- 1State Key Laboratory for Functions and Applications of Medicinal Plants, Guizhou Medical University, Guiyang, China
- 2The Key Laboratory of Chemistry for Natural Products of Guizhou Province and Chinese Academic of Sciences, Guiyang, China
- 3School of Pharmaceutical Sciences, Guizhou Medical University, Guiyang, China
Chronic myeloid leukemia (CML) accounts for a major cause of death in adult leukemia patients due to mutations or other reasons for dysfunction in the ABL proto-oncogene. The ubiquitous BCR–ABL expression stimulates CML by activating CDK1 and cyclin B1, promoting pro-apoptotic, and inhibiting antiapoptotic marker expression along with regulations in RAS pathway activation. Thus, inhibitors of cyclins and the RAS pathway by ERK are of great interest in antileukemic treatments. Mikanolide is a sesquiterpene dilactone isolated from several Asteraceae family Mikania sp. plants. Sesquiterpene dilactone is a traditional medicine for treating ailments, such as flu, cardiovascular diseases, bacterial infections, and other blood disorders. It is used as a cytotoxic agent as well. The need of the hour is potent chemotherapeutic agents with cytotoxic effects inhibition of proliferation and activation of apoptotic machinery. Recently, ERK inhibitors are used in clinics as anticancer agents. Thus, in this study, we synthesized 22-mikanolide derivatives that elucidated to be potent antileukemic agents in vitro. However, a bioactive mikanolide derivative, 3g, was found with potent antileukemic activity, through the Ras/Raf/MEK/ERK pathway. It can arrest the cell cycle by inhibiting phosphorylation of CDC25C, triggering apoptosis, and promoting DNA and mitochondrial damage, thus suggesting it as a potential chemotherapeutic agent for leukemia patients.
Introduction
Chronic myeloid leukemia (CML) is a myeloproliferative syndrome of primitive hematopoietic progenitor cells. It accounts for around 20% of leukemic cases reported in adults. The major triggers for this type of leukemia are the BCR–ABL tyrosine kinase dysfunctions due to mutations or other causes (Ben-Neriah et al., 1986; Jackson and Baltimore, 1989; Branford et al., 2002). The ABL is a proto-oncogene that coordinates many cellular activities, such as cell differentiation, adhesion, DNA damage response, and apoptosis. This BCR–ABL gene, when expressed, monitors the expression of cyclin-dependent kinase 1 (CDK1). Therefore, the mutations in this gene cause DNA damage and the activation of RAS pathways. BCR–ABL expression promotes CML due to damaged cell adhesion capacity cells (Overduin et al., 1992; Musacchio et al., 1994; Willis et al., 2005; Vaidya et al., 2015), causing uncontrolled blood cells’ growth in CML patients. Thus, recently the chemotherapeutic drugs such as imatinib and several tyrosine inhibitors (Wan et al., 2016; Liu et al., 2020; Maiti et al., 2020) focusing on BCR–ABL gene regulation and related pathways are of great attraction for leukemia treatment researchers.
Deregulation of the ERK pathway usually happens due to genetic adjustments of several crucial molecules of this pathway. In contrast, uncontrolled leukemic proliferation can occur due to diminished compassion to apoptosis-initiating factors or due to chemo-resistance correlated to pro-survival molecules activation. Nevertheless, inactivated ERK pathway molecules could considerably alter reactions toward small molecule inhibitors. Moreover, the ERK pathway is inhibited by many usual chemotherapeutic drugs used for leukemia (McCubrey et al., 2007; McCubrey et al., 2008a; Steelman et al., 2008). Mikanolide is a sesquiterpene dilactone isolated from many kinds of Mikania plants. It has been used as a folk medicine for a long time and has been paid attention by researchers due to its antibacterial, antitumor, antimicrobial, and cytotoxic functions (Bohlmann et al., 1984; Gutierrez et al., 1985; Pickman, 1986; Ysrael and Croft, 1990; Facey et al., 1999; Ahmed et al., 2001; Zhuang et al., 2010). Sesquiterpenes are biosynthesized in the plant endoplasmic reticulum by farnesyl pyrophosphate as colorless lipophilic compounds. Sesquiterpenes are a 15-carbon backbone molecule with diverse structural orientations and some functional cyclic ones. The most bioactive compounds having cardiovascular effects inhibit iNOS and NF-κB (Giordano et al., 1990). Another study revealed parthenolide— a germacranolide sesquiterpene lactone—inhibits JNK activation in CNE1, COLO205, HELA, HBL-100, and MDA-MB-231 cells. Parthenolide sensitizes TNF-related apoptosis-inducing ligand (TRAIL) proteins offering anticancer action (Nakshatri et al., 2004; Zhang et al., 2004; Guzman et al., 2005). Consequently, as treatment regimens for CML, novel mikanolide derivatives that can act as inhibitors of abnormal cell growth with reduced side effects are investigated. Likewise, the ERK pathway inhibitors can control the sensitivity as well as resistance in leukemia treatment. Thus, in this study, we synthesized 22-mikanolide derivatives that can be antileukemic agents by targeting inhibition of cell proliferation signaling molecules, thus suggesting them as a potential chemotherapeutic agent for leukemia patients.
Materials and Methods
Synthesis of Compounds
Proton nuclear magnetic resonance (1H NMR) spectra were recorded using Bruker AV 400 MHz or 700 MHz spectrometers. Proton chemical shifts are reported in parts per million (d scale) and are referenced using residual protium in the NMR solvent [CDCl3: δ 7.26 (CHCl3), DMSO-d6: d 2.54 (DMSO)]. Data are reported as follows: chemical shift [multiplicity (s = singlet, d = doublet, dd = doublet of doublets, ddd = doublet of doublet of doublets, t = triplet, q = quartet, m = multiplet, and br s = broad singlet), coupling constant(s) (Hz), and integration]. Carbon-13 nuclear magnetic resonance (13C NMR) spectra were recorded using Bruker AV 100 MHz or 151 MHz or 176 MHz spectrometers. Carbon chemical shifts are reported in parts per million (d scale), and are referenced using the carbon resonances of the solvent [CDCl3: δ 77.0 (CHCl3), DMSO-d6: δ 40.45 (DMSO)]. Data are reported as follows: chemical shift [multiplicity (if not singlet) and assignment (Cq = fully substituted carbon)]. High resolution mass spectra (HRMS) were documented on a Waters SYNAPT G2 using an electrospray (ESI) ionization source. Column chromatography was performed on silica gel (400–500 mesh) eluting with ethyl acetate and petroleum ether. TLC was performed on glass-backed silica plates, and visualized by UV light and I2 products.
A mixture of mikanolide or deoxymikanolide (1.0 mmol) and an appropriate aromatic iodide (1.1 mmol) was refluxed at 115°C using palladium (II) ferrocene (0.01 mmol) and DIPEA (3.0 mmol) in dry toluene (1 ml) under air for 6–12 h. A TLC monitor was used until the reaction was complete, then the reaction mixture was allowed to cool to room temperature, water (10 ml) was added, and the resultant mixture was extracted with ethyl acetate (15 ml × 3). The separated organics were dried over Na2SO4. Later it was filtered. The filtrate was concentrated under reduced pressure. The obtained crude residue was purified by silica flash chromatography (500:1 to 100:1, dichloromethane/methanol) to afford the corresponding aryl-substituted parthenolide as a solid (65–195 mg) in 15–45% isolated yield.
Reagents
Roswell Park Memorial Institute 1640 (RPMI-1640, Gibco) was purchased from Thermo Fisher Scientific (Shanghai, China). Fetal bovine serum (FBS) was purchased from the VACCA Biologics LLC. (United States). Dimethyl sulfoxide (D8371), bovine serum albumin (A8020), BCA protein assay kit (PC0020), thiazolyl blue tetrazolium bromide (M8180), and color mixed protein marker (PR 1920) were obtained from Solarbio Life Sciences (Beijing, China). Annexin V-FITC apoptosis detection kit was purchased from BD (United States, Cat. No. 556547). JC-1 mitochondrial membrane potential detection kit (C2006), Hoechst staining kit (C0003), reactive oxygen species assay kit (S0033), SDS-PAGE gel preparation kit (P0012AC), cell lysis buffer (P0013), SDS-PAGE sample loading buffer, 5X (P0015L), and transfer buffer (P0021B) were purchased from Beyotime Biotechnology (Shanghai, China). Ras (ab52939), Bim (ab32158), ERK (ab184699), p-ERK (ab32538), p-PKCδ (ab76181), PKCδ (ab182126), cyclin B1 (ab32053), CDK1 (ab133327), p90RSK(ab32413), caspase 8 (ab25901), Bcl-2 (ab32124), Bcl-xl (ab2568), and p-γH2AX (ab11174) were purchased from Abcam (Abcam, Cambridge, United Kingdom); BID (#2002), P-B-Raf (#2696), P-MEK1/2 (#9154), MEK1/2 (#8727), PARP (#9542), P-CDC25C (#4901), CDC25C (#4688), p21 (#2947), Bad (#9292), Phospho-CDC2 (#4539), caspase 3 (#9662), cleaved caspase 3 (#9661), cleaved caspase 9 (#7237), and c-FLIP (#56343S) were purchased from Cell Signaling Technology; p27 (380960), caspase 9 (381336), and GAPDH (301341) were purchased from Zen Bioscience, China; B-Raf (AF6171) was purchased from Affinity Biosciences, United States .
Cell Culture
HEL (erythroleukemia), K562 (chronic myeloid leukemia), CEM-C7H2 (T-cell acute lymphoblastic leukemia), and HL7702 (normal hepatic cell line) were purchased from ATCC, Manassas, VA, United States . The cells were cultured in RPMI 1640 medium supplemented with 5% FBS at 37°C in a CO2 incubator (5% CO2 and 95% air, 95% humidity), as per standard conditions of passage.
Cell Viability Assays
The cell viability assay was performed by treating different concentrations of compounds on HEL, K562, and CEM-C7H2 cells to find their IC50 values. The control groups were treated with 0.1% DMSO or compounds. After 72 h treatment, the cells were added with an MTT solution of 20 µl (5 mg/mL) for 4 h. Later, the 96 well-plates were centrifuged at 2,500 rpm for 20 min and discarded the medium, then DMSO (160 µl) was used to dissolve formazan crystals. The resulting solution was determined using absorbance at 490 nm (BioTek, Winooski, VT, United States). The cell morphology was analyzed under an inverted microscope (Nikon) and photographed (Gajendran et al., 2020a).
Flow Cytometric Analysis
Cells were seeded at 3 × 106 cells per 60 mm dish in 3 ml medium with different concentrations of 3g (0.3, 0.5, and 1 µM). After growing for 24 and 48 h, the cells were harvested, washed twice with cold PBS, and then the cells were resuspended in 1X binding buffer at a concentration of 1 × 106 cells/ml. Later, 100 µl of cells (1 × 105) were transferred to a 2 ml Eppendorf tube, added with 5 µL of FITC Annexin V and PI (BD FITC Annexin V apoptosis detection kit I) in each tube. After gently vortexing the cells and incubating for 15 min at RT (25°C) in the dark, 400 µl of 1X binding buffer to each tube was added and analyzed by flow cytometry within 1 h (ACEA Biosciences Inc. San Diego, CA, United States) and compared with controls (Varier and Sumathi, 2019).
Cell Cycle Measurement
Cells were seeded in a 6-well culture plate at a density of 1 × 106 cells/ml in 2 ml medium and were treated with 3g at different concentrations (0.3, 0.5, and 1 µM) for 24 and 48 h. After the incubation period, the cells were harvested and transferred into a sterile centrifuge tube for cell cycle analysis (Gajendran et al., 2020b). Cells were washed with pre-cool PBS and suspended in 70% ice ethanol, incubated for 4 h at 4°C, and preserved in a refrigerator at −20°C overnight. To remove the stationary liquid, the cells were centrifuged and washed twice with cold PBS. Then, 500 µL mix dye solution (RNaseA 100 μg/mL, PI 50 μg/mL, and Triton X-100 0.2%) were added into each tube, gently mixed, and incubated for 10 min at room temperature in the dark. Before analysis by flow cytometry, the cells were washed with cold PBS, and 200 µl suspensions were used for analysis by a NovoCyte flow cytometer (ACEA Biosciences, Inc. San Diego, CA, United States).
Measurement of Mitochondrial Membrane Potential (∆Ψm)
Cells (1 × 106) were seeded in a 3 ml medium with different concentrations of 3g (0.3, 0.5, and 1 µM) to detect the change in the membrane potential of treated cells concerning control cells. After being treated with 3g for 48 h, the cells were preincubated with JC-1 working solution for 20 min at 37°C, 5% CO2. After incubation, the dye was removed, and the cells were washed two times with JC-1 (Ma et al., 2021) and investigated using a Nikon fluorescent microscope.
Hoechst 33258 Staining
K562 cells (1 × 106) were seeded into a 6-well plate and treated with different concentrations of 3g (0.3, 0.5, and 1 µM) for 72 h (Ma et al., 2021). Cells were consequently collected, rinsed twice with PBS, and stained with Hoechst 33258 (Beyotime, Jiangsu, China) for 10 min and examined by using a Nikon fluorescence microscope.
Western Blot Analysis
Cells were treated with 3g in different concentrations (0.3, 0.5, and 1 µM) for 24 h, cells were extracted, and total protein was collected from cell lysis buffer. Protein concentration was determined by the BCA test kit (Solarbio Life Sciences, China), and proteins were separated by 10% SDS-PAGE, then blotted onto the PVDF membrane (0.22 µm, Merck KGaA, Germany). The membranes were incubated in solution with 5% milk (dissolved in TBST) at room temperature for 1 h and probed with primary antibodies and GAPDH at 4°C overnight. After TBST washing, the membrane was developed with a secondary antibody (Hu et al., 2021). The antibodies were diluted according to the manufacturer’s instructions. Last, immunoreactive protein signals were spotted by the Odyssey Infrared Imaging System. GAPDH served as an internal loading control.
AutoDock Analysis
The molecular docking simulation was performed via the AutoDock Vina program (Morris et al., 2009; Varier et al., 2017). ERK (PDB code: 5V60) interaction with U0126 (PubChem ID: 3006531) and 3g were carefully analyzed.
Statistical Analysis
All statistical analyses by two-way ANOVA test were performed, followed by Tukey’s post hoc test analysis by GraphPad Prism 8 software (San Diego, CA, United States). All the experiments were organized in triplicates. The data were expressed as mean ± SD with significant p values (*p < 0.05, **p < 0.01, ***p < 0.001, and ****p < 0.0001).
Results
Chemistry
Mikanolide and deoxymikanolide were isolated according to the reported procedures (Li et al., 2013; Li et al., 2013), and 22 novel E-olefinic coupling products of mikanolides (3a-u and 4) were prepared in 15–45% yields under heck reaction conditions utilizing palladium ferrocene [Pd (dppf)Cl2] as a catalyst in toluene and heating the mixture with an appropriate iodo-aromatic or iodo-heteroaromatic compound in the presence of di-isopropylethyl-amine (DIPEA) as the base (Figure 1A; Supplementary Material).
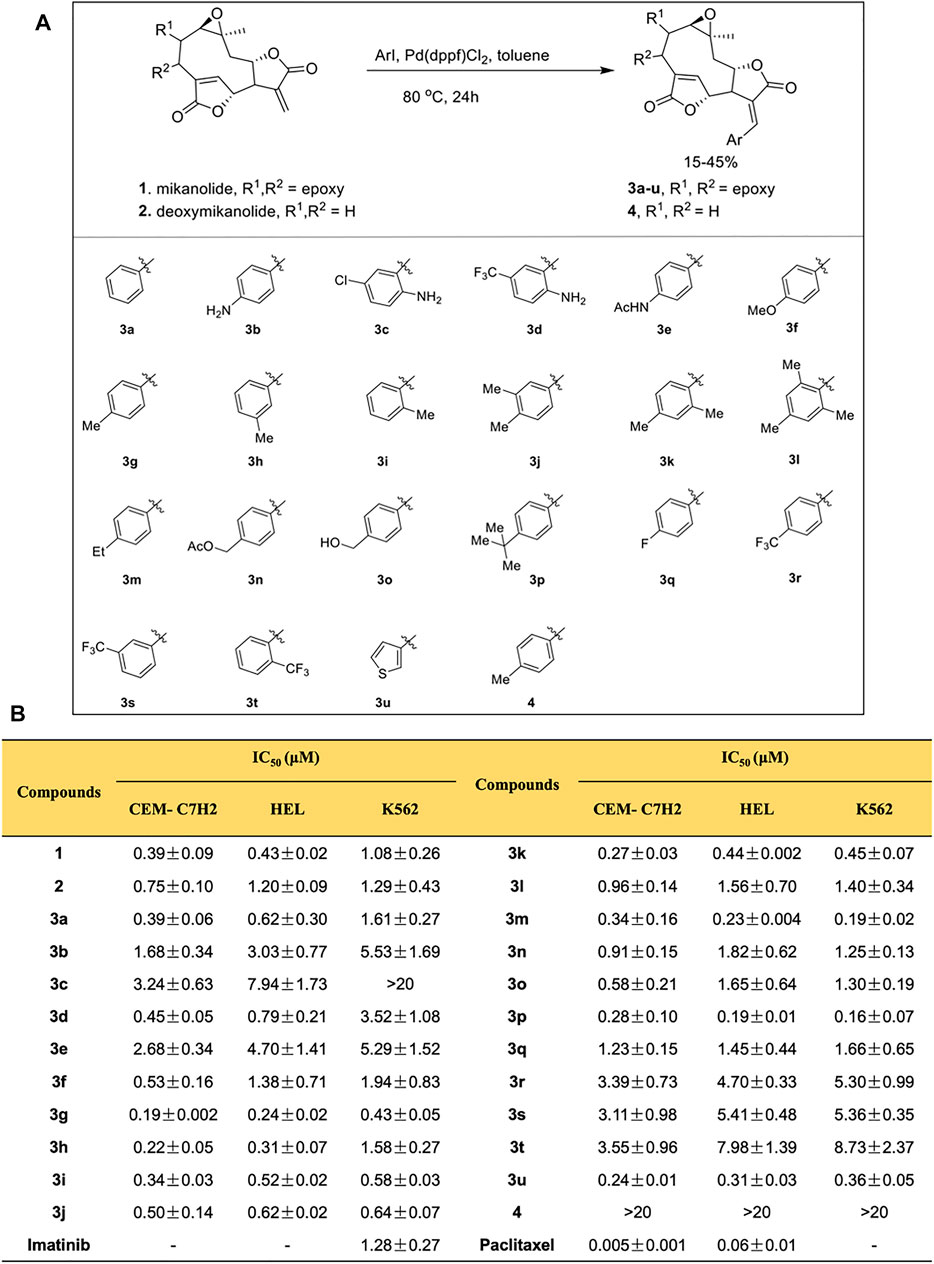
FIGURE 1. Mikanolide derivatives offer antileukemic action. (A) Schematic diagram explaining the procedure of synthesis of mikanolide derivatives. (B) Cytotoxicity (IC50) of mikanolide derivatives against indicated cancer cells. Data represented as means ± SD of three independent experiments.
Compound 3a (1aR,1bR,2aR,6R,6aR,9aS,10aS,Z)-7-[(E)-benzylidene]-10a-methyl-1a,1b,2a,6a,7,9a,10,10a-octahydro-4H-3,6-(metheno) furo [3,2-c] bis (oxireno) [2,3-f:2',3'-h] [1] oxacycloundecine-4,8 (6H)-dione: 149.5 mg; isolated yield 39.5%; white solid; IR (KBr): 2,926.49, 2,359.84, 1,755.88, 1,645.58, 1,645.58, 1,195.91, 1,079.00, 1,021.15, 804.95, 696.93, and 472.58 cm−1; 1H NMR (600 MHz, CDCl3): δ 7.87 (d, J = 3.0 Hz, 1H), 7.62 (d, J = 7.8 Hz, 2H), 7.52 (t, J = 7.5 Hz, 2H), 7.47 (t, J = 7.5 Hz, 1H), 7.11 (s, 1H), 5.20 (s, 1H), 4.37 (m, 1H), 4.05 (s, 1H), 3.83 (m, 1H), 3.38 (d, J = 3.6 Hz, 1H), 2.94 (s, 1H), 2.21 (m, 2H), and 1.24 (m, 3H) ppm; 13C NMR (125 MHz, CDCl3): δ 170.0, 169.0, 146.2, 141.3, 131.8, 131.4, 130.9, 130.5, 129.0, 124.6, 79.7, 78.5, 58.4, 57.8, 54.9, 51.0, 50.6, 43.4, and 21.1 ppm. HRMS calculated for C21H19NO6, (M + H)+: 367.1176 found 367.1168.
Compound 3b (1aR,1bR,2aR,6R,6aR,9aS,10aS,Z)-7-[(E)-4-aminobenzylidene]-10a-methyl-1a,1b,2a,6a,7,9a,10,10a-octahydro-4H-3,6-(metheno) furo [3,2-c] bis (oxireno) [2,3-f:2',3'-h] [1] oxacycloundecine-4,8 (6H)-dione: 161.6 mg; isolated yield 41.0%; light yellow solid; IR (KBr): 3,473.59, 3,365.14, 3,239.04, 2,935.41, 2,360.15, 1744.47, 1,519.48, 1,365.13, 1,309.06, 1,261.56, 1,199.75, 1,177.24, 1,106.54, 1,078.05, 1,035.83, 1,012.46, 928.06, 831.10, and 626.06 cm−1; 1H NMR (600 MHz, CDCl3): δ 7.67 (d, J = 3.6 Hz, 1H), 7.47 (d, J = 8.4 Hz, 2H), 7.17 (s, 1H), 6.75 (d, J = 8.4 Hz, 2H), 5.30 (s, 1H), 4.59 (d, J = 7.8 Hz, 1H), 3.88 (dd, J = 10.8, 3.0 Hz, 1H), 2.70 (m, 2H), 2.56 (m, 1H), 2.18 (s, 2H), and 1.32 (s, 3H) ppm; 13C NMR (125 MHz, CDCl3): δ 171.6, 170.3, 149.3, 147.6, 140.9, 133.1, 133.0, 121.8, 118.7, 114.7, 78.5, 78.2, 61.8, 57.0, 50.5, 44.0, 22.8, 22.3, and 19.9 ppm. HRMS calculated for C21H20NO6, (M + H)+: 382.1285 found 382.1275.
Compound 3c (1aR,1bR,2aR,6R,6aR,9aS,10aS,Z)-7-[(E)-2-amino-5-chlorobenzylidene]-10a-methyl-1a,1b,2a,6a,7,9a,10,10a-octahydro-4H-3,6-(metheno) furo [3,2-c] bis (oxireno) [2,3-f:2',3'-h] [1] oxacycloundecine-4,8 (6H)-dione: 180.3 mg; isolated yield 42.0%; light yellow solid: IR (KBr): 3,367.06, 2,935.41, 2,360.36, 1748.95, 1,635.78, 1,487.85, 1,418.17, 1,364.27, 1,300.64, 1,265.09, 1,228.68, 1,188.36, 1,105.23, 1,035.56, 1,012.47, 930.06, and 635.04 cm−1; 1H NMR (600 MHz, CDCl3): δ 7.70 (d, J = 3.6 Hz, 1H), 7.36 (d, J = 1.8 Hz, 1H), 7.20 (dd, J = 10.8, 2.4 Hz, 1H), 7.09 (s, 1H), 6.73 (d, J = 9.0 Hz, 1H), 5.05 (s, 1H), 4.58 (m, 1H), 3.88 (m, 1H), 2.69 (m, 2H), 2.52 (m, 1H), 2.20 (m, 2H), and 1.32 (s, 3H) ppm; 13C NMR (125 MHz, CDCl3): δ 171.3, 169.3, 146.9, 144.0, 134.2, 133.3, 131.8, 128.6, 126.3, 123.6, 118.4, 117.9, 78.4, 77.9, 61.8, 56.9, 50.5, 44.0, 22.7, 22.1, and 20.0 ppm. HRMS calculated for C21H19ClNO6, (M + H)+: 416.0895 found 416.0886.
Compound 3 d (1aR,1bR,2aR,6R,6aR,9aS,10aS,Z)-7-[(E)-2-amino-5-(trifluoromethyl)benzylidene]-10a-methyl-1a,1b,2a,6a,7,9a,10,10a-octahydro-4H-3,6-(metheno) furo [3,2-c] bis (oxireno) [2,3-f:2',3'-h] [1] oxacycloundecine-4,8 (6H)-dione: 180.3 mg; isolated yield 42.0%; white solid; IR (KBr): 3,400.39, 2,359.86, 1775.83, 1,634.46, 1,510.61, 1,333.31, 1,266.66, 1,230.21, 1,111.72, 1,017.45, 843.14, 802.66, 730.89, and 602.09 cm−1; 1H NMR (600 MHz, DMSO-d6): δ 7.68 (s, 1H), 7.54 (m, 2H), 7.38 (dd, J = 10.2, 1.8Hz, 1H), 6.82 (d, J = 8.4 Hz, 1H), 6.16 (s, 2H), 5.01 (s, 1H), 4.77 (m, 1H), 4.00 (m, 1H), 3.93 (d, J = 3.0 Hz, 1H), 3.38 (d, J = 3.6 Hz, 1H), 3.20 (s, 1H), 2.30 (t, J = 12.0 Hz, 1H), 1.84 (dd, J = 17.4, 4.2 Hz, 1H), and 1.05 (s, 3H) ppm, 13C NMR (125 MHz, DMSO-d6): δ 170.4, 169.2, 150.8, 149.6, 133.4, 128.7, 128.4, 127.6, 127.5, 125.9, 124.1, 115.6, 114.9, 81.1, 77.4, 57.5, 57.3, 54.9, 50.2, 48.9, 42.1, and 20.6 ppm; 19F NMR (125 MHz, DMSO-d6): 59.2 (3) ppm. HRMS calculated for C22H19F3NO6, (M + H)+: 450.1159 found 450.1160.
Compound 3e N-{4-(E)-[(1aR,1bR,2aR,6R,6aR,9aS,10aS,Z)-10a-methyl-4,8-dioxo-1a,1b,2a,6,6a,9a,10,10a-octahydro-4H-3,6-(metheno) furo [3,2-c] bis (oxireno) [2,3-f:2',3'-h] [1] oxacycloundecin-7 (8H)-ylidene)methyl] phenyl} acetamide: 155.3 mg; isolated yield 35.5%; light yellow solid: IR (KBr): 3,577.39, 2,359.53, 1769.28, 1748.68, 1,683.73, 1,635.05, 1,592.76, 1,514.66, 1,415.59, 1,371.33, 1,313.69, 1,259.48, 1,199.65, 1,179.40, 1,079.17, 1,024.75, 840.01, 803.69, and 679.07 cm−1; 1H NMR (600 MHz, DMSO-d6): δ 10.2 (s, 1H), 7.69 (m, 4H), 7.56 (t, J = 6.0,Hz, 2H), 5.13 (s, 1H), 4.73 (m, 1H), 4.11 (t, J = 3.6 Hz, 1H), 3.98 (d, J = 3.0 Hz, 1H), 3.40 (d, J = 3.0 Hz, 1H), 3.16 (s, 1H), 2.24 (t, J = 12.6 Hz, 1H), 2.08 (d, J = 6.0 Hz, 3H), 1.84 (dd, J = 16.8, 3.0 Hz, 1H), and 1.07 (s, 3H) ppm; 13C NMR (125 MHz, DMSO-d6): δ 170.7, 169.6, 168.7, 150.0, 141.0, 137.6, 131.4, 128.6, 127.0, 125.8, 118.3, 81.2, 77.8, 57.7, 57.4, 54.7, 50.3, 48.8, 42.0, 24.2, and 20.5 ppm. HRMS calculated for C23H22NO7, (M + H)+: 424.1391 found 424.1393.
Compound 3f (1aR,1bR,2aR,6R,6aR,9aS,10aS,Z)-7-[(E)-4-methoxybenzylidene]-10a-methyl-1a,1b,2a,6a,7,9a,10,10a-octahydro-4H-3,6-(metheno) furo [3,2-c] bis (oxireno) [2,3-f:2',3'-h] [1] oxacycloundecine-4,8 (6H)-dione: 163.8 mg; isolated yield 41.4%; white solid; IR (KBr): 2,358.91, 1751.23, 1,644.06, 1,604.30, 1,514.57, 1,307.14, 1,216.08, 1,199.72, 1,176.07, 1,021.69, and 805.33 cm−1; 1H NMR (600 MHz, CDCl3): δ 7.78 (d, J = 3.0 Hz, 1H), 7.62 (d, J = 8.4 Hz, 2H), 7.16 (s, 1H), 7.03 (d, J = 8.4 Hz, 2H), 5.28 (d, J = 1.8 Hz, 1H), 4.38 (m, 1H), 4.06 (d, J = 3.0 Hz, 1H), 3.88 (s, 3H), 3.80 (m, 1H), 3.38 (d, J = 3.0 Hz, 1H), 2.95 (s, 1H), 2.22 (m, 2H), and 1.25 (s, 3H) ppm; 13C NMR (125 MHz, CDCl3): δ 170.1, 169.5, 161.8, 146.6, 141.1, 132.7, 131.2, 124.3, 121.5, 114.5, 79.7, 78.5, 58.3, 57.9, 55.5, 54.9, 51.0, 50.7, 43.3, and 21.1 ppm. HRMS calculated for C22H21O7, (M + H)+: 397.1282 found 397.1283.
Compound 3g (1aR,1bR,2aR,6R,6aR,9aS, 10aS,Z)-10a-methyl-7-[(E)-4-methylbenzylidene]-1a,1b,2a,6a,7,9a, 10,10a-octahydro-4H-3,6-(metheno) furo [3,2-c] bis (oxireno) [2,3-f:2',3'-h] [1] oxacycloundecine-4,8(6H)-dione: 78.0 mg; isolated yield 39.7%; white solid; IR (KBr):2,959.55, 2,359.98, 1764.47, 1,651.30, 1,608.26, 1,510.49, 1,353.13, 1,261.73, 1,197.54, 1,081.93, 1,022.75, 896.31, 736.61, and 677.40 cm−1; 1H NMR (600 MHz, CDCl3): δ 7.82 (d, J = 3.6 Hz, 1H), 7.53 (d, J = 7.8 Hz, 2H), 7.33 (d, J = 8.4 Hz, 2H), 7.13 (s, 1H), 5.24 (d, J = 1.8 Hz, 2H), 4.37 (m, 1H), 4.05 (d, J = 2.4 Hz, 1H), 3.82 (m, 1H), 3.38 (d, J = 3.6 Hz, 1H), 2.94 (s, 1H), 2.42 (s, 3H), 2.22 (m, 2H), and 1.25 (s, 3H) ppm; 13C NMR (125 MHz, CDCl3): δ 170.0, 169.3, 146.6, 141.6, 141.4, 131.3, 130.6, 129.7, 128.9, 123.3, 79.7, 78.5, 58.3, 57.9, 54.9, 51.0, 50.7, 43.3, 21.6, and 21.1 ppm. HRMS calculated for C22H20O6Na, (M + Na)+: 403.1152 found 403.1144.
Compound 3h (1aR,1bR,2aR,6R,6aR,9aS, 10aS,Z)-10a-methyl-7-[(E)-3-methylbenzylidene]-1a,1b,2a,6a,7,9a, 10,10a-octahydro-4H-3,6-(metheno) furo [3,2-c] bis (oxireno) [2,3-f:2',3'-h] [1] oxacycloundecine-4,8 (6H)-dione: 149.4 mg; isolated yield 38.0%; white solid; IR (KBr): 2,929.12, 2,359.41, 1764.42, 1,372.25, 1,347.99, 1,264.94, 1,209.79, 1,165.25, 1,084.03, 1,022.65, 892.60, 790.61, and 697.10 cm−1; 1H NMR (600 MHz, DMSO-d6): δ 7.84 (d, J = 3.6 Hz, 1H), 7.46 (s, 1H), 7.41 (d, J = 5.4 Hz, 2H), 7.28 (m, 1H), 7.10 (s, 1H), 5.20 (d, J = 1.8 Hz, 2H), 4.37 (m, 1H), 4.05 (d, J = 3.0 Hz, 1H), 3.82 (m, 1H), 3.38 (d, J = 3.0 Hz, 1H), 2.94 (s, 1H), 2.46 (s, 3H), 2.18 (m, 2H), and 1.25 (s, 3H) ppm; 13C NMR (125 MHz, DMSO-d6): δ 169.9, 169.1, 146.2, 141.5, 138.8, 131.7, 131.6, 131.3, 131.0, 128.8, 127.7, 124.3, 79.6, 78.4, 58.3, 57.8, 54.9, 51.0, 50.7, 43.4, 21.1, and 18.4 ppm. HRMS calculated for C22H21O6, (M + H)+: 381.1333 found 381.1323.
Compound 3i (1aR,1bR,2aR,6R,6aR,9aS,10aS,Z)-10a-methyl-7-[(E)-2-methylbenzylidene] 1a,1b,2a,6a,7,9a,10,10a-octahydro-4H-3,6-(metheno) furo [3,2-c] bis (oxireno) [2,3-f:2',3'-h] [1] oxacycloundecine-4,8 (6H)-dione: 143.5 mg; isolated yield 36.5%; white solid; IR (KBr): 2,920.46, 2,359.93, 1,743.78, 1,662.12, 1,457.26, 1,376.24, 1,349.31, 1,262.43, 1,212.20, 1,083.32, 1,024.69, 804.54, 755.47, and 678.90 cm−1; 1H NMR (600 MHz, CDCl3): δ 7.68 (d, J = 3.6 Hz, 1H), 7.63 (d, J = 7.8 Hz, 1H), 7.47 (d, J = 1.2 Hz, 1H), 7.32 (m, 1H), 7.29 (m, 2H), 4.96 (s, 1H), 4.74 (m, 1H), 4.02 (m, 1H), 3.93 (d, J = 3.6 Hz, 1H), 3.37 (dd, J = 4.8, 1.2 Hz, 1H), 3.18 (s, 1H), 2.35 (s, 3H), 2.26 (t, J = 12.3 Hz, 1H), 1.85 (dd, J = 16.8, 3.6 Hz, 1H), and 1.02 (s, 3H) ppm; 13C NMR (125 MHz, CDCl3): δ 170.6, 169.2, 149.7, 137.3, 136.3, 131.9, 129.8, 129.7, 129.3, 129.1, 128.6, 125.7, 81.4, 77.7, 57.5, 57.3, 54.8, 50.2, 48.4, 42.1, 20.6, and 19.6 ppm. HRMS calculated for C22H21O6, (M + H)+: 381.1333 found 381.1323.
Compound 3j (1aR,1bR,2aR,6R,6aR,9aS,10aS,Z)-7-[(E)-3,4-dimethylbenzylidene]-10a-methyl-1a,1b,2a,6a,7,9a,10,10a-octahydro-4H-3,6-(metheno) furo [3,2-c] bis (oxireno) [2,3-f:2',3'-h] [1] oxacycloundecine-4,8(6H)-dione: 144.3 mg; isolated yield 35.4%; white solid; IR (KBr): 2,918.19, 2,359.93, 1766.70, 1747.04, 1,652.51, 1,454.87, 1,373.63, 1,348.69, 1,265.66, 1,205.77, 1,163.64, 1,022.18, and 669.13 cm−1; 1H NMR (600 MHz, DMSO-d6): δ 7.55 (m, 3H), 7.44 (d, J = 7.8 Hz, 1H), 7.24 (d, J = 7.8 Hz, 1H), 5.10 (s, 1H), 4.74 (m, 1H), 4.10 (m, 1H), 3.97 (d, J = 3.0 Hz, 1H), 3.16 (s, 1H), 2.27 (d, J = 9.0 Hz, 6H), 1.82 (m, 2H), and 1.07 (s, 3H) ppm; 13C NMR (125 MHz, DMSO-d6): δ 170.6, 169.6, 149.9, 138.9, 138.0, 136.4, 131.4, 129.9, 129.6, 128.6, 128.2, 126.4, 81.1, 77.9, 57.7, 57.4, 50.4, 50.2, 48.9, 42.0, 20.6, 19.4, and 19.1 ppm. HRMS calculated for C23H23O6, (M + H)+: 395.1489 found 395.1491.
Compound 3k (1aR,1bR,2aR,6R,6aR,9aS,10aS,Z)-7-[(E)-2,4-dimethylbenzylidene]-10a-methyl-1a,1b,2a,6a,7,9a,10,10a-octahydro-4H-3,6-(metheno) furo [3,2-c] bis (oxireno) [2,3-f:2',3'-h] [1] oxacycloundecine-4,8(6H)-dione: 142.6 mg; isolated yield 35.0%; white solid; IR (KBr): 2,989.49, 2,360.13, 1740.93, 1,632.70, 1,612.44, 1,496.93, 1,450.06, 1,407.02, 1,365.09, 1,297.59, 1,263.76, 1,226.20, 1,201.65, 1,123.63, 1,072.60, 1,025.97, 894.11, 803.86, and 747.24 cm−1; 1H NMR (600 MHz, DMSO-d6): δ 7.64 (d, J = 3.6 Hz, 1H), 7.56 (d, J = 7.2 Hz, 1H), 7.49 (d, J = 1.2 Hz, 1H), 7.12 (m, 2H), 5.00 (t, J = 1.2 Hz, 1H), 4.73 (m, 1H), 4.05 (m, 1H), 3.94 (d, J = 3.0 Hz, 1H), 3.37 (d, J = 3.6 Hz, 1H), 3.17 (s, 1H), 2.32 (d, J = 5.4 Hz, 6H), 2.25 (m, 1H), 1.84 (dd, J = 17.4, 3.6 Hz, 1H), and 1.04 (s, 3H) ppm; 13C NMR (125 MHz, DMSO-d6): δ 170.7, 169.4, 149.8, 139.5, 137.5, 136.1, 130.7, 129.5, 128.9, 128.6, 127.9, 126.3, 81.3, 77.7, 57.5, 57.3, 54.8, 50.3, 48.5, 42.1, 21.0, 20.6, and 19.5 ppm. HRMS calculated for C23H23O6, (M + H)+: 395.1489 found 395.1491.
Compound 3l (1aR,1bR,2aR,6R,6aR,9aS,10aS,Z)-10a-methyl-7-[(E)-2,4,6-trimethylbenzylidene]-1a,1b,2a,6a,7,9a,10,10a-octahydro-4H-3,6-(metheno) furo [3,2-c] bis (oxireno) [2,3-f:2',3'-h] [1] oxacycloundecine-4,8 (6H)-dione: 164.6 mg; isolated yield 39.0%; white solid; IR (KBr): 2,970.53, 2,359.70, 1759.72, 1,668.57, 1,342.95, 1,225.26, 1,202.19, 1,080.08, 1,018.43, 903.49, 848.07, 803.99, and 678.79 cm−1; 1H NMR (600 MHz, DMSO-d6): δ 7.60 (d, J = 3.0 Hz, 1H), 7.43 (s, 1H), 6.97 (s, 1H), 6.87 (s, 1H), 4.73 (m, 2H), 3.88 (d, J = 3.0 Hz, 1H), 3.35 (d, J = 3.0 Hz, 1H), 3.24 (m, 1H), 3.20 (s, 1H), 2.33 (t, J = 12.3 Hz, 1H), 2.22 (m, 9H), 1.85 (dd, J = 17.4, 3.6 Hz, 1H), and 0.98 (s, 3H) ppm; 13C NMR (125 MHz, DMSO-d6): δ 170.6, 169.5, 149.8, 146.1, 137.9, 130.5, 129.9, 128.6, 127.9, 126.7, 81.1, 77.9, 57.6, 57.4, 54.7, 50.3, 48.8, 42.0, 28.1, 20.5, and 15.2 ppm. HRMS calculated for C24H25O6, (M + H)+: 409.1646 found 409.1647.
Compound 3m (1aR,1bR,2aR,6R,6aR,9aS,10aS,Z)-7-[(E)-4-ethylbenzylidene]-10a-methyl-1a,1b,2a,6a,7,9a,10,10a-octahydro-4H-3,6-(metheno) furo [3,2-c] bis (oxireno) [2,3-f:2',3'-h] [1] oxacycloundecine-4,8 (6H)-dione: 152.0 mg; isolated yield 37.3%; white solid; IR (KBr): 2,968.14, 2,360.02, 1741.34, 1,637.90, 1,606.81, 1,508.35, 1,420.09, 1,370.07, 1,260.38, 1,225.74, 1,200.81, 1,181.62, 1,073.20, 1,023.79, 896.50, 834.37, 796.24, and 679.52 cm−1; 1H NMR (600 MHz, DMSO-d6): δ 7.66 (d, J = 7.8 Hz, 2H), 7.60 (s, J = 3.6 Hz, 1H), 7.54 (s, 1H), 7.33 (d, J = 8.4 Hz, 2H), 5.13 (s, 1H), 4.73 (m, 1H), 4.12 (m, 1H), 3.98 (d, J = 3.0 Hz, 1H), 3.40 (d, J = 3.6 Hz, 1H), 3.16 (s, 1H), 2.67 (q, J = 7.6 Hz, 2H), 2.26 (t, J = 12.6 Hz, 1H), 1.84 (dd, J = 16.8, 3.6 Hz, 1H), 1.21 (t, J = 7.5 Hz, 3H), and 1.07 (s, 3H) ppm; 13C NMR (125 MHz, DMSO-d6): δ 170.6, 169.5, 149.8, 146.1, 137.8, 130.5, 129.9, 128.6, 127.9, 126.7, 81.1, 77.9, 57.6, 57.4, 54.7, 50.3, 48.8, 42.0, 28.1, 20.5, and 15.2 ppm. HRMS calculated for C23H23O6, (M + H)+: 395.1489 found 395.1491.
Compound 3n 4-{(E)-[(1aR,1bR,2aR,6R,6aR,9aS,10aS,Z)-10a-methyl-4,8-dioxo-1a,1b,2a,6,6a,9a, 10,10a-octahydro-4H-3,6-(metheno) furo [3,2-c] bis (oxireno) [2,3-f:2',3'-h] [1] oxacycloundecin-7 (8H)-ylidene]methyl}benzyl acetate: 140.4 mg; isolated yield 30.1%; white solid; IR (KBr): 3,409.25, 2,359.98, 1740.10, 1,635.00, 1,115.80, and 668.60 cm−1; 1H NMR (600 MHz, DMSO-d6): δ 7.74 (d, J = 8.4 Hz, 2H), 7.63 (s, J = 3.6 Hz, 1H), 7.53 (d, J = 1.2 Hz, 1H), 7.48 (d, J = 8.4 Hz, 2H), 5.14 (s, 2H), 5.11 (s, 1H), 4.75 (m, 1H), 4.15 (m, 1H), 3.98 (d, J = 3.6 Hz, 1H), 3.40 (dd, J = 4.2, 0.6 Hz, 1H), 3.17 (s, 1H), 2.25 (t, J = 12.3 Hz, 1H), 2.10 (s, 3H), 1.85 (dd, J = 16.8, 3.6 Hz, 1H), and 1.07 (s, 3H) ppm; 13C NMR (125 MHz, DMSO-d6): δ 171.1, 170.7, 169.9, 150.3, 138.6, 137.7, 132.6, 130.9, 129.1, 128.5, 128.2, 81.7, 78.4, 65.5, 58.1, 57.8, 55.2, 50.8, 49.1, 42.5, 21.2, and 20.0 ppm. HRMS calculated for C24H23O8, (M + H)+: 439.1387 found 439.1389.
Compound 3o (1aR,1bR,2aR,6R,6aR,9aS,10aS,Z)-7-[(E)-4-(hydroxymethyl)benzylidene]-10a-methyl-1a,1b,2a,6a,7,9a,10,10a-octahydro-4H-3,6-(metheno) furo [3,2-c] bis (oxireno) [2,3-f:2',3'-h] [1] oxacycloundecine-4,8 (6H)-dione: 154.8 mg; isolated yield 37.8%; white solid; IR (KBr): 3,123.86, 2,975.22, 2,359.69, 1753.75, 1,644.59, 1,519.60, 1,376.80, 1,338.73, 1,257.45, 1,215.98, 1,167.84, 1,071.03, 1,020.90, 800.01, 738.41, and 645.26 cm−1; 1H NMR (600 MHz, DMSO-d6): δ 7.70 (d, J = 7.8 Hz, 2H), 7.62 (d, J = 3.6 Hz, 1H), 7.54 (s, 1H), 7.43 (d, J = 8.4 Hz, 2H), 5.32 (t, J = 5.4 Hz, 1H), 5.11 (s, 1H), 4.74 (m, 1H), 4.56 (d, J = 5.4 Hz, 2H), 4.14 (m, 1H), 3.97 (d, J = 3.0 Hz, 1H), 3.40 (d, J = 3.0 Hz, 1H), 3.33 (s, 1H), 2.25 (t, J = 12.3 Hz, 1H), 1.85 (dd, J = 16.8, 3.6 Hz, 1H), and 1.08 (s, 3H) ppm; 13C NMR (125 MHz, DMSO-d6): δ 170.6, 169.5, 149.8, 144.8, 137.8, 130.8, 130.3, 128.6, 127.1, 126.2, 81.1, 77.9, 62.6, 57.6, 57.4, 54.7, 50.3, 48.8, 42.0, and 20.5 ppm. HRMS calculated for C22H20O7Na, (M + Na)+: 419.1101 found 419.1094.
Compound 3p (1aR,1bR,2aR,6R,6aR,9aS, 10aS,Z)-7-[(E)-4-(tert-butyl)benzylidene]-10a-methyl-1a,1b,2a,6a,7,9a, 10,10a-octahydro-4H-3,6-(metheno) furo [3,2-c] bis (oxireno) [2,3-f:2',3'-h] [1] oxacycloundecine-4,8 (6H)-dione: 196.8 mg; isolated yield 45.1%; white solid; IR (KBr): 2,966.76, 2,359.91, 1741.72, 1,639.05, 1,607.81, 1,508.23, 1,458.24, 1,418.31, 1,364.24, 1,318.52, 1,287.23, 1,260.75, 1,188.70, 1,106.62, 1,072.97, 1,023.76, 895.73, 837.84, 795.91, and 752.83 cm−1; 1H NMR (600 MHz, DMSO-d6): δ 7.69 (d, J = 8.4 Hz, 2H), 7.60 (d, J = 3.6 Hz, 1H), 7.56 (s, 1H), 7.50 (d, J = 7.8 Hz, 2H), 5.18 (s, 1H), 4.74 (m, 1H), 4.01 (m, 1H), 3.98 (d, J = 3.0 Hz, 1H), 3.39 (d, J = 3.6 Hz, 1H), 3.17 (s, 1H), 2.27 (t, J = 12.6 Hz, 1H), 1.85 (dd, J = 16.8, 3.0 Hz, 1H), 1.31 (s, 9H), and 1.08 (s, 3H) ppm; 13C NMR (125 MHz, DMSO-d6): δ 170.7, 169.6, 152.9, 149.8, 137.7, 130.4, 129.7, 128.6, 126.6, 125.3, 81.2, 77.9, 57.6, 57.4, 54.7, 50.3, 48.8, 42.0, 34.7, 30.9, and 20.5 ppm. HRMS calculated for C25H27O6, (M + H)+: 423.1802 found 423.1804.
Compound 3q (1aR,1bR,2aR,6R,6aR,9aS, 10aS,Z)-7-[(E)-4-fluorobenzylidene]-10a-methyl-1a,1b,2a,6a,7,9a, 10,10a-octahydro-4H-3,6-(metheno) furo [3,2-c] bis (oxireno) [2,3-f:2',3'-h] [1] oxacycloundecine-4,8(6H)-dione:142.2 mg; isolated yield 35.8%; white solid; IR (KBr): 3,496.40, 2,977.92, 2,359.48, 1748.93, 1,655.73, 1,600.37, 1,508.59, 1,390.63, 1,349.85, 1,262.30, 1,218.84, 1,193.94, 1,159.43, 1,082.65, 1,022.77, 786.88, 737.41, and 678.17 cm−1; 1H NMR (600 MHz, DMSO-d6): δ 7.74 (t, J = 9.9 Hz, 2H), 7.58 (d, J = 4.8 Hz, 1H), 7.48 (s, 1H), 7.27 (t, J = 12.9 Hz, 2H), 5.03 (s, 1H), 4.70 (m, 1H), 4.06 (t, J = 5.4 Hz, 1H), 3.92 (d, J = 3.0 Hz, 1H), 3.33 (d, J = 4.8 Hz, 1H), 3.13 (s, 1H), 2.18 (t, J = 18.6 Hz, 1H), 1.80 (dd, J = 23.4, 4.2 Hz, 1H), and 1.08 (s, 3H) ppm; 13C NMR (125 MHz, DMSO-d6): δ 170.6, 169.3, 163.6, 162.0, 149.8, 136.7, 132.7, 129.2, 128.6, 128.0, 115.5, 115.4, 81.3, 77.8, 57.6, 57.3, 54.8, 50.3, 48.5, 42.0, and 20.5 ppm; 19F NMR (125 MHz, DMSO-d6): 110.3 ppm. HRMS calculated for C21H18FO6, (M + H)+: 385.1082 found 385.1083.
Compound 3r (1aR,1bR,2aR,6R,6aR,9aS,10aS,Z)-10a-methyl-7-[(E)-4-(trifluoromethyl)benzylidene]-1a,1b,2a,6a,7,9a,10,10a-octahydro-4H-3,6-(metheno) furo [3,2-c] bis (oxireno) [2,3-f:2',3'-h] [1] oxacycloundecine-4,8(6H)-dione (3r): 125.7 mg; isolated yield 28.0%; white solid; IR (KBr): 3,396.13, 2,922.14, 2,851.70, 2,359.82, 1748.13, 1,644.99, 1,420.16, 1,373.28, 1,324.64, 1,261.53, 1,225.56, 1,202.77, 1,166.46, 1,130.27, 1,067.68, 1,023.34, 840.75, 797.18, 744.85, 677.19, and 601.81 cm−1; 1H NMR (600 MHz, DMSO-d6): δ 7.93 (d, J = 7.8 Hz, 2H), 7.84 (d, J = 7.8 Hz, 2H), 7.71 (d, J = 3.0 Hz, 1H), 7.50 (s, 1H), 5.06 (s, 1H), 4.77 (m, 1H), 4.16 (t, J = 3.3 Hz, 1H), 3.96 (d, J = 2.4 Hz, 1H), 3.39 (d, J = 3.6 Hz, 1H), 3.20 (s, 1H), 2.25 (t, J = 12.3 Hz, 1H), 1.86 (dd, J = 16.2, 3.0 Hz, 1H), and 1.07 (s, 3H) ppm; 13C NMR (125 MHz, DMSO-d6): δ 170.6, 169.1, 149.8, 137.0, 135.8, 130.9, 130.8, 129.6, 128.7, 125.2, 123.2, 81.6, 77.9, 57.5, 57.3, 54.8, 50.3, 48.4, 42.0, 29.8, and 20.6 ppm; 19F NMR (125 MHz, DMSO-d6): 61.1 3) ppm. HRMS calculated for C22H18F3O6, (M + H)+: 435.1050 found 435.1051.
Compound 3s (1aR,1bR,2aR,6R,6aR,9aS,10aS,Z)-10a-methyl-7-[(E)-3-(trifluoromethyl)benzylidene]-1a,1b,2a,6a,7,9a,10,10a-octahydro-4H-3,6-(metheno) furo [3,2-c] bis (oxireno) [2,3-f:2',3'-h] [1] oxacycloundecine-4,8 (6H)-dione: 121.2 mg; isolated yield 27.0%; white solid; IR (KBr): 3,399.81, 2,918.51, 2,849.79, 2,360.10, 1748.57, 1,635.68, 1,558.17, 1,541.00, 1,507.72, 1,457.24, 1,130.23, and 668.88 cm−1; 1H NMR (600 MHz, DMSO-d6): δ 8.10 (s, 1H), 7.99 (d, J = 7.8 Hz, 1H), 7.79 (d, J = 7.2 Hz, 1H), 7.71 (m, 2H), 7.49 (s, 1H), 5.05 (s, 1H), 4.77 (m, 1H), 4.16 (m, 1H), 3.95 (s, 1H), 3.19 (s, 1H), 2.25 (t, J = 12.0 Hz, 1H), 1.86 (dd, J = 15.6, 3.0 Hz, 1H), and 1.07 (s, 3H) ppm; 13C NMR (125 MHz, DMSO-d6): δ 170.3, 169.1, 149.7, 135.9, 134.0, 133.8, 130.3, 129.4, 128.7, 126.7, 126.3, 124.9, 123.1, 81.4, 77.8, 57.5, 57.3, 54.8, 50.3, 48.4, 42.0, and 20.6 ppm; 19F NMR (125 MHz, DMSO-d6): 61.2 (3) ppm. HRMS calculated for C22H18F3O6, (M + H)+: 435.1050 found 435.1051.
Compound 3t (1aR,1bR,2aR,6R,6aR,9aS,10aS,Z)-10a-methyl-7-[(E)-2-(trifluoromethyl)benzylidene]-1a,1b,2a,6a,7,9a,10,10a-octahydro-4H-3,6-(metheno) furo [3,2-c] bis (oxireno) [2,3-f:2',3'-h] [1] oxacycloundecine-4,8 (6H)-dione: 67.3 mg; isolated yield 15.0%; white solid; IR (KBr): 3,566.03, 2,988.58, 2,360.09, 1773.14, 1,665.68, 1,488.49, 1,455.59, 1,352.39, 1,318.49, 1,229.70, 1,203.73, 1,165.16, 1,109.05, 1,021.95, 770.30, and 679.70 cm−1; 1H NMR (600 MHz, DMSO-d6): δ 7.95 (d, J = 7.8 Hz, 1H), 7.82 (m, 2H), 7.67 (m, 2H), 7.47 (d, J = 1.2 Hz, 1H), 4.89 (m, 1H), 4.81 (m, 1H), 4.00 (m, 1H), 3.91 (d, J = 3.6 Hz, 1H), 3.37 (dd, J = 4.8, 1.2 Hz, 1H), 3.18 (s, 1H), 2.25 (t, J = 12.3 Hz, 1H), 1.86 (dd, J = 17.4, 4.2 Hz, 1H), and 1.02 (s, 3H) ppm; 13C NMR (125 MHz, DMSO-d6): δ 170.4, 168.7, 149.8, 132.8, 132.7, 132.3, 131.2, 130.0, 128.7, 127.0, 125.9, 125.0, 123.7, 81.7, 77.6, 57.3, 57.2, 54.9, 50.1, 48.1, 42.0, and 20.6 ppm; 19F NMR (125 MHz, DMSO-d6): 58.9 (3) ppm. HRMS calculated for C22H18F3O6, (M + H)+: 435.1050 found 435.1051.
Compound 3u (1aR,1bR,2aR,3Z,6R,6aR,7E,9aS,10aS)-10a-methyl-7-(thiophen-3-ylmethylene)-1a,1b,2a,6a,7,9a,10,10a-octahydro-4H-3,6-(metheno) furo [3,2-c] bis (oxireno) [2,3-f:2',3'-h] [1] oxacycloundecine-4,8 (6H)-dione: 145.5 mg; isolated yield 37.8%; white solid; IR (KBr): 3,564.51, 2,932.29, 2,359.82, 1750.94, 1,654.39, 1,508.66, 1,418.21, 1,390.71, 1,262.38, 1,231.78, 1,198.39, 1,083.17, 1,022.15, 898.23, 805.15, and 677.67 cm−1; 1H NMR (600 MHz, DMSO-d6): δ 8.16 (d, J = 3.0 Hz, 1H), 7.71 (q, J = 2.6 Hz, 1H), 7.63 (d, J = 3.6 Hz, 1H), 7.60 (m, 2H), 5.27 (m, 1H), 4.73 (m, 1H), 3.99 (m, 2H), 3.40 (d, J = 3.6 Hz, 1H), 3.17 (s, 1H), 2.23 (t, J = 12.6 Hz, 1H), 1.83 (dd, J = 16.8, 3.6 Hz, 1H), and 1.08 (s, 3H) ppm; 13C NMR (125 MHz, DMSO-d6): δ 170.5, 169.7, 149.9, 134.4, 131.8, 131.1, 128.8, 128.6, 126.9, 125.6, 81.4, 77.8, 57.7, 57.4, 54.7, 50.3, 48.8, 42.0, and 20.5 ppm. HRMS calculated for C19 H17O6S, (M + H)+: 373.0740 found 373.0742.
Compound 4 (3aR,4R,9aR,10aS, 11aS,Z)-10a-methyl-3-[(E)-4-methylbenzylidene]-3a,4,8,9,9a,10a, 11,11a-octahydro-6H-4,7-(metheno) furo [3,2-c] oxireno [2,3-f] [1] oxacycloundecine-2,6 (3H)-dione: 110.0 mg; isolated yield 41.5%; white solid; IR (KBr): 3,083.01, 2,967.20, 2,359.81, 1749.00, 1,636.71, 1,603.52, 1,509.84, 1,452.20, 1,360.58, 1,314.44, 1,193.36, 1,178.89,1,106.03, 1,033.82, 1,012.46, 817.58, and 473.69 cm−1; 1H NMR (600 MHz, DMSO-d6): δ 7.73 (s, 1H), 7.64 (d, J = 8.4 Hz, 2H), 7.57 (d, J = 3.6 Hz, 1H), 7.33 (d, J = 7.8 Hz, 2H), 5.01 (s, 1H), 4.80 (m, 1H), 4.16 (m, 1H), 2.87 (dd, J = 13.8, 2.4 Hz, 1H), 2.54 (m, 1H), 2.45 (m, 1H), 2.36 (s, 3H), 2.22 (t, J = 12.6 Hz, 1H), 2.02 (m, 1H), 1.82 (dd, J = 17.4, 4.2 Hz, 1H), 1.29 (m, 1H), and 1.14 (s, 3H) ppm; 13C NMR (125 MHz, DMSO-d6): δ 172.5, 170.1, 151.7, 140.6, 137.6, 131.0, 130.9, 130.3, 129.8, 126.8, 80.0, 78.2, 61.0, 57.2, 49.1, 43.5, 23.3, 21.7, 21.5, and 20.0 ppm. HRMS calculated for C22 H23O5, (M + H)+: 367.1540 found 367.1541.
Mikanolide Derivatives Possess Antileukemic Action
Using MTT assay, the cell viabilities of mikanolide derivatives were assessed. Paclitaxel and imatinib served as positive controls. Mikanolide derivatives were treated at different concentrations (0.0075–10 μmol/L) in CEM-C7H2, HEL, and K562 cells. The results exhibited significant antileukemic activity in the selected human leukemic cells. Notably, the IC50 value of mikanolide derivatives in 72 h showed good antiproliferative activity against the three leukemia cell lines (Figure 1B). However, among the 22 mikanolide derivatives, the compound 3g exhibited improved activity against the three leukemic cell lines.
Compound 3g Potently Impedes Cell Proliferation in Leukemia Cells
The inhibition rate of 3g in K562 cells was effective in a dose- and time-dependent manner. Furthermore, 3g was assessed vs. normal hepatocyte cells, HL7702 (Figure 2A). When normal cells HL7702 were incubated with the 3g compound for 72 h, there were no variations in the cellular viability. But this compound showed a prominent decrease in cell viability to the human chronic myeloid leukemia cells K562 at the concentration of 0.43 ± 0.05 µM in 72 h. The results represented that 3g had a considerable antileukemic property in K562 cells (Figures 1B, 2A, Supplementary Figure S1), compared to the positive controls, imatinib (Supplementary Figure S2), and paclitaxel.
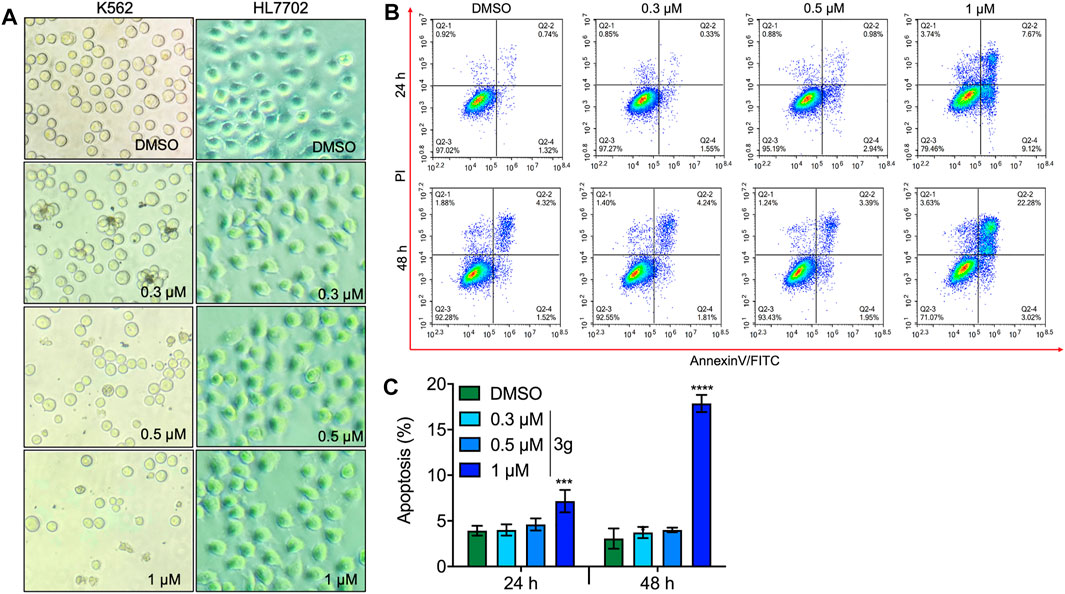
FIGURE 2. 3g induces apoptosis in both dose- and time-dependent manners. (A) Dose-dependent cell morphology variations in K562 and normal hepatocyte cells at the indicated 3g concentrations (40X), for 72 h. (B) Flow cytometric analysis of apoptosis caused by indicated doses of 3g treatment at 24 and 48 h in K562 cells. (C) Densitometry of the apoptotic cells percentage at an indicated time interval of 3g incubation. Data are represented as mean ± SD (n = 3; ***p < 0.001 and ****p < 0.0001 vs. control).
3g Triggers Apoptosis in Leukemia Cells
Different concentrations of 3g were treated in K562, HEL, and CEM-C7H2 cells after 24 and 48 h of incubation, to analyze the apoptotic effect using flow cytometry. The study revealed a significant apoptotic rate at higher concentrations in 24 and 48 h treatment of 3g (Figures 2B,C; Supplementary Figures S3, S4), compared to non-tumor lineage cells (Supplementary Figure S5).
3g Induces Cell Cycle Arrest in Leukemia Cells Both Dose- and Time-Dependently
The cell cycle was analyzed to facilitate the apoptosis study. The study revealed a significant effect on the G2/M phase of the cell cycle, both dose- and time-dependently after 3g treatment in K562 cells (Figures 3A,B) and in the G1/G2 and S phases in HEL and CEM-C7H2 cells (Supplementary Figures S6, S7), respectively. There were no significant cell arrests in non-tumor lineage cells (Supplementary Figure S8). To reconfirm the hypothesis, Western blot was performed for cell cycle markers. There was a dose-dependent significant reduction in protein expressions of p-CDC25C and cyclin B1, with increased expression of P27, and P21, after 3g post-treatment (Figure 3C).
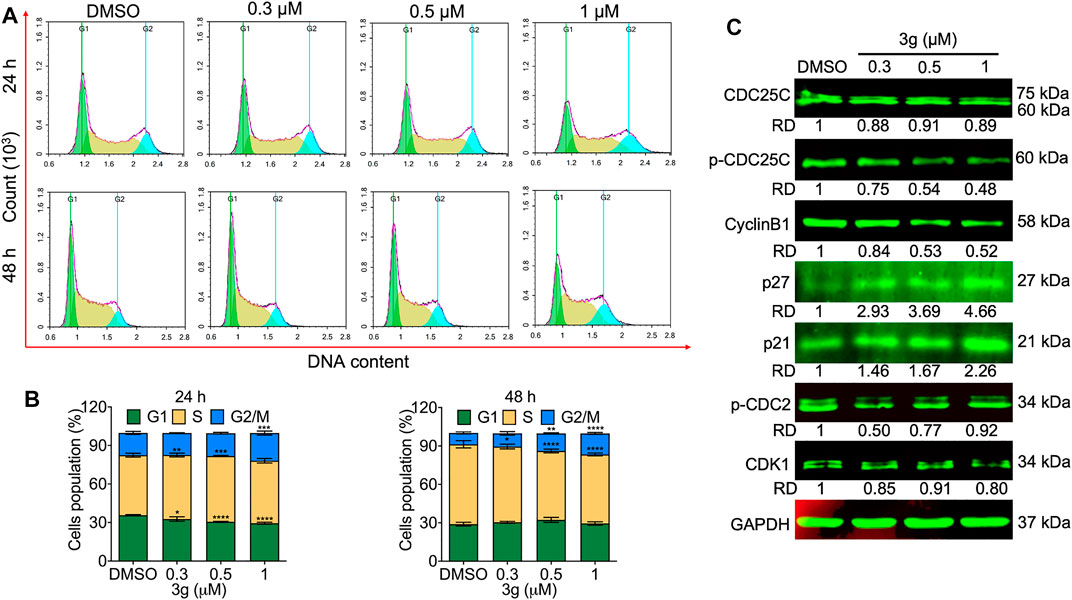
FIGURE 3. 3g arrests the cell cycle in K562 cells. (A) Cell cycle arrest evaluation by flow cytometry after 24 and 48 h of 3g treatment. (B) Densitometry plots depicting the variations in the stages of cell cycle arrest. (C) Protein expression levels of indicated proteins after 48 h of 3g incubation. Data are represented as mean ± SD (n = 3; *p < 0.05, **p < 0.01, ***p < 0.001, and ****p < 0.0001 vs. control).
3g Engenders Mitochondrial and DNA Damage in K562 Cells
To study whether the K562 cell’s mitochondria were affected by 3g treatment, we analyzed the loss of mitochondrial membrane potential (MMP). The results showed an increased green fluorescence after 3g treatment compared with control cells (Figures 4A,B). The reduced MMP suggests the damage to the chondriosome, by releasing cytochrome C and mitochondrial damage. Excitingly, the Western blot results further confirmed that the mitochondrial damage was accomplished by the 3g treatment in a dose-dependent manner, by decreased expression of Bcl-2, Bcl-XL, caspase 3, caspase 9, c-FLIPL, c-FLIPS, and cleaved BID. While, there was an increased expression of caspase 8, cleaved caspase 3, cleaved caspase 9, Bim, BAD, and BID proteins (Figure 4C). However, the Hoechst 33,258 staining revealed DNA damage, detected after K562 cells were treated with different concentrations of 3g. As shown in Figures 5A,B, the DNA damage levels were increased in a dose-dependent manner, suggesting cellular damage.
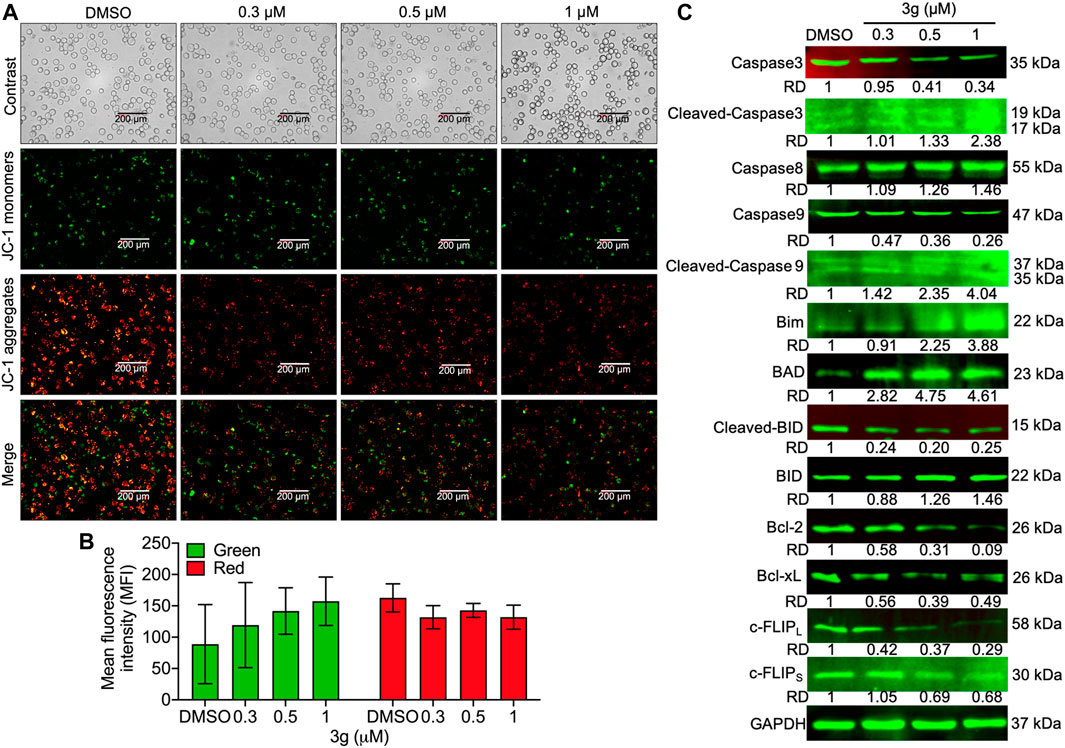
FIGURE 4. Mitochondrial damage after 3g treatment in K562 cells. (A) Photomicrograph of JC-1 monomers and aggregates after 3g incubation at 24 h. (B) Quantification of the red/green levels showing extent of mitochondrial damage. (C) Protein expression levels of indicated proteins at 48 h of 3g treatment.
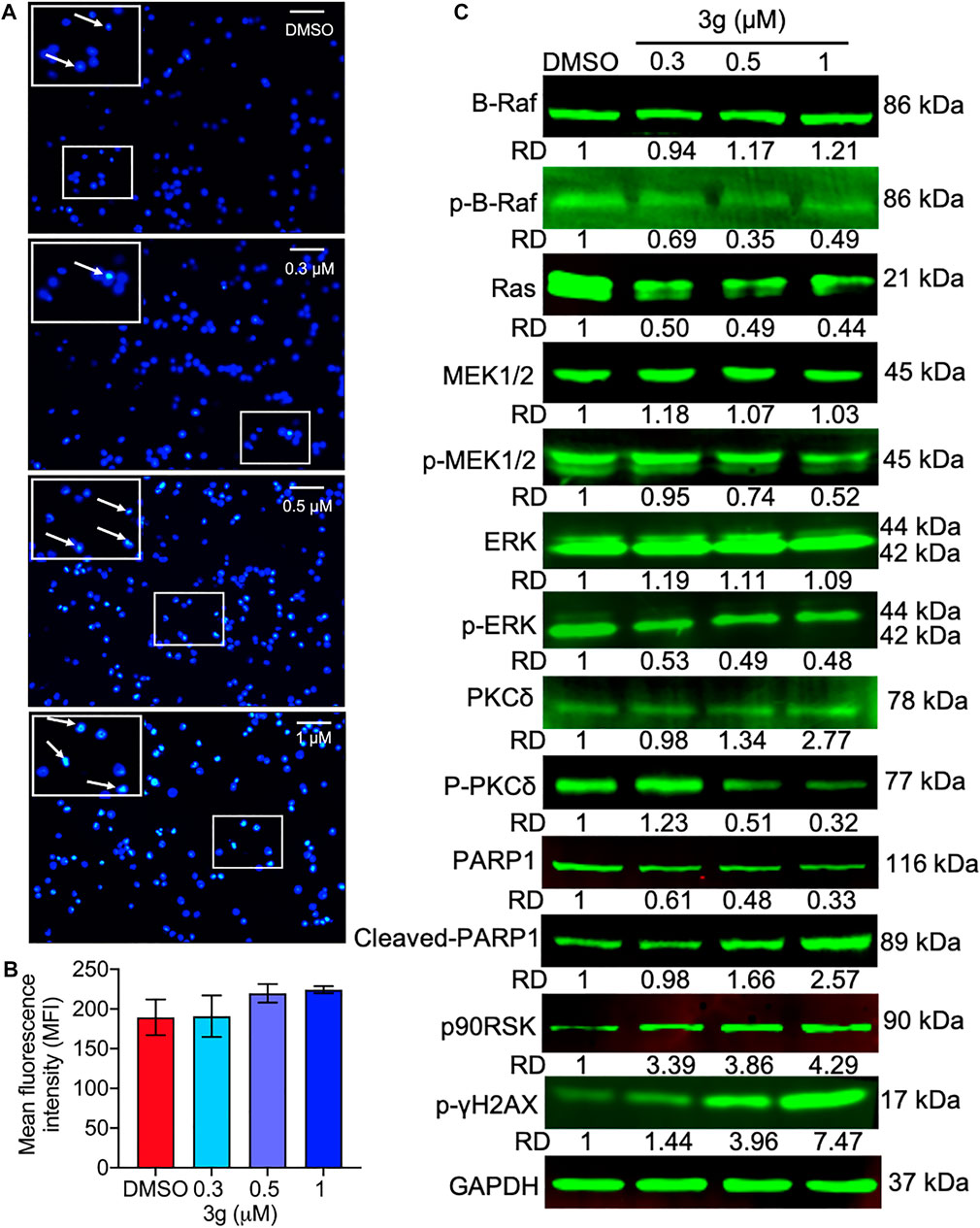
FIGURE 5. DNA damage and the Ras/Raf/Mek/Erk pathway alterations induced by 3g in K562 cells. (A) Photomicrograph of DNA damage (in boxes) at indicated concentrations of 3g treatment. (B) Quantification of the fluorescence levels showing extent of DNA damage. (C) Protein expression levels of indicated proteins after 3g treatment at 48 h.
3g Selectively Targets the Ras/Raf/MEK/ERK Pathway in K562 Cells
The effect of 3g on Ras- and Raf-related molecules was analyzed by Western blot. The protein expressions of B-Raf, PKCδ, cleaved PARP1, p90RSK, and p-γH2AX were significantly increased. On the other hand, the expression levels of p-B-Raf, RAS, p-MEK1/2, p-PKCδ, PARP1, and p-ERK were significantly reduced, compared to the control. There were no variations in the expression levels of other molecules (Figure 5C). To further facilitate the specific molecular target of 3g, AutoDock was performed. The in silico study revealed that the 3g compound is a potent inhibitor of ERK (Figures 6A–D). With a considerable amount of binding energy and ligand efficiency such as U0126, 3g proved to inhibit functional p-ERK, which was elucidated by AutoDock (Figure 6E).
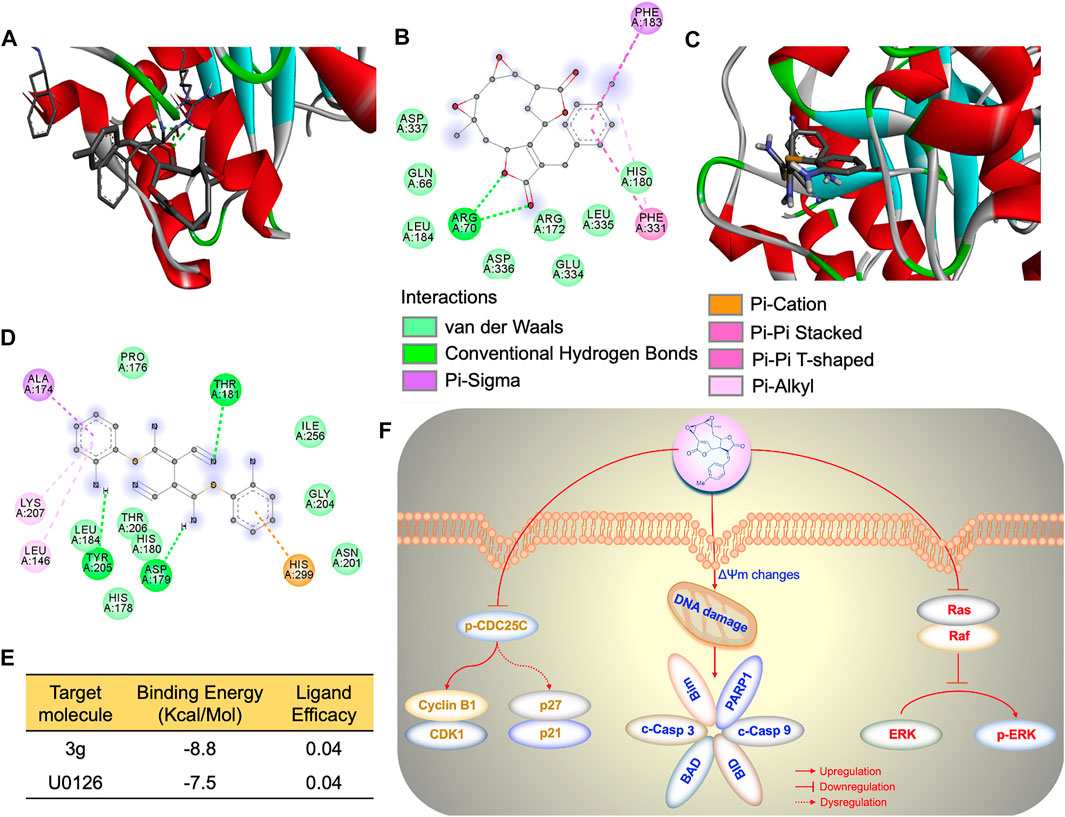
FIGURE 6. 3g cause p-ERK inhibition. (A) AutoDock 3D image depicting the interactive site between U0126 and ERK. (B) AutoDock 2D amino acid interactive site image between U0126 and ERK. (C) AutoDock 3D image depicting the interactive site between 3g and ERK. (D) AutoDock 2D amino acid interactive site image between 3g and ERK. (E) Binding energy and ligand efficiency between ERK and ligands. (F) Molecular mechanism of 3g as an antileukemic agent.
Discussion
Leukemia is described by uncontrolled proliferation of blood cells, with limited cell death. The existence of leukemia is severe, generally affecting almost all ages of people. Thus, a strategy or drug of choice to treat the disease is in high demand. In this investigation, we prepared 22 novel mikanolide derivatives using heck reaction to study their antileukemic activities in human leukemia cell lines.
Biological agents and their derivatives are widely in the preclinical and clinical sectors for cancer chemotherapies. These agents have mainly been investigated for their properties to initiate apoptosis (Hanahan and Weinberg, 2000). Our investigation has indicated that mikanolide derivatives have cytotoxic effects on leukemic cells such as CEM-C7H2, HEL, and K562. The present study has proven that mikanolide derivatives have appropriate antileukemic activities. The selected mikanolide derivative compound, 3g inspired us to focus on K562 cells to predict the specific molecular antileukemic mechanism.
Apoptosis is a multi-stage process. Antiapoptosis is a key factor in many types of cancer. Protein kinases participate in the management of early stages of apoptosis by phosphorylating key apoptotic proteins or in later events by enacting downstream caspases. CDC25C can monitor G2/M phase progression and DNA damage repair. It is a specific phosphatase family cyclin that can trigger the cyclin B1/CDK1 complexes. These complexes can manage the cells inflowing with mitosis by regulating G2/M progression. They play as a checkpoint for protein regulation and DNA damage. The regulation of CDC25C is closely associated with tumorigenesis and is considered a possible target for cancer conduct (Liu et al., 2020). Likewise, dysfunction of p27 has been reported in human cancers, resulting from p27 phosphorylation-dependent protein degradation and cell cycle arrest (Sun et al., 2016). The p21 can regulate p53-dependent and independent cancer pathways, consenting to DNA repair and sponsoring tumorigenesis (Al Bitar and Gali-Muhtasib, 2019). In our investigation, a most effective mikanolide derivative, 3g arrested the K652 cells by entering the G2/M phase by increasing the expression of CDK1/cyclin B1. This blockage entry of the M phase by the mikanolide derivative is through inhibiting the activity of CDK1/cyclin B1 viz decreased phosphorylation of CDC25C or increased expression of p27 and p21 as explained earlier.
However, cancer cells have defective or uncontrolled cellular proliferation (Fulda et al., 2010; Indran et al., 2011). This apoptotic machinery may be either mitochondrial-dependent or -independent (Adams, 2003; Redza-Dutordoir and Averill-Bates, 2016). In our study, mikanolide derivative induced DNA and mitochondrial damage. So, disturbances in mitochondrial function can lead to cell impairment (Dai et al., 2014), 3g could downregulate cleaved caspase 3 and 9 and upregulate the Bim, BAD, and BID proteins, ensuring cellular mortality. Moreover, the cells treated with 3g decreased the expression of c-FLIP, Bcl-2, and Bcl-xL. c-FLIP regulates the apoptosis by attenuating autophagy by directly acting on the autophagy machinery by inhibiting autophagosome formation. The upregulation of c-FLIP has been found in various tumor types, and its silencing has been shown to restore apoptosis triggered by cytokines and various chemotherapeutic agents (Bagnoli et al., 2010). Similarly, 3g reduced the expression levels of c-FLIP dose-dependently. The nascent DNA responds to its damage by disrupting the histone phosphorylation in H2AX on its ser4 residue of C-terminus establishing γH2AX (Rogakou et al., 1998). This cellular response gets augmented about 20 min after the initiation of DNA damage (Redon et al., 2002). Thus, this molecule is considered as a powerful maker of DNA that breaks in the cancer cells experimentally. Likewise, the study elucidated that the drug 3g conferred DNA damage of K562 cell lysates by increasing the expression of γH2AX.
ERK is responsible for the broadcast of antiapoptotic signals from membrane-bound receptors (James et al., 2011). Chemotherapeutic drugs, frequently used in leukemia therapy, often inactivate this pathway. Inhibition of Ras (or Ras-related molecules), Raf, MEK, and ERK may prove useful in leukemia treatment. These observations have boosted the pharmaceutical industry to improve inhibitors that direct key factors of this pathway and are currently in clinical trials (McCubrey et al., 2007; McCubrey et al., 2008b; McCubrey et al., 2008b; Steelman et al., 2008; Steelman et al., 2010). The protein kinase C family of serine-threonine kinases is activated by diverse stimuli and participates in cellular processes such as growth, differentiation, and apoptosis (Abrams et al., 2010). The 90 kDa ribosomal S6 kinase (RSK) family of proteins is highly conserved Ser/Thr kinases that can regulate the downstream effectors of the ERK/MAPK trail (Hug and Sarre, 1993). Likewise, the 3g treatment, dose-dependently reduced the protein expression of p-ERK molecules, thus availing as a possible treatment for leukemia.
The MEK/ERK cascade sends signals to cell surface receptors and transcription factors that control gene expression. Moreover, additional signal transduction pathways usually interrelate with the Raf/MEK/ERK pathway to regulate its action, positively or negatively, by varying the phosphorylation of downstream targets (Anjum and Blenis, 2008), including phosphorylation of ERK (Chang et al., 2003; Ahearn et al., 2011). Thus, inhibitors of ERK pathway molecules are currently in clinical trials as anticancer agents (Knight and Irving, 2014). In line with it, in our study, mikanolide derivative 3g could significantly downregulate p-ERK. MEK is associated with apoptosis and cell cycle and the activation of P38, which could cause the p-CDC25C to dephosphorylate (Xiaofei and Kowalik, 2014), arresting K562 cells in the G2/M phase. These studies elucidated the inhibition of the pathway that could revive apoptosis, suggesting its capability to act as an apoptotic agent via p-ERK inhibition. Moreover, the in silico AutoDock studies discovered that ERK is inhibited by significant binding energies that might appeal for the downregulation and effect of 3g in the pathway.
Conclusion
To conclude, our study revealed that mikanolide derivatives, especially 3g, could cross the K562 cells plasma membrane when administered an ideal dose. The 3g reduced the phosphorylation of CDC25C by significantly diminishing the expression of cyclin B1/CDK1, causing dysregulation of P27/P21, triggering cell cycle arrest, and leading to cellular apoptosis. Moreover, 3g could induce mitochondrial/DNA damage, leading to activation of caspase 3/9 and increased expression of cellular Bim, BAD, and BID proteins-mediated apoptosis. Similarly, 3g could inhibit phosphorylation of ERK, causing the dysregulation of the Ras/Raf/MEK/ERK pathway, promoting cell death of K562 cells (Figure 6F). These results thus suggest 3g as a novel potent chemotherapeutic agent for leukemia.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Author Contributions
QR, KX, KV, LH, JS, JY, JQ, and YH contributed to the conception, design of the study, as well as data acquisition and interpretation. QR, BG, and KV were involved in data and statistical analysis. QR drafted the manuscript. YL, BG, YML, and SL reviewed the manuscript critically. YML supervised, conceived, and designed the study. All authors contributed to the interpretation of the findings, reviewed, edited, and approved the final manuscript.
Funding
This research was supported financially by the grants from the National Natural Science Foundation of China (81872772, 81960546, U1812403, 82160808, 82160813, 81760630), Guizhou Provincial Natural Science Foundation (QKHPTRC[2020]5008, QKHJC-ZK[2022]YB293, QKHPTRC[2019]5627, QKHJC-ZK[2021]YB569, QKHZC[2019]2762, QKHJC-ZK[2022]YB297, QKHJC[2020]1Y033), Foundation of State Key Laboratory of Functions and Applications of Medicinal Plants (FAMP201901K, QZYY-019-022), the State Key Laboratory of Drug Research (SIMM2105KF-15), and the Guizhou Medical University Research Grants N21025 (fund for BG) and RN21024 (fund for KV).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors, and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fphar.2022.809551/full#supplementary-material
References
Abrams, S. L., Steelman, L. S., Shelton, J. G., Wong, E. W., Chappell, W. H., Bäsecke, J., et al. (2010). The Raf/MEK/ERK Pathway Can Govern Drug Resistance, Apoptosis and Sensitivity to Targeted Therapy. Cell Cycle 9 (9), 1781–1791. doi:10.4161/cc.9.9.11483
Adams, J. M. (2003). Ways of Dying: Multiple Pathways to Apoptosis. Genes Dev. 17 (20), 2481–2495. doi:10.1101/gad.1126903
Ahearn, I. M., Haigis, K., Bar-Sagi, D., and Philips, M. R. (2011). Regulating the Regulator: post-translational Modification of RAS. Nat. Rev. Mol. Cell Biol 13 (1), 39–51. doi:10.1038/nrm3255
Ahmed, M., Rahman, M. T., Alimuzzaman, M., and Shilpi, J. A. (2001). Analgesic Sesquiterpene Dilactone from Mikania Cordata. Fitoterapia 72 (8), 919–921. doi:10.1016/s0367-326x(01)00318-5
Al Bitar, S., and Gali-Muhtasib, H. (2019). The Role of the Cyclin Dependent Kinase Inhibitor P21cip1/waf1 in Targeting Cancer: Molecular Mechanisms and Novel Therapeutics. Cancers (Basel) 11 (10), 1475. doi:10.3390/cancers11101475
Anjum, R., and Blenis, J. (2008). The RSK Family of Kinases: Emerging Roles in Cellular Signalling. Nat. Rev. Mol. Cell Biol 9 (10), 747–758. doi:10.1038/nrm2509
Bagnoli, M., Canevari, S., and Mezzanzanica, D. (2010). Cellular FLICE-Inhibitory Protein (C-FLIP) Signalling: a Key Regulator of Receptor-Mediated Apoptosis in Physiologic Context and in Cancer. Int. J. Biochem. Cell Biol 42, 210–213. doi:10.1016/j.biocel.2009.11.015
Ben-Neriah, Y., Daley, G. Q., Mes-Masson, A. M., Witte, O. N., and Baltimore, D. (1986). The Chronic Myelogenous Leukemia-specific P210 Protein Is the Product of the Bcr/abl Hybrid Gene. Science 233 (4760), 212–214. doi:10.1126/science.3460176
Bohlmann, F., Tsankova, E., King, R. M., and Robinson, H. (1984). Five Mikanolide Derivatives from Mikania Cynanchifolia and Their Biogenetic Relationships. Phytochemistry 23 (5), 1099–1101. doi:10.1016/S0031-9422(00)82617-5
Branford, S., Rudzki, Z., Walsh, S., Grigg, A., Arthur, C., Taylor, K., et al. (2002). High Frequency of point Mutations Clustered within the Adenosine Triphosphate-Binding Region of BCR/ABL in Patients with Chronic Myeloid Leukemia or Ph-Positive Acute Lymphoblastic Leukemia Who Develop Imatinib (STI571) Resistance. Blood 99 (9), 3472–3475. doi:10.1182/blood.v99.9.3472
Chang, F., Steelman, L. S., Lee, J. T., Shelton, J. G., Navolanic, P. M., Blalock, W. L., et al. (2003). Signal Transduction Mediated by the Ras/Raf/MEK/ERK Pathway from Cytokine Receptors to Transcription Factors: Potential Targeting for Therapeutic Intervention. Leukemia 17 (7), 1263–1293. doi:10.1038/sj.leu.2402945
Dai, D. F., Chiao, Y. A., Marcinek, D. J., Szeto, H. H., and Rabinovitch, P. S. (2014). Mitochondrial Oxidative Stress in Aging and Healthspan. Longev Healthspan 3, 6. doi:10.1186/2046-2395-3-6
Facey, P. C., Pascoe, K. O., Porter, R. B., and Jones, A. D. (1999). Investigation of Plants Used in Jamaican Folk Medicine for Anti-bacterial Activity. J. Pharm. Pharmacol. 51 (12), 1455–1460. doi:10.1211/0022357991777119
Fulda, S., Galluzzi, L., and Kroemer, G. (2010). Targeting Mitochondria for Cancer Therapy. Nat. Rev. Drug Discov. 9 (6), 447–464. doi:10.1038/nrd3137
Gajendran, B., Durai, P., Madhu Varier, K., and Chinnasamy, A. (2020a). A Novel Phytosterol Isolated from Datura Inoxia, RinoxiaB Is a Potential Cure colon Cancer Agent by Targeting BAX/Bcl2 Pathway. Bioorg. Med. Chem. 28 (2), 115242. doi:10.1016/j.bmc.2019.115242
Gajendran, B., Varier, K. M., Liu, W., Wang, C., Sample, K. M., Zacksenhaus, E., et al. (2020b). A C21-Steroidal Derivative Suppresses T-Cell Lymphoma in Mice by Inhibiting SIRT3 via SAP18-SIN3. Commun. Biol. 3 (1), 732. doi:10.1038/s42003-020-01458-3
Giordano, O. S., Guerreiro, E., Pestchanker, M. J., Guzman, J., Pastor, D., and Guardia, T. (1990). The Gastric Cytoprotective Effect of Several Sesquiterpene Lactones. J. Nat. Prod. 53 (4), 803–809. doi:10.1021/np50070a004
Gutierrez, A. B., Oberti, J. C., Kulanthaivel, P., and Herz, W. (1985). Sesquiterpene Lactones and Diterpenes from Mikania Periplocifolia. Phytochemistry 24, 2967–2971. doi:10.1016/0031-9422(85)80037-6
Guzman, M. L., Rossi, R. M., Karnischky, L., Li, X., Peterson, D. R., Howard, D. S., et al. (2005). The Sesquiterpene Lactone Parthenolide Induces Apoptosis of Human Acute Myelogenous Leukemia Stem and Progenitor Cells. Blood 105 (11), 4163–4169. doi:10.1182/blood-2004-10-4135
Hanahan, D., and Weinberg, R. A. (2000). The Hallmarks of Cancer. Cell 100 (1), 57–70. doi:10.1016/s0092-8674(00)81683-9
Hu, M., Varier, K. M., Li, Z., Qin, X., Rao, Q., Song, J., et al. (2021). A Natural Acylphloroglucinol Triggered Antiproliferative Possessions in HEL Cells by Impeding STAT3 Signaling and Attenuating Angiogenesis in Transgenic Zebrafish Model. Biomed. Pharmacother. 141, 111877. doi:10.1016/j.biopha.2021.111877
Hug, H., and Sarre, T. F. (1993). Protein Kinase C Isoenzymes: Divergence in Signal Transduction? Biochem. J. 291 (Pt 2), 329–343. doi:10.1042/bj2910329
Indran, I. R., Tufo, G., Pervaiz, S., and Brenner, C. (2011). Recent Advances in Apoptosis, Mitochondria and Drug Resistance in Cancer Cells. Biochim. Biophys. Acta 1807 (6), 735–745. doi:10.1016/j.bbabio.2011.03.010
Jackson, P., and Baltimore, D. (1989). N-terminal Mutations Activate the Leukemogenic Potential of the Myristoylated Form of C-Abl. EMBO J. 8 (2), 449–456. doi:10.1002/j.1460-2075.1989.tb03397.x
Knight, T., and Irving, J. A. (2014). Ras/Raf/MEK/ERK Pathway Activation in Childhood Acute Lymphoblastic Leukemia and its Therapeutic Targeting. Front. Oncol. 4, 160. doi:10.3389/fonc.2014.00160
Li, Y., Li, J., Li, Y., Wang, X. X., and Cao, A. C. (2013). Antimicrobial Constituents of the Leaves of Mikania Micrantha H. B. K. PLoS One 8 (10), e76725. doi:10.1371/journal.pone.0076725
Liu, J., Zhang, Y., Huang, H., Lei, X., Tang, G., Cao, X., et al. (2021). Recent Advances in Bcr‐Abl Tyrosine Kinase Inhibitors for Overriding T315I Mutation. Chem. Biol. Drug Des. 97 (3), 649–664. doi:10.1111/cbdd.13801
Liu, K., Zheng, M., Lu, R., Du, J., Zhao, Q., Li, Z., et al. (2020). The Role of CDC25C in Cell Cycle Regulation and Clinical Cancer Therapy: a Systematic Review. Cancer Cell Int 20, 213. doi:10.1186/s12935-020-01304-w
Ma, L., Zhang, J., Wang, X., Yang, J., Guo, L., Wang, X., et al. (2021). Design and Synthesis of Diosgenin Derivatives as Apoptosis Inducers through Mitochondria-Related Pathways. Eur. J. Med. Chem. 217, 113361. doi:10.1016/j.ejmech.2021.113361
Maiti, A., Franquiz, M. J., Ravandi, F., Cortes, J. E., Jabbour, E. J., Sasaki, K., et al. (2020). Venetoclax and BCR-ABL Tyrosine Kinase Inhibitor Combinations: Outcome in Patients with Philadelphia Chromosome-Positive Advanced Myeloid Leukemias. Acta Haematol. 143 (6), 567–573. doi:10.1159/000506346
McCubrey, J. A., Abrams, S. L., Ligresti, G., Misaghian, N., Wong, E. W., Steelman, L. S., et al. (2008a). Involvement of P53 and Raf/MEK/ERK Pathways in Hematopoietic Drug Resistance. Leukemia 22 (11), 2080–2090. doi:10.1038/leu.2008.207
McCubrey, J. A., Steelman, L. S., Abrams, S. L., Bertrand, F. E., Ludwig, D. E., Bäsecke, J., et al. (2008b). Targeting Survival Cascades Induced by Activation of Ras/Raf/MEK/ERK, PI3K/PTEN/Akt/mTOR and Jak/STAT Pathways for Effective Leukemia Therapy. Leukemia 22 (4), 708–722. doi:10.1038/leu.2008.27
McCubrey, J. A., Steelman, L. S., Chappell, W. H., Abrams, S. L., Wong, E. W., Chang, F., et al. (2007). Roles of the Raf/MEK/ERK Pathway in Cell Growth, Malignant Transformation and Drug Resistance. Biochim. Biophys. Acta 1773 (8), 1263–1284. doi:10.1016/j.bbamcr.2006.10.001
McCubrey, J. A., Steelman, L. S., Kempf, C. R., Chappell, W. H., Abrams, S. L., Stivala, F., et al. (2011). Therapeutic Resistance Resulting from Mutations in Raf/MEK/ERK and PI3K/PTEN/Akt/mTOR Signaling Pathways. J. Cell Physiol 226 (11), 2762–2781. doi:10.1002/jcp.22647
Morris, G. M., Huey, R., Lindstrom, W., Sanner, M. F., Belew, R. K., Goodsell, D. S., et al. (2009). AutoDock4 and AutoDockTools4: Automated Docking with Selective Receptor Flexibility. J. Comput. Chem. 30 (16), 2785–2791. doi:10.1002/jcc.21256
Musacchio, A., Saraste, M., and Wilmanns, M. (1994). High-resolution crystal Structures of Tyrosine Kinase SH3 Domains Complexed with Proline-Rich Peptides. Nat. Struct. Biol. 1 (8), 546–551. doi:10.1038/nsb0894-546
Nakshatri, H., Rice, S. E., and Bhat-Nakshatri, P. (2004). Antitumor Agent Parthenolide Reverses Resistance of Breast Cancer Cells to Tumor Necrosis Factor-Related Apoptosis-Inducing Ligand through Sustained Activation of C-Jun N-Terminal Kinase. Oncogene 23 (44), 7330–7344. doi:10.1038/sj.onc.1207995
Overduin, M., Rios, C. B., Mayer, B. J., Baltimore, D., and Cowburn, D. (1992). Three-dimensional Solution Structure of the Src Homology 2 Domain of C-Abl. Cell 70 (4), 697–704. doi:10.1016/0092-8674(92)90437-h
Pickman, A. K. (1986). Biological Activity of Sesquiterpene Lactones. Biochem. Syst. Ecol. 14, 255–281. doi:10.1016/0305-1978(86)90101-8
Redon, C., Pilch, D., Rogakou, E., Sedelnikova, O., Newrock, K., and Bonner, W. (2002). Histone H2A Variants H2AX and H2AZ. Curr. Opin. Genet. Dev. 12 (2), 162–169. doi:10.1016/s0959-437x(02)00282-4
Redza-Dutordoir, M., and Averill-Bates, D. A. (2016). Activation of Apoptosis Signalling Pathways by Reactive Oxygen Species. Biochim. Biophys. Acta 1863 (12), 2977–2992. doi:10.1016/j.bbamcr.2016.09.012
Rogakou, E. P., Pilch, D. R., Orr, A. H., Ivanova, V. S., and Bonner, W. M. (1998). DNA Double-Stranded Breaks Induce Histone H2AX Phosphorylation on Serine 139. J. Biol. Chem. 273 (10), 5858–5868. doi:10.1074/jbc.273.10.5858
Steelman, L. S., Abrams, S. L., Shelton, J. G., Chappell, W. H., Bäsecke, J., Stivala, F., et al. (2010). Dominant Roles of the Raf/MEK/ERK Pathway in Cell Cycle Progression, Prevention of Apoptosis and Sensitivity to Chemotherapeutic Drugs. Cell Cycle 9 (8), 1629–1638. doi:10.4161/cc.9.8.11487
Steelman, L. S., Abrams, S. L., Whelan, J., Bertrand, F. E., Ludwig, D. E., Bäsecke, J., et al. (2008). Contributions of the Raf/MEK/ERK, PI3K/PTEN/Akt/mTOR and Jak/STAT Pathways to Leukemia. Leukemia 22 (4), 686–707. doi:10.1038/leu.2008.26
Sun, C., Wang, G., Wrighton, K. H., Lin, H., Songyang, Z., Feng, X. H., et al. (2016). Regulation of p27Kip1 Phosphorylation and G1 Cell Cycle Progression by Protein Phosphatase PPM1G. Am. J. Cancer Res. 6 (10), 2207–2220.
Vaidya, S., Vundinti, B. R., Shanmukhaiah, C., Chakrabarti, P., and Ghosh, K. (2015). Evolution of BCR/ABL Gene Mutation in CML Is Time Dependent and Dependent on the Pressure Exerted by Tyrosine Kinase Inhibitor. PloS one 10 (1), e0114828. doi:10.1371/journal.pone.0114828
Varier, K. M., and Sumathi, T. (2019). Hinokitiol Offers Neuroprotection against 6-OHDA-Induced Toxicity in SH-SY5Y Neuroblastoma Cells by Downregulating mRNA Expression of MAO/α-Synuclein/LRRK2/PARK7/PINK1/PTEN Genes. Neurotox Res. 35 (4), 945–954. doi:10.1007/s12640-018-9988-x
Varier, K. M., Thangarajan, S., Chinnasamy, A., Balakrishnan, G., and Paulose, R. (2017). Comparative Analysis of Potentiality of Esculin and Hinokitol (Β-Thujaplicin) as Anti-parkinsonism Drugs: a Pilot In Silico Study. Int. J. Pharm. Pharm. Sci. 9, 108–115. doi:10.22159/ijpps.2017v9i1.15340
Wan, Y. L., Wang, Y., Liu, B. C., Liu, X., Gong, X. Y., Zhao, X. L., et al. (2016). Zhonghua xue ye xue za zhi = Zhonghua xueyexue zazhi 37 (10), 886–891. doi:10.3760/cma.j.issn.0253-2727.2016.10.014
Willis, S. G., Lange, T., Demehri, S., Otto, S., Crossman, L., Niederwieser, D., et al. (2005). High-sensitivity Detection of BCR-ABL Kinase Domain Mutations in Imatinib-Naive Patients: Correlation with Clonal Cytogenetic Evolution but Not Response to Therapy. Blood 106 (6), 2128–2137. doi:10.1182/blood-2005-03-1036
Xiaofei, E., and Kowalik, T. F. (2014). The DNA Damage Response Induced by Infection with Human Cytomegalovirus and Other Viruses. Viruses 6 (5), 2155–2185. doi:10.3390/v6052155
Ysrael, M. C., and Croft, K. D. (1990). Inhibition of Leukotriene and Platelet Activating Factor Synthesis in Leukocytes by the Sesquiterpene Lactone Scandenolide. Planta Med. 56 (3), 268–270. doi:10.1055/s-2006-960953
Zhang, S., Lin, Z. N., Yang, C. F., Shi, X., Ong, C. N., and Shen, H. M. (2004). Suppressed NF-kappaB and Sustained JNK Activation Contribute to the Sensitization Effect of Parthenolide to TNF-Alpha-Induced Apoptosis in Human Cancer Cells. Carcinogenesis 25 (11), 2191–2199. doi:10.1093/carcin/bgh234
Keywords: mikanolide derivatives, 3G, sesquiterpene, leukemia, apoptosis, cell cycle, K562
Citation: Rao Q, Xie K, Varier KM, Huang L, Song J, Yang J, Qiu J, Huang Y, Li Y, Gajendran B, Li Y and Liu S (2022) Design, Synthesis, and Antileukemic Evaluation of a Novel Mikanolide Derivative Through the Ras/Raf/MEK/ERK Pathway. Front. Pharmacol. 13:809551. doi: 10.3389/fphar.2022.809551
Received: 18 November 2021; Accepted: 06 April 2022;
Published: 20 May 2022.
Edited by:
Jiang-Jiang Qin, Institute of Cancer and Basic Medicine, (CAS), ChinaReviewed by:
Sebastian Halbach, University of Freiburg, GermanySung-Gook Cho, Korea National University of Transportation, South Korea
Copyright © 2022 Rao, Xie, Varier, Huang, Song, Yang, Qiu, Huang, Li, Gajendran, Li and Liu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yan Li, bGl5YW4xNjEyQDE2My5jb20=; Babu Gajendran, YmFidUBnbWMuZWR1LmNu; Yanmei Li, bGl5YW5tZWk1MThAaG90bWFpbC5jb20=; Sheng Liu, bHNoZW5nQDEyNi5jb20=
†These authors have contributed equally to this work
 Qing Rao
Qing Rao Kaiqiang Xie1,2†
Kaiqiang Xie1,2† Jue Yang
Jue Yang Babu Gajendran
Babu Gajendran Yanmei Li
Yanmei Li