- 1Institut für Biochemie, Fachbereich Biologie, Universität des Saarlandes, Naturwissenschaftlich-Technische Fakultät, Saarbrücken, Germany
- 2Institut für Klinische Bioinformatik, Universität des Saarlandes, Saarbrücken, Germany
- 3Institut für Medizinische Biometrie, Epidemiologie und Medizinische Informatik, Universität des Saarlandes, Homburg, Germany
- 4KLinik für Neurologie, Fachbereich Klinische Medizin, Universität des Saarlandes, Homburg, Germany
- 5Klinik für Neurologie, SHG Kliniken Sonnenberg, Saarbrücken, Germany
Genetic and environmental factors lead to the manifestation of Parkinson’s disease (PD) but related mechanisms are only rudimentarily understood. Cytochromes P450 (P450s) are involved in the biotransformation of toxic compounds and in many physiological processes and thus predestinated to be involved in PD. However, so far only SNPs (single nucleotide polymorphisms) in CYP2D6 and CYP2E1 have been associated with the susceptibility of PD. Our aim was to evaluate the role of all 57 human P450s and their redox partners for the etiology and pathophysiology of PD and to identify novel potential players which may lead to the identification of new biomarkers and to a causative treatment of PD. The PPMI (Parkinson’s Progression Markers Initiative) database was used to extract the gene sequences of all 57 P450s and their three redox partners to analyze the association of SNPs with the occurrence of PD. Applying statistical analyses of the data, corresponding odds ratios (OR) and confidence intervals (CI) were calculated. We identified SNPs significantly over-represented in patients with a genetic predisposition for PD (GPD patients) or in idiopathic PD (IPD patients) compared to HC (healthy controls). Xenobiotic-metabolizing P450s show a significant accumulation of SNPs in PD patients compared with HC supporting the role of toxic compounds in the pathogenesis of PD. Moreover, SNPs with high OR values (>5) in P450s catalyzing the degradation of cholesterol (CYP46A1, CY7B1, CYP39A1) indicate a prominent role of cholesterol metabolism in the brain for PD risk. Finally, P450s participating in the metabolism of eicosanoids show a strong over-representation of SNPs in PD patients underlining the effect of inflammation on the pathogenesis of PD. Also, the redox partners of P450 show SNPs with OR > 5 in PD patients. Taken together, we demonstrate that SNPs in 26 out of 57 P450s are at least 5-fold over-represented in PD patients suggesting these P450s as new potential players in the pathogenesis of PD. For the first time exceptionally high OR values (up to 12.9) were found. This will lead to deeper insight into the origin and development of PD and may be applied to develop novel strategies for a causative treatment of this disease.
Introduction
Parkinson’s disease (PD) is the second most common neurodegenerative disorder. It was shown that the number of people with PD doubled to over six million from 1990 to 2015 and is further increasing (Dorsey et al., 2018). The main characteristic for PD is the degeneration of the Substantia nigra pars compacta and the connected loss of dopaminergic neurons. However, so far due to lack in understanding the reason of this degeneration, causative treatment is not possible. Therefore, intense research is going on trying to decipher the pathogenesis of PD, to find new biomarkers for early detection and novel candidates and approaches for treatment. While the majority of PD cases are idiopathic (IPD), about 5%–10% are familial and linked to mutations in several genes (GPD) (Bandres-Ciga et al., 2020). Mutations in more than 20 genes have been associated with this disease during the past years and 90 more were found in a recent genome-wide association study (Nalls et al., 2019). On the other hand, it is assumed that in the case of IPD the disease is caused and modulated by a complex interplay of genetic and environmental factors (Kalia and Lang, 2015).
A starting point for our investigations were reports on a higher susceptibility of patients for PD in the case of activity-decreasing mutations of the cytochrome P450 (P450) CYP2D6. P450s are hemo-proteins and in humans they are involved in the biotransformation of drugs, the conversion and bioactivation as well as degradation of environmental substances as well as the biosynthesis of endogenous compounds like steroid hormones, prostaglandins, vitamin D and others (Bernhardt, 2021). P450s are localized on the cytoplasmic side of the endoplasmic reticulum and on the luminal side of the inner mitochondrial membrane. They are monooxygenases and obtain electrons from NAD(P)H via a short redox chain, a NADPH-dependent FAD- and FMN-containing reductase (POR) in case of microsomal P450s and an iron-sulfur protein of the [2Fe-2S] type, adrenodoxin (Adx), and a FAD-containing reductase, adrenodoxin reductase (AdR) in case of mitochondrial P450s (Omura, 2010). In humans, 50 out of 57 P450s are located in the endoplasmic reticulum, while seven are mitochondrial (Omura, 2006). P450s have been found in many organs, including the brain (Ghosh et al., 2016). Much less is known on their specific functions in the brain. CYP2D6, an important drug metabolizer (Tyndale et al., 1991; Rendic and Guengerich, 2015), as well as CYP2E1 (Vaglini et al., 2004) were found to convert the neurotoxin 1-methyl-4-phenyl-1,2,3,6-tetrahydropyridine (MPTP) which was shown to induce PD-like symptoms in mouse models of PD. Later, a correlation between the incidence of PD and the occurrence of so-called poor metabolizers (PMs) was described (Smith et al., 1992; Deng et al., 2004; Elbaz et al., 2004; Anwarullah et al., 2017). However, the results are conflicting (Riedl et al., 1998; Nicholl et al., 1999; Ur Rasheed et al., 2017) and need further research. A pooled odds ratio (OR) from ten published studies of 1.47 was calculated for PMs for PD susceptibility by (McCann et al., 1997).
We used the well-accepted PPMI (The Parkinson’s Progression Markers Initiative) database to analyze possible relations between mutations in the genes of selected proteins and the origin and development of PD. The association of individual SNPs in the genes of all 57 P450s as well as their three redox partners (POR, Adx, AdR) with PD was analyzed. We observed statistically significant changes in patients suffering from IPD versus healthy controls (HC) and in patients with a genetic predisposition (mutations of the LRRK2, SNCA, PRKN and other genes) for PD (GPD) versus HC and identified several P450s displaying a significant over-representation of selected SNPs in PD patients compared with the HC.
Materials and methods
Extraction of the data on 57 P450s, POR, Adx and AdR from the PPMI database
Lists for the variants were obtained by downloading the whole-genome sequencing (WGS) VCF files of the Parkinson’s Progression Markers Initiative (PPMI; July 2018 release). The PPMI study is an ongoing international multicenter cohort study to identify biomarkers of progression in PD (Marek et al., 2018). Briefly, these were generated based on whole-blood extracted DNA samples and processed with the Centers for Common Disease Genomics standard pipeline, with the human hg38 assembly as reference genome (Regier et al., 2018). Subsequently, we annotated the called variants with SnpEff 5.0 (Cingolani, et al., 2012b) and SnpSift 5.0 (Cingolani et al., 2012a). Variants were further processed with the R programming language and aggregated per diagnosis group at the allele level. An overview of the workflow is given in Supplementary Figure S1.
Besides genomic data, information on group assignment (HC, IPD, GPD) for the investigated subjects were extracted from the PPMI database (https://ida.loni.usc.edu/login.jsp?project=ppmi). Genomic data as well as information on group assignment were merged into one data set for analyses.
Statistical analysis of the obtained data
Data analysis was performed using IBM-SPSS Version 26. Any p values reported are two-sided and subject to a significance level of 0.05. Due to the explorative nature of the study, we did not account for the issue of multiple statistical testing. Thus, we report raw 2-sided p-values without adjustment. Risk factors were identified with logistic regression and reported as odds ratios (OR) with 95% confidence intervals (CI).
Results
Bioinformatic approach
We extracted the variants from the whole-genome sequencing data of the PPMI cohort for the 57 cytochromes of the P450 family as well as their three redox partners. We thereby obtained allele information for 40,514 genomic positions, of which 3,256 were located in protein coding regions. The potential effect of the SNPs was forecasted using the available programs SnpEff and SnpSift, which combine data on functional consequences of SNPs from the GWAS Catalog, SIFT, Mutation Taster as well as ClinVar (Supplementary Figure S1). Most of the found SNPs are new and have been classified as “modifiers” of the corresponding gene function by the programs used. It has to be mentioned that in few cases the SNPs in individual P450s (or the three redox partners) were not found with highest frequency in the reference genome but in the studied individuals from the PPMI database. This might be due either to mistakes in the assignment of the corresponding SNP in the reference genome or to adaptation leading to an over-representation of the corresponding SNP in the studied PPMI cohort. Where necessary, we will mention this point for individual SNPs.
SNPs in the genes of human P450 systems and Parkinson’s disease
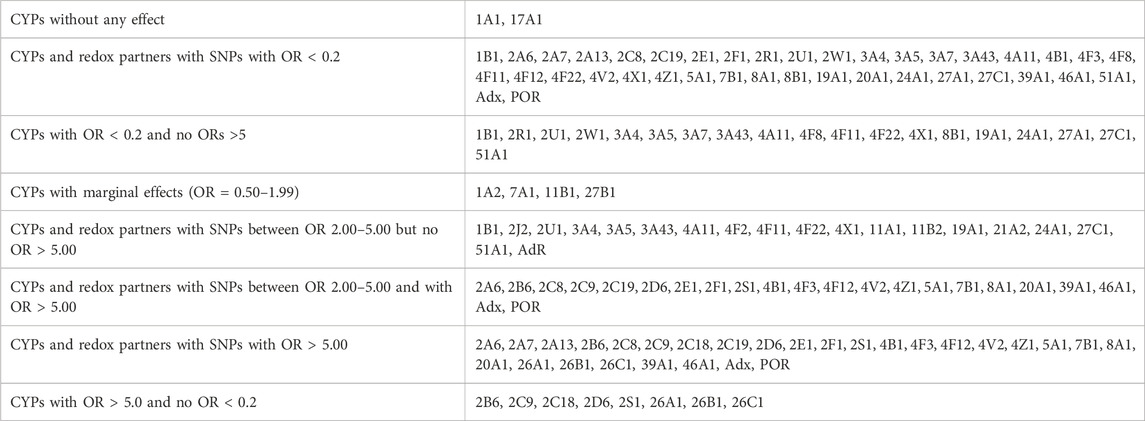
To get an overview on the P450 species, which display an over-representation of SNPs in their genes in PD patients, we categorized them into three groups: HC, IPD, GPD. Data from 193 HC representing the control group, 362 IPD and 317 GPD, representing the two classes of patients, were extracted from the PPMI database. Based on this data, we performed statistical analyses to find out how many SNPs in the genes encoding the investigated 60 proteins are significantly different between IPD and HC or GPD and HC. Since we considered only one gene at a time in these three groups, we did not perform corrections for multiple testing to get a direct impression about the occurrence of certain SNPs in different individuals. As a starting point, we focused on the association between certain SNPs and the occurrence of IPD or GPD. OR values showing the corresponding over- or under-representation of a certain SNP in a gene were calculated. Only data where the statistically obtained probability values (p-values) were below 0.05 were taken into account and are discussed in this paper. When the OR value is >1, an over-representation (higher odds) of the corresponding SNP occurs in PD patients. If OR is < 1, then the corresponding SNP is under-represented (lower odds) in PD patients and over-represented in HC this way suggesting a protective effect. Since we identified for some P450s many different SNPs with very high or high statistical significance, we grouped the data into five main groups: SNPs with OR values > 5, showing a significant over-representation of the SNP in PD patients, SNPs with over-representation between 2 and 5, groups with OR values between 0.5 and 2.0 demonstrating a medium effect, SNPs with more than 5-fold over-representation in HC (OR<0.2) and SNPs with 2-5-fold over-representation in HC (OR between 0.2 and 0.5). As demonstrated in Tables 1, 2, only two P450s, CYP1A1 and CYP17A1, did not show any SNPs which were significantly different between PD patients and HC although a total of 86 and 99 SNPs was found in CYP1A1 and CYP17A1, respectively. Four P450s (CYPs 1A2, 7A1, 11B1, 27B1) displayed differences with OR values of 0.5–2.0. Intriguingly, 26 P450s showed SNPs with OR values > 5. It is of importance to mention that most SNPs with OR values > 5 (and also between 2 and 5) are found in GPD patients suggesting that they may potentiate the effect of a genetic predisposition caused by known PD-inducing mutations to a considerable extent. Only SNPs in CYP46A1, Adx and POR were found in IPD patients with OR values > 5, while SNPs in 20 out of the 56 remaining P450s displayed OR values between two and five in IPD patients. The identification of so many significant SNPs in P450s was completely unexpected but suggests a significant contribution of these enzymes to the pathogenesis or development of PD. It is also of interest that the two immediate redox partners of P450s (Adx, POR) contain SNPs which were found > 5-fold over-represented in PD patients compared with HC. POR also shows two SNPs with OR<0.2 in IPD and one in GPD patients while Adx has three SNPs with OR<0.2 in GPD patients. Moreover, 21 and 34 P450s display SNPs with OR values < 0.2 in IPD and GPD patients (Table 2), respectively, with CYP51A1 and CYP4B1 having the highest numbers of SNPs with OR <0.2. SNPs with OR values < 0.2 indicate a protective effect on the pathogenesis of PD. In the following paragraphs we discuss the putative association of SNPs in different P450s with PD for the various classes of P450s as divided according to their major substrate class by (Guengerich, 2017).

TABLE 2. Summary of significant SNPs in 57 cytochrome P450s and three redox partners. CYP: cytochrome P450 gene; GPD and IPD display the numbers of significant SNPs found in patients suffering from GPD or IPD, respectively; number of total SNPs shows the number of all SNPs (significant and non-significant) found in the corresponding gene. Significant means with p < 0.01 or 0.05. The data are subdivided according to the function of the individual SNPs as described by Guengerich (Guengerich, 2017).
Association of SNPs in the genes of various P450s with PD
Due to the large quantity of obtained data (Table 2), we will focus in this paper mainly on SNPs in genes which display OR>5 or <0.2 and discuss possible effects on the derived P450 function.
CYP2D6 and CYP2E1
Based on initial reports on an increased occurrence of PD in CYP2D6 PMs as well as a higher incidence of activity-decreasing SNPs in CYP2E1 of PD patients (McCann et al., 1997; Vaglini et al., 2004), we first analyzed the distribution of SNPs in these two genes in PD patients and HC. In CYP2D6, 13 out of 153 total SNPs were found to be statistically significantly different compared to HC, 11 in GPD and two in IPD patients (Table 2).
Three SNPs in the gene of CYP2D6 were found with OR values of 9.54, 9.54 and 9.49 in patients suffering from GPD (compared to HC) (Table 2; Table 3) demonstrating that GPD patients have an up to 9.5-fold over-representation (higher odds) of one of these gene variations compared with HC. This means that only patients which have a predisposition for PD due to mutations in one of the genes like LRRK2, SNCA etc. are prone to develop PD when the CYP2D6 gene is mutated, whereas none of the SNPs alone (OR values in IPD patients of 0.228 and 0.229 were found) seems to be sufficient to cause symptoms of PD. SNP rs76015180 (OR = 9.49) was found in intron 5, while the other two SNPs with OR values > 5 are in the downstream region. The functional effect for the corresponding CYP2D6 gene products was classified as “modifier” by the used programs and no other information was found in the literature with regard to these SNPs.
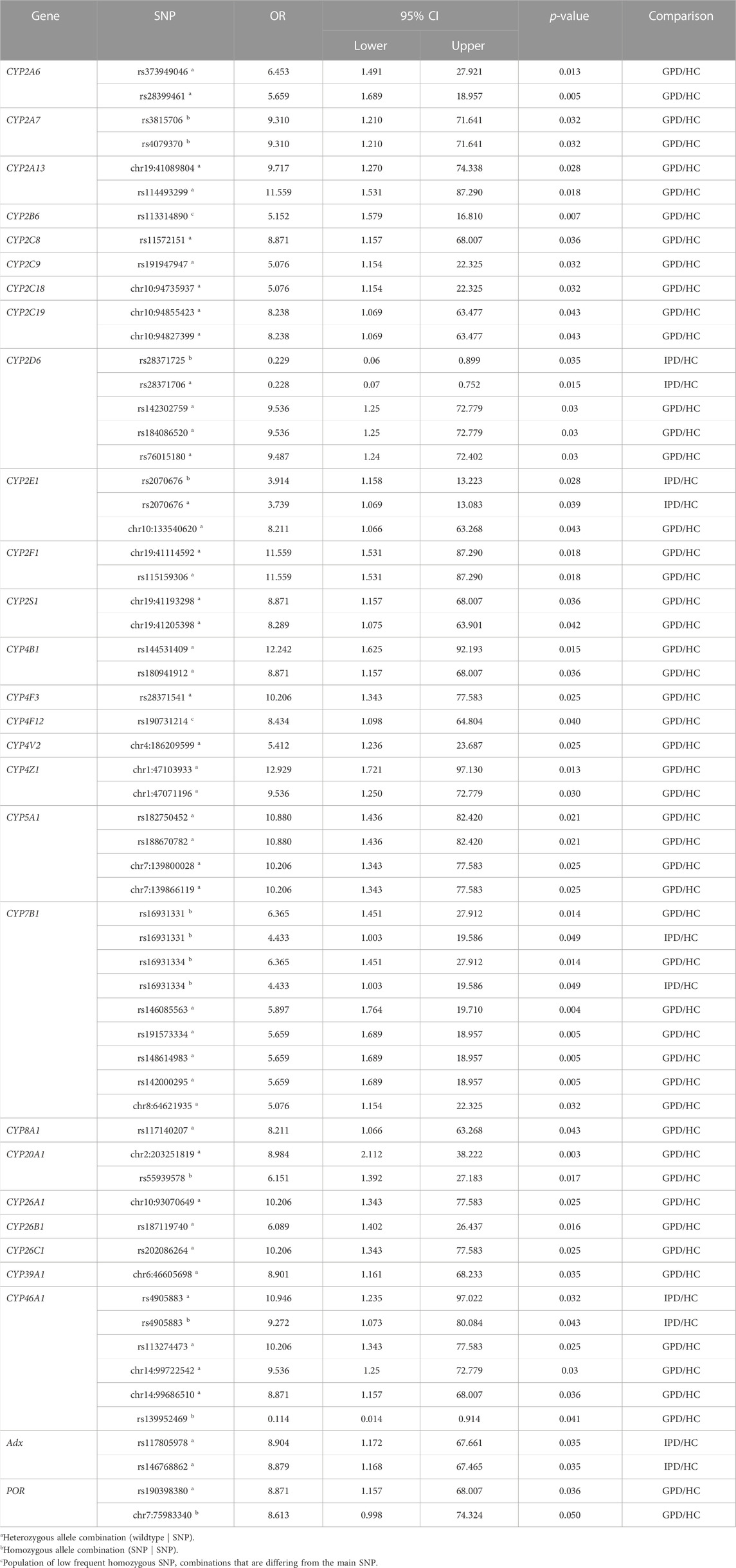
TABLE 3. Summary of selected single nucleotide polymorphisms (SNP) with statistically significant association (p-value <0.05) to Parkinson´s Disease (PD) for various P450s. The strength of association is represented by the odds ratio (OR) for patients with genetic PD (GPD) or idiopathic PD (IPD) and the healthy control group (HC), respectively. Only SNPs discussed in the paper are shown. The confidence interval (CI) of the OR is indicated by the lower and upper limit for a confidence level of 95%.
In CYP2E1 a total of 965 SNPs was identified in patients and HC from the data base, 114 of them being statistically different among patients and controls (Table 2). Only one SNP (chr10133540620) had an OR value > 5 (OR = 8.21) and was found in GPD patients, while three SNPs were found with OR = 0.15 in GPD patients and one SNP with OR = 0.10 is present in IPD patients suggesting a protective role of these latter SNPs. These five SNPs are located in a regulatory genomic position and described as modifiers of the function. The high number of SNPs with OR values between two and five in IPD patients (Table 2) indicates that CYP2E1 might have a more pronounced effect on the development of PD than CYP2D6. In a recent study on PD progression also changes in the CYP2E1 gene expression have been described (Kern et al., 2021). This might be due to its prominent role in the conversion of xenobiotics (acetaminophen, anesthetics), alcohols, acrylamide, carcinogens and halogenated alkanes (Guengerich, 2020). Interestingly, the SNP with OR>5 belonged to a group of SNPs which were not most frequently found in the reference gene but in homozygous patients (see above). Among the 77 SNPs over-represented in IPD patients, SNP rs2070676 (OR = 3.74 and 3.91 in heterozygous and homozygous form, respectively) which is located in intron 7, was before shown to be associated with PD (Shahabi et al., 2009). Interestingly, these authors also observed a susceptibility for PD by the C-allele being much more prominent than the G-allele (as in our case), but only in patients without relatives having PD (corresponding to IPD). It may be speculated that SNPs in CYP2E1 could affect ROS production or its capability to detoxify potential neurotoxins this way promoting the development of PD and supporting the importance of environmental factors for the risk to develop PD.
Association of SNPs in other xenobiotic-metabolizing P450s with PD
After supporting the role of the xenobiotic-metabolizing P450s CYP2D6 and CYP2E1 for the development of PD and identifying new SNPs being over-represented in GDP and IPD patients, we decided to look at the other 14 xenobiotic-metabolizing P450s (Guengerich, 2017) to find out whether other ones play a prominent role in PD. Interestingly, CYP1A1 and CYP1A2 are not or barely related to PD (Table 1; Table 2), although a total of 86 and 130 SNPs was observed in CYP1A1 and CYP1A2, respectively. When it comes to the members of the CYP2 family involved in the metabolism of xenobiotics, it turns out that all of them display SNPs with OR values >5 in case of GPD patients. This again suggests the importance of toxic compounds and due to this to environmental factors on the development of PD. In addition to this, (besides CYP2E1, which is discussed above) CYP2A6, CYP2B6, CYP2C8, CYP2C9, CYP2C19 and CYP2F1 show SNPs which are 2–5 times over-represented in GPD patients, while SNPs with similar OR in CYP2C8, CYP2C9 and CYP2F1 genes were found to be over-represented in IPD patients. In contrast to this, members of the CYP3A family do not show such a strong variation in their genes in PD patients compared to HC. None of them (CYP3A4, CYP3A5, CYP3A7, CYP3A43) displays a SNP with an OR value > 5 in PD patients. When looking at SNPs with OR values < 0.2 it becomes clear that seven out of 15 P450 genes (CYP2D6 and 2E1 are being discussed in the previous paragraph) display one or two of these protecting SNPs with OR<0.2 in IPD patients and eight in GPD patients.
Taken together, our data indicates the importance of environmental toxins for the risk to develop PD and on a close relationship between endogenous (genetic) and exogenous (environmental) factors for the pathogenesis of this disease.
Analysis of SNPs in P450s involved in fatty acid conversion
As shown in Table 2, only three P450 genes (CYP4B1, CYP4F12 and CYP4V2) contain SNPs with OR values > 5 in GPD patients but no such SNP was found in IPD patients. However, five of eight P450s display OR values between two and five in IPD patients and six of eight in GPD patients. This demonstrates that fatty acids also seem to be of importance for the development of IPD. When taking a closer look onto the substrates of the most prominent P450s of this group, it turns out that the predominant reaction catalyzed by members of the CYP4 family is the ω-hydroxylation of fatty acids and eicosanoids which leads to a removal of excess free fatty acids and the production of bioactive metabolites like 20-HETE (20-hydroxyeicosatetraenoic acid) synthesized from arachidonic acid (Hsu et al., 2007). CYP4B1 seems to be involved in the production of 12-HETE (12-hydroxy-5,8,11,14-eicosatetraenoic acid) (Mastyugin et al., 1999). In addition, its participation in endogenous as well as xenobiotic metabolism has been described (Thesseling et al., 2020) and it was demonstrated to catalyze the production of cytotoxic metabolites (Kowalski et al., 2019). When looking at protective SNPs, it turns out that CYP4B1 shows the highest numbers with four SNPs having OR values < 0.2 in IPD and 20 in GPD patients. This is not surprising when taking into account that in the genes of this P450 a total of 1218 SNPs was found, whereas this number varied between 238 (CYP4A11) and 877 (CYP4F22) for the other CYP4 genes (Table 2).
Analysis of SNPs in P450s involved in eicosanoid metabolism
Eicosanoids are generated from arachidonic acid and are oxidized derivatives of this and other polyunsaturated fatty acids (PUFAs). Eicosanoids play a pivotal role in inflammation and immunity. Besides cyclooxygenase and lipoxygenase, P450s play an essential role in their biosynthesis (Calder, 2020). CYP8A1 catalyzes the isomerization of prostaglandin H2 to prostacyclin (Wada et al., 2004), while CYP5A1 converts the cyclooxygenase product prostaglandin H2 to thromboxane A2 (Hsu et al., 1999). CYP5A1 is highly polymorphic. With 4.394 SNPs found in the individuals of the PPMI database it shows the highest number of SNPs in a P450. Out of the 4.394 SNPs 339 are significantly different between PD patients and HC. Four SNPs were found with OR values > 10 in GPD patients (Table 3). All of them are attributed to regulatory genomic positions. Also, in CYP8A1 and CYP4F3 SNPs with OR values > 5 have been described in GPD patients. CYP8A1 displays one SNP with OR = 8.21 in GPD patients in the regulatory region, described as modifier (Table 3). The SNP in CYP4F3 with OR = 10.21 comprises also a variation in a regulatory genomic position suggested to be a modifier. It has been found that CYP4F3 is a leukotriene B4 ω-hydroxylase which leads to a powerful proinflammatory mediator (Kikuta et al., 2002). This observation indicates that P450s involved in eicosanoid metabolism are of special importance for the development and progression of PD in patients, which have already a predisposition due to another PD-relevant mutation in the genome and thus may play an outstanding role in the pathogenesis of PD.
Analysis of SNPs in P450s involved in vitamin metabolism
Seven P450s (CYPs 2R1, 24A1, 26A1, 26B1, 26C1, 27B1, 27C1) were shown to be involved in the metabolism of vitamins (Guengerich, 2017). Only three of them (all being members of the CYP26 family) show one SNP each with an OR value > 5 (between 6 and 10) in GPD patients (Table 3). All three members of the CYP26 family are involved in retinoic acid (RA) metabolism (Guengerich, 2017) thus indicating a possible effect of retinoids on the development of PD. This coincides well with previous observations that retinoic acid is involved in neuronal differentiation (Janesick et al., 2015). In addition, it was very recently discussed that altered vitamin A metabolism and bioavailability contributes to oxidative stress, neuroinflammation and disturbance in biological rhythms indicating on multiple ways how retinoic acid may be associated with the pathogenesis of PD (Marie et al., 2021). Also, CYP27C1 is involved in the desaturation of retinoids (Glass et al., 2021), but demonstrates only a rather moderate association with PD with only three SNPs with OR values between two and five in GPD patients. However, one SNP was found with OR<0.2 in GPD patients suggesting a protecting effect. Interestingly, SNPs for genes involved in the biosynthesis and degradation of vitamin D (CYP2R1, CYP27B1, CYP24A) (Schuster, 2011) do not correlate to a high extent (OR values > 5 or <0.2) with the occurrence of PD. This indicates that the biosynthesis of vitamin D is obviously not related in a very significant manner to PD, but rather may have a moderate effect.
Analysis of SNPs in sterol-metabolizing and steroid-synthesizing P450s
The 14 P450s shown in Table 2 in the sterol section can be divided into those being involved in steroid hormone biosynthesis (CYP11A1, CYP11B1, CYP11B2, CYP17A1, CYP19A1, CYP21A2), those involved in the conversion of cholesterol and oxy-cholesterol (CYP7A1, CYP7B1, CYP27A1, CYP39A1, CYP46A1) and those involved in the metabolism of sterols like estradiol and bile acids (CYP1B1, CYP8B1) (Lorbek et al., 2012; Niwa et al., 2015; Guengerich, 2017). A special case is CYP51A1 catalyzing the removal of the C14 methyl group of lanosterol leading to cholesterol, which is the evolutionary most conserved P450 enzyme and is expressed ubiquitously. As shown in Table 2, P450s catalyzing steroid hormone biosynthesis (reactions reviewed in (Bernhardt and Neunzig, 2021)) demonstrate no or only moderate association to PD. In contrast, among the P450s involved in oxy-sterol metabolism, CYP7B1, CYP39A1 and CYP46A1 have SNPs with OR>5 in GPD patients (Table 2; Table 3). CYP46A1 is located specifically in the central nervous system and catalyzes the conversion of cholesterol to 24OH-cholesterol this way promoting its excretion from the brain (Petrov and Pikuleva, 2019). As shown, CYP46A1 displayed 734 total SNPs in the investigated PPMI cohort and is the only P450 that shows SNPs with OR >5 in IPD patients (Table 2; Table 3). However, the corresponding base in the reference group is not the most frequent one. In addition to this, three SNPs with OR>5 (being 10.21, 9.54 and 8.87) were discovered in GPD patients. The three SNPs are located in regulatory regions and described as modifiers. One SNP is found with an OR = 0.11 in GPD patients and is located in a regulatory genomic position. This data demonstrates a very significant association between SNPs in CYP46A1 and the occurrence of PD with one mutation being effective without predisposition and others enhancing the negative effect of the predisposition to a significant extent. CYP7B1, being a 7α-hydroxylase of 24OH-cholesterol, shows seven SNPs with OR>5 which are located in regulatory genomic positions and classified as modifiers. The SNP in the CYP39A1 gene is a variant located in a regulatory region as well. Interestingly, SNPs rs16931331 and rs16931334 in CYP7B1 show OR values of 6.36 in GPD patients and 4.43 in IPD patients thus underlining their special impact on the pathogenesis of PD. In contrast, CYP7A1, described to be a cholesterol 7α-hydroxylase (Wu et al., 1999), is not significantly associated with PD, while CYP27A1 being a human sterol 27-hydroxylase (Ali et al., 2013) displays several SNPs with OR<0.2 indicating a more protective effect on the development of PD. CYP8B1, which like CYP1B1 is involved in the metabolism of sterols, also does not display a strong association with the occurrence of PD. Finally, CYP51A1 demonstrates no SNPs with OR>5, but has 81 SNPs with OR values between two and five in GPD. This suggests that out of the 169 SNPs present in the corresponding gene, about half of them is over-represented in PD. Together this data indicates a very important association of sterol, especially cholesterol metabolism, and the pathogenesis of PD.
Analysis of SNPs in P450s with so far unidentified function (orphan P450s)
Seven P450s have been classified as P450s with unknown function by Guengerich (Guengerich, 2017), but for some of them substrates have been described more recently although detailed studies are still missing. CYP2A7 shows 97% sequence identity in the exons with CYP2A6 and a connection to nicotine metabolism has been found (Gao et al., 2020). A role of CYP2S1 in the ω-1 hydroxylation of polyunsaturated fatty acids has been described in an untargeted metabolomic study (Fekry et al., 2019). The enzyme is induced by dioxin and seems to play also an important role in in-situ toxicity, primarily in extra-hepatic tissues. It is highly expressed in the brain (Yan et al., 2018). CYP2W1 is expressed in a cancer-specific way and converts pro-dugs as well as fatty acids like arachidonic acid (Pan and Ong, 2017). CYP20A1 is highly expressed in the brain, especially in the Substantia nigra (Stark et al., 2008; Lemaire et al., 2016), and a connection to methotrexate toxicity has been found (Papapetropoulos et al., 2004; Aslibekyan et al., 2014) although this finding is controversial (Koponen et al., 2022). Interestingly, for CYPs of the CYP4 family (CYP4A22, CYP4X1 and CYP4Z1) a participation in fatty acid conversion and epoxide formation of eicosatrienoic acids has been described (Zöllner et al., 2009; Carver et al., 2014; McDonald et al., 2017; Durairaj et al., 2019). When looking at the association of SNPs in these genes (CYPs with unidentified function) with PD, it becomes clear that the largest effects owing six SNPs with OR values > 5 in GPD patients are identified in CYP2A7 (Table 2). Five of these SNPs were found to be homozygous and also five are intron variants with a described modifier effect. One homozygous SNP was found as a missense mutation leading to an amino acid exchange Met204Val. No information on the functional consequences of this amino acid replacement has been described in the literature so far. Also, CYP2S1, CYP4Z1 and CYP20A1 have two SNPs each with OR values > 5 in GPD patients. Most of them are located in regulatory regions and described by our bioinformatic tool as having a moderate impact. It needs to be mentioned that CYP4Z1 demonstrates 781 total SNPs in our PPMI cohort with only very few being significantly higher in PD patients compared to HC. The two SNPs discussed here have OR values of 9.54 and 12.93 suggesting that they are significantly over-represented in PD. Also, CYP20A1 shows many total SNPs (1011) but only 2 with OR>5. Both are described as variants in regulatory genomic positions and modifiers. CYP4A22, which so far has not been found in the brain, does not show any SNPs with OR values ≥ 2. Together this indicates on a special importance of CYP2A7, CYP20A1, CYP2S1 and CYP4Z1 for the pathogenesis of PD.
Association of SNPs in the genes of P450 redox partners with PD
POR
As most of the human P450s, POR is located on the cytoplasmic side of the endoplasmic reticulum. We found one SNP associated with IPD (with OR = 6.86) and nine associated with GPD displaying OR values of >5 (Table 2). This indicates that changes in POR also seem to be strongly associated with the occurrence of PD. When looking at the genome position of the SNP with OR>5 in IPD patients, it turns out that it is located in a regulatory genomic position of heterozygous patients. The same SNP is also found in different heterozygous GPD patients with an OR value between 2.84 and 5.36 as well as in heterozygous IPD patients with OR values between 3.08 and 3.27. Bioinformatic analyses described this SNP as modifier of activity. However, the reference genome does not show the most frequent base at that place (see above). Unfortunately, no more information is available concerning the effect of this SNP on the expression and activity of POR. Some significant SNPs in GPD patients seem to be clustered in the regulatory region with five SNPs displaying exactly the same OR of 8.57 and two with OR = 8.61 and 8.87, respectively. The SNPs with OR = 8.57 and 8.61 again do not display the “original” base in the reference gene. In contrast, SNP rs190398380 (OR = 8.87) which is also described as being located in the regulatory region shows the base defined as “original” one in the reference gene. When looking at the SNPs, which show a strong protective effect on PD (OR<0.2), it becomes clear that two of them can be found in IPD patients and one in GPD patients. All three coincide with the reference gene. The two SNPs found in the genome of IPD patients with OR<0.2 are homozygous and described to be located in regulatory genomic positions. To both SNPs a modifier function has been prescribed. The SNP with OR = 0.197 in GPD patients is also located in a regulatory region and described as modifier. Interestingly, SNP rs1057868 (also known as POR*28) has not been found as a significant SNP in our PD patient cohort.
Adx
Adx (assigned as Fdx1 in the database) is the immediate electron transfer protein to mitochondrial P450s. Interestingly, two SNPs were found with an OR value > 5 (8.90 and 8.88) in IPD patients (Table 2). These two SNPs are heterozygous, located in intron regions and described as modifiers. In GPD patients three protective SNPs with OR values of 0.11, 0.15 and 0.20 were found. All of them are intron variants and also described as modifiers. This data indicates that Adx may have a significant effect on the pathogenesis of PD, especially since high OR values have been obtained for IPD patients.
AdR
AdR (assigned as FDXR in the database) transfers the electrons from NADPH in two one-electron steps to Adx. No SNPs were found displaying statistically significant differences between IPD patients and HC, but one heterozygous SNP displayed an OR value of 2.24 in GPD patients (Table 2). This indicates a low or moderate effect of AdR on the etiology of PD.
Discussion
Although PD is the second most frequent neurodegenerative disease world-wide, its etiology is still unknown. To create novel approaches for the causative treatment of this disease, new biomarkers and targets have to be identified and characterized. Based on early studies on the interrelation between polymorphisms in the CYP2D6 gene, leading to decreased elimination of neurotoxins, and the occurrence of PD, we aimed to investigate the contribution of all 57 members of the P450 superfamily to the development of PD. Members of the P450 family play a prominent role not only in the detoxification and bioactivation of xenobiotics, but also in endogenous pathways like steroid hormone metabolism, biosynthesis of fatty acids, vitamins and eicosanoids etc. And they are, therefore, involved in many physiological and pathophysiological processes (Bernhardt, 2021).
Surprisingly, their contribution to the risk of developing PD has barely been studied so far (Breckenridge et al., 2016) although besides CYP2D6 15 more P450s are involved in the biotransformation of drugs and xenobiotics so that it is challenging to investigate their potential as a risk factor for PD. Furthermore, the effect of P450-catalyzed endogenous reactions on the pathogenesis of PD has not been investigated so far.
Our data clearly indicate a significant contribution of xenobiotic-converting P450s as a cause or promoting factor of PD and for the first time identify important SNPs in P450s involved in drug (and xenobiotic) metabolism associated with PD. As shown, all P450s belonging to the CYP2 family display an over-representation of SNPs in the genes of GPD patients with OR values > 5. Eight of these 10 P450s are described for the first time as linked to PD (Table 1; Table 2; Table 3 and Figure 1). CYP2C19 and CYP2F1 seem to play a prominent role for the pathology of GPD with eight and six SNPs displaying OR values > 5, respectively. In contrast, CYP1A1 and CYP1A2 are of no or low importance, while CYP3A7 has three significant SNPs, however, with OR values <0.5 suggesting a protective role against PD. Our data on CYP2D6 also show a clear and significant over-representation of several SNPs in the CYP2D6 gene of patients suffering from GPD with OR values of up to 9.5 (Table 3). This result is especially remarkable when taking into account that previous studies on a correlation of SNPs in so-called “poor metabolizers” with PD reported an average OR value of 1.47 (McCann et al., 1997), which is more than 6-fold lower than the value we found in our studies. Although the OR values for these SNPs is very high in GPD patients, no association of CYP2D6 with PD-causing genes has been found in GWAS studies (Edwards et al., 2010; Li et al., 2022). However, associations between CYP2D6 and pesticides on PD risk have been described in some studies and are also found between LRRK2 und PINK1 and pesticides (Dardiotis et al., 2013). Moreover, our data may shed light onto the partially conflicting results obtained when studying the correlation of PD with the occurrence of the “poor metabolizer” type. While the previously published studies did not differentiate between GPD and IPD patients, our investigations, taking this fact into account, clearly demonstrate that only GPD patients have a significant over-representation of several SNPs compared with HC, while in IPD patients only two under-represented SNPs have been found suggesting a protective effect of these SNPs. This data may explain why the various studies in the literature came to inconsistent findings. It seems that when the amount of GPD patients in the studied cohort is high, a correlation with PD becomes detectable, while it can barely be observed when the portion of GPD patients is too low to be statistically relevant. The bioinformatic tools used in our study classified the observed SNPs in CYP2D6 mostly as modifiers. Literature data concerning their function are not available. Experimental investigations, which are outside the scope of this study are necessary and certainly will contribute to a deeper understanding of the relation between the observed SNPs and the manifestation of PD. This is particularly interesting, since CYP2D6 has also been shown to catalyze the formation of dopamine from p-tyramine (Hiroi et al., 1998). This suggests that a lower expression or activity of CYP2D6 could also reduce the production of dopamine via this pathway. It is of special interest that SNPs in CYP2E1 and CYP2C8 also exert a strong influence on the development of IPD. While a connection between CYP2E1 and PD has been described in previous studies (Shahabi et al., 2009; Kern et al., 2021), the contribution of CYP2C8 as a risk factor for IPD is completely new. The effect of CYP2E1 can be mainly traced to ROS production, whereas the effect of CYP2C8 SNPs on the metabolism is not clear yet. However, it has been shown that CYP2C8 expression can be increased in human brain cells by the organophosphorous pesticide moncrotophos and that exposure to this pesticide on the other hand causes an increase in ROS production leading to oxidative damage (Tripathi et al., 2017).
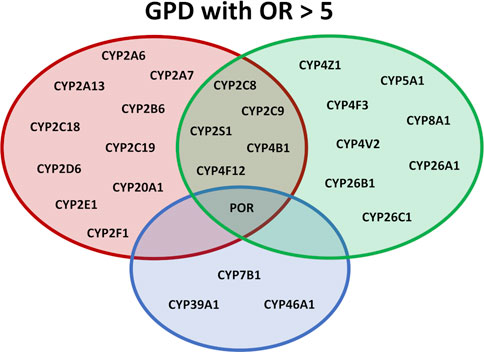
FIGURE 1. Overview of the effect of different SNPs in cytochromes P450 on various physiological pathways. P450s are shown which display OR values >5.0 in GPD patients. In the red circle P450s are listed participating in the biotransformation of xenobiotica, in the green circle P450s are listed participating in immune response and inflammation and in the blue circle P450s are shown involved in sterol metabolism or the degradation of cholesterol.
Taken together, the data found here demonstrate an association between PD and SNPs in drug-metabolizing and xenobiotic-converting enzymes. This strongly supports the hypothesis that toxic substances may have a devasting effect on the initiation and/or development of PD underlining the importance of environment-gene interaction as a significant factor for the pathology of PD (Figure 1).
The second group of P450s displaying SNPs with high OR values is connected to fatty acid metabolism, especially biosynthesis and metabolism of eicosanoids (Figure 1). As can be seen from Table 2 and Table 3, especially CYP5A1 displays a high variability of the gene with 10 SNPs having OR values > 5 and 4 with OR>10. Moreover, besides CYP5A1, which catalyzes the biosynthesis of thromboxane A2, CYP8A1, a prostacyclin synthase, shows a SNP with an OR value of 8.21. Both products, thromboxane and prostacyclin, are involved in inflammatory processes. Another P450, CYP4F3, catalyzes the production of leukotriene B4, a strong proinflammatory mediator, whereas CYP4F12, CYP4V2 and CYP4Z1 participate in fatty acid hydroxylation which can lead to the biosynthesis of bioactive HETE metabolites also playing a role in inflammation (Zöllner et al., 2009; Carver et al., 2014; McDonald et al., 2017; Durairaj et al., 2019). Furthermore, CYP4B1, which, besides participating in the formation of 12-HETE, is also involved in the metabolism of xenobiotics, displays four SNPs that are 6.67–12.24 over-represented in GPD patients compared to HC. In addition to CYP4B1 also CYPs 2C8, 2C9, 2S1 and 4F12 are involved in both, xenobiotic as well as fatty acid/eicosanoid metabolism, thus being on the interface of both metabolic pathways and possibly affecting the development of PD from both sides (Figure 1). This indicates for the first time that SNPs in 19 P450s involved in the biosynthesis of fatty acids and eicosanoids are closely linked to PD, even in cases where no predisposition by mutations of known PD causing genes exists (Table 2) and underlines the outstanding importance of eicosanoids and of inflammation (Sarparast et al., 2020) for neurodegenerative diseases and as a risk factor of PD (Figure 1).
In addition to P450s participating in the conversion of toxic substances and the production of immune modulators, a third group of P450s shows high numbers of SNPs with significant OR values. They belong to the group of sterol-converting enzymes. Here, CYP46A1, which is highly expressed in various regions of the brain (Petrov and Pikuleva, 2019; Pikuleva and Cartier, 2021), plays an outstanding role (Table 2; Figure 1). It is the only P450 with a SNP with OR>5 (10.94 and 9.27 in heterozygous and homozygous patients, respectively) found in IPD patients and three SNPs with OR>5 in GPD patients. As discussed above, it has, however, to be taken into account that the SNP in IPD patients as well as in three of the four SNPs with OR values < 0.2 show the most frequent base in the SNP and not in the reference genome. Interestingly, the occurrence of Alzheimer’s disease was also related to a SNP in one of the introns of CYP46A1. An OR value of 1.69, about 5-fold lower than for the association of some of the SNPs with PD found here, has been described for this disease (He et al., 2012). Besides CYP46A1, also CYP7B1 displays OR values > 5 in GPD patients as does CYP39A1. The three P450s are involved in the conversion of cholesterol. While CYP46A1 catalyzes the conversion of cholesterol to 24OH-cholesterol, CYP39A1 converts this substance to a 7α product to finally synthesize bile acids. Interestingly, an increase in the level of 24OH-cholesterol has been observed in the cerebrospinal fluid of PD patients (Björkhem et al., 2013; Björkhem et al., 2018). CYP7B1 is also highly expressed in the brain and a potent oxysterol 7α-hydroxylase with 27OH-cholesterol as main substrate, which is formed by CYP27A1. This data suggests that the metabolism, especially the degradation, of cholesterol in the brain is of pivotal importance for the pathogenesis of PD. This conclusion is further supported when looking at SNPs in IPD patients with OR values between two and five displaying one, four and three SNPs in CYP46A1, CYP39A1 and CYP7B1, respectively.
Since P450s are monooxygenases, they need electrons to perform oxygen activation and the following substrate hydroxylation. Mutations in the redox proteins may cause varying effects on different P450s (Ewen et al., 2011; Pandey and Flück, 2013). With SNPs displaying OR values > 5 in IPD patients, POR and Adx are significantly linked to PD. This is supported by the finding that for POR nine SNPs with OR>5 are found in GPD patients. The SNPs found are located in regulatory regions and no detailed information about their influence on the activities of POR or Adx with various P450s is available.
There are also some limitations of the investigations presented here. First of all, the data do not allow individual characterization of the two alleles of a studied person. Secondly, our analyses considered individual SNPs but did not analyze the multitude of various combinations of SNPs from different P450s in individual patients, since this was outside the focus of this paper, is very time-consuming and requires additional independent investigation. Finally, although suggestions concerning the functional consequences of the observed SNPs were drawn, experimental studies are missing. This is, however, common to most of the identified PD risk factors so that only very few mechanistic details are available to explain the pathogenesis of PD. Analysis of the data obtained in our studies reveal that most of the 40,514 SNPs in P450s were found in regulatory genomic positions (introns, UTR) and only 3,256 are located in exons. This is not surprising when taking into account that only 1%–2% of the human genome represent protein-coding structures and that 22,000 genes produce a proteome with over 200,000 distinct proteins (Annalora et al., 2017). It supports the importance of non-coding regions for the regulation of physiological processes and the pathogenesis of many diseases as well as the identification of new targets for drug treatment (Eisenstein, 2021).
Summarizing our findings, the data demonstrate for the first time a significant contribution of cytochrome P450 systems for the development of PD. CYP46A1 as well as the direct redox partners of P450s, POR and Adx, have been found to possess SNPs in IPD and GPD patients with OR>5. This is a very strong association, especially when taking into account that previous associations between SNPs of CYP2D6 (one of the only two studied P450s concerning their possible contribution to the occurrence of PD) and PD had a mean OR of 1.47, more than 6-fold lower than observed in the present paper. The strongly affected P450s were classified into three main groups concerning their function (Figure 1): (i) P450s involved in the biotransformation of xenobiotics, (ii) P450s involved in fatty acid and eicosanoid metabolism and (iii) P450s participating in the degradation of cholesterol. Our data for the first time give detailed information on enzymes, especially P450s and their redox partners, contributing to the development of PD by three mechanisms: toxicity, inflammation, and cholesterol degradation. We thus were able to identify novel potential players involved in the pathogenesis of this disease, but extensive and time-consuming experimental studies are needed to get insight into mechanisms of PD development, especially since most of the SNPs are located in regulatory regions.
Data availability statement
The original contributions presented in the study are included in the article/Supplementary Materials, further inquiries can be directed to the corresponding authors.
Author contributions
RB and MU designed the project. RB wrote the paper and participated in the analyses and discussion of results. PH performed statistical analyses of the data and participated in the data analyses, writing and preparation of the manuscript. MU participated in analyses of the data and writing of the manuscript. TF performed extraction and classification of the data from the PPMI database and contributed to the writing of the manuscript. GW performed statistical analyses of the data. All authors read and approved the manuscript.
Funding
The work was supported by a research grant from the “Dr. Rolf M. Schwiete Stiftung” Mannheim/Germany.
Acknowledgments
The work was made possible by accession to the Parkinson’s Progression Markers Initiative (PPMI) database. The authors thank Prof. Dr. Andreas Keller for his help in extraction and classification of the data from the PPMI database. We thank Stephan Baas for critical reading of the manuscript.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fphar.2022.1094265/full#supplementary-material
Abbreviations
CYP, cytochrome P450; P450, cytochrome P450; Adx, adrenodoxin; POR, NADPH-dependent cytochrome P450 reductase; CYP, P450; Adx; AdR and POR designates thecorresponding genes of P450, Adx, AdR and POR; PD, Parkinson’s disease; IPD, idiopathic (sporadic) PD; GPD, PD with a genetic predisposition; PPMI, The Parkinson’s Progression Markers Initiative; OR, odds ratio; CI, confidence interval.
References
Ali, Z., Heverin, M., Olin, M., Acimovic, J., Lövgren-Sandblom, A., Shafaati, M., et al. (2013). On the regulatory role of side-chain hydroxylated oxysterols in the brain. Lessons from CYP27A1 transgenic and Cyp27a1(-/-) mice. J. Lipid Res. 54, 1033–1043. doi:10.1194/jlr.M034124
Annalora, A. J., Marcus, C. B., and Iversen, P. L. (2017). Alternative splicing in the cytochrome P450 superfamily expands protein diversity to augment gene function and redirect human drug metabolism. Drug Metab. Dispos. 45, 375–389. doi:10.1124/dmd.116.073254
Anwarullah, A M., Badshah, M., Abbasi, R., Sultan, A., Khan, K., Ahmad, N., et al. (2017). Further evidence for the association of CYP2D6*4 gene polymorphism with Parkinson’s disease: A case control study. Genes Environ. 39, 18. doi:10.1186/s41021-017-0078-8
Aslibekyan, S., Brown, E. E., Reynolds, R. J., Redden, D. T., Morgan, S., Baggott, J. E., et al. (2014). Genetic variants associated with methotrexate efficacy and toxicity in early rheumatoid arthritis: Results from the treatment of early aggressive rheumatoid arthritis trial. Pharmacogenomics J. 14, 48–53. doi:10.1038/tpj.2013.11
Bandres-Ciga, S., Diez-Fairen, M., Kim, J. J., and Singleton, A. B. (2020). Genetics of Parkinson’s disease: An introspection of its journey towards precision medicine. Neurobiol. Dis. 137, 104782. doi:10.1016/j.nbd.2020.10478210.1016/j.nbd.2020.104782
Bernhardt, R. (2021). “Bioenergetics theory and components | cytochrome P450,” in Encyclopedia of biological chemistry III (Amsterdam, Netherlands: Elsevier), 123–129. doi:10.1016/B978-0-12-809633-8.21391-5
Bernhardt, R., and Neunzig, J. (2021). Underestimated reactions and regulation patterns of adrenal cytochromes P450. Mol. Cell Endocrinol. 530, 111237. doi:10.1016/j.mce.2021.111237
Björkhem, I., Lövgren-Sandblom, A., Leoni, V., Meaney, S., Brodin, L., Salveson, L., et al. (2013). Oxysterols and Parkinson’s disease: Evidence that levels of 24S-hydroxycholesterol in cerebrospinal fluid correlates with the duration of the disease. Neurosci. Lett. 555, 102–105. doi:10.1016/j.neulet.2013.09.003
Björkhem, I., Patra, K., Boxer, A. L., and Svenningsson, P. (2018). 24S-Hydroxycholesterol correlates with tau and is increased in cerebrospinal fluid in Parkinson’s disease and corticobasal syndrome. Front. Neurol. 9, 756. doi:10.3389/fneur.2018.00756
Breckenridge, C. B., Berry, C., Chang, E. T., Sielken, R. L., and Mandel, J. S. (2016). Association between Parkinson’s disease and cigarette smoking, rural living, well-water consumption, farming and pesticide use: Systematic review and meta-analysis. PloS One 11, e0151841. doi:10.1371/journal.pone.0151841
Carver, K. A., Lourim, D., Tryba, A. K., and Harder, D. R. (2014). Rhythmic expression of cytochrome P450 epoxygenases CYP4x1 and CYP2c11 in the rat brain and vasculature. Am. J. Physiol. Cell Physiol. 307, C989–C998. doi:10.1152/ajpcell.00401.2013
Cingolani, P., Patel, V. M., Coon, M., Nguyen, T., Land, S. J., Ruden, D. M., et al. (2012a). Using Drosophila melanogaster as a model for genotoxic chemical mutational studies with a new program, SnpSift. Front. Genet. 3, 35. doi:10.3389/fgene.2012.00035
Cingolani, P., Platts, A., Wang, L. L., Coon, M., Nguyen, T., Wang, L., et al. (2012b). A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff: SNPs in the genome of Drosophila melanogaster strain w1118; iso-2; iso-3. Fly. (Austin) 6, 80–92. doi:10.4161/fly.19695
Dardiotis, E., Xiromerisiou, G., Hadjichristodoulou, C., Tsatsakis, A. M., Wilks, M. F., and Hadjigeorgiou, G. M. (2013). The interplay between environmental and genetic factors in Parkinson's disease susceptibility: The evidence for pesticides. Toxicology 307, 17–23. doi:10.1016/j.tox.2012.12.016
Deng, Y., Newman, B., Dunne, M. P., Silburn, P. A., and Mellick, G. D. (2004). Further evidence that interactions between CYP2D6 and pesticide exposure increase risk for Parkinson’s disease. Ann. Neurol. 55, 897. doi:10.1002/ana.20143
Dorsey, E. R., Sherer, T., Okun, M. S., and Bloem, B. R. (2018). The emerging evidence of the Parkinson pandemic. J. Park. Dis. 8, S3–S8. doi:10.3233/JPD-181474
Durairaj, P., Fan, L., Du, W., Ahmad, S., Mebrahtu, D., Sharma, S., et al. (2019). Functional expression and activity screening of all human cytochrome P450 enzymes in fission yeast. FEBS Lett. 593, 1372–1380. doi:10.1002/1873-3468.13441
Edwards, T. L., Scott, W. K., Almonte, C., Burt, A., Powell, E. H., Beecham, G. W., et al. (2010). Genome-wide association study confirms SNPs in SNCA and the MAPT region as common risk factors for Parkinson disease. Ann. Hum. Genet. 74, 97–109. doi:10.1111/j.1469-1809.2009.00560.x
Eisenstein, M. (2021). Drug hunters uncloak the non-coding ‘hidden’ genome. Nat. Biotechnol. 39, 1169–1171. doi:10.1038/s41587-021-01088-y
Elbaz, A., Levecque, C., Clavel, J., Vidal, J-S., Richard, F., Amouyel, P., et al. (2004). CYP2D6 polymorphism, pesticide exposure, and Parkinson’s disease: CYP2D6, pesticides, and PD. Ann. Neurol. 55, 430–434. doi:10.1002/ana.20051
Ewen, K. M., Hannemann, F., Iametti, S., Morleo, A., and Bernhardt, R. (2011). Functional characterization of Fdx1: Evidence for an evolutionary relationship between P450-type and ISC-type ferredoxins. J. Mol. Biol. 413, 940–951. doi:10.1016/j.jmb.2011.09.010
Fekry, M. I., Xiao, Y., Berg, J. Z., and Guengerich, F. P. (2019). A role for the orphan human cytochrome P450 2S1 in polyunsaturated fatty acid ω-1 hydroxylation using an untargeted metabolomic approach. Drug Metab. Dispos. 47, 1325–1332. doi:10.1124/dmd.119.089086
Gao, Y., Miksys, S., Palmour, R. M., and Tyndale, R. F. (2020). The influence of tobacco smoke/nicotine on CYP2A expression in human and african green monkey lungs. Mol. Pharmacol. 98, 658–668. doi:10.1124/molpharm.120.000100
Ghosh, C., Hossain, M., Solanki, J., Dadas, A., Marchi, N., and Janigro, D. (2016). Pathophysiological implications of neurovascular P450 in brain disorders. Drug Discov. Today 21, 1609–1619. doi:10.1016/j.drudis.2016.06.004
Glass, S. M., Webb, S. N., and Guengerich, F. P. (2021). Binding of cytochrome P450 27C1, a retinoid desaturase, to its accessory protein adrenodoxin. Arch. Biochem. Biophys. 714, 109076. doi:10.1016/j.abb.2021.109076
Guengerich, F. P. (2020). Cytochrome P450 2E1 and its roles in disease. Chem. Biol. Interact. 322, 109056. doi:10.1016/j.cbi.2020.109056
Guengerich, F. P. (2017) Intersection of the roles of cytochrome P450 enzymes with xenobiotic and endogenous substrates: Relevance to toxicity and drug interactions, Chem. Res. Toxicol., 30(1):2, doi:10.1021/acs.chemrestox.6b00226
He, X., Zhang, Z., Zhang, J., Zhou, Y., Wu, C., Tang, M., et al. (2012). An intronic CYP46A1 polymorphism is associated with alzheimer disease in a Chinese han population. J. Mol. Neurosci. 47, 514–518. doi:10.1007/s12031-012-9778-5
Hiroi, T., Imaoka, S., and Funae, Y. (1998). Dopamine formation from tyramine by CYP2D6. Biochem. Biophys. Res. Commun. 249, 838–843. doi:10.1006/bbrc.1998.9232
Hsu, M-H., Savas, Ü., Griffin, K. J., and Johnson, E. F. (2007). Human cytochrome P450 family 4 enzymes: Function, genetic variation and regulation. Drug Metab. Rev. 39, 515–538. doi:10.1080/03602530701468573
Hsu, P-Y., Tsai, A-L., Kulmacz, R. J., and Wang, L-H. (1999). Expression, purification, and spectroscopic characterization of human thromboxane synthase. J. Biol. Chem. 274, 762–769. doi:10.1074/jbc.274.2.762
Janesick, A., Wu, S. C., and Blumberg, B. (2015). Retinoic acid signaling and neuronal differentiation. Cell Mol. Life Sci. 72, 1559–1576. doi:10.1007/s00018-014-1815-9
Kalia, L. V., and Lang, A. E. (2015). Parkinson’s disease. Lancet 386, 896–912. doi:10.1016/S0140-6736(14)61393-3
Kern, F., Fehlmann, T., Violich, I., Alsop, E., Hutchins, E., Kahraman, M., et al. (2021). Deep sequencing of sncRNAs reveals hallmarks and regulatory modules of the transcriptome during Parkinson’s disease progression. Nat. Aging 1, 309–322. doi:10.1038/s43587-021-00042-6
Kikuta, Y., Kusunose, E., and Kusunose, M. (2002). Prostaglandin and leukotriene ω-hydroxylases. Prostagl. Other Lipid Mediat 68–69, 345–362. doi:10.1016/S0090-6980(02)00039-4
Koponen, M., Paakinaho, A., Lin, J., Hartikainen, S., and Tolppanen, A. M. (2022). Identification of drugs associated with lower risk of Parkinson's disease using a systematic screening approach in a nationwide nested case-control study. Clin. Epidemiol. 14, 1217–1227. doi:10.2147/CLEP.S381289
Kowalski, J. P., McDonald, M. G., Whittington, D., Guttman, M., Scian, M., Girhard, M., et al. (2019). Structure-activity relationships for CYP4B1 bioactivation of 4-ipomeanol congeners: Direct correlation between cytotoxicity and trapped reactive intermediates. Chem. Res. Toxicol. 32, 2488–2498. doi:10.1021/acs.chemrestox.9b00330
Lemaire, B., Kubota, A., O’Meara, C. M., Lamb, D. C., Tanguay, R. L., Goldstone, J. V., et al. (2016). Cytochrome P450 20A1 in zebrafish: Cloning, regulation and potential involvement in hyperactivity disorders. Toxicol. Appl. Pharmacol. 296, 73–84. doi:10.1016/j.taap.2016.02.001
Li, J., Amoh, B. K., McCormick, E., Tarkunde, A., Zhu, K. F., Perez, A., et al. (2022). Integration of transcriptome-wide association study with neuronal dysfunction assays provides functional genomics evidence for Parkinson's disease genes. Hum. Mol. Genet., doi:10.1093/hmg/ddac230
Lorbek, G., Lewinska, M., and Rozman, D. (2012). Cytochrome P450s in the synthesis of cholesterol and bile acids - from mouse models to human diseases: CYPs in cholesterol and BA synthesis. FEBS J. 279, 1516–1533. doi:10.1111/j.1742-4658.2011.08432.x
Marek, K., Chowdhury, S., Siderowf, A., Lasch, S., Coffey, C. S., Caspell-Garcia, C., et al. (2018). The Parkinson's progression markers initiative (PPMI) - establishing a PD biomarker cohort. Ann. Clin. Transl. Neurol. 5, 1460–1477. doi:10.1002/acn3.644
Marie, A., Darricau, M., Touyarot, K., Parr-Brownlie, L. C., and Bosch-Bouju, C. (2021). Role and mechanism of vitamin A metabolism in the pathophysiology of Parkinson’s disease. J. Park. Dis. 11, 949–970. doi:10.3233/JPD-212671
Mastyugin, V., Aversa, E., Bonazzi, A., Vafaes, C., Mieyal, P., and Schwartzman, M. L. (1999). Hypoxia-induced production of 12-hydroxyeicosanoids in the corneal epithelium: Involvement of a cytochrome P-4504B1 isoform. J. Pharmacol. Exp. Ther. 289, 1611–1619.
McCann, S. J., Pond, S. M., James, K. M., and Le Couteur, D. G. (1997). The association between polymorphisms in the cytochrome P-450 2D6 gene and Parkinson’s disease: A case-control study and meta-analysis. J. Neurol. Sci. 153, 50–53. doi:10.1016/S0022-510X(97)00179-2
McDonald, M. G., Ray, S., Amorosi, C. J., Sitko, K. A., Kowalski, J. P., Paco, L., et al. (2017). Expression and functional characterization of breast cancer-associated cytochrome P450 4Z1 in Saccharomyces cerevisiae. Drug Metab. Dispos. 45, 1364–1371. doi:10.1124/dmd.117.078188
Nalls, M. A., Blauwendraat, C., Vallerga, C. L., Heilbron, K., Bandres-Ciga, S., Chang, D., Tan, M., Kia, D. A., Noyce, A. J., Xue, A., Bras, J., Young, E., von Coelln, R., Simón-Sánchez, J., Schulte, C., Sharma, M., Krohn, L., Pihlstrøm, L., Siitonen, A., Iwaki, H., Leonard, H., Faghri, F., Gibbs, J. R., Hernandez, D. G., Scholz, S. W., Botia, J. A., Martinez, M., Corvol, J-C., Lesage, S., Jankovic, J., Shulman, L. M., Sutherland, M., Tienari, P., Majamaa, K., Toft, M., Andreassen, O. A., Bangale, T., Brice, A., Yang, J., Gan-Or, Z., Gasser, T., Heutink, P., Shulman, J. M., Wood, N. W., Hinds, D. A., Hardy, J. A., Morris, H. R., Gratten, J., Visscher, P. M., Graham, R. R., and Singleton, A. B., 23 and Me Research Team et al. (2019). System genomics of Parkinson’s disease consortium, international Parkinson’s disease genomics ConsortiumIdentification of novel risk loci, causal insights, and heritable risk for Parkinson’s disease: A meta-analysis of genome-wide association studies. Lancet Neurol. 18, 1091–1102. doi:10.1016/S1474-4422(19)30320-5
Nicholl, D. J., Bennett, P., Hiller, L., Bonifati, V., Vanacore, N., Fabbrini, G., et al. (1999). A study of five candidate genes in Parkinson’s disease and related neurodegenerative disorders. European Study Group on Atypical Parkinsonism. Neurology 53, 1415–1421. doi:10.1212/wnl.53.7.1415
Niwa, T., Murayama, N., Imagawa, Y., and Yamazaki, H. (2015). Regioselective hydroxylation of steroid hormones by human cytochromes P450. Drug Metab. Rev. 47, 89–110. doi:10.3109/03602532.2015.1011658
Omura, T. (2006). Mitochondrial P450s. Chem. Biol. Interact. 163, 86–93. doi:10.1016/j.cbi.2006.06.008
Omura, T. (2010). Structural diversity of cytochrome P450 enzyme system. J. Biochem. 147, 297–306. doi:10.1093/jb/mvq001
Pan, Y., and Ong, E. C. (2017). Cytochrome P450 2W1 (CYP2W1) - ready for use as the biomarker and drug target for cancer? Xenobiotica 47, 923–932. doi:10.1080/00498254.2016.1244370
Pandey, A. V., and Flück, C. E. (2013). NADPH P450 oxidoreductase: Structure, function, and pathology of diseases. Pharmacol. Ther. 138, 229–254. doi:10.1016/j.pharmthera.2013.01.010
Papapetropoulos, S., Argyriou, A. A., Liossis, S. N., Daousis, D., Katsoulas, G., Apostolopoulos, D., et al. (2004). A case of L-dopa-responsive parkinsonian syndrome after low-dose oral methotrexate intake. Clin. Neuropharmacol. 27, 95–98. doi:10.1097/00002826-200403000-00011
Petrov, A. M., and Pikuleva, I. A. (2019). Cholesterol 24-hydroxylation by CYP46A1: Benefits of modulation for brain diseases. Neurotherapeutics 16, 635–648. doi:10.1007/s13311-019-00731-6
Pikuleva, I. A., and Cartier, N. (2021). Cholesterol hydroxylating cytochrome P450 46A1: From mechanisms of action to clinical applications. Front. Aging Neurosci. 13, 696778. doi:10.3389/fnagi.2021.696778
Regier, A. A., Farjoun, Y., Larson, D. E., Krasheninina, O., Kang, H. M., Howrigan, D. P., et al. (2018). Functional equivalence of genome sequencing analysis pipelines enables harmonized variant calling across human genetics projects. Nat. Commun. 9, 4038. doi:10.1038/s41467-018-06159-4
Rendic, S., and Guengerich, F. P. (2015). Survey of human oxidoreductases and cytochrome P450 enzymes involved in the metabolism of xenobiotic and natural chemicals. Chem. Res. Toxicol. 28, 38–42. doi:10.1021/tx500444e
Riedl, A. G., Watts, P. M., Jenner, P., and Marsden, C. D. (1998). P450 enzymes and Parkinson’s disease: The story so far. Mov. Disord. 13, 212–220. doi:10.1002/mds.870130204
Sarparast, M., Dattmore, D., Alan, J., and Lee, K. S. S. (2020). Cytochrome P450 metabolism of polyunsaturated fatty acids and neurodegeneration. Nutrients 12, E3523. doi:10.3390/nu12113523
Schuster, I. (2011). Cytochromes P450 are essential players in the vitamin D signaling system. Biochim. Biophys. Acta 1814, 186–199. doi:10.1016/j.bbapap.2010.06.022
Shahabi, H. N., Westberg, L., Melke, J., Håkansson, A., Belin, A. C., Sydow, O., et al. (2009). Cytochrome P450 2E1 gene polymorphisms/haplotypes and Parkinson’s disease in a Swedish population. J. Neural Transm. (Vienna) 116, 567–573. doi:10.1007/s00702-009-0221-1
Smith, C. A., Gough, A. C., Leigh, P. N., Summers, B. A., Harding, A. E., Maraganore, D. M., et al. (1992). Debrisoquine hydroxylase gene polymorphism and susceptibility to Parkinson’s disease. Lancet 339, 1375–1377. doi:10.1016/0140-6736(92)91196-f
Stark, K., Wu, Z-L., Bartleson, C. J., and Guengerich, F. P. (2008). mRNA distribution and heterologous expression of orphan cytochrome P450 20A1. Drug Metab. Dispos. 36, 1930–1937. doi:10.1124/dmd.108.022020
Thesseling, F. A., Hutter, M. C., Wiek, C., Kowalski, J. P., Rettie, A. E., and Girhard, M. (2020). Novel insights into oxidation of fatty acids and fatty alcohols by cytochrome P450 monooxygenase CYP4B1. Arch. Biochem. Biophys. 679, 108216. doi:10.1016/j.abb.2019.108216
Tripathi, V. K., Kumar, V., Pandey, A., Vatsa, P., Dhasmana, A., Singh, R. P., et al. (2017). Monocrotophos induces the expression of xenobiotic metabolizing cytochrome P450s (CYP2C8 and CYP3A4) and neurotoxicity in human brain cells. Mol. Neurobiol. 54, 3633–3651. doi:10.1007/s12035-016-9938-7
Tyndale, R., Aoyama, T., Broly, F., Matsunaga, T., Inaba, T., Kalow, W., et al. (1991). Identification of a new variant CYP2D6 allele lacking the codon encoding lys-281: Possible association with the poor metabolizer phenotype. Pharmacogenetics 1, 26–32. doi:10.1097/00008571-199110000-00005
Ur Rasheed, M. S., Mishra, A. K., and Singh, M. P. (2017). Cytochrome P450 2D6 and Parkinson’s disease: Polymorphism, metabolic role, risk and protection. Neurochem. Res. 42, 3353–3361. doi:10.1007/s11064-017-2384-8
Vaglini, F., Pardini, C., Viaggi, C., Bartoli, C., Dinucci, D., and Corsini, G. U. (2004). Involvement of cytochrome P450 2E1 in the 1-methyl-4-phenyl-1, 2, 3, 6-tetrahydropyridine-induced mouse model of Parkinson’s disease. J. Neurochem. 91, 285–298. doi:10.1111/j.1471-4159.2004.02720.x
Wada, M., Yokoyama, C., Hatae, T., Shimonishi, M., Nakamura, M., Imai, Y., et al. (2004). Purification and characterization of recombinant human prostacyclin synthase. J. Biochem. 135, 455–463. doi:10.1093/jb/mvh059
Wu, Z., Martin, K. O., Javitt, N. B., and Chiang, J. Y. L. (1999). Structure and functions of human oxysterol 7α-hydroxylase cDNAs and gene CYP7B1. J. Lipid Res. 40, 2195–2203. doi:10.1016/S0022-2275(20)32094-0
Yan, P., Eng, O. C., and Yu, C. J. (2018). A review on the expression and metabolic features of orphan human cytochrome P450 2S1 (CYP2S1). Curr. Drug Metab. 19, 917–929. doi:10.2174/1389200219666180528090237
Keywords: cytochrome P450, Parkinson’s disease, CYP46A1, POR, Adx, xenobiotica, cholesterol, PPMI
Citation: Hartz P, Fehlmann T, Wagenpfeil G, Unger MM and Bernhardt R (2023) A CYPome-wide study reveals new potential players in the pathogenesis of Parkinson’s disease. Front. Pharmacol. 13:1094265. doi: 10.3389/fphar.2022.1094265
Received: 09 November 2022; Accepted: 22 December 2022;
Published: 19 January 2023.
Edited by:
Vita Dolzan, University of Ljubljana, SloveniaReviewed by:
Irina A. Pikuleva, Case Western Reserve University, United StatesAnastazja Gorecki, Perron Institute for Neurological and Translational Science, Australia
Copyright © 2023 Hartz, Fehlmann, Wagenpfeil, Unger and Bernhardt. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Marcus Michael Unger, bS51bmdlckBzYi5zaGcta2xpbmlrZW4uZGU=; Rita Bernhardt, cml0YWJlcm5AbXgudW5pLXNhYXJsYW5kLmRl
 Philip Hartz1
Philip Hartz1 Rita Bernhardt
Rita Bernhardt