- 1Department of Biological Pharmacy, Biotechnology Pharmacy Laboratory, Faculty of Pharmacy, Universitas Padjadjaran, Sumedang, Indonesia
- 2Unit of Clinical Pharmacy and Community, Faculty of Pharmacy, Universitas Mulawarman, Samarinda, Indonesia
- 3Center of Excellence for Pharmaceutical Care Innovation, Universitas Padjadjaran, Sumedang, Indonesia
- 4Division of Respirology and Critical Care, Department of Internal Medicine, Faculty of Medicine, Universitas Padjadjaran-Hasan Sadikin Hospital, Bandung, Indonesia
- 5Division of Pharmacology and Therapy, Department of Biomedical Sciences, Faculty of Medicine, Universitas Padjadjaran, Bandung, Indonesia
Tuberculosis (TB) is an infectious disease that occurs globally. Treatment of TB has been hindered by problems with multidrug-resistant strains (MDR-TB). Fluoroquinolones are one of the main drugs used for the treatment of MDR-TB. The success of therapy can be influenced by genetic factors and their impact on pharmacokinetic parameters. This review was conducted by searching the PubMed database with keywords polymorphism and fluoroquinolones. The presence of gene polymorphisms, including UGT1A1, UGT1A9, SLCO1B1, and ABCB1, can affect fluoroquinolones pharmacokinetic parameters such as area under the curve (AUC), creatinine clearance (CCr), maximum plasma concentration (Cmax), half-life (t1/2) and peak time (tmax) of fluoroquinolones.
Introduction
Multidrug Resistant-Tuberculosis (MDR-TB) is a severe problem in various parts of the world, and Tuberculosis (TB) cases are particularly prevalent in India and Indonesia (WHO, 2020a). However, the use of pharmacogenomic and pharmacokinetic aspects as therapeutic parameters were expected to have a positive impact on treatment and may achieve an 80% reduction in TB incidence rates by 2030 as described in the WHO End TB strategy.
Drug responses of individual patients can be determined by the drug pharmacokinetic parameters, and these responses can be affected by single nucleotide polymorphisms (SNPs) in genes that encode drug-metabolizing enzymes and transporters; the influence of these SNPs on drug response is called pharmacogenetics. Therefore, comprehensive molecular understanding and clinical information for precise treatment of individuals are needed to improve the outcome therapy (Roden and Tyndale, 2011). Genetic polymorphism is due to naturally existing variants in genes that occur in more than 1% of the population. Polymorphism may influence the action of a drug by changing the pharmacokinetic or pharmacodynamic profile (Belle and Singh, 2008).
Transport proteins or transporters are membrane channels and molecular pumps that facilitate the movement of ions, small molecules, macromolecules, and drugs across membranes (Nelson et al., 2008). The movement of biochemical compounds through biological membranes is critical to the absorption, distribution, metabolism, and excretion of nutrients, neurotransmitters, and drugs (Overington et al., 2006; Ware, 2006; Giacomini et al., 2010; Yan, 2010; Rask-Andersen et al., 2011). The dynamic partnerships of transporters with other signaling molecules in subcellular locations are regarded as essential processes for cellular function. The attenuation of transporter gene sequence by polymorphisms often contributes to complex human diseases and individual drug responses (Ware, 2006; Cardoso et al., 2010; Yan, 2010; Longo et al., 2011; Rask-Andersen et al., 2011; Ueda, 2011).
In shorter MDR-TB treatment regimens, fluoroquinolones such as levofloxacin (L-isomer ofloxacin) and moxifloxacin (8-methoxy fluoroquinolone) are an important class of drugs (group A) used in the initial treatment phase for 4–6 months and the continuation phase for 5 months (WHO, 2020b). Fluoroquinolones such as levofloxacin and moxifloxacin are effective against gram-positive and gram-negative anaerobic bacteria through bactericidal action that acts via inhibition of the topoisomerase II (DNA gyrase) and topoisomerase IV enzymes required for bacterial DNA replication, transcription, repair, and recombination (FDA, 2008; Fàbrega et al., 2009; FDA, 2016).
Fluoroquinolone antibiotics play a significant role in the elimination of bacteria in the treatment of infections, necessitating accuracy in drug administration to maintain their efficacy and safety, which can be influenced by several circumstances. Therefore, the objective of this study was to determine explicitly how human genetic variation can alter the pharmacokinetic profile of fluoroquinolone antibiotic and how it also impact in clinical implication.
Materials and methods
This narrative review used articles published in PubMed obtained using the combination of “polymorphism” OR “single nucleotide polymorphism” OR “SNP” AND “fluoroquinolone” AND “pharmacokinetic” as keywords and found 387 articles. Finally, 6 out of 387 studies were reviewed to identify gene polymorphisms and their effect on the pharmacokinetic parameters of fluoroquinolones. Due to the limitations of published studies, we also searched several other studies that have been performed on other drugs for more comprehensive approaches to gene polymorphisms.
Result and discussion
Metabolic processes in the liver play an important role in influencing drug levels in the body. Drug metabolism in the liver can result in the formation of drugs that are more hydrophilic and that are then excreted through the liver, kidneys, and/or intestines. Drug metabolism involves the chemical biotransformation of drug molecules by enzymes present in the body. In addition, drug transporters facilitate the movement of drugs and metabolites in and out of cells and organs (Taxax and Bhartam, 2014).
Drug metabolism pathways consist of phases I and II and may include phase III. The phase I pathway is generally controlled by the Cytochrome P450 (CYP450) family, the main group of enzymes that chemically modify drugs so that they are more soluble in water and are then easily excreted by the kidneys and/or liver. The phase II pathway of drugs/metabolites involves enzymatic conjugation with endogenous hydrophilic compounds assisted by transferase enzymes; phase II metabolic enzymes are UDP-glucuronosyltransferase (UGT), Sulfotransferases, N-acyl transferases, Glutathione S-transferases, N-acetyl transferases, and Methyl transferases (Taxax and Bhartam, 2014). Phase III pathways are classified into two main superfamilies: ATP-binding cassette (ABC) proteins and solute carrier (SLC) transporters. The phase III pathway is facilitated by drug transporters that in general are transmembrane proteins that facilitate the transport of large molecules and/or ionized molecules into or out of the cell. ABC transporters require energy (ATP) to actively absorb or efflux the drug from one side of the cell membrane to the other, whereas SLCs enable the passage of certain solutes (e.g., sugars and amino acids) across the membrane while actively transporting other solutes despite their electrochemical gradients by coupling the process with another solute or ion (Almazroo et al., 2017). Fifty-two percent of moxifloxacin’s oral or intravenous dose is metabolized by glucuronide and sulfate conjugation (phase II metabolism). While the CYP450 system is not involved in the metabolic process of the moxifloxacin (FDA, 2016).
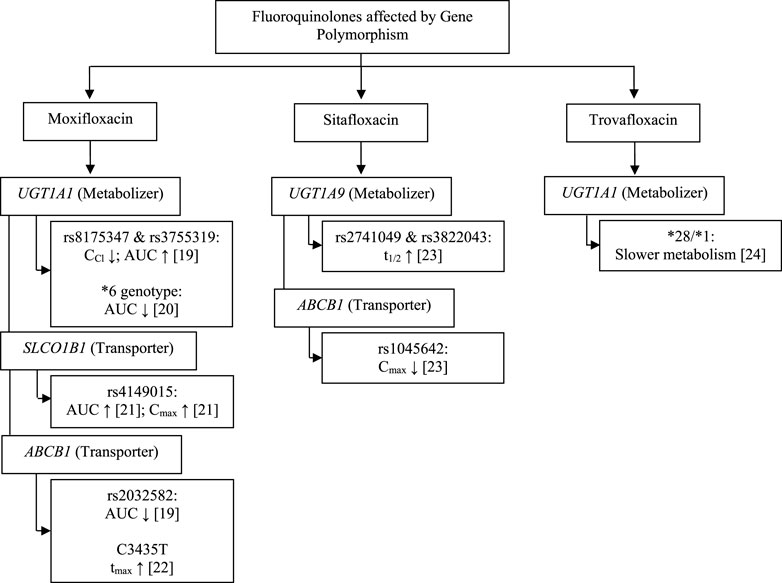
The pharmacokinetic profile of a drug can be influenced by internal factors that cannot be modified, such as genetic components, and several genes are known to be involved in the pharmacokinetic profile of fluoroquinolones. We found the study in fluoroquinolones antibiotics group are moxifloxacin, sitafloxacin, and trovafloxacin as fourth-generation fluoroquinolones. The gene found were uridine 5′-diphospho-glucuronosyltransferase family 1 member A1 (UGT1A1), 5′-diphospho-glucuronosyltransferase family 1 member A9 (UGT1A9), solute carrier organic anion transporter family member 1B1 (SLCO1B1), and ATP-binding cassette subfamily B member 1 (ABCB1) may affect fluoroquinolones pharmacokinetic parameters, including creatinine clearance (CCr), area under the curve (AUC), maximum plasma concentration (Cmax), half-life (t1/2) and time to the maximum plasma concentration (tmax).
Thus, the pharmacokinetic profile of fluoroquinolones may be affected by proteins expressed by the UGT1A1, UGT1A9, SLCO1B1, and ABCB1 genes (Figure 1; Tables 1, 2). UGT functions as a drug metabolizer, while the roles of SLC and ABC as transporters will certainly affect the pharmacokinetic profile. The metabolism of moxifloxacin by UGT with SNPs at rs8175347 and rs3755319 reduces CCr and increases AUC in healthy subjects, in contrast to studies that show a reduction in AUC with the UGT1A1*6 genotype. SLCO1B1 encodes organic anion transporting polypeptide 1B1 (OATP1B1), which acts as a moxifloxacin drug transporter. The rs4149015 SNP of SLCO1B1 increases the AUC and Cmax of moxifloxacin, whereas the p-glycoprotein drug transporter encoded by ABCB1 decreases the AUC, Cmax, and increases Tmax. Studies have shown a similar decrease in drug exposure and a prolonged time to reach the peak drug level in the body (Weiner et al., 2007; Hasunuma et al., 2016; Naidoo et al., 2018; Weiner et al., 2018).

TABLE 1. The effect of transporter and metabolizer human gene polymorphism on moxifloxacin pharmacokinetic parameter.
TABLE 2. The effect of transporter and metabolizer human gene polymorphism on sitafloxacin and trovafloxacin pharmacokinetic parameter.
Polymorphism of UGT1A1, UGT1A9, and ABCB1 also affects the pharmacokinetic profile in sitafloxacin and trovafloxacin (Figure 1; Table 2). A study on sitafloxacin showed that there was an association of genetic polymorphism of the human drug transporter ABCB1 rs1045642 in subjects with heterozygous or homozygous genotype variants with a lower Cmax of sitafloxacin than that in subjects without variants (p < 0.05) (Sun et al., 2021). SNPs in UGT1A9 rs2741049 and rs3832043 have a longer half-life (t1/2) than those in subjects without the variant; this was thought to occur due to decreased metabolism and disposition of sitafloxacin. However, this study did not show significant changes in AUC in either the ABCB1 or UGT1A9 groups (Sun et al., 2021). Acyl glucuronidation of trovafloxacin has been studied in human liver microsomes and also in UGT isoform-expressing systems in human liver microsomes. UGT1A1*28/*28 carriers were significantly slower metabolism than that in microsomes from UGT1A1*1/*1, suggesting that UGT1A1 is the main contributor to the glucuronidation of trovafloxacin (Fujiwara et al., 2015). At the molecular level, an influx transporter implicated in the membrane transport of quinolone antibacterial drugs (levofloxacin) has been identified for the first time as a result of the current work, in conclusion. OATP1A2, the discovered transporter molecule, is expressed in several tissues, including the small intestine, blood-brain barrier, liver, lung, and testis. As a result, it may play a part in regulating the intestinal absorption, tissue distribution, and hepatic excretion of these substances (Maeda et al., 2007).
Covariates can have an impact on the pharmacokinetic profile of fluoroquinolones in addition to genetic variables. According to a study conducted in Africa, gender, height, and body size (fat-free mass) may affect the pharmacokinetic characteristics of moxifloxacin, but these variables were adjusted before the pharmacokinetic analysis (Naidoo et al., 2018). Studies on the body weight, body mass index (BMI), and CCr of subjects of Japanese, Chinese, Korean, and Caucasian ancestry have been adjusted for pharmacokinetic moxifloxacin outcomes at a body weight of 70 kg (Hasunuma et al., 2016). Age, race, and weight were all correlated with geographic origin (African and American). The moxifloxacin AUC0-24 and Cmax were significantly increased by the moxifloxacin milligrams per kilogram dosage and genotype of variant g.11187G.A in the SLCO1B1 gene (rs4149015), but not by geographic location (Weiner et al., 2018). To adjust factors that can impact the pharmacokinetic profile of moxifloxacin, values from monitored univariate tests, sex, ethnicity (Black, White non-Hispanic, White Hispanic, and Asian), and univariate test results were employed (Weiner et al., 2007). Sitafloxacin can reduce Cmax by 50% when consumed by subjects who are fasting or having high fat foods intake, but it does not affect tmax, t1/2 elimination, or total exposure (AUC0-t and AUC0-∞). Despite only being possible in one dosing group of 10 participants, it is possible to affect the pharmacokinetic properties of ciprofloxacin, including t1/2 and Cmax (Sun et al., 2021).
UGT1A1 and UGT1A9
UGT1A1 is a member of the UGT family and encodes a UDP-glucuronosyltransferase, an enzyme of the glucuronidation pathway (Genecards, 2022), (National Library of Medicines, 2022). This enzyme catalyzes glucuronidation during phase II of drug metabolism, especially in conjugate reactions (Guillemette et al., 2000; Balram et al., 2002; Zhang et al., 2007). Moxifloxacin and trovafloxacin are metabolized via glucuronide and sulfate conjugation by glucuronosyltransferase and sulfotransferase (Moise et al., 2000), (Vincent et al., 1998). Therefore, genetic variations of UGT1A1 may affect the pharmacokinetic profile and clinical response of moxifloxacin and trovafloxacin.
Studies in South African patients (of black African ethnicity) with TB (Naidoo et al., 2018), UGT1A1 rs8175347, and rs3755319 were significantly associated with the alteration of pharmacokinetic parameters for moxifloxacin. After controlling for other factors, it was shown that having the TA 5/6 repeat in rs8175347 was linked to a 20.6% poorer clearance and roughly a 26% higher AUC (p = 0.001). Subjects with the AC and AA rs3755319 genotypes had 11.6% higher clearance than those with the CC genotype in the model (p = 0.032) (Naidoo et al., 2018). Other studies showed that UGT1A1*6 (211G > A; G71R) carriers had a significantly lower total exposure to the drug (AUCinf) for the glucuronide conjugate (M2 metabolite) moxifloxacin than that in the other genotypes (p < 0.0001), although the metabolism of moxifloxacin itself was not influenced by variation of the UGT1A1 genotype. There were significant differences between Japanese, Chinese, Korean, and Caucasian populations in Cmax, AUCinf, and CCr of moxifloxacin. The average AUCinf and Cmax compared to the parent compound in this study showed significant differences between the Japanese and Korean groups. AUCinf M2 in Caucasian races showed a higher value than in East Asian population groups (Japanese, Chinese and Korean) and was associated with differences in the frequency of UGT1A1*6 genotypes. These studies showed that ethnic differences may affect the pharmacokinetic parameters of drugs (Hasunuma et al., 2016). In a study using human liver microsomes, trovafloxacin glucuronidation was substantially slower in liver microsomes from UGT1A1*28/*28 carriers than it was in UGT1A1*1/*1 carriers (Fujiwara et al., 2015). According to these findings, UGT1A1*28/*28, rs8175347, and rs3755319 carriers have a poor metabolizer phenotype, while carriers of the *6 genotype have a hyper metabolizer phenotype (Naidoo et al., 2018), (Hasunuma et al., 2016), (Fujiwara et al., 2015).
The genetic variations of UGT1A1 also affected the pharmacokinetic profile of other drugs, such as telmisartan, irinotecan, dolutegravir, letermovir, and axitinib (Table 3). In general, the effect of the UGT1A1 polymorphism on other drugs also has an influence on the pharmacokinetic profile (AUC and CCr) and moxifloxacin.
TABLE 3. UGT1A1 gene polymorphism on telmisartan, irinotecan, dolutegavir, latermovir, axitinib pharmacokinetic parameter.
A study on the hypertension drug telmisartan showed that there was a genotype difference (rs4124874) associated with decreased clearance and increased bioavailability, and studies of dolutegravir and letermovir showed similar results with an increase in AUC (Huang et al., 2019; Kobie et al., 2019; Elliot et al., 2020). Studies on irinotecan (UGT1A1*6 and *8) and axitinib (UGT1A1*6 221GA and *27 686CA) showed a reduction in AUC (Minami et al., 2007), (Igarashi et al., 2018).
Similarly with UGT1A1, UGT1A9 also encodes a UDP-glucuronosyltransferase, an enzyme of the glucuronidation pathway that transforms small lipophilic molecules. UDP-glucuronosyltransferases (UGT) as a catalyst for the phase II biotransformation reaction in a lipophilic substrate conjugated with glucuronic acid to increase the polarity of the metabolite, which in turn can facilitate excretion in urine or bile. In addition, it has an important role in the elimination of drugs, xenobiotics, and endogenous compounds (Gene Cards, 2022a). Studies on sitafloxacin at rs2741049 and rs3832043 showed a significant effect on longer drug half-life t1/2 (p < 0.05). Inferring a poor metabolizer, rs2741049 and rs3822043 may play a part in the pharmacokinetic profile of the drug (Sun et al., 2021), another study at rs3832043 showed a possible effect on acetaminophen metabolism in neonates (Linakis et al., 2018). Thus, UGT1A9 may have a role in the pharmacokinetic profile of the drug.
It can be summarized the effect of UGT1A1 and UGT1A9 can affect metabolic processes that have an impact on decreasing CCr, decreasing t1/2, and increasing AUC in the fluoroquinolone antibiotics (moxifloxacin, sitafloxacin, and trovafloxacin) (Naidoo et al., 2018), (Sun et al., 2021), (Fujiwara et al., 2015). An increase in AUC needs to be considered to avoid drug side effects and can be considered in treatment interventions such as lowering drug doses for the safety of drug use and the effectiveness of therapy.
SLCO1B1
The SLCO1B1 gene is located on chromosome 12p12.1, 796 base pairs, and is an important pharmacokinetic gene. The function of the OATP1B1 protein, encoded by SLCO1B1, is to enable the transport of several compounds (hormones, toxins, and drugs) from the blood to the liver for elimination (Genetic Home Reference, 2022), (Ensembl Asia, 2022). OATP1B1 is located on the sinusoidal membrane of human hepatocytes, where it mediates the uptake of its substrates from portal blood into the hepatocytes (Takaaki et al., 1999), (Oshiro et al., 2010).
4 (8%) persons with the SLCO1B1 g.11187 AG genotype had a median moxifloxacin AUC0-24 that was 46% higher and a median Cmax that was 30% higher than 45 participants with the wild-type GG genotype (median AUC0–24 from the model [p 0.005, ANCOVA]; median Cmax from the model [p 0.009, ANCOVA]) (Weiner et al., 2018). This suggests that subject with g.-11187G>A shows poor metabolizer phenotype. Elevated levels of moxifloxacin can lead to risk for blood and lymphatic system orders (anemia), gastrointestinal disorders (nausea, diarrhea, vomiting, constipation, abdominal pain, dyspepsia), metabolic and nutritional disorders (hypokalemia), nervous system disorders (headache, dizziness) and QT prolongation (FDA, 2016). Increased AUC and Cmax need to be seen to avoid drug side effects and can be considered in treatment interventions such as lowering doses for the safety of drug use.
SNPs of SLCO1B1 may affect the pharmacokinetic profiles of other drugs (Table 4). Studies on atorvastatin polymorphisms in (SLCO1B1*15) and fluvastatin (c.521TA>G) showed an AUC-enhancing effect. Similarly, studies on methotrexate (388A>G and wild-type (AA) for 388A>G) showed the largest decrease in clearance in this genotype (Lee et al., 2010; Hirvensalo et al., 2019; Schulte et al., 2021). Conversely, studies on rifampin (c.388AA) and repaglinide/nateglinide (SLCO1B1*1B/*1B) in the Finnish ethnic group showed a decrease in drug concentrations in the body. In addition, the same study on repaglinide (SLCO1B1*1B/*1B) in ethnic Chinese was consistent with previous studies showing decreased AUC (Kalliokoski et al., 2008; He et al., 2011; Dompreh et al., 2018). Several studies also showed an effect of increasing the AUC that was similar to that of moxifloxacin.
TABLE 4. SLCO1B1 gene polymorphism on atorvastatin, 2-hydroatorvastatin, rifampin, repaglinide, nateglinide, methotrexate, fluvastatin & repaglinide pharmacokinetic parameter.
ABCB1
ABCB1 is a member of the ABC family. ABCB1 is a member of the ABC family. ABC proteins are divided into seven subfamilies, namely, ATP binding cassette subfamily A (ABC1), ATP binding cassette subfamily B (MDR/TAP), ATP binding cassette subfamily C (ABCC), ATP binding cassette subfamily D (ABCD), ATP binding cassette subfamily E (ABCE), ATP binding cassette subfamily F (GCN20), and ATP binding cassette subfamily G (WHITE) (Gene Cards, 2022b), (Human Genome Organisation, 2022). ABC proteins are found on chromosome 7q21.12, 323 base pairs that span 209.6 kb with 29 exons (Gene Cards, 2022b). The p-glycoprotein encoded by ABCB1 plays a key role in the elimination of drugs in the first pass of orally administered drugs, thereby limiting their bioavailability by excreting the drug through the epithelium that faces the lumen of the small intestine and colon and from the canaliculi facing the hepatic bile. The drug substrate will be removed from the systemic circulation through urine via the proximal renal tubule and through biliary excretion (Fromm, 2004). The expression and function of p-glycoprotein are ABCB1 SNPs-dependent. Changes in p-glycoprotein expression and function will affect the absorption, tissue distribution, and excretion of drug substances. Therefore, transporters genetic variations potentially affected the fate of drugs as well as the effectiveness of therapy (Marzolini et al., 2004).
For the ABCB1 SNP rs2032582, only one person in the cohort under study had the CA genotype. The patient with the CA genotype had a 40% lower prehepatic bioavailability and a comparable decrease in AUC when the effect of the rs2032582 SNP was taken into account in the population PK model (p = 0.01) (Naidoo et al., 2018). However, in another study, in univariate analyses of the pharmacogenetic data obtained with moxifloxacin plus rifampin, cases with the MDR1 3435CC genotype showed a significant increase in the time to the peak concentration of moxifloxacin (Tmax) compared to cases with the other genotypes, but there were no differences in the mean peak concentration of moxifloxacin [a 23% lower geometric mean for the 3435 CC genotype (p = 0.08)] (Weiner et al., 2007).
Two studies showed in-line results in the ABCB1 gene. A decrease in AUC and an increase in Tmax occurred with moxifloxacin, whereas in sitafloxacin there was a decrease in Cmax (p < 0.05) (Naidoo et al., 2018), (Weiner et al., 2007), (Sun et al., 2021). According to the moxifloxacin pharmacokinetics results, ABCB1 rs2032582 and C3435T suggest a hyper metabolizer phenotype, while rs1045642 on sitafloxacin suggests a poor metabolizer. Low levels of drugs in the blood in the use of antibiotics will risk the occurrence of drug resistance and failed treatment. This data can be taken into consideration in treatment interventions such as increasing or adjusting the drug dose until it reaches the expected level.
Genetic variations of ABCB1 may also affect the pharmacokinetic profiles of other drugs (Table 5). ABCB1 polymorphism studies have been performed using other drugs and show that lower drug levels are in line with those used for moxifloxacin and sitafloxacin (decreased Cmax). Studies on other drugs showed similar results where variations in pharmacokinetic parameters tended to decrease the blood levels of the drug. Studies on aripiprazole showed that the 1236TT genotype compared with the CC genotype had a lower clearance of aripiprazole, and also lower AUC and Cmax of dehydro-aripiprazole (it is an active metabolite). The azithromycin study showed that individuals with a heterozygous genotype of 2677GT/3435CC could inhibit intermediate levels of AUC and Cmax. In the sunitinib study, the mutant genotype (CT/TT) had a greater Cl/F compared with that of the wild genotype (CC). Clopidogrel (C3435) and tacrolimus (3435T) studies have revealed decreased plasma drug levels (Belmonte et al., 2018; Nazir et al., 2020; Woo et al., 2016; Wang et al., 2015; Naito et al., 2015).
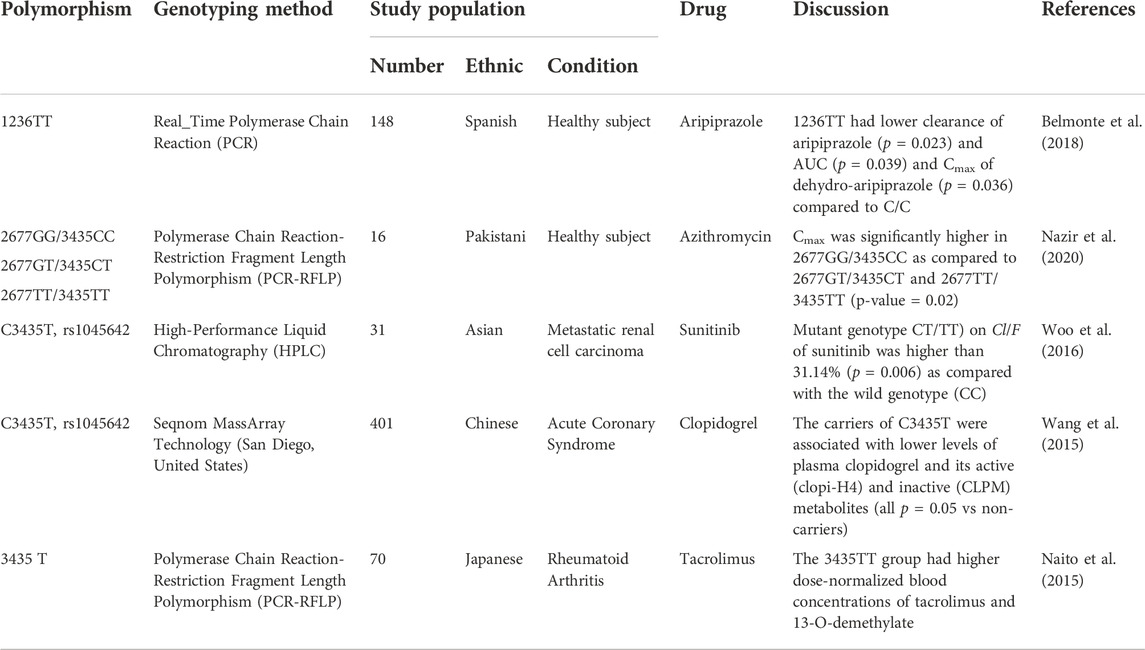
TABLE 5. ABCB1 gene polymorphism on aripiprazole, azithromycin, sunitinib, clopidogrel, and tacrolimus pharmacokinetic parameter.
Clinical implications
Treatment of MDR-TB is critical to eradicating tuberculosis. Genetic variations (SNPs) in genes that encode drug metabolizers or transporter proteins may affect the response to treatment. Identification of these SNPs is expected to provide specific information on the alteration of pharmacokinetic profiles, especially that of fluoroquinolones, a key drug in MDR-TB therapy (group A). Therefore, dose adjustments for the phenotype that appears (hyper metabolizer or poor metabolizer) may be required. In addition, it is also possible to consider changing the drug, because in antibiotic therapy a certain dose must be reached for efficacy without causing toxic effects. Therefore, it is deemed necessary to carry out pharmacogenetic studies to see the genetic profile of a population. Hence, genetic profile information can be used as a database for genetic screening for personalized treatment recommendations.
Limitation of the review
A limitation of this review is that studies on relevant SNPs and their effect on the pharmacokinetic profile of fluoroquinolones are limited. Hence, the number of subjects in those included studies are also limited and this may not be representative enough for the population. We also discovered that SNPs variation in the same gene can influence the pharmacokinetic profile of other drugs. This information also enriched the description of altered functions of transporters (ABCB1 and SLCO1B1) and metabolizers (UGT1A1 and UGT1A9) although their effect on fluoroquinolones still needs further study.
Conclusion
SNP polymorphisms that have been known to affect the pharmacokinetic parameters of fluoroquinolone as an important drug in the treatment of MDR-TB are found in the UGT1A1, UGT1A9, SLCO1B1, and ABCB1 genes. UGT1A1 and UGT1A9, genes that encode enzyme metabolizers for fluoroquinolones, can reduce CCr, t1/2, and influence AUC. SLCO1B1, a gene that encodes the OATP1B1 protein as a drug transporter for moxifloxacin, can increase the AUC and Cmax. ABCB1, a gene that encodes p-glycoprotein as a drug transporter for fluoroquinolones, has the effect of decreasing AUC and Cmax, and increasing Tmax.
Author contributions
NA writing original draft; NA and MB conceptualization and design; PS and MB editing draft manuscript; RR critical revision and supervision.
Funding
This research was funded by grants-in-aid from the Ministry of Research, Technology, and the Higher Education Republic of Indonesia for MB [1827/UN6.3.1/LT/2020].
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Almazroo, O. A., Miah, M. K., and Venkataramanan, R. (2017). Drug metabolism in the liver. Clin. Liver Dis. 21, 1–20. doi:10.1016/j.cld.2016.08.001
Balram, C., Sabapathy, K., Fei, G., Khoo, K. S., and Lee, E. J. D. (2002). Genetic polymorphisms of UDP-glucuronosyltransferase in asians: UGT1A1*28 is a common allele in Indians. Pharmacogenetics 12 (1), 81–83. doi:10.1097/00008571-200201000-00012
Belle, D. J., and Singh, H. (2008). Genetic factors in drug metabolism. Am. Fam. Physician 77 (11), 1553–1560.
Belmonte, C., Ochoa, D., Roman, M., Saiz-Rodriguez, M., Wojnicz, A., Gomez-Sanchez, C. I., et al. (2018). Influence of CYP2D6, CYP3A4, CYP3A5 and ABCB1 polymorphisms on pharmacokinetics and safety of aripiprazole in healthy volunteers. Basic Clin. Pharmacol. Toxicol. 122 (6), 596–605. doi:10.1111/bcpt.12960
Cardoso, A. R., Queliconi, B. B., and Kowaltowski, A. J. (2010). Mitochondrial ion transport pathways: Role in metabolic diseases. Biochim. Biophys. Acta 1797 6 (7), 832–838. doi:10.1016/j.bbabio.2009.12.017
Dompreh, A., Tang, X., Zhou, J., Yang, H., Topletz, A., Adu Ahwireng, E., et al. (2018). Effect of genetic variation of NAT2 on isoniazid and SLCO1B1 and CES2 on rifampin pharmacokinetics in Ghanaian children with tuberculosis. Antimicrob. Agents Chemother. 62 (3), e02099–e02117. doi:10.1128/AAC.02099-17
Elliot, E. R., Neary, M., Else, L., Khoo, S., Moyle, G., Carr, D. F., et al. (2020). Genetic influence of ABCG2, UGT1A1 and NR1I2 on dolutegravir plasma pharmacokinetics. J. Antimicrob. Chemother. 75 (5), 1259–1266. doi:10.1093/jac/dkz558
Ensembl Asia (2022). Ensembl Asia, rs4149015 SNP. [Online]. Available: http://asia.ensembl.org/Homo_sapiens/Variation/Population?db=core;r=12:21129888-21130888;v=rs4149015;vdb=variation;vf=418101832.
Fàbrega, A., Madurga, S., Giralt, E., and Vila, J. (2009). Mechanism of action of and resistance to quinolones. Microb. Biotechnol. 2 (1), 40–61. doi:10.1111/j.1751-7915.2008.00063.x
FDA (2016). Highlights of prescribing information. Avelox® (moxifloxacin hydrocloride). [Online]. Available: https://www.accessdata.fda.gov/drugsatfda_docs/label/2016/021085s063lbl.pdf.
FDA (2008). Highlights of prescribing information. Levaquin® (levofloxacin). [Online]. Available:https://www.accessdata.fda.gov/drugsatfda_docs/label/2006/020634s040,020635s043,021721s007lbl.pdf.
Fromm, M. F. (2004). Importance of P-glycoprotein at blood-tissue barriers. Trends Pharmacol. Sci. 25 (8), 423–429. doi:10.1016/j.tips.2004.06.002
Fujiwara, R., Sumida, K., Kutsuno, Y., Sakamoto, M., and Itoh, T. (2015). UDP-glucuronosyltransferase (UGT) 1A1 mainly contributes to the glucuronidation of trovafloxacin. Drug Metab. Pharmacokinet. 30 (1), 82–88. doi:10.1016/j.dmpk.2014.09.003
Gene Cards (2022). Human gene database index: ABCB1 gene. [Online]. Available: https://www.genecards.org/cgi-bin/carddisp.pl?gene=ABCB1.
Gene Cards (2022). Human gene database: UGT1A9. [Online]. Available: https://www.genecards.org/cgi-bin/carddisp.pl?gene=UGT1A9.
Genecards (2022). Human gene database index: UGT1A1. [Online]. Available: https://www.genecards.org/cgi-bin/carddisp.pl?gene=UGT1A1.
Genetic Home Reference (2022). SLCO1B1 gene. [Online]. Available:https://ghr.nlm.nih.gov/gene/SLCO1B1.
Giacomini, K. M., Huang, S. M., Tweedie, D. J., Benet, L. Z., Brouwer, K. L. R., Chu, X., et al. (2010). Membrane transporters in drug development. Nat. Rev. Drug Discov. 9 (3), 215–236. doi:10.1038/nrd3028
Guillemette, C., Millikan, R. C., Newman, B., and Housman, D. E. (2000). Genetic polymorphisms in uridine diphospho-glucuronosyltransferase 1A1 and association with breast cancer among African Americans. Cancer Res. 60 (4), 950–956.
Huang, L., Yang, L., Huang, J., Tan, H. Y., Liu, S. K., Guo, C. X., et al. (2019). Effects of UGT1A1 polymorphism, gender and triglyceride on the pharmacokinetics of telmisartan in Chinese patients with hypertension: A population pharmacokinetic analysis. Eur. J. Drug Metab. Pharmacokinet. 44 (6), 797–806. doi:10.1007/s13318-019-00567-7
Hasunuma, T., Tohkin, M., Kaniwa, N., Jang, I. J., Yimin, C., Kaneko, M., et al. (2016). Absence of ethnic differences in the pharmacokinetics of moxifloxacin, simvastatin, and meloxicam among three East Asian populations and Caucasians. Br. J. Clin. Pharmacol. 81 (6), 1078–1090. doi:10.1111/bcp.12884
He, J., Qiu, Z., Li, N., Yu, Y., Lu, Y., Han, D., et al. (2011). Effects of SLCO1B1 polymorphisms on the pharmacokinetics and pharmacodynamics of repaglinide in healthy Chinese volunteers. Eur. J. Clin. Pharmacol. 67 (7), 701–707. doi:10.1007/s00228-011-0994-7
Hirvensalo, P., Tornio, A., Neuvonen, M., Kiander, W., Kidron, H., Paile-Hyvarinen, M., et al. (2019). Enantiospecific pharmacogenomics of fluvastatin. Clin. Pharmacol. Ther. 106 (3), 668–680. doi:10.1002/cpt.1463
Human Genome Organisation (2022). HUGO gene nomenclature. [Online]. Available: https://www.hugo-international.org/standards/.
Igarashi, R., Inoue, T., Fujiyama, N., Tsuchiya, N., Numakura, K., Kagaya, H., et al. (2018). Contribution of UGT1A1 genetic polymorphisms related to axitinib pharmacokinetics to safety and efficacy in patients with renal cell carcinoma. Med. Oncol. 35, 51. doi:10.1007/s12032-018-1113-8
Kalliokoski, A., Backman, J. T., Neuvonen, P. J., and Niemi, M. (2008). Effects of the SLCO1B1*1B haplotype on the pharmacokinetics and pharmacodynamics of repaglinide and nateglinide. Pharmacogenet. Genomics 18 (11), 937–942. doi:10.1097/FPC.0b013e32830d733e
Kobie, J., Guo, Z., Cho, C. R., Menzel, K., McCrea, J. B., Blanchard, R., et al. (2019). Pharmacogenetic analysis of OATP1B1, UGT1A1, and BCRP variants in relation to the pharmacokinetics of letermovir in previously conducted clinical studies. J. Clin. Pharmacol. 59 (9), 1236–1243. doi:10.1002/jcph.1420
Lee, Y. J., Lee, M. G., Lim, L. A., Jang, S. B., and Chung, J. Y. (2010). Effects of SLCO1B1 and ABCB1 genotypes on the pharmacokinetics of atorvastatin and 2-hydroxyatorvastatin in healthy Korean subjects. Int. J. Clin. Pharmacol. Ther. 48 (1), 36–45. doi:10.5414/cpp48036
Linakis, M. W., Cook, S. F., Kumar, S. S., Liu, X., Wilkins, D. G., Gaedigk, R., et al. (2018). Polymorphic expression of UGT1A9 is associated with variable acetaminophen glucuronidation in neonates: A population pharmacokinetic and pharmacogenetic study. Clin. Pharmacokinet. 57 (10), 1325–1336. doi:10.1007/s40262-018-0634-9
Longo, N., Ardon, O., Vanzo, R., Schwartz, E., and Pasquali, M. (2011). Disorders of creatine transport and metabolism. Am. J. Med. Genet. C Semin. Med. Genet. 157 (1), 72–78. doi:10.1002/ajmg.c.30292
Maeda, T., Takahashi, K., Ohtsu, N., Oguma, T., Ohnishi, T., Atsumi, R., et al. (2007). Identification of influx transporter for the quinolone antibacterial agent levofloxacin. Mol. Pharm. 4 (1), 85–94. doi:10.1021/mp060082j
Marzolini, C., Paus, E., Buclin, T., and Kim, R. B. (2004). Polymorphisms in human MDR1 (P-glycoprotein): Recent advances and clinical relevance. Clin. Pharmacol. Ther. 75 (1), 13–33. doi:10.1016/j.clpt.2003.09.012
Minami, H., Sai, K., Saeki, M., Saito, Y., Ozawa, S., Suzuki, K., et al. (2007). Irinotecan pharmacokinetics/pharmacodynamics and UGT1A genetic polymorphisms in Japanese: Roles of UGT1A1*6 and *28. Pharmacogenet. Genomics 17 (7), 497–504. doi:10.1097/FPC.0b013e328014341f
Moise, P., Birmingham, M., and Schentang, J. (2000). Pharmacokinetics and metabolism of moxifloxacin. Metabolism Moxifloxacin Drugs Today 36 (4), 229–244. doi:10.1358/dot.2000.36.4.570201
Naidoo, A., Ramsuran, V., Chirehwa, M., Denti, P., McIlleron, H., Naidoo, K., et al. (2018). Effect of genetic variation in UGT1A and ABCB1 on moxifloxacin pharmacokinetics in South African patients with tuberculosis. Pharmacogenomics 19 (1), 17–29. doi:10.2217/pgs-2017-0144
Naito, T., Mino, Y., Aoki, Y., Hirano, K., Shimoyama, K., Ogawa, N., et al. (2015). ABCB1 genetic variant and its associated tacrolimus pharmacokinetics affect renal function in patients with rheumatoid arthritis. Clin. Chim. Acta. 445, 79–84. doi:10.1016/j.cca.2015.03.021
National Library of Medicines (2022). Gene summary: UGT1A1-UDP gluconosyltransferase family 1 member A1 (human). Available at: https://www.ncbi.nlm.nih.gov/gene/54658.
Nazir, S., Adnan, K., Gul, R., Ali, G., Saleha, S., and Khan, A. (2020). The effect of gender and ABCB1 gene polymorphism on the pharmacokinetics of azithromycin in healthy male and female Pakistani subjects. Can. J. Physiol. Pharmacol. 98 (8), 506–510. doi:10.1139/cjpp-2019-0569
Nelson, D., Lehninger, A., and Cox, M. (2008). Lehninger principles of biochemistry. New York: W. H. Freeman.
Oshiro, C., Mangravite, L., Klein, T., and Altman, R. (2010). PharmGKB very important pharmacogene: SLCO1B1. Pharmacogenet. Genomics 20 (3), 211–216. doi:10.1097/FPC.0b013e328333b99c
Overington, J. P., Al-Lazikani, B., and Hopkins, A. L. (2006). How many drug targets are there? Nat. Rev. Drug Discov. 5 (12), 993–996. doi:10.1038/nrd2199
Rask-Andersen, M., Almén, M. S., and Schiöth, H. B. (2011). Trends in the exploitation of novel drug targets. Nat. Rev. Drug Discov. 10 (8), 579–590. doi:10.1038/nrd3478
Roden, D. M., and Tyndale, R. F. (2011). Pharmacogenomics at the Tipping Point: Challenges and Opportunities. Clinical Pharmacology and Therapeutics 89 (3), 323–327. doi:10.1038/clpt.2010.340
Schulte, R. R., Choi, L., Utreja, N., Van Driest, S. L., Stein, C. M., and Ho, R. H. (2021). Effect of SLCO1B1 polymorphisms on high-dose methotrexate clearance in children and young adults with leukemia and lymphoblastic lymphoma. Clin. Transl. Sci. 14 (1), 343–353. doi:10.1111/cts.12879
Sun, L. N., Sun, G. X., Yang, Y. Q., Shen, Y., Huang, F. R., Xie, L. J., et al. (2021). Effects of ABCB1, UGT1A1, and UGT1A9 genetic polymorphisms on the pharmacokinetics of sitafloxacin granules in healthy subjects. Clin. Pharmacol. Drug Dev. 10 (1), 57–67. doi:10.1002/cpdd.848
Takaaki, A., KakyoM., , Tokui, T., Nakagomi, R., Nishio, T., Nakai, D., et al. (1999). Identification of a novel gene family encoding human liver-specific organic anion transporter LST-1. J. Biol. Chem. 274 (24), 17159–17163. doi:10.1074/jbc.274.24.17159
Taxax, N., and Bhartam, P. (2014). Drug metabolism: A fascinating link between chemistry and biology resonance. Reson. 19, 259–282. doi:10.1007/s12045-014-0031-0
Ueda, K. (2011). ABC proteins protect the human body and maintain optimal health. Biosci. Biotechnol. Biochem. 75 (3), 401–409. doi:10.1271/bbb.100816
Vincent, J. T., R Dalvie, D. K., and Friedman, H. L. (1998). Pharmacokinetics and metabolism of single oral doses of trovafloxacin. Am. J. Surg. 176 (6), 8S–13S. doi:10.1016/s0002-9610(98)00213-x
Wang, X. Q., Shen, C. L., Wang, B. N., Huang, X. H., Hu, Z. L., and Li, J. (2015). Genetic polymorphisms of CYP2C19*2 and ABCB1 C3435T affect the pharmacokinetic and pharmacodynamic responses to clopidogrel in 401 patients with acute coronary syndrome. Gene 558 (2), 200–207. doi:10.1016/j.gene.2014.12.051
Ware, J. A. (2006). Membrane transporters in drug discovery and development: A new mechanistic adme era. Mol. Pharm. 3 (1), 1–2. doi:10.1021/mp058084b
Weiner, M., Burman, W., Luo, C. C., Peloquin, C. A., Engle, M., Goldberg, S., et al. (2007). Effects of rifampin and multidrug resistance gene polymorphism on concentrations of moxifloxacin. Antimicrob. Agents Chemother. 51 (8), 2861–2866. doi:10.1128/AAC.01621-06
Weiner, M., Gelfond, J., Johnson-Pais, T. L., Engle, M., Peloquin, C. A., Johnson, J. L., et al. (2018). Elevated plasma moxifloxacin concentrations and SLCO1B1 g.-11187G>A polymorphism in adults with pulmonary tuberculosis. Antimicrob. Agents Chemother. 62 (5), e01802–e01817. doi:10.1128/AAC.01802-17
WHO (2020a). Global tuberculosis report 2020. Geneva: World Health Organization. Licence: CC BY-NC-SA 3.0 IGO.
WHO (2020b). WHO operational handbook on tuberculosis. Module 4: treatment -drug-resistant tuberculosis treatment. Geneva: World Health Organization. Licence: CC BY-NC-SA 3.0 IGO.
Woo, C. J., Teo, Y. L., Ho, H. K., Lee, J., Back, H. M., Yun, H. Y., et al. (2016). BSA and ABCB1 polymorphism affect the pharmacokinetics of sunitinib and its active metabolite in Asian mRCC patients receiving an attenuated sunitinib dosing regimen. Cancer Chemother. Pharmacol. 78 (3), 623–632. doi:10.1007/s00280-016-3104-9
Yan, Q. (2010). Membrane transporters and drug development: Relevance to pharmacogenomics, nutrigenomics, epigenetics, and systems biology. Methods Mol. Biol. 637, 1–21. doi:10.1007/978-1-60761-700-6_1
Keywords: pharmacogenetics, UGT1A1, UGT1A9, SLCO1B1, ABCB1, moxifloxacin, sitafloxacin, trovafloxacin
Citation: Annisa N, Barliana MI, Santoso P and Ruslami R (2022) Transporter and metabolizer gene polymorphisms affect fluoroquinolone pharmacokinetic parameters. Front. Pharmacol. 13:1063413. doi: 10.3389/fphar.2022.1063413
Received: 07 October 2022; Accepted: 28 November 2022;
Published: 12 December 2022.
Edited by:
Junmin Zhang, Lanzhou University, ChinaReviewed by:
Olga Butranova, Peoples’ Friendship University of Russia, RussiaRamazan Rezaei, Shahid Beheshti University of Medical Sciences, Iran
Lian-Sheng Wang, Nanjing Medical University, China
Copyright © 2022 Annisa, Barliana, Santoso and Ruslami. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Melisa I. Barliana, bWVsaXNhLmJhcmxpYW5hQHVucGFkLmFjLmlk
 Nurul Annisa
Nurul Annisa Melisa I. Barliana
Melisa I. Barliana Prayudi Santoso4
Prayudi Santoso4