- 1Department of Medical Science, Chungnam National University College of Medicine, Daejeon, South Korea
- 2Department of Microbiology, Chungnam National University College of Medicine, Daejeon, South Korea
- 3Infection Control Convergence Research Center, Chungnam National University College of Medicine, Daejeon, South Korea
The nucleoside inosine is an essential metabolite for purine biosynthesis and degradation; it also acts as a bioactive molecule that regulates RNA editing, metabolic enzyme activity, and signaling pathways. As a result, inosine is emerging as a highly versatile bioactive compound and second messenger of signal transduction in cells with diverse functional abilities in different pathological states. Gut microbiota remodeling is closely associated with human disease pathogenesis and responses to dietary and medical supplementation. Recent studies have revealed a critical link between inosine and gut microbiota impacting anti-tumor, anti-inflammatory, and antimicrobial responses in a context-dependent manner. In this review, we summarize the latest progress in our understanding of the mechanistic function of inosine, to unravel its immunomodulatory actions in pathological settings such as cancer, infection, inflammation, and cardiovascular and neurological diseases. We also highlight the role of gut microbiota in connection with inosine metabolism in different pathophysiological conditions. A more thorough understanding of the mechanistic roles of inosine and how it regulates disease pathologies will pave the way for future development of therapeutic and preventive modalities for various human diseases.
Introduction
Inosine, an inert purine nucleoside, is formed by breakdown (deamination) of adenosine both intracellularly and extracellularly; it also generated by the action of 5′-nucleotidase on inosine monophosphate (IMP) (Conway and Cooke, 1939; Itoh et al., 1967; Chen et al., 1996). Recent studies have revealed that inosine is also produced by several species found in the gut microbiome and modulates host immune and inflammatory functions (Wang et al., 2020; Brown et al., 2021). Inosine can be metabolized into hypoxanthine, xanthine, and uric acid (Sorensen, 1970; Zoref-Shani et al., 1988; Doyle et al., 2018; Garcia-Gil et al., 2021). Cell membrane transport of inosine is mediated by equilibrative and concentrative nucleoside transporters (Körber et al., 1975; Belt et al., 1993; Cass et al., 1999; Miller et al., 2021). Inosine functions are mediated in receptor-dependent or–independent manners. The receptor-mediated function of inosine is thought to be related to adenosine receptor family members including A1, A2A, A2B, and A3 G-protein coupled receptors (Kypson and Hait, 1976; Fredholm et al., 2001; Fredholm et al., 2011; Welihinda et al., 2018). Compared with the known role of adenosine as a signaling molecule, the function of inosine in the context of physiological and pathological responses in human health and diseases remain poorly understood.
Earlier studies suggested that the inert purine nucleoside inosine has neuroprotective, cardioprotective, and immunomodulatory effects in different experimental models (Aviado, 1983; Haskó et al., 2004; Guinzberg et al., 2006). The beneficial function of inosine has been thought to be mediated through modulation of oxidative stress and inflammatory responses (Haskó et al., 2004). More recent studies have revealed therapeutic effects of inosine in motor function improvement during neurologic injury or stroke (Benowitz et al., 1999; Kuricova et al., 2014), learning and memory (Ruhal and Dhingra, 2018), and Parkinson’s disease (Schwarzschild et al., 2014). Inosine treatment also results in the activation of anti-tumor and anti-inflammatory responses in different disease models (Haskó et al., 2000; Mabley et al., 2003; Panebianco et al., 2018; Tobólska et al., 2018; Kovachev, 2020; Mager et al., 2020). Importantly, the beneficial effects of inosine have been demonstrated with the preclinical and clinical use of Isoprinosine (inosine pranobex), formed by inosine with the immunostimulatory dimepranol acedoben (acetamidobenzoic acid and dimethylaminoisopropanol), for treatment of neurological disorders and acute respiratory viral infections (Beran et al., 2016; Sliva et al., 2019; Teixeira et al., 2020; Beran et al., 2021; Nascimento et al., 2021; Teixeira et al., 2022a; Yang et al., 2022). In this Review, we discuss recent updates on the regulation of pathological responses by inosine and its association with gut microbiota remodeling in different contexts. Furthermore, we discuss the functions of exogenous inosine in terms of cancers, inflammation, infection, and cardiovascular and neurological diseases through its immunomodulatory roles (Table 1).
Overview of inosine biology
Inosine, an intermediate in purine metabolism, consists of hypoxanthine and ribose. Three enzymatic reactions can form inosine endogenously (Figure 1). First, adenosine deaminase (ADA) irreversibly removes the amine group of adenine ring intracellularly and extracellularly (Akedo et al., 1972; Van Heukelom et al., 1976; Van der Weyden and Kelley, 1977; Akeson et al., 1988). In particular, double-stranded RNA (dsRNA)-specific adenosine deaminases (ADARs) mediate adenosine-to-inosine editing (Liddicoat et al., 2015; George et al., 2016; Eisenberg and Levanon, 2018; Nakahama and Kawahara, 2020; Pfaller et al., 2021). ADAR1 and ADAR2 maintain balanced immune activation and self-tolerance through inhibiting dsRNA-binding proteins, such as RIG-I-like receptors, protein kinase R, and oligoadenylate synthases-RNAse L (Yang et al., 2014; Liddicoat et al., 2015; George et al., 2016; Jain et al., 2019; Lamers et al., 2019; Schaffer et al., 2020; Pfaller et al., 2021; Li et al., 2022). In a recent study, the severe autoinflammatory disease Aicardi–Goutières syndrome mouse model with ADAR1 mutation reveals immunopathology associated with type I interferon signaling and Z-binding protein 1 (Rice et al., 2012; de Reuver et al., 2022). Contrary to ADAR1 and ADAR2, ADAR3 suppresses A-to-I editing by binding to RNA-binding domains (Chen et al., 2000; Oakes et al., 2017; Raghava Kurup et al., 2022). Second, 5′-nucleotidase catalyzes the reversible dephosphorylation of IMP both inside and outside cells (Barsotti et al., 2005; Ipata and Balestri, 2013). Cytosolic 5′-nucleotidase (NT5C2) is correlated with chemotherapy resistance in acute lymphoblastic leukemia (Tzoneva et al., 2013). Ecto-5′-nucleotidase (NT5E, CD73) mediates immune suppression (Romio et al., 2011; Lovászi et al., 2021). Third, purine nucleoside phosphorylase (PNP) converts hypoxanthine and ribose-1-phosphate (R-1-P) into inosine and thermodynamically favors this enzymatic synthesis over phosphorolysis (Kalckar, 1947; Friedmin, 1950; Tozzi et al., 2006). However, the PNP reaction equilibrium is biased toward inosine degradation due to the more significant concentration of inorganic phosphate than base and R-1-P (Traut, 1994), and the linked reaction of hypoxanthine catalyzed by hypoxanthine phosphoribosyl transferase and xanthine oxidase (Pugmire and Ealick, 2002; Il’icheva et al., 2020). Xanthine oxidase catabolizes hypoxanthine into uric acid via xanthine (Heinz et al., 1980; Moriwaki et al., 1999; Doyle et al., 2018). Humans and higher primates excrete uric acid in their urine, but other mammals convert uric acid to allantoin by uricase and then excrete it in urine (Heinz et al., 1980; Kurtz et al., 1986; Moriwaki et al., 1999). Uric acid, the end product of human purine metabolism, is one of the major antioxidants (Ames et al., 1981; Whiteman et al., 2002; Muraoka and Miura, 2003) and protects against neurological and intestinal diseases (Hooper et al., 1998; Hooper et al., 2000; Toncev et al., 2002; Ascherio et al., 2009; Matsuo et al., 2015; Yasutake et al., 2017). Consequently, inosine-related metabolism should be extensively explored for its multidimensional effects on human illnesses.
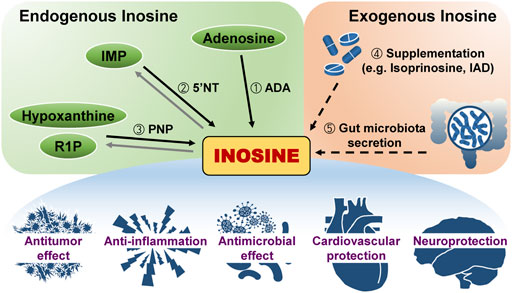
FIGURE 1. Inosine generation in the body and its effect. Inosine originates either within or outside the body. Three different reactions produce endogenous inosine: ①Hydrolytic deamination of adenosine by adenosine deaminase (ADA), ② dephosphorylation of inosine monophosphate (IMP) by 5′-nucleotidase (5′NT), and ③ reaction of hypoxanthine and ribose-1-phosphate (R1P) by purine nucleoside phosphorylase (PNP). Exogenous inosine is from two ways: ④ Supplementation such as isoprinosine or inosine acedoben dimepranol (IAD) or ⑤ secretion from gut microbiota. Inosine has various effects on the body.
In the human gut, 1013–1014 microorganisms are found and are emerging as important players in the pathogenesis and therapeutics of a variety of human diseases (Wardman et al., 2022). Recently, a mechanistic link between microbiota and inosine has been revealed. For example, the microbiome-derived Bxa, an abundant ADP-ribosyltransferase (ADPRT) of Bacteroides, induced secretion of inosine as a carbon source, thus acting as a bacterial fitness factor (Brown et al., 2021). Gut microbiota remodeling enriched Bifidobacterium pseudolongum and supplementation with its metabolite inosine increased the anti-tumor effects of immune checkpoint blockade and functioned as a carbon source for CD8+ effector T-cell function (Wang et al., 2020). In a more recent study, inosine was found to be a microbiota-derived immunostimulatory metabolite that enhanced immunotherapeutic effects and antitumor T-cell responses (Kroemer and Zitvogel, 2020; Mager et al., 2020). Indeed, gut microbiota remodeling enriched B. pseudolongum significantly increased the anticancer immunotherapy response through the generation of inosine via A2A receptors (Mager et al., 2020).
A multifaceted role for inosine in cancer
Multiple studies have suggested that inosine functions as a crucial biomarker metabolite associated with cancer metastasis, drug resistance and/or treatment, and tumor progression (Li et al., 2021a; Lee et al., 2021; Stockard et al., 2021). A recent study showed that inosine can predict the metastatic potential of lung squamous cell carcinoma (Lee et al., 2021). Inosine has also been shown to be associated with acute myeloid leukemia with cytarabine resistance (Stockard et al., 2021). In esophageal squamous cell carcinoma, inosine has been associated with cancer progression (Li et al., 2021a). In colorectal cancer organoids, inosine was elevated following treatment with 5-fluorouracil (Neef et al., 2020), although the underlying mechanism is unclear. In head and neck squamous cell carcinoma, inosine and adenosine are abundant purine metabolites present in exosomes from the supernatant of UMSCC47 cells (Ludwig et al., 2020). Inosine is a crucial serum metabolite that can differentiate between low- and high-grade bladder cancer patients (Tan et al., 2017). Moreover, urine and serum levels of inosine as well as other metabolites can be used to characterize hepatocellular carcinoma patients (Chen et al., 2011; Ladep et al., 2014). These data suggest that inosine has a promising role in the diagnosis and grading of tumors as a signature metabolite. By contrast, inosine levels are generally decreased in pancreatic cancer patients, and elevated by a low carbohydrate ketogenic diet, compared to a general hospital diet (Kang et al., 2019). These data collectively suggest that inosine functions as a potential biomarker for prediction of cancer risk, drug response, and early detection of metastasis of various tumors.
Several recent studies have indicated the role of inosine in anti-tumor responses in various cancers. In a xenograft model of pancreatic ductal adenocarcinoma, gemcitabine-mediated chemotherapy significantly induced activation of nuclear factor (NF)-κB-mediated inflammatory responses and decreased the proportion of Firmicutes and Bacteroides. The chemotherapy also suppressed serum levels of inosine and xanthine (Panebianco et al., 2018), suggesting chemotherapy may influence gut microbiota remodeling and changes in serum metabolites. Moreover, inosine supplementation promoted immune checkpoint blockade and provided a carbon source for CD8+ effector T-cell function (Wang et al., 2020). In a more recent study, inosine was found to be a microbiota-derived immunostimulatory metabolite that enhanced immunotherapeutic effects and antitumor T-cell responses (Kroemer and Zitvogel, 2020; Mager et al., 2020). Indeed, gut microbiota remodeling enriched B. pseudolongum and significantly increased the effects of immunotherapy through the generation of inosine via A2A receptors (Mager et al., 2020).
Although inosine pranobex (isoprinosin) has been shown to exhibit significant antiviral effects (Beran et al., 2021), few studies have investigated its effects in cancer. Treatment with isoprinosin significantly increased the clearance of cervical human papillomavirus (HPV) infection during postoperative immunotherapy in women receiving surgical treatment for high-grade squamous intraepithelial cervical lesions (Kovachev, 2020). Addtionally, a previous study revealed increased cytotoxicity of fibroblasts and hepatocellular carcinoma HepG2 cells following inosine pranobex treatment (Tobólska et al., 2018). Because inosine pranobex is useful in viral infections and has remarkable immunomodulating functions in both cellular and humoral immune responses (Kovachev, 2021), the therapeutic application of inosine pranobex is challenging in terms of virus-related tumors accompanied with chronic inflammation and immunosuppression. Although it would have been shown to be therapeutically beneficial in animal studies, the use of inosine pranobex in anticancer regimens should be considered following more data from clinical trials.
Inosine and antimicrobial immunity
In nonhuman primate models infected with human immunodeficiency virus type 1/simian immunodeficiency virus (HIV-1/SIV), elevated inosine levels have been related to immune activation and disease progression markers (He et al., 2015). The disease-progressive model reveals higher ADA activity and CD26 expression on intestinal T cells than the nonprogressive model, suggesting that adenosine degradation stimulates T cells (He et al., 2015). In addition, several recent studies have suggested a link between inosine and antimicrobial function during bacterial infection. Our recent study showed that inosine, as a metabolite of B. pseudolongum, contributes to antimicrobial responses in mouse models with Mycobacteroides abscessus (Mabc) infection (Kim et al., 2022). Interestingly, ʟ-arginine administration of Mabc-infected mice led to gut microbiota remodeling toward enrichment of B. pseudolongum, which promoted effector T-cell responses with IFN-γ activation and inducible nitric oxide synthase (iNOS) expression, indicating a Th1-mediated M1 shift (Kim et al., 2022). Importantly, ʟ-arginine administration upregulated the serum level of inosine in mice infected with Mabc, and inosine treatment exhibited a similar protective phenotype as observed in ʟ-arginine-treated conditions in the context of NTM infections (Sun et al., 2020; Kim et al., 2022). It was previously reported suberic acid is produced by B. pseudolongum (Sun et al., 2020); however, treatment of Mabc-infected mice with suberic acid did not show any protective immune responses during Mabc infection. These data strongly suggest that inosine, produced by B. pseudolongum, plays a distinct role in the enhancement of antimicrobial responses during NTM infection.
Isoprinosine appears to be an effective treatment for various viral infections through pleiotropic immunomodulatory roles including T-cell activation and proinflammatory cytokine-mediated functions (Sliva et al., 2019; Beran et al., 2021). Isoprinosine combined with antiviral and antioxidant drugs has been shown to produce a protective effect, with increased survival and decreased lung pathologies, in influenza H3N2 virus-infected mice (Pavlova et al., 2018). In addition, a Phase 4 clinical study showed that isoprinosine is safe for treatment of patients with acute respiratory viral infections and is effective in the resolution of influenza-like symptoms in individuals less than 50 years of age (Beran et al., 2016). A preliminary report emphasized the effectiveness of inosine pranobex in the reduction of the case-fatality rate in older COVID-19 patients in the Czech Republic (Beran et al., 2020), although larger-scale trials are warranted to clarify the therapeutic effects of inosine pranobex against COVID-19. Inosine acedoben dimepranol (IAD), another licensed inosine-based drug, has been shown to boost NK cell numbers in clinical trials (Rumel Ahmed et al., 2017). Future clinical studies are needed to determine whether isoprinosine or IAD can be effectively used as adjunctive drugs in antiviral therapeutics without overt complications.
Inosine and therapeutic implications for inflammation
A recent report showed that inosine, metabolized from IMP, suppressed tumor necrosis factor (TNF)-α in vitro and in vivo, although A2A, A2B, and A3 receptors were not involved (Lovászi et al., 2021). In addition, inosine augmented IL-β production in response to NLRP3 inflammasome stimuli in macrophages (Lovászi et al., 2021). These data suggest that inosine is involved in the activation or suppression of inflammatory responses, depending on the stimuli.
Several studies have shown the combined beneficial effects of inosine and probiotics in experimental inflammatory disease models. For example, Lactobacillus rhamnosus GG (LGG) combined with inosine has been shown to ameliorate hepatic inflammation and restore regulatory T-cell function in an alcohol-induced liver injury model (Zhu et al., 2022). Importantly, combined treatment of LGG and inosine significantly improved immune homeostasis and intestinal microecology with amelioration of gut dysbiosis during liver injury (Zhu et al., 2022). Interestingly, inosine treatment can alter gut microbiota toward an abundance of Bifidobacterium and Lachnospiraceae UCG-006 and suppress TLR4 signaling, thus negatively regulating hepatic inflammation and damage (Guo et al., 2021).
In the context of colitis, dietary barley leaf supplementation has been shown to produce anti-inflammatory effects in dextran sulfate sodium (DSS)-induced colitis and dysbiosis of the gut microbiome. Mechanistically, dietary barley leaf elicits inosine accumulation in colonic epithelial cells through the alteration of gut bacterial composition, such as an abundance of Lactobacillus (Li et al., 2021b). In addition, the exogenous inosine administration showed a similar protective effect on colitis through the activation of A2A receptor and peroxisome proliferator-activated receptor (PPAR)-γ signaling (Li et al., 2021b). Furthermore, indomethacin-induced enteropathy is ameliorated by inosinic acid (1,000 mg/kg, i.p.) in mice. Inosinic acid treatment leads to increased serum levels of uric acid, resulting in protection against intestinal injury via antioxidative effects (Yasutake et al., 2017).
A recent study showed that NAD + precursor nicotinamide riboside (NR) suppresses lipopolysaccharide-induced IFN-β production and autophagy activation in myeloid cells at least partly through inosine-mediated signaling (Wu et al., 2022). Importantly, NR indicated anti-inflammatory effects in monocytes from patients with the autoimmune disorder systemic lupus erythematosus (SLE), showing dysregulated type I IFN production (Wu et al., 2022). Plasma inosine increase via Limosilactobacillus reuteri (LR) administration exhibits a protective effect in the scurfy mouse, lacking regulatory T cells, that mimics human IPEX syndrome (immune dysregulation, polyendocrinopathy, and enteropathy, with X-linked inheritance) (He et al., 2017; Liu et al., 2021). LR reverses decreased inosine levels in plasma from scurfy mice and impacts the amelioration of disease pathologies and multiorgan inflammation through A2A receptors (He et al., 2017; Liu et al., 2021). Interestingly, the ability of LR to increase plasma inosine is unique compared to the probiotic LGG (Liu et al., 2021). These data suggest inosine may act as a bioactive molecule responsible for distinct anti-inflammatory properties in immunodeficiency and autoimmune diseases.
Inosine and other diseases
Inosine and cardiovascular diseases
Inosine is thought to be as “a coronary dilator” through relaxation of the coronary artery and inotropic action, although the mechanisms of action remain unclear (Aviado, 1983). Alternatively, the role of ADA competitive inhibition by inosine cannot be excluded. In addition, accumulating evidence suggests that adenosine has a protective role against endothelial dysfunction and vascular inflammation (Kutryb-Zajac et al., 2020). Dysregulated ADA activity leads to cardiovascular pathologies such as atherosclerosis, thrombosis, and myocardial ischemia-reperfusion injury (Kutryb-Zajac et al., 2020), suggesting that ADA may be a critical target for treatment of cardiovascular disease. Interestingly, co-administration of febuxostat and inosine exhibited a favorable response in two patients with mitochondrial diseases, cardiomyopathy and diabetes (Kamatani et al., 2019).
In a hypercholesterolemic rat model, inosine treatment improved endothelium-mediated vasodilatation and antiplatelet function (Lima et al., 2020). A recent study suggested that urinary inosine is inversely associated with coronary heart disease risk in men (Yoon et al., 2020). Several metabolites including inosine are prominent biomarkers in Kawasaki disease patients with coronary artery lesions (Qian et al., 2021). The existence of brown adipose tissue (BAT) is an independent factor for lowering cardiovascular and metabolic disease (Becher et al., 2021). Extracellular inosine enhances the energy expenditure of BAT in mice. In addition, loss of function mutation of human SLC29A1, which decreases extracellular inosine levels, is strongly correlated with lean body mass and non-obese (Niemann et al., 2022). These data underpin the protective effects of inosine on the cardiovascular system.
Inosine in psychiatric and neurological pathologies
Recent studies have shown neuroprotective and neuromodulatory functions of inosine in a variety of neurological and psychiatric diseases (Nascimento et al., 2021). Inosine treatment ameliorated 3-nitropropionic acid (3-NP)-induced neurotoxicity and boosted brain-derived neurotrophic factor (BDNF) levels, p-cAMP response element-binding protein (CREB) expression, and glutathione content (El-Shamarka et al., 2022). In addition, metformin-mediated amelioration of methamphetamine withdrawal syndrome is at least partly mediated through altered bacterial composition and metabolite changes such as abundance of Rikenellaceae and inosine (Yang et al., 2022). Interestingly, inosine supplementation of mice improved methamphetamine withdrawal-mediated anxiety and depression-like symptoms (Yang et al., 2022), indicating that metformin effects may depend on microbiota-derived inosine.
In rat models of Alzheimer’s disease, inosine treatment has been shown to prevent memory deficits, suppress immunoreactivity via brain A2A receptor, and enhance anti-inflammatory cytokine levels and oxidative alterations in the brain (Teixeira et al., 2020; Teixeira et al., 2022a). In addition, inosine administration had beneficial effects in a rat model of diabetic peripheral neuropathy, resulting in a hypoglycemic effect and enhanced myelination (Abdelkader et al., 2022). Inosine-induced neuroprotective function depends on Nrf2 expression, downstream HO-1, and suppression of PKC and TRPV1 (Abdelkader et al., 2022). Inosine treatment also resulted in elevated memory acquisition and consolidation in a rat model of scopolamine-induced cognitive impairment. These effects are partly mediated through modulation of brain redox status, cholinesterase function, and ion pump activity, suggesting promising approaches for neurodegenerative diseases (Teixeira et al., 2022b). In a ketamine-induced rat mania model, inosine administration may prevent hyperlocomotion and attenuation of maniac phase symptoms (Camerini et al., 2020). In an experimental model of sciatic nerve crush injury in mice, inosine treatment resulted in accelerated axonal regeneration and a recovery of motor and sensory functions (Soares Dos Santos Cardoso et al., 2019).
A clinical trial with co-administration of febuxostat and inosine showed improvement of the symptoms of Parkinson’s disease (Watanabe et al., 2020). However, another recent clinical trial showed that inosine treatment does not modify progression of early Parkinson’s disease (Schwarzschild et al., 2021). In a clinical trial for use of IMP in multiple system atrophy (MSA), IMP raised serum levels of uric acids, and was considered to be tolerable and safe during a 24-week treatment (Jung Lee et al., 2021), although a more long-term follow-up study is warranted.
Concluding remarks
Recent advances have contributed to our understanding of the role of inosine as a biomarker or a regulator of immunity, infection, inflammation, cancer, and other pathological conditions. In addition, accumulating reports suggest that the levels of inosine or inosine-related enzymes are dysregulated in diseases such as cancers. More studies are needed to identify how inosine levels are regulated in different tissues under various physiological and pathological conditions. The information available on the impact of inosine on immunomodulatory and protective functions has identified this metabolite as a possible therapeutic target for tumors, inflammation, infection, and cardiovascular and neurological disorders. Future studies with preclinical models and clinical research are required to bring the inosine-based therapeutic approach into the clinic.
Several microbes from the intestinal microbiota have been reported to produce inosine, exhibiting therapeutic efficacy in various disease models. Future molecular studies will be important to reveal how the link between the microbiota and inosine regulates immune and inflammatory responses in specific human diseases.
The use of isoprinosine, a synthetic agent of inosine with an immunostimulant, in several clinical trials has suggested that inosine or inosine-based drugs may be therapeutically useful in neurological and autoimmune diseases. However, most data regarding the physiological and therapeutic potential of inosine have been derived from experimental studies. There are also concerns for the possibility of uric acid stones, which can be produced by inosine metabolism. Therefore, another strategy based on the regulation of inosine-degrading enzymes (i.e., PNP) could be also useful for potential treatment targeting inosine-based therapeutics. Future preclinical and clinical trials are needed to assess the safety and adverse effects associated with long-term use of inosine or inosine-related enzyme modulation.
To date, there is limited information on the molecular mechanisms through which inosine exerts its biological functions as a signaling mediator. Evidence suggests that inosine may function through multiple subtypes of adenosine receptors. However, it remains unclear how inosine targets adenosine receptor subtypes or whether there are alternative inosine receptors in different cell types. Understanding the pathways responsible for the dysregulation of inosine production and finding inosine-specific effectors will facilitate the development of therapeutic strategies against various disorders.
Author contributions
E-KJ and IK contributed to the conception and critically revised the manuscript. Both authors contributed to the article and approved the submitted version.
Funding
This research was supported by the National Research Foundation of Korea (NRF) grant funded by the Korea government (MSIT) (No. 2017R1A5A2015385) and two grants of the Korea Health Technology R&D Project through the Korea Health Industry Development Institute (KHIDI) funded by the Ministry of Health & Welfare, Republic of Korea (grant numbers: HR20C0025, HI22C1361).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Abdelkader, N. F., Ibrahim, S. M., Moustafa, P. E., and Elbaset, M. A. (2022). Inosine mitigated diabetic peripheral neuropathy via modulating GLO1/AGEs/RAGE/NF-κB/Nrf2 and TFG-β/PKC/TRPV1 signaling pathways. Biomed. Pharmacother. 145, 112395. doi:10.1016/j.biopha.2021.112395
Akedo, H., Nishihara, H., Shinkai, K., Komatsu, K., and Ishikawa, S. (1972). Multiple forms of human adenosine deaminase. I. Purification and characterization of two molecular species. Biochim. Biophys. Acta 276 (1), 257–271. doi:10.1016/0005-2744(72)90028-9
Akeson, A. L., Wiginton, D. A., Dusing, M. R., States, J. C., and Hutton, J. J. (1988). Mutant human adenosine deaminase alleles and their expression by transfection into fibroblasts. J. Biol. Chem. 263 (31), 16291–16296. doi:10.1016/s0021-9258(18)37591-4
Ames, B. N., Cathcart, R., Schwiers, E., and Hochstein, P. (1981). Uric acid provides an antioxidant defense in humans against oxidant- and radical-caused aging and cancer: A hypothesis. Proc. Natl. Acad. Sci. U. S. A. 78 (11), 6858–6862. doi:10.1073/pnas.78.11.6858
Ascherio, A., LeWitt, P. A., Xu, K., Eberly, S., Watts, A., Matson, W. R., et al. (2009). Urate as a predictor of the rate of clinical decline in Parkinson disease. Arch. Neurol. 66 (12), 1460–1468. doi:10.1001/archneurol.2009.247
Barsotti, C., Pesi, R., Giannecchini, M., and Ipata, P. L. (2005). Evidence for the involvement of cytosolic 5'-nucleotidase (cN-II) in the synthesis of guanine nucleotides from xanthosine. J. Biol. Chem. 280 (14), 13465–13469. doi:10.1074/jbc.M413347200
Becher, T., Palanisamy, S., Kramer, D. J., Eljalby, M., Marx, S. J., Wibmer, A. G., et al. (2021). Brown adipose tissue is associated with cardiometabolic health. Nat. Med. 27 (1), 58–65. doi:10.1038/s41591-020-1126-7
Belt, J. A., Marina, N. M., Phelps, D. A., and Crawford, C. R. (1993). Nucleoside transport in normal and neoplastic cells. Adv. Enzyme Regul. 33, 235–252. doi:10.1016/0065-2571(93)90021-5
Benowitz, L. I., Goldberg, D. E., Madsen, J. R., Soni, D., and Irwin, N. (1999). Inosine stimulates extensive axon collateral growth in the rat corticospinal tract after injury. Proc. Natl. Acad. Sci. U. S. A. 96 (23), 13486–13490. doi:10.1073/pnas.96.23.13486
Beran, J., Šalapová, E., and Špajdel, M. (2016). Inosine pranobex is safe and effective for the treatment of subjects with confirmed acute respiratory viral infections: Analysis and subgroup Analysis from a phase 4, randomised, placebo-controlled, double-blind study. BMC Infect. Dis. 16 (1), 648. doi:10.1186/s12879-016-1965-5
Beran, J., Špajdel, M., Katzerová, V., Holoušová, A., Malyš, J., Finger Rousková, J., et al. (2020). Inosine pranobex significantly decreased the case-fatality rate among PCR positive elderly with SARS-CoV-2 at three nursing homes in the Czech Republic. Pathogens 9 (12), E1055. doi:10.3390/pathogens9121055
Beran, J., Špajdel, M., and Slíva, J. (2021). Inosine pranobex deserves attention as a potential immunomodulator to achieve early alteration of the COVID-19 disease course. Viruses 13 (11), 2246. doi:10.3390/v13112246
Brown, E. M., Arellano-Santoyo, H., Temple, E. R., Costliow, Z. A., Pichaud, M., Hall, A. B., et al. (2021). Gut microbiome ADP-ribosyltransferases are widespread phage-encoded fitness factors. Cell Host Microbe 29 (9), 1351–1365.e11. doi:10.1016/j.chom.2021.07.011
Camerini, L., Ardais, A. P., Xavier, J., Bastos, C. R., Oliveira, S., Soares, M. S. P., et al. (2020). Inosine prevents hyperlocomotion in a ketamine-induced model of mania in rats. Brain Res. 1733, 146721. doi:10.1016/j.brainres.2020.146721
Cass, C. E., Young, J. D., Baldwin, S. A., Cabrita, M. A., Graham, K. A., Griffiths, M., et al. (1999). Nucleoside transporters of mammalian cells. Pharm. Biotechnol. 12, 313–352. doi:10.1007/0-306-46812-3_12
Chen, C. X., Cho, D. S., Wang, Q., Lai, F., Carter, K. C., and Nishikura, K. (2000). A third member of the RNA-specific adenosine deaminase gene family, ADAR3, contains both single- and double-stranded RNA binding domains. Rna 6 (5), 755–767. doi:10.1017/s1355838200000170
Chen, T., Xie, G., Wang, X., Fan, J., Qiu, Y., Zheng, X., et al. (2011). Serum and urine metabolite profiling reveals potential biomarkers of human hepatocellular carcinoma. Mol. Cell. Proteomics 10 (7), M110.004945004945. doi:10.1074/mcp.M110.004945
Chen, W., Hoerter, J., and Guéron, M. (1996). A comparison of AMP degradation in the perfused rat heart during 2-deoxy-D-glucose perfusion and anoxia. Part I: The release of adenosine and inosine. J. Mol. Cell. Cardiol. 28 (10), 2163–2174. doi:10.1006/jmcc.1996.0208
Conway, E. J., and Cooke, R. (1939). The deaminases of adenosine and adenylic acid in blood and tissues. Biochem. J. 33 (4), 479–492. doi:10.1042/bj0330479
de Reuver, R., Verdonck, S., Dierick, E., Nemegeer, J., Hessmann, E., Ahmad, S., et al. (2022). ADAR1 prevents autoinflammation by suppressing spontaneous ZBP1 activation. Nature 607 (7920), 784–789. doi:10.1038/s41586-022-04974-w
Doyle, C., Cristofaro, V., Sullivan, M. P., and Adam, R. M. (2018). Inosine - a multifunctional treatment for complications of neurologic injury. Cell. Physiol. biochem. 49 (6), 2293–2303. doi:10.1159/000493831
Eisenberg, E., and Levanon, E. Y. (2018). A-to-I RNA editing - immune protector and transcriptome diversifier. Nat. Rev. Genet. 19 (8), 473–490. doi:10.1038/s41576-018-0006-1
El-Shamarka, M. E., El-Sahar, A. E., Saad, M. A., Assaf, N., and Sayed, R. H. (2022). Inosine attenuates 3-nitropropionic acid-induced huntington's disease-like symptoms in rats via the activation of the A2AR/BDNF/TrKB/ERK/CREB signaling pathway. Life Sci. 300, 120569. doi:10.1016/j.lfs.2022.120569
Fredholm, B. B., Ijzerman, A. P., Jacobson, K. A., Klotz, K. N., and Linden, J. (2001). International union of Pharmacology. Xxv. Nomenclature and classification of adenosine receptors. Pharmacol. Rev. 53 (4), 527–552.
Fredholm, B. B., Ijzerman, A. P., Jacobson, K. A., Linden, J., and Müller, C. E. (2011). International union of basic and clinical Pharmacology. LXXXI. Nomenclature and classification of adenosine receptors-an update. Pharmacol. Rev. 63 (1), 1–34. doi:10.1124/pr.110.003285
Friedmin, M. (1950). Desoxyribose-1-Phosphate. II. The isolation of crystalline desoxyribose-1-phosphate. J. Biol. Chem. 184 (2), 449–459.
Garcia-Gil, M., Camici, M., Allegrini, S., Pesi, R., and Tozzi, M. G. (2021). Metabolic aspects of adenosine functions in the brain. Front. Pharmacol. 12, 672182. doi:10.3389/fphar.2021.672182
George, C. X., Ramaswami, G., Li, J. B., and Samuel, C. E. (2016). Editing of cellular self-RNAs by adenosine deaminase ADAR1 suppresses innate immune stress responses. J. Biol. Chem. 291 (12), 6158–6168. doi:10.1074/jbc.M115.709014
Guinzberg, R., Cortés, D., Díaz-Cruz, A., Riveros-Rosas, H., Villalobos-Molina, R., and Piña, E. (2006). Inosine released after hypoxia activates hepatic glucose liberation through A3 adenosine receptors. Am. J. Physiol. Endocrinol. Metab. 290 (5), E940–E951. doi:10.1152/ajpendo.00173.2005
Guo, W., Xiang, Q., Mao, B., Tang, X., Cui, S., Li, X., et al. (2021). Protective effects of microbiome-derived inosine on lipopolysaccharide-induced acute liver damage and inflammation in mice via mediating the TLR4/NF-κB pathway. J. Agric. Food Chem. 69 (27), 7619–7628. doi:10.1021/acs.jafc.1c01781
Haskó, G., Kuhel, D. G., Németh, Z. H., Mabley, J. G., Stachlewitz, R. F., Virág, L., et al. (2000). Inosine inhibits inflammatory cytokine production by a posttranscriptional mechanism and protects against endotoxin-induced shock. J. Immunol. 164 (2), 1013–1019. doi:10.4049/jimmunol.164.2.1013
Haskó, G., Sitkovsky, M. V., and Szabó, C. (2004). Immunomodulatory and neuroprotective effects of inosine. Trends Pharmacol. Sci. 25 (3), 152–157. doi:10.1016/j.tips.2004.01.006
He, B., Hoang, T. K., Wang, T., Ferris, M., Taylor, C. M., Tian, X., et al. (2017). Resetting microbiota by Lactobacillus reuteri inhibits T reg deficiency-induced autoimmunity via adenosine A2A receptors. J. Exp. Med. 214 (1), 107–123. doi:10.1084/jem.20160961
He, T., Brocca-Cofano, E., Gillespie, D. G., Xu, C., Stock, J. L., Ma, D., et al. (2015). Critical role for the adenosine pathway in controlling simian immunodeficiency virus-related immune activation and inflammation in gut mucosal tissues. J. Virol. 89 (18), 9616–9630. doi:10.1128/jvi.01196-15
Heinz, F., Reckel, S., Pilz, R., and Kalden, J. R. (1980). A new spectrophotometric assay for enzymes of purine metabolism. IV. Determination of adenosine deaminase. Enzyme 25 (1), 50–55. doi:10.1159/000459215
Hooper, D. C., Scott, G. S., Zborek, A., Mikheeva, T., Kean, R. B., Koprowski, H., et al. (2000). Uric acid, a peroxynitrite scavenger, inhibits CNS inflammation, blood-CNS barrier permeability changes, and tissue damage in a mouse model of multiple sclerosis. Faseb J. 14 (5), 691–698. doi:10.1096/fasebj.14.5.691
Hooper, D. C., Spitsin, S., Kean, R. B., Champion, J. M., Dickson, G. M., Chaudhry, I., et al. (1998). Uric acid, a natural scavenger of peroxynitrite, in experimental allergic encephalomyelitis and multiple sclerosis. Proc. Natl. Acad. Sci. U. S. A. 95 (2), 675–680. doi:10.1073/pnas.95.2.675
Il'icheva, I. A., Polyakov, K. M., and Mikhailov, S. N. (2020). Strained conformations of nucleosides in active sites of nucleoside phosphorylases. Biomolecules 10 (4), E552. doi:10.3390/biom10040552
Ipata, P. L., and Balestri, F. (2013). The functional logic of cytosolic 5'-nucleotidases. Curr. Med. Chem. 20 (34), 4205–4216. doi:10.2174/0929867311320340002
Itoh, R., Mitsui, A., and Tsushima, K. (1967). 5'-Nucleotidase of chicken liver. Biochim. Biophys. Acta 146 (1), 151–159. doi:10.1016/0005-2744(67)90081-2
Jain, M., Jantsch, M. F., and Licht, K. (2019). The editor's I on disease development. Trends Genet. 35 (12), 903–913. doi:10.1016/j.tig.2019.09.004
Jung Lee, J., Han Yoon, J., Jin Kim, S., Soo Yoo, H., Jong Chung, S., Hyun Lee, Y., et al. (2021). Inosine 5'-monophosphate to raise serum uric acid level in multiple system Atrophy (IMPROVE-MSA study). Clin. Pharmacol. Ther. 109 (5), 1274–1281. doi:10.1002/cpt.2082
Kalckar, H. M. (1947). The enzymatic synthesis of purine ribosides. J. Biol. Chem. 167 (2), 477–486. doi:10.1016/s0021-9258(17)31000-1
Kamatani, N., Kushiyama, A., Toyo-Oka, L., and Toyo-Oka, T. (2019). Treatment of two mitochondrial disease patients with a combination of febuxostat and inosine that enhances cellular ATP. J. Hum. Genet. 64 (4), 351–353. doi:10.1038/s10038-018-0558-0
Kang, C. M., Yun, B., Kim, M., Song, M., Kim, Y. H., Lee, S. H., et al. (2019). Postoperative serum metabolites of patients on a low carbohydrate ketogenic diet after pancreatectomy for pancreatobiliary cancer: A nontargeted metabolomics pilot study. Sci. Rep. 9 (1), 16820. doi:10.1038/s41598-019-53287-y
Kim, Y. J., Lee, J. Y., Lee, J. J., Jeon, S. M., Silwal, P., Kim, I. S., et al. (2022). Arginine-mediated gut microbiome remodeling promotes host pulmonary immune defense against nontuberculous mycobacterial infection. Gut Microbes 14 (1), 2073132. doi:10.1080/19490976.2022.2073132
Körber, W., Meisterernst, E. B., and Hermann, G. (1975). Quantitative measurement of adenosine deaminase from human erythrocytes. Clin. Chim. Acta. 63 (3), 323–333. doi:10.1016/0009-8981(75)90054-6
Kovachev, S. M. (2021). A review on inosine pranobex immunotherapy for cervical HPV-positive patients. Infect. Drug Resist. 14, 2039–2049. doi:10.2147/idr.S296709
Kovachev, S. M. (2020). Immunotherapy in patients with local HPV infection and high-grade squamous intraepithelial lesion following uterine cervical conization. Immunopharmacol. Immunotoxicol. 42 (4), 314–318. doi:10.1080/08923973.2020.1765374
Kroemer, G., and Zitvogel, L. (2020). Inosine: Novel microbiota-derived immunostimulatory metabolite. Cell Res. 30 (11), 942–943. doi:10.1038/s41422-020-00417-1
Kuricova, M., Ledecky, V., Liptak, T., Madari, A., Grulova, I., Slovinska, L., et al. (2014). Oral administration of inosine promotes recovery after experimental spinal cord injury in rat. Neurol. Sci. 35 (11), 1785–1791. doi:10.1007/s10072-014-1840-3
Kurtz, T. W., Kabra, P. M., Booth, B. E., Al-Bander, H. A., Portale, A. A., Serena, B. G., et al. (1986). Liquid-chromatographic measurements of inosine, hypoxanthine, and xanthine in studies of fructose-induced degradation of adenine nucleotides in humans and rats. Clin. Chem. 32 (5), 782–786. doi:10.1093/clinchem/32.5.782
Kutryb-Zajac, B., Mierzejewska, P., Slominska, E. M., and Smolenski, R. T. (2020). Therapeutic perspectives of adenosine deaminase inhibition in cardiovascular diseases. Molecules 25 (20), E4652. doi:10.3390/molecules25204652
Kypson, J., and Hait, G. (1976). Effects of uridine and inosine on glucose metabolism in skeletal muscle and activated lipolysis in adipose tissue. J. Pharmacol. Exp. Ther. 199 (3), 565–574.
Ladep, N. G., Dona, A. C., Lewis, M. R., Crossey, M. M., Lemoine, M., Okeke, E., et al. (2014). Discovery and validation of urinary metabotypes for the diagnosis of hepatocellular carcinoma in West Africans. Hepatology 60 (4), 1291–1301. doi:10.1002/hep.27264
Lamers, M. M., van den Hoogen, B. G., and Haagmans, B. L. (2019). Adar1: "Editor-in-Chief" of cytoplasmic innate immunity. Front. Immunol. 10, 1763. doi:10.3389/fimmu.2019.01763
Lee, H., Lee, H., Park, S., Kim, M., Park, J. Y., Jin, H., et al. (2021). Integrative metabolomic and lipidomic profiling of lung squamous cell carcinoma for characterization of metabolites and intact lipid species related to the metastatic potential. Cancers (Basel) 13 (16), 4179. doi:10.3390/cancers13164179
Li, D., Feng, Y., Tian, M., Ji, J., Hu, X., and Chen, F. (2021). Gut microbiota-derived inosine from dietary barley leaf supplementation attenuates colitis through PPARγ signaling activation. Microbiome 9 (1), 83. doi:10.1186/s40168-021-01028-7
Li, Q., Gloudemans, M. J., Geisinger, J. M., Fan, B., Aguet, F., Sun, T., et al. (2022). RNA editing underlies genetic risk of common inflammatory diseases. Nature 608 (7923), 569–577. doi:10.1038/s41586-022-05052-x
Li, X., Zhao, L., Wei, M., Lv, J., Sun, Y., Shen, X., et al. (2021). Serum metabolomics analysis for the progression of esophageal squamous cell carcinoma. J. Cancer 12 (11), 3190–3197. doi:10.7150/jca.54429
Liddicoat, B. J., Piskol, R., Chalk, A. M., Ramaswami, G., Higuchi, M., Hartner, J. C., et al. (2015). RNA editing by ADAR1 prevents MDA5 sensing of endogenous dsRNA as nonself. Science 349 (6252), 1115–1120. doi:10.1126/science.aac7049
Lima, G. F., Lopes, R. O., Mendes, A. B. A., Brazão, S. C., Autran, L. J., Motta, N. A. V., et al. (2020). Inosine, an endogenous purine nucleoside, avoids early stages of atherosclerosis development associated to eNOS activation and P38 MAPK/NF-κB inhibition in rats. Eur. J. Pharmacol. 882, 173289. doi:10.1016/j.ejphar.2020.173289
Liu, Y., Hoang, T. K., Taylor, C. M., Park, E. S., Freeborn, J., Luo, M., et al. (2021). Limosilactobacillus reuteri and Lacticaseibacillus rhamnosus GG differentially affect gut microbes and metabolites in mice with treg deficiency. Am. J. Physiol. Gastrointest. Liver Physiol. 320 (6), G969–G981. doi:10.1152/ajpgi.00072.2021
Lovászi, M., Németh, Z. H., Gause, W. C., Beesley, J., Pacher, P., and Haskó, G. (2021). Inosine monophosphate and inosine differentially regulate endotoxemia and bacterial sepsis. Faseb J. 35 (11), e21935. doi:10.1096/fj.202100862R
Ludwig, N., Gillespie, D. G., Reichert, T. E., Jackson, E. K., and Whiteside, T. L. (2020). Purine metabolites in tumor-derived exosomes may facilitate immune escape of head and neck squamous cell carcinoma. Cancers (Basel) 12 (6), E1602. doi:10.3390/cancers12061602
Mabley, J. G., Pacher, P., Liaudet, L., Soriano, F. G., Haskó, G., Marton, A., et al. (2003). Inosine reduces inflammation and improves survival in a murine model of colitis. Am. J. Physiol. Gastrointest. Liver Physiol. 284 (1), G138–G144. doi:10.1152/ajpgi.00060.2002
Mager, L. F., Burkhard, R., Pett, N., Cooke, N. C. A., Brown, K., Ramay, H., et al. (2020). Microbiome-derived inosine modulates response to checkpoint inhibitor immunotherapy. Science 369 (6510), 1481–1489. doi:10.1126/science.abc3421
Matsuo, H., Tomiyama, H., Satake, W., Chiba, T., Onoue, H., Kawamura, Y., et al. (2015). ABCG2 variant has opposing effects on onset ages of Parkinson's disease and gout. Ann. Clin. Transl. Neurol. 2 (3), 302–306. doi:10.1002/acn3.167
Miller, S. R., Lane, T. R., Zorn, K. M., Ekins, S., Wright, S. H., and Cherrington, N. J. (2021). Multiple computational approaches for predicting drug interactions with human equilibrative nucleoside transporter 1. Drug Metab. Dispos. 49 (7), 479–489. doi:10.1124/dmd.121.000423
Moriwaki, Y., Yamamoto, T., and Higashino, K. (1999). Enzymes involved in purine metabolism-a review of histochemical localization and functional implications. Histol. Histopathol. 14 (4), 1321–1340. doi:10.14670/hh-14.1321
Muraoka, S., and Miura, T. (2003). Inhibition by uric acid of free radicals that damage biological molecules. Pharmacol. Toxicol. 93 (6), 284–289. doi:10.1111/j.1600-0773.2003.pto930606.x
Nakahama, T., and Kawahara, Y. (2020). Adenosine-to-Inosine RNA editing in the immune system: Friend or foe? Cell. Mol. Life Sci. 77 (15), 2931–2948. doi:10.1007/s00018-020-03466-2
Nascimento, F. P., Macedo-Júnior, S. J., Lapa-Costa, F. R., Cezar-Dos-Santos, F., and Santos, A. R. S. (2021). Inosine as a tool to understand and treat central nervous system disorders: A neglected actor? Front. Neurosci. 15, 703783. doi:10.3389/fnins.2021.703783
Neef, S. K., Janssen, N., Winter, S., Wallisch, S. K., Hofmann, U., Dahlke, M. H., et al. (2020). Metabolic drug response phenotyping in colorectal cancer organoids by LC-QTOF-MS. Metabolites 10 (12), E494. doi:10.3390/metabo10120494
Niemann, B., Haufs-Brusberg, S., Puetz, L., Feickert, M., Jaeckstein, M. Y., Hoffmann, A., et al. (2022). Apoptotic Brown adipocytes enhance energy expenditure via extracellular inosine. Nature 609 (7926), 361–368. doi:10.1038/s41586-022-05041-0
Oakes, E., Anderson, A., Cohen-Gadol, A., and Hundley, H. A. (2017). Adenosine deaminase that acts on RNA 3 (ADAR3) binding to glutamate receptor subunit B pre-mRNA inhibits RNA editing in glioblastoma. J. Biol. Chem. 292 (10), 4326–4335. doi:10.1074/jbc.M117.779868
Panebianco, C., Adamberg, K., Jaagura, M., Copetti, M., Fontana, A., Adamberg, S., et al. (2018). Influence of gemcitabine chemotherapy on the microbiota of pancreatic cancer xenografted mice. Cancer Chemother. Pharmacol. 81 (4), 773–782. doi:10.1007/s00280-018-3549-0
Pavlova, E. L., Simeonova, L. S., and Gegova, G. A. (2018). Combined efficacy of oseltamivir, isoprinosine and ellagic acid in influenza A(H3N2)-infected mice. Biomed. Pharmacother. 98, 29–35. doi:10.1016/j.biopha.2017.12.014
Pfaller, C. K., George, C. X., and Samuel, C. E. (2021). Adenosine deaminases acting on RNA (ADARs) and viral infections. Annu. Rev. Virol. 8 (1), 239–264. doi:10.1146/annurev-virology-091919-065320
Pugmire, M. J., and Ealick, S. E. (2002). Structural analyses reveal two distinct families of nucleoside phosphorylases. Biochem. J. 361, 1–25. doi:10.1042/0264-6021:3610001
Qian, G., Xu, L., Qin, J., Huang, H., Zhu, L., Tang, Y., et al. (2021). Leukocyte proteomics coupled with serum metabolomics identifies novel biomarkers and abnormal amino acid metabolism in Kawasaki disease. J. Proteomics 239, 104183. doi:10.1016/j.jprot.2021.104183
Raghava Kurup, R., Oakes, E. K., Manning, A. C., Mukherjee, P., Vadlamani, P., and Hundley, H. A. (2022). RNA binding by ADAR3 inhibits adenosine-to-inosine editing and promotes expression of immune response protein MAVS. J. Biol. Chem. 298 (9), 102267. doi:10.1016/j.jbc.2022.102267
Rice, G. I., Kasher, P. R., Forte, G. M., Mannion, N. M., Greenwood, S. M., Szynkiewicz, M., et al. (2012). Mutations in ADAR1 cause Aicardi-Goutières syndrome associated with a type I interferon signature. Nat. Genet. 44 (11), 1243–1248. doi:10.1038/ng.2414
Romio, M., Reinbeck, B., Bongardt, S., Hüls, S., Burghoff, S., and Schrader, J. (2011). Extracellular purine metabolism and signaling of CD73-derived adenosine in murine Treg and Teff cells. Am. J. Physiol. Cell Physiol. 301 (2), C530–C539. doi:10.1152/ajpcell.00385.2010
Ruhal, P., and Dhingra, D. (2018). Inosine improves cognitive function and decreases aging-induced oxidative stress and neuroinflammation in aged female rats. Inflammopharmacology 26 (5), 1317–1329. doi:10.1007/s10787-018-0476-y
Rumel Ahmed, S., Newman, A. S., O'Daly, J., Duffy, S., Grafton, G., Brady, C. A., et al. (2017). Inosine acedoben dimepranol promotes an early and sustained increase in the natural killer cell component of circulating lymphocytes: A clinical trial supporting anti-viral indications. Int. Immunopharmacol. 42, 108–114. doi:10.1016/j.intimp.2016.11.023
Schaffer, A. A., Kopel, E., Hendel, A., Picardi, E., Levanon, E. Y., and Eisenberg, E. (2020). The cell line A-to-I rna editing catalogue. Nucleic Acids Res. 48 (11), 5849–5858. doi:10.1093/nar/gkaa305
Schwarzschild, M. A., Ascherio, A., Beal, M. F., Cudkowicz, M. E., Curhan, G. C., Hare, J. M., et al. (2014). Inosine to increase serum and cerebrospinal fluid urate in Parkinson disease: A randomized clinical trial. JAMA Neurol. 71 (2), 141–150. doi:10.1001/jamaneurol.2013.5528
Schwarzschild, M. A., Ascherio, A., Casaceli, C., Curhan, G. C., Fitzgerald, R., Kamp, C., et al. (2021). Effect of urate-elevating inosine on early Parkinson disease progression: The sure-Pd3 randomized clinical trial. Jama 326 (10), 926–939. doi:10.1001/jama.2021.10207
Sliva, J., Pantzartzi, C. N., and Votava, M. (2019). Inosine pranobex: A key player in the game against a wide range of viral infections and non-infectious diseases. Adv. Ther. 36 (8), 1878–1905. doi:10.1007/s12325-019-00995-6
Soares Dos Santos Cardoso, F., Cardoso, R., Dos Santos Ramalho, B., Bastos Taboada, T., Dos Santos Nogueira, A. C., Blanco Martinez, A. M., et al. (2019). Inosine accelerates the regeneration and anticipates the functional recovery after sciatic nerve crush injury in mice. Neuroscience 423, 206–215. doi:10.1016/j.neuroscience.2019.09.023
Sorensen, L. B. (1970). Mechanism of excessive purine biosynthesis in hypoxanthine-guanine phosphoribosyltransferase deficiency. J. Clin. Invest. 49 (5), 968–978. doi:10.1172/jci106316
Stockard, B., Bhise, N., Shin, M., Guingab-Cagmat, J., Garrett, T. J., Pounds, S., et al. (2021). Cellular metabolomics profiles associated with drug chemosensitivity in AML. Front. Oncol. 11, 678008. doi:10.3389/fonc.2021.678008
Sun, S., Luo, L., Liang, W., Yin, Q., Guo, J., Rush, A. M., et al. (2020). Bifidobacterium alters the gut microbiota and modulates the functional metabolism of T regulatory cells in the context of immune checkpoint blockade. Proc. Natl. Acad. Sci. U. S. A. 117 (44), 27509–27515. doi:10.1073/pnas.1921223117
Tan, G., Wang, H., Yuan, J., Qin, W., Dong, X., Wu, H., et al. (2017). Three serum metabolite signatures for diagnosing low-grade and high-grade bladder cancer. Sci. Rep. 7, 46176. doi:10.1038/srep46176
Teixeira, F. C., de Mattos, B. D. S., Mello, J. E., Cardoso, J., Spohr, L., Luduvico, K. P., et al. (2022). Protective effects of inosine on memory consolidation in a rat model of scopolamine-induced cognitive impairment: Involvement of cholinergic signaling, redox status, and ion pump activities. Neurochem. Res. 47 (2), 446–460. doi:10.1007/s11064-021-03460-5
Teixeira, F. C., Gutierres, J. M., Soares, M. S. P., da Siveira de Mattos, B., Spohr, L., do Couto, C. A. T., et al. (2020). Inosine protects against impairment of memory induced by experimental model of Alzheimer disease: A nucleoside with multitarget brain actions. Psychopharmacol. Berl. 237 (3), 811–823. doi:10.1007/s00213-019-05419-5
Teixeira, F. C., Soares, M. S. P., Blödorn, E. B., Domingues, W. B., Reichert, K. P., Zago, A. M., et al. (2022). Investigating the effect of inosine on brain purinergic receptors and neurotrophic and neuroinflammatory parameters in an experimental model of Alzheimer’s disease. Mol. Neurobiol. 59 (2), 841–855. doi:10.1007/s12035-021-02627-z
Tobólska, S., Terpiłowska, S., Jaroszewski, J., and Siwicki, A. K. (2018). Influence of inosine pranobex on cell viability in normal fibroblasts and liver cancer cells. J. Vet. Res. 62 (2), 215–220. doi:10.2478/jvetres-2018-0031
Toncev, G., Milicic, B., Toncev, S., and Samardzic, G. (2002). Serum uric acid levels in multiple sclerosis patients correlate with activity of disease and blood-brain barrier dysfunction. Eur. J. Neurol. 9 (3), 221–226. doi:10.1046/j.1468-1331.2002.00384.x
Tozzi, M. G., Camici, M., Mascia, L., Sgarrella, F., and Ipata, P. L. (2006). Pentose phosphates in nucleoside interconversion and catabolism. Febs J. 273 (6), 1089–1101. doi:10.1111/j.1742-4658.2006.05155.x
Traut, T. W. (1994). Physiological concentrations of purines and pyrimidines. Mol. Cell. Biochem. 140 (1), 1–22. doi:10.1007/bf00928361
Tzoneva, G., Perez-Garcia, A., Carpenter, Z., Khiabanian, H., Tosello, V., Allegretta, M., et al. (2013). Activating mutations in the NT5C2 nucleotidase gene drive chemotherapy resistance in relapsed ALL. Nat. Med. 19 (3), 368–371. doi:10.1038/nm.3078
Van der Weyden, M. B., and Kelley, W. N. (1977). Adenosine deaminase: Characterization of the molecular heterogeneity of the enzyme in human tissue. Adv. Exp. Med. Biol. 76a, 235–248. doi:10.1007/978-1-4613-4223-6_29
Van Heukelom, L. H., Boom, A., Bartstra, H. A., and Staal, G. E. (1976). Characterization of adenosine deaminase isozymes from normal human erythrocytes. Clin. Chim. Acta. 72 (1), 109–115. doi:10.1016/0009-8981(76)90041-3
Wang, T., Gnanaprakasam, J. N. R., Chen, X., Kang, S., Xu, X., Sun, H., et al. (2020). Inosine is an alternative carbon source for CD8(+)-T-cell function under glucose restriction. Nat. Metab. 2 (7), 635–647. doi:10.1038/s42255-020-0219-4
Wardman, J. F., Bains, R. K., Rahfeld, P., and Withers, S. G. (2022). Carbohydrate-active enzymes (CAZymes) in the gut microbiome. Nat. Rev. Microbiol. 20 (9), 542–556. doi:10.1038/s41579-022-00712-1
Watanabe, H., Hattori, T., Kume, A., Misu, K., Ito, T., Koike, Y., et al. (2020). Improved Parkinsons disease motor score in a single-arm open-label trial of febuxostat and inosine. Med. Baltim. 99 (35), e21576. doi:10.1097/md.0000000000021576
Welihinda, A. A., Kaur, M., Raveendran, K. S., and Amento, E. P. (2018). Enhancement of inosine-mediated A2AR signaling through positive allosteric modulation. Cell. Signal. 42, 227–235. doi:10.1016/j.cellsig.2017.11.002
Whiteman, M., Ketsawatsakul, U., and Halliwell, B. (2002). A reassessment of the peroxynitrite scavenging activity of uric acid. Ann. N. Y. Acad. Sci. 962, 242–259. doi:10.1111/j.1749-6632.2002.tb04072.x
Wu, J., Singh, K., Lin, A., Meadows, A. M., Wu, K., Shing, V., et al. (2022). Boosting NAD(+) blunts TLR4-induced type I IFN in control and systemic lupus erythematosus monocytes. J. Clin. Invest. 132 (5), e139828. doi:10.1172/jci139828
Yang, J., Zhang, Z., Xie, Z., Bai, L., Xiong, P., Chen, F., et al. (2022). Metformin modulates microbiota-derived inosine and ameliorates methamphetamine-induced anxiety and depression-like withdrawal symptoms in mice. Biomed. Pharmacother. 149, 112837. doi:10.1016/j.biopha.2022.112837
Yang, S., Deng, P., Zhu, Z., Zhu, J., Wang, G., Zhang, L., et al. (2014). Adenosine deaminase acting on RNA 1 limits RIG-I RNA detection and suppresses IFN production responding to viral and endogenous RNAs. J. Immunol. 193 (7), 3436–3445. doi:10.4049/jimmunol.1401136
Yasutake, Y., Tomita, K., Higashiyama, M., Furuhashi, H., Shirakabe, K., Takajo, T., et al. (2017). Uric acid ameliorates indomethacin-induced enteropathy in mice through its antioxidant activity. J. Gastroenterol. Hepatol. 32 (11), 1839–1845. doi:10.1111/jgh.13785
Yoon, H. S., Jeong Yang, J., Rivera, E. S., Shu, X. O., Xiang, Y. B., Calcutt, M. W., et al. (2020). Urinary metabolites and risk of coronary heart disease: A prospective investigation among urban Chinese adults. Nutr. Metab. Cardiovasc. Dis. 30 (3), 467–473. doi:10.1016/j.numecd.2019.10.011
Zhu, Y., Wang, X., Zhu, L., Tu, Y., Chen, W., Gong, L., et al. (2022). Lactobacillus rhamnosus GG combined with inosine ameliorates alcohol-induced liver injury through regulation of intestinal barrier and Treg/Th1 cells. Toxicol. Appl. Pharmacol. 439, 115923. doi:10.1016/j.taap.2022.115923
Keywords: inosine, cancer, inflammation, infection, cardiovasclar disease, neuroprotection
Citation: Kim IS and Jo E-K (2022) Inosine: A bioactive metabolite with multimodal actions in human diseases. Front. Pharmacol. 13:1043970. doi: 10.3389/fphar.2022.1043970
Received: 20 September 2022; Accepted: 02 November 2022;
Published: 16 November 2022.
Edited by:
Jianbo Xiao, University of Vigo, SpainCopyright © 2022 Kim and Jo. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Eun-Kyoung Jo, aGF5b3VuZ2pAY251LmFjLmty
 In Soo Kim
In Soo Kim Eun-Kyoung Jo
Eun-Kyoung Jo