- Changzhou Key Laboratory of Respiratory Medical Engineering, Institute of Biomedical Engineering and Health Sciences, School of Medical and Health Engineering, Changzhou University, Changzhou, Jiangsu, China
The ongoing COVID-19 pandemic caused by severe acute respiratory syndrome coronavirus-2 (SARS-CoV-2) poses a never before seen challenge to human health and the world economy. However, it is difficult to widely use conventional animal and cell culture models in understanding the underlying pathological mechanisms of COVID-19, which in turn hinders the development of relevant therapeutic treatments, including drugs. To overcome this challenge, various three-dimensional (3D) pulmonary cell culture models such as organoids are emerging as an innovative toolset for simulating the pathophysiology occurring in the respiratory system, including bronchial airways, alveoli, capillary network, and pulmonary interstitium, which provide a robust and powerful platform for studying the process and underlying mechanisms of SARS-CoV-2 infection among the potential primary targets in the lung. This review introduces the key features of some of these recently developed tools, including organoid, lung-on-a-chip, and 3D bioprinting, which can recapitulate different structural compartments of the lung and lung function, in particular, accurately resembling the human-relevant pathophysiology of SARS-CoV-2 infection in vivo. In addition, the recent progress in developing organoids for alveolar and airway disease modeling and their applications for discovering drugs against SARS-CoV-2 infection are highlighted. These innovative 3D cell culture models together may hold the promise to fully understand the pathogenesis and eventually eradicate the pandemic of COVID-19.
1 Introduction
The world has been facing coronavirus 2019 (COVID-19) pandemic for 3 years, caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) (Wang C. et al., 2020; Castiello et al., 2022; Luo et al., 2022). Although most COVID-19 patients present mild or moderate symptoms including cough, fever, fatigue, shortness of breath, and pharyngodynia, critically ill patients of COVID-19 always present severe respiratory complications such as pulmonary edema, acute respiratory distress syndrome (ARDS), and even death (Chen et al., 2020; Guan et al., 2020; Huang et al., 2020). Therefore, COVID-19 which has led to such a global health crisis with huge economic and social impact requires urgent and thorough investigations to elucidate its mechanisms of pathogenesis and explore corresponding therapeutic treatments as well as potential post-disease implications.
However, these investigations so far have been limited by the scarcity of suitable models to mimic the pathophysiological processes of SARS-CoV-2 infection that occur during COVID-19 in vivo. Conventional models used to experimentally investigate pathophysiology of human diseases include animal-based models (de Oliveira et al., 2021; Lee and Lowen, 2021). Several kinds of animals including mice (Dinnon et al., 2020), hamsters (Tostanoski et al., 2020), ferrets (Cox et al., 2021), and non-human primates (NHPs) (Corbett et al., 2020; Maisonnasse et al., 2020) have been used to study pathogenesis and host responses associated with COVID-19. It is however difficult to use animal models to recapitulate human physiology and at the same time decipher fundamental molecular mechanisms of host-pathogen interactions, viral replication kinetics, and virus tropism. These animal models are also not very suitable for early-stage drug screening since they are time-consuming and often fail to translate to human trials due to the species difference (Knight, 2008; Konar et al., 2016).
Alternatively, two-dimensional (2D) cell culture systems can be used as experimental models in the study of COVID-19, which are both relatively inexpensive and highly efficient (Duval et al., 2017). But increasing evidence suggests that 2D cell culture systems have inherent drawbacks. For example, 2D cell culture cannot correctly mimic the organ’s in vivo architecture and microenvironments (Elsdale and Bard, 1972; Korff and Augustin, 1999; Ghajar et al., 2008). More specifically, the lung is a complex organ including 23 generations of branching airways, multiple cell types, specified tubular three-dimensional (3D) geometry, and cyclic stretch stimulation, which limit the simulating effect using 2D cell culture (Weibel and Gomez, 1962). A recent report has shown that SARS-CoV-2 infection arises from the proximal airways (containing basal, secretory, and ciliated cells) and induces associated inflammation in distal alveoli (containing type I and type II cells) (Morrisey and Hogan, 2010; Borczuk et al., 2020; Tay et al., 2020), which are largely infeasible to be recapitulated by the traditional 2D cell culture systems.
Fortunately, many novel 3D cell culture toolsets such as organoid, lung-on-a-chip, and 3D bioprinting have been developed during the past decade, which provide new platforms for exploring complex pathophysiology of lung diseases such as COVID-19 (de Melo et al., 2021). These 3D cell culture models are indeed gaining increasing momentum to be the primary choice of experimental methods due to their physiological relevance and operational flexibility as well as high-throughput adaptability (Bircsak et al., 2021). Cells cultured in these 3D models exhibit features close to the complex lung conditions in vivo, mimic cell-cell and cell-matrix interactions, and reproduce the morphology and function of the lung (Baker and Chen, 2012). Thus, it is evident that 3D cell culture models are advantageous in the investigation of COVID-19 pathogenesis and the development of therapeutic agents to combat the pandemic disease.
In this review, we first describe the characteristics of SARS-CoV-2 and immuno-inflammatory responses related to the viral infection and pathogenesis of COVID-19, and then summarize the 3D cell culture models including the traditional air-liquid interface (ALI) culture and spheroid, and the emerging organoid, lung-on-a-chip (containing organoplate), and 3D bioprinting which may be suitable ex vivo models to mimic various vital lung functions in a cell culture dish. We also highlight the possibility of building new robust models to recapitulate different structural compartments of the lung and lung function, in particular, accurately resembling the pathophysiology of COVID-19 in vivo. This information may hopefully help investigators to select and/or develop suitable 3D cell culture models for the pursuit of mechanistic understanding and therapeutic treatment of COVID-19.
2 Characteristics and immuno-inflammatory responses of SARS-CoV-2
SARS-CoV-2 is a virus with a genome of nearly 30 kb, with 11 open reading frames (ORFs) and 27 viral encoding proteins (Lu et al., 2020). Among them, an array of ORFs, i.e., ORF 3, 6, 7a, 7b, 8, and 10 are the accessory proteins, and the main structural viral proteins include spike glycoprotein (S), envelope glycoprotein (E), membrane glycoprotein (M), and nucleocapsid proteins (N) (Zhou P. et al., 2020; Wu et al., 2020). SARS-CoV-2 enters host cells (ciliated, club, alveolar epithelial type 2 (AT2) cells, vascular endothelial cells, and alveolar macrophages) by endocytosis mediated by the interaction of the S proteins with host receptors such as angiotensin-converting enzyme 2 (ACE2). The S proteins on the envelope of SARS-CoV-2 are cleaved into S1 and S2 subunits (Ke et al., 2020). But only S1 consists of the receptor-binding domain (RBD), which directly binds to the peptidase domain (PD) of ACE2 to gain entry into host cells (Yan et al., 2020). Therefore, the S1 protein/receptor interaction is the critical determinant for the virus to infect host cells. According to various mutation sites in the S protein, six main variants of SARS-CoV-2 viruses have been identified including Alpha, Beta, Gamma, Delta, Lambda, and Omicron. Some variants are more transmissible or easier to escape from immunity than others, which leads to increased transmissibility and a higher viral load in the human body. Garrett and others (Garrett et al., 2022) evaluated asymptomatic carriage in a sub-study of the Sisonke vaccine trial and found that 2.6% of the asymptomatic carriage during the Beta and Delta outbreaks rose to 16% during the Omicron period.
Additionally, some host factors can enable and/or facilitate viral entry. For example, transmembrane protease serine 2 (TMPRSS2) is widely expressed in epithelial cells of the respiratory tract and could activate SARS-CoV-2 in Calu-3 cells (Bugge et al., 2009; Bestle et al., 2020). Furin also plays a critical role in the cleavage activation of SARS-CoV-2 spike proteins (Johnson et al., 2021). Neuropilin-1 (NRP1), which regulates pleiotropic biological processes, facilitates SARS-CoV-2 cell entry and infectivity (Cantuti-Castelvetri et al., 2020; Daly et al., 2020). In brief, these host factors provide an essential mechanism for SARS-CoV-2 infectivity and a scientific basis for targeting infected cells to develop antiviral drugs. Therefore, these host factors’ expression levels in 3D cell culture models may be an important indicator for selecting suitable models for SARS-CoV-2 infection study.
Once SARS-CoV-2 infects the host, both innate and adaptive immune systems initiate to counteract the virus infection. The innate immune response provides the first line of defense against SARS-CoV-2 infection in the airways, via various mechanisms for rapid sensing and suppressing of the viral infection. For example, the viral infection is detected by endosomal Toll-like receptor 3&7 (TLR3, TLR7), and melanoma differentiation-associated gene 5 (MDA5) of the innate immune cells in the airways (Totura et al., 2015; Zhou et al., 2021). This subsequently triggers the release of a series of pro-inflammatory factors such as tumor necrosis factor-alpha (TNF-α), and interleukin 1&6 (IL-1, IL-6), which together facilitate the early controlling of the viral infection.
On the other hand, the adaptive immune response provides the second line of defense against SARS-CoV-2 infection, which is enabled by a broader and more finely tuned repertoire of recognition mechanisms for viral infection, involving antigen presenting cells (APCs), CD4+ T cells, CD8+ T cells and B cells (Sette and Crotty, 2021). More specifically, following SARS-CoV-2 infection, the APCs present viral particles to CD8+/CD4+ T cells via interaction of TCR-MHC I or II, respectively. When exposed to antigens, CD8+ T cells release cytotoxic granules that are critical for clearance of virus-infected cells, and CD4+ T cells polarize towards Th1 and Th2 cells. Then Th1 cells release IFN-γ to eliminate the virus, and Th2 cells activate humoral immunity (such as B cells) to generate antibodies that neutralize SARS-CoV-2 (Toor et al., 2021). Unfortunately, in severe cases of COVID-19 the potential of this mechanism is significantly limited because the number of APCs is largely reduced (Zhou R. et al., 2020; Qin et al., 2021).
It is obvious that a deficiency of the immune responses would give an opportunity for viruses such as SARS-CoV-2 to freely complete their RNA replication process and subsequent release of the genetic materials, and ultimately result in reassembling and release of large amounts viruses (Jiang et al., 2021). However, an excessive immune response may also trigger excessive production of inflammatory cytokines, a phenomenon known as the cytokine storm. For example, in critically ill patients of COVID-19 the expression of inflammatory cytokines including IL-2, IL-6, IL-7, IL-10, IP-10, MCP-1, TNF-α, and IFN-γ has been shown to be excessively elevated, and this kind of cytokine storm is thought as the main cause of multi-organ failure and death in COVID-19 patients (Castelli et al., 2020; Hu et al., 2021; Luo et al., 2022).
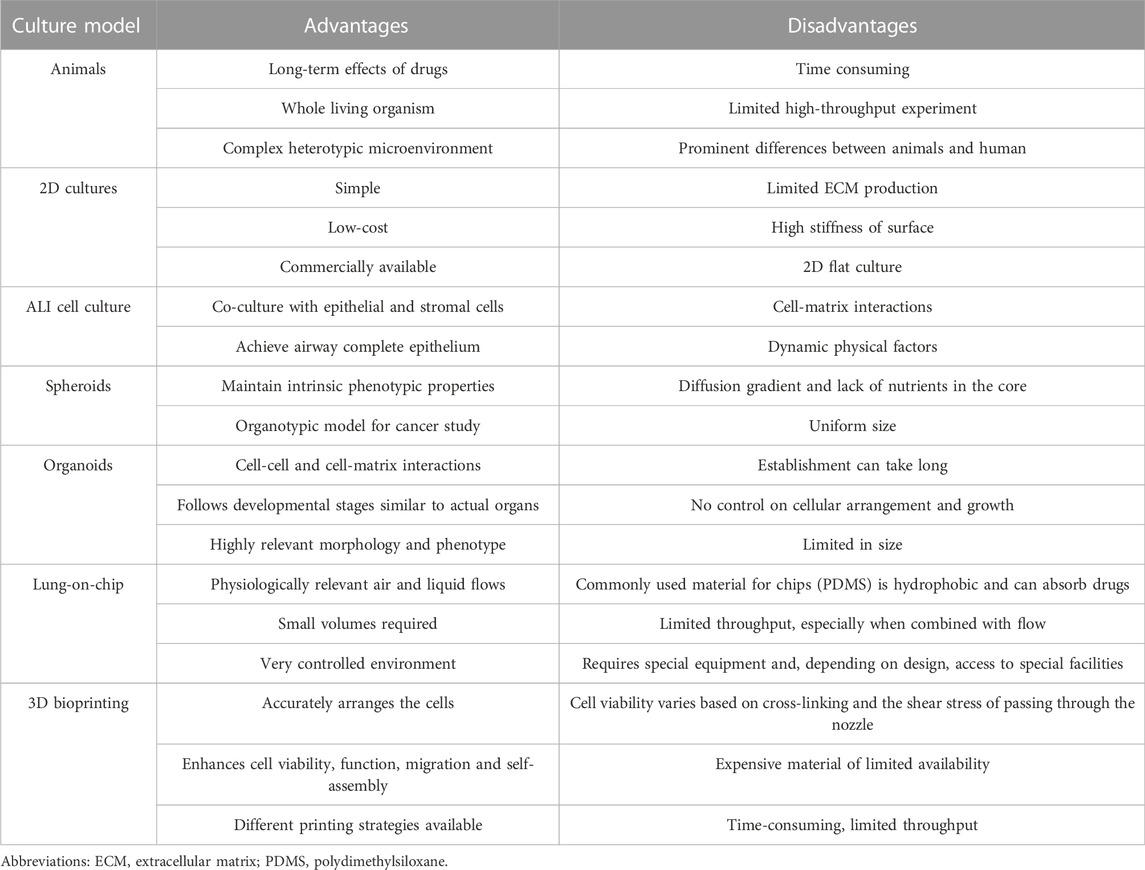
3 3D cell culture models to mimic lung in study of SARS-CoV-2 infection
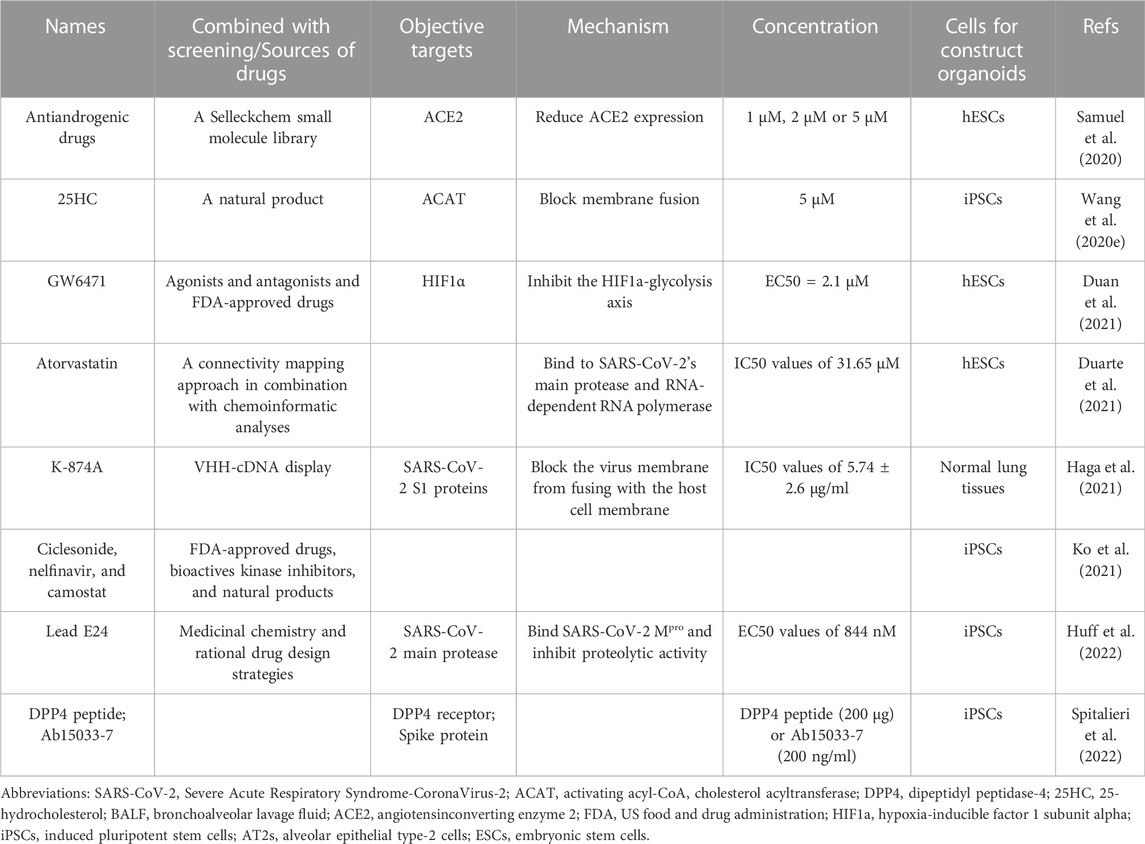
With increasing studies of COVID-19, the importance is acknowledged that different lung regions (airway, alveolus, a thin epithelial-endothelial barrier, and pulmonary interstitium) play different essential roles in the pathogenesis and development of COVID-19. Therefore, 3D cell culture models (ALI culture, spheroid, organoid, organ-on-a-chip including organoplate, and 3D bioprinting) which could mimic different structural compartments of the lung and lung function may be more suitable for studying different physiopathological processes of COVID-19 (Figure 1). In addition, these 3D cell culture models have different advantages in mimicking the physiology and pathology of diseases (Table 1). For example, the ALI approach is primarily used to mimic the epithelial air-liquid interface environment of respiratory tract in the lung. Spheroids are mainly used in study of tumor growth such as in lung cancer. Organoids are self-assembled constructs that can “freely” grow in resemblance to natural development. Organ-on-a-chip has bridged microfluidic technology and living cells, resulting in a dynamic biomimetic device (de Oliveira et al., 2021). 3D bioprinting models can print more complex constructs and mimic complex anatomical structures.
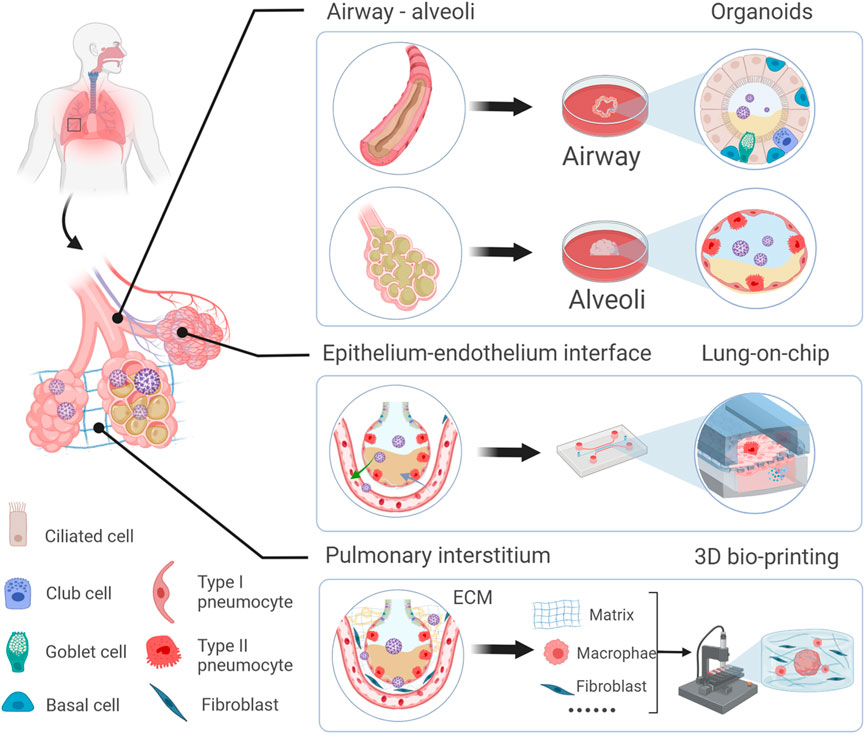
FIGURE 1. Schematic representation of novel 3D models to mimic in vivo the physiology and pathophysiology of different lung regions. Different 3D cell culture models (organoid, organ-on-a-chip, and 3D bioprinting) have their own advantages which could mimic the structure and function of different lung regions (airway, alveoli, a thin epithelial-endothelial barrier, and pulmonary interstitium).
3.1 3D cell culture to mimic ALI of the lung
The respiratory system provides an ALI interface to protect the body from invasion by inhaled pathogens that are commonly encountered in the environments. This interface can not be mimicked by the submerged 2D cell culture models, until the development of 3D ALI culture models (Whitcutt et al., 1988). 3D ALI culture mainly consists of an outer plastic culture dish and an inner porous membrane insert (usually in a 6-well plate format with a pore size of 0.4 μm). Cells are cultured on the insert, and once confluent (often in 2–4 days), the medium is removed from the apical aspect of the insert, forming an ALI. Consequently, this cell culture method has been used to mimic ALI in the lung with different types of airway epithelial cells, including ciliated cells, club cells, goblet cells, and basal cells, to self-assemble into a pseudo-stratified columnar epithelium (Ghio et al., 2013). These functional epithelia provide crucial functions for maintaining airway tissue integrity and homeostasis by secreting inflammatory mediators and antimicrobial peptides (Tam et al., 2011).
By using this traditional 3D cell culture, Mulay et al. (Mulay et al., 2021) discovered that SARS-CoV-2 infected ciliated cells at a significantly higher rate than goblet cells in the proximal airway epithelium. In addition, Mullen and others (Mullen et al., 2021) found in ALI cultures that SARS-CoV-2 infection increased pyruvate carboxylase (PC) and mTORC1 activity and inhibition of mTORC1 could reduce SARS-CoV-2 replication. These studies collectively demonstrate that ALI culture system is an important platform for screening therapeutics targeting airway cells of SARS-CoV-2 and related antivirals for COVID-19.
3.2 Spheroids culture to mimic alveoli of the lung
Spheroids allow cell colonies to self-assemble and form aggregates of 3D microtissues (Ryu et al., 2019). The process of spheroid formation is affected by adhesion and differentiation of cells and various factors, including gradients of nutrients, oxygen, and growth factors in cell culture medium.
Spheroids are mainly used for cancer studies because it shows some characteristics similar to tumor microenvironments such as hypoxia and tumoral cell-to-cell interaction (Nigjeh et al., 2018). But it has also been used as 3D cell cultures for recapitulating the anti-viral drug responses in airway cells. For example, Ebisudani et al. (Ebisudani et al., 2021) established and validated a long-term culture of alveolospheres that could be used as an efficient drug testing platform for the development of therapeutic agents to combat viruses such as SARS-CoV-2.
3.3 Organoids to mimic different regions of the lung
Novel 3D cell culture systems such as organoids have emerged recently with great potential in biomedical research. Organoids are 3D structures derived from stem/progenitor cells in specific biomaterials that can be differentiated to generate 3D structures containing multiple cell types and assembles that resemble the organization and functions of specific tissue/organs. Therefore, the development of stem cell technology has been central to the formation and progression of organoids. For example, Takahashi and Yamanaka (Takahashi and Yamanaka, 2006) demonstrated the creation of pluripotent cells directly from mouse embryonic or adult fibroblasts by introducing four transcription factors. Soon afterward, Sato et al. (Sato et al., 2009) reported the formation of 3D structures of single-sorted Lgr5+ stem cells in Matrigel. Ever since, there have been many organoids successfully generated and used in different fields.
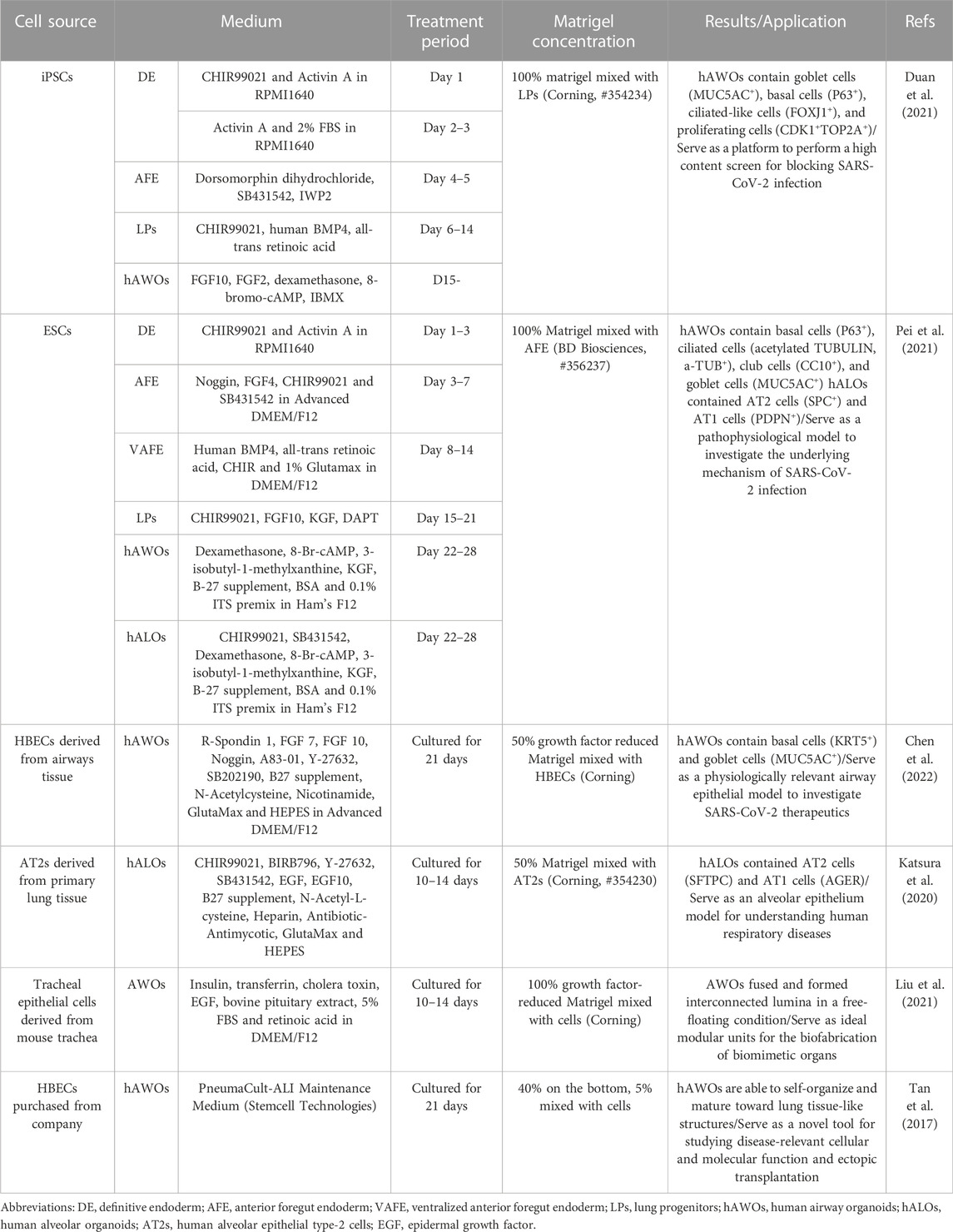
Lung organoids can be established from induced pluripotent stem cells (iPSCs) or airway epithelial progenitor cells such as AT2 cells or basal cells under optimal conditions. The processes of different cell sources to generate lung organoids are described in Table 2. iPSCs-derived organoids contain AT1, AT2, and epithelial cells, and recapitulate the structure and function of the alveolus or airway when exposed to suitable induction signals (Takahashi et al., 2007; Leibel et al., 2020).
AT2 cells reside in the alveoli which are characterized by the production of pulmonary surfactant proteins and can behave as alveolar stem cells during repair after injury, repopulating both AT1 and AT2 cells (Diem et al., 2020). Basal cells, characterized by the marker of the transcription factor Trp63, the cytokeratin Krt5, and integrin alpha 6, are one type of proximal airway epithelium (accounting for ∼30%), which can differentiate into secretory and ciliated cells (Lambrecht and Hammad, 2012; Barkauskas et al., 2017; Bilodeau et al., 2021). Compared to iPSCs, alveolar or airway organoids based on AT2 cells and basal cells from adults present the physiological dynamic consistent with normal in vivo or human patients, especially for COVID-19 (Wang et al., 2021).
Due to the need to mimic the effects in different lung regions, alveolar and airway organoids have been developed to mimic the airway and alveoli, respectively. And a simplified visual description of the main steps to generate lung organoids of different origin cells is in Figure 2.
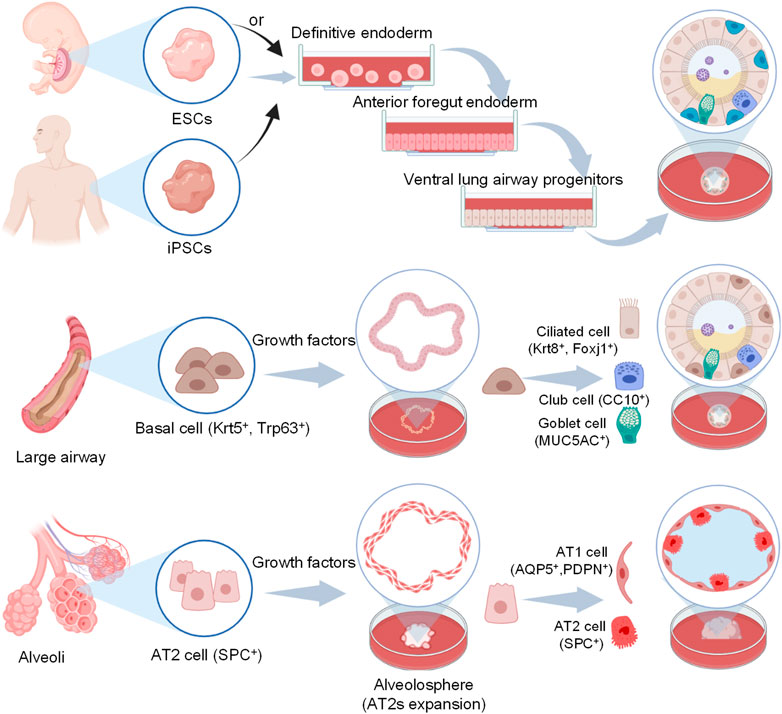
FIGURE 2. Schematic representation of human airway and alveoli organoids developed from different origin cells and cultivation include particular steps. For iPSCs, they first generate definitive endoderm (DE), then anterior foregut endoderm (AFE) and lung progenitors (LPs) with different additives, finally yield lung organoids via 3D culture using Matrigel (Barkauskas et al., 2017). For basal cells and AT2 cells, they maintain under suitable conditions and self-organize in a more natural manner with Matrigel.
3.3.1 Alveolar organoids
Though the SARS-CoV-2 virus typically initiates in the proximal airways, severe symptoms of COVID-19 arise from infection and associated inflammation in the distal alveoli, which has strikingly different physiology from that of the proximal airways (Katsura et al., 2020; Mulay et al., 2021). Previous studies have demonstrated that ACE2 is essential for SARS-CoV-2 entry, indicating that AT2 cells with a high expression of ACE2 proteins are significant targets of SARS-CoV-2 infection (Zhao B. et al., 2020). To explore the response of lung cells (particularly AT2 cells) to the SARS-CoV-2 infection, Han and collaborators (Han et al., 2021) developed an alveolar organoid model derived from iPSCs. Similar to what is seen in COVID-19 patients’ lung autopsy tissues (Blanco-Melo et al., 2020), their results revealed that robust induction of chemokines (such as rheumatoid arthritis, TNF signaling, and IL-17 signaling) upon SARS-CoV-2 infection. Also, these AT2-derived organoids were able to differentiate into AT1 cells and could facilitate diverse investigations of pulmonary pathogens, including SARS-CoV-2 infection (Salahudeen et al., 2020).
In addition, the expression of ACE2 could be modulated by the activation of different pathways. When treated with particulate matter (PM2.5), Kim et al. (Kim et al., 2020) proved that ACE2 significantly upregulated in alveolar organoids. On the contrary, androgen signaling inhibition reduced ACE2 expression and protected lung organoids against SARS-CoV-2 infection (Samuel et al., 2020).
3.3.2 Airway organoids
Since the proximal airway regions are the first target of SARS-CoV-2 infection, airway organoids are suitable for exploring the interaction of SARS-CoV-2 with proximal airway cells. To accurately mimic proximal airway physiological conditions during SARS-CoV-2 infection, (Chen et al., 2022) used human airway basal cells to generate airway organoids, in which basal cells differentiate into ciliated cells, goblet cells, and club cells. Furthermore, they compared ALI cultures and airway organoid cultures and acknowledged that the latter expressed high levels of ACE2 and TMPRSS2, which are highly susceptible to SARS-CoV-2 infection and promote inflammatory cytokine responses (Marescotti et al., 2019; Wang J. et al., 2020; Chen et al., 2022).
Furthermore, to observe cellular dynamic changes similar to clinical features in COVID-19 patients, (Xu et al., 2021) developed airway organoids derived from patient biopsy residues. Therefore, these airway organoids can be used to investigate the tissue-specific SARS-CoV-2 infection, host responses, and viral infection inhibitors.
To investigate whether multibasic cleavage site (MBCS) can alter protease usage during entry and which entry pathway is taken by SARS-CoV-2, (Mykytyn et al., 2021) found that SARS-CoV-2 spike MBCS increases infectivity and serine protease usage on human airway organoids based on collagen-coated transwell inserts. In addition, (Kastenhuber et al., 2022) demonstrated that coagulation factors, including factor Xa and thrombin, increase SARS-CoV-2 infection in human lung organoids derived from iPSCs. These data indicate that these hose factors can directly cleave SARS-CoV-2 spike, which is important to promote viral entry into airway epithelia.
Together, these data demonstrate that organoid models could accurately mimic alveolar and airway cellular composition to provide a valuable platform for screening new drugs to identify candidate COVID-19 therapeutics.
3.3.3 Lung organoids
iPSCs-derived lung organoids are indisputably a fast-moving field due to the unique property of unlimited self-renewal capacity (Kolagar et al., 2020; Sharma et al., 2020). To date, the most classical differentiation protocols first generate definitive endoderm (DE), then anterior foregut endoderm (AFE) and lung progenitors (LPs), and finally yield lung organoids using 3D Matrigel. Tiwari et al. (2021) developed human lung organoids derived from iPSCs to investigate viral pathogenesis. iPSCs were differentiated into definitive endoderm, lung progenitor cells, then an epithelial-like structure with surrounding mesenchymal cells (labeled with smooth muscle actin and acetylated tubulin) by day 60, and subsequently a pseudostratified epithelial structure with P63+ basal-like cells, FOXJ1+ ciliated cells, and structural alveolar type 1 & 2 cells.
In addition, 3D cell culture models such as organoids are suitable for further understanding immuno-inflammatory responses associated with SARS-CoV-2 infection, which is crucial for effective control and clearance of the virus. For example, iPSCs-derived lung organoids have been used to determine the early cellular response to SARS-CoV-2 infection, particularly the change in the expression level of inflammatory factors with 48 h infection (Pei et al., 2021). The results of RNA-sequencing analysis show that several inflammatory factors, including IL-6, TNF, CXCL8, CXCL2, CXCL3, CXCL10, CXCL11, and NF-κB were upregulated, which is consistent with the clinically observed phenomenon in COVID-19 patients (Huang et al., 2020; Wilk et al., 2020).
Therefore, iPSCs-derived lung organoids containing the component and structure of proximal airways and distal alveoli, can be used for revealing cell/tissue-specific SARS-CoV-2 Infection and host responses in the whole lung (Dye et al., 2015).
3.4 Lung-on-a-chip models to mimic epithelium-endothelium interface
Although lung organoids are promising tools to elucidate the pathophysiological mechanisms of COVID-19, a significant limitation of them is the absence of vasculature, and could not mimic the interaction of alveoli-capillary networks and related gas exchange in the lung (Barkauskas et al., 2017). The gas exchange process in the human body depends on the direct interaction between a monolayer alveolar epithelia lining the alveoli and a monolayer endothelial cell lining the capillary network, which allows for diffusive gas exchange and prevents plasma fluid entry into the alveoli (Weibel, 2017; Bernard et al., 2020). With growing interest in COVID-19, the importance of crosstalk between alveolar epithelial cells and the capillary network gets more attention. Fortunately, the lung-on-a-chip model could replicate this alveolar-capillary interaction by integrating tissue–tissue interfaces and may be crucial for the systemic understanding of COVID-19.
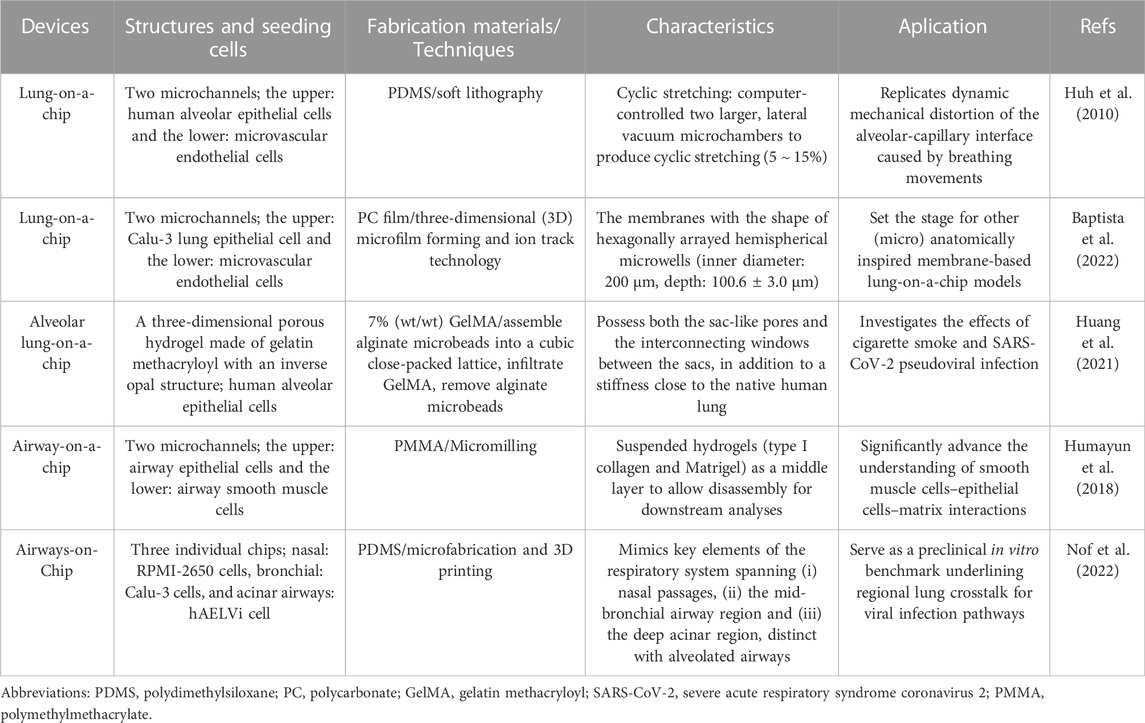
Organ-on-a-chip is a novel 3D cell culture tool based on the integration of the techniques of bioengineering and microfluidics disciplines (Shrestha et al., 2020). It always consists of upper and lower microchannels separated by a thin, flexible, extracellular matrix (ECM)-coated membrane, which is very suitable for mimicking the alveolar-capillary interface (Huh et al., 2010; Kızılkurtlu et al., 2018). For example, when human alveolar epithelial cells and pulmonary microvascular endothelial cells are cultured on the opposite sides of the membrane and grown to confluence, the upper channel is introduced into the air to create an air-liquid interface. In addition, a computer-controlled vacuum in these chambers can be used to produce cyclic strain ranging from 5% to 15% to match normal levels of strain to mimic physiopathological breathing movements (Birukov et al., 2003). Therefore, organ-on-a-chip can manipulate not only biochemical factors (such as cytokines, oxygen, and nutrients) but also dynamic physical factors (such as shear stress and cyclic strain), both of which are critical in understanding lung organ-level functions and permit researchers to mimic disease pathogenesis of COVID-19 (Huh et al., 2010; Benam et al., 2016; Novak et al., 2021; Wang et al., 2022).
In addition to dynamic mechanical distortion of the alveolar-capillary interface, there is increasing evidence that the microscale curved surfaces affect the spatiotemporal organization and behaviors of cells (Callens et al., 2020; Jin et al., 2021). To recreate the mainly spherical geometry of the cells’ native microenvironment, Baptista et al. (2022) made the membranes the shape of hexagonally arrayed hemispherical microwells by using a combination of 3D microfilm forming and ion track technology. Each microwell has a little bit more than 200 μm maximum inner diameter and an average maximum depth of 100.6 ± 3.0 μm. With integration in microfluidic chips, the microcurved membranes were seeded with Calu-3 lung epithelial cell line and human lung microvascular endothelial cells, respectively. Also, Huang et al. (2021) successfully designed an alveolar lung-on-a-chip platform with the alveoli-like 3D gelatin methacryloyl (GelMA) inverse opal structure. Significantly, this device also provided an air-liquid interface and cyclic strain, which was better able to maintain the functions of human alveolar epithelial cells. A list of characteristics and applications of current lung-on-a-chip devices is given in Table 3. A visual representation of these classifications is found in Figure 3.
FIGURE 3. Schematic representation of functional lung-on-a-chip based on epithelium-endothelium interface physiology characteristic. During normal inspiration, pressure pulls air into the lungs, resulting in stretching of the alveolar epithelium. For mimicking this phenomenon, pressure-driven stretching by incorporating two larger, lateral microchambers into the device design (Huh et al., 2010). Also, microscale curved surfaces affect the spatiotemporal organization and behaviors of cells. The membrane was given the shape of hexagonally arrayed hemispherical microwells into the device design (Baptista et al., 2022).
Multiple clinical trials have found that microvascular thrombotic and inflammatory processes may be crucial in exacerbating ARDS and increasing lung damage (Ackermann et al., 2020; Jung et al., 2020; Sardu et al., 2020). The vascular damage, and whether it is a direct consequence of endothelial infection or an indirect consequence of immune cells-mediated cytokine storm remain unclear. By using a vascularized lung-on-a-chip model, Thacker et al. (2021) found that infection of alveolar epithelial cells leads to the limited apical release of virions, and viral RNA and proteins are rapidly detected in underlying endothelial cells, which are themselves refractory to apical infection in monocultures. In addition, endothelial cells infected by SARS-CoV-2 lose expression of tight junction markers and adopt a pro-coagulatory phenotype. These results indicate that the dynamics of vascular damage are a direct consequence of endothelial infection independently of a cytokine storm.
To accurately resemble human-relevant responses to viral infection, Zhang M. et al. (2020) also created an alveoli-on-a-chip consisting of the upper human alveolar epithelium channel, lower microvascular endothelium, and circulating immune cells channel. They found a higher susceptibility to SARS-CoV-2 infection in the epithelium than in endothelium upon SARS-CoV-2 infection. Furthermore, they used RNA-seq analysis of immune responses to SARS-CoV-2 infection in this alveolus chip, and the results suggested the crucial role of immune cells involved in alveolar barrier injury and exacerbated inflammation. These results might explain the pathogenesis of the lung microvascular thrombosis and endotheliitis that existed in severe cases of SARS-CoV-2 infection.
With the advances in microfabrication technology, microfluidics, and tissue engineering (Jin et al., 2021), new approaches to the development of lung-on-a-chip models enable the production of more robust and high-throughput human in vitro respiratory tract models. The organoplate is an organ-on-a-chip platform comprising 96 microchambers that can be used for 3D cell culture (Trietsch et al., 2013). These microchambers are incorporated into a standard 384-well microtiter plate that is pipette-operatable and fully compatible with industrial readout and liquid handling equipment. Each microchamber consists of adjacent microchannels separated by phase-guides and four wells are linked together by microfluidic channels (Junaid and Hankemeier, 2021). van Duinen et al. (2017) used such organoplate to assess the vascular barrier function of 96 perfusable blood vessels which have a size-selective permeability with data from in vivo studies, and found that cytokines such VEGF and TNFα have dose-dependent effects on the vascular permeability.
In short, these studies show that lung-on-a-chip contributes to the exploration of the intricate cross-talk between vascular networks and alveolar epithelial cells.
3.5 3D bioprinting to mimic pulmonary interstitium
Although organoid and lung-on-a-chip models are suitable for mimicking the 3D structure and function of alveoli and airway as well as the interaction of alveoli and capillary networks, they cannot mimic the heterogenic pulmonary interstitium structures with complex components (consisting of collagen, elastin, fibronectin, glycoproteins, proteoglycans, 60 different kinds of cells) and certain arrangement (airways tree, alveoli network, and the vascular tree), which play an important role in controlling and guiding cellular behaviors to ultimately define tissue architecture (Suki et al., 2011; Barreiro Carpio et al., 2021; Novak et al., 2021).
3D bioprinting, an emerging novel 3D cell culture tool, deposits layer-by-layer cells and biomaterials in an organized and automatized manner (Zhang et al., 2017). Due to the capability of delivering cells and biomaterials with precise control over spatial distributions, it is possible to rapidly recreate engineered constructs with accurate architecture and composition of targeting tissues such as lungs, which are suitable for drug testing and virus infection (Malda et al., 2013; Murphy and Atala, 2014; Matai et al., 2020).
The general process of 3D bioprinting contains three steps: acquisition of a 3D computer-aided design (CAD) model, automated deposition of biomaterials (referred to biomaterial inks) or the mixture of cells and biomaterials (referred to bioinks), and maturation of cell-laden constructs to reinforce the development of desired tissue constructs (Figure 4) (Mironov et al., 2003; Mironov et al., 2008; Zhang et al., 2017; Barreiro Carpio et al., 2021). There are different 3D printing strategies including extrusion-based bioprinting, inkjet/drop-on-demand, laser-assisted, stereolithography, and electronspinning-based bioprinting (Table 4). The most commonly used bioprinting is the extrusion-based technique, due to its ease of handling and low cost. The success of 3D bioprinting chiefly depends on the ability to formulate complex, cell-laden 3D structures. To date, there is no single bioprinting technique that enables the production of all scales and complexities of synthetic tissues and organs (Matai et al., 2020).
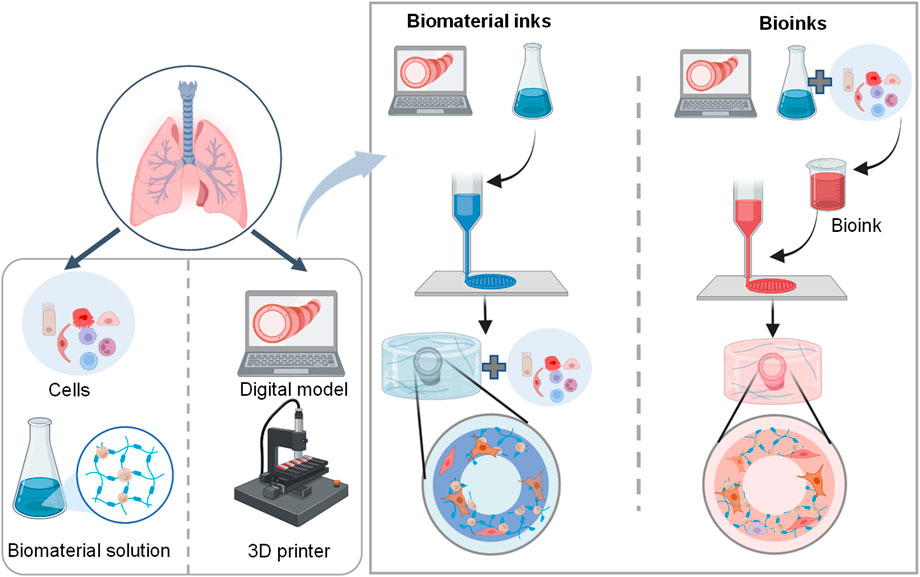
FIGURE 4. Schematic representation of the 3D lung bioprinting process. The general process of 3D bioprinting contains three steps: acquisition of a 3D CAD model, automated deposition of biomaterials (referred to biomaterial inks) or the mixture of cells and biomaterials (referred to bioinks), and maturation of cell-laden constructs to mimic desired tissue constructs (Barreiro Carpio et al., 2021).
To get high mm-sized high-precision scaffolds, Erben and co-workers (Erben et al., 2020) adopted two-photon stereolithography to print 3D cell scaffolds with varying Young’s moduli ranging from 7–300 kPa. The dynamics of colonizing primary human lung fibroblast cells are observed by modifying scaffold geometry. Also, Jing et al. (2021) successfully fabricated fibrous scaffolds via electrohydrodynamic printing. Based on drop-on-demand inkjet-printing, high-resolution deposition of alveolar cells enables to fabricate a three-layered alveolar barrier model with an unprecedented thickness of ≈10 μm, which better recapitulates the structure, morphologies, and functions of the lung tissue and could reproduce practical tissue-level responses to influenza infection (Kang et al., 2021).
Nowadays, more than 100 types of biomaterials are currently being used as bioinks for 3D bioprinting. The most important features of bioinks are bioactivity, biocompatibility, biodegradability, and mechanical properties which provide mechanical strength, physical stability, and biological features (Petersen et al., 2010; O’Brien, 2011; Nichols et al., 2014). Therefore, these bioinks are composed of many types of biocompatible materials from either natural or synthetic materials. Hydrogels such as collagen and GelMA are the most common scaffold to encapsulate cells since they are akin to the native ECM to enable cell differentiation and proper function (Zhao Z. et al., 2020). Studies suggested that some patients with severe COVID-19 develop fibrotic lung disease, an interstitial lung disease characterized by the excessive deposition of ECM proteins in the lung (Bharat et al., 2020; Tale et al., 2020). When the virus binds to the ECM components, it may significantly affect cellular responses to assist its infection in the host (de Melo et al., 2021). A good selection of bioink is of great significance for cell growth and the establishment of disease models. For example, Falcones et al. (2021) successfully designed a new bioink with tunable stiffness based on decellularized porcine lung ECM hydrogels for 3D culturing of lung-resident MSCs. This bioink doesn’t need additional chemical or physical crosslinking and improves preconditioning MSCs for therapeutic purposes.
These elements provide a basis for mimicking the heterogenic pulmonary interstitium structures. Horvath et al. (2015) reported the 3D bioprinting of an alveolar barrier by using a micro-extrusion bioprinter. The biofabricated structure was morphologically similar to the native tissue, being highly organized in a thin layer. On the other hand, cells manually mixed to Matrigel formed multi-layered clusters with tick ECM between the epithelial and endothelial cells, which can affect the permeability of biomolecules. Using 3D bioprinting via photopolymerizable hydrogels, Grigoryan et al. (2019) successfully designed the most complex alveolar models, which contain elaborate entangled vascular networks from space-filling mathematical topologies and can be used to explore the oxygenation and flow of human red blood cells during tidal ventilation and distension of a proximate airway.
To study the mechanisms of SARS-CoV-2 infection in the lung, the alveolar tissue can be mimicked using a 3D bioprinting model with high repeatability and reliability. Ng et al. (2021) reported the fabrication of human triple-layered alveolar lung models using the drop-on-demand 3D bioprinting. These 3D bioprinted human triple-layered alveolar lung models consisted of human lung epithelial cells, human endothelial cells, and human lung fibroblasts and showed high survivability rates over a long-term period of at least 14 days. This would help to address the demand for highly repeatable and scalable fabrication of 3D in-vitro alveolar lung models for studying global respiratory diseases caused by infectious pathogens. Also, in order to clarify virus detection and characterization, Koban et al. (2020) designed easy-to-handle 3D bioprinting platforms based on Wellbrick matrix containing gelatin and collagen additives. This will provide a promising way for the characterization of virus infections due to sensitive monitoring virus-host interactions and replication of different viruses under physiologically relevant conditions.
4 Application of 3D cell culture models in study of COVID-19 treatments
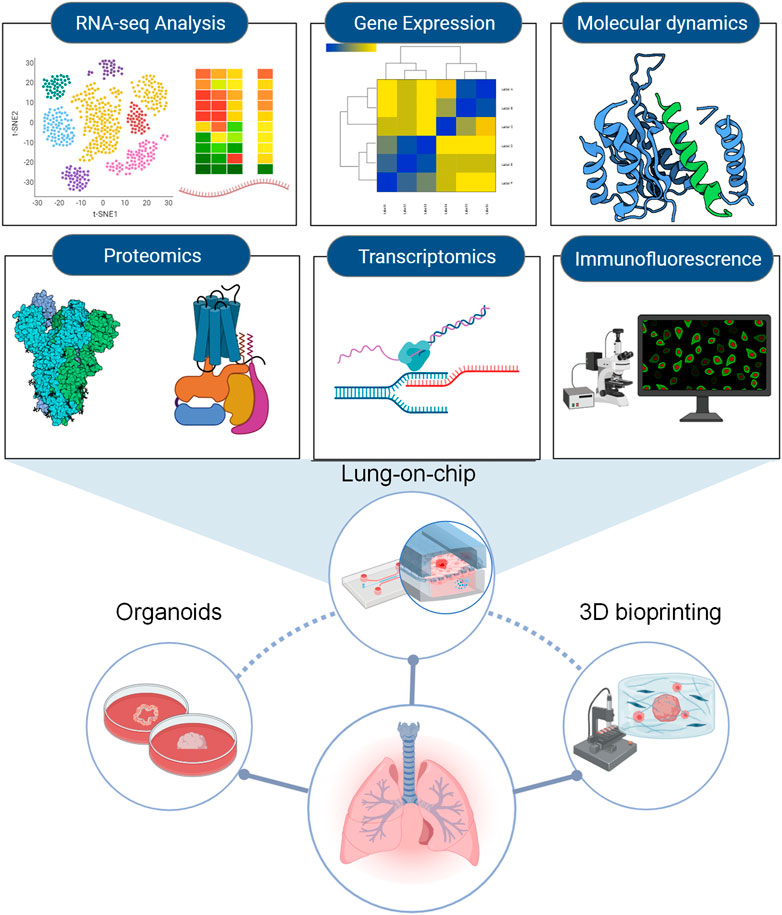
3D cell culture models can provide robust support to simulate SARS-CoV-2 infection, replication, and related immuno-inflammatory responses in humans, which is crucial to understanding the virus’s biology and developing antiviral drugs. For example, in addition to investigating viral effects on human pulmonary tissue, lung organoids are very helpful for high-throughput assays to screen therapeutic drugs, such as RNA-seq analyses, proteomics, phosphoproteomics, transcriptomics, and molecular docking analysis (Elbadawi and Efferth, 2020; Hekman et al., 2020). Moreover, organoids derived from patient lung tissue may lead to a promising resource of potentially effective drugs for treating and managing the disease (Xu et al., 2021). This process from the organoid establishment to drug testing is concise so that the organoid models will be a valuable platform for screening patient-specific drugs (Zhou et al., 2018; Sachs et al., 2019). Apart from organoid, a spheroid culture system for human alveolus was established and this platform has been used for accurate pre-clinical testing of candidate drugs for the treatment of COVID-19 (Ebisudani et al., 2021).
ACE2 is the canonical entry receptor used by SARS-CoV-2 yet is expressed in only a small fraction of airway epithelial cells, predominantly AT2 (Zhao B. et al., 2020). For example, by using hESC-derived cardiac and lung organoids, 1,443 FDA-approved drugs were used to search for modulators of ACE2 levels and identified that inhibitors of 5 alpha reductases, which inhibit androgen signaling, can reduce ACE2 levels in the target cells and thereby decrease SARS-CoV-2 infectivity (Samuel et al., 2020).
However, ACE2 alone cannot explain the multi-organ tropism of SARS-CoV-2, and some analyses of patients with COVID-19 revealed many virus-positive cells without ACE2 expression (Chua et al., 2020; Hikmet et al., 2020; Ren et al., 2021). This evidence indicates other receptors involved in SARS-CoV-2 host interactions. By using lung organoids that provide more physiological conditions, KREMEN1/ASGR1 and Tetraspanin 8 (TSPAN8) were identified as alternative functional receptors of SARS-CoV-2 (Hysenaj et al., 2021; Gu et al., 2022). Adult human stem cell-derived alveolosphere has been developed to provide long-term expansion and differentiation of human alveolar type 2 cells/pneumocytes, which express ACE2 and TMPRSS2. These organoids respond to SARS-CoV-2 infection with upregulation of IFN and downregulation of surfactant, and low-dose IFN blocks SARS-CoV-2 replication, which mimics the features of COVID-19 lungs (Katsura et al., 2020). These newly discovered host receptors play essential roles in ACE2-independent virus entry and potential therapeutic targets for COVID-19.
COVID-19 patients who are hospitalized frequently manifest lymphopenia, which suggests suppression of cellular immune responses (Zhang J. J. et al., 2020; Wang D. et al., 2020). 3D models mimicking the lung structure and function have great advantages to investigate airway immune responses to viral infection. For example, Nelli et al. (2016) developed an ALI culture system that closely mimics the natural airway epithelium to characterize the innate immune response of feline herpesvirus-1 (FHV-1). In addition, Purwada et al. (2015) described a B cell follicle organoid to control the rate of immune reaction through tunable design parameters. The next major development may be organoids of the thymus and lymph node because the thymus and bone marrow are the central immune organs for development, differentiation, and maturation of human immune cells.
COVID-19 patients in severe cases may also develop complications such as hypercoagulopathy, systemic endotheliitis, and even multi-organ failure, which are concomitant with a sustained release of inflammatory factors caused by an excessive inflammatory response. Such phenomenon of elevated levels of inflammatory factors has been detected in organoid models following SARS-CoV-2 infection (Mills et al., 2021; Pei et al., 2021). SARS-CoV-2 is also likely to enter vascular endothelial cells through infected airway epithelial cells, which may lead to endothelial dysfunction (Harrison et al., 2020). More recently, Hashimoto et al. (2022) used a lung-on-a-chip model consisting of two microchannels to mimic the interactions between epithelial, endothelial, and immune cells, and they demonstrated in this model that Claudin-5 is a key factor in disrupting vascular endothelial cadherin-mediated adherent junctions.
In clinical trials different strategies have been adopted to deal with an excessive inflammatory response. They either boost the early type I or III interferon responses in order to help patient get rid of the virus before it causes hyper-inflammation, or suppress immune response by inhibiting specific pro-inflammatory pathways such as IL-6. However, it is still in question how to balance the immune status so that the viral replication can be effectively suppressed without cytokine storm to cause organ failure. Correct use of 3D models may help better understand this question and thus accelerate optimization of the immune therapeutic approach for SARS-CoV-2 infection.
Furthermore, 3D lung models serve as a powerful platform for drug screening and safety assays against SARS-CoV-2, targeting host cells factors (ACE2, TMPRSS2, ACAT and HIF1α) and the virus itself (Tiwari et al., 2021; Kastenhuber et al., 2022). By combining iPSCs-derived lung organoids and high-throughput screening techniques from United States Food and Drug Administration (FDA)-approved drugs, GW6471 has been identified to block SARS-CoV-2 infection (Duan et al., 2021) and imatinib, mycophenolic acid (MPA), and quinacrine dihydrochloride (QNHC) have been identified to block SARS-CoV-2 entry (Han et al., 2021). Combining iPSCs-derived lung organoids with differentiated AT2 cells and a connectivity mapping approach, atorvastatin was predicted to be the most promising candidate from 20,000 small compounds for blocking SARS-CoV-2 (Duarte et al., 2021).
Remdesivir has been recently recognized as a promising antiviral drug against many RNA viruses (e.g., SARS, MERS-CoV), including SARS-CoV-2 (Wang M. et al., 2020). By using an air-liquid interface model, remdesivir has been reported to decrease the colony-forming efficiency (CFE) of club cells, but promote the growth of club organoids (Wang J. et al., 2020). Also, by using an alveolar chip, remdesivir has been proven to inhibit viral replication and alleviate the disruption of the alveolar-capillary barrier (Zhang M. et al., 2020).
A systematic example of 3D cell culture models with some fast-track approaches used in screening drugs against COVID-19 is listed in Figure 5. A summary of the aforementioned drugs suitable for COVID-19 treatment can be seen in Table 5.
5 Future direction and perspectives
Since the complexity of SARS-CoV-2 infection and the pathogenesis of COVID-19 and the timeliness for high-throughput of drug screening for therapeutic treatment, the use of 3D cell culture toolsets provides increasing similarity to the in vivo pathophysiology and may give us valuable insights and more reliable tools for drug development and testing in the treatments of COVID-19 (Ravi et al., 2015).
Recent advances in organoids have enabled the modeling of SARS-CoV-2 infection using human airway epithelial and alveolar cells. These culture models, however, lack airway smooth muscle cells (ASMCs). ASMCs are known to switch between contractile and proliferative phenotypes in response to various physical and chemical cues, which play essential roles in lung development and respiratory diseases (Kim et al., 2015). Therefore, organoids containing ASMCs may better recapitulate the airway structure and function in vivo, and if co-cultured with immune cells, could be used to reveal more detailed mechanisms of the COVID-19-associated immunology.
Appropriate culture media should also be developed for efficient generation and maintenance of lung organoids. Such media should contain not only basal medium, but also a variety of growth factors and small-molecules. For example, TGF-β has an anti-inflammatory effect on cell response induced by influenza H1N1 virus (BustosRivera-Bahena et al., 2021). TGF-β also inhibits cell proliferation and promotes apoptosis and differentiation. So far, effect of TGF-β in the culture medium on efficiency of organoid formation has been evaluated (Shi and Massague, 2003), and evaluation may be required of other signaling pathways such as Wnt/β regarding their effects on organoid culture medium.
Lung-on-a-chip is always restricted to mimic the alveolar-capillary barrier and cannot be used to resemble the airway tubular structure. In addition, lung-on-a-chip is typically limited by the number of cells as compared to that in vivo, which may lead to altered cellular function such as the metabolic rate. Such issues need to be addressed in the development of next-generation lung-on-a-chip systems.
3D bioprinting airway or alveoli is always expensive, which leads that it is almost impossible to set up high-throughput approaches for drug screening using these 3D cell culture models. In addition, despite the extensive use of 3D cell culture models in basic research, their translational biomedical application is restricted since the lack of robust, reproducible, and scalable methods of production in compliance with current pharmaceutical standards. The reproducibility, accuracy, and scalability of the methodologies proposed still need to be improved (Vives and Batlle-Morera, 2020). Therefore, easy-to-use, rapid, and low-cost strategies in the fabrication of 3D cell culture models with more complex cytoarchitecture and a more physiological microenvironment still need to develop.
Despite of their potential, 3D models still entail considerable technical problems that may compromise their application, which include but are not limited to sample collection, high-quality imaging, and cost of substrate materials such as Matrigel. For example, it is technically challenging to directly collect samples from Matrigel-based 3D models such as organoids or 3D bioprinting because direct sample collection would disrupt the integrity of Matrigel and thus the structure of the 3D model, leading to inaccurate results. It is also difficult to obtain high-quality imaging of these 3D cultures by using traditional imaging techniques such as paraffin embedding and tissue slicing (Rios and Clevers, 2018). Although non-invasive microscopy methods such as multi-photon and light-sheet microscopy can be used to visualize cellular details of 3D cultures, the thick sample can cause light scattering because of mismatched refractive indices. To reduce scattering within the sample, various optical clearing methods have been developed. For example, Dekkers et al. (2019) successfully designed a simple optical clearing method utilizing a homemade fructose-glycerol clearing agent to improve the light penetration through fixed organoids, whereas Boothe et al. (2017) used another agent named Iodixanol for the same purpose.
3D models commonly use Matrigel (Corning) or Cultrex BME (Trevigen) as substrate materials. These materials are too expensive to be used in large-scale studies such as high-throughput screening, regenerative medicine and diagnostics (Curvello et al., 2020). This high cost may be reduced by replacing Matrigel/Cultrex BME with an alternative in the 3D models. For example, gels made of decellularized porcine small intestine mucosa/submucosa were used to generate human gastric organoids (Giobbe et al., 2019). Recently, a novel engineered plant-based nanocellulose hydrogel was developed to provide the required microenvironment for small intestinal organoid growth and budding (Curvello et al., 2020).
Materials properties and fluid dynamics are also important issues to consider in developing 3D cultures. For example, Matrigel needs to be mixed with agarose to achieve proper mechanical properties for long time maintenance of a tubular organoid structure (Güney et al., 2021). In order to provide the low shear environment suitable for iPSCs differentiation, organoids can be cultured in spinner flasks, which can be further combined with microfluidic design to achieve both chemical and mechanical stimulation in tunable fashions.
Last but not least, each of the 3D models has its own advantages and disadvantages, and it may be feasible to combine some of these models as a novel platform with enhanced functionality. For example, although lung organoids always automatedly generate target tissue structure and function, they lack typical epithelium-endothelium tissue interfaces in vivo and are time-consuming to fabricate. On the other hand, the advantage of 3D bioprinting is the possibility to precisely define the composition and arrangement of a culture which can be realized rapidly. Combining the organoid and 3D bioprinting techniques may establish a new cell culture model to reflect the near-physiological cross-talk among airway and vascular vessels after SARS-CoV-2 infection. Advances in bioink preparation including the incorporation of bioactive matrices, and induced pluripotent stem cells, and suitable induction factors may open up broad applications for 3D cell culture models in disease diagnostics, and regenerative medicine.
6 Conclusion
The COVID-19 pandemic has resulted in global health and economic burden, but the underlying pathogenesis and therapeutic treatments remain to be further explored. Novel 3D cell culture models representing the lung structure and function in a dish that is in good agreement with reports from animal models and clinical disease are undoubtedly a valuable toolset for these studies, which may provide a promising alternative for animal models and 2D cell culture models. This review of the strategies to fabricate useful 3D pulmonary cell culture models including organoids, lab-on-a-chip, and bioprinting, although by no means comprehensive, indeed provide new insights for understanding how SARS-CoV-2 infects human lung cells, which is essential for elaborating the virus-induced human responses and helping further development of novel therapeutics and prophylactics for COVID-19.
Author contributions
ML and LD conceived and designed the study. KN and BC wrote the manuscript. CY, YQ, RG, CW collected some data for this manuscript. ML and LD edited and revised the manuscript. All authors read and approved the final version of the manuscript.
Funding
This work was supported by NSF of China (Nos.12072048, 12272063, and 31670950), the Key Program of NSF of China (No.11532003), the science and technology innovation leading plan of high tech industry in Hunan Province (2020SK2018), and the International Science and Technology Cooperation Project of Changzhou (CZ20180017).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Ackermann, M., Verleden, S. E., Kuehnel, M., Haverich, A., Welte, T., Laenger, F., et al. (2020). Pulmonary vascular endothelialitis, thrombosis, and angiogenesis in covid-19. N. Engl. J. Med. 383 (2), 120–128. doi:10.1056/NEJMoa2015432
Baker, B. M., and Chen, C. S. (2012). Deconstructing the third dimension: How 3D culture microenvironments alter cellular cues. J. Cell Sci. 125, 3015–3024. doi:10.1242/jcs.079509
Baptista, D., Moreira Teixeira, L., Barata, D., Tahmasebi Birgani, Z., King, J., van Riet, S., et al. (2022). 3D lung-on-chip model based on biomimetically microcurved culture membranes. ACS Biomater. Sci. Eng. 8 (6), 2684–2699. doi:10.1021/acsbiomaterials.1c01463
Barkauskas, C. E., Chung, M. I., Fioret, B., Gao, X., Katsura, H., and Hogan, B. L. (2017). Lung organoids: Current uses and future promise. Development 144 (6), 986–997. doi:10.1242/dev.140103
Barreiro Carpio, M., Dabaghi, M., Ungureanu, J., Kolb, M. R., Hirota, J. A., and Moran-Mirabal, J. M. (2021). 3D bioprinting strategies, challenges, and opportunities to model the lung tissue microenvironment and its function. Front. Bioeng. Biotechnol. 9, 773511. doi:10.3389/fbioe.2021.773511
Benam, K. H., Villenave, R., Lucchesi, C., Varone, A., Hubeau, C., Lee, H.-H., et al. (2016). Small airway-on-a-chip enables analysis of human lung inflammation and drug responses in vitro. Nat. Methods 13 (2), 151–157. doi:10.1038/nmeth.3697
Bernard, I., Limonta, D., Mahal, L. K., and Hobman, T. C. (2020). Endothelium infection and dysregulation by SARS-CoV-2: Evidence and caveats in COVID-19. Viruses 13 (1), 29. doi:10.3390/v13010029
Bestle, D., Heindl, M. R., Limburg, H., Van Lam van, T., Pilgram, O., Moulton, H., et al. (2020). TMPRSS2 and furin are both essential for proteolytic activation of SARS-CoV-2 in human airway cells. Life Sci. Alliance 3 (9), e202000786. doi:10.26508/lsa.202000786
Bharat, A., Querrey, M., Markov, N. S., Kim, S., Kurihara, C., Garza-Castillon, R., et al. (2020). Lung transplantation for patients with severe COVID-19. Sci. Transl. Med. 12 (574), eabe4282. doi:10.1126/scitranslmed.abe4282
Bilodeau, C., Shojaie, S., Goltsis, O., Wang, J., Luo, D., Ackerley, C., et al. (2021). TP63 basal cells are indispensable during endoderm differentiation into proximal airway cells on acellular lung scaffolds. NPJ Regen. Med. 6 (1), 12. doi:10.1038/s41536-021-00124-4
Bircsak, K. M., DeBiasio, R., Miedel, M., Alsebahi, A., Reddinger, R., Saleh, A., et al. (2021). A 3D microfluidic liver model for high throughput compound toxicity screening in the OrganoPlate®. Toxicology 450, 1879–3185. doi:10.1016/j.tox.2020.152667
Birukov, K. G., Jacobson, J. R., Flores, A. A., Ye, S. Q., Birukova, A. A., Verin, A. D., et al. (2003). Magnitude-dependent regulation of pulmonary endothelial cell barrier function by cyclic stretch. Am. J. Physiol. Lung Cell. Mol. Physiol. 285 (4), L785–L797. doi:10.1152/ajplung.00336.2002
Blanco-Melo, D., Nilsson-Payant, B. E., Liu, W. C., Uhl, S., Hoagland, D., Møller, R., et al. (2020). Imbalanced host response to SARS-CoV-2 drives development of COVID-19. Cell 181 (5), 1036–1045. doi:10.1016/j.cell.2020.04.026
Boothe, T., Hilbert, L., Heide, M., Berninger, L., Huttner, W. B., Zaburdaev, V., et al. (2017). A tunable refractive index matching medium for live imaging cells, tissues and model organisms. Elife 6, e27240. doi:10.7554/eLife.27240
Borczuk, A. C., Salvatore, S. P., Seshan, S. V., Patel, S. S., Bussel, J. B., Mostyka, M., et al. (2020). COVID-19 pulmonary pathology: A multi-institutional autopsy cohort from Italy and New York city. Mod. Pathol. 33 (11), 2156–2168. doi:10.1038/s41379-020-00661-1
Bugge, T. H., Antalis, T. M., and Wu, Q. (2009). Type II transmembrane serine proteases. J. Biol. Chem. 284 (35), 23177–23181. doi:10.1074/jbc.R109.021006
BustosRivera-Bahena, G., Lopez-Guerrero, D. V., Marquez-Bandala, A. H., Esquivel-Guadarrama, F. R., and Montiel-Hernandez, J. L. (2021). TGF-β1 signaling inhibit the in vitro apoptotic, infection and stimulatory cell response induced by influenza H1N1 virus infection on A549 cells. Virus Res. 297, 198337. doi:10.1016/j.virusres.2021.198337
Callens, S. J. P., Uyttendaele, R. J. C., Fratila-Apachitei, L. E., and Zadpoor, A. A. (2020). Substrate curvature as a cue to guide spatiotemporal cell and tissue organization. Biomaterials 232, 119739. doi:10.1016/j.biomaterials.2019.119739
Cantuti-Castelvetri, L., Ojha, R., Pedro, L. D., Djannatian, M., Franz, J., Kuivanen, S., et al. (2020). Neuropilin-1 facilitates SARS-CoV-2 cell entry and infectivity. Science 370 (6518), 856–860. doi:10.1126/science.abd2985
Castelli, V., Cimini, A., and Ferri, C. (2020). Cytokine storm in COVID-19: "When you come out of the storm, you won't Be the same person who walked in. Front. Immunol. 11, 2132. doi:10.3389/fimmu.2020.02132
Castiello, T., Georgiopoulos, G., Finocchiaro, G., Claudia, M., Gianatti, A., Delialis, D., et al. (2022). COVID-19 and myocarditis: A systematic review and overview of current challenges. Heart fail. Rev. 27 (1), 251–261. doi:10.1007/s10741-021-10087-9
Chen, N., Zhou, M., Dong, X., Qu, J., Gong, F., Han, Y., et al. (2020). Epidemiological and clinical characteristics of 99 cases of 2019 novel coronavirus pneumonia in wuhan, China: A descriptive study. Lancet 395 (10223), 507–513. doi:10.1016/S0140-6736(20)30211-7
Chen, Q., Langenbach, S., Li, M., Xia, Y. C., Gao, X., Gartner, M. J., et al. (2022). ACE2 expression in organotypic human airway epithelial cultures and airway biopsies. Front. Pharmacol. 13, 813087. doi:10.3389/fphar.2022.813087
Chua, R. L., Lukassen, S., Trump, S., Hennig, B. P., Wendisch, D., Pott, F., et al. (2020). COVID-19 severity correlates with airway epithelium-immune cell interactions identified by single-cell analysis. Nat. Biotechnol. 38 (8), 970–979. doi:10.1038/s41587-020-0602-4
Corbett, K. S., Flynn, B., Foulds, K. E., Francica, J. R., Boyoglu-Barnum, S., Werner, A. P., et al. (2020). Evaluation of the mRNA-1273 vaccine against SARS-CoV-2 in nonhuman primates. N. Engl. J. Med. 383 (16), 1544–1555. doi:10.1056/NEJMoa2024671
Cox, R. M., Wolf, J. D., and Plemper, R. K. (2021). Therapeutically administered ribonucleoside analogue MK-4482/EIDD-2801 blocks SARS-CoV-2 transmission in ferrets. Nat. Microbiol. 6 (1), 11–18. doi:10.1038/s41564-020-00835-2
Curvello, R., Kerr, G., Micati, D. J., Chan, W. H., Raghuwanshi, V. S., Rosenbluh, J., et al. (2020). Engineered plant-based nanocellulose hydrogel for small intestinal organoid growth. Adv. Sci. 8 (1), 2002135. doi:10.1002/advs.202002135
Daly, J. L., Simonetti, B., Klein, K., Chen, K. E., Williamson, M. K., Antón-Plágaro, C., et al. (2020). Neuropilin-1 is a host factor for SARS-CoV-2 infection. Science 370 (6518), 861–865. doi:10.1126/science.abd3072
de Melo, B. A. G., Benincasa, J. C., Cruz, E. M., Maricato, J. T., and Porcionatto, M. A. (2021). 3D culture models to study SARS-CoV-2 infectivity and antiviral candidates: From spheroids to bioprinting. Biomed. J. 44 (1), 31–42. doi:10.1016/j.bj.2020.11.009
de Oliveira, M., De Sibio, M. T., Costa, F. A. S., and Sakalem, M. E. (2021). Airway and alveoli organoids as valuable research tools in COVID-19. ACS Biomater. Sci. Eng. 7 (8), 3487–3502. doi:10.1021/acsbiomaterials.1c00306
Dekkers, J. F., Alieva, M., Wellens, L. M., Ariese, H. C. R., Jamieson, P. R., Vonk, A. M., et al. (2019). High-resolution 3D imaging of fixed and cleared organoids. Nat. Protoc. 14 (6), 1756–1771. doi:10.1038/s41596-019-0160-8
Diem, K., Fauler, M., Fois, G., Hellmann, A., Winokurow, N., Schumacher, S., et al. (2020). Mechanical stretch activates piezo1 in caveolae of alveolar type I cells to trigger ATP release and paracrine stimulation of surfactant secretion from alveolar type II cells. FASEB J. 34 (9), 12785–12804. doi:10.1096/fj.202000613RRR
Dinnon, K. H., Leist, S. R., Schäfer, A., Edwards, C. E., Martinez, D. R., Montgomery, S. A., et al. (2020). A mouse-adapted model of SARS-CoV-2 to test COVID-19 countermeasures. Nature 586 (7830), 560–566. doi:10.1038/s41586-020-2708-8
Duan, X., Tang, X., Nair, M. S., Zhang, T., Qiu, Y., Zhang, W., et al. (2021). An airway organoid-based screen identifies a role for the HIF1α-glycolysis axis in SARS-CoV-2 infection. Cell Rep. 37 (6), 109920. doi:10.1016/j.celrep.2021.109920
Duarte, R. R. R., Copertino, D. C., Iñiguez, L. P., Marston, J. L., Bram, Y., Han, Y., et al. (2021). Identifying FDA-approved drugs with multimodal properties against COVID-19 using a data-driven approach and a lung organoid model of SARS-CoV-2 entry. Mol. Med. 27 (1), 105. doi:10.1186/s10020-021-00356-6
Duval, K., Grover, H., Han, L. H., Mou, Y., Pegoraro, A. F., Fredberg, J., et al. (2017). Modeling physiological events in 2D vs. 3D cell culture. Physiol. (Bethesda) 32 (4), 266–277. doi:10.1152/physiol.00036.2016
Dye, B. R., Hill, D. R., Ferguson, M. A., Tsai, Y. H., Nagy, M. S., Dyal, R., et al. (2015). In vitro generation of human pluripotent stem cell derived lung organoids. Elife 4, e05098. doi:10.7554/eLife.05098
Ebisudani, T., Sugimoto, S., Haga, K., Mitsuishi, A., Takai-Todaka, R., Fujii, M., et al. (2021). Direct derivation of human alveolospheres for SARS-CoV-2 infection modeling and drug screening. Cell Rep. 35 (10), 109218. doi:10.1016/j.celrep.2021.109218
Elbadawi, M., and Efferth, T. (2020). Organoids of human airways to study infectivity and cytopathy of SARS-CoV-2. Lancet. Respir. Med. 8 (7), e55–e56. doi:10.1016/S2213-2600(20)30238-1
Elsdale, T., and Bard, J. (1972). Collagen substrata for studies on cell behavior. J. Cell Biol. 54 (3), 626–637. doi:10.1083/jcb.54.3.626
Erben, A., Horning, M., Hartmann, B., Becke, T., Eisler, S. A., Southan, A., et al. (2020). Precision 3D-printed cell scaffolds mimicking native tissue composition and mechanics. Adv. Healthc. Mat. 9 (24), e2000918. doi:10.1002/adhm.202000918
Falcones, B., Sanz-Fraile, H., Marhuenda, E., Mendizabal, I., Cabrera-Aguilera, I., Malandain, N., et al. (2021). Bioprintable lung extracellular matrix hydrogel scaffolds for 3D culture of mesenchymal stromal cells. Polym. (Basel) 13 (14), 2350. doi:10.3390/polym13142350
Garrett, N., Tapley, A., Andriesen, J., Seocharan, I., Fisher, L. H., Bunts, L., et al. (2022). High rate of asymptomatic carriage associated with variant strain Omicron. medRxiv., 2021.2012.2021.12.20.21268130.2021.12.20.21268130. doi: doi:10.1101/2021.12.20.21268130
Ghajar, C. M., Chen, X., Harris, J. W., Suresh, V., Hughes, C. C., Jeon, N. L., et al. (2008). The effect of matrix density on the regulation of 3-D capillary morphogenesis. Biophys. J. 94 (5), 1930–1941. doi:10.1529/biophysj.107.120774
Ghio, A. J., Dailey, L. A., Soukup, J. M., Stonehuerner, J., Richards, J. H., and Devlin, R. B. (2013). Growth of human bronchial epithelial cells at an air-liquid interface alters the response to particle exposure. Part. Fibre Toxicol. 10, 25. doi:10.1186/1743-8977-10-25
Giobbe, G. G., Crowley, C., Luni, C., Campinoti, S., Khedr, M., Kretzschmar, K., et al. (2019). Extracellular matrix hydrogel derived from decellularized tissues enables endodermal organoid culture. Nat. Commun. 10 (1), 5658. doi:10.1038/s41467-019-13605-4
Grigoryan, B., Paulsen, S. J., Corbett, D. C., Sazer, D. W., Fortin, C. L., Zaita, A. J., et al. (2019). Multivascular networks and functional intravascular topologies within biocompatible hydrogels. Science 364 (6439), 458–464. doi:10.1126/science.aav9750
Gu, Y., Cao, J., Zhang, X., Gao, H., Wang, Y., Wang, J., et al. (2022). Receptome profiling identifies KREMEN1 and ASGR1 as alternative functional receptors of SARS-CoV-2. Cell Res. 32 (1), 24–37. doi:10.1038/s41422-021-00595-6
Guan, W. J., Ni, Z. Y., Hu, Y., Liang, W. H., Ou, C. Q., He, J. X., et al. (2020). Clinical characteristics of coronavirus disease 2019 in China. N. Engl. J. Med. 382 (18), 1708–1720. doi:10.1056/NEJMoa2002032
Güney, T. G., Herranz, A. M., Mumby, S., Dunlop, I. E., and Adcock, I. M. (2021). Epithelial-stromal cell interactions and extracellular matrix mechanics drive the formation of airway-mimetic tubular morphology in lung organoids. iScience 24 (9), 103061. doi:10.1016/j.isci.2021.103061
Haga, K., Takai-Todaka, R., Matsumura, Y., Song, C., Takano, T., Tojo, T., et al. (2021). Nasal delivery of single-domain antibody improves symptoms of SARS-CoV-2 infection in an animal model. PLoS Pathog. 17 (10), e1009542. doi:10.1371/journal.ppat.1009542
Hakobyan, D., Medina, C., Dusserre, N., Stachowicz, M. L., Handschin, C., Fricain, J. C., et al. (2020). Laser-assisted 3D bioprinting of exocrine pancreas spheroid models for cancer initiation study. Biofabrication 12 (3), 035001. doi:10.1088/1758-5090/ab7cb8
Han, Y., Duan, X., Yang, L., Nilsson-Payant, B. E., Wang, P., Duan, F., et al. (2021). Identification of SARS-CoV-2 inhibitors using lung and colonic organoids. Nature 589 (7841), 270–275. doi:10.1038/s41586-020-2901-9
Harrison, A. G., Lin, T., and Wang, P. (2020). Mechanisms of SARS-CoV-2 transmission and pathogenesis. Trends Immunol. 41 (12), 1100–1115. doi:10.1016/j.it.2020.10.004
Hashimoto, R., Takahashi, J., Shirakura, K., Funatsu, R., Kosugi, K., Deguchi, S., et al. (2022). SARS-CoV-2 disrupts respiratory vascular barriers by suppressing Claudin-5 expression. Sci. Adv. 8 (38), eabo6783. doi:10.1126/sciadv.abo6783
Hekman, R. M., Hume, A. J., Goel, R. K., Abo, K. M., Huang, J., Blum, B. C., et al. (2020). Actionable cytopathogenic host responses of human alveolar type 2 cells to SARS-CoV-2. Mol. Cell 80 (6), 1104–1122.e9. doi:10.1016/j.molcel.2020.11.028
Hikmet, F., Mear, L., Edvinsson, A., Micke, P., Uhlen, M., and Lindskog, C. (2020). The protein expression profile of ACE2 in human tissues. Mol. Syst. Biol. 16 (7), e9610. doi:10.15252/msb.20209610
Horvath, L., Umehara, Y., Jud, C., Blank, F., Petri-Fink, A., and Rothen-Rutishauser, B. (2015). Engineering an in vitro air-blood barrier by 3D bioprinting. Sci. Rep. 5, 7974. doi:10.1038/srep07974
Hu, B., Huang, S., and Yin, L. (2021). The cytokine storm and COVID-19. J. Med. Virol. 93 (1), 250–256. doi:10.1002/jmv.26232
Huang, C., Wang, Y., Li, X., Ren, L., Zhao, J., Hu, Y., et al. (2020). Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet 395 (10223), 497–506. doi:10.1016/s0140-6736(20)30183-5
Huang, D., Liu, T., Liao, J., Maharjan, S., Xie, X., Perez, M., et al. (2021). Reversed-engineered human alveolar lung-on-a-chip model. Proc. Natl. Acad. Sci. U. S. A. 118 (19), e2016146118. doi:10.1073/pnas.2016146118
Huff, S., Kummetha, I. R., Tiwari, S. K., Huante, M. B., Clark, A. E., Wang, S., et al. (2022). Discovery and mechanism of SARS-CoV-2 main protease inhibitors. J. Med. Chem. 65 (4), 2866–2879. doi:10.1021/acs.jmedchem.1c00566
Huh, D., Matthews, B. D., Mammoto, A., Montoya-Zavala, M., Hsin, H. Y., and Ingber, D. E. (2010). Reconstituting organ-level lung functions on a chip. Science 328 (5986), 1662–1668. doi:10.1126/science.1188302
Humayun, M., Chow, C. W., and Young, E. W. K. (2018). Microfluidic lung airway-on-a-chip with arrayable suspended gels for studying epithelial and smooth muscle cell interactions. Lab. Chip 18 (9), 1298–1309. doi:10.1039/c7lc01357d
Hysenaj, L., Little, S., Kulhanek, K., Gbenedio, O. M., Rodriguez, L., Shen, A., et al. (2021). SARS-CoV-2 infection studies in lung organoids identify TSPAN8 as novel mediator. bioRxiv., 2021.2006.2001.2021.06.01.446640. doi: doi:10.1101/2021.06.01.446640
Jiang, Y., Wu, Q., Song, P., and You, C. (2021). The variation of SARS-CoV-2 and advanced research on current vaccines. Front. Med. 8, 806641. doi:10.3389/fmed.2021.806641
Jin, Y., Liu, L., Yu, P., Lin, F., Shi, X., Guo, J., et al. (2021). Emergent differential organization of airway smooth muscle cells on concave and convex tubular surface. Front. Mol. Biosci. 8, 717771. doi:10.3389/fmolb.2021.717771
Jing, L., Wang, X., Leng, B., Zhan, N., Liu, H., Wang, S., et al. (2021). Engineered nanotopography on the microfibers of 3D-printed PCL scaffolds to modulate cellular responses and establish an in vitro tumor model. ACS Appl. Bio Mat. 4 (2), 1381–1394. doi:10.1021/acsabm.0c01243
Johnson, B. A., Xie, X., Bailey, A. L., Kalveram, B., Lokugamage, K. G., Muruato, A., et al. (2021). Loss of furin cleavage site attenuates SARS-CoV-2 pathogenesis. Nature 591 (7849), 293–299. doi:10.1038/s41586-021-03237-4
Junaid, A., and Hankemeier, T. (2021). OrganoPlate micro-fluidic microvessel culture and analysis. Bio. Protoc. 11 (13), e4070. doi:10.21769/BioProtoc.4070
Jung, F., Kruger-Genge, A., Franke, R. P., Hufert, F., and Kupper, J. H. (2020). COVID-19 and the endothelium. Clin. Hemorheol. Microcirc. 75 (1), 7–11. doi:10.3233/CH-209007
Kang, D., Park, J. A., Kim, W., Kim, S., Lee, H. R., Kim, W. J., et al. (2021). All-inkjet-printed 3D alveolar barrier model with physiologically relevant microarchitecture. Adv. Sci. 8 (10), 2004990. doi:10.1002/advs.202004990
Kastenhuber, E. R., Mercadante, M., Nilsson-Payant, B., Johnson, J. L., Jaimes, J. A., Muecksch, F., et al. (2022). Coagulation factors directly cleave SARS-CoV-2 spike and enhance viral entry. Elife 11, e77444. doi:10.7554/eLife.77444
Katsura, H., Sontake, V., Tata, A., Kobayashi, Y., Edwards, C. E., Heaton, B. E., et al. (2020). Human lung stem cell-based alveolospheres provide insights into SARS-CoV-2-mediated interferon responses and pneumocyte dysfunction. Cell Stem Cell 27 (6), 890–904. doi:10.1016/j.stem.2020.10.005
Ke, Z., Oton, J., Qu, K., Cortese, M., Zila, V., McKeane, L., et al. (2020). Structures and distributions of SARS-CoV-2 spike proteins on intact virions. Nature 588 (7838), 498–502. doi:10.1038/s41586-020-2665-2
Kim, H. Y., Pang, M. F., Varner, V. D., Kojima, L., Miller, E., Radisky, D. C., et al. (2015). Localized smooth muscle differentiation is essential for epithelial bifurcation during branching morphogenesis of the mammalian lung. Dev. Cell 34 (6), 719–726. doi:10.1016/j.devcel.2015.08.012
Kim, J.-H., Kim, J., Kim, W. J., Choi, Y. H., Yang, S.-R., and Hong, S.-H. (2020). Diesel particulate matter 2.5 induces epithelial-to-mesenchymal transition and upregulation of SARS-CoV-2 receptor during human pluripotent stem cell-derived alveolar organoid development. Int. J. Environ. Res. Public Health 17 (22), 8410. doi:10.3390/ijerph17228410
Kızılkurtlu, A. A., Polat, T., Aydın, G. B., and Akpek, A. (2018). Lung on a chip for drug screening and design. Curr. Pharm. Des. 24 (45), 5386–5396. doi:10.2174/1381612825666190208122204
Knight, A. (2008). Systematic reviews of animal experiments demonstrate poor contributions toward human healthcare. Rev. Recent Clin. Trials 3 (2), 89–96. doi:10.2174/157488708784223844
Ko, M., Chang, S. Y., Byun, S. Y., Ianevski, A., Choi, I., Pham, H., et al. (2021). Screening of FDA-approved drugs using a MERS-CoV clinical isolate from South Korea identifies potential therapeutic options for COVID-19. Viruses 13 (4), 651. doi:10.3390/v13040651
Koban, R., Lam, T., Schwarz, F., Kloke, L., Bürge, S., Ellerbrok, H., et al. (2020). Simplified bioprinting-based 3D cell culture infection models for virus detection. Viruses 12 (11), 1298. doi:10.3390/v12111298
Kolagar, T. A., Farzaneh, M., Nikkar, N., and Khoshnam, S. E. (2020). Human pluripotent stem cells in neurodegenerative diseases: Potentials, advances and limitations. Curr. Stem Cell Res. Ther. 15 (2), 102–110. doi:10.2174/1574888x14666190823142911
Konar, D., Devarasetty, M., Yildiz, D. V., Atala, A., and Murphy, S. V. (2016). Lung-on-A-chip technologies for disease modeling and drug development. Biomed. Eng. Comput. Biol. 7, 17–27. doi:10.4137/BECB.S34252
Korff, T., and Augustin, H. G. (1999). Tensional forces in fibrillar extracellular matrices control directional capillary sprouting. J. Cell Sci. 112, 3249–3258. doi:10.1242/jcs.112.19.3249
Lambrecht, B. N., and Hammad, H. (2012). The airway epithelium in asthma. Nat. Med. 18 (5), 684–692. doi:10.1038/nm.2737
Lee, C. Y., and Lowen, A. C. (2021). Animal models for SARS-CoV-2. Curr. Opin. Virol. 48, 73–81. doi:10.1016/j.coviro.2021.03.009
Leibel, S. L., McVicar, R. N., Winquist, A. M., Niles, W. D., and Snyder, E. Y. (2020). Generation of complete multi-cell type lung organoids from human embryonic and patient-specific induced pluripotent stem cells for infectious disease modeling and therapeutics validation. Curr. Protoc. Stem Cell Biol. 54 (1), e118. doi:10.1002/cpsc.118
Liu, Y., Dabrowska, C., Mavousian, A., Strauss, B., Meng, F., Mazzaglia, C., et al. (2021). Bio-assembling macro-scale, lumenized airway tubes of defined shape via multi-organoid patterning and fusion. Adv. Sci. 8 (9), 2003332. doi:10.1002/advs.202003332
Lu, R., Zhao, X., Li, J., Niu, P., Yang, B., Wu, H., et al. (2020). Genomic characterisation and epidemiology of 2019 novel coronavirus: Implications for virus origins and receptor binding. Lancet 395 (10224), 565–574. doi:10.1016/S0140-6736(20)30251-8
Luo, M., Ni, K., Sun, Y., Guo, J., Wen, K., and Deng, L. (2022). Toward an optimized strategy of using various airway mucus clearance techniques to treat critically ill COVID-19 patients. Biocell 46 (4), 855–871. doi:10.32604/biocell.2022.017520
Maisonnasse, P., Guedj, J., Contreras, V., Behillil, S., Solas, C., Marlin, R., et al. (2020). Hydroxychloroquine use against SARS-CoV-2 infection in non-human primates. Nature 585 (7826), 584–587. doi:10.1038/s41586-020-2558-4
Malda, J., Visser, J., Melchels, F. P., Jungst, T., Hennink, W. E., Dhert, W. J., et al. (2013). 25th anniversary article: Engineering hydrogels for biofabrication. Adv. Mat. 25 (36), 5011–5028. doi:10.1002/adma.201302042
Marescotti, D., Serchi, T., Luettich, K., Xiang, Y., Moschini, E., Talikka, M., et al. (2019). How complex should an in vitro model be? Evaluation of complex 3D alveolar model with transcriptomic data and computational biological network models. ALTEX 36 (3), 388–402. doi:10.14573/altex.1811221
Matai, I., Kaur, G., Seyedsalehi, A., McClinton, A., and Laurencin, C. T. (2020). Progress in 3D bioprinting technology for tissue/organ regenerative engineering. Biomaterials 226, 119536. doi:10.1016/j.biomaterials.2019.119536
Mills, R. J., Humphrey, S. J., Fortuna, P. R. J., Lor, M., Foster, S. R., Quaife-Ryan, G. A., et al. (2021). BET inhibition blocks inflammation-induced cardiac dysfunction and SARS-CoV-2 infection. Cell 184 (8), 2167–2182.e22. doi:10.1016/j.cell.2021.03.026
Mironov, V., Boland, T., Trusk, T., Forgacs, G., and Markwald, R. R. (2003). Organ printing: Computer-aided jet-based 3D tissue engineering. Trends Biotechnol. 21 (4), 157–161. doi:10.1016/S0167-7799(03)00033-7
Mironov, V., Kasyanov, V., Drake, C., and Markwald, R. R. (2008). Organ printing: Promises and challenges. Regen. Med. 3 (1), 93–103. doi:10.2217/17460751.3.1.93
Morrisey, E. E., and Hogan, B. L. (2010). Preparing for the first breath: Genetic and cellular mechanisms in lung development. Dev. Cell 18 (1), 8–23. doi:10.1016/j.devcel.2009.12.010
Mulay, A., Konda, B., Garcia, G., Yao, C., Beil, S., Villalba, J. M., et al. (2021). SARS-CoV-2 infection of primary human lung epithelium for COVID-19 modeling and drug discovery. Cell Rep. 35 (5), 109055. doi:10.1016/j.celrep.2021.109055
Mullen, P. J., Garcia, G., Purkayastha, A., Matulionis, N., Schmid, E. W., Momcilovic, M., et al. (2021). SARS-CoV-2 infection rewires host cell metabolism and is potentially susceptible to mTORC1 inhibition. Nat. Commun. 12 (1), 1876. doi:10.1038/s41467-021-22166-4
Murphy, S. V., and Atala, A. (2014). 3D bioprinting of tissues and organs. Nat. Biotechnol. 32 (8), 773–785. doi:10.1038/nbt.2958
Mykytyn, A. Z., Breugem, T. I., Riesebosch, S., Schipper, D., van den Doel, P. B., Rottier, R. J., et al. (2021). SARS-CoV-2 entry into human airway organoids is serine protease-mediated and facilitated by the multibasic cleavage site. Elife 10, e64508. doi:10.7554/eLife.64508
Nelli, R. K., Maes, R., Kiupel, M., and Hussey, G. S. (2016). Use of a feline respiratory epithelial cell culture system grown at the air-liquid interface to characterize the innate immune response following feline herpesvirus 1 infection. Virus Res. 214, 39–48. doi:10.1016/j.virusres.2016.01.006
Ng, W. L., Ayi, T. C., Liu, Y. C., Sing, S. L., Yeong, W. Y., and Tan, B. H. (2021). Fabrication and characterization of 3D bioprinted triple-layered human alveolar lung models. Int. J. Bioprint. 7 (2), 332. doi:10.18063/ijb.v7i2.332
Nichols, J. E., Niles, J. A., Vega, S. P., Argueta, L. B., Eastaway, A., and Cortiella, J. (2014). Modeling the lung: Design and development of tissue engineered macro- and micro-physiologic lung models for research use. Exp. Biol. Med. 239 (9), 1135–1169. doi:10.1177/1535370214536679
Nigjeh, S. E., Yeap, S. K., Nordin, N., Kamalideghan, B., Ky, H., and Rosli, R. (2018). Citral induced apoptosis in MDA-MB-231 spheroid cells. BMC Complement. Altern. Med. 18 (1), 56. doi:10.1186/s12906-018-2115-y
Nof, E., Zidan, H., Artzy-Schnirman, A., Mouhadeb, O., Beckerman, M., Bhardwaj, S., et al. (2022). Human multi-compartment airways-on-chip platform for emulating respiratory airborne transmission: From nose to pulmonary acini. Front. Physiol. 13, 853317. doi:10.3389/fphys.2022.853317
Novak, C., Ballinger, M. N., and Ghadiali, S. (2021). Mechanobiology of pulmonary diseases: A review of engineering tools to understand lung mechanotransduction. J. Biomech. Eng. 143 (11), 110801. doi:10.1115/1.4051118
O'Brien, F. J. (2011). Biomaterials & scaffolds for tissue engineering. Mater. Today 14(3), 88–95. doi: Doi doi:10.1016/S1369-7021(11)70058-X
Pei, R., Feng, J., Zhang, Y., Sun, H., Li, L., Yang, X., et al. (2021). Host metabolism dysregulation and cell tropism identification in human airway and alveolar organoids upon SARS-CoV-2 infection. Protein Cell 12 (9), 717–733. doi:10.1007/s13238-020-00811-w
Petersen, T. H., Calle, E. A., Zhao, L., Lee, E. J., Gui, L., Raredon, M. B., et al. (2010). Tissue-engineered lungs for in vivo implantation. Science 329 (5991), 538–541. doi:10.1126/science.1189345
Purwada, A., Jaiswal, M. K., Ahn, H., Nojima, T., Kitamura, D., Gaharwar, A. K., et al. (2015). Ex vivo engineered immune organoids for controlled germinal center reactions. Biomaterials 63, 24–34. doi:10.1016/j.biomaterials.2015.06.002
Qin, S., Jiang, Y., Wei, X., Liu, X., Guan, J., Chen, Y., et al. (2021). Dynamic changes in monocytes subsets in COVID-19 patients. Hum. Immunol. 82 (3), 170–176. doi:10.1016/j.humimm.2020.12.010
Ravi, M., Paramesh, V., Kaviya, S. R., Anuradha, E., and Solomon, F. D. (2015). 3D cell culture systems: Advantages and applications. J. Cell. Physiol. 230 (1), 16–26. doi:10.1002/jcp.24683
Ren, X., Wen, W., Fan, X., Hou, W., Su, B., Cai, P., et al. (2021). COVID-19 immune features revealed by a large-scale single-cell transcriptome atlas. Cell 184 (7), 1895–1913.e19. doi:10.1016/j.cell.2021.01.053
Rios, A. C., and Clevers, H. (2018). Imaging organoids: A bright future ahead. Nat. Methods 15 (1), 24–26. doi:10.1038/nmeth.4537
Ryu, N. E., Lee, S. H., and Park, H. (2019). Spheroid culture system methods and applications for mesenchymal stem cells. Cells 8 (12), 1620. doi:10.3390/cells8121620
Sachs, N., Papaspyropoulos, A., Zomer-van Ommen, D. D., Heo, I., Bottinger, L., Klay, D., et al. (2019). Long-term expanding human airway organoids for disease modeling. EMBO J. 38 (4), e100300. doi:10.15252/embj.2018100300
Salahudeen, A. A., Choi, S. S., Rustagi, A., Zhu, J., van Unen, V., de la, O. S., et al. (2020). Progenitor identification and SARS-CoV-2 infection in human distal lung organoids. Nature 588 (7839), 670–675. doi:10.1038/s41586-020-3014-1
Samuel, R. M., Majd, H., Richter, M. N., Ghazizadeh, Z., Zekavat, S. M., Navickas, A., et al. (2020). Androgen signaling regulates SARS-CoV-2 receptor levels and is associated with severe COVID-19 symptoms in men. Cell Stem Cell 27 (6), 876–889. doi:10.1016/j.stem.2020.11.009
Sardu, C., Gambardella, J., Morelli, M. B., Wang, X., Marfella, R., and Santulli, G. (2020). Hypertension, thrombosis, kidney failure, and diabetes: Is COVID-19 an endothelial disease? A comprehensive evaluation of clinical and basic evidence. J. Clin. Med. 9 (5), 1417. doi:10.3390/jcm9051417
Sato, T., Vries, R. G., Snippert, H. J., van de Wetering, M., Barker, N., Stange, D. E., et al. (2009). Single Lgr5 stem cells build crypt-villus structures in vitro without a mesenchymal niche. Nature 459 (7244), 262–265. doi:10.1038/nature07935
Sette, A., and Crotty, S. (2021). Adaptive immunity to SARS-CoV-2 and COVID-19. Cell 184 (4), 861–880. doi:10.1016/j.cell.2021.01.007
Sharma, A., Sances, S., Workman, M. J., and Svendsen, C. N. (2020). Multi-lineage human iPSC-derived platforms for disease modeling and drug discovery. Cell Stem Cell 26 (3), 309–329. doi:10.1016/j.stem.2020.02.011
Shi, Y., and Massague, J. (2003). Mechanisms of TGF-beta signaling from cell membrane to the nucleus. Cell 113 (6), 685–700. doi:10.1016/s0092-8674(03)00432-x
Shrestha, J., Razavi Bazaz, S., Aboulkheyr Es, H., Yaghobian Azari, D., Thierry, B., Ebrahimi Warkiani, M., et al. (2020). Lung-on-a-chip: The future of respiratory disease models and pharmacological studies. Crit. Rev. Biotechnol. 40 (2), 213–230. doi:10.1080/07388551.2019.1710458
Spitalieri, P., Centofanti, F., Murdocca, M., Scioli, M. G., Latini, A., Di Cesare, S., et al. (2022). Two different therapeutic approaches for SARS-CoV-2 in hiPSCs-derived lung organoids. Cells 11 (7), 1235. doi:10.3390/cells11071235
Suki, B., Stamenovic, D., and Hubmayr, R. (2011). Lung parenchymal mechanics. Compr. Physiol. 1 (3), 1317–1351. doi:10.1002/cphy.c100033
Takahashi, K., Tanabe, K., Ohnuki, M., Narita, M., Ichisaka, T., Tomoda, K., et al. (2007). Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell 131 (5), 861–872. doi:10.1016/j.cell.2007.11.019
Takahashi, K., and Yamanaka, S. (2006). Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell 126 (4), 663–676. doi:10.1016/j.cell.2006.07.024
Tale, S., Ghosh, S., Meitei, S. P., Kolli, M., Garbhapu, A. K., and Pudi, S. (2020). Post-COVID-19 pneumonia pulmonary fibrosis. QJM Mon. J. Assoc. Physicians 113 (11), 837–838. doi:10.1093/qjmed/hcaa255
Tam, A., Wadsworth, S., Dorscheid, D., Man, S. F., and Sin, D. D. (2011). The airway epithelium: More than just a structural barrier. Ther. Adv. Respir. Dis. 5 (4), 255–273. doi:10.1177/1753465810396539
Tan, Q., Choi, K. M., Sicard, D., and Tschumperlin, D. J. (2017). Human airway organoid engineering as a step toward lung regeneration and disease modeling. Biomaterials 113, 118–132. doi:10.1016/j.biomaterials.2016.10.046
Tay, M. Z., Poh, C. M., Rénia, L., MacAry, P. A., and Ng, L. F. P. (2020). The trinity of COVID-19: Immunity, inflammation and intervention. Nat. Rev. Immunol. 20 (6), 363–374. doi:10.1038/s41577-020-0311-8
Thacker, V. V., Sharma, K., Dhar, N., Mancini, G. F., Sordet-Dessimoz, J., and McKinney, J. D. (2021). Rapid endotheliitis and vascular damage characterize SARS-CoV-2 infection in a human lung-on-chip model. EMBO Rep. 22 (6), e52744. doi:10.15252/embr.202152744
Tiwari, S. K., Wang, S., Smith, D., Carlin, A. F., and Rana, T. M. (2021). Revealing tissue-specific SARS-CoV-2 infection and host responses using human stem cell-derived lung and cerebral organoids. Stem Cell Rep. 16 (3), 437–445. doi:10.1016/j.stemcr.2021.02.005
Toor, S. M., Saleh, R., Sasidharan Nair, V., Taha, R. Z., and Elkord, E. (2021). T-cell responses and therapies against SARS-CoV-2 infection. Immunology 162 (1), 30–43. doi:10.1111/imm.13262
Tostanoski, L. H., Wegmann, F., Martinot, A. J., Loos, C., McMahan, K., Mercado, N. B., et al. (2020). Ad26 vaccine protects against SARS-CoV-2 severe clinical disease in hamsters. Nat. Med. 26 (11), 1694–1700. doi:10.1038/s41591-020-1070-6
Totura, A. L., Whitmore, A., Agnihothram, S., Schafer, A., Katze, M. G., Heise, M. T., et al. (2015). Toll-like receptor 3 signaling via TRIF contributes to a protective innate immune response to severe acute respiratory syndrome coronavirus infection. mBio 6 (3), e00638–e00615. doi:10.1128/mBio.00638-15
Trietsch, S. J., Israels, G. D., Joore, J., Hankemeier, T., and Vulto, P. (2013). Microfluidic titer plate for stratified 3D cell culture. Lab. Chip 13 (18), 3548–3554. doi:10.1039/c3lc50210d
van Duinen, V., van den Heuvel, A., Trietsch, S. J., Lanz, H. L., van Gils, J. M., van Zonneveld, A. J., et al. (2017). 96 perfusable blood vessels to study vascular permeability in vitro. Sci. Rep. 7 (1), 18071. doi:10.1038/s41598-017-14716-y
Vives, J., and Batlle-Morera, L. (2020). The challenge of developing human 3D organoids into medicines. Stem Cell Res. Ther. 11 (1), 72. doi:10.1186/s13287-020-1586-1
Wang, C., Horby, P. W., Hayden, F. G., and Gao, G. F. (2020a). A novel coronavirus outbreak of global health concern. Lancet 395 (10223), 470–473. doi:10.1016/s0140-6736(20)30185-9
Wang, D., Hu, B., Hu, C., Zhu, F., Liu, X., Zhang, J., et al. (2020b). Clinical characteristics of 138 hospitalized patients with 2019 novel coronavirus-infected pneumonia in wuhan, China. JAMA 323 (11), 1061–1069. doi:10.1001/jama.2020.1585
Wang, J., Li, X., Wang, A., Zhao, F., Wu, Q., Li, L., et al. (2020c). Organoid technology demonstrates effects of potential drugs for COVID-19 on the lung regeneration. Cell Prolif. 53 (12), e12928. doi:10.1111/cpr.12928
Wang, M., Cao, R., Zhang, L., Yang, X., Liu, J., Xu, M., et al. (2020d). Remdesivir and chloroquine effectively inhibit the recently emerged novel coronavirus (2019-nCoV) in vitro. Cell Res. 30 (3), 269–271. doi:10.1038/s41422-020-0282-0
Wang, S., Li, W., Hui, H., Tiwari, S. K., Zhang, Q., Croker, B. A., et al. (2020e). Cholesterol 25-Hydroxylase inhibits SARS-CoV-2 and other coronaviruses by depleting membrane cholesterol. EMBO J. 39 (21), e106057. doi:10.15252/embj.2020106057
Wang, T., Zhang, N., Fan, S., Zhao, L., Song, W., Gong, Y., et al. (2021). Establishment of human distal lung organoids for SARS-CoV-2 infection. Cell Discov. 7 (1), 108. doi:10.1038/s41421-021-00346-2
Wang, Y., Wang, P., and Qin, J. (2022). Human organoids and organs-on-chips for addressing COVID-19 challenges. Adv. Sci. 9 (10), e2105187. doi:10.1002/advs.202105187
Weibel, E. R., and Gomez, D. M. (1962). Architecture of the human lung. Use of quantitative methods establishes fundamental relations between size and number of lung structures. Science 137 (3530), 577–585. doi:10.1126/science.137.3530.577
Weibel, E. R. (2017). Lung morphometry: The link between structure and function. Cell Tissue Res. 367 (3), 413–426. doi:10.1007/s00441-016-2541-4
Whitcutt, M. J., Adler, K. B., and Wu, R. (1988). A biphasic chamber system for maintaining polarity of differentiation of cultured respiratory tract epithelial cells. Vitro Cell. Dev. Biol. 24 (5), 420–428. doi:10.1007/BF02628493
Wilk, A. J., Rustagi, A., Zhao, N. Q., Roque, J., Martinez-Colon, G. J., McKechnie, J. L., et al. (2020). A single-cell atlas of the peripheral immune response in patients with severe COVID-19. Nat. Med. 26 (7), 1070–1076. doi:10.1038/s41591-020-0944-y
Wu, F., Zhao, S., Yu, B., Chen, Y.-M., Wang, W., Song, Z.-G., et al. (2020). A new coronavirus associated with human respiratory disease in China. Nature 579 (7798), 265–269. doi:10.1038/s41586-020-2008-3
Xu, G., Li, Y., Zhang, S., Peng, H., Wang, Y., Li, D., et al. (2021). SARS-CoV-2 promotes RIPK1 activation to facilitate viral propagation. Cell Res. 31 (12), 1230–1243. doi:10.1038/s41422-021-00578-7
Yan, R., Zhang, Y., Li, Y., Xia, L., Guo, Y., and Zhou, Q. (2020). Structural basis for the recognition of SARS-CoV-2 by full-length human ACE2. Science 367 (6485), 1444–1448. doi:10.1126/science.abb2762
Zhang, J. J., Dong, X., Cao, Y. Y., Yuan, Y. D., Yang, Y. B., Yan, Y. Q., et al. (2020a). Clinical characteristics of 140 patients infected with SARS-CoV-2 in Wuhan, China. Allergy 75 (7), 1730–1741. doi:10.1111/all.14238
Zhang, M., Wang, P., Luo, R., Wang, Y., Li, Z., Guo, Y., et al. (2020b). Biomimetic human disease model of SARS-CoV-2 induced lung injury and immune responses on organ chip system. Adv. Sci. 8 (3), 2002928. doi:10.1002/advs.202002928
Zhang, Y. S., Yue, K., Aleman, J., Moghaddam, K. M., Bakht, S. M., Yang, J., et al. (2017). 3D bioprinting for tissue and organ fabrication. Ann. Biomed. Eng. 45 (1), 148–163. doi:10.1007/s10439-016-1612-8
Zhao, B., Ni, C., Gao, R., Wang, Y., Yang, L., Wei, J., et al. (2020a). Recapitulation of SARS-CoV-2 infection and cholangiocyte damage with human liver ductal organoids. Protein Cell 11 (10), 771–775. doi:10.1007/s13238-020-00718-6
Zhao, Z., Vizetto-Duarte, C., Moay, Z. K., Setyawati, M. I., Rakshit, M., Kathawala, M. H., et al. (2020b). Composite hydrogels in three-dimensional in vitro models. Front. Bioeng. Biotechnol. 8, 611. doi:10.3389/fbioe.2020.00611
Zhou, J., Li, C., Sachs, N., Chiu, M. C., Wong, B. H., Chu, H., et al. (2018). Differentiated human airway organoids to assess infectivity of emerging influenza virus. Proc. Natl. Acad. Sci. U. S. A. 115 (26), 6822–6827. doi:10.1073/pnas.1806308115
Zhou, P., Yang, X.-L., Wang, X.-G., Hu, B., Zhang, L., Zhang, W., et al. (2020a). A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature 579 (7798), 270–273. doi:10.1038/s41586-020-2012-7
Zhou, R., To, K. K., Wong, Y. C., Liu, L., Zhou, B., Li, X., et al. (2020b). Acute SARS-CoV-2 infection impairs dendritic cell and T cell responses. Immunity 53 (4), 864–877. doi:10.1016/j.immuni.2020.07.026
Zhou, Z., Zhang, X., Lei, X., Xiao, X., Jiao, T., Ma, R., et al. (2021). Sensing of cytoplasmic chromatin by cGAS activates innate immune response in SARS-CoV-2 infection. Signal Transduct. Target. Ther. 6 (1), 382. doi:10.1038/s41392-021-00800-3
Glossary
2D Two-dimensional
3D Three-dimensional
ACE2 Angiotensin-converting enzyme 2
AFE Anterior foregut endoderm
ALI Air liquid interface
APCs Antigen presenting cells
ARDS Acute respiratory distress syndrome
ASMCs Airway smooth muscle cells
AT2 Alveolar epithelial type 2
CAD Computer-aided design
CFE Colony-forming efficiency
COVID-19 Coronavirus disease 2019
DE Definitive endoderm
ECM Extracellular matrix
ESCs Embryonic stem cells
FDA Food and Drug Administration
FHV-1 Feline herpesvirus-1
GelMA Gelatin methacryloyl
IL Interleukin
iPSCs Induced pluripotent stem cells
LPs Lung progenitors
MDA5 Melanoma differentiation-associated gene 5
MBCS Multibasic cleavage site
MPA Mycophenolic acid
NHPs Non-human primates
NRP1 Neuropilin-1
ORFs Open reading frames
PC Pyruvate carboxylase
PD Peptidase domain
PM2.5 Fine particulate matter
QNHC Quinacrine dihydrochloride
RBD Receptor-binding domain
SARS-CoV-2 Severe acute respiratory syndrome coronavirus-2
TLR3 Toll-like receptor 3
TLR7 Toll-like receptor 7
TMPRSS2 Transmembrane protease serine 2
TNF-a Tumor necrosis factor alpha
TSPAN8 Tetraspanin 8
Keywords: COVID-19, lung organoid, lung-on-a-chip, 3D bioprinting, drug discovery
Citation: Ni K, Che B, Yang C, Qin Y, Gu R, Wang C, Luo M and Deng L (2022) Emerging toolset of three-dimensional pulmonary cell culture models for simulating lung pathophysiology towards mechanistic elucidation and therapeutic treatment of SARS-COV-2 infection. Front. Pharmacol. 13:1033043. doi: 10.3389/fphar.2022.1033043
Received: 31 August 2022; Accepted: 30 November 2022;
Published: 12 December 2022.
Edited by:
Rebecca L. Heise, Virginia Commonwealth University, United StatesReviewed by:
Manash K. Paul, California State University, Los Angeles, United StatesRahul K. Nelli, Iowa State University, United States
Copyright © 2022 Ni, Che, Yang, Qin, Gu, Wang, Luo and Deng. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Mingzhi Luo, bHVvbWluZ3poaUBjY3p1LmVkdS5jbg==; Linhong Deng, ZGxoQGNjenUuZWR1LmNu
†These authors have contributed equally to this work
 Kai Ni
Kai Ni Bo Che†
Bo Che† Mingzhi Luo
Mingzhi Luo