
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Pharmacol. , 07 October 2022
Sec. Inflammation Pharmacology
Volume 13 - 2022 | https://doi.org/10.3389/fphar.2022.1023878
This article is part of the Research Topic Natural Compounds Regulating Epigenetics for Treating Chronic Inflammatory Diseases View all 8 articles
Background: Astragalus mongholicus polysaccharides (APS) have anti-inflammatory, antioxidant and immunomodulatory effects. Recent studies have demonstrated the epigenetic regulation of N6-methyladenosine (m6A) in the development of inflammation. However, the effect of APS on m6A modification is unclear. Here, for the first time, we investigate the mechanism of m6A modification in APS regulation of THP-1 macrophage inflammation.
Methods: We treated LPS-induced THP-1 macrophages with APS at different concentrations and times, and detected IL-6 mRNA and protein levels by quantitative real-time PCR (qRT-PCR) and western blot, respectively. The m6A modification level was detected by m6A quantification kit. The proteins that regulate m6A modification were screened by western blot. Wilms’ tumor 1-associating protein (WTAP) was overexpressed in APS-treated THP-1 macrophages and the m6A modification level and IL-6 expressions were detected.
Results: These findings confirmed that APS significantly abolished LPS-induced IL-6 levels in THP-1 macrophages. Meanwhile, APS reduced m6A modification levels and WTAP gene expression in THP-1 macrophages. Further overexpression of WTAP can significantly reverse APS-induced m6A modification level and IL-6 expression. Mechanistically, APS regulates IL-6 expression through WTAP-mediated p65 nuclear translocation.
Conclusion: Overall, our study suggested that WTAP mediates the anti-inflammatory effect of APS by regulating m6A modification levels in THP-1 macrophages. This study reveals a new dimension of APS regulation of inflammation at the epigenetic level.
N6-Methyladenine (m6A) modification is an epigenetic modification and is the most common and abundant modification of RNA molecules in eukaryotes (Marnef and Legube, 2020). In mammalian cells, m6A methylation was shown to be enriched in mRNAs containing 3–5 m6 A modifications per mRNA (Wang et al., 2014). RNA m6A modification has been shown to play important roles in regulating RNA splicing, translation, stability, translocation and high-level structure (Jiang et al., 2021). Studies have shown that m6A methylation of RNA is mediated by a methyltransferase complex consisting of methyltransferase 3 and 14 (METTL3, METTL14) and wilms’ tumor 1-associating protein (WTAP). M6A methylation is reversed by two demethylases: fat mass and obesity-associated (FTO) and alkylation repair homolog 5 (ALKBH5) (Xu et al., 2020). Recently, research has shown that m6A modifications occur in most species and play important roles in the development of many diseases, including tumors (Liu et al., 2020), neurological (Li et al., 2019) and cardiovascular diseases (Paramasivam et al., 2020), etc.
Monocytes and macrophages are major players in many chronic inflammatory diseases (Aguzzi et al., 2020). Macrophages initiate and maintain inflammation, produce inflammatory cytokines (TNF-α, IL-1β and IL-6) (Tabas and Bornfeldt, 2016). At present, multiple studies have reported that defects in m6A modification affect the development of inflammation in mouse and human macrophages (Li et al., 2022). However, no targeted therapy has been shown to ameliorate macrophage inflammatory responses through m6A modifications.
Astragalus mongholicus polysaccharide (APS) is a water-soluble heteropolysaccharide obtained by extracting, concentrating and purifying the dried roots of leguminous plants Astragalus mongolica or Astragalus membranaceus. The molecular formula of APS is C10H7CIN2O2S. APS is one of the most well-known herbal medicines that is widely used in China (Yuan et al., 2009). In addition to its antioxidant (Jia et al., 2019), antitumor (Liao et al., 2020) and immunomodulatory (Shao et al., 2006) effects, several studies have shown that APS can be used to treat chronic inflammatory diseases, including atherosclerosis (Chen et al., 2015). However, whether APS can exert anti-inflammatory effects by regulating the m6A modification remains unclear.
In this study, we investigated the effect of APS on RNA m6A modification during LPS-induced inflammation in THP-1 macrophages. It was further shown that APS affected the m6A modification level through the m6A “writer” protein WTAP. Overall, our data suggest that WTAP mediates the anti-inflammatory effects of APS by regulating m6A modification levels in THP-1 macrophages. This study reveals a new mechanism by which APS regulates inflammation at the epigenetic level.
THP-1 was purchased from ATCC (TIB-202) and maintained in RPMI 1640 medium in 10% FBS with penicillin and streptomycin at 37 °C in humidified 5% CO2. The macrophage-like state was obtained by treating the THP-1 monocytes for 48 h with 100 nM PMA (MCE, United States). The culture medium was removed, and then the cells were exposed to various concentrations of APS (50–500 μg/ml, Solarbio, CHN) diluted in culture medium for different time intervals at 37°C. Subsequently, the cells were stimulated with 10 μg/ml lipopolysaccharide (LPS, Sigma, United States) for 4 h until further analysis.
Cell toxicity indicated as the percentage of cell viability was determined using the standard MTT assay (Sigma-Aldrich, Germany). THP-1 macrophages were seeded in 96-well (1 × 105 cells/well) plates for 48 h and then treated with APS (0–500 μg/ml) for 24 h. After treatment, 20 μL MTT solution was added to each well and incubated for 4 h. Next, 150 μL of dimethyl sulfoxide was added to each well, and then the absorbance of each well was measured at 490 nm using a microplate reader. Cell viability was normalized to the control group.
Total RNA was extracted from cultured cells with TRizol reagent (TAKARA BIO INC., Osaka, Japan), followed by reverse transcription into cDNA with the PrimeScript RT reagent Kit with gDNA Eraser (TAKARA BIO INC., Osaka, Japan). qRT-PCR was performed with a SYBR Green Premix Pro Taq HS qPCR Kit (ACCURATE BIOLOGY, Hunan, China). Relative changes in mRNA levels were determined by the ΔΔCT method, using GAPDH levels as controls. Primer pair sequences are shown in Supplementary Table S1.
Cells were washed in cold PBS and harvested as following. Whole cell lysates were prepared by lysing the cells for 30 min on ice with a modified RIPA lysis buffer (Beyotime, Shanghai, China) containing 1:100 phenylmethanesulfonyl fluoride (PMSF, Beyotime, Shanghai, China). Cytosolic and nuclear fractions were obtained using the Nuclear/Cytosol Fractionation Kit (Biovision, United States), according to the manufacturer’s instructions. Protein amounts were measured by a protein assay kit (BCA; Beyotime, Shanghai, China).
Cellular proteins were resolved by SDS-PAGE and transferred onto nitrocellulose membranes (Merck Millipore Ltd., Melbourne, Australia). After being blocked by 5% non-fat milk in TBST for 1 h, membranes were incubated with corresponding primary antibodies at 4°C overnight. Washed by TBST for three times, they would be incubated with HRP-conjugated secondary antibodies for 1 h at room temperature. The immunoblots were detected with an imaging system (Bio-Rad, United States) using enhanced Immobilon Wrstern Chemiluminescent HRP Substran (Millipore, Boston, United States). GAPDH were selected to be the loading controls. Primary antibodies are shown in Supplementary Table S2.
Briefly, THP‐1 cells were seeded in 6-well plates, and 48 h later, WTAP siRNA (150 pM) was introduced to the cells using GP-transfect-Mate (Genepharma, Shanghai, China) according to the manufacturer’s instructions. Non‐silencing siRNA and siRNAs specific for WTAP were transfected. Cells were harvested after 24 h for qRT-PCR examination or after 48 h for western blotting. The selected sequences for knockdown as follow: si-WTAP sense were 5′-GCUUGGACUCAAGUGUAAATT-3′, si-WTAP antisense were5′-UUUACACUUGAGUCCAAGCTT-3′.
All new plasmids were generated using standard molecular biology methods and were verified by sequencing. Overexpressing plasmid (1.2 μg) (WeiZhen Biosciences, Shangdong, China) of indicated genes were transfected into THP-1 macrophages using Attractene Transfection Reagent (Qiagen, Germany) according to the manufacturer’s instructions. For each well (6-well plates), plasmid DNA was mixed with Attractene Transfection Reagent (with 1.2 μg–4.5 μL ratio) in 100 μL serum-free medium. After incubating the mixture at room temperature for 15 min, 1.9 ml per well of fully supplemented RPMI 1640 medium was added. 100 μL of the resulting mixture was added to the wells that had 1.9 ml of cell medium. The plate was gently shaken to ensure that the reagents were well mixed. The efficiency of WTAP plasmid transfection using Attractene Transfection Reagent was as high as about 80%.
Total RNA was isolated using TRIzol reagent (TAKARA BIO INC., Osaka, Japan) according to the manufacturer’s instructions. The RNA quality was analyzed using the NanoDrop2000 system. The change in global m6A levels in the mRNA was measured using the EpiQuik™ m6A RNA Methylation Quantification Kit (Colorimetric; Epigentek, P-9005–48) following the manufacturer’s protocol. For total RNA, 200 ng of RNA per sample was transferred to corresponding wells in a 96-well plate. All samples were performed in triplicate. Both negative and positive controls as well as a standard curve in the range of 0.02 ng–1 ng of m6A were included as recommended by the manufacturer. The capture antibody solution and detection antibody solution were then added to the assay wells separately using a suitable diluted concentration. The m6A levels were colorimetrically quantified by reading the absorbance at a wavelength of 450 nm, and then calculations were performed based on the standard curve.
To evaluate mRNA stability, THP-1 macrophages were treated with 4 μM Actinomycin D (Act D, MedChemExpress, United States) during indicated times and harvested. Then mRNA expression of IL-6 was determined by qRT-RCR analysis.
To evaluate protein stability, THP-1 macrophages were treated with 10 μM cycloheximide (CHX, MedChemExpress, United States) during indicated times and harvested. Then protein expression of IL-6 was determined by western blot analysis.
THP-1 macrophages were cultured at 37°C, and the cells were washed with PBS and fixed in 500 μL 4% paraformaldehyde for 20 min at room temperature (RT). The cells were washed three times with PBS for 5 min each, permeabilized in 0.1% Triton X-100 in PBS for 30 min at RT. The cells were washed three times with PBS for 5 min each, and blocked with diluted goat serum (1:10) for 1 h with gentle shaking at RT. The cells were then incubated in blocking buffer containing diluted primary antibodies (anti - P65) overnight at 4°C and washed three times with PBS for 5 min each time. The cells were incubated with secondary antibodies in blocking buffer for 1 h at 37°C in the dark, and then washed three times with PBS for 5 min each time. The cells were incubated with DAPI (0.1 μg/ml) for 10 min, washed three times with PBS for 5 min, and fixed. Images were acquired using a confocal microscope (Zeiss, German).
The results are presented as means ± SEM. Statistical analyses were performed using the GraphPad Prism version 8.0.1. Two-group comparisons were performed by unpaired two-tailed Student’s t test. p-values <0.05 were considered statistically significant. All experiments were performed at least three times.
It has since been clarified that IL-6 is a critical factor in inflammation. To assess the effect of APS on IL-6 gene expression in THP-1 macrophages, we pretreated THP-1 macrophages with different concentrations (50, 100, 200 and 500 μg/ml) of APS for 24 h and stimulated them with LPS (10 μg/ml) for 4 h. First, we performed the MTT assay and found that 50–500 μg/ml APS did not show any cytotoxicity to THP-1 cells over 24 h (Supplementary Figure S1), which allowed us to rule out nonspecific cytotoxicity as a possible explanation for the decrease in cytokine output. Second, relevant inflammatory factor expression was measured by qRT-PCR and Western blotting. The results showed that 500 μg/ml APS significantly inhibited the LPS-induced increase of IL-6 mRNA and protein in THP-1 macrophages (Figures 1A–C and Supplementary Figure S2). We further examined the effects of APS treatment time (12, 24 and 48 h) on LPS-induced IL-6 expression in THP-1 macrophages. The results showed that APS treatment for 24 h had the most obvious inhibitory effect on IL-6 (Figures 1D–F). These results indicated that the inhibitory effect of APS on IL-6 expression in THP-1 macrophages was concentration- and time-dependent.
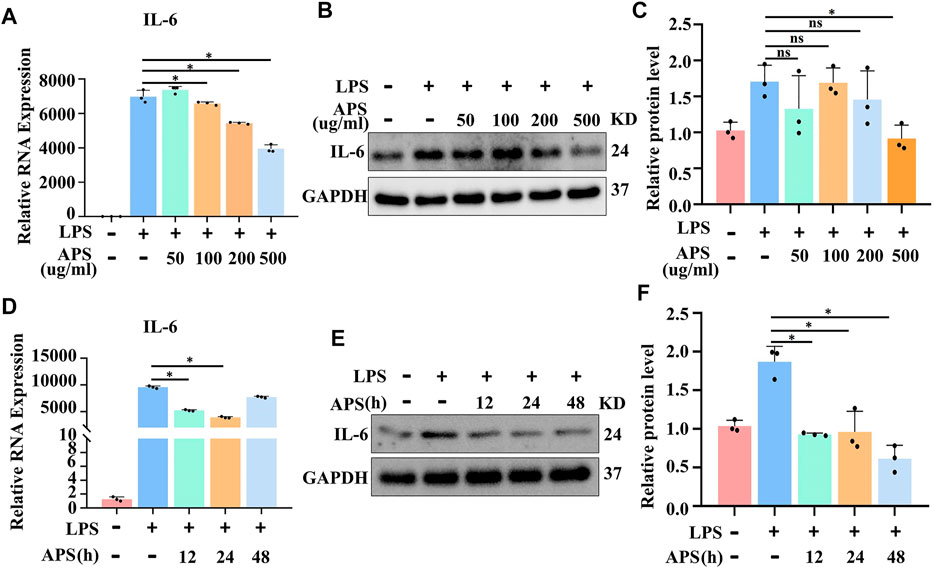
FIGURE 1. APS abrogated LPS-induced IL-6 gene expression in THP-1 macrophages. THP-1 macrophages were treated with APS (50, 100, 200, 500 μg/ml) for 24 h and then induced with LPS (10μg/ml) for 4 h (A) IL-6 mRNA was detected by qRT-PCR. (B) IL-6 protein was detected by western blot. (C) Densitometry quantification of protein. THP-1 macrophages were treated with APS (500 μg/ml) for different time (0, 12, 24, 48 h) and then induced with LPS (10 μg/ml) for 4 h (D) IL-6 mRNA was detected by qRT-PCR. (E) IL-6 protein was detected by western blot. (F) Densitometry quantification of protein. *p < 0.05 compared with LPS group. Data represent the mean ± SEM. n = 3 per group.
Recent evidence suggests that m6A methylation plays a crucial role in regulating THP-1 macrophage inflammation (Wang et al., 2019), but whether APS plays a role in the m6A modification is unclear. To investigate the effect of APS on m6A methylation, m6A levels in APS-treated THP-1 macrophages were measured by an m6A quantification kit. The results showed that LPS-induced m6A methylation was decreased with increasing APS concentrations (Figure 2A). Furthermore, we examined the m6A modification level after stimulation with APS for 12, 24 and 48 h, and the results indicated that the level of m6A was significantly lower than that in the LPS-treated group (Figure 2B). These results suggest that APS regulates m6A modification levels in THP-1 macrophages. Then, we treated LPS-induced THP-1 macrophages with 500 μg/ml APS for 24 h for the following mechanistic studies.
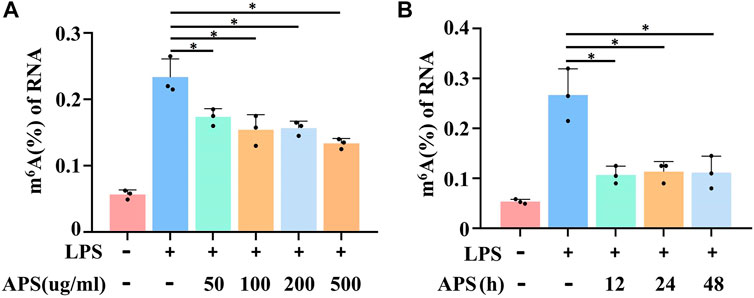
FIGURE 2. APS abrogated LPS-induced m6A level in THP-1 macrophages. (A) THP-1 macrophages were treated with APS (50, 100, 200, 500 μg/ml) for 24 h and then induced with LPS (10 μg/ml) for 4 h m6A level was detected in the cells by m6A RNA methylation quantification. (B) THP-1 macrophages were treated with APS (500 μg/ml) for different time (0, 12, 24, 48 h) and then induced with LPS (10 μg/ml) for 4 h. m6A level was detected in the cells by m6A RNA methylation quantification. *p < 0.05 compared with LPS group. Data represent the mean ± SEM. n = 3 per group.
To examine the mechanism by which APS regulates m6A modification levels, we examined whether APS treatment altered m6A modification system-related protein expression in LPS-induced THP-1 macrophages. As shown in Figure 3A, WTAP mRNA expression was markedly decreased after treatment with APS for 24 h. The protein level of WTAP was also significantly decreased compared with that of the other related proteins (Figures 3B,C). WTAP is one of the essential proteins in the m6A methyltransferase complex and plays an important role in m6A writing. To verified the relationship between WTAP and m6A levels. We knocked down WTAP in LPS-treated THP-1 macrophages with siRNA. The results showed that WTAP knockdown suppressed the LPS-induced increase in total m6A levels in THP-1 macrophages. Taken together, these data suggest that APS can affect m6A modification levels by regulating WTAP expression (Figure 3D).
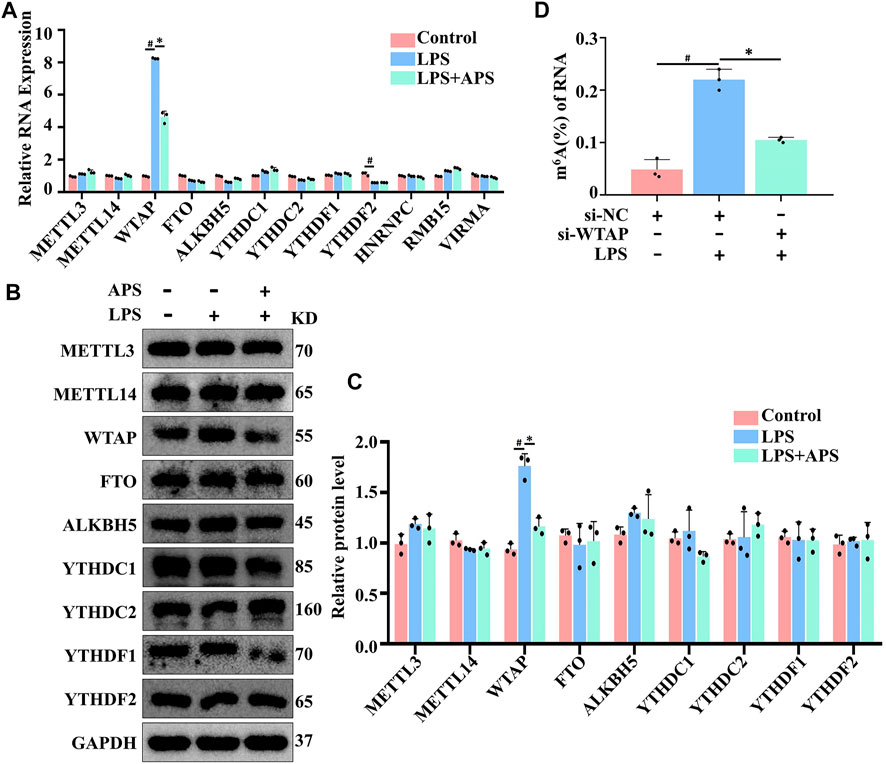
FIGURE 3. APS decreased LPS-induced WTAP gene in THP-1 macrophages. THP-1 macrophages were treated with APS (500 μg/ml) for 24 h and then induced with LPS (10 μg/ml) for 4 h. m6A modification system-related proteins were detected by (A) qRT-PCR and (B) western blot. (C) Densitometry quantification of protein. (D) THP-1 macrophages were transfected with si-WTAP and then induced with LPS (10 μg/ml) for 4 h. Overall m6A level in RNA is detected by m6A RNA methylation quantification. #p < 0.05 compared with Control group. *p < 0.05 compared with LPS group. Data represent the mean ± SEM. n = 3 per group.
To verify that APS affects the expression of IL-6 through WTAP, we examined the mRNA and protein expression of IL-6 by qRT‒PCR and western blotting, respectively, after WTAP overexpression. WTAP overexpression ameliorated the low expression of IL-6 in APS-treated THP-1 macrophages (Figures 4A–C). Next, we identified the mechanism by which WTAP regulates IL-6. Because WTAP is one of the essential proteins in the m6A writer, we hypothesized that WTAP-mediated regulation of IL-6 expression may be dependent on the m6A modification. Interestingly, we predicted the m6A modification sites of IL-6 with SRAMP website (http://www.cuilab.cn/sramp) and found that there were multiple m6A modification sites (GGAC) in the IL-6 mRNA sequence (Supplementary Table S3), suggesting that IL-6 mRNA can undergo m6A modification. The main mechanism by which m6A modification regulates protein expression is by regulating mRNA stability, translation shearing and nuclear transport. Among these, m6A modification most commonly regulates the stability of mRNA and protein. Therefore, we first examined the stability of IL-6 mRNA. As shown in Figure 4D, LPS stimulation and WTAP knockdown had no effect on the stability of IL-6 mRNA. We further examined protein levels after treatment with CHX to stop translation. The results revealed that knockdown of WTAP had no effect on the stability of IL-6 protein in THP-1 macrophages (Figure 4E). Therefore, these results indicated that WTAP does not regulate IL-6 through m6A modifications.
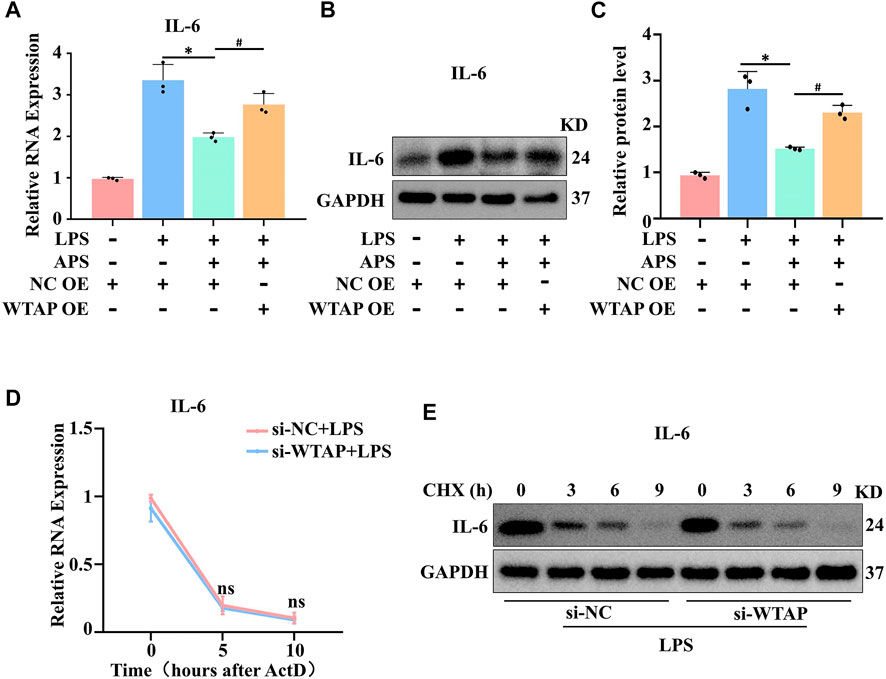
FIGURE 4. APS exerted an anti-inflammatory effect by regulating WTAP expression. IL-6 expression in APS-treated THP-1 macrophages after WTAP overexpression was detected by (A) qRT-PCR and (B) western blot. (C) Densitometry quantification of protein. (D) IL-6 mRNA stability following treatment with actinomycin D (ActD, 2 μM) was detected by qRT-PCR. (E) THP-1 macrophages were treated with cycloheximide (CHX, 10 μM) after si-WTAP transfection. IL-6 protein was detected by western blot. *p < 0.05 compared with NC OE + LPS group. #p < 0.05 compared with NC OE + LPS + APS group. Data represent the mean ± SEM. n = 3 per group.
Therefore, how does WTAP regulate the expression of IL-6? Multiple studies have shown that NF-κB p65 can regulate transcription factors associated with IL-6 expression (Wang et al., 2019; Jiang et al., 2022). We hypothesized that WTAP regulates IL-6 through the NF-κB pathway and examined the expression level of p65 in APS-treated THP-1 macrophages by western blotting. The results showed that APS did not affect the expression of p65 (Figures 5A,B). WTAP overexpression did not affect the overall expression level of p65 in THP-1 macrophages (Figures 5A,B, Supplementary Figure S3). We further examined the nuclear transfer of p65. Immunofluorescence staining showed that APS prevented LPS-induced translocation of p65 from the cytoplasm to the nucleus in THP-1 macrophages (Figure 5C). However, WTAP overexpression promoted the translocation of p65 from the cytoplasm to the nucleus in APS-treated THP-1 macrophages (Figure 5C). To better examine the nuclear and cytoplasmic distribution of p65, we extracted nuclear and cytoplasmic proteins from THP-1 macrophages. The results showed that APS-mediated inhibition of p65 translocation to the nucleus was reversed in THP-1 macrophages that were transfected with the WTAP overexpression plasmid (Figures 5D,E). These results suggest that APS regulates IL-6 expression through WTAP-mediated p65 nuclear translocation.
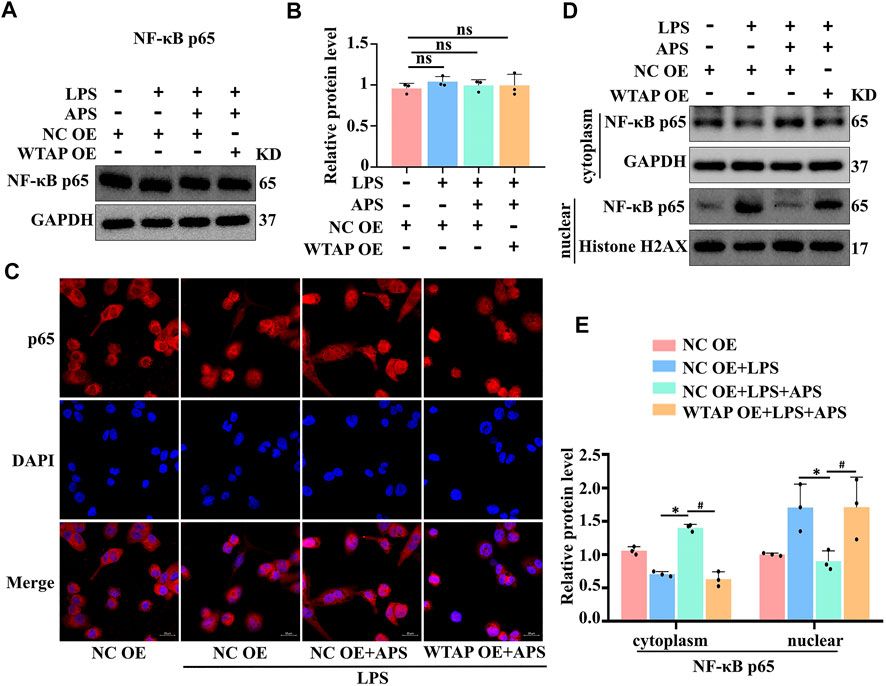
FIGURE 5. APS regulates IL-6 expression through WTAP-mediated p65 nuclear translocation. THP-1 macrophages were transfected with WTAP overexpression plasmid prior to treatment with APS. (A) NF-κb p65 protein expression was detected by western blot. (B) Densitometry quantification of protein. (C) The localization of NF-κb p65 (red) was observed with immunofuorescence, and the nuclei (blue) were stained with DAPI, scale bar, 20 μm. (D) The nuclear translocation of p65 was analyzed, and its localization in the cytosol and nucleus was separately quantified by western blot. (E) Densitometry quantification of protein. *p < 0.05 compared with NC OE + LPS group. #p < 0.05 compared with NC OE + LPS + APS group. Data represent the mean ± SEM. n = 3 per group.
APS has attracted the attention of researchers because of its anti-inflammatory, antioxidant and immunomodulatory effects. Studies have shown that APS exerts its anti-inflammatory effect by inhibiting LPS-induced production of TNF-α and IL-8 in intestinal epithelial cells by blocking mitogen-activated protein kinase (MAPK) transcriptional activity (Yuan et al., 2009). In addition to identifying the role of APS through its anti-inflammatory and antioxidant properties, recent studies have demonstrated a link between APS and the DNA methylome (Liu et al., 2021). In this study, we verified that APS exerts its anti-inflammatory effect on THP-1 macrophages through m6A modifications.
The m6A is the most widely studied RNA modification and have been recognized as a new layer of epigenetic regulation, which is closely related to the occurrence and development of macrophage inflammation. Recent studies have shown that the level of the m6A modification is significantly increased in LPS-stimulated THP-1 macrophages, and knocking down the key m6A modification enzyme METTL14 inhibits the occurrence of macrophage inflammation (Zheng et al., 2022). In our study, we also found that the m6A modification level was significantly increased in LPS-stimulated THP-1 macrophages, and the m6A modification level decreased with the concentration increasing of APS. These results suggest that APS can regulate the m6A modification.
In recent clinical trials, ziltivekimab, a novel IL-6 ligand inhibitor, was highly effective in reducing inflammatory responses and biomarkers of atherosclerosis, suggesting that IL-6 is an effective therapeutic target for atherosclerosis (Ridker and Rane, 2021). Our findings suggested that IL-6 is a target of WTAP in APS-treated THP-1 macrophages. Since WTAP is one of the essential proteins for m6A writing, we hypothesize that WTAP-mediated regulation of IL-6 expression may depend on m6A modifications. However, our results demonstrated that the stability of IL-6 mRNA and protein were not significantly changed in WTAP knockdown cells. The m6A modifications on mRNA are mainly written by the METTL3, METTL14 and WTAP complexes (Xu et al., 2022), and the evidence shown that METTL14 does not regulate IL-6 through m6A modification (Zheng et al., 2022). This evidence shows that WTAP does not directly regulate IL-6 through m6A modification.
Multiple studies have shown that NF-κB p65 can regulate the transcription of IL-6. As expected, WTAP overexpression reversed the distribution of p65 in the cytoplasm in APS-treated THP-1 macrophages, which regulated the transcription of IL-6. Moreover, we examined the expression of other m6A enzymes upon overexpression of WTAP, and the results suggested that WTAP could not induce complementary effects of other m6A writers (Supplementary Figure S4).In conclusion, the therapeutic mechanism of APS involves regulating IL-6 expression through WTAP-mediated p65 nuclear translocation.
This study has certain limitations. First, the underlying mechanism by which WTAP regulates the nucleocytoplasmic distribution of p65 needs to be further examined. Second, our entire study was performed at the cellular level, and further validation of the results in mice is needed.
In conclusion, our findings confirmed that APS could exert an anti-inflammatory effect in THP-1 macrophages. We demonstrated that APS could reduce the m6A modification level. To the best of our knowledge, this is the first study on the m6A modification in the anti-inflammatory mechanism of APS. Mechanistically, we showed that WTAP regulated the expression of IL-6 by regulating p65 nuclear translocation. Our findings reveal a novel role for APS in the regulation of the m6A modification during macrophage inflammation, and this has great potential for macrophage inflammation therapy.
The datasets presented in this article are not readily available because N.A. Requests to access the datasets should be directed to emhhb2d1b2p1bkBnemhtdS5lZHUuY24=.
Study design: GZ and HjL; Experiments: PZ, HyL, SL, LH and MZ; Data analysis: GZ, KY, and HjL; Writing and Revising: GZ, KY, HjL, and PZ; All authors read and approved the final version of the manuscript.
This work was supported by grants from National Natural Science Foundation of China (81870337) and Natural Science Foundation of Guangdong Province (2021A1515010717).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fphar.2022.1023878/full#supplementary-material
Aguzzi, C., Marinelli, O., Zeppa, L., Santoni, G., Maggi, F., and Nabissi, M. (2020). Evaluation of anti-inflammatory and immunoregulatory activities of Stimunex® and Stimunex D3® in human monocytes/macrophages stimulated with LPS or IL-4/IL-13. Biomed. Pharmacother. 132, 110845. doi:10.1016/j.biopha.2020.110845
Chen, C., Fu, Q., Li, Y., and Li, Z. (2015). Effect of Astragalus mongholicus polysaccharides on gene expression profiles of dendritic cells isolated from healthy donors. J. South. Med. Univ. 35 (12), 1802–1805.
Jia, N., Qiao, H., Zhu, W., Zhu, M., Meng, Q., Lu, Q., et al. (2019). Antioxidant, immunomodulatory, oxidative stress inhibitory and iron supplementation effect of Astragalus membranaceus polysaccharide-iron (III) complex on iron-deficiency anemia mouse model. Int. J. Biol. Macromol. 132, 213–221. doi:10.1016/j.ijbiomac.2019.03.196
Jiang, R., Tang, J., Zhang, X., He, Y., Yu, Z., Chen, S., et al. (2022). CCN1 promotes inflammation by inducing IL-6 production via α6β1/PI3K/akt/NF-κB pathway in autoimmune hepatitis. Front. Immunol. 13, 810671. doi:10.3389/fimmu.2022.810671
Jiang, X., Liu, B., Nie, Z., Duan, L., Xiong, Q., Jin, Z., et al. (2021). The role of m6A modification in the biological functions and diseases. Signal Transduct. Target. Ther. 6 (1), 74. doi:10.1038/s41392-020-00450-x
Li, J., Yang, X., Qi, Z., Sang, Y., Liu, Y., Xu, B., et al. (2019). The role of mRNA m6A methylation in the nervous system. Cell Biosci. 9, 66. doi:10.1186/s13578-019-0330-y
Li, Z., Teng, M., Jiang, Y., Zhang, L., Luo, X., Liao, Y., et al. (2022). YTHDF1 negatively regulates Treponema pallidum-induced inflammation in THP-1 macrophages by promoting SOCS3 translation in an m6A-dependent manner. Front. Immunol. 13, 857727. doi:10.3389/fimmu.2022.857727
Liao, C. H., Yong, C. Y., Lai, G. M., Chow, J. M., Cheng, C. F., Fang, C. L., et al. (2020). Astragalus polysaccharide (PG2) suppresses macrophage migration inhibitory factor and aggressiveness of lung adenocarcinoma cells. Am. J. Chin. Med. 48 (6), 1491–1509. doi:10.1142/S0192415X20500731
Liu, J., Liu, J., Liu, L., Zhang, G., and Peng, X. (2021). Reprogrammed intestinal functions in Astragalus polysaccharide-alleviated osteoporosis: Combined analysis of transcriptomics and DNA methylomics demonstrates the significance of the gut-bone axis in treating osteoporosis. Food Funct. 12 (10), 4458–4470. doi:10.1039/d1fo00113b
Liu, T., Wei, Q., Jin, J., Luo, Q., Liu, Y., Yang, Y., et al. (2020). The m6A reader YTHDF1 promotes ovarian cancer progression via augmenting EIF3C translation. Nucleic Acids Res. 48 (7), 3816–3831. doi:10.1093/nar/gkaa048
Marnef, A., and Legube, G. (2020). m6A RNA modification as a new player in R-loop regulation. Nat. Genet. 52 (1), 27–28. doi:10.1038/s41588-019-0563-z
Paramasivam, A., Vijayashree Priyadharsini, J., and Raghunandhakumar, S. (2020). N6-adenosine methylation (m6A): A promising new molecular target in hypertension and cardiovascular diseases. Hypertens. Res. 43 (2), 153–154. doi:10.1038/s41440-019-0338-z
Ridker, P. M., and Rane, M. (2021). Interleukin-6 signaling and anti-interleukin-6 therapeutics in cardiovascular disease. Circ. Res. 128 (11), 1728–1746. doi:10.1161/CIRCRESAHA.121.319077
Shao, P., Zhao, L. H., and Zhi-Chen Pan, J. P. (2006). Regulation on maturation and function of dendritic cells by Astragalus mongholicus polysaccharides. Int. Immunopharmacol. 6 (7), 1161–1166. doi:10.1016/j.intimp.2006.02.009
Tabas, I., and Bornfeldt, K. E. (2016). Macrophage phenotype and function in different stages of atherosclerosis. Circ. Res. 118 (4), 653–667. doi:10.1161/CIRCRESAHA.115.306256
Wang, J., Yan, S., Lu, H., Wang, S., and Xu, D. (2019). METTL3 attenuates LPS-induced inflammatory response in macrophages via NF-κB signaling pathway. Mediat. Inflamm. 2019, 3120391. doi:10.1155/2019/3120391
Wang, X., Lu, Z., Gomez, A., Hon, G. C., Yue, Y., Han, D., et al. (2014). N6-methyladenosine-dependent regulation of messenger RNA stability. Nature 505 (7481), 117–120. doi:10.1038/nature12730
Xu, K., Mo, Y., Li, D., Yu, Q., Wang, L., Lin, F., et al. (2020). N6-methyladenosine demethylases Alkbh5/Fto regulate cerebral ischemia-reperfusion injury. Ther. Adv. Chronic Dis. 11, 2040622320916024. doi:10.1177/2040622320916024
Xu, W., He, C., Kaye, E. G., Li, J., Mu, M., Nelson, G. M., et al. (2022). Dynamic control of chromatin-associated m6A methylation regulates nascent RNA synthesis. Mol. Cell 82 (6), 1156–1168.e7. e7. doi:10.1016/j.molcel.2022.02.006
Yuan, Y., Sun, M., and Li, K. S. (2009). Astragalus mongholicus polysaccharide inhibits lipopolysaccharide-induced production of TNF-alpha and interleukin-8. World J. Gastroenterol. 15 (29), 3676–3680. doi:10.3748/wjg.15.3676
Keywords: APS, M6A, WTAP, macrophage inflammation, IL-6
Citation: Long H, Lin H, Zheng P, Hou L, Zhang M, Lin S, Yin K and Zhao G (2022) WTAP mediates the anti-inflammatory effect of Astragalus mongholicus polysaccharide on THP-1 macrophages. Front. Pharmacol. 13:1023878. doi: 10.3389/fphar.2022.1023878
Received: 20 August 2022; Accepted: 23 September 2022;
Published: 07 October 2022.
Edited by:
Qianqian Wang, Dalian University, ChinaReviewed by:
Shoudong Guo, Weifang Medical University, ChinaCopyright © 2022 Long, Lin, Zheng, Hou, Zhang, Lin, Yin and Zhao. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Kai Yin, S2FpeWluYnlAcXEuY29t; Guojun Zhao, emhhb2d1b2p1bkBnemhtdS5lZHUuY24=
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.