
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Pharmacol., 23 September 2022
Sec. Experimental Pharmacology and Drug Discovery
Volume 13 - 2022 | https://doi.org/10.3389/fphar.2022.1011561
This article is part of the Research TopicPharmacological Mechanisms of Drugs Affecting Bone Formation and Bone ResorptionView all 16 articles
Osteoporosis is the most common metabolic disease of skeleton with reduced bone density and weaker bone. Qianggu Capsule as a traditional chinese medicine has been widely used to treat osteoporosis. The potential pharmacological mechanism of its active ingredient Gusuibu is not well understood. The purpose of this work is to analyze the anti-osteoporosis function of Gusuibu based on network pharmacology, and further explore the potential mechanism of Qianggu Capsule. The active compounds and their corresponding targets of Gusuibu were obtained from TCMSP, TCMID, and BATMAN-TCM databases. Potential therapeutic targets for osteoporosis were obtained through DisGeNET, TTD, GeneCards, MalaCards, CTD, and OMIM databases. The overlapping targets of Gusuibu and osteoporosis were obtained. GO and KEGG pathway enrichment analysis were performed. The “Gusuibu-active compounds-target genes-osteoporosis” network and protein-protein interaction (PPI) network were constructed, and the top hub genes were screened by using the plug-in CytoHubba. Molecular docking was used to verify the binding activity of hub genes and key compounds. We identified 21 active compounds and 140 potential therapeutic targets that may be related to Gusuibu and 10 hub genes (AKT1, IL6, JUN, TNF, MAPK3, VEGFA, EGFR, MAPK1, CASP3, PTGS2). Molecular docking analysis demonstrated that four key active small molecules in Gusuibu (including Luteolin, Naringenin, Kaempferol, and Beta-sitosterol) have excellent binding affinity to the target proteins encoded by the top 10 hub genes. Our new findings indicated that one key active compound kaempferol activated the expression of osteoblast specific transcription factor OSX through JNK kinase pathway.
Osteoporosis is the most common metabolic disease of skeleton with reduced bone density and weaker bone structure. Patients are more likely to have fractures with the pain and other complications, and the quality of life is dramatically affected (Zheng et al., 2020). Postmenopausal osteoporosis is one common form of primary osteoporosis. Osteoporosis is classified into the categories of “Gu Bi” and “Gu Lou” in the “Huang Di Nei Jing” of Traditional Chinese Medicine (TCM) theory (CSoOaBM, 2019). Bone metabolism is a continuous process, and bone mass will be gradually lost after the age of 35. People especially postmenopausal women and the elderly should pay more attention to osteoporosis prevention (Li et al., 2020a; Saul and Kosinsky, 2021). The fragility fractures induced by osteoporosis will lead to increased disability and mortality, resulting in a heavy family, social and economic burden (Kanis, 2002; Xiong et al., 2021). The commonly used anti-osteoporosis drugs include estrogen, calcium, vitamin D, bisphosphonates, and denosumab; however, the consequent adverse effects are still a challenge to be solved due to the increased duration of use and dose (Jackson and Mysiw, 2014). Therefore, it is needed to explore potential anti-osteoporosis drugs with high efficacy and safety.
It is well known that TCM has a long history of practice in China and other Asian countries to treat a wide range of diseases, including osteoporosis (Li et al., 2017a; Wei et al., 2017; Wu et al., 2017; Zhang et al., 2017; Ge et al., 2018; Li et al., 2020a; Li et al., 2020b; Liu et al., 2020; Zheng et al., 2020). Among those to treat osteoporosis, the clinically commonly used Traditional Chinese Medicine prescriptions include Qianggu Capsule, XianlingGubao Capsule, DuhuoJisheng Decoction, LiuweiDihuang Pill, and Erxian Decoction (Li et al., 2017a; Wei et al., 2017; Wu et al., 2017; Ge et al., 2018; Li et al., 2020a). Qianggu Capsule (Drug approval number: Z20030007, Qi-Huang Pharmaceutical CO. LTD., Beijing, China) is a China Food and Drug Administration (CFDA)-approved TCM for the treatment of osteoporosis and bone loss. The active ingredients in Qianggu Capsule are total flavonoids extracted from Rhizoma Drynariae (English name is Fortune’s Drynaria Rhizome, and Chinese name is Gusuibu), the dried rhizome of Drynaria fortune (Kunze) J. SM. (Wang et al., 2008; CSoOaBM, 2019). The active ingredient of Qianggu Capsules is the herbal Gusuibu. Gusuibu is still a kind of raw material, and there are many compounds in Gusuibu, including flavonoids. Studies have shown that the flavonoids in Gusuibu may treat osteoporosis by improving bone density and reducing bone loss (Zhang et al., 2017; Shen et al., 2020). However, the mechanism of function of many TCM cannot be elucidated due to the complexity of the ingredients in TCM.
In recent years, many researchers have started to use bioinformatics and network pharmacology to analyze the multi-component, multi-target and multi-pathway characteristics of TCM in order to elucidate mechanisms of its action, thus providing directions for further research (Shuai et al., 2020). Gusuibu contains a variety of components. Although some studies have suggested that total flavonoids play a key role in anti-osteoporosis (Wang et al., 2008; Zhang et al., 2017), the molecular mechanism has not been elucidated yet.
In this study we used a network pharmacology approach to analyze the key genes, active compounds and pathways in the anti-osteoporosis of Gusuibu, and further explored possible underlying molecular mechanisms. We identified one key active compound kaempferol in Gusuibu as an upregulator of osteoblast specific transcription factor OSX.
Using TCM Systems Pharmacology (TCMSP, Version: 2.3, https://tcmspw.com/tcmsp.php) database (Ru et al., 2014), BATMAN-TCM platform (http://bionet.ncpsb.org/batman-tcm/) (Liu et al., 2016) and TCM Integrated Database (TCMID, http://www.megabionet.org/tcmid/) (Huang et al., 2018), the corresponding compounds of Gusuibu and related information were obtained. The compounds retrieved in the previous step were screened for active compounds according to the absorption, distribution, metabolism and excretion (ADME) protocol (oral bioavailability (OB) ≥ 30 and drug-likeness (DL) ≥ 0.18) (Tsaioun et al., 2016; Sheng et al., 2020). The databases were then used to mine potential targets corresponding to the active compounds. The target proteins were then converted to the corresponding gene names and UniProt ID using the UniProt database (https://www.uniprot.org/) for “Homo sapiens” (UniProt Consortium, 2018).
The targets related to osteoporosis were obtained through retrieving GeneCards (https://www.genecards.org/) (Relevance score ≥10) (Stelzer et al., 2016), MalaCards (https://www.malacards.org/) (Rappaport et al., 2017), DisGeNet database (https://www.disgenet.org/, v7.0) (Score ≥ 0.01) (Piñero et al., 2020), Therapeutic Target Database (TTD) (http://db.idrblab.net/ttd/, Last update by 1 June 2020) (Wang et al., 2020), Comparative Toxicogenomics Database (CTD) (http://ctdbase.org/, Last update by June 2020) (Inference score ≥ 15) (Davis et al., 2020), and Online Mendelian Inheritance in Man (OMIM) (https://omim.org/, updated 25 November 2020) (Amberger and Hamosh, 2017) using the keyword “osteoporosis or postmenopausal osteoporosis”. The potential targets obtained from the these 6 databases were integrated and deduplicated to generate the osteoporosis-related target set.
Gusuibu-related targets and osteoporosis-related targets were input to the Vennonline tool (http://www.bioinformatics.com.cn/) to obtain the common targets. These were the candidate targets of Gusuibu to treat osteoporosis. Cytoscape software (version 3.7.2) was used to construct the network diagram of “Gusuibu-active compounds-target genes-osteoporosis” (Shannon et al., 2003).
PPI is the basis of most biological processes and is important to understand cell physiology in both normal and disease states (Xiao et al., 2020). The STRING database (http://string-db.org/; version 11) was used to perform a PPI network analysis on the common targets (Szklarczyk et al., 2017). The species is limited to “Homo sapiens” with a confidence level greater than 0.4. Cytoscape software (version 3.7.2) was used to construct the PPI network, and the plug-in 12 CytoHubba algorithms [Degree, Maximal Clique Centrality (MCC), Clustering Coefficient, Density of Maximum Neighborhood Component (DMNC), BottleNeck, Maximum Neighborhood Component (MNC), Radiality, Edge Percolated Component (EPC), EcCentricity, Closeness, Betweenness, Stress] were used to find the top 10 hub genes (Chin et al., 2014; Xu et al., 2020).
The cluster Profiler package in R (R 4.0.2 for Windows) was used to perform Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) analysis to explore biological processes and signaling pathways in Gusuibu for osteoporosis (Kanehisa et al., 2008; Yu et al., 2012; Consortium, 2015). An adjusted p-value of less than 0.05 was used to determine the enrichment term.
The Sankey diagram (http://sankeymatic.com/) reveals the correspondence between herbs, components, and targets by constructing the interrelationship between the active compounds of Gusuibu and the top 10 hub genes. AutoDock Vina was used to perform molecular docking between the top 10 hub genes and key active ingredients to predict their binding affinity (Trott and Olson, 2010). The PDB format of the target proteins and the MOL2 format of active compounds could be obtained from the RCSB protein data (http://www.rcsb.org/) and the PubChem database (https://pubchem.ncbi.nlm.nih.gov/) (Kanis, 2002). The smaller the molecular docking score, the more stable the binding affinity between target proteins and active compounds (Chang et al., 2020).
C2C12 mesenchymal stem cells (ATCC) were cultured in DMEM with 10% fetal bovine serum (FBS, Gibco), 100U/mL penicillin G sodium and 100 μg/ml streptomycin sulfate in a humidified atmosphere at 37°C. Cells were plated in 6-well plates, cultured to 60–80% confluence and treated with kaempferol.
Kaempferol (1,000 ng/ml; Sigma) was used to stimulate C2C12 cells for 24 h before harvest. TRIzol (Invitrogen Life Technologies, Carlsbad, CA, United States) was used to extract total RNA from cultured cells as previously described (Tang et al., 2012). RNA purity and integrity were determined by the RNA 6000 Nano assay with an Agilent Bioanalyzer 2,100 (Agilent Technologies, Santa Clara, CA). 1 μg total RNA was reverse transcribed into cDNA using the GoScript Reverse Transcription System (Promega Corp., Madison, WI, United States) based on the manufacturer protocol. qPCR was performed in triplicate using SYBR-Green SuperReal PreMix Plus [Tiangen Biotech (Beijing) Co., Ltd., China] and the iQ5 PCR system (Bio-Rad Laboratories, Inc., Hercules, CA, United States). The reaction conditions were as follows: 95°C for 30 s, and 40 cycles of 95°C for 10 s and 60°C for 30 s. Data were reported as cycle threshold (Ct) values. The 2−ΔΔCt method was used to compare the RNA expressions. RNA levels were normalized to glyceraldehyde-3-phosphate dehydrogenase (GAPDH) levels.
All qPCR experiments were performed in triplicate. Data were expressed as the mean ± SD. Comparisons were made between groups by Student’s t test with p < 0.05 being considered as statistically significant.
A total of 131 compounds in Gusuibu (Rhizoma Drynariae) were obtained through the TCMSP database (Number: 71), TCMID database (Number: 53), and BATMAN-TCM platform (Number: 7). Based on the respective standards of TCMSP database (OB ≥ 30% and DL ≥ 0.18) and BATMAN-TCM platform (Score cutoff>20, adjusted p-value < 0.05) along with the existence of target proteins, a total of 24 active compounds of Gusuibu were screened. The corresponding target proteins were deduplicated, and a total of 331 target proteins were obtained. The UniProt database was used to convert the target proteins predicted by Gusuibu’s bioactive compounds into gene names. Table 1 includes the basic information of active compounds in Gusuibu.
By searching the GeneCard, TTD, MalaCard, CTD, DisGeNET, and OMIM databases, a total of 1,578 potential therapeutic targets for osteoporosis were obtained. By constructing the Venn diagram of the targets regulated by Gusuibu’s active ingredients and the potential targets of osteoporosis, a total of 140 intersection targets were obtained, which are potential therapeutic targets of Gusuibu against osteoporosis as shown in Figure 1. Cytoscape software was used to construct the “Gusuibu-active compounds-target genes-osteoporosis” network for Gusuibu against osteoporosis as shown in Figure 2. There are three compounds (Cycloartenone, Narirutin, Naringin) corresponding to targets that are not intersecting genes, so there are 21 active compounds in the network.
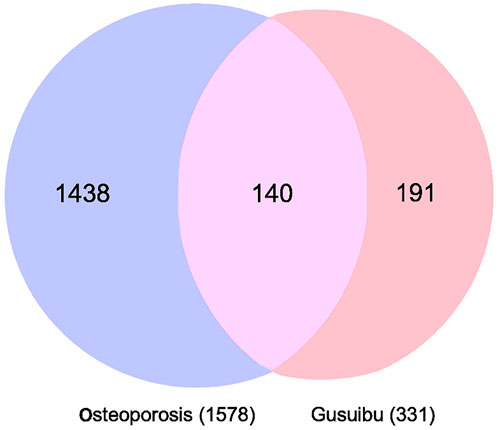
FIGURE 1. Venn diagram for the integrated analysis of the related targets of Gusuibu and osteoporosis.
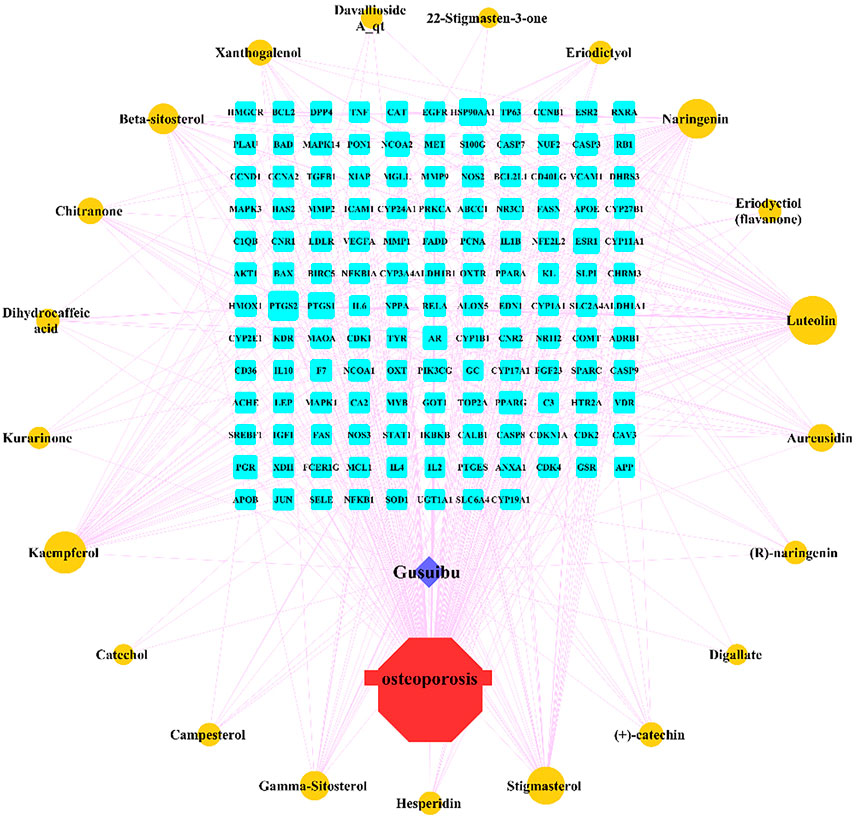
FIGURE 2. “Gusuibu-active compounds-target genes-osteoporosis” network. The red octagon represents osteoporosis; the blue diamond represents Gusuibu; the cyan rectangle represents the potential target; the brown ellipse represents the active compound contained in Gusuibu. The line between two nodes indicates that there is a relationship, and the size of each node indicates the number of relationships.
A total of 140 potential target genes of Gusuibu for treating osteoporosis were entered into the STRING database to obtain a PPI network, which involved 140 nodes and 2,199 edges (Figure 3). The data obtained was imported into Cytoscape software (version 3.7.2) for further visualization (Figure 4A). According to the 12 CytoHubba algorithms of Cytoscape software, the top 10 hub genes of Gusuibu for the treatment of osteoporosis were screened. A total of 37 different genes and the number of algorithms to which these genes belong were sorted in Table 2. The top 10 hub genes were consistent with the results of the Degree algorithm (Figure 4B). The Sankey diagram was constructed by using the top 10 ranked hub genes (AKT1, IL6, JUN, TNF, MAPK3, VEGFA, EGFR, MAPK1, CASP3, PTGS2) and the corresponding active compounds of Gusuibu. Luteolin targeted most of the hub genes while PTGS2 targeted most of the active compounds (Figure 5).
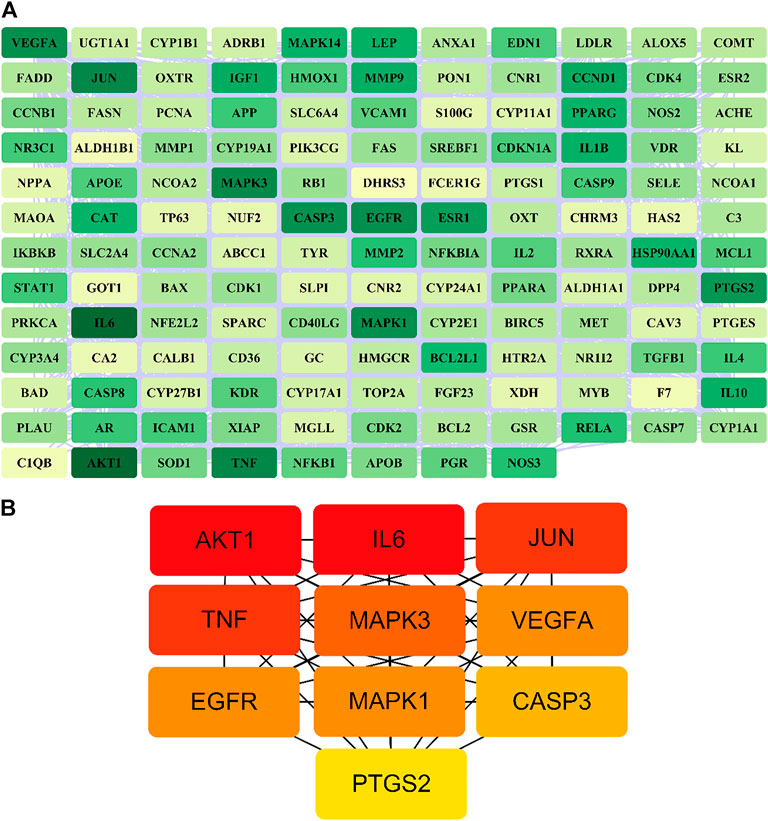
FIGURE 4. PPI network of potential target genes and top 10 hub genes for Gusuibu against osteoporosis. (A) PPI network constructed by using Cytoscape software. (B) The top 10 hub genes was identified by the Degree algorithm.
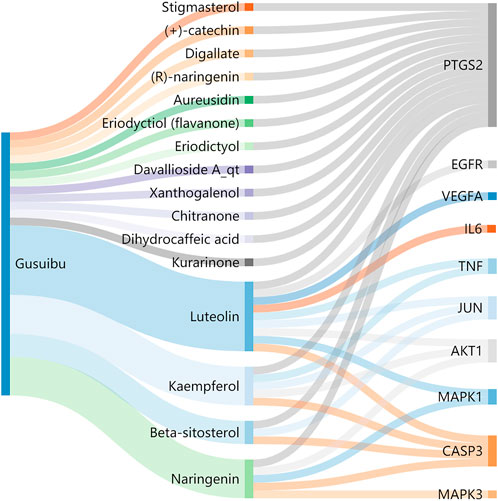
FIGURE 5. Sankey diagram. The blue vertical bar on the left represents Gusuibu, the middle bars represent the active ingredients of Gusuibu, and the right bars represent the top 10 hub genes. The size of the bars represents the number of correlations.
The Cluster Profiler package in R was used to perform GO enrichment analysis on 140 potential targets of Gusuibu in the treatment of osteoporosis. 2601 GO items (adjusted, p < 0.05) were obtained, including 2,398 biological process (BP) items, 75 cellular component (CC) terms and 128 molecular function (MF) items. As shown in Figure 6, the top 10 enrichment results of GO-BP, GO-CC, and GO-MF were visualized by bubble chart.
A total of 166 KEGG enriched pathways (adjusted, p < 0.05) were obtained through the above R package. As shown in Figure 7, the top 20 KEGG pathway enrichment terms were visualized by bubble chart.
We searched for osteoporosis in the KEGG database, and compared the retrieved relevant pathways with the enriched 166 pathways. A total of 50 pathways that may be related to osteoporosis were obtained, and we used Cytoscape software to construct a network diagram of potential target genes and pathways as shown in Figure 8.
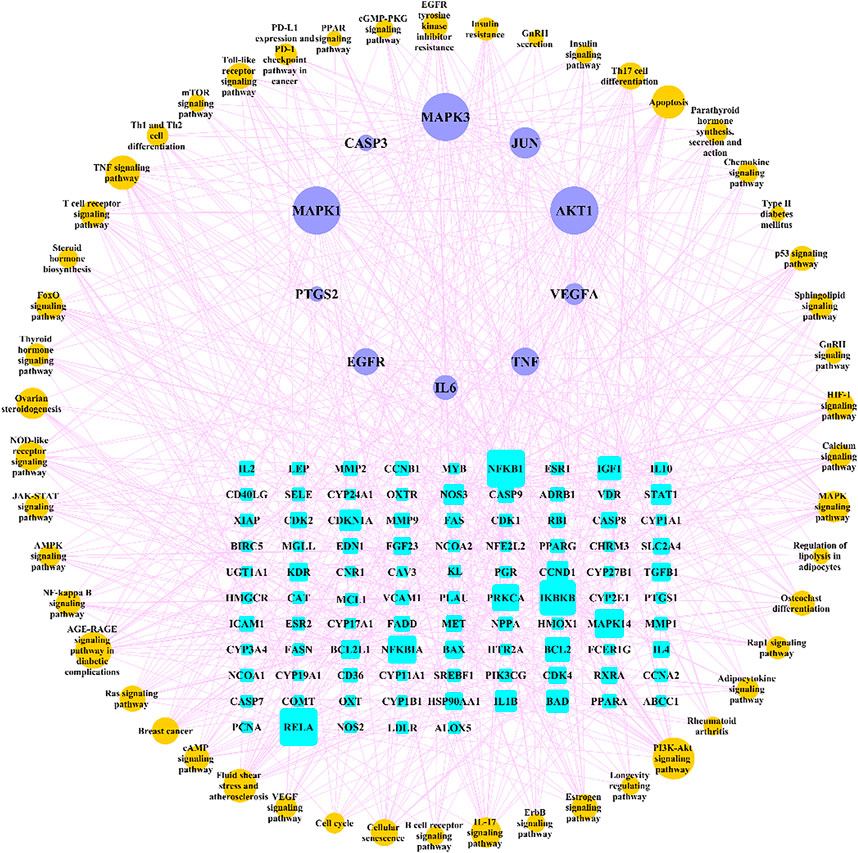
FIGURE 8. “Target genes-pathways” network. The cyan rectangle represents the potential target genes of Gusuibu against osteoporosis; the light blue ellipse represents the top 10 hub genes; the brown ellipse represents potential osteoporosis-related pathways.
Based on the Sankey diagram results, molecular docking was performed between the top 10 target proteins (AKT1, IL6, JUN, TNF, MAPK3, VEGFA, EGFR, MAPK1, CASP3, PTGS2) and four key active compounds (Luteolin, Naringenin, Kaempferol, and Beta-sitosterol) using AutoDock Vina. As shown in Figure 9, a heatmap was used to visualize the strongest affinity docking scores of top 10 target proteins and 4 key active compounds. The binding energy between the top 10 target proteins and the active compound is about −6.4 to −10.8 kcal mol−1, indicating that the key active compounds in Gusuibu bind well to the top 10 target proteins.
To explore the possible role of one key active compound kaempferol in osteoblast genes, we examined the effect of kaempferol on osteoblast differentiation related gene expressions. C2C12 cells were cultured and treated as indicated with kaempferol (1,000 ng/ml, Sigma) for 24 h. Total RNA was isolated and measured by real time RT-PCR. As shown in Figure 10, after kaempferol stimulation osteoblast specific transcription factor Osterix (OSX) expression was upregulated by about 6.1 folds, while the expression of OSX downstream osteoblastic gene osteocalcin (OCN) increased by 5.9 fold. These observations demonstrated that kaempferol induced OSX gene expression.

FIGURE 10. Kaempferol induced the expression of osteoblast genes. C2C12 cells were treated with 1,000 ng/ml kaempferol for 24 h. Total RNA was isolated and measured by real time RT-PCR. The RNA level from the control group was normalized to a value of 1. Values were presented as the mean ± S.D. *: A star indicates statistical significance compared to control group with p < 0.05. (A) Effect of kaempferol on OSX expression; (B) Effect of kaempferol on OCN expression.
We asked which pathway was involved in kaempferol induced OSX gene expression. To explore molecular mechanisms of kaempferol effect on OSX expression, we used a loss-of-function approach to examine possible pathways involved. The selective inhibitors were used: SB203580 as a specific inhibitor for p38 MAPK pathway, and SP600125 is a specific inhibitor for JNK kinase pathway. C2C12 cells were treated with kaempferol. Different inhibitors were added in the culture medium. As shown in Figure 11, kaempferol treatment led to OSX expression increase by 10.8-fold. Addition of 50 μM SP600125 almost abolished the increment of OSX expression induced by kaempferol. Kaempferol-induced OSX activation remained unchanged after treatment with 10 μM SB203580. The data indicate that kaempferol-induced OSX activation is mediated through JNK kinase pathway.
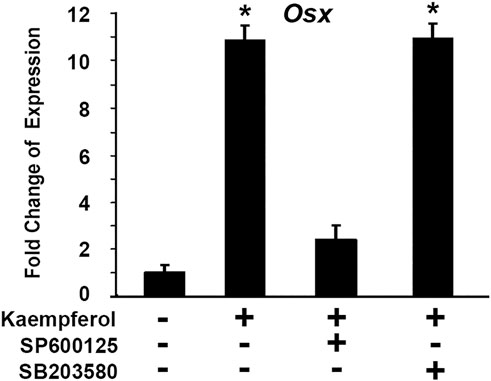
FIGURE 11. Kaempferol-induced OSX activation is mediated through JNK kinase pathway. Gene expressions were determined via qRT-PCR with or without kaempferol stimulation. The RNA level from the control group was normalized to a value of 1. *: A star indicates statistical significance compared to control group with p < 0.05. Specific inhibitors (50 μM SP600125 or 10 μM SB203580) were added as indicated.
Osteoporosis has become an important health issue with aging (Zheng et al., 2020). For example, postmenopausal osteoporosis as a very common form of primary osteoporosis affects most postmenopausal women (Jackson and Mysiw, 2014). Due to severe side effects of current clinical osteoporosis drugs, more and more attention has been paid to possible drugs for the prevention and treatment of osteoporosis from natural products (Li et al., 2020a). Qianggu Capsule is a proprietary Traditional Chinese Medicine approved by CFDA to treat osteoporosis, and its active ingredient is total flavonoids of Rhizoma Drynariae (Gusuibu) (CSoOaBM, 2019). Studies have shown that Qianggu Capsule may prevent and treat osteoporosis by improving bone density and reducing bone loss (Wei et al., 2017). The raw material of Qianggu Capsules is the flavonoids of the herbal Gusuibu, but the mixture is not a single compound at all, and the underlying mechanism is not clear yet. Based on the network pharmacological approaches such as databases mining, PPI network construction, GO and KEGG enrichment analysis and molecular docking validation, this study aims to analyze the active ingredients and potential targets of Gusuibu for osteoporosis treatment and to explore possible mechanisms of Qianggu Capsule active ingredients.
Based on the principle of ADME (setting OB ≥ 30 and DL ≥ 0.18) (Tsaioun et al., 2016), we searched three herbal databases, including TCMSP, TCMID and BATMAN-TCM platform, and screened 24 active compounds from Gusuibu (131 initially, after removing duplicates and null targets), and finally 331 corresponding target proteins were found. The protein name of the target protein obtained was converted into a gene ID through the UniProt database. Subsequently, we searched 6 disease-related databases (GeneCards, MalaCards, DisGeNET, TTD, CTD, and OMIM) and obtained a total of 1,578 potential therapeutic targets for osteoporosis. After constructing Venn diagrams with potential targets of Gusuibu, we obtained 140 potential target genes of Gusuibu for osteoporosis treatment. “Gusuibu-active compounds-target genes-osteoporosis” network was established, which involved 163 nodes and 432 interactions. The PPI network of potential therapeutic targets had also been constructed, involving 140 nodes and 2,199 edges. Based on the CytoHubba 12 algorithms of Cytoscape software (Degree, Clustering Coefficient, DMNC, Bottle Neck, MCC, MNC, Radiality, EPC, EcCentricity, Closeness, Betweenness, Stress), the top 10 hub genes were obtained, including AKT1, IL6, JUN, TNF, MAPK3, VEGFA, EGFR, MAPK1, CASP3, PTGS2. The physiological processes regulated by the proteins encoded by these core target genes mainly include inflammatory response, cell proliferation and differentiation, apoptosis, migration, cell cycle progression, angiogenesis, immune response, endocrine metabolism, bone metabolism, reproductive function, growth and nutrition. Therefore, based on these results, it may be speculated that Gusuibu may play a role in resisting osteoporosis through the multiple processes.
Sankey diagram showed the one-to-one correspondence between the top 10 hub genes and the corresponding active compounds contained in Gusuibu [Luteolin, Naringenin, Kaempferol, Beta-sitosterol, (+)-catechin, (R)-naringenin, Aureusidin, Digallate, Eriodyctiol (flavanone), Eriodictyol, DavalliosideA_qt, Dihydrocaffeic acid, Kurarinone, Chitranone, Stigmasterol, Xanthogalenol]. Among them, Luteolin targeted nine hub genes, Naringenin targeted five hub genes, Kaempferol targeted five hub genes, Beta-sitosterol targeted three hub genes, and the others targeted one hub gene each. Luteolin is a flavonoid found in many herbal extracts with antioxidant, anti-inflammatory and anti-tumor activities, and has been shown to reduce glucocorticoid-induced osteoporosis by modulating the ERK/Lrp-5/GSK-3β signaling pathway in vitro and in vivo (Jing et al., 2019). We summarized the relevant literature exploring the possible mechanisms of Qianggu Capsule and its compounds for osteoporosis treatment (Liu et al., 2001; Wong et al., 2007; Wang et al., 2008; Chen et al., 2009; Hung et al., 2010; Guo et al., 2011; Huang et al., 2011; Ouyang et al., 2012; Li et al., 2017b; Wei et al., 2017; Zhang et al., 2017; Jing et al., 2019; Dong et al., 2020; Ruangsuriya et al., 2020; Shen et al., 2020; Liu et al., 2021) in Table 3.
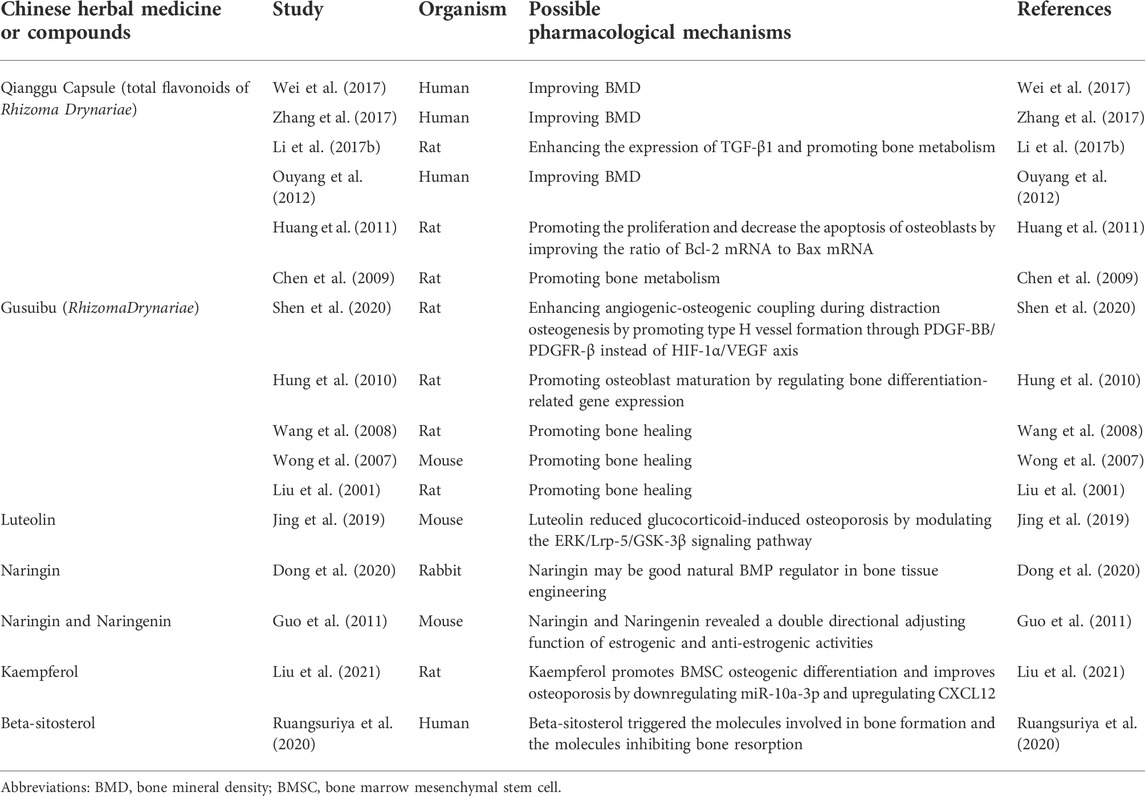
TABLE 3. Studies related to the regulation of bone metabolism by Qianggu Capsule and its active compounds.
We further analyzed the potential therapeutic genes of Gusuibu against osteoporosis by GO and KEGG pathway enrichment analysis. We used osteoporosis as a search term in the KEGG database and obtained ten directly related pathways, including osteoclast differentiation (hsa04380), AGE-RAGE signaling pathway in diabetic complications (hsa04933), Endocrine and other factor-regulated calcium reabsorption (hsa04961), Mineral absorption (hsa04978), Wnt pathway (hsa04310), MAPK pathway (hsa04010), Apoptosis (hsa04210), Chemokine pathway (hsa04062), T cell receptor pathway (hsa04660), and B cell receptor pathway (hsa04662). Five of the top 10 hub genes (AKT1, TNF, MAPK3, JUN, MAPK1) are enriched in osteoclast differentiation. Eight of the top 10 hub genes (AKT1, IL6, CASP3, TNF, VEGFA, MAPK3, JUN, MAPK1) are enriched in AGE-RAGE pathway in diabetic complications. Eight of the top 10 hub genes (AKT1, CASP3, TNF, VEGFA, MAPK3, JUN, EGFR, MAPK1) are enriched in MAPK pathway. Six of the top 10 hub genes (AKT1, CASP3, TNF, MAPK3, JUN, MAPK1) are enriched in Apoptosis. Three of the top 10 hub genes (AKT1, MAPK3, MAPK1) are enriched in Chemokine pathway. Five of the top 10 hub genes (AKT1, TNF, MAPK3, JUN, MAPK1) are enriched in T cell receptor pathway. Four of the top 10 hub genes (AKT1, MAPK3, JUN, MAPK1) are enriched in B cell receptor pathway. The three pathways are not enriched including Endocrine and other factor-regulated calcium reabsorption, Mineral absorption, and Wnt pathway.
To search for potentially relevant pathways, we obtained 50 enriched pathways as osteoporosis-related pathways and used Cytoscape software to construct the “pathway-gene” network. Studies have shown that there are many pathways associated with osteoporosis, which are mainly related to bone metabolism, inflammatory response, immune response, endocrine metabolism and apoptosis (Liu et al., 2020). Based on these analyses, Gusuibu may exert anti-osteoporosis effects through many pathways and hub genes.
As an osteoblast-specific transcription factor OSX is required for bone formation and osteoblast differentiation (Nakashima et al., 2002; Zhang et al., 2008). OSX was originally discovered as a BMP2 inducible gene in C2C12 mesenchymal stem cells, and OSX knock-out mice lack bone formation (Nakashima et al., 2002). OCN is a well-known downstream osteoblastic gene of OSX. There are three critical transcription factors required for osteoblast differentiation and bone formation: IHH, RUNX2 and OSX. OSX is the only osteoblast-specific transcription factor identified so far, and it is only expressed in cells in the bone matrix and the inner (endosteum) and outer (periosteum) bone surfaces. OSX discovery as a master regulator of bone formation and osteoblast differentiation opens a new window to bone biology. As far as we know, this is the first study to discover that one active compound kaempferol in Gusuibu activated the expression of osteoblast specific transcription factor OSX through JNK kinase pathway.
In summary, through network pharmacology, we explored the potential mechanisms by which Gusuibu exerts its anti-osteoporosis effects with a multi-component, multi-target, and multi-pathway profile. Potential therapeutic targets for Gusuibu to exert anti-osteoporosis effects include AKT1, IL6, JUN, TNF, MAPK3, VEGFA, EGFR, MAPK1, CASP3, PTGS2. Most of the key active compounds in Gusuibu are flavonoids, mainly including Luteolin, Naringenin, Kaempferol, and Beta-sitosterol. Interestingly, kaempferol was identified as an upregulator of OSX.
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author.
CZ designed the study. AH, ZX, KL, YC, and LS performed the experiments. GG and CZ analyzed the data. CZ supervised the study. AH and ZX drafted the text. CZ corrected the manuscript.
This work was supported by the Beijing Municipal Science and Technology Commission (Grant Number: Z181100001818006).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Amberger, J., and Hamosh, A. (2017). Searching online mendelian inheritance in man (OMIM): A knowledgebase of human genes and genetic phenotypes. Curr. Protoc. Bioinforma. 581, 1–2. doi:10.1002/cpbi.27
Chang, J., Liu, L., Wang, Y., Sui, Y., Li, H., and Feng, L. (2020). Investigating the multitarget mechanism of traditional Chinese medicine prescription for cancer-related pain by using network pharmacology and molecular docking approach. Evid. Based. Complement. Altern. Med. 2020, 7617261. doi:10.1155/2020/7617261
Chen, L., Chen, L., Chen, H., Guo, X., Xu, H., Zhang, G., et al. (2009). Skeletal biomechanical effectiveness of retinoic acid on induction of osteoporotic rats treated by alendronate and qianggu capsules. Zhonghua yi xue za zhi 89 (27), 1930–1933.
Chin, C., Chen, S., Wu, H., Ho, C., Ko, M., and Lin, C. (2014). cytoHubba: identifying hub objects and sub-networks from complex interactome. BMC Syst. Biol. S11, S11. doi:10.1186/1752-0509-8-S4-S11
Consortium, T. G. O. (2015). Gene Ontology Consortium: Going forward. Nucleic Acids Res. 43, D1049–D1056. doi:10.1093/nar/gku1179
CSoOaBM, Research (2019). Guidelines for the diagnosis and management of primary osteoporosis (2017). Chin. J. Osteoporos. 25 (3), 281.
Davis, A., Grondin, C., Johnson, R., Sciaky, D., Wiegers, J., Wiegers, T., et al. (2020). Comparative Toxicogenomics database (CTD): Update 2021. Nucleic acids Res. 49, D1138–D1143. doi:10.1093/nar/gkaa891
Dong, G., Ma, T., Li, C., Chi, C., Su, C., Huang, C., et al. (2020). A study of Drynaria fortunei in modulation of BMP–2 signalling by bone tissue engineering. Turk. J. Med. Sci. 50 (5), 1444–1453. doi:10.3906/sag-2001-148
Ge, J., Xie, L., Chen, J., Li, S., Xu, H., Lai, Y., et al. (2018). Liuwei dihuang Pill treats postmenopausal osteoporosis with shen (kidney) yin deficiency via janus kinase/signal transducer and activator of transcription signal pathway by up-regulating cardiotrophin-like cytokine factor 1 expression. Chin. J. Integr. Med. 24 (6), 415–422. doi:10.1007/s11655-016-2744-2
Guo, D., Wang, J., Wang, X., Luo, H., Zhang, H., Cao, D., et al. (2011). Double directional adjusting estrogenic effect of naringin from Rhizoma drynariae (Gusuibu). J. Ethnopharmacol. 138 (2), 451–457. doi:10.1016/j.jep.2011.09.034
Huang, L., Xie, D., Yu, Y., Liu, H., Shi, Y., Shi, T., et al. (2018). Tcmid 2.0: A comprehensive resource for TCM. Nucleic Acids Res. 46, D1117–D1120. doi:10.1093/nar/gkx1028
Huang, Z., Ouyang, G., Xiao, L., Li, N., Gao, H., He, Y., et al. (2011). Effects of Drynaria total flavonoids on apoptosis of osteoblasts mediated by tumor necrosis factor-α. Zhong xi yi jie he xue bao 9 (2), 173–178. doi:10.3736/jcim20110210
Hung, T., Chen, T., Liao, M., Ho, W., Liu, D., Chuang, W., et al. (2010). Drynaria fortunei J. Sm. promotes osteoblast maturation by inducing differentiation-related gene expression and protecting against oxidative stress-induced apoptotic insults. J. Ethnopharmacol. 131 (1), 70–77. doi:10.1016/j.jep.2010.05.063
Jackson, R., and Mysiw, W. (2014). Insights into the epidemiology of postmenopausal osteoporosis: The women's health initiative. Semin. Reprod. Med. 32 (6), 454–462. doi:10.1055/s-0034-1384629
Jing, Z., Wang, C., Yang, Q., Wei, X., Jin, Y., Meng, Q., et al. (2019). Luteolin attenuates glucocorticoid-induced osteoporosis by regulating ERK/Lrp-5/GSK-3β signaling pathway in vivo and in vitro. J. Cell. Physiol. 234 (4), 4472–4490. doi:10.1002/jcp.27252
Kanehisa, M., Araki, M., Goto, S., Hattori, M., Hirakawa, M., Itoh, M., et al. (2008). KEGG for linking genomes to life and the environment. Nucleic Acids Res. 36, D480–D484. doi:10.1093/nar/gkm882
Kanis, J. A. (2002). Diagnosis of osteoporosis and assessment of fracture risk. Lancet (London, Engl. 359–1936. doi:10.1016/S0140-6736(02)08761-5
Li, G., Xu, Q., Han, K., Yan, W., and Huang, C. (2020). Experimental evidence and network pharmacology-based analysis reveal the molecular mechanism of Tongxinluo capsule administered in coronary heart diseases. Biosci. Rep. 40 (10), BSR20201349. doi:10.1042/BSR20201349
Li, J., Jia, Y., Chai, L., Mu, X., Ma, S., Xu, L., et al. (2017). Effects of Chinese herbal formula erxian decoction for treating osteoporosis: A systematic review. Clin. Interv. Aging 12, 45–53. doi:10.2147/CIA.S117597
Li, J., Wang, W., Feng, G., Du, J., Kang, S., Li, Z., et al. (2020). Efficacy and safety of duhuo jisheng decoction for postmenopausal osteoporosis: A systematic review and meta-analysis. Evid. Based. Complement. Altern. Med. 2020, 6957825. doi:10.1155/2020/6957825
Li, M., Wang, Y., Liao, N., Li, J., and Dong, Q. (2017). Changes of TGF-β1 expression during orthodontic tooth movement in rats with osteoporosis. Shanghai kou qiang yi xue 26 (1), 17–20.
Liu, H., Chen, R., Jian, W., and Lin, Y. (2001). Cytotoxic and antioxidant effects of the water extract of the traditional Chinese herb gusuibu (Drynaria fortunei) on rat osteoblasts. J. Formos. Med. Assoc. = Taiwan yi zhi 100 (6), 383–388.
Liu, H., Yi, X., Tu, S., Cheng, C., and Luo, J. (2021). Kaempferol promotes BMSC osteogenic differentiation and improves osteoporosis by downregulating miR-10a-3p and upregulating CXCL12. Mol. Cell. Endocrinol. 520, 111074. doi:10.1016/j.mce.2020.111074
Liu, W., Jiang, Z., Chen, Y., Xiao, P., Wang, Z., Huang, T., et al. (2020). Network pharmacology approach to elucidate possible action mechanisms of Sinomenii Caulis for treating osteoporosis. J. Ethnopharmacol. 257, 112871. doi:10.1016/j.jep.2020.112871
Liu, Z., Guo, F., Wang, Y., Li, C., Zhang, X., Li, H., et al. (2016). BATMAN-TCM: A bioinformatics analysis tool for molecular mechANism of traditional Chinese medicine. Sci. Rep. 6, 21146. doi:10.1038/srep21146
Nakashima, K., Zhou, X., Kunkel, G., Zhang, Z., Deng, J. M., Behringer, R. R., et al. (2002). The novel zinc finger-containing transcription factor osterix is required for osteoblast differentiation and bone formation. Cell. 108, 17–29. doi:10.1016/s0092-8674(01)00622-5
Ouyang, G., Feng, X., Xiao, L., Huang, Z., Xia, Q., and Zhu, F. (2012). Effects of Chinese herbal medicine qianggu capsule on patients with rheumatoid arthritis-induced osteoporosis: A report of 82 cases. Zhong Xi Yi Jie He Xue Bao 10 (12), 1394–1399. doi:10.3736/jcim20121210
Piñero, J., Ramírez-Anguita, J., Saüch-Pitarch, J., Ronzano, F., Centeno, E., Sanz, F., et al. (2020). The DisGeNET knowledge platform for disease genomics: 2019 update. Nucleic Acids Res. 48, D845–D855. doi:10.1093/nar/gkz1021
Rappaport, N., Twik, M., Plaschkes, I., Nudel, R., Iny Stein, T., Levitt, J., et al. (2017). MalaCards: An amalgamated human disease compendium with diverse clinical and genetic annotation and structured search. Nucleic Acids Res. 45, D877–D887. doi:10.1093/nar/gkw1012
Ru, J., Li, P., Wang, J., Zhou, W., Li, B., Huang, C., et al. (2014). Tcmsp: A database of systems pharmacology for drug discovery from herbal medicines. J. Cheminform. 6 (13). doi:10.1186/1758-2946-6-13
Ruangsuriya, J., Charumanee, S., Jiranusornkul, S., Sirisa-Ard, P., Sirithunyalug, B., Sirithunyalug, J., et al. (2020). Depletion of β-sitosterol and enrichment of quercetin and rutin in Cissus quadrangularis Linn fraction enhanced osteogenic but reduced osteoclastogenic marker expression. BMC Complement. Med. Ther. 20 (1), 105. doi:10.1186/s12906-020-02892-w
Saul, D., and Kosinsky, R. (2021). Epigenetics of aging and aging-associated diseases. Int. J. Mol. Sci. 22 (1), E401. doi:10.3390/ijms22010401
Shannon, P., Markiel, A., Ozier, O., Baliga, N., Wang, J., Ramage, D., et al. (2003). Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 13 (11), 2498–2504. doi:10.1101/gr.1239303
Shen, Z., Chen, Z., Li, Z., Zhang, Y., Jiang, T., Lin, H., et al. (2020). Total flavonoids of rhizoma drynariae enhances angiogenic-osteogenic coupling during distraction osteogenesis by promoting type H vessel formation through PDGF-BB/PDGFR-β instead of HIF-1α/VEGF Axis. Front. Pharmacol. 11, 503524. doi:10.3389/fphar.2020.503524
Sheng, S., Yang, Z., Xu, F., and Huang, Y. (2020). Network pharmacology-based exploration of synergistic mechanism of guanxin II formula (II) for coronary heart disease. Chin. J. Integr. Med. 27, 106–114. doi:10.1007/s11655-020-3199-z
Shuai, Y., Jiang, Z., Yuan, Q., Tu, S., and Zeng, F. (2020). Deciphering the underlying mechanism of eucommiae cortex against osteoporotic fracture by network pharmacology. Evid. Based. Complement. Altern. Med. 2020, 7049812. doi:10.1155/2020/7049812
Stelzer, G., Rosen, N., Plaschkes, I., Zimmerman, S., Twik, M., Fishilevich, S., et al. (2016). The GeneCards suite: From gene data mining to disease genome sequence analyses. Curr. Protoc. Bioinforma. 541, 1–30. doi:10.1002/cpbi.5
Szklarczyk, D., Morris, J., Cook, H., Kuhn, M., Wyder, S., Simonovic, M., et al. (2017). The STRING database in 2017: Quality-controlled protein-protein association networks, made broadly accessible. Nucleic Acids Res. 45, D362–D368. doi:10.1093/nar/gkw937
Tang, W., Yang, F., Li, Y., de Crombrugghe, B., Jiao, H., Xiao, G., et al. (2012). Transcriptional regulation of vascular endothelial growth factor (VEGF) by osteoblast-specific transcription factor osterix (osx) in osteoblasts. J. Biol. Chem. 287, 1671–1678. doi:10.1074/jbc.M111.288472
Trott, O., and Olson, A. (2010). AutoDock Vina: Improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J. Comput. Chem. 31 (2), 455–461. doi:10.1002/jcc.21334
Tsaioun, K., Blaauboer, B., and Hartung, T. (2016). Evidence-based absorption, distribution, metabolism, excretion (ADME) and its interplay with alternative toxicity methods. ALTEX 33 (4), 343–358. doi:10.14573/altex.1610101
UniProt Consortium, T. (2018). UniProt: The universal protein knowledgebase. Nucleic Acids Res. 46 (5), 2699. doi:10.1093/nar/gky092
Wang, X., Wang, N., Zhang, Y., Gao, H., Pang, W., Wong, M., et al. (2008). Effects of eleven flavonoids from the osteoprotective fraction of Drynaria fortunei (KUNZE) J. SM. on osteoblastic proliferation using an osteoblast-like cell line. Chem. Pharm. Bull. 56 (1), 46–51. doi:10.1248/cpb.56.46
Wang, Y., Zhang, S., Li, F., Zhou, Y., Zhang, Y., Wang, Z., et al. (2020). Therapeutic target database 2020: Enriched resource for facilitating research and early development of targeted therapeutics. Nucleic Acids Res. 48, D1031–D1041. doi:10.1093/nar/gkz981
Wei, X., Xu, A., Shen, H., and Xie, Y. (2017). Qianggu capsule for the treatment of primary osteoporosis: Evidence from a Chinese patent medicine. BMC Complement. Altern. Med. 17 (1), 108. doi:10.1186/s12906-017-1617-3
Wong, R., Rabie, B., Bendeus, M., and Hägg, U. (2007). The effects of rhizoma curculiginis and rhizoma drynariae extracts on bones. Chin. Med. 2, 13. doi:10.1186/1749-8546-2-13
Wu, Z., Zhu, X., Xu, C., Chen, Y., Zhang, L., and Zhang, C. (2017). Effect of Xianling Gubao capsules on bone mineral density in osteoporosis patients. J. Biol. Regul. Homeost. Agents 31 (2), 359–363.
Xiao, W., Sun, W., Lian, H., and Shen, J. (2020). Integrated network and experimental pharmacology for deciphering the medicinal substances and multiple mechanisms of duhuo jisheng decoction in osteoarthritis therapy. Evid. Based. Complement. Altern. Med. 2020, 7275057. doi:10.1155/2020/7275057
Xiong, Z., Zheng, C., Chang, Y., Liu, K., Shu, L., and Zhang, C. (2021). Exploring the pharmacological mechanism of duhuo jisheng decoction in treating osteoporosis based on network pharmacology. Evidence-based complementary Altern. Med. 5510290, 2021–21. doi:10.1155/2021/5510290
Xu, C., Li, R., and Wu, J. (2020). Effects of Yuanhu- Zhitong tablets on alcohol-induced conditioned place preference in mice. Biomed. Pharmacother. = Biomedecine Pharmacother. 133, 110962. doi:10.1016/j.biopha.2020.110962
Yu, G., Wang, L., Han, Y., and He, Q. (2012). clusterProfiler: an R package for comparing biological themes among gene clusters. Omics a J. Integr. Biol. 16 (5), 284–287. doi:10.1089/omi.2011.0118
Zhang, C., Cho, K., Huang, Y., Lyons, J. P., Zhou, X., Sinha, K., et al. (2008). Inhibition of Wnt signaling by the osteoblast-specific transcription factor Osterix. Proc. Natl. Acad. Sci. U. S. A. 105, 6936–6941. doi:10.1073/pnas.0710831105
Zhang, Y., Jiang, J., Shen, H., Chai, Y., Wei, X., and Xie, Y. (2017). Total flavonoids from rhizoma drynariae (gusuibu) for treating osteoporotic fractures: Implication in clinical practice. Drug Des. devel. Ther. 11–1890. doi:10.2147/DDDT.S139804
Keywords: kaempferol, osteoporosis, qianggu capsule, gusuibu, OSX, network pharmacology
Citation: Huang AY, Xiong Z, Liu K, Chang Y, Shu L, Gao G and Zhang C (2022) Identification of kaempferol as an OSX upregulator by network pharmacology-based analysis of qianggu Capsule for osteoporosis. Front. Pharmacol. 13:1011561. doi: 10.3389/fphar.2022.1011561
Received: 04 August 2022; Accepted: 31 August 2022;
Published: 23 September 2022.
Edited by:
Xiaofeng Zhu, Jinan University, ChinaReviewed by:
Liusheng Huang, University of California, San Francisco, United StatesCopyright © 2022 Huang, Xiong, Liu, Chang, Shu, Gao and Zhang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Chi Zhang, Y2hpLnpoYW5nQGNhc2UuZWR1
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.