- 1Natural Medicine Research Center, College of Veterinary Medicine, Sichuan Agricultural University, Chengdu, China
- 2Key Laboratory of Animal Disease and Human Health of Sichuan Province, Sichuan Agricultural University, Chengdu, China
- 3Yibin university Sichuan Oil Cinnamon Engineering Technology Research Center, Yibin University, Yibin, China
Fungal-infections are mostly due to fungi in an adhering, biofilm-mode of growth and not due to planktonically growing, suspended-fungi. 1, 8-cineole is a natural product, which has been shown to possess antifungal effect. However, the anti-biofilm effect and mechanism of 1,8-cineole against Fusarium solani species complex has not reported previously. In this study, we found that 1,8-cineole has a good antifungal activity against F. solani with an MIC value of 46.1 μg/ml. Notably, 1,8-cineole showed good anti-biofilm formation activity against F. solani via inhibiting cell adhesion, hypha formation and decreasing the secretion of extracellular matrix at the concentration of ≥5.76 μg/ml. In addition, transcriptome sequencing analysis results showed that F. solani species complex genes related to ECM, protein synthesis and energy metabolism were down-expressed in the biofilms formation process treated with 1,8-cineole. In conclusion, these results show that 1,8-cineole has good anti-biofilm formation activity against F. solani species complex, and it exerts its anti-biofilm formation activity by downregulating of ergosterol biosynthetic genes, inhibiting adhesion, hindering the synthesis of ECM and interfering mitochondrial activity. This study suggests that 1,8-cineole is a promising anti-biofilm agent against F. solani species complex.
1 Introduction
Fusarium spp. can cause infections not only in plants but also in humans and animals, especially members of the Fusarium oxysporum and F. solani species complex, which have been implicated in human infections worldwide (Dalyan Cilo et al., 2015; van Diepeningen and de Hoog, 2016). The number of human infections by Fusarium spp. is on the rise worldwide due to an increase in the number of susceptible and immunocompromised people (Batista et al., 2020). Studies showed that up to 10% of onychomycosis is caused by Fusarium spp. (van Diepeningen et al., 2015). Eye infections associated with plants or soil commonly resulted in keratomycosis. Moreover, it could cause life-threatening infections in patients with severe immunodeficiency, especially leukaemia (van Diepeningen et al., 2015). The biofilm of filamentous fungi is one of its pathogenic factors. Infections associated with biofilm formation have been considered as a significant and increasing clinical problem.
Fungal biofilm is a community of microorganisms that are embedded in a polymeric matrix attached to an object’s surface. Biofilm formation is a very complicated process involving initial adhesion, hyphae formation, maturation and dispersion (Liu et al., 2021). It has been reported that a dramatic change will happen in biofilms of filamentous fungi from planktonic organisms to cells, such as the metabolism and biosynthesis (O’Toole et al., 2000). The extracellular matrix (ECM) of biofilm normally includes proteins, polysaccharides, lipids, and extracellular DNA (eDNA) (Chiang et al., 2013), which is one of the main protective component of microbial biofilms.
Although there are many kinds of antifungal drugs for treating fusariosis, such as amphotericin B and voriconazole, the cure rate is low in clinical trials (Guarro, 2013). Moreover, these drugs have serious toxicity concerns, such as voriconazole can increase risk of cutaneous malignancies (Levine Miriam, T., and Chandrasekar Pranatharthi, H. (2016))and amphotericin B is related to nephrotoxicity (Hamill, 2013). Therefore, it is urgently needed to discover novel and effective medicines to treat fusariosis. Natural products from plants are one of the important resources for drug discovery. 1,8-cineole, a kind of cyclic-ether monoterpene, which is widely exist in plants, including Eucalyptus, tea, cinnamon, bay and sage (Rodenak-Kladniew et al., 2020; Cai et al., 2021). 1,8-cineole has been shown with multiple biological activities in the treatment of cancers (Sampath et al., 2018), neuropathic pain (Zhang et al., 2018), cardiovascular illnesses (Wang et al., 2021), digestive disorders (Santos et al., 2004), nervous system disease (Figuêiredo et al., 2019) and antimicrobial (De Sousa et al., 2012). However, its activity against Fusarium solani species complex biofilms has not yet been intensively investigated.
In this study, we investigated the anti-biofilm formation activity of 1,8-cineole against F. solani. Moreover, the effect of 1,8-cineole on F. solani species complex biofilm architecture was visualized using optical microscopy, scanning electron microscopy (SEM), and confocal laser scanning microscopy (CLSM). Furthermore, to investigate the influence of 1,8-cineole on the biofilm formation of F. solani at the gene expression level, transcriptome sequencing analysis was performed on the F. solani biofilm treated with 1,8-cineole. To the best of our knowledge, this is the first study to estimate the anti-biofilm formation activity of 1,8-cineole against F. solani, and also the first time RNA-seq analysis was used to explore the anti-biofilm formation mechanism of 1,8-cineole.
2 Materials and methods
A clinical strain of Fusarium solani species complex preserved by Sichuan Agricultural University was used in this study. Frozen stock of the strain was revived by subculture on sabouraud dextrose agar (SDA; Qingdao Hope Bio-Technology Co., Ltd. Qingdao, China) plates at 27°C for 4–6 days, and the conidia were subsequently harvested in PBS (0.02 M phosphate, 0.15 M, pH 7.2). The resulting F. solani suspensions counted on a hemocytometer. For all experiments, the final sporangiospore concentration was 2 × 106 CFU/ml planktonic cells. 1,8-cineole was dissolved in 40% acetone (XILONG SCIENTIFIC CO., LTD. Sichuan, China) to a concentration of 461 μg/ml, and 1,8-cineole was further diluted to 92.2 μg/ml in RPMI 1640 medium (Hyclone, Logan, UT, United States) and used to prepare a series of 2-fold dilutions ranging from 2.88 to 92.2 μg/ml.
2.1 Antifungal susceptibility testing
The minimal inhibitory concentrations (MICs) of 1,8-cineole were determined by the microdilution method using the approach of the Clinical and Laboratory Standards Institute (CLSI) guideline in 96-well microplates (Fothergill, 2012). 100 μl of F. solani species complex suspension (2 × 106 CFU/ml) and 100 μl of 1,8-cineole (in the range of 92.2 to 2.88 μg/ml in the above described medium) were added in each well and incubated at 27°C for 24 h. Wells containing only 40% acetone served as controls, and wells without test compounds served as controls blank control. To determine the minimal fungicidal concentrations (MFC), fungal cells from MIC assays were collected and rinsed with fresh RPMI 1640 medium, and inoculated on SDA plates. Colonies were counted after 3 days of incubation. The MFC was defined as the lowest concentration of testing compound at which 99.9% of the microorganisms were killed.
2.2 In vitro biofilm inhibition assay
The biofilm formation and the viability of the cells were tested after 1,8-cineole treatments (Zhong et al., 2017). Briefly, 100 μl F solani species complex suspensions (2 × 106 CFU/ml) were added to a 96-well microplate to adhesion for 90 min at 27°C. After pre adhesion, the unattached cells were washed by using sterile PBS, then add 100 μl 1,8-cineole (in the range of 23.05 to 2.88 μg/ml in the above described medium), and incubated at 27°C for 72 h to evaluate the effect of 1,8-cineole on biofilm formation.
The inhibitory effect of 1,8-cineole on F. solani species complex biofilm was evaluated using the 2,3-bis-(2-methoxy-4-nitro-5-sulfophenyl)-2H-tetrazolium-5-carboxanilide (XTT) (Shanghai yuanye Bi-Technology Co., Ltd. Shanghai, China) reduction assay according to the method described in a previous study (Liu et al., 2017). After incubation at 27 C for 72 h, the biofilms were washed twice with PBS to remove unattached cells and 200 μl XTT solution containing 1 mM menadione was added. After incubation at 37 C for 2 h in dark, 100 μl of each supernatant were transferred to new 96-well plates and the absorbance of each solution was measured at 490 nm using SPECTROstar Nano plate reader (BMG LabTech, Offenburg, Germany).
2.3 Microscopic observations on biofilms
Three microscopic techniques were used to investigate the structure of biofilms: optical microscopy, SEM, and CLSM. Incubation of F. solani cells and 1,8-cineole for 72 h without shake, to observe the effect on biofilms, and the final concentrations of 1,8-cineole were adjusted to 5.76, 11.52, 23.05 μg/ml, respectively.
The expolysaccharides was detected using the argentation method as described in a previous study (Bai and Cui, 2019). Briefly, put the coverslips containing biofilms into 2.5% glutaraldehyde solution and fixation for 2 h, and then, saturated calcium chloride solution was added to coverslips for 15 min 5% silver nitrate solution was added to coverslips and incubated for 15 min. After that, the biofilm was treated with 1% hydroquinone solution for 2 min, and washed with sterilized water for three times. The coverslips containing biofilms were placed in 5% sodium thiosulfate for 2 min, and washed with sterilized water for three times. Finally, the samples were dried and observed under an optical microscope for photography.
For SEM analysis, biofilms were washed and placed in 2.5% glutaraldehyde at 4°C overnight to fix samples (Wu et al., 2020). Following, the samples were dehydrated with ethanol in ascending concentrations (30, 50, 70, 80, 90, 95, and 100% ethanol) for 10 min at each concentration, and dried overnight. After drying, the samples were covered with a gold layer and observed with an Inspect FEI 50 scanning electron microscope at 20 kV. The images were processed by Photoshop software (Adobe Systems, San Jose, CA, United States).
For CLSM analysis, the coverslips containing biofilms were washed with sterile PBS, and the biofilms were stained with the Live/Dead TM viability/Cytotoxicity Assay Kit to distinguish live and dead cells. NucGreen is a green fluorescent dye and viable cells with intact membranes were stained green, while EthD-Ⅲ is a red fluorescent dye, which is not permeable by the viable cell membrane, and its fluorescence during microscopy studies indicates the cell death. The biofilms were visualized by STELLARIS STED/EM CPD 300 confocal laser scanning microscopy (Brilhante et al., 2019; Xie et al., 2020).
2.4 Adhesion assay
100 μl F solani species complex suspensions and 100 μl 1,8-cineole (in the range of 23.05 to 2.88 μg/ml in the above described medium) were added and incubated at 96-well microplates at 27°C for 4 h. Following the initial incubation, the medium was aspirated and unattached cells were washed by using sterile PBS thrice. Adhesion assay was measured by XTT-reduction assay as described previously (Zhong et al., 2017). The colorimetric change at 490 nm was measured with a SPECTROstar Nano plate reader (BMG LabTech, Offenburg, Germany).
2.5 Hyphae growth inhibition assay
In a 6-well microplate, 1 ml cell suspension (2 × 106 CFU/ml) was treated with different concentrations of 1,8-cineole (in the range of 2.88–23.05 μg/ml in the above described medium) and incubated at 27°C for 6 h. After 6 h of incubation, the culture plate was washed three times with sterile PBS, and then all samples were visualized under bright field using inverted microscope and photographed.
2.6 Composition of F. solani species complex biofilm and biomass quantification
1 ml cell suspension (2 × 106 CFU/ml) and 1 ml 1,8-cineole (in the range of 46.1 to 11.52 μg/ml) were added in 6-well microplates and incubated in RPMI 1640 medium 27°C for 72 h. Firstly, the contents of extracellular protein, extracellular polysaccharide and eDNA in biofilm of F. solani species complex were determined by enzymatic hydrolysis method. The samples were treated with protease K (100 μg/ml), DNaseI (2 mg/ml) or sodium periodate (10 μM) at 27°C for 2 h. After that, the biofilm was stained with crystal violet (CV), decolorized with 33% glacial acetic acid, and the OD595 value was determined with a SPECTROstar Nano plate reader (Sacks et al., 2018).
The extraction of ECM was performed as the method described in a previous study (Gupta et al., 2021). The biofilm was treated with 1,8-cineole in 6-well plates according to the above method. After that, the wells were washed to remove the media and 1,8-cineole and the biofilm was scrapped in PBS. The biofilm was sonicated at 35W for five cycles (30 s each) on ice bath and then the suspension was centrifuged for 5 min at 12,000 rpm. The cell pellet was visualized under light microscope for any cell lysis or breakage. The supernatant was used for extracelluar protein, extracellular polysaccharide (EPS), eDNA.
The EPS in F. solani species complex biofilm was estimated by a congored binding assay. The Congo red was added to 1 ml suspension to a final concentration of 40 μg/ml, and incubated at 37°C for 2 h at 200 r/min in a constant temperature shaker. Then the incubated solution was centrifuged at 5,000 g for 5 min to precipitate the cells, and the absorbance of supernatant was measured at 490 nm (Desai et al., 2019).
The Bradford method was used to estimate the protein content in the biofilm. According to manufacturer’s instructions, the protein concentration of samples can obtain by standard protein curves.
The eDNA was estimated by phenol-chloroform method. Briefly, The DNA of the ECM fraction was extracted with an equal volume of phenol: trichloromethane: isoamyl alcohol (25:24:1, v/v) (Invitrogen, Carlsbad, CA, United States) and then precipitated with trichloromethane: isoamyl alcohol (24:1). After centrifugation, 1/10 equivalent (v/v) of 3M sodium acetate and 2.5 equivalent (v/v) of anhydrous ethanol were added to the separated upper water layer and stored overnight at −20°C. The next day, the solution was centrifuged at 12,000×g for 10 min to obtain the precipitate, the precipitate was dissolved in 100 μl sterile distilled water, and the absorbance was measured at 260 nm.
2.7 Transcriptomic analysis
F. solani species complex was incubated in RPMI-1640 with or without 5.76 μg/ml of 1,8-cineole (MIC = 46.1 μg/ml) for 72 h. After removing the planktonic cells, the biofilms were collected and subsequently frozen in liquid nitrogen. After mRNA purification and sequencing library construction, the samples were sequenced by next-generation sequencing (NGS) based on the illumine platform. The threshold value of DEGs (differential expression genes) is a fold change of >2 and a p value of <0.05. RNA extraction, mRNA purification, and cDNA synthesis and sequencing were performed by Beijing Novogene (China). Three independent biological replicates were performed for each treatment. The RNA-Seq data have been deposited in the NCBI Gene Expression Omnibus with the accession number PRJNA847035.
2.8 Reverse transcription quantitative PCR (qRT-PCR)
The samples were collected and frozen using liquid nitrogen. Total RNA was extracted from mycelia using the TrIzol REAgent (Biomed, Beijing, China) according to the manufacturer’s instructions. Reverse transcription of cDNA was performed by reverse transcription kits according to the manufacturer’s instructions (Thermo Fisher Scientific, MA). The qRT-PCR assays were performed on seven genes detected as highly differentially expressed via RNA-seq. The seven selected genes are Atg3 (which can catalyze conserved reactions in glycolysis) (Graham et al., 2002), CLAH10 (a stress response enzymes) (Mukherjee et al., 2019), nuo-51 (a ubiquinone oxidoreductase) (Ryčovská et al., 2000), SDH2 (a component of the tricarboxylic acid cycle) (Huang and Millar, 2013), msh-2 (which is necessary for microsatellite stability and maintenance of genome integrity) (Kunkel and Erie, 2015) ATPeV1A (an ATP-driven enzyme) (Beyenbach and Wieczorek., 2006). gpi17 (a subunit of GPI-Transamidase) (Gamage et al., 2017). The reaction system of qRT-PCR included 5 μl SYBR Green Mix, 3 μl RNA-free water, 1 μl a pair of specific primers (0.5 μl forward primer and 0.5 μl reverse primer) and 1 μl cDNA templates. All the sequences of forward and reverse primers are shown in Table 1. The qRT-PCR procedure was set as follows: 95°C for 5 min, followed 40 cycles of 95°C for 10 s and 60°C for 30 s, with a final extension of 72°C for 60 s. According to the fluorescence threshold results (Ct values), all data were normalized to the housekeeping gene Actin (Shen et al., 2020) as the internal reference gene, and expression levels were calculated using the ΔΔCt method, as previously described (Schmittgen and Livak, 2001; Chen et al., 2020).
2.9 Statistics
The datas were analyzed using parametric statistical tests. Three independent biological replicates were performed for each assay. One-way analysis of variance (ANOVA) was used to analyze differences between the 1,8-cineole and control group. Statistical Product and Service Solutions (SPSS) 18.0 software (SPSS Inc., Chicago, IL, United States) was used for statistical analysis. p < 0.05 was considered statistically significant.
3 Results
3.1 1,8-cineole inhibits F. solani species complex biofilm
In this study, we firstly evaluated the antifungal effect of 1,8-cineole against F. solani species complex. Antifungal susceptibility testing results showed that the 1,8-cineole has a same MIC and MFC against F. solani species complex at 46.1 μg/ml (Figure 1A). To determine the inhibitory effect of 1,8-cineole against F. solani species complex biofilm, a XTT reduction assay was performed. The results showed that 1,8-cineole has a good anti-biofilm activity against F. solani species complex biofilm in a dose-dependent-manner (Figure 1B). Notably, 1,8-cineole inhibited 98.24% (p < 0.01) biofilm formation at a concentration of 11.52 μg/ml.
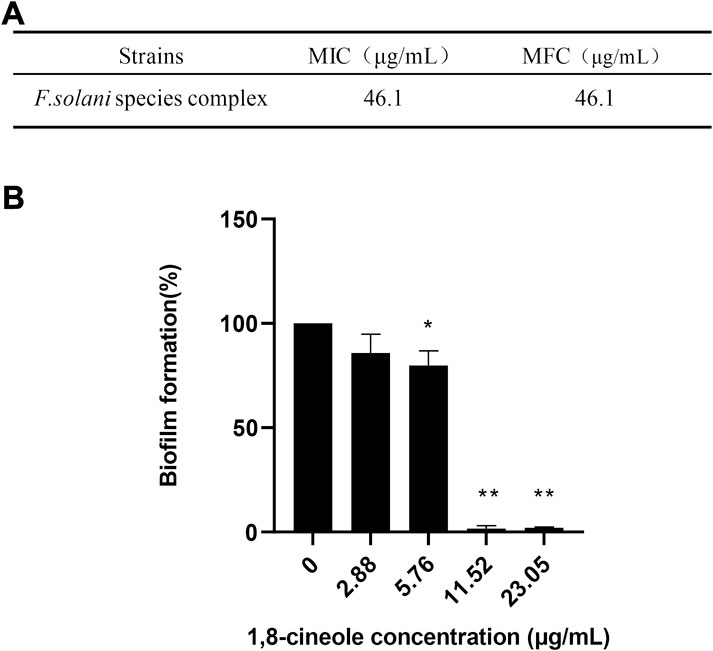
FIGURE 1. 1,8-cineole inhibits F. solani species complex biofilm formation in vitro. Biofilm formation was evaluated using an XTT reduction assay, and the results are presented as the percent of 1,8-cineole treated biofilm relative to control biofilm formed without treatment. (A) The MICs and MFCs of 1,8-cineole against F. solani species complex (46.1–2.88 μg/ml). (B) Effects of different concentrations of 1,8-cineole on biofilm formation. The results represent means ± standard deviations for three independent experiments. *, p < 0.05; **, p < 0.01.
3.2 1,8-cineole inhibits the formation of F. solani species complex biofilm structure
Optical microscope, SEM, CLSM techniques were used to observe F. solani species complex biofilms treated with 23.05–5.76 μg/ml 1,8-cineole as well as untreated biofilms (Figure 2). Optical microscope, with low magnifications (Figure 2A), provided an overview and preliminary inspection of F. solani species complex biofilms were affected by different concentrations of 1,8-cineole treatment, and showed untreated F. solani species complex biofilm with dense fungal growth (Figure 2Aa). The biofilm gradually decreased after treating with 1,8-cineole. Notably, 23.05 μg/ml 1,8-cineole could completely inhibit the growth of biofilm (Figure 2Ab, Ac, Ad).
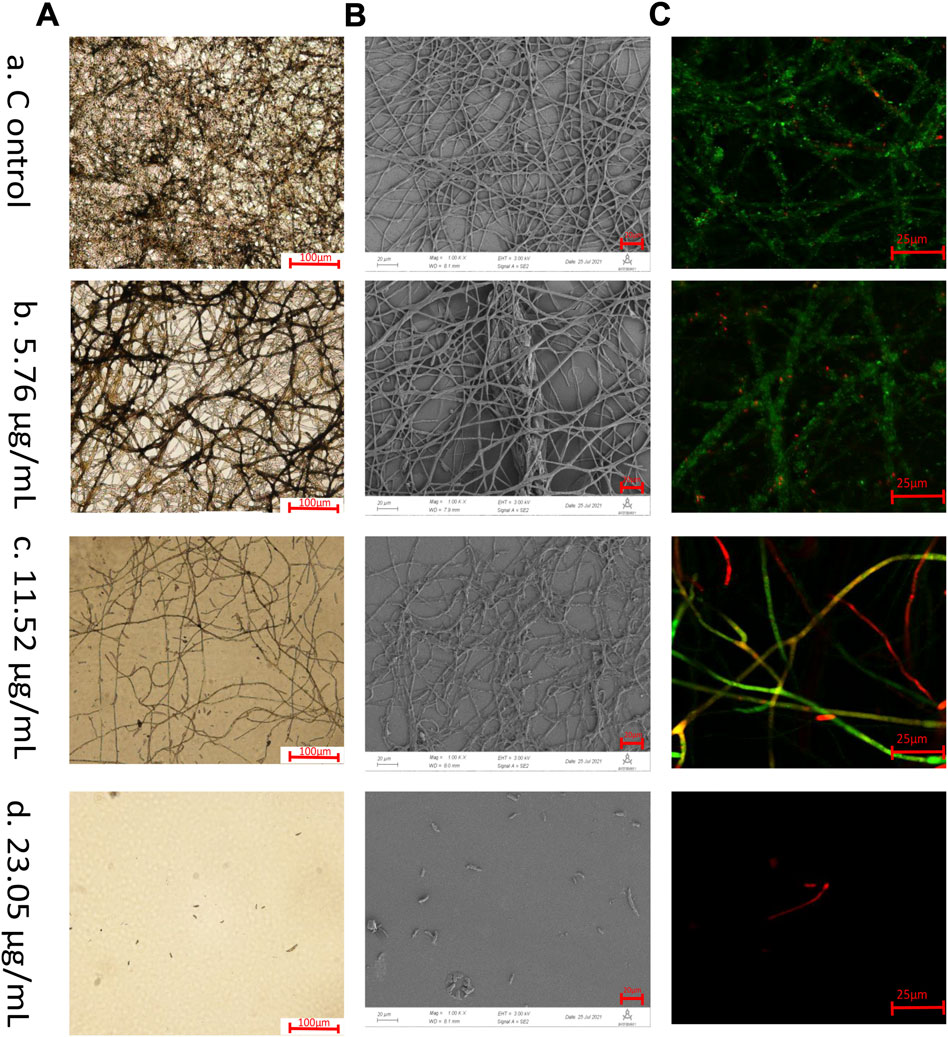
FIGURE 2. Microscopic observation. Biofilms of F. solani species complex are visualized by different microscopic techniques. The F. solani species complex cells were transferred to RPMI 1640 media with different concentrations of 1,8-cineole at 27°C for 72 h. The samples were treated at (A) 5.76 μg/ml, (B) 11.52 μg/ml, (C) 23.05 μg/ml, and (D) 46.1 μg/ml of 1,8-cineole, respectively.(A) Optical microscopic images of biofilm formation under 1,8-cineole treatment. Scale bar is 100 μm (100x). (B) Scanning electron microscopy images of biofilm formation under 1,8-cineole treatment. Scale bar is 20 μm (1,000x). The arrow indicated that the biofilm has multilayer mycelium structure. (C) CLSM images of biofilm formation under 1,8-cineole treatment. NucGreen is green nucleic acid staining and EthD-III is red nucleic acid staining. Red represents dead cells, green represents living cells, and yellow represents weakened cells Scale bar is 25 μm (200x).
SEM with a higher magnification and larger depth of focus, which can provide three-dimensional close up inspection of biofilms, allowing a better assessment of the inhibition of F. solani species complex biofilms formation that treated with 1,8-cineole (Figure 2B). After 72 h incubation, the untreated F. solani species complex formed a mature biofilm, fusion of hyphae, interconnection or anastomosis with well-development channels between several layers of hyphae and production of ECM (Figure 2Ba). After 11.52 μg/ml 1,8-cineole treatment, the morphological arrangement of the hyphae was significantly different from that of the control group, with significantly reduced ECM, damaged hyphae with flattened structures, and reduced hyphal density (Figure 2Bc).
To evaluate the 1,8-cineole killing efficiency against F. solani species complex biofilm cells during biofilm formation, NucGreen - EthD-III dual staining was performed and observed by CLSM (Figure 2C). In the growth control group, it was observed that the cells grew well, with green fluorescence. The number of cell death gradually increased with the increase of 1,8-cineole concentration (Figure 2Ca, Cb, Cc), with an increasing red fluorescent signal. At 23.05 μg/ml of 1,8-cineole, there was no cell survived (Figure 2Cd).
The hyphae in F. solani species complex biofilms were inhibited by 1,8-cineole in a dose-dependent manner, and more specifically, after 23.05 μg/ml of 1,8-cineole treatment, F. solani species complex cells maintained the state of spore and criss-crossing true hyphae could not be observed (Figure 2Ad, 2Bd, 2Cd).
3.3 1,8-cineole inhibits adhesion of F. solani species complex
A cell adhesion assay was performed to determine the effect of 1,8-cineole on attachment of F. solani species complex strain (Figure 3A). Compared with the control cells, a significant reduction (p < 0.05) in the adhesion of cells to the plastic surfaces was observed under the treatment of 1,8-cineole. The results showed that F. solani species complex adhesion was reduced 98% at the 11.52 μg/ml concentration of 1,8-cineole treatment (p < 0.01).
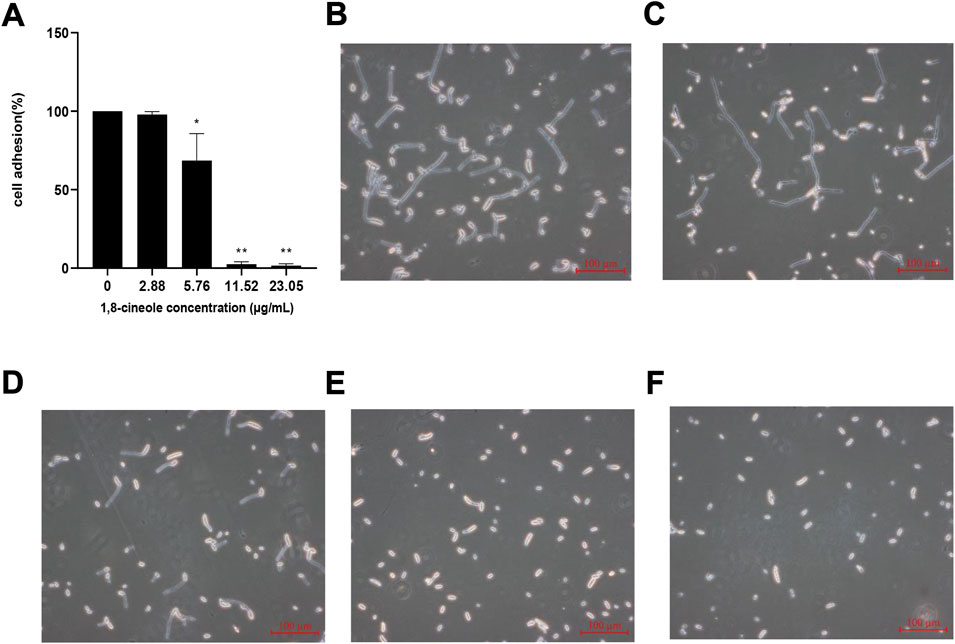
FIGURE 3. Effect of different concentrations of 1,8-cineole on adhesion and hypha formation. (A) Effects of different concentrations of 1,8-cineole on adhesion. Exponentially growing F. solani species complex cells were transferred to RPMI 1640 media with different concentrations of 1,8-cineole at 27°C for 4 h. In adhesion assay, exponentially growing F. solani species complex cells were incubated with different concentrations of 1,8-cineole for 4 h and was evaluated using an XTT reduction assay. (C ∼ F) Effects of different concentrations of 1,8-cineole on hypha formation. Exponentially growing F. solani species complex cells were transferred to RPMI 1640 media with different concentrations of 1,8-cineole at 27°C for 6 h. The group of control (B) and the concentration of 2.88 μg/ml (C), 5.76 μg/ml (D), 11.52 μg/ml (E), and 23.05 μg/ml (F) 1,8-cineole, respectively. The results represent means ± standard deviations for three independent experiments. *, p < 0.05; **, p < 0.01.
3.4 1,8-cineole inhibits hypha formation of F. solani species complex
The effect of 1,8-cineole on F. solani species complex hypha formation was further evaluated by incubating F. solani species complex in RPMI 1640 media for 72 h (Figure 3B∼ 3F). 1,8-cineole inhibited hypha formation in a dose-dependent manner. After treatment with 1,8-cineole at a concentration of 5.76 μg/ml, the hypha of the F. solani species complex began to be shorten. Interestingly, hypha formation was completely inhibited by 23.05 μg/ml of 1,8-cineole in RPMI 1640 medium and all F. solani species complex cells were maintained as spores.
3.5 1,8-cineole decreases the formation of extracellular matrix of F. solani species complex biofilm
To corroborate the molecules involved in the formation of biofilm of F. solani species complex, it was subjected to different treatments by detachment assay: protein degradation, polysaccharide degradation and DNA degradation. The biofilm was quantified by CV assay, in which the biomass formed by F. solani species complex was reduced by 28.44% with sodium periodate, 36.38% with Proteinase K, and 16.56% with DNaseⅠ (Figure 4A).
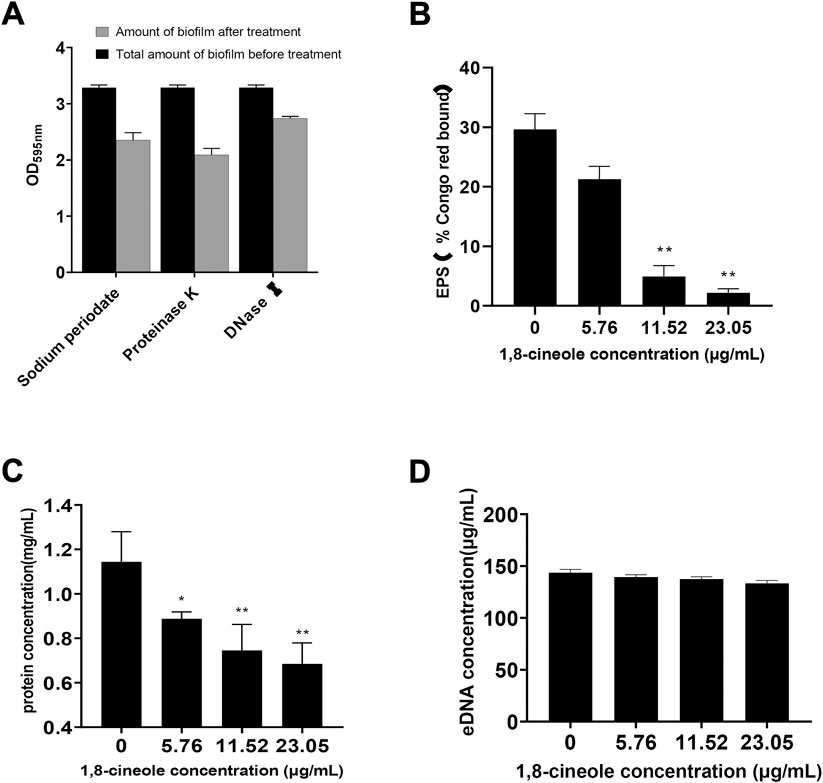
FIGURE 4. Effect of different concentrations of 1,8-cineole on Extracellular matrix. (A) The major components of F. solani species complex extracellular matrix were analyzed by adding to the preformed biofilm protease K to degrade proteins, DNase I to degrade DNA and sodium periodate to degrade β-1,6-linked polysaccharides. Biofilms were developed in 24-well plates incubated at 27 C for 72 h in RPMI 1640 medium and quantified by crystal violet assay. (B) The effect of 1,8-cineole on the extracellular polysaccharide (EPS) was evaluated using Congo red test. (C) Effect of 1,8-cineole on extracellular proteins was determined by Coomassie Brilliant Blue staining method. (D) The impact of various concentrations of 1,8-cineole on eDNA was evaluated by using Phenol chloroform method. Comparisons between absorbance levels revealed significant differences. *, p < 0.05; **, p < 0.01.
Compared with the control group, the extracellular polysaccharide and protein formation was reduced in 1,8-cineole treatment group (Figures 4B,C). When the concentration of 1,8-cineole reached to 11.52 μg/ml, extracellular polysaccharide and protein content were decreased by 83.36% and 35.01%, respectively. However, no significant differences in the leakage of eDNA were observed for control group and 1,8-cineole treated groups, at the experimental dose (Figure 4D).
3.6 1,8-cineole inhibited biofilms formation exhibit a novel transcriptome
3.6.1 Illuminate sequencing and de novo transcriptome assembly
In order to avoid the influence of cell growth inhibition on the expression of mRNA, 1,8-cineole at 5.76 μg/ml was chosen for RNA-seq analysis. F. solani species complex biofilm with 5.76 μg/ml 1,8-cineole treatment was named “treatment group”, and F. solani species complex biofilm without 1,8-cineole treatment was named “control group”.
To obtain a comprehensive overview of F. solani species complex biofilm transcriptome, a cDNA library from F. solani species complex biofilm was constructed and sequenced using Illumina RNA-seq. A total of 23233245 and 21189058 reads were generated from the control and treatment group, respectively (Table 2). The average length of raw reads was 3,028 nucleotides (Table 3). After removing of adapter sequences, ambiguous reads and low quality reads, 22161005 and 19733412 high-quality clean reads were obtained with an average GC content of 56.16% and 56.74% (Table 2). Finally, all short sequences were assembled into 29766246 unigenes with an average length of 2,185 bp, a maximum length of 26,965 bp and an N50 of 3,524 bp. The length distribution of the unigenes was illustrated in Figure 5A. The sequences ranging from 300 bp to 1,000 bp in length accounted for nearly 36.99% of the total. Up to 3,261 unigenes (23.94%) and 5,322 unigenes (39.07%) were 1,000 bp to 2000 bp and >2000 bp in length, respectively (Table 4).

TABLE 2. De novo transcriptome analysis in F. solani species complex biofilm and identification of critical genes involved in 1,8-cineole treatment.
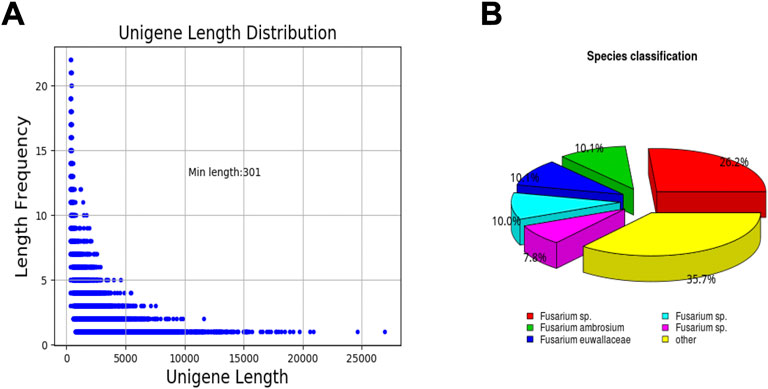
FIGURE 5. Unigene length distribution and species classification. (A) unigene length distribution. (B) species classification.
3.6.2 Transcriptome annotation and functional classification
After eliminating low-quality and short-length sequences, 13,621 unigenes were subjected to annotation analysis by matching sequences against Nr, Pfam and Swiss-prot databases. 11,190 unigenes (82.15% of the total) can be matched in Nr database, 8,276 unigenes (60.75% of the total) matched Pfam, and 7,200 unigenes (52.85% of the total) matched in Swiss-prot (Table 5). By comparing and annotating with the Nr database, the similarity between the gene sequence of this species with its relatives and the functional information can be obtained. The results showed that the gene sequences covered 44% of the Fusarium species. In addition, 10.1% and 10.1% of the gene sequences were matched to Fusarium ambrosium and Fusarium euwallaceae, respectively (Figure 5B).
Significant transcriptional changes were observed in the F. solani species complex biofilms after a 72 h treatment with 1,8-cineole (Figure 6). The raw expression levels of each sample were shown in the boxplots, and little variability was observed between the sample (Figure 6A). A total of 2,979 differentially expressed genes (DEGs) were seen in the 1,8-cineole-treated biofilms group, compared to the control group (Figure 6B), of which 1,203 were upregulated and 1776 downregulated (Figure 6C) (log2 fold change ≥1, p < 0.05).
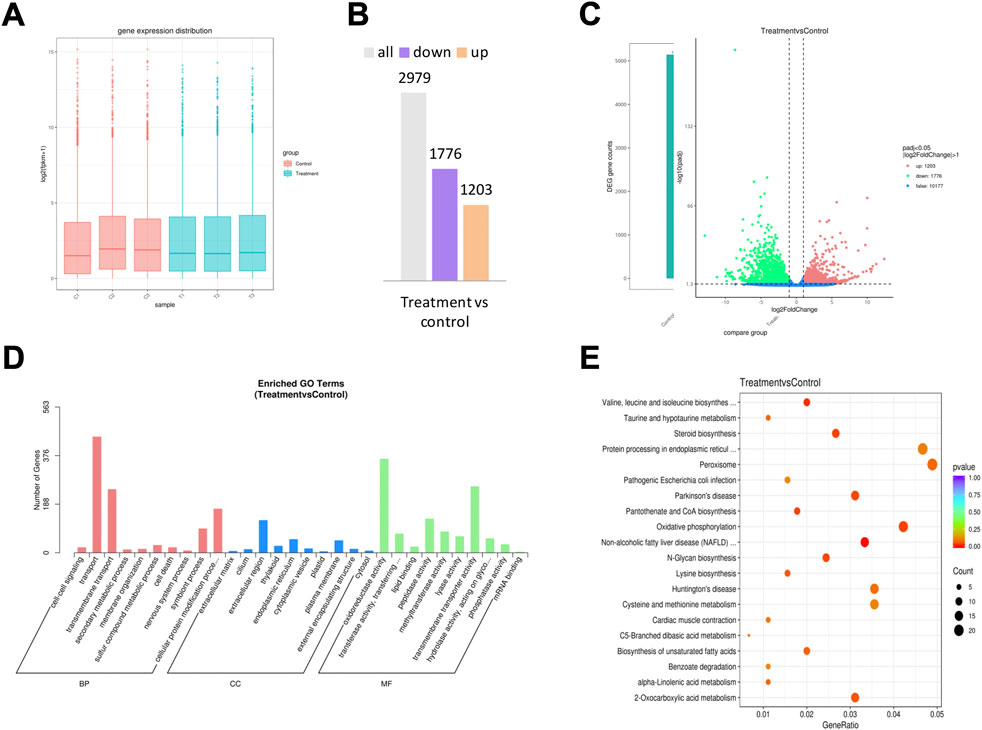
FIGURE 6. RNA sequencing analysis. F. solani species complex cells under treatment with 5.76 μg/ml of 1,8-cineole at 27 C for 72 h. (A) The box plot shows the variation in mRNA expression. (B) Statistical chart of DEGs. (C) Volcano plots of DEGs for F. solani species complex biofilm formed in response to 5.76 μg/ml 1,8-cineole; the red dots represent the up-regulated genes, the green dots represent the down-regulated genes, and the blue dotted line represents the threshold line of differential gene screening criteria. (D) GO analysis and (E) KEGG pathway classification analysis of upregulated and downregulated genes.
GO terms were assigned to assemble unigenes and provided defined ontologies to express gene product properties. GO function classification analysis was performed to identify alterations in the biological processes (BP), cellular composition (CC), and molecular functions (MF). GO revealed that the DEGs were mainly related to transport, transmembrane transport, extracellular region, oxidoreductase activity and transmembrane transporter activity (Figure 6D).
KEGG analysis demonstrated the biological pathways in which the unigenes are involved. Assembled unigenes were compared with the KEGG database using BLASTx and the corresponding pathways were established. KEGG pathway classification analysis results showed that the DEGs were mainly enriched in the pathways of valine, leucine and isoleucine biosynthesis, pantothenate and COA biosynthesis, 2- oxocarboxylic acid metabolism and oxidative phosphorylation (Figure 6E).
3.7 Validation of RNA-Sequencing data using qRT-PCR
To validate the DEGs expression levels of F. solani species complex that identified by RNA-Seq analysis, 7 genes (5 up-regulated and 2 down-regulated) with highest fold changes were selected from stages of biofilm matura and qRT-PCR was performed. The results of qRT-PCR confirmed the gene expression trend observed in RNA-seq data (Figure 7).
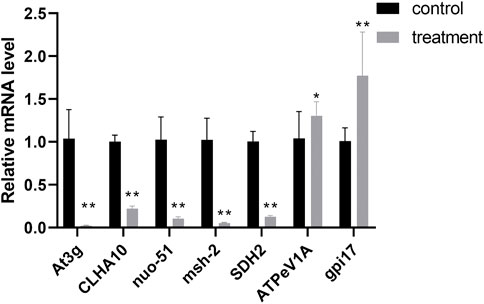
FIGURE 7. Correlation of expression level analyzed by qRT-PCR with data obtained using RNA-Seq platform. The data represent mean ± SD (n = 4). *, p < 0.05; **, p < 0.01.
4 Discussion
1,8-cineole has a good inhibitory effect on many fungi, such as Fusarium oxysporum, Fusarium sporotrichioides, Aspergillus tubingensis (Morcia et al., 2012). In this study, we demonstrated that 1,8-cineole can effectively inhibit F. solani species complex biofilm formation in vitro. To get more deep insight in the inhibition biofilm formation activity of 1,8-cineole against F. solani species complex, different microscopy assays were performed. The results of silver staining and SEM showed that the biofilm and ECM decreased with 1,8-cineole treatment. We observed that most of the ECM wrapped the mycelium, causing adhesion between the mycelium, while a small part of the extracellular matrix was “blanket” shaped, covering the mycelium (as shown in Figure 2B). This morphology of ECM was observed in this experiment has a similar structure with Chen. B (Chen et al., 2019). The formation of ECM will greatly increase the drug resistance of fungus in the biofilm state. Noteworthy, a large quantity of well-organized hyphae can be seen, which was consistent with images illustrating the biofilm of F. oxysporum (Veiga et al., 2021).
Since 1,8-cineole has a good inhibitory effect on the formation of biofilm, we further investigated the effect of 1,8-cineole on the formation stage of biofilm. The cycle of biofilm is adhesion, hyphal formation, maturation and dispersion (Pereira et al., 2021). Among them, adhesion and hyphal formation are key factors for biofilm formation and maturation. Adhesion is the first step for biofilm formation, which makes planktonic cells adhere to form colonies. The spores adhered to the surfaces, continuously grow and multiplying, and then further forming hyphae. The transformation from spores to hyphal phases at this stage is considered to be the main pathogenic factor (Huang et al., 2020). Our results demonstrated that 5.76 μg/ml 1,8-cineole could effectively reduce the adhesion of F. solani species complex and the formation of hypha (p < 0.05). These results indicated that the effect of 1,8-cineole on the biofilm may be attributed to inhibiting adhesion and hypha formation.
In addition, ECM is produced during the formation of biofilm, which is a complex mixture of macromolecules including proteins, exopolysaccharides and eDNA. Previous studies have shown that the extracellular matrix (ECM) formed by biofilm is the direct reason of fungal resistance to commonly used antifungal drugs (Gupta et al., 2016; Nogueira Brilhante et al., 2017). Saima Muzammil found that the content of protein, extracellular polysaccharide and eDNA in extracellular matrix of Acinetobacter baumannii were reduced after Aluminium oxide nanoparticles treatment (Muzammil et al., 2020). Therefore, we hypothesised that 1,8-cineole could affect the formation of biofilm by affecting the composition of ECM. Interestingly, the results showed that 1,8-cineole could inhibit extracellular protein and extracellular polysaccharide.
To investigate the effect of 1,8-cineole on gene expression of F. solani biofilm, transcriptomics was then used. KEGG and GO enrichment analyses were performed to determine the effect of biofilm formation while 1,8-cineole was added. There were different changes of several pathways associated with substance and energy metabolism and genetic information.
In view of the lipophilicity of essential oil, microbial cell plasma membrane is considered as the target of essential oil or its volatile components (Wang et al., 2019; Nunes et al., 2021). Essential oils can affect the expression of genes involved in cell membrane related pathways, such as fatty acid biosynthesis (FAS2) and sterol biosynthesis (ERG), thereby damaging cell membrane integrity (OuYang et al., 2016). ERG genes were upregulated in C. albicans after exposure to the itraconazole. (De Backer et al., 2001). Our results showed that genes related to steroid biosynthesis, such as sterol 14 α-demethylase (CYP51), sterol synthase (ERG7) and sterol reductase (ERG4) were up-regulated in F. solani. Genes involved in phosphatidylic acid biosynthesis were down regulated 6.2 times after treatment with 1,8-cineole, and phosphatidylserine decarboxylase (PSD) was down regulated 2.7 times. However, diacylglycerol kinase (DGK1) and choline kinase (CKI1) were up-regulated. Based on this phenomenon, the results indicated 1,8-cineole may affect membrane fluidity by changing the expression of membrane component related genes. In addition, the results indicated 1,8-cineole may penetrate the cell membrane and destroy the organelles without damaging the membrane.
Energy and substance metabolisms are the basis of life activity, especially under stress conditions. M. alternifolia essential oil can affect the respiration and metabolism of Sitophilus zeamais (Liao et al., 2016). This study found that almost all of differential genes encoding carbohydrate metabolism, amino acid metabolism and fatty acid metabolism were down regulated after 72 h of treatment with 1,8-cineole (Supplementary Table S1). Mitochondria are one of the important potential targets of essential oils (Wu et al., 2009). Turmeric EO inhibited the activities of ergosterol synthesis, mitochondrial ATPase, succinate dehydrogenase, and malate dehydrogenase (Hu et al., 2017). Transcriptome sequencing results showed that the genes encoding glycolysis related enzymes, including PGK, GPI, eno, and TPI were down regulated. Moreover, the genes encoding the TCA cycle including malate dehydrogenase (mdh2) and aconitase (ACO) were inhibited. Glucose is metabolized through TCA cycle to produce ATP and reduce NADH (Fernie et al., 2004). The glycolytic pathway and oxidative phosphorylation (Song et al., 2019) create the majority of the ATP necessary for fungal growth, reproduction and infection. Oxidative phosphorylation in mitochondria is the main source of energy required for life activities. ATP synthesized in this way accounts for about 95% of the total (Chinopoulos and Seyfried, 2018). Electrons produce a proton gradient across the membrane through the electron transport chain and produce ATP to provide energy for cells. Ruopeng Yang made cinnamaldehyde, eugenol or carvacrol into a compound nanoemulsion, which exerts antifungal activity by influencing the cellular respiration and proton transmembrane biological processes (Yang et al., 2021). Our data showed that the oxidative phosphorylation pathway was inhibited, in which NADH dehydrogenase (ubiquinone) (Complex I), succinate dehydrogenase (SDHB) (Complex II), cytochrome c oxidase (cox1) (Complex III), Fe-S protein 1 (NDUFS1) and other enzymes were down regulated. The results indicated 1,8-cineole caused the transmembrane transport of protons and electrons in mitochondria affects the production of ATP and then made F. solani starved of energy and dead. Meanwhile, the decline of the complex I, II, III and IV activity also led to the destruction of mitochondria.
Essential oil treatment can affect genes related to genetic information processing (transcription, translation, replication and repair (Lv et al., 2018). Interestingly, in this study, transcriptional regulatory-related genes (RPAC2 and RPC3) were downregulated in biofilm cells exposed to 1,8-cineole (Supplementary Table S2). Therefore, the results indicated that 1,8-cineole may interfere with the regulatory network during biofilm formation. Besides, the downregulated enolase gene with 1,8-cineole treatment suggested fungal adhesion was inhibited. Moreover, 1,8-cineole had reduced the activity of proteinase (glnS, asps and cycS) which made the formation of ECM of the F. solani biofilm after 1,8-cineole treatment was blocked. This means that the anti-biofilm formation ability of 1,8-cineole could not only interfere with cellular pathway, but also target the ECM of biofilm.
Collectively, the probable anti-biofilm mechanism of 1,8-cineole against F. solani biofilm formation involves multiple pathways and components. In the process of biofilm formation, 1,8-cineole first interacts with different biomolecules in ECM and inactivates various enzymes, which inhibits the formation of ECM. And then 1,8-cineole pass through ECM. After reaching the cell surface, 1,8-cineole penetrated the cell through the membrane. It can inhibit ergosterol synthesis in the cell membrane and change the permeability of the cell membrane. In the cytoplasm, 1,8-cineole mediates antifungal activity by attacking multiple pathways at the same time. Firstly, 1,8-cineole interferes with the metabolism of ability, including oxidative phosphorylation, glycolysis/gluconeogenesis and other energy metabolism pathways. In addtition, it can affect the production of ATP by changing the activity of mitochondrial complex I, II, III and IV; secondly, 1,8-cineole interferes with the transcriptional expression of biosynthetic pathway genes and inhibits the activity of protease.
In conclusion, the formation of F. solani species complex biofilm was greatly inhibited by 1,8-cineole. Further, the findings in our study suggested 1,8-cineole downregulated of ergosterol biosynthetic genes, inhibited adhesion, hindered the synthesis of ECM and interfered mitochondrial activity. This further demonstrated the value of 1,8-cineole as an anti-biofilm formation agent against F. solani. To our best knowledge, this research is the first study to report the phenotype and expression profiles of F. solani in biofilms exposed to 1,8-cineole.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://www.ncbi.nlm.nih.gov/, PRJNA847035.
Author contributions
YZ, ZY and XZ contributed to the conception and design of the study. YW, LL and XR performed the experiments and wrote the draft. XS, YFZ, LXL, HW, RJ, LY, XL and CH contributed to the methodology, investigation and visualization of the study. ZY, XZ, and QW provieded the fund support. All authors have read and approved the manuscript.
Funding
This research was financially supported by the program Sichuan Veterinary Medicine and Drug Innovation Group of China Agricultural Research System (SCCXTD-2020-18); the Science and Technology Project of Sichuan Province (2021NZZJ0021); Application and Industrialization Plan of Scientific and Technological Achievements of Guizhou Province (202221481317030275); the Science and Technology Project of Sichuan Province (2022YFH0057); the Bureau of Science and technology(2021YGC03).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fphar.2022.1010593/full#supplementary-material
References
Bai, M., and Cui, H. (2019) Inhibition mechanism of Litsea cubeba essential oil on Staphylococcus aureus and its biofilm (in Chinese) [D]. Jiangsu University.
Batista, B. G., de Chaves, M. A., Reginatto, P., Saraiva, O. J., and Fuentefria, A. M. (2020). Human fusariosis: An emerging infection that is difficult to treat. Rev. Soc. Bras. Med. Trop. 53, e20200013. doi:10.1590/0037-8682-0013-2020
Beyenbach, K., W., and Wieczorek, H. (2006). The V-type H+ ATPase: Molecular structure and function, physiological roles and regulation. J. Exp. Biol. 209, 577–589. doi:10.1242/jeb.02014
Brilhante, R. S. N., Aguiar, L. de, Sales, J. A., Araújo, G., dos, S., Pereira, V. S., et al. (2019). Ex vivo biofilm-forming ability of dermatophytes using dog and cat hair: An ethically viable approach for an infection model. Biofouling 35, 392–400. doi:10.1080/08927014.2019.1599361
Cai, Z. M., Peng, J. Q., Chen, Y., Tao, L., Zhang, Y. Y., Fu, L. Y., et al. (2021). 1, 8-cineole: A review of source, biological activities, and application. J. Asian Nat. Prod. Res. 23, 938–954. doi:10.1080/10286020.2020.1839432
Chen, B., Sun, Y., Zhang, J., Chen, R., Zhong, X., Wu, X., et al. (2019). In vitro evaluation of photodynamic effects against biofilms of dermatophytes involved in onychomycosis. Front. Microbiol. 10, 1228. doi:10.3389/fmicb.2019.01228
Chen, Y., Le Mauff, F., Wang, Y., Lu, R., Sheppard, D. C., Lu, L., et al. (2020). The transcription factor SomA synchronously regulates biofilm formation and cell wall homeostasis in Aspergillus fumigatus. MBio 11, 023299–e2420. doi:10.1128/mBio.02329-20
Chiang, W. C., Nilsson, M., Jensen, P. Ø., Høiby, N., Nielsen, T. E., Givskov, M., et al. (2013). Extracellular DNA shields against aminoglycosides in Pseudomonas aeruginosa biofilms. Antimicrob. Agents Chemother. 57, 2352–2361. doi:10.1128/AAC.00001-13
Chinopoulos, C., and Seyfried, T. N. (2018). Mitochondrial substrate-level phosphorylation as energy source for glioblastoma: Review and hypothesis. ASN Neuro 10, 1759091418818261. doi:10.1177/1759091418818261
Dalyan Cilo, B., Al-Hatmi, A. M. S., Seyedmousavi, S., Rijs, A. J. M. M., Verweij, P. E., Ener, B., et al. (2015). Emergence of fusarioses in a University hospital in Turkey during a 20-year period. Eur. J. Clin. Microbiol. Infect. Dis. 34, 1683–1691. doi:10.1007/s10096-015-2405-y
De Backer, M. D., Ilyina, T., Ma, X. J., Vandoninck, S., Luyten, W. H., and Vanden Bossche, H. (2001). Genomic profiling of the response of Candida albicans to itraconazole treatment using a DNA microarray. Antimicrob. Agents Chemother. 45, 1660–1670. doi:10.1128/AAC.45.6.1660-1670.2001
De Sousa, J. P., De Azerêdo, G. A., De Araújo Torres, R., Da Silva Vasconcelos, M. A., Da Conceição, M. L., and De Souza, E. L. (2012). Synergies of carvacrol and 1, 8-cineole to inhibit bacteria associated with minimally processed vegetables. Int. J. Food Microbiol. 154, 145–151. doi:10.1016/j.ijfoodmicro.2011.12.026
Desai, S., Sanghrajka, K., and Gajjar, D. (2019). High adhesion and increased cell death contribute to strong biofilm formation in Klebsiella pneumoniae. Pathogens 8, E277. doi:10.3390/pathogens8040277
Fernie, A. R., Carrari, F., and Sweetlove, L. J. (2004). Respiratory metabolism: Glycolysis, the TCA cycle and mitochondrial electron transport. Curr. Opin. Plant Biol. 7, 254–261. doi:10.1016/j.pbi.2004.03.007
Figuêiredo, F. R. S. D. N. de, Lbm, A., Iradm, B., Vdss, A., Epdn, A., Ckdsr, A., et al. (2019). Effects of the Hyptis martiusii Benth. leaf essential oil and 1, 8-cineole (eucalyptol) on the central nervous system of mice - ScienceDirect. Food Chem. Toxicol. 133, 110802. doi:10.1016/j.fct.2019.110802
Fothergill, A. W. (2012). Antifungal susceptibility testing: Clinical laboratory and standards Institute (CLSI) methods. Interact. Yeasts, Moulds, Antifung. Agents How Detect Resist. 2012, 65–74. doi:10.1007/978-1-59745-134-5_2
Gamage, D. G., Varma, Y., Meitzler, J. L., Morissette, R., Ness, T. J., and Hendrickson, T. L. (2017). The soluble domains of Gpi8 and Gaa1, two subunits of glycosylphosphatidylinositol transamidase (GPI-T), assemble into a complex. Arch. Biochem. Biophys. 633, 58–67. doi:10.1016/j.abb.2017.09.006
Graham, D. E., Xu, H., and White, R. H. (2002). A divergent archaeal member of the alkaline phosphatase binuclear metalloenzyme superfamily has phosphoglycerate mutase activity. FEBS Lett. 517, 190–194. doi:10.1016/s0014-5793(02)02619-4
Guarro, J. (2013). Fusariosis, a complex infection caused by a high diversity of fungal species refractory to treatment. Eur. J. Clin. Microbiol. Infect. Dis. 32, 1491–1500. doi:10.1007/s10096-013-1924-7
Gupta, A. K., Daigle, D., and Carviel, J. L. (2016). The role of biofilms in onychomycosis. J. Am. Acad. Dermatol. 74, 1241–1246. doi:10.1016/j.jaad.2016.01.008
Gupta, P., Goel, A., Singh, K. R., Meher, M. K., Gulati, K., and Poluri, K. M. (2021). Dissecting the anti-biofilm potency of kappa-carrageenan capped silver nanoparticles against Candida species. Int. J. Biol. Macromol. 172, 30–40. doi:10.1016/j.ijbiomac.2021.01.035
Hamill, R. J. (2013). Amphotericin B formulations: A comparative review of efficacy and toxicity. Drugs 73, 919–934. doi:10.1007/s40265-013-0069-4
Hu, Y., Zhang, J., Kong, W., Zhao, G., and Yang, M. (2017). Mechanisms of antifungal and anti-aflatoxigenic properties of essential oil derived from turmeric (Curcuma longa L.) on Aspergillus flavus. Food Chem. 220, 1–8. doi:10.1016/j.foodchem.2016.09.179
Huang, S., and Millar, A. H. (2013). Succinate dehydrogenase: The complex roles of a simple enzyme. Curr. Opin. Plant Biol. 16, 344–349. doi:10.1016/j.pbi.2013.02.007
Huang, X., Zheng, M., Yi, Y., Patel, A., and Li, Y. (2020). Inhibition of berberine hydrochloride on Candida albicans biofilm formation. Biotechnol. Lett. 42, 2263–2269. doi:10.1007/s10529-020-02938-6
Kunkel, T. A., and Erie, D. A. (2015). Eukaryotic mismatch repair in relation to DNA replication. Annu. Rev. Genet. 49, 291–313. doi:10.1146/annurev-genet-112414-054722
Levine Miriam, T., and Chandrasekar Pranatharthi, H. (2016). Adverse effects of voriconazole: Over a decade of use. Clin. Transpl. 30, 1377–1386. doi:10.1111/ctr.12834
Liao, M., Xiao, J.-J., Zhou, L.-J., Liu, Y., Wu, X.-W., Hua, R.-M., et al. (2016). Insecticidal activity of melaleuca alternifolia essential oil and RNA-seq analysis of Sitophilus zeamais transcriptome in response to oil fumigation. PLoS One 11, e0167748. doi:10.1371/journal.pone.0167748
Liu, Y., Xu, Y., Song, Q., Wang, F., Sun, L., Liu, L., et al. (2017). Anti-biofilm Activities from Bergenia crassifolia leaves against Streptococcus mutans. Front. Microbiol. 8, 1738. doi:10.3389/fmicb.2017.01738
Liu, Z., Li, L., Fang, Z., Lee, Y., Zhao, J., Zhang, H., et al. (2021). Integration of transcriptome and metabolome reveals the genes and metabolites involved in bifidobacterium bifidum biofilm formation. Int. J. Mol. Sci. 22, 7596. doi:10.3390/ijms22147596
Lv, C., Wang, P., Ma, L., Zheng, M., Liu, Y., and Xing, F. (2018). Large-scale comparative analysis of eugenol-induced/repressed genes expression in Aspergillus flavus using RNA-seq. Front. Microbiol. 9, 1116. doi:10.3389/fmicb.2018.01116
Morcia, C., Malnati, M., and Terzi, V. (2012). In vitro antifungal activity of terpinen-4-ol, eugenol, carvone, 1, 8-cineole (eucalyptol) and thymol against mycotoxigenic plant pathogens. Food Addit. Contam. Part A Chem. Anal. Control Expo. Risk Assess. 29, 415–422. doi:10.1080/19440049.2011.643458
Mukherjee, A., Yadav, R., Marmeisse, R., Fraissinet-Tachet, L., and Reddy, M. S. (2019). Heavy metal hypertolerant eukaryotic aldehyde dehydrogenase isolated from metal contaminated soil by metatranscriptomics approach. Biochimie 160, 183–192. doi:10.1016/j.biochi.2019.03.010
Muzammil, S., Khurshid, M., Nawaz, I., Siddique, M. H., Zubair, M., Nisar, M. A., et al. (2020). Aluminium oxide nanoparticles inhibit EPS production, adhesion and biofilm formation by multidrug resistant Acinetobacter baumannii. Biofouling 36, 492–504. doi:10.1080/08927014.2020.1776856
Nogueira Brilhante, R. S., Correia, E. E. M., De Melo Guedes, G. M., Pereira, V. S., De Oliveira, J. S., Bandeira, S. P., et al. (2017). Quantitative and structural analyses of the in vitro and ex vivo biofilm-forming ability of dermatophytes. J. Med. Microbiol. 66, 1045–1052. doi:10.1099/jmm.0.000528
Nunes, T. A. de L., Costa, L. H., De Sousa, J. M. S., De Souza, V. M. R., Rodrigues, R. R. L., Val, M., et al. (2021). Eugenia piauhiensis Vellaff. essential oil and γ-elemene its major constituent exhibit antileishmanial activity, promoting cell membrane damage and in vitro immunomodulation. Chem. Biol. Interact. 339, 109429. doi:10.1016/j.cbi.2021.109429
O’Toole, G., Kaplan, H. B., and Kolter, R. (2000). Biofilm formation as microbial development. Annu. Rev. Microbiol. 54, 49–79. doi:10.1146/annurev.micro.54.1.49
OuYang, Q., Tao, N., and Jing, G. (2016). Transcriptional profiling analysis of Penicillium digitatum, the causal agent of citrus green mold, unravels an inhibited ergosterol biosynthesis pathway in response to citral. BMC Genomics 17, 599. doi:10.1186/s12864-016-2943-4
Pereira, R., dos Santos Fontenelle, R. O., de Brito, E. H. S., and de Morais, S. M. (2021). Biofilm of Candida albicans: Formation, regulation and resistance. J. Appl. Microbiol. 131, 11–22. doi:10.1111/jam.14949
Rodenak-Kladniew, B., Castro, A., Stärkel, P., Galle, M., and Crespo, R. (2020). 1, 8-Cineole promotes G0/G1 cell cycle arrest and oxidative stress-induced senescence in HepG2 cells and sensitizes cells to anti-senescence drugs. Life Sci. 243, 117271. doi:10.1016/j.lfs.2020.117271
Ryčovská, A., Szabo, R., Tomáška, L., Nosek, J., and Rycovska, A. (2000). The respiratory complex I in yeast: Isolation of a gene NUO51 coding for the nucleotide-binding subunit of NADH:ubiquinone oxidoreductase from the obligately aerobic yeast yarrowia lipolytica. Folia Microbiol. 45, 429–433. doi:10.1007/BF02817616
Sampath, S., Subramani, S., Janardhanam, S., Subramani, P., Yuvaraj, A., and Chellan, R. (2018). Bioactive compound 1, 8-Cineole selectively induces G2/M arrest in A431 cells through the upregulation of the p53 signaling pathway and molecular docking studies. Phytomedicine 46, 57–68. doi:10.1016/j.phymed.2018.04.007
Santos, F. A., Silva, R. M., Campos, A. R., De Araújo, R. P., Lima Júnior, R. C. P., and Rao, V. S. N. (2004). 1, 8-Cineole (eucalyptol), a monoterpene oxide attenuates the colonic damage in rats on acute TNBS-colitis. Food Chem. Toxicol. 42, 579–584. doi:10.1016/j.fct.2003.11.001
Schmittgen, T. D., and Livak, K. J. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 25, 402–408. doi:10.1006/meth.2001.1262
Shen, T., Wang, Q., Li, C., Zhou, B., Li, Y., and Liu, Y. (2020). Transcriptome sequencing analysis reveals silver nanoparticles antifungal molecular mechanism of the soil fungi Fusarium solani species complex. J. Hazard. Mat. 388, 122063. doi:10.1016/j.jhazmat.2020.122063
Song, Y., Li, S., Zhao, Y., Zhang, Y., Lv, Y., Jiang, Y., et al. (2019). ADH1 promotes Candida albicans pathogenicity by stimulating oxidative phosphorylation. Int. J. Med. Microbiol. 309, 151330. doi:10.1016/j.ijmm.2019.151330
van Diepeningen, A. D., and de Hoog, G. S. (2016). Challenges in Fusarium, a trans-kingdom pathogen. Mycopathologia 181, 161–163. doi:10.1007/s11046-016-9993-7
van Diepeningen, A. D., Feng, P., Ahmed, S., Sudhadham, M., Bunyaratavej, S., and de Hoog, G. S. (2015). Spectrum of Fusarium infections in tropical dermatology evidenced by multilocus sequencing typing diagnostics. Mycoses 58, 48–57. doi:10.1111/myc.12273
Veiga, F. F., Castro-Hoshino, L., Sato, F., Baesso, M. L., Svidzinski, T., Negri, M., et al. (2021). Characterization of a biofilm formed by Fusarium oxysporum on the human nails. Int. J. Dermatol. 61, 191–198. doi:10.1111/ijd.15747
Wang, Y., Wei, K., Han, X., Zhao, D., Zheng, Y., Chao, J., et al. (2019). The antifungal effect of garlic essential oil on phytophthora nicotianae and the inhibitory component involved. Biomolecules 9, E632. doi:10.3390/biom9100632
Wang, Y., Zhen, D., Fu, D., Fu, Y., Zhang, X., Gong, G., et al. (2021). 1, 8-cineole attenuates cardiac hypertrophy in heart failure by inhibiting the miR-206-3p/SERP1 pathway. Phytomedicine. 91, 153672. doi:10.1016/j.phymed.2021.153672
Wu, R., Tao, Y., Cao, Y., Zhou, Y., and Lin, H. (2020). Streptococcus mutans membrane vesicles harboring glucosyltransferases augment Candida albicans biofilm development. Front. Microbiol. 11, 581184. doi:10.3389/fmicb.2020.581184
Wu, X.-Z., Cheng, A.-X., Sun, L.-M., Sun, S.-J., and Lou, H.-X. (2009). Plagiochin E, an antifungal bis(bibenzyl), exerts its antifungal activity through mitochondrial dysfunction-induced reactive oxygen species accumulation in Candida albicans. Biochim. Biophys. Acta 1790, 770–777. doi:10.1016/j.bbagen.2009.05.002
Xie, Y., Liu, X., and Zhou, P. (2020). In vitro antifungal effects of berberine against candida spp. in planktonic and biofilm conditions. Drug Des. devel. Ther. 14, 87–101. doi:10.2147/DDDT.S230857
Yang, R., Chen, X., Huang, Q., Chen, C., Rengasamy, K. R. R., Chen, J., et al. (2021). Mining RNA-seq data to depict how Penicillium digitatum shapes its transcriptome in response to nanoemulsion. Front. Nutr. 8, 724419. doi:10.3389/fnut.2021.724419
Zhang, Y. L., Liu, Y. G., Li, Q., Wang, X. D., Zheng, X. B., Yang, B. L., et al. (2018). 1, 8-cineole decreases neuropathic pain probably via a mechanism mediating P2X3 receptor in the dorsal root ganglion. Neurochem. Int. 121, 69–74. doi:10.1016/j.neuint.2018.09.007
Keywords: Fusarium solani species complex, biofilm, 1, 8-cineole, adhesion, mitochondrial, transcriptome sequencing
Citation: Zhang Y, Wang Y, Zhao X, Liu L, Xing R, Song X, Zou Y, Li L, Wan H, Jia R, Yin L, Liang X, He C, Wei Q and Yin Z (2022) Study on the anti-biofilm mechanism of 1,8-cineole against Fusarium solani species complex. Front. Pharmacol. 13:1010593. doi: 10.3389/fphar.2022.1010593
Received: 03 August 2022; Accepted: 28 September 2022;
Published: 14 October 2022.
Edited by:
Krishna Mohan Poluri, Indian Institute of Technology Roorkee, IndiaReviewed by:
Anne Debourgogne, Université de Lorraine, FrancePayal Gupta, Graphic Era University, India
Mohd Sajjad Ahmad Khan, Imam Abdulrahman Bin Faisal University, Saudi Arabia
Copyright © 2022 Zhang, Wang, Zhao, Liu, Xing, Song, Zou, Li, Wan, Jia, Yin, Liang, He, Wei and Yin. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Qin Wei, V2VpcWluMjAwMS02N0AxNjMuY29t; Zhongqiong Yin, eWluemhvbmdxQDE2My5jb20=
†These authors have contributed equally to this work
 Yu Zhang1†
Yu Zhang1† Yiming Wang
Yiming Wang Xinghong Zhao
Xinghong Zhao Xu Song
Xu Song Yuanfeng Zou
Yuanfeng Zou Lizi Yin
Lizi Yin Zhongqiong Yin
Zhongqiong Yin


