- 1School of Life Sciences, Chongqing University, Chongqing, China
- 2Center of Plant Functional Genomics, Institute of Advanced Interdisciplinary Studies, Chongqing University, Chongqing, China
- 3Department of Anesthesiology, First Affiliated Hospital of Kunming Medical University, Kunming, China
- 4Department of Pharmacy, West China Hospital, Sichuan University, Chengdu, China
- 5Department of Anesthesiology, The First People’s Hospital of Yunnan Province, Kunming, China
- 6Department of Anesthesiology, The Affiliated Hospital of Kunming University of Science and Technology, Kunming, China
Circular RNAs (circRNAs) are a class of covalently closed single-stranded RNA molecules. Four types of circRNAs have been reported in animal cells, and they have typical characteristics in their biogenesis, nuclear export and degradation. Advances in our understanding of the molecular functions of circRNAs in sponging microRNAs, modulating transcription, regulating RNA-binding proteins, as well as encoding proteins have been made very recently. Dysregulated circRNAs are associated with human diseases such as acute myeloid leukemia (AML). In this review, we focus on the recently described mechanisms, role and clinical significance of circRNAs in AML. Although great progress of circRNAs in AML has been achieved, substantial efforts are still required to explore whether circRNAs exert their biological function by other mechanisms such as regulation of gene transcription or serving as translation template in AML. It is also urgent that researchers study the machineries regulating circRNAs fate, the downstream effectors of circRNAs modulatory networks, and the clinical application of circRNAs in AML.
Introduction
Acute myeloid leukemia (AML) is a hematological malignancy characterized by clonal expansion of myeloid blasts cells with uncontrolled proliferation in the bone marrow and peripheral blood (Almatani et al., 2021; Aung et al., 2021; Andreozzi et al., 2022). AML has become a central research focus because it is the most common type of acute leukemia in adults worldwide, with rising morbidity and mortality (Gallipoli et al., 2015; Liu et al., 2019; Bhattacharya and Gutti 2022). The key therapeutic strategies for AML include chemotherapy, allogenic hematopoietic stem cell transplantation and palliative care (Xiang et al., 2022). However, although these advancements in the treatment of AML, the overall prognosis is poor (5-yeal overall survival only 28.7%) (Singh V. et al., 2021). Thus, new biomarker and precision therapy method are urgent to be found for the treatment of AML.
By the splicing machinery in linear order, most eukaryotic genes were divided by introns which must be removed from precursor message RNA via linking an upstream 5′ splicing site to a downstream 3′ splicing site (Wahl et al., 2009; Wilusz 2018) (Figure 1A). However, it has been reported that the splicing event can also occur in a non-canonical way to make the backsplicing reaction by linking a downstream 5′ splicing site to an upstream 3′ splicing site, thereby producing circular RNAs (Nigro et al., 1991; Cocquerelle et al., 1993; Chen et al., 2015; Wilusz 2017; Chen 2020). Circular RNAs (circRNAs), typically produced from protein-encoding genes through backsplicing, are a large class of covalently closed single-stranded RNA molecules, without a 5′ end or a 3’ poly (A) tail (Wilusz 2018; Kristensen et al., 2019; Papatsirou et al., 2021; Zhou et al., 2021) (Figure 1B). There are four types circRNAs in animal cells, including exonic circRNAs (EcircRNAs), exon-intron circRNAs (EIciRNAs), and intronic circRNAs (ciRNAs) (Jeck et al., 2013; Chen et al., 2015; Wilusz 2018; Chen 2020; Chen L. et al., 2022) (Figure 1B). Recently another type has been reported as mitochondria-encoded circRNAs (mecciRNA) (Liu et al., 2020) (Figure 1C). Although some circRNAs (such as EIciRNAs and ciRNAs) have been found in the nucleus (Li Z. et al., 2015; Conn et al., 2017), most circRNAs (EcircRNAs) are export to the cytoplasm (Salzman et al., 2012; Jeck et al., 2013). With the development of high-throughput RNA sequencing, many circRNAs have been identified from protein-coding genes across different species, tissues and cell lines (Memczak et al., 2013; Chen et al., 2015; Wang et al., 2016; Wilusz 2018; Zheng et al., 2021). CircRNAs have been found to play vital roles in different molecular and cellular events through different mechanisms including acting as microRNA sponges, regulation of transcription, interacting with RNA binding proteins and serving as translation template (Kristensen et al., 2019; Li et al., 2020b; Ma et al., 2020; Singh D. et al., 2021; Zheng et al., 2021; Zhou et al., 2021).
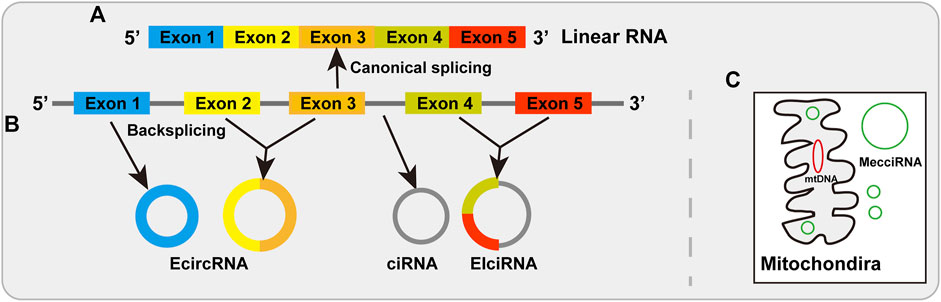
FIGURE 1. Biogenesis of linear RNAs and circRNAs. (A) Linear RNAs are produced by canonical splicing via joining an upstream 5′ splicing site to a downstream 3′ splicing site. (B) The biogenesis of circRNAs. EcircRNAs are produced by backsplicing and distribute predominantly in the cytoplasm. EIciRNAs are generated by intronic sequences retained between the backsplicing exons that are distributed mainly in the nucleus. CiRNAs from intronic lariat RNA precursors that distribute in the nucleus. (C) MecciRNAs are mitochondria-encoded circRNAs that are found in the cytosol and mitochondria.
With the development of study methods, our understanding of the general characteristics of circRNAs and their functions in normal physiology and most human diseases has been improved. Here, we focus on the recently described functional relevance of individual circRNAs to leukemia and their clinical significance. We first offer a brief introduction to the mechanisms of circRNAs, and then focus on recently described their roles and clinical significance in AML.
Mechanisms of circRNAs
Although the functions of most circRNAs are not fully explored, emerging evidence is beginning to uncover that dysregulated circRNAs play invital roles in many biological processes as regulatory noncoding RNAs, such as acting as microRNAs, regulating transcription, interacting with RNA binding proteins (Figures 2A–C). A part of circRNAs are also recognized as regulatory coding RNAs encoding small functional peptides (Figure 2D).
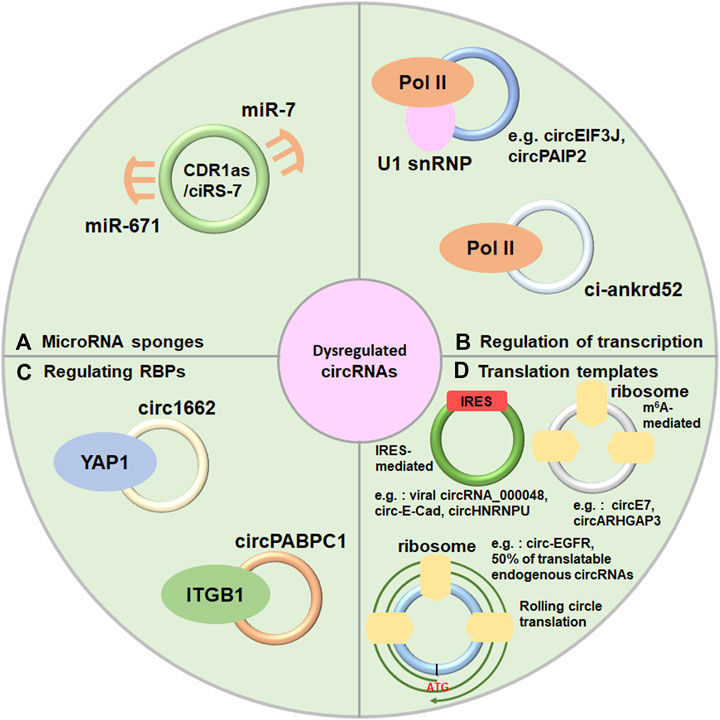
FIGURE 2. Diverse mechanisms of circRNAs. (A) Acting as microRNAs sponges, (B) Regulation of transcription. (C) Interacting with RNA binding proteins. (D) Serving as translation templates.
Acting as microRNA sponges
Numerous researches have indicated that circRNAs have one or more microRNA binding sites and serve as microRNA sponges to prevent microRNAs away from their target genes which have been well-documented by researchers (D'Ambra et al., 2019; Kristensen et al., 2019; Zhou et al., 2021). For instance, CDR1as/ciRS-7 is one of the classic and most researched circRNAs which contains one miR-671 binding site and over 60 conserved miR-7 binding sites (Memczak et al., 2013) (Figure 2A). In addition, circBIRC6 contains miR-145 and miR-34a binding sites to regulate human cell pluripotency (Yu et al., 2017). Some other circRNAs also can sponge microRNAs, despite the majority of them only have a limited amount of microRNA binding sites (Panda 2018; Qiu et al., 2021; Shen et al., 2022). Further researches are required to identify in what extend circRNAs could sponge microRNAs due to the lower expression of circRNAs.
Regulation of transcription
Nucleus-localized circRNAs (EIciRNAs and ciRNAs) and some EcircRNAs were proposed to play important role in transcriptional regulation. EIcircRNAs such as circEIF3J and circPAIP2, facilitate transcription initiation by RNA polymerasemer II (Pol II) at the promoter of host gene through recruiting U1 small nuclear ribonucleoprotein (U1 snRNP) in human cells (Li Z. et al., 2015) (Figure 2B). Furthermore, ci-ankrd52, a ciRNA, produces an R-loop in cis and facilitates transcription elongation via Pol II (Zhang et al., 2013; Li X. et al., 2021) (Figure 2B). Furthermore, some EcircRNAs also modulate transcription through interacting with chromatin. For instance, circFECR1 promotes FLI1 transcription in cis through recruting the TET1 (a demethylase) to result in DNA demethylation (Chen N. F. et al., 2018), and circSMARCA5 forms an R-loop by binding to its parent gene locus, resulting in transcriptional pausing at exon 15 of SMARCA5 (Xu et al., 2020).
Interacting with RNA binding proteins
Numerous studies indicated that circRNAs were also found to reveal different functions through directly interacting with different proteins (Holdt et al., 2016; Abdelmohsen et al., 2017; Li X. et al., 2017; Wu et al., 2019; Shen et al., 2020; Chen C. et al., 2021; Shi et al., 2021). For instance, Circ1662 accelerated the nuclear transport of TAP1 by restraining YAP1 phosphorylation (Chen C. et al., 2021) (Figure 2C). In a ubiquitination-dependent manner, circPABPC1 directly linked ITGB1 to the 26S proteasome for degradation in liver cancer (Shi et al., 2021) (Figure 2C). Furthermore, circECE1 prevented speckle-type POZ-mediated c-Myc ubiquitination and degradation through interacting with c-Myc in osteosarcoma (Shen et al., 2020). In addition, circYAP negatively modulated YAP expression by inhibiting the assembly of the YAP translation inititaion marinery in breast cancer cells (Wu et al., 2019).
Serving as translation template
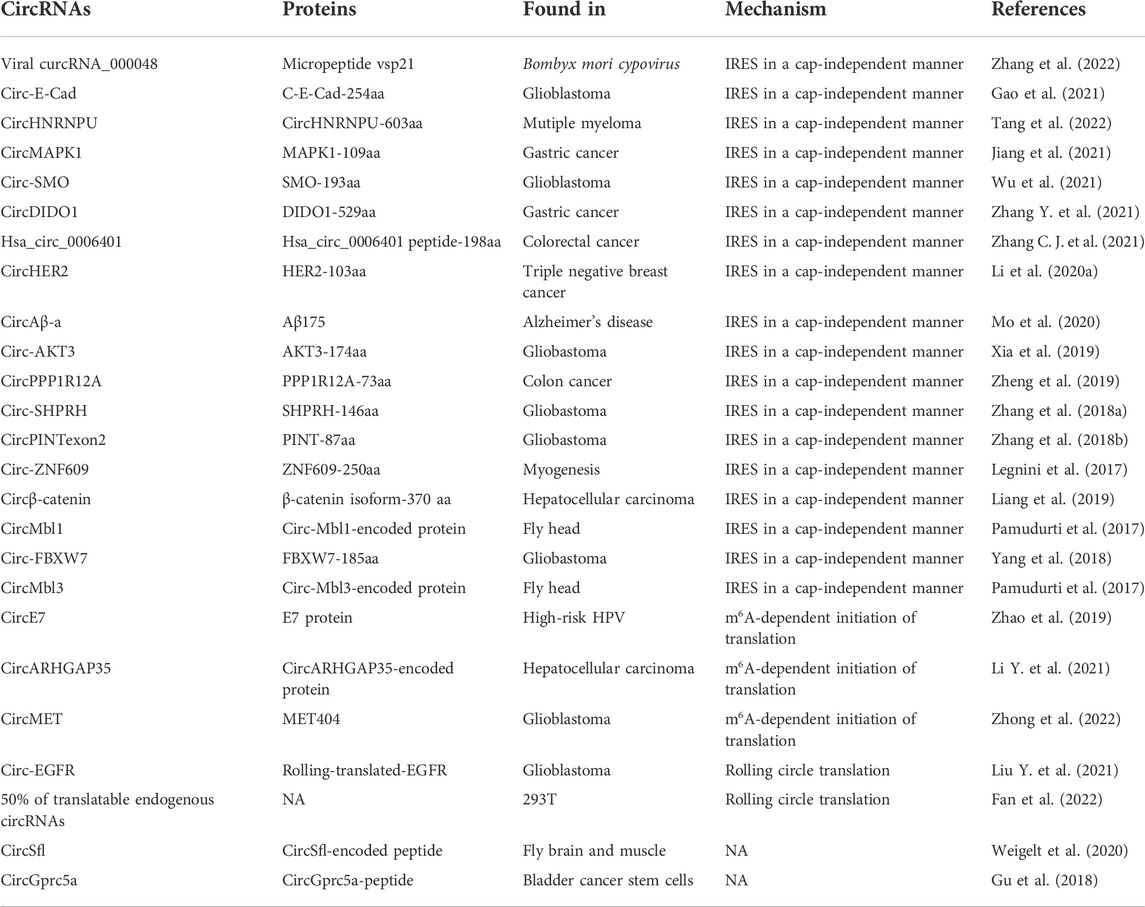
CircRNAs have been regarded as a class of non-coding RNAs for long periods of time. However, recent researches have revealed that circRNA can be translated into functional peptides (Legnini et al., 2017; Liu Y. et al., 2021; Wu et al., 2021; Liu et al., 2022; Zhang et al., 2022) (Table 1). CircRNAs could be translated into proteins through internal ribosome entry site (IRES) in a cap-independent manner, m6A-dependent initiation of translation and rolling circle translation (Figure 2D). For instance, endogenous circRNAs, such as viral circRNA_000048, circ-E-Cad and circHNRNPU, can translate to a micropeptide vsp21, C-E-Cad-254aa and MAPK1-109aa through IRES-dependent manner (Gao et al., 2021; Tang et al., 2022; Zhang et al., 2022) (Figure 2D), respectively. Moreover, circE7 and circARHGAP3 can translate to E7 protein and circARHGAP35-encoded protein by m6A-dependent initiation of translation (Zhao et al., 2019; Li Y. et al., 2021) (Figure 2D), respectively. Furthermore, circ-EGFR can translate to rolling-translated-EGFR through rolling circle translation (Liu Y. et al., 2021) (Figure 2D), respectively. In addition, Fan et al. recently reported that 50% of translatable endogenous circRNAs experience rolling circle translation, several of which are experimentally verfied (Fan et al., 2022) (Figure 2D). For instance, mutation of the IRES-like element (AAGAAG) in circPFAS decrease its translation (Fan et al., 2022).
The role of circRNAs in acute myeloid leukemia
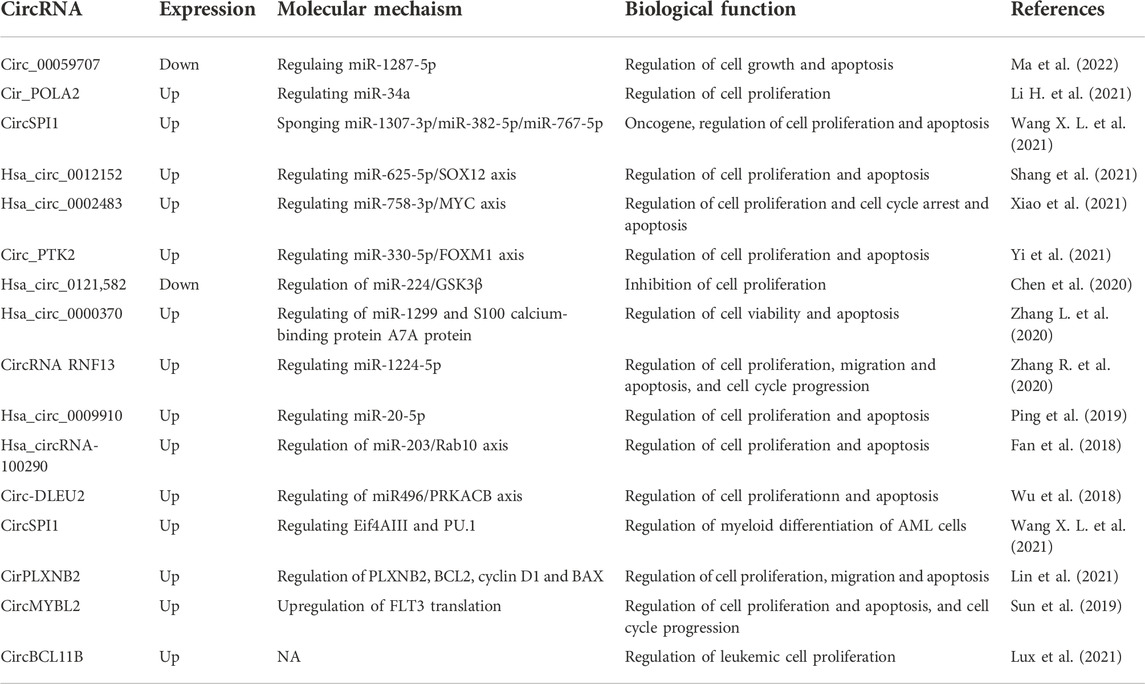
The role of circRNAs in AML biology and pathogenesis has been investigated (Table 2). Increasing evidenence shows that circRNAs play important role in gene expression and regulate distinct steps of leukemogenesis, such as differentiation, cell cycle progress, proliferation and apoptosis (Jamal et al., 2019; Singh V. et al., 2021). They also involve in drug resistance in AML chemotherapy (Shang et al., 2019; Li M. et al., 2020; Ding et al., 2021). The role of circRNAs in AML will be discussed in the following sections.
Dysregulation of circRNAs in acute myeloid leukemia and their association with acute myeloid leukemia phenotype
CircRNAs was first reported in viroids by Sanger et al., in 1976 (Sanger et al., 1976). With the development of high-throughput sequencing and increased research interest in circRNAs, many bioinformatics tools have been improved to study circRNAs in the past few years (Wang Y. et al., 2020; Rbbani et al., 2021; Yang et al., 2021). Especially, accumulating evidence demonstrates that circRNA expression is deregulated in AML compared with healthy control and reveals AML subgroup-specific signatures (Li W. et al., 2017; Chen H. L. et al., 2018; Lv et al., 2018; Lux et al., 2021; Wang J. H. et al., 2021). For instance, Lux et al. reported that hundreds of circRNAs were differentially expressed between 61 AML patients (including 20 NPM1mut patients, 25 CBF leukemias and 16 patients with mutations in splicing factors (PMSF)) and 16 healthy hematopoietic stem and progenitor cell samples (HSPCs) through using ribosomla RNA-depleted RNA sequencing (Lux et al., 2021) (Figure 3A). Their results showed that circRNA expression patterns are distinct in AML subgroups compared with healthy HSPCs. Many circRNA isoforms were deregulated in only one of the AML subgroups with 40%, 51% and 24% of the differentially expressed circRNAs in NPM1mut, CBF leukemia and PMSF, repecstively. Their results also showed that AML-related circRNA expression patterns are enriched for leukemia-relevant genes, such as JAATINEN_HEMATOPOIETIC_STEM_CELL_UP gene set, VERHAAK_AML_WITH_NPM1_MUTATED gene set and ROSS_AML_CBF gene set (Figure 3A).
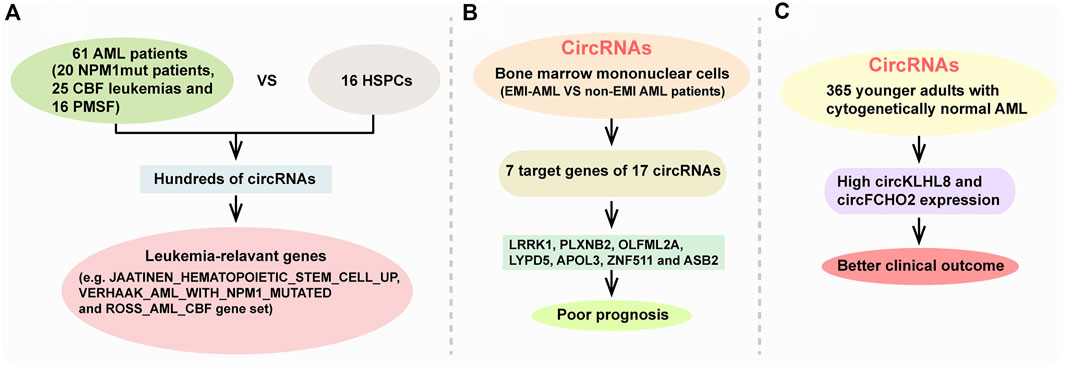
FIGURE 3. CircRNAs in AML and their association with AML phenotype. (A) AML-related circRNA expression patterns are enriched for leukemia-relevant gene. (B) seven target genes of 17 circRNAs (LRRK1, PLXNB2, OLFML2A, LYPD5, APOL3, ZNF511, and ASB2) indicated a poor prognosis. (C) High circKLHL8 and circFCHO2 expression were independently associated with better clinical outcome of cytogenetically normal AML patient.
AML can develop as myobasts infiltrate into organs and tissues anywhere other than the bone marrow, which called extramedullary infiltration (EMI), revealing a poor prognosis. Through comparing differentially expressed circRNAs in bone marrow mononuclear cells between EMI-AML and non-EMI AML patients, they found that seven target genes of 17 circRNAs (LRRK1, PLXNB2, OLFML2A, LYPD5, APOL3, ZNF511, and ASB2) revealed a poor prognosis (Lv et al., 2018) (Figure 3B). Through analyzing whole-transcriptome profiling of 365 younger adults with cytogenetically normal AML, another study identified three different circRNA expression-based clusters with distinct clinical and molecular characristics such as somatic mutations, differences in age and white blood cell count. They found that high circKLHL8 and circFCHO2 expression were independently associated with better clinical outcome of cytogenetically normal AML patients (Papaioannou et al., 2020) (Figure 3C). Above all, these circRNAs sequencing results highlight a potential involvement of circRNAs in the pathogenesis of AML. However, most researches have applied bone marrow samples, and only a few used peripheral blood samples. Correlative researches between bone marrow samples and peripheral blood are also limited.
CircRNAs regulte cell differentiation, cell cycle progression, and cell proliferation
AML is characterized by aberrant differentiation and abnormal clonal expansion of myeloid blasts (Newell and Cook 2021; Xiang et al., 2022). It has been reported that dysregulated circRNAs can regulate cell differentiation, cell cycle progression and cell proliferation through acting as microRNA sponges in various diseases (Xiao et al., 2021; Shi et al., 2022; Wang et al., 2022). The myelocytomatosis oncogene (MYC) is a typical leukemia-associated transcription factor and plays important role in leukemic cell growth, AML cell proliferation and apoptosis (Beyer et al., 2019; Li et al., 2020c). Hsa_circ_0002483 (circ_0002483) expression was increased in AML patients and cells (Table 2). Knockdown of circ_0002483 inhibited AML cell proliferation and facilitated cell cycle arrest and apoptosis by regulating miR-758-3p/MYC axis (Xiao et al., 2021) (Figure 4A). Shang et al. found that the expression of circ_0012152 was enhanced in AML tissues and cells, circ_0012152 knockdown inhibited cell proliferation, induced cell apoptosis and facilitated death in AML cells by regulating miR-625-5p/SOX12 axis (Shang et al., 2021) (Figure 4A). In addition, Lux et al. reported that circBCL11B exclusively expressed in AML patients but not detected in healthy hematopoietic stem and progenitor cell samples, inhibition of circBCL11B suppressed leukemic cell proliferation and led to enhanced cell death of leukemic cells (Lux et al., 2021). However, the molecular mechanisms of circBCL118-mediated function in AML need to be further investigated in future studies. Except for acting as microRNA sponges, circRNAs interacting with RNA binding proteins functionally to exhibit their roles in various diseases (Sun et al., 2019; Shen et al., 2020; Zhang Y. N. et al., 2020). While, the study about circRNAs interacting with RNA binding proteins to exert their functions in AML was limited. Sun et al. reported that circMYBL2 expression to be about 5-fold higher in AML patient samples with FLT3-ITD mutations (FLT3-ITD+) compared with those without FLT3-ITD mutation (FLT3-ITD-). CircMYBL2 suppressed cell apoptosis, increased cell proliferation, and promoted cell-cycle progression in FLT3-ITD+ leukemic cells but not FLT3-ITD- cells. Mechanistically, it increased translation of FLT3 kinase by promoting the PTBP1 binding to FLT3 messenger RNA (Sun et al., 2019) (Figure 4B). In addition, some circRNAs exert different biological functions through different mechanisms in various diseases (Xing et al., 2020; Wang X. L. et al., 2021; Yang Z. G. et al., 2022). Overexpression of circ-FOXO3 suppressed cell growth, migration and invasion through sponging miR-23 in esophageal squamous cell cancer (Xing et al., 2020), while circ-FOXO3 relieved blood-brain barrier by sequestering mTOR and E2F1 in ischemia/reperfusion injury (Yang Z. G. et al., 2022). While in AML, Wang et al. reported that silencing circSPI1 decreased myeloid differentiation of AML cells through interacting with the translation initiation factor eIF4AIII to inhibit PU.1 expression at the translation level. While, knockdown of it specially reduced cell proliferation and apoptosis through interacting with miR-1307-3p, miR-382-5p, and miR-767-5p (Wang X. L. et al., 2021) (Figure 4C).
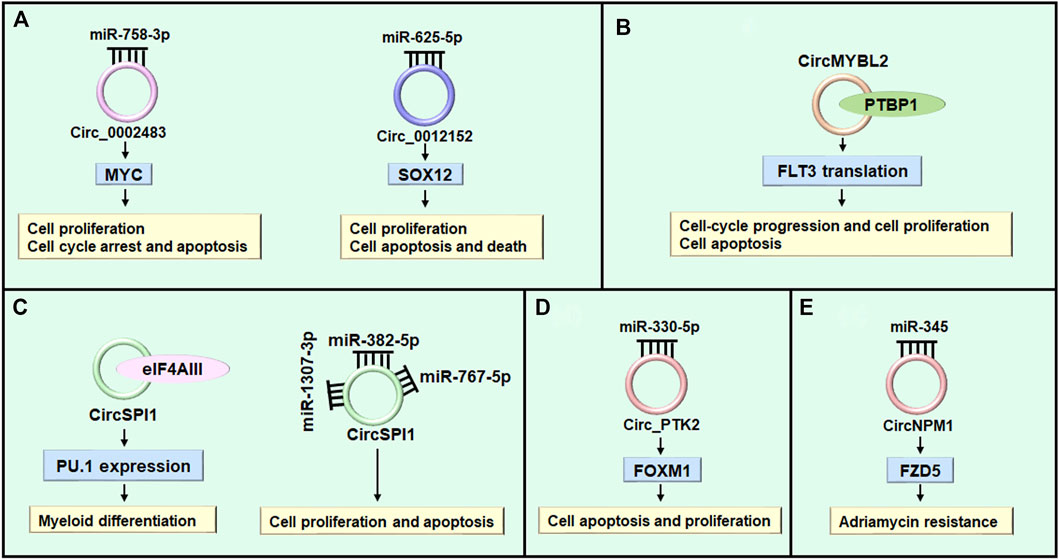
FIGURE 4. The role of circRNAs in AML. (A) Circ_0002483 and circ_0012152 regulate cell proliferation through sponging miRNAs. (B) CircMYBL2 modulates cell proliferation through regulating RBP (e.g. PTBP1). (C) CircSPI1 regulates myeloid differentiation of AML cells through interacting with the translation initiation factor eIF4AIII to inhibit PU.1 expression at the translation level. While, it regulates cell proliferation and apoptosis through sponging miR-1307-3p, miR-382-5p, and miR-767-5p. (D) CircPTK2 regulates cell apoptosis through modulating miR-330-5p/FOXM1 axis. (E) CircNPM1 reveals adriamycin resistance through regulating miR-345/FZD5 axis.
CircRNAs regulte cell apoptosis
Apoptosis, or programmed cell death, plays a key role in the development and homeostasis of the hematopoietic system (Testa and Riccioni 2007; Testa 2010). Although there are many factors contributing to the hematopoetic cell homeostasis, apoptotic machinery seems to have an important role (Droin et al., 2013). Recent studies indicated that circRNAs play vital role in cell apoptosis through sponging micrioRNAs in AML (Fan et al., 2018; Wu et al., 2018; Zhang L. et al., 2020; Wang X. L. et al., 2021; Xiao et al., 2021; Yi et al., 2021). Forkhead box M1 (FOXM1) functioned as an oncogene in cancers and can be regulated by multiple microRNAs in mang maliganancies (Gartel 2017; Hamurcu et al., 2021; Xing et al., 2021). While in AML, suppression of highly expressed-circ_PTK2 induced apoptosis and inhibited proliferation of AML cell by regulating miR-330-5p/FOXM1 axis (Yi et al., 2021) (Table 2) (Figure 4D). Hsa_circ-0000370 facilitated cell viability and inhibited apoptosis of FLT3-ITD-positive AML cells via modulating miR-1299 and S100 calcium-binding protein A7A expression (Zhang L. et al., 2020). CircRNA_100290 promoted cell proliferation and suppressed apoptosis in AML cells by regulating miR203/Rab10 axis (Fan et al., 2018). Furthermore, circRNA-DLEU2 was upregulated in AML tissues and cell, which accelerated AML cell proliferation and suppressed cell apoptosis through inhibiting miR-496 and facilitating PRKACB expression (Wu et al., 2018). In addition, Wang et al. reported that circ_0009910-containing exosomes regulated proliferation, apoptosis and cell cycle progression of AML cells partially by miR-5195-3p and GRB10 (Wang D. et al., 2021).
Relation between circRNAs and drug resistance in acute myeloid leukemia
Drug resistance is one of the key factors that lead to the failure of AML chemotherapy (Bester et al., 2018). Various genes and non-coding RNAs are participated in the development of drug resistance in AML (Tian et al., 2017; Xu et al., 2017; Hu et al., 2018; Gebru and Wang 2020; Chen X. et al., 2021; Kirtonia et al., 2022). Non-coding RNAs, such as microRNAs and lncRNAs, are regarding as vital players in regulating drug resistance, and their targeting provides avenues for the development of new treatment choices (Tian et al., 2017; Bester et al., 2018; Wang C. et al., 2020; Kirtonia et al., 2022). Nevertheless, studies on the potential involvement of aberrant expressed circRNAs in drug resistance of AML are just appearing. Ding et al. reported that circNPM1 increased adriamycin resistance in AML through regulating the miR-345/FZD5 pathway (Ding et al., 2021) (Figure 4E). Similarly, Shang et al. found that circPAN3 was increased in refractory and recurrent AML patient tissues and doxorubicin-resistant THP-1 AML cell lines than non-transformed tissue and THP-1 AML cell lines. Mechanistically, circPAN3 could be an important mediator of chemoresistance in AML cells by regulating miR-153-5p/miR-183-5p-XIAP (X-linked inhibitor of apoptosis) axis (Shang et al., 2019). Moreover, miR-153-5p and miR-183-5p were revealed to interact with XIAP, which has been indicated as a drug resistance gene in AML (Katragadda et al., 2013). In addition, overexpression of circPVT1 has also been found to involve in resistance to vincristine in AML (L'Abbate et al., 2018), and knockdown of fusion circM9 revealed enhanced sensitivity to anti-leukemic drugs (Guarnerio et al., 2016). These results suggest that circRNAs can potentially be applied to reverse drug resistance. However, the relation between circRNAs and other drugs in AML needs to be further investigated.
In conclusion, circRNAs play important role in regulating cell differentiation, cell cycle progress, proliferation and apoptosis, as well as involve in drug resistance through acting as microRNA sponges or interacting with RNA binding proteins in AML. As discussed in above, circRNAs also can regulate gene transcription and serve as translation template to exert their function. However, whether circRNAs exert their function through regulating gene transcription and serving as translation template in AML need to be further explored.
Clinical significance of circRNAs in acute myeloid leukemia
CircRNAs have the potential to be diagostic and prognostic biomarkers, and therapeutic targets because that they are highly stable, cell- and tissue-specific expressed, and their expression levels often associated with clinical and pathological characteristics (D'Ambra et al., 2019; Li et al., 2020b; Wang Y. et al., 2020). Different molecular-based biomarkers such as cytogenetics, epigenetics, genetics, noncoding RNAs and protemocis have been well-documented in AML (Trino et al., 2018; Thakral et al., 2020; Ribeiro et al., 2021; Kirtonia et al., 2022; Wiatrowski et al., 2022). CircRNAs act as tumor suppressors or oncogenes to involve in the development of various diseases such as AML and are becoming new diagnostic and prognostic biomarkers (Zhou et al., 2020; Issah et al., 2021; Singh V. et al., 2021) (Table 3). Li et al. reported that hsa_circ0004277 might be a potential diagnostic marker through evaluating its expression in 115 AML patients samples and increasing level of hsa_circ0004277 was associated with successful chemotherapy (Li W. et al., 2017) (Figure 5A). Lin et al. found that enhanced circPLXNB2 levels were related to an obviously shorter overall survival and leukaemia-free survival of patients with AML. Their study highlights the potential of circPLXNB2 as a novel prognostic marker and therapeutic target for AML in the future (Lin et al., 2021) (Figure 5A). In other studies, they found that hsa_circ_0075451, circ-VIM and circ_0009910 can serve as important prognostic factor in AML, respectively (Ping et al., 2019; Yi et al., 2019; Wang J. H. et al., 2021). Furthermore, Zhou et al. reported that circ-Foxo3 and Foxo3 expressed low in AML patients compared to control group and patients with high expression of Foxo3 often revealed a trend of better prognosis (Zhou et al., 2019) (Figure 5A). In addition, Liu et al. recently found that circRNF220 was specifically enriched in the peripheral blood and bone marrow of pediatric patients with AML. CircRNF220 could distinguish AML from acute lymphoblastic leukemia and other hematological malignancies with high sensitivity and specificity (Figure 5A). CircRNF220 expression independently predicted prognosis, while high expression of circRNF220 was unsuitable prognostic marker for relapse (Liu X. et al., 2021) (Figure 5A).
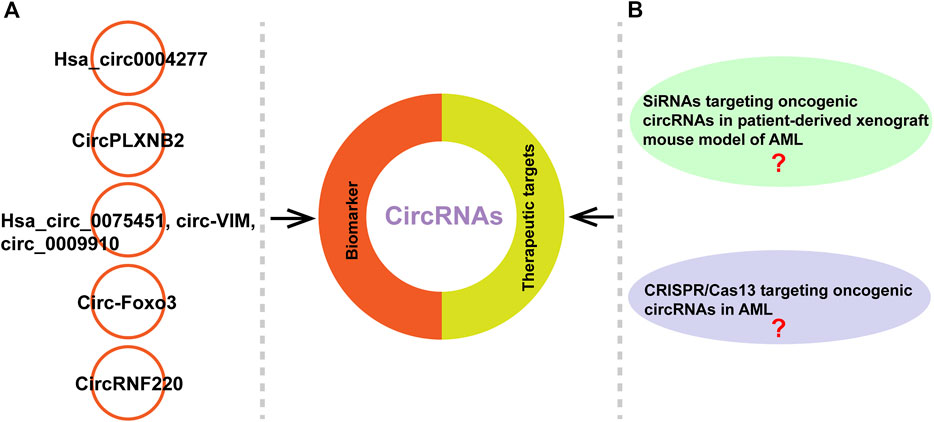
FIGURE 5. CircRNAs can serve as biomarkers or therapeutic targets. (A) Examples of circRNAs as potential biomarkers in AML. (B) The studies about siRNAs targeting oncogenic circRNAs in patient-derived xenograft mouse model of AML and CRISPR/Cas13 targeting oncogenic circRNAs in AML remain unclear.
Increasing reports in patient-derived xenograft mouse model indicated that the siRNAs specifically targeting oncogenic circRNAs can effectively suppress tumor growth (Meng et al., 2018; Zhang et al., 2019; Du et al., 2022). Meng et al. found that silencing of si-circ-10720 via intravenous injection inhibited the promotive effect on tumor growth and metastasis in a mouse hepatocellilar carcinoma model (Meng et al., 2018). Another study reported that knockdown of circNRIP1 using siRNA suppressed the proliferation, migration and invasion of gastric cancer (GC) cells in vitro and blocked tumor growth in GC-patient-derived xgenograft mouse model (Zhang et al., 2019). Recently, Du et al. reported that poly (β-amino esters)-delivered circMDK siRNA significantly inhibited the growth of hepatocellular carinoma through reducing the expression of ATG16L1 in patient-derived xenograft mouse model, suggesting that the oncogenic circMDK may be a potential treatment targtet for hepatocellular carinoma (Du et al., 2022). An interference RNA can be designed to precisely target the unique back-spliced junction of oncogenc circRNA in cancers in order to eliminate the possibility of interference with the expression of parent linear mRNA. In AML, currently, some circRNAs have been reported as oncogenic circRNAs (Ping et al., 2019; Zhang R. et al., 2020; Wang X. L. et al., 2021). Ping et al. found that circ_0009910 acting as oncogene in AML patients and knockdown of it suppressed AML cell proliferation and resulted in cell apoptosis (Ping et al., 2019). Another study reported that circRNF12 as an oncogene in blood of AML patients and interference of it reduced the migration and invasion ability of AML cells (Zhang R. et al., 2020). Wang et al. found another circRNA, circSPl1 also as an oncogene in AML, evidenced by the results that knockdown of circSPl1 induced apoptosis of AML cells (Wang X. L. et al., 2021). Although some circRNAs acting as oncogene has been reported in AML, the study about siRNAs targeting oncogenic circRNAs in patient-derived xenograft mouse model of AML needs to be explored in the future researches (Figure 5B). In addition, it has been reported that CRISPR-Cas13 system can be applied to knock down circRNAs, without any influence on related mRNAs (Koch 2021). This method has been used to few studies. For instance, Li et al. reported that knockdown circFAM120A (oncogenic circRNA) by CRISPR-RfxCas13d system promoted cell proliferation by inhibiting FAM120A from binding the translation inhibitor IGF2BP2 in 293FT cells (Li S. Q. et al., 2021). Ishola et al. found that knockdown of hsa_circ_0000190 using CRISPR/Cas13a inhibited tumor growth in vivo non-small cell lung cancer xenograft model (Ishola et al., 2022). However, the study about CRISPR/Cas13 targeting oncogenic circRNAs in AML needs to be investigated in the future (Figure 5B).
Conclusion and perspective
AML is a malignant tumor characterized by the accumulation and clonal expansion of the immature myeloid hematopoietic cells in the bone marrow, with rising morbidity and mortality (Liu et al., 2019). Although advances in AML molecular characterization and targeted methods, most AML cases still lack therapeutically actionable targets and long-term survival remains low (Decroocq et al., 2022; Pabon et al., 2022). Therefore, it is necessary to discover new biomarkers for prognostication, diagnosis, and therapeutic targets of AML to explore more effective surveillance and treatment programs. It has been reported that circRNAs could regulate cell differentiation, cell cycle progress, proliferation and apoptosis, as well as involve in drug resistance in AML through acting as microRNA sponges or interacting with RNA binding proteins. However, whether circRNAs exhibit their biological function through regulating gene transcription or serving as translation template in AML need to be further investigated. Moreover, the accurate mechanism of modulation of circRNAs expression in AML is not well researched. It is not clear if abnormal circRNA expression is central event in leukemogenesis or an epiphenomenon. Most researches have applied bone marrow samples, and only a few used peripheral blood samples. Correlative researches between bone marrow samples and peripheral blood are also limited.
Recently, Qu et al. reported that circRNA vaccine successfully elicited potent neutralizing antibodies and T cell response by encoding the trimeric receptor-binding domain of SARS-CoV-2 spike protein (Qu et al., 2022). Their results suggested that the synthesis of translatable circRNAs is of great value in the field of biomedicine. Moreover, Chen et al. recently found high-efficiency method to enhance circRNA protein yields by several hundred-fold by optimizing five functional elements controlling circRNA translation including IRES, 5′ and 3’ UTRs, vector topology and synthetic aptamers (Chen R. et al., 2022). Their results enable potent and durable protein production by translatable circRNA in vivo. However, whether translatable circRNAs could applied to the treatment of AML required to be further investigated. Furthermore, increasing evidence indicates the siRNAs specifically targeting oncogenic circRNAs can effectively suppress tumor growth in patient-derived xenograft mouse model (Huang et al., 2020; Yang et al., 2020; Liang et al., 2021). However, siRNAs targeting oncogenic circRNAs in patient-derived xenograft mouse model of AML needs to be explored in the future study. In addition, it has been reported that CRISPR-Cas13 sysrem can be used to knock down circRNAs to explore the function of circRNAs (Li S. Q. et al., 2021; Ishola et al., 2022). However, the study about CRISPR/Cas13 targeting oncogenic circRNAs in AML needs to be investigated in the future.
Our understanding of the metabolism and transport of circRNA within and outside the cell is also lacking. It has been reported that excessive circRNAs are transported out of the cell in exosomes (Li Y. et al., 2015). This is also of great interest because it is well documented in other cancers (Wang et al., 2019; Pan et al., 2022; Yang C. et al., 2022). However, in AML, exosomal circRNAs are few been explored. The use of exosomal cirRNAs in regulating bone marrow microenvironment and extreamedullary infiltration of leukemia cells can be an interest field to research.
In summary, although great progress of circRNAs in AML has been achieved, substantial efforts are still needed to find whether circRNAs exert their biological function by other mechainsms such as regulation of gene transcription or serving as translation template in AML. It is also urgent that scientists study the machineries regulating circRNAs fate, the downstream effectors of circRNAs modulatory networks, and the clinical application of circRNAs in AML. Better understanding of these will promote our knowledge of circRNAs in AML biology and the development of circRNAs-based diagnosis, prognosis and therapeutic methods for AML.
Author contributions
All authors listed have made a substantial, direct, and intellectual contribution to the work and approved it for publication.
Funding
This research was funded by the Natural Science Foundation of Chongqing, China, Grant number: cstc2021jcyj-bshX0121.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Abdelmohsen, K., Panda, A. C., Munk, R., Grammatikakis, I., Dudekula, D. B., De, S., et al. (2017). Identification of HuR target circular RNAs uncovers suppression of PABPN1 translation by CircPABPN1. RNA Biol. 14, 361–369. doi:10.1080/15476286.2017.1279788
Almatani, M. F., Ali, A., Onyemaechi, S., Zhao, Y., Gutierrez, L., Vaikari, V. P., et al. (2021). Strategies targeting FLT3 beyond the kinase inhibitors. Pharmacol. Ther. 225, 107844. doi:10.1016/j.pharmthera.2021.107844
Andreozzi, F., Massaro, F., Wittnebel, S., Spilleboudt, C., Lewalle, P., and Salaroli, A. (2022). New perspectives in treating acute myeloid leukemia: Driving towards a patient-tailored strategy. Int. J. Mol. Sci. 23, 3887. doi:10.3390/Ijms23073887
Aung, M. M. K., Mills, M. L., Bittencourt-Silvestre, J., and Keeshan, K. (2021). Insights into the molecular profiles of adult and paediatric acute myeloid leukaemia. Mol. Oncol. 15, 2253–2272. doi:10.1002/1878-0261.12899
Bester, A. C., Lee, J. D., Chavez, A., Lee, Y. R., Nachmani, D., Vora, S., et al. (2018). An integrated genome-wide CRISPRa approach to functionalize lncRNAs in drug resistance. Cell 173, 649–664. doi:10.1016/j.cell.2018.03.052
Beyer, M., Romanski, A., Mustafa, A. H. M., Pons, M., Buchler, I., Vogel, A., et al. (2019). HDAC3 activity is essential for human leukemic cell growth and the expression of beta-catenin, MYC, and WT1. Cancers 11, E1436. doi:10.3390/Cancers11101436
Bhattacharya, M., and Gutti, R. K. (2022). Non-coding RNAs: Are they the protagonist or antagonist in the regulation of leukemia? Am. J. Transl. Res. 14, 1406–1432.
Chen, C., Yuan, W. T., Zhou, Q. B., Shao, B., Guo, Y., Wang, W., et al. (2021). N6-methyladenosine-induced circ1662 promotes metastasis of colorectal cancer by accelerating YAP1 nuclear localization. Theranostics 11, 4298–4315. doi:10.7150/thno.51342
Chen, H. L., Liu, T., Liu, J., Feng, Y., Wang, B., Wang, J., et al. (2018). Circ-ANAPC7 is upregulated in acute myeloid leukemia and appears to target the miR-181 family. Cell. Physiol. biochem. 47, 1998–2007. doi:10.1159/000491468
Chen, J. J., Lei, P., and Zhou, M. (2020). hsa_circ_0121582 inhibits leukemia growth by dampening Wnt/β-catenin signaling. Clin. Transl. Oncol. 22, 2293–2302. doi:10.1007/s12094-020-02377-9
Chen, L., Huang, C., and Shan, G. (2022). Circular RNAs in physiology and non-immunological diseases. Trends biochem. Sci. 47, 250–264. doi:10.1016/j.tibs.2021.11.004
Chen, L., Huang, C., Wang, X., and Shan, G. (2015). Circular RNAs in eukaryotic cells. Curr. Genomics 16, 312–318. doi:10.2174/1389202916666150707161554
Chen, L. L. (2020). The expanding regulatory mechanisms and cellular functions of circular RNAs. Nat. Rev. Mol. Cell Biol. 21, 475–490. doi:10.1038/s41580-020-0243-y
Chen, N. F., Zhao, G., Yan, X., Lv, Z., Yin, H., Zhang, S., et al. (2018). A novel FLI1 exonic circular RNA promotes metastasis in breast cancer by coordinately regulating TET1 and DNMT1. Genome Biol. 19, 218. doi:10.1186/S13059-018-1594-Y
Chen, R., Wang, S. K., Belk, J. A., Amaya, L., Li, Z., Cardenas, A., et al. (2022). Engineering circular RNA for enhanced protein production. Nat. Biotechnol. Online ahead of print. doi:10.1038/s41587-022-01393-0
Chen, X., Chen, X., Huang, Y., Lin, J., Wu, Y., and Chen, Y. (2021). TCP1 increases drug resistance in acute myeloid leukemia by suppressing autophagy via activating AKT/mTOR signaling. Cell Death Dis. 12, 1058. doi:10.1038/s41419-021-04336-w
Cocquerelle, C., Mascrez, B., Hetuin, D., and Bailleul, B. (1993). Mis-splicing yields circular RNA molecules. FASEB J. 7, 155–160. doi:10.1096/fasebj.7.1.7678559
Conn, V. M., Hugouvieux, V., Nayak, A., Conos, S. A., Capovilla, G., Cildir, G., et al. (2017). A circRNA from SEPALLATA3 regulates splicing of its cognate mRNA through R-loop formation. Nat. Plants 3, 17053. doi:10.1038/Nplants.2017.53
D'Ambra, E., Capauto, D., and Morlando, M. (2019). Exploring the regulatory role of circular RNAs in neurodegenerative disorders. Int. J. Mol. Sci. 20, E5477. doi:10.3390/Ijms20215477
Decroocq, J., Birsen, R., Montersino, C., Chaskar, P., Mano, J., Poulain, L., et al. (2022). RAS activation induces synthetic lethality of MEK inhibition with mitochondrial oxidative metabolism in acute myeloid leukemia. Leukemia 36, 1237–1252. doi:10.1038/s41375-022-01541-0
Ding, J., Zhang, X. C., Xue, J. A., Fang, L., Ban, C., Song, B., et al. (2021). CircNPM1 strengthens Adriamycin resistance in acute myeloid leukemia by mediating the miR-345-5p/FZD5 pathway. Cent. Eur. J. Immunol. 46, 162–182. doi:10.5114/ceji.2021.108175
Droin, N., Guery, L., Benikhlef, N., and Solary, E. (2013). Targeting apoptosis proteins in hematological malignancies. Cancer Lett. 332, 325–334. doi:10.1016/j.canlet.2011.06.016
Du, A. S., Li, S. Q., Zhou, Y. Z., Disoma, C., Liao, Y., Zhang, Y., et al. (2022). M6A-mediated upregulation of circMDK promotes tumorigenesis and acts as a nanotherapeutic target in hepatocellular carcinoma. Mol. Cancer 21, 109. doi:10.1186/s12943-022-01575-z
Fan, H., Li, Y., Liu, C., Bai, J., and Li, W. (2018). Circular RNA-100290 promotes cell proliferation and inhibits apoptosis in acute myeloid leukemia cells via sponging miR-203. Biochem. Biophys. Res. Commun. 507, 178–184. doi:10.1016/j.bbrc.2018.11.002
Fan, X., Yang, Y., Chen, C., and Wang, Z. (2022). Pervasive translation of circular RNAs driven by short IRES-like elements. Nat. Commun. 13, 3751. doi:10.1038/s41467-022-31327-y
Gallipoli, P., Giotopoulos, G., and Huntly, B. J. (2015). Epigenetic regulators as promising therapeutic targets in acute myeloid leukemia. Ther. Adv. Hematol. 6, 103–119. doi:10.1177/2040620715577614
Gao, X. Y., Xia, X., Li, F. Y., Zhang, M., Zhou, H., Wu, X., et al. (2021). Circular RNA-encoded oncogenic E-cadherin variant promotes glioblastoma tumorigenicity through activation of EGFR-STAT3 signalling. Nat. Cell Biol. 23, 278–291. doi:10.1038/s41556-021-00639-4
Gartel, A. L. (2017). FOXM1 in cancer: Interactions and vulnerabilities. Cancer Res. 77, 3135–3139. doi:10.1158/0008-5472.CAN-16-3566
Gebru, M. T., and Wang, H. G. (2020). Therapeutic targeting of FLT3 and associated drug resistance in acute myeloid leukemia. J. Hematol. Oncol. 13, 155. doi:10.1186/s13045-020-00992-1
Gu, C. H., Zhou, N. C., Wang, Z. Y., Li, G., Kou, Y., Yu, S., et al. (2018). circGprc5a promoted bladder oncogenesis and metastasis through Gprc5a-targeting peptide. Mol. Ther. Nucleic Acids 13, 633–641. doi:10.1016/j.omtn.2018.10.008
Guarnerio, J., Bezzi, M., Jeong, J. C., Paffenholz, S. V., Berry, K., Naldini, M. M., et al. (2016). Oncogenic role of fusion-circRNAs derived from cancer-associated chromosomal translocations. Cell 165, 289–302. doi:10.1016/j.cell.2016.03.020
Hamurcu, Z., Sener, E. F., Taheri, S., Nalbantoglu, U., Kokcu, N. D., Tahtasakal, R., et al. (2021). MicroRNA profiling identifies Forkhead box transcription factor M1 (FOXM1) regulated miR-186 and miR-200b alterations in triple negative breast cancer. Cell. Signal. 83, 109979. doi:10.1016/j.cellsig.2021.109979
Holdt, L. M., Stahringer, A., Sass, K., Pichler, G., Kulak, N. A., Wilfert, W., et al. (2016). Circular non-coding RNA ANRIL modulates ribosomal RNA maturation and atherosclerosis in humans. Nat. Commun. 7, 12429. doi:10.1038/ncomms12429
Hu, X., Mei, S., Meng, W., Xue, S., Jiang, L., Yang, Y., et al. (2018). CXCR4-mediated signaling regulates autophagy and influences acute myeloid leukemia cell survival and drug resistance. Cancer Lett. 425, 1–12. doi:10.1016/j.canlet.2018.03.024
Huang, Q., Guo, H., Wang, S., Ma, Y., Chen, H., Li, H., et al. (2020). A novel circular RNA, circXPO1, promotes lung adenocarcinoma progression by interacting with IGF2BP1. Cell Death Dis. 11, 1031. doi:10.1038/s41419-020-03237-8
Ishola, A. A., Chien, C. S., Yang, Y. P., Chien, Y., Yarmishyn, A. A., Tsai, P. H., et al. (2022). Oncogenic circRNA C190 promotes non-small cell lung cancer via modulation of the EGFR/ERK pathway. Cancer Res. 82, 75–89. doi:10.1158/0008-5472.CAN-21-1473
Issah, M. A., Wu, D., Zhang, F., Zheng, W., Liu, Y., Fu, H., et al. (2021). Epigenetic modifications in acute myeloid leukemia: The emerging role of circular RNAs (Review). Int. J. Oncol. 59, 107. doi:10.3892/ijo.2021.5287
Jamal, M., Song, T., Chen, B., Faisal, M., Hong, Z., Xie, T., et al. (2019). Recent progress on circular RNA research in acute myeloid leukemia. Front. Oncol. 9, 1108. doi:10.3389/fonc.2019.01108
Jeck, W. R., Sorrentino, J. A., Wang, K., Slevin, M. K., Burd, C. E., Liu, J., et al. (2013). Circular RNAs are abundant, conserved, and associated with ALU repeats. RNA 19, 141–157. doi:10.1261/rna.035667.112
Jiang, T. L., Xia, Y. W., Lv, J. L., Li, B., Li, Y., Wang, S., et al. (2021). A novel protein encoded by circMAPK1 inhibits progression of gastric cancer by suppressing activation of MAPK signaling. Mol. Cancer 20, 66. doi:10.1186/S12943-021-01358-Y
Katragadda, L., Carter, B. Z., and Borthakur, G. (2013). XIAP antisense therapy with AEG 35156 in acute myeloid leukemia. Expert Opin. Investig. Drugs 22, 663–670. doi:10.1517/13543784.2013.789498
Kirtonia, A., Ashrafizadeh, M., Zarrabi, A., Hushmandi, K., Zabolian, A., Bejandi, A. K., et al. (2022). Long noncoding RNAs: A novel insight in the leukemogenesis and drug resistance in acute myeloid leukemia. J. Cell. Physiol. 237, 450–465. doi:10.1002/jcp.30590
Koch, L. (2021). CRISPR-Cas13 targets circRNAs. Nat. Rev. Genet. 22, 68. doi:10.1038/s41576-020-00318-4
Kristensen, L. S., Andersen, M. S., Stagsted, L. V. W., Ebbesen, K. K., Hansen, T. B., and Kjems, J. (2019). The biogenesis, biology and characterization of circular RNAs. Nat. Rev. Genet. 20, 675–691. doi:10.1038/s41576-019-0158-7
L'Abbate, A., Tolomeo, D., Cifola, I., Severgnini, M., Turchiano, A., Augello, B., et al. (2018). MYC-Containing amplicons in acute myeloid leukemia: Genomic structures, evolution, and transcriptional consequences. Leukemia 32, 2152–2166. doi:10.1038/s41375-018-0033-0
Legnini, I., Di Timoteo, G., Rossi, F., Morlando, M., Briganti, F., Sthandier, O., et al. (2017). Circ-ZNF609 is a circular RNA that can be translated and functions in myogenesis. Mol. Cell 66, 22–37. doi:10.1016/j.molcel.2017.02.017
Li, H., Bi, K. H., Feng, S. R., Wang, Y., and Zhu, C. (2021). CircRNA circ_POLA2 is upregulated in acute myeloid leukemia (AML) and promotes cell proliferation by suppressing the production of mature miR-34a. Cancer Manag. Res. 13, 3629–3637. doi:10.2147/Cmar.S281690
Li, J., Ma, M., Yang, X., Zhang, M., Luo, J., Zhou, H., et al. (2020a). Circular HER2 RNA positive triple negative breast cancer is sensitive to Pertuzumab. Mol. Cancer 19, 142. doi:10.1186/s12943-020-01259-6
Li, J., Sun, D., Pu, W., Wang, J., and Peng, Y. (2020b). Circular RNAs in cancer: Biogenesis, function, and clinical significance. Trends Cancer 6, 319–336. doi:10.1016/j.trecan.2020.01.012
Li, J., Wang, M., and Chen, X. (2020c). Long non-coding RNA UCA1 modulates cell proliferation and apoptosis by regulating miR-296-3p/Myc axis in acute myeloid leukemia. Cell Cycle 19, 1454–1465. doi:10.1080/15384101.2020.1750814
Li, M., Meng, F., and Lu, Q. (2020). Expression profile screening and bioinformatics analysis of circRNA, LncRNA, and mRNA in acute myeloid leukemia drug-resistant cells. Turk. J. Haematol. 37, 104–110. doi:10.4274/tjh.galenos.2019.2019.0312
Li, S. Q., Li, X., Xue, W., Zhang, L., Yang, L. Z., Cao, S. M., et al. (2021). Screening for functional circular RNAs using the CRISPR-Cas13 system. Nat. Methods 18: 51–59. doi:10.1038/s41592-020-01011-4
Li, W., Zhong, C., Jiao, J., Li, P., Cui, B., Ji, C., et al. (2017). Characterization of hsa_circ_0004277 as a new biomarker for acute myeloid leukemia via circular RNA profile and bioinformatics analysis. Int. J. Mol. Sci. 18, E597. doi:10.3390/Ijms18030597
Li, X., Liu, C. X., Xue, W., Zhang, Y., Jiang, S., Yin, Q. F., et al. (2017). Coordinated circRNA biogenesis and function with NF90/NF110 in viral infection. Mol. Cell 67, 214–227. doi:10.1016/j.molcel.2017.05.023
Li, X., Zhang, J. L., Lei, Y. N., Liu, X. Q., Xue, W., Zhang, Y., et al. (2021). Linking circular intronic RNA degradation and function in transcription by RNase H1. Sci. China. Life Sci. 64, 1795–1809. doi:10.1007/s11427-021-1993-6
Li, Y., Chen, B., Zhao, J., Li, Q., Chen, S., Guo, T., et al. (2021). HNRNPL circularizes ARHGAP35 to produce an oncogenic protein. Adv. Sci. 8, 2001701. doi:10.1002/advs.202001701
Li, Y., Zheng, Q., Bao, C., Li, S., Guo, W., Zhao, J., et al. (2015). Circular RNA is enriched and stable in exosomes: A promising biomarker for cancer diagnosis. Cell Res. 25, 981–984. doi:10.1038/cr.2015.82
Li, Z., Huang, C., Bao, C., Chen, L., Lin, M., Wang, X., et al. (2015). Exon-intron circular RNAs regulate transcription in the nucleus. Nat. Struct. Mol. Biol. 22, 256–264. doi:10.1038/nsmb.2959
Liang, W. C., Wong, C. W., Liang, P. P., Shi, M., Cao, Y., Rao, S. T., et al. (2019). Translation of the circular RNA circβ-catenin promotes liver cancer cell growth through activation of the Wnt pathway. Genome Biol. 20, 84. doi:10.1186/s13059-019-1685-4
Liang, Z. X., Liu, H. S., Xiong, L., Yang, X., Wang, F. W., Zeng, Z. W., et al. (2021). A novel NF-κB regulator encoded by circPLCE1 inhibits colorectal carcinoma progression by promoting RPS3 ubiquitin-dependent degradation. Mol. Cancer 20, 103. doi:10.1186/s12943-021-01404-9
Lin, L. L., Wang, Y., Bian, S. C., Sun, L., Guo, Z., Kong, D., et al. (2021). A circular RNA derived from PLXNB2 as a valuable predictor of the prognosis of patients with acute myeloid leukaemia. J. Transl. Med. 19, 123. doi:10.1186/s12967-021-02793-7
Liu, C., Wu, X., Gokulnath, P., Li, G., and Xiao, J. (2022). The functions and mechanisms of translatable circRNAs. J. Pharmacol. Exp. Ther. JPET-MR, 2022-001085. doi:10.1124/jpet.122.001085
Liu, X., Liu, X., Cai, M., Luo, A., He, Y., Liu, S., et al. (2021). CircRNF220, not its linear cognate gene RNF220, regulates cell growth and is associated with relapse in pediatric acute myeloid leukemia. Mol. Cancer 20, 139. doi:10.1186/s12943-021-01395-7
Liu, X., Wang, X. L., Li, J. X., Hu, S., Deng, Y., Yin, H., et al. (2020). Identification of mecciRNAs and their roles in the mitochondrial entry of proteins. Sci. China. Life Sci. 63, 1429–1449. doi:10.1007/s11427-020-1631-9
Liu, Y., Cheng, Z. H., Pang, Y. F., Cui, L., Qian, T., Quan, L., et al. (2019). Role of microRNAs, circRNAs and long noncoding RNAs in acute myeloid leukemia. J. Hematol. Oncol. 12, 51. doi:10.1186/s13045-019-0734-5
Liu, Y., Li, Z., Zhang, M., Zhou, H., Wu, X., Zhong, J., et al. (2021). Rolling-translated EGFR variants sustain EGFR signaling and promote glioblastoma tumorigenicity. Neuro. Oncol. 23, 743–756. doi:10.1093/neuonc/noaa279
Lux, S., Blatte, T. J., Gillissen, B., Richter, A., Cocciardi, S., Skambraks, S., et al. (2021). Deregulated expression of circular RNAs in acute myeloid leukemia. Blood Adv. 5, 1490–1503. doi:10.1182/bloodadvances.2020003230
Lv, C. F., Sun, L. L., Guo, Z. B., Li, H., Kong, D., Xu, B., et al. (2018). Circular RNA regulatory network reveals cell-cell crosstalk in acute myeloid leukemia extramedullary infiltration. J. Transl. Med. 16, 361. doi:10.1186/S12967-018-1726-X
Ma, J., Wen, X., Xu, Z., Xia, P., Jin, Y., Lin, J., et al. (2022). The down-regulation of circ_0059707 in acute myeloid leukemia promotes cell growth and inhibits apoptosis by regulating miR-1287-5p. Curr. Oncol. 29, 6688–6699. doi:10.3390/curroncol29090525
Ma, S., Kong, S., Wang, F., and Ju, S. (2020). CircRNAs: Biogenesis, functions, and role in drug-resistant tumours. Mol. Cancer 19, 119. doi:10.1186/s12943-020-01231-4
Memczak, S., Jens, M., Elefsinioti, A., Torti, F., Krueger, J., Rybak, A., et al. (2013). Circular RNAs are a large class of animal RNAs with regulatory potency. Nature 495, 333–338. doi:10.1038/nature11928
Meng, J., Chen, S., Han, J. X., Qian, B., Wang, X. R., Zhong, W. L., et al. (2018). Twist1 regulates vimentin through Cul2 circular RNA to promote EMT in hepatocellular carcinoma. Cancer Res. 78, 4150–4162. doi:10.1158/0008-5472.can-17-3009
Mo, D., Li, X., Raabe, C. A., Rozhdestvensky, T. S., Skryabin, B. V., and Brosius, J. (2020). Circular RNA encoded amyloid beta peptides-a novel putative player in Alzheimer's disease. Cells 9, 2196. doi:10.3390/Cells9102196
Newell, L. F., and Cook, R. J. (2021). Advances in acute myeloid leukemia. BMJ 375, n2026. doi:10.1136/bmj.n2026
Nigro, J. M., Cho, K. R., Fearon, E. R., Kern, S. E., Ruppert, J. M., Oliner, J. D., et al. (1991). Scrambled exons. Cell 64: 607–613. doi:10.1016/0092-8674(91)90244-S
Pabon, C. M., Abbas, H. A., and Konopleva, M. (2022). Acute myeloid leukemia: Therapeutic targeting of stem cells. Expert Opin. Ther. Targets 26, 547–556. doi:10.1080/14728222.2022.2083957
Pamudurti, N. R., Bartok, O., Jens, M., Ashwal-Fluss, R., Stottmeister, C., Ruhe, L., et al. (2017). Translation of circRNAs. Mol. Cell 66, 9–21. doi:10.1016/j.molcel.2017.02.021
Pan, Z., Zhao, R., Li, B., Qi, Y., Qiu, W., Guo, Q., et al. (2022). EWSR1-induced circNEIL3 promotes glioma progression and exosome-mediated macrophage immunosuppressive polarization via stabilizing IGF2BP3. Mol. Cancer 21, 16. doi:10.1186/s12943-021-01485-6
Panda, A. C. (2018). Circular RNAs act as miRNA sponges. Adv. Exp. Med. Biol. 1087, 67–79. doi:10.1007/978-981-13-1426-1_6
Papaioannou, D., Volinia, S., Nicolet, D., Swierniak, M., Petri, A., Mrozek, K., et al. (2020). Clinical and functional significance of circular RNAs in cytogenetically normal AML. Blood Adv. 4, 239–251. doi:10.1182/bloodadvances.2019000568
Papatsirou, M., Artemaki, P. I., Karousi, P., Scorilas, A., and Kontos, C. K. (2021). Circular RNAs: Emerging regulators of the major signaling pathways involved in cancer progression. Cancers (Basel) 13, 2744. doi:10.3390/cancers13112744
Ping, L., Chen, J. J., Liao, C. S., Guang-Hua, L., and Ming, Z. (2019). Silencing of circ_0009910 inhibits acute myeloid leukemia cell growth through increasing miR-20a-5p. Blood Cells Mol. Dis. 75, 41–47. doi:10.1016/j.bcmd.2018.12.006
Qiu, Y., Yu, Y., Qin, X. M., Jiang, T., Tan, Y. F., Ouyang, W. X., et al. (2021). CircTLK1 modulates sepsis-induced cardiomyocyte apoptosis via enhancing PARP1/HMGB1 axis-mediated mitochondrial DNA damage by sponging miR-17-5p. J. Cell. Mol. Med. 25, 8244–8260. doi:10.1111/jcmm.16738
Qu, L., Yi, Z., Shen, Y., Lin, L., Chen, F., Xu, Y., et al. (2022). Circular RNA vaccines against SARS-CoV-2 and emerging variants. Cell 185, 1728–1744. e16. doi:10.1016/j.cell.2022.03.044
Rbbani, G., Nedoluzhko, A., Galindo-Villegas, J., and Fernandes, J. M. O. (2021). Function of circular RNAs in fish and their potential application as biomarkers. Int. J. Mol. Sci. 22, 7119. doi:10.3390/Ijms22137119
Ribeiro, S., Eiring, A. M., and Khorashad, J. S. (2021). Genomic abnormalities as biomarkers and therapeutic targets in acute myeloid leukemia. Cancers 13, 5055. doi:10.3390/Cancers13205055
Salzman, J., Gawad, C., Wang, P. L., Lacayo, N., and Brown, P. O. (2012). Circular RNAs are the predominant transcript isoform from hundreds of human genes in diverse cell types. PLoS One 7, e30733. doi:10.1371/journal.pone.0030733
Sanger, H. L., Klotz, G., Riesner, D., Gross, H. J., and Kleinschmidt, A. K. (1976). Viroids are single-stranded covalently closed circular RNA molecules existing as highly base-paired rod-like structures. Proc. Natl. Acad. Sci. U. S. A. 73, 3852–3856. doi:10.1073/pnas.73.11.3852
Shang, J., Chen, W. M., Wang, Z. H., Wei, T. N., Chen, Z. Z., and Wu, W. B. (2019). CircPAN3 mediates drug resistance in acute myeloid leukemia through the miR-153-5p/miR-183-5p-XIAP axis. Exp. Hematol. 70, 42–54. doi:10.1016/j.exphem.2018.10.011
Shang, Z., Ming, X., Wu, J. Y., and Xiao, Y. (2021). Downregulation of circ_0012152 inhibits proliferation and induces apoptosis in acute myeloid leukemia cells through the miR-625-5p/SOX12 axis. Hematol. Oncol. 39, 539–548. doi:10.1002/hon.2895
Shen, M. J., Yan, S. T., Zhang, X. Y., Li, W., Chen, X., Zheng, X. X., et al. (2022). The circular RNA hsa_circ_0003091 regulates sepsis-induced lung injury by sponging the miR-149/Smad2 axis. Aging (Albany NY) 14, 5059–5074. doi:10.18632/aging.204125
Shen, S. Y., Yao, T., Xu, Y. N., Zhang, D., Fan, S., and Ma, J. (2020). CircECE1 activates energy metabolism in osteosarcoma by stabilizing c-Myc. Mol. Cancer 19, 151. doi:10.1186/s12943-020-01269-4
Shi, L., Liu, B., Shen, D. D., Yan, P., Zhang, Y., Tian, Y., et al. (2021). A tumor-suppressive circular RNA mediates uncanonical integrin degradation by the proteasome in liver cancer. Sci. Adv. 7, eabe5043. doi:10.1126/sciadv.abe5043
Shi, Y. B., Tian, Y. Y., Sun, X. R., Qiu, Y., and Zhao, Y. (2022). Silencing circOMA1 inhibits osteosarcoma progression by sponging miR-1294 to regulate c-Myc expression. Front. Oncol. 12, 889583. doi:10.3389/Fonc.2022.889583
Singh, D., Kesharwani, P., Alhakamy, N. A., and Siddique, H. R. (2021). Accentuating circRNA-miRNA-transcription factors axis: A conundrum in cancer research. Front. Pharmacol. 12, 784801. doi:10.3389/fphar.2021.784801
Singh, V., Uddin, M. H., Zonder, J. A., Azmi, A. S., and Balasubramanian, S. K. (2021). Circular RNAs in acute myeloid leukemia. Mol. Cancer 20, 149. doi:10.1186/s12943-021-01446-z
Sun, Y. M., Wang, W. T., Zeng, Z. C., Chen, T. Q., Han, C., Pan, Q., et al. (2019). circMYBL2, a circRNA from MYBL2, regulates FLT3 translation by recruiting PTBP1 to promote FLT3-ITD AML progression. Blood 134, 1533–1546. doi:10.1182/blood.2019000802
Tang, X. Z., Deng, Z. D., Ding, P. G., Qiang, W., Lu, Y., Gao, S., et al. (2022). A novel protein encoded by circHNRNPU promotes multiple myeloma progression by regulating the bone marrow microenvironment and alternative splicing. J. Exp. Clin. Cancer Res. 41, 85. doi:10.1186/S13046-022-02276-7
Testa, U., and Riccioni, R. (2007). Deregulation of apoptosis in acute myeloid leukemia. Haematologica 92, 81–94. doi:10.3324/haematol.10279
Testa, U. (2010). TRAIL/TRAIL-R in hematologic malignancies. J. Cell. Biochem. 110, 21–34. doi:10.1002/jcb.22549
Thakral, D., Gupta, R., Sahoo, R. K., Verma, P., Kumar, I., and Vashishtha, S. (2020). Real-time molecular monitoring in acute myeloid leukemia with circulating tumor DNA. Front. Cell Dev. Biol. 8, 604391. doi:10.3389/Fcell.2020.604391
Tian, C., Zheng, G. G., Zhuang, H. Q., Li, X., Hu, D., Zhu, L., et al. (2017). MicroRNA-494 activation suppresses bone marrow stromal cell-mediated drug resistance in acute myeloid leukemia cells. J. Cell. Physiol. 232, 1387–1395. doi:10.1002/jcp.25628
Trino, S., Lamorte, D., Caivano, A., Laurenzana, I., Tagliaferri, D., Falco, G., et al. (2018). MicroRNAs as new biomarkers for diagnosis and prognosis, and as potential therapeutic targets in acute myeloid leukemia. Int. J. Mol. Sci. 19, E460. doi:10.3390/Ijms19020460
Wahl, M. C., Will, C. L., and Luhrmann, R. (2009). The spliceosome: Design principles of a dynamic RNP machine. Cell 136, 701–718. doi:10.1016/j.cell.2009.02.009
Wang, C., Li, L. L., Li, M. Y., Wang, W., Liu, Y., and Wang, S. (2020). Silencing long non-coding RNA XIST suppresses drug resistance in acute myeloid leukemia through down-regulation of MYC by elevating microRNA-29a expression. Mol. Med. 26, 114. doi:10.1186/s10020-020-00229-4
Wang, D., Ming, X., Xu, J., and Xiao, Y. (2021). Circ_0009910 shuttled by exosomes regulates proliferation, cell cycle and apoptosis of acute myeloid leukemia cells by regulating miR-5195-3p/GRB10 axis. Hematol. Oncol. 39, 390–400. doi:10.1002/hon.2874
Wang, F., Nazarali, A. J., and Ji, S. (2016). Circular RNAs as potential biomarkers for cancer diagnosis and therapy. Am. J. Cancer Res. 6, 1167–1176.
Wang, J., Pan, J., Huang, S., Li, F., Huang, J., Li, X., et al. (2021). Development and validation of a novel circular RNA as an independent prognostic factor in acute myeloid leukemia. BMC Med. 19, 28. doi:10.1186/S12916-020-01898-Y
Wang, X. L., Jin, P., Zhang, Y., and Wang, K. (2021). CircSPI1 acts as an oncogene in acute myeloid leukemia through antagonizing SPI1 and interacting with microRNAs. Cell Death Dis. 12, 297. doi:10.1038/s41419-021-03566-2
Wang, Y., Yan, Q., Mo, Y., Liu, Y., Wang, Y., Zhang, S., et al. (2022). Splicing factor derived circular RNA circCAMSAP1 accelerates nasopharyngeal carcinoma tumorigenesis via a SERPINH1/c-Myc positive feedback loop. Mol. Cancer 21, 62. doi:10.1186/S12943-022-01502-2
Wang, Y., Liu, J., Ma, J., Sun, T., Zhou, Q., Wang, W., et al. (2019). Exosomal circRNAs: Biogenesis, effect and application in human diseases. Mol. Cancer 18, 116. doi:10.1186/s12943-019-1041-z
Wang, Y., Zhang, Y., Wang, P., Fu, X., and Lin, W. (2020). Circular RNAs in renal cell carcinoma: Implications for tumorigenesis, diagnosis, and therapy. Mol. Cancer 19, 149. doi:10.1186/s12943-020-01266-7
Weigelt, C. M., Sehgal, R., Tain, L. S., Cheng, J., Eber, J., Pahl, A., et al. (2020). An insulin-sensitive circular RNA that regulates lifespan in Drosophila. Mol. Cell 79, 268–279. doi:10.1016/j.molcel.2020.06.011
Wiatrowski, K., Kim, T. H., and Przespolewski, A. (2022). Cellular and molecular biomarkers predictive of response to immunotherapy in acute myeloid leukemia. Front. Oncol. 12, 826768. doi:10.3389/Fonc.2022.826768
Wilusz, J. E. (2018). A 360 degrees view of circular RNAs: From biogenesis to functions. Wiley Interdiscip. Rev. RNA 9, e1478. doi:10.1002/wrna.1478
Wilusz, J. E. (2017). Circular RNAs: Unexpected outputs of many protein-coding genes. RNA Biol. 14, 1007–1017. doi:10.1080/15476286.2016.1227905
Wu, D. M., Wen, X., Han, X. R., Wang, S., Wang, Y. J., Shen, M., et al. (2018). Role of circular RNA DLEU2 in human acute myeloid leukemia. Mol. Cell. Biol. 38, 002599-18. doi:10.1128/MCB.00259-18
Wu, N., Yuan, Z. D., Du, K. Y., Fang, L., Lyu, J., Zhang, C., et al. (2019). Translation of yes-associated protein (YAP) was antagonized by its circular RNA via suppressing the assembly of the translation initiation machinery. Cell Death Differ. 26, 2758–2773. doi:10.1038/s41418-019-0337-2
Wu, X., Xiao, S., Zhang, M., Yang, L., Zhong, J., Li, B., et al. (2021). A novel protein encoded by circular SMO RNA is essential for Hedgehog signaling activation and glioblastoma tumorigenicity. Genome Biol. 22, 33. doi:10.1186/s13059-020-02250-6
Xia, X., Li, X. X., Li, F. Y., Wu, X., Zhang, M., Zhou, H., et al. (2019). A novel tumor suppressor protein encoded by circular AKT3 RNA inhibits glioblastoma tumorigenicity by competing with active phosphoinositide-dependent Kinase-1. Mol. Cancer 18, 131. doi:10.1186/s12943-019-1056-5
Xiang, W., Lam, Y. H., Periyasamy, G., and Chuah, C. (2022). Application of high throughput technologies in the development of acute myeloid leukemia therapy: Challenges and progress. Int. J. Mol. Sci. 23, 2863. doi:10.3390/Ijms23052863
Xiao, Y., Ming, X., and Wu, J. Y. (2021). Hsa_circ_0002483 regulates miR-758-3p/MYC axis to promote acute myeloid leukemia progression. Hematol. Oncol. 39, 243–253. doi:10.1002/hon.2829
Xing, S., Tian, Z., Zheng, W., Yang, W., Du, N., Gu, Y., et al. (2021). Hypoxia downregulated miR-4521 suppresses gastric carcinoma progression through regulation of IGF2 and FOXM1. Mol. Cancer 20, 9. doi:10.1186/s12943-020-01295-2
Xing, Y., Zha, W. J., Li, X. M., Li, H., Gao, F., Ye, T., et al. (2020). Circular RNA circ-Foxo3 inhibits esophageal squamous cell cancer progression via the miR-23a/PTEN axis. J. Cell. Biochem. 121, 2595–2605. doi:10.1002/jcb.29481
Xu, B., Zhao, Y., Wang, X., Gong, P., and Ge, W. (2017). MZH29 is a novel potent inhibitor that overcomes drug resistance FLT3 mutations in acute myeloid leukemia. Leukemia 31, 913–921. doi:10.1038/leu.2016.297
Xu, X. L., Zhang, J. W., Tian, Y. H., Gao, Y., Dong, X., Chen, W., et al. (2020). CircRNA inhibits DNA damage repair by interacting with host gene. Mol. Cancer 19, 128. doi:10.1186/s12943-020-01246-x
Yang, C., Wu, S., Mou, Z., Zhou, Q., Dai, X., Ou, Y., et al. (2022). Exosome-derived circTRPS1 promotes malignant phenotype and CD8+ T cell exhaustion in bladder cancer microenvironments. Mol. Ther. 30, 1054–1070. doi:10.1016/j.ymthe.2022.01.022
Yang, H. B., Li, X. B., Meng, Q. T., Sun, H., Wu, S., Hu, W., et al. (2020). CircPTK2 (hsa_circ_0005273) as a novel therapeutic target for metastatic colorectal cancer. Mol. Cancer 19, 13. doi:10.1186/s12943-020-1139-3
Yang, X. Q., Ye, T., Liu, H. R., Lv, P., Duan, C., Wu, X., et al. (2021). Expression profiles, biological functions and clinical significance of circRNAs in bladder cancer. Mol. Cancer 20, 4. doi:10.1186/s12943-020-01300-8
Yang, Y. B., Gao, X. Y., Zhang, M. L., Yan, S., Sun, C., Xiao, F., et al. (2018). Novel role of FBXW7 circular RNA in repressing glioma tumorigenesis. J. Natl. Cancer Inst. 110, 304–315. doi:10.1093/jnci/djx166
Yang, Z. G., Huang, C., Wen, X. Y., Liu, W., Huang, X., Li, Y., et al. (2022). Circular RNA circ-FoxO3 attenuates blood-brain barrier damage by inducing autophagy during ischemia/reperfusion. Mol. Ther. 30, 1275–1287. doi:10.1016/j.ymthe.2021.11.004
Yi, L., Zhou, L. B., Luo, J. X., and Yang, Q. (2021). Circ-PTK2 promotes the proliferation and suppressed the apoptosis of acute myeloid leukemia cells through targeting miR-330-5p/FOXM1 axis. Blood Cells Mol. Dis. 86, 102506. doi:10.1016/J.Bcmd.2020.102506
Yi, Y. Y., Yi, J., Zhu, X., Zhang, J., Zhou, J., Tang, X., et al. (2019). Circular RNA of vimentin expression as a valuable predictor for acute myeloid leukemia development and prognosis. J. Cell. Physiol. 234, 3711–3719. doi:10.1002/jcp.27145
Yu, C. Y., Li, T. C., Wu, Y. Y., Yeh, C. H., Chiang, W., Chuang, C. Y., et al. (2017). The circular RNA circBIRC6 participates in the molecular circuitry controlling human pluripotency. Nat. Commun. 8, 1149. doi:10.1038/s41467-017-01216-w
Zhang, C. J., Zhou, X. L., Geng, X. G., Zhang, Y., Wang, J., Wang, Y., et al. (2021). Circular RNA hsa_circ_0006401 promotes proliferation and metastasis in colorectal carcinoma. Cell Death Dis. 12, 443. doi:10.1038/s41419-021-03714-8
Zhang, L., Bu, Z., Shen, J., Shang, L., Chen, Y., and Wang, Y. (2020). A novel circular RNA (hsa_circ_0000370) increases cell viability and inhibits apoptosis of FLT3-ITD-positive acute myeloid leukemia cells by regulating miR-1299 and S100A7A. Biomed. Pharmacother. 122, 109619. doi:10.1016/j.biopha.2019.109619
Zhang, M. L., Huang, N. N., Yang, X. S., Luo, J., Yan, S., Xiao, F., et al. (2018a). A novel protein encoded by the circular form of the SHPRH gene suppresses glioma tumorigenesis. Oncogene 37, 1805–1814. doi:10.1038/s41388-017-0019-9
Zhang, M. L., Zhao, K., Xu, X. P., Yang, Y., Yan, S., Wei, P., et al. (2018b). A peptide encoded by circular form of LINC-PINT suppresses oncogenic transcriptional elongation in glioblastoma. Nat. Commun. 9, 4475. doi:10.1038/S41467-018-06862-2
Zhang, R., Li, Y., Wang, H., Zhu, K., and Zhang, G. (2020). The regulation of circRNA RNF13/miRNA-1224-5p Axis promotes the malignant evolution in acute myeloid leukemia. Biomed. Res. Int. 2020, 5654380. doi:10.1155/2020/5654380
Zhang, X., Wang, S., Wang, H. X., Cao, J., Huang, X., Chen, Z., et al. (2019). Circular RNA circNRIP1 acts as a microRNA-149-5p sponge to promote gastric cancer progression via the AKT1/mTOR pathway. Mol. Cancer 18, 20. doi:10.1186/s12943-018-0935-5
Zhang, Y., Jiang, J. J., Zhang, J. Y., Shen, H., Wang, M., Guo, Z., et al. (2021). CircDIDO1 inhibits gastric cancer progression by encoding a novel DIDO1-529aa protein and regulating PRDX2 protein stability. Mol. Cancer 20, 101. doi:10.1186/s12943-021-01390-y
Zhang, Y. N., Zhao, L., Yang, S. Z., Cen, Y., Zhu, T., Wang, L., et al. (2020). CircCDKN2B-AS1 interacts with IMP3 to stabilize hexokinase 2 mRNA and facilitate cervical squamous cell carcinoma aerobic glycolysis progression. J. Exp. Clin. Cancer Res. 39, 281. doi:10.1186/s13046-020-01793-7
Zhang, Y., Zhang, X. O., Chen, T., Xiang, J. F., Yin, Q. F., Xing, Y. H., et al. (2013). Circular intronic long noncoding RNAs. Mol. Cell 51, 792–806. doi:10.1016/j.molcel.2013.08.017
Zhang, Y., Zhu, M., Zhang, X., Dai, K., Liang, Z., Pan, J., et al. (2022). Micropeptide vsp21 translated by Reovirus circular RNA 000048 attenuates viral replication. Int. J. Biol. Macromol. 209, 1179–1187. doi:10.1016/j.ijbiomac.2022.04.136
Zhao, J., Lee, E. E., Kim, J., Yang, R., Chamseddin, B., Ni, C., et al. (2019). Transforming activity of an oncoprotein-encoding circular RNA from human papillomavirus. Nat. Commun. 10, 2300. doi:10.1038/S41467-019-10246-5
Zheng, S. L., Zhang, X. J., Odame, E., Xu, X., Chen, Y., Ye, J., et al. (2021). CircRNA-protein interactions in muscle development and diseases. Int. J. Mol. Sci. 22, 3262. doi:10.3390/Ijms22063262
Zheng, X., Chen, L. J., Zhou, Y., Wang, Q., Zheng, Z., Xu, B., et al. (2019). A novel protein encoded by a circular RNA circPPP1R12A promotes tumor pathogenesis and metastasis of colon cancer via Hippo-YAP signaling. Mol. Cancer 18, 47. doi:10.1186/s12943-019-1010-6
Zhong, J., Wu, X., Chen, J., Gao, Y., Zhang, M., Zhou, H., et al. (2022). Circular RNA-encoded MET variant is a targetable factor in glioblastoma. Res. Square. Preprint. doi:10.21203/rs.3.rs-1730428/v1
Zhou, J., Zhou, L. Y., Tang, X., Zhang, J., Zhai, L. L., Yi, Y. Y., et al. (2019). Circ-Foxo3 is positively associated with the Foxo3 gene and leads to better prognosis of acute myeloid leukemia patients. Bmc Cancer 19, 930. doi:10.1186/S12885-019-5967-8
Zhou, M., Xiao, M. S., Li, Z. G., and Huang, C. (2021). New progresses of circular RNA biology: From nuclear export to degradation. RNA Biol. 18, 1365–1373. doi:10.1080/15476286.2020.1853977
Keywords: circular RNAs, acute myeloid leukemia, molecular functions, clinical significance, role
Citation: Zhou M, Gao X, Zheng X and Luo J (2022) Functions and clinical significance of circular RNAs in acute myeloid leukemia. Front. Pharmacol. 13:1010579. doi: 10.3389/fphar.2022.1010579
Received: 03 August 2022; Accepted: 08 November 2022;
Published: 24 November 2022.
Edited by:
Stefania Bortoluzzi, University of Padua, ItalyReviewed by:
Paola Guglielmelli, Università degli Studi di Firenze, ItalyAnke Van Den Berg, University Medical Center Groningen, Netherlands
Copyright © 2022 Zhou, Gao, Zheng and Luo. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Min Zhou, bWluX3pob3VAY3F1LmVkdS5jbg==; Jing Luo, bGoyMjkyOTQwNjcwQDE2My5jb20=
†These authors have contributed equally to this work and share first authorship
 Min Zhou
Min Zhou Xianling Gao3†
Xianling Gao3†