- 1Shanghai Key Laboratory of Molecular Imaging, Jiading District Central Hospital Affiliated Shanghai University of Medicine and Health Sciences, Shanghai, China
- 2Department of Nuclear Medicine, Shanghai Chest Hospital, Shanghai Jiao Tong University, Shanghai, China
Warburg effect is characterized by excessive consumption of glucose by the tumor cells under both aerobic and hypoxic conditions. This metabolic reprogramming allows the tumor cells to adapt to the unique microenvironment and proliferate rapidly, and also promotes tumor metastasis and therapy resistance. Metabolic reprogramming of tumor cells is driven by the aberrant expression and activity of metabolic enzymes, which results in the accumulation of oncometabolites, and the hyperactivation of intracellular growth signals. Recent studies suggest that tumor-associated metabolic remodeling also depends on intercellular communication within the tumor microenvironment (TME). Small extracellular vesicles (sEVs), also known as exosomes, are smaller than 200 nm in diameter and are formed by the fusion of multivesicular bodies with the plasma membrane. The sEVs are instrumental in transporting cargoes such as proteins, nucleic acids or metabolites between the tumor, stromal and immune cells of the TME, and are thus involved in reprogramming the glucose metabolism of recipient cells. In this review, we have summarized the biogenesis and functions of sEVs and metabolic cargos, and the mechanisms through they drive the Warburg effect. Furthermore, the potential applications of targeting sEV-mediated metabolic pathways in tumor liquid biopsy, imaging diagnosis and drug development have also been discussed.
1 Introduction
Tumor cells can rapidly utilize glucose through “aerobic glycolysis” and produce high levels of lactate—a phenomenon known as the Warburg effect. Hanahan and Weinberg characterized this metabolic reprogramming of tumor cells as one of the hallmarks of cancer, and a potential therapeutic target (Hanahan and Weinberg, 2011; Hanahan, 2022). The Warburg effect is primarily driven by the aberrant expression of key metabolic enzymes, production of oncometabolites, and overactivation of growth signals (Vander Heiden et al., 2009; Liberti and Locasale, 2016; Poff et al., 2019). The key enzymes of glycolysis, including hexokinase 2 (HK2), 6-phosphofructo-2-kinase/fructose-2,6-bisphosphatase-3 (PFKFB3) and pyruvate kinase M2 (PKM2), promote the conversion of glucose to lactate instead of pyruvate in the tumor cells, which in turn impairs mitochondrial oxidative phosphorylation (Minchenko et al., 2002; Yang et al., 2014; Xu and Herschman, 2019). Furthermore, glucose 6-phosphate dehydrogenase (G6PD) produces ribose and NADPH in the tumor cells through the pentose phosphate pathway (PPP) as a source of energy for tumor growth (Jiang et al., 2011; Deng et al., 2021). Therefore, understanding the complex mechanisms underlying glucose uptake and metabolism by tumors, and the role of the tumor microenvironment (TME), can improve diagnostic accuracy and aid in drug development.
According to the classification criteria of the International Society for Extracellular Vesicles (ISEV), small extracellular vesicles (sEVs) or exosomes measure less than 200 nm in diameter, and are formed by the fusion of multivesicular bodies with the plasma membrane. They are typically secreted by tumor cells and other cells in the TME into various body fluids (Théry et al., 2018; Armacki et al., 2020; Möller and Lobb, 2020; Wu et al., 2021). The sEVs contain a large number of functional proteins, nucleic acid fragments (including DNA, mRNA or non-coding RNA) and other biologically active substances from the parent cells, and thus mediate transport and information exchange between cells. Most tumor cells release higher amounts of sEVs compared to normal cells. The tumor-derived sEVs promote angiogenesis, induce chemoresistance and differentiation of stromal cells in the TME, and regulate immune responses and the pre-metastatic microenvironment (Li and Nabet, 2019; Mashouri et al., 2019; Yousefi et al., 2020). Studies show that the sEVs in TME can also regulate tumor metabolism by directly delivering metabolic enzymes, metabolites or other factors to the recipient cells (Zhao et al., 2016; Wang et al., 2020b). Furthermore, these metabolically reprogrammed sEVs also modulate glucose uptake by tumor cells and other TME components, including stromal cells, immune cells and fibroblasts (Zhao et al., 2016; Yan et al., 2018; Rai et al., 2019; Morrissey et al., 2021). In this review, we have summarized the mechanisms through which the sEVs mediate the crosstalk between tumor cells and the TME, and propose possible applications of targeting sEV-related metabolic pathways for precise diagnosis and effective treatment of tumors.
2 Sorting of metabolic cargoes into sEVs and secretion
Tumor-specific sEVs can directly trigger the Warburg effect by transporting metabolites, enzymes, and the regulatory proteins and RNAs throughout the TME. The different intracellular components are loaded into sEVs through distinct sorting mechanisms (Figure 1). The biogenesis of sEVs is initiated following the inward budding of the membranes of multivesicular bodies (MVBs) to form intraluminal vesicles (ILVs). The mature MVBs fuse either with the plasma membrane and are expelled as sEVs, or with the lysosomes and are broken down (Anand et al., 2019). Rab GTPases act as the molecular switches that control ILV formation and transport within MVBs. The protein cargo is first sorted into the ILVs by tetraspanins and the endosomal sorting complex required for transport (ESCRT). VAMP2, VAMP3, VAMP7 and VAMP8 on the surface of MVBs form a SNARE complex with SNAP-23 on the plasma membrane, which releases the ILVs as sEVs (Clancy et al., 2019; Kumar et al., 2020; Zhang et al., 2021b). Pyruvate kinase M2 (PKM2) is co-sorted into the sEVs with the SNARE complex by phosphorylating SANP-23. In addition, the intra-vesicular PKM2 also promotes the exocytosis of sEVs from tumor cells (Wei et al., 2017; Fan et al., 2022). Lipids such as phospholipids and cholesterol are sorted into the sEVs by forming lipid rafts that are incorporated into sEV membranes (Zebrowska et al., 2019; Kumar and Deep, 2020). The sorting mechanism of other metabolites, including sugars, amino acids, nucleotides or vitamins, are not completely understood.
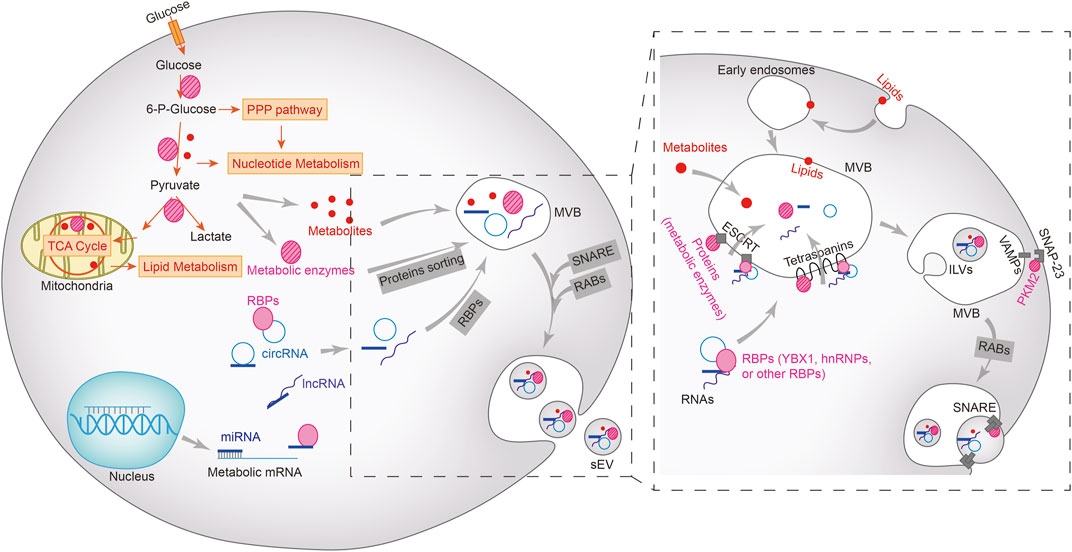
FIGURE 1. Metabolic cargoes in sEVs and their packaging, sorting and biogenesis. Metabolic enzymes and other regulatory proteins, metabolites, miRNAs, lncRNAs, and circRNAs are sorted into multivesicular bodies (MVBs) in different ways and subsequently excretes from the cells via the vesicle secretion pathways. Lipids (including phospholipids, sphingolipids, or triglycerides) on the plasma membrane enter MVBs via the early endosomes, but the sorting mechanism of other metabolites remains unclear. Proteins, including metabolic enzymes as well as RNA-bound RNA binding proteins (RBPs) enter the MVBs through ESCRT-dependent or independent mechanisms. The intraluminal vesicles (ILVs) in MVBs bind to the plasma membrane to form SNARE complex, which facilitates the extracellular release of sEV.
The mechanism underlying exosomal sorting of RNAs has been partially revealed (Liu et al., 2021). The RNA-binding Y-box protein I (YBX1) and Lupus La protein are required for loading miR-223 and miR-122 into sEVs, and is dependent on the high affinity of miRNAs for these RNA-binding proteins (Shurtleff et al., 2016; Temoche-Diaz et al., 2019). In fact, miRNAs that are sorted into sEVs possess similar motifs, and are enriched in the vesicles by specific RNA-binding proteins (Garcia-Martin et al., 2022b). For long RNAs such as mRNAs or lncRNAs, hnRNP A2B1 is responsible for binding to the corresponding motifs and sorting them into sEVs (O'Grady et al., 2022). The exact mechanism through which circRNAs are sorted into sEVs is still being explored, and studies suggest that it is likely accomplished through a miRNA binding-dependent manner or via direct interaction with RNA-binding proteins (Bao et al., 2016; Xu et al., 2020).
3 sEVs as drivers of Warburg effect
As already mentioned, the sEVs derived from tumor cells and TME components (fibroblasts, epithelial cells, macrophages, NK cells and other immune cells) reprogram glucose metabolism in tumor tissues by transporting proteins (metabolic enzymes, membrane proteins and other growth signaling proteins), nucleic acids (miRNAs, lncRNAs, circRNAs and mRNAs) and metabolites. The resulting dysregulation in metabolic pathways promote tumor progression, metastasis, and therapy resistance (Göran Ronquist, 2019; Zhang et al., 2021a). The contents of sEVs that regulate the Warburg effect are listed in Table 1.
3.1 Metabolic enzymes in sEV
The hypoxic environment within solid tumors is a major inducer of Warburg Effect. Ovarian cancer cell-derived sEVs load higher levels of hexokinase (HK), UDP-glucuronosyltransferase, 6-phosphogluconolactonase and CTP synthase 1, which are the key enzymes in glucose, phospholipid and nucleic acid metabolism, when exposed to hypoxic conditions (Alharbi et al., 2021). These sEVs loaded with metabolic enzymes are critical for the development of platinum resistance in ovarian cancer. In a previous study, we showed that cisplatin-resistant non-small cell lung cancer (NSCLC) cells transmitted drug resistance to sensitive cells by secreting PKM2-loaded exosomes in response to hypoxic stimulation (Wang et al., 2021). Thus, inhibiting glucose metabolism could potentially reduce sEV-mediated chemoresistance in cancer cells. For example, pancreatic cancer cells with autophagy blockade exhibited low glucose uptake and expressed low levels of ATP-binding cassette sub-family G member 2 (ABCG2) in the secreted exosomes, which restored gemcitabine sensitivity (Bhattacharya et al., 2014). Although oxidative phosphorylation is attenuated in tumor cells, some enzymes of the Krebs cycle are also released within sEVs. We previously observed that exosomal release of cytoplasmic IDH1 promoted 5FU resistance in colorectal cancer cells (Yang et al., 2021). In addition to chemoresistance, exosomal metabolic enzymes can also enhance other malignant features such as invasion, metastasis, angiogenesis and immune escape. For example, exosomal angiopoietin-like protein 7 (ANGPTL7) secreted by the human ovarian cancer cells can promote tumor angiogenesis by regulating glucose and lipid metabolism, and oxidative stress (Parri et al., 2014). Several studies in recent years have established the presence of metabolic enzymes in sEVs and their impact on oncogenic signaling networks. Retinoblastoma-derived exosomes are enriched in multiple metabolic enzymes, including enolase 3 (ENO3), galactokinase-1 (GALK1), asparagine synthase (ASNS) and saccharopine dehydrogenase (SCCPDH) (Galardi et al., 2020). Taken together, transmission of metabolic enzymes via sEVs affects tumor progression through metabolic or non-metabolic signaling pathways.
3.2 Metabolites in sEV
Tumor or TME-derived sEVs can modulate tumor progression by transporting specific metabolites to distant tissues, immune cells, etc. For instance, colon cancer cells expressing CD63, CD9 and A33 are known to deliver exosomal phosphatidylcholine to liver tissues via the portal vein (Kumar et al., 2021). Furthermore, lung adenocarcinoma-derived exosomes are enriched in linoleic acid (LA) compared to those derived from normal lung epithelial cells, and promote tumor metastasis by reprogramming macrophage glycolysis and PD-L1-dependent immunosuppression (Morrissey et al., 2021). Metabolomics have been used to identify the species, content and signaling pathways of metabolites in sEVs and other vesicles. Metabolite analysis of CAF-derived exosomes by gas chromatography-mass spectrometer (GC-MS) and ultra-high performance liquid chromatography (UPLC) revealed high levels of intermediates produced during glucose metabolism (such as citric acid and pyruvate), lipids (stearic and palmitic) and most amino acids (Zhao et al., 2016). However, the exact functions of these exosomal metabolites in tumor progression remains to be explored.
3.3 Nucleic acids regulate tumor metabolism via sEV
Exosomal nucleic acids, especially non-coding RNAs, are established diagnostic biomarkers of various tumors. Non-coding RNAs can promote or impede the Warburg effect by targeting specific metabolic enzymes or regulatory genes. For example, colorectal cancer cell-derived exosomal circ-RNF121 enhanced glycolysis and reduced ATP/ADP production in the tumor cells (Jiang et al., 2021). Furthermore, circ-0008928 is highly expressed in NSCLC cell-derived sEVs, and confers cisplatin resistance by acting as a “sponge” for miR-488, which enhances HK2 activity and glycolysis (Shi et al., 2021). NSCLC cells also secrete the lncRNA MALAT1 via sEVs, which is known to promote lactate dehydrogenase A (LDHA) expression and glycolysis (Wang et al., 2020a). Oxaliplatin-resistant colorectal cancer cells can transmit chemoresistance to the sensitive cells through exosomal circ-0005963, which sponges miR-122 and upregulates PKM2 (Wang et al., 2020c). Exosomal RNA-mediated decrease in glucose uptake in the normal tissues also contributes to tumor development. For instance, breast cancer cell-derived exosomal miR-122 drives brain/lung metastasis by downregulating PKM2 and inhibiting glucose uptake in distant non-tumor tissues (Fong et al., 2015). In addition to tumor cells, the stromal and immune cells of the TME also secrete sEVs that reprogram tumor metabolism. There is evidence that exosomal lncRNA TUG1 secreted by cancer-associated fibroblasts (CAFs) in HCC tissues increases LDH activity and lactate production in the tumor cells (Lu et al., 2022).
Interestingly, the metabolic reprograming induced by exosomal components exert a positive feedback by increasing cargo loading into the sEVs. For example, exosomal circ_0072083 derived from temozolomide (TMZ)-resistant glioma cells activates glycolysis and increases the levels of glucose transporter 1 (GLUT1), LDHA and PKM2, which further induce the expression of the circRNA and amplify TMZ resistance (Ding et al., 2021). Furthermore, the dissemination of sEVs in the TME also depends on the extracellular glucose status. Under energy-deficient low-glucose conditions, gastric cancer-derived exosomal miR-451 is more inclined to polarize T cells to the immunosuppressive Th17 cells, which leads to poor prognosis (Liu et al., 2018). In fact, tumor-derived sEVs can also trigger metabolic disorders, as exemplified by the interaction of pancreatic cancer and diabetes (Sah et al., 2019). Pancreatic cancer cell-derived exosomes delivered miR-19a to islet β cells and disrupted normal insulin secretion (Pang et al., 2021; Su et al., 2022), and reprogramed enteroendocrine cell function via multiple exosomal miRNAs (Zhang et al., 2018). Thus, TME-derived sEV RNA reprograms tumor glucose metabolism and creates favorable conditions for tumor cell growth and metastasis, while the sEV RNA cargo secreted by metabolically abnormal tumor cells also enhances the malignant potential.
3.4 Resident proteins in sEV
The sEVs derived from different tissues may express similar marker proteins, including CD9, CD63, CD81 or TSG101, which are used for their identification. However, most of these ubiquitous resident proteins of sEVs, most of which are membrane proteins, show differential expression across cell types during proteomics examination (Kowal et al., 2016; Rai et al., 2021; Garcia-Martin et al., 2022a). Some resident exosomal proteins are transferred to recipient cells, wherein they reprogram glucose metabolism. For example, nasopharyngeal carcinoma cell-derived sEVs can transform normal fibroblasts to a tumor-promoting phenotype by delivering latent membrane protein 1 (LMP1), which enhances glycolysis and inhibits mitochondrial oxidative phosphorylation via activation of the NF-κB pathway (Wu et al., 2020). Integrin beta 4 (ITGB4) is highly expressed on triple-negative breast cancer (TNBC) cells and their derived exosomes, and promotes glycolysis in CAFs through mitophagy (Sung et al., 2020). High mobility group box-1 (HMGB-1) is highly expressed in lung adenocarcinoma exosomes and promotes glycolysis in the tumor macrophages by upregulating PD-L1 expression (Morrissey et al., 2021). The 78 kDa glucose-regulated protein (GRP78) is translocated from the endoplasmic reticulum to the plasma membrane, and released via exosomes (Li et al., 2016; Gonzalez-Gronow et al., 2021). High expression of exosomal GRP78 in colorectal and breast tumors is associated with increased glycolysis and metastasis (Zheng et al., 2019; Li et al., 2022). Therefore, proteins that are differentially expressed in the tumor-derived exosomes relative to that secreted by normal cells should be considered as potential as biomarkers. Overall, tumor cell-derived sEV-resident proteins act as “villain messengers” by promoting malignant behaviors such as invasion, metastasis and immunosuppression through metabolic reprograming.
4 Application of targeting metabolic sEVs
Given their unique protein profiles, sEVs are promising biomarkers for early, non-invasive tumor detection, and for monitoring therapy (Liang et al., 2021). Tumor-derived exosomal protein or RNA cargoes can be easily detected in liquid biopsies for rapid diagnostic and prognostic evaluation. Furthermore, inhibitors of sEV secretion or the loaded cargos potential drug candidates for tumor therapy. In addition, in vivo imaging of sEVs can be used as a surrogate for tracking tumor progression and therapeutic efficacy.
4.1 Metabolic biomarkers for liquid biopsy
Proteomic mass spectrometry analysis of salivary exosomes from human papillomavirus (HPV)-driven oropharyngeal cancer patients revealed significantly elevated aldolase, glyceraldehyde dehydrogenase-3-phosphate dehydrogenase (GAPDH), LDHA/B, phosphoglycerate kinase 1 (PGK1) and PKM1/2 (Tang et al., 2021). Compared to healthy volunteers, serum-derived exosomal circARHGAP10 was upregulated in NSCLC patients, and associated with increased the expression of GLUT1 and LDH, which are drivers of NSCLC progression (Fang et al., 2022). Furthermore, presence of serum-derived exosomes with low levels of miR-543 in patients with epithelial ovarian cancer indicates favorable prognosis since downregulation of this miRNA inhibits glucose uptake by tumor cells (Zhang et al., 2022). Cancer stem cells (CSCs) drive tumor initiation, drug resistance and metastasis, and are the primary cause of tumor recurrence. Metabolomics analysis of CSCs-enriched primary melanoma tissues and serum-derived exosomes from melanoma patients revealed aberrant expression of multiple metabolites (Palacios-Ferrer et al., 2021). Thus, exosomal metabolites in liquid biopsies are ideal markers of tumor diagnosis, therapeutic efficacy and prognosis.
4.2 Therapeutic agents targeting metabolic regulators
Some small molecule drugs and natural compounds can inhibit tumor-associated sEV secretion or metabolic pathways, and are thus potential candidates for tumor therapy. The AMPK inhibitor compound C and the c-Jun inhibitor SP600125 blocked the delivery of exosomes from the TNBC cells into CAFs, which inhibited glycolysis and tumor cell proliferation (Sung et al., 2020). The sEV-mediated immunosuppression is partly attributed to the metabolic reprogramming of TME, and drugs targeting sEV-related tumor immunity are suitable for tumor therapy, such as immune checkpoint inhibitors (Marshall and Djamgoz, 2018; Bader et al., 2020; Yang et al., 2020). As already described, tumor cell-derived exosomes increased glucose uptake and PD-L1 expression in CAFs in an NF-kB-dependent manner, which was inhibited by the glycolysis inhibitor 2-deoxyglucose. Furthermore, tumor sEVs also inhibited oxidative phosphorylation in tumor-associated macrophages by upregulating NOS2 expression, and administration of the NOS2 inhibitor S-ethylisothiourea hydrobromide restored oxygen utilization (Kuruppu et al., 2014; Morrissey et al., 2021). In addition, coptisine disrupted the secretion of exosomal circCCT3 from CAFs and reduced the expression of HK2 in liver cancer cells (Lv et al., 2020), which has potential therapeutic value. The natural compound oleanolic acid reduced the secretion of exosomes from TMZ-resistant glioma cells, and shikonin inhibited resistant cells-derived exosomal circ_0072083 levels and the Warburg effect (Ding et al., 2021). These compounds have potential sensitizing effects on TMZ treatment of glioma. Therapeutic agents targeting sEV-related communications have also been shown to multidrug resistance -associated protein (MRP) -mediated resistance. The calmodulin-dependent protein kinase inhibitor KN-93 was used to reverse MSC sEV-mediated 5-FU resistance in gastric cancer (Ji et al., 2015). Mechanistically, these sEVs enhanced the expression of MRP and P-glycoprotein by regulating miR-301b and ERK kinase pathways (Ji et al., 2015; Zhu et al., 2022), which are inducers of glucose metabolism and malignant tumorigenesis (Asl et al., 2021; Jandova and Wondrak, 2021). However, the limitation lies in the lack of the effect of sEV from other TME components (other than MSC) on MRP and chemotherapy resistance. More studies are needed to explore the mechanism of chemotherapy resistance of metabolic sEV in order to develop therapeutic agents with relevant targets.
4.3 Metabolic imaging of sEVs in nuclear medicine
Due to the sentinel role of exosomes in early TME shaping and formation of the pre-metastatic niche, in vivo sEV imaging can be helpful for early tumor monitoring. At present, sEV imaging mainly focuses on tracking the fluorescently-labeled sEVs. For example, plasma membrane-specific fluorescent dyes such as DIR and PKH26 are commonly used to label exosomes and track them at the cellular level (Pužar Dominkuš et al., 2018; Sun et al., 2019; Kumar et al., 2021). Nuclear imaging, including 18F-deoxyglucose (FDG)-positron emission tomography/computed tomography (PET/CT), is currently the main diagnostic method for evaluating tumor glucose uptake and metabolic activity, and offers the possibility for isotopic labeling and imaging of exosomes. Engineered exosomes labeled with radioisotopes (including 99mTc, 111I, 125I, 131I, 64Cu, 68Ga) have achieved non-invasive PET or SPECT detection in animal models, and show promise for clinical translation (Morishita et al., 2015; Gangadaran et al., 2018; Rashid et al., 2019; Jung et al., 2020; Lázaro-Ibáñez et al., 2021).
Radio-labeled exosomes have excellent intra-tumoral homing ability and biosafety (Shi et al., 2019). FDG-PET imaging has been used to assess the function of exosomes in Alzheimer’s disease development since glucose metabolism in the affected tissues correlates with the secretion of plasma-specific exosomes (Coumans et al., 2017; Chanteloup et al., 2019). In addition, FDG-PET imaging has also demonstrated the activation of glucose metabolism in breast tumor cells through EVs uptake (Kang et al., 2021). Furthermore, the enriched miRNA levels in plasma EVs from Hodgkin lymphoma patients matched the FDG PET imaging results, suggesting that EVs can be used for metabolic tracing in tumor patients (van Eijndhoven et al., 2016). In an in vitro study, 13C isotope-labeled glucose was used to evaluate the effect of CAF-derived exosomes on the metabolites of glycolysis (Zhao et al., 2016). Although isotopic tracing of exosomes can be used for tumor imaging, very few studies have reported direct radiolabeling of metabolites in EVs for in vivo imaging.
5 Conclusion and perspectives
The Warburg effect is manifested as overactive glycolysis and aberrant activity of metabolic enzymes, and is instrumental for the rapid proliferation, invasiveness, drug resistance and immune escape of tumor cells. Extracellular vesicles (EVs) mediate intercellular communication in the TME, and the sEVs or exosomes have been studied the most. In this review, we outlined the mechanisms through which proteins, RNAs and metabolites are sorted into the sEVs within cells, as well as the pathways regulated by the exosomal cargo to remodel tumor metabolism and promote tumor progression (Figure 2). Furthermore, we have also discussed the diagnostic and therapeutic potential of tumor-derived sEVs.
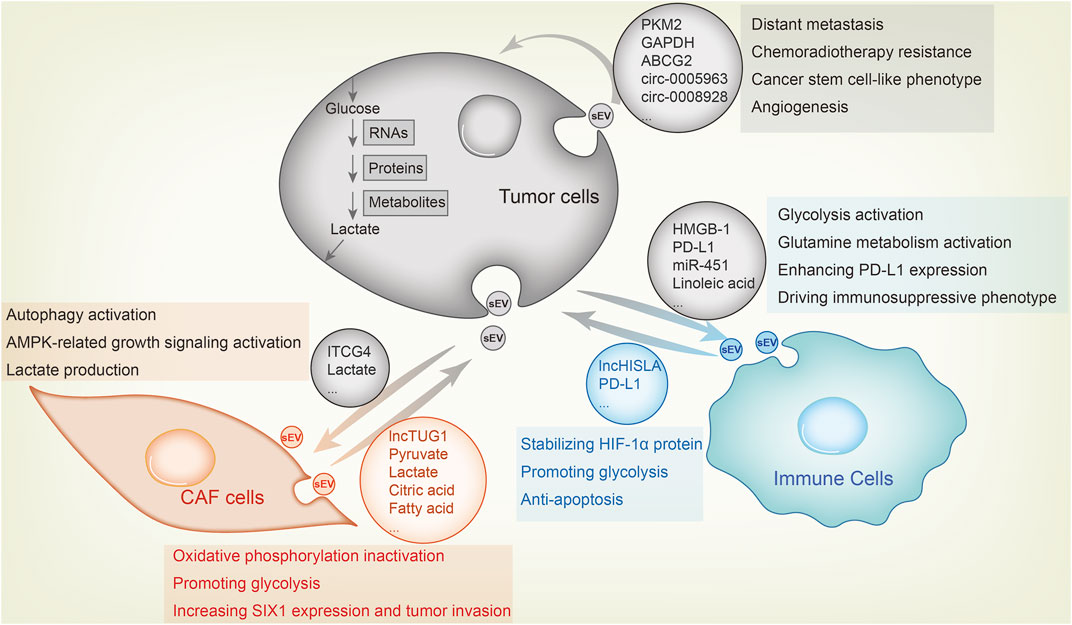
FIGURE 2. Metabolic sEV exchange between tumor cells and other components in the TME. The uptake of tumor-derived sEVs by CAFs or immune cells facilitates tumor growth, whereas tumor cell fusion of metabolic sEVs in TME leads to its malignant transformation. For example, CAFs-derived sEV lncRNA TUG1 delivers to liver cancer cells, increasing SIX1 expression and promoting glycolysis and tumor invasion. CAFs-derived sEVs inhibit oxidative phosphorylation and activates glycolysis by delivering lactate, pyruvate, citric acid, fatty acids and amino acids to pancreatic cancer cells. As feedback, breast cancer cells sEVs carry ITGB4 proteins that can be transmitted to CAFs, inducing autophagy, AMPK activation and lactate production in CAFs, which contribute to tumor invasion. As for the effect of sEV on immune cells, lung cancer-derived sEV HMGB1, PD-L1 and linoleic acid enhance glycolysis, glutamine metabolism and PD-L1 expression in macrophages and T cells through NF-κB signaling, endowing these immune cells with an immunosuppressive phenotype. MiR-451 in sEV of gastric cancer transmits to T cells and enhances their infiltration, causing them to differentiate to Th17 cells and forming malignant transformation of tumor microenvironment. Macrophages also secrete sEVs, which enhance aerobic glycolysis and anti-apoptosis of breast cancer cells by delivering lncRNA HISLA and stabilizing HIF-1α protein. Among tumor cells, highly metastatic cells, chemotherapy or radiotherapy resistant cells and cancer stem cell-like cells transfer sEV to sensitive cells to confer corresponding malignant phenotype.
Analysis of exosomal protein mass spectrometry and sequencing data form EV databases such as ExoCarta, exoRBase or Vesiclepedia have also indicated that the resident metabolic enzymes or RNAs may be involved in regulating tumor metabolism (Kalra et al., 2012; Keerthikumar et al., 2016; Lai et al., 2021). Therefore, more sEV cargoes and functions may be discovered in the future, and provide new insights into tumor metabolic remodeling. Although some carbohydrate-metabolizing enzymes (such as HK2, ENO3 and PKM2) delivered via sEVs are known to remodel the TME, given the opposing functions of glycolytic and mitochondrial metabolic enzymes in the Warburg effect, it remains to be ascertained whether these enzymes exert different functions when sorted into sEVs and in the recipient cells. In addition, since tetraspanins may potentially regulate nutrient metabolism in tumor cells, their use as a biomarker of sEV protein cargo also needs to be considered (Wang et al., 2019; Najy et al., 2021).
The diagnostic, therapeutic and imaging applications of tumor-derived sEVs has gained considerable attention in recent years. Tumor patient-derived sEVs containing metabolic enzymes or miRNAs are diagnostic biomarkers, which opens up the possibility of developing rapid sEV isolation and microfluidic chip analysis of the cargo. 18F-deoxyglucose (FDG) is routinely used as a tracer for the radiometabolic imaging of tumors during diagnosis and treatment evaluation (Jacobson and Chen, 2013; Yang et al., 2017; Sheikhbahaei et al., 2020). However, the clinical utility is somewhat limited. For example, FDG PET/CT is prone to inflammatory interference in cervical cancer, resulting in low diagnostic specificity (Dejanovic et al., 2021). In addition, tumors such as hepatocellular carcinoma (HCC) have a lower capacity for FDG uptake (Li et al., 2018). Therefore, isotope probes targeting tumor-specific sEVs offer more possibilities for the diagnosis of heterogeneous tumors than FDG. Although sEVs can be labeled with fluorophores or isotopes for in vivo tumor imaging, the metabolic components of sEVs cannot be directly labeled as yet. The development of metabolic enzyme-targeted probes or radionuclide-labeled metabolites will be a significant progress in tumor imaging and clinical decision-making. Finally, more therapeutic or engineered agents targeting the metabolic cargo in sEVs need to be developed as drug delivery systems. In conclusion, targeting metabolic sEVs will play an important role in tumor diagnosis and therapy.
Author contributions
Preparing the original manuscripts, HY and JW; writing and revising manuscripts, HY, JW, and GH. All authors have read and agreed to the published version of the manuscript.
Funding
This study was supported by grants from the National Natural Science Foundation of China (Grant Nos 82127807 and 81903065) and construction project of Shanghai Key Laboratory of Molecular Imaging (18DZ2260400).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Aga, M., Bentz, G. L., Raffa, S., Torrisi, M. R., Kondo, S., Wakisaka, N., et al. (2014). Exosomal HIF1α supports invasive potential of nasopharyngeal carcinoma-associated LMP1-positive exosomes. Oncogene 33 (37), 4613–4622. doi:10.1038/onc.2014.66
Alharbi, M., Lai, A., Sharma, S., Kalita-de Croft, P., Godbole, N., Campos, A., et al. (2021). Extracellular vesicle transmission of chemoresistance to ovarian cancer cells is associated with hypoxia-induced expression of glycolytic pathway proteins, and prediction of epithelial ovarian cancer disease recurrence. Cancers (Basel) 13 (14), 3388. doi:10.3390/cancers13143388
Anand, S., Samuel, M., Kumar, S., and Mathivanan, S. (2019). Ticket to a bubble ride: Cargo sorting into exosomes and extracellular vesicles. Biochim. Biophys. Acta. Proteins Proteom. 1867 (12), 140203. doi:10.1016/j.bbapap.2019.02.005
Armacki, M., Polaschek, S., Waldenmaier, M., Morawe, M., Ruhland, C., Schmid, R., et al. (2020). Protein kinase D1, reduced in human pancreatic tumors, increases secretion of small extracellular vesicles from cancer cells that promote metastasis to lung in mice. Gastroenterology 159 (3), 10191019–10191035. doi:10.1053/j.gastro.2020.05.052
Asl, E. R., Amini, M., Najafi, S., Mansoori, B., Mokhtarzadeh, A., Mohammadi, A., et al. (2021). Interplay between MAPK/ERK signaling pathway and MicroRNAs: A crucial mechanism regulating cancer cell metabolism and tumor progression. Life Sci. 278, 119499. doi:10.1016/j.lfs.2021.119499
Bader, J. E., Voss, K., and Rathmell, J. C. (2020). Targeting metabolism to improve the tumor microenvironment for cancer immunotherapy. Mol. Cell 78 (6), 1019–1033. doi:10.1016/j.molcel.2020.05.034
Bao, C., Lyu, D., and Huang, S. (2016). Circular RNA expands its territory. Mol. Cell. Oncol. 3 (2), e1084443. doi:10.1080/23723556.2015.1084443
Bhattacharya, S., Pal, K., Sharma, A. K., Dutta, S. K., Lau, J. S., Yan, I. K., et al. (2014). GAIP interacting protein C-terminus regulates autophagy and exosome biogenesis of pancreatic cancer through metabolic pathways. PLoS One 9 (12), e114409. doi:10.1371/journal.pone.0114409
Chanteloup, G., Cordonnier, M., Moreno-Ramos, T., Pytel, V., Matías-Guiu, J., Gobbo, J., et al. (2019). Exosomal HSP70 for monitoring of frontotemporal dementia and alzheimer's disease: Clinical and FDG-PET correlation. J. Alzheimers Dis. 71 (4), 1263–1269. doi:10.3233/jad-190545
Chen, F., Chen, J., Yang, L., Liu, J., Zhang, X., Zhang, Y., et al. (2019). Extracellular vesicle-packaged HIF-1α-stabilizing lncRNA from tumour-associated macrophages regulates aerobic glycolysis of breast cancer cells. Nat. Cell Biol. 21 (4), 498–510. doi:10.1038/s41556-019-0299-0
Clancy, J. W., Zhang, Y., Sheehan, C., and D'Souza-Schorey, C. (2019). An ARF6-Exportin-5 axis delivers pre-miRNA cargo to tumour microvesicles. Nat. Cell Biol. 21 (7), 856–866. doi:10.1038/s41556-019-0345-y
Coumans, F. A. W., Brisson, A. R., Buzas, E. I., Dignat-George, F., Drees, E. E. E., El-Andaloussi, S., et al. (2017). Methodological guidelines to study extracellular vesicles. Circ. Res. 120 (10), 1632–1648. doi:10.1161/circresaha.117.309417
Dai, Y., Liu, Y., Li, J., Jin, M., Yang, H., and Huang, G. (2022). Shikonin inhibited glycolysis and sensitized cisplatin treatment in non-small cell lung cancer cells via the exosomal pyruvate kinase M2 pathway. Bioengineered 13 (5), 13906–13918. doi:10.1080/21655979.2022.2086378
Dejanovic, D., Hansen, N. L., and Loft, A. (2021). PET/CT variants and pitfalls in gynecological cancers. Semin. Nucl. Med. 51 (6), 593–610. doi:10.1053/j.semnuclmed.2021.06.006
Deng, P., Li, K., Gu, F., Zhang, T., Zhao, W., Sun, M., et al. (2021). LINC00242/miR-1-3p/G6PD axis regulates Warburg effect and affects gastric cancer proliferation and apoptosis. Mol. Med. 27 (1), 9. doi:10.1186/s10020-020-00259-y
Ding, C., Yi, X., Chen, X., Wu, Z., You, H., Chen, X., et al. (2021). Warburg effect-promoted exosomal circ_0072083 releasing up-regulates NANGO expression through multiple pathways and enhances temozolomide resistance in glioma. J. Exp. Clin. Cancer Res. 40 (1), 164. doi:10.1186/s13046-021-01942-6
Fan, M., Sun, W., Gu, X., Lu, S., Shen, Q., Liu, X., et al. (2022). The critical role of STAT3 in biogenesis of tumor-derived exosomes with potency of inducing cancer cachexia in vitro and in vivo. Oncogene 41 (7), 1050–1062. doi:10.1038/s41388-021-02151-3
Fang, K., Chen, X., Qiu, F., Xu, J., Xiong, H., and Zhang, Z. (2022). Serum-derived exosomes-mediated circular RNA ARHGAP10 modulates the progression of non-small cell lung cancer through the miR-638/FAM83F Axis. Cancer biother. Radiopharm. 37 (2), 96–110. doi:10.1089/cbr.2019.3534
Fong, M. Y., Zhou, W., Liu, L., Alontaga, A. Y., Chandra, M., Ashby, J., et al. (2015). Breast-cancer-secreted miR-122 reprograms glucose metabolism in premetastatic niche to promote metastasis. Nat. Cell Biol. 17 (2), 183–194. doi:10.1038/ncb3094
Galardi, A., Colletti, M., Lavarello, C., Di Paolo, V., Mascio, P., Russo, I., et al. (2020). Proteomic profiling of retinoblastoma-derived exosomes reveals potential biomarkers of vitreous seeding. Cancers (Basel) 12 (6), 1555. doi:10.3390/cancers12061555
Gangadaran, P., Hong, C. M., Oh, J. M., Rajendran, R. L., Kalimuthu, S., Son, S. H., et al. (2018). In vivo non-invasive imaging of radio-labeled exosome-mimetics derived from red blood cells in mice. Front. Pharmacol. 9, 817. doi:10.3389/fphar.2018.00817
Garcia-Martin, R., Brandao, B. B., Thomou, T., Altindis, E., and Kahn, C. R. (2022a). Tissue differences in the exosomal/small extracellular vesicle proteome and their potential as indicators of altered tissue metabolism. Cell Rep. 38 (3), 110277. doi:10.1016/j.celrep.2021.110277
Garcia-Martin, R., Wang, G., Brandão, B. B., Zanotto, T. M., Shah, S., Kumar Patel, S., et al. (2022b). MicroRNA sequence codes for small extracellular vesicle release and cellular retention. Nature 601 (7893), 446–451. doi:10.1038/s41586-021-04234-3
Giovannetti, E., van der Borden, C. L., Frampton, A. E., Ali, A., Firuzi, O., and Peters, G. J. (2017). Never let it go: Stopping key mechanisms underlying metastasis to fight pancreatic cancer. Semin. Cancer Biol. 44, 43–59. doi:10.1016/j.semcancer.2017.04.006
Gonzalez-Gronow, M., Gopal, U., Austin, R. C., and Pizzo, S. V. (2021). Glucose-regulated protein (GRP78) is an important cell surface receptor for viral invasion, cancers, and neurological disorders. IUBMB Life 73 (6), 843–854. doi:10.1002/iub.2502
Göran Ronquist, K. (2019). Extracellular vesicles and energy metabolism. Clin. Chim. Acta. 488, 116–121. doi:10.1016/j.cca.2018.10.044
Hanahan, D. (2022). Hallmarks of cancer: New dimensions. Cancer Discov. 12 (1), 31–46. doi:10.1158/2159-8290.cd-21-1059
Hanahan, D., and Weinberg, R. A. (2011). Hallmarks of cancer: The next generation. Cell 144 (5), 646–674. doi:10.1016/j.cell.2011.02.013
Jacobson, O., and Chen, X. (2013). Interrogating tumor metabolism and tumor microenvironments using molecular positron emission tomography imaging. Theranostic approaches to improve therapeutics. Pharmacol. Rev. 65 (4), 1214–1256. doi:10.1124/pr.113.007625
Jandova, J., and Wondrak, G. T. (2021). Genomic GLO1 deletion modulates TXNIP expression, glucose metabolism, and redox homeostasis while accelerating human A375 malignant melanoma tumor growth. Redox Biol. 39, 101838. doi:10.1016/j.redox.2020.101838
Ji, R., Zhang, B., Zhang, X., Xue, J., Yuan, X., Yan, Y., et al. (2015). Exosomes derived from human mesenchymal stem cells confer drug resistance in gastric cancer. Cell Cycle 14 (15), 2473–2483. doi:10.1080/15384101.2015.1005530
Jiang, P., Du, W., Wang, X., Mancuso, A., Gao, X., Wu, M., et al. (2011). p53 regulates biosynthesis through direct inactivation of glucose-6-phosphate dehydrogenase. Nat. Cell Biol. 13 (3), 310–316. doi:10.1038/ncb2172
Jiang, Z., Hu, H., Hu, W., Hou, Z., Liu, W., Yu, Z., et al. (2021). Circ-RNF121 regulates tumor progression and glucose metabolism by miR-1224-5p/FOXM1 axis in colorectal cancer. Cancer Cell Int. 21 (1), 596. doi:10.1186/s12935-021-02290-3
Jung, K. O., Kim, Y.-H., Chung, S.-J., Lee, C.-H., Rhee, S., Pratx, G., et al. (2020). Identification of lymphatic and hematogenous routes of rapidly labeled radioactive and fluorescent exosomes through highly sensitive multimodal imaging. Int. J. Mol. Sci. 21 (21), 7850. doi:10.3390/ijms21217850
Kalra, H., Simpson, R. J., Ji, H., Aikawa, E., Altevogt, P., Askenase, P., et al. (2012). Vesiclepedia: A compendium for extracellular vesicles with continuous community annotation. PLoS Biol. 10 (12), e1001450. doi:10.1371/journal.pbio.1001450
Kang, S. Y., Lee, E. J., Byun, J. W., Han, D., Choi, Y., Hwang, D. W., et al. (2021). Extracellular vesicles induce an aggressive phenotype in luminal breast cancer cells via PKM2 phosphorylation. Front. Oncol. 11, 785450. doi:10.3389/fonc.2021.785450
Keerthikumar, S., Chisanga, D., Ariyaratne, D., Al Saffar, H., Anand, S., Zhao, K., et al. (2016). ExoCarta: A web-based compendium of exosomal cargo. J. Mol. Biol. 428 (4), 688–692. doi:10.1016/j.jmb.2015.09.019
Kowal, J., Arras, G., Colombo, M., Jouve, M., Morath, J. P., Primdal-Bengtson, B., et al. (2016). Proteomic comparison defines novel markers to characterize heterogeneous populations of extracellular vesicle subtypes. Proc. Natl. Acad. Sci. U. S. A. 113(8), E968–E977. doi: doi:doi:10.1073/pnas.1521230113
Kumar, A., and Deep, G. (2020). Hypoxia in tumor microenvironment regulates exosome biogenesis: Molecular mechanisms and translational opportunities. Cancer Lett. 479, 23–30. doi:10.1016/j.canlet.2020.03.017
Kumar, A., Sundaram, K., Mu, J., Dryden, G. W., Sriwastva, M. K., Lei, C., et al. (2021). High-fat diet-induced upregulation of exosomal phosphatidylcholine contributes to insulin resistance. Nat. Commun. 12 (1), 213. doi:10.1038/s41467-020-20500-w
Kumar, R., Tang, Q., Müller, S. A., Gao, P., Mahlstedt, D., Zampagni, S., et al. (2020). Fibroblast growth factor 2-mediated regulation of neuronal exosome release depends on VAMP3/cellubrevin in hippocampal neurons. Adv. Sci. 7 (6), 1902372. doi:10.1002/advs.201902372
Kuruppu, S., Rajapakse, N. W., Dunstan, R. A., and Smith, A. I. (2014). Nitric oxide inhibits the production of soluble endothelin converting enzyme-1. Mol. Cell. Biochem. 396 (1-2), 49–54. doi:10.1007/s11010-014-2141-0
Lai, H., Li, Y., Zhang, H., Hu, J., Liao, J., Su, Y., et al. (2021). exoRBase 2.0: an atlas of mRNA, lncRNA and circRNA in extracellular vesicles from human biofluids. Nucleic Acids Res. 50 (D1), D118–D128. doi:10.1093/nar/gkab1085
Lázaro-Ibáñez, E., Faruqu, F. N., Saleh, A. F., Silva, A. M., Tzu-Wen Wang, J., Rak, J., et al. (2021). Selection of fluorescent, bioluminescent, and radioactive tracers to accurately reflect extracellular vesicle biodistribution in vivo. ACS Nano 15 (2), 3212–3227. doi:10.1021/acsnano.0c09873
Li, B., Cao, Y., Sun, M., and Feng, H. (2021). Expression, regulation, and function of exosome-derived miRNAs in cancer progression and therapy. FASEB J. 35 (10), e21916. doi:10.1096/fj.202100294RR
Li, I., and Nabet, B. Y. (2019). Exosomes in the tumor microenvironment as mediators of cancer therapy resistance. Mol. Cancer 18 (1), 32. doi:10.1186/s12943-019-0975-5
Li, M., Zhao, X., Yong, H., Xu, J., Qu, P., Qiao, S., et al. (2022). Transketolase promotes colorectal cancer metastasis through regulating AKT phosphorylation. Cell Death Dis. 13 (2), 99. doi:10.1038/s41419-022-04575-5
Li, M., Zou, X., Xia, T., Wang, T., Liu, P., Zhou, X., et al. (2019). A five-miRNA panel in plasma was identified for breast cancer diagnosis. Cancer Med. 8 (16), 7006–7017. doi:10.1002/cam4.2572
Li, Y. C., Yang, C. S., Zhou, W. L., Li, H. S., Han, Y. J., Wang, Q. S., et al. (2018). Low glucose metabolism in hepatocellular carcinoma with GPC3 expression. World J. Gastroenterol. 24 (4), 494–503. doi:10.3748/wjg.v24.i4.494
Li, Z., Zhuang, M., Zhang, L., Zheng, X., Yang, P., and Li, Z. (2016). Acetylation modification regulates GRP78 secretion in colon cancer cells. Sci. Rep. 6, 30406. doi:10.1038/srep30406
Liang, Y., Lehrich, B. M., Zheng, S., and Lu, M. (2021). Emerging methods in biomarker identification for extracellular vesicle-based liquid biopsy. J. Extracell. Vesicles 10 (7), e12090. doi:10.1002/jev2.12090
Liberti, M. V., and Locasale, J. W. (2016). The Warburg effect: How does it benefit cancer cells? Trends biochem. Sci. 41 (3), 211–218. doi:10.1016/j.tibs.2015.12.001
Liu, F., Bu, Z., Zhao, F., and Xiao, D. (2018). Increased T-helper 17 cell differentiation mediated by exosome-mediated microRNA-451 redistribution in gastric cancer infiltrated T cells. Cancer Sci. 109 (1), 65–73. doi:10.1111/cas.13429
Liu, J., Ren, L., Li, S., Li, W., Zheng, X., Yang, Y., et al. (2021). The biology, function, and applications of exosomes in cancer. Acta Pharm. Sin. B 11 (9), 2783–2797. doi:10.1016/j.apsb.2021.01.001
Lu, L., Huang, J., Mo, J., Da, X., Li, Q., Fan, M., et al. (2022). Exosomal lncRNA TUG1 from cancer-associated fibroblasts promotes liver cancer cell migration, invasion, and glycolysis by regulating the miR-524-5p/SIX1 axis. Cell. Mol. Biol. Lett. 27 (1), 17. doi:10.1186/s11658-022-00309-9
Lv, B., Zhu, W., and Feng, C. (2020). Coptisine blocks secretion of exosomal circCCT3 from cancer-associated fibroblasts to reprogram glucose metabolism in hepatocellular carcinoma. DNA Cell Biol. 39, 2281–2288. doi:10.1089/dna.2020.6058
Marshall, H. T., and Djamgoz, M. B. A. (2018). Immuno-Oncology: Emerging targets and combination therapies. Front. Oncol. 8, 315. doi:10.3389/fonc.2018.00315
Mashouri, L., Yousefi, H., Aref, A. R., Ahadi, A. M., Molaei, F., and Alahari, S. K. (2019). Exosomes: Composition, biogenesis, and mechanisms in cancer metastasis and drug resistance. Mol. Cancer 18 (1), 75. doi:10.1186/s12943-019-0991-5
Minchenko, A., Leshchinsky, I., Opentanova, I., Sang, N., Srinivas, V., Armstead, V., et al. (2002). Hypoxia-inducible factor-1-mediated expression of the 6-phosphofructo-2-kinase/fructose-2, 6-bisphosphatase-3 (PFKFB3) gene. Its possible role in the Warburg effect. J. Biol. Chem. 277 (8), 6183–6187. doi:10.1074/jbc.M110978200
Möller, A., and Lobb, R. J. (2020). The evolving translational potential of small extracellular vesicles in cancer. Nat. Rev. Cancer 20 (12), 697–709. doi:10.1038/s41568-020-00299-w
Morishita, M., Takahashi, Y., Nishikawa, M., Sano, K., Kato, K., Yamashita, T., et al. (2015). Quantitative analysis of tissue distribution of the B16BL6-derived exosomes using a streptavidin-lactadherin fusion protein and iodine-125-labeled biotin derivative after intravenous injection in mice. J. Pharm. Sci. 104 (2), 705–713. doi:10.1002/jps.24251
Morrissey, S. M., Zhang, F., Ding, C., Montoya-Durango, D. E., Hu, X., Yang, C., et al. (2021). Tumor-derived exosomes drive immunosuppressive macrophages in a pre-metastatic niche through glycolytic dominant metabolic reprogramming. Cell Metab. 33 (10), 20402040–20402058.e10. doi:10.1016/j.cmet.2021.09.002
Najy, A. J., Jung, Y.-S., Kim, S., Fridman, R., and Kim, H.-R. C. (2021). Regulation of tumor metabolism and extracellular acidosis by the TIMP-10–CD63 Axis in breast carcinoma. Cells 10 (10), 2721. doi:10.3390/cells10102721
O'Grady, T., Njock, M. S., Lion, M., Bruyr, J., Mariavelle, E., Galvan, B., et al. (2022). Sorting and packaging of RNA into extracellular vesicles shape intracellular transcript levels. BMC Biol. 20 (1), 72. doi:10.1186/s12915-022-01277-4
Palacios-Ferrer, J. L., García-Ortega, M. B., Gallardo-Gómez, M., García, M., Díaz, C., Boulaiz, H., et al. (2021). Metabolomic profile of cancer stem cell-derived exosomes from patients with malignant melanoma. Mol. Oncol. 15 (2), 407–428. doi:10.1002/1878-0261.12823
Pang, W., Yao, W., Dai, X., Zhang, A., Hou, L., Wang, L., et al. (2021). Pancreatic cancer-derived exosomal microRNA-19a induces β-cell dysfunction by targeting ADCY1 and EPAC2. Int. J. Biol. Sci. 17 (13), 3622–3633. doi:10.7150/ijbs.56271
Parri, M., Pietrovito, L., Grandi, A., Campagnoli, S., De Camilli, E., Bianchini, F., et al. (2014). Angiopoietin-like 7, a novel pro-angiogenetic factor over-expressed in cancer. Angiogenesis 17 (4), 881–896. doi:10.1007/s10456-014-9435-4
Poff, A., Koutnik, A. P., Egan, K. M., Sahebjam, S., D'Agostino, D., and Kumar, N. B. (2019). Targeting the Warburg effect for cancer treatment: Ketogenic diets for management of glioma. Semin. Cancer Biol. 56, 135–148. doi:10.1016/j.semcancer.2017.12.011
Pužar Dominkuš, P., Stenovec, M., Sitar, S., Lasič, E., Zorec, R., Plemenitaš, A., et al. (2018). PKH26 labeling of extracellular vesicles: Characterization and cellular internalization of contaminating PKH26 nanoparticles. Biochim. Biophys. Acta. Biomembr. 1860 (6), 1350–1361. doi:10.1016/j.bbamem.2018.03.013
Rai, A., Fang, H., Claridge, B., Simpson, R. J., and Greening, D. W. (2021). Proteomic dissection of large extracellular vesicle surfaceome unravels interactive surface platform. J. Extracell. Vesicles 10 (13), e12164. doi:10.1002/jev2.12164
Rai, A., Greening, D. W., Chen, M., Xu, R., Ji, H., and Simpson, R. J. (2019). Exosomes derived from human primary and metastatic colorectal cancer cells contribute to functional heterogeneity of activated fibroblasts by reprogramming their proteome. Proteomics 19 (8), e1800148. doi:10.1002/pmic.201800148
Rashid, M. H., Borin, T. F., Ara, R., Angara, K., Cai, J., Achyut, B. R., et al. (2019). Differential in vivo biodistribution of (131)I-labeled exosomes from diverse cellular origins and its implication for theranostic application. Nanomedicine 21, 102072. doi:10.1016/j.nano.2019.102072
Sah, R. P., Sharma, A., Nagpal, S., Patlolla, S. H., Sharma, A., Kandlakunta, H., et al. (2019). Phases of metabolic and soft tissue changes in months preceding a diagnosis of pancreatic ductal adenocarcinoma. Gastroenterology 156 (6), 1742–1752. doi:10.1053/j.gastro.2019.01.039
Sheikhbahaei, S., Verde, F., Hales, R. K., Rowe, S. P., and Solnes, L. B. (2020). Imaging in therapy response assessment and surveillance of lung cancer: Evidenced-based review with focus on the utility of (18)F-FDG PET/CT. Clin. Lung Cancer 21 (6), 485–497. doi:10.1016/j.cllc.2020.06.020
Shi, Q., Ji, T., Ma, Z., Tan, Q., and Liang, J. (2021). Serum exosomes-based biomarker circ_0008928 regulates cisplatin sensitivity, tumor progression, and glycolysis metabolism by miR-488/HK2 Axis in cisplatin-resistant nonsmall cell lung carcinoma. Cancer Biotherapy Radiopharm. doi:10.1089/cbr.2020.4490
Shi, S., Li, T., Wen, X., Wu, S. Y., Xiong, C., Zhao, J., et al. (2019). Copper-64 labeled PEGylated exosomes for in vivo positron emission tomography and enhanced tumor retention. Bioconjug. Chem. 30 (10), 2675–2683. doi:10.1021/acs.bioconjchem.9b00587
Shurtleff, M. J., Temoche-Diaz, M. M., Karfilis, K. V., Ri, S., and Schekman, R. (2016). Y-box protein 1 is required to sort microRNAs into exosomes in cells and in a cell-free reaction. Elife 5, e19276. doi:10.7554/eLife.19276
Su, J., Pang, W., Zhang, A., Li, L., Yao, W., and Dai, X. (2022). Exosomal miR-19a decreases insulin production by targeting Neurod1 in pancreatic cancer associated diabetes. Mol. Biol. Rep. 49 (3), 1711–1720. doi:10.1007/s11033-021-06980-z
Sun, W., Li, Z., Zhou, X., Yang, G., and Yuan, L. (2019). Efficient exosome delivery in refractory tissues assisted by ultrasound-targeted microbubble destruction. Drug Deliv. 26 (1), 45–50. doi:10.1080/10717544.2018.1534898
Sung, J. S., Kang, C. W., Kang, S., Jang, Y., Chae, Y. C., Kim, B. G., et al. (2020). ITGB4-mediated metabolic reprogramming of cancer-associated fibroblasts. Oncogene 39 (3), 664–676. doi:10.1038/s41388-019-1014-0
Tang, K. D., Wan, Y., Zhang, X., Bozyk, N., Vasani, S., Kenny, L., et al. (2021). Proteomic alterations in salivary exosomes derived from human papillomavirus-driven oropharyngeal cancer. Mol. Diagn. Ther. 25 (4), 505–515. doi:10.1007/s40291-021-00538-2
Temoche-Diaz, M. M., Shurtleff, M. J., Nottingham, R. M., Yao, J., Fadadu, R. P., Lambowitz, A. M., et al. (2019). Distinct mechanisms of microRNA sorting into cancer cell-derived extracellular vesicle subtypes. Elife 8, e47544. doi:10.7554/eLife.47544
Théry, C., Witwer, K. W., Aikawa, E., Alcaraz, M. J., Anderson, J. D., Andriantsitohaina, R., et al. (2018). Minimal information for studies of extracellular vesicles 2018 (MISEV2018): A position statement of the international society for extracellular vesicles and update of the MISEV2014 guidelines. J. Extracell. Vesicles 7 (1), 1535750. doi:10.1080/20013078.2018.1535750
van Eijndhoven, M. A., Zijlstra, J. M., Groenewegen, N. J., Drees, E. E., van Niele, S., Baglio, S. R., et al. (2016). Plasma vesicle miRNAs for therapy response monitoring in Hodgkin lymphoma patients. JCI Insight 1 (19), e89631. doi:10.1172/jci.insight.89631
Vander Heiden, M. G., Cantley, L. C., and Thompson, C. B. (2009). Understanding the Warburg effect: The metabolic requirements of cell proliferation. Science 324 (5930), 1029–1033. doi:10.1126/science.1160809
Wang, D., Zhao, C., Xu, F., Zhang, A., Jin, M., Zhang, K., et al. (2021). Cisplatin-resistant NSCLC cells induced by hypoxia transmit resistance to sensitive cells through exosomal PKM2. Theranostics 11 (6), 2860–2875. doi:10.7150/thno.51797
Wang, S., Wang, T., Liu, D., and Kong, H. (2020a). LncRNA MALAT1 aggravates the progression of non-small cell lung cancer by stimulating the expression of COMMD8 via targeting miR-613. Cancer Manag. Res. 12, 10735–10747. doi:10.2147/cmar.s263538
Wang, V. M. Y., Ferreira, R. M. M., Almagro, J., Evan, T., Legrave, N., Zaw Thin, M., et al. (2019). CD9 identifies pancreatic cancer stem cells and modulates glutamine metabolism to fuel tumour growth. Nat. Cell Biol. 21 (11), 1425–1435. doi:10.1038/s41556-019-0407-1
Wang, W., Zhu, N., Yan, T., Shi, Y. N., Chen, J., Zhang, C. J., et al. (2020b). The crosstalk: Exosomes and lipid metabolism. Cell Commun. Signal. 18 (1), 119. doi:10.1186/s12964-020-00581-2
Wang, X., Zhang, H., Yang, H., Bai, M., Ning, T., Deng, T., et al. (2020c). Exosome-delivered circRNA promotes glycolysis to induce chemoresistance through the miR-122-PKM2 axis in colorectal cancer. Mol. Oncol. 14 (3), 539–555. doi:10.1002/1878-0261.12629
Wei, Y., Tang, X., Ren, Y., Yang, Y., Song, F., Fu, J., et al. (2021). An RNA–RNA crosstalk network involving HMGB1 and RICTOR facilitates hepatocellular carcinoma tumorigenesis by promoting glutamine metabolism and impedes immunotherapy by PD-L1+ exosomes activity. Signal Transduct. Target. Ther. 6 (1), 421. doi:10.1038/s41392-021-00801-2
Wei, Y., Wang, D., Jin, F., Bian, Z., Li, L., Liang, H., et al. (2017). Pyruvate kinase type M2 promotes tumour cell exosome release via phosphorylating synaptosome-associated protein 23. Nat. Commun. 8, 14041. doi:10.1038/ncomms14041
Wu, H., Fu, M., Liu, J., Chong, W., Fang, Z., Du, F., et al. (2021). The role and application of small extracellular vesicles in gastric cancer. Mol. Cancer 20 (1), 71. doi:10.1186/s12943-021-01365-z
Wu, J., Yuan, M., Shen, J., Chen, Y., Zhang, R., Chen, X., et al. (2022). Effect of modified Jianpi Yangzheng on regulating content of PKM2 in gastric cancer cells-derived exosomes. Phytomedicine. 103, 154229. doi:10.1016/j.phymed.2022.154229
Wu, X., Zhou, Z., Xu, S., Liao, C., Chen, X., Li, B., et al. (2020). Extracellular vesicle packaged LMP1-activated fibroblasts promote tumor progression via autophagy and stroma-tumor metabolism coupling. Cancer Lett. 478, 93–106. doi:10.1016/j.canlet.2020.03.004
Xu, S., and Herschman, H. R. (2019). A tumor agnostic therapeutic strategy for hexokinase 1-null/hexokinase 2-positive cancers. Cancer Res. 79 (23), 5907–5914. doi:10.1158/0008-5472.can-19-1789
Xu, Y., Kong, S., Qin, S., Shen, X., and Ju, S. (2020). Exosomal circRNAs: Sorting mechanisms, roles and clinical applications in tumors. Front. Cell Dev. Biol. 8, 581558. doi:10.3389/fcell.2020.581558
Yan, W., Wu, X., Zhou, W., Fong, M. Y., Cao, M., Liu, J., et al. (2018). Cancer-cell-secreted exosomal miR-105 promotes tumour growth through the MYC-dependent metabolic reprogramming of stromal cells. Nat. Cell Biol. 20 (5), 597–609. doi:10.1038/s41556-018-0083-6
Yang, H., Xie, S., Liang, B., Tang, Q., Liu, H., Wang, D., et al. (2021). Exosomal IDH1 increases the resistance of colorectal cancer cells to 5-Fluorouracil. J. Cancer 12 (16), 4862–4872. doi:10.7150/jca.58846
Yang, H. Y., Wu, C. Y., Powell, J. D., and Lu, K. L. (2020). Manipulation of metabolic pathways and its consequences for anti-tumor immunity: A clinical perspective. Int. J. Mol. Sci. 21 (11), 4030. doi:10.3390/ijms21114030
Yang, L., Xia, L., Wang, Y., He, S., Chen, H., Liang, S., et al. (2017). Development and external validation of nomograms to predict the risk of skeletal metastasis at the time of diagnosis and skeletal metastasis-free survival in nasopharyngeal carcinoma. BMC Cancer 17 (1), 628. doi:10.1186/s12885-017-3630-9
Yang, L., Xie, M., Yang, M., Yu, Y., Zhu, S., Hou, W., et al. (2014). PKM2 regulates the Warburg effect and promotes HMGB1 release in sepsis. Nat. Commun. 5, 4436. doi:10.1038/ncomms5436
Yousefi, H., Maheronnaghsh, M., Molaei, F., Mashouri, L., Reza Aref, A., Momeny, M., et al. (2020). Long noncoding RNAs and exosomal lncRNAs: Classification, and mechanisms in breast cancer metastasis and drug resistance. Oncogene 39 (5), 953–974. doi:10.1038/s41388-019-1040-y
Zebrowska, A., Skowronek, A., Wojakowska, A., Widlak, P., and Pietrowska, M. (2019). Metabolome of exosomes: Focus on vesicles released by cancer cells and present in human body fluids. Int. J. Mol. Sci. 20 (14), 3461. doi:10.3390/ijms20143461
Zhang, Q., Yang, X., and Liu, H. (2021a). Extracellular vesicles in cancer metabolism: Implications for cancer diagnosis and treatment. Technol. Cancer Res. Treat. 20, 15330338211037821. doi:10.1177/15330338211037821
Zhang, R., Xia, Y., Wang, Z., Zheng, J., Chen, Y., Li, X., et al. (2017). Serum long non coding RNA MALAT-1 protected by exosomes is up-regulated and promotes cell proliferation and migration in non-small cell lung cancer. Biochem. Biophys. Res. Commun. 490 (2), 406–414. doi:10.1016/j.bbrc.2017.06.055
Zhang, S., Pan, D., Zhang, S., Wu, Q., Zhen, L., Liu, S., et al. (2022). Exosomal miR-543 inhibits the proliferation of ovarian cancer by targeting IGF2. J. Immunol. Res. 2022, 2003739. doi:10.1155/2022/2003739
Zhang, Y., Huang, S., Li, P., Chen, Q., Li, Y., Zhou, Y., et al. (2018). Pancreatic cancer-derived exosomes suppress the production of GIP and GLP-1 from STC-1 cells in vitro by down-regulating the PCSK1/3. Cancer Lett. 431, 190–200. doi:10.1016/j.canlet.2018.05.027
Zhang, Y., Li, Y., Liu, P., Gong, D., Zhou, H., Li, W., et al. (2021b). Phosphatase Shp2 regulates biogenesis of small extracellular vesicles by dephosphorylating Syntenin. J. Extracell. Vesicles 10 (5), e12078. doi:10.1002/jev2.12078
Zhao, H., Yang, L., Baddour, J., Achreja, A., Bernard, V., Moss, T., et al. (2016). Tumor microenvironment derived exosomes pleiotropically modulate cancer cell metabolism. Elife 5, e10250. doi:10.7554/eLife.10250
Zheng, Y., Liu, P., Wang, N., Wang, S., Yang, B., Li, M., et al. (2019). Betulinic acid suppresses breast cancer metastasis by targeting GRP78-mediated glycolysis and ER stress apoptotic pathway. Oxid. Med. Cell. Longev. 2019, 8781690. doi:10.1155/2019/8781690
Zhou, S., Lan, Y., Li, Y., Li, Z., Pu, J., and Wei, L. (2022). Hypoxic tumor-derived exosomes induce M2 macrophage polarization via PKM2/AMPK to promote lung cancer progression. Cell Transpl. 31, 9636897221106998. doi:10.1177/09636897221106998
Keywords: small extracellular vesicles, exosomes, warburg effect, tumor metabolism, liquid biopsy, glycolysis, drug development
Citation: Yang H, Wang J and Huang G (2022) Small extracellular vesicles in metabolic remodeling of tumor cells: Cargos and translational application. Front. Pharmacol. 13:1009952. doi: 10.3389/fphar.2022.1009952
Received: 02 August 2022; Accepted: 07 December 2022;
Published: 16 December 2022.
Edited by:
Hai-Long Piao, Dalian Institute of Chemical Physics (CAS), ChinaReviewed by:
Zhiyang Li, Nanjing Drum Tower Hospital, ChinaBilikere S. Dwarakanath, Sri Ramachandra Institute of Higher Education and Research, India
Copyright © 2022 Yang, Wang and Huang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Gang Huang, aHVhbmdnYW5nQHN1bWhzLmVkdS5jbg==; Hao Yang, eWFuZ2hAc3VtaHMuZWR1LmNu
 Hao Yang
Hao Yang Jingyi Wang2
Jingyi Wang2 Gang Huang
Gang Huang