
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Pharmacol., 27 January 2022
Sec. Experimental Pharmacology and Drug Discovery
Volume 12 - 2021 | https://doi.org/10.3389/fphar.2021.785981
This article is part of the Research TopicEpigenetic Mechanisms and Post-translational Modifications as Novel Therapeutic Targets in CancerView all 12 articles
 Jing Yang1†
Jing Yang1† Zimeng Wu1†
Zimeng Wu1† Li Yang1,2,3*
Li Yang1,2,3* Ji-Hak Jeong4
Ji-Hak Jeong4 Yuanhang Zhu1
Yuanhang Zhu1 Jie Lu1
Jie Lu1 Baojin Wang1,2
Baojin Wang1,2 Nannan Wang1
Nannan Wang1 Yan Wang1
Yan Wang1 Ke Shen1
Ke Shen1 Ruiqing Li1
Ruiqing Li1Kinesin family member 2C (KIF2C) is known as an oncogenic gene to regulate tumor progression and metastasis. However, its pan-cancer analysis has not been reported. In this study, we comprehensively analyzed the characteristics of KIF2C in various cancers. We found that KIF2C was highly expressed and corresponded to a poor prognosis in various cancers. We also found a significant correlation between KIF2C and clinicopathological characteristics, particularly in cervical cancer, which is the most common gynecological malignancy and is the second leading cause of cancer-related deaths among women worldwide. KIF2C mutation is strongly associated with the survival rate of cervical cancer, and KIF2C expression was significantly upregulated in cervical cancer tissues and cervical cancer cells. Moreover, KIF2C promoted cervical cancer cells proliferation, invasion, and migration in vitro and as well increased tumor growth in vivo. KIF2C knockdown promotes the activation of the p53 signaling pathway by regulating the expression of related proteins. The rescue assay with KIF2C and p53 double knockdown partially reversed the inhibitory influence of KIF2C silencing on cervical cancer processes. In summary, our study provided a relatively comprehensive description of KIF2C as an oncogenic gene and suggested KIF2C as a therapeutic target for cervical cancer.
Kinesin family member 2C (KIF2C), also known as mitotic centromere associated kinesin (MCAK), is the most representative member of the kinesin family-13 (Kinesin-13) (Ritter et al., 2015). It participates in the functions of microtubules and regulates mitosis and cell cycle, involving in the disaggregation and formation of secondary spindles and the separation of chromosomes (DeLuca et al., 2006; Ganguly et al., 2008; Hedrick et al., 2008; Huang et al., 2009). Because KIF2C is involved in the regulation of tumor development, proliferation, and metastasis, it has been considered as a candidate promoting factor for breast cancer, liver cancer, lung cancer, bladder cancer, colon cancer, and other cancers (Bai et al., 2019; Wei et al., 2020; Yang et al., 2020). Shi et al. reported that KIF2C is activated by the Wnt/β-catenin signaling pathway and plays a key role in mTORC1 signal transduction and hepatocellular carcinoma progression through its interaction with TbC1D7 (Wei et al., 2020). Gan et al. (2019) revealed that KIF2C has a carcinogenic effect in non-small cell lung cancer and is negatively regulated by miR-325-3p. Yang et al. (2020) showed that circRGNEF promotes the occurrence and development of bladder cancer by directly targeting miR-548 and upregulating the expression of KIF2C.
Cervical cancer is one of the most common gynecological malignancies with high morbidity and mortality, and it is the second leading cause of cancer-related deaths among all female malignant tumors (Siegel et al., 2021). The most common pathological type of cervical cancer is cervical squamous cell carcinoma (Zhao et al., 2019). When cervical cancer patients are diagnosed and treated in the early tumor stage, the chance of survival can be increased and may push the 5-year survival rate to reach as high as 90% (Das, 2021). However, the early symptoms of cervical cancer are not obvious, and 70% of cervical cancer patients are in the middle and late stages of the disease when they are diagnosed with cancer before receiving treatment. Additionally, approximately 31% of cervical cancers relapse after the treatment, and most of the recurrence occurs within 2 years after the initial diagnosis (Zhao and Qiao, 2019; Brisson et al., 2020). The treatment of recurrent cervical cancer is very difficult, and its prognosis is very poor, causing the 1-year survival rate to be only 8–12% (Cohen et al., 2019). Therefore, early treatment provides life-saving, and more and more attention has been paid to finding meaningful therapy targets for cervical cancer.
In the present study, we used The Cancer Genome Atlas (TCGA) project and Gene Expression Omnibus databases to conduct a pan-cancer analysis of KIF2C for the first time, including gene expression, survival status, and genetic alteration. Interestingly, we identified KIF2C that has a significant value for the research in cervical cancer. Hence, we experimentally confirm that KIF2C expression was increased in cervical cancer tissues and cervical cancer cells. Through in vivo experiments, KIF2C was found to play an important role in the development of cervical cancer. In addition, RNA sequencing analysis of cervical cancer cells with KIF2C knockdown showed that differentially expressed genes were enriched in the p53 signaling pathway. Further analysis of the expression of proteins in the p53 signaling pathway showed that the knockdown of KIF2C could induce the activation of the p53 signaling pathway. Beyond this, rescue experiments demonstrated that knockdown of p53 partially reversed the inhibitory influence of knockdown KIF2C silencing on cervical cancer processes. Our findings provide new insights into the functional and mechanistic link between the KIF2C and P53 signaling pathways in cervical cancer, which might help discover adjuvant therapy for patients with cervical cancer.
The KIF2C expression of tumor and adjacent normal tissues profile was analyzed using the TIMER 2.0 database (https://cistrome.shinyapps.io/timer/), a bioinformatics tool for analyzing and visualizing TCGA (Li et al., 2020a). The 33 TCGA cancer types analyzed are shown in Table 1. The gene expression levels are represented as log2 transcript per million (TPM) values. For some tumor tissues without normal tissues, normal expression comparison analyses, the TPM of the given genes from TCGA, and matched GETx (Genotype-Tissue Expression) normal tissue RNA sequencing (RNA-seq) datasets were visualized by GEPIA2 (http://gepia2.cancer-pku.cn/#analysis) (Tang et al., 2017). The cutoff for significance was chosen at p = 0.01. The tumor pathological staging map was also obtained from GEPIA2. TPM values were calculated, and the expression levels of genes were presented using the Log2 (TPM + 1) scale.
Protein expression of KIF2C was evaluated using the UALCAN online analysis software (http://ualcan.path.uab.edu/index.html), which is a comprehensive web resource for analyzing cancer-omics data including Clinical Proteomic Tumor Analysis Consortium analysis in various tumor types (Chen et al., 2019).
The correlation between KIF2C messenger RNA (mRNA) expression and overall survival (OS) and disease-free survival (DFS) was predicted using the PrognoScan database (http://www.abren.net/PrognoScan/) and GEPIA2. PrognoScan database is also a collection of publicly available cancer microarray datasets and a tool for assessing the biological relationships between gene expression and prognosis, providing a convenient platform to evaluate potential tumor markers and therapeutic targets (Mizuno et al., 2009). The threshold was set as a Cox p-value < 0.05.
Genomic alteration data were analyzed through the cBio Cancer Genomics Portal (http://cbioportal.org) (Cerami et al., 2012; Gao et al., 2013), which collects multidimensional cancer genomics dataset. We observed the alteration frequency and mutation type of all 33 tumors. We also obtain the data on the overall disease-free, progression-free, and DFS differences for TCGA cancer cases with or without KIF2C genetic alteration.
HPA provides tissue and cell distribution information of 26,000 human proteins (Uhlén et al., 2015). In this database, researchers used highly specific antibodies and immunoassay techniques to detect the expression of each protein in 64 cell lines, 48 human normal tissues, and 20 tumor tissues in detail. In this study, we conducted HPA to observe the localization of KIF2C in cells and compare the protein expression of KIF2C between normal tissue and carcinoma tissue.
The LinkedOmics database 8 (http://linkedomics.org) (Vasaikar et al., 2018) is an online open-access powerful bioinformatics platform, which includes multi-omics information and clinical data involving 11,158 patients and 32 cancer types in TCGA project. We used the Linkedomics platform to analyze the relationship between the expression level of KIF2C in TCGA database and the clinical–pathological characteristics of patients.
Tissue specimens were obtained from patients who underwent hysterectomy and pathological diagnosis in the Department of Pathology of The Third Affiliated Hospital of Zhengzhou University (Zhengzhou, China) from 2017 to 2021. Paraffin-embedded histological specimens of 35 cervical squamous tissue samples, 25 cervical adenocarcinoma tissue samples, and 40 normal cervical tissues samples were sectioned (thickness 5 μm). All the patients had not received any antitumor therapy before their diagnoses. The project was approved by the Medical Ethics Committee of the Third Affiliated Hospital of Zhengzhou University (ethics approval number: 2021-080-01) and performed in accordance with the principles of the Declaration of Helsinki.
Immunohistochemistry and scoring criteria were performed as described (Soumaoro et al., 2004). Briefly, the tissue sections were deparaffinized in xylene, rehydrated in graded ethanol, and soaked in distilled water. After heat-mediated citric acid antigen retrieval, the tissue sections were incubated with the primary antibody for KIF2C (DF7348, rabbit antihuman, 1:100, Affinity) and Ki-67 (ab92742, rabbit antihuman, 1:500, Abcam) at 4°C overnight and incubated with horseradish peroxidase-conjugated antibody for 60 min at room temperature. Staining was developed by incubating with diaminobenzidine. Finally, these processed tissue sections were observed under a microscope and then counterstained with hematoxylin. The results were scored independently by two pathologists. A total score of >3 points was considered a high expression, and ≤3 points was considered low expression.
Cervical cancer cell lines SiHa, HeLa, C33a, and Caski and normal cervical cell line HCerEpic were borrowed from the relevant research group with our laboratory. All cells were cultured in Dulbecco’s modified Eagle’s medium (Solarbio, SuoLaibao Technology Co., Ltd.) supplemented with 10% fetal bovine serum (Gibco, Thermo Fisher Scientific, Inc.) and incubated at 37°C in a moist atmosphere containing 5% CO2.
When C33a and SiHa cells reached 20–30% confluence on the day of transfection, two lentiviral stocks (named GV248-shKIF2C and GV248-shCon, purchased from Genechem, Shanghai, China) were transduced into the cells with HitransG A, respectively. Stable cell lines were screened with 0.25 μg/ml puromycin. The cells were collected when 80–90% of the cells with green fluorescence were grown under a fluorescence microscope, then quantitative real-time polymerase chain reaction (qRT-PCR) and Western blot analysis were used to analyze the level of KIF2C expression.
CRISPR/Cas9 p53 knockdown and its control plasmids were kindly provided by Dr. Qi Zhang; then, the plasmids were transfected into SiHa-sh-KIF2C and SiHa-sh-NC cervical cancer cells, respectively. The cells were transfected using JetPrime transfection reagent (Polyplus transfection, Illkirch, France) according to the manufacturer’s protocol; then, Western blot analysis was used to analyze the level of p53 expression.
For overexpression, the plasmid (vector name: GV657) for human KIF2C and negative control were synthesized by Genechem (Shanghai, China), then transfected into Caski cell using JetPrime transfection reagent (Polyplus transfection, Illkirch, France) according to the manufacturer’s protocol using 1=µl of JetPrime for 1 µg of plasmid DNA. After being transfected with 24 h, the protein was collected, and Western blotting was used to verify the transfection efficiency.
According to the supplier’s instructions, synthesis of complementary DNA by extracting 1 μg total RNA with Trizol (Cwbio, Beijing, China) and SYBR reverse transcription reagent (US Everbright Inc., Suzhou, China). The ratio of target glyceraldehyde 3-phosphate dehydrogenase expressed the relative expression of target gene mRNA. The sequences for the primers are as follows:
GADPH, Forword, 5′-TGACTTCAACAGCGACACCCA-3′;
Reverse, 5′-CACCCTGTTGCTGTAGCCAAA-3′;
KIF2C, Forword, 5′-GGAGGAGAAGGCTATGGAAGA-3′;
Reverse, 5′-TCGCAGGGCTGAGAAATG-3′.
The relative gene expression was normalized to control by the 2−ΔΔCT method.
Total proteins from cells were extracted with radioimmunoprecipitation assay lysis buffer (Solarbio, SuoLaibao Technology Co., Ltd.). Western blot analysis was carried out by first electrophoresing the proteins through a sodium dodecyl sulfate–polyacrylamide gel electrophoresis gel and subsequently transferring to 0.45-µm polyvinylidene fluoride membrane (Millipore, Ireland). Immunoblotting was performed using antibodies against indicated proteins: KIF2C (DF7348, rabbit antihuman, 1:1,000, Affinity), p21 (10355-1-AP, rabbit antihuman, 1:1,000, Proteintech), p53 (sc126, mouse antihuman, 1:, Santa), Bax (50599-2-Ig, rabbit antihuman, 1:5,000, Proteintech), bcl-2 (60178-1-Ig, mouse antihuman, 1:2,000, Proteintech), and GADPH (bs2188R, rabbit antihuman, 1:5,000, Bioss). Glyceraldehyde 3-phosphate dehydrogenase was used as an endogenous control. The transferred membranes were blocked with 5% milk for 2 h and then incubated with the indicated primary antibody overnight at 4°C. Then, the membranes were incubated with the appropriate secondary antibodies (1:9,000) at room temperature for 1 h. Finally, the electrogenerated chemiluminescence solution (A:B = 1:1) was added onto the membrane, and the front of the membrane was placed on the glass plate. The GelView 6000Plus was used to process the pictures after exposure.
The successfully transfected cervical cancer cells (C33a, SiHa, and Caski) were inoculated in 96-well plates with 4 × 103 cells per well. A 10-µl CCK-8 solution (US Everbright Inc., Suzhou, China) was added to each well after plating for 12, 24, 48, 72, and 96 h. When the cells were further incubated for 3 h, a microboard reader (Thermo Scientific) was used to measure the spectrometric absorbance at the wavelengths of 450 nm.
After transfection, the cells were inoculated in six-well plates with 600 per well and cultured 2 weeks. Colonies were fixed with methanol for 20 min and subsequently stained with 0.1% crystal violet for 30 min. Colonies with more than 60 cells were defined as positive.
For this assay, serum-free C33a, SiHa, and Caski cells were seeded to the upper chamber with 5 × 104 cells in 100 µl, and the medium containing 20% fetal bovine serum was used as a chemoattractant in the lower chamber. After incubating for 36 h, migrated cells were fixed by formaldehyde and stained with crystal violet. For invasion assay, cells were also suspended in serum-free medium and counted and seeded in the upper chamber at a concentration of 5 × 104 cells in 100 µl; the only difference was spreading the Matrigel Basement Membrane Matrix (BD Bioscience, United States) on the upper chamber before adding the cells. The number of migrated and invaded cells was captured and calculated using a microscope.
Total RNAs were extracted from successfully transfected cervical cancer cells using Trizol reagent (Cwbio, Beijing, China). After a primary test for RNA quality and integrity, the sequencing samples were finally sequenced on Novaseq 6000 sequencer (Illumina) with PE150 model. The raw sequencing data were mapped to the human genome using STRA 2.5 software. Reads mapped to the exon regions of each gene were counted by featureCounts, and then reads per kilobase of transcript per million reads mapped was calculated. Differentially expressed (p-value cutoff of 0.05; the fold-change cutoff of 2) genes between groups were identified using the edgeR package. For further precise analysis, Gene Ontology (GO) analysis and Kyoto Encyclopedia of Genes and Genomes (KEGG) enrichment analysis for differentially expressed genes were conducted by KOBAS 2.1 software (p-value < 0.05; false discovery rate < 0.05). The RNA sequencing analysis and data analysis were performed with the help of Seqhealth cooperation (Wuhan, China). RNA-seq data have been deposited to National Center for Biotechnology Information Sequence Read Archive (Sequence Read Archive accession: PRJNA767113).
Four- to 5-week-old female BALB/c nude mice (SPF Biotechnology Co., Ltd., Beijing, China) were used in this study. The control and experimental cervical cancer cells (2 × 106) were transplanted to establish the subcutaneous xenograft model. Tumor volumes were measured every 7 days. After 4 weeks, the mice were killed, and tumor weight was examined. The tumor volumes were calculated by the formula: tumor volume = length × width2/2. Animal research was approved by the Animal Care and Use Committee of Zhengzhou University and was conducted in accordance with the Animal Care Guidelines of Zhengzhou University.
Statistical analysis of all data was done using SPSS 26.0 software. The measurement data with normal distribution was exhibited as the mean ± standard deviation. The statistical significance between the two groups was compared with the two-tailed Student’s t-test. Pearson’s chi-square test analyzed the association between KIF2C expression and clinicopathological features. p < 0.05 was considered statistically significant. All cell experiments had three repetitions.
In the TIMER database, we investigated the expression of KIF2C in pan-cancer. The GEPIA2 database supplemented part of cancers without normal samples. The results are shown in Figures 1A,B. As expected, KIF2C expression was significantly increased in most tumor tissues compared with normal tissues, including TCGT and LAML. However, there was no significant difference between tumor and normal tissues in ACC and LGG. As there were no normal samples studied in either dataset, an analysis could not be performed for MESO and UVM. In addition, the protein expression level of KIF2C was also shown in a similar manner. Clinical Proteomic Tumor Analysis Consortium analysis showed that the protein expression of KIF2C was higher in primary tissues of BRAC, OV, COAD, KIRC, UCEC, and LUAD compared with those in normal tissues (Figure 1C). All the results were statistically significant (p < 0.05). Next, we used the UALCAN database to analyze the correlation between KIF2C mRNA expression and clinicopathological stages. Expression of KIF2C was highly associated with advanced clinical stage (Figure 1D) of ACC (p < 0.001), BRCA (p < 0.001), KICH (p < 0.001), KIRC (p < 0.001), KIRP (p < 0.001), SKCM (p = 0.04), LUAD (p = 0.001), and LIHC (p < 0.001) but not others.
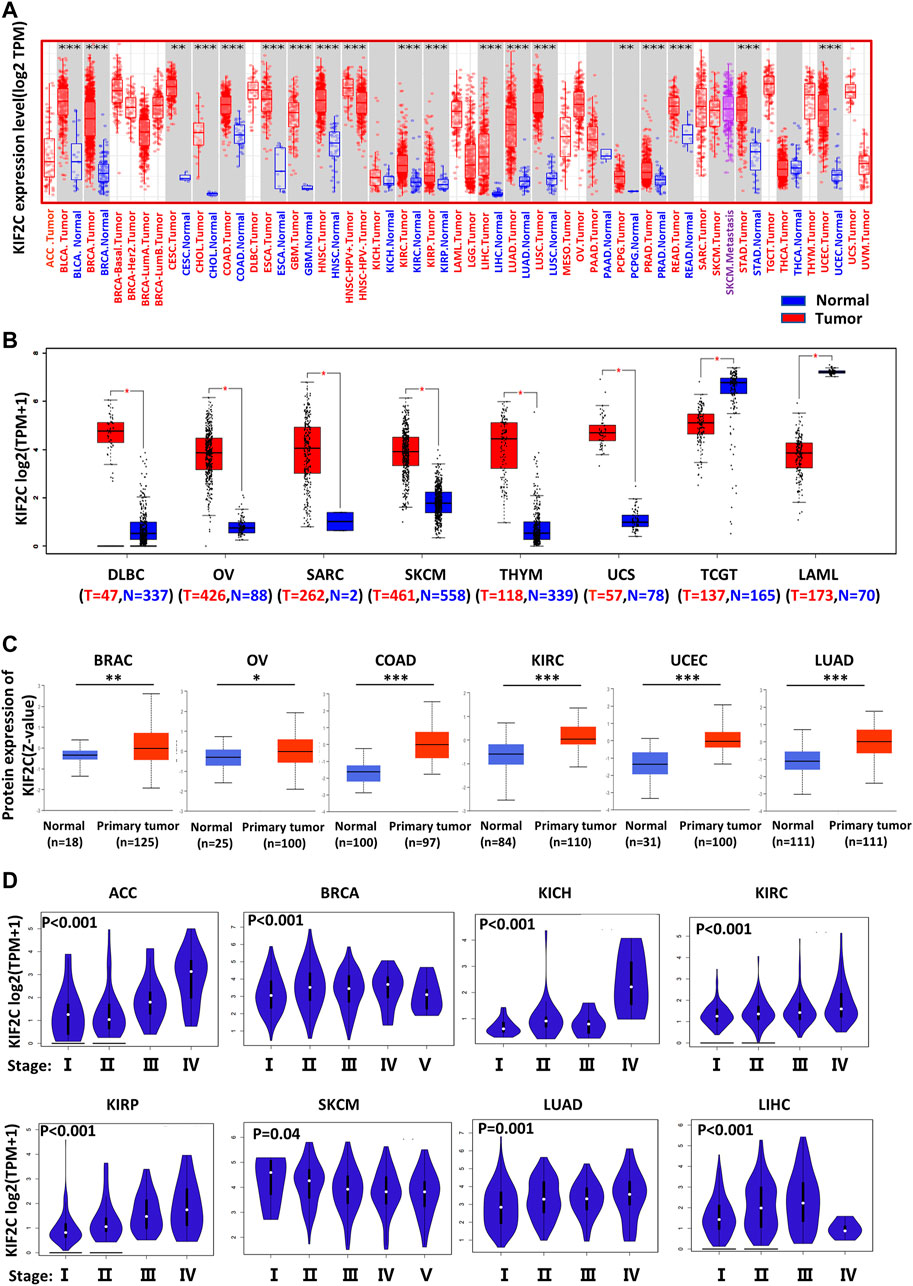
FIGURE 1. Different expression of KIF2C in various tumors and pathological stages. (A) KIF2C expression in 33 tumor types and paracancer tissues determined by TIMER. (B) Tumor types of DLBC, OV, SARC, SKCM, THYM, UCS, TGCT, and LAML were analyzed by box plot, for which corresponding normal tissues of GTEx database were included as controls. (C) Diagrams of quantified proteins analyzed by Clinical Proteomic Tumor Analysis Consortium in BRAC, OV, COAD, KIRC, UCEC, and LUAD. (D) Expression of KIF2C in different tumor stages. Log2 (TPM+1) was applied for log-scale. *p < 0.05; **p < 0.01; ***p < 0.001.
Next, we investigated KIF2C prognostic value using PrognoScan and GEPIA2 database. In PrognoScan, high KIF2C expression is associated with worse OS [hazard ratio (HR) = 1.45, Cox p = 0.000964] and disease-specific survival (DSS, HR = 2.2, Cox p = 0.000025) in bladder cancer (Figures 2A,B), with worse OS (HR = 4.35, Cox p = 0.000473) in brain cancer (Figure 2C), with worse OS (HR = 1.39, COX p = 0.047079), distant metastasis-free survival (HR = 4.98, COX p = 0.003309), DFS (HR = 6.19, COX p = 0.001105), and recurrence-free survival (RFS, HR = 4.98, COX p = 0.000259) in breast cancer (Figures 2D–G), with worse distant relapse-free survival (HR = 2.33, COX p = 0.000006) in soft tissue cancer (Figure 2H), with worse OS (HR = 3.6, COX p = 0.042603), RFS (HR = 2.41, COX p = 0.000001) in lung cancer (Figures 2I,J) and with worse OS (HR = 9.01, COX p = 0.000058) in skin cancer (Figure 2K). Conversely, KIF2C high expression is associated with good DFS (HR = 0.53, COX p = 0.013566) in colorectal cancer (Figure 2L). For blood cancer (Figures 2M,N), KIF2C presented the opposite manner in OS (HR = 0.53, COX p = 0.017) and DSS (HR = 2.53, COX p = 0.000363). For other cancers, no statistically significant associations were observed.
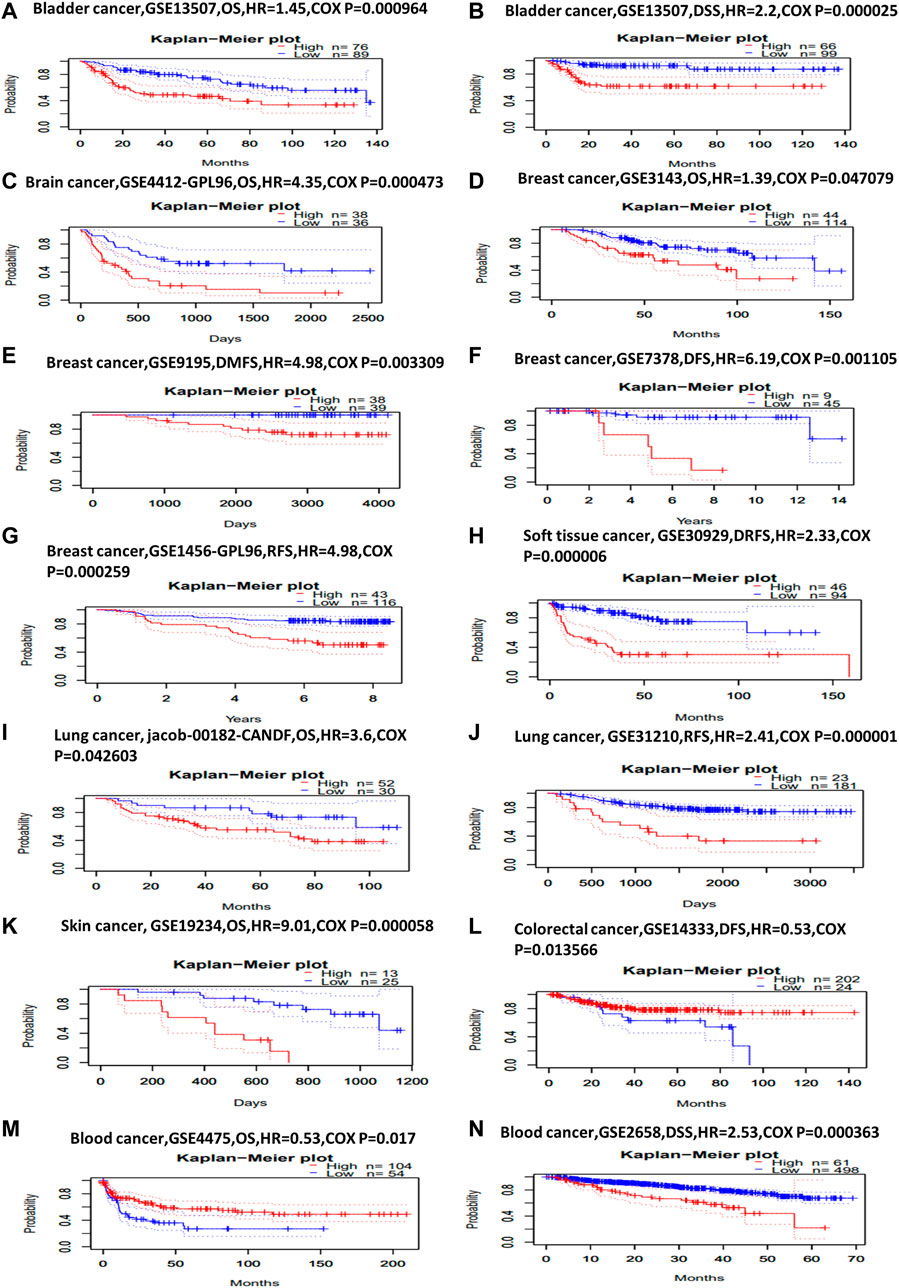
FIGURE 2. Kaplan–Meier survival curves comparing high and low expression of KIF2C in different cancer types in PrognoScan. (A) OS (n = 165) in bladder cancer cohort GSE13507. (B) DSS (n = 165) in bladder cancer cohort GSE13507. (C) OS (n = 74) in brain cancer cohort GSE4412-GPL96. (D) OS (n = 158) in breast cancer cohort GSE3143. (E) Distant metastasis-free survival (n = 77) in breast cancer cohort GSE9195. (F) DFS (n = 54) in breast cancer cohort GSE7378. (G) RFS (n = 159) in breast cancer cohort GSE1456-GPL96. (H) Distant relapse-free survival (n = 140) in soft-tissue cancer cohort GSE30929. (I) OS (n = 82) in lung cancer cohort jacob-00182-CANDF. (J) RFS (n = 204) in lung cancer cohort GSE31210. (K) OS (n = 38) in skin cancer cohort GSE19234. (L) DFS (n = 226) in colorectal cancer cohort GSE14333. (M) OS (n = 158) in blood cancer cohort GSE4475. (N) DSS (n = 559) in blood cancer cohort GSE2658.
To further explore the prognostic value of KIF2C, we performed the survival analysis using Kaplan–Meier curves from the GEPIA2. As shown in Figure 3A, higher KIF2C expression was associated with poor OS in ACC (p < 0.001), KICH (p = 0.018), KIRC (p = 0.006), KIRP (p = 0.0017), LGG (p < 0.001), LIHC (p < 0.001), LUAD (p = 0.016), MESO (p < 0.001), PAAD (p = 0.02), SKCM (p = 0.01), and PRAD (p = 0.049), whereas higher KIF2C expression was linked to better prognosis in THYM (p = 0.034). As for DFS (Figure 3B), high KIF2C expression was significantly associated with worse DFS in ACC (p < 0.001), KIRC (p = 0.0018), KIRP (p < 0.001), LGG (p < 0.001), PAAD (p < 0.001), LIHC (p < 0.001), PRAD (p = 0.00011), THCA (p = 0.03), and MESO (p = 0.016). These results suggested that higher KIF2C was associated with worse prognosis.
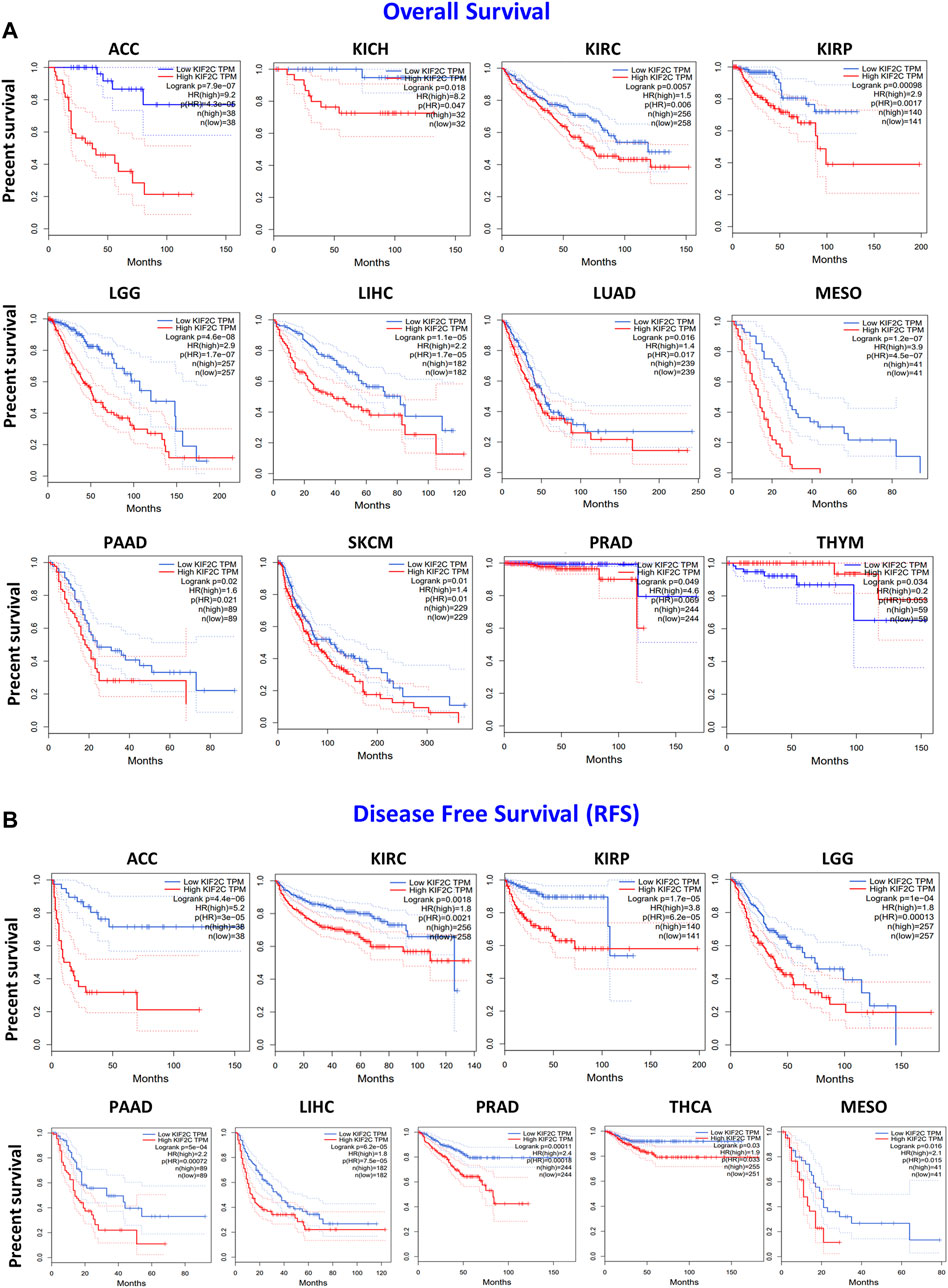
FIGURE 3. KIF2C expression and its association with survival in cancers based on TCGA data analysis. GEPIA2 software was used to analyze (A) overall survival rate and (B) disease-free survival rate of different tumors in TCGA. Survival plots and Kaplan–Meier curves showing positive results.
KIF2C mutations and copy number alterations were investigated using the cBioPortal database. The genetic alteration includes mutation, amplification, deep deletion, and multiple alterations. Notably, the highest mutation frequency (>6%) was observed in UCEC (Figure 4A). The amplification type of CNA was the major type in the OV, which showed an alteration frequency of ∼4% (Figure 4A). The predominant mutation type in PGPC was a deep deletion that resulted in a frameshift (Figure 4A). All COAD and LAML cases with genetic alteration had a mutation of KIF2C (Figure 4A). In addition, the amplification of KIF2C was present in all SARC, MESO, and LIHC cases (Figure 4A). The information of KIF2C genetic alteration is presented in Figure 4B. The missense mutation was the only type of genetic alteration of KIF2C. In addition, we also explored the potential association between KIF2C genetic alteration and clinical survival rate in various cancers. The CESC patients with KIF2C genetic alteration had poor prognosis in OS (Figure 4C, p = 0.047), progression-free survival (Figure 4C, p < 0.001), DFS (Figure 4C, p < 0.001), and DSS (Figure 4C, p = 0.0167). We also used the HPA to investigate the intracellular location and tissue protein expression levels of KIF2C. We found that KIF2C was mostly localized in both cellular nucleus and cytosol (Figure 4D), and it was only detected in CESC tissues (Figure 4D). These results suggested that KIF2C could be a potential therapeutic target in CESC.
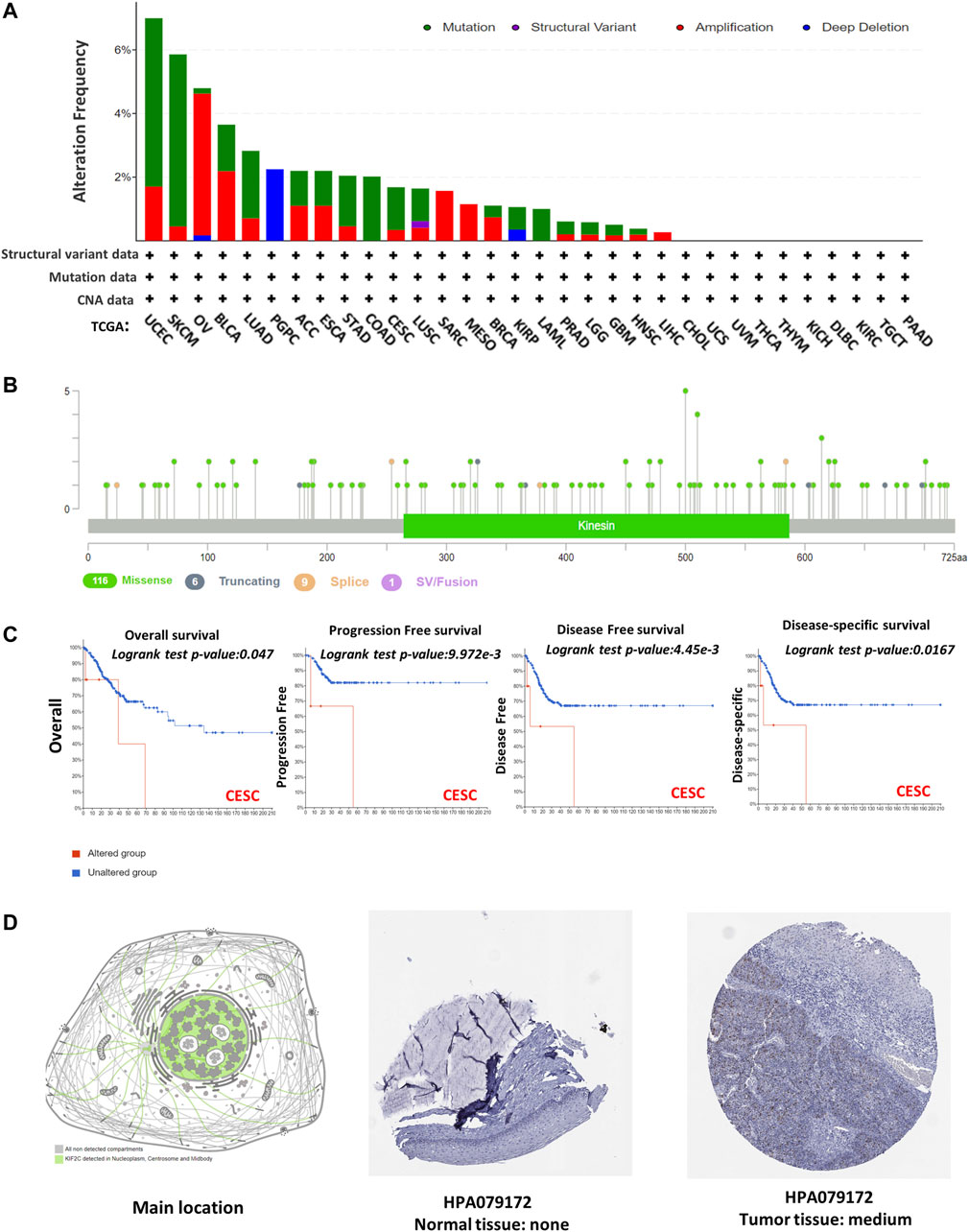
FIGURE 4. Mutation feature and tissue protein expression of KIF2C in different tumors of TCGA. Mutation features data were obtained using cBioPortal. Alteration frequencies with (A) mutation type and (B) mutation site are shown. (C) Using cBioPortal tool, we analyzed potential correlation between mutation status and overall, disease-specific, disease-free, and progression-free survivals of CESC. Nuclear localization of KIF2C and immunohistochemical results in CESC are shown in (D).
To explore the possibility of KIF2C as a therapeutic target of cervical cancer, we first detected its expression in human tissues. Findings showed that KIF2C was upregulated in cervical cancer tissues compared with normal tissues (Figures 5A,C,E). Not only that, the expression of KIF2C was higher in squamous cervical carcinoma than cervical adenocarcinoma. All three differences were statistically significant (p < 0.05, Figure 5G). Besides, we also found that the expression of KIF2C has a similar expression pattern with Ki-67 in cervical tissues (Figures 5B,D,F). This suggested that the expression of KIF2C may be related to the active proliferation of cancer cells.
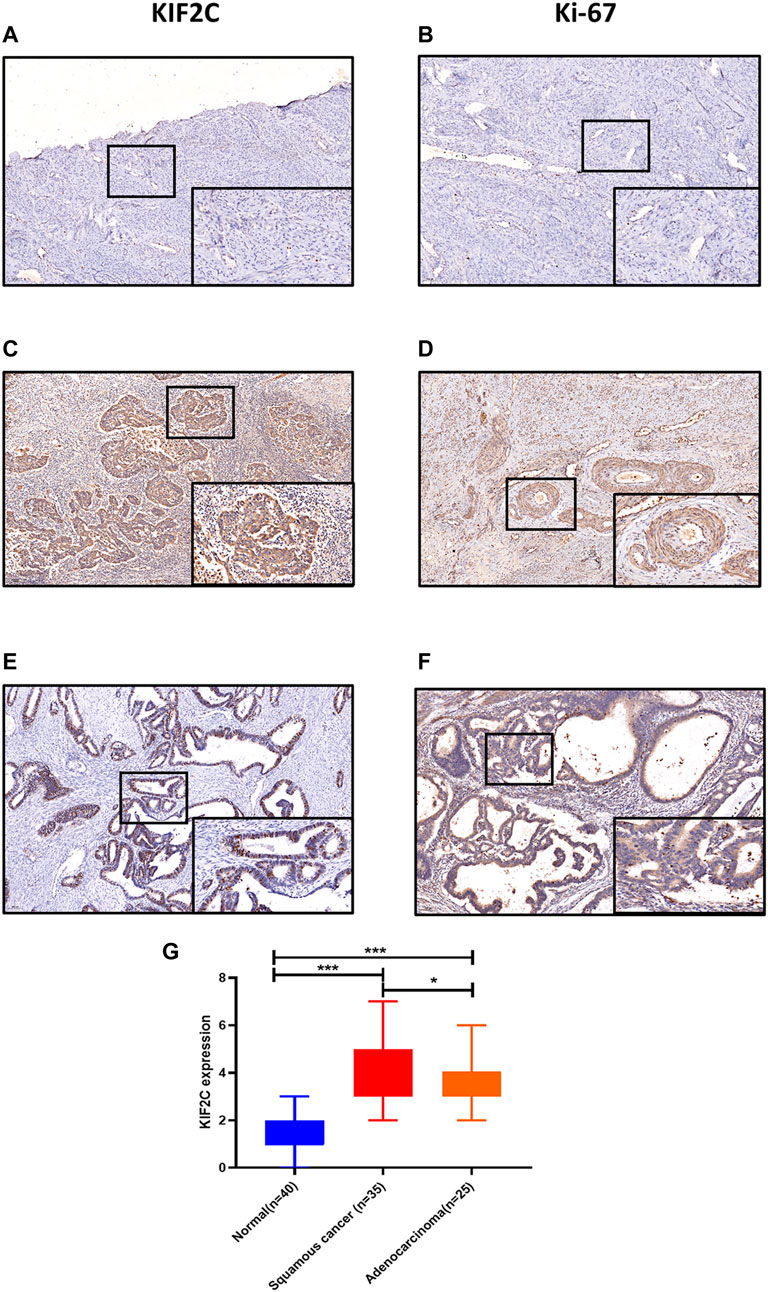
FIGURE 5. KIF2C was highly expressed in cervical cancer tissues. (A, B) Immunohistochemical staining of KIF2C and Ki-67 in normal cervical tissues. (C, D) Immunohistochemical staining of KIF2C and Ki-67 in cervical squamous cell carcinoma tissues. (E,F) Immunohistochemical staining of KIF2C and Ki-67 in cervical adenocarcinoma tissues. (Insets) Regions with higher magnification. (G) Immunohistochemical intensity scores of KIF2C staining in 100 specimens. *p < 0.05, **p < 0.01; ***p < 0.001.
To investigate the clinical relevance of KIF2C in cervical cancer, 304 cervical cancer cases from TCGA were analyzed. The results showed that KIF2C expression was correlated with the type of cervical cancer (p < 0.05), whereas it was not directly correlated with age, tumor–nodes–metastases stage, 2018 International Federation of Gynecology and Obstetrics stage, lymphatic metastasis, and distant metastasis (Table 2).
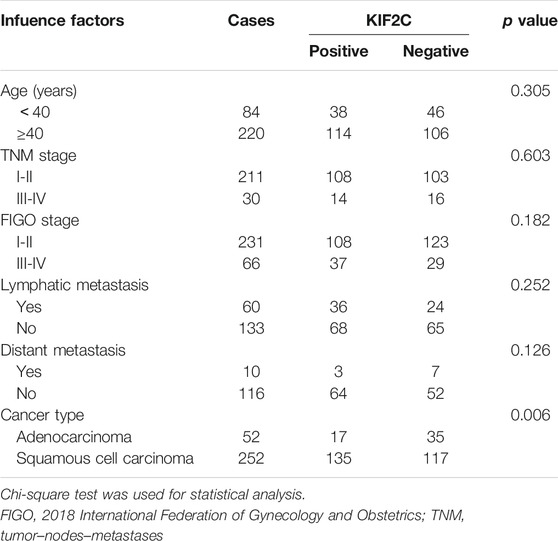
TABLE 2. Relationship between KIF2C and clinicopathological characteristics in cervical cancer patients.
Because we found that KIF2C was clinically relevant in cervical cancer in the discussion earlier, we examined the KIF2C expression in various cervical cancer and normal cell lines, including C33a, HeLa, SiHa, Caski, and HCerEpic cell lines. We analyzed mRNA and protein levels of KIF2C using qRT-PCR and Western blot assay in those cells. The results showed that the mRNA or protein levels of KIF2C in C33a, HeLa, and SiHa cells were higher than those in HCerEpic cells (Figures 6A–C). The two cell lines (SiHa and C33a) with the highest KIF2C expression and Caski cell, which had a relatively low level of KIF2C expression, were used for subsequent experiments. To further investigate the role of KIF2C in cervical cancer, we established the stable KIF2C knockdown cell lines using the short hairpin RNA lentiviral system. We confirmed the knockdown efficiency using Western blot, qRT-PCR (Figures 6D–I), and green fluorescent protein (Figure 6J) in SiHa and C33a cells. For overexpression, the Caski cell was transfected with KIF2C plasmid, and the transfection efficiency was verified with Western blot (Figure 6K,L).
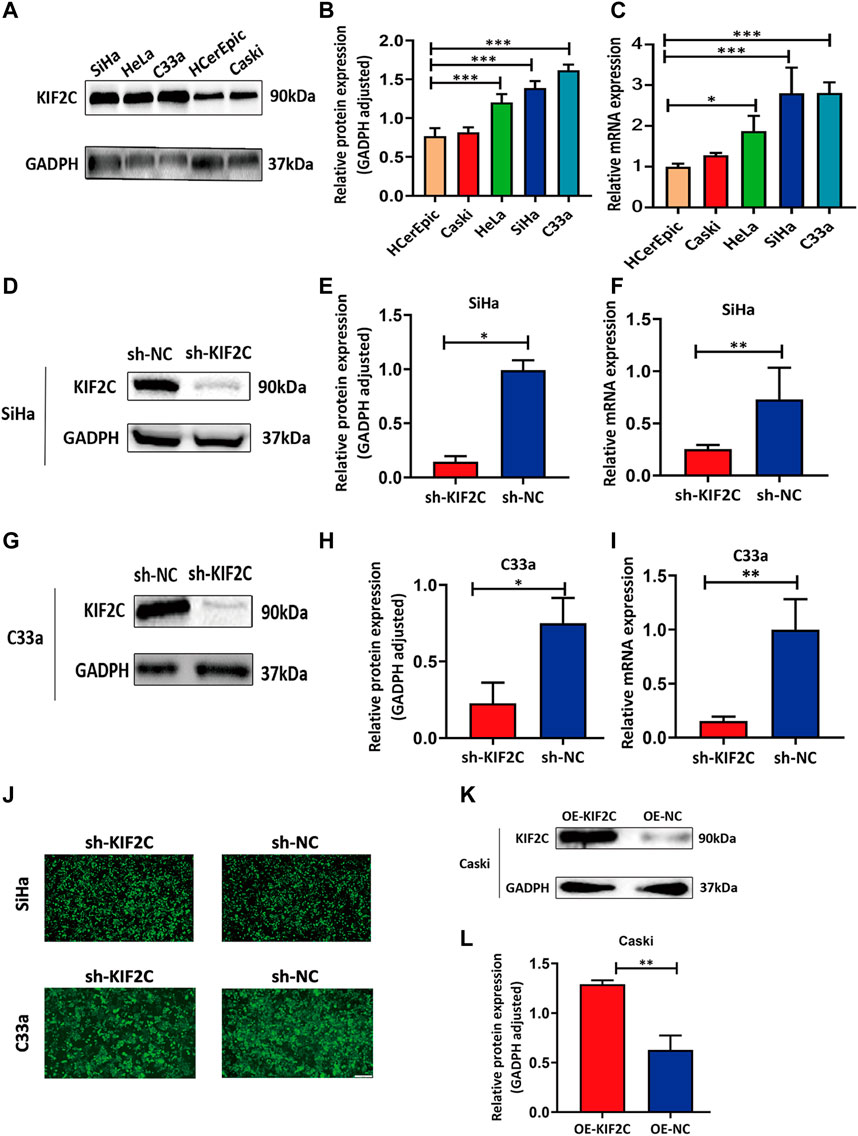
FIGURE 6. Establishment of KIF2C knockdown and overexpression cell lines using cervical cancer cell lines. (A) Gray value of KIF2C protein band in SiHa, HeLa, Caski, C33a and HCerEpic; (B) relative protein expression of KIF2C in SiHa, HeLa, Caski, C33a, and HCerEpic; (C) relative mRNA expression of KIF2C in SiHa, HeLa, Caski, C33a, and HCerEpic; (D) gray value of KIF2C protein band in SiHa after transfection; (E) relative protein expression of KIF2C in SiHa after transfection; (F) relative mRNA expression of KIF2C in SiHa after transfection; gray value, protein and relative mRNA expression of KIF2C in C33a after transfection are shown in Figures 5G–I; (J) green fluorescence indicated a transfection efficiency greater than 80%; (K) gray value of KIF2C protein band in Caski after transfecting KIF2C overexpressed plasmid; (L) relative protein expression of KIF2C in Caski after transfecting overexpressed plasmid. *p < 0.05; **p < 0.01; ***p < 0.001.
To explore the role of KIF2C in cervical cancer cells, we examined the cell viability using CCK-8 assay and clone formation assay in SiHa or C33a cells. The viability of sh-KIF2C SiHa or C33a cells was significantly decreased compared with that of control cells (Figure 7A). These results indicated that the KIF2C knockdown inhibited the proliferation of cervical cancer cells. Next, we investigated the role of KIF2C on in vitro tumorigenicity in SiHa and C33a cells. The clonal colonies of sh-KIF2C SiHa and C33a cells were also significantly decreased compared with those of control cells (Figure 7B). We then examined whether KIF2C knockdown could affect the migration and invasion in SiHa and C33a cells. Results showed that the numbers of migrated or invaded cells in KIF2C knockdown groups were significantly lower than those in control groups (Figure 7C). These results indicated that KIF2C knockdown suppresses the migration and invasion of cervical cancer cells.
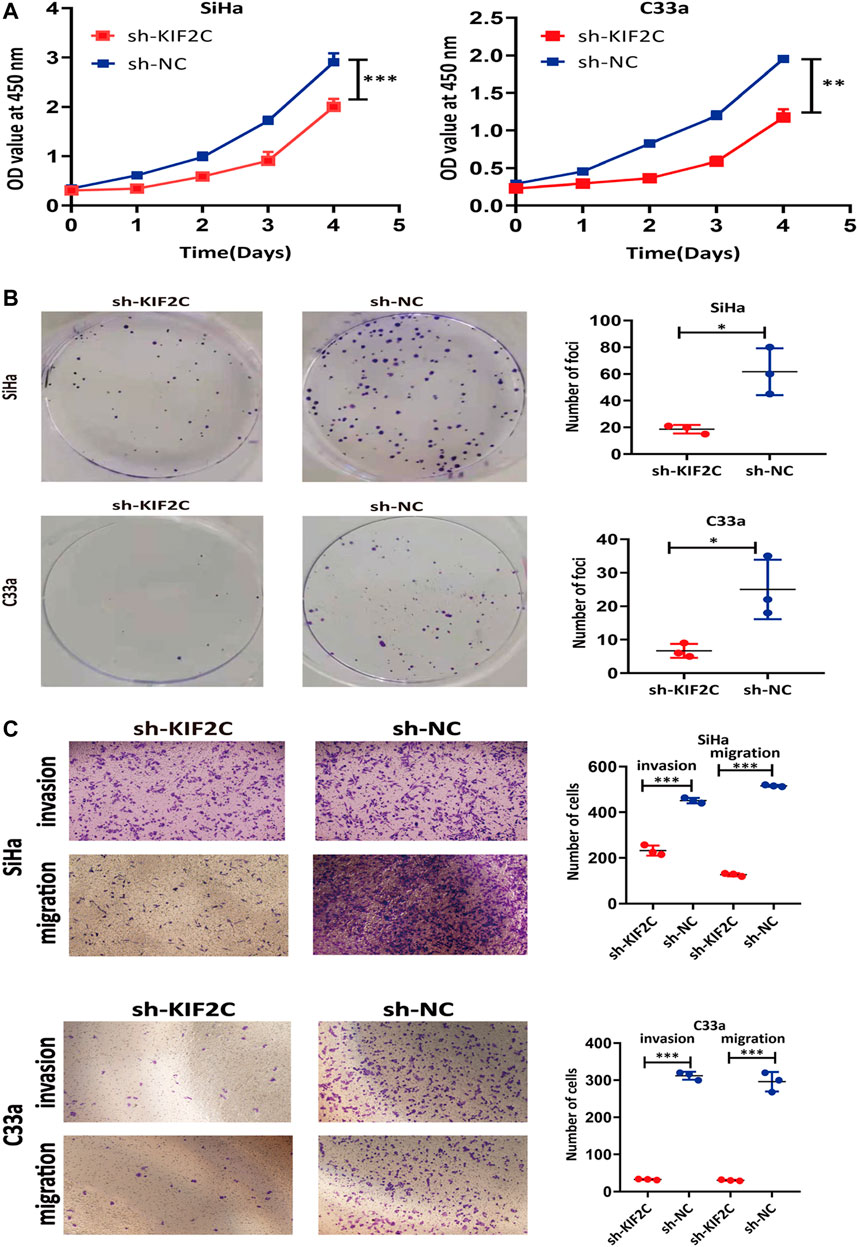
FIGURE 7. KIF2C knockdown inhibited tumorigenicity of cervical cancer cells. CCK-8 assay (A) and colony formation assay (B) were used to evaluate ability of cell proliferation in cervical cancer cells with KIF2C knockdown; dot plots in right showed results of quantitative analyses. (C) Migration and invasion of cervical cancer cells were determined after KIF2C knockdown and quantification of number of cells in migration and invasion. *p < 0.05; **p < 0.01; ***p < 0.001.
To further probe the role of KIF2C in cervical cancer cells, we transfected the KIF2C overexpression plasmid into the Caski cell line. CCK-8 assay and clone formation experiment were used to detect the cell viability and clone formation ability. For the CCK-8 assay, the results showed the cell viability was significantly increased in the KIF2C overexpression groups compared with the control groups (Figure 8A). For the clone formation experiment, the clonogenic ability of Caski cells, which overexpressed KIF2C, was significantly increased (Figure 8B). All findings indicated that KIF2C overexpression could promote cell proliferation. By the same token, a transwell assay was performed to examine cell migration and invasion ability, and the results demonstrated that KIF2C overexpression can remarkably improve the migration and invasion ability of cervical cancer cells (Figure 8C).
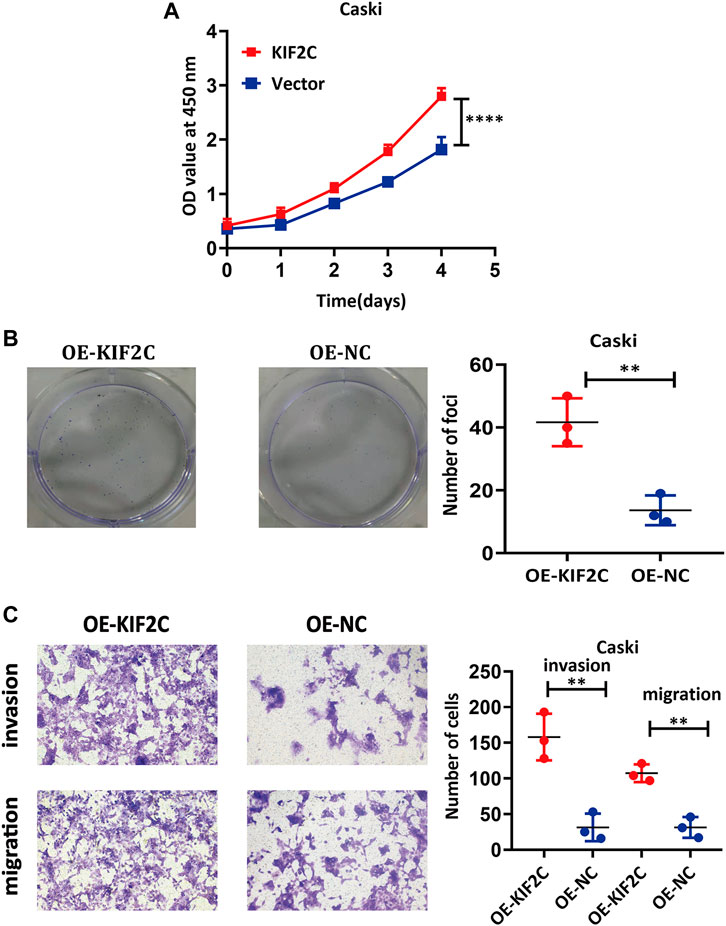
FIGURE 8. KIF2C overexpression promoted tumorigenicity of cervical cancer cells. CCK-8 assay (A) and colony formation assay (B) were used to evaluate ability of cell proliferation in cervical cancer cells with KIF2C overexpression; dot plots in right showed results of quantitative analyses. (C) Migration and invasion of cervical cancer cells were determined after KIF2C overexpression and quantification of number of cells in migration and invasion. **p < 0.01; ***p < 0.001.
To gain a deeper insight into the mechanism of KIF2C expression, we used RNA-seq analysis. We screened the differentially expressed genes using sh-KIF2C SiHa and sh-NC SiHa. Under the screening criteria, which were false discovery rate < 0.05 and |log2FC| >1.5, a total of 349 differentially expressed genes (104 upregulated and 245 downregulated genes) were analyzed (Figure 9A). The results were presented as a heat map, in which blue was the downregulated genes and red was the upregulated genes (Figure 9B). Then, we subjected the 349 genes to GO and KEGG analyses. The results of the GO analysis showed the main pathways in which the upregulated genes were involved in protein transport into membrane raft, protein transport within the lipid bilayer, negative regulation of signal transduction, negative regulation of signaling, etc. (Figure 9C). In addition, it also showed the main pathways in which the downregulated genes were involved in xenobiotic glucuronidation, negative regulation of cellular glucuronidation, regulation of glucuronosyltransferase activity, immune system process, and other signaling pathways (Figure 9D). The results of the KEGG analysis revealed that the upregulated genes were mainly enriched in mucin-type O-glycan biosynthesis, leukocyte transendothelial migration, cell adhesion molecules (CAMs), phagosome, etc. (Figure 9E), whereas the downregulated genes were mainly enriched in drug metabolism-other enzymes, ascorbate and aldarate metabolism, pentose and glucuronate interconversions, and other related pathways (Figure 9F).
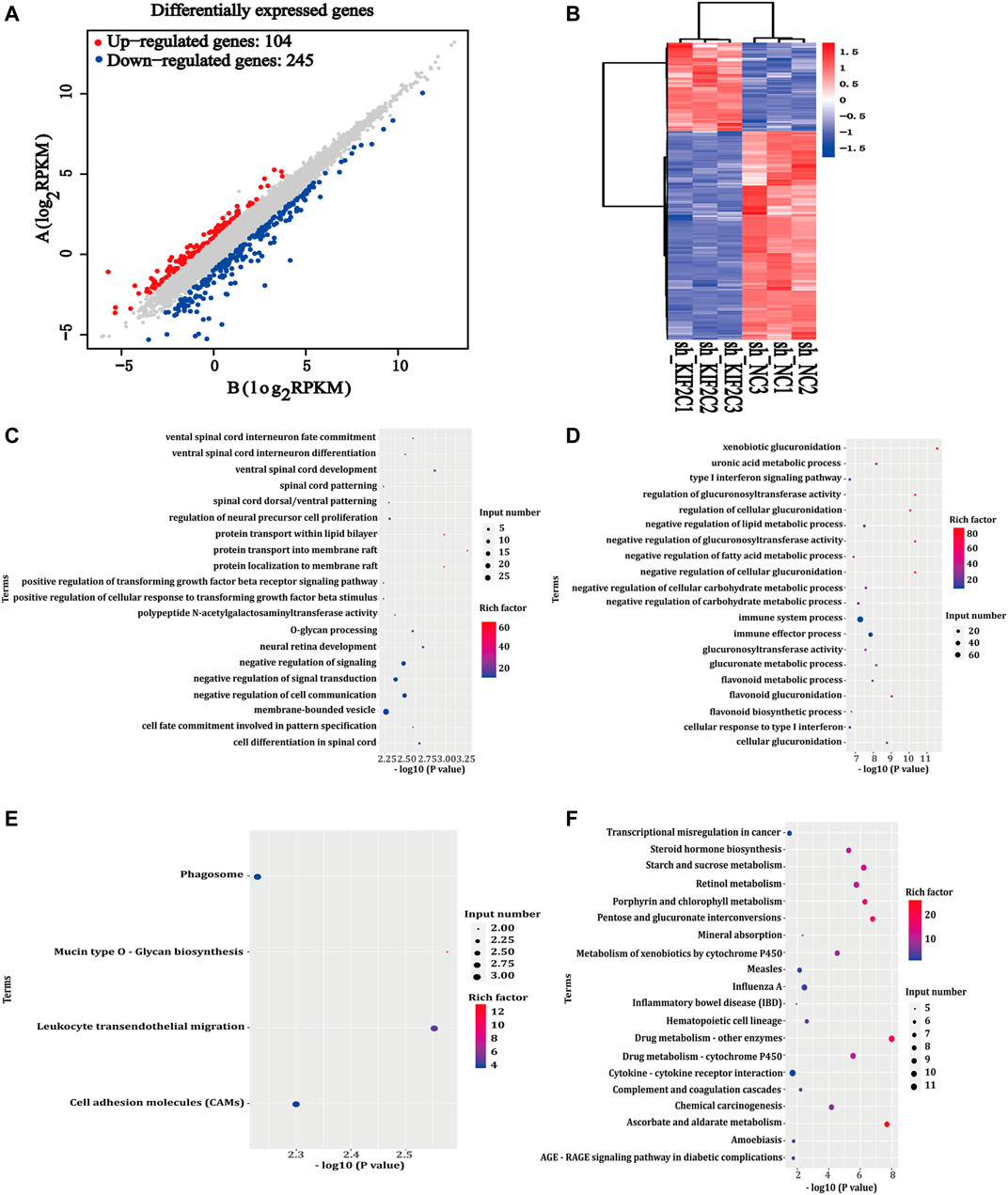
FIGURE 9. Differentially expressed genes of KIF2C. (A) Scatter plot of differentially expressed genes between comparison groups; gray dots represent not differentially expressed genes, blue dots represent differentially downregulated genes, and red dots represent differentially upregulated genes; (B) leftmost image is overall hierarchical clustering diagram of all differentially expressed genes in all comparison groups, clustered by reads per kilobase of transcript per million reads mapped value, red represents high-expressed genes, and blue represents low-expressed genes. x-axis represents different samples, and y-axis represents gene names; (C,D) GO enrichment map of upregulated/downregulated genes; (E,F) KEGG enrichment map of upregulated/downregulated genes.
To further experimentally validate the specific mechanism by which KIF2C regulates cervical carcinogenesis, we deeply analyzed the KEGG pathway and found that its differentially expressed genes were enriched in the p53 signaling pathway (Supplementary Table S1). There have been some studies showing that alteration of the p53 signaling pathway is involved in the development of cervical cancer. Given that, we further analyzed the expression of proteins involved in the p53 signaling pathways, including p21, p53, bcl-2, and bax. In SiHa and C33a cells with KIF2C knockdown, the expression of p21, p53, and bax proteins was significantly increased, whereas the expression of bcl-2 protein significantly decreased (Figures 10A–D). However, the expression of the proteins mentioned earlier exhibited an opposite trend in the presence of the overexpressed KIF2C (Figures 10E,F). These results indicated that KIF2C might promote cervical cancer progression through the p53 signaling pathway.
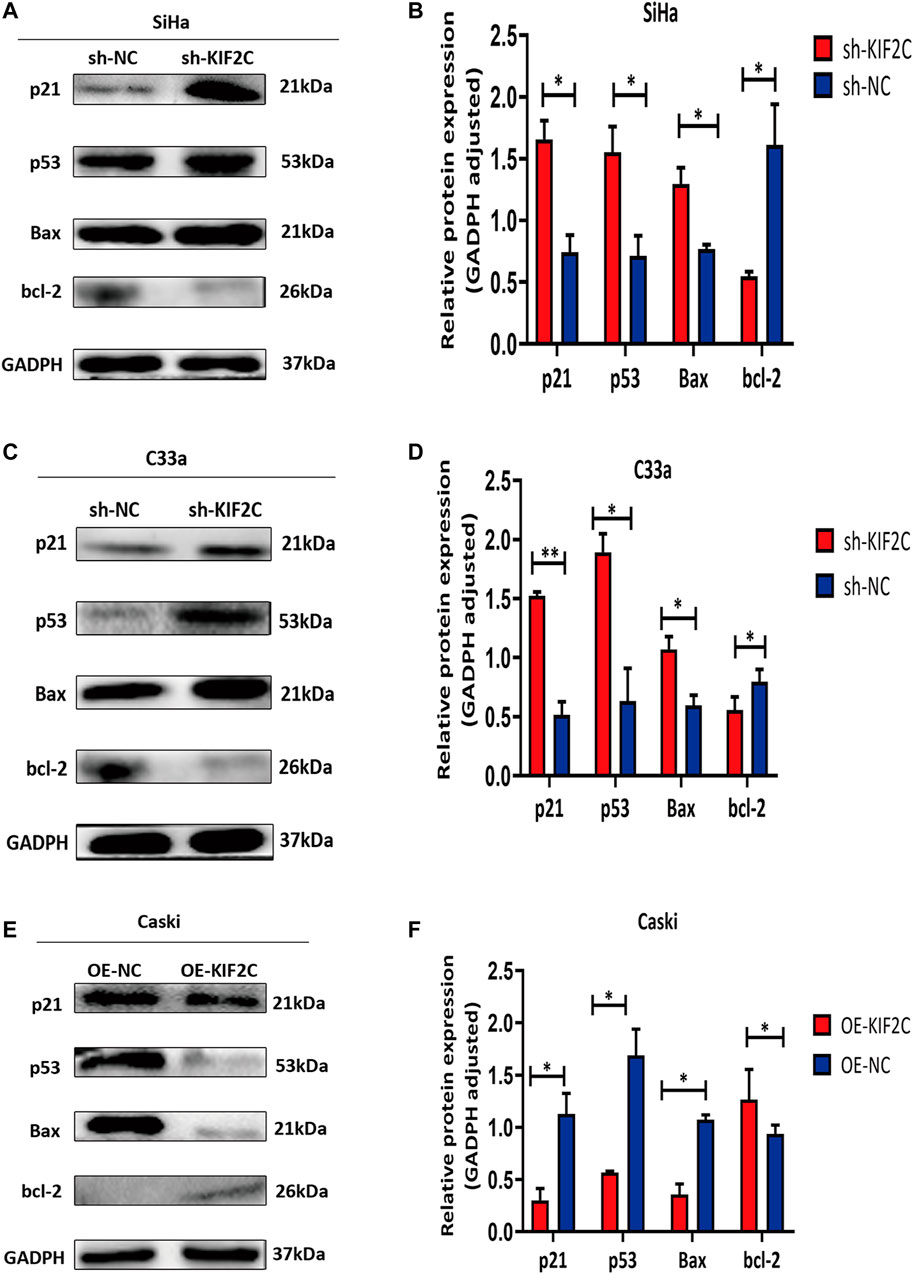
FIGURE 10. KIF2C knockdown promoted activation of p53 signaling pathway. Relative protein expression levels of p21, p53, and Bax were significantly increased, whereas those of bcl-2 was significantly decreased in SiHa (A,B) and C33a (C,D) cells with KIF2 knockdown. However, expression of proteins mentioned earlier exhibited an opposite trend in presence of KIF2C overexpression (E,F). *p < 0.05, **p < 0.01.
For additionally exploring the associations of KIF2C with p53 signaling pathways, the p53 CRISPR/Cas9 knockout plasmid and control CRISPR/Cas9 plasmid were transfected into the SiHa cell with KIF2C stable knockdown. Transfection efficiency was verified using Western blot assay (Figures 11A,B). The outcome revealed that p53 knockdown reversed the prohibiting effect of KIF2C-silenced on cell proliferation (Figures 11C–E), migration, and invasion (Figures 11F–H). Besides, we also found that the tendency of p21 and Bax showed the same trend with p53; the level of bcl-2 showed the opposite results (Figures 11I,J). All these data detailed that KIF2C might induce cervical cancer cells growth, invasion, and migration by regulating the p53 signaling pathway.
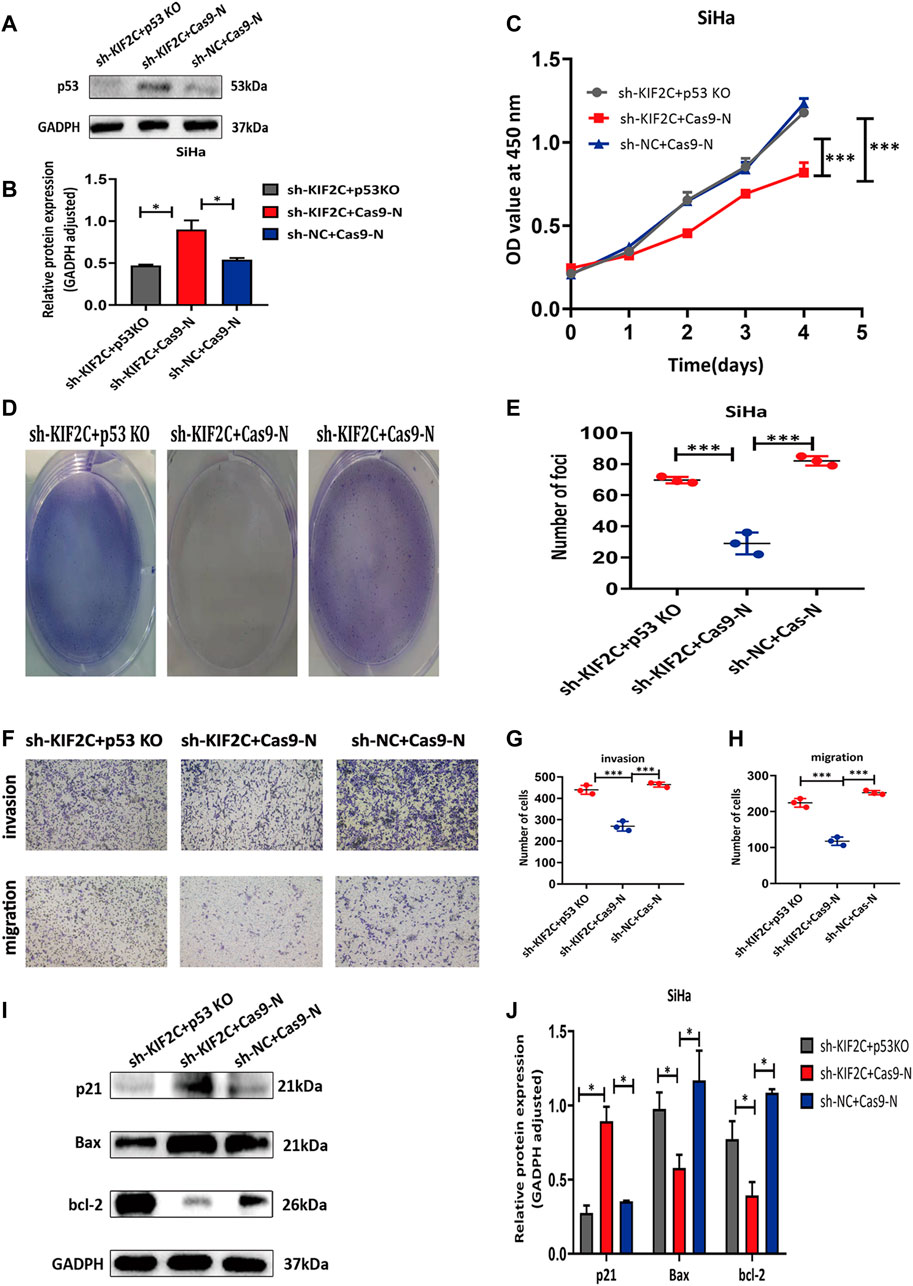
FIGURE 11. p53 knockdown rescued inhibitory effects of KIF2C in cervical cancer. (A) Gray value of p53 protein band in SiHa after transfection; (B) relative protein expression of p53 in SiHa after transfection; knockdown of p53 partially reversed KIF2C induced inhibition of proliferation in SiHa cells determined by CCK-8 assay (C) and colony formation assay (D,E); knockdown of p53 partially reversed KIF2C induced suppression of invasion and migration (F); bar graphs in (G,H) show results of quantitative analyses. (I) Gray value of p21, Bax, and bcl-2 protein band in SiHa after transfection; (J) relative protein expression of p21, bax, and bcl-2 in SiHa after transfection. *p < 0.05; **p < 0.01; ***p < 0.001.
To assess the role of KIF2C in mouse xenograft models, we subcutaneously inoculated SiHa and C33a cells in the flanks of nude mice. We continuously monitored the tumor growth and measured the volume and weight of tumors for 4 weeks. We found that tumor growth of the sh-KIF2C group was significantly decreased compared with that of the control group (Figure 12A). In addition, the volume and weight of tumors of the sh-KIF2C group were lower than those of the control group (Figures 12B,C). These results indicated that KIF2C knockdown played a tumor-suppressive role in vivo.
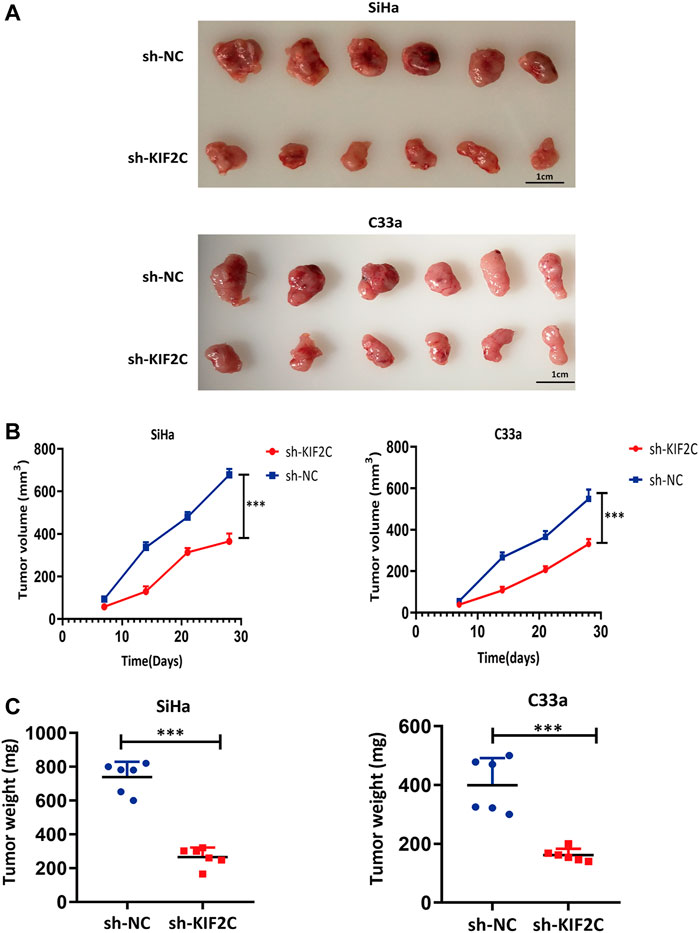
FIGURE 12. KIF2C knockdown suppressed tumor growth in vivo. (A) Photos of tumor samples from mice; (B) curve of tumor volume in mice; (C) scatter graph of tumor weight. ***p < 0.001.
KIF2C belongs to the kinesin superfamily proteins, which participate in microtubule depolymerization, microfilament binding to centromere sites, and chromosome segregation, thus regulating the mitotic process of cells (Ritter et al., 2015). Based on its biological function, KIF2C has been suggested to have tumor-promoting properties. Previous studies have reported that KIF2C is overexpressed in a variety of cancer types. Shimo et al. (2008) used complementary DNA microarray coupled with laser microbeam microdissection gene chip and laser microbeam technology to detect the genomic expression of 81 breast cancers, from which they found that KIF2C is a type of trans-activated gene. Their further research also showed that KIF2C is highly expressed in breast cancer tissues and other cell lines. Through bioinformatics analysis, Li et al. (2020b) revealed that KIF2C is highly expressed in breast cancer and is involved in the worst OS, RFS, and distant metastasis-free survival of breast cancer. In addition to breast cancer, KIF2C is also found to be highly expressed in gastric cancer. Nakamura et al. (2007) reported that the expression level of KIF2C in gastric cancer tissues is significantly higher than that in adjacent tissues and is significantly correlated with lymph node invasion, lymph node metastasis, and poor prognosis. Moreover, gastric cancer cell lines transfected with the KIF2C gene were verified to have a higher proliferation rate and migration capabilities in vitro.
In this study, we performed a series of bioinformatics analyses to identify KIF2C gene features in pan-cancer. We analyzed the expression characteristics of KIF2C according to the TIMER dataset, TCGA dataset, and GEPIA2 dataset. We found that KIF2C was frequently abnormally expressed in various types of cancer. KIF2C expression was upregulated in many tumor tissues (BLCA, BRCA, CESC, CHOL, COAD, DLBC, ESCA, GBM, HNSC, KIRC, KIRP, LIHC, LUAD, LUSC, OV, PAAD, PCPG, PRAD, READ, SARC, SKCM, STAD, THYM, UCEC, and UCS) compared with the corresponding normal tissues. However, KIF2C expression was downregulated in tumor tissues (e.g., TCGT, LAML). Thus, the difference in KIF2C expression in various cancers suggested that it might have different biological functions in different types of cancers. Moreover, high expression levels of KIF2C were associated with the poor prognosis of most cancers, which strongly suggested that KIF2C was a potential prognostic biomarker for tumor treatment. Tumorigenesis is a complex phenomenon, which is often caused by gene mutations (Fan et al., 2021). Based on this, we then studied the characteristics of KIF2C gene mutation. The results demonstrated that the main mutation type of KIF2C was missense mutation, which was also the most common oncogenic mutation type (Brown et al., 2014). This may indicate that KIF2C was closely related to the development of tumors to some extent. Interestingly, we found that the mutation of KIF2C was closely related to the survival rate of CESC, which suggested that KIF2C might be a potential therapeutic target of CESC. At the same time, immunohistochemical results in public datasets also showed that KIF2C was only highly expressed in cervical cancer tissue. This prompted us to investigate further the biological function of KIF2C in cervical cancer, which might provide new druggable targets and novel possibilities for cervical cancer therapy.
To explore these possibilities, we first studied the expression of KIF2C in human tissues. Immunohistochemical staining showed that KIF2C was highly expressed in cervical cancer tissues. More than that, there were some similarities in expression levels between Ki-67 and KIF2C. This suggested a latent relationship for KIF2C with the proliferation of cervical cancer. To further investigate the correlation between KIF2C and clinicopathological characteristics, we performed a statistical analysis using the raw data from TCGA database. We found that the expression of KIF2C was dependent on the tumor types, whereas it had no direct correlation with tumor stage, lymph node invasion, and distant metastasis. Therefore, we assumed that KIF2C might be involved in the occurrence and development of cervical carcinoma. KIF2C was highly expressed in most cervical cancer cells. Downregulation of the KIF2C expression could significantly inhibit the biological functions of cervical cancer cell lines both in vitro and in vivo. In contrast, the overexpression of KIF2C had the opposite effects. These results also suggest that KIF2C may be a potential gene contributing to cervical carcinoma development.
The p53 gene is a tumor suppressor gene that has the highest correlation with human tumorigenesis among genes that have been discovered so far (Levine, 2020). The gene exerts its growth inhibitory effect by activating and interacting with a variety of signaling pathways (Kasteri et al., 2018). As a key transcription factor, p53 has been found to be inactive at both gene and protein levels in a variety of human cancers. Its upregulation in cancer cells may prevent the proliferation of cancer cells by promoting cell cycle arrest and apoptosis (Wang et al., 2015). For example, SNRPB can promote the occurrence and development of cervical cancer by inhibiting the expression of p53 (Zhu et al., 2020). In addition, in cervical cancer, ISG15 can upregulate and activate p53, which in turn causes the inhibition of proliferation and growth of cancer cells (Zhou et al., 2017). Kong et al. (2015) have reported that VTRNA2-1-5p is a direct regulator of p53; the inhibition of VTRNA2-1-5p can reduce the invasion, proliferation, and tumorigenicity of cervical cancer cells and can increase cell apoptosis and p53 expression. Furthermore, because p53 plays a central role in cellular emergency response, deletion or mutation of the p53 gene can lead to the dysfunctions of upstream regulators or downstream effectors in cancer cells (Miedl et al., 2020). The development of therapy based on the p53 pathway has always faced a huge challenge. Many strategies targeting the p53 pathway have been developed, including the restoration of p53 function, the inhibition of p53-MDM2 interaction, and the conversion of mutant p53 into wild-type p53 by gene therapy, the targeting of p53 family proteins, the elimination of mutant p53, and the development of p53-based vaccines (Ladds and Laín, 2019). Herein, we found that the downregulation of KIF2C causes the activation of the p53 signaling pathway, whereas the upregulation of KIF2C inhibits the activation of the p53 signaling pathway. To further confirm the mechanisms by which KIF2C regulates cervical cancer formation and development, we applied p53 CRISPR/Cas9 knockout plasmid in Siha cells to rescue the phenotype induced by KIF2C. Indeed, the knockdown of p53 partially reversed the oncogenic effects of KIF2C. Thus, we assumed that KIF2C might promote the development of cervical cancer by inhibiting the activation of the p53 signaling pathway. Whether or not this finding can lead to the development of a therapeutic strategy targeting the p53 pathway of cervical cancer, it provides clues and directions for targeted therapy and combined radiotherapy and chemotherapy of cervical cancer, which will be the main topics for our future research.
In summary, the results of this pan-cancer analysis showed that the abnormal expression of KIF2C was associated with poor prognosis of different cancer types. We also found that KIF2C promoted cervical cancer cell proliferation by suppressing the p53 signaling pathway. These findings suggest the potential use of KIF2C as a therapeutic target for the treatment of cervical cancer.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
The studies involving human participants were reviewed and approved by the Medical Ethics Committee of the Third Affiliated Hospital of Zhengzhou University (ethics approval number: 2021-080-01). Written informed consent for participation was not required for this study in accordance with the national legislation and the institutional requirements. The animal study was reviewed and approved by the Animal Center of Zhengzhou University.
LY designed the study. JY and ZW performed the experiments and wrote the manuscript. J-HJ and BW modified the language. YW, NW, KS, and RL contributed to the analysis of the data. YZ and JL reviewed and edited the manuscript. All authors read and approved the manuscript.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fphar.2021.785981/full#supplementary-material
Bai, Y., Xiong, L., Zhu, M., Yang, Z., Zhao, J., and Tang, H. (2019). Co-Expression Network Analysis Identified KIF2C in Association with Progression and Prognosis in Lung Adenocarcinoma. Cancer Biomark 24 (3), 371–382. doi:10.3233/cbm-181512
Brisson, M., Kim, J. J., Canfell, K., Drolet, M., Gingras, G., Burger, E. A., et al. (2020). Impact of HPV Vaccination and Cervical Screening on Cervical Cancer Elimination: a Comparative Modelling Analysis in 78 Low-Income and Lower-Middle-Income Countries. Lancet 395 (10224), 575–590. doi:10.1016/s0140-6736(20)30068-4
Brown, S. D., Warren, R. L., Gibb, E. A., Martin, S. D., Spinelli, J. J., Nelson, B. H., et al. (2014). Neo-antigens Predicted by Tumor Genome Meta-Analysis Correlate with Increased Patient Survival. Genome Res. 24 (5), 743–750. doi:10.1101/gr.165985.113
Cerami, E., Gao, J., Dogrusoz, U., Gross, B. E., Sumer, S. O., Aksoy, B. A., et al. (2012). The cBio Cancer Genomics portal: an Open Platform for Exploring Multidimensional Cancer Genomics Data. Cancer Discov. 2 (5), 401–404. doi:10.1158/2159-8290.Cd-12-0095
Chen, F., Chandrashekar, D. S., Varambally, S., and Creighton, C. J. (2019). Pan-Cancer Molecular Subtypes Revealed by Mass-Spectrometry-Based Proteomic Characterization of More Than 500 Human Cancers. Nat. Commun. 10 (1), 5679. doi:10.1038/s41467-019-13528-0
Cohen, P. A., Jhingran, A., Oaknin, A., and Denny, L. (2019). Cervical Cancer. Lancet 393 (10167), 169–182. doi:10.1016/s0140-6736(18)32470-x
Das, M. (2021). WHO Launches Strategy to Accelerate Elimination of Cervical Cancer. Lancet Oncol. 22 (1), 20–21. doi:10.1016/s1470-2045(20)30729-4
DeLuca, J. G., Gall, W. E., Ciferri, C., Cimini, D., Musacchio, A., and Salmon, E. D. (2006). Kinetochore Microtubule Dynamics and Attachment Stability Are Regulated by Hec1. Cell 127 (5), 969–982. doi:10.1016/j.cell.2006.09.047
Fan, T., Lu, Z., Liu, Y., Wang, L., Tian, H., Zheng, Y., et al. (2021). A Novel Immune-Related Seventeen-Gene Signature for Predicting Early Stage Lung Squamous Cell Carcinoma Prognosis. Front. Immunol. 12, 665407. doi:10.3389/fimmu.2021.665407
Gan, H., Lin, L., Hu, N., Yang, Y., Gao, Y., Pei, Y., et al. (2019). KIF2C Exerts an Oncogenic Role in Nonsmall Cell Lung Cancer and Is Negatively Regulated by miR-325-3p. Cell Biochem. Funct. 37 (6), 424–431. doi:10.1002/cbf.3420
Ganguly, A., Bhattacharya, R., and Cabral, F. (2008). Cell Cycle Dependent Degradation of MCAK: Evidence against a Role in Anaphase Chromosome Movement. Cell Cycle 7 (20), 3187–3193. doi:10.4161/cc.7.20.6814
Gao, J., Aksoy, B. A., Dogrusoz, U., Dresdner, G., Gross, B., Sumer, S. O., et al. (2013). Integrative Analysis of Complex Cancer Genomics and Clinical Profiles Using the cBioPortal. Sci. Signal. 6 (269), pl1. doi:10.1126/scisignal.2004088
Hedrick, D. G., Stout, J. R., and Walczak, C. E. (2008). Effects of Anti-microtubule Agents on Microtubule Organization in Cells Lacking the Kinesin-13 MCAK. Cell Cycle 7 (14), 2146–2156. doi:10.4161/cc.7.14.6239
Huang, S. M., Mishina, Y. M., Liu, S., Cheung, A., Stegmeier, F., Michaud, G. A., et al. (2009). Tankyrase Inhibition Stabilizes Axin and Antagonizes Wnt Signalling. Nature 461 (7264), 614–620. doi:10.1038/nature08356
Kasteri, J., Das, D., Zhong, X., Persaud, L., Francis, A., Muharam, H., et al. (2018). Translation Control by P53. Cancers (Basel) 10 (5), 133. doi:10.3390/cancers10050133
Kong, L., Hao, Q., Wang, Y., Zhou, P., Zou, B., and Zhang, Y. X. (2015). Regulation of P53 Expression and Apoptosis by Vault RNA2-1-5p in Cervical Cancer Cells. Oncotarget 6 (29), 28371–28388. doi:10.18632/oncotarget.4948
Ladds, M. J. G. W., and Laín, S. (2019). Small Molecule Activators of the P53 Response. J. Mol. Cel. Biol. 11 (3), 245–254. doi:10.1093/jmcb/mjz006
Levine, A. J. (2020). p53: 800 Million Years of Evolution and 40 Years of Discovery. Nat. Rev. Cancer 20 (8), 471–480. doi:10.1038/s41568-020-0262-1
Li, T., Fu, J., Zeng, Z., Cohen, D., Li, J., Chen, Q., et al. (2020a). TIMER2.0 for Analysis of Tumor-Infiltrating Immune Cells. Nucleic Acids Res. 48, W509–W514. doi:10.1093/nar/gkaa407
Li, T. F., Zeng, H. J., Shan, Z., Ye, R. Y., Cheang, T. Y., Zhang, Y. J., et al. (2020b). Overexpression of Kinesin Superfamily Members as Prognostic Biomarkers of Breast Cancer. Cancer Cel Int 20, 123. doi:10.1186/s12935-020-01191-1
Miedl, H., Dietrich, B., Kaserer, K., and Schreiber, M. (2020). The 40bp Indel Polymorphism Rs150550023 in the MDM2 Promoter Is Associated with Intriguing Shifts in Gene Expression in the P53-MDM2 Regulatory Hub. Cancers (Basel) 12 (11), 3363. doi:10.3390/cancers12113363
Mizuno, H., Kitada, K., Nakai, K., and Sarai, A. (2009). PrognoScan: a New Database for Meta-Analysis of the Prognostic Value of Genes. BMC Med. Genomics 2, 18. doi:10.1186/1755-8794-2-18
Nakamura, Y., Tanaka, F., Haraguchi, N., Mimori, K., Matsumoto, T., Inoue, H., et al. (2007). Clinicopathological and Biological Significance of Mitotic Centromere-Associated Kinesin Overexpression in Human Gastric Cancer. Br. J. Cancer 97 (4), 543–549. doi:10.1038/sj.bjc.6603905
Ritter, A., Kreis, N. N., Louwen, F., Wordeman, L., and Yuan, J. (2015). Molecular Insight into the Regulation and Function of MCAK. Crit. Rev. Biochem. Mol. Biol. 51 (4), 228–245. doi:10.1080/10409238.2016.1178705
Shimo, A., Tanikawa, C., Nishidate, T., Lin, M. L., Matsuda, K., Park, J. H., et al. (2008). Involvement of Kinesin Family Member 2C/mitotic Centromere-Associated Kinesin Overexpression in Mammary Carcinogenesis. Cancer Sci. 99 (1), 62–70. doi:10.1111/j.1349-7006.2007.00635.x
Miller, K. D., Ortiz, A. P., Pinheiro, P. S., Bandi, P., Minihan, A., Fuchs, H. E., et al. (2021). Cancer Statistics for the US Hispanic/Latino Population, 2021. CA Cancer J. Clin. 71 (1), 466–487. doi:10.3322/caac.21695
Soumaoro, L. T., Uetake, H., Higuchi, T., Takagi, Y., Enomoto, M., and Sugihara, K. (2004). Cyclooxygenase-2 Expression: a Significant Prognostic Indicator for Patients with Colorectal Cancer. Clin. Cancer Res. 10 (24), 8465–8471. doi:10.1158/1078-0432.Ccr-04-0653
Tang, Z., Li, C., Kang, B., Gao, G., Li, C., and Zhang, Z. (2017). GEPIA: a Web Server for Cancer and normal Gene Expression Profiling and Interactive Analyses. Nucleic Acids Res. 45, W98–W102. doi:10.1093/nar/gkx247
Uhlén, M., Fagerberg, L., Hallström, B., Lindskog, C., Oksvold, P., Mardinoglu, A., et al. (2015). Proteomics. Tissue-Based Map of the Human Proteome. Science (New York, N.Y.) 347 (6220), 1260419. doi:10.1126/science.1260419
Vasaikar, S. V., Straub, P., Wang, J., and Zhang, B. (2018). LinkedOmics: Analyzing Multi-Omics Data within and across 32 Cancer Types. Nucleic Acids Res. 46, D956–D963. doi:10.1093/nar/gkx1090
Wang, X., Simpson, E. R., and Brown, K. A. (2015). p53: Protection against Tumor Growth beyond Effects on Cell Cycle and Apoptosis. Cancer Res. 75 (23), 5001–5007. doi:10.1158/0008-5472.Can-15-0563
Wei, S., Dai, M., Zhang, C., Teng, K., Wang, F., Li, H., et al. (2020). KIF2C: a Novel Link between Wnt/β-Catenin and mTORC1 Signaling in the Pathogenesis of Hepatocellular Carcinoma. Protein Cell 12, 788–809. doi:10.1007/s13238-020-00766-y
Yang, C., Li, Q., Chen, X., Zhang, Z., Mou, Z., Ye, F., et al. (2020). Circular RNA circRGNEF Promotes Bladder Cancer Progression via miR-548/KIF2C axis Regulation. Aging (Albany NY) 12 (8), 6865–6879. doi:10.18632/aging.103047
Zhao, F., and Qiao, Y. (2019). Cervical Cancer Prevention in China: a Key to Cancer Control. Lancet 393 (10175), 969–970. doi:10.1016/s0140-6736(18)32849-6
Zhao, Z., Zhou, S., Li, W., Zhong, F., Zhang, H., Sheng, L., et al. (2019). AIB1 Predicts Tumor Response To Definitive Chemoradiotherapy And Prognosis In Cervical Squamous Cell Carcinoma. J. Cancer 10 (21), 5212–5222. doi:10.7150/jca.31697
Zhou, M. J., Chen, F. Z., Chen, H. C., Wan, X. X., Zhou, X., Fang, Q., et al. (2017). ISG15 Inhibits Cancer Cell Growth and Promotes Apoptosis. Int. J. Mol. Med. 39 (2), 446–452. doi:10.3892/ijmm.2016.2845
Keywords: KIF2C, cervical cancer, P53 signaling pathway, molecularly targeted therapy, pan-cancer analysis
Citation: Yang J, Wu Z, Yang L, Jeong J-H, Zhu Y, Lu J, Wang B, Wang N, Wang Y, Shen K and Li R (2022) Characterization of Kinesin Family Member 2C as a Proto-Oncogene in Cervical Cancer. Front. Pharmacol. 12:785981. doi: 10.3389/fphar.2021.785981
Received: 29 September 2021; Accepted: 21 December 2021;
Published: 27 January 2022.
Edited by:
Yingjie Zhang, Shandong University, ChinaReviewed by:
Haiyan Zhu, First Affiliated Hospital of Wenzhou Medical University, ChinaCopyright © 2022 Yang, Wu, Yang, Jeong, Zhu, Lu, Wang, Wang, Wang, Shen and Li. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Li Yang, emRzZnl5YW5nbGlAMTYzLmNvbQ==
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.