
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
METHODS article
Front. Pharmacol. , 30 November 2021
Sec. Experimental Pharmacology and Drug Discovery
Volume 12 - 2021 | https://doi.org/10.3389/fphar.2021.771808
This article is part of the Research Topic Artificial Intelligence for Drug Design, Development and Therapy View all 20 articles
Drug targets are biological macromolecules or biomolecule structures capable of specifically binding a therapeutic effect with a particular drug or regulating physiological functions. Due to the important value and role of drug targets in recent years, the prediction of potential drug targets has become a research hotspot. The key to the research and development of modern new drugs is first to identify potential drug targets. In this paper, a new predictor, DrugHybrid_BS, is developed based on hybrid features and Bagging-SVM to identify potentially druggable proteins. This method combines the three features of monoDiKGap (k = 2), cross-covariance, and grouped amino acid composition. It removes redundant features and analyses key features through MRMD and MRMD2.0. The cross-validation results show that 96.9944% of the potentially druggable proteins can be accurately identified, and the accuracy of the independent test set has reached 96.5665%. This all means that DrugHybrid_BS has the potential to become a useful predictive tool for druggable proteins. In addition, the hybrid key features can identify 80.0343% of the potentially druggable proteins combined with Bagging-SVM, which indicates the significance of this part of the features for research.
Drug targets refer to the binding sites of drugs in the body. To date, there are approximately 130 protein families as therapeutic drug targets, which usually include enzymes (Liu et al., 2019a; Meng et al., 2020; Xu et al., 2021a; Wang et al., 2021), G protein-coupled receptors (Ru et al., 2020), ion channels and transporters (Han et al., 2019), nuclear hormone receptors, etc (Li and Lai, 2007). These drug targets are of great significance for disease treatment and drug research and development (Ding et al., 2019a; Ding et al., 2019b; Shi et al., 2019; Ding et al., 2020a; Wang et al., 2020a; Ding et al., 2020b; Shang et al., 2021; Zhuang et al., 2021). However, the discovery and development of modern drugs is usually a time-consuming and laborious process. It is estimated that it takes an average of 10–15 years to bring a drug to the market, which costs approximately US $2,558 million (Zhong et al., 2018). Therefore, predicting whether a protein can potentially be used as a drug target has significant value in disease treatment and reducing the time and cost of drug development, which greatly accelerates the drug development process for the protein (Wang et al., 2020b; Yu et al., 2021).
The discovery of drug targets has attracted extensive attention in both academia and the pharmaceutical industry. The commonly used methods for drug target prediction can be roughly divided into three types. The first type is to analyse known drug targets at the genome level based on sequence homology and to find potential drug targets from protein families (Hopkins and Groom, 2002; Russ and Lampel, 2005; Munir et al., 2019; Ao et al., 2021). Not all members of the same protein family can be used as therapeutic drug targets. The second type predicts whether the new target is druggable based on several chemical properties, molecular drug similarity, and target properties (Gayvert et al., 2016). This method is usually limited by experimental cost. The third type is discovering drug targets based on protein structure, which predicts the protein’s drug properties by searching for the binding site and binding affinity of the target protein (Salmaso and Moro, 2018). However, this method has limitations because the three-dimensional structure of most proteins is not easy to obtain.
With the advent of the genome era, revolutionary changes have taken place in the field of drug research and development. Many computing methods were used for effective drug target prediction. To better find potential drug targets and provide new options for drug redirection, Cheng et al. (Cheng et al., 2021) established the GraphMS model. They fused heterogeneous graph information using mutual information in the heterogeneous graph to obtain effective node information and substructure information. The experimental results show that the area under the receiver operating characteristic curve (AUROC) was 0.959, and the area under the precision-recall curve (AUPR) was 0.847. Dezső et al. (Dezső and Ceccarelli, 2020) developed a machine learning model for tumour drug targets. A variety of protein features, including features from sequences, features that characterize protein functions, and network features from protein-protein interaction networks, were included in the model. It has achieved high accuracy on the drug target of independent clinical trial drug targets, with an area under the curve of 0.89. In order to establish a high-quality environment-specific metabolic model that can be used for drug target prediction, Pacheco et al. (Pacheco et al., 2019) developed a metabolic model FASTCORMICS RNA-seq workflow (rFASTCORMICS) based on RNA-seq data. The genes and response characteristics of 13 different types of cancer were extracted. At the same time, 17 new colon cancer candidate drugs were predicted, of which 3 drugs were verified in vitro in colon cancer cell lines. Ji et al. (Ji et al., 2019) proposed a DTINet method based on network propagation, starting from the diffusion component analysis of potential drug targets and disease networks. The DTINet performed well under the receiver operating characteristic curve (AUROC = 0.86 ± 0.008). To achieve the rapid identification of novel targets, Li et al. (Li and Lai, 2007) constructed a simple model extraction characteristics from known drug target protein sequences. Using this model, drug targets and nondrug targets can be distinguished with 84% accuracy. Jamali et al. (Jamali et al., 2016) based on the protein features derived from 443 sequences, the accuracy of predicting drug targets through neural network models reached 89.98%.
This paper selected three feature extraction methods: monoDiKGap (k = 2), cross covariance (CC) and grouped amino acid composition (GAAC) (Zuo et al., 2017). The three individual features were mixed in different combinations through the hybrid feature method. The MRMD was used to remove redundant hybrid features, and the integrated method bagging was used to improve the classification performance of potentially druggable proteins. We performed the importance analysis on the best feature combination and selected the key features that distinguish potentially druggable proteins. The results show that the hybrid features of the three feature extraction methods can predict the potentially druggable proteins well by the integrated method bagging, and can correctly predict 96.9944% of the druggable target proteins. This model was conducive to better promotion of drug development. Furthermore, the potential drug targets screened out can provide references for new drug targets.
This paper mainly studied the following parts, and the step flow chart was shown in Figure 1:
1. Establishment of dataset.
2. Use three single feature extraction methods, monoDiKGap, Cross Covariance, and Grouped Amino Acid Composition, to represent the features of dataset.
3. Combine three single feature methods to obtain hybrid features.
4. The MRMD was used to remove redundant features, and the MRMD2.0 obtained key features.
5. The feature subset predicted the potentially druggable proteins through the optimized Bagging-SVM model.
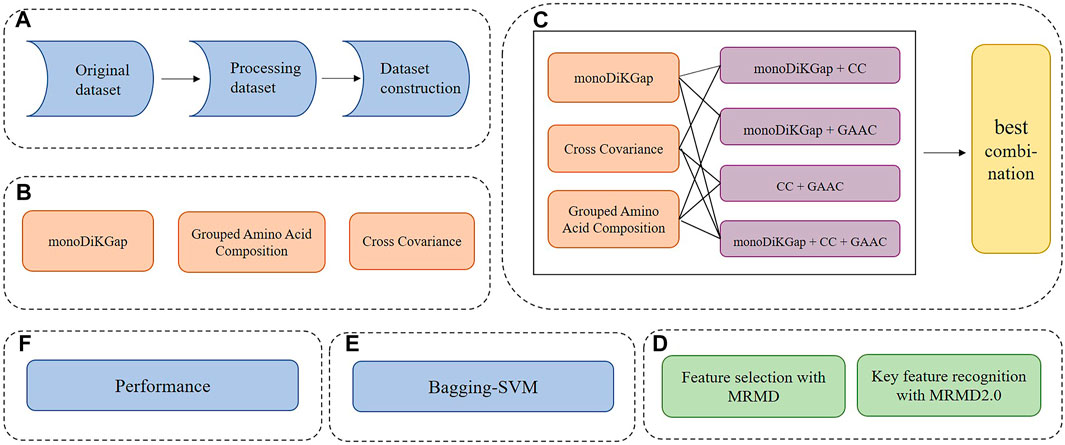
FIGURE 1. Flow chart of DrugHybrid_BS model (A) Process the referenced dataset (B) Three single feature representation methods were used to extract features (C) Combine three single feature representation methods and select the best hybrid feature (D) Use MRMD to remove redundant features and MRMD2.0 to obtain key features (E) Feature subsets were used to predict potentially druggable proteins through the optimized Bagging-SVM model (F) Evaluate model prediction effects based on performance indicators.
This research was carried out under the software python 3.7.4. By comparing the new method DrugHybrid_BS with other machine learning models, the study found that the classification effect of DrugHybrid_BS was better, which was helpful for the prediction of potentially druggable proteins.
This paper cited the dataset proposed by Lin et al. (Lin et al., 2019), in which the drug target dataset was downloaded from the DrugBank (Wishart et al., 2006) database. In the original dataset, 1,224 druggable protein sequences were selected as the positive sample set, and 1,319 non-druggable proteins were selected as the negative sample set. We further processed the dataset by removing the protein sequences containing non-standard amino acid characters “B", “J", “O", “U", “X" and “Z". For the remaining sequences, the CD-Hit program (Fu et al., 2012) was used to set a critical value of 60% sequence identity to delete highly similar sequences to avoid overfitting caused by homologous deviation and noise in training (Zou et al., 2020).
The processed dataset was represented by D, which is the combination of
where
The monoDiKGap feature is a variant of the kmer feature extraction method in the PyFeat package. Kmer, as our common feature extraction method, is also called k-tuples (Liu et al., 2019b; Lv et al., 2020; Niu et al., 2021a). MonoDiKGap refers to the combination of subsequences with KGap used to describe the sequence. While monoDiKGap generates all feature sets, it can also use the AdaBoost (Zhu et al., 2006) classification model to reduce redundant features to generate the optimal feature set. The generated optimal feature set will not only reduce the feature dimension but also ensure a good prediction. In this study, we set KGap to 2. At this time, the monoDiKGap feature can be expressed as:
where
CC is the correlation between two different attributes separated by lag (Guo et al., 2008). For this study, the CC variable described the average interaction between two fragments with different physical and chemical properties separated by lag fragments. Suppose that the protein sequence P has L residues,
where
CC converts protein sequences of different lengths into feature vectors of the same length. The calculation formula of the CC feature representation method is as follows:
In the GAAC code, twenty amino acid types are divided into five categories based on their physical and chemical properties (Lee et al., 2011; Zheng et al., 2019; Zheng et al., 2021). These five categories include the aliphatic group
The GAAC descriptor refers to the frequency of each amino acid group, which is calculated as follows:
where
As an example, for the sequence
In this study, predicting druggable proteins was a typical binary classification problem. To better explore prediction models and analysis features, we mainly used four machine learning algorithms for prediction tasks, namely, support vector machine, K-nearest neighbour, bagging integrated learning, and random forest.
The k-nearest neighbour algorithm is a classic machine learning algorithm (Liao and Vemuri, 2002; Samanthula et al., 2014). The principle of the k-nearest neighbour algorithm is straightforward: a sample in the feature space will always find the k data closest to it, that is, the nearest sample in the feature space. If most of the k data belong to a specific category, the sample also belongs to this category. In this study, the default parameters of the prediction model were selected, and the value of k was 3.
Although the support vector machine has only a short development history of more than 20 years. It shows strong energy in classification problems (Ding et al., 2017; Wei et al., 2018a; Wang et al., 2019; Wang et al., 2020c; Huo et al., 2020). It has become the mainstream technology of machine learning from the end of the 20th century to the beginning of the 21st century, applied to many fields (Jiang et al., 2013; Xu et al., 2018; Zhang et al., 2018; Wei et al., 2019a; Liu et al., 2021). The support vector machine uses the maximum classification interval to determine the optimal partitioning hyperplane to obtain good generalization. For the binary classification problem in this study, when we obtain a feature dataset containing category information:
where n was the number of samples, the feature dimension of each sample was d, and the samples were divided into positive categories (
We used
In addition, the kernel function is a unique feature of the support vector model. For the same dataset, different kernel function choices will have different prediction effects. Appropriate kernel functions can improve prediction performance. The commonly used functions include the linear, Gaussian, and polynomial kernel functions. In this study, a linear kernel function was selected as the kernel of the support vector machine by comparing different kernel functions.
Bagging is one of the common ensemble learning models (Dudoit and Fridlyand, 2003; Jin et al., 2019; Jin et al., 2021; Wu and Yu, 2021). The ensemble learning model uses a series of weak learners (also called basic models) for learning and integrates the results of each weak learner to obtain a better learning effect than individual learners.
The bagging algorithm uses the simplest combination strategy to obtain the integration model. For the classification problem, the majority voting method is adopted. Each weak learner has one vote, and the final prediction result is generated according to the votes of all weak learners. The process of the bagging method is as follows: suppose we have a training set containing N samples and randomly put back the data to form a new training set. Because there is a way to put back sampling, a sample may be selected multiple times, or a sample may not be selected once. Hence, the size of the sampled data samples is the same as that of the original training data samples, but they contain different data. In this way, after T groups of data are extracted, T weak learners trained by different training sets can be obtained at the end of training. According to the prediction results of T weak learners, the most voting method is adopted to obtain a more accurate and reasonable prediction model.
Random forest is a representative bagging algorithm based on decision trees. Because random forest has good performance in regression and classification prediction, it has attracted great attention. It has been widely used in many practical problems, such as genome data analysis and disease risk prediction. When making classification prediction, each decision tree will make classification judgment on the data according to the characteristics of the data. Through the majority voting method, the category with the most votes is the prediction result of the random forest.
In the feature extraction section, we introduced three feature representation methods. The optimal feature subset of the dataset sample generated by the monoDiKGap method had 466 features. The CC feature representation method generated 12 features, and the GAAC feature method generated five features. Different feature extraction methods were combined to obtain hybrid features. However, the hybrid of features may lead to feature redundancy and affect the predictive effect of potentially druggable proteins. Therefore, we used MRMD and MRMD2.0 to select features and used fewer features to distinguish between potentially druggable and non-druggable proteins better.
In this study, the MRMD (Quan et al., 2016) was used to remove redundant features in hybrid features. The MRMD will leave the optimal feature subset after automatic feature selection. The main principle of this method is to use the Euclidean distance, cosine distance, and the Tanimoto coefficient to calculate the redundancy between features and use the Pearson correlation coefficient to calculate the correlation between dataset features and class labels to generate feature subsets with low redundancy and strong correlation automatically. When we analyse the hybrid feature subset that can accurately predict potentially druggable proteins, we also need to analyse the importance of different features. MRMD2.0 (He et al., 2021) combined seven algorithms, such as ANOVA, MIC, LASSO, mRMR, and chi-square test, through the PageRank strategy algorithm to rank different algorithm lists to form a directed graph, and each feature obtained a score. According to the ranking information, we analyse the importance of features and obtain key features that influence the prediction of potentially druggable proteins.
To intuitively measure the quality of the model, we evaluated the predictive effect of the model. This study used common evaluation indicators, including TP rate (TPR), FP rate (FPR), precision (Su et al., 2018), F-score (Sokolova et al., 2006), and accuracy (ACC) (Wei et al., 2017a; Wei et al., 2017b; Wei et al., 2018b; Wei et al., 2019b; Huang et al., 2020; Liang et al., 2020; Zhang et al., 2020; Xu et al., 2021b; Zhu et al., 2021). The calculation method of each measurement index was as follows:
Here, TP represents the classification number of correct positive samples, and TN represents the classification number of correct negative samples. FP represents the classification number of false positive samples. FN represents the classification number of false negative samples. In addition, this study also used 5-fold cross-validation to predict and evaluate the model.
Because the monoDiKGap feature extraction method gradually increases with the value of KGap, the number of corresponding generated feature vectors increases exponentially. In this study, the total feature set generated by monoDiKGap(k = 2) has 16,000 features, but in fact many small fragments appear very rarely, and some even appear 0 or 1 times. At this time, a large number of feature vectors composed of 0 or one also have no meaning already. In order to avoid high-dimensional feature vectors introducing dimensional disasters for subsequent machine learning algorithms, resulting in a significant decline in predictive classification performance. Therefore, this study used AdaBoost to automatically generate a more discriminative 466-dimensional feature subset, and compared the ACC values of the full feature set and the feature subset of monoDiKGap(k = 2) under different classifiers, as shown in Figure 3.
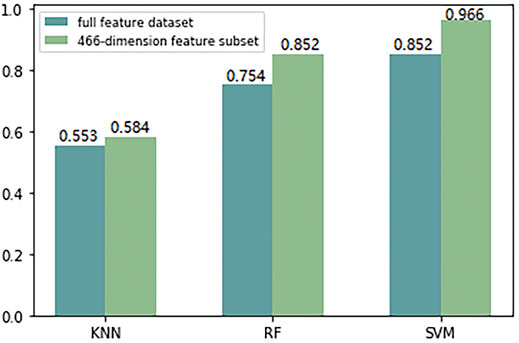
FIGURE 3. Comparison the ACC values of the full feature set and 466-dimensional feature subset extracted by monoDiKGap(k = 2) under different classifiers.
In this paper, three single feature extraction methods, monoDiKGap(k = 2), CC and GAAC, were used to represent the features of the dataset. Three single feature representation methods extracted 466-dimensional, 12-dimensional, and 5-dimensional features. The prediction performance of each extraction method under SVM, KNN, and RF was shown in Table 1. The data in Table 1 showed that the accuracy of the monoDiKGap(k = 2) feature representation method in predicting potentially druggable proteins through the SVM classification algorithm was higher than that of KNN and RF. The model can accurately predict 96.608% of the potentially druggable proteins. At this time, the TPR value reached 0.965, the FPR value reached 0.033, the F-score reached 0.962, and the ROC curve area was 0.966. The GAAC feature extraction method had an accuracy of 77.2864% in predicting potentially druggable proteins under SVM, which was 1.20 and 2.40% higher than that of the RF and KNN classification models, respectively. The accuracy of the CC feature extraction method to predict proteins through the SVM feature representation method was only 1.07% lower than that of the KNN algorithm. Therefore, considering the performance evaluation of the three feature representation methods under different classifiers, the SVM classification algorithm was more suitable for accurately predicting potentially druggable proteins.
To explore the prediction performance of hybrid features, we combined the above three feature representation methods and obtained new feature vectors of different combinations. After the combination of three single feature extraction methods, four new feature vectors were obtained: monoDiKGap + CC, monoDiKGap + GAAC, CC + GAAC and monoDiKGap + CC + GAAC. Table 2 showed the evaluation performance of different combinations of hybrid features using the SVM classification algorithm. Table 2 indicated that compared with the single feature representation method, the hybrid feature showed higher performance. The accuracy of the combination of monoDiKGap and other feature representation methods was more than 96%. In addition, the prediction performance of the CC + GAAC feature combination was also higher than that of the single feature representation method. Importantly, we found that the combination of monoDiKGap, CC, and GAAC features showed the best prediction performance, and the hybrid feature could accurately predict 96.6509% of potentially druggable proteins.
The kernel function is an important feature of support vector machines. The kernel function choice of the support vector machine affects the prediction performance of the model. For the monoDiKGap, CC, and GAAC hybrid features to represent the dataset features, we used different kernel functions and 5-fold cross-validation to select the appropriate kernel function. We compared the performance of the linear kernel function, quadratic polynomial kernel function, and radial basis kernel function. The ROC curves of different kernel functions were shown in Figure 4. The ROC values were 0.966, 0.955, and 0.846. The evaluation indicators of the three kernel functions were shown in Table 3. We can see that the prediction effect of the hybrid feature using the linear kernel function was better than the quadratic kernel function and the radial basis function. At this time, the three kernel functions predicted 96.6509, 95.5346, and 85.745% of the potentially druggable proteins, respectively. Therefore, this paper chose a linear kernel function as the kernel of the support vector machine.
For the linear kernel of the support vector machine, the penalty parameter C is an important parameter. The larger the value of C is, the easier it is to overfit, while the smaller the value of C is, the easier it is to underfit. The most commonly used C values are 1, 10, 100, and 1,000. We selected the appropriate C value with the help of grid search. Table 4 showed the prediction performance of different penalty parameters. When the C value was 1, the support vector machine classification algorithm achieved a better prediction effect and shortened the running time.
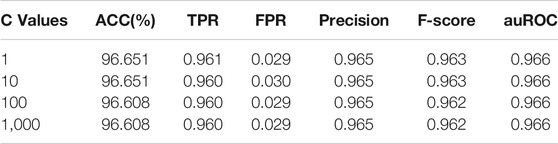
TABLE 4. Performance comparison of hybrid features with different penalty parameter C values under linear kernel.
The best hybrid features are 483-dimensional features mixed by the monoDiKGap, CC, and GAAC feature representation methods. These features may contain redundancy and affect the performance. Since the monoDiKGap feature extraction method automatically generated the optimal feature subset, we also need to remove redundant features from the CC and GAAC feature extraction methods. We used MRMD to filter the feature sets extracted by CC and GAAC and generated the optimal feature subset with low redundancy and strong correlation. Finally, we combined the feature subsets to obtain the filtered new hybrid features. These hybrid features not only reduced the feature dimensions but also had more expressiveness (Table 5).
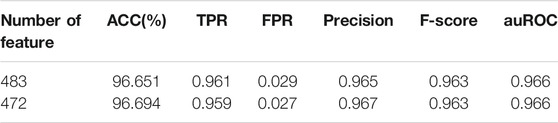
TABLE 5. Comparison of classification performance of hybrid features before and after using MRMD feature selection.
The expressive ability of a single support vector machine classification model may be limited so that the bagging ensemble algorithm based on a support vector machine has room for improvement. Compared with a single model, the bagging integration method can enhance the expressive ability of the model and reduce the error. When it is difficult for a single model to correctly distinguish the two types of data, the ensemble algorithm can often improve the model’s prediction performance by constructing multiple independent base models.
In this study, a support vector machine with a penalty coefficient of one and a linear kernel function was used as the basic model, and the number of optimal basic models was selected to construct a Bagging-SVM classification algorithm. The hybrid features of monoDiKGap, CC, and GAAC, which removed the cumbersome features, were shown in Figure 5 under the Bagging-SVM classification algorithm where the number of base models was 1–20. The accuracy of combining hybrid features and Bagging-SVM to predict potentially druggable proteins was basically more than 96.73%, and the highest prediction accuracy was 96.9944% when the number of base models was 12.
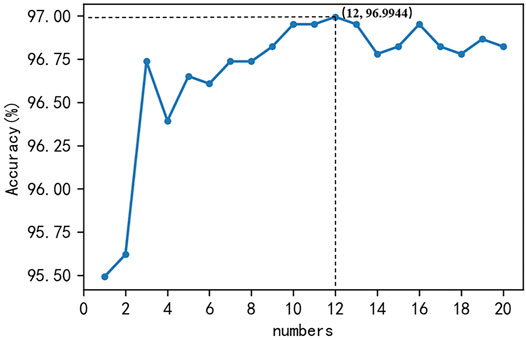
FIGURE 5. The accuracy of hybrid features in predicting potential druggable proteins under the Bagging-SVM classification algorithm where the number of base models was 1–20.
Based on the hybrid features of monoDiKGap, CC, GAAC, and Bagging-SVM, a new predictive model, DrugHybrid_BS, was constructed. To further explore the prediction model, we evaluated the performance of SVM, RF, and KNN using the same hybrid feature set. Table 6 showed that the DrugHybrid_BS model can better predict potentially druggable proteins. At this time, the TPR value reached 0.970, the F-score reached 0.967, and the AUC value reached 0.992. In addition, Table 6 showed the prediction performance comparison between the DrugHybrid_BS model and the previous model when using the same dataset as Lin et al. (Lin et al., 2019) and Jamali et al. (Jamali et al., 2016). The study found that the accuracy of the original data set using the DrugHybrid_BS model reached 100%, which shows that the original data does have redundancy, and it also reflects the significance of the initial data preprocessing in this article.
The accuracy of the classification and prediction model in predicting the training set cannot well reflect the future performance of the prediction model. To effectively judge the performance of a predictive model, we divided 80% of the dataset as the training set and 20% as the test set. The detailed information was shown in Figure 6. The independent test set using the DrugHybrid_BS model can accurately predict 96.5665% of the potentially druggable protein. The TPR value was 0.948, the FPR value was 0.02, the precision value was 0.975, and the AUC value was 0.990.
From the DrugHybrid_BS model, we obtained the following: after combining the single feature representation methods, the hybrid features of monoDiKGap, CC and GAAC combined with Bagging-SVM can improve the accuracy of predicting druggable proteins. This part further explored the features that play a key role in the DrugHybrid_BS model, that is, the importance of these features.
First, we used the MRMD2.0 to sort the feature sets extracted by three single feature representation methods and simultaneously obtained the relationship between the number of features and the accuracy of predicting potential druggable proteins (Figures 7A–C). Figure 7A showed that when the number of features of the CC feature extraction method was more than eight, the accuracy rate reached more than 60% and continued to grow. Therefore, we selected the top eight features as the key features of the CC feature representation method. Figure 7B showed the GAAC feature representation method. When the number of features was two, the accuracy rate reached more than 70%, and the accuracy rate continued to increase as the number increased. Therefore, we selected the top two features as the key features of the GAAC feature extraction method. Figure 7C showed the monoDiKGap feature extraction method. When the number of features was twenty-six, the accuracy of predicting potentially druggable proteins was significantly improved, and then the accuracy increased steadily as the number of features increased. Therefore, we chose the top twenty-six features as the key features of the monoDiKGap feature extraction method. Second, we combined the key features of the single feature extraction methods to obtain the hybrid key features. The detailed information was shown in Table 7. Finally, the number of base models suitable for the hybrid key features was selected through the Bagging-SVM classification model.
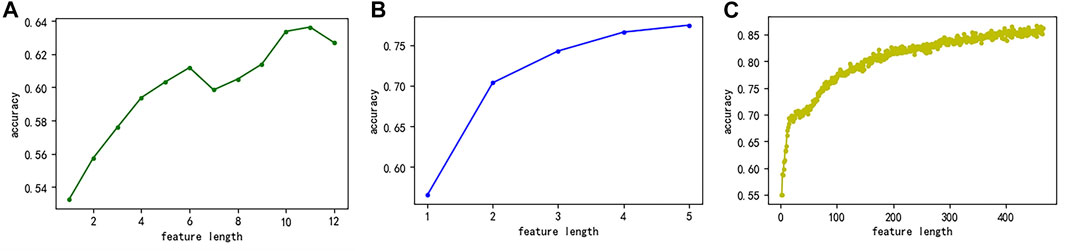
FIGURE 7. The relationship between the number of features extracted by the three methods and the accuracy of predicting potentially druggable proteins (A) CC (B) GAAC, and (C) monoDiKGap.
After research, we obtained that the hybrid key features can accurately predict 80.0343% of the potentially druggable proteins under the bagging algorithm based on the integration of fifteen SVMs. These hybrid key features combined with Bagging-SVM have achieved good prediction results, which fully demonstrated the importance of this part of the feature for the new method DrugHybrid_BS for predicting potentially druggable proteins.
Research on potentially druggable proteins is of great significance in the field of drug development and disease treatment. However, identifying potentially druggable proteins is the first step in research. This research focused on combining hybrid features and Bagging-SVM to predict potentially druggable proteins. The hybrid features included three feature extraction methods: monoDiKGap, CC, and GAAC, which were based on sequence information, physiochemical properties, and correlation. Through the three single feature representation methods of monoDiKGap, CC, GAAC, and the comparison of combined feature prediction, it was found that the hybrid features of monoDiKGap, CC, and GAAC can accurately predict 96.9944% of the potentially druggable proteins under Bagging-SVM. In addition, the accuracy of the independent test set using the new method DrugHybrid_BS reached 96.5665%. Therefore, the DrugHybrid_BS model used in this study could be a powerful method to study potentially druggable proteins and provide a reference value for other studies. In the future, we will try more deep learning techniques (Zou et al., 2019; Guo et al., 2020; Zeng et al., 2020; Niu et al., 2021b; Zhang et al., 2021) for this problem.
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author.
Conceptualization, BL and QZ; data collection or analysis, YG and PW; validation, YG; writing—original draft preparation, YG; writing—review and editing, YG. and QZ All authors have read and agreed to the published version of the manuscript.
This work was supported by the National Nature Science Foundation of China (Grant Nos 61863010, 11926205, 11926412, and 61873076), National Key R&D Program of China (No.2020YFB2104400), Natural Science Foundation of Hainan, China(Grant Nos. 119MS036 and 120RC588), Hainan Normal University 2020 Graduate Student Innovation Research Project (hsyx 2020-40) and the Special Science Foundation of Quzhou (2020D003).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fphar.2021.771808/full#supplementary-material
Ao, C., Yu, L., and Zou, Q. (2021). RFhy-m2G: Identification of RNA N2-Methylguanosine Modification Sites Based on Random Forest and Hybrid Features. Methods. doi:10.1016/j.ymeth.2021.05.016
Cheng, S., Zhang, L., Jin, B., Zhang, Q., and Lu, X. (2021). Drug Target Prediction Using Graph Representation Learning via Substructures Contrast, Appl. Sci., 11, 3239. doi:10.3390/app11073239
Dezső, Z., and Ceccarelli, M. (2020). Machine Learning Prediction of Oncology Drug Targets Based on Protein and Network Properties. BMC Bioinformatics 21, 104. doi:10.1186/s12859-020-3442-9
Ding, Y., Jijun, T., and Guo, F. (2020a). Identification of Drug-Target Interactions via Dual Laplacian Regularized Least Squares with Multiple Kernel Fusion. Knowledge-Based Syst. 204, 106254. doi:10.1016/j.knosys.2020.106254
Ding, Y., Tang, J., and Guo, F. (2019a). Identification of Drug-Side Effect Association via Semisupervised Model and Multiple Kernel Learning. IEEE J. Biomed. Health Inform. 23, 2619–2632. doi:10.1109/jbhi.2018.2883834
Ding, Y., Tang, J., and Guo, F. (2019b). Identification of Drug-Side Effect Association via Multiple Information Integration with Centered Kernel Alignment. Neurocomputing 325, 211–224. doi:10.1016/j.neucom.2018.10.028
Ding, Y., Tang, J., and Guo, F. (2020b). Identification of Drug-Target Interactions via Fuzzy Bipartite Local Model. Neural Comput. Applic 32, 10303–10319. doi:10.1007/s00521-019-04569-z
Ding, Y., Tang, J., and Guo, F. (2017). Identification of Drug-Target Interactions via Multiple Information Integration. Inf. Sci. 418-419, 546–560. doi:10.1016/j.ins.2017.08.045
Dudoit, S., and Fridlyand, J. (2003). Bagging to Improve the Accuracy of a Clustering Procedure. Bioinformatics 19, 1090–1099. doi:10.1093/bioinformatics/btg038
Fu, L., Niu, B., Zhu, Z., Wu, S., and Li, W. (2012). CD-HIT: Accelerated for Clustering the Next-Generation Sequencing Data. Bioinformatics 28, 3150–3152. doi:10.1093/bioinformatics/bts565
Gayvert, K. M., Madhukar, N. S., and Elemento, O. (2016). A Data-Driven Approach to Predicting Successes and Failures of Clinical Trials. Cell Chem Biol 23, 1294–1301. doi:10.1016/j.chembiol.2016.07.023
Guo, L., Jiang, Q., Jin, X., Liu, L., Zhou, W., Yao, S., et al. (2020). A Deep Convolutional Neural Network to Improve the Prediction of Protein Secondary Structure. Curr. Bioinformatics 15, 767–777. doi:10.2174/1574893615666200120103050
Guo, Y., Yu, L., Wen, Z., and Li, M. (2008). Using Support Vector Machine Combined with Auto Covariance to Predict Protein-Protein Interactions from Protein Sequences. Nucleic Acids Res. 36, 3025–3030. doi:10.1093/nar/gkn159
Han, K., Wang, M., Zhang, L., Wang, Y., Guo, M., Zhao, M., et al. (2019). Predicting Ion Channels Genes and Their Types with Machine Learning Techniques. Front. Genet. 10, 399. doi:10.3389/fgene.2019.00399
He, S., Guo, F., Zou, Q., and HuiDing, H. (2021). MRMD2.0: A Python Tool for Machine Learning with Feature Ranking and Reduction. Curr. Bioinformatics 15, 1213–1221. doi:10.2174/1574893615999200503030350
Hopkins, A. L., and Groom, C. R. (2002). The Druggable Genome. Nat. Rev. Drug Discov. 1, 727–730. doi:10.1038/nrd892
Huang, Y., Zhou, D., Wang, Y., Zhang, X., Su, M., Wang, C., et al. (2020). Prediction of Transcription Factors Binding Events Based on Epigenetic Modifications in Different Human Cells. Epigenomics 12, 1443–1456. doi:10.2217/epi-2019-0321
Huo, Y., Xin, L., Kang, C., Wang, M., Ma, Q., and Yu, B. (2020). SGL-SVM: A Novel Method for Tumor Classification via Support Vector Machine with Sparse Group Lasso. J. Theor. Biol. 486, 110098. doi:10.1016/j.jtbi.2019.110098
Jamali, A. A., Ferdousi, R., Razzaghi, S., Li, J., Safdari, R., and Ebrahimie, E. (2016). DrugMiner: Comparative Analysis of Machine Learning Algorithms for Prediction of Potential Druggable Proteins. Drug Discov. Today 21, 718–724. doi:10.1016/j.drudis.2016.01.007
Ji, X., Freudenberg, J. M., and Agarwal, P. (2019). Integrating Biological Networks for Drug Target Prediction and Prioritization. Methods Mol. Biol. 1903, 203–218. doi:10.1007/978-1-4939-8955-3_12
Jiang, Q., Wang, G., Jin, S., Li, Y., and Wang, Y. (2013). Predicting Human microRNA-Disease Associations Based on Support Vector Machine. Int. J. Data Min Bioinform 8, 282–293. doi:10.1504/ijdmb.2013.056078
Jin, Q., Cui, H., Sun, C., Meng, Z., and Su, R. (2021). Free-form Tumor Synthesis in Computed Tomography Images via Richer Generative Adversarial Network. Knowledge-Based Syst. 218, 106753. doi:10.1016/j.knosys.2021.106753
Jin, Q., Meng, Z., Pham, T. D., Chen, Q., Wei, L., and Su, R. (2019). DUNet: A Deformable Network for Retinal Vessel Segmentation. Knowledge-Based Syst. 178, 149–162. doi:10.1016/j.knosys.2019.04.025
Lee, T. Y., Lin, Z. Q., Hsieh, S. J., Bretaña, N. A., and Lu, C. T. (2011). Exploiting Maximal Dependence Decomposition to Identify Conserved Motifs from a Group of Aligned Signal Sequences. Bioinformatics 27, 1780–1787. doi:10.1093/bioinformatics/btr291
Li, Q., and Lai, L. (2007). Prediction of Potential Drug Targets Based on Simple Sequence Properties. BMC Bioinformatics 8, 353. doi:10.1186/1471-2105-8-353
Liang, X., Zhu, W., Liao, B., Wang, B., Yang, J., Mo, X., et al. (2020). A Machine Learning Approach for Tracing Tumor Original Sites with Gene Expression Profiles. Front. Bioeng. Biotechnol. 8, 607126. doi:10.3389/fbioe.2020.607126
Liao, Y., and Vemuri, V. R. (2002). Use of K-Nearest Neighbor Classifier for Intrusion Detection. Comput. Security 21, 439–448. doi:10.1016/s0167-4048(02)00514-x
Lin, J., Chen, H., Li, S., Liu, Y., Li, X., and Yu, B. (2019). Accurate Prediction of Potential Druggable Proteins Based on Genetic Algorithm and Bagging-SVM Ensemble Classifier. Artif. Intell. Med. 98, 35–47. doi:10.1016/j.artmed.2019.07.005
Liu, D., Li, G., and Zuo, Y. (2019). Function Determinants of TET Proteins: the Arrangements of Sequence Motifs with Specific Codes. Brief Bioinform 20, 1826–1835. doi:10.1093/bib/bby053
Liu, B., Gao, X., and Zhang, H. (2019). BioSeq-Analysis2.0: an Updated Platform for Analyzing DNA, RNA and Protein Sequences at Sequence Level and Residue Level Based on Machine Learning Approaches. Nucleic Acids Res. 47, e127. doi:10.1093/nar/gkz740
Liu, J., Su, R., Zhang, J., and Wei, L. (2021). Classification and Gene Selection of Triple-Negative Breast Cancer Subtype Embedding Gene Connectivity Matrix in Deep Neural Network. Brief. Bioinform. 22, bbaa395. LID - bbaa395 [pii] LID - 10.1093/bib/bbaa395 [doi]. doi:10.1093/bib/bbaa395
Lv, H., Zhang, Z. M., Li, S. H., Tan, J. X., Chen, W., and Lin, H. (2020). Evaluation of Different Computational Methods on 5-methylcytosine Sites Identification. Brief Bioinform 21, 982–995. doi:10.1093/bib/bbz048
Meng, C., Guo, F., and Zou, Q. (2020). CWLy-SVM: A Support Vector Machine-Based Tool for Identifying Cell wall Lytic Enzymes. Comput. Biol. Chem. 87, 107304. doi:10.1016/j.compbiolchem.2020.107304
Munir, A., Malik, S. I., and Malik, K. A. (2019). Proteome Mining for the Identification of Putative Drug Targets for Human Pathogen Clostridium tetani. Curr. Bioinformatics 14, 532–540. doi:10.2174/1574893613666181114095736
Niu, M., Lin, Y., and Zou, Q. (2021). sgRNACNN: Identifying sgRNA On-Target Activity in Four Crops Using Ensembles of Convolutional Neural Networks. Plant Mol. Biol. 105, 483–495. doi:10.1007/s11103-020-01102-y
Niu, M., Wu, J., Zou, Q., Liu, Z., and Xu, L. (2021). rBPDL:Predicting RNA-Binding Proteins Using Deep Learning. IEEE J. Biomed. Health Inform. 25, 3668–3676. doi:10.1109/jbhi.2021.3069259
Pacheco, M. P., Bintener, T., Ternes, D., Kulms, D., Haan, S., Letellier, E., et al. (2019). Identifying and Targeting Cancer-specific Metabolism with Network-Based Drug Target Prediction. EBioMedicine 43, 98–106. doi:10.1016/j.ebiom.2019.04.046
Platt, J. C. (1998). Sequential Minimal Optimization: A Fast Algorithm for Training Support Vector Machines. Technical Report MSR-TR-98-14.
Quan, Z., Zeng, J., Cao, L., and Ji, R. (2016). A Novel Features Ranking Metric with Application to Scalable Visual and Bioinformatics Data Classification. Neurocomputing 173, 346–354. doi:10.1016/j.neucom.2014.12.123
Ru, X., Wang, L., Li, L., Ding, H., Ye, X., and Zou, Q. (2020). Exploration of the Correlation between GPCRs and Drugs Based on a Learning to Rank Algorithm. Comput. Biol. Med. 119, 103660. doi:10.1016/j.compbiomed.2020.103660
Russ, A. P., and Lampel, S. (2005). The Druggable Genome: an Update. Drug Discov. Today 10, 1607–1610. doi:10.1016/s1359-6446(05)03666-4
Salmaso, V., and Moro, S. (2018). Bridging Molecular Docking to Molecular Dynamics in Exploring Ligand-Protein Recognition Process: An Overview. Front. Pharmacol. 9, 923. doi:10.3389/fphar.2018.00923
Samanthula, B. K., Elmehdwi, Y., and Jiang, W. (2014). K-Nearest Neighbor Classification over Semantically Secure Encrypted Relational Data. IEEE Trans. Knowledge Data Eng. 27, 1261–1273. doi:10.1109/TKDE.2014.2364027
Shang, Y., Gao, L., Zou, Q., and Yu, L. (2021). Prediction of Drug-Target Interactions Based on Multi-Layer Network Representation Learning. Neurocomputing 434, 80–89. doi:10.1016/j.neucom.2020.12.068
Shi, H., Liu, S., Chen, J., Li, X., Ma, Q., and Yu, B. (2019). Predicting Drug-Target Interactions Using Lasso with Random forest Based on Evolutionary Information and Chemical Structure. Genomics 111, 1839–1852. doi:10.1016/j.ygeno.2018.12.007
Sokolova, M., Japkowicz, N., and Szpakowicz, S. (2006). Beyond Accuracy, F-Score and ROC: A Family of Discriminant Measures for Performance Evaluation. Berlin, Heidelberg: Springer.
Su, R., Wu, H., Xu, B., Liu, X., and Wei, L. (2018). Developing a Multi-Dose Computational Model for Drug-Induced Hepatotoxicity Prediction Based on Toxicogenomics Data. Ieee/acm Trans. Comput. Biol. Bioinform 16, 1231–1239. doi:10.1109/TCBB.2018.2858756
Wang, J., Wang, H., Wang, X., and Chang, H. (2020). Predicting Drug-Target Interactions via FM-DNN Learning. Curr. Bioinformatics 15, 68–76. doi:10.2174/1574893614666190227160538
Wang, J., Shi, Y., Wang, X., and Chang, H. (2020). A Drug Target Interaction Prediction Based on LINE-RF Learning. Curr. Bioinformatics 15, 750–757. doi:10.2174/1574893615666191227092453
Wang, H., Ding, Y., Tang, J., and Guo, F. (2020c). Identification of Membrane Protein Types via Multivariate Information Fusion with Hilbert-Schmidt Independence Criterion. Neurocomputing 383, 257–269. doi:10.1016/j.neucom.2019.11.103
Wang, Y., Liu, K., Ma, Q., Tan, Y., Du, W., Lv, Y., et al. (2019). Pancreatic Cancer Biomarker Detection by Two Support Vector Strategies for Recursive Feature Elimination. Biomark Med. 13, 105–121. doi:10.2217/bmm-2018-0273
Wang, Z., Liu, D., Xu, B., Tian, R., and Zuo, Y. (2021). Modular Arrangements of Sequence Motifs Determine the Functional Diversity of KDM Proteins. Brief. Bioinformatics 22, bbaa215. doi:10.1093/bib/bbaa215
Wei, L., Luan, S., Nagai, L. A. E., Su, R., and Zou, Q. (2019). Exploring Sequence-Based Features for the Improved Prediction of DNA N4-Methylcytosine Sites in Multiple Species. Bioinformatics 35, 1326–1333. doi:10.1093/bioinformatics/bty824
Wei, L., Xing, P., Shi, G., Ji, Z., and Zou, Q. (2019). Fast Prediction of Protein Methylation Sites Using a Sequence-Based Feature Selection Technique. Ieee/acm Trans. Comput. Biol. Bioinform 16, 1264–1273. doi:10.1109/tcbb.2017.2670558
Wei, L., Ding, Y., Su, R., Tang, J., and Zou, Q. (2018). Prediction of Human Protein Subcellular Localization Using Deep Learning. J. Parallel Distributed Comput. 117, 212–217. doi:10.1016/j.jpdc.2017.08.009
Wei, L., Zhou, C., Chen, H., Song, J., and Su, R. (2018). ACPred-FL: a Sequence-Based Predictor Using Effective Feature Representation to Improve the Prediction of Anti-cancer Peptides. Bioinformatics 34, 4007–4016. doi:10.1093/bioinformatics/bty451
Wei, L., Tang, J., and Zou, Q. (2017). Local-DPP: An Improved DNA-Binding Protein Prediction Method by Exploring Local Evolutionary Information. Inf. Sci. 384, 135–144. doi:10.1016/j.ins.2016.06.026
Wei, L., Xing, P., Zeng, J., Chen, J., Su, R., and Guo, F. (2017). Improved Prediction of Protein-Protein Interactions Using Novel Negative Samples, Features, and an Ensemble Classifier. Artif. Intell. Med. 83, 67–74. doi:10.1016/j.artmed.2017.03.001
Wishart, D. S., Knox, C., Guo, A. C., Shrivastava, S., Hassanali, M., Stothard, P., et al. (2006). DrugBank: a Comprehensive Resource for In Silico Drug Discovery and Exploration. Nucleic Acids Res. 34, D668–D672. doi:10.1093/nar/gkj067
Wu, X., and Yu, L. (2021). EPSOL: Sequence-Based Protein Solubility Prediction Using Multidimensional Embedding. Bioinformatics (Oxford, England), btab463. doi:10.1093/bioinformatics/btab463
Xu, B., Liu, D., Wang, Z., Tian, R., and Zuo, Y. (2021). Multi-substrate Selectivity Based on Key Loops and Non-homologous Domains: New Insight into ALKBH Family. Cell Mol Life Sci 78, 129–141. doi:10.1007/s00018-020-03594-9
Xu, L., Liang, G., Shi, S., and Liao, C. (2018). SeqSVM: A Sequence-Based Support Vector Machine Method for Identifying Antioxidant Proteins. Int. J. Mol. Sci. 19. doi:10.3390/ijms19061773
Xu, Z., Luo, M., Lin, W., Xue, G., Wang, P., Jin, X., et al. (2021). DLpTCR: an Ensemble Deep Learning Framework for Predicting Immunogenic Peptide Recognized by T Cell Receptor. Brief Bioinform 22, bbab335. doi:10.1093/bib/bbab335
Yu, L., Wang, M., Yang, Y., Xu, F., Zhang, X., Xie, F., et al. (2021). Predicting Therapeutic Drugs for Hepatocellular Carcinoma Based on Tissue-specific Pathways. Plos Comput. Biol. 17, e1008696. doi:10.1371/journal.pcbi.1008696
Zeng, X., Zhong, Y., Lin, W., and Zou, Q. (2020). Predicting Disease-Associated Circular RNAs Using Deep Forests Combined with Positive-Unlabeled Learning Methods. Brief Bioinform 21, 1425–1436. doi:10.1093/bib/bbz080
Zhang, L., Xiao, X., and Xu, Z. C. (2020). iPromoter-5mC: A Novel Fusion Decision Predictor for the Identification of 5-Methylcytosine Sites in Genome-wide DNA Promoters. Front Cel Dev Biol 8, 614. doi:10.3389/fcell.2020.00614
Zhang, N., Sa, Y., Guo, Y., Lin, W., Wang, P., and Feng, Y. (2018). Discriminating Ramos and Jurkat Cells with Image Textures from Diffraction Imaging Flow Cytometry Based on a Support Vector Machine. Curr. Bioinformatics 11, 1. doi:10.2174/1574893611666160608102537
Zhang, Y., Yan, J., Chen, S., Gong, M., Gao, D., Zhu, M., et al. (2021). Review of the Applications of Deep Learning in Bioinformatics. Curr. Bioinformatics 15, 898–911. doi:10.2174/1574893615999200711165743
Zheng, L., Huang, S., Mu, N., Zhang, H., Zhang, J., Chang, Y., et al. (2019). RAACBook: a Web Server of Reduced Amino Acid Alphabet for Sequence-dependent Inference by Using Chou's Five-step Rule. Database (Oxford) 2019, baz131. doi:10.1093/database/baz131
Zheng, L., Liu, D., Yang, W., Yang, L., and Zuo, Y. (2021). RaacLogo: a New Sequence Logo Generator by Using Reduced Amino Acid Clusters. Brief. Bioinformatics 22, bbaa096. doi:10.1093/bib/bbaa096
Zhong, F., Xing, J., Li, X., Liu, X., Fu, Z., Xiong, Z., et al. (2018). Artificial Intelligence in Drug Design. Sci. China Life Sci. 61, 1191–1204. doi:10.1007/s11427-018-9342-2
Zhu, J., Arbor, A., and Hastie, T. (2006). Multi-class AdaBoost. Stat. Its Interf. 2, 349–360. doi:10.4310/SII.2009.v2.n3.a8
Zhu, Y., Li, F., Xiang, D., Akutsu, T., Song, J., and Jia, C. (2021). Computational Identification of Eukaryotic Promoters Based on Cascaded Deep Capsule Neural Networks. Brief Bioinform 22, bbaa299. doi:10.1093/bib/bbaa299
Zhuang, J., Dai, S., Zhang, L., Gao, P., Han, Y., Tian, G., et al. (2021). Identifying Breast Cancer-Induced Gene Perturbations and its Application in Guiding Drug Repurposing. Curr. Bioinformatics 15, 1075–1089. doi:10.2174/1574893615666200203104214
Zou, Q., Xing, P., Wei, L., and Liu, B. (2019). Gene2vec: Gene Subsequence Embedding for Prediction of Mammalian N 6-methyladenosine Sites from mRNA. RNA 25, 205–218. doi:10.1261/rna.069112.118
Zou, Q., Lin, G., Jiang, X., Liu, X., and Zeng, X. (2020). Sequence Clustering in Bioinformatics: an Empirical Study. Brief. Bioinform. 21, 1–10. doi:10.1093/bib/bby090
Keywords: monoDiKGap, CC, GAAC, bagging, support vector machine
Citation: Gong Y, Liao B, Wang P and Zou Q (2021) DrugHybrid_BS: Using Hybrid Feature Combined With Bagging-SVM to Predict Potentially Druggable Proteins. Front. Pharmacol. 12:771808. doi: 10.3389/fphar.2021.771808
Received: 07 September 2021; Accepted: 15 November 2021;
Published: 30 November 2021.
Edited by:
Xiujuan Lei, Shaanxi Normal University, ChinaReviewed by:
Jiawei Luo, Hunan University, ChinaCopyright © 2021 Gong, Liao, Wang and Zou. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Bo Liao, ZHJhZ29uYndAMTYzLmNvbQ==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.