
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Pharmacol., 25 October 2021
Sec. Translational Pharmacology
Volume 12 - 2021 | https://doi.org/10.3389/fphar.2021.768210
This article is part of the Research TopicVenoms and Toxins: Functional Omics and Pharmacological InsightsView all 7 articles
The Andaman and Nicobar Islands are an abode to a diversity of flora and fauna, including the many endemic species of snakes, such as the elusive Andaman cobra (Naja sagittifera). However, the ecology and evolution of venomous snakes inhabiting these islands have remained entirely uninvestigated. This study aims to bridge this knowledge gap by investigating the evolutionary history of N. sagittifera and its venom proteomic, biochemical and toxicity profile. Phylogenetic reconstructions confirmed the close relationship between N. sagittifera and the Southeast Asian monocellate cobra (N. kaouthia). Overlooking this evolutionary history, a polyvalent antivenom manufactured using the venom of the spectacled cobra (N. naja) from mainland India is used for treating N. sagittifera envenomations. Comparative evaluation of venoms of these congeners revealed significant differences in their composition, functions and potencies. Given the close phylogenetic relatedness between N. sagittifera and N. kaouthia, we further assessed the cross-neutralising efficacy of Thai monovalent N. kaouthia antivenom against N. sagittifera venoms. Our findings revealed the inadequate preclinical performance of the Indian polyvalent and Thai monovalent antivenoms in neutralising N. sagittifera venoms. Moreover, the poor efficacy of the polyvalent antivenom against N. naja venom from southern India further revealed the critical need to manufacture region-specific Indian antivenoms.
Located 1,400 km off mainland India in the Bay of Bengal, the Andaman and Nicobar Islands are among India’s ten biogeographic zones. Over 85% of the total land area being covered by forests and only 38 out of the 572 islands are home to nearly 400,000 human inhabitants (Sawhney, 2019). Among these are the indigenous Andamanese people, the aboriginal residents belonging to many tribes, such as the Jarawa and Sentinelese. These islands are also an abode to many endemic plants and animals, including many species of enigmatic snakes. The Andaman cobra (Naja sagittifera), Andaman krait (Bungarus andamanensis), king cobra (Ophiophagus hannah) and several endemic pit vipers (Trimeresurus spp.) on the island are amongst the venomous snakes that are capable of inflicting severe clinical symptoms in humans. The Andaman cobra, in particular, is primarily distributed across the islands, but the ecology and evolution of venom of this enigmatic snake remain entirely uninvestigated.
Unlike in mainland India, the impact of snakebite to populations of Andaman and Nicobar Islands is unclear. While a study examining the Health Management Information System data from 2017 to 2019 estimated that between 23 and 28 people per 100,000 suffer snake envenoming on these islands (Salve et al., 2020), a rigorous snakebite epidemiological survey is warranted to precisely understand the burden of this socioeconomic disease. Moreover, completely neglecting the distinct biogeographic histories of these islands and the phylogenetic histories of snakes inhabiting them, a polyvalent antivenom manufactured against the “big four” Indian snakes [i.e., spectacled cobra (N. naja), common krait (B. caeruleus), Russell’s viper (Daboia russelii) and saw-scaled viper (Echis carinatus)] from Tamil Nadu is used for the treatment of snakebites. However, the effectiveness of this antivenom in treating bites from the phylogenetically distant snakes of these islands remains to be evaluated.
In this study, we unveil the venom composition, biochemistry, pharmacological activity, and potency of the Andaman cobra (N. sagittifera). We comparatively evaluate the venom of this enigmatic snake with its congener from mainland India: the spectacled cobra (N. naja). Phylogenetic reconstructions using mitochondrial markers revealed the distant evolutionary relationship between these Naja species. Consistent with previous report (Kazemi et al., 2021), N. sagittifera was found to have originated from the common ancestor of Southeast Asian monocled cobra (N. kaouthia). Furthermore, we assessed the effectiveness of the “big four” polyvalent Indian antivenom in cross-neutralising the venom of this phylogenetically distant elapid snake. Considering its close phylogenetic relationship with N. kaouthia from Southeast Asia, we also evaluated the potential use of Thai Red Cross Society’s N. kaouthia monovalent antivenom for the treatment of N. sagittifera bites. Our findings unravel the preclinical ineffectiveness of commercial antivenoms against N. sagittifera and underscore the necessity to manufacture a specific antivenom product for therapeutic use on these islands.
Venom samples were sourced from an adult N. naja from Bannerghatta, Karnataka in mainland India, and a subadult N. sagittifera from Port Blair, Andaman Island. Since encountering N. sagittifera in the wild is extremely rare, we could not find full grown adult individuals of this species during our sampling trips. Following collection, N. sagittifera venom sample was immediately flash frozen and transported back to the lab in a vapor shipper. The snake was released back into the wild after sampling. Samples were collected with permissions from the Karnataka [PCCF(WL)/CR-06/2018-19)] and Andaman and Nicobar Islands [CWLW/WL/134(A)/253] Forest Departments. The venoms were flash-frozen immediately after extraction, lyophilised and stored at −80°C. Details of the commercial antivenoms and venoms tested in this study are provided in Table 1 and Supplementary Table S1, respectively.
Venom toxicity and neutralisation experiments were conducted in the murine model following WHO guidelines. Animal ethical approval was granted by the Institutional Animal Ethics Committee (IAEC), Indian Institute of Science (IISc), Bangalore (CAF/Ethics/770/2020 and CAF/Ethics/814/2021). Guidelines issued by the Committee for the Purpose of Control and Supervision of Experiments on Animals (CPCSEA) were followed while conducting these experiments. Collection of fresh blood for coagulation assay was obtained from healthy adult volunteers with written consent and approved by the Institutional Human Ethical Committee (IHEC No: 5-24072019 and 18/20201216).
Scale samples were harvested from the ventral side of the snake using microscissors and preserved in molecular grade absolute ethanol. The genomic DNA was isolated using Xpress DNA Tissue kit following the manufacturer’s protocol (MagGenome, United Kingdom) and the purity of the isolate was determined by measuring the 260/280 nm wavelength absorbance ratio with an Epoch 2 microplate spectrophotometer (BioTek Instruments, Inc., United States). Mitochondrial markers (ND4 and cyt b) were amplified by performing polymerase chain reaction (PCR) on a ProFlex PCR System (Thermo Fisher Scientific, MA, United States) using universal primers (Arevalo et al., 1994; Palumbi, 1996; Harvey et al., 2000) (Supplementary Table S2). Components of PCR reaction mixtures for a total volume of 50 μl were: 2 μl of template DNA (∼50 ng), 25 μl of Taq DNA Polymerase Mastermix [Tris-HCl pH 8.5, (NH4)2SO4, 3 mM MgCl2, 0.2% Tween 20, 3 μl of 25 mM MgCl2, 0.4 mM dNTPs, Amplicon Taq polymerase], 2.5 μl each of forward and reverse primers and 18 μl nuclease-free water. The PCR program was set as follows: initial denaturation at 94°C for 5 min, followed by 35–40 cycles of denaturation (94°C for 30 s), annealing (TA for 35 s) and extension (72°C for 2 min) and a final extension step for 10 min at 72°C. QIAquick PCR and Gel Cleanup Kit (Qiagen) were used for gel purification, and the amplicons were sequenced on an Applied Biosystems 3730xl platform (Thermo Fisher Scientific, MA, United States). The sequence data were acquired with Sequence Scanner Software v2.0. Datasets used for phylogenetic reconstructions, in addition to the cyt b and ND4 sequences generated in this study, have been provided in Supplementary Files S1, S2.
Homologues of the sequenced mitochondrial markers were retrieved from NCBI’s GenBank repository using BLAST searches, and the nucleotide datasets were compiled and manually curated. Clustal-Omega algorithm was used for generating multiple sequence alignments (Sievers et al., 2011), and the phylogenetic histories of Naja spp. were reconstructed using a maximum likelihood approach implemented in the PhyML package (Guindon et al., 2010). Smart Model Selection (SMS) tool on the ATGC server (Lefort et al., 2017) was used to determine the best nucleotide substitution scheme and the phylogenetic trees were subsequently built using the Subtree-Pruning-Regrafting (SPR) method of tree topology search, and the node support was derived with 100 bootstrapping replicates. Additionally, Bayesian inference-based phylogenies were estimated using MrBayes 3.2.7 (Altekar et al., 2004; Ronquist et al., 2012). The analyses, which were parallelised on four independent runs, each executing eleven simultaneous Markov chain simulations, were programmed to terminate after 10 million generations or when the standard deviation of split frequencies converged to 0.01. At every 100th generation, trees and the corresponding parameter estimates were sampled, and the first 25% of these were discarded as burn-in. Finally, a majority-rule consensus tree was generated with the posterior probability for each node. The evolutionary divergence between sequences for each gene was determined by estimating p-distances using MEGA X (Kumar et al., 2018) with 100 bootstrap replicates.
The total protein content of venoms and antivenoms were estimated using the Bradford method, wherein crude venom was incubated with 250 µl of Bradford reagent (Thermo Fisher Scientific, United States), and absorbance was recorded at 595 nm on an Epoch 2 microplate reader (BioTek Instruments, Inc., United States). Bovine Serum Albumin (BSA) and Bovine Gamma Globulin (BGG) served as standards for protein estimation of venoms and antivenoms, respectively (Bradford, 1976).
N. sagittifera and N. naja venoms were subjected to reducing SDS-PAGE to evaluate the qualitative and quantitative differences in venoms. Briefly, venom samples (12 µg) were loaded into 12.5% gel, followed by an electrophoretic separation in Tris-Glycine-SDS buffer at 80 V (Smith, 1984). The Precision Plus Protein Dual Color Xtra Standard (Bio-Rad Laboratories, United States) was used as a marker, and the gel was stained with Coomassie Brilliant Blue R-250 (Sisco Research Laboratories Pvt. Ltd., India). iBright CL1000 gel documentation system (Thermo Fisher Scientific, United States) was then used for visualising the gel.
Snake venom samples (500 µg) were fractionated by RP-HPLC using a Shimadzu LC-20AD series system (Kyoto, Japan) equipped with Shimadzu C18 (25 cm × 4.6 mm, 5 µm particle size, 300 Å pore size) column equilibrated with solution A (0.1% trifluoroacetic acid in HPLC grade water) and a diode array detector (DAD). The elution was carried out at a flow rate of 1 ml/min with the following concentration gradients of solution B (0.1% trifluoroacetic acid in 100% acetonitrile): 5–15% for 10 min, 15–45% over 60 min, 45–70% over 10 min and 70% for 9 min (Lomonte and Calvete, 2017; Senji Laxme R. R. et al., 2021). The absorbance was monitored at 215 nm and venom fractions (1 ml/min) were collected semi-automatically using a fraction collector (Shimadzu FRC-10A, Japan).
The RP-HPLC fractions of N. sagittifera and N. naja venom were subjected to in-solution trypsin digestion, followed by lyophilisation and storage at −80°C. The samples were then reconstituted moments before injection and characterisation with tandem mass spectrometry (Rashmi et al., 2021). Samples (40 µg) were first reduced with 10 mM dithiothreitol (DTT), alkylated using 30 mM iodoacetamide (IAA) and further digested with trypsin (0.2 µg/µl) overnight at 37°C. The digested samples were run through a C18 nano-LC column (50 cm × 75 µm, 3 µm particle size and 100 Å pore size) with a Thermo EASY nLC 1200 series system (Thermo Fisher Scientific, MA, United States). Elution was carried out at a constant flow rate of 300 nl/min for 120 min with buffer A (0.1% formic acid in HPLC grade water) and gradient buffer B (0.1% formic acid in 80% acetonitrile) as follows: 10–45% over 98 min, 45–95% over 4 min and 95% over 18 min. A Thermo Orbitrap Fusion Mass Spectrometer (Thermo Fisher Scientific, MA, United States) was used for mass spectrometric analyses of the samples. MS scans were performed using the following parameters: scan range (m/z) of 375–1700 with a resolution of 120,000 and maximum injection time of 50 ms. Fragment scans (MS/MS) were performed using an ion trap detector with high collision energy fragmentation (30%), scan range (m/z) of 100–2000, and maximum injection time of 35 ms.
For the identification of various toxin families in the proteomic profiles of venom fractions, the raw MS/MS spectra were searched against the NCBI-NR Serpentes databases (taxid: 8570) using PEAKS Studio X Plus (Bioinformatics Solutions Inc., ON, Canada) with the following parameters: parent and fragment mass error tolerance limits of 10 ppm and 0.6 Da, respectively; “monoisotopic” precursor ion search type; “semispecific” trypsin digestion; cysteine carbamidomethylation (+57.02) as a fixed modification; methionine oxidation (+15.99) as a variable modification and False Discovery Rate (FDR) of 0.1. Mass spectrometry data has been deposited to the ProteomeXchange Consortium via the PRIDE partner repository (Perez-Riverol et al., 2019) with data identifier: PXD025362. Protein families with at least one unique matching peptide were considered for downstream analyses. The redundant protein hits from each protein family were removed manually, and their relative abundance in a fraction was determined by calculating its area under the spectral intensity curve (AUC) relative to the overall AUC for all protein families in that fraction. AUC values obtained from PEAKS Studio analyses, representing the mean spectral intensities, were normalised across fractions using the percentage of peak areas for the respective RP-HPLC fractions (Tan et al., 2017; Rashmi et al., 2021). The relative abundance of a protein family hit (X) was estimated using the equation below, where “N” indicates the total number of fractions obtained from RP-HPLC and “n” corresponds to the fraction number.
PLA2 activity of N. sagittifera and N. naja venoms was evaluated using modifications to a previously described protocol (Marinetti, 1965; Joubert and Taljaard, 1980; Senji Laxme R. R. et al., 2021). In brief, a substrate solution containing chicken egg yolk dissolved in 0.9% NaCl was freshly prepared with an absorbance of 1 at 740 nm. Time-dependent kinetics of varying amounts of Naja venoms (0.01, 0.1, 0.5, and 1 µg) were assayed for their PLA2 activity in triplicates. 250 µl of the egg yolk substrate solution was added, and the absorbance was monitored at 1 min intervals for 60 min at 740 nm using an Epoch 2 microplate spectrophotometer (BioTek Instruments, Inc., United States). The unit PLA2 activity was estimated as the amount of crude venom that reduces the absorbance of the substrate by 0.01 OD unit per minute (Joubert and Taljaard, 1980).
N. sagittifera and N. naja venoms were analysed for their proteolytic activities using previously published protocols (Chowdhury et al., 1990). 10 µl of the crude venom was incubated with 80 µg azocasein (Sigma-Aldrich, United States) substrate for 90 min at 37°C. Following the incubation period, the enzymatic reaction was quenched by the addition of 200 µl of trichloroacetic acid. The supernatant of this mixture, obtained by centrifuging at 1,000 × g for 5 min, was mixed with equal volumes of 0.5 M NaOH and the absorbance was measured at 440 nm in an Epoch 2 microplate spectrophotometer (BioTek Instruments, Inc., United States). Purified bovine pancreatic protease (Sigma-Aldrich, United States) was used as a positive control to estimate the relative proteolytic activity of Naja venoms.
The LAAO activity of N. sagittifera and N. naja venoms was evaluated with a previously described endpoint assay (Kishimoto and Takahashi, 2001; Senji Laxme R. R. et al., 2021). 90 µl of the L-leucine substrate solution (5 mM L-leucine, 50 nM Tris-HCl buffer, 5 IU/ml horseradish peroxidase, 2 mM o-phenylenediamine dihydrochloride) was incubated with the crude venom (10 µl) at 37°C for 60 min. Following incubation, 2 M H2SO4 solution was added to terminate the reaction, and the absorbance was recorded at 492 nm using an Epoch 2 microplate spectrophotometer.
0.05 µg/µl of crude venom in 1X phosphate buffered saline (PBS) was mixed with purified calf thymus DNA (Sigma-Aldrich, United States) and incubated at 37°C for 60 min to assess the DNase activity of N. sagittifera and N. naja venoms (Gerceker et al., 2009; Senji Laxme R. R. et al., 2021). Post-incubation, these reaction mixtures were subjected to 0.8% agarose gel electrophoresis. GelRed (Sigma-Aldrich, United States) nucleic acid stain was used to visualise DNA in an iBright CL1000 gel documentation system (Thermo Fisher Scientific, United States). The percentage DNase activity of the venom samples, and that of the positive control, was calculated by densitometric analysis of the images using the ImageJ software (Schneider et al., 2012)
The fibrinogenolytic activity of N. sagittifera and N. naja venoms was elucidated using an electrophoresis-based protocol (Teng et al., 1985). A predefined concentration of venom (1.5 µg) based on protein content was incubated with human fibrinogen (15 µg) (Sigma-Aldrich, United States) in 1X PBS (pH 7.4) at 37°C for 60 min. An equal volume of the loading dye solution (1 M Tris-HCl, pH 6.8; 50% Glycerol; 0.5% Bromophenol blue; 10% SDS; and 20% β-mercaptoethanol) was added to the reaction mixture, and the resulting mixture was then heated at 70°C for 10 min. Samples were subjected to 15% SDS-PAGE, and the gel was subsequently stained with Coomassie Brilliant Blue R-250. An iBright CL1000 gel documentation system (Thermo Fisher Scientific, United States) was used to visualise the stained gels. A negative control containing human fibrinogen alone was used to compare and interpret the results.
The abilities of N. sagittifera and N. naja venoms in altering the two central blood coagulation cascades (i.e., the intrinsic and extrinsic pathway) was assessed by measuring the activated partial thromboplastin time (aPTT) and prothrombin time (PT). Whole blood from healthy human volunteers was collected in 2% Sodium citrate tubes (BD Vacutainer®) after obtaining written consent. Platelet-poor plasma (PPP) was obtained by centrifuging the whole blood at 3,000 × g for 10 min at 4°C. Reagents, specific for PT and aPTT tests [i.e., prewarmed calcium thromboplastin reagent (200 µl of Uniplastin; Tulip diagnostics, Mumbai) and activated cephaloplastin (100 µl of Liquicelin-E; Tulip diagnostics, Mumbai), along with 0.02 M calcium chloride (100 µl of CaCl2), respectively], was mixed with 50 µl PPP. This mixture was then treated with four distinct amounts of Naja venoms (5, 10, 20, and 40 µg), and the time taken for the formation of the first fibrin clot was measured on a Hemostar XF 2.0 coagulometer (Tulip Diagnostics).
The extent of snake venom-induced haemolysis was measured by assessing the destruction of human red blood cells (RBCs) using a previously described method (Maisano et al., 2013; Senji Laxme et al., 2019). By centrifugation at 3,000 × g for 10 min at 4°C, RBCs were separated from the whole blood and resuspended in 1X PBS (pH 7.4). This step was repeated five times to remove undesired clotting factors and debris. An RBC solution (1%) was then prepared and mixed with varying concentrations of N. naja and N. sagittifera venoms (5, 10, 20, and 40 µg). These mixtures were then incubated at 37°C for 24 h and followed with centrifugation at 3,000 × g for 10 min. The absorbance of the supernatant was measured at 540 nm in an Epoch 2 microplate spectrophotometer (BioTek Instruments, Inc., United States). Triton X (0.5%) was used as the positive control, and the relative haemolytic activity of venoms was calculated with respect to this control.
Interspecific venom variation was assessed by estimating the median lethal dose or LD50 of the venom (the minimum amount of venom that can kill 50% of the test population) using the WHO-recommended murine model of envenoming (WHO, 2018). Five distinct concentrations of N. naja and N. sagittifera venoms, prepared in physiological saline (0.9% NaCl), were administered intravenously into the caudal vein of male CD-1 mice (200 µl/mouse). The death and survival patterns were recorded for each venom dose group (n = 5) 24 h post venom injection. Finally, using Probit analysis, the LD50 values were calculated with 95% confidence intervals (Finney, 1971).
The in vitro venom recognition of the major Indian polyvalent and Thai N. kaouthia monovalent antivenoms were elucidated using a previously described ELISA protocol (Casewell et al., 2010; Senji Laxme et al., 2019). Venom samples (100 ng per well) were diluted in sodium carbonate-bicarbonate buffer (pH 9.6) and coated onto 96-well plates. Overnight incubation of these coated plates at 4°C was followed by washing with Tris-buffered saline and 1% Tween 20 (TBST). A blocking buffer (5% skimmed milk in TBST) was then added to the plates, and followed by incubation at room temperature (RT) for 3 hours. This was followed by another round of washing with TBST to remove the blocking solution. Various dilutions of antivenoms (Premium Serums, VINS, Bharat, Haffkine and QSMI) were added, and plates were incubated overnight at 4°C. Unbound antibodies were removed the next day by washing the plates with a TBST solution. Horseradish peroxidase (HRP)-conjugated rabbit anti-horse secondary antibody (Sigma-Aldrich, United States), diluted in PBS (1:1000), was then added to these plates, followed by an incubation period of 2 h at RT. The ABTS [2,2'-azino-bis(3-ethylbenzothiazoline-6-sulfonic acid)] substrate solution (Sigma-Aldrich, United States) was added to the plates post-incubation, and the absorbance was measured at a wavelength of 405 nm for 40 min in an Epoch 2 microplate reader. In addition, purified antibodies from naive horses (Bio-Rad Laboratories, United States) were used as a negative control.
Venom and antivenom immunoblotting experiments were performed by following a previously described protocol (Casewell et al., 2010). Firstly, crude venoms (20 µg) were subjected to SDS-PAGE in a 12.5% gel, followed by electrotransfer of the separated proteins onto a nitrocellulose membrane, as per the manufacturer’s instructions (Bio-Rad Laboratories, United States). Ponceau S reversible stain was used to verify the transfer efficacy, and the membrane was incubated overnight with a blocking buffer at 4°C. After washing with the TBST, the membrane was incubated overnight with a known concentration (1:200) of commercial antivenom at 4°C. On the following day, an HRP-conjugated rabbit anti-horse secondary antibody was added at a dilution of 1:2000, after six TBST washes to remove the unbound antivenom. Finally, following the manufacturer’s instructions (Thermo Fisher Scientific, United States), an enhanced chemiluminescence substrate was used to visualise the binding efficacy of commercial antivenoms to venom, and the membrane was imaged in an iBright CL1000 (Thermo Fisher Scientific, United States). Densitometric analyses of high- (>50 kDa), mid- (15–50 kDa) and low-molecular-weight bands (<15 kDa) in the immunoblots of all antivenoms against N. naja and N. sagittifera venoms were performed using ImageJ software (Schneider et al., 2012).
A best binding antivenom (Bharat Serums) was selected for the in vivo venom neutralisation assays based on the results of the in vitro binding experiments (ELISA and western blotting). Furthermore, to evaluate whether the increased binding efficiency under in vitro conditions translates into effective in vivo neutralisation, we also performed these experiments with the antivenom that exhibited relatively poor binding (Premium Serums). The median effective dose (ED50), or the minimum amount of antivenom required to save 50% of the test population injected with the challenge dose of venom (five times the LD50), was determined for each of these antivenoms (WHO, 2018). Briefly, four dilutions of the antivenom were prepared in the physiological saline (0.9% NaCl) and mixed with the challenge dose. These mixtures were incubated for 30 min at 37°C, and followed with intravenous administration into their respective group male CD-1 mice (n = 5). The animals were kept under observation for 24 h, and the number of dead and surviving animals were documented. When the antivenom was found to be ineffective in neutralising 5X LD50 of the snake venom (N. sagittifera: 2.375 mg/kg; N. naja: 4.2 mg/kg), the experiment was repeated with a 3X LD50 challenge dose (N. sagittifera: 1.425 mg/kg; N. naja: 2.52 mg/kg). Probit analysis was used for calculating the ED50 values with 95% confidence intervals (Finney, 1971). Neutralisation potency of the antivenoms against N. naja and N. sagittifera venoms was calculated using the formula below, where ‘n’ indicates the number of LD50 used as the challenge dose (Mendonca-da-Silva et al., 2017; Senji Laxme et al., 2019).
Antivenom potencies were also described in terms of µl of antivenom per mg of the venom (Supplementary Table S8), following a previously described method (Ainsworth et al., 2020).
Statistical differences in the results of ELISA and biochemical assays were determined using two-way ANOVA and unpaired t-tests, respectively. These comparisons were made using GraphPad Prism (GraphPad Software 9.0, San Diego, California, United States, www.graphpad.com).
Phylogenetic reconstructions of the evolutionary histories of mitochondrial markers (cyt b and ND4) provided fascinating insights into the evolution of Asiatic cobras (Figure 1 and Supplementary Figures S1–S4). The overall topology of the Naja phylogeny was in agreement with the previously reported mitochondrial species tree (Kazemi et al., 2021). Consistent with the literature, N. sagittifera was recovered as a sister lineage to N. kaouthia (Figure 1 and Supplementary Figures S1–S4) with significant node support [Bayesian Posterior Probability (BPP): 1; bootstrap (BS): 100]. However, the N. kaouthia clade was found to be polyphyletic with two distinct lineages: one consisting of N. kaouthia and N. atra from China, and the other with the Southeast Asian N. kaouthia and N. sagittifera (Figure 1 and Supplementary Figures S1–S4). The estimation of evolutionary divergences further supported these findings (Supplementary Tables S3, S4). Limited pairwise differences were observed between cyt b (2.45–2.91%) and ND4 (1.51–1.85%) sequences of N. sagittifera and N. kaouthia from Southeast Asia, while significant differences were found when N. sagittifera sequences were compared to N. kaouthia from China (6.45 and 5.21%, respectively, for cyt b and ND4).
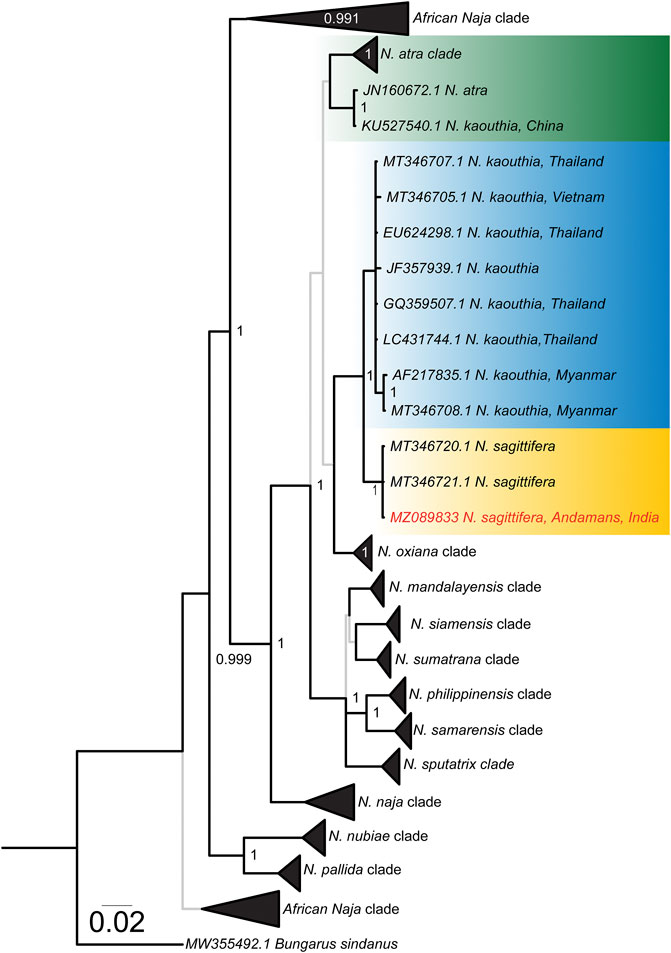
FIGURE 1. Bayesian cyt b phylogeny of Naja species. Lineages of interest have been shown in uniquely coloured boxes and the accession number of the individual sequenced in this study has been highlighted in red. Branches with superior (BPP ≥ 0.95) and relatively inferior (BPP ≤ 0.95) node support are shown in thick black and thin grey lines, respectively. Branch lengths are scaled by the number of nucleotide substitutions per site.
The SDS-PAGE electrophoretic profiles of N. naja and N. sagittifera unveiled significant differences in band patterns and intensities, highlighting the considerable extent of venom variation in these congeners (Figure 2B).
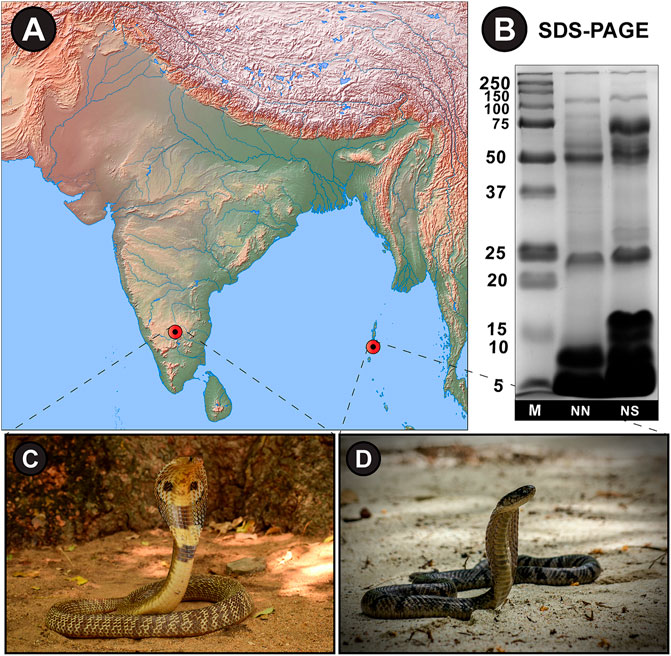
FIGURE 2. (A) Sampling locations from mainland India and Andaman and Nicobar Islands have been shown on a map of India prepared using (QGIS, 2019). (B) SDS-PAGE venom profiles and photographs of (C) N. naja and (D) N. sagittifera. M: Protein marker (units in kDa); NN: N. naja; NS: N. sagittifera.
RP-HPLC profiles revealed remarkable differences in peak areas and intensities, particularly between the retention time of 30–60 min, highlighting the significant compositional differences in the venom proteomes of these snakes (Figure 3A). Furthermore, mass spectrometric analysis of individual RP-HPLC fractions of N. naja and N. sagittifera venoms against NCBI-NR Serpentes databases (taxid: 8570) identified 101 and 103 non-redundant protein families, respectively (Figure 3B; Supplementary Tables S5, S6; Supplementary File S3). The identified toxin proteins belonged to 20 toxin families, namely three-finger toxin [3FTx; neurotoxic-3FTx (N-3FTx) and cytotoxic-3FTx (C-3FTx)], cysteine-rich secretory proteins (CRISP), disintegrin-like, vespryn, phospholipase A2 (PLA2), cobra venom factor (CVF), cystatin, vascular endothelial growth factor (VEGF), L-amino-acid oxidase (LAAO), nerve growth factor (NGF), Kunitz-type serine protease inhibitor (Kunitz), calreticulin, 5’-nucleotidase (5’-NT), natriuretic peptide (NP), C-type lectin (CTL), phosphodiesterase (PDE), hyaluronidase, phospholipase B (PLB), acetylcholinesterase (AChE) and snake venom serine protease (SVSP) (Figure 3B; Supplementary Tables S5, S6). Interestingly, cathelicidin antimicrobial peptides (de Barros et al., 2019) were discovered in N. naja venom from Karnataka, emphasising the biodiscovery potential of Indian snake venoms. PLA2 inhibitors (PLI) were also detected in both N. naja and N. sagittifera venoms, which have been theorised to play a role in preventing self-envenomation in snakes (Kochva, 1978). These anti-toxins have also been previously reported from N. naja and D. russelii venoms from various parts of India (Senji Laxme RR. et al., 2021; Senji Laxme R. R. et al., 2021).
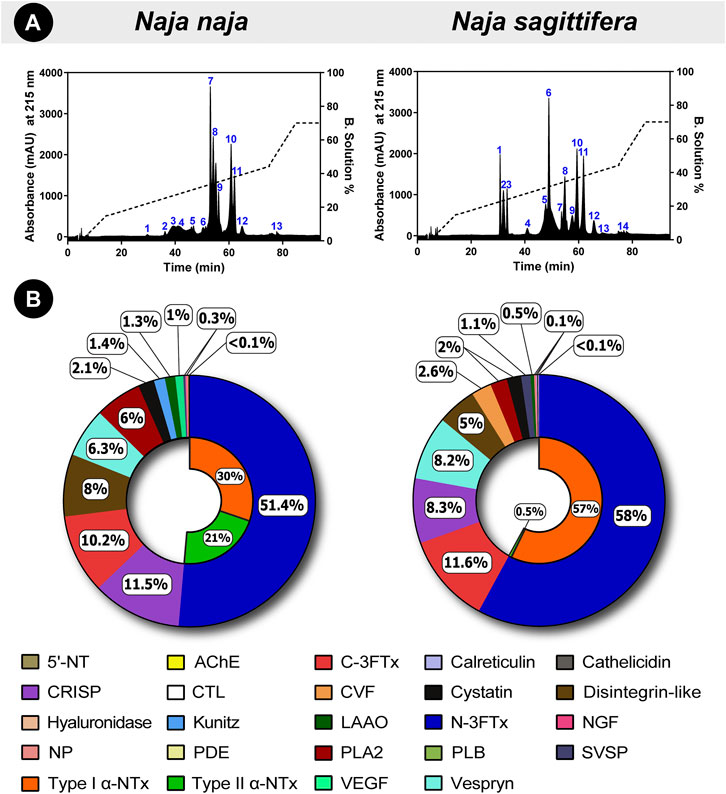
FIGURE 3. Comparative venom proteomics of N. naja and N. sagittifera. In this figure, (A) RP-HPLC profiles and (B) venom proteomes of Naja species are shown. Uniquely colour-coded individual fractions in RP-HPLC profiles are numbered from 1–14, whereas the relative abundance of each toxin family is indicated in the doughnut charts as percentages. The relative abundance of N-3FTx and C-3FTx in the two venoms has also been shown.
Further analysis of mass spectrometry data revealed that N-3FTx are the major toxic components in the venoms and they comprised 51% of N. naja and 58% of N. sagittifera protein content. However, considerable differences were noted in the relative abundance of α-neurotoxins (Figure 3B; Supplementary Tables S5, S6). Type I (or short) neurotoxins were found to be more prevalent in N. sagittifera venom (57%) than in the N. naja (30%) venom, whereas Type II (or long) neurotoxins were significantly more in N. naja venom (21%), in comparison to the trace amounts detected in N. sagittifera venom (0.5%). In contrast to the abundance of N-3FTx, C-3FTxs were nearly equally abundant in the N. sagittifera (12%) and N. naja (10%) venoms. Slight differences in the abundances of PLA2s were also observed. While 6% of the N. naja venom consisted of this toxin type, N. sagittifera venom contained only 2% (Figure 3B; Supplementary Tables S5, S6). Similarly, minor differences in the abundances of disintegrin, CRISP, vespryn, LAAO, Kunitz, SVSP and CVF were documented. Interestingly, certain toxin families, including NP, CTL, hyaluronidase, serpin, and calreticulin, were only detected in the venom of N. sagittifera (Figure 3B; Supplementary Tables S5, S6). Thus, our findings reveal that the major distinction between the proteomic composition of N. naja and N. sagittifera venom is in the relative abundance of α-neurotoxins.
PLA2s are amongst the most important and widespread snake venom superfamilies and are known to be responsible for various pharmacological manifestations in bite victims (Kini, 2003; Tasoulis and Isbister, 2017). Taking the clinical significance of this toxin into account, we assessed the PLA2 activity of N. naja and N. sagittifera venoms. Despite the relatively lower abundance of PLA2s in the venom proteome of N. sagittifera (2%) in comparison to N. naja (6%), the former exhibited slightly increased PLA2 activity (p < 0.05; Table 2; Supplementary Figure S5A).
Elapid venom proteases, such as snake venom serine proteases and disintegrin-like toxins, are known to inflict many coagulopathies, including the inhibition of platelet aggregation, and thrombin-, plasminogen-, and bradykinin-like effects (Kalogeropoulos et al., 2019; Senji Laxme R. R. et al., 2021). Therefore, we assayed the protease activities of N. naja and N. sagittifera venoms, in comparison to that of the bovine pancreatic protease (positive control). Both Naja venoms were found to exhibit limited ability in cleaving the azocasein substrate (p > 0.05; Table 2; Supplementary Figure S5B), which corresponds to the lower abundance of these proteases in the venoms of these snakes.
LAAOs in snake venoms have been shown to exhibit cytotoxic effects and induce or inhibit platelet aggregation. In the presence of LAAO, L-amino acids undergo oxidative deamination to form α-keto acids and hydrogen peroxide (H2O2). H2O2 results in the accumulation of reactive oxygen species (ROS), leading to oxidative stress and cell death (Hiu and Yap, 2020). Upon assessing the LAAO activities of Naja venoms, it was found that the venom of N. naja has significantly higher LAAO activity (p < 0.05) compared to that of its island-counterpart (Table 2; Supplementary Figure S5C), which correlates with the relative abundance of LAAOs in the two venoms.
DNases in elapid snake venoms are shown to work synergistically with other venom toxins and cause clinically severe manifestations in snakebite victims (Katkar et al., 2016). When the venoms of N. naja and N. sagittifera were tested for their ability to cleave the purified DNA from calf thymus, both venoms exhibited significant DNase activity (93.10 and 98.83%, respectively), which was comparable to that of the positive control: purified DNase I from bovine pancreas (94.11%; Table 2; Supplementary Figures S5D, S6).
Fibrinogen, also known as clotting factor I, is a soluble glycoprotein complex comprising of Aα, Bβ, and γ subunits. Upon infliction of a wound, fibrinogen undergoes enzymatic conversion in the presence of thrombin to form an insoluble fibrin clot, maintaining haemostasis (Weisel and Litvinov, 2017). Certain snake venom toxins, such as CTLs, SVSPs and snake venom metalloproteinases (SVMPs), have been documented to perturb haemostasis in bite victims by exhibiting fibrinogenolytic activity (Yamazaki and Morita, 2007). When the fibrinogenolytic activities of N. naja and N. sagittifera venoms were evaluated, both venoms were found to completely degrade the Aα subunit of the human fibrinogen, while both Bβ and γ subunits remained intact (Supplementary Figure S7).
Snake venoms are known to perturb the coagulation cascade of snakebite victims (Kumar et al., 2010; Gowtham et al., 2012; Das et al., 2013; Senji Laxme et al., 2019). Hence, we evaluated the abilities of N. naja and N. sagittifera venoms on the intrinsic and extrinsic coagulation cascades via PT and aPTT experiments, respectively. In these assays, the N. naja venom was found to affect the intrinsic coagulation pathway (aPTT) considerably and exhibited strong anticoagulatory properties (delayed coagulation by 422 s) under in vitro conditions at the highest tested concentration of the venom (40 µg; Figure 4B). Moreover, its congener from the island, N. sagittifera (40 µg), altered the intrinsic pathway in a relatively limited manner (delayed coagulation by 83 s), highlighting the significant difference between the two venoms in inflicting these effects (Figure 4B). Similarly, N. naja venom (40 µg) affected the extrinsic (PT) coagulation cascade to a greater extent and more than doubled the time required for blood coagulation (a delay of coagulation by 20 s), when compared to N. sagittifera venom (a delay of coagulation by 4 s) which had limited effect on the extrinsic cascade (Figure 4A).
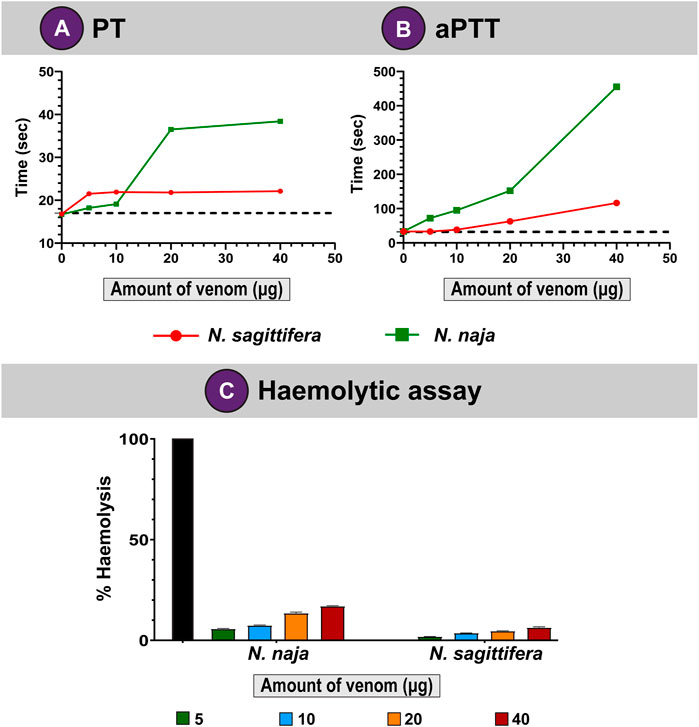
FIGURE 4. Coagulopathies induced by N. naja and N. sagittifera venoms. Effects of N. naja and N. sagittifera venoms on (A) extrinsic and (B) intrinsic pathways are depicted as line graphs. (C) Haemolytic abilities of N. naja and N. sagittifera venoms have also been shown. The horizontal dotted line in (A,B) indicate the plasma clotting time in seconds for the control sample.
Snake venom toxins such as PLA2s have been shown to hydrolyse the phospholipid membranes of red blood cells (RBCs) and disrupt haemostasis (Stoykova et al., 2013). When we evaluated the haemolytic potential of N. naja and N. sagittifera venoms on human RBCs and compared these with the Triton X-100 positive control, the N. naja venom (40 µg) showed appreciable haemolytic effects (17%). In contrast, N. sagittifera venom (40 µg) was found to exhibit limited haemolytic activity (7%) (Figure 4C). These results are consistent with the relative abundance of venom PLA2s in the two snake species (Figure 3B; Supplementary Tables S5, S6).
We evaluated the abilities of commercial Indian polyvalent antivenoms in recognising N. naja and N. sagittifera venoms using indirect ELISA assays. In these experiments, a fixed amount of the venom (100 ng) was incubated with varying concentrations of commercial antivenoms, and the absorbance, which corresponds to the venom binding capabilities of antivenoms, was measured at 405 nm. While all tested antivenoms exhibited a titre of 1:2500, Bharat Serums was found to be the best binding antivenom against both N. naja and N. sagittifera venoms (Figure 5). Bharat Serums exhibited between 1.4 to nearly two times better binding against the N. sagittifera venom and between 1.2 and 1.75 times better binding against the N. naja venom when compared to its Indian counterparts. Bharat Serums was closely followed by VINS and Premium Serums antivenoms, whereas the Haffkine antivenom exhibited the worst venom recognition capability. Considering their close phylogenetic relationship with N. kaouthia snakes, we also evaluated the cross-neutralising potential of Thai monovalent N. kaouthia antivenom against N. sagittifera venoms from the Andaman and Nicobar Islands. Similar to the Bharat Serums polyvalent antivenom, Thai monovalent N. kaouthia antivenom was very effective in recognising the N. sagittifera venom (Figure 5).
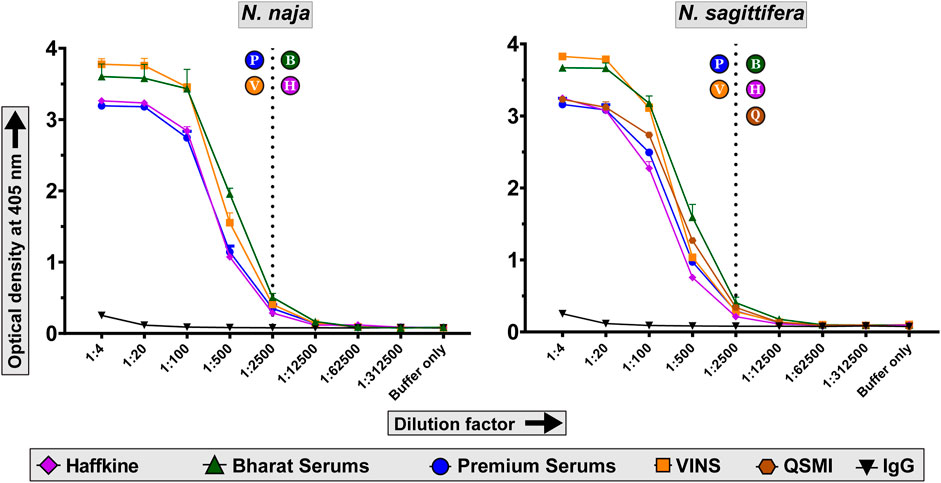
FIGURE 5. Venom recognition capabilities of commercial antivenoms. The line graphs in this figure indicate the in vitro venom binding of naive horse IgG and commercial Indian polyvalent and Thai monovalent antivenoms against N. naja and N. sagittifera. Multiple dilutions of antivenoms were evaluated in these indirect ELISA experiments. Dotted lines indicate the titre value of antivenoms against the two venoms. The alphabets in circles next to the dotted lines indicate the respective titres of antivenoms: P—Premium Serums and Vaccines Pvt. Ltd.; V—VINS Bioproducts Ltd.; B—Bharat Serums and Vaccines Ltd.; H—Haffkine Bio-Pharmaceutical Corporation Ltd. and Q—Queen Saovabha Memorial Institute monovalent N. kaouthia antivenom.
Moreover, when the binding efficacy of antivenoms to venom toxins was assessed using western blotting experiments, it was found that the Bharat Serums antivenom bound to many low-, mid-, and high-molecular-weight toxins in the venoms of N. naja and N. sagittifera (Supplementary Figures S8, S9). Further, Thai QSMI monovalent antivenom notably exhibited high binding to low-molecular-weight toxins (<15 kDa) in the N. sagittifera venom (Supplementary Figures S8, S9). In contrast, the antivenom manufactured by Premium Serums exhibited poor immunorecognition against both the Naja venoms, predominantly binding to high-molecular-weight toxins (>50 kDa), while poorly recognising their low-molecular-weight counterparts (Supplementary Figures S8, S9). Therefore, both in vitro binding experiments identified Bharat Serum and Premium Serums as the best- and worst-binding Indian antivenoms, respectively. Furthermore, similarly to the Bharat Serums antivenom, Thai QSMI monovalent antivenom exhibited increased in vitro binding to N. sagittifera venom (Supplementary Figures S8, S9).
The venom potencies of the endemic Andaman cobra and its congener from mainland India were evaluated in a murine model of envenoming using WHO-recommended protocols. The N. sagittifera venom (0.475 mg/kg) was found to be nearly two times as potent as the N. naja venom (0.84 mg/kg) from mainland India (Figure 6A; Supplementary Table S7). Although N-3FTxs dominated the venoms of both species (>50%), the significant difference in venom toxicities could be attributed to the differences in the abundance of Type I and Type II α-neurotoxins and variations in their sequence compositions. Type I α-neurotoxins were significantly more abundant in N. sagittifera venom (57%) than N. naja venom (30%).
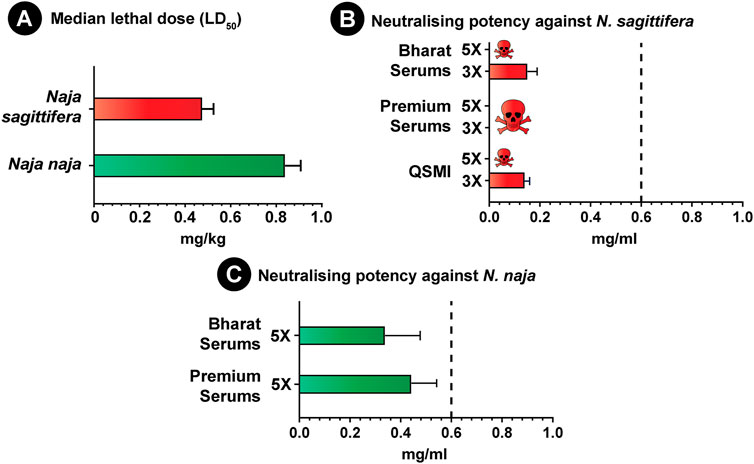
FIGURE 6. Venom toxicities of N. naja and N. sagittifera and the neutralisation potential of commercial antivenoms. (A) in this figure depicts the venom potencies (mg/kg) of N. naja and N. sagittifera, while the neutralising potencies (mg/ml) of Indian polyvalent (Premium Serums and Bharat Serums) and Thai monovalent antivenoms against the Naja venoms (3X and 5X LD50 challenge dose) are shown in (B,C). The dotted vertical line in (B,C) represents the marketed claim of neutralisation (0.60 mg/ml) against N. naja and N. kaouthia for the Indian polyvalent and Thai monovalent antivenoms, whereas the error bars represent 95% confidence intervals.
Given the ethical considerations in animal usage, and based on the results of the in vitro binding studies (ELISA and western blotting), a single best binding polyvalent antivenom (Bharat Serums) was selected for the in vivo preclinical assessment. However, to evaluate whether the in vitro binding potential of antivenoms corresponds to their in vivo venom neutralisation efficacy, we also estimated the neutralisation potency of the relatively poor binding Premium Serums antivenom against the venoms of both Naja species. The outcomes of these experiments revealed the inadequacy of both the best and worst binding Indian polyvalent antivenoms in neutralising the N. sagittifera venom, indicating that the in vitro binding does not necessarily correlate with in vivo venom neutralisation (Figure 6B; Supplementary Table S8). Furthermore, as test animals rapidly (∼5 min) succumbed to the fatal effects of the venom in experiments with 5X LD50 of the challenge dose, neutralisation experiments were repeated by lowering the challenge dose to 3X LD50. In these experiments, Bharat Serums antivenom exhibited a meagre 0.151 mg/ml potency (Figure 6B; Supplementary Table S8), while Premium Serums was again completely ineffective in saving mice from the lethal effects of N. sagittifera envenoming. These results are suggestive of the preclinical ineffectiveness of Indian antivenoms in treating bites from the Andaman cobra.
As N. sagittifera is closely related to N. kaouthia from Southeast Asia, we further explored the cross-neutralisation effectiveness of Thai N. kaouthia monovalent antivenom manufactured by QSMI in treating N. sagittifera bites. Similar to its Indian polyvalent counterparts, Thai monovalent antivenom could not protect the experimental animals injected with 5X LD50 of venom. However, when the challenge dose was reduced to 3X LD50, this antivenom exhibited a very poor neutralisation potential (0.140 mg/ml; Figure 6B; Supplementary Table S8), highlighting the negative impact of geographical and phylogenetic divergence between the two species on antivenom’s effectiveness.
Both Indian antivenoms were also severely limited in their ability to neutralise the venom of N. naja from mainland India, as they were found to exhibit a neutralisation potential (Premium Serums: 0.442 mg/ml; Bharat Serums: 0.338 mg/ml; Figure 6C; Supplementary Table S8) that was significantly lower than the marketed value (0.6 mg/ml) when tested at 5X LD50 challenge dose. These results are congruent with a previous study, wherein the Indian polyvalent antivenom showed poor neutralising efficacy against N. naja venoms from various biogeographical zones of the Indian subcontinent (Senji Laxme R. R. et al., 2021).
The Andaman and Nicobar Islands, which extend over an area of 800 km2, are separated from Southeast Asia and mainland India by the Andaman Sea and Bay of Bengal, respectively. The archipelago, comprising 572 islands, was once a part of the Asian mainland that later got detached due to geological events during the Upper Mesozoic and became submerged underwater [100 million years ago (MYA)] (Sivaperuman and Deepak, 2013). The archipelago finally re-emerged in the Late Miocene (10 MYA) (Lee and Lawver, 1995). Interestingly, the Andaman and Nicobar Islands were never a contiguous landmass and are geographically separated by a narrow water canal: the Ten-Degree (10°) channel (Ripley and Beehler, 1989). These islands are a part of two biodiversity hotspots: the Indo-Burma region consisting of the Andaman Islands, and Sundaland comprising the Nicobar Islands (Myers et al., 2000). It is believed that before its separation from the Asian mainland, the contiguous landmass of Andaman and Nicobar Islands may have had an influx of organisms from the mainland (Ripley and Beehler, 1989). This proposition is further corroborated by the phylogenetic affinity of Andaman and Nicobar herpetofauna with their counterparts in the Indo-China region (Das, 1999). However, as a consequence of geographic distance and saltwater barrier, species found on these islands experienced reduced gene flow that resulted in highly divergent populations/species (MacArthur and Wilson, 2001).
The Andaman and Nicobar Islands are an abode to a vast floral and faunal biodiversity due to the conducive tropical climatic conditions. In addition to mammalian and marine fauna, the archipelago harbours diverse herpetofauna with over 64 reptilian species, including 31 endemics (Tikader, 1992; Srivastava and Ambast, 2009). These include several species of snakes, such as Anderson’s pit viper (T. andersonii), Andaman krait (B. andamanensis) and Andaman cobra (N. sagittifera), that can potentially inflict medically significant envenomation in humans (Smith, 1941). Among these, the Andaman cobra probably causes a large number of snakebites, primarily due to its occurrence in the vicinity of human settlements and agricultural cultivations. Despite their clinical importance, N. sagittifera has received limited research attention. Though initially considered a conspecific of the monocled cobra (N. kaouthia), these snakes were elevated to species rank based on molecular phylogenetics (Wüster et al., 1995). Consistent with the literature (Kazemi et al., 2021), mitochondrial phylogeny of the genus Naja recovered N. sagittifera as a sister lineage to one of the branches of the polyphyletic N. kaouthia clades (Figure 1).
As a consequence of local adaptation to shifts in ecology and environment, stark inter and intraspecific differences in venom composition and toxicity profiles have been observed in snakes (Daltry et al., 1996; Sunagar et al., 2014; Rokyta et al., 2017; Casewell et al., 2020; Senji Laxme RR. et al., 2021; Senji Laxme R. R. et al., 2021), even within a restricted geographic locale (Rashmi et al., 2021). Consistently, the characterisation of venom proteomes in this study revealed considerable differences in the venoms of N. naja and N. sagittifera. While RP-HPLC profiling unravelled distinct differences in intensities and areas of peaks, tandem mass spectrometry of these fractions revealed the domination of N-3FTxs (>50%) in the venoms of both Naja species. However, a prominent difference in the relative abundance of Type I and Type II α-neurotoxins was noted. Perhaps, this variation in the relative abundance of α-neurotoxins, as well as sequence variations in venom toxins, underpins the increased toxicity of N. sagittifera venom to mice (0.475 mg/kg) when compared to N. naja (0.84 mg/kg).
Moreover, as snake venoms are complex concoctions of various macromolecules, we also subjected these venoms to in vitro biochemical and pharmacological assays, including PLA2, protease, LAAO, fibrinogenolytic, blood coagulation, and haemolytic assays. Considerable differences in PLA2 (Supplementary Figure S5A), LAAO (Supplementary Figure S5C), PT and aPTT (Figure 4) activities of N. naja and N. sagittifera venoms were noted. Additional experiments are necessary to elucidate the biological and clinical relevance of these differences in activities. However, the significant variations documented in proteomic composition, toxicity profile and functional activity between the mainland N. naja and island-endemic N. sagittifera venoms are very likely to result in varied clinical manifestations and therapeutic implications in snakebite victims.
In vitro and in vivo experiments in this study revealed alarming preclinical deficiencies of the Indian polyvalent “big four” and Thai monovalent N. kaouthia antivenoms in neutralising the venoms of N. naja and N. sagittifera. Indirect ELISA and western blotting experiments identified Bharat Serums antivenom as the best binding Indian antivenom against both N. naja and N. sagittifera venoms, while VINS and Haffkine antivenoms showed intermediary in vitro venom recognition. In contrast, the Premium Serums antivenom exhibited the worst venom binding against both Naja venoms. In contrast, Thai N. kaouthia monovalent antivenom was also found to exhibit significant immunological cross-reactivity against the N. sagittifera venom.
Following the tenets of the three R’s in animal ethics (reduction, replacement and refinement), we utilised the results of the in vitro binding experiments to downselect the best binding antivenom for the in vivo preclinical assessment in mice. A statistically significant correlation between the in vitro binding effectiveness of antivenoms and their in vivo venom neutralising potency has been shown in the past (Theakston and Reid, 1979; Rungsiwongse and Ratanabanangkoon, 1991). To test whether this correlation holds in the case of the Naja species, we evaluated the neutralising efficacies of antivenoms that exhibited the best (Bharat Serum) and worst (Premium Serums) binding in ELISA and western blotting experiments. While Bharat Serum exhibited significantly better binding efficacy in our in vitro assays compared to its Indian antivenom counterparts, its preclinical neutralisation in the murine model was extremely poor against both N. sagittifera (0.151 mg/ml) and N. naja (0.338 mg/ml) and was on par with the neutralising potency of the worst binding Premium Serums antivenom (0.442 mg/ml against N. naja; 0 mg/ml against N. sagittifera). The plausible explanation for this discrepancy could be the presence of a higher abundance of antibodies against medically unimportant toxins or non-toxic proteins in snake venoms that do not inflict clinical manifestations in envenomed victims (Casewell et al., 2010). This could also be explained by the presence of antibodies against regions in toxins that are not involved in binding interactions with the receptor.
Due to the presence of many shared toxin families across snakes, antivenoms may exhibit cross-neutralisation against non-target species (de Roodt et al., 1998; Tan et al., 2015; Whiteley et al., 2019; Casewell et al., 2020; Mora-Obando et al., 2021). This cross-neutralisation potential of antivenoms has been previously exploited for the treatment of snakebites. For example, Thai monovalent N. kaouthia antivenom, and Thai neuro polyvalent snake antivenom manufactured using the venoms of N. kaouthia, O. hannah, Malayan krait (B. candidus) and banded krait (B. fasciatus), have been shown to cross-neutralise the Malaysian sea snake (Hydrophis spp.) venoms (Tan et al., 2015). Similarly, commercial polyvalent antivenoms raised against the Indian “big four” snake species have been used to treat snakebites in regions where these snakes are absent. Until recently, antivenoms marketed by VINS BioProducts for Central Africa were formulated by hyperimmunising equines with the venoms of Indian Russell’s (D. russelii) and saw-scaled (E. carinatus) vipers. Other antivenom manufacturers have also adopted similar practices. For example, Premium Serums continues to market Indian “big four” antivenoms to treat hump-nosed pit viper (Hypnale hypnale) bites in Sri Lanka.
Considering the preclinical ineffectiveness of the “big four” Indian polyvalent antivenoms and the close phylogenetic relationship between N. kaouthia and N. sagittifera, we evaluated the potential use of N. kaouthia specific Thai monovalent antivenoms for the treatment of N. sagittifera envenomation in the Andaman and Nicobar Islands. Unfortunately, although in vitro experiments revealed that the QSMI monovalent antivenom binds effectively to N. sagittifera venom, our subsequent in vivo assessments revealed its limited neutralisation potential (0.14 mg/ml) against the N. sagittifera venom, falling well short of the marketed claim of neutralisation of 0.60 mg/ml against the Southeast Asian N. kaouthia. This disconcerting lack of neutralising efficacy exhibited by the Indian polyvalent and Thai monovalent antivenoms against N. sagittifera further highlights the pressing need to develop regional antivenoms tailored for the treatment of snakebites on the Andaman and Nicobar Islands.
In conclusion, comparative proteomics, in vitro biochemical and pharmacological characterisation, and in vivo toxicity assessments revealed significant differences in the venoms of N. naja from mainland India (Karnataka) and N. sagittifera from Andaman and Nicobar Islands. When the Indian polyvalent antivenoms manufactured using the venoms of the “big four” snakes were tested for their in vitro immunorecognition capabilities, Bharat Serums and Premium Serums antivenoms were identified as the best- and worst-performing, respectively. Thai monovalent antivenom manufactured by QSMI using the phylogenetically relevant Southeast Asian N. kaouthia venoms too exhibited increased immunorecognition towards the N. sagittifera venom. Furthermore, in vivo venom neutralisation experiments in the murine model revealed that both the Indian polyvalent (Bharat Serums and Premium Serums) and Thai monovalent (QSMI) antivenoms are inefficacious in neutralising the lethal effects of N. sagittifera from Andaman and Nicobar Islands. Unfortunately, the Indian antivenoms were also found to lack desirable potency against the venoms of the Southern Indian population of N. naja from Karnataka. It should be noted that extensive sampling efforts across islands from individuals of various age groups of N. sagittifera are needed to reveal the extent of intraspecific and ontogenetic venom variation in this species, and its consequent impact on the effectiveness of snakebite therapy in Andaman and Nicobar Islands.
Overall, our results indicate the importance of preclinical assays in evaluating the efficacies of antivenoms and suggest the need to identify key toxins that are medically important. Immunisation of animals with the medically important toxins or toxin fractions, instead of the crude venom, is not only anticipated to reduce the abundance of therapeutically irrelevant antibodies in the finished product, but will also reduce the number of vials required to effect cure by enhancing antivenom’s dose effectiveness. Furthermore, in line with our previous findings (Senji Laxme RR. et al., 2021; Senji Laxme R. R. et al., 2021), the results of this study highlight the significance of phylogenetic and biogeographic considerations for the manufacture of commercial antivenoms in the Indian subcontinent. As our in vivo venom toxicity and neutralisation experiments revealed that N. sagittifera is more toxic than N. naja and tested antivenoms failed to reverse lethality of the venoms, it demonstrates the necessity to investigate the venoms of the medically important yet neglected species of Indian snakes [a.k.a. the “neglected many”; (Senji Laxme et al., 2019)]. Finally, while the preclinical effectiveness of the “big four” antivenom in treating bites of the other medically important snakes on the Andaman and Nicobar Islands is yet to be evaluated, our results demonstrate the need for a N. sagittifera specific antivenom product on these islands.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
The studies involving human participants were reviewed and approved by the Institutional Human Ethical Committee, Indian Institute of Science (IISc), Bangalore. The patients/participants provided their written informed consent to participate in this study. The animal study was reviewed and approved by the Institutional Animal Ethics Committee (IAEC), Indian Institute of Science (IISc), Bangalore.
Conceptualisation: KS; Formal analysis: SA, SK, PD, AI, and KS; Funding acquisition: KS; Investigation: SA, SK, PD, AI, GM, and KS; Supervision: KS; Visualisation: SA and KS; original draft: SA, AI, PD, SK, and KS; Review and editing: KS.
This work was supported by the DBT/Wellcome Trust India Alliance Fellowship (grant number IA/I/19/2/504647) awarded to KS. This research was also supported by DST-FIST (SR/FST/LS-II/2018/233). PD was supported by the Ministry of Human Resource Development (MHRD) research fellowship.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The authors are thankful to the Forest Departments of Karnataka and Andaman and Nicobar Islands for the sampling permits and the logistic support during venom collection. In addition, the authors are thankful to Vivek Suranse (IISc), Ajay Kartik (MCBT), Lohith Kumar (Liana Trust), Manjeet Singh (Andaman Forest Department), Aaron Fernandes, Anil Khaire (Indian Herpetological Society) and Aditya Maladi (IISc) for assistance with sampling. The authors are also thankful to Ajay Kartik (N. naja) and Aaron Fernandes (N. sagittifera) for contributing snake photographs in Figure 2.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fphar.2021.768210/full#supplementary-material
Ainsworth, S., Menzies, S. K., Casewell, N. R., and Harrison, R. A. (2020). An Analysis of Preclinical Efficacy Testing of Antivenoms for Sub-saharan Africa: Inadequate Independent Scrutiny and Poor-Quality Reporting Are Barriers to Improving Snakebite Treatment and Management. Plos Negl. Trop. Dis. 14 (8), e0008579. doi:10.1371/journal.pntd.0008579
Altekar, G., Dwarkadas, S., Huelsenbeck, J. P., and Ronquist, F. (2004). Parallel metropolis Coupled Markov Chain Monte Carlo for Bayesian Phylogenetic Inference. Bioinformatics 20 (3), 407–415. doi:10.1093/bioinformatics/btg427
Arevalo, E., Davis, S. K., and Sites, J. W. (1994). Mitochondrial DNA Sequence Divergence and Phylogenetic Relationships Among Eight Chromosome Races of the Sceloporus Grammicus Complex (Phrynosomatidae) in central Mexico. Syst. Biol. 43 (3), 387–418. doi:10.1093/sysbio/43.3.387
Bradford, M. M. (1976). A Rapid and Sensitive Method for the Quantitation of Microgram Quantities of Protein Utilizing the Principle of Protein-Dye Binding. Anal. Biochem. 72, 248–254. doi:10.1006/abio.1976.9999
Casewell, N. R., Cook, D. A., Wagstaff, S. C., Nasidi, A., Durfa, N., Wüster, W., et al. (2010). Pre-clinical Assays Predict Pan-African Echis viper Efficacy for a Species-specific Antivenom. Plos Negl. Trop. Dis. 4 (10), e851. doi:10.1371/journal.pntd.0000851
Casewell, N. R., Jackson, T. N. W., Laustsen, A. H., and Sunagar, K. (2020). Causes and Consequences of Snake Venom Variation. Trends Pharmacol. Sci. 41 (8), 570–581. doi:10.1016/j.tips.2020.05.006
Chowdhury, M. A., Miyoshi, S., and Shinoda, S. (1990). Purification and Characterization of a Protease Produced by Vibrio Mimicus. Infect. Immun. 58 (12), 4159–4162. doi:10.1128/IAI.58.12.4159-4162.1990
Daltry, J. C., Wüster, W., and Thorpe, R. S. (1996). Diet and Snake Venom Evolution. Nature 379 (6565), 537–540. doi:10.1038/379537a0
Das, D., Urs, N., Hiremath, V., Vishwanath, B. S., and Doley, R. (2013). Biochemical and Biological Characterization of Naja Kaouthia Venom from North-East India and its Neutralization by Polyvalent Antivenom. J. Venom Res. 4, 31–38.
Das, I. (1999). “Biogeography of the Amphibians and Reptiles of the Andaman and Nicobar Islands, India,” in Tropical Island Herpetofauna. Origin, Current Diversity and Current Status (New York, United States: Elsevier), 43–77.
de Barros, E., Gonçalves, R. M., Cardoso, M. H., Santos, N. C., Franco, O. L., and Cândido, E. S. (2019). Snake Venom Cathelicidins as Natural Antimicrobial Peptides. Front. Pharmacol. 10, 1415. doi:10.3389/fphar.2019.01415
de Roodt, A. R., Dolab, J. A., Fernández, T., Segre, L., and Hajos, S. E. (1998). Cross-reactivity and Heterologous Neutralization of Crotaline Antivenoms Used in Argentina. Toxicon 36 (7), 1025–1038. doi:10.1016/s0041-0101(97)00111-6
Gerceker, D., Karasartova, D., Elyürek, E., Barkar, S., Kiyan, M., Ozsan, T. M., et al. (2009). A New, Simple, Rapid Test for Detection of DNase Activity of Microorganisms: DNase Tube Test. J. Gen. Appl. Microbiol. 55 (4), 291–294. doi:10.2323/jgam.55.291
Gowtham, Y. J., Kumar, M. S., Girish, K. S., and Kemparaju, K. (2012). Hemostatic Interference of Indian king Cobra (Ophiophagus Hannah) Venom. Comparison with Three Other Snake Venoms of the Subcontinent. Biochemistry (Mosc) 77 (6), 639–647. doi:10.1134/S0006297912060119
Guindon, S., Dufayard, J. F., Lefort, V., Anisimova, M., Hordijk, W., and Gascuel, O. (2010). New Algorithms and Methods to Estimate Maximum-Likelihood Phylogenies: Assessing the Performance of PhyML 3.0. Syst. Biol. 59 (3), 307–321. doi:10.1093/sysbio/syq010
Harvey, M. B., Barker, D. G., Ammerman, L. K., and Chippindale, P. T. (2000). Systematics of Pythons of the Morelia Amethistina Complex (Serpentes: Boidae) with the Description of Three New Species. Herpetol. Monogr. 14, 139–185. doi:10.2307/1467047
Hiu, J. J., and Yap, M. K. K. (2020). Cytotoxicity of Snake Venom Enzymatic Toxins: Phospholipase A2 and L-Amino Acid Oxidase. Biochem. Soc. Trans. 48 (2), 719–731. doi:10.1042/BST20200110
Joubert, F. J., and Taljaard, N. (1980). Purification, Some Properties and Amino-Acid Sequences of Two Phospholipases A (CM-II and CM-III) from Naja naja Kaouthia Venom. Eur. J. Biochem. 112 (3), 493–499. doi:10.1111/j.1432-1033.1980.tb06112.x
Kalogeropoulos, K., Treschow, A., auf dem Keller, U., Escalante, T., Rucavado, A., Gutiérrez, J., et al. (2019). Protease Activity Profiling of Snake Venoms Using High-Throughput Peptide Screening. Toxins 11 (3), 170. doi:10.3390/toxins11030170
Katkar, G. D., Sundaram, M. S., NaveenKumar, S. K., Swethakumar, B., Sharma, R. D., Paul, M., et al. (2016). NETosis and Lack of DNase Activity Are Key Factors in Echis carinatus Venom-Induced Tissue Destruction. Nat. Commun. 7, 11361. doi:10.1038/ncomms11361
Kazemi, E., Nazarizadeh, M., Fatemizadeh, F., Khani, A., and Kaboli, M. (2021). The Phylogeny, Phylogeography, and Diversification History of the Westernmost Asian Cobra (Serpentes: Elapidae: Naja Oxiana) in the Trans-caspian Region. Ecol. Evol. 11 (5), 2024–2039. doi:10.1002/ece3.7144
Kini, R. M. (2003). Excitement Ahead: Structure, Function and Mechanism of Snake Venom Phospholipase A2 Enzymes. Toxicon 42 (8), 827–840. doi:10.1016/j.toxicon.2003.11.002
Kishimoto, M., and Takahashi, T. (2001). A Spectrophotometric Microplate Assay for L-Amino Acid Oxidase. Anal. Biochem. 298 (1), 136–139. doi:10.1006/abio.2001.5381
Kochva, E. (1978). Evolution and Secretion of Venom and its Antidotes in Snakes. New York, United States: Academic Press.
Kumar, M. S., Devaraj, V. R., Vishwanath, B. S., and Kemparaju, K. (2010). Anti-coagulant Activity of a Metalloprotease: Further Characterization from the Indian Cobra (Naja naja) Venom. J. Thromb. Thrombolysis 29 (3), 340–348. doi:10.1007/s11239-009-0379-2
Kumar, S., Stecher, G., Li, M., Knyaz, C., and Tamura, K. (2018). MEGA X: Molecular Evolutionary Genetics Analysis across Computing Platforms. Mol. Biol. Evol. 35 (6), 1547–1549. doi:10.1093/molbev/msy096
Lee, T.-Y., and Lawver, L. A. (1995). Cenozoic Plate Reconstruction of Southeast Asia. Tectonophysics 251 (1-4), 85–138. doi:10.1016/0040-1951(95)00023-2
Lefort, V., Longueville, J. E., and Gascuel, O. (2017). SMS: Smart Model Selection in PhyML. Mol. Biol. Evol. 34 (9), 2422–2424. doi:10.1093/molbev/msx149
Lomonte, B., and Calvete, J. J. (2017). Strategies in 'snake Venomics' Aiming at an Integrative View of Compositional, Functional, and Immunological Characteristics of Venoms. J. Venom Anim. Toxins Incl Trop. Dis. 23, 26. doi:10.1186/s40409-017-0117-8
MacArthur, R. H., and Wilson, E. O. (2001). The Theory of Island Biogeography. Princeton, United States: Princeton university press.
Maisano, M., Trapani, M. R., Parrino, V., Parisi, M. G., Cappello, T., D'Agata, A., et al. (2013). Haemolytic Activity and Characterization of Nematocyst Venom fromPelagia noctiluca(Cnidaria: Scyphozoa). Ital. J. Zoolog. 80 (2), 168–176. doi:10.1080/11250003.2012.758782
Marinetti, G. V. (1965). The Action of Phospholipase A on Lipoproteins. Biochim. Biophys. Acta 98 (3), 554–565. doi:10.1016/0005-2760(65)90152-9
Mendonça-da-Silva, I., Magela Tavares, A., Sachett, J., Sardinha, J. F., Zaparolli, L., Gomes Santos, M. F., et al. (2017). Safety and Efficacy of a Freeze-Dried Trivalent Antivenom for Snakebites in the Brazilian Amazon: An Open Randomized Controlled Phase IIb Clinical Trial. Plos Negl. Trop. Dis. 11 (11), e0006068. doi:10.1371/journal.pntd.0006068
Mora-Obando, D., Pla, D., Lomonte, B., Guerrero-Vargas, J. A., Ayerbe, S., and Calvete, J. J. (2021). Antivenomics and In Vivo Preclinical Efficacy of Six Latin American Antivenoms towards South-Western Colombian Bothrops asper Lineage Venoms. Plos Negl. Trop. Dis. 15 (2), e0009073. doi:10.1371/journal.pntd.0009073
Myers, N., Mittermeier, R. A., Mittermeier, C. G., Da Fonseca, G. A., and Kent, J. (2000). Biodiversity Hotspots for Conservation Priorities. Nature 403 (6772), 853–858. doi:10.1038/35002501
Palumbi, S. R. (1996). “Nucleic Acids II: The Polymerase Chain Reaction,” in Molecular Systematics (Sunderland, MA, United States: Sinauer Ass..), 205–247.
Perez-Riverol, Y., Csordas, A., Bai, J., Bernal-Llinares, M., Hewapathirana, S., Kundu, D. J., et al. (2019). The PRIDE Database and Related Tools and Resources in 2019: Improving Support for Quantification Data. Nucleic Acids Res. 47 (D1), D442–D450. doi:10.1093/nar/gky1106
QGIS (2019). Q.D.T. "QGIS Geographic Information System. Open Source Geospatial Foundation Project. Available at: http://qgis.osgeo.org (Accessed June 10, 2021).
Rashmi, U., Khochare, S., Attarde, S., Laxme, R. S., Suranse, V., Martin, G., et al. (2021). Remarkable Intrapopulation Venom Variability in the Monocellate Cobra (Naja Kaouthia) Unveils Neglected Aspects of India's Snakebite Problem. J. Proteomics 242, 104256. doi:10.1016/j.jprot.2021.104256
Ripley, S. D., and Beehler, B. M. (1989). Ornithogeographic Affinities of the Andaman and Nicobar Islands. J. Biogeogr. 16, 323–332. doi:10.2307/2845224
Rokyta, D. R., Margres, M. J., Ward, M. J., and Sanchez, E. E. (2017). The Genetics of Venom Ontogeny in the Eastern Diamondback Rattlesnake (Crotalus adamanteus). PeerJ 5, e3249. doi:10.7717/peerj.3249
Ronquist, F., Teslenko, M., Van Der Mark, P., Ayres, D. L., Darling, A., Höhna, S., et al. (2012). MrBayes 3.2: Efficient Bayesian Phylogenetic Inference and Model Choice across a Large Model Space. Syst. Biol. 61 (3), 539–542. doi:10.1093/sysbio/sys029
Rungsiwongse, J., and Ratanabanangkoon, K. (1991). Development of an ELISA to Assess the Potency of Horse Therapeutic Antivenom against Thai Cobra Venom. J. Immunol. Methods 136 (1), 37–43. doi:10.1016/0022-1759(91)90247-d
Salve, P. S., Vatavati, S., and Hallad, J. (2020). Clustering the Envenoming of Snakebite in India: The District Level Analysis Using Health Management Information System Data. Clin. Epidemiol. Glob. Health 8 (3), 733–738. doi:10.1016/j.cegh.2020.01.011
Schneider, C. A., Rasband, W. S., and Eliceiri, K. W. (2012). NIH Image to ImageJ: 25 Years of Image Analysis. Nat. Methods 9 (7), 671–675. doi:10.1038/nmeth.2089
Senji Laxme, R. R., Khochare, S., de Souza, H. F., Ahuja, B., Suranse, V., Martin, G., et al. (2019). Beyond the 'big Four': Venom Profiling of the Medically Important yet Neglected Indian Snakes Reveals Disturbing Antivenom Deficiencies. Plos Negl. Trop. Dis. 13 (12), e0007899. doi:10.1371/journal.pntd.0007899
Senji Laxme, R. R., Khochare, S., Attarde, S., Suranse, V., Iyer, A., Casewell, N. R., et al. (2021a). Biogeographic Venom Variation in Russell's viper (Daboia russelii) and the Preclinical Inefficacy of Antivenom Therapy in Snakebite Hotspots. Plos Negl. Trop. Dis. 15 (3), e0009247. doi:10.1371/journal.pntd.0009247
Senji Laxme, R. R., Attarde, S., Khochare, S., Suranse, V., Martin, G., Casewell, N. R., et al. (2021b). Biogeographical Venom Variation in the Indian Spectacled Cobra (Naja naja) Underscores the Pressing Need for Pan-India Efficacious Snakebite Therapy. Plos Negl. Trop. Dis. 15 (2), e0009150. doi:10.1371/journal.pntd.0009150
Sievers, F., Wilm, A., Dineen, D., Gibson, T. J., Karplus, K., Li, W., et al. (2011). Fast, Scalable Generation of High-Quality Protein Multiple Sequence Alignments Using Clustal Omega. Mol. Syst. Biol. 7 (1), 539. doi:10.1038/msb.2011.75
Sivaperuman, C., and Deepak, R. (2013). Diversity and Distribution of Amphibians and Reptiles in the Ritchie’s Archipelago, Andaman and Nicobar Islands. J. Andaman Sci. Assoc. 18 (1), 32–38.
Smith, M. A. (1941). “The Herpetology of the Andaman and Nicobar Islands,” in Proceedings of the Linnean Society of London, November, 1941 (London: Wiley Online Library), 150–158.
Smith, B. J. (1984). SDS Polyacrylamide Gel Electrophoresis of Proteins. Methods Mol. Biol. 1, 41–55. doi:10.1385/0-89603-062-8:41
Srivastava, R. C., and Ambast, S. K. (2009). Water Policy for Andaman & Nicobar Islands: A Scientific Perspective. Port Blair: CARI, 1–18.
Stoykova, S., Goranova, Y., Pantcheva, I., Atanasov, V., Danchev, D., and Petrova, S. (2013). Hemolytic Activity and Platelet Aggregation Inhibitory Effect of Vipoxin's Basic sPLA2 Subunit. Interdiscip. Toxicol. 6 (3), 136–140. doi:10.2478/intox-2013-0021
Sunagar, K., Undheim, E. A., Scheib, H., Gren, E. C., Cochran, C., Person, C. E., et al. (2014). Intraspecific Venom Variation in the Medically Significant Southern Pacific Rattlesnake (Crotalus Oreganus Helleri): Biodiscovery, Clinical and Evolutionary Implications. J. Proteomics 99, 68–83. doi:10.1016/j.jprot.2014.01.013
Tan, C. H., Tan, N. H., Tan, K. Y., and Kwong, K. O. (2015). Antivenom Cross-Neutralization of the Venoms of Hydrophis Schistosus and Hydrophis Curtus, Two Common Sea Snakes in Malaysian Waters. Toxins (Basel) 7 (2), 572–581. doi:10.3390/toxins7020572
Tan, N. H., Wong, K. Y., and Tan, C. H. (2017). Venomics of Naja Sputatrix, the Javan Spitting Cobra: A Short Neurotoxin-Driven Venom Needing Improved Antivenom Neutralization. J. Proteomics 157, 18–32. doi:10.1016/j.jprot.2017.01.018
Tasoulis, T., and Isbister, G. K. (2017). A Review and Database of Snake Venom Proteomes. Toxins (Basel) 9 (9), 290. doi:10.3390/toxins9090290
Teng, C. M., Ouyang, C., and Lin, S. C. (1985). Species Difference in the Fibrinogenolytic Effects of Alpha- and Beta-Fibrinogenases from Trimeresurus Mucrosquamatus Snake Venom. Toxicon 23 (5), 777–782. doi:10.1016/0041-0101(85)90008-x
Theakston, R. D., and Reid, H. A. (1979). Enzyme-linked Immunosorbent Assay (ELISA) in Assessing Antivenom Potency. Toxicon 17 (5), 511–515. doi:10.1016/0041-0101(79)90284-8
Weisel, J. W., and Litvinov, R. I. (2017). Fibrin Formation, Structure and Properties. Fibrous proteins: structures Mech., 405–456. doi:10.1007/978-3-319-49674-0_13
Whiteley, G., Casewell, N. R., Pla, D., Quesada-Bernat, S., Logan, R. A. E., Bolton, F. M. S., et al. (2019). Defining the Pathogenic Threat of Envenoming by South African Shield-Nosed and Coral Snakes (Genus Aspidelaps), and Revealing the Likely Efficacy of Available Antivenom. J. Proteomics 198, 186–198. doi:10.1016/j.jprot.2018.09.019
WHO (2018). World Health Organisation Guidelines for the Production, Control and Regulation of Snake Antivenom Immunoglobulins. Geneva, Switzerland: World Health Organisation.
Wüster, W., Thorpe, R. S., J. Cox, M. M., Jintakune, P., and Nabhitabhata, J. (1995). Population Systematics of the Snake Genus Naja (Reptilia: Serpentes: Elapidae) in Indochina: Multivariate Morphometrics and Comparative Mitochondrial DNA Sequencing (Cytochrome Oxidase I). J. Evol. Biol 8 (4), 493–510. doi:10.1046/j.1420-9101.1995.8040493.x
Keywords: Naja sagittifera, Naja naja, venomics, Andaman and Nicobar Islands, antivenoms
Citation: Attarde S, Khochare S, Iyer A, Dam P, Martin G and Sunagar K (2021) Venomics of the Enigmatic Andaman Cobra (Naja sagittifera) and the Preclinical Failure of Indian Antivenoms in Andaman and Nicobar Islands. Front. Pharmacol. 12:768210. doi: 10.3389/fphar.2021.768210
Received: 31 August 2021; Accepted: 05 October 2021;
Published: 25 October 2021.
Edited by:
Kae Yi Tan, University of Malaya, MalaysiaReviewed by:
Dong-Zong Hung, China Medical University Hospital, TaiwanCopyright © 2021 Attarde, Khochare, Iyer, Dam, Martin and Sunagar. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Kartik Sunagar, a3N1bmFnYXJAaWlzYy5hYy5pbg==
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.