
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Pharmacol. , 23 November 2021
Sec. Drug Metabolism and Transport
Volume 12 - 2021 | https://doi.org/10.3389/fphar.2021.764124
CYP2B10 is responsible for metabolism and detoxification of many clinical drugs. Here, we aimed to investigate a potential role of Period 2 (PER2) in regulating expression of hepatic CYP2B10. Regulatory effects of PER2 on hepatic expression of CYP2B10 and other enzymes were determined using Per2-deficient mice with exons 4-6 deleted (named Per2Del4-6 mice). In vitro and in vivo metabolic activities of CYP2B10 were probed using cyclophosphamide (CPA) as a specific substrate. Regulatory mechanism was investigated using luciferase reporter assays. Genotyping and Western blotting demonstrated loss of wild-type Per2 transcript and markedly reduced PER2 protein in Per2Del4-6 mice. Hepatic expression of a plenty of drug-metabolizing genes (including Cyp2a4/2a5, Cyp2b10, Ugt1a1, Ugt1a9, Ugt2b36, Sult1a1 and Sult1e1) were altered (and majority were down-regulated) in Per2Del4-6 mice. Of note, Cyp2b10, Ugt1a9 and Sult1a1 were three genes considerably affected with reduced expression. Decreased expression of CYP2B10 was translated to reduced metabolism and altered pharmacokinetics of CPA as well as attenuated CPA hepatotoxicity in Per2Del4-6 mice. Positive regulation of CYP2B10 by PER2 was further confirmed in both Hepa-1c1c7 and AML-12 cells. Based on luciferase reporter assays, it was shown that PER2 regulated Cyp2b10 transcription in a REV-ERBα-dependent manner. REV-ERBα was negatively regulated by PER2 (increased REV-ERBα expression in Per2Del4-6 mice) and itself was also a repressor of CYP2B10. In conclusion, PER2 positively regulates CYP2B10 expression and activity in mouse liver through inhibiting its repressor REV-ERBα.
Drug metabolism (biotransformation) is a main component of pharmacokinetics, profoundly affecting drug efficacy and toxicity (Yan et al., 2018). Drug metabolism reactions, mediated by drug-metabolizing enzymes (DMEs), have been historically divided into two types, namely, phase I and phase II reactions. In phase I reactions (also known as functionalization reactions), DMEs such as cytochromes P450 (CYPs) introduce polar chemical moieties to drug molecules. The modified molecules are then converted to more hydrophilic and excretable metabolites in phase II reactions catalyzed by conjugating DMEs such as UDP-glucuronosyltransferases (UGTs) and sulfotransferases (SULTs). Human CYP superfamily contains 18 families consisting of 57 members (Elfaki et al., 2018). Human UGTs include 22 isoenzymes and are classified into four families (i.e., UGT1, UGT2, UGT3, and UGT8) (Mano et al., 2018). Enzymes from UGT1A and 2B sunfamilies (with a total of 16 members) are the main contributors to drug glucuronidation (Rowland et al., 2013). To date, there are four families of SULT enzymes in humans, namely SULT1, SULT2, SULT4, and SULT6. Enzymes of SULT1 and SULT2 (with a total of 12 members) play a leading role in drug sulfonation (Allali-Hassani et al., 2007).
DMEs such as CYP enzymes are distributed in many tissue and organs, particularly in the drug-metabolizing organs liver, kidney, and small intestine (Renaud et al., 2011). Of CYPs, the members from CYP1 to CYP4 families are of most importance as they metabolize >55% of FDA-approved drugs (Saravanakumar et al., 2019). Mouse CYP2B10 (CYP2B6 in humans) is a member of CYP2B subfamily and primarily expressed in the liver, where it accounts for about 10% of the total microsomal CYP pool (Ekins et al., 1998; Ekins et al., 1999). CYP2B10 is responsible for the metabolism and detoxification of many clinical drugs including cyclophosphamide (CPA) and bupropion (Turpeinen and Zanger, 2012). Expression of CYP2B10 is known to be regulated by a number of nuclear receptors such as pregnane X receptor (PXR), constitutive androstane receptor (CAR), glucocorticoid receptors (GR), and vitamin D receptor (VDR) (Beigneux et al., 2002; Pascussi et al., 2004).
Period 2 (PER2) is a central component of the mammalian circadian clock machinery. It acts as a negative regulator of circadian clock through a direct interaction with the transcriptional activator CLOCK/BMAL1 heterodimer (Langmesser et al., 2008). Phosphorylation by casein kinase 1δ/ε (CK1δ/ε) is a key step that determines the stability of the PER2 protein and therefore the period of the circadian rhythms in mammals (Narasimamurthy et al., 2018). A mutation in CK1-phosphorylating site of PER2 (S662G) has been identified as a determiner of human familial advanced sleep phase syndrome (Vanselow et al., 2006; Xu et al., 2007). PER2 is also involved in the regulation of many physiological and pathological processes such as neurobiological activities (Kim et al., 2018), metabolic homeostasis (Zani et al., 2013), thermogenesis (Chappuis et al., 2013), and tumourigenesis (Mteyrek et al., 2016). Moreover, it has been found that PER2 regulates the hepatotoxicity of drugs such as acetaminophen in mice probably via modulating the expression of CYP1A2 and CYP2E1 (Kakan et al., 2011; Ge et al., 2021). However, whether and how PER2 regulates CYP2B10 remain unknown.
Interestingly, Per2−/− mice show decreases in serum triglycerides and free fatty acids coupled with increases in hepatic triglycerides and free fatty acids, indicating that PER2 is involved in lipid metabolism (Kettner et al., 2016). In addition, Heintza’s work illustrates that the repression or inhibition of CYP2B (e.g., CYP2B10) may exacerbate metabolic disorders and cause obesity by perturbing fatty acid metabolism, suggesting a role of CYP2B in lipid homeostasis (Heintz et al., 2019). These findings indicate a potential link between PER2 and CYP2B, particularly CYP2B10.
In the present study, we aimed to investigate a potential role of PER2 in regulating expression of hepatic CYP2B10. Regulatory effects of PER2 on hepatic expression of CYP2B10 and other DMEs were determined using Per2-deficient mice with exons 4-6 deleted. mRNAs and proteins were quantified by qPCR and Western blotting, respectively. In vitro and in vivo metabolic activities of CYP2B10 were probed using CPA as a specific substrate. Regulation of CYP2B10 by PER2 was validated in Hepa-1c1c7 and AML-12 cells. Regulatory mechanism was investigated using luciferase reporter assays. We for the first time demonstrated that PER2 positively regulates CYP2B10 expression and activity in mouse liver through inhibiting its repressor REV-ERBα.
Cyclophosphamide (CPA) was purchased from J&K Scientific (Beijing, China). O-methylhydroxylamine (OMHA) was obtained from Steraloids (Wilton, NH). Nicotinamide adenine dinucleotide phosphate (NADPH), uridine diphosphoglucuronic acid (UDPGA), alamethicin, and 3′-phosphoadenosine-5′-phosphosulfate (PAPS) were purchased from Sigma-Aldrich (St. Louis, MO). Propofol and propofol glucuronide were purchased from TargetMol (Shanghai, China). Galangin was obtained from Weikeqi Biotech (Sichuan, China). Assay kits for ALT (alanine aminotransferase), AST (aspartate aminotransferase) and GSH (glutathione) were purchased from Jiancheng Bioengineering Institute (Nanjing, China). Anti-PER2 (ab180655) and anti-GAPDH antibodies (ab9485) were purchased from Abcam (Cambridge, MA). Anti-CYP2B10 (TA504328) and anti-SULT1A1 (TA501951) antibodies were obtained from OriGene (Rockville, MD). Anti-UGT1A9 (bs-4224R) antibody was purchased from Bioss (Beijing, China). Anti-REV-ERBα antibody (AB10130) was purchased from Sigma-Aldrich (St Louis, MO). All primary antibodies were diluted with 5% BSA at a ratio of 1:1,000. The secondary antibody was purchased from Huaan Biotechnology (Hangzhou, China) and diluted with 5% skim milk at a ratio of 1:5,000. siPer2 (siRNA targeting Per2) and siNC (a negative control for siPer2) were obtained from TranSheep Biotech (Shanghai, China). pRL-TK vector was purchased from Promega (Madison, WI).
Per2 heterozygotes (on a C57BL/6 background) were obtained from Cyagen Biosciences (Guangzhou, China) (Supplementary Material S1). Homozygotes with exons 4-6 deleted (named Per2Del4-6 mice) were generated by inter-crossing heterozygous mice. Per2-deficient mice and their wild-type littermates were housed and maintained under a 12 h light/12 h dark cycle (lights on at 6:00 AM (= Zeitgeber time 0/ZT0) and lights off at 6:00 PM (= ZT12)), with free access to food and water. Animal experimental procedures were approved by Institutional Animal Care and Use Committee of Guangzhou University of Chinese Medicine (Appr. date: 2020-11-19; IACUC Issue No: ZYD-2020-111) and were performed in accordance with the NIH Guide for the Care and Use of Laboratory Animals. Efforts were made to minimize suffering and the number of mice used in the experiments.
DNA was extracted from mouse tail (0.5–1 mm). PCR reactions were performed with 400 ng DNA template. Amplification program consisted of an initial denaturation at 94°C for 3 min, 35 cycles of denaturation at 94°C for 30 s, annealing at 60°C for 35 s, and extension at 72°C for 35 s. The products were subjected to 2% agarose gel electrophoresis, and bands were imaged using the Omega Lum G imaging system (Aplegen). The primers are listed in Table 1.
Per2Del4-6 and control wild-type mice (8–12 weeks, male) were treated with a single dose of CPA (100 mg/kg) by intraperitoneal injection at ZT14. At predetermined time points (15, 30, 60 and 120 min), mice (n = 5 per time point) were rendered unconscious with isoflurane, and plasma samples were collected by retro-orbital bleeding. Plasma samples were processed for UPLC-QTOF/MS analysis as previously described (Sadagopan et al., 2001; Zhao et al., 2019). Of note, the CPA metabolite, 4-hydroxycyclophosphamide (4-OH-CPA) is unstable in the plasma (Sadagopan et al., 2001). To quantify 4-OH-CPA, plasma samples were treated with O-methylhydroxylamine (OMHA) to transform 4-OH-CPA into a stable product OH-CPA O-methyloxime (Sadagopan et al., 2001). Additionally, liver samples were collected at two time points (i.e., 30 and 120 min), and processed to measure the drug/metabolite concentrations in the livers as previously described (Zhang et al., 2018a).
Per2Del4-6 and control wild-type mice (8–12 weeks, male, n = 5 per group) were injected (i.p.) with CPA at 100 mg/kg or with vehicle at ZT14. Four hours after drug administration, mice were sacrificed to collect plasma and liver samples, followed by biochemical analyses.
ALT, AST, and GSH were measured with their assay kits according to manufacturer’s instructions (Jiancheng Bioengineering Institute, Nanjing, China).
Livers were collected from Per2Del4-6 and control wild-type mice. Liver microsomes were prepared by sequential ultracentrifugation, first at 9,000 g for 10 min and then at 100,000 g for 1 h as previously described (Van et al., 1992). CPA was used as a specific substrate to determine the microsomal activity of CYP2B10 (Pass et al., 2005; Zhao et al., 2019). In brief, liver microsomes (4 mg/ml) was incubated with CPA (20 μM), NADPH (1.5 mM), and MgCl2 (5 mM) in potassium phosphate (50 mM, pH 7.4) at 37°C for 2 h. The reaction was terminated by adding ice-cold acetonitrile (containing an internal standard). To quantify 4-OH-CPA, the resultant mixture was treated with OMHA (1.5 mg/ml) to transform 4-OH-CPA into a stable product OH-CPA O-methyloxime. The reaction mixture was centrifuged at 13,000 g for 15 min and the supernatant was subjected to UPLC-QTOF/MS analysis.
Glucuronidation assays were performed by incubating liver microsomes with propofol. The incubation mixture contained liver microsomes (4 mg/ml), propofol (100 μM), MgCl2 (0.88 mM), saccharolactone (4.4 mM), alamethicin (11.2 μM) and UDPGA (3.5 mM) in 50 mM potassium phosphate (pH 7.4). After 30 min, the metabolic reaction was terminated by adding 200 μl ice-cold water/acetonitrile (50:50, v/v) (containing an internal standard). The resulting mixture was centrifuged at 2,000 g for 10 min, and the supernatant was subjected to UPLC-QTOF/MS analysis.
Livers were collected from Per2Del4-6 and control wild-type mice. Liver S9 fraction was prepared by centrifugation at 9,000 g for 10 min (Richardson et al., 2016). Sulfation activity was determined using an incubation method with liver S9 fraction as previously described (Guo et al., 2018). Briefly, liver S9 fraction (4 mg/ml), PAPS (50 μM) and galangin (6 μM) were incubated in potassium phosphate buffer (50 mM, pH 7.4) at 37°C for 3 h. The reactions were terminated by adding 200 μl ice-cold acetonitrile (containing an internal standard), followed by vortex and centrifugation at 13,000 g for 15 min. The supernatant was subjected to UPCL-QTOF/MS analysis.
Drugs and metabolites were quantified using an UPLC-QTOF/MS system (AB SCIEX, CA) and a Phenomenex C18 column (2 × 100 mm, 1.6 μm; Phenomenex, Torrance). The mobile phases were 0.1% formic acid in acetonitrile (mobile phase A) and 0.1% formic acid in water (mobile phase B). For determination of CPA and OH-CPA O-methyloxime, the gradient elution program was as follows: 0–2 min, 90% B; 2–6 min, 90-10% B; and 6–8 min, 10–90% B. The mass spectrometer was operated at the positive ion full scan mode. For determination of propofol glucuronide, the gradient elution program was as follows: 0–2 min, 80% B; 2–4 min, 80-30% B; and 4–5 min, 30-80% B. For determination of galangin sulfate, the gradient elution program was as follows: 0–1 min, 90%B; 1–3.5 min; 90-10% B; 3.5–4.5 min, 10% B; and 4.5–5 min, 10–90% B. The mass spectrometer was operated at the negative ion full scan mode for propofol glucuronide and galangin sulfate. The flow rate was set at 0.3 ml/min. Peak areas were recorded with extract masses as follows: m/z 261.03 ± 0.05 Da for CPA, 306.09 ± 0.05 Da for OH-CPA O-methyloxime; m/z 353.16 ± 0.05 Da for propofol glucuronide; and m/z 348.9 ± 0.05 Da for galangin sulfate.
Hepa-1c1c7 cells were cultured in DMEM containing 10% fetal bovine serum (FBS). AML-12 cells were cultured in DMEM/F12 supplemented with 0.1% dexamethasone, 1% ITS (i.e., insulin, transferrin and selenium) and 10% FBS. Cells were seeded into 6-well plates. Once growing to a density of 60–70%, cells were co-transfected with Per2 expression plasmid (2 μg) or siPer2 (50 nM) or control using jetPRIME transfection reagent (Polyplus Transfection, Illkirch, France) according to the manufacturer’s protocol. After 24 or 48 h, cells were collected for mRNA and protein quantification.
Total RNA was extracted from mouse liver and cell samples with TRIzol reagent (Accurate Biotech, Hunan, China) following the manufacturer’s instructions, and used as a template for cDNA synthesis. qPCR reactions were performed with SYBR Green Master Mix (Vazyme, Nanjing, China) using a Biometra Toptical Thermocycler (Analytik Jena, Goettingen, Germany) as previously described (Yamamura et al., 2010). Amplification reactions consisted of an initial denaturation at 95°C for 5 min, followed by 40 cycles of denaturation at 95°C for 15 s, annealing at 60°C for 30 s, and extension at 72°C for 30 s. Peptidylprolyl isomerase B (Ppib) was used as an internal control. Relative mRNA expression was calculated using the 2-∆∆Ct method and normalized to the control group. Primer sequences are shown in Table 1.
Mouse tissues and cell samples were lysed in RIPA buffer supplemented with a protease inhibitor cocktail (HY-K0010, Monmouth Junction, NJ). Protein concentration was detected by a BCA assay kit (Beyotime, Shanghai, China). After denaturing at 95°C for 10 min, protein samples (40 μg) were subjected to SDS-polyacrylamide gel electrophoresis (10% gel) and transferred to polyvinylidene fluoride membranes (Millipore, Bedford, MA). The membranes were sequentially incubated with primary antibody and HRP-conjugated secondary antibody. Protein bands were visualized using the Omega Lum G imaging system (Aplegen), and band density was analyzed by using FlourChem software (Bio-Rad). Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) was used as a loading control. Protein expression was normalized to GAPDH.
HEK293T cells were cultured in DMEM supplemented with 10% FBS and seeded into 48-well plates. Cells were transfected with Cyp2b10 luciferase (firefly) reporter plasmid (50 ng), pRL-TK (15 ng) and overexpression plasmid (Per2 or Rev-erbα, 200 ng) or blank pcDNA3.1 (control) using jetPRIME transfection reagent (PolyplusTransfection, Illkirch, France). After 24 h, cells were lysed in 45 μl passive lysis buffer (Promega, Madison, WI), and cell lysate was collected to determine the luciferase activities using the Dual-Luciferase Reporter Assay system on a GloMax 20/20 luminometer (Promega). Firefly luciferase activities were normalized to renilla luciferase values, and expressed as relative luciferase units.
Data are presented as mean ± SD (standard deviation). Sample sizes are provided in figure legends. Student’s t-test was used to analyze a statistical difference between two groups. One-way ANOVA followed by Bonferroni post hoc test was used for multiple group comparisons. Statistical analyses were performed with GraphPad Prism 7 (GraphPad Software Inc., San Diego, CA). Pharmacokinetic analysis was performed using WinNonlin software (Pharsight Corp, St. Louis, MO). The level of significance was set at p < 0.05 (*).
Per2-deficient mice with exons 4-6 deleted (named Per2Del4-6 mice) were obtained from Cyagen Biosciences (Guangzhou, China) (Figure 1A). PCR genotyping of mice was performed using tail biopsies with the primers F1, F2 and R1/2, resulting in a 707 bp fragment for Per2Del4-6 mice and a 514 bp fragment for wild-type allele (Figure 1B). qPCR assay with primer set 1 (targeting the knockout sequence) confirmed the absence of wild-type Per2 transcript in mouse liver (Figure 1C). Western blotting showed that hepatic PER2 protein was markedly reduced in Per2Del4-6 mice, however, it was not completely lost (Figure 1D). qPCR assay with primer set 2 targeting non-knockout sequence suggested the presence of a new version of Per2 transcript in Per2Del4-6 mice albeit at a reduced level (Figure 1C). This new transcript may be translated to a PER2-related protein that retains a reactivity with the commercial PER2 antibody, thereby explaining why PER2 protein was detected in Per2Del4-6 mice (Figure 1D). Taken together, Per2Del4-6 mice were characterized with loss of wild-type Per2 transcript and markedly reduced PER2 protein.
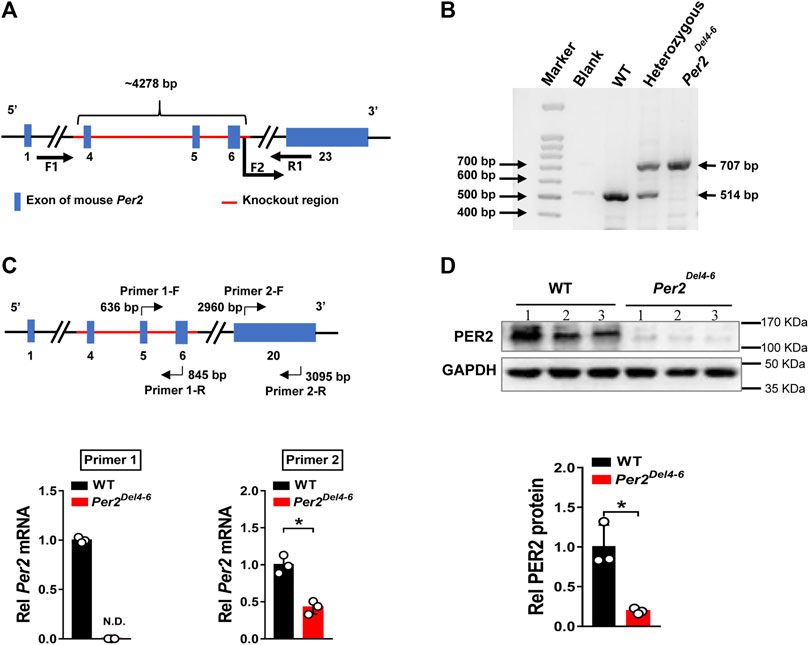
FIGURE 1. Characterization of Per2-deficient mice. (A) Schematic diagram showing target site (exons 4-6) deleted for establishment of Per2Del4-6 mice. (B) PCR genotyping using mouse tails from wild-type (WT), heterozygous and homozygous (Per2Del4-6) mice. (C) qPCR analyses of Per2 in Per2Del4-6 and WT mice. The top panel shows the target sites of primer sets 1 and 2. (D) Western blotting analyses of PER2 protein in Per2Del4-6 and WT mice. Western blot strips (a target protein and a loading control) were cut from one gel. Data are mean ± SD (n = 3). *p < 0.05 (t test).
Per2 expression displays a robust diurnal oscillation with the nadir at ZT2 and the zenith at ZT14 (Yamamura et al., 2010; Yoo et al., 2017). Therefore, the impact of Per2 on expression of drug-metabolizing genes was assessed at two diurnal time points (i.e., ZT2 and ZT14) using Per2Del4-6 mice. According to qPCR analyses, hepatic expression of several drug-metabolizing genes (including Cyp2a4/2a5, Cyp2b10, Ugt1a1, Ugt1a9, Ugt2b36, Sult1a1, and Sult1e1) was altered (and majority were down-regulated) at both ZT2 and ZT14 in Per2Del4-6 mice (Figure 2A). Of note, Cyp2b10, Ugt1a9, and Sult1a1 were three genes considerably affected, independent of diurnal times (Figure 2B). Consistent with the mRNA changes, the proteins of CYP2B10, UGT1A9 and SULT1A1 in the liver were significantly reduced in Per2Del4-6 mice (Figure 2C). In addition, Per2Del4-6 mice showed decreased enzymatic activities of CYP2B10, UGT1A9 and SULT1A1 against their specific substrates (CPA for CYP2B10, propofol for UGT1A9, and galangin for SULT1A1) based on in vitro incubation assays (Figure 3). Taken together, these data revealed a role of Per2 in regulation of drug-metabolizing enzymes.
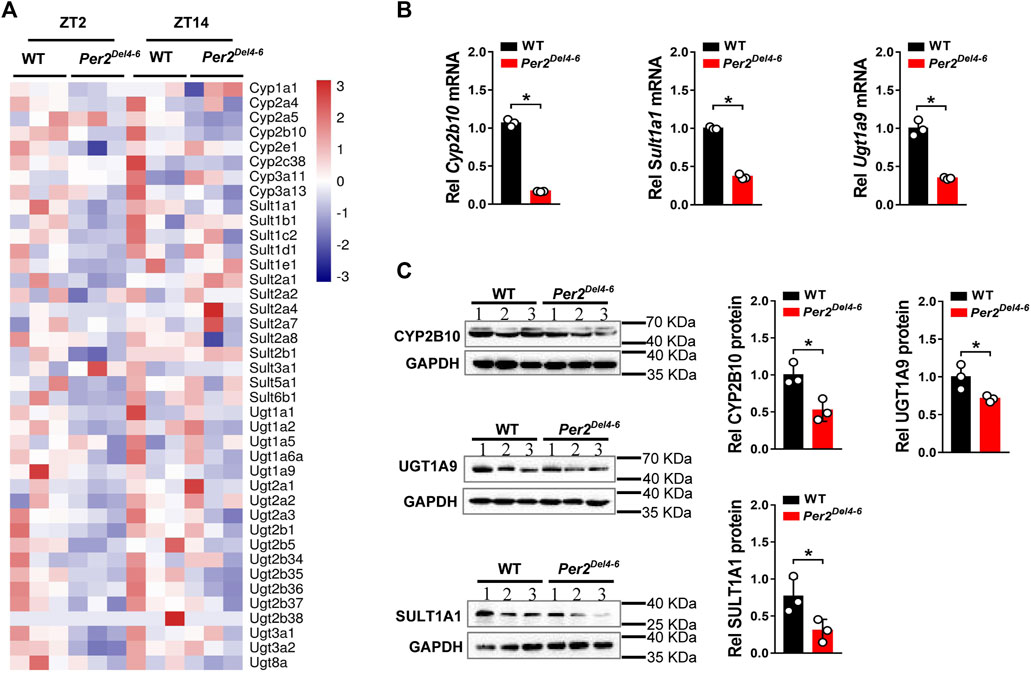
FIGURE 2. Expression of selected drug-metabolizing enzymes in Per2Del4-6 mice. (A) Heatmap showing relative mRNA expression of drug-metabolizing enzymes at ZT2 and ZT14 in Per2Del4-6 versus in WT (wild-type) mice. (B) mRNA expression levels of three drug-metabolizing genes (Cyp2b10, Ugt1a9 and Sult1a1) in the livers of Per2Del4-6 and WT mice. (C) Protein expression levels of CYP2B10, UGT1A9 and SULT1A1 in the livers of Per2Del4-6 and WT mice. Western blot strips (a target protein and a loading control) were cut from one gel. Data are mean ± SD (n = 3). *p < 0.05 (t test). Rel, Relative. ZT, zeitgeber time.
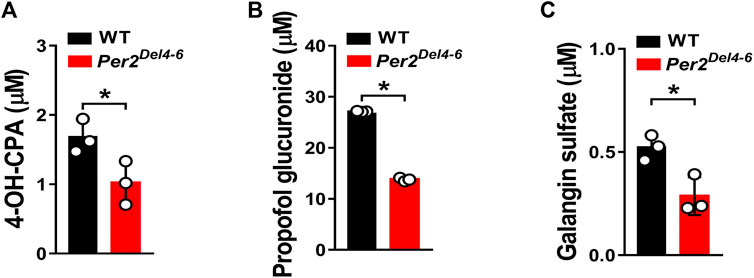
FIGURE 3. Metabolic activities of CYP2B10, UGT1A9 and SULT1A1 in Per2Del4-6 mice based on in vitro metabolism assays. (A) Liver CYP microsomal metabolism assays showing attenuated metabolism of CPA in Per2Del4-6 mice. (B) Reduced glucuronidation of propofol in liver microsomes of Per2Del4-6 mice. (C) Sulfation assay showing reduced hepatic metabolism of galangin in Per2Del4-6 mice. Data are mean ± SD (n = 3). *p < 0.05 (t test).
Since CYP2B10 expression was altered in the liver of Per2Del4-6 mice, we next tried to test whether this affects the metabolism and pharmacokinetics of substrate drugs. To this end, we performed pharmacokinetic experiments of CPA with Per2Del4-6 and control mice. Per2Del4-6 mice showed increases in plasma CPA concentrations (Figure 4A). Accordingly, AUC (area under the curve, a measure of systemic exposure) of CPA was significantly increased in Per2Del4-6 mice (Table 2). In the meantime, liver CPA concentrations were increased in Per2Del4-6 mice (Figure 4B). By contrast, the plasma and hepatic levels of the metabolite 4-hydroxycyclophosphamide (4-OH-CPA) were lower in Per2Del4-6 mice, suggesting reduced metabolism of CPA (Figures 4C,D). Therefore, Per2 ablation altered the pharmacokinetics of CPA in mice by down-regulating drug metabolism.
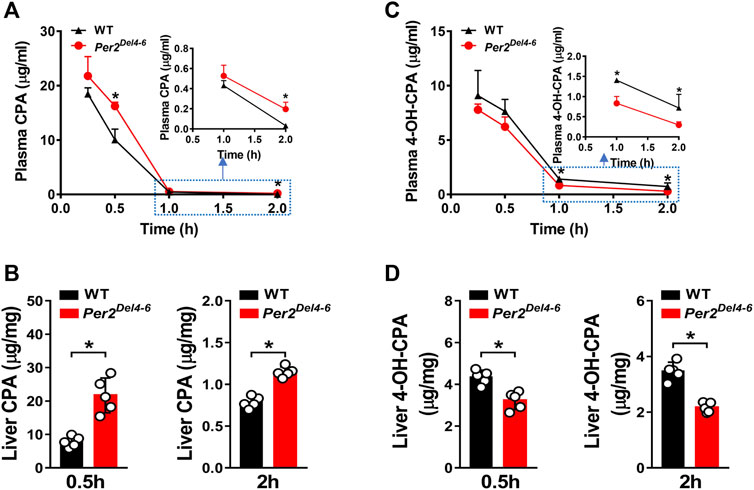
FIGURE 4. Characterization of CPA metabolism and pharmacokinetics in Per2Del4-6 mice. (A) Plasma concentrations of CPA in Per2Del4-6 and WT mice at different time points after CPA treatment (i.p., 100 mg/kg). (B) Plasma concentrations of 4-OH-CPA in Per2Del4-6 and WT mice at different time points after CPA treatment. (C) Liver concentrations of CPA in Per2Del4-6 and WT mice at 0.5 and 2 h after CPA treatment. (D) Liver concentrations of CPA in Per2Del4-6 and WT mice at 0.5 and 2 h after CPA treatment. Data are mean ± SD (n = 5). *p < 0.05 (t test).
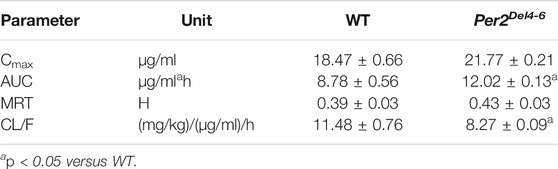
TABLE 2. Pharmacokinetic parameters for CPA (100 mg/kg, i.p.) after drug administration to Per2Del4-6 and wild-type (WT) mice.
CPA is an anticancer prodrug whose cytotoxic effects depend on metabolic activation to its metabolites such as 4-OH-CPA (Richardson et al., 2016). Our previous study shows that CPA treatment induces hepatotoxicity in mice, which is positively related to formation of 4-OH-CPA (Zhao et al., 2019). We thus examined whether CPA-induced hepatotoxicity is affected in Per2Del4-6 mice. We found lower levels of ALT and AST activities, but a higher GSH level in the liver of Per2Del4-6 mice, indicating reduced hepatotoxicity in the genetically modified mice (Figure 5). This was consistent with reduced metabolism of CPA and lowered formation of 4-OH-CPA (Figure 4). Altogether, CPA hepatotoxicity is reduced in Per2Del4-6 mice due to decreased metabolism.
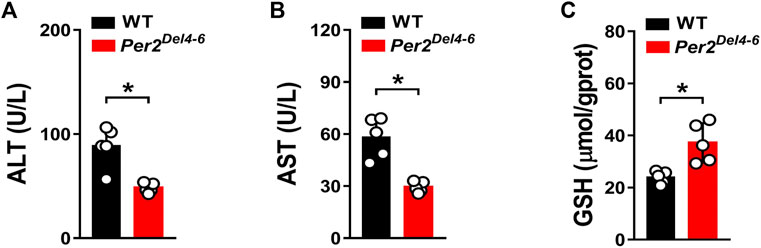
FIGURE 5. Characterization of CPA-induced hepatotoxicity in Per2Del4-6 mice. (A) Plasma ALT levels in Per2Del4-6 and WT mice at 4 h after CPA administration (i.p., 100 mg/kg, n = 5). (B) Plasma AST levels in Per2Del4-6 and WT mice at 4 h after CPA administration. (C) Hepatic GSH levels in Per2Del4-6 and WT mice at 4 h after CPA administration. Data are mean ± SD (n = 5). *p < 0.05 (t test).
Next, we assessed the regulatory effects of Per2 on CYP2B10 expression in mouse Hepa-1c1c7 hepatoma cells by performing overexpression and knockdown assays. We confirmed that overexpression plasmid significantly elevated the levels of PER2 mRNA and protein, whereas siRNA decreased PER2 expression, in Hepa-1c1c7 cells (Figure 6A). Per2 overexpression significantly increased cellular levels of CYP2B10 mRNA and protein (Figure 6B). By contrast, Per2 knockdown reduced cellular expression of CYP2B10 (Figure 6B). In an identical manner, we examined the effects of Per2 on CYP2B10 expression in mouse AML-12 hepatocytes (Figure 7). Per2 overexpression significantly increased CYP2B10 expression, whereas Per2 knockdown deceased CYP2B10 expression, in AML-12 cells (Figures 7A,B). Taken together, Per2 positively regulates CYP2B10 expression in both Hepa-1c1c7 and AML-12 cells, consistent with our in vivo findings (Figure 2).
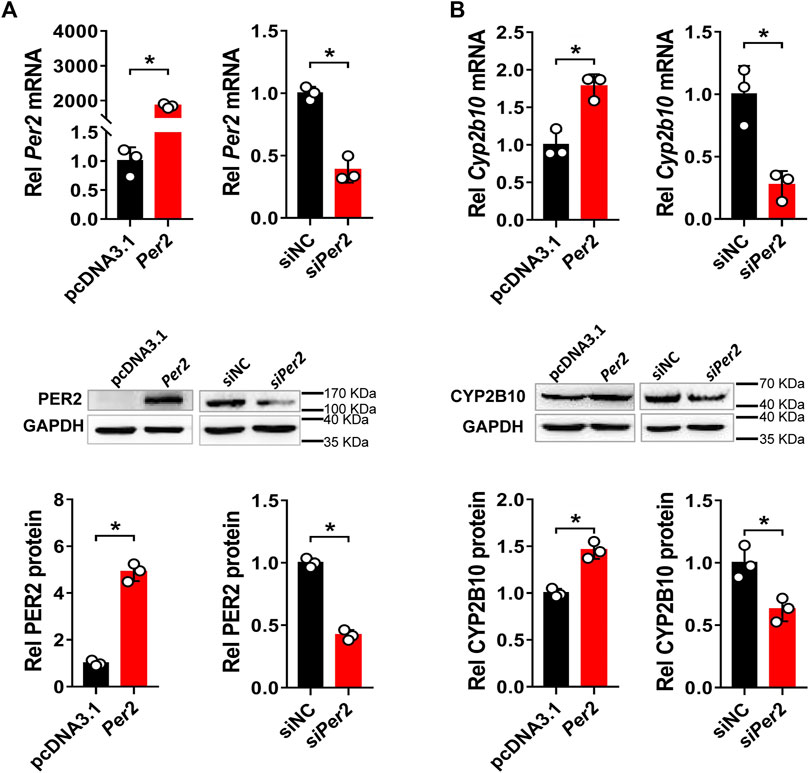
FIGURE 6. Per2 regulates CYP2B10 expression in Hepa-1c1c7 cells. (A) Efficacy validation of Per2 overexpression (by overexpression plasmid) and knockdown (by siRNA). (B) Effects of Per2 overexpression or knockdown on the expression of CYP2B10 mRNA and protein. Western blot strips (a target protein and a loading control) were cut from one gel. Data are mean ± SD from three independent experiments. *p < 0.05 (t test). Rel, Relative.
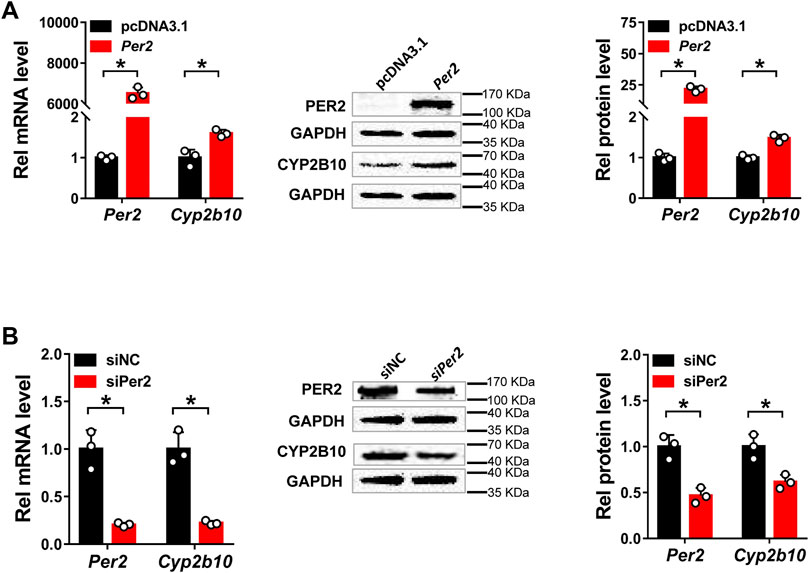
FIGURE 7. Per2 regulates Cyp2b10 expression in AML-12 cells. (A) Effects of Per2 overexpression on the expression of CYP2B10 mRNA and protein. (B) Effects of Per2 knockdown on the expression of CYP2B10 mRNA and protein. Western blot strips (a target protein and a loading control) were cut from one gel. Data are mean ± SD from three independent experiments. *p < 0.05 (t test). Rel, Relative.
PER2 is an integral component of circadian clock, and functions as a co-repressor to repress the transcriptional activity of the BMAL1/CLOCK heterodimer, a well-known activator of many clock-controlled genes (Langmesser et al., 2008). Thus, it was speculated that PER2 may regulate CYP2B10 expression through an indirect mechanism that involves BMAL1/CLOCK and an intermediate regulator. This intermediate regulator should be a target of BMAL1/CLOCK and a repressor of CYP2B10. A survey of the literature suggested REV-ERBα as a candidate for such intermediate regulator because it transcriptionally inhibits CYP2B10 and the expression itself is directly driven by BMAL1/CLOCK (Dunlap, 1999; Crumbley and Burris, 2011; Zhang et al., 2018b). We therefore tested whether REV-ERBα mediates PER2 regulation of CYP2B10. We found that REV-ERBα mRNA and protein were significantly increased in the liver of Per2Del4-6 mice (Figure 8A). This was expected because PER2 can repress the expression of REV-ERBα by inhibiting the transcriptional activity of BMAL1/CLOCK (Liu et al., 2008). We confirmed that REV-ERBα is a transcriptional repressor of Cyp2b10 in HEK293T cells based on luciferase reporter assays (Figure 8B). Intriguingly, PER2 dose-dependently induced the transcription of Cyp2b10 (Figure 8C). However, the induction effect of PER2 was attenuated in Rev-erbα silenced cells (Figure 8D). Moreover, overexpressing Rev-erbα reversed the effect of PER2 on Cyp2b10 transcription (Figure 8E). Taken together, our findings suggest that PER2 regulates CYP2B10 expression by down-regulating REV-ERBα.
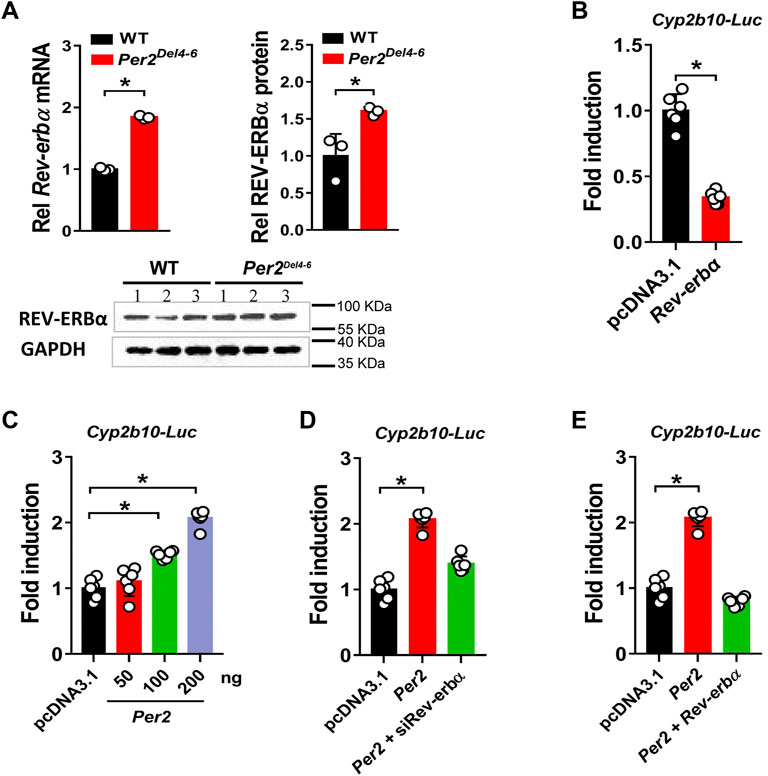
FIGURE 8. Per2 regulates Cyp2b10 transcription through down-regulating REV-ERBα. (A) Increased expression of REV-ERBα in Per2Del4-6 mice. Western blot strips (a target protein and a loading control) were cut from one gel. (B) Effects of Rev-erbα overexpression Cyp2b10-Luc reporter activity in HEK293T cells. (C) Per2 dose-dependently induces Cyp2b10-Luc reporter activity in HEK293T cells. (D) Regulatory effects of Per2 overexpression on Cyp2b10 are attenuated in Rev-erbα-deficient HEK293T cells. (E) Rev-erbα reverses the effects of Per2 on Cyp2b10 transcription in HEK293T cells. Data are mean ± SD (n = 3 or 6). In panels A/B, *p < 0.05 (t test). In panels (C–E), *p < 0.05 (one-way ANOVA followed by Bonferroni post hoc test). Rel, Relative.
In this study, we have revealed that many hepatic DMEs including CYP2B10, UGT1A9 and SULT1A1 are under the control of PER2 in mice (Figures 1–3). We further showed that decreased expression of CYP2B10 was translated to reduced metabolism and altered pharmacokinetics of its substrate drug CPA as well as attenuated CPA hepatotoxicity in Per2-deficient (Per2Del4-6) mice (Figures 4, 5). Positive regulation of CYP2B10 by PER2 was confirmed in both Hepa-1c1c7 and AML-12 cells (Figures 6, 7). Based on luciferase reporter assays, it was shown that PER2 regulated Cyp2b10 transcription in a REV-ERBα-dependent manner (Figure 8). REV-ERBα is negatively regulated by PER2 (decreased REV-ERBα in Per2Del4-6 mice, Figure 8) and itself is also a repressor of CYP2B10. Therefore, we propose that PER2 regulates CYP2B10 expression and activity through down-regulating REV-ERBα, thereby impacting the metabolism and pharmacokinetics of substrate drugs.
CYP2B10/CYP2B6 activity is a critical determinant to the pharmacokinetics and efficacy of many drugs including CPA, ifosfamide, ketamine, pethidine, methadone, nevirapine and efavirenz (Ekins et al., 2008). For instance, CYP2B6*6 allele carriers have been linked to increased ifosfamide plasma levels and exacerbated toxicities (Mo et al., 2009). Induction of CYP2B6 by a selective activator (e.g., CITCO) facilitates the bioactivation of CPA to 4-OH-CPA and improves the therapeutic outcome of CHOP (cyclophosphamide–doxorubicin–vincristine–prednisone) chemotherapy against non-Hodgkin lymphoma (Shu et al., 2017). Therefore, identification of PER2 as a novel pharmacokinetic determinant of CYP2B10 substrates may facilitate an increased understanding of varied pharmacokinetics and possibly varied drug toxicity and efficacy in a circadian fashion.
A previous study has revealed Cyp2b10 as a circadian gene that is under the control of the Clock gene (Zhao et al., 2019). It is noteworthy that Per2 is a circadian clock gene (whose expression oscillates with time of the day) and involved in regulation of circadian rhythms (Yamamura et al., 2010). Therefore, there is a high possibility that Per2 may be involved in regulation of circadian expression of Cyp2b10. However, of clock genes, Rev-erbα seems to be a direct transcriptional regulator of Cyp2b10 and thus a direct key regulator of circadian Cyp2b10 (Zhang et al., 2018a). Circadian regulation of Cyp2b10 by other clock genes such as Per2 and Clock is possible, but is attained through modulating Rev-erbα (Figure 9). In addition, CAR is another direct transcriptional regulator of Cyp2b10 (Kettner et al., 2016). However, CAR should not be involved in PER2 regulation of Cyp2b10 as it positively regulates Cyp2b10 expression (Kettner et al., 2016). Our findings may provide increased understanding of the complex regulatory pathways for CYP2B10, and highlight the role of clock genes in regulating drug metabolism and pharmacokinetics.
It is noteworthy that we detected a new version of Per2 transcript in Per2Del4-6 mice albeit at a reduced level (Figure 1C). This new Per2 transcript most likely is a truncated version lacking exons 4-6 transcribed region, and may be translated to a PER2-related protein that retains a reactivity with the commercial PER2 antibody, thereby explaining why PER2 protein was detected in Per2Del4-6 mice (Figure 1D). It is acknowledged that whether or not this PER2-related protein is functional remains unknown. However, it was of no concern that Per2Del4-6 mice can be used to determine the regulatory effects of Per2 on hepatic DME in vivo as they were characterized with loss of wild-type Per2 transcript and markedly reduced PER2 protein in the liver (Figure 1).
CPA is a prodrug and bioactivated by CYP2B10 in mice to the hydroxylated metabolite 4-OH-CPA, one active form with cytotoxic effects and hepatotoxicity (Sadagopan et al., 2001). 4-OH-CPA is unstable in biological samples (Sadagopan et al., 2001). To quantify 4-OH-CPA, the samples were treated with OMHA to transform 4-OH-CPA into an equivalent stable product OH-CPA O-methyloxime for mass spectrometric quantification as previously noted (Sadagopan et al., 2001). Alleviated hepatotoxicity of CPA in Per2Del4-6 mice supported reduced metabolism of CPA to 4-OH-CPA caused by decreased expression of CYP2B10 (Figures 4, 5). Also, reduced CPA metabolism was consistent with decreased systemic clearance (CL/F) according to the pharmacokinetic analysis (Table 2).
Current study focused on determination of the regulatory effects of PER2 on hepatic CYP2B10 and drug pharmacokinetics. It is noteworthy that CYP2B10 expression in other drug-metabolizing organs such as the kidney and intestine besides the liver may be also under the control of PER2 in mice, however, a potential role of renal and intestinal CYP2B10 in altered CPA pharmacokinetics remains unresolved. In addition, we have provided in vitro and in vivo evidence that hepatic PER2 regulated CYP2B10 in mice to alter pharmacokinetics and drug toxicity. However, whether PER2 regulates CYP2B6-mediated pharmacokinetics and drug toxicity in humans as its counterpart does in mouse liver awaits further investigations.
In summary, CYP2B10 was down-regulated at the mRNA, protein, and enzymatic levels in Per2-deficient mice. Consistently, PER2 positively regulated CYP2B10 expression in both Hepa-1c1c7 and AML-12 cells. Moreover, PER2 regulated Cyp2b10 transcription in a REV-ERBα-dependent manner. Therefore, PER2 regulates CYP2B10 expression and activity through REV-ERBα, impacting the metabolism and pharmacokinetics of substrate drugs.
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding authors.
The animal study was reviewed and approved by the Animal experimental procedures were approved by Institutional Animal Care and Use Committee of Guangzhou University of Chinese Medicine (Appr. date: 2020-11-19; IACUC Issue No: ZYD-2020-111) and were performed in accordance with the NIH Guide for the Care and Use of Laboratory Animals.
MLC, MC, ZW, and BW designed the study; MLC, MC, YW, and LZ performed experiments; MLC, MC, YW, DL, and ZW collected and analyzed data; BW wrote the manuscript.
This work was supported by the Guangdong Basic and Applied Basic Research Foundation (2020A1515010682 and 2021A1515011256), and the Project of Administration of Traditional Chinese Medicine of Guangdong Province of China (20212047).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fphar.2021.764124/full#supplementary-material
Allali-Hassani, A., Pan, P. W., Dombrovski, L., Najmanovich, R., Tempel, W., Dong, A., et al. (2007). Structural and Chemical Profiling of the Human Cytosolic Sulfotransferases. Plos Biol. 5 (5), e97. doi:10.1371/journal.pbio.0050097
Beigneux, A. P., Moser, A. H., Shigenaga, J. K., Grunfeld, C., and Feingold, K. R. (2002). Reduction in Cytochrome P-450 Enzyme Expression Is Associated with Repression of CAR (Constitutive Androstane Receptor) and PXR (Pregnane X Receptor) in Mouse Liver during the Acute Phase Response. Biochem. Biophys. Res. Commun. 293 (1), 145–149. doi:10.1016/S0006-291X(02)00196-1
Chappuis, S., Ripperger, J. A., Schnell, A., Rando, G., Jud, C., Wahli, W., et al. (2013). Role of the Circadian Clock Gene Per2 in Adaptation to Cold Temperature. Mol. Metab. 2 (3), 184–193. doi:10.1016/j.molmet.2013.05.002
Crumbley, C., and Burris, T. P. (2011). Direct Regulation of CLOCK Expression by REV-ERB. PloS one 6 (3), e17290. doi:10.1371/journal.pone.0017290
Dunlap, J. C. (1999). Molecular Bases for Circadian Clocks. Cell 96 (2), 271–290. doi:10.1016/s0092-8674(00)80566-8
Ekins, S., Bravi, G., Ring, B. J., Gillespie, T. A., Gillespie, J. S., Vandenbranden, M., et al. (1999). Three-dimensional Quantitative Structure Activity Relationship Analyses of Substrates for CYP2B6. J. Pharmacol. Exp. Ther. 288 (1), 21–29.
Ekins, S., Iyer, M., Krasowski, M. D., and Kharasch, E. D. (2008). Molecular Characterization of CYP2B6 Substrates. Curr. Drug Metab. 9 (5), 363–373. doi:10.2174/138920008784746346
Ekins, S., Vandenbranden, M., Ring, B. J., Gillespie, J. S., Yang, T. J., Gelboin, H. V., et al. (1998). Further Characterization of the Expression in Liver and Catalytic Activity of CYP2B6. J. Pharmacol. Exp. Ther. 286 (3), 1253–1259.
Elfaki, I., Mir, R., Almutairi, F. M., and Duhier, F. M. A. (2018). Cytochrome P450: Polymorphisms and Roles in Cancer, Diabetes and Atherosclerosis. Asian Pac. J. Cancer Prev. 19 (8), 2057–2070. doi:10.22034/APJCP.2018.19.8.2057
Ge, W., Wang, T., Zhao, Y., Yang, Y., Sun, Q., Yang, X., et al. (2021). Period1 Mediates Rhythmic Metabolism of Toxins by Interacting with CYP2E1. Cell Death Dis 12 (1), 76. doi:10.1038/s41419-020-03343-7
Guo, L., Yu, F., Zhang, T., and Wu, B. (2018). The Clock Protein Bmal1 Regulates Circadian Expression and Activity of Sulfotransferase 1a1 in Mice. Drug Metab. Dispos 46 (10), 1403–1410. doi:10.1124/dmd.118.082503
Heintz, M. M., Kumar, R., Rutledge, M. M., and Baldwin, W. S. (2019). Cyp2b-null Male Mice Are Susceptible to Diet-Induced Obesity and Perturbations in Lipid Homeostasis. J. Nutr. Biochem. 70, 125–137. doi:10.1016/j.jnutbio.2019.05.004
Kakan, X., Chen, P., and Zhang, J. (2011). Clock Gene mPer2 Functions in Diurnal Variation of Acetaminophen Induced Hepatotoxicity in Mice. Exp. Toxicol. Pathol. 63 (6), 581–585. doi:10.1016/j.etp.2010.04.011
Kettner, N. M., Voicu, H., Finegold, M. J., Coarfa, C., Sreekumar, A., Putluri, N., et al. (2016). Circadian Homeostasis of Liver Metabolism Suppresses Hepatocarcinogenesis. Cancer Cell 30 (6), 909–924. doi:10.1016/j.ccell.2016.10.007
Kim, M., de la Peña, J. B., Cheong, J. H., and Kim, H. J. (2018). Neurobiological Functions of the Period Circadian Clock 2 Gene, Per2. Biomol. Ther. (Seoul) 26 (4), 358–367. doi:10.4062/biomolther.2017.131
Langmesser, S., Tallone, T., Bordon, A., Rusconi, S., and Albrecht, U. (2008). Interaction of Circadian Clock Proteins PER2 and CRY with BMAL1 and CLOCK. BMC Mol. Biol. 9, 41. doi:10.1186/1471-2199-9-41
Liu, A. C., Tran, H. G., Zhang, E. E., Priest, A. A., Welsh, D. K., and Kay, S. A. (2008). Redundant Function of REV-ERBalpha and Beta and Non-essential Role for Bmal1 Cycling in Transcriptional Regulation of Intracellular Circadian Rhythms. Plos Genet. 4 (2), e1000023. doi:10.1371/journal.pgen.1000023
Mano, E. C. C., Scott, A. L., and Honorio, K. M. (2018). UDP-glucuronosyltransferases: Structure, Function and Drug Design Studies. Curr. Med. Chem. 25 (27), 3247–3255. doi:10.2174/0929867325666180226111311
Mo, S. L., Liu, Y. H., Duan, W., Wei, M. Q., Kanwar, J. R., and Zhou, S. F. (2009). Substrate Specificity, Regulation, and Polymorphism of Human Cytochrome P450 2B6. Curr. Drug Metab. 10 (7), 730–753. doi:10.2174/138920009789895534
Mteyrek, A., Filipski, E., Guettier, C., Okyar, A., and Lévi, F. (2016). Clock Gene Per2 as a Controller of Liver Carcinogenesis. Oncotarget 7 (52), 85832–85847. doi:10.18632/oncotarget.11037
Narasimamurthy, R., Hunt, S. R., Lu, Y., Fustin, J. M., Okamura, H., Partch, C. L., et al. (2018). CK1δ/ε Protein Kinase Primes the PER2 Circadian Phosphoswitch. Proc. Natl. Acad. Sci. U S A. 115 (23), 5986–5991. doi:10.1073/pnas.1721076115
Pascussi, J. M., Dvorák, Z., Gerbal-Chaloin, S., Assenat, E., Drocourt, L., Maurel, P., et al. (2004). “Regulation of Xenobiotic Detoxification by PXR, CAR, GR, VDR and SHP Receptors: Consequences in Physiology,” in Transcription Factors. Editors M. Gossen, J. Kaufmann, and S. J. Triezenberg (Heidelberg: Springer, Berlin), Vol. 166, 409–435. doi:10.1007/978-3-642-18932-6_13
Pass, G. J., Carrie, D., Boylan, M., Lorimore, S., Wright, E., Houston, B., et al. (2005). Role of Hepatic Cytochrome P450s in the Pharmacokinetics and Toxicity of Cyclophosphamide: Studies with the Hepatic Cytochrome P450 Reductase Null Mouse. Cancer Res. 65 (10), 4211–4217. doi:10.1158/0008-5472.CAN-04-4103
Renaud, H. J., Cui, J. Y., Khan, M., and Klaassen, C. D. (2011). Tissue Distribution and Gender-Divergent Expression of 78 Cytochrome P450 mRNAs in Mice. Toxicol. Sci. 124 (2), 261–277. doi:10.1093/toxsci/kfr240
Richardson, S. J., Bai, A., Kulkarni, A. A., and Moghaddam, M. F. (2016). Efficiency in Drug Discovery: Liver S9 Fraction Assay as a Screen for Metabolic Stability. Drug Metab. Lett. 10 (2), 83–90. doi:10.2174/1872312810666160223121836
Rowland, A., Miners, J. O., and Mackenzie, P. I. (2013). The UDP-Glucuronosyltransferases: Their Role in Drug Metabolism and Detoxification. Int. J. Biochem. Cel Biol 45 (6), 1121–1132. doi:10.1016/j.biocel.2013.02.019
Sadagopan, N., Cohen, L., Roberts, B., Collard, W., and Omer, C. (2001). Liquid Chromatography-Tandem Mass Spectrometric Quantitation of Cyclophosphamide and its Hydroxy Metabolite in Plasma and Tissue for Determination of Tissue Distribution. J. Chromatogr. B Biomed. Sci. Appl. 759 (2), 277–284. doi:10.1016/s0378-4347(01)00243-2
Saravanakumar, A., Sadighi, A., Ryu, R., and Akhlaghi, F. (2019). Physicochemical Properties, Biotransformation, and Transport Pathways of Established and Newly Approved Medications: A Systematic Review of the Top 200 Most Prescribed Drugs vs. The FDA-Approved Drugs between 2005 and 2016. Clin. Pharmacokinet. 58 (10), 1281–1294. doi:10.1007/s40262-019-00750-8
Shu, W., Chen, L., Hu, X., Zhang, M., Chen, W., Ma, L., et al. (2017). Cytochrome P450 Genetic Variations Can Predict mRNA Expression, Cyclophosphamide 4-Hydroxylation, and Treatment Outcomes in Chinese Patients with Non-hodgkin's Lymphoma. J. Clin. Pharmacol. 57 (7), 886–898. doi:10.1002/jcph.878
Turpeinen, M., and Zanger, U. M. (2012). Cytochrome P450 2B6: Function, Genetics, and Clinical Relevance. Drug Metabol Drug Interact 27 (4), 185–197. doi:10.1515/dmdi-2012-0027
Van der Hoeven, T., Lefevre, R., and Mankes, R. (1992). Effects of Intrauterine Position on the Hepatic Microsomal Polysubstrate Monooxygenase and Cytosolic Glutathione S-Transferase Activity, Plasma Sex Steroids and Relative Organ Weights in Adult Male and Female Long-Evans Rats. J. Pharmacol. Exp. Ther. 263 (1), 32–39.
Vanselow, K., Vanselow, J. T., Westermark, P. O., Reischl, S., Maier, B., Korte, T., et al. (2006). Differential Effects of PER2 Phosphorylation: Molecular Basis for the Human Familial Advanced Sleep Phase Syndrome (FASPS). Genes Dev. 20 (19), 2660–2672. doi:10.1101/gad.397006
Xu, Y., Toh, K. L., Jones, C. R., Shin, J. Y., Fu, Y. H., and Ptácek, L. J. (2007). Modeling of a Human Circadian Mutation Yields Insights into Clock Regulation by PER2. Cell 128 (1), 59–70. doi:10.1016/j.cell.2006.11.043
Yamamura, Y., Yano, I., Kudo, T., and Shibata, S. (2010). Time-dependent Inhibitory Effect of Lipopolysaccharide Injection on Per1 and Per2 Gene Expression in the Mouse Heart and Liver. Chronobiol Int. 27 (2), 213–232. doi:10.3109/07420521003769111
Yan, R., Yang, Y., and Chen, Y. (2018). Pharmacokinetics of Chinese Medicines: Strategies and Perspectives. Chin. Med. 13, 24. doi:10.1186/s13020-018-0183-z
Yoo, S. H., Kojima, S., Shimomura, K., Koike, N., Buhr, E. D., Furukawa, T., et al. (2017). Period2 3'-UTR and microRNA-24 Regulate Circadian Rhythms by Repressing PERIOD2 Protein Accumulation. Proc. Natl. Acad. Sci. U S A. 114 (42), E8855–E8864. doi:10.1073/pnas.1706611114
Zani, F., Breasson, L., Becattini, B., Vukolic, A., Montani, J. P., Albrecht, U., et al. (2013). PER2 Promotes Glucose Storage to Liver Glycogen during Feeding and Acute Fasting by Inducing Gys2 PTG and G L Expression. Mol. Metab. 2 (3), 292–305. doi:10.1016/j.molmet.2013.06.006
Zhang, T., Yu, F., Guo, L., Chen, M., Yuan, X., and Wu, B. (2018a). Small Heterodimer Partner Regulates Circadian Cytochromes P450 and Drug-Induced Hepatotoxicity. Theranostics 8 (19), 5246–5258. doi:10.7150/thno.28676
Zhang, T., Zhao, M., Lu, D., Wang, S., Yu, F., Guo, L., et al. (2018b). REV-ERBα Regulates CYP7A1 through Repression of Liver Receptor Homolog-1. Drug Metab. Dispos 46 (3), 248–258. doi:10.1124/dmd.117.078105
Keywords: PER2, Cyp2b10, REV-ERBα, cyclophosphamide, drug metabolism
Citation: Chen M, Chen M, Lu D, Wang Y, Zhang L, Wang Z and Wu B (2021) Period 2 Regulates CYP2B10 Expression and Activity in Mouse Liver. Front. Pharmacol. 12:764124. doi: 10.3389/fphar.2021.764124
Received: 25 August 2021; Accepted: 03 November 2021;
Published: 23 November 2021.
Edited by:
David E. Stec, University of Mississippi Medical Center, United StatesReviewed by:
William Baldwin, Clemson University, United StatesCopyright © 2021 Chen, Chen, Lu, Wang, Zhang, Wang and Wu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Zhigang Wang, ZHJ3YW5nemdAc2luYS5jb20=; Baojian Wu, Ymoud3VAaG90bWFpbC5jb20=
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.