
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Pharmacol. , 19 November 2021
Sec. Ethnopharmacology
Volume 12 - 2021 | https://doi.org/10.3389/fphar.2021.761980
 Minfeng Zhou1†
Minfeng Zhou1† Jinxiao Li1†
Jinxiao Li1† Dan Luo2
Dan Luo2 Haiming Zhang3
Haiming Zhang3 Zhaomin Yu4
Zhaomin Yu4 Youlin Chen5
Youlin Chen5 Qiumeng Li6
Qiumeng Li6 Fengxia Liang7*
Fengxia Liang7* Rui Chen1*
Rui Chen1*Colorectal cancer (CRC) is one of the most pervasive cancers in the human disease spectrum worldwide, ranked the second most common cause of cancer death by the end of 2020. Prunus mume (PM) is an essential traditional Chinese medicine for the adjuvant treatment of solid tumors, including CRC. In the current study, we utilize means of network pharmacology, molecular docking, and multilayer experimental verification to research mechanism. The five bioactive compounds and a total of eight critical differentially expressed genes are screened out using the bioinformatics approaches of Cytoscape software, String database, Gene Ontology analysis, Kyoto Encyclopedia of Genes and Genomes pathways, and molecular docking. RelA has been proven to be highly expressed in CRC. Experiments in vitro have shown that kaempferol, the main active component of PM, dramatically inhibited the growth, migration, and invasion of CRC cells, and experiments in vivo have shown that PM effectively delays CRC formation and improves the survival cycle of mice. Further analysis shows that PM inhibits the CRC progression by down-regulating the expression level of RelA, Bax, caspase 3, caspase 9, and EGFR in CRC. PM and its extract are potentially effective therapeutics for the treatment of CRC via the RelA/nuclear factor κB signaling pathway.
1. Components of Prunus mume (PM) and differentially expressed genes (DEGs) of colorectal cancer (CRC) screening by intelligent network pharmacology and molecular docking.
2. Bioinformatics approaches of Cytoscape software, String database, Gene Ontology analysis, Kyoto Encyclopedia of Genes and Genomes pathways, and molecular docking applying to select the critical genes of PM-DEGs.
3. RelA proved to be highly expressed in CRC in human CRC tissue in vitro and in vivo.
4. First report on the comprehensive study to explore the molecular mechanism of PM in the treatment of CRC.
Colorectal cancer (CRC) without obvious symptoms expresses early the main symptoms of occult or overt rectal bleeding, anemia, change in defecation habits, or abdominal pain in an advanced period (Dekker et al., 2019). CRC had been ranked the second most common cause of cancer death in America by 2020. Morbidity and mortality rates were increasing recently by 1% and 1.3% per year among the population younger than 65 years (Siegel et al., 2020). As screening programs and colonoscopies increase, CRC morbidity and mortality are stabilized and decline in developed countries. Still, with progress in developing countries, the global incidence of CRC will continue to increase (Bray et al., 2018; Dekker et al., 2019). The CRC incidence continues to rise in countries with medium to high human development index (HDI) (Wong et al., 2021). Unreasonable diet structure, overweight, physical inactivity, smoking, ultraviolet radiation, and cancer-related infections are modifiable factors leading to the development of CRC (Islami et al., 2018). Endoscopic and surgical resection, preoperative chemotherapy, local metastatic ablation, palliative chemotherapy, local radiotherapy, targeted therapy, biologic therapy, and immunotherapy are current treatments for CRC treatment (Dekker et al., 2019). However, the risk of recurrence, metastasis, and severe side effects is inevitable. As adjunctive therapy, traditional Chinese herbal medicine (CHM) is expected to play a role in reducing toxicity and enhancing efficacy in the treatment of CRC.
Traditional Chinese medicine (TCM) is natural medicine, including plants, animals, and minerals. As most TCM are plants, TCM is also called CHM (Qi et al., 2020). CHM and their active components played a positive role in the treatment of multiple cancers (Wang et al., 2021), including combination therapy after first-line treatment for the non–small cell lung cancer (Wang et al., 2018), triple-negative breast cancer (Yang et al., 2021), liver cancer (Liu et al., 2018a), gastric cancer (Zhou et al., 2019), and CRC (Wang et al., 2021). As a representative herb vested in the homology of medicine and food, the safety of Prunus mume (PM) can be guaranteed. PM has the function of inhibiting a variety of inflammatory factors, anti-inflammation and antioxidation, repairing gastrointestinal tract, and regulating gastrointestinal movement and intestinal secretion (Xing et al., 2018). PM treated diarrhea and improved intestinal symptoms caused by lapatinib combined with capecitabine. PM plays anti-inflammatory effects by inhibiting MBP degradation and activating intracellular signals, including TLR4 and P38 MAPK, thus improving brain injury and becoming a potential drug for treating vascular dementia (Lee et al., 2015). Mumefural (MF), a component of PM, improved cognitive impairment in BCCAO rats by enhancing neurological function and inhibiting neuroinflammation (Bang et al., 2019). PM inhibits the proliferation and angiogenesis of endothelial cells in CRC (Cho et al., 2019). Isoquercitrin (IQ) is the main flavonol in PM fruits, which effectively inhibits the proliferation of SK-MEL-2 skin cancer cells. At the same time, the same concentration of IQ has no cytotoxic effect on human keratinocytes HaCaT (Won et al., 2020). However, very few studies have investigated the treatment of CRC with PM and its active ingredients roundly. Therefore, this article aims to explore the mechanism of PM in the treatment of CRC.
RelA/P65 is a member of the nuclear factor κB (NF-κB) transcription factor family, which is a class of proteins with specific DNA-binding activity that regulates a variety of functional target genes (Hoesel and Schmid 2013). NF-κB is a class of nuclear proteins that combine with the response element of immunoglobulin K enhancer to regulate the biological function of κB light chain in B cells (Sen and Baltimore 2006). External stimuli gather together to activate the upstream IκB kinase (IKK) complex, resulting in the release of NF-κBs into the nucleus to play a role in transcriptional regulation. Dimerization of NF-κB family members mediated by Rel homologous domain (RHD) is a necessary condition for DNA binding, and the up-regulation of gene expression depends on the transcriptional activation domain of RelA, C-REL, and RelB (Lu and Yarbrough 2015). Thus, the posttranslational modification of RelA determines the functional activity of NF-κB. Increased NF-κB activity can be attributed to activation pathways treated by mutated NF-κB genes or oncogenes and increased stimulators in the tumor immune microenvironment (Ben-Neriah and Karin 2011). Various adverse factors promote the high expression of NF-κB, which is a negative signal to the body, leading to tissue damage, immune disorders, and cancer (Staudt 2010). Activated NF-κB promotes CRC by accelerating cell proliferation and angiogenesis, inhibiting apoptosis, and promoting cell invasion and metastasis. The function of NF-κB in CRC is further complicated by the interaction of other signaling pathways. In short, NF-κB is involved in all stages of CRC development, from early adenoma to advanced cancer invasion and metastasis (Patel et al., 2018). It is also demonstrated that RelA is engaged in the whole process of CRC.
In this study, the mechanism of and its bioactive component in the treatment of CRC was investigated by means of network pharmacology, molecular docking, and experimental validation. Figure 1 is the flowchart of this study. The function of RelA and the role of the NF-κB pathway in the treatment of CRC were investigated. PM and its extract are expected to be an effective drug in the treatment of CRC.
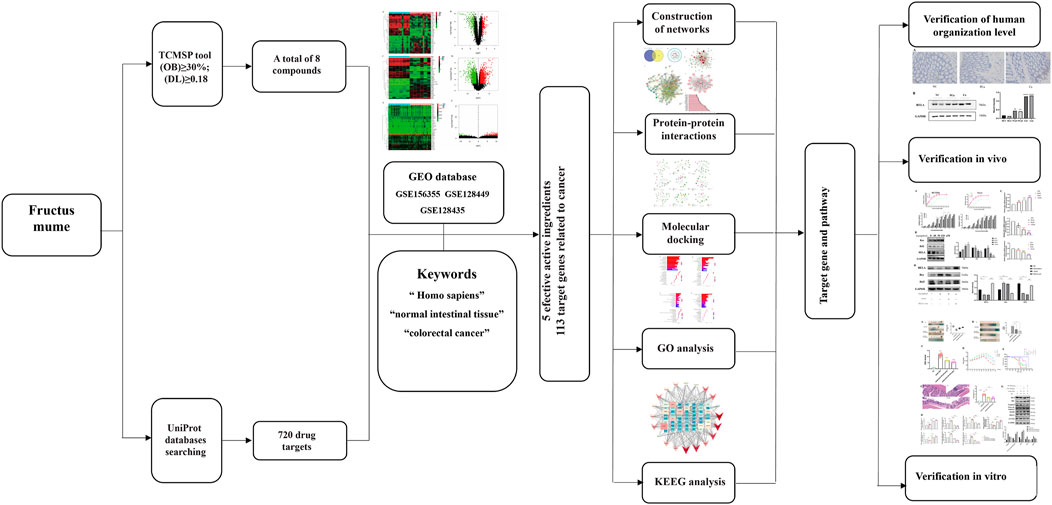
FIGURE 1. Network pharmacological, molecular docking, and experience verification workflow for identifying PM targets in the treatment of colorectal cancer.
We searched the critical word “Prunus mume” in the Traditional Chinese Medicine Database and Analysis Platform (TCMSP) website and the TCM Integrative Database (www.megabionet.org/tcmid/). The bioavailability (OB) >30% and drug similarity (DL) >0.18, main pharmacokinetic parameters, were used to screen the active components of PM using the pharmacokinetic indexes of absorption, distribution, metabolism, and excretion (ADME) (Liu et al., 2013; Shen et al., 2017; He R. et al., 2020). The OB value mainly reflects the proportion that a drug can overcome a series of barriers to be absorbed by the body and enter the blood circulation. The DL value mainly reflects the degree of similarity between the target molecule and the proven drug in composition, which can remove the components that are not suitable for the drug in chemical composition (Shen et al., 2017; He T. et al., 2020). The combination of the two values is used to screen drug components. Then, the target genes corresponding to the active ingredients of the drug were retrieved using the TCMSP (https://tcmsp-e.com/).
The index words, “Homo sapiens,” “normal intestinal tissue,” and “colorectal cancer,” were used as keywords in the GEO database. Genes expressed differently between CRC tissues and normal tissues were sorted out and classified into functional groups with high expression and low expression (GSE156355, GSE128449, and GSE128435). The obtained gene list was imported into the Uniprot database (http://www.uniprot.org/) to bring the name and gene ID (He R. et al., 2020).
Drugs’ active ingredients, the corresponding target genes of active ingredients, and disease screening target genes form the building “drug–disease” common target genes together, which build plum-visual networks of CRC target genes by Cytoscape 3.7.0 (http://www.cytoscape.org/). In the network diagram, nodes represent the active components or gene/proteins of PM, and interactions between nodes are represented by lines.
The “drug–disease” target genes were imported into the STRING biological database (https://string-db.org/) to study the target protein interaction relationship, also known as protein–protein interaction (PPI). All targets were limited to the species H. sapiens. In the PPI diagram, each solid circle depicts a gene, and the middle of the circle represents the protein structure. The circles are connected by different-color lines, and the strength of the interaction relationship is indicated by the number of connecting lines. At the same time, the bioinformatics technology was used to verify the protein interaction relationship again and draw the PPI interaction graph.
By bioinformatics technology, with its programming packets and Bioconductor high-throughput genomic data analysis (http://www.bioconductor.org/), Gene Ontology (GO) enrichment analysis and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analysis were performed to extract the biological functions of target proteins and cancer-related pathways. PM plays an anti-CRC role by regulating the biological functions of target proteins or related signaling pathways. At the same time, the histogram and bubble graph are drawn to visualize the data.
Using molecular docking technology to study the active components of PM and their related targets for CRC treatment can explain the mechanism of action and binding activity between the active components and target proteins to some extent. Compound structures in SDF format downloaded from the PubChem database (https://pubchem.ncbi.nlm.nih.gov/) were converted to a mol2 format file using Chem 3D software. The PDB format structures of BAX (PDB ID: 4S0O), BCL2 (PDB ID: 4LXD), CASP3 (PDB ID: 4PS0), CCND1 (PDB ID: 2W96), EGFR (PDB ID: 4I24), MAPK8 (PDB ID: 2XRW), RELA (PDB ID: 3QXY), and VEGFA (PDB ID: 1MKK) were downloaded from the RCSB Database (HTTPS://www.rcsb.org/), using Pymol software to remove solvent molecules and ligands, using AutoDock Tools 1.5.6 software to hydrogenate, calculate charge, assign atomic types, and so on and saving them as PDBQT format. Finally, Autodock Vina 1.1.2 was run for molecular docking, and Discovery Studio 2020 was used to analyze the conformation of docking visually. According to the data of intermolecular binding energy, hydrophilic and hydrophobic interaction, and electrostatic interaction, the score was given comprehensively, and then the intermolecular interaction was verified by next experiments.
The experiments involving human tissues have been carried out in accordance with The Code of Ethics of the World Medical Association (Declaration of Helsinki) for experiments involving humans and uniform requirements for manuscripts submitted to biomedical journals. All animal experiments should be carried out in accordance with the National Institutes of Health guide for the Care and Use of Laboratory animals (NIH publication no. 8023, revised 1978). All animal studies complied with the ARRIVE guidelines. All of the following tissue and animal experiments have passed the ethical review of Huazhong University of Science and Technology and Wuhan Union Hospital.
Plant extract takes plants as raw materials, and according to the needs of the final product extracted, it is obtained and concentrated through the physical and chemical extraction and separation process. A product formed by one or more of the active ingredients in a substance without changing the structure of the active ingredients. Wumei is extracted on a proportion, which is the powder made after the extraction and concentration of plants raw materials. The means of proportion is the mathematical proportion of the amount of raw materials before extraction and the product after extraction and concentration. Proportional extracts generally do not have a very specific composition and content. Wumei was extracted from 10 kg of raw material and condensed into 1 kg of powder product. PM was purchased from Chengdu Effa Biological Technology Co., Ltd. (Chengdu, China). Certificate of analysis is shown in Supplementary Figure 2. The species was identified by Prof. Bo Liu from Changchun University of Chinese Medicine (Changchun, China). PM was dried and extracted in 75% ethanol in a shake incubator (Shanghai Jiecheng Experimental Instrument Co., Ltd., Shanghai, China) overnight at room temperature and filtered with Whatman Quantitative Filter Paper. The extracts of PM (EPM) were concentrated using a rotary vacuum evaporator (GongyiYuhua instrument Co., Ltd., Gongyi, China).
The CRC cell lines of HCT116 and Lovo were purchased from the American Type Culture Collection (ATCC, Manassas, VA, United States). The culture media was Dulbecco modified eagle medium, supplemented with 10% fetal bovine serum. The cells were put in an incubator (37°C, 5% CO2). Cell lines were authenticated by short tandem repeat analysis using the GenePrint 10 System at the Australian Genome Research Facility and confirmed to be Mycoplasma-free using the Lookout Mycoplasma PCR Detection Kit.
The overexpression plasmid transfection of RelA and the negative control plasmid were designed and synthesized by Nanjing Norvizan Biological Technology Co., Ltd. (Nanjing, China). When the cells reached 70%–80% confluence, we performed siRNA transfection using Lipofectamine® RNAiMAX Transfection Reagen (Invitrogen, United States) strictly under the manufacturer’s instructions.
Sixty C57BL/6 wild-type age-matched (8–9 weeks) male mice (The Beijing Vitong Lihua Experimental Animal Technology Co. Ltd.) were used for this study. Forty-five mice were injected intraperitoneally (i.p.) with 12 mg/kg azoxymethane (AOM) and 3% dextran sulfate sodium (DSS). Starting at week 2, the mice were given 3% DSS three times a week (Tuesdays, Thursdays, and Saturdays) by intragastric administration for 1 week, followed by regular water for 2 weeks and repeated three cycles for 9 weeks. The rest of the 15 mice were injected i.p. with only 0.9% normal saline in week 1 served as untreated healthy controls. The AOM-injected mice were further subdivided into three groups of 15 mice. Group 1 (model group): Except for the above operation, the mice were given a regular diet; group 2 (PM 200 mg/kg group): except for the above operation, the mice were given 200 mg/kg PM three times a week (Monday, Wednesday, Friday) by intragastric administration for 9 weeks; group 3 (PM 400 mg/kg group): except for the above operation, the mice were given 400 mg/kg PM three times a week (Monday, Wednesday, Friday) by intragastric administration for 9 weeks. All mice were killed by cervical dislocation in week 10, and the number of macroscopic colonic tumors and the length of colorectum of the four groups were analyzed. Animal experiments were conducted following internationally recognized experimental animal management standards and approved by the Animal Ethics Committee of Huazhong University of Science and Technology (Wuhan, China) with ad libitum access to water and standard rodent diet.
The mice were maintained under controlled temperature and a 12-h light–dark cycle. All animal procedures were strictly carried out under the approved guidelines.
Cell viability was analyzed by Cell Counting Kit-8 (CCK8; Beyotime, Shanghai, China), according to the manufacturer’s protocols. Cells were seeded and cultured at a density of 5 × 103/well in 100 μL of the medium into 96-well microplates (Corning, United States). Then, the cells were treated with various concentrations of kaempferol (0, 15, 30, 60, 90, 120, 150, 240 μM). After treatment for 6, 12, 24, and 48 h; 10 μL of CCK-8 reagent was added to each well and then cultured for 2 h. All experiments were performed in triplicate. The absorbance was analyzed at 450 nm using a microplate reader (Bio-Rad, Hercules, CA, United States) using wells without cells as blanks. The proliferation of cells was expressed by absorbance.
Total protein was extracted from the tissues and cell lines using tissue extraction reagent and cell extraction buffer (Beyotime) purified and assessed qualitatively by Western blot (WB) analysis. Total protein extracts (30 µg) were shifted to 12% gradient sodium dodecyl sulfate–polyacrylamide gel electrophoresis gels to separate the proteins by different molecular weights and transferred onto nitrocellulose membranes for antigen–antibody reaction. The membranes were then blocked with 5% skimmed milk in TBST for 1 h and mixed with primary antibodies against RELA (1:1,000; ABclonal), Bax (1:1,500; ABclonal), Bcl2 (1:1,000; ABclonal), caspase 3 (1:1,000; ABclonal), cleaved caspase 3 (1:1,000; ABclonal), EGFR (1:1,000; ABclonal), CCND1(1:1,000; ABclonal), and GAPDH (1:1,000; ABclonal) and then incubated with goat anti-rabbit secondary antibodies (ABclonal) for 2 h at 37°C (Jiang et al., 2018).
Total RNA was extracted from the cancer tissues, matched normal tissues and cell lines. Total RNA was prepared using Trizol (Takara), and the cDNAs were generated by PrimeScript™ RT reagent kit according to the manufacturer’s instructions. The relative mRNA expression of GAPDH (mouse), RELA (mouse), Bax (mouse), Bcl2 (mouse), caspase 3 (mouse), cleaved caspase 3 (mouse), EGFR (mouse), CCND1(mouse), GAPDH (human), RELA (human), Bax (human) and Bcl2 (human) were executed using specific primers and the QuantiTest SYBR-Green polymerase chain reaction (PCR) kit (Norvizan Biological Technology Co., Ltd., Nanjing, China) on an Applied Biosystems 7500 real-time PCR machine (Applied Biosystems, Foster City, CA, United States). The GAPDH acted as a normalization control for all of the mRNAs listed above. The primers for qRT-PCR are shown in Table 1.
Formalin-fixed tissues were embedded in paraffin and sliced. The 4-mm-thick sections were then incubated with monoclonal antibodies against RELA (1:100; ABclonal) overnight at 4°C and then incubated with goat anti-rabbit secondary antibodies (Abcam, United States). 3,3′-Diaminobenzidine was used as chromogen. The sections were then dyed using hematoxylin and mounted. RELA expression was evaluated qualitatively by two independent laboratory technicians.
Based on three independent experiments, data are expressed as mean ± SEM, and statistical analyses were performed using SPSS 18.0 software, R software version 3.5.0 (R Foundation for Statistical Computing, Vienna, Austria), and GraphPad Prism version 8.0.1 (GraphPad Software Inc., United States). Significant differences were considered if the probability of the difference was <5 in 100 (p < 0.05).
Data of PM and differential gene expression (DGE) in CRC were obtained from TCMSP (https://old.tcmsp-e.com/tcmsp.php) and GEO (https://www.ncbi.nlm.nih.gov/geo/) databases. The 40 known components and 720 drug target indexes of PM were obtained by TCMSP database retrieval. A total of eight compounds were obtained from the TCMSP database under the screening conditions of OB ≥30% and DL ≥0.18 (Table 2). The corresponding genes were converted into GeneSymbol through the Uniprot database, and then duplicated and nonhuman targets were deleted; 170 drug targets were finally obtained. The two-dimensional and three-dimensional structures of protein molecules were retrieved by the PubChem database (https://pubchem.ncbi.nlm.nih.gov/) (Table 2). Differential genes were screened out by processing of three datasets, GSE164191, GSE110225, and GSE156355 in the GEO database. DGE analysis was conducted on the three datasets respectively, and the results are shown in Figure 2. A total of 10,009 genes were screened, among which 8,981 target genes were screened by removing duplicate, meaningless, and blank data.
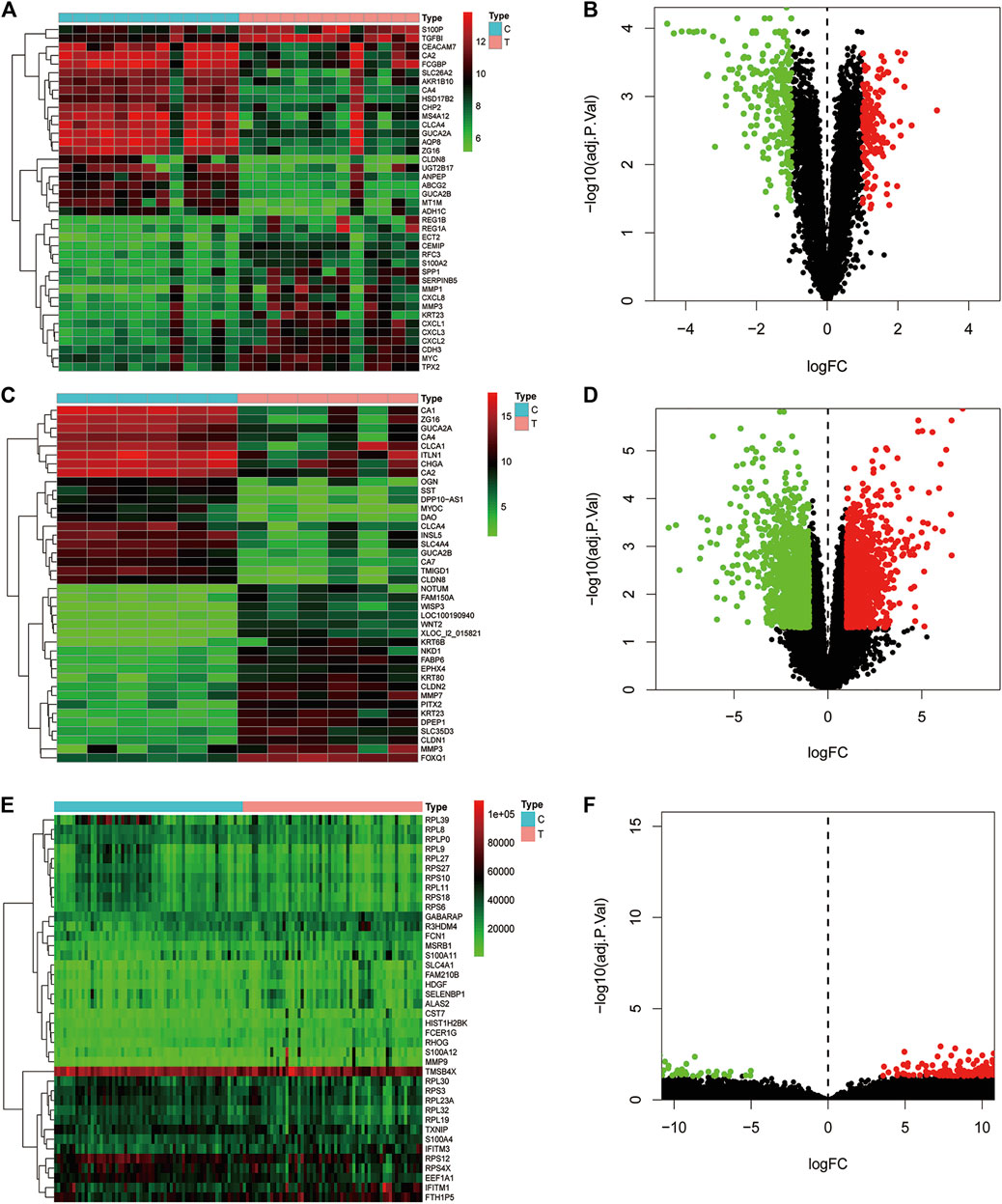
FIGURE 2. Analysis of differential gene expression in three datasets associated with colorectal cancer and normal tissue (GSE164191, GSE110225, and GSE156355) in GEO database. A total of 6,649, 406, 2954 differentially expressed genes were screened from normal tissues and colorectal cancer tissues, respectively. Heat maps and scatter maps were drawn according to different expression levels of differentially expressed genes, and some differentially expressed genes were selected for display. In the figure, red represents high expression, and green represents low expression. (A, B) Heat map and scatter map of differential genes in GSE164191 dataset. (C, D) Heat map and scatter diagram of differential genes in GSE110225 dataset. (E, F) Heat map and scatter diagram of differential genes in GSE156355 dataset.
A total of 170 PM targets and 8,981 CRC DGEs were comapped, common targets were obtained through the Venny 2.1 website, and Venn diagrams were drawn (Figure 3A). Through the docking and integration of drug targets and disease-related targets, a total of 87 drug–disease key targets were screened out. Eight active ingredients and 87 drug–disease targets were imported into Cytoscape 3.8.2 software to visualize the gene network relationship between PM active ingredients and target genes (Figure 3B). As shown in Figure 3B, there are 93 nodes and 130 relation lines; The red rhomboid nodules are the active components of PM. The light blue oval node is the target gene of drug disease. Each active ingredient acts on at least one target gene, and each gene is regulated by at least two active ingredients, indicating the multitarget action characteristics of multiactive components of PM. The active ingredients with the most target genes in PM are Mol000098 (quercetin), Mol000422 (kaempferol), Mol000358 (β-sitosterol), and MOL000449 (stigmasterol). This study speculated that these active ingredients might be the fundamental constituents for PM treatment of CRC.

FIGURE 3. Construction of PM active ingredient—target gene network in colorectal cancer and screening of key molecules. (A) Venn diagrams of drug and disease targets. After screening and repeating, blue represents only the drug target, yellow represents only the disease target, and the middle color represents the intersection of the two targets. (B) Construction of drug active components—target gene network. (C) Target protein interaction network analysis (PPI) in STRING database. (D) Target protein interaction network analysis (PPI) in Cytoscape 3.8.2 software. (E) The top 25 genes with higher expression differences were obtained by PPI screening. (F) Protein interaction relationship histogram of PM.
The STRING database was utilized to construct PM target–CRC DGE network. PM-CRC target genes were imported, the species selected was H. sapiens with a combined score of ≥0.4, and the network diagram of protein interaction was obtained (Figure 3C). As shown in Figure 3C, there are 166 different-color circles, each representing all the proteins encoded by a target gene, 2,361 interacting relationship lines. Each line represents a protein–protein association, representing that two proteins interact functionally, but not necessarily bind structurally. Meanwhile, Cytoscape 3.8.2 software was used to analyze further and score the PM target–CRC DGE network, and the molecular network diagram as shown in Figure 3D was obtained. Statistical analysis was carried out on each target gene, and the top 30 proteins with the most significant number of adjacent genes were selected for statistical analysis (Figure 3E). The proteins whose connection degree is for more than 40 contain VEGFA, CASP3, EGF, EGFR, MYC, MAPK8, FOS, CCND1, and MMP9 (Figure 3F). Other molecules with high connectivity obtained HIF1A, RelA, and Bcl2 through PPI screening (Figure 3E). Genes with more linked proteins play a more critical role in the network, play a more pronounced role in PM-CRC correlation, and are more likely to become therapeutic targets of drugs acting on diseases.
GO annotation analysis showed that drug–disease target proteins were mainly involved in protein heterodimerization activity, DNA-binding transcription factor binding, and oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen and ubiquitin-like protein ligase–binding functional activities (Figures 4A–C). Meanwhile, KEGG enrichment analysis showed many target genes were closely related to lipid and atherosclerosis, chemical carcinogenesis-receptor activation, human cytomegalovirus infection, Kaposi sarcoma–associated herpesvirus infection, and otitis B (Figure 4D). Drug therapy often works at the cellular and molecular levels through a network of multiple pathways and multiple targets. Among these potential targets, RELA (RELA proto-oncogene, NF-κB subunit), CASP3, CASP9, BAX, BCL2, CCND1, and CDKNIA were screened as relatively high-level targets (Figure 5), which played an essential role in the inhibition effect of PM on CRC. We hypothesized that PM plays an inhibitory role in CRC by regulating RELA and above other molecules. That NF-κB pathway is involved in the treatment of CRC by PM, which may play an antitumor role by promoting the apoptosis of tumor cells.
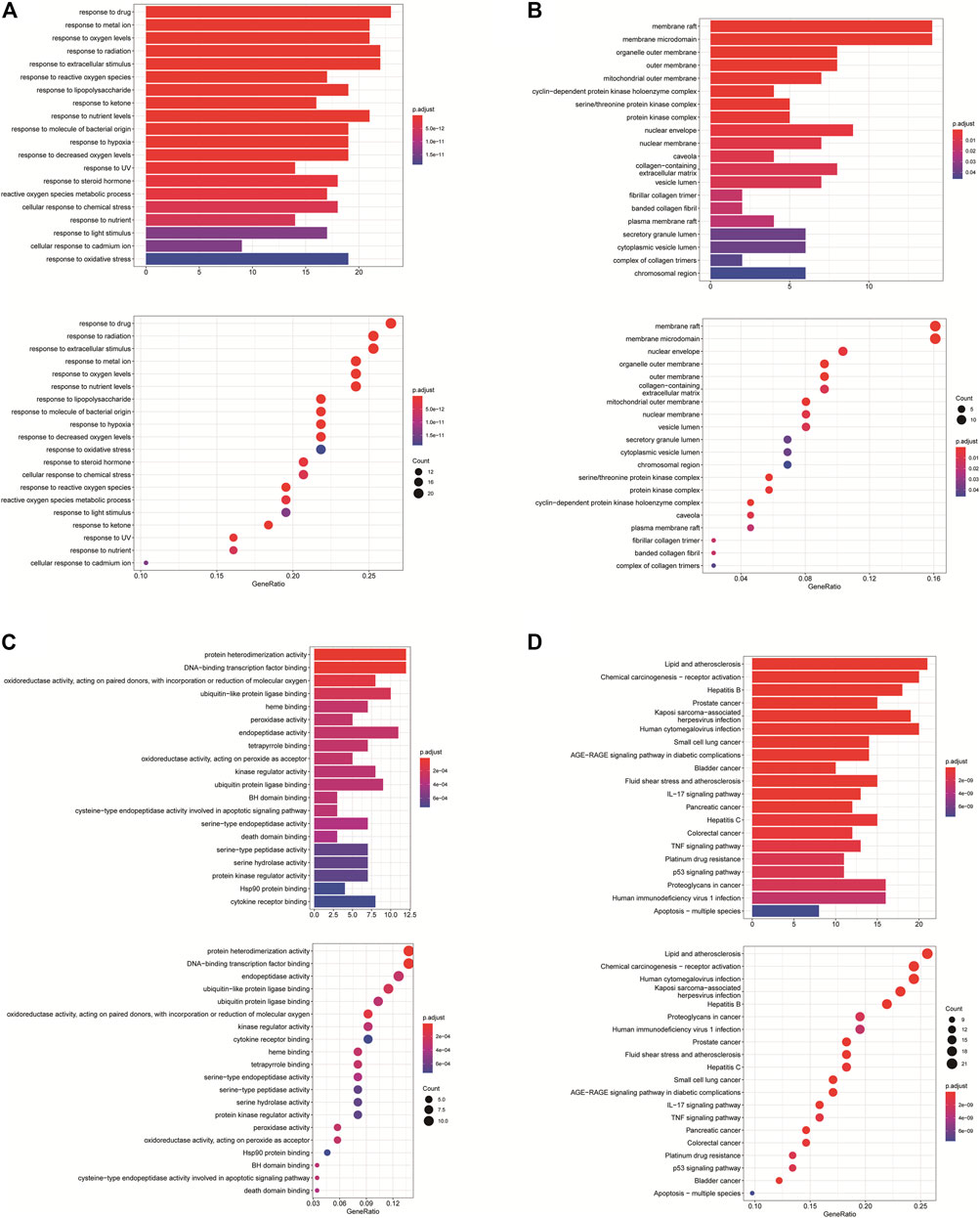
FIGURE 4. Enrichment analysis of Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway biological process of anticolorectal cancer targets gene from active ingredients of PM. The color scale indicates the p value, and the dot size indicates the gene count in each term. (A) GO analysis of multicellular organismal development (BP), mitotic G1 DNA damage checkpoint, G1 DNA damage checkpoint, and mitotic G1/S transition checkpoint were screened out. (B) GO analysis of intracellular membrane-bounded organelles (CC), cyclin-dependent protein kinase holoenzyme complex, serine/threonine kinase complex and protein kinase complex were screened out; (C) GO analysis of protein binding (MF). (D) KEGG analysis, CXCR chemokine receptor binding, and sulfur compound binding were screened out.
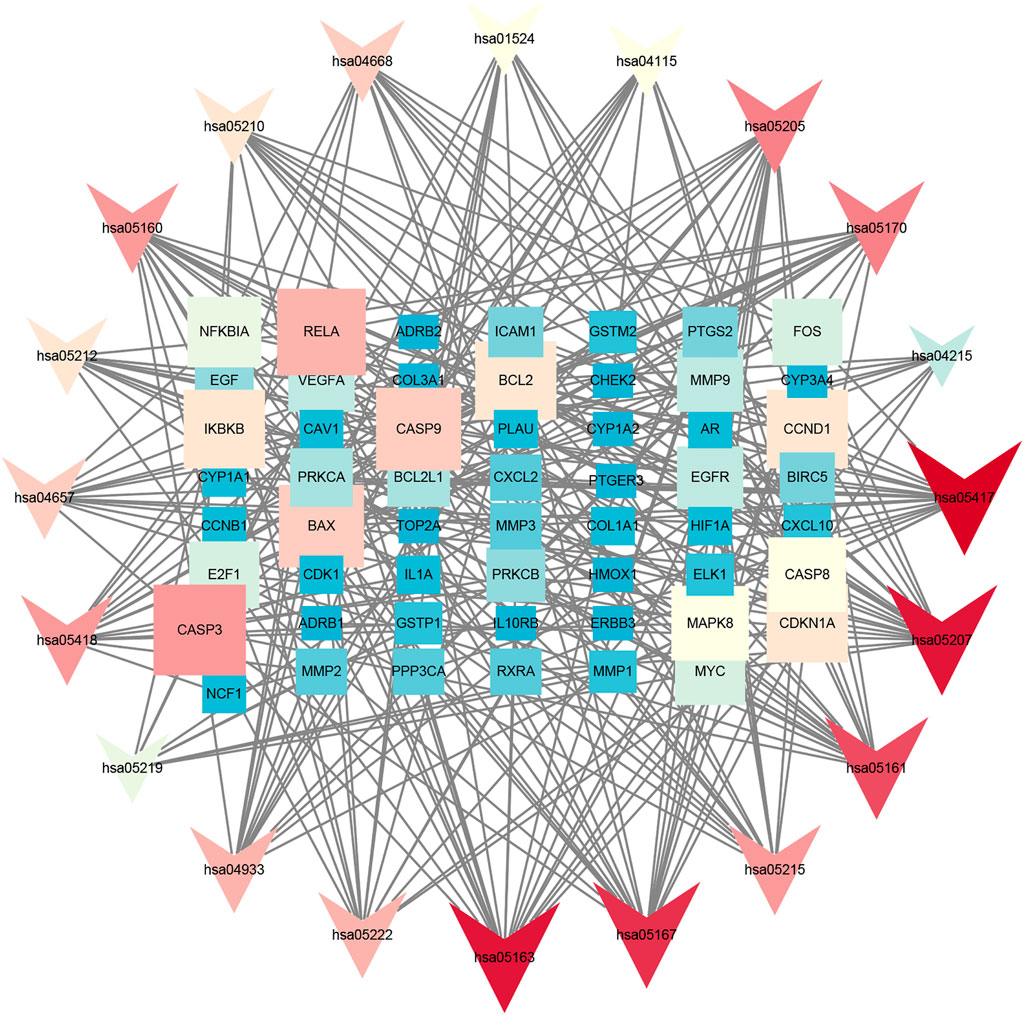
FIGURE 5. Kyoto Encyclopedia of Genes and Genomes (KEGG) analysis in Cytoscape 3.8.2 software. The KEGG screening was conducted using Cytoscape 3.8.2 software. The association network between target genes and pathways was constructed. The target genes and KEGG pathways were scored according to the connectivity degree. A total of eight high expression of target protein include CASP3, BAX, RELA, EGFR, BCL2, CCND1, MAPK8, and VEGFA. The pathway involved in this process mainly includes the apoptosis pathway by KEGG analysis.
Molecular docking was used to verify if the top five compounds had a significant role in regulating above eight DGEs. The interaction was scored, and the results are shown in Table 3. According to the scoring results, stigmasterol-BAX, stigmasterol-CASP3, stigmasterol-EGFR, stigmasterol-MAPK8, kaempferol-RELA, kaempferol-CCND1, quercetin-CCND1, quercetin-VEGFA, and CLR-BCL2 had a strong correlation, among which kaempferol-RELA had the highest correlation score. Further experiments are needed to verify the strong relationship in CRC. The molecular docking results are shown in Figure 6, which shows that all the essential compounds in the network had a strong affinity with corresponding DGEs.
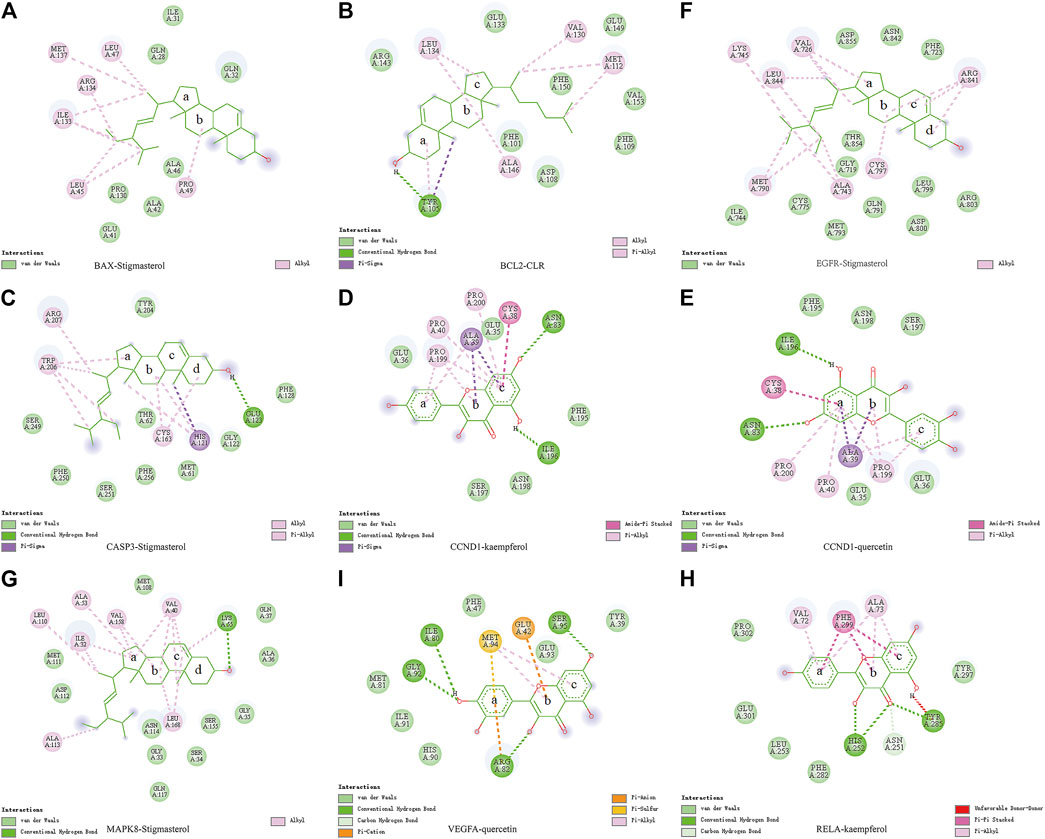
FIGURE 6. Binding of PM compounds in interaction with target proteins. The bingding mode of PM compounds, including stigmasterol, CLR, kaempferol, and quercetin in the proteins (A) BAX, (B) BCL2, (C) CASP3, (D,E) CCND1, (F) EGFR, (G) MAPK8, (H) RELA, and (I) VEGFA.
NF-κB is a crucial transcription factor in the inflammatory tumor microenvironment, and RelA/P65 is center to the activation of Wnt signaling by NF-κB, which is one of the center mechanisms of tumor cell initiation in CRC (Schwitalla et al., 2013). To verify the expression of RelA in colon cancer tissue, we conducted immunohistochemical analyses in average, paracancer, and colon cancer tissue samples from the patients. The presentation of RelA in colon cancer specimens was significantly higher than that in paracancer and standard specimens (Figure 7A). We randomly selected two groups from 16 groups of tissue samples provided by the Department of Gastrointestinal Surgery of Wuhan Union Hospital for a WB test. WB result and gray value statistics of RELA in human colon tissue samples showed that RELA is highly expressed in colonic adenocarcinoma tissues (Figure 7B). The above results indicate that RELA is highly expressed in human colon tissue, and RELA can be used as a marker of CRC formation.
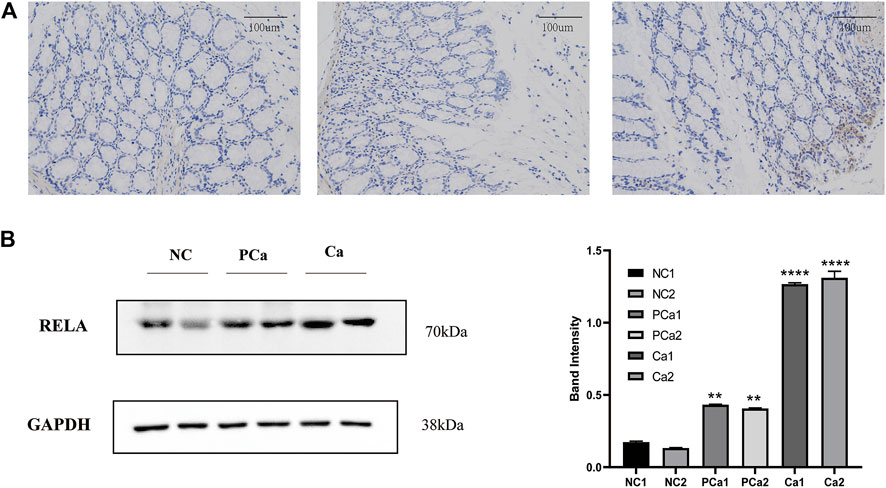
FIGURE 7. The RELA molecule was highly expressed in human colorectal cancer tissue samples. (A) Images depicting immunohistochemical (IHC) staining of RelA conducted in tissue sections derived from human normal colon, paracarcinoma tissues, and colon cancer samples. Images were captured at ×200 magnification. (B) Western blot and gray value statistics of RelA in human colon tissue samples. NC, normal colon of the patient; PCa, paracarcinoma tissues of the patient; Ca, patient colon cancer tissue. *p < 0.05 vs. negative control (NC) group, **p < 0.02 vs. NC group, ***p < 0.01 vs. NC group, ****p < 0.001 vs. NC group.
To further explore the effect of PM extract on CRC in vivo, we established the AOM/DSS–induced mouse CRC model and treated the mice with PM simultaneously. After 11 weeks, the mice in each group were sacrificed to obtain the entire colorectal tissue from the cecum to the end of the anus, conducting experimental validation at the organizational level. The results showed that the length of the CRC was significantly shortened in the AOM/DSS–induced model, and the shortening of the intestinal tract was improved considerably in the PM-treated group compared to that of the untreated model group. However, the improvement of colonic length was not apparent with the increasing concentration of PM extract (Figure 8A). The number of tumor formation per unit area of intestinal tissue is the indicator to measure the effect of modeling and treatment. PM extract significantly reduced the number of tumor formation per unit area (p < 0.05) and inhibited tumor formations in macroscopic perspective (Figure 8B). At the same time, the higher the concentration of PM extract, the more pronounced the inhibition of tumor formation, but the safety of a high concentration of PM extract for the body needs to be further verified. Compared with the control group, DAI (Disease Activity Index) score was significantly decreased (p < 0.01) (Figure 8C), weight was significantly increased (p < 0.01) (Figure 8D), and survival period was prolonged (p < 0.01) (Figure 8E) in the PM extract treatment group, which had not improved in the high-concentration group. The colon tissues were taken for hematoxylin-eosin staining. The results showed that the model group lost the normal tissue morphology, the intestinal villi were destroyed, and there were apparent nuclear enlargement, nuclear aggregation, and nuclear shrinkage. Treatment with PM extract restored part of the damaged intestinal structure and improved the above pathological changes (Figure 8F). Western blotting and quantitative PCR (qPCR) were performed to examine the expression levels of key regulators responsible for apoptosis and core molecules screened through network pharmacology. As shown in Figures 8G,H , PM extract inhibited the expression of RelA, Bcl2, caspase 3, and CCND1 and promoted apoptosis-related proteins Bax, cleaved caspase 3, and EGFR. These results confirmed that PM extract is mainly involved in inhibiting CRC through apoptosis-related pathways, especially the NF-κB pathway in which RelA is engaged. RelA is a vital molecule regulated by PM, and the NF-κB pathway involved by RelA is a momentous mechanism of PM’s treatment of CRC.
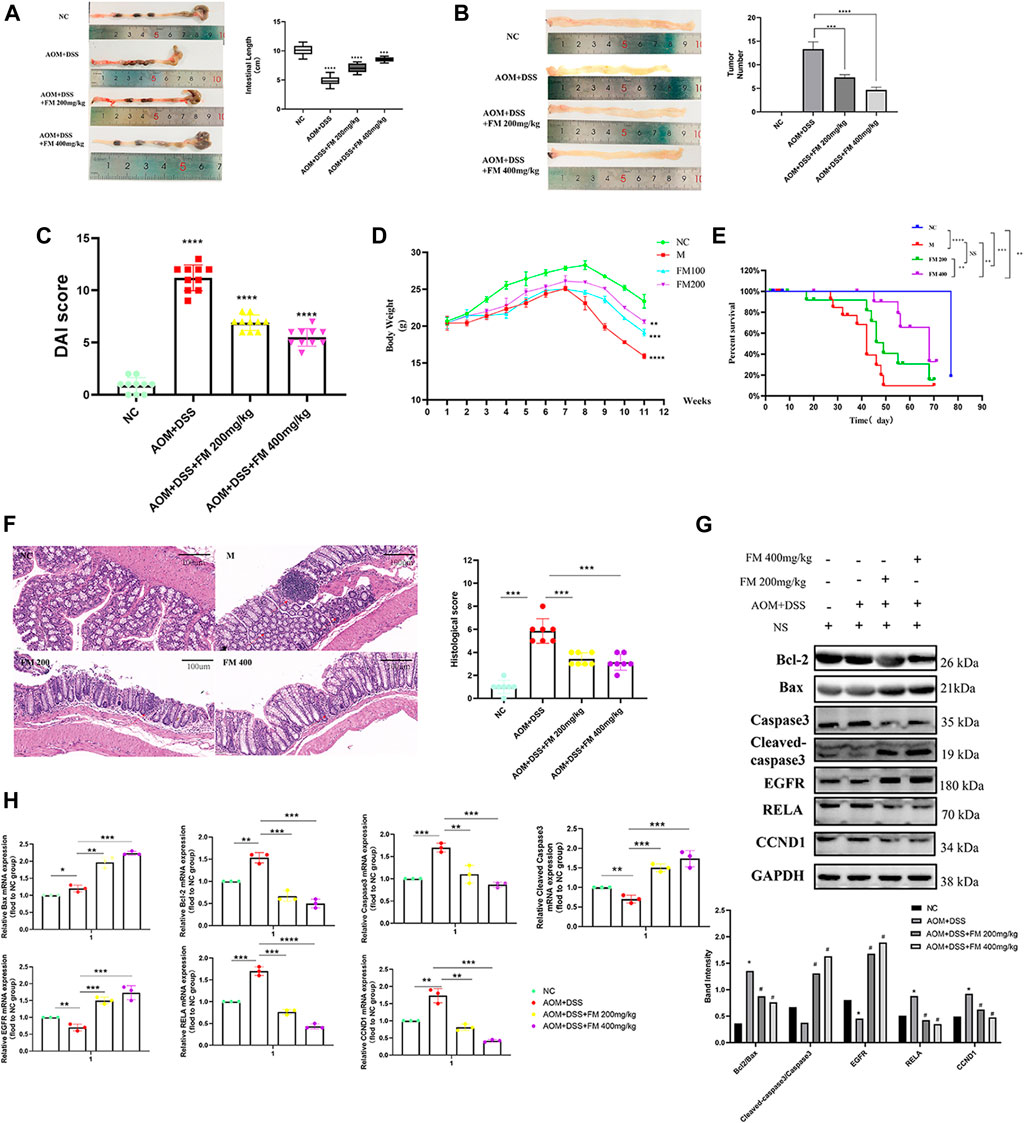
FIGURE 8. PM inhibits the progression of colorectal cancer in mice by promoting apoptosis. (A) PM improved intestinal shortening in colon cancer model mice. (B) PM inhibits intestinal tumorigenesis in colon cancer model mice. (C) PM improved intestinal DAI score in colon cancer model mice. (D) PM slowed weight loss in colon cancer model mice. (E) In survival analysis, PM improved survival in colon cancer model mice. (F) Hematoxylin-eosin staining experiment verified that PM improved the intestinal structure destroyed by AOM and DSS and inhibited the development of tumor tissues. The tumor inhibition effect of 400 mg/kg was higher than 200 mg/kg. (G) Western blot and (H) qPCR assays confirmed that PM inhibition of colon cancer in mice was mediated by apoptosis-related pathways. NC, normal group; M, model group, AOM + DSS; PM2, AOM + DSS + PM 200 mg/kg; PM4, AOM + DSS + PM 400 mg/kg. *p < 0.05 vs. negative control (NC) group, **p < 0.02 vs. NC group, ***p < 0.01 vs. NC group, ****p < 0.001 vs. NC group.
Based on the above molecular docking results and previous literature reports, RelA was a potential target gene of kaempferol, one of the active components of PM, for the treatment of ulcerative colitis (Wei et al., 2020). Kaempferol could improve virus-induced acute lung injury through the NF-κB signaling pathway (Zhang et al., 2017). Therefore, we hypothesized that kaempferol might be involved in PM regulation of RelA, thus affecting the activity of CRC cells. Cell viability assays were performed on two human colon cancer cell lines HCT116 and Lovo, as shown in Figure 9A. These findings demonstrated that the activity of kaempferol on colon cancer cells was time- and dose-dependent. There was no difference between the two cell lines in IC50 value, which after 24 h of treatment was approximately 60 µM for the two cell lines. Moreover, after treating colon cancer cell HCT116 at concentrations of 60, 90, and 120 µM for 24 h by kaempferol, the expressions of RelA and apoptosis-related proteins Bax and Bcl2 were detected by WB. The results showed that kaempferol could inhibit the expression of RelA in colon cancer cell HCT116 (Figure 9B). At the same time, the effect of promoting apoptosis of tumor cells was more evident with the increase of the concentration. At the gene expression level, the expression of RelA and apoptosis-related proteins Bax and Bcl2 was detected by qPCR, and similar protein levels were obtained (Figure 9C). The above results only proved that kaempferol could change the expression of RELA in colon cancer cells, but did not indicate that kaempferol had a regulatory relationship with RELA. Therefore, we completed the cell transfection experiment to transfect the RELA-specific overexpression plasmid into HCT116 cells, and the results showed that in the case of RelA overexpression, the inhibitory effect of kaempferol on RelA was decreased, and the ability to induce apoptosis of tumor cells was weakened, which proved that kaempferol could regulate the expression of RelA, and RelA was the downstream target of kaempferol (Figure 9D). Kaempferol inhibits the expression of RelA, a crucial molecule in the NF-κB pathway, thereby promoting the apoptosis of colon cancer cells and inhibiting tumor growth.
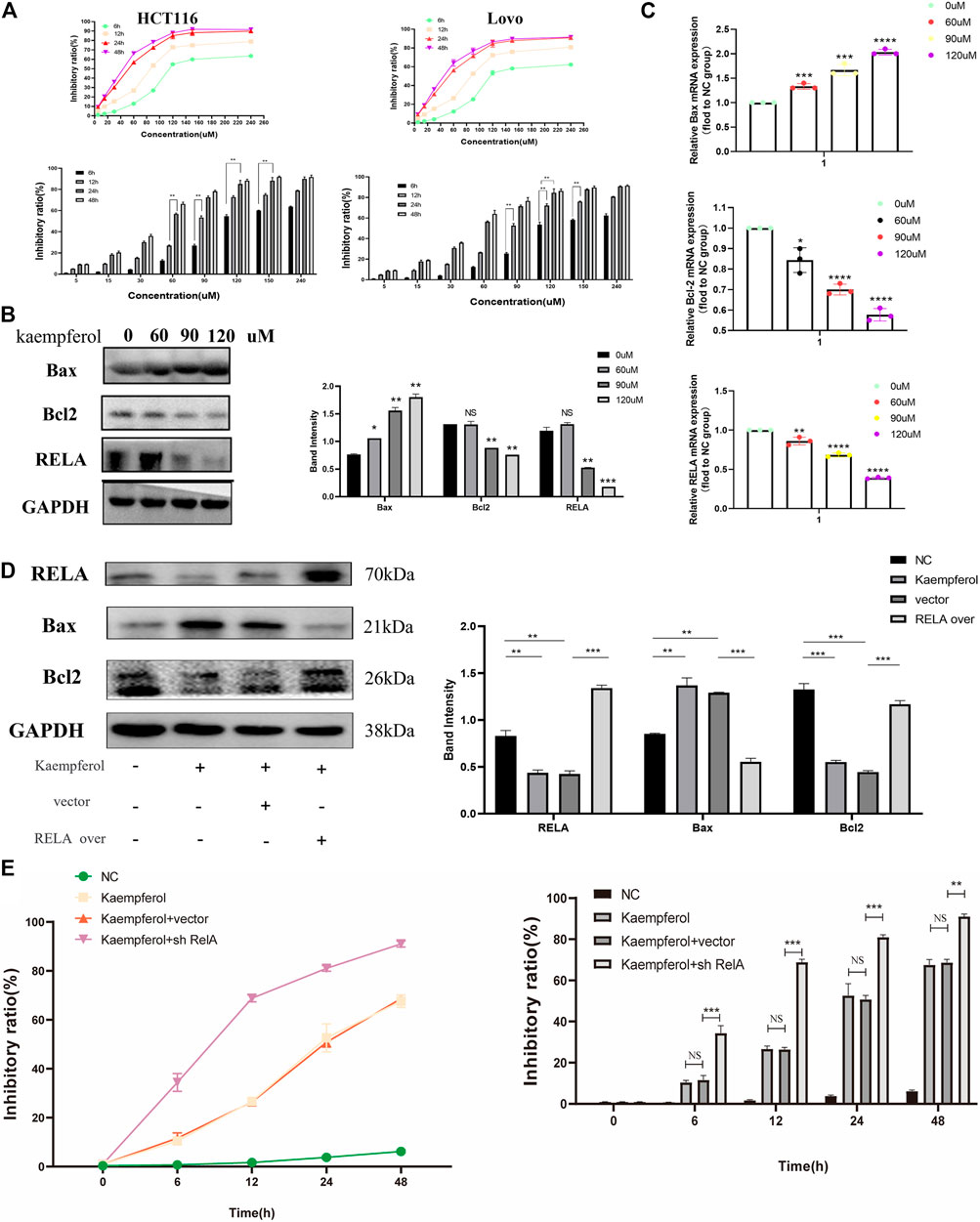
FIGURE 9. Kaempferol, an important active component in PM, inhibits the growth of colon cancer cells by promoting RELA-related apoptotic pathways. (A) CCK8 assay verified the effect of kaempferol on HCT116 and Lovo human colon cancer cell lines. Eight concentration gradients of 5, 15, 30, 60, 90, 120, 150, and 240 µM were implemented, and the results showed that the inhibition rates of the two cell lines were similar. The cells were significantly inhibited at the concentrations of 60, 90, and 120 µM. (B) Western blot assay and (C) qPCR assay verified that kaempferol regulated the expression of Bax, Bcl2, and RELA in HCT116 cells at concentrations of 60, 90, and 120 µM, respectively. RELA was significantly inhibited with the increase of the concentration. Apoptosis was increased. (D) After overexpressed RELA plasmid and no-loaded plasmid were transfected, the effects of kaempferol on the expression of RELA, Bax, and Bcl2 in HCT116 cell lines were confirmed by Western blotting. (E) After sh-RELA plasmid and no-loaded plasmid were transfected, the effects of kaempferol on the cell viability of HCT116 cell lines were confirmed by CCK8. *p < 0.05 vs. negative control (NC) group, **p < 0.02 vs. NC group, ***p < 0.01 vs. NC group, ****p < 0.001 vs. NC group.
In this study, we found that PM possesses an adequate therapeutic effect on CRC, primarily by inhibiting RelA expression and NF-κB pathway from promoting tumor apoptosis. On the GEO database, differentially expressed genes (DEGs) in CRC were identified. Simultaneously, we detected the bioactive components (β-sitosterol, kaempferol, stigmasterol, methyl arachidonate, and quercetin) and main target proteins related to CRC in PM using the TCMSP database. Subsequently, we further screened essential genes and core-dependent pathways through the PPI network, GO analysis, and KEGG analysis. Ultimately, the eight genes (RELA, CASP3, BAX, EGFR, BCL2, CCND1, MAPK8, and VEGFA) with the highest differential expression within the constraints and the apoptosis-associated pathway were identified. Then, we performed molecular fitting scoring and molecular docking for the eight genes and five bioactive components. The outcomes indicated that RELA and kaempferol were genes and bioactive compounds, respectively, with the best affinity and reasonable degree in PM for CRC treatment. The NF-κB transcription factors are known to consist of five homologous subunits (RelA/p65, C-rel, RelB, P50/NF-κB1, and P52/NF-κB2), which are usually dimerized and controlled by NF-κB inhibitors (IκBs) in the cytoplasm (Naugler and Karin 2008). Nuclear localization of RelA/P50 dimer is one of the regulatory factors of NF-κB canonical pathways, which is closely related to inflammatory tumorigenia (Annunziata et al., 2007; Keats et al., 2007). RelA had been used as an evaluator to evaluate the role of NF-κB activation in various tumors, including lung adenocarcinoma (He T. et al., 2020), breast cancer (Li et al., 2020), liver cancer (Moles et al., 2016), CRC (Liu et al., 2018b; Xu et al., 2019; Wang S. et al., 2020), and so on. In intestinal diseases, NF-κB is involved in the early formation of colorectal adenoma and may contribute to adenoma canceration (Shaked et al., 2012). At the mechanic level, RelA/NF-κB plays a crucial role in colorectal tumor–related processes such as angiogenesis, cell proliferation, apoptosis, autophagy, and metastasis (Ben-Neriah and Karin 2011). NF-κB represented antiapoptotic activity mediated by cFLIP, cIAP2, Bcl-XL, and Bcl2 and other antiapoptotic genes (Naugler and Karin 2008). Iκβ-mediated NF-κB activity had a cell-specific role in the development of colitis-associated cancers. NF-κB played a role in the early development of precancerous adenomas, which may be a potential target for clinical drug development and treatment (Greten et al., 2004; Hayakawa et al., 2009). Consequently, we hypothesized that PM inhibited the occurrence and growth of tumors by inhibiting the expression of RELA/NF-κB and the apoptosis-associated pathway.
The feasibility of PM in the treatment of human CRC from the perspective of morphology and protein expression was further proven (Figure 7), by verifying that the expression of RelA in human colon cancer tissues was indeed higher than that in normal tissues and paracancer tissues. Then we explored the potential mechanism of PM in the treatment of CRC, as it is well-known that inducing tumor cell apoptosis is a crucial therapeutic approach in the treatment of tumors. After applying kaempferol, apoptosis of HCT116 cells was significantly increased, and the expression of apoptosis-related proteins was increased.
The comparison results between the kaempferol treatment group and the RelA over with kaempferol treatment group showed that PM mainly affected apoptosis by inhibiting RelA. Our study showed that RelA overexpression could reduce the therapeutic effect of kaempferol, inhibit the expression of Bax, and promote the expression of Bcl2. Therefore, we can demonstrate that kaempferol, the bioactive component of PM, can promote the apoptosis of colon cancer cells by inhibiting RelA, thus playing a role in treating cancer. Simultaneously, we conducted experiments in mice to verify that PM improved the symptoms of CRC model mice and participated in regulating the expression of RelA and apoptosis-related proteins. PM extract inhibited the expression of RelA, Bcl2, caspase 3, and CCND1 and promoted apoptosis-related proteins Bax, cleaved caspase 3, and EGFR. These results were consistent with the predictions of network pharmacology. In addition, the binding force between a drug and its target protein is an important indicator to evaluate its mechanism of action on disease (Schmidt et al., 2010). We investigated the docking of five bioactive ingredients and eight proteins through molecular docking, and the results showed that kaempferol-RelA had the best docking effect. These results confirmed that PM extract is mainly involved in inhibiting CRC through apoptosis-related pathways, especially the NF-κB pathway in which RelA is engaged.
CHM was also verified as a regulator to improve the function of the immune system and inhibit tumor progression (Wang Z. et al., 2020). Many scholars believe that the bioactive components play a major role in anticancer. Among bioactive compounds of PM, β-sitosterol, kaempferol, stigmasterol, methyl arachidonate, and quercetin showed strong pharmacological effects. β-Sitosterol, kaempferol, and stigmasterol had obvious antioxidation, anti-inflammatory, antitumor, immunoregulation, and metabolic regulation activities (Wilt et al., 2000; Bin Sayeed and Ameen 2015; Devi et al., 2015; Eid and Haddad 2017; Imran et al., 2019; Reyes-Farias and Carrasco-Pozo 2019; Babu and Jayaraman 2020; Bae et al., 2020; Vo et al., 2020). β-Sitosterol interferes with multiple signaling pathways such as cell cycle, proliferation, apoptosis, inflammation, invasion, metastasis, and angiogenesis (Bin Sayeed and Ameen 2015). In terms of antitumor, β-sitosterol induced apoptosis by activating caspase-3 and caspase-9 and showed cytotoxic effects on HepG2 and Huh7 cells, but did not have an effect on human primary hepatic fibroblasts (Vo et al., 2020). Kaempferol reduced the risk of skin cancer, liver cancer, and colon cancer by blocking the cell cycle in the G2/M phase, down-regulating the related markers of epithelial–mesenchymal transition and accelerating cell necrosis and apoptosis (Imran et al., 2019). Stigmasterol had toxic effects on various tumors and restrained ES2 and OV90 cells by inhibiting cell migration and angiogenic genes in human ovarian cancer cells (Bae et al., 2020). Quercetin played an anticancer role by inducing autophagy, apoptosis, and inactivation of cells through MAPK/Erk1/2, PI3K/Akt/mTOR and Wnt/-catenin pathways (Reyes-Farias and Carrasco-Pozo 2019). Therefore, there is a theoretical basis for PM and its bioactive ingredients to inhibit the occurrence and development of CRC.
Combined with the results of network pharmacology, molecular docking, and experimental verification of cell and animal and human tissue samples at three levels, PM promotes colorectal cell apoptosis and inhibits the development of CRC mainly by inhibiting the expression of RelA. Among the five bioactive ingredients associated with CRC in PM, kaempferol and RelA had the highest molecular docking level. We also verified the effect of kaempferol on RelA and CRC cells. However, there are also limitations. First, PM is a natural botanical medicine containing many components, so pharmaceutical composition identification should be conducted. Second, other bioactive ingredients of PM should also be verified by experiments. Finally, it is necessary to further explore the direct regulation of bioactive ingredients on RelA.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://www.ncbi.nlm.nih.gov/, GSE110225; https://www.ncbi.nlm.nih.gov/, GSE156355; https://www.ncbi.nlm.nih.gov/, GSE164191.
The animal study was reviewed and approved by Ethics Committee of Union Hospital affiliated to Tongji Medical College of Huazhong University of Science and Technology.
ZMF designed the study and completed the bioinformatics part, tissue experiment, and the cell experiment part, while ZMF and LJX completed the animal experiment. All authors were involved in drafting the manuscript and revising it before final submission.
This work was supported by Natural Science Foundation of China, Grant No. 81774401.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
We would like to thank everyone who contributed to this article. We also would like to acknowledge the reviewers for their helpful comments on this paper.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fphar.2021.761980/full#supplementary-material
TCM, Chinese traditional medicine; CRC, colorectal cancer; PM, Prunus mume; TCMSP, TCM Systematic Pharmacology Database; GO, biological functions of gene ontology; KEGG, Kyoto Encyclopedia of Genes and Genomes; CHM, Chinese herbal medicine; MF, mumefural; IQ, isoquercitrin; IKK, the upstream IκB kinase; RHD, Rel homologous domain.
Annunziata, C. M., Davis, R. E., Demchenko, Y., Bellamy, W., Gabrea, A., Zhan, F., et al. (2007). Frequent Engagement of the Classical and Alternative NF-kappaB Pathways by Diverse Genetic Abnormalities in Multiple Myeloma. Cancer Cell 12 (2), 115–130. doi:10.1016/j.ccr.2007.07.004
Babu, S., and Jayaraman, S. (2020). An Update on β-sitosterol: A Potential Herbal Nutraceutical for Diabetic Management. Biomed. Pharmacother. 131, 110702. doi:10.1016/j.biopha.2020.110702
Bae, H., Song, G., and Lim, W. (2020). Stigmasterol Causes Ovarian Cancer Cell Apoptosis by Inducing Endoplasmic Reticulum and Mitochondrial Dysfunction. Pharmaceutics 12 (6). doi:10.3390/pharmaceutics12060488
Bang, J., Kim, M. S., and Jeon, W. K. (2019). Mumefural Ameliorates Cognitive Impairment in Chronic Cerebral Hypoperfusion via Regulating the Septohippocampal Cholinergic System and Neuroinflammation. Nutrients 11 (11). doi:10.3390/nu11112755
Ben-Neriah, Y., and Karin, M. (2011). Inflammation Meets Cancer, with NF-Κb as the Matchmaker. Nat. Immunol. 12 (8), 715–723. doi:10.1038/ni.2060
Bin Sayeed, M. S., and Ameen, S. S. (2015). Beta-Sitosterol: A Promising but Orphan Nutraceutical to Fight against Cancer. Nutr. Cancer 67 (8), 1214–1220. doi:10.1080/01635581.2015.1087042
Bray, F., Ferlay, J., Soerjomataram, I., Siegel, R. L., Torre, L. A., and Jemal, A. (2018). Global Cancer Statistics 2018: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J. Clin. 68 (6), 394–424. doi:10.3322/caac.21492
Cho, H. D., Kim, J. H., Won, Y. S., Moon, K. D., and Seo, K. I. (2019). Inhibitory Effects of Pectinase-Treated Prunus Mume Fruit Concentrate on Colorectal Cancer Proliferation and Angiogenesis of Endothelial Cells. J. Food Sci. 84 (11), 3284–3295. doi:10.1111/1750-3841.14824
Dekker, E., Tanis, P. J., Vleugels, J. L. A., Kasi, P. M., and Wallace, M. B. (2019). Colorectal Cancer. Lancet 394 (10207), 1467–1480. doi:10.1016/s0140-6736(19)32319-0
Devi, K. P., Malar, D. S., Nabavi, S. F., Sureda, A., Xiao, J., Nabavi, S. M., et al. (2015). Kaempferol and Inflammation: From Chemistry to Medicine. Pharmacol. Res. 99, 1–10. doi:10.1016/j.phrs.2015.05.002
Eid, H. M., and Haddad, P. S. (2017). The Antidiabetic Potential of Quercetin: Underlying Mechanisms. Curr. Med. Chem. 24 (4), 355–364. doi:10.2174/0929867323666160909153707
Greten, F. R., Eckmann, L., Greten, T. F., Park, J. M., Li, Z. W., Egan, L. J., et al. (2004). IKKbeta Links Inflammation and Tumorigenesis in a Mouse Model of Colitis-Associated Cancer. Cell 118 (3), 285–296. doi:10.1016/j.cell.2004.07.013
Hayakawa, Y., Maeda, S., Nakagawa, H., Hikiba, Y., Shibata, W., Sakamoto, K., et al. (2009). Effectiveness of IkappaB Kinase Inhibitors in Murine Colitis-Associated Tumorigenesis. J. Gastroenterol. 44 (9), 935–943. doi:10.1007/s00535-009-0098-7
He, R., Ou, S., Chen, S., and Ding, S. (2020a). Network Pharmacology-Based Study on the Molecular Biological Mechanism of Action for Compound Kushen Injection in Anti-cancer Effect. Med. Sci. Monit. 26, e918520. doi:10.12659/MSM.918520
He, T., Shen, H., Wang, S., Wang, Y., He, Z., Zhu, L., et al. (2020b). MicroRNA-3613-5p Promotes Lung Adenocarcinoma Cell Proliferation through a RELA and AKT/MAPK Positive Feedback Loop. Mol. Ther. Nucleic Acids 22, 572–583. doi:10.1016/j.omtn.2020.09.024
Hoesel, B., and Schmid, J. A. (2013). The Complexity of NF-Κb Signaling in Inflammation and Cancer. Mol. Cancer 12, 86. doi:10.1186/1476-4598-12-86
Imran, M., Salehi, B., Sharifi-Rad, J., Aslam Gondal, T., Saeed, F., Imran, A., et al. (2019). Kaempferol: A Key Emphasis to its Anticancer Potential. Molecules 24 (12). doi:10.3390/molecules24122277
Islami, F., Goding Sauer, A., Miller, K. D., Siegel, R. L., Fedewa, S. A., Jacobs, E. J., et al. (2018). Proportion and Number of Cancer Cases and Deaths Attributable to Potentially Modifiable Risk Factors in the United States. CA Cancer J. Clin. 68 (1), 31–54. doi:10.3322/caac.21440
Jiang, J., Liu, H. L., Tao, L., Lin, X. Y., Yang, Y. D., Tan, S. W., et al. (2018). Let-7d I-nhibits C-olorectal C-ancer C-ell P-roliferation through the CST1/p65 P-athway. Int. J. Oncol. 53 (2), 781–790. doi:10.3892/ijo.2018.4419
Keats, J. J., Fonseca, R., Chesi, M., Schop, R., Baker, A., Chng, W. J., et al. (2007). Promiscuous Mutations Activate the Noncanonical NF-kappaB Pathway in Multiple Myeloma. Cancer Cell 12 (2), 131–144. doi:10.1016/j.ccr.2007.07.003
Lee, K. M., Bang, J., Kim, B. Y., Lee, I. S., Han, J. S., Hwang, B. Y., et al. (2015). Fructus Mume Alleviates Chronic Cerebral Hypoperfusion-Induced white Matter and Hippocampal Damage via Inhibition of Inflammation and Downregulation of TLR4 and P38 MAPK Signaling. BMC Complement. Altern. Med. 15, 125. doi:10.1186/s12906-015-0652-1
Li, M., Pan, M., Wang, J., You, C., Zhao, F., Zheng, D., et al. (2020). miR-7 Reduces Breast Cancer Stem Cell Metastasis via Inhibiting RELA to Decrease ESAM Expression. Mol. Ther. Oncolytics 18, 70–82. doi:10.1016/j.omto.2020.06.002
Liu, H., Wang, J., Zhou, W., Wang, Y., and Yang, L. (2013). Systems Approaches and Polypharmacology for Drug Discovery from Herbal Medicines: an Example Using Licorice. J. Ethnopharmacol 146 (3), 773–793. doi:10.1016/j.jep.2013.02.004
Liu, W., Wang, S., Sun, Q., Yang, Z., Liu, M., and Tang, H. (2018a). DCLK1 Promotes Epithelial-Mesenchymal Transition via the PI3K/Akt/NF-Κb Pathway in Colorectal Cancer. Int. J. Cancer 142 (10), 2068–2079. doi:10.1002/ijc.31232
Liu, X., Wang, S., Xu, J., Kou, B., Chen, D., Wang, Y., et al. (2018b). Extract of Stellerachamaejasme L(ESC) Inhibits Growth and Metastasis of Human Hepatocellular Carcinoma via Regulating microRNA Expression. BMC Complement. Altern. Med. 18 (1), 99. doi:10.1186/s12906-018-2123-y
Lu, X., and Yarbrough, W. G. (2015). Negative Regulation of RelA Phosphorylation: Emerging Players and Their Roles in Cancer. Cytokine Growth Factor. Rev. 26 (1), 7–13. doi:10.1016/j.cytogfr.2014.09.003
Moles, A., Butterworth, J. A., Sanchez, A., Hunter, J. E., Leslie, J., Sellier, H., et al. (2016). A RelA(p65) Thr505 Phospho-Site Mutation Reveals an Important Mechanism Regulating NF-κb-dependent Liver Regeneration and Cancer. Oncogene 35 (35), 4623–4632. doi:10.1038/onc.2015.526
Naugler, W. E., and Karin, M. (2008). NF-kappaB and Cancer-Identifying Targets and Mechanisms. Curr. Opin. Genet. Dev. 18 (1), 19–26. doi:10.1016/j.gde.2008.01.020
Patel, M., Horgan, P. G., McMillan, D. C., and Edwards, J. (2018). NF-κB Pathways in the Development and Progression of Colorectal Cancer. Transl Res. 197, 43–56. doi:10.1016/j.trsl.2018.02.002
Qi, L., Zhang, Y., Song, F., and Ding, Y. (2020). Chinese Herbal Medicine Promote Tissue Differentiation in Colorectal Cancer by Activating HSD11B2. Arch. Biochem. Biophys. 695, 108644. doi:10.1016/j.abb.2020.108644
Reyes-Farias, M., and Carrasco-Pozo, C. (2019). The Anti-cancer Effect of Quercetin: Molecular Implications in Cancer Metabolism. Int. J. Mol. Sci. 20 (13). doi:10.3390/ijms20133177
Schmidt, S., Gonzalez, D., and Derendorf, H. (2010). Significance of Protein Binding in Pharmacokinetics and Pharmacodynamics. J. Pharm. Sci. 99 (3), 1107–1122. doi:10.1002/jps.21916
Schwitalla, S., Fingerle, A. A., Cammareri, P., Nebelsiek, T., Göktuna, S. I., Ziegler, P. K., et al. (2013). Intestinal Tumorigenesis Initiated by Dedifferentiation and Acquisition of Stem-cell-like Properties. Cell 152 (1-2), 25–38. doi:10.1016/j.cell.2012.12.012
Sen, R., and Baltimore, D. (2006). Multiple Nuclear Factors Interact with the Immunoglobulin Enhancer Sequences (Reprinted from Cell, Vol 46, Pg 705-716, 1986). J. Immunol. 177 (11), 7485–7496.
Shaked, H., Hofseth, L. J., Chumanevich, A., Chumanevich, A. A., Wang, J., Wang, Y., et al. (2012). Chronic Epithelial NF-Κb Activation Accelerates APC Loss and Intestinal Tumor Initiation through iNOS Up-Regulation. Proc. Natl. Acad. Sci. U S A. 109 (35), 14007–14012. doi:10.1073/pnas.1211509109
Shen, X., Zhao, Z., Wang, H., Guo, Z., Hu, B., and Zhang, G. (20172017). Elucidation of the Anti-inflammatory Mechanisms of Bupleuri and Scutellariae Radix Using System Pharmacological Analyses. Mediators Inflamm. 2017, 3709874. doi:10.1155/2017/3709874
Siegel, R. L., Miller, K. D., Goding Sauer, A., Fedewa, S. A., Butterly, L. F., Anderson, J. C., et al. (2020). Colorectal Cancer Statistics, 2020. CA Cancer J. Clin. 70 (3), 145–164. doi:10.3322/caac.21601
Staudt, L. M. (2010). Oncogenic Activation of NF-kappaB. Cold Spring Harb Perspect. Biol. 2 (6), a000109. doi:10.1101/cshperspect.a000109
Vo, T. K., Ta, Q. T. H., Chu, Q. T., Nguyen, T. T., and Vo, V. G. (2020). Anti-Hepatocellular-Cancer Activity Exerted by β-Sitosterol and β-Sitosterol-Glucoside from Indigofera Zollingeriana Miq. Molecules 25 (13). doi:10.3390/molecules25133021
Wang, K., Chen, Q., Shao, Y., Yin, S., Liu, C., Liu, Y., et al. (2021). Anticancer Activities of TCM and Their Active Components against Tumor Metastasis. Biomed. Pharmacother. 133, 111044. doi:10.1016/j.biopha.2020.111044
Wang, Q., Jiao, L., Wang, S., Chen, P., Bi, L., Zhou, D., et al. (2018). Maintenance Chemotherapy with Chinese Herb Medicine Formulas vs. With Placebo in Patients with Advanced Non-small Cell Lung Cancer after First-Line Chemotherapy: A Multicenter, Randomized, Double-Blind Trial. Front. Pharmacol. 9, 1233. doi:10.3389/fphar.2018.01233
Wang, S., Long, S., Deng, Z., and Wu, W. (2020a). Positive Role of Chinese Herbal Medicine in Cancer Immune Regulation. Am. J. Chin. Med. 48 (7), 1577–1592. doi:10.1142/S0192415X20500780
Wang, Z., Sheng, C., Kan, G., Yao, C., Geng, R., and Chen, S. (2020b). RNAi Screening Identifies that TEX10 Promotes the Proliferation of Colorectal Cancer Cells by Increasing NF-Κb Activation. Adv. Sci. (Weinh) 7 (17), 2000593. doi:10.1002/advs.202000593
Wei, M., Li, H., Li, Q., Qiao, Y., Ma, Q., Xie, R., et al. (2020). Based on Network Pharmacology to Explore the Molecular Targets and Mechanisms of Gegen Qinlian Decoction for the Treatment of Ulcerative Colitis. Biomed. Res. Int. 2020, 5217405. doi:10.1155/2020/5217405
Wilt, T., Ishani, A., MacDonald, R., Stark, G., Mulrow, C., and Lau, J. (2000). Beta-sitosterols for Benign Prostatic Hyperplasia. Cochrane Database Syst. Rev. (2), CD001043. doi:10.1002/14651858.CD001043
Won, Y. S., Kim, J. H., Lizardo, R. C. M., Min, H. J., Cho, H. D., Hong, S. M., et al. (2020). The Flavonol Isoquercitrin Promotes Mitochondrial-dependent Apoptosis in SK-Mel-2 Melanoma Cell via the PI3K/AKT/mTOR Pathway. Nutrients 12 (12). doi:10.3390/nu12123683
Wong, M. C. S., Huang, J., Lok, V., Wang, J., Fung, F., Ding, H., et al. (2021). Differences in Incidence and Mortality Trends of Colorectal Cancer Worldwide Based on Sex, Age, and Anatomic Location. Clin. Gastroenterol. Hepatol. 19 (5), 955–966. doi:10.1016/j.cgh.2020.02.026
Xing, H., Zhang, L., Ma, J., Liu, Z., Song, C., and Liu, Y. (2018). Fructus Mume Extracts Alleviate Diarrhea in Breast Cancer Patients Receiving the Combination Therapy of Lapatinib and Capecitabine. Front. Pharmacol. 9, 516. doi:10.3389/fphar.2018.00516
Xu, J., Meng, Q., Li, X., Yang, H., Xu, J., Gao, N., et al. (2019). Long Noncoding RNA MIR17HG Promotes Colorectal Cancer Progression via miR-17-5p. Cancer Res. 79 (19), 4882–4895. doi:10.1158/0008-5472.CAN-18-3880
Yang, Z., Zhang, Q., Yu, L., Zhu, J., Cao, Y., and Gao, X. (2021). The Signaling Pathways and Targets of Traditional Chinese Medicine and Natural Medicine in Triple-Negative Breast Cancer. J. Ethnopharmacol 264, 113249. doi:10.1016/j.jep.2020.113249
Zhang, R., Ai, X., Duan, Y., Xue, M., He, W., Wang, C., et al. (2017). Kaempferol Ameliorates H9N2 Swine Influenza Virus-Induced Acute Lung Injury by Inactivation of TLR4/MyD88-Mediated NF-Κb and MAPK Signaling Pathways. Biomed. Pharmacother. 89, 660–672. doi:10.1016/j.biopha.2017.02.081
Keywords: Prunus mume, colorectal cancer, network pharmacology, moleculardocking, RelA, apoptosis
Citation: Zhou M, Li J, Luo D, Zhang H, Yu Z, Chen Y, Li Q, Liang F and Chen R (2021) Network Pharmacology and Molecular Docking–Based Investigation: Prunus mume Against Colorectal Cancer via Silencing RelA Expression. Front. Pharmacol. 12:761980. doi: 10.3389/fphar.2021.761980
Received: 20 August 2021; Accepted: 13 October 2021;
Published: 19 November 2021.
Edited by:
Juei-Tang Cheng, Chang Jung Christian University, TaiwanReviewed by:
Eswar Shankar, The Ohio State University, United StatesCopyright © 2021 Zhou, Li, Luo, Zhang, Yu, Chen, Li, Liang and Chen. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Rui Chen, MjAwMVhIMDY2OUBodXN0LmVkdS5jbg==; Fengxia Liang, ZnhsaWFuZzVAaG90bWFpbC5jb20=
†These authors share first authorship
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.