- 1Department of Pharmacy, Chengdu Second People’s Hospital, Chengdu, China
- 2Department of Pharmacy, Sichuan Academy of Medical Sciences and Sichuan Provincial People’s Hospital, School of Medicine, University of Electronic Science and Technology of China, Chengdu, China
- 3Department of Pharmacy, Sichuan Vocational College of Health and Rehabilitation, Zigong, China
- 4Department of Pharmacy, The First People’s Hospital of Zigong, Zigong, China
Systemic lupus erythematosus (SLE) is a kind of chronic diffuse connective tissue illness characterized by multisystem and multiorgan involvement, repeated recurrence and remission, and the presence of a large pool of autoantibodies in the body. Although the exact cause of SLE is not thoroughly revealed, accumulating evidence has manifested that intake of probiotics alters the composition of the gut microbiome, regulating the immunomodulatory and inflammatory response, which may be linked to the disease pathogenesis. Particularly, documented experiments demonstrated that SLE patients have remarkable changes in gut microbiota compared to healthy controls, indicating that the alteration of microbiota may be implicated in different phases of SLE. In this review, the alteration of microbiota in the development of SLE is summarized, and the mechanism of intestinal microbiota on the progression of immune and inflammatory responses in SLE is also discussed. Due to limited reports on the effects of probiotics supplementation in SLE patients, we emphasize advancements made in the last few years on the function and mechanisms of probiotics in the development of SLE animal models. Besides, we follow through literature to survey whether probiotics supplements can be an adjuvant therapy for comprehensive treatment of SLE. Research has indicated that intake of probiotics alters the composition of the gut microbiome, contributing to prevent the progression of SLE. Adjustment of the gut microbiome through probiotics supplementation seems to alleviate SLE symptoms and their cardiovascular and renal complications in animal models, marking this treatment as a potentially novel approach.
Introduction
Systemic lupus erythematosus (SLE) is a chronic autoimmune disease, where a large pool of autoantibodies are produced, causing the immune system to attack its tissues, resulting in damage to multiple organs and systems throughout the body (Mu et al., 2015; Yacoub et al., 2018). Its main clinical features are multiple systems and organs involvement, repeated relapse and remission, and the development of a large pool of autoantibodies against double-stranded (ds) DNA (Lisnevskaia et al., 2014; Durcan et al., 2019; Fava and Petri, 2019). Individuals affected by SLE have extensive symptoms and course of the disease, the most frequent of which are fever, fatigue, facial butterfly erythema, photosensitivity, muscle or joint pain, arthritis, and renal symptoms (Goldblatt and O’Neill, 2013). Moreover, patients with SLE have an increased incidence of atherosclerosis, thrombosis, arteritis, embolization, and vascular spasm (Kasselman et al., 2018). The most lethal outcomes in SLE patients are infection, severe multiple organ injury, especially damage to the nervous system and kidney (Lee et al., 2016; Yen et al., 2017). Production of the immune response in SLE is distinguished by an overreaction of B cell and T cell responses, and impaired self-tolerance to autoantigens. (Lisnevskaia et al., 2014; Durcan et al., 2019; Fava and Petri, 2019; Kiriakidou and Ching, 2020). The prevalence of SLE varies widely from region to region, with the current global prevalence approaching or even above 50 to 241 per 100,000 adults, among which the prevalence rates of African Americans, American Indians, and Alaska Natives are higher (Helmick et al., 2008; Ferucci et al., 2014; Somers et al., 2014; Rees et al., 2017; Nikolopoulos et al., 2020). The prevalence of SLE is higher among African Americans and Europe, which is less prevalent in Africa (Symmons, 1995; Pons-Estel et al., 2010). The severity of the disease may also vary by ethnic background, with patients of African and Latin American descent commonly more severe (Carter et al., 2016). Particularly, SLE is strikingly dominated by women of childbearing age, with nearly 10 female patients for every male suffering from the disease (Carter et al., 2016; Durcan et al., 2019; Fava and Petri, 2019; Fanouriakis et al., 2021). Among women aged 15 to 44, the ratio of women to men is 13:1 while the ratio is only 2:1 between children and the elderly (Petri, 2002; Danchenko et al., 2006).
A variety of medications are applied to SLE therapy, including glucocorticoids (GCs), antimalarial agents, nonsteroidal anti-inflammatory drugs (NSAIDs), immunosuppressive agents, and B cell–targeting biologics. NSAIDs block the synthesis of prostaglandins by attenuating the enzymes cyclooxygenases (COX-1 and COX-2) to counter inflammation and pain. The adverse reactions with the highest incidence of this drug are gastrointestinal gastritis, nephrotoxicity, fluid retention (Kiriakidou and Ching, 2020). Hydroxychloroquine is the cornerstone of lupus treatment (Kiriakidou and Ching, 2020). Hydroxychloroquine or other antimalarial agents have the effect of immunomodulatory and antithrombotic (Chrisman et al., 1976; Espinola et al., 2002; Wang et al., 2019). But the drugs can induce retinopathy, skin pigmentation, and rare cases of neuromuscular or cardiac toxicity (Marmor et al., 2016). GCs are the most commonly used agents in SLE-induced remission therapy and are consistently recommended by the guidelines as first-line agents for the control of SLE. (Gordon et al., 2018; Fanouriakis et al., 2019; Kiriakidou and Ching, 2020). The side effects that may occur after taking GCs are gastrointestinal adverse reaction, metabolic disorders, infections, weight gain, hypertension, psychiatric disorders, lipodystrophy, fractures, and adrenal suppression, which are mainly dose and time-dependent (Saag et al., 1994; van Staa et al., 2002; Wei et al., 2004; Da Silva et al., 2006; Warrington and Bostwick, 2006; Dixon et al., 2011; Sarnes et al., 2011). Furthermore, immunosuppressant therapy is suggested for SLE patients who continue to respond poorly to GCs and hydroxychloroquine combination therapy, or who cannot adjust the dose of GCs to a relatively safe dose. Immunosuppressive agents such as methotrexate inhibit DNA synthesis and increase the release of adenosine, but some patients were forced to stop using the drug because of intolerance to adverse reactions (Kiriakidou and Ching, 2020). What occurs most after the application of methotrexate are gastrointestinal side effects (nausea, vomiting, diarrhea), hepatotoxicity, and blood-related toxicity (anemia, leucopenia) (Sakthiswary and Suresh, 2014; Andreoli et al., 2017). For patients with refractory (ineffective after conventional treatment) or recurrent SLE, the administration of biological agents can reduce disease activity, disease recurrence rate and reduce hormone dosage (Wei et al., 2016; Alshaiki et al., 2018). Belimumab targets B-lymphocyte stimulator inhibits B-lymphocyte proliferation and activation (van Vollenhoven et al., 2012), but its common adverse effects are hypersensitivity reaction, gastrointestinal toxicity, myalgias, depression, migraine, infection (Lee and Song, 2018; Peterknecht et al., 2018). Rituximab depletes CD20-expressing B lymphocytes, but patients may occur an infusion reaction, infection, progressive multifocal leukoencephalopathy (rare) (Alshaiki et al., 2018; Peterknecht et al., 2018). In addition, other measures can be used to treat SLE. When SLE develops to severe or refractory levels, the addition of plasma exchange or DNA immunosorbent as adjunct therapy may be considered, which may ameliorate clinical symptoms rapidly but cannot improve the outcome (Kronbichler et al., 2016). In summary, all the medicines used in the treatment of SLE induce adverse reactions, whereas, probiotics supplementation appears to have no significant side effects clinically. Hence, it is necessary to further investigate probiotics for the exploration of theoretical basis as adjuvant therapy in SLE patients.
The precise pathogenesis of SLE is not entirely revealed, it is believed to be caused by the human immune system attacking self-tissues after being abnormally activated. The pathogenesis of SLE may be related to genetic, hormones and, environmental factors (infection, drugs, UVA light) (De Luca and Shoenfeld, 2019). Nevertheless, with research on intestinal flora dysregulation going depth recently, dysbiosis as a vital internal environmental factor has also been shown to be concerned with SLE (Meng et al., 2019). In 1994, Apperloo-Renkema et al. first demonstrated experimentally that alterations in intestinal microbiota composition can cause SLE in animal models, possibly due to a weakened defense of native gut microbes against foreign bacteria (Apperloo-Renkema et al., 1994). Repeated antigen stimulation may lead to changes in intestinal microecology, confusion of the immune system, and the body subsequently attacks its tissue by producing antibodies or sensitized lymphocytes (Zhang and Reichlin, 2008). The production of these antibodies is exacerbated by extensive inflammatory responses, which leads to a range of clinical symptoms and further complications associated with SLE (Zhang and Reichlin, 2008). Long-term use of probiotics is believed to neutralize an imbalance in the gut microbiota that results in the reduced antibody production and suppressed inflammatory response, leading to attenuation of severity, signs and, manifestation of SLE (Esmaeili et al., 2017). In a study assessing the resistance of intestinal microbiota to pathogen colonization in SLE patients and healthy controls, the colonization resistance of patients with active SLE is lower than that of healthy people, which indicates that the incomplete normal intestinal microbiota may lead to more intestinal transfection of pathogenic bacteria (Apperloo-Renkema et al., 1994). The study presented above confirmed that the balance of intestinal microbiota is associated with the pathogenesis of SLE, but it is still unclear whether administering probiotics to restore normal intestinal flora and reduce inflammation may have therapeutic benefits for SLE patients or not (van der Meulen et al., 2016).
In brief, raising an understanding of how to ameliorate gut dysbiosis could help explore an alternative approach to prevent or alleviate SLE. Therefore, the alteration of microbiota associated with SLE was reviewed and the function and mechanisms of probiotics in the development of SLE animal models were also discussed.
An Inflammatory Pathway of SLE
SLE is a multifactorial caused disease, and the pathogenesis is considered to be related to hormonal, environmental and genetic factors that lead to an intolerance to autoantigens (Rahman and Isenberg, 2008; De Luca and Shoenfeld, 2019). SLE is featured by the generation of autoantibodies, aggregation of autoreactive and inflammatory T cells, and abnormal production of inflammatory cells and pro-inflammatory cytokines (Buckner, 2010; Tsokos, 2011; Rastin et al., 2013). Autoantibodies produced by autoimmune B cells bring about the generation and accumulation of immunocomplex which do harm to multiple organs, containing the skin, joints, heart, kidneys, and brain (Zhang and Reichlin, 2008; Tsokos, 2011). Although SLE is regarded as mainly B cells mediated disease, there is some evidence indicating the significance of unbalanced regulatory T (Treg) cells in the development of SLE (Buckner, 2010; Ma et al., 2010; Talaat et al., 2015). In addition, it has been proven that improved T helper cell 17 (Th17) amount and effect play a crucial role by secreting pro-inflammatory cytokines, such as interleukin (IL)-17 and IL-23), as the primary trigger of autoimmune response, and these cytokines are related to the inflammatory formation and tissue damage in SLE (Crispín et al., 2008; Doreau et al., 2009; Chen et al., 2010; Pan et al., 2013). Studies have demonstrated that strengthening Treg cells restrain abnormal reactions of effector T cells, which can steadfastly alleviate autoimmune and inflammatory responses (Shevach, 2009; Lavi Arab et al., 2015; Reihani et al., 2015). It has been identified that patients with SLE have decreased numbers and functional deficiencies of Tregs as well as the resistance of effector T cells to the inhibitory effects of Tregs, which exert significant effects in the pathogenesis of SLE (Lyssuk et al., 2007; Valencia et al., 2007; Gómez et al., 2009; Esmaeili et al., 2017). It is reported that anti-inflammatory cytokines, such as transforming growth factor β (TGF-β) and pro-inflammatory cytokines, including IL-6, IFN-γ, and IL-23/IL-17 are drastic in every developmental stage of SLE (Su et al., 2012). Therefore, restoration of unbalanced cytokines and defective immune cells may be a potential remedial strategy for alleviating SLE manifestations (Esmaeili et al., 2017).
The Microbiome and Probiotics
The Gut Microbiota
The intestine contains the largest complex mic-ecosystem in humans, which can be regarded as an independent organ in the body (Van de Wiele et al., 2016). According to high-throughput culture-independent sequencing analysis, the microbiome of the gut tract is more complicated than those of other parts of the body, with over 1,000 microorganisms identified so far, and the total biomass is close to 1,000 colony-forming units 1013∼1014(CUF) (Claesson et al., 2009; Sankar et al., 2015). The intestine microbiota is dominated mainly by two phyla (approximately 90%) Firmicutes and Bacteroidetes, and the rest is involved in Actinobacteria, Proteobacteria, Synergistetes, Verrucomicrobia, Fusobacteria, and so on (Eckburg et al., 2005; de la Visitación et al., 2019). The existing methanogenic archaea, yeasts, and viruses (mainly phages) increase the complexity of the gut microbiota (Lozupone et al., 2012). Although only two phyla have predominance in the gut microbiota, there are striking differences in the intestinal microecology between people and people across different life cycles (Van de Wiele et al., 2016). Individual diversity in host genes, mode of delivery and lactation, geographic origin, age, diet, disease, drug uptake, and lifestyle contribute to differences in intestine microbiota composition (Ley, 2015). With the development of the functional characteristics of individual microbiota, growing evidence shows that the gut microbiota participated in critical activities related to disease and health. It has been proven that the functions of the human gut bacteria are to affect digestion, provide nutrients, form intestinal barriers and produce colonization resistance, regulate the development of intestinal epithelium, as well as to modulate the activation and progression of the immune system (Van de Wiele et al., 2016).
Therefore, any factors that disrupt the host-microbial balance (such as acute alters in dietary behavior; malnutrition; pathogen infection; inflammation; administration of anti-biological drugs; gastrointestinal surgery; etc.) may influence the homeostasis of microbiota which exerts a significant impact on the regulation of host immune functions (Ogura et al., 2003; Cho, 2008; De Filippo et al., 2010; Delzenne et al., 2011).
Probiotics
Probiotics are living organisms that regulate the gut microbiome in various ways to improve intestinal health. Probiotics can affect immune homeostasis by keeping a healthy microbial balance, and can also adjust mucus secretion through intestinal epithelial cells, thereby contributing to maintaining the stability of the mucus barrier and providing resistance to pathogen colonization (Bron et al., 2017; de Oliveira et al., 2017). Besides, Probiotics can promote the generation of multiple nutrients such as SCFAs and vitamins, which contribute to form the entire host intestinal microbiome (Yadav et al., 2013; de Oliveira et al., 2017). Moreover, probiotics participate in the degradation of toxic compounds and the production of antimicrobial compounds, like bacteriocins (de Oliveira et al., 2017). Therefore, these probiotics can be used as a treatment option for immune-related diseases (Balakrishnan and Taneja, 2018).
Probiotics have been widely evaluated for their benefits in preventing or treating extensive diseases, including infection, inflammation, cancer, and autoimmune diseases in animal and human trails (Borchers et al., 2009). The recorded probiotic-inducing impacts include suppression of infection, immune regulation, prolonged remission of patients with ulcerative colitis, treating or preventing infective or antibiotic-associated diarrhea in both adults and infants, assisting in the eradication of Helicobacter pylori, improving nonalcoholic fatty liver disease and metabolic diseases, reducing the recurrence rate of colorectal cancer and alleviating lactose intolerance symptoms (de Vrese et al., 2001; Van Niel et al., 2002; Bengmark, 2003; Gill, 2003; Gionchetti et al., 2003; Kalliomäki et al., 2003; Tamboli et al., 2003). Probiotics have been reported to exert their beneficial effects mainly in three ways, containing competitive exclusion, antibacterial action, and regulation of immune responses. It has been found that the administration of immunoregulatory probiotics in the prevention or treatment of autoimmune diseases is mainly attributable to improving the inflammatory responses and modulating tolerance in the host to pathogens (Esmaeili et al., 2017).
Mechanism of Action of Probiotics
It has been discovered that probiotics influence each segment of the intestine, containing the intraluminal microbiota, the epithelial microbial, mucosal barrier, the lamina propria rich in lymphocytes and plasma cells, the blood vessels and nerves of lamina propria components, the underlying smooth muscles commanding movement and the mesenteric lymph nodes associated with systemic immunity (Liu et al., 2018). In mechanism, immunomodulatory probiotics are known to prevent inflammation and modulate immunity to improve SLE symptoms (Liu et al., 2018). As Liu et al. (Liu et al., 2018) summarized that short-chain fatty acids (SCFAs) generated by bifidobacterium, lactobacillus, and symbiotic bacteria combine and activate receptors (FFAR2, FFAR3, or GPR109a) on enterocytes to inhibit inflammatory responses by blocking nuclear factor-κ-light chain enhancer of B cells activation pathway. SCFAs also suppress histone deacetylases to facilitate amassing of Tregs and discharge glucagon-like protein-1/peptide tyrosine to respond to the enteric and central nervous system, thereby affecting intestinal homeostasis and motion. They also induce tolerogenic dendritic cells (DCs), which induce immature CD4+ T cells to differentiate into Tregs. The above response restraint the generation of cytokines via neutrophils and macrophages by binding to receptors. Adenosine and its derivative inosine interact with adenosine receptor-2A expressed on T cells to enhance Treg effects and suppress TH1 and TH17 subsets inflammation. Histamine generated by L. reuteri 6475 reacts on H2 receptor located in intestinal epithelial cells and macrophages to decrease the secretion of proinflammatory cytokines, containing tumor necrosis factor (TNF)-α, MCP (monocyte chemoattractant protein)-1, and IL-12. In conclusion, the pivotal metabolites generated by probiotics exhibit anti-inflammatory properties and improvement of the intestinal barrier function during the disease.
The Microbiota Studies in SLE
Studies that described the microbiota of SLE are relatively limited, although the increasing prevalence of Crohn’s disease (CD) in patients with SLE (Shor et al., 2016) has sparked interest in its involvement. Although the pathogenesis of SLE is not completely understood, an imbalance in the microbiome has been manifested to be associated with the establishment of SLE (Hevia et al., 2014; De Luca and Shoenfeld, 2019). Until now, human studies that investigate the connection between the microbiome and SLE initiation are observational case-control studies that compare differences of the human microbiome in areas like the gut or buccal cavity between SLE patients and controls (Arron et al., 2014; Hevia et al., 2014; Zhang et al., 2015; López et al., 2016). Therefore, revealing the microbial composition and possible function of these microbes in SLE patients may illuminate the cause and development, and may even find diagnostic biomarkers.
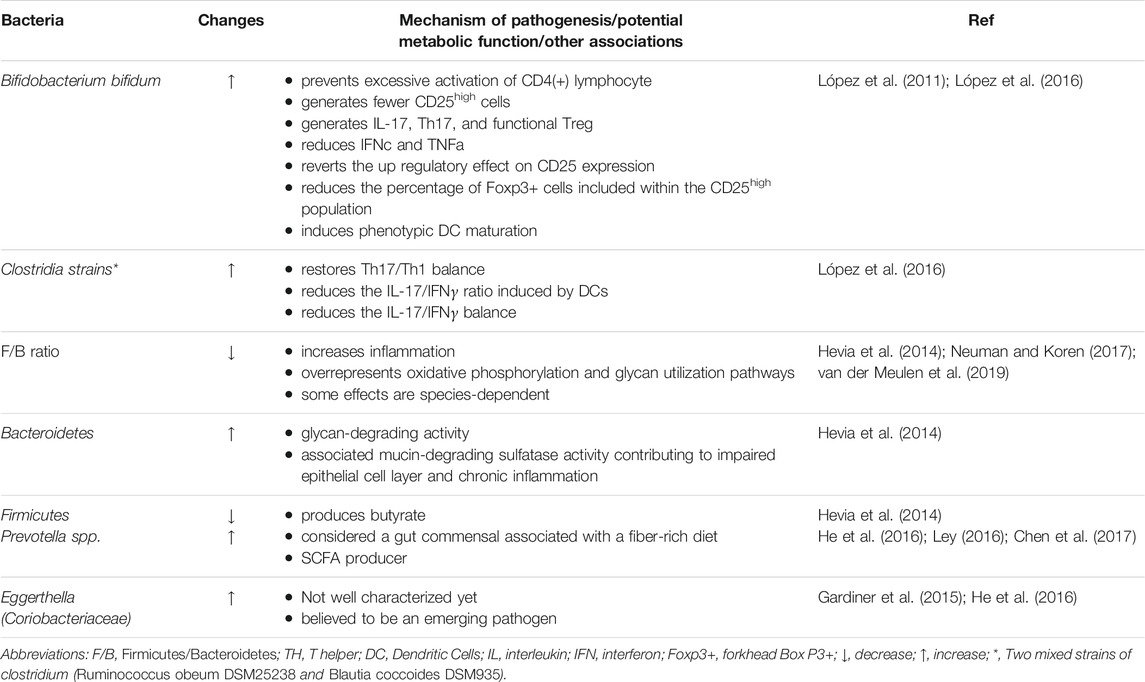
SLE patients have compositional and functional alterations in gut microbiota, possibly due to a weakened defense of native gut microbes against foreign bacteria (Apperloo-Renkema et al., 1994). Noticeable increase of several genera, including Rhodococcus, Klebsiella, Eggerthella, Prevotella, Eubacterium, and Flavonifractor, has been found in patients with SLE, whereas, Dialister and Pseudobutyrivibrio decreased (Hevia et al., 2014; He et al., 2016; Chen et al., 2017). Quantitative polymerase chain reaction analysis confirmed that the ratio of Firmicutes to Bacteroidetes was lower in patients with SLE, and the abundance of some families of Firmicutes was decreased (López et al., 2016) (Hevia et al., 2014; Neuman and Koren, 2017; van der Meulen et al., 2019) (Table 1). Such alterations are also present in other diseases, such as Crohn’s disease and type 2 diabetes mellitus (Man et al., 2011), suggesting that an overall imbalanced microbiota state is not specific to SLE (Larsen et al., 2010). A similar study on the composition of gut microbiota in 45 Chinese patients with SLE was in accordance with the results mentioned above, showing lower Firmicutes and higher Bacteroidetes in SLE patients (He et al., 2016). Downregulating inflammation can be achieved in several ways, such as elimination of apoptotic cells and cell debris, clearance of oxidized lipids, and blocking the stimulation of mitogen-activated protein kinase (MAPK) and other pro-inflammatory cytokines (Grönwall et al., 2012; López et al., 2016). As the anti-dsDNA titer increased, the frequency of the Synergistetes, which was positively associated with the rate of Firmicutes to Bacteroidetes in healthy controls, verged to decrease in SLE patients and was present a significantly negative association with the level of proinflammatory cytokines IL-6 in serum, meanwhile, correlating positively with natural protective IgM anti-phosphorylcholine secreted by B1 cells (López et al., 2016).
In female SLE patients, studies have shown the abundances of Lactobacillaceae decreases, while the levels of Lachnospiraceae increase, both of which belong to the Firmicutes phylum (Zhang et al., 2014; Mu et al., 2015; Neuman and Koren, 2017). Notably, comthe level of abutyrate-producing bacterium Lachnospiraceae was augmenter in SLE patients compared with healthy controls, thus Lachnospiraceae or any bacterium that produces butyrate may be unable to inhibit inflammation in SLE cases (Kakiyama et al., 2013; Zhang et al., 2014; Kasselman et al., 2018; Luo et al., 2018). Further, patients with SLE tend to generate more CD25high cells, whereas Bifidobacterium bifidum (B. bifidum) strain can revert to the up regulatory effect (López et al., 2016). On the other hand, the microbiota isolated from the feces of SLE patients has been found to accelerate the activation of lymphocytes and the differentiation of Th17 from primitive CD4+ lymphocytes (López et al., 2016). Additionally, B. bifidum may prevent lymphocyte activation whereas mixtures of two Clostridia strains supplementation, including Ruminococcus obeum DSM25238 and Blautia coccoides DSM935, restore Th17/Th1 balance (Atarashi et al., 2011; Atarashi et al., 2013; López et al., 2016). Alternatively, increased numbers of Selenomonas, Veillonella, T. denticola, and Leptotrichia are directly associated with raised concentrations of inflammatory factors like IL-6, IL-17, and IL-33, which are indicative of a decline in oral microbial species diversity in SLE patients (Corrêa et al., 2017).
Roles of Probiotics Against SLE
As a result of the above-mentioned findings, researchers believe that SLE treatment with probiotics (Table 2), has already presented some benefits like in other autoimmune diseases, can help ameliorate the symptomatology of disease (Schiffer et al., 2011; Zamani et al., 2016). Studies in animal and human trials have identified the potential benefits of probiotics in the alleviation and suppression of inflammation and autoimmune responses (Liu et al., 2018). Research in the SLE animal model has demonstrated that certain probiotic strains, including B. bifidum, Lactobacillus, Ruminococcus obeum, and Blautia coccoides, contribute to regulating excessive inflammation and restore tolerances (Esmaeili et al., 2017).
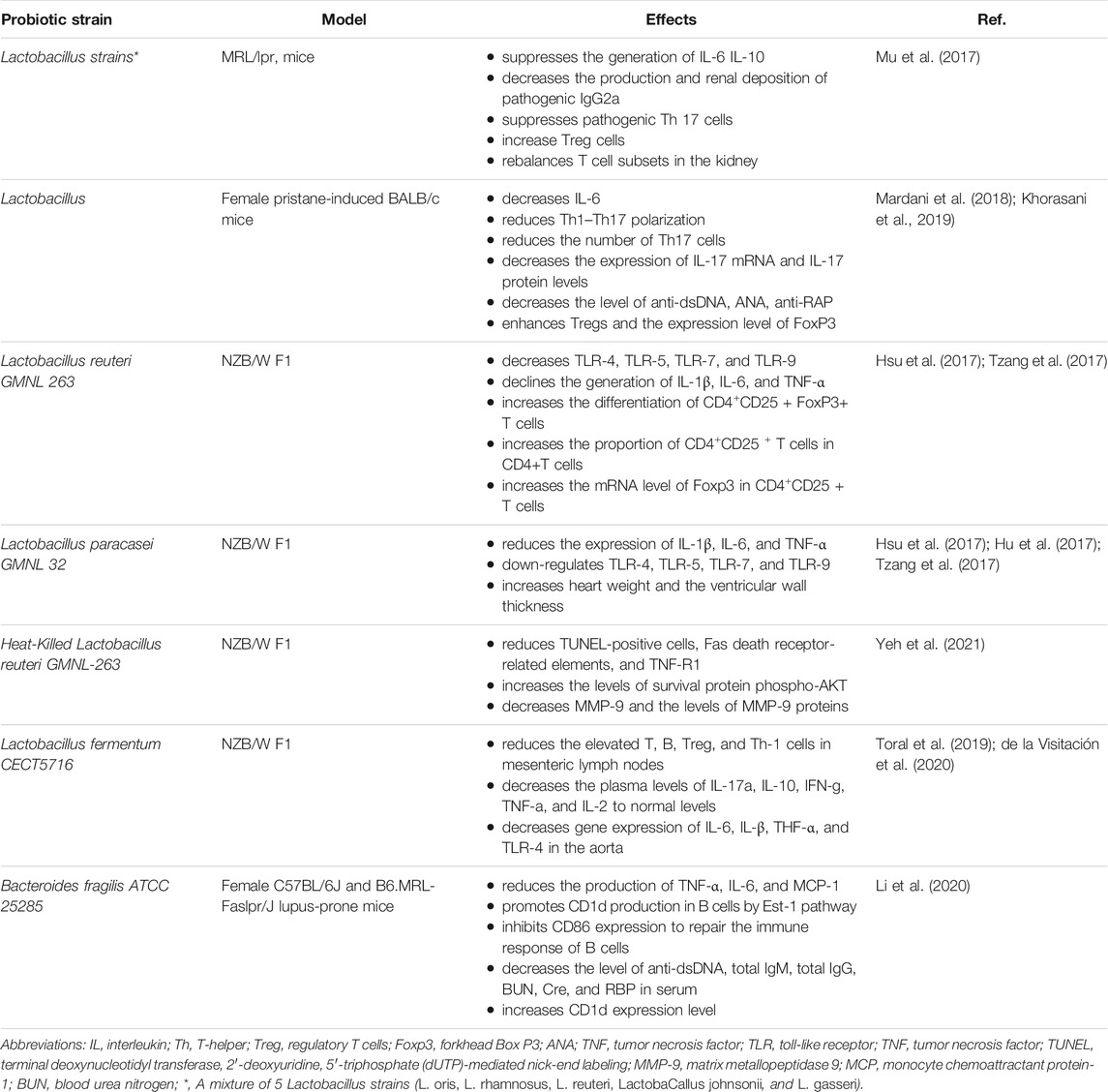
Researchers found that enteral administration of combinations of lactobacilli or L. reuteri alone in MRL/LPR mice, SLE mouse models, can skew the balance of Treg–Th17 toward Treg cell advantage in the kidneys, reduce endotoxemia, decrease the concentrations of dsDNA-reactive IgG, decrease urinary protein, and ameliorate the survival rate of patients (Mu et al., 2017). These outcomes were related to a shift in intestinal microbiota and extension of Lactobacilli, Clostridiales, and Desulfovibrionales. Mu et al. (2017) found that Lactobacillus spp. supplementation plays an anti-inflammatory role through reducing IL-6 and enhancing IL-10 generation in the intestine. The supplement of therapeutic Lactobacillus enhanced circulating IL-10 and declined IgG2a, which is regarded as a main immune deposition in MRL/lpr mice kidney. These benefits were observed in female and unsexed male mice, rather than in male functional mice, indicating that intestinal flora may regulate inflammation in a sex hormone-dependent pattern. According to Mardani et al.(2018), the consumption of Lactobacillus delbrueckii and actobacillus rhamnosusin to Female pristane-induced BALB/c mice improved the symptoms of SLE, exhibit anti-inflammatory properties by attenuating the generation of Th7 and down-regulating its major cytokines of IL-17a, one of the critical mediators in the formation and progression of inflammation.
L. reuteri GMNL 263 can down-regulate cytokine levels and repaired Tregs in NZB/W F1 mouse model, which is distinguished by oxidative stress and reduction of regulatory Tregs levels in circulation (Tzang et al., 2017; Liu et al., 2018). Alternatively, L. GMNL 263(GMNL 263) showed a diverse mechanism in the NZB/W F1mouse model of SLE (Tzang et al., 2017). These probiotics strains can enhance the production of Treg lymphocytes and the levels of transcription factor fork head box P3 (FoxP3), which is the hallmark of a natural Treg. These cells are in charge of regulating these pro-inflammatory lymphocytes and have significant anti-inflammatory characteristics. Besides, TLR-4, TLR-5, TLR-7, and TLR-9, which are the common pathogen-associated molecular pattern receptors that mediate the inflammation progression in the liver, were decreased and the antioxidant activity was increased under probiotics treatment (Hsu et al., 2017; Tzang et al., 2017). In addition, GMNL 263 also promoted the differentiation of CD4+CD25+FoxP3+ T cells and the proportion of CD4+CD25+ T cells number in CD4+T cells of spleen and enhanced the expression of Foxp3 mRNA in CD4+CD25+ T cells (Hsu et al., 2017).
In similar trials, the above alterations of TLRs and oxidative stress were also found using probiotics L. paracasei GMNL 32(GMNL-32) and L. reuteri GMNL 89, although GMNL 263 presented an effect on Treg expression in those cases (Hsu et al., 2017). The SLE-associated inflammation was also decreased with the administration of these probiotics, through enhancing the activity of antioxidation in serum and levels of CD4+CD25 + regulatory T cells in NZB/W F1 mice (Tzang et al., 2017). Moreover, in the treatment of these three probiotics strains, pro-inflammatory cytokines IL-1β, TNF-α, and IL-6 declined in the liver by inhibiting nuclear factor kB (NF-κB) and the signaling pathway of mitogen-activated protein kinase (Hsu et al., 2017; Hu et al., 2017). Specifically, GMNL-32 supplement attenuated left ventricular hypertrophy and the cardiac cell apoptosis in this genetic model of lupus (Hu et al., 2017; Tzang et al., 2017). These results indicated that oral supplement of several probiotic strains, such as GMNL32, L. reuteri GMNL-89, and L. reuteri GMNL263, to NZB/W F1 mice can not only mitigates hepatic inflammation and apoptosis caused by SLE, but also presents a protective function on cardiac cells of lupus-prone mice (Hsu et al., 2017; Hu et al., 2017; Tzang et al., 2017).
Yeh et al. (2021) first revealed the preventive effect of Heat-killed L. reuteri GMNL-263 on expanded interstitial spaces and abnormal myocardial structures in the hearts of NZB/W F1 mice, and lowered area of fibrosis and rescues cardiomyocyte arrangement, which demonstrate the clinical applications of the Lactobacillus in SLE-related cardiovascular diseases therapy. Because Heat-Killed L. reuteri GMNL263 prevented the development of the proinflammatory response, cardiac and renal hypertrophy complications in SLE were averted (Yeh et al., 2021). Compared with the controls, the anti-apoptotic effects were observed in the NZB/W F1 mice, and the significant declines of TUNEL-positive cells, Fas death receptor-related elements, and apoptosis were also detected after the consumption of GMNL-263. Additionally, administration of L. reuteri GMNL-263 to NZB/W F1mice present markedly higher levels of phospho-AKT (survival protein) than in NZB/W F1 control group. In fact, feeding L. Paracasei GMNL 32 was also detected to exhibit a similar protective pathway and prevent cardiac complications associated with SLE in NZB/W F1 mice. The Heat-killed L. reuteri GMNL-263 was a kind of dead bacteria, which was killed after heat treatment at 121°C for 5 min in 0.9% sterile NaCl and were made into powder freeze-dried. During the experiment, the powder was dissolved into a probiotic solution and fed to mice. Live probiotics provide barrier protection and immune system modulation; while components of dead cells exert an anti-inflammatory response in the gastrointestinal tract. Both live and dead probiotics can exert specific actions. To sum up, both L. reuteri GMNL-263 and L. paracasei GMNL 32 have been observed to exert cardioprotective properties by reducing TNF-R1, Fas-associated protein with death domain (FADD) and fibrosis proteins matrix metallopeptidase 9 (MMP-9).
Research has found L. fermentum CECT5716(LC40) protects the kidney and cardiovascular complications as well as disease activity in a female mouse model of SLE (Toral et al., 2019; de la Visitación et al., 2020). The administration of the immune-modulatory bacterium LC40 could increase the number of Bifidobacterium in the intestine of female NZB/WF1 mice. LC40 can reduce the activity of lupus and splenomegaly in SLE mice, improving the integrity of the intestinal barrier, reducing the plasma level of lipopolysaccharide (LPS), and subsequently decreasing the immune activation, which was characterized by reduced T and B cells in mesenteric lymph nodes (MLNs) and declined plasma pro-inflammatory factors, containing TNF-α, IFN-γ, IL-17a, and IL-21. Since probiotics prevented the progression of proinflammatory responses, complications related to SLE, such as cardiac and renal hyperplasia, were prevented (Toral et al., 2019). Another research reported that treatment with LC40 decreased the enhanced plasma anti-dsDNA, endotoxemia, and hypertension in NZB/WF1 mice. Meanwhile, LC40 also protected lupus mice from deterioration in renal function and kidney damage, as well as suppressing immune-complex deposition and inflammatory infiltration in glomerular, tubulointerstitial, and vascular lesions (de la Visitación et al., 2020).
In one recent experiment conducted by Li et al. (2020), oral supplementation of Bacteroides fragilis (B. fragilis) ATCC 25285 reduced autoantibodies levels and symptoms of lupus nephritis in MRL/lpr mice. The results confirmed that B. fragilis ATCC 25285 could improve the expression CD1d in B cells through Est-1 pathway, but suppress the expression of CD86 through SHP-2 signaling pathway to restore the immune response of B cells. Furthermore, levels of anti-dsDNA, total IgG and total IgM, as well levels of BUN, CRE, and RBP in serum decreased in MRL/lpr mice. In parallel, B. fragilis ATCC 25285 was found to play a role in restoring the balance of Th17/Treg in MRL/lpr mice, as it does in other autoimmune diseases (Li et al., 2020).
Through the above research, we have an overall understanding of the beneficial role of probiotics in adjuvant therapy of SLE, particularly the regulatory function of Treg and Th17 (de la Visitación et al., 2019). Dendritic and T Treg cells, cytokines like IL-6, IFN-γ, IL-17, and IL-23 are currently regarded as the most dominant mediators of dysregulation in the tolerated condition (Esmaeili et al., 2017). Different strains of probiotics may exhibit different beneficial functions but still fall within the same species. In this regard, live versus heat-killed probiotics present different properties, such as L. reuteri GMNL-263 and Heat-killed L. reuteri GMNL-263 (Adams, 2010). Therefore, further exploration of the potential mechanisms of probiotics is necessary, which will not only contribute to the cause and progression of SLE but may also support an alternative strategy for the comprehensive treatment of SLE, such as renal, cardiovascular, and hepatic complications.
Discussion
It is well known that there is a connection between microbes and autoimmune diseases. Alterations of the microbiome, namely “dysbiosis” can cause autoimmune disease influenced by the factors of certain genetic backgrounds and environments (De Luca and Shoenfeld, 2019). Dysbiosis can occur with the following three situations: reduction of beneficial microorganisms, overgrowth of potentially harmful microorganisms, and decrease of microbial diversity (DeGruttola et al., 2016). In order to ameliorate the adverse effects produced by microbial imbalances during the course of the disease, it may be possible to reestablish a healthy microbiota by supplement multiple probiotic strains, such as Bifidobacteriaum spp., Lactobacillus spp., Lactococcus spp., Pediococcus spp., or more varieties (Solis et al., 2002; Homayouni et al., 2014). Furthermore, fecal microbiota transplantation (FMT) is the transplantation of healthy fecal fluids into the gut of the recipient to restore a stable intestinal flora, which affects both the endogenous and host microbes (Gough et al., 2011).
Theoretical risks of probiotics have been illustrated in case reports, including systemic infections, harmful metabolic activities, gene transfer, extreme immune activation in susceptible populations, and gastrointestinal adverse reactions (Doron and Snydman, 2015). Notably, the most frequently reported single event is fungemia caused by consumption of Lactobacillus acidophilus and Lactobacillus casei (Barton et al., 2001; De Groote et al., 2005; Ledoux et al., 2006; Vahabnezhad et al., 2013). Meanwhile, incidents of endocarditis caused by both Lactobacillus and Streptococcus probiotics have also been reported (Mackay et al., 1999; Doron and Snydman, 2015). Although probiotics supplements appear to have no clinically significant side effects, the administration of probiotics in susceptible individuals should be treated with caution.
In conclusion, the existing evidence manifests that some probiotics, such as Lactobacillus, which can restore dysbiosis and enhance intestinal barrier function may prevent the occurrence of cardiovascular and renal complications of SLE and alleviate its symptoms. The mechanism of intestinal microflora imbalance inducing the occurrence and development of SLE may be associated with the abnormal T cell subsets, particularly the abnormal levels of Naïve CD4+T, γδT, Tfh, Treg, and Th17 cells. Further exploration of the mechanism by which the probiotics influence the disease state of SLE, most likely through inflammation and the immune system, may contribute to the progression of future clinical treatments. Therefore, it is significant to shed light on the variation of intestine microbiota to exhibit anti-inflammatory properties, and potentially they can be considered as biomarkers to reflecting disease status. Particularly important is that more animal trials combined with clinical studies are needed to further elucidate the mechanisms for the effect of probiotics, meanwhile, to unravel whether specific probiotics bacteria have a positive impact on the treatment or prevention of SLE to develop novel therapeutic targets.
Author Contributions
XG, XY, QL, and XS searched and reviewed literature. XG defined the structure of the paper and wrote the original draft. HZ and YY supervised the study and revised the paper. All authors reviewed and approved the final format.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
We acknowledge the support from Chengdu Second People’s Hospital and Sichuan Academy of Medical Sciences and Sichuan Provincial People’s Hospital, also express gratitude to the contributions of YY.
References
Adams, C. A. (2010). The Probiotic Paradox: Live and Dead Cells Are Biological Response Modifiers. Nutr. Res. Rev. 23 (1), 37–46. doi:10.1017/S0954422410000090
Alshaiki, F., Obaid, E., Almuallim, A., Taha, R., El-Haddad, H., and Almoallim, H. (2018). Outcomes of Rituximab Therapy in Refractory Lupus: A Meta-Analysis. Eur. J. Rheumatol. 5 (2), 118–126. doi:10.5152/eurjrheum.2018.17096
Andreoli, L., Bertsias, G. K., Agmon-Levin, N., Brown, S., Cervera, R., Costedoat-Chalumeau, N., et al. (2017). EULAR Recommendations for Women's Health and the Management of Family Planning, Assisted Reproduction, Pregnancy and Menopause in Patients with Systemic Lupus Erythematosus And/or Antiphospholipid Syndrome. Ann. Rheum. Dis. 76 (3), 476–485. doi:10.1136/annrheumdis-2016-209770
Apperloo-Renkema, H. Z., Bootsma, H., Mulder, B. I., Kallenberg, C. G., and van der Waaij, D. (1994). Host-microflora Interaction in Systemic Lupus Erythematosus (SLE): Colonization Resistance of the Indigenous Bacteria of the Intestinal Tract. Epidemiol. Infect. 112 (2), 367–373. doi:10.1017/s0950268800057770
Arron, S. T., Dimon, M. T., Li, Z., Johnson, M. E., Wood, T. A., Feeney, L., et al. (2014). High Rhodotorula Sequences in Skin Transcriptome of Patients with Diffuse Systemic Sclerosis. J. Invest. Dermatol. 134 (8), 2138–2145. doi:10.1038/jid.2014.127
Atarashi, K., Tanoue, T., Shima, T., Imaoka, A., Kuwahara, T., Momose, Y., et al. (2011). Induction of Colonic Regulatory T Cells by Indigenous Clostridium Species. Science 331 (6015), 337–341. doi:10.1126/science.1198469
Atarashi, K., Tanoue, T., Oshima, K., Suda, W., Nagano, Y., Nishikawa, H., et al. (2013). Treg Induction by a Rationally Selected Mixture of Clostridia Strains from the Human Microbiota. Nature 500 (7461), 232–236. doi:10.1038/nature12331
Balakrishnan, B., and Taneja, V. (2018). Microbial Modulation of the Gut Microbiome for Treating Autoimmune Diseases. Expert Rev. Gastroenterol. Hepatol. 12 (10), 985–996. doi:10.1080/17474124.2018.1517044
Barton, L. L., Rider, E. D., and Coen, R. W. (2001). Bacteremic Infection with Pediococcus: Vancomycin-Resistant Opportunist. Pediatrics 107 (4), 775–776. doi:10.1542/peds.107.4.775
Bengmark, S. (2003). Use of Some Pre-, Pro- and Synbiotics in Critically Ill Patients. Best Pract. Res. Clin. Gastroenterol. 17 (5), 833–848. doi:10.1016/s1521-6918(03)00073-8
Borchers, A. T., Selmi, C., Meyers, F. J., Keen, C. L., and Gershwin, M. E. (2009). Probiotics and Immunity. J. Gastroenterol. 44 (1), 26–46. doi:10.1007/s00535-008-2296-0
Bron, P. A., Kleerebezem, M., Brummer, R. J., Cani, P. D., Mercenier, A., MacDonald, T. T., et al. (2017). Can Probiotics Modulate Human Disease by Impacting Intestinal Barrier Function. Br. J. Nutr. 117 (1), 93–107. doi:10.1017/S0007114516004037
Buckner, J. H. (2010). Mechanisms of Impaired Regulation by CD4(+)CD25(+)FOXP3(+) Regulatory T Cells in Human Autoimmune Diseases. Nat. Rev. Immunol. 10 (12), 849–859. doi:10.1038/nri2889
Carter, E. E., Barr, S. G., and Clarke, A. E. (2016). The Global burden of SLE: Prevalence, Health Disparities and Socioeconomic Impact. Nat. Rev. Rheumatol. 12 (10), 605–620. doi:10.1038/nrrheum.2016.137
Chen, X. Q., Yu, Y. C., Deng, H. H., Sun, J. Z., Dai, Z., Wu, Y. W., et al. (2010). Plasma IL-17A Is Increased in New-Onset SLE Patients and Associated with Disease Activity. J. Clin. Immunol. 30 (2), 221–225. doi:10.1007/s10875-009-9365-x
Chen, B., Sun, L., and Zhang, X. (2017). Integration of Microbiome and Epigenome to Decipher the Pathogenesis of Autoimmune Diseases. J. Autoimmun. 83, 31–42. doi:10.1016/j.jaut.2017.03.009
Cho, J. H. (2008). The Genetics and Immunopathogenesis of Inflammatory Bowel Disease. Nat. Rev. Immunol. 8 (6), 458–466. doi:10.1038/nri2340
Chrisman, O. D., Snook, G. A., Wilson, T. C., and Short, J. Y. (1976). Prevention of Venous Thromboembolism by Administration of Hydroxychloroquine. A Preliminary Report. J. Bone Jt. Surg Am 58 (7), 918–920. doi:10.2106/00004623-197658070-00003
Claesson, M. J., O'Sullivan, O., Wang, Q., Nikkilä, J., Marchesi, J. R., Smidt, H., et al. (2009). Comparative Analysis of Pyrosequencing and a Phylogenetic Microarray for Exploring Microbial Community Structures in the Human Distal Intestine. PloS one 4 (8), e6669. doi:10.1371/journal.pone.0006669
Corrêa, J. D., Calderaro, D. C., Ferreira, G. A., Mendonça, S. M., Fernandes, G. R., Xiao, E., et al. (2017). Subgingival Microbiota Dysbiosis in Systemic Lupus Erythematosus: Association with Periodontal Status. Microbiome 5 (1), 34. doi:10.1186/s40168-017-0252-z
Crispín, J. C., Oukka, M., Bayliss, G., Cohen, R. A., Van Beek, C. A., Stillman, I. E., et al. (2008). Expanded Double Negative T Cells in Patients with Systemic Lupus Erythematosus Produce IL-17 and Infiltrate the Kidneys. J. Immunol. 181 (12), 8761–8766. Baltimore, Md. : 1950. doi:10.4049/jimmunol.181.12.8761
Da Silva, J. A., Jacobs, J. W., Kirwan, J. R., Boers, M., Saag, K. G., Inês, L. B., et al. (2006). Safety of Low Dose Glucocorticoid Treatment in Rheumatoid Arthritis: Published Evidence and Prospective Trial Data. Ann. Rheum. Dis. 65 (3), 285–293. doi:10.1136/ard.2005.038638
Danchenko, N., Satia, J. A., and Anthony, M. S. (2006). Epidemiology of Systemic Lupus Erythematosus: a Comparison of Worldwide Disease burden. Lupus 15 (5), 308–318. doi:10.1191/0961203306lu2305xx
De Filippo, C., Cavalieri, D., Di Paola, M., Ramazzotti, M., Poullet, J. B., Massart, S., et al. (2010). Impact of Diet in Shaping Gut Microbiota Revealed by a Comparative Study in Children from Europe and Rural Africa. Proc. Natl. Acad. Sci. U S A. 107 (33), 14691–14696. doi:10.1073/pnas.1005963107
De Groote, M. A., Frank, D. N., Dowell, E., Glode, M. P., and Pace, N. R. (2005). Lactobacillus Rhamnosus GG Bacteremia Associated with Probiotic Use in a Child with Short Gut Syndrome. Pediatr. Infect. Dis. J. 24 (3), 278–280. doi:10.1097/01.inf.0000154588.79356.e6
de la Visitación, N., Robles-Vera, I., Toral, M., and Duarte, J. (2019). Protective Effects of Probiotic Consumption in Cardiovascular Disease in Systemic Lupus Erythematosus. Nutrients 11 (11), 2676. doi:10.3390/nu11112676
de la Visitación, N., Robles-Vera, I., Toral, M., O'Valle, F., Moleon, J., Gómez-Guzmán, M., et al. (2020). Lactobacillus Fermentum CECT5716 Prevents Renal Damage in the NZBWF1 Mouse Model of Systemic Lupus Erythematosus. Food Funct. 11 (6), 5266–5274. doi:10.1039/d0fo00578a
De Luca, F., and Shoenfeld, Y. (2019). The Microbiome in Autoimmune Diseases. Clin. Exp. Immunol. 195 (1), 74–85. doi:10.1111/cei.13158
de Oliveira, G. L. V., Leite, A. Z., Higuchi, B. S., Gonzaga, M. I., and Mariano, V. S. (2017). Intestinal Dysbiosis and Probiotic Applications in Autoimmune Diseases. Immunology 152 (1), 1–12. doi:10.1111/imm.12765
de Vrese, M., Stegelmann, A., Richter, B., Fenselau, S., Laue, C., and Schrezenmeir, J. (2001). Probiotics--compensation for Lactase Insufficiency. Am. J. Clin. Nutr. 73 (2 Suppl. l), 421S–429S. doi:10.1093/ajcn/73.2.421s
DeGruttola, A. K., Low, D., Mizoguchi, A., and Mizoguchi, E. (2016). Current Understanding of Dysbiosis in Disease in Human and Animal Models. Inflamm. Bowel Dis. 22 (5), 1137–1150. doi:10.1097/MIB.0000000000000750
Delzenne, N. M., Neyrinck, A. M., Bäckhed, F., and Cani, P. D. (2011). Targeting Gut Microbiota in Obesity: Effects of Prebiotics and Probiotics. Nat. Rev. Endocrinol. 7 (11), 639–646. doi:10.1038/nrendo.2011.126
Dixon, W. G., Kezouh, A., Bernatsky, S., and Suissa, S. (2011). The Influence of Systemic Glucocorticoid Therapy upon the Risk of Non-serious Infection in Older Patients with Rheumatoid Arthritis: a Nested Case-Control Study. Ann. Rheum. Dis. 70 (6), 956–960. doi:10.1136/ard.2010.144741
Doreau, A., Belot, A., Bastid, J., Riche, B., Trescol-Biemont, M. C., Ranchin, B., et al. (2009). Interleukin 17 Acts in Synergy with B Cell-Activating Factor to Influence B Cell Biology and the Pathophysiology of Systemic Lupus Erythematosus. Nat. Immunol. 10 (7), 778–785. doi:10.1038/ni.1741
Doron, S., and Snydman, D. R. (2015). Risk and Safety of Probiotics. Clin. Infect. Dis. 60 (Suppl. 2), S129–S134. doi:10.1093/cid/civ085
Durcan, L., O'Dwyer, T., and Petri, M. (2019). Management Strategies and Future Directions for Systemic Lupus Erythematosus in Adults. Lancet 393 (10188), 2332–2343. doi:10.1016/S0140-6736(19)30237-5
Eckburg, P. B., Bik, E. M., Bernstein, C. N., Purdom, E., Dethlefsen, L., Sargent, M., et al. (2005). Diversity of the Human Intestinal Microbial flora. Science 308 (5728), 1635–1638. doi:10.1126/science.1110591
Esmaeili, S. A., Mahmoudi, M., Momtazi, A. A., Sahebkar, A., Doulabi, H., and Rastin, M. (2017). Tolerogenic Probiotics: Potential Immunoregulators in Systemic Lupus Erythematosus. J. Cel Physiol 232 (8), 1994–2007. doi:10.1002/jcp.25748
Espinola, R. G., Pierangeli, S. S., Gharavi, A. E., Harris, E. N., and Ghara, A. E. (2002). Hydroxychloroquine Reverses Platelet Activation Induced by Human IgG Antiphospholipid Antibodies. Thromb. Haemost. 87 (3), 518–522. doi:10.1055/s-0037-1613033
Fanouriakis, A., Kostopoulou, M., Alunno, A., Aringer, M., Bajema, I., Boletis, J. N., et al. (2019). 2019 Update of the EULAR Recommendations for the Management of Systemic Lupus Erythematosus. Ann. Rheum. Dis. 78 (6), 736–745. doi:10.1136/annrheumdis-2019-215089
Fanouriakis, A., Tziolos, N., Bertsias, G., and Boumpas, D. T. (2021). Update οn the Diagnosis and Management of Systemic Lupus Erythematosus. Ann. Rheum. Dis. 80 (1), 14–25. doi:10.1136/annrheumdis-2020-218272
Fava, A., and Petri, M. (2019). Systemic Lupus Erythematosus: Diagnosis and Clinical Management. J. Autoimmun. 96, 1–13. doi:10.1016/j.jaut.2018.11.001
Ferucci, E. D., Johnston, J. M., Gaddy, J. R., Sumner, L., Posever, J. O., Choromanski, T. L., et al. (2014). Prevalence and Incidence of Systemic Lupus Erythematosus in a Population-Based Registry of American Indian and Alaska Native People, 2007-2009. Arthritis Rheumatol. 66 (9), 2494–2502. doi:10.1002/art.38720
Gardiner, B. J., Tai, A. Y., Kotsanas, D., Francis, M. J., Roberts, S. A., Ballard, S. A., et al. (2015). Clinical and Microbiological Characteristics of Eggerthella Lenta Bacteremia. J. Clin. Microbiol. 53 (2), 626–635. doi:10.1128/JCM.02926-14
Gill, H. S. (2003). Probiotics to Enhance Anti-infective Defences in the Gastrointestinal Tract. Best Pract. Res. Clin. Gastroenterol. 17 (5), 755–773. doi:10.1016/s1521-6918(03)00074-x
Gionchetti, P., Amadini, C., Rizzello, F., Venturi, A., Poggioli, G., and Campieri, M. (2003). Probiotics for the Treatment of Postoperative Complications Following Intestinal Surgery. Best Pract. Res. Clin. Gastroenterol. 17 (5), 821–831. doi:10.1016/s1521-6918(03)00071-4
Gómez, J., Prado, C., López, P., Suárez, A., and Gutiérrez, C. (2009). Conserved Anti-proliferative Effect and Poor Inhibition of TNFalpha Secretion by Regulatory CD4+CD25+ T Cells in Patients with Systemic Lupus Erythematosus. Clin. Immunol. 132 (3), 385–392. doi:10.1016/j.clim.2009.05.012
Goldblatt, F., and O'Neill, S. G. (2013). Clinical Aspects of Autoimmune Rheumatic Diseases. Lancet 382 (9894), 797–808. doi:10.1016/S0140-6736(13)61499-3
Gordon, C., Amissah-Arthur, M. B., Gayed, M., Brown, S., Bruce, I. N., D'Cruz, D., et al. (2018). The British Society for Rheumatology Guideline for the Management of Systemic Lupus Erythematosus in Adults. Rheumatology (Oxford) 57 (1), e1–e45. doi:10.1093/rheumatology/kex286
Gough, E., Shaikh, H., and Manges, A. R. (2011). Systematic Review of Intestinal Microbiota Transplantation (Fecal Bacteriotherapy) for Recurrent Clostridium difficile Infection. Clin. Infect. Dis. 53 (10), 994–1002. doi:10.1093/cid/cir632
Grönwall, C., Chen, Y., Vas, J., Khanna, S., Thiel, S., Corr, M., et al. (2012). MAPK Phosphatase-1 Is Required for Regulatory Natural Autoantibody-Mediated Inhibition of TLR Responses. Proc. Natl. Acad. Sci. U S A. 109 (48), 19745–19750. doi:10.1073/pnas.1211868109
He, Z., Shao, T., Li, H., Xie, Z., and Wen, C. (2016). Alterations of the Gut Microbiome in Chinese Patients with Systemic Lupus Erythematosus. Gut Pathog. 8, 64. doi:10.1186/s13099-016-0146-9
Helmick, C. G., Felson, D. T., Lawrence, R. C., Gabriel, S., Hirsch, R., Kwoh, C. K., et al. (2008). Estimates of the Prevalence of Arthritis and Other Rheumatic Conditions in the United States. Part I. Arthritis Rheum. 58 (1), 15–25. doi:10.1002/art.23177
Hevia, A., Milani, C., López, P., Cuervo, A., Arboleya, S., Duranti, S., et al. (2014). Intestinal Dysbiosis Associated with Systemic Lupus Erythematosus. mBio 5 (5), e01548–14. doi:10.1128/mBio.01548-14
Homayouni, A., Bastani, P., Ziyadi, S., Mohammad-Alizadeh-Charandabi, S., Ghalibaf, M., Mortazavian, A. M., et al. (2014). Effects of Probiotics on the Recurrence of Bacterial Vaginosis: a Review. J. Low Genit Tract Dis. 18 (1), 79–86. doi:10.1097/LGT.0b013e31829156ec
Hsu, T. C., Huang, C. Y., Liu, C. H., Hsu, K. C., Chen, Y. H., and Tzang, B. S. (2017). Lactobacillus Paracasei GMNL-32, Lactobacillus Reuteri GMNL-89 and L. Reuteri GMNL-263 Ameliorate Hepatic Injuries in Lupus-Prone Mice. Br. J. Nutr. 117 (8), 1066–1074. doi:10.1017/S0007114517001039
Hu, W. S., Rajendran, P., Tzang, B. S., Yeh, Y. L., Shen, C. Y., Chen, R. J., et al. (2017). Lactobacillus Paracasei GMNL-32 Exerts a Therapeutic Effect on Cardiac Abnormalities in NZB/W F1 Mice. PloS one 12 (9), e0185098. doi:10.1371/journal.pone.0185098
Kakiyama, G., Pandak, W. M., Gillevet, P. M., Hylemon, P. B., Heuman, D. M., Daita, K., et al. (2013). Modulation of the Fecal Bile Acid Profile by Gut Microbiota in Cirrhosis. J. Hepatol. 58 (5), 949–955. doi:10.1016/j.jhep.2013.01.003
Kalliomäki, M., Salminen, S., Poussa, T., Arvilommi, H., and Isolauri, E. (2003). Probiotics and Prevention of Atopic Disease: 4-year Follow-Up of a Randomised Placebo-Controlled Trial. Lancet 361 (9372), 1869–1871. doi:10.1016/S0140-6736(03)13490-3
Kasselman, L. J., Vernice, N. A., DeLeon, J., and Reiss, A. B. (2018). The Gut Microbiome and Elevated Cardiovascular Risk in Obesity and Autoimmunity. Atherosclerosis 271, 203–213. doi:10.1016/j.atherosclerosis.2018.02.036
Khorasani, S., Mahmoudi, M., Kalantari, M. R., Lavi Arab, F., Esmaeili, S. A., Mardani, F., et al. (2019). Amelioration of Regulatory T Cells by Lactobacillus Delbrueckii and Lactobacillus Rhamnosus in Pristane-Induced Lupus Mice Model. J. Cel Physiol 234 (6), 9778–9786. doi:10.1002/jcp.27663
Kiriakidou, M., and Ching, C. L. (2020). Systemic Lupus Erythematosus. Ann. Intern. Med. 172 (11), ITC81–ITC96. doi:10.7326/AITC202006020
Kronbichler, A., Brezina, B., Quintana, L. F., and Jayne, D. R. (2016). Efficacy of Plasma Exchange and Immunoadsorption in Systemic Lupus Erythematosus and Antiphospholipid Syndrome: A Systematic Review. Autoimmun. Rev. 15 (1), 38–49. doi:10.1016/j.autrev.2015.08.010
Larsen, N., Vogensen, F. K., van den Berg, F. W., Nielsen, D. S., Andreasen, A. S., Pedersen, B. K., et al. (2010). Gut Microbiota in Human Adults with Type 2 Diabetes Differs from Non-diabetic Adults. PloS one 5 (2), e9085. doi:10.1371/journal.pone.0009085
Lavi Arab, F., Rastin, M., Faraji, F., Zamani Taghizadeh Rabe, S., Tabasi, N., Khazaee, M., et al. (2015). Assessment of 1,25-dihydroxyvitamin D3 Effects on Treg Cells in a Mouse Model of Systemic Lupus Erythematosus. Immunopharmacol. Immunotoxicol. 37 (1), 12–18. doi:10.3109/08923973.2014.968255
Ledoux, D., Labombardi, V. J., and Karter, D. (2006). Lactobacillus Acidophilus Bacteraemia after Use of a Probiotic in a Patient with AIDS and Hodgkin's Disease. Int. J. STD AIDS 17 (4), 280–282. doi:10.1258/095646206776253507
Lee, Y. H., and Song, G. G. (2018). Comparative Efficacy and Safety of Intravenous or Subcutaneous Belimumab in Combination with Standard Therapy in Patients with Active Systemic Lupus Erythematosus: a Bayesian Network Meta-Analysis of Randomized Controlled Trials. Lupus 27 (1), 112–119. doi:10.1177/0961203317713143
Lee, Y. H., Choi, S. J., Ji, J. D., and Song, G. G. (2016). Overall and Cause-specific Mortality in Systemic Lupus Erythematosus: an Updated Meta-Analysis. Lupus 25 (7), 727–734. doi:10.1177/0961203315627202
Ley, R. E. (2016). Gut Microbiota in 2015: Prevotella in the Gut: Choose Carefully. Nat. Rev. Gastroenterol. Hepatol. 13 (2), 69–70. doi:10.1038/nrgastro.2016.4
Li, D., Pan, Y., Xia, X., Liang, J., Liu, F., Dou, H., et al. (2020). Bacteroides Fragilis Alleviates the Symptoms of Lupus Nephritis via Regulating CD1d and CD86 Expressions in B Cells. Eur. J. Pharmacol. 884, 173421. doi:10.1016/j.ejphar.2020.173421
Lisnevskaia, L., Murphy, G., and Isenberg, D. (2014). Systemic Lupus Erythematosus. Lancet 384 (9957), 1878–1888. doi:10.1016/S0140-6736(14)60128-8
Liu, Y., Alookaran, J. J., and Rhoads, J. M. (2018). Probiotics in Autoimmune and Inflammatory Disorders. Nutrients 10 (10), 1537. doi:10.3390/nu10101537
López, P., González-Rodríguez, I., Gueimonde, M., Margolles, A., and Suárez, A. (2011). Immune Response to Bifidobacterium Bifidum Strains Support Treg/Th17 Plasticity. PloS one 6 (9), e24776. doi:10.1371/journal.pone.0024776
López, P., de Paz, B., Rodríguez-Carrio, J., Hevia, A., Sánchez, B., Margolles, A., et al. (2016). Th17 Responses and Natural IgM Antibodies Are Related to Gut Microbiota Composition in Systemic Lupus Erythematosus Patients. Sci. Rep. 6, 24072. doi:10.1038/srep24072
Lozupone, C. A., Stombaugh, J. I., Gordon, J. I., Jansson, J. K., and Knight, R. (2012). Diversity, Stability and Resilience of the Human Gut Microbiota. Nature 489 (7415), 220–230. doi:10.1038/nature11550
Luo, X. M., Edwards, M. R., Mu, Q., Yu, Y., Vieson, M. D., Reilly, C. M., et al. (2018). Gut Microbiota in Human Systemic Lupus Erythematosus and a Mouse Model of Lupus. Appl. Environ. Microbiol. 84 (4), e02288–17. doi:10.1128/AEM.02288-17
Lyssuk, E. Y., Torgashina, A. V., Soloviev, S. K., Nassonov, E. L., and Bykovskaia, S. N. (2007). Reduced Number and Function of CD4+CD25highFoxP3+ Regulatory T Cells in Patients with Systemic Lupus Erythematosus. Adv. Exp. Med. Biol. 601, 113–119. doi:10.1007/978-0-387-72005-0_12
Ma, J., Yu, J., Tao, X., Cai, L., Wang, J., and Zheng, S. G. (2010). The Imbalance between Regulatory and IL-17-secreting CD4+ T Cells in Lupus Patients. Clin. Rheumatol. 29 (11), 1251–1258. doi:10.1007/s10067-010-1510-7
Mackay, A. D., Taylor, M. B., Kibbler, C. C., and Hamilton-Miller, J. M. (1999). Lactobacillus Endocarditis Caused by a Probiotic Organism. Clin. Microbiol. Infect. 5 (5), 290–292. doi:10.1111/j.1469-0691.1999.tb00144.x
Man, S. M., Kaakoush, N. O., and Mitchell, H. M. (2011). The Role of Bacteria and Pattern-Recognition Receptors in Crohn's Disease. Nat. Rev. Gastroenterol. Hepatol. Gastroenterol. Hepatol. 8 (3), 152–168. doi:10.1038/nrgastro.2011.3
Mardani, F., Mahmoudi, M., Esmaeili, S. A., Khorasani, S., Tabasi, N., and Rastin, M. (2018). In Vivo study: Th1-Th17 Reduction in Pristane-Induced Systemic Lupus Erythematosus Mice after Treatment with Tolerogenic Lactobacillus Probiotics. J. Cel Physiol 234 (1), 642–649. doi:10.1002/jcp.26819
Marmor, M. F., Kellner, U., Lai, T. Y., Melles, R. B., and Mieler, W. F. (2016). Recommendations on Screening for Chloroquine and Hydroxychloroquine Retinopathy (2016 Revision). Ophthalmology 123 (6), 1386–1394. doi:10.1016/j.ophtha.2016.01.058
Meng, X., Zhou, H. Y., Shen, H. H., Lufumpa, E., Li, X. M., Guo, B., et al. (2019). Microbe-metabolite-host axis, Two-Way Action in the Pathogenesis and Treatment of Human Autoimmunity. Autoimmun. Rev. 18 (5), 455–475. doi:10.1016/j.autrev.2019.03.006
Mu, Q., Zhang, H., and Luo, X. M. (2015). SLE: Another Autoimmune Disorder Influenced by Microbes and Diet. Front. Immunol. 6, 608. doi:10.3389/fimmu.2015.00608
Mu, Q., Zhang, H., Liao, X., Lin, K., Liu, H., Edwards, M. R., et al. (2017). Control of Lupus Nephritis by Changes of Gut Microbiota. Microbiome 5 (1), 73. doi:10.1186/s40168-017-0300-8
Neuman, H., and Koren, O. (2017). The Gut Microbiota: a Possible Factor Influencing Systemic Lupus Erythematosus. Curr. Opin. Rheumatol. 29 (4), 374–377. doi:10.1097/BOR.0000000000000395
Nikolopoulos, D. S., Kostopoulou, M., Pieta, A., Flouda, S., Chavatza, K., Banos, A., et al. (2020). Transition to Severe Phenotype in Systemic Lupus Erythematosus Initially Presenting with Non-severe Disease: Implications for the Management of Early Disease. Lupus Sci. Med. 7 (1), e000394. doi:10.1136/lupus-2020-000394
Ogura, Y., Lala, S., Xin, W., Smith, E., Dowds, T. A., Chen, F. F., et al. (2003). Expression of NOD2 in Paneth Cells: a Possible Link to Crohn's Ileitis. Gut 52 (11), 1591–1597. doi:10.1136/gut.52.11.1591
Pan, H. F., Leng, R. X., Feng, C. C., Li, X. P., Chen, G. M., Li, B. Z., et al. (2013). Expression Profiles of Th17 Pathway Related Genes in Human Systemic Lupus Erythematosus. Mol. Biol. Rep. 40 (1), 391–399. doi:10.1007/s11033-012-2073-2
Peterknecht, E., Keasey, M. P., and Beresford, M. W. (2018). The Effectiveness and Safety of Biological Therapeutics in Juvenile-Onset Systemic Lupus Erythematosus (JSLE): a Systematic Review. Lupus 27 (13), 2135–2145. doi:10.1177/0961203318804879
Petri, M. (2002). Epidemiology of Systemic Lupus Erythematosus. Best Pract. Res. Clin. Rheumatol. 16 (5), 847–858. doi:10.1053/berh.2002.0259
Pons-Estel, G. J., Alarcón, G. S., Scofield, L., Reinlib, L., and Cooper, G. S. (2010). Understanding the Epidemiology and Progression of Systemic Lupus Erythematosus. Semin. Arthritis Rheum. 39 (4), 257–268. doi:10.1016/j.semarthrit.2008.10.007
Rahman, A., and Isenberg, D. A. (2008). Systemic Lupus Erythematosus. N. Engl. J. Med. 358 (9), 929–939. doi:10.1056/NEJMra071297
Rastin, M., Mahmoudi, M., Hatef, M., Sahebari, M., Tabasi, N., Haghmorad, D., et al. (2013). T Lymphocyte Apoptosis in Systemic Lupus Erythematosus Patients. Iran J. Basic Med. Sci. 16 (8), 936–941. doi:10.22038/IJBMS.2013.1353
Rees, F., Doherty, M., Grainge, M. J., Lanyon, P., and Zhang, W. (2017). The Worldwide Incidence and Prevalence of Systemic Lupus Erythematosus: a Systematic Review of Epidemiological Studies. Rheumatology (Oxford) 56 (11), 1945–1961. doi:10.1093/rheumatology/kex260
Reihani, H., Rastin, M., Mahmoudi, M., Ghoryani, M., Abdollahi, N., Tabasi, N. S., et al. (2015). Influence of 1 Alpha, 25-Dihydroxyvitamin D3 on T Helper 17 Cells and Related Cytokines in Systemic Lupus Erythematosus. Iran J. Immunol. 12 (2), 82–93. IJIv12i2A1.IJIv12i2A1.
Saag, K. G., Koehnke, R., Caldwell, J. R., Brasington, R., Burmeister, L. F., Zimmerman, B., et al. (1994). Low Dose Long-Term Corticosteroid Therapy in Rheumatoid Arthritis: an Analysis of Serious Adverse Events. Am. J. Med. 96 (2), 115–123. doi:10.1016/0002-9343(94)90131-7
Sakthiswary, R., and Suresh, E. (2014). Methotrexate in Systemic Lupus Erythematosus: a Systematic Review of its Efficacy. Lupus 23 (3), 225–235. doi:10.1177/0961203313519159
Sankar, S. A., Lagier, J. C., Pontarotti, P., Raoult, D., and Fournier, P. E. (2015). The Human Gut Microbiome, a Taxonomic Conundrum. Syst. Appl. Microbiol. 38 (4), 276–286. doi:10.1016/j.syapm.2015.03.004
Sarnes, E., Crofford, L., Watson, M., Dennis, G., Kan, H., and Bass, D. (2011). Incidence and US Costs of Corticosteroid-Associated Adverse Events: a Systematic Literature Review. Clin. Ther. 33 (10), 1413–1432. doi:10.1016/j.clinthera.2011.09.009
Schiffer, C., Lalanne, A. I., Cassard, L., Mancardi, D. A., Malbec, O., Bruhns, P., et al. (2011). A Strain of Lactobacillus Casei Inhibits the Effector Phase of Immune Inflammation. J. Immunol. 187 (5), 2646–2655. Baltimore, Md. : 1950. doi:10.4049/jimmunol.1002415
Shevach, E. M. (2009). Mechanisms of Foxp3+ T Regulatory Cell-Mediated Suppression. Immunity 30 (5), 636–645. doi:10.1016/j.immuni.2009.04.010
Shor, D. B., Dahan, S., Comaneshter, D., Cohen, A. D., and Amital, H. (2016). Does Inflammatory Bowel Disease Coexist with Systemic Lupus Erythematosus. Autoimmun. Rev. 15 (11), 1034–1037. doi:10.1016/j.autrev.2016.07.027
Solis, B., Samartín, S., Gómez, S., Nova, E., de la Rosa, B., and Marcos, A. (2002). Probiotics as a Help in Children Suffering from Malnutrition and Diarrhoea. Eur. J. Clin. Nutr. 56 (Suppl. 3), S57–S59. doi:10.1038/sj.ejcn.1601488
Somers, E. C., Marder, W., Cagnoli, P., Lewis, E. E., DeGuire, P., Gordon, C., et al. (2014). Population-based Incidence and Prevalence of Systemic Lupus Erythematosus: the Michigan Lupus Epidemiology and Surveillance Program. Arthritis Rheumatol. 66 (2), 369–378. doi:10.1002/art.38238
Su, D. L., Lu, Z. M., Shen, M. N., Li, X., and Sun, L. Y. (2012). Roles of Pro- and Anti-inflammatory Cytokines in the Pathogenesis of SLE. J. Biomed. Biotechnol. 2012, 347141. doi:10.1155/2012/347141
Symmons, D. P. (1995). Frequency of Lupus in People of African Origin. Lupus 4 (3), 176–178. doi:10.1177/096120339500400303
Talaat, R. M., Mohamed, S. F., Bassyouni, I. H., and Raouf, A. A. (2015). Th1/Th2/Th17/Treg Cytokine Imbalance in Systemic Lupus Erythematosus (SLE) Patients: Correlation with Disease Activity. Cytokine 72 (2), 146–153. doi:10.1016/j.cyto.2014.12.027
Tamboli, C. P., Caucheteux, C., Cortot, A., Colombel, J. F., and Desreumaux, P. (2003). Probiotics in Inflammatory Bowel Disease: a Critical Review. Best Pract. Res. Clin. Gastroenterol. 17 (5), 805–820. doi:10.1016/s1521-6918(03)00076-3
Toral, M., Robles-Vera, I., Romero, M., de la Visitación, N., Sánchez, M., O'Valle, F., et al. (2019). Lactobacillus Fermentum CECT5716: a Novel Alternative for the Prevention of Vascular Disorders in a Mouse Model of Systemic Lupus Erythematosus. FASEB J. 33 (9), 10005–10018. doi:10.1096/fj.201900545RR
Tsokos, G. C. (2011). Systemic Lupus Erythematosus. N. Engl. J. Med. 365 (22), 2110–2121. doi:10.1056/NEJMra1100359
Tzang, B. S., Liu, C. H., Hsu, K. C., Chen, Y. H., Huang, C. Y., and Hsu, T. C. (2017). Effects of Oral Lactobacillus Administration on Antioxidant Activities and CD4+CD25+forkhead Box P3 (FoxP3)+ T Cells in NZB/W F1 Mice. Br. J. Nutr. 118 (5), 333–342. doi:10.1017/S0007114517002112
Vahabnezhad, E., Mochon, A. B., Wozniak, L. J., and Ziring, D. A. (2013). Lactobacillus Bacteremia Associated with Probiotic Use in a Pediatric Patient with Ulcerative Colitis. J. Clin. Gastroenterol. 47 (5), 437–439. doi:10.1097/MCG.0b013e318279abf0
Valencia, X., Yarboro, C., Illei, G., and Lipsky, P. E. (2007). Deficient CD4+CD25high T Regulatory Cell Function in Patients with Active Systemic Lupus Erythematosus. J. Immunol. 178 (4), 2579–2588. doi:10.4049/jimmunol.178.4.2579
Van de Wiele, T., Van Praet, J. T., Marzorati, M., Drennan, M. B., and Elewaut, D. (2016). How the Microbiota Shapes Rheumatic Diseases. Nat. Rev. Rheumatol. 12 (7), 398–411. doi:10.1038/nrrheum.2016.85
van der Meulen, T. A., Harmsen, H., Bootsma, H., Spijkervet, F., Kroese, F., and Vissink, A. (2016). The Microbiome-Systemic Diseases Connection. Oral Dis. 22 (8), 719–734. doi:10.1111/odi.12472
van der Meulen, T. A., Harmsen, H. J. M., Vila, A. V., Kurilshikov, A., Liefers, S. C., Zhernakova, A., et al. (2019). Shared Gut, but Distinct Oral Microbiota Composition in Primary Sjögren's Syndrome and Systemic Lupus Erythematosus. J. Autoimmun. 97, 77–87. doi:10.1016/j.jaut.2018.10.009
Van Niel, C. W., Feudtner, C., Garrison, M. M., and Christakis, D. A. (2002). Lactobacillus Therapy for Acute Infectious Diarrhea in Children: a Meta-Analysis. Pediatrics 109 (4), 678–684. doi:10.1542/peds.109.4.678
van Staa, T. P., Leufkens, H. G., and Cooper, C. (2002). The Epidemiology of Corticosteroid-Induced Osteoporosis: a Meta-Analysis. Osteoporos. Int. 13 (10), 777–787. doi:10.1007/s001980200108
van Vollenhoven, R. F., Petri, M. A., Cervera, R., Roth, D. A., Ji, B. N., Kleoudis, C. S., et al. (2012). Belimumab in the Treatment of Systemic Lupus Erythematosus: High Disease Activity Predictors of Response. Ann. Rheum. Dis. 71 (8), 1343–1349. doi:10.1136/annrheumdis-2011-200937
Wang, F., Zhang, W., Wang, S., Pan, W., Liu, L., Wu, M., et al. (2019). Protective Effects of Antimalarials in Chinese Patients with Systemic Lupus Erythematosus. Ann. Rheum. Dis. 78 (8), e80. doi:10.1136/annrheumdis-2018-213819
Warrington, T. P., and Bostwick, J. M. (2006). Psychiatric Adverse Effects of Corticosteroids. Mayo Clin. Proc. 81 (10), 1361–1367. doi:10.4065/81.10.1361
Wei, L., MacDonald, T. M., and Walker, B. R. (2004). Taking Glucocorticoids by Prescription Is Associated with Subsequent Cardiovascular Disease. Ann. Intern. Med. 141 (10), 764–770. doi:10.7326/0003-4819-141-10-200411160-00007
Wei, L. Q., Liang, Y. G., Zhao, Y., Liang, H. T., Qin, D. C., and She, M. C. (2016). Efficacy and Safety of Belimumab Plus Standard Therapy in Patients with Systemic Lupus Erythematosus: A Meta-Analysis. Clin. Ther. 38 (5), 1134–1140. doi:10.1016/j.clinthera.2016.02.022
Yacoub, R., Jacob, A., Wlaschin, J., McGregor, M., Quigg, R. J., and Alexander, J. J. (2018). Lupus: The Microbiome Angle. Immunobiology 223 (6-7), 460–465. doi:10.1016/j.imbio.2017.11.004
Yadav, H., Lee, J. H., Lloyd, J., Walter, P., and Rane, S. G. (2013). Beneficial Metabolic Effects of a Probiotic via Butyrate-Induced GLP-1 Hormone Secretion. J. Biol. Chem. 288 (35), 25088–25097. doi:10.1074/jbc.M113.452516
Yeh, Y. L., Lu, M. C., Tsai, B. C., Tzang, B. S., Cheng, S. M., Zhang, X., et al. (2021). Heat-Killed Lactobacillus Reuteri GMNL-263 Inhibits Systemic Lupus Erythematosus-Induced Cardiomyopathy in NZB/W F1 Mice. Probiotics Antimicrob. Proteins 13 (1), 51–59. doi:10.1007/s12602-020-09668-1
Yen, E. Y., Shaheen, M., Woo, J. M. P., Mercer, N., Li, N., McCurdy, D. K., et al. (2017). 46-Year Trends in Systemic Lupus Erythematosus Mortality in the United States, 1968 to 2013: A Nationwide Population-Based Study. Ann. Intern. Med. 167 (11), 777–785. doi:10.7326/M17-0102
Zamani, B., Golkar, H. R., Farshbaf, S., Emadi-Baygi, M., Tajabadi-Ebrahimi, M., Jafari, P., et al. (2016). Clinical and Metabolic Response to Probiotic Supplementation in Patients with Rheumatoid Arthritis: a Randomized, Double-Blind, Placebo-Controlled Trial. Int. J. Rheum. Dis. 19 (9), 869–879. doi:10.1111/1756-185X.12888
Zhang, W., and Reichlin, M. (2008). A Possible Link between Infection with Burkholderia Bacteria and Systemic Lupus Erythematosus Based on Epitope Mimicry. Clin. Dev. Immunol. 2008, 683489. doi:10.1155/2008/683489
Zhang, H., Liao, X., Sparks, J. B., and Luo, X. M. (2014). Dynamics of Gut Microbiota in Autoimmune Lupus. Appl. Environ. Microbiol. 80 (24), 7551–7560. doi:10.1128/AEM.02676-14
Keywords: autoimmunity, microbiota, probiotics, systemic lupus erythematosus, inflammation
Citation: Guo X, Yang X, Li Q, Shen X, Zhong H and Yang Y (2021) The Microbiota in Systemic Lupus Erythematosus: An Update on the Potential Function of Probiotics. Front. Pharmacol. 12:759095. doi: 10.3389/fphar.2021.759095
Received: 15 August 2021; Accepted: 01 November 2021;
Published: 23 November 2021.
Edited by:
Siomar De Castro Soares, Universidade Federal do Triângulo Mineiro, BrazilReviewed by:
Yehuda Julyus Shoenfeld, Sheba Medical Center, IsraelJian Gao, Shanghai Children’s Medical Center, China
Copyright © 2021 Guo, Yang, Li, Shen, Zhong and Yang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Huiyun Zhong, emhvbmdodWl5dW5AMTI2LmNvbQ==; Yong Yang, eXl4cG93ZXJAMTYzLmNvbQ==
 Xirui Guo
Xirui Guo Xuerong Yang
Xuerong Yang Qi Li
Qi Li Xiaoyan Shen2
Xiaoyan Shen2