
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Pharmacol. , 02 September 2021
Sec. Pharmacogenetics and Pharmacogenomics
Volume 12 - 2021 | https://doi.org/10.3389/fphar.2021.736511
This article is part of the Research Topic Developing Medicines for Infectious Diseases – How Can We Better Use The Potential of Local and Traditional Knowledge? View all 9 articles
 Ochuko L. Erukainure1
Ochuko L. Erukainure1 Motlalepula G. Matsabisa1*
Motlalepula G. Matsabisa1* Aliyu Muhammad2
Aliyu Muhammad2 Musa M. Abarshi2
Musa M. Abarshi2 James F. Amaku3
James F. Amaku3 Sanusi B. Katsayal2
Sanusi B. Katsayal2 Adeline Lum Nde1
Adeline Lum Nde1The possible evolutionary trend of COVID-19 in South Africa was investigated by comparing the genome of SARS-CoV-2 isolated from a patient in KwaZulu-Natal, South Africa with those isolated from China, Spain, Italy, and United States, as well as the genomes of Bat SARS CoV, Middle East Respiratory Syndrome Coronavirus (MERS-CoV), Mouse Hepatitis Virus (MHV), and Infectious Bronchitis Virus (IBV). Phylogenetic analysis revealed a strong homology (96%) between the genomes of SARS-CoV-2 isolated from KwaZulu-Natal, South Africa and those isolated from the study countries as well as those isolated from bat SARS CoV, MERS-CoV, MHV and IBV. The ability of phytocannabinoids from Cannabis sativa infusion to interact with gene segments (mRNAs) coding for proteins implicated in viral replication, assembly and release were also investiagted using computational tools. Hot water infusion of C. sativa leaves was freeze-dried and subjected to Gas Chromatography-Mass Spectroscopy analysis which revealed the presence of tetrahydrocannabivarin, cannabispiran, cannabidiol tetrahydrocannabinol, cannabigerol, and cannabinol. Molecular docking analysis revealed strong binding affinities and interactions between the phytocannabinoids and codon mRNAs for ORF1ab, Surface glycoprotein, Envelope protein and Nucleocapsid phosphoprotein from SARS-CoV-2 whole genome which may be due to chemico-biological interactions as a result of nucleophilic/electrophilic attacks between viral nucleotides and cannabinoids. These results depict the spread of SARS-CoV-2 is intercontinental and might have evolved from other coronaviruses. The results also portray the phytocannabinoids of C. sativa infusion as potential therapies against COVID-19 as depicted by their ability to molecularly interact with codon mRNAs of proteins implicated in the replication, translation, assembly, and release of SARS-CoV-2. However, further studies are needed to verify these activities in pre-clinical and clinical studies.
The outbreak of the corona virus disease 2019 (COVID-19) pandemic has led to an unprecedented pressure on the global health sector, with the sub-Saharan African countries as well as other developing countries being among the most hit (Chen et al., 2020; Willan et al., 2020). The disease which originated from Wuhan China in December 2019 as cluster crises of pneumonia, is caused by the novel β-coronavirus, Severe Acute Respiratory Syndrome Coronavirus 2 popularly regarded as SARS-CoV-2.
Italy, the United Kingdom and Spain were the epicenters for the pandemic in Europe, United States for North America, Brazil for South America, and South Africa for Africa. Although the cases of COVID-19 and mortality rate is lower in African countries compared to other continents, the pandemic has caused a lot of pressure on the poor health systems and economies of these countries which were already burdened with existing non-communicable diseases particularly malaria, AIDS and tuberculosis (Hogan et al., 2020; Sands, 2020; Sherrard-Smith et al., 2020). These setbacks coupled with an uncontrolled growing population exacerbates the continent’s vulnerability to the pandemic with major consequences. Egypt was the first to announce a case in the continent on February 14, 2020, this was followed by Nigeria on February 27, 2020, while South Africa announced her index case on March 5, 2020 (Lone and Ahmad, 2020; Zhao et al., 2020). These figures have so far grown to over 6 digits with South Africa presently having the highest cases. It is pertinent to mention that the virus was brought into the continent by travelers from Europe and has so far led to community spread.
The absence of a substantial standard protocol treatment and vaccine for the disease have led to continuous efforts by scientists and policy makers to curb the disease spread through mainly non-pharmaceutical interventions. These efforts include public sensitization on the virus and the disease, testing, patient isolation and contact tracing, as well as implementation of government policing such as national lockdown, social and physical distancing, mandatory public use of face mask, and constant washing of hands (Mitjà and Clotet, 2020). Despite South Africa passing some of the world’s stringent lockdown rules to curb the transmission of the virus, it has experienced an exacerbated growth in the number of cases which have caused tremendous strain on her health care system and economy. This has led to the search of novel and cheaper alternatives for the treatment of the diseases, with much attention given to indigenous medicinal plants. This has also led to the African stance through the WHO afro, Africa-CDC and the AU Commission in setting up an expert committee on traditional medicines for COVID-19 which aims at strengthening African countries by finding traditional medicines for the disease.
Several studies have reported the potentials of medicinal plants in the treatment and management of COVID-19 and its complications (Benarba and Pandiella, 2020; Erukainure et al., 2020; Vellingiri et al., 2020). These therapeutic properties have been attributed to the phytochemical constituents of the plants. Amongst these plants is Cannabis sativa.
Cannabis sativa is a member of the Cannabis genus and the Cannabaceae family. It is an annual herbaceous plant commonly known as weed, marijuana, Indian hemp, weed and dagga in different parts of the world. Its utilization cuts across religious and recreational purposes, as well as traditional medicine and food (Kuddus et al., 2013; Bonini et al., 2018). The plant is now the most reported for its pharmaceutical medical uses and it is regarded as a global specie as it is widely distributed across the world (UNODC, 2012). Although its utilization is under strict rules in most countries, C. sativa has over the years been reported to be effective in the treatment and management of several ailments such as diabetes mellitus (Ren et al., 2016), different cancer types (Guzman, 2003), insomnia (Ramar et al., 2018), epilepsy (Fusar-Poli et al., 2009), severe pains (Whiting et al., 2015) and neurodegeneration (Aso and Ferrer, 2016). These medicinal properties have been attributed to the phytochemical constituents of C. sativa, particularly the phytocannabinoids. Cannabidiol (CBD), tetrahydrocannabinol (THC) and cannabinol (CBN) constitute the predominant phytocannabinoids in C. sativa (Andre et al., 2016), and have been studied widely for their medicinal applications.
Recently, C. sativa has been gaining interests as a possible therapy for the treatment of COVID-19 and its complications (Byrareddy and Mohan, 2020; Mabou Tagne et al., 2020). This has been attributed to the antiviral activities of its phytocannabinoids such as CBD against hepatitis C and Kaposi sarcoma (Mabou Tagne et al., 2020), as well as its potent anti-inflammatory properties as depicted by its ability to suppress the production of TNF-α, MIP-1a, IL-2, 6, 1α and 1β, MCP-1, and interferon gamma (Nichols and Kaplan, 2020). These proinflammatory cytokines have been implicated in the pathogenesis and progression of COVID-19 induced by SARS-CoV-2 via the cytokine release syndrome (CRS) (Mehta et al., 2020; Nichols and Kaplan, 2020; Shi et al., 2020) commonly regarded as the cytokine storm. Therefore, studying the potentials of C. sativa and its phytocannabinoids as a possible therapy in the treatment and management of COVID-19 will contribute to the curbing of the disease.
This study aimed at decoding the possible evolutionary trend of COVID-19 in South Africa by comparing the genome of SARS-CoV-2 isolated from KwaZulu-Natal, South Africa with those isolated from China, Spain, Italy and United States, as well as the genomes of Bat SARS CoV, Middle East Respiratory Syndrome Coronavirus (MERS-CoV), Mouse Hepatitis Virus (MHV), and Infectious Bronchitis Virus (IBV). The study also investigated the ability of identified compounds from C. sativa to interact with gene segments (mRNAs) coding for proteins implicated in viral replication, assembly and release vis-à-vis ORF1ab, Surface glycoprotein, Envelope protein and Nucleocapsid phosphoprotein from the whole genome of SARS-CoV-2 isolated from KwaZulu-Natal, South Africa using computational tools.
Different complete genome sequences (MT324062, MT291830, MT198652, MT077125, MT371047, FJ588686.1, MG923481.1, MF618252.1, and NC_001451.1) of SARS-CoV-2 (KwaZulu-Natal, South Africa), SARS-CoV-2 (Wuhan, China), SARS-CoV-2 (Valencia, Spain), SARS-CoV-2 (Rome, Italy), SARS-CoV-2 (New York, United States), Bat SARS CoV Rs672/2006, MERS-CoV, MHV and IBV respectively (Table 1), were retrieved from the National Center for Biotechnology Information (NCBI) database and thereafter subjected to alignment using CLUSTALW X Software, version 10.1.8 (Kumar et al., 2018) (Supplementary Figure S1). The aligned sequences were then used to plot phylogenetic tree.
The evolutionary history was inferred using the UPGMA method (Sneath and Sokal, 1973). The optimal tree with the sum of branch length = 1.90049281 is shown. The tree was drawn to scale, with branch lengths in the same units as those of the evolutionary distances used to infer the phylogenetic tree. The evolutionary distances were computed using the Maximum Composite Likelihood method (Tamura et al., 2004) and are in the units of the number of base substitutions per site. This analysis involved 9 nucleotide sequences of the aforementioned viruses. All ambiguous positions were removed for each sequence pair (pairwise deletion option). There was a total of 31,304 positions in the final dataset. Evolutionary analyses were conducted using MEGA X (Kumar et al., 2018).
The use of C. sativa was approved by the South African Health Products Regulatory Authority to conduct, collect, posses, transport and store cannabis plant, plant parts and products for research purposes (Permit No. POS 248/2019/2020; permit issued to the University of Free State).
Cannabis sativa leaves were collected from Mohale’s Hoek District, Lesotho (GPS coordinates: 30.333776″S and 27.651201″E) under the permit (Permit #: 01/LS/2019/10/02–01). The leaves were deposited with the voucher number: BLFU MGM 0018 following its identification and authentication by the Geo Potts Herbarium at the University of the Free State, Bloemfontein 9300, South Africa.
The leaves were air-dried and blended to smooth powder. About 30 g of the powdered samples were infused in boiled water and allowed to extract for 2 h. The infusion was decanted into plastic bowls and allowed to freeze at −80°C before freeze-drying to yield about 9 g of concentrated infusion. The sample was stored in glass vials and stored at 2°C until further analysis.
The concentrated sample was subjected to GC-MS analysis in order to identify the compounds. This was carried out with an Agilent technologies 6890 Series GC coupled with (an Agilent) 5973 Mass Selective detector, which is driven by Agilent Chemstation software. The operating parameters were:
Column: HP-5MS capillary column (30 × 0.25 mm ID, 0.25 μm film thickness, 5% phenylmethylsiloxane); Carrier gas: ultra-pure helium; Gas flow rate: 1.0 ml/min at a linear velocity of 37 cm/s; Programmed oven temperature: 280°C (at the rate of 10 °C/min with a hold time of 3 min); Injector temperature: 250°C; Ion source temperature: 230°C; Quadrupole temperature: 150°C; Electron ionization mode: 70 eV; Electron multiplier voltage: 1859 V; Solvent delay: 4 min; Scan range: 50–70 amu. An inbuilt NIST data library was used in identifying the compounds via comparison of retention time and mass spectral data. The GC-MS spectra is shown in Supplementary Figure S2
SARS-CoV-2 (KwaZulu-Natal, South Africa) whole genome mRNA sequence was obtained from PubMed database from corona virus whole genome (MT324062.1). Gene segment coding for ORF1ab, Surface glycoprotein, Envelope protein and Nucleocapsid phosphoprotein were identified. Possible initiation and termination sequence of the gene were selected based on the identification of regions carrying initiation and termination codons as shown in Table 2. About 45–50 nucleotide sequence from both initiation and termination codons were converted into 3D single mRNA strand using Discovery studio (v19.1.0.18287) and saved as protein data bank file (Biovia, 2015). The structures were prepared by removing all solvent molecules and optimized to simulate physiological conditions using Chimera v 1.1. Polar hydrogens were added, and partial charges were assigned to the standard residue using Gasteiger partial charge.
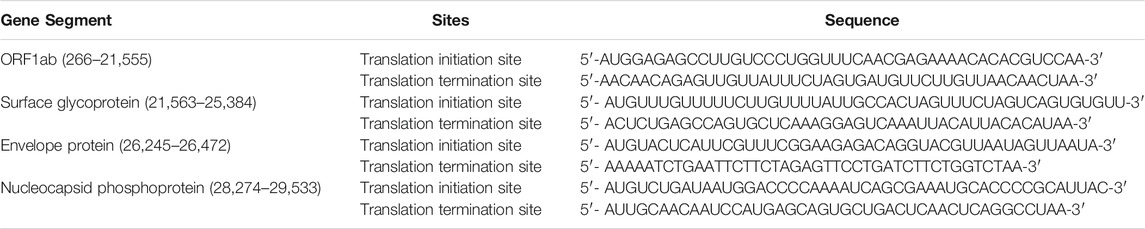
TABLE 2. Selected sequence of initiation and termination sites of target protein mRNAs from SARS-CoV-2 whole genome.
The chemical structure of tetrahydrocannabivarin, cannabispiran, cannabidiol tetrahydrocannabinol, cannabigerol, cannabinol, hydroxychloroquine and remdesivir were retrieved from PubChem online webserver. Gaussian 09 equipped with Gaussview 5.0 was used to optimize the chemical structures (ligands). The optimize ligands were then converted to PDB files using UCSF Chimera tool. The crystal structure of the mRNA (receptors) was built from selected sequence of initiation and termination sites obtained from SARS-COV 2 proteins by making use of UCSF Chimera tool. Hydrogen and Gasteiger partial charges were added to the receptor using the UCSF Chimera tool. Molecular docking simulation was performed on a nucleic acid docking server (HNADOCK). Finally, the ligand-mRNA interaction was assessed using Maestro 11.5 a Schrodinger 2018–1 suite package.
Hydroxychloroquine and remdesivir were chosen as the control antiviral drugs owing to previous reports on their use in the treatment and management of COVID-19 (Al-Tannak et al., 2020; Wang et al., 2020).
The South African SARS-CoV-2 isolate evidently clustered with other SARS-CoV-2 isolates from other countries (China, Italy, United States and Spain) with 96% homology as shown in Figure 1. Interestingly, it also showed 93% homology with the isolate from Bat. This was followed by 92 and 94% homology with MERS-CoV isolate and MHV/IBV, respectively.
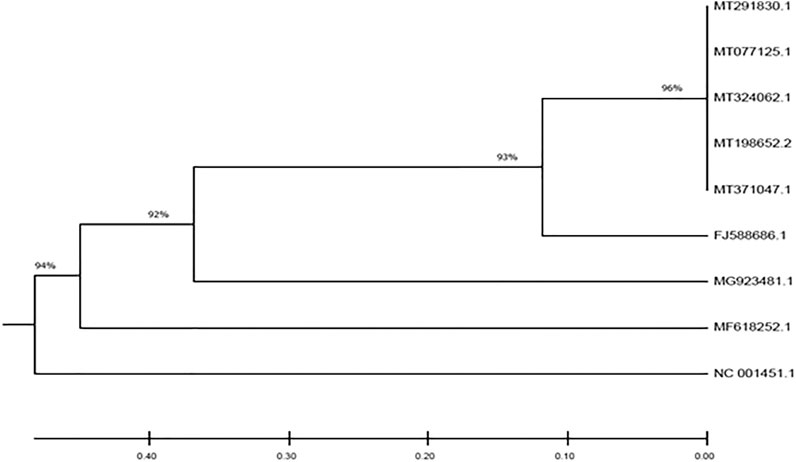
FIGURE 1. Phylogenetic tree depicting the evolutionary relationships between the studied viral genomes.
As depicted in Figure 2 and Table 3, GC-MS analysis of the infusion revealed the presence of tetrahydrocannabivarin, cannabispiran, CBD, THC, cannabigerol and cannabinol. The highest concentration were THC and cannabinol, with 73.84 and 21.74% abundance, respectively as shown in Table 3.
The structure of the compounds and their best docked conformations within the binding site of the selected receptors are shown in Figures 3–8. These favorable interactions between the molecules are depicted by the binding energy as indicated in Table 4. All the compounds bounded to the target sequence reasonably with similar binding pattern. The bindings were seen within the beginning, middle and end of the initiation and termination codons of the various viral proteins respectively. These interactions include both polar and nonpolar through hydrogen, hydrophobic and other non-conventional interactions.
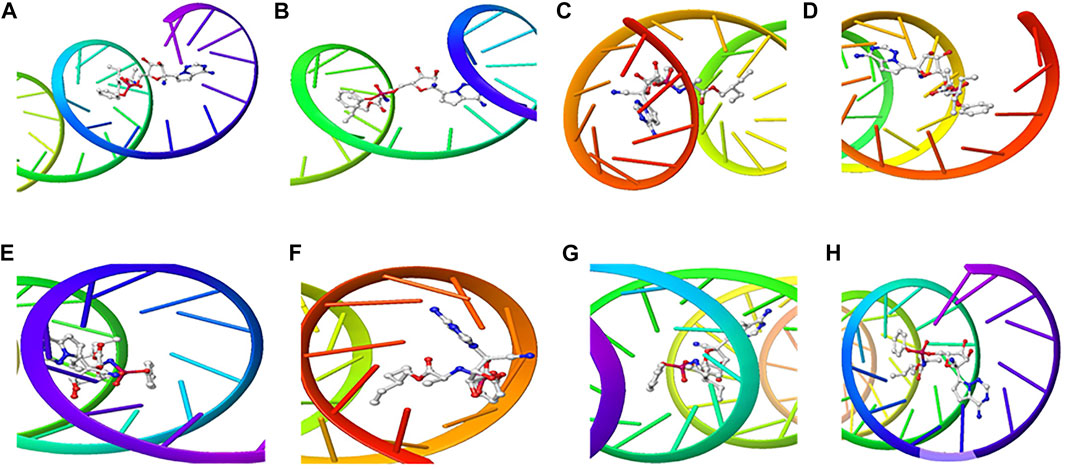
FIGURE 3. 3D Interaction of Remdesivir with Initiation and Termination Sequence of the Viral Proteins. A = Nucleocapsid Phosphoprotein Translation Initiation Site; B = Envelope Protein Translation Initiation Site; C = Envelope Protein Translation Termination Site; D = Nucleocapsid Phosphoprotein Translation Termination Site; E = ORF1ab Translation Initiation Site; F = ORF1ab Translation Termination Site; G = Surface Glycoprotein Translation Termination Site; H = Surface Glycoprotein Translation Initiation Site.
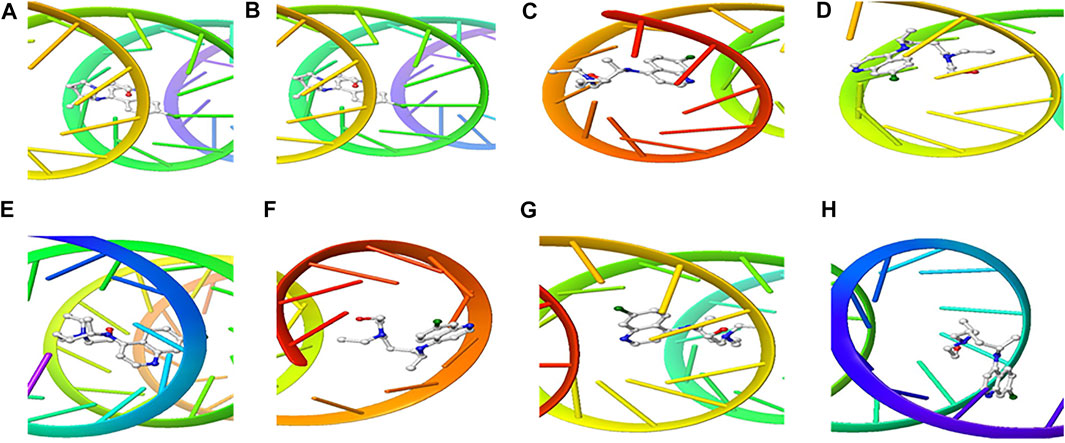
FIGURE 4. 3D Interaction of hydroxychloroquine with Initiation and Termination Sequence of the Viral Proteins. A = Nucleocapsid Phosphoprotein Translation Initiation Site; B = Envelope Protein Translation Initiation Site; C = Envelope Protein Translation Termination Site; D = Nucleocapsid Phosphoprotein Translation Termination Site; E = ORF1ab Translation Initiation Site; F = ORF1ab Translation Termination Site; G = Surface Glycoprotein Translation Termination Site; H = Surface Glycoprotein Translation Initiation Site.
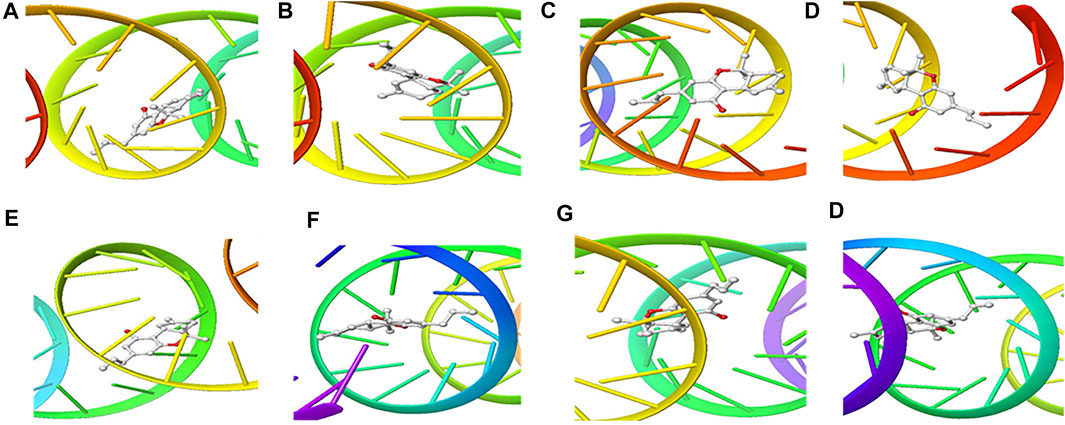
FIGURE 5. 3D Interaction of tetraydrocannabivarin with Initiation and Termination Sequence of the Viral Proteins. A = Nucleocapsid Phosphoprotein Translation Initiation Site; B = Envelope Protein Translation Initiation Site; C = Envelope Protein Translation Termination Site; D = Nucleocapsid Phosphoprotein Translation Termination Site; E = ORF1ab Translation Initiation Site; F = ORF1ab Translation Termination Site; G = Surface Glycoprotein Translation Termination Site; H = Surface Glycoprotein Translation Initiation Site.
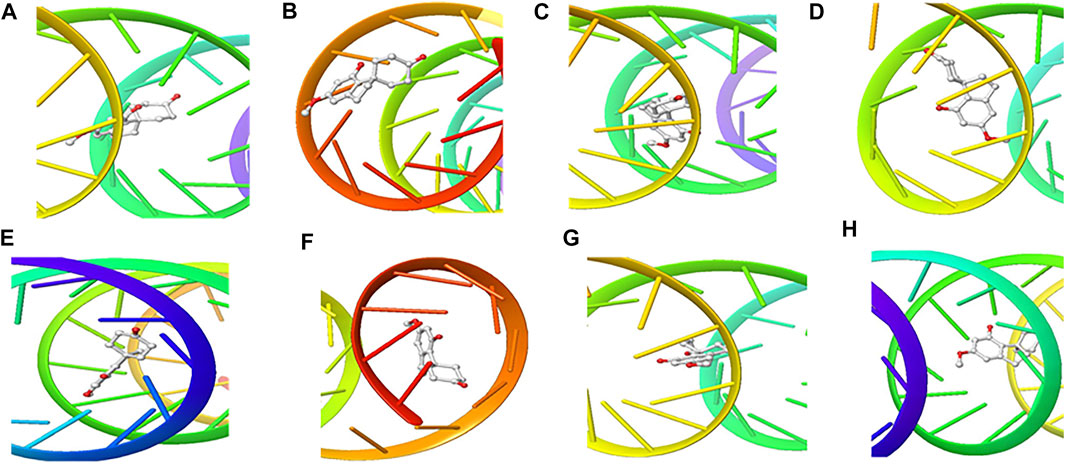
FIGURE 6. 3D Interaction of Cannabispiran with Initiation and Termination Sequence of the Viral Proteins. A = Nucleocapsid Phosphoprotein Translation Initiation Site; B = Envelope Protein Translation Initiation Site; C = Envelope Protein Translation Termination Site; D = Nucleocapsid Phosphoprotein Translation Termination Site; E = ORF1ab Translation Initiation Site; F = ORF1ab Translation Termination Site; G = Surface Glycoprotein Translation Termination Site; H = Surface Glycoprotein Translation Initiation Site.
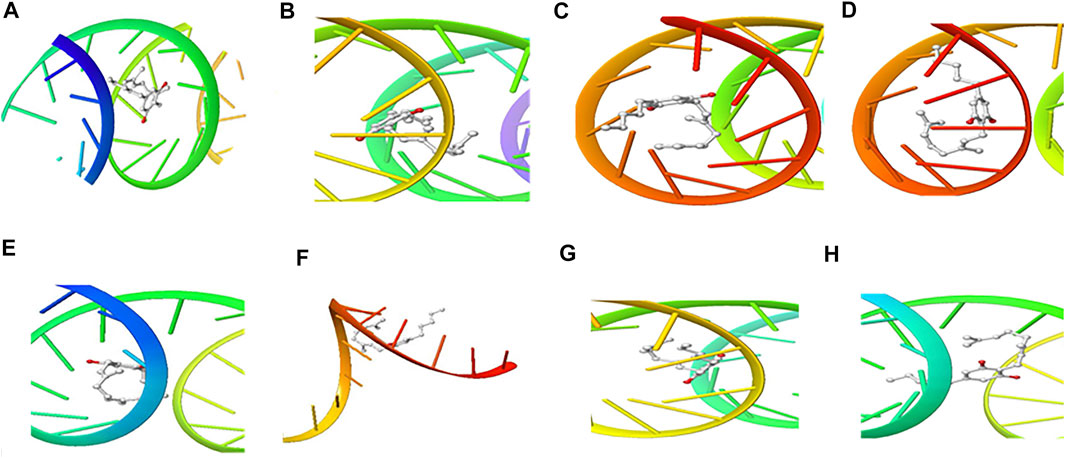
FIGURE 7. 3D Interaction of Cannabigerol with Initiation and Termination Sequence of the Viral Proteins. A = Nucleocapsid Phosphoprotein Translation Initiation Site; B = Envelope Protein Translation Initiation Site; C = Envelope Protein Translation Termination Site; D = Nucleocapsid Phosphoprotein Translation Termination Site; E = ORF1ab Translation Initiation Site; F = ORF1ab Translation Termination Site; G = Surface Glycoprotein Translation Termination Site; H = Surface Glycoprotein Translation Initiation Site.
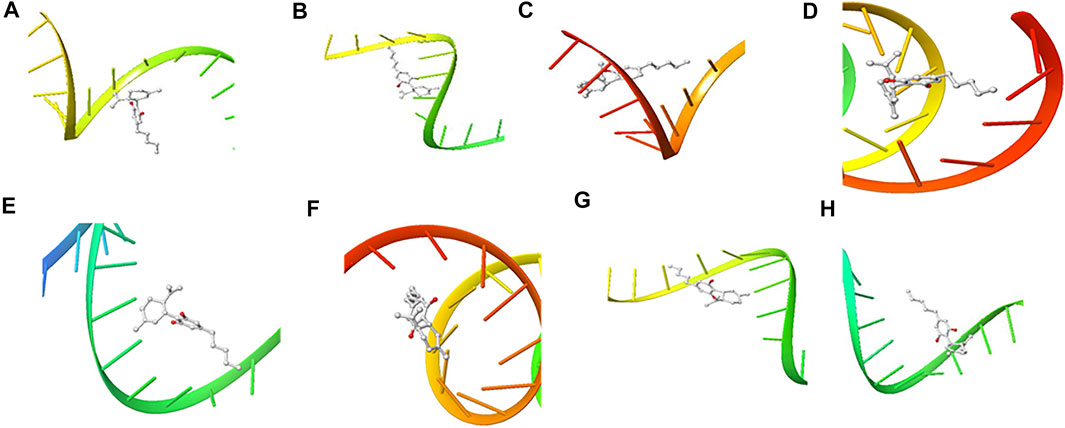
FIGURE 8. 3D Interaction of Cannabidiol with Initiation and Termination Sequence of the Viral Proteins. A = Nucleocapsid Phosphoprotein Translation Initiation Site; B = Envelope Protein Translation Initiation Site; C = Envelope Protein Translation Termination Site; D = Nucleocapsid Phosphoprotein Translation Termination Site; E = ORF1ab Translation Initiation Site; F = ORF1ab Translation Termination Site; G = Surface Glycoprotein Translation Termination Site; H = Surface Glycoprotein Translation Initiation Site.
Since its outbreak in Wuhan China, SARS-CoV-2 has spread across the globe leading to the COVID-19 pandemic. The spread of the disease has been attributed mostly to infected travelers who act as carrier of the virus, and then leading to community spread. In the present study, we decoded the origin of the virus in South Africa by comparing the genome of SARS-CoV-2 isolated from KwaZulu-Natal, South Africa with those of countries with reported high cases (Table 1). The 96% homology with genomes from China, Spain Italy and United States suggests SARS-CoV-2 in South Africa originated from outside the continent and corroborates reports that her index COVID-19 case was a male citizen who tested positive to the virus after a trip to Italy (SAG, 2020). This also corroborates the global spread of the virus and the disease (Lee, 2020; Macintyre, 2020). Furthermore, the 93% homology of the genome of the South African isolate with bat SARS COVs, as well as the 92 and 94% homology with MERS-COV and MHV/IBV isolates, respectively insinuates an evolutionary conservation among the whole genomes of the studied coronaviruses. These evolutionary trends corroborate reports that suggest that coronaviruses are of bat origin (Lu et al., 2020; York, 2020; Zhou et al., 2020).
Coronaviruses are enveloped viruses with a single strand, positive-sense RNA genome of 27–32 kb in length (Liu et al., 2007). Several studies have reported more than 100 full-length or partial genomic sequences for SARS-COV-2 (Wang et al., 2020). The genome contains about six to nine sorts of mRNA, comprising the genome-length mRNA and five to eight sub-genomic mRNA which code for structural protein and many other for non-structural proteins (Liu et al., 2007). In an infected cell, coronaviruses induce the formation of double-membrane vesicles and convoluted membranes that harbor the nonstructural proteins (Verheije et al., 2010). The structural proteins include transmembrane spike glycoprotein, nucleocapsid phosphoprotein, envelope, and surface glycoproteins (Baudoux et al., 1998), whereas the nonstructural includes ORF1ab, ORF3, ORF6, ORF7a, ORF8, and ORF10 which contains information for genome RNA synthesis and replication (Tsai et al., 2020). The entry mechanism of SARS-CoV-2 starts with its transmembrane spike glycoprotein attachment to host cell membrane receptor (angiotensin-converting enzyme 2) and induce its entry through membrane endocytosis. ORF1 stimulates the virus replication and synthesis of the subgenomic RNAs, while nucleocapsid phosphoprotein packages the viral genome to form the helical nucleocapsid that is unified into the budding particle and can also function as an RNA chaperone. Envelope and surface glycoproteins induce virion assembly and morphogenesis leading to the formation of virus-like particles and the final release of the virions by exocytosis (Ho et al., 2004; Zúñiga et al., 2007; Andersen et al., 2020). Any attempt to interfere with the expression of one of these proteins could be tantamount to disrupting the overall SARS-COV-2 transmission cycle. Therefore, targeting these structural and nonstructural protein components especially at translational level of mRNAs being converted to proteins, could serve as a viable therapeutic strategy against SARS-COV-2. This is corroborated by studies which reported ORF1ab, surface glycoprotein, nucleocapsid phosphoprotein, envelope, membrane, spike protein, protease, hemagglutinin esterase and helicase as possible therapeutic target for coronaviruses including SARS-CoV-2 (Mcbride et al., 2014; Bhatia et al., 2020). The strong binding affinities of the identified compounds from C. sativa (Figure 2 and Table 3) with both the initiation and termination codons of ORF1ab, Surface glycoprotein, Envelope protein and Nucleocapsid phosphoprotein mRNAs from the whole genome of SARS-CoV-2 isolated from KwaZulu-Natal, South Africa (Figures 3–8) therefore, portray their high potential as possible therapy for treatment of COVID-19 and its complication by curbing the replication, translation, assembly, and release of the virus. The mechanism of these strong binding affinities of the identified compounds from C. sativa againts the identified mRNA initiation and termination codons could be due some chemico-biological interactions based on the chemistry of the said bioactive compounds versus that of nucleotides. Literally, the constituents of mRNA include adenine, guanine, cytosine, uracil and a phosphate terminal (Elliott and Ladomery, 2017). Meanwhile, these chemical compounds are classified as purine and pyrimidines and are known to constitute basic functional group (amine and carbonyl) with the capacity to undergo nucleophilic or electrophilic reactions (Ulbricht, 2016). In the biological system, natural product dissociates, hence, the tendency to interact with purines and pyrimidines via electrostatic, hydrophobic, intermolecular force or covalent bond is certain. GC-MS analysis of Cannabis sativa extract revealed the presence of tetrahydrocannabivarin, cannabispiran, cannabidiol tetrahydrocannabinol, cannabigerol, and cannabinol. These compounds contain free hydroxyl and aromatic rings which confers on them the ease to interact with the purines, pyrimidines, or phosphate terminal of SARS-CoV-2 genome mRNA sequence via hydrogen bonding or intermolecular interaction. Hydrogen bond formation between the oxygen at carbon 2 of the purines on the genome of SARS-CoV-2 mRNA may be responsible for its interaction with natural product. On the other hand, the free amine attached to carbon 2 and 6 of guanine and adenine respectively may account for the mechanism of interactions (electrostatic) between pyrimidines and the natural product obtained from Cannabis sativa extract. Similarly, the phosphate terminal of the SARS-CoV-2 mRNA could electrostatically interact with the polar region of electron deficient natural product that may have dissociated with the biochemical environment. Hence, at a favourable pH within the biochemical system, natural product obtained from Cannabis sativa extract can dissociate to impart a possible therapeutic function against COVID-19. Furthermore, flavonoid-based molecules from C. sativa have also been shown to bind with high affinity to the spike protein, helicase, and protease sites on the ACE2 receptor used by the coronavirus 2 to infect cells and cause COVID-19 (Attia et al., 2020; Khanna et al., 2020).
Interestingly, the viral proteins have also been implicated in manipulating the host’s innate immune system (Alexander et al., 2019). Conversely, C. sativa and its phytocannabinoids have been proposed as a possible natural products for the treatment of COVID-19 (El Biali et al., 2020; Mckernan et al., 2020). This is also coupled with their reported potent anti-inflammatory activities esepecially CBD, tetrahydrocannabivarin and THC (Bolognini et al., 2010; Anil et al., 2020; Nichols and Kaplan, 2020) as the pathologenesis and progression of the virus have been linked to excessive producition of pro-inflammatory cytokines often termed as cytokine storm syndrome leading to suppression of the immune system (Ragab et al., 2020; Vaninov, 2020).
These results further indicate that phytochemicals from C. sativa can respond to the COVID-19 at different levels of its mRNAs and different mechanisms. They corroborate previous studies that tentatively indicated that CBD and THC may be beneficial in the treatment of patients whose bodies’ inflammatory response has become pathogenic and therefore respond to the cytokine storm (Nagarkatti et al., 2009; Onaivi and Sharma, 2020; Rossi et al., 2020). Much research in this field has focused on the ability of cannabinoids and terpenes to lower the immune system’s response without suppressing.
Taken together, the results from this study indicates a homology between the genome of SARS-CoV-2 isolated from KwaZulu-Natal, South Africa and those isolated from Europe, Asia and North America, as well as those isolated from bat SARS COV, MERS-CoV, MHV and IBV. Thus, depicting the spread of the virus is intercontinental and might have evolved from other coronaviruses. The results also indicate the phytocannabinoids of C. sativa infusion as potential therapies against COVID-19 as depicted by their ability to molecularly interact with codon mRNAs of proteins implicated in the replication, translation, assembly, and release of SARS-CoV-2. However, further studies are needed to verify these activities pre-clinically and clinically. It is also recommended that LC-MS analysis should be carried out on the infusion to further identify its polar constituents.
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author.
Concepts: OE and MM; Research Design: OE, AM, and JA; Execution and Interpretation of Results: AM, MA, SK; AN; JA and OE; Original Manuscript Draft: OE, AM, MA, and JA; Manuscript Revision and Approval: all authors; Supervision: MM.
Prof. MG is thankful to the IKS Based Technology Innovation Unit of DSI South Africa, for financial support (Grant contracts: DST/CON 0162/201 and DST/CON 0206/2019/2020). OE is thankful to the University of the Free State, Bloemfontein, South Africa for Incentives for Rated Researchers (2019060769); and the National Research Foundation (NRF) for Scarce Skills Postdoctoral Research Grant (UID: 132822).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The authors are thankful for to the University of the Free State for technical support and postdoctoral support for OE, Sunday O. Oyedemi and Omolola R. Oyenihi. The authors are also thankful for the assistance and support from the Free State Provincial Police and the Police at Port St Johns and Lusikisiki in the Eastern Cape. The authors are further thankful to Mirranda Javu, senior community liaison officer, Department of Pharmacology UFS for community engagement and negotiations on the cannabis collections. Similar appreciation goes to L R Mafura and S E Molomo both from the Ministry of Forestry, Range, Soil & Water Conservation, Mafeteng, Lesotho for the assistance with the cannabis project and cannabis wild collections.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fphar.2021.736511/full#supplementary-material
Al-Tannak, N. F., Novotny, L., and Alhunayan, A. (2020). Remdesivir-Bringing Hope for COVID-19 Treatment. Sci. Pharm. 88, 29. doi:10.3390/scipharm88020029
Alexander, S. P. H., Kelly, E., Mathie, A., Peters, J. A., Veale, E. L., Armstrong, J. F., et al. (2019). The Concise Guide to PHARMACOLOGY 2019/20: Introduction and Other Protein Targets. Br. J. Pharmacol. 176 (Suppl. 1), S1–S20. doi:10.1111/bph.14747
Andersen, K. G., Rambaut, A., Lipkin, W. I., Holmes, E. C., and Garry, R. F. (2020). The Proximal Origin of SARS-CoV-2. Nat. Med. 26, 450–452. doi:10.1038/s41591-020-0820-9
Andre, C. M., Hausman, J. F., and Guerriero, G. (2016). Cannabis Sativa: The Plant of the Thousand and One Molecules. Front. Plant Sci. 7, 19. doi:10.3389/fpls.2016.00019
Anil, S. M., Shalev, N., Vinayaka, A. C., Nadarajan, S., Namdar, D., Belausov, E., et al. (2020). Cannabis Compounds Have Anti-Inflammatory Activity in Lung Epithelial Cells but Pro-Inflammatory Activity in Macrophages while Increasing Phagocytosis In Vitro. Res. Square. doi:10.21203/rs.3.rs-89166/v1
Aso, E., and Ferrer, I. (2016). CB2 Cannabinoid Receptor as Potential Target against Alzheimer's Disease. Front. Neurosci. 10, 243. doi:10.3389/fnins.2016.00243
Attia, Y. A., Alagawany, M. M., Farag, M. R., Alkhatib, F. M., Khafaga, A. F., Abdel-Moneim, A. E., et al. (2020). Phytogenic Products and Phytochemicals as a Candidate Strategy to Improve Tolerance to Coronavirus. Front. Vet. Sci. 7, 573159. doi:10.3389/fvets.2020.573159
Baudoux, P., Carrat, C., Besnardeau, L., Charley, B., and Laude, H. (1998). Coronavirus Pseudoparticles Formed with Recombinant M and E Proteins Induce Alpha Interferon Synthesis by Leukocytes. J. Virol. 72, 8636–8643. doi:10.1128/JVI.72.11.8636-8643.1998
Benarba, B., and Pandiella, A. (2020). Medicinal Plants as Sources of Active Molecules against COVID-19. Front. Pharmacol. 11, 1189. doi:10.3389/fphar.2020.01189
Bhatia, R., Narang, R. K., and Rawal, R. K. (2020). A Summary of Viral Targets and Recently Released PDB IDs of SARS-CoV-2. Tovj 14, 7–8. doi:10.2174/1874357902014010007
Bolognini, D., Costa, B., Maione, S., Comelli, F., Marini, P., Di Marzo, V., et al. (2010). The Plant Cannabinoid Delta9-Tetrahydrocannabivarin Can Decrease Signs of Inflammation and Inflammatory Pain in Mice. Br. J. Pharmacol. 160, 677–687. doi:10.1111/j.1476-5381.2010.00756.x
Bonini, S. A., Premoli, M., Tambaro, S., Kumar, A., Maccarinelli, G., Memo, M., et al. (2018). Cannabis Sativa: A Comprehensive Ethnopharmacological Review of a Medicinal Plant with a Long History. J. Ethnopharmacol. 227, 300–315. doi:10.1016/j.jep.2018.09.004
Byrareddy, S. N., and Mohan, M. (2020). SARS-CoV2 Induced Respiratory Distress: Can Cannabinoids Be Added to Anti-Viral Therapies to Reduce Lung Inflammation? Brain Behav. Immun. 87, 120–121. doi:10.1016/j.bbi.2020.04.079
Chen, Q., Liang, M., Li, Y., Guo, J., Fei, D., Wang, L., et al. (2020). Mental Health Care for Medical Staff in China during the COVID-19 Outbreak. Lancet Psychiatry 7, e15–e16. doi:10.1016/S2215-0366(20)30078-X
El Biali, M., Broers, B., Besson, M., and Demeules, J. (2020). Cannabinoids and COVID-19. Med. Cannabis Cannabinoids. 3, 1–5. doi:10.1159/000510799
Elliott, D., and Ladomery, M. (2017). Molecular Biology of RNA. Oxford, United Kingdom: Oxford University Press.
Erukainure, O. L., Atolani, O., Muhammad, A., Katsayal, S. B., Ebhuoma, O. O., Ibeji, C. U., et al. (2020). Targeting the Initiation and Termination Codons of SARS-CoV-2 Spike Protein as Possible Therapy Against COVID-19: The Role of Novel Harpagide 5-O-β-D-Glucopyranoside from Clerodendrum Volubile P Beauv.(Labiatae). J. Biomol. Struct. Dyn., 1–14. doi:10.1080/07391102.2020.1840439
Fusar-Poli, P., Crippa, J. A., Bhattacharyya, S., Borgwardt, S. J., Allen, P., Martin-Santos, R., et al. (2009). Distinct Effects of {delta}9-Tetrahydrocannabinol and Cannabidiol on Neural Activation during Emotional Processing. Arch. Gen. Psychiatry 66, 95–105. doi:10.1001/archgenpsychiatry.2008.519
Guzmán, M. (2003). Cannabinoids: Potential Anticancer Agents. Nat. Rev. Cancer 3, 745–755. doi:10.1038/nrc1188
Ho, Y., Lin, P. H., Liu, C. Y., Lee, S. P., and Chao, Y. C. (2004). Assembly of Human Severe Acute Respiratory Syndrome Coronavirus-like Particles. Biochem. Biophys. Res. Commun. 318, 833–838. doi:10.1016/j.bbrc.2004.04.111
Hogan, A. B., Jewell, B. L., Sherrard-Smith, E., Vesga, J. F., Watson, O. J., Whittaker, C., et al. (2020). Potential Impact of the COVID-19 Pandemic on HIV, Tuberculosis, and Malaria in Low-Income and Middle-Income Countries: A Modelling Study. Lancet Glob. Health 8, e1132–e1141. doi:10.1016/S2214-109X(20)30288-6
Khanna, K., Kohli, S. K., Kaur, R., Bhardwaj, A., Bhardwaj, V., Ohri, P., et al. (2021). Herbal Immune-Boosters: Substantial Warriors of Pandemic COVID-19 Battle. Phytomedicine 85, 153361. doi:10.1016/j.phymed.2020.153361
Kuddus, M., Ginawi, I., and AlHazimi, A. (2013). Cannabis Sativa: An Ancient Wild Edible Plant of India. Emir. J. Food Agric. 25, 736–745. doi:10.9755/ejfa.v25i10.16400
Kumar, S., Stecher, G., Li, M., Knyaz, C., and Tamura, K. (2018). MEGA X: Molecular Evolutionary Genetics Analysis across Computing Platforms. Mol. Biol. Evol. 35, 1547–1549. doi:10.1093/molbev/msy096
Lee, A. (2020). Wuhan Novel Coronavirus (COVID-19): Why Global Control Is Challenging? Public health 179, A1. doi:10.1016/j.puhe.2020.02.001
Liu, D. X., Yuan, Q., and Liao, Y. (2007). Coronavirus Envelope Protein: a Small Membrane Protein with Multiple Functions. Cell. Mol. Life Sci. 64, 2043–2048. doi:10.1007/s00018-007-7103-1
Lone, S. A., and Ahmad, A. (2020). COVID-19 Pandemic - an African Perspective. Emerg. Microbes Infect. 9, 1300–1308. doi:10.1080/22221751.2020.1775132
Lu, R., Zhao, X., Li, J., Niu, P., Yang, B., Wu, H., et al. (2020). Genomic Characterisation and Epidemiology of 2019 Novel Coronavirus: Implications for Virus Origins and Receptor Binding. Lancet 395, 565–574. doi:10.1016/S0140-6736(20)30251-8
Mabou Tagne, A., Pacchetti, B., Sodergren, M., Cosentino, M., and Marino, F. (2020). Cannabidiol for Viral Diseases: Hype or Hope? Cannabis Cannabinoid Res. 5, 121–131. doi:10.1089/can.2019.0060
Macintyre, C. R. (2020). Global Spread of COVID-19 and Pandemic Potential. Glob. Biosecur. 1, 3. doi:10.31646/gbio.55
Mcbride, R., Van Zyl, M., and Fielding, B. C. (2014). The Coronavirus Nucleocapsid Is a Multifunctional Protein. Viruses 6, 2991–3018. doi:10.3390/v6082991
Mckernan, K., Kane, L. T., and Helbert, Y. (2020). Quantitative PCR for Cannabis Flower Containing SARs-CoV-2. BioRxiv.
Mehta, P., Mcauley, D. F., Brown, M., Sanchez, E., Tattersall, R. S., and Manson, J. J. (2020). COVID-19: Consider Cytokine Storm Syndromes and Immunosuppression. Lancet 395, 1033–1034. doi:10.1016/S0140-6736(20)30628-0
Mitjà, O., and Clotet, B. (2020). Use of Antiviral Drugs to Reduce COVID-19 Transmission. Lancet Glob. Health 8, e639-e640. doi:10.1016/s2214-109x(20)30114-5
Nagarkatti, P., Pandey, R., Rieder, S. A., Hegde, V. L., and Nagarkatti, M. (2009). Cannabinoids as Novel Anti-Inflammatory Drugs. Future Med. Chem. 1, 1333–1349. doi:10.4155/fmc.09.93
Nichols, J. M., and Kaplan, B. L. F. (2020). Immune Responses Regulated by Cannabidiol. Cannabis Cannabinoid Res. 5, 12–31. doi:10.1089/can.2018.0073
Onaivi, E. S., and Sharma, V. (2020). Cannabis for COVID-19: Can Cannabinoids Quell the Cytokine Storm? Future Sci. OA 6, FSO625. doi:10.2144/fsoa-2020-0124
Ragab, D., Salah Eldin, H., Taeimah, M., Khattab, R., and Salem, R. (2020). The COVID-19 Cytokine Storm; what We Know So Far. Front. Immunol. 11, 1446. doi:10.3389/fimmu.2020.01446
Ramar, K., Rosen, I. M., Kirsch, D. B., Chervin, R. D., Carden, K. A., Aurora, R. N., et al. (2018). Medical Cannabis and the Treatment of Obstructive Sleep Apnea: An American Academy of Sleep Medicine Position Statement. J. Clin. Sleep Med. 14, 679–681. doi:10.5664/jcsm.7070
Ren, Y., Liang, K., Jin, Y., Zhang, M., Chen, Y., Wu, H., et al. (2016). Identification and Characterization of Two Novel α-Glucosidase Inhibitory Oligopeptides from Hemp ( Cannabis Sativa L.) Seed Protein. J. Funct. Foods 26, 439–450. doi:10.1016/j.jff.2016.07.024
Rossi, F., Tortora, C., Argenziano, M., Di Paola, A., and Punzo, F. (2020). Cannabinoid Receptor Type 2: A Possible Target in SARS-CoV-2 (CoV-19) Infection?. Int. J. Mol. Sci. 21, 3809. doi:10.3390/ijms21113809
SAG (2020). Minister Zweli Mkhize Reports First Case of Coronavirus Covid-19 [Online]. Available: https://www.gov.za/speeches/health-reports-first-case-covid-19-coronavirus-5-mar-2020-0000 (Accessed November 29, 2020).
Sands, P. (2020). HIV, Tuberculosis, and Malaria: How Can the Impact of COVID-19 Be Minimised? Lancet Glob. Health 8, e1102–e1103. doi:10.1016/S2214-109X(20)30317-X
Sherrard-Smith, E., Hogan, A. B., Hamlet, A., Watson, O. J., Whittaker, C., Winskill, P., et al. (2020). The Potential Public Health Consequences of COVID-19 on Malaria in Africa. Nat. Med. 26, 1411–1416. doi:10.1038/s41591-020-1025-y
Shi, Y., Wang, Y., Shao, C., Huang, J., Gan, J., Huang, X., et al. (2020). COVID-19 Infection: The Perspectives on Immune Responses. Cell Death Differ 27, 1451–1454. doi:10.1038/s41418-020-0530-3
Sneath, P. H., and Sokal, R. R. (1973). Numerical Taxonomy. The Principles and Practice of Numerical Classification. San Francisco: Freeman.
Tamura, K., Nei, M., and Kumar, S. (2004). Prospects for Inferring Very Large Phylogenies by Using the Neighbor-Joining Method. Proc. Natl. Acad. Sci. U S A. 101, 11030–11035. doi:10.1073/pnas.0404206101
Tsai, P. H., Wang, M. L., Yang, D. M., Liang, K. H., Chou, S. J., Chiou, S. H., et al. (2020). Genomic Variance of Open Reading Frames (ORFs) and Spike Protein in Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2). J. Chin. Med. Assoc. 83, 725–732. doi:10.1097/JCMA.0000000000000387
Ulbricht, T. L. V. (2016). Purines, Pyrimidines and Nucleotides and the Chemistry of Nucleic Acids. Elsevier.
UNODC (2012). Cannabis: A Short Review [Online]. Available: https://www.unodc.org/documents/drug-prevention-and-treatment/cannabis_review.pdf (Accessed November 12, 2019).
Vaninov, N. (2020). In the Eye of the COVID-19 Cytokine Storm. Nat. Rev. Immunol. 20, 277. doi:10.1038/s41577-020-0305-6
Vellingiri, B., Jayaramayya, K., Iyer, M., Narayanasamy, A., Govindasamy, V., Giridharan, B., et al. (2020). COVID-19: A Promising Cure for the Global Panic. Sci. Total Environ. 725, 138277. doi:10.1016/j.scitotenv.2020.138277
Verheije, M. H., Hagemeijer, M. C., Ulasli, M., Reggiori, F., Rottier, P. J., Masters, P. S., et al. (2010). The Coronavirus Nucleocapsid Protein Is Dynamically Associated with the Replication-Transcription Complexes. J. Virol. 84, 11575–11579. doi:10.1128/JVI.00569-10
Wang, C., Liu, Z., Chen, Z., Huang, X., Xu, M., He, T., et al. (2020a). The Establishment of Reference Sequence for SARS-CoV-2 and Variation Analysis. J. Med. Virol. 92, 667–674. doi:10.1002/jmv.25762
Wang, S., Mulier, S., Jonscher, C., Ye, S., Chen, L., Feng, Y., et al. (2020b). Hypothesis: What Is the Best We Can Do with Hydroxychloroquine for COVID-19? Clin. Epidemiol. 12, 1139–1144. doi:10.2147/CLEP.S277889
Whiting, P. F., Wolff, R. F., Deshpande, S., Di Nisio, M., Duffy, S., Hernandez, A. V., et al. (2015). Cannabinoids for Medical Use: A Systematic Review and Meta-Analysis. JAMA 313, 2456–2473. doi:10.1001/jama.2015.6358
Willan, J., King, A. J., Jeffery, K., and Bienz, N. (2020). Challenges for NHS Hospitals during Covid-19 Epidemic. BMJ 368, m1117. doi:10.1136/bmj.m1117
York, A. (2020). Novel Coronavirus Takes Flight from Bats? Nat. Rev. Microbiol. 18, 191. doi:10.1038/s41579-020-0336-9
Zhao, Z., Li, X., Liu, F., Zhu, G., Ma, C., and Wang, L. (2020). Prediction of the COVID-19 Spread in African Countries and Implications for Prevention and Control: A Case Study in South Africa, Egypt, Algeria, Nigeria, Senegal and Kenya. Sci. Total Environ. 729, 138959. doi:10.1016/j.scitotenv.2020.138959
Zhou, P., Yang, X. L., Wang, X. G., Hu, B., Zhang, L., Zhang, W., et al. (2020). A Pneumonia Outbreak Associated with a New Coronavirus of Probable Bat Origin. Nature 579, 270–273. doi:10.1038/s41586-020-2012-7
Keywords: Cannabis sativa, COVID-19, SARS-CoV-2, phytocannabinoids, South Africa
Citation: Erukainure OL, Matsabisa MG, Muhammad A, Abarshi MM, Amaku JF, Katsayal SB and Nde AL (2021) Targeting of Protein’s Messenger RNA for Viral Replication, Assembly and Release in SARS-CoV-2 Using Whole Genomic Data From South Africa: Therapeutic Potentials of Cannabis Sativa L. Front. Pharmacol. 12:736511. doi: 10.3389/fphar.2021.736511
Received: 06 July 2021; Accepted: 17 August 2021;
Published: 02 September 2021.
Edited by:
Joao Lago, Federal University of ABC, BrazilReviewed by:
Roberto Carlos Campos Martins, Federal University of Rio de Janeiro, BrazilCopyright © 2021 Erukainure, Matsabisa, Muhammad, Abarshi, Amaku, Katsayal and Nde. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Motlalepula G. Matsabisa, bWF0c2FiaXNhbWdAdWZzLmFjLnph
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.