
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Pharmacol., 14 July 2021
Sec. Pharmacogenetics and Pharmacogenomics
Volume 12 - 2021 | https://doi.org/10.3389/fphar.2021.711940
This article is part of the Research TopicPersonalized Medicine in Neuropsychiatric Disorders - From Preclinical Studies to Clinical ApplicationsView all 5 articles
 Paula Soria-Chacartegui1†
Paula Soria-Chacartegui1† Gonzalo Villapalos-García1†
Gonzalo Villapalos-García1† Pablo Zubiaur1,2
Pablo Zubiaur1,2 Francisco Abad-Santos1,2,3*
Francisco Abad-Santos1,2,3* Dora Koller4*
Dora Koller4*Olanzapine, aripiprazole and risperidone are atypical antipsychotics or neuroleptics widely used for schizophrenia treatment. They induce various adverse drug reactions depending on their mechanisms of action: metabolic effects, such as weight gain and alterations of glucose and lipid metabolism; hyperprolactinemia and extrapyramidal effects, such as tremor, akathisia, dystonia, anxiety and distress. In this review, we listed polymorphisms associated with individual response variability to olanzapine, aripiprazole and risperidone. Olanzapine is mainly metabolized by cytochrome P450 enzymes, CYP1A2 and CYP2D6, whereas aripiprazole and risperidone metabolism is mainly mediated by CYP2D6 and CYP3A4. Polymorphisms in these genes and other enzymes and transporters, such as enzymes from the uridine 5'-diphospho-glucuronosyltransferase (UGT) family and ATP-binding cassette sub-family B member 1 (ABCB1), are associated to differences in pharmacokinetics. The three antipsychotics act on dopamine and serotonin receptors, among others, and several studies found associations between polymorphisms in these genes and variations in the incidence of adverse effects and in the response to the drug. Since olanzapine is metabolized by CYP1A2, a lower starting dose should be considered in patients treated with fluvoxamine or other CYP1A2 inhibitors. Regarding aripiprazole, a reduced dose should be administered in CYP2D6 poor metabolizers (PMs). Additionally, a reduction to a quarter of the normal dose is recommended if the patient is treated with concomitant CYP3A4 inhibitors. Risperidone dosage should be reduced for CYP2D6 PMs and titrated for CYPD6 ultrarapid metabolizers (UMs). Moreover, risperidone dose should be evaluated when a CYP2D6, CYP3A4 or ABCB1 inhibitor is administered concomitantly.
Schizophrenia is a debilitating mental illness characterized by a distortion of thinking, perceptions, emotions and behavior. It is a complex syndrome that begins in adolescence and early adulthood, affecting daily functioning and educational and work performance. One percent of the world's population suffers from schizophrenia, establishing an urgent problem for healthcare systems worldwide (Schultz et al., 2007). It is a polygenic disorder influenced by environmental factors. The symptoms of this illness are classified into positive, negative and cognitive. Positive symptoms, also called psychotic symptoms, include hallucinations and delusions. The main negative symptoms include flattened affect, anhedonia, apathy, social withdrawal and loss of will. Cognitive impairment includes attention, memory and language deficit and disorientation (Mueser and McGurk, 2004).
Schizophrenia is developed by alterations in different neurotransmitters (Laruelle, 2014). The main hypothesis is the dopaminergic hypothesis, which proposes a chemical imbalance of dopamine in the brain (Brisch et al., 2014). Nevertheless, alternative hypotheses were developed that complement the dopaminergic hypothesis and that are related to other neurotransmitters, such as serotonin or glutamate (Laruelle, 2014; Stahl, 2018). Consequently, the main targets of antipsychotic drugs which are used for schizophrenia treatment are dopamine and serotonin receptors. These antipsychotics, also referred as neuroleptics, are classified into typical and atypical, and differ in their mechanism of action, therefore the side effects they induce (Meltzer, 2013). Typical antipsychotics, e.g., haloperidol, which act mainly as dopamine receptor (DRD) antagonists, have little or no effect on negative and cognitive symptoms and frequently cause extrapyramidal adverse effects (Maric et al., 2016). Atypical antipsychotics, also called second generation antipsychotics (SGAs), act as antagonists to DRD, serotonin (5-HTR), histamine, α-adrenergic and muscarinic receptors. The most common SGAs are olanzapine, quetiapine, clozapine, risperidone and paliperidone. These antipsychotics are associated with metabolic adverse effects and weight gain. In addition, some authors propose that aripiprazole, a SGA with unique mechanism of action, is a third generation antipsychotic as it is a partial agonist of DRD2 (Maric et al., 2016). Moreover, antipsychotic drugs are also used for the treatment of acute manic or mixed episodes associated with bipolar disorder, for the major depression disorder and for autistic disorder (FDA, 1993, 1996, 2014).
This review provides a comprehensive summary of pharmacogenetic biomarkers associated with the metabolic effects of three representative atypical antipsychotics: risperidone, olanzapine and aripiprazole. Aripiprazole was selected since it has a unique mechanism of action. Risperidone and olanzapine were selected for being representative of two of the main types of atypical antipsychotics.
Olanzapine and risperidone are atypical antipsychotics that act as antagonists at different neurotransmitters receptors, such as DRD, 5-HTR, histamine, α-adrenergic and muscarinic receptors. Aripiprazole acts as a partial agonist of DRD2 and 5-HTR (Maric et al., 2016). The complete mechanism of action of olanzapine, risperidone and aripiprazole is shown in Table 1. However, the mechanism of action of antipsychotics is not completely understood. Therefore, the barrier between side effects and adverse events is questionable as their mechanism of action is poorly described.
Olanzapine has a 60% of oral bioavailability and displays linear pharmacokinetics over the clinical dosing range. Its bioavailability is not influenced by food intake (Callaghan et al., 1999). It is extensively eliminated by first pass metabolism, with approximately 40% of the dose metabolized before reaching the systemic circulation. Its maximum plasma concentration (Cmax) is reached in 5 h (Tmax) following an oral dose (Tamminga and Kane, 1997). When administered once daily, it requires approximately 1 week to reach steady-state concentrations. Olanzapine is extensively distributed throughout the body, with a volume of distribution (Vd) of approximately 1533 L or 21 L/kg 93% of the drug binds to plasma proteins, primarily to albumin and α1-acid glycoprotein (Callaghan et al., 1999). It is mainly excreted in urine (57%) and feces (30%) (Callaghan et al., 1999). Olanzapine is highly metabolized, since 7% of the drug is excreted as unchanged drug in urine (FDA, 1996). Its half-life (T1/2) ranges from 21 to 54 h and its clearance (Cl), from 12 to 47 L/h. Sex, smoking and age affect olanzapine plasma concentrations, Cl and T1/2 (Callaghan et al., 1999).
The metabolism of olanzapine is extensive and complex (Figure 1). It generates different metabolites with a lower pharmacological activity compared to the parent compound, therefore, they do not significantly contribute to the clinical effects of olanzapine (Söderberg and Dahl, 2013). The main reaction olanzapine undergoes is N-glucuronidation, which results in the 10 N-glucuronide and, to a lesser extent, 4 N-glucuronide metabolites. The 10 N-glucuronide metabolite is the main metabolite circulating in plasma, present at steady state at 44% of olanzapine concentration (FDA, 1996). These reactions are mainly catalyzed by uridine diphosphate glucuronosyl transferase UGT1A4 and UGT2B10 (Söderberg and Dahl, 2013). In addition, olanzapine undergoes oxidation reactions catalyzed by different isoforms of the cytochrome P450 enzymes (CYP). The 1A2 isoform (CYP1A2) produces the N-desmethylolanzapine metabolite (DMO), which is present at 31% of the concentration of olanzapine at steady state. DMO is also produced by CYP2D6 and CYP2C8, although to a lesser extent. CYP2D6 and secondarily, CYP3A4 and CYP2C9, produce the 2-hydroximethylolanzapine metabolite (Söderberg and Dahl, 2013; Korprasertthaworn et al., 2015). However, CYP2D6 metabolism seems to occur marginally in vivo (FDA, 1996). In addition, olanzapine undergoes N-oxidation reactions, catalyzed by the flavin monooxygenase 3 (FMO3) enzyme, which produces N-oxide olanzapine, also generated by CYP2D6 (Figure 1) (Söderberg and Dahl, 2013; Okubo et al., 2016).
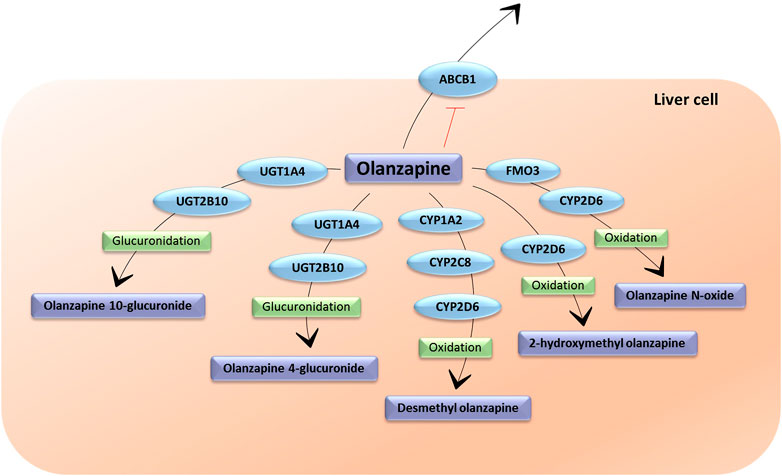
FIGURE 1. The metabolic pathways of olanzapine. UGT1A4: Uridine 5′-diphospho-glucuronosyltransferase Family 1 Subfamily A Member 4; UGT2B10: Uridine 5′ diphospho-glucuronosyltransferase Family 2 Subfamily B Member 10; FMO3: flavin monooxigenase 3; ABCB1: ATP-binding cassette sub-family B member 1; CYP1A2: cytochrome P450 Family 1 Subfamily A Member 2; CYP2C8: cytochrome P450 Family 2 Subfamily C Member 8; CYP2D6: cytochrome P450 Family 2 Subfamily D Member 6 (Figure inspired by PharmGKB olanzapine pharmacokinetic pathway).
The prevalence of smokers is higher in schizophrenic patients than in general population (52.9% and 40.1%, respectively, p = 0.031) (Ohi et al., 2019). Tobacco induces CYP1A2 activity, what leads to reduced olanzapine plasma concentration. Thus, smoking affects olanzapine Cl, however, dosage modifications are not routinely recommended (FDA, 1996; Tsuda et al., 2014).
Olanzapine is transported by and acts as inhibitor of the ATP-binding cassette sub-family B member 1 transporter (ABCB1). This transporter, also called P-glycoprotein (P–gp), is an efflux pump involved in the excretion of several antipsychotics in the blood–brain barrier (BBB), limiting their bioavailability in the brain (Wang et al., 2006). Moreover, it is located in other organs, such as the gastrointestinal tract and the liver, influencing olanzapine location (Wang et al., 2006).
Aripiprazole has an absolute oral bioavailability of 87%. Depending on the dose, the Cmax is reached between 2.8 and 6.8 h after drug intake, with a median Tmax of 3–4 h. It presents linear pharmacokinetics. Its Vd is of 404 L or 4.9 L/kg and it has high affinity for plasma proteins with 99% binding. Cl is 0.7 L/min/kg and is eliminated in feces (60%) and urine (27%) (Shapiro et al., 2003).
Aripiprazole is mainly transformed in the liver through dehydrogenation, hydroxylation and N-dealkylation by CYP2D6 and CYP3A4 (Figure 2) (Kinghorn and McEvoy, 2005). Its main and only active metabolite is dehydro-aripiprazole, which amounts to 40% of aripiprazole concentration at steady state, reached after 14 days of treatment (Kinghorn and McEvoy, 2005). The T1/2 varies between 58 and 78 h and can reach 140 h in CYP2D6 poor metabolizers (PMs) (Kinghorn and McEvoy, 2005). Both aripiprazole and dehydro-aripiprazole are possible substrates of P–gp (Belmonte et al., 2018).
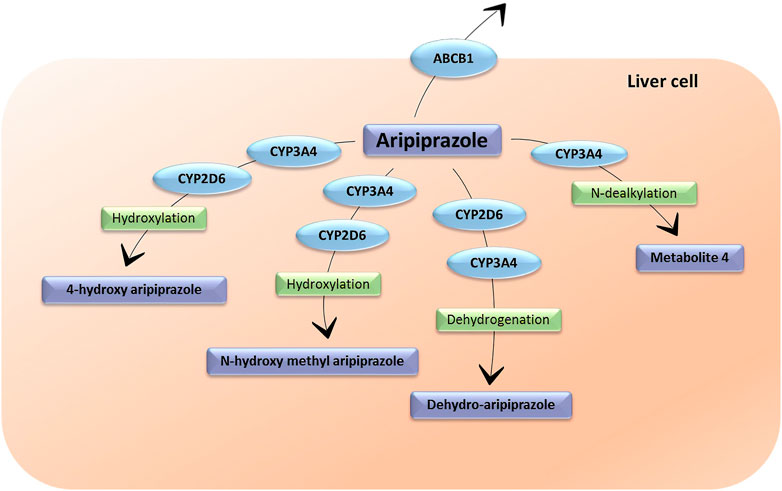
FIGURE 2. The metabolic pathways of aripiprazole. CYP3A4: cytochrome P450 Family 3 Subfamily A Member 4; CYP2D6: cytochrome P450 Family 2 Subfamily D Member 6. ABCB1: ATP-binding cassette sub-family B member 1. Metabolite 4 does not have an assigned name.
Risperidone has an absolute oral bioavailability of 70%. Plasma concentrations of risperidone, its major metabolite and both together are dose proportional over the dosing range (0.5–8 mg twice daily) (FDA, 1993). Following administration, risperidone Cmax is reached after about 1 h 9-hydroxyrisperidone Tmax depends on CYP2D6 phenotype: 3 h in normal metabolizers (NM), and 13 h in PM (FDA, 1993). The Vd is 1–2 L/kg. In plasma, 90% of risperidone and 77.4% of 9-hydroxirisperidone is bound to albumin and α1-acid glycoprotein. Risperidone and its metabolites are eliminated via the urine and, to a much lesser extent, via the feces (Germann et al., 2012). The pharmacokinetics of risperidone and 9-hydroxyrisperidone combined, after single and multiple doses, are similar in NM and PM, with an overall mean T1/2 of about 20 h (FDA, 1993).
Risperidone suffers a 9-hydroxylation in the liver catalyzed by CYP2D6, CYP3A4 and CYP3A5 (Fang et al., 1999). This hydroxylation produces 9-hydroxyrisperidone, which is an active metabolite that was commercialized as paliperidone (Corena-McLeod, 2015). The sum of risperidone and 9-hydroxyrisperidone represents the active moiety of this antipsychotic. A minor metabolic pathway is through N-dealkylation through CYP3A4 and CYP3A5 (Germann et al., 2012; Ganoci et al., 2021) and 7-hydroxylation (Berecz et al., 2004), both resulting in inactive metabolites. In vitro studies showed that risperidone is a substrate and an inhibitor of P-gp (Ganoci et al., 2021) (Figure 3).
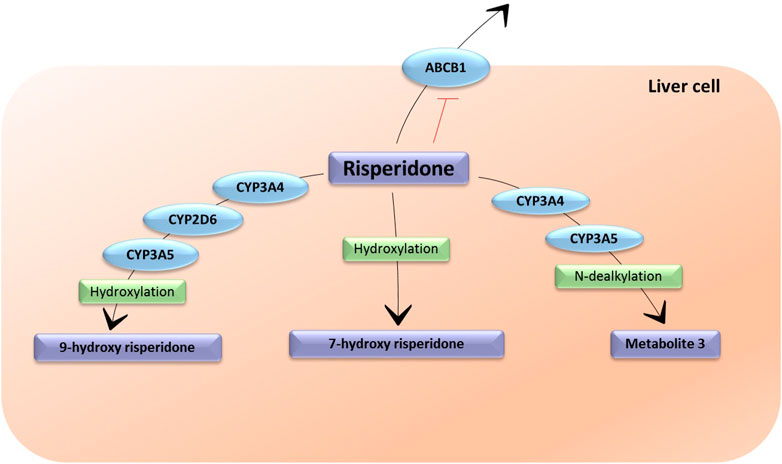
FIGURE 3. The metabolic pathways of risperidone. CYP3A4: Cytochrome P450 Family 3 Subfamily A Member 4; CYP3A5: Cytochrome P450 Family 3 Subfamily A Member 5; CYP2D6: Cytochrome P450 Family 2 Subfamily D Member 6; ABCB1: ATP-binding cassette sub-family B member 1. Metabolite 3 does not have an assigned name. The enzymes responsible for the formulation of 7-hydroxy risperidone are unknown.
Olanzapine causes metabolic adverse effects to the greatest extent among antipsychotics e.g., weight gain and alterations in glucose and lipid metabolism (Rummel-Kluge et al., 2010; Roerig et al., 2011; Barton et al., 2020). The metabolic effects of risperidone are also notable, but less frequent compared to olanzapine (FDA, 1993; Rummel-Kluge et al., 2010). On the contrary, aripiprazole barely causes these effects (Bak et al., 2014; Barton et al., 2020). The mechanisms by which olanzapine and risperidone cause increased intake, weight gain and fat deposition metabolism are not completely known. It seems to involve different peptide, neurotransmitter and receptor systems in the appetite and reward systems in the brain (Roerig et al., 2011; Aringhieri et al., 2018). A clinical trial reported that olanzapine- and risperidone-induced long term weight gain was not correlated with appetite alterations in chronically treated patients. Nonetheless, the authors did not exclude that weight gain in first time treated patients could be produced by increased appetite (Smith et al., 2012). It is hypothesized that additional drug-mediated mechanisms are partially responsible for antipsychotic-induced weight gain (Himmerich et al., 2015). Olanzapine-induced weight gain was observed even after only 5 days acute treatment in healthy volunteers (Koller et al., 2021). Other studies observed a 0.9 kg per month weight gain in schizophrenic patients treated with olanzapine (Lieberman et al., 2005) or a weight gain of 13.9 kg in patients after 1 year of olanzapine treatment (Kahn et al., 2008).
Risperidone-induced weight gain or metabolic impairments are less frequent and severe compared to olanzapine, but greater compared to aripiprazole (Komossa et al., 2009, 2011; Aringhieri et al., 2018). Clinical trials demonstrated that the same phenomenon occurs in different populations. In 107 adult schizophrenic stable outpatients, olanzapine induced greater weight gain than risperidone, however, risperidone was associated with more hospitalizations (Noordsy et al., 2017). In 144 antipsychotic naïve young patients, adverse changes in adiposity and insulin sensitivity were observed during 12 weeks of antipsychotic treatment with olanzapine, risperidone or aripiprazole, and olanzapine-related fat increase was the most significant (Nicol et al., 2018). An overweight (body mass index (BMI) superior to 26 kg/m2) adult cohort of patients presented reduced prevalence of overweight in the risperidone group compared to those receiving olanzapine (Meyer et al., 2005). In a Chinese cohort of first episode schizophrenia patients, aripiprazole was the less weight-gain inducer, as opposed to olanzapine, which was the greater weight gain inducer (Cheng et al., 2019). In contrast to this, other studies reported no significant differences in metabolic syndrome development induced by olanzapine or risperidone. In a clinical trial of 46 overweight patients, olanzapine and risperidone showed no difference in the prevalence of diabetes or prediabetic glucose or glycohemoglobin levels after 5 months of treatment. Nonetheless, olanzapine induced higher insulin resistance and consequently, increased insulin levels (Smith et al., 2009). In the same population, no difference was found in weight-gain after 5 months, although at 2 months of treatment olanzapine induced greater increase in triglycerides, and very low density (VLDL) cholesterol and triglycerides (Smith et al., 2010). Despite these facts, risperidone was found to induce metabolic side effects such as lipid metabolism imbalance, cholesterol and VLDL level increase, hyperglycemia and insulin resistance in other studies (Robinson et al., 2015; Nicol et al., 2018).
One of the receptors that might be involved in the development of metabolic effects is the 5-HT2A serotonin receptor. The blockade of this receptor by olanzapine causes an increase in food intake and weight, by altering the secretion of the neuropeptide PY (Ruaño et al., 2007). In addition, this receptor is involved in the regulation of glucose intake by the muscle, so that its inhibition favors the development of hyperglycemia and insulin resistance (Roerig et al., 2011). On the other hand, blockage of the 5-HT2C receptor causes alterations in satiety and food intake by altering the regulation of leptin (Reynolds et al., 2006). Olanzapine and risperidone are antagonists to 5-HTR and aripiprazole is a partial agonist (Roerig et al., 2011; Aringhieri et al., 2018); therefore the signaling cascade that leads to weight gain via 5-HT receptors may be less stimulated by aripiprazole. Thus, the different affinity profile of aripiprazole could explain the reduced number of metabolic adverse events compared to other antipsychotics (Shapiro et al., 2003; Kane et al., 2009; Rummel-Kluge et al., 2010).
The role of DRD1, DRD2 and DRD3 in weight gain is not clearly known. Nonetheless, presumably olanzapine’s DRD2 antagonism alters feeding behavior since this receptor is involved in the reward system (Rivera-Iñiguez et al., 2019). The blockage of H1 histamine receptor and α1 and α2 adrenergic receptors by olanzapine may alter feeding and glucose control and regulation of body energy balance, playing a role in weight gain and metabolism (Roerig et al., 2011). Aripiprazole and risperidone present moderate affinity for H1 histamine receptors, which may explain their moderate effect on weight gain (Shapiro et al., 2003; Aringhieri et al., 2018).
Olanzapine is also known to increase the risk of developing type II diabetes (Lambert et al., 2006; Koller et al., 2021). Its M3 receptor antagonism causes an alteration in glucose control and insulin response, and the blockade of the H1 receptor favors the development of insulin resistance. Inhibition of 5-HT receptors, in particular 5-HT1A, leads to alterations in glucose metabolism favoring hyperglycemia (Jeon and Kim, 2017). Moreover, atypical antipsychotics can also provoke dyslipemia or hyperlipidemia, however, the mechanisms by which this occurs are not clearly known (Jeon and Kim, 2017). It is proposed that olanzapine alters hepatic lipid metabolism by interfering with lipogenesis, lipoprotein internalization and cholesterol clearance, favoring the accumulation of lipids and the development of hyperlipidemia (Chen et al., 2018).
The most common (≥5% and at least twice than placebo) adverse events to olanzapine in schizophrenic patients are constipation, weight gain, dizziness, personality disorder, akathisia, postural hypotension, sedation, headache, increased appetite, fatigue, dry mouth, hyperprolactinemia and abdominal pain. Olanzapine was associated to leukopenia and thrombocytopenia (FDA, 1996). Apart from metabolic effects, olanzapine causes short and long-term movement disorders, such as bradykinesia, akathisia; Parkinsonism and dystonia. These are known as extrapyramidal adverse drug reactions.
The most frequently reported adverse events during risperidone treatment are of greater prevalence (more than 10%) than metabolic effects, and are the following: insomnia, anxiety, headache, upper respiratory tract infection, Parkinsonism, depression and akathisia. The less frequent, but still common (1–10% of cases) adverse events are pneumonia, bronchitis, sinusitis, urinary tract infection, influenza; anemia, sleep disorders, agitation, decreased libido, sedation or sleepiness, dystonia, dizziness, dyskinesia, tremor, blurred vision, tachycardia, hypotension or hypertension, abdominal pain, abdominal discomfort, vomiting, nausea, constipation, gastroenteritis, diarrhea, dyspepsia, dry mouth, hyperprolactinemia, toothache, exanthema, muscle spasms, musculoskeletal pain, back pain, arthralgia (FDA, 1993).
The most common adverse drug reactions (>10% patients) to aripiprazole are mostly extrapyramidal effects, headache, agitation, insomnia, anxiety, nausea and vomiting, akathisia, light-headedness, and constipation (Prommer, 2017).
Dizziness seems to happen due to the antagonism on α1-adrenergic receptors, as well as tachycardia, bradycardia and syncope (FDA, 1996). Antagonism on muscarinic receptors causes anticholinergic effects, such as dry mouth and constipation (Bhana et al., 2001). Antagonism on 5-HT receptors seems also to be in involved in the appearance of constipation, as serotonin participates in the activation of colonic smooth muscle contraction (Zhang et al., 2016). Headache and somnolence are thought to be caused by the antagonism on 5-HT, H1 histamine and α1-adrenergic receptors, however, the molecular mechanisms involved are not completely known (Fang et al., 2016). Olanzapine seems to induce the least neurological adverse events compared to risperidone and aripiprazole (Cheng et al., 2019).
During antipsychotic treatment, the follow up of several adverse events is highly recommended, e.g. QTc interval monitoring, since some antipsychotics may be associated with QTc prolongation, such as thioridazine or ziprasidone, which might be a signal of increased risk of arrhythmia (Marder et al., 2004). Olanzapine, risperidone and aripiprazole do not usually alter the QTc length (Czekalla et al., 2001; Glassman and Bigger, 2001; Ozeki et al., 2010; Spellmann et al., 2018). Risperidone can cause hypotension (Wilson et al., 2017), while olanzapine and aripiprazole usually do not affect blood pressure (Woo et al., 2009; Seven et al., 2017). Some studies observed a lower systolic and diastolic blood pressure, heart rate and QTc lowering effect on the first day of olanzapine treatment, however these effects progressively diminished on the following days probably due to developing tolerance (Bever and Perry, 1998; Koller et al., 2021).
Antipsychotics, including olanzapine, risperidone and aripiprazole, are also known to cause neuroleptic malignant syndrome, a rare life threatening adverse drug reaction (FDA, 1993, 1996, 2014). The four main symptoms of this syndrome are altered mental status, muscle rigidity, hyperthermia, and autonomic instability (Velamoor, 2017). It is thought to be related with D2 receptor blockage, among others (Velamoor, 2017).
Personalized medicine is an emergent approach with the objective to prevent, diagnose and treat diseases using specific information about genes, proteins and environment of each patient individually (Di Sanzo et al., 2017). This field evolved rapidly during the last decade thanks to the continuous evolution of technology and to the development of genetic analyses (Di Sanzo et al., 2017).
Pharmacogenetics studies the influence of genetic variability on drug response. Its aim is to select "the right drug, at the right dose, for the right patient," predicting the tolerance and effectiveness of the drug in each patient, considering the individual's genetic information (Daudén Tello, 2006). To date, two of the most important groups that develop pharmacogenetic guidelines are the Clinical Pharmacogenetics Implementation Consortium (CPIC) (Caudle et al., 2014) and the Dutch Pharmacogenetics Working Group (DPWG) (Bank et al., 2018). The CPIC was established in 2009 as a joint project between the Pharmacogenomics Research Network (PGRN) and the Pharmacogenomics Knowledge Base (PharmGKB) (Caudle et al., 2014). These guidelines classify individuals into different metabolizer phenotypes for different enzymes based on the genetic variants in the gene of interest. The main phenotypes are PM, when individuals carry non function or decreased function variants; intermediate metabolizers (IM), when individuals carry decreased function variants or non-function variants with normal function variants, presenting higher activity than PMs; NM, when individuals carry normal function variants and ultrarapid metabolizers (UM), when individuals carry increased function variants. Moreover, the phenotype of specific patients can be modified if they are treated with inhibitors or inducers of the enzyme of interest. Pharmacogene Variation Consortium also gathers information about the polymorphisms identified in each pharmacogene (Pharmacogene Variation Consortium, 2017). Regulatory agencies such as FDA (Food and Drug Administration) and EMA (European Medicines Agency) already include pharmacogenetic dosing recommendations for different drugs (FDA, 2019; EMA, 2020).
Antipsychotic treatment shows a hardly replicable response that cannot be predicted, leading to a trial-and-error prescribing strategy to choose the ideal treatment for each patient (Lally and MacCabe, 2015). Genetic factors influencing pharmacokinetics and pharmacodynamics may be the cause of part of this heterogeneity (Eum et al., 2016). The DPWG published guidelines in favor of CYP2D6 genotyping for patients under aripiprazole and risperidone treatment, whereas no recommendation has been published for olanzapine to date (Bank et al., 2018; PharmGKB guidelines, 2020). Regarding aripiprazole, the DPWG proposed that a reduced dose should be administered for CYP2D6 PMs (Swen et al., 2011). Additionally, an adjustment of the normal dose is recommended if the patient is treated with concomitant CYP3A4 inhibitors, inducers or CYP2D6 inhibitors (FDA, 2014). There are no dosage recommendations for olanzapine based on CYP2D6 phenotype. Since olanzapine is metabolized by CYP1A2, a lower starting dose should be considered in patients treated with fluvoxamine or other CYP1A2 inhibitors (FDA, 1996). Regarding risperidone, the DPWG recommends a reduction of risperidone dose in CYP2D6 PMs since May 2020. Additionally, it recommends changing treatment to an alternative drug or titration of the dose according to the maximum concentration for the active metabolite for CYP2D6 UMs (PharmGKB guidelines, 2020). Moreover, the proper dose for the patient has to be evaluated in case of coadministration of other drugs, such as CYP3A4 and ABCB1 inducers (carbamazepine, phenobarbital, etc.), CYP2D6 inhibitors (fluoxetine, paroxetine) or ABCB1 inhibitors (e. g. Verapamil) (FDA, 1993).
The findings of several studies show that pharmacogenetics may be useful in the clinical practice, mainly to reduce the adverse drug reactions to these drugs, and thus achieve greater adherence to treatment (Brandl et al., 2014; Zai et al., 2018).
Several approaches allow investigating if genetic variants are associated with pharmacokinetics and pharmacodynamics of antipsychotics. One approach is genome-wide association studies (GWAS), which can discover new genes without any prior hypothesis on the complex biology of the disease. The main drawbacks of GWAS are its high price and complexity. Moreover, careful statistical analysis must be performed in other to avoid false positives resulting from the massive amount of statistical tests. Additionally, the selection of the individuals included should be done carefully to avoid insufficient sample size and population stratification. GWAS are used to discover new genes involved in a disease or in a drug response. However, the findings made in GWAS have to be confirmed in candidate genes studies. Candidate gene studies are association studies with the objective to identify relationships between polymorphisms in candidate genes, also called pharmacogenes, and different clinical events, such as efficacy and safety in patients or pharmacokinetics and safety in healthy volunteers. Candidate genes are previously selected based on a suspected role or a previous association in the development of a specific disease or phenotype. The advantages of candidate gene studies are its simplicity and affordability. The main disadvantages are the considerable chance of not finding associations derived from the limited gene selection and no possible gene discovery. The majority of the data presented here was collected from candidate gene studies.
The discovery of pharmacogenetic biomarkers could improve antipsychotic treatment. The administration of the right drug at the right dose for each patient facilitates the achievement of the desired efficacy while lowering the risks of adverse effects, thus achieving a greater adherence to therapy and a better control of the disease.
Among the polymorphisms found in CYP1A2, only some of them were associated with variation in the expression or inducibility of the enzyme. Five alleles were defined that can affect olanzapine metabolism: *1C (rs2069514, −3860G > A), *1D (rs35694136, 22467delT), *1E (rs2069526), *1F (rs762551, −163C > A) and *1K (rs2069526, rs12720461, and rs762551). However, only the *1D and *1F alleles were associated with increased and reduced olanzapine plasma concentration, respectively, in some candidate gene studies (Söderberg and Dahl, 2013; Czerwensky et al., 2015), although this association could not be found in other candidate gene studies (Cabaleiro et al., 2013; Koller et al., 2020). A polygenic risk score model was proposed for CYP1A2, which classifies individuals into PMs, NMs and UMs (Saiz-Rodríguez et al., 2019). However, associations between this risk score model and pharmacokinetic parameters were not found in other candidate gene studies (Koller et al., 2020; Zubiaur et al., 2021) (Table 2). The rs2472297 polymorphism, located between the CYP1A1 and CYP1A2 genes, was associated with variations in olanzapine plasma concentration in a GWAS, explaining 2% of the variability (Söderberg and Dahl, 2013). The lack of agreement among studies means that there are still no recommendations for olanzapine dosing based on CYP1A2 genotype (Table 2).
No-function and reduced-function were defined in CYP2D6 (Caudle et al., 2020). These alleles allow the classification of individuals into different phenotypes: PM, IM, NM and UM. Although this cytochrome is involved in the metabolism of olanzapine, numerous candidate gene studies did not find associations between phenotypes and plasma concentration of olanzapine (Nozawa et al., 2008; Söderberg and Dahl, 2013; Koller et al., 2020; Zubiaur et al., 2021). This is consistent with its secondary role on the metabolism of this drug (Table 2).
CYP3A4 is known to be in involved in olanzapine metabolism (Söderberg and Dahl, 2013). CYP3A4 presents high allelic variance (26 alleles), however, only a few of these alleles possess altered function (Pharmacogene Variation Consortium, 2017). Its most studied no-function alleles are CYP3A4*6 (rs4646438) and CYP3A4*20 (rs67666821). However, there is no clear evidence for their effects to date (Sanchez Spitman et al., 2017). Regardless the presence of no function variants, CYP3A4 could compensate for this effect by being overexpressed, as it is highly inducible (Sanchez Spitman et al., 2017). Its high inducibility makes it difficult to find pharmacogenetic associations with drugs (Sanchez Spitman et al., 2017). Due to the similar substrate specificity between CYP3A4 and CYP3A5, some drugs metabolized by CYP3A4 are also metabolized by CYP3A5. CYP3A5*3 (defined by rs776746) is a non-function allele (Wong, 2004). A study found significantly lower AUC in individuals *3/*3 for CYP3A5 (Cabaleiro et al., 2013). A different candidate gene study did not find differences in olanzapine pharmacokinetics regarding CYP3A4/5 (Zubiaur et al., 2021). Therefore, results are contradictory, and additional studies are needed to evaluate its impact on olanzapine metabolism (Table 2).
Additionally, two candidate gene studies found a higher T1/2 and Vd in CYP2C9 PM (*2/*3 and *3/*3 carriers) compared to IM and NM, and a higher incidence of adverse drug reactions in *3 carriers (Cabaleiro et al., 2013; Zubiaur et al., 2021) (Table 2).
Several polymorphisms were identified in the ABCB1 gene; three of them were related to olanzapine pharmacokinetics in different candidate gene studies. 3435T > C (rs1045642) and 2677T > G/A (rs2032582) T allele carriers presented a higher plasma area under the curve (AUC) of olanzapine, as well as a better response in olanzapine treatment (Skogh et al., 2011; Saiz-Rodríguez et al., 2018). In addition, one study observed that 1236T > C (rs1128503) T allele carriers were associated with a higher serum concentration of olanzapine (Skogh et al., 2011). In a different candidate gene study, ABCB1 rs3842 CC individuals showed a greater olanzapine exposure (Zubiaur et al., 2021). Other polymorphisms were related to a variation of olanzapine T1/2 (Koller et al., 2020) (Table 2).
Two alleles were identified in the UGT1A4 gene: *2 (p.P24T, rs6755571) and *3 (p.L48V, rs2011425). The *3 allele was associated with increased glucuronidation of olanzapine in vivo. UGT2B10 *2 (p.D67Y, rs61750900) was associated with a decreased olanzapine glucuronidation in vitro (Erickson-Ridout et al., 2011). A candidate gene study found that UGT1A1*80 (rs887829) *80 homozygotes had higher Tmax than *1/*80 and *1/*1 individuals (Koller et al., 2020) (Table 2).
The three most studied FM O 3 polymorphisms are p. E158K (rs2266782), p. V257M (rs1736557) and p. E308G (rs2266780). p. E158K and p. E308G constitute the allele K158-G308 (Söderberg and Dahl, 2013). This allele was associated with a reduction of olanzapine N-oxidation in vitro. The *6 allele (rs12720462) in FM O 1 gene seems to be associated with higher olanzapine plasma concentrations (Söderberg et al., 2013) (Table 2).
The only existing pharmacogenetic guideline in aripiprazole treatment is for CYP2D6. The DPWG and the Food and Drug Administration (FDA) recommend dose reduction for CYP2D6 PM (Swen et al., 2011; FDA, 2014, 2016). This group presents higher Cmax and T1/2 (Kinghorn and McEvoy, 2005; FDA, 2014; Belmonte et al., 2018) (Table 3).
CYP3A4 is aripiprazole’s secondary metabolizing enzyme. Patients should receive a reduced dose when concomitant CYP3A4 inhibitors are prescribed (FDA, 2014). Since it is highly inducible (Sanchez Spitman et al., 2017), it is difficult to find associations between polymorphisms and drug pharmacodynamics or pharmacokinetics.
Due to similarity between CYP3A4 and CYP3A5 families (Huang et al., 2004), other genes could be involved in aripiprazole metabolism, e.g., CYP3A5. CYP3A5 *3/*3 carriers do not express CYP3A5, therefore, higher aripiprazole concentrations could be expected (Lamba et al., 2012). However, results are contradictory. Some candidate gene studies reveal no effect (Suzuki et al., 2014), while other reports its involvement in the development of adverse drug reactions (Belmonte et al., 2018) (Table 3).
Other gene studied in aripiprazole pharmacokinetics is ABCB1 (Kirschbaum et al., 2010; Saiz-Rodríguez et al., 2018; Koller et al., 2020). Among the different mutations of ABCB1, carriers of the two mutations 2677T > G/A (rs2032582) and 3435T > C (rs1045642) presented lower plasma concentration (Rafaniello et al., 2018). Nonetheless, these results are controversial since other candidate gene studies reported no differences between ABCB1 variants (Suzuki et al., 2014; Belmonte et al., 2018; Koller et al., 2018). In another study, carriers of the rs2032582 and rs1128503 polymorphisms affected aripiprazole Cl (Belmonte et al., 2018) (Table 3).
PMs for CYP2D6 showed a higher risperidone Cmax, AUC and T1/2 and a lower Cl (Jovanović et al., 2010; Cabaleiro et al., 2014) and a lower 9-hydroxyrisperidone AUC, Cmax and a higher Tmax (Cabaleiro et al., 2014). A different candidate gene study observed a higher plasma concentration risperidone/9-hydroxyrisperidone ratio in IM compared to NM and UM (Vanwong et al., 2016). Similar results were found in healthy volunteers (Novalbos et al., 2010). According to these results, children who are PM or IM for CYP2D6 have higher risk of suffering from adverse effects when treated with risperidone (Oshikoya et al., 2019) and extrapyramidal effects are more probable in IM patients (Ito et al., 2018). However, the active moiety levels are not always affected by CYP2D6 phenotype (Kang et al., 2009) (Table 4).
CYP3A4 and CYP3A5 are also involved in risperidone metabolism. CYP3A5 non-expressors exhibited higher plasma concentrations of both risperidone and 9-hydroxyrisperidone and a higher active moiety than its expressors (Kang et al., 2009). However, two different candidate gene studies did not find differences in concentrations of risperidone, 9-hydroxyrisperidone and the active moiety regarding the CYP3A phenotype (Vandenberghe et al., 2015; Ganoci et al., 2016). Further studies are needed to estimate the influence of CYP3A phenotype on risperidone pharmacokinetics (Table 4).
A candidate gene study showed that schizophrenic women who were G for rs2032582 (2677T > G/A) and C for rs1045642 (3435T > C) for ABCB1 were less represented in the group of patients who suffered a greatest weight gain in risperidone treatment (Kuzman et al., 2008). In a different study, rs1045642T carriers and T carriers in rs2032582 and rs1045642 suffered from extrapyramidal symptoms with a higher frequency (Jovanović et al., 2010). For this reason, it was proposed that rs1045642T and rs2032582T carriers reach a higher risperidone concentration in the brain, allowing stronger effects of this antipsychotic, including its adverse effects. T carriers in the rs1045642 single nucleotide polymorphism (SNP) were found to have a higher risperidone and 9-hydroxyrisperidone Cl (Saiz-Rodríguez et al., 2018). A different candidate gene study did not find differences in risperidone and 9-hydroxyrisperidone plasma concentrations depending on rs2032582 and rs1045642 ABCB1 polymorphisms (Yasui-Furukori et al., 2007), and another one did not find differences in risperidone response (Xing et al., 2006). However, this last study found a better response to risperidone in T/T rs1128503 (1236T > C) patients (Xing et al., 2006). Different candidate gene studies obtained different results regarding these polymorphisms and risperidone pharmacokinetics. For this reason, further studies should be conducted to clarify the impact of these SNPs in risperidone pharmacokinetics (Saiz-Rodríguez et al., 2018) (Table 4).
The catechol-O-methyl transferase (COMT) enzyme is involved in the regulation of dopamine availability in the brain. Some studies found that some COMT polymorphisms influenced antipsychotics response. In the treatment with risperidone, rs9606186 was associated with clinical efficacy in male patients (Zhao et al., 2012), while rs165599 was significantly associated with risperidone efficacy on negative symptoms (Han et al., 2017) (Table 4).
Several studies showed that the HTR2C −759C > T (rs3813929) T allele has a protective effect against olanzapine-produced weight gain (Templeman et al., 2005). The C allele of the rs1414334 intronic polymorphism is associated with an increased risk for developing metabolic syndrome and a higher weight gain after olanzapine treatment (Ma et al., 2014; Koller et al., 2021). This SNP is in complete linkage disequilibrium with the rs6318 (p.Cys23Ser) polymorphism, where the Ser allele is constitutively more active (Ma et al., 2014). Carriers of T, C and Ser alleles for the −759C > T, −697G > C (rs518147) and Cys23Ser, presented lower weight gain and body mass index (BMI) (Sicard et al., 2010) compared to the other alleles (Table 2).
Genetic variations in the DRD2 gene caused different responses to olanzapine treatment (Houston et al., 2011). The TaqIA (rs1800497) T allele seems to be associated with increased prolactin levels during olanzapine treatment (López-Rodríguez et al., 2011; Brennan, 2014). The InDel polymorphism rs1799732 is located in the gene promoter. Individuals who presented the deletion suffered greater weight gain when treated with olanzapine than those without the deletion (Lencz et al., 2010). This variation in the DRD2 promoter region may produce differences in the sensitivity of the receptor to the effects of antipsychotics on reward signals associated with food intake and satiety (Lencz et al., 2010). A different candidate gene study also found a higher incidence of adverse reactions in mutant individuals for TaqIA and rs1799732 (Zubiaur et al., 2021) (Table 2).
The presence of Gly in DRD3 rs6280 (p.Ser9Gly) is associated with a better response to olanzapine (Brennan, 2014). Moreover, the Gly allele was related to a higher increase in prolactin levels compared to Ser allele (Koller et al., 2021) (Table 2).
The apolipoprotein C3 (APOC3) rs4520 C/C genotype was associated with higher triglyceride concentrations after olanzapine administration in a candidate gene study (Koller et al., 2021). Polymorphisms in this gene influence plasma triglyceride levels since the APOC3 protein increases plasma triglyceride levels by the inhibition of lipoprotein lipase, stimulates low-density lipoprotein secretion and intestinal triglyceride trafficking modulation (Koller et al., 2021). A different study observed lower T1/2 in G/G rs5128 APOC3 (Zubiaur et al., 2021) (Table 2).
There is no extensive evidence on the influence of polymorphisms in aripiprazole pharmacodynamics. One of the most studied variants in antipsychotic pharmacodynamics is the DRD2 TaqIA polymorphism. The *A1 allele corresponds with lower density of DRD2 receptors in the striatum (Jönsson et al., 1999). Some groups reported better aripiprazole response for *A1 allele carriers (Kwon et al., 2008). Furthermore, C/C homozygotes for rs6277 in DRD2 gene present poorer response to aripiprazole (Shen et al., 2009). Also, carriers of C allele in rs6277 and *A1 allele in Taq1A had poorer cognitive performance (Ramsay et al., 2015) (Table 3).
Other reported genetic variants are mutations of serotonin receptor such as 5-HTR2A. The different variants of this receptor present different binding affinities for aripiprazole (Davies et al., 2011). Concretely, subjects with the G/G C/C genotype of HTR2A rs6311 (1438G > A/T) and rs6313 (102T > C) polymorphisms showed poorer aripiprazole response for negative symptoms in a candidate gene study (Chen et al., 2009) (Table 3).
DRD2 rs1801028 (p.Ser311Cys) was significantly associated with risperidone efficacy on the treatment of negative symptoms (Han et al., 2017). In addition, being heterozygous for the rs1801028 variant in DRD2 was associated with a better response to risperidone compared to Ser homozygous (Lane et al., 2004). Taq1A and rs1799732 genetic variants were not associated with antipsychotic to risperidone in one candidate gene study (Alladi et al., 2019). However, a different candidate gene study found an association between *A1/*A1 genotype for TaqIA and A/A genotype for rs1799978 SNP and a higher improvement in PANSS scale (Ikeda et al., 2008). The TaqIA*A1 allele seemed to be associated with increased prolactin levels during risperidone treatment (López-Rodríguez et al., 2011; Brennan, 2014) (Table 4).
There are controversial results regarding the influence of the rs6280 polymorphism on DRD3 on risperidone response. A candidate gene study with autistic children observed a better risperidone response in Gly allele carriers (Firouzabadi et al., 2017). However, a different candidate gene study with schizophrenic patients did not find this association (Ikeda et al., 2008) (Table 4).
Another candidate gene study observed that HTR2A rs6313 C/C patients showed a superior risperidone response particularly for negative symptoms (Lane et al., 2002). However, a different candidate gene study did not find associations between HTR2A rs6311 and rs6313 polymorphisms and risperidone response (Alladi et al., 2019). In a candidate gene study carried in schizophrenic women, HTR2C rs3813929 (−759C > T) polymorphism was not found to be associated to a higher weight gain (Kuzman et al., 2008) or to a better response (Ikeda et al., 2008; Alladi et al., 2019). Neither was the −697C > G SNP associated with a better risperidone response (Ikeda et al., 2008) (Table 4).
Based on our review, olanzapine induces more metabolic adverse drug reactions than risperidone and aripiprazole, being aripiprazole the drug that induces the least metabolic adverse reactions. The goal of personalized medicine is to predict the side effects of each drug in each patient individually. In order to reach this goal, specific biomarkers, i.e. genetic polymorphisms should be tested before starting the treatment. Only a few polymorphisms were found to be associated to olanzapine, aripiprazole and risperidone treatment outcomes. Since olanzapine is metabolized by CYP1A2, a lower starting dose should be considered in patients treated with fluvoxamine or other CYP1A2 inhibitors. Regarding aripiprazole, a reduced dose should be administered for CYP2D6 PMs. Additionally, a reduction to a quarter of the normal dose is recommended if the patient is treated with concomitant CYP3A4 inhibitors. For risperidone, a dose adjustment should be evaluated in case of coadministration of CYP3A, CYP2D6 and ABCB1 inhibitors. Although several studies found possible associations with the metabolic adverse drug reactions of these drugs, to date, no genetic polymorphism is recommended for testing. Consequently, this deficit should be addressed to offer better treatment to patients under antipsychotic therapy. This review lists all of the possible targets which have been found to date.
Author Contributions: writing—original draft preparation, PS-C, GV-G, DK; writing—review and editing, DK, FA-S, PZ.
GV-G is financed by Instituto de Salud Carlos III (ISCIII) and the European Social Fund (PFIS predoctoral grant, number FI20/00090). PS-C is financed by the scholarship “Ayuda para el fomento de la investigación en estudios de máster 2020” granted by Universidad Autónoma de Madrid.
FA-S has been consultant or investigator in clinical trials sponsored by the following pharmaceutical companies: Abbott, Alter, Chemo, Cinfa, FAES, Farmalider, Ferrer, GlaxoSmithKline, Galenicum, Gilead, Janssen-Cilag, Kern, Normon, Novartis, Servier, Silverpharma, Teva and Zambon. The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Alladi, C. G., RajKumar, R. P., Adithan, S., Marie‐Claire, C., Bellivier, F., and Shewade, D. G. (2019). Dopamine (DRD 2) and Serotonin ( HTR 2A, 2C) Receptor Gene Polymorphisms Do Not Influence Early Response to Risperidone in South Indian Patients with Schizophrenia. Fundam. Clin. Pharmacol. 33, 355–364. doi:10.1111/fcp.12424
Aringhieri, S., Carli, M., Kolachalam, S., Verdesca, V., Cini, E., Rossi, M., et al. (2018). Molecular Targets of Atypical Antipsychotics: From Mechanism of Action to Clinical Differences. Pharmacol. Ther. 192, 20–41. doi:10.1016/j.pharmthera.2018.06.012
Bak, M., Fransen, A., Janssen, J., van Os, J., and Drukker, M. (2014). Almost All Antipsychotics Result in Weight Gain: A Meta-Analysis. PLoS ONE 9, e94112. doi:10.1371/journal.pone.0094112
Bank, P., Caudle, K., Swen, J., Gammal, R., Whirl‐Carrillo, M., Klein, T., et al. (2018). Comparison of the Guidelines of the Clinical Pharmacogenetics Implementation Consortium and the Dutch Pharmacogenetics Working Group. Clin. Pharmacol. Ther. 103, 599–618. doi:10.1002/cpt.762
Barton, B. B., Segger, F., Fischer, K., Obermeier, M., and Musil, R. (2020). Update on Weight-Gain Caused by Antipsychotics: a Systematic Review and Meta-Analysis. Expert Opin. Drug Saf. 19, 295–314. doi:10.1080/14740338.2020.1713091
Belmonte, C., Ochoa, D., Román, M., Saiz-Rodríguez, M., Wojnicz, A., Gómez-Sánchez, C. I., et al. (2018). Influence of CYP2D6, CYP3A4, CYP3A5 and ABCB1 Polymorphisms on Pharmacokinetics and Safety of Aripiprazole in Healthy Volunteers. Basic Clin. Pharmacol. Toxicol. 122, 596–605. doi:10.1111/bcpt.12960
Berecz, R., Dorado, P., Rubia, A., Caceres, M., Degrell, I., and LLerena, A. (2004). The Role of Cytochrome P450 Enzymes in the Metabolism of Risperidone and its Clinical Relevance for Drug Interactions. CDT 5, 573–579. doi:10.2174/1389450043345263
Bever, K. A., and Perry, P. J. (1998). Olanzapine: a Serotonin-Dopamine-Receptor Antagonist for Antipsychotic Therapy. Am. J. Health-System Pharm. 55, 1003–1016. doi:10.1093/ajhp/55.10.1003
Bhana, N., Foster, R. H., Olney, R., and Plosker, G. L. (2001). Olanzapine. Drugs 61, 111–161. doi:10.2165/00003495-200161010-00011
Brandl, E. J., Kennedy, J. L., and Müller, D. J. (2014). Pharmacogenetics of Antipsychotics. Can. J. Psychiatry 59, 76–88. doi:10.1177/070674371405900203
Brennan, M. D. (2014). Pharmacogenetics of Second-Generation Antipsychotics. Pharmacogenomics 15, 869–884. doi:10.2217/pgs.14.50
Brisch, R., Saniotis, A., Wolf, R., Bielau, H., Bernstein, H.-G., Steiner, J., et al. (2014). The Role of Dopamine in Schizophrenia from a Neurobiological and Evolutionary Perspective: Old Fashioned, but Still in Vogue. Front. Psychiatry 5, 47. doi:10.3389/fpsyt.2014.00047
Cabaleiro, T., López-Rodríguez, R., Ochoa, D., Román, M., Novalbos, J., and Abad-Santos, F. (2013). Polymorphisms Influencing Olanzapine Metabolism and Adverse Effects in Healthy Subjects. Hum. Psychopharmacol. Clin. Exp. 28, 205–214. doi:10.1002/hup.2308
Cabaleiro, T., Ochoa, D., López-Rodríguez, R., Román, M., Novalbos, J., Ayuso, C., et al. (2014). Effect of Polymorphisms on the Pharmacokinetics, Pharmacodynamics, and Safety of Risperidone in Healthy Volunteers. Hum. Psychopharmacol. Clin. Exp. 29, 459–469. doi:10.1002/hup.2420
Callaghan, J. T., Bergstrom, R. F., Ptak, L. R., and Beasley, C. M. (1999). Olanzapine. Clin. Pharmacokinet. 37, 177–193. doi:10.2165/00003088-199937030-00001
Caudle, K. E., Sangkuhl, K., Whirl‐Carrillo, M., Swen, J. J., Haidar, C. E., Klein, T. E., et al. (2020). Standardizing CYP 2D6 Genotype to Phenotype Translation: Consensus Recommendations from the Clinical Pharmacogenetics Implementation Consortium and Dutch Pharmacogenetics Working Group. Clin. Transl Sci. 13, 116–124. doi:10.1111/cts.12692
Caudle, K., Klein, T., Hoffman, J., Muller, D., Whirl-Carrillo, M., Gong, L., et al. (2014). Incorporation of Pharmacogenomics into Routine Clinical Practice: the Clinical Pharmacogenetics Implementation Consortium (CPIC) Guideline Development Process. CDM 15, 209–217. doi:10.2174/1389200215666140130124910
Chen, C.-H., Shyue, S.-K., Hsu, C.-P., and Lee, T.-S. (2018). Atypical Antipsychotic Drug Olanzapine Deregulates Hepatic Lipid Metabolism and Aortic Inflammation and Aggravates Atherosclerosis. Cell Physiol Biochem 50, 1216–1229. doi:10.1159/000494573
Chen, S.-F., Shen, Y.-C., and Chen, C.-H. (2009). HTR2A A-1438G/T102C Polymorphisms Predict Negative Symptoms Performance upon Aripiprazole Treatment in Schizophrenic Patients. Psychopharmacology 205, 285–292. doi:10.1007/s00213-009-1538-z
Cheng, Z., Yuan, Y., Han, X., Yang, L., Cai, S., Yang, F., et al. (2019). An Open-Label Randomised Comparison of Aripiprazole, Olanzapine and Risperidone for the Acute Treatment of First-Episode Schizophrenia: Eight-Week Outcomes. J. Psychopharmacol. 33, 1227–1236. doi:10.1177/0269881119872193
Corena-McLeod, M. (2015). Comparative Pharmacology of Risperidone and Paliperidone. Drugs R. D 15, 163–174. doi:10.1007/s40268-015-0092-x
Czekalla, J., Kollack-Walker, S., and Beasley, C. M. (2001). Cardiac Safety Parameters of Olanzapine: Comparison with Other Atypical and Typical Antipsychotics. J. Clin. Psychiatry 62 (Suppl. 2), 35–40. doi:10.4088/jcp.v62n0310
Czerwensky, F., Leucht, S., and Steimer, W. (2015). CYP1A2*1D and *1F Polymorphisms Have a Significant Impact on Olanzapine Serum Concentrations. Ther. Drug Monit. 37, 152–160. doi:10.1097/FTD.0000000000000119
Daudén Tello, E. (2006). Farmacogenética I. Concepto, historia, objetivos y áreas de estudio. Actas Dermo-Sifiliográficas 97, 623–629. doi:10.1016/S0001-7310(06)73482-2
Davies, M. A., Conley, Y., and Roth, B. L. (2011). Functional SNPs in Genes Encoding the 5-HT2A Receptor Modify the Affinity and Potency of Several Atypical Antipsychotic Drugs. Biol. Res. Nurs. 13, 55–60. doi:10.1177/1099800409358760
Di Sanzo, M., Cipolloni, L., Borro, M., La Russa, R., Santurro, A., Scopetti, M., et al. (2017). Clinical Applications of Personalized Medicine: A New Paradigm and Challenge. CPB 18, 194–203. doi:10.2174/1389201018666170224105600
Ema, (2020). EMA Recommendations on DPD Testing Prior to Treatment with Fluorouracil, Capecitabine, Tegafur and Flucytosine. Available at: https://www.ema.europa.eu/en/news/ema-recommendations-dpd-testing-prior-treatment-fluorouracil-capecitabine-tegafur-flucytosine (Accessed April 27, 2021).
Erickson-Ridout, K. K., Zhu, J., and Lazarus, P. (2011). Olanzapine Metabolism and the Significance of UGT1A448V and UGT2B1067Y Variants. Pharmacogenetics and Genomics 21, 539–551. doi:10.1097/FPC.0b013e328348c76b
Eum, S., Lee, A. M., and Bishop, J. R. (2016). Pharmacogenetic Tests for Antipsychotic Medications: Clinical Implications and Considerations. Dialogues Clin. Neurosci. 18, 323–337.
Fang, F., Sun, H., Wang, Z., Ren, M., Calabrese, J. R., and Gao, K. (2016). Antipsychotic Drug-Induced Somnolence: Incidence, Mechanisms, and Management. CNS Drugs 30, 845–867. doi:10.1007/s40263-016-0352-5
Fang, J., Bourin, M., and Baker, G. B. (1999). Metabolism of Risperidone to 9-hydroxyrisperidone by Human Cytochromes P450 2D6 and 3A4. Naunyn-schmiedeberg's Arch. Pharmacol. 359, 147–151. doi:10.1007/PL00005334
FDA (2014). ABILIFY (Aripiprazole). HIGHLIGHTS of PRESCRIBING INFORMATION. Available at: https://www.accessdata.fda.gov/drugsatfda_docs/label/2016/021436s041,021713s032,021729s024,021866s026lbl.pdf.
FDA (2016). ABILIFY MAINTENA (Aripiprazole). HIGHLIGHTS of PRESCRIBING INFORMATION. Available at: https://www.accessdata.fda.gov/drugsatfda_docs/label/2016/202971s008lbl.pdf (Accessed April 28, 2021).
FDA (1993). RISPERIDAL (Risperidone). HIGHLIGHTS of PRESCRIBING INFORMATION. Available at: https://www.accessdata.fda.gov/drugsatfda_docs/label/2009/020272s056,020588s044,021346s033,021444s03lbl.pdf (Accessed December 29, 2020).
FDA (2019). ZIAGEN (Abacavir Sulfate). HIGHLIGHTS of PRESCRIBING INFORMATION. Available at: https://www.accessdata.fda.gov/drugsatfda_docs/pepfar/077844PI.pdf (Accessed April 27, 2021).
FDA (1996). ZYPREXA (Olanzapine). HIGHLIGHTS of PRESCRIBING INFORMATION. Available at: https://www.accessdata.fda.gov/drugsatfda_docs/label/2009/020592s051,021086s030,021253s036lbl.pdf (Accessed November 15, 2020).
Firouzabadi, N., Nazariat, A., and Zomorrodian, K. (2017). DRD3 Ser9Gly Polymorphism and its Influence on Risperidone Response in Autistic Children. J. Pharm. Pharm. Sci. 20, 445. doi:10.18433/J3H63T
Ganoci, L., Lovrić, M., Živković, M., Šagud, M., Klarica Domjanović, I., and Božina, N. (2016). The Role of Cyp2d6, Cyp3a4/5, and Abcb1 Polymorphisms in Patients Using Long-Acting Injectable Risperidone. Clin. Ther. 38, e10–e11. doi:10.1016/j.clinthera.2016.07.110
Ganoci, L., Trkulja, V., Živković, M., Božina, T., Šagud, M., Lovrić, M., et al. (2021). ABCB1, ABCG2 and CYP2D6 Polymorphism Effects on Disposition and Response to Long-Acting Risperidone. Prog. Neuro-Psychopharmacology Biol. Psychiatry 104, 110042. doi:10.1016/j.pnpbp.2020.110042
Germann, D., Kurylo, N., and Han, F. (2012). ““Risperidone,”,” in Profiles of Drug Substances, Excipients and Related Methodology, 37, 313–361. doi:10.1016/B978-0-12-397220-0.00008-8
Glassman, A. H., and Bigger, J. T. (2001). Antipsychotic Drugs: Prolonged QTc Interval, Torsade de Pointes, and Sudden Death. AJP 158, 1774–1782. doi:10.1176/appi.ajp.158.11.1774
Han, J., Li, Y., and Wang, X. (2017). Potential Link between Genetic Polymorphisms of Catechol-O-Methyltransferase and Dopamine Receptors and Treatment Efficacy of Risperidone on Schizophrenia. NDT Vol. 13, 2935–2943. doi:10.2147/NDT.S148824
Himmerich, H., Minkwitz, J., and Kirkby, K. (2015). Weight Gain and Metabolic Changes during Treatment with Antipsychotics and Antidepressants. EMIDDT 15, 252–260. doi:10.2174/1871530315666150623092031
Houston, J., Dharia, S., Bishop, J. R., Ellingrod, V. L., Fijal, B., Jacobson, J. G., et al. (2011). Association of DRD2 and ANKK1 Polymorphisms with Prolactin Increase in Olanzapine-Treated Women. Psychiatry Res. 187, 74–79. doi:10.1016/j.psychres.2010.10.020
Huang, W., Lin, Y. S., McConn, D. J., Calamia, J. C., Totah, R. A., Isoherranen, N., et al. (2004). EVIDENCE OF SIGNIFICANT CONTRIBUTION FROM CYP3A5 TO HEPATIC DRUG METABOLISM. Drug Metab. Dispos 32, 1434–1445. doi:10.1124/dmd.104.001313
Ikeda, M., Yamanouchi, Y., Kinoshita, Y., Kitajima, T., Yoshimura, R., Hashimoto, S., et al. (2008). Variants of Dopamine and Serotonin Candidate Genes as Predictors of Response to Risperidone Treatment in First-Episode Schizophrenia. Pharmacogenomics 9, 1437–1443. doi:10.2217/14622416.9.10.1437
Ito, T., Yamamoto, K., Ohsawa, F., Otsuka, I., Hishimoto, A., Sora, I., et al. (2018). Association of CYP2D6 Polymorphisms and Extrapyramidal Symptoms in Schizophrenia Patients Receiving Risperidone: a Retrospective Study. J. Pharm. Health Care Sci. 4, 28. doi:10.1186/s40780-018-0126-y
Jeon, S., and Kim, Y.-K. (2017). Unresolved Issues for Utilization of Atypical Antipsychotics in Schizophrenia: Antipsychotic Polypharmacy and Metabolic Syndrome. IJMS 18, 2174. doi:10.3390/ijms18102174
Jönsson, E. G., Nöthen, M. M., Grünhage, F., Farde, L., Nakashima, Y., Propping, P., et al. (1999). Polymorphisms in the Dopamine D2 Receptor Gene and Their Relationships to Striatal Dopamine Receptor Density of Healthy Volunteers. Mol. Psychiatry 4, 290–296. doi:10.1038/sj.mp.4000532
Jovanović, N., Božina, N., Lovrić, M., Medved, V., Jakovljević, M., and Peleš, A. M. (2010). The Role of CYP2D6 and ABCB1 Pharmacogenetics in Drug-Naïve Patients with First-Episode Schizophrenia Treated with Risperidone. Eur. J. Clin. Pharmacol. 66, 1109–1117. doi:10.1007/s00228-010-0850-1
Kahn, R. S., Fleischhacker, W. W., Boter, H., Davidson, M., Vergouwe, Y., Keet, I. P., et al. (2008). Effectiveness of Antipsychotic Drugs in First-Episode Schizophrenia and Schizophreniform Disorder: an Open Randomised Clinical Trial. The Lancet 371, 1085–1097. doi:10.1016/S0140-6736(08)60486-9
Kane, J. M., Osuntokun, O., Kryzhanovskaya, L. A., Xu, W., Stauffer, V. L., Watson, S. B., et al. (2009). A 28-Week, Randomized, Double-Blind Study of Olanzapine versus Aripiprazole in the Treatment of Schizophrenia. J. Clin. Psychiatry 70, 572–581. doi:10.4088/JCP.08m04421
Kang, R.-H., Jung, S.-M., Kim, K.-A., Lee, D.-K., Cho, H.-K., Jung, B.-J., et al. (2009). Effects of CYP2D6 and CYP3A5 Genotypes on the Plasma Concentrations of Risperidone and 9-Hydroxyrisperidone in Korean Schizophrenic Patients. J. Clin. Psychopharmacol. 29, 272–277. doi:10.1097/JCP.0b013e3181a289e0
Kinghorn, W. A., and McEvoy, J. P. (2005). Aripiprazole: Pharmacology, Efficacy, Safety and Tolerability. Expert Rev. Neurotherapeutics 5, 297–307. doi:10.1586/14737175.5.3.297
Kirschbaum, K. M., Uhr, M., Holthoewer, D., Namendorf, C., Pietrzik, C., Hiemke, C., et al. (2010). Pharmacokinetics of Acute and Sub-chronic Aripiprazole in P-Glycoprotein Deficient Mice. Neuropharmacology 59, 474–479. doi:10.1016/j.neuropharm.2010.06.010
Koller, D., Almenara, S., Mejía, G., Saiz-Rodríguez, M., Zubiaur, P., Román, M., et al. (2021). Metabolic Effects of Aripiprazole and Olanzapine Multiple-Dose Treatment in a Randomised Crossover Clinical Trial in Healthy Volunteers: Association with Pharmacogenetics. Adv. Ther. 38, 1035–1054. doi:10.1007/s12325-020-01566-w
Koller, D., Belmonte, C., Lubomirov, R., Saiz-Rodríguez, M., Zubiaur, P., Román, M., et al. (2018). Effects of Aripiprazole on Pupillometric Parameters Related to Pharmacokinetics and Pharmacogenetics after Single Oral Administration to Healthy Subjects. J. Psychopharmacol. 32, 1212–1222. doi:10.1177/0269881118798605
Koller, D., Saiz‐Rodríguez, M., Zubiaur, P., Ochoa, D., Almenara, S., Román, M., et al. (2020). The Effects of Aripiprazole and Olanzapine on Pupillary Light Reflex and its Relationship with Pharmacogenetics in a Randomized Multiple‐dose Trial. Br. J. Clin. Pharmacol. 86, 2051–2062. doi:10.1111/bcp.14300
Komossa, K., Rummel-Kluge, C., Schmid, F., Hunger, H., Schwarz, S., El-Sayeh, H. G. G., et al. (2009). “Aripiprazole versus Other Atypical Antipsychotics for Schizophrenia,” in In Cochrane Database Of Systematic Reviews The Cochrane Collaboration, (Chichester, UK: John Wiley & Sons, Ltd), CD006569. doi:10.1002/14651858.CD006569.pub3
Komossa, K., Rummel-Kluge, C., Schwarz, S., Schmid, F., Hunger, H., Kissling, W., et al. (2011). Risperidone versus Other Atypical Antipsychotics for Schizophrenia. Cochrane Database Syst. Rev. doi:10.1002/14651858.CD006626.pub2
Korprasertthaworn, P., Polasek, T. M., Sorich, M. J., McLachlan, A. J., Miners, J. O., Tucker, G. T., et al. (2015). In Vitro Characterization of the Human Liver Microsomal Kinetics and Reaction Phenotyping of Olanzapine Metabolism. Drug Metab. Dispos 43, 1806–1814. doi:10.1124/dmd.115.064790
Kuzman, M. R., Medved, V., Bozina, N., Hotujac, L., Sain, I., and Bilusic, H. (2008). The Influence of 5-HT2C and MDR1 Genetic Polymorphisms on Antipsychotic-Induced Weight Gain in Female Schizophrenic Patients. Psychiatry Res. 160, 308–315. doi:10.1016/j.psychres.2007.06.006
Kwon, J. S., Kim, E., Kang, D.-H., Choi, J. S., Yu, K.-S., Jang, I.-J., et al. (2008). Taq1A Polymorphism in the Dopamine D2 Receptor Gene as a Predictor of Clinical Response to Aripiprazole. Eur. Neuropsychopharmacol. 18, 897–907. doi:10.1016/j.euroneuro.2008.07.010
Lally, J., and MacCabe, J. H. (2015). Antipsychotic Medication in Schizophrenia: a Review. Br. Med. Bull. 114, 169–179. doi:10.1093/bmb/ldv017
Lamba, J., Hebert, J. M., Schuetz, E. G., Klein, T. E., and Altman, R. B. (2012). PharmGKB Summary. Pharmacogenetics and Genomics 22, 555–558. doi:10.1097/FPC.0b013e328351d47f
Lambert, B. L., Cunningham, F. E., Miller, D. R., Dalack, G. W., and Hur, K. (2006). Diabetes Risk Associated with Use of Olanzapine, Quetiapine, and Risperidone in Veterans Health Administration Patients with Schizophrenia. Am. J. Epidemiol. 164, 672–681. doi:10.1093/aje/kwj289
Lane, H.-Y., Chang, Y.-C., Chiu, C.-C., Chen, M.-L., Hsieh, M.-H., and Chang, W.-H. (2002). Association of Risperidone Treatment Response with a Polymorphism in the 5-HT2AReceptor Gene. AJP 159, 1593–1595. doi:10.1176/appi.ajp.159.9.1593
Lane, H.-Y., Lee, C.-C., Chang, Y.-C., Lu, C.-T., Huang, C.-H., and Chang, W.-H. (2004). Effects of Dopamine D2 Receptor Ser311Cys Polymorphism and Clinical Factors on Risperidone Efficacy for Positive and Negative Symptoms and Social Function. Int. J. Neuropsychopharm. 7, 461–470. doi:10.1017/S1461145704004389
Laruelle, M. (2014). Schizophrenia: from Dopaminergic to Glutamatergic Interventions. Curr. Opin. Pharmacol. 14, 97–102. doi:10.1016/j.coph.2014.01.001
Lencz, T., Robinson, D. G., Napolitano, B., Sevy, S., Kane, J. M., Goldman, D., et al. (2010). DRD2 Promoter Region Variation Predicts Antipsychotic-Induced Weight Gain in First Episode Schizophrenia. Pharmacogenetics and Genomics 20, 569–572. doi:10.1097/FPC.0b013e32833ca24b
Lieberman, J. A., Stroup, T. S., McEvoy, J. P., Swartz, M. S., Rosenheck, R. A., Perkins, D. O., et al. (2005). Effectiveness of Antipsychotic Drugs in Patients with Chronic Schizophrenia. N. Engl. J. Med. 353, 1209–1223. doi:10.1056/NEJMoa051688
López-Rodríguez, R., Román, M., Novalbos, J., Pelegrina, M. L., Ochoa, D., and Abad-Santos, F. (2011). DRD2 Taq1A Polymorphism Modulates Prolactin Secretion Induced by Atypical Antipsychotics in Healthy Volunteers. J. Clin. Psychopharmacol. 31, 555–562. doi:10.1097/JCP.0b013e31822cfff2
Ma, X., Maimaitirexiati, T., Zhang, R., Gui, X., Zhang, W., Xu, G., et al. (2014). HTR2Cpolymorphisms, Olanzapine-Induced Weight Gain and Antipsychotic-Induced Metabolic Syndrome in Schizophrenia Patients: A Meta-Analysis. Int. J. Psychiatry Clin. Pract. 18, 229–242. doi:10.3109/13651501.2014.957705
Marder, S. R., Essock, S. M., Miller, A. L., Buchanan, R. W., Casey, D. E., Davis, J. M., et al. (2004). Physical Health Monitoring of Patients with Schizophrenia. AJP 161, 1334–1349. doi:10.1176/appi.ajp.161.8.1334
Maric, N. P., Jovicic, M. J., Mihaljevic, M., and Miljevic, C. (2016). Improving Current Treatments for Schizophrenia. Drug Dev. Res. 77, 357–367. doi:10.1002/ddr.21337
Meltzer, H. Y. (2013). Update on Typical and Atypical Antipsychotic Drugs. Annu. Rev. Med. 64, 393–406. doi:10.1146/annurev-med-050911-161504
Meyer, J. M., Pandina, G., Bossie, C. A., Turkoz, I., and Greenspan, A. (2005). Effects of Switching from Olanzapine to Risperidone on the Prevalence of the Metabolic Syndrome in Overweight or Obese Patients with Schizophrenia or Schizoaffective Disorder: Analysis of a Multicenter, Rater-Blinded, Open-Label Study. Clin. Ther. 27, 1930–1941. doi:10.1016/j.clinthera.2005.12.005
Mueser, K. T., and McGurk, S. R. (2004). Schizophrenia. The Lancet 363, 2063–2072. doi:10.1016/S0140-6736(04)16458-1
Nicol, G. E., Yingling, M. D., Flavin, K. S., Schweiger, J. A., Patterson, B. W., Schechtman, K. B., et al. (2018). Metabolic Effects of Antipsychotics on Adiposity and Insulin Sensitivity in Youths. JAMA Psychiatry 75, 788. doi:10.1001/jamapsychiatry.2018.1088
Noordsy, D. L., Glynn, S. M., Sugar, C. A., O'Keefe, C. D., and Marder, S. R. (2017). Risperidone versus Olanzapine Among Patients with Schizophrenia Participating in Supported Employment: Eighteen-Month Outcomes. J. Psychiatr. Res. 95, 299–307. doi:10.1016/j.jpsychires.2017.09.008
Novalbos, J., López-Rodríguez, R., Román, M., Gallego-Sandín, S., Ochoa, D., and Abad-Santos, F. (2010). Effects of CYP2D6 Genotype on the Pharmacokinetics, Pharmacodynamics, and Safety of Risperidone in Healthy Volunteers. J. Clin. Psychopharmacol. 30, 504–511. doi:10.1097/JCP.0b013e3181ee84c7
Nozawa, M., Ohnuma, T., Matsubara, Y., Sakai, Y., Hatano, T., Hanzawa, R., et al. (2008). The Relationship between the Response of Clinical Symptoms and Plasma Olanzapine Concentration, Based on Pharmacogenetics. Ther. Drug Monit. 30, 35–40. doi:10.1097/FTD.0b013e31816336fd
Ohi, K., Shimada, T., Kuwata, A., Kataoka, Y., Okubo, H., Kimura, K., et al. (2019). Smoking Rates and Number of Cigarettes Smoked Per Day in Schizophrenia: A Large Cohort Meta-Analysis in a Japanese Population. Int. J. Neuropsychopharmacol. 22, 19–27. doi:10.1093/ijnp/pyy061
Okubo, M., Narita, M., Murayama, N., Akimoto, Y., Goto, A., and Yamazaki, H. (2016). Individual Differences Inin Vitroandin Vivometabolic Clearances of the Antipsychotic Drug Olanzapine from Non-smoking and Smoking Japanese Subjects Genotyped for Cytochrome P4502D6 and Flavincontaining Monooxygenase 3. Hum. Psychopharmacol. Clin. Exp. 31, 83–92. doi:10.1002/hup.2515
Oshikoya, K. A., Neely, K. M., Carroll, R. J., Aka, I. T., Maxwell-Horn, A. C., Roden, D. M., et al. (2019). CYP2D6 Genotype and Adverse Events to Risperidone in Children and Adolescents. Pediatr. Res. 85, 602–606. doi:10.1038/s41390-019-0305-z
Ozeki, Y., Fujii, K., Kurimoto, N., Yamada, N., Okawa, M., Aoki, T., et al. (2010). QTc Prolongation and Antipsychotic Medications in a Sample of 1017 Patients with Schizophrenia. Prog. Neuro-Psychopharmacology Biol. Psychiatry 34, 401–405. doi:10.1016/j.pnpbp.2010.01.008
Pharmacogene Variation Consortium (2017). PharmVar. Available at: https://www.pharmvar.org/gene/CYP3A4.
PharmGKB guidelines (2020). Annotation of DPWG Guideline for Risperidone and CYP2D6. Available at: https://www.pharmgkb.org/guidelineAnnotation/PA166104943.
Prommer, E. (2017). Aripiprazole. Am. J. Hosp. Palliat. Care 34, 180–185. doi:10.1177/1049909115612800
Rafaniello, C., Sessa, M., Bernardi, F. F., Pozzi, M., Cheli, S., Cattaneo, D., et al. (2018). The Predictive Value of ABCB1, ABCG2, CYP3A4/5 and CYP2D6 Polymorphisms for Risperidone and Aripiprazole Plasma Concentrations and the Occurrence of Adverse Drug Reactions. Pharmacogenomics J. 18, 422–430. doi:10.1038/tpj.2017.38
Ramsay, H., Barnett, J. H., Miettunen, J., Mukkala, S., Mäki, P., Liuhanen, J., et al. (2015). Association between Dopamine Receptor D2 (DRD2) Variations Rs6277 and Rs1800497 and Cognitive Performance According to Risk Type for Psychosis: A Nested Case Control Study in a Finnish Population Sample. PLoS ONE 10, e0127602. doi:10.1371/journal.pone.0127602
Reynolds, G. P., Hill, M. J., and Kirk, S. L. (2006). The 5-HT2C Receptor and Antipsychoticinduced Weight Gain - Mechanisms and Genetics. J. Psychopharmacol. 20, 15–18. doi:10.1177/1359786806066040
Rivera-Iñiguez, I., Panduro, A., Ramos-Lopez, O., Villaseñor-Bayardo, S. J., and Roman, S. (2019). DRD2/ANKK1 TaqI A1 Polymorphism Associates with Overconsumption of Unhealthy Foods and Biochemical Abnormalities in a Mexican Population. Eat. Weight Disord. 24, 835–844. doi:10.1007/s40519-018-0596-9
Robinson, D. G., Gallego, J. A., John, M., Petrides, G., Hassoun, Y., Zhang, J.-P., et al. (2015). A Randomized Comparison of Aripiprazole and Risperidone for the Acute Treatment of First-Episode Schizophrenia and Related Disorders: 3-Month Outcomes. SCHBUL 41, 1227–1236. doi:10.1093/schbul/sbv125
Roerig, J. L., Steffen, K. J., and Mitchell, J. E. (2011). Atypical Antipsychotic-Induced Weight Gain. CNS Drugs 25, 1035–1059. doi:10.2165/11596300-000000000-00000
Ruaño, G., Goethe, J. W., Caley, C., Woolley, S., Holford, T. R., Kocherla, M., et al. (2007). Physiogenomic Comparison of Weight Profiles of Olanzapine- and Risperidone-Treated Patients. Mol. Psychiatry 12, 474–482. doi:10.1038/sj.mp.4001944
Rummel-Kluge, C., Komossa, K., Schwarz, S., Hunger, H., Schmid, F., Lobos, C. A., et al. (2010). Head-to-head Comparisons of Metabolic Side Effects of Second Generation Antipsychotics in the Treatment of Schizophrenia: A Systematic Review and Meta-Analysis. Schizophrenia Res. 123, 225–233. doi:10.1016/j.schres.2010.07.012
Saiz-Rodríguez, M., Belmonte, C., Román, M., Ochoa, D., Jiang-Zheng, C., Koller, D., et al. (2018). Effect of ABCB1 C3435T Polymorphism on Pharmacokinetics of Antipsychotics and Antidepressants. Basic Clin. Pharmacol. Toxicol. 123, 474–485. doi:10.1111/bcpt.13031
Saiz-Rodríguez, M., Ochoa, D., Belmonte, C., Román, M., Vieira de Lara, D., Zubiaur, P., et al. (2019). Polymorphisms in CYP1A2, CYP2C9 and ABCB1 Affect Agomelatine Pharmacokinetics. J. Psychopharmacol. 33, 522–531. doi:10.1177/0269881119827959
Sanchez Spitman, A. B., Moes, D. J. A. R., Gelderblom, H., Dezentje, V. O., Swen, J. J., and Guchelaar, H. J. (2017). Effect of CYP3A4*22, CYP3A5*3, and CYP3A Combined Genotypes on Tamoxifen Metabolism. Eur. J. Clin. Pharmacol. 73, 1589–1598. doi:10.1007/s00228-017-2323-2
Schultz, S. H., North, S. W., and Shields, C. G. (2007). Schizophrenia: a Review. Am. Fam. Physician 75, 1821–1829.
Seven, H., Ayhan, M. G., Kürkcü, A., Özbek, S., and Eren, İ. (2017). Aripiprazole-induced Asymptomatic Hypertension: A Case Report. Psychopharmacol. Bull. 47, 53–56.
Shapiro, D. A., Renock, S., Arrington, E., Chiodo, L. A., Liu, L.-X., Sibley, D. R., et al. (2003). Aripiprazole, A Novel Atypical Antipsychotic Drug with a Unique and Robust Pharmacology. Neuropsychopharmacol 28, 1400–1411. doi:10.1038/sj.npp.1300203
Shen, Y.-C., Chen, S.-F., Chen, C.-H., Lin, C. C. H., Chen, S.-J., Chen, Y.-J., et al. (2009). Effects of DRD2/ANKK1 Gene Variations and Clinical Factors on Aripiprazole Efficacy in Schizophrenic Patients. J. Psychiatr. Res. 43, 600–606. doi:10.1016/j.jpsychires.2008.09.005
Sicard, M. N., Zai, C. C., Tiwari, A. K., Souza, R. P., Meltzer, H. Y., Lieberman, J. A., et al. (2010). Polymorphisms of theHTR2Cgene and Antipsychotic-Induced Weight Gain: an Update and Meta-Analysis. Pharmacogenomics 11, 1561–1571. doi:10.2217/pgs.10.123
Skogh, E., Sjödin, I., Josefsson, M., and Dahl, M.-L. (2011). High Correlation between Serum and Cerebrospinal Fluid Olanzapine Concentrations in Patients with Schizophrenia or Schizoaffective Disorder Medicating with Oral Olanzapine as the Only Antipsychotic Drug. J. Clin. Psychopharmacol. 31, 4–9. doi:10.1097/JCP.0b013e318204d9e2
Smith, R. C., Lindenmayer, J.-P., Davis, J. M., Kelly, E., Viviano, T. F., Cornwell, J., et al. (2009). Effects of Olanzapine and Risperidone on Glucose Metabolism and Insulin Sensitivity in Chronic Schizophrenic Patients with Long-Term Antipsychotic Treatment. J. Clin. Psychiatry 70, 1501–1513. doi:10.4088/JCP.08m04446yel
Smith, R. C., Lindenmayer, J.-P., Hu, Q., Kelly, E., Viviano, T. F., Cornwell, J., et al. (2010). Effects of Olanzapine and Risperidone on Lipid Metabolism in Chronic Schizophrenic Patients with Long-Term Antipsychotic Treatment: A Randomized Five Month Study. Schizophrenia Res. 120, 204–209. doi:10.1016/j.schres.2010.04.001
Smith, R. C., Rachakonda, S., Dwivedi, S., and Davis, J. M. (2012). Olanzapine and Risperidone Effects on Appetite and Ghrelin in Chronic Schizophrenic Patients. Psychiatry Res. 199, 159–163. doi:10.1016/j.psychres.2012.03.011
Söderberg, M. M., and Dahl, M.-L. (2013). Pharmacogenetics of Olanzapine Metabolism. Pharmacogenomics 14, 1319–1336. doi:10.2217/pgs.13.120
Söderberg, M. M., Haslemo, T., Molden, E., and Dahl, M.-L. (2013). Influence of FMO1 and 3 Polymorphisms on Serum Olanzapine and its N-Oxide Metabolite in Psychiatric Patients. Pharmacogenomics J. 13, 544–550. doi:10.1038/tpj.2012.47
Spellmann, I., Reinhard, M. A., Veverka, D., Zill, P., Obermeier, M., Dehning, S., et al. (2018). QTc Prolongation in Short-Term Treatment of Schizophrenia Patients: Effects of Different Antipsychotics and Genetic Factors. Eur. Arch. Psychiatry Clin. Neurosci. 268, 383–390. doi:10.1007/s00406-018-0880-8
Stahl, S. M. (2018). Beyond the Dopamine Hypothesis of Schizophrenia to Three Neural Networks of Psychosis: Dopamine, Serotonin, and Glutamate. CNS Spectr. 23, 187–191. doi:10.1017/S1092852918001013
Suzuki, T., Mihara, K., Nakamura, A., Kagawa, S., Nagai, G., Nemoto, K., et al. (2014). Effects of Genetic Polymorphisms of CYP2D6, CYP3A5, and ABCB1 on the Steady-State Plasma Concentrations of Aripiprazole and its Active Metabolite, Dehydroaripiprazole, in Japanese Patients with Schizophrenia. Ther. Drug Monit. 36, 651–655. doi:10.1097/FTD.0000000000000070
Swen, J. J., Nijenhuis, M., de Boer, A., Grandia, L., Maitland-van der Zee, A. H., Mulder, H., et al. (2011). Pharmacogenetics: From Bench to Byte- an Update of Guidelines. Clin. Pharmacol. Ther. 89, 662–673. doi:10.1038/clpt.2011.34
Tamminga, C. A., and Kane, J. M. (1997). Olanzapine (Zyprexa):charactirestics of a New Antipsychotic. Expert Opin. Investig. Drugs 6, 1743–1752. doi:10.1517/13543784.6.11.1743
Templeman, L. A., Reynolds, G. P., Arranz, B., and San, L. (2005). Polymorphisms of the 5-HT2C Receptor and Leptin Genes Are Associated with Antipsychotic Drug-Induced Weight Gain in Caucasian Subjects with a First-Episode Psychosis. Pharmacogenetics and Genomics 15, 195–200. doi:10.1097/01213011-200504000-00002
Tsuda, Y., Saruwatari, J., and Yasui-Furukori, N. (2014). Meta-analysis: the Effects of Smoking on the Disposition of Two Commonly Used Antipsychotic Agents, Olanzapine and Clozapine. BMJ Open 4, e004216. doi:10.1136/bmjopen-2013-004216
Vandenberghe, F., Guidi, M., Choong, E., von Gunten, A., Conus, P., Csajka, C., et al. (2015). Genetics-Based Population Pharmacokinetics and Pharmacodynamics of Risperidone in a Psychiatric Cohort. Clin. Pharmacokinet. 54, 1259–1272. doi:10.1007/s40262-015-0289-8
Vanwong, N., Ngamsamut, N., Hongkaew, Y., Nuntamool, N., Puangpetch, A., Chamnanphon, M., et al. (2016). Detection of CYP2D6 Polymorphism Using Luminex xTAG Technology in Autism Spectrum Disorder: CYP2D6 Activity Score and its Association with Risperidone Levels. Drug Metab. Pharmacokinet. 31, 156–162. doi:10.1016/j.dmpk.2016.01.005
Velamoor, R. (2017). Neuroleptic Malignant Syndrome: A Neuro-Psychiatric Emergency: Recognition, Prevention, and Management. Asian J. Psychiatry 29, 106–109. doi:10.1016/j.ajp.2017.05.004
Wang, J.-S., Zhu, H.-J., Markowitz, J. S., Donovan, J. L., and DeVane, C. L. (2006). Evaluation of Antipsychotic Drugs as Inhibitors of Multidrug Resistance Transporter P-Glycoprotein. Psychopharmacology 187, 415–423. doi:10.1007/s00213-006-0437-9
Wilson, M. P., Nordstrom, K., Hopper, A., Porter, A., Castillo, E. M., and Vilke, G. M. (2017). Risperidone in the Emergency Setting Is Associated with More Hypotension in Elderly Patients. J. Emerg. Med. 53, 735–739. doi:10.1016/j.jemermed.2017.06.026
Wong, M. (2004). CYP3A5 Genotype and Midazolam Clearance in Australian Patients Receiving Chemotherapy*1. Clin. Pharmacol. Ther. 75, 529–538. doi:10.1016/j.clpt.2004.02.005
Woo, Y. S., Kim, W., Chae, J.-H., Yoon, B.-H., and Bahk, W.-M. (2009). Blood Pressure Changes during Clozapine or Olanzapine Treatment in Korean Schizophrenic Patients. World J. Biol. Psychiatry 10, 420–425. doi:10.1080/15622970801910399
Xing, Q., Gao, R., Li, H., Feng, G., Xu, M., Duan, S., et al. (2006). Polymorphisms of theABCB1gene Are Associated with the Therapeutic Response to Risperidone in Chinese Schizophrenia Patients. Pharmacogenomics 7, 987–993. doi:10.2217/14622416.7.7.987
Yasui-Furukori, N., Tsuchimine, S., Saito, M., Nakagami, T., Sato, Y., and Kaneko, S. (2007). Association between Major Multidrug Resistance 1 (MDR1) Gene Polymorphisms and Plasma Concentration of Prolactin during Risperidone Treatment in Schizophrenic Patients. Prog. Neuro-Psychopharmacology Biol. Psychiatry 31, 1230–1234. doi:10.1016/j.pnpbp.2007.04.021
Zai, C. C., Tiwari, A. K., Zai, G. C., Maes, M. S., and Kennedy, J. L. (2018). New Findings in Pharmacogenetics of Schizophrenia. Curr. Opin. Psychiatry 31, 200–212. doi:10.1097/YCO.0000000000000417
Zhang, J., Qiao, Y., Le, J., Sun, D., Guan, Y., and Li, Z. (2016). Olanzapine May Inhibit Colonic Motility Associated with the 5-HT Receptor and Myosin Light Chain Kinase. Psychiatry Investig. 13, 232. doi:10.4306/pi.2016.13.2.232
Zhao, Q.-Z., Liu, B.-C., Zhang, J., Wang, L., Li, X.-W., Wang, Y., et al. (2012). Association between a COMT Polymorphism and Clinical Response to Risperidone Treatment. Psychiatr. Genet. 22, 298–299. doi:10.1097/YPG.0b013e328358629a
Zubiaur, P., Soria-Chacartegui, P., Koller, D., Navares-Gómez, M., Ochoa, D., Almenara, S., et al. (2021). Impact of Polymorphisms in Transporter and Metabolizing Enzyme Genes on Olanzapine Pharmacokinetics and Safety in Healthy Volunteers. Biomed. Pharmacother. 133, 111087. doi:10.1016/j.biopha.2020.111087
Keywords: antipsychotics, pharmacogenetics, metabolism, genetic polymorphism, pharmacokinetics
Citation: Soria-Chacartegui P, Villapalos-García G, Zubiaur P, Abad-Santos F and Koller D (2021) Genetic Polymorphisms Associated With the Pharmacokinetics, Pharmacodynamics and Adverse Effects of Olanzapine, Aripiprazole and Risperidone. Front. Pharmacol. 12:711940. doi: 10.3389/fphar.2021.711940
Received: 19 May 2021; Accepted: 28 June 2021;
Published: 14 July 2021.
Edited by:
Jatinder K. Lamba, University of Florida, United StatesReviewed by:
Elena De Mattia, Centro di Riferimento per l'oncologia di Aviano (IRCCS), ItalyCopyright © 2021 Soria-Chacartegui, Villapalos-García, Zubiaur, Abad-Santos and Koller. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Francisco Abad-Santos, ZnJhbmNpc2NvLmFiYWRAdWFtLmVz; Dora Koller, ZG9yYS5rb2xsZXJAeWFsZS5lZHU=
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.