- 1School of Pharmacy, Chengdu University of Traditional Chinese Medicine, Chengdu, China
- 2School of Health Preservation and Rehabilitation, Chengdu University of Traditional Chinese Medicine, Chengdu, China
- 3Center for Health Policy & Drug Affairs Operation Management, Chengdu University of Traditional Chinese Medicine, Chengdu, China
Ulcerative colitis (UC) is a chronic non-specific inflammatory bowel disease, which usually manifests as abdominal pain, diarrhea and hematochezia. The disease often recurs and is difficult to cure. At present, the pathogenesis is not clear, but it is believed that the disease is caused by a complex interaction among immunity, heredity, environment and intestinal microflora disorders. MicroRNA (miRNA) is endogenous single-stranded non-coding RNA of 17–25 nucleotides (nts). They target the 3'Untranslated Region of a target gene and inhibit or degrade the target gene according to the extent of complementary bases. As important gene expression regulators, miRNAs are involved in regulating the expression of most human genes, and play an important role in the pathogenesis of many autoimmune diseases including UC. Studies in recent years have illustrated that abnormal expression of miRNA occurs very early in disease pathogenesis. Moreover, this abnormal expression is highly related to disease activity of UC and colitis-associated cancer, and involves virtually all key UC-related mechanisms, such as immunity and intestinal microbiota dysregulation. Recently, it was discovered that miRNA is highly stable outside the cell in the form of microvesicles, exosomes or apoptotic vesicles, which raises the possibility that miRNA may serve as a novel diagnostic marker for UC. In this review, we summarize the biosynthetic pathway and the function of miRNA, and summarize the usefulness of miRNA for diagnosis, monitoring and prognosis of UC. Then, we described four types of miRNAs involved in regulating the mechanisms of UC occurrence and development: 1) miRNAs are involved in regulating immune cells; 2) affect the intestinal epithelial cells barrier; 3) regulate the homeostasis between gut microbiota and the host; and 4) participate in the formation of tumor in UC. Altogether, we aim to emphasize the close relationship between miRNA and UC as well as to propose that the field has value for developing potential biomarkers as well as therapeutic targets for UC.
Introduction
Ulcerative colitis (UC) is one of two main inflammatory bowel diseases (IBD), which is a chronic nonspecific inflammation, mainly manifested as long-term inflammation and ulcer of the rectum and colon. The inflammation caused by UC is usually confined to the mucosa and submucosa, and is usually distributed continuously from the rectum to the proximal colon (Rubin et al., 2019). The course of UC is intermittent, with active and inactive periods appearing alternately. Patients in the active stage usually suffer from abdominal pain, diarrhea, bloody stools and weight loss. It is believed that UC is caused by a complex interaction among environmental factors, genetic susceptibility, altered gut microbiota, and immune disorders. UC often recurs, which may lead to intestinal perforation, toxic megacolon, colorectal cancer (CRC) and other complications (Du and Ha, 2020). UC is incurable. At present, treatment is used to achieve clinical symptom relief, mucosal healing and histological relief (Hindryckx et al., 2015; Ng et al., 2017; Ungaro et al., 2017). At present, the diagnosis of UC is through endoscopic examination combined with clinical symptoms, histological analysis, laboratory examination and imaging research (Kobayashi et al., 2020). However, 15–30% of IBD patients can’t be diagnosed with UC or Crohn’s disease (CD), which is called uncertain colitis (Ballard and M'Koma, 2015), which has a negative impact on the future treatment of patients (Soubières and Poullis, 2016). With the increasing incidence and prevalence rate of UC, it has become a global medical burden (Ng et al., 2017). It has become an urgent clinical demand to seek more effective clinical diagnosis, monitoring and treatment of UC.
MicroRNA (miRNA) is a single-stranded non-coding RNA of 17–25 nucleotides (nts), which is a kind of small RNA and highly conserved in different organisms. miRNA participates in regulating various physiological processes, such as cell growth, differentiation, apoptosis and carcinogenesis, including DNA double-strand breaks (DSBs) (Chowdhury et al., 2013; Hu et al., 2017). Unlike small interfering RNAs (siRNA) and Piwi-associated RNAs (piRNA) (Table 1), piRNA is currently found to be associated with the development of human germ cells, but no piRNA has been found to exist in somatic cells (Slack and Chinnaiyan, 2019; Chen et al., 2021). The action patterns of miRNA and siRNA in the human body are similar, but the difference is that miRNA is mostly endogenous RNA while siRNA is mostly exogenous (Senapati et al., 2019). More than 60% of human protein-coding genes contain at least one conserved miRNA binding site (Lam et al., 2015). miRNA has been found to have significant relevance to a variety of diseases, including UC. In this paper, the physiological synthesis and function of miRNAs were reviewed, and the differential expression of miRNAs in UC was summarized. miRNA as a biomarker for UC was discussed. Finally, the action mechanisms of miRNA were discussed, which involved intestinal immune disorder, barrier dysfunction, intestinal flora imbalance and UC canceration.
Overview of miRNAS
miRNA was first discovered and reported in 1993 (Lee et al., 1993), and found in introns, exons or regulatory sequences of the genome, mainly located in the intergenic regions (Galatenko et al., 2018). It exists in various forms, such as single copy, multiple copies or gene clusters (Cantini et al., 2019). More than 2,500 miRNAs have been found in the human genome (Alles et al., 2019). miRNA participates in almost all biological processes in eukaryotic cells (Friedman et al., 2009) and stably exists in almost all body fluids through extracellular vesicles, thus playing a remote regulatory role (Schaefer et al., 2015; Glinge et al., 2017; Yang et al., 2017; Fredsøe et al., 2019; Zamanillo et al., 2019).
In the canonical biogenesis pathway of miRNAs (Figure 1), RNA polymerase II or III transcribes the miRNA gene into the primary transcript pri-miRNA. Then, DGCR8, an RNA-binding protein, binds to Drosha (a type of RNase III) to form the larger Drosha-DGCR8 complex, the microprocessor (Ameres and Zamore, 2013; Kwon et al., 2016). It is a heterotrimeric complex containing one Drosha and two DGCR8 proteins (Nguyen et al., 2015). The Drosha-DGCR8 complex cleaves the pri-miRNA into the intermediate pre-miRNA (Rawat et al., 2019). In this complex, Drosha acts as the catalytic subunit and determines the cleavage site (Ha and Kim, 2014; Nguyen et al., 2015), and DGCR8 stabilizes Drosha through protein-protein interactions, increasing the affinity of Drosha for the substrate and the accuracy of the cleavage site (Han et al., 2009; Ameres and Zamore, 2013; Ha and Kim, 2014; Quick-Cleveland et al., 2014; Nguyen et al., 2015; Rawat et al., 2019). After exportin-5 (XPO-5) and GTP-binding protein (Ran-GTP) form a complex, pre-miRNA is transported to cytoplasm by the complex (Yamazawa et al., 2018). In the cytoplasm, after binding of Dicer and transactivating response RNA binding protein (TRBP), the pre-miRNA is cut into mature double-stranded miRNA of 17–25 nts (Fareh et al., 2016; Takahashi et al., 2018). Then, through loading and strand selection, mature single-stranded miRNA was transferred into Argonaute (AGO) protein by the Dicer-TRBP complex to form an RNA-induced silence complex (RISC). In the past several years, non-canonical biogenesis pathways of miRNAs such as the mirtron (Kim et al., 2016) synthesis pathway continue to be discovered, but most pathways still require Dicer enzymes. The mirtron pathway is the first non-classical pathway discovered. This pathway does not require the Drosha/Dgcr8 complex to generate pre-miRNA, but still requires the transport of XPO-5 and the cleavage of Dicer enzyme (Figure 1) (Abdelfattah et al., 2014; Stavast and Erkeland, 2019).
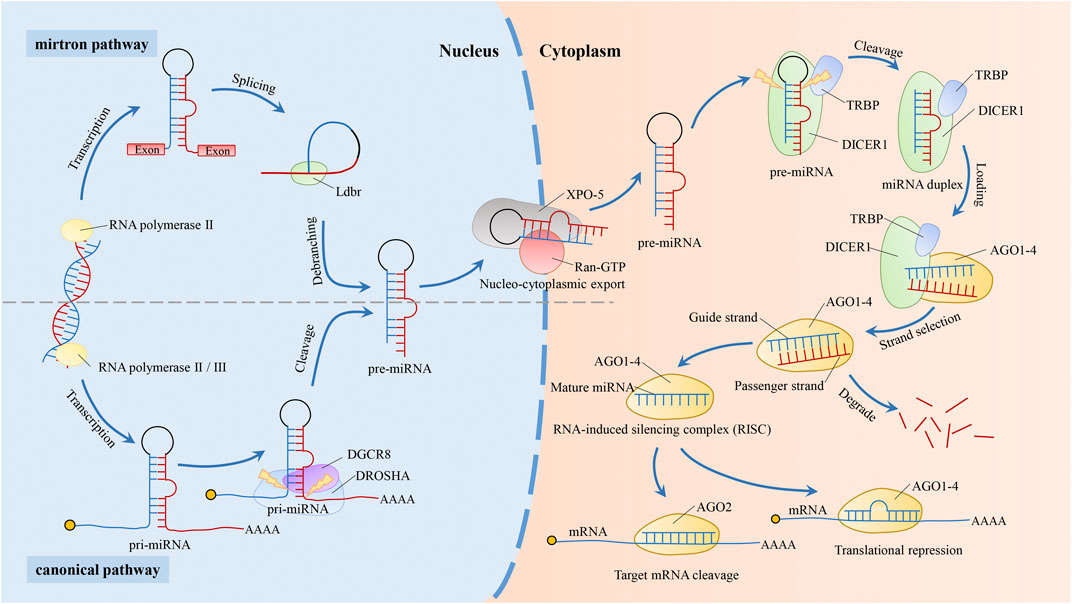
FIGURE 1. Biogenesis pathway of miRNAs. In the canonical biogenesis pathway of miRNAs. After the miRNA gene is transcribed by RNA polymerase II or III into pri-miRNA, it is sheared by a microprocessor consisting of Drosha and DGCR8 into a pre-miRNA of about 70–100 nucleotides (Rawat et al., 2019). The pre-miRNA is then transported to the cytoplasm by XPO-5 (Yamazawa et al., 2018) and sheared to miRNA duplex by the Dicer-TRBP complex (Fareh et al., 2016; Takahashi et al., 2018). Dicer-TRBP complex binds to AGO1-4 and dissociates after transferring the mature miRNA duplex into AGO1-4 to form RISC. RISC recognizes and binds to target genes by the second to eighth nucleotide counted from 5′-end of the miRNAs (Meijer et al., 2014; Gebert and MacRae, 2019), which is degraded or suppressed the target gene based on the extent of sequence complementarity. Target genes are degraded when completely matched, while inhibited when incompletely matched. During RISC loading, one of the mature miRNAs (the “guide strand”) is retained and forms RISC, while the other is degraded (the “passenger strand”) (Matsuyama and Suzuki, 2019). The non-canonical pathway, mirtron, does not require Drosha. introns are spliced and debranched by the lariat debranching enzyme (Ldbr) to produce pre-miRNA. Similar to the canonical miRNA pathway, this pre-miRNA is transported outside the nucleus via XPO-5. And the mirtron pathway merges with the canonical pathway during this transport stage (Abdelfattah et al., 2014; Stavast and Erkeland, 2019).
RISC recognizes and binds to target genes via the seed region (second to eighth nucleotide counted from 5′-end) of miRNA. The canonical pathway of miRNAs mechanism is to recognize target genes by targeting the 3′Untranslated Region (3′-UTR) of mRNA (Kim et al., 2017; Gruszka and Zakrzewska, 2018; Perconti et al., 2019), which is suppressed or degraded based on the extent of sequence complementarity between the target gene and miRNAs (Bartel, 2004). In addition, some studies have reported that a few miRNAs regulate the transcription of target genes through non-canonical pathways. For example, some miRNAs can target the 5′Untranslated Region (5′-UTR) (Kalantari et al., 2017), coding sequences (CDS) (Tang et al., 2017), promoter regions (Matsui et al., 2013), and pri-miRNAs (Tang et al., 2012). miRNAs are widely present in a variety of organisms, including animals, plants and viruses, and most miRNA sequences are highly conserved across organisms and usually have similar functions (Fromm et al., 2015; Bartel, 2018). Of the more than 500 canonical miRNA genes identified in human genome, 296 are conserved among placental mammals (Bartel, 2018). However, for the hundreds of other human miRNAs that are not conserved for mammals, they can not be studied using mouse models. And for miRNAs that are conserved for mice, the mechanisms still need to be validated in human cells (Bartel, 2018).
High-throughput microarray analysis is mainly used to measure changes in miRNA expression profiles, and reverse transcriptase quantitative real-time polymerase chain reaction (RT-qPCR) is used to verify the results of miRNA expression in screening experiments, and to determine the changes of miRNA expression in specific groups. miRNA is abnormally expressed in many diseases including UC, and has been proved to be involved in regulating the immune response of UC and the occurrence and development of colon tumors. Recent studies have also found that miRNA participates in the mutual regulation between host and intestinal flora (Aguilar et al., 2019; Dong et al., 2019; Zhao et al., 2021). These studies of miRNA have provided new insights into the molecular mechanism of UC.
miRNAs Are Involved in Regulating the Pathogenesis of Ulcerative Colitis
miRNAs Are Involved in Regulating Immune Cells
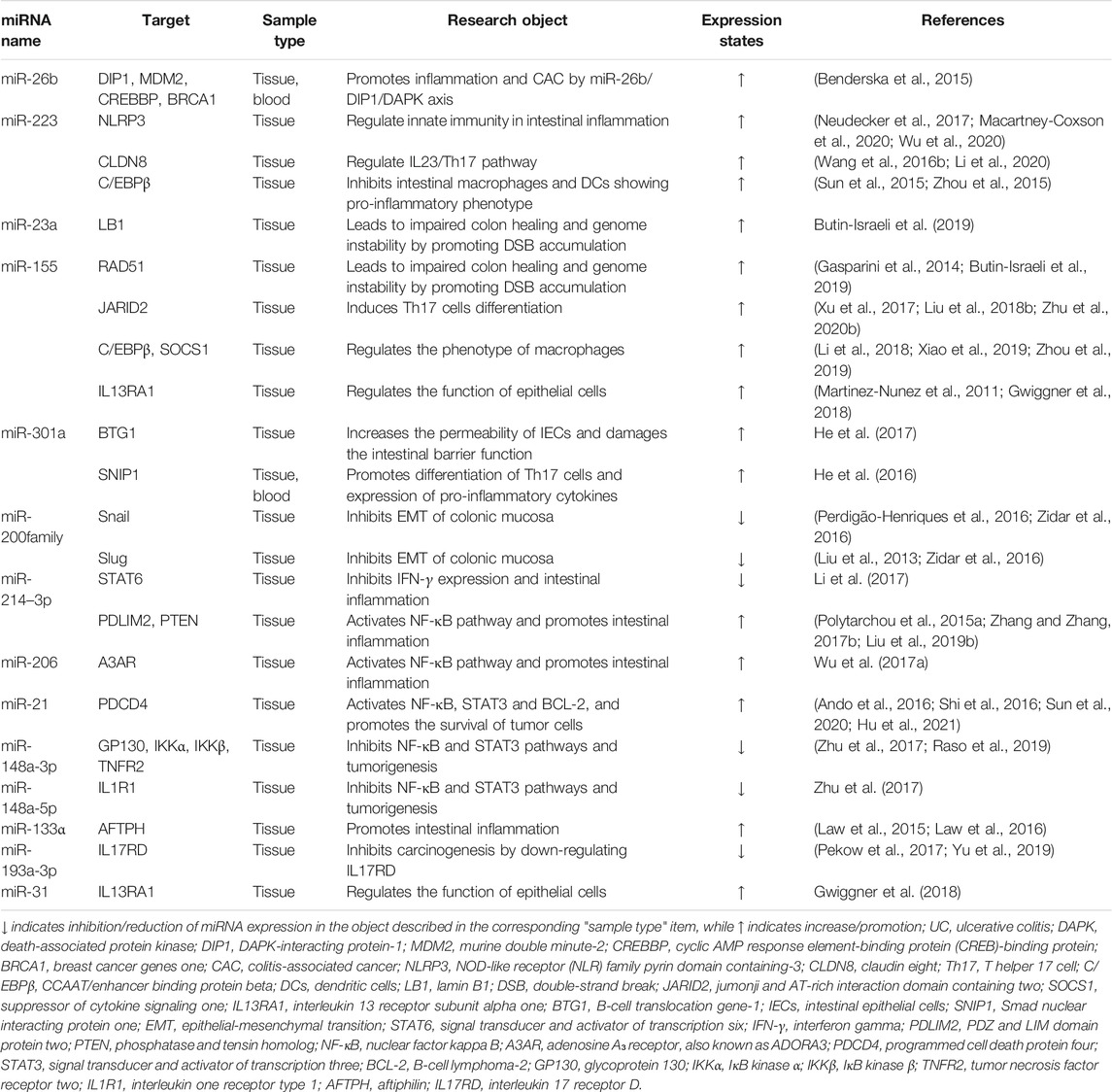
Generally, tissues derived from similar anatomical locations in healthy individuals showed no significant differences in the expression of miRNAs (Landgraf et al., 2007; Liang et al., 2007; Martínez et al., 2017). However, the expression of miRNAs appears significantly dysregulated would lead to activation or inhibition of signaling pathways and result in disease (Pallante et al., 2014; Bracken et al., 2016; Hwang et al., 2018). Dysregulation of miRNAs is strongly associated with UC development (Table 2). miRNAs are crucial regulators of intestinal immunity and are involved in the innate and adaptive immune. Thus, they respond to inflammation, and influence the maturation, differentiation, and infiltration of immune cells (Figure 2). miR-141–3p and let-7b/c/f/g-5p are significantly up-regulated in colon intestinal epithelial cells (IECs) of UC mice models and may affect inflammatory cell infiltration by targeting the chemokines CXCL9 or CXCL16 (Lee et al., 2015). miR-223 is significantly up-regulated in patients with active UC and is a potential biomarker closely related to disease activity (see section miRNAs as Biomarkers for Diagnosis, Monitoring and Prognosis of Ulcerative Colitis). However, it is reported that miR-223 has an anti-inflammatory role in UC. Myeloid-derived miR-223 reduces IL-1β release via repressing Nlrp3 inflammasome to attenuate experimental colitis (Neudecker et al., 2017). Intestinal macrophages and dendritic cells (DCs) lacking miR-223 exhibit a pro-inflammatory phenotype, and monocytes deficiency of miR-223 promotes an increase in monocyte-derived DCs, resulting in more severe colitis (Zhou et al., 2015). Besides, a high-fat diet promotes the release of exosomes with pro-inflammatory factors (e.g., miR-155) from visceral adipose into the intestine to promote macrophage M1 polarization and aggravate colitis (Wei et al., 2020). Erobic exercise can significantly reduce the expression of miR-155 and miR-146a, and increase miR-126 expression in mouse vascular tissue (Wu et al., 2014). In addition, pro-inflammatory miRNAs, such as miR-23a and miR-155, can be released by activated tissue-infiltrating neutrophils via microparticles (Butin-Israeli et al., 2019). miR-23a and miR-155 can promote the accumulation of DNA double-strand breaks (DSBs) in IECs by downregulating nuclear envelope protein Lamin-B1 and RAD51 (a key homologous recombination regulator), leading to genomic instability and ultimately tumorigenesis (Butin-Israeli et al., 2019).

FIGURE 2. miRNAs are involved in the occurrence and development of UC. miRNAs regulate the generation, differentiation, and function of multiple immune cells (e.g., macrophages, DCs, T-cell) (Murugaiyan et al., 2015; Zhou et al., 2015; Liu et al., 2016a; Wang et al., 2016a; Hou et al., 2017; Neudecker et al., 2017; Xu et al., 2017; Li et al., 2018; Wei et al., 2020). miRNAs also affect the physical barrier of intestinal tract by regulating IEC's tight junctions and apoptosis (Bian et al., 2011; Van der Goten et al., 2014; Zhang et al., 2015; Wang et al., 2016b; He et al., 2017). In addition, miRNAs are secreted by IECs into the intestinal lumen through exosomes and regulate the bacterial growth (Liu et al., 2016b).
In adaptive immunity, miRNAs regulate the differentiation and function of regulatory T-cell (Treg), T helper 17 cell (Th17), cluster of differentiation eight positive (CD8+) T-cell and CD8+ B-cell (Xu and Zhang, 2016; Yang et al., 2016). miR-155 could induce Th17 differentiation via targeting Jarid2 (Xu et al., 2017) and is essential for the generation and function of T follicular helper cells (Tfh) (Liu et al., 2016a). Macrophages lacking miR-155 are polarized to the M2 phenotype, attenuating intestinal immune cell proliferation, and inhibiting cluster of differentiation four positive (CD4+) T-cell polarization to Th1 and Th17 (Hou et al., 2017; Li et al., 2018). miR-21 also promotes Th2 cell differentiation and is involved in Th2-type inflammation development (Murugaiyan et al., 2015; Wang et al., 2016a).
The above studies indicated that the regulation of miRNAs to suppress inflammation is an effective way to treat UC. It should be noted that since a single miRNA can regulate hundreds of target genes, the effect of anti-inflammatory miRNAs is a combined result presented after acting on various immune cells, rather than on a certain type of immune cells. The therapeutic use of miRNAs mainly includes both inhibition of pro-inflammatory miRNAs and overexpression of anti-inflammatory miRNAs. Viral vectors, lipids, polymers, inorganic and extracellular vesicles are usually used to deliver therapeutic miRNAs (Dasgupta and Chatterjee, 2021). However, due to the wide regulation of miRNA (Lewis et al., 2003; Broughton and Pasquinelli, 2016), how to reduce the regulation unrelated to the therapeutic purpose is the main problem at present.
miRNAs Affect the IEC’s Barrier
miRNAs are key regulators of the IEC's barrier, regulating the growth and apoptosis of IECs, as well as the tight junctions between IECs (Figure 2). IEC's barrier is the foremost component of the intestinal mucosal barrier, and the intestinal mucosal barrier enables epithelial cells to maintain immune tolerance to more than 10 trillion intestinal microorganisms (Kanneganti, 2017). When the intestinal mucosal barrier is disrupted, intestinal microbes invade the IECs, triggering intestinal inflammation (Jostins et al., 2012). miR-223 is induced by the IL23/Th17 pathway and disrupts the tight junctions between IECs via targeting Claudin-8 (Wang et al., 2016b). miR-21 may also regulate IEC's permeability but by promoting protein kinase B (AKT) phosphorylation and inhibiting PTEN expression through the PTEN/PI3K/AKT signaling pathway. And the knockout of miR-21 reduces intestinal permeability (Zhang et al., 2015). miR-301a is up-regulated in IECs of active IBD patients. It decreases the expression of cadherin-1 and increases cell permeability to disrupt intestinal barrier function and promotes inflammation and tumorigenesis via targeting BTG anti-proliferation factor 1 (BTG1) (He et al., 2017).
In IBD, the miR-200 family may maintain the integrity of IECs and inhibit intestinal fibrosis by inhibiting epithelial-mesenchymal transition (EMT) (Zidar et al., 2016). miR-200b-3p is significantly up-regulated in cancerous epithelial cells and dysplastic tissues of UC patients, which indicates that miR-200b-3p may be involved in the carcinogenesis of UC (Lewis et al., 2017). miR-200c-3p is down-regulated in the mucosa of UC patients in the active stage, the inhibition of its target IL-8 and cadherin-11 (related to the barrier function of IEC) subsequently decreased (Van der Goten et al., 2014). miR-150 is up-regulated in the mucosa of UC patients, and inhibits the expression of target transcription factor c-Myb, resulting in a decrease in the expression of Bcl-2. This pattern suggests that miR-150 may disrupt epithelial barrier function through an apoptotic mechanism (Bian et al., 2011). But miR-150 can also induce apoptosis and inhibit tumor cell migration and invasion, which correlates with CRC patient prognosis (Ma et al., 2012).
In general, miRNA plays a key role in maintaining the function of intestinal tissue barriers. miRNA can enhance the tight junctions between IECs and reduce their permeability, and is related to apoptosis. miRNA not only can be used as potential therapeutic targets for UC, but also has a good applications prospect in maintaining the stability of the intestinal barriers in healthy individuals.
miRNAs Regulate the Homeostasis Between Gut Microbiota and the Host
Recent studies have reported that miRNAs are involved in the cross-regulation between gut microbiota and host (Figure 2). Intestinal microflora is an important part of the gut microenvironment. UC is usually associated with the proliferation of pathogenic intestinal bacteria (such as Escherichia coli, Salmonella and Clostridium difficile) (Mead et al., 1999).
The expression of some miRNAs in B-cell can be promoted by short-chain fatty acids (a metabolite of intestinal microorganisms). These miRNAs inhibit Aicda and Prdm1 in B-cell, and thus regulate the differentiation of B-cell (Sanchez et al., 2020). Seth and his colleagues found that nitric oxide produced by gut microbes in Caenorhabditis elegans can cause S-nitrosylation modification of AGO, a key protein of the miRNA pathway, thereby inhibiting miRNA activity and affecting C. elegans development (Seth et al., 2019). Moreover, nitric oxide can modify the S-nitrosylation of AGO proteins in mammals (Seth et al., 2019). In addition, the reduction of intestinal microbial community abundance in specific pathogen-free (SPF) mice mediated by broad-spectrum antibiotics promoted tumor lung metastasis through circRNA/miRNA networks, while tumor metastasis was effectively inhibited by transplantation of fecal bacteria into germ-free (GF) mice using SPF mice feces (Zhu et al., 2020a). Fusobacterium nucleatum inhibits the expression of miR-18a* and miR-4802 by activating the TLR4/MYD88 pathway. This reduces the inhibition of miR-18a* and miR-4802 on their target genes ATG7 and ULK1, resulting in activation of the autophagic pathway and alteration of CRC chemotherapy response (Yu et al., 2017).
Conversely, the host can regulate the development of intestinal microbes via miRNAs. Liu and his colleagues found that miRNAs are secreted into the intestinal lumen by IECs via extracellular vesicles and enter the microbes to target mRNAs and regulate microbial development (Liu et al., 2016b). And fecal miRNA from wild-type mice could alleviate dextran sulfate sodium (DSS)-induced colitis in miRNA-deficient mice (Liu et al., 2016b). Subsequently, Liu et al. found that fecal miR-30d targeted Akkermansia muciniphila and increased its abundance in the intestine by upregulating lactase expression (Liu et al., 2019a). Ji et al. screened fecal miRNAs that are differentially expressed in IBD, and found that four of these miRNAs (miR-199a-5p, miR-548ab, miR-1226 and miR-515–5p) could target and regulate the proliferation of Fusobacterium nucleatum, Escherichia coli and segmental filamentous bacteria (Ji et al., 2018). In addition, food-derived miRNAs can also regulate the development of gut microbiota. miRNAs from ginger exosomes reduce colitis by promoting the multiplication of Lactobacillus rhamnosus and the production of ligands for aryl hydrocarbon receptors (Teng et al., 2018).
miRNAs can also indirectly participate in the regulation of intestinal homeostasis by affecting the expression of intestinal immunoglobulins. Some immunoglobulins promote host-microbiota interactions by binding symbiotic bacteria, which are important for maintaining intestinal homeostasis (Pabst, 2012; Magri et al., 2017; Castro-Dopico et al., 2019). For example, immunoglobulin A (IgA) secretion-deficient mice and humans show increased susceptibility to inflammatory bowel disease, celiac disease and allergy. (Moon et al., 2015). The study showed that miR-221–5p could target polymeric immunoglobulin receptor (pIgR), which is closely related to the sustained supply of secretory IgA, reducing the intestinal secretory IgA level (Bruno et al., 2011; Zeng et al., 2021). Activation-induced cytidine deaminase (AID), a key enzyme of intestinal immune antibody, was targeted by miR-155. Inhibition of miR-155 leads to upregulation of AID and ameliorates the disruption of the intestinal immune barrier caused by IgA and IgM dysfunction (Fairfax et al., 2015; Zhang et al., 2018). Crk-like protein (CRKL) is found to be related to the regulation of B-cell and immunoglobulin G (IgG) levels. miR-29a targets and inhibits CRKL level, and regulates IgG expression through miR-29a/CRKL axis (Shi et al., 2020). In addition, miR-17–92 is reported to be necessary for the production of immunoglobulin G2c (IgG2c) isoforms in B-cell. The IgG generation of miR-17–92-deficient B-cell is significantly reduced in the intestine (Xu et al., 2015; Wu et al., 2018). Currently, we know little about the effects of miRNAs on immunoglobulins in UC, but how miRNAs participate in the interaction between these immunoglobulins and the gut microbiota will be a promising field.
These findings reveal the role of miRNAs in host-microbe interactions, and provide a new method to maintain gut ecological stability.
miRNAs Participate in the Formation of Tumor in Ulcerative Colitis
Patients with UC for more than 8 years have an increased risk of colitis-associated cancer (CAC) (Chen et al., 2016), and it is an urgent clinical need to prevent UC from developing into CRC. At present, it is believed that the sustained activation of carcinogenic signaling pathways, such as NF-κB, STAT3 (Li et al., 2017), and PI3K (Zhang et al., 2015), the release of pro-inflammatory mediators and the increase of local levels of reactive oxygen species and nitrogen substances, contribute to the development of CAC. Now it has been found that some miRNAs can regulate the carcinogenesis of UC (Figure 3). miR-301a promotes Th17 differentiation and decreases cadherin-1 expression to disrupt intestinal barrier function by targeting SNIP1 and BTG1. And knockout of miR-301a reduced inflammation and inhibited tumor occurrence (He et al., 2016; He et al., 2017).
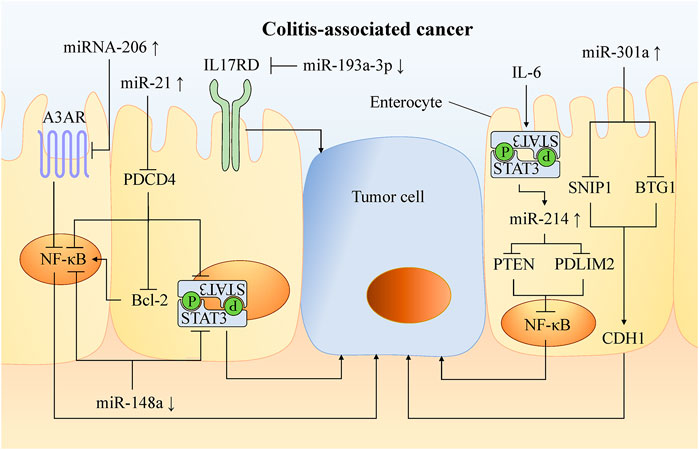
FIGURE 3. Mechanism of miRNA involved in carcinogenesis of UC. In UC, chronic activation of carcinogenic pathways such as NF-κB and STAT3 leads to a higher risk of CAC. A variety of miRNAs can promote or inhibit the occurrence of cancer by targeting molecules in these pathways. Increasing the abundance of anti-oncogenic miRNAs and decreasing the abundance of oncogenic miRNAs can inhibit tumor formation and progression in experimental colitis. This provides a new therapeutic target for the preventing and treatng CAC.
NF-κB is an essential pathway that promotes tissues with inflammation to transform into cancer. Through molecules targeting this pathway, a variety of miRNAs can be involved in cancer formation. For example, miR-206 is up-regulated in the mucosa of UC patients, and promotes intestinal inflammation by targeting the adenosine A3 receptor (A3AR) to reduce the inhibition of NF-κB pathway (Wu et al., 2017a). miR-214 is highly expressed in the colonic tissues of active UC and CAC patients by STAT3 pathway. It activates the NF-κB pathway by targeting and inhibiting PDLIM2 and PTEN expression. miR-214 inhibitors significantly inhibit (>90%) NF-κB phosphorylation, thus reducing the severity of colitis and the number and size of tumors in mice (Polytarchou et al., 2015a).
miR-21 is significantly up-regulated in tissues of UC (Yan et al., 2020), CRC (Wu et al., 2017b) and CAC, activates the NF-κB, STAT3, and Bcl-2 by targeting PDCD4 (Figure 3). Then, it reduced the apoptosis of mouse tumor cells (Shi et al., 2016). Blocking miR-21 reduced pro-inflammatory and pro-cancer factors, and then decreased the number and size of colon tumors in mice (Shi et al., 2016).
miR-148a is down-regulated in the colon tissues of UC and CRC patients, and reduces the activation of NF-κB and STAT3 in macrophages of colon tissues by directly targeting upstream regulators of NF-κB and STAT3 pathway (including GP130, IKKα, IKKβ, IL1R1 and TNFR2). miR-148a-deficient mice are more susceptible to UC and CAC (Zhu et al., 2017).
miR-155 is up-regulated in a variety of malignant tumors including CRC, which promotes proliferation and metastasis of colon tumor cells, and is related to tumor location, tumor grade, metastasis and TNM stage (Qu et al., 2015). In CRC, miR-155 targets protein tyrosine phosphatase receptor J-type (PTPRJ) mRNA, inhibits the anti-proliferation effects of PTPRJ, promotes the proliferation and migration of tumor cells, and activates the AKT pathway (Zhang et al., 2017a). Al-Haidari et al. found that miR-155–5p upregulates the human antigen R (HuR) by targeting the AU-rich elements (AREs) in the 3′-UTR region of HuR mRNA, thus promoting the metastasis of colon cancer cells. The migration of colon cancer cells can be inhibited by blocking the binding of miR-155–5p and ARE in HuR (Al-Haidari et al., 2018).
These studies prove that miRNA is related to the degree of inflammation and canceration in UC. The pro-inflammatory miRNAs can promote the occurrence and development of cancer, whereas anti-inflammatory miRNAs can also resist cancer. miRNA is involved in the transformation of inflammation to cancer in epigenetics, and thus provides a potential therapeutic target for UC and CRC.
miRNAs Are Specifically Expressed in Ulcerative Colitis
miRNAs as Biomarkers for Diagnosis, Monitoring and Prognosis of Ulcerative Colitis
The early diagnosis of UC can provide more accurate drug treatment and prevent complications. At present, colonoscopy is the gold standard in the diagnosis of UC, supplemented by blood tests and biomarker analysis. However, colonoscopy is invasive, which brings considerable physical and economic burden to patients, accompanied by serious complications such as intestinal perforation and death (Hagel et al., 2012; Carmona et al., 2013; Adler et al., 2014; Borgaonkar et al., 2016; Niikura et al., 2016; Vatandoost et al., 2016). We summarized the promising miRNA biomarkers in UC (Table 3) and discussed the prospect of miRNA biomarkers in the diagnosis, surveillance, and prognosis of UC, as well as in the screening for CAC (Figure 4).
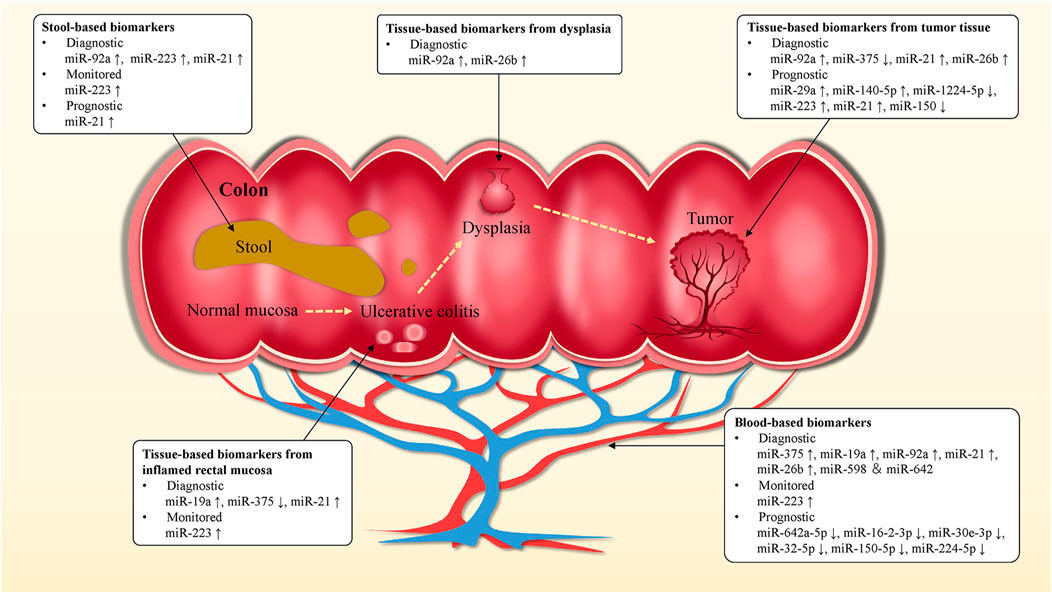
FIGURE 4. miRNA biomarkers in UC. Blood, stool and tissue-based miRNA biomarkers might be used for the diagnosis, monitoring, prognosis of UC and the prediction of early stage CAC, as well as to guide clinical treatment and medication strategies. miRNA biomarkers could be of value in improving survival and cure rates in UC and CAC patients.
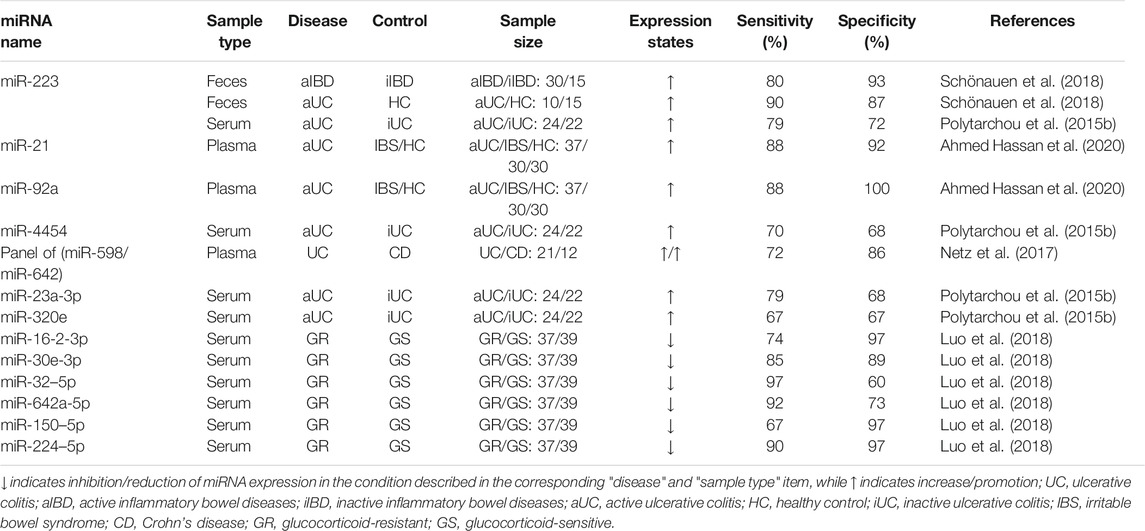
Tissue, blood and stool samples were mainly used in the study of miRNAs in UC. For example, plasma miR-21 and miR-92a are up-regulated in UC and could distinguish active UC (n = 37) from non-IBD (30 healthy individuals and 30 patients with irritable bowel syndrome (IBS)), with the specificity of 92 and 100%, respectively, and sensitivity of 88% (Ahmed Hassan et al., 2020). Among them, miR-21 level is positively correlated with histologically assessed disease severity, which is closely related to the development of inflammation. And miR-21 increases IEC permeability and promotes Th2-type inflammation (Murugaiyan et al., 2015; Wang et al., 2016a). Recent studies have shown that circulating miR-375 is significantly up-regulated in UC patients compared to healthy controls and CD patients, which is a potential biomarker for the diagnosis of UC (Schaefer et al., 2015). Further, it may be a potential early biomarker for CAC. Patel et al. reported the analysis of plasma miR-375 in UC patients (n = 37), UC patients with dysplasia (n = 2) and CAC patients (n = 6) (Patel et al., 2015). They found that plasma miR-375 in CAC patients was significantly up-regulated. Notably, miR-375 has low expression in colon tissues of UC and CRC patients, and participates in the regulation of epidermal growth factor receptor signal pathway by targeting the connective tissue growth factor (Alam et al., 2017). miR-375 affects the growth and invasion of colon cells, and overexpression of it can enhance apoptosis and necrosis (Alam et al., 2017; Garrido-Mesa et al., 2018).
High recurrence rate is still one of the main characteristics of UC, and the evaluation of patients' clinical symptoms has a great impact on treatment decision-making and nursing management. At present, the indicators used to monitor the activity of UC all have their own limitations, which makes the nursing care of patients with UC complicated (Tsilidis et al., 2008; Henderson et al., 2015; Mosli et al., 2015; D'Angelo et al., 2017; Fabian et al., 2019). Recently, miRNA in circulation and feces has become a potential non-invasive biomarker for monitoring disease activity of IBD patients, and it is significantly related to endoscopic activity. For example, miRNA-146a is down-regulated in UC patients, which is negatively correlated with Sutherland Disease Activity Index (DAI) score, clinical activity index and endoscopy index (Feng et al., 2020). The research showed that miRNA-146a could relieve colitis and CRC by targeting TRAF6, PTGES2, and TLR4/MyD88/NF-κB signaling pathways (Wang et al., 2019; Garo et al., 2021). Compared to healthy controls, the expression of serum miR-146b-5p in UC is up-regulated by 2.72 fold and significantly correlates with disease activity (Chen et al., 2019). This may be related to miR-146b promoting colonic mucosal repair. miR-146b can strongly inhibit the activation of M1 macrophages by suppressing the Toll-like receptor 4 (TLR4) signaling pathway, thus inhibiting the induction of pro-inflammatory cytokines. (Deng et al., 2019). In addition, miR-223 increased significantly in feces, tissue and serum samples of UC patients with the active stage (Wang et al., 2016c; Schönauen et al., 2018; Mohammadi et al., 2019; Verdier et al., 2020). And it is significantly up-regulated compared with non-inflammatory areas in colonoscopy biopsy of UC patients (Valmiki et al., 2017). Previous studies have shown that miR-223 helps to alleviate colonic inflammation in UC patients (Zhou et al., 2015; Neudecker et al., 2017). However, the studies of biomarkers show that the expression level of miR-223 is positively correlated with disease activity in UC patients. The sensitivity and specificity of fecal-derived miR-223 in distinguishing active IBD patients (fecal calprotectin >500 μg/g and active disease in colonoscopy, n = 30) from remission IBD patients (fecal calprotectin <100 μg/g and remission in colonoscopy, n = 15) was 80 and 93%, respectively (Schönauen et al., 2018). Wang et al. (2016c) analyzed serum samples from 50 UC patients, 50 CD patients and 50 healthy controls, and found that serum miR-223 level in UC patients was positively correlated with Mayo Endoscopic Score (MES), erythrocyte sedimentation rate (ESR), high-sensitivity C-reactive protein (hs-CRP) and Ulcerative Colitis Endoscopic Index of Severity (UCEIS). And miR-223 (r = 0.481) has a higher Spearman r value than ESR (r = 0.334) when correlated with UCEIS. Likewise, Polytarchou et al. (Polytarchou et al., 2015b) analyzed serum miR-223 levels in 21 healthy controls and 46 UC patients (22 in remission, 10 with moderately active disease and 14 with severe active disease) and found that miR-223 was positively correlated with MES and C-reactive protein (CRP). The Spearman r value of miR-223 (r = 0.44) is higher than that of CRP (r = 0.30) when it is related to MES. But miR-223 is highly expressed in platelets, and improper treatment during serum separation can lead to the introduction of miR-223 into platelets, thus these may affect the experimental results (De Guire et al., 2013).
Currently, some studies have reported the use of miRNA biomarkers for predicting response to treatment, disease recurrence, and the severity of disease over time or something else in UC. Although this area has not been adequately studied, it does represent a valuable direction. Malham et al. found that the expression level of miR-21 was significantly lower in pediatric UC patients than in adult UC patients, and was greater in male patients than in female patients in the adult group (Malham et al., 2021). Batra et al. screened for biomarkers associated with clinical response to anti-TNF-α therapy and glucocorticoid (GC) therapy in pediatric IBD patients and identified five serum miRNAs (miR-146a, miR-146b, miR-320a, miR-126, and let-7c), which expression correlated not only with disease mucosal biopsy but also with treatment response and are potential non-invasive biomarkers for clinical monitoring (Batra et al., 2020). It was found that miR-206 levels were elevated in UC and associated with inhibition of anti-inflammatory A3 adenosine receptor (A3AR) expression, and miR-206 expression was reduced in mesalazine-treated colon cells as well as in colon tissue from patients treated with mesalazine (Minacapelli et al., 2019). A prospective study was conducted by Kalla et al., which found that miR-3615 and miR-4792 in T-cell in the blood contributed to the prognosis of UC patients. When patients meet at least three criteria of the four biomarkers [relative miR-3615 expression <0.95, miR-4792 >2.26, albumin <39 g/dl and extensive colitis (logrank p = 6.93 × 107)], there was a 90% probability that the patient would require treatment escalation within one year (Kalla et al., 2020). Luo et al. reported that six miRNAs were significantly down-regulated in the serum of GCs-resistant UC patients and these could be used to identify GCs-resistant UC patients. In particular, miR-224–5p is a strong predictor with an area under the curve (AUC) = 0.99, specificity = 97.30%, and sensitivity = 89.70% (Luo et al., 2018). Similarly, Morilla et al. used a panel of miRNA biomarkers (has-miR-3934, hsa-miR-100, hsa-miR-718, hsa-miR-193b, hsa-miR-3150a-5p, hsa-miR-1260b, hsa-miR-938, has-miR-518b, and hsa-miR-1468) to distinguish steroid-resistant acute severe ulcerative colitis (ASUC) patients with high accuracy (AUC = 0.87) (Morilla et al., 2019). Jabandziev et al. analyzed miRNA expression in colonic tissue of 60 pediatric UC patients and found four panels of miRNAs, which were used to distinguish pediatric UC patients from healthy controls (let-7i-5p, miR-223–3p and miR-4284, AUC = 0.99), to detect severity in pediatric UC patients (miR-375–3p, miR-146a-5p, miR-223–3p and miR-200b-3p, ACU = 0.73), to detect disease relapse within one year of treatment (miR-21–5p, miR-192–5p and miR-194–5p, AUC = 0.73) and to diagnose UC patients with primary sclerosing cholangitis (miR-142–3p, miR-146a-5p, miR-223–3p, let-7i-5p, miR-192–5p and miR-194–5p, AUC = 0.858) (Jabandziev et al., 2021).
The above studies demonstrate that miRNAs are promising potential biomarkers of UC. The quantitative detection of miRNA has high sensitivity and accuracy, and is related to the activity of UC. However, at present, most studies only detect the changing trend of miRNA expression, but do not quantitatively analyze miRNA. In future research, it is necessary to standardize the sample detection scheme, and detect the abundance of miRNA in UC, so as to determine the accurate expression range of potential miRNA biomarkers. In addition, at present, there are few studies on how the drugs commonly used in UC, the length of duration of the disease, the age of patients, and other factors affect miRNA expression, but this is indeed a promising field.
miRNAs as Biomarkers for the Diagnosis of Colitis-Associated Cancer
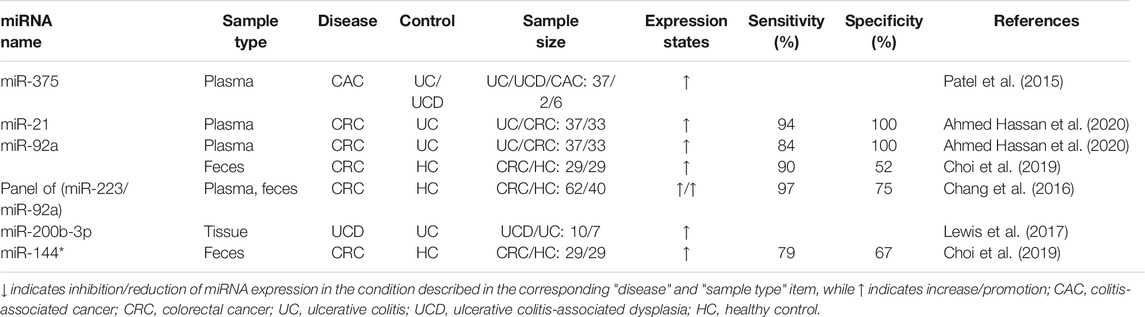
Investigations have shown that patients with long-term UC have an increased risk of tumor complications (Chen et al., 2016), while patients with CAC have a poor prognosis and higher mortality than patients with sporadic CRC (Dugum et al., 2017). In addition, the five-year survival rate for diagnosis at early stages of CRC is 90%, compared to 13% for stage IV (Bray et al., 2018). As important post-transcriptional regulators, some miRNAs are abnormally expressed in the carcinogenesis of UC (Table 4), which can be used for the diagnosis of early canceration.
miR-21, one of the most studied miRNA in UC, is up-regulated in blood (Ahmed Hassan et al., 2020), feces (Schönauen et al., 2018), and colonic tissues (Yan et al., 2020) of UC patients and promotes the development of intestinal inflammation (Murugaiyan et al., 2015; Wang et al., 2016a). miR-21 can activate NF-κB, STAT3 and bcl-2 signaling pathways by targeting PDCD4, thus reducing apoptosis in tumor cells (Figure 3). Compared to the expression in UC patients, miR-21 further increased in CRC (Wu et al., 2017b; Choi et al., 2019; Ahmed Hassan et al., 2020) where it regulated the physiological function of tumor cells (Wu et al., 2017b). Therefore, detection of miR-21 level might be an effective method to assess the risk of carcinogenesis in patients with UC. A meta-analysis confirmed that miR-21 is the most reliable potential fecal-based miRNA biomarker of CRC, with an AUC of 0.843, sensitivity of 59.3% and specificity of 85.6% (Yau et al., 2019). Ahmed Hassan et al. (Ahmed Hassan et al., 2020) measured plasma miR-21 expression in 37 patients with active UC, 33 with CRC, and 30 with IBS as well as 30 healthy controls. The results showed that the sensitivity and specificity of distinguishing UC from IBS and healthy controls were 87.5 and 91.7% (AUC = 0.844). The sensitivity and specificity of miR-21 in distinguishing CRC from UC were 93.5 and 100%, respectively. In addition, miR-26b is up-regulated in the progression of UC to CAC and down-regulated in sporadic CRC, thus distinguishing CAC from UC and sporadic CRC (Benderska et al., 2015). Furthermore, research has shown that miR-26b inhibits tumor cell proliferation and invasion, and induces apoptosis of tumor cells. Compared with paired primary CRC tissues, the expression level of miR-26b was almost 4-fold higher in lung metastases, which may be associated with lung metastasis (Cristóbal et al., 2015).
Detection of miRNA methylation, rather than expression, is another accurate diagnostic method for CAC. Methylation changes the stability and specific recognition of miRNA (Konno et al., 2019), and the methylation level is related to the age and course of UC patients (Toiyama et al., 2017). miRNA methylation in UC patients with dysplasia or CRC increased significantly (Toiyama et al., 2017). Toiyama et al. accurately diagnosed UC patients with dysplasia and CRC by detecting a panel of methylated miRNA (evaluation cohort AUC = 0.81; validation cohort AUC = 0.78) (Toiyama et al., 2017).
Compared with the traditional blood-based biomarkers (CEA or CA 19–9), miRNA expression has been found to significantly different in every stage of CRC and has a higher diagnostic value for early diagnosis, disease monitoring and prognosis of CRC (Inoue et al., 2017; Liu et al., 2018a; El-Daly et al., 2019; Baassiri et al., 2020).
Discussion
Currently, miRNA, as a biomarker of UC, has the advantage of early diagnosis and better correlation with disease activity (Wang et al., 2016c; Schönauen et al., 2018). However, because miRNA disorders involve many diseases, how to ensure miRNA biomarkers in UC are not interfered by other diseases or therapies is a challenge in the future (Kalla et al., 2015). In future studies of miRNA in UC, it is necessary to study the role of miRNA in specific tissues and cell types (Whiteoak et al., 2015). In addition, some immunoglobulins, such as sIgA and IgG, are not only important molecules of the intestinal mucosal immune barrier, but also the main effective antibodies of the humoral immune response, which play an important role in the pathological mechanism of UC. Castro-Dopico et al. reported that IgG drives intestinal inflammation and Th17-related immunity in the mucosal immune hyperactivation caused by genetic variants of UC (Castro-Dopico et al., 2019). sIgA has been reported to bind intestinal microbes and regulate the composition of the intestinal microbiota (Okai et al., 2016). As post-transcriptional regulators, the interaction between miRNAs and immunoglobulins in intestinal immunity is also a field of interest in the future.
At present, miRNA biomarkers in UC are mainly studied for clinical diagnosis and monitoring, however, miRNA biomarkers seem to have other values. Due to the broad regulatory effects of miRNA, it is associated with almost all clinical reactions, such as in predicting disease recurrence (Jabandziev et al., 2021), therapeutic escalation (Kalla et al., 2020), and response to drug therapy (Batra et al., 2020). Although this aspect is currently under-researched, they do represent a valuable direction of research. The current studies show that miR-21 seems to be the most promising potential miRNA biomarker for UC. miR-21 is up-regulated in blood, feces, and colonic tissues of UC patients and further increased in CRC (Wu et al., 2017b; Schönauen et al., 2018; Choi et al., 2019; Ahmed Hassan et al., 2020; Yan et al., 2020), which allows it to differentiate UC from non-IBD (healthy individuals and IBS) and also for the diagnosis of CAC. And the expression of miR-21 was correlated with the age and gender of UC patients (Malham et al., 2021). Moreover, fecal-derived miR-21 has no significant changes in CD, which increases the possibility that miR-21 may be used to distinguish UC from CD (Zhou et al., 2021). miR-223 has a higher correlation with disease activity than ESR and CRP, and is the most promising potential biomarker for monitoring disease activity in UC (Polytarchou et al., 2015b; Wang et al., 2016c). In addition, since most miRNA biomarkers are tested in small sample sizes, they must be verified in large-scale longitudinal studies before being applied to clinical practice. On the other hand, there is no uniform quantitative standard for the detection of miRNA, which is one of the reasons limiting the development of miRNA biomarkers. Especially for complex samples, such as feces, there is a need for more effective and standardized measurement method (Niu et al., 2019). Measuring exosomal-derived miRNAs in blood may reduce the interference of other components, but the technology and cost of isolating exosomes are also challenging issues (Vasconcelos et al., 2019). Detection of fecal exosomal-derived miRNAs may be a better approach. Fecal exosome-derived miRNAs have been proved to be secreted predominantly by IECs and Hopx-positive cells (Liu et al., 2016b), thus allowing a more accurate assessment of disease activity or mucosal healing in UC.
miRNA is regarded as a potential target for the treatment of UC. But one miRNA can regulate hundreds of proteins, then how to reduce the influence on non-target proteins is the main problem of miRNA-based treatment. At present, this problem is mainly solved by improving drug carriers or finding more specific targets. It was found that plant-derived exosomes can be preferentially absorbed by specific intestinal microorganisms (Teng et al., 2018). Gut microbiota dysbiosis contributes to the pathogenesis of UC, and targeted inhibition of specific pathogenic bacteria using exosomal-derived miRNAs may be a novel approach to reduce off-target effects. Of course, local medication may be the most effective method to reduce systemic exposure. For example, Alicaforsen is currently in clinical trial stage, which is an oligonucleotide therapeutic drug, targeting mRNA and then inhibits the generation of protein. A phase IV clinical trial showed that it is equivalent to mesalazine enema (Miner et al., 2004; Miner et al., 2006a; Miner et al., 2006b). On the whole, although there are still some problems to be solved, miRNA is a promising target in the treatment and diagnosis of UC.
Author Contributions
SC and LS contributed to conception and design of the study. JZ are the first authors and responsible for collecting materials and writing the paper. JL, YG and SL helped organizing the information and edited the article pictures. All authors contributed to manuscript revision, read, and approved the submitted version.
Funding
This work was supported by the National Natural Science Foundation of China (No. 81503325) and the Xinglin Scholar Research Promotion Project of Chengdu University of TCM (No. CXTD2018021).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We are very grateful to our alma mater, Chengdu University of Traditional Chinese Medicine, for its convenience in collecting documents. Thanks to all the people in our laboratory for their help.
References
Abdelfattah, A. M., Park, C., and Choi, M. Y. (2014). Update on Non-canonical microRNAs. Biomol. Concepts 5 (4), 275–287. doi:10.1515/bmc-2014-0012
Adler, A., Geiger, S., Keil, A., Bias, H., Schatz, P., deVos, T., et al. (2014). Improving Compliance to Colorectal Cancer Screening Using Blood and Stool Based Tests in Patients Refusing Screening Colonoscopy in Germany. BMC Gastroenterol. 14, 183. doi:10.1186/1471-230X-14-183
Aguilar, C., Mano, M., and Eulalio, A. (2019). MicroRNAs at the Host-Bacteria Interface: Host Defense or Bacterial Offense. Trends Microbiol. 27 (3), 206–218. doi:10.1016/j.tim.2018.10.011
Ahmed Hassan, E., El-Din Abd El-Rehim, A. S., Mohammed Kholef, E. F., and Abd-Elgwad Elsewify, W. (2020). Potential Role of Plasma miR-21 and miR-92a in Distinguishing between Irritable Bowel Syndrome, Ulcerative Colitis, and Colorectal Cancer. Gastroenterol. Hepatol. Bed Bench 13 (2), 147–154.
Al-Haidari, A., Algaber, A., Madhi, R., Syk, I., and Thorlacius, H. (2018). MiR-155-5p Controls colon Cancer Cell Migration via post-transcriptional Regulation of Human Antigen R (HuR). Cancer Lett. 421, 145–151. doi:10.1016/j.canlet.2018.02.026
Alam, K. J., Mo, J. S., Han, S. H., Park, W. C., Kim, H. S., Yun, K. J., et al. (2017). MicroRNA 375 Regulates Proliferation and Migration of colon Cancer Cells by Suppressing the CTGF-EGFR Signaling Pathway. Int. J. Cancer 141 (8), 1614–1629. doi:10.1002/ijc.30861
Alles, J., Fehlmann, T., Fischer, U., Backes, C., Galata, V., Minet, M., et al. (2019). An Estimate of the Total Number of True Human miRNAs. Nucleic Acids Res. 47 (7), 3353–3364. doi:10.1093/nar/gkz097
Ameres, S. L., and Zamore, P. D. (2013). Diversifying microRNA Sequence and Function. Nat. Rev. Mol. Cel Biol. 14 (8), 475–488. doi:10.1038/nrm3611
Ando, Y., Mazzurana, L., Forkel, M., Okazaki, K., Aoi, M., Schmidt, P. T., et al. (2016). Downregulation of MicroRNA-21 in Colonic CD3+ T Cells in UC Remission. Inflamm. Bowel Dis. 22 (12), 2788–2793. doi:10.1097/MIB.0000000000000969
Baassiri, A., Nassar, F., Mukherji, D., Shamseddine, A., Nasr, R., and Temraz, S. (2020). Exosomal Non Coding RNA in LIQUID Biopsies as a Promising Biomarker for Colorectal Cancer. Ijms 21 (4), 1398. doi:10.3390/ijms21041398
Ballard, B. R., and M'Koma, A. E. (2015). Gastrointestinal Endoscopy Biopsy Derived Proteomic Patterns Predict Indeterminate Colitis into Ulcerative Colitis and Crohn's Colitis. World J. Gastrointest. Endosc. 7 (7), 670–674. doi:10.4253/wjge.v7.i7.670
Bartel, D. P. (2004). MicroRNAs: Genomics, Biogenesis, Mechanism, and Function. Cell 116 (2), 281–297. doi:10.1016/s0092-8674(04)00045-5
Batra, S. K., Heier, C. R., Diaz-Calderon, L., Tully, C. B., Fiorillo, A. A., van den Anker, J., et al. (2020). Serum miRNAs Are Pharmacodynamic Biomarkers Associated with Therapeutic Response in Pediatric Inflammatory Bowel Disease. Inflamm. Bowel Dis. 26 (10), 1597–1606. doi:10.1093/ibd/izaa209
Benderska, N., Dittrich, A. L., Knaup, S., Rau, T. T., Neufert, C., Wach, S., et al. (2015). miRNA-26b Overexpression in Ulcerative Colitis-Associated Carcinogenesis. Inflamm. Bowel Dis. 21 (9), 2039–2051. doi:10.1097/MIB.0000000000000453
Bian, Z., Li, L., Cui, J., Zhang, H., Liu, Y., Zhang, C. Y., et al. (2011). Role of miR-150-Targeting C-Myb in Colonic Epithelial Disruption during Dextran Sulphate Sodium-Induced Murine Experimental Colitis and Human Ulcerative Colitis. J. Pathol. 225 (4), 544–553. doi:10.1002/path.2907
Borgaonkar, M. R., Pace, D., Lougheed, M., Marcoux, C., Evans, B., Hickey, N., et al. (2016). Canadian Association of Gastroenterology Indicators of Safety Compromise Following Colonoscopy in Clinical Practice. Can. J. Gastroenterol. Hepatol. 2016, 2729871. doi:10.1155/2016/2729871
Bracken, C. P., Scott, H. S., and Goodall, G. J. (2016). A Network-Biology Perspective of microRNA Function and Dysfunction in Cancer. Nat. Rev. Genet. 17 (12), 719–732. doi:10.1038/nrg.2016.134
Bray, F., Ferlay, J., Soerjomataram, I., Siegel, R. L., Torre, L. A., and Jemal, A. (2018). Global Cancer Statistics 2018: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J. Clin. 68 (6), 394–424. doi:10.3322/caac.21492
Broughton, J. P., and Pasquinelli, A. E. (2016). A Tale of Two Sequences: microRNA-Target Chimeric Reads. Genet. Sel. Evol. 48, 31. doi:10.1186/s12711-016-0209-x
Bruno, M. E., Frantz, A. L., Rogier, E. W., Johansen, F. E., and Kaetzel, C. S. (2011). Regulation of the Polymeric Immunoglobulin Receptor by the Classical and Alternative NF-Κb Pathways in Intestinal Epithelial Cells. Mucosal Immunol. 4 (4), 468–478. doi:10.1038/mi.2011.8
Butin-Israeli, V., Bui, T. M., Wiesolek, H. L., Mascarenhas, L., Lee, J. J., Mehl, L. C., et al. (2019). Neutrophil-induced Genomic Instability Impedes Resolution of Inflammation and Wound Healing. J. Clin. Invest. 129 (2), 712–726. doi:10.1172/JCI122085
Cantini, L., Bertoli, G., Cava, C., Dubois, T., Zinovyev, A., Caselle, M., et al. (2019). Identification of microRNA Clusters Cooperatively Acting on Epithelial to Mesenchymal Transition in Triple Negative Breast Cancer. Nucleic Acids Res. 47 (5), 2205–2215. doi:10.1093/nar/gkz016
Carmona, F. J., Azuara, D., Berenguer-Llergo, A., Fernández, A. F., Biondo, S., de Oca, J., et al. (2013). DNA Methylation Biomarkers for Noninvasive Diagnosis of Colorectal Cancer. Cancer Prev. Res. (Phila) 6 (7), 656–665. doi:10.1158/1940-6207.CAPR-12-0501
Castro-Dopico, T., Dennison, T. W., Ferdinand, J. R., Mathews, R. J., Fleming, A., Clift, D., et al. (2019). Anti-commensal IgG Drives Intestinal Inflammation and Type 17 Immunity in Ulcerative Colitis. Immunity 50 (4), 1099–e10. doi:10.1016/j.immuni.2019.02.006
Chang, P. Y., Chen, C. C., Chang, Y. S., Tsai, W. S., You, J. F., Lin, G. P., et al. (2016). MicroRNA-223 and microRNA-92a in Stool and Plasma Samples Act as Complementary Biomarkers to Increase Colorectal Cancer Detection. Oncotarget 7 (9), 10663–10675. doi:10.18632/oncotarget.7119
Chen, P., Li, Y., Li, L., Yu, Q., Chao, K., Zhou, G., et al. (2019). Circulating microRNA146b-5p Is superior to C-Reactive Protein as a Novel Biomarker for Monitoring Inflammatory Bowel Disease. Aliment. Pharmacol. Ther. 49 (6), 733–743. doi:10.1111/apt.15159
Chen, R., Lai, L. A., Brentnall, T. A., and Pan, S. (2016). Biomarkers for Colitis-Associated Colorectal Cancer. World J. Gastroenterol. 22 (35), 7882–7891. doi:10.3748/wjg.v22.i35.7882
Chen, S., Ben, S., Xin, J., Li, S., Zheng, R., Wang, H., et al. (2021). The Biogenesis and Biological Function of PIWI-Interacting RNA in Cancer. J. Hematol. Oncol. 14 (1), 93. doi:10.1186/s13045-021-01104-3
Choi, H. H., Cho, Y. S., Choi, J. H., Kim, H. K., Kim, S. S., and Chae, H. S. (2019). Stool-based miR-92a and miR-144* as Noninvasive Biomarkers for Colorectal Cancer Screening. Oncology 97 (3), 173–179. doi:10.1159/000500639
Chowdhury, D., Choi, Y. E., and Brault, M. E. (2013). Charity Begins at home: Non-coding RNA Functions in DNA Repair. Nat. Rev. Mol. Cel Biol. 14 (3), 181–189. doi:10.1038/nrm3523
Cristóbal, I., Manso, R., Gónzález-Alonso, P., Madoz-Gúrpide, J., Rojo, F., and García-Foncillas, J. (2015). Clinical Value of miR-26b Discriminating Ulcerative Colitis-Associated Colorectal Cancer in the Subgroup of Patients with Metastatic Disease. Inflamm. Bowel Dis. 21 (10), 1–E25. doi:10.1097/MIB.0000000000000572
D'Angelo, F., Felley, C., and Frossard, J. L. (2017). Calprotectin in Daily Practice: where Do We Stand in 2017?. Digestion 95 (4), 293–301. doi:10.1159/000476062
Dasgupta, I., and Chatterjee, A. (2021). Recent Advances in miRNA Delivery Systems. Methods Protoc. 4 (1), 10. doi:10.3390/mps4010010
De Guire, V., Robitaille, R., Tétreault, N., Guérin, R., Ménard, C., Bambace, N., et al. (2013). Circulating miRNAs as Sensitive and Specific Biomarkers for the Diagnosis and Monitoring of Human Diseases: Promises and Challenges. Clin. Biochem. 46 (10-11), 846–860. doi:10.1016/j.clinbiochem.2013.03.015
Del Pozo-Acebo, L., López de Las Hazas, M. C., Margollés, A., Dávalos, A., and García-Ruiz, A. (2021). Eating microRNAs: Pharmacological Opportunities for Cross-Kingdom Regulation and Implications in Host Gene and Gut Microbiota Modulation. Br. J. Pharmacol. 178 (11), 2218–2245. doi:10.1111/bph.15421
Deng, F., He, S., Cui, S., Shi, Y., Tan, Y., Li, Z., et al. (2019). A Molecular Targeted Immunotherapeutic Strategy for Ulcerative Colitis via Dual-Targeting Nanoparticles Delivering miR-146b to Intestinal Macrophages. J. Crohns. Colitis 13 (4), 482–494. doi:10.1093/ecco-jcc/jjy181
Dong, J., Tai, J. W., and Lu, L. F. (2019). miRNA-Microbiota Interaction in Gut Homeostasis and Colorectal Cancer. Trends Cancer 5 (11), 666–669. doi:10.1016/j.trecan.2019.08.003
Du, L., and Ha, C. (2020). Epidemiology and Pathogenesis of Ulcerative Colitis. Gastroenterol. Clin. North. Am. 49 (4), 643–654. doi:10.1016/j.gtc.2020.07.005
Dugum, M., Lin, J., Lopez, R., Estfan, B., Manilich, E., Stocchi, L., et al. (2017). Recurrence and Survival Rates of Inflammatory Bowel Disease-Associated Colorectal Cancer Following Postoperative Chemotherapy: a Comparative Study. Gastroenterol. Rep. (Oxf) 5 (1), 57–61. doi:10.1093/gastro/gow016
El-Daly, S. M., Morsy, S. M., Medhat, D., El-Bana, M. A., Latif, Y. A., Omara, E. A., et al. (2019). The Diagnostic Efficacy of Circulating miRNAs in Monitoring the Early Development of Colitis-Induced Colorectal Cancer. J. Cel. Biochem. 120 (10), 16668–16680. doi:10.1002/jcb.28925
Fabian, O., Hradsky, O., Lerchova, T., Mikus, F., Zamecnik, J., and Bronsky, J. (2019). Limited Clinical Significance of Tissue Calprotectin Levels in Bowel Mucosa for the Prediction of Complicated Course of the Disease in Children with Ulcerative Colitis. Pathol. Res. Pract. 215 (12), 152689. doi:10.1016/j.prp.2019.152689
Fairfax, K. A., Gantier, M. P., Mackay, F., Williams, B. R., and McCoy, C. E. (2015). IL-10 Regulates Aicda Expression through miR-155. J. Leukoc. Biol. 97 (1), 71–78. doi:10.1189/jlb.2A0314-178R
Fareh, M., Yeom, K. H., Haagsma, A. C., Chauhan, S., Heo, I., and Joo, C. (2016). TRBP Ensures Efficient Dicer Processing of Precursor microRNA in RNA-Crowded Environments. Nat. Commun. 7, 13694. doi:10.1038/ncomms13694
Feng, J., Zhu, Y., Chen, L., and Wang, M. (2020). Clinical Significance of microRNA-146a in Patients with Ulcerative Colitis. Ann. Clin. Lab. Sci. 50 (4), 463–467.
Fredsøe, J., Rasmussen, A. K. I., Mouritzen, P., Borre, M., Ørntoft, T., and Sørensen, K. D. (2019). A Five-microRNA Model (pCaP) for Predicting Prostate Cancer Aggressiveness Using Cell-free Urine. Int. J. Cancer 145 (9), 2558–2567. doi:10.1002/ijc.32296
Friedman, R. C., Farh, K. K.-H., Burge, C. B., and Bartel, D. P. (2009). Most Mammalian mRNAs Are Conserved Targets of microRNAs. Genome Res. 19 (1), 92–105. doi:10.1101/gr.082701.108
Fromm, B., Billipp, T., Peck, L. E., Johansen, M., Tarver, J. E., King, B. L., et al. (2015). A Uniform System for the Annotation of Vertebrate microRNA Genes and the Evolution of the Human microRNAome. Annu. Rev. Genet. 49, 213–242. doi:10.1146/annurev-genet-120213-092023
Galatenko, V. V., Galatenko, A. V., Samatov, T. R., Turchinovich, A. A., Shkurnikov, M. Y., Makarova, J. A., et al. (2018). Comprehensive Network of miRNA-Induced Intergenic Interactions and a Biological Role of its Core in Cancer. Sci. Rep. 8 (1), 2418. doi:10.1038/s41598-018-20215-5
Garo, L. P., Ajay, A. K., Fujiwara, M., Gabriely, G., Raheja, R., Kuhn, C., et al. (2021). MicroRNA-146a Limits Tumorigenic Inflammation in Colorectal Cancer. Nat. Commun. 12 (1), 2419. doi:10.1038/s41467-021-22641-y
Garrido-Mesa, J., Rodríguez-Nogales, A., Algieri, F., Vezza, T., Hidalgo-Garcia, L., Garrido-Barros, M., et al. (2018). Immunomodulatory Tetracyclines Shape the Intestinal Inflammatory Response Inducing Mucosal Healing and Resolution. Br. J. Pharmacol. 175 (23), 4353–4370. doi:10.1111/bph.14494
Gasparini, P., Lovat, F., Fassan, M., Casadei, L., Cascione, L., Jacob, N. K., et al. (2014). Protective Role of miR-155 in Breast Cancer through RAD51 Targeting Impairs Homologous Recombination after Irradiation. Proc. Natl. Acad. Sci. U. S. A. 111 (12), 4536–4541. doi:10.1073/pnas.1402604111
Gebert, L. F. R., and MacRae, I. J. (2019). Regulation of microRNA Function in Animals. Nat. Rev. Mol. Cel Biol. 20 (1), 21–37. doi:10.1038/s41580-018-0045-7
Glinge, C., Clauss, S., Boddum, K., Jabbari, R., Jabbari, J., Risgaard, B., et al. (2017). Stability of Circulating Blood-Based microRNAs - Pre-analytic Methodological Considerations. PLoS One 12 (2), e0167969. doi:10.1371/journal.pone.0167969
Gruszka, R., and Zakrzewska, M. (2018). The Oncogenic Relevance of miR-17-92 Cluster and its Paralogous miR-106b-25 and miR-106a-363 Clusters in Brain Tumors. Ijms 19 (3), 879. doi:10.3390/ijms19030879
Gwiggner, M., Martinez-Nunez, R., Whiteoak, S., Bondanese, V., Claridge, A., Collins, J., et al. (2018). MicroRNA-31 and MicroRNA-155 Are Overexpressed in Ulcerative Colitis and Regulate IL-13 Signaling by Targeting Interleukin 13 Receptor α-1. Genes 9 (2), 85. doi:10.3390/genes9020085
Ha, M., and Kim, V. N. (2014). Regulation of microRNA Biogenesis. Nat. Rev. Mol. Cel Biol. 15 (8), 509–524. doi:10.1038/nrm3838
Hagel, A. F., Boxberger, F., Dauth, W., Kessler, H. P., Neurath, M. F., and Raithel, M. (2012). Colonoscopy-associated Perforation: a 7-year Survey of In-Hospital Frequency, Treatment and Outcome in a German university Hospital. Colorectal Dis. 14 (9), 1121–1125. doi:10.1111/j.1463-1318.2011.02899.x
Han, J., Pedersen, J. S., Kwon, S. C., Belair, C. D., Kim, Y. K., Yeom, K. H., et al. (2009). Posttranscriptional Crossregulation between Drosha and DGCR8. Cell 136 (1), 75–84. doi:10.1016/j.cell.2008.10.053
He, C., Shi, Y., Wu, R., Sun, M., Fang, L., Wu, W., et al. (2016). miR-301a Promotes Intestinal Mucosal Inflammation through Induction of IL-17A and TNF-α in IBD. Gut 65 (12), 1938–1950. doi:10.1136/gutjnl-2015-309389
He, C., Yu, T., Shi, Y., Ma, C., Yang, W., Fang, L., et al. (2017). MicroRNA 301A Promotes Intestinal Inflammation and Colitis-Associated Cancer Development by Inhibiting BTG1. Gastroenterology 152 (6), 1434–e15. doi:10.1053/j.gastro.2017.01.049
Henderson, P., Kennedy, N. A., Van Limbergen, J. E., Cameron, F. L., Satsangi, J., Russell, R. K., et al. (2015). Serum C-Reactive Protein and CRP Genotype in Pediatric Inflammatory Bowel Disease: Influence on Phenotype, Natural History, and Response to Therapy. Inflamm. Bowel Dis. 21 (3), 596–605. doi:10.1097/MIB.0000000000000296
Hindryckx, P., Baert, F., Hart, A., Magro, F., Armuzzi, A., Peyrin-Biroulet, L., et al. (2015). Clinical Trials in Ulcerative Colitis: a Historical Perspective. J. Crohns. Colitis 9 (7), 580–588. doi:10.1093/ecco-jcc/jjv074
Hou, J., Hu, X., Chen, B., Chen, X., Zhao, L., Chen, Z., et al. (2017). miR-155 Targets Est-1 and Induces Ulcerative Colitis via the IL-23/17/6-mediated Th17 Pathway. Pathol. Res. Pract. 213 (10), 1289–1295. doi:10.1016/j.prp.2017.08.001
Hu, B., Wang, X., Hu, S., Ying, X., Wang, P., Zhang, X., et al. (2017). miR-21-mediated Radioresistance Occurs via Promoting Repair of DNA Double Strand Breaks. J. Biol. Chem. 292 (8), 3531–3540. doi:10.1074/jbc.M116.772392
Hu, M., Lu, Y., Zeng, H., Zhang, Z., Chen, S., Qi, Y., et al. (2021). MicroRNA-21 Maintains Hematopoietic Stem Cell Homeostasis through Sustaining the NF-Κb Signaling Pathway in Mice. Haematologica 106 (2), 412–423. doi:10.3324/haematol.2019.236927
Hwang, J., Min, B. H., Jang, J., Kang, S. Y., Bae, H., Jang, S. S., et al. (2018). MicroRNA Expression Profiles in Gastric Carcinogenesis. Sci. Rep. 8 (1), 14393. doi:10.1038/s41598-018-32782-8
Inoue, E., Hata, K., Kimura, H., Yamaguchi, K., Nojima, M., Endo, I., et al. (2017). Altered Expression of microRNAs in Patients with Pouchitis after Restorative Proctocolectomy. Surg. Today 47 (12), 1484–1491. doi:10.1007/s00595-017-1550-6
Jabandziev, P., Kakisaka, T., Bohosova, J., Pinkasova, T., Kunovsky, L., Slaby, O., et al. (2021). MicroRNAs in Colon Tissue of Pediatric Ulcerative Pancolitis Patients Allow Detection and Prognostic Stratification. J. Clin. Med. 10 (6), 1325. doi:10.3390/jcm10061325
Ji, Y., Li, X., Zhu, Y., Li, N., Zhang, N., and Niu, M. (2018). Faecal microRNA as a Biomarker of the Activity and Prognosis of Inflammatory Bowel Diseases. Biochem. Biophys. Res. Commun. 503 (4), 2443–2450. doi:10.1016/j.bbrc.2018.06.174
Jostins, L., Ripke, S., Weersma, R. K., Duerr, R. H., McGovern, D. P., Hui, K. Y., et al. (2012). Host-microbe Interactions Have Shaped the Genetic Architecture of Inflammatory Bowel Disease. Nature 491 (7422), 119–124. doi:10.1038/nature11582
Kalantari, P., Harandi, O. F., Agarwal, S., Rus, F., Kurt-Jones, E. A., Fitzgerald, K. A., et al. (2017). miR-718 Represses Proinflammatory Cytokine Production through Targeting Phosphatase and Tensin Homolog (PTEN). J. Biol. Chem. 292 (14), 5634–5644. doi:10.1074/jbc.M116.749325
Kalla, R., Adams, A. T., Ventham, N. T., Kennedy, N. A., White, R., Clarke, C., et al. (2020). Whole Blood Profiling of T-Cell Derived miRNA Allows the Development of Prognostic Models in Inflammatory Bowel Disease. J. Crohns. Colitis 2020, jjaa134. doi:10.1093/ecco-jcc/jjaa134
Kalla, R., Ventham, N. T., Kennedy, N. A., Quintana, J. F., Nimmo, E. R., Buck, A. H., et al. (2015). MicroRNAs: New Players in IBD. Gut 64 (3), 504–517. doi:10.1136/gutjnl-2014-307891
Kanneganti, T. D. (2017). Inflammatory Bowel Disease and the NLRP3 Inflammasome. N. Engl. J. Med. 377 (7), 694–696. doi:10.1056/NEJMcibr1706536
Kim, B., Jeong, K., and Kim, V. N. (2017). Genome-wide Mapping of DROSHA Cleavage Sites on Primary microRNAs and Noncanonical Substrates. Mol. Cel 66 (2), 258–e5. doi:10.1016/j.molcel.2017.03.013
Kim, Y. K., Kim, B., and Kim, V. N. (2016). Re-evaluation of the Roles of DROSHA, Export in 5, and DICER in microRNA Biogenesis. Proc. Natl. Acad. Sci. U. S. A. 113 (13), E1881–E1889. doi:10.1073/pnas.1602532113
Kobayashi, T., Siegmund, B., Le Berre, C., Wei, S. C., Ferrante, M., Shen, B., et al. (2020). Ulcerative Colitis. Nat. Rev. Dis. Primers 6 (1), 74. doi:10.1038/s41572-020-0205-x
Konno, M., Koseki, J., Asai, A., Yamagata, A., Shimamura, T., Motooka, D., et al. (2019). Distinct Methylation Levels of Mature microRNAs in Gastrointestinal Cancers. Nat. Commun. 10 (1), 3888. doi:10.1038/s41467-019-11826-1
Kwon, S. C., Nguyen, T. A., Choi, Y. G., Jo, M. H., Hohng, S., Kim, V. N., et al. (2016). Structure of Human DROSHA. Cell 164 (1-2), 81–90. doi:10.1016/j.cell.2015.12.019
Lam, J. K., Chow, M. Y., Zhang, Y., and Leung, S. W. (2015). siRNA versus miRNA as Therapeutics for Gene Silencing. Mol. Ther. Nucleic Acids 4 (9), e252. doi:10.1038/mtna.2015.23
Landgraf, P., Rusu, M., Sheridan, R., Sewer, A., Iovino, N., Aravin, A., et al. (2007). A Mammalian microRNA Expression Atlas Based on Small RNA Library Sequencing. Cell 129 (7), 1401–1414. doi:10.1016/j.cell.2007.04.040
Law, I. K., Bakirtzi, K., Polytarchou, C., Oikonomopoulos, A., Hommes, D., Iliopoulos, D., et al. (2015). Neurotensin--regulated miR-133α Is Involved in Proinflammatory Signalling in Human Colonic Epithelial Cells and in Experimental Colitis. Gut 64 (7), 1095–1104. doi:10.1136/gutjnl-2014-307329
Law, I. K., Jensen, D., Bunnett, N. W., and Pothoulakis, C. (2016). Neurotensin-induced miR-133α Expression Regulates Neurotensin Receptor 1 Recycling through its Downstream Target Aftiphilin. Sci. Rep. 6, 22195. doi:10.1038/srep22195
Lee, J., Park, E. J., Yuki, Y., Ahmad, S., Mizuguchi, K., Ishii, K. J., et al. (2015). Profiles of microRNA Networks in Intestinal Epithelial Cells in a Mouse Model of Colitis. Sci. Rep. 5, 18174. doi:10.1038/srep18174
Lee, R. C., Feinbaum, R. L., and Ambros, V. (1993). The C. elegans Heterochronic Gene Lin-4 Encodes Small RNAs with Antisense Complementarity to Lin-14. Cell 75 (5), 843–854. doi:10.1016/0092-8674(93)90529-y
Lewis, A., Felice, C., Kumagai, T., Lai, C., Singh, K., Jeffery, R. R., et al. (2017). The miR-200 Family Is Increased in Dysplastic Lesions in Ulcerative Colitis Patients. PLoS One 12 (3), e0173664. doi:10.1371/journal.pone.0173664
Lewis, B. P., Shih, I. H., Jones-Rhoades, M. W., Bartel, D. P., and Burge, C. B. (2003). Prediction of Mammalian microRNA Targets. Cell 115 (7), 787–798. doi:10.1016/s0092-8674(03)01018-3
Li, J., Zhang, J., Guo, H., Yang, S., Fan, W., Ye, N., et al. (2018). Critical Role of Alternative M2 Skewing in miR-155 Deletion-Mediated protection of Colitis. Front. Immunol. 9, 904. doi:10.3389/fimmu.2018.00904
Li, J. A., Wang, Y. D., Wang, K., Wang, Z. L., Jia, D. Y., Yang, B. Y., et al. (2017). Downregulation of miR-214-3p May Contribute to Pathogenesis of Ulcerative Colitis via Targeting STAT6. Biomed. Res. Int. 2017, 8524972. doi:10.1155/2017/8524972
Li, M., Zhao, J., Cao, M., Liu, R., Chen, G., Li, S., et al. (2020). Mast Cells-Derived MiR-223 Destroys Intestinal Barrier Function by Inhibition of CLDN8 Expression in Intestinal Epithelial Cells. Biol. Res. 53 (1), 12. doi:10.1186/s40659-020-00279-2
Liang, Y., Ridzon, D., Wong, L., and Chen, C. (2007). Characterization of microRNA Expression Profiles in normal Human Tissues. BMC Genomics 8, 166. doi:10.1186/1471-2164-8-166
Liu, H. N., Liu, T. T., Wu, H., Chen, Y. J., Tseng, Y. J., Yao, C., et al. (2018a). Serum microRNA Signatures and Metabolomics Have High Diagnostic Value in Colorectal Cancer Using Two Novel Methods. Cancer Sci. 109 (4), 1185–1194. doi:10.1111/cas.13514
Liu, S., Rezende, R. M., Moreira, T. G., Tankou, S. K., Cox, L. M., Wu, M., et al. (2019a). Oral Administration of miR-30d from Feces of MS Patients Suppresses MS-like Symptoms in Mice by Expanding Akkermansia Muciniphila. Cell Host Microbe 26 (6), 779–e8. doi:10.1016/j.chom.2019.10.008
Liu, S., Zhang, S., Lv, X., Lu, J., Ren, C., Zeng, Z., et al. (2019b). Limonin Ameliorates Ulcerative Colitis by Regulating STAT3/miR-214 Signaling Pathway. Int. Immunopharmacol. 75, 105768. doi:10.1016/j.intimp.2019.105768
Liu, W. H., Kang, S. G., Huang, Z., Wu, C. J., Jin, H. Y., Maine, C. J., et al. (2016a). A miR-155-Peli1-C-Rel Pathway Controls the Generation and Function of T Follicular Helper Cells. J. Exp. Med. 213 (9), 1901–1919. doi:10.1084/jem.20160204
Liu, S., da Cunha, A. P., Rezende, R. M., Cialic, R., Wei, Z., Bry, L., et al. (2016b). The Host Shapes the Gut Microbiota via Fecal microRNA. Cell Host & Microbe 19 (1), 32–43. doi:10.1016/j.chom.2015.12.005
Liu, Y., Dong, Y., Zhu, X., Fan, H., Xu, M., Chen, Q., et al. (2018b). miR-155 Inhibition Ameliorates 2, 4, 6-Trinitrobenzenesulfonic Acid (TNBS)-induced Experimental Colitis in Rat via Influencing the Differentiation of Th17 Cells by Jarid2. Int. Immunopharmacol. 64, 401–410. doi:10.1016/j.intimp.2018.09.007
Liu, Y. N., Yin, J. J., Abou-Kheir, W., Hynes, P. G., Casey, O. M., Fang, L., et al. (2013). miR-1 and miR-200 Inhibit EMT via Slug-dependent and Tumorigenesis via Slug-independent Mechanisms. Oncogene 32 (3), 296–306. doi:10.1038/onc.2012.58
Luo, J., Wang, Y., Lan, D., Niu, J., Miao, J., Dong, X., et al. (2018). Differential Expression of Serum microRNAs in Glucocorticoid-Resistant Patients with Ulcerative Colitis. Int. J. Clin. Exp. Pathol. 11 (2), 936–946.
Ma, Y., Zhang, P., Wang, F., Zhang, H., Yang, J., Peng, J., et al. (2012). miR-150 as a Potential Biomarker Associated with Prognosis and Therapeutic Outcome in Colorectal Cancer. Gut 61 (10), 1447–1453. doi:10.1136/gutjnl-2011-301122
Macartney-Coxson, D., Danielson, K., Clapham, J., Benton, M. C., Johnston, A., Jones, A., et al. (2020). MicroRNA Profiling in Adipose before and after Weight Loss Highlights the Role of miR-223-3p and the NLRP3 Inflammasome. Obesity (Silver Spring) 28 (3), 570–580. doi:10.1002/oby.22722
Magri, G., Comerma, L., Pybus, M., Sintes, J., Lligé, D., Segura-Garzón, D., et al. (2017). Human Secretory IgM Emerges from Plasma Cells Clonally Related to Gut Memory B Cells and Targets Highly Diverse Commensals. Immunity 47 (1), 118–e8. doi:10.1016/j.immuni.2017.06.013
Malham, M., James, J. P., Jakobsen, C., Hoegdall, E., Holmstroem, K., Wewer, V., et al. (2021). Mucosal microRNAs Relate to Age and Severity of Disease in Ulcerative Colitis. Aging (Albany NY) 13 (5), 6359–6374. doi:10.18632/aging.202715
Martínez, C., Rodiño-Janeiro, B. K., Lobo, B., Stanifer, M. L., Klaus, B., Granzow, M., et al. (2017). miR-16 and miR-125b Are Involved in Barrier Function Dysregulation through the Modulation of Claudin-2 and Cingulin Expression in the Jejunum in IBS with Diarrhoea. Gut 66 (9), 1–1538. doi:10.1136/gutjnl-2016-311477
Martinez-Nunez, R. T., Louafi, F., and Sanchez-Elsner, T. (2011). The Interleukin 13 (IL-13) Pathway in Human Macrophages Is Modulated by microRNA-155 via Direct Targeting of Interleukin 13 Receptor Alpha1 (IL13Ralpha1). J. Biol. Chem. 286 (3), 1786–1794. doi:10.1074/jbc.M110.169367
Matsui, M., Chu, Y., Zhang, H., Gagnon, K. T., Shaikh, S., Kuchimanchi, S., et al. (2013). Promoter RNA Links Transcriptional Regulation of Inflammatory Pathway Genes. Nucleic Acids Res. 41 (22), 10086–10109. doi:10.1093/nar/gkt777
Matsuyama, H., and Suzuki, H. I. (2019). Systems and Synthetic microRNA Biology: From Biogenesis to Disease Pathogenesis. Ijms 21 (1), 132. doi:10.3390/ijms21010132
Mead, P. S., Slutsker, L., Dietz, V., McCaig, L. F., Bresee, J. S., Shapiro, C., et al. (1999). Food-related Illness and Death in the United States. Emerg. Infect. Dis. 5 (5), 607–625. doi:10.3201/eid0505.990502
Meijer, H. A., Smith, E. M., and Bushell, M. (2014). Regulation of miRNA Strand Selection: Follow the Leader?. Biochem. Soc. Trans. 42 (4), 1135–1140. doi:10.1042/BST20140142
Minacapelli, C. D., Bajpai, M., Geng, X., Van Gurp, J., Poplin, E., Amenta, P. S., et al. (2019). miR-206 as a Biomarker for Response to Mesalamine Treatment in Ulcerative Colitis. Inflamm. Bowel Dis. 25 (1), 78–84. doi:10.1093/ibd/izy279
Miner, P. B., Geary, R. S., Matson, J., Chuang, E., Xia, S., Baker, B. F., et al. (2006a). Bioavailability and Therapeutic Activity of Alicaforsen (ISIS 2302) Administered as a Rectal Retention Enema to Subjects with Active Ulcerative Colitis. Aliment. Pharmacol. Ther. 23 (10), 1427–1434. doi:10.1111/j.1365-2036.2006.02909.x
Miner, P. B., Wedel, M. K., Xia, S., and Baker, B. F. (2006b). Safety and Efficacy of Two Dose Formulations of Alicaforsen Enema Compared with Mesalazine Enema for Treatment of Mild to Moderate Left-Sided Ulcerative Colitis: a Randomized, Double-Blind, Active-Controlled Trial. Aliment. Pharmacol. Ther. 23 (10), 1403–1413. doi:10.1111/j.1365-2036.2006.02837.x
Miner, P., Wedel, M., Bane, B., and Bradley, J. (2004). An Enema Formulation of Alicaforsen, an Antisense Inhibitor of Intercellular Adhesion Molecule-1, in the Treatment of Chronic, Unremitting Pouchitis. Aliment. Pharmacol. Ther. 19 (3), 281–286. doi:10.1111/j.1365-2036.2004.01863.x
Mohammadi, A., Kelly, O. B., Smith, M. I., Kabakchiev, B., and Silverberg, M. S. (2019). Differential miRNA Expression in Ileal and Colonic Tissues Reveals an Altered Immunoregulatory Molecular Profile in Individuals with Crohn's Disease versus Healthy Subjects. J. Crohns. Colitis 13 (11), 1459–1469. doi:10.1093/ecco-jcc/jjz076
Moon, C., Baldridge, M. T., Wallace, M. A., Burnham, C-A.D., Virgin, H. W., Stappenbeck, T. S., et al. (2015). Vertically Transmitted Faecal IgA Levels Determine Extra-chromosomal Phenotypic Variation. Nature 521(7550), 90–93. doi:10.1038/nature14139
Morilla, I., Uzzan, M., Laharie, D., Cazals-Hatem, D., Denost, Q., Daniel, F., et al. (2019). Colonic microRNA Profiles, Identified by a Deep Learning Algorithm, that Predict Responses to Therapy of Patients with Acute Severe Ulcerative Colitis. Clin. Gastroenterol. Hepatol. 17 (5), 905–913. doi:10.1016/j.cgh.2018.08.068
Mosli, M. H., Zou, G., Garg, S. K., Feagan, S. G., MacDonald, J. K., Chande, N., et al. (2015). C-reactive Protein, Fecal Calprotectin, and Stool Lactoferrin for Detection of Endoscopic Activity in Symptomatic Inflammatory Bowel Disease Patients: a Systematic Review and Meta-Analysis. Am. J. Gastroenterol. 110 (6), 802–820. doi:10.1038/ajg.2015.120
Murugaiyan, G., Garo, L. P., and Weiner, H. L. (2015). MicroRNA-21, T Helper Lineage and Autoimmunity. Oncotarget 6 (12), 9644–9645. doi:10.18632/oncotarget.3928
Netz, U., Carter, J., Eichenberger, M. R., Feagins, K., Galbraith, N. J., Dryden, G. W., et al. (2017). Plasma microRNA Profile Differentiates Crohn's Colitis from Ulcerative Colitis. Inflamm. Bowel Dis. 24 (1), 159–165. doi:10.1093/ibd/izx009
Neudecker, V., Haneklaus, M., Jensen, O., Khailova, L., Masterson, J. C., Tye, H., et al. (2017). Myeloid-derived miR-223 Regulates Intestinal Inflammation via Repression of the NLRP3 Inflammasome. J. Exp. Med. 214 (6), 1737–1752. doi:10.1084/jem.20160462
Ng, S. C., Shi, H. Y., Hamidi, N., Underwood, F. E., Tang, W., Benchimol, E. I., et al. (2017). Worldwide Incidence and Prevalence of Inflammatory Bowel Disease in the 21st century: a Systematic Review of Population-Based Studies. Lancet 390 (10114), 2769–2778. doi:10.1016/S0140-6736(17)32448-0
Nguyen, T. A., Jo, M. H., Choi, Y. G., Park, J., Kwon, S. C., Hohng, S., et al. (2015). Functional Anatomy of the Human Microprocessor. Cell 161 (6), 1374–1387. doi:10.1016/j.cell.2015.05.010
Niikura, R., Yasunaga, H., Yamada, A., Matsui, H., Fushimi, K., Hirata, Y., et al. (2016). Factors Predicting Adverse Events Associated with Therapeutic Colonoscopy for Colorectal Neoplasia: a Retrospective Nationwide Study in Japan. Gastrointest. Endosc. 84 (6), 971–982.e6. doi:10.1016/j.gie.2016.05.013
Niu, Y., Xia, S., Su, M., Dang, Q., Kang, K., Li, L., et al. (2019). Direct S-Poly(T) Plus Assay in Quantification of microRNAs without RNA Extraction and its Implications in Colorectal Cancer Biomarker Studies. J. Transl. Med. 17 (1), 316. doi:10.1186/s12967-019-2061-6
Okai, S., Usui, F., Yokota, S., Hori-I, Y., Hasegawa, M., Nakamura, T., et al. (2016). High-affinity Monoclonal IgA Regulates Gut Microbiota and Prevents Colitis in Mice. Nat. Microbiol. 1 (9), 16103. doi:10.1038/nmicrobiol.2016.103
Pabst, O. (2012). New Concepts in the Generation and Functions of IgA. Nat. Rev. Immunol. 12 (12), 821–832. doi:10.1038/nri3322
Pallante, P., Battista, S., Pierantoni, G. M., and Fusco, A. (2014). Deregulation of microRNA Expression in Thyroid Neoplasias. Nat. Rev. Endocrinol. 10 (2), 88–101. doi:10.1038/nrendo.2013.223
Patel, M., Verma, A., Aslam, I., Pringle, H., and Singh, B. (2015). Novel Plasma microRNA Biomarkers for the Identification of Colitis-Associated Carcinoma. Lancet 385 (Suppl. 1), S78. doi:10.1016/S0140-6736(15)60393-2
Pekow, J., Meckel, K., Dougherty, U., Huang, Y., Chen, X., Almoghrabi, A., et al. (2017). miR-193a-3p Is a Key Tumor Suppressor in Ulcerative Colitis-Associated colon Cancer and Promotes Carcinogenesis through Upregulation of Il17rd. Clin. Cancer Res. 23 (17), 5281–5291. doi:10.1158/1078-0432.CCR-17-0171
Perconti, G., Rubino, P., Contino, F., Bivona, S., Bertolazzi, G., Tumminello, M., et al. (2019). RIP-chip Analysis Supports Different Roles for AGO2 and GW182 Proteins in Recruiting and Processing microRNA Targets. BMC Bioinformatics 20 (Suppl. 4), 120. doi:10.1186/s12859-019-2683-y
Perdigão-Henriques, R., Petrocca, F., Altschuler, G., Thomas, M. P., Le, M. T., Tan, S. M., et al. (2016). miR-200 Promotes the Mesenchymal to Epithelial Transition by Suppressing Multiple Members of the Zeb2 and Snail1 Transcriptional Repressor Complexes. Oncogene 35 (2), 158–172. doi:10.1038/onc.2015.69
Polytarchou, C., Hommes, D. W., Palumbo, T., Hatziapostolou, M., Koutsioumpa, M., Koukos, G., et al. (2015a). MicroRNA214 Is Associated with Progression of Ulcerative Colitis, and Inhibition Reduces Development of Colitis and Colitis-Associated Cancer in Mice. Gastroenterology 149 (4), 981–992.e11. doi:10.1053/j.gastro.2015.05.057
Polytarchou, C., Oikonomopoulos, A., Mahurkar, S., Touroutoglou, A., Koukos, G., Hommes, D. W., et al. (2015b). Assessment of Circulating microRNAs for the Diagnosis and Disease Activity Evaluation in Patients with Ulcerative Colitis by Using the NanoString Technology. Inflamm. Bowel Dis. 21 (11), 2533–2539. doi:10.1097/MIB.0000000000000547
Qu, Y. L., Wang, H. F., Sun, Z. Q., Tang, Y., Han, X. N., Yu, X. B., et al. (2015). Up-regulated miR-155-5p Promotes Cell Proliferation, Invasion and Metastasis in Colorectal Carcinoma. Int. J. Clin. Exp. Pathol. 8 (6), 6988–6994.
Quick-Cleveland, J., Jacob, J. P., Weitz, S. H., Shoffner, G., Senturia, R., and Guo, F. (2014). The DGCR8 RNA-Binding Heme Domain Recognizes Primary microRNAs by Clamping the Hairpin. Cell Rep 7 (6), 1994–2005. doi:10.1016/j.celrep.2014.05.013
Raso, A., Dirkx, E., Philippen, L. E., Fernandez-Celis, A., De Majo, F., Sampaio-Pinto, V., et al. (2019). Therapeutic Delivery of Mir-148a Suppresses Ventricular Dilation in Heart Failure. Mol. Ther. 27 (3), 584–599. doi:10.1016/j.ymthe.2018.11.011
Rawat, M., Kadian, K., Gupta, Y., Kumar, A., Chain, P., Kovbasnjuk, O., et al. (2019). MicroRNA in Pancreatic Cancer: from Biology to Therapeutic Potential. Genes 10 (10), 752. doi:10.3390/genes10100752
Riahi Rad, Z., Riahi Rad, Z., Goudarzi, H., Goudarzi, M., Mahmoudi, M., Yasbolaghi Sharahi, J., et al. (2021). MicroRNAs in the Interaction between Host-Bacterial Pathogens: A New Perspective. J. Cel. Physiol. 236 (9), 6249–6270. In press. doi:10.1002/jcp.30333
Rubin, D. T., Ananthakrishnan, A. N., Siegel, C. A., Sauer, B. G., and Long, M. D. (2019). ACG Clinical Guideline: Ulcerative Colitis in Adults. Am. J. Gastroenterol. 114 (3), 384–413. doi:10.14309/ajg.0000000000000152
Sanchez, H. N., Moroney, J. B., Gan, H., Shen, T., Im, J. L., Li, T., et al. (2020). B Cell-Intrinsic Epigenetic Modulation of Antibody Responses by Dietary Fiber-Derived Short-Chain Fatty Acids. Nat. Commun. 11 (1), 60. doi:10.1038/s41467-019-13603-6
Schaefer, J. S., Attumi, T., Opekun, A. R., Abraham, B., Hou, J., Shelby, H., et al. (2015). MicroRNA Signatures Differentiate Crohn's Disease from Ulcerative Colitis. BMC Immunol. 16, 5. doi:10.1186/s12865-015-0069-0
Schönauen, K., Le, N., von Arnim, U., Schulz, C., Malfertheiner, P., and Link, A. (2018). Circulating and Fecal microRNAs as Biomarkers for Inflammatory Bowel Diseases. Inflamm. Bowel Dis. 24 (7), 1547–1557. doi:10.1093/ibd/izy046
Senapati, D., Patra, B. C., Kar, A., Chini, D. S., Ghosh, S., Patra, S., et al. (2019). Promising Approaches of Small Interfering RNAs (siRNAs) Mediated Cancer Gene Therapy. Gene 719, 144071. doi:10.1016/j.gene.2019.144071
Seth, P., Hsieh, P. N., Jamal, S., Wang, L., Gygi, S. P., Jain, M. K., et al. (2019). Regulation of microRNA Machinery and Development by Interspecies S-Nitrosylation. Cell 176 (5), 1014–1025.e12. doi:10.1016/j.cell.2019.01.037
Shi, C., Yang, Y., Xia, Y., Okugawa, Y., Yang, J., Liang, Y., et al. (2016). Novel Evidence for an Oncogenic Role of microRNA-21 in Colitis-Associated Colorectal Cancer. Gut 65 (9), 1470–1481. doi:10.1136/gutjnl-2014-308455
Shi, X., Ye, L., Xu, S., Guo, G., Zuo, Z., Ye, M., et al. (2020). Downregulated miR-29a Promotes B Cell Overactivation by Upregulating Crk-like Protein in Systemic Lupus Erythematosus. Mol. Med. Rep. 22 (2), 841–849. doi:10.3892/mmr.2020.11166
Slack, F. J., and Chinnaiyan, A. M. (2019). The Role of Non-coding RNAs in Oncology. Cell 179 (5), 1033–1055. doi:10.1016/j.cell.2019.10.017
Soubières, A. A., and Poullis, A. (2016). Emerging Biomarkers for the Diagnosis and Monitoring of Inflammatory Bowel Diseases. Inflamm. Bowel Dis. 22 (8), 2016–2022. doi:10.1097/MIB.0000000000000836
Spinler, J. K., Karri, V., and Hirschi, K. D. (2019). Planting the Microbiome. Trends Microbiol. 27 (2), 90–93. doi:10.1016/j.tim.2018.12.001
Stavast, C. J., and Erkeland, S. J. (2019). The Non-canonical Aspects of MicroRNAs: Many Roads to Gene Regulation. Cells 8 (11), 1465. doi:10.3390/cells8111465
Sun, D., Wang, C., Long, S., Ma, Y., Guo, Y., Huang, Z., et al. (2015). C/EBP-β-activated microRNA-223 Promotes Tumour Growth through Targeting RASA1 in Human Colorectal Cancer. Br. J. Cancer 112 (9), 1491–1500. doi:10.1038/bjc.2015.107
Sun, L. H., Tian, D., Yang, Z. C., and Li, J. L. (2020). Exosomal miR-21 Promotes Proliferation, Invasion and Therapy Resistance of colon Adenocarcinoma Cells through its Target PDCD4. Sci. Rep. 10 (1), 8271. doi:10.1038/s41598-020-65207-6
Takahashi, T., Nakano, Y., Onomoto, K., Murakami, F., Komori, C., Suzuki, Y., et al. (2018). LGP2 Virus Sensor Regulates Gene Expression Network Mediated by TRBP-Bound microRNAs. Nucleic Acids Res. 46 (17), 9134–9147. doi:10.1093/nar/gky575
Tang, B., Li, X., Ren, Y., Wang, J., Xu, D., Hang, Y., et al. (2017). MicroRNA-29a Regulates Lipopolysaccharide (LPS)-induced Inflammatory Responses in Murine Macrophages through the Akt1/NF-Κb Pathway. Exp. Cel Res. 360 (2), 74–80. doi:10.1016/j.yexcr.2017.08.013
Tang, R., Li, L., Zhu, D., Hou, D., Cao, T., Gu, H., et al. (2012). Mouse miRNA-709 Directly Regulates miRNA-15a/16-1 Biogenesis at the Posttranscriptional Level in the Nucleus: Evidence for a microRNA Hierarchy System. Cell Res 22 (3), 504–515. doi:10.1038/cr.2011.137
Teng, Y., Ren, Y., Sayed, M., Hu, X., Lei, C., Kumar, A., et al. (2018). Plant-derived Exosomal Micrornas Shape the Gut Microbiota. Cell Host Microbe 24 (5), 637–652.e8. doi:10.1016/j.chom.2018.10.001
Toiyama, Y., Okugawa, Y., Tanaka, K., Araki, T., Uchida, K., Hishida, A., et al. (2017). A Panel of Methylated microRNA Biomarkers for Identifying High-Risk Patients with Ulcerative Colitis-Associated Colorectal Cancer. Gastroenterology 153 (6), 1634–1646.e8. doi:10.1053/j.gastro.2017.08.037
Tsilidis, K. K., Branchini, C., Guallar, E., Helzlsouer, K. J., Erlinger, T. P., and Platz, E. A. (2008). C-reactive Protein and Colorectal Cancer Risk: a Systematic Review of Prospective Studies. Int. J. Cancer 123 (5), 1133–1140. doi:10.1002/ijc.23606
Ungaro, R., Mehandru, S., Allen, P. B., Peyrin-Biroulet, L., and Colombel, J. F. (2017). Ulcerative Colitis. Lancet 389 (10080), 1756–1770. doi:10.1016/S0140-6736(16)32126-2
Valmiki, S., Ahuja, V., and Paul, J. (2017). MicroRNA Exhibit Altered Expression in the Inflamed Colonic Mucosa of Ulcerative Colitis Patients. World J. Gastroenterol. 23 (29), 5324–5332. doi:10.3748/wjg.v23.i29.5324
Van der Goten, J., Vanhove, W., Lemaire, K., Van Lommel, L., Machiels, K., Wollants, W. J., et al. (2014). Integrated miRNA and mRNA Expression Profiling in Inflamed colon of Patients with Ulcerative Colitis. PLoS One 9 (12), e116117. doi:10.1371/journal.pone.0116117
Vasconcelos, M. H., Caires, H. R., Ābols, A., Xavier, C., and Linē, A. (2019). Extracellular Vesicles as a Novel Source of Biomarkers in Liquid Biopsies for Monitoring Cancer Progression and Drug Resistance. Drug Resist. Updat. 47, 100647. doi:10.1016/j.drup.2019.100647
Vatandoost, N., Ghanbari, J., Mojaver, M., Avan, A., Ghayour-Mobarhan, M., Nedaeinia, R., et al. (2016). Early Detection of Colorectal Cancer: from Conventional Methods to Novel Biomarkers. J. Cancer Res. Clin. Oncol. 142 (2), 341–351. doi:10.1007/s00432-015-1928-z
Verdier, J., Breunig, I. R., Ohse, M. C., Roubrocks, S., Kleinfeld, S., Roy, S., et al. (2020). Faecal Micro-RNAs in Inflammatory Bowel Diseases. J. Crohns. Colitis 14 (1), 110–117. doi:10.1093/ecco-jcc/jjz120
Wang, S., Wan, X., and Ruan, Q. (2016a). The microRNA-21 in Autoimmune Diseases. Int. J. Mol. Sci. 17 (6), 864. doi:10.3390/ijms17060864
Wang, H., Chao, K., Ng, S. C., Bai, A. H., Yu, Q., Yu, J., et al. (2016b). Pro-inflammatory miR-223 Mediates the Cross-Talk between the IL23 Pathway and the Intestinal Barrier in Inflammatory Bowel Disease. Genome Biol. 17, 58. doi:10.1186/s13059-016-0901-8
Wang, J. P., Dong, L. N., Wang, M., Guo, J., and Zhao, Y. Q. (2019). MiR-146a Regulates the Development of Ulcerative Colitis via Mediating the TLR4/MyD88/NF-Κb Signaling Pathway. Eur. Rev. Med. Pharmacol. Sci. 23 (5), 2151–2157. doi:10.26355/eurrev_201903_17260
Wang, H., Zhang, S., Yu, Q., Yang, G., Guo, J., Li, M., et al. (2016c). Circulating microRNA223 Is a New Biomarker for Inflammatory Bowel Disease. Medicine 95 (5), e2703. doi:10.1097/MD.0000000000002703
Wei, M., Gao, X., Liu, L., Li, Z., Wan, Z., Dong, Y., et al. (2020). Visceral Adipose Tissue Derived Exosomes Exacerbate Colitis Severity via Pro-inflammatory miRNAs in High Fat Diet Fed Mice. ACS Nano 14 (4), 5099–5110. doi:10.1021/acsnano.0c01860
Whiteoak, S. R., Felwick, R., Sanchez-Elsner, T., and Fraser Cummings, J. R. (2015). MicroRNAs in Inflammatory Bowel Diseases: Paradoxes and Possibilities. Inflamm. Bowel Dis. 21 (5), 1160–1165. doi:10.1097/MIB.0000000000000288
Wu, W., He, Y., Feng, X., Ye, S., Wang, H., Tan, W., et al. (2017a). MicroRNA-206 Is Involved in the Pathogenesis of Ulcerative Colitis via Regulation of Adenosine A3 Receptor. Oncotarget 8 (1), 705–721. doi:10.18632/oncotarget.13525
Wu, X. D., Zeng, K., Liu, W. L., Gao, Y. G., Gong, C. S., Zhang, C. X., et al. (2014). Effect of Aerobic Exercise on miRNA-TLR4 Signaling in Atherosclerosis. Int. J. Sports Med. 35 (4), 344–350. doi:10.1055/s-0033-1349075
Wu, X., Pan, S., Luo, W., Shen, Z., Meng, X., Xiao, M., et al. (2020). Roseburia Intestinalis-Derived Flagellin Ameliorates Colitis by Targeting miR-223-3p-Mediated Activation of NLRP3 Inflammasome and Pyroptosis. Mol. Med. Rep. 22 (4), 2695–2704. doi:10.3892/mmr.2020.11351
Wu, Y., Schutt, S., Paz, K., Zhang, M., Flynn, R. P., Bastian, D., et al. (2018). MicroRNA-17-92 Is Required for T-Cell and B-Cell Pathogenicity in Chronic Graft-Versus-Host Disease in Mice. Blood 131 (17), 1974–1986. doi:10.1182/blood-2017-06-789321
Wu, Y., Song, Y., Xiong, Y., Wang, X., Xu, K., Han, B., et al. (2017b). MicroRNA-21 (Mir-21) Promotes Cell Growth and Invasion by Repressing Tumor Suppressor PTEN in Colorectal Cancer. Cell. Physiol. Biochem. 43 (3), 945–958. doi:10.1159/000481648
Xiao, T., Ling, M., Xu, H., Luo, F., Xue, J., Chen, C., et al. (2019). NF-κB-regulation of miR-155, via SOCS1/STAT3, Is Involved in the PM2.5-accelerated Cell Cycle and Proliferation of Human Bronchial Epithelial Cells. Toxicol. Appl. Pharmacol. 377, 114616. doi:10.1016/j.taap.2019.114616
Xu, M., Zuo, D., Liu, X., Fan, H., Chen, Q., Deng, S., et al. (2017). MiR-155 Contributes to Th17 Cells Differentiation in Dextran Sulfate Sodium (DSS)-induced Colitis Mice via Jarid2. Biochem. Biophys. Res. Commun. 488 (1), 6–14. doi:10.1016/j.bbrc.2017.04.143
Xu, S., Ou, X., Huo, J., Lim, K., Huang, Y., Chee, S., et al. (2015). Mir-17-92 Regulates Bone Marrow Homing of Plasma Cells and Production of Immunoglobulin G2c. Nat. Commun. 6, 6764. doi:10.1038/ncomms7764
Xu, X. M., and Zhang, H. J. (2016). miRNAs as New Molecular Insights into Inflammatory Bowel Disease: Crucial Regulators in Autoimmunity and Inflammation. World J. Gastroenterol. 22 (7), 2206–2218. doi:10.3748/wjg.v22.i7.2206
Yamazawa, R., Jiko, C., Choi, S., Park, I. Y., Nakagawa, A., Yamashita, E., et al. (2018). Structural Basis for Selective Binding of export Cargoes by Exportin-5. Structure 26 (10), 1393–1398.e2. doi:10.1016/j.str.2018.06.014
Yan, H., Zhang, X., and Xu, Y. (2020). Aberrant Expression of miR-21 in Patients with Inflammatory Bowel Disease: A Protocol for Systematic Review and Meta Analysis. Medicine 99 (17), e19693. doi:10.1097/MD.0000000000019693
Yang, F., Ning, Z., Ma, L., Liu, W., Shao, C., Shu, Y., et al. (2017). Exosomal miRNAs and miRNA Dysregulation in Cancer-Associated Fibroblasts. Mol. Cancer 16 (1), 148. doi:10.1186/s12943-017-0718-4
Yang, H. Y., Barbi, J., Wu, C. Y., Zheng, Y., Vignali, P. D., Wu, X., et al. (2016). MicroRNA-17 Modulates Regulatory T Cell Function by Targeting Co-regulators of the Foxp3 Transcription Factor. Immunity 45 (1), 83–93. doi:10.1016/j.immuni.2016.06.022
Yau, T. O., Tang, C. M., Harriss, E. K., Dickins, B., and Polytarchou, C. (2019). Faecal microRNAs as a Non-invasive Tool in the Diagnosis of Colonic Adenomas and Colorectal Cancer: A Meta-Analysis. Sci. Rep. 9 (1), 9491. doi:10.1038/s41598-019-45570-9
Yu, H. M., Wang, C., Yuan, Z., Chen, G. L., Ye, T., and Yang, B. W. (2019). LncRNA NEAT1 Promotes the Tumorigenesis of Colorectal Cancer by Sponging miR-193a-3p. Cell Prolif 52 (1), e12526. doi:10.1111/cpr.12526
Yu, T., Guo, F., Yu, Y., Sun, T., Ma, D., Han, J., et al. (2017). Fusobacterium Nucleatum Promotes Chemoresistance to Colorectal Cancer by Modulating Autophagy. Cell 170 (3), 548–563.e16. doi:10.1016/j.cell.2017.07.008
Zamanillo, R., Sánchez, J., Serra, F., and Palou, A. (2019). Breast Milk Supply of microRNA Associated with Leptin and Adiponectin Is Affected by Maternal Overweight/obesity and Influences Infancy BMI. Nutrients 11 (11), 2589. doi:10.3390/nu11112589
Zeng, B., Wang, H., Luo, J., Xie, M., Zhao, Z., Chen, X., et al. (2021). Porcine Milk-Derived Small Extracellular Vesicles Promote Intestinal Immunoglobulin Production through pIgR. Animals 11 (6), 1522. doi:10.3390/ani11061522
Zhang, L., Shen, J., Cheng, J., and Fan, X. (2015). MicroRNA-21 Regulates Intestinal Epithelial Tight junction Permeability. Cell Biochem. Funct. 33 (4), 235–240. doi:10.1002/cbf.3109
Zhang, X. F., Tu, R., Li, K., Ye, P., and Cui, X. (2017a). Tumor Suppressor PTPRJ Is a Target of miR-155 in Colorectal Cancer. J. Cel. Biochem. 118 (10), 3391–3400. doi:10.1002/jcb.25995
Zhang, Q., and Zhang, S. (2017b). miR-214 Promotes Radioresistance in Human Ovarian Cancer Cells by Targeting PETN. Biosci. Rep. 37 (4), BSR20170327. doi:10.1042/BSR20170327
Zhang, X. Y., Guan, S., Zhang, H. F., Li, R. Y., and Liu, Z. M. (2018). Activation of PD-1 Protects Intestinal Immune Defense through IL-10/miR-155 Pathway after Intestinal Ischemia Reperfusion. Dig. Dis. Sci. 63 (12), 3307–3316. doi:10.1007/s10620-018-5282-2
Zhao, Y., Zeng, Y., Zeng, D., Wang, H., Zhou, M., Sun, N., et al. (2021). Probiotics and microRNA: Their Roles in the Host-Microbe Interactions. Front. Microbiol. 11, 604462. doi:10.3389/fmicb.2020.604462
Zhou, H., Xiao, J., Wu, N., Liu, C., Xu, J., Liu, F., et al. (2015). MicroRNA-223 Regulates the Differentiation and Function of Intestinal Dendritic Cells and Macrophages by Targeting C/EBPβ. Cel Rep 13 (6), 1149–1160. doi:10.1016/j.celrep.2015.09.073
Zhou, J., Liang, A., Hong, J., Sun, J., Lin, X., Peng, Y., et al. (2019). MicroRNA-155 Suppresses the Catabolic Effect Induced by TNF-α and IL-1β by Targeting C/EBPβ in Rat Nucleus Pulposus Cells. Connect. Tissue Res. 60 (2), 165–177. doi:10.1080/03008207.2018.1483356
Zhou, R., Qiu, P., Wang, H., Yang, H., Yang, X., Ye, M., et al. (2021). Identification of microRNA-16-5p and microRNA-21-5p in Feces as Potential Noninvasive Biomarkers for Inflammatory Bowel Disease. Aging 13 (3), 4634–4646. doi:10.18632/aging.202428
Zhu, Z., Huang, J., Li, X., Xing, J., Chen, Q., Liu, R., et al. (2020a). Gut Microbiota Regulate Tumor Metastasis via circRNA/miRNA Networks. Gut Microbes 12 (1), 1788891. doi:10.1080/19490976.2020.1788891
Zhu, Y., Gu, L., Li, Y., Lin, X., Shen, H., Cui, K., et al. (2017). miR-148a Inhibits Colitis and Colitis-Associated Tumorigenesis in Mice. Cell Death Differ 24 (12), 2199–2209. doi:10.1038/cdd.2017.151
Zhu, F., Li, H., Liu, Y., Tan, C., Liu, X., Fan, H., et al. (2020b). miR-155 Antagomir Protect against DSS-Induced Colitis in Mice through Regulating Th17/Treg Cell Balance by Jarid2/Wnt/β-Catenin. Biomed. Pharmacother. 126, 109909. doi:10.1016/j.biopha.2020.109909
Zidar, N., Boštjančič, E., Jerala, M., Kojc, N., Drobne, D., Štabuc, B., et al. (2016). Down-regulation of microRNAs of the miR-200 Family and Up-Regulation of Snail and Slug in Inflammatory Bowel Diseases - Hallmark of Epithelial-Mesenchymal Transition. J. Cel. Mol. Med. 20 (10), 1813–1820. doi:10.1111/jcmm.12869
Keywords: ulcerative colitis, microRNA, colitis-associated cancer, inflammatory bowel diseases, gut microbiota
Citation: Zhou J, Liu J, Gao Y, Shen L, Li S and Chen S (2021) miRNA-Based Potential Biomarkers and New Molecular Insights in Ulcerative Colitis. Front. Pharmacol. 12:707776. doi: 10.3389/fphar.2021.707776
Received: 10 May 2021; Accepted: 29 June 2021;
Published: 09 July 2021.
Edited by:
Tao Xu, Anhui Medical University, ChinaReviewed by:
Celia Escudero-Hernández, University Medical Center Schleswig-Holstein, GermanyJinxia Zhu, Capital Medical University, China
Copyright © 2021 Zhou, Liu, Gao, Shen, Li and Chen. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Simin Chen, bWluZ21pbmcyMTk5QGFsaXl1bi5jb20=; Liwei Shen, MTkzMzM3MzY2NkBxcS5jb20=
 Jing Zhou
Jing Zhou Jialing Liu
Jialing Liu Yangyang Gao1
Yangyang Gao1