- 1Department of Neurology, The First Affiliated Hospital of Kunming Medical University, Kunming, China
- 2Department of Clinical Laboratory, The First Affiliated Hospital of Kunming Medical University, Kunming, China
- 3Biomedicine Engineering Research Center, Kunming Medical University, Kunming, China
Background: Epilepsy is a debilitating brain disease with complex inheritance and frequent treatment resistance. However, the role of STX1B single nucleotide polymorphisms (SNPs) in epilepsy treatment remains unknown.
Objective: This study aimed to explore the genetic association of STX1B SNPs with treatment response in patients with epilepsy in a Han Chinese population.
Methods: We first examined the associations between STX1B SNPs and epilepsy in 1000 Han Chinese and the associations between STX1B SNPs and drug-resistant epilepsy in 450 subjects. Expression quantitative trait loci analysis was then conducted using 16 drug-resistant epileptic brain tissue samples and results from the BrainCloud database (http://eqtl.brainseq.org).
Results: The allelic frequencies of rs140820592 were different between the epilepsy and control groups (p = 0.002) after Bonferroni correction. The rs140820592 was associated with significantly lower epilepsy risk among 1,000 subjects in the dominant model after adjusting for gender and age and Bonferroni correction (OR = 0.542, 95%CI = 0.358–0.819, p = 0.004). The rs140820592 also conferred significantly lower risk of drug-resistant epilepsy among 450 subjects using the same dominant model after adjusting for gender and age and Bonferroni correction (OR = 0.260, 95%CI = 0.103–0.653, p = 0.004). Expression quantitative trait loci analysis revealed that rs140820592 was associated with STX1B expression level in drug-resistant epileptic brain tissues (p = 0.012), and this result was further verified in the BrainCloud database (http://eqtl.brainseq.org) (p = 2.3214 × 10–5).
Conclusion: The STX1B rs140820592 may influence the risks of epilepsy and drug-resistant epilepsy by regulating STX1B expression in brain tissues.
Introduction
Epilepsy is one of the most prevalent chronic neurological disorders worldwide, with an estimated globe prevalence > 0.5% (Shorvon, 1990; Fiest et al., 2017). The disease is characterized by episodes of hyper-synchronized neuronal activity leading to recurrent seizures. It is estimated that more than half of all epilepsy cases are associated with genetic factors (Pal et al., 2010). Mutations in many genes have been reported to cause epilepsy (Helbig et al., 2008; Poduri and Lowenstein, 2011; Ponnala et al., 2012; Abou El Ella et al., 2018; Butilă et al., 2018), and epilepsies associated with different mutations exhibit substantial heterogeneity in disease course, clinical manifestations, and treatment response (Wang et al., 2017), presenting significant challenges for diagnosis and management.
The STX1B gene (16p11) encodes Syntaxin 1B, a protein of the SNARE complex mediating calcium-dependent synaptic vesicle release (Südhof, 2013). Synaptic dysfunctions are associated with a myriad of neurological disorders, including epilepsy (Steinlein, 2004; Luscher and Isaac, 2009; van Spronsen and Hoogenraad, 2010; Lepeta et al., 2016; Torres et al., 2017). Recent studies suggest that STX1B is involved in epilepsy (Schubert et al., 2014; Wolking et al., 2019). However, the role of STX1B SNPs in epilepsy treatment remains unknown, so it is necessary to explore the genetic association of STX1B SNPs with treatment response in patients with epilepsy in a Han Chinese population.
In this study, we investigated the associations between seven STX1B tagging SNPs and treatment response in patients with epilepsy in Han Chinese, and then conducted brain expression quantitative trait loci (eQTL) analysis. The STX1B rs140820592 was associated with reduced risks for epilepsy and drug-resistant epilepsy, likely by regulating STX1B expression in brain tissues.
Materials and Methods
Subjects
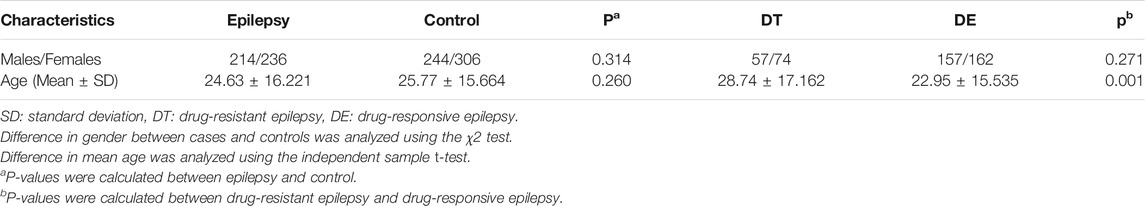
A case‒control study was performed to investigate the associations between STX1B tagging SNPs and drug-resistant epilepsy. Clinical and demographic characteristics of the study cohort are summarized in Table 1. All blood samples were collected at the First Affiliated Hospital of Kunming Medical University, and stored at −80°C in the biobank of the First Affiliated Hospital of Kunming Medical University. All brain tissue samples were collected at the First Affiliated Hospital of Kunming Medical University and Xinqiao Hospital and stored at −80°C in the biobank of Kunming Medical University. All subjects included in this study were of Han Chinese ancestry. The epilepsy patients were diagnosed according to 2017 International League Against Epilepsy (ILAE) criteria, and drug-resistant epilepsy patients were diagnosed according to 2010 ILAE criteria. Carbamazepine, valproic acid, levetiracetam and Lamotrigine were prescribed to the epilepsy patients. In order to improve epilepsy care and research, the ILAE defined drug-resistant epilepsy as failure of adequate trials of two tolerated, appropriately chosen and used antiepileptic drug schedules (whether as monotherapies or in combination) to achieve sustained seizure freedom in 2010 (Kwan et al., 2010), and developed the new classification of seizures and epilepsy relevant to clinical practice in 2017 (Fisher et al., 2017; Scheffer et al., 2017). Symptomatic epilepsy was excluded through auxiliary examination and disease history review. The healthy controls had no individual or family history of epilepsy and were neurologically normal. All participates or legal representatives provided written informed consent in accordance with the tenets of the Declaration of Helsinki. All study protocols were approved by the Ethics Committee of the First Affiliated Hospital of Kunming Medical University (No.2020-L-40).
Selection and Genotyping of SNPs
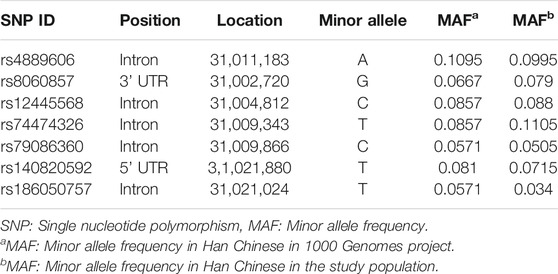
We first identified all STX1B SNPs in the Chinese Han South (CHS) population recorded in the 1000 Genomes database (http://www.internationalgenome.org/), and then used Haploview software (Barrett et al., 2005) to pick seven tagging SNPs (r2 > 0.8) with minor allele frequency > 5%. Basic information for seven STX1B SNPs is summarized in Table 2. Genomic DNA was extracted from human brain tissues using the Tissue DNA Kit (OMEGA, United States) and from peripheral blood using the Blood DNA Mini Kit (OMEGA, United States). SNP genotyping was conducted using the Bio-Rad CFX96 (BioRad, United States) platform. Primers for PCR were designed using Primer Premier V6.0 (Premier Biosoft Inc., United States). Details on PCR primers are provided in Supplementary Table 1.
Association Analysis of SNPs
SPSS V23.0 (IBM Corp, Armonk, NY) was used for all analyses. Haplotypes were constructed and analyzed using SHEsis software (http://analysis.bio-x.cn/myAnalysis. php). Hardy‒Weinberg equilibrium and allele frequencies of all SNPs were analyzed using the Chi-square (χ2) test or Fisher’s exact test. Difference in genotype frequencies of all SNPs between cases and controls was analyzed using binary logistic regression. In order to calculate the statistical power, type I error rate of 0.05, dominant mode, and 0.007 of baseline risk were assumed with QUANTO V1.2.4 (Written by John Morrison and W.James Gauderman at the University of Southern California), so the dominant model was also used to evaluate the associations between the genotypes of all SNPs and the risks of epilepsy and drug-resistant epilepsy.
eQTL Analysis
To examine if the tagging SNPs associated with epilepsy and drug-resistant epilepsy were also associated with STX1B expression level in drug-resistant epileptic brain tissues, we conducted eQTL analysis using temporal lobe samples from 16 patients (nine males and seven females) with drug-resistant epilepsy. Genomic RNA was extracted using the Tissue RNA Kit (OMEGA, United States), and reverse transcribed into cDNA using the FastQuant cDNA kit (Tiangen, China). Quantitative PCR was conducted using SYBR®Green I (Vazyme, China) and the ABI QuantStudio six Flex™ (ABI, United States) analyzer. Primers were designed using Primer Premier V6.0 (Premier Biosoft Inc., United States) Relative expression levels were determined using the 2−ΔΔCT method, and differences in STX1B mRNA expression between genotypes were analyzed by independent sample t-test using Graphpad Prism9.0 (www.graphpad- prism.cn). We also conducted eQTL analysis using results from the BrainCloud database (http://eqtl.brainseq.org) to validate eQTL findings from epileptic brain tissues. Details on qPCR primers are provided in Supplementary Table 2.
Statistical Analysis
Data are expressed as number, frequency, or mean ± standard deviation (SD) as appropriate. Continuous variables were compared between groups by independent samples t-test and categorical variables by χ2 test, Fisher’s exact test and logistic regression analysis using SPSS V23.0 (IBM Corp, Armonk, NY) and Graphpad Prism9.0 (www.graphpad-prism.cn) A p < 0.05 (two-tailed) was considered significant. For Bonferroni correction, A p < 0.007 (0.05/7) was considered significant.
Results
Subject Characteristics
Neither average age nor gender ratio differed significantly between the epilepsy group and control group. However, average age is significantly older in the drug-resistant epilepsy group than drug-responsive epilepsy group. Subject information is summarized in Table 1.
STX1B SNPs Associated Epilepsy Treatment Response in Han Chinese
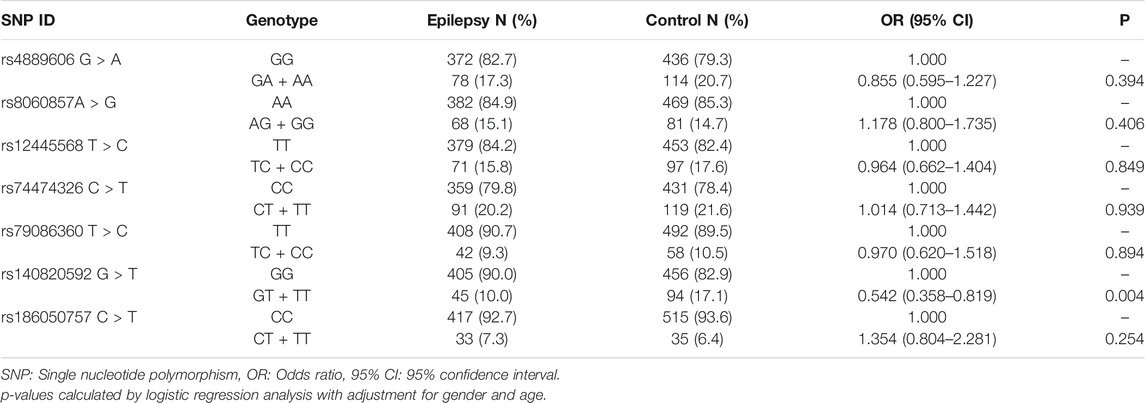
To comprehensively evaluate the relationships between the seven selected SNPs and epilepsy treatment response in Han Chinese, we performed a case‒control study involving 1,000 individuals (450 cases, 550 healthy controls). All seven tagging SNPs were in Hardy‒Weinberg equilibrium among control participants (p > 0.05) (Table 3).
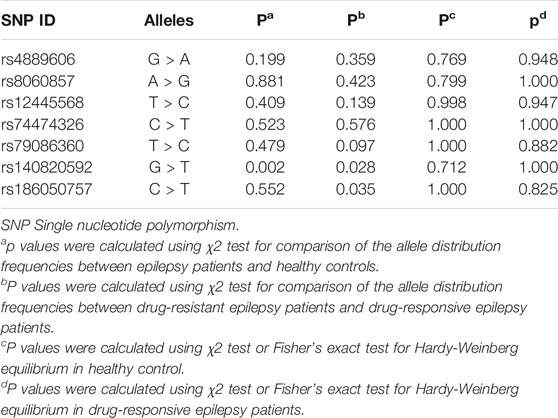
TABLE 3. Comparison of allele frequency distributions for seven tagging SNPs between cases and controls.
In the Stage I study, we found a significant difference in rs140820592 allele distribution between the epilepsy group and the control group (p = 0.002) (Table 3), and the association remained significant after adjusting for age and gender and Bonferroni correction (OR = 0.542, 95%CI = 0.358–0.819, p = 0.004) in the dominant model (GT + TT vs. GG) (Table 4).
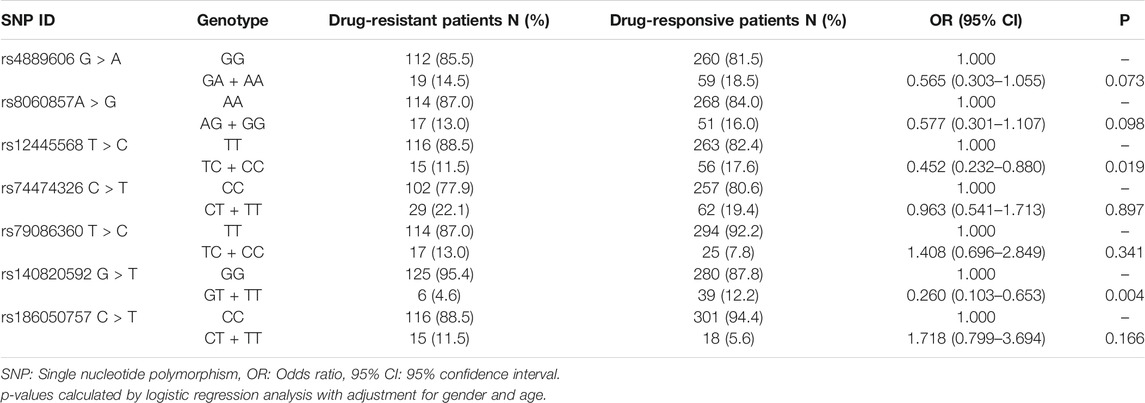
In Stage II, we further explored the relationships between STX1B SNPs and drug-resistant epilepsy among 450 Han Chinese individuals (131 drug-resistant epilepsy patients, and 319 drug-responsive epilepsy patients), and again found this association remained after adjusting for age and gender and Bonferroni correction (OR = 0.260, 95%CI = 0.103–0.653, p = 0.004) in the dominant model (GT + TT vs. GG) (Table 5).
The rs8060857, rs12445568, and rs74474326 polymorphisms have strong linkage disequilibrium for both epilepsy vs. control groups and drug-resistant epilepsy vs. drug-responsive epilepsy groups (Figure 1). There were no significant difference in the distributions of haplotypes A-T-C, A-T-T, and G-C-C between epilepsy and control groups (p = 0.812, p = 0.726, and p = 0.938, respectively) (Table 6) or between drug-resistant epilepsy vs. drug-responsive epilepsy groups (p = 0.936, p = 0.636, and p = 0.478, respectively) (Table 7).
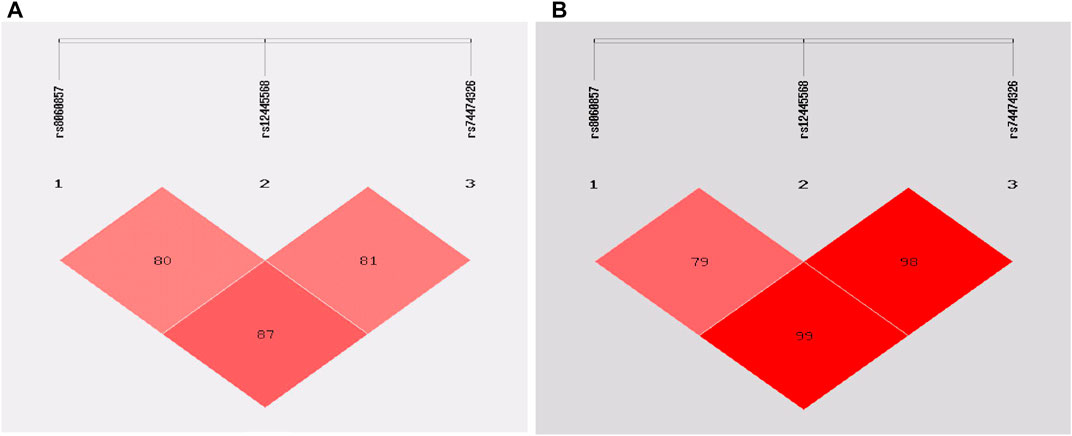
FIGURE 1. Linkage disequilibrium (LD) analysis of the STX1B SNPs in cases and controls: (A) in epilepsy patients and controls (B) in drug-resistant epilepsy patients and drug-responsive patients.
eQTL Analysis
We then conducted eQTL analysis based on brain tissue samples from 16 epilepsy patients with drug-resistant epilepsy and found that T allele carriers of rs140820592 exhibited higher STX1B expression than GG carriers (p = 0.012) (Figure 2), a finding consistent with records from the BrainCloud database (http://eqtl.brainseq.org) (Figure 3).
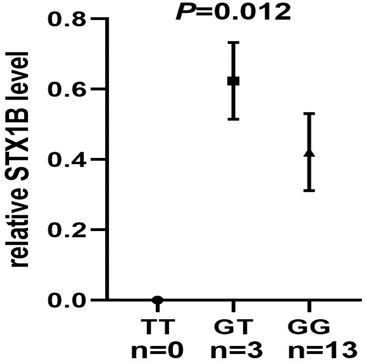
FIGURE 2. The rs140820592 is an eQTL in temporal lobe tissue of drug-resistant epilepsy patients. The carriers of the T allele exhibited upregulated STX1B gene expression.
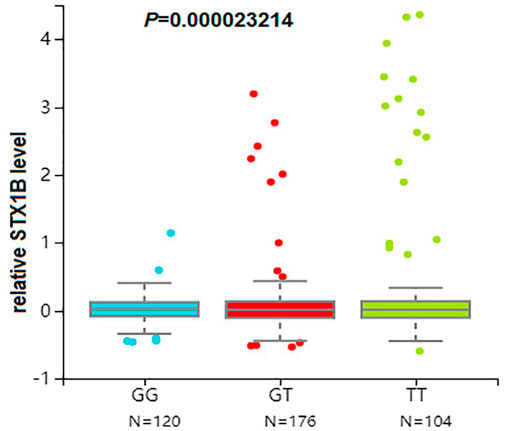
FIGURE 3. The rs140820592 is an eQTL in the dorsolateral prefrontal cortex. Data were retrieved from the brain tissue database Braincloud (http://eqtl.brainseq.org).
Discussion
Syntaxin 1B together with its binding partner Syntaxin binding protein 1 (Hata et al., 1993; Ogawa et al., 1998) is a critical component of the minimal presynaptic transmitter release machinery (Jahn and Sheller, 2006; Rizo and Sudhof, 2012). Here we showed that rs140820592 in the Syntaxin 1B-coding gene STX1B was significantly associated with the risks of epilepsy (drug-resistant epilepsy + drug-responsive epilepsy) and drug-resistant epilepsy. This is a study combining genetic association and eQTL analyses in brain tissues and blood samples to comprehensively evaluate the relationship between the STX1B gene and epilepsy treatment.
Among the seven tagging SNPs, the SNP rs140820592 was significantly associated with epilepsy. Specifically, risk of epilepsy was lower in rs140820592 GT + TT genotype carriers in the dominant model after adjusting for gender and age, and significance was maintained after Bonferroni correction (Table 4). Furthermore, eQTL analysis showed that the T allele carriers of rs140820592 was associated with increased STX1B expression in drug-resistant epileptic brain tissues (Figure 1), suggesting rs140820592 is a functional SNP.
In addition to epilepsy (drug-resistant epilepsy + drug-responsive epilepsy), the rs140820592 GT + TT genotype decreased the risk of drug-resistant epilepsy in the dominant model after adjusting for gender and age, and significance remained after Bonferroni correction (Table 5). To our knowledge, there are no other reports on the relationship between STX1B SNPs and drug-resistant epilepsy. In summary, rs140820592 may decrease the risks of epilepsy and drug-resistant epilepsy by regulating STX1B gene expression in human brain tissues. However, further larger sample studies are required for confirmation.
Haplotype analysis showed a strong LD for rs8060857, rs12445568, and rs74474326 (D’ = 0.79–0.99) in this study. However, the frequencies of these haplotypes, including A-T-C, A-T-T and G-C-C, did not differ between case and control groups for either epilepsy or drug-resistant epilepsy (Table 6 and Table 7), at least in part because these SNPs were not associated with epilepsy or drug-resistant epilepsy in this study cohort. We did not find a strong LD between rs140820592 and other tagging SNPs, suggesting that rs140820592 may be an independent factor reducing the risks of epilepsy and drug-resistant epilepsy in Han Chinese.
Syntaxin 1B is mainly expressed in neurons, and has similar physiological functions to STX1A (Inoue et al., 1992; Kushima et al., 1997). The STX1B gene is highly expressed in temporal lobe according to BioGPS (http://biogps.org), so this region was chosen for eQTL analysis. Indeed, this analysis showed that rs140820592 is an eQTL in drug-resistant epileptic brain tissues. As reported in previous studies, disease-associated SNPs often influence the risk of disease by regulating gene expression (Nicolae et al., 2010; Maurano et al., 2012; Ward and Kellis, 2012; Bi et al., 2018; Guo et al., 2018; Li et al., 2018; Wang et al., 2021), and this result further verified the association between rs140820592 and epilepsy treatment response. These findings may aid in the development of new diagnostic methods and therapeutic targets for epilepsy.
Genetic factors have been found to affect drug response in many studies (Franco and Perucca, 2015; Balestrini and Sisodiya. 2018). Understanding the associations between STX1B SNPs and drug response will aid in translation of genetic findings to clinical treatment. We found that the STX1B rs140820592 was significantly associated with the risk of drug-resistant epilepsy at the genetic and expressional levels, suggesting a role of STX1B in epilepsy treatment. STX1B plays an important role in the regulation of synaptic vesicle exocytosis, including release of glutamatergic and GABAergic synaptic transmission (Mishima et al., 2014). Furthermore, some important neurotransmitters, such as glutamate and GABA have been found to be involved in epilepsy and drug treatment (Teichgräber et al., 2009; Ngomba and van Luijtelaar, 2018; Chun et al., 2019; Alcoreza et al., 2021). Therefore, STX1B may be involved in epilepsy and drug responsiveness by regulating synaptic vesicle exocytosis.
However, our study in Han Chinese failed to replicate rs140820592 minor allele (T) as a increased risk for epilepsy in the United Kingdom biobank database. The opposite direction of the effect can be explained as follows: First, the difference in the distribution of alleles among different populations should be considered. Second, study subjects for different types of epilepsy may also draw inconsistent conclusions.
Although we have provided important evidence that STX1B gene is involved in the development of epilepsy and drug-resistant epilepsy at the genetic and expressional levels in Han Chinese, some limitations of this study should also be noted: First, our study only included the Han population, and the sample size was relatively small (We estimated that our study had 80% power to detect significant SNPs of MAF = 0.034 with relative risk >1.75.), so a larger sample study including different ethnic groups should be conducted to further verify our findings. Second, we performed functional analysis only in human brain tissues, and further experiments are necessary. Third, in this study, only STX1B gene was included, and other important epilepsy genes were not included in our study.
Conclusion
We provide multiple lines of evidence that the STX1B rs140820592 may decrease the risks of epilepsy (drug-resistant epilepsy + drug-responsive epilepsy) and drug-resistant epilepsy by regulating STX1B expression. STX1B gene may be a new therapeutic target for epilepsy. However, It is necessary to further verify our conclusions through studies with large samples in the future.
Data Availability Statement
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author.
Ethics Statement
The studies involving human participants were reviewed and approved by the Ethics Committee of the First Affiliated Hospital of Kunming Medical University. Written informed consent to participate in this study was provided by the participants' legal guardian/next of kin.
Author Contributions
YH and SW conceived and designed this research; DW, XC, YY, CH, and LC collected blood and brain tissue samples; DL, LB, and QW analyzed the data; YH, SW, and LZ wrote the manuscript; YY, LC, and SD performed the experiments. All authors read and approved the final manuscript.
Funding
This work was supported by the National Natural Science Foundation of China (81260199, 81660228, and 81601134), the Yunnan Province Talent Training Program (2017HB048, L-2019019, and H-2018056), and the Yunnan Science and Research Funding Program (2016NS029).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank all the participants for providing blood and brain tissue samples. We also appreciate all the professionals and databases managers who helped us in this study.
References
Abou El Ella, S. S., Tawfik, M. A., Abo El Fotoh, W. M. M., and Soliman, O. A. M. (2018). The Genetic Variant "C588T" of GABARG2 Is Linked to Childhood Idiopathic Generalized Epilepsy and Resistance to Antiepileptic Drugs. Seizure 60, 39–43. doi:10.1016/j.seizure.2018.06.004
Alcoreza, O. B., Patel, D. C., Tewari, B. P., and Sontheimer, H. (2021). Dysregulation of Ambient Glutamate and Glutamate Receptors in Epilepsy: An Astrocytic Perspective. Front. Neurol. 12, 652159. doi:10.3389/fneur.2021.652159
Balestrini, S., and Sisodiya, S. M. (2018). Pharmacogenomics in Epilepsy. Neurosci. Lett. 667, 27–39. doi:10.1016/j.neulet.2017.01.014
Barrett, J. C., Fry, B., Maller, J., and Daly, M. J. (2005). Haploview: Analysis and Visualization of LD and Haplotype Maps. Bioinformatics 21 (2), 263–265. doi:10.1093/bioinformatics/bth457
Bi, R., Zhang, W., Zhang, D. F., Xu, M., Fan, Y., Hu, Q. X., et al. (2018). Genetic Association of the Cytochrome Coxidase-Related Genes with Alzheimer’s Disease in Han Chinese. Neuropsychopharmacology 43 (11), 2264–2276. doi:10.1038/s41386-018-0144-3
Butilă, A. T., Zazgyva, A., Sin, A. I., Szabo, E. R., and Tilinca, M. C. (2018). GABRG2 C588T Gene Polymorphisms Might Be a Predictive Genetic Marker of Febrile Seizures and Generalized Recurrent Seizures: a Case-Control Study in a Romanian Pediatric Population. Arch. Med. Sci. 14 (1), 157–166. doi:10.5114/aoms.2016.63739
Chun, E., Bumanglag, A. V., Burke, S. N., and Sloviter, R. S. (2019). Targeted Hippocampal GABA Neuron Ablation by Stable Substance P-Saporin Causes Hippocampal Sclerosis and Chronic Epilepsy in Rats. Epilepsia 60 (5), e52–e57. doi:10.1111/epi.14723
Fiest, K. M., Sauro, K. M., Wiebe, S., Patten, S. B., Kwon, C. S., Dykeman, J., et al. (2017). Prevalence and Incidence of Epilepsy: A Systematic Review and Meta-Analysis of International Studies. Neurology 88 (3), 296–303. doi:10.1212/WNL.0000000000003509
Fisher, R. S., Cross, J. H., French, J. A., Higurashi, N., Hirsch, E., Jansen, F. E., et al. (2017). Operational Classification of Seizure Types by the International League against Epilepsy: Position Paper of the ILAE Commission for Classification and Terminology. Epilepsia 58 (4), 522–530. doi:10.1111/epi.13670
Franco, V., and Perucca, E. (2015). The Pharmacogenomics of Epilepsy. Expert Rev. Neurother. 15 (10), 1161–1170. doi:10.1586/14737175.2015.1083424
Guo, X., Lin, W., Bao, J., Cai, Q., Pan, X., Bai, M., et al. (2018). A Comprehensive Cis-eQTL Analysis Revealed Target Genes in Breast Cancer Susceptibility Loci Identified in Genome-wide Association Studies. Am. J. Hum. Genet. 102, 890–903. doi:10.1016/j.ajhg.2018.03.016
Hata, Y., Slaughter, C. A., and Sudhof, T. C. (1993). Synaptic Vesicle Fusion Complex Contains Unc-18 Homologue Bound to Syntaxin. Nature 366 (6453), 347–351. doi:10.1038/366347a0
Helbig, I., Scheffer, I. E., Mulley, J. C., and Berkovic, S. F. (2008). Navigating the Channels and beyond: Unravelling the Genetics of the Epilepsies. Lancet Neurol. 7 (3), 231–245. doi:10.1016/S1474-4422(08)70039-5
Inoue, A., Obata, K., and Akagawa, K. (1992). Cloning and Sequence Analysis of cDNA for a Neuronal Cell Membrane Antigen, HPC-1. J. Biol. Chem. 267 (15), 10613–10619. doi:10.1016/s0021-9258(19)50061-8
Jahn, R., and Scheller, R. H. (2006). SNAREs–Engines for Membrane Fusion. Nat. Rev. Mol. Cel Biol. 7 (9), 631–643. doi:10.1038/nrm2002
Kushima, Y., Fujiwara, T., Sanada, M., and Akagawa, K. (1997). Characterization of HPC-1 Antigen, an Isoform of Syntaxin-1, with the Isoform-specific Monoclonal Antibody, 14D8. J. Mol. Neurosci. 8 (1), 19–27. doi:10.1007/BF02736860
Kwan, P., Arzimanoglou, A., Berg, A. T., Brodie, M. J., Allen Hauser, W., Mathern, G., et al. (2010). Definition of Drug Resistant Epilepsy: Consensus Proposal by the Ad Hoc Task Force of the ILAE Commission on Therapeutic Strategies. Epilepsia 51 (6), 1069–1077. doi:10.1111/j.1528-1167.2009.02397.x
Lepeta, K., Lourenco, M. V., Schweitzer, B. C., Martino, Adami, P. V., Banerjee, P., Catuara-Solarz, S., et al. (2016). Synaptopathies: Synaptic Dysfunction in Neurological Disorders - A Review from Students to Students. J. Neurochem. 138 (6), 785–805. doi:10.1111/jnc.13713
Li, X., Luo, Z., Gu, C., Hall, L. S., McIntosh, A. M., Zeng, Y., et al. (2018). Common Variants on 6q16.2, 12q24.31 and 16p13.3 Are Associated with Major Depressive Disorder. Neuropsychopharmacology 43 (10), 2146–2153. doi:10.1038/s41386-018-0078-9
Luscher, C., and Isaac, J. T. (2009). The Synapse: center Stage for many Brain Diseases. J. Physiol. 587 (Pt4), 727–729. doi:10.1113/jphysiol.2008.167742
Maurano, M. T., Humbert, R., Rynes, E., Thurman, R. E., Haugen, E., Wang, H., et al. (2012). Systematic Localization of Common Diseaseassociated Variation in Regulatory DNA. Science 337 (6099), 1190–1195. doi:10.1126/science.1222794
Ngomba, R. T., and van Luijtelaar, G. (2018). Metabotropic Glutamate Receptors as Drug Targets for the Treatment of Absence Epilepsy. Curr. Opin. Pharmacol. 38, 43–50. doi:10.1016/j.coph.2018.01.012
Nicolae, D. L., Gamazon, E., Zhang, W., Duan, S., Dolan, M. E., and Cox, N. J. (2010). Trait-associated SNPs Are More Likely to Be eQTLs: Annotation to Enhance Discovery from GWAS. Plos Genet. 6 (4), e1000888. doi:10.1371/journal.pgen.1000888
Ogawa, H., Harada, S., Sassa, T., Yamamoto, H., and Hosono, R. (1998). Functional Properties of the Unc-64 Gene Encoding a Caenorhabditis elegans Syntaxin. J. Biol. Chem. 273 (4), 2192–2198. doi:10.1074/jbc.273.4.2192
Pal, D. K., Pong, A. W., and Chung, W. K. (2010). Genetic Evaluation and Counseling for Epilepsy. Nat. Rev. Neurol. 6 (8), 445–453. doi:10.1038/nrneurol.2010.92
Poduri, A., and Lowenstein, D. (2011). Epilepsy Genetics—Past, Present, and Future. Curr. Opin. Genet. Dev. 2011 (3), 21325–21332. doi:10.1016/j.gde.2011.01.005
Ponnala, S., Chaudhari, J. R., Jaleel, M. A., Bhiladvala, D., Kaipa, P. R., Das, U. N., et al. (2012). Role of MDR1 C3435T and GABRG2 C588T Gene Polymorphisms in Seizure Occurrence and MDR1 Effect on Anti-epileptic Drug (Phenytoin) Absorption. Genet. Test. Mol. Biomarkers. 16 (6), 550–557. doi:10.1089/gtmb.2011.0225
Rizo, J., and Sudhof, T. C. (2012). The Membrane Fusion enigma: SNAREs, Sec1/Munc18 Proteins, and Their Accomplices–Guilty as Charged?. Annu. Rev. Cel Dev. Biol. 28, 279–308. doi:10.1146/annurev-cellbio-101011-155818
Scheffer, I. E., Berkovic, S., Capovilla, G., Connolly, M. B., French, J., Guilhoto, L., et al. (2017). ILAE Classification of the Epilepsies: Position Paper of the ILAE Commission for Classification and Terminology. Epilepsia 58 (4), 512–521. doi:10.1111/epi.13709
Schubert, J., Siekierska, A., Langlois, M., May, P., Huneau, C., Becker, F., et al. (2014). Mutations in STX1B, Encoding a Presynaptic Protein, Cause Fever-Associated Epilepsy Syndromes. Nat. Genet. 46 (12), 1327–1332. doi:10.1038/ng.3130
Shorvon, S. D. (1990). Epidemiology, Classification, Natural History, and Genetics of Epilepsy. Lancet 336 (8707), 93–96. doi:10.1016/0140-6736(90)91603-8
Steinlein, O. K. (2004). Genetic Mechanisms that Underlie Epilepsy. Nat. Rev. Neurosci. 5 (5), 400–408. doi:10.1038/nrn1388
Südhof, T. C. (2013). Neurotransmitter Release: The Last Millisecond in the Life of a Synaptic Vesicle. Neuron 80 (3), 675–690. doi:10.1016/j.neuron.2013.10.022
Teichgräber, L. A., Lehmann, T. N., Meencke, H. J., Weiss, T., Nitsch, R., and Deisz, R. A. (2009). Impaired Function of GABA(B) Receptors in Tissues from Pharmacoresistant Epilepsy Patients. Epilepsia 50 (7), 1697–1716. doi:10.1111/j.1528-1167.2009.02094.x
Torres, V. I., Vallejo, D., and Inestrosa, N. C. (2017). Emerging Synaptic Molecules as Candidates in the Etiology of Neurological Disorders. Neural Plast. 2017, 8081758. doi:10.1155/2017/8081758
van Spronsen, M., and Hoogenraad, C. C. (2010). Synapse Pathology in Psychiatric and Neurologic Disease. Curr. Neurol. Neurosci. Rep. 10 (3), 207–214. doi:10.1007/s11910-010-0104-8
Wang, J., Lin, Z. J., Liu, L., Xu, H. Q., Shi, Y. W., Yi, Y. H., et al. (2017). Epilepsy-associated Genes. Seizure 44, 11–20. doi:10.1016/j.seizure.2016.11.030
Wang, S., Zhang, X., Zhou, L., Wu, Q., and Han, Y. (2021). Analysis of GABRG2 C588T Polymorphism in Genetic Epilepsy and Evaluation of GABRG2 in Drug Treatment. Clin. Transl. Sci. 00, 1–9. doi:10.1111/cts.12997
Ward, L. D., and Kellis, M. (2012). Interpreting Noncoding Genetic Variation in Complex Traits and Human Disease. Nat. Biotechnol. 30 (11), 1095–1106. doi:10.1038/nbt.2422
Keywords: STX1B, epilepsy, polymorphism, association, treatment
Citation: Wang S, Zhou L, He C, Wang D, Cai X, Yu Y, Chen L, Lu D, Bian L, Du S, Wu Q and Han Y (2021) The Association Between STX1B Polymorphisms and Treatment Response in Patients With Epilepsy. Front. Pharmacol. 12:701575. doi: 10.3389/fphar.2021.701575
Received: 28 April 2021; Accepted: 29 June 2021;
Published: 09 July 2021.
Edited by:
Youssef Daali, Geneva University Hospitals (HUG), SwitzerlandReviewed by:
Yanfei Zhang, Geisinger Health System, United StatesStanley Hooker, City of Hope National Medical Center, United States
Copyright © 2021 Wang, Zhou, He, Wang, Cai, Yu, Chen, Lu, Bian, Du, Wu and Han. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yanbing Han, eW5oeWJAMTYzLmNvbQ==
 Shitao Wang1
Shitao Wang1 Yanbing Han
Yanbing Han