- 1Key Laboratory of Molecular Target & Clinical Pharmacology and the State Key Laboratory of Respiratory Disease, School of Pharmaceutical Sciences & the Fifth Affiliated Hospital, Guangzhou Medical University, Guangzhou, China
- 2Department of Cellular and Molecular Biology, Beijing Chest Hospital, Capital Medical University/Beijing Tuberculosis and Thoracic Tumor Research Institute, Beijing, China
- 3BioMatter Unit, École Polytechnique de Bruxelles, Université Libre de Bruxelles (ULB), Brussels, Belgium
- 4Department of Biochemistry and Molecular Biology, School of Integrated Chinese and Western Medicine, Anhui University of Chinese Medicine, Hefei, China
Cancer is a major global health challenge for our health system, despite the important pharmacological and therapeutic discoveries we have seen since past 5 decades. The increasing prevalence and mortality of cancer may be closely related to smoking, exposure to environmental pollution, dietary and genetic factors. Despite significant promising discoveries and developments such as cell and biotechnological therapies a new breakthrough in the medical field is needed to develop specific and effective drugs for cancer treatment. On the development of cell therapies, anti-tumor vaccines, and new biotechnological drugs that have already shown promising effects in preclinical studies. With the continuous enrichment and development of chromatin immunoprecipitation sequencing (ChIP-seq) and its derivative technologies, epigenetic modification has gradually become a research hotspot. As key ingredients of epigenetic modification, Writers, Readers, Erasers have been gradually unveiled. Cancer has been associated with epigenetic modification especially methylation and therefore different epigenetic drugs have been developed and some of those are already undergoing clinical phase I or phase II trials, and it is believed that these drugs will certainly assist the treatment in the near future. With respect to this, an overview of anti-tumor drugs targeting modified enzymes and de-modified enzymes will be performed in order to contribute to future research.
Introduction
The epigenetic modification will affect gene expression. The genetic changes in gene expression or cell phenotype can happen without the sequence change of DNA. The phenomenon is called the epigenetic phenomenon, which can be stably inherited and may be reversible during embryonic development and cell proliferation (Harvey et al., 2018). The core of epigenetics includes various covalent modifications of histones and nucleic acids, which coordinately regulate chromatin structure and gene expression. Disorders of the epigenetic processes will drive abnormal transcription programs and promote the occurrence and progression of cancer (Sapienza and Issa, 2016; Nebbioso et al., 2018). According to the current research results, the regulatory mechanisms of epigenetic modification mainly include DNA methylation (Skvortsova et al., 2019), RNA interference (Holoch and Moazed, 2015), histone modification (Stoll et al., 2018), chromatin remodeling (Dawson and Kouzarides, 2012), and nucleosome localization (Allis and Jenuwein, 2016).
Currently, DNA methylation is the most fully studied form of epigenetic modification, which mainly involves adding a methyl group to the C5 position of the cytosine base to produce 5-methylcytosine. Normal methylation is necessary for maintaining cell growth and metabolism, while abnormal DNA methylation can cause diseases (such as cancers). The reason for the occurrence of this event may be that, on the one hand, abnormal methylation may prevent the transcription of tumor suppressor genes, and on the other hand, it may cause genome instability. Therefore, the study of DNA methylation is very helpful for understanding biological growth and disease treatment (Koch et al., 2018). In the cell nucleus, DNA is wrapped around histones and packaged to form a chromatin structure, and the tightness of chromatin packaging determines the activity of gene expression. Studies have confirmed that chromatin can switch between the “on and off” states by regulating the chemical modification of histones (Bannister and Kouzarides, 2011; Calo and Wysocka, 2013). Non-coding RNA refers to a functional RNA molecule that cannot be translated into protein and has a regulatory effect, and it plays a very important role in regulating gene expression. The regulation of non-coding RNA is the regulation of gene transcription through certain mechanisms, such as RNA interference (Holoch and Moazed, 2015).
Chromatin remodeling involves a series of biological processes mediated by the chromatin remodeling complex, which is characterized by nucleosome changes in chromatin, and it was considered to be an important epigenetic mechanism (Nacev et al., 2020). Studies have suggested that nucleosomes were barriers to gene transcription, and DNA tightly wound on histones cannot bind to many transcription factors and activation factors. In each type of cell, several specific genes are activated while others inhibited; thus, resulting in a variety of gene expression patterns. Therefore, changes in the position of nucleosomes in the genome have an important impact on regulating gene expression. With the development of life cycle activities, such as DNA replication, recombination, repair, and transcriptional regulation, the positioning of nucleosomes in chromatin has undergone dynamic changes. This continuous change requires participation of a series of chromatin remodeling complexes (Dawson and Kouzarides, 2012).
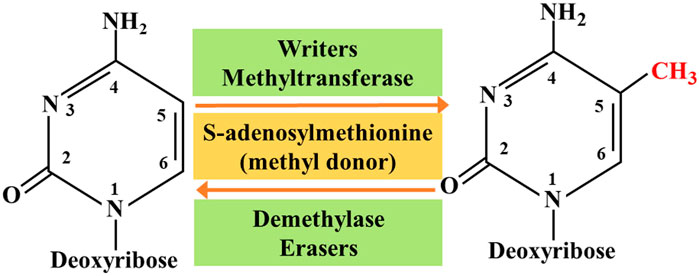
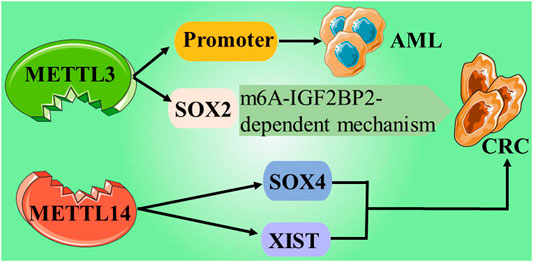
So far, researchers have identified four different types of cytosine residue modifications in DNA, including methylation, hydroxymethylation, formylation, and carboxylation (Biswas and Rao, 2018). In addition, more than ten different types of histone modifications have been identified; and among them, methylation and acetylation are the most stable and suitable. With continuous progress in the research on epigenetics, the key roles associated with these modifications are gradually being deciphered. They are as follows: 1) the writers-a large number of enzymes that can modify nucleotide bases and specific amino acid residues in histones, 2) the erasers-a group of enzymes proficient in removing these markers, and 3) the readers -a series of different proteins that possess specific domains that can recognize specific epigenetic marks in a locus. Together, these enzymes and protein domains constitute epigenetic tools (Biswas and Rao, 2018; Zaccara et al., 2019). The modification of DNA and histones occurs by adding various chemical groups using a variety of enzymes. We will focus on the most extensive study of epigenetic modifications, namely methylation. Both DNA and histones are prone to methylation, and this modification usually controls gene expression in cells through changes in transcription activation or inhibition. Here, the epigenetic writers, which we intend to focus on include DNA methyltransferase, histone lysine methyltransferase, and protein arginine methyltransferase (Biswas and Rao, 2018). Depending on the requirements of the cell to modify the expression state of the locus, epigenetic marks formed by histone post-translational modification and DNA covalent modification can be removed. To achieve this purpose, a group of enzymes called erasers can counter the writer's activities. The eraser can catalyze the removal of epigenetic marks, thereby reducing the effect of epigenetic marks on transcription; thus, leading to regulation of gene expression. The key difference between genetic mutation and epigenetic modification is that epigenetic changes are reversible, which makes them attractive drug targets. On the whole, the ultimate goal of research on tumor-related epigenetic mechanisms is to apply them to clinical diagnosis and preventive treatment (Figure 1).
Writers
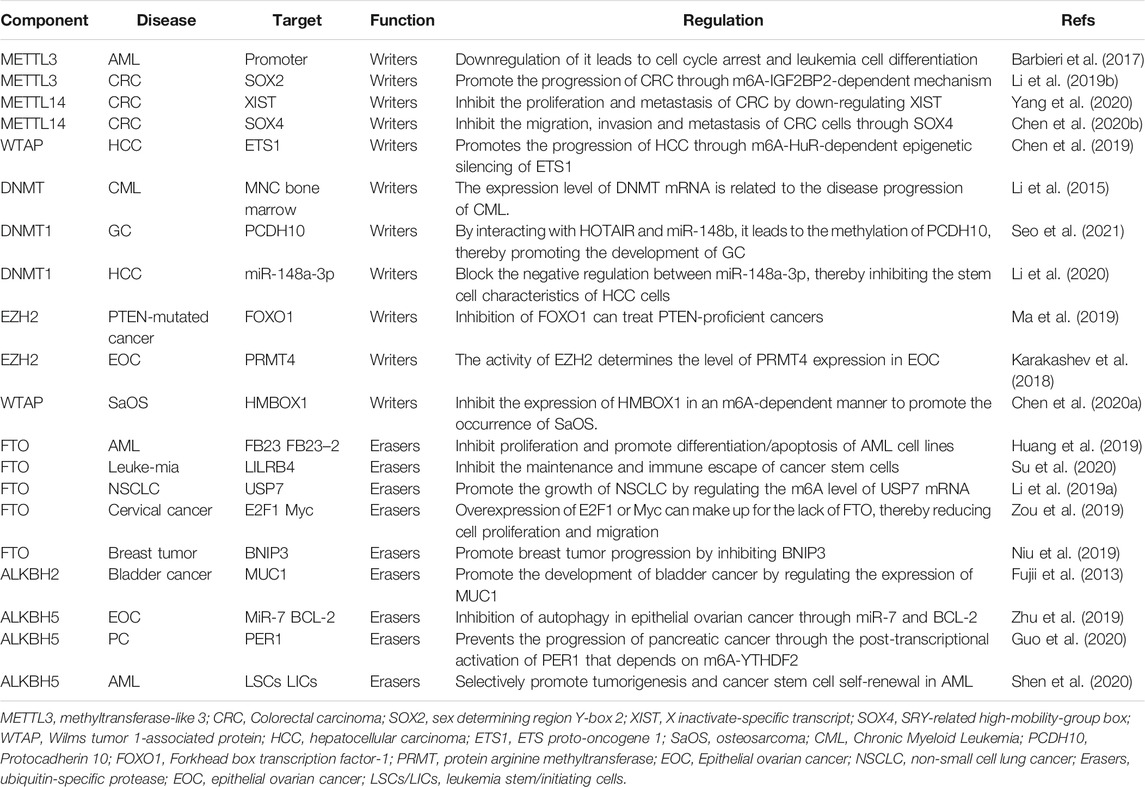
Modifications of DNA and histone proteins occur through the additions of various chemical groups utilizing numerous enzymes. Although a plethora of modifications are possible, we have focused on the two most widely studied epigenetic alterations, methylation and acetylation. Both DNA and histone proteins are prone to methylation, while acetylation is associated only with histones. These two modifications frequently govern the gene expression pattern in a cell by switching between transcriptional activation or repression. Changes in global DNA methylation and individual gene methylation patterns are distinguishing features of cancer cells, which are governed by DNMT: DNA methyltransferase. Global DNA hypomethylation and hypermethylation of the promoter regions of tumor suppressor genes have been reported in malignant cells (Kulis and Esteller, 2010). These modifications are caused by DNMT providing a viable target for developing drugs against these enzymes.
Abnormal DNA methylation at the C5 position of the cytosine catalyzed by DNMTs is not only related to silencing of many tumor suppressor genes, but also to other diseases. Small molecule inhibitors of DNMTs are the most widely used epigenetic therapies for cancer treatment, mainly for the treatment of myelodysplastic syndrome (MDS) and acute myeloid leukemia (AML) (Khan et al., 2013; Du et al., 2020). DNMT is usually overexpressed in various cancer tissues and cell lines. Since DNA methylation is reversible, DNMT is considered an important epigenetic target for drug development. DNMT inhibitors are categorized into nucleosides and non-nucleosides. Among these inhibitors, the nucleoside analogue azacytidine and its deoxy derivative decitabine are both irreversible DNMT inhibitors and have been approved for the treatment of bone marrow hyperplastic syndrome (Zhou et al., 2018b). Studies have shown that DNMTs can up-regulate the immune signal in ovarian cancer through the viral defense pathway (Chiappinelli et al., 2015).
Inhibitors of poly (ADP-ribose) polymerase 1 (PARP1) have shown promise for targeting cancer cells harboring mutations in the double-strand break (DSB) repair breast cancer genes, BRCA1 and BRCA2, where these drugs induce synthetic lethality. Studies have shown that PARP1 and DNMTs have shown an unexpected benefit in the combined treatment of non-small cell lung cancer (NSCLC). Chemoradiation is the main treatment method for NSCLC. Interestingly, combined treatment with PARP1 and DNMTs can make NSCLC cells very sensitive to ionizing radiation both in vitro and in vivo. With respect to the key mechanism, DNMTs may produce a BRCA-like phenotype by down-regulating the expression of key homologous recombination and non-homologous end joining genes (Abbotts et al., 2019).
The enhancer of Zeste homolog 2 (EZH2) is histone methyltransferase, and it catalyzes the methylation of histone 3 lysine 27, which is a sign of transcriptional inhibition. Many studies have clarified the complex role of EZH2 in normal biology and tumorigenesis. The effect of EZH2 on tumorigenesis is attributed to direct gene overexpression and point mutations leading to gain or loss of function. To a large extent, point mutations can increase the function of lymphoma and inactivate mutations in myeloid malignancies, and overexpression is the main manifestation of epithelial malignancies and specific lymphoma subtypes. Interestingly, EZH2 may have a dual function, and based on the dynamic expression of EZH2 in cell differentiation and cell cycle progression, it has the ability to act as both an oncogene and a tumor suppressor. Tazemetostat is the first EZH2 inhibitor, and it has shown enhanced clinical activity in mutant follicular lymphoma and diffuse large B-cell lymphoma. A new treatment strategy is required not only for the treatment of lymphoma, but it may also be beneficial in the treatment of many other malignant tumors (Lue and Amengual, 2018).
Another in vivo clinical study on tazemetostat published in 2018 reported that it showed good safety and anti-tumor activity in refractory B-cell non-Hodgkin’s lymphoma and advanced solid tumors, including epithelioid sarcoma. Further clinical studies of tazemetostat monotherapy are ongoing in phase 2 trials in adults and phase 1 trials in children. A total of 64 patients (21 cases of B-cell non-Hodgkin's lymphoma and 43 cases of advanced solid tumors) received treatment with tazemetostat. No treatment-related deaths occurred. A durable objective response was observed in 21 patients with B-cell non-Hodgkin's lymphoma (Italiano et al., 2018) (NCT01897571).
Excellent efficacy of EZH2 inhibitors in cancer is likely to cause the problem of drug resistance. Studies have confirmed that the role of Forkhead box transcription factor-1 (FOXO1) is linked to the role of EZH2 inhibitors in cancer. It is believed that the FOXO1 gene is a new inhibitory target of EZH2 and FOXO1 is a key mediator of EZH2 inhibition and induction of prostate cancer cell death. Phosphatase and tension homolog (PTEN) is a well-known tumor suppressor and FOXO1 is a key downstream effector of PTEN in inhibiting cell growth and survival. Further studies have proved that EZH2 inhibitors cannot effectively induce PTEN-deficient cancer cell death, but they can be overcome by combination therapy with taxanes. The following mechanism is likely: EZH2 inhibits FOXO1 expression and may serve as a target of EZH2 inhibitors, which can be used to overcome EZH2 inhibitor resistance in PTEN mutant cancers or to treat PTEN-deficient prostate cancer in combination with taxane. A new inhibitory target and FOXO1 is a key mediator of EZH2 inhibition to induce prostate cancer cell death. Further studies have proved that EZH2 inhibitors cannot effectively induce PTEN-deficient cancer cell death, but they can be overcome by combination therapy with taxanes (Ma et al., 2019). Furthermore, Bitler et al. believe that inhibition of EZH2 methyltransferase can lead to regression of ARID1A mutant tumors in mouse models of ovarian cancer. This is because PIK3IPI is the target of ARID1A and EZH2, which up-regulates PIK3IPI by inhibiting EZH2, and ultimately inhibits the oncogenic PI3K/Akt signal (Bitler et al., 2015).
DOT1L is the only known histone 3 lysine 79 (H3K79) methyltransferase, while AML or acute lymphocytic leukemia (ALL) is a malignant clonal disease of hematopoietic stem cells, and without any special treatment, these patients can only survive for about 3 months. Some of these patients can even die within a few days after the diagnosis. Both these diseases are associated with aberration in mixed lineage leukemia (MLL) gene (also known as KMT2A) translocation at chromosome 11q23, and both show a poor prognosis. If DOT1L can become an attractive target for the treatment of acute leukemia, it will be of great benefit to the medical field. Pinometostat (EPZ-5676) is a small molecule inhibitor of DOT1L histone methyltransferase interference. In an in vivo clinical trial, the DOT1L inhibitor pinometostat can reduce H3K79 methylation and has moderate clinical activity in adult acute leukemia. This study demonstrates the therapeutic potential of DOT1L in the mixed lineage leukemia (MLL) gene rearrangement of leukemia and lays the foundation for future combination therapy in this patient population (Stein et al., 2018) (NCT01684150).
In another study, convincing evidence was obtained by using DOT1L inhibitors as targeted therapy for MLL. EPZ004777 is a potent and selective inhibitor of DOT1L. According to the reports, MLL cells treated with EPZ004777 can selectively inhibit H3K79 methylation and can effectively block the expression of leukemia-associated genes. In addition, in vitro experiments showed that EPZ004777 has a selective killing effect on leukemia cells translocated by the MLL gene, and it has minimal effect on non-MLL translocated cells. In vivo experiments also showed a good performance; i.e., by administering EPZ004777 to a mouse xenograft model of MLL, the survival period can be significantly extended (Daigle et al., 2011).
It is undeniable that effective inhibitors of DOT1L have achieved many surprising results in targeting leukemia with MLL gene rearrangement. Research on this subject will continue in the future. In a document published in 2019, it has been confirmed that pinometostat as a DOT1L inhibitor has entered a phase 1 clinical trial to treat children with relapsed/refractory leukemia with MLL gene rearrangement Patient (NCT02141828). It has also been confirmed that pinometostat is undergoing a phase 1/2 clinical trial for evaluating the combination of pinometostat and standard chemotherapy for the treatment of newly diagnosed MLL rearranged leukemia in children and adults (NCT03724084) (Lonetti et al., 2019).
Different from the leukemia mechanism of MLL gene translocation, DOT1L is also believed to play an important role in the occurrence and development of breast cancer. Studies have suggested that DOT1L can target the gene expression of epithelial-mesenchymal transition (EMT) promoters by cooperating with the c-Myc/p300 transcriptional activity complex, thereby playing an important role in the occurrence and development of breast cancer, which suggests that DOT1L is a potential therapeutic target for invasive breast cancer (Lee and Kong, 2015).
Similarly, the new psammaplin A analogue (PsA-3091) is considered as a new template for DOT1L inhibitors, and it shows an effective inhibitory effect on DOT1L-mediated H3K79 methylation. As already known, triple-negative breast cancer (TNBC) is the most difficult disease to treat among women and has a high risk of metastasis. According to reports, PsA-3091 has a significant inhibitory effect on the proliferation, migration, and invasion of TNBC cells, and it can significantly enhance the expression of E-cadherin and inhibit the expression of N-cadherin, ZEB1, and vimentin. In addition, by developing an orthotopic mouse model, it has been suggested that PsA-3091 can effectively inhibit lung metastasis and tumor growth by regulating DOT1L activity and EMT biomarkers. In vivo and in vitro studies have confirmed that PsA-3091 exhibits excellent anti-tumor and anti-metastasis effects, and it can be speculated as a promising potential target for the treatment of patients with metastatic breast cancer (Byun et al., 2019). In addition, DOT1L inhibitors have also shown surprising performance in neuroblastoma (Wong et al., 2017) and colorectal cancer (Huang et al., 2017).
In recent years, abundant research evidence has shown that PRMT5, as an oncogene, can play an indispensable regulatory role in the pathological progression of various human cancers by promoting the proliferation, invasion, and metastasis of cancer cells. This indicates that PRMT5 may become a potential biomarker or therapeutic target for cancer. Regulation of epigenetics may be one of the main mechanisms by which PRMTs affect cell activities. Currently, PRMT1 and PRMT5 inhibitors have entered the first phase of clinical trials; thus, opening up a new path for the treatment of solid tumors and hematological malignancies (Jarrold and Davies, 2019) (NCT03666988) (Table 1 and Figure 2).
Erasers
In recent years, the regulatory role of epigenetic modifications in the occurrence and development of malignant tumors has received extensive attention. It is obvious that some epigenetic markers, whether in the form of post-translational modifications of histones or equivalent modifications of DNA, have proven to be non-permanent; i.e., removable. An enzyme called eraser appears immediately, which can remove these genetic markers, thereby resisting the writer's activity and leading to the regulation of gene expression (Biswas and Rao, 2018).
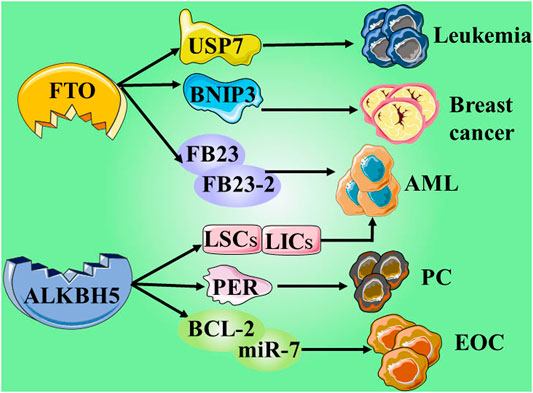
In the past few decades, with the rise of RNA epigenetics, the focus of N6-methyladenosine (m6A) RNA modification has gradually become a research hotspot in the field of biological sciences. In the early days, due to the incompatibility of technology and information, research on epigenetic modification was focused on the DNA and protein levels. Currently, the RNA epigenetic modification based on m6A has dynamic and reversible characteristics, and it is the most common mRNA epigenetic modification that exists in all higher eukaryotes. It involves fine regulation of many complex cellular processes, such as RNA processing, transportation, localization, translation, and degradation. Studies have shown that m6A can dynamically regulate RNA transport, localization, translation, and degradation through the abnormal expression of “writing”, “erasing”, and “reading” related factors. It can play a role in tumor development through a variety of mechanisms, including promotion or inhibition. Here, we mainly introduce the eraser, which can encode m6A demethylase to remove the m6A modification in the RNA molecule, which is the key to the reversibility of the m6A modification process (Roundtree et al., 2017). Currently, the identified “erasers” are mainly fat mass and obesity associated (FTO) protein and AlkB homolog 5 (ALKBH5) (Zou et al., 2016; Zhang et al., 2017). Although these two erasers have similar functions, they have different processes.
Studies have proved that FTO is not only limited to playing a major role in obesity-related diseases, but is also involved in the occurrence, development, and prognosis of a variety of cancers, including AML, glioblastoma, and breast cancer (Chen and Du, 2019; Lan et al., 2020). Long-term studies have shown that m6A modification is related to tumorigenesis, proliferation, invasion, and metastasis, and it acts as an oncogene or tumor suppressor gene in malignant tumors (Sun et al., 2019). Rhein, meclofenamic acid (MA), MO-I-500, fluorescein, and R-2-hydroxyglutarate (R-2HG) belong to a group of specific or non-specific FTO inhibitors that have been identified. Briefly, these small molecules are limited in their clinical applications due to their weak biological functions and low sensitivity and/or specificity (Huang et al., 2019).
Su et al. developed two highly effective FTO inhibitors, CS1 and CS2. Compared with the two previously reported FTO inhibitors (FB23–2 and MO-I-500), CS1 and CS2 showed better performance in inhibiting the viability of AML cells. Su et al. obtained a set of up- or down-regulated pathways through global gene set enrichment analysis (GSEA). Surprisingly, in the up-regulated pathway, CS1, CS2, and FTO shared most of the abundant signaling pathways and core genes. Among the down-regulated pathways, the pathways inhibited by FTO are also widely present in the pathways inhibited by CS1 or CS2. The results showed that CS1 and CS2 may be able to exhibit high-efficiency performance by regulating the basic signal pathway of FTO. Interestingly, further in vivo experiments showed that CS2 treatment significantly reduced leukemic infiltration and doubled overall survival. However, CS1 treatment did not exert any significant effect. Further analysis showed that poor solubility and absorption of CS1 may be the reason for its inability to function in the body, and Su et al. subsequently solved this problem. However, it is undeniable that the FTO inhibitors CS1 and CS2 with anti-tumor and low side effects derived from the data have great potential in clinical applications (Su et al., 2020). Su et al. also found that R-2HG can display anti-tumor activity by targeting the FTO/m6A/MYC/CEPPA signaling pathway. In terms of the mechanism, R-2HG inhibits the increase in fat mass and the expression of FTO, thereby increasing the modification of m6A RNA in leukemia cells sensitive to R-2HG; thus, leading to further reduction in the stability of MYC/CEPPA transcripts, and ultimately suppressing the related pathways (Su et al., 2018).
It is recognized that ubiquitin-specific protease 7 (USP7) has a wide range of substrates, and most of the substrates, such as p53, PTEN, and FOXO4, are related to tumor suppression, DNA repair, or immune response. Therefore, USP7 is a potential cancer treatment target. Studies have found that FTO can promote the expression of USP7 through demethylation and can increase the stability of USP7, which is expected to become a potential target for the treatment of human lung cancer (Li J. et al., 2019). FTO can directly interact with oncogenic factors E2F1 and Myc. Experiments showed that by inhibiting FTO, the translation function of the two factors can be disrupted, thereby inhibiting the proliferation and migration of cervical cancer cells. But the study also revealed an interesting point, i.e., the overexpression of oncogenic factors E2F1 and Myc can also counteract the proliferation or migration of cervical cancer cells induced by the lack of FTO, and weaken its inhibitory effect (Zou et al., 2019). Similarly, some studies have shown that the expression of FTO is related to the occurrence and prognosis of gastric cancer. Experimental data have shown that FTO in a low expression state has the ability to inhibit the proliferation, migration, and invasion of gastric cancer. However, high expression of FTO exerts an opposite result, which promotes the activity of gastric cancer cells (Xu et al., 2017). FTO also provided deep insights in AML (Li Z. et al., 2017; Huang et al., 2019), breast cancer (Niu et al., 2019; Xu et al., 2020), and melanoma (Yang et al., 2019).
ALKBH5, another m6A demethylase of messenger RNA (mRNA) in higher eukaryotes, has also shown impressive performance. However, compared with the research on FTO, the research on ALKBH5 is still not thorough and abundant. In a study published in 2021, it was reported that the expression of ALKBH5 can be activated by Myc, and that the expression can reduce the m6A level in some selected Myc suppressor genes. Further experiments have also found that inhibiting the overexpression of ALKBH5 or selected Myc inhibitor genes can effectively inhibit the growth of Myc-mediated B-cell lymphoma in vivo and in vitro (Wu et al., 2021). In another study on osteosarcoma, ALKBH5 was also found to inhibit the occurrence and development of tumors through m6A-dependent epigenetic silencing of the pre-miR-181b-1/YAP signal axis present in osteosarcoma (Yuan et al., 2021). It is worth mentioning a recent novel discovery that ALKBH5 is not involved in DNA repair, but studies have found that demethylation of ALKBH5 may play a supporting role in maintaining genome integrity. In terms of the mechanism, the overexpression of ALKBH5 reduces the level of 3-methylcytosine in genomic DNA and the cytotoxic effect of the DNA-damaging alkylating agent methyl methanesulfonate (Akula et al., 2021) (Table 1 and Figure 3; ; Fujii et al., 2013; Li et al., 2015; Barbieri et al., 2017; Karakashev et al., 2018; Ma et al., 2019; Chen et al. (2019); Li et al. (2019b); Niu et al. (2019); Zhu et al., 2019; Li et al. (2019a); Zou et al. (2019); Yang et al., 2020; Chen et al., 2020b; Li et al., 2020; Chen et al., 2020a; Huang et al., 2019; Su et al., 2020; Guo et al., 2020; Shen et al., 2020; Seo et al., 2021)
Interaction
Epigenetic modification is not a static entity, but it develops in a way dependent on the cellular environment and dynamically changes to cope with a complex environment. It can be simply understood as follows: epigenetic modifications undergo dynamic changes. Under a certain physiological environment, certain modifications will be added, and when external factors or physiological environments change, these newly added modifications will be removed. As mentioned earlier, epigenetic writers and erasers have proven to be ubiquitous in cancer, and epigenetic changes at different types and sites usually represent different meanings. This also provides many potential targets for cancer research and treatment. A large number of small molecule drugs are currently being developed mainly to target these epigenetic regulatory factors. Although only a few epigenetic drugs have been certified by the FDA for cancer treatment, a large number of excellent epigenetic drugs have gone through clinical research on cancer treatment and have achieved remarkable results (Ganesan et al., 2019). The above discussion has already been involved. However, there is no research linking epigenetic drugs and cancer treatment to prove whether there is a certain synergy. This point will be elaborated subsequently.
Enhancement of Chemotherapy Sensitivity
Chemotherapy is one of the most important methods for the treatment of malignant tumors. However, the resistance of tumor cells to chemotherapeutic drugs often leads to failure of chemotherapy. Therefore, it is particularly important to resensitize the patients to chemotherapy-resistant drugs. Among the existing writers, DNMT decitabine can resensitize the patients with ovarian cancer to platinum-based drugs, and its mechanism may involve demethylation and re-expression in DNA repair and immune activation pathways (Fang et al., 2018). Sara et al. also strongly agreed with this finding and believed that epigenetic therapy has great potential in ovarian cancer (Moufarrij et al., 2019).
In breast cancer, Ye et al. found that the restoration of Spalt-like transcription factor 2 (SALL2) is mediated by DNMT inhibition, which can make tamoxifen-resistant breast cancer sensitive to tamoxifen therapy again. This discovery reveals and represents a potential clinical target that can be used for the treatment of tamoxifen-resistant breast cancer. These patients can benefit from the combination therapy of tamoxifen and DNMT inhibitors (Ye et al., 2019).
In colorectal cancer, Wei et al. confirmed that zebularine, a DNMT inhibitor with low toxicity, overcomes hypoxia-induced resistance to oxaliplatin in HCT116 cells by down-regulating the expression of hypoxia-inducible factor-1α (HIF-1α). It also provides a new strategy for the treatment of colorectal cancer, which is to overcome the resistance to oxaliplatin by increasing the hydroxylation of HIF-1α (Wei et al., 2020).
Similarly, in gefitinib-resistant PC9/AB2 cells, the combination of the EZH2 inhibitor GSK343 and gefitinib could significantly inhibit the activity of drug-resistant cells, reduce the migration ability of the cells, and induce the apoptosis of drug-resistant cells (Gong et al., 2019). EZH2 also showed surprising results in reversing the chemotherapy resistance of cervical cancer cells (Cai et al., 2016), cisplatin resistance in epithelial ovarian cancer (Liu et al., 2014), melanoma resistance to immunotherapy (Emran et al., 2019), and resistance of prostate cancer (Shankar et al., 2020), and it can be described as the gospel for patients.
Not only the writer, but the eraser is also excellent at improving the resistance to chemicals (Xiang et al., 2020). Zhou et al. demonstrated for the first time that FTO with elevated gene levels in cervical squamous cell carcinoma (CSCC) tissues can enhance the resistance of CSCC to radiotherapy and chemotherapy. The mechanism may be upregulation of β-catenin and subsequent activation of excision repair cross-complementation group 1 (ERCC1), and ultimately it was attributed to the demethylation of FTO (Zhou et al., 2018a). According to a report in 2020, malignant glioma is one of the deadly primary brain tumors in adults. Li et al. found that the inhibition of FTO can enhance the anti-tumor effect of the chemotherapeutic drug temozolomide in malignant glioma, and its mechanism may involve the MYC-miR-155/23a cluster-MXI1 feedback loop (Xiao et al., 2020). Compared with the well-researched FTO, there are relatively few studies on the resistance of ALKBH. Alkylating agents have broad-spectrum effects. It has been reported that it has anti-tumor, immunosuppressive, and other therapeutic effects, as well as carcinogenic, teratogenic, mutagenic, and bone marrow suppressive effects. Although alkylating agents have high toxicity and carcinogenic potential, they are still important first-line anti-tumor drugs for many highly aggressive and metastatic cancers (Sauter and Gillingham, 2020). Tran et al. inhibited the DNA repair activity of the ALKBH enzyme by targeting glutamine metabolism, leading to the accumulation of DNA alkylation damage, thereby increasing the sensitivity of cells to alkylating agents (Tran et al., 2017). The combination of drugs has achieved the effect of improving the efficacy and reducing the side effects of alkylating agents, which can be considered as a new strategy. In addition, the glutaminase inhibitor CB-839 has entered clinical trials for determining its safety and activity (Riess et al., 2021).
Inhibition of Proliferation and Promotion of Apoptosis
As a DNMT inhibitor, RG108 has been proven to effectively inhibit the proliferation of endometrial cancer cells and block the G2/M phase of the cell cycle to induce apoptosis, and it can be regarded as a promising drug candidate for endometrial cancer patients (Yang et al., 2017). Coincidentally, zebularine, another DNMT inhibitor, is thought to induce apoptosis in gastric cancer cells through the mitochondrial pathway (Tan et al., 2013). In addition, zebularine has also been shown to inhibit the proliferation of lung cancer A549 cells and HeLa cervical cancer cells through cell cycle arrest and apoptosis (You and Park, 2012; You and Park, 2014). Zhang et al. also stated that the combined application of DNMT and mTOR signals can inhibit the formation and proliferation of colorectal cancer (Zhang et al., 2009).
Kazuya Ishiguro et al. discussed the role of DOT1L in the development of multiple myeloma, and they believed that the inhibitory effect of DOT1L may be a new therapy for myeloma. Further experiments showed that the use of DOT1L inhibitors in multiple myeloma can induce cell cycle arrest and apoptosis, and can strongly inhibit cell proliferation in vitro (Ishiguro et al., 2019). Li et al. explored the effects of DOT1L inhibitor EPZ-5676 in combination with chemotherapy drugs on the proliferation and apoptosis of human ALL cells, and they concluded that the combination of these drugs has a synergistic inhibitory effect on proliferation, and low concentrations of EPZ-5676 combined with different chemotherapy drugs cause synergistic induction of apoptosis and have pro-apoptotic effects (Li LH. et al., 2017). However, the achievements of EZH2 in inhibiting proliferation and promoting apoptosis are countless, and they have been observed in colorectal cancer (Xu et al., 2018; Li X. et al., 2019), breast cancer (Han et al., 2018), and bladder cancer (Chen et al., 2019).
Similarly, FTO and ALKBH, the only known erasers, also have great potential in inhibiting proliferation and promoting apoptosis. In the study by Tang et al., it was suggested that FTO is necessary for the proliferation of pancreatic cancer cells by using RNA interference and knocking out FTO. With respect to its basic mechanism, Tang et al. believed that FTO is related to the interaction of MYC proto-oncogene and BHLH transcription factor (Tang et al., 2019). In the review of FTO by Wang et al., they believed that FTO is involved in a variety of biological processes, such as cancer cell apoptosis, proliferation, migration, metastasis, and stem cell self-renewal in human cancer. These modulations mainly rely on the communication between FTO and specific signal pathways, including PI3K/AKT, MAPK and mTOR signal pathways (Wang et al., 2020).
With respect to another type of eraser, ALKBH5, studies have found that silencing of ALKBH5 can significantly increase the proliferation, migration, and invasion of pancreatic cancer cells, and its overexpression will cause the opposite effect (Tang et al., 2020). In the study by Jin et al., it is believed that effective inhibition of the m6A modification of ALKBH5 may constitute a potential treatment strategy for lung cancer. Further experiments showed that ALKBH5 inhibited tumor proliferation and metastasis by reducing YTHDFs-mediated YAP expression and inhibiting miR-107/LATS2-mediated YAP activity in NSCLC (Jin et al., 2020). In general, both the writer and the eraser have their own unique advantages, attracting the attention of researchers.
Conclusion
Epigenetic modification is becoming a significant topic for research. With the development of next-generation sequencing technology, it will be possible explore more unknown areas for the world. As an important part of it. The resulting drugs that target epigenetic modifications will also be closely related to diseases, especially cancer. The change of epigenetic factors in cancer provides new targets for the development of cancer drugs, not only limited to a single therapy, but combined drugs that can achieve the desired efficacy. As with the emergence of any other new therapies, the fever of epigenetic modification is also essential to bring about adverse reactions. In the study of Allen et al., epigenetics affects gene expression, which may lead to adverse reactions of statins and have long-term effects on the health of statin users (Allen and Mamotte, 2017). In addition, the general public’s attitude toward new therapies is not very positive. Paradoxically, any new thing is also pursued by the public. In the endless research on epigenetic modification, a large number of options will bring the dawn on cancer and even more refractory diseases.
Author Contributions
J-yZ and HS designed and managed this article. HS, W-mZ and BL wrote the original manuscript. LL, HS, AS, and J-yZ reviewed this manuscript.
Funding
This work is supported by the National Natural Science Foundation of China (U1903126, 81773888 and 81802103), the Natural Science Foundation of Guangdong Province (2020A1515010605) and Project of High-Level Talents in AHUTCM (2019rcZD001).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fphar.2021.690057/full#supplementary-material
References
Abbotts, R., Topper, M. J., Biondi, C., Fontaine, D., Goswami, R., Stojanovic, L., et al. (2019). DNA Methyltransferase Inhibitors Induce a BRCAness Phenotype that Sensitizes NSCLC to PARP Inhibitor and Ionizing Radiation. Proc. Natl. Acad. Sci. USA 116 (45), 22609–22618. doi:10.1073/pnas.1903765116
Akula, D., O’Connor, T. R., and Anindya, R. (2021). Oxidative Demethylase ALKBH5 Repairs DNA Alkylation Damage and Protects against Alkylation-Induced Toxicity. Biochem. Biophysical Res. Commun. 534, 114–120. doi:10.1016/j.bbrc.2020.12.017
Allen, S. C., and Mamotte, C. D. S. (2017). Pleiotropic and Adverse Effects of Statins-Do Epigenetics Play a Role? J. Pharmacol. Exp. Ther. 362 (2), 319–326. doi:10.1124/jpet.117.242081
Allis, C. D., and Jenuwein, T. (2016). The Molecular Hallmarks of Epigenetic Control. Nat. Rev. Genet. 17 (8), 487–500. doi:10.1038/nrg.2016.59
Bannister, A. J., and Kouzarides, T. (2011). Regulation of Chromatin by Histone Modifications. Cell Res 21 (3), 381–395. doi:10.1038/cr.2011.22
Barbieri, I., Tzelepis, K., Pandolfini, L., Shi, J., Millán-Zambrano, G., and Robson, S. C. (2017). Promoter-Bound METTL3 Maintains Myeloid Leukaemia by m(6)A-Dependent Translation Control. Nature 552, 126–131. doi:10.1038/nature24678
Biswas, S., and Rao, C. M. (2018). Epigenetic Tools (The Writers, the Readers and the Erasers) and Their Implications in Cancer Therapy. Eur. J. Pharmacol. 837, 8–24. doi:10.1016/j.ejphar.2018.08.021
Bitler, B. G., Aird, K. M., Garipov, A., Li, H., Amatangelo, M., Kossenkov, A. V., et al. (2015). Synthetic Lethality by Targeting EZH2 Methyltransferase Activity in ARID1A-Mutated Cancers. Nat. Med. 21 (3), 231–238. doi:10.1038/nm.3799
Byun, W. S., Kim, W. K., Han, H. J., Chung, H.-J., Jang, K., Kim, H. S., et al. (2019). Targeting Histone Methyltransferase DOT1L by a Novel Psammaplin A Analog Inhibits Growth and Metastasis of Triple-Negative Breast Cancer. Mol. Ther. - Oncolytics 15, 140–152. doi:10.1016/j.omto.2019.09.005
Cai, L., Wang, Z., and Liu, D. (2016). Interference with Endogenous EZH2 Reverses the Chemotherapy Drug Resistance in Cervical Cancer Cells Partly by Up-Regulating Dicer Expression. Tumor Biol. 37 (5), 6359–6369. doi:10.1007/s13277-015-4416-9
Calo, E., and Wysocka, J. (2013). Modification of Enhancer Chromatin: what, How, and Why? Mol. Cel 49 (5), 825–837. doi:10.1016/j.molcel.2013.01.038
Chen, J., and Du, B. (2019). Novel Positioning from Obesity to Cancer: FTO, an m6A RNA Demethylase, Regulates Tumour Progression. J. Cancer Res. Clin. Oncol. 145 (1), 19–29. doi:10.1007/s00432-018-2796-0
Chen, Z., Du, Y., Liu, X., Chen, H., Weng, X., Guo, J., et al. (2019). EZH2 Inhibition Suppresses Bladder Cancer Cell Growth and Metastasis via the JAK2/STAT3 Signaling Pathway. Oncol. Lett. 18 (1), 907–915. doi:10.3892/ol.2019.10359
Chen, S., Li, Y., Zhi, S., Ding, Z., Wang, W., and Peng, Y. (2020a). WTAP Promotes Osteosarcoma Tumorigenesis by Repressing HMBOX1 Expression in an m(6)A-Dependent Manner. Cell Death Dis. 11, 659. doi:10.1038/s41419-020-02847-6
Chen, X., Xu, M., Xu, X., Zeng, K., Liu, X., and Pan, B. (2020b). METTL14-Mediated N6-Methyladenosine Modification of SOX4 mRNA Inhibits Tumor Metastasis in Colorectal Cancer. Mol. Cancer 19, 106. doi:10.1186/s12943-020-01220-7
Chiappinelli, K. B., Strissel, P. L., Desrichard, A., Li, H., Henke, C., Akman, B., et al. (2015). Inhibiting DNA Methylation Causes an Interferon Response in Cancer via dsRNA Including Endogenous Retroviruses. Cell 162 (5), 974–986. doi:10.1016/j.cell.2015.07.011
Daigle, S. R., Olhava, E. J., Therkelsen, C. A., Majer, C. R., Sneeringer, C. J., Song, J., et al. (2011). Selective Killing of Mixed Lineage Leukemia Cells by a Potent Small-Molecule DOT1L Inhibitor. Cancer Cell 20 (1), 53–65. doi:10.1016/j.ccr.2011.06.009
Dawson, M. A., and Kouzarides, T. (2012). Cancer Epigenetics: from Mechanism to Therapy. Cell 150 (1), 12–27. doi:10.1016/j.cell.2012.06.013
Du, W., Xu, A., Huang, Y., Cao, J., Zhu, H., Yang, B., et al. (2020). The Role of Autophagy in Targeted Therapy for Acute Myeloid Leukemia. Autophagy, 1–15. doi:10.1080/15548627.2020.1822628
Emran, A. A., Chatterjee, A., Rodger, E. J., Tiffen, J. C., Gallagher, S. J., Eccles, M. R., et al. (2019). Targeting DNA Methylation and EZH2 Activity to Overcome Melanoma Resistance to Immunotherapy. Trends Immunol. 40 (4), 328–344. doi:10.1016/j.it.2019.02.004
Fang, F., Cardenas, H., Huang, H., Jiang, G., Perkins, S. M., Zhang, C., et al. (2018). Genomic and Epigenomic Signatures in Ovarian Cancer Associated with Resensitization to Platinum Drugs. Cancer Res. 78 (3), 631–644. doi:10.1158/0008-5472.Can-17-1492
Fujii, T., Shimada, K., Anai, S., Fujimoto, K., Konishi, N., et al. (2013). ALKBH2, a Novel AlkB Homologue, Contributes to Human Bladder Cancer Progression by Regulating MUC1 Expression. Cancer Sci. 104, 321–327. doi:10.1111/cas.12089
Ganesan, A., Arimondo, P. B., Rots, M. G., Jeronimo, C., and Berdasco, M. (2019). The Timeline of Epigenetic Drug Discovery: from Reality to Dreams. Clin. Epigenet 11 (1), 174. doi:10.1186/s13148-019-0776-0
Gong, H., Yuan, Y., Li, Y., Zhang, H., Li, Y., Li, W., et al. (2019). [Role of EZH2 Inhibitor Combined with Gefitinib in EGFR-TKIs Resistant Lung Cancer Cells]. Zhongguo Fei Ai Za Zhi 22 (5), 255–263. doi:10.3779/j.issn.1009-3419.2019.05.01
Guo, X., Li, K., Jiang, W., Hu, Y., Xiao, W., Huang, Y., et al. (2020). RNA demethylase ALKBH5 Prevents Pancreatic Cancer Progression by Posttranscriptional Activation of PER1 in an m6A-YTHDF2-Dependent Manner. Mol. Cancer 19, 91. doi:10.1186/s12943-020-01158-w
Han, L., Zhang, H.-C., Li, L., Li, C.-X., Di, X., and Qu, X. (2018). Downregulation of Long Noncoding RNA HOTAIR andEZH2Induces Apoptosis and Inhibits Proliferation, Invasion, and Migration of Human Breast Cancer Cells. Cancer Biother. Radiopharm. 33 (6), 241–251. doi:10.1089/cbr.2017.2432
Harvey, Z. H., Chen, Y., and Jarosz, D. F. (2018). Protein-Based Inheritance: Epigenetics beyond the Chromosome. Mol. Cel 69 (2), 195–202. doi:10.1016/j.molcel.2017.10.030
Holoch, D., and Moazed, D. (2015). RNA-mediated Epigenetic Regulation of Gene Expression. Nat. Rev. Genet. 16 (2), 71–84. doi:10.1038/nrg3863
Huang, T., Lin, C., Zhong, L. L. D., Zhao, L., Zhang, G., Lu, A., et al. (2017). Targeting Histone Methylation for Colorectal Cancer. Therap Adv. Gastroenterol. 10 (1), 114–131. doi:10.1177/1756283x16671287
Huang, Y., Su, R., Sheng, Y., Dong, L., Dong, Z., Xu, H., et al. (2019). Small-Molecule Targeting of Oncogenic FTO Demethylase in Acute Myeloid Leukemia. Cancer Cell 35 (4), 677–691. doi:10.1016/j.ccell.2019.03.006
Ishiguro, K., Kitajima, H., Niinuma, T., Ishida, T., Maruyama, R., Ikeda, H., et al. (2019). DOT1L Inhibition Blocks Multiple Myeloma Cell Proliferation by Suppressing IRF4-MYC Signaling. Haematologica 104 (1), 155–165. doi:10.3324/haematol.2018.191262
Italiano, A., Soria, J.-C., Toulmonde, M., Michot, J.-M., Lucchesi, C., Varga, A., et al. (2018). Tazemetostat, an EZH2 Inhibitor, in Relapsed or Refractory B-Cell Non-hodgkin Lymphoma and Advanced Solid Tumours: a First-In-Human, Open-Label, Phase 1 Study. Lancet Oncol. 19 (5), 649–659. doi:10.1016/s1470-2045(18)30145-1
Jarrold, J., and Davies, C. C. (2019). PRMTs and Arginine Methylation: Cancer's Best-Kept Secret? Trends Mol. Med. 25 (11), 993–1009. doi:10.1016/j.molmed.2019.05.007
Jin, D., Guo, J., Wu, Y., Yang, L., Wang, X., Du, J., et al. (2020). m6A Demethylase ALKBH5 Inhibits Tumor Growth and Metastasis by Reducing YTHDFs-Mediated YAP Expression and Inhibiting miR-107/lats2-Mediated YAP Activity in NSCLC. Mol. Cancer 40 (1), 1–24. doi:10.1186/s12943-020-01161-1
Karakashev, S., Zhu, H., Wu, S., Yokoyama, Y., Bitler, B. G., Park, P. H., et al. (2018). CARM1-Expressing Ovarian Cancer Depends on the Histone Methyltransferase EZH2 Activity. Nat. Commun. 9, 631. doi:10.1038/s41467-018-03031-3
Khan, H., Vale, C., Bhagat, T., and Verma, A. (2013). Role of DNA Methylation in the Pathogenesis and Treatment of Myelodysplastic Syndromes. Semin. Hematol. 50 (1), 16–37. doi:10.1053/j.seminhematol.2013.01.001
Koch, A., Joosten, S. C., Feng, Z., de Ruijter, T. C., Draht, M. X., Melotte, V., et al. (2018). Analysis of DNA Methylation in Cancer: Location Revisited. Nat. Rev. Clin. Oncol. 15 (7), 459–466. doi:10.1038/s41571-018-0004-4
Kulis, M., and Esteller, M. (2010). DNA Methylation and Cancer. Adv. Genet. 70, 27–56. doi:10.1016/b978-0-12-380866-0.60002-2
Lan, N., Lu, Y., Zhang, Y., Pu, S., Xi, H., Nie, X., et al. (2020). FTO - A Common Genetic Basis for Obesity and Cancer. Front. Genet. 559138, 1–12. doi:10.3389/fgene.2020.559138
Lee, J.-Y., and Kong, G. (2015). DOT1L: a New Therapeutic Target for Aggressive Breast Cancer. Oncotarget 6 (31), 30451–30452. doi:10.18632/oncotarget.5860
Li, J., Han, Y., Zhang, H., Qian, Z., Jia, W., Gao, Y., et al. (2019a). The m6A Demethylase FTO Promotes the Growth of Lung Cancer Cells by Regulating the m6A Level of USP7 mRNA. Biochem. Biophysical Res. Commun. 512 (3), 479–485. doi:10.1016/j.bbrc.2019.03.093
Li, L. H., Wang, J., and Ke, X. Y. (2017a). [Effects of DOT1L Inhibitor EPZ-5676 Combined with Chemotherapeutic Drugs on Prolifiration and Apoptosis of RS 4;11 Cells]. Zhongguo Shi Yan Xue Ye Xue Za Zhi 25 (5), 1334–1341. doi:10.7534/j.issn.1009-2137.2017.05.010
li, X., Wang, L., Cao, X., Zhou, L., Xu, C., Cui, Y., et al. (2020). Casticin Inhibits Stemness of Hepatocellular Carcinoma Cells Via Disrupting The Reciprocal Negative Regulation Between DNMT1 and miR-148a-3p. Toxicol. Appl. Pharmacol. 396, 114998. doi:10.1016/j.taap.2020.114998
Li, X., Xing, J., Wang, H., and Yu, E. (2019b). The SLC34A2-ROS-HIF-1-Induced Up-Regulation of EZH2 Expression Promotes Proliferation and Chemo-Resistance to Apoptosis in Colorectal Cancer. Biosci. Rep. 112 (5), 1–15. doi:10.1042/bsr20180268
Li, L. H., Liu, X. D., Guo, X. F., Liu, X., Luo, J. M., and Zhang, Y. X. (2015). Expression and Clinical Significance of DNMT in Patients With Chronic Myeloid Leukemia. Zhongguo Shi Yan Xue Ye Xue Za Zhi 23, 1547–1550. doi:10.7534/j.issn.1009-2137.2015.06.003
Li, Z., Weng, H., Su, R., Weng, X., Zuo, Z., Li, C., et al. (2017b). FTO Plays an Oncogenic Role in Acute Myeloid Leukemia as a N 6 -Methyladenosine RNA Demethylase. Cancer Cell 31 (1), 127–141. doi:10.1016/j.ccell.2016.11.017
Liu, L., Guo, J., Yu, L., Cai, J., Gui, T., Tang, H., et al. (2014). miR-101 Regulates Expression of EZH2 and Contributes to Progression of and Cisplatin Resistance in Epithelial Ovarian Cancer. Tumor Biol. 35 (12), 12619–12626. doi:10.1007/s13277-014-2585-6
Lonetti, A., Pession, A., and Masetti, R. (2019). Targeted Therapies for Pediatric AML: Gaps and Perspective. Front. Pediatr. 7, 463. doi:10.3389/fped.2019.00463
Lue, J. K., and Amengual, J. E. (2018). Emerging EZH2 Inhibitors and Their Application in Lymphoma. Curr. Hematol. Malig Rep. 13 (5), 369–382. doi:10.1007/s11899-018-0466-6
Ma, L., Yan, Y., Bai, Y., Yang, Y., Pan, Y., Gang, X., et al. (2019). Overcoming EZH2 Inhibitor Resistance by Taxane in PTEN-Mutated Cancer. Theranostics 9 (17), 5020–5034. doi:10.7150/thno.34700
Moufarrij, S., Dandapani, M., Arthofer, E., Gomez, S., Srivastava, A., Lopez-Acevedo, M., et al. (2019). Epigenetic Therapy for Ovarian Cancer: Promise and Progress. Clin. Epigenet 11 (1), 7. doi:10.1186/s13148-018-0602-0
Nacev, B. A., Jones, K. B., Intlekofer, A. M., Yu, J. S. E., Allis, C. D., Tap, W. D., et al. (2020). The Epigenomics of Sarcoma. Nat. Rev. Cancer 20 (10), 608–623. doi:10.1038/s41568-020-0288-4
Nebbioso, A., Tambaro, F. P., Dell’Aversana, C., and Altucci, L. (2018). Cancer Epigenetics: Moving Forward. Plos Genet. 14 (6), e1007362. doi:10.1371/journal.pgen.1007362
Niu, Y., Lin, Z., Wan, A., Chen, H., Liang, H., Sun, L., et al. (2019). RNA N6-Methyladenosine Demethylase FTO Promotes Breast Tumor Progression through Inhibiting BNIP3. Mol. Cancer 18 (1), 46. doi:10.1186/s12943-019-1004-4
Riess, J. W., Frankel, P., Shackelford, D., Dunphy, M., Badawi, R. D., Nardo, L., et al. (2021). Phase 1 Trial of MLN0128 (Sapanisertib) and CB-839 HCl (Telaglenastat) in Patients with Advanced NSCLC (NCI 10327): Rationale and Study Design. Clin. Lung Cancer 22 (1), 67–70. doi:10.1016/j.cllc.2020.10.006
Roundtree, I. A., Evans, M. E., Pan, T., and He, C. (2017). Dynamic RNA Modifications in Gene Expression Regulation. Cell 169 (7), 1187–1200. doi:10.1016/j.cell.2017.05.045
Sapienza, C., and Issa, J.-P. (2016). Diet, Nutrition, and Cancer Epigenetics. Annu. Rev. Nutr. 36, 665–681. doi:10.1146/annurev-nutr-121415-112634
Sauter, B., and Gillingham, D. (2020). DNA Damaging Agents in Chemical Biology and Cancer. chimia (aarau) 74 (9), 693–698. doi:10.2533/chimia.2020.693
Seo, S. I., Yoon, J. H., Byun, H. J., Lee, S. K., et al. (2021). HOTAIR Induces Methylation of PCDH10, a Tumor Suppressor Gene, by Regulating DNMT1 and Sponging with miR-148b in Gastric Adenocarcinoma. Yonsei Med J 62, 118–128. doi:10.3349/ymj.2021.62.2.118
Shankar, E., Franco, D., Iqbal, O., Moreton, S., Kanwal, R., and Gupta, S. (2020). Dual Targeting of EZH2 and Androgen Receptor as a Novel Therapy for Castration-Resistant Prostate Cancer. Toxicol. Appl. Pharmacol. 404, 115200. doi:10.1016/j.taap.2020.115200
Shen, C., Sheng, Y., Zhu, A. C., Robinson, S., Jiang, X., Dong, L., et al. (2020). RNA Demethylase ALKBH5 Selectively Promotes Tumorigenesis and Cancer Stem Cell Self-Renewal in Acute Myeloid Leukemia. Cell Stem Cell 27, 64–80.e69. doi:10.1016/j.stem.2020.04.009
Skvortsova, K., Stirzaker, C., and Taberlay, P. (2019). The DNA Methylation Landscape in Cancer. Essays Biochem. 63 (6), 797–811. doi:10.1042/ebc20190037
Stein, E. M., Garcia-Manero, G., Rizzieri, D. A., Tibes, R., Berdeja, J. G., Savona, M. R., et al. (2018). The DOT1L Inhibitor Pinometostat Reduces H3K79 Methylation and Has Modest Clinical Activity in Adult Acute Leukemia. Blood 131 (24), 2661–2669. doi:10.1182/blood-2017-12-818948
Stoll, S., Wang, C., and Qiu, H. (2018). DNA Methylation and Histone Modification in Hypertension. Ijms 19 (4), 1174. doi:10.3390/ijms19041174
Su, R., Dong, L., Li, C., Nachtergaele, S., Wunderlich, M., Qing, Y., et al. (2018). R-2HG Exhibits Anti-tumor Activity by Targeting FTO/m6A/MYC/CEBPA Signaling. Cell 172 (1-2), 90–105. doi:10.1016/j.cell.2017.11.031
Su, R., Dong, L., Li, Y., Gao, M., Han, L., Wunderlich, M., et al. (2020). Targeting FTO Suppresses Cancer Stem Cell Maintenance and Immune Evasion. Cancer Cell 38 (1), 79–96. doi:10.1016/j.ccell.2020.04.017
Sun, T., Wu, R., and Ming, L. (2019). The Role of m6A RNA Methylation in Cancer. Biomed. Pharmacother. 112, 108613. doi:10.1016/j.biopha.2019.108613
Tan, W., Zhou, W., Yu, H.-g., Luo, H.-S., and Shen, L. (2013). The DNA Methyltransferase Inhibitor Zebularine Induces Mitochondria-Mediated Apoptosis in Gastric Cancer Cells In Vitro and In Vivo. Biochem. Biophysical Res. Commun. 430 (1), 250–255. doi:10.1016/j.bbrc.2012.10.143
Tang, B., Yang, Y., Kang, M., Wang, Y., Wang, Y., Bi, Y., et al. (2020). m6A Demethylase ALKBH5 Inhibits Pancreatic Cancer Tumorigenesis by Decreasing WIF-1 RNA Methylation and Mediating Wnt Signaling. Mol. Cancer 19 (1), 3. doi:10.1186/s12943-019-1128-6
Tang, X., Liu, S., Chen, D., Zhao, Z., and Zhou, J. (2019). The Role of the Fat Mass and Obesity-associated P-rotein in the P-roliferation of P-ancreatic C-ancer C-ells. Oncol. Lett. 17 (2), 2473–2478. doi:10.3892/ol.2018.9873
Tran, T. Q., Ishak Gabra, M. B., Lowman, X. H., Yang, Y., Reid, M. A., Pan, M., et al. (2017). Glutamine Deficiency Induces DNA Alkylation Damage and Sensitizes Cancer Cells to Alkylating Agents through Inhibition of ALKBH Enzymes. Plos Biol. 15 (11), e2002810. doi:10.1371/journal.pbio.2002810
Wang, J.-y., Chen, L.-j., and Qiang, P. (2020). The Potential Role of N6-Methyladenosine (m6A) Demethylase Fat Mass and Obesity-Associated Gene (FTO) in Human Cancers. Ott 13, 12845–12856. doi:10.2147/ott.S283417
Wei, T.-T., Lin, Y.-T., Tang, S.-P., Luo, C.-K., Tsai, C.-T., Shun, C.-T., et al. (2020). Metabolic Targeting of HIF-1α Potentiates the Therapeutic Efficacy of Oxaliplatin in Colorectal Cancer. Oncogene 39 (2), 414–427. doi:10.1038/s41388-019-0999-8
Wong, M., Tee, A. E. L., Milazzo, G., Bell, J. L., Poulos, R. C., Atmadibrata, B., et al. (2017). The Histone Methyltransferase DOT1L Promotes Neuroblastoma by Regulating Gene Transcription. Cancer Res. 77 (9), 2522–2533. doi:10.1158/0008-5472.Can-16-1663
Wu, G., Suo, C., Yang, Y., Shen, S., Sun, L., Li, S. T., et al. (2021). MYC Promotes Cancer Progression by Modulating M 6 A Modifications to Suppress Target Gene Translation. EMBO Rep. 22, e51519. doi:10.15252/embr.202051519
Xiang, M., Liu, W., Tian, W., You, A., and Deng, D. (2020). RNA N-6-Methyladenosine Enzymes and Resistance of Cancer Cells to Chemotherapy and Radiotherapy. Epigenomics 12 (9), 801–809. doi:10.2217/epi-2019-0358
Xiao, L., Li, X., Mu, Z., Zhou, J., Zhou, P., Xie, C., et al. (2020). FTO Inhibition Enhances the Antitumor Effect of Temozolomide by Targeting MYC-miR-155/23a Cluster-MXI1 Feedback Circuit in Glioma. Cancer Res. 80 (18), 3945–3958. doi:10.1158/0008-5472.Can-20-0132
Xu, D., Shao, W., Jiang, Y., Wang, X., Liu, Y., and Liu, X. (2017). FTO Expression Is Associated with the Occurrence of Gastric Cancer and Prognosis. Oncol. Rep. 38 (4), 2285–2292. doi:10.3892/or.2017.5904
Xu, M., Chen, X., Lin, K., Zeng, K., Liu, X., Pan, B., et al. (2018). The Long Noncoding RNA SNHG1 Regulates Colorectal Cancer Cell Growth through Interactions with EZH2 and miR-154-5p. Mol. Cancer 17 (1), 141. doi:10.1186/s12943-018-0894-x
Xu, Y., Ye, S., Zhang, N., Zheng, S., Liu, H., Zhou, K., et al. (2020). The FTO/miR‐181b‐3p/ARL5B Signaling Pathway Regulates Cell Migration and Invasion in Breast Cancer. Cancer Commun. 40 (10), 484–500. doi:10.1002/cac2.12075
Yang, L., Hou, J., Cui, X. H., Suo, L. N., and Lv, Y. W. (2017). RG108 Induces the Apoptosis of Endometrial Cancer Ishikawa Cell Lines by Inhibiting the Expression of DNMT3B and Demethylation of HMLH1. Eur. Rev. Med. Pharmacol. Sci. 21 (22), 5056–5064. doi:10.26355/eurrev_201711_13818
Yang, S., Wei, J., Cui, Y.-H., Park, G., Shah, P., Deng, Y., et al. (2019). m6A mRNA Demethylase FTO Regulates Melanoma Tumorigenicity and Response to Anti-PD-1 Blockade. Nat. Commun. 10 (1), 2782. doi:10.1038/s41467-019-10669-0
Yang, X., Zhang, S., He, C., Xue, P., Zhang, L., and He, Z. (2020). METTL14 Suppresses Proliferation and Metastasis of Colorectal Cancer by Down-Regulating Oncogenic Long Non-Coding RNA XIST. Mol. Cancer 19, 46. doi:10.1186/s12943-020-1146-4
Ye, L., Lin, C., Wang, X., Li, Q., Li, Y., Wang, M., et al. (2019). Epigenetic Silencing of SALL 2 Confers Tamoxifen Resistance in Breast Cancer. EMBO Mol. Med. 11 (12), e10638. doi:10.15252/emmm.201910638
You, B. R., and Park, W. H. (2014). Zebularine Inhibits the Growth of A549 Lung Cancer Cells via Cell Cycle Arrest and Apoptosis. Mol. Carcinog. 53 (11), 847–857. doi:10.1002/mc.22042
You, B. R., and Park, W. H. (2012). Zebularine Inhibits the Growth of HeLa Cervical Cancer Cells via Cell Cycle Arrest and Caspase-dependent Apoptosis. Mol. Biol. Rep. 39 (10), 9723–9731. doi:10.1007/s11033-012-1837-z
Yuan, Y., Yan, G., He, M., Lei, H., Li, L., Wang, Y., et al. (2021). ALKBH5 Suppresses Tumor Progression via an m6A-dependent Epigenetic Silencing of Pre-miR-181b-1/YAP Signaling axis in Osteosarcoma. Cell Death Dis 12 (1), 60. doi:10.1038/s41419-020-03315-x
Zaccara, S., Ries, R. J., and Jaffrey, S. R. (2019). Reading, Writing and Erasing mRNA Methylation. Nat. Rev. Mol. Cel Biol 20 (10), 608–624. doi:10.1038/s41580-019-0168-5
Zhang, S., Zhao, B. S., Zhou, A., Lin, K., Zheng, S., Lu, Z., et al. (2017). m 6 A Demethylase ALKBH5 Maintains Tumorigenicity of Glioblastoma Stem-like Cells by Sustaining FOXM1 Expression and Cell Proliferation Program. Cancer Cell 31 (4), 591–606. doi:10.1016/j.ccell.2017.02.013
Zhang, Y.-J., Zhao, S.-L., Tian, X.-Q., Sun, D.-F., Xiong, H., Dai, Q., et al. (2009). Combined Inhibition of Dnmt and mTOR Signaling Inhibits Formation and Growth of Colorectal Cancer. Int. J. Colorectal Dis. 24 (6), 629–639. doi:10.1007/s00384-009-0664-8
Zhou, S., Bai, Z.-L., Xia, D., Zhao, Z.-J., Zhao, R., Wang, Y.-Y., et al. (2018a). FTO Regulates the Chemo-Radiotherapy Resistance of Cervical Squamous Cell Carcinoma (CSCC) by Targeting β-catenin through mRNA Demethylation. Mol. Carcinogenesis 57 (5), 590–597. doi:10.1002/mc.22782
Zhou, Z., Li, H.-Q., and Liu, F. (2019b). DNA Methyltransferase Inhibitors and Their Therapeutic Potential. Ctmc 18 (28), 2448–2457. doi:10.2174/1568026619666181120150122
Zhu, H., Gan, X., Jiang, X., Diao, S., Wu, H., Hu, J., et al. (2019). ALKBH5 Inhibited Autophagy of Epithelial Ovarian Cancer Through miR-7 and BCL-2. J Exp. Clin. Cancer 38, 163. doi:10.1186/s13046-019-1159-2
Zou, D., Dong, L., Li, C., Yin, Z., Rao, S., and Zhou, Q. (2019). The m6A Eraser FTO Facilitates Proliferation and Migration of Human Cervical Cancer Cells. Cancer Cel Int 19, 321. doi:10.1186/s12935-019-1045-1
Keywords: epigenetics, writers, readers, erasers, cancer, epigenetic drugs
Citation: Zhou W-m, Liu B, Shavandi A, Li L, Song H and Zhang J-y (2021) Methylation Landscape: Targeting Writer or Eraser to Discover Anti-Cancer Drug. Front. Pharmacol. 12:690057. doi: 10.3389/fphar.2021.690057
Received: 02 April 2021; Accepted: 18 May 2021;
Published: 03 June 2021.
Edited by:
Yi-Chao Zheng, Zhengzhou University, ChinaReviewed by:
Faming Wang, Blood Center of Wisconsin, United StatesFazhi Yu, University of Science and Technology of China, China
Copyright © 2021 Zhou, Liu, Shavandi, Li, Song and Zhang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Lu Li, ZGVlcmx5ZWVAaG90bWFpbC5jb20=; Hang Song, dG9zb25naGFuZ0BzaW5hLmNvbQ==; Jian-ye Zhang, amlhbnllekAxNjMuY29t
†These authors have contributed equally to this work
 Wen-min Zhou1†
Wen-min Zhou1† Bin Liu
Bin Liu Lu Li
Lu Li Hang Song
Hang Song Jian-ye Zhang
Jian-ye Zhang