- 1Affiliated Hospital of Integrated Traditional Chinese and Western Medicine, Nanjing University of Chinese Medicine, Nanjing, China
- 2Department of Pharmacology, School of Pharmacy, Nanjing University of Chinese Medicine, Nanjing, China
- 3Department of General Surgery, Changshu No. 1 People’s Hospital, Affiliated Changshu Hospital of Soochow University, Changshu, China
- 4Jiangsu Key Laboratory for Pharmacology and Safety Evaluation of Chinese Materia Medica, Nanjing University of Chinese Medicine, Nanjing, China
- 5Yangtze River Pharmaceutical Group, Taizhou, China
Colorectal cancer is a multifactorial disease involving genetic, environmental, and lifestyle risk factors. Intestinal microbiota plays an important role in the occurrence and development of colorectal cancer. Studies have shown that the behavior of intestinal microbiota can lead to pathological changes in the host intestine, which can be divided into epigenetic changes and carcinogenic changes at the gene level, and ultimately promote the formation and development of colorectal cancer. Intestinal microbiota is mainly distributed in the intestinal epithelium, which is composed of a large number of microorganisms interacting with the host intestinal cells. It can affect the immune-inflammation and metabolism of the gastrointestinal tract, and may be used as a biomarker for disease diagnosis. Regulation of gut microbiota is a promising strategy for the prevention and treatment of colorectal cancer. This article reviews the role of intestinal microbiota in the development of colorectal cancer, including the related mechanisms of intestinal microbiota promoting colorectal cancer, the use of intestinal microbiota in the diagnosis of colorectal cancer, and the regulation of intestinal microbiota in the prevention or treatment of colorectal cancer.
Introduction
Colorectal cancer (CRC) accounts for about 10% of the new cancer cases worldwide. Its incidence rate is third among all cancers worldwide, and its mortality rate ranks second among all cancers (Bray et al., 2018). The formation of CRC involves complex changes of multiple genes, steps, and stages, and various genetic and environmental factors are related to the occurrence and development of CRC (Theodoratou et al., 2017). Recently, more and more attention has been paid to the role of microorganisms in cancer. Researchers have begun to study the impact of changes in microbial communities on cancer. These microbial communities have become an important influencing factor of some cancers, including CRC, liver cancer, breast cancer, and so on. The human intestinal tract is one of the most complex organs of the human body, which hosts tens of thousands of microorganisms, including bacteria, archaea, fungi, protozoa, and viruses, of which bacteria account for the majority (Gill et al., 2006). Intestinal microflora mainly act on intestinal epithelial cells, interact with intestinal cells, maintain intestinal environment, and play an important role in human health, such as energy metabolism (Turnbaugh et al., 2006) and immune regulation (Chung et al., 2012). The change in its relative abundance will disrupt the balance of the intestinal microenvironment, thus causing some diseases inside and outside the intestine. This review provides an overview of the relationship between intestinal microbiota and CRC, focusing on the mechanism of intestinal microbiota in CRC, and finally discusses the potential strategies for the prevention or treatment of CRC based on the regulation of intestinal microbiota.
Intestinal Microbiota and Colorectal Cancer
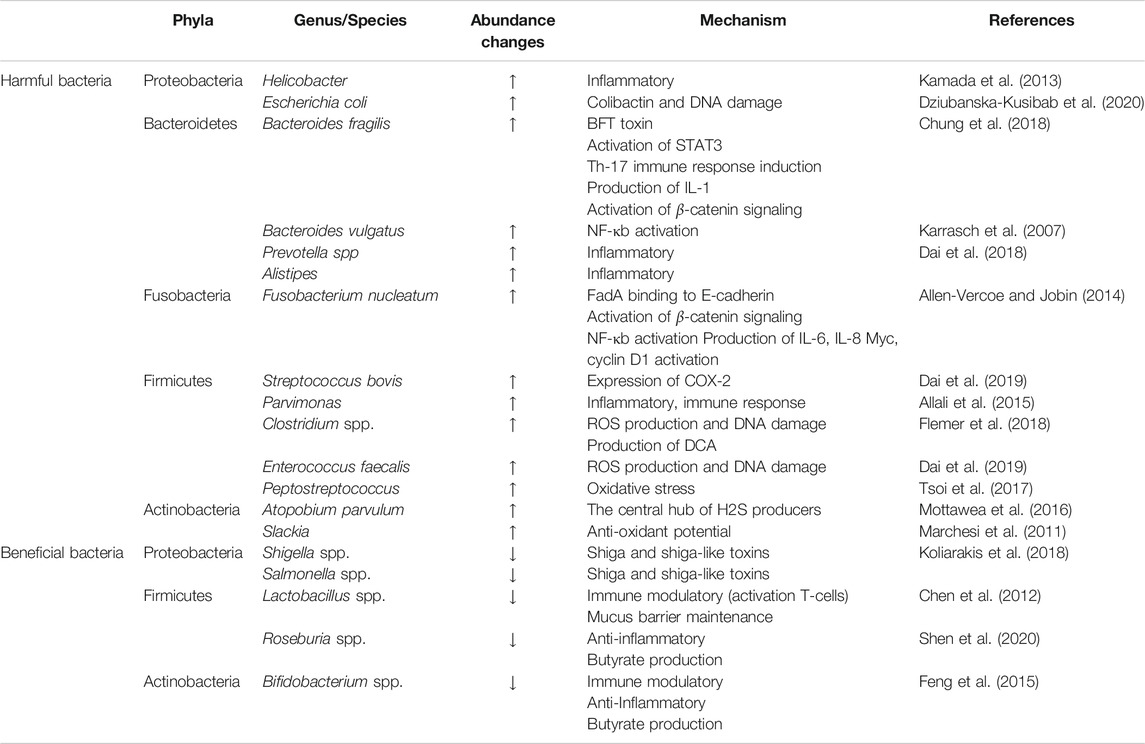
There are hundreds of microorganisms in the intestine, which form a symbiotic system with intestinal cells to maintain a dynamic balance and maintain the intestinal environment. Once the balance is destroyed, the intestinal flora will be out of balance, causing a series of intestinal diseases. As early as the 1960s, the relationship between intestinal flora and CRC was studied. In this study, through the induction of conventional rats and sterile rats, it can be found that Cycas has carcinogenic effect on conventional rats, but failed to cause cancer in sterile rats, which indicates that intestinal microorganisms play an important role in the carcinogenesis mediated by Cycas (Laqueur et al., 1967). Recently, through a case-control study of CRC patients, a research team analyzed the characteristics of fecal flora and blood inflammatory factors in various stages of colorectal tumorigenesis (benign polyps to advanced adenoma), identified 24 CRC-related bacteria, and revealed that the flora is involved in promoting the formation of CRC microenvironment in the process of gradual malignant transformation (Zhang et al., 2018). At present, the known flora associated with CRC mainly includes Fusobacterium nucleatum (F. nucleatum), Escherichia coli (E. coli), Bacteroides fragilis (B. fragilis), Campylobacter jejuni (C. jejuni), etc. (Dai et al., 2019) (Figure 1).
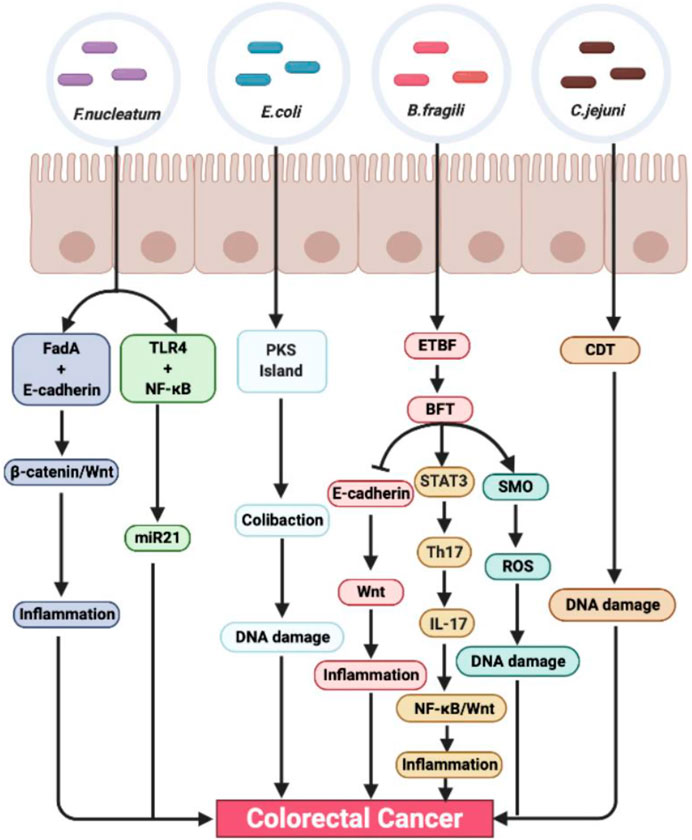
FIGURE 1. The role and mechanism of intestinal microbiota promoting colorectal cancer. Fusobacterium nucleatum, E. coli, B. fragili, and C. jejuni are closely related to the occurrence and development of colorectal cancer. It can stimulate inflammation and promote tumor formation through many inflammation-related signaling pathways, such as NF-κB, STAT3, and Wnt/β-catenin. At the same time, intestinal flora can also cause DNA damage, lead to gene mutation, and promote tumor formation through its own toxins.
Fusobacterium nucleatum
Fusobacterium nucleatum is a common symbiotic anaerobic Gram-negative bacteria in oral cavity. It is unique to human oral cavity and plays an important role in periodontal disease. It can bridge different microorganisms in biofilm through a variety of adhesins. More and more evidence show that F. nucleatum affects many stages of CRC development. Fusobacterium nucleatum can increase the proliferation of cancer cells through two different mechanisms: 1) The binding of FadA and E-cadherin drives the activation of β-catenin and Wnt pathway (Rubinstein et al., 2013). 2) The activation of TLR4 and NF-κB leads to the increased expression of carcinogenic miR21 (Yang et al., 2017). The researchers verified the above conclusion by constructing an intestinal tumor model in Apcmin/+ mice, and found that compared with the control group, F. nucleatum can cause Apcmin/+ mice to exhibit high levels of proliferating cell nuclear antigen, promote cell proliferation, and increase the level of inflammatory mediators in mouse serum and myeloid cells infiltrating into the tumor, thus promoting the development of tumor (Kostic et al., 2013). In addition, F. nucleatum may also affect the metastatic transmission. It can work together with other species and genera of bacteria such as Bacteroides, Selenomonas, and Prevotella to promote the metastasis of CRC, and can be isolated from liver and lymph node metastasis (Bullman et al., 2017). After CRC treatment, F. nucleatum can inhibit mir-18a* and mir-4802 involved in autophagy, thus increasing the risk of CRC recurrence and chemotherapy resistance (Yu et al., 2017). In addition, F. nucleatum also promotes the glycolysis and carcinogenesis of CRC by upregulating the histone modification of ENO1 (a key component in the glycolysis pathway) and other genes by upregulating the long non-coding RNA enolase 1 (ENO1)-intron transcript (ENO1-IT1)] (Hong et al., 2020).
Escherichia coli
Escherichia coli is a kind of anaerobic Gram-negative symbiotic bacteria, which colonizes the human intestinal tract shortly after birth. However, some virulent strains of E. coli with pathogenicity island can infect human gastrointestinal system and induce diseases. E. coli strains are divided into four main phylogenetic groups: A, B1, B2, and D. Fecal strains often belong to groups A and B1, while the most common pathogenic strains carrying virulence factors belong to groups B2 and D (Smati et al., 2013). Some pathogenic strains in groups B2 and D are associated with inflammatory bowel disease (IBD), and IBD is a risk factor for CRC. Colibactin is a bacterial gene toxin first mentioned by Nougayrede et al., in 2006. It is mainly produced by E. coli. Some studies have shown that colibactin is closely related to CRC, which can cause double-strand DNA breakage, eukaryotic cell cycle arrest, and chromosome aberration, and induce CRC. In the CRC mouse model, colibactin-producing E. coli can promote the occurrence of colon cancer (Arthur et al., 2012). In human CRC cells, colibactin preferentially destroys DNA rich in AT-enriched hexamer sequence motifs, which is related to different DNA shape characteristics and electrostatic potential. Somatic mutations of colibactin targets were analyzed in thousands of cancer genomes, and it was found that colibactin-binding sequences aggregated in CRC (Dziubanska-Kusibab et al., 2020).
Bacteroides fragilis
Bacteroides fragilis is a kind of common bacteria in the intestinal tract, which is symbiotic with the host. Most bacteria help the human body digest food and maintain intestinal health. However, in some cases, these bacteria produce a toxin that disrupts the cells on the surface of the gut, thereby promoting CRC. There are two subtypes of B. fragilis, namely, nontoxic B. fragilis and enterotoxigenic B. fragilis (ETBF), which produce a kind of B. fragilis toxin (BFT). BFT can lead to the destruction of epithelial barrier and the cleavage of E-cadherin. At the same time, the cleavage of E-cadherin can activate the Wnt signal transduction pathway, stimulate mucosal inflammation, and promote the formation of colon tumor. In addition, the STAT3 pathway is the target of colon transformation, and it is also necessary for the development of Th17 cells. ETBF promotes the production of IL-17 b y Th17 cells through the rapid activation of the STAT3 pathway, which further promotes the activation of the NF-κB and Wnt pathways and the production of intestinal inflammatory tumor microenvironment. This process depends on the production of IL-17, and injection of IL-17 blocking antibody in mice can inhibit tumor formation (Dejea et al., 2013). ETBF can also upregulate the expression of spermine oxidase (SMO) in colonic epithelial cells, thus increasing the SMO-dependent reactive oxygen species (ROS), promoting the release of inflammatory cytokines and causing DNA damage, and ultimately promoting the development of CRC.
Campylobacter jejuni
As early as 20 years previously, it was reported that some Campylobacter, including C. jejuni, had cytolethal distending toxin (CDT) (Lara-Tejero and Galan, 2000). CDT has dnase activity, leading to DNA double-strand breaks. Zheng He et al. (Yusuf et al., 2019) found that C. jejuni 81–176 isolated from human body was colonized in sterile Apcmin/+ mice, which could significantly increase the number and size of tumor, and the macro transcriptome and intestinal flora of mice were also significantly changed. C. jejuni has a mutated CDTb subunit, which makes the bacteria unable to produce CDT, thus inhibiting its carcinogenesis in vivo and reducing the DNA damage response of cells and intestinal organs.
Mechanism of Intestinal Microbiota Promoting Colorectal Cancer
The changes in intestinal microbiota abundance and intestinal microecological structure will lead to an imbalance of intestinal microbiota. Once the intestinal microbiota is unbalanced, the number of probiotics in the intestine will decrease, and the number of pathogenic bacteria will increase. Pathogenic bacteria will secrete a variety of toxic factors, damage intestinal epithelial cells, and cause chronic inflammatory reaction. Meanwhile, in the process of inflammation, a variety of cytokines and chemokines will be released to activate inflammation-related signaling pathways, thus promoting CRC. In addition, epigenetic modification enables host cells to alter gene expression and sequence. Microorganisms can interact with host genomes dynamically through the interface of epigenetic modification. Epigenetics also plays an important role in the occurrence and development of CRC.
Intestinal Microbiota and Immune Response
Intestinal microbiota promotes the maturation and regulation of mucosal and systemic immune systems through innate and adaptive immune cells. For example, the activation of intestinal cells, macrophages, pattern recognition receptors, and highly specific receptors on the surface of T cells and B cells in the mucosa is highly regulated by intestinal microorganisms (Lucas et al., 2017). Studies have shown that intestinal microflora can activate mucosal macrophages through intestinal mucosal M cells and trigger innate immune response, thus promoting the occurrence and development of CRC (Grivennikov et al., 2012). In adaptive immune response, including humoral response, soluble tumor antigen is recognized by specific B cells, matures, and produces tumor antigen-specific antibody with the help of CD4+ T cells (Vesely et al., 2011). Adaptive immunity plays an important role in regulating the composition of intestinal microflora, and the intestinal microbiota of mice lacking adaptive immunity will change accordingly (Kato et al., 2014; Zhang et al., 2015). Helper T cells, T cells, and B cells secrete immunoglobulin A (IgA) and participate in tumorigenesis through adaptive immune response (Palm et al., 2015). The main mechanism of T cells affecting the composition of intestinal microorganisms is to assist B cells to produce IgA in the intestine, and IgA is mainly secreted into the intestinal cavity, which can bind and cover the intestinal microorganisms and participate in shaping the intestinal microbiota.
Intestinal Microbiota and Immune Signal Pathway
IBD is an inflammatory bowel disease, including ulcerative colitis and Crohn's disease. CRC is closely related to IBD. Intestinal microbiota can induce mucosal lesions through pro-inflammatory cytokines, thus promoting IBD and increasing the risk of CRC (Ullman and Itzkowitz, 2011; Manichanh et al., 2012). Inflammatory cells release cytokines, such as IL-6, IL-8, IL-17, and TNF-α, which play an important role in the development of CRC. Studies have shown that Enterococcus faecalis (E. faecalis) infection can cause colitis and express TGF-β in intestinal epithelial cells, thus activating the Smad4 signaling pathway (Szigeti et al., 2012). E. faecalis can also produce ROS and H2O2, leading to DNA damage, thus activating NF-κB signaling pathway, stimulating inflammation, and affecting the occurrence and development of CRC. At the same time, it seems to be involved in the bystander effect of COX-2, that is, macrophages release TNF-α, causing chromosome instability and cell transformation, leading to CRC (Wang et al., 2013). In addition, Helicobacter pylori (H. pylori) can cause colorectal epithelial damage through IL-8-mediated inflammatory response, thus promoting the occurrence and development of CRC (Rubin et al., 2012). IL-17 is an important cytokine produced by Th17 cells, and the activation of STAT3 is required for Th17 cell growth. Two transcription factors, NF-κB and STAT3, are necessary for inflammation to promote the occurrence and development of cancer. ETBF can rapidly activate the STAT3 signaling pathway and promote Th17 cells to secrete IL-17; thus, IL-17-dependent NF-κB and Wnt pathways are activated to establish inflammatory tumor microenvironment in the gut (Goktuna et al., 2016; Housseau et al., 2016).
Intestinal Microbiota and Epigenetic Modification
Epigenetic modification is heritable and potentially reversible. It is the central mechanism of transcriptional response to environmental cues and regulates gene expression through DNA modification, histone modification, and non-coding RNA. Studies have shown that intestinal flora can directly or indirectly regulate epigenetic modification and affect the occurrence and development of CRC. Epigenetic mechanisms that play an important role in the occurrence and development of CRC include DNA methylation, histone modification, miRNA and non-coding RNA, and nucleosome localization (Sharma et al., 2010). miRNA-21 and miRNA-200 b are two carcinogenic miRNAs, which are often upregulated in colorectal cancer cells. However, the expression levels of miRNA-21 and miRNA-200 b were significantly decreased in HT-29 cells treated with Leuconostoc mesenteroides (Zununi Vahed et al., 2017). In addition, intestinal microbiota metabolites can regulate cell differentiation through epigenetics, such as T cell development. Symbiotic bacteria, such as Clostridium, can promote the production of colon ptreg cells through its fermentation product butyric acid, which is an HDAC inhibitor and can promote the acetylation of Foxp3 promoter and histone H3 in CNS1 (Furusawa et al., 2013). At the same time, acetic acid and propionic acid promote the accumulation of colon Treg cells by activating GPR43 (Smith et al., 2013). Both induce Foxp3+ and CD4+ Treg cells, which play a key role in limiting the inflammatory response during carcinogenesis. In addition, butyric acid can increase the histone methylation of NF-κB1 promoter, thereby downregulating the expression of NF-κB1. Short-chain fatty acids (SCFAs) induce the expression of anti-inflammatory IL-10 R A (IL-10 receptor alpha subunit mRNA) and antimicrobial peptides through HDAC inhibitor function. Therefore, with the downregulation of NF-κB1, SCFAs can inhibit colitis through epigenetic modification. In addition, the fecal bacteria transplantation experiment found that the intestinal flora can also affect the expression of tumor-related genes by increasing DNA methylation of multiple CpG sites in CRC cells (Yu et al., 2015).
Intestinal Microbiota and Diagnosis of Colorectal Cancer
Intestinal flora plays an important role in the diagnosis of CRC. It can be used as a biomarker in the diagnosis and prediction of CRC. Some studies revealed the characteristics of intestinal microflora that can predict CRC through metagenomic analysis of feces (Table 1). In this study, through metagenomic analysis of feces of people from different countries, different living habits and eating habits, bacteria associated with CRC were identified, such as Fusobacterium, Porphyromonas, Parvimonas, etc., which can be used as biomarkers for the diagnosis of CRC. In addition, they have also obtained two important research results: 1) The discovery of a special bacterium in CRC patients, F. nucleatum, which is common in the oral cavity and respiratory tract. Compared with healthy individuals, the level of F. nucleatum in CRC patients is higher. 2) The discovery of the correlation between CRC and microbial enzyme genes, which is an important microorganism in the feces of CRC patients Choline trimethylamine lyase (cutC) gene is a biological enzyme, which can degrade choline in meat and release acetaldehyde (Wirbel et al., 2019). This finding was also confirmed by Thomas et al. (Thomas et al., 2019), which indicates that intestinal bacteria in feces may be able to predict CRC. In addition, the assessment of gene changes in stool samples can accurately reflect the status of intestinal microbiota, which may provide important clues for the diagnosis and cause and effect of diseases and contribute to the early diagnosis of cancer (Yachida et al., 2019).
Intestinal Microbiota and Prevention of Colorectal Cancer
Diet
CRC is a multifactorial disease, and changes in diet and lifestyle can aggravate the incidence and mortality rates of CRC. Excessive intake of animal protein and fat produces excessive secondary bile acids and hydrogen sulfide, which leads to barrier dysfunction, inflammation, DNA damage, genotoxicity, etc., and increases the risk of CRC, whereas dietary fiber produces SCFAs, such as butyrate, which plays anti-inflammatory and anti-tumor roles through cell metabolism, bacterial homeostasis, anti-proliferation, immune regulation, and epigenetics (Olagnier et al., 2018). In a follow-up study of more than 170,000 people with an average of 5.7 years, it was found that eating more red meat and drinking alcohol may increase the risk of CRC, while eating more fiber in breakfast may reduce the risk of CRC (Bradbury et al., 2020). In addition, in a case-control study, the correlation between flavonoids from different dietary sources and the risk of CRC was evaluated, and it was found that specific flavonoids, especially flavonoids from vegetables and fruits, were negatively correlated with the risk of colorectal cancer (Xu et al., 2016). Other studies have shown that a high-fiber diet can increase the diversity of SCFA-producing bacteria and the expression of butyrate receptor in mice, thereby inhibiting colon cancer (Bishehsari et al., 2018).
Fecal Microbiota Transplantation
In recent years, more and more studies have reported that fecal microbiota transplantation (FMT) has a good therapeutic effect on IBD, CRC, immune-related diseases, and other diseases. Routy et al. (Routy et al., 2018) found that intestinal flora improves the efficacy of anti-tumor immunotherapy based on PD-1, indicating that FMT can help fight tumors. The patients who responded to anti-PD-1 therapy had a varied species composition, such as Bifidobacterium, E. faecalis, etc. After anti-PD-1 treatment, the fecal bacteria from the patients with reaction can enhance the T cell effect and inhibit the development of tumor.
Probiotics, Prebiotics, Synbiotics, and Postbiotics
At present, the use of probiotics in the treatment and prevention of cancer has become a research hotspot. Probiotics are living microorganisms that, when given in sufficient quantity, will bring health benefits to the host (Hill et al., 2014). The most common probiotic strains are Bifidobacterium and Lactobacillus. The study found that Lactobacillus acidophilus and Lactobacillus plantarum have a potential preventive effect in patients with polyps or CRC. The study used 16 S rRNA to conduct preliminary gene detection on 77 samples of CRC, polyps, and healthy subjects, and used absolute real-time PCR to determine the copy number of bacteria per Gram of feces. The results confirmed that taking L. acidophilus and L. plantarum in people with a family history of CRC and patients with polyps may be a method to prevent, treat, or alleviate CRC (Zinatizadeh et al., 2018). In addition, Lactobacillus casei BL23, a probiotic-specific strain, can inhibit colitis-associated CRC, which can be used as a potential new strategy for the prevention and treatment of CRC. In the CRC mouse model induced by azoxymethane/dextran sulfate sodium (AOM/DSS), L. casei BL23 can reduce the histological score and proliferation parameters; reduce the level of cytokine IL-22; mediate the immunoregulation; upregulate the expression of caspase-7, caspase-9, and Bik; mediate the antiproliferative effect; and counteract the fecal flora imbalance induced by CRC in mice (Jacouton et al., 2017).
In addition, prebiotics are also a useful strategy to prevent CRC. Prebiotics are substrates that can be used by intestinal microorganisms and are beneficial to host health. The common prebiotics are fructans, inulin, fructooligosaccharides and galactooligosaccharides (Gibson et al., 2004; Bruno-Barcena and Azcarate-Peril, 2015). Galactooligosaccharides derived from lactulose (GOS-Lu) is a prebiotic preparation. In order to test its preventive effect on CRC, the Rattus norvegicus F344 animal model was established. The results showed that compared with the control group, the number of colon tumors in the GOS-Lu group was significantly reduced. At the same time, through the metagenomic sequencing of intestinal flora, it was found that the pro-inflammatory bacteria in the GOS-Lu group decreased significantly, while the beneficial bacteria increased significantly (Fernandez et al., 2018).
The combination of probiotics and prebiotics is called synbiotics, which have synergistic effects. Roller et al. found that the synbiotic combination of oligofructose-enriched inulin-based Lactobacillus rhamnosus and Bifidobacterium lactis can reduce the number of tumors by regulating the immune function in Peyer's patches, thus inhibiting the occurrence of CRC (Roller et al., 2004).
Postbiotics are soluble byproducts and metabolites secreted by intestinal microbiota, which have biological activity and thus interact with the host. Postbiotics have a protective effect on intestinal epithelial cells. Studies have shown that P40, a soluble protein derived from L. rhamnosus GG, can inhibit cytokine-induced epithelial cell apoptosis and intestinal barrier damage (Yan and Polk, 2012).
Conclusions
CRC, as a disease induced by many factors, is closely related to diet, the environment, and lifestyle. Intestinal microbiota plays an important role in the development of CRC by destroying the homeostasis of the microenvironment, changing immune response, producing toxic metabolites, and directly or indirectly affecting epigenetic modification. Several types of bacteria have been proven to promote CRC in various ways and mechanisms. As a biomarker, intestinal microbiota provides a new method for the early diagnosis of noninvasive CRC. At the same time, intestinal flora in feces can also help predict CRC, thus reducing the occurrence and development of CRC. However, due to the complexity of carcinogenic mechanism, the best biomarker for disease diagnosis has not yet been determined. CRC can be managed through healthy diet, the use of probiotics, and fecal transplantation; however, each method has certain limitations, such as in the use of probiotics, there will be individual differences in probiotics, by which under different pathogenic conditions, its role will change accordingly, so the use of probiotics must be targeted. Intestinal microbiota can ferment dietary fiber to produce SCFAs. Some bacteria have been identified as potential butyric acid producers, including FN (Vital et al., 2014). Butyrate is one of the most produced SCFAs. It can pass through the intestinal epithelium and reach the lamina propria, directly forming the mucosal immune response (Tilg et al., 2018). Most studies have shown that butyrate can inhibit inflammation and canceration of the colon, but some studies have found contrasting results (butyrate paradox) in which the beneficial or harmful effects of butyrate on the mucosal immune system depend on its concentration and immune environment (De Almeida et al., 2019). In addition, dietary fiber intake is not enough to produce high levels of butyrate for the prevention of inflammation and cancer. Therefore, it is necessary to conduct more research on interventions involving high-level dietary fiber intake in the near future. With the deepening of various studies, the etiology and pathogenesis of CRC will be further revealed, and the role of microorganisms in the formation, progress, and therapeutic response of CRC will be further confirmed. Intestinal microorganisms is anticipated to become an important part of the prevention and treatment of CRC in the future.
Author Contributions
YY and LR were in charge of searching all the relative papers and writing this manuscript. JY was in charge of drawing the picture. PC gave her valuable and professional suggestions and guide in organizing and drafting this manuscript. JS helped to revise the manuscript. BZ was in charge of summarizing and drawing the table.
Funding
This work was supported by the National Natural Science Foundation of China (Grant Numbers 81973498 and 81774283), the Joint Open Project of Jiangsu Key Laboratory for Pharmacology and Safety Evaluation of Chinese Materia Medica and Yangtze River Pharmaceutical Group (No. JKLPSE202003), Suzhou Clinical Key disease Diagnosis and Treatment Technology foundation (LCZX201823), Suzhou Science and Technology Development Program (SYS2020058) and Changshu Science and Technology Development Program (CS202029).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank the two reviewers for significantly improving this manuscript.
Abbreviations
AMPS, antimicrobial peptides; AOM, azoxymethane; BFT, Bacteroides fragilis toxin; CDT, cytolethal distending toxin; CRC, colorectal cancer; CutC, choline trimethylamine lyase; DSS, dextran sulfate sodium; ETBF, enterotoxigenic Bacteroides fragilis; IgA, immunoglobulin A; IBD, inflammatory bowel disease; IL-10 R A, IL-10 receptor alpha subunit mRNA; ROS, reactive oxygen species; SCFAs, short-chain fatty acids; SMO, spermine oxidase.
References
Allali, I., Delgado, S., Marron, P. I., Astudillo, A., Yeh, J. J., Ghazal, H., et al. (2015). Gut microbiome compositional and functional differences between tumor and non-tumor adjacent tissues from cohorts from the US and Spain. Gut Microbes 6 (3), 161–172. doi:10.1080/19490976.2015.1039223
Allen-Vercoe, E., and Jobin, C. (2014). Fusobacterium and Enterobacteriaceae: important players for CRC?. Immunol. Lett. 162 (2 Pt A), 54–61. doi:10.1016/j.imlet.2014.05.014
Almeida, C. V. D., Camargo, M. R. d., Russo, E., and Amedei, A. (2018). Role of diet and gut microbiota on colorectal cancer immunomodulation. Wjg 25 (2), 151–162. doi:10.3748/wjg.v25.i2.151
Arthur, J. C., Perez-Chanona, E., Mühlbauer, M., Tomkovich, S., Uronis, J. M., Fan, T.-J., et al. (2012). Intestinal inflammation targets cancer-inducing activity of the microbiota. Science 338 (6103), 120–123. doi:10.1126/science.1224820
Bishehsari, F., Engen, P., Preite, N., Tuncil, Y., Naqib, A., Shaikh, M., et al. (2018). Dietary fiber treatment corrects the composition of gut microbiota, promotes SCFA production, and suppresses colon carcinogenesis. Genes 9 (2), 102. doi:10.3390/genes9020102
Bradbury, K. E., Murphy, N., and Key, T. J. (2020). Diet and colorectal cancer in UK Biobank: a prospective study. Int. J. Epidemiol. 49 (1), 246–258. doi:10.1093/ije/dyz064
Bray, F., Ferlay, J., Soerjomataram, I., Siegel, R. L., Torre, L. A., and Jemal, A. (2018). Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA: A Cancer J. Clinicians 68 (6), 394–424. doi:10.3322/caac.21492
Bruno-Barcena, J. M., and Azcarate-Peril, M. A. (2015). Galacto-oligosaccharides and colorectal cancer: feeding our intestinal probiome. J. Funct. Foods 12, 92–108. doi:10.1016/j.jff.2014.10.029
Bullman, S., Pedamallu, C. S., Sicinska, E., Clancy, T. E., Zhang, X., Cai, D., et al. (2017). Analysis ofFusobacteriumpersistence and antibiotic response in colorectal cancer. Science 358 (6369), 1443–1448. doi:10.1126/science.aal5240
Chen, W., Liu, F., Ling, Z., Tong, X., and Xiang, C. (2012). Human intestinal lumen and mucosa-associated microbiota in patients with colorectal cancer. PLoS One 7 (6), e39743, doi:10.1371/journal.pone.0039743
Chung, H., Pamp, S. J., Hill, J. A., Surana, N. K., Edelman, S. M., Troy, E. B., et al. (2012). Gut immune maturation depends on colonization with a host-specific microbiota. Cell 149 (7), 1578–1593. doi:10.1016/j.cell.2012.04.037
Chung, L., Thiele Orberg, E., Geis, A. L., Chan, J. L., Fu, K., DeStefano Shields, C. E., et al. (2018). Bacteroides fragilis toxin coordinates a pro-carcinogenic inflammatory cascade via targeting of colonic epithelial cells. Cell Host Microbe 23 (2), 203–214. doi:10.1016/j.chom.2018.01.007
Dai, Z., Coker, O. O., Nakatsu, G., Wu, W. K. K., Zhao, L., Chen, Z., et al. (2018). Multi-cohort analysis of colorectal cancer metagenome identified altered bacteria across populations and universal bacterial markers. Microbiome 6 (1), 70. doi:10.1186/s40168-018-0451-2
Dai, Z., Zhang, J., Wu, Q., Chen, J., Liu, J., Wang, L., et al. (2019). The role of microbiota in the development of colorectal cancer. Int. J. Cancer 145 (8), 2032–2041. doi:10.1002/ijc.32017
Dejea, C., Wick, E., and Sears, C. L. (2013). Bacterial oncogenesis in the colon. Future Microbiol. 8 (4), 445–460. doi:10.2217/fmb.13.17
Dziubańska-Kusibab, P. J., Berger, H., Battistini, F., Bouwman, B. A. M., Iftekhar, A., Katainen, R., et al. (2020). Colibactin DNA-damage signature indicates mutational impact in colorectal cancer. Nat. Med. 26 (7), 1063–1069. doi:10.1038/s41591-020-0908-2
Feng, Q., Liang, S., Jia, H., Stadlmayr, A., Tang, L., Lan, Z., et al. (2015). Gut microbiome development along the colorectal adenoma-carcinoma sequence. Nat. Commun. 6, 6528. doi:10.1038/ncomms7528
Fernández, J., Moreno, F. J., Olano, A., Clemente, A., Villar, C. J., and Lombó, F. (2018). A galacto-oligosaccharides preparation derived from lactulose protects against colorectal cancer development in an animal model. Front. Microbiol. 9, 2004. doi:10.3389/fmicb.2018.02004
Flemer, B., Herlihy, M., O'Riordain, M., Shanahan, F., and O'Toole, P. W. (2018). Tumour-associated and non-tumour-associated microbiota: Addendum. Gut Microbes 9 (4), 1–5. doi:10.1080/19490976.2018.1435246
Furusawa, Y., Obata, Y., Fukuda, S., Endo, T. A., Nakato, G., Takahashi, D., et al. (2013). Commensal microbe-derived butyrate induces the differentiation of colonic regulatory T cells. Nature 504 (7480), 446–450. doi:10.1038/nature12721
Gibson, G. R., Probert, H. M., Loo, J. V., Rastall, R. A., and Roberfroid, M. B. (2004). Dietary modulation of the human colonic microbiota: updating the concept of prebiotics. Nutr. Res. Rev. 17 (2), 259–275. doi:10.1079/NRR200479
Gill, S. R., Pop, M., Deboy, R. T., Eckburg, P. B., Turnbaugh, P. J., Samuel, B. S., et al. (2006). Metagenomic analysis of the human distal gut microbiome. Science 312 (5778), 1355–1359. doi:10.1126/science.1124234
Göktuna, S. I., Shostak, K., Chau, T.-L., Heukamp, L. C., Hennuy, B., Duong, H.-Q., et al. (2016). The prosurvival IKK-related kinase IKKε integrates LPS and IL17A signaling cascades to promote wnt-dependent tumor development in the intestine. Cancer Res. 76 (9), 2587–2599. doi:10.1158/0008-5472.CAN-15-1473
Grivennikov, S. I., Wang, K., Mucida, D., Stewart, C. A., Schnabl, B., Jauch, D., et al. (2012). Adenoma-linked barrier defects and microbial products drive IL-23/IL-17-mediated tumour growth. Nature 491 (7423), 254–258. doi:10.1038/nature11465
Hill, C., Guarner, F., Reid, G., Gibson, G. R., Merenstein, D. J., Pot, B., et al. (2014). The International Scientific Association for Probiotics and Prebiotics consensus statement on the scope and appropriate use of the term probiotic. Nat. Rev. Gastroenterol. Hepatol. 11 (8), 506–514. doi:10.1038/nrgastro.2014.66
Hong, J., Guo, F., Lu, S.-Y., Shen, C., Ma, D., Zhang, X., et al. (2020). F. nucleatum targets lncRNA ENO1-IT1 to promote glycolysis and oncogenesis in colorectal cancer. Gut gutjnl-2020, 322780. doi:10.1136/gutjnl-2020-322780
Housseau, F., Wu, S., Wick, E. C., Fan, H., Wu, X., Llosa, N. J., et al. (2016). Redundant innate and adaptive sources of IL17 production drive colon tumorigenesis. Cancer Res. 76 (8), 2115–2124. doi:10.1158/0008-5472.CAN-15-0749
Jacouton, E., Chain, F., Sokol, H., Langella, P., and Bermúdez-Humarán, L. G. (2017). Probiotic strain Lactobacillus casei BL23 prevents colitis-associated colorectal cancer. Front. Immunol. 8, 1553. doi:10.3389/fimmu.2017.01553
Kamada, N., Seo, S.-U., Chen, G. Y., and Núñez, G. (2013). Role of the gut microbiota in immunity and inflammatory disease. Nat. Rev. Immunol. 13 (5), 321–335. doi:10.1038/nri3430
Karrasch, T., Kim, J.-S., Muhlbauer, M., Magness, S. T., and Jobin, C. (2007). Gnotobiotic IL-10−/−;NF-κBEGFPMice reveal the critical role of TLR/NF-κB signaling in commensal bacteria-induced colitis. J. Immunol. 178 (10), 6522–6532. doi:10.4049/jimmunol.178.10.6522
Kato, L. M., Kawamoto, S., Maruya, M., and Fagarasan, S. (2014). The role of the adaptive immune system in regulation of gut microbiota. Immunol. Rev. 260 (1), 67–75. doi:10.1111/imr.12185
Koliarakis, I., Psaroulaki, A., Nikolouzakis, T. K., Kokkinakis, M., Sgantzos, M. N., Goulielmos, G., et al. (2018). Intestinal microbiota and colorectal cancer: a new aspect of research. J. BUON 23 (5), 1216–1234.
Kostic, A. D., Chun, E., Robertson, L., Glickman, J. N., Gallini, C. A., Michaud, M., et al. (2013). Fusobacterium nucleatum potentiates intestinal tumorigenesis and modulates the tumor-immune microenvironment. Cell Host Microbe 14 (2), 207–215. doi:10.1016/j.chom.2013.07.007
Laqueur, G. L., McDaniel, E. G., and Matsumoto, H. (1967). Tumor induction in germfree rats with methylazoxymethanol (MAM) and synthetic MAM acetate. J. Natl. Cancer Inst. 39, 355–371.
Lara-Tejero, M., and Galan, J. E. (2000). A bacterial toxin that controls cell cycle progression as a deoxyribonuclease I-like protein. Science 290 (5490), 354–357. doi:10.1126/science.290.5490.354
Lucas, C., Barnich, N., and Nguyen, H. T. T. (2017). Microbiota, inflammation and colorectal cancer. Int. J. Mol. Sci. 18 (6). doi:10.3390/ijms18061310
Manichanh, C., Borruel, N., Casellas, F., and Guarner, F. (2012). The gut microbiota in IBD. Nat. Rev. Gastroenterol. Hepatol. 9 (10), 599–608. doi:10.1038/nrgastro.2012.152
Marchesi, J. R., Dutilh, B. E., Hall, N., Peters, W. H. M., Roelofs, R., Boleij, A., et al. (2011). Towards the human colorectal cancer microbiome. PLoS One 6 (5), e20447, doi:10.1371/journal.pone.0020447
Mottawea, W., Chiang, C.-K., Mühlbauer, M., Starr, A. E., Butcher, J., Abujamel, T., et al. (2016). Altered intestinal microbiota-host mitochondria crosstalk in new onset Crohn's disease. Nat. Commun. 7, 13419. doi:10.1038/ncomms13419
Olagnier, D., Brandtoft, A. M., Gunderstofte, C., Villadsen, N. L., Krapp, C., Thielke, A. L., et al. (2018). Nrf2 negatively regulates STING indicating a link between antiviral sensing and metabolic reprogramming. Nat. Commun. 9 (1), 3506. doi:10.1038/s41467-018-05861-7
Palm, N. W., de Zoete, M. R., and Flavell, R. A. (2015). Immune-microbiota interactions in health and disease. Clin. Immunol. 159 (2), 122–127. doi:10.1016/j.clim.2015.05.014
Roller, M., Femia, A. P., Caderni, G., Rechkemmer, G., and Watzl, B. (2004). Intestinal immunity of rats with colon cancer is modulated by oligofructose-enriched inulin combined with Lactobacillus rhamnosus and Bifidobacterium lactis. Br. J. Nutr. 92 (6), 931–938. doi:10.1079/bjn20041289
Routy, B., Le Chatelier, E., Derosa, L., Duong, C. P. M., Alou, M. T., Daillère, R., et al. (2018). Gut microbiome influences efficacy of PD-1-based immunotherapy against epithelial tumors. Science 359 (6371), 91–97. doi:10.1126/science.aan3706
Rubin, D. C., Shaker, A., and Levin, M. S. (2012). Chronic intestinal inflammation: inflammatory bowel disease and colitis-associated colon cancer. Front. Immun. 3, 107. doi:10.3389/fimmu.2012.00107
Rubinstein, M. R., Wang, X., Liu, W., Hao, Y., Cai, G., and Han, Y. W. (2013). Fusobacterium nucleatum promotes colorectal carcinogenesis by modulating E-cadherin/β-catenin signaling via its FadA adhesin. Cell Host Microbe 14 (2), 195–206. doi:10.1016/j.chom.2013.07.012
Sharma, S., Kelly, T. K., and Jones, P. A. (2010). Epigenetics in cancer. Carcinogenesis 31 (1), 27–36. doi:10.1093/carcin/bgp220
Shen, J., Li, P., Liu, S., Liu, Q., Li, Y., Zhang, Z., et al. (2020). The chemopreventive effects of Huangqin-tea against AOM-induced preneoplastic colonic aberrant crypt foci in rats and omics analysis. Food Funct. 11 (11), 9634–9650. doi:10.1039/d0fo01731k
Smati, M., Clermont, O., Le Gal, F., Schichmanoff, O., Jaureguy, F., Eddi, A., et al. (2013). Real-time PCR for quantitative analysis of human commensal Escherichia coli populations reveals a high frequency of subdominant phylogroups. Appl. Environ. Microbiol. 79 (16), 5005–5012. doi:10.1128/AEM.01423-13
Smith, P. M., Howitt, M. R., Panikov, N., Michaud, M., Gallini, C. A., Bohlooly-Y, M., et al. (2013). The microbial metabolites, short-chain fatty acids, regulate colonic Treg cell homeostasis. Science 341 (6145), 569–573. doi:10.1126/science.1241165
Szigeti, R., Pangas, S. A., Nagy-Szakal, D., Dowd, S. E., Shulman, R. J., Olive, A. P., et al. (2012). SMAD4 haploinsufficiency associates with augmented colonic inflammation in select humans and mice. Ann. Clin. Lab. Sci. 42 (4), 401–408.
Theodoratou, E., Timofeeva, M., Li, X., Meng, X., and Ioannidis, J. P. A. (2017). Nature, nurture, and cancer risks: genetic and nutritional contributions to cancer. Annu. Rev. Nutr. 37, 293–320. doi:10.1146/annurev-nutr-071715-051004
Thomas, A. M., Manghi, P., Asnicar, F., Pasolli, E., Armanini, F., Zolfo, M., et al. (2019). Author Correction: metagenomic analysis of colorectal cancer datasets identifies cross-cohort microbial diagnostic signatures and a link with choline degradation. Nat. Med. 25 (12), 1948. doi:10.1038/s41591-019-0663-4
Tilg, H., Adolph, T. E., Gerner, R. R., and Moschen, A. R. (2018). The intestinal microbiota in colorectal cancer. Cancer Cell 33 (6), 954–964. doi:10.1016/j.ccell.2018.03.004
Tsoi, H., Chu, E. S. H., Zhang, X., Sheng, J., Nakatsu, G., Ng, S. C., et al. (2017). Peptostreptococcus anaerobius induces intracellular cholesterol biosynthesis in colon cells to induce proliferation and causes dysplasia in mice. Gastroenterology 152 (6), 1419–1433. doi:10.1053/j.gastro.2017.01.009
Turnbaugh, P. J., Ley, R. E., Mahowald, M. A., Magrini, V., Mardis, E. R., and Gordon, J. I. (2006). An obesity-associated gut microbiome with increased capacity for energy harvest. Nature 444 (7122), 1027–1031. doi:10.1038/nature05414
Ullman, T. A., and Itzkowitz, S. H. (2011). Intestinal inflammation and cancer. Gastroenterology 140 (6), 1807–1816. doi:10.1053/j.gastro.2011.01.057
Vesely, M. D., Kershaw, M. H., Schreiber, R. D., and Smyth, M. J. (2011). Natural innate and adaptive immunity to cancer. Annu. Rev. Immunol. 29, 235–271. doi:10.1146/annurev-immunol-031210-101324
Vital, M., Howe, A. C., and Tiedje, J. M. (2014). Revealing the bacterial butyrate synthesis pathways by analyzing (meta)genomic data. mBio 5 (2), e00889. doi:10.1128/mBio.00889-14
Wang, X., Allen, T. D., Yang, Y., Moore, D. R., and Huycke, M. M. (2013). Cyclooxygenase-2 generates the endogenous mutagen trans-4-hydroxy-2-nonenal in Enterococcus faecalis-infected macrophages. Cancer Prev. Res. 6 (3), 206–216. doi:10.1158/1940-6207.CAPR-12-0350
Wirbel, J., Pyl, P. T., Kartal, E., Zych, K., Kashani, A., Milanese, A., et al. (2019). Meta-analysis of fecal metagenomes reveals global microbial signatures that are specific for colorectal cancer. Nat. Med. 25 (4), 679–689. doi:10.1038/s41591-019-0406-6
Xu, M., Chen, Y.-M., Huang, J., Fang, Y.-J., Huang, W.-Q., Yan, B., et al. (2016). Flavonoid intake from vegetables and fruits is inversely associated with colorectal cancer risk: a case-control study in China. Br. J. Nutr. 116 (7), 1275–1287. doi:10.1017/S0007114516003196
Yachida, S., Mizutani, S., Shiroma, H., Shiba, S., Nakajima, T., Sakamoto, T., et al. (2019). Metagenomic and metabolomic analyses reveal distinct stage-specific phenotypes of the gut microbiota in colorectal cancer. Nat. Med. 25 (6), 968–976. doi:10.1038/s41591-019-0458-7
Yan, F., and Polk, D. B. (2012). Characterization of a probiotic-derived soluble protein which reveals a mechanism of preventive and treatment effects of probiotics on intestinal inflammatory diseases. Gut Microbes 3 (1), 25–28. doi:10.4161/gmic.19245
Yang, Y., Weng, W., Peng, J., Hong, L., Yang, L., Toiyama, Y., et al. (2017). Fusobacterium nucleatum increases proliferation of colorectal cancer cells and tumor development in mice by activating toll-like receptor 4 signaling to nuclear Factor−κB, and up-regulating expression of MicroRNA-21. Gastroenterology 152 (4), 851–866. doi:10.1053/j.gastro.2016.11.018
Yu, D.-H., Gadkari, M., Zhou, Q., Yu, S., Gao, N., Guan, Y., et al. (2015). Postnatal epigenetic regulation of intestinal stem cells requires DNA methylation and is guided by the microbiome. Genome Biol. 16, 211. doi:10.1186/s13059-015-0763-5
Yu, T., Guo, F., Yu, Y., Sun, T., Ma, D., Han, J., et al. (2017). Fusobacterium nucleatum promotes chemoresistance to colorectal cancer by modulating autophagy. Cell 170 (3), 548–563. doi:10.1016/j.cell.2017.07.008
Yusuf, I. O., Chen, H.-M., Cheng, P.-H., Chang, C.-Y., Tsai, S.-J., Chuang, J.-I., et al. (2019). Fibroblast growth factor 9 activates anti-oxidative functions of Nrf2 through ERK signalling in striatal cell models of Huntington's disease. Free Radic. Biol. Med. 130, 256–266. doi:10.1016/j.freeradbiomed.2018.10.455
Zhang, H., Sparks, J. B., Karyala, S. V., Settlage, R., and Luo, X. M. (2015). Host adaptive immunity alters gut microbiota. ISME J. 9 (3), 770–781. doi:10.1038/ismej.2014.165
Zhang, Y., Yu, X., Yu, E., Wang, N., Cai, Q., Shuai, Q., et al. (2018). Changes in gut microbiota and plasma inflammatory factors across the stages of colorectal tumorigenesis: a case-control study. BMC Microbiol. 18 (1), 92. doi:10.1186/s12866-018-1232-6
Zinatizadeh, N., Khalili, F., Fallah, P., Farid, M., Geravand, M., and Yaslianifard, S. (2018). Potential preventive effect of Lactobacillus acidophilus and Lactobacillus plantarum in patients with polyps or colorectal câncer. Arq. Gastroenterol. 55 (4), 407–411. doi:10.1590/S0004-2803.201800000-87
Zununi Vahed, S., Barzegari, A., Rahbar Saadat, Y., Goreyshi, A., and Omidi, Y. (2017). Leuconostoc mesenteroides-derived anticancer pharmaceuticals hinder inflammation and cell survival in colon cancer cells by modulating NF-κB/AKT/PTEN/MAPK pathways. Biomed. Pharmacother. 94, 1094–1100. doi:10.1016/j.biopha.2017.08.033
Keywords: colorectal cancer, intestinal flora, immune-inflammation, metabolism, prevention and treatment of colorectal cancer
Citation: Ren L, Ye J, Zhao B, Sun J, Cao P and Yang Y (2021) The Role of Intestinal Microbiota in Colorectal Cancer. Front. Pharmacol. 12:674807. doi: 10.3389/fphar.2021.674807
Received: 02 March 2021; Accepted: 23 March 2021;
Published: 19 April 2021.
Edited by:
Jian Gao, Second Affiliated Hospital of Dalian Medical University, ChinaReviewed by:
Sizhi Paul Gao, Memorial Sloan Kettering Cancer Center, United StatesJing Li, China Pharmaceutical University, China
Copyright © 2021 Ren, Ye, Zhao, Sun, Cao and Yang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yang Yang, eW91bmcxNTcwQDEyNi5jb20=; Peng Cao, Y2FvX3BlbmdAbmp1Y20uZWR1LmNu
†These authors contributed equally to this work as co-first authors
 Lingli Ren1,2†
Lingli Ren1,2† Juan Ye
Juan Ye Peng Cao
Peng Cao Yang Yang
Yang Yang