- 1Department of Plant Molecular Biology and Bioinformatics, Tamil Nadu Agricultural University, Coimbatore, India
- 2Department of Plant Biotechnology, Tamil Nadu Agricultural University, Coimbatore, India
- 3Centre for Plant Molecular Biology and Biotechnology, Tamil Nadu Agricultural University, Coimbatore, India
- 4Tamil Nadu Agricultural University, Coimbatore, India
Severe Acute Respiratory Syndrome Corona Virus 2 (SARS-CoV-2) being a causative agent for global pandemic disease nCOVID’19, has acquired much scientific attention for the development of effective vaccines and drugs. Several attempts have been made to explore repurposing existing drugs known for their anti-viral activities, and test the traditional herbal medicines known for their health benefiting and immune-boosting activity against SARS-CoV-2. In this study, efforts were made to examine the potential of 605 phytochemicals from 37 plant species (of which 14 plants were endemic to India) and 139 antiviral molecules (Pubchem and Drug bank) in inhibiting SARS-CoV-2 multiple protein targets through a virtual screening approach. Results of our experiments revealed that SARS-CoV-2 MPro shared significant disimilarities against SARS-CoV MPro and MERS-CoV MPro indicating the need for discovering novel drugs. This study has screened the phytochemical cyanin (Zingiber officinale) which may exhibit broad-spectrum inhibitory activity against main proteases of SARS-CoV-2, SARS-CoV and MERS-CoV with binding energies of (−) 8.3 kcal/mol (−) 8.2 kcal/mol and (−) 7.7 kcal/mol respectively. Amentoflavone, agathisflavone, catechin-7-o-gallate and chlorogenin were shown to exhibit multi-target inhibitory activity. Further, Mangifera indica, Anacardium occidentale, Vitex negundo, Solanum nigrum, Pedalium murex, Terminalia chebula, Azadirachta indica, Cissus quadrangularis, Clerodendrum serratum and Ocimum basilicumaree reported as potential sources of phytochemicals for combating nCOVID’19. More interestingly, this study has highlighted the anti-viral properties of the traditional herbal formulation “Kabasura kudineer” recommended by AYUSH, a unit of Government of India. Short listed phytochemicals could be used as leads for future drug design and development. Genomic analysis of identified herbal plants will help in unraveling molecular complexity of therapeutic and anti-viral properties which proffer lot of chance in the pharmaceutical field for researchers to scout new drugs in drug discovery.
Introduction
Novel Coronavirus disease (nCOVID’19) caused by SARS-CoV-2 virus has become a global threat and WHO has declared it as a pandemic. nCOVID’19 is the third life threatening virus in the SARS family of viruses after SARS-CoV occurred during 2002–03 and MERS-CoV during 2012 (Lee et al., 2003; Cheng et al., 2007; Zaki et al., 2012; De Groot et al., 2013). It is named as a novel Coronavirus as it shares significant dissimilarity against other members of the SARS family of viruses viz., SARS-CoV (30%) and MERS-CoV (60%) (Xu et al., 2020). Its unique genetic makeup has made it not responsive to available medical treatments and necessitated search for novel targets for vaccine development and drugs for effective prevention and treatment of nCOVID’19.
Exploding increase in the nCOVID’19 affected cases has brought this globe to a halt. Scientific community is trying to unravel genome complexity of nCOVID’19 for identifying novel targets for development of vaccines, screen available anti-viral drugs for effective management and short listing effective botanicals for therapeutic interventions. This has resulted in the accumulation enormous genomic information of nCOVID’19 in the public domain (https://www.ncbi.nlm.nih.gov/genbank/sars-cov-2-seqs/). Genomic analysis of nCOVID’19 revealed that it is approximately 30 kb in size (NCBI Accession # NC_045512) and further investigations identified three key genes viz., 1) coronavirus main protease (3CLpro)/papain-like protease (PLpro); 2) RNA-dependent RNA polymerase (RdRp) and 3) spike glycoprotein (S protein) as potential targets for drug designing (Elfiky, 2020; Sampangi-Ramaiah et al., 2020).
Screening of existing antiviral drugs including interferon α (IFN-α), lopinavir/ritonavir, chloroquine phosphate, ribavirin, chloroquine, hydroxychloroquine and arbidol is in progress and many of these experiments require pre-clinical and clinical validation (Elfiky, 2020). Ineffectiveness of existing anti-viral drugs have made the doctors to resort using traditional medicines in nCOVID’19 treatments (Yang et al., 2014). Several attempts have been made to exploit the potential of several herbal products having potential to inhibit the main protease (Mpro)/chymotrypsin-like protease (3CLpro) using molecular modeling and docking studies (Colson et al., 2020; Dong et al., 2020; Touret and de Lamballerie, 2020). Elfiky (2020) made an attempt to screen 27 different ligands present in commonly used herbals of Indian cuisines against SARS-CoV-2 Main protease and identified 15 different ligands effective in binding the viral protease. Yang et al. (2014) made a systematic review of herbal drugs used in the effective treatment of SARS-CoV and MERS-CoV and emphasized the urgent need for evolving procedures involving complementary and alternative treatments in managing nCOVID’19. Studies conducted so far have made attempts by using limited number of ligands which may hinder discovery of effective viral inhibitor in the herbal gene pool. In this context, short listing potential herbal drugs effective against nCOVID’19 through in silico docking of globally available ligands and validating them through laboratory and clinical trials is one of the viable approaches in managing this pandemic. India is one of the richest biodiversity centers in the world and known for its vast repository of medicinal plants. Considering India’s richest biodiversity of herbal medicinal plants and regular use of such medicinal plants in Indian health care system, the present study was undertaken to screen about 744 ligands including small molecules and phytochemicals from 37 different Indian medicinal plants against seven different protein targets of nCOVID’19 through molecular docking. Protein-Ligand interactions were analyzed carefully to shortlist potential small molecules and phytochemicals for drug development.
Materials and Methods
Phylogenetic Analysis of Main Protease of nCOVID’19
Protein sequence of SARS-CoV-2 encoding for main protease (PDB ID: 5R81) was used for PSI BLAST (NCBI) (Altschul et al., 1997) search to identify its homologs for understanding the evolutionary relationship with main proteases of other viruses. Multiple sequence alignment and phylogenetic analysis of SARS-CoV-2 main protease with other viral proteins was done using MAFFT server (Katoh et al., 2019).
Virtual Screening of Herbal Ligands Against Potential Targets of nCOVID’19
Protein Targets
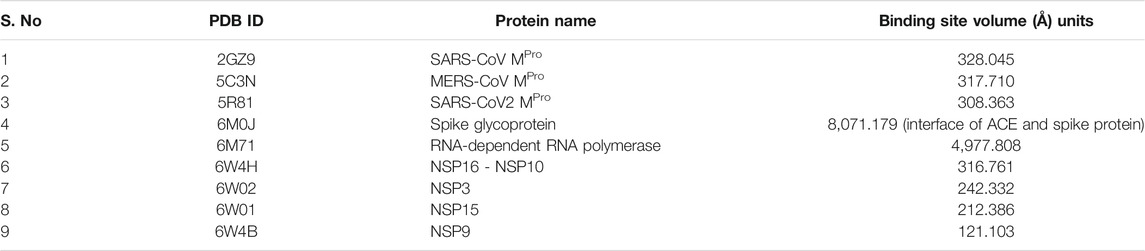
Coronavirus genome was reported to encode for 29 proteins, out of which the main protease is considered to be an important drug target. ORF1ab of the coronavirus genome contains 15 polypeptide chains encoding for non-structural proteins (NS proteins). Another part of the genome encodes for envelope and coat proteins. Availability of X-ray crystal structures for most of the proteins in the coronavirus two genome facilitates virtual screening to search for potential inhibitors. We have performed molecular docking of 744 ligands against seven different target proteins of the nCOVID’19 genome (Table 1). In addition, virtual screening was also performed against MPro of SARS CoV (PDB ID: 2GZ9) and MERS CoV (PDB ID: 5C3N) for identifying inhibitors against main protease of all three viruses and inhibitors specific to nCOVID’19.
Ligand Library Preparation
Chemical structures of all the small molecules were retrieved from Dukes database (1992–2016), PubChem (Kim et al., 2019) and DrugBank (Wishart et al., 2018). From the DrugBank database, 99 chemical structures of drugs approved for the treatment of respiratory diseases and compounds exhibiting antiviral activity were collected. Forty chemical compounds with COVID 19 antiviral property retrieved from pubchem database were also included in the ligand dataset. Information regarding the origin, traditional use and protocol were obtained from literature, Indian medicinal plants database (http://www.medicinalplants.in/) and Indian Medicinal Plants, Phytochemistry and Therapeutics (https://cb.imsc.res.in/imppat/home) database. Structures of 605 phytochemicals belonging to 37 different herbals and spices used in South Indian Traditional Medicine were also used for virtual screening (Supplementary Table S1). Among the 37 herbals, 14 herbals were found native to India (Table 2) namely, Abutilon indicum (Thuthi), Ocimum tenuiflorum (Tulsi), Pedalium murex (Peru nerinji), Anacyclus pyrethrum (Akara), Andrographis paniculata (Nilavembu), Pepper nigrum (Pepper), Azadirachta indica (Neem), Phyllanthus niruri (Keelanelli), Cissus_quadrangularis_L (Perandai), Plectranthus amboinicus (Karppuravalli), Clerodendrum serratum (Kanduparangi), Solanum torvum (sundakai), Solanum trilobatum (Thoothuvalai) and Terminalia chebula (Kadukai). Phytochemicals from different parts of the plant such as Root, Rhizome, pericarp leaves, Flower, Seed essential oil, Stem and Fruits was curated in order to find bioactive molecule that may have role in the inhibition of SARS CoV two targets.
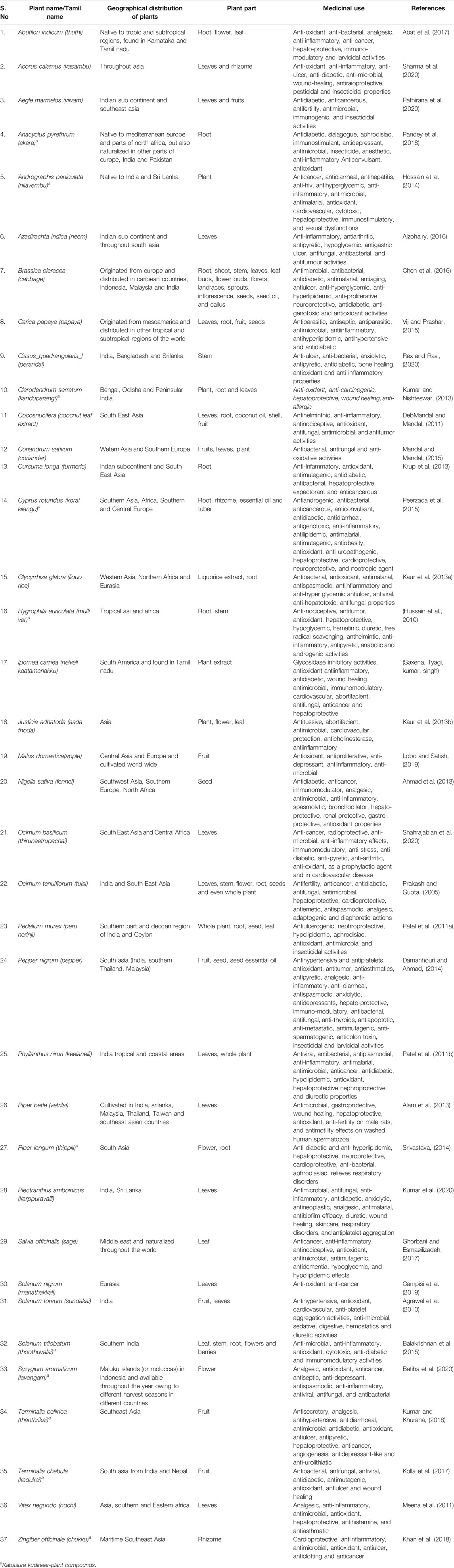
TABLE 2. List of herbal plants used in the study for screening antiviral compounds and their distribution and medicinal uses.
Known active ingredients of eight herbal plants included in the Tamil traditional medicine “Kabasura Kudineer” (meaning water capable of boosting immunity) were also included in the screening. Overall, a total of 744 small molecules/ligands (Supplementary Table S1: a-c) were used for virtual screening against seven different protein targets. Interactions of phytochemicals were compared with drugs such as hydroxycholoroquine (Pubchem ID: 3652), chloroquine (Pubchem ID: 2719) and ivermectin (Pubchem ID: 6427057) retrieved from pubchem database.
Virtual Screening
Virtual screening was performed using Python Prescription Virtual Screening tool (PyRx 0.8) containing AutoDock Vina module (Dallakyan and Olson, 2015). Protein structure was prepared using SWISS PDB Viewer by adding hydrogen atoms and energy minimization. Prepared protein structure was fed into the PyRx tool along with the structure of 744 ligands. Both the ligands and protein molecules were converted to pdbqt file using the AutoDock module of PyRx tool. Inhibitors are expected to bind in the active site/binding site of the protein to inhibit the function of the protein target. In the present study, binding sites were predicted using CASTP server (Tian et al., 2018) and the predicted sites were used for setting grid (XYZ dimensions: 25*25*25) in the AutoDock Vina for virtual screening experiment with the exhaustiveness value of 8. Predicted binding sites were also verified with protein-ligand binding sites of experimental structures for accuracy. Furthermore, phylogenetic analysis of SARS-CoV-2 MPro was carried out using PSI-BLAST (NCBI) (Altschul et al., 1997) and MAFFT server (Katoh et al., 2019). Top 10 ligand hits against each of the seven protein targets were taken for further analysis. 2D and 3D interactions between the protein-ligand were analyzed using Schrodinger Maestro visualizer and BIOVIA discovery studio visualizer 2020 (Accelrys Inc. San Diego, CA, United States) software. ADME properties of top 10 ligands screened against individual protein targets were predicted using SWISSADME server (http://www.swissadme.ch) (Supplementary Table S2).
Results and Discussion
Phylogenetic Analysis on Coronavirus Main Proteases
Main protease (Mpro, also called 3CLpro) is considered as one of the important molecular targets for designing novel drugs against coronaviruses (Anand et al., 2003). With a view to design drugs/inhibitors specifically targeting main protease of nCoVID’19, in-silico analysis was performed using main protease sequences of SARS-CoV-2, SARS-CoV and MERS-CoV. Multiple sequence alignment identified 12 significant differences between main proteases of SARS-CoV and SARS-CoV-2 (Figure 1). Out of the 12 differences, S45 to A45 was found to reside within in the binding site of SARS-CoV-2 main protease. This may play a crucial role in determining differential binding affinity of the two proteases.
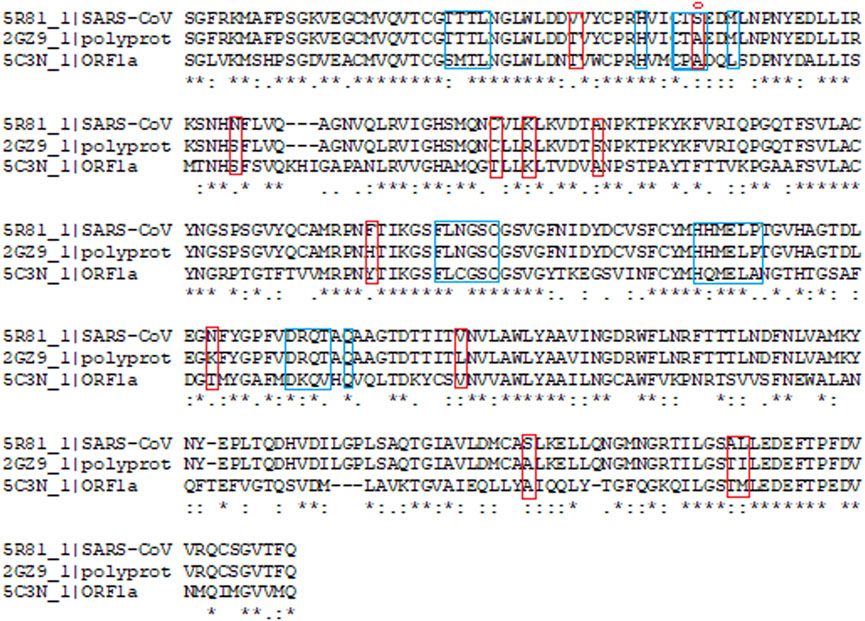
FIGURE 1. Multiple sequence alignment of SARS-CoV-2 (PDB ID: 5R81), SARS-CoV (PDB ID: 2GZ9) and MERS-CoV (PDB ID: 5C3N) main proteases. Binding site residues are marked with blue box. Red marked residues indicate the change observed between SARS-CoV-2 and SARS CoV. * denotes the conserved regions with identical residues. Serine to Alanine change at the 45th position observed in the binding site is marked with circle.
Phylogenetic analysis of SARS-CoV2MPro with other CoV MPro sequences sharing >50 percentage similarity revealed its significant genetic relatedness with SARS CoV (96.08% similarity) and bat CoV (76.84% similarity) (Figure 2). Also it shared significant similarity with ORF1ab of Rousettus bat coronavirus. Main protease of nCoVID’19 shared only 50.65% similarity against main proteases of MERS-CoV. Above results clearly indicated the need for a highly specific novel drug specifically inhibiting main proteases of SARS-CoV-2.
Virtual Screening of Potential Herbal Ligands Against Major Protein Targets of SARS-CoV-2
Virtual screening of 744 ligands belonging to small molecules and active compounds from 37 medicinal herbs against seven major protein targets of nCOVID’19 predicted probable SARS- CoV-2 inhibitors. Information regarding the binding site residues predicted using CASTp server is provided in Table 3. Binding affinity of drugs such as hydroxychloroquine, chloroquine and ivermectin are provided in Table 4 which was used as reference for comparison of phytochemicals. Top 10 hits reported with higher binding affinity for each target protein is considered for downstream analysis (Table 5). Seven molecular targets of SARS-CoV-2 include, Main protease (PDB ID: 5R81), RNA-dependent RNA polymerase (RdRp) (PDB ID: 6M71), NSP3 (PDB ID: 6W02), NSP9 (PDB ID: 6W4B), NSP10-NSP16 (PDB ID: 6W4H), NSP15 (PDB ID: 6W01) and Spike protein (PDB ID: 6M0J).

TABLE 4. Binding affinity value of Hydroxychloroquine, Cholroquine and Ivermectin against SARS-COV-2 targets.

TABLE 5. Details of shortlisted top 10 natural compounds/small molecules screened against various targets of coronaviruses.
Small Molecules/Herbal Compounds Exhibiting Significant Binding Affinity Against Main Protease
Despite of significant structural (RMSD: 0.71 Å) and binding site volume similarity of MPro between SARS-CoV (PDB ID: 2GZ9) and SARS-CoV-2 (PDB ID: 5R81), they showed differential binding affinity against different inhibitors (Table 1). Virtual screening of small molecules against MPro identified agathisflavone as the best hit exhibiting the binding affinity value of −8.2 kcal/mol.
Out of 744 ligands screened against SARS-CoV main protease, a ligand namely rutin abundantly found in Terminalia chebula, Azadirachta indica and Ocimum basilicum exhibited highest binding affinity value of −9.0 kcal/mol. In case of MERS-CoV main protease (PDB ID: 5C3N), amentoflavone predominantly found in Mangifera indica and Garcinia species showed the maximum binding affinity value of -8.6 kcal/mol. Interestingly, a cytotoxic biflavonoid agathisflavone found in cashew nut (Anacardium occidentale) was shown to exhibit significant binding affinity with −8.0 kcal/mol against the SARS-CoV-2 MPro (Supplementary Table S3). Agathisflavone is a biflavanoid derived from plant source and has been found to possess several biological activities (De Amorim et al., 2018). Various studies have found that agathisflavone possesses antioxidant, anti-inflammatory, antiviral, antiparasitic, cytotoxic, neuroprotective, and hepatoprotective activities. It has also been suggested that agathisflavone could be used in the treatment of oxidative stress, inflammatory diseases, microbial infection, hepatic and neurological diseases and cancer (Islam et al., 2019). Also, Agathisflavones have been reported for their cytotoxicity against malignant cell lines in earlier studies (Konan et al., 2012). This compound was found to involve in the formation of three hydrogen bonds with residues such as ASP 187, PRO 52 and ARG 40. This was followed by Rubusic acid (−8.1 kcal/mol) (Pedalium murex), solanocapsine (−7.9 kcal/mol)) (Solanum nigrum), chlorogenin (−7.7 kcal/mol) (Solanum torvum), Lupeol (−7.7 kcal/mol) (Carica papaya and Azadirachta indica), Cyanin (−7.7 kcal/mol) (Zingiber officinale), 3-O-trans-cafeoyltormentic acid (−7.7 kcal/mol) (antiviral compound), Luteolin-7-O-(6″-malonylglucoside) (−7.7 kcal/mol) (Vitex negundo), Agnuside (−7.6 kcal/mol) (Vitex negundo) and Luteolin 7-O-beta-D-glucoside (−7.6 kcal/mol) (Vitex negundo) exhibiting significant binding affinity in the decreasing order against MPro.
The phytochemical cyanin found in Zingiber officinale was found to have higher binding affinity with the main proteases of all three coronaviruses (Figures 3A–C) and placed in the top 10 hits list. It showed a binding affinity value of −8.3 kcal/mol, −8.2 kcal/mol and −7.7 kcal/mol against SARS-CoV-2(PDB ID: 5R81), SARS CoV (PDB ID: 2GZ9) and MERS CoV (PDB ID: 5C3N) main proteases respectively. Hydrogen bond interactions for SARS-CoV-2 (THR 26, SER 46 and GLU 166), SARS CoV (THR 26, LEU 141 and CYS 145) and MERS CoV (THR 26, CYS 149 and GLU 169) were observed in the binding sites respectively.
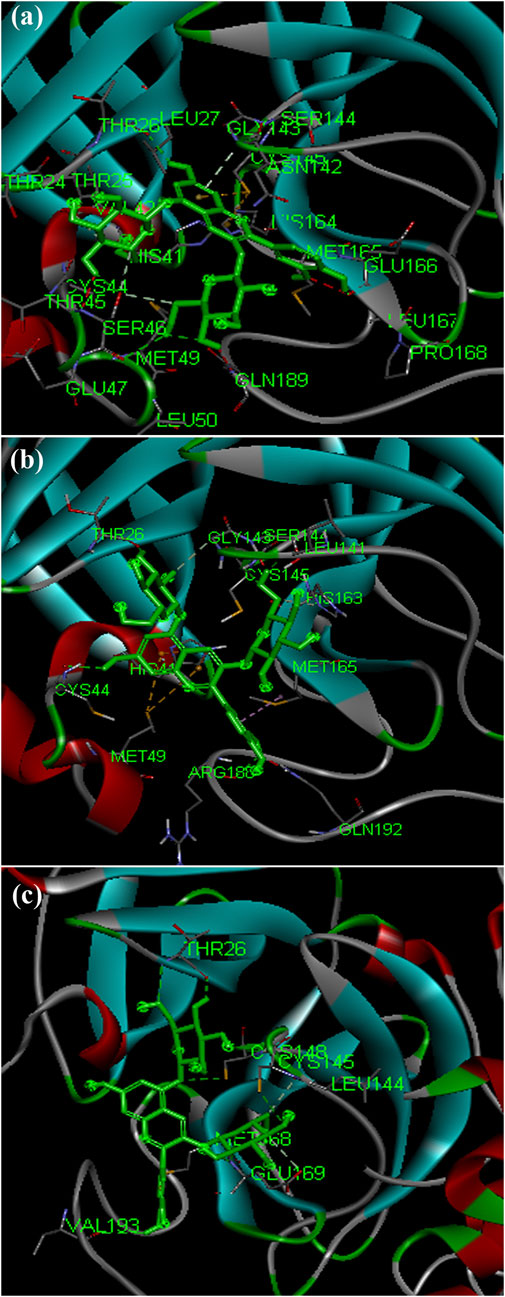
FIGURE 3. Complex structure of Cyanin with (A) SARS-CoV-2 MPro (PDB ID: 5R81) (B) SARS-CoV MPro (PDB ID: 2GZ9) (C) MERS-CoVMPro (PDB ID: 5C3N).
Effect of Suggested FDA Drugs on SARS-CoV-2 Protein Targets
Hydroxycholoroquine, chloroquine and ivermectin drugs were selected as positive controls to compare the binding interaction of phytochemicals for the assessment of anti-viral activity (Caly et al., 2020; Principi and Esposito, 2020). Hydroxychloroquine was reported to show promising inhibitory activity against nCOVID’19 spike protein (Gautret et al., 2020; Liu et al., 2020). Our results revealed that hydroxycholorquine and chloroquine showed less binding affinity against all the seven targets of nCOVID’19 compared to ivermectin (Table 4). Ivermectin exhibited significant binding affinity value of −9.4 kcal/mol and −8.2 kcal/mol against RNA - dependent RNA polymerase (RdRp) (PDB ID: 6M71) and spike protein (PDB ID: 6M0J) respectively (Figures 4A,D). Ivermection also exhibited significant binding affinity against NSP3 (PDB ID: 6W02) (-6.7 kcal/mol) and NSP9 (PDB ID: 6W4B) (-7.5 kcal/mol) (Figures 5A–D) (Supplementary Table S4). Analysis of hydrogen bond interaction with ivermectin, showed three hydrogen bonds for RdRp target (ARG 403, TYR 453 and TYR 489) and two hydrogen bonds for spike protein (ASN 497 and SER 814), each one hydrogen bond for NSP3 (TYR 161) and NSP9 (ARG 40) respectively. It is also important to note that the ivermectin was reported as best hits for the above mentioned targets compared to hydroxychloroquine and chloroquine against SARS-CoV-2 targets. In the following section, binding of ivermectin was compared with amentoflavone and agasthisflavone phytochemicals as they were predicted to inhibit multi-target SARS-CoV-2 proteins based on the docking score.
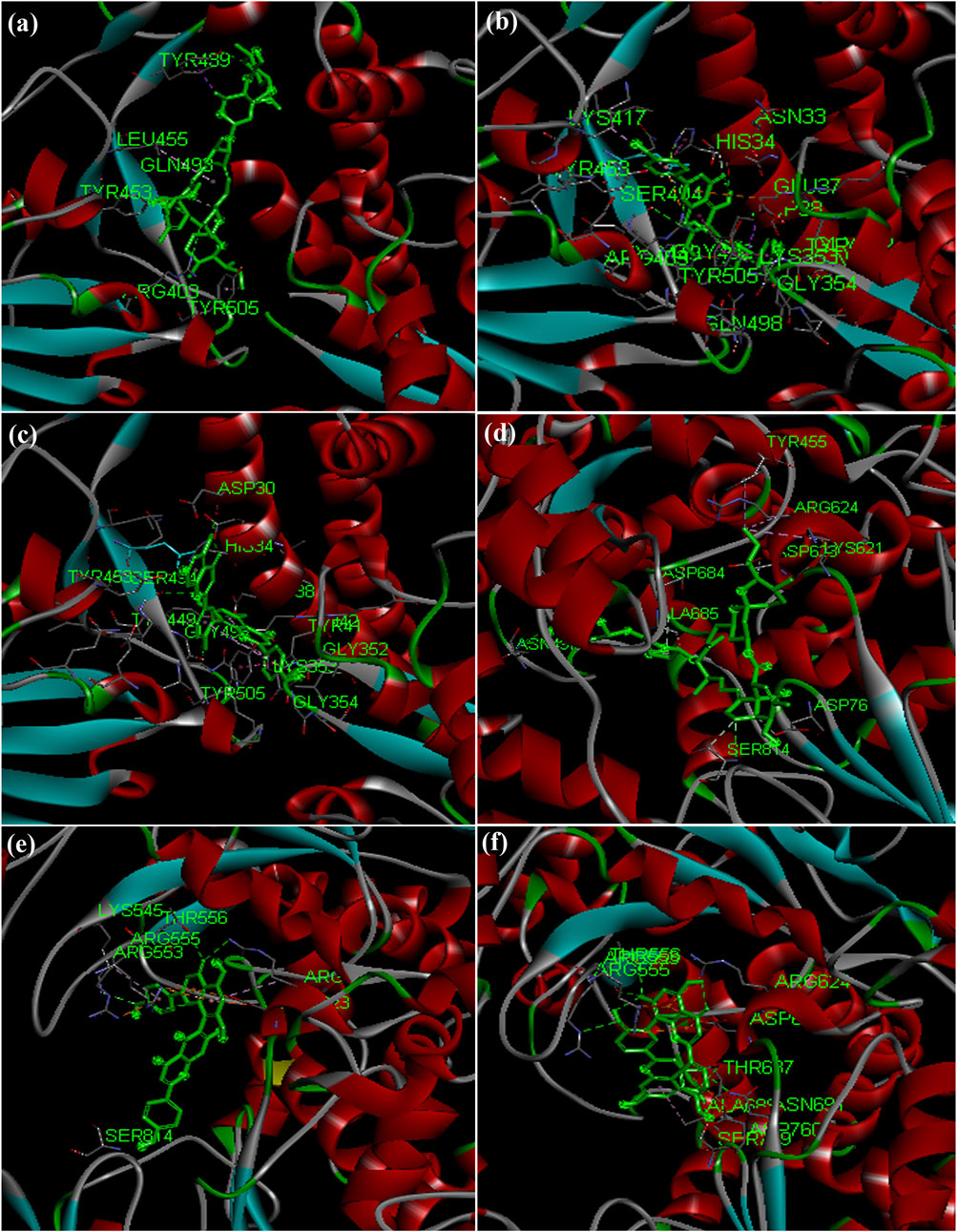
FIGURE 4. Docked complex structures of spike protein with (A) ivermectin, (B) agasthisflavone, (C) amentoflavone, and RdRp with (D) ivermectin (E) agasthisflavone (F) amentoflavone.
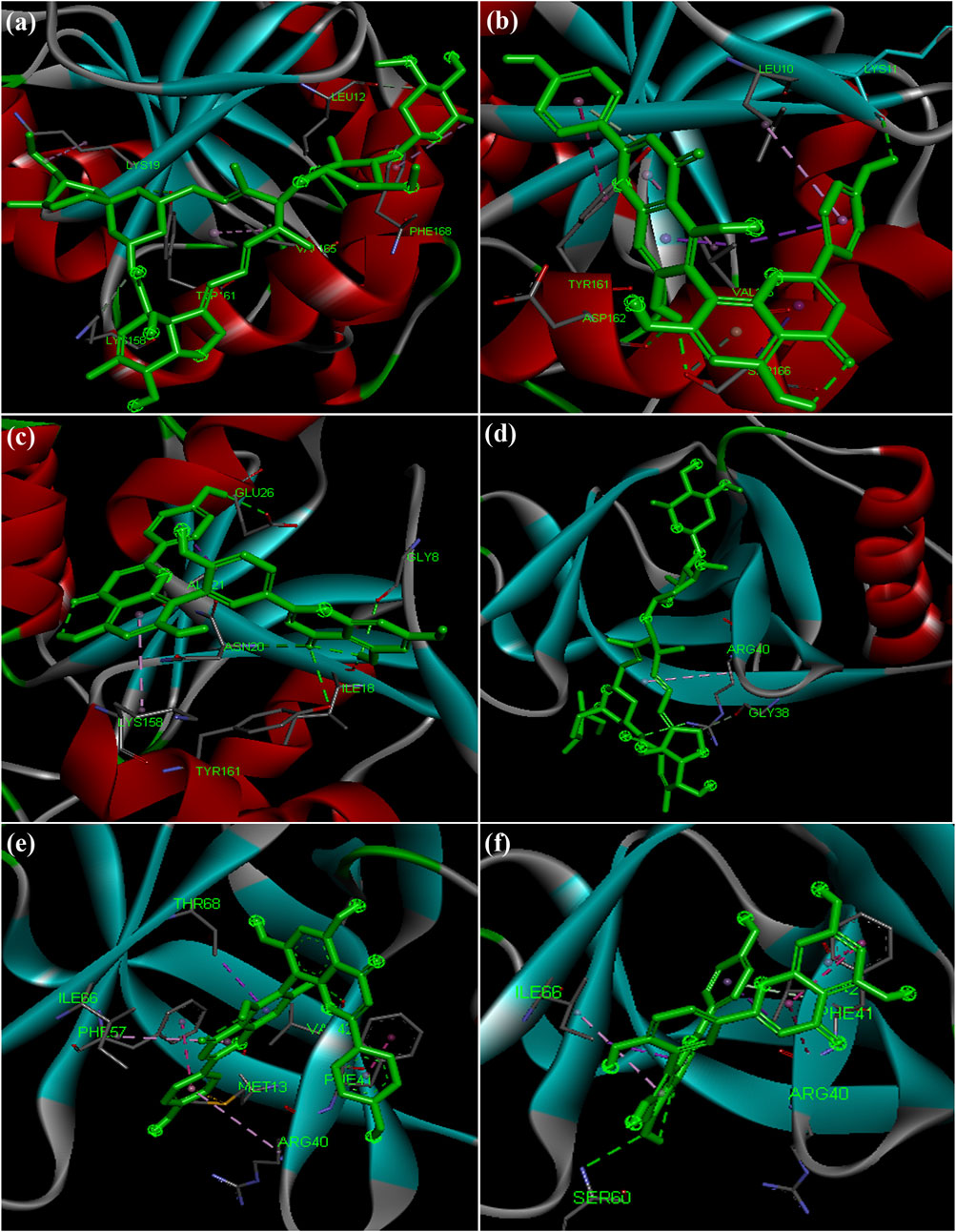
FIGURE 5. Docked complex structures of NSP3 protein with (A) ivermectin (B) agasthisflavone (C) amentoflavone, and NSP9 with (D) ivermectin (E) agasthisflavone (F) amentoflavone.
Spike Protein
X-ray crystal structure of spike glycoprotein (PDB ID: 6M0J) was chosen for performing virtual screening. Virtual screening was performed by choosing ACE interacting region as the binding site (Figure 6A). 1,8-Dichloro-9,10-diphenylanthracene-9,10-diol from Carica papaya was found to exhibit significant binding affinity against spike glycoprotein (−8.2 kcal/mol). GLY 496 residue was found to be involved in the formation of hydrogen bond with the 1, 8-Dichloro-9, 10-diphenylanthracene-9,10-diol. Earlier, leaf extracts of Carica papaya was reported to have significant effect in combating dengue virus infection (Rajapakse et al., 2019) and its exact role in increasing platelet counts is not clear. 1,8-Dichloro-9,10-diphenylanthracene-9,10-diol was found buried in the binding site of spike glycoprotein exhibiting hydrophobic interactions with residues such as LEU 39, TYR 41, TYR 449, TYR 453, TYR 495, PHE 497 and TYR 505 (Figure 7). This was followed by other small molecules viz., agasthisflavone (−8.2 kcal/mol) (Figure 4B), amentoflavone (−8.4 kcal/mol) (Figure 4C), ivermectin (−8.2 kcal/mol), three o caffeoyltormentic acid (−8.1 kcal/mol), agnuside (−7.5 kcal/mol) (Vitex negundo), taraxerol (−7.3 kcal/mol) (Cissus quadrangularis), nimbinene (−7.3 kcal/mol) (Azadirachta indica) and β-amyrin (−7.3 kcal/mol) (Cissus quadrangularis) exhibiting significantly higher level of binding affinity toward spike protein of nCOVID’19. Comparison of hydrogen bond interactions at the binding site for ivermectin (3 hydrogen bonds), amentoflavone (4 hydrogen bonds) and agasthisflavone (3 hydrogen bonds) showed higher number of interactions for amentoflavone involving residues TYR 449, SER 494, GLY 496 and ASP 30 respectively.

FIGURE 6. (A) PDB ID: 6M0J - Residue interaction between ACE-2 (chain A) and spike glycoprotein (chain E). (B) PDB ID: 6W4H - Interface of NSP10-NSP16 complex.
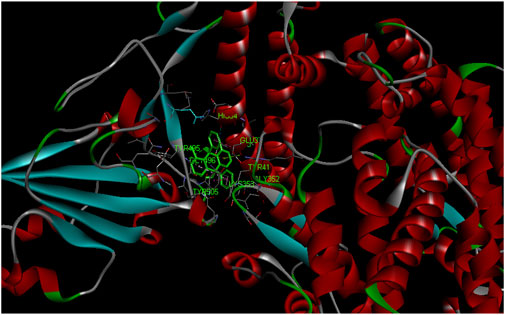
FIGURE 7. Spike protein (PDB ID: 6M0J) complex with 1,8-Dichloro-9,10-diphenylanthracene-9,10-diol (binding affinity: 8.2 kcal/mol).
Residues are colored based on their physiochemical properties.
Ribonucleic Acid-dependent Ribonucleic Acid Polymerase (RdRp)
RdRp (PDB ID: 6M71) is responsible for replication of COVID-19 genome inside the host. Among the ligands tested, ivermectin (Figure 4D) showed the higher binding affinity value of −9.4 kcal/mol compared to the 744 small molecules screened in the present study. Amentoflavone stood at second position with the binding affinity value of −9.3 kcal/mol (Figure 4F). This was found to have interaction with three amino acid residues namely ARG 553, THR 556 and ASN 691. Ligands viz., Corilagin (−9.0 kcal/mol) (Terminalia bellirica and Terminalia chebula), Agasthisflavone (−8.9 kcal/mol) (Anacardium occidentale) (Figure 4E), 3-O-trans-caffeoyltormentic acid (−8.6 kcal/mol), Arjungenin (−8.2 kcal/mol) (Terminalia chebula), Crateogolic acid (−8.2 kcal/mol) (Syzygium aromaticum), 3.8’-biapigenin (−8.0 kcal/mol), cyanin (−7.9 kcal/mol) (Zingiber officinale) were found to exhibit significant binding affinity. Interaction analysis showed higher number of vander waals interactions for ivermectin compared to hydrogen bond interactions. Comparison of hydrogen bond interactions exhibited toward ivermectin (3 hydrogen bonds), agasthisflavone (4 hydrogen bond interactions) and amentoflavone (6 hydrogen bond interactions–ARG 553, ARG 555, THR 556, SER 759, ASN 691 and THR 687).
Non-structural Proteins
Apart from the four major structural proteins (S, E, M, and N proteins), non-structural proteins namely NSP3 (cleavage of N-terminal replicase poly protein) (PDB ID: 6W02), NSP9 (ssRNA binding) (PDB ID: 6W4B), NSP10-NSP16 (co-factor in activating replicating enzyme) (PDB ID: 6W4H) and NSP15 (PDB ID: 6W01) involved in the transcription and replication of nCoVID’19 can also serve as potential targets for containing the virus using inhibitory herbal molecules (Prajapat et al., 2020).
Virtual screening of small molecules against NSP3 identified amentoflavone (Mangifera indica) as the top scored ligand with binding affinity of −7.5 kcal/mol (Figure 5C). In the decreasing order of binding affinity, luteolin 7-O-(6''-malonylglucoside) (−7.1 kcal/mol) (Vitex negundo), rubusic acid (−6.8 kcal/mol) (Pedalium murex), acteoside (−6.7 kcal/mol) (Clerodendrum serratum), ivermectin (−6.7 kcal/mol), taraxerol acetate (−6.7 kcal/mol) (Cissus_quadrangularis), catechin 7-O-gallate (-6.6 kcal/mol), luteolin 7-O-beta-D-glucoside (−6.6 kcal/mol) (Vitex negundo), agathisflavone (−6.6 kcal/mol) (Anacardium occidentale) (Figure 5B) and luteolin-7-o-beta-d-glucopyranoside (−6.6 kcal/mol) (Vitex negundo) were found to have higher binding affinity next to amentoflavone. It was observed that three inhibitors from Vitex negundo were reported with the highest binding scores. Vitex negundo belongs to Verbenaceae family known for its effects for ailments like ophthalmia, deafness, indigestion, piles and jaundice (Venkateswarlu, 2012). Results of earlier experiments conducted by Wu et al. (2020) also revealed similar findings of vitexin from Vitex negundo exhibiting significant binding affinity toward NSP3.
Another small molecule, friedelin from Vitex negundo and Acorus calamus was also found to exhibit significant binding affinity of −9.6 kcal/mol against NSP9 (PDB ID: 6W4B) (Table 5). Even though, many hydrophobic interactions were observed, no hydrogen bond interaction was found in the binding site of NSP9. It is very interesting to observe that five out of the top ten inhibitors are from a single plant source Solanum nigrum. As evidenced from other studies, Solanum nigrum is one of the traditionally known medicinal plants known for its use in treatment ofseizure, pain, ulcer, inflammation, diarrhea, eye infections, jaundice and oxidative stresses (Jain et al., 2011; Javed et al., 2011; Wang et al., 2013; Zaidi et al., 2014).
Virtual screening against NSP15 (PDB ID:6W01) identified oleonolic acid and urosolic acid as molecules exhibiting highest binding affinity values of −9.2 kcal/mol. Oleonolic acid and urosolic acid are known for their anti-cancerous and anti-inflammatory activities (Arshad Qamar et al., 2010; Kashyap et al., 2016; Yu et al., 2017). Both the phytochemicals are reported to be enriched in Ocimum basilicum and Ocimum tenuiflorum (Table 5). This was followed by crategolic acid (−8.9 kcal/mol) (Syzygium aromaticum), arjungenin (−8.8 kcal/mol) (Terminalia chebula), hederagenin (−8.7 kcal/mol) (Nigella sativa), triterpenoid (−8.6 kcal/mol) (Abutilon indicum), beta-amyrin (−8.6 kcal/mol) (Cissus quadrangularis), friedelin (−8.6 kcal/mol) (Vitex negundo, Acorus calamus), catechin 7-O-gallate (−8.5 kcal/mol) (Camellia sinensis) and arjunolic acid (−8.5 kcal/mol) (Terminalia chebula).
In the case of NSP10-NSP16 (PDB ID: 6W4H) protein complex, interface of the complex (Figure 6B) was chosen as the binding site for performing virtual screening. Compounds such as amentoflavone (−8.5 kcal/mol), 10-methoxycamptothecin (−8.3 kcal/mol), 3.8'-biapigenin (−8.2 kcal/mol), taraxerol acetate (−8.0 kcal/mol) (Cissus_quadrangularis), corilagin (−7.8 kcal/mol) (Terminalia bellirica and Terminalia chebula), lupeol acetate (−7.8 kcal/mol) (Pedalium murex), emetine (−7.6 kcal/mol), chlorogenin (−7.6 kcal/mol) (Solanum torvum), spirostan-3-ol (−7.6 kcal/mol) (Solanum torvum) and 5,7-Dihydroxy-4'-methoxy-8.3'-di-C-prenylflavanone (−7.6 kcal/mol) (Azadirachta indica) were ranked among the top 10 molecules exhibiting highest binding affinity.
Molecules Exhibiting Inhibitory Activity Against Multiple Protein Targets of nCOVID’19
Phytochemicals showing strong interactions against multiple targets of viruses are expected to confer durable protection to the patients. This will be more beneficial in situations where the virus is developing mutations in one of the targets. Small molecules namely, amentoflavone, agathisflavone, catechin-o-gallate and chlorogenin exhibited significant binding affinity toward multiple targets of nCOVID’19.
Amentoflavone showed docking score of RdRp (−9.3 kcal/mol) (Figure 4C), NSP9 (−8.3 kcal/mol) (Figure 5F), NSP3 (−7.4 kcal/mol) (Figure 5C), NSP10-NSP16 (−8.5 kcal/mol) and spike glycoprotein (−8.2 kcal/mol). In all the docked complexes, the target - ligand binding affinity was greater than (−8.0 kcal/mol) except NSP3. 3D and 2D Protein-ligand interactions exhibited by the small molecules amentoflavone and agathisflavone are shown in Figure 4, Figure 5 and Supplementary Table S3. Molecular interactions such as hydrogen bond, vander waals interaction, pi interaction were observed to be higher for amentoflavone incomparison with the drug ivermectin. Amentoflavone is a naturally occurring biflavonoid reported to be found in more than 120 plants (Yu et al., 2017). Many of these plants have been used in traditional medicine for several thousand years in different parts of the world. Several studies have reported that amentoflavone possess anti-inflammatory, anti-oxidative, anti-diabetic, anti-tumor, anti-viral and anti-fungal activities (Yu et al., 2017). Evidences have been reported for amentoflavone exhibiting anti-senescence activity in the cardiovascular and central nervous system (Park et al., 2011). Further, Amentoflavone isolated from Torreya nucifera was demonstrated to possess inhibitory activity against SARS-CoV3CLPro (Ryu et al., 2010).
Similarly, agathisflavone was found to exhibit significant interaction against four different protein targets viz., RNA-dependent RNA polymerase (−8.9 kcal/mol), SARS-CoV-2 main protease (−8.2 kcal/mol), spike glycoprotein (−8.2 kcal/mol) and NSP 3 (−6.6 kcal/mol) (Supplementary Table. S3). Catechin-o-gallate was also found to possess significant binding affinity toward spike glycoprotein (−7.3 kcal/mol), NSP3 (−6.6 kcal/mol), RNA-dependent RNA polymerase (−7.9 kcal/mol) and NSP15 (−8.5 kcal/mol). Role of catechins for antiviral activity against the number of viruses due to the increased affinity for cellular membranes has been reported in many articles (Kaihatsu et al., 2018). Chlorogenin from Solanum torvum exhibited binding affinity value of −8.2 kcal/mol (NSP9), −7.7 kcal/mol (MPro) and −7.6 kcal mol (NSP16-NSP10) of SARS-CoV-2. Solanum torvum is well known for its pharmacological properties for the treatment of human ailments and its phytoconstituent chlorogenin also has antiviral effect on influenza viruses (Agrawal et al., 2010; Lim, 2013; Shi et al., 2020).
Fifteen different compounds viz., 3.8'-biapigenin, 3-O-trans-caffeoyltormentic acid, agnuside, arjungenin, corilagin, crategolic acid, cyanin, friedelin, luteolin 7-O-(6''-, malonylglucoside), N-methylsolasodine, rubusic acid, solanocapsine, spirostan-3-ol, taraxerol and taraxerol acetate were found to possess significant interactions with at least two protein targets of SARS-CoV-2 (Table 6) (Figure 8). Among the above reported 15 compounds, cyanin targeted all the three Coronavirus MPro. 2D representation of MPro-cyanin interaction of all the coronaviruses is provided in the Supplementary Table S5.

TABLE 6. Phytochemicals having strong interaction based on the top 10 hits screened for SARS-CoV-2targets.
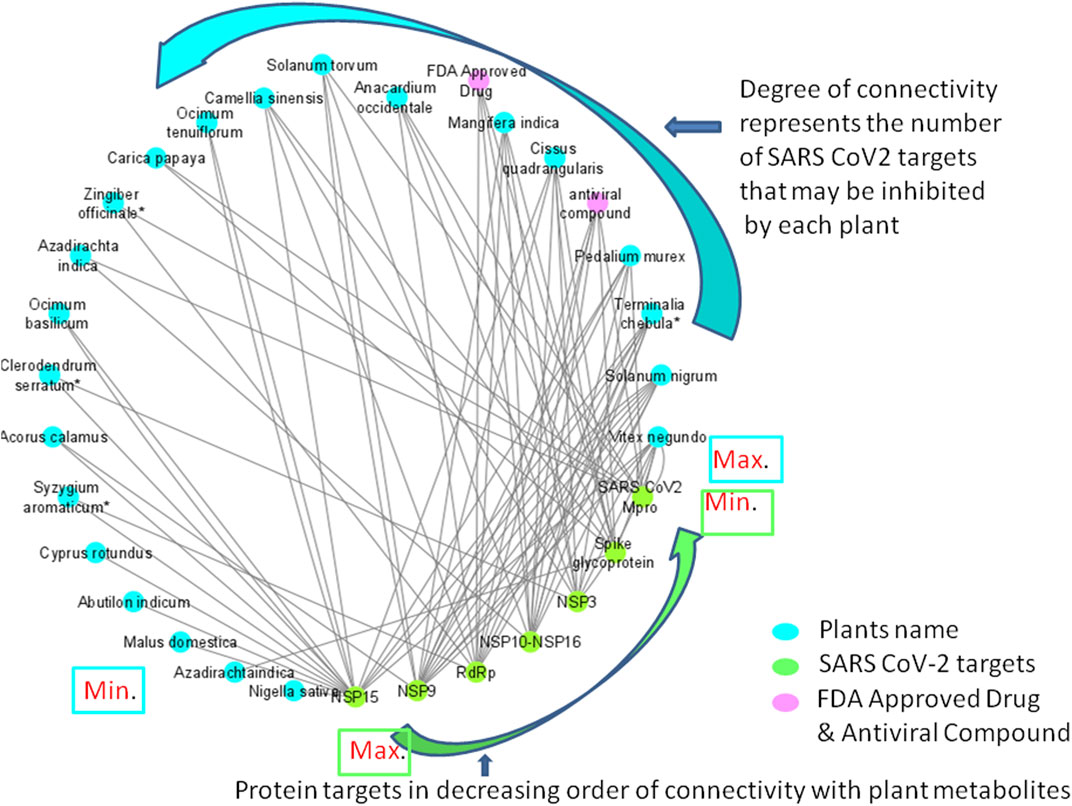
FIGURE 8. Network diagram showing the interaction of plant phytochemcials with SARS-CoV-2 protein targets. Degree of connectivity represents number of SARS-CoV-2 that may be inhibited by each plant. In this network, NSP 15 stands first in the order where many plants connected (degree of connectivity) which indicates that metabolites from the connected plants has shown highest binding affinity (top ten screened compounds). Likewise, Vitex negundo was predicted to inhibit highest number of SARS-CoV-2 targets in the virtual screening. Plants name with * indicates their presence in Kabasura kudineer.
Discussion
Among the 605 phytochemicals originating from 37 plant species, 33 (6% approx.) phytochemicals from 22 plants (Figure 8) were found to be the best hits with higher binding affinities against all the seven targets (Table 5). Among those 22 plants, four plants were found to be the ingredients of a traditional siddha herbal formulation namely “Kabasura kudineer” recommended by AYUSH Board of Government of India for boosting immunity. Vitex negundo was reported to possess 32 different phytochemicals that were included in this study. Out of the 33, five different compounds namely, luteolin 7-O-beta-D-glucoside, luteolin 7-O-(6'-malonylglucoside), agnuside, luteolin-7-o-beta-d-glucopyranoside and friedelin were found to exhibit significant binding affinity against five different protein targets of SARS-CoV-2 namely, spike glycoprotein, SARS-CoV-2 main protease, NSP3, NSP9, NSP15 in the SARS-CoV-2.
Solanocapsine, Spirostan-3-ol, N-methylsolasodine, Diosgenin and Solasodine are the phytochemicals reported in Solanum nigrum which effectively inhibited three SARS-CoV-2 targets (SARS-CoV-2MPro, NSP9 and NSP16-NSP10).
Pedalium murex was reported to have Diosgenin (NSP9), Lupeol acetate (NSP16-NSP10), Urosolic acid (NSP15) and Rubusic acid (SARS-CoV-2 Mpro, NSP3) which might be probable SARS-CoV-2inhibitors. It is noteworthy that maximum of five targets were predicted to be inhibited by the compounds from Pedalium murex. In spite of its role as anti-ulcerogenic, nephroprotective, hypolipidemic, aphrodisiac, anti-oxidant, anti-microbial and insecticidal activities, Pedalium murex has been traditionally used in treating ailments like cough and cold as a regular practice (Patel et al., 2011a).
Medicinal plants namely, Azadirachta indica, Terminalia chebula, Cissus quadrangularis, Clerodendrum serratum and Ocimum basilicum reported more than two phytochemicals that might possibly inhibit SARS-CoV-2 targets. These herbal plants might be the potential targets for future research toward developing herbal formulations against SARS-CoV-2. Intensive genomics and proteomics research may lead to identification of novel drugs against this pandemic disease.
Knowledge of the phytochemical composition in the reported plants is important to understand the role of bioactive molecule in the traditional medicine. Phytochemical composition of the some of the screened plants quantified using various analytical techniques are given as follows: Cyanin (Zingiber officinale) (20%) (Ghasemzadeh et al., 2010), amentoflavone (Mangifera indica) (90.92 ± 4.91 mg/100 g) (Ma et al., 2011), agathisflavone (Anacardium occidentale) (4.39 ± 0.01 (mg/g)) (Onuh et al., 2017), Corilagin (Terminalia chebula) (13.05 mg/g) (Juang et al., 2004) luteolin derivatives (Vitex negundo) (123.776 – 127.774 mg/g) (Koirala et al., 2020), Solanocapsine (Solanum nigrum) (>50%) (Almoulah et al., 2017), Solasodine (Solanum nigrum) (75.89%) (Jasim et al., 2015), Diosgenin (Pedalium murex) (0.06%) (Patel et al., 2011a), Quercetrin (Azadirachta indica) (0.016%) (Sarkar et al., 2020), Rutin (Azadirachta indica) (0.14%) (Sarkar et al., 2020), Nimbinene (Azadirachta indica) (1.2%) (Sadeghian and Mortazaienezhad, 2007), Rutin (Terminalia chebula) (0.17%) (Kumar et al., 2010), Rutin (Ocimum basilicum) (425.71 ± 2.15 μg/g) (Vlase et al., 2014) and Isoquercetin (Ocimum basilicum) (179.19 ± 1.93 μg/g) (Vlase et al., 2014).
Antiviral activity of the plants highlighted in this study, whose phytochemicals are predicted to have inhibitory effect on SARS-CoV-2 targets have proven antiviral role on many other human pathogenic viruses. Antiviral activity of Vitex negundo L. against Chikungunya virus and HIV-1 Reverse transcriptase (RT) have been previously reported by Kothandan and Swaminathan (2014) and Nair and Ramachandran (2012) (Nair, 2012; Kothandan and Swaminathan, 2014). Solanum nigrum was reported to have antiviral effect against Hepatitis C Virus chronic infection (Javed et al., 2011). Similarily, the antiviral and virucidal effect of neem leaves was studied regarding its activity and possible mechanism of action against Coxsackie B group of viruses (Badam et al., 1999). Sivagami and Sailaja 2021 (Sivagami and Sailaja, 2021) have reported the antiviral activity of Clerodendrum serratum against yellow fever virus. Likewise, antiviral effect of Terminalia chebula on swine influenza A virus infection has also been proved (Ma et al., 2010). Phytochemicals like apigenin and ursolic acid from Ocimum basilicum has shown broad-spectrum of antiviral activity against many of the DNA and RNA viruses (Chiang et al., 2005). Zingiber officinale has been proved to have antiviral activity against human respiratory syncytial virus (Chang et al., 2013). Also, the plants like Mangifera indica and Anacardium occidentale L. inhibits Herpes simplex virus two and influenza virus neuraminidase respectively (Parvez, 2016; de Freitas et al., 2020).
ADME properties are important to understand the pharmacokinetics and physio-chemical properties of phytochemicals which further provides insight on drug design and formulation research. Analysis on solubility and bioavailability based on the SWISSADME prediction for the screened inhibitors showed different level of classifications as given in Supplementary Table S2. According to the Silicos-IT LogSw score, 41% of the screened ligands were classified soluble, 31% (moderately soluble), 27% (poorly soluble) and one molecule (1,8-Dichloro-9,10-diphenylanthracene-9,10-diol) alone as insoluble. Bioavailability score predicted on the probability of a compound to have at least 10% oral bioavailability (Martin, 2005) in rat has been reported in which one compound (uteolin 7-O-(6''-malonylglucoside)) has less score (0.11 score), 17 compounds have (0.17 score), 27 compounds have (0.55 score) and three compounds (ursolic acid, betulinic acid and oleanolic acid) with 0.85 score respectively.
Conclusion
Generation of improved knowledge and understanding biochemical and molecular basis of herbals used in traditional Ayurveda and siddha medicine will accelerate development of effective drugs in controlling emerging diseases. In this study, comparative analysis of main proteases of MERS-CoV (PDB ID: 5C3N), SARS-CoV (PDB ID: 2GCZ9) and SARS-CoV-2 (PDB ID: 5R81) revealed significant differences between the three homologs which were confirmed by differential binding affinity exhibited by 744 phytochemicals/small molecules against the three main proteases. Cyanin from Zingiber officinale was screened as the best phytochemical with highest binding energy against main proteases of all the three viruses. Popular fruit tree, mango (Mangifera indica) and, cashew nut (Anacardium occidentale) rich in amentoflavone and agathisflavone were showing possible inhibitory activity against multiple targets of SARS-CoV-2. Vitex negundo, Solanum nigrum, Pedalium Murex, Terminalia chebula, Azadirachta indica, Cissus quadrangularis, Clerodendrum serratum and Ocimum basilicum were also found to contain phytochemicals that may have possible inhibitory activity against SARS-CoV-2 proteins. More interestingly, this study has picked up Carica Papaya that may possess inhibitory activity against spike glycoprotein and MPro of SARS-CoV-2 which was known for its protective role against dengue virus in humans (Rajapakse et al., 2019). Overall, this study has shortlisted potential phytochemicals that may have inhibitory activity against SARS-CoV-2 which could be taken for further testing, formulation and discovery of novel drugs.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Author Contributions
SN, JM, CR, KA, and BN performed experimental, data analysis, and drafting of original manuscript. KN, GR, RM, MS, and KN conceptualized the experiment, refined the manuscript, funded the project, and helped in revising the manuscript. All authors read and approved the final version of the manuscript.
Funding
This research was carried out in the Bioinformatics Laboratory, Center for Plant Molecular Biology and Biotechnology, Tamil Nadu Agricultural University supported by Biotechnology Information System (BTIS) program of Department of Biotechnology (DBT), Government of India, New Delhi.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fphar.2021.667704/full#supplementary-material
References
Abat, J. K., Kumar, S., and Mohanty, A. (2017). Ethnomedicinal, Phytochemical and Ethnopharmacological Aspects of Four Medicinal Plants of Malvaceae Used in Indian Traditional Medicines: a Review. Medicines 4 (4), 75. doi:10.3390/medicines4040075
Agrawal, A. D., Bajpei, P. S., Patil, A. A., and Bavaskar, S. R. (2010). Solanum Torvum Sw.—A Phytopharmacological Review. Der pharmacia lettre 2 (4), 403–407.
Ahmad, A., Husain, A., Mujeeb, M., Khan, S. A., Najmi, A. K., Siddique, N. A., et al. (2013). A Review on Therapeutic Potential of Nigella Sativa: A Miracle Herb. Asian Pac. J. Trop. Biomed. 3 (5), 337–352. doi:10.1016/s2221-1691(13)60075-1
Alam, B., Akter, F., Parvin, N., Sharmin Pia, R., Akter, S., Chowdhury, J., et al. (2013). Antioxidant, Analgesic and Anti-inflammatory Activities of the Methanolic Extract of Piper Betle Leaves. Avicenna J. Phytomed 3 (2), 112–125.
Almoulah, N. F., Voynikov, Y., Gevrenova, R., Schohn, H., Tzanova, T., Yagi, S., et al. (2017). Antibacterial, Antiproliferative and Antioxidant Activity of Leaf Extracts of Selected Solanaceae Species. South Afr. J. Bot. 112, 368–374. doi:10.1016/j.sajb.2017.06.016
Altschul, S., Madden, T. L., Schäffer, A. A., Zhang, J., Zhang, Z., Miller, W., et al. (1997). Gapped BLAST and PSI-BLAST: a New Generation of Protein Database Search Programs. Nucleic Acids Res. 25 (17), 3389–3402. doi:10.1093/nar/25.17.3389
Alzohairy, M. A. (2016). Therapeutics Role of Azadirachta indica (Neem) and Their Active Constituents in Diseases Prevention and Treatment. Evidence-Based Complement. Altern. Med. 2016, 7382506. doi:10.1155/2016/7382506
Anand, K., Ziebuhr, J., Wadhwani, P., Mesters, J. R., and Hilgenfeld, R. (2003). Coronavirus Main Proteinase (3CLpro) Structure: Basis for Design of Anti-SARS Drugs. Science 300 (5626), 1763–1767. doi:10.1126/science.1085658
Arshad Qamar, K., Dar, A., Siddiqui, B. S., Kabir, N., and Aslam, H., Ahmed, S., et al. (2010). Anticancer Activity of Ocimum Basilicum and the Effect of Ursolic Acid on the Cytoskeleton of MCF-7 Human Breast Cancer Cells. Lett. Drug. Des. Discov. 7 (10), 726–736. doi:10.2174/1570180811007010726
Badam, L., Joshi, S. P., and Bedekar, S. S. (1999). 'In Vitro' Antiviral Activity of Neem (Azadirachta indica. A. Juss) Leaf Extract against Group B Coxsackieviruses. J. Commun. Dis. 31 (2), 79–90.
Balakrishnan, P., Ansari, T., Gani, M., Subrahmanyam, S., and Shanmugam, K. (2015). A Perspective on Bioactive Compounds from Solanum Trilobatum. J. Chem. Pharm. Res. 7 (8), 507–512.
Batiha, G. E-S., Alkazmi, L. M., Wasef, L. G., Beshbishy, A. M., Nadwa, E. H., and Rashwan, E. K. (2020). Syzygium Aromaticum L.(Myrtaceae): Traditional Uses, Bioactive Chemical Constituents, Pharmacological and Toxicological Activities. Biomolecules 10 (2), 202. doi:10.3390/biom10020202
Caly, L., Druce, J. D., Catton, M. G., Jans, D. A., and Wagstaff, K. M. (2020). The FDA-Approved Drug Ivermectin Inhibits the Replication of SARS-CoV-2 In Vitro. Antivir. Res. 178, 104787. doi:10.1016/j.antiviral.2020.104787
Campisi, A., Acquaviva, R., Raciti, G., Duro, A., Rizzo, M., and Santagati, N. A. (2019). Antioxidant Activities of Solanum nigrum L. Leaf Extracts Determined in In Vitro Cellular Models. Foods 8 (2), 63. doi:10.3390/foods8020063
Chang, J. S., Wang, K. C., Yeh, C. F., Shieh, D. E., and Chiang, L. C. (2013). Fresh Ginger (Zingiber Officinale) Has Anti-viral Activity against Human Respiratory Syncytial Virus in Human Respiratory Tract Cell Lines. J. ethnopharmacology 145 (1), 146–151. doi:10.1016/j.jep.2012.10.043
Chen, J., Zhang, J., Xiang, Y., Xiang, L., Liu, Y., He, X., et al. (2016). Extracts of Tsai Tai (Brassica Chinensis): Enhanced Antioxidant Activity and Anti-aging Effects Both In Vitro and in Caenorhabditis elegans. Food Funct. 7 (2), 943–952. doi:10.1039/c5fo01241d
Cheng, V. C. C., Lau, S. K. P., Woo, P. C. Y., and Yuen, K. Y. (2007). Severe Acute Respiratory Syndrome Coronavirus as an Agent of Emerging and Reemerging Infection. Clin. Microbiol. Rev. 20 (4), 660–694. doi:10.1128/cmr.00023-07
Chiang, L.-C., Ng, L.-T., Cheng, P.-W., Chiang, W., and Lin, C.-C. (2005). Antiviral Activities of Extracts and Selected Pure Constituents of Ocimum Basilicum. Clin. Exp. Pharmacol. Physiol. 32 (10), 811–816. doi:10.1111/j.1440-1681.2005.04270.x
Colson, P., Rolain, J.-M., Lagier, J.-C., Brouqui, P., and Raoult, D. (2020). Chloroquine and Hydroxychloroquine as Available Weapons to Fight COVID-19. Int. J. Antimicrob. Agents 55 (4), 105932. doi:10.1016/j.ijantimicag.2020.105932
Dallakyan, S., and Olson, A. J. (2015). Small-molecule Library Screening by Docking with PyRx. Chem. Biol. 1263, 243–250. doi:10.1007/978-1-4939-2269-7_19
Damanhouri, Z. A., and Ahmad, A. (2014). A Review on Therapeutic Potential of Piper Nigrum L. Black Pepper) The King of Spices. Med. Aromat Plants 3, 161. doi:10.4172/2167-0412.1000161
De Amorim, V. C. M., Júnior, M. S. O., Bastos, E. M. S., Da Silva, V. D. A., and Costa, S. L. (2018). Research on the Scientific Evolution of the Flavonoid Agathisflavone. J. Pharm. Pharm. Sci. 21, 376–385. doi:10.18433/jpps30103
de Freitas, C. S., Rocha, M. E. N., Sacramento, C. Q., Marttorelli, A., Ferreira, A. C., Rocha, N., et al. (2020). Agathisflavone, a Biflavonoid from Anacardium Occidentale L., Inhibits Influenza Virus Neuraminidase. Curr. Top. Med. Chem. 20 (2), 111–120. doi:10.2174/1568026620666191219150738
De Groot, R. J., Baker, S. C., Baric, R. S., Brown, C. S., Drosten, C., Enjuanes, L., et al. (2013). Middle East Respiratory Syndrome Coronavirus (MERS-CoV): Announcement of the Coronavirus Study Group. J. Virol. 87 (14), 7790–7792. doi:10.1128/jvi.01244-13
DebMandal, M., and Mandal, S. (2011). Coconut (Cocos Nucifera L.: Arecaceae): in Health Promotion and Disease Prevention. Asian Pac. J. Trop. Med. 4 (3), 241–247. doi:10.1016/s1995-7645(11)60078-3
Dong, L., Hu, S., and Gao, J. (2020). Discovering Drugs to Treat Coronavirus Disease 2019 (COVID-19). Drug. Discov. Ther. 14 (1), 58–60. doi:10.5582/ddt.2020.01012
Elfiky, A. A. (2020). Anti-HCV, Nucleotide Inhibitors, Repurposing against COVID-19. Life Sci. 248, 117477. doi:10.1016/j.lfs.2020.117477
Gautret, P., Lagier, J.-C., Parola, P., Hoang, V. T., Meddeb, L., Mailhe, M., et al. (2020). Hydroxychloroquine and Azithromycin as a Treatment of COVID-19: Results of an Open-Label Non-randomized Clinical Trial. Int. J. Antimicrob. Agents 56 (1), 105949. doi:10.1016/j.ijantimicag.2020.105949
Ghasemzadeh, A., Jaafar, H. Z. E., and Rahmat, A. (2010). Antioxidant Activities, Total Phenolics and Flavonoids Content in Two Varieties of Malaysia Young Ginger (Zingiber Officinale Roscoe). Molecules 15 (6), 4324–4333. doi:10.3390/molecules15064324
Ghorbani, A., and Esmaeilizadeh, M. (2017). Pharmacological Properties of Salvia Officinalis and its Components. J. Traditional Complement. Med. 7 (4), 433–440. doi:10.1016/j.jtcme.2016.12.014
Hossain, M., Urbi, Z., Sule, A., and Rahman, K. (2014). Andrographis Paniculata (Burm. f.) Wall. Ex Nees: a Review of Ethnobotany, Phytochemistry, and Pharmacology. Scientific World J. 2014, 274905. doi:10.1155/2014/274905
Hussain, M. S., Fareed, S., and Ali, M. (2010). Hygrophila Auriculata (K. Schum) Heine: Ethnobotany, Phytochemistry and Pharmacology. Asian J. Traditional Medicines 5 (4), 121–131.
Islam, M. T., Zihad, S. M. N. K., Rahman, M. S., Sifat, N., Khan, M. R., Uddin, S. J., et al. (2019). Agathisflavone: Botanical Sources, Therapeutic Promises, and Molecular Docking Study. IUBMB life 71 (9), 1192–1200. doi:10.1002/iub.2053
Jain, R., Sharma, A., Gupta, S., Sarethy, I. P., and Gabrani, R. (2011). Solanum nigrum: Current Perspectives on Therapeutic Properties. Altern. Med. Rev. 16 (1), 78–85.
Jasim, H., Hussein, A. O., Hameed, I. H., and Kareem, M. A. (2015). Characterization of Alkaloid Constitution and Evaluation of Antimicrobial Activity of Solanum nigrum Using Gas Chromatography Mass Spectrometry (GC-MS). J. Pharmacognosy phytotherapy 7 (4), 56–72.
Javed, T., Ashfaq, U. A., Riaz, S., Rehman, S., and Riazuddin, S. (2011). In-vitro Antiviral Activity of Solanum nigrum against Hepatitis C Virus. Virol. J. 8 (1), 1–7. doi:10.1186/1743-422x-8-26
Juang, L.-J., Sheu, S.-J., and Lin, T.-C. (2004). Determination of Hydrolyzable Tannins in the Fruit ofTerminalia Chebula Retz. By High-Performance Liquid Chromatography and Capillary Electrophoresis. J. Sep. Sci. 27 (9), 718–724. doi:10.1002/jssc.200401741
Kaihatsu, K., Yamabe, M., and Ebara, Y. (2018). Antiviral Mechanism of Action of Epigallocatechin-3-O-Gallate and its Fatty Acid Esters. Molecules 23 (10), 2475. doi:10.3390/molecules23102475
Kashyap, D., Sharma, A., Tuli, S. H., Punia, S., and Sharma, K. A. (2016). Ursolic Acid and Oleanolic Acid: Pentacyclic Terpenoids with Promising Anti-inflammatory Activities. Iad 10 (1), 21–33. doi:10.2174/1872213x10666160711143904
Katoh, K., Rozewicki, J., and Yamada, K. D. (2019). MAFFT Online Service: Multiple Sequence Alignment, Interactive Sequence Choice and Visualization. Brief. Bioinformatics 20 (4), 1160–1166. doi:10.1093/bib/bbx108
Kaur, R., Kaur, H., and Dhindsa, A. S. (2013). Glycyrrhiza Glabra: a Phytopharmacological Review. Int. J. Pharm. Sci. Res. 4 (7), 2470. doi:10.13040/IJPSR.0975-8232.4(7).2470-77
Kaur, R., Ruhil, S., Balhara, M., Dhankhar, S., and Chhillar, A. (2013). A Review on Justicia Adhatoda: A Potential Source of Natural Medicine. Afr. J. Plant Sci. 5 (11), 620–627. doi:10.5897/AJPS.9000004
Khan, M. A. A., Ansair, M., and Maheshwari, R. K. (2018). Phytochemistry and Pharmacological Properties of Ginger (Zingiber Officinale). Delhi, India: Lenin Media Pvt. Ltd.
Kim, S., Chen, J., Cheng, T., Gindulyte, A., He, J., He, S., et al. (2019). PubChem 2019 Update: Improved Access to Chemical Data. Nucleic Acids Res. 47 (D1), D1102–D1109. doi:10.1093/nar/gky1033
Koirala, N., Dhakal, C., Munankarmi, N. N., Ali, S. W., Hameed, A., Martins, N., et al. (2020). Vitex Negundo Linn.: Phytochemical Composition, Nutritional Analysis, and Antioxidant and Antimicrobial Activity. Cel Mol Biol. (Noisy-le-grand) 66 (4), 1–7. doi:10.14715/cmb/2020.66.4.1
Kolla, J. N., Kulkarni, N. M., Kura, R. R., and Theepireddy, S. K. R. (2017). Terminalia Chebula Retz.-an Important Medicinal Plant. Herba Pol. 63 (4), 45–56. doi:10.1515/hepo-2017-0024
Konan, N. A., Lincopan, N., Collantes Díaz, I. E., de Fátima Jacysyn, J., Tanae Tiba, M. M., Pessini Amarante Mendes, J. G., et al. (2012). Cytotoxicity of Cashew Flavonoids towards Malignant Cell Lines. Exp. Toxicologic Pathol. 64 (5), 435–440. doi:10.1016/j.etp.2010.10.010
Kothandan, S., and Swaminathan, R. (2014). Evaluation of In Vitro Antiviral Activity of Vitex Negundo L., Hyptis Suaveolens (L) poit., Decalepis Hamiltonii Wight & Arn., to Chikungunya Virus. Asian Pac. J. Trop. Dis. 4, S111–S115. doi:10.1016/s2222-1808(14)60424-2
Krup, V., Prakash, L., and Harini, A. (2013). Pharmacological Activities of Turmeric (Curcuma Longa Linn): a Review 1000133. J. Homeop Ayurv Med. 2 (133), 2167–1206.
Kumar, A., Lakshman, K., Jayaveera, K., Tripathi, S. M., and Satish, K. (2010). Estimation of Gallic Acid, Rutin and Quercetin in Terminalia Chebula by HPTLC. Jordan J. Pharm. Sci. 3 (1), 63–68.
Kumar, N., and Khurana, S. (2018). Phytochemistry and Medicinal Potential of the Terminalia Bellirica Roxb.(Bahera). Indian J. Nat. Prod. Resour. (IJNPR)[Formerly Nat. Product. Radiance (NPR)] 9 (2), 97–107.
Kumar, P., and Nishteswar, K. (2013). Phytochemical and Pharmacological Profiles of Clerodendrum Serratum Linn.(Bharngi): A Review. Int. J. Res. Ayurveda Pharm. 4 (2), 276–278.
Kumar, P., Singh, S., and Kumar, N. (2020). Plectranthus Amboinicus: a Review on its Pharmacological and, Pharmacognostical Studies. A. J. Physiol. Biochem. Pharmacol. 10 (2), 55–62. doi:10.5455/ajpbp.20190928091007
Lee, N., Hui, D., Wu, A., Chan, P., Cameron, P., Joynt, G. M., et al. (2003). A Major Outbreak of Severe Acute Respiratory Syndrome in Hong Kong. N. Engl. J. Med. 348 (20), 1986–1994. doi:10.1056/nejmoa030685
Lim, T. K. (2013). Solanum Torvum. Edible Medicinal and Non-medicinal Plants. Dordrecht: Springer, 429–441. doi:10.1007/978-94-007-5628-1_48
Liu, J., Cao, R., Xu, M., Wang, X., Zhang, H., Hu, H., et al. (2020). Hydroxychloroquine, a Less Toxic Derivative of Chloroquine, Is Effective in Inhibiting SARS-CoV-2 Infection In Vitro. Cel Discov. 6 (1), 1–4. doi:10.1038/s41421-020-0156-0
Lobo, A. R., and Satish, S. (2019). An Investigation on Anti-depressant Activity of Fresh Fruit Juice of Malus Domestica in Experimental Animal Models. Int. J. Res. Pharm. Pharm. Sci. 4 (1), 19–23.
Ma, H., Diao, Y., Zhao, D., Li, K., and Kang, T. (2010). A New Alternative to Treat Swine Influenza A Virus Infection: Extracts from Terminalia Chebula Retz. Afr. J. Microbiol. Res. 4 (6), 497–499.
Ma, X., Wu, H., Liu, L., Yao, Q., Wang, S., Zhan, R., et al. (2011). Polyphenolic Compounds and Antioxidant Properties in Mango Fruits. Scientia Horticulturae 129 (1), 102–107. doi:10.1016/j.scienta.2011.03.015
Mandal, S., and Mandal, M. (2015). Coriander (Coriandrum Sativum L.) Essential Oil: Chemistry and Biological Activity. Asian Pac. J. Trop. Biomed. 5 (6), 421–428. doi:10.1016/j.apjtb.2015.04.001
Martin, Y. C. (2005). A Bioavailability Score. J. Med. Chem. 48 (9), 3164–3170. doi:10.1021/jm0492002
Meena, A. K., Niranjan, U., Rao, M., Padhi, M., and Babu, R. (2011). A Review of the Important Chemical Constituents and Medicinal Uses of Vitex Genus. Asian J. Traditional Medicines 6 (2), 54–60.
Nair, R. (2012). HIV-1 Reverse Transcriptase Inhibition by Vitex Negundo L. Leaf Extract and Quantification of Flavonoids in Relation to Anti-HIV Activity. J. Cel Mol Biol. 10, 53–59.
Onuh, J. O., Idoko, G., Yusufu, P., and Onuh, F. (2017). Comparative Studies of the Phytochemical, Antioxidant and Antimicrobial Properties of Cashew Leaf, Bark and Fruits Extracts. Am. J. Food Nutr. 5 (4), 115–120.
Pandey, S., Kushwaha, G. R., Singh, A., and Singh, A. (2018). Chemical Composition and Medicinal Uses of Anacyclus Pyrethrum. Pharma Sci. Monitor 9 (1), 551–560.
Park, N.-H., Lee, C.-W., Bae, J.-h., and Na, Y. J. (2011). Protective Effects of Amentoflavone on Lamin A-dependent UVB-Induced Nuclear Aberration in normal Human Fibroblasts. Bioorg. Med. Chem. Lett. 21 (21), 6482–6484. doi:10.1016/j.bmcl.2011.08.067
Parvez, G. M. (2016). Pharmacological Activities of Mango (Mangifera Indica): A Review. J. Pharmacognosy Phytochemistry 5 (3), 1.
Patel, D. K., Laloo, D., Kumar, R., and Hemalatha, S. (2011a). Pedalium Murex Linn.: an Overview of its Phytopharmacological Aspects. Asian Pac. J. Trop. Med. 4 (9), 748–755. doi:10.1016/s1995-7645(11)60186-7
Patel, J. R., Tripathi, P., Sharma, V., Chauhan, N. S., and Dixit, V. K. (2011b). Phyllanthus Amarus: Ethnomedicinal Uses, Phytochemistry and Pharmacology: a Review. J. ethnopharmacology 138 (2), 286–313. doi:10.1016/j.jep.2011.09.040
Pathirana, C. K., Madhujith, T., and Eeswara, J. (2020). Bael (Aegle Marmelos L. Corrêa), a Medicinal Tree with Immense Economic Potentials. Adv. Agric. 2020, 8814018. doi:10.1155/2020/8814018
Peerzada, A. M., Ali, H. H., Naeem, M., Latif, M., Bukhari, A. H., and Tanveer, A. (2015). Cyperus Rotundus L.: Traditional Uses, Phytochemistry, and Pharmacological Activities. J. ethnopharmacology 174, 540–560. doi:10.1016/j.jep.2015.08.012
Prajapat, M., Sarma, P., Shekhar, N., Avti, P., Sinha, S., Kaur, H., et al. (2020). Drug Targets for corona Virus: A Systematic Review. Indian J. Pharmacol. 52 (1), 56–65. doi:10.4103/ijp.IJP_115_20
Prakash, P., and Gupta, N. (2005). Therapeutic Uses of Ocimum Sanctum Linn (Tulsi) with a Note on Eugenol and its Pharmacological Actions: a Short Review. Indian J. Physiol. Pharmacol. 49 (2), 125–131.
Principi, N., and Esposito, S. (2020). Chloroquine or Hydroxychloroquine for Prophylaxis of COVID-19. Lancet Infect. Dis. 20 (10), 1118. doi:10.1016/S1473-3099(20)30296-6
Rajapakse, S., de Silva, N. L., Weeratunga, P., Rodrigo, C., Sigera, C., and Fernando, S. D. (2019). Carica Papaya Extract in Dengue: a Systematic Review and Meta-Analysis. BMC Complement. Altern. Med. 19 (1), 1–8. doi:10.1186/s12906-019-2678-2
Rex, M. C., and Ravi, L. (2020). A Review on Cissus Quadrangularis L. As Herbal Medicine. Indian J. Nat. Prod. Resour. (IJNPR)[Formerly Nat. Product. Radiance (NPR)] 11 (3), 155–164.
Ryu, Y. B., Jeong, H. J., Kim, J. H., Kim, Y. M., Park, J.-Y., Kim, D., et al. (2010). Biflavonoids from Torreya Nucifera Displaying SARS-CoV 3CLpro Inhibition. Bioorg. Med. Chem. 18 (22), 7940–7947. doi:10.1016/j.bmc.2010.09.035
Sadeghian, M. M., and Mortazaienezhad, F. (2007). Investigation of Compounds from Azadirachta indica (Neem). Asian J. Plant Sci. 6, 444-445. doi:10.3923/ajps.2007.444.445
Sampangi-Ramaiah, M. H., Vishwakarma, R., and Shaanker, R. U. (2020). Molecular Docking Analysis of Selected Natural Products from Plants for Inhibition of SARS-CoV-2 Main Protease. Curr. Sci. 118 (7), 1087–1092.
Sarkar, S., Pal, A., Chouni, A., and Paul, S. (2020). A Novel Compound β-sitosterol-3-O-β-D-glucoside Isolated from Azadirachta indica Effectively Induces Apoptosis in Leukemic Cells by Targeting G0/G1 Populations. Indian J. Biochem. Biophys. (Ijbb) 57 (1), 27–32.
Shahrajabian, M. H., Sun, W., and Cheng, Q. (2020). Chemical Components and Pharmacological Benefits of Basil (Ocimum Basilicum): a Review. Int. J. Food Properties 23 (1), 1961–1970. doi:10.1080/10942912.2020.1828456
Sharma, V., Sharma, R., Gautam, D., Kuca, K., Nepovimova, E., and Martins, N. (2020). Role of Vacha (Acorus calamus Linn.) in Neurological and Metabolic Disorders: Evidence from Ethnopharmacology, Phytochemistry, Pharmacology and Clinical Study. Jcm 9 (4), 1176. doi:10.3390/jcm9041176
Shi, W.-Z., Jiang, L.-Z., Song, G.-P., Wang, S., Xiong, P., and Ke, C.-W. (2020). Study on the Antiviral Activities and Hemagglutinin-Based Molecular Mechanism of Novel Chlorogenin 3-O-β-Chacotrioside Derivatives against H5N1 Subtype Viruses. Viruses 12 (3), 304. doi:10.3390/v12030304
Sivagami, B., and Sailaja, B. (2021). A Review on Analytical Methods for Antiviral Phytoconstituents. J. Young Pharm. 13 (1), 7.
Srivastava, P. (2014). Therapeutic Potential of Piper Longum L. For Disease Management-A Review. Int. J. Pharma Sci. 4 (4), 692–696.
Tian, W., Chen, C., Lei, X., Zhao, J., and Liang, J. (2018). CASTp 3.0: Computed Atlas of Surface Topography of Proteins. Nucleic Acids Res. 46 (W1), W363–W367. doi:10.1093/nar/gky473
Touret, F., and de Lamballerie, X. (2020). Of Chloroquine and COVID-19. Antivir. Res. 177, 104762. doi:10.1016/j.antiviral.2020.104762
Venkateswarlu, K. (2012). Vitex Negundo: Medicinal Values, Biological Activities, Toxicity Studies and Phytopharmacological Actions. Int. J. Pharm. Phytopharmacological Res. 2 (2), 126–133.
Vij, T., and Prashar, Y. (2015). A Review on Medicinal Properties of Carica Papaya Linn. Asian Pac. J. Trop. Dis. 5 (1), 1–6. doi:10.1016/s2222-1808(14)60617-4
Vlase, L., Benedec, D., Hanganu, D., Damian, G., Csillag, I., Sevastre, B., et al. (2014). Evaluation of Antioxidant and Antimicrobial Activities and Phenolic Profile for Hyssopus Officinalis, Ocimum Basilicum and Teucrium Chamaedrys. Molecules 19 (5), 5490–5507. doi:10.3390/molecules19055490
Wang, Z., Li, J., Ji, Y., An, P., Zhang, S., and Li, Z. (2013). Traditional Herbal Medicine: a Review of Potential of Inhibitory Hepatocellular Carcinoma in Basic Research and Clinical Trial. Evid. Based Complement. Alternat Med. 2013, 268963. doi:10.1155/2013/268963
Wishart, D. S., Feunang, Y. D., Guo, A. C., Lo, E. J., Marcu, A., Grant, J. R., et al. (2018). DrugBank 5.0: a Major Update to the DrugBank Database for 2018. Nucleic Acids Res. 46 (D1), D1074–D1082. doi:10.1093/nar/gkx1037
Wu, C., Liu, Y., Yang, Y., Zhang, P., Zhong, W., Wang, Y., et al. (2020). Analysis of Therapeutic Targets for SARS-CoV-2 and Discovery of Potential Drugs by Computational Methods. Acta Pharmaceutica Sinica B 10 (5), 766–788. doi:10.1016/j.apsb.2020.02.008
Xu, X., Chen, P., Wang, J., Feng, J., Zhou, H., Li, X., et al. (2020). Evolution of the Novel Coronavirus from the Ongoing Wuhan Outbreak and Modeling of its Spike Protein for Risk of Human Transmission. Sci. China Life Sci. 63 (3), 457–460. doi:10.1007/s11427-020-1637-5
Yang, X., Chen, X., Bian, G., Tu, J., Xing, Y., Wang, Y., et al. (2014). Proteolytic Processing, Deubiquitinase and Interferon Antagonist Activities of Middle East Respiratory Syndrome Coronavirus Papain-like Protease. J. Gen. Virol. 95 (3), 614–626. doi:10.1099/vir.0.059014-0
Yu, S., Yan, H., Zhang, L., Shan, M., Chen, P., Ding, A., et al. (2017). A Review on the Phytochemistry, Pharmacology, and Pharmacokinetics of Amentoflavone, a Naturally-Occurring Biflavonoid. Molecules 22 (2), 299. doi:10.3390/molecules22020299
Zaidi, S. K., Hoda, M., Tabrez, S., Ansari, S. A., Jafri, M. A., Shahnawaz Khan, M., et al. (2014). Protective Effect of Solanum nigrum Leaves Extract on Immobilization Stress Induced Changes in Rat’s Brain. Evidence-Based Complement. Altern. Med. 2014, 912450. doi:10.1155/2014/912450
Keywords: nCOVID-19, SARS-CoV-2, molecular docking, cyanin, amentoflavone, agathisflavone
Citation: Nallusamy S, Mannu J, Ravikumar C, Angamuthu K, Nathan B, Nachimuthu K, Ramasamy G, Muthurajan R, Subbarayalu M and Neelakandan K (2021) Exploring Phytochemicals of Traditional Medicinal Plants Exhibiting Inhibitory Activity Against Main Protease, Spike Glycoprotein, RNA-dependent RNA Polymerase and Non-Structural Proteins of SARS-CoV-2 Through Virtual Screening. Front. Pharmacol. 12:667704. doi: 10.3389/fphar.2021.667704
Received: 14 February 2021; Accepted: 24 June 2021;
Published: 08 July 2021.
Edited by:
Christophe F. Hano, INRA EA1207 Laboratoire de Biologie des Ligneux et des Grandes Cultures, FranceReviewed by:
José Blanco-Salas, University of Extremadura, SpainJen-Tsung Chen, National University of Kaohsiung, Taiwan
Copyright © 2021 Nallusamy, Mannu, Ravikumar, Angamuthu, Nathan, Nachimuthu, Ramasamy, Muthurajan, Subbarayalu and Neelakandan. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Mohankumar Subbarayalu, bW9oYW5rdW1hci5zQHRuYXUuYWMuaW4=
 Saranya Nallusamy
Saranya Nallusamy Jayakanthan Mannu
Jayakanthan Mannu Caroline Ravikumar
Caroline Ravikumar Kandavelmani Angamuthu1
Kandavelmani Angamuthu1 Bharathi Nathan
Bharathi Nathan