
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Pharmacol. , 12 May 2021
Sec. Ethnopharmacology
Volume 12 - 2021 | https://doi.org/10.3389/fphar.2021.652335
This article is part of the Research Topic Biomolecules Against Coronaviruses: Molecular Aspects, Multi-omics and Systems Pharmacology View all 7 articles
 Rachana Singh1
Rachana Singh1 Pradhyumna Kumar Singh2
Pradhyumna Kumar Singh2 Rajnish Kumar1*
Rajnish Kumar1* Md. Tanvir Kabir3
Md. Tanvir Kabir3 Mohammad Amjad Kamal4,5,6
Mohammad Amjad Kamal4,5,6 Abdur Rauf7
Abdur Rauf7 Ghadeer M. Albadrani8
Ghadeer M. Albadrani8 Amany A. Sayed9
Amany A. Sayed9 Shaker A. Mousa10
Shaker A. Mousa10 Mohamed M. Abdel-Daim11
Mohamed M. Abdel-Daim11 Md. Sahab Uddin12,13*†
Md. Sahab Uddin12,13*†COVID-19 is caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2). It has a disastrous effect on mankind due to the contagious and rapid nature of its spread. Although vaccines for SARS-CoV-2 have been successfully developed, the proven, effective, and specific therapeutic molecules are yet to be identified for the treatment. The repurposing of existing drugs and recognition of new medicines are continuously in progress. Efforts are being made to single out plant-based novel therapeutic compounds. As a result, some of these biomolecules are in their testing phase. During these efforts, the whole-genome sequencing of SARS-CoV-2 has given the direction to explore the omics systems and approaches to overcome this unprecedented health challenge globally. Genome, proteome, and metagenome sequence analyses have helped identify virus nature, thereby assisting in understanding the molecular mechanism, structural understanding, and disease propagation. The multi-omics approaches offer various tools and strategies for identifying potential therapeutic biomolecules for COVID-19 and exploring the plants producing biomolecules that can be used as biopharmaceutical products. This review explores the available multi-omics approaches and their scope to investigate the therapeutic promises of plant-based biomolecules in treating SARS-CoV-2 infection.
The world has faced several chronic diseases, a few epidemics, and pandemics that were disastrous and eliminated a predominant human population throughout history. Among all these, the present-day COVID-19 is potentially contagious and has left the most devastating effect on humankind (Prasad and Prasad, 2020; Shahriar et al., 2021). The virulent nature of COVID-19 and the rise of its different strains have forced researchers worldwide to search for rapid diagnostic methods for detection, development of new vaccines, various therapeutic drugs, and immunization options. Success has been achieved in developing different diagnosis methods, but no drug has been developed to efficiently provide treatment to the infected patient (Hossain et al., 2020; Lurie et al., 2020; Loeffelholz and Tang, 2020).
Coronavirus belongs to the family Coronaviridae in the order Nidovirales. This group of viruses is classified into four genera such as Alpha coronavirus (α-CoV), Beta coronavirus (β-CoV), Gamma coronavirus (γ-CoV), and Delta coronavirus (δ-CoV) (Woo et al., 2010). Both the alpha- and beta-coronaviruses tend to infect mammals. Among them, β-CoV has become the utmost concern of the world due to its ability to cause serious illness in the human population, like the Middle East respiratory syndrome–related coronavirus (MERS-CoV), SARS-CoV, and SARS-CoV-2, which cause fatal respiratory tract infection in humans (Lu et al., 2020). The structural and nonstructural information of SARS-CoV-2 has already been explained and explored by many studies available in the literature (Hillen et al., 2020; Jin et al., 2020; Clark et al., 2021; Hasana et al., 2021; Semper et al., 2021).
Although many vaccines such as Comirnaty (mRNA based), Moderna (mRNA based), AstraZeneca/Covishield (adenovirus vaccine), Sputnik V (nonreplicating viral vector), CoronaVac (inactivated vaccine), BBIBP-CorV (inactivated vaccine), EpiVacCorona (peptide vaccine), Convidicea (adenovirus type 5 vector-recombinant vaccine), and Covaxin (inactivated vaccine) have been approved in various countries, a specific approved drug for COVID-19 is still missing. The significant challenges in mass immunization include large-scale production, cost-effectiveness, and global allocation. Moreover, until the vaccination preferably drives herd immunity, social distancing is still the most effective measure adopted all over the world to prevent the spread of the disease (McKibbin and Fernando, 2020; Sohrabi et al., 2020). Mostly, preexisting antiviral drugs are still being used in the management of SARS-CoV-2–infected patients. Simultaneously, the repurposing of drugs is also recommended for testing and application. Some preexisting antiviral drugs like remdesivir and lopinavir/ritonavir are currently being used in the management of COVID-19 (Kumar et al., 2020a; Choy et al., 2020; Wang et al., 2020f; Zhu et al., 2020).
However, there is an urgent need to investigate and discover potential therapeutic compounds that could restrict viral replication and its assembly in the human body. Finding a permissive cell and delivering genetic information into its cytoplasm are essential steps for viral infection into the host cell. From a therapeutic point of view, there are at least two important ways. The first is that blocking viral entry stops infection early on and the second is preventing viral replication. The antiviral drugs can be developed by targeting different viral entry stages, either by blocking virus-specific interactions or by inhibiting conserved cellular mechanisms that viruses exploit to enter cells (Mazzon and Marsh, 2019).
Advancements in biotechnology and computational analysis have taken place in the past decade, and they have provided massive amounts of new data that have given an optimistic outlook for intensifying disease treatment developments (Belfiore et al., 2020; Infusino et al., 2020; Stebbing et al., 2020). Nearly every disease and clinical research area has exploded with enormous data (Lin et al., 2020). Computational analysis of the available diverse omics data could provide an in-depth understanding of molecular mechanisms and associated transitions of the diseases (Kumar et al., 2020b; Muthuramalingam et al., 2020; Su et al., 2020). Clinical data management, genome and proteome analyses, next-generation sequencing data mining, machine learning, and deep learning algorithms have progressed significantly for mining patterns from such enormous data (Kumar et al., 2016; Kumar et al., 2018; Hossain et al., 2021; Overmyer et al., 2021).
Omics represent the collective technologies that help to investigate the different molecules’ roles and actions that make up an organism’s cells. These include genomics (gene), transcriptomics (mRNA), proteomics (proteins), and metabolomics (metabolites). These techniques have already played a significant role in vaccine development and repurposing of drugs, as shown in Figure 1. Omics technologies’ main importance lies in detecting and verifying all gene products (transcripts, proteins, and metabolites) available in any given biological sample. Omics analysis has been extensively applied in drug discovery (Horgan and Kenny, 2011; Chen B. et al., 2020; Aaron et al., 2020) and estimation of their effectiveness and toxicity (Figure 2). These are high-throughput technologies that have been significantly assisting in describing gene/protein expression profiles and their complex effects for SARS-CoV-2 (Wang et al., 2020a; Bojkova et al., 2020; Kim et al., 2020; Li et al., 2020). With their immense possibilities, the powerful omics techniques seem like they will continue supporting researchers and healthcare professionals in exploring and exploiting SARS-CoV-2 pathophysiology for a deeper understanding of its processes and understanding the disease for diagnosis, screening, and prognosis (Wang et al., 2020b; Hart and Halden, 2020; Robson, 2020).
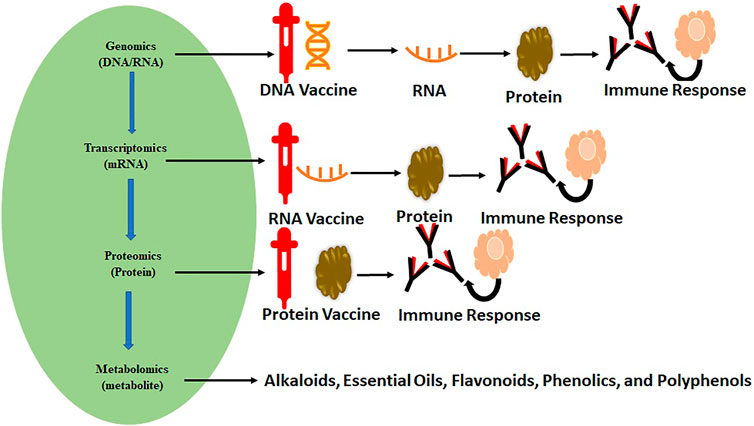
FIGURE 1. Omics science and flow of biological information assisting the vaccine development and drug repurposing for various diseases.
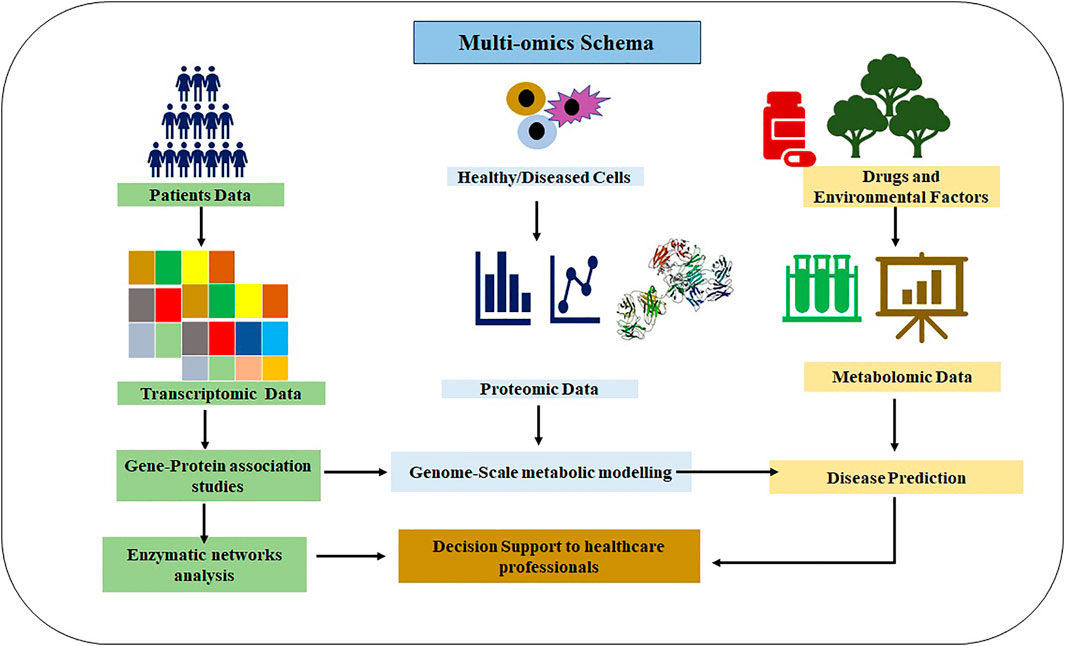
FIGURE 2. Multi-omics schema integrating different types of omics data to support the healthcare professionals in decision-making.
The reports on the uses of preexisting drugs suggest that 3CLpro, Spike, RNA-dependent RNA polymerase (RdRp), PLpro, and human angiotensin-converting enzyme 2 (human ACE2) are potential drug targets for SARS-CoV-2 for further in vitro and in vivo studies (Xu et al., 2020).
Since ancient times, plant preparations have been utilized as herbal medicines, which probably also contain active antiviral extracts/compounds, to cure and control infectious diseases (Israt and Ahmet, 2020; Yang et al., 2020; Al-Ishaq et al., 2020). Screening of plants to find the potential antiviral compounds was conducted long ago, during 1952. A total of 288 extracts from plant sources were experimented to check their role against the influenza A virus (Chantrill et al., 1952). Thereafter, many scientists have worked on utilizing the potential of plant extracts with different viral diseases. Debiaggi et al. (1988) worked on the antiviral activity of Chamaecyparis lawsoniana (A. Murr bis) Parl. against herpes simplex virus type 2. Another study found that Geranium sanguineum L. has an antiviral activity for the influenza A virus. It was discovered that polyphenolic extract derived from Geranium sanguineum L. effectively inhibits the influenza virus’s reproduction (Serkedjieva, 2003). Medicinal plants contain a variety of secondary metabolites with the potential to inhibit viral proteins and their enzymes, which consequently stop/suppress the viral entry and replication into the host (Semple et al., 1998; Li and Peng, 2013; Arbab et al., 2017; Akram et al., 2018; Dhama et al., 2018). Kotwal and coworkers studied the acidic extract of Trifolium species that showed an antiviral activity (broad-spectrum) (Kotwal et al., 2005). Several studies have been carried out, and they have confirmed that bioactive natural compounds are potential candidates for the treatment of SARS-CoV-2 because of their effective antiviral activity (Figure 3) (Behl et al., 2020; Kabir et al., 2020; Qamar et al., 2020; Zhang L. et al., 2020; Kumar et al., 2021).
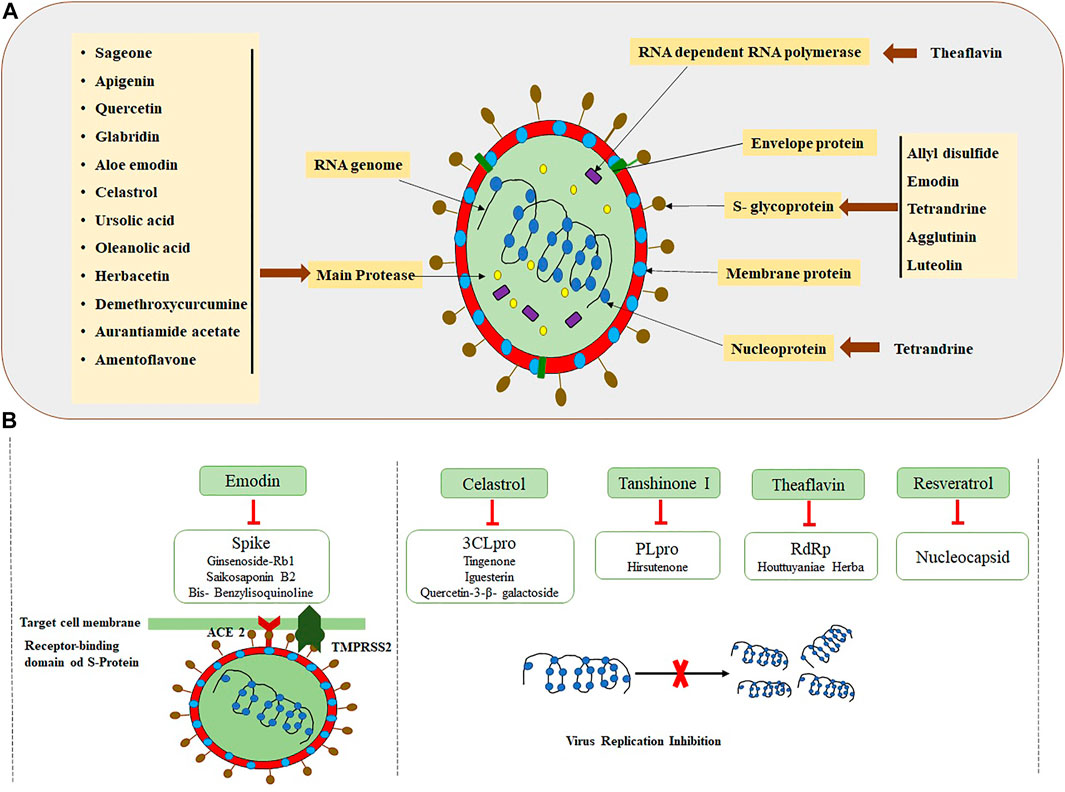
FIGURE 3. Interaction of SARS-CoV-2 structural proteins with the potential plant-based biomolecules. (A) Structure of SARS-CoV-2 and available biomolecules. (B) Rich resources of Chinese herbal medicines targeting SARS-CoV-2 (Sharma N. et al., 2020; Xian et al., 2020).
These natural biomolecules are economical and deemed safe with minimum side effects compared to synthetic compounds (Enmozhi et al., 2020). Although the use of natural biomolecules generally involves minimal or no side effects, sometimes, they can potentially be toxic. This may be due to misidentification of the biomolecules in the form that they are sold, wrong preparation, dispensing, or administration by inadequately trained personnel (Karimi et al., 2015). Moreover, if a potential biomolecule could be found to regulate COVID-19 infection, it would be a boon in the treatment and could be used immediately after acquiring compulsory ethical clearances. Despite sufficient evidence that plant-derived biomolecules are effective as an antiviral agent, their use is still limited because these compounds are either available at minimal levels or might not be stable outside the plants (Anand et al., 2021; Balkrishna et al., 2021). Therefore, a comprehensive strategy is needed to properly identify potential biomolecules’ biosynthetic pathways and analyze the genes underlying those pathways (Shree et al., 2020). As the identification of plant-based moieties with anti–SARS-CoV-2 action is in a devolving phase, it will take a long time to explore their full potential using the traditional drug development method. The omics techniques used in plant molecular biology could accelerate this procedure using a strategically planned study design and provide a platform to researchers and the bio-manufacturer of these biomolecules to manage the COVID-19 pandemic (Sharma N. et al., 2020; Chojnacka et al., 2020). This article has explored the plant-based compounds that contain antiviral activities to assess the impact of the multi-omics approach for plant biomolecular research and its potential application against SARS-CoV-2.
There is enough evidence that herbal medicines have been used in different parts of the world since prehistoric times. Asian countries like India, Japan, China, and Pakistan, and countries in Africa are using medicinal plants for herbal treatment. Herbal plants are supplemented with various phytochemicals like alkaloids, flavonoids, phenolic acids, lignins, and terpenoids. All have shown their potential against infectious diseases (Kim et al., 2019; Lalani and Poh, 2020). It was demonstrated that biomolecules of plants exhibit inhibitory activity on hepatitis-B virus (Huang et al., 2006), herpes simplex virus type-2 (Debiaggi et al., 1988), human immunodeficiency viruses (Asres and Bucar, 2005; Lubbe et al., 2012), dengue virus, rotavirus, Zika virus (Oliveira et al., 2017; Akram et al., 2018), and SARS coronavirus (Prasad et al., 2020; Tsai et al., 2020; Yang et al., 2020). Different computational studies on the antiviral promises of herbal plants indicated that secondary metabolites present in plant extracts could interact with different targets of the SARS-CoV-2 virus. Table 1 summarizes some of the recently published plant-based secondary metabolites that have shown some therapeutic promises against SARS-CoV-2 in computational studies.
Due to homology in SARS-CoV and SARS-CoV-2, the previous studies may also endorse the potential of naturally occurring compounds to inhibit SARS-CoV-2. Various studies are currently using traditional medicines and extracting their therapeutic potential against SARS-CoV-2 (Ang et al., 2020; Ren et al., 2020; Silveira et al., 2020). A well-studied traditional Chinese medicine (TCM) is reported to treat SARS-CoV and various other diseases (Tong et al., 2004; Cao et al., 2020; Huang et al., 2020; Zhang Q. et al., 2020; Zhang et al., 2021). This medicine is also applied to treat the patients of SARS-CoV-2, with approximately 85% success (Yang et al., 2020). The plants used in TCM contain active biomolecules like flavonoids (herbacetin, isobavaschacone, rhoifolin, quercetin 3-b-D-glucoside, epigallocatechin gallate, and pectolinarin) that regulate virus accumulation by restricting the multiplication. In a recent report, Sehailia and Chemat (2020) studied the infection mechanism of SARS-CoV-2, compared it with that of plasmodium, and reported that each pathogen causes lung infection by crystallizing carbon dioxide. Based on this finding, artemisinin, isolated from Artemisia annua L., has been proposed to treat SARS-CoV-2–infected patients. Artemisinin is commonly used to treat malaria and against various viral infections like MERS-CoV and SARS-CoV (D’Alessandro et al., 2020; Hahn et al., 2018). Collaborative research between the Max-Plank Research Center, Germany, and Mateon Therapeutics, California, is being performed using Artemisia annua L. to treat COVID-19–infected patients. This research is supported by the World Health Organization (WHO).
3-Chymotrypsin–like protease (3CLpro) is vital for replicating the virus, and thus represents a promising drug target for the development of therapeutic agents for SARS-CoV and other human coronaviruses, including SARS-CoV-2 (Yang et al., 2020). The following reports suggest that Chinese rhubarb extracts (Luo et al., 2009), Houttuynia cordata (Lau et al., 2008; Fung et al., 2011), flavonoids (Jo et al., 2019; Jo et al., 2020), beta-sitosterol extracted from the root extract of Isatis indigotica, and herbal extracts of TCM could inhibit the enzymatic activity of SARS 3CLpro. Besides that, the flavonoids, namely, herbacetin, isobavaschalcone, quercetin 3-β-D-glucoside, and helichrysetin, had shown their capability to block the enzymatic activity of MERS-CoV 3CL protease (Jo et al., 2019). The RNA-dependent RNA polymerase (RdRp) is a key enzyme responsible for both positive- and negative-strand RNA synthesis, and it is another potential target for a drug. It was reported by Fung et al. (2011) that the extracts of Kang Du Bu Fei Tang, Sinomenium acutum, Coriolus versicolor, and Ganoderma lucidum, inhibited SARS-CoV RdRp when tested in different doses (Fung et al., 2011).
The applications of essential oils, extracted from various medicinal plants like Citrus spp, Mentha spp, ginger, Hyssopus officinalis L., Illicium spp, and Santalum spp with antiviral effects, have been well studied by numerous researchers (Li and Peng, 2013; Akram et al., 2018; Dhama et al., 2018; Wink 2020). Ben-Shabat and coworkers reported that these essential oils alter the viral-envelope lipid-bilayer membrane's fluidity (Ben-Shabat et al., 2020). In 2003, Schnitzler reported that monoterpenes, oxygenated sesquiterpenes, and phenylpropanoids of essential oils could disrupt the phospholipid bilayer membrane of human coronavirus that interferes with the envelope protein structure of the virus during infection (Schnitzler et al., 2008).
A major study showed that eucalyptol (essential oil from gum trees, Eucalyptus spp.) is effective against SARS-CoV-2 and other coronaviruses because its major component, eucalyptus oil, consists of ketone, ether, and hydroxyl groups that play a crucial role in the inhibition of SARS-CoV-2 (Sharma, 2020). Another compound named jensenone, obtained from eucalyptus essential oil, has also shown antiviral potential to inhibit Mpro of COVID-19 (Sharma and Kaur, 2020).
Chloroquine is an alkaloid that is obtained from the cinchona tree’s bark. It has DNA intercalating properties and is identified as a potential candidate for developing an effective drug for SARS-CoV-2 (Devaux et al., 2020). Another alkaloid, Resochin, used in malaria treatment, has been thought to be an effective antiviral compound as it can interfere with the replication, transcription, and protein synthesis of viral RNA (Wink, 2020). The isoquinolines, for example, palmatine and chelidonine, are also promising biomolecules that could be potential drug candidates against COVID-19 (Ho et al., 2019; Wink, 2020).
Kim et al. (2019) studied the important bis-benzylisoquinoline alkaloids extracted from Stephania tetrandra S. Moore and related species of Menispermaceae, such as cepharanthine (CEP), tetrandrine (TET), and fangchinoline (OFAN). They investigated the antiviral activity of these alkaloids against HCoV-OC43 in human coronavirus–infected MRC-5 human lung cells (Kim et al., 2019). The result showed that all three of these alkaloids could decrease the replication of HCoV-OC43 inside host cells. Apart from that, they also inhibited the viral spike and nucleocapsid protein expression.
Flavonoids, phenolic compounds, steroids, polyphenols, and terpenoids, and their derivatives are commonly found in secondary metabolites of plants and consist of aromatic rings with one or many hydroxyl groups (Vieira et al., 2010; Wink, 2020). Wink's study revealed that polyphenols could bind with the lipoprotein of the virus envelope that checks the viral invasion in host cells. Various other studies confirmed the antiviral activity of phenolic compounds like curcumin, catechin, bavachinin, gallate, silvestrol, and tomentin (Ryu et al., 2010; Khaerunnisa et al., 2020; Wink, 2020; Yang et al., 2020). Besides phenolic compounds, flavonoids are also potential candidates against SARS 3CLpro enzymes of human coronavirus. Various antiviral studies on flavonoids have shown promising results (Table 2).
TABLE 2. List of the promising phytochemicals used worldwide for antiviral studies that could play a crucial role in the treatment of COVID-19.
In recent research on biomolecules, Letko et al. (2020) reported that SARS-CoV-2 associates with host cells using angiotensin-converting enzyme 2 (hACE2), that is, a host receptor. Therefore, plant-based biomolecules that can inhibit the interaction with this receptor could become an excellent pharmaceutical candidate to fight against SARS-CoV-2. For further analysis, molecular docking was performed with cannabinoids and different phytochemicals to establish these biomolecules’ binding positions with viral spike protein (S) (Ho et al., 2007; Tallei et al., 2020).
Before discussing the application of the multi-omics approach in the different research fields, it is essential to understand how all these technologies work individually and how information could be combined to generate a more in-depth understanding. Various methods can apply all the omics knowledge depending on the availability of data and requirements (Zhou et al., 2020; Zoppi et al., 2021). The data available in the public domain related to all these omics technologies are extracted for complex analysis, and attempts are made to link all markers at the different levels (genomic, proteomic, and metabolomic) back to annotated genes (Figure 4; Subramanian et al., 2020). Usually, this method works appropriately because well-curated and interpreted databases with a complete description of genes and their biological functions can be acquired from several data sources. However, it is a big challenge to collect all the information separately and make a common analysis. Multi-omics data are usually not cross-referenced between repositories. A web application, that is, MOD-Finder, searches for multi-omics datasets related to a user-defined chemical of interest (Canzler et al., 2020).
FIGURE 4. Schema representing steps to discover therapeutics (plant-based biomolecules and vaccines) for COVID-19. The biomolecules/plant metabolites can be screened and tested following the mentioned steps for potential antiviral activity against SARS-CoV-2. The figure also depicts the steps to develop vaccines by exploring the genetic material of the virus.
The commonly used and readily available databases are the gene database of NCBI (http://www.ncbi.nlm.nih.gov/gene), Gene Ontology (http://geneontology.org) (Carbon et al., 2019), Ensembl (http://useast.ensembl.org) (Andrew et al., 2020), KEGG (http://genome.jp/kegg/pathway.html), HMDB (http://hmdb.ca/), MetaCyc (http://metacyc.org/), Wikipathways (http://wikipathways.org/index.php/WikiPathways), and DAVID (http://david.abcc.ncifcrf.gov/P). Extensive work is underway on the genome using its different aspects. The forecasting or designating genotypes that cannot be assayed directly in individuals’ samples are known as genotype imputation (Ashburner et al., 2000; Naj, 2019; Chen S. F. et al., 2020). In recent years, the studies on genome-wide association (GWA) have revealed many significantly replicated associations for various complicated diseases (McGuire et al., 2020). In the analysis of GWA studies, genotype imputation has been widely used to enhance power, fine-map connections, and expedite the integration of results throughout the studies using meta-analysis. For genotype imputation, a resource like 1000 Genomes (http://www.1000genomes.org) (Marchini and Howie, 2010; Howie et al., 2011; Belsare et al., 2019) is available that facilitated combining various genotyping platforms and consequently enhanced the capability to interpret the genomics data and execute meta-analyses. Although some forms of data cannot be easily mapped to annotated genes, such limitations of annotation are addressed with the newest omics technologies.
Metabolomics is seen with considerable gaps in the annotation, restricting the efficacy of pathway-based and integrative method approaches (Kilk, 2020). Metabolomic datasets are mostly deciphered in the form of metabolic pathways. The KEGG database consists of metabolic pathways and contains information about both enzymes and metabolites. This database is categorized into groups related to cellular processes, metabolism, human diseases, etc. (Kanehisa and Sato, 2020). However, the annotated metabolites are very limited, suggesting that there is a lot to be learned regarding the role of several metabolites in human health.
Due to the availability and storage of the extensive amount of molecular data, an urgent demand has developed as a new branch of science: “system biology,” which unravels the basic functional properties of living beings originating from the interaction of macromolecules (Zupanic et al., 2020). The increased ability to elucidate genetic variations and their role in downstream molecular changes, like metabolite levels, would play a crucial role in interpreting and combining various data types.
Multi-omics approaches can be used to identify and screen plant-based biomolecules. They also provide deep insights into the effect of these biomolecules on COVID-19. The addition of proteomic datasets to genomic and transcriptomic data helps to understand the role of plant-based biomolecules against COVID-19. The integration of proteomics data complements genomics in the identification of multiple pathways. As soon as any cell is given exposure to biomolecules, it triggers a series of effects at the regulatory pathway level, which involve changes of levels and interactions of different types of biomolecules. Transcriptomics can detect biomolecules of one type and can only capture changes in a small subset of the biological cascade.
How to employ multi-omics data for the study is solely dependent on integration strategies. Various methods are available for the integrative analysis of multi-omics data. This review focuses on implementing these approaches for screening potential therapeutic biomolecules, which can be efficiently used against COVID-19. The comprehensive reviews on multi-omics integration methods are provided by various researchers (Cavill et al., 2016; Huang et al., 2017; Tarazona et al., 2018; Subramanian et al., 2020).
Plant biotechnology provides a comprehensive platform with several strategies to facilitate the synthesis of biomolecules on a large scale, viz. hairy root culture, cell suspension culture, etc. The advancement of genomics can accelerate the gene manipulation and pathway triggering the biosynthesis of therapeutically active compounds. Genetic manipulation of genes and genomes in plants can be achieved by various methods, including transformation (development of transgenic), inducible and transient expression systems, gene-silencing methodologies, knockout, knockdown, and the most advanced genome-editing (Wang et al., 2020c). In pandemic situations, the best vaccine production method is transient expression because it is time-saving and could be advanced for large-scale manufacturing. The most advanced next-generation sequencing offers detailed information about the genes and complete genomes, including the noncoding region and regulatory elements that facilitate the identification of genes associated with biomolecules’ biosynthesis (Peška et al., 2017). Gene cloning helps in the identification and molecular characterization of the genes that encode biomolecules. Furthermore, it can be transformed into a plant system for the expression and optimization of the product. For example, Nicotiana benthamiana Domin extends a systematic and effective system and is used to express VLP of SARS-CoV-2 to produce the plant-based vaccine (Rattanapisit et al., 2020). This system could yield the required biomolecules that can be scaled up, isolated, and purified by downstream approaches (Sharma N. et al., 2020).
The CRISPER/Cas9 (genome editing) approach facilitates accurate editing of genes (Dangi et al., 2019). Specifically, this can be applied to knock out the enzymes that involve the target biomolecule as a precursor. It accelerates the processes with the overproduction of required metabolites. The higher level of biomolecule production and fine-tuning of biosynthetic machinery could be attained by using transient methods like virus-induced gene silencing or RNA interference. A genetic transformation like Agrobacterium-mediated transformation provides the stable and reliable change and expression of genes in the plants (Ma et al., 2020). This method could easily be customized to develop target biomolecules in different plants, which can further undergo downstream processing and convenient purifications. Some of the useful phytochemicals are found in endangered species viz. Chinese medicinal plants Panax ginseng C. A. Mey. and Magnolia officinalis Rehder and E. H. Wilson, but their use in therapeutics is prohibited. Genetic manipulation and transformation methods offer model plant species as an alternative. The biosynthetic pathway can be studied and engineered into these, such as tomato, tobacco, rice, and maize. This has the advantage of producing beneficial compounds without putting any pressure on the original medicinal plant (Sassi et al., 2008; Moon et al., 2019). An excellent example of the application of genomics and biotechnology is the saponin glycyrrhizin. It is a naturally occurring class of compounds and is reported to inhibit coronavirus replication, and its antiviral activity augments ten folds by modification in its glycosidic chain (Hoever et al., 2005). The successful whole nucleic acid sequencing of SARS-CoV-2 from a different population of patients has given a new vision regarding the pathogen and its nonuniformity worldwide. As sequence data on the virus’s proteomics and metabolomics are available, they could help study it more precisely (Gordon et al., 2020; Shen et al., 2020; Wang M. et al., 2020).
Proteomics is the most powerful tool for studying total expressed proteins in an organism or cell type at a particular time. This provides the methodologies used for identification, detection, sample preparation, separation methods, and quantification of proteins. Proteins are responsible for the cells’ function, and the expression, localization, and activity of proteins differ in various conditions. Hence, the study of protein expression in cell types or different conditions helps to identify and understand their biological information. All plant biomolecules derive from specific biosynthetic pathways. The comprehensive study of those pathways, starting from the analysis of the genes underlying them and biosynthetic enzymes and their regulation (Song et al., 2015), is another challenging task that can be achieved by deploying different proteomics methods. Mass spectrometry (MS) is an important technique that enables the analysis of proteomes and identification of proteins present in the biological system. The separation of proteomes can be performed by gel chromatography or liquid chromatography before analysis. Production of allergens and toxins while deriving a specific plant biomolecule should also be monitored to check if any toxic by-product is also produced. All such allergens and toxins are identified and systematized in www.allergenonline.org/ and www.allergen.org (Ahsan et al., 2016; Croote and Quake, 2016). Identification of accurate therapeutic plant biomolecules is the critical and key step. Once they are identified, their interaction and effect on cells and cellular activities in another aspect of proteomics and proteomics research can help identify medicinal plant biomolecules.
Proteomics relates proteins’ functional role to host and pathogen (Zheng and Perlman, 2018). Zheng and Perlman (2018) studied proteomics’ role in the host immune system and its responses to the respiratory virus interactions. Other researchers also studied G-protein–coupled receptors (Sriram and Insel, 2018), enzymes (Ding et al., 2017), and ion channels (Duncan et al., 2020). Recently, various protein-based analyses of coronavirus have been done to identify structural proteins that include SDS-PAGE analysis, Western blot, protein categorization, protein identification, separation, and quantification. The proteome microarray of SARS-CoV-2 was demonstrated by Wang et al., which helps in mapping COVID-19 antibody interactions (Wang X. et al., 2020). A detailed report was given by Gordon et al. on the human protein–protein interaction map of SARS-CoV-2, where they defined 332 human protein–protein interactions, and out of them, 66 were targeted by several preexisting FDA-approved drugs or under trial drugs (Gordon et al., 2020).
Transcriptomics studies primarily deal with gene expression profiles, that is, by RNA sequencing (Depledge et al., 2019), ribosome profiling, and high-throughput DNA microarray studies (Wang et al., 2009). Transcriptomics is an approach for exhaustive study and detection of RNA in the cell. In transcriptomics, a pathway response is mainly detected via a known set of target genes of the pathway expressed differentially. The information on association with a particular pathway is mainly available for protein-coding RNAs (mRNAs). Earlier, transcriptomics was dependent on microarrays as a measurement technique. Microarrays constitute a targeted detection approach; that is, they require prior selection and knowledge of the sequence of the interrogated RNAs. Recently, transcriptomics has switched to transcriptome sequencing (RNA-seq), which provides a platform for simultaneous identification of transcripts, isoform detection, and quantification (Canzler et al., 2020). The dose–response models were generated to determine the factors affecting gene expressions (Hashem et al., 2019) at various viral protein concentrations (Haas, 2020). Studies were also conducted to check the concentration of mRNA at different infection stages and its propagation (Albarino et al., 2018).
The metabolome is a collection of chemically highly heterogeneous molecules. It can be defined as the complete complement of all small molecule metabolites found in a specific cell, organ, or organism (Wishart, 2007). Different metabolic enzymes run cellular metabolism (Ahmed et al., 2020), and this is reported by a fierce study of genomics, proteomics, and transcriptomics (Fanos et al., 2020). These are directly connected with pathways available in metabolomics (Haas et al., 2016). The primary importance of metabolomics is related to diagnostics (Debnath et al., 2010; To et al., 2016). The concentration of metabolites is observed and identified by high-performance liquid chromatography/mass spectrometry (HPLC/MS) and nuclear magnetic resonance (NMR) (Peng and Liu, 2017). The early studies concluded that metabolic profile analysis reveals the inactivation and binding of metabolites with the therapeutic compounds (Eisfeld et al., 2017).
Artificial intelligence (AI) is the computational design, development, and application of computer programs and algorithms that perform cognitive functions based on human intelligence traits, for example, anticipating, problem-solving, and learning (Saxena et al., 2019; Sharma A. et al., 2020). AI techniques have the potential to accelerate the virtual screening, lead discovery and validation, etc. (Kumar et al., 2011; Kumar et al., 2017), thereby assisting in drug design and repurposing, and they can complement the traditional drug development methods for COVID-19. AI technique–based Benevolent AI has been successfully applied to identify baricitinib as a potential drug against COVID-19 (Favalli et al., 2020; Randhawa et al., 2020). PolypharmDB (Redka et al., 2020) and inclProject IDentif.AI (Abdulla et al., 2020) have already been successful in identifying potential drug candidates against SARS-Cov-2. A supervised learning–based Vaxign reverse vaccinology-machine learning platform has been developed for assisting the development of vaccine candidates against COVID-19 (Ong et al., 2020). Figure 5 summarizes the applications of biotechnology and multi-omics approaches for screening and profiling of plant-based biomolecules against SARS-CoV-2.
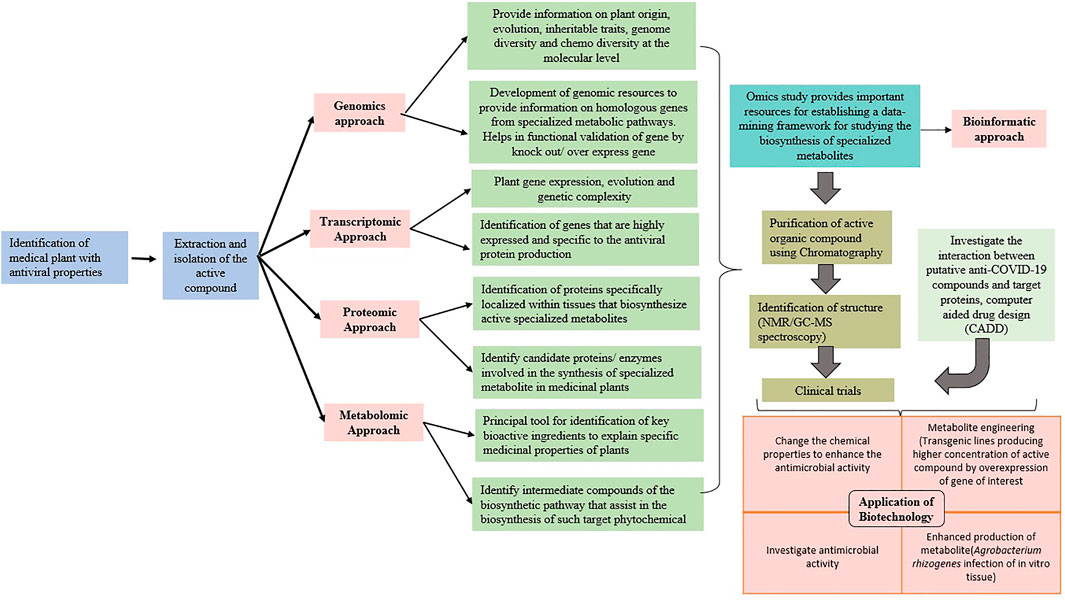
FIGURE 5. Use of biotechnology and multi-omics approaches for screening and profiling of plant-based biomolecules against SARS-CoV-2.
Wang et al. (2020d) developed a deep learning and ontology-based side effect prediction framework to evaluate and assess traditional Chinese medicines against COVID-19 treatment. Moreover, AI techniques are efficiently applied in SARS-CoV-2 protein structure prediction (Zhang et al., 2004; Senior et al., 2020). Apart from assisting the drug design, drug repurposing, and vaccine candidate development, AI has also been instrumental in spreading awareness, curbing misinformation (Hung et al., 2020; Miner et al., 2020; Rashid and Wang, 2020), assisting in early diagnosis (Xu et al., 2020), and decreasing the burden in healthcare professions by providing accurate decision support (Iwendi et al., 2020) during the COVID-19 pandemic.
The present review gives an insight into the applicability of multi-omics tools and different omics approaches in identifying potential therapeutic plant biomolecules. These tools could explore the immense potential of plant-based biomolecules for the prevention, mitigation, or cure of SARS-CoV-2–infected patients. There is a need to reduce the gaps between the conventional treatment from plant extracts and herbs with an updated understanding of biomolecules/phytochemicals present in plant extracts using omics technologies. Advancement in technology and discovery of different omics approaches to explore and analyze the genomic, proteomic, and metabolomic data can generate the profile of plant biomolecules and identify the potential antiviral compounds that could be used against SARS-CoV-2.
Due to the highly contagious nature of SARS-CoV-2, handling of clinical samples in omics research facilities is often restricted. This has made the implementation of system-level molecular research extremely challenging. With this limitation, it is helpful for academicians, scientists, and health professionals from this field to be aware of the recent trends in omics approaches to address issues related to COVID-19. Various plant-based biomolecules have already been identified and studied at different levels of omics research. Now, screening those potential therapeutic compounds to treat SAR-CoV-2–infected patients and devising a relevant strategy to optimize the production and purification of those biomolecules using multi-omics approaches are the urgent needs of the situation. Multi-omics techniques are anticipated to play a crucial role in the identification of potential therapeutic plant biomolecules and effective clinical management of COVID-19. Once these approaches have been applied successfully, the screened repository of plant-based therapeutic biomolecules could be used for future health emergencies like the emergence of a new strain or mutation in the virus. We hope that this document may help future researchers to quickly get an overview and understand the applications of omics approaches to find out therapeutically active plant-based biomolecules in infectious outbreaks or pandemics.
All authors contributed to the article and approved the submitted version.
This work was funded by the Deanship of Scientific Research at Princess Nourah bint Abdulrahman University through the Fast-Track Research Funding Program.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Aanouz, I., Belhassan, A., El-Khatabi, K., Lakhlifi, T., El-ldrissi, M., and Bouachrine, M. (2020). Moroccan Medicinal Plants as Inhibitors against SARS-CoV-2 Main Protease: Computational Investigations. J. Biomol. Struct. Dyn. 2020, 1–9. doi:10.1080/07391102.2020.1758790
Aaron, G., Daire, C., Leticia, M. W., and Simon, J. W. (2020). Multi-Omics Technologies Applied to Tuberculosis Drug Discovery. Appl. Sci. 10, 4629. doi:10.3390/app10134629
Abdelli, I., Hassani, F., Bekkel Brikci, S., and Ghalem, S. (2020). In Silico study the Inhibition of Angiotensin Converting Enzyme 2 Receptor of COVID-19 by Ammoides Verticillata Components Harvested from Western Algeria. J. Biomol. Struct. Dyn. 2020, 1–14. doi:10.1080/07391102.2020.1763199
Abdulla, A., Wang, B., Qian, F., Kee, T., Blasiak, A., Ong, Y. H., et al. (2020). Project IDentif.AI: Harnessing Artificial Intelligence to Rapidly Optimize Combination Therapy Development for Infectious Disease Intervention. Adv. Therap. 3, 2000034. doi:10.1002/adtp.202000034
Ahmed, I., Hyder, S., and Altayb, N. (2020). The Important Role of Fever during Infection and the Use of Fever for Treatment of Coronavirus Patients. SSRN Elec J. doi:10.2139/ssrn.3556215
Ahsan, N., Rao, R. S. P., Gruppuso, P. A., Ramratnam, B., and Salomon, A. R. (2016). Targeted Proteomics: Current Status and Future Perspectives for Quantification of Food Allergens. J. Proteomics 143, 15–23. doi:10.1016/j.jprot.2016.04.018
Akram, M., Tahir, I. M., Shah, S. M. A., Mahmood, Z., Altaf, A., Ahmad, K., et al. (2018). Antiviral Potential of Medicinal Plants against HIV, HSV, Influenza, Hepatitis, and Coxsackievirus: a Systematic Review. Phytotherapy Res. 32, 811–822. doi:10.1002/ptr.6024
Al-Ishaq, R. K., Overy, A. J., and Büsselberg, D. (2020). Phytochemicals and Gastrointestinal Cancer: Cellular Mechanisms and Effects to Change Cancer Progression. Biomolecules 10 (1), 105. doi:10.3390/biom10010105
Al-Jabir, K., and Insel, P. A. (2018). G Protein-Coupled Receptors as Targets for Approved Drugs: How Many Targets and How Many Drugs?. Mol. Pharmacol. 93, 251–258. doi:10.1124/mol.117.111062
Albarino, C. G., Guerrero, L. W., Chakrabarti, A. K., and Nichol, S. T. (2018). Transcriptional Analysis of Viral mRNAs Reveals Common Transcription Patterns in Cells Infected by Five Different Filoviruses. PLoS One 13, e0201827. doi:10.1371/journal.pone.0201827
Anand, A. V., Balamuralikrishnan, B., Kaviya, M., Bharathi, K., Parithathvi, A., Arun, M., et al. (2021). Medicinal Plants, Phytochemicals, and Herbs to Combat Viral Pathogens Including SARS-CoV-2. Molecules 26 (6), 1775. doi:10.3390/molecules26061775
Andrew, D. Y., Premanand, A., Wasiu, A., James, A., Jamie, A., Jorge, A., et al. (2020). Ensembl 2020. Nucleic Acids Res. 48, D682–D688. doi:10.1093/nar/gkz966
Ang, L., Song, E., Lee, H. W., and Lee, M. S. (2020). Herbal Medicine for the Treatment of Coronavirus Disease 2019 (COVID-19): A Systematic Review and Meta-Analysis of Randomized Controlled Trials. J Clin Med. 9 (5), 1583. doi:10.3390/jcm9051583
Arbab, A. H., Parvez, M. K., Al-Dosari, M. S., and Al-Rehaily, A. J. (2017). In vitro evaluation of Novel Antiviral Activities of 60 Medicinal Plants Extracts against Hepatitis B Virus. Exp. Ther. Med 14, 626–634. doi:10.3892/etm.2017.4530
Ashburner, M., Ball, C. A., Blake, J. A., Botstein, D., Butler, H., Cherry, J. M., et al. (2000). Gene Ontology: Tool for the Unification of Biology. Nat. Genet. 25, 25–29. doi:10.1038/75556
Asres, K., and Bucar, F. (2005). Anti-HIV Activity against Immunodeficiency Virus Type 1 (HIV-I) and Type II (HIV-II) of Compounds Isolated from the Stem Bark of Combretum Molle. Ethiop Med. J. 43, 15–20.
Balkrishna, A., Haldar, S., Singh, H., Roy, P., and Varshney, A. (2021). Coronil, a Tri-herbal Formulation, Attenuates Spike-Protein-Mediated SARS-CoV-2 Viral Entry into Human Alveolar Epithelial Cells and Pro-Inflammatory Cytokines Production by Inhibiting Spike Protein-ACE-2 Interaction. J. Inflamm. Res. 14, 869–884. doi:10.2147/JIR.S298242
Basu, A., Sarkar, A., and Maulik, U. (2020). Computational Approach for the Design of Potential Spike Protein Binding Natural Compounds in SARS- CoV2. Pharmacodynamics. doi:10.21203/rs.3.rs-33181/v1
Behl, T., Kaur, I., Bungau, S., Kumar, A., Uddin, M. S., Kumar, C., et al. (2020). The Dual Impact of ACE2 in COVID-19 and Ironical Actions in Geriatrics and Pediatrics with Possible Therapeutic Solutions. Life Sci. 257, 118075. doi:10.1016/j.lfs.2020.118075
Belfiore, M. P., Urraro, F., Grassi, R., Giacobbe, G., Patelli, G., Cappabianca, S., et al. (2020). Artificial Intelligence to Codify Lung CT in Covid-19 Patients. Radiol. Med. 125 (5), 500–504. doi:10.1007/s11547-020-01195-x
Belsare, S., Levy-Sakin, M., Mostovoy, Y., Durinck, S., Chaudhuri, S., Xiao, M., et al. (2019). Evaluating the Quality of the 1000 Genomes Project Data. BMC genomics 20 (1), 620. doi:10.1186/s12864-019-5957-x
Ben-Shabat, S., Yarmolinsky, L., Porat, D., and Dahan, A. (2020). Antiviral Effect of Phytochemicals from Medicinal Plants: Applications and Drug Delivery Strategies. Drug Deliv. Transl. Res. 10, 354–367. doi:10.1007/s13346-019-00691-6
Bhandari, M., Bhandari, A., and Bhandari, A. (2011). Recent Updates on Codeine. Pharm. Methods 2, 3–8. doi:10.4103/2229-4708.81082
Bojkova, D., Klann, K., Koch, B., Widera, M., Krause, D., Ciesek, S., et al. (2020). Proteomics of SARS-CoV-2-Infected Host Cells Reveals Therapy Targets. Nature 583 (7816), 469–472. doi:10.1038/s41586-020-2332-7
Borkotoky, S., and Banerjee, M. (2020). A Computational Prediction of SARS-CoV-2 Structural Protein Inhibitors from Azadirachta indica (Neem). J. Biomol. Struct. Dyn. 2020, 1–11. doi:10.1080/07391102.2020.1774419
Canzler, S., Schor, J., Busch, W., Schubert, K., Rolle-Kampczyk, U. E., Seitz, H., et al. (2020). Prospects and Challenges of Multi-Omics Data Integration in Toxicology. Arch. Toxicol. 94, 371–388. doi:10.1007/s00204-020-02656-y
Cao, P., Wu, S., Wu, T., Deng, Y., Zhang, Q., Wang, K., et al. (2020). The Important Role of Polysaccharides from a Traditional Chinese Medicine-Lung Cleansing and Detoxifying Decoction against the COVID-19 Pandemic. Carbohydr. Polym. 240, 116346. doi:10.1016/j.carbpol.2020.116346
Carbon, S., Douglass, E., Dunn, N., Good, B., Harris, N. L., Lewis, S. E., et al. (2019). The Gene Ontology Resource: 20 Years and Still GOing Strong. Nucleic Acids Res. 47 (D1), D330–D338. doi:10.1093/nar/gky1055
Cavill, R., Jennen, D., Kleinjans, J., and Briedé, J. J. (2016). Transcriptomic and Metabolomic Data Integration. Br. Bioinform. 17 (5), 891–901. doi:10.1093/bib/bbv090
Chantrill, B. H., Coulthard, C. E., Dickinson, L., Inkley, G. W., Morris, W., and Pyle, A. H. (1952). The Action of Plant Extracts on a Bacteriophage of Pseudomonas Pyocyanea and on Influenza A Virus. J. Gen. Microbiol. 6, 74–84. doi:10.1099/00221287-6-1-2-74
Chen, B., Garmire, L., Calvisi, D. F., Chua, M.-S., Kelley, R. K., and Chen, X. (2020). Harnessing Big 'omics' Data and AI for Drug Discovery in Hepatocellular Carcinoma. Nat. Rev. Gastroenterol. Hepatol. 17, 238–251. doi:10.1038/s41575-019-0240-9
Chen, S. F., Dias, R., Evans, D., Salfati, E. L., Liu, S., Wineinger, N. E., et al. (2020). Genotype Imputation and Variability in Polygenic Risk Score Estimation. Genome Med. 12 (1), 100. doi:10.1186/s13073-020-00801-x
Cho, J. K., Curtis-Long, M. J., Lee, K. H., Kim, D. W., Ryu, H. W., Yuk, H. J., et al. (2013). Geranylated Flavonoids Displaying SARS-CoV Papain-like Protease Inhibition from the Fruits of Paulownia Tomentosa. Bioorg. Med. Chem. 21, 3051–3057. doi:10.1016/j.bmc.2013.03.027
Chojnacka, K., Witek-Krowiak, A., Skrzypczak, D., Mikula, K., and Młynarz, P. (2020). Phytochemicals Containing Biologically Active Polyphenols as an Effective Agent against Covid-19-Inducing Coronavirus. J. Funct. Foods 73, 104146. doi:10.1016/j.jff.2020.104146
Chong, L., Zhang, W., Nie, Y., Yu, G., Liu, L., Lin, L., et al. (2014). Protective Effect of Curcumin on Acute Airway Inflammation of Allergic Asthma in Mice through Notch1-GATA3 Signaling Pathway. Inflammation 37, 1476–1485. doi:10.1007/s10753-014-9873-6
Clark, L. K., Green, T. J., and Petit, C. M. (2021). Structure of Nonstructural Protein 1 from SARS-CoV-2. J. Virol. 95 (4), e02019–20. doi:10.1128/JVI.02019-20
Croote, D., and Quake, S. R. (2016). Food Allergen Detection by Mass Spectrometry: the Role of Systems Biology. Npj Syst. Biol. Appl. 2, 16022. doi:10.1038/npjsba.2016.22
D’Alessandro, S., Scaccabarozzi, D., Signorini, L., Perego, F., Ilboudo, D. P., Ferrante, P., et al. (2020). The Use of Antimalarial Drugs against Viral Infection. Microorganisms 8, 85. doi:10.3390/microorganisms8010085
Dangi, A., Yu, S., and Luo, X. (2019). Emerging Approaches and Technologies in Transplantation: the Potential Game Changers. Cell Mol Immunol 16 (4), 334–342. doi:10.1038/s41423-019-0207-3
Debiaggi, M., Pagani, L., Cereda, P. M., Landini, P., and Romero, E. (1988). Antiviral Activity of Chamaecyparis Lawsoniana Extract: Study with Herpes Simplex Virus Type 2. Microbiologica 11, 55–61.
Debnath, M., Prasad, G. B. K. S., and Bisen, P. S. (2010). Molecular Diagnostics: Promises and Possibilities Dordrech Heidelberg. London: Springer, 11–31.
Depledge, D. P., Srinivas, K. P., Sadaoka, T., Bready, D., Mori, Y., Placantonakis, D. G., et al. (2019). Direct RNA Sequencing on Nanopore Arrays Redefines the Transcriptional Complexity of a Viral Pathogen. Nat. Commun. 10, 1–13. doi:10.1038/s41467-019-08734-9
Devaux, C. A., Rolain, J.-M., Colson, P., and Raoult, D. (2020). New Insights on the Antiviral Effects of Chloroquine against Coronavirus: what to Expect for COVID-19?. Int. J. Antimicrob. Agents 55, 105938. doi:10.1016/j.ijantimicag.2020.105938
Dhama, K., Karthik, K., Khandia, R., Munjal, A., Tiwari, R., Rana, R., et al. (2018). Medicinal and Therapeutic Potential of Herbs and Plant Metabolites/ Extracts Countering Viral Pathogens - Current Knowledge and Future Prospects. Curr. Drug Metab. 19, 236–263. doi:10.2174/1389200219666180129145252
Ding, Y., Tang, J., and Guo, F. (2017). Identification of Drug-Target Interactions via Multiple Information Integration. Inf. Sci. 418-419, 546–560. doi:10.1016/j.ins.2017.08.04510.1016/j.ins.2017.08.045
Drake, K. B., Ganeshpurkar, A., and Mishra, M. K. (2020). Natural RNA Dependent RNA Polymerase Inhibitors: Molecular Docking Studies of Some Biologically Active Alkaloids of Argemone Mexicana. Med. Hypotheses 144, 109905. doi:10.1016/j.mehy.2020.109905
Duncan, A. L., Song, W., and Sansom, M. S. P. (2020). Lipid-dependent Regulation of Ion Channels and G Protein-Coupled Receptors: Insights from Structures and Simulations. Annu. Rev. Pharmacol. Toxicol. 60, 31–50. doi:10.1146/annurev-pharmtox-010919-023411
Eisfeld, A. J., Halfmann, P. J., Wendler, J. P., Kyle, J. E., Burnum-Johnson, K. E., Peralta, Z., et al. (2017). Multi-platform 'Omics Analysis of Human Ebola Virus Disease Pathogenesis. Cell Host & Microbe 22, 817–829. doi:10.1016/j.chom.2017.10.011
Enmozhi, S. K., Raja, K., Sebastine, I., and Joseph, J. (2020). Andrographolide as a Potential Inhibitor of SARS-CoV-2 Main Protease: an In Silico Approach. J. Biomol. Struct. Dyn. 5, 1–7. doi:10.1080/07391102.2020.1760136
Fanos, V., Pintus, M. C., Pintus, R., and Marcialis, M. A. (2020). Lung Microbiota in the Acute Respiratory Disease: from Coronavirus to Metabolomics. J. Pediatr. Neonatal. Individual. Med. 9, e090139. doi:10.7363/090139
Favalli, E. G., Biggioggero, M., Maioli, G., and Caporali, R. (2020). Baricitinib for COVID-19: a Suitable Treatment?. Lancet Infect. Dis. 20, 1012–1013. doi:10.1016/S1473-3099(20)30262-0
Fung, K. P., Leung, P. C., Tsui, K. W., Wan, C. C., Wong, K. B., Waye, M. Y., et al. (2011). Immunomodulatory Activities of the Herbal Formula Kwan Du Bu Fei Dang in Healthy Subjects: a Randomised, Double-Blind, Placebo-Controlled Study. Hong Kong Med. J. 17 (Suppl. 2), 41–43.
Gordon, D. E., Jang, G. M., Bouhaddou, M., Xu, J., Obernier, K., White, K. M., et al. (2020). A SARS-CoV-2 Protein Interaction Map Reveals Targets for Drug Repurposing. Nature 583 (7816), 459–468. doi:10.1038/s41586-020-2286-9
Greiner, J. F.-W., Müller, J., Zeuner, M.-T., Hauser, S., Seidel, T., Klenke, C., et al. (2013). 1,8-Cineol Inhibits Nuclear Translocation of NF-κB P65 and NF-κB-dependent Transcriptional Activity. Biochim. Biophys. Acta. 1833, 2866–2878. doi:10.1016/j.bbamcr.2013.07.001
Gupta, M. K., Vemula, S., Donde, R., Gouda, G., Behera, L., and Vadde, R. (2020). In-silico Approaches to Detect Inhibitors of the Human Severe Acute Respiratory Syndrome Coronavirus Envelope Protein Ion Channel. J. Biomol. Struct. Dyn. 39, 2617–2627. doi:10.1080/07391102.2020.1751300
Gyebi, G. A., Ogunro, O. B., Adegunloye, A. P., Ogunyemi, O. M., and Afolabi, S. O. (2020). Potential Inhibitors of Coronavirus 3-chymotrypsin-like Protease (3CLpro): an In Silico Screening of Alkaloids and Terpenoids from African Medicinal Plants. J. Biomol. Struct. Dyn. 2020, 1–13. doi:10.1080/07391102.2020.1764868
Haas, C. T., Roe, J. K., Pollara, G., Mehta, M., and Noursadeghi, M. (2016). Diagnostic ‘omics’ for Active Tuberculosis. BMC Med. 14, 37. doi:10.1186/s12916-016-0583-9
Hahn, F., Fröhlich, T., Frank, T., Bertzbach, L. D., Kohrt, S., Kaufer, B. B., et al. (2018). Artesunate-derived Monomeric, Dimeric and Trimeric Experimental Drugs - Their Unique Mechanistic Basis and Pronounced Antiherpesviral Activity. Antiviral Res. 152, 104–110. doi:10.1016/j.antiviral.2018.02.013
Hart, O. E., and Halden, R. U. (2020). Computational Analysis of SARS-CoV-2/COVID-19 Surveillance by Wastewater-Based Epidemiology Locally and Globally: Feasibility, Economy, Opportunities and Challenges. Sci. Total Environ. 730, 138875. doi:10.1016/j.scitotenv.2020.138875
Hasana, S., Hossain, M. F., Jalouli, M., Kabir, M. T., Uddin, M. G., Wahed, M. I. I., et al. (2021). Genetic Diversity of SARS-CoV2 and Environmental Settings: Possible Association with Neurological Disorders. Mol. Neurobiol. 58, 1917–1931. Epub ahead of print. PMID: 33404977. doi:10.1007/s12035-020-02239-z
Hashem, A. M., Algaissi, A., Agrawal, A. S., Al-Amri, S. S., Alhabbab, R. Y., Sohrab, S. S., et al. (2019). A Highly Immunogenic, Protective, and Safe Adenovirus-Based Vaccine Expressing Middle East Respiratory Syndrome Coronavirus S1-CD40L Fusion Protein in a Transgenic Human Dipeptidyl Peptidase 4 Mouse Model. J. Infect. Dis. 220, 1558–1567. doi:10.1093/infdis/jiz13710.1093/infdis/jiz137
Hillen, H. S., Kokic, G., Farnung, L., Dienemann, C., Tegunov, D., and Cramer, P. (2020). Structure of Replicating SARS-CoV-2 Polymerase. Nature 584 (7819), 154–156. doi:10.1038/s41586-020-2368-8
Ho, T., Wu, S., Chen, J., Li, C., and Hsiang, C. (2007). Emodin Blocks the SARS Coronavirus Spike Protein and Angiotensin-Converting Enzyme 2 Interaction. Antiviral Res. 74, 92–101. doi:10.1016/j.antiviral.2006.04.014
Ho, Y.-J., Lu, J.-W., Huang, Y.-L., and Lai, Z.-Z. (2019). Palmatine Inhibits Zika Virus Infection by Disrupting Virus Binding, Entry, and Stability. Biochem. Biophysical Res. Commun. 518, 732–738. doi:10.1016/j.bbrc.2019.08.120
Hoever, G., Baltina, L., Michaelis, M., Kondratenko, R., Baltina, L., Tolstikov, G. P., et al. (2005). Antiviral Activity of Glycyrrhizic Acid Derivatives against SARS−Coronavirus. J. Med. Chem. 48, 1256–1259. doi:10.1021/jm0493008
Horgan, R. P., and Kenny, L. C. (2011). 'Omic' Technologies: Genomics, Transcriptomics, Proteomics and Metabolomics. Obstet. Gynaecol. 13, 189–195. doi:10.1576/toag.13.3.189.27672
Hossain, M. F., Hasana, S., Mamun, A. A., Uddin, M. S., Wahed, M. I. I., Sarker, S., et al. (2020). COVID-19 Outbreak: Pathogenesis, Current Therapies, and Potentials for Future Management. Front. Pharmacol. 11, 563478. doi:10.3389/fphar.2020.563478
Hossain, M. U., Bhattacharjee, A., Emon, M. T. H., Chowdhury, Z. M., Mosaib, M. G., Mourin, M., et al. (2021). Recognition of Plausible Therapeutic Agents to Combat COVID-19: An Omics Data Based Combined Approach. Gene 771, 145368. doi:10.1016/j.gene.2020.145368
Howie, B., Marchini, J., and Stephens, M. (2011). Genotype Imputation with Thousands of Genomes. G3. Genes, Genomes, Genet. 1, 457–470. doi:10.1534/g3.111.001198
Huang, K.-L., Lai, Y. K., Lin, C. C., and Chang, J. M. (2006). Inhibition of Hepatitis B Virus Production byBoehmeria Nivearoot Extract in HepG2 2.2.15 Cells. World J Gastroenterol. 12, 5721–5725. doi:10.3748/wjg.v12.i35.5721
Huang, S., Chaudhary, K., and Garmire, L. X. (2017). More is Better: Recent Progress in Multi-Omics Data Integration Methods. Front. Genet. 8, 84. doi:10.3389/fgene.2017.00084
Huang, Y.-F., Bai, C., He, F., Xie, Y., and Zhou, H. (2020). Review on the Potential Action Mechanisms of Chinese Medicines in Treating Coronavirus Disease 2019 (COVID-19). Pharmacol. Res. 158, 104939. doi:10.1016/j.phrs.2020.104939
Hung, M., Lauren, E., Hon, E. S., Birmingham, W. C., Xu, J., Su, S., et al. (2020). Social Network Analysis of COVID-19 Sentiments: Application of Artificial Intelligence. J. Med. Internet Res. 22, e22590. doi:10.2196/22590
Infusino, F., Marazzato, M., Mancone, M., Fedele, F., Mastroianni, C. M., Severino, P., et al. (2020). Diet Supplementation, Probiotics, and Nutraceuticals in SARS-CoV-2 Infection: A Scoping Review. Nutrients 12 (6), 1718. doi:10.3390/nu12061718
Islam, M. T., Sarkar, C., El‐Kersh, D. M., Jamaddar, S., Uddin, S. J., Shilpi, J. A., et al. (2020). Natural Products and Their Derivatives against Coronavirus: A Review of the Non‐clinical and Pre‐clinical Data. Phytotherapy Res. 34, 2471–2492. doi:10.1002/ptr.6700
Israt, J., and Ahmet, O. (2020). Potentials of Plant-Based Substance to Inhabit and Probable Cure for the COVID-19. Turkish J. Biol. 44, 228–241. doi:10.3906/biy-2005-114
Iwendi, C., Bashir, A. K., Peshkar, A., Sujatha, R., Chatterjee, J. M., Pasupuleti, S., et al. (2020). COVID-19 Patient Health Prediction Using Boosted Random Forest Algorithm. Front. Public Health 8, 357. doi:10.3389/fpubh.2020.00357
Jin, Z., Du, X., Xu, Y., Deng, Y., Liu, M., Zhao, Y., et al. (2020). Structure of Mpro from SARS-CoV-2 and Discovery of its Inhibitors. Nature 582 (7811), 289–293. doi:10.1038/s41586-020-2223-y
Jo, S., Kim, H., Kim, S., Shin, D. H., and Kim, M. S. (2019). Characteristics of Flavonoids as Potent MERS-CoV 3C-like Protease Inhibitors. Chem. Biol. Drug Des. 94, 2023. doi:10.1111/cbdd.13604
Jo, S., Kim, S., Shin, D. H., and Kim, M.-S. (2020). Inhibition of SARS-CoV 3CL Protease by Flavonoids. J. Enzyme Inhib. Med. Chem. 35, 145–151. doi:10.1080/14756366.2019.1690480
Kabir, M. T., Uddin, M. S., Hossain, M. F., Abdulhakim, J. A., Alam, M. A., Ashraf, G. M., et al. (2020). nCOVID-19 Pandemic: From Molecular Pathogenesis to Potential Investigational Therapeutics. Front. Cel Dev. Biol. 8, 616. doi:10.3389/fcell.2020.00616
Kanehisa, M., and Sato, Y. (2020). KEGG Mapper for Inferring Cellular Functions from Protein Sequences. Protein Sci. 29 (1), 28–35. doi:10.1002/pro.3711
Karimi, A., Rahim, K. A., and Rafieian-Kopaei, M. (2015). Classification of External Stakeholders Pressures in Green Supply Chain Management. Proced. Environ. Sci. 30 (1), 27–32. doi:10.1016/j.proenv.2015.10.005
Khaerunnisa, S., Kurniawan, H., Awaluddin, R., Suhartati, S., and Soetjipto, S. (2020). Potential Inhibitor of COVID-19 Main Protease (Mpro) from Several Medicinal Plant Compounds by Molecular Docking Study. Preprints 2020, 2020030226. doi:10.20944/preprints202003.0226.v1
Khan, S. A., Zia, K., Ashraf, S., Uddin, R., and Ul-Haq, Z. (2020). Identification of Chymotrypsin-like Protease Inhibitors of SARS-CoV-2 via Integrated Computational Approach. J. Biomol. Struct. Dyn. 39, 2607–2616. doi:10.1080/07391102.2020.1751298
Kilk, K. (2020). Metabolomics Horizon Scanning for 2020: Three Actionable Challenges. Omics: a J. Integr. Biol. 24 (1), 51–52. doi:10.1089/omi.2019.0194
Kim, S., Kim, D., Cho, S. W., Kim, J., and Kim, J. S. (2014). Highly Efficient RNA-Guided Genome Editing in Human Cells Via Delivery of Purified Cas9 Ribonucleoproteins. Genome Res. 24 (6), 1012–1019. doi:10.1101/gr.171322.113
Kim, D., Lee, J.-Y., Yang, J.-S., Kim, J. W., Kim, V. N., and Chang, H. (2020). The Architecture of SARS-CoV-2 Transcriptome. Cell 181 (4), 914–921.e10. doi:10.1016/j.cell.2020.04.011
Kim, D., Min, J., Jang, M., Lee, J., Shin, Y., Park, C., et al. (2019). Natural Bis-Benzylisoquinoline Alkaloids-Tetrandrine, Fangchinoline, and Cepharanthine, Inhibit Human Coronavirus OC43 Infection of MRC-5 Human Lung Cells. Biomolecules 9, 696. doi:10.3390/biom9110696
Kiran, G., Karthik, L., Shree Devi, M. S., Sathiyarajeswaran, P., Kanakavalli, K., Kumar, K. M., et al. (2020). In Silico computational Screening of Kabasura Kudineer - Official Siddha Formulation and JACOM against SARS-CoV-2 Spike Protein. J. Ayurveda Integr. Med. doi:10.1016/j.jaim.2020.05.009
Klenke, C. N. (2020). Coronavirus and Environmental Engineering Science. Environ. Eng. Sci. 37, 233–234. doi:10.1089/ees.2020.0096
Kodaira, H., and Spector, S. (1988). Transformation of Thebaine to Oripavine, Codeine, and Morphine by Rat Liver, Kidney, and Brain Microsomes. Proc. Natl. Acad. Sci. 85, 1267–1271. doi:10.1073/pnas.85.4.1267
Kotwal, G. J., Kaczmarek, J. N., Leivers, S., Ghebremariam, Y. T., Kulkarni, A. P., Bauer, G., et al. (2005). Anti-HIV, Anti-poxvirus, and Anti-SARS Activity of a Nontoxic, Acidic Plant Extract from the Trifollium Species Secomet-V/anti-Vac Suggests that it Contains a Novel Broad-Spectrum Antiviral. Ann. N.Y Acad. Sci. 1056, 293–302. doi:10.1196/annals.1352.014
Krishnasamy, R., T, A., Baba, M., Bharath, M. V., Phuntsho, J., Arunachalam, D., et al. (2020). In Silico Analysis of Active Compounds from Siddha Herbal Infusion of Ammaiyar Koondhal Kudineer (Akk) against SARS-CoV-2 Spike Protein and Its ACE2 Receptor Complex. SSRN J. doi:10.2139/ssrn.3578294
Kumar, R., Sharma, A., Haris Siddiqui, M., and Kumar Tiwari, R. (2016). Prediction of Metabolism of Drugs Using Artificial Intelligence: How Far Have We Reached?. Curr. Drug. Metab. 17, 129–141. doi:10.2174/1389200216666151103121352
Kumar, R., Sharma, A., Iqbal, M. S., and Srivastava, J. K. (2020b). Therapeutic Promises of Chlorogenic Acid with Special Emphasis on its Anti-obesity Property. Curr. Mol. Pharmacol. 13 (1), 7–16. doi:10.2174/1874467212666190716145210
Kumar, R., Sharma, A., Siddiqui, M. H., and Tiwari, R. K. (2017). Prediction of Human Intestinal Absorption of Compounds Using Artificial Intelligence Techniques. Curr. Drug. Discov. Technol. 14, 244–254. doi:10.2174/1570163814666170404160911
Kumar, R., Sharma, A., Siddiqui, M. H., and Tiwari, R. K. (2018). Promises of Machine Learning Approaches in Prediction of Absorption of Compounds. Mini. Rev. Med. Chem. 18, 196–207. doi:10.2174/1389557517666170315150116
Kumar, R., Sharma, A., Srivastava, J. K., Siddiqui, M. H., Uddin, M. S., and Aleya, L. (2021). Hydroxychloroquine in COVID-19: Therapeutic Promises, Current Status, and Environmental Implications. Environ. Sci. Pollut. Res. 1–14. doi:10.1007/s11356-020-12200-1
Kumar, R., Sharma, A., Varadwaj, P., Ahmad, A., and Ashraf, G. M. (2012). Classification of Oral Bioavailability of Drugs by Machine Learning Approaches: a Comparative Study. Jcis 2, 1–18. doi:10.6062/jcis.2011.02.03.0045
Kumar, R., Srivastava, J. K., Singh, R., Siddiqui, M. H., Mansouri, R. A., Abdulhakim, J. A., et al. (2020a). Available Compounds with Therapeutic Potential against COVID-19: Antimicrobial Therapies, Supportive Care, and Probable Vaccines. Front. Pharmacol. 11, 1592. doi:10.3389/fphar.2020.582025
Lalani, S., and Poh, C. L. (2020). Flavonoids as Antiviral Agents for Enterovirus A71 (EV-A71). Viruses 12 (2), 184. doi:10.3390/v12020184
Lau, K.-M., Lee, K.-M., Koon, C.-M., Cheung, C. S.-F., Lau, C.-P., Ho, H.-M., et al. (2008). Immunomodulatory and Anti-SARS Activities of Houttuynia Cordata. J. Ethnopharmacology 118, 79–85. doi:10.1016/j.jep.2008.03.018
Letko, M., Marzi, A., and Munster, V. (2020). Functional Assessment of Cell Entry and Receptor Usage for SARS-CoV-2 and Other Lineage B Betacoronaviruses. Nat. Microbiol. 5, 562–569. doi:10.1038/s41564-020-0688-y
Li, M. Y., Li, L., Zhang, Y., and Wang, X. S. (2020). Expression of the SARS-CoV-2 Cell Receptor Gene ACE2 in a Wide Variety of Human Tissues. Infect. Dis. poverty 9 (1), 45. doi:10.1186/s40249-020-00662-x
Li, T., and Peng, T. (2013). Traditional Chinese Herbal Medicine as a Source of Molecules with Antiviral Activity. Antiviral Res. 97, 1–9. doi:10.1016/j.antiviral.2012.10.006
Lin, B., Liu, J., Liu, Y., and Qin, X. (2020). Progress in Understanding COVID-19: Insights from the Omics Approach. Crit. Rev. Clin. Lab. Sci. 1–18. Advance online publicationdoi:10.1080/10408363.2020.1851167
Lin, K.-T., Wong, A. Y.-L., Kaewpreedee, P., Sia, S. F., Chen, D., Hui, K. P. Y., et al. (2020). Remdesivir, Lopinavir, Emetine, and Homoharringtonine Inhibit SARS-CoV-2 Replication in vitro. Antiviral Res. 178, 104786. doi:10.1016/j.antiviral.2020.104786
Loeffelholz, M. J., and Tang, Y.-W. (2020). Laboratory Diagnosis of Emerging Human Coronavirus Infections - the State of the Art. Emerging Microbes & Infections 9, 747–756. doi:10.1080/22221751.2020.1745095
Lu, R., Zhao, X., Li, J., Niu, P., Yang, B., Wu, H., et al. (2020). Genomic Characterisation and Epidemiology of 2019 Novel Coronavirus: Implications for Virus Origins and Receptor Binding. The Lancet 395, 565–574. doi:10.1016/S0140-6736(20)30251-8
Lubbe, A., Verpoorte, R., and Choi, Y. H. (2012). Effects of Fungicides on Galanthamine and Metabolite Profiles in Narcissus Bulbs. Plant Physiol. Biochem. 94, 371–388. doi:10.1007/s00204-020-02656-y
Luo, W., Su, X., Gong, S., Qin, Y., Liu, W., and Li, J. (2009). Anti-SARS Coronavirus 3C-like Protease Effects of Rheum Palmatum L. Extracts. Biosci. Trends 3, 124–126.
Lurie, N., Saville, M., Hatchett, R., and Halton, J. (2020). Developing Covid-19 Vaccines at Pandemic Speed. N. Engl. J. Med. 382, 1969–1973. doi:10.1056/NEJMp2005630
Ma, R., Yu, Z., Cai, Q., Li, H., Dong, Y., Oksman-Caldentey, K.-M., et al. (2020). Agrobacterium-mediated Genetic Transformation of the Medicinal Plant Veratrum Dahuricum. Plants 9, 191. doi:10.3390/plants9020191
Marchini, J., and Howie, B. (2010). Genotype Imputation for Genome-wide Association Studies. Nat. Rev. Genet. 11, 499–511. doi:10.1038/nrg2796
Maurya, V. K., Kumar, S., Bhatt, M. L. B., and Saxena, S. K. (2020). Therapeutic Development and Drugs for the Treatment of COVID-19. Nat. Public Health Emerg. Collec 2019, 109–126. doi:10.1007/978-981-15-4814-7_10
Mazzon, M., and Marsh, M. (2019). Targeting Viral Entry as a Strategy for Broad-Spectrum Antivirals, F1000Res 8, 1628. Faculty Rev-1628doi:10.12688/f1000research.19694.1
McGuire, A. L., Gabriel, S., Tishkoff, S. A., Wonkam, A., Chakravarti, A., Furlong, E. E. M., et al. (2020). The Road Ahead in Genetics and Genomics. Nat. Rev. Genet. 21 (10), 581–596. doi:10.1038/s41576-020-0272-6
McKibbin, W. J., and Fernando, R. (2020). The Global Macroeconomic Impacts of COVID-19: Seven Scenarios. SSRN J. doi:10.2139/ssrn.3547729
Michaelis, M., Doerr, H. W., and Cinatl, J. (2011). Investigation of the Influence of EPs 7630, a Herbal Drug Preparation from Pelargonium Sidoides, on Replication of a Broad Panel of Respiratory Viruses. Phytomedicine 18, 384–386. doi:10.1016/j.phymed.2010.09.008
Miner, A. S., Laranjo, L., and Kocaballi, A. B. (2020). Chatbots in the Fight against the COVID-19 Pandemic. Npj Digit. Med. 3, 65. doi:10.1038/s41746-020-0280-0
Moon, K.-B., Park, J.-S., Park, Y.-I., Song, I.-J., Lee, H.-J., Cho, H.-S., et al. (2019). Development of Systems for the Production of Plant-Derived Biopharmaceuticals. Plants 9, 30. doi:10.3390/plants9010030
Murray, A., Ravichandran, K., Singh, S. V., Sankar, M., Thomas, P., Dhama, K., et al. (2020). In Silico Molecular Docking Analysis Targeting SARS-CoV-2 Spike Protein and Selected Herbal Constituents. J. Pure Appl. Microbiol. 14, 989–998. doi:10.22207/JPAM.14.SPL1.37
Muthuramalingam, P., Jeyasri, R., Valliammai, A., Selvaraj, A., Karthika, C., Gowrishankar, S., et al. (2020). Global Multi-Omics and Systems Pharmacological Strategy Unravel the Multi-Targeted Therapeutic Potential of Natural Bioactive Molecules against COVID-19: An In Silico Approach. Genomics 112, 4486–4504. doi:10.1016/j.ygeno.2020.08.003
Muller, C., Schulte, F. W., Lange-Grünweller, K., Obermann, W., Madhugiri, R., Pleschka, S., et al. (2018). Broad-spectrum Antiviral Activity of the eIF4A Inhibitor Silve Strol against Corona- and Picornaviruses. Antivir. Res 150, 123–129. doi:10.1016/j.antiviral.2017.12.010
Naj, A. C. (2019). Genotype Imputation in Genome‐Wide Association Studies. Curr. Protoc. Hum. Genet. 102 (1), e84. doi:10.1002/cphg.84
Oliveira, R. V., Oliveira, M. C., and Pelli, A. (2017). Disease Infection by Enterobacteriaceae Family in Fishes: A Review. J. Microbiol. Exp. 4 (5), 00128. doi:10.15406/jmen.2017.04.00128
Ong, E., Wong, M. U., Huffman, A., and He, Y. (2020). COVID-19 Coronavirus Vaccine Design Using Reverse Vaccinology and Machine Learning. Front. Immunol. 11, 1581. doi:10.3389/fimmu.2020.01581
Overmyer, K. A., Shishkova, E., Miller, I. J., Balnis, J., Bernstein, M. N., Peters-Clarke, T. M., et al. (2021). Large-Scale Multi-Omic Analysis of COVID-19 Severity. Cel Syst. 12 (1), 23–40. e7. doi:10.1016/j.cels.2020.10.003
Pandit, M., and Latha, N. (2020). In Silico studies Reveal Potential Antiviral Activity of Phytochemicals from Medicinal Plants for the Treatment of COVID-19 Infection. Res. Sq. [Epub ahead of print]. doi:10.21203/rs.3.rs-22687/v1
Park, J.-Y., Yuk, H. J., Ryu, H. W., Lim, S. H., Kim, K. S., Park, K. H., et al. (2017). Evaluation of Polyphenols from Broussonetia Papyrifera as Coronavirus Protease Inhibitors. J. Enzyme Inhib. Med. Chem. 32, 504–512. doi:10.1080/14756366.2016.1265519
Peng, D., and Liu, X. (2017). Research Advances in Biomarker for Sepsis. Advanced Trauma and Surgery. Editors X. Fu, and L. Liu (Singapore:Elsevier), doi:10.1007/978-981-10-2425-2_15
Peška, V., Sitová, Z., Fajkus, P., and Fajkus, J. (2017). BAL31-NGS Approach for Identification of Telomeres De Novo in Large Genomes. Methods (San Diego, Calif.) 114, 16–27. doi:10.1016/j.ymeth.2016.08.017
Prasad, A., Muthamilarasan, M., and Prasad, M. (2020). Synergistic Antiviral Effects against SARS-CoV-2 by Plant-Based Molecules. Plant Cel Rep 39 (9), 1109–1114. doi:10.1007/s00299-020-02560-w
Prasad, A., and Prasad, M. (2020). SARS-CoV-2: the Emergence of a Viral Pathogen Causing Havoc on Human Existence. J. Genet. 99, 37. doi:10.1007/s12041-020-01205-x
Qamar, U. l., Alqahtani, M. T., Alamri, S. M., and Chen, M. A. (2020). Structural Basis of SARS-CoV-2 3CLpro and Anti-COVID-19 Drug Discovery from Medicinal Plants. J. Pharm. Anal. 10, 313–319. doi:10.1016/j.jpha.2020.03.009
Randhawa, G. S., Soltysiak, M. P. M., El Roz, H., de Souza, C. P. E., Hill, K. A., and Kari, L. (2020). Machine Learning Using Intrinsic Genomic Signatures for Rapid Classification of Novel Pathogens: COVID-19 Case Study. PLoS One 15, e0232391. doi:10.1371/journal.pone.0232391
Rashid, M. T., and Wang, D. (2020). CovidSens: a Vision on Reliable Social Sensing for COVID-19. Artif. Intell. Rev. 54, 1–25. doi:10.1007/s10462-020-09852-3
Rattanapisit, K., Shanmugaraj, B., Manopwisedjaroen, S., Purwono, P. B., Siriwattananon, K., Khorattanakulchai, N., et al. (2020). Rapid Production of SARS-CoV-2 Receptor Binding Domain (RBD) and Spike Specific Monoclonal Antibody CR3022 in Nicotiana Benthamiana. Sci. Rep. 10 (1), 17698. doi:10.1038/s41598-020-74904-1
Redka, D. y. S., MacKinnon, S. S., Landon, M., Windemuth, A., Kurji, N., and Shahani, V. (2020). PolypharmDB, a Deep Learning-Based Resource, Quickly Identifies Repurposed Drug Candidates for COVID-19. ChemRxiv. doi:10.26434/chemrxiv.12071271.v1
Ren, J.-l., Zhang, A.-H., and Wang, X.-J. (2020). Traditional Chinese Medicine for COVID-19 Treatment. Pharmacol. Res. 155, 104743. doi:10.1016/j.phrs.2020.104743
Robson, B. (2020). COVID-19 Coronavirus Spike Protein Analysis for Synthetic Vaccines, a Peptidomimetic Antagonist, and Therapeutic Drugs, and Analysis of a Proposed Achilles' Heel Conserved Region to Minimize Probability of Escape Mutations and Drug Resistance. Comput. Biol. Med. 121, 103749. doi:10.1016/j.compbiomed.2020.103749
Roh, C. (2012). A Facile Inhibitor Screening of SARS Coronavirus N Protein Using Nanoparticle-Based RNA Oligonucleotide. Int. J. Nanomedicine 7, 2173–2179. doi:10.2147/IJN.S31379
Rothan, H. A., and Byrareddy, S. N. (2020). The Epidemiology and Pathogenesis of Coronavirus Disease (COVID-19) Outbreak. J. Autoimmun. 109, 102433. doi:10.1016/j.jaut.2020.102433
Ryu, Y. B., Jeong, H. J., Kim, J. H., Kim, Y. M., Park, J.-Y., Kim, D., et al. (2010). Biflavonoids from Torreya Nucifera Displaying SARS-CoV 3CLpro Inhibition. Bioorg. Med. Chem. 18, 7940–7947. doi:10.1016/j.bmc.2010.09.035
Sampangi-Ramaiah, M. H., Vishwakarma, R., and Shaanker, R. U. (2020). Molecular Docking Analysis of Selected Natural Products from Plants for Inhibition of SARS-CoV-2 Main Protease. Curr. Sci. 118, 1087–1092.
Sassi, B. A., Harzallah-Skhiri, F., Bourgougnon, N., and Aouni, M. (2008). Antiviral Activity of Some Tunisian Medicinal Plants against Herpes Simplex Virus Type 1. Nat. Prod. Res. 22, 53–65. doi:10.1080/14786410701589790
Saxena, D., Sharma, A., Siddiqui, M. H., and Kumar, R. (2019). Blood Brain Barrier Permeability Prediction Using Machine Learning Techniques: An Update. Curr. Pharm. Biotechnol. 20, 1163–1171. doi:10.2174/1389201020666190821145346
Schnitzler, P., Schuhmacher, A., Astani, A., and Reichling, J. (2008). Melissa Officinalis Oil Affects Infectivity of Enveloped Herpesviruses. Phytomedicine 15, 734–740. doi:10.1016/j.phymed.2008.04.01810.1016/j.phymed.2008.04.018
Sehailia, M., and Chemat, S. (2020). In-silico Studies of Antimalarial-Agent Artemisinin and Derivatives Portray More Potent Binding to Lys353 and Lys31-Binding Hotspots of SARS-CoV-2 Spike Protein Than Hydroxychloroquine: Potential Repurposing of Artenimol for COVID-19. ChemRxiv. doi:10.26434/chemrxiv.12098652.v1
Semper, C., Watanabe, N., and Savchenko, A. (2021). Structural Characterization of Nonstructural Protein 1 from SARS-CoV-2. iScience 24 (1), 101903. doi:10.1016/j.isci.2020.101903
Semple, S. J., Reynolds, G. D., O'Leary, M. C., and Flower, R. L. P. (1998). Screening of Australian Medicinal Plants for Antiviral Activity. J. Ethnopharmacology 60, 163–172. doi:10.1016/s0378-8741(97)00152-9
Senior, A. W., Evans, R., Jumper, J., Kirkpatrick, J., Sifre, L., Green, T., et al. (2020). Improved Protein Structure Prediction Using Potentials from Deep Learning. Nature 577, 706–710. doi:10.1038/s41586-019-1923-7
Sepay, N., Sepay, N., Al Hoque, A., Mondal, R., Halder, U. C., and Muddassir, M. (2020). In Silico fight against Novel Coronavirus by Finding Chromone Derivatives as Inhibitor of Coronavirus Main Proteases Enzyme. Struct. Chem., 31, 1831, 1840. doi:10.1007/s11224-020-01537-5
Serkedjieva, J. (2003). Influenza Virus Variants with Reduced Susceptibility to Inhibition by a Polyphenol Extract from Geranium Sanguineum L. Pharmazie 58, 53–57.
Serseg, T., Benarous, K., and Yousfi, M. (2020). Hispidin and Lepidine E: Two Natural Compounds and Folic Acid as Potential Inhibitors of 2019-novel Coronavirus Main Protease (2019-nCoVMpro), Molecular Docking and SAR Study. Curr. Comput. Aided Drug Des. 16. doi:10.2174/1573409916666200422075440
Shahriar, S., Rana, M. S., Hossain, M. S., Karim, A., Mredula, T. N., Nourin, N., et al. (2021). COVID-19: Epidemiology, Pathology, Diagnosis, Treatment, and Impact. Curr. Pharm. Des. [Epub ahead of print]. doi:10.2174/1381612827666210224142446
Sharma, A. D., and kaur, i. (2020). Eucalyptol (1,8 Cineole) from Eucalyptus Essential Oil a Potential Inhibitor of COVID 19 Corona Virus Infection by Molecular Docking Studies. Preprints. [Epub ahead of print]. doi:10.20944/preprints202003.0455.v1
Sharma, A. D., and Kaur, I. (2020). Eucalyptol (1,8 Cineole) from Eucalyptus Essential Oil a Potential Inhibitor of COVID 19 Corona Virus Infection by Molecular Docking Studies. Res. Rev. Biotech. Biosci. 7, 59–66. [Epub ahead of print]. doi:10.20944/preprints202003.0455.v1
Sharma, A., Kumar, R., Semwal, R., Aier, I., Tyagi, P., and Varadwaj, P. (2020). DeepOlf: Deep Neural Network Based Architecture for Predicting Odorants and Their Interacting Olfactory Receptors. IEEE/ACM Trans. Comput. Biol. Bioinf., 1. doi:10.1109/TCBB.2020.3002154
Sharma, N., Muthamilarasan, M., Prasad, A., and Prasad, M. (2020). Genomics Approaches to Synthesize Plant-Based Biomolecules for Therapeutic Applications to Combat SARS-CoV-2. Genomics 112, 4322–4331. doi:10.1016/j.ygeno.2020.07.033
Shen, L., Niu, J., Wang, C., Huang, B., Wang, W., Zhu, N., et al. (2019). High-Throughput Screening and Identification of Potent Broad-Spectrum Inhibitors of Coronaviruses. J. Virol. 93, e00023–e00019. doi:10.1128/JVI.00023-19
Shen, F., Fu, Z. Y., Wu, Y. R., Li, L., Zhao, Y. D., Xia, Y., et al. (2020). Based on Network Pharmacology and High-Throughput Molecular Docking to Study the Potential Molecular Mechanism of Active Compounds That Bind SARS-Cov-2 Specific Target Protein in Jinhua Qingan Granules to Interfere with CovID-19, Modernization of Traditional Chinese Medicine and Materia Materia-World Science and Technology. Available at: http://kns.cnki.net/kcms/detail/11.5699.R.20200421.0949.004.html (Accessed December 20, 2020).
Shree, P., Mishra, P., Selvaraj, C., Singh, S. K., Chaube, R., Garg, N., et al. (2020). Targeting COVID-19 (SARS-CoV-2) Main Protease through Active Phytochemicals of Ayurvedic Medicinal Plants - Withania somnifera (Ashwagandha), Tinospora cordifolia (Giloy) and Ocimum sanctum (Tulsi) - a Molecular Docking Study. J. Biomol. Struct. Dyn. 1, 1–14. Advance online publication doi:10.1080/07391102.2020.1810778
Silveira, D., Prieto-Garcia, J. M., Boylan, F., Estrada, O., Fonseca-Bazzo, Y. M., Jamal, C. M., et al. (2020). COVID-19: Is There Evidence for the Use of Herbal Medicines as Adjuvant Symptomatic Therapy? Front. Pharmacol. 11, 581840. doi:10.3389/fphar.2020.581840
Singh, A., and Mishra, A. (2020). Leucoefdin a Potential Inhibitor against SARS CoV-2 Mpro. J. Biomol. Struct. Dyn. 2020, 1–6. doi:10.1080/07391102.2020.1777903
Sohrabi, C., Alsafi, Z., O'Neill, N., Khan, M., Kerwan, A., Al-Jabir, A., et al. (2020). World Health Organization Declares Global Emergency: a Review of the 2019 Novel Coronavirus (COVID-19). Int. J. Surg. 76, 71–76. doi:10.1016/j.ijsu.2020.02.034
Song, J., Du, L., Li, L., Palmer, L. C., Forney, C. F., Fillmore, S., et al. (2015). Targeted Quantitative Proteomic Investigation Employing Multiple Reaction Monitoring on Quantitative Changes in Proteins that Regulate Volatile Biosynthesis of Strawberry Fruit at Different Ripening Stages. J. Proteomics 126, 288–295. doi:10.1016/j.jprot.2015.06.004
Stebbing, J., Krishnan, V., de Bono, S., Ottaviani, S., Casalini, G., Richardson, P. J., et al. (2020). Mechanism of Baricitinib Supports Artificial Intelligence-Predicted Testing in COVID-19 Patients. EMBO Mol. Med. 12 (8), e12697. doi:10.15252/emmm.202012697
Su, Y., Chen, D., Yuan, D., Lausted, C., Choi, J., Dai, C. L., et al. (2020). Multi-Omics Resolves a Sharp Disease-State Shift between Mild and Moderate COVID-19. Cell 183 (6), 1479–1495 e20. doi:10.1016/j.cell.2020.10.037
Subramanian, I., Verma, S., Kumar, S., Jere, A., and Anamika, K. (2020). Multi-omics Data Integration, Interpretation, and Its Application. Bioinformatics Biol. insights 14, 1177932219899051. doi:10.1177/1177932219899051
Suryanarayana, L., and Banavath, D. (2020). A Review on Identification of Antiviral Potential Medicinal Plant Compounds against with COVID-19. Int. J. Res. Eng. Sci. Manage. 3, 675–679.
Tallei, T. E., Tumilaar, S. G., Niode, N. J., Fatimawali, F., Kepel, B. J., Idroes, R., et al. (2020). Potential of Plant Bioactive Compounds as SARS-CoV-2 Main Protease (Mpro) and Spike (S) Glycoprotein Inhibitors: a Molecular Docking Study. Preprints 2020, 2020040102. doi:10.20944/preprints202004.0102.v2
Tarazona, S., Balzano-Nogueira, L., and Conesa, A. (2018). “Multiomics Data Integration in Time Series Experiments,” in Data Analysis for Omic Sciences: Methods and Applications, Comprehensive Analytical Chemistry. Editors J. Jaumot, C. Bedia, and R. Tauler (Amsterdam, Netherlands:Elsevier) 82, 502–532. doi:10.1016/bs.coac.2018.06.005
To, K. K. W., Lee, K.-C., Wong, S. S. Y., Sze, K.-H., Ke, Y.-H., Lui, Y.-M., et al. (2016). Lipid Metabolites as Potential Diagnostic and Prognostic Biomarkers for Acute Community Acquired Pneumonia. Diagn. Microbiol. Infect. Dis. 85, 249–254. doi:10.1016/j.diagmicrobio.2016.03.012
Tong, X., Li, A., Zhang, Z., Duan, J., Chen, X., Hua, C., et al. (2004). TCM Treatment of Infectious Atypical Pneumonia-Aa Report of 16 Cases. J. Tradit Chin. Med. 24, 266–269.
Tsai, Y.-C., Lee, C.-L., Yen, H.-R., Chang, Y.-S., Lin, Y.-P., Huang, S.-H., et al. (2020). Antiviral Action of Tryptanthrin Isolated from Strobilanthes Cusia Leaf against Human Coronavirus NL63. Biomolecules 10 (3), 366. doi:10.3390/biom10030366
Ulasli, M., Gurses, S. A., Bayraktar, R., Yumrutas, O., Oztuzcu, S., Igci, M., et al. (2014). The Effects of Nigella Sativa (Ns), Anthemis Hyalina (Ah) and Citrus Sinensis (Cs) Extracts on the Replication of Coronavirus and the Expression of TRP Genes Family. Mol. Biol. Rep. 41, 1703–1711. doi:10.1007/s11033-014-3019-7
Umesh, U., Kundu, D., Selvaraj, C., Singh, S. K., and Dubey, V. K. (2020). Identification of New Anti-nCoV Drug Chemical Compounds from Indian Spices Exploiting SARS-CoV-2 Main Protease as Target. J. Biomol. Struct. Dyn. 2020, 1–9. doi:10.1080/07391102.2020.1763202
Vieira, R. F., Bizzo, H. R., and Deschamps, C. (2010). Genetic Resources of Aromatic Plants from Brazil. Isr. J. Plant Sci. 58, 263–271. doi:10.1560/IJPS.58.2.263
Wahedi, H. M., Ahmad, S., and Abbasi, S. W. (2020). Stilbene-based Natural Compounds as Promising Drug Candidates against COVID-19. J. Biomol. Struct. Dyn. 2020, 1–10. doi:10.1080/07391102.2020.1762743
Wang, C., Liu, Z., Chen, Z., Huang, X., Xu, M., He, T., et al. (2020a). The Establishment of Reference Sequence for SARS‐CoV‐2 and Variation Analysis. J. Med. Virol. 92 (6), 667–674. doi:10.1002/jmv.25762
Wang, D., Yang, J., Fan, J., Chen, W., Nikolic‐Paterson, D. J., and Li, J. (2020b). Omics Technologies for Kidney Disease Research. Anat. Rec. 303 (10), 2729–2742. doi:10.1002/ar.24413
Wang, D., Zhang, F., and Gao, G. (2020c). CRISPR-Based Therapeutic Genome Editing: Strategies and In Vivo Delivery by AAV Vectors. Cell 181 (1), 136–150. doi:10.1016/j.cell.2020.03.023
Wang, M., Cao, R., Zhang, L., Yang, X., Liu, J., Xu, M., et al. (2020d). Remdesivir and Chloroquine Effectively Inhibit the Recently Emerged Novel Coronavirus (2019-nCoV) in vitro. Cell Res 30, 269–271. doi:10.1038/s41422-020-0282-0
Wang, X., Dhindsa, R., Povysil, G., Zoghbi, A., Motelow, J., Hostyk, J., et al. (2020e). Transcriptional Inhibition of Host Viral Entry Proteins as a Therapeutic Strategy for SARS-CoV-2. Preprints 2020, 030360. doi:10.20944/preprints202003.0360.v1
Wang, Z., Gerstein, M., and Snyder, M. (2009). RNA-Seq: a Revolutionary Tool for Transcriptomics. Nat. Rev. Genet. 10, 57–63. doi:10.1038/nrg2484
Wang, Z., Li, L., Yan, J., and Yao, Y. (2020f). Evaluating the Traditional Chinese Medicine (TCM) Officially Recommended in China for COVID-19 Using Ontology-Based Side-Effect Prediction Framework (OSPF) and Deep Learning. Preprints. [Epub ahead of print]. doi:10.20944/preprints202002.0230.v1
Wink, M. (2020). Potential of DNA Intercalating Alkaloids and Other Plant Secondary Metabolites against SARS-CoV-2 Causing COVID-19. Diversity 12, 175. doi:10.3390/d12050175
Wishart, D. S. (2007). Human Metabolome Database: Completing the 'human Parts List'. Pharmacogenomics 8, 683–686. doi:10.2217/14622416.8.7.683
Woo, P. C., Huang, Y., Lau, S. K., and Yuen, K. Y. (2010). Coronavirus Genomics and Bioinformatics Analysis. Viruses 2 (8), 1804–1820. doi:10.3390/v2081803
Xian, Y., Zhang, J., Bian, Z., Zhou, H., Zhang, Z., Lin, Z., et al. (2020). Bioactive Natural Compounds against Human Coronaviruses: a Review and Perspective. Acta Pharmaceutica Sinica. B 10 (7), 1163–1174. doi:10.1016/j.apsb.2020.06.002
Xu, Z., Shi, L., Wang, Y., Zhang, J., Huang, L., Zhang, C., et al. (2020). Pathological Findings of COVID-19 Associated with Acute Respiratory Distress Syndrome. Lancet Respir. Med. 8, 420–422. doi:10.1016/S2213-2600(20)30076-X
Yang, Y., Islam, M. S., Wang, J., Li, Y., and Chen, X. (2020). Traditional Chinese Medicine in the Treatment of Patients Infected with 2019-new Coronavirus (SARS-CoV-2): a Review and Perspective. Int. J. Biol. Sci. 16, 1708–1717. doi:10.7150/ijbs.45538
Yu, M.-S., Lee, J., Lee, J. M., Kim, Y., Chin, Y.-W., Jee, J.-G., et al. (2012). Identification of Myricetin and Scutellarein as Novel Chemical Inhibitors of the SARS Coronavirus Helicase, nsP13. Bioorg. Med. Chem. Lett. 22, 4049–4054. doi:10.1016/j.bmcl.2012.04.08110.1016/j.bmcl.2012.04.081
Zhang, L., Yu, J., Zhou, Y., Shen, M., and Sun, L. (2020). Becoming a Faithful Defender: Traditional Chinese Medicine against Coronavirus Disease 2019 (COVID-19). Am. J. Chin. Med. 48 (4), 763–777. doi:10.1142/s0192415x2050038x
Zhang, Q., Wang, Y., Qi, C., Shen, L., and Li, J. (2020). Clinical Trial Analysis of 2019‐nCoV Therapy Registered in China. J. Med. Virol. 92, 540–545. doi:10.1002/jmv.25733
Zhang, M.-M., Liu, X. M., and He, L. (2004). Effect of Integrated Traditional Chinese and Western Medicine on SARS: a Review of Clinical Evidence. World. J. Gastroenterol. 10, 3500–3505. doi:10.3748/wjg.v10.i23.3500
Zhang, T., Zhang, L., Payne, P. R. O., and Li, F. (2021). Synergistic Drug Combination Prediction by Integrating Multiomics Data in Deep Learning Models. Methods Mol. Biol. (Clifton, N.J.) 2194, 223–238. doi:10.1007/978-1-0716-0849-4_12
Zheng, J., and Perlman, S. (2018). Immune Responses in Influenza A Virus and Human Coronavirus Infections: an Ongoing Battle between the Virus and Host. Curr. Opin. Virol. 28, 43–52. doi:10.1016/j.coviro.2017.11.002
Zhou, G., Li, S., and Xia, J. (2020). Network-Based Approaches for Multi-Omics Integration. Methods Mol. Biol. (Clifton, N.J.) 2104, 469–487. doi:10.1007/978-1-0716-0239-3_23
Zhu, N., Zhang, D., Wang, W., Li, X., Yang, B., Song, J., et al. (2020). A Novel Coronavirus from Patients with Pneumonia in China, 2019. N. Engl. J. Med. 382, 727–733. doi:10.1056/NEJMoa2001017
Zoppi, J., Guillaume, J. F., Neunlist, M., and Chaffron, S. (2021). MiBiOmics: an Interactive Web Application for Multi-Omics Data Exploration and Integration. BMC bioinformatics 22 (1), 6. doi:10.1186/s12859-020-03921-8
Keywords: COVID-19, SARS-CoV-2, coronavirus, multi-omics, biomolecules, therapeutic molecules
Citation: Singh R, Singh PK, Kumar R, Kabir MT, Kamal MA, Rauf A, Albadrani GM, Sayed AA, Mousa SA, Abdel-Daim MM and Uddin MS (2021) Multi-Omics Approach in the Identification of Potential Therapeutic Biomolecule for COVID-19. Front. Pharmacol. 12:652335. doi: 10.3389/fphar.2021.652335
Received: 12 January 2021; Accepted: 21 April 2021;
Published: 12 May 2021.
Edited by:
Christophe F. Hano, INRA EA1207 Laboratoire De Biologie Des Ligneux et Des Grandes Cultures, FranceReviewed by:
Giuseppe Biondi-Zoccai, Sapienza University of Rome, ItalyCopyright © 2021 Singh, Singh, Kumar, Kabir, Kamal, Rauf, Albadrani, Sayed, Mousa, Abdel-Daim and Uddin. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Rajnish Kumar, cmt1bWFyMkBsa28uYW1pdHkuZWR1; Md. Sahab Uddin, bXN1LW5ldXJvcGhhcm1hQGhvdG1haWwuY29t
†ORCID: Md. Sahab Uddin, orcid.org/0000-0002-0805-7840
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.