
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Pharmacol., 22 February 2021
Sec. Respiratory Pharmacology
Volume 12 - 2021 | https://doi.org/10.3389/fphar.2021.636989
The outbreak of a new coronavirus (SARS-CoV-2), which is responsible for the COVID-19 disease and is spreading rapidly around the world, urgently requires effective therapeutic treatments. In this context, drug repurposing represents a valuable strategy, as it enables accelerating the identification of drug candidates with already known safety profiles, possibly aiding in the late stages of clinical evaluation. Moreover, therapeutic treatments based on drugs with beneficial multi-target activities (polypharmacology) may show an increased antiviral activity or help to counteract severe complications concurrently affecting COVID-19 patients. In this study, we present the results of a computational drug repurposing campaign that aimed at identifying potential inhibitors of the main protease (Mpro) of the SARS-CoV-2. The performed in silico screening allowed the identification of 22 candidates with putative SARS-CoV-2 Mpro inhibitory activity. Interestingly, some of the identified compounds have recently entered clinical trials for COVID-19 treatment, albeit not being assayed for their SARS-CoV-2 antiviral activity. Some candidates present a polypharmacology profile that may be beneficial for COVID-19 treatment and, to the best of our knowledge, have never been considered in clinical trials. For each repurposed compound, its therapeutic relevance and potential beneficial polypharmacological effects that may arise due to its original therapeutic indication are thoroughly discussed.
At present, we are faced with one of the most devastating pandemic crises in human history, the coronavirus disease 2019 (COVID-19), which has affected around 56 million people to date and is responsible for more than 1.38 million deaths (Who.int1). Unfortunately, effective therapeutic options are not currently available to prevent or cure this disease, which is caused by the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) (Who.int1). Only very recently, Pfizer, in partnership with BioNTech Manufacturing GmbH, has filed their clinically evaluated vaccine (Comirnaty) to Food and Drug Administration (FDA), for an Emergency Use Authorization (EUA) (Pfizer Inc.2) and received a Conditional Marketing Authorization from the European Medicines Agency (EMA3). However, it should be noted that the efficacy of vaccines for this disease have yet to be assessed on a large scale, and challenges in vaccine dose supply worldwide should be overcome. Moreover, the approval by worldwide regulatory agencies of effective and un-restricted treatments for patients already suffering from the disease is urgently needed.
SARS-CoV-2 is a positive-sense single-stranded RNA virus belonging to the β-genus of the Coronaviridae family (Wan et al., 2020) and is closely related to coronaviruses responsible for the SARS and MERS outbreaks that spread in 2003 and 2012, respectively (Zhu et al., 2020). Although outbreaks deriving from coronavirus infections appear to be recurrent (Zhu et al., 2020), no specific antiviral drugs are currently available to treat these diseases. Therefore, major efforts are currently focused on developing vaccines, as well as effective drugs to treat infected patients (Clinicaltrials.gov4). Indeed, the first COVID-19 vaccine has been approved after the submission of the present study. In the meanwhile, many biological targets are now under investigation to develop SARS-CoV-2-specific antiviral compounds, the most studied being the RNA-dependent RNA polymerase, the spike protein, and the main protease (Morse et al., 2020). Moreover, the number of clinical trials based on antivirals developed for other infections or drugs meant to reduce inflammation and severe respiratory complications are now increasing at previously unseen rates (Clinicaltrials.gov4). Currently investigated antivirals include favipiravir, an anti-influenza drug, which resulted effective also against the Ebola virus (ClinicalTrials Identifiers: NCT04303299, NCT04310228, NCT04349241), and the RNA-dependent RNA polymerase prodrug inhibitor remdesivir, which was shown to be effective also against MERS and SARS (Clinicaltrials.gov)5. Indeed, remdesivir has very recently been reported to significantly reduce the median recovery time of COVID-19 patients and has been approved by FDA as a therapeutic option in hospital settings (Beigel et al., 2020). Moreover, boceprevir, which is a NS3/4A serine protease inhibitor used in the treatment of chronic Hepatitis C, has also very recently gained the attention of the research community for COVID-19 treatment (Fu et al., 2020). Hydroxychloroquine was also investigated for the treatment of COVID-19, but clinical trials on this compound were stopped by the World Health Organization in June 2020 (Who.int6). Moreover, hydroxychloroquine did not show significant activity on COVID-19 hospitalized patients, according to recently retrieved clinical data (RECOVERY Collaborative Group et al., 2020b). The monoclonal antibodies tocilizumab and sarilumab directed against the interleukin-6 receptor (IL-6R) have entered clinical trials (ClinicalTrials Identifiers: NCT04322773, NCT04332913) for the same purpose. Finally, the use of dexamethasone was endorsed by the European Medicines Agency to treat hospitalized patients with COVID-19 (RECOVERY Collaborative Group et al., 2020a).
The rapid spread of the SARS-CoV-2 pandemic and the lack of specific antiviral drugs suggest that drug repurposing should be the preferred way for rapidly selecting suitable candidates for clinical testing (Senanayake, 2020). Indeed, these compounds are well characterized and already possess optimized pharmacokinetics and safety profiles. In this scenario, computational approaches can provide new opportunities for drug repurposing (March-Vila et al., 2017), allowing the identification of valuable drug candidates to be used either alone or in combination. These approaches take advantage of curated databases reporting chemical, structural and activity information on different protein targets and already known therapeutic effects (e.g., PubChem, ChEMBL, DrugBank), or other repositories specifically focusing on repurposing data (e.g., ReframeDB and Drug Repurposing Hub) and COVID-19 databases (e.g., NIH7), which are currently available for already approved or under clinical evaluation COVID-19 drugs (Corsello et al., 2017; Gaulton et al., 2017; Janes et al., 2018; Wishart et al., 2018; Kim et al., 2019). Moreover, considering the severe life-threatening disease complications, polypharmacological drugs, i.e., single drug molecules that combine antiviral activity with e.g., anti-inflammatory or antithrombotic activity, may be particularly useful (Anighoro et al., 2014). To this end, in this study we performed an extensive structure-based virtual screening campaign and identified 22 top-candidate approved or experimental drugs as potential inhibitors of the SARS-CoV-2 main protease (Mpro). The DrugBank database (Wishart et al., 2018), including drug metabolites, was docked to the Mpro enzyme and the results were post-processed with BEAR (Rastelli et al., 2009), an in-house developed screening tool with a well-documented ability to refine virtual screening results (Rastelli and Pinzi, 2019). Moreover, retrospective structure-based analyses were performed on a set of compounds with already known SARS-CoV-2 Mpro activity, to further validate the adopted in silico structure-based protocol. The candidate selection process took into special consideration the analysis of drug annotations and biological activity information reported in the literature to attain possible favorable polypharmacological effects arising from the original therapeutic indication. Because of their fitting to the SARS-CoV-2 main protease active site, the reported drugs could be readily repurposed to elicit an antiviral response.
The recently reported 6LU7 crystal structure of the SARS-CoV-2 main protease (Jin et al., 2020) was first collected from the Protein Data Bank (accessed on March 17th, 2020) and then prepared for the in silico screening process using the Protein Preparation Wizard (Protein Preparation Wizard, Schrödinger, LLC, New York, NY, 2020). Defaults parameters were used during the protein preparation. Atom types and connectivity issues were fixed, hydrogen atoms were added, and interaction geometries were optimized. The co-crystallized water molecules and the peptide-like PRD_002214 inhibitor (compound N3) were retained during the protein preparation process, while they were removed in the following docking and post-processing phases.
Docking calculations were performed in the active site of the prepared 6LU7 crystal structure by using Glide (Glide, Schrödinger, LLC, New York, NY, 2020; Friesner et al., 2004) with the Standard Precision (SP) protocol. Specifically, the receptor grid was first generated on the coordinates of the co-crystallized PRD_002214 ligand, with a box of (10 Å × 10 Å × 10 Å) dimensions (default settings). Then, the docking protocol was validated by redocking the co-crystallized ligand into its parent crystal structure, with satisfactory results.
Approved drugs, clinical and preclinical candidates, and metabolites were first downloaded from the DrugBank database (www.drugbank.ca, accessed on March 17th, 2020), and then prepared for the structure-based calculations using the LigPrep utility (LigPrep, Schrödinger, LLC, New York, NY, 2020). Specifically, ionization states and tautomers at pH values equal to 7 ± 2 were first generated for each ligand in the screening database and then minimized according to the OPLS3e force field. Stereoisomers were also generated for the DrugBank ligands with undefined chiralities. Subsequently, the pre-treated compounds were screened with the validated docking protocol. Finally, the predicted poses were visually inspected, and the first 2000 top-scoring ligand-protein complexes were further post-processed with the BEAR post-docking tool (Rastelli et al., 2009).
The BEAR protocol consists of three steps based on molecular mechanics (MM) minimization and molecular dynamics cycles, followed by more accurate binding free energy estimation of the refined complex with the MM-PBSA and MM-GBSA methods (Rastelli et al., 2009). Further details on the BEAR post-processing procedure, which was shown to considerably increase the prediction performances in several virtual screening campaigns, are reported by (Rastelli et al., 2009).
A final step of visual inspection of the refined complexes and their comparison with the corresponding poses predicted by Glide aided in the final selection of the potential candidates for the SARS-CoV-2 main protease inhibition. Analysis of data annotation and literature searches returned a set of top candidates with a potentially beneficial polypharmacology profile.
Retrospective structure-based calculations were also performed with the previously described protocol on a set of compounds (Supplementary Table S1) with recently reported activity data on the SARS-CoV-2 Mpro (Dai et al., 2020; Jin et al., 2020; Ma et al., 2020). Finally, results were visually inspected and the obtained scores compared with those from the performed virtual screening.
The SARS-CoV-2 main protease (Mpro), also known as 3C-like protease, is a relevant target for drug repurposing because it plays a crucial role in the maturation of the viral particle (Jin et al., 2020). Indeed, this cysteine protease presents a highly conserved active site in several coronaviruses, such as SARS-CoV and MERS-CoV, and plays a key role in the cleavage of precursor polyproteins translated from viral RNA.
A computational screening workflow (Figure 1) was devised to identify drug candidates able to bind and inhibit the SARS-CoV-2 main protease. To this end, the complete DrugBank database (Release Version 5.1.5, 13,227 compounds, accessed on March 17th, 2020), including drug metabolites, was docked to the crystal structure of the SARS-CoV-2 main protease (PDB ID: 6LU7) (Jin et al., 2020) using Glide (Glide, Schrödinger, LLC, New York, NY, 2020; Friesner et al., 2004), and the results were ranked according to the Glide “Docking score” scoring function.
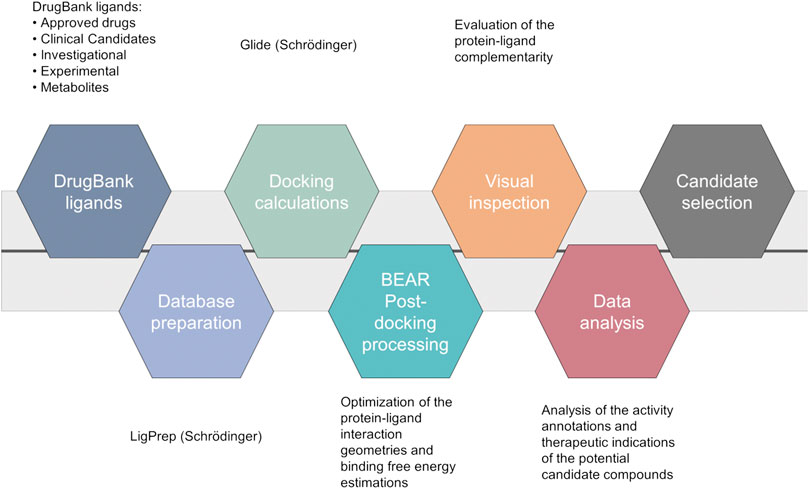
FIGURE 1. Computational workflow of the repurposing screening for the identification of SARS-CoV-2 main protease inhibitors.
The 2000 top ranking compounds were post-processed with BEAR (Rastelli et al., 2009; Rastelli and Pinzi, 2019). BEAR is an automated procedure that combines the structural refinement of docking poses through molecular dynamics, which accounts for protein flexibility, with the re-ranking of ligands on the basis of binding free-energies. Binding free energies of the refined complexes are calculated through the molecular mechanics generalized Born surface area (MM-GBSA) and molecular mechanics Poisson Boltzmann surface area (MM-PBSA) scoring functions, which implicitly account for solvation effects (Rastelli et al., 2009). Then, the best candidates were selected according to i) docking and post-docking scores, which provide an estimation of the binding affinity of a ligand to a target, ii) visual inspection of the protein-ligand complexes, and iii) analysis of drug annotations and literature information to repurpose known drugs, clinical candidates or experimental compounds as Mpro inhibitors that may benefit from their original therapeutic indications to reduce severe COVID-19 complications, while inhibiting viral particle maturation (beneficial polypharmacology (Anighoro et al., 2014)). Interestingly, several ligands already present in the DrugBank database, which were very recently confirmed as inhibitors of the SARS-CoV-2 Mpro enzyme, turned out to be among the top-scored compounds in our virtual screening campaign (see Supplementary Table S1), thus demonstrating that the used protocol is able to identify active compounds within the screened database. Moreover, to further validate the ability of the used in silico workflow to aid in the identification of candidates with potential SARS-CoV-2 Mpro activity, retrospective analyses were also performed on a set of inhibitors extracted from the literature and not included in the DrugBank database. In particular, ligand structures were first collected from their reference publications (Dai et al., 2020; Jin et al., 2020; Ma et al., 2020), built and prepared for structure-based calculations. Then, docking calculations followed by BEAR binding-free energy estimations were performed, as detailed in the “Materials and Methods” section. Notably, the majority of retrospectively investigated compounds with known activity data (i.e., 16 out of 23; ∼70% of the validation dataset) provided comparable docking scores to those of the 2,000 top ranking compounds (Glide “Docking scores” lower than −7.18 Kcal/mol), therefore, these compounds would have been selected for further investigations in our workflow. Moreover, it should be noted that the seven known SARS-CoV-2 Mpro inhibitors discarded at this stage by our workflow present a chemical structure significantly different to that of the crystallographic ligand, and it is widely acknowledged that docking results can be strongly affected by the selected receptor conformation (Broccatelli and Brown, 2014). Finally, results of the BEAR calculations on the retrospectively investigated compounds further strengthen the soundness of the implemented workflow. Indeed, the most active compounds in the validation dataset provided BEAR GB and PB scores comparable to those of the best-ranking ligands in the screened database (Supplementary Table S1).
The most promising compounds resulting from the virtual screening are reported in Table 1, along with their DrugBank IDs, the predicted docking and binding free energy scores, the rank position in the screening outcome, the therapeutic targets and original indications, and the PDB IDs of the crystal structure of each drug in complex with its original target, when available. The 2D chemical structures of the selected candidates are reported in Supplementary Material S1. A detailed discussion of the selected drug candidates divided into candidates for drug repurposing, candidates for drug repurposing with beneficial polypharmacology, and candidates for drug repurposing based on active drug metabolites, is reported in the following paragraphs.
The drugs herein reported (either approved or investigational) are excellent examples of potential candidates for repurposing as SARS-CoV-2 Mpro inhibitors. Indeed, all compounds are well accommodated in the SARS-CoV-2 Mpro catalytic site. Moreover, as evidence of the reliability of our results, several of the proposed candidates are viral protease inhibitors, which in some cases are already under study as COVID-19 therapeutic agents (see below).
The hydroxymethyl ketones EXPT02467 (cruz-2, DB02128) and EXPT02989 (cruz-1, DB01871) are two reversible inhibitors of cruzipain, a cysteine-type endopeptidase of Trypanosoma cruzi (Cazzulo et al., 1990). The structures of their complexes with the cruzipain target are available under the PDB IDs 1ME3 and 1ME4, respectively. Another interesting cruzipain inhibitor that emerged from our screening is WRR-204 (EXPT03235, DB04502), which is an irreversible inhibitor (PDB ID: 1EWO). To the best of our knowledge, these compounds have never been investigated for drug repurposing, including on COVID-19 related therapeutic targets.
MMI-175 (DB02378) is an experimental drug that inhibits β-secretase (BACE-1) (Ghosh et al., 2005), one of the two aspartic proteases responsible for the generation of amyloid-β peptides in neurons. As such, drugs blocking this enzyme may aid in slowing down Alzheimer’s disease progression. According to the predicted docking pose (Figure 2, panel (A)) and binding affinities, this compound is expected to efficiently bind to the SARS-CoV-2 Mpro enzyme. The ability of this compound to cross the blood-brain barrier would be of great interest for COVID-19 treatment, as previous studies have reported the presence of coronavirus particles in the CNS and their potential association with neurologic manifestations in patients (Baig et al., 2020). Also in this case, the compound emerged for the first time as a putative candidate for drug repurposing, according to the available literature data.
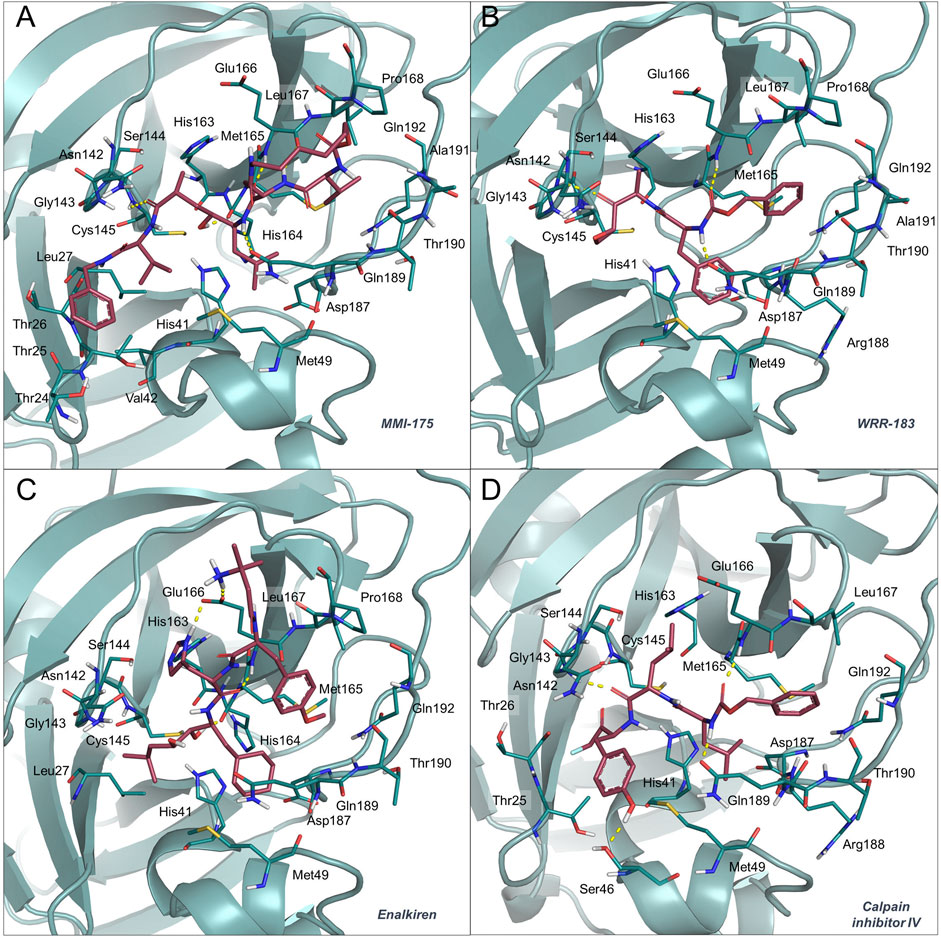
FIGURE 2. Binding mode of MMI-175 (A), WRR-183 (B), enalkiren (C) and calpain inhibitor IV (D) to the 6LU7 crystal structure. The SARS-CoV-2 Mpro binding site residues and the potential repurposed compounds are represented in deepteal and raspberry sticks, respectively. The image was created with PyMol (The PyMOL Molecular Graphics System, Version 2.1.1, Schrödinger, LLC).
EH58 (EXPT01332, DB03063) is a potent inhibitor of plasmepsin 2, an aspartic protease in the food vacuole of Plasmodium falciparum (PDB ID: 1LF3), and exhibits antimalarial activity (Ki = 100 nM) (Asojo et al., 2003). Another plasmepsin 2 inhibitor worthy of consideration, even if predicted scores are lower than those of EH58, is RS370 (DrugBank ID: DB04378, PDB ID: 1LF2), with a reported Ki of 30 nM on its original target (Asojo et al., 2002). EXPT00713 (DB03648) is a P. falciparum formylmethionine deformylase inhibitor with a reported IC50 of 130 nM (Robien et al., 2004). Interestingly, several antimalarial drugs are under the spotlight of COVID-19 investigations (Gendrot et al., 2020). The possibility of repurposing these compounds for the COVID-19 treatment would provide great benefits for cases of malaria and SARS-CoV-2 co-infections. This is especially important in those countries where malaria is endemic, considering that COVID-19 early symptoms (e.g., fatigue and fever) might result in misleading clinical diagnoses (Di Gennaro et al., 2020).
QF34 (EXPT02729, DB04353) is a pseudopeptide inhibitor of several variants of HIV-1 and HIV-2 proteases (Weber et al., 2002), including those of some highly resistant mutants. The compound was also crystallized with the HIV-1 protease (PDB IDs: 1IZH, 1IZI). Interestingly, the identification of ligands with already optimized safety profiles and reported HIV activity might be particularly effective for HIV/SARS-CoV-2 co-infected patients, which, unfortunately, seem to have poorer prognoses (Bhaskaran et al., 2020).
Compounds I2 (DB04692), N1 (DB04710) and N3 (PRD_002214, DB04595), three experimental inhibitors of the SARS-CoV Mpro reported in 2005, may also be valuable candidate inhibitors of the SARS-CoV-2 Mpro. Compounds I2, N1, and N3 were co-crystallized in complex with the SARS-CoV Mpro (PDB IDs: 2D2D, 1WOF, and 2AMQ, respectively). Notably, compound N3 has been very recently confirmed to bind and inhibit the SARS-CoV-2 Mpro, and the crystal structure of its complex was the starting point of our virtual screening (PDB ID: 6LU7) (Jin et al., 2020).
Compound WRR-183 (DB08732) is an α,β-epoxyketone that irreversibly inhibits the SARS-CoV Mpro (PDB ID 2OP9) (Goetz et al., 2007). According to our docking results, this compound was predicted to bind to the SARS-CoV-2 Mpro tightly and with a similar binding mode, where the epoxide is in close proximity to the Cys residue (Figure 2, panel (B)). Moreover, WRR-183 and especially its C-2 (R) epoxide isomer WRR-182 are highly active against the SARS-CoV spike-mediated entry (Zhou et al., 2011). Therefore, they show the potential to block SARS-CoV and, hopefully, SARS-CoV-2 at two different steps of the replication cycle, i.e., viral entry and particle assembly (Zhou et al., 2011).
Difelikefalin (formerly known as CR-845, DB11938) is a highly selective agonist of the κ-opioid receptor (Keppel Hesselink, 2017). This analgesic opioid peptide acts peripherally and is under investigation for the treatment of acute and post-operative pain and, more recently, chronic pruritus (Keppel Hesselink, 2017). Difelikefalin is currently in two Phase II clinical trials for the treatment of pruritus in atopic dermatitis and biliary cholangitis (Clinical Trials Identifiers: NCT04018027 and NCT03995212). The peripheral analgesic activity of the compound, together with its potential SARS-CoV-2 Mpro activity, may prove beneficial to COVID-19 patients experiencing peripheral neurologic symptoms and pain.
Polypharmacological ligands are extremely interesting in drug repurposing, because they offer the potential for higher efficacy and a combination of synergistic effects (Anighoro et al., 2014). Therefore, for each top-ranking compound, we carefully investigated whether a possible beneficial polypharmacological effect may arise owing to the reported biological activities and original therapeutic indications.
Enalkiren (DB03395) belongs to the class of direct renin inhibitors. By mimicking the transition state of angiotensin, enalkiren is able to block the first step of the renin-angiotensin system (Glassman et al., 1990). Interestingly, hypertension is among the most common comorbidities in hospitalized COVID-19 patients, and is often treated with angiotensin II receptor blockers or angiotensin-converting enzyme (ACE) inhibitors (Ran et al., 2020). Unfortunately, it has been recently reported that SARS-CoV-2 binds to the widespread angiotensin-converting enzyme 2 receptor to enter target cells (Bavishi et al., 2020) and that levels of serum angiotensin II are considerably increased in COVID-19 patients (Liu et al., 2020b). Moreover, the use of drugs lowering blood pressure through such mechanisms might lead to overexpression of ACE2, thus potentially increasing the susceptibility to SARS-CoV-2 in patients (Fang et al., 2020). Based on these premises, the possibility of enalkiren to modulate the renin-angiotensin system without altering patient susceptibility to the virus, together with inhibition of the SARS-CoV-2 Mpro, may exhibit beneficial effects for COVID-19 treatment. According to our docking results, enalkiren is well accommodated within the SARS-CoV-2 Mpro binding site (Figure 2, panel (C)). Interestingly, another very recent computational study based on a different workflow also identified enalkiren as a potential candidate to target the SARS-CoV-2 Mpro, further supporting its selection as a promising candidate for COVID-19 treatment (Liu et al., 2020a).
The calpain inhibitor IV (ZLLYCH2F, DB04653) is a covalent inhibitor of the calpain-1 cysteine protease (PDB ID: 1ZCM) that regulates the activity of proteins involved in processes influencing neuronal plasticity, cognition and neurodegeneration (Vosler et al., 2008) and is a potential target for intervention in inflammatory diseases (Cuzzocrea et al., 2000). In our docked structure (Figure 2, panel (D)), the reactive methylene group of the compound is in close proximity to the Cys residue of the SARS-CoV-2 Mpro active site. This result indicates that a covalent bond can potentially be formed that would provide specificity and higher affinity over other proteases. Interestingly, in 2004, calpain inhibitor IV was shown to be an active agent against SARS-CoV (Barnard et al., 2004) and its potential use against SARS-CoV-2 has also very recently been proposed in other repurposing studies (Abhithaj et al., 2020). Moreover, other calpain inhibitors have recently been tested for inhibition of the SARS-CoV-2 Mpro and among them the calpain inhibitor XII was shown to be the most active with an IC50 of 0.45 μM (Ma et al., 2020). Importantly, calpain inhibitor IV is reported to also act as an inhibitor of cathepsin L (Angliker et al., 1992), which is a necessary factor for the SARS-CoV-2 entry into host cells (Ou et al., 2020). Based on these premises, calpain inhibitor IV represents a valuable multi-target candidate for further clinical investigation to treat COVID-19.
Ethylsulfonamide-D-Trp-Gln-p-aminobenzamidine (DB04758) was designed to potently inhibit factor VIIa (FVIIa), which forms a complex with tissue factor (TF) to initiate the extrinsic coagulation cascade (Kadono et al., 2005). Compared with other anti-thrombotic agents, the specific targeting of the extrinsic coagulation provides a decreased risk of bleeding. SARS-CoV-2 infection often has dramatic consequences for the circulatory system (Tang et al., 2020b), with preliminary reports including thrombocytopenia, elevated d-dimer levels, prolonged prothrombin time, and disseminated intravascular coagulation (Han et al., 2020). Interestingly, research findings suggest that inhibition of TF-FVIIa complex may reduce the cytokine storm responsible for the increased coagulation and multi-organ failure, and thus the mortality rate in COVID-19 patients (Geisbert et al., 2003; Eslamifar et al., 2020). Our in silico findings suggested that DB04758 could also bind with high affinity to the SARS-CoV-2 Mpro. Therefore, this molecule might exhibit a dual activity against two crucial aspects of the SARS-CoV-2 infection.
Z-LY-CMK (DB07571) is a covalent inhibitor of the ATP-dependent Clp protease proteolytic subunit (ClpP), an enzyme that has recently gained attention as a promising drug target for antibiotic development (Bhandari et al., 2018). If confirmed, the potential of this compound to act as both an antimicrobial agent and a SARS-CoV-2 inhibitor would be particularly useful to treat secondary bacterial infections, potentially affecting COVID-19 patients. Moreover, a structurally similar compound (Z LVG CHN2 in the original paper), previously shown to inhibit the herpes simplex virus cysteine protease (Björck et al., 1990), has recently been tested on Vero E6 cells infected with SARS-CoV-2. Its antiviral activity (EC50 = 0.19 µM) was suggested to be due to the inhibition of the SARS-CoV-2 Mpro (Riva et al., 2020).
The thrombin inhibitor BM51.1011 (DB07934) is another candidate with a promising multi-target activity profile. Indeed, this compound has been originally studied as a low molecular weight inhibitor of thrombin, a class of therapeutics with anticoagulant effects (Engh et al., 1996). The structure of BM51.1011 in complex with the original target (thrombin) is reported in PDB ID: 1UVS. Notably, thrombotic complications and, in particular, coagulopathies appear to be an important issue in patients affected by COVID-19 (Han et al., 2020). Moreover, reduced levels of antithrombin, a protein involved in the regulation of the coagulation cascade, have been reported in COVID-19 patients (Arachchillage et al., 2020). Interestingly, according to recent findings, treatment with Argatroban, a direct thrombin inhibitor with anticoagulant effects independent from antithrombin, seems to provide significant therapeutic effects in patients (Arachchillage et al., 2020; Sagardia and Daniels, 2020), albeit this drug was shown to be unable to inhibit SARS-CoV-2 replication (NIH8). As the pandemic is spreading, the reported coagulation disorders in COVID-19 patients as in previous SARS and MERS patients should be carefully addressed. Based on these premises, the possibility of BM51.1011 to exert anticoagulant and Mpro inhibitory activity, if confirmed, would certainly provide a valuable therapeutic advantage for COVID-19 patients.
Delparantag (formerly known as PMX-60056, DB12955) is a top scoring candidate for all the scoring functions. This molecule reverses the anticoagulation effects of heparin by binding to the pentasaccharide group of unfractionated heparin (UFH) and low-molecular-weight heparins (LMWH) (Kuziej et al., 2010). Heparin has gained increasing attention for its ability to prevent blood coagulation in COVID-19 patients affected by severe pneumonia and its concomitant anti-inflammatory effects that result in reduced IL-6 levels (Tang et al., 2020a). However, recent studies have shown that patients with COVID-19 can rapidly develop severe or critical vascular diseases, which may result in venous thromboembolism and bleeding status (Xu et al., 2020). As a close monitoring of venous thromboembolism and bleeding risks is essential in such patients, delparantag may be an effective tool to mitigate bleeding risks while eliciting antiviral activity due to the potential inhibition of the SARS-CoV-2 Mpro. To the best of our knowledge, this compound has never been investigated in repurposing campaigns.
A unique feature of our repurposing strategy involves the inclusion of drug metabolites among screened compounds. Although they are typically discarded in drug repurposing studies, major metabolites can provide extremely interesting results. For this reason, here we describe some of the top scoring metabolites from our screening. Remarkably, many of these metabolites were shown to bind to the active site of the SARS-Cov-2 Mpro with higher affinity than their parent drug.
Ritonavir and lopinavir, two HIV-1 protease inhibitors that are administered in combination, are currently being clinically investigated against COVID-19 (Cao et al., 2020). Unfortunately, recent research findings showed that lopinavir or its combination with ritonavir are not able to significantly inhibit the SARS-CoV-2 Mpro up to a concentration of 100 µg/m (Jang et al., 2020). However, they were shown to be slightly active on other SARS-CoV-2 related targets, as reported in the COVID-19 open-data portal of the National Institutes of Health (NIH9; NIH10). Based on our in silico analyses, neither of these drugs appears to interact favourably with the SARS-CoV-2 Mpro, although the N-desmethyl metabolite M7 of ritonavir (DBMET00084) was demonstrated to better interact with the protein owing to its free urea group that establishes favourable hydrogen bonds with the SARS-CoV-2 Mpro.
The saquinavir decahydroisoquinoline metabolites M2 (DBMET01550 and DBMET01549) and t-butyl hydroxyl M10 (DBMET01548) were predicted to strongly bind to the SARS-CoV-2 Mpro active site according to the docking scores. Interestingly, while saquinavir metabolites are inactive against the HIV-1 protease (Noble and Faulds, 1996), they appear to bind to the SARS-CoV-2 Mpro binding site with a more favourable predicted binding energy than saquinavir. This finding may be relevant for the pharmacokinetics and dosing of this antiviral for the treatment of COVID-19. Indeed, saquinavir was shown to be inactive against the Mpro, as recently reported in the COVID-19 open-data portal of the National Institutes of Health (NIH11). However, it was identified as a putative inhibitor of the spike glycoprotein (Ramírez-Salinas et al., 2020), and lately confirmed as active (NIH11). If experimentally confirmed, the potential activity of saquinavir metabolites on both the spike glycoprotein and the Mpro would result in beneficial effects on COVID-19 patients.
The goal of the present study was to perform a systematic drug repurposing screening of compounds from the DrugBank database for their ability to bind to the SARS-CoV-2 main protease. The performed analyses allowed the selection of 22 drugs or experimental compounds, which, to the best of our knowledge, can be considered “novel”, as they had not yet been included in COVID-19 clinical trials at the time of manuscript submission. Notably, several DrugBank compounds already reported as active against the SARS-CoV-2 Mpro scored among the top-ranked ligands, suggesting that the implemented protocol is able to correctly identify active ligands. Moreover, retrospective analyses were also performed on an additional set of known SARS-CoV-2 Mpro inhibitors, not included in the screened database, thus allowing to further validate the in silico workflow. Interestingly, many of the identified candidates could be able to combine a potential antiviral activity with other activities, e.g., antithrombotic or anti-inflammatory activity, due to their activity on the primary target and therapeutic indication. Such “polypharmacological” behavior, if confirmed, would make the identified candidate drugs extremely attractive to be further evaluated for COVID-19 treatment. Indeed, a single molecule would be able to concurrently exert an antiviral activity and mitigate or abolish COVID-19 comorbidities, the severity of which often leads to patient death. Therefore, in this study the importance of these repurposed molecules is also discussed from a polypharmacologic perspective. Furthermore, we identified a number of drug metabolites that appeared to be stronger binders of the SARS-CoV-2 Mpro than the parent drugs (e.g., saquinavir and ritonavir metabolites). To the best of our knowledge, drug repurposing based on drug (major) metabolites represents a novel approach that may offer additional and valuable opportunities to repurpose candidate drugs through the modulation of in vivo pharmacokinetics. In addition, the in-depth analysis of currently available literature data, including cell-based results on SARS-CoV-2 infected cells, further substantiate the relevance of the potential Mpro inhibitors selected in this work. Given the drastic need for therapeutic options for COVID-19, our results can suggest some key drugs for repurposing.
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author.
LP performed all calculations. All authors participated in data analysis, compound selection and manuscript editing. GR conceived and coordinated the study, and wrote the initial draft.
This work was supported by an Investigator Grant from the Associazione Italiana per la Ricerca sul Cancro (AIRC IG 23635) to GR, by a FIRC-AIRC fellowship (AIRC Rif. 24096) to LP, and a Ph.D. fellowship from the Regione Emilia Romagna on Data driven technologies for drug repurposing to AT.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fphar.2021.636989/full#supplementary-material.
1World Health Organization (2020) Coronavirus disease (COVID-19) pandemic. https://www.who.int/emergencies/diseases/novel-coronavirus-2019 [Accessed November 23, 2020].
2Pfizer Inc. (2020) Pfizer and Biontech to submit emergency use authorization request today to the U.S. FDA for Covid-19 vaccine. https://www.pfizer.com/news/press-release/press-release-detail/pfizer-and-biontech-submit-emergency-use-authorization [Accessed November 23, 2020].
3EMA (2020) https://www.ema.europa.eu/en/news/ema-recommends-first-covid-19-vaccine-authorisation-eu [Accessed December 21, 2020].
4Clinicaltrials.gov (2020) COVID-19. https://clinicaltrials.gov/ct2/results?cond=COVID-19&term=&cntry=&state=&city=&dist= [Accessed November 6, 2020].
5Clinicaltrials.gov (2020) Remdesivir. https://www.clinicaltrials.gov/ct2/results?term=remdesivir&cond=covid-19&Search=Apply&recrs=b&recrs=a&recrs=f&recrs=d&age_v=&gndr=&type=&rslt= [Accessed November 25, 2020].
6World Health Organization (2020) Coronavirus disease (COVID-19): Hydroxychloroquine. https://www.who.int/news-room/q-a-detail/coronavirus-disease-covid-19-hydroxychloroquine [Accessed November 6, 2020].
7National Institutes of Health (2020) COVID-19 - OpenData Portal. https://opendata.ncats.nih.gov/covid19/databrowser [Accessed December 21, 2020].
8National Institutes of Health (2020) COVID-19 - OpenData Portal – Argatroban. https://opendata.ncats.nih.gov/covid19/databrowser?q=argatroban [Accessed December 22, 2020].
9National Institutes of Health (2020) COVID-19 - OpenData Portal – Ritonavir. https://opendata.ncats.nih.gov/covid19/databrowser?q=Ritonavir [Accessed December 22, 2020].
10National Institutes of Health (2020) COVID-19 - OpenData Portal – Lopinavir. https://opendata.ncats.nih.gov/covid19/databrowser?q=lopinavir [Accessed December 22, 2020].
11National Institutes of Health (2020) COVID-19 - OpenData Portal – Saquinavir. https://opendata.ncats.nih.gov/covid19/databrowser?q=saquinavir [Accessed December 22, 2020].
Abhithaj, J., Dileep, F., Sharanya, C. S., Arun, K. G., Sadasivan, C., and Variyar Jayadevi, E. (2020). Repurposing simeprevir, calpain inhibitor IV and a cathepsin F inhibitor against SARS-CoV-2 and insights into their interactions with Mpro. J. Biomol. Struct. Dyn. 1, 23–35. doi:10.1080/07391102.2020.1813200
Angliker, H., Anagli, J., and Shaw, E. (1992). Inactivation of calpain by peptidyl fluoromethyl ketones. J. Med. Chem. 35, 216–220. doi:10.1021/jm00080a003
Anighoro, A., Bajorath, J., and Rastelli, G. (2014). Polypharmacology: challenges and opportunities in drug discovery. J. Med. Chem. 57, 7874–7887. doi:10.1021/jm5006463
Arachchillage, D. J., Remmington, C., Rosenberg, A., Xu, T., Passariello, M., Hall, D., et al. (2020). Anticoagulation with argatroban in patients with acute antithrombin deficiency in severe COVID-19. Br. J. Haematol. 190, e286–e288. doi:10.1111/bjh.16927
Asojo, O. A., Afonina, E., Gulnik, S. V., Yu, B., Erickson, J. W., Randad, R., et al. (2002). Structures of Ser205 mutant plasmepsin II from Plasmodium falciparum at 1.8 Å in complex with the inhibitors rs367 and rs370. Acta Crystallogr. Sect. D Biol. Crystallogr. 58, 2001–2008. doi:10.1107/S0907444902014695
Asojo, O. A., Gulnik, S. V., Afonina, E., Yu, B., Ellman, J. A., Haque, T. S., et al. (2003). Novel uncomplexed and complexed structures of plasmepsin II, an aspartic protease from Plasmodium falciparum. J. Mol. Biol. 327, 173–181. doi:10.1016/S0022-2836(03)00036-6
Baig, A. M., Khaleeq, A., Ali, U., and Syeda, H. (2020). Evidence of the COVID-19 virus targeting the CNS: tissue distribution, host–virus interaction, and proposed neurotropic mechanisms. ACS Chem. Neurosci. 11, 995–998. doi:10.1021/acschemneuro.0c00122
Barnard, D. L., Hubbard, V. D., Burton, J., Smee, D. F., Morrey, J. D., Otto, M. J., et al. (2004). Inhibition of severe acute respiratory syndrome-associated coronavirus (SARSCoV) by calpain inhibitors and beta-D-N4-hydroxycytidine. Antivir. Chem. Chemother. JAMA Cardiol. 5 (7), 15–22. doi:10.1177/095632020401500102.Bavishi10.1001/jamacardio.2020.1282
Bavishi, C., Maddox, T. M., and Messerli, F. H. (2020). Coronavirus disease 2019 (COVID-19) infection andrenin angiotensin system blockers. JAMA Cardiol. 5(7), 745–747. doi:10.1001/jamacardio.2020.1282
Beigel, J. H., Tomashek, K. M., Dodd, L. E., Mehta, A. K., Zingman, B. S., Kalil, A. C., et al. (2020). Remdesivir for the treatment of Covid-19—final report. N. Engl. J. Med. 383, 1813–1826. doi:10.1056/NEJMoa2007764
Bhandari, V., Wong, K. S., Zhou, J. L., Mabanglo, M. F., Batey, R. A., and Houry, W. A. (2018). The role of ClpP protease in bacterial pathogenesis and human diseases. ACS Chem. Biol. 13, 1413–1425. doi:10.1021/acschembio.8b00124
Bhaskaran, K., Rentsch, C. T., MacKenna, B., Schultze, A., Mehrkar, A., Bates, C. J., et al. (2020). HIV infection and COVID-19 death: a population-based cohort analysis of UK primary care data and linked national death registrations within the OpenSAFELY platform. Lancet HIV 14, 33–39. doi:10.1016/S2352-3018(20)30305-2
Björck, L., Grubb, A., and Kjellén, L. (1990). Cystatin C, a human proteinase inhibitor, blocks replication of herpes simplex virus. J. Virol. 64, 941–943. doi:10.1128/JVI.64.2.941-943.1990
Broccatelli, F., and Brown, N. (2014). Best of both worlds: on the complementarity of ligand-based and structure-based virtual screening. J. Chem. Inf. Model. 54, 1634–1641. doi:10.1021/ci5001604
Cao, B., Wang, Y., Wen, D., Liu, W., Wang, J., Fan, G., et al. (2020). A trial of lopinavir–ritonavir in adults hospitalized with severe Covid-19. N. Engl. J. Med. 382, 1787–1799. doi:10.1056/NEJMoa2001282
Cazzulo, J. J., Cazzulo Franke, M. C., Martínez, J., and Franke de Cazzulo, B. M. (1990). Some kinetic properties of a cysteine proteinase (cruzipain) from Trypanosoma cruzi. Biochim. Biophys. Acta (BBA) Protein Struct. Mol. 1037, 186–191. doi:10.1016/0167-4838(90)90166-D
Corsello, S. M., Bittker, J. A., Liu, Z., Gould, J., McCarren, P., Hirschman, J. E., et al. (2017). The Drug Repurposing Hub: a next-generation drug library and information resource. Nat. Med. 23, 405–408. doi:10.1038/nm.4306
Cuzzocrea, S., McDonald, M. C., Mazzon, E., Siriwardena, D., Serraino, I., Dugo, L., et al. (2000). Calpain inhibitor I reduces the development of acute and chronic inflammation. Am. J. Pathol. 157, 2065–2079. doi:10.1016/S0002-9440(10)64845-6
Dai, W., Zhang, B., Jiang, X.-M., Su, H., Li, J., Zhao, Y., et al. (2020). Structure-based design of antiviral drug candidates targeting the SARS-CoV-2 main protease. Science 368, 1331–1335. doi:10.1126/science.abb4489
Di Gennaro, F., Marotta, C., Locantore, P., Pizzol, D., and Putoto, G. (2020). Malaria and COVID-19: common and different findings. Trav. Med. Infect. Dis. 5, 141. doi:10.3390/tropicalmed5030141
Engh, R. A., Brandstetter, H., Sucher, G., Eichinger, A., Baumann, U., Bode, W., et al. (1996). Enzyme flexibility, solvent and “weak” interactions characterize thrombin-ligand interactions: implications for drug design. Structure 4, 1353–1362. doi:10.1016/s0969-2126(96)00142-6
Eslamifar, Z., Behzadifard, M., Soleimani, M., and Behzadifard, S. (2020). Coagulation abnormalities in SARS-CoV-2 infection: overexpression tissue factor. Thromb. J. 18, 38. doi:10.1186/s12959-020-00250-x
Fang, L., Karakiulakis, G., and Roth, M. (2020). Are patients with hypertension and diabetes mellitus at increased risk for COVID-19 infection? Lancet Respir. Med. 8, e21. doi:10.1016/S2213-2600(20)30116-8
Friesner, R. A., Banks, J. L., Murphy, R. B., Halgren, T. A., Klicic, J. J., Mainz, D. T., et al. (2004). Glide: a new approach for rapid, accurate docking and scoring. 1. Method and assessment of docking accuracy. J. Med. Chem. 47, 1739–1749. doi:10.1021/jm0306430
Fu, L., Ye, F., Feng, Y., Yu, F., Wang, Q., Wu, Y., et al. (2020). Both Boceprevir and GC376 efficaciously inhibit SARS-CoV-2 by targeting its main protease. Nat. Commun. 11, 4417. doi:10.1038/s41467-020-18233-x
Gaulton, A., Hersey, A., Nowotka, M., Bento, A. P., Chambers, J., Mendez, D., et al. (2017). The ChEMBL database in 2017. Nucleic Acids Res. 45, D945–D954. doi:10.1093/nar/gkw1074
Geisbert, T. W., Hensley, L. E., Jahrling, P. B., Larsen, T., Geisbert, J. B., Paragas, J., et al. (2003). Treatment of Ebola virus infection with a recombinant inhibitor of factor VIIa/tissue factor: a study in rhesus monkeys. Lancet 362, 1953–1958. doi:10.1016/S0140-6736(03)15012-X
Gendrot, M., Andreani, J., Boxberger, M., Jardot, P., Fonta, I., Le Bideau, M., et al. (2020). Antimalarial drugs inhibit the replication of SARS-CoV-2: an in vitro evaluation. Trav. Med. Infect. Dis. 37, 101873. doi:10.1016/j.tmaid.2020.101873
Ghosh, A. K., Devasamudram, T., Hong, L., Dezutter, C., Xu, X., Weerasena, V., et al. (2005). Structure-based design of cycloamide-urethane-derived novel inhibitors of human brain memapsin 2 (β-secretase). Bioorg. Med. Chem. Lett 15, 15–20. doi:10.1016/j.bmcl.2004.10.084
Glassman, H. N., Kleinert, H. D., Boger, R. S., Moyse, D. M., Griffiths, A. N., and Luther, R. R. (1990). Clinical pharmacology of enalkiren, a novel, dipeptide renin inhibitor. J. Cardiovasc. Pharmacol. 16, S76–S81. doi:10.1097/00005344-199016004-00016
Glide, Schrödinger, LLC, New York, NY (2020). Schrödinger Release 2020-1: Glide. New York, NY: Schrödinger, LLC.
Goetz, D. H., Choe, Y., Hansell, E., Chen, Y. T., McDowell, M., Jonsson, C. B., et al. (2007). Substrate specificity profiling and identification of a new class of inhibitor for the major protease of the SARS Coronavirus. Biochemistry 46, 8744–8752. doi:10.1021/bi0621415
Han, H., Yang, L., Liu, R., Liu, F., Wu, K. L., Li, J., et al. (2020). Prominent changes in blood coagulation of patients with SARS-CoV-2 infection. Clin. Chem. Lab. Med. 58, 1116–1120. doi:10.1515/cclm-2020-0188
RECOVERY Collaborative Group, Horby, P., Lim, W., Emberson, J., Mafham, M., Bell, J., et al. (2020a). Dexamethasone in hospitalized patients with Covid-19—preliminary report. N. Engl. J. Med. 11, 31–49. doi:10.1056/NEJMoa2021436
RECOVERY Collaborative Group, Horby, P., Mafham, M., Linsell, L., Bell, J. L., Staplin, N., et al. (2020b). Effect of hydroxychloroquine in hospitalized patients with Covid-19. N. Engl. J. Med. 7, 133–137. doi:10.1056/NEJMoa2022926
Janes, J., Young, M. E., Chen, E., Rogers, N. H., Burgstaller-Muehlbacher, S., Hughes, L. D., et al. (2018). The ReFRAME library as a comprehensive drug repurposing library and its application to the treatment of cryptosporidiosis. Proc. Natl. Acad. Sci. U.S.A. 115, 10750. doi:10.1073/pnas.1810137115
Jang, M., Park, Y.-I., Park, R., Cha, Y.-E., Namkoong, S., Lee, J. I., et al. (2020). Lopinavir-ritonavir is not an effective inhibitor of the main protease activity of SARS-CoV-2 in vitro. Biorxiv [Preprint] Available at: https://www.biorxiv.org/content/10.1101/2020.09.16.299800v1 (Accessed: November 6, 2020).
Jin, Z., Du, X., Xu, Y., Deng, Y., Liu, M., Zhao, Y., et al. (2020). Structure of Mpro from SARS-CoV-2 and discovery of its inhibitors. Nature 582, 289–293. doi:10.1038/s41586-020-2223-y
Kadono, S., Sakamoto, A., Kikuchi, Y., Oh-eda, M., Yabuta, N., Yoshihashi, K., et al. (2005). Structure-based design of P3 moieties in the peptide mimetic factor VIIa inhibitor. Biochem. Biophys. Res. Commun. 327, 589–596. doi:10.1016/j.bbrc.2004.12.042
Keppel Hesselink, J. M. (2017). Kappa receptors agonist in postoperative pain. J. Anesth. Intensive Care Med. 3 (4), 555620. doi:10.19080/jaicm.2017.03.555620
Kim, S., Chen, J., Cheng, T., Gindulyte, A., He, J., He, S., et al. (2019). PubChem 2019 update: improved access to chemical data. Nucleic Acids Res. 47, D1102–D1109. doi:10.1093/nar/gky1033
Kuziej, J., Litinas, E., Hoppensteadt, D. A., Liu, Dahui, Walenga, J. M., Fareed, J., et al. (2010). In vivo neutralization of unfractionated heparin and low-molecular-weight heparin by a novel salicylamide derivative. Clin. Appl. Thromb. Hemost. 16, 377–386. doi:10.1177/1076029610366439
LigPrep, Schrödinger, LLC, New York, NY (2020). Schrödinger Release 2020-1. New York, NY: LigPrep, Schrödinger, LLC.
Liu, H., Jiang, T., Liu, W., and Zheng, Z. (2020a). Computational evaluation of the COVID-19 3c-like protease inhibition mechanism, and drug repurposing screening. Chemrxiv [Preprint]. Available at: https://chemrxiv.org/articles/Computational_Evaluation_of_the_COVID-19_3c-like_Protease_Inhibition_Mechanism_and_Drug_Repurposing_Screening/12090426/1 (Accessed: November 6, 2020).
Liu, Y., Yang, Y., Zhang, C., Huang, F., Wang, F., Yuan, J., et al. (2020b). Clinical and biochemical indexes from 2019-nCoV infected patients linked to viral loads and lung injury. Sci. China Life Sci. 63, 364–374. doi:10.1007/s11427-020-1643-8
Ma, C., Sacco, M. D., Hurst, B., Townsend, J. A., Hu, Y., Szeto, T., et al. (2020). Boceprevir, GC-376, and calpain inhibitors II, XII inhibit SARS-CoV-2 viral replication by targeting the viral main protease. Cell Res. 30, 678–692. doi:10.1038/s41422-020-0356-z
March-Vila, E., Pinzi, L., Sturm, N., Tinivella, A., Engkvist, O., Chen, H., et al. (2017). On the integration of in silico drug design methods for drug repurposing. Front. Pharmacol. 8, 298. doi:10.3389/fphar.2017.00298
Morse, J. S., Lalonde, T., Xu, S., and Liu, W. R. (2020). Learning from the past: possible urgent prevention and treatment options for severe acute respiratory infections caused by 2019‐nCoV. Chembiochem. 21, 730–738. doi:10.1002/cbic.202000047
Noble, S., and Faulds, D. (1996). Saquinavir: a review of its pharmacology and clinical potential in the management of HIV infection. Drugs 52, 93–112. doi:10.2165/00003495-199652010-00007
Ou, X., Liu, Y., Lei, X., Li, P., Mi, D., Ren, L., et al. (2020). Characterization of spike glycoprotein of SARS-CoV-2 on virus entry and its immune cross-reactivity with SARS-CoV. Nat. Commun. 11, 1620. doi:10.1038/s41467-020-15562-9
Protein Preparation Wizard, Schrödinger, LLC, New York, NY (2020). Schrödinger Release 2020-1: protein preparation Wizard. New York, NY: Schrödinger, LLC.
Ramírez-Salinas, G. L., Martínez-Archundia, M., Correa-Basurto, J., and García-Machorro, J. (2020). Repositioning of ligands that target the spike glycoprotein as potential drugs for SARS-CoV-2 in an in silico study. Molecules 25, 121–129. doi:10.3390/molecules25235615
Ran, J., Song, Y., Zhuang, Z., Han, L., Zhao, S., Cao, P., et al. (2020). Blood pressure control and adverse outcomes of COVID-19 infection in patients with concomitant hypertension in Wuhan, China. Hypertens. Res. 43, 1267–1276. doi:10.1038/s41440-020-00541-w
Rastelli, G., Degliesposti, G., Del Rio, A., and Sgobba, M. (2009). Binding estimation after refinement, a new automated procedure for the refinement and rescoring of docked ligands in virtual screening. Chem. Biol. Drug Des. 73, 283–286. doi:10.1111/j.1747-0285.2009.00780.x
Rastelli, G., and Pinzi, L. (2019). Refinement and rescoring of virtual screening results. Front. Chem. 7, 498. doi:10.3389/fchem.2019.00498
Riva, L., Yuan, S., Yin, X., Martin-Sancho, L., Matsunaga, N., Burgstaller-Muehlbacher, S., et al. (2020). A large-scale drug repositioning survey for SARS-CoV-2 antivirals. BioRxiv [Preprint] Available at: https://www.biorxiv.org/content/10.1101/2020.04.16.044016v1 (Accessed: November 6, 2020).
Robien, M. A., Nguyen, K. T., Kumar, A., Hirsh, I., Turley, S., Pei, D., et al. (2004). An improved crystal form of Plasmodium falciparum peptide deformylase. Protein Sci. 13, 1155–1163. doi:10.1110/ps.03456404
Sagardia, L. M., and Daniels, L. M. (2020). Thrombolysis and use of argatroban for the treatment of massive pulmonary embolism following anticoagulation failure in a patient with COVID-19. Am. J. Health Syst. Pharm. 77, 1961–1964. doi:10.1093/ajhp/zxaa287
Senanayake, S. L. (2020). Drug repurposing strategies for COVID-19. Futur. Drug Discov. 2, 2. doi:10.4155/fdd-2020-0010
Tang, N., Bai, H., Chen, X., Gong, J., Li, D., and Sun, Z. (2020a). Anticoagulant treatment is associated with decreased mortality in severe coronavirus disease 2019 patients with coagulopathy. J. Thromb. Haemostasis 18, 1094–1099. doi:10.1111/jth.14817
Tang, N., Li, D., Wang, X., and Sun, Z. (2020b). Abnormal coagulation parameters are associated with poor prognosis in patients with novel coronavirus pneumonia. J. Thromb. Haemostasis 18, 844–847. doi:10.1111/jth.14768
Vosler, P. S., Brennan, C. S., and Chen, J. (2008). Calpain-mediated signaling mechanisms in neuronal injury and neurodegeneration. Mol. Neurobiol. 38, 78–100. doi:10.1007/s12035-008-8036-x
Wan, Y., Shang, J., Graham, R., Baric, R. S., and Li, F. (2020). Receptor recognition by the novel coronavirus from Wuhan: an analysis based on decade-long structural studies of SARS coronavirus. J. Virol. 94, 127–147. doi:10.1128/jvi.00127-20
Weber, J., Mesters, J. R., Lepšík, M., Prejdová, J., Švec, M., Šponarová, J., et al. (2002). Unusual binding mode of an HIV-1 protease inhibitor explains its potency against multi-drug-resistant virus strains. J. Mol. Biol. 324, 739–754. doi:10.1016/S0022-2836(02)01139-7
Wishart, D. S., Feunang, Y. D., Guo, A. C., Lo, E. J., Marcu, A., Grant, J. R., et al. (2018). DrugBank 5.0: a major update to the DrugBank database for 2018. Nucleic Acids Res. 46, D1074–D1082. doi:10.1093/nar/gkx1037
Xu, J., Wang, L., Zhao, L., Li, F., Liu, J., Zhang, L., et al. (2020). Risk assessment of venous thromboembolism and bleeding in COVID-19 patients. Researchsquare [Preprint]. Available at: https://www.researchsquare.com/article/rs-18340/v1 (Accessed: November 6, 2020).
Zhou, Y., Agudelo, J., Lu, K., Goetz, D. H., Hansell, E., Chen, Y. T., et al. (2011). Inhibitors of SARS-CoV entry - identification using an internally-controlled dual envelope pseudovirion assay. Antivir. Res. 92, 187–194. doi:10.1016/j.antiviral.2011.07.016
Keywords: COVID-19, SARS-2-CoV-2, drug repurposing, polypharmacology, structure-based, molecular docking, BEAR
Citation: Pinzi L, Tinivella A, Caporuscio F and Rastelli G (2021) Drug Repurposing and Polypharmacology to Fight SARS-CoV-2 Through Inhibition of the Main Protease. Front. Pharmacol. 12:636989. doi: 10.3389/fphar.2021.636989
Received: 02 December 2020; Accepted: 12 January 2021;
Published: 22 February 2021.
Edited by:
Robert Wallace Malone, Independent Researcher, Madison, VA, United StatesReviewed by:
Arnab K. Chatterjee, Calibr at Scripps Research, United StatesCopyright © 2021 Pinzi, Tinivella, Caporuscio and Rastelli. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Giulio Rastelli, Z2l1bGlvLnJhc3RlbGxpQHVuaW1vcmUuaXQ=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.