- Molecular and Cellular Glycobiology Unit, Department of Biological Sciences, Sungkyunkhwan University, Suwon, South Korea
Severe acute respiratory syndrome–related coronavirus-2 (SARS-CoV-2), a β-coronavirus, is the cause of the recently emerged pandemic and worldwide outbreak of respiratory disease. Researchers exchange information on COVID-19 to enable collaborative searches. Although there is as yet no effective antiviral agent, like tamiflu against influenza, to block SARS-CoV-2 infection to its host cells, various candidates to mitigate or treat the disease are currently being investigated. Several drugs are being screened for the ability to block virus entry on cell surfaces and/or block intracellular replication in host cells. Vaccine development is being pursued, invoking a better elucidation of the life cycle of the virus. SARS-CoV-2 recognizes O-acetylated neuraminic acids and also several membrane proteins, such as ACE2, as the result of evolutionary switches of O-Ac SA recognition specificities. To provide information related to the current development of possible anti–SARS-COV-2 viral agents, the current review deals with the known inhibitory compounds with low molecular weight. The molecules are mainly derived from natural products of plant sources by screening or chemical synthesis via molecular simulations. Artificial intelligence–based computational simulation for drug designation and large-scale inhibitor screening have recently been performed. Structure–activity relationship of the anti–SARS-CoV-2 natural compounds is discussed.
Introduction
General Virology of Coronaviruses
The coronaviruses (CoVs) target humans and animals with exchangeable infectivity, causing a zoonotic outbreak. SARS-CoV-2 or 2019-nCoV spreads and causes the human life crisis of COVID-19 by infecting the human respiratory tract and causing pneumonia (Zhou et al., 2020). In addition, SARS-CoV-2 spreads by easy transmission among people, and COVID-19 patients exhibit flu-like symptoms such as fever and cough. Enveloped CoVs contain positive ssRNA genomes with relatively small RNAs (approximately 30 kb). They are classified into the Riboviria–Nidovirales–Cornidovirineae–Coronaviridae–Orthocoronavirinae–CoV genus (α-, β-, γ-, and δ-CoV). Most mammals are infected by α-CoV and β-CoV only, while avians and some mammals are infected by δ-CoV and γ-CoV. SARS-CoV-2, belonging to the β-CoV genus, and bat SARS-like CoV-ZXC-21 are similar in their RNA genomes. The COVID-19–causing CoV isolates exhibit 79% identity with the previously named SARS-CoV and 50% identity with the Middle East respiratory syndrome (MERS) virus (Chan et al., 2020).
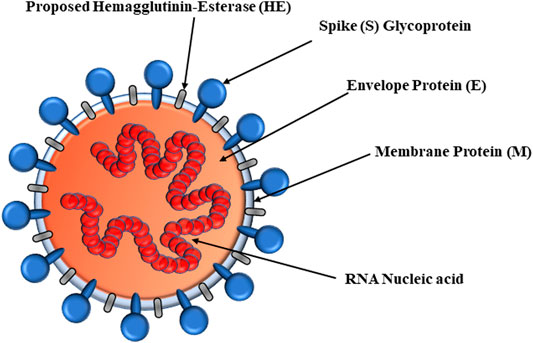
SARS-CoV-2 viral proteins include RNA-dependent RNA polymerase (RdRp) and hemagglutinin-esterase (HE) enzymes as well as proteins including spike (S), envelope (E), membrane (M), and nucleocapsid (N) (Figure 1) (Tu et al., 2020). Nonstructural protein 3 (Nsp3), Nsp5, Nsp9, and Nsp12 RdRp are enzymes. The E-, S-, and M-proteins are embedded into the endoplasmic reticulum (ER) membrane and translocated to the ER–Golgi intermediate compartment (ERGIC). As the first step, the S-glycoprotein of SARS-CoV-2 binds to surfaced O-acetyl (Ac)-neuraminic acids of host cells. The neuraminic acid-O-Ac-esterase of HE evolved from the influenza C virus, nidoviruses, and salmon anemia virus (teleost orthomyxovirus). Fusion of S-glycoprotein and HE is important for CoV attachment to neuraminic acid–bearing host receptors (Tortorici et al., 2019). Therefore, the HE found in the β-CoV genus mediates viral attachment to O-Ac-neuraminic acids. The glycoproteins of the HA, HE, S, and HA-esterase-fusion protein (HEF) bind to the host receptor. However, certain α-CoV and γ-CoV are deficient of the neuraminic acid-O-Ac-esterases but bind to Ac-neuraminic acids or glycolyl-neuraminic acids. Murine CoVs esterize the C4-O-Ac (Smits et al., 2005). The SARS-CoV-2 S-glycoprotein N-terminal domain recognizes the surface entry site, binding to 9-O-Ac-neuraminic acid in a similar manner to CoV HEs as well as influenza C and D HEFs. Bovine CoV (BCoV) and human CoV (HCoV)-OC43 can recognize the 5,9-Ac2-neuraminic acids (Vlasak et al., 1988) and bear neuraminic acid 9-O-Ac-esterase. Most β-CoVs bind to the 9-O-Ac-neuraminic acids, but mutant strains target 4-O-Ac-neuraminic acids. Specifically, the HEs of β-CoVs recognize the 9-O-Ac-neuraminic acids, although certain species bind to the 4-O-Ac forms (Kim, 2020). In fact, 9-O-Ac-neuraminic acid is the recognition site for HCoV-OC43, β1-CoV, and SARS-CoV-2 S-glycoproteins, but MERS-CoV recognizes the α2,3-linkage. The CoV glycoproteins, BCoV HEs, and influenza virus HEFs are specific for 9-O-Ac-neuraminic acid (Mani et al., 2020; McKee et al., 2020), but the influenza HA is specific for glycolylneuraminic acids (Vogel et al., 2006). Upon interaction with neuraminic acid, host furin proteases cleave the S-glycoprotein to potentiate entry into host cells (Oliveira et al., 2017).
The current global COVID-19 pandemic is threatening the daily lives of human beings. The disease biology is a topic of interest. To overcome the disease, the academic society urgently needs to exchange the pandemic CoV-controlling drugs, but no truly effective agent has yet been discovered. In this review, antiviral candidate agents and the availability of natural compounds are discussed.
Natural Products to Target and Inhibit Infection of Coronaviruses
Recently, natural phytochemicals that exhibit anti-CoV activity have been extensively summarized (Li et al., 2005). LMW molecules exhibit antiviral activity. Recently, development of anti-CoV drugs has also been applied for molecular docking via simulation approaches. Computer-based artificial intelligence technology contributes to the development of anti-CoV agents. Human angiotensin-converting enzyme (ACE)-2, papain-like protease (PLpro), main 3C-like protease (3CLpro), RdRp, helicase, N7 methyltransferase, human DDP4, receptor-binding domain (RBD), cathepsin L, type II transmembrane (TM) Ser-protease, or transmembrane protease serine (TMPRSS)-2 is mainly targeted. CoV 3CLpro and PLpro are polyprotein-specific viral proteases. RdRp is a complementary RNA strand synthetic replicase. Remdesivir inhibits RdRp in the ssRNA genome of CoV, where the RdRp mediates RNA replication and remdesivir acts as an ATP analog and thus inhibits RdRp.
Currently, effective anti-CoV agents are not available, although several drugs have been prescribed and some natural compounds exhibit antiviral activity. Natural resources are a tremendous treasure trove of chemical compounds that are applicable for various viral infections. Natural products are produced by the metabolic pathways of a given organism, but humans utilize them for their benefits from the modern view of pharmacology. Therefore, phytochemicals have been screened to test their effectiveness against viruses, and some natural products inhibit the infection and amplification of viruses with a broad antiviral spectrum (Pour et al., 2019). Naturally occurring compounds such as artemisinin, baicalin, curcumin, rutin, glycyrrhizic acid, hesperidin, hesperetin, and quercetin have been examined for their anti-CoV activity by various assay based on the viral life cycle. However, none of the natural compounds have direct antiviral activity against CoVs or other RNA viruses. Only GA has been frequently described to be the most active component in several previous articles. Indeed, the molecular action mechanisms of the natural products are not specific because current candidates of natural antiviral agents are mainly examined by using in vitro cell-based assays or computer modeling through docking simulation before application to animal and clinical studies (Li et al., 2005; Packer and Cadenas, 2011). Conventional approaches to natural products were a mix of chemical analysis and structure–function relationship analysis. Recent AI-aided approaches combined with in silico computational simulation is cost-effective for the prediction of chemical compounds (Müller et al., 2018). Currently, new concepts of AI-aided in silico computational approaches have evolved for drug prediction based on drug candidate–ligand/receptor interaction. These approaches utilize known structures of the molecules to predict in silico docking molecules. Fundamental limitations of these AI in silico approaches have also been identified. In fact, in blocking or inhibiting viral proteases, S-glycoprotein, and the entry of SARS-CoV-2, several plant compounds have been suggested through computer simulation techniques (Li et al., 2005).
Natural compounds including chemoenzymatic modified molecules can be used in ethnopharmacology to modulate SARS-CoV-2/nCoV-19 infections due to current limited therapeutic options. The efficacy of natural products depends on the CoV strains. Several natural products inhibit viral replication (Müller et al., 2020), implying antiviral properties. (Kalhori et al., 2021) For example, several compounds exhibit promising prospects for CoV treatment in human patients, as described above for lycorine, scutellarein, silvestrol, tryptanthrin, saikosaponin B2, and polyphenolic compounds such as caffeic acid, isobavachalcone, myricetin, psoralidin, and quercetin, as well as lectins such as griffithsin. For example, Lycoris radiata (L’Hér.) Herb lycorine shows cytopathogenic and antiviral activities against SARS-associated CoV (Yu et al., 2012). Currently known natural products that are pharmacologically effective for SARS-CoV-2 inhibition are shown in Figure 2, with the synthetic compounds previously utilized for other targets in humans.
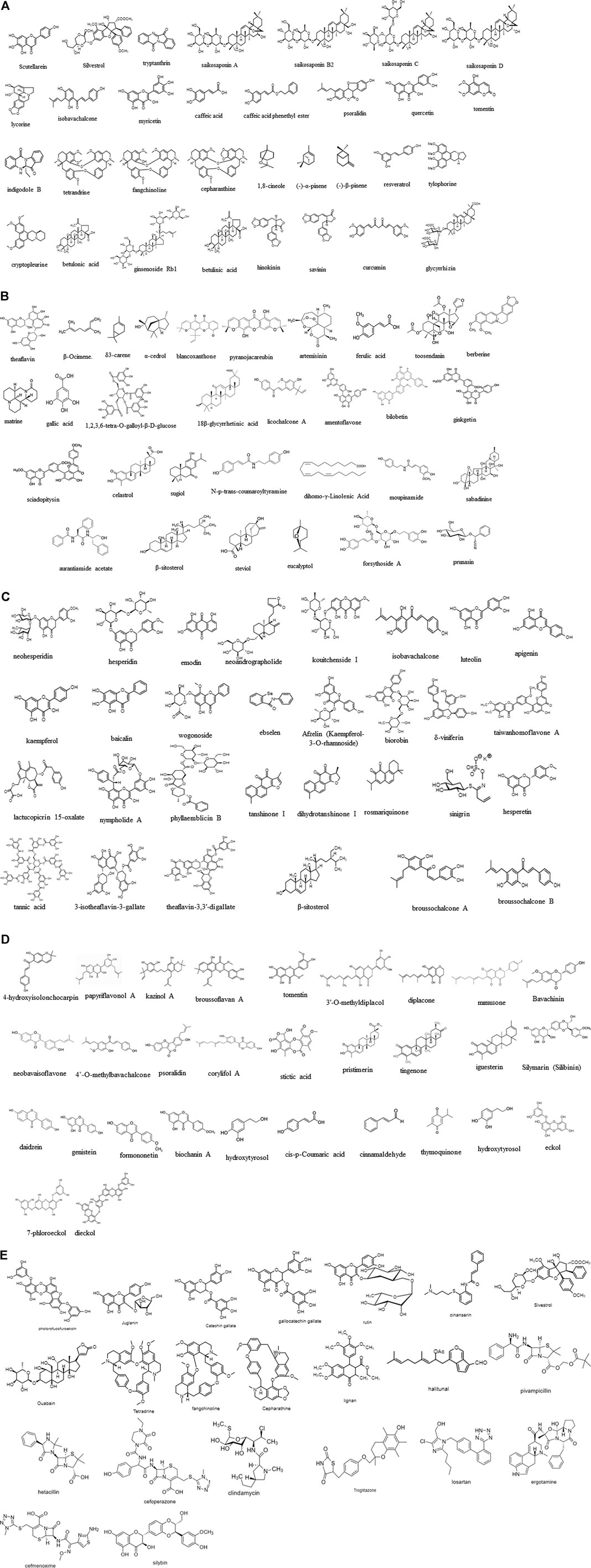
FIGURE 2. (A–E) Molecular structures experimentally effective for SARS-CoV therapeutic target. Scutellarein (CAS No: 529–53-3), silvestrol (CAS No: 697235-38-4), tryptanthrin (CAS No: 13220-57-0), saikosaponin A (CAS No: 20736-09-8), saikosaponin B2 (CAS No: 58316-41-9), saikosaponin C (CAS No: 20736-08-7), saikosaponin D (CAS No: 20874-52-6), lycorine (CAS No: 476-28-8), isobavachalcone (CAS No: 20784-50-3), myricetin (CAS No: 529-44-2), caffeic acid (CAS No: 331-39-5), caffeic acid phenethyl ester (CAS No: 104594-70-9), psoralidin (CAS No: 18642-23-4), quercetin (CAS No: 117-39-5), tomentin (CAS No: 28449-62-9), indigodole B, theaflavin (CAS No: 4,670–05-7), β-ocimene (CAS No: 3,338–55-4), δ3-carene (CAS No: 13,466–78-9), α-cedrol (CAS No: 77–53-2), blancoxanthone (PubChem CID: 11703574), pyranojacareubin (CAS No:78,343–62-1), artemisinin (CAS No: 63,968–64-9), ferulic acid (CAS No: 1,135–24-6), toosendanin (CAS No: 58,812–37-6), berberine (CAS No: 633–65-8), matrine (CAS No: 519–02-8), gallic acid (CAS No:149–91-7), 1.2,3,6-tetra-O-galloyl-β-d-glucose (CAS No: 79,886–50-3), 18β-glycyrrhetinic acid (CAS No: 471–53-4), licochalcone A (CAS No: 58,749–22-7), amentoflavone (CAS No:1,617–53-4), bilobetin (CAS No: 521–32-4), ginkgetin (CAS No: 481–46-9), sciadopitysin (CAS No: 521–34-6), celastrol (CAS No: 34,157–83-0), sugiol (CAS No: 511–05-7), N-p-trans-coumaroyltyramine (CAS No: 36,417–86-4), dihomo-γ-linolenic acid (CAS No: 1783–84-2), moupinamide (CAS No: 66,648–43-9), sabadinine (CAS No: 5,876–23-3), aurantiamide acetate (CAS No: 56,121–42-7), β-sitosterol (CAS No: 83–46-5), steviol (CAS No: 471–80-7), eucalyptol (CAS No: 470–82-6), forsythoside A (CAS No: 79,916–77-1), prunasin (CAS No: 99–18-3), tetrandrine (CAS No: 518–34-3), fangchinoline (CAS No: 436–77-1), cepharanthine (CAS No: 481–49-2), 1,8-cineole (CAS No: 470–82-6) (-)-α-pinene (CAS No: 7,785–70-8), (-)-β-pinene (CAS No: 18,172–67-3), ginsenoside Rb1 (CAS No: 41,753–43-9), resveratrol (trans-3,5,4′-trihydroxystilbene), homoharringtonine (CAS No: 501–36-0), tylophorine (CAS No: 482–20-2), cryptopleurine (CAS No: 482–22-4), betulonic acid (CAS No: 4,481–62-3), betulinic acid (CAS No: 472–15-1), hinokinin (CAS No: 26,543–89-5), savinin (CAS No: 493–95-8), curcumin (CAS No: 458–37-7), glycyrrhizin (CAS No: 1,405–86-3), neohesperidin (CAS No: 13,241–33-3), hesperidin (CAS No: 520–26-3), emodin (6-methyl-1,3,8-trihydroxyanthraquinone) (CAS No: 518–82-1), neoandrographolide (CAS No: 27,215–14-1), kouitchenside I (CAS No: 1444411-79-3), isobavachalcone (CAS No: 20,784–50-3), luteolin (CAS No: 491–70-3), 7-methylluteolin, apigenin (CAS No: 520–36-5), kaempferol (CAS No: 520–18-3), baicalin (CAS No: 491–67-8), wogonoside (CAS No: 51,059–44-0), ebselen (CAS No: 60,940–34-3), afzelin (CAS No: 482–39-3), biorobin (CAS No: 17,297–56-2), δ-viniferin (CAS No: 253,435–07-3), taiwanhomoflavone A (CAS No: 265,120–00-1), lactucopicrin 15-oxalate (CAS No: 303,130–75-8), nympholide A (CAS No: 604,004–58-2), phyllaemblicin B (CAS No: 307,504–07-0), tanshinone I (CAS No: 568–73-0), cryptotanshinone (CAS No: 35,825–57-1), dihydrotanshinone I (CAS No: 87,205–99-0), rosmariquinone (CAS No: 27,210–57-7), tannic acid (CAS No: 1,401–55-4), 3-isotheaflavin-3-gallate (CAS No: 30,462–34-1), theaflavin-3,3′-digallate (CAS No: 33,377–72-9), sinigrin (CAS No: 3,952–98-5), hesperetin (CAS No: 520–33-2), β-sitosterol (CAS No: 83–46-5), bavachalcone (CAS No: 28,448–85-30), broussochalcone A (CAS No: 99,217–68-2), broussochalcone B (CAS No: 28,448–85-3), 4-hydroxyisolonchocarpin (CAS No: 41,743–38-8), papyriflavonol A (CAS No: 363,134–28-5), 3′-(3-methylbut-2-enyl)-3′,4,7-trihydroxyflavane, kazinol A (CAS No: 99,624–28-9), kazinol B (CAS No: 99,624–27-8), kazinol F (CAS No: 104,494–35-1), kazinol J (CAS No: 104,778–05-4), broussoflavan A (CAS No: 99,217–69-3), tomentin A/B/C/D/E (CAS No: 36,034–36-3), 3′-O-methyldiplacol, 4′-O-methyldiplacol, 3′-O-methyldiplacone (CID No: 14,539,951), 4′-O-methyldiplacone (CID No: 24,854,122), mimulone (CAS No: 97,126–57-3), diplacone (CAS No: 73,676–38-7), bavachinin (CAS No: 19,879–30-2), neobavaisoflavone (CAS No: 41,060–15-5), isobavachalcone (CAS No: 20,784–50-3), 4′-O-methylbavachalcone (CAS No: 0,784–60-5), psoralidin (CAS No: 18,642–23-4), corylifol A (CAS No: 775,351–88-7), stictic acid (CAS No: 549–06-4), pristimerin (CAS No: 1,258–84-0), tingenone (CAS No: 50,802–21-6), iguesterin (CAS No: 53,527–47-2), silymarin (silibinin) (CAS No: 22,888–70-6), daidzein (CAS No: 486–66-8), genistein (CAS No: 446–72-0), formononetin (CAS No: 485–72-3), biochanin A (CAS No: 491–80-5), linolenic acid (CAS No: 463–40-1), palmitic acid (CAS No: 57–10-3), hydroxytyrosol (CAS No: 10,597–60-1), cis-p-coumaric acid (CAS No: 501–98-4), cinnamaldehyde (CAS No: 14,371–10-9), thymoquinone (CAS No: 490–91-5), hydroxytyrosol (CAS No: 10,597–60-1), eckol (CAS No: 88,798–74-7), 7-phloroeckol, dieckol (CAS No: 88,095–77-6), phlorofucofuroeckoln (CAS No: 128,129–56-6), juglanin (CAS No: 5,041–67-8), catechin gallate (CAS No: 1,257–08-5), (−)-gallocatechin gallate (CAS No: 5,127–64-0), rutin (CAS No: 153–18-4), cinanserin (CAS No: 1,166–34-3), sivestrol (CAS No: 697,235–38-4), ouabain (CAS No: 630–60-4), tetrandrine (CAS No: 518–34-3), fangchinoline (CAS No: 33,889–68-8), cepharanthine (CAS No: 481–49-2), diterpene (CAS No: 28,957–10-0), diterpene aldehyde, sesquiterpene (CAS No: 72,826–63-2), triterpene (CAS No: 125,343–14-8), lignan (CAS No: 6,549–68-4), halitunal (CAS No: 133,076–08-1), antibacterial agents such as pivampicillin (CAS No: 33,817–20-8), hetacillin (CAS No: 3,511–16-8), cefoperazone (CAS No: 62,893–19-0), clindamycin (CAS No: 18,323–44-9), antidiabetic drug troglitazone (CAS No: 97,322–87-7), antihypertensive drug losartan (CAS No: 114,798–26-4), analgesia drug ergotamine (CAS No: 113–15-5), antibacterial drug cefmenoxime (CAS No: 65,085–01-0), and hepatoprotective drug silybin (CAS No: 22,888–70-6).
Natural products that show viral inhibitory activity are promising candidates as anti-CoV agents. Natural products to combat the CoVs are reviewed in this article, focusing on the general properties of CoVs and suggesting applicable drugs and natural compounds effective against several CoV species. Viral proteins such as 3CLpro, PLpro, N, S, and ACE2 have been targeted for antiviral replication or anti-infection. Some limited antiviral agents as inhibitors specific for proteases and RNA synthases are known to block viral replication (Häkkinen et al., 1999). CoV bioactive natural products can also enhance and strengthen host immunity. Vitamins A and C lower susceptibility to infections and help in the prevention of viral infections through host immune function (Häkkinen et al., 1999).
Inhibition of Ribonucleic Acid Helicase eIF4A and Protein Expression
SARS-CoV helicase, a virus replication enzyme, is involved in the unwinding of RNA. Helicases in protein sequences are commonly conserved during evolution in CoVs and other related nidoviruses. CoV helicase is an important therapeutic target because it hydrolyzes all deoxyribonucleotide and ribonucleotide triphosphates in SARS-CoV. Therefore, SARS-CoV-2 helicase has been targeted to screen for inhibitors. The 420-amino acid–long helicase is phylogenetically homologous to the helicases of other CoVs. Favipiravir or hydroxychloroquine, described later in further detail, recognizes SARS-CoV-2 helicase with weak affinities.
Aglaia sp. silvestrol inhibits the replication of MERS types with an EC50 value of 1.3 nM, acting as an inhibitor of RNA helicase eIF4A and protein expression via blocking replication/transcription complex formation (Miean and Mohamed, 2001). Silvestrol inhibits HcoV-229E protein synthesis with an EC50 of 3 nM. Silvestrol also inhibits HCoV-229E ex vivo in bronchial epithelial cells via RNA helicase eIF4A inhibition (Lau et al., 2008). The polyphenolic compounds myricetin and scutellarein inhibit the helicase activity of SARS-CoVs. Phenolic compounds including myricetin and scutellarein of Isatis indigotica Fort. and Torreya nucifera L. inhibit SARS-CoV helicases including nsP13 helicase (Cho et al., 2013). Scutellaria baicalensis Georgi (Scutellaria radix) myricetin and scutellarein inhibit the ATPase activity of the SARS-CoV helicase Nsp-13 (Yu et al., 2012). Myricetin is enriched in fruits such as cranberry Vaccinium oxycoccos L. (Mikulic-Petkovsek et al., 2012) and in vegetables such as Calamus scipionum Lam. and garlic (Qing et al., 2016). Scutellarein from S. baicalensis is a strong inhibitor of SARS-CoV helicase because it inhibits SARS-CoV helicase Nsp13 via ATPase activity inhibition but not via direct inhibition of helicase activity. The flavonoid quercetin is structurally similar to other polyphenolics such as myricetin and scutellarein and shows similar inhibitory activity of SARS-CoV helicase (Yang et al., 2010). In addition, naturally occurring tomentins of Paulownia tomentosa (Thunb.) Steud., belonging to Scrophulariaceae, reversibly and allosterically inhibit the PLpro activity of SARS-CoV (Lung et al., 2020).
Inhibition of Ribonucleic Acid Genome Synthesis and Replication
RdRp, also known as nsp12, synthesizes a complementary RNA strand by using the original virus RNA genome as template. Inhibition of the SARS-CoV-2 RdRp enzyme is a potential therapeutic option for COVID-19 patients. Tryptanthrin inhibits viral RNA synthesis and PLpro-2 enzyme activity, important for the early, late, and post-entry step of HCoV replication. Strobilanthes cusia (Nees) Kuntze tryptanthrin, an indoloquinazoline moiety–carrying alkaloid, and the indigodole B (5aR-ethyltryptanthrin) alkaloid (Wen et al., 2007) have anti–HCoV-NL63 activity. S. cusia tryptanthrin and indigodole B also block RNA synthesis and PLpro-2 enzyme activity. Tryptanthrin is effective against SARS-CoV-2 and other HCoVs. The antiviral EC50 values were 1.52 µM for tryptanthrin and 2.60 µM for indigodole B. Tryptanthrin also has multiple pharmacological activities (Chen et al., 2008). In addition, tryptanthrin and indigodole B have direct antiviral activities to HCoV-NL63. Houttuynia cordata Thunb. aqueous extracts inhibit the enzyme activities of the viral 3CL protease and viral RdRp of SARS-CoV (Hoever et al., 2005). In computer modeling, Camellia sinensis (L.) Kuntze theaflavin (TF), 3,4,5-trihydroxy-1,8-bis [(2R,3R)-3,5,7-trihydroxy-2-chromanyl]-6-benzo (Kim, 2020) annulenone (C29H24O12), has been demonstrated to interact with the SARS-CoV-2 RdRp (Zhang et al., 2020). The TFs, known as antioxidant polyphenols, are formed from the precursor flavan-3-ols, which occur in green tea leaves, via condensation and enzymatic oxidation. Several derivatives, including TF-3′-gallate, TF-3-gallate, and TF-3–3′-di-gallate, are known. The TFs belong to the thearubigins, polymeric polyphenols, which show a red color, with a tropolone moiety.
Similarly, redwood Sequoia sempervirens (D.Don) Endl. natural phenol ferruginol compounds have various terpenoid substructures, such as betulonic acid [C30H46O3; 3-oxolup-20 (29)-en-28-oic acid; CAS No: 4481-62-3; CID 122844] and betulinic acid [C30H48O3; (3β)-3-hydroxy-lup-20 (29)-en-28-oic acid; CAS No: 472-15-1; CID 64971]; 8β-hydroxyabieta-9 (11),13-dien-12-one; 3β,12-diacetoxyabieta-6,8,11,13-tetraene; curcumin; hinokinin; and savinin. These compounds inhibit the replication of SARS-CoV (Loizzo et al., 2008). Toona sinensis (Juss.) M.Roem. extracts inhibit SARS-CoV replication (Jeong et al., 2014). Glycyrrhizin inhibits the replication of SARS-CoV after viral entrance into Vero cells, inhibiting virus attachment and entry (Shen et al., 2005). Several glycyrrhizin-derived compounds inhibit SARS-CoV replication more effectively (EC50 of 5–50 μM), but they are highly cytotoxic to Vero cells (Hoever et al., 2005). Lignin, betulinic acid, and desmethoxyreserpine inhibit viral replication as well as 3CLpro (Cheng et al., 2006). Especially desmethoxyreserpine blocks virus entry. Laurus nobilis L. and Thuja orientalis (L.) Franco essential oils are also inhibitors of viral replication. For example, L. nobilis β-ocimene (3,7-dimethyl-1,3,6-octatriene; CAS 502-99-8), 1,8-cineole (1,3,3-trimethyl-2-oxabicyclo-2.2.2-octane; CAS No: 470-82-6), α-pinene, and β-pinene, as well as T. orientalis α-pinene, δ3-carene, and α-cedrol inhibit viral replication (McDonagh et al., 2014).
β-Ocimene acts as a plant defense and antifungal agent and is derived from the plant genus Ocimum. Pinene (C10H16) is a bicyclic monoterpene. 1,8-Cineole oil, known as eucalyptol, is a cyclic ether and a monoterpenoid. Its synonyms are cajeputol; 1,8-epoxy-p-menthane; 1,8-oxido-p-menthane; and 1,3,3-trimethyl-2-oxabicyclo octane (Chan et al., 2020; Chan et al., 2020; Chan et al., 2020). a-Pinene is also a major constituent of the essential oil of Rosmarinus officinalis L (rosemary). Two enantiomers (1S,5S)- or (−)-α-pinene and (1R,5R)- or (+)-α-isomer are present. δ3-Carene or 3-carene is also a bicyclic monoterpene and has a pungent odor. α-Cedrol is a sesquiterpene alcohol and an essential oil component. It is an antioxidant with antiseptic, anti-inflammatory, anti-spasmodic, tonic, astringent, diuretic, sedative, insecticidal, and antifungal activities (Kim et al., 2008) and has been used in traditional medicine. Calophyllum blancoi Planch. & Triana pyranoxanthones such as blancoxanthone (C23H22O5; 5,10-dihydroxy-2,2-dimethyl-12-(2-methylbut-3-en-2-yl)-2H,6H-pyrano[3,2-b]xanthen-6-one) and pyranojacareubin inhibit HCoV-229E–infected host cell growth (Kim et al., 2010). The pyranoxanthones tested against HCoV-229E reverse in vitro virus-induced cytopathic effects. Blancoxanthone exhibits viral inhibition at 3 ug/ml in MRC-5 cells. Bupleurum sp., Heteromorpha sp., and Scrophularia scorodonia L. saikosaponin compounds interfere with the early replication step and entrance of HCoV-229E (Ulasli et al., 2014).
For animal CoVs such as feline CoVs, LMW molecules including artemisinin, baicalin, curcumin, quercetin, rutin, glycyrrhizic acid, hesperidin, and hesperitin inhibit replication of feline viruses (FCoVs) such as feline infectious peritonitis virus (FIPV)-1146 and FECV1683 via cytotoxicity of the virus-infected cells (O’Flaherty et al., 2019). For murine hepatitis virus (MHV) CoVs, ferulic acid, isoferulic acid, toosendanin, berberine, protoberberine alkaloids, matrine, oxymatrine, sophoranone, and sophocarpine isolated from Cimicifuga racemosa L., Melia sp., Coptis sp., Phellodendron sp., and Sophora subprostrata Chun & T. Chen. (Fabaceae) inhibit the replication of the murine hepatitis virus (MHV)-A59 strain (Song et al., 2014; Jin et al., 2019; Sun et al., 2019). Methanol extracts from the plants of Sophorae sp., Acanthopanacis sp., Sanguisorbae sp., and Torilis sp. possibly inhibit RdRp or other protease activity of MHV-A59. Nigella sativa L. and Citrus sinensis L. ethanol extract inhibits viral replication of MHV-A59 via an undetermined mechanism (Jin et al., 2019).
Artemisinin, isolated from Artemisia annua L. in 1972 by Dr. Tu Youyou, co-recipient of the 2015 Nobel Prize in Medicine, is an anti–Plasmodium falciparum malaria drug. It is a sesquiterpene lactone with an endoperoxide 1,2,4-trioxane ring, which is necessary to exert its activity. Artemisinin is a potential therapeutic candidate for certain RNA viruses (de Vries et al., 1997). Ferulic acid, as a hydroxycinnamic acid and a component of lignin, is a major metabolite of chlorogenic acid along with caffeic acid (CA) and isoferulic acid. Ferulic acid and its derivatives, including caffeoyltyramine, feruloyltyramine, and feruloyloctopamine, also inhibit SARS-CoV PLpro activity (Li, 2015). Recently, Adem et al. (2020) described that CA derivatives such as khainaosides, 6-O-caffeoyl-arbutin, and vitexfolin have been suggested to be inhibitory candidates with higher binding activities than that of nelfinavir against SARS CoV-2 S-protein as well as Nsp15 and Mpro enzymes by using molecular docking simulation via Web engines named Toxtree and www.swissadme.ch (Adem et al., 2021). Toosendanin (C₃₀H₃₈O₁₁), a triterpenoid isolated from the bark of Melia azedarach L., has analgesic, insecticidal, anti-botulinum, antimicrobial, and anti-inflammatory activities, and antiviral RNA polymerase complex activities (Simmons et al., 2013). Matrine, an alkaloid of Sophora flavescens Aiton, inhibits IL-1β expression and MyD88/NF-κB and NLRP3 inflammasome in the inflammatory response in porcine respiratory syndrome virus–infected pigs (Hulswit et al., 2019). Using a structure- and activity-based computational approach, natural products have been analyzed for the Nsp-9 (PDB ID-6W4B) enzyme inhibition of SARS-CoV-2 RNA replication and S-protein binding (Chandel et al., 2020). Baicalin exhibits binding affinity to both S-protein and Nsp9 enzyme.
Inhibition of N-Protein and S-Glycoprotein Synthesis and Replication
Modulation of S-Glycoprotein
S-glycoprotein recognizes the host cell receptor to enter the cells through endosomal fusion, after which the S-glycoprotein is cleaved, endosomal membranes released, and RNA liberated into the cytosol (Langereis et al., 2012). S-glycoprotein interacts with its receptors via its RBD and plays a role in host tropism and pathogenicity, and in proposing some therapeutic clues (Xiong et al., 2013). Therefore, modulation of the S-glycoprotein is a potential target to control SARS-CoV-2 propagation. Tetrandrine, fangchinoline, and cepharanthine as bis-benzylisoquinoline alkaloids from Stephania tetrandra var. glabra Maxim. protect cells from virus-induced cell death. In addition, they inhibit viral replication, as well as CoV S-glycoprotein and N-protein synthesis. Also, they induce the virus-induced host response by the p38MAPK pathway. Terpenoid compounds such as α- and β-pinene as well as cineole interact with the infectious bronchitis virus (IBV) N-protein to inhibit the N-protein–RNA interaction and block IBV replication. The terpenoids bind to the N-terminal active site of the N-protein. The active site is composed of five amino acid residues (Yang et al., 2011; Müller et al., 2020). These conserved amino acids in the active sites are commonly located in various IBVs. CA from Sambucus javanica subsp. chinensis Fukuoka (elderberry) extract inhibits the HCoV strain HCoV-NL63 (Weng et al., 2019). CA inhibits HCoV S-glycoprotein attachment to host cells. Chlorogenic acid and gallic acid (3,4,5-trihydroxybenzoic acid) also exhibit the same activities as CA. Gallic acid is a trihydroxybenzoic acid and forms dimeric ellagic acid. Tannins are hydrolyzed to glucose and gallic acid (gallotannin), or glucose and ellagic acid (ellagitannin). CA also inhibits the hepatitis B virus (Wang et al., 2009). Sambucus nigra L. extract (black elderberry) has been used for treating cold and flu symptoms. The adsorption, bioavailability, metabolism, and delivery mechanism of the extracts are documented for therapeutic plasma concentrations (Wittemer et al., 2005).
Four saikosaponins inhibit human CoV-229E infectivity. These saikosaponin pentacyclic triterpenoid glycoside derivatives purified from Bupleurum spp., Heteromorpha spp., and S. scorodonia L. also have anti-HIV and anti-HCoV-22E9 activities in vitro (Ushio and Abe, 1992; Chiang et al., 2003; Ulasli et al., 2014). Saikosaponins show anti-CoV activity, and saikosaponin B2 also shows the highest potency with an EC50 of 1.7 µM and inhibits the early stage of CoV viral attachment to host receptors via S-glycoprotein and penetration into the cells. The Streptomyces parvulus actinomycin D antibiotic also inhibits CoV attachment and penetration stages (O’Flaherty et al., 2019). Panax ginseng (T.Nees) C.A. Mey. ginsenoside Rb1 (gynosaponin C) as steroidal glycosides and triterpene saponins exhibit antiviral activity (Wu et al., 2004). Stephania tetrandra var. glabra bis-benzylisoquinoline alkaloid compounds such as tetrandrine, fangchinoline, and cepharanthine exhibit antiviral activity on HCoV-OC43 (Kim et al., 2019). They inhibit virus-induced cell death via blocking of virus replication and S-glycoprotein and N-protein synthesis with the host response. Resveratrol (CAS number: 501–36-0), a stilbenoid, inhibits MERS-CoV replication and infection in a cell-based system by inhibition of MERS-CoV N-protein expression and MERS-CoV–induced host cell death (Lin et al., 2017). Resveratrol inhibits SARS-CoV-2 infection.
The NIH clinical collection of 727 tested antiviral compounds showed that the alkaloid omacetaxine (homoharringtonine) shows a nonomolar IC50 level (Cao et al., 2015). Two alkaloids of Tylophora indica (Burm. f.) Merr., tylophorine and 7-methoxycryptopleurine, inhibit transmissible gastroenteritis CoV replication (Yang et al., 2010). The T. indica alkaloids tylophorine and 7-methoxycryptopleurine block replication in CoV-infected cells of swine testicular tissues (Cho et al., 2006). 7-Methoxycryptopleurine (IC50 of 20 nM) is rather more efficient than tylophorine (IC50 of 58 nM). Tylophorine also blocks virus RNA replication and NF-κB activation mediated by cellular JAK phosphorylation in CoV (Yang et al., 2017). Tylophorine and 7-methoxycryptopleurine inhibit N- and S-glycoprotein activity. Dihydrotanshinone recognizes the S-glycoprotein of SARS-CoV-2 to block its entry (Zhang et al., 2020). Rhus chinensis Mill. luteolin and tetra-O-galloyl-β-d-glucose (TGG) specifically recognize the S2 subunit and prevent viral entry of SARS-CoV (Yi et al., 2004). Luteolin also binds to the S2 protein to exert its antiviral capacity by interfering with virus–cell attachment and consequent fusion. TGG and luteolin exhibit anti–SARS-CoV activities. Therefore, LMW natural products, which bind to the SARS-CoV S-glycoprotein, can block virus infection in its host cells.
For animal CoVs such as avian IBV, Alstonia scholaris (L.) R. Br. alstotide-1 and -3 interfere with membrane proteins and S-glycoproteins but not the nucleocapsid proteins of avian IBV (Nguyen et al., 2015). These peptide-derived drugs are potentially applicable for therapeutic characteristics, although they are poor in oral bioavailability. However, alstotides are suggested to be permeable to cells, stable, and nontoxic with anti-IBV activities. Alstotide-1 interacts with the IBV M-protein during the assembly and budding of virus particles. M-protein is a glycosylated and membrane-spanning protein. The alstotide-1 and M-protein interaction implicates that alstotide-1 inhibits the assembly and budding of virus particles. Punica granatum L. polyphenols also interact with the surface S-glycoprotein of murine CoV, MHV-A59 (Sundararajan et al., 2010).
Inhibition of Interaction of S-Glycoprotein With ACE2
The β-CoV SARS-CoV recognizes ACE2 in respiratory epithelial or type I and II alveolar epithelial cells of the lung in membrane-bound and soluble forms (Alifano et al., 2020). ACE2 is a type I membrane-anchored carboxypeptidase with an N-terminal signal peptide. The host SARS-CoV-2 receptor ACE2 in the renin–angiotensin system (RAS) plays a role in lung infection through removal of the barrier. The SARS-CoV-2 S-glycoprotein recognizes ACE2. ACE2 is necessary for a virus receptor. The receptor-binding motif (RBM) recognizes human ACE2 (Li, 2015; Gheblawi et al., 2020). The α-CoV HCoV-NL63 and the lineage B β-CoV SARS-CoV S-glycoproteins are well known to bind to ACE2, but β-CoV MERS virus is not specific for the ACE2 recognition, while the α-CoV HCoV-NL63 is specific for the ACE2 recognition. Thus, the S viral protein drives the first attachment step on respiratory cell surfaces. This is a therapeutic target. Host ACE2 is the known host site for the S-glycoprotein RBD. The RBD sequence of the SARS-CoV-2 S-glycoprotein is homologous to the RBD of the SARS-CoV S-glycoprotein. ACE2 is also a SARS-CoV-2 drug target. To date, the ACE2 protein can be recognized by the antidiabetic troglitazone, antihypertensive losartan, anti-analgesic ergotamine, antibacterial cefmenoxime, and hepatic-protective silybin. Phyllanthus emblica L. phyllaemblicin G7, the genus Swertia, Citrus aurantium L. xanthones, neohesperidin, and hesperidin bind to the ACE2 protein, but not to the ACE2–S-protein RBD interface. A flavonoid hesperidin isolated from citrus peel interacts with the SARS-CoV-2 receptors (Meneguzzo et al., 2020). In molecular docking analysis, flavonoids and anthraquinones exhibit binding capacities to ACE2. Their binding sites of ACE2 protein are different from that of the viral S-protein. For example, the flavone chrysin (CID: 5281607) isolated from the medicinal plant Oroxylum indicum binds to the ACE2 protein in silico (Basu et al., 2020).
The S-glycoprotein cleavage TMPRSS2 enzyme potentiates SARS- and MERS-CoVs infections. Several antibacterial agents such as pivampicillin, hetacillin, cefoperazone, and clindamycin, and antiviral kouitchenside I potentially inhibit the TMPRSS2 enzyme (Wu et al., 2020). The anthraquinone emodin (CAS No: 518-82-1) blocks the SARS-CoV S-glycoprotein and ACE2 interaction (Ho et al., 2007). Glycyrrhizin-modified compounds such as 18β-glycyrrhetinic acid are known to be anti–SARS-CoV agents due to their cytopathogenic effects (Haiying et al., 2003; Hoever et al., 2005). 18β-Glycyrrhetinic acid is a glycyrrhizin metabolite that is converted by intestinal microbes in humans and is an inhibitor of the complement cascade. 18β-Glycyrrhetinic acid inhibits DNA polymerases and suppresses TNF-α expression. Glycoside chain modification of glycyrrhizin with 2-acetamido-β-d-glucopyranosylamine increased its antiviral activity by 10-fold, through increased interaction with the S-glycoprotein. Glycyrrhizin and its derivatives, such as 18β-glycyrrhetinic acid and licochalcone A, are constituents of Glycyrrhiza uralensis Fisch. (licorice), G. glabra L., or G. inflata Bat. (Fu et al., 2016).
In addition, 18β-glycyrrhetinic acid and licochalcone A bind to the ssRNA virus nucleoprotein (NP), a target candidate for therapeutic development, because these natural ligands influence the RNA-binding property of NP. The two agents specifically recognize the RNA-binding groove of NP (PDB code 4Z9P) and disrupt the NP–viral ssRNA interaction through a conformational shift of NP oligomers to impair ssRNA assembly. Glycyrrhizin and glycyrrhetinic acid also block SARS-CoV replication (Cinatl et al., 2003). In addition, glycyrrhizin inhibits H5N1 influenza A virus replication (Michaelis et al., 2011). Glycyrrhizin and glycyrrhetinic acid are also anti-inflammatory, antiviral, and anti-allergic agents. Licochalcone A is a natural phenolic chalconoid found in the Glycyrrhiza species and exhibits antimalarial and antiviral activities. It inhibits influenza neuraminidases (NAs) of influenza subtypes such as H1N1, H9N2, and oseltamivir-resistant novel H1N1strains (Chen et al., 1994; Dao et al., 2011).
Inhibition of NLRP3 Inflammasome Signaling
SARS-CoV protein domains modulate NLRP3 inflammasome–triggered pulmonary inflammation via chemokines. Therefore, the NLRP3 inflammasome is a potential candidate for therapeutic agents against CoV-mediated inflammatory diseases. Natural products such as flavonoids interfere with signaling mediated by the NLRP3 inflammasome. Respiratory inflammatory SARS-CoVs induce the NLRP3 inflammasome in macrophages and Th1 cells. Several flavonoids inhibit NLRP3 inflammasome–related inflammatory response to SARS-CoVs. Such flavonoids include isobavachalcone, saikosaponin B2, silvestrol, tryptanthrin, CA, quercetin, myricetin, psoralidin, scutellarein, luteolin, apigenin, kaempferol, baicalin, and wogonoside (Sun et al., 2015; Fu et al., 2016; Choe and Kim, 2017; Lim et al., 2018; Zhang et al., 2018; Chen et al., 2019; Chen et al., 2019; Yamagata et al., 2019).
Inhibition of SARS-CoV Mpro, PLpro, 3CLpro, and Related Proteases
The CoV genomes encode a polypeptide which contains a protease region. Two cysteine proteases, PLpro and 3CLpro, are directly associated with RNA virus replication. PLpro and 3CLpro cleave the viral polyprotein and produce nonstructural proteins for viral replication at the commonly conserved 11 substrate-recognition sites. Structure-based information on PLpro from SARS-CoV or other CoVs is limited. 3CLpro is also called the CoV main protease (MPro) (MW 34 kDa). Thus, Mpro is used as a target for anti-CoV drugs. Mpro controls overall RNA replication and transcription. Therefore, it is a target protease, and computational in silico simulation enables the discovery of SARS-CoV-2 Mpro-specific inhibitors (Jin et al., 2020). 3CLpro has 100% identity with other SARS-CoV genomic RNA sequences. The 3CLpro of bat and human SARS-CoV-2 exhibits 99.02% amino acid sequence homology. The SARS-CoV-2 3CLpro protein is homologous with the known SARS-CoV, HCoV, MERS-CoV, and BCoV.
Among the virus targets, CoV Mpro and ACE2 are the main targets to screen. An Mpro inhibitor N3 was designed by AI-driven drug simulation and docking. Michael acceptor inhibitor or N3 inhibits the SARS- and MERS-CoV Mpros through an inhibitory mechanism of irreversible covalent bond formation with Mpro (Jin et al., 2020). Additionally, another SARS-CoV-2 Mpro inhibitor, ebselen, was designed, with N3 and ebselen exhibiting antiviral activity against SARS-CoV-2. Also, multiple natural compounds including afzelin, biorobin, hesperidin, δ-viniferin, myricitrin, taiwanhomoflavone A, lactucopicrin 15-oxalate, nympholide A, and phyllaemblicin B recognize SARS-CoV-2 Mpro with additional binding activities to hACE-2 and RdRp (Rasool et al., 2020). In the molecular interaction of natural products with the Mpro docking pocket, Psorothamnus arborescens var. simplifolius (Parish) Barneby 5,7,3′,4′-tetrahydroxy-2’-(3,3-dimethylallyl) isoflavone (PubChem 11610052) showed a high binding affinity via rigid hydrogen bonds to the catalytic dyad amino acid residues, and binding to the RBD at the amino acid residues. Similarly, Myrica cerifera L. myricitrin (PubChem 5281673) and methyl rosmarinate showed RBD to and receptor binding affinities to SARS-CoV-2 Mpro in a stability assay of ligand–protein complex (Joshi et al., 2020). Withanolide derivatives such as withaferin isolated from Ashwagandha species and the CA derivative CA phenethyl ester (CAPE) exhibit binding capacities to Mpro enzyme. CAPE and withaferin recognize the SARS-CoV-2 Mpro SBD with equivalent potentials to the known N3 protease inhibitor, as analyzed using dynamic simulation (Kumar et al., 2020). As potential COVID-19 Mpro inhibitors, flavonoid derivatives including curcumin derivatives, apigenin derivatives, oleuropein, catechin derivatives, and kaempferol have been suggested as candidates in the docking analysis (Khaerunnisa et al., 2020). In a recent report using the docking analysis (Erlina et al., 2020), ermanin compound known as kaempferol-di-O-methyl ether, myricetin glycosides, peonidin arabinosides, quercetin rhamnosides, rhamnetin mannosyllsides, and hesperidin have also been suggested to exhibit SARS-CoV-2 protease inhibitory activities. A cyclic ether and monoterpenoid component, jensenone, which is found in eucalyptus plant oil, potentially inhibits Mpro enzyme activity (Sharma and Kaur, 2020). Similarly, phytochemicals such as Allium cepa L. oleanolic acids, Cocos nucifera L. α-tocotrienols, Psidium guajava L. asiatic acids, and Eucalyptus globulus culinosides exhibit anti–SARS-CoV-2 activity in molecular dynamic docking analysis. Oleanolic acid specifically binds to the Mpro enzyme (Fitriani et al., 2020).
The SARS-CoV PLpro cleaves junctions spanning Nsp1–Nsp4. PLpro also deubiquitinates proteins and helps the virus to evade the innate immune response (Ratia et al., 2008). Therefore, PLpro is a target for drug development against disease-associated deubiquitinating enzymes (Ghosh et al., 2010). Cinnamic amides isolated from Tribulus terrestris L. fruits inhibit PLpro activity (Song et al., 2014). Some plants including Cassia tora L., Cibotium barometz (L.) J. Sm., Dioscorea polystachya Turcz., Gentiana scabra Bunge, and Taxillus chinensis (DC.) Danser showed SARS-CoV 3CLpro enzyme activity (Wen et al., 2011). Natural compounds such as lignins, tannins, and coumarins have also been found to exhibit CoV inhibitory activities (Islam et al., 2020). The diterpenoid 8β-hydroxyabieta-9 (11),13-dien-12-one and a lignin compound savinin inhibited SARS-CoV 3CLpro activity. Salvia miltiorrhiza Bunge tanshinone I, IIA, IIB; dihydrotanshinone; methyl tanshinonate; cryptotanshinone; and rosmariquinone inhibited 3CLpro and PLpro with anti-infection and anti-replication activities, where tanshinone I and dihydrotanshinone I are strong 3CLpro and PLpro inhibitors (Park et al., 2012). The above S. miltiorrhiza Bunge tanshinone derivatives are noncompetitive inhibitors of protease enzyme isomerization. Especially, rosmariquinone reversibly inhibits the slow binding during cysteine protease isomerization (Park et al., 2012).
Black tea phenolic components such as tannic acid, 3-isotheaflavin-3-gallate, and theaflavin-3,3′-digallate also exhibited 3CLpro inhibition (Chen et al., 2005). Isatis indigotica Fortune ex Lindl. phenolic compounds including sinigrin, indigo, emodin, hesperetin, and β-sitosterol inhibited 3CLpro activity (Lin et al., 2005). Torreya nucifera (L.) Siebold & Zucc. flavones, biflavones, amentoflavone, apigenin, luteolin, bilobetin, ginkgetin, sciadopitysin, and quercetin also inhibit SARS-CoV 3CLpro (Ryu et al., 2010). Amentoflavone, bilobetin, ginkgetin, and sciadopitysin biflavonoids are constituents of Ginkgo biloba L. Other plants such as Chamaecyparis obtusa (Siebold & Zucc.) Endl. and Hypericum perforatum L. are also known to contain these compounds. They inhibit the cathepsin B inhibitor and influenza virus NA. Amentoflavone, bilobetin, ginkgetin, and sciadopitysin are noncompetitive inhibitors of CoV 3CLpro (Ryu et al., 2010). Myricetin and scutellarein inhibit SARS-CoV 3CLpro activity (Cho et al., 2013). Broussonetia papyrifera (L.) L'Hér. ex Vent. broussochalcone A/B, kazinol A/B/F/J, broussoflavan A, 4-hydroxyisolonchocarpin, papyriflavonol A, and 3′-(3-methylbut-2-enyl)-3′,4,7-trihydroxyflavane inhibit the 3CLpro and PLpro enzymes, where papyriflavonol A is the best inhibitor of the PLpro enzyme (Ryu et al., 2010). B. papyrifera 3′-(3-methylbut-2-enyl)3′,4,7-trihydroxyflavane noncompetitively inhibits PLpro activity (Park et al., 2017). However, these compounds do not inhibit PLpro of MERs-CoV, indicating strain dependence. Other polyphenolic compounds such as kazinol F and broussochalcone A of the same B. papyrifera inhibit MERS-CoV PLpro.
Paulownia tomentosa tomentins 3′-O-/4′-O-methyldiplacol, 3′-O-/4′-O-methyldiplacone, mimulone, diplacone, and 6-geranyl-4′,5,7-trihydroxy-3′,5′-dimethoxyflavone inhibit PLpro of SARS-CoV (Lung et al., 2020). Psoralea corylifolia L. polyphenolics including bavachinin, neobavaisoflavone, isobavachalcone, 4′-O-methylbavachalcone, psoralidin, and corylifol A inhibit PLpro activity. Among these, isobavachalcone and psoralidin exhibit the highest antiviral activity with reversible inhibitory activity against PLpro via a type I mechanism (Kim et al., 2014). Quercetin inhibits PLpro and 3CLpro proteases (Ryu et al., 2010). For porcine epidemic diarrhea virus (PEDV), quercetin-7-rhamnoside, a disaccharide glucoside, inhibits viral activity rather than quercetin alone. In a computer-based simulation for a protease 3CLpro inhibitor of feline CoVs, naturally occurring compounds such as 7-methylluteolin, stictic acid, and quercetin-7-rhamnoside showed inhibition. However, only stictic acid prevented virus-induced death and virus attachment to the host cells, while 7-benzyl luteolin and steviol showed no inhibitory effects (Theerawatanasirikul et al., 2020). Similarly, quercetin-7-rhamnoside exhibits higher antiviral activity against animal CoVs than quercetin (Choi et al., 2009). Psoralea corylifolia bavachinin, neobavaisoflavone, isobavachalcone, 4′-O-methylbavachalcone, psoralidin, and corylifol inhibited PLpro of SARS-CoV (Lin et al., 2005). Interestingly, psoralidin strongly inhibited the protease activity of SARS-CoV. Using an in vitro cell-based assay of Vero E6 cells, terpenoids and lignoids were shown to block 50% cell growth of Vero E6 cells infected with SARS-CoV. Betulinic acid and savinin competitively inhibit SARS-CoV 3CLpro with a Ki of 8.2 and 9.1 μM, respectively (Loizzo et al., 2008). Quinone-methide triterpenes such as celastrol (tripterine), pristimerin, tingenone, and iguesterin of Triterygium regelii Sprague & Takeda inhibit the 3CLpro activity (IC50 = 5.5, 9.9, and 2.6 μM) as competitive inhibitors (Ryu et al., 2010). Celastrol as a pentacyclic triterpenoid in quinone methides also inhibits the RNA of hepatitis C and dengue viruses (Tseng et al., 2017; Yu et al., 2017). Torreya nucifera ethanolic extract contains SARS-CoV 3CLpro inhibitors. Biflavone amentoflavone was identified as a potent 3CLpro inhibitor via molecular docking (Ryu et al., 2010). Geranylated flavonoids are the strongest inhibitors of PLpro activity (Lung et al., 2020).
Sugiol, coumaroyltyramine, N-cis-feruloyltyramine, kaempferol, quercetin, cryptotanshinone, and tanshinone IIA inhibit PLpro and 3CLpro. Dihomo-γ-linolenic acid and moupinamide (feruloyltyramine) inhibit 3CLpro and PLpro, respectively (Cheng et al., 2006). Through computer docking modeling of SARS-CoV 3CLpro, Veratrum sabadilla Retz. sabadinine inhibits CoV protease (Toney et al., 2004). Artemisia annua aurantiamide acetate inhibits the active pocket of the CoV cathepsin-L protease (Wang et al., 2007). Isatis indigotica sinigrin, indigo, β-sitosterol, aloe-emodin, and hesperetin (Islam et al., 2020) as well as Rheum palmatum L. anthraquinones inhibit 3CLpro (Luo et al., 2009). Houttuynia cordata Thunb. water extract inhibits 3CLpro protease (Yang et al., 2010). For animal CoVs, stictic acid, 7-methylluteolin, quercetin-7-rhamnoside, 7-benzyl luteolin, and steviol, which exist in plants such as lichen, inhibit the 3CLpro protease activity of FIPV1146 (FCoV) (Theerawatanasirikul et al., 2020). Uncaria tomentosa (Willd. ex Schult.) DC, known as cat’s claw, exhibits 3CLPro inhibitory activity as found by molecular docking analysis (Yepes-Pérez et al., 2020). Isolated phytochemicals such as cadambine, speciophylline, and proanthocyanidin of Uncaria tomentosa effectively interacted with 3CLpro.
As described above, CoV proteases are considered antiviral targets for the reduction of virus replication and host pathogenicity. However, nM affinity–leveled compounds are not developed for the targets. Apart from the conventional discovery from natural products, computer-aided methods to design drugs have been applied by using chemical databases for the inhibitor screening of SARS-CoV 3CLpro activity. Moreover, the known crystal structure of HCoV-229E 3CLpro facilitates design of inhibitors (Anand et al., 2003). Currently, SARS-CoV 3CLpro (PDB: 1Q2W and 1UK4) and SARS-CoV 3CLpro are elucidated for their 3D structures (Yang et al., 2003).
ADAM17 and TMPRSS2 Serine Protease Inhibitors
SARS-CoV-2 also utilizes TMPRSS2 for infection. A disintegrin and metallopeptidase domain (ADAM) family comprises Zn-metalloproteinases, and ADAM17 is a specific TNF-α–converting enzyme (TACE), thus also named TNF-α sheddase. ADAMs include ADAM-9, -10, and -12. ADAM17 sheds the ACE2 enzyme and catalyzes the formation of the soluble ACE2 N-terminal carboxypeptidase domain from ACE2 (Tipnis et al., 2000) and also converts pro-TNF-α to soluble TNF-α. Thus, ADAM17 is an anti-inflammatory target. Interaction of SARS S-glycoprotein and ACE2 cleaves ACE2 via ADAM17/TACE, facilitating its shedding and virus entry (Chen et al., 2008). TMPRSS2, human airway trypsin-like protease (HAT), TM protease, serine 13, serine protease DESC1, furin, factor Xa, and endosomal cathepsin L/B can cleave the SARS-CoV S-protein, facilitating SARS-CoV infection (Heurich et al., 2014). However, only TMPRSS2 allows SARS-CoV infection (Haga et al., 2010; Park et al., 2016; Zuniga et al., 2020). ACE2 interaction and TMPRSS2 activation potentiate the viral attachment to host cells. Thus, TMPRSS2 is a target for therapeutic agents (Figure 3).
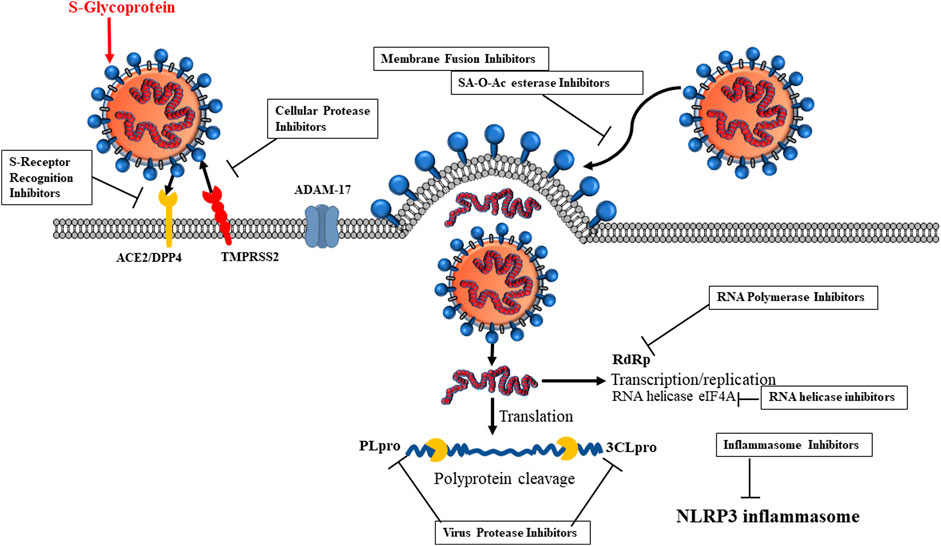
FIGURE 3. Inhibitor candidates for SARS-CoV-2 life cycle. 1) S-glycoprotein receptor recognition, 2) cellular protease, 3) virus proteases, 4) RNA polymerase, 5) inflammasome, 6) RNA helicase, 7) SA-O-Ac-esterase.
SARS-CoV S-cellular TNF-α–converting enzyme activation facilitates virus entry, and thus this enzyme is an antiviral target. The inhibitor TAPI-2 inhibits virus entry of SARS-CoV into host cells. TAPI-2 inhibits SARS S-glycoprotein–mediated ACE2 shedding and TNF-α synthesis in the lung (Haga et al., 2010). ADAM17 inhibitors are widely beneficial for various diseases related to tumor immunosurveillance, cancer, and inflammatory diseases. As described previously, ADAM17 inhibitors reduce TNF-α–induced proinflammatory diseases and are attractive target candidates for the inflammatory diseases involved in SARS-CoVs. For example, a dual and selective small molecular inhibitor of ADAM17 and ADAM10, named INCB7839, is currently under combined usage with rituximab for B-cell non-Hodgkin lymphoma therapy (Witters et al., 2008). Although ADAM17 inhibitors such as matrix metalloproteinase (MMP) inhibitors marimastat and prinomastat inhibit ADAM17 activity (Packer and Cadenas, 2011), they are not clinically applicable due to ADAM17 sequence homology with the MMP enzymes and physiological problems. In this context, naturally occurring molecules have been used to attempt to develop selective ADAM17 inhibitors by using in silico approaches toward ligands and targets. Through the binding of ADAM17 to ligands, silymarin has been purified as an ADAM17-specific inhibitor that binds to the active amino acid residues in the ADAM17 protein. The inhibiting capacity has been compared with a previously known inhibitor, IK682. Silymarin is found in Silybum marianum (L.) Gaertn., known as milk thistle; and Cynara cardunculus L., known as wild artichokes; Curcuma longa L. turmeric rhizome; and Coriandrum sativum L. coriander seeds (Borah et al., 2016).
Cryptotanshinone, a natural compound isolated from S. miltiorrhiza, modulates androgen receptor (AR) transcriptional regulation and downregulates TMPRSS2 gene expression as an AR target gene in androgen-responsive tumor cells. Interestingly, cryptotanshinone selectively inhibits the AR gene and thus has potential as anti-AR or SARS-CoV therapy (Xu et al., 2012).
Inhibition of GRP78 (HSPA5) Interaction in Silico
MERS-CoV spikes also recognize a 78-kDa glucose-regulated protein (GRP78) known as Byun1, heat shock 70-kDa protein 5 (HSPA5), and binding immunoglobulin protein (BiP). HSP5A is an ER-resident unfolded protein response (UPR) protein and acts as an alternative entry site via S-protein interaction for human viruses including papillomavirus, Ebola virus, Zika virus, and HCoVs, as well as the fungus Rhizopus oryzae (Pujhari et al., 2019; Elfiky, 2020; Elfiky, 2020; Ibrahim et al., 2020). Viral infection increases HSPA5 translocation to the plasma membrane (PM) and forms a membrane protein complex. In addition, GRP78 regulates MERS-CoV entry in the presence of DPP4. Lineage D β-CoV and bat CoV HKU also recognize the GRP78 (Ho et al., 2007; Yang et al., 2010), as simulated by molecular modeling and docking (Rao et al., 2002). Other ER molecules such as activating transcription factor 6 (ATF6), inositol-requiring enzyme 1 (IRE1), and protein kinase RNA (PKR)–like ER kinase (PERK) (Ibrahim et al., 2019) are involved. GRP78 releases IRE1, ATF6, and PERK activation, contributing to translation and refolding. GRP78 translocates to the membrane and recognizes the virus by its substrate-binding domain β (SBDβ), which is bound by the RBD. The binding region is molecularly targeted for COVID-19–specific drugs. Therefore, natural products can inhibit the HSPA5 binding to the S-glycoprotein. Small natural products prevent the S-glycoprotein–HSPA5 SBDβ interaction in silico. The effects of natural products that cause HSPA5 SDBβ dysfunction prevent SARS-CoV-2 S recognition because the HSPA5 SBDβ is the binding site for the SARS-CoV-2 S-glycoprotein. During viral infection, the HSPA5 (GRP78) translocated to the cell PM recognizes the SARS-CoV-2 S-protein. In in silico AI computer-aided simulation, several natural products recognize HSPA5 SBDβ. HSPA5 SBDβ-binding natural products can block virus attachment to the host cells if they are stressed. Thus, anti–COVID-19 agents specific for HSPA5 SBDβ recognition can be beneficial for elderly humans with cell stress. Therefore, approaches using AI computer-based molecular docking simulation yielded some natural products that bind to HSPA5 SBDβ (Elfiky, 2020). Four Cicer arietinum L. phytoestrogens, daidzein, genistein, formononetin, and biochanin A, recognize HSPA5 SBDβ. In addition, other natural compounds such as chlorogenic acid, linolenic acid, palmitic acid, CA, CA-phenethyl ester (CAPE), hydroxytyrosol, cis-p-coumaric acid, cinnamaldehyde, and thymoquinone showed moderate binding affinities to HSPA5 SBDβ. Phytoestrogens bear the same recognition affinity to HSPA5 SBDβ. Estrogenic hormones such as estrogens, progesterone, testosterone, and cholesterol have also binding affinities to HSPA5 SBDβ. From the binding affinity, phytoestrogens and estrogens are found to be the most feasible ligands to bind to HSPA5. Phytoestrogens such as daidzein, genistein, formononetin, and biochanin A also bind to estrogen receptors (ER) of humans and murines in silico and act like estrogen-like molecules (Sayed and Elfiky, 2018). Olive leaf hydroxytyrosol has moderate binding affinity to HSPA5 SBDβ. CA and p-coumaric acid also have average binding affinities to surface HSPA5 SBDβ and compete for recognition by the S-glycoprotein. The CAPE has a medium binding affinity to HSPA5 SBDβ. Cinnamaldehyde and thymoquinone have average binding affinity to HSPA5 SBDβ.
Inhibition of SARS-CoV by Plant Lectins
Plant lectins are potent inhibitors of CoV infection of host cells. Plant lectin-like protein interacts with virus surface proteins. Agglutinins, including mannose-specific lectin, inhibit the attachment and replication of SARS-CoV. Lectins can inhibit SARS-CoV-2 infection. Lectins such as griffithsin exert anti-CoV activity by multiple mechanisms (Moghaddam et al., 2014; Dai et al., 2019). Of the 33 plant lectins screened for the inhibition of SARS-CoV, L. radiata agglutinin was found to be effective (Keyaerts et al., 2007). GlcNAc-specific lectin, (GlcNAc)n-specific lectin, Gal-specific lectin, Man/Glc-specific lectin, Gal/GalNAc-specific lectin, GalNAcα(1.3)Gal > GalNAc > Gal-specific lectin, and Man/GalNAc-specific lectins inhibit the viral attachment to host cells and replication in host cells (Keyaerts et al., 2007). For example, Urtica dioica L. agglutinin inhibits viral replication in the penetration stages and binds to the S-glycoprotein and GlcNAc-like residues on the envelope glycan (Kumaki et al., 2011). Lectins from Allium porrum L., Nicotiana tabacum L., and U. dioica inhibit the virus propagation (EC50) (Yonesi and Rezazadeh, 2020).
Plant lectins are promising antiviral agents against influenza, herpes simplex virus (Hwang et al., 2020), and Ebola (Michelow et al., 2011; Covés-Datson et al., 2019). Galanthus nivalis L. agglutinin recognizes the S-glycoprotein and membrane proteins of feline CoV. Red alga Griffithsia sp. griffithsin directly interacts with S-glycoprotein (O’Keefe et al., 2010) and MERS-CoV (Millet et al., 2016). Griffithsin lectin purified from the Griffithsia sp. has three identical glycan-binding domains (GBDs) (O’Keefe et al., 2010). Different inhibition spectrums of griffithsin against different strains may be caused by genomic differences of the S-glycoproteins between SARS-CoV strains, potentiating different binding to the GBDs and affinity to the S-glycoproteins. Griffithsin lectin is relatively a small molecule and classified to be a MERS-CoV and HCoV inhibitor (EC50 of 0.0032–0.33 µM) (Millet et al., 2016). The three carbohydrate-binding domains specific for S-glycoprotein glycans inhibit MERS-CoV viral attachment to host cells (EC50 of 0.125 µM) (O’Keefe et al., 2010). Griffithsin has low toxicity and is a candidate agent against SARS-CoV-2. Griffithsin was effective for the SARS-CoV Urbani/Tor-II strains and not effective for the Frank strain. Human mannose-binding lectin (MBL) protects mice from fatal Ebola infections (Michelow et al., 2011). The legume Jack bean, Canavalia ensiformis (L.) DC., lectin concanavalin A (Con-A) is a phytagglutinin that hemagglutinates the hemagglutinating encephalomyelitis CoV, via binding to glycoconjugates (Greig and Bouillant, 1977). The therapeutic utility of Con-A is limited due to its hepatotoxic side effects. Leguminous Dioclea lasiocarpa Mart. ex Benth. lectin DLasiL inhibits feline CoV at an EC50 of 5 nM. Interestingly, Galanthus nivalis L. lectin, agglutinin, recognizes the S-glycoprotein and feline coronavirus (FCoV) NTU156 (Hsieh et al., 2010). Griffithsia sp. griffithsin blocks PEDV (NJPEDV) attachment to host cells (Li et al., 2019).
Virus Entry Inhibitors via Nonspecific Inhibition
Lycorine, emetine, berbamine, and mycophenolate mofetil are known to inhibit several CoV strains including MHV-A59, HCoV-OC43/-NL63, and MERS-CoV (Shen et al., 2019). Additionally, mycophenolate mofetil showed immunosuppressive activity on the related virus-infected cells. Similarly, marine brown alga species Ecklonia cava Kjellman eckols, 7-phloroeckol, phlorofucofuroeckoln, and dieckol, blocked virus binding to porcine epidemic cells (Kwon et al., 2013). Cinnamomum cassia (L.) J. Presl. cortex procyanidin A2/B1 and cinnamtannin B1 inhibited SARS-CoV infection (Zhuang et al., 2009). Among these, procyanidin A2 inhibits the early stage of virus entry by blocking the clathrin-dependent endocytosis pathway. As virus entry inhibitors, tetra-O-galloyl-beta-d-glucose and luteolin prevent SARS-CoV entry into host cells (Yi et al., 2004). Upon interaction with ACE2, SARS-CoVs are incorporated into vesicle forms to facilitate entry into the cells. Juglanin inhibits SARS-CoV channel 3a (Schwarz et al., 2014). (−)-Catechin gallate and (−)-gallocatechin gallate block the nanoparticle-based RNA oligomer of SARS-CoV (Roh, 2012). Houttuynia cordata Thunb. quercetin, quercetrin, rutin, and cinanserin inhibit murine CoV (Chiow et al., 2016).
Aglaia foveolata Pannell sivestrol blocks Cap-dependent translation of HCoV-229E mRNA genome (Müller et al., 2018). Ouabain reduces the viral titers, yields, and viral RNA copy numbers (Yang et al., 2018). Its carboxylic amide derivatives exhibit specific SARS-CoV antiviral activity (Kim et al., 2019). Plant alkaloids such as cepharanthine, tetrandrine, and fangchinoline protected HCoV-OC43–infected human lung MRC-5 cells from cell death (Kim et al., 2019; Majnooni et al., 2020). Cepharanthine also blocks the SARS-CoV protease enzyme (Zhang et al., 2005). Some diterpenes, sesquiterpenes, triterpenes, lignans, and curcumin also exhibited antiviral activities against SARS-CoV (Yamagata et al., 2019). The marine algae Halimeda tuna (Ulvophyceae, Chlorophyta) diterpene aldehyde, halituna, shows antiviral activity against murine CoV A59 (Koehn et al., 1991). Some compounds inhibit SARS-CoV S-protein RBD interaction with ACE2. For example, the cathepsin L inhibitor inhibits fusion of viral membrane with host cell PM, blocking virus entry (Adedeji et al., 2013). On the other hand, Aglaia sp. silvestrol specifically inhibits the RNA helicase eIF4A of MERS-CoV (Müller et al., 2018). The Boenninghausenia sessilicarpa H. Lév. bioactive coumarin, leptodactylone, exhibits cytopathogenic effects on SARS-CoV–infected cells (Yang et al., 2007). Pelargonium sidoides DC. 11% ethanol extract interferes with the virus surface and causes inactivation of respiratory viruses (Michaelis et al., 2011).
For animal CoVs, Sambucus nigra L. lectins and flavonols also disrupt virion structure, compromising virus membrane integrity of avian IBV (Chen et al., 2014). Mentha piperita L., Thymus vulgaris L., and Desmodium canadense (L.) DC. 40% ethanol extracts directly inactivate the virus envelope structure of avian IBV (Lelesius et al., 2019). Houttuynia cordata essential oils and methyl-nonyl-ketone inhibit the release of avian IBV (Yin et al., 2011). Plant eucalyptol blocks the interaction of RNA with the nucleocapsid protein of avian IBV (Yang et al., 2010), and α-/β-pinene suppresses the N-protein function, hindering the interaction of avian IBV RNA and N-protein (Yang et al., 2011). Forsythia suspensa (Thunb.) Vahl forsythoside A affects cell signaling of avian IBV-infected avian cells (Li et al., 2011). For bovine CoVs, Rosa nutkana C. Presl and Amelanchier alnifolia (Nutt.) Nutt. ex M. Roem. prunasin exhibits cytotoxicity against BCoV (McCutcheon et al., 1995). Ziziphus jujuba Mill. jubanine G and H as well as nummularine B exhibit cytotoxicity in PEDV-infected cells (Kang et al., 2015). Ginkgo biloba polysaccharides dose-dependently inhibit viral attachment and the entry steps of PEDV CoV-777 (Lee et al., 2015). Houttuynia cordata quercetin 7-rhamnoside interacts directly with PEDV (Song et al., 2011). H. cordata quercetin 7-rhamnoside, quercetin, apigenin, and luteolin exhibit cytotoxicity in PEDV-infected host cells (Choi et al., 2009). Prunus serrulata var. spontanea (Maxim.) E.H. Wilson polyphenols exhibit cytotoxicity in PEDV (KPEDV9)-infected host cells (Yook et al., 2010).
Carcinoembryonic Antigen Cell Adhesion Molecule Receptor
The N-terminal domain of S1 recognizes CEACAM1. S-glycoprotein–CEACAM receptor binding leads to S-glycoprotein–mediated fusion of membrane. For example, MHV recognizes the CEACAM expressed on BHK cell cultures (Heino et al., 2000). In MERS-CoV, CEACAM5 isoforms are associated with attachment (Naskalska et al., 2019). Therefore, MERS-CoV recognizes CEACAM5 as the attachment and entry site (Chan et al., 2016). In the structural aspect, the S1 N-terminal domain exhibits an identical tertiary structure compared with human galectins which recognize Gal-residues. The S1 N-terminal domain of the MHV recognizes mouse CEACAM1a and that of BCoV recognizes carbohydrate residues (Peng et al., 2011; Peng et al., 2012; Walls et al., 2016). Because CEACAM1a mRNA is alternatively spliced, HCoVs have been suggested to be evolutionarily recombinant between the host galectin and the S1-glycoprotein genes. However, the BCoV S1-glycoprotein gene is not subjected to such recombination but bears the glycan-binding lectin activity. MHV S1-glycoprotein has also been suggested to acquire mouse CEACAM1a-binding capacity (Peng et al., 2017), suggesting that CoVs receive evolutionary pressure to acquire the interaction capacity with host receptors over cross-species (Li, 2015; Li, 2016). Moreover, soluble forms of CEACAM directly involve in S-glycoprotein–mediated PM fusion, inducing conformational shifts (Matsuyama and Taguchi, 2002; Taguchi and Matsuyama, 2002). On the host side, host organisms have also evolved to escape the lethal pressure from coronavirus infections. The acquired geno- and phenotypes of such hosts are expressed for SA-recognizing proteins. For example, Siglecs are representatively expressed to utilize the innate responses of host immune cells.
Major Histocompatibility Complex Class I (MHC-I) C and DC-SIGN (CD209) for Coronavirus Attachment Site
CoV-HKU1 spikes additionally bind to MHC-I C (Chan et al., 2009). HCoV-HKU1 S-glycoprotein also binds to MHC-IC known as HLA-C (Song et al., 2011). SARS-CoV utilizes dendritic cell (DC)–specific intercellular adhesion molecule (ICAM)-3–grabbing nonintegrin (DC-SIGN) (Marzi et al., 2004). SARS-CoV also uses the C-type lectins of DC-SIGN and DC-L-SIGN. DC/L-SIGN recognizes the S-glycoprotein glycans, where seven N-glycan sites are known to enable DC/L-SIGN–mediated infection (Marzi et al., 2004; Han et al., 2007).
Dipeptidyl Peptidase-4, Aminopeptidase N, and Tetraspanin CD9
Ser exopeptidase dipeptidyl peptidase-4 (DPP-4)/human CD26 is the MERS-CoV receptor. DPP4 is a ubiquitous membrane-type aminopeptidase in the PM. The MERS-CoV S1 N-terminal domain binds to DPP4 (Raj et al., 2013; Gheblawi et al., 2020; Letko et al., 2020). The CD9 tetraspanin, but not the CD81 tetraspanin, interacts with DPP4 and TMPRSS2 (Earnest et al., 2017). These CD9–DPP4–TMPRSS2 receptors and proteases permit entrance of the MERS-CoV pseudovirus into the host cells. Tetraspanin CD9 binds to the DPP4–TMPRSS2 complex, and this triggers the S-glycoprotein. The α-CoV HCoV-229E S-glycoprotein binds to human aminopeptidase N (hAPN) (Yeager et al., 1992). hAPN (CD13) is a TM alanyl aminopeptidase and Zn-dependent metalloprotease (EC 3.4.11.2) with a MW of 150 kDa and made up of 967 amino acids. The C-terminal domain has zinc–MMP-related pentapeptides. The S1 C-terminal region is the APN-binding domain (Deng et al., 2016). Porcine APN (pAPN) and hAPN exhibit about 80% protein similarity. PEDV can bind to hAPN and neuraminic acid as its co-receptors, as human cytomegalovirus, pCoV, FIPV, feline enteric virus (FeCV), and canine CoVs recognize them (Delmas et al., 1992; Söderberg et al., 1993; Tresnan et al., 1996; Nomura et al., 2004). APN is the functional receptor for HCoV-229E (Yeager et al., 1992; Zhu et al., 2018). Bestatin, an APN inhibitor, binds to its catalytic site (Milewska et al., 2014).
Heparan Sulfate as Human Coronavirus Entry Site
MHV and HCoV-NL63 are known to interact with heparan sulfate (HS) (Watanabe et al., 2007; Milewska et al., 2014). The HS proteoglycans (HSPGs) are recognized by M-protein in the absence of the S-glycoprotein in the HCoV-NL63 entry into host cells. Then, the M-protein and S-glycoprotein enhance virus entry into the host cells (Milewska et al., 2014; Naskalska et al., 2019). In general, ACE2, APN, HSPA5, furin, O-Ac-neuraminic acid, and HSPGs are the CoV-binding molecules. Apart from the precise targeting of the molecules, several medicinal plant resources also exhibit antiviral activities against respiratory and influenza virus diseases. For example, Panax ginseng can prevent viral respiratory diseases and influenza virus diseases (Cheng et al., 2020). Pelargonium sidoides also prevents respiratory viral infections (Im et al., 2015). Astragalus mongholicus Bunge can treat common cold and upper respiratory infections and also prevent influenza virus infections (Kolodziej, 2011; Liang et al., 2019). Compounds and extracts with anti-CoV activities are summarized in Table 1.
Relationship Between Structures and Activities of Natural Products
The anti–SARS-CoV-2 natural compounds have been subjected to screening for understanding their structure–activity relationships (SARs). A possible approach to understand the SARs and inhibitory mechanism(s) is to resolve the inhibitor–target complex by using analytic tools. For example, crystallized complexes of the natural products and target proteins such as enzymes, surface proteins, and host receptors can be instrumentally analyzed. However, information on the successful SARs and the inhibitory mechanism(s) are currently limited. Instead, using molecular in silico ducking simulation and computational analysis, the SAR results have been reported. Using molecular modeling and docking techniques, potential binding abilities of the compounds to the pocket sites, interface sites, or catalytic sites of targets including proteases and the ACE2–S-glycoprotein complex have been suggested. The functional groups of the binding pocket interact with targets in van der Waals, hydrophilic, hydrophobic, and H-bond interactions.
As regards natural anthraquinones, rings and substituted glycosides differentially inhibit SARS-CoV-2 targets (Li and Jiang, 2018). For example, dihydroxyanthraquinone with C1 and C2−OH groups differently inhibit SARS-CoV-2 infection (Li and Jiang, 2018). Anthocyanins interact with the active site pockets of Mpro and human ACE2, where the active site of Mpro is polar in its chemical property, having affordable binding energies. Delphinidin, an anthocyanin derivative, forms H-bond in the binding site and π stacking with the hydrophobic pocket. A diglycosidic anthocyanin, delphinidin 3,5-diglucoside binds to the Mpro and ACE2 (Sharma and Shanavas, 2020), where it recognizes the flavylium nucleus ring and the Mpro catalytic site. In addition, the −OH groups of the phenyl ring recognize the Mpro S1 catalytic site through H-bonds (Sharma and Shanavas, 2020). The benzene ring and the Hie41 of the hydrophobic Mpro S2 domain form the p–π interaction. The −OH group of the flavylium nucleus binds to the Mpro S4, while the −OH groups of the benzoyl moiety of non-glycosidic 3,5-di-O-galloylshikimic acid form H-bonds with the Mpro cavity site. The OH- group and oxygen atoms of the benzoyl groups bind to the Mpro cavity site (Sharma and Shanavas, 2020). In contrast, for ACE2, oxygen of the COOH of 5-di-O-galloylshikimic acid binds to the Mpro cavity site via H-bonds, and non-covalent and ionic interactions. The −OH groups of the benzoyl moiety form H-bonds with the Mpro cavity site, where the side chain groups bind to benzoyl rings via the p–π stacking interaction. Therefore, the −OH groups are crucial for the SAR.
Flavones such as apigenin and quercetin inhibit 3CLpro activity, which coincides with the enzyme-inhibitory data. The 3CLpro inhibitory potential of biflavone with apigenin residue at the flavone C-3ʹ is enhanced, indicating that the 3CLpro inhibitory activity is upregulated by the additional apigenin residue at C-3′. In fact, the biflavonoid amentoflavone inhibits the 3CLpro activity. Quercetin recognizes the S-glycoprotein–ACE2 interface site (Smith and Smith, 2020; Williamson and Kerimi, 2020). A quercetin derivative, avicularin (Fukunaga et al., 1989) has also the Mpro-binding affinity. A similar scutellarein glucoside has the Mpro- and ACE2-binding affinities, where the −OH groups of glycoside form the H-bonds with the Mpro catalytic site. Another −OH group of the phenyl ring also forms H-bond with the Mpro. The phenyl ring also forms the π–π stacking interaction. The carbonyl oxygen and −OH group of the chromone nucleus form the H-bonds (Sharma and Shanavas, 2020). Similar to delphinidin diglucoside and scutellarein glucoside, l-arabinoside of avicularin binds to the catalytic site through H-bonds. The benzene ring involves in the π–π stacking with the hydrophobic subsite. Also, the −OH group of the chromone nucleus and benzene ring recognize the active site through H-bonds. The −OH groups of the arabinoside and phenyl ring recognize Mpro domain 1. Multiple π–π stacking interactions are formed between the chromone nucleus and the Mpro domain. The carbonyl group of the main nucleus forms the H-bonds with the Mpro. Likely, a flavanone glycoside, hesperidin, forms multiple H-bonds with the Mpro.
The flavonoid myricetin binds to both nsp13 and anti-3CLpro (Ananda Silva et al., 2020) as well as the TMPRSS2 active pocket through the 3 H-bonds, van der Waals forces, and π-anion (Pooja et al., 2021). Similarly, baicalein interacts with TMPRSS2 via 3 H-bonds, and van der Waals and π-stacking interactions. Aesculitannin B (Pooja et al., 2021) and proanthocyanidin bind to the TMPRSS2 active site via 5 H-bonds, and van der Waals and amide–π stacking interaction. Hydrocinnamic caffeic acid and ferulic acid recognize the Mpro active site via the H-bonds. Caffeic acid forms the H-bonds with both E- and N-proteins (Bhowmik et al., 2020). A bioflavonoid rutin also forms H-bonds with M- and N-proteins. Theaflavin interacts with the catalytic pocket groove near the RdRp active site through H-bonds and π–cation interaction, resulting in low docking score (Lung et al., 2020). Membrane binding of the alkyl gallates depends on alkyl chain lengths (Stefaniu et al., 2020) by high polarity–triggered reactivity. Therefore, the position of the −OH groups on the benzoic acid ring seems to be essential, compared with the number or type of ester, −OH, and methoxy groups.
For the SAR of glycyrrhizin and glycyrrhetinic acid, the free −OH (C-3), carbonyl (C-11), and COOH (C-30) groups influence the antiviral activity, while esterification of the −OH group on C-3 or the COOH group on C-30 decreases the activity. In addition, the dual esterification in the C-3 and C-30 decreases the activity, while substitution of the C-30 increases the activity (Wang et al., 2012). Betulonic acid, a triterpenoid, has an anti–SARS-CoV activity through the ketoxime backbone (Kazakova et al., 2011). The betulonic acid has a −OH and a COOH with a double bond at position C-20, 3-OH and 28-COOH groups (Regueiro-Ren et al., 2018), and C-3 and C-17 positions are crucial for the activity. Polyphenolic tannins show different binding capacities to the 3CLpro due to their SAR activities. The tannins recognize the receptor-binding spot and putative catalytic dyad of the 3CLpro. Tannic acid is a specific polyphenolic form of tannin with weak acidic properties due to the grouped phenols, where −OH groups, ketone groups (=O), and phenolic rings involve in binding to the 3CLpro through H-bonds and other forces (Khalifa et al., 2020). For example, hydrolyzable tannins including pedunculagin directly recognize the catalytic dyads and 3CLpro receptor–binding site with 5 H-bonds. Similarly, castalin and tercatain recognize the 3CLpro receptor–binding site via H-bonds and arene–arene interactions, influencing the catalytic dyad residues. Other hydrolyzable tannins including punicalin and isoterchebin secondarily recognize the catalytic dyad residues of the 3CLpro. Thymoquinone also interacts with the catalytic site of the 3CLpro via multiple H-bonds and π–H interactions (Kadil. et al., 2020). The −OH and carbonyl groups interact with the targets via H-bonds. For example, the −OH group binds to the Mpro, Nsp15, and S-glycoprotein (Kodchakorn et al., 2020).
Conclusion
The COVID-19 outbreak is a global pandemic health problem. The SARS-CoV-2 RNA sequence has been known to be highly homologous with those of the CoVs. For the present crisis of pandemic SARS-CoV-2 infections, therapeutic and preventive approaches are simultaneously required to overcome the current life-threatening disease. Because development of blockers and inhibitors of viral entry and replication is urgent, computational AI has been incorporated to accelerate drug designation. Natural resources contain promising ligands for the development of therapeutic targets. Naturally occurring compounds are potentially promising resources for their antiviral properties. SARS-targeting agents can be effective against related CoV strains due to their similar life cycles. LMW compounds can be generated, discovered, and simulated with AI assistance for target molecules. Chemical derivative modification of known structures by AI-based technologies can enhance such drug activities. Thus, natural products may be useful for use in medical therapy of SARS-CoV-2 infections.
Author Contributions
Conceptualization, C-HK; writing-draft preparation, C-HK; review and editing, C-HK.
Funding
This study has been in part supported by a grant (NRF-2018R1D1A1A09081927 to C-HK) of the Basic Science Research Program through the National Research Foundation (NRF) funded by the Ministry of Education, Science and Technology (MEST) and the Korea Institute of Planning and Evaluation for Technology in Food, Agriculture and Forestry (IPET 119010032HD050 derived from the IPET No.119010032CG000) during 2019–2021.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fphar.2021.590509/full#supplementary-material.
Glossary
COVID-19 coronavirus disease 2019
SARS-CoV severe acute respiratory syndrome-related coronavirus
MERS-V Middle East respiratory syndrome virus
HCoV human CoV
HE hemagglutinin-esterase
RdRp RNA-dependent RNA polymerase
E envelope
N nucleocapsid
S spike glycoprotein
M membrane matrix glycoprotein
ER endoplasmic reticulum
ERGIC endoplasmic reticulum–Golgi intermediate compartment
Nsp nonstructural protein
TM transmembrane
PLpro Papain-like protease
RBD receptor-binding domain
ssRNA single-stranded RNA
NA neuraminidase
Neu5Ac N-acetylneuraminic acid
Neu5,9Ac2 9-O-acetyl-N-acetylneuraminic acid
Neu5Ac9NAc 9-acetamido-9-deoxy-N-acetylneuraminic acid
HEF HE fusion protein
BCoV Bovine CoV
MHV murine hepatitis virus
CEACAM carcinoembryonic antigen cell adhesion molecule
9-O-Ac-SA 9-O-acetylated SA
IBV infectious bronchitis virus
pAPN porcine aminopeptidase N
GRP78 membrane-associated 78-kDa glucose-regulated protein
ACE2 angiotensin-converting enzyme 2
DPP4 dipeptidyl peptidase-4 dipeptidyl peptidase-4
hAPN human APN
MHC-I major histocompatibility complex class I
DC-SIGN dendritic cell–specific intercellular adhesion molecule-3-grabbing nonintegrin
HSPG heparan sulfate proteoglycan
HSPA5 heat shock protein A5
RAS renin–angiotensin system
ADAM A disintegrin and metallopeptidase domain
TACE TNF-α–converting enzyme
TMPRSS transmembrane protease serine
HAT human airway trypsin-like protease
MSPL serine 13
DPP4 dipeptidyl peptidase-4 dipeptidyl peptidase-4
BiP immunoglobulin protein
UPR unfolded protein response
ATF6 activating transcription factor 6
IRE1 inositol-requiring enzyme 1
PERK protein kinase RNA (PKR)–like ER kinase
SBD substrate-binding domain
PEDV porcine epidemic diarrhea coronavirus virus
FIPV feline infectious peritonitis virus
FeCV feline enteric virus
GAG glycosaminoglycan
RBM receptor-binding motif
3CLpro 3-chymotrypsin–like protease
References
Adedeji, A. O., Severson, W., Jonsson, C., Singh, K., Weiss, S. R., and Sarafianos, S. G. (2013). Novel Inhibitors of Severe Acute Respiratory Syndrome Coronavirus Entry that Act by Three Distinct Mechanisms. J. Virol. 87, 8017–8028. doi:10.1128/jvi.00998-13
Adem, Ş., Eyupoglu, V., Sarfraz, I., Rasul, A., Zahoor, A. F., Ali, M., et al. (2021). Caffeic Acid Derivatives (CAFDs) as Inhibitors of SARS-CoV-2: CAFDs-Based Functional Foods as a Potential Alternative Approach to Combat COVID-19. Phytomedicine. 85, 153310. doi:10.1016/j.phymed.2020.153310
Alifano, M., Alifano, P., Forgez, P., and Iannelli, A. (2020). Renin-angiotensin System at the Heart of COVID-19 Pandemic, Biochimie, 174, 30, 33. doi:10.1016/j.biochi.2020.04.008
Anand, K., Ziebuhr, J., Wadhwani, P., Mesters, J. R., and Hilgenfeld, R. (2003). Coronavirus Main Proteinase (3CLpro) Structure: Basis for Design of Anti-SARS Drugs. Science 300(5626), 1763–1767. doi:10.1126/science.1085658
Ananda Silva, A. A., Wiedemann, L. S. M., and Veiga-Junior, V. F. (2020). Natural Products' Role against COVID-19. RSC Adv. 10(39), 23379–23393. doi:10.1039/D0RA03774E
Basu, A., Sarkar, A., and Maulik, U. (2020). Molecular Docking Study of Potential Phytochemicals and Their Effects on the Complex of SARS-CoV2 Spike Protein and Human ACE2. Sci. Rep. 10(1), 17699. doi:10.1038/s41598-020-74715-4
Bhowmik, D., Nandi, R., Jagadeesan, R., Kumar, N., Prakash, A., and Kumar, D. (2020).Identification of Potential Inhibitors against SARS-CoV-2 by Targeting Proteins Responsible for Envelope Formation and Virion Assembly Using Docking Based Virtual Screening, and Pharmacokinetics Approaches. Infect. Genet. Evol. 84, 104451. doi:10.1016/j.meegid.2020.104451
Borah, P. K., Chakraborty, S., Jha, A. N., Rajkhowa, S., and Duary, R. K. (2016). In Silico approaches and Proportional Odds Model towards Identifying Selective ADAM17 Inhibitors from Anti-inflammatory Natural Molecules. J. Mol. Graphics Model. 70, 129–139. doi:10.1016/j.jmgm.2016.10.003
Cao, J., Forrest, J. C., and Zhang, X. (2015). A Screen of the NIH Clinical Collection Small Molecule Library Identifies Potential Anti-coronavirus Drugs. Antiviral Res. 114, 1–10. doi:10.1016/j.antiviral.2014.11.010
Chan, C.-M., Chu, H., Wang, Y., Wong, B. H.-Y., Zhao, X., Zhou, J., et al. (2016). Carcinoembryonic Antigen-Related Cell Adhesion Molecule 5 Is an Important Surface Attachment Factor that Facilitates Entry of Middle East Respiratory Syndrome Coronavirus. J. Virol. 90, 9114–9127. doi:10.1128/jvi.01133-16
Chan, C. M., Lau, S. K. P., Woo, P. C. Y., Tse, H., Zheng, B.-J., Chen, L., et al. (2009). Identification of Major Histocompatibility Complex Class I C Molecule as an Attachment Factor that Facilitates Coronavirus HKU1 Spike-Mediated Infection. Jvi 83, 1026–1035. doi:10.1128/jvi.01387-08
Chan, J. F.-W., Kok, K.-H., Zhu, Z., Chu, H., To, K. K.-W., Yuan, S., et al. (2020). Genomic Characterization of the 2019 Novel Human-Pathogenic Coronavirus Isolated from a Patient with Atypical Pneumonia after Visiting Wuhan. Emerging Microbes & Infections. 9(1), 221–236. doi:10.1080/22221751.2020.1719902
Chandel, V., Sharma, P. P., Raj, S., Choudhari, R., Rathi, B., and Kumar, D. (2020). Structure-based Drug Repurposing for Targeting Nsp9 Replicase and Spike Proteins of Severe Acute Respiratory Syndrome Coronavirus 2. J. Biomol. Struct. Dyn. 38, 1–14. doi:10.1080/07391102.2020.1811773
Chen, C.-J., Michaelis, M., Hsu, H.-K., Tsai, C.-C., Yang, K. D., Wu, Y.-C., et al. (2008). Toona Sinensis Roem Tender Leaf Extract Inhibits SARS Coronavirus Replication. J. Ethnopharmacology 120, 108–111. doi:10.1016/j.jep.2008.07.048
Chen, C.-N., Lin, C. P. C., Huang, K.-K., Chen, W.-C., Hsieh, H.-P., Liang, P.-H., et al. (2005). Inhibition of SARS-CoV 3C-like Protease Activity by Theaflavin-3,3'-Digallate (TF3). Evidence-Based Complement. Altern. Med. 2(2), 209–215. doi:10.1093/ecam/neh081
Chen, C., Zuckerman, D. M., Brantley, S., Sharpe, M., Childress, K., Hoiczyk, E., et al. (2014). Sambucus Nigra Extracts Inhibit Infectious Bronchitis Virus at an Early Point during Replication. BMC Vet. Res. 10, 24. doi:10.1186/1746-6148-10-24
Chen, H., Lin, H., Xie, S., Huang, B., Qian, Y., Chen, K., et al. 2019). Myricetin Inhibits NLRP3 Inflammasome Activation via Reduction of ROS-dependent Ubiquitination of ASC and Promotion of ROS-independent NLRP3 Ubiquitination. Toxicol. Appl. Pharmacol. 365, 19–29. doi:10.1016/j.taap.2018.12.019
Chen, I.-Y., Moriyama, M., Chang, M.-F., and Ichinohe, T. (2019). Severe Acute Respiratory Syndrome Coronavirus Viroporin 3a Activates the NLRP3 Inflammasome. Front. Microbiol. 10(50), doi:10.3389/fmicb.2019.00050
Chen, M., Theander, T. G., Christensen, S. B., Hviid, L., Zhai, L., and Kharazmi, A. (1994). Licochalcone A, a New Antimalarial Agent, Inhibits In Vitro Growth of the Human Malaria Parasite Plasmodium Falciparum and Protects Mice from P. Yoelii Infection. Antimicrob. Agents Chemother. 38 (7), 1470–1475. doi:10.1128/aac.38.7.1470
Cheng, L., Zheng, W., Li, M., Huang, J., Bao, S., Xu, Q., et al. (2020). Citrus Fruits Are Rich in Flavonoids for Immunoregulation and Potential Targeting ACE2. Preprints 2020020313
Cheng, P.-W., Ng, L.-T., Chiang, L.-C., and Lin, C.-C. (2006). Antiviral Effects of Saikosaponins on Human Coronavirus 229E In Vitro. Clin. Exp. Pharmacol. Physiol. 33(7), 612–616. doi:10.1111/j.1440-1681.2006.04415.x
Chiang, L. C., Ng, L. T., Liu, L. T., Shieh, D. E., and Lin, C. C. (2003). Cytotoxicity and Anti-hepatitis B Virus Activities of Saikosaponins from Bupleurum Species. Planta Med. 69(08), 705–709. doi:10.1055/s-2003-42797
Chiow, K. H., Phoon, M. C., Putti, T., Tan, B. K. H., and Chow, V. T. (2016). Evaluation of Antiviral Activities of Houttuynia Cordata Thunb. Extract, Quercetin, Quercetrin and Cinanserin on Murine Coronavirus and Dengue Virus Infection. Asian Pac. J. Trop. Med. 9(1), 1–7. doi:10.1016/j.apjtm.2015.12.002
Cho, J. H., Bernard, D. L., Sidwell, R. W., Kern, E. R., and Chu, C. K. (2006). Synthesis of Cyclopentenyl Carbocyclic Nucleosides as Potential Antiviral Agents against Orthopoxviruses and SARS. J. Med. Chem. 49(3), 1140–1148. doi:10.1021/jm0509750
Cho, J. K., Curtis-Long, M. J., Lee, K. H., Kim, D. W., Ryu, H. W., Yuk, H. J., et al. (2013). Geranylated Flavonoids Displaying SARS-CoV Papain-like Protease Inhibition from the Fruits of Paulownia Tomentosa. Bioorg. Med. Chem. 21(11), 3051–3057. doi:10.1016/j.bmc.2013.03.027
Choe, J.-Y., and Kim, S.-K. (2017). Quercetin and Ascorbic Acid Suppress Fructose-Induced NLRP3 Inflammasome Activation by Blocking Intracellular Shuttling of TXNIP in Human Macrophage Cell Lines. Inflammation 40(3), 980–994. doi:10.1007/s10753-017-0542-4
Choi, H.-J., Kim, J.-H., Lee, C.-H., Ahn, Y.-J., Song, J.-H., Baek, S.-H., et al. (2009). Antiviral Activity of Quercetin 7-rhamnoside against Porcine Epidemic Diarrhea Virus. Antiviral Res. 81(1), 77–81. doi:10.1016/j.antiviral.2008.10.002
Cinatl, J., Morgenstern, B., Bauer, G., Chandra, P., Rabenau, H., and Doerr, H. (2003). Glycyrrhizin, an Active Component of Liquorice Roots, and Replication of SARS-Associated Coronavirus. The Lancet 361, 2045–2046. doi:10.1016/s0140-6736(03)13615-x
Covés-Datson, E. M., Dyall, J., DeWald, L. E., King, S. R., Dube, D., Legendre, M., et al. (2019). Inhibition of Ebola Virus by a Molecularly Engineered Banana Lectin. Plos Negl. Trop. Dis. 13(7), e0007595. doi:10.1371/journal.pntd.0007595
Dai, W., Bi, J., Li, F., Wang, S., Huang, X., Meng, X., et al. 2019). Antiviral Efficacy of Flavonoids against Enterovirus 71 Infection In Vitro and in Newborn Mice. Viruses 11(7), 625. doi:10.3390/v11070625
Dao, T. T., Nguyen, P. H., Lee, H. S., Kim, E., Park, J., Lim, S. I., et al. (2011). "Chalcones as Novel Influenza A (H1N1) Neuraminidase Inhibitors from Glycyrrhiza Inflata". Bioorg. Med. Chem. Lett. 21 (1), 294–298. doi:10.1016/j.bmcl.2010.11.016
de Vries, A. A. F., Horzinek, M. C., Rottier, P. J. M., and de Groot, R. J. (1997). The Genome Organization of the Nidovirales: Similarities and Differences between Arteri-, Toro-, and Coronaviruses. Semin. Virol. 8, 33–47. doi:10.1006/smvy.1997.0104
Delmas, B., Gelfi, J., L'Haridon, R., Vogel, L. K., Sjöström, H., Norén, O., et al. (1992). Aminopeptidase N Is a Major Receptor for the Enteropathogenic Coronavirus TGEV. Nature 357(6377), 417–420. doi:10.1038/357417a0
Deng, F., Ye, G., Liu, Q., Navid, M., Zhong, X., Li, Y., et al. (2016). Identification and Comparison of Receptor Binding Characteristics of the Spike Protein of Two Porcine Epidemic Diarrhea Virus Strains. Viruses 8(3):55. doi:10.3390/v8030055
Earnest, J. T., Hantak, M. P., Li, K., McCray, P. B., Perlman, S., and Gallagher, T. (2017). The Tetraspanin CD9 Facilitates MERS-Coronavirus Entry by Scaffolding Host Cell Receptors and Proteases. Plos Pathog. 13(7), e1006546. doi:10.1371/journal.ppat.1006546
Elfiky, A. A. (2020). Ebola Virus Glycoprotein GP1-Host Cell-Surface HSPA5 Binding Site Prediction. Cell Stress and Chaperones. 25(3), 541–548. doi:10.1007/s12192-020-01106-z
Elfiky, A. A. (2020). Natural Products May Interfere with SARS-CoV-2 Attachment to the Host Cell. J. Biomol. Struct. Dyn. 1, 1–10. doi:10.1080/07391102.2020.1761881
Erlina, L., Paramita, R. I., Kusuma, W. A., Fadilah, F., Tedjo, A., Pratomo, I. P., et al. (2020). Virtual Screening on Indonesian Herbal Compounds as COVID-19 Supportive Therapy: Machine Learning and Pharmacophore Modeling Approaches. BMC Med. Inform. Decis. Making. doi:10.21203/rs.3.rs-29119/v1
Fitriani, I. N., Utami, W., Zikri, A. T., and Santoso, P. (2020), In Silico Approach of Potential Phytochemical Inhibitor from Moringa Oleifera, Cocos Nucifera, Allium cepa, Psidium Guajava, and Eucalyptus Globulus for the Treatment of COVID-19 by Molecular Docking. doi:10.21203/rs.3.rs-42747/v1
Fu, S., Xu, L., Li, S., Qiu, Y., Liu, Y., and Wu, Z. (2016). Baicalin Suppresses NLRP3 Inflammasome and Nuclear Factor-Kappa B (NF-Κb) Signaling during Haemophilus Parasuis Infection. Vet. Res. 47(1), 80. doi:10.1186/s13567-016-0359-4
Fu, X., Wang, Z., Li, L., Dong, S., Li, Z., Jiang, Z., et al. (2016). Novel Chemical Ligands to Ebola Virus and Marburg Virus Nucleoproteins Identified by Combining Affinity Mass Spectrometry and Metabolomics Approaches. Sci. Rep. 6, 29680. doi:10.1038/srep29680
Fukunaga, T., Nishiya, K., Kajikawa, I., Takeya, K., and Itokawa, H. (1989). Studies on the Constituents of Japanese Mistletoes from Different Host Trees, and Their Antimicrobial and Hypotensive Properties. Chem. Pharm. Bull. Bulletin, 37, 1543–1546. doi:10.1248/cpb.37.1543
Gheblawi, M., Wang, K., Viveiros, A., Nguyen, Q., Zhong, J.-C., Turner, A. J., et al. (2020). Angiotensin-Converting Enzyme 2: SARS-CoV-2 Receptor and Regulator of the Renin-Angiotensin System. Circ. Res. 126(10), 1456–1474. doi:10.1161/circresaha.120.317015
Ghosh, A. K., Takayama, J., Rao, K. V., Ratia, K., Chaudhuri, R., Mulhearn, D. C., et al. (2010). Severe Acute Respiratory Syndrome Coronavirus Papain-like Novel Protease Inhibitors: Design, Synthesis, Protein−Ligand X-Ray Structure and Biological Evaluation. J. Med. Chem., 53, 4968–4979. doi:10.1021/jm1004489
Greig, A. S., and Bouillant, A. M. (1977). Binding Effects of Concanavalin A on a Coronavirus. Can. J. Comp. Med. 41(1), 122–126.
Haga, S., Nagata, N., Okamura, T., Yamamoto, N., Sata, T., Yamamoto, N., et al. (2010). TACE Antagonists Blocking ACE2 Shedding Caused by the Spike Protein of SARS-CoV Are Candidate Antiviral Compounds. Antiviral Res. 85, 551–555. doi:10.1016/j.antiviral.2009.12.001
Haiying, L., Na, H., and Xiaoyuan, X. (2003). The Curative Effects of Glycyrrhizin on Patients with SARS. Annual Meeting of The Society of Infectious and Parasitic Diseases, Chinese Medical Association, Wuhan, China, pp. 18–22.
Häkkinen, S. H., Kärenlampi, S. O., Heinonen, I. M., Mykkänen, H. M., and Törrönen, A. R. (1999). Content of the Flavonols Quercetin, Myricetin, and Kaempferol in 25 Edible Berries. J. Agric. Food Chem. 47(6), 2274–2279. doi:10.1021/jf9811065
Han, D. P., Lohani, M., and Cho, M. W. (2007). Specific Asparagine-Linked Glycosylation Sites Are Critical for DC-SIGN- and L-SIGN-Mediated Severe Acute Respiratory Syndrome Coronavirus Entry. J. Virol. 81(21), 12029–12039. doi:10.1128/jvi.00315-07
Heino, S., Lusa, S., Somerharju, P., Ehnholm, C., Olkkonen, V. M., and Ikonen, E. (2000). Dissecting the Role of the Golgi Complex and Lipid Rafts in Biosynthetic Transport of Cholesterol to the Cell Surface. Proc. Natl. Acad. Sci. 97, 8375–8380. doi:10.1073/pnas.140218797
Heurich, A., Hofmann-Winkler, H., Gierer, S., Liepold, T., Jahn, O., and Pohlmann, S. (2014). TMPRSS2 and ADAM17 Cleave ACE2 Differentially and Only Proteolysis by TMPRSS2 Augments Entry Driven by the Severe Acute Respiratory Syndrome Coronavirus Spike Protein. J. Virol. 88(2), 1293–1307. doi:10.1128/jvi.02202-13
Ho, T., Wu, S., Chen, J., Li, C., and Hsiang, C. (2007). Emodin Blocks the SARS Coronavirus Spike Protein and Angiotensin-Converting Enzyme 2 Interaction. Antiviral Res. 74(2), 92–101. doi:10.1016/j.antiviral.2006.04.014
Hoever, G., Baltina, L., Michaelis, M., Kondratenko, R., Baltina, L., Tolstikov, G. A., et al. (2005). Antiviral Activity of Glycyrrhizic Acid Derivatives against SARS−Coronavirus. J. Med. Chem. 48, 1256–1259. doi:10.1021/jm0493008
Hsieh, L.-E., Lin, C.-N., Su, B.-L., Jan, T.-R., Chen, C.-M., Wang, C.-H., et al. (2010). Synergistic Antiviral Effect of Galanthus Nivalis Agglutinin and Nelfinavir against Feline Coronavirus. Antiviral Res. 88(1), 25–30. doi:10.1016/j.antiviral.2010.06.010
Hulswit, R. J. G., Lang, Y., Bakkers, M. J. G., Li, W., Li, Z., Schouten, A., et al. (2019). Human Coronaviruses OC43 and HKU1 Bind to 9-O-Acetylated Sialic Acids via a Conserved Receptor-Binding Site in Spike Protein Domain A. Proc. Natl. Acad. Sci. USA. 116:2681–2690. doi:10.1073/pnas.1809667116
Hwang, H.-J., Han, J.-W., Jeon, H., Cho, K., Kim, J.-h., Lee, D.-S., et al. (2020). Characterization of a Novel Mannose-Binding Lectin with Antiviral Activities from Red Alga, Grateloupia Chiangii. Biomolecules 10(2), 333. doi:10.3390/biom10020333
Ibrahim, I. M., Abdelmalek, D. H., and Elfiky, A. A. (2019). GRP78: a Cell's Response to Stress. Life Sci. 226, 156–163. doi:10.1016/j.lfs.2019.04.022
Ibrahim, I. M., Abdelmalek, D. H., Elshahat, M. E., and Elfiky, A. A. (2020). COVID-19 Spike-Host Cell Receptor GRP78 Binding Site Prediction. J. Infect. 80(5), 554–562. doi:10.1016/j.jinf.2020.02.026
Im, K., Kim, J., and Min, H. (2015). Ginseng, the Natural Effectual Antiviral: Protective Effects of Koran Red Ginseng against Viral Infection 40(4):309–314. doi:10.1016/j.jgr.2015.09.002Available at: https://www.ncbi.nlm.nih.gov/pmc/articles/PMC5052424/
Islam, M. T., Sarkar, C., El‐Kersh, D. M., Jamaddar, S., Uddin, S. J., Shilpi, J. A., et al. (2020). Natural Products and Their Derivatives against Coronavirus: A Review of the Non‐clinical and Pre‐clinical Data. Phytotherapy Res. 34(10), 2471–2492. doi:10.1002/ptr.6700
Jeong, H.-U., Kwon, S.-S., Kong, T. Y., Kim, J. H., and Lee, H. S. (2014). "Inhibitory Effects of Cedrol, β-Cedrene, and Thujopsene on Cytochrome P450 Enzyme Activities in Human Liver Microsomes". J. Toxicol. Environ. Health A 77 (22–24): 1522–1532. doi:10.1080/15287394.2014.955906
Jin, Y. H., Kwon, S., Choi, J. G., Cho, W. K., Lee, B., and Ma, J. Y. (2019). Toosendanin from Melia Fructus Suppresses Influenza A Virus Infection by Altering Nuclear Localization of Viral Polymerase PA Protein. Front. Pharmacol. 10, 1025. doi:10.3389/fphar.2019.01025
Jin, Z., Du, X., Xu, Y., Deng, Y., Liu, M., Zhao, Y., et al. 2020). Structure of Mpro from SARS-CoV-2 and Discovery of its Inhibitors. Nature, 582, 289, 293., doi:10.1038/s41586-020-2223-y
Joshi, R. S., Jagdale, S. S., Bansode, S. B., Shankar, S. S., Tellis, M. B., Pandya, V. K., et al. (2020). Discovery of Potential Multi-Target-Directed Ligands by Targeting Host-specific SARS-CoV-2 Structurally Conserved Main Protease. J. Biomol. Struct. Dyn. 2020, 1–16. doi:10.1080/07391102.2020.1760137
Kadil, Y., Mouhcine, M., and Filali, H. (2020). In Silico Investigation of the SARS CoV2 Protease with Thymoquinone Major Constituent of Nigella Sativa. Cddt, 17, 12. doi:10.2174/1570163817666200712164406
Kang, K. B., Ming, G., Kim, G. J., Ha, T.-K. -Q., Choi, H., Oh, W. K., et al. (2015). Jubanines F-J, Cyclopeptide Alkaloids from the Roots of Ziziphus Jujuba. Phytochemistry 119, 90–95. doi:10.1016/j.phytochem.2015.09.001
Kalhori, M. R., Saadatpour, F., Arefian, E., Soleiman, M., Farzaei, M. H., Aneva, I. Y., et al. (2021). The Potential Therapeutic Effect of RNA Interference and Natural Products on COVID-19: A Review of the Coronaviruses Infection. Front. Pharmacol. 12, 616993. doi:10.3389/fphar.2021.616993
Kazakova, O. B., Giniiatullina, G. V., and Tolstikov, G. A. (2011). Synthesis of A-Secomethylenamino- and Substituted Amidoximotriterpenoids. Bioorg. Khim. 37(5), 690–696. doi:10.1134/s1068162011050086
Keyaerts, E., Vijgen, L., Pannecouque, C., Van Damme, E., Peumans, W., Egberink, H., et al. (2007). Plant Lectins Are Potent Inhibitors of Coronaviruses by Interfering with Two Targets in the Viral Replication Cycle. Antiviral Res. 75(3), 179–187. doi:10.1016/j.antiviral.2007.03.003
Khaerunnisa, S., Kurniawan, H., Awaluddin, R., Suhartati, S., and Soetjipto, S. (2020). Potential Inhibitor of COVID-19 Main Protease (Mpro) from Several Medicinal Plant Compounds by Molecular Docking Study. Preprints 2020, 2020030226. doi:10.20944/preprints202003.0226.v1
Khalifa, I., Zhu, W., Mohammed, H. H. H., Dutta, K., and Li, C. (2020). Tannins Inhibit SARS‐CoV‐2 through Binding with Catalytic Dyad Residues of 3CL Pro : An In Silico Approach with 19 Structural Different Hydrolysable Tannins. J. Food Biochem., 44 e13432. doi:10.1111/jfbc.13432
Kim, C.-H. (2020). SARS-CoV-2 Evolutionary Adaptation toward Host Entry and Recognition of Receptor O-Acetyl Sialylation in Virus-Host Interaction. Ijms 21(12), 4549. doi:10.3390/ijms21124549
Kim, D., Min, J., Jang, M., Lee, J., Shin, Y., Park, C., et al. (2019). Natural Bis-Benzylisoquinoline Alkaloids-Tetrandrine, Fangchinoline, and Cepharanthine, Inhibit Human Coronavirus OC43 Infection of MRC-5 Human Lung Cells. Biomolecules 9(11), 696. doi:10.3390/biom9110696
Kim, D. W., Seo, K. H., Curtis-Long, M. J., Oh, K. Y., Oh, J.-W., Cho, J. K., et al. (2014). Phenolic Phytochemical Displaying SARS-CoV Papain-like Protease Inhibition from the Seeds of Psoralea Corylifolia. J. Enzyme Inhib. Med. Chem. 29(1), 59–63. doi:10.3109/14756366.2012.753591
Kim, H.-Y., Eo, E.-Y., Park, H., Kim, Y.-C., Park, S., Shin, H.-J., et al. (2010). Medicinal Herbal Extracts of Sophorae Radix, Acanthopanacis Cortex, Sanguisorbae Radix and Torilis Fructus Inhibit Coronavirus Replication In Vitro. Antivir. Ther. 15(5), 697–709. doi:10.3851/imp1615
Kim, H.-Y., Shin, H.-S., Park, H., Kim, Y.-C., Yun, Y. G., Park, S., et al. (2008). In vitro inhibition of Coronavirus Replications by the Traditionally Used Medicinal Herbal Extracts, Cimicifuga Rhizoma, Meliae Cortex, Coptidis Rhizoma, and Phellodendron Cortex. J. Clin. Virol. 41(2), 122–128. doi:10.1016/j.jcv.2007.10.011
Kodchakorn, K., Poovorawan, Y., Suwannakarn, K., and Kongtawelert, P. (2020). Molecular Modelling Investigation for Drugs and Nutraceuticals against Protease of SARS-CoV-2. J. Mol. Graphics Model. 101, 107717. doi:10.1016/j.jmgm.2020.107717
Koehn, F. E., Sarath, G. P., Neil, D. N., and Cross, S. S. (1991). Halitunal, an Unusual Diterpene Aldehyde from the Marine Alga Halimeda Tuna. Tetrahedron Lett. 32(2), 169–172. doi:10.1016/0040-4039(91)80845-w
Kolodziej, H. (2011). Antimicrobial, Antiviral and Immunomodulatory Activity Studies of Pelargonium Sidoides (EPs 7630) in the Context of Health Promotion. Pharmaceuticals, 4(10), 1295–1314. doi:10.3390/ph4101295
Kumaki, Y., Wandersee, M. K., Smith, A. J., Zhou, Y., Simmons, G., Nelson, N. M., et al. (2011). Inhibition of Severe Acute Respiratory Syndrome Coronavirus Replication in a Lethal SARS-CoV BALB/c Mouse Model by Stinging Nettle Lectin, Urtica Dioica Agglutinin. Antiviral Res. 90(1), 22–32. doi:10.1016/j.antiviral.2011.02.003
Kumar, V., Dhanjal, J. K., Kaul, S. C., Wadhwa, R., and Sundar, D. (2020). Withanone and Caffeic Acid Phenethyl Ester Are Predicted to Interact with Main Protease (Mpro) of SARS-CoV-2 and Inhibit its Activity. J. Biomol. Struct. Dyn. 39, 1–13. doi:10.1080/07391102.2020.1772108
Kwon, H.-J., Ryu, Y. B., Kim, Y.-M., Song, N., Kim, C. Y., Rho, M.-C., et al. (2013). In vitro antiviral Activity of Phlorotannins Isolated from Ecklonia Cava against Porcine Epidemic Diarrhea Coronavirus Infection and Hemagglutination. Bioorg. Med. Chem. 21(15), 4706–4713. doi:10.1016/j.bmc.2013.04.085
Langereis, M. A., Zeng, Q., Heesters, B. A., Huizinga, E. G., and de Groot, R. J. (2012). The Murine Coronavirus Hemagglutinin-Esterase Receptor-Binding Site: a Major Shift in Ligand Specificity through Modest Changes in Architecture. Plos Pathog. 8, e1002492. doi:10.1371/journal.ppat.1002492
Lau, K.-M., Lee, K.-M., Koon, C.-M., Cheung, C. S.-F., Lau, C.-P., Ho, H.-M., et al. (2008). Immunomodulatory and Anti-SARS Activities of Houttuynia Cordata. J. Ethnopharmacology 118, 79–85. doi:10.1016/j.jep.2008.03.018
Lee, J.-H., Park, J.-S., Lee, S.-W., Hwang, S.-Y., Young, B.-E., and Choi, H.-J. (2015). Porcine Epidemic Diarrhea Virus Infection: Inhibition by Polysaccharide from Ginkgo Biloba Exocarp and Mode of its Action. Virus. Res. 195, 148–152. doi:10.1016/j.virusres.2014.09.013
Lelesius, R., Karpovaite, A., Mickiene, R., Drevinskas, T., Tiso, N., Ragazinskiene, O., et al. (2019). In vitro antiviral Activity of Fifteen Plant Extracts against Avian Infectious Bronchitis Virus. BMC Vet. Res. 15(1), 178.
Letko, M., Marzi, A., and Munster, V. (2020). Functional Assessment of Cell Entry and Receptor Usage for SARS-CoV-2 and Other Lineage B Betacoronaviruses. Nat. Microbiol. 5(4), 562–569. doi:10.1038/s41564-020-0688-y
Li, F. (2015). Receptor Recognition Mechanisms of Coronaviruses: a Decade of Structural Studies. J. Virol. 89, 1954–1964. doi:10.1128/JVI.02615-14
Li, F. (2016). Structure, Function, and Evolution of Coronavirus Spike Proteins. Annu. Rev. Virol. 3, 237–261. doi:10.1146/annurev-virology-110615-042301
Li, H., Wu, J., Zhang, Z., Ma, Y., Liao, F., Zhang, Y., et al. (2011). Forsythoside a Inhibits the Avian Infectious Bronchitis Virus in Cell Culture. Phytother. Res. 25(3), 338–342. doi:10.1002/ptr.3260
Li, L., Yu, X., Zhang, H., Cheng, H., Hou, L., Zheng, Q., et al. (2019). In vitro antiviral Activity of Griffithsin against Porcine Epidemic Diarrhea Virus. Virus Genes 55(2), 174–181. doi:10.1007/s11262-019-01633-7
Li, S., Chen, C., Zhang, H., Guo, H., Wang, H., Wang, L., et al. (2005). Identification of Natural Compounds with Antiviral Activities against SARS-Associated Coronavirus. Antiviral Res. 67(1), 18–23. doi:10.1016/j.antiviral.2005.02.007
Li, Y., and Jiang, J.-G. (2018). Health Functions and Structure-Activity Relationships of Natural Anthraquinones from Plants. Food Funct. 9(12), 6063–6080. doi:10.1039/c8fo01569d
Liang, Y., Zhang, Q., Zhang, L., Wang, R., Xu, X., and Hu, X. (2019). Astragalus Membranaceus Treatment Protects Raw264.7 Cells from Influenza Virus by Regulating G1 Phase and the TLR3-Mediated Signaling Pathway. Evid. Based Complement. Alternat. Med. 2019, 2971604. doi:10.1155/2019/2971604
Lim, H., Min, D. S., Park, H., and Kim, H. P. (2018). Flavonoids Interfere with NLRP3 Inflammasome Activation. Toxicol. Appl. Pharmacol. 355, 93–102. doi:10.1016/j.taap.2018.06.022
Lin, C.-W., Tsai, F.-J., Tsai, C.-H., Lai, C.-C., Wan, L., Ho, T.-Y., et al. (2005). Anti-SARS Coronavirus 3C-like Protease Effects of Isatis Indigotica Root and Plant-Derived Phenolic Compounds. Antiviral Res. 68, 36–42. doi:10.1016/j.antiviral.2005.07.002
Lin, S. C., Ho, C. T., Chuo, W. H., Li, S., Wang, T. T., and Lin, C. C. (2017). Effective Inhibition of MERS-CoV Infection by Resveratrol. BMC Infect. Dis. 17(1), 144. doi:10.1186/s12879-017-2253-8
Loizzo, M. R., Saab, A. M., Tundis, R., Statti, G. A., Menichini, F., Lampronti, I., et al. (2008). Phytochemical Analysis Andin Vitro Antiviral Activities of the Essential Oils of Seven Lebanon Species. C&B. 5(3), 461–470. doi:10.1002/cbdv.200890045
Lung, J., Lin, Y. S., Yang, Y. H., Chou, Y. L., Shu, L. H., Cheng, Y. C., et al. (2020). The Potential Chemical Structure of anti‐SARS‐CoV‐2 RNA‐dependent RNA Polymerase. J. Med. Virol. 92(6), 693–697. doi:10.1002/jmv.25761
Luo, W., Su, X., Gong, S., Qin, Y., Liu, W., Li, J., et al. (2009). Anti-SARS Coronavirus 3C-like Protease Effects of Rheum Palmatum L. Extracts. Biosci. Trends 3(4), 124–126.
Mani, J. S., Johnson, J. B., Steel, J. C., Broszczak, D. A., Neilsen, P. M., Walsh, K. B., et al. (2020). Natural Product-Derived Phytochemicals as Potential Agents against Coronaviruses: a Review. Virus. Res. 284, 197989. doi:10.1016/j.virusres.2020.197989
Majnooni, M. B., Fakhri, S., Shokoohinia, Y., Kiyani, N., Stage, K., Mohammadi, P., et al. (2020). Phytochemicals: Potential Therapeutic Interventions Against Coronavirus-Associated Lung Injury. Front. Pharmacol. 11, 1744. doi:10.3389/fphar.2020.588467
Marzi, A., Gramberg, T., Simmons, G., Moller, P., Rennekamp, A. J., Krumbiegel, M., et al. (2004). DC-SIGN and DC-SIGNR Interact with the Glycoprotein of Marburg Virus and the S Protein of Severe Acute Respiratory Syndrome Coronavirus. Jvi 78, 12090–12095. doi:10.1128/jvi.78.21.12090-12095.2004
Matsuyama, S., and Taguchi, F. (2002). Receptor-induced Conformational Changes of Murine Coronavirus Spike Protein. J. Virol. 76, 11819–11826. doi:10.1128/jvi.76.23.11819-11826.2002
McCutcheon, A. R., Roberts, T. E., Gibbons, E., Ellis, S. M., Babiuk, L. A., Hancock, R. E. W., et al. (1995). Antiviral Screening of British Columbian Medicinal Plants. J. Ethnopharmacology 49(2), 101–110. doi:10.1016/0378-8741(95)90037-3
McDonagh, P., Sheehy, P. A., and Norris, J. M. (2014). Identification and Characterisation of Small Molecule Inhibitors of Feline Coronavirus Replication. Vet. Microbiol. 174(3-4), 438–447. doi:10.1016/j.vetmic.2014.10.030
McKee, D. L., Sternberg, A., Stange, U., Laufer, S., and Naujokat, C. (2020). Candidate Drugs against SARS-CoV-2 and COVID-19. Pharmacol. Res., 157, 104859. 104859. doi:10.1016/j.phrs.2020.104859
Meneguzzo, F., Ciriminna, R., Zabini, F., and Pagliaro, M. (2020). Review of Evidence Available on Hesperidin-Rich Products as Potential Tools against COVID-19 and Hydrodynamic Cavitation-Based Extraction as a Method of Increasing Their Production. Processes 8, 549. doi:10.3390/pr8050549
Michaelis, M., Doerr, H. W., and Cinatl, J. (2011). Investigation of the Influence of EPs 7630, a Herbal Drug Preparation from Pelargonium Sidoides, on Replication of a Broad Panel of Respiratory Viruses. Phytomedicine 18(5), 384–386. doi:10.1016/j.phymed.2010.09.008
Michaelis, M., Geiler, J., Naczk, P., Sithisarn, P., Leutz, A., Doerr, H. W., et al. (2011). Glycyrrhizin Exerts Antioxidative Effects in H5N1 Influenza A Virus-Infected Cells and Inhibits Virus Replication and Pro-inflammatory Gene Expression. PLoS One 6, e19705. doi:10.1371/journal.pone.0019705
Michelow, I. C., Lear, C., Scully, C., Prugar, L. I., Longley, C. B., Yantosca, L. M., et al. (2011). High-dose Mannose-Binding Lectin Therapy for Ebola Virus Infection. J. Infect. Dis. 203(2), 175–179. doi:10.1093/infdis/jiq025
Miean, K. H., and Mohamed, S. (2001). Flavonoid (Myricetin, Quercetin, Kaempferol, Luteolin, and Apigenin) Content of Edible Tropical Plants. J. Agric. Food Chem. 49(6), 3106–3112. doi:10.1021/jf000892m
Mikulic-Petkovsek, M., Slatnar, A., Stampar, F., and Veberic, R. (2012). HPLC-MSn Identification and Quantification of Flavonol Glycosides in 28 Wild and Cultivated Berry Species. Food Chem. 135(4), 2138–2146. doi:10.1016/j.foodchem.2012.06.115
Milewska, A., Zarebski, M., Nowak, P., Stozek, K., Potempa, J., and Pyrc, K. (2014). Human Coronavirus NL63 Utilizes Heparan Sulfate Proteoglycans for Attachment to Target Cells. J. Virol. 88, 13221–13230. doi:10.1128/jvi.02078-14
Millet, J. K., Séron, K., Labitt, R. N., Danneels, A., Palmer, K. E., Whittaker, G. R., et al. (2016). Middle East Respiratory Syndrome Coronavirus Infection Is Inhibited by Griffithsin. Antiviral Res. 133, 1–8. doi:10.1016/j.antiviral.2016.07.011
Moghaddam, E., Teoh, B. T., Sam, S. S., Lani, R., Hassandarvish, P., and Chik, Z. (2014). Baicalin, a Metabolite of Baicalein with Antiviral Activity against Dengue Virus. Sci. Rep. 4, 5452. doi:10.1038/srep05452
Müller, C., Obermann, W., Schulte, F. W., Lange-Grünweller, K., Oestereich, L., Elgner, F., et al. (2020). Comparison of Broad-Spectrum Antiviral Activities of the Synthetic Rocaglate CR-31-B (−) and the eIF4A-Inhibitor Silvestrol. Antiviral Res. 175, 104706. doi:10.1016/j.antiviral.2020.104706
Müller, C., Schulte, F. W., Lange-Grünweller, K., Obermann, W., Madhugiri, R., Pleschka, S., et al. (2018). Broad-spectrum Antiviral Activity of the eIF4A Inhibitor Silvestrol against Corona- and Picornaviruses. Antiviral Res. 150, 123–129. doi:10.1016/j.antiviral.2017.12.010
Naskalska, A., Dabrowska, A., Szczepanski, A., Milewska, A., Jasik, K. P., and Pyrc, K. (2019). Membrane Protein of Human Coronavirus NL63 Is Responsible for Interaction with the Adhesion Receptor. J. Virol. 93(19), e00355-19. doi:10.1128/JVI.00355-19
Nguyen, P. Q. T., Ooi, J. S. G., Nguyen, N. T. K., Wang, S., Huang, M., Liu, D. X., et al. (2015). Antiviral Cystine Knot α-Amylase Inhibitors from Alstonia scholaris. J. Biol. Chem. 290(52), 31138–31150. doi:10.1074/jbc.m115.654855
Nomura, R., Kiyota, A., Suzaki, E., Kataoka, K., Ohe, Y., Miyamoto, K., et al. 2004). Human Coronavirus 229E Binds to CD13 in Rafts and Enters the Cell through Caveolae. Jvi 78(16), 8701–8708. doi:10.1128/jvi.78.16.8701-8708.2004
O’Flaherty, K., Ataíde, R., Zaloumis, S. G., Ashley, E. A., Powell, R., Feng, L., et al. (2019). Contribution of Functional Antimalarial Immunity to Measures of Parasite Clearance in Therapeutic Efficacy Studies of Artemisinin Derivatives. J. Infect. Dis. 220, 1178–1187. doi:10.1093/infdis/jiz247
O'Keefe, B. R., Giomarelli, B., Barnard, D. L., Shenoy, S. R., Chan, P. K. S., McMahon, J. B., et al. (2010). Broad-Spectrum In Vitro Activity and In Vivo Efficacy of the Antiviral Protein Griffithsin against Emerging Viruses of the Family Coronaviridae. Jvi 84(5), 2511–2521. doi:10.1128/jvi.02322-09
Oliveira, A. F., Teixeira, R. R., Oliveira, A. S., Souza, A. P., Silva, M. L., and Paula, S. O. (2017). Potential Antivirals: Natural Products Targeting Replication Enzymes of Dengue and Chikungunya Viruses. Molecules, 22(3): 505. doi:10.3390/molecules22030505
Packer, L., and Cadenas, E. (2011). Lipoic Acid: Energy Metabolism and Redox Regulation of Transcription and Cell Signaling. J. Clin. Biochem. Nutr. 48(1), 26–32. doi:10.3164/jcbn.11-005FR
Park, J.-E., Li, K., Barlan, A., Fehr, A. R., Perlman, S., McCray, P. B., et al. (2016). Proteolytic Processing of Middle East Respiratory Syndrome Coronavirus Spikes Expands Virus Tropism. Proc. Natl. Acad. Sci. USA 113, 12262–12267. doi:10.1073/pnas.1608147113
Park, J.-Y., Kim, J. H., Kim, Y. M., Jeong, H. J., Kim, D. W., Park, K. H., et al. (2012). Tanshinones as Selective and Slow-Binding Inhibitors for SARS-CoV Cysteine Proteases. Bioorg. Med. Chem. 20(19), 5928–5935. doi:10.1016/j.bmc.2012.07.038
Park, J.-Y., Yuk, H. J., Ryu, H. W., Lim, S. H., Kim, K. S., Park, K. H., et al. (2017). Evaluation of Polyphenols from Broussonetia Papyrifera as Coronavirus Protease Inhibitors. J. Enzyme Inhib. Med. Chem. 32(1), 504–512. doi:10.1080/14756366.2016.1265519
Peng, G., Sun, D., Rajashankar, K. R., Qian, Z., Holmes, K. V., and Li, F. (2011). Crystal Structure of Mouse Coronavirus Receptor-Binding Domain Complexed with its Murine Receptor. Proc. Natl. Acad. Sci. 108(26), 10696–10701. doi:10.1073/pnas.1104306108
Peng, G., Xu, L., Lin, Y.-L., Chen, L., Pasquarella, J. R., Holmes, K. V., et al. (2012). Crystal Structure of Bovine Coronavirus Spike Protein Lectin Domain*. J. Biol. Chem. 287(50), 41931–41938. doi:10.1074/jbc.m112.418210
Peng, G., Yang, Y., Pasquarella, J. R., Xu, L., Qian, Z., Holmes, K. V., et al. (2017). Structural and Molecular Evidence Suggesting Coronavirus-Driven Evolution of Mouse Receptor. J. Biol. Chem. 292(6), 2174–2181. doi:10.1074/jbc.m116.764266
Pooja, M., Reddy, G. J., Hema, K., Dodoala, S., and Koganti, B. (2021). Unravelling High-Affinity Binding Compounds towards Transmembrane Protease Serine 2 Enzyme in Treating SARS-CoV-2 Infection Using Molecular Modelling and Docking Studies. Eur. J. Pharmacol. 890, 173688. doi:10.1016/j.ejphar.2020.173688
Pour, P. M., Fakhri, S., Asgary, S., Farzaei, M. H., and Echeverría, J. (2019). The Signaling Pathways, and Therapeutic Targets of Antiviral Agents: Focusing on the Antiviral Approaches and Clinical Perspectives of Anthocyanins in the Management of Viral Diseases. Front. Pharmacol. 10, 1207. doi:10.3389/fphar.2019.01207
Pujhari, S., Brustolin, M., Macias, V. M., Nissly, R. H., Nomura, M., Kuchipudi, S. V., et al. (2019). Heat Shock Protein 70 (Hsp70) Mediates Zika Virus Entry, Replication, and Egress from Host Cells. Emerging Microbes & Infections 8(1), 8–16. doi:10.1080/22221751.2018.1557988
Qing, Z., Chen, X. Y., and Martin, C. (2016). Scutellaria Baicalensis, the Golden Herb from the Garden of Chinese Medicinal Plants. Sci. Bull. 61(18), 1391–1398. doi:10.1007/s11434-016-1136-5
Raj, V. S., Mou, H., Smits, S. L., Dekkers, D. H. W., Müller, M. A., Dijkman, R., et al. (2013). Dipeptidyl Peptidase 4 Is a Functional Receptor for the Emerging Human Coronavirus-EMC. Nature 495, 251–254. doi:10.1038/nature12005
Rao, R. V., Peel, A., Logvinova, A., del Rio, G., Hermel, E., Yokota, T., et al. 2002). Coupling Endoplasmic Reticulum Stress to the Cell Death Program: Role of the ER Chaperone GRP78. FEBS Lett. 514(2–3), 122–128. doi:10.1016/s0014-5793(02)02289-5
Rasool, N., Akhtar, A., and Hussain, W. (2020). Insights into the Inhibitory Potential of Selective Phytochemicals against Mpro of 2019-nCoV: a Computer-Aided Study. Struct. Chem. 31, 1777–1783. doi:10.1007/s11224-020-01536-6
Ratia, K., Pegan, S., Takayama, J., Sleeman, K., Coughlin, M., Baliji, S., et al. (2008). A Noncovalent Class of Papain-like Protease/deubiquitinase Inhibitors Blocks SARS Virus Replication. Proc. Natl. Acad. Sci. 105, 16119–16124. doi:10.1073/pnas.0805240105
Regueiro-Ren, A., Swidorski, J. J., Liu, Z., Chen, Y., Sin, N., Sit, S. Y., et al. (2018). Design, Synthesis, and SAR of C-3 Benzoic Acid, C-17 Triterpenoid Derivatives. Identification of the HIV-1 Maturation Inhibitor 4-((1 R,3a S,5a R,5b R,7a R,11a S,11b R,13a R,13b R)-3a-((2-(1,1-Dioxidothiomorpholino)ethyl)amino)-5a,5b,8,8,11a-pentamethyl-1-(prop-1-en-2-yl)-2,3,3a,4,5,5a,5b,6,7,7a,8,11,11a,11b,12,13,13a,13b-octadecahydro-1 H-Cyclopenta[ A]chrysen-9-Yl)benzoic Acid (GSK3532795, BMS-955176). J. Med. Chem. 61(16), 7289–7313. doi:10.1021/acs.jmedchem.8b00854
Roh, C. (2012). A Facile Inhibitor Screening of SARS Coronavirus N Protein Using Nanoparticle-Based RNA Oligonucleotide. Ijn 7, 2173. doi:10.2147/IJN.S31379
Ryu, Y. B., Jeong, H. J., Kim, J. H., Kim, Y. M., Park, J.-Y., Kim, D., et al. (2010). Biflavonoids from Torreya Nucifera Displaying SARS-CoV 3CLpro Inhibition. Bioorg. Med. Chem. 18, 7940–7947. doi:10.1016/j.bmc.2010.09.035
Ryu, Y. B., Park, S.-J., Kim, Y. M., Lee, J.-Y., Seo, W. D., Chang, J. S., et al. (2010). SARS-CoV 3CLpro Inhibitory Effects of Quinone-Methide Triterpenes from Tripterygium Regelii. Bioorg. Med. Chem. Lett. 20, 1873–1876. doi:10.1016/j.bmcl.2010.01.152
Sayed, A. A., and Elfiky, A. A. (2018). In Silico estrogen-like Activity and In Vivo Osteoclastogenesis Inhibitory Effect of Cicer Arietinum Extract. Cel Mol Biol (Noisy-le-grand) 64(5), 29–39. doi:10.14715/cmb/2018.64.5.5
Schwarz, S., Sauter, D., Wang, K., Zhang, R., Sun, B., Karioti, A., et al. (2014). Kaempferol Derivatives as Antiviral Drugs against the 3a Channel Protein of Coronavirus. Planta Med. 80(02/03), 177–182. doi:10.1055/s-0033-1360277
Sharma, A. D., and Kaur, I. (2020). Jensenone from eucalyptus Essential Oil as a Potential Inhibitor of COVID 19 Corona Virus Infection. Res. Rev. Biotech. Biosci., 7, 59–66. doi:10.5281/zenodo.3748477
Sharma, P., and Shanavas, A. (2020). Natural Derivatives with Dual Binding Potential against SARS-CoV-2 Main Protease and Human ACE2 Possess Low Oral Bioavailability: a Brief Computational Analysis. J. Biomol. Struct. Dyn. 21, 1–12. doi:10.1080/07391102.2020.1794970
Shen, L., Niu, J., Wang, C., Huang, B., Wang, W., Zhu, N., et al. (2019). High-throughput Screening and Identification of Potent Broad-spectrum Inhibitors of Coronaviruses. J. Virol. 93(12), e00023–e00019. doi:10.1128/jvi.00023-19
Shen, Y.-C., Wang, L.-T., Khalil, A. T., Chiang, L. C., and Cheng, P.-W. (2005). Bioactive Pyranoxanthones from the Roots of Calophyllum Blancoi. Chem. Pharm. Bull. 53(2), 244–247. doi:10.1248/cpb.53.244
Simmons, G., Zmora, P., Gierer, S., Heurich, A., and Pöhlmann, S. (2013). Proteolytic Activation of the SARS-Coronavirus Spike Protein: Cutting Enzymes at the Cutting Edge of Antiviral Research. Antiviral Res. 100, 605–614. doi:10.1016/j.antiviral.2013.09.028
Smith, M., and Smith, J. C. (2020). Repurposing Therapeutics for COVID-19: Supercomputer-Based Docking to the SARS-CoV-2 Viral Spike Protein and Viral Spike Protein-Human ACE2 Interface, ChemRxiv doi:10.26434/chemrxiv.11871402.v4
Smits, S. L., Gerwig, G. J., van Vliet, A. L. W., Lissenberg, A., Briza, P., Kamerling, J. P., et al. (2005). Nidovirus Sialate-O-Acetylesterases. J. Biol. Chem. 280, 6933–6941. doi:10.1074/jbc.m409683200
Söderberg, C., Giugni, T. D., Zaia, J. A., Larsson, S., Wahlberg, J. M., and Möller, E. (1993). CD13 (Human Aminopeptidase N) Mediates Human Cytomegalovirus Infection. J. Virol. 67 (11), 6576–6585. doi:10.1128/jvi.67.11.6576-6585.1993
Song, J., Shim, J., and Choi, H. (2011). Quercetin 7-rhamnoside Reduces Porcine Epidemic Diarrhea Virus Replication via Independent Pathway of Viral Induced Reactive Oxygen Species. Virol. J. 8, 460. doi:10.1186/1743-422x-8-460
Song, Y. H., Kim, D. W., Curtis-Long, M. J., Yuk, H. J., Wang, Y., Zhuang, N., et al. (2014). Papain-like Protease (PLpro) Inhibitory Effects of Cinnamic Amides from Tribulus Terrestris Fruits. Biol. Pharm. Bull. 37(6), 1021–1028. doi:10.1248/bpb.b14-00026
Stefaniu, A., Pirvu, L., Albu, B., and Pintilie, L. (2020). Molecular Docking Study on Several Benzoic Acid Derivatives against SARS-CoV-2. Molecules 25(24), 5828. doi:10.3390/molecules25245828
Sun, P., Sun, N., Yin, W., Sun, Y., Fan, K., Guo, J., et al. (2019). Matrine Inhibits IL-1β Secretion in Primary Porcine Alveolar Macrophages through the MyD88/NF-Κb Pathway and NLRP3 Inflammasome. Vet. Res. 50(1), 53. doi:10.1186/s13567-019-0671-x
Sun, Y., Zhao, Y., Yao, J., Zhao, L., Wu, Z., Wang, Y., et al. (2015). Wogonoside Protects against Dextran Sulfate Sodium-Induced Experimental Colitis in Mice by Inhibiting NF-Κb and NLRP3 Inflammasome Activation. Biochem. Pharmacol. 94(2), 142–154. doi:10.1016/j.bcp.2015.02.002
Sundararajan, A., Ganapathy, R., Huan, L., Dunlap, J. R., Webby, R. J., Kotwal, G. J., et al. (2010). Influenza Virus Variation in Susceptibility to Inactivation by Pomegranate Polyphenols Is Determined by Envelope Glycoproteins. Antiviral Res. 88(1), 1–9. doi:10.1016/j.antiviral.2010.06.014
Taguchi, F., and Matsuyama, S. (2002). Soluble Receptor Potentiates Receptor-independent Infection by Murine Coronavirus. Jvi 76, 950–958. doi:10.1128/jvi.76.3.950-958.2002
Theerawatanasirikul, S., Kuo, C. J., Phetcharat, N., and Lekcharoensuk, P. (2020). In Silico and In Vitro Analysis of Small Molecules and Natural Compounds Targeting the 3CL Protease of Feline Infectious Peritonitis Virus. Antiviral Res. 174, 104697. doi:10.1016/j.antiviral.2019.104697
Tipnis, S. R., Hooper, N. M., Hyde, R., Karran, E., Christie, G., and Turner, A. J. (2000). A Human Homolog of Angiotensin-Converting Enzyme. J. Biol. Chem. 275(43), 33238–33243. doi:10.1074/jbc.m002615200
Toney, J. H., Navas-Martín, S., Weiss, S. R., and Koeller, A. (2004). Sabadinine: A Potential Non-peptide Anti-severe Acute-Respiratory-Syndrome Agent Identified Using Structure-Aided Design. J. Med. Chem. 47(5), 1079–1080. doi:10.1021/jm034137m
Tortorici, M. A., Walls, A. C., Lang, Y., Wang, C., Li, Z., Koerhuis, D., et al. (2019). Structural Basis for Human Coronavirus Attachment to Sialic Acid Receptors. Nat. Struct. Mol. Biol. 26(6), 481–489. doi:10.1038/s41594-019-0233-y
Tresnan, D. B., Levis, R., and Holmes, K. V. (1996). Feline Aminopeptidase N Serves as a Receptor for Feline, Canine, Porcine, and Human Coronaviruses in Serogroup I. J. Virol. 70(12), 8669–8674. doi:10.1128/jvi.70.12.8669-8674.1996
Tseng, C.-K., Hsu, S.-P., Lin, C.-K., Wu, Y.-H., Lee, J.-C., and Young, K.-C. (2017). Celastrol Inhibits Hepatitis C Virus Replication by Upregulating Heme Oxygenase-1 via the JNK MAPK/Nrf2 Pathway in Human Hepatoma Cells. Antiviral Res. 146, 191–200. doi:10.1016/j.antiviral.2017.09.010
Tu, Y.-F., Chien, C.-S., Yarmishyn, A. A., Lin, Y.-Y., Luo, Y.-H., Lin, Y.-T., et al. (2020). A Review of SARS-CoV-2 and the Ongoing Clinical Trials. Ijms 21(7), 2657. doi:10.3390/ijms21072657
Ulasli, M., Gurses, S. A., Bayraktar, R., Yumrutas, O., Oztuzcu, S., Igci, M., et al. (2014). The Effects of Nigella Sativa (Ns), Anthemis Hyalina (Ah) and Citrus Sinensis (Cs) Extracts on the Replication of Coronavirus and the Expression of TRP Genes Family. Mol. Biol. Rep. 41(3), 1703–1711. doi:10.1007/s11033-014-3019-7
Ushio, Y., and Abe, H. (1992). Inactivation of Measles Virus and Herpes Simplex Virus by Saikosaponin D. Planta Med. 58(02), 171–173. doi:10.1055/s-2006-961422
Vlasak, R., Luytjes, W., Spaan, W., and Palese, P. (1988). Human and Bovine Coronaviruses Recognize Sialic Acid-Containing Receptors Similar to Those of Influenza C Viruses. Proc. Natl. Acad. Sci. 85, 4526–4529. doi:10.1073/pnas.85.12.4526
Vogel, H. G., Maas, J., and Mayer, D. (2006). Drug Discovery and Evaluation: Safety and Pharmacokinetic Assays; with 125 Tables. Springer Science & Business Media. 2006 – 889.
Walls, A. C., Tortorici, M. A., Bosch, B.-J., Frenz, B., Rottier, P. J. M., DiMaio, F., et al. (2016). Cryo-electron Microscopy Structure of a Coronavirus Spike Glycoprotein Trimer. Nature 531(7592), 114–117. doi:10.1038/nature16988
Wang, G.-F., Shi, L.-P., Ren, Y.-D., Liu, Q.-F., Liu, H.-F., Zhang, R.-J., et al. (2009). Anti-hepatitis B Virus Activity of Chlorogenic Acid, Quinic Acid and Caffeic Acid In Vivo and In Vitro. Antiviral Res. 83(2), 186–190. doi:10.1016/j.antiviral.2009.05.002
Wang, L.-J., Geng, C.-A., Ma, Y.-B., Huang, X.-Y., Luo, J., Chen, H., et al. (2012). Synthesis, Biological Evaluation and Structure-Activity Relationships of Glycyrrhetinic Acid Derivatives as Novel Anti-hepatitis B Virus Agents. Bioorg. Med. Chem. Lett. 22(10), 3473–3479. doi:10.1016/j.bmcl.2012.03.081
Wang, S.-Q., Du, Q.-S., Zhao, K., Li, A.-X., Wei, D.-Q., and Chou, K.-C. (2007). Virtual Screening for Finding Natural Inhibitor against Cathepsin-L for SARS Therapy. Amino Acids 33(1), 129–135. doi:10.1007/s00726-006-0403-1
Watanabe, R., Sawicki, S. G., and Taguchi, F. (2007). Heparan Sulfate Is a Binding Molecule but Not a Receptor for CEACAM1-independent Infection of Murine Coronavirus. Virology 366, 16–22. doi:10.1016/j.virol.2007.06.034
Wen, C.-C., Kuo, Y.-H., Jan, J.-T., Liang, P.-H., Wang, S.-Y., Liu, H.-G., et al. (2007). Specific Plant Terpenoids and Lignoids Possess Potent Antiviral Activities against Severe Acute Respiratory Syndrome Coronavirus. J. Med. Chem. 50, 4087–4095. doi:10.1021/jm070295s
Wen, C.-C., Shyur, L.-F., Jan, J.-T., Liang, P.-H., Kuo, C.-J., Arulselvan, P., et al. (2011). Traditional Chinese Medicine Herbal Extracts of Cibotium Barometz, Gentiana Scabra, Dioscorea Batatas, Cassia Tora, and Taxillus Chinensis Inhibit SARS-CoV Replication. J. Traditional Complement. Med. 1(1), 41–50. doi:10.1016/s2225-4110(16)30055-4
Weng, J.-R., Lin, C.-S., Lai, H.-C., Lin, Y.-P., Wang, C.-Y., Tsai, Y.-C., et al. (2019). Antiviral Activity of Sambucus FormosanaNakai Ethanol Extract and Related Phenolic Acid Constituents against Human Coronavirus NL63. Virus. Res. 273, 197767 doi:10.1016/j.virusres.2019.197767
Williamson, G., and Kerimi, A. (2020). Testing of Natural Products in Clinical Trials Targeting the SARS-CoV-2 (Covid-19) Viral Spike Protein-Angiotensin Converting Enzyme-2 (ACE2) Interaction. Biochem. Pharmacol. 178, 114123–123. doi:10.1016/j.bcp.2020.114123
Wittemer, S. M., Ploch, M., Windeck, T., Müller, S. C., Drewelow, B., Derendorf, H., et al. (2005). Bioavailability and Pharmacokinetics of Caffeoylquinic Acids and Flavonoids after Oral Administration of Artichoke Leaf Extracts in Humans. Phytomedicine 12(1), 28–38. doi:10.1016/j.phymed.2003.11.002
Witters, L., Scherle, P., Friedman, S., Fridman, J., Caulder, E., Newton, R., et al. (2008). Synergistic Inhibition with a Dual Epidermal Growth Factor receptor/HER-2/neu Tyrosine Kinase Inhibitor and a Disintegrin and Metalloprotease Inhibitor. Cancer Res. 68(17), 7083–7089. doi:10.1158/0008-5472.can-08-0739
Wu, C.-Y., Jan, J.-T., Ma, S.-H., Kuo, C.-J., Juan, H.-F., Cheng, Y.-S. E., et al. (2004). Small Molecules Targeting Severe Acute Respiratory Syndrome Human Coronavirus. Proc. Natl. Acad. Sci. 101(27), 10012–10017. doi:10.1073/pnas.0403596101
Wu, C., Liu, Y., Yang, Y., Zhang, P., Zhong, W., Wang, Y., et al. (2020). Analysis of Therapeutic Targets for SARS-CoV-2 and Discovery of Potential Drugs by Computational Methods. Acta Pharmaceutica Sinica B, 10, 766, 788. doi:10.1016/j.apsb.2020.02.008
Xiong, X., Coombs, P. J., Martin, S. R., Liu, J., Xiao, H., McCauley, J. W., et al. (2013). Receptor Binding by a Ferret-Transmissible H5 Avian Influenza Virus. Nature 497, 392–396. doi:10.1038/nature12144
Xu, D., Lin, T.-H., Li, S., Da, J., Wen, X.-Q., Ding, J., et al. (2012). Cryptotanshinone Suppresses Androgen Receptor-Mediated Growth in Androgen Dependent and Castration Resistant Prostate Cancer Cells. Cancer Lett. 316(1), 11–22. doi:10.1016/j.canlet.2011.10.006
Yamagata, K., Hashiguchi, K., Yamamoto, H., and Tagami, M. (2019). Dietary Apigenin Reduces Induction of LOX-1 and NLRP3 Expression, Leukocyte Adhesion, and Acetylated Low-Density Lipoprotein Uptake in Human Endothelial Cells Exposed to Trimethylamine-N-Oxide. J. Cardiovasc. Pharmacol. 74(6), 558–565. doi:10.1097/fjc.0000000000000747
Yang, C.-W., Chang, H.-Y., Lee, Y.-Z., Hsu, H.-Y., and Lee, S.-J. (2018). The Cardenolide Ouabain Suppresses Coronaviral Replication via Augmenting a Na+/K+-ATPase-dependent PI3K_PDK1 axis Signaling. Toxicol. Appl. Pharmacol. 356, 90–97. doi:10.1016/j.taap.2018.07.028
Yang, C.-W., Lee, Y.-Z., Hsu, H.-Y., Shih, C., Chao, Y.-S., Chang, H.-Y., et al. (2017). Targeting Coronaviral Replication and Cellular JAK2 Mediated Dominant NF-Κb Activation for Comprehensive and Ultimate Inhibition of Coronaviral Activity. Sci. Rep. 7(1), 4105. doi:10.1038/s41598-017-04203-9
Yang, C.-W., Lee, Y.-Z., Kang, I.-J., Barnard, D. L., Jan, J.-T., Lin, D., et al. (2010). Identification of Phenanthroindolizines and Phenanthroquinolizidines as Novel Potent Anti-coronaviral Agents for Porcine Enteropathogenic Coronavirus Transmissible Gastroenteritis Virus and Human Severe Acute Respiratory Syndrome Coronavirus. Antiviral Res. 88(2), 160–168. doi:10.1016/j.antiviral.2010.08.009
Yang, H., Yang, M., Ding, Y., Liu, Y., Lou, Z., Zhou, Z., et al. (2003). The Crystal Structures of Severe Acute Respiratory Syndrome Virus Main Protease and its Complex with an Inhibitor. Proc. Natl. Acad. Sci. 100(23), 13190–13195. doi:10.1073/pnas.1835675100
Yang, Q.-Y., Tian, X.-Y., and Fang, W.-S. (2007). Bioactive Coumarins from Boenninghausenia Sessilicarpa. J. Asian Nat. Prod. Res. 9(1), 59–65. doi:10.1080/10286020500382397
Yang, Z., Wu, N., Fu, Y., Yang, G., Wang, W., Zu, Y., et al. (2010). Anti-infectious Bronchitis Virus (IBV) Activity of 1,8-cineole: Effect on Nucleocapsid (N) Protein. J. Biomol. Struct. Dyn. 28(3), 323–330. doi:10.1080/07391102.2010.10507362
Yang, Z., Wu, N., Zu, Y., and Fu, Y. (2011). Comparative Anti-infectious Bronchitis Virus (IBV) Activity of (-)-pinene: Effect on Nucleocapsid (N) Protein. Molecules 16(2), 1044–1054. doi:10.3390/molecules16021044
Yang, Z., Wu, N., Zu, Y., and Fu, Y. (2011). Comparative Anti-infectious Bronchitis Virus (IBV) Activity of (-)-Pinene: Effect on Nucleocapsid (N) Protein. Molecules 16(2), 1044–1054. doi:10.3390/molecules16021044
Yeager, C. L., Ashmun, R. A., Williams, R. K., Cardellichio, C. B., Shapiro, L. H., Look, A. T., et al. (1992). Human Aminopeptidase N Is a Receptor for Human Coronavirus 229E. Nature 357 (6377), 420–422. doi:10.1038/357420a0
Yeager, C. L., Ashmun, R. A., Williams, R. K., Cardellichio, C. B., Shapiro, L. H., Look, A. T., et al. (1992). Human Aminopeptidase N Is a Receptor for Human Coronavirus 229E. Nature 357, 420–422. doi:10.1038/357420a0
Yepes-Pérez, A. F., Herrera-Calderon, O., Sánchez-Aparicio, J.-E., Tiessler-Sala, L., Maréchal, J.-D., and Cardona-G, W. (2020). Investigating Potential Inhibitory Effect of Uncaria Tomentosa (Cat's Claw) against the Main Protease 3CLPro of SARS-CoV-2 by Molecular Modeling. Preprints 2020, 2020060326. doi:10.20944/preprints202006.0326.v1
Yi, L., Li, Z., Yuan, K., Qu, X., Chen, J., Wang, G., et al. (2004). Small Molecules Blocking the Entry of Severe Acute Respiratory Syndrome Coronavirus into Host Cells. Jvi 78(20), 11334–11339. doi:10.1128/jvi.78.20.11334-11339.2004
Yin, J., Li, G., Li, J., Yang, Q., and Ren, X. (2011). In Vitroandin Vivoeffects ofHouttuynia Cordataon Infectious Bronchitis Virus. Avian Pathol. 40(5), 491–498. doi:10.1080/03079457.2011.605107
Yonesi, M., and Rezazadeh, A. (2020). Plants as a Prospective Source of Natural Anti-viral Compounds and Oral Vaccines against COVID-19 Coronavirus. Preprints 2020040321:0321. doi:10.20944/preprints202004.0321.v1
Yook, H.-S., Kim, K.-H., Park, J.-E., and Shin, H.-J. (2010). Antioxidative and Antiviral Properties of Flowering Cherry Fruits (Prunus serrulataL. var.Spontanea). Am. J. Chin. Med. 38(5), 937–948. doi:10.1142/s0192415x10008366
Yu, J.-S., Tseng, C.-K., Lin, C.-K., Hsu, Y.-C., Wu, Y.-H., Hsieh, C.-L., et al. (2017). Celastrol Inhibits Dengue Virus Replication via Up-Regulating Type I Interferon and Downstream Interferon-Stimulated Responses. Antiviral Res. 137, 49–57. doi:10.1016/j.antiviral.2016.11.010
Yu, M.-S., Lee, J., Lee, J. M., Kim, Y., Chin, Y.-W., Jee, J.-G., et al. (2012). Identification of Myricetin and Scutellarein as Novel Chemical Inhibitors of the SARS Coronavirus Helicase, nsP13. Bioorg. Med. Chem. Lett. 22, 4049–4054. doi:10.1016/j.bmcl.2012.04.081
Zhang, B. C., Li, Z., Xu, W., Xiang, C. H., and Ma, Y. F. (2018). Luteolin Alleviates NLRP3 Inflammasome Activation and Directs Macrophage Polarization in Lipopolysaccharide-Stimulated RAW264.7 Cells. Am. J. Transl. Res. 10(1), 265–273.
Zhang, C. H., Wang, Y. F., Liu, X. J., Lu, J. H., Qian, C. W., Wan, Z. Y., et al. (2005). Antiviral Activity of Cepharanthine against Severe Acute Respiratory Syndrome Coronavirus In Vitro. Chin. Med. J. (Engl) 118(6), 493–496.
Zhang, D.-h., Wu, K.-l., Zhang, X., Deng, S.-q., and Peng, B. (2020). In Silico screening of Chinese Herbal Medicines with the Potential to Directly Inhibit 2019 Novel Coronavirus. J. Integr. Med. 18(2), 152–158. doi:10.1016/j.joim.2020.02.005
Zhou, P., Yang, X.-L., Wang, X.-G., Hu, B., Zhang, L., Zhang, W., et al. (2020). A Pneumonia Outbreak Associated with a New Coronavirus of Probable Bat Origin. Nature 579(7798), 270–273. doi:10.1038/s41586-020-2012-7
Zhu, X., Liu, S., Wang, X., Luo, Z., Shi, Y., Wang, D., et al. (2018). Contribution of Porcine Aminopeptidase N to Porcine Deltacoronavirus Infection. Emerging Microbes & Infections 7(1), 1, 13. doi:10.1038/s41426-018-0068-3
Zhuang, M., Jiang, H., Suzuki, Y., Li, X., Xiao, P., Tanaka, T., et al. (2009). Procyanidins and Butanol Extract of Cinnamomi Cortex Inhibit SARS-CoV Infection. Antiviral Res. 82(1), 73–81. doi:10.1016/j.antiviral.2009.02.001
Zuniga, S., Cruz, J. L., Sola, I., Mateos-Gómez, P. A., Palacio, L., and Enjuanes, L. (2020). COVID-19 and the Cardiovascular System. Nat. Rev. Cardiol. 17(5), 259–260. doi:10.1038/s41569-020-0360-5
Keywords: SARS-CoV-2, ACE2 inhibitor, natural products, replication inhibitor, virus entry blocker
Citation: Kim C-H (2021) Anti–SARS-CoV-2 Natural Products as Potentially Therapeutic Agents. Front. Pharmacol. 12:590509. doi: 10.3389/fphar.2021.590509
Received: 01 August 2020; Accepted: 19 April 2021;
Published: 27 May 2021.
Edited by:
Jon Wardle, Southern Cross University, AustraliaReviewed by:
Armando Caceres, Universidad De San Carlos De Guatemala, GuatemalaRuchi Tiwari, Pranveer Singh Institute of Technology PSIT, India
Copyright © 2021 Kim. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Cheorl-Ho Kim, Y2hraW1iaW9Ac2trdS5lZHU=
 Cheorl-Ho Kim
Cheorl-Ho Kim