- 1Department of Pharmacology, Manipal College of Pharmaceutical Sciences, Manipal Academy of Higher Education, Manipal, India
- 2Department of Biomedical Engineering, School of Engineering and Technology, Central University of Rajasthan, Kishangarh, India
- 3Department of Pharmacology, All India Institute of Medical Sciences (AIIMS), Bathinda, India
Sirtuins, NAD + dependent proteins belonging to class III histone deacetylases, are involved in regulating numerous cellular processes including cellular stress, insulin resistance, inflammation, mitochondrial biogenesis, chromatin silencing, cell cycle regulation, transcription, and apoptosis. Of the seven mammalian sirtuins present in humans, Sirt6 is an essential nuclear sirtuin. Until recently, Sirt6 was thought to regulate chromatin silencing, but new research indicates its role in aging, diabetes, cardiovascular disease, lipid metabolism, neurodegenerative diseases, and cancer. Various murine models demonstrate that Sirt6 activation is beneficial in alleviating many disease conditions and increasing lifespan, showing that Sirt6 is a critical therapeutic target in the treatment of various disease conditions in humans. Sirt6 also regulates the pathogenesis of multiple diseases by acting on histone proteins and non-histone proteins. Endogenous and non-endogenous modulators regulate both activation and inhibition of Sirt6. Few Sirt6 specific non-endogenous modulators have been identified. Hence the identification of Sirt6 specific modulators may have potential therapeutic roles in the diseases described above. In this review, we describe the development of Sirt6, the role it plays in the human condition, the functional role and therapeutic importance in disease processes, and specific modulators and molecular mechanism of Sirt6 in the regulation of metabolic homeostasis, cardiovascular disease, aging, and neurodegenerative disease.
Introduction
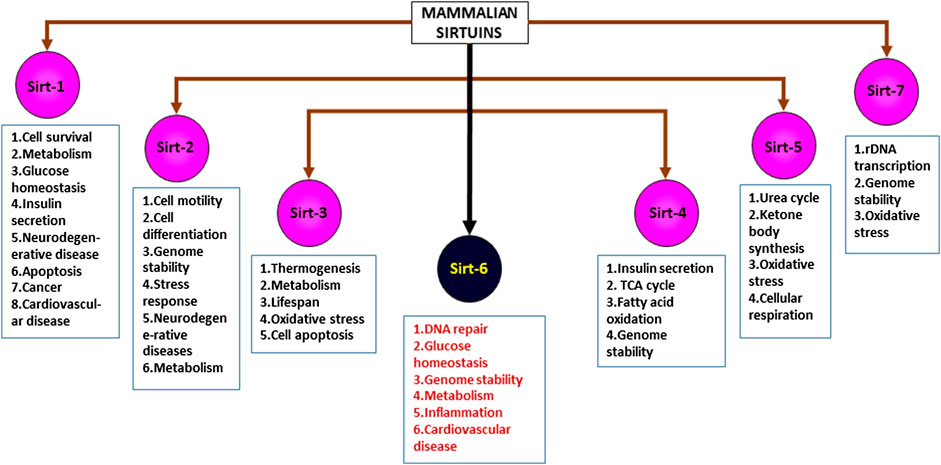
Sirtuins are energy linked NAD + dependent proteins that are activated during calorie restriction (Corbi et al., 2012; Rack et al., 2015; Bheda et al., 2016; Yu et al., 2018), mainly altering the acetylation/deacetylation status of histones, thus regulating chromatin silencing. Proteomic studies suggest that human cells possess histones with various lysine acyl modifications and these sirtuins have specific deacetylating activities that remove acyl moieties from lysine, thus modulating gene expression, without altering the gene sequence itself (Kupis et al., 2016). This unique property of sirtuins and their ability to bind to various substrates and other moieties bearing acyl modifications allow us to examine different mechanisms involved in multiple diseases, thus showing the importance of sirtuins (Carafa et al., 2012). Sirtuins are epigenetic regulators, belonging to class III Histone Deacetylases (HDACs) (Rack et al., 2015) which are known to affect multiple pathways involved in various disease conditions including cancer, diabetes, cardiac failure, hypertrophy, cachexia, pulmonary fibrosis, aging etc. (Dryden et al., 2003; Vassilopoulos et al., 2011; Lee and Goldberg, 2013; Lee and Gu, 2013; Bheda et al., 2016; Kupis et al., 2016; Graham et al., 2018; Kanwal and Dsouza, 2019) These energy linked proteins were first discovered in the yeast cells, as silencing information regulator-2 (Sir-2) (Haigis and Sinclair, 2010; Grabowska et al., 2017; Graham et al., 2018; Rajabi et al., 2018). Orthologues of Sir-2 in mammals are known as sirtuins (Polito et al., 2010). In humans, sirtuins are divided into seven types (Figure 1, Mammalian sirtuins and their functions) (Sirt1- Sirt7), localized in different cellular components, that has crucial role in various cellular processes (Kobayashi et al., 2005; Haigis and Sinclair, 2010; Vassilopoulos et al., 2011; Hoffmann et al., 2014; Wu et al., 2015; Tang, 2016; Dai et al., 2018; Graham et al., 2018; Wang et al., 2018; Kanwal, 2018). Significant differences exist between sirtuin homologues in their variable N and C terminal extensions with conserved C-terminal extensions. Among the 7 types of sirtuins, Sirt6 is an essential sirtuin in humans that is localized in the nucleus. Like other sirtuins, Sirt6 is also a stress responsive protein deacetylase, but unlike other sirtuins, in addition to its deacetylase activity, it also transfers ADP ribosyl via mono-ADP ribosyltransferase enzyme. In humans, Sirt6 has a plethora of functions, including DNA repair, telomerase function, genomic stability, cellular senescence, and metabolic homeostasis (Figure 2: Clinical significance of Sirt6 in various diseases) (Michishita et al., 2008; Watroba and Szukiewicz, 2016). Until recently, Sirt6 was mainly known for chromatin signaling. Recent data indicate that Sirt6 is involved in various disease conditions mentioned above (Khan R. I. et al., 2018); due to their action on multiple substrates and catalytic sites (Tasselli et al., 2017). Studies indicate that they control activities of p53, FOXO proteins, NF-KB, PGC-1α, PARP1, TNFα, GCN5, HIF1α, which are involved in the pathogenesis of various diseases (Kawahara et al., 2009; Dominy et al., 2012; Zwaans and Lombard, 2014). This review focuses on Sirt6, its mechanism(s) in multiple disease states, its significance in human health, and as a therapeutic target in drug discovery.
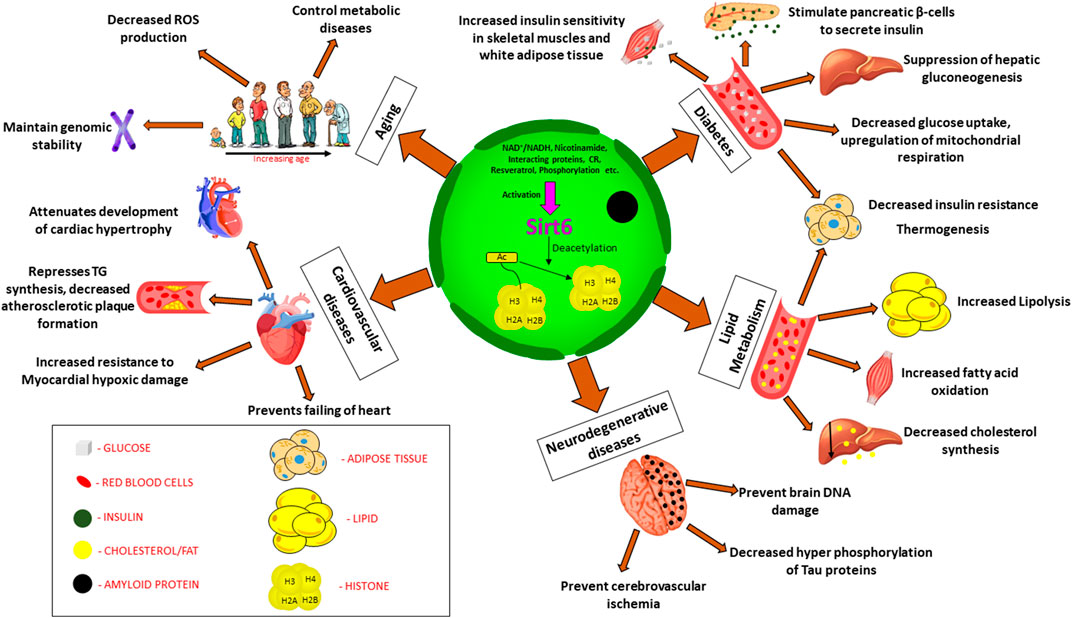
FIGURE 2. Clinical significance of Sirt6 in various diseases. Sirt6 is a nuclear sirtuin activated by CR, NAD+, resveratrol etc., promoting deacetylation of histone which plays a major role in (a) Diabetes: increased insulin sensitivity, insulin secretion, suppress gluconeogenesis, decrease glucose uptake. (b) Lipid metabolism: increased lipolysis, increased fatty acid oxidation, decreased cholesterol synthesis. (c) Neurodegenerative diseases: decreased phosphorylation of Tau proteins, prevent cerebrovascular ischemia and brain DNA damage (d) Cardiovascular diseases: decrease TG synthesis, attenuates cardiac hypertrophy, prevents failing of heart, resistance to hypoxic damage (e) Aging: decreased ROS production, maintain genomic stability, control metabolic diseases. Abbreviation: Ac, Acetylated; CR, calorie restriction; TG, triglycerides; ROS, reactive oxygen species.
Sirt6: Importance and Recent Development
Sirt6 is a NAD+ dependent nuclear histone deacetylase, having deacetylase, deacylase and mono-ADP ribosyltransferase activity. In the past 5 years, the development of Sirt6 suggests a crucial role in a broad spectrum of metabolic processes. Sirt6 has a significant role in maintaining genetic stability and DNA repair, by activating several DNA-repair genes (Mao et al., 2011; Khan R. I. et al., 2018). It was identified as a suppressor of genomic instability due to its association with chromatin through its ability to modulate base excision-pair repair and double strand break repair (Kugel and Mostoslavsky, 2014). In addition to maintaining genetic stability, it also plays a crucial role in intermediary metabolism including glucose metabolism, lipid metabolism, circadian metabolism, etc. (Watroba and Szukiewicz, 2016; Tasselli et al., 2017). Studies in the recent past have also shown implications of Sirt6 in various diseases including dyslipidemia, diabetes, heart disease, cancer, neurodegenerative diseases, brain aging etc. (Roichman et al., 2016; Xiwen et al., 2016; D’Onofrio et al., 2017). Sirt6 is a longevity protein; various studies have demonstrated its involvement in increasing life expectancy (Schumacher, 2011; Kanfi et al., 2012; Giblin and Lombard, 2016; Schumacher, 2017). Overexpression of Sirt6 in transgenic mice resulted in significantly longer half-life compared to wild-types, and that IGF-1 plays a crucial role in regulating lifespan (Kanfi et al., 2012). Sirt6 blocks the action of IGF-1, which is responsible for increasing lifespan (Sundaresan et al., 2012). There are two main theories that postulate how Sirt6 regulates longevity, they are: maintenance of genetic stability and regulation of metabolism, and various studies show that either of these theories contribute to Sirt6’s function in longevity (Mostoslavsky et al., 2006; Michishita et al., 2008; Kanfi et al., 2012). Sirt6 knockout mice died within a month of birth due to the development of significant metabolic abnormalities due to deficiency of IGF-1 (Watroba and Szukiewicz, 2016). Lack of IGF-1 was correlated with a decrease in adipose tissue, lordokyphosis, and severe hypoglycemia (Lombard et al., 2008; Peshti et al., 2017; Ferrer et al., 2018). Another study reveals the role of Sirt6 in longevity by the maintenance of genetic stability (Tian et al., 2019). The mechanism involved is double-strand break repair of DNA; an activity controlled by Sirt6 resulted in an increase in lifespan in animal models (Mostoslavsky et al., 2006; Lombard et al., 2008; Tian et al., 2019). Macaque monkeys that lacked the gene for Sirt6 died a few hours after the birth and exhibited prenatal developmental retardation (Zhang W. et al., 2018). In this study, it was observed that the brain was underdeveloped, suggesting low levels of Sirt6 can lead to neurodegenerative diseases and brain aging (Zhang W. et al., 2018; Naiman and Cohen, 2018; Niu, 2019). Sirt6 is necessary in human development as shown by a study that the deletion of Sirt6 in humans can cause perinatal lethality (Ferrer et al., 2018). Several other studies have highlighted telomeres’ role in aging and its association with Sirt6 (Aubert and Lansdorp, 2008; Tennen et al., 2011). With aging, the length of the telomere declines, but Sirt6 by its deacetylation activity, maintain the length of telomeres, thus preventing telomere sequence loss (Aubert and Lansdorp, 2008; Cacchione et al., 2019; Cao et al., 2019). Cells lacking Sirt6 had malformed telomere structure and also replication associated sequence loss of telomeres (Aubert and Lansdorp, 2008). Apart from telomeres, mammalian aging and aging related disorders are also associated with abnormal IGF-Akt signaling, which in turn is controlled by Sirt6 (Pillai et al., 2014). Heart failure is one of the age-related disorder in mammals, where the IGF-Akt on sustained activation promotes hypertrophy and heart failure (Wang et al., 2015; D’Onofrio et al., 2017; Lee and Kim, 2018), whereas Sirt6 impedes IGF-Akt signaling via c-Jun by deacetylation of H3K9 (Sundaresan et al., 2012). Few studies found that failing hearts of humans and mice showed decreased levels of Sirt6, suggesting that low levels of Sirt6 increase the activity of IGF-Akt, leading to initiation and progression of cardiac hypertrophy and heart failure (Sundaresan et al., 2012; Zhang D. et al., 2018). Osteoporosis is yet another age-related disorder in mammals, and recent studies have found that Sirt6 prevents osteoporosis, but the mechanism remains unclear. Sirt6 knockout mice suffering from osteopenia, exhibited more significant bone loss than the non-mutant mice (Zhang D. et al., 2018), indicating that Sirt6 was involved in decreased osteoclast activation (Zhang D. et al., 2018; Wang and Mbalaviele, 2019).
Substrates for Sirt6
Sirt6 functions are diversified (Pan et al., 2011; Kuang et al., 2018). They include different molecular pathways associated with glycolysis, DNA repair, gluconeogenesis, cardiac hypertrophic responses, neurodegeneration, and tumorigenesis due to its activity on varied number of substrates, which include PARP1, TNFα, GCN5, HIF1α, etc. (Liu et al., 2012; Sundaresan et al., 2012; Li et al., 2017; Kuang et al., 2018; Khan R. I. et al., 2018; Yang H. et al., 2019). HIFs are transcription factors that are expressed as regulators of genes during cellular deprivation of oxygen. Overexpression of HIFs is implicated with tumor growth and metastasis, and is involved in initiating angiogenesis (Jun et al., 2017; Pezzuto and Carico, 2018). Sirt6 can inhibit these activities of HIF. Thus activation of Sirt6 controls tumor growth and metastasis by regulating the overexpression of HIF1α (Zhong et al., 2010; Zwaans and Lombard, 2014; Yang Z. et al., 2019).
TNF-α, a Sirt6 substrate, is a proinflammatory cytokine that is involved in various inflammatory pathological processes (Ravussin and Smith, 2016; Josephs et al., 2018), and increased expression of Sirt6 inhibits TNF-α (He et al., 2017). Although Sirt6 is essential in the deacetylation process, the de-fatty acylation, specifically the hydrolysis of lysine units at the 19 and 20 positions of H3 histone, regulates TNF-α secretion (Jiang et al., 2016). Catalysis of fatty acyl lysine hydrolysis by Sirt6 is more efficient when compared to deacetylation (Jiang et al., 2013). Sirt6 has a significant role in chronic inflammation due to its association with TNF-α. Cells treated with TNF-α had decreased levels of Sirt6 in a dose dependent manner, identifying a symbiotic relationship between TNF-α and Sirt6 (Yeo et al., 2017). In addition to the above, Sirt6 attenuates inflammatory response through inhibition of NF-κB signaling (Li Z. et al., 2018; Santos-Barriopedro et al., 2018; Santos-Barriopedro and Vaquero, 2018).
Sirt6 binds to and activates GCN5, thereby inhibiting the acetylation of PGC-1α (Peroxisome Proliferator activated Receptor coactivator one alpha) and decreasing gluconeogenic gene expression (Giblin and Lombard, 2016; D’Onofrio et al., 2017; Kuang et al., 2018; De Céu Teixeira et al., 2019). This correlation between Sirt6 levels and glucose metabolism is a key in understanding involvement of Sirt6 in the pathogenesis in diabetes mellitus and a potential therapeutic target in the management of the disease (Dominy et al., 2012; D’Onofrio et al., 2017; Kuang et al., 2018).
Sirt6 deacetylase, along with FoxO3a, can reduce LDL-cholesterol levels by regulating the expression of PCSK9 (Proprotein Convertase Subtilisin/Kexin Type 9) (Chaudhary et al., 2017). PCSK9 is a gene that regulates LDL receptors, which in turn is involved in decreased LDL clearance from the circulation in the hepatocytes (Spolitu et al., 2019). FoxO3 recruits NAD+ -dependent Sirt6 deacetylase, to the promoter region of the PCSK9 gene for the deacetylating histone H3 at lysine 9 and 56, which resulted in suppression of the expression of PCSK9 gene, thereby promoting LDL receptors and subsequent hepatic clearance of pathogenic LDL (Tao et al., 2013a; Gao et al., 2018). Activation of Sirt6 inhibited PCSK9, thus positively regulating hypercholesterolemia. Therefore Sirt6 could be a potential target in the treatment of dyslipidemia (Tao et al., 2013a; Chaudhary et al., 2017; Glerup et al., 2017; Handelsman and Lepor Norman, 2018; Spolitu et al., 2019).
The activity of Poly (ADP-ribose) polymerase 1 (PARP1) is increased when acted upon by Sirt6 where it undergoes mono (ADP-ribosylation). This enzyme is involved in the modification of nuclear proteins, responsible for DNA repair and proliferation, differentiation and tumor transformation (Xu et al., 2015; Rizzo et al., 2016; Van Meter et al., 2016; Tang, 2017; Fujimoto et al., 2017; Yang et al., 2018). PARP-1 is also an essential regulator of Apoptosis Inducing Factor (AIF) mediated cell death (Song Y. et al., 2016). The expression levels of this protein were low under Sirt6 deficient conditions. Levels of PARP-1 were reduced faster under Sirt6 defective conditions (Zhang Q. et al., 2019).
Recently attention has been focused on the role of Thioredoxin-interacting protein (TXNIP), as overexpression of this protein is negatively associated with the insulin secretion by β-cells of the pancreas (Alhawiti et al., 2017; Nagaraj et al., 2018). Sirt6 influences the expression of TXNIP i.e. overexpression of Sirt6 inhibits TXNIP, thus making it an important therapeutic target (Kuang et al., 2018; Qin et al., 2018). In addition to the above, Sirt6 has been reported to act on various other substrates that could potentially be implicated in multiple other diseases for which a better understanding of the molecular mechanism is required (Tasselli et al., 2017; Khan R. I. et al., 2018; Gertman et al., 2018).
Sirt6 and Diseases
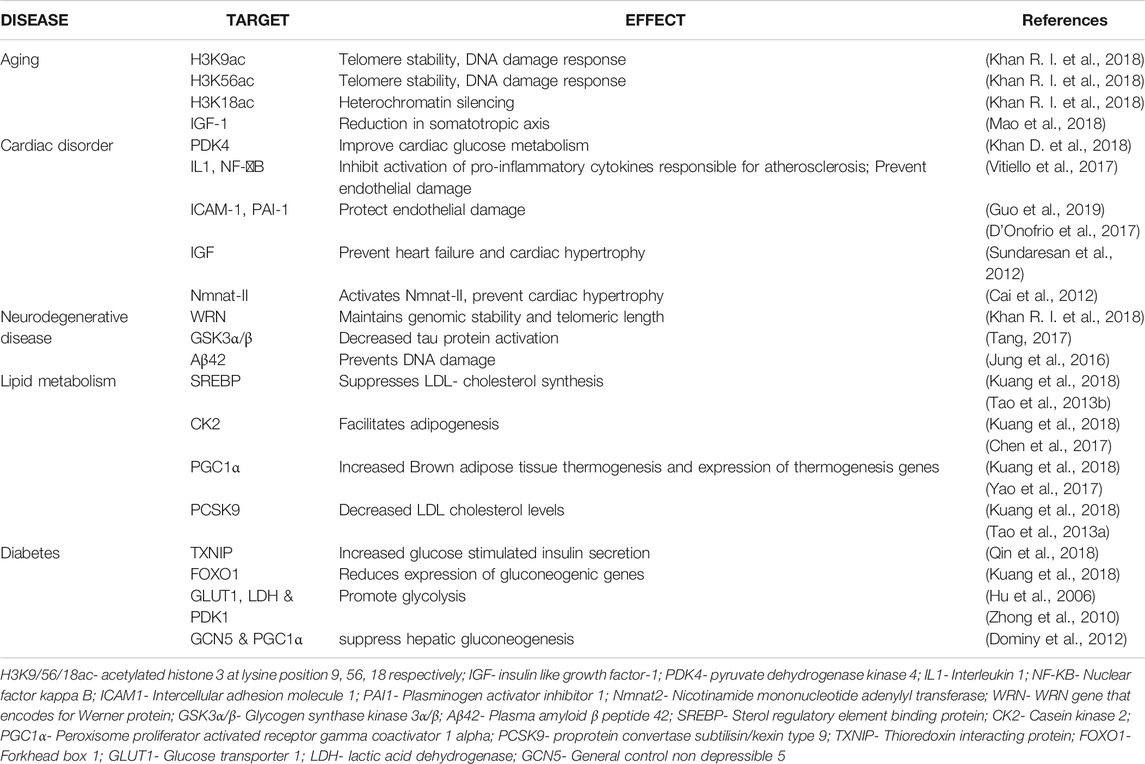
As we delve into the cellular level, Sirt6 deprivation leads to several changes in sensitivity to reactive oxygen species, glucose metabolism, and genomic stability (Mostoslavsky et al., 2006; Kanfi et al., 2012; Li et al., 2017; Peshti et al., 2017; Xu et al., 2019; Yepuri and Ramasamy, 2019). Mice placed on caloric restriction or Sirt6 activators overexpress Sirt6, improving cancer, and age-related disorders in animal models (Zhang et al., 2016a; Kuang et al., 2018; Iachettini et al., 2018; Rahnasto-Rilla et al., 2018), on the contrary, lower Sirt6 levels in mice showed shorter life expectancy, cancer occurrence, diabetes and other metabolic disorders increased. The animals also exhibited other complications such as curved spines, decreased subcutaneous fat, hypoglycemia, and lowered levels of IGF-1 (Peshti et al., 2017; Khan R. I. et al., 2018; Ghosh et al., 2018; Simon et al., 2019). This suggests that Sirt6 is a therapeutic target in aging, cardiac disorders, neurodegenerative disorders, and metabolic disorders (Rodgers and Puigserver, 2006; Serravallo et al., 2013; Demir et al., 2017; Harlan et al., 2019). Table 1.
Sirt6 in Aging
Aging is a complex, multifactorial process resulting in the accumulation of diverse harmful changes in the cell, increasing risk of disease and death. Multiple theories can explain the aging process but none can be considered absolute (Tosato et al., 2007; Jin, 2010; Sergiev et al., 2015). These include evolutionary theory, Free radical theory, Mitochondrial theory, Gene regulation theory, Telomere theory, Inflammation hypothesis, Immunity theory, Neuroendocrine theory, Neuroendocrine- immune method theory, and Caloric restriction (Wei et al., 2001; Gavrilov and Gavrilova, 2002; Tosato et al., 2007; López-Lluch and Navas, 2016). In addition to these theories, sirtuins too contribute to aging. Initially, sirtuins were first identified in yeast, which led them to discover its life prolonging activity. Studies in the worm flies confirmed the link between sirtuins and aging (Finkel et al., 2007; Guarente, 2007). Of the seven mammalian sirtuins, Sirt6 is pivotal in regulating lifespan (Kanfi et al., 2012; Hirvonen et al., 2017; Peshti et al., 2017). It promotes chromatin changes essential for DNA repair and maintenance of telomere structure, preventing genomic instability, and cellular senescence (Tasselli et al., 2017). The DNA repair mechanism in longevity is Double-Strand Break repair, where Sirt6 performs this function more efficiently (Tian et al., 2019). Sirt6 is thought to have a protective action on telomeres, as deficiency of Sirt6 causes a loss of telomere sequence associated with replication, leading to genomic instability and early cell death (Watroba and Szukiewicz, 2016; Naiman and Cohen, 2018). Furthermore, Sirt6 maintains redox homeostasis in mesenchymal stem cells, thus suggesting that it regulates longevity (Liao and Kennedy, 2016; Pan et al., 2016). IGF-1 is yet another factor that is related to aging. Lower levels of IGF-1 delayed the process of aging. Transgenic mice models that overexpressed the Sirt6 gene had low levels of IGF-1, slowing the aging process in mice (Kanfi et al., 2012; Ravi et al., 2019). Thus, Sirt6 can be targeted for therapeutic interventions in aging and aging related diseases (Khan R. I. et al., 2018).
Sirt6 in Cardiac Disorders
Congestive heart failure and coronary artery diseases are the two most common types of cardiac complications associated with Sirt6 activity (Bindu et al., 2016; D’Onofrio et al., 2017; Li et al., 2017; Guo et al., 2019; Yepuri and Ramasamy, 2019). Deficiency of Sirt6 in cardiomyocytes results in the accumulation of lactate due to impaired glucose oxidation, leading to various comorbidities relating to the heart, including heart failure (Khan D. et al., 2018). Sirt6 heterozygous mice were used to show lactic acid accumulation in mice hearts. Sirt6 deficiency increased FOXO1 localization in heart, upregulating PDK4, reducing oxygen consumption and ATP production, thereby demonstrating the protective role of Sirt6 in maintaining cardiac homeostasis (Khan D. et al., 2018). Another factor involved in the development of cardiac disorders is the formation of atherosclerotic plaque. No evidence suggests a direct link between Sirt6 and atherosclerotic plaque formation (Zi et al., 2019). However, Sirt6 deficiency enhances the expression of pro-inflammatory cytokines like Interleukin-1 and transcriptional signaling of NF-KB (Lappas, 2012; Yu et al., 2013), which are indirectly linked to the pathogenesis of atherosclerosis (Xu et al., 2016). In addition, Sirt6 depletion increased expression of ICAM-1 and PAI-1 and upregulation of p21 gene and reduction of eNOS (Figure 3, Schematic representation of molecular mechanism of Sirt6 in various diseases) further leading to atherosclerotic vascular disease (Yepuri and Ramasamy, 2019; Zi et al., 2019). Sirt6 is also involved in the pathogenesis of cardiomyocyte hypertrophy (Yu et al., 2013; Lu et al., 2016; Ravi et al., 2019; Kanwal et al., 2019b), which includes angiotensin-II induced and IGF-Akt signaling induced cardiac hypertrophy (Li et al., 2017). Increased expression of Sirt6 in cardiomyocytes decreased angiotensin-II action on cardiomyocytes (Yu et al., 2013; Zhang et al., 2017; Eguchi et al., 2018; Ianni et al., 2018). Angiotensin-II induced cardiac hypertrophy is blocked by over-expression of nicotinamide mononucleotide adenylyltransferase-II (Nmnat-II) that is responsible for activation of Sirt6 (Vitiello et al., 2017). Another pathway involved in cardiac hypertrophy is mediated through increased activation of IGF-Akt pathway (Yan et al., 2019) (Figure 3: Schematic representation of molecular mechanism of Sirt6 in various diseases), which was shown in Sirt6 deficient mice (Sundaresan et al., 2012; Vitiello et al., 2017).
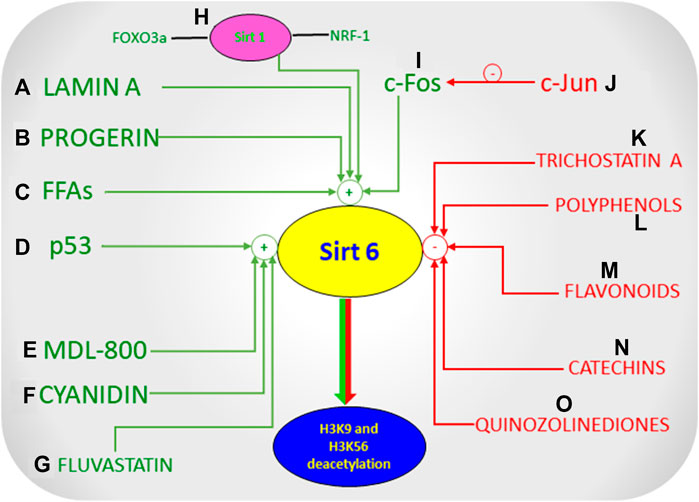
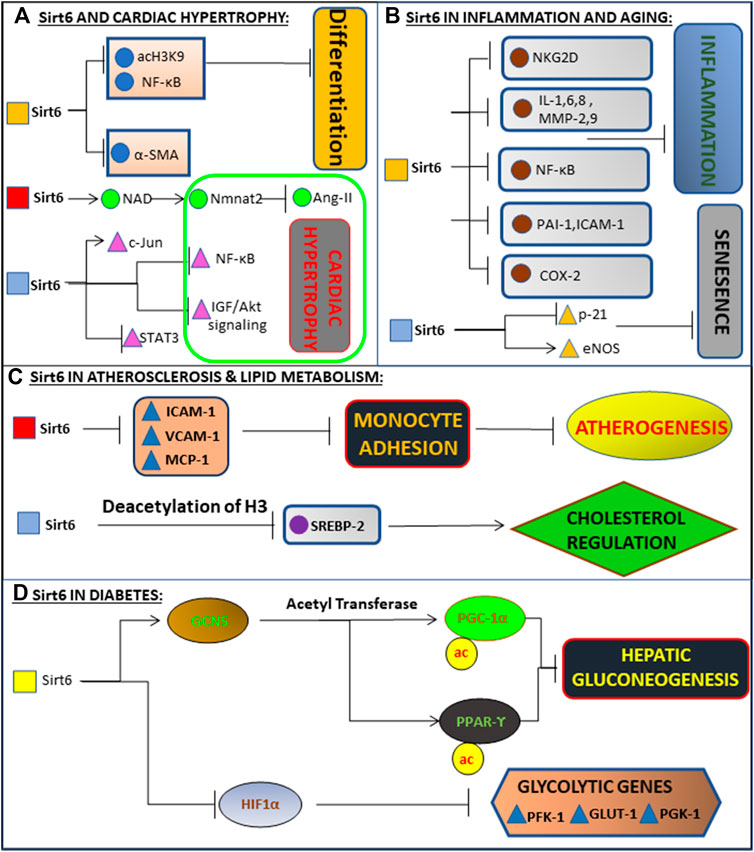
FIGURE 3. Schematic representation of molecular mechanism of Sirt6 in various diseases. (A) Deacetylates H3K9, inhibiting NF-kB and differentiation of cardiac cells into myofibroblasts. Also binds to c-Jun, promoting deacetylation of H3K9, inhibiting the expression of NF-kB and IGF-Akt signalling. Inhibits activation of Ang 2 and STAT3 via Nmnat1 and NAD, together preventing Cardiac Hypertrophy and subsequently Heart Failure. (B) Exerts anti-inflammatory action in the endothelial cells via blockade of NF-kB, cytokines (i.e., IL-1, IL-6, IL-8), metalloproteinases (MMP-2, MMP-9), PAI-1, ICAM-1, and COX-2. Delays senescence by inhibiting p21 signalling and maintaining high levels of eNOS. (C) Prevents monocyte adhesion and Atherogenesis by inhibiting pro inflammatory mediators (ICAM-1, VCAM-1, MCP-1). Regulates Cholesterol levels by deacetylating H3K9 and inhibiting SREBP gene. (D) Enhances activity of GCN5, leading to down-regulation of gluconeogenesis-related enzymes and inhibiting gluconeogenesis via acetylation of PGC-1α and activation of PPARγ. It inhibits glycolysis by inhibiting glycolytic genes (PFK-1, GLUT-1, PGK-1) via inhibition of HIF-1α.
Sirt6 in Neurodegenerative Diseases and Brain Aging
Alzheimer’s disease (AD) is involved in neurodegeneration, and characterized by dementia. AD is pathologically characterized by formation of beta amyloid plaques and neurofibrillary tangles known as tau proteins (Kocahan and Doğan, 2017; Takahashi et al., 2017). Oxidative stress, cell senescence, and aging are the major risk factors for the progression of this disease (Markesbery, 1999; Huang et al., 2016; Kerchner and Wyss-Coray, 2016; Xia X. et al., 2018; Butterfield and Boyd-Kimball, 2018; Trevisan et al., 2019). During the pathogenesis of AD, telomere length has a causal role. As cells divide, telomere length shortens, and this shortening is associated with cognitive impairment, amyloid plaque deposition and hyper-phosphorylation of tau protein that are characteristics of AD (Cai et al., 2013; Forero et al., 2016; Liu et al., 2016). Oxidative stress affects telomeres as they contain guanine, which undergoes oxidation to produce 8-oxo-7,8-dihydro-2-deoxyguanosine (8-oxodG) (Kawanishi and Oikawa, 2004; Cai et al., 2013). Sirt6 maintains telomere function as it prevents telomere dysfunction through WRN protein stabilization at telomeric chromatin (Mohamad Nasir et al., 2018). In addition to telomere maintenance, Sirt6 regulates tau protein stabilization during AD oxidative stress. Activation of Sirt6 maintains both genomic stability in the brain and leads to loss of tau protein stability via inhibition of GSK3α/β (Kaluski et al., 2017; Stein and Toiber, 2017; Kim H. et al., 2018). Sirt6 reduction alters DNA repair in the brain of an AD mouse model, and Sirt6 overexpression prevents Amyloid beta protein (Aβ42) induced DNA damage, thereby proving the beneficial effects of Sirt6 in AD (Jung et al., 2016; Tang, 2017). This is confirmed by the fact that the levels of Sirt6 are reduced in the brains of both AD containing mice as well as AD patients. In addition, Aβ42 decreased Sirt6 levels (Kaluski et al., 2017; Cacabelos et al., 2019). Sirt6 is associated with age related disorders. Activation of Sirt6 is protective in AD and other neurodegenerative disorders involving brain aging, thus proving to be an essential therapeutic target in the treatment of neurodegenerative disorders (Naiman and Cohen, 2018).
Sirt6 in Lipid Metabolism
Lipid metabolism disorders are pervasive and play a vital role in the pathogenesis of atherosclerosis, leading to cardiovascular diseases (Parhofer, 2016; Schofield et al., 2016). Recently Sirt6 has been shown to have a critical role in lipid metabolism (Ye et al., 2017; Assadi-Porter et al., 2018; Kuang et al., 2018) as it is involved in the regulation of fatty acid synthesis, triglyceride synthesis, cholesterol synthesis, fatty acid beta oxidation, lipolysis, adipogenesis and thermogenesis (Liu et al., 2012; Elhanati et al., 2013; Tao et al., 2013a; Tao et al., 2013b; Chen et al., 2017; Yao et al., 2017; Kuang et al., 2018; Gao et al., 2019). Lipid homeostasis and cholesterol biosynthesis are regulated by a transcription factor called Sterol regulatory binding proteins (SREBP), and the expression of SREBP is controlled by Sirt6 (Eberle et al., 2004). SREBP activation is also implicated in inflammation, autophagy, endoplasmic reticulum stress (Shimano and Sato, 2017). Sirt6 inhibits the expression of SREBP, which is indirectly linked with cholesterol biosynthesis, hence regulating Cholesterol homeostasis (Tao et al., 2013b; Ye et al., 2017). Sirt6 negatively regulates cholesterol biosynthesis by various pathways involving SREBP. It causes downregulation of SREBP by reducing mRNA expression (Kuang et al., 2018). Sirt6 is also recruited to the Srebp gene promoter (Figure 3: Schematic representation of molecular mechanism of Sirt6 in various diseases) by FOXO3, where it deacetylates H3K9 and H3K56 in the promoter regions of Srebp and suppresses the transcription levels of Srebp and its target genes (Tao et al., 2013b; Elhanati et al., 2013; Kugel and Mostoslavsky, 2014). It also inhibits SREBP’s subsequent conversion into active forms (Elhanati et al., 2013; Kugel and Mostoslavsky, 2014) and activates AMPK, leading to the phosphorylation and inactivation of SREBP (Kugel and Mostoslavsky, 2014; Kuang et al., 2018). The absence of Sirt6 leads to increased production of Triglycerides due to increased expression of genes involved in Triglyceride synthesis (Ye et al., 2017). The genes responsible for Fatty acid metabolism by beta oxidation is downregulated (Ye et al., 2017; Kuang et al., 2018). Sirt6 also regulates adipogenesis and thermogenesis, it is an essential factor in adipogenesis, by enhancing casein kinase 2 (CK2) activity (Chen et al., 2017; Kuang et al., 2017; Kuang et al., 2018). Similarly, expression of PGC-1α is a central regulator in thermogenesis. Overexpression of PGC-1α causes mitochondrial oxidative phosphorylation and expression of thermogenic genes. Sirt6 controls expression of PGC-1α, as depletion of Sirt6 decreases the appearance of PGC-1α, thus resulting in decreased thermogenesis (Yao et al., 2017; Kuang et al., 2018; Singh et al., 2020).
Sirt6 and Diabetes
The prevalence of Type 2 Diabetes Mellitus (T2DM) is increasing at an alarming rate worldwide. With an increased understanding of the pathogenesis of T2DM various new therapeutic approaches are being developed to target the fundamental cause of T2DM, Sirt6 being one among them (Kitada et al., 2013; Bae, 2017; Chellappan et al., 2018). The pathophysiology of T2DM is characterized by many causes (Mahler and Adler, 1999). The primary cause of T2DM is decreased sensitivity of beta cell functioning to the levels of glucose, but recent studies have proven the crucial role Sirt6 in glucose stimulated insulin secretion (GSIS), enhancing the release of insulin (Mahler and Adler, 1999; Song M. Y. et al., 2016; Xiwen et al., 2016) (Figure 3: Schematic representation of molecular mechanism of Sirt6 in various diseases). Various studies show different mechanisms of how Sirt6 is involved in increasing beta cell function. One such pathway is Sirt6 suppresses expression of the thioredoxin-interacting protein (TXNIP), which is engaged in β-cell apoptosis (Shalev, 2014; Qin et al., 2018). Thus, Sirt6 maintains the functioning of beta cells. Another pathway where Sirt6 supports GSIS functioning of beta cells is via regulation of FOXO1 expression. Sirt6 inhibits FOXO1, maintaining the glucose-sensing ability of pancreatic β-cell and systemic glucose tolerance (Song M. Y. et al., 2016). Hence Sirt6 has proven to be a chief regulator in glucose homeostasis (Zhong et al., 2010; Gertman et al., 2018). Studies done on mice have been conclusive in showing that knockdown of Sirt6 can lead to complications, namely severe hypoglycemia leading to death (Lee et al., 2017; Kuang et al., 2018). The primary reason for this were increased uptake from the muscle and adipose tissue rather than intestinal uptake of glucose or increased secretion from the kidneys (Kuang et al., 2018). There is both in vitro and in vivo evidence, showing that increased glucose uptake may be due to deficiency of Sirt6 (Zhong et al., 2010; Zhong and Mostoslavsky, 2010). Sirt6 suppresses HIF1α (hypoxia inducible factor-1α), which is responsible for suppressing several genes like GLUT-1, LDH, and PDK-1, which coordinate various processes involving glucose metabolism such as glycolysis (Kim et al., 2006; Zhong et al., 2010; Laemmle et al., 2012; Khan D. et al., 2018; Kuang et al., 2018). Growth hormone and IGF-1 signaling alter the metabolism of glucose (Sundaresan et al., 2012; Takasaka et al., 2014). Sirt6 controls gluconeogenesis by the receptor PGC-1α and p53/FOXO1 signaling. Inhibition of PGC-1α activity by Sirt6 occurs via deacetylation of GCN5, increasing its acetyltransferase activity, which is a form of histone acetylation and thereby increases the acetylation of PGC-1α which leads to inhibition of hepatic gluconeogenesis and thereby hyperglycemia (Figure 3: Schematic representation of molecular mechanism of Sirt6 in various diseases) (Jeninga et al., 2010; Satoh and Imai, 2014; Sharabi et al., 2017; Kanwal and Dsouza, 2019; Singh et al., 2019).
Sirt6 Modulators
Sirt6 has a plethora of biological activities, making it a vital molecule, allowing researchers to identify Sirt6 modulators (Figure 4, Modulators of Sirt6) in developing effective therapeutic approaches to a broad spectrum of diseases. Some endogenous activators of Sirt6 include Lamin A and long chain free fatty acids (Feldman et al., 2013; Rahnasto-Rilla et al., 2016; Ghosh, 2019). Lamin A increases the deacetylation activity of Sirt6 and Sirt1 by directly interacting with the deacetylating proteins (Ghosh et al., 2013; Ghosh et al., 2015). Free fatty acids were also seen to increase the deacetylate ing activity of Sirt6. Free fatty acids stimulated Sirt6 deacetylase activity, where acyl group binding pocket binds to free fatty acids, splaying Sirt6 subdomains, thus stimulating deacetylase activity (Feldman et al., 2013). In addition to the above endogenous activators, Sirt6 is activated by CR (Caloric restriction), c-Fos protein, p53 and increased intracellular levels of NAD+ (Kugel and Mostoslavsky, 2014; Zhang et al., 2014; Zhang et al., 2016b; Li M. et al., 2018; Khan R. I. et al., 2018; Kuang et al., 2018; De Céu Teixeira et al., 2019). Although CR and NAD + activates all isoforms of sirtuins, free-fatty acids and c-Fos protein selectively activates Sirt6. Furthermore, other endogenous activators that indirectly activate Sirt6 include increased Sirt1, FoxO3a, and Nrf-1 (Tosato et al., 2007; D’Onofrio et al., 2017). Pyrrolo (1,2-α) quinoxaline derivatives are among the first discovered Sirt6 activators (Hassanieh and Mostoslavsky, 2018). UBCS039 is another activator of Sirt6 which is one of the first synthetic and specific activators of the same. It has been known to cause Sirt6 specific Histone H3 deacetylation and accentuate autophagy in various types of cancer cells thereby showing its tumor suppressor effects (Iachettini et al., 2018). Another novel Sirt6 activator, CL5D also regulates the process of Histone deacetylation but its exact mechanism and clinical relevance has not yet been clearly elucidated (Klein et al., 2020).
Polyphenols, such as quercetin and luteolin at higher concentrations activate Sirt6, whereas inhibit at lower levels. Thus polyphenols modulate Sirt6 activity in a concentration dependent manner (Rahnasto-Rilla et al., 2016; Rahnasto-Rilla et al., 2018; Heger et al., 2019). Cyanidin, another polyphenol, is a potent activator of Sirt6 producing 5–15 folds increase in Sirt6 activity compared to other polyphenols (Rahnasto-Rilla et al., 2018). These compounds are non-specific modulators of Sirt6, and are able to modulate the activities of other sirtuin isoforms. Selective small molecule activators of Sirt6 like MDL-800 bind to the allosteric site, increasing Sirt6’s deacetylase activity (Klein et al., 2017; You et al., 2017; Huang et al., 2018). This binding led to a significant and overall increase in deacetylation of H3K9ac and H3K56ac in HCC (human-hepatocellular-carcinoma cells) which prevented the proliferation and differentiation of HCC cells through cell cycle arrest thus proving that the activation of Sirt6 is crucial in the treatment of Hepatocellular Carcinoma (Huang et al., 2018). Fluvastatin, which competitively inhibits HMG CoA reductase, reduces cholesterol synthesis and is yet another activator of Sirt6 (Kim J. H. et al., 2018). Exposure of fluvastatin to HepG2 cells increased Sirt6 expression (Kim J. H. et al., 2018; Zhang C. et al., 2019). The mechanism underlying cholesterol regulation when fluvastatin increased Sirt6 expression was via phosphorylation of AMPKα and SREBP-1 pathway (Kim J. H. et al., 2018). Apart from activators, many small molecule Sirt6 inhibitors have been developed over time that directly acts on Sirt6 (Liu and Zheng, 2016). 2,4-dioxo-N-(4-(pyridin-3-yloxy) phenyl)-1,2,3,4-tetrahydroquinazoline-6-sulfonamide, a Sirt6 inhibitor, improved glucose tolerance in mice, and reduced insulin, triglycerides, and cholesterol levels indicating that a Sirt6 inhibitor could improve glycemic control in T2DM (Sociali et al., 2017; Khan R. I. et al., 2018). Trichostatin A (TSA) is an inhibitor of Sirt6 that selectively inhibits Sirt6 and no other mammalian sirtuins (Parenti et al., 2014; Wood et al., 2018; You and Steegborn, 2018). A study showed that TSA inhibited Sirt6 thus inhibiting deacetylation of p53 at lysine 382 (Wood et al., 2018), thus providing a lead compound in development of Sirt6 specific inhibitors in regulating apoptosis and stress resistance (Zhao et al., 2019). In addition to the above inhibitors, specific peptides and pseudo peptides including SDK (thioAc)TM21, HKK(thioAc)LM21, AKK(thioAc)LM21 were also studied for their ability to inhibit Sirt6 activity (Kokkonen et al., 2012; Rahnasto-Rilla et al., 2018). Derivatives of Quinazolinedione were recently discovered to inhibit Sirt6 activity, and these compounds were seen to sensitise the tumour cells to chemotherapeutic agents (Sociali et al., 2015; Rahnasto-Rilla et al., 2018). Their activity of inhibition of Sirt6 would have represented a potential therapeutic approach in the treatment of cancer, but this is not entirely true because Sirt6 acts as a double-edged sword in cancer, as activation of Sirt6 has shown to act as a tumor suppressor in many forms of cancer such as Colon, Ovarian, Prostate, Breast Cancer etc. (Lerrer et al., 2015; Lee et al., 2016; Xia Y. Q. et al., 2018) Although various Sirt6 modulators show a promising therapeutic intervention, Sirt6 under certain circumstances seems to play contradictory roles, the reason for this discrepancy is unknown, hence making it difficult to develop Sirt6 modulators (Gomes, Leal et al. 2019).
Conclusion
In an era where the sheer number of cases of metabolic and cardiovascular diseases is progressively escalating, resulting in a significant health challenge. Scientists are on a continuous search to discover novel molecular targets. One such target is Sirt6, an NAD + dependent histone deacetylase that regulates the expression of several essential genes. As a regulator of gene expression, Sirt6 has been implicated in cancer, neurodegenerative diseases, heart diseases, diabetes, and aging-related processes. Popularly known as “longevity protein,” Sirt6 plays a critical role in aging by controlling cellular processes including genomic stability, DNA-repair, maintenance of telomere length, thereby increasing lifespan. Sirt6 involvement in increasing lifespan requires a better understanding of the molecular mechanisms of Sirt6 in humans relating to aging. In cardiovascular diseases, Sirt6 is involved in regulating the heart's multiple pathophysiological conditions, including hypertrophy, atherosclerotic vascular disease, coronary artery disease, and heart failure. Abnormal lipid metabolism also plays a crucial role in the pathogenesis of heart disease. Sirt6 activation is beneficial in alleviating various factors involved in heart disease. The ubiquity of neurodegenerative diseases is increasing, and decreased levels of Sirt6 in degenerative disease animal models have given a clear understanding of its role, especially in AD. The purpose of Sirt6 in T2DM has been extensively studied, as it improves major pathophysiological defects in pancreatic β-cells, skeletal muscle, and tissues impaired during T2DM. Therefore, Sirt6 plays a crucial role in metabolic diseases and neurodegenerative diseases; future studies should be directed in developing genetic and pharmacologic activation of Sirt6. Various activators and inhibitors that directly and indirectly modulate the activity of Sirt6 have been discovered. Further studies on these Sirt6 modulators may generate potential therapeutic targets that may enhance therapy. Sirt6 has the potential to be a critical therapeutic target in clinical approaches to treating a broad spectrum of disease states. Due to the diversified effects of Sirt6, it poses both a challenge as well as a ray of hope for extensive studies to understand its precise mechanism and functioning, further as a potential therapeutic target, it can enhance or even substitute existing lines of therapy. Apart from the development of new modulators, extensive research must be done in concluding the roles of Sirt6 in various other diseases.
Author Contributions
AK and SPS has designed, edited, and revised the manuscript. SR and LAD have written the manuscript. Final proof reading is done by by SPS and AK. All the authors have agreed the ultimate version.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
All authors have read and agree with the manuscript as written. We thank Professor Nader G. Abraham (1R56HL139561 NA) Departments of Internal medicine and Pharmacology from New York Medical College New York, United States, for his editorial assistance and suggestions for preparing the manuscript.
References
Alhawiti, N. M., Al Mahri, S., Aziz, M. A., Malik, S. S., and Mohammad, S. (2017). TXNIP in metabolic regulation: physiological role and therapeutic outlook. Curr. Drug Targets 18, 1095–1103. doi:10.2174/1389450118666170130145514
Assadi-Porter, M. F., Reiland, H., Sabatini, M., Lorenzini, L., Carnicelli, V., Rogowski, M., et al. (2018). Metabolic reprogramming by 3-iodothyronamine (T1AM): a new perspective to reverse obesity through Co-regulation of sirtuin 4 and 6 expression. Int. J. Mol. Sci. 19 (5), 1535. doi:10.3390/ijms19051535
Aubert, G., and Lansdorp, P. M. (2008). Telomeres and aging. Physiol. Rev. 88, 557–579. doi:10.1152/physrev.00026.2007
Bae, E. J. (2017). Sirtuin 6, a possible therapeutic target for type 2 diabetes. Arch. Pharm. Res. 40, 1380–1389. doi:10.1007/s12272-017-0989-8
Bheda, P., Jing, H., Wolberger, C., and Lin, H. (2016). The substrate specificity of sirtuins. Annu. Rev. Biochem. 85, 405–429. doi:10.1146/annurev-biochem-060815-014537
Bindu, S., Pillai, V. B., and Gupta, M. P. (2016). Role of sirtuins in regulating pathophysiology of the heart. Trends Endocrinol. Metab. 27, 563–573. doi:10.1016/j.tem.2016.04.015
Butterfield, D. A., and Boyd-Kimball, D. (2018). Oxidative stress, amyloid-β peptide, and altered key molecular pathways in the pathogenesis and progression of alzheimer's disease. J. Alzheimers Dis. 62, 1345–1367. doi:10.3233/JAD-170543
Cacabelos, R., Carril, J. C., Cacabelos, N., Kazantsev, A. G., Vostrov, A. V., Corzo, L., et al. (2019). Sirtuins in alzheimer's disease: SIRT2-related GenoPhenotypes and implications for PharmacoEpiGenetics. Int. J. Mol. Sci. 20, 1249. doi:10.3390/ijms20051249
Cacchione, S., Biroccio, A., and Rizzo, A. (2019). Emerging roles of telomeric chromatin alterations in cancer. J. Exp. Clin. Cancer Res. 38, 21. doi:10.1186/s13046-019-1030-5
Cai, Y., Yu, S. S., Chen, S. R., Pi, R. B., Gao, S., Li, H., et al. (2012). Nmnat2 protects cardiomyocytes from hypertrophy via activation of SIRT6. FEBS Lett. 586, 866–874. doi:10.1016/j.febslet.2012.02.014
Cai, Z., Yan, L. J., and Ratka, A. (2013). Telomere shortening and Alzheimer's disease. Neuromolecular Med. 15, 25–48. doi:10.1007/s12017-012-8207-9
Cao, D., Zhao, J., Nguyan, L. N., Nguyen, L. N. T., Khanal, S., Dang, X., et al. (2019). Disruption of telomere integrity and DNA repair machineries by KML001 induces T cell senescence, apoptosis, and cellular dysfunctions. Front. Immunol. 10, 1152. doi:10.3389/fimmu.2019.01152
Carafa, V., Nebbioso, A., and Altucci, L. (2012). Sirtuins and disease: the road ahead. Front. Pharmacol. 3, 4. doi:10.3389/fphar.2012.00004
Chaudhary, R., Garg, J., Shah, N., and Sumner, A. (2017). PCSK9 inhibitors: a new era of lipid lowering therapy. World J. Cardiol. 9, 76–91. doi:10.4330/wjc.v9.i2.76
Chellappan, D. K., Yap, W. S., Bt Ahmad Suhaimi, N. A., Gupta, G., and Dua, K. (2018). Current therapies and targets for type 2 diabetes mellitus. Panminerva Med. 60, 117–131. doi:10.23736/S0031-0808.18.03455-9
Chen, Q., Hao, W., Xiao, C., Wang, R., Xu, X., Lu, H., et al. (2017). SIRT6 is essential for adipocyte differentiation by regulating mitotic clonal expansion. Cell Rep. 18, 3155–3166. doi:10.1016/j.celrep.2017.03.006
Corbi, G., Conti, V., Scapagnini, G., Filippelli, A., and Ferrara, N. (2012). Role of sirtuins, calorie restriction and physical activity in aging. Front. Biosci. (Elite Ed.) 4, 768–778. doi:10.2741/417
D’onofrio, N., Servillo, L., and Balestrieri, M. L. (2017). SIRT1 and SIRT6 signaling pathways in cardiovascular disease protection. Antioxid. Redox Signal. 28, 711–732. doi:10.1089/ars.2017.7178
Dai, H., Sinclair, D. A., Ellis, J. L., and Steegborn, C. (2018). Sirtuin activators and inhibitors: promises, achievements, and challenges. Pharmacol. Ther. 188, 140–154. doi:10.1016/j.pharmthera.2018.03.004
De Céu Teixeira, M., Sanchez-Lopez, E., Espina, M., Garcia, M. L., Durazzo, A., Lucarini, M., et al. (2019). Sirtuins and SIRT6 in carcinogenesis and in diet. Int. J. Mol. Sci. 20, 4945. doi:10.3390/ijms20194945
Demir, I. E., Ceyhan, G. O., and Friess, H. (2017). Epigenomic therapies: the potential of targeting SIRT6 for the treatment of pancreatic cancer. Expert Opin. Ther. Targets 21, 1–3. doi:10.1080/14728222.2017.1265507
Dominy, J. E., Lee, Y., Jedrychowski, M. P., Chim, H., Jurczak, M. J., Camporez, J. P., et al. (2012). The deacetylase Sirt6 activates the acetyltransferase GCN5 and suppresses hepatic gluconeogenesis. Mol. Cell 48, 900–913. doi:10.1016/j.molcel.2012.09.030
Dryden, S. C., Nahhas, F. A., Nowak, J. E., Goustin, A. S., and Tainsky, M. A. (2003). Role for human SIRT2 NAD-dependent deacetylase activity in control of mitotic exit in the cell cycle. Mol. Cell Biol. 23, 3173–3185. doi:10.1128/mcb.23.9.3173-3185.2003
Eberle, D., Hegarty, B., Bossard, P., Ferré, P., and Foufelle, F. (2004). SREBP transcription factors: master regulators of lipid homeostasis. Biochimie 86, 839–848. doi:10.1016/j.biochi.2004.09.018
Eguchi, S., Kawai, T., Scalia, R., and Rizzo, V. (2018). Understanding angiotensin II type 1 receptor signaling in vascular pathophysiology. Hypertension 71, 804–810. doi:10.1161/HYPERTENSIONAHA.118.10266
Elhanati, S., Kanfi, Y., Varvak, A., Roichman, A., Barth, S., Gibor, G., et al. (2013). Carmel-gross, I.multiple regulatory layers of SREBP1/2 by SIRT6. Cell Rep. 4, 905–912. doi:10.1016/j.celrep.2013.08.006
Feldman, J. L., Baeza, J., and Denu, J. M. (2013). Activation of the protein deacetylase SIRT6 by long-chain fatty acids and widespread deacylation by mammalian sirtuins. J. Biol. Chem. 288, 31350–31356. doi:10.1074/jbc.C113.511261
Ferrer, C. M., Alders, M., Postma, A. V., Park, S., Klein, M. A., Cetinbas, M., et al. (2018). An inactivating mutation in the histone deacetylase SIRT6 causes human perinatal lethality. Genes Dev. 32, 373–388. doi:10.1101/gad.307330.117
Finkel, T., Serrano, M., and Blasco, M. A. (2007). The common biology of cancer and ageing. Nature 448, 767–774. doi:10.1038/nature05985
Forero, D. A., González-Giraldo, Y., López-Quintero, C., Castro-Vega, L. J., Barreto, G. E., and Perry, G. (2016). Meta-analysis of telomere length in alzheimer’s disease. J. Gerontol. A Biol. Sci. Med. Sci. 71, 1069–1073. doi:10.1093/gerona/glw053
Fujimoto, M., Takii, R., Takaki, E., Katiyar, A., Nakato, R., Shirahige, K., et al. (2017). The HSF1-PARP13-PARP1 complex facilitates DNA repair and promotes mammary tumorigenesis, Nat. Commun. 8, 1638. doi:10.1038/s41467-017-01807-7
Gao, T., Li, M., Mu, G., Hou, T., Zhu, W.-G., and Yang, Y. (2019). PKCζ phosphorylates SIRT6 to mediate fatty acid β-oxidation in colon cancer cells. Neoplasia 21, 61–73. doi:10.1016/j.neo.2018.11.008
Gao, W. Y., Chen, P. Y., Chen, S. F., and Wu, M. J. (2018). Pinostrobin inhibits proprotein convertase subtilisin/kexin-type 9 (PCSK9) gene expression through the modulation of FoxO3a protein in HepG2 cells, J. Agric. Food Chem. 66, 6083–6093. doi:10.1021/acs.jafc.8b02559
Gavrilov, L. A., and Gavrilova, N. S. (2002). Evolutionary theories of aging and longevity. ScientificWorldJournal 2, 339–356. doi:10.1100/tsw.2002.96
Gertman, O., Omer, D., Hendler, A., Stein, D., Onn, L., Khukhin, Y., et al. (2018). Directed evolution of SIRT6 for improved deacylation and glucose homeostasis maintenance. Sci. Rep. 8, 3538. doi:10.1038/s41598-018-21887-9
Ghosh, S., Liu, B., and Zhou, Z. (2013). Resveratrol activates SIRT1 in a Lamin A-dependent manner.Cell Cycle 12 (6), 872–876. doi:10.4161/cc.24061
Ghosh, S., Liu, B., Wang, Y., Hao, Q., and Zhou, Z. (2015). Lamin A is an endogenous SIRT6 activator and promotes SIRT6-mediated DNA repair. Cell Rep. 13, 1396–1406. doi:10.1016/j.celrep.2015.10.006
Ghosh, S. (2019). “Results-I. Lamin A is an endogenous activator of SIRT6 in DNA damage repair process,” in SIRT6 activities in DNA damage repair and premature aging: functions of SIRT6. Editor S. Ghosh (Singapore: Springer Singapore), 73–95.
Ghosh, S., Wong, S. K., Jiang, Z., and Liu, B. (2018). Haploinsufficiency of Trp53 dramatically extends the lifespan of Sirt6-deficient mice. Elife 7, e32127. doi:10.7554/eLife.32127
Giblin, W., and Lombard, D. B. (2016). “Chapter 3 - sirtuins, healthspan, and longevity in mammals,” in Handbook of the biology of aging. 8th Edn, Editors M.R. Kaeberlein and G.M. Martin (San Diego: Academic Press), 83–132.
Glerup, S., Schulz, R., Laufs, U., and Schlüter, K. D. (2017). Physiological and therapeutic regulation of PCSK9 activity in cardiovascular disease. Basic Res. Cardiol. 112, 32. doi:10.1007/s00395-017-0619-0
Gomes, P., Leal, H., Mendes, A. F., Reis, F., and Cavadas, C. (2019). Dichotomous sirtuins: implications for drug discovery in neurodegenerative and cardiometabolic diseases. Trends Pharmacol. Sci. 40 (12), 1021–1039. doi:10.1016/j.tips.2019.09.003
Grabowska, W., Sikora, E., and Bielak-Zmijewska, A. (2017). Sirtuins, a promising target in slowing down the ageing process, Biogerontology 18, 447–476. doi:10.1007/s10522-017-9685-9
Graham, E., Rymarchyk, S., Wood, M., and Cen, Y. (2018). Development of activity-based chemical probes for human sirtuins, ACS Chem. Biol. 13, 782–792. doi:10.1021/acschembio.7b00754
Guarente, L. (2007). Sirtuins in aging and disease. Cold Spring Harb. Symp. Quant. Biol. 72, 483–488. doi:10.1101/sqb.2007.72.024
Guo, J., Wang, Z., Wu, J., Liu, M., Li, M., Sun, Y., et al. (2019). Endothelial SIRT6 is vital to prevent hypertension and associated cardiorenal injury through targeting nkx3.2-GATA5 signaling. Circ. Res. 124, 1448–1461. doi:10.1161/CIRCRESAHA.118.314032
Haigis, M. C., and Sinclair, D. A. (2010). Mammalian sirtuins: biological insights and disease relevance. Annu. Rev. Pathol. 5, 253–295. doi:10.1146/annurev.pathol.4.110807.092250
Handelsman, Y., and Lepor Norman, E. (2018). PCSK9 inhibitors in lipid management of patients with diabetes mellitus and high cardiovascular risk: a review. J. Am. Heart Assoc. 7, e008953. doi:10.1161/JAHA.118.008953
Harlan, B. A., Pehar, M., Killoy, K. M., and Vargas, M. R. (2019). Enhanced SIRT6 activity abrogates the neurotoxic phenotype of astrocytes expressing ALS-linked mutant SOD1. FASEB J. 33, 7084–7091. doi:10.1096/fj.201802752R
Hassanieh, S., and Mostoslavsky, R. (2018). “Chapter 9 - multitasking roles of the mammalian deacetylase SIRT6,” in Introductory review on sirtuins in biology, aging, and disease. Editors L. Guarente, R. Mostoslavsky, and A. Kazantsev (Cambridge, MA: Academic Press), 117–130.
He, Y., Xiao, Y., Yang, X., Li, Y., Wang, B., Yao, F., et al. (2017). SIRT6 inhibits TNF-α-induced inflammation of vascular adventitial fibroblasts through ROS and Akt signaling pathway. Exp. Cell Res. 357, 88–97. doi:10.1016/j.yexcr.2017.05.001
Heger, V., Tyni, J., Hunyadi, A., Horáková, L., Lahtela-Kakkonen, M., and Rahnasto-Rilla, M. (2019). Quercetin based derivatives as sirtuin inhibitors. Biomed. Pharmacother. 111, 1326–1333. doi:10.1016/j.biopha.2019.01.035
Hirvonen, K., Laivuori, H., Lahti, J., Strandberg, T., Eriksson, J. G., and Hackman, P. (2017). SIRT6 polymorphism rs117385980 is associated with longevity and healthy aging in Finnish men. BMC Med. Genet. 18, 41. doi:10.1186/s12881-017-0401-z
Hoffmann, G., Breitenbucher, F., Schuler, M., and Ehrenhofer-Murray, A. E. (2014). A novel sirtuin 2 (SIRT2) inhibitor with p53-dependent pro-apoptotic activity in non-small cell lung cancer. J. Biol. Chem. 289, 5208–5216. doi:10.1074/jbc.M113.487736
Hu, C. J., Iyer, S., Sataur, A., Covello, K. L., Chodosh, L. A., and Simon, M. C. (2006). Differential regulation of the transcriptional activities of hypoxia-inducible factor 1 alpha (HIF-1alpha) and HIF-2alpha in stem cells. Mol. Cell Biol. 26, 3514–3526. doi:10.1128/MCB.26.9.3514-3526.2006
Huang, W. J., Zhang, X., and Chen, W. W. (2016). Role of oxidative stress in Alzheimer’s disease. Biomed. Rep. 4, 519–522. doi:10.3892/br.2016.630
Huang, Z., Zhao, J., Deng, W., Chen, Y., Shang, J., Song, K., et al. (2018). Identification of a cellularly active SIRT6 allosteric activator, Nat. Chem. Biol. 14, 1118–1126. doi:10.1038/s41589-018-0150-0
Ianni, A., Yuan, X., Bober, E., and Braun, T. (2018). Sirtuins in the cardiovascular system: potential targets in pediatric cardiology. Pediatr. Cardiol. 39, 983–992. doi:10.1007/s00246-018-1848-1
Jeninga, E. H., Schoonjans, K., and Auwerx, J. (2010). Reversible acetylation of PGC-1: connecting energy sensors and effectors to guarantee metabolic flexibility. Oncogene 29, 4617–4624. doi:10.1038/onc.2010.206
Jiang, H., Khan, S., Wang, Y., Charron, G., He, B., Sebastian, C., et al. (2013). SIRT6 regulates TNF-α secretion through hydrolysis of long-chain fatty acyl lysine. Nature 496, 110–113. doi:10.1038/nature12038
Jiang, H., Zhang, X., and Lin, H. (2016). Lysine fatty acylation promotes lysosomal targeting of TNF-α. Sci. Rep. 6, 24371. doi:10.1038/srep24371
Josephs, S. F., Ichim, T. E., Prince, S. M., Kesari, S., Marincola, F. M., Escobedo, A. R., et al. (2018). Unleashing endogenous TNF-alpha as a cancer immunotherapeutic. J. Transl. Med. 16, 242. doi:10.1186/s12967-018-1611-7
Jun, J. C., Rathore, A., Younas, H., Gilkes, D., and Polotsky, V. Y. (2017). Hypoxia-inducible factors and cancer. Curr. Sleep Med. Rep. 3, 1–10. doi:10.1007/s40675-017-0062-7
Jung, E. S., Choi, H., Song, H., Hwang, Y. J., Kim, A., Ryu, H., et al. (2016). p53-dependent SIRT6 expression protects Aβ42-induced DNA damage. Sci. Rep. 6, 25628. doi:10.1038/srep25628
Kaluski, S., Portillo, M., Besnard, A., Stein, D., Einav, M., Zhong, L., et al. (2017). Neuroprotective functions for the histone deacetylase SIRT6. Cell Rep. 18, 3052–3062. doi:10.1016/j.celrep.2017.03.008
Kanfi, Y., Naiman, S., Amir, G., Peshti, V., Zinman, G., Nahum, L., et al. (2012). The sirtuin SIRT6 regulates lifespan in male mice. Nature 483, 218–221. doi:10.1038/nature10815
Kanwal, A. (2018). Functional and therapeutic potential of mitochondrial SIRT3 deacetylase in disease conditions. Expert Rev. Clin. Pharmacol. 11, 1151–1155. doi:10.1080/17512433.2018.1546119
Kanwal, A., Pillai, V. B., Samant, S., Gupta, M., and Gupta, M. P. (2019b). The nuclear and mitochondrial sirtuins, Sirt6 and Sirt3, regulate each other's activity and protect the heart from developing obesity-mediated diabetic cardiomyopathy. FASEB J. 33, 10872–10888. doi:10.1096/fj.201900767R
Kanwal, A., and Dsouza, L. A. (2019). Sirtuins and diabetes: optimizing the sweetness in the blood. Transl. Med. Commun. 4, 3. doi:10.1186/s41231-019-0034-7
Kawahara, T. L., Michishita, E., Adler, A. S., Damian, M., Berber, E., Lin, M., et al. (2009). SIRT6 links histone H3 lysine 9 deacetylation to NF-kappaB-dependent gene expression and organismal life span. Cell 136, 62–74. doi:10.1016/j.cell.2008.10.052
Kawanishi, S., and Oikawa, S. (2004). Mechanism of telomere shortening by oxidative stress. Ann. N. Y. Acad. Sci. 1019, 278–284. doi:10.1196/annals.1297.047
Kerchner, G. A., and Wyss-Coray, T. (2016). “The role of aging in alzheimer’s disease,” in Advances in geroscience. Editors F. Sierra, and R. Kohanski (Cham: Springer International Publishing), 197–227.
Khan, D., Sarikhani, M., Dasgupta, S., Maniyadath, B., Pandit, A. S., Mishra, S., et al. (2018). SIRT6 deacetylase transcriptionally regulates glucose metabolism in heart, J. Cell. Physiol. 233, 5478–5489. doi:10.1002/jcp.26434
Khan, R. I., Nirzhor, S. S. R., and Akter, R. (2018). A review of the recent advances made with SIRT6 and its implications on aging related processes, major human diseases, and possible therapeutic targets. Biomolecules 8, 44. doi:10.3390/biom8030044
Kim, H., Kim, H.-S., and Kaang, B.-K. (2018). Elevated contextual fear memory by SIRT6 depletion in excitatory neurons of mouse forebrain. Mol. Brain 11, 49. doi:10.1186/s13041-018-0391-6
Kim, J. H., Lee, J. M., Kim, J. H., and Kim, K. R. (2018). Fluvastatin activates sirtuin 6 to regulate sterol regulatory element-binding proteins and AMP-activated protein kinase in HepG2 cells. Biochem. Biophys. Res. Commun. 503, 1415–1421. doi:10.1016/j.bbrc.2018.07.057
Kim, J. W., Tchernyshyov, I., Semenza, G. L., and Dang, C. V. (2006). HIF-1-mediated expression of pyruvate dehydrogenase kinase: a metabolic switch required for cellular adaptation to hypoxia. Cell Metab. 3, 177–185. doi:10.1016/j.cmet.2006.02.002
Kitada, M., Kume, S., Kanasaki, K., Takeda-Watanabe, A., and Koya, D. (2013). Sirtuins as possible drug targets in type 2 diabetes. Curr. Drug Targets 14, 622–636. doi:10.2174/1389450111314060002
Klein, M. A., Liu, C., Kuznetsov, V. I., Feltenberger, J. B., Tang, W., and Denu, J. M. (2020). Mechanism of activation for the sirtuin 6 protein deacylase. J. Biol. Chem. 295 (5), 1385–1399. doi:10.1074/jbc.RA119.011285
Klein, M., Liu, C., Camacho, B., Tang, W., and Denu, J. M. (2017). Development and mechanism of small-molecule SIRT6 activators. FAESB J. 31, 921.9. doi:10.1096/fasebj.31.1_supplement.921.9
Kobayashi, Y., Furukawa-Hibi, Y., Chen, C., Horio, Y., Isobe, K., Ikeda, K., et al. (2005). SIRT1 is critical regulator of FOXO-mediated transcription in response to oxidative stress. Int. J. Mol. Med. 16, 237–243. doi:10.3892/ijmm.16.2.237
Kocahan, S., and Doğan, Z. (2017). Mechanisms of alzheimer’s disease pathogenesis and prevention: the brain, neural pathology, N-methyl-D-aspartate receptors, tau protein and other risk factors. Clin. Psychopharmacol. Neurosci. 15, 1–8. doi:10.9758/cpn.2017.15.1.1
Kokkonen, P., Rahnasto-Rilla, M., Kiviranta, P. H., Huhtiniemi, T., Laitinen, T., Poso, A., et al. (2012). Peptides and pseudopeptides as SIRT6 deacetylation inhibitors. ACS Med. Chem. Lett. 3, 969–974. doi:10.1021/ml300139n
Kuang, J., Zhang, Y., Liu, Q., Shen, J., Pu, S., Cheng, S., et al. (2017). Fat-specific Sirt6 ablation sensitizes mice to high-fat diet-induced obesity and insulin resistance by inhibiting lipolysis. Diabetes 66, 1159. doi:10.2337/db16-1225
Kuang, J., Chen, L., Tang, Q., Zhang, J., Li, Y., and He, J. (2018). The role of Sirt6 in obesity and diabetes. Front. Physiol. 9, 135. doi:10.3389/fphys.2018.00135
Kugel, S., and Mostoslavsky, R. (2014). Chromatin and beyond: the multitasking roles for SIRT6. Trends Biochem. Sci. 39, 72–81. doi:10.1016/j.tibs.2013.12.002
Kupis, W., Pałyga, J., Tomal, E., and Niewiadomska, E. (2016). The role of sirtuins in cellular homeostasis. J. Physiol. Biochem. 72, 371–380. doi:10.1007/s13105-016-0492-6
Laemmle, A., Lechleiter, A., Roh, V., Schwarz, C., Portmann, S., Furer, C., et al. (2012). Inhibition of SIRT1 impairs the accumulation and transcriptional activity of HIF-1α protein under hypoxic conditions. PLoS One 7, e33433. doi:10.1371/journal.pone.0033433
Lappas, M. (2012). Anti-inflammatory properties of sirtuin 6 in human umbilical vein endothelial cells. Mediators Inflamm. 2012, 597514. doi:10.1155/2012/597514
Lee, D., and Goldberg, A. L. (2013). SIRT1 protein, by blocking the activities of transcription factors FoxO1 and FoxO3, inhibits muscle atrophy and promotes muscle growth. J. Biol. Chem. 288, 30515–30526. doi:10.1074/jbc.M113.489716
Lee, J. T., and Gu, W. (2013). SIRT1: regulator of p53 deacetylation. Genes Cancer 4, 112–117. doi:10.1177/1947601913484496
Lee, N., Ryu, H. G., Kwon, J. H., Kim, D. K., Kim, S. R., Wang, H. J., et al. (2016). SIRT6 depletion suppresses tumor growth by promoting cellular senescence induced by DNA damage in HCC. PLoS One 11, e0165835. doi:10.1371/journal.pone.0165835
Lee, W.-S., and Kim, J. (2018). Insulin-like growth factor-1 signaling in cardiac aging. Biochim. Biophys. Acta Mol. Basis Dis. 1864, 1931–1938. doi:10.1016/j.bbadis.2017.08.029
Lee, Y., Ka, S. O., Cha, H. N., Chae, Y. N., Kim, M. K., Park, S. Y., et al. (2017). Myeloid sirtuin 6 deficiency causes insulin resistance in high-fat diet-fed mice by eliciting macrophage polarization toward an M1 phenotype, Diabetes 66, 2659–2668. doi:10.2337/db16-1446
Lerrer, B., Gertler, A. A., and Cohen, H. Y. (2015). The complex role of SIRT6 in carcinogenesis. Carcinogenesis 37, 108–118. doi:10.1093/carcin/bgv167
Li, M., Hou, T., Gao, T., Lu, X., Yang, Q., Zhu, Q., et al. (2018). p53 cooperates with SIRT6 to regulate cardiolipin de novo biosynthesis. Cell Death Dis. 9, 941. doi:10.1038/s41419-018-0984-0
Li, Z., Xu, K., Zhang, N., Amador, G., Wang, Y., Zhao, S., et al. (2018). Overexpressed SIRT6 attenuates cisplatin-induced acute kidney injury by inhibiting ERK1/2 signaling. Kidney Int. 93, 881–892. doi:10.1016/j.kint.2017.10.021
Li, Y., Meng, X., Wang, W., Liu, F., Hao, Z., Yang, Y., et al. (2017). Cardioprotective effects of SIRT6 in a mouse model of transverse aortic constriction-induced heart failure. Front. Physiol. 8, 394. doi:10.3389/fphys.2017.00394
Liao, C. Y., and Kennedy, B. K. (2016). SIRT6, oxidative stress, and aging. Cell Res. 26, 143–144. doi:10.1038/cr.2016.8
Liu, J., and Zheng, W. (2016). Cyclic peptide-based potent human SIRT6 inhibitors. Org. Biomol. Chem. 14, 5928–5935. doi:10.1039/c5ob02339d
Liu, M., Huo, Y. R., Wang, J., Wang, C., Liu, S., Liu, S., et al. (2016). Telomere shortening in alzheimer's disease patients. Ann. Clin. Lab. Sci. 46, 260–265
Liu, T. F., Vachharajani, V. T., Yoza, B. K., and Mccall, C. E. (2012). NAD+-dependent sirtuin 1 and 6 proteins coordinate a switch from glucose to fatty acid oxidation during the acute inflammatory response. J. Biol. Chem. 287, 25758–25769. doi:10.1074/jbc.M112.362343
Lombard, D. B., Schwer, B., Alt, F. W., and Mostoslavsky, R. (2008). SIRT6 in DNA repair, metabolism and ageing. J. Intern. Med. 263, 128–141. doi:10.1111/j.1365-2796.2007.01902.x
López-Lluch, G., and Navas, P. (2016). Calorie restriction as an intervention in ageing. J. Physiol. 594, 2043–2060. doi:10.1113/JP270543
Lu, J., Sun, D., Liu, Z., Li, M., Hong, H., Liu, C., et al. (2016). SIRT6 suppresses isoproterenol-induced cardiac hypertrophy through activation of autophagy. Transl. Res. 172, 96. doi:10.1016/j.trsl.2016.03.002
Mahler, R. J., and Adler, M. L. (1999). Clinical review 102: type 2 diabetes mellitus: update on diagnosis, pathophysiology, and treatment. J. Clin. Endocrinol. Metab. 84, 1165–1171. doi:10.1210/jcem.84.4.5612
Mao, K., Quipildor, G. F., Tabrizian, T., Novaj, A., Guan, F., Walters, R. O., et al. (2018). Late-life targeting of the IGF-1 receptor improves healthspan and lifespan in female mice. Nat. Commun. 9, 2394. doi:10.1038/s41467-018-04805-5
Mao, Z., Hine, C., Tian, X., Van Meter, M., Au, M., Vaidya, A., et al. (2011). SIRT6 promotes DNA repair under stress by activating PARP1. Science 332, 1443–1446. doi:10.1126/science.1202723
Markesbery, W. R. (1999). The role of oxidative stress in alzheimer disease. Arch. Neurol. 56, 1449–1452. doi:10.1001/archneur.56.12.1449
Michishita, E., Mccord, R. A., Berber, E., Kioi, M., Padilla-Nash, H., Damian, M., et al. (2008). SIRT6 is a histone H3 lysine 9 deacetylase that modulates telomeric chromatin. Nature 452, 492–496. doi:10.1038/nature06736
Mohamad Nasir, N. F., Zainuddin, A., and Shamsuddin, S. (2018). Emerging roles of sirtuin 6 in alzheimer's disease. J. Mol. Neurosci. 64, 157–161. doi:10.1007/s12031-017-1005-y
Mostoslavsky, R., Chua, K. F., Lombard, D. B., Pang, W. W., Fischer, M. R., Gellon, L., et al. (2006). Genomic instability and aging-like phenotype in the absence of mammalian SIRT6. Cell 124, 315–329. doi:10.1016/j.cell.2005.11.044
Nagaraj, K., Lapkina-Gendler, L., Sarfstein, R., Gurwitz, D., Pasmanik-Chor, M., Laron, Z., et al. (2018). Identification of thioredoxin-interacting protein (TXNIP) as a downstream target for IGF1 action. Proc. Natl. Acad. Sci. U.S.A. 115, 1045. doi:10.1073/pnas.1715930115
Naiman, S., and Cohen, H. Y. (2018). Role for the longevity protein SIRT6 in primate development. Nature 560, 559–560. doi:10.1038/d41586-018-05970-9
Niu, Y. (2019). Genetic monkeys reveal a new role for a longevity protein in embryonic development. Natl. Sci. Rev. 6, 392. doi:10.1093/nsr/nwz051
Pan, H., Guan, D., Liu, X., Li, J., Wang, L., Wu, J., et al. (2016). SIRT6 safeguards human mesenchymal stem cells from oxidative stress by coactivating NRF2. Cell Res. 26, 190–205. doi:10.1038/cr.2016.4
Pan, P. W., Feldman, J. L., Devries, M. K., Dong, A., Edwards, A. M., and Denu, J. M. (2011). Structure and biochemical functions of SIRT6. J. Biol. Chem. 286, 14575–14587. doi:10.1074/jbc.M111.218990
Parenti, M. D., Grozio, A., Bauer, I., Galeno, L., Damonte, P., Millo, E., et al. (2014). Discovery of novel and selective SIRT6 inhibitors. J. Med. Chem. 57, 4796–4804. doi:10.1021/jm500487d
Parhofer, K. G. (2016). The treatment of disorders of lipid metabolism. Dtsch. Arztebl. Int. 113, 261–268. doi:10.3238/arztebl.2016.0261
Peshti, V., Obolensky, A., Nahum, L., Kanfi, Y., Rathaus, M., Avraham, M., et al. (2017). Characterization of physiological defects in adult SIRT6-/- mice. PLoS One 12, e0176371. doi:10.1371/journal.pone.0176371
Pezzuto, A., and Carico, E. (2018). Role of HIF-1 in cancer progression: novel insights. A review. Curr. Mol. Med. 18, 343–351. doi:10.2174/1566524018666181109121849
Pillai, V. B., Sundaresan, N. R., and Gupta, M. P. (2014). Regulation of Akt signaling by sirtuins: its implication in cardiac hypertrophy and aging. Circ. Res. 114, 368–378. doi:10.1161/CIRCRESAHA.113.300536
Polito, L., Kehoe, P. G., Forloni, G., and Albani, D. (2010). The molecular genetics of sirtuins: association with human longevity and age-related diseases. Int. J. Mol. Epidemiol. Genet. 1, 214–225
Qin, K., Zhang, N., Zhang, Z., Nipper, M., Zhu, Z., Leighton, J., et al. (2018). SIRT6-mediated transcriptional suppression of Txnip is critical for pancreatic beta cell function and survival in mice. Diabetologia 61, 906–918. doi:10.1007/s00125-017-4542-6
Rack, J. G., Morra, R., Barkauskaite, E., Kraehenbuehl, R., Ariza, A., Qu, Y., et al. (2015). Identification of a class of protein ADP-ribosylating sirtuins in microbial pathogens. Mol. Cell 59, 309–320. doi:10.1016/j.molcel.2015.06.013
Rahnasto-Rilla, M., Kokkola, T., Jarho, E., Lahtela-Kakkonen, M., and Moaddel, R. (2016). N-acylethanolamines bind to SIRT6. Chembiochem 17, 77–81. doi:10.1002/cbic.201500482
Rahnasto-Rilla, M., Tyni, J., Huovinen, M., Jarho, E., Kulikowicz, T., Ravichandran, S., et al. (2018). Natural polyphenols as sirtuin 6 modulators. Sci. Rep. 8, 4163. doi:10.1038/s41598-018-22388-5
Rajabi, N., Galleano, I., Madsen, A. S., and Olsen, C. A. (2018). Targeting sirtuins: substrate specificity and inhibitor design. Prog. Mol. Biol. Transl. Sci. 154, 25–69. doi:10.1016/bs.pmbts.2017.11.003
Ravi, V., Jain, A., Khan, D., Ahamed, F., Mishra, S., Giri, M., et al. (2019). SIRT6 transcriptionally regulates global protein synthesis through transcription factor Sp1 independent of its deacetylase activity. Nucleic Acids Res. 47, 9115–9131. doi:10.1093/nar/gkz648
Ravussin, E., and Smith, S. R. (2016). “Chapter 36 - role of the adipocyte in metabolism and endocrine function,” in Endocrinology: adult and pediatric. 7th Edn, Editors J. L. Jameson, L. J. De Groot, D. M. De Kretser, L. C. Giudice, A. B. Grossman, S. Melmed, et al. (Philadelphia: W.B. Saunders), 627–647.e629.
Rizzo, A., Iachettini, S., Salvati, E., Zizza, P., Maresca, C., D'angelo, C., et al. (2016). SIRT6 interacts with TRF2 and promotes its degradation in response to DNA damage. Nucleic Acids Res. 45, 1820–1834. doi:10.1093/nar/gkw1202
Rodgers, J. T., and Puigserver, P. (2006). Certainly can't live without this: SIRT6. Cell Metab. 3, 77–78. doi:10.1016/j.cmet.2006.01.009
Roichman, A., Kanfi, Y., Glazz, R., Naiman, S., Amit, U., Landa, N., et al. (2016). SIRT6 overexpression improves various aspects of mouse healthspan. J. Gerontol. A Biol. Sci. Med. Sci. 72, 603–615. doi:10.1093/gerona/glw152
Santos-Barriopedro, I., Bosch-Presegué, L., Marazuela-Duque, A., De La Torre, C., Colomer, C., Vazquez, B. N., et al. (2018). SIRT6-dependent cysteine monoubiquitination in the PRE-SET domain of Suv39h1 regulates the NF-κB pathway. Nat. Commun. 9, 101. doi:10.1038/s41467-017-02586-x
Santos-Barriopedro, I., and Vaquero, A. (2018). Complex role of SIRT6 in NF-κB pathway regulation. Mol. Cell. Oncol. 5, e1445942. doi:10.1080/23723556.2018.1445942
Satoh, A., and Imai, S. (2014). Systemic regulation of mammalian ageing and longevity by brain sirtuins. Nat. Commun. 5, 4211. doi:10.1038/ncomms5211
Schofield, J. D., Liu, Y., Rao-Balakrishna, P., Malik, R. A., and Soran, H. (2016). Diabetes dyslipidemia. Diabetes Ther. 7, 203–219. doi:10.1007/s13300-016-0167-x
Schumacher, A. (2011). “Chapter 25 - aging epigenetics,” in Handbook of epigenetics. Editor T. Tollefsbol (San Diego: Academic Press), 405–422.
Schumacher, A. (2017). “Chapter 25–aging epigenetics,” in Handbook of epigenetics. 2nd Edn, Editor T. O. Tollefsbol (Academic Press), 371–388.
Sergiev, P. V., Dontsova, O. A., and Berezkin, G. V. (2015). Theories of aging: an ever-evolving field. Acta Naturae 7, 9–18. doi:10.32607/20758251-2015-7-1-9-18
Serravallo, M., Jagdeo, J., Glick, S., Siegel, D., and Brody, N. (2013). Sirtuins in dermatology: applications for future research and therapeutics. Arch. Dermatol. Res. 305, 269. doi:10.1007/s00403-013-1320-2
Shalev, A. (2014). Minireview: thioredoxin-interacting protein: regulation and function in the pancreatic β-cell. Mol. Endocrinol. 28, 1211–1220. doi:10.1210/me.2014-1095
Sharabi, K., Lin, H., Tavares, C. D., Dominy, J. E., Camporez, J. P., Perry, R. J., et al. (2017). Selective chemical inhibition of PGC-1α gluconeogenic activity ameliorates type 2 diabetes. Cell 169, 148–e15. doi:10.1016/j.cell.2017.03.001
Shimano, H., and Sato, R. (2017). SREBP-regulated lipid metabolism: convergent physiology - divergent pathophysiology. Nat. Rev. Endocrinol. 13, 710. doi:10.1038/nrendo.2017.91
Simon, M., Van Meter, M., Ablaeva, J., Ke, Z., Gonzalez, R. S., Taguchi, T., et al. (2019). LINE1 derepression in aged wild-type and SIRT6-deficient mice drives inflammation. Cell Metab. 29, 871–e875. doi:10.1016/j.cmet.2019.02.014
Singh, S. P., Mcclung, J. A., Thompson, E., Glick, Y., Greenberg, M., Acosta-Baez, G., et al. (2019). Cardioprotective heme oxygenase-1-pgc1α signaling in epicardial fat attenuates cardiovascular risk in humans as in obese mice. Obesity (Silver Spring) 27, 1634–1643. doi:10.1002/oby.22608
Singh, S. P., Greenberg, M., Glick, Y., Bellner, L., Favero, G., Rezzani, R., et al. (2020). Adipocyte specific HO-1 gene therapy is effective in antioxidant treatment of insulin resistance and vascular function in an obese mice model. Antioxidants (Basel) 9, 40. doi:10.3390/antiox9010040
Sociali, G., Galeno, L., Parenti, M. D., Grozio, A., Bauer, I., Passalacqua, M., et al. (2015). Quinazolinedione SIRT6 inhibitors sensitize cancer cells to chemotherapeutics. Eur. J. Med. Chem. 102, 530–539. doi:10.1016/j.ejmech.2015.08.024
Sociali, G., Magnone, M., Ravera, S., Damonte, P., Vigliarolo, T., Von Holtey, M., et al. (2017). Pharmacological Sirt6 inhibition improves glucose tolerance in a type 2 diabetes mouse model. FASEB J. 31, 3138–3149. doi:10.1096/fj.201601294R
Song, M. Y., Wang, J., Ka, S. O., Bae, E. J., and Park, B. H. (2016). Insulin secretion impairment in Sirt6 knockout pancreatic β cells is mediated by suppression of the FoxO1-Pdx1-Glut2 pathway. Sci. Rep. 6, 30321. doi:10.1038/srep30321
Song, Y., Fan, Z., Bai, X., Liu, W., Han, Y., Xu, L., et al. (2016). PARP-1-modulated AIF translocation is involved in streptomycin-induced cochlear hair cell death. Acta Otolaryngol. 136, 545–550. doi:10.3109/00016489.2016.1143968
Spolitu, S., Okamoto, H., Dai, W., Zadroga John, A., Wittchen Erika, S., Gromada, J., et al. (2019). Hepatic glucagon signaling regulates PCSK9 and low-density lipoprotein cholesterol. Circ. Res. 124, 38–51. doi:10.1161/CIRCRESAHA.118.313648
Stein, D., and Toiber, D. (2017). DNA damage and neurodegeneration: the unusual suspect. Neural Regen. Res. 12, 1441–1442. doi:10.4103/1673-5374.215254
Sundaresan, N. R., Vasudevan, P., Zhong, L., Kim, G., Samant, S., Parekh, V., et al. (2012). The sirtuin SIRT6 blocks IGF-Akt signaling and development of cardiac hypertrophy by targeting c-Jun. Nat. Med. 18, 1643–1650. doi:10.1038/nm.2961
Takahashi, R. H., Nagao, T., and Gouras, G. K. (2017). Plaque formation and the intraneuronal accumulation of β-amyloid in Alzheimer's disease. Pathol. Int. 67, 185–193. doi:10.1111/pin.12520
Takasaka, N., Araya, J., Hara, H., Ito, S., Kobayashi, K., Kurita, Y., et al. (2014). Autophagy induction by SIRT6 through attenuation of insulin-like growth factor signaling is involved in the regulation of human bronchial epithelial cell senescence. J. Immunol. 192, 958. doi:10.4049/jimmunol.1302341
Tang, B. L. (2016). Sirt1 and the mitochondria. Mol. Cells 39, 87–95. doi:10.14348/molcells.2016.2318
Tang, B. L. (2017). Is SIRT6 activity neuroprotective and how does it differ from SIRT1 in this regard? Front. Cell. Neurosci. 11, 165. doi:10.3389/fncel.2017.00165
Tao, R., Xiong, X., Depinho, R. A., Deng, C. X., and Dong, X. C. (2013a). FoxO3 transcription factor and Sirt6 deacetylase regulate low density lipoprotein (LDL)-cholesterol homeostasis via control of the proprotein convertase subtilisin/kexin type 9 (Pcsk9) gene expression. J. Biol. Chem. 288, 29252–29259. doi:10.1074/jbc.M113.481473
Tao, R., Xiong, X., Depinho, R. A., Deng, C. X., and Dong, X. C. (2013b). Hepatic SREBP-2 and cholesterol biosynthesis are regulated by FoxO3 and Sirt6. J. Lipid Res. 54, 2745–2753. doi:10.1194/jlr.M039339
Tasselli, L., Zheng, W., and Chua, K. F. (2017). SIRT6: novel mechanisms and links to aging and disease. Trends Endocrinol. Metab. 28, 168–185. doi:10.1016/j.tem.2016.10.002
Tennen, R. I., Bua, D. J., Wright, W. E., and Chua, K. F. (2011). SIRT6 is required for maintenance of telomere position effect in human cells. Nat. Commun. 2, 433. doi:10.1038/ncomms1443
Tian, X., Firsanov, D., Zhang, Z., Cheng, Y., Luo, L., Tombline, G., et al. (2019). SIRT6 is responsible for more efficient DNA double-strand break repair in long-lived species. Cell 177, 622–e22. doi:10.1016/j.cell.2019.03.043
Tosato, M., Zamboni, V., Ferrini, A., and Cesari, M. (2007). The aging process and potential interventions to extend life expectancy. Clin. Interv. Aging 2, 401–412
Trevisan, K., Cristina-Pereira, R., Silva-Amaral, D., and Aversi-Ferreira, T. A. (2019). Theories of aging and the prevalence of alzheimer’s disease. BioMed. Res. Int. 2019, 9171424. doi:10.1155/2019/9171424
Van meter, M., Simon, M., Tombline, G., May, A., Morello, T. D., Hubbard, B. P., et al. (2016). JNK phosphorylates SIRT6 to stimulate DNA double-strand break repair in response to oxidative stress by recruiting PARP1 to DNA breaks. Cell Rep. 16, 2641–2650. doi:10.1016/j.celrep.2016.08.006
Vassilopoulos, A., Fritz, K. S., Petersen, D. R., and Gius, D. (2011). The human sirtuin family: evolutionary divergences and functions. Hum. Genomics. 5, 485–496. doi:10.1186/1479-7364-5-5-485
Vitiello, M., Zullo, A., Servillo, L., Mancini, F. P., Borriello, A., Giovane, A., et al. (2017). Multiple pathways of SIRT6 at the crossroads in the control of longevity, cancer, and cardiovascular diseases. Ageing Res. Rev. 35, 301–311. doi:10.1016/j.arr.2016.10.008
Wang, C., and Mbalaviele, G. (2019). Role of APD-ribosylation in bone health and disease. Cells 8, 1201. doi:10.3390/cells8101201
Wang, G., Meyer, J. G., Cai, W., Li, M. E., Softic, S., and Kahn, C. R. (2018). Sirt5 plays a critical role in mitochondrial protein acylation and mitochondrial metabolic homeostasis in brown fat. Diabetes 67, 274. doi:10.2337/db18-274-OR
Wang, K. C., Tosh, D. N., Zhang, S., Mcmillen, I. C., Duffield, J. A., Brooks, D. A., et al. (2015). IGF-2R-Gαq signaling and cardiac hypertrophy in the low-birth-weight lamb. Am. J. Physiol. Regul. Integr. Comp. Physiol. 308, R627–R635. doi:10.1152/ajpregu.00346.2014
Watroba, M., and Szukiewicz, D. (2016). The role of sirtuins in aging and age-related diseases. Adv. Med. Sci. 61, 52–62. doi:10.1016/j.advms.2015.09.003
Wei, Y. H., Ma, Y. S., Lee, H. C., Lee, C. F., and Lu, C. Y. (2001). Mitochondrial theory of aging matures–roles of mtDNA mutation and oxidative stress in human aging. Zhonghua Yixue Zazhi (Taipei) 64, 259–270
Wood, M., Rymarchyk, S., Zheng, S., and Cen, Y. (2018). Trichostatin A inhibits deacetylation of histone H3 and p53 by SIRT6. Arch. Biochem. Biophys. 638, 8–17. doi:10.1016/j.abb.2017.12.009
Wu, Y., Chen, L., Wang, Y., Li, W., Lin, Y., Yu, D., et al. (2015). Overexpression of Sirtuin 6 suppresses cellular senescence and NF-κB mediated inflammatory responses in osteoarthritis development. Sci. Rep. 5, 17602. doi:10.1038/srep17602
Xia, X., Jiang, Q., Mcdermott, J., and Han, J. J. (2018). Aging and Alzheimer's disease: comparison and associations from molecular to system level. Aging Cell 17, e12802. doi:10.1111/acel.12802
Xia, Y. Q., Hua, R. J., Juan, C., Zhong, Z. H., Tao, C. S., Fang, R., et al. (2018). SIRT6 depletion sensitizes human hepatoma cells to chemotherapeutics by downregulating MDR1 expression. Front. Pharmacol. 9, 194. doi:10.3389/fphar.2018.00194
Xiwen, X., Xupeng, S., Qingzhi, W., Xinlai, Q., Yang, Z., Xiaoyan, P., et al. (2016). SIRT6 protects against palmitate-induced pancreatic β-cell dysfunction and apoptosis. J. Endocrinol. 231, 159–165. doi:10.1530/JOE-16-0317
Xu, P., Wang, T.-T., Liu, X.-Z., Wang, N.-Y., Sun, L.-H., Zhang, Z.-Q., et al. (2019). Sirt6 regulates efficiency of mouse somatic reprogramming and maintenance of pluripotency. Stem Cell Res. Ther. 10, 9. doi:10.1186/s13287-018-1109-5
Xu, S., Yin, M., Koroleva, M., Mastrangelo, M. A., Zhang, W., Bai, P., et al. (2016). SIRT6 protects against endothelial dysfunction and atherosclerosis in mice. Aging (Albany NY) 8, 1064–1082. doi:10.18632/aging.100975
Xu, Z., Zhang, L., Zhang, W., Meng, D., Zhang, H., Jiang, Y., et al. (2015). SIRT6 rescues the age related decline in base excision repair in a PARP1-dependent manner. Cell Cycle 14, 269–276. doi:10.4161/15384101.2014.980641
Yan, K., Wang, K., and Li, P. (2019). The role of post-translational modifications in cardiac hypertrophy. J. Cell Mol. Med. 23, 3795–3807. doi:10.1111/jcmm.14330
Yang, G., Liu, C., Chen, S. H., Kassab, M. A., Hoff, J. D., Walter, N. G., et al. (2018). Super-resolution imaging identifies PARP1 and the Ku complex acting as DNA double-strand break sensors. Nucleic Acids Res. 46, 3446–3457. doi:10.1093/nar/gky088
Yang, H., Zhu, R., Zhao, X., Liu, L., Zhou, Z., Zhao, L., et al. (2019). Sirtuin-mediated deacetylation of hnRNP A1 suppresses glycolysis and growth in hepatocellular carcinoma. Oncogene 38, 4915–4931. doi:10.1038/s41388-019-0764-z
Yang, Z., Yu, W., Huang, R., Ye, M., and Min, Z. (2019). SIRT6/HIF-1α axis promotes papillary thyroid cancer progression by inducing epithelial–mesenchymal transition. Cancer Cell Int. 19, 17. doi:10.1186/s12935-019-0730-4
Yao, L., Cui, X., Chen, Q., Yang, X., Fang, F., Zhang, J., et al. (2017). Cold-inducible SIRT6 regulates thermogenesis of Brown and beige fat. Cell Rep. 20, 641–654. doi:10.1016/j.celrep.2017.06.069
Ye, X., Li, M., Hou, T., Gao, T., Zhu, W. G., and Yang, Y. (2017). Sirtuins in glucose and lipid metabolism. Oncotarget 8, 1845–1859. doi:10.18632/oncotarget.12157
Yeo, D., Kang, C., and Ji, L. (2017). Overexpression of SIRT6 suppresses TNFα-induced muscle cell atrophy via NFκB inhibition. FASEB J. 31, doi:1022.10161022.1016
Yepuri, G., and Ramasamy, R. (2019). Significance and mechanistic relevance of SIRT6-mediated endothelial dysfunction in cardiovascular disease progression. Circ. Res. 124, 1408–1410. doi:10.1161/CIRCRESAHA.119.315098
You, W., Rotili, D., Li, T. M., Kambach, C., Meleshin, M., Schutkowski, M., et al. (2017). Structural basis of sirtuin 6 activation by synthetic small molecules. Angew Chem. Int. Ed. Engl. 56, 1007–1011. doi:10.1002/anie.201610082
You, W., and Steegborn, C. (2018). Structural basis of sirtuin 6 inhibition by the hydroxamate trichostatin A: implications for protein deacylase drug development J. Med. Chem. 61, 10922–10928. doi:10.1021/acs.jmedchem.8b01455
Yu, S. S., Cai, Y., Ye, J. T., Pi, R. B., Chen, S. R., Liu, P. Q., et al. (2013). Sirtuin 6 protects cardiomyocytes from hypertrophy in vitro via inhibition of NF-κB-dependent transcriptional activity. Br. J. Pharmacol. 168, 117–128. doi:10.1111/j.1476-5381.2012.01903.x
Yu, W., Qin, J., Chen, C., Fu, Y., and Wang, W. (2018). Moderate calorie restriction attenuates age‑associated alterations and improves cardiac function by increasing SIRT1 and SIRT3 expression. Mol. Med. Rep. 18, 4087–4094. doi:10.3892/mmr.2018.9390
Zhang, C., Yu, Y., Huang, Q., and Tang, K. (2019). SIRT6 regulates the proliferation and apoptosis of hepatocellular carcinoma via the ERK1/2 signaling pathway. Mol. Med. Rep. 20, 1575–1582. doi:10.3892/mmr.2019.10398
Zhang, Q., Tu, W., Tian, K., Han, L., Wang, Q., Chen, P., et al. (2019). Sirtuin 6 inhibits myofibroblast differentiation via inactivating transforming growth factor-beta1/Smad2 and nuclear factor-kappaB signaling pathways in human fetal lung fibroblasts. J. Cell Biochem. 120, 93–104. doi:10.1002/jcb.27128
Zhang, D., Jing, J., Lou, F., Li, R., Ping, Y., Yu, F., et al. (2018). Evidence for excessive osteoclast activation in SIRT6 null mice. Sci. Rep. 8, 10992. doi:10.1038/s41598-018-28716-z
Zhang, W., Wan, H., Feng, G., Qu, J., Wang, J., Jing, Y., et al. (2018). SIRT6 deficiency results in developmental retardation in cynomolgus monkeys. Nature 560, 661–665. doi:10.1038/s41586-018-0437-z
Zhang, N., Li, Z., Mu, W., Li, L., Liang, Y., Lu, M., et al. (2016a). Calorie restriction-induced SIRT6 activation delays aging by suppressing NF-κB signaling. Cell Cycle 15, 1009–1018. doi:10.1080/15384101.2016.1152427
Zhang, N., Li, Z., Mu, W., Li, L., Liang, Y., Lu, M., et al. (2016b). Calorie restriction-induced SIRT6 activation delays aging by suppressing NF-κB signaling. Cell Cycle 15, 1009–1018. doi:10.1080/15384101.2016.1152427
Zhang, P., Tu, B., Wang, H., Cao, Z., Tang, M., Zhang, C., et al. (2014). Tumor suppressor p53 cooperates with SIRT6 to regulate gluconeogenesis by promoting FoxO1 nuclear exclusion. Proc. Natl. Acad. Sci. U.S.A. 111, 10684–10689. doi:10.1073/pnas.1411026111
Iachettini, S., Trisciuoglio, D., Rotili, D., Lucidi, A., Salvati, E., Zizza, P., et al. (2018). Pharmacological activation of SIRT6 triggers lethal autophagy in human cancer cells. Cell Death Dis. 9, 996. doi:10.1038/s41419-018-1065-0
Zhang, Z. Z., Cheng, Y. W., Jin, H. Y., Chang, Q., Shang, Q. H., Xu, Y. L., et al. (2017). The sirtuin 6 prevents angiotensin II-mediated myocardial fibrosis and injury by targeting AMPK-ACE2 signaling. Oncotarget 8, 72302–72314. doi:10.18632/oncotarget.20305
Zhao, J., Wozniak, A., Adams, A., Cox, J., Vittal, A., Voss, J., et al. (2019). SIRT7 regulates hepatocellular carcinoma response to therapy by altering the p53-dependent cell death pathway. J. Exp. Clin. Cancer Res. 38, 252. doi:10.1186/s13046-019-1246-4
Zhong, L., D’urso, A., Toiber, D., Sebastian, C., Henry, R. E., Vadysirisack, D. D., et al. (2010). The histone deacetylase Sirt6 regulates glucose homeostasis via Hif1alpha. Cell 140, 280–293. doi:10.1016/j.cell.2009.12.041
Zhong, L., and Mostoslavsky, R. (2010). SIRT6: a master epigenetic gatekeeper of glucose metabolism. Transcription 1, 17–21. doi:10.4161/trns.1.1.12143
Zi, Y., Yi-An, Y., Bing, J., Yan, L., Jing, T., Chun-Yu, G., et al. (2019). Sirt6-induced autophagy restricted TREM-1-mediated pyroptosis in ox-LDL-treated endothelial cells: relevance to prognostication of patients with acute myocardial infarction. Cell Death Discov. 5, 88. doi:10.1038/s41420-019-0168-4
Keywords: drug discovery, epigenetics, diseases, modulators, Sirt6, sirtuins
Citation: Raj S, Dsouza LA, Singh SP and Kanwal A (2020) Sirt6 Deacetylase: A Potential Key Regulator in the Prevention of Obesity, Diabetes and Neurodegenerative Disease. Front. Pharmacol. 11:598326. doi: 10.3389/fphar.2020.598326
Received: 24 August 2020; Accepted: 27 October 2020;
Published: 07 December 2020.
Edited by:
Terry D. Hinds Jr., University of Toledo, United StatesReviewed by:
Dante Rotili, Sapienza University of Rome, ItalyPedro Gomes, University of Coimbra, Portugal
Copyright © 2020 Raj, Dsouza, Singh and Kanwal. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Abhinav Kanwal, YWJoaW5hdmthbndhbEBnbWFpbC5jb20=; Shailendra Pratap Singh, c3BzaW5naEBjdXJhai5hYy5pbg==
†These authors have contributed equally to this work
 Swapnil Raj
Swapnil Raj Liston Augustine Dsouza
Liston Augustine Dsouza Shailendra Pratap Singh
Shailendra Pratap Singh Abhinav Kanwal
Abhinav Kanwal