- 1Department of Pharmacy, Huadong Hospital, Fudan University, Shanghai, China
- 2Shanghai Key Laboratory of Clinical Geriatric Medicine, Huadong Hospital, Fudan University, Shanghai, China
- 3Department of Biological Medicines & Shanghai Engineering Research Center of Immunotherapeutics, Fudan University School of Pharmacy, Shanghai, China
Recently, there has been renewed interest in metabolic therapy for cancer, particularly in amino acid deprivation by enzymes. L-asparaginase was approved for the treatment of acute lymphoblastic leukemia by the U.S. Food and Drug Administration. Arginine deiminase and recombinant human arginase have been developed into clinical trials as potential cancer therapeutic agents for the treatment of arginine-auxotrophic tumors. Moreover, other novel amino acid degrading enzymes, such as glutaminase, methionase, lysine oxidase, phenylalanine ammonia lyase, have been developed for the treatment of malignant cancers. One of the greatest obstacles faced by anticancer drugs is the development of drug resistance, which is reported to be associated with autophagy. Autophagy is an evolutionarily conserved catabolic process that is responsible for the degradation of dysfunctional proteins and organelles. There is a growing body of literature revealing that, in response to metabolism stress, autophagy could be induced by amino acid deprivation. The manipulation of autophagy in combination with amino acid degrading enzymes is actively being investigated as a potential therapeutic approach in preclinical studies. Importantly, shedding light on how autophagy fuels tumor metabolism during amino acid deprivation will enable more potential combinational therapeutic strategies. This study summarizes recent advances, discussing several potential anticancer enzymes, and highlighting the promising combined therapeutic strategy of amino acid degrading enzymes and autophagy modulators in tumors
Introduction
In recent years, there has been increasing interest in targeting tumor amino acid metabolism as a strategy to treat cancer (Dang et al., 2011; Koppenol et al., 2011). It has been recognized that tumor cells often undergo metabolic reprogramming to support the high metabolic demands that are required for tumorigenesis (Nicholatos et al., 2019; Souder and Anderson, 2019). Cancer cells attempt to utilize various fuel sources to maintain rapid and abnormal proliferation (Vander Heiden and DeBerardinis, 2017). However, certain tumor cells become auxotrophic for specific amino acids, such as asparagine, arginine, and methionine (Cheng et al., 2007; Rytting, 2010; Gao et al., 2019). Therefore, depleting these amino acids by amino acid degrading enzymes inhibits and impairs tumor growth. Whereas, normal cells are kept in good condition due to their capability to synthesize these specific amino acids via endogenous synthesis. The difference between normal and cancer cells in terms of nutritional demand makes tumor tissue vulnerable to certain amino acid deprivation treatments (Fung and Chan, 2017; Dhankhar et al., 2020). Thus, degrading amino acids by enzymes may be an excellent therapeutic approach for the treatment of malignant cancer. L-Asparaginase was the first amino acid degrading enzyme approved by the U.S. Food and Drug Administration (FDA) in 1978, used to treat acute lymphoblastic leukemia (ALL) (Truelove et al., 2013). Since then, many other amino acid depriving enzymes have been developed through preclinical or clinical evaluations (Stasyk et al., 2015; Fernandes et al., 2017).
There is a growing body of literature indicating that cellular metabolism stress, for example, nutrient deprivation, could induce macroautophagy (Xie et al., 2015; Fulop et al., 2018; Nacarelli et al., 2018). Macroautophagy is an evolutionarily conserved catabolic process in which damaged cellular organelles and proteins are engulfed into double-membraned vesicles named autophagosomes, and then delivered to lysosomes for degradation (Nishida et al., 2009; Honda et al., 2014). Besides macroautophagy, there are other categories of autophagy, including macroautophagy, microautophagy, and chaperone-mediated autophagy (Mony et al., 2016). Macroautophagy (hereafter referred to as autophagy) plays a crucial role not only in normal cells and tissues but also in pathological environments. Amino acid starvation initiates autophagy by inhibiting the mammalian target of rapamycin (mTOR) and activating adenosine 5′-monophosphate (AMP)-activated protein kinase (AMPK). In the initial stage, the ULK1 and VPS34 complexes are most essential in recruiting and assembling other components that are needed for autophagy. There are more autophagy-related genes (ATGs) involved in the whole process of autophagy (Hsu et al., 2018; Chang, 2020; Gu et al., 2020). A large number of studies have been published on the complicated and context-dependent role of autophagy in cancer (White et al., 2015; Levy et al., 2017). Although autophagy served as a double-edged sword in the carcinogenesis, progression, treatment, and resistance of tumors (White, 2015; Limpert et al., 2018; Monkkonen and Debnath, 2018), most studies have suggested that autophagy is vital to promote tumor growth and survival. Nowadays, blocking autophagy as a potential anticancer therapy is currently undergoing clinical trials (Bortnik and Gorski, 2017; Chude and Amaravadi, 2017). Autophagy serves a cytoprotective role in cancer through its capability to support cancer metabolism. Given that autophagy can degrade various substrates, it is not surprising that autophagy provides cancer cells with abundant metabolic plasticity, for example, degradation of protein or peptide into amino acid could fuel the tricarboxylic acid (TCA) cycle (Kimmelman and White, 2017).
Importantly, the combination of amino acid degrading enzymes and autophagy regulators has been demonstrated to show marvelous synergistic anticancer effects in preclinical and clinical studies (Kim et al., 2009b; Zeng et al., 2013; Song et al., 2015). This review will highlight recent advances in the development of amino acid depriving enzymes and the combinational employment of autophagy regulators and enzymes which have been successfully used as potential therapeutic approaches in the therapy of cancer.
L-Asparaginase and Autophagy
L-asparaginase, the first bacterial enzyme approved for cancer therapy, hydrolyzes L-asparagine (ASN) and yields aspartic acid and ammonia (Pieters et al., 2011). ASN plays an important role in glycoproteins biosynthesis, regulating the functions of the immune and nervous systems (Wu, 2013; Knott et al., 2018). Importantly, several types of tumors, particularly leukemia cells, cannot synthesize ASN due to lack of or low expression of asparagine synthetase (Willems et al., 2013), which renders the tumors sensitive to asparaginase. The first commercial drug of L-asparaginase is a native E. coli-derived asparaginase, an indication of which is ALL. Although L-asparaginase derived from E. coli achieved great therapeutic improvements, it is subject to hypersensitivity and other toxicities, such as hepatic and renal dysfunction (Spiers and Wade, 1979; Salzer et al., 2014). A more stable and efficient form of L-asparaginase derived from E. coli was PEGylated to reduce the allergy to foreign proteins and prolong half-life (Dinndorf et al., 2007). Nowadays, L-asparaginase derived from E. coli has been applied as first-line therapy and L-asparaginase derived from Erwinia chrysanthemi has been used for the treatment of ALL patients when hypersensitivity to E. coli-derived L-asparaginase happens (Keating, 2013). Apart from hypersensitivity, glutamine depletion is another clinical problem of L-asparaginase due to its dual asparaginase and glutaminase activity, which can cause hepatotoxicity, thrombotic complication, and neurotoxicity (Reinert et al., 2006). Researchers have explored solutions by modifying L-asparaginase via replacing amino acid residues (Derst et al., 2000).
One of the greatest obstacles faced by L-asparaginase in clinical applications is the development of drug resistance. We reported that L-asparaginase not only induced caspase 3-dependent apoptosis but also triggered obvious autophagy in chronic myeloid leukemia (CML) cells, accompanied by inhibition of Akt/mTOR and activation of the ERK signaling pathway (Song et al., 2015), as illustrated in Figure 1. The blocking of autophagy by LY294002, chloroquine (CQ), and quinacrine enhanced apoptosis is triggered by L-asparaginase, suggesting the pro-survival role of autophagy in L-asparaginase-treated CML cells. Moreover, the ROS-p53 feedback loop played an important role in the combinational treatment of L-asparaginase and CQ. In addition to CML, ALL, glioblastoma, laryngeal squamous cell carcinoma, and pulmonary adenocarcinoma showed sensitivity to L-asparaginase, and autophagy was demonstrated to be activated through autophagosomes formation and the conversion of cytoplasmic LC3-I to membranal LC3-II (Zhang et al., 2016; Chen et al., 2017; Ji et al., 2017; Takahashi et al., 2017; Polak et al., 2019). Moreover, the combination of CQ and L-asparaginase significantly enhanced the antitumor effect of L-asparaginase. Based on the studies mentioned, autophagy played a cytoprotective role in most cancer therapy of L-asparaginase, which indicated that both targeting asparagine metabolism and autophagy was a new promising therapeutic strategy for malignant tumors. More studies and evaluations of the combinational treatment of amino acid degrading enzymes and autophagy regulators are listed in Table 1.
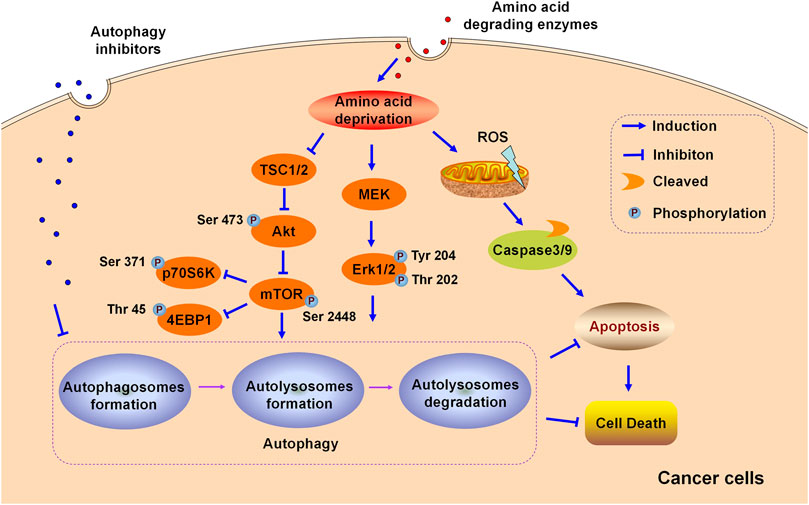
FIGURE 1. Schematic illustration of the cytoprotective role of autophagy in the cytotoxicity induced by amino acid degrading enzymes (L-Asparaginase, ADI, and rhArg) in cancer cells.
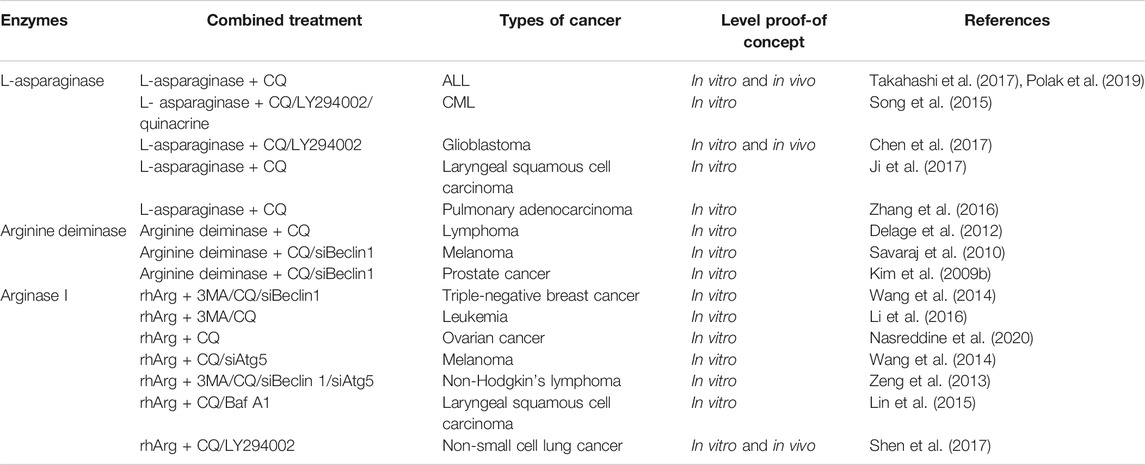
TABLE 1. Current developments of the combination of amino acid degrading enzymes and autophagy inhibitors in pre-clinical studies.
It is noteworthy that there is a close relationship between autophagy and the immune system (Gonzalo and Coll-Bonfill, 2019), which is vital for efficient cancer therapy (Jin et al., 2017; Yamamoto et al., 2020). L-asparaginase is demonstrated to not only suppress autophagy in macrophages but also inhibit phagocytosis, MHC-II expression, secretion of cytokine IL-6, and TNF-α in activated macrophages. Importantly, activating autophagy could overcome the immune suppression triggered by L-asparaginase in macrophages (Song et al., 2017).
Arginine Deiminase and Autophagy
Arginine is a semi-essential amino acid that cannot be adequately supplied by endogenous biosynthesis. Arginine metabolism is complicated, as arginine plays an essential role in several biological functions, such as precursors for cell signaling molecules as well as intermediates of the urea cycle and TCA cycle (Shen and Shen, 2006; Ni et al., 2008). Arginine can be produced through sucargininosuccinate synthetase (ASS), ornithine transcarbamylase (OTC), and argininosuccinate lyase in the urea cycle. ASS and OTC are the key enzymes for arginine synthesis (Fultang et al., 2020).
Arginine deiminase (ADI), an arginine-depleting enzyme, is regarded as a novel anticancer candidate (Synakiewicz et al., 2014) and hydrolyzes arginine into citrulline and ammonia. For ADI, ASS-deficient tumors are arginine-auxotrophic and its efficiency is highly dependent on the ASS deficiency of cancer patients (Lam et al., 2009). ASS silencing in cholangiocarcinoma cells (Roeksomtawin et al., 2018) and glioblastoma multiforme cells (Przystal et al., 2018) significantly enhanced their sensitivity to ADI-PEG20 treatment. It is reported that ADI-PEG20 significantly reduced tumor growth in ASSlow sarcoma models. However, significantly increased expression of ASS was observed in tumor tissue over time (Delage et al., 2012; Bean et al., 2016). In randomized clinical trials, neutralizing antibodies against ADI-PEG20 and enhanced ASS expression was found in the latter treatment, which caused drug resistance (Szlosarek et al., 2017). Pegylation significantly reduces antigenicity of ADI and ADI-PEG20 has been used in clinical trials in patients with hepatocellular carcinoma (HCC) (in phase III), melanoma (in phase I/II), small cell lung cancer (SCLC) (in phase II), mesothelioma (in phase I/II) and other arginine-auxotrophic advanced tumors.
In addition to neutralizing antibodies and enhanced ASS expression, autophagy is reported to be associated with ADI-resistance. Arginine deprivation by ADI could induce obvious autophagy and autophagy inhibitors potentiated the cytotoxicity of ADI-PEG20 in lymphoma cells, indicating the cytoprotective role of autophagy triggered by ADI-PEG20 in lymphoma. Furthermore, blocking autophagy by CQ or silencing autophagy-related Beclin1 mRNA accelerated and enhanced the antitumor effect of ADI in melanoma (Savaraj et al., 2010) and prostate cancer (Kim et al., 2009a; Kim et al., 2009b), suggesting that both targeting autophagy and arginine metabolism may provide novel potential avenues for cancer therapy. ADI activated MEK and ERK signaling pathways but suppressed the Akt/mTOR pathway in melanoma cells, as shown in Figure 1. In general, Akt/mTOR and ERK signaling pathways are vital in manipulating autophagy in eukaryotic cells (Dai et al., 2019; Farias Quipildor et al., 2019). Nutritional starvation triggers autophagy via inhibiting mTOR, which is a vital negative regulator of autophagy. The ERK signaling pathway is documented to regulate the induction of autophagy by interacting with LC3 and the expression of autophagy as well as lysosomal genes.
It was observed that depriving arginine by ADI triggered a cytotoxic excessive autophagy which contributed to cell death in ASS-deficient prostate cancer cells. Representative micrographs exhibited atypical autophagy with large autophagosomes formation, nucleus membrane rupture, and DNA/chromatin leakage was captured by autophagosomes, which was referred to as chromatin autophagy (Changou et al., 2014; Li et al., 2019). ADI triggered regular-sized autophagosomes during the first 24 h and giant autophagosomes after 48 h in prostate cancer cells. Moreover, ADI triggered mitochondrial dysfunction, for example, mitochondrial membrane potential depolarization (Kung et al., 2015).
Arginase and Autophagy
Another arginine degrading enzyme used for cancer therapy is arginase I. Previous studies have suggested that cancers with deficiencies in either ASS and/or OTC expression are sensitive to arginine-auxotrophy induced by arginase I (Cheng et al., 2007; Tsui et al., 2009). Recombinant human arginase I (rhArg) is a modified enzyme, which contains cobalt (II) ion or manganese (II) ion (Cheng et al., 2007; Lam et al., 2009; Yau et al., 2013). During a preclinical evaluation, rhArg exhibited significant antitumor activity in many ASS and/or OTC deficient cancer cells, namely HCC (Cheng et al., 2007), melanoma (Wang et al., 2014b), breast cancer (Leung et al., 2019), keratinocytic carcinoma (Bobak et al., 2010), SCLC (Xu et al., 2018) and Merkel cell carcinoma (Agnello et al., 2020). Pegylated rhArg has a remarkable advantage over the native arginase I on account of the extended half-life, from several hours to 72–96 h, due to its enhanced stability (Lam et al., 2011). According to the records on ClinicalTrials.gov, PEG-rhArg has been studied in clinical trials for a variety of malignant cancers, including HCC, pediatric AML, pediatric ALL, and prostate adenocarcinoma.
Arginase I is mainly expressed in the liver. Blocking autophagy by liver-specific deletion of the important autophagy genes Atg7 and Atg5, which generated circulating arginase I and inhibited tumor growth and identifies a metabolic vulnerability of cancer. Moreover, supplementation with arginine in Atg7-deficient mice model partially relieved arginine reduction and tumor growth inhibition. Whole-body deletion of Atg7 in a mice model triggered a bigger regression of KRAS-driven tumors than the knockdown of cancer-specific autophagy, suggesting that basal autophagy facilitates tumor growth (Poillet-Perez et al., 2018). Previously, we reported that rhArg suppressed cell growth of triple-negative breast cancer (TNBC), which lacks an effective druggable target, resulting in poor prognosis. Also, autophagic flux was observed in TNBC cells. Blocking autophagy by CQ, 3-MA and silencing Beclin1 enhanced the antitumor effect of rhArg in TNBC (Wang et al., 2014a). Until now, rhArg was also found to have an inhibitory effect on melanoma cells (Wang et al., 2014b), non-Hodgkin’s lymphoma cells (Zeng et al., 2013), laryngeal squamous cell carcinoma cells (Lin et al., 2015), leukemia cells (Li et al., 2016), ovarian cancer cells (Nasreddine et al., 2020), and non-small-cell lung cancer (NSCLC) cells (Shen et al., 2017). Autophagy inhibitors enhanced the antitumor effect of rhArg in these tumors, indicating that autophagy is pro-survival in the treatment of cancer (as shown in Table 1). Notably, arginase I was reported to contribute to tumor-driven immune suppression which is a major obstacle for the immunotherapy of cancer (Czystowska-Kuzmicz et al., 2019; Wang et al., 2019).
Other Amino Acid Enzymes and Autophagy
Apart from asparaginase, arginine deiminase, arginase, some other amino acid enzymes have been recently developed for cancer therapy, including methionase, lysine oxidase, phenylalanine ammonia lyase, and glutaminase. These amino acid degrading enzymes and their related autophagy studies are relatively fewer than the three enzymes discussed above.
Glutaminase is a vital enzyme that breaks down glutamine into glutamate. Glutaminase is not regarded as a potential drug for cancer therapy, but, instead, as a druggable target (Masisi et al., 2020). Cancers with high glutaminase expression are related to poor prognosis. Recently, the strategy of cancer therapy in glutamine metabolism inhibition has begun to concentrate on glutamine deprivation, glutaminase blocking, and membrane glutamine transporter inhibition (Chiu et al., 2014; Gross et al., 2014; Lee et al., 2014; Song et al., 2018). Among glutaminase inhibitors, CB-839, one of the most successful drug candidates, is under clinical trials for NSCLC, melanoma, and leukemia (NCT03965845, NCT02771626, and NCT02071927 respectively). It was reported that glutamine deprivation was synthetically lethal for autophagy inhibition in colorectal cancer (Li et al., 2017). Autophagy is an essential process that provides glutamine for anaplerosis of the TCA cycle in pancreatic ductal adenocarcinoma. Therefore, targeting glutamine metabolism and autophagy simultaneously to completely inhibit glutamine uptake offers a novel therapeutic approach for treating refractory cancers (Seo et al., 2016).
Methionase, also named L-Methionine-γ-lyase, converts methionine into ammonia, α-ketobutyrate, and methanethiol (Cellarier et al., 2003; Thivat et al., 2007; Ho et al., 2011). Methionine-dependent cancer cannot generate or generate low levels of methionine. Methionase was regarded as a potential anticancer candidate for Lewis lung, human colon carcinoma (Tan et al., 1998), neuroblastoma (Hu and Cheung, 2009), and glioblastoma (Kokkinakis et al., 2001). PEGylated recombinant methionase has been developed into phase I clinical trials, in which recombinant methionase showed no significant toxicity (Tan et al., 1996; Tan et al., 1997). However, the antitumor activity of PEGylated recombinant methionase was not reported. Notably, methionine acts as a signal for amino acid which could suppress autophagy induced by nitrogen starvation via methylation of PP2A (a protein phosphatase enzyme), also depleting methionine and cystine induced autophagy and suppressed tumor growth in glioma cells in vivo.
Lysine oxidase, one of the most studied amino acid oxidases, showed considerable cytotoxicity against a wide variety of cancers, including leukemia, colorectal adenocarcinoma, prostate cancer, pheochromocytoma (Pokrovsky et al., 2013; Lukasheva et al., 2015). The short half-life of lysine oxidase restricted its development and commercialization (Krupyanko et al., 2017). Moreover, a few studies have shown that lysine oxidase supports the growth of some tumors (Wang et al., 2016), which makes the role of lysine oxidase in antitumor therapy controversial and, therefore, demands more preclinical data.
Phenylalanine ammonia lyase converts phenylalanine to trans-cinnamic acid and ammonia. Like other enzymes. The antitumor mechanism of phenylalanine ammonia lyase is associated with a reduced level and disability of synthesis of phenylalanine. Phenylalanine ammonia lyase showed to be effective against colorectal cancer in vivo (Yang et al., 2019) and leukemic lymphoblasts in vitro (Stith et al., 1973).
Conclusion
There exist several advantages of amino acid degrading enzymes over conventional anticancer therapeutics. Firstly, amino acid enzymes have strong effects against specific amino acid auxotrophic tumors. Secondly, the side effect pattern of the enzymes is unique, which is significant for drug combinational therapy. Lastly, there exist key synthetases as biomarkers to forecast the therapeutic effect (Timosenko et al., 2017; Pokrovsky et al., 2019). Clinical trials of amino acid-degrading enzymes have shown that enzyme treatment is a safe and effective therapeutic approach. Despite the advantages of amino acid in depleting enzymes, a few weaknesses still affect clinical applications. The high immunogenicity and shorter half-life may be the greatest obstacles in the development of drugs (Schiffmann et al., 2019; Thisted et al., 2019). Chemical modification, construction of fusion protein, and encapsulation of enzymes are some of the existing solutions to overcome those obstacles and increase the bioavailability of amino acid degrading enzymes (Veronese, 2001; Li et al., 2007; Chen and Zeng, 2016; Bilal et al., 2018; Sinha and Shukla, 2019).
Recently, both targeting autophagy and amino acid metabolism have entered into clinical studies on the basis of preclinical experiments (as shown in Table 1) and synergistic drug effects in cancer therapy. Combinational therapy is a great opportunity for cancer patients. Although the context-dependent role of autophagy during tumor treatment has attracted great attention, amino acid degrading enzyme induced pro-survival autophagy in the majority of tumors. Therefore, manipulating autophagy provides a chance to make a tumor more sensitive to subsequent therapeutics. Among them, CQ is one of the most used autophagy inhibitors. CQ inhibits autophagosome fusing with lysosome, and significantly improves the expression level of LC3-II. Furthermore, there is a growing body of literature that recognizes the importance of potential applications of autophagy related proteins, including LC3, ATG7, ATG5, Beclin1, and SH3GLB1, as prognostic biomarkers in some tumors, like glioma, breast cancer, and colon cancer (Park et al., 2013; Lebovitz et al., 2015). Under the right conditions, in the future, a co-targeting autophagy and amino acid metabolism may become a potential cancer therapy.
Despite the advances mentioned in this study, patients still have a poor prognosis. Hence, further studies are required to provide a deeper understanding of the underlying molecular mechanisms and more clinical trials are needed to collect evidence-based data with respect to the efficacy and safety of these therapeutics.
Author Contributions
ZW made the draft. QX, JS, and ZS revised the manuscript. HZ and MZ analyzed the scientific literature. DJ designed the study and revised the manuscript.
Funding
This work was supported by grants from the National Natural Science Foundation of China (No. 81773620, 31872746), the Shanghai Sailing Program (17YF1405100) and Key Innovative Team of Shanghai Top-Level University Capacity Building in Clinical Pharmacy and Regulatory Science at Shanghai Medical College, Fudan University (HJW-R-2019-66-19).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Abbreviations
3MA, 3-methyladenine; ALL, acute lymphoblastic leukemia; AML, acute myeloid leukemia; Baf A1, bafilomycin A1; CQ, chloroquine; rhArg, recombinant human arginase I; siAtg5, siRNA targeting Atg5; siBeclin, siRNA targeting Beclin1.
References
Agnello, G., Alters, S. E., and Rowlinson, S. W. (2020). Preclinical safety and antitumor activity of the arginine-degrading therapeutic enzyme pegzilarginase, a PEGylated, cobalt-substituted recombinant human arginase 1. Transl. Res. 217, 11–22. doi:10.1016/j.trsl.2019.12.005
Bean, G. R., Kremer, J. C., Prudner, B. C., Schenone, A. D., Yao, J. C., Schultze, M. B., et al. (2016). A metabolic synthetic lethal strategy with arginine deprivation and chloroquine leads to cell death in ASS1-deficient sarcomas. Cell Death Dis. 7, e2406. doi:10.1038/cddis.2016.232
Bilal, M., Iqbal, H. M. N., Guo, S., Hu, H., Wang, W., and Zhang, X. (2018). State-of-the-art protein engineering approaches using biological macromolecules: a review from immobilization to implementation view point. Int. J. Biol. Macromol. 108, 893–901. doi:10.1016/j.ijbiomac.2017.10.182
Bobak, Y. P., Vynnytska, B. O., Kurlishchuk, Y. V., Sibirny, A. A., and Stasyk, O. V. (2010). Cancer cell sensitivity to arginine deprivation in vitro is not determined by endogenous levels of arginine metabolic enzymes. Cell Biol. Int. 34 (11), 1085–1089. doi:10.1042/cbi20100451
Bortnik, S., and Gorski, S. M. (2017). Clinical applications of autophagy proteins in cancer: from potential targets to biomarkers. Int. J. Mol. Sci. 18, 1496. doi:10.3390/ijms18071496
Cellarier, E., Durando, X., Vasson, M. P., Farges, M. C., Demiden, A., Maurizis, J. C., et al. (2003). Methionine dependency and cancer treatment. Canc. Treat Rev. 29, 489–499. doi:10.1016/s0305-7372(03)00118-x
Chang, N. C. (2020). Autophagy and stem cells: self-eating for self-renewal. Front Cell Dev Biol 8, 138. doi:10.3389/fcell.2020.00138
Changou, C. A., Chen, Y. R., Xing, L., Yen, Y., Chuang, F. Y., et al. (2014). Arginine starvation-associated atypical cellular death involves mitochondrial dysfunction, nuclear DNA leakage, and chromatin autophagy. Proc. Natl. Acad. Sci. U.S.A. 111, 14147–14152. doi:10.1073/pnas.1404171111
Chen, Q., Ye, L., Fan, J., Zhang, X., Wang, H., Liao, S., et al. (2017). Autophagy suppression potentiates the anti-glioblastoma effect of asparaginase in vitro and in vivo. Oncotarget 8, 91052–91066. doi:10.18632/oncotarget.19409
Chen, Z., and Zeng, A. P. (2016). Protein engineering approaches to chemical biotechnology. Curr. Opin. Biotechnol. 42, 198–205. doi:10.1016/j.copbio.2016.07.007
Cheng, P. N., Lam, T. L., Lam, W. M., Tsui, S. M., Cheng, A. W., Lo, W. H., et al. (2007). Pegylated recombinant human arginase (rhArg-peg5,000mw) inhibits the in vitro and in vivo proliferation of human hepatocellular carcinoma through arginine depletion. Canc. Res. 67, 309–317. doi:10.1158/0008-5472.can-06-1945
Chiu, M., Tardito, S., Pillozzi, S., Arcangeli, A., Armento, A., Uggeri, J., et al. (2014). Glutamine depletion by crisantaspase hinders the growth of human hepatocellular carcinoma xenografts. Br. J. Canc. 111, 1159–1167. doi:10.1038/bjc.2014.425
Chude, C. I., and Amaravadi, R. K. (2017). Targeting autophagy in cancer: update on clinical trials and novel inhibitors. Int. J. Mol. Sci. 18, 1279. doi:10.3390/ijms18061279
Czystowska-Kuzmicz, M., Sosnowska, A., Nowis, D., Ramji, K., Szajnik, M., Chlebowska-Tuz, J., et al. (2019). Small extracellular vesicles containing arginase-1 suppress T-cell responses and promote tumor growth in ovarian carcinoma. Nat. Commun. 10 (1), 3000. doi:10.1038/s41467-019-10979-3
Dai, D. F., Liu, Y., Basisty, N., Karunadharma, P., Dastidar, S. G., Chiao, Y. A., et al. (2019). Differential effects of various genetic mouse models of the mechanistic target of rapamycin complex I inhibition on heart failure. Geroscience 41 (6), 847–860. doi:10.1007/s11357-019-00119-6
Dang, C. V., Hamaker, M., Sun, P., Le, A., and Gao, P. (2011). Therapeutic targeting of cancer cell metabolism. J. Mol. Med. 89, 205–212. doi:10.1007/s00109-011-0730-x
Delage, B., Luong, P., Maharaj, L., O’Riain, C., Syed, N., Crook, T., et al. (2012). Promoter methylation of argininosuccinate synthetase-1 sensitises lymphomas to arginine deiminase treatment, autophagy and caspase-dependent apoptosis. Cell Death Dis. 3, e342. doi:10.1038/cddis.2012.83
Derst, C., Henseling, J., and Röhm, K. H. (2000). Engineering the substrate specificity of Escherichia coli asparaginase. II. Selective reduction of glutaminase activity by amino acid replacements at position 248. Protein Sci. 9 (10), 2009–2017. doi:10.1110/ps.9.10.2009
Dhankhar, R., Gupta, V., Kumar, S., Kapoor, R. K., and Gulati, P. (2020). Microbial enzymes for deprivation of amino acid metabolism in malignant cells: biological strategy for cancer treatment. Appl. Microbiol. Biotechnol. 104 (7), 2857–2869. doi:10.1007/s00253-020-10432-2
Dinndorf, P. A., Gootenberg, J., Cohen, M. H., Keegan, P., and Pazdur, R. (2007). FDA drug approval summary: pegaspargase (oncaspar) for the first-line treatment of children with acute lymphoblastic leukemia (ALL). Oncol. 12, 991–998. doi:10.1634/theoncologist.12-8-991
Farias Quipildor, G. E., Mao, K., Hu, Z., Novaj, A., Cui, M. H., Gulinello, M., et al. (2019). Central IGF-1 protects against features of cognitive and sensorimotor decline with aging in male mice. Geroscience 41 (2), 185–208. doi:10.1007/s11357-019-00065-3
Fernandes, H. S., Silva Teixeira, C. S., Fernandes, P. A., Ramos, M. J., and Cerqueira, N. M. (2017). Amino acid deprivation using enzymes as a targeted therapy for cancer and viral infections. Expert Opin. Ther. Pat. 27, 283–297. doi:10.1080/13543776.2017.1254194
Fulop, G. A., Kiss, T., Tarantini, S., Balasubramanian, P., Yabluchanskiy, A., Farkas, E., et al. (2018). Nrf2 deficiency in aged mice exacerbates cellular senescence promoting cerebrovascular inflammation. Geroscience 40 (5–6), 513–521. doi:10.1007/s11357-018-0047-6
Fultang, L., Booth, S., Yogev, O., Martins da Costa, B., Tubb, V., Panetti, S., et al. (2020). Metabolic engineering against the arginine microenvironment enhances CAR-T cell proliferation and therapeutic activity. Blood 136, 1155–1160. doi:10.1182/blood.2019004500
Fung, M. K. L., and Chan, G. C. (2017). Drug-induced amino acid deprivation as strategy for cancer therapy. J. Hematol. Oncol. 10, 144. doi:10.1186/s13045-017-0509-9
Gao, X., Sanderson, S. M., Dai, Z., Reid, M. A., Cooper, D. E., Lu, M., et al. (2019). Dietary methionine influences therapy in mouse cancer models and alters human metabolism. Nature 572, 397–401. doi:10.1038/s41586-019-1437-3
Gonzalo, S., and Coll-Bonfill, N. (2019). Genomic instability and innate immune responses to self-DNA in progeria. Geroscience 41 (3), 255–266. doi:10.1007/s11357-019-00082-2
Gross, M. I., Demo, S. D., Dennison, J. B., Chen, L., Chernov-Rogan, T., Goyal, B., et al. (2014). Antitumor activity of the glutaminase inhibitor CB-839 in triple-negative breast cancer. Mol. Canc. Therapeut. 13, 890–901. doi:10.1158/1535-7163.mct-13-0870
Gu, Y., Han, J., Jiang, C., and Zhang, Y. (2020). Biomarkers, oxidative stress and autophagy in skin aging. Ageing Res. Rev. 59, 101036. doi:10.1016/j.arr.2020.101036
Ho, E., Beaver, L. M., Williams, D. E., and Dashwood, R. H. (2011). Dietary factors and epigenetic regulation for prostate cancer prevention. Adv. Nutr. 2, 497–510. doi:10.3945/an.111.001032
Honda, S., Arakawa, S., Nishida, Y., Yamaguchi, H., Ishii, E., and Shimizu, S. (2014). Ulk1-mediated Atg5-independent macroautophagy mediates elimination of mitochondria from embryonic reticulocytes. Nat. Commun. 5, 4004. doi:10.1038/ncomms5004
Hsu, C. L., Lee, E. X., Gordon, K. L., Paz, E. A., Shen, W. C., Ohnishi, K., et al. (2018). MAP4K3 mediates amino acid-dependent regulation of autophagy via phosphorylation of TFEB. Nat. Commun. 9 (1), 942. doi:10.1038/s41467-018-03340-7
Hu, J., and Cheung, N. K. (2009). Methionine depletion with recombinant methioninase: in vitro and in vivo efficacy against neuroblastoma and its synergism with chemotherapeutic drugs. Int. J. Canc. 124, 1700–1706. doi:10.1002/ijc.24104
Ji, Y., Li, L., Tao, Q., Zhang, X., Luan, J., Zhao, S., Liu, H., and Ju, D. (2017). Deprivation of asparagine triggers cytoprotective autophagy in laryngeal squamous cell carcinoma. Appl. Microbiol. Biotechnol. 101, 4951–4961. doi:10.1007/s00253-017-8221-9
Jin, Y., Hong, Y., Park, C. Y., and Hong, Y. (2017). Molecular interactions of autophagy with the immune system and cancer. Int. J. Mol. Sci. 18, 1694. doi:10.3390/ijms18081694
Keating, G. M. (2013). Asparaginase Erwinia chrysanthemi (Erwinaze®): a guide to its use in acute lymphoblastic leukemia in the USA. BioDrugs 27, 413–418. doi:10.1007/s40259-013-0051-4
Kim, R. H., Bold, R. J., and Kung, H. J. (2009a). ADI, autophagy and apoptosis: metabolic stress as a therapeutic option for prostate cancer. Autophagy 5, 567–568. doi:10.4161/auto.5.4.8252
Kim, R. H., Coates, J. M., Bowles, T. L., McNerney, G. P., Sutcliffe, J., Jung, J. U., et al. (2009b). Arginine deiminase as a novel therapy for prostate cancer induces autophagy and caspase-independent apoptosis. Canc. Res. 69, 700–708. doi:10.1158/0008-5472.can-08-3157
Kimmelman, A. C., and White, E. (2017). Autophagy and tumor metabolism. Cell Metabol. 25, 1037–1043. doi:10.1016/j.cmet.2017.04.004
Knott, S. R. V., Wagenblast, E., Khan, S., Kim, S. Y., Soto, M., Wagner, M., et al. (2018). Asparagine bioavailability governs metastasis in a model of breast cancer. Nature 554, 378–381. doi:10.1038/nature25465
Kokkinakis, D. M., Hoffman, R. M., Frenkel, E. P., Wick, J. B., Han, Q., Xu, M., et al. (2001). Synergy between methionine stress and chemotherapy in the treatment of brain tumor xenografts in athymic mice. Canc. Res. 61, 4017–4023.
Koppenol, W. H., Bounds, P. L., and Dang, C. V. (2011). Otto Warburg’s contributions to current concepts of cancer metabolism. Nat. Rev. Canc. 11, 325–337. doi:10.1038/nrc3038
Krupyanko, V. I., Medentsev, A. G., Lukasheva, E. V., and Arinbasarova, A. Y. (2017). Kinetic characteristics of L-lysine α- oxidase from Trichoderma cf. aureoviride Rifai VKM F-4268D: substrate specificity and allosteric effects. Biochem. Biophys. Rep. 9, 9–12. doi:10.1016/j.bbrep.2016.11.003
Kung, H. J., Changou, C. A., Li, C. F., and Ann, D. K. (2015). Chromatophagy: autophagy goes nuclear and captures broken chromatin during arginine-starvation. Autophagy 11, 419–421. doi:10.1080/15548627.2015.1009789
Lam, T. L., Wong, G. K., Chong, H. C., Cheng, P. N., Choi, S. C., Chow, T. L., et al. (2009). Recombinant human arginase inhibits proliferation of human hepatocellular carcinoma by inducing cell cycle arrest. Canc. Lett. 277, 91–100. doi:10.1016/j.canlet.2008.11.031
Lam, T. L., Wong, G. K., Chow, H. Y., Chong, H. C., Chow, T. L., Kwok, S. Y., et al. (2011). Recombinant human arginase inhibits the in vitro and in vivo proliferation of human melanoma by inducing cell cycle arrest and apoptosis. Pigment Cell Melanoma Res. 24, 366–376. doi:10.1111/j.1755-148X.2010.00798.x
Lebovitz, C. B., DeVorkin, L., Bosc, D., Rothe, K., Singh, J., Bally, M., et al. (2015). Precision autophagy: will the next wave of selective autophagy markers and specific autophagy inhibitors feed clinical pipelines? Autophagy 11, 1949–1952. doi:10.1080/15548627.2015.1078962
Lee, Y. Z., Yang, C. W., Chang, H. Y., Hsu, H. Y., Chen, I. S., Chang, H. S., et al. (2014). Discovery of selective inhibitors of Glutaminase-2, which inhibit mTORC1, activate autophagy and inhibit proliferation in cancer cells. Oncotarget 5, 6087–6101. doi:10.18632/oncotarget.2173
Leung, S. L., Ho, M. K., Lam, Y. M., Chow, H. Y., So, Y. H., and Leung, Y. C. (2019). PEGylated recombinant human arginase as a drug for breast cancer. Hong Kong Med. J. 25 (Suppl. 9), 28–31
Levy, J. M. M., Towers, C. G., and Thorburn, A. (2017). Targeting autophagy in cancer. Nat. Rev. Canc. 17, 528–542. doi:10.1038/nrc.2017.53
Li, I. T., Pham, E., and Truong, K. (2007). Current approaches for engineering proteins with diverse biological properties. Adv. Exp. Med. Biol. 620, 18–33. doi:10.1007/978-0-387-76713-0_2
Li, Y., Zeng, X., Wang, S., Fan, J., Wang, Z., Song, P., et al. (2016). Blocking autophagy enhanced leukemia cell death induced by recombinant human arginase. Tumour Biol 37, 6627–6635. doi:10.1007/s13277-015-4253-x
Li, J., Song, P., Zhu, L., Aziz, N., Zhou, Q., Zhang, Y., et al. (2017). Synthetic lethality of glutaminolysis inhibition, autophagy inactivation and asparagine depletion in colon cancer. Oncotarget 8, 42664–42672. doi:10.18632/oncotarget.16844
Li, Y., Jiang, X., Zhang, Y., Gao, Z., Liu, Y., Hu, J., et al. (2019). Nuclear accumulation of UBC9 contributes to SUMOylation of lamin A/C and nucleophagy in response to DNA damage. J. Exp. Clin. Canc. Res. 38, 67. doi:10.1186/s13046-019-1048-8
Limpert, A. S., Lambert, L. J., Bakas, N. A., Bata, N., Brun, S. N., Shaw, R. J., et al. (2018). Autophagy in cancer: regulation by small molecules. Trends Pharmacol. Sci. 39, 1021–1032. doi:10.1016/j.tips.2018.10.004
Lin, C., Wang, Z., Li, L., He, Y., Fan, J., Liu, Z., et al. (2015). The role of autophagy in the cytotoxicity induced by recombinant human arginase in laryngeal squamous cell carcinoma. Appl. Microbiol. Biotechnol. 99, 8487–8494. doi:10.1007/s00253-015-6565-6
Lukasheva, E. V., Ribakova, Y. S., Fedorova, T. N., Makletsova, M. G., Arinbasarova, A. Y., Medentzev, A. G., et al. (2015). [Effect of L-lysine alpha-oxidase from Trichoderma cf. aureoviride Rifai ВКМF-4268D on pheochromocytoma PC12 cell line]. Biomed Khim 61, 99–104. doi:10.18097/pbmc20156101099
Masisi, B. K., El Ansari, R., Alfarsi, L., Rakha, E. A., Green, A. R., and Craze, M. L. 2020). The role of glutaminase in cancer. Histopathology 76 (4), 498–508. doi:10.1111/his.14014
Monkkonen, T., and Debnath, J. (2018). Inflammatory signaling cascades and autophagy in cancer. Autophagy 14, 190–198. doi:10.1080/15548627.2017.1345412
Mony, V. K., Benjamin, S., and O’Rourke, E. J. (2016). A lysosome-centered view of nutrient homeostasis. Autophagy 12, 619–631. doi:10.1080/15548627.2016.1147671
Nacarelli, T., Azar, A., Altinok, O., Orynbayeva, Z., and Sell, C. (2018). Rapamycin increases oxidative metabolism and enhances metabolic flexibility in human cardiac fibroblasts. Geroscience 40, 243–256. doi:10.1007/s11357-018-0030-2
Nasreddine, G., El-Sibai, M., and Abi-Habib, R. J. (2020). Cytotoxicity of [HuArgI (co)-PEG5000]-induced arginine deprivation to ovarian cancer cells is autophagy dependent. Invest. N. Drugs 38 (1), 10–19. doi:10.1007/s10637-019-00756-w
Ni, Y., Schwaneberg, U., and Sun, Z. H. (2008). Arginine deiminase, a potential anti-tumor drug. Canc. Lett. 261, 1–11. doi:10.1016/j.canlet.2007.11.038
Nicholatos, J. W., Robinette, T. M., Tata, S. V. P., Yordy, J. D., Francisco, A. B., Platov, M., et al. (2019). Cellular energetics and mitochondrial uncoupling in canine aging. Geroscience 41 (2), 229–242. doi:10.1007/s11357-019-00062-6
Nishida, Y., Arakawa, S., Fujitani, K., Yamaguchi, H., Mizuta, T., Kanaseki, T., et al. (2009). Discovery of Atg5/Atg7-independent alternative macroautophagy. Nature 461, 654–658. doi:10.1038/nature08455
Park, J. M., Huang, S., Wu, T. T., Foster, N. R., and Sinicrope, F. A. (2013). Prognostic impact of Beclin 1, p62/sequestosome 1 and LC3 protein expression in colon carcinomas from patients receiving 5-fluorouracil as adjuvant chemotherapy. Canc. Biol. Ther. 14, 100–107. doi:10.4161/cbt.22954
Pieters, R., Hunger, S. P., Boos, J., Rizzari, C., Silverman, L., Baruchel, A., et al. (2011). L-asparaginase treatment in acute lymphoblastic leukemia: a focus on Erwinia asparaginase. Cancer 117, 238–249. doi:10.1002/cncr.25489
Poillet-Perez, L., Xie, X., Zhan, L., Yang, Y., Sharp, D. W., Hu, Z. S., et al. (2018). Autophagy maintains tumour growth through circulating arginine. Nature 563, 569–573. doi:10.1038/s41586-018-0697-7
Pokrovsky, V. S., Chepikova, O. E., Davydov, D. Z., Zamyatnin, A. A., Lukashev, A. N., and Lukasheva, E. V. (2019). Amino acid degrading enzymes and their application in cancer therapy. Curr. Med. Chem. 26, 446–464. doi:10.2174/0929867324666171006132729
Pokrovsky, V. S., Treshalina, H. M., Lukasheva, E. V., Sedakova, L. A., Medentzev, A. G., Arinbasarova, A. Y., et al. (2013). Enzymatic properties and anticancer activity of L-lysine α-oxidase from Trichoderma cf. aureoviride Rifai BKMF-4268D. Anti Canc. Drugs 24, 846–851. doi:10.1097/CAD.0b013e328362fbe2
Polak, R., Bierings, M. B., van der Leije, C. S., Sanders, M. A., Roovers, O., Marchante, J. R. M., et al. (2019). Autophagy inhibition as a potential future targeted therapy for ETV6-RUNX1-driven B-cell precursor acute lymphoblastic leukemia. Haematologica 104, 738–748. doi:10.3324/haematol.2018.193631
Przystal, J. M., Hajji, N., Khozoie, C., Renziehausen, A., Zeng, Q., Abaitua, F., et al. (2018). Efficacy of arginine depletion by ADI-PEG20 in an intracranial model of GBM. Cell Death Dis. 9 (12), 1192. doi:10.1038/s41419-018-1195-4
Reinert, R. B., Oberle, L. M., Wek, S. A., Bunpo, P., Wang, X. P., Mileva, I., et al. (2006). Role of glutamine depletion in directing tissue-specific nutrient stress responses to L-asparaginase. J. Biol. Chem. 281 (42), 31222–31233. doi:10.1074/jbc.M604511200
Roeksomtawin, S., Navasumrit, P., Waraprasit, S., Parnlob, V., Sricharunrat, T., Bhudhisawasdi, V., et al. (2018). Decreased argininosuccinate synthetase expression in Thai patients with cholangiocarcinoma and the effects of ADI-PEG20 treatment in CCA cell lines. Oncol. Lett. 16 (2), 1529–1538. doi:10.3892/ol.2018.8807
Rytting, M. (2010). Peg-asparaginase for acute lymphoblastic leukemia. Expet Opin. Biol. Ther. 10, 833–839. doi:10.1517/14712591003769808
Salzer, W. L., Asselin, B. L., Plourde, P. V., Corn, T., and Hunger, S. P. (2014). Development of asparaginase Erwinia chrysanthemi for the treatment of acute lymphoblastic leukemia. Ann. N. Y. Acad. Sci. 1329, 81–92. doi:10.1111/nyas.12496
Savaraj, N., You, M., Wu, C., Wangpaichitr, M., Kuo, M. T., and Feun, L. G. (2010). Arginine deprivation, autophagy, apoptosis (AAA) for the treatment of melanoma. Curr. Mol. Med. 10, 405–412. doi:10.2174/156652410791316995
Schiffmann, R., Goker-Alpan, O., Holida, M., Giraldo, P., Barisoni, L., Colvin, R. B., et al. (2019). Pegunigalsidase alfa, a novel PEGylated enzyme replacement therapy for Fabry disease, provides sustained plasma concentrations and favorable pharmacodynamics: a 1-year Phase 1/2 clinical trial. J. Inherit. Metab. Dis. 42, 534–544. doi:10.1002/jimd.12080
Seo, J. W., Choi, J., Lee, S. Y., Sung, S., Yoo, H. J., Kang, M. J., et al. (2016). Autophagy is required for PDAC glutamine metabolism. Sci. Rep. 6, 37594. doi:10.1038/srep37594
Shen, L. J., and Shen, W. C. (2006). Drug evaluation: ADI-PEG-20--a PEGylated arginine deiminase for arginine-auxotrophic cancers. Curr. Opin. Mol. Therapeut. 8, 240–248.
Shen, W., Zhang, X., Fu, X., Fan, J., Luan, J., Cao, Z., et al. (2017). A novel and promising therapeutic approach for NSCLC: recombinant human arginase alone or combined with autophagy inhibitor. Cell Death Dis. 8, e2720. doi:10.1038/cddis.2017.137
Sinha, R., and Shukla, P. (2019). Current trends in protein engineering: updates and progress. Curr. Protein Pept. Sci. 20 (5), 398–407. doi:10.2174/1389203720666181119120120
Song, M., Kim, S. H., Im, C. Y., and Hwang, H. J. (2018). Recent development of small molecule glutaminase inhibitors. Curr. Top. Med. Chem. 18, 432–443. doi:10.2174/1568026618666180525100830
Song, P., Wang, Z., Zhang, X., Fan, J., Li, Y., Chen, Q., et al. (2017). The role of autophagy in asparaginase-induced immune suppression of macrophages. Cell Death Dis. 8, e2721. doi:10.1038/cddis.2017.144
Song, P., Ye, L., Fan, J., Li, Y., Zeng, X., Wang, Z., et al. (2015). Asparaginase induces apoptosis and cytoprotective autophagy in chronic myeloid leukemia cells. Oncotarget 6, 3861–3873. doi:10.18632/oncotarget.2869
Souder, D. C., and Anderson, R. M. (2019). An expanding GSK3 network: implications for aging research. Geroscience 41 (4), 369–382. doi:10.1007/s11357-019-00085-z
Spiers, A. S., and Wade, H. E. (1979). Achromobacter L-glutaminase-L-asparaginase: human pharmacology, toxicology, and activity in acute leukemias. Canc. Treat Rep. 63, 1019–1024.
Stasyk, O. V., Boretsky, Y. R., Gonchar, M. V., and Sibirny, A. A. (2015). Recombinant arginine-degrading enzymes in metabolic anticancer therapy and bioanalytics. Cell Biol. Int. 39, 246–252. doi:10.1002/cbin.10383
Stith, W. J., Hodgins, D. S., and Abell, C. W. (1973). Effects of phenylalanine amonia-lyase and phenylalanine deprivation on murine leukemic lymphoblasts in vitro. Canc. Res. 33, 966–971.
Synakiewicz, A., Stachowicz-Stencel, T., and Adamkiewicz-Drozynska, E. (2014). The role of arginine and the modified arginine deiminase enzyme ADI-PEG 20 in cancer therapy with special emphasis on phase I/II clinical trials. Expet Opin. Invest. Drugs 23, 1517–1529. doi:10.1517/13543784.2014.934808
Szlosarek, P. W., Steele, J. P., Nolan, L., Gilligan, D., Taylor, P., Spicer, J., et al. (2017). Arginine deprivation with pegylated arginine deiminase in patients with argininosuccinate synthetase 1-deficient malignant pleural mesothelioma: a randomized clinical trial. JAMA Oncol. 3, 58–66. doi:10.1001/jamaoncol.2016.3049
Takahashi, H., Inoue, J., Sakaguchi, K., Takagi, M., Mizutani, S., and Inazawa, J. (2017). Autophagy is required for cell survival under L-asparaginase-induced metabolic stress in acute lymphoblastic leukemia cells. Oncogene 36, 4267–4276. doi:10.1038/onc.2017.59
Tan, Y., Sun, X., Xu, M., An, Z., Tan, X., Han, Q., et al. (1998). Polyethylene glycol conjugation of recombinant methioninase for cancer therapy. Protein Expr. Purif. 12, 45–52. doi:10.1006/prep.1997.0805
Tan, Y., Zavala, J., Han, Q., Xu, M., Sun, X., Tan, X., et al. (1997). Recombinant methioninase infusion reduces the biochemical endpoint of serum methionine with minimal toxicity in high-stage cancer patients. Anticancer Res. 17 (5B), 3857–3860.
Tan, Y., Zavala, J., Xu, M., Zavala, J., and Hoffman, R. M. (1996). Serum methionine depletion without side effects by methioninase in metastatic breast cancer patients. Anticancer Res. 16 (6c), 3937–3942.
Thisted, T., Biesova, Z., Walmacq, C., Stone, E., Rodnick-Smith, M., Ahmed, S. S., et al. (2019). Optimization of a nicotine degrading enzyme for potential use in treatment of nicotine addiction. BMC Biotechnol. 19, 56. doi:10.1186/s12896-019-0551-5
Thivat, E., Durando, X., Demidem, A., Farges, M. C., Rapp, M., Cellarier, E., et al. (2007). A methionine-free diet associated with nitrosourea treatment down-regulates methylguanine-DNA methyl transferase activity in patients with metastatic cancer. Anticancer Res. 27, 2779–2783.
Timosenko, E., Hadjinicolaou, A. V., and Cerundolo, V. (2017). Modulation of cancer-specific immune responses by amino acid degrading enzymes. Immunotherapy 9, 83–97. doi:10.2217/imt-2016-0118
Truelove, E., Fielding, A. K., and Hunt, B. J. (2013). The coagulopathy and thrombotic risk associated with L-asparaginase treatment in adults with acute lymphoblastic leukaemia. Leukemia 27, 553–559. doi:10.1038/leu.2012.290
Tsui, S. M., Lam, W. M., Lam, T. L., Chong, H. C., So, P. K., Kwok, S. Y., et al. (2009). Pegylated derivatives of recombinant human arginase (rhArg1) for sustained in vivo activity in cancer therapy: preparation, characterization and analysis of their pharmacodynamics in vivo and in vitro and action upon hepatocellular carcinoma cell (HCC). Canc. Cell Int. 9, 9. doi:10.1186/1475-2867-9-9
Vander Heiden, M. G., and DeBerardinis, R. J. (2017). Understanding the intersections between metabolism and cancer Biology. Cell 168, 657–669. doi:10.1016/j.cell.2016.12.039
Veronese, F. M. (2001). Peptide and protein PEGylation: a review of problems and solutions. Biomaterials 22, 405–417. doi:10.1016/s0142-9612(00)00193-9
Wang, T. H., Hsia, S. M., and Shieh, T. M. (2016). Lysyl oxidase and the tumor microenvironment. Int. J. Mol. Sci. 18. doi:10.3390/ijms18010062
Wang, Z., Shi, X., Li, Y., Fan, J., Zeng, X., Xian, Z., et al. (2014a). Blocking autophagy enhanced cytotoxicity induced by recombinant human arginase in triple-negative breast cancer cells. Cell Death Dis. 5, e1563. doi:10.1038/cddis.2014.503
Wang, Z., Shi, X., Li, Y., Zeng, X., Fan, J., Sun, Y., et al. (2014b). Involvement of autophagy in recombinant human arginase-induced cell apoptosis and growth inhibition of malignant melanoma cells. Appl. Microbiol. Biotechnol. 98, 2485–2494. doi:10.1007/s00253-013-5118-0
Wang, Z., Song, P., Li, Y., Wang, S., Fan, J., Zhang, X., et al. (2019). Recombinant human arginase I elicited immunosuppression in activated macrophages through inhibiting autophagy. Appl. Microbiol. Biotechnol. 103 (12), 4825–4838. doi:10.1007/s00253-019-09832-w
White, E. (2015). The role for autophagy in cancer. J. Clin. Invest. 125, 42–46. doi:10.1172/jci73941
White, E., Mehnert, J. M., and Chan, C. S. (2015). Autophagy, metabolism, and cancer. Clin. Canc. Res. 21, 5037–5046. doi:10.1158/1078-0432.ccr-15-0490
Willems, L., Jacque, N., Jacquel, A., Neveux, N., Maciel, T. T., Lambert, M., et al. (2013). Inhibiting glutamine uptake represents an attractive new strategy for treating acute myeloid leukemia. Blood 122, 3521–3532. doi:10.1182/blood-2013-03-493163
Wu, G. (2013). Functional amino acids in nutrition and health. Amino Acids 45, 407–411. doi:10.1007/s00726-013-1500-6
Xie, Y., Kang, R., Sun, X., Zhong, M., Huang, J., Klionsky, D. J., et al. (2015). Posttranslational modification of autophagy-related proteins in macroautophagy. Autophagy 11, 28–45. doi:10.4161/15548627.2014.984267
Xu, S., Lam, S. K., Cheng, P. N., and Ho, J. C. (2018). Recombinant human arginase induces apoptosis through oxidative stress and cell cycle arrest in small cell lung cancer. Canc. Sci. 109 (11), 3471–3482. doi:10.1111/cas.13782
Yamamoto, K., Venida, A., Yano, J., Biancur, D. E., Kakiuchi, M., Gupta, S., et al. (2020). Autophagy promotes immune evasion of pancreatic cancer by degrading MHC-I. Nature 581 (7806), 100–105. doi:10.1038/s41586-020-2229-5.2020
Yang, J., Tao, R., Wang, L., Song, L., Wang, Y., Gong, C., et al. (2019). Thermosensitive micelles encapsulating phenylalanine ammonia lyase act as a sustained and efficacious therapy against colorectal cancer. J. Biomed. Nanotechnol. 15, 717–727. doi:10.1166/jbn.2019.2734
Yau, T., Cheng, P. N., Chan, P., Chan, W., Chen, L., Yuen, J., et al. (2013). A phase 1 dose-escalating study of pegylated recombinant human arginase 1 (Peg-rhArg1) in patients with advanced hepatocellular carcinoma. Invest. N. Drugs 31, 99–107. doi:10.1007/s10637-012-9807-9
Zeng, X., Li, Y., Fan, J., Zhao, H., Xian, Z., Sun, Y., et al. (2013). Recombinant human arginase induced caspase-dependent apoptosis and autophagy in non-Hodgkin’s lymphoma cells. Cell Death Dis. 4, e840. doi:10.1038/cddis.2013.359
Keywords: autophagy, amino acid degrading enzymes, cancer therapy, L-asparaginase, arginine deiminase, recombinant human arginase, chloroquine
Citation: Wang Z, Xie Q, Zhou H, Zhang M, Song Z, Shen J and Ju D (2021) Amino Acid Degrading Enzymes and Autophagy in Cancer Therapy. Front. Pharmacol. 11:582587. doi: 10.3389/fphar.2020.582587
Received: 12 July 2020; Accepted: 20 October 2020;
Published: 11 January 2021.
Edited by:
Marco Cordani, IMDEA Nanociencia, SpainReviewed by:
Oleh Stasyk, National Academy of Sciences of Ukraine (NAN Ukraine), UkraineZhijie Xu, Central South University, China
Copyright © 2021 Wang, Xie, Zhou, Zhang, Shen and Ju. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jie Shen, c2hqNDIxQDEyNi5jb20=; Dianwen Ju, ZGlhbndlbmp1QGZ1ZGFuLmVkdS5jbg==
 Ziyu Wang1,2
Ziyu Wang1,2 Dianwen Ju
Dianwen Ju