- 1Department of Psychiatry, Universidad Autónoma de Nuevo León, Monterrey, México
- 2Department of Psychiatry and Psychology, Mayo Clinic Depression Center, Mayo Clinic, Rochester, MN, United States
Background: Epimutations secondary to gene-environment interactions have a key role in the pathophysiology of major psychiatric disorders. In vivo and in vitro evidence suggest that mood stabilizers can potentially reverse epigenetic deregulations found in patients with schizophrenia or mood disorders through mechanisms that are not yet fully understood. However, their activity on epigenetic processes has made them a research target for therapeutic approaches.
Methods: We conducted a comprehensive literature search of PubMed and EMBASE for studies investigating the specific epigenetic changes induced by non-antipsychotic mood stabilizers (valproate, lithium, lamotrigine, and carbamazepine) in animal models, human cell lines, or patients with schizophrenia, bipolar disorder, or major depressive disorder. Each paper was reviewed for the nature of research, the species and tissue examined, sample size, mood stabilizer, targeted gene, epigenetic changes found, and associated psychiatric disorder. Every article was appraised for quality using a modified published process and those who met a quality score of moderate or high were included.
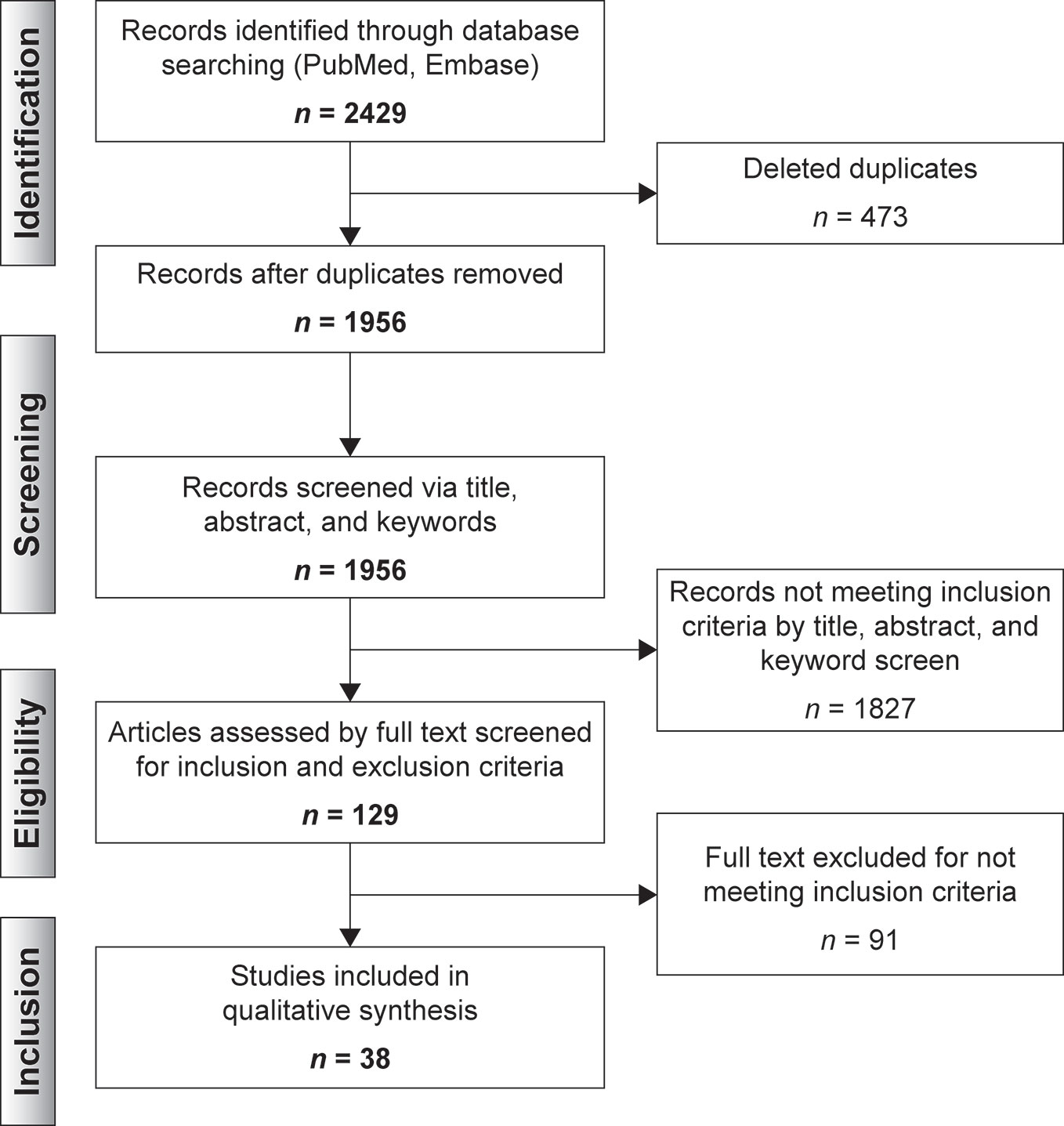
Results: A total of 2,429 records were identified; 1,956 records remained after duplicates were removed and were screened via title, abstract and keywords; 129 records were selected for full-text screening and a remaining of 38 articles were included in the qualitative synthesis. Valproate and lithium were found to induce broader epigenetic changes through different mechanisms, mainly DNA demethylation and histones acetylation. There was less literature and hence smaller effects attributable to lamotrigine and carbamazepine could be associated overall with the small number of studies on these agents. Findings were congruent across sample types.
Conclusions: An advanced understanding of the specific epigenetic changes induced by classic mood stabilizers in patients with major psychiatric disorders will facilitate personalized interventions. Further related drug discovery should target the induction of selective chromatin remodeling and gene-specific expression effects.
Introduction
Rationale
Psychiatric illness entails a severe disease burden that is associated with reduced productivity, quality of life, and life expectancy (Vigo et al., 2016). Globally, bipolar disorder (BD), major depressive disorder (MDD), and schizophrenia (SCZ) affect >1%, 6%, and ≈1% of the population, respectively (Malhi and Mann, 2018; Vieta et al., 2018; Marder and Cannon, 2019). Differentiation between these three major psychiatric diseases disorders is typically based on symptoms and course patterns. However, overlapping foundations, heterogeneous clinical features, and common underlying genetic vulnerabilities challenge traditional definitions of related nosological boundaries (Cross-Disorder Group of the Psychiatric Genomics, 2013; Cheng et al., 2018). The pleiotropic nature and the influence of gene-environment interactions on pathophysiology has focused attention towards a more comprehensive understanding of psychiatric illness and a pharmacogenomics approach of existing treatment strategies (Lopez-Leon et al., 2008; Serretti and Mandelli, 2008; Gatt et al., 2015; Harrison, 2015).
Epigenetic changes involve reversible chromatin rearrangements that induce mitotically heritable, stable, long-term, and reversible gene expression patterns without altering the DNA sequence (Figure 1) (Bernstein et al., 2010; Allis and Jenuwein, 2016). DNA methylation and histone modifications are the most studied epigenetic marks in physiological and pathological contexts (Schenkel et al., 2017). In DNA methylation, DNA methyltransferases (DNMT) transfer a methyl group to 5′-cytosine residues at cytosine-guanine sequences (CpG) which are clustered in CpG islands, often unmethylated and located within gene promoters of active transcription and tumor suppressor genes (Inbar-Feigenberg et al., 2013; Houtepen et al., 2016). Acetylation and deacetylation of histones through histone deacetylases (HDACs) and histone acetyltransferases (HATs) activity regulate chromatin structure and gene expression (Eyal et al., 2004; Machado-Vieira et al., 2011; Inbar-Feigenberg et al., 2013; Schenkel et al., 2017). Whilst increased histone acetylation and decreased DNA methylation are associated with active transcription and gene expression, histone deacetylation and DNA hypermethylation are indicators of heterochromatin (condensed state of chromatin) and gene silencing (Machado-Vieira et al., 2011; Inbar-Feigenberg et al., 2013). Similarly, some studies on epigenetic modifications are based on the hypotheses that abnormal RNA expression is linked with altered epigenetics at gene promoter regions and regulatory sequences (Houston et al., 2013).
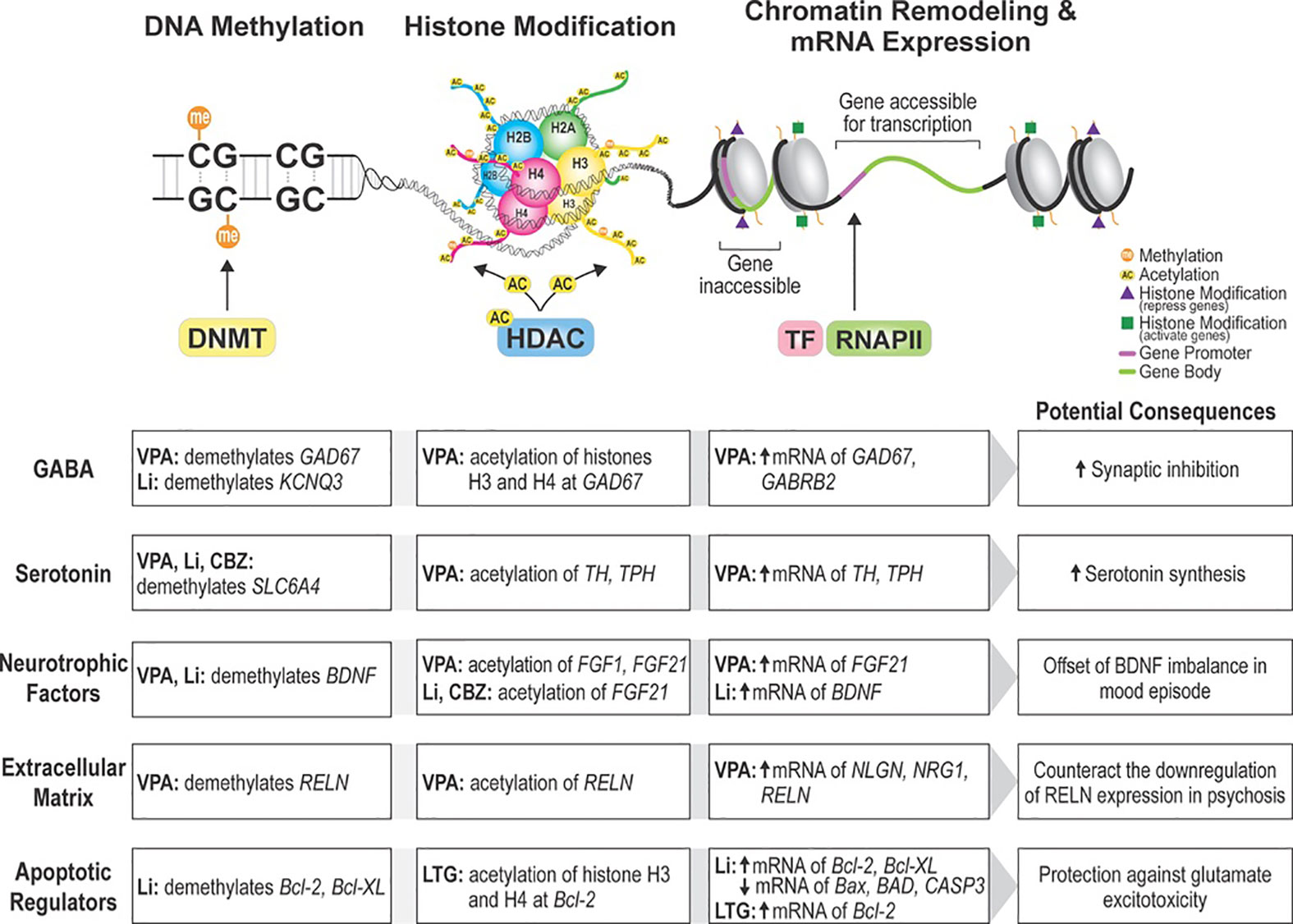
Figure 1 Pathways epigenetically impacted by mood stabilizers. Non-antipsychotic mood stabilizers alter the epigenetic expression of a variety of candidate genes in bipolar disorder and schizophrenia. Findings from our systematic review suggest that epigenetic changes induced by mood stabilizers produce neuroprotective effects through different pathways. DNMT, DNA methyltransferase; HDAC, histone deacetylase; TF, transcription factor; RNAPII, RNA polymerase II; VPA, valproic acid; Li, lithium; CBZ, carbamazepine; LTG, lamotrigine; GAD67, glutamate decarboxylase 67; KCNQ3, potassium voltage-gated channel subfamily Q member 3; GABRB2, gamma-aminobutyric acid receptor subunit beta-2; SLC6A4, sodium-dependent serotonin transporter and solute carrier family 6 member 4; TH, tyrosine hydroxylase; TPH, tryptophan hydroxylase; FGF21, fibroblast growth factor 21; BDNF, brain-derived neurotrophic factor; RELN, reelin; NLGN, neuroligin; NRG1, neuregulin 1; Bcl-2, Bcl-2 apoptotic regulator Bcl-XL- BCL2 Like 1; Bax, BCL2 associated X, apoptosis regulator; BAD, BCL2 associated agonist of cell death; CASP3, caspase 3.
In physiological circumstances, epigenetic mechanisms control neurobiological processes but deregulation in these mechanisms can translate into an increased risk of disease development (Bernstein et al., 2010; Davies et al., 2012). Epimutations secondary to gene-environment interactions have been described to have a key role in the pathophysiology of major psychiatric disorders (Abdolmaleky et al., 2019) where aberrant DNA methylation and histone modification patterns have been identified. Histone acetylation levels are significantly altered in individuals with BD and, to a lesser extent, in individuals with SCZ (Gavin and Sharma, 2010; Machado-Vieira et al., 2011).There is also a reduction in global and specific DNA methylation levels in individuals with BD, MDD, and SCZ (Alladi et al., 2018). In vitro and in vivo studies have suggested the potential of mood stabilizers to reverse epimutations in major psychiatric disorders (Pisanu et al., 2018) making them a target for further research.
Classic mood stabilizers comprising of lithium, valproate, lamotrigine, and carbamazepine, which show antimanic, antidepressant and prophylactic effects, have been characterized as the mainstay of treatment for BD and as aides in MDD and SCZ (Bauer and Mitchner, 2004; Goodwin and Malhi, 2007). While mechanisms of action of valproic acid (VPA), carbamazepine (CBZ), lamotrigine (LTG), and lithium (Li) are not completely understood, there is robust evidence on their ability to target altered epigenetic functions (Seo et al., 2014; Houtepen et al., 2016; Pisanu et al., 2018) involved in the pathophysiology of BD, MDD and SCZ (Higuchi et al., 2011; Ludwig and Dwivedi, 2016). The putative neuroprotective and neurotrophic actions of Li are thought to be induced through epigenetic mechanisms that enhance the expression of molecules involved in neuroplasticity and cytoprotective proteins (Chuang et al., 2002; Schloesser et al., 2012). Likewise, identification of VPA as a class I and IIa HDAC inhibitor (Gottlicher et al., 2001; Phiel et al., 2001) suggests that associated reversion of HDAC-dependent transcriptional repression and histone hyperacetylation could be involved in its mood-stabilizing properties (Gavin and Sharma, 2010; Machado-Vieira et al., 2011). Less studied are the mechanisms of action of LTG and CBZ; neuroprotective effects of LTG exerted through upregulation of excitatory amino acid transporter activity (Schloesser et al., 2012; Leng et al., 2013) and increased global DNA methylation induced by CBZ (Pisanu et al., 2018) are the best described epigenetic changes. The histone deacetylase inhibitory properties of anticonvulsants (Eyal et al., 2004) and the potent antioxidant effects of lithium (Leng et al., 2008; Dwivedi and Zhang, 2015) have been postulated as potential pathways to reverse dysfunctional epigenetic regulation and variability in treatment response (Machado-Vieira et al., 2011).
Objectives and Research Question
Studies on the epigenetic impact on candidate genes of mood stabilizers, especially Li and VPA, have consistently increased in the past decade but attempts to summarize the findings have been scarce (Alladi et al., 2018; Pisanu et al., 2018). This systematic review provides a qualitative summary of the current state of knowledge of the epigenetic effects of non-antipsychotic mood stabilizers in MDD, BD, and SCZ in an attempt to define the specific mechanisms through which these agents act at the epigenomic level.
Methods
Study Design
We developed the systematic review protocol based on the Preferred Reporting Items for Systematic Review and Meta-Analysis Protocols (PRISMA-P) 2015 (Shamseer et al., 2015) and conducted a comprehensive literature search of PubMed and EMBASE from their inception through 30 September 2019.
Search Strategy
The following search string was used: (“epigenetic” OR “epigenomic” OR “DNA methylation” OR “DNA hydroxymethylation” OR “histone acetylation” OR “histone deacetylation” OR “histone methylation”) AND (“lithium” OR “carbamazepine” OR “lamotrigine” OR “mood stabilizer” OR “valproic acid”) NOT “cancer”. Search strategy for valproic acid was narrowed using the Boolean operator NOT, to exclude studies related to use of VPA as an epigenetic cancer drug.
Articles were collated in Rayaan QCRI (Ouzzani et al., 2016). Duplicates were eliminated by the software. Each abstract was reviewed, through a blinded process, for eligibility by two independent reviewers (M.G.R. and M.U.K.). Disagreements were resolved via discussion and a senior reviewer (M. V.) assessing the relevance of the selected data. A total of 129 articles were selected for full-text review (Figure 2).
Participants, Interventions, Comparators
Literature eligible for inclusion were (a) published articles with original data in English; (b) about gene-specific studies with patients, human-tissue, or mammalian models involving assessment of epigenetic changes with use of lithium, divalproex, carbamazepine, or lamotrigine for major psychiatric disorders (psychotic disorders, bipolar disorder, and depression) that (c) met a minimum quality standard of moderate or higher. The three selected major psychiatric disorders were chosen due to phenotypic similarities and shared symptomatology. Exclusion criteria were (a) reviews or non-experimental papers, (b) papers that did not report specific epigenetic changes, (c) papers not focused on the aforementioned psychiatric disorders, or (d) literature only published as abstracts or conference summaries.
Data Sources, Studies Sections and Data Extraction
Each paper was reviewed for the nature of research, the species and tissue examined, sample size, mood stabilizer, targeted gene, epigenetic changes found, and associated psychiatric disorder.
Results
Study Selection and Characteristics
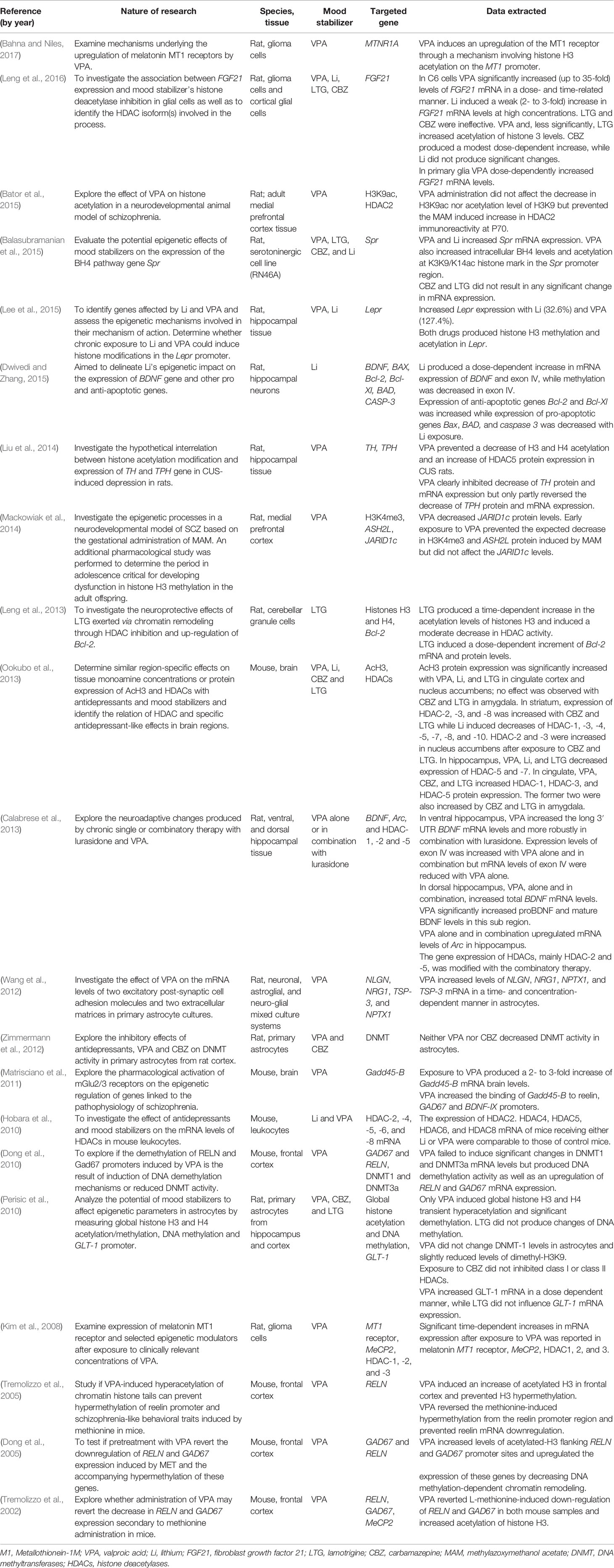
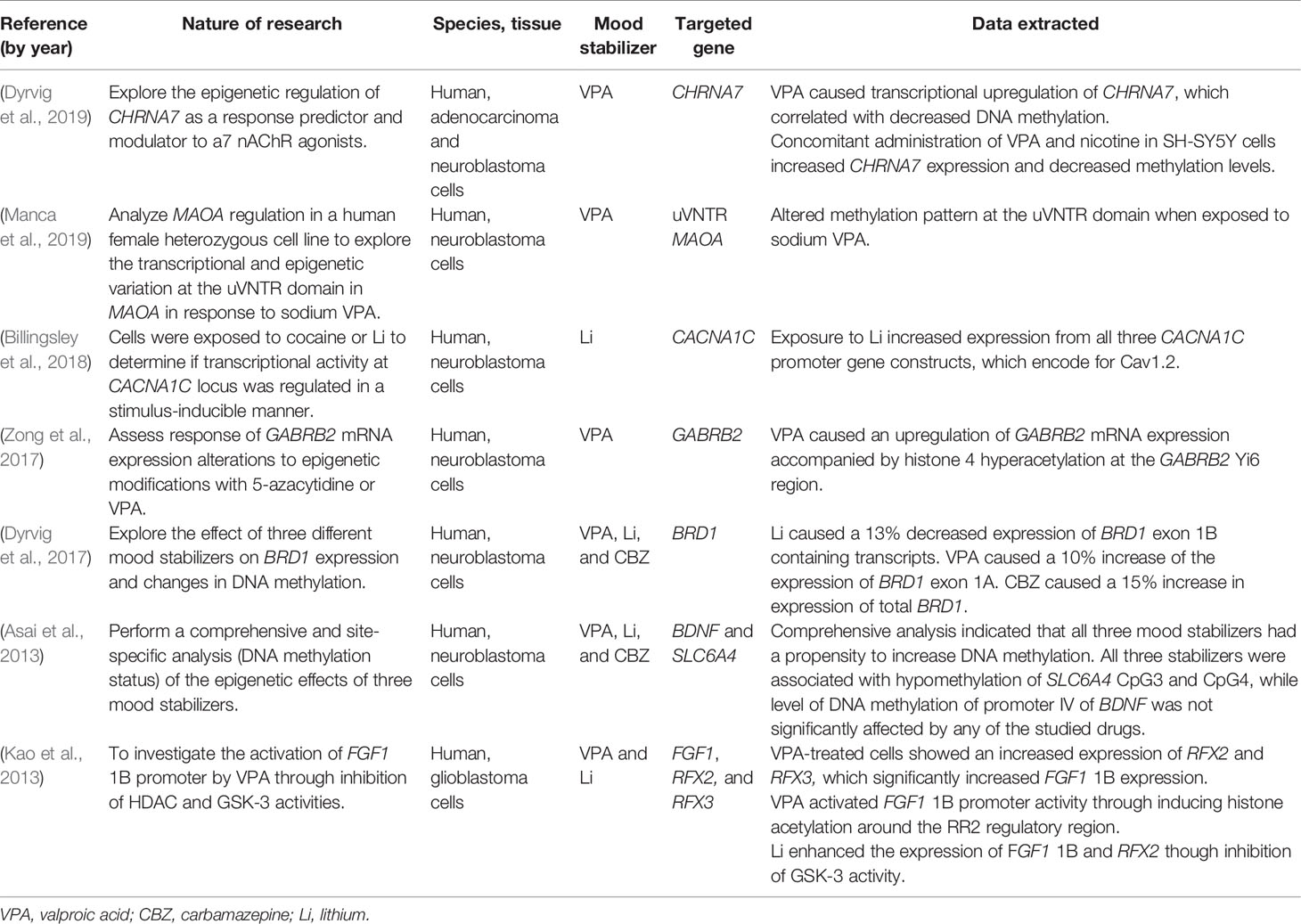
Thirty-eight articles met all the inclusion criteria; studies retrieved for the review were organized in a flow diagram (Figure 2). Twenty-one described studies of rodent samples (Tremolizzo et al., 2002; Dong et al., 2005; Tremolizzo et al., 2005; Kim et al., 2008; Dong et al., 2010; Hobara et al., 2010; Perisic et al., 2010; Matrisciano et al., 2011; Wang et al., 2012; Zimmermann et al., 2012; Calabrese et al., 2013; Leng et al., 2013; Ookubo et al., 2013; Liu et al., 2014; Mackowiak et al., 2014; Balasubramanian et al., 2015; Bator et al., 2015; Dwivedi and Zhang, 2015; Lee et al., 2015; Leng et al., 2016; Bahna and Niles, 2017). Seven articles studied human cell lines (Asai et al., 2013; Kao et al., 2013; Dyrvig et al., 2017; Zong et al., 2017; Billingsley et al., 2018; Dyrvig et al., 2019; Manca et al., 2019) and eight focused on blood samples of subjects diagnosed with either of the selected psychiatric disorders (Gavin et al., 2009; D’Addario et al., 2012; Dell’Osso et al., 2014; Huzayyin et al., 2014; Burghardt et al., 2015; Burghardt et al., 2016; Houtepen et al., 2016; Bengesser et al., 2018). One article studied rodents and human cell lines and subjects (Kaminsky et al., 2015) and one focused on patients and human cell lines (Kakiuchi et al., 2003). The papers are summarized in Tables 1–4.
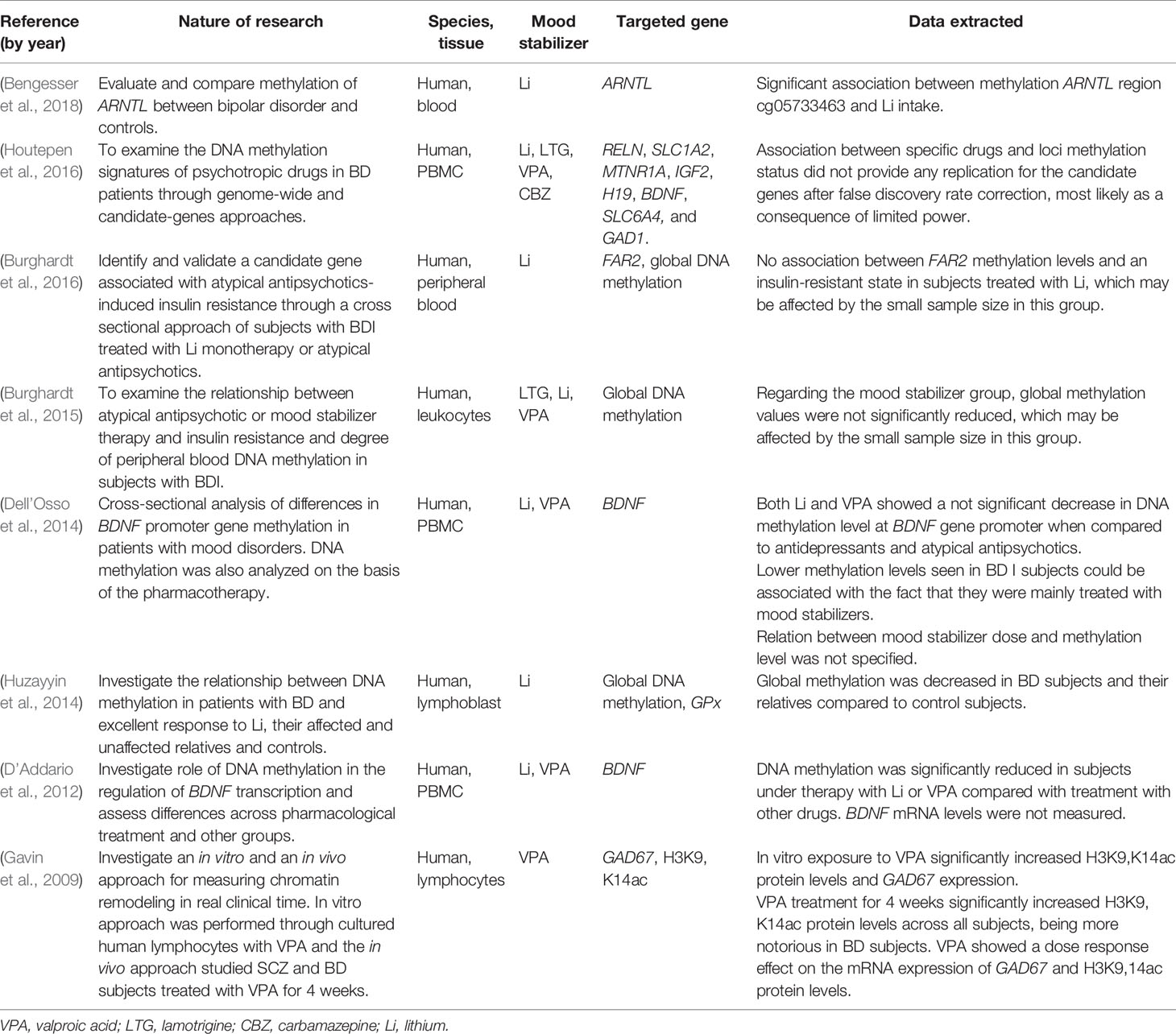
Table 3 Summaries of studies on human subjects affected by a major psychiatric disorder included in the systematic analysis.
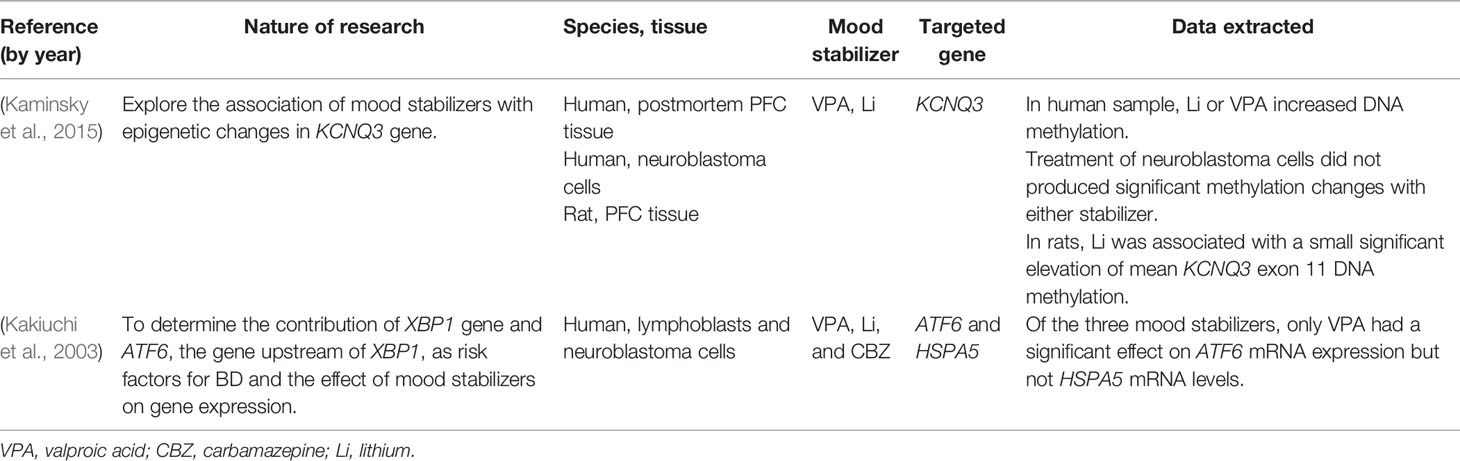
Table 4 Summaries of studies on more than one type of populations included in the systematic analysis.
Synthesized Findings
Rodent Models
SCZ-like epigenetic modifications and symptoms, namely positive, negative, and cognitive, have been replicated in animal samples through the administration of L-methionine (MET) and methylazoxymethanol acetate (MAM) to obtain models for SCZ. Both VPA and antipsychotics have shown a potential reversibility effect of symptoms and epigenetic markers on MET or MAM treated rodents (Tremolizzo et al., 2002; Lodge, 2013; Wang et al., 2015).
Five papers examined the impact of mood stabilizers on genes involved in the regulation of histone activity (Kim et al., 2008; Hobara et al., 2010; Ookubo et al., 2013; Mackowiak et al., 2014; Bator et al., 2015). Kim et al. (Kim et al., 2008) exposed rat C6 glioma cells to prolonged treatment with low clinically relevant doses of VPA (0.5 mM or 1 mM), reporting significant increases in HDAC-1, -2, and -3 mRNA expression (class I HDAC isoforms), as well as increases in methyl-CpG binding protein 2 (MeCP2) and Metallothionein-1M (MT1) mRNA expression. These findings suggest a compensatory temporal upregulation following the HDAC inhibitory effects of VPA as well as a relation between HDACs involvement in the melatonin MT1 receptor induction. Similarly, Hobara et al. (2010) investigated the effect of Li and VPA in class I and II HDACs (HDAC-2, -4, -5, -6, and -8) in lymphocytes from male C57BL/6 and BALB/c mice exposed to a forced swim test, a highly reliable model of depressive-like behavior (Yankelevitch-Yahav et al., 2015). After 21 days of treatment, expression of HDAC-2, -4, -5, -6, and -8 mRNA levels were comparable to those of stress-free control mice, suggesting a non-selective nature of Li and VPA as HDAC inhibitors, and that they do not have a causal role in the altered expression of HDACs in mood disorder patients (Hobara et al., 2010). The region-specific changes in histone deacetylase expression and histone H3 acetylation (AcH3) induced by VPA, Li, CBZ, and LTG in C57BL/6 mice was investigated in another study (Ookubo et al., 2013). Treatment with VPA, Li, and LTG significantly increased AcH3 expression in nucleus accumbens and cingulate cortex, while CBZ and LTG increased HDAC-2 and -3 in striatum; VPA, CBZ, and LTG increased HDAC-3 in cingulate cortex and HDAC-5 in the amygdala (Ookubo et al., 2013). The latter shows epigenetic changes associated with modified HDAC expression and that transcriptional activation includes an alternating recruitment of histone acetyltransferases and HDACs, rather than a process of increasing acetylation only (Shahbazian and Grunstein, 2007). Impact on histone H3 methylation in the prefrontal cortex was studied in MAM-injected Wistar male rats through measurements of ASH2L levels, which encodes for Set1/Ash2 histone methyltransferase complex subunit ASH2, and lysine-specific demethylase 5C enzyme, JARID1c (Mackowiak et al., 2014). Use of VPA in early adolescence prevented disturbances in ASH2L and H3K4me3 but did not affect the levels of JARID1c, a key protein in the demethylase activity on H3K4me3 (Mackowiak et al., 2014; Rondinelli et al., 2015). The same group reported that VPA prevented an increase in HDAC-2 levels evoked by MAM but not H3K9 acetylation levels (Bator et al., 2015). An additional paper studied the impact of CBZ and VPA on DNMT1 activity in primary astrocytes from rat cortex, reporting that neither of the agents altered the expression levels of DNMT (Zimmermann et al., 2012).
Chronic unpredictable stress (CUS)–induced depression in rats was used in a study of epigenetic histone modification and gene expression induced by VPA on tyrosine hydroxylase (TH) and tryptophan hydroxylase (TPH), enzymes involved in the biosynthesis of L-DOPA (a precursor to norepinephrine) and serotonin respectively (Liu et al., 2014). In controls, CUS led to decreased acetylation of H3K9 and H4K12, and reduced expression of TH and TPH. Administration of VPA reversed these downregulations and prevented an increase of HDAC-5 in the hippocampus. Regulation of another gene involved in the monoamine neurotransmitter formation, Spr, associated with increased susceptibility to mood disorders, was studied by evaluating the epigenetic changes after exposure to VPA, Li, LTG, and CBZ (Balasubramanian et al., 2015). Exposing RN46A cells (derived from embryonic rat medullary raphe nucleus) to VPA and Li showed increased Spr mRNA expression, protein levels, and histone acetylation in the Spr promoter. The increases in Spr resulted in augmented levels of tetrahydrobioptherin (BH4), a cofactor involved in the biosynthesis of diverse neurotransmitters. Conversely, treatment with LTG and CBZ did not result in significant changes in Spr mRNA expression (Balasubramanian et al., 2015). A fourth study analyzed the potential of mood stabilizers VPA’s, CBZ’s, and LTG’s affect on global DNA methylation, histone acetylation, and methylation of the GLT-1 promoter, which encodes for the excitatory amino acid transporter 2 (EAAT2), a research target for MDD and BD, in astroglial cultures of Sprague-Dawley rats (Perisic et al., 2010; Blacker et al., 2019). Only VPA induced histone hyperacetylation of H3 and H4, and DNA methylation, both reversible after drug removal. VPA also led to reduced GLT-1promoter methylation at three CpG sites, enriched acetylation of H3 and H4, and a dose dependent increase of GLT-1 mRNA. LTG and CBZ failed to produce significant changes (Perisic et al., 2010).
A preliminary study by Wang et al. (2012) examined rat astrocytes and the effect of VPA on the synaptic excitatory/inhibitory balance through modulation of cell adhesion molecules (CAM) and extracellular matrices (ECM), which are known to be involved in the formation and maturation of synapses (Dalva et al., 2007). This paper evaluated VPA-induced changes on mRNA levels of neuroligin-1 (NLGN) and neuregulin-1 (NRG1), which are CAMs involved in the regulation of glutamatergic and GABAergic synapses (Craig and Kang, 2007), and neuronal pentraxin-1 (NPTX1) and thrombospondin-3 (TSP3), which are ECMs that promote synaptogenesis (Christopherson et al., 2005; Koch and Ullian, 2010). Exposure to VPA significantly increased the mRNA levels of the four molecules in a time- and concentration-dependent manner in astrocytes (Wang et al., 2012).
Activity on genes encoding for pro- and anti-apoptotic proteins have also been reported, highlighting them as a potential therapeutic target in BD, a disorder were neuronal resilience and plasticity cascades are altered (Schloesser et al., 2008). A novel study in Sprague Dawley rats identified that LTG had protective effects against glutamate excitotoxicity through HDAC inhibition and Bcl-2 upregulation in primary neuronal cerebellar granule cells (CGC) (Leng et al., 2013). LTG pretreatment provided a dose- and time-dependent protection against glutamate toxicity and decrease in HDAC activity, sparing histone acetyltransferase (HAT) activity (Leng et al., 2013). LTG treatment caused moderate time-dependent increase of histones H3 and H4 acetylation levels. Inhibition of HDAC activity by LTG was less robust, however, than VPA-related inhibition. In this study, the anti-apoptotic Bcl-2 gene was positively impacted with LTG treatment. Concentration-dependent increases in Bcl-2 mRNA levels with an associated increase of Bcl-2 protein were observed after exposure of CGC cultures to incremental doses of LTG (0-100 µM) for 2 days (Leng et al., 2013).
The relevance of Bcl-2 has risen since recent links between Bcl-2 dysregulation and mood disorders were identified (Schloesser et al., 2008), and the mechanisms through which Li and VPA impact cellular plasticity cascades. One study described the influence of lithium on the expression of neuroprotective genes BDNF, Bcl-2, and Bcl-XL and pro-apoptotic genes Bax, BAD and caspase-3 in rat hippocampal neurons (Dwivedi and Zhang, 2015). After exposure to Li, expression of Bcl-2 and Bcl-XL genes were increased in a dose-dependent manner, while expression of Bax, BAD, and caspase-3 was decreased by both doses of Li (1 mM and 2 mM), with magnitude of change being higher at 2 mM (Dwivedi and Zhang, 2015). The mRNA expression of BDNF was increased by 67% when exposed to Li at 1 mM and was fully sustained at 2 mM, especially on specific BDNF promoter exon IV, where methylation was decreased by Li; protein levels of BDNF were increased by 53% and 89% for 1 mM and 2 mM Li, respectively (Dwivedi and Zhang, 2015). One additional study focused on changes in BDNF, Arc, and epigenetic regulators HDAC-1, -2, and -5 in Sprague-Dawley rats’ hippocampi after exposure to VPA, lurasidone, or a combination of both (Calabrese et al., 2013). Regarding treatment with VPA only, an increase in BDNF mRNA levels in ventral and dorsal hippocampus was reported, as well as proBDNF and mature BDNF levels. Arc mRNA levels were also upregulated after exposure to VPA. Arc and BDNF expression increased more consistently with combination therapy, and HDAC isoform mRNA levels were significantly decreased (Calabrese et al., 2013).
Four papers examined the epigenetic changes secondary to VPA administration on RELN (reelin) and GAD67 in MET-induced mouse models of SCZ (Tremolizzo et al., 2002; Dong et al., 2005; Tremolizzo et al., 2005; Dong et al., 2010). RELN expression, synthetized by GABAergic interneurons, is reduced in neocortex of SCZ and BD, paralleled by a down-regulation of GAD67 levels, responsible for regulating cortical GABA levels (Guidotti et al., 2000; Guidotti et al., 2016). When comparing randomly sampled MET-injected populations of heterozygous reeler mice (reelin-deficient) versus wild type mice, VPA increased H3 histone acetylation, and reversed decreased mRNA expression of RELN and GAD67 induced by L-methionine in prefrontal cortices of both populations. VPA failed to modify H4 acetylation and GAD65 expression (Tremolizzo et al., 2002). Likewise, in Swiss albino mice’s frontal cortices, VPA prevented MET-induced RELN promoter hypermethylation and reduced the MeCP2 binding to RELN and GAD67 in frontal cortices (Dong et al., 2005). Upregulated MeCP2 binding is associated with hypermethylation of CpG island promoters and increased recruitment of HDACs which favors gene transcriptional repression (Dong et al., 2005). VPA action on histone acetylation and methylation dynamics in RELN promoter was confirmed in MET-treated B6C3Fe male mice in which enhanced acetylation of H3 histone and prevention of MET-induced RELN hypermethylation and mRNA downregulation was observed after exposure to VPA (Tremolizzo et al., 2005). In this study, correction of L-methionine-related behavioral deficits was also observed after VPA treatment. More recently, the association between inhibition of RELN expression and DNA methyltransferases (DNMT) was explored in Swiss albino mice to better determine the pathway through which VPA induces demethylation of RELN and GAD67 (Dong et al., 2010). Although this study confirmed previous evidence of the association between overexpression of DNMT1 and DNMT3a in SCZ cortex and hypermethylation of the promoters of RELN and GAD67 (Veldic et al., 2004), promoter demethylation induced by VPA was not caused by a decrease of DNMT activity or levels, but by increased acetylation of histones by HDAC inhibition (Dong et al., 2010). A fifth study examined the impact of VPA on growth arrest and DNA-damage-inducible beta (Gadd45-β) expression and binding properties (Matrisciano et al., 2011). In mice hippocampi, frontal cortices, and cerebellums, VPA was reported to robustly upregulate Gadd45-β expression by activation of DNA demethylation, a 2- to 3-fold increase of Gadd45-β mRNA levels, and an increased binding to reelin, GAD67, and BDNF-IX promoters (Matrisciano et al., 2011).
VPA- and Li-mediated epigenetic effects on Lepr, leptin receptor gene, implicated in regulation of mood and satiety, were explored in rats’ hippocampi (Lee et al., 2015). Both agents produced an augment in the expression of Lepr, although VPA showed a more significant increase (127.4% vs. 32.6%). Histone H3 methylation and acetylation were induced by VPA and Li; for Li, a more pronounced demethylation of H3 was observed. Another gene involved in metabolism, fibroblast growth factor 21 (FGF21), and its effect after exposure to VPA, CBZ, LTG, and Li was studied by Leng et al. (2016) in rat primary cortical glial cells and C6 glioma cells. C6 and glial cells treated with either VPA or Li experienced a dose-dependent increase in FGF21 mRNA levels, while LTG and CBZ were ineffective. VPA, LTG and, less potently, CBZ significantly increased H3 acetylation levels (Leng et al., 2016). In another study, further examination of the mechanisms underlying melatonin MT1 receptors’ upregulation by VPA was performed in rat glioma cells (Bahna and Niles, 2017). Exposure to VPA increased expression of MT1 mRNA and acetylation of H3K9 across the receptor’s promoter.
In Vitro Human Cell Line Model
Study of human neuroblastoma and glioblastoma cells offer a window of what might happen in vivo and might be better models than HeLa adenocarcinoma cells to investigate the effects of mood stabilizers, due to their neuronal characteristics (Dyrvig et al., 2017).
CHRNA7, encoding the α7 nicotinic acetylcholine receptor (nAChR) has been linked to SCZ (Stephens et al., 2009). In one study, VPA was found to cause an upregulation of CHRNA7 expression, correlating with decreased DNA methylation on HeLa cells, suggesting a potential reversing effect of pharmacotherapy on an epigenetic mechanism (Dyrvig et al., 2019). A dose-dependent effect was observed; while treatment with 0.3mM VPA produced a 4-fold upregulation of the expression, 0.6 mM VPA resulted in an 8.5-fold increase. Concomitant exposure to VPA and nicotine did not produce additional effects of CHRNA7 in HeLa cells but increased CHRNA7 expression, and decreased DNA methylation, in SH-SY5Y cells.
Necessary for acetylation of H3K14, dysregulations of bromodomain-containing protein 1 (BRD1), and its encoding gene BRD1 have been linked to SCZ and BD and show stress-related changes in expression (Christensen et al., 2012; Dyrvig et al., 2017). The effects of CBZ, VPA, and Li on the BRD1 gene was studied in cultured neuroblastoma cells (SH-SY5Y) providing evidence that these mood stabilizers altered the expression of BRD1 by mechanisms other than DNA methylation, which remained unchanged after exposure to these agents (Christensen et al., 2012; Dyrvig et al., 2017). Increased expression of total BRD1 with CBZ was observed, while VPA increased the expression of exon 1C, and Li treatment decreased the expression of exon 1B.
Two papers examined epigenetic changes induced by mood stabilizers on genes involved in synaptic processes, GABRB2 (encoding a GABA receptor subunit), and CACNA1C (encoding a calcium channel) (Zong et al., 2017; Billingsley et al., 2018). GABAA receptor β2-subunit gene (GABRB2) has been linked to SCZ and BD in multiple studies (Chen et al., 2009; Zhao et al., 2012). Zong et al. (Zong et al., 2017) assessed the response of deregulated GABRB2 mRNA expression to epigenetic modifications with VPA. Treatment of neuroblastoma cells (IMR-32) with VPA not only induced hyperacetylation of histone H4 and promoter regions, but also had a five-fold increase in mRNA expression providing further evidence of GABAergic dysfunction in psychotic disorders. Billingsley et al. (2018) focused on the effect of lithium and cocaine exposure in the transcriptional activity on CACNA1C gene, encoding for the L-type a1c sub-unit of the voltage-dependent calcium channel, Cav1.2, the most abundant human neuronal L-type calcium channel and a significant risk gene for BD and SCZ (Moon et al., 2018; Zhu et al., 2019). Treatment of neuroblastoma cells (SH-SY5Y) with Li resulted in an increased expression of all three studied CACNA1C promoters (Billingsley et al., 2018). A gene-specific analysis of DNA methylation status of BDNF and SLC6A4 genes after exposure to VPA, Li, and CBZ showed that all three mood stabilizers decreased, at different concentrations, methylation of CpG sites of SLC6A4 but not BDNF promoter IV in neuroblastoma cells (Asai et al., 2013). This study was limited by the lack of testing of treatment duration but provided, nonetheless, insights into epigenetic regulation exerted by mood stabilizers in two genes involved in synaptic regulation and associated with mood disorders.
Epigenetic variations secondary to VPA in the monoamine system were explored by Manca et al. (Manca et al., 2019) through analysis of MAOA gene regulation. Neuroblastoma cells (SH-SY5Y), which is a female cell line, showed a decreased methylation pattern at the uVNTR domain of MAOA after exposure to VPA. This study is particularly relevant due to the heterozygous nature of the MAOA gene, located on the X chromosome, in females and because findings showed a different pattern of binding of transcription factors on each X chromosome allele.
Two studies provided insights on the influence of mood stabilizers on neuroprotective- and transcription-related genes (Asai et al., 2013; Kao et al., 2013). Activation of FGF-1B gene promoter, member of the fibroblast growth factor family, by VPA- and Li-related inhibition of HDACs and GSK-3 pathway in glioblastoma cells was investigated (Kao et al., 2013). VPA-treated cells showed increased expression levels of transcriptional factors RFX2 and RFX3, which bind to FGF-1B promoter, as well as activation of FGF-1B promoter through acetylation of histone H3 secondary to HDAC inhibition. Through a different pathway, Li, a well-known GSK-3 inhibitor, enhanced the expression levels of FGF-1B and RFX2 (Kao et al., 2013). This study suggests that FGF1 is an important target of mood stabilizers through different pathways. Asai et al. performed an analysis of the DNA methylation status of BDNF and SLC6A4 after exposure to VPA, CBZ, and Li. Findings suggested a propensity of all three mood stabilizers to alter global DNA methylation and were associated with decreased methylation of SLC6A4 CpG3 and CpG4. DNA methylation status of BDN promoter IV was not significantly altered by any of the studied stabilizers (Asai et al., 2013).
Human Studies
Studies comparing the effects of Li and VPA, alone or in combination with other medications, on DNA methylation of BDNF gene of subjects with BD or MDD were similar. A more significant reduction in DNA methylation level was observed with VPA and Li when compared to other medications (D’Addario et al., 2012). Lower methylation levels during manic states observed in this study could be attributed to the use of higher doses of mood stabilizers than during depressive or euthymic states. More recently, this same group generated data suggesting a decrease in DNA methylation level at BDNF gene promoter in patients under treatment with VPA or Li, although due to lack of reporting on used doses, a dose-dependent effect on methylation levels remains uncertain (Dell’Osso et al., 2014).
Lithium was also associated with reduced methylation at one site in the 5′ regulatory region of ARNTL (cg05733463) when compared to individuals with BD not taking lithium, suggesting an activation of epigenetic marks with an associated increased gene expression (Bengesser et al., 2018). ARNTL, a core component of the circadian clock, is involved in the expression of the MAOA gene, therefore imbalances in the relation between circadian rhythms and neurotransmitter degradation may contribute towards an increased BD susceptibility (Geoffroy, 2018). Effects of Li on FAR2 (fatty acyl CoA reductase 2) methylation and insulin resistance was done in another paper, although the main focus of the research was on second generation antipsychotics (Burghardt et al., 2016). Only 25% of the 72 subjects included were on Li monotherapy, so lack of association between FAR2 methylation levels and Li treatment could be a consequence of the small sample size. A previous study by Burghardt et al. (Burghardt et al., 2015) also compared global DNA methylation induced second generation antipsychotics versus Li, VPA, and LTG and insulin resistance. No change in global methylation was observed with mood stabilizers but, as in their subsequent study, the small size of the sample under mood stabilizer treatment (31 of 115 BD1 patients) was a significant limitation. Another study measured global DNA methylation and glutathione peroxidase (GPx) activity in subjects with BD and excellent response to Li monotherapy, in unaffected and affected relatives, and in healthy controls (Huzayyin et al., 2014). Interestingly, GPx activity was increased in affected relatives after treatment with Li and a significant, negative correlation between GPx activity and DNA methylation was observed. GPx levels have been reported to be reduced in postmortem prefrontal cortices in SCZ, MDD, and BD (Gawryluk et al., 2011).
Studies indicate that GAD67 mRNA levels are reduced in postmortem brains of subjects diagnosed with SCZ and BD (Akbarian and Huang, 2006). Similar to findings in previously described animal studies, a dual in vivo and in vitro approach in humans consisting of 4-week treatment with VPA was found to upregulate, in response to dosage, the expression of GAD67 in individuals with SCZ and BD, as well as H3K9 and H3K14 acetylation levels (Gavin et al., 2009). Cultured lymphocytes exposed to 0.7 mM VPA showed a 383% increase in GAD67 mRNA and an 89% increase in H3K9ac and H3K14 while lymphocytes from SCZ and BD patients before and after treatment with VPA showed a significant increase in GAD67 mRNA expression (Gavin et al., 2009). H3K9Ac is an epigenetic mark that typically indicates transcriptionally active chromatin (Qiao et al., 2015), therefore the 482% increase in H3K9ac/H3K14ac attachment to the GAD67 promoter could predict a further increase in GAD67 expression (Gavin et al., 2009).
DNA methylation signatures of psychotropic drugs on candidate-genes in 172 Dutch patients with BD were studied (Houtepen et al., 2016). Impact of the classic mood stabilizers VPA, CBZ, LTG, and Li were measured on RELN, SLCQA2, MTNR1A, IGF2, H19, BDNF, SLC6A4, and GAD1. No specific methylated CpG sites survived multiple testing correction; Q-Q (quantile-quantile) plot analysis and trend level results suggested that it was likely the result of limited statistical power, although VPA was significantly associated with altered methylation signatures (Houtepen et al., 2016).
It is worth mentioning that all studies on human subjects were performed on peripheral blood mononuclear cells (PBMC), widely used for DNA methylation studies and a model of epigenetic gene regulation in the brain (Gavin and Sharma, 2010; Dell’Osso et al., 2014). Despite DNA methylation being subject to tissue-specific variations in brain and blood, PBMC is considered a reliable biomarker of brain activity (Davies et al., 2012).
Studies With Animal Models, Human Cell Lines, and Psychiatric Populations
Two studies performed gene-specific analysis with sample-specific approaches on rodents, human cell lines, and/or patients with BD (Kakiuchi et al., 2003; Kaminsky et al., 2015). Kakiuchi et al. studied the effect of mood stabilizers (Li, VPA, and CBZ) on activating transcription factor gene (ATF6), and heat shock protein family A member 5 gene (HSPA5), on lymphoblastoid cells in two pairs of monozygotic twins affected with BD, a pair of healthy twins, and in neuroblastoma (SHSY5Y) cells (Kakiuchi et al., 2003). Only VPA significantly increased mRNA expression of ATF6 and induced HSPA5 gene expression through upregulation of XBP1 gene. The second study explored the association of mood stabilizers, VPA and Li, with epigenetic changes in KCNQ3 gene, which encodes for potassium voltage-gated channel, subfamily Q member 3 (Kaminsky et al., 2015). DNA methylation and gene expression levels were tested in four different samples 1) a group comparing post-mortem prefrontal cortex tissue from 12 BD subjects versus 2) 10 control subjects, 3) a sample of human neuroblastoma cell line, and 4) prefrontal cortices of Brown-Norway rats. On the first and fourth groups, increased DNA methylation was observed after treatment with VPA and Li, while no changes were observed in neuroblastoma cells (Kaminsky et al., 2015).
Risk of Bias
Every article was appraised for quality using a modified published process (du Prel et al., 2009); all papers met a quality score of moderate or high.
Discussion
Summary of Main Findings
This systematic review explored the epigenetic changes induced by VPA, Li, CBZ, and LTG in major psychiatric disorders. Despite the genetic complexity of mental illnesses, evidence suggest that epigenome can be targeted by mood stabilizers in psychiatric disorders which can potentially translate into long-term illness trajectory changes (Ludwig and Dwivedi, 2016), although, aside from VPA and Li, the evidence found was limited. Currently, only VPA and Li have shown to consistently induce epigenetic changes while LTG and CBZ have shown less consistent evidence, in part due to the lack of studies or to small sample sizes. Mood stabilizers exert their actions not only through histone deacetylase inhibition, but also through downregulation of methylation of cytosines in CpG dinucleotides, increased histone activity, and induction of RNA, which leads to an augmentation of gene expression (Machado-Vieira et al., 2011; Houston et al., 2013). Previously identified disease-associated alterations in epigenetic markings of specific genes were susceptible to mood stabilizers, according to our findings.
We found few studies primarily focusing on the epigenetic effects of CBZ and LTG. This is relevant because both agents have Level 1 evidence for treatment of different phases of BD and are used in multiple psychiatric disorders (Reid et al., 2013; Yatham et al., 2018). While CBZ has been linked to moderate effects on global and gene-specific DNA methylation and HDAC inhibition, no clear conclusions can be drawn with respect to its epigenetic effects from available literature. Neuroprotective effects of LTG are believed to occur via inhibition of glutamate excitotoxicity, HDAC inhibition, histone acetylation, and activity on neuronal survival, and plasticity cascades. Up-regulation of anti-apoptotic gene Bcl-2 has been studied as a potential mechanisms through which LTG exerts its epigenetic effects (Leng et al., 2013; Reid et al., 2013). It has been previously noted that perturbations in the apoptotic pathway could be involved in the pathogenesis of mood disorders (Zhang et al., 2014; Scaini et al., 2017) and that apoptotic regulator genes are targeted by VPA, Li, and LTG (Leng et al., 2013; Dwivedi and Zhang, 2015). Hence, the therapeutic potential of a synergistic neuroprotection from combined mood stabilizer therapy is a target for future investigation.
VPA is the prototypical HDAC inhibitor in major psychiatric disorders research. VPA relieves class I and II HDAC-dependent repression of transcription factors and increases histones H3 and H4 acetylation thereby enhancing activation of gene expression (Gottlicher et al., 2001). However, evidence suggests that VPA also exerts an effect on DNA methylation signatures, suggesting that VPA’s activity over chromatin remodeling is not limited to HDACs inhibition (Houtepen et al., 2016; Pisanu et al., 2018). It is worth mentioning that HDAC inhibition and associated hyperacetylation by mood stabilizers is not absolute and that increased gene expression is a process that entails high levels of acetylation turnover and recruitment of HATs and HDACs to the active gene, suggesting that HDACs have both inhibitory and activating effects on transcription (Shahbazian and Grunstein, 2007). The latter may explain reports of increased HDAC expression after exposure to mood stabilizers described in several of the included studies (Ookubo et al., 2013; Liu et al., 2014; Bator et al., 2015). Likewise, since DNA methylation and histone modification pathways are not completely independent from each other (Cedar and Bergman, 2009), one mechanism by VPA can partially affect activity in the other. VPA-induced augmentation of methylcytosine-binding protein MeCP2 reported in animal models (Tremolizzo et al., 2002; Kim et al., 2008) suggests a relationship between DNA methylation and histone modification through HDACs recruitment to the methylated region by MeCP2 (Cedar and Bergman, 2009). VPA has also been reported to reduce methylation of gene promoters, thereby upregulating mRNA/expression of genes linked to SCZ and BD, indicating other mechanisms of action additional to HDAC inhibition (Dong et al., 2010). Animal and human studies generally reported that prominent epigenetic activity induced by valproic acid were exerted through different types of epigenetic mechanisms which led to a significant rise in the expression of silenced genes and regulation of altered transcriptional processes (Pisanu et al., 2018).
Although the precise mechanisms through which Li induces neuroprotective and neurotrophic activity are not yet fully elucidated, evidence suggests that activity on synapsis strength, cellular resilience, and glial function at a molecular level has a key role (Machado-Vieira, 2018). Overlapping activity on different pathways is characteristic of Li; however, epigenetic contributions on neurotrophins, apoptosis pathways, and glycogen synthase kinase-3 (GSK-3β) stand out, among the included papers, as central targets of lithium (D’Addario et al., 2012; Kao et al., 2013; Dell’Osso et al., 2014; Dwivedi and Zhang, 2015). Several studies have identified a negative correlation between Bcl-2 levels and positive symptoms in SCZ and manic symptoms in BD (Tsai et al., 2013; Chen et al., 2015), implying a dysregulation of apoptotic pathways in these disorders. Findings from our search reflect that Li can reverse not only the decreased levels of Bcl-2 and Bcl-XL but also inhibit the expression of pro-apoptotic molecules such as BAD, BAX and caspase-3 (Dwivedi and Zhang, 2015), highlighting the anti-apoptotic properties of this mood stabilizer. Similarly, expression of neuroprotective and anti-apoptotic protein BDNF has been shown to be downregulated in BD, MDD, and SCZ (Machado-Vieira, 2018) but also epigenetically susceptible to mood stabilizers, including Li which exerts this activity mainly through DNA demethylation (D’Addario et al., 2012; Asai et al., 2013; Dwivedi and Zhang, 2015; Houtepen et al., 2016). Inhibition of GSK-3β activity on the intracellular signaling cascade has been one of the most well-defined mechanisms of action of Li (Beurel et al., 2015). One study reported an upregulation of the expression of plasticity proteins secondary to Li inhibition of GSK-3 activity (Kao et al., 2013). This is an especially important target for future research. Additionally, our systematic process corroborated the already defined role of Li as an HDAC inhibitor (Hobara et al., 2010; Ookubo et al., 2013).
Strengths and Limitations
While the epigenetic effects of mood stabilizers were evident, there are limitations to our results. First, most studies were limited by relatively small group samples and focused on cohort-based effects. This small group-based approach could hinder the identification of disease-relevant signals and be underpowered (Houston et al., 2013). Although subject to availability of samples and resources, future research should aim to increase the size of the studied groups. Second, studies were heterogeneous in methodology and studied population. Third, the majority of papers were focused on VPA, highlighting an urgent need to study the activity at an epigenetic level of other mood stabilizers. As we previously mentioned, although partly comparable, epigenetic patterns in peripheral blood cells are not an exact reflection of those in brain tissue (Davies et al., 2012; Pisanu et al., 2018), neuroblastoma cells, or animal models; however they are widely accepted as models for research of neuropsychiatric disorders in the absence of cell type-specific epigenome mappings (Houston et al., 2013). Finally, despite the measures taken to reduce publication bias through well-defined inclusion criteria, selective reporting cannot be excluded.
Several strengths were identified in our study. We reviewed a large body of evidence using criteria to capture a specific feature of mood stabilizers in SCZ, BD, and MDD and the quality of scientific publications were evaluated. Since we focused mainly on gene-specific effects of mood stabilizers, we identified small regulatory networks of epigenetic modulators of gene expression and neuroplasticity; consistency across studies was also found. To our knowledge, this systematic review is the first to attempt to summarize epigenetic changes secondary to non-antipsychotic mood stabilizers in three major psychiatric disorders.
Conclusions
In conclusion, available data confirms the effects of VPA and Li on the epigenome of genes associated with major psychiatric disorders and suggest that LTG and CBZ might have a greater role in the manipulation of epigenetic mechanisms, although further studies are required to elucidate their mechanisms of action. Our systematic review not only reflects the genetic complexity of major psychiatric disorders but, by summarizing the existing evidence, also underscores the ability of these classic mood stabilizers to target genes involved in synaptic plasticity, neuroplasticity, and transcription factors. A better understanding of the specific epigenetic changes induced by classic mood stabilizers in patients with major psychiatric disorders can lead towards more personalized interventions and to the development of new agents able to induce selective chromatin remodeling and gene-specific expression effects.
Data Availability Statement
The original contributions presented in the study are included in the article/supplementary material, further inquiries can be directed to the corresponding author.
Author Contributions
MG-R and MV conceived the research idea. MG-R, MV, and CB designed the search strategy. MG-R and MK performed the data collection. AH co-wrote and reviewed the paper. PC, MF, and CB reviewed and edited the paper. MV supervised the research.
Conflict of Interest
MF’s potential conflicts include previous grant support from Assurex Health, Mayo Foundation, and Medibio; consultancy from Actify Neurotherapies, Allergan, Intra-Cellular Therapies, Inc., Janssen, Myriad, Neuralstem Inc., Takeda, and Teva Pharmaceuticals; CME/Travel/Honoraria from American Physician Institute, CME Outfitters, and Global Academy for Medical Education.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
Abdolmaleky, H. M., Gower, A. C., Wong, C. K., Cox, J. W., Zhang, X., Thiagalingam, A., et al. (2019). Aberrant transcriptomes and DNA methylomes define pathways that drive pathogenesis and loss of brain laterality/asymmetry in schizophrenia and bipolar disorder. Am. J. Med. Genet. B. Neuropsychiatr. Genet. 180 (2), 138–149. doi: 10.1002/ajmg.b.32691
Akbarian, S., Huang, H. S. (2006). Molecular and cellular mechanisms of altered GAD1/GAD67 expression in schizophrenia and related disorders. Brain Res. Rev. 52 (2), 293–304. doi: 10.1016/j.brainresrev.2006.04.001
Alladi, C. G., Etain, B., Bellivier, F., Marie-Claire, C. (2018). DNA Methylation as a Biomarker of Treatment Response Variability in Serious Mental Illnesses: A Systematic Review Focused on Bipolar Disorder, Schizophrenia, and Major Depressive Disorder. Int. J. Mol. Sci. 19 (10), 1–19. doi: 10.3390/ijms19103026
Allis, C. D., Jenuwein, T. (2016). The molecular hallmarks of epigenetic control. Nat. Rev. Genet. 17 (8), 487–500. doi: 10.1038/nrg.2016.59
Asai, T., Bundo, M., Sugawara, H., Sunaga, F., Ueda, J., Tanaka, G., et al. (2013). Effect of mood stabilizers on DNA methylation in human neuroblastoma cells. Int. J. Neuropsychopharmacol. 16 (10), 2285–2294. doi: 10.1017/S1461145713000710
Bahna, S. G., Niles, L. P. (2017). Epigenetic induction of melatonin MT1 receptors by valproate: Neurotherapeutic implications. Eur. Neuropsychopharmacol. 27 (8), 828–832. doi: 10.1016/j.euroneuro.2017.06.002
Balasubramanian, D., Deng, A. X., Doudney, K., Hampton, M. B., Kennedy, M. A. (2015). Valproic acid exposure leads to upregulation and increased promoter histone acetylation of sepiapterin reductase in a serotonergic cell line. Neuropharmacology 99, 79–88. doi: 10.1016/j.neuropharm.2015.06.018
Bator, E., Latusz, J., Radaszkiewicz, A., Wedzony, K., Mackowiak, M. (2015). Valproic acid (VPA) reduces sensorimotor gating deficits and HDAC2 overexpression in the MAM animal model of schizophrenia. Pharmacol. Rep. 67 (6), 1124–1129. doi: 10.1016/j.pharep.2015.04.004
Bauer, M. S., Mitchner, L. (2004). What is a “mood stabilizer”? An evidence-based response. Am. J. Psychiatry 161 (1), 3–18. doi: 10.1176/appi.ajp.161.1.3
Bengesser, S. A., Reininghaus, E. Z., Lackner, N., Birner, A., Fellendorf, F. T., Platzer, M., et al. (2018). Is the molecular clock ticking differently in bipolar disorder? Methylation analysis of the clock gene ARNTL. World J. Biol. Psychiatry 19 (sup2), S21–S29. doi: 10.1080/15622975.2016.1231421
Bernstein, B. E., Stamatoyannopoulos, J. A., Costello, J. F., Ren, B., Milosavljevic, A., Meissner, A., et al. (2010). The NIH Roadmap Epigenomics Mapping Consortium. Nat. Biotechnol. 28 (10), 1045–1048. doi: 10.1038/nbt1010-1045
Beurel, E., Grieco, S. F., Jope, R. S. (2015). Glycogen synthase kinase-3 (GSK3): regulation, actions, and diseases. Pharmacol. Ther. 148, 114–131. doi: 10.1016/j.pharmthera.2014.11.016
Billingsley, K. J., Manca, M., Gianfrancesco, O., Collier, D. A., Sharp, H., Bubb, V. J., et al. (2018). Regulatory characterisation of the schizophrenia-associated CACNA1C proximal promoter and the potential role for the transcription factor EZH2 in schizophrenia aetiology. Schizophr. Res. 199, 168–175. doi: 10.1016/j.schres.2018.02.036
Blacker, C. J., Millischer, V., Webb, L. M., Ho, A. M. C., Schalling, M., Frye, M. A., et al. (2019). EAAT2 as a Research Target in Bipolar Disorder and Unipolar Depression: A Systematic Review. Mol. Neuropsychiatry 1–16. doi: 10.1159/000501885
Burghardt, K. J., Goodrich, J. M., Dolinoy, D. C., Ellingrod, V. L. (2015). DNA methylation, insulin resistance and second-generation antipsychotics in bipolar disorder. Epigenomics 7 (3), 343–352. doi: 10.2217/epi.15.5
Burghardt, K. J., Goodrich, J. M., Dolinoy, D. C., Ellingrod, V. L. (2016). Gene-specific DNA methylation may mediate atypical antipsychotic-induced insulin resistance. Bipolar Disord. 18 (5), 423–432. doi: 10.1111/bdi.12422
Calabrese, F., Luoni, A., Guidotti, G., Racagni, G., Fumagalli, F., Riva, M. A. (2013). Modulation of neuronal plasticity following chronic concomitant administration of the novel antipsychotic lurasidone with the mood stabilizer valproic acid. Psychopharmacology 226 (1), 101–112. doi: 10.1007/s00213-012-2900-0
Cedar, H., Bergman, Y. (2009). Linking DNA methylation and histone modification: patterns and paradigms. Nat. Rev. Genet. 10 (5), 295–304. doi: 10.1038/nrg2540
Chen, J. H., Tsang, S. Y., Zhao, C. Y., Pun, F. W., Yu, Z. L., Mei, L. L., et al. (2009). GABRB2 in schizophrenia and bipolar disorder: disease association, gene expression and clinical correlations. Biochem. Soc. Trans. 37, 1415–1418. doi: 10.1042/Bst0371415
Chen, W. T., Huang, T. L., Tsai, M. C. (2015). Bcl-2 associated with severity of manic symptoms in bipolar patients in a manic phase. Psychiatry Res. 225 (3), 305–308. doi: 10.1016/j.psychres.2014.12.020
Cheng, C. M., Chang, W. H., Chen, M. H., Tsai, C. F., Su, T. P., Li, C. T., et al. (2018). Co-aggregation of major psychiatric disorders in individuals with first-degree relatives with schizophrenia: a nationwide population-based study. Mol. Psychiatry 23 (8), 1756–1763. doi: 10.1038/mp.2017.217
Christensen, J. H., Elfving, B., Muller, H. K., Fryland, T., Nyegaard, M., Corydon, T. J., et al. (2012). The Schizophrenia and Bipolar Disorder associated BRD1 gene is regulated upon chronic restraint stress. Eur. Neuropsychopharmacol. 22 (9), 651–656. doi: 10.1016/j.euroneuro.2012.01.005
Christopherson, K. S., Ullian, E. M., Stokes, C. C. A., Mullowney, C. E., Hell, J. W., Agah, A., et al. (2005). Thrombospondins are astrocyte-secreted proteins that promote CNS synaptogenesis. Cell 120 (3), 421–433. doi: 10.1016/j.cell.2004.12.020
Chuang, D. M., Chen, R. W., Chalecka-Franaszek, E., Ren, M., Hashimoto, R., Senatorov, V., et al. (2002). Neuroprotective effects of lithium in cultured cells and animal models of diseases. Bipolar Disord. 4 (2), 129–136. doi: 10.1034/j.1399-5618.2002.01179.x
Craig, A. M., Kang, Y. (2007). Neurexin-neuroligin signaling in synapse development. Curr. Opin. Neurobiol. 17 (1), 43–52. doi: 10.1016/j.conb.2007.01.011
Cross-Disorder Group of the Psychiatric Genomics, C (2013). Identification of risk loci with shared effects on five major psychiatric disorders: a genome-wide analysis. Lancet (London Engl.) 381 (9875), 1371–1379. doi: 10.1016/S0140-6736(12)62129-1
D’Addario, C., Dell’Osso, B., Palazzo, M. C., Benatti, B., Lietti, L., Cattaneo, E., et al. (2012). Selective DNA Methylation of BDNF Promoter in Bipolar Disorder: Differences Among Patients with BDI and BDII. Neuropsychopharmacology 37 (7), 1647–1655. doi: 10.1038/npp.2012.10
Dalva, M. B., McClelland, A. C., Kayser, M. S. (2007). Cell adhesion molecules: signalling functions at the synapse. Nat. Rev. Neurosci. 8 (3), 206–220. doi: 10.1038/nrn2075
Davies, M. N., Volta, M., Pidsley, R., Lunnon, K., Dixit, A., Lovestone, S., et al. (2012). Functional annotation of the human brain methylome identifies tissue-specific epigenetic variation across brain and blood. Genome Biol. 13 (6), 1–14. doi: 10.1186/gb-2012-13-6-r43
Dell’Osso, B., D’Addario, C., Palazzo, M. C., Benatti, B., Camuri, G., Galimberti, D., et al. (2014). Epigenetic modulation of BDNF gene: differences in DNA methylation between unipolar and bipolar patients. J. Affect. Disord. 166, 330–333. doi: 10.1016/j.jad.2014.05.020
Dong, E., Agis-Balboa, R. C., Simonini, M. V., Grayson, D. R., Costa, E., Guidotti, A. (2005). Reelin and glutamic acid decarboxylase(67) promoter remodeling in an epigenetic methionine-induced mouse model of schizophrenia. Proc. Natl. Acad. Sci. U. States America 102 (35), 12578–12583. doi: 10.1073/pnas.0505394102
Dong, E., Chen, Y., Gavin, D. P., Grayson, D. R., Guidotti, A. (2010). Valproate induces DNA demethylation in nuclear extracts from adult mouse brain. Epigenetics 5 (8), 730–735. doi: 10.4161/epi.5.8.13053
du Prel, J. B., Rohrig, B., Blettner, M. (2009). Critical appraisal of scientific articles: part 1 of a series on evaluation of scientific publications. Dtsch Arztebl Int. 106 (7), 100–105. doi: 10.3238/arztebl.2009.0100
Dwivedi, T., Zhang, H. (2015). Lithium-induced neuroprotection is associated with epigenetic modification of specific BDNF gene promoter and altered expression of apoptotic-regulatory proteins. Front. Neurosci. 8, 1–8. doi: 10.3389/fnins.2014.00457
Dyrvig, M., Qvist, P., Lichota, J., Larsen, K., Nyegaard, M., Borglum, A. D., et al. (2017). DNA Methylation Analysis of BRD1 Promoter Regions and the Schizophrenia rs138880 Risk Allele. PloS One 12 (1), 1–19. doi: 10.1371/journal.pone.0170121
Dyrvig, M., Mikkelsen, J. D., Lichota, J. (2019). DNA methylation regulates CHRNA7 transcription and can be modulated by valproate. Neurosci. Lett. 704, 145–152. doi: 10.1016/j.neulet.2019.04.015
Eyal, S., Yagen, B., Sobol, E., Altschuler, Y., Shmuel, M., Bialer, M. (2004). The activity of antiepileptic drugs as histone deacetylase inhibitors. Epilepsia 45 (7), 737–744. doi: 10.1111/j.0013-9580.2004.00104.x
Gatt, J. M., Burton, K. L. O., Williams, L. M., Schofield, P. R. (2015). Specific and common genes implicated across major mental disorders: A review of meta-analysis studies. J. Psychiatr. Res. 60, 1–13. doi: 10.1016/j.jpsychires.2014.09.014
Gavin, D. P., Sharma, R. P. (2010). Histone modifications, DNA methylation, and Schizophrenia. Neurosci. Biobehav. Rev. 34 (6), 882–888. doi: 10.1016/j.neubiorev.2009.10.010
Gavin, D. P., Kartan, S., Chase, K., Jayaraman, S., Sharma, R. P. (2009). Histone deacetylase inhibitors and candidate gene expression: An in vivo and in vitro approach to studying chromatin remodeling in a clinical population. J. Psychiatr. Res. 43 (9), 870–876. doi: 10.1016/j.jpsychires.2008.12.006
Gawryluk, J. W., Wang, J. F., Andreazza, A. C., Shao, L., Young, L. T. (2011). Decreased levels of glutathione, the major brain antioxidant, in post-mortem prefrontal cortex from patients with psychiatric disorders. Int. J. Neuropsychopharmacol. 14 (1), 123–130. doi: 10.1017/S1461145710000805
Geoffroy, P. A. (2018). Clock Genes and Light Signaling Alterations in Bipolar Disorder: When the Biological Clock Is Off. Biol. Psychiatry 84 (11), 775–777. doi: 10.1016/j.biopsych.2018.09.006
Goodwin, G. M., Malhi, G. S. (2007). What is a mood stabilizer? psychol. Med. 37 (5), 609–614. doi: 10.1017/S0033291706009305
Gottlicher, M., Minucci, S., Zhu, P., Kramer, O. H., Schimpf, A., Giavara, S., et al. (2001). Valproic acid defines a novel class of HDAC inhibitors inducing differentiation of transformed cells. EMBO J. 20 (24), 6969–6978. doi: 10.1093/emboj/20.24.6969
Guidotti, A., Auta, J., Davis, J. M., Di-Giorgi-Gerevini, V., Dwivedi, Y., Grayson, D. R., et al. (2000). Decrease in reelin and glutamic acid decarboxylase67 (GAD67) expression in schizophrenia and bipolar disorder: a postmortem brain study. Arch. Gen. Psychiatry 57 (11), 1061–1069. doi: 10.1001/archpsyc.57.11.1061
Guidotti, A., Grayson, D. R., Caruncho, H. J. (2016). Epigenetic RELN Dysfunction in Schizophrenia and Related Neuropsychiatric Disorders. Front. Cell. Neurosci. 10, 55–60. doi: 10.3389/fncel.2016.00089
Harrison, P. J. (2015). Recent genetic findings in schizophrenia and their therapeutic relevance. J. Psychopharmacol. 29 (2), 85–96. doi: 10.1177/0269881114553647
Higuchi, F., Uchida, S., Yamagata, H., Otsuki, K., Hobara, T., Abe, N., et al. (2011). State-dependent changes in the expression of DNA methyltransferases in mood disorder patients. J. Psychiatr. Res. 45 (10), 1295–1300. doi: 10.1016/j.jpsychires.2011.04.008
Hobara, T., Uchida, S., Otsuki, K., Matsubara, T., Funato, H., Matsuo, K., et al. (2010). Altered gene expression of histone deacetylases in mood disorder patients. J. Psychiatr. Res. 44 (5), 263–270. doi: 10.1016/j.jpsychires.2009.08.015
Houston, I., Peter, C. J., Mitchell, A., Straubhaar, J., Rogaev, E., Akbarian, S. (2013). Epigenetics in the Human Brain. Neuropsychopharmacology 38 (1), 183–197. doi: 10.1038/npp.2012.78
Houtepen, L. C., van Bergen, A. H., Vinkers, C. H., Boks, M. P. M. (2016). DNA methylation signatures of mood stabilizers and antipsychotics in bipolar disorder. Epigenomics 8 (2), 197–208. doi: 10.2217/epi.15.98
Huzayyin, A. A., Andreazza, A. C., Turecki, G., Cruceanu, C., Rouleau, G. A., Alda, M., et al. (2014). Decreased global methylation in patients with bipolar disorder who respond to lithium. Int. J. Neuropsychopharmacol. 17 (4), 561–569. doi: 10.1017/S1461145713001569
Inbar-Feigenberg, M., Choufani, S., Butcher, D. T., Roifman, M., Weksberg, R. (2013). Basic concepts of epigenetics. Fertil. Sterility 99 (3), 607–615. doi: 10.1016/j.fertnstert.2013.01.117
Kakiuchi, C., Iwamoto, K., Ishiwata, M., Bundo, M., Kasahara, T., Kusumi, I., et al. (2003). Impaired feedback regulation of XBP1 as a genetic risk factor for bipolar disorder. Nat. Genet. 35 (2), 171–175. doi: 10.1038/ng1235
Kaminsky, Z., Jones, I., Verma, R., Saleh, L., Trivedi, H., Guintivano, J., et al. (2015). DNA methylation and expression of KCNQ3 in bipolar disorder. Bipolar Disord. 17 (2), 150–159. doi: 10.1111/bdi.12230
Kao, C. Y., Hsu, Y. C., Liu, J. W., Lee, D. C., Chung, Y. F., Chiu, I. M. (2013). The mood stabilizer valproate activates human FGF1 gene promoter through inhibiting HDAC and GSK-3 activities. J. Neurochem. 126 (1), 4–18. doi: 10.1111/jnc.12292
Kim, B., Rincon Castro, L. M., Jawed, S., Niles, L. P. (2008). Clinically relevant concentrations of valproic acid modulate melatonin MT(1) receptor, HDAC and MeCP2 mRNA expression in C6 glioma cells. Eur. J. Pharmacol. 589 (1-3), 45–48. doi: 10.1016/j.ejphar.2008.04.058
Koch, S. M., Ullian, E. M. (2010). Neuronal Pentraxins Mediate Silent Synapse Conversion in the Developing Visual System. J. Neurosci. 30 (15), 5404–5414. doi: 10.1523/Jneurosci.4893-09.2010
Lee, R. S., Pirooznia, M., Guintivano, J., Ly, M., Ewald, E. R., Tamashiro, K. L., et al. (2015). Search for common targets of lithium and valproic acid identifies novel epigenetic effects of lithium on the rat leptin receptor gene. Trans. Psychiatry 5, 1–10. doi: 10.1038/tp.2015.90
Leng, Y., Liang, M. H., Ren, M., Marinova, Z., Leeds, P., Chuang, D. M. (2008). Synergistic neuroprotective effects of lithium and valproic acid or other histone deacetylase inhibitors in neurons: Roles of glycogen synthase kinase-3 inhibition. J. Neurosci. 28 (10), 2576–2588. doi: 10.1523/Jneurosci.5467-07.2008
Leng, Y., Fessler, E. B., Chuang, D. M. (2013). Neuroprotective effects of the mood stabilizer lamotrigine against glutamate excitotoxicity: roles of chromatin remodelling and Bcl-2 induction. Int. J. Neuropsychopharmacol. 16 (3), 607–620. doi: 10.1017/S1461145712000429
Leng, Y., Wang, J. Y., Wang, Z. F., Liao, H. M., Wei, M., Leeds, P., et al. (2016). Valproic Acid and Other HDAC Inhibitors Upregulate FGF21 Gene Expression and Promote Process Elongation in Glia by Inhibiting HDAC2 and 3. Int. J. Neuropsychopharmacol. 19 (8), 1–13. doi: 10.1093/ijnp/pyw035
Liu, D., Qiu, H. M., Fei, H. Z., Hu, X. Y., Xia, H. J., Wang, L. J., et al. (2014). Histone acetylation and expression of mono-aminergic transmitters synthetases involved in CUS-induced depressive rats. Exp. Biol. Med. 239 (3), 330–336. doi: 10.1177/1535370213513987
Lodge, D. J. (2013). The MAM rodent model of schizophrenia. Curr. Protoc. Neurosci. 9, 1–8. doi: 10.1002/0471142301.ns0943s63. Chapter 9, Unit9 43.
Lopez-Leon, S., Janssens, A. C. J. W., Ladd, A. M. G. Z., Del-Favero, J., Claes, S. J., Oostra, B. A., et al. (2008). Meta-analyses of genetic studies on major depressive disorder. Mol. Psychiatry 13 (8), 772–785. doi: 10.1038/sj.mp.4002088
Ludwig, B., Dwivedi, Y. (2016). Dissecting bipolar disorder complexity through epigenomic approach. Mol. Psychiatry 21 (11), 1490–1498. doi: 10.1038/mp.2016.123
Machado-Vieira, R., Ibrahim, L., Zarate, C. A., Jr. (2011). Histone deacetylases and mood disorders: epigenetic programming in gene-environment interactions. CNS Neurosci. Ther. 17 (6), 699–704. doi: 10.1111/j.1755-5949.2010.00203.x
Machado-Vieira, R. (2018). Lithium, Stress, and Resilience in Bipolar Disorder: Deciphering this key homeostatic synaptic plasticity regulator. J. Affect. Disord. 233, 92–99. doi: 10.1016/j.jad.2017.12.026
Mackowiak, M., Bator, E., Latusz, J., Mordalska, P., Wedzony, K. (2014). Prenatal MAM administration affects histone H3 methylation in postnatal life in the rat medial prefrontal cortex. Eur. Neuropsychopharmacol. 24 (2), 271–289. doi: 10.1016/j.euroneuro.2013.05.013
Malhi, G. S., Mann, J. J. (2018). Depression. Lancet 392 (10161), 2299–2312. doi: 10.1016/S0140-6736(18)31948-2
Manca, M., Pessoa, V., Myers, P., Pickles, A., Hill, J., Sharp, H., et al. (2019). Distinct chromatin structures at the monoamine oxidase-A promoter correlate with allele-specific expression in SH-SY5Y cells. Genes Brain Behav. 18 (6), e12483. doi: 10.1111/gbb.12483
Marder, S. R., Cannon, T. D. (2019). Schizophrenia. N. Engl. J. Med. 381 (18), 1753–1761. doi: 10.1056/NEJMra1808803
Matrisciano, F., Dong, E., Gavin, D. P., Nicoletti, F., Guidotti, A. (2011). Activation of Group II Metabotropic Glutamate Receptors Promotes DNA Demethylation in the Mouse Brain. Mol. Pharmacol. 80 (1), 174–182. doi: 10.1124/mol.110.070896
Moon, A. L., Haan, N., Wilkinson, L. S., Thomas, K. L., Hall, J. (2018). CACNA1C: Association With Psychiatric Disorders, Behavior, and Neurogenesis. Schizophr. Bull. 44 (5), 958–965. doi: 10.1093/schbul/sby096
Ookubo, M., Kanai, H., Aoki, H., Yamada, N. (2013). Antidepressants and mood stabilizers effects on histone deacetylase expression in C57BL/6 mice: Brain region specific changes. J. Psychiatr. Res. 47 (9), 1204–1214. doi: 10.1016/j.jpsychires.2013.05.028
Ouzzani, M., Hammady, H., Fedorowicz, Z., Elmagarmid, A. (2016). Rayyan-a web and mobile app for systematic reviews. Syst. Rev. 5, 1–10. doi: 10.1186/s13643-016-0384-4
Perisic, T., Zimmermann, N., Kirmeier, T., Asmus, M., Tuorto, F., Uhr, M., et al. (2010). Valproate and Amitriptyline Exert Common and Divergent Influences on Global and Gene Promoter-Specific Chromatin Modifications in Rat Primary Astrocytes. Neuropsychopharmacology 35 (3), 792–805. doi: 10.1038/npp.2009.188
Phiel, C. J., Zhang, F., Huang, E. Y., Guenther, M. G., Lazar, M. A., Klein, P. S. (2001). Histone deacetylase is a direct target of valproic acid, a potent anticonvulsant, mood stabilizer, and teratogen. J. Biol. Chem. 276 (39), 36734–36741. doi: 10.1074/jbc.M101287200
Pisanu, C., Papadima, E. M., Del Zompo, M., Squassina, A. (2018). Understanding the molecular mechanisms underlying mood stabilizer treatments in bipolar disorder: Potential involvement of epigenetics. Neurosci. Lett. 669, 24–31. doi: 10.1016/j.neulet.2016.06.045
Qiao, Y. B., Wang, R., Yang, X. F., Tang, K., Jing, N. H. (2015). Dual Roles of Histone H3 Lysine 9 Acetylation in Human Embryonic Stem Cell Pluripotency and Neural Differentiation. J. Biol. Chem. 290 (4), 2508–2520. doi: 10.1074/jbc.M114.603761
Reid, J. G., Gitlin, M. J., Altshuler, L. L. (2013). Lamotrigine in Psychiatric Disorders. J. Clin. Psychiatry 74 (7), 675–684. doi: 10.4088/JCP.12r08046
Rondinelli, B., Schwerer, H., Antonini, E., Gaviraghi, M., Lupi, A., Frenquelli, M., et al. (2015). H3K4me3 demethylation by the histone demethylase KDM5C/JARID1C promotes DNA replication origin firing. Nucleic Acids Res. 43 (5), 2560–2574. doi: 10.1093/nar/gkv090
Scaini, G., Fries, G. R., Valvassori, S. S., Zeni, C. P., Zunta-Soares, G., Berk, M., et al. (2017). Perturbations in the apoptotic pathway and mitochondrial network dynamics in peripheral blood mononuclear cells from bipolar disorder patients. Trans. Psychiatry 7, 1–10. doi: 10.1038/tp.2017.83
Schenkel, L. C., Rodenhiser, D., Siu, V., McCready, E., Ainsworth, P., Sadikovic, B. (2017). Constitutional Epi/Genetic Conditions: Genetic, Epigenetic, and Environmental Factors. J. Pediatr. Genet. 6 (1), 30–41. doi: 10.1055/s-0036-1593849
Schloesser, R. J., Huang, J., Klein, P. S., Manji, H. K. (2008). Cellular plasticity cascades in the pathophysiology and treatment of bipolar disorder. Neuropsychopharmacology 33 (1), 110–133. doi: 10.1038/sj.npp.1301575
Schloesser, R. J., Martinowich, K., Manji, H. K. (2012). Mood-stabilizing drugs: mechanisms of action. Trends Neurosci. 35 (1), 36–46. doi: 10.1016/j.tins.2011.11.009
Seo, M. S., Scarr, E., Lai, C. Y., Dean, B. (2014). Potential Molecular and Cellular Mechanism of Psychotropic Drugs. Clin. Psychopharmacol. Neurosci. 12 (2), 94–110. doi: 10.9758/cpn.2014.12.2.94
Serretti, A., Mandelli, L. (2008). The genetics of bipolar disorder: genome ‘hot regions,’ genes, new potential candidates and future directions. Mol. Psychiatry 13 (8), 742–771. doi: 10.1038/mp.2008.29
Shahbazian, M. D., Grunstein, M. (2007). Functions of Site-Specific Histone Acetylation and Deacetylation. Annu. Rev. Biochem. 76, 75–100. doi: 10.1146/annurev.biochem.76.052705.162114
Shamseer, L., Moher, D., Clarke, M., Ghersi, D., Liberati, A., Petticrew, M., et al. (2015). Preferred reporting items for systematic review and meta-analysis protocols (PRISMA-P) 2015: elaboration and explanation. BMJ 350, g7647. doi: 10.1136/bmj.g7647
Stephens, S. H., Logel, J., Barton, A., Franks, A., Schultz, J., Short, M., et al. (2009). Association of the 5′-upstream regulatory region of the alpha7 nicotinic acetylcholine receptor subunit gene (CHRNA7) with schizophrenia. Schizophr. Res. 109 (1-3), 102–112. doi: 10.1016/j.schres.2008.12.017
Tremolizzo, L., Carboni, G., Ruzicka, W. B., Mitchell, C. P., Sugaya, I., Tueting, P., et al. (2002). An epigenetic mouse model for molecular and behavioral neuropathologies related to schizophrenia vulnerability. Proc. Natl. Acad. Sci. U. States America 99 (26), 17095–17100. doi: 10.1073/pnas.262658999
Tremolizzo, L., Doueiri, M. S., Dong, E., Grayson, D. R., Davis, J., Pinna, G., et al. (2005). Valproate corrects the schizophrenia-like epigenetic behavioral modifications induced by methionine in mice. Biol. Psychiatry 57 (5), 500–509. doi: 10.1016/j.biopsych.2004.11.046
Tsai, M. C., Liou, C. W., Lin, T. K., Lin, I. M., Huang, T. L. (2013). Bcl-2 associated with positive symptoms of schizophrenic patients in an acute phase. Psychiatry Res. 210 (3), 735–738. doi: 10.1016/j.psychres.2013.08.032
Veldic, M., Caruncho, H. J., Liu, W. S., Davis, J., Satta, R., Grayson, D. R., et al. (2004). DNA-methyltransferase 1 mRNA is selectively overexpressed in telencephalic GABAergic interneurons of schizophrenia brains. Proc. Natl. Acad. Sci. U. States America 101 (1), 348–353. doi: 10.1073/pnas.2637013100
Vieta, E., Berk, M., Schulze, T. G., Carvalho, A. F., Suppes, T., Calabrese, J. R., et al. (2018). Bipolar disorders. Nat. Rev. Dis. Primers 4, 18008. doi: 10.1038/nrdp.2018.8
Vigo, D., Thornicroft, G., Atun, R. (2016). Estimating the true global burden of mental illness. Lancet Psychiatry 3 (2), 171–178. doi: 10.1016/S2215-0366(15)00505-2
Wang, C.-C., Chen, P., Hsu, C.-W., Wu, S.-J., Lin, C.-T., Gean, P.-W. (2012). Valproic acid mediates the synaptic excitatory/inhibitory balance through astrocytes - A preliminary study. Prog. Neuropsychopharmacol. Biol. Psychiatry 37, 111–120. doi: 10.1016/j.pnpbp.2012.01.017
Wang, L., Alachkar, A., Sanathara, N., Belluzzi, J. D., Wang, Z. W., Civelli, O. (2015). A Methionine-Induced Animal Model of Schizophrenia: Face and Predictive Validity. Int. J. Neuropsychopharmacol. 18 (12), 1–11. doi: 10.1093/ijnp/pyv054
Yankelevitch-Yahav, R., Franko, M., Huly, A., Doron, R. (2015). The forced swim test as a model of depressive-like behavior. J. Vis. Exp. 97, 1–7. doi: 10.3791/52587
Yatham, L. N., Kennedy, S. H., Parikh, S. V., Schaffer, A., Bond, D. J., Frey, B. N., et al. (2018). Canadian Network for Mood and Anxiety Treatments (CANMAT) and International Society for Bipolar Disorders (ISBD) 2018 guidelines for the management of patients with bipolar disorder. Bipolar Disord. 20 (2), 97–170. doi: 10.1111/bdi.12609
Zhang, C., Wu, Z. G., Hong, W., Wang, Z. W., Peng, D. H., Chen, J., et al. (2014). Influence of BCL2 gene in major depression susceptibility and antidepressant treatment outcome. J. Affect. Disord. 155, 288–294. doi: 10.1016/j.jad.2013.11.010
Zhao, C. Y., Wang, F., Pun, F. W., Mei, L. L., Ren, L. H., Yu, Z. L., et al. (2012). Epigenetic regulation on GABRB2 isoforms expression: Developmental variations and disruptions in psychotic disorders. Schizophr. Res. 134 (2-3), 260–266. doi: 10.1016/j.schres.2011.11.029
Zhu, D. J., Yin, J. W., Liang, C. M., Luo, X. D., Lv, D., Dai, Z., et al. (2019). CACNA1C (rs1006737) may be a susceptibility gene for schizophrenia: An updated meta-analysis. Brain Behav. 9 (6), 1–8. doi: 10.1002/brb3.1292
Zimmermann, N., Zschocke, J., Perisic, T., Yu, S., Holsboer, F., Rein, T. (2012). Antidepressants inhibit DNA methyltransferase 1 through reducing G9a levels. Biochem. J. 448 (1), 93–102. doi: 10.1042/bj20120674
Keywords: epigenetics, mood disorders, schizophrenia, lithium carbonate, valproic acid, anticonvulsants, histone deacetylase inhibitors, DNA methylation
Citation: Gardea-Resendez M, Kucuker MU, Blacker CJ, Ho AM-C, Croarkin PE, Frye MA and Veldic M (2020) Dissecting the Epigenetic Changes Induced by Non-Antipsychotic Mood Stabilizers on Schizophrenia and Affective Disorders: A Systematic Review. Front. Pharmacol. 11:467. doi: 10.3389/fphar.2020.00467
Received: 03 January 2020; Accepted: 25 March 2020;
Published: 22 April 2020.
Edited by:
Hector J. Caruncho, University of Victoria, CanadaReviewed by:
Shota Nishitani, University of Fukui, JapanRaquel Romay-Tallon, University of Illinois at Chicago, United States
Carlos Spuch, Instituto de Investigación Sanitaria Galicia Sur (IISGS), Spain
Copyright © 2020 Gardea-Resendez, Kucuker, Blacker, Ho, Croarkin, Frye and Veldic. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Marin Veldic, VmVsZGljLk1hcmluQG1heW8uZWR1
 Manuel Gardea-Resendez
Manuel Gardea-Resendez Mehmet Utku Kucuker
Mehmet Utku Kucuker Caren J. Blacker
Caren J. Blacker Ada M.-C. Ho
Ada M.-C. Ho Paul E. Croarkin
Paul E. Croarkin Mark A. Frye
Mark A. Frye Marin Veldic
Marin Veldic