
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Pharmacol. , 07 November 2019
Sec. Pharmacogenetics and Pharmacogenomics
Volume 10 - 2019 | https://doi.org/10.3389/fphar.2019.01318
The objective of this study was to investigate whether MTNR1B gene variants influence repaglinide response in Chinese patients with newly diagnosed type 2 diabetes mellitus (T2DM). A total of 300 patients with T2DM and 200 control subjects were enrolled to identify MTNR1B rs10830963 and rs1387153 genotypes by real-time polymerase chain reaction (PCR), with subsequent high-resolution melting (HRM) analysis. Ninety-five patients with newly diagnosed T2DM were randomly selected to undergo 8 weeks of repaglinide treatment (3 mg/day). After 8-week repaglinide monotherapy, patients with at least one G allele of MTNR1B rs10830963 showed a smaller decrease in fasting plasma glucose (FPG) (P = 0.031) and a smaller increase in homeostasis model assessment for beta cell function (HOMA-B) (P = 0.002) levels than those with the CC genotype did. The T allele carriers at rs1387153 exhibited a smaller decrease in FPG (P = 0.007) and smaller increases in postprandial serum insulin (PINS) (P = 0.016) and HOMA-B (P < 0.001) levels compared to individuals with the CC genotype. These data suggest that the MTNR1B rs10830963 and rs1387153 polymorphisms are associated with repaglinide monotherapy efficacy in Chinese patients with T2DM.
The incidence of type 2 diabetes mellitus (T2DM) has increased at an alarming rate worldwide over the recent decades (Persaud et al., 2016). It is well established that T2DM is a complex multifactorial and polygenic metabolic disorder characterized by impaired insulin secretion and reduced sensitivity to the peripheral actions of insulin (Pal and McCarthy, 2013; Mannino et al., 2019). Genome-wide association studies (GWAS) have identified more than 150 so-called risk alleles that increase a person’s susceptibility to T2DM, including a common variant in the melatonin receptor 1B gene (MTNR1B) (Persaud et al., 2016). The MTNR1B gene encodes the melatonin receptor MT2, a G protein-coupled receptor, that is expressed in many tissues, including pancreatic islets (Lyssenko et al., 2008). In 2008, three GWAS in European cohort studies reported that the MTNR1B gene was a new susceptibility gene for T2DM (Bouatia-Naji et al., 2008; Lyssenko et al., 2008; Prokopenko et al., 2008). Meanwhile, human and animal studies already pointed to a significant impact of MTNR1B on the regulation of glucose homeostasis (Tuomi et al., 2016; Patel et al., 2018; Karamitri and Jockers, 2019). It has been reported that two common variants (rs1387153 and rs10830963) near/in MTNR1B were associated with elevated plasma glucose levels, a reduction of the early insulin response to both oral and intravenous glucose, and a faster deterioration of insulin secretion over time (Bouatia-Naji et al., 2008; Lyssenko et al., 2008; Prokopenko et al., 2008; Rönn et al., 2009; Kan et al., 2010). These association studies have demonstrated a link between MTNR1B and T2DM, but the fundamental mechanism underlying the association remains far from clear.
Repaglinide is an oral antidiabetic agent that is widely used in the treatment of T2DM. It triggers insulin secretion by closing the ATP-dependent potassium channels and activating Ca2+ channels on the surface of pancreatic beta cells, thus lowering plasma glucose levels and especially the postprandial glucose level (Scott, 2012). However, some studies have shown that there were marked interindividual differences in therapeutic response and adverse drug reactions in patients with T2DM treated with repaglinide monotherapy (Eichelbaum et al., 2006; Maruthur et al., 2014; Chen et al., 2016). These differences were partly due to genetic factors related to drug absorption, distribution, metabolism, and targets and genes involved in the causal pathway of T2DM (Tomalik-Scharte et al., 2011; Chakraborty et al., 2015; Chen et al., 2015; Sanchez-Ibarra et al., 2018). Many variants of T2DM susceptibility genes, such as KCNJ11, ABCC8, and KCNQ1, were associated with pharmacogenomic effects of repaglinide in previous studies (Maruthur et al., 2014). Therefore, polymorphisms in certain susceptibility genes associated with T2DM may potentially affect repaglinide efficacy. However, it remains unknown whether MTNR1B single-nucleotide polymorphisms (SNPs) have the same influence on the therapeutic effects of repaglinide. Thus, in this study, we further explored the association of MTNR1B polymorphisms with the therapeutic effect of repaglinide in Chinese patients with T2DM.
A total of 300 patients with T2DM and 200 healthy controls were recruited for MTNR1B polymorphism analysis in this study. All participants were evaluated by analysis of medical histories, physical examinations, and routine clinical laboratory tests. The basic clinical and biochemical characteristics of subjects have previously been described in detail (Wang et al., 2014). T2DM was diagnosed according to the 1999 World Health Organization criteria (Gabir et al., 2000). The inclusion criteria were that the patients had a body mass index (BMI) of 18.5–30 kg/m2 and had not received any insulin secretagogue or any agonist or inhibitor of CYP2C8, CYP3A4, and OATP1B1 in the past 3 months. Patients with serious diseases such as acute myocardial infarction, cerebral vascular accident, trauma, and kidney or liver diseases; patients receiving insulin treatment; and pregnant or lactating women were excluded from this study. A total of 95 patients with T2DM (54 male and 41 female) with different MTNR1B rs10830963 and rs1387153 genotypes were randomly selected to receive repaglinide (3 mg/day) for 8 consecutive weeks. The study was registered in the Chinese Clinical Trial Registry (registration number: ChiCTR-CCC13003536), and the protocol was approved by the Ethics Committee of the Affiliated Hospital of Xuzhou Medical University. Written informed consent was obtained from each participant before taking part in the study.
The general anthropometric parameters considered for this study were height (in meters), weight (in kilograms), and waist and hip circumferences (in centimeters). After an overnight fast, venous blood samples were obtained both in the fasting state and 2 h later during a standard 75-g oral glucose tolerance test. Parameters were measured at the end of weeks 0 and 8 after administration of repaglinide. Plasma glucose and serum lipids were assessed using standard laboratory methods using a Roche Cobas 8000 analyzer (Roche, Basel, Switzerland). Levels of plasma insulin and hemoglobin A1c (HbA1c) were measured by electro-chemiluminescence assay (Roche, Shanghai, China) and high-performance liquid chromatography assay, respectively. Insulin resistance and beta cell function were calculated with the following formulae: homeostasis model assessment for insulin resistance (HOMA-IR) = fasting insulin level (mU/L) × fasting glucose level (mmol/L)/22.5; homeostasis model assessment for beta cell function (HOMA-B) = 20 × fasting serum insulin (FINS)/(FPG-3.5) (Kong et al., 2017).
Genomic DNA was extracted from peripheral blood leucocytes. Genotypes for the MTNR1B polymorphisms were analyzed using a HRM assay. For the rs10830963 locus, we used the primer pair 5’-GAGGATTTGCTTGCTGAACA-3’ (forward) and 5’-CCCAGGCAGTTACTGGTTCT-3’ (reverse). For rs1387153, we used the primer pair 5’-AACACATGGAAAATGCTTGG-3’ (forward) and 5’-CTGGTTCCAAACCCAACTCT-3’ (reverse). The HRM analyses were performed in a 10 µl reaction system containing 5 µl LC480 HRM Master Mix (Roche Applied Science, Mannheim, Germany), which comprises FastStart Taq DNA Polymerase and the High Resolution Melting Dye in a reaction buffer; 1.2 µl of 25 mM MgCl2, 0.2 µl of each primer, 2.5 µl of target DNA sample, 0.5 µl of one known genotype DNA sample (wide type), and 0.4 µl deionized water. The thermal cycling profile for the real-time PCR comprised an initial denaturation at 95°C for 10 min, followed by 55 cycles of 10 s denaturation at 95°C, 10 s annealing at 65°C, and 15 s extension at 72°C. The subsequent melt curve program comprised: 95°C for 1 min, 40°C for 1 min, 70°C for 1 s, 95°C for 1 min, and then cooling at 40°C for 30 s. To confirm the HRM assay results, 5.0% of all samples were directly sequenced.
Patients were categorized into two cohorts based on glycemic response to repaglinide use: responder and nonresponder. Previous studies have reported that T2DM patients undergoing repaglinide treatment experience a decline of HbA1c by 15%–30% (Abbink et al., 2004; Fang et al., 2014; Jia et al., 2018; Wang et al., 2018). In the current study, levels of HbA1c were reduced by 28% on average. Therefore, responders were defined as those patients with an HbA1c decrease of 25% or more after 8 weeks of repaglinide treatment, whereas nonresponders were defined as patients who failed to achieve this level of decrease.
Statistical analyses were performed using SPSS software (version 13.0 for Windows; SPSS Inc., Chicago, IL, USA). All data were expressed as mean ± standard deviation (SD), median [interquartile range (IQR)], or percentages as appropriate. The Hardy-Weinberg equilibrium and allelic frequencies in different groups were assessed using χ2 tests. A comparison of baseline characteristics in patients with T2DM and healthy controls was carried out using independent samples t tests. Differences among three genotypes were analyzed using the one-way ANOVA when appropriate. Paired t tests and independent samples t tests were used to estimate the effects of repaglinide on biochemical index among genotypes. Parameters with nonnormal distribution were analyzed by the Mann-Whitney U-test or the Kruskal-Wallis test. Genotype distribution differences between responders and nonresponders were compared using the χ2 test or Fisher’s exact test. Logistic regression analysis was used to test the major determinants of repaglinide response rate. The odds ratio (OR) values were presented with 95% confidence intervals (CIs). Linkage disequilibrium (LD) between SNPs was estimated in control subjects using Haploview version 3.2. Statistical power was calculated by power calculator software (http://www.ncss.com). A value of P < 0.05 was considered statistically significant.
The distribution of genotypes and allele frequencies of the MTNR1B rs10830963 and rs1387153 polymorphisms in T2DM patients and control subjects are summarized in Table 1. The frequency of the G allele at the MTNR1B rs10830963 locus was higher in patients with T2DM than in healthy controls (44.17 vs. 35.75%, P = 0.008). For the MTNR1B rs1387153 polymorphism, there was no significant difference in allele frequencies between T2DM patients and control subjects. The genotype distributions of rs10830963 and rs1387153 SNPs were in Hardy-Weinberg equilibrium (P > 0.05). Assessment of the LD between the polymorphisms using our control subjects revealed a relatively high disequilibrium between rs10830963 and rs1387153 (r2 = 0.630, P < 0.05).
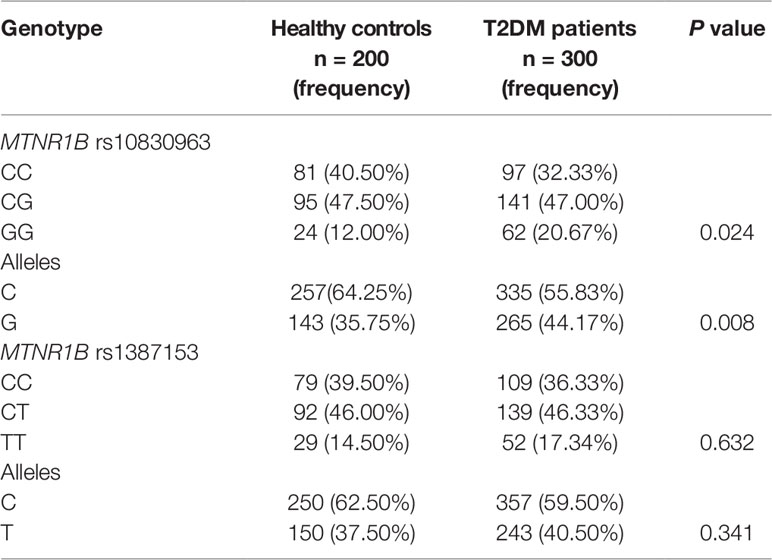
Table 1 Genotypes and frequencies of MTNR1B polymorphisms in patients with type 2 diabetes mellitus (T2DM) (n = 300) and healthy controls (n = 200).
The baseline clinical characteristics of 300 T2DM patients with different rs10830963 and rs1387153 genotypes are listed in Tables 2 and 3. There were no significant differences in sex, age, BMI, waist to hip ratio (WHR), postprandial plasma glucose (PPG), HOMA-IR, triglyceride, total cholesterol, high-density lipoprotein-cholesterol (HDL-C), and low-density lipoprotein-cholesterol (LDL-C) between different genotype groups. However, patients with the GG genotype at MTNR1B rs10830963 had higher levels of FPG (P < 0.001) but lower levels of FINS (P = 0.022), postprandial serum insulin (PINS) (P = 0.003), and HOMA-B (P < 0.001) than those patients with genotypes CC and CG (Table 2, Figure S1). The MTNR1B rs1387153 polymorphism was markedly associated with levels of FPG and HbA1c; patients with the rs1387153 risk T allele had noticeably higher FPG (P = 0.013) and HbA1c (P = 0.005) levels compared to patients with the C allele (Table 3, Figure S2). In addition, the association of MTNR1B gene polymorphisms with baseline clinical characteristics was still significant when stratified by age and gender (Table S1).
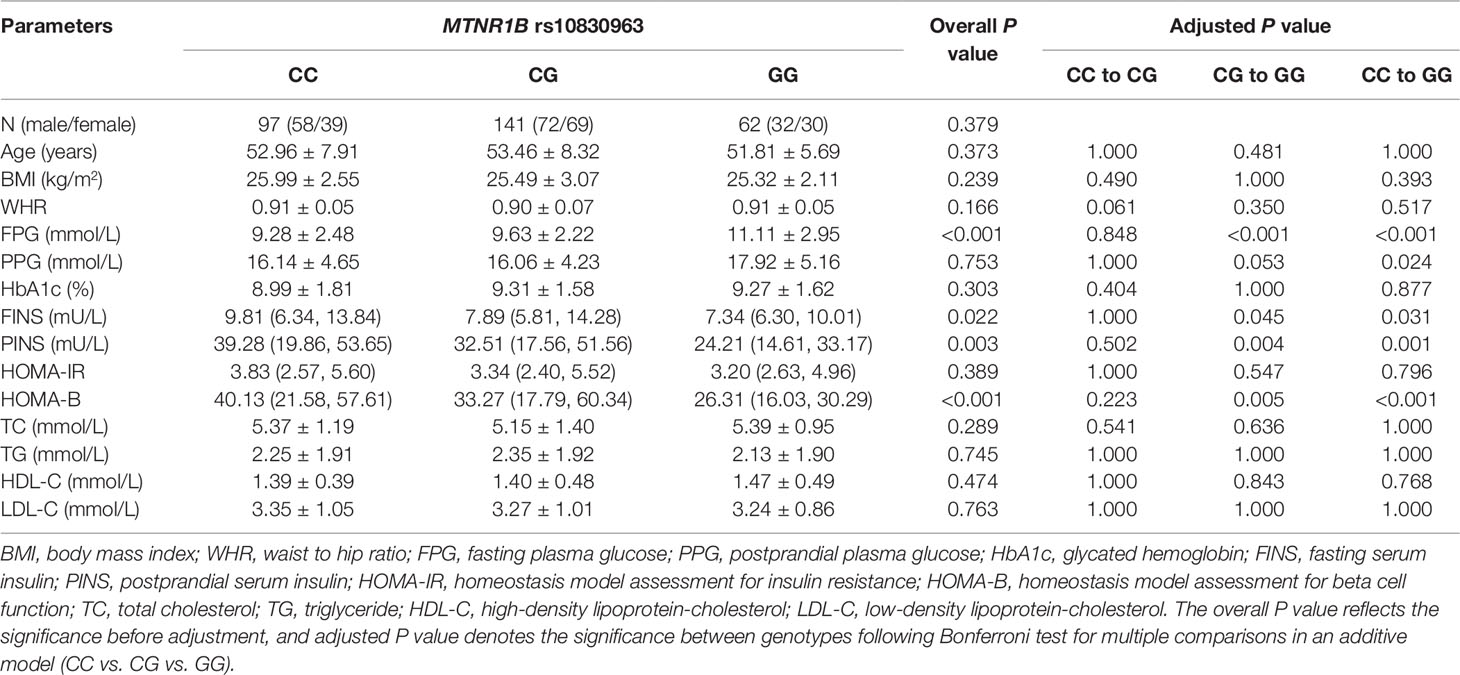
Table 2 Baseline characteristics of different genotypes of MTNR1B rs10830963 in 300 patients with type 2 diabetes mellitus (T2DM) before treatment with repaglinide.
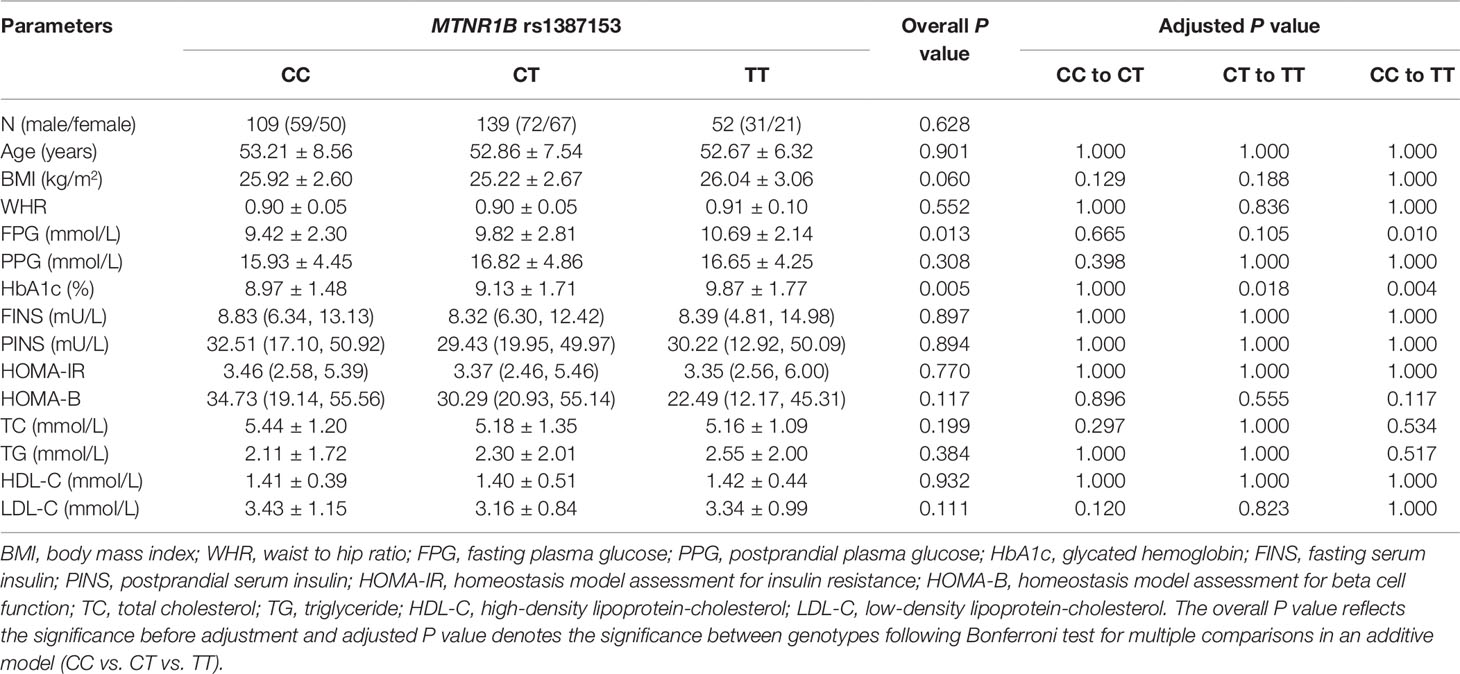
Table 3 Baseline characteristics of different genotypes of MTNR1B rs1387153 in 300 patients with type 2 diabetes mellitus (T2DM) before treatment with repaglinide.
We observed the effects of MTNR1B rs1387153 and rs10830963 on the therapeutic efficacy of repaglinide monotherapy in 95 patients with T2DM. After treatment with 3 mg repaglinide daily for 8 consecutive weeks, the levels of FPG, PPG, HbA1c, HOMA-IR, triglycerides, and total cholesterol decreased significantly, and the levels of FINS, PINS, and HOMA-B markedly increased in all patients with T2DM (Table S2). Our data also showed that patients with genotype CC at MTNR1B rs10830963 had better efficacy of repaglinide monotherapy with respect to FPG and HOMA-B than CG + GG genotype carriers (Table S3, Figure 1). Moreover, patients with MTNR1B rs1387153 CT + TT genotypes had attenuated efficacy of repaglinide monotherapy with respect to FPG, PINS, and HOMA-B compared with CC genotype carriers (Table S3, Figure 2). When the patients were stratified by age and gender, the association of MTNR1B gene polymorphisms with repaglinide monotherapy efficacy, as measured by FPG, PINS, and HOMA-B, did not vary significantly (Table S4).
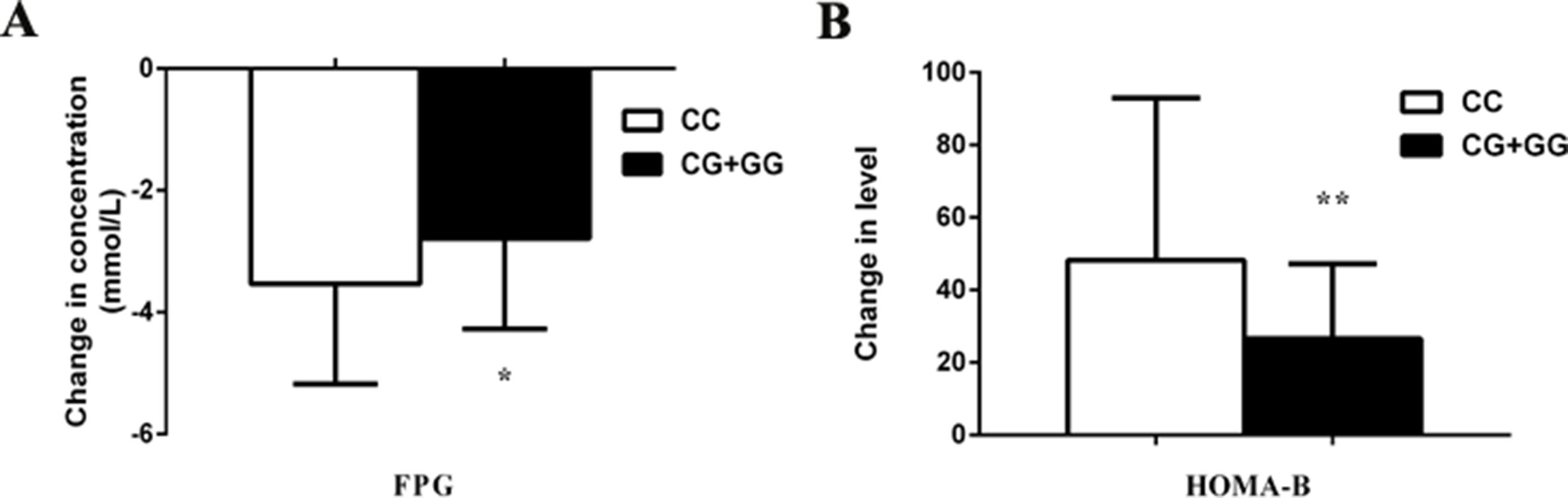
Figure 1 Comparisons of differential values (post-administration levels minus pre-administration levels) of fasting plasma glucose (FPG) (A) and homeostasis model assessment for beta cell function (HOMA-B) (B) between type 2 diabetes mellitus (T2DM) patients with the CC (n = 44) genotype and those with CG (n = 39) and GG (n = 12) genotypes of MTNR1B rs10830963 in T2DM patients after treatment with repaglinide. *P < 0.05 and **P < 0.01 compared with the CC genotype group (n = 95).
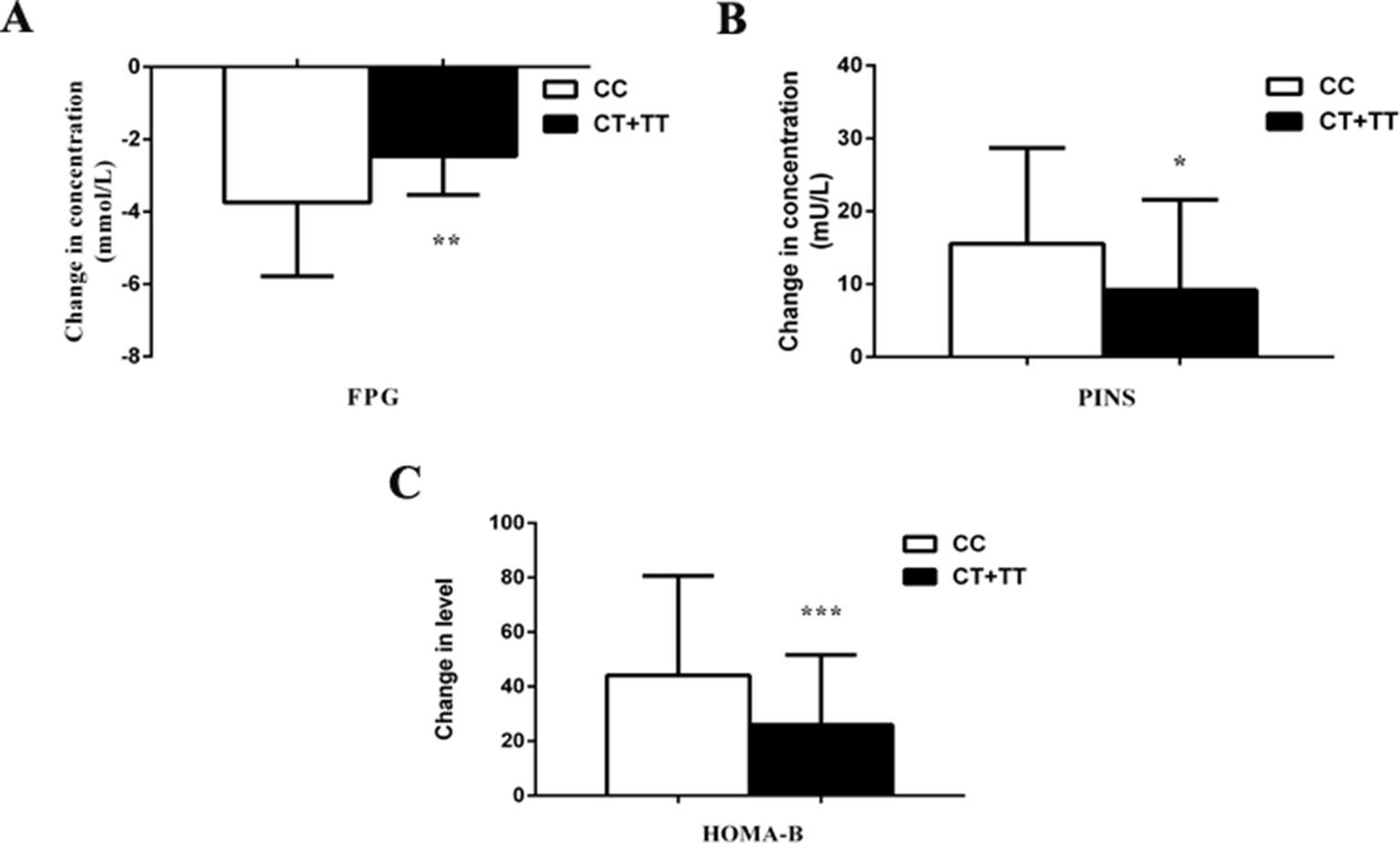
Figure 2 Comparisons of differential values (post-administration levels minus pre-administration levels) of fasting plasma glucose (FPG) (A), postprandial serum insulin (PINS) (B), and homeostasis model assessment for beta cell function (HOMA-B) (C) between type 2 diabetes mellitus (T2DM) patients with the CC (n = 47) genotype and those with CT (n = 36) and TT (n = 12) genotypes of MTNR1B rs1387153 after treatment with repaglinide. *P < 0.05, **P < 0.01, and ***P < 0.001 compared with the CC genotype group (n = 95).
To assess the association of the MTNR1B genetic polymorphisms with the response rate to repaglinide treatment in the present study, the genotype and allele frequencies are summarized according to the therapeutic responses in Table 4. According to predetermined criteria, the carriers of allele C at the MTNR1B rs1387153 locus exhibited good responses to repaglinide therapy; however, there were more responders among the CC allele homozygotes: approximately 70.21% of the CC homozygotes responded compared with only 44.44% of CT heterozygotes and 33.33% of the TT homozygotes (P = 0.016). No significant effects of the variation in MTNR1B rs10830963 on repaglinide treatment were detected. After adjusting for age, sex, BMI, WHR, FPG, PPG, HbA1c, and blood lipid at baseline, the logistic regression analysis indicated that the MTNR1B rs1387153 variant was the only covariate for the response rate to repaglinide treatment. Compared with CC homozygotes, the OR for treatment success was 0.277 for CT (95% CI: 0.088–0.865, P = 0.027) and 0.137 for TT (95% CI: 0.027–0.684, P = 0.015) genotype carriers.
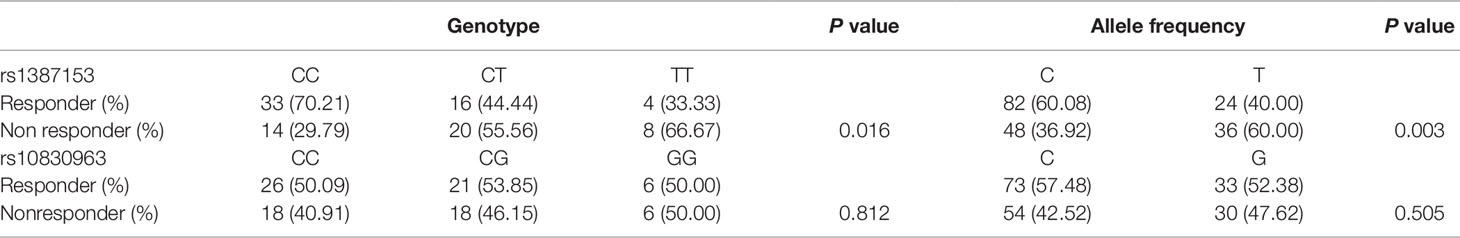
Table 4 Genotype and allele distributions between responders and nonresponders of MTNR1B rs1387153 and rs10830963 variants (n = 95).
In the present study, we investigated the potential effects of two SNPs (rs1387153 and rs10830963) near/in MTNR1B on the therapeutic efficacy of repaglinide in Chinese patients with T2DM. Our data demonstrated that patients with the G allele of MTNR1B rs10830963 or the T allele of rs1387153 may be less responsive to treatment with repaglinide, indicating that the MTNR1B genotype may serve as repaglinide response predictor. We therefore suggest that prior genotyping and identification of these individuals may be beneficial for T2DM patients who need treatment with repaglinide.
The common MTNR1B gene variant is associated with an increased risk of T2DM across populations of different ethnic backgrounds including Europeans, African-Americans, East Asians, and South Asians (Kim et al., 2011; Ramos et al., 2011). Our study on genetic variants of MTNR1B showed that the frequency of the risk G allele of rs10830963 (44.17%) was higher in patients with T2DM than in the control group (P < 0.01) and was higher than that in the African (2.70%), European (28.80%), East Asian (42.30%), South Asian (43.00%), and American (19.00%) populations (Auton and Abecasis, 2015). In contrast, rs1387153, which has been identified as a risk factor in European populations, did not show a significant association with T2DM in our subjects, but the frequency of the risk T allele of rs1387153 (40.50%) was lower than that in the East Asian (44.40%) populations and higher than that in the African (38.80%), European (28.70%), South Asian (37.00%), and American (21.00%) populations (Auton and Abecasis, 2015). Our data showed that MTNR1B rs10830963 and rs1387153 have dramatically different allele frequencies in Chinese and the other ethnic populations. The major reasons for this discrepancy may be the differences in specific ethnic groups and exposure to environmental factors. The data from this study also showed that patients carrying the rs10830963 risk G allele had higher levels of FPG but lower levels of FINS, PINS, and HOMA-B than those carrying the C allele (Table 2, Figure S1). In contrast, FPG and HbA1c values were higher in patients with the rs1387153 risk T allele compared to those with the C allele (Table 3, Figure S2). Robust associations have been identified between genetic variants of the MTNR1B gene and increased risk of T2DM along with a higher FPG level and reduced beta-cell function.
The fact that MTNR1B is directly or indirectly involved in glucose homeostasis and insulin secretion suggests that MTNR1B gene polymorphisms may contribute to interindividual differences in repaglinide response. No previous studies have proven that a patient with a specific MTNR1B gene variant shows a better or poorer response to repaglinide. To exclude the effects of OATP1B1 and CYP2C8 genetic polymorphisms on the response to repaglinide, 95 patients with different MTNR1B genotypes, but the same OATP1B1 521TT and CYP2C8*3 139Arg genotypes, were randomly selected from the 300 T2DM patients. In this study, we observed that subjects with at least one G allele of the MTNR1B rs10830963 showed a smaller decrease in FPG and a smaller increase in HOMA-B levels than those with the CC genotype, which suggested that the MTNR1B rs10830963 G allele confers the poor repaglinide response through a reduction of beta cell function, as measured by HOMA-B. Lyssenko et al. have confirmed the presence of MTNR1B in human pancreatic islets and an increased MTNR1B mRNA expression in carriers of the rs10830963 risk genotype, reporting a negative correlation between MTNR1B mRNA levels and insulin secretion (Lyssenko et al., 2008). Accordingly, overexpression of MTNR1B in clonal beta cells results in a stronger inhibition of insulin secretion (Lyssenko et al., 2008). Tuomi and colleagues found that mice with a global deletion of MTNR1B exhibit increased insulin release during an intravenous glucose tolerance test when compared to wild-type mice (Tuomi et al., 2016). The proposed mechanism by which MTNR1B rs10830963 polymorphism may influence the therapeutic efficacy of repaglinide involves altering the expression of MTNR1B in pancreatic beta cells leading to impaired insulin secretion and resulting in deteriorated beta cell function. However, rs10830963 is in the intronic region of MTNR1B, and the precise mechanism by which the SNP influences the therapeutic effects of repaglinide is not yet completely clear. Further detailed studies to elucidate the causal variant by fine mapping of the noncoding association signals are warranted.
Regarding rs1387153, which was located 28 kb upstream of the 5’ region of MTNR1B at chromosome 11q21-q22, Bouatia et al. confirmed that the rs1387153 T allele was associated with both increased FPG and risk of T2DM in Europeans (Bouatia-Naji et al., 2008). In the present study, the carriers of T alleles at rs1387153 exhibited a smaller decrease in FPG and smaller increases in PINS and HOMA-B levels compared to individuals with the CC genotype following 8 weeks of repaglinide treatment, potentially accounting for the lower failure rate of repaglinide therapy among the CC homozygotes. Further analysis of metabolic traits indicated that the T allele of rs1387153 had negative impacts on FPG and HOMA-B, suggesting that an increased FPG may result, at least in part, from a reduction of beta cell function. The global expression data available from human lymphoblastoid cell lines and human cortical gene expression data have not shown that rs1387153 directly influences MTNR1B expression (Dixon et al., 2007; Myers et al., 2007); instead, Lyssenko et al. found that individuals carrying the G allele showed higher expression of MTNR1B rs10830963 than carriers of the C allele in human pancreatic islets (Lyssenko et al., 2008). Furthermore, MTNR1B rs10830963 was in high LD with rs1387153 (r2 = 0.630, P < 0.05) in our study, as observed in previous studies (Prokopenko et al., 2008; Kan et al., 2010). Therefore, the association of rs1387153 variant with biochemical index and repaglinide efficacy might be involved in the observed LD with rs10830963. Ultimately, detailed functional analyses in pancreatic islets would be necessary to define the causal allele and to confirm that this effect is mediated through altered expression or function of MTNR1B.
In interpreting the results of our study, several limitations must be addressed. First, the sample size of included participants was relatively small, although consistent with that used in pilot studies. However, the statistical power of our study (77%–98%) may have been restricted by the sample size. Further studies with a larger sample size and comparisons at multiple time points within 8 weeks are required to confirm the effects of MTNR1B polymorphisms on the therapeutic efficacy of repaglinide. Second, the different locations of rs10830963 (in the intronic region of MTNR1B) and rs1387153 (in the 5′ region) have made functional studies to elucidate their effects more difficult. The mechanisms by which the two SNPs in MTNR1B affect the therapeutic efficacy of repaglinide are not fully understood. Third, incomplete data for healthy controls may have affected a comprehensive exploration of association of MTNR1B polymorphisms with clinical characteristics. Fourth, we focused only on the effect of MTNR1B gene variants on repaglinide efficacy. However, the possibility still exists that other susceptibility loci for T2DM may affect the therapeutic efficacy of repaglinide. Hence, we will evaluate the joint effects of multiple loci on the efficacy of repaglinide in future studies. Last, although individual differences are the product of the interactions between multiple genetic and environmental factors, it is naturally presumed that some, but not all, patients can benefit from taking certain drugs. We observed distinct responses to repaglinide in different patients with T2DM, which may result from genetic and environmental factors and perhaps other susceptible genetic variations involved in T2DM.
In conclusion, the MTNR1B polymorphisms appear to be associated with the therapeutic response to repaglinide in Chinese patients who are newly diagnosed with T2DM. Therefore, the MTNR1B risk variants may serve as repaglinide response predictors based on MTNR1B mediated beta cell dysfunction. Further pharmacogenetic and functional studies to elucidate the exact effects of MTNR1B variants on repaglinide therapeutic efficacy are necessary to lay the foundation for tailoring of a more precise therapy for T2DM.
All datasets generated for this study are included in the article/Supplementary Material.
This study was carried out in accordance with the recommendations of the Ethics Committee of the Affiliated Hospital of Xuzhou Medical University with written informed consent from all subjects. All subjects gave written informed consent in accordance with the Declaration of Helsinki. The protocol was approved by the Ethics Committee of the Affiliated Hospital of Xuzhou Medical University.
TW contributed to conception of the article, data acquisition, statistical analysis, result interpretation, manuscript drafting and approved the final version. X-XY contributed to conception of the article, results interpretation, manuscript drafting and approved the final version. X-TW, RL, and H-WL contributed to conception of the article, data acquisition, and approved the final version. FZ contributed to statistical analysis and approved the final version. QL and D-ML contributed to conception of the article, manuscript revising, and approved the final version.
This work was supported by the Project Funded by Xuzhou Science and Technology Bureau (KC15SH005).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The authors are very grateful to all the volunteers and patients for their participation in this study. Our thanks also to the physicians of the Department of Endocrinology in the Affiliated Hospital of Xuzhou Medical University for their support.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fphar.2019.01318/full#supplementary-material
Figure S1 | Baseline levels of FPG, FINS, PINS, and HOMA-B in T2DM patients with CC (n = 97), CG (n = 141) and GG (n = 62) genotypes of MTNR1B rs10830963. *, P< 0.05 compared with the CG and CC genotype groups (n = 300).
Figure S2 | Baseline levels of FPG and HbA1c in T2DM patients with CC (n = 109), CT (n = 139) or TT (n = 52) genotypes of MTNR1B rs1387153. *, P< 0.05 compared with the CT and CC genotype groups (n = 300).
Abbink, E. J., van der Wal, P. S., Sweep, C. G. J., Smits, P., Tack, C. J. (2004). Compared to glibenclamide, repaglinide treatment results in a more rapid fall in glucose level and beta-cell secretion after glucose stimulation. Diabetes/Metab. Res. Rev. 20 (6), 466–471. doi: 10.1002/dmrr.474
Auton, A., Abecasis, G. R. (2015). A global reference for human genetic variation. Nature 526 (7571), 68–74. doi: 10.1038/nature15393
Bouatia-Naji, N., Bonnefond, A., Cavalcanti-Proença, C., Sparsø, T., Holmkvist, J., Marchand, M., et al. (2008). A variant near MTNR1B is associated with increased fasting plasma glucose levels and type 2 diabetes risk. Nat. Genet. 41 (1), 89–94. doi: 10.1038/ng.277
Chakraborty, C., Hsu, M. J., Agoramoorthy, G. (2015).Drug metabolizing enzymes in type II diabetes and their pharmacogenetics during therapy of anti-diabetes drugs. Curr. Drug Metab. 16 (10), 864–876. doi: 10.2174/138920021610151210183821
Chen, M., Hu, C., Zhang, R., Jiang, F., Wang, J., Peng, D., et al. (2015). A variant of PSMD6 is associated with the therapeutic efficacy of oral antidiabetic drugs in Chinese type 2 diabetes patients. Sci. Rep. 5 (1), 1–7. doi: 10.1038/srep10701
Chen, M., Zhang, R., Jiang, F., Wang, J., Peng, D., Yan, J., et al. (2016). Joint effects of diabetic-related genomic loci on the therapeutic efficacy of oral anti-diabetic drugs in Chinese type 2 diabetes patients. Sci. Rep. 6 (1), 1–7. doi: 10.1038/srep23266
Dixon, A. L., Liang, L., Moffatt, M. F., Chen, W., Heath, S., Wong, K. C. C., et al. (2007). A genome-wide association study of global gene expression. Nat. Genet. 39 (10), 1202–1207. doi: 10.1038/ng2109
Eichelbaum, M., Ingelman-Sundberg, M., Evans, W. E. (2006). Pharmacogenomics and individualized drug therapy. Annu. Rev. Med. 57 (1), 119–137. doi: 10.1146/annurev.med.56.082103.104724
Fang, F.-S., Gong, Y.-P., Li, C.-L., Li, J., Tian, H., Huang, W., et al. (2014). Comparison of repaglinide and metformin monotherapy as an initial therapy in Chinese patients with newly diagnosed type 2 diabetes mellitus. Eur. J. Endocrinol. 170 (6), 901–908. doi: 10.1530/eje-14-0052
Gabir, M., Hanson, R., Dabelea, D., Imperatore, G., Roumain, J., Bennett, P., et al. (2000). The 1997 American Diabetes Association and 1999 World Health Organization criteria for hyperglycemia in the diagnosis and prediction of diabetes. Diabetes Care 23 (8), 1108–1112. doi: 10.2337/diacare.23.8.1113
Jia, Y., Lao, Y., Zhu, H., Li, N., Leung, S. W. (2018). Is metformin still the most efficacious first-line oral hypoglycaemic drug in treating type 2 diabetes? A network meta-analysis of randomized controlled trials. Obesity Rev. 20 (1), 1–12. doi: 10.1111/obr.12753
Kan, M. Y., Zhou, D. Z., Zhang, D., Zhang, Z., Chen, Z., Yang, Y. F., et al. (2010). Two susceptible diabetogenic variants near/inMTNR1Bare associated with fasting plasma glucose in a Han Chinese cohort. Diabetic Med. 27 (5), 598–602. doi: 10.1111/j.1464-5491.2010.02975.x
Karamitri, A., Jockers, R. (2019). Melatonin in type 2 diabetes mellitus and obesity. Nat. Rev. Endocrinol. 15 (2), 105–125. doi: 10.1038/s41574-018-0130-1
Kim, Y. J., Go, M. J., Hu, C., Hong, C. B., Kim, Y. K., Lee, J. Y., et al. (2011). Large-scale genome-wide association studies in east Asians identify new genetic loci influencing metabolic traits. Nat. Genet. 43 (10), 990–995. doi: 10.1038/ng.939
Kong, X., Xing, X., Hong, J., Zhang, X., Yang, W. (2017). Association of a type 2 diabetes genetic risk score with insulin secretion modulated by insulin sensitivity among Chinese Hans. Clin. Genet. 91 (6), 832–842. doi: 10.1111/cge.12817
Lyssenko, V., Nagorny, C. L. F., Erdos, M. R., Wierup, N., Jonsson, A., Spégel, P., et al. (2008). Common variant in MTNR1B associated with increased risk of type 2 diabetes and impaired early insulin secretion. Nat. Genet. 41 (1), 82–88. doi: 10.1038/ng.288
Mannino, G. C., Andreozzi, F., Sesti, G. (2019). Pharmacogenetics of type 2 diabetes mellitus, the route toward tailored medicine. Diabetes/Metab. Res. Rev. 35 (3), e3109. doi: 10.1002/dmrr.3109
Maruthur, N. M., Gribble., M. O., Bennett., W. L., Bolen., S., Wilson, L. M., Balakrishnan, P., et al. (2014). The Pharmacogenetics of Type 2 diabetes: a systematic review. Diabetes Care 37 (3), 876–886. doi: 10.2337/dc13-1276/-/DC1
Myers, A. J., Gibbs, J. R., Webster, J. A., Rohrer, K., Zhao, A., Marlowe, L., et al. (2007). A survey of genetic human cortical gene expression. Nat. Genet. 39 (12), 1494–1499. doi: 10.1038/ng.2007.16
Pal, A., McCarthy, M. (2013). The genetics of type 2 diabetes and its clinical relevance. Clin. Genet. 83 (4), 297–306. doi: 10.1111/cge.12055
Patel, R., Rathwa, N., Palit, S. P., Ramachandran, A. V., Begum, R. (2018). Association of melatonin & MTNR1B variants with type 2 diabetes in Gujarat population. Biomed. Pharmacother. 103 (2018), 429–434. doi: 10.1016/j.biopha.2018.04.058
Persaud, S. J., Phimister, E. G., Jones, P. M. (2016). A wake-up call for type 2 diabetes? New Engl. J. Med. 375 (11), 1090–1092. doi: 10.1056/NEJMcibr1607950
Prokopenko, I., Langenberg, C., Florez, J. C., Saxena, R., Soranzo, N., Thorleifsson, G., et al. (2008). Variants in MTNR1B influence fasting glucose levels. Nat. Genet. 41 (1), 77–81. doi: 10.1038/ng.290
Rönn, T., Wen, J., Yang, Z., Lu, B., Du, Y., Groop, L., et al. (2009). A common variant in MTNR1B, encoding melatonin receptor 1B, is associated with type 2 diabetes and fasting plasma glucose in Han Chinese individuals. Diabetologia 52 (5), 830–833. doi: 10.1007/s00125-009-1297-8
Ramos., E., Chen., G., Shriner., D., Doumatey., A., Gerry., N. P., Herbert., A., et al. (2011). Replication of genome-wide association studies (GWAS) loci for fasting plasma glucose in African-Americans. Diabetologia 54 (4), 783–788. doi: 10.1007/s00125-010-2002-7
Sanchez-Ibarra, H. E., Reyes-Cortes, L. M., Jiang, X.-L., Luna-Aguirre, C. M., Aguirre-Trevino, D., Morales-Alvarado, I. A., et al. (2018). Genotypic and phenotypic factors influencing drug response in mexican patients with type 2 diabetes mellitus. Front. Pharmacol. 9, 320. doi: 10.3389/fphar.2018.00320
Scott, L. J. (2012). Repaglinide: a review of its use in type 2 diabetes mellitus. Drugs 72 (2), 249–272. doi: 10.2165/11207600-000000000-00000
Tomalik-Scharte, D., Fuhr, U., Hellmich, M., Frank, D., Doroshyenko, O., Jetter, A., et al. (2011). Effect of the CYP2C8 genotype on the pharmacokinetics and pharmacodynamics of repaglinide. Drug Metab. Disposition 39 (5), 927–932. doi: 10.1124/dmd.110.036921
Tuomi, T., Nagorny, C. L. F., Singh, P., Bennet, H., Yu, Q., Alenkvist, I., et al. (2016). Increased melatonin signaling is a risk factor for Type 2 Diabetes. Cell Metab. 23 (6), 1067–1077. doi: 10.1016/j.cmet.2016.04.009
Wang, L.-C., Fang, F.-S., Gong, Y.-P., Yang, G., Li, C.-L. (2018). Characteristics of repaglinide and its mechanism of action on insulin secretion in patients with newly diagnosed type-2 diabetes mellitus. Medicine 97 (38), e12476. doi: 10.1097/md.0000000000012476
Keywords: MTNR1B, rs10830963, rs1387153, genetic polymorphism, type 2 diabetes mellitus, repaglinide, pharmacogenetics
Citation: Wang T, Wang X-t, Lai R, Ling H-w, Zhang F, Lu Q, Lv D-m and Yin X-x (2019) MTNR1B Gene Polymorphisms Are Associated With the Therapeutic Responses to Repaglinide in Chinese Patients With Type 2 Diabetes Mellitus. Front. Pharmacol. 10:1318. doi: 10.3389/fphar.2019.01318
Received: 29 April 2019; Accepted: 15 October 2019;
Published: 07 November 2019.
Edited by:
Amit V. Pandey, University of Bern, SwitzerlandReviewed by:
Wei Zhang, Central South University, ChinaCopyright © 2019 Wang, Wang, Lai, Ling, Zhang, Lu, Lv and Yin. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xiao-xing Yin, eWlueHhAeHptYy5lZHUuY24=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.