
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Pharmacol., 31 May 2019
Sec. Drug Metabolism and Transport
Volume 10 - 2019 | https://doi.org/10.3389/fphar.2019.00591
Cytochrome P450 3A4 (CYP3A4) enzyme activity is known to show considerable ethnic heterogeneity and inter-individual differences, affecting the outcome of drug treatment. CYP3A4 genetic polymorphisms are believed to be one of the important causes, leading to inter-individual variability in drug metabolism. Quinine is an antipyretic drug with antimalarial properties that is metabolized primarily by CYP3A4. Quinine 3-hydroxylation has been proven as a biomarker reaction for evaluating CYP3A4 ability. Quinine has frequent adverse effects and there are distinct inter-individual differences in quinine sensitivity. The open reading frame for 30 CYP3A4 allelic variants were constructed from wild-type CYP3A4*1A by an overlap extension polymerase chain reaction. Recombinant CYP3A4 variants were expressed using baculovirus-insect cell expression system, and their catalytic activities towards quinine hydroxylation were determined and evaluated. Of the 30 CYP3A4 allelic variants, 23 variants exhibited significantly reduced intrinsic clearance towards quinine, 2 variants showed increased intrinsic clearance for quinine, 2 variants possessed no significant differences towards quinine, compared with CYP3A4*1A, and 3 variants had no detected expression and enzyme activity. Our assessment on the enzymatic activities of CYP3A4 variants towards quinine may contribute to laying an experimental foundation for further clinical studies so as to accelerate the process of determining the associations between genetic variations and clinical phenotypes.
Cytochrome P450 3A4 (CYP3A4) contributes to the metabolism of ∼50% of drugs used therapeutically (Rendic, 2002; Guengerich, 2005) and also to the metabolism of endogenous substrates, such as retinoic acid, bile acids, and steroid hormones (Ingelman-Sundberg et al., 2007). It has been reported that CYP3A4 expression differs between people, with 43.4-fold differences in expression observed in human liver microsomes from different individuals (Ohtsuki et al., 2012). Further, CYP3A4 enzyme activity shows considerable ethnic heterogeneity and inter-individual differences, which have a major impact on the outcome of drug treatment (Zanger and Schwab, 2013). Inter-individual variability in CYP3A4 activity is due to environmental factors (e.g., environmental exposures, drug interactions), as well as genetic variation in the CYP3A4 gene, and results in variability in drug efficacy and adverse events (Ma and Lu, 2011; Rahmioglu et al., 2011). Evaluation of CYP3A4 activity using probe drugs is important to elucidate the impact of genetic variation on inter-individual variability in CYP3A4 activity, relative to environmental factors.
Quinine, a component of the bark of the Cinchona tree, has been used as an important drug for the treatment of malaria for several hundred years (Achan et al., 2011). Quinine is recommended as the treatment for uncomplicated malaria, particularly in the first trimester of pregnancy (W. H. Organization, 2015), or is used to treat severe malaria if the first-line drug fails or is not available (W. H. Organization, 2015). Further, quinine is used as a flavor modifier in beverages such as bitter lemon and tonic water (Horie et al., 2006; Bel et al., 2009), and is an ingredient of some lotions and shampoos. Quinine is eliminated over 80% via hepatic biotransformation and is excreted unchanged about 20% by the kidneys. Zhang et al. (1997) reported that the formation of the major metabolite of quinine, 3-hydroxyquinine, was highly correlated with CYP3A4 apoprotein levels as determined by western blotting. In vivo CYP3A4 has also been proven to be important for the 3-hydroxylation of quinine (Mirghani et al., 1999; Mirghani et al., 2003). Quinine has been used as a reliable probe substrate of CYP3A4 for association studies between enzymatic activities and genotypes because CYP3A4 is the major metabolic enzyme involved in quinine 3-hydroxylation (Mirghani et al., 2002; Mirghani et al., 2003; Rodriguez-Antona et al., 2005) both in vivo and in vitro.
So far, 30 CYP3A4 variants, with amino-acid changes located in coding regions, have been identified among different ethnic population (www.pharmvar.org/gene/CYP3A4), including seven new variants, CYP3A4*28–*34, detected in our recent study (Hu et al., 2017). Along with the new allelic variants added, the demand for functional studies of CYP3A4 variants has been increasing. In view of substrate specificity for a genetic variant, our group tested various CYP3A4 substrates, including testosterone, lidocaine (Fang et al., 2017), and amiodarone (Yang et al., 2019), with these variants. In the present study, we attempted to evaluate the enzymatic activities of 31 CYP3A4 alleles towards quinine in vitro using high expression of CYP3A4 holoenzymes in insect cells.
Quinine, 3-hydroxyquinine (3-OH quinine), and carbamazepine (internal standard [IS]) were purchased from Toronto Research Chemicals (Toronto, ON, Canada). Cells from the fall armyworm (Spodoptera frugiperda [Sf]21), Sf-900™ III SFM insect culture medium, fetal bovine serum, Cellfectin® II Reagent, and the Bac-to-Bac™ baculovirus cell expression system were obtained from Invitrogen (Carlsbad, CA, USA). PrimeSTAR® HS DNA polymerase, restriction enzymes, and a DNA ligation kit were purchased from TaKaRa Bio (Shiga, Japan). A human cDNA clone for oxidoreductase (OR) (catalog number SC100401) and a wild-type CYP3A4 cDNA clone (catalog number SC125488) were obtained from Origene (Rockville, MD, USA). Mouse monoclonal anti-OR antibody, anti-CYP3A4 antibody, and anti-CYP3 antibody were from Santa Cruz Biotechnology (sc-25270, sc-53850, and sc-365415; Santa Cruz, CA, USA). Rabbit monoclonal anti-CYP3A4 antibody was purchased from Abcam (ab 3572; Cambridge, UK). A regenerating system for the reduced form of nicotinamide adenine dinucleotide phosphate (NADPH) was obtained from Promega (Madison, WI, USA). High-performance liquid chromatography-grade organic solvents and liquid chromatography–mass spectrometry-grade acetonitrile were purchased from Merck (Darmstadt, Germany). All other chemicals and reagents used were of the highest purity available.
The dual-expression baculovirus plasmid pFastBac™ Dual was used to express the CYP3A4 and OR proteins in Sf21 cells simultaneously. First, the open reading frame (ORF) of OR was obtained and inserted into cloning sites downstream to the P10 promoter in the baculovirus vector pFastBac Dual to create the intermediate plasmid pFastBac-OR. Second, ORF fragment of CYP3A4 *1A was isolated from wild-type CYP3A4 cDNA (catalog number SC125488) vector using 3A4*1A-F (5′-ACCA
Recombinant baculoviruses expressing CYP3A4 protein and OR protein were obtained from Sf21 cells according to manufacturer instructions. Then, Sf21 cells were infected with these viruses and cultivated in Sf-900 II SFM insect culture medium containing 10% fetal bovine serum, 1× penicillin (100 U/ml)–streptomycin (0.1 mg/ml), and 4 ng/μl hemin at 27°C. After 4 days of transfection, cells were harvested and resuspended in 100 mM KPO4 containing 1 mM ethylenediamine tetra-acetic acid, 1 mM phenylmethane sulfonyl fluoride, and 0.25 M sucrose. After centrifugation at 1,600 × g for 5 min, pellets were resuspended and sonicated for 30 s on ice at 25% of its full power (750 W) using a Vibracell™ sonicator (Sonics & Materials, Newtown, CT, USA). Then, the homogenate was centrifuged at 13,000 × g for 20 min at 4°C to remove the precipitate. Finally, the microsomal fraction was collected by ultracentrifugation at 100,000 × g for 1 h at 4°C and the pellet resuspended in 100 mM KPO4 (pH = 7.4) containing 20% glycerol for storage at −80°C.
Expression of CYP3A4 apoenzyme and OR protein was determined by Western blotting according to standard procedures. Microsomal proteins were quantified using a bicinchoninic acid protein assay kit according to manufacturer (Pierce, Rockford, IL, USA) instructions. Then, 2 μg of microsomal protein was separated by electrophoresis on 10% sodium dodecyl sulfate-polyacrylamide gels, and transferred onto polyvinylidene fluoride (PVDF) membranes. The CYP3A4 apoprotein and OR protein were detected using a rabbit anti-CYP3A4 monoclonal antibody (1:2,000 dilution) and mouse anti-OR monoclonal antibody (1:8,000 dilution) as the primary antibody, respectively. After incubation overnight at 4°C, PVDF membranes were washed four times (5 min each time). Horseradish peroxidase-conjugated goat anti-rabbit and goat anti-mouse immunoglobulin (Ig) G (1:5,000 dilution) were used as secondary antibodies. Signals were visualized with a SuperSignal™ West Pico Trial Kit (Pierce).
Levels of recombinant CYP3A4 holoenzymes were evaluated using reduced carbon monoxide difference spectroscopy (RCODS) (Omura and Sato, 1964) employing an Evolution 201 system (Thermo Scientific, Rockford, IL, USA). Microsomal preparations suspended in 0.1 M potassium phosphate (pH = 7.0) were divided equally into a sample cuvette and reference cuvette, and a baseline determined. After recording the baseline, carbon monoxide was bubbled carefully through the sample for ∼40 s in the sample cuvette. Then, a few milligrams of solid Na2S2O4 were used for the chemical reduction of samples in the sample cuvette. Accordingly, the spectral difference in microsomal preparations was recorded by ultraviolet–visible absorption spectrometry. The change in absorbance at 450 nm relative to that at 490 nm was converted to a concentration of cytochrome P450 by using a millimolar difference extinction coefficient of 91 (Omura and Sato, 1964; Estabrook and Werringloer, 1978).
The hydroxylation of quinine by recombinant CYP3A4 variants derived from Sf21 cells was assayed according to a specific method. According to previous experiences (Lee et al., 1995; Dai et al., 2013; Zhou et al., 2018), we performed optimization of reaction conditions including amount of enzyme, incubation time, and reaction buffer, etc. The conditions were finally controlled and selected under the linear range of the enzymatic reaction. Briefly, the incubation mixture comprised of 100 mM phosphate buffer (pH = 7.4), purified cytochrome b5, 0.7–1.2 pmol of a recombinant CYP3A4 microsomal fraction obtained from Sf21 cells (CYP3A4:b5 = 1:1), and quinine (25–1000 μM). This incubation mixture was pre-incubated at 37°C for 5 min. Then, 50 µl of 4 mM NADPH was added to start the reaction in a final volume of 200 μl. The reaction was carried out at 37°C for 45 min with gentle shaking and terminated by cooling to −80°C for 15 min. Finally, the mixture was precipitated with 400 μl acetonitrile and 30 μl carbamazepine (50 μg/ml, IS). The supernatant was diluted 1:9 with water and used for subsequent measurements after centrifugation at 13,000 × g for 10 min at 4°C. Data are the mean ± SD of three independent experiments.
The levels of 3-OH quinine and carbamazepine (IS) were measured using ultra-high-performance liquid chromatography–tandem mass spectrometry (UPLC-MS/MS). Detection was carried out with a 1290 LC system coupled to a 6490 triple quadrupole mass spectrometer (Agilent Technologies, Santa Clara, CA, USA), with a 1.8 µm Rapid Resolution HT C18 column (3.0 × 100 mm, Agilent Technologies). The mobile phase consisted of water (A) and methanol (B). The gradient elution program was 0–2.00 min (90–10% A), 2.00–4.40 min (10% A), 4.40–4.41 min (10–90% A), and 4.41–6.00 min (90% A). The flow rate was maintained at 0.4 ml/min and the injection volume was 3 µl. The total run time was 6 min. Target compounds were detected using positive electrospray ionization with multiple reaction monitoring transitions of mass/charge ratio (m/z) 341.1–160.1 for 3-OH quinine and m/z 237.1–194.1 for carbamazepine (IS). Under the conditions mentioned above, 3-OH quinine and carbamazepine (IS) separated well and their retention times were 3.47 and 3.19 min, respectively. The capillary voltage was set to 2.0 kV in positive mode and the nebulizer pressure was set to 30 psi. The fragment voltage and collision energy were set at 380 V and 35 eV for 3-OH quinine, and 380 V and 20 eV for carbamazepine, respectively. The gas temperature was set to 200°C at a flow rate of 16 L/min.
The 3-hydroxyquinine formation rate was calculated as follows: the amount of 3-hydroxyquinine formed/incubation time/amount of CYP3A4 variant used (units: pmol/min per pmol of CYP3A4 enzyme).
Kinetic parameters were estimated using Prism 5 (GraphPad, San Diego, CA, USA) based on non-linear regression analyses. All values of enzyme parameters, Michaelis–Menten constant (apparent Km), maximum reaction velocity (Vmax), and intrinsic clearance (CLint, which is Vmax/apparent Km), are presented as the mean ± SD from three separate experiments. Comparisons of enzymatic activity between the wild-type and variant were performed by one-way analysis of variance using the post hoc Dunnett method. p < 0.05 was considered to represent statistical significance.
It is known that Baculovirus host cells are deficient in all electron-transport components required for human CYP expression (Gonzalez and Korzekwa, 1995). As shown in previous studies, coexpression of CYP3A4 and OR proteins could make increase of the catalytic activity of recombinant CYP3A4 microsome, catalytically similar to human liver microsomal CYP3A4 (Buters et al., 1994; Lee et al., 1995). In the current study, the dual-expression baculovirus vectors p-FastBac-OR-CYP3A4 were constructed. Consequently, recombinant human CYP3A4 and OR proteins were highly co-expressed in the microsomes of Sf21 cells to assess the enzymatic activities of CYP3A4 variants towards quinine 3-hydroxylation in vitro.
Expression of recombinant CYP3A4 apoenzymes was measured by immunoblotting (Figure 1A). The results of the relative intensity of CYP3A4/OR indicated that CYP3A4 protein expression levels were similar across most variants (Figure 1B). Compared with wild-type CYP3A4*1A, several recombinant CYP3A4 variants exhibited a remarkable variation in expression. Apoprotein expression of five variants, CYP3A4*11 (T363M), CYP3A4*12 (L373F), CYP3A4*13 (P416L), CYP3A4*20 (488 frameshift), and CYP3A4*23 (R162W), was particularly lower than that of wild-type CYP3A4*1A. Conversely, one variant, CYP3A4*18 (L293P), had obviously higher expression than CYP3A4*1A. CYP3A4 apoproteins were not detected in Sf21 cells transfected with the allelic variants CYP3A4*6 (277 frameshift), CYP3A4*21 (Y319C), CYP3A4*26 (R268STOP), or CYP3A4*30 (R130STOP).
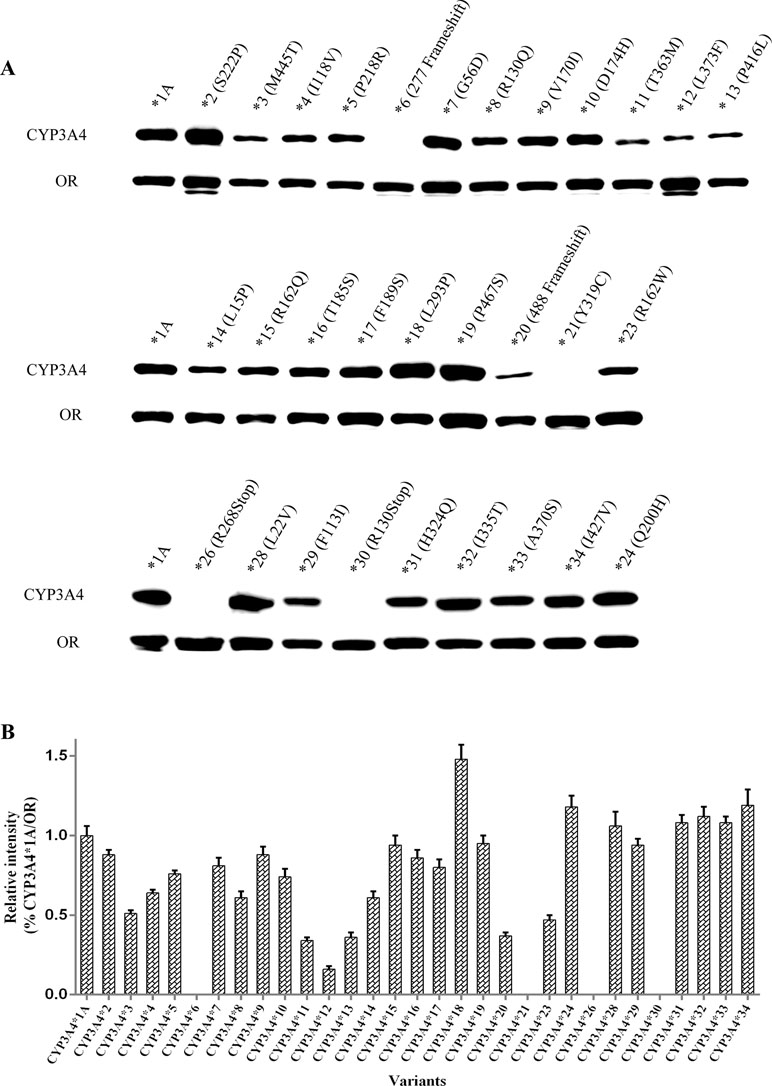
Figure 1 Expression of 31 CYP3A4 alleles in Sf21 cells. The expressed OR and CYP3A4 proteins were detected by western blotting with antibodies against OR and CYP3A4, respectively (A). Relative intensities (% of CYP3A4*1A/OR) (B). Each bar represents the mean ± SD from three experiments.
Recombinant CYP3A4 holoenzymes were quantified using RCODS. CYP3A4 holoenzyme was not detected for three CYP3A4 variants: CYP3A4*6 (277 frameshift), CYP3A4*26 (R268STOP), and CYP3A4*30 (R130STOP). For the other CYP3A4 variants, the concentrations of recombinant CYP3A4 holoenzymes ranged from 0.07 pmol/μl to 1.06 pmol/μl.
The enzymatic properties of 31 CYP3A4 holoenzymes towards quinine were identified and enzymatic kinetic parameters are shown in Table 2 and Figure 2. The apparent Km, Vmax, and CLint for the 3-hydroxylation of quinine by wild-type CYP3A4*1A was 16 μM, 3.5 pmol/min per pmol CYP3A4, and 217.5 μl/min per nmol CYP3A4, respectively. Most CYP3A4 variants exhibited significantly reduced Vmax and increased apparent Km. Consequently, most of the recombinant CYP3A4 holoenzymes exhibited decreased CLint towards quinine, especially for the variants CYP3A4*8 (R130Q), CYP3A4*12 (L373F), CYP3A4*13 (P416L), CYP3A4*17 (F189S), CYP3A4*20 (488 frameshift), and CYP3A4*21 (Y319C), which exhibited a reduction in CLint of quinine of >90% compared with that of wild-type CYP3A4*1A. However, CYP3A4*15 (R162Q), and CYP3A4*29 (F113I) proteins displayed increased CLint, with higher Vmax and similar apparent Km, than those of wild-type CYP3A4*1A protein. In contrast, CYP3A4*28 (L22V) and CYP3A4*34 (I427V) had comparable CLint to the wild-type CYP3A4 enzyme. In addition, three recombinant microsomes, CYP3A4*6 (277 frameshift), CYP3A4*26 (R268STOP), and CYP3A4*30 (R130STOP), exhibited no detectable enzymatic activity towards quinine hydroxylation, which was probably because active holoenzymes derived from these recombinant variants could not be detected (Table 2).
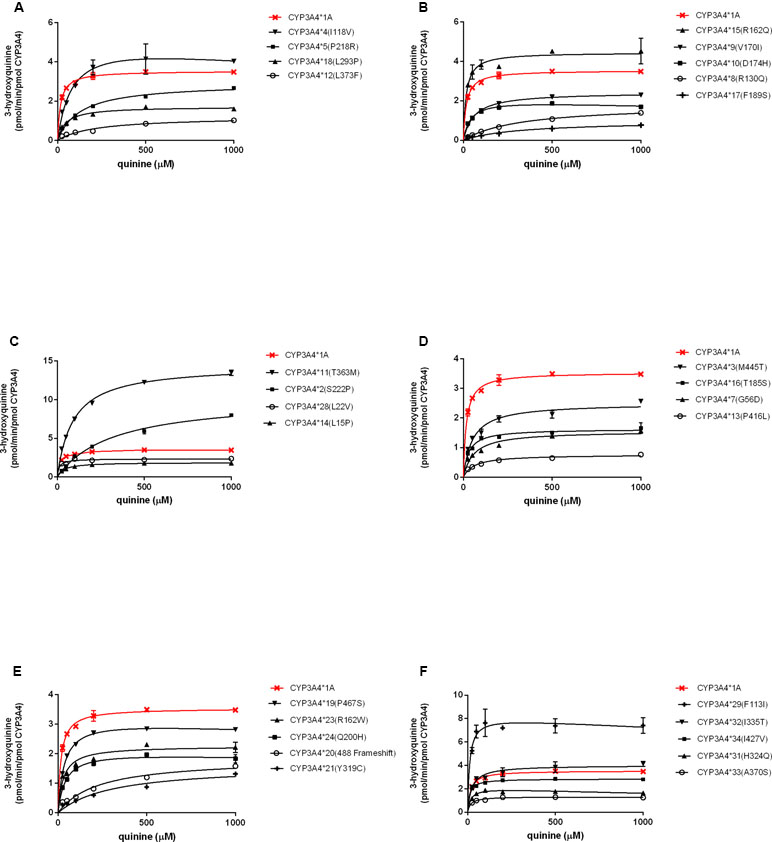
Figure 2 Michaelis–Menten kinetics for quinine hydroxylation by recombinant CYP3A4 variants. Each point is the mean ± SD from three experiments (A–F).
Paradoxically, CYP3A4*21 (Y319C) showed detectable microsomal holoenzyme by RCODS and exhibited extremely low Vmax and high apparent Km towards quinine, with relative clearance of 2.44%; however, no detectable apoenzyme expression was found by Western blotting (Figure 1A, Table 2). We also used other commercial antibodies (sc-53850 & sc-365415, Santa Cruz, USA) to detect the expression of CYP3A4*21 but no detectable expression was identified either (results were not shown).
It is important to relate gene variations to clinical phenotypes using proper phenotyping tools of CYP3A4 activity. The 3-hydroxylation of quinine had been proven as a biomarker reaction for CYP3A4 activity because 3-hydroxylation of quinine is catalyzed by CYP3A4 both in vivo and in in vitro (Mirghani et al., 2003; Rahmioglu et al., 2011). Therefore, the kinetic parameters of enzymatic catalysis on quinine 3-hydroxylation were used as a measure of CYP3A4 activity in the present study.
Twenty-three of the recombinant CYP3A4 holoenzymes showed significantly decreased CLint towards quinine. Six variants, CYP3A4*8 (R130Q), CYP3A4*12 (L373F), CYP3A4*13 (P416L), CYP3A4*17 (F189S), CYP3A4*20 (488 frameshift), and CYP3A4*21 (Y319C), exhibited much lower CLint for quinine (1.44–5.80%, Table 2) than wild-type CYP3A4*1A. If our in vitro findings are replicated in clinical studies, patients that are carriers of these alleles may be potentially CYP3A4 poor-metabolizers and may: i) require lower quinine doses to reach therapeutic plasma concentrations; and ii) be at increased risk for adverse effects and toxicity when conventional doses of quinine are used.
CYP3A4*2 (S222P) exhibited a reduced CLint of more than sixfold (14.76%, Table 2) for quinine compared with wild-type CYP3A4*1A. This decreased enzymatic activity might be because a serine→proline amino-acid substitution could cause a major change in the three-dimensional structure of the enzyme (proline is known to break alpha-helices). Likewise, CYP3A4*2 has been reported to possess a lower CLint for midazolam (Miyazaki et al., 2008), nifedipine (Sata et al., 2000; Miyazaki et al., 2008), ibrutinib (Xu et al., 2018), and lidocaine (Fang et al., 2017), than that of the wild-type enzyme. In contrast, CYP3A4*2 showed a higher CLint for amiodarone (Yang et al., 2019). For testosterone, CYP3A4*2 had a decreased intrinsic clearance when expressed using Escherichia coli express system in vitro (Miyazaki et al., 2008), but the activity was comparable to the wild-type enzyme when expressed in baculovirus expression system (Sata et al., 2000) (Table 3).
CYP3A4*3 (M445T) displayed lower enzymatic activity towards quinine, compared with the wild-type allele (Table 2). The amino-acid residue 445 is located within the conserved heme-binding region, just two amino-acid C-terminals to the absolutely conserved Cys at position 442 in the CYP3A4 protein (Sata et al., 2000); therefore, a Met445Thr change may exert an unfavorable effect on heme binding, resulting in an alteration in catalytic activity of the enzyme, especially when binding a suitable substrate. But as noted in Table 3, the enzymatic activity of CYP3A4*3 was similar to that of the wild-type CYP3A4 for most substrates tested. The discrepancy of results might arise from different heterologous expression methods used, and due to differing substrate specificity for the variant.
CYP3A4*4 (I118V) exhibited lower CLint towards quinine, relative to wild-type CYP3A4*1A. Studies of site-directed mutagenesis suggest that ser119 is a key amino acid implicated as an active-site residue (Yano et al., 2004). Hence, the Ile118Val mutation is likely to affect the conformation of the active site, resulting in decreased catalytic activity. Similarly, an in vivo study showed that the ratio of the urinary level of 6β-hydroxycortisol/free cortisol in these heterozygous individuals was lower than that in healthy controls, suggesting decreased enzymatic activities (Hsieh et al., 2001).
Both CYP3A4*8 and *13 variants displayed detectable, albeit lower, levels of CYP3A4 holoproteins and had extremely low catalytic activities towards quinine. Contrary to our results, CYP3A4*8 and *13 have been reported to exhibit no detectable CYP holoproteins when expressed in bacterial systems (Eiselt et al., 2001). We speculate that the use of bacterial vs. insect expression systems may explain the differing results. The bacterial expression system lacks an intracellular membrane environment (Gonzalez and Korzekwa, 1995) required for high expression of protein. In contrast, insect cells can process and modify proteins in a manner similar to human cells, permitting proper folding of the enzyme and incorporation of the heme cofactor, resulting in higher expression of CYP3A4 proteins.
CYP3A4*11 (T363M) was expressed at significantly lower levels than wild-type CYP3A4 (Figure 2), a finding that is consistent with studies using bacterial or mammalian expression systems (Eiselt et al., 2001; Murayama et al., 2002). Threonine at residue 363 within the substrate recognition site (SRS)-5 may contribute to the hydrogen bonding network formation. A substitution from threonine to methionine might result in the loss of catalytic activity (Murayama et al., 2002). So CYP3A4*11 displayed reduced quinine hydroxylation activity in current study. In addition, CYP3A4*11 showed reduced testosterone hydroxylation activity (Murayama et al., 2002), had remarkably increased CLint towards lidocaine (Fang et al., 2017) and amiodarone (Yang et al., 2019), but was not associated with noticeable changes in metabolic activities for ibrutinib (Xu et al., 2018) (Table 3).
CYP3A4*12 (L373F) displayed a dramatically decreased CLint towards quinine, similarly with a previous research result towards midazolam (Eiselt et al., 2001). CYP3A4*12 possesses an amino-acid change of Leu373Phe; the mutated residue is located near Arg-372 and Glu-374, which participate in formation of a hydrogen-bonding network and are involved in active sites according to studies involving site-directed mutagenesis (Yano et al., 2004). Hence, the mutational residue (L373F) is likely to affect the stability of the hydrogen-bonding network and spatial conformation of the active-site cavity, resulting in altered catalytic activity of CYP3A4*12.
CYP3A4*17 (F189S) was found in Caucasians (Dai et al., 2001), Moroccans (Fernandez-Santander et al., 2016), Libyans (Fernandez-Santander et al., 2016), and Chinese (Du et al., 2006) with a frequency of 2.1%, 1.8%, 1%, and 2%, respectively. CYP3A4*17 displayed an especially low CLint towards quinine compared with the wild-type. Similarly, it also exhibited extremely low catalytic activities towards testosterone (Dai et al., 2001), chlorpyrifos (Dai et al., 2001), amiodarone (Yang et al., 2019), and nifedipine (Lee et al., 2005) (Table 3).
CYP3A4*18 (L293P) exhibited decreased CLint towards quinine (17.62%, Table 2). The Leu293Pro substitution has an influence on the overall protein structure and leads to the modification of the arrangement of SRS regions, the important sites for substrate recognition and substrate access to the active site (Kang et al., 2009). As described in in vitro studies, CYP3A4*18 displayed decreased metabolic activities towards midazolam (Lee et al., 2007; Kang et al., 2009; Maekawa et al., 2010), paclitaxel (Maekawa et al., 2010), docetaxel (Maekawa et al., 2010), and irinotecan (Maekawa et al., 2010), had increased catalytic activities towards lidocaine (Fang et al., 2017), amiodarone (Yang et al., 2019), testosterone (Dai et al., 2001; Kang et al., 2009), chlorpyrifos (Dai et al., 2001), and estrogens (Kang et al., 2009), but exhibited unchanged relative activities for nifedipine (Lee et al., 2005; Miyazaki et al., 2008), carbamazepine (Maekawa et al., 2010), and ibrutinib (Xu et al., 2018). There were also conflicting results when regarding midazolam (Miyazaki et al., 2008) or testosterone (Murayama et al., 2002; Miyazaki et al., 2008) as substrates (Table 3). An in vivo study indicated that CYP3A4*1*18-heterozygous subjects did not appear to have a significant change in midazolam clearance compared with CYP3A4*1*1 wild-type subjects (Lee et al., 2007). However, another in vivo study indicated that CYP3A4*18 carriers, including 52 of heterozygotes and a homozygote, showed diminished midazolam clearance (Kang et al., 2009), which is consistent with the results obtained from in vitro experiments (Lee et al., 2007; Kang et al., 2009; Maekawa et al., 2010).
CYP3A4*20 (488 frameshift) showed remarkably diminished apoprotein expression (Figure 1) and dramatically reduced CLint towards quinine. In contrast, no catalytic activities of midazolam 1′- and 4-hydroxylation have been reported in yeast or HEK 293 microsomes expressing CYP3A4*20 (Westlind-Johnsson et al., 2006). These conflicting results may be attributable to the different heterologous expression systems and substrates used in the experiments. In addition, a CYP3A4*20 heterozygote, identified in Brazilian individuals, has been reported to exhibit reduced systemic clearance of ∼1.9-fold for midazolam in vivo (Westlind-Johnsson et al., 2006). It was hypothesized previously that this rare mutation could influence protein folding, heme incorporation (Westlind-Johnsson et al., 2006), and the spatial conformation of active-site cavity, giving rise to a decrease or loss of catalytic activity associated with the substrate used.
CYP3A4*21 (Y319C), which was identified in Chinese population (Zhou et al., 2011), displayed weak catalytic activity towards quinine in our study. Interestingly, no detectable apoenzyme expression was identified in our results. Tyr319 is a highly conserved residue in the cytochrome P450 family throughout eukaryote evolution from nematode to human (Zhou et al., 2011). According to in silico functional predictions (Zhou et al., 2011), Tyr319 residue lies on an important domain and a substitution of Tyr with Cys at position 319 of the CYP3A4*21 protein may induce dramatic alterations in active-site cavity configuration. Therefore, the CYP3A4*21 may produce a malfunctional protein with reduced catalytic activity towards quinine. Additionally, we speculated that these antibodies we used to detect CYP3A4 recombinant apoproteins may not be suitable to detect the variant CYP3A4*21. That is, the mutation of Tyr319Cys in CYP3A4*21 may prevent these antibodies from recognizing the specific epitopes in CYP3A4.
In contrast, two of the recombinant CYP3A4 holoenzymes, CYP3A4*15 (R162Q) and CYP3A4*29 (F113I), displayed significantly increased CLint for quinine. Similarly, both CYP3A4*15 and CYP3A4*29 exhibited increased CLint towards lidocaine (Fang et al., 2017), but showed decreased CLint for ibrutinib (Xu et al., 2018). CYP3A4*15 was detected first in different ethnic populations with small samples for each ethnic group (Lamba et al., 2002). CYP3A4*29 was identified and named in our recent study (Hu et al., 2017). The two variants are related to rapid metabolism of quinine in vitro. If confirmation of increased quinine clearance is found in individuals who are homozygous or heterozygous carriers of CYP3A4*29 or CYP3A4*15, these individuals may be CYP3A4 ultra-rapid metabolizers of quinine. Patients with these genotypes may: i) have poorer therapeutic efficacy when usual clinical doses of quinine are administered; and ii) require higher doses to reach therapeutic plasma concentrations.
No active holoenzymes of the CYP3A4*6 (277 frameshift), CYP3A4*26 (R268Stop), and CYP3A4*30 (R130Stop) variants were detected and, accordingly, they exhibited no metabolic activity upon quinine (Table 2).
CYP3A4*6 has an insertion of an adenine residue in exon 9 (830–831 insA); this mutation causes a frameshift and leads to the translation of a truncated protein that cannot incorporate heme (Hsieh et al., 2001). Consequently, the catalytic activity of CYP3A4*6 towards quinine was not identified in our study. As shown previously in vivo, a Chinese person was found to be heterozygous for CYP3A4*6 with a much lower ratio of urinary level of 6β-hydroxycortisol:free cortisol than wild-type individuals, suggesting decreased CYP3A4 activity (Hsieh et al., 2001). Similarly, a patient who is heterozygous for CYP3A4*6, undergoing organ transplantation, had a tacrolimus concentration-to-adjusted dose ratio 4.3-fold higher than that of wild-type patients (Jun et al., 2009).
CYP3A4*26 has a C→T nucleotide substitution in exon 5. This causes a change from arginine to a premature stop codon at position 268, and results in a putative shortened protein lacking catalytic domain and therefore activity (Werk et al., 2014). CYP3A4*26 may not be expressed due to the premature stop codon at position 268 or if it is, the protein may be marked for rapid degradation (Werk et al., 2014). Accordingly, we were unable to detect the holoprotein of the CYP3A4*26 variant expressed in Sf21 cells. Likewise, a 19-year-old kidney-transplanted patient, identified as a homozygote for CYP3A4*26 and meanwhile also a homozygote for CYP3A5*3, exhibited an unexpectedly high plasma level of tacrolimus as a result of extremely diminished tacrolimus clearance (Werk et al., 2014).
Similar to CYP3A4*26, CYP3A4*30 has a change from arginine to a premature stop codon at position 130 (Hu et al., 2017). The Arg130 is considered important for heme incorporation (Eiselt et al., 2001) and thus mutation of Arg130 would disrupt heme incorporation and so affect holoenzyme expression of CYP3A4.
In combination with previous studies (Table 3), some CYP3A4 allelic variants showed the different preferences for the substrates unlike those of the wild type enzymes. Namely, some variants exhibit altered catalytic activities with a substrate-dependent profile. As discussed above, conflicting results deriving from a specific substrate in studies may arise from many factors such as discrepancies from in vitro or in vivo method and different heterologous expression systems.
Quinine has frequent adverse effects, because of its narrow therapeutic index (Bateman and Dyson, 1986). These adverse reactions were not only induced by drugs; they were also caused by common quinine-containing beverages. Based on a systematic review on clinical data from one hundred-fourteen articles, adverse reactions, which were resulted from quinine-containing beverages, accounted for 20% of the total adverse reactions to quinine (Liles et al., 2016). Surprisingly, even with minute exposure from common beverages, some individuals experienced severe adverse reactions involving multiple organ systems. Given obvious inter-individual diversity in quinine sensitivity, variation in CYP3A4 may be one possible mechanism underlying quinine sensitive and adverse effects. In addition, regulations for quinine use have not been established in some countries, such as, pills containing 50 mg or less of quinine are regarded as a natural health product and are available without a prescription in Canada (Liles et al., 2016); quinine-containing lotions and shampoos, as well as quinine-containing beverages, remain commonly available and supervision is not required. Thus, increased awareness of inter-individual differences in quinine sensitivity and adverse reactions to quinine among physicians and the public is believed to be necessary.
In conclusion, 31 CYP3A4 alleles were expressed using the baculovirus insect cell expression system and their catalytic abilities towards quinine were evaluated. Most recombinant CYP3A4 variants exhibited significantly decreased intrinsic clearance towards quinine. To our knowledge, this is the first study to describe functional activities of all reported CYP3A4 variants with amino acid substitutions, in quinine 3-hydroxylation. Our results may provide one possible mechanism underlying the inter-individual differences in quinine sensitivity. Further clinical studies are needed to confirm whether these CYP3A4 variants mentioned above are relevant to the variability in quinine metabolism in vivo. The study may contribute to providing an experimental basis for further clinical studies on quinine and so accelerate the process for establishing a genotype-phenotype connection.
The raw data supporting the conclusions of this manuscript will be made available by the authors, without undue reservation, to any qualified researcher.
All authors contributed to this work and agreed to submit the final version. X-YZ, G-XH, and J-PC organized and designed the study. X-YZ, X-XH, C-CW, X-RL, ZC, and QL conducted the experiments. X-YZ performed data analysis and wrote the draft of the manuscript. G-XH and J-PC contributed to the final version of the manuscript.
This work was supported by a grant from the Ministry of Science and Technology of the People’s Republic of China (2017ZX09304026).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
CYP3A4, Cytochrome P450 3A4; RCODS, reduced carbon monoxide difference spectroscopy; URM, ultra-rapid-metabolizer; IS, internal standard; UPLC-MS/MS, ultra-high-performance liquid chromatography–tandem mass spectrometry.
Achan, J., Talisuna, A. O., Erhart, A., Yeka, A., Tibenderana, J. K., Baliraine, F. N., et al. (2011). Quinine, an old anti-malarial drug in a modern world: role in the treatment of malaria. Malar. J. 10, 144. doi: 10.1186/1475-2875-10-144
Bateman, D. N., Dyson, E. H. (1986). Quinine toxicity. Adverse Drug React Acute Poisoning Rev. 5, 215–233.
Bel, B., Jeudy, G., Bouilly, D., Dalac, S., Vabres, P., Collet, E. (2009). Fixed eruption due to quinine contained in tonic water: positive patch-testing. Contact Derm. 61, 242–244. doi: 10.1111/j.1600-0536.2009.01617.x
Buters, J. T., Korzekwa, K. R., Kunze, K. L., Omata, Y., Hardwick, J. P., Gonzalez, F. J. (1994). cDNA-directed expression of human cytochrome P450 CYP3A4 using baculovirus. Drug Metab. Dispos. 22, 688–692.
Dai, D., Tang, J., Rose, R., Hodgson, E., Bienstock, R. J., Mohrenweiser, H. W., et al. (2001). Identification of variants of CYP3A4 and characterization of their abilities to metabolize testosterone and chlorpyrifos. J. Pharmacol. Exp. Ther. 299, 825–8231.
Dai, D. P., Wang, Y. H., Wang, S. H., Geng, P. W., Hu, L. M., Hu, G. X., et al. (2013). In vitro functional characterization of 37 CYP2C9 allelic isoforms found in Chinese Han population. Acta. Pharmacol. Sin. 34, 1449–1456. doi: 10.1038/aps.2013.123
Du, J., Xing, Q., Xu, L., Xu, M., Shu, A., Shi, Y., et al. (2006). Systematic screening for polymorphisms in the CYP3A4 gene in the Chinese population. Pharmacogenomics 7, 831–841. doi: 10.2217/14622416.7.6.831
Eiselt, R., Domanski, T. L., Zibat, A., Mueller, R., Presecan-Siedel, E., Hustert, E., et al. (2001). Identification and functional characterization of eight CYP3A4 protein variants. Pharmacogenetics 11, 447–458. doi: 10.1097/00008571-200107000-00008
Estabrook, R. W., Werringloer, J. (1978). The measurement of difference spectra: application to the cytochromes of microsomes. Meth. Enzymol. 52, 212–220. doi: 10.1016/S0076-6879(78)52024-7
Fang, P., Tang, P. F., Xu, R. A., Zheng, X., Wen, J., Bao, S. S., et al. (2017). Functional assessment of CYP3A4 allelic variants on lidocaine metabolism in vitro. Drug Des. Devel. Ther. 11, 3503–3510. doi: 10.2147/DDDT.S152366
Fernandez-Santander, A., Novillo, A., Gaibar, M., Romero-Lorca, A., Moral, P., Sanchez-Cuenca, D., et al. (2016). Cytochrome and sulfotransferase gene variation in north African populations. Pharmacogenomics 17, 1415–1423. doi: 10.2217/pgs-2016-0016
Gonzalez, F. J., Korzekwa, K. R. (1995). Cytochromes P450 expression systems. Annu. Rev. Pharmacol. Toxicol. 35, 369–390. doi: 10.1146/annurev.pa.35.040195.002101
Guengerich, F. P. (2005). CytochromeP450: Structure, Mechanism, and Biochemistry. New York, N.Y: Kluwer Academic/Plenum Publishers, 10013.
Horie, M., Oishi, M., Ishikawa, F., Shindo, T., Yasui, A., Ogino, S., et al. (2006). Liquid chromatographic analysis of cinchona alkaloids in beverages. J. AOAC Int. 89, 1042–1047.
Hsieh, K. P., Lin, Y. Y., Cheng, C. L., Lai, M. L., Lin, M. S., Siest, J. P., et al. (2001). Novel mutations of CYP3A4 in Chinese. Drug Metab. Dispos. 29, 268–273.
Hu, G. X., D. D., Wang, H., Huang, X. X., Zhou, X. Y., Cai, J., Chen, H., et al. (2017). Systematic screening for CYP3A4 genetic polymorphisms in a Han Chinese population. Pharmacogenomics 18, 369–379. doi: 10.2217/pgs-2016-0179
Ingelman-Sundberg, M., Sim, S. C., Gomez, A., Rodriguez-Antona, C. (2007). Influence of cytochrome P450 polymorphisms on drug therapies: pharmacogenetic, pharmacoepigenetic and clinical aspects. Pharmacol. Ther. 116, 496–526. doi: 10.1016/j.pharmthera.2007.09.004
Jun, K. R., Lee, W., Jang, M. S., Chun, S., Song, G. W., Park, K. T., et al. (2009). Tacrolimus concentrations in relation to CYP3A and ABCB1 polymorphisms among solid organ transplant recipients in Korea. Transplantation 87, 1225–1231. doi: 10.1097/TP.0b013e31819f117e
Kang, Y. S., Park, S. Y., Yim, C. H., Kwak, H. S., Gajendrarao, P., Krishnamoorthy, N., et al. (2009). The CYP3A4*18 genotype in the cytochrome P450 3A4 gene, a rapid metabolizer of sex steroids, is associated with low bone mineral density. Clin. Pharmacol. Ther. 85, 312–318. doi: 10.1038/clpt.2008.215
Lamba, J. K., Lin, Y. S., Thummel, K., Daly, A., Watkins, P. B., Strom, S., et al. (2002). Common allelic variants of cytochrome P4503A4 and their prevalence in different populations. Pharmacogenetics 12, 121–132. doi: 10.1097/00008571-200203000-00006
Lee, C. A., Kadwell, S. H., Kost, T. A., Serabjit-Singh, C. J. (1995). CYP3A4 expressed by insect cells infected with a recombinant baculovirus containing both CYP3A4 and human NADPH-cytochrome P450 reductase is catalytically similar to human liver microsomal CYP3A4. Arch. Biochem. Biophys. 319, 157–167. doi: 10.1006/abbi.1995.1278
Lee, S. J., Bell, D. A., Coulter, S. J., Ghanayem, B., Goldstein, J. A. (2005). Recombinant CYP3A4*17 is defective in metabolizing the hypertensive drug nifedipine, and the CYP3A4*17 allele may occur on the same chromosome as CYP3A5*3, representing a new putative defective CYP3A haplotype. J. Pharmacol. Exp. Ther. 313, 302–309. doi: 10.1124/jpet.104.078758
Lee, S. J., Lee, S. S., Jeong, H. E., Shon, J. H., Ryu, J. Y., Sunwoo, Y. E., et al. (2007). The CYP3A4*18 allele, the most frequent coding variant in asian populations, does not significantly affect the midazolam disposition in heterozygous individuals. Drug Metab. Dispos. 35, 2095–2101. doi: 10.1124/dmd.107.016733
Liles, N. W., Page, E. E., Liles, A. L., Vesely, S. K., Raskob, G. E., George, J. N. (2016). Diversity and severity of adverse reactions to quinine: a systematic review. Am. J. Hematol. 91, 461–466. doi: 10.1002/ajh.24314
Ma, Q., Lu, A. Y. (2011). Pharmacogenetics, pharmacogenomics, and individualized medicine. Pharmacol. Rev. 63, 437–459. doi: 10.1124/pr.110.003533
Maekawa, K., Yoshimura, T., Saito, Y., Fujimura, Y., Aohara, F., Emoto, C., et al. (2009). Functional characterization of CYP3A4.16: catalytic activities toward midazolam and carbamazepine. Xenobiotica 39, 140–147. doi: 10.1080/00498250802617746
Maekawa, K., Harakawa, N., Yoshimura, T., Kim, S. R., Fujimura, Y., Aohara, F., et al. (2010). CYP3A4*16 and CYP3A4*18 alleles found in East Asians exhibit differential catalytic activities for seven CYP3A4 substrate drugs. Drug Metab. Dispos. 38, 2100–2104. doi: 10.1124/dmd.110.034140
Mirghani, R. A., Hellgren, U., Westerberg, P. A., Ericsson, O., Bertilsson, L., Gustafsson, L. L. (1999). The roles of cytochrome P450 3A4 and 1A2 in the 3-hydroxylation of quinine in vivo. Clin. Pharmacol. Ther. 66, 454–460. doi: 10.1016/S0009-9236(99)70008-1
Mirghani, R. A., Yasar, U., Zheng, T., Cook, J. M., Gustafsson, L. L., Tybring, G., et al. (2002). Enzyme kinetics for the formation of 3-hydroxyquinine and three new metabolites of quinine in vitro; 3-hydroxylation by CYP3A4 is indeed the major metabolic pathway. Drug Metab. Dispos. 30, 1368–1371. doi: 10.1124/dmd.30.12.1368
Mirghani, R. A., Ericsson, O., Tybring, G., Gustafsson, L. L., Bertilsson, L. (2003). Quinine 3-hydroxylation as a biomarker reaction for the activity of CYP3A4 in man. Eur. J. Clin. Pharmacol. 59, 23–28. doi: 10.1007/s00228-003-0575-5
Miyazaki, M., Nakamura, K., Fujita, Y., Guengerich, F. P., Horiuchi, R., Yamamoto, K. (2008). Defective activity of recombinant cytochromes P450 3A4.2 and 3A4.16 in oxidation of midazolam, nifedipine, and testosterone. Drug Metab. Dispos. 36, 2287–2291. doi: 10.1124/dmd.108.021816
Murayama, N., Nakamura, T., Saeki, M., Soyama, A., Saito, Y., Sai, K., et al. (2002). CYP3A4 gene polymorphisms influence testosterone 6beta-hydroxylation. Drug Metab. Pharmacokinet. 17, 150–156. doi: 10.2133/dmpk.17.150
Ohtsuki, S., Schaefer, O., Kawakami, H., Inoue, T., Liehner, S., Saito, A., et al. (2012). Simultaneous absolute protein quantification of transporters, cytochromes P450, and UDP-glucuronosyltransferases as a novel approach for the characterization of individual human liver: comparison with mRNA levels and activities. Drug Metab. Dispos. 40, 83–92. doi: 10.1124/dmd.111.042259
Omura, T., Sato, R. (1964). The carbon monoxide-binding pigment of liver microsomes. J. Biol. Chem. 239, 2370–2378.
Rahmioglu, N., Heaton, J., Clement, G., Gill, R., Surdulescu, G., Zlobecka, K., et al. (2011). Genetic epidemiology of induced CYP3A4 activity. Pharmacogenet. Genomics 21, 642–651. doi: 10.1097/FPC.0b013e3283498ecf
Rendic, S. (2002). Summary of information on human CYP enzymes: human P450 metabolism data. Drug Metab. Rev. 34, 83–448. doi: 10.1081/DMR-120001392
Rodriguez-Antona, C., Sayi, J. G., Gustafsson, L. L., Bertilsson, L., Ingelman-Sundberg, M. (2005). Phenotype-genotype variability in the human CYP3A locus as assessed by the probe drug quinine and analyses of variant CYP3A4 alleles. Biochem. Biophys. Res. Commun. 338, 299–305. doi: 10.1016/j.bbrc.2005.09.020
Sata, F., Sapone, A., Elizondo, G., Stocker, P., Miller, V. P., Zheng, W., et al. (2000). CYP3A4 allelic variants with amino acid substitutions in exons 7 and 12: evidence for an allelic variant with altered catalytic activity. Clin. Pharmacol. Ther. 67, 48–56. doi: 10.1067/mcp.2000.104391
Werk, A. N., Lefeldt, S., Bruckmueller, H., Hemmrich-Stanisak, G., Franke, A., Roos, M., et al. (2014). Identification and characterization of a defective CYP3A4 genotype in a kidney transplant patient with severely diminished tacrolimus clearance. Clin. Pharmacol. Ther. 95, 416–422. doi: 10.1038/clpt.2013.210
Westlind-Johnsson, A., Hermann, R., Huennemeyer, A., Hauns, B., Lahu, G., Nassr, N., et al. (2006). Identification and characterization of CYP3A4*20, a novel rare CYP3A4 allele without functional activity. Clin. Pharmacol. Ther. 79, 339–349. doi: 10.1016/j.clpt.2005.11.015
W. H. Organization. (2015). Guidelines for the Treatment of Malaria. 3rd edition. Geneva, Switzerland: WHO Press, World Health Organization.
Xu, R. A., Wen, J., Tang, P., Wang, C., Xie, S., Zhang, B. W., et al. (2018). Functional characterization of 22 CYP3A4 protein variants to metabolize ibrutinib in vitro. Basic Clin. Pharmacol. Toxicol. 122, 383–387. doi: 10.1111/bcpt.12934
Yang, C. C., Zheng, X., Liu, T. H., Wang, C. C., Tang, P. F., Chen, Z., et al. (2019). Functional characterization of 21 CYP3A4 variants on amiodarone metabolism in vitro. Xenobiotica 49, 120–126. doi: 10.1080/00498254.2017.1414971
Yano, J. K., Wester, M. R., Schoch, G. A., Griffin, K. J., Stout, C. D., Johnson, E. F. (2004). The structure of human microsomal cytochrome P450 3A4 determined by X-ray crystallography to 2.05-A resolution. J. Biol. Chem. 279, 38091–38094. doi: 10.1074/jbc.C400293200
Zanger, U. M., Schwab, M. (2013). Cytochrome P450 enzymes in drug metabolism: regulation of gene expression, enzyme activities, and impact of genetic variation. Pharmacol. Ther. 138, 103–141. doi: 10.1016/j.pharmthera.2012.12.007
Zhang, H., Coville, P. F., Walker, R. J., Miners, J. O., Birkett, D. J., Wanwimolruk, S. (1997). Evidence for involvement of human CYP3A in the 3-hydroxylation of quinine. Br. J. Clin. Pharmacol. 43, 245–252. doi: 10.1046/j.1365-2125.1997.00556.x
Zhou, Q., Yu, X., Shu, C., Cai, Y., Gong, W., Wang, X., et al. (2011). Analysis of CYP3A4 genetic polymorphisms in Han Chinese. J. Hum. Genet. 56, 415–422. doi: 10.1038/jhg.2011.30
Keywords: cytochrome P450 3A4, drug metabolism, quinine, 3-hydroxylation, polymorphism
Citation: Zhou X-Y, Hu X-X, Wang C-C, Lu X-R, Chen Z, Liu Q, Hu G-X and Cai J-P (2019) Enzymatic Activities of CYP3A4 Allelic Variants on Quinine 3-Hydroxylation In Vitro. Front. Pharmacol. 10:591. doi: 10.3389/fphar.2019.00591
Received: 16 February 2019; Accepted: 08 May 2019;
Published: 31 May 2019.
Edited by:
Yurong Lai, Gilead, United StatesReviewed by:
Hyunyoung Jeong, University of Illinois at Chicago, United StatesCopyright © 2019 Zhou, Hu, Wang, Lu, Chen, Liu, Hu and Cai. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jian-Ping Cai, Y2FpanA2MUB2aXAuc2luYS5jb20=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.