
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Pharmacol. , 19 March 2019
Sec. Ethnopharmacology
Volume 10 - 2019 | https://doi.org/10.3389/fphar.2019.00266
This article is part of the Research Topic Network Pharmacology and Traditional Medicine View all 31 articles
 Yu Dong1,2†
Yu Dong1,2† Ping Qiu2†
Ping Qiu2† Rui Zhu2
Rui Zhu2 Lisha Zhao1
Lisha Zhao1 Pinghu Zhang3
Pinghu Zhang3 Yiqi Wang2
Yiqi Wang2 Changyu Li2
Changyu Li2 Kequn Chai1,2,4
Kequn Chai1,2,4 Dan Shou1*
Dan Shou1* Huajun Zhao2*
Huajun Zhao2*Phellinus igniarius (P. igniarius) is a medicinal fungus that is widely used in East Asia for the adjuvant treatment of cancer. To elucidate the antitumor effective substances and mechanism of P. igniarius, we designed an approach incorporating cytotoxicity screening, phytochemical analysis, network pharmacology construction, and cellular and molecular experiments. The dichloromethane extract of P. igniarius (DCMPI) was identified as the active portion in HT-29 cells. Nineteen constituents were identified, and 5 were quantified by UPLC-ESI-Q/TOF-MS. Eight ingredients were obtained in the network pharmacology study. In total, 473 putative targets associated with DCMPI and 350 putative targets related to colon cancer were derived from online databases and target prediction tools. Protein-protein interaction networks of drug and disease putative targets were constructed, and 84 candidate targets were identified based on topological features. Pathway enrichment analysis showed that the candidate targets were mostly related to reactive oxygen species (ROS) metabolic processes and intrinsic apoptotic pathways. Then, a cellular experiment was used to validate the drug-target mechanisms predicted by the system pharmacology analysis. Experimental results showed that DCMPI increased intracellular ROS levels and induced HT-29 cell apoptosis. Molecular biology experiments indicated that DCMPI not only increased Bax and Bad protein expression and promoted PARP and caspase-3/9 cleavage but also down-regulated Bcl-2 and Bcl-xl protein levels to induce apoptosis in HT-29 cells. In conclusion, our study provides knowledge on the chemical composition and antitumor mechanism of P. igniarius, which may be exploited as a promising therapeutic option for colon cancer.
Cancer serves as a major public health problem worldwide and is expected to surpass heart disease as the leading cause of death in the next few years (Siegel et al., 2016). In the past decade, numerous significant advances in cancer research have revealed the genetics and pathologies of malignant tumors, which, in turn, facilitates the development of novel anticancer agents (Hare et al., 2017). Interestingly, multiple bioactive phytochemicals in consumed edible and medicinal fungi have recently attracted considerable attention as potential candidates for anticancer agents (Li et al., 2015).
Phellinus igniarius (P. igniarius), a well-known mushroom belonging to the genus Phellinus in the polyporaceae family, is a physiologically functional food and exemplary source of natural medicine that has been widely used in China, Japan, Korea, and other countries (Ma et al., 2016). P. igniarius possesses high anti-inflammatory, antioxidant and antitumor biological activities due to its accumulation of various secondary metabolites, including polysaccharides, flavonoids, polyphenols, steroids and organic acids (Dong et al., 2015; Shou et al., 2016). Notably, recent studies have creatively focused on the potential functions of P. igniarius extracts and their constituent compounds in the prevention and treatment of cancer (Zhou et al., 2014; Sangdee et al., 2017). A considerable amount of evidence indicates that P. igniarius extracts resulting from water, alcohol, ethyl acetate and other solvent extractions have a significant inhibitory effect on various tumor cells, such as S180, PC3, SK-HEP-1, and HT-29 cells (Yang et al., 2006; Song et al., 2008; Jeon et al., 2013). To date, no prior system reports on the chemical composition of P. igniarius contribute to its antitumor activity or functional mechanisms.
Network pharmacology is now popularly utilized to discover the basis of pharmacodynamic substances, explore their molecular mechanisms of action, and elucidate their scientific connotations (Kim et al., 2017). Especially, TCM network pharmacology focuses on the wholeness and systemicity of the interactions between components, targets and diseases of TCM (Cao et al., 2018; Huang et al., 2018), and is crucial to select the beneficial therapeutic targets of TCM, typical TCM syndromes and corresponding classic formulas (Zhang et al., 2017). At the same time, TCM network pharmacology substantially reduces the workload of follow-up experimental studies on TCM.
In the present study, we designed an approach incorporating cytotoxicity screening, phytochemical analysis, network pharmacology construction, and cellular and molecular biology validation to clarify the antitumor effective substance and mechanism of P. igniarius. Notably, this is the first integral study using multiple methods in combination to elucidate the antitumor efficacy substances and mechanism of this large medicinal fungus.
Phellinus igniarius sporocarps were purchased from Zhejiang Qingzheng Biotechnology Co. Ltd. (Hangzhou, China). Reference standards (purity > 98%) of protocatechuic aldehyde (671E-QHX2) and osmundacetone (RC5E-FH31) were purchased from the National Institute for the Control of Pharmaceutical and Biological Products (Beijing, China), and naringenin (170124), eriodictycol (170309) and sakuranetin (170124) were purchased from Beijing Century Aoke Biology Research Co., Ltd. (Beijing, China). HPLC grade methanol and acetonitrile were purchased from Merck (Darmstadt, Germany). Distilled water was purchased from Watson’s Food & Beverage Co., Ltd. (Guangzhou, China). Leucine enkephalin and formic acid were purchased from Sigma-Aldrich (Darmstadt, Germany).
Human hepatoma carcinoma (HepG2, SMMC7721), human gastric carcinoma (BGC-823, SGC790, AGS), human colon carcinoma (HT-29) and human lung carcinoma (A549) cells were purchased from American Type Culture Collection (Rockefeller, MD, United States). DMEM and RPMI 1640 cell culture mediums were purchased from HyClone Corporation (Logan, UT, United States). Fetal bovine serum was purchased from Gibco Corporation (Grand Island, NE, United States). MTT was purchased from Sigma-Aldrich (Darmstadt, Germany). An AnnexinV-FITC/PI apoptosis detection kit was purchased from BD Biosciences (Franklin lakes, NJ, United States). A ROSs assay kit and JC-1 dye were purchased from Beyotime Biosciences (Nanjing, China). Antibodies against PARP (#9532), Caspase-3 (#9662), Caspase-8 (#6790), Caspase-9 (#9508), Bax (#5023), Bcl-2 (#2870), Bcl-xl (#2764), and β-actin (#4970) were purchased from Cell Signaling Technology (Boston, MA, United States). Antibody against Bad (ab32445) was purchased from Abcam (Cambridge, MA, United States). HRP-conjugated secondary antibody was purchased from Bio-Rad (Hercules, CA, United States).
The dried P. igniarius sporocarps were crushed into a powder (with an approximately 100 mesh screen) using a plant disintegrator. In total, 600 g of powder was weighed, immersed in 3 L of 95% (v/v) ethanol for 30 min, and extracted in an ultrasonic bath 3 times for 1 h each time. The extracted solution was merged and evaporated by a rotary evaporator, and distilled water was added to 1 L of the suspension (containing 20% ethanol). The suspension was then subjected to liquid-liquid extraction with petroleum ether, dichloromethane, ethyl acetate and n-butanol successively. Each extract fraction was merged, evaporated and then freeze-dried to obtain the petroleum ether extract (0.32 g), dichloromethane extract (0.51 g), ethyl acetate extract (2.23 g), and n-butanol extract (1.28 g).
Human hepatoma carcinoma (HepG2) and human gastric carcinoma (BGC-823) cells were cultured in DMEM. Human gastric carcinoma (SGC790, AGS), human colon carcinoma (HT-29) and human lung carcinoma (A549) cells were maintained in RPMI 1640 medium. Human hepatoma carcinoma cells (SMMC7721) were cultured in DMEM/F12 medium. Then, the three media were supplemented with 10% fetal bovine serum, 100 units/mL penicillin and 100 μg/mL streptomycin. The cultures were maintained in a humidified incubator containing 5% (v/v) CO2 at 37°C.
The cytotoxic effects of the P. igniarius extracts were detected using the MTT calorimetric method. Briefly, cells (3 × 103 cells/well) were seeded in 96-well plates and incubated for 24 h. The supernatant was subsequently aspirated, and the cells were exposed to various concentrations of extracts and 0.05% DMSO (v/v). After 72 h of incubation, MTT solution (0.5 mg/mL) was added to each well and incubated for 4 h. Thereafter, the medium containing MTT was removed and replaced with 120 μL of DMSO to dissolve the formazan crystals. Next, the plates were shaken for 20 min, and the absorbance at 570 nm was recorded using an enzyme-linked immunosorbent assay reader (TECAN, Switzerland).
In addition, HT-29 cells (5 × 105 cells/well) were seeded into a six-well plate and incubated for 24 h. Then, the medium was replaced with RPMI-1640 medium containing different concentrations of dichloromethane extract of P. igniarius (DCMPI). After incubation for 72 h, the cytomorphology of the cells was observed and digitally photographed using a phase contrast microscope (Olympus, Japan).
The UPLC-ESI-Q/TOF-MS system consisted of an AcquityTM ultra performance liquid chromatography (UPLC) system (Waters Corporation, Milford, MA, United States) and a Synapt G2 mass spectrometer (MS) (Waters MS-Technologies, Manchester, United Kingdom) equipped with an electrospray ion (ESI) source. The system and data were controlled by MassLynx (V4.1) software. Chromatography was performed on an Acquity UPLC BEH C18 column (2.1 × 150 mm, 1.7 μm, Waters Corporation, Milford, MA, United States) at a flow rate of 0.3 mL/min and a 40°C column temperature. The optimal mobile phases consisted of A (HCOOH: CH3CN = 0.1: 100, v/v) and B (HCOOH: H2O = 0.1: 100, v/v): 0–1 min, 1–8% A; 1–2.5 min, 8–15% A; 2.5–4.5 min, 15–20% A; 4.5–5.5 min, maintained at 20% A; 5.5–7.5 min, 20–30% A; 7.5–13 min, 30–60%; 13–14 min, 60–99% A; and 14–15 min, maintained at 99% A. The extracts were dissolved in methanol to a concentration of 1 mg/mL and filtered through a 0.22 μm membrane. A 2 μL aliquot of sample solution was injected for analysis.
The full-scan LC-MS data were acquired in both positive and negative ion modes from 50 to 1500 Da with a 0.3 s scan time. The optimal Q/TOF-MS conditions were as follows: capillary voltage 3.0 kV, sampling cone voltage 40.0 V and extraction cone voltage 5.0 V for positive ion mode; capillary voltage 2.4 kV, sampling cone voltage 35.0 V and extraction cone voltage 3.5 V for negative ion mode. The source temperature and desolvation gas temperature were set to 150 and 350°C, respectively, and the cone gas flow and desolvation gas flow were set to 60 L/h and 550 L/h, respectively. The collision energy was set to 4 eV for positive ion mode and 2 eV for negative ion mode. Sodium formate solution was used to calibrate the mass spectrometer prior to the experiment. Leucine-enkephalin was used as an external reference (Lock-SprayTM) to correct the mass during data acquisition via a LockSpray interface, generating reference ions at m/z 556.2771 Da ([M+H]+) and m/z 554.2615 Da ([M-H]−) in the positive and negative ion modes, respectively.
All LC-MS and MS/MS data were processed with MassLynxTM (V4.1) software. Molecular formula speculations of the compounds were determined with Elemental Composition software. Structural identification of the main compounds, which included the chemical structure, accurate molecular mass and potential molecular fragmentation pathways, was determined with Mass Fragment software. Previously published compounds were identified by comprehensively searching databases, such as PubMed1, Chemspider2, HMDB3, and Metlin4. Compounds with standard materials were validated using reference standards.
DCMPI was dissolved in methanol to a concentration of 1 mg/mL as a sample solution. Mother stock solutions (approximately 1 mg/mL) of protocatechuic aldehyde, osmundacetone, eriodictyol, naringenin and sakuranetin were separately prepared in MeOH. Further, combined spiking stock solutions of the five reference substances were prepared in MeOH from the mother stock solutions by stepwise dilution in the ranges of 4.61–73.15 μg/mL for protocatechuic aldehyde, 3.97–63.44 μg/mL for osmundacetone, 3.91–62.50 μg/mL for eriodictyol, 4.20–67.19 μg/mL for naringenin and 4.04–64.69 μg/mL for sakuranetin. The sample solution and each combined spiking stock solution were filtered through a 0.22 μm membrane, and a 2 μL volume was injected into the UPLC-ESI-Q/TOF-MS system for analysis. The LC-MS conditions were the same as those previously described.
Eight major DCMPI components, including five validated components (protocatechuic aldehyde, osmundacetone, eriodictyol, naringenin and sakuranetin) and three identified components (inoscavin A, phelligrin A and phelligrin B) with a peak area greater than 5% were included in the network pharmacology study. By searching for the relevant predictive genes of the 8 main components at the websites PharmMapper Server5, SEA Search Server6, STITCH7 and TCMSP8, the DCMPI prediction genes were obtained.
Two main methods were used to identify colon cancer-related targets. To identify the main DEGs between normal human colon samples and colon cancer samples, GDS4382 microarray data were downloaded from the GEO9. The data set consisted of 34 human samples; GEO analysis was performed using 17 normal colon samples and 17 colon cancer samples. DEGs were defined by the Bioconductor/R limma package. P < 0.05 and a fold change ≥ 2 were applied. The known targets associated with colon cancer were derived from five databases, GAD, OMIM10, TTD11, DrugBank12 and CoolGen13, using ‘colon cancer’ as the key word.
Protein–protein interaction data from the six currently available PPI databases, the Human Protein Reference Database, the Biomolecular Interaction Network Database, the Biological General Repository for Interaction Datasets, the Database of Interacting Proteins and the Molecular INTeraction Database, were searched using the Cytoscape plugin BisoGenet. An interactive network of DCMPI drug targets and colon cancer-related targets was constructed based on interaction data, and the network was visualized utilizing Cytoscape software (version 3.2.1).
By calculating the ‘betweenness centrality,’ ‘degree centrality,’ ‘eigenvector centrality,’ ‘closeness centrality,’ ‘network centrality’ and ‘local average connectivity’ using CytoNCA, the topological nature of each node in the interactive network was analyzed. The definitions and calculation formulas of these six parameters represent the topological importance of the nodes in the network, with more important nodes yielding higher quantitative values in the network.
DAVID Bioinformatics Resources 6.814 was used to perform GO enrichment analysis of the differentially expressed targets to explore their roles in many BPs. Twenty significantly enriched terms in the CC, BP, and MF categories are shown. ClueGO is a Cytoscape plugin for visualizing the non-redundant features of a large number of gene clusters in a functional grouping network to assess the enrichment of DCMPI candidate targets. The ClueGO network was created using kappa statistics, reflecting the relationship between terms based on the similarity of related genes. The significances of terms and groups are automatically calculated.
HT-29 colon cancer cells were seeded into 24-well plates (5 × 104 cells/well) and treated with 30 and 60 μg/mL DCMPI for 24 h. Then, the cells were rinsed with PBS, stained with DAPI (0.5 μg/mL) and further incubated for 20 min in the dark. The slides were observed under a fluorescence microscope (Nikon, Japan), and the cells with nucleus condensation or fragmentation were considered apoptotic.
HT-29 colon cancer cells were seeded into 6-well plates (5 × 105 cells/well) and cultured overnight prior to exposure to different concentrations of DCMPI (30 μg/mL, 60 μg/mL) for 24 h. Then, the percentage of apoptotic cells was determined by an Annexin V-FITC/PI staining kit following the manufacturer’s protocol. Cells were then analyzed with the Guava Easy cytometer (Merck Millipore Co. Ltd., Darmstadt, Germany) within 1 h.
Mitochondrial membrane potential (Δψm) was monitored utilizing the JC-1 cationic dye (Molecular Probes) as recommended by the manufacturer. HT-29 cells were seeded into 6-well plates (5 × 105 cells/well) and cultured overnight prior to exposure to different concentrations of DCMPI (30 μg/mL, 60 μg/mL) for 24 h and then resuspended in a buffer solution containing 10 μg/mL JC-1 cationic dye. After 20 min of incubation in the dark at 37°C, the samples were rinsed with PBS to remove unreacted dye. The fluorescence intensity was read with the Guava Easy cytometer within 1 h.
The cell-permeable dye DCFH-DA (Molecular Probes) was used to assay intracellular ROS levels. This dye diffuses into cells and becomes trapped inside by de-esterification. After a reaction with peroxides, the fluorescent product, 5-chloromethyl-2′-7′-dichlorofluorescein (DCF), is formed. HT-29 cells were seeded into 6-well plates (5 × 105 cells/well) and cultured overnight prior to exposure to different concentrations of DCMPI (30 μg/mL, 60 μg/mL) for 24 h and then resuspended in DMEM/F-12 medium containing 10 μM DCFH-DA. Thereafter, the samples were incubated in the dark at 37°C for 20 min and then rinsed with PBS to remove unreacted dye. The fluorescence intensity was read with the Guava Easy cytometer within 1 h.
Protein extraction was performed as follows: after treatment with DCMPI (30 μg/mL, 60 μg/mL) for 24 h, HT-29 cells were cultured, collected, rinsed twice with ice-cold PBS, lysed, incubated in RIPA buffer containing a 1% protease inhibitor cocktail for 30 min on ice, and then centrifuged at 12,000 × g for 15 min. The supernatants were harvested and prepared by mixing with 5× sample buffer for subsequent Western blot analysis. The protein samples were then separated by 10% SDS-PAGE and transferred onto 0.22 μm polyvinylidene fluoride membranes (Millipore, Bedford, MA, United States). The membranes were blocked with 5% non-fat skim milk for 1 h at room temperature and then incubated with the appropriate primary antibodies overnight at 4°C. The membranes were incubated with a horseradish peroxidase-conjugated secondary antibody at room temperature for 2 h, followed by rinsing three times with 1× TBST. The protein bands were visualized utilizing ECL detection reagents (Bio-Rad, United States).
All data were analyzed with GraphPad Prism 5.0 software (GraphPad Software, Inc., La Jolla, CA, United States) and SPSS 20.0 Software (SPSS, Inc., Chicago, IL, United States). Furthermore, all statistical comparisons were performed using one-way analysis of variance followed by a post hoc Dunnett’s test. Values of P < 0.01 were considered statistically significant.
The cell viability inhibition induced by P. igniarius extracts, including the petroleum ether fraction, dichloromethane fraction, ethyl acetate fraction, n-butanol fraction and ethanol extract of P. igniarius, was evaluated in vitro with an MTT assay. Moreover, each cancer cell line was incubated with different concentrations of P. igniarius extracts for 72 h to compare their cytotoxicities. Notably, DCMPI as the strongest inhibitor decreased the viabilities of all the tested cell lines in a concentration-dependent manner, and especially significant cytotoxicity was observed in HT-29 cells (Figure 1A). Normal HT-29 cells appeared whole, healthy, and polygonal in shape under a microscope; however, after administering DCMPI, the HT-29 cells exhibited a number of morphological alterations, including cell shrinkage, condensed chromatin, membrane blebbing, and an increased density of apoptotic cells (Figure 1B).
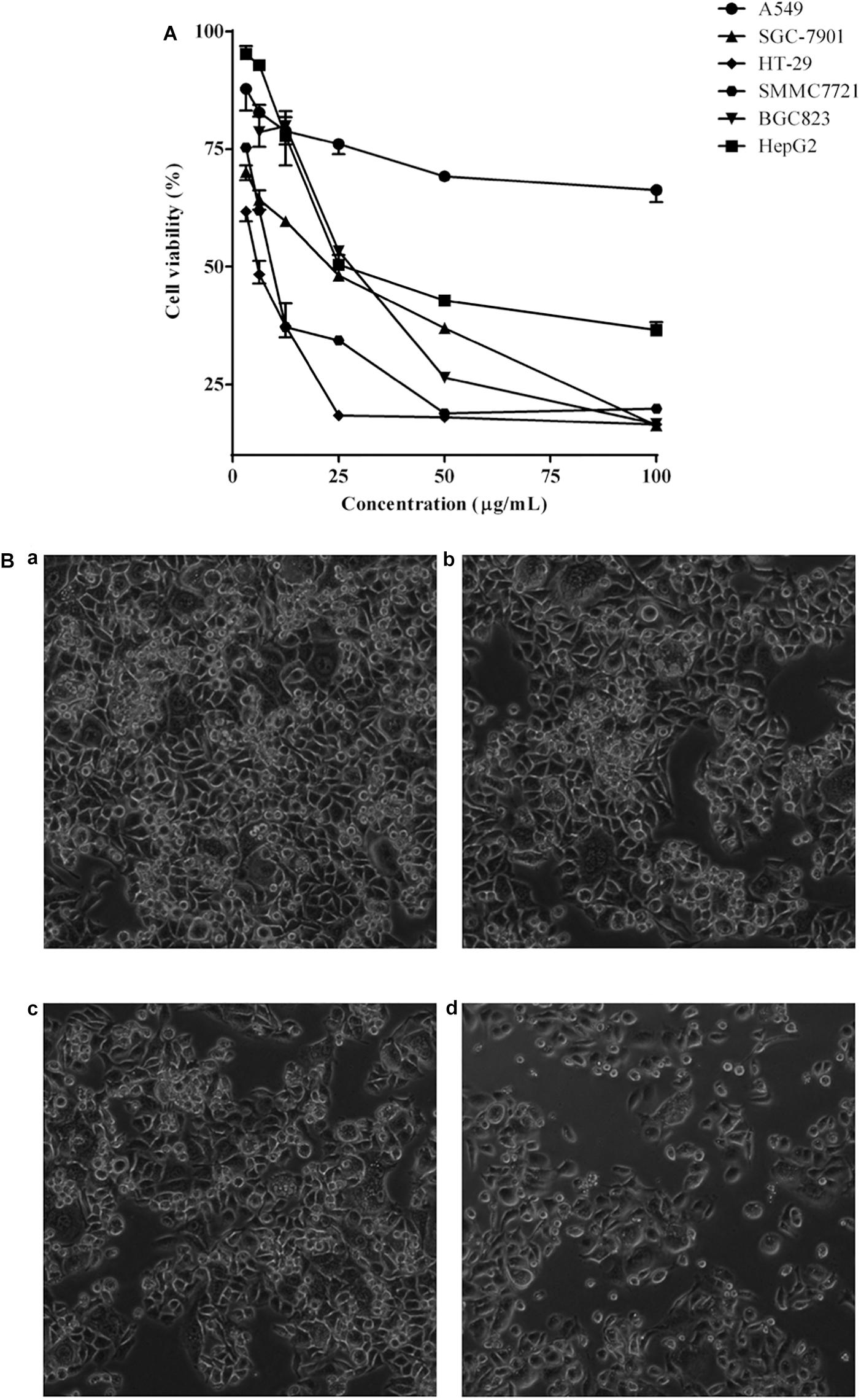
Figure 1. (A) Cytotoxic effects of DCMPI on the A549, SGC-7901, HT-29, SMMC7721, BGC-823 and HepG2 cell lines as determined by the MTT assay for 72 h. (B) Microscopic observation of HT-29 cells after administrating DCMPI for 72 h (a): untreated; (b): treated with 12.5 μg/mL DCMPI; (c): treated with 25 μg/mL DCMPI; (d): treated with 50 μg/mL DCMPI).
The chemical profiles of DCMPI were analyzed by UPLC-ESI-Q/TOF-MS. All LC-MS data, including retention times, accurate molecular masses, and MS/MS data, are necessary for the structural analysis of compounds. The element compositions were calculated and clearly confirmed by combining with a mass accuracy (ppm) less than 5.0 using MarkerLynx (4.1) software. The majority of the components detected in the positive ion mode were also detected in the negative ion mode; therefore, data analysis was performed in only the negative ion mode. Using the optimal UPLC and Q/TOF-MS conditions described above, the BPI chromatogram was obtained in the negative ion mode (Figure 2A). A total of 20 peaks were obtained from the BPI chromatogram, and 19 of these peaks were identified or characterized (Table 1). Peak 15 was tentatively characterized as phelligrin A based on the mass spectrum and fragmentation pathway illustrated in Figures 2B,C.
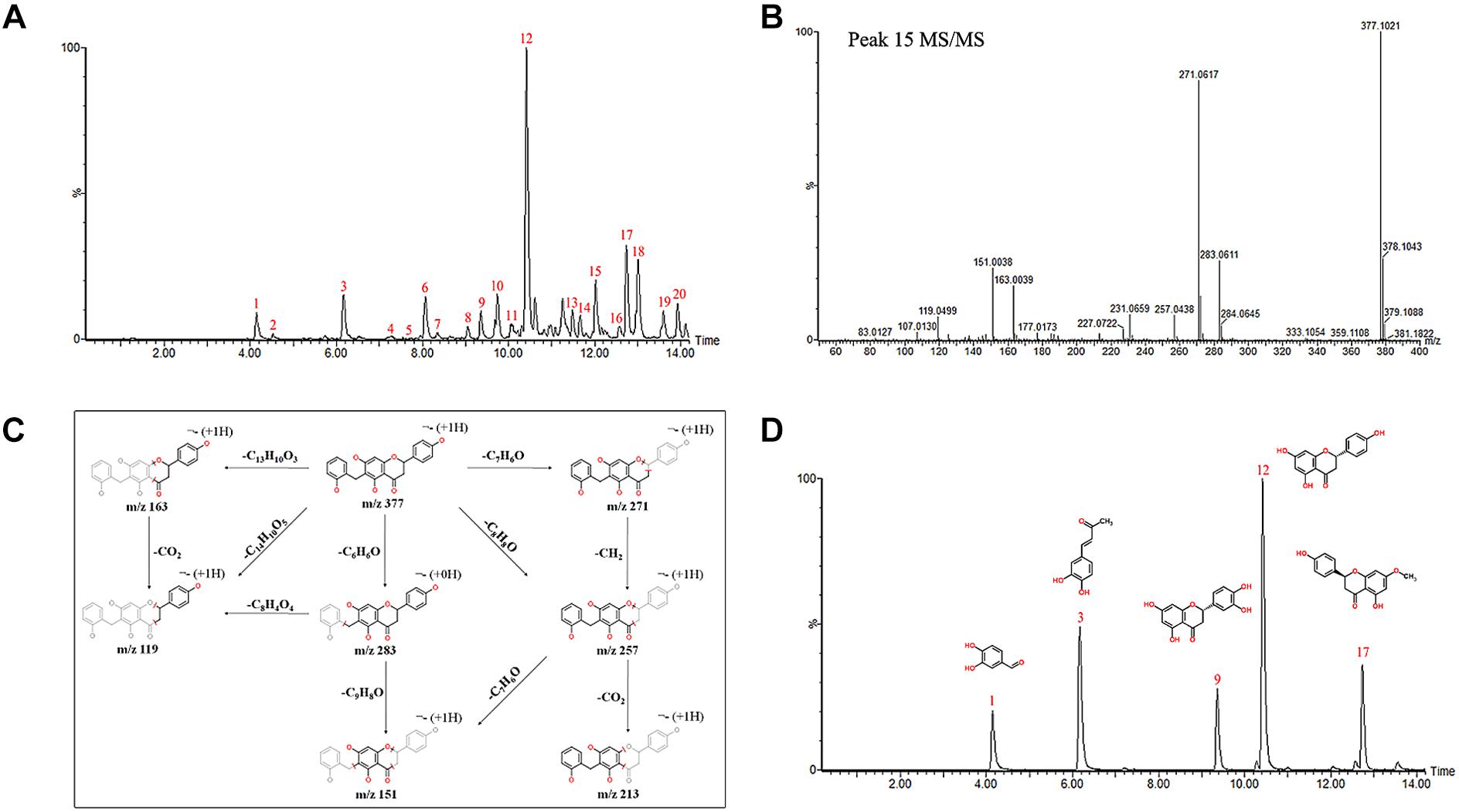
Figure 2. (A) UPLC-ESI-Q/TOF-MS BPI chromatogram of DCMPI. (B) Mass fragment spectrum of Phelligrin A in the negative ion mode. (C) Possible fragmentation mechanistic pathway of Phelligrin A. (D) Typical mixed standard substance chromatograms of protocatechuic aldehyde, osmundacetone, eriodictyol, naringenin, and sakuranetin.
The established quantitative method was applied to determine the contents of protocatechuic aldehyde, osmundacetone, eriodictyol, naringenin, and sakuranetin in DCMPI. The linear parameters of the five ingredients are listed in Table 2. Typical chromatograms of the five ingredients are shown in Figure 2D. DCMPI contained approximately 1.17% protocatechuic aldehyde, 0.62% osmundacetone, 1.10% eriodictyol, 3.59% naringenin, and 3.71% sakuranetin.
By screening the eight main components of DCMPI (the structures are shown in Figure 3A) in the PharmMapper Server, SEA Search Server, STITCH and TCMSP databases, potential targets of DCMPI were obtained. A total of 1727 potential targets were predicted for the 8 main compounds (196 for protocatechuic aldehyde, 149 for osmundacetone, 284 for eriodictyol, 293 for naringenin, 285 for sakuranetin, 250 for inoscavin A, 270 for phelligrin A, and 264 for phelligrin B), and 473 targets remained after deleting duplicates.
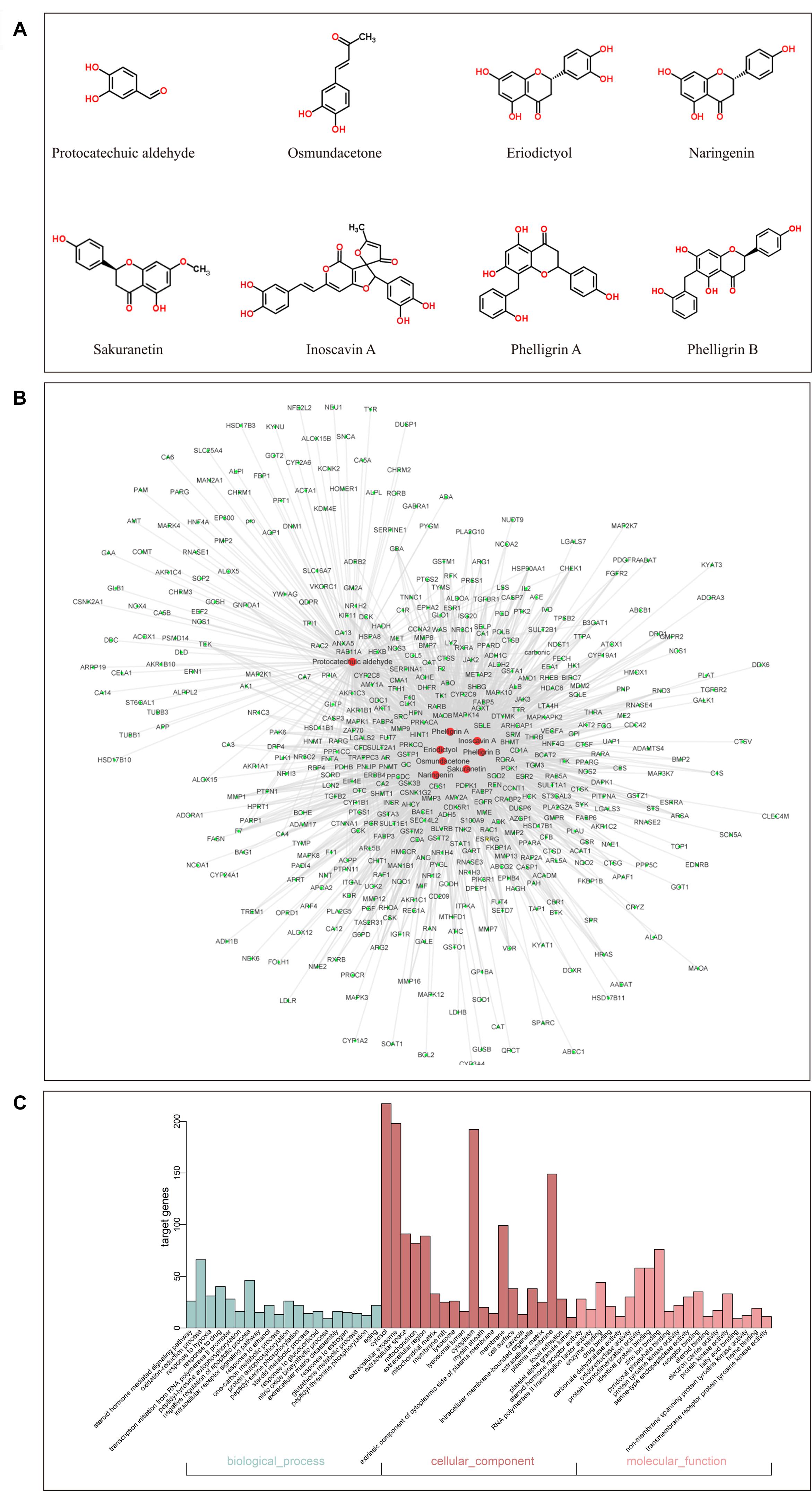
Figure 3. GO analysis of putative DCMPI targets and compound-target network construction. (A) Chemical structures of the eight main ingredients. (B) GO analysis of BP, cell component, and MF terms was performed on putative DCMPI targets, and the top 20 terms with P < 0.05 are shown. (C) The compound-target network was constructed by linking the 8 main compounds (red dots) and their potential targets (green dots).
Traditional Chinese medicine always exhibit versatile biological and pharmacological activities with complex chemical compositions by acting on multiple targets. Studying the complicated interactions between compounds and their targets at the system level may help comprehensively understand the mechanisms underlying TCM effects. We constructed a compound-target network based on the 8 candidate DCMPI compounds and their 473 potential targets. In total, 480 nodes and 1988 compound-target interactions are embodied in this network (Figure 3B).
GO analysis of the putative DCMPI targets was employed based on DAVID software for the visualization, annotation, and integrated discovery described by the BP, CC, and MF terms. In total, 671 BPs, 73 CCs, and 192 MFs that were enriched for this dataset were identified, of which 140 BPs, 55 CCs, and 140 MFs had P-values < 0.05. Figure 3C exhibits an overview of the GO analysis, and 20 remarkably enriched terms in the BP, CC, and MF categories are shown.
We herein collected colon cancer-related targets from two main sources: DEGs obtained from publicly available microarray data and disease-related databases. As shown Figure 4, 200 DEGs were identified from the GEO repository microarray data, while other targets were obtained from five databases: 163 from GAD, 54 from OMIM, 25 from TTD, 66 from DrugBank, and 246 from CoolGen. After removing redundant genes, 349 colon cancer-related targets were collected. Of these, 84 genes were also the targets of DCMPI, which suggests an obvious therapeutic potential for P. igniarius.
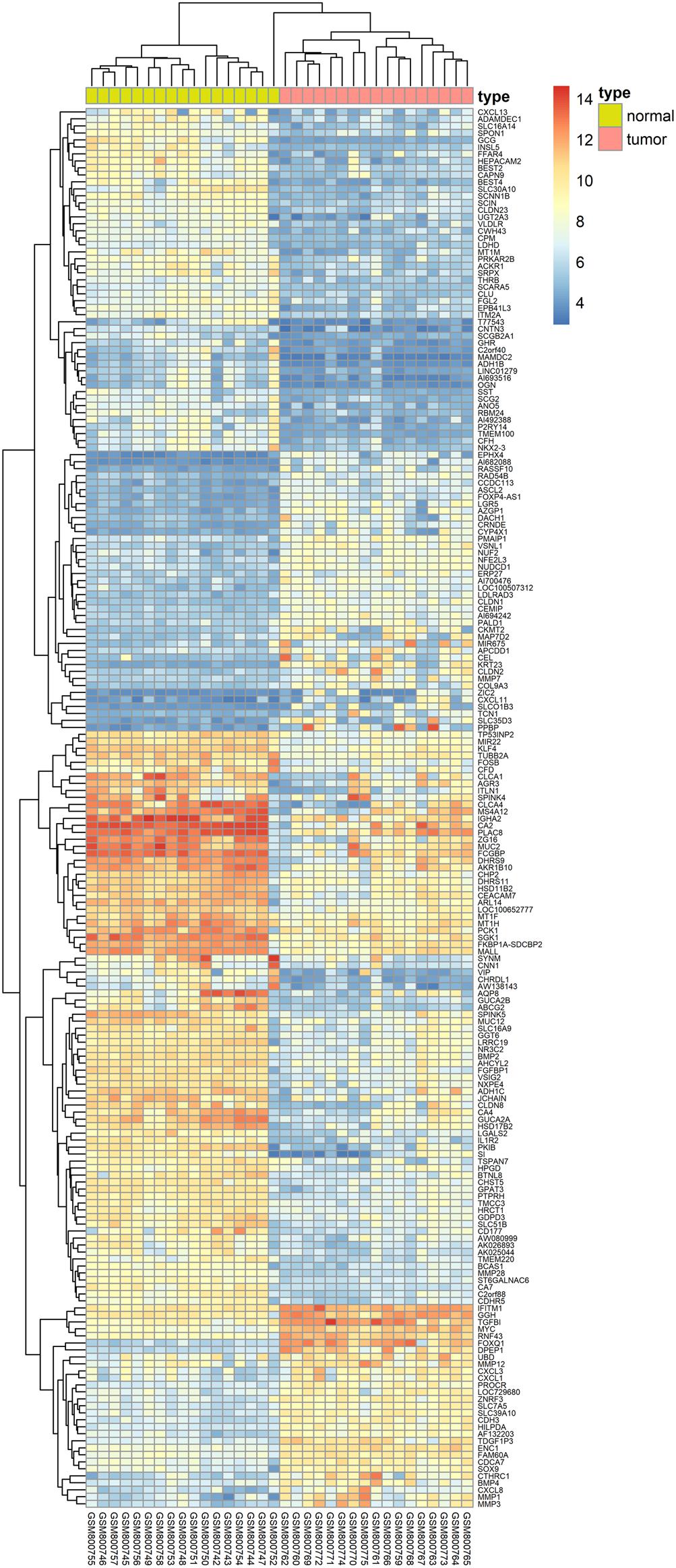
Figure 4. Identification of colon cancer-related targets using pre-existing microarray data. Two hundred DEGs identified by the limma package were highly related to colon cancer. P < 0.05 and FC > 2 were considered cut-off values.
Numerous evidence in network biology has shown that genes and proteins exert their functions via interactions. Thus, we selected proteins as nodes for constructing the network. To elaborate the pharmacological mechanism by which DCMPI ameliorates colon cancer, we established PPI networks that may reflect the behavior and properties of biological molecules. First, a putative target PPI network of DCMPI-related genes was obtained using Cytoscape 3.2.1 software with the plugin BisoGenet (8933 nodes and 195,493 edges) and database retrieval of the PPI network of colon cancer-related targets (4615 nodes and 113,727 edges). We then merged these two networks to obtain a core PPI (CPPI) network that consisted of 4614 nodes and 113,727 edges. Subsequently, colon cancer targets of DCMPI were screened using the topological features of CPPI. Then, utilizing a Cytoscape plugin (CytoNCA), the main hubs of the network were screened by calculating the topological features for each hub, which were identified when their degree exceeded twice the median degree of all nodes in the network. A flow chart depicting the core target screening is presented in Figures 5A–C.
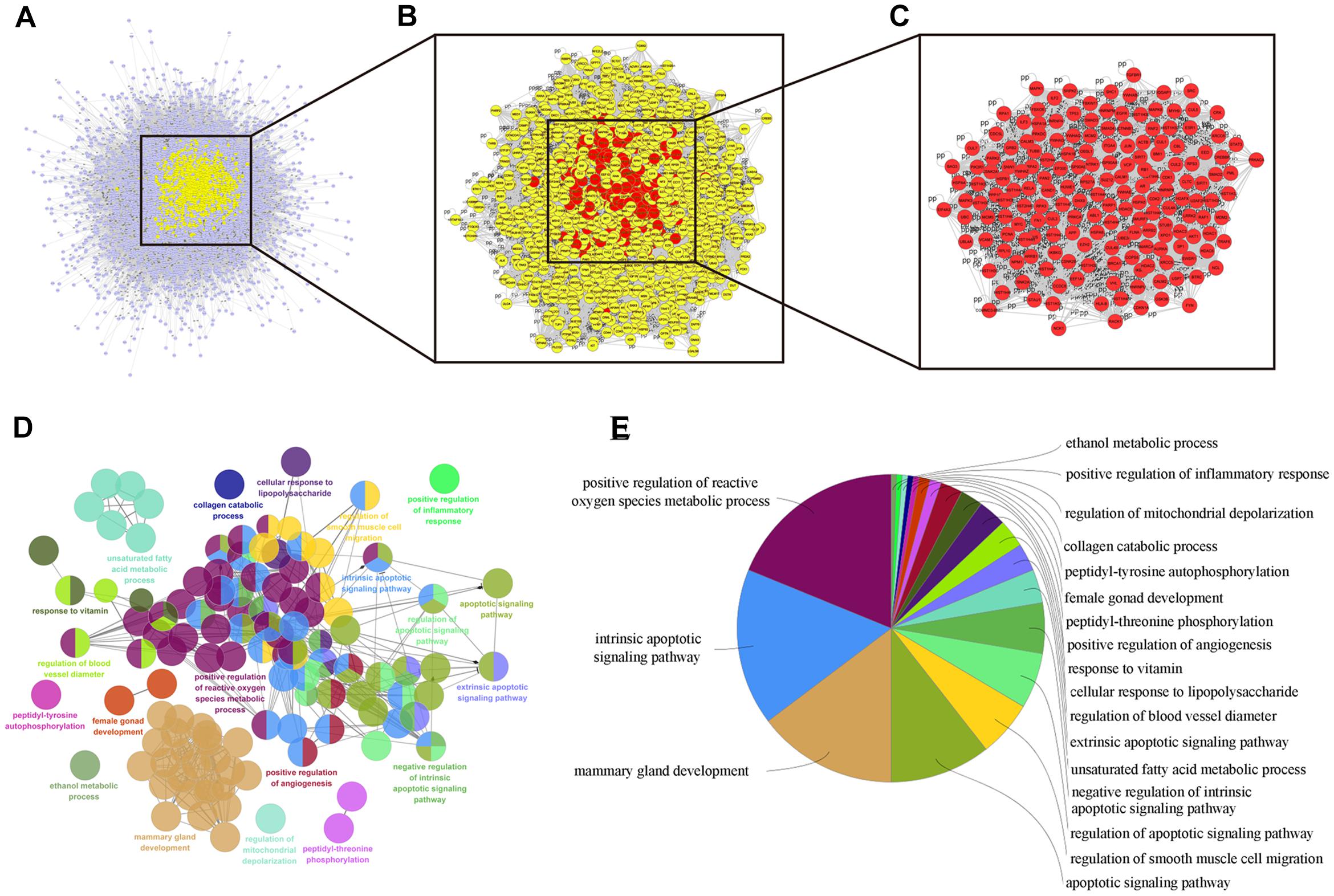
Figure 5. Candidate target identification and ClueGO pathway analysis. (A) Core CPPI network of DCMPI targets. (B) Large hubs of the DCMPI CPPI network extracted from (A) whose degrees were more than twice the median degree of all nodes in the network. (C) PPI network of the main DCMPI targets extracted from (B) constructed by calculating 6 topological features. (D,E) A functionally grouped network of enriched categories was generated for the target genes. GO terms are represented as nodes, and the node size represents the term’s enrichment significance. Functionally related groups partially overlap. Only the most significant term in the group is labeled. Representative enriched pathway (P < 0.05) interactions among the main DCMPI targets.
A Cytoscape plugin (ClueGO) was applied to further define the pathways involved in the biological networks identified above. As shown in Figures 5D,E, the biological networks consisted of 189 nodes and 1503 edges, and the potential targets were mainly assigned to positive regulation of ROS metabolic processes, intrinsic apoptotic pathways and mammary gland development. Of these, the first two pathways have well-established roles in cell apoptosis, and the results suggested that DCMPI is highly likely to exert its antitumor effect via the apoptotic pathway.
In this study, HT-29 cells were monitored by DAPI staining to determine the pro-apoptotic effect of DCMPI. The administration of DCMPI at doses of 30 and 60 μg/mL for 24 h dramatically induced nuclear morphological changes, such as apoptotic bodies and nuclear fragmentation, compared with the control group (Figure 6A).
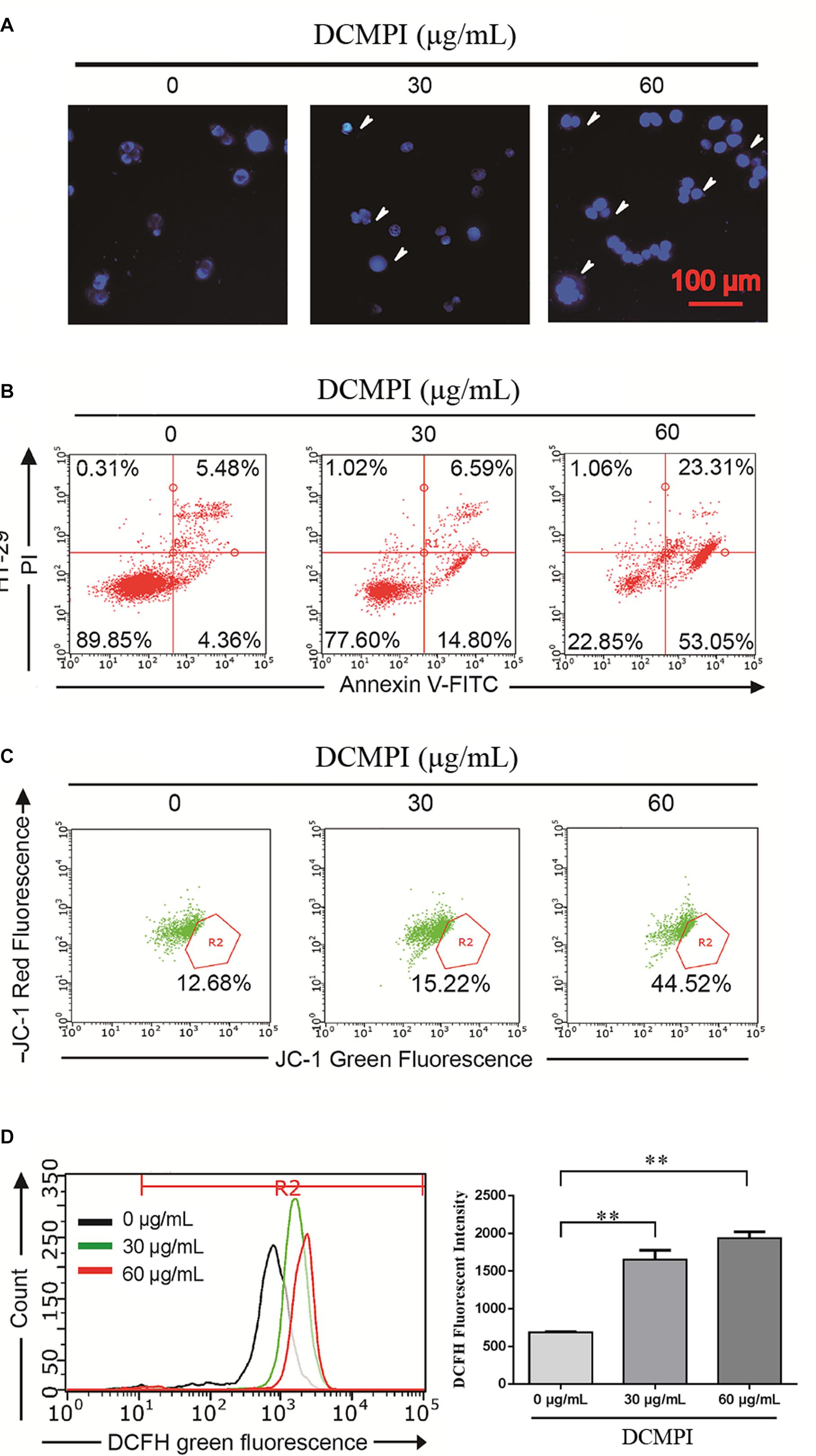
Figure 6. DCMPI induced apoptosis in HT-29 colon cancer cells. (A) Nuclear morphological changes were observed in DCMPI-exposed HT-29 cells by DAPI staining. (B) The percentage of apoptotic cells induced by DCMPI treatment was detected by Annexin V/PI double-staining analysis. (C) The effect of DCMPI on the mitochondrial membrane potential, which was determined using a JC-1 staining assay and flow cytometry. (D) The effect of DCMPI on the intracellular ROS, which was detected using DCFH-DA probes and flow cytometry. ∗∗P < 0.01. The data are presented as the mean ± SD from at least three independent experiments.
Next, the percentage of apoptotic cells induced by DCMPI treatment was detected by Annexin V-FITC/PI staining. As shown in Figure 6B, the numbers of HT-29 cells in both early- and late-stage apoptotic populations were remarkably increased in response to DCMPI (30 and 60 μg/mL). Therefore, this finding was consistent with the DAPI staining results and confirmed that DCMPI induced HT-29 cell apoptosis in a concentration-dependent manner.
The loss of Δψm, which occurs prior to caspase activation, is regarded as a hallmark of apoptosis. To determine whether DCMPI treatment activated the mitochondria-mediated apoptotic pathway, we utilized the JC-1 fluorescent probe to evaluate alterations in Δψm by flow cytometry. In apoptotic cells with low Δψm, JC-1 remains in the monomeric form, and the loss of Δψm is followed by a red-to-green shift. The percentage of green JC-1 was significantly elevated by treatment with DCMPI (Figure 6C), and DCMPI thus markedly increased the mitochondrial membrane permeability and induced Δψm collapse.
Reactive oxygen species generation was closely associated with the cancer cell death caused by mitochondrial dysfunction and subsequent mitochondria-induced apoptosis. To identify the roles of ROS in DCMPI-induced colon cancer cell death, we measured intracellular ROS production by DCFH-DA staining with flow cytometry. DCMPI significantly increased the accumulation of fluorescent dye in HT-29 cells in a concentration-dependent manner (Figure 6D).
DCMPI not only significantly up-regulated the protein expression of Bax and Bad but also simultaneously attenuated the protein levels of Bcl-2 and Bcl-xl. These results revealed that DCMPI alters the Bax/Bcl-2 ratio in HT-29 cells, which subsequently stimulates the mitochondrial-mediated apoptosis pathway. Furthermore, DCMPI (30 and 60 μg/mL) induced both PARP and caspase-3/9 cleavage to aggravate HT-29 cell apoptosis (Figure 7).
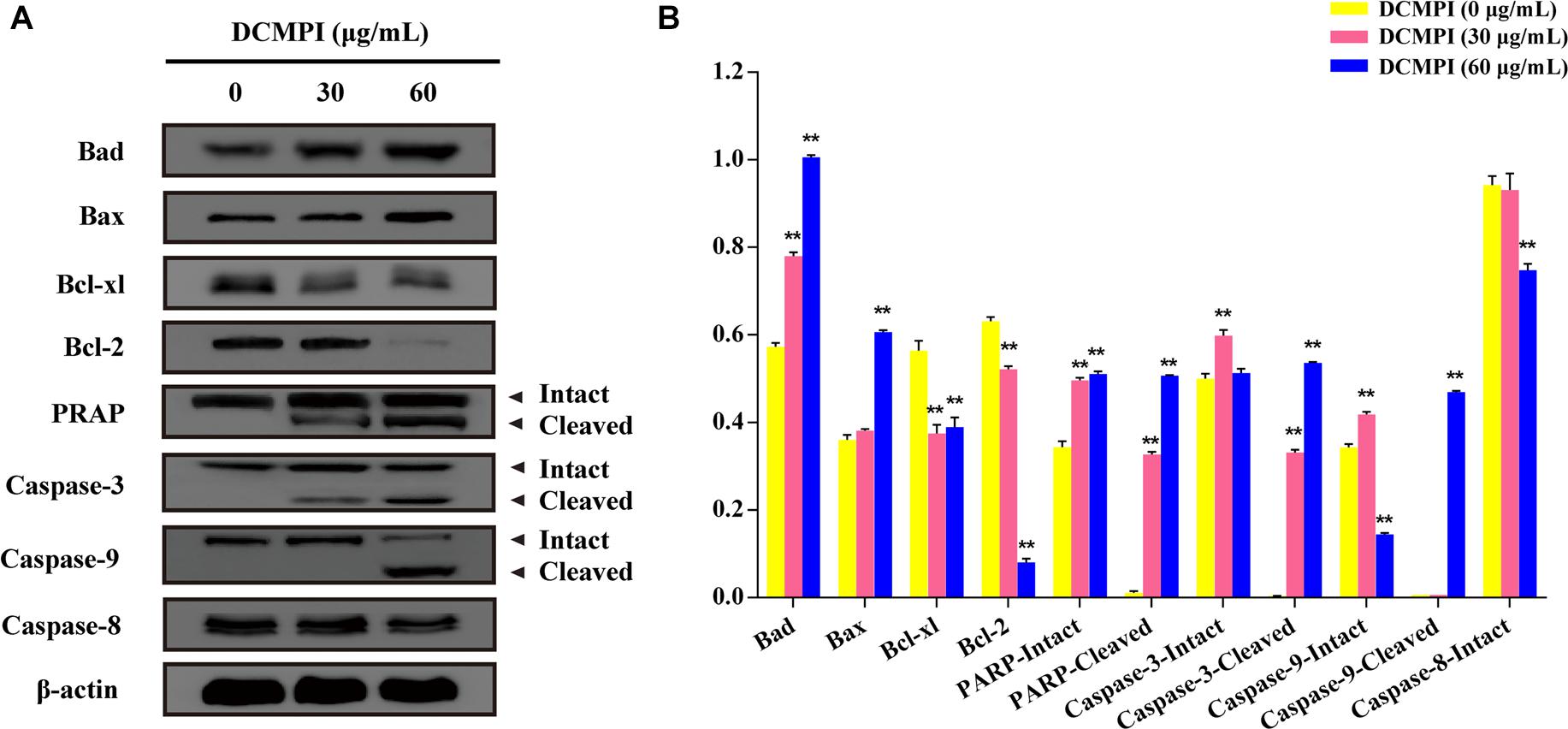
Figure 7. Western blot analysis of apoptosis-related protein expression. (A) Bad, Bax, Bcl-xl, Bcl-2, PARP, caspase-3, caspase-9, and caspase-8 protein expression in HT-29 cells after DCMPI treatment (30 and 60 μg/mL) for 24 h. (B) Densitometric analysis of Bad, Bax, Bcl-xl, Bcl-2, PARP, caspase-3, caspase-9, and caspase-8 expression. ∗∗P < 0.01. The data are presented as the mean ± SD from at least three independent experiments.
To clarify whether apoptosis is initiated by caspase cascade activation, we adopted the caspase inhibitor Z-VAD-FMK. Western blot analysis revealed that the cleavage of PARP and caspase 3/9 caused by DCMPI was dramatically inhibited by Z-VAD-FMK (50 μM) pretreatment (Figure 8). Together, these results clearly support that DCMPI induces HT-29 cell apoptosis via the mitochondrial apoptosis pathway.
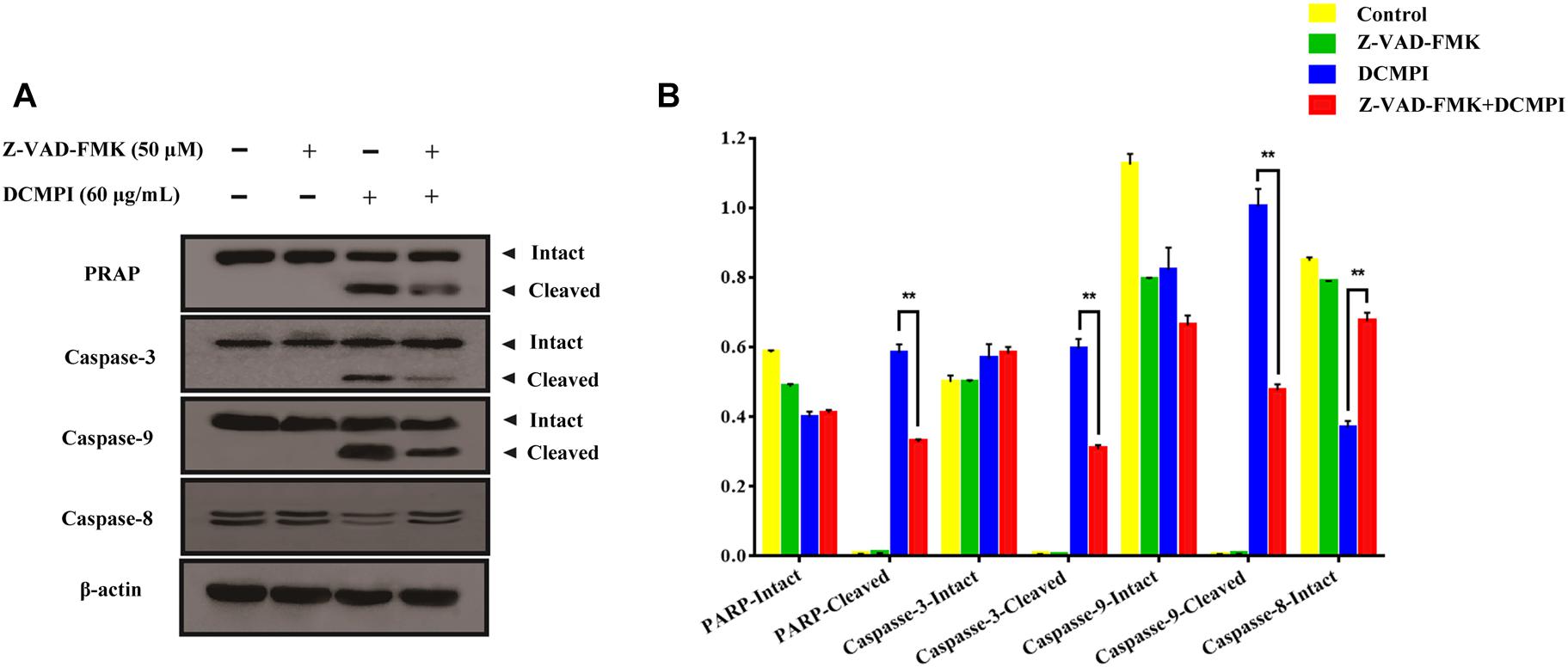
Figure 8. The effect of Z-VAD-FMK (50 μM) pretreatment and DCMPI (60 μg/mL) treatment on the cleavage of PARP and caspase-3/9 in HT-29 cells. (A) PARP, caspase-3, caspase-9, and caspase-8 protein expression in HT-29 cells after treatment for 24 h. (B) Densitometric analysis of PARP, caspase-3, caspase-9, and caspase-8 expression. ∗∗P < 0.01. The data are presented as the mean ± SD from at least three independent experiments.
Recently, medicinal fungi were shown to be ubiquitously present in nature, recognized as a promising source of food and medicine with multi-targeted effects and to have low toxicity (Gargano et al., 2017). Intriguingly, numerous preclinical and clinical trials have demonstrated that medicinal fungi contain a variety of structurally unique metabolites, which most likely possess significant activities like anti-inflammatory and antitumor (Piska et al., 2017). For instance, P. igniarius has been proposed as a prevalent type of edible and medicinal mushroom that has multiple highly effective protective properties (Dong et al., 2016). More importantly, P. igniarius attracts overwhelming attention as a potential candidate for anticancer therapeutics (De Silva et al., 2012). Cancer is a complex disease and tumorigenesis is related to inflammation (Guo et al., 2017). P. igniarius has potential anti-inflammatory and antitumor activities, hopeful to be developed a functional food for the prevention and control of inflammation-induced tumorigenesis in the future. Previous studies had established that the ethanol extract of P. igniarius suppresses SK-Hep-1 and RHE cell proliferation in a dose-dependent manner (Song et al., 2008). Furthermore, the hot water extract of P. igniarius directly inhibits S180 tumors and simultaneously exhibits an immune-regulatory effect on S180-bearing mice (Yang et al., 2006). However, to date, these studies largely emphasize the pharmacological effects of P. igniarius extracts, and no studies have characterized the chemical composition P. igniarius extracts or clarified their underlying mechanisms.
Traditional Chinese medicine network pharmacology was a new research strategy for translating TCM from an experience-based medicine to an evidence-based medicine system, which predicted the target profiles and pharmacological actions of herbal compounds, and revealed drug-gene-disease co-module associations to interpret the combinatorial rules and network regulation effects of herbal formulae (Liang et al., 2014; Li and Zhang, 2013). It provided a new paradigm for revealing the pharmacodynamics substance basis and mechanisms of TCM and revealing the problem of the effectiveness of TCM (Wang et al., 2017).
Therefore, we herein utilized a combination method that included a cytotoxicity test, phytochemical analysis, TCM network pharmacology, and cell and molecular biology experiments to elucidate the antitumor substances and potential mechanisms of P. igniarius. First, cytotoxicity experiments were used to show that DCMPI had a certain inhibitory effect on each tumor strain. Among the strains, the inhibitory effects on the BGC-823, SMMC7721 and HT-29 cell lines were higher than those on the others. Second, we elucidated the systemic phytochemical composition of DCMPI via UPLC-ESI-Q/TOF-MS. As previously described, 19 constituents were identified or tentatively characterized, and 5 of them were quantified by standard substances. Numerous ingredients in DCMPI have been reported to possess antitumor activity or cytotoxicity. For example, naringenin, a flavanone compound, has highly effective, diverse bioactivities, including anticancer activity (Chang et al., 2017). Eriodictyol also exerts multiple bioactive effects, and it especially inhibits cell proliferation and transformation (Zhu et al., 2015). Dihydrokaempferol serves as a flavonol extensively in citrus fruits and provides anticancer effects via suppressing cell migration and invasion (Liu X. et al., 2017). The flavonoids phelligrin A, phelligrin B, methylphelligrin A and methylphelligrin B were first discovered in the Phellinus family and shown to possess multiple bioactivities (Wu et al., 2011). Furthermore, several studies revealed that phelligrin A and phelligrin B provide selective cytotoxicity against cancer cells (Lee and Yun, 2011). In summary, the main compounds in DCMPI are attributed to the flavone family, and most have anticancer activity and likely account for the antitumor activity of P. igniarius.
Because of their complex compositions and mechanisms of action, investigating the pharmacology of TCM is a daunting task. Here, to explore the antitumor mechanism of DCMPI, we selected eight major components of DCMPI and utilized TCM network pharmacology approach to clarify their potential mechanisms. The network pharmacological analysis showed that DCMPI exerted its anticolon cancer function mainly via the positive regulation of ROS metabolic processes and intrinsic apoptotic pathways.
Apoptosis induction has been recognized as an essential mechanism in antitumor therapeutics (Janssen, 2001). Notably, numerous studies have demonstrated that anticancer agents exert their anti-proliferative effects mainly via two different apoptosis pathways involving mitochondria or death receptors (Liu B. et al., 2017). In particular, the mitochondria-associated pathway is a classic intrinsic pathway that is initiated by ROS overproduction, resulting in the depletion of Δψm (Kuranaga, 2012). In general, a variety of protein molecules are involved in regulating the mitochondrial apoptotic pathway, including pro-apoptotic members (e.g., Bax and Bad) and anti-apoptotic members (e.g., Bcl-2 and Bcl-xl) (Czabotar et al., 2014). Furthermore, the pathway activates the specific pivotal proteinases as initiator caspase-9 and effector caspase-3, which subsequently leads to DNA fragmentation and nuclear PARP degradation during apoptosis (Kuranaga, 2012).
To verify the network pharmacology prediction results, in vitro cell experiments were carried out. DAPI and Annexin V-FITC/PI staining were performed to determine the apoptotic alterations associated with the cytotoxicity of HT-29 cells. Apoptotic bodies, nuclear fragmentation and early- and late-stage apoptotic populations were increased by DCMPI. Therefore, the cytotoxic effect of DCMPI on HT-29 cells appeared to be mediated by apoptosis. Apoptosis is considered to be a regulated form of cell death, and dysregulated apoptosis leads to a variety of aberrant pathological conditions and subsequent diseases, especially cancer (Sankari et al., 2012; Goldar et al., 2015). Remarkably, two main signaling pathways involving caspase-mediated apoptosis have been identified: the mitochondrial-mediated intrinsic pathway and the death receptor-mediated extrinsic pathway (Khan et al., 2014). Compelling evidence has indicated that increased levels of intracellular ROS result in mitochondrial swelling and MMP loss, which, in turn, activates the intrinsic apoptotic pathway. Because ROS overproduction not only causes severe DNA damage but also suppresses tumor growth, aggravated cell structures break down, and cancer cells die (Renschler, 2004). In this study, DCMPI stimulated the generation of intracellular ROS in a dose-dependent manner, induced MMP collapse and ultimately contributed to mitochondria-mediated apoptosis.
Bcl-2 family members, including Bcl-2, Bcl-xl, Bax, and Bad, are major regulators located in the mitochondria (Borkan, 2016). As anti-apoptotic proteins, Bcl-2 and Bcl-xl intervene in various apoptotic signals, whereas Bax and Bad serve as pro-apoptotic proteins and trigger the activation of caspases by mitochondrial dysfunction. In the present study, DCMPI down-regulated anti-apoptotic Bcl-xl and Bcl-2 expression and simultaneously enhanced the translocation of the activated pro-apoptotic proteins Bax and Bad. More importantly, subsequent activation of the caspase-dependent pathway was recognized as a pivotal step in the apoptotic process, and a series of initiator caspases was involved in the progression. Briefly, activation of the initiator caspase as caspase-9 activates downstream or executioner caspases undergoing apoptosis, such as caspase-3 (Nuñez et al., 1998; Kim and Hong, 2011). Furthermore, executioner caspase activation was responsible for a series of apoptotic biochemical characteristics, especially the cleavage of PARP, which facilitated cellular disassembly and resulted in DNA fragmentation. Herein, our findings demonstrated that DCMPI induced apoptosis in HT-29 cells by up-regulating cleaved caspase-3/9 and PARP. In addition, pretreatment with the pan-caspase inhibitor Z-VAD-FMK inhibited caspase and PARP cleavage, which confirmed that the pro-apoptotic effect of DCMPI in HT-29 cells was exerted via a caspase-dependent pathway (Figure 9).
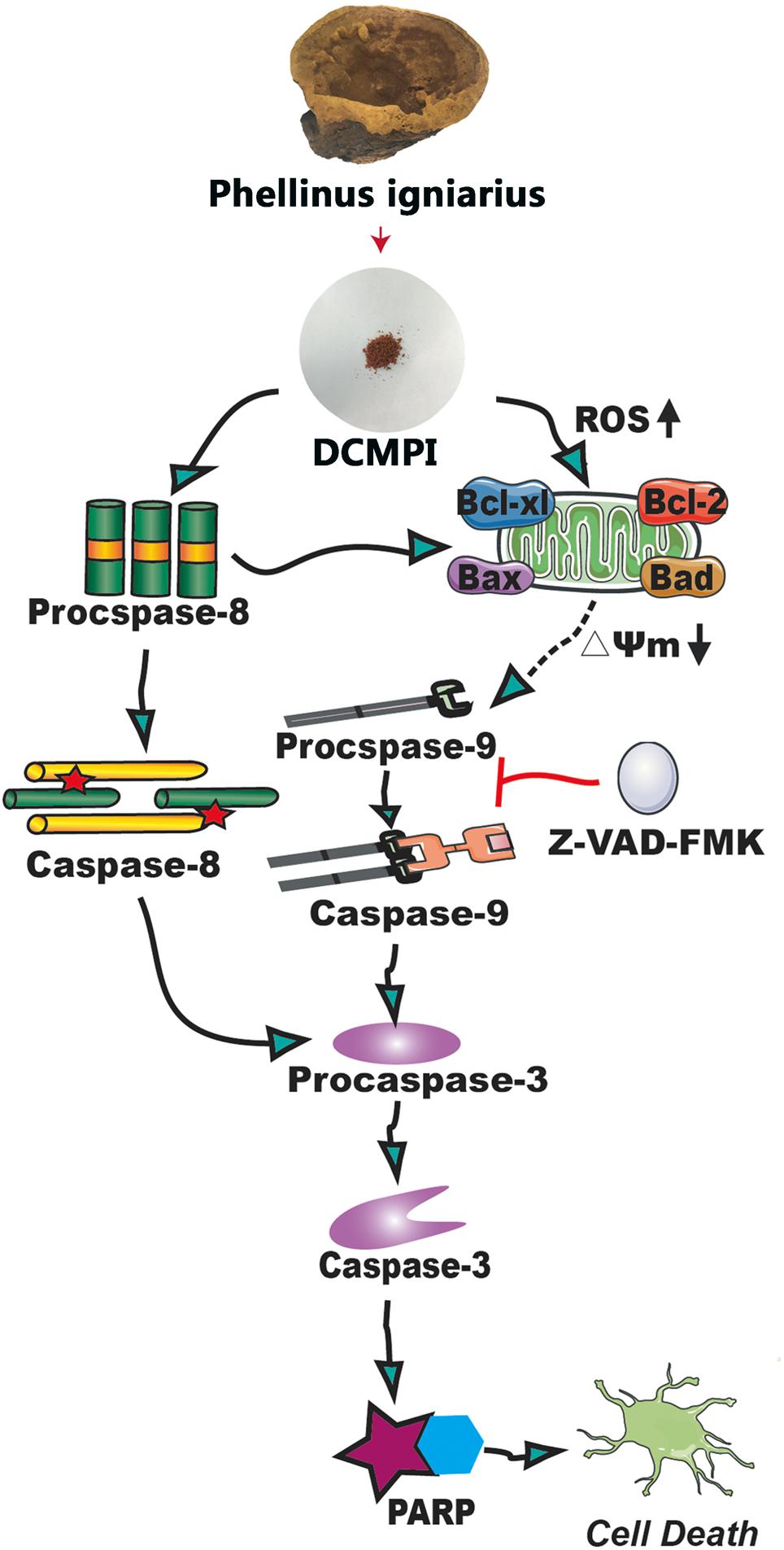
Figure 9. Molecular mechanisms underlying the antitumor activity of DCMPI. DCMPI increased intracellular ROS levels and induced HT-29 cell apoptosis mainly mediated via the mitochondrial apoptosis pathway.
In the current study, we first demonstrated that the DCMPI dramatically suppressed HT-29 colon cancer cell proliferation. Then, we characterized 19 main constituents of DCMPI by UPLC-ESI-Q/TOF-MS, and quantified 5 of them. Furthermore, network pharmacology analysis focusing on the 8 high content compounds of DCMPI (different from other fractions) was performed, revealing that the potential targets were mainly associated with the positive regulation of ROS metabolic processes and intrinsic apoptotic pathways. Finally, cellular tests verified that DCMPI increased intracellular ROS levels to induce HT-29 cell apoptosis. Taken together, our results provide the antitumor chemical composition of and mechanistic insights into P. igniarius, which may be exploited as a promising therapeutic option for colon cancer.
YD and PQ wrote the manuscript. YD, PQ, RZ, LZ, PZ, and YW conducted the research and analyzed the data. CL, KC, DS, and HZ conceived or designed the studies. All authors read and approved the final manuscript.
This research was funded by National Natural Science Foundation of China (Nos. 81603349 and 81673809) and Key Research and Development Plan of Zhejiang Province (No. 2015C03033).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The reviewer YZhu declared a shared affiliation, with no collaboration, with several of the authors to the handling Editor at the time of review.
BP, biological process; BPI, base peak intensity; CC, cellular component; DAPI, 4’,6-diamidino-2-phenylindole; DEGs, differentially expressed genes; DMSO, dimethyl sulfoxide; ECL, enhanced chemiluminescence; GAD, genetic association database; GEO, gene expression omnibus database; GO, gene ontology; HMDB, Human Metablome Database; MF, molecular function; MMP, mitochondrial membrane potential; MTT, 3-[4,5-dimethylthiazol-2-yl]-2,5-diphenyltetrazolium bromide; OMIM, online mendelian inheritance in man; PARP, poly ADP-ribose polymerase; PBS, phosphate buffer saline; P. igniarius, Phellinus igniarius; P. igniarius DCM, dichloromethane extract of Phellinus igniarius; PPI, protein–protein interaction; ROS, reactive oxygen species; SDS-PAGE, sodium dodecyl sulfate polyacrylamide gel electrophoresis; SEA, similarity ensemble approach; TCM, traditional Chinese medicine; TCMSP, traditional Chinese medicine systems pharmacology; TTD, therapeutic target database; UPLC-ESI-Q/TOF-MS, ultra performance liquid chromatography-electrospray ionization- quadrupole/time-of-flight mass spectrometry.
Borkan, S. C. (2016). The role of BCL-2 family members in acute kidney injury. Semin. Nephrol. 36, 237–250. doi: 10.1016/j.semnephrol.2016.04.001
Cao, H., Li, S., Xie, R., Xu, N., Qian, Y., Chen, H., et al. (2018). Exploring the mechanism of dangguiliuhuang decoction against hepatic fibrosis by network pharmacology and experimental validation. Front. Pharmacol. 9:187. doi: 10.3389/fphar.2018.00187
Chang, H. L., Chang, Y. M., Lai, S. C., Chen, K. M., Wang, K. C., Chiu, T. T., et al. (2017). Naringenin inhibits migration of lung cancer cells via the inhibition of matrix metalloproteinases-2 and -9. Exp. Ther. Med. 13, 739–744. doi: 10.3892/etm.2016.3994
Czabotar, P. E., Lessene, G., Strasser, A., and Adams, J. M. (2014). Control of apoptosis by the BCL-2 protein family: implications for physiology and therapy. Nat. Rev. Mol. Cell Biol. 15, 49–63. doi: 10.1038/nrm3722
De Silva, D. D., Rapior, S., Fons, F., Bahkali, A. H., and Hyde, K. D. (2012). Medicinal mushrooms in supportive cancer therapies: an approach to anti-cancer effects and putative mechanisms of action. Fungal Divers. 55, 1–35. doi: 10.1007/s13225-012-0151-3
Dong, Y., He, Y., Yu, Z. M., Zhang, Y., Wang, N. N., Shou, D., et al. (2016). Metabolomic investigation of rat serum following oral administration of the willow bracket medicinal mushroom, phellinus igniarius (Agaricomycetes), by UPLC-HDMS. Int. J. Med. Mushrooms 18, 699–711. doi: 10.1615/IntJMedMushrooms.v18.i8.60
Dong, Y., Wang, H., Zhang, Y., An, N., Zhang, Y., and Shou, D. (2015). Ultra high performance liquid chromatography with synapt high-definition mass spectrometry and a pattern recognition approach to characterize chemical constituents and rat metabolites after the oral administration of Phellinus igniarius. J. Sep. Sci. 38, 1137–1148. doi: 10.1002/jssc.201401293
Gargano, M. L., van Griensven, L. J. L. D., Isikhuemhen, O. S., Lindequist, U., Venturella, G., Wasser, S. P., et al. (2017). Medicinal mushrooms: valuable biological resources of high exploitation potential. Plant Biosyst. 151, 548–565. doi: 10.1080/11263504.2017.1301590
Goldar, S., Khaniani, M. S., Derakhshan, S. M., and Baradaran, B. (2015). Molecular mechanisms of apoptosis and roles in cancer development and treatment. Asian Pac. J. Cancer Prev. 16, 2129–2144. doi: 10.7314/APJCP.2015.16.6.2129
Guo, Y., Nie, Q., Maclean, A. L., Li, Y., Lei, J., and Li, S. (2017). Multiscale modeling of inflammation-induced tumorigenesis reveals competing oncogenic and oncoprotective roles for inflammation. Cancer Res. 77, 6429–6441. doi: 10.1158/0008-5472.CAN-17-1662
Hare, J. I., Lammers, T., Ashford, M. B., Puri, S., Storm, G., and Barry, S. T. (2017). Challenges and strategies in anti-cancer nanomedicine development: an industry perspective. Adv. Drug Deliver. Rev. 108, 25–38. doi: 10.1016/j.addr.2016.04.025
Huang, T., Ning, Z., Hu, D., Zhang, M., Zhao, L., Lin, C., et al. (2018). Uncovering the mechanisms of chinese herbal medicine (mazirenwan) for functional constipation by focused network pharmacology approach. Front. Pharmacol. 9:270. doi: 10.3389/fphar.2018.00270
Janssen, O. (2001). Staurosporine and conventional anticancer drugs induce overlapping, yet distinct pathways of apoptosis and caspase activation. Oncogene 20, 1193–1202. doi: 10.1038/sj.onc.1204221
Jeon, T. I., Jung, C. H., Cho, J. Y., Park, D. K., and Moon, J. H. (2013). Identification of an anticancer compound against HT-29 cells from Phellinus linteus grown on germinated brown rice. Asian Pac. J. Trop. Biomed. 3, 785–789. doi: 10.1016/S2221-1691(13)60156-2
Khan, K. H., Blancocodesido, M., and Molife, L. R. (2014). Cancer therapeutics: targeting the apoptotic pathway. Crit. Rev. Oncol. Hemat. 90, 200–219. doi: 10.1016/j.critrevonc.2013.12.012
Kim, B. M., and Hong, S. H. (2011). Sequential caspase-2 and caspase-8 activation is essential for saikosaponin a-induced apoptosis of human colon carcinoma cell lines. Apoptosis 16, 184–197. doi: 10.1007/s10495-010-0557-x
Kim, B. Y., Song, K. H., Lim, C. Y., and Cho, S. I. (2017). Therapeutic properties of scutellaria baicalensis in db/db mice evaluated using connectivity map and network pharmacology. Sci. Rep. 7:41711. doi: 10.1038/srep41711
Kuranaga, E. (2012). Beyond apoptosis: caspase regulatory mechanisms and functions in vivo. Genes Cells 17, 83–97. doi: 10.1111/j.1365-2443.2011.01579.x
Lee, I. K., and Yun, B. S. (2011). Styrylpyrone-class compounds from medicinal fungi phellinus and inonotus spp. and their medicinal importance. J. Antibiot. 64, 349–359. doi: 10.1038/ja.2011.2
Li, S., and Zhang, B. (2013). Traditional chinese medicine network pharmacology: theory, methodology and application. Chin. J. Nat. Med. 11, 110–120. doi: 10.3724/SP.J.1009.2013.00110
Li, X., Wu, Q., Xie, Y., Ding, Y. R., Du, W. W., Sdiri, M., et al. (2015). Ergosterol purified from medicinal mushroom Amauroderma rude inhibits cancer growth in vitro and in vivo by up-regulating multiple tumor suppressors. Oncotarget 6, 17832–17846. doi: 10.18632/oncotarget.4026
Liang, X., Li, H., and Li, S. (2014). A novel network pharmacology approach to analyse traditional herbal formulae: the Liu-Wei-Di-Huang pill as a case study. Mol. BioSyst. 10, 1014–1022. doi: 10.1039/c3mb70507b
Liu, B., Sun, R., Luo, H., Liu, X., Jiang, M., Yuan, C., et al. (2017). Both intrinsic and extrinsic apoptotic pathways are involved in Toll-like receptor 4 (TLR4)-induced cell death in monocytic THP-1 cells. Immunobiology 222, 198–205. doi: 10.1016/j.imbio.2016.10.002
Liu, X., Xiang, M., Fan, Y., Yang, C., Zeng, L., Zhang, Q., et al. (2017). A root-preferential DFR-Like gene encoding dihydrokaempferol reductase involved in anthocyanin biosynthesis of purple-fleshed sweet potato. Front. Plant Sci. 8:279. doi: 10.3389/fpls.2017.00279
Ma, X., dan Guo, D., Peterson, E. C., Dun, Y., and Li, D. Y. (2016). Structural characterization and anti-aging activity of a novel extracellular polysaccharide from fungus Phellinus sp. in a mammalian system. Food Funct. 7, 3468–3479. doi: 10.1039/c6fo00422a
Nuñez, G., Benedict, M. A., Hu, Y., and Inohara, N. (1998). Caspases: the proteases of the apoptotic pathway. Oncogene 17, 3237–3245. doi: 10.1038/sj.onc.1202581
Piska, K., Sułkowska-Ziaja, K., and Muszyńska, B. (2017). Edible mushroom pleurotus ostreatus (Oyster mushroom)-Its dietary significance and biological activity. Acta Sci. Pol Hortoru. 16, 151–161.
Renschler, M. F. (2004). The emerging role of reactive oxygen species in cancer therapy. Eur. J. Cancer 40, 1934–1940. doi: 10.1016/j.ejca.2004.02.031
Sangdee, K., Buranrat, B., Naksuwankul, K., Jaihan, P., and Sangdee, A. (2017). Antibacterial and anti-breast cancer cell line activities of Sanghuangporus sp. 1 extracts. Trop. J. Pharm. Res. 16, 613–620. doi: 10.4314/tjpr.v16i3.16
Sankari, S. L., Masthan, K. M., Babu, N. A., Bhattacharjee, T., and Elumalai, M. (2012). Apoptosis in cancer–an update. Asian Pac. J. Cancer Prev. 13, 4873–4878. doi: 10.7314/APJCP.2012.13.10.4873
Shou, D., Dong, Y., Wang, N. N., Li, H. Y., Zhang, Y., and Zhu, Y. (2016). Simultaneous quantification of antioxidant compounds in phellinus igniarius using ultra performance liquid chromatography-photodiode array detection-electrospray ionization tandem mass spectrometry. PLoS One 11:e0163797. doi: 10.1371/journal.pone.0163797
Siegel, R. L., Miller, K. D., and Jemal, A. (2016). Cancer statistics, 2016. CA Cancer J. Clin. 66, 7–30. doi: 10.3322/caac.21332
Song, T. Y., Lin, H. C., Yang, N. C., and Hu, M. L. (2008). Antiproliferative and antimetastatic effects of the ethanolic extract of Phellinus igniarius (Linnearus: Fries) quelet. J. Ethnopharmacol. 115, 50–56. doi: 10.1016/j.jep.2007.09.001
Wang, X., Yu, S., Jia, Q., Chen, L., Zhong, J., Pan, Y., et al. (2017). Niaoduqing granules relieve chronic kidney disease symptoms by decreasing renal fibrosis and anemia. Oncotarget 8, 55920–55937. doi: 10.18632/oncotarget.18473
Wu, C. S., Lin, Z. M., Wang, L. N., Guo, D. X., Wang, S. Q., Liu, Y. Q., et al. (2011). Phenolic compounds with NF-κB inhibitory effects from the fungus phellinus baumii. Bioorg. Med. Chem. Lett. 21, 3261–3267. doi: 10.1016/j.bmcl.2011.04.025
Yang, Q., Hu, X. G., Wan, Q., and Wang, W. Q. (2006). Study on anti-tumor effect of medicinal fungi Phellinus igniarius extracts. Chin. J. Chinese Mater. Med. 31, 1713–1715.
Zhang, Q., Yu, H., Qi, J., Tang, D., Chen, X., Wan, J. B., et al. (2017). Natural formulas and the nature of formulas: exploring potential therapeutic targets based on traditional Chinese herbal formulas. PLoS One 12:e0171628. doi: 10.1371/journal.pone.0171628
Zhou, C., Jiang, S. S., Wang, C. Y., Li, R., and Che, H. L. (2014). Different immunology mechanisms of Phellinus igniarius in inhibiting growth of liver cancer and melanoma cells. Asian Pac. J. Cancer Prev. 15, 3659–3665. doi: 10.7314/APJCP.2014.15.8.3659
Keywords: Phellinus igniarius, antitumor, phytochemistry, network pharmacology, effective substances, mitochondrial apoptosis pathway
Citation: Dong Y, Qiu P, Zhu R, Zhao L, Zhang P, Wang Y, Li C, Chai K, Shou D and Zhao H (2019) A Combined Phytochemistry and Network Pharmacology Approach to Reveal the Potential Antitumor Effective Substances and Mechanism of Phellinus igniarius. Front. Pharmacol. 10:266. doi: 10.3389/fphar.2019.00266
Received: 28 June 2018; Accepted: 04 March 2019;
Published: 19 March 2019.
Edited by:
Shao Li, Tsinghua University, ChinaReviewed by:
Wentzel Christoffel Gelderblom, Cape Peninsula University of Technology, South AfricaCopyright © 2019 Dong, Qiu, Zhu, Zhao, Zhang, Wang, Li, Chai, Shou and Zhao. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Dan Shou, c2hvdWRhbm9rQDE2My5jb20= Huajun Zhao, emhqQHpjbXUuZWR1LmNu
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.