- 1Institute of Medicinal Plant Development, Chinese Academy of Medical Sciences, Peking Union Medical College, Beijing, China
- 2Key Laboratory of Bioactive Substances and Resources Utilization of Chinese Herbal Medicine (Peking Union Medical College), Ministry of Education, Beijing, China
- 3Engineering Research Center of Traditional Chinese Medicine Resources, Ministry of Education, Beijing, China
The stems and roots of Kadsura species have been used as the folk medicine in Traditional Chinese medicine (TCM) and have good traditional efficacy and medicinal application with a long history. Among these species, K. coccinea, K. heteroclita and K. longipedunculata are the most widely distributed species in the regions of south and southwest China. Owing to their similar appearance, the crude drugs are often confusedly used by some folk doctors, even some pharmaceutical factories. To discriminate the crude drugs, haplotype analysis based on cpDNA markers and ITS was firstly employed in this study. Generic delimitation, interspecific interrelationships, and the identification of medicinal materials between K. longipedunculata and K. heteroclita remained unresolved by the existing molecular fragments. The original plant could be identified through the morphological character of flower, fruit and leaf. However, in most situation collectors have no chance to find out these characters due to lack of reproductive organs, and have no experience with the minor difference and transitional variation of leaf morphology. The chemical characterization show that the chemometric of chemical composition owned higher resolution to discriminate three herbs of Kadsura species. In conclusion, this integrative approach involving molecular phylogeny, morphology and chemical characterization could be applied for authentication of the Kadusra. Our study suggests the use of this comprehensive approach for accurate characterization of this closely related taxa as well as identifying the source plant and confused herbs of TCM.
Introduction
Traditional Chinese medicine (TCM) is widely accepted in the health care system, and has made a significant contribution to prevention and treatment of human diseases. This extensive use warrants safety measures and so TCM drug safety monitoring and quality control are becoming increasingly important tasks to guarantee the safety and efficacy of TCM treatments (Chen et al., 1999). However, the use of substitute products and confused materials still aggravate the chaotic situation in clinical application. It is important to find a reliable way for distinguishing them from each other (Li et al., 2015).
Kadsura belongs to the economically and medicinally important family Schisandraceae and eight species mainly distributed in the southwest and southeast in China (Saunders, 1998; Wu et al., 2008). In China, the stems and roots of genus Kadsura are commonly used as folk medicines and 5 species of genus Kadsura are documented in the official Pharmacopoeia and folk record (GuangXi Zhuang Autonomous Region Health Department, 1992; Guangdong Food and Drug Administration, 2004; FuJian Food and Drug Administration, 2006; Chinese Pharmacopeia Commission, 2015). The stems of K. heteroclita, K. longipedunculata and K. coccinea are the most widely used in south China which have good traditional efficacy and medicinal application with a long history, and often used confusedly in clinical application. Numerous phytochemical and pharmacological studies have been carried out and focus on its health benefits (Liu et al., 2014). There are differences in clinical efficacy between the three crude drugs. K. heteroclita was used for the treatment of rheumatic arthralgia. K. longipedunculata was used for the treatment of irregular menstruation. K. coccinea was used for the treatment of gastric and duodenal ulcer. Owing to their similar appearance, these crude drugs are often confusedly used by some folk doctors, even some pharmaceutical factories. There is an urgent need to find a reliable, accurate way for distinguishing three Kadsura crude drugs (Liu et al., 2012).
In order to identify this kind of medicine herbs by molecular sequences, Zhou et al. chose psbA-trnH for distinguishing eight species. Although they found a stabilized single nucleotide polymorphism (SNP), SNPs as potential tool to distinguish Kadsrua crude drugs could not be further analyzed because of poor samples of K. heteroclite (Zhou et al., 2016). Combination of ITS + psbA-trnH + matK + rbcL as the most ideal DNA barcode for discriminating the medicinal plants of Schisandra and Kadsura, nonetheless, degree of species resolution was lower among the closely related species, and exposed K. heteroclita and K. longipedunculata could not be discriminated by four commonly used DNA barcodes (Zhang et al., 2015). Molecular identification of TCM is objective, more accurate, and easier to perform than traditional identification methods, and has successfully been applied to identify medicinal plants (Kress et al., 2005; Chen et al., 2010; de Boer et al., 2015). However, previous studies of DNA barcoding have not effectively resolved the problem of identifying three crude drugs of the Kadsura. In this study, we try to identify three crude drugs by haplotype analysis based on cpDNA and ITS markers.
The high selectivity and sensitivity of UPLC-QTOF/MS has been successfully applied to the metabolite analysis and identification of complex compounds in herbal materials (Song et al., 2013; Yao et al., 2013; Cubero-Leon et al., 2014). There are few researches in investigated chemical profiles of Kadsura species for the safety and efficacy, and the comparative analysis on chemical composition of these Kadsura herbs is needed. Bioactivity-based characteristics are good quality indicators too, as they are pharmacologically relevant (Liu et al., 2017). Analysis on main chemical components disparity in three medicinal materials could guarantee the clinical uses the medicine the rationality, the security and the validity.
Adequately considering samples representativeness and experiments economy, we therefore started multi-populations survey in twenty populations covering five provinces including Hunan, Guangxi, Guizhou, Chongqing and Sichuan provinces and in reproductive stage at summer and autumn from June 2016 to December 2017. The overall aim of this study was to explore the usefulness of an authentication approach to three crude drugs of the Kadsura complex using cpDNA and ITS markers, morphology, and UPLC-QTOF/MS chemical profiling. We want to compare the genetic polymorphism of haplotypes and analyze their population difference for the taxa in the Kadsura species, and distinguish chemotypes of the species complex by comparing their UPLC-QTOF/MS chemical profiles using chemometric data analysis.
Materials and Methods
Plant Materials
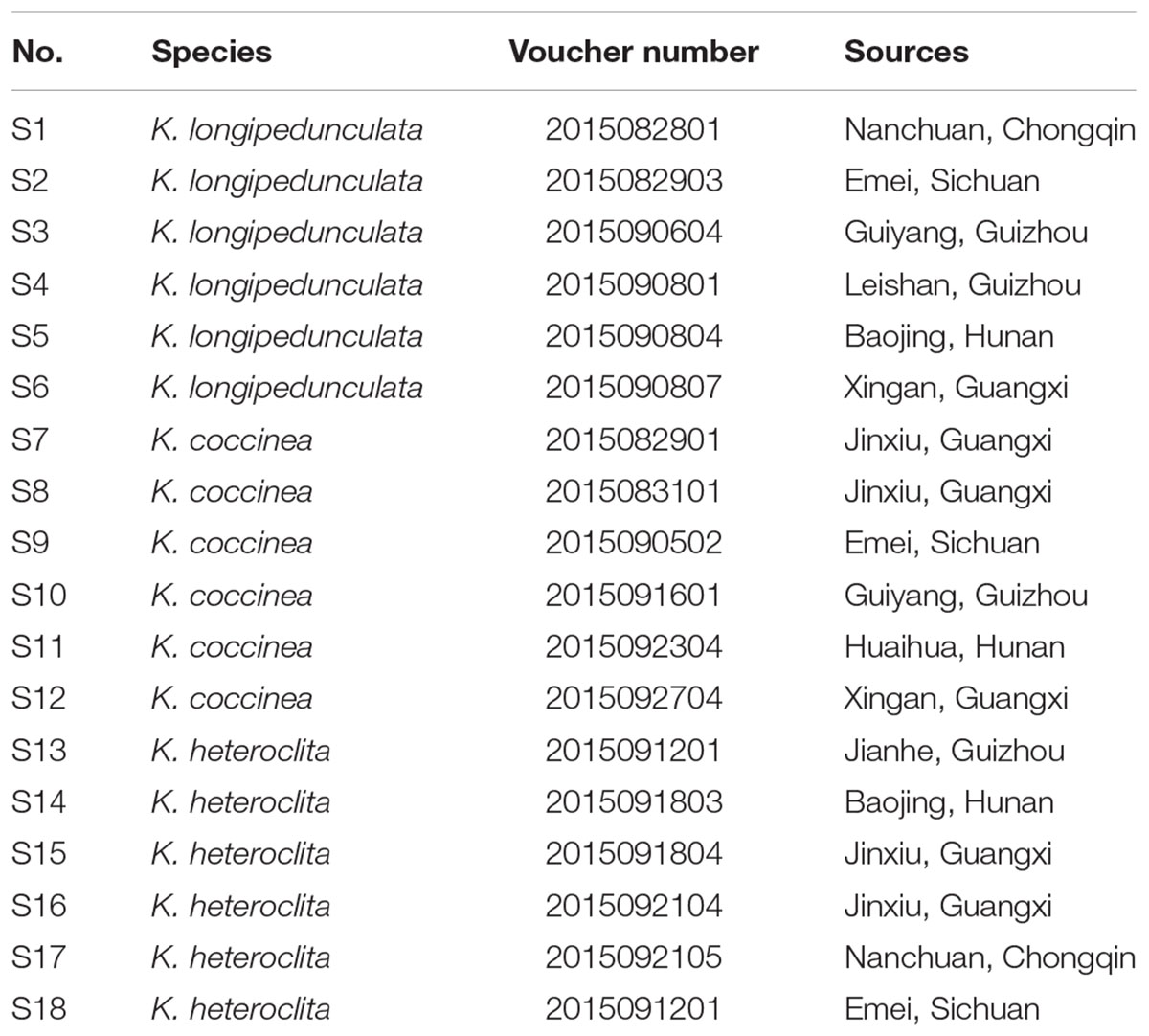
There are morphological differences between the three species to discriminate them during flower or fruit stages of life cycles. For the sake of sampling accuracy, we therefore started multi-populations survey in reproductive stage at summer and autumn from June 2016 to December 2017. The Kadsura samples were collected in the main areas in China: Hunan, Guangxi, Guizhou, Chongqing and Sichuan provinces. In total 52 samples of K. coccinea, K. heteroclita and K. longipedunculata were collected directly from wild region. The 52 leaves dried using silica gel for DNA extraction and stored at 4°C until use. And 18 the stems dried in the shade for UPLC-MS analysis (Table 1).
DNA Extraction, PCR Amplification and Sequencing
Total genomic DNA was extracted from silica gel-dried leaves by using the Plant Genomic DNA Kit (Tiangen Biotech, Beijing, China) following the manufacturer’s instructions. Three cpDNA gene markers, matK,rbcL,psbA-trnH and one nrDNA ITS, were separately amplified for each individual by using the primers and protocol of Guo et al. (2015). Sanger sequence reactions were carried out using the DYEnamic ETDye Terminator Cycle Sequencing Kit (Amersham Pharmacia Biotech) and sequenced on ABI 3730XL genetic analyzer (Applied Biosystems, CA, United States).
Network Analysis of Haplotypes
The DNA sequences were aligned using the program Clustal X v.1.83 (Thompson et al., 1997) and manually adjusted in BioEdit v. 7.0.9 (Hall, 1999). Voucher and GenBank accession numbers were listed in the Supplementary Table S1. A network of the cpDNA haplotypes (chlorotypes) was constructed using NETWORK 5.0.0.1 (Bandelt et al., 1999), with a default parsimony connection limit of 95% and each insertion/deletion (indel) treated as a single mutation event.
Sample Preparation and UPLC-QTOF/MS Conditions
HPLC-grade acetonitrile (Merck KGaA, Darmstadt, Germany) and formic acid (Fisher Scientific, NH, United States) were utilized for UPLC analysis. Pure water (18.2 MΩ) for UPLC analysis was obtained from a Milli-Q system (Millipore, MA, United States). All other chemicals were of analytical grade.
Kadsura samples (0.5000 g, 65-mesh) were accurately weighed and extracted with 25 mL methanol by ultrasonication (35 kHz) for 30 min. After centrifugation at 10,000 ×g for 10 min, the supernatant was stored at 4°C and filtered through 0.22 μm membrane prior to injection into the UPLC system.
A Thermo ScientificTM DionexTM UltiMateTM 3000 Rapid Separation LC (RSLC) system performed UHPLC separations using the gradient conditions as follows. Mobile phase A was water and mobile phase B was acetonitrile; both A and B contained 0.1% formic acid. The conditions were optimized as follows: 0–3 min, 2–20% B; 3–4.5 min, 20–75% B; 4.5–6.5 min, 75–100% B; 6.5–15 min, 100% B; 15–15.5 min, 100–5% B; 15.5–17 min, 5% B. The column was a HSS T3 column (2.1 mm × 100 mm, 1.7 μm, waters) operated at 45°C. The flow rate was 300 μL/min and the injection volume was 2 μL.
A Thermo ScientificTM Q ExactiveTM hybrid quadrupole Orbitrap mass spectrometer equipped with a HESI-II probe was employed. The HESI-II spray voltages were 3.7 kV for positive mode, the heated capillary temperature was 320°C, the sheath gas pressure was 30 psi, the auxiliary gas setting was 10 psi, and the heated vaporizer temperature was 300°C. Both the sheath gas and the auxiliary gas were nitrogen. The collision gas was argon at a pressure of 1.5 mTorr. The parameters of the full mass scan were as follows: a resolution of 70,000, an auto gain control target under 1 × 106, a maximum isolation time of 50 ms, and an m/z range 150–1500. The calibration was customized for the analysis of Q Exactive to keep the mass tolerance of 5 ppm. The LC-MS system was controlled using Xcalibur 2.2 SP1.48 software (Thermo Fisher Scientific), and data were collected and processed with the same software.
UPLC-QTOF/MS Data Analysis
UPLC-QTOF/MS data for Kadsura samples were analyzed to identify potential discriminant variables. Peak finding, alignment and filtering of ES raw data were carried out using Xcalibur 2.2 SP1.48 software (Thermo Fisher Scientific). The parameters used were as follows: retention time (tR) of 0.5–10.5 min, mass of 150–800 Da, retention time tolerance of 0.05 min, and mass tolerance of 0.02 Da. Three replicate samples collected from each geographic location were used (n = 3). A total of three, 114 variables were used to create the model. The resulting data was analyzed by heatmap analysis with MetaboAnalyst, which is a web-based tool for visualization of chemometrics (Deng et al., 2014). And principal component analysis (PCA) and partial least squares discriminant analysis (PLS-DA) were applied to discriminate three Kadsura species by the EZinfo 2.0 software (Masssart et al., 1998; Xia et al., 2012).
Results
Morphology
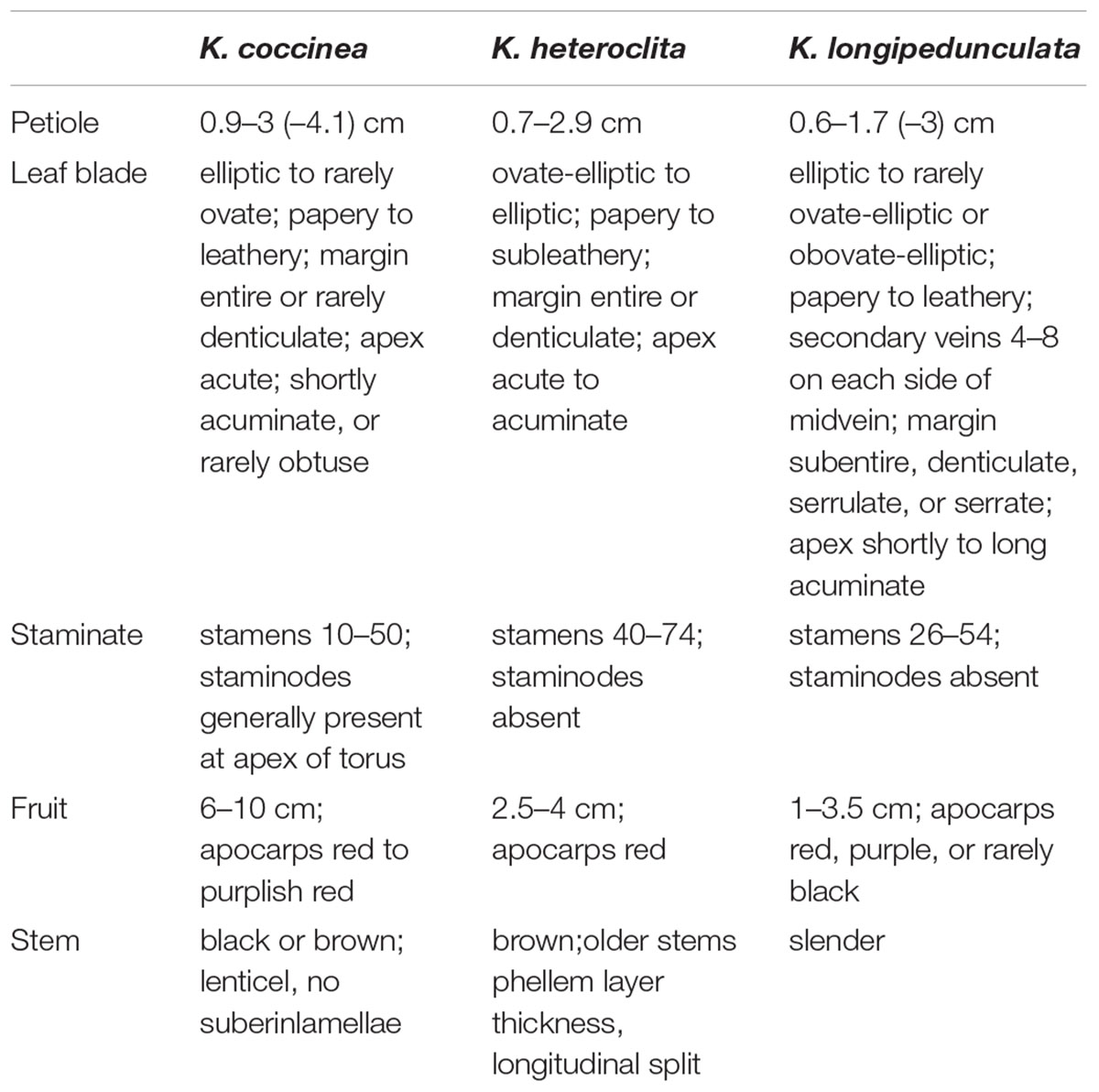
The variation of morphological traits in the Kadsura is relatively complex and the Kadsura in relationship was near with each other. There are still some morphological differences among three species. We observed main morphology characters (male flower, fruit shape, stem and leaf listed in Table 2) of K. coccinea, K. heteroclita and K. longipedunculata by specimens and natural populations, and established morphological basis to identify three Kadsura species. The morphology of male flower, fruit shape, stem and leaf are an important basis of identifying Kadsura (Figure 1). In most situations, collectors have no chance to find out these characters because of comparatively short flower and fruit time or lack of these organs in some habits and young individuals. Minor difference and transitional variation of leaf morphology make it difficult for those inexperienced collectors to identify. It is important to find a reliable, accurate way for distinguishing three crude drugs.
Haplotypes Network
The alignment of the combined three cpDNA fragments were designated 13 haplotypes (C1–C13) including 52 variable characters, and ITS were designated 29 haplotypes (H1–H29) with 49 variable characters. K. coccinea occupied eight private chloroplastic haplotypes (Figure 2A; C1–C8), while K. longipedunculata and K. heteroclita shared the three main haplotypes (C9, C11, and C12), two rare haplotypes C10 and C13 were fixed by K. heteroclita and K. longipedunculata, respectively. In the ITS haplotypes network (Figure 2B), K. coccinea occupied four private haplotypes which were quite different from the others (H1–H4). 17 haplotypes were fixed in K. longipedunculata, and seven haplotypes in K. heteroclita. Only one H17 was shared by K. longipedunculata and K. heteroclita. H17 was one of the main haplotypes of ITS and it was the center of the “star-like” haplotypes network of K. longipedunculata and K. heteroclita. It was obviously to see that both cpDNA markers and ITS can distinguish K. coccinea from K. longipedunculata and K. heteroclita clearly. However, the phylogenetic relationship between K. longipedunculata and K. heteroclita are quite closely related, their haplotypes are star-like and they shared the main haploptypes in both cpDNA markers and ITS. (Supplementary Table S2)
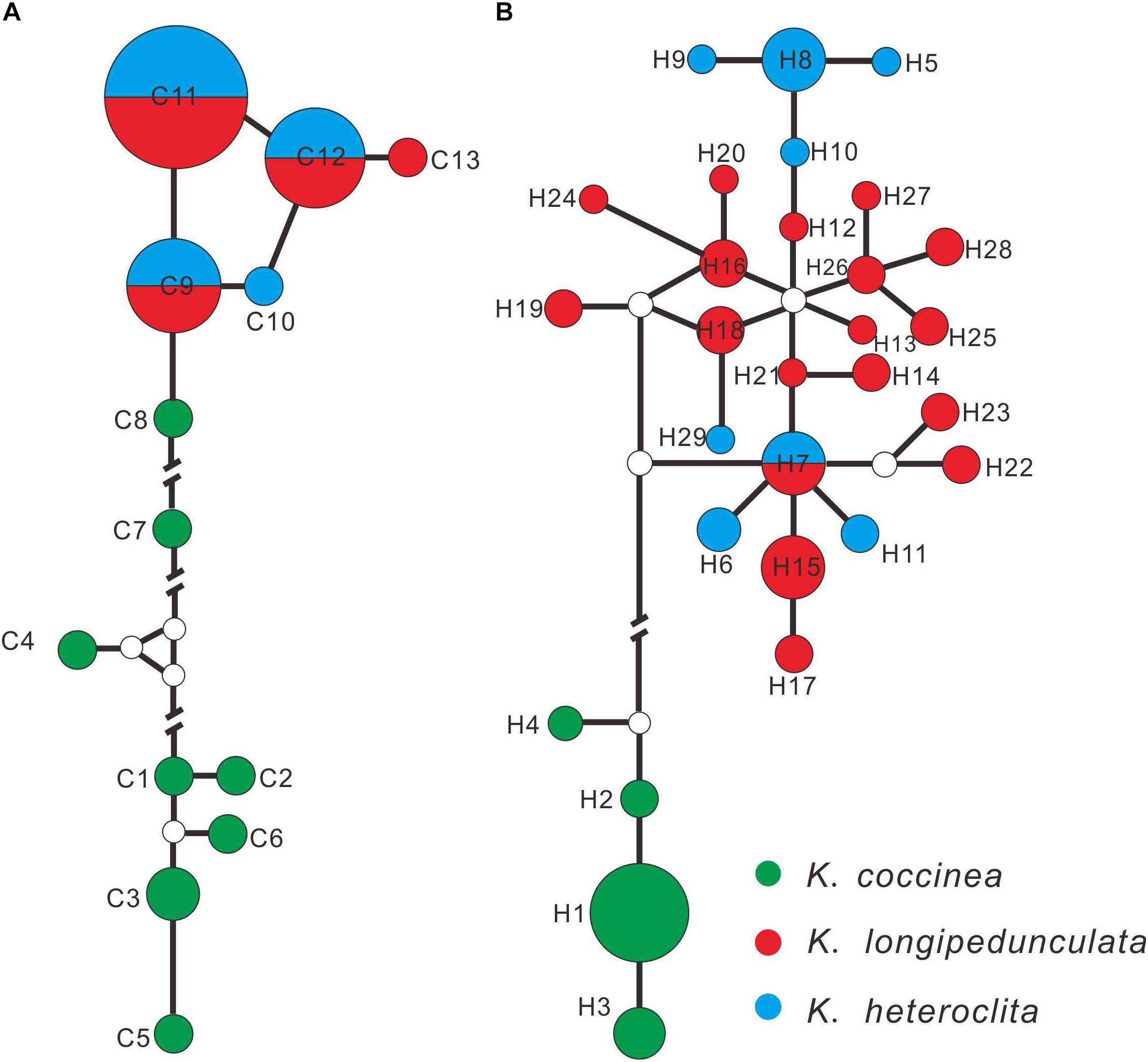
Figure 2. Networks of the cpDNA (A) and ITS (B) haplotypes constructed by using NETWORK 5.0.0.1. The sizes of the circles in the network are proportional to the observed frequencies of the haplotypes.
UPLC-QTOF/MS Analysis
Twenty compounds were tentatively identified by elucidating the retention time (min), parentions [M+H]+, MS/MS fragmentation pattern and calculated molecular formula of each peak, and by matching above data with those reported previously (Table 3). For example, schisandrin, in the low energy spectrum, the protonated adduct ion [M+H]+ at m/z 433.1866. Further confirmation of schisandrin was provided by the high-energy function. At m/z 415.2115 was detected the fragment identified as [M-H2O+H]+ and at m/z 384.1932 we assigned a fragment due to the further loss of methoxy group corresponding to [M-H2O-OCH3+H]+. It was identified to schisandrin based on the parent and characteristic fragmentions information.
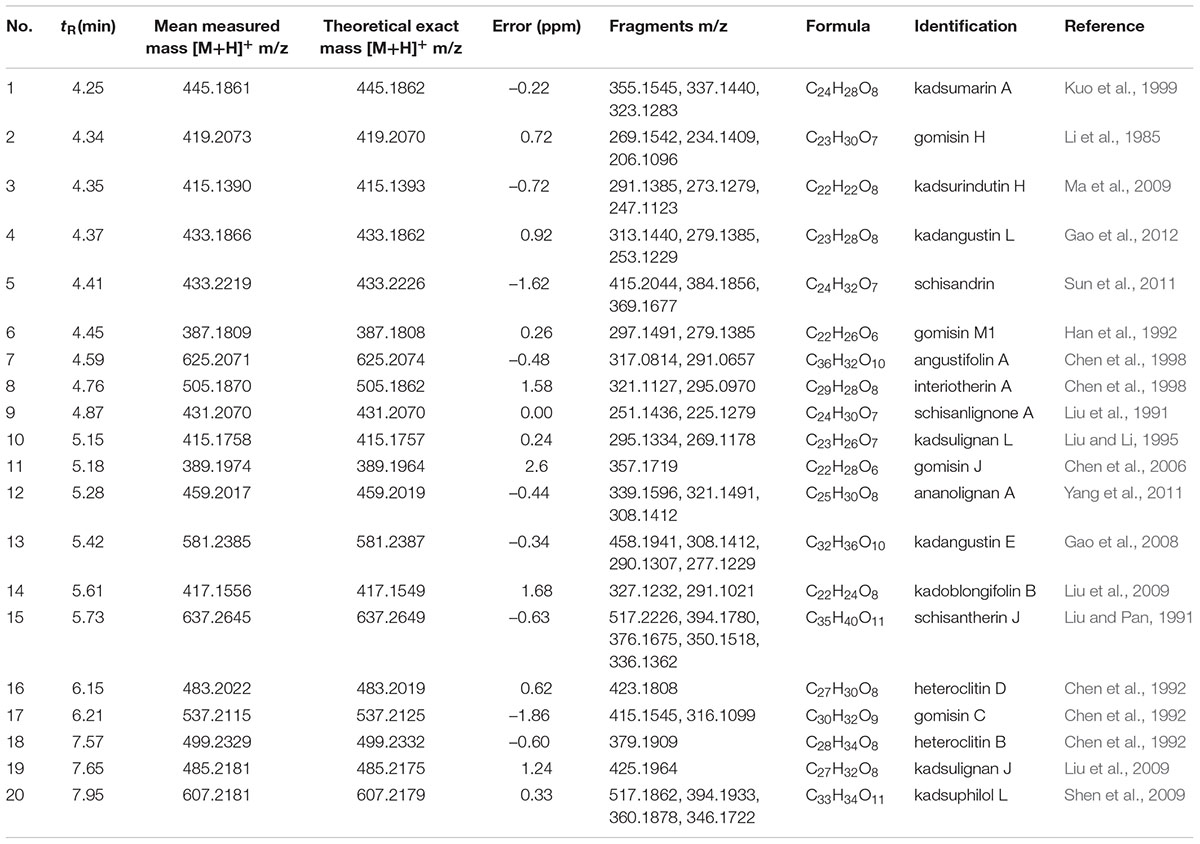
Table 3. Tentatively identified compounds from K. longipedunculata, K. heteroclita, and K. coccinea.
As depicted in Figure 3, we can observe that: (a) the chemical components of three Kadsura species were very different by the heatmaps, and the components of K. longipedunculata were close to K. heteroclita. (b) Among all the identified compounds, kadangustin L, gomisin H, and ananolignan A have a relatively high concentration in K. coccinea, while showing low levels in the K. heteroclita and K. longipedunculata. While kadangustin E, kadsumarin A, interiotherin A and kadoblongifolin B are present mainly in the K. longipedunculata, followed by the K. heteroclita and limited concentratins in the K. coccinea. Previous studies have suggested that these compounds are found in high concentrations in K. heteroclita, which is supported by our results (Liu et al., 2014).
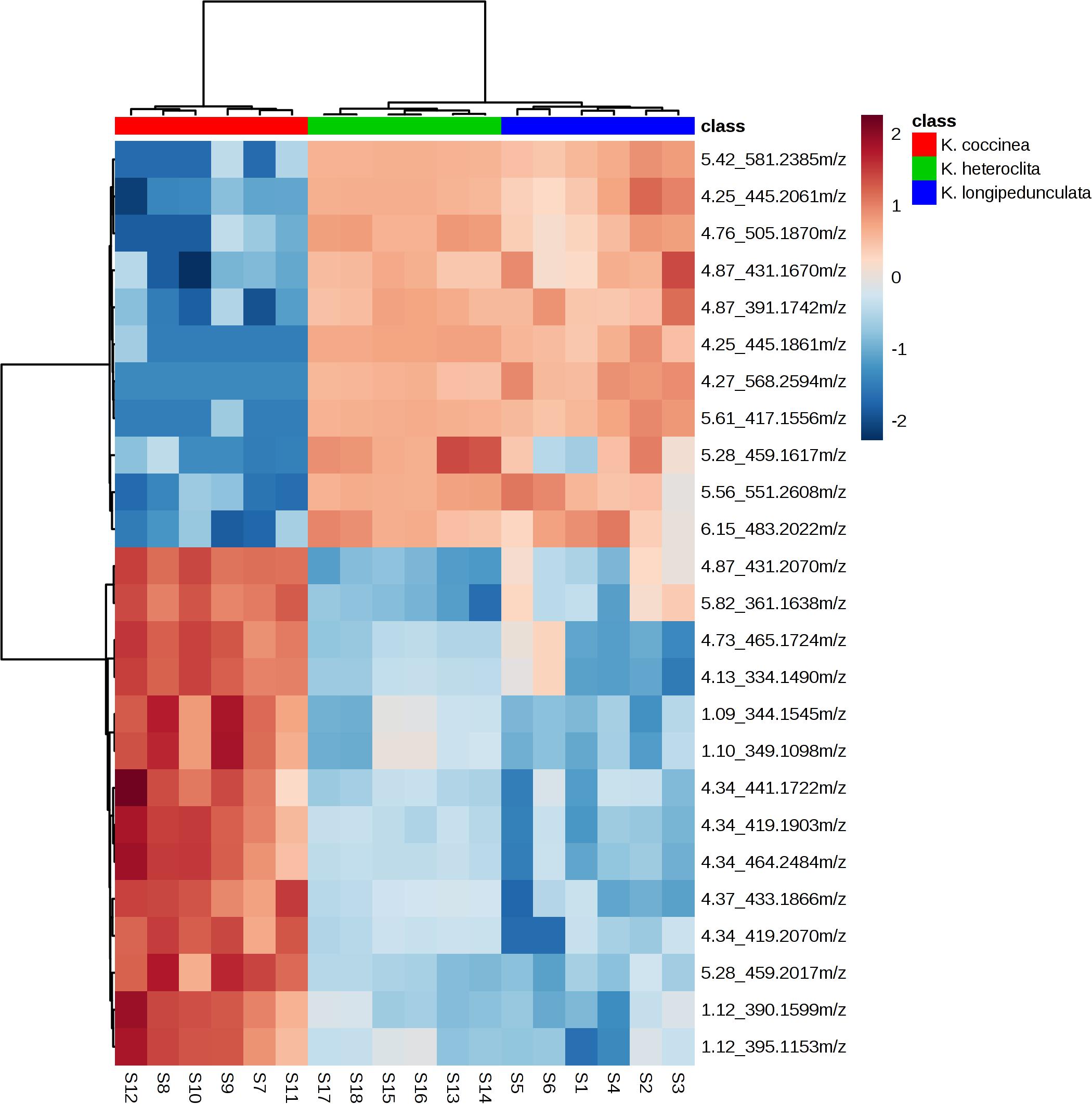
Figure 3. The heatmap presenting the distribution of constituents in K. longipedunculata, K. coccinea and K. heteroclita.
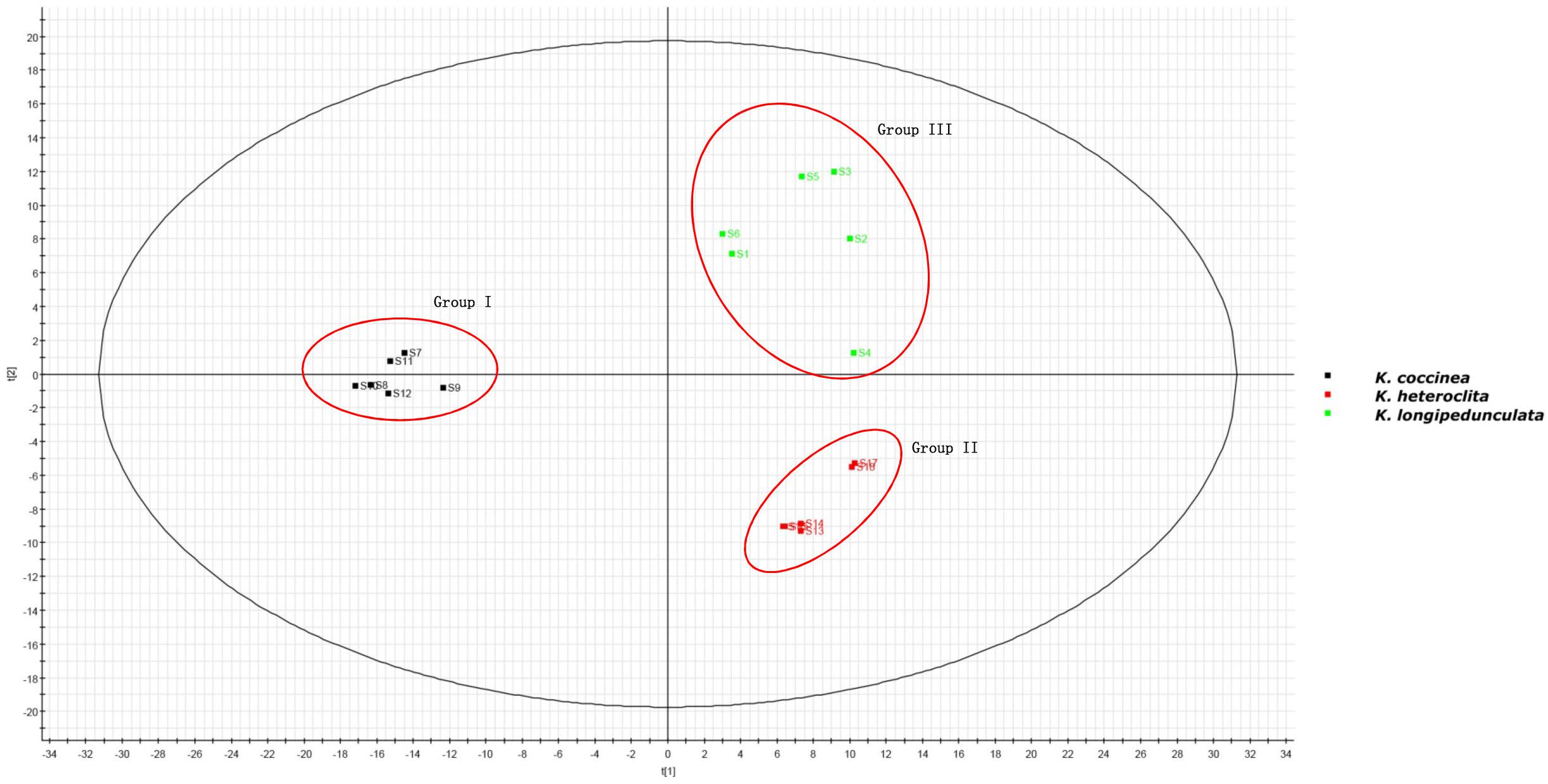
The two-component PCA model cumulatively accounted for 46.04% of the variation (PC1, 36.43%; PC2, 9.61%). The PCA score plot shows that these three species were obtained the very good separation. Group I was formed by K. coccinea, Group II consisted of K. heteroclita and Group III was formed by K. longipedunculata (Figure 4).
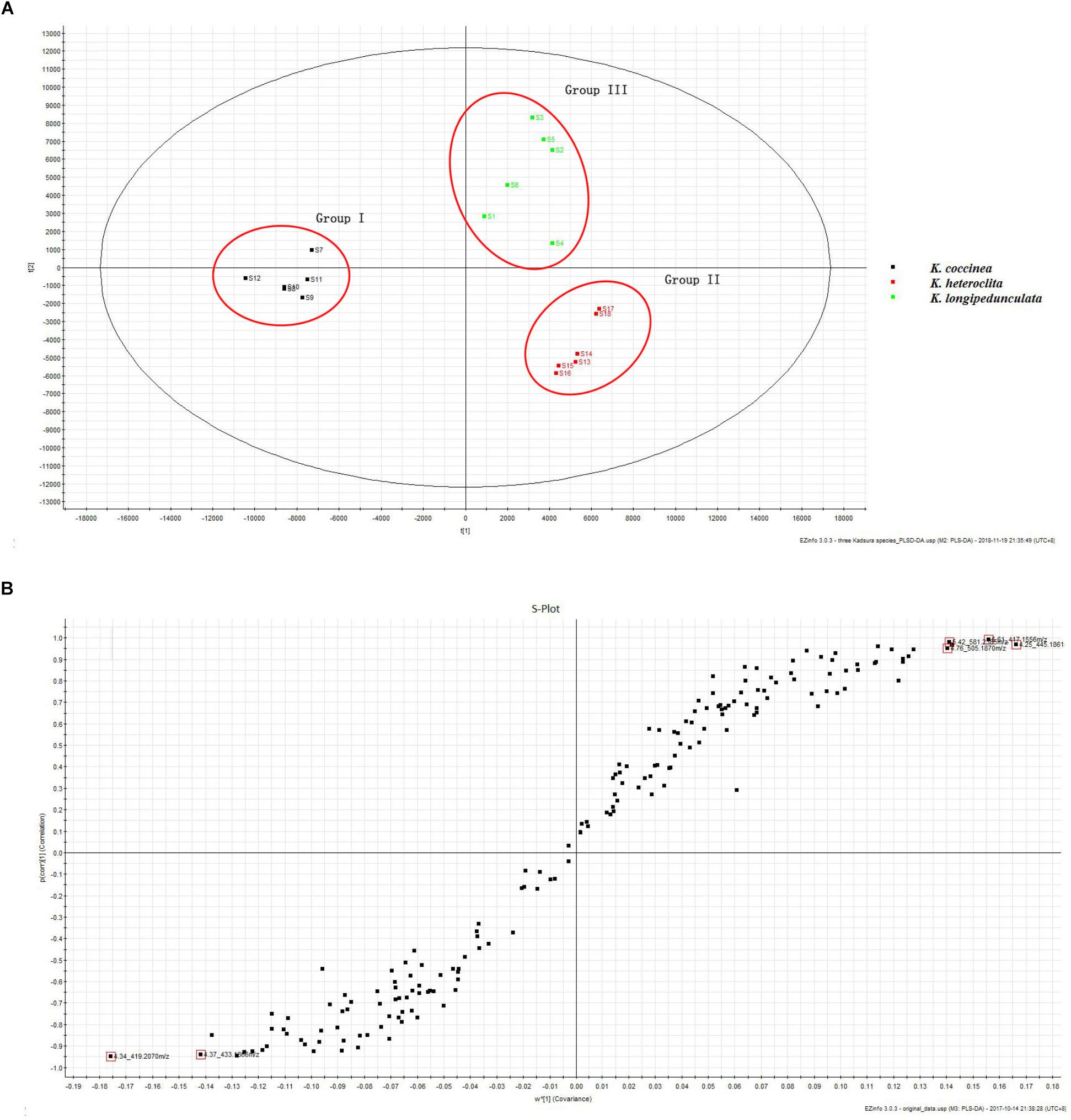
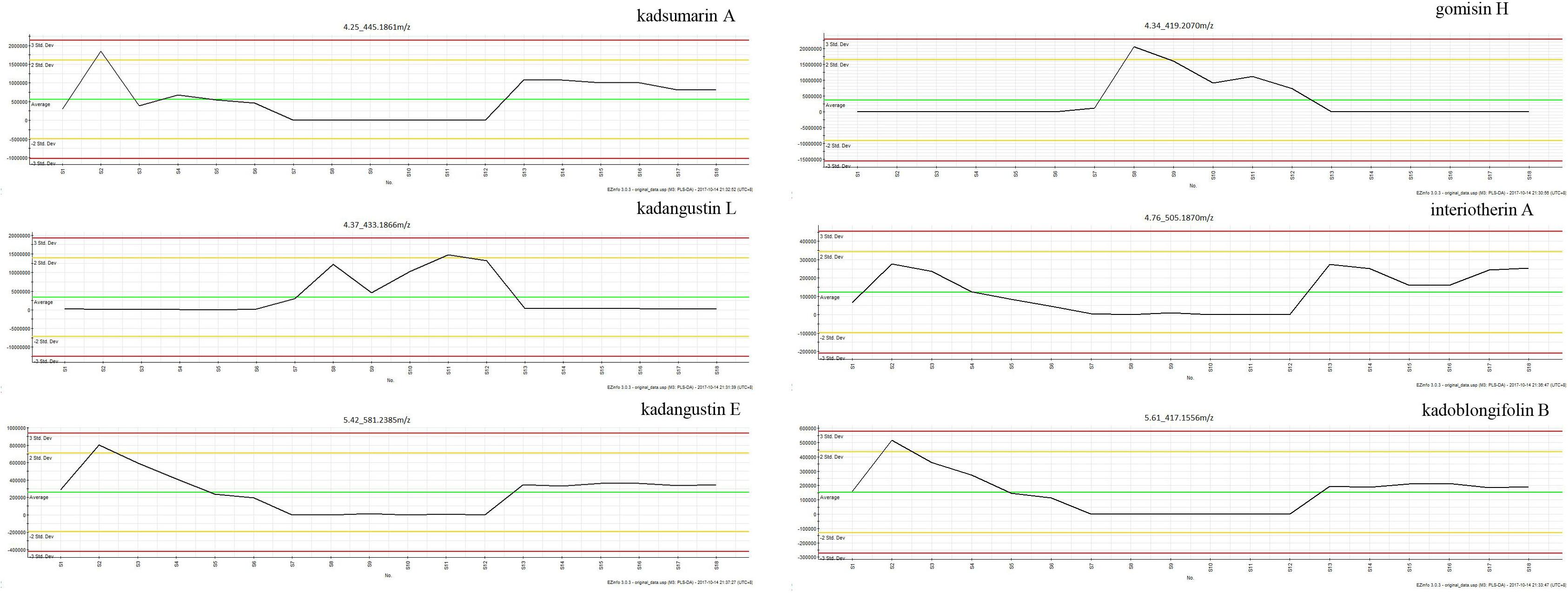
The PLS–DA model performed well in classifying the three species of Kadsura, and group I was far away from the group II and III (Figure 5A). A total of six credible and significant markers were determined to facilitate discrimination of these groups by the S-plot of PLS-DA. The identities of six potential markers were tentatively assigned (Figure 5B). The components correlated with these six ions were tentatively identified as isomers of kadsumarin A, gomisin H, kadangustin L, interiotherin A, kadangustin E, and kadoblongifolin B. The marker compounds could be used to distinguish the three plant species, as the ion intensities of kadsumarin A, kadoblongifolin B, and interiotherin A in K. heteroclita and K. longipedunculata was higher than in K. coccinea. Marker gomisin H and kadangustin L could be detected in K. coccinea, which was higher than in the other two species (Figure 6 and Supplementary Data Sheet S1).
Discussion
The three Kadsura species distribute widely in tropical or subtropical evergreen forests of south of Yangtze River in China. In fact, there are morphological differences between the three species to discriminate them during flower or fruit stages of life cycles. For the sake of sampling accuracy, we therefore started multi-populations survey in reproductive stage at summer and autumn from 2016 June to December 2017. Adequately considering samples representativeness and experiments economy, we chose following survey and sampling strategy. 80 individuals were observed in twenty populations covering five provinces including Hunan, Guangxi, Guizhou, Chongqing and Sichuan provinces, while 45 leaves samples were collected for DNA barcoding experiment and 18 stems samples were used for metabolites analysis, in which samples of each species included ten individuals (DNA test) and six individuals (Chemical test) from different populations.
Morphology
As mentioned above, we usually discriminate the three species by the shape of staminate flower torus, the size and shape of fruits, the length of fruit stalk and leaf shape. For example, the shape of staminate flower torus of K. coccinea is conical, K. heteroclite is elliptical and K. longipedunculata is spherical. The size of fruit is K. coccinea (6–10 cm) > K. heteroclita (2.5–4 cm) > K. longipedunculata (1–3.5 cm). These identifying characteristics also were record in FRPS and FOC (Academiae Sinicae Edita, 2004; Flora of China Edita, 2013). When we surveyed in wild populations, these morphological characters were very valuable to discriminate them. In spite of obvious differences between reproductive organs, in most situation collectors have no chance to find out these characters due to comparatively short flower and fruit time or lack of these organs in some habits and young individuals. In addition, leaf morphology may be observed through whole growth period, minor difference and transitional variation between species make difficult for those inexperienced collectors. Consequently this leads to collection uncertainty for the three crude drugs, and confusedly mixed application often occurs in present research and clinic use. Nowadays a popular solution is to extract DNA fragments from dried materials, and then conducts DNA barcoding or SNPs analysis (Kress et al., 2005; Chen et al., 2010).
DNA Sequence
In our study, haplotype analysis based on cpDNA and ITS markers can distinguish clearly K. coccinea from K. longipedunculata and K. heteroclita, but can’t distinguish K. longipedunculata and K. heteroclite. Haplotype analysis is suitable for the study of closely related species and genetic diversity of intraspecific species by molecular biology methods. However, it does not show any advantage to delimit the boundary between K. longipendunculata and K. heteroclita. Haplotypes of K. longipedunculata and K. heteroclita shared the main haploptypes in both cpDNA markers and ITS. MatK,rbcL,psbA-trnH and ITS are the four suggested DNA barcode in plant (CBOL Plant Working Group, 2009). The cpDNA is characterized by its evolutionary conservatism, matrilineal inheritance, and lack of recombination (Wolfe et al., 1987). However, the complicated relationship such as the potential hybridization, reticulate evolution and gene introgression may further intersify the difficulty of species identification in closely related species of the Kadsura. The existing DNA barcode have not effectively resolved the problem of identifying K. longipedunculata and K. heteroclita.
Chemical Characteristics
Three herbs differ from their metabolite profiles including lignan based chemometric analysis. Heatmap analysis, PCA analysis and PLS-DA showed the chemical constituents of three kinds of medicinal materials differed significantly. We summarized the chemical constituents of Kadsura and found that a lot of spirobenzofuranoid dibenzocyclooctadiene compounds have been found in K. heteroclita. Tetrahydrofura compounds have been found in K. longipedunculata. 18 (13→12)-abeo-lanostane and nortriterpenoid compounds have been found only in K. coccinea (Liu et al., 2014). The different chemical constituents can influence the curative effect and security. The chemometrics analysis can make up for the shortage of molecular identification and has successfully been applied to identify the three Kadsura crude drug.
In this study, the DNA sequence analyzes, the recheck of morphology and chemical characteristics applied to identify the three Kadsura crude drug. The identification of medicinal materials between K. longipedunculata and K. heteroclita remained unresolved by the existing molecular fragments. The chemical characterization shows that the chemometric of chemical composition owned higher resolution to discriminate three crude drugs of the Kadsura and helpful to differentiate the source of samples and judge the consistency of three Kadsura species which make up for the shortage of molecular identification. This paper conducts a comprehensive analysis on three Kadsura crude drugs and provides a new research route for the confused herbs by molecular phylogeny, morphology and chemical composition.
Author Contributions
JL involved field survey, performed operation of the whole experiments, and wrote the manuscript. XZ and XW assisted with JL in the experiments. YQ and HL responsible for provided the technical guidance and designed the experiments. BZ and PX improved the manuscript.
Funding
The authors are grateful for the financial support provided by the National Natural Sciences Foundation of China (Nos. 81373913 and 81703650) and CAMS Initiative for Innovative Medicine (CAMS-I2M-1-010).
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank the reviewers for carefully reviewing our manuscript and making many valuable suggestions.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fphar.2018.01576/full#supplementary-material
References
Bandelt, H.-J., Forster, P., and Röhl, A. (1999). Median-joining networks for inferring intraspecific phylogenies. Mol. Boil. Evol. 16, 37–48. doi: 10.1093/oxfordjournals.molbev.a026036
CBOL Plant Working Group (2009). A DNA barcode for land plants. Proc. Natl. Acad. Sci. U.S.A. 106, 12794–12797. doi: 10.1073/pnas.0905845106
Chen, D. F., Xu, G. J., Yang, X. W., Hattori, M., Tezuka, Y., Kikuchi, T., et al. (1992). Dibenzocyclooctadiene lignans from Kadsura heteroclita. Phytochemistry 31, 629–632. doi: 10.1016/0031-9422(92)90049-V
Chen, M., Jia, Z. W., and Chen, D. F. (2006). Heteroclitin H, a new lignan from Kadsura heteroclita. J. Asian Natl. Prod. Res. 8, 643–648. doi: 10.1080/10286020500209053
Chen, P., Zhengou, W., and Peiping, X. (1999). History and Development of Traditional Chinese Medicine. Beijing: Science Press.
Chen, S. L., Yao, H., Han, J. P., Liu, C., Song, J. Y., Shi, L. C., et al. (2010). Validation of the ITS2 Region as a novel dna barcode for identifying medicinal plant species. PLoS One 5:e8613. doi: 10.1371/journal.pone.0008613
Chen, Y. G., Wang, P., Lin, Z. W., Sun, H. D., Qin, G. W., and Xie, Y. Y. (1998). Dibenzocyclooctadiene lignans from Kadsura angustifolia. Phytochemistry 48, 1059–1062. doi: 10.1016/S0031-9422(97)00996-5
Chinese Pharmacopeia Commission (2015). Pharmacopoeia of the People’s Republicof China Version (2015). Beijing: Chinese Medical Science Press.
Cubero-Leon, E., Peñalver, R., and Maquet, A. (2014). Review on metabolomics for food authentication. Food Res. Int. 60, 95–107. doi: 10.1016/j.foodres.2013.11.041
de Boer, H. J., Ichim, M. C., and Newmaster, S. G. (2015). DNA barcoding and pharmacovigilance of herbal medicines. Drug Safety 38, 611–620. doi: 10.1007/s40264-015-0306-8
Deng, W., Wang, Y., Liu, Z., Cheng, H., and Xue, Y. (2014). HemI: a toolkit for illustrating heatmaps. PLoS One 9:e111988. doi: 10.1371/journal.pone.0111988
FuJian Food and Drug Administration (2006). Chinese Materia Medica Standards of FuJian Province. Fujian: HaiFeng Press.
Gao, X. M., Pu, J. X., Xiao, W. L., Huang, S. X., Lou, L. G., and Sun, H. D. (2008). Kadcoccilactones K-R, triterpenoids from Kadsura coccinea. Tetrahedron 64, 11673–11679. doi: 10.1016/j.tet.2008.10.011
Gao, X. M., Pu, J. X., Zhao, Y., Yang, L. B., and Sun, H. D. (2012). Lignans from Kadsura angustifolia and Kadsura coccinea. J. Asian Natl. Prod. Res. 14, 129–134. doi: 10.1080/10286020.2011.637922
Guangdong Food and Drug Administration (2004). Chinese Materia Medica Standards of Guang Dong Province. Guangzhou: Guangdong Science and Technology Press.
GuangXi Zhuang Autonomous Region Health Department (1992). Chinese Materia Medica Standards of Guang Xi Province. Nanning: Guangxi Science and Technology Press.
Guo, Y. J., Guo, H. J., Liu, J. S., Li, X. W., and Qi, Y. D. (2015). Advances in studies on chemical constituents in Kadsura longipedunculata and their pharmacological activities. Mod. Chin. Med. 17, 1350–1358.
Hall, T. A. (1999). BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucl. Acids Symp. Ser. 41, 95–98.
Han, G. Q., Dai, P., Xue, R., Arison, B. H., Lankin, D. C., and Hwang, S. B. (1992). Dibenzocyclooctadiene lignans with platelet-activating factor (PAF) antagonist activity from Kadsura heteroclita. J. Chin. Pharm. Sci. 1, 20–27.
Kress, W. J., Wurdack, K. J., Zimmer, E. A., Weigt, L. A., and Janzen, D. H. (2005). Use of DNA barcodes to identify flowering plants. Proc. Natl. Acad. Sci. 102, 8369–8374. doi: 10.1073/pnas.0503123102
Kuo, Y. H., Li, S. Y., Wu, M. D., Huang, R. L., Yang, K. L., and Chen, C. F. (1999). A new anti-HBeAg lignan, kadsumarin A, from Kadsura matsudai and Schizandra arisanensis. Chem. Pharm. Bull. 47, 1047–1048. doi: 10.1248/cpb.47.1047
Li, L. N., Xue, H., and Tan, R. (1985). Dibenzocyclooctadiene lignans from roots and stems of Kadsura coccinea. Planta Med. 51, 297–300. doi: 10.1055/s-2007-969495
Li, Y., Wang, Y., Tai, W., Yang, L., Chen, Y., Chen, C., et al. (2015). Challenges and solutions of pharmacokinetics for efficacy and safety of traditional Chinese medicine. Curr. Drug Metab. 16, 756–776. doi: 10.2174/138920021609151201114223
Liu, H. T., Qi, Y. D., Xu, L. J., Peng, Y., Zhang, B. G., and Xiao, P. G. (2012). Ethnopharmacological investigation of Schisandraceae plants in China. China J. Chin. Mater. Med. 37, 1353–1359.
Liu, H. T., Xu, L. J., Peng, Y., Li, R. T., and Xiao, P. G. (2009). Chemical study on ethyl acetate soluble portion of Kadsura oblongifolia. China J. Chin. Mater. Med. 34, 864–866.
Liu, J. S., Huang, M. F., and Zhou, H. X. (1991). Kadsulignan C and D, two novel lignans from Kadsura longipedunculata. Can. J. Chem. 69, 1403–1407. doi: 10.1139/v91-207
Liu, J. S., and Li, L. (1995). Kadsulignans L-N, three dibenzocyclooctadiene lignans from Kadsura coccinea. Phytochemistry 38, 241–245. doi: 10.1016/0031-9422(94)00557-A
Liu, J. S., Qi, Y. D., Lai, H. W., Zhang, J., Jia, X. G., Liu, H., et al. (2014). Genus Kadsura, a good source with considerable characteristic chemical constituents and potential bioactivities. Phytomedicine 21, 1092–1097. doi: 10.1016/j.phymed.2014.01.015
Liu, J. X., and Pan, Y. P. (1991). Isolation and structures of schisantherin J and schisanlactone F. Yaoxue Xuebao 49, 308–312.
Liu, Z., Wang, D., Li, D., and Zhang, S. (2017). Quality evaluation of Juniperus rigida sieb. et zucc. based on phenolic profiles, bioactivity, and hplc fingerprint combined with chemometrics. Front. Pharmacol. 8:198. doi: 10.3389/fphar.2017.00198
Ma, W. H., Ma, X. L., Lu, Y., and Chen, D. F. (2009). Lignans and triterpenoids from the stems of Kadsura induta. Helv. Chim. Acta 92, 709–715. doi: 10.1002/hlca.200800363
Masssart, D. L., Vandeginste, B. G. M., Deming, S. N., Michotte, Y., and Kaufman, L. (1998). Data Handling in Science and Technology. Chemometrics: A Textbook. Amsterdam: Elsevier.
Saunders, R. M. (1998). Monograph of Kadsura (Schisandraceae). Syst. Bot. Monogr. 1:106. doi: 10.2307/25096646
Shen, Y. C., Lin, Y. C., Cheng, Y. B., Chiang, M. Y., Liou, S. S., and Khalil, A. T. (2009). Dibenzocyclooctadiene lignans from Kadsura philippinensis. Phytochemistry 70, 114–120. doi: 10.1016/j.phytochem.2008.11.005
Song, H. H., Kim, D. Y., Woo, S., Lee, H. K., and Oh, S. R. (2013). An approach for simultaneous determination for geographical origins of Korean Panax ginseng by UPLC-QTOF/MS coupled with OPLS-DA models. J. Gins. Res. 37:341. doi: 10.5142/jgr.2013.37.341
Sun, R., Song, H. C., Wang, C. R., Shen, K. Z., Xu, Y. B., Gao, Y. X., et al. (2011). Compounds from Kadsura angustifolia with anti-HIV activity. Bioorg. Med. Chem. Lett. 21, 961–965. doi: 10.1016/j.bmcl.2010.12.055
Thompson, J. D., Gibson, T. J., Plewniak, F., Jeanmougin, F., and Higgins, D. G. (1997). The Clustal_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 25, 4876–4882. doi: 10.1093/nar/25.24.4876
Wolfe, K. H., Li, W. H., and Sharp, P. M. (1987). Rates of nucleotide substitution vary greatly among plant mitochondrial, chloroplast, and nuclear DNAs. Proc. Natl. Acad. Sci. 84, 9054–9058. doi: 10.1073/pnas.84.24.9054
Xia, J. G., Mandal, R. I, Sinelnikov, V., Broadhurst, D., and Wishart, D. S. (2012). MetaboAnalyst 2.0-a comprehensive server for metabonomic data analysis. Nucleic Acids Res. 40, 127–133. doi: 10.1093/nar/gks374
Yang, J. H., Zhang, H. Y., Wen, J., Du, X., Chen, J. H., Zhang, H. B., et al. (2011). Dibenzocyclooctadiene lignans with antineurodegenerative potential from Kadsura ananosma. J. Nat. Prod. 74, 1028–1035. doi: 10.1021/np1009288
Yao, Y., Chen, H., Xie, L., and Rao, X. (2013). Assessing the temperature influence on the soluble solids content of watermelon juice as measured by visible and near-infrared spectroscopy and chemometrics. J. Food Eng. 119, 22–27. doi: 10.1016/j.jfoodeng.2013.04.033
Keywords: Kadsura, morphology, molecular markers, chemical characterization, identification
Citation: Liu J, Wei X, Zhang X, Qi Y, Zhang B, Liu H and Xiao P (2019) A Comprehensive Comparative Study for the Authentication of the Kadsura Crude Drug. Front. Pharmacol. 9:1576. doi: 10.3389/fphar.2018.01576
Received: 25 June 2018; Accepted: 31 December 2018;
Published: 22 January 2019.
Edited by:
Karl Tsim, Hong Kong University of Science and Technology, Hong KongReviewed by:
Shuai Ji, Xuzhou Medical University, ChinaJianping Chen, Shenzhen Traditional Chinese Medicine Hospital, China
Copyright © 2019 Liu, Wei, Zhang, Qi, Zhang, Liu and Xiao. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Haitao Liu, aHRsaXUwNzE4QDEyNi5jb20=
 Jiushi Liu
Jiushi Liu Xueping Wei
Xueping Wei Xiaoyi Zhang1,2
Xiaoyi Zhang1,2