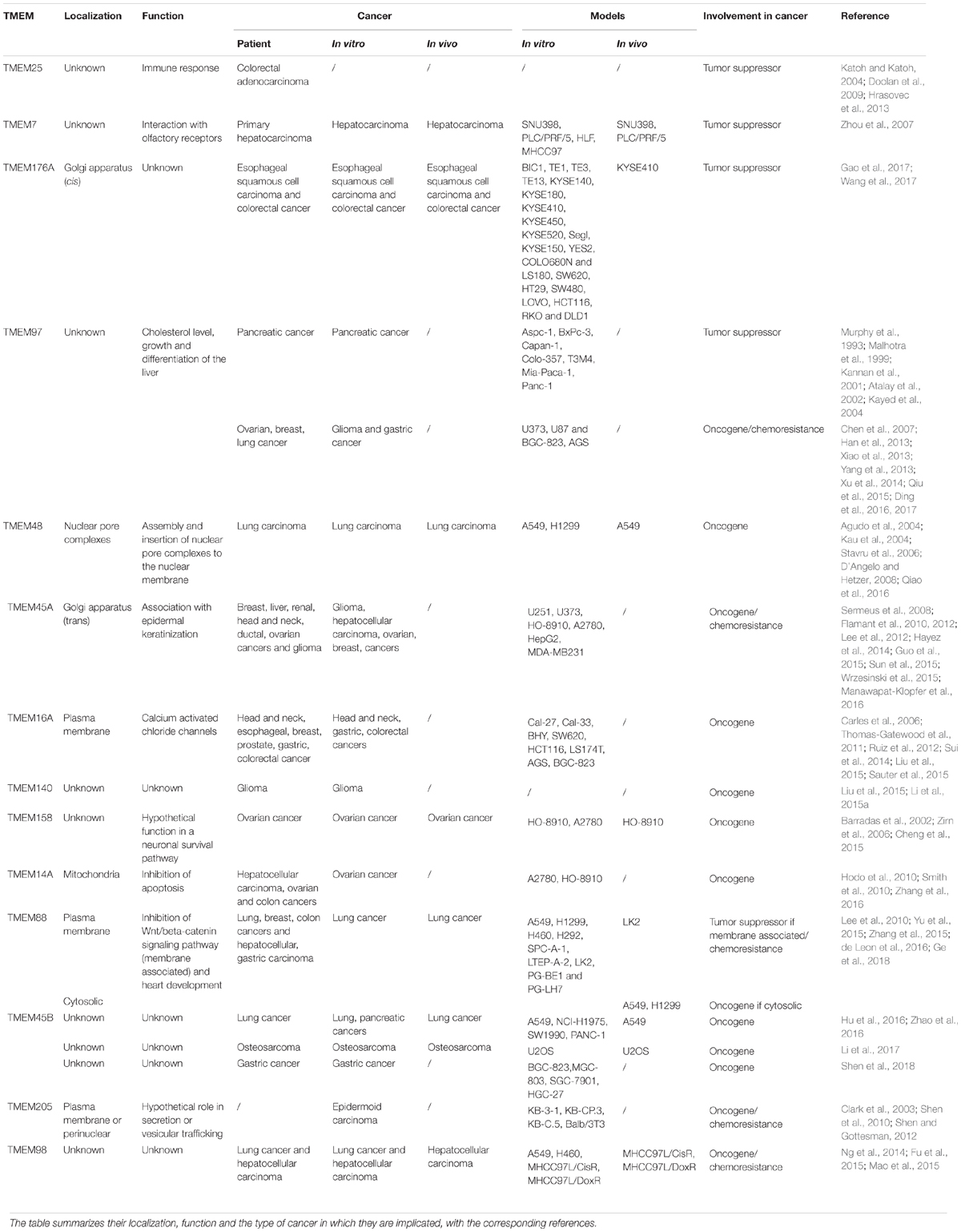
- URBC-NARILIS, University of Namur, Namur, Belgium
A transmembrane protein (TMEM) is a type of protein that spans biological membranes. Many of them extend through the lipid bilayer of the plasma membrane but others are located to the membrane of organelles. The TMEM family gathers proteins of mostly unknown functions. Many studies showed that TMEM expression can be down- or up-regulated in tumor tissues compared to adjacent healthy tissues. Indeed, some TMEMs such as TMEM48 or TMEM97 are defined as potential prognostic biomarkers for lung cancer. Furthermore, experimental evidence suggests that TMEM proteins can be described as tumor suppressors or oncogenes. TMEMs, such as TMEM45A and TMEM205, have also been implicated in tumor progression and invasion but also in chemoresistance. Thus, a better characterization of these proteins could help to better understand their implication in cancer and to allow the development of improved therapy strategies in the future. This review gives an overview of the implication of TMEM proteins in cancer.
Introduction
A TMEM is a type of protein that spans the entire width of the lipid bilayer and to which it is permanently anchored. Many TMEMs function as channels to permit the transport of specific substances across the biological membranes. But the biological functions of many of them remain unknown mainly due to difficulties in the extraction and purification of these proteins. There are two ways to classify the TMEMs. The first one is according to their structure. Indeed there are two basic types of TMEMs, alpha-helical proteins and the beta-barrel proteins (Vinothkumar and Henderson, 2010). The second classification is according to their topology, this classification refers to the position of the N- and C-terminal domains (von Heijne, 2006).
Among TMEMs is the TMEM family. The proteins of this family are predicted to be components of various cell membranes, such as mitochondrial, endoplasmic reticulum, lysosome, and Golgi membranes. TMEMs are present in many cell types and fulfill important physiological functions such as epidermal keratinization (TMEM45A) (Hayez et al., 2014), autophagy, smooth muscle contraction (TMEM16) (Thomas-Gatewood et al., 2011), protein glycosylation (TMEM165) (Foulquier et al., 2012) and development and differentiation of the liver (TMEM97) (Malhotra et al., 1999). Among them, some members play a primordial role in immune response (TMEM9B) (Dodeller et al., 2008). Indeed, TMEM9B is a key component of inflammatory signaling pathways through the enhancement of the production of pro-inflammatory cytokines induced by TNF, IL1β, and TLR ligands.
In many cancers, differential regulation of the expression of TMEMs has been observed, such as in lymphomas (TMEM176) (Cuajungco et al., 2012), colorectal cancer (TMEM25) (Hrasovec et al., 2013), hepatic cancer (TMEM7) (Zhou et al., 2007), and lung cancer (TMEM48) (Qiao et al., 2016). Some of them are used as prognostic biomarkers. For example, in renal cancers, many TMEMs with predicted ER localization have been shown to be potential classifiers of cancer grade (e.g., TMEM45A, TMEM116, TMEM207, TMEM213…) (Wrzesinski et al., 2015). A large number of TMEMs have also been implicated in cancer development and in drug resistance, suggesting that the TMEM family is a prominent group for cancer research. Furthermore, some of these proteins act as tumor suppressors (e.g., TMEM25, TMEM7) (Zhou et al., 2007; Doolan et al., 2009) while others act as pro-oncogenes (e.g., TMEM158, TMEM14A…) (Cheng et al., 2015; Zhang et al., 2016). This review aims to describe the implication of the TMEM proteins in cancer.
Part 1: TMEMs as Tumor Suppressors
Some TMEMs have been described in the literature to act as tumor suppressors. A downregulation of their expression is generally observed in tumor tissue compared to adjacent healthy tissue. It is for example the case for TMEM25. This protein is a member of the immunoglobulin super-family and is involved in immune response, growth factor signaling and cell adhesion (Katoh and Katoh, 2004). The expression of this protein has been studied in fresh tumor samples collected during surgical colectomy from patients who had been diagnosed with primary colorectal adenocarcinoma. TMEM25 mRNA expression was significantly decreased in 68% of tumor tissues in comparison to corresponding normal tissues. This downregulation has been correlated with the hypermethylation of a specific CpG site in the 5′ UTR region of TMEM25 gene in a high proportion of tumor samples (Hrasovec et al., 2013). Another study revealed that TMEM25 expression in the tumor tissues was lower than the one in normal healthy tissues in 50% of tumor samples in human breast tumor biopsies. The expression of TMEM25 was correlated with a better overall survival and associated with a longer survival time for patients who received adjuvant chemotherapy. Furthermore, in triple-negative breast tumors, TMEM25 was generally not expressed (Doolan et al., 2009). All together these findings suggest that TMEM25 may be used as a tumor biomarker of favorable prognosis.
Another example is TMEM7. This protein of 232 amino acids has a single transmembrane domain and is expressed in the liver. The gene coding for TMEM7 is localized in the short arm of chromosome 3, which is commonly deleted in cancer cells (Huebner, 2001). Chromosomal regions that are deleted in cancer are generally the loci of tumor suppressor genes, suggesting that TMEM7 is a candidate suppressor gene. This protein has been studied in 18 hepatocellular carcinoma cell lines but also in primary tumors obtained from surgical resection of hepatocellular carcinoma from 17 patients. Each tumor sample was matched with its corresponding healthy liver tissue. In the absence of homozygous deletion, TMEM7 is down regulated in 33% of the cell lines and 85% of the tumor samples compared to healthy tissue. Tumor suppressor genes located at chromosomal regions deleted in some cancer cells are found to be silenced by promoter methylation in other cell lines. In two lines of the latter that displayed TMEM7 downregulation, 5-aza-2′-deoxycytidine, a DNA methylation inhibitor and trichostatin A, a HDAC inhibitor, increased TMEM7 expression suggesting that aberrant methylation and histone deacetylation are responsible for the transcriptional silencing of this gene. The study of this protein also showed that INF-α induced TMEM7 mRNA expression and the restoration of its expression by overexpression or by induction with IFN-α decreased the proliferation and the invasion of hepatocellular carcinoma cell lines (SNU398 and PLC/PRF/5 or HLF and MHCC97 respectively). These data have also been validated in vivo. Indeed, ectopic expression of TMEM7 in two TMEM7 deficient hepatocarcinoma cell lines decreased tumor growth in nude mice (Zhou et al., 2007). All these data highlight the tumor suppressor role of TMEM7 in hepatocellular carcinoma.
Two recent studies also showed that TMEM176A could act as tumor suppressor. The first one was performed in esophageal squamous cell carcinoma. Wang et al. analyzed the methylation profile of TMEM176A promoter in 13 cell lines (BIC1, TE1, TE3, TE13, KYSE140, KYSE180, KYSE410, KYSE450, KYSE520, Segl, KYSE150, YES2, and COLO680N) and 267 primary esophageal squamous cell carcinoma. The results showed the loss of TMEM176 expression in 12 cell lines (TE1, TE3, TE13, KYSE140, KYSE180, KYSE410, KYSE450, KYSE520, Segl, KYSE150, YES2, and COLO680N) in association with a complete methylation of its promoter. It also revealed that 66% of primary tumors presented TMEM176A promoter methylation. This methylation and TMEM176A decreased expression were correlated with poor overall survival. The restoration in two cell lines, KYSE410 and KYSE150, of TMEM176A expression with 5′-aza-2′-deoxycytidine treatment and the downregulation of TMEM176A in BIC1 cells showed that TMEM176A inhibited cell invasion and migration and induced apoptosis. Furthermore, TMEM176A inhibited cell growth both in vitro and in vivo with a decrease in tumor volume when TMEM176A was re-expressed (Wang et al., 2017). A very similar study has been performed in colorectal cancer. It revealed that 50% of the primary tumors presented methylation of TMEM176 promoter. The results also showed a normal expression of TMEM176A in LS180 and SW620 cell lines, a decreased expression in HT29 and SW480 cell lines and a total loss of expression in LOVO, HCT116, RKO, and DLD1 cell lines respectively associated with no methylation, partial methylation and total methylation of TMEM176A promoter. In colorectal cancer as well as in esophageal squamous cell carcinoma, TMEM176A overexpression inhibited cell migration and invasion, induced apoptosis and inhibited cell growth both in vitro and in vivo (Gao et al., 2017). These two studies together presented TMEM176A as tumor suppressor of esophageal squamous cell carcinoma and colorectal cancer.
The last protein described in this part is TMEM97. This protein, also named MAC30, is a member of the insulin-like growth factor binding proteins (Murphy et al., 1993). TMEM97 mRNA is expressed in the fetal liver but not in adult liver suggesting a role in development and differentiation of the liver (Malhotra et al., 1999). In 2001 and 2002, two studies showed that the expression of TMEM97 can be induced by other genes like BRCA1 but also be downregulated by others like p53 suggesting that the expression of this gene can be deregulated in cancers (Kannan et al., 2001; Atalay et al., 2002). Indeed, the expression of TMEM97 is increased in several types of cancer as described later in this review, except in pancreatic and renal cancers that both display a low expression level of TMEM97 protein and mRNA. In 2004, 30 pancreatic cancer tissues obtained from patients after tumor resection and 19 non-cancerous pancreatic tissues obtained through an organ donor program have been used to analyze the expression level of TMEM97 in pancreatic cancer both at the mRNA level by RT-qPCR and at the protein level by histochemistry. 50% of pancreatic cancer biopsies displayed a lower TMEM97 mRNA expression compared to normal pancreatic tissue, 20% displayed no change and 30% presented higher TMEM97 mRNA levels. These results highlighted a high variability regarding TMEM97 expression levels in pancreatic cancer. A high variation in mRNA level expression was also observed in different pancreatic cancer cell lines (Aspc-1, BxPc-3, Capan-1, Colo-357, T3M4, Mia-PaCa-2 and Panc-1 cells). The protein expression and localization of TMEM97 were also analyzed: TMEM97 protein was strongly expression in the cytoplasm of islet cells and moderately in acinar cells. Cancer cells in pancreatic cancer tissues displayed weak or no expression of this protein in more than 75% of cases. But at low levels in pancreatic cancer cells (Aspc-1, BxPc-3, Capan-1, Colo-357, T3M4, Mia-PaCa-2 and Panc-1 cells). Knowing that tubular complexes are considered as potential pre-neoplastic lesions, The observed reduction of TMEM97 expression in pancreatic cancer suggests that this gene might act as a tumor suppressor in this disease (Kayed et al., 2004). This hypothesis may also be true for prostate cancer since miR-152-3p downregulation and promoter methylation were found to be prevalent in primary prostate cancers. TMEM97, which is overexpressed in this type of cancer, is a target of miR-152-3p (Ramalho-Carvalho et al., 2018).
Part 2: TMEMs as Oncogenes
Many TMEMs are up regulated in cancer. Some of them are implicated in tumor progression, invasion and in the formation of metastasis while others are associated with poor prognosis and can be used as prognostic biomarker. The studies behind these conclusions are summarized here under.
TMEMs as Prognostic Biomarkers
TMEM48, also named NDC1 is localized to the nuclear pore complexes. This nucleoporin has six membrane-spanning segments and is crucial for nuclear pore complexes and nuclear envelope assembly (Stavru et al., 2006). The integrity of the nuclear envelope and a correct nucleocytoplasmic transport are important for many cellular processes such as genome stability, DNA replication, or DNA repair (D’Angelo and Hetzer, 2008). Nucleoporin deregulation has been implicated in several malignancies such as breast cancers (Agudo et al., 2004; Kau et al., 2004) in multiple tumors including melanoma, pancreatic, colon, gastric, prostate, esophageal, lung cancer, and lymphomas (Mahipal and Malafa, 2016). A study based on 60 patients with NSCLC showed that TMEM48 expression was significantly higher in cancer tissues compared to healthy tissues. This overexpression was associated with poor prognosis, lymph node metastasis, increased tumor size and short survival (Qiao et al., 2016). All together these results suggest that, since TMEM48 mRNA expression is increased in non-small lung carcinoma in association with advanced tumor stage, TMEM48 may be a potential prognostic factor for NSCLC.
TMEM45A is a TMEM of 275 amino acids, predicted to have five to seven transmembrane domains and localized in the trans Golgi apparatus. Very little is known about this protein except that TMEM45A is highly expressed in the skin and is associated with epiderm keratinization (Hayez et al., 2014). This protein is overexpressed in many cancers: breast cancer, liver cancer, renal cancer, glioma, head and neck cancer, ductal cancer, and ovarian cancer (Flamant et al., 2012; Lee et al., 2012; Guo et al., 2015; Sun et al., 2015; Wrzesinski et al., 2015; Manawapat-Klopfer et al., 2016). In the cases of breast cancer and cervical lesions, a higher expression level of TMEM45A has been correlated with a lower patient overall survival suggesting that TMEM45A is a potential biomarker for aggressiveness of breast cancer and cervical lesions (Flamant et al., 2012; Manawapat-Klopfer et al., 2016).
Despite the putative tumor suppressor role of TMEM97 in pancreatic and prostate cancers, this protein is overexpressed in different types of cancer and associated with tumor progression, recurrence and poor survival. It is the case in breast, gastric, colon, epithelial ovarian, oral squamous, and NSCLC. Indeed, the expression of TMEM97 has been analyzed in 20 cases of NSCLC compared to adjacent healthy tissue: 65% of patients showed a higher expression level of TMEM97 in tumor tissue compared to healthy tissue. Furthermore, the expression of this protein has been correlated with poor tumor differentiation and a shorter patient survival (Han et al., 2013). A similar study performed in human SQCLC showed TMEM97 overexpression in 26 of the 32 tumor samples in comparison to corresponding non-tumor tissues. TMEM97 overexpression was associated with poor tumor differentiation and shorter overall patient survival (Ding et al., 2016). Another study in breast cancer revealed that 59.7% of tumor samples displayed a higher expression level of TMEM97 compared to healthy tissue and that this overexpression correlated with larger tumor size and tumor recurrences. One study on ovarian cancer showed that high expression of TMEM97 was correlated with high histological grade and tumor recurrence (Xiao et al., 2013; Yang et al., 2013). All these studies demonstrated that TMEM97 expression could affect the prognosis of NSCLC, SQCLC, ovarian and breast cancer patients.
Another important TMEM protein is TMEM16A. TMEM16A, also known as anoctamin-1, is expressed in cerebral artery smooth muscle cells and is predicted to have eight transmembrane domains. This protein is a TMEM that functions as a calcium-activated Cl- channel (Thomas-Gatewood et al., 2011). TMEM16A has recently been shown to be upregulated in several cancers including HNSCC, esophageal, breast and gastric cancers. In HNSCC, the expression of TMEM16A has been studied by fluorescence in situ hybridization and immunohistochemistry on several primary tumors. The results demonstrated that TMEM16A was highly expressed in 4–19% of the samples and that higher TMEM16A expression strongly correlated with poor prognosis of HNSCC patients (Ruiz et al., 2012). In another study in HNSCC, TMEM16A has been shown to be overexpressed in 84% of tumor samples (Carles et al., 2006). In the context of gastric cancer, the expression of TMEM16A has been evidenced to be higher in tumor tissue than in adjacent non-tumor tissue. Furthermore, the expression of this protein has been correlated with the tumor stage and negatively correlated with patient survival in this cancer type (Liu et al., 2015). TMEM16A is thus proposed to be a negative prognostic factor.
Two other TMEMs have been described as prognosis biomarker. In glioma, TMEM140 expression has been analyzed in 47 of the 70 glioma samples by immunohistochemistry. The results showed a higher expression in tumor tissue than in the control brain tissue and a correlation with poor prognosis in this cancer (Li et al., 2015a,b). In lung cancer, TMEM45B expression has been analyzed in 110 tumor tissue samples and 35 non-tumor tissue samples. TMEM45B was shown to be upregulated in lung cancer and its expression was negatively correlated with overall survival (Hu et al., 2016).
TMEMs Involved in Tumor Growth
Besides the evidence for a correlation between TMEM expression and patient survival, some of these proteins have been shown to be directly involved in tumor growth but the mechanisms by which they act are not always known.
With an Identified Pathway
The first protein described in this part is TMEM158, also called Ras-induced senescence 1 protein (RIS1). The gene coding for this protein is known to be upregulated during Ras-induced senescence in human diploid fibroblasts infected with rasV12-containing retrovirus (Barradas et al., 2002). TMEM158 is overexpressed in Wilms tumors (also known as nephroblastoma) with somatic mutations in catenin beta-1 gene suggesting a relationship between the Ras and Wnt signaling pathways (Zirn et al., 2006). TMEM158 is also overexpressed in ovarian cancer in 84% of the 25 tumor samples which were analyzed. The involvement of TMEM158 in tumor growth has been studied in two ovarian cancer cell lines, HO-8910 and A2780. This protein was evidenced to regulate cell proliferation, adhesion, and invasion. Furthermore, TMEM158 knockdown inhibited tumor growth of HO-8910 cell line in nude mice highlighting the role of this protein in tumorigenicity. TMEM158 silencing led to the deregulation of the expression of different genes, including a downregulation of ICAM1 and VCAM1 expression. These two proteins are involved in cell adhesion. TMEM158 silencing also impaired the TGF-β signaling pathway (Cheng et al., 2015). All these results showed that TMEM158 may work as an oncogene in ovarian cancer.
The implication of TMEM48 in NSCLC progression has been studied in two cell lines that overexpressed this protein, A549 and H1299. The results suggested a role of TMEM48 in cell proliferation, migration and invasion. Indeed, the silencing of this gene impaired cell proliferation, induced cell cycle arrest and decreased the migration and invasive ability of NSCLC cells. The downregulation of TMEM48 also induced cell apoptosis in association with a decrease or an increase in anti- or pro-apoptotic gene expression respectively. One of these two cell lines (A549) was also used to study the involvement of TMEM48 in tumorigenicity in vivo and the data revealed that TMEM48 is involved in tumor formation from A549 cells in nude mice. A marked decrease in tumor weight (50%) was evidenced when TMEM48 was silenced. All these evidences showed a role of TMEM48 in lung cancer progression (Qiao et al., 2016). A recent study demonstrated that TMEM48 suppression by miR-421 increased the expression of the apoptotic and tumor suppressor proteins caspase 3, PTEN and p53 in A549 cells (Akkafa et al., 2018). These results suggest that TMEM48 modulates the apoptotic pathway.
TMEM14A is a TMEM with three transmembrane domains, localized in mitochondria. This protein is deregulated in different types of cancer such as ovarian cancer, colon cancer and hepatocellular carcinoma (Hodo et al., 2010; Smith et al., 2010; Zhang et al., 2016). In the context of ovarian cancer, TMEM14A is involved in cell proliferation as shown by a cell cycle arrest when TMEM14A was invalidated in two ovarian cancer cell lines, A2780 and HO-8910. TMEM14A up regulation also increased the cell invasive ability of ovarian cancer cells highlighting a potential role of this protein to promote metastasis. Further investigations showed that TMEM14A knockdown may down-regulate the protein expression of PCNA, cyclins and MMPs. It may also downregulate TGF-β signaling (Zhang et al., 2016). These results could explain the decrease in cell proliferation and invasiveness in ovarian cancer cell lines when TMEM14A was invalidated.
TMEM97 is found deregulated in several types of cancer but this protein has been particularly involved in the tumor growth of two cancers: glioma and gastric cancer. Indeed, the silencing of TMEM97 expression in glioma U373 and U87 cells inhibited cell proliferation and cell cycle progression associated with a decrease in cyclin B1, E, CDK2 and CDK4 expression, but also in cell invasiveness. TMEM97 silencing also induced the deregulation of the expression of EMT markers like β-catenin, Twist and E-cadherin (Qiu et al., 2015). The downregulation of TMEM97 in gastric cancer BGC-823 and AGS cell lines inhibited the cell proliferation and mobility with a decrease in Akt phosphorylation, hence suggesting that Akt may mediate the TMEM97-induced inhibition of proliferation (Xu et al., 2014). The invalidation of TMEM97 also induced an inhibition of cell migration and invasion by reducing the expression of cyclin B1 and WAVE2. These data showed that TMEM97 plays an important role in tumor growth and aggressiveness in glioma and gastric cancer.
Another TMEM protein involved in tumor growth is TMEM16A. In human colorectal cancer cells, the mRNA and protein expression of TMEM16A has been reported in several cell lines like SW620, HCT116 and LS174T but not in HCT8 and SW480. TMEM16A knockdown in SW620 cell line inhibited cell proliferation, migration and invasion. These effects were mediated through a decrease in the expression of cyclin D1 and in the phosphorylation of MEK and ERK1/2. Furthermore, invalidation of TMEM16A expression led to a delay in cell cycle progression (Sui et al., 2014). TMEM16A expression is also regulated epigenetically. Indeed, inhibition of HDAC class I and II by siRNA or pharmacological agents decreased the expression of TMEM16A. HDAC3 seems to be the most important one in this regard. Hence, the inhibition of HDAC3 may exert suppressive effect on cancer cell viability via the downregulation of TMEM16A in prostate or breast cancer (Matsuba et al., 2014). TMEM16A has also been well-studied in gastric cancer. Knockdown in AGS and BGC-823 gastric cancer cell lines inhibited cell migration and invasion via a downregulation of E-cadherin expression (EMT marker) probably via a decrease in TGF-β secretion since the supplementation of exogenous TGF-β restored E-cadherin expression and cell migration and invasion (Liu et al., 2015). TMEM16A silencing was also shown to induce apoptosis in human prostate cancer PC3 cells by upregulating TGF-β signaling (Song et al., 2018). In pancreatic ductal adenocarcinoma, TMEM16A is overexpressed in several cancer cell lines (Mia PaCa-2, Panc-1, BxPC-3, and AsPC-1) in comparison to HPDE-derived cells. The invalidation of TMEM16A expression in these cell lines using siRNA showed an implication of this protein in cell migration but not in the proliferation illustrating that TMEM16A modulates the metastatic potential of pancreatic cancer cells. Contrary to colorectal cancer, the molecular mechanism underlying this effect is still unknown (Sauter et al., 2015).
The last TMEM described in this part is a very peculiar TMEM protein, TMEM88. This protein is a potential 2-transmembrane type protein that interacts with an important component of Wnt signaling pathway: DVL1 (Lee et al., 2010). According to the localization of its partner DVL1, TMEM88 may be localized in the cytoplasm or to the plasma membrane. This protein is overexpressed in cancer tissue compared to non-cancerous tissue in different types of cancer such as in lung, colon, gastric, breast cancer (Yu et al., 2015; Zhang et al., 2015) and can be involved in the tumor initiation and progression through Wnt signaling pathway (Ge et al., 2018). For the majority of these cancer types, immunohistochemistry analysis demonstrated a cytosolic localization. But in the context of NSCLC, two different subcellular localizations for TMEM88 have been reported, suggesting different roles in tumor development depending on its localization. Indeed, an in vitro analysis on nine lung cancer cell lines (A549, H1299, H460, H292, SPC-A-1, LTEP-A-2, LK2, PG-BE1, and PG-LH7) showed that the overexpression of membrane-associated TMEM88 led to the inhibition of the canonical Wnt pathway through the downregulation of the expression of effectors like cyclin D1, MMP-7, and c-Myc. The increase in membrane-associated TMEM88 expression also led to a decrease of proliferation, colony formation, migration and invasion and to a decrease in tumor growth in vivo highlighting the tumor suppressor role of TMEM88 when it is localized to the membrane of the cell. Furthermore, TMEM88 promoter methylation is associated with unfavorable prognosis in NSCLC (Ma et al., 2017). On the contrary, its cytosolic localization is correlated with a low level of differentiation of the tumor and poor prognosis of patients with NSCLC. Furthermore in vitro analysis demonstrated that the overexpression or downregulation of this protein respectively enhanced or suppressed NSCLC cell migration and invasion through a deregulation of the EMT signaling pathway. Indeed, the TMEM88-DVL complex increased p38 and GSK3β phosphorylation leading to a stabilization of the protein SNAIL and hence to a decreased occludin and zonula occludens-1 (ZO-1) expression. Moreover, in vivo analysis showed that the number of lung metastatic nodules increased in the mice transplanted with cell lines expressing cytosolic TMEM88 (Zhang et al., 2015). Very similar results have also been observed in triple-negative breast cancer (Yu et al., 2015). These data confirmed that, in NSCLC and breast cancer, the cytosolic localization of TMEM88 conferred an oncogenic role to the protein.
Depending of cancer stage, TGF-β signaling can have different impact on tumor growth. Indeed, in early stage TGF-β plays a tumor suppressor role whereas in advanced stage, cancer cells benefit from TGF-β to initiate proliferation, invasion, and metastasis dissemination. It seems that several TMEM proteins are involved in tumor growth through TGF-β pathway modulation in order to facilitate malignant progression (Figure 1). Indeed, TMEM16A, TMEM158, TMEM14A, TMEM97, TMEM88 and probably TMEM45A interacts with several components of the TGF-β-induced signal transduction.
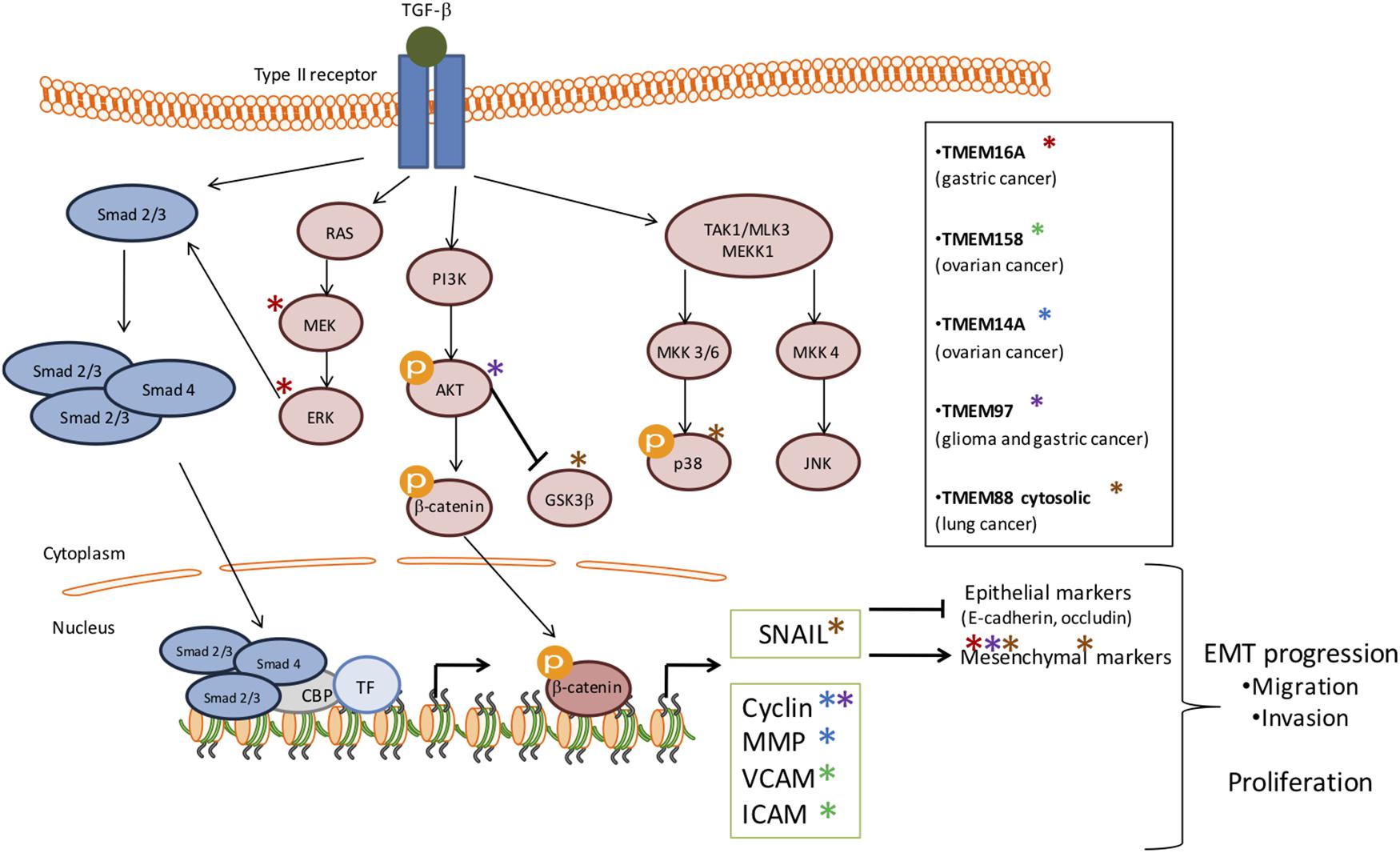
Figure 1. Schematic representation of the involvement of several TMEMs in tumor growth through the TGF-β signaling pathway. The activation of TGF-β signaling pathway has been implicated in many cellular processes and in tumor growth. This activation is induced by its ligand which then activates the phosphorylation of serine/threonine residues and triggers phosphorylation of the intracellular effectors, SMADs (blue). TGF-β receptors can also activate Smad-independent pathways (pink). In early stage TGF-β plays a tumor suppressor role whereas, in advanced stage, cancer cells benefit from TGF-β to initiate proliferation, invasion and metastasis dissemination. It seems that several TMEM proteins are involved in tumor growth through TGF-β activation in order to facilitate malignant progression and EMT progression. The stars represent the effectors deregulated by TMEMs.
Through an Unknown Pathway
Other TMEMs have also an impact on tumor growth but the mechanisms by which they act are still unknown. Such an example is TMEM140 that is up regulated in cancer tissue compared to healthy tissue. TMEM140 has been involved in the regulation of the growth of glioma in vitro and in vivo. Indeed, when TMEM140 is silenced in two glioma cell lines in vitro, U87 and U373, the proliferation decreased with a higher proportion of cells in G1 phase and the cell viability decreased due to the activation of the apoptotic pathway. Furthermore, the knockdown of TMEM140 led to a decreased cell adhesion, migration and invasion. It has also been shown that the invalidation of this protein inhibited tumor growth in vivo with a decrease in the size and the weight of tumors in the invalidated group compared to the control group (Li et al., 2015a,b). These findings demonstrate that TMEM140 can be used as a prognosis biomarker but also as a therapeutic target.
Two other TMEM proteins have been involved in tumor progression, TMEM45A and TMEM45B, already described above. TMEM45A is implicated in cell proliferation, migration, and invasion of different cancers like glioma (U251 and U373 cells) and ovarian cancer (HO-8910 and A2780 cells) (Guo et al., 2015; Sun et al., 2015). In the context of ovarian cancer, TMEM45A protein expression has been positively correlated to TGF-β signaling pathway and this data could explain the impact of TMEM45A invalidation in this cancer (Guo et al., 2015). On the other hand, TMEM45B is up-regulated in human lung cancer and promotes tumorigenicity in vivo. Invalidation of TMEM45B in A549 and NCI-H1975 cells led to the inhibition of cell proliferation, migration, and invasion highlighting its role in tumor growth in lung cancer (Hu et al., 2016). In the case of pancreatic cancer, TMEM45B had also been involved in proliferation, invasion, and migration since its silencing in SW1990 and PANC-1 cell lines induced an inhibition of cell proliferation associated with cell cycle arrest. It also led to a decrease in cell mobility and invasiveness. Conversely, the overexpression of TMEM45B in CFPAC-1 cells promoted cell proliferation, invasion and migration (Zhao et al., 2016). TMEM45B is also upregulated in osteosarcoma cell lines. Its knockdown suppressed the prolifreation, migration, and invasion of U2OS cells in vitro as well as tumor growth in nude mice. These effects were associated with a decrease in the expression of β-catenin, cyclin D1 and c-Myc (Li et al., 2017). Similar results were obtained in gastric cancer cells, in which TMEM45B silencing was associated with a decrease in the abundance of p-STAT3 and p-JAK2 (Shen et al., 2018). These two proteins can be described as potential prognosis markers but also as regulators of tumor growth in several types of cancer.
Part 3: TMEMs Involved in Chemoresistance
Although mutagenic alterations have long been associated with cancer development or drug resistance, epigenetic modifications and tumor microenvironment have also been linked to chemoresistance. Both epigenetic modifications and the tumor microenvironment can impact the expression or the localization of several TMEMs leading to a deregulation of treatment responses. The first example is hypoxia, one component of the tumor microenvironment. Indeed, in hypoxic condition (<1% of O2), hepatocellular carcinoma cells (HepG2) (Sermeus et al., 2008) and breast cancer cells (MDA-MB-231) (Flamant et al., 2010) were protected against cell death normally induced by chemotherapeutic drugs. In this condition, TMEM45A was shown to be upregulated and its silencing led to a decrease in this protective effect conferred by hypoxia against cell death induced by chemotherapeutic agents. These results suggest that, in hypoxic condition, TMEM45A is involved in the chemoresistance of breast and liver cancers. However, the mechanism underlying this protection is still unknown (Flamant et al., 2012).
The second example is related to epigenetic modifications, in particular DNA methylation. Indeed, in ovarian cancer, it has been shown in vivo, that the methylation profile of some promoters was different in xenografts resistant to cisplatin compared to control ones. This observation has been associated with a differential expression profile of the genes whose expression is regulated by these promoters. It is the case for TMEM88, which is a DNA methylation-regulated gene. The hypomethylation of TMEM88 promoter observed in ovarian cancer led to an increased expression of the protein and to platinum resistance. Indeed, knowing that TMEM88 was involved in Wnt signaling pathway, De Leon et al investigated the possible association of Wnt pathway and the observed phenotype. First of all, TMEM88 downregulation led to an increase in Wnt target gene expression such as β-catenin or Jun, validating the interaction between TMEM88 and Wnt pathway in ovarian cancer. Then, they studied the link between this interaction and the observed chemoresistance. TMEM88 overexpression in resistant cells inhibited the Wnt signaling pathway associated with a decrease in target gene expression while the activation of the Wnt pathway in resistant cells increased the chemosensitivity of the cells to cisplatin. Furthermore, the invalidation of TMEM88 in cisplatin resistant cells increased the sensitivity of the cells to the chemotherapeutic drug. This increase in chemosensitivity was associated to a decrease in cell proliferation allowing the escape of the cells from the genotoxic effects of cisplatin (de Leon et al., 2016).
Another TMEM involved in chemoresistance is TMEM205, also known as MBC3205. This protein of 21 kDa has four transmembrane domains and belongs to the group of secreted proteins (Clark et al., 2003). In 2011, a study revealed that TMEM205 is highly expressed in the pancreas, adrenal gland, liver, mammary gland and kidney (Shen et al., 2010). This study also showed that, in epidermoid carcinoma, this protein had the particularity to translocate in the presence of cisplatin. Indeed, TMEM205 is located at the cell surface but in the presence of the chemotherapeutic drug, the protein is translocated in an intracellular compartment at the periphery of the nucleus. Furthermore, its expression is increased in a cell line resistant to cisplatin and TMEM205 overexpression conferred resistance to cisplatin (Shen et al., 2010). Another study demonstrated that TMEM205 colocalized with RAB8, a marker of recycling endosomes. Interestingly, TMEM205 also colocalized with syntaxin 6 (STXR6), a regulator of protein trafficking, which is translocated at the same subcellular localization that TMEM205 in the presence of cisplatin. Then, the translocation of TMEM205 may allow the exocytosis of platinum containing vesicles, which thus results in the accumulation of the drug outside the cell (Shen and Gottesman, 2012).
In the tumor microenvironment, the immune system plays a crucial role that modulates tumor growth. Furthermore, cancer-associated inflammation also plays a role in chemoresistance (Chen et al., 2007). In this context, TMEM98, which has immune-related properties, mainly regarding the differentiation of T helper (Th) 1 cells, may be proposed as a novel chemoresistance-conferring gene (Fu et al., 2015). There are two RNA splicing forms of TMEM98 reported in the NCBI database, TMEM98-v1 and TMEM98-v2 respectively. Although there is a slight difference between them in the 5′ UTR sequence, their coding products are almost the same, which consists of 226 amino acids and a molecular weight of 24.6 kDa. In lung cancer, TMEM98 mRNA expression is higher in cancer tissues compared to healthy tissues. Furthermore, in two lung cancer cell lines, A549 and H460, the silencing of TMEM98 inhibited cell proliferation and suppressed the invasion and the migration of cancer cells meaning that this protein can have an impact in tumor growth (Mao et al., 2015). Knowing that tumor progression and chemoresistance can be accompanied with inflammation injuries and the link between TMEM98 and inflammation, this protein is a very interesting target for further investigations on anti-cancer drug resistance. In the case of hepatocellular carcinoma, TMEM98 has been identified as a chemoresistance-associated gene. Indeed, its expression is increased in two chemoresistant cell lines, MHCC97L/CisR and MHCC97L/DoxR resistant to cisplatin and doxorubicin respectively. Furthermore, the level of the upregulation increased with the degree of chemoresistance. This study also showed that TMEM98 mRNA expression was higher in tumor tissue of patients who received a transarterial chemoembolization treatment. Moreover, the patients who did not respond well to the treatment had higher TMEM98 expression level. These data demonstrated that this protein is involved in chemoresistance of hepatocellular carcinoma. In order to identify the mechanism of TMEM88 in chemoresistance, further investigation had been performed. In the absence of TMEM88 in resistant cell lines, a repression of activation of AKT in association with a repression of its downstream targets had been observed. Furthermore, the silencing of TMEM88 restored p53 phosphorylation and activation under cisplatin or doxorubicin treatment. These data showed that the chemoresistance induced by TMEM88 is associated with AKT activation and the repression of p53 activation (Ng et al., 2014).
The platinum-based chemotherapy is used for the treatment of several cancers such as lung cancer. In this model, the high expression level of TMEM97 has been correlated with the resistance of cancer to platinum-based treatment but also with poor patient survival (Chen et al., 2016; Ding et al., 2017). Indeed, Chen et al. (2016), showed that only 4% of patients with elevated expression of TMEM97 showed responses to therapy while 65% of patients with low expression of TMEM97 responded to the treatment. This study proposed TMEM97 as a biomarker of prognosis but also of the responses of NSCLC patients to chemotherapies.
Two other TMEMs could have an impact in chemoresistance via the immune system, TMEM176A and TMEM176B. These two proteins can physically interact one with the other and are both localized in the plasma membrane and vesicular intracellular compartments (Cuajungco et al., 2012). The expression of these two proteins is increased in lymphoma, which may allow the cancer cells to evade the immune system or negatively impact their detection by immune system (Cuajungco et al., 2012).
Knowing that many chemotherapeutic drugs induced cancer cell death, several TMEMs could also have an impact in chemoresistance by exerting an anti-apoptotic function. TMEM48 is such an example for lung cancer (Qiao et al., 2016), TMEM14A for ovarian cancer (Zhang et al., 2016) and TMEM45B for lung and pancreatic cancers (Hu et al., 2016; Zhao et al., 2016).
The resistance to chemotherapy is not only due to the adaptation of cancer cells themselves but can involve tumor microenvironment. Furthermore, the mechanisms underlying the resistance to treatment can differ according to the cancer type and to the chemotherapeutic drug. The studies reported in this review showed that some TMEM proteins are involved in resistance to treatment and so can be used as new therapeutic targets (Figure 2). Finally, since TGF-β-induced quiescence renders cancer cells resistant to some anticancer agents (Brown et al., 2017; Tamai et al., 2017) and since many TMEM proteins interfer with TGF-β-induced intracellular signaling, TGF-β pathway is probably one of the key mechanisms through which TMEM proteins exert their effects.
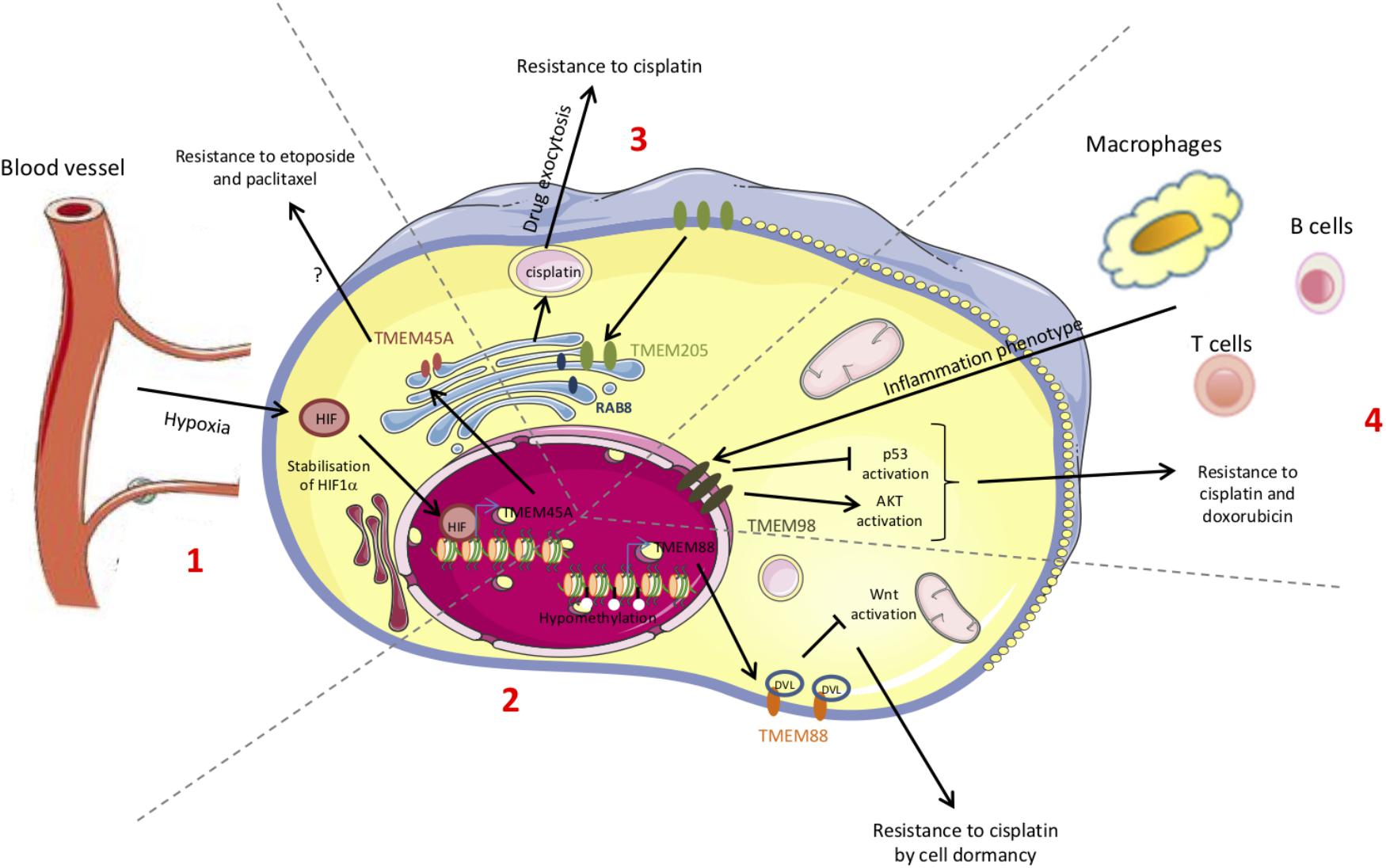
Figure 2. Schematic representation of the chemoresistance mechanisms conferred by TMEM proteins. The chemoresistance can be due to an adaptation of the cancer cells themselves (mutations, DNA methylation, proteins, translocation…) but can also be provided from the interactions with the microenvironment. Some of these chemoresistance mechanisms involve TMEM proteins. (1) Hypoxia leads to HIF-1α stabilization and to the expression of several target genes such as TMEM45A. (2) Methylation or acetylation of promoters leads to the transcriptional regulation of genes such as TMEM88. (3) The increase in TMEM205 expression and its translocation modify its partners. (4) The immune system induces the expression of several genes such as TMEM98.
Conclusion
Despite the different role and localization of TMEM proteins, many of them are implicated in cancer (Table 1). Some of them can be correlated with stages and patient survival and so be used as biomarkers and/or classifiers. Others have a role in carcinogenesis and tumor progression, but for most of them, the mechanism involved is still unknown. A better characterization of these proteins could help to better understand their implication in cancer. A few of them are even involved in chemoresistance and could be used as new therapeutic targets to enhance the efficiency of chemotherapies.
Author Contributions
KS wrote the review and designed the figures and the table. CM supervised the whole work, contributed to writing, and critically revised the paper.
Funding
KS was recipient of a Télévie grant (FNRS, Belgium).
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Abbreviations
BRCA1, breast cancer type 1 susceptibility protein; CDK, cyclin-dependent kinase; DVL1, disheveled 1; EMT, epithelial–mesenchymal transition; ERK, extracellular signal-regulated kinase; GSKβ, glycogen synthase kinase 3β; HDAC, histone deacetylase; HNSCC, head and neck squamous cell carcinoma; HPDE, normal pancreatic ductal epithelium; ICAM, intercellular adhesion molecule 1; IFN, interferon; IL, interleukin; MAC30, meningioma-associated protein; MEK, mitogen-activated protein kinase kinase; MMP, matrix metalloproteinase; mRNA, messenger ribonucleic acid; NDC1, transmembrane Nucleoporin; NSCLC, non-small cell lung cancer; PCNA, proliferating cell nuclear antigen; RAB8, Ras-related protein; SQCLC, squamous cell lung carcinoma; STXR6, syntaxin 6; TLR, toll-like receptor; TGF-β, transforming growth factor-β; TNF, tumor necrosis factor; TMEM, transmembrane protein; UTR, untranslated region; VCAM, vascular cell adhesion molecule 1; ZO-1, zona occludens 1.
References
Agudo, D., Gomez-Esquer, F., Martinez-Arribas, F., Nunez-Villar, M. J., Pollan, M., and Schneider, J. (2004). Nup88 mRNA overexpression is associated with high aggressiveness of breast cancer. Int. J. Cancer 109, 717–720. doi: 10.1002/ijc.20034
Akkafa, F., Koyuncu, I., Temiz, E., Dagli, H., Dilmec, F., and Akbas, H. (2018). miRNA-mediated apoptosis activation through TMEM 48 inhibition in A549 cell line. Biochem. Biophys. Res. Commun. 503, 323–329. doi: 10.1016/j.bbrc.2018.06.023
Atalay, A., Crook, T., Ozturk, M., and Yulug, I. G. (2002). Identification of genes induced by BRCA1 in breast cancer cells. Biochem. Biophys. Res. Commun. 299, 839–846. doi: 10.1016/S0006-291X(02)02751-1
Barradas, M., Gonos, E. S., Zebedee, Z., Kolettas, E., Petropoulou, C., Delgado, M. D., et al. (2002). Identification of a candidate tumor-suppressor gene specifically activated during Ras-induced senescence. Exp. Cell Res. 273, 127–137. doi: 10.1006/excr.2001.5434
Brown, J. A., Yonekubo, Y., Hanson, N., Sastre-Perona, A., Basin, A., Rytlewski, J. A., et al. (2017). TGF-beta-induced quiescence mediates chemoresistance of tumor-propagating cells in squamous cell carcinoma. Cell Stem Cell 21, 650.e8–664.e8. doi: 10.1016/j.stem.2017.10.001
Carles, A., Millon, R., Cromer, A., Ganguli, G., Lemaire, F., Young, J., et al. (2006). Head and neck squamous cell carcinoma transcriptome analysis by comprehensive validated differential display. Oncogene 25, 1821–1831. doi: 10.1038/sj.onc.1209203
Chen, R., Alvero, A. B., Silasi, D. A., and Mor, G. (2007). Inflammation, cancer and chemoresistance: taking advantage of the toll-like receptor signaling pathway. Am. J. Reprod. Immunol. 57, 93–107. doi: 10.1111/j.1600-0897.2006.00441.x
Chen, R., Fen, Y., Lin, X., Ma, T., Cai, H., and Ding, H. (2016). Overexpression of MAC30 is resistant to platinum-based chemotherapy in patients with non-small cell lung cancer. Technol. Cancer Res. Treat. 15, 815–820. doi: 10.1177/1533034615605208
Cheng, Z., Guo, J., Chen, L., Luo, N., Yang, W., and Qu, X. (2015). Overexpression of TMEM158 contributes to ovarian carcinogenesis. J. Exp. Clin. Cancer Res. 34:75. doi: 10.1186/s13046-015-0193-y
Clark, H. F., Gurney, A. L., Abaya, E., Baker, K., Baldwin, D., Brush, J., et al. (2003). The secreted protein discovery initiative (SPDI), a large-scale effort to identify novel human secreted and transmembrane proteins: a bioinformatics assessment. Genome Res. 13, 2265–2270. doi: 10.1101/gr.1293003
Cuajungco, M. P., Podevin, W., Valluri, V. K., Bui, Q., Nguyen, V. H., and Taylor, K. (2012). Abnormal accumulation of human transmembrane (TMEM)-176A and 176B proteins is associated with cancer pathology. Acta Histochem. 114, 705–712. doi: 10.1016/j.acthis.2011.12.006
D’Angelo, M. A., and Hetzer, M. W. (2008). Structure, dynamics and function of nuclear pore complexes. Trends Cell Biol. 18, 456–466. doi: 10.1016/j.tcb.2008.07.009
de Leon, M., Cardenas, H., Vieth, E., Emerson, R., Segar, M., Liu, Y., et al. (2016). Transmembrane protein 88 (TMEM88) promoter hypomethylation is associated with platinum resistance in ovarian cancer. Gynecol. Oncol. 142, 539–547. doi: 10.1016/j.ygyno.2016.06.017
Ding, H., Gui, X., Lin, X., Chen, R., Ma, T., Sheng, Y., et al. (2017). The prognostic effect of MAC30 expression on patients with non-small cell lung cancer receiving adjuvant chemotherapy. Technol. Cancer Res. Treat. 16, 645–653. doi: 10.1177/1533034616670443
Ding, H., Gui, X. H., Lin, X. B., Chen, R. H., Cai, H. R., Fen, Y., et al. (2016). Prognostic value of MAC30 expression in human pure squamous cell carcinomas of the lung. Asian Pac. J. Cancer Prev. 17, 2705–2710.
Dodeller, F., Gottar, M., Huesken, D., Iourgenko, V., and Cenni, B. (2008). The lysosomal transmembrane protein 9B regulates the activity of inflammatory signaling pathways. J. Biol. Chem. 283, 21487–21494. doi: 10.1074/jbc.M801908200
Doolan, P., Clynes, M., Kennedy, S., Mehta, J. P., Germano, S., Ehrhardt, C., et al. (2009). TMEM25, REPS2 and Meis 1: favourable prognostic and predictive biomarkers for breast cancer. Tumour Biol. 30, 200–209. doi: 10.1159/000239795
Flamant, L., Notte, A., Ninane, N., Raes, M., and Michiels, C. (2010). Anti-apoptotic role of HIF-1 and AP-1 in paclitaxel exposed breast cancer cells under hypoxia. Mol. Cancer 9:191. doi: 10.1186/1476-4598-9-191
Flamant, L., Roegiers, E., Pierre, M., Hayez, A., Sterpin, C., De Backer, O., et al. (2012). TMEM45A is essential for hypoxia-induced chemoresistance in breast and liver cancer cells. BMC Cancer 12:391. doi: 10.1186/1471-2407-12-391
Foulquier, F., Amyere, M., Jaeken, J., Zeevaert, R., Schollen, E., Race, V., et al. (2012). TMEM165 deficiency causes a congenital disorder of glycosylation. Am. J. Hum. Genet. 91, 15–26. doi: 10.1016/j.ajhg.2012.05.002
Fu, W., Cheng, Y., Zhang, Y., Mo, X., Li, T., Liu, Y., et al. (2015). The secreted form of transmembrane protein 98 promotes the differentiation of T helper 1 cells. J. Interferon Cytokine Res. 35, 720–733. doi: 10.1089/jir.2014.0110
Gao, D., Han, Y., Yang, Y., Herman, J. G., Linghu, E., Zhan, Q., et al. (2017). Methylation of TMEM176A is an independent prognostic marker and is involved in human colorectal cancer development. Epigenetics 12, 575–583. doi: 10.1080/15592294.2017.1341027
Ge, Y. X., Wang, C. H., Hu, F. Y., Pan, L. X., Min, J., Niu, K. Y., et al. (2018). New advances of TMEM88 in cancer initiation and progression, with special emphasis on Wnt signaling pathway. J. Cell Physiol. 233, 79–87. doi: 10.1002/jcp.25853
Guo, J., Chen, L., Luo, N., Yang, W., Qu, X., and Cheng, Z. (2015). Inhibition of TMEM45A suppresses proliferation, induces cell cycle arrest and reduces cell invasion in human ovarian cancer cells. Oncol. Rep. 33, 3124–3130. doi: 10.3892/or.2015.3902
Han, K. Y., Gu, X., Wang, H. R., Liu, D., Lv, F. Z., and Li, J. N. (2013). Overexpression of MAC30 is associated with poor clinical outcome in human non-small-cell lung cancer. Tumour Biol. 34, 821–825. doi: 10.1007/s13277-012-0612-z
Hayez, A., Malaisse, J., Roegiers, E., Reynier, M., Renard, C., Haftek, M., et al. (2014). High TMEM45A expression is correlated to epidermal keratinization. Exp. Dermatol. 23, 339–344. doi: 10.1111/exd.12403
Hodo, Y., Hashimoto, S., Honda, M., Yamashita, T., Suzuki, Y., Sugano, S., et al. (2010). Comprehensive gene expression analysis of 5′-end of mRNA identified novel intronic transcripts associated with hepatocellular carcinoma. Genomics 95, 217–223. doi: 10.1016/j.ygeno.2010.01.004
Hrasovec, S., Hauptman, N., Glavac, D., Jelenc, F., and Ravnik-Glavac, M. (2013). TMEM25 is a candidate biomarker methylated and down-regulated in colorectal cancer. Dis. Markers 34, 93–104. doi: 10.1155/2013/427890
Hu, R., Hu, F., Xie, X., Wang, L., Li, G., Qiao, T., et al. (2016). TMEM45B, up-regulated in human lung cancer, enhances tumorigenicity of lung cancer cells. Tumour Biol. 37, 12181–12191. doi: 10.1007/s13277-016-5063-5
Huebner, K. (2001). Tumor suppressors on 3p: a neoclassic quartet. Proc. Natl. Acad. Sci. U.S.A. 98, 14763–14765. doi: 10.1073/pnas.261586598
Kannan, K., Amariglio, N., Rechavi, G., Jakob-Hirsch, J., Kela, I., Kaminski, N., et al. (2001). DNA microarrays identification of primary and secondary target genes regulated by p53. Oncogene 20, 2225–2234. doi: 10.1038/sj.onc.1204319
Katoh, M., and Katoh, M. (2004). Identification and characterization of human TMEM25 and mouse Tmem25 genes in silico. Oncol. Rep. 12, 429–433. doi: 10.3892/or.12.2.429
Kau, T. R., Way, J. C., and Silver, P. A. (2004). Nuclear transport and cancer: from mechanism to intervention. Nat. Rev. Cancer 4, 106–117. doi: 10.1038/nrc1274
Kayed, H., Kleeff, J., Ding, J., Hammer, J., Giese, T., Zentgraf, H., et al. (2004). Expression analysis of MAC30 in human pancreatic cancer and tumors of the gastrointestinal tract. Histol. Histopathol. 19, 1021–1031.
Lee, H. J., Finkelstein, D., Li, X., Wu, D., Shi, D. L., and Zheng, J. J. (2010). Identification of transmembrane protein 88 (TMEM88) as a dishevelled-binding protein. J. Biol. Chem. 285, 41549–41556. doi: 10.1074/jbc.M110.193383
Lee, S., Stewart, S., Nagtegaal, I., Luo, J., Wu, Y., Colditz, G., et al. (2012). Differentially expressed genes regulating the progression of ductal carcinoma in situ to invasive breast cancer. Cancer Res. 72, 4574–4586. doi: 10.1158/0008-5472.CAN-12-0636
Li, Y., Guo, W., Liu, S., Zhang, B., Yu, B. B., Yang, B., et al. (2017). Silencing transmembrane protein 45B (TNEM45B) inhibits proliferation, invasion, and tumorigenesis in osteosarcoma cells. Oncol. Res. 25, 1021–1026. doi: 10.3727/096504016X14821477992177
Li, B., Huang, M. Z., Wang, X. Q., Tao, B. B., Zhong, J., Wang, X. H., et al. (2015a). Erratum: TMEM140 is associated with the prognosis of glioma by promoting cell viability and invasion. J. Hematol. Oncol. 8:101. doi: 10.1186/s13045-015-0199-0
Li, B., Huang, M. Z., Wang, X. Q., Tao, B. B., Zhong, J., Wang, X. H., et al. (2015b). TMEM140 is associated with the prognosis of glioma by promoting cell viability and invasion. J. Hematol. Oncol. 8:101. doi: 10.1186/s13045-015-0199-0
Liu, F., Cao, Q. H., Lu, D. J., Luo, B., Lu, X. F., Luo, R. C., et al. (2015). TMEM16A overexpression contributes to tumor invasion and poor prognosis of human gastric cancer through TGF-beta signaling. Oncotarget 6, 11585–11599.
Ma, R., Feng, N., Yu, X., Lin, H., Zhang, X., Shi, O., et al. (2017). Promoter methylation of Wnt/beta-Catenin signal inhibitor TMEM88 is associated with unfavorable prognosis of non-small cell lung cancer. Cancer Biol. Med. 14, 377–386. doi: 10.20892/j.issn.2095-3941.2017.0061
Mahipal, A., and Malafa, M. (2016). Importins and exportins as therapeutic targets in cancer. Pharmacol. Ther. 164, 135–143. doi: 10.1016/j.pharmthera.2016.03.020
Malhotra, K., Luehrsen, K. R., Costello, L. L., Raich, T. J., Sim, K., Foltz, L., et al. (1999). Identification of differentially expressed mRNAs in human fetal liver across gestation. Nucleic Acids Res. 27, 839–847. doi: 10.1093/nar/27.3.839
Manawapat-Klopfer, A., Thomsen, L. T., Martus, P., Munk, C., Russ, R., Gmuender, H., et al. (2016). TMEM45A, SERPINB5 and p16INK4A transcript levels are predictive for development of high-grade cervical lesions. Am. J. Cancer Res. 6, 1524–1536.
Mao, M., Chen, J., Li, X., and Wu, Z. (2015). siRNA-TMEM98 inhibits the invasion and migration of lung cancer cells. Int. J. Clin. Exp. Pathol. 8, 15661–15669.
Matsuba, S., Niwa, S., Muraki, K., Kanatsuka, S., Nakazono, Y., Hatano, N., et al. (2014). Downregulation of Ca2(-activated Cl- channel TMEM16A by the inhibition of histone deacetylase in TMEM16A-expressing cancer cells. J. Pharmacol. Exp. Ther. 351, 510–518. doi: 10.1124/jpet.114.217315
Murphy, M., Pykett, M. J., Harnish, P., Zang, K. D., and George, D. L. (1993). Identification and characterization of genes differentially expressed in meningiomas. Cell Growth Differ. 1993, 715–722.
Ng, K. T., Lo, C. M., Guo, D. Y., Qi, X., Li, C. X., Geng, W., et al. (2014). Identification of transmembrane protein 98 as a novel chemoresistance-conferring gene in hepatocellular carcinoma. Mol. Cancer Ther. 13, 1285–1297. doi: 10.1158/1535-7163.MCT-13-0806
Qiao, W., Han, Y., Jin, W., Tian, M., Chen, P., Min, J., et al. (2016). Overexpression and biological function of TMEM48 in non-small cell lung carcinoma. Tumour Biol. 37, 2575–2586. doi: 10.1007/s13277-015-4014-x
Qiu, G., Sun, W., Zou, Y., Cai, Z., Wang, P., Lin, X., et al. (2015). RNA interference against TMEM97 inhibits cell proliferation, migration, and invasion in glioma cells. Tumour Biol. 36, 8231–8238. doi: 10.1007/s13277-015-3552-6
Ramalho-Carvalho, J., Goncalves, C. S., Graca, I., Bidarra, D., Pereira-Silva, E., Salta, S., et al. (2018). multiplatform approach identifies miR-152-3p as a common epigenetically regulated onco-suppressor in prostate cancer targeting TMEM97. Clin. Epigenet. 10:40. doi: 10.1186/s13148-018-0475-2
Ruiz, C., Martins, J. R., Rudin, F., Schneider, S., Dietsche, T., Fischer, C. A., et al. (2012). Enhanced expression of ANO1 in head and neck squamous cell carcinoma causes cell migration and correlates with poor prognosis. PLoS One 7:e43265. doi: 10.1371/journal.pone.0043265
Sauter, D. R. P., Novak, I., Pedersen, S. F., Larsen, E. H., and Hoffmann, E. K. (2015). ANO1 (TMEM16A) in pancreatic ductal adenocarcinoma (PDAC). Pflugers Arch. 467, 1495–1508. doi: 10.1007/s00424-014-1598-8
Sermeus, A., Cosse, J. P., Crespin, M., Mainfroid, V., de Longueville, F., Ninane, N., et al. (2008). Hypoxia induces protection against etoposide-induced apoptosis: molecular profiling of changes in gene expression and transcription factor activity. Mol. Cancer 7:27. doi: 10.1186/1476-4598-7-27
Shen, D. W., and Gottesman, M. M. (2012). RAB8 enhances TMEM205-mediated cisplatin resistance. Pharm. Res. 29, 643–650. doi: 10.1007/s11095-011-0562-y
Shen, D. W., Ma, J., Okabe, M., Zhang, G., Xia, D., and Gottesman, M. M. (2010). Elevated expression of TMEM205, a hypothetical membrane protein, is associated with cisplatin resistance. J. Cell Physiol. 225, 822–828. doi: 10.1002/jcp.22287
Shen, K., Yu, W., Yu, Y., Liu, X., and Cui, X. (2018). Knockdown of TMEM45B inhibits cell proliferation and invasion in gastric cancer. Biomed. Pharmacother. 104, 576–581. doi: 10.1016/j.biopha.2018.05.016
Smith, J. J., Deane, N. G., Wu, F., Merchant, N. B., Zhang, B., Jiang, A., et al. (2010). Experimentally derived metastasis gene expression profile predicts recurrence and death in patients with colon cancer. Gastroenterology 138, 958–968. doi: 10.1053/j.gastro.2009.11.005
Song, Y., Gao, J., Guan, L., Chen, X., Gao, J., and Wang, K. (2018). Inhibition of ANO1/TMEM16A induces apoptosis in human prostate carcinoma cells by activating TNF-alpha signaling. Cell Death Dis. 9:703. doi: 10.1038/s41419-018-0735-2
Stavru, F., Hulsmann, B. B., Spang, A., Hartmann, E., Cordes, V. C., and Gorlich, D. (2006). NDC1: a crucial membrane-integral nucleoporin of metazoan nuclear pore complexes. J. Cell Biol. 173, 509–519. doi: 10.1083/jcb.200601001
Sui, Y., Sun, M., Wu, F., Yang, L., Di, W., Zhang, G., et al. (2014). Inhibition of TMEM16A expression suppresses growth and invasion in human colorectal cancer cells. PLoS One 9:e115443. doi: 10.1371/journal.pone.0115443
Sun, W., Qiu, G., Zou, Y., Cai, Z., Wang, P., Lin, X., et al. (2015). Knockdown of TMEM45A inhibits the proliferation, migration and invasion of glioma cells. Int. J. Clin. Exp. Pathol. 8, 12657–12667.
Tamai, M., Furuichi, Y., Kasai, S., Ando, N., Harama, D., Goi, K., et al. (2017). TGFbeta1 synergizes with FLT3 ligand to induce chemoresistant quiescence in acute lymphoblastic leukemia with MLL gene rearrangements. Leuk Res. 61, 68–76. doi: 10.1016/j.leukres.2017.08.013
Thomas-Gatewood, C., Neeb, Z. P., Bulley, S., Adebiyi, A., Bannister, J. P., Leo, M. D., et al. (2011). TMEM16A channels generate Ca(2)(()-activated Cl(-) currents in cerebral artery smooth muscle cells. Am. J. Physiol. Heart Circ Physiol. 301, H1819–H1827. doi: 10.1152/ajpheart.00404.2011
Vinothkumar, K. R., and Henderson, R. (2010). Structures of membrane proteins. Quart. Rev. Biophys. 43, 65–158. doi: 10.1017/S0033583510000041
von Heijne, G. (2006). Membrane-protein topology. Nat. Rev. Mol. Cell Biol. 7, 909–918. doi: 10.1038/nrm2063
Wang, Y., Zhang, Y., Herman, J. G., Linghu, E., and Guo, M. (2017). Epigenetic silencing of TMEM176A promotes esophageal squamous cell cancer development. Oncotarget 8, 70035–70048. doi: 10.18632/oncotarget.19550
Wrzesinski, T., Szelag, M., Cieslikowski, W. A., Ida, A., Giles, R., Zodro, E., et al. (2015). Expression of pre-selected TMEMs with predicted ER localization as potential classifiers of ccRCC tumors. BMC Cancer 15:518. doi: 10.1186/s12885-015-1530-4
Xiao, M., Li, H., Yang, S., Huang, Y., Jia, S., Wang, H., et al. (2013). Expression of MAC30 protein is related to survival and clinicopathological variables in breast cancer. J. Surg. Oncol. 107, 456–462. doi: 10.1002/jso.23269
Xu, X. Y., Zhang, L. J., Yu, Y. Q., Zhang, X. T., Huang, W. J., Nie, X. C., et al. (2014). Down-regulated MAC30 expression inhibits proliferation and mobility of human gastric cancer cells. Cell Physiol. Biochem. 33, 1359–1368. doi: 10.1159/000358703
Yang, S., Li, H., Liu, Y., Ning, X., Meng, F., Xiao, M., et al. (2013). Elevated expression of MAC30 predicts lymph node metastasis and unfavorable prognosis in patients with epithelial ovarian cancer. Med. Oncol. 30:324. doi: 10.1007/s12032-012-0324-7
Yu, X., Zhang, X., Zhang, Y., Jiang, G., Mao, X., and Jin, F. (2015). Cytosolic TMEM88 promotes triple-negative breast cancer by interacting with Dvl. Oncotarget 6, 25034–25045. doi: 10.18632/oncotarget.4379
Zhang, Q., Chen, X., Zhang, X., Zhan, J., and Chen, J. (2016). Knockdown of TMEM14A expression by RNAi inhibits the proliferation and invasion of human ovarian cancer cells. Biosci. Rep. 36:e00298. doi: 10.1042/BSR20150258
Zhang, X., Yu, X., Jiang, G., Miao, Y., Wang, L., Zhang, Y., et al. (2015). Cytosolic TMEM88 promotes invasion and metastasis in lung cancer cells by binding DVLS. Cancer Res. 75, 4527–4537. doi: 10.1158/0008-5472.CAN-14-3828
Zhao, L. C., Shen, B. Y., Deng, X. X., Chen, H., Zhu, Z. G., and Peng, C. H. (2016). TMEM45B promotes proliferation, invasion and migration and inhibits apoptosis in pancreatic cancer cells. Mol. Biosyst. 12, 1860–1870. doi: 10.1039/C6MB00203J
Zhou, X., Popescu, N. C., Klein, G., and Imreh, S. (2007). The interferon-alpha responsive gene TMEM7 suppresses cell proliferation and is downregulated in human hepatocellular carcinoma. Cancer Genet. Cytogenet. 177, 6–15. doi: 10.1016/j.cancergencyto.2007.04.007
Keywords: cancer, TMEM proteins, biomarkers, tumor suppressors, oncogenes, chemoresistance
Citation: Schmit K and Michiels C (2018) TMEM Proteins in Cancer: A Review. Front. Pharmacol. 9:1345. doi: 10.3389/fphar.2018.01345
Received: 17 June 2018; Accepted: 31 October 2018;
Published: 06 December 2018.
Edited by:
Agnès Noël, Université de Liège, BelgiumReviewed by:
Zhijie Xu, Central South University, ChinaKaren Maegley, Pfizer (United States), United States
Copyright © 2018 Schmit and Michiels. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Carine Michiels, Y2FyaW5lLm1pY2hpZWxzQHVuYW11ci5iZQ==
 Kathleen Schmit
Kathleen Schmit Carine Michiels
Carine Michiels