- 1Division of Bacterial Diseases, State Key Laboratory of Veterinary Biotechnology, Harbin Veterinary Research Institute, Chinese Academy of Agricultural Sciences, Harbin, China
- 2Tianjin Animal Science and Veterinary Research Institute, Tianjin, China
- 3Institute of Microbiology and Epizootics, Department of Veterinary Medicine, Freie Universität Berlin, Berlin, Germany
This study aimed at investigating the antibacterial activity of aspidinol, an extract from Dryopteris fragrans (L.) Schott, against methicillin-resistant Staphylococcus aureus (MRSA). MRSA isolates were treated with aspidinol to determine the differential expression of genes and associated pathways following the drug treatment. Aspidinol displayed significant anti-MRSA activity, both in vivo (minimum inhibitory concentration = 2 μg/mL) and in vitro, and achieved an antibacterial effect comparable to that of vancomycin. In the lethal septicemic mouse study, a dose of 50 mg/kg of either aspidinol or vancomycin provided significant protection from mortality. In the non-lethal septicemic mouse study, aspidinol and vancomycin produced a significant reduction in mean bacterial load in murine organs, including the spleen, lung, and liver. After treatment with aspidinol, we found through RNA-seq and RT-PCR experiments that the inhibition of the formation of ribosomes was the primary S. aureus cell-killing mechanism, and the inhibition of amino acid synthesis and the reduction of virulence factors might play a secondary role.
Introduction
Methicillin-resistant Staphylococcus aureus (MRSA) is the most common pathogen and is associated with high morbidity and mortality in both humans and animals (VanEperen and Segreti, 2016). MRSA causes nosocomial infections, pneumonia (Woods and Colice, 2014), sepsis (Taylor, 2013), and skin infections (Mihu et al., 2015). In January 2017, MRSA was officially ranked as one of the 12 deadliest drug-resistant bacteria by WHO (2017). Antibiotics are the most effective tool to combat infections due to pathogenic bacteria. While new antimicrobial agents are becoming increasingly difficult to develop, medicine is currently unable to keep pace with the emergence of resistant bacteria (Rennie, 2012). Therefore, it is imperative to develop new agents to fight resistant bacteria, especially MRSA.
Natural products are alternative tools in the fight against drug-resistant bacteria, and synthetic approaches are unable to replace natural products (Guzman et al., 2010; Vandal et al., 2015; Farah et al., 2016). Chinese herb Dryopteris fragrans (L.) Schott is a deciduous perennial herbaceous plant of Dryopteris, it is widely distributed in many countries and is mainly grown in northeast of China (Zhong et al., 2017). As a traditional Chinese herb medicine, D. fragrans was used to treat a variety of diseases, especially skin diseases such as psoriasis, rashes, dermatitis, and beriberi (Huang et al., 2014). Recently, there are also reports of the antibacterial (Gao et al., 2016), anti-tumor (Zhang et al., 2012), anti-inflammatory, and antifungal activity of its extracts (Huang et al., 2014; Peng et al., 2016). Aspidinol is a phloroglucinol derivative from the stem and leaf of traditional D. fragrans (Zhao et al., 2014; Gao et al., 2016). To date, the research on aspidinol has been rather limited; only one publication has reported that it has anti-cancer activity (Zhao et al., 2014).
In this study, aspidinol was found to be an effective anti-MRSA agent. We have verified that aspidinol efficiently cleared intracellular bacteria and exhibited excellent in vivo activity, which indicated its potential for application as an antibacterial agent to treat systemic MRSA infections. Building upon this seminal work, we investigated the anti-MRSA mechanism of aspidinol. RNA-seq demonstrated that aspidinol appears to interfere with multiple biological pathways in S. aureus and that this interference ultimately results in the killing of MRSA.
Herein, we report for the first time the validation process of the anti-MRSA activity of aspidinol and identify its mode of action. All of the findings strongly verified the potential therapeutic application of aspidinol as a novel antibacterial agent.
Materials and Methods
Strains and Growth Conditions
The S. aureus strains ATCC 29213 and ATCC 33591 (MRSA) were obtained from the American Type Culture Collection. The clinical MSSA and MRSA isolates used in this study were provided by the First Affiliated Hospital of Harbin Medical University, Harbin, China. A complete list of bacterial strains used in this study can be found in Supplementary Table S1. All strains were maintained in Mueller-Hinton broth (MHB, Oxoid, Basingstoke, United Kingdom), frozen at -80°C until use, and cultured in MHB at 37°C under aerobic conditions.
Antimicrobial Agents
Aspidinol, vancomycin, and linezolid were purchased from Sigma-Aldrich (Bornem, Belgium). Aspidinol was stored in DMSO at -20°C. All antibiotics were dissolved in ultrapure water.
Minimum Inhibitory Concentration and Minimum Bactericidal Concentration
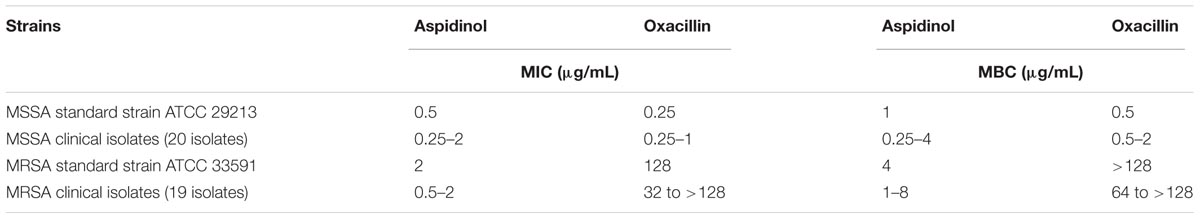
Broth microdilution was used to determine the minimum inhibitory concentration (MIC) according to CLSI standards M07-A9. The test medium used for most species was MHB, and the cell concentration was adjusted to approximately 5 × 105 cells/mL. After 20 h of incubation at 35 ± 2°C, the MIC was read as the lowest concentration of antibiotic that inhibited visible growth of the bacteria. The minimum bactericidal concentration (MBC) was defined as the lowest concentration of aspidinol to kill S. aureus ATCC 33591 cells. S. aureus ATCC 33591 cells from the MIC assays were resuspended in fresh media and plated onto Mueller-Hinton agar (MHA). After incubating for 24 h at 37°C, the colonies were enumerated. All experiments were performed with three replicates.
Time-Dependent Killing
An overnight culture of cells (S. aureus ATCC 33591) was diluted 1:5,000 in MHB and incubated at 37°C at 220 r.p.m. for 2 h after overnight culture. The bacteria cells were then treated with aspidinol, linezolid, or vancomycin at a concentration of 10 × MIC. One milliliter aliquots were removed at 2, 4, 6, 8, 12, 24, and 48 h, and 1 mL of MHB was added as a replacement. The suspension was centrifuged at 10,000 g for 1 min, and the pellet was resuspended in 100 mL of sterile PBS buffer. The diluted suspensions were plated onto MHA plates and incubated at 37°C overnight for CFU calculation. All experiments were performed with three replicates.
Cytotoxicity Assay
Macrophage cells (RAW264.7) were seeded at a density of 10,000 cells per well into a 96-well cell culture plate (NEST, Nest Biotech Co. Ltd, NJ, United States) and incubated overnight at 37°C in DMEM media containing 10% fetal bovine serum (FBS). Cells were then treated with aspidinol for 24 h at different concentrations ranging from 0 to 128 μg/mL. The treated cells were washed four times with PBS, and DMEM media containing the MTS assay reagent, 3-(4,5-dimethylthiazol-2-yl)-5-(3carboxymethoxyphenyl)-2-(4-sulfophenyl)-2H-tetrazolium) (Promega, Madison, WI, United States) was then added. After 4 h of incubation at 37°C, the absorbance was measured using an ELISA microplate reader (Molecular Devices, Sunnyvale, CA, United States). The percent cell viability of the aspidinol-treated cells was calculated.
Intracellular Infection Assay
After overnight incubation, macrophage cells (RAW 264.7) were infected with MRSA ATCC 33591 for 30 min at a multiplicity of infection (MOI) ratio of 100:1. The infected cells were then washed three times with DMEM medium containing 10 IU lysostaphin. Aspidinol (20 μg/mL), vancomycin (20 μg/mL), or linezolid (10 μg/mL) were added in complete DMEM medium containing 4 IU lysostaphin, which was then used to incubate the infected cells at 37°C (with 5% CO2). After 24 h, the cells were washed three times with PBS and lysed with 0.1% Triton X-100 (Sigma-Aldrich). The cell lysates were plated onto MHA plates and incubated for 24 h at 37°C, followed by CFU determination.
Biofilm Assay
Staphylococcus aureus ATCC 33591 was cultured in tryptic soy broth containing 1% glucose. A biofilm formed after 24 h of incubation at 37°C. The medium was then removed, and the biofilm washed with PBS. The drugs at a concentration of 10 × MIC were added and incubated for an additional 24 h at 37°C. The 96-well plates were washed again, and the biofilms were stained with 0.1% (wt/vol) crystal violet. The 96-well plates were washed and air-dried, and finally, the biofilm mass was dissolved using 95% ethanol. A microplate reader (Bio-Tek Instruments, Inc., United States) was used to measure the intensity of crystal violet. The data were presented as the percent of biofilm mass reduction in the treated groups in relation to the control group.
Scanning Electron Microscopy
The method of biofilm formation was carried out as described above on a glass coverslip in 24-well plates. The formed biofilm was fixed with 2.5% glutaraldehyde in 0.1 M sodium cacodylate buffer (pH 7.2) at 4°C for 10 min and then washed three times with PBS. Next, the biofilm was fixed with 1% osmic acid at room temperature for 10 min. Gradual dehydration was then carried out with ethyl alcohol (60, 70, 80, 90, 95, and 100%), and tertiary butanol was used as a displacement liquid (60, 70, 80, 90, 95, and 100%). Finally, the samples were freeze-dried overnight. The specimens were then sputter coated with gold for observation using a JSM 7500 (JEOL, Tokyo, Japan).
Mouse Studies
Eight-week-old female BALB/c mice (Vital River, Beijing, China) were used in all of the mouse studies. The animal experiments were performed in accordance with the animal ethics guidelines and approved protocols. The animal experiment was approved by the Animal Ethics Committee of Harbin Veterinary Research Institute of the Chinese Academy of Agricultural Sciences.
Systemic Lethal Infection
An overnight culture of S. aureus ATCC 33591 cells was washed and resuspended in PBS. Each mouse received an intraperitoneal (i.p.) injection (200 μL) containing the bacterial suspension (9 × 109 CFU). One hour after infection, the mice were divided into seven groups (10 mice per group). The mice were intravenously injected with either aspidinol or vancomycin (5, 15, or 25 mg/kg) or the vehicle alone (10% ethanol). The treatment was provided once daily for 3 days following infection. Mortality was monitored daily for 5 days, and the moribund mice were euthanized using CO2 asphyxiation.
Systemic Non-lethal Infection
The infection protocol was carried out as described above (systemic lethal infection) with the following exceptions. Each mouse received an i.p. injection containing 2 × 107 CFU S. aureus ATCC 33591. The mice were divided into three groups (16 mice per group) and intravenously injected with either aspidinol, vancomycin (25 mg/kg), or vehicle (10% ethanol). The mice were treated once daily for 6 days. The mice were euthanized 24 h after the last administration, and the organs (including heart, lung, kidney, spleen, and liver) were excised, homogenized in MHA, and finally incubated at 37°C for 24 h for MRSA CFU counting. For evaluation of the extent of tissue damage and cellular response, histological analyses were also performed. Mice from the control group, vancomycin group, and aspidinol group that underwent the same infection protocol as the systemic non-lethal infection were submitted to histopathology examination after 6 days. The following organs were collected from each animal: heart, liver, spleen, lung, and kidneys. The tissues were fixed with 4% paraformaldehyde and then stained with hematoxylin–eosin.
RNA-Seq Transcriptomics
Staphylococcus aureus ATCC 33591 was grown to an OD600 of 0.4 from an initial absorbance of 0.01, and aspidinol was added to a final sub-MIC concentration (1 μg/mL). Samples were collected at 1-h post treatment and preserved with RNA protect (Qiagen, United States) following the manufacturer’s instructions. Cells were pelleted by centrifugation at 5,000 g for 10 min at 4°C. The RNA was isolated using an RNeasy minikit (Qiagen, United States) in accordance with the manufacturer’s instructions, with the following additions. The cell pellets were homogenized in 1 mL Tris-buffered saline (TBS) (20 mM Tris, pH 7.5) containing 0.4 mg of lysostaphin and incubated at 37°C for 15 min. Subsequently, 20 mg of lysozyme in TE buffer (20 mM Tris, pH 7.5, 2 mM EDTA, pH 7.8) was added, and the sample was incubated at 25°C for 10 min. Control samples were collected from an antibiotic-free culture, and each experiment was repeated three times.
Three independently prepared RNA samples from each strain were used for RNA-Seq. Illumina sequencing was performed by Shanghai Majorbio Bio-pharm Technology Co., Ltd. (Shanghai, China) using the Illumina Hiseq2000 Truseq SBS Kit v3-HS (200 cycles) and Miseq Reagent Kit V2 (500 cycle/600 cycle) (Illumina, Inc.). The data analyses were performed using edgeR software. Genes exhibiting twofold changes in expression, which were statistically significant as determined by Student’s t-test (p < 0.05), were considered to be differentially expressed under the conditions indicated.
Real-Time PCR
To confirm the RNA-Seq data, we selected some genes that were downregulated and assessed their relative expression levels by real-time PCR. S. aureus ATCC 33591 cells were cultured in the same conditions as the RNA-Seq transcriptomics experiments. qRT-PCR was performed by a two-step process. These reactions were performed using the Applied qTOWER 2.2 (Analytik Jena, Jena, Germany) Real-Time PCR System by using the following cycle parameters: 95°C for 5 min, followed by 40 cycles of 95°C for 15 s, 55°C for 15 s, and 72°C for 15 s; and one dissociation step of 95°C for 1 min, 55°C for 30 s, and 95°C for 30 s. All measurements were independently conducted three times on two separate biological isolates. All of the primers and sequences are listed in Supplementary Table S2.
The melting curve analysis was performed immediately after amplification to verify the specificity of the PCR amplification products. Fluorescence was measured at the end of the annealing-extension phase of each cycle. A threshold value for the fluorescence of all samples was manually set. The reaction cycle at which the PCR product exceeds this fluorescence threshold was identified as the threshold cycle (CT). The relative quantitation was calculated by the 2-ΔΔCT method.
Statistical Analysis
Statistical analyses were carried out using GraphPad Prism 5.0 (Graph Pad Software, LaJolla, CA). One-way model ANOVA was performed between the groups. In ANOVA, the observed variance is partitioned into components due to different explanatory variables. A level of p < 0.05 was considered to be statistically significant.
Results
Antibacterial Activity of Aspidinol
The antimicrobial activity of aspidinol was tested against clinical isolates of S. aureus, including drug-resistant strains, the MICs and MBCs of which are given in Table 1. Aspidinol showed potent bactericidal activity against MSSA and MRSA, with MICs and MBCs ranging from 0.25 to 2 μg/mL and 0.5 to 4 μg/mL, respectively.
After confirming that aspidinol possessed excellent antimicrobial activity against S. aureus, we next assessed the killing kinetics of aspidinol. As depicted in Figure 1A, aspidinol also showed excellent bactericidal activity against S. aureus.
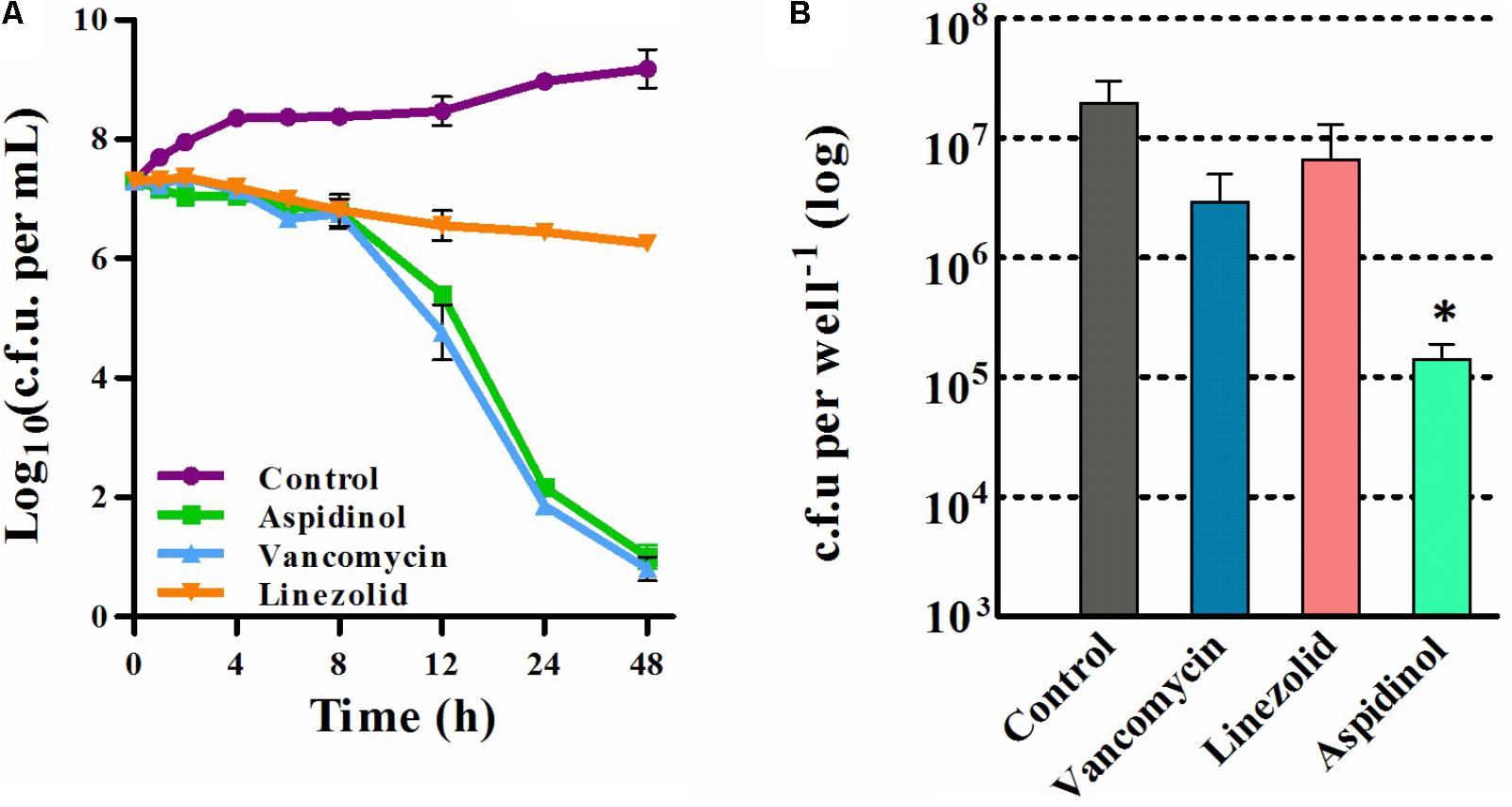
FIGURE 1. Time-kill analysis and the intracellular bacterial killing activity of aspidinol. (A) Time-kill kinetics of aspidinol against S. aureus ATCC 33591. The error bars show the standard deviations of the results from three independent biological repeats. (B) S. aureus ATCC 33591-infected RAW264.7 cells were treated with aspidinol and control antibiotics (vancomycin or linezolid) for 24 h, and the percent bacterial reduction was calculated compared to that of the untreated control groups. The results are given as the mean ± SD (n = 3). Two-tailed Student’s t-test was employed, and ∗p-values of ≤0.05 are considered to be significant.
Cytotoxicity of Aspidinol
Cytotoxicity tests indicated that the toxicity of aspidinol toward macrophage cells (RAW264.7) was negligible. At 128 μg/mL, i.e., the highest dose tested, aspidinol showed no toxicity to RAW264.7 cells. The dose response curve of aspidinol in the cytotoxicity assay can be found in Supplementary Figure S2.
Aspidinol Efficiently Clears Intracellular Bacteria
Eliminating intercellular S. aureus has long been considered to be key to clinical success because S. aureus is capable of entering multiple mammalian cells, thereby evading the antibiotic therapy. The ability to internalize into mammalian cells might result in the long-term colonization of the host and explain clinical failures. Lehar et al. have confirmed this hypothesis, showing that three major antibiotics commonly used for clinical standard care (vancomycin, daptomycin, and linezolid) failed to kill the highly virulent community-acquired S. aureus ATCC 33591 strain (Lehar et al., 2015).
As aspidinol exhibited potent anti-MRSA activity, we explored the ability of aspidinol to permeate cellular membranes and kill MRSA located inside eukaryotic cells. To investigate the efficacy of aspidinol in eliminating intracellular MRSA, macrophage cells (RAW264.7) infected with S. aureus ATCC 33591 were employed. As depicted in Figure 1B, intracellular S. aureus ATCC 33591 exposed to aspidinol (20 μg/mL) were efficiently killed, and the intracellular bacteria decreased by 100-fold. In contrast, conventional antibiotics such as linezolid (20 μg/mL) and vancomycin (10 μg/mL) were shown to reduce the bacterial burden inside the infected macrophages by 5- to 10-fold. From the above data, aspidinol was confirmed to be capable of eradicating MRSA infection in macrophage cells.
Inhibitory Effect of Aspidinol on MRSA Biofilm Formation
The biofilms were stained with crystal violet after treatment with aspidinol, and the absorbance was measured at 570 nm for mass estimation. Compared to linezolid and vancomycin, aspidinol was not able to significantly reduce adherent biofilms of S. aureus ATCC 33591. At a concentration of 256 μg/mL, which corresponds to the 128-fold MIC, aspidinol reduced the biofilm mass by approximately by 30%. The control antibiotics, linezolid (256 μg/mL, i.e., 128-fold MIC) and vancomycin (128 μg/mL, i.e., 128-fold MIC), were able to reduce the biofilm mass by only 20% (Figure 2A).
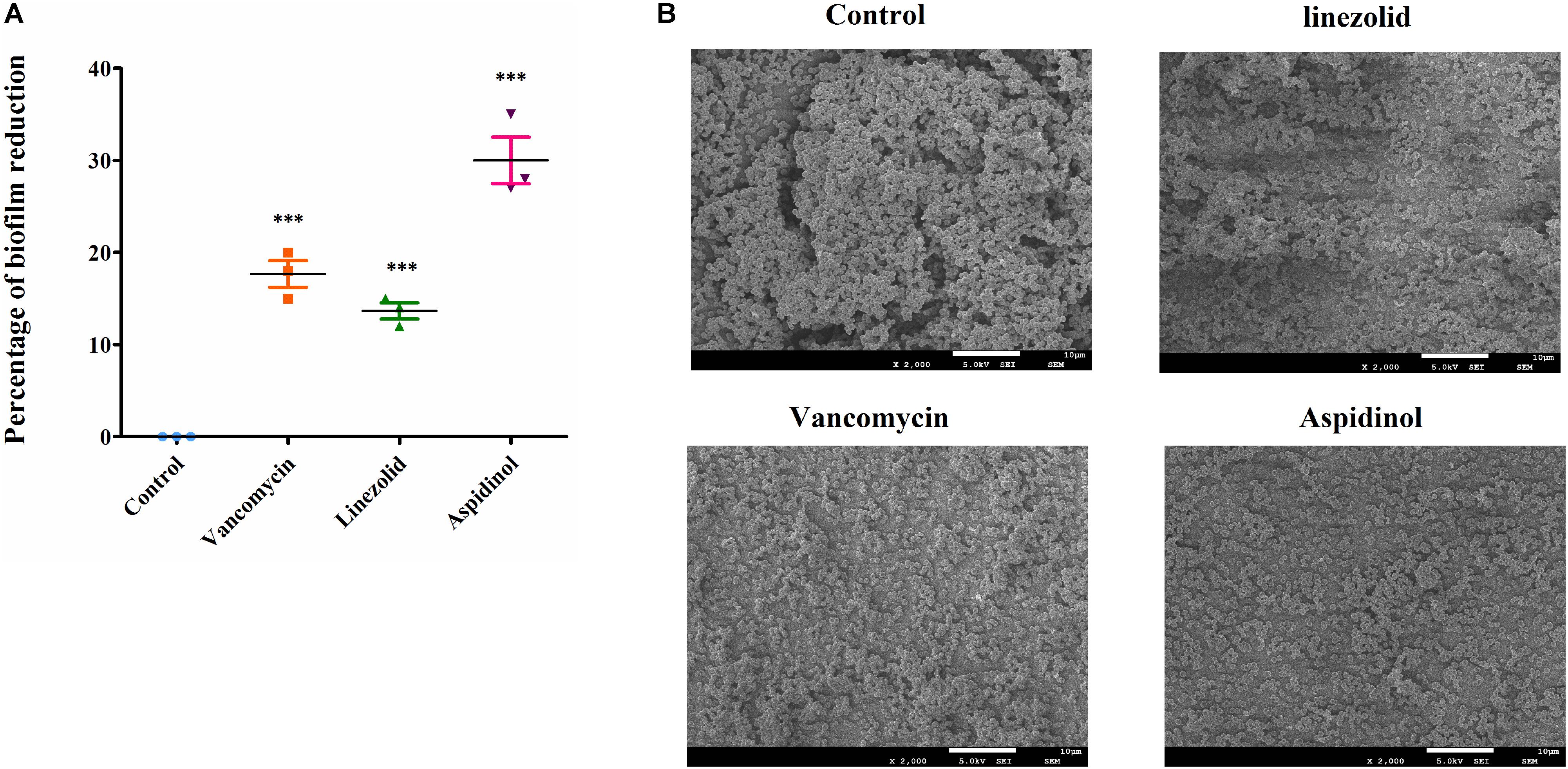
FIGURE 2. Anti-biofilm activity of aspidinol. (A) The effects of aspidinol and antibiotics (linezolid and vancomycin) on established biofilms of S. aureus ATCC 33591 were evaluated. The pre-formed biofilms were treated with linezolid, vancomycin, or aspidinol and then stained with crystal violet. The optical density of the dissolved crystal violet was measured using a spectrophotometer. The values are the mean of the triplicate samples with standard deviation bars. The results are given as the mean ± SD (n = 3). Two-tailed Student’s t-test was employed, and ∗∗∗ means p-values ≤ 0.005. (B) Scanning electron microscopy images showing the structure of MRSA biofilms treated with aspidinol and antibiotics at 24 h. Magnifications, ×2000.
The morphology of S. aureus ATCC 33591 biofilms on the surface of coverslips was observed using scanning electron microscope (SEM). Under a 2,000-fold magnification, the biofilm was shown to be composed of many multilayered MRSA colonies. The SEM analysis results were consistent with those of the crystal violet staining observations. At a concentration of 256 μg/mL of aspidinol, linezolid, and vancomycin, we observed a decrease in density of the MRSA biofilms (Figure 2B).
In Vivo Efficacy of Aspidinol
To determine the role of aspidinol against MRSA in vivo, both a lethal and a non-lethal systemic MRSA infection model were used. In the lethal systemic study, mice were infected intraperitoneally with MRSA ATCC 33591 (2.95 × 109 CFU per mouse). Both aspidinol and vancomycin provided significant protection from mortality (Figure 3A). The survival rate of infected mice varied in a concentration-dependent manner, with the survival rate improving dramatically when the dose of aspidinol increased. Approximately 80% of mice that received a higher dose of aspidinol (25 mg/kg) survived for 5 days. These results exhibited the in vivo activity of aspidinol in protecting mice from septicemic MRSA infection.
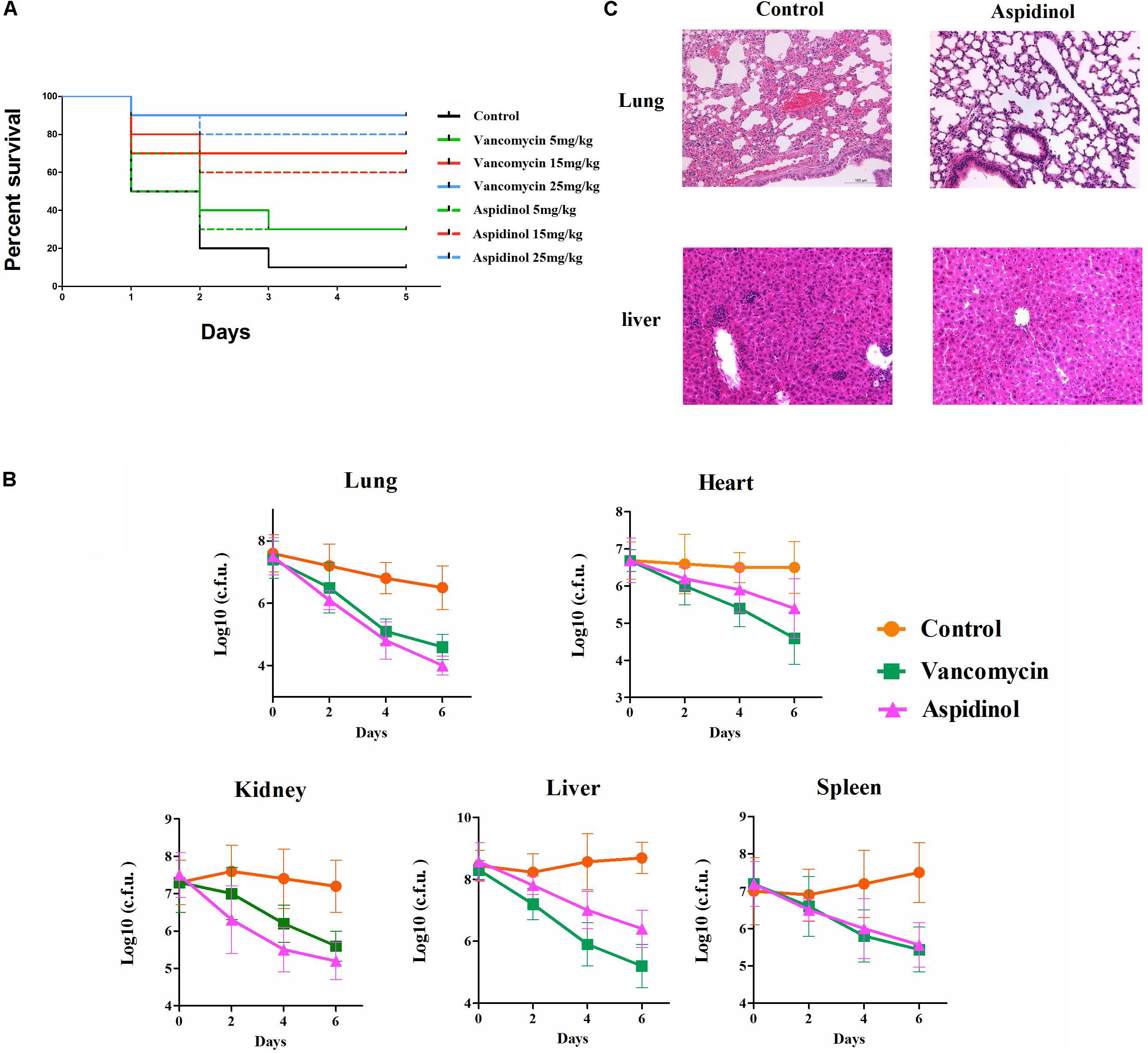
FIGURE 3. Aspidinol is effective in a mouse model of MRSA septicemic infection. (A) Ten mice per group were infected (i.p.) with a lethal dose of S. aureus ATCC 33591 and intravenously injected with aspidinol, vancomycin (5, 15, or 25 mg/kg), or the vehicle alone for 5 days (one dose per day). Mice were monitored for 5 days and the percentage survival was calculated. The statistical significance was calculated in order to compare treated to control groups. (B) Six mice per group were infected (i.p.) with a non-lethal dose of S. aureus ATCC 33591 and intravenously injected with aspidinol, vancomycin (25 mg/kg) or the vehicle alone for 6 days (one dose per day). Twenty-four hours after the last treatment, the mice were euthanized, and their organs were excised and homogenized in TSB to count viable MRSA colonies. The number of CFUs from each mouse is plotted as individual points. Values are the mean of triplicate samples with standard deviation bars. (C) Histological evaluation of lung and liver of mice infected with S. aureus ATCC 33591 receiving no treatment or a treatment with aspidinol. Both lung and liver in the control group demonstrated acute inflammation; no apparent pathological changes were observed in the treatment group.
Next, the efficacy of aspidinol against MRSA ATCC 33591 in a non-lethal model was investigated. In brief, mice were first infected with a non-lethal dose of MRSA ATCC 33951 (4.20 × 107 CFU/mouse), and then each group of mice received two oral doses of aspidinol, vancomycin (25 mg/kg), or the vehicle alone. As depicted in Figure 3B, aspidinol and vancomycin produced a significant reduction in mean bacterial load in different organs. In particular, both treatments reduced the mean bacterial load by more than 1000-fold in the lung. The histopathological inspection 6 days after infection with the non-lethal dose of MRSA ATCC 33951 revealed no changes in the heart, spleen, and kidney. While the animals showed moderate histopathological alterations in lung and liver in the control group, after treatment with aspidinol or vancomycin, there were no obvious histopathological alterations (Figure 3C).
RNA-Seq
To better understand the mechanism of the anti-MRSA effect of aspidinol, the underlying differential expression of MRSA genes was analyzed after treatment with aspidinol.
A heatmap and volcano plot analyses revealed the differential gene expression for both untreated and treated MRSA (Figures 4A,B). A total of 2952 genes were identified; among these, 1147 genes were upregulated and 1183 were downregulated during aspidinol treatment. After quantile normalization of the FPKM values followed by Student’s t-test at p = 0.05 and the selection of DEGs with at least a twofold change in their expression in response to aspidinol treatment, we identified 949 DEGs (456 upregulated and 493 downregulated) with highly significant expression patterns before and after the treatment. All the sequencing reads have been submitted to the NCBI short-read archive (SRA) with accession number SRP129492.
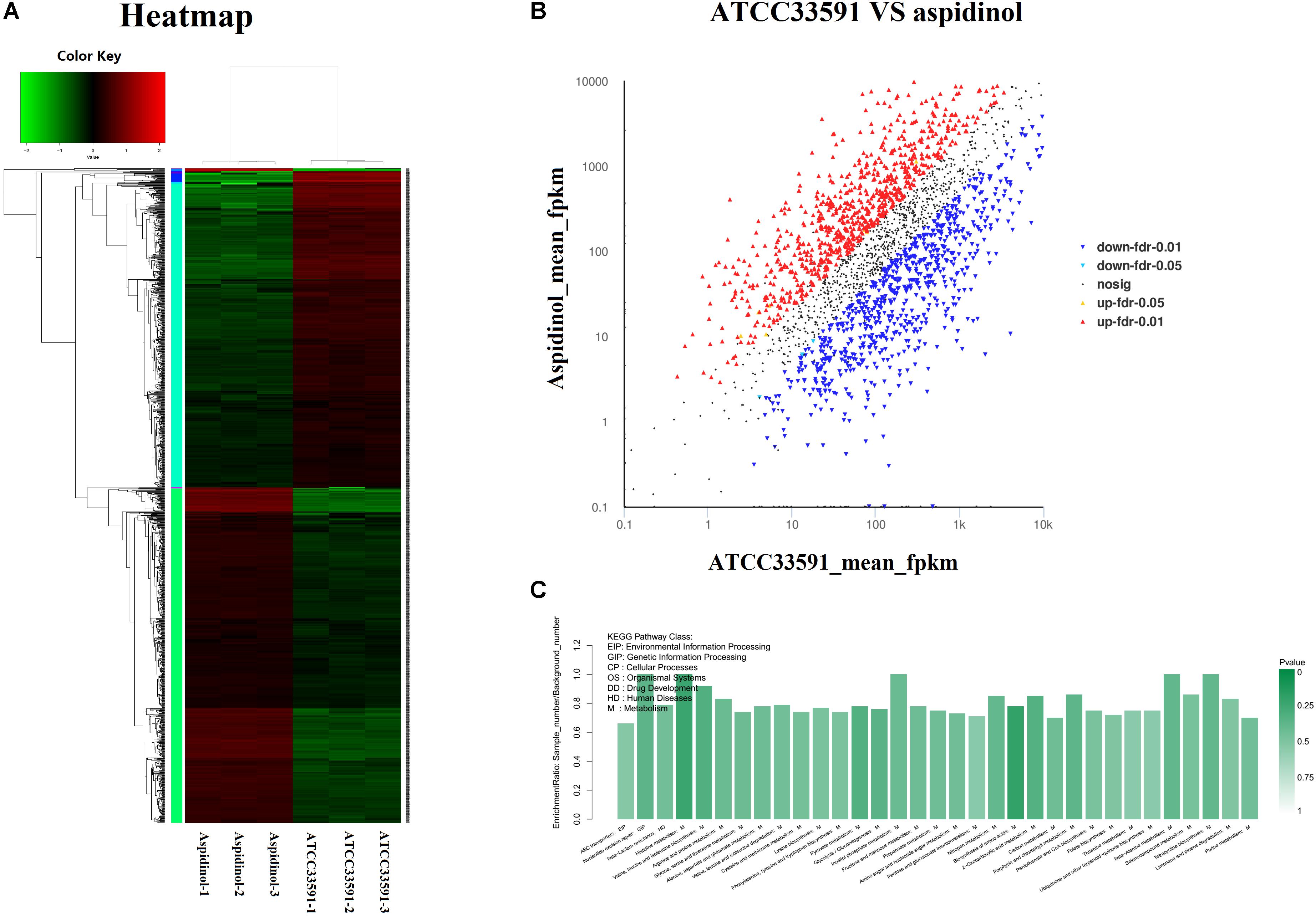
FIGURE 4. RNA-Seq gene expression results for S. aureus ATCC 33591 cells treated and not treated with aspidinol. (A) A heatmap generated comparing aspidinol-treated cells to untreated control S. aureus ATCC 33591 cells is shown. Triplicate samples were used for each group. (B) Volcano plot analyses of unigenes in aspidinol and the ATCC 33591 group. (C) Differentially expressed genes enriched in the KEGG pathway.
KEGG Pathway Analysis of the DEGs
KEGG pathway analysis was used to further clarify the biological function of aspidinol. Overall, DEGs were significantly enriched in 55 KEGG pathways, meeting the criteria of p-values <0.05 (Figure 4C, more clearer pictures can be found in Supplementary Figure S1). The KEGG pathways that showed the highest levels of significance were the lysine biosynthesis pathway (ko00300), the valine, leucine, and isoleucine biosynthesis pathways (ko00290), nitrogen metabolism (ko00910), galactose metabolism (ko00052), carbon metabolism (ko01200), and amino sugar and nucleotide sugar metabolism (ko00520), suggesting that these pathways may play important roles in the antibacterial processes of aspidinol.
Genes Involved in the Synthesis of Amino Acid
The analysis of genes downregulated by aspidinol in S. aureus ATCC 33591 showed a significant decrease in the expression of the genes involved in the synthesis of amino acids. The synthesis of amino acids is important for the survival of bacteria. Related genes that act in the synthesis of valine, leucine, isoleucine, and lysine were downregulated after treatment with aspidinol. Genes encoding important intermediates in amino acid synthesis, such as ilvC and ilvD, were also downregulated 3.47- and 4.73-fold, respectively. In addition, the leucine synthesis genes leuABCD were also downregulated 2.6-, 3.08-, 2.58-, and 2.49-fold, respectively. The gene lysC, which encodes an aspartate kinase, was downregulated 5.77-fold. In addition to amino acid biosynthesis, genes involved in peptide import were also significantly downregulated. The genes oppABCDF, encoding a peptide transport system, were downregulated 1.15-, 7.37-, 3.68-, 2.30-, and 1.40-fold, respectively.
Genes Involved in Ribosome Synthesis
Four genes involved in structural components of ribosomes, 30S ribosomal protein S20 rpsT (-4.01-fold), 50S ribosomal protein L1 rplA (-4.34-fold), L20 rplT (-4.11-fold), and L32 (-2.33-fold), were downregulated after treatment with aspidinol. Nevertheless, three genes involved in structural components of ribosomes, 30S ribosomal protein S11 rpsK (2.3-fold), 50S ribosomal protein L17 rplQ (2.33-fold), and L7/L12 SAV1268 (2.22-fold) were upregulated. In addition, some genes involved in ribosome synthesis were significantly changed. The gene expression of the ribosomal large subunit pseudouridine synthase D rluD, ribosomal-protein-serine N-acetyltransferase rimL, and ribosome-binding factor A rbfA decreased by 4.66-, 2.82-, and 2.9-fold, respectively. The ribosomal subunit interface protein SAV0752 and ribosomal large subunit pseudouridine synthase B SAV1493 were increased by 4.21- and 2.32-fold, respectively.
Repression of Genes Involved in Iron Transport and Key Virulence Factors
Aspidinol also significantly repressed genes involved in iron ABC transporter synthesis. The constituents of the ferrichrome ABC transporter system comprising fhuABG were downregulated by aspidinol 4.93-, 4.12-, and 3.87-fold, respectively. The genes encoding metal iron ABC transport mtsABC were downregulated 4.06-, 5.31-, and 8.59-fold, respectively. Heme transporter IsdA was downregulated 4.34-fold. After treatment with aspidinol, the repression of key virulence factors, exfoliative toxins eta and IgG-binding protein sbi, were downregulated 4.82- and 2.82-fold, respectively.
Genes Involved in the Metabolism of Energy
Aspidinol-treated S. aureus ATCC 700699 showed an increase in the expression of genes involved in basic metabolic processes, including metabolism of nitrogen, galactose, and carbon. The genes involved in nitrogen metabolism, including nirBD, narGHJ, and arc, were upregulated 5.36-, 4.68-, 4.68-, 3.81-, and 9.91-fold, respectively. The lacABCDEFG genes encoding a galactose-6-phosphate isomerase were upregulated 5.68-, 4.28-, 4.22-, 5.72-, 3.6-, 6.22-, and 5.54-fold, respectively. In addition, the carbon metabolism genes sdaA, sdaB, lpd, prsA, and tdcB were also upregulated 4.29-, 3.67-, 2.84-, 2.75-, and 5.1-fold, respectively.
Genes Involved in β-Lactam Resistance
Most genes related to the β-lactam resistance pathway were significantly downregulated after treatment with aspidinol, including beta-lactamase repressor blaI, beta-lactamase regulatory gene blaR1, and methicillin resistance regulatory genes mecI and mecR1, which are key genes attributed to drug resistance of β-lactam antibiotics. After treatment, blaR1, blaI, mecR1, mecI were downregulated 5.6-, 4.49-, 3.06-, and 4.35-fold, respectively, relative to their expression in the untreated control.
RNA-Seq Results Were Verified by qRT-PCR
A qRT-PCR assay was conducted to further validate transcriptional changes induced by aspidinol and observed in the RNA-seq experiments. After treatment with sub-MIC (1 μg/mL) of aspidinol, 30 different genes were chosen, including oppABCDF, fhuABG, and ilvBCD among others (Figure 5), to determine changes in their transcript accumulation. There was no obvious variation between qRT-PCR and transcriptomic data in the expression patterns except for mtsA. Although the exact fold change difference in expression for each gene by qRT-PCR was different from the RNA-seq data, similar trends were observed, which suggests that there is a relatively high consistency between the RNA-seq and qRT-PCR results.
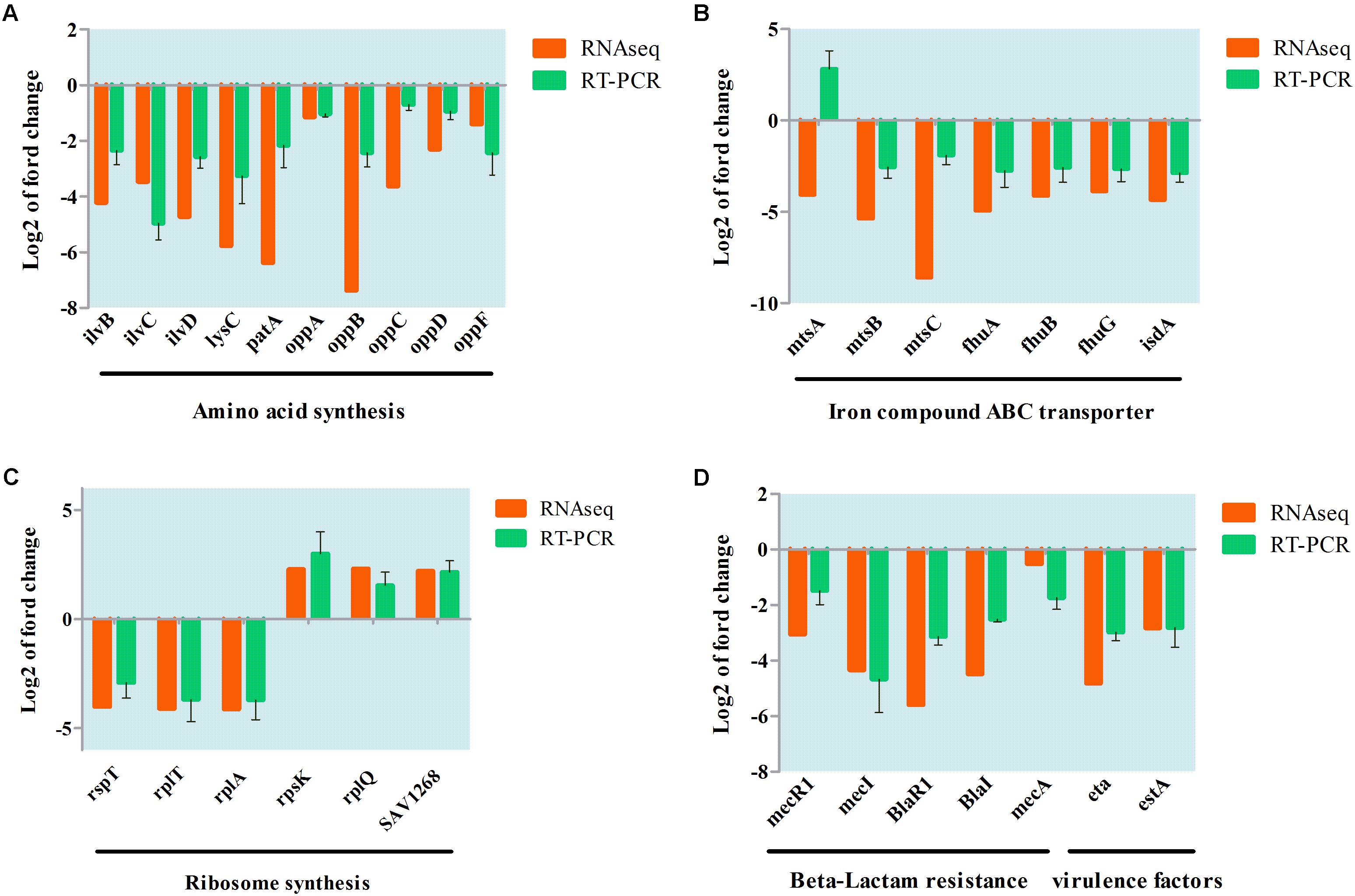
FIGURE 5. Validation of RNA-seq data for selected genes by real-time PCR. (A) Differentially expressed genes involved in amino acid synthesis; (B) differentially expressed genes involved in iron ABC transporter synthesis; (C) differentially expressed genes involved in ribosome synthesis; (D) differentially expressed genes involved in beta-lactam resistance and virulence factor.
Discussion
Staphylococcus aureus, in particular MRSA, has acquired numerous antimicrobial resistance genes, rendering these isolates multi-resistant. This development of resistance and multi-resistance is likely to have the effect that the currently available antibiotics will not be as efficacious in the near future (Rodvold and McConeghy, 2014; Shenoy et al., 2014; Stahlmann, 2014). Unfortunately, the development of novel antibiotics and their introduction into clinical use cannot keep pace with the emergence of resistant pathogens. Relatedly, pharmaceutical companies have increasingly withdrawn investment in the development of new antimicrobial agents. Despite this situation and the associated challenges to healthcare, new strategies, and alternative methods for anti-infective therapy need to be explored.
In preliminary experiments, aspidinol was chosen as a study object through antibacterial activity screening. Aspidinol exhibited an excellent anti-MRSA activity in vitro and no cytotoxicity. Time-kill assays revealed that the anti-MRSA effect of aspidinol was nearly identical to that seen with vancomycin and linezolid. In previous reports of Mu, two analogs of aspidinol, aspidinol C and D, were reported to have significant anti-MRSA activity. The MICs of aspidinol C and D against MRSA were 2 and 4 μg/mL, respectively (The structures of aspidinol, aspidinol C and D can be found in Supplementary Figure S3). However, there was no further in-depth study of aspidinol C and D except for determining their MIC value (Mu et al., 2012). Therefore, it could be speculated that aspidinol may also have excellent anti-MRSA activity due to it similar chemical structure. Indeed, compared with aspirinol C and D, aspirinol shows stronger anti-MRSA activity.
Once MRSA internalizes into eukaryotic cells, intracellular persistence will prolong the presence of the pathogen in the host and result in a weakening of the host defenses (Lemaire et al., 2008). Because of their inability to penetrate the cell membranes, most antibiotics, including vancomycin, cannot effectively combat intracellular MRSA (Garzoni and Kelley, 2009; Seleem et al., 2009; Tenover and Goering, 2009; Thangamani et al., 2015). Vancomycin has been reported to have a failure rate of greater than 40% in treating S. aureus pneumonia (Rubinstein et al., 2008). In this study, aspidinol exhibited excellent anti-MRSA activity inside macrophages; this activity was 100 times better than that of vancomycin. However, compared to vancomycin and linezolid, aspidinol did not significantly reduce adherent MRSA biofilms. This outcome suggests that aspidinol may be more suitable for systemic MRSA infectious diseases, such as septicemia, rather than skin infections.
The in vivo efficacy of aspidinol was assessed in two murine MRSA systemic infection models (non-lethal and lethal). Both the non-lethal and lethal mouse studies confirmed that aspidinol retains its antibacterial activity in vivo. The results were consistent with the results of the anti-MRSA activity in vitro, proving that aspidinol was able to effectively prevent and treat systemic infections caused by MRSA.
To better understand the pathway of the anti-MRSA effect of aspidinol, RNA-seq experiments were conducted. The analysis of transcriptomic changes caused by aspidinol treatment showed that the expression of a number of genes involved in ribosome synthesis was up- and downregulated upon treatment with aspidinol. The results indicated that ribosome synthesis was more likely to be inhibited after treatment with aspidinol and result in the inhibition of protein synthesis in bacteria. The increase in the expression of genes involved in ribosome synthesis may therefore be in response to stress. The inhibition of the formation of the ribosome, which is composed of the 30S and 50S subunits, is an important antibacterial mechanism, such as in linezolid (Colca et al., 2003; Ippolito et al., 2008; Wilson et al., 2008). Thus, the changes of genes involved in ribosome synthesis may contribute to the antimicrobial activity of aspidinol.
Iron transport was also found to be suppressed. With the possible exception of lactobacilli, all bacteria require iron for growth because it plays a vital role in the biological processes of bacteria (Sebulsky et al., 2000). MtsABC proteins were reported to be associated with metal binding and transmembrane transport. A metal solute-binding lipoprotein and an ATP-binding protein were encoded by mtsA and mtsB, respectively, while mtsC encodes a transmembrane permease protein (Sun et al., 2008, 2009; Zou et al., 2010). Thus, the mtsABC operon is also essential for the survival of bacteria (Janulczyk et al., 2003); mtsA, as a metal ion transporter, is responsible for iron release and transmembrane uptake and interacts with membrane components. The Fhu system is the best-characterized system in S. aureus for ferric hydroxamate uptake and is composed of up to five genes (Sebulsky et al., 2000). The genes fhuC, fhuB, and fhuG reside in an operon that encodes an ATP-binding cassette (ABC) transporter; fhuC encodes an ATPase at the inner side of the cytoplasmic membrane, and fhuB and fhuG are membrane-spanning proteins (Pramanik and Braun, 2006; Speziali et al., 2006). When treated with aspidinol, mtsABC and fhuABCDF are downregulated, resulting in decreased iron transport and further reducing the energy supply of ribosome synthesis, which also might contribute to MRSA killing.
Moreover, amino acid biosynthesis was downregulated after aspidinol treatment, including the leucine, isoleucine, and valine synthesis. Pathway analysis using the transcriptome data showed that the expression of the valine, leucine, and isoleucine biosynthesis system was decreased. The downregulated ilv and leu operons have been reported to encode leucine, isoleucine, and valine intermediates in the synthesis of these amino acids in E. coli (Miyanoiri et al., 2016). According to our results, we speculate that these operons may play the same role, although there are no reports regarding their function in S. aureus. Components of other amino acid pathways, such as lysC, were apparently downregulated by aspidinol; lysC is the primary gene for lysine biosynthesis and is involved in the biosynthesis of lysine from aspartic acid (Wiltshire and Foster, 2001; Nishi et al., 2004). In addition to amino acid biosynthesis, the oppABCDF genes were also downregulated in response to aspidinol. The Opp system is essential for the uptake of nutrients by bacteria (Gardan et al., 2009) and has been shown to supply bacteria with exogenous peptides that serve as an amino acid resource (Zhao et al., 2016). In this study, downregulation of amino acid biosynthesis pathways after treatment with aspidinol could be attributed to the killing of MRSA.
Aspidinol also significantly repressed genes involved in the virulence of MRSA. Following aspidinol treatment at sub-inhibitory concentrations, exfoliative toxin eta and IgG-binding protein sbi were measurably downregulated. Our data suggest that in addition to killing ATCC 33591, it is very likely that aspidinol reduced the virulence of the infecting bacterial cells as a secondary effect.
According to the data analysis, genes associated with β-lactam resistance were significantly reduced after treatment with aspidinol. Through the analysis of the transcription data, we found that the expression levels of genes associated with β-lactam resistance (mecI, mecR1, blaZ, and blaR1) were significantly reduced after treatment with aspidinol; mecA coding for PBP2a (a low affinity penicillin binding protein) is responsible for methicillin resistance in S. aureus and other staphylococci (Wilke et al., 2004; Staude et al., 2015). While transcription of mecA is regulated by mecI and mecR1 (Wilke et al., 2004; Aedo and Tomasz, 2016), blaR1 and blaI are involved in the regulation of blaZ (Thumanu et al., 2006). Based on our current understanding, blocking of blaR1 and mecR1 is helpful to restore the susceptibility of MRSA to β-lactam antibiotics. Hou et al. (2011) have designed specific anti-mecR1 and anti-blaR1 deoxyribozymes and confirmed their function in restoring the β-lactam susceptibility of MRSA.
According to the transcriptome data, the expression of mecA was decreased 0.52-fold, but there were no statistically significant differences. However, the RT-PCR results showed that mecA was downregulated 1.75-fold after treatment with aspidinol. Subsequently, a combination administration experiment was conducted to test whether a sub-MIC concentration of aspidinol was able to restore the susceptibility to β-lactams. In this experiment, oxacillin was co-administered with 1 μg/mL aspidinol, and the oxacillin MIC decreased from 256 to 0.5 μg/mL. This observation might provide a first hint toward the use of aspidinol as a combination drug to restore the β-lactam susceptibility. We theorize that aspidinol is able to inhibit the expression of mecA-related genes to restore susceptibility to β-lactam antibiotics.
Genes involved in some metabolic pathways, such as carbon metabolism and nitrogen metabolism, were upregulated after treatment with aspidinol, which could be attributed to metabolic disorders of bacteria in the process of dying.
Although the validation of the effects of aspidinol on metabolic pathways has been carried out by RT-PCR, further studies are still needed to more thoroughly understand the antibacterial mechanisms of aspidinol.
However, these data were not sufficient to fully elucidate the antibacterial mechanisms of aspidinol against MRSA. In future studies, we will focus on the identification of potential drug targets to clearly reveal the mechanism of action.
Conclusion
In this study, the anti-MRSA activity of aspidinol, both in vitro and in vivo, was reported for the first time. The mechanism of aspidinol’s anti-MRSA effect was also investigated. In particular, RNA-seq was applied for the analysis of differentially expressed genes after treatment with aspidinol. When combined with verification experiments, the results suggest that the inhibition of the formation of ribosomes was the primary S. aureus cell-killing mechanism, and the inhibition of amino acid synthesis and the reduction of virulence factors were considered to be a secondary effect.
Author Contributions
XH and SL responsible for experimental design. XH, QY, and ZD responsible for experimental operation. XH, WZ, and SY responsible for writing the manuscript. SS responsible for reviewing the manuscript.
Funding
The authors gratefully acknowledge the financial support from the Program for Natural Science Foundation of Heilongjiang Province of China (C2016063), China Postdoctoral Science Foundation (2016M591311), National Natural Science Foundation of China (31602100), and Central Public-interest Scientific Institution Basal Research Fund (1610302017007).
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fphar.2018.00619/full#supplementary-material
FIGURE S1 | Differentially expressed genes enriched in the KEGG pathway for S. aureus ATCC 33591 cells treated and not treated with aspidinol.
FIGURE S2 | Cell survival curve after treated with aspidinol.
FIGURE S3 | Chemical structure of aspidinol, aspidinol C and aspidinol D.
TABLE S1 | Strain list.
TABLE S2 | Primers used in this study.
References
Aedo, S., and Tomasz, A. (2016). Role of the stringent stress response in the antibiotic resistance phenotype of methicillin-resistant Staphylococcus aureus. Antimicrob. Agents Chemother. 60, 2311–2317. doi: 10.1128/AAC.02697-15
Colca, J. R., Mcdonald, W. G., Waldon, D. J., Thomasco, L. M., Gadwood, R. C., Lund, E. T., et al. (2003). Cross-linking in the living cell locates the site of action of oxazolidinone antibiotics. J. Biol. Chem. 278, 21972–21979. doi: 10.1074/jbc.M302109200
Farah, S. I., Abdelrahman, A., North, E. J., and Chauhan, H. (2016). Opportunities and challenges for natural products as novel antituberculosis agents. Assay Drug Dev. Technol. 14, 29–38. doi: 10.1089/adt.2015.673
Gao, C., Guo, N., Li, N., Peng, X., Wang, P., Wang, W., et al. (2016). Investigation of antibacterial activity of aspidin BB against Propionibacterium acnes. Arch. Dermatol. Res. 308, 79–86. doi: 10.1007/s00403-015-1603-x
Gardan, R., Besset, C., Guillot, A., Gitton, C., and Monnet, V. (2009). The oligopeptide transport system is essential for the development of natural competence in Streptococcus thermophilus strain LMD-9. J. Bacteriol. 191, 4647–4655. doi: 10.1128/JB.00257-09
Garzoni, C., and Kelley, W. L. (2009). Staphylococcus aureus: new evidence for intracellular persistence. Trends Microbiol. 17, 59–65. doi: 10.1016/j.tim.2008.11.005
Guzman, J. D., Gupta, A., Evangelopoulos, D., Basavannacharya, C., Pabon, L. C., Plazas, E. A., et al. (2010). Anti-tubercular screening of natural products from Colombian plants: 3-methoxynordomesticine, an inhibitor of MurE ligase of Mycobacterium tuberculosis. J. Antimicrob. Chemother. 65, 2101–2107. doi: 10.1093/jac/dkq313
Hou, Z., Zhou, Y., Wang, H., Bai, H., Meng, J., Xue, X., et al. (2011). Co-blockade of mecR1/blaR1 signal pathway to restore antibiotic susceptibility in clinical isolates of methicillin-resistant Staphylococcus aureus. Arch. Med. Sci. 7, 414–422. doi: 10.5114/aoms.2011.23404
Huang, Y. H., Zeng, W. M., Li, G. Y., Liu, G. Q., Zhao, D. D., Wang, J., et al. (2014). Characterization of a new sesquiterpene and antifungal activities of chemical constituents from Dryopteris fragrans (L.) Schott. Molecules 19, 507–513. doi: 10.3390/molecules19010507
Ippolito, J. A., Kanyo, Z. F., Wang, D., Franceschi, F. J., Moore, P. B., Steitz, T. A., et al. (2008). Crystal structure of the oxazolidinone antibiotic linezolid bound to the 50S ribosomal subunit. J. Med. Chem. 51, 3353–3356. doi: 10.1021/jm800379d
Janulczyk, R., Ricci, S., and Bjorck, L. (2003). MtsABC is important for manganese and iron transport, oxidative stress resistance, and virulence of Streptococcus pyogenes. Infect. Immun. 71, 2656–2664. doi: 10.1128/IAI.71.5.2656-2664.2003
Lehar, S. M., Pillow, T., Xu, M., Staben, L., Kajihara, K. K., Vandlen, R., et al. (2015). Novel antibody-antibiotic conjugate eliminates intracellular S. aureus. Nature 527, 323–328. doi: 10.1038/nature16057
Lemaire, S., Olivier, A., Van Bambeke, F., Tulkens, P. M., Appelbaum, P. C., and Glupczynski, Y. (2008). Restoration of susceptibility of intracellular methicillin-resistant Staphylococcus aureus to beta-lactams: comparison of strains, cells, and antibiotics. Antimicrob. Agents Chemother. 52, 2797–2805. doi: 10.1128/AAC.00123-08
Mihu, M. R., Roman-Sosa, J., Varshney, A. K., Eugenin, E. A., Shah, B. P., Lee, H. H., et al. (2015). Methamphetamine alters the antimicrobial efficacy of phagocytic cells during methicillin-resistant Staphylococcus aureus skin infection. MBio 6:e1622-15. doi: 10.1128/mBio.01622-15
Miyanoiri, Y., Ishida, Y., Takeda, M., Terauchi, T., Inouye, M., and Kainosho, M. (2016). Highly efficient residue-selective labeling with isotope-labeled Ile, Leu, and Val using a new auxotrophic E. coli strain. J. Biomol. NMR 65, 109–119. doi: 10.1007/s10858-016-0042-0
Mu, Q., Zeng, C. H., Gibbons, S., and Osman, C. (2012). Aspidin Phenolic Compounds and Their Use in the Orparation of Anti-drug Resistant. CN Patent No. 102464578 A. Peking: State Intellectual Property Office of the People’s Republic of China.
Nishi, H., Komatsuzawa, H., Fujiwara, T., Mccallum, N., and Sugai, M. (2004). Reduced content of lysyl-phosphatidylglycerol in the cytoplasmic membrane affects susceptibility to moenomycin, as well as vancomycin, gentamicin, and antimicrobial peptides, in Staphylococcus aureus. Antimicrob. Agents Chemother. 48, 4800–4807. doi: 10.1128/AAC.48.12.4800-4807.2004
Peng, B., Bai, R. F., Li, P., Han, X. Y., Wang, H., Zhu, C. C., et al. (2016). Two new glycosides from Dryopteris fragrans with anti-inflammatory activities. J. Asian Nat. Prod. Res. 18, 59–64. doi: 10.1080/10286020.2015.1121853
Pramanik, A., and Braun, V. (2006). Albomycin uptake via a ferric hydroxamate transport system of Streptococcus pneumoniae R6. J. Bacteriol. 188, 3878–3886. doi: 10.1128/JB.00205-06
Rennie, R. P. (2012). Current and future challenges in the development of antimicrobial agents. Handb. Exp. Pharmacol. 211, 45–65. doi: 10.1007/978-3-642-28951-4_4
Rodvold, K. A., and McConeghy, K. W. (2014). Methicillin-resistant Staphylococcus aureus therapy: past, present, and future. Clin. Infect. Dis. 58(Suppl. 1), S20–S27. doi: 10.1093/cid/cit614
Rubinstein, E., Kollef, M. H., and Nathwani, D. (2008). Pneumonia caused by methicillin-resistant Staphylococcus aureus. Clin. Infect. Dis. 46, S378–S385. doi: 10.1086/533594
Sebulsky, M. T., Hohnstein, D., Hunter, M. D., and Heinrichs, D. E. (2000). Identification and characterization of a membrane permease involved in iron-hydroxamate transport in Staphylococcus aureus. J. Bacteriol. 182, 4394–4400. doi: 10.1128/JB.182.16.4394-4400.2000
Seleem, M. N., Jain, N., Pothayee, N., Ranjan, A., Riffle, J. S., and Sriranganathan, N. (2009). Targeting Brucella melitensis with polymeric nanoparticles containing streptomycin and doxycycline. FEMS Microbiol. Lett. 294, 24–31. doi: 10.1111/j.1574-6968.2009.01530.x
Shenoy, E. S., Paras, M. L., Noubary, F., Walensky, R. P., and Hooper, D. C. (2014). Natural history of colonization with methicillin-resistant Staphylococcus aureus (MRSA) and vancomycin-resistant Enterococcus (VRE): a systematic review. BMC Infect. Dis. 14:177. doi: 10.1186/1471-2334-14-177
Speziali, C. D., Dale, S. E., Henderson, J. A., Vines, E. D., and Heinrichs, D. E. (2006). Requirement of Staphylococcus aureus ATP-binding cassette-ATPase FhuC for iron-restricted growth and evidence that it functions with more than one iron transporter. J. Bacteriol. 188, 2048–2055. doi: 10.1128/JB.188.6.2048-2055.2006
Stahlmann, R. (2014). Antibiotics for treatment of infections by methicillin-resistant Staphylococcus aureus (MRSA). Pneumologie 68, 676–684. doi: 10.1055/s-0034-1377747
Staude, M. W., Frederick, T. E., Natarajan, S. V., Wilson, B. D., Tanner, C. E., Ruggiero, S. T., et al. (2015). Investigation of signal transduction routes within the sensor/transducer protein BlaR1 of Staphylococcus aureus. Biochemistry 54, 1600–1610. doi: 10.1021/bi501463k
Sun, X., Baker, H. M., Ge, R., Sun, H., He, Q. Y., and Baker, E. N. (2009). Crystal structure and metal binding properties of the lipoprotein MtsA, responsible for iron transport in Streptococcus pyogenes. Biochemistry 48, 6184–6190. doi: 10.1021/bi900552c
Sun, X., Ge, R., Chiu, J. F., Sun, H., and He, Q. Y. (2008). Lipoprotein MtsA of MtsABC in Streptococcus pyogenes primarily binds ferrous ion with bicarbonate as a synergistic anion. FEBS Lett. 582, 1351–1354. doi: 10.1016/j.febslet.2008.03.020
Taylor, A. R. (2013). Methicillin-resistant Staphylococcus aureus infections. Prim. Care 40, 637–654. doi: 10.1016/j.pop.2013.06.002
Tenover, F. C., and Goering, R. V. (2009). Methicillin-resistant Staphylococcus aureus strain USA300: origin and epidemiology. J. Antimicrob. Chemother. 64, 441–446. doi: 10.1093/jac/dkp241
Thangamani, S., Younis, W., and Seleem, M. N. (2015). Repurposing clinical molecule ebselen to combat drug resistant pathogens. PLoS One 10:e0133877. doi: 10.1371/journal.pone.0133877
Thumanu, K., Cha, J., Fisher, J. F., Perrins, R., Mobashery, S., and Wharton, C. (2006). Discrete steps in sensing of beta-lactam antibiotics by the BlaR1 protein of the methicillin-resistant Staphylococcus aureus bacterium. Proc. Natl. Acad. Sci. U.S.A. 103, 10630–10635. doi: 10.1073/pnas.0601971103
Vandal, J., Abou-Zaid, M. M., Ferroni, G., and Leduc, L. G. (2015). Antimicrobial activity of natural products from the flora of Northern Ontario, Canada. Pharm. Biol. 53, 800–806. doi: 10.3109/13880209.2014.942867
VanEperen, A. S., and Segreti, J. (2016). Empirical therapy in methicillin-resistant Staphylococcus aureus infections: an up-to-date approach. J. Infect. Chemother. 22, 351–359. doi: 10.1016/j.jiac.2016.02.012
WHO (2017). Global Priority List of Antibiotic-Resistant Bacteria to Guide Research, Discovery, and Development of New Antibiotics. Available at: http://www.who.int/medicines/publications/WHO-PPL-Short_Summary_25Feb-ET_NM_WHO.pdf
Wilke, M. S., Hills, T. L., Zhang, H. Z., Chambers, H. F., and Strynadka, N. C. (2004). Crystal structures of the Apo and penicillin-acylated forms of the BlaR1 beta-lactam sensor of Staphylococcus aureus. J. Biol. Chem. 279, 47278–47287. doi: 10.1074/jbc.M407054200
Wilson, D. N., Schluenzen, F., Harms, J. M., Starosta, A. L., Connell, S. R., and Fucini, P. (2008). The oxazolidinone antibiotics perturb the ribosomal peptidyl-transferase center and effect tRNA positioning. Proc. Natl. Acad. Sci. U.S.A. 105, 13339–13344. doi: 10.1073/pnas.0804276105
Wiltshire, M. D., and Foster, S. J. (2001). Identification and analysis of Staphylococcus aureus components expressed by a model system of growth in serum. Infect. Immun. 69, 5198–5202. doi: 10.1128/IAI.69.8.5198-5202.2001
Woods, C., and Colice, G. (2014). Methicillin-resistant Staphylococcus aureus pneumonia in adults. Expert Rev. Respir. Med. 8, 641–651. doi: 10.1586/17476348.2014.940323
Zhang, Y., Luo, M., Zu, Y., Fu, Y., Gu, C., Wang, W., et al. (2012). Dryofragin, a phloroglucinol derivative, induces apoptosis in human breast cancer MCF-7 cells through ROS-mediated mitochondrial pathway. Chem. Biol. Interact. 199, 129–136. doi: 10.1016/j.cbi.2012.06.007
Zhao, D. D., Zhao, Q. S., Liu, L., Chen, Z. Q., Zeng, W. M., Lei, H., et al. (2014). Compounds from Dryopteris fragrans (L.) Schott with cytotoxic activity. Molecules 19, 3345–3355. doi: 10.3390/molecules19033345
Zhao, J., Cheah, S.-E., Roberts, K. D., Nation, R. L., Thompson, P. E., Velkov, T., et al. (2016). Transcriptomic analysis of the activity of a novel polymyxin against Staphylococcus aureus. mSphere 1:e00119-16. doi: 10.1128/mSphere.00119-16
Zhong, Z.-C., Zhao, D.-D., Liu, Z.-D., Jiang, S., and Zhang, Y.-L. (2017). A new human cancer cell proliferation inhibition sesquiterpene, dryofraterpene a, from medicinal plant Dryopteris fragrans (L.) Schott. Molecules 22:180. doi: 10.3390/molecules22010180
Keywords: aspidinol, anti-MRSA activity, antimicrobial mechanism, RNA-seq, inhibit ribosomes synthesis
Citation: Hua X, Yang Q, Zhang W, Dong Z, Yu S, Schwarz S and Liu S (2018) Antibacterial Activity and Mechanism of Action of Aspidinol Against Multi-Drug-Resistant Methicillin-Resistant Staphylococcus aureus. Front. Pharmacol. 9:619. doi: 10.3389/fphar.2018.00619
Received: 11 January 2018; Accepted: 23 May 2018;
Published: 13 June 2018.
Edited by:
Jiang Bo Li, Second People’s Hospital of Wuhu, ChinaReviewed by:
Hongjie Fan, Nanjing Agricultural University, ChinaCassandra L. Quave, Emory University School of Medicine, United States
Copyright © 2018 Hua, Yang, Zhang, Dong, Yu, Schwarz and Liu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Siguo Liu, bGl1c2lndW9AY2Fhcy5jbg==
†These authors have contributed equally to this work.
 Xin Hua
Xin Hua Qin Yang1†
Qin Yang1† Stefan Schwarz
Stefan Schwarz