- 1First Faculty of Medicine, Institute of Physiology, Charles University, Prague, Czechia
- 2Institute of Experimental Botany, Academy of Sciences of the Czech Republic, Prague, Czechia
Autoradiography helps to determine the distribution and density of muscarinic receptor (MR) binding sites in the brain. However, it relies on the selectivity of radioligands toward their target. 3H-Pirenzepine is commonly believed to label predominantly M1MR, 3H-AFDX-384 is considered as M2MR selective ligand. Here we performed series of autoradiographies with 3H-AFDX-384 (2 nM), and 3H-pirenzepine (5 nM) in WT, M1KO, M2KO, and M4KO mice to address the ligand selectivity. Labeling with 3H-pirenzepine using M1KO, M2KO, and M4KO brain sections showed the high selectivity toward M1MR. Selectivity of 3H-AFDX-384 toward M2MR varies among brain regions and depends on individual MR subtype proportion. All binding sites in the medulla oblongata and pons, correspond to M2MR. In caudate putamen, nucleus accumbens and olfactory tubercle, 77.7, 74.2, and 74.6% of 3H-AFDX-384 binding sites, respectively, are represented by M4MR and M2MR constitute only a minor portion. In cortex and hippocampus, 3H-AFDX-384 labels almost similar amounts of M2MR and M4MR alongside significant amounts of non-M2/non-M4MR. In cortex, the proportion of 3H-AFDX-384 binding sites attributable to M2MR can be increased by blocking M4MR with MT3 toxin without affecting non-M4MR. PD102807, which is considered as a highly selective M4MR antagonist failed to improve the discrimination of M2MR. Autoradiography with 3H-QNB showed genotype specific loss of binding sites. In conclusion: while 3H-pirenzepine showed the high selectivity toward M1MR, 3H-AFDX-384 binding sites represent different populations of MR subtypes in a brain-region-specific manner. This finding has to be taken into account when interpreting the binding data.
Introduction
Muscarinic receptors (MR) are typical members of G protein coupled receptors family (Kruse et al., 2013) and can be divided into 5 subtypes (M1–M5) (Eglen, 2012), which activate different G proteins (Gq, Gi). Odd-numbered subtypes activate Gq, even-numbered activate Gi protein (Eglen, 2012; Kow and Nathanson, 2012; Reiner and Nathanson, 2012).
Respective MR subtypes have been assigned to different functions in CNS (Wess et al., 2007; Thomsen et al., 2017). Odd-numbered receptors are considered to be localized primarily post-synaptically, however, both M2MR and M4MR are localized both pre- and post-synaptically. As cholinergic autoreceptors, M2 and M4 provide feedback control of acetylcholine release (Zhang et al., 2002; Shin et al., 2015).
One of the means to determine MR density in different CNS areas is autoradiography. In vitro autoradiography has high sensitivity allowing to explore brain regions even with few MR. The use of very thin tissue sections in vitro autoradiography provides several advantages over the large tissue blocks used in binding studies in homogenates/membrane fractions. Brain sectioning allows analyzing MR density in virtually all brain areas of a single animal greatly reducing the number of experimental animals. Moreover, sectioning of a single brain generates sufficient number of tissue sections to explore the binding of multiple radioligands in a particular brain area of the same animal. This further reduces the number of experimental animals and allows comparing the effect of treatment on multiple targets (receptors, transporters) in a single animal (Farar and Myslivecek, 2016).
An important issue is the selectivity of radioligand used in experiments. A general problem in identification of MR subtypes present in specific regions of the central nervous system is the lack of highly subtype-selective muscarinic antagonists. The MR subtypes affinities for pirenzepine and AFDX-384, the most commonly used ones for discrimination of M1 and M2MR, respectively, are shown in Table 1. It can be deduced from this table that both pirenzepine and AFDX-384 have not only high affinity for M1, and M2 MRs, respectively, but also for M4 MR subtype. In radioligand binding studies, it is therefore necessary to use a combination of various antagonists. However, for autoradiography detection this approach is not suitable because of evaluation limitations of such changed “binding.” Thus, the present protocols for M1 and M2 MR subtypes identification should be considered as method for detection of M1 (or M2) and also yet unidentified portion of M4 MRs. Unfortunately, only few papers report these binding sites as M2/M4 MRs (e.g., Zavitsanou et al., 2003; Wang et al., 2014).
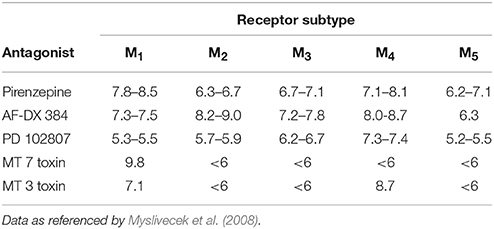
Table 1. Muscarinic antagonist affinity constants (log affinity or pKi values) for mammalian muscarinic receptor subtypes.
The lack of MR subtype-selective ligands constitutes a significant obstacle in anatomical localization studies of MR, which are fundamental to our understanding of MR neuronal circuits. Three key strategies were developed to address regional distribution and relative abundance of particular MR subtype in the central nervous system.
First, in situ hybridization studies have identified neurons that synthetize MR (Buckley et al., 1988; Vilaró et al., 1990; Weiner et al., 1990). These and several other studies have shown that all five MR subtypes are expressed in the brain and mRNA for an individual MR subtype is distributed in a brain-region specific manner. However, while in situ hybridization studies provide valuable information about the sites of MR expression they do not address the real distribution of final proteins.
Second, MR subtype selective antibodies have been developed to map and quantify the distribution of individual MR by means of immunocytochemistry and immunoprecipitation (Levey et al., 1991; Yasuda et al., 1993). A body of reports have provided information about the distribution and quantities of individual MR. However, the reliability and selectivity of commonly used antibodies against MR have been questioned by their testing in specimens devoidof particular MR subtypes. This knockout-proof specific labeling has shown that the vast majority of tested antisera have identical labeling patterns in wild-type and knockout mice tissues (Jositsch et al., 2009).
Finally, anatomical localization and relative quantification of MR can be done by means of in vitro radioligand binding studies. This includes direct radioligand binding studies in tissue homogenates or plasma membrane preparation and indirect in vitro autoradiographic assays. In vitro autoradiography offers several advantages over direct binding studies. These includes tissue saving, high precision and reproducibility of results, high sensitivity and high degree of anatomical resolution. Moreover several ligands can be applied on consecutive sections derived from the same animal, further reducing numbers of experimental animals (Farar and Myslivecek, 2016).
Historically, tritiated pirenzepine was used as ligand that binds to MR with distinct binding in specific brain areas (Yamamura et al., 1983). Further, distinct distribution was found in the central nervous system. 3H-pirenzepine labels regions of the cerebral cortex, hippocampus, striatum and dorsal horn of the spinal cord, while sites in the cerebellum, nucleus tractus solitarius, facial nucleus and ventral horn of the spinal cord are labeled with 3H-QNB (non-specific muscarinic ligand) and not by 3H-pirenzepine (Wamsley et al., 1984). These observations indicated binding to different subtypes of MR. This was further expanded to definition of binding sites as M1 MR (Villiger and Faull, 1985). In the middle of eighties pirenzepine binding sites were considered as M1 MR (Buckley and Burnstock, 1986; Cortes and Palacios, 1986). On the other hand, 3H-AFDX-384 was considered from the beginning as a M2 MR specific ligand (Aubert et al., 1992) and some authors were aware of limited selectivity (e.g., Mulugeta et al., 2003). In many cases3H-AFDX-384 and 3H-pirenzepine are still considered as selective ligands (Tien et al., 2004; Wolff et al., 2008).
Here we performed series of in vitro autoradiographies with 3H-AFDX-384 (2 nM, concentration below KD and the most commonly used one), and 3H-pirenzepine (5 nM, concentration below KD and the most used one). We took advantage of knockout mice models in the standard autoradiography procedures to compare binding in WT, M1 KO, M2 KO and M4 KO mice. With this approach we addressed the selectivity of in vitro 3H-AFDX-384 autoradiography and provide a guide on the interpretation of results.
Moreover, we tried to block M4MR using three concentration of MT3 toxin isolated from Dendroaspis angusticeps venom (1, 10, and 100 nmol/l). Another method to disable the binding to M4MR was co-incubation with specific M4MR antagonist PD102807 (10, 100, and 1 μmol/l). With an aim to block M1MR we used MT7 toxin isolated from Dendroaspis angusticeps venom (1, 10, or 100 nmol/l) or pirenzepine (10 and 100 nmol/l).
In another of our experiment, using radiolabeled non-selective anatagonist, which binds all five MR subtypes (3H-QNB), we determined the contribution of M1, M2, and M4 MRs to the total expression of MRs in the mouse brain.
We can conclude that 3H-pirenzepine showed high selectivity toward M1MR. In contrast, 3H-AFDX-384 binding sites represent different populations of MR subtypes in a brain-region-specific manner. This finding has to be taken into account when interpreting the binding data not only in the autoradiographies but also when these antagonists (pirenzepine, AFDX-384, PD102807 or MT3 toxin) are used as a mean to detect M1MR, M2MR, or M4MR effects in functional studies. Experiments with 3H-QNB binding decrease in M1, , and M4 KO animals showed the highest proportion (usually above 50%) of M1MR in virtually all studied brain areas. M2MR take up to 20% in cortical areas and 34% in thalamus. M4MR were abundant (40% approximately) in thalamus, striatum and ventral striatum (NAc and OT), about 20% of M4MR can be found in cortical structures.
Methods
Drugs
Atropine sulfate and pirenzepine dihydrochloride were purchased from Sigma-Aldrich (Sigma-Aldrich Co, St. Louis, MO, USA). PD102807 was purchased from Tocris Bioscience (Tocris Bioscience, Bristol, United Kingdom). MT3 toxin and MT7 toxin were purchased from Peptide Institute (Peptide Institute, Inc., Osaka, Japan). Pirenzepine [N-methyl-3H] (83.4 Ci/mmol), and AFDX-384 [2,3-dipropylamino-3H] (100.0 Ci/mmol) were from American Radiolabeled Chemicals (ARC, Inc.), Qinuclidinyl benzilate L-[benzilic-4,4′-3H] (50.5 Ci/mmol) was purchased from Perkin Elmer (Perkin Elmer Inc., USA).
Animals
Mice were treated in accordance with the legislature of the Czech Republic and EU legislature [European Convention for the Protection of Vertebrate Animals used for Experimental and other Scientific Purposes (Council of Europe No 123, Strasbourg 1985)], and the experimental protocol was approved by the Committee for the Protection of Experimental Animals of the 1st Medical Faculty, Charles University, Prague under N° MSMT-6316/2014-39.
Mice were maintained (3 per cage) under controlled environmental conditions (12 h/12 h light/dark cycle, 22±1°C, light on at 06:00 a.m.). Food and water were available ad libitum. Knockout mice and their WT counterparts of both genders (weighting 20–25 g, 11–13 weeks old), were used in the study. We studied fully backcrossed (at least 10 generations) muscarinic KO and WT littermates.
M1 KO Mice
Mice lacking M1 MR subtype were generated in the Wess laboratory (Bymaster et al., 2003) and then bred in our animal facility (Prague, Czech Republic). Their genetic background was C57Bl6/NTac. WT and KO genotypes were confirmed using polymerase chain reaction (PCR) analysis as previously described (Cea-del Rio et al., 2010).
M2 KO Mice
Mice lacking M2 MR subtype was generated in the Wess laboratory (Gomeza et al., 1999a) and then bred in our animal facility (Prague, Czech Republic). The genetic background was maintained on C57Bl6/NTac mouse line. WT and KO genotypes were confirmed using PCR analysis as previously described (Cea-del Rio et al., 2010).
M4 KO Mice
Mice lacking M4 MR subtype were generated in Wess laboratory (Gomeza et al., 1999b) and then bred in our animal facility (Prague, Czech Republic). Their genetic background was C57Bl6/NTac. WT and KO genotypes were confirmed using PCR analysis as previously described (Cea-del Rio et al., 2010).
Receptor Autoradiography
Tissue Preparation
For receptor determination, autoradiography was performed in several brain areas [motor cortex (MOCx), somatosensory cortex (SSCx), visual cortex (VisCx), striatum (Caudatum-Putamen, CPu), nucleus accumbens (NAc), thalamus (TH), hippocampus (Hipp) and its specific areas CA1, CA3 and dentate gyrus (DG), olfactory tubercle (OT), pons (Pons), and medulla oblongata (MY)] on sagittal brain sections of M1KO, M2KO, M4KO mice and their WT littermates. Brains were rapidly removed (4–6 brains per group), frozen on dry ice, and then stored at −80°C until cryostat sectioning. Sixteen-micrometer thick sagittal sections were cut on a cryostat at −20°C and thaw-mounted on Superfrost® Plus glass slides (Carl Roth GmbH & Co. KG, Karlsruhe, Germany) and stored in storage boxes at −80°C until use. For autoradiography experiments brain sections were allowed to thaw and dry for 30 min at 22°C.
Autoradiography of Muscarinic Receptors on M1KO, M2KO, M4KO, and WT Mice (3H-Pirenzepine, 3H-AFDX-384, and 3H-QNB Radioligand Binding)
Brain sections were allowed to thaw and dry for 30 min at 22°C and density of receptors was determined as previously described (Farar et al., 2012; Farar and Myslivecek, 2016). Dry M1KO M2KO, M4KO, and WT sagittal brain sections were pre-incubated for 30 min in 50 mM potassium phosphate buffer (pH 7.4) at room temperature (RT). Following pre-incubation, sections were transferred into fresh 50 mM potassium phosphate buffer containing 2 nM 3H-AFDX-384, or 5 nM 3H-pirenzepine, or 2 nM 3H-QNB, and incubated for 60 min at RT. Non-specific binding was assessed on adjacent sections in the presence of 10 μM atropine sulfate. After incubation, sections were washed two times for 5 min each in ice-cold buffer and dipped for 2 s in ice-cold water to remove buffer salts. Wet sections were immediately dried by gentle stream of cold air. Dry sections were opposed to tritium sensitive Fuji BAS imaging plates (GE Healthcare Europe GmbH, Freiburg, Germany) in Kodak Biomax autoradiographic cassettes (Carestream Health, Inc., Rochester, NY).
Autoradiography of M2 Muscarinic Receptors Labeled with 3H-AFDX-384 and Simultaneous Blocking of M4 Receptors with PD102807
The procedure was similar as described above. However, the sections were transferred into buffer containing 2 nM 3H-AFDX-384 together with 10; 100 or 1,000 nM PD102807 or without PD102807 but with addition of DMSO which serves as dissolving agent for PD102807 and incubated for 60 min at RT. The final concentration of DMSO in incubation buffer must be less than 1%, in our case it was 0.1%. The non-specific binding was assessed as described above. This specific experiment was conducted on M2 KO and M4 KO brain sections only. After incubation, sections were treated as stated above.
Autoradiography of M2 Muscarinic Receptors Labeled with 3H-AFDX-384 and Simultaneous Blocking of M4 Receptors with MT3 Toxin Isolated from Dendroaspis Angusticeps Venom
The procedure was similar as described above. However, the sections were transferred into buffer containing 2 nM 3H-AFDX-384 together with 1; 10 or 100 nM MT3 toxin or without MT3 toxin and incubated for 60 min at RT. Non-specific binding was assessed as described above. After incubation, sections were treated as described above.
Autoradiography of M2 Muscarinic Receptors Labeled with 3H-AFDX-384 and Simultaneous Blocking of M1 Receptors with MT7 Toxin Isolated from Dendroaspis Angusticeps Venom or with Pirenzepine
The procedure was similar as described above. However, the sections were transferred into buffer containing 2 nM 3H-AFDX-384 together with 10 or 100 nM pirenzepine or without pirenzepine (serves as control) and incubated for 60 min at RT. Blocking of M1 MR with MT7 toxin was conducted as following: the brain sections were at first pre-incubated with different concentrations of MT7 toxin (1; 10 or 100 nM) for 45 min. After pre-incubation with MT7, 3H-AFDX-384 with final concentration 2 nM was added into the buffer containing MT7 toxin and incubated for another 45 min. Non-specific binding as mentioned earlier. After incubation, sections were processed as described above.
Quantification of Receptor Density
To assure linearity of the signal, autoradiographic standards (American Radiolabeled Chemicals, Inc., St. Louis, MO, USA) were exposed along with the samples to the screens. Imaging plates were processed in Fuji Bioimaging Analyzer BAS-5000 (FUJIFILM corporation, Tokio, Japan) and digitized images analyzed with MCID analysis software (InterFocus GmbH, Mering, Germany). Measurements were taken and averaged from three sections for each animal and brain region.
Statistical Analysis
Statistical significance between groups was determined using 1-way ANOVA with Sidak post-hoc analysis. Student's t-test was used for comparison between two groups (typically WT vs. KO only).
Results
The Selectivity of 3H-Pirenzepine Toward M1MR
Autoradiography with 3H-pirenzepine showed high selectivity of this ligand toward M1MR (see Figure 1). In many brain areas, except in CPu and OT the binding in M1 KO animals decreased by 90–99% (see Table 2). Especially, in hippocampal areas (dorsal hippocampus, CA1, CA3 area, and DG) 3H-pirenzepine binding sites were almost completely abolished. Please note very low binding in TH. The selectivity of pirenzepine was confirmed in M2 KO animals (see Table 3) where no significant difference was shown in M2 KO animals when compared to WT animals. Similar results were obtained when using M4 KO animals (see Table 4). However, in some brain areas, we found small, but significant decrease in 3H-pirenzepine binding in M4 KO animals (MOCx, SSCx, VisCx, CPu; 7, 13, 9, and 11%, respectively).
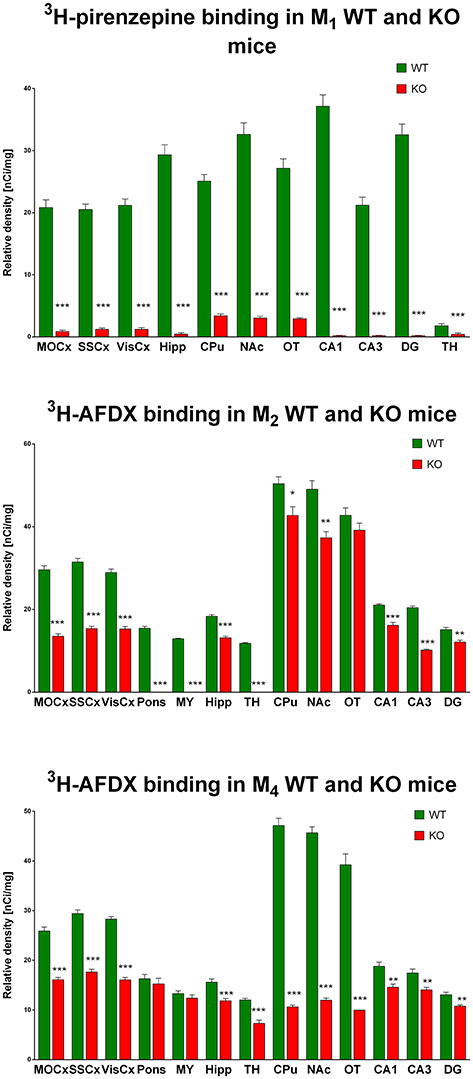
Figure 1. Changes in 3H-pirenzepine specific binding in M1KO mice (top) and 3H-AFDX binding in M2KO (middle) and M4KO mice (bottom) when compared to WT animals. Non-specific binding was determined in the presence of 10 μM atropine sulfate. Ordinate: brain areas [motor cortex (MOCx), somatosensory cortex (SSCx), visual cortex (VisCx), striatum (Caudatum-Putamen, CPu), nucleus accumbens (NAc), thalamus (TH), hippocampus (Hipp) and its specific areas CA1, CA3, and dentate gyrus (DG), olfactory tubercle (OT)], abscissa: relative density [nCi/mg]. WT, wild type; KO, knockout mice. *p < 0.05, **p < 0.01, ***p < 0.001.
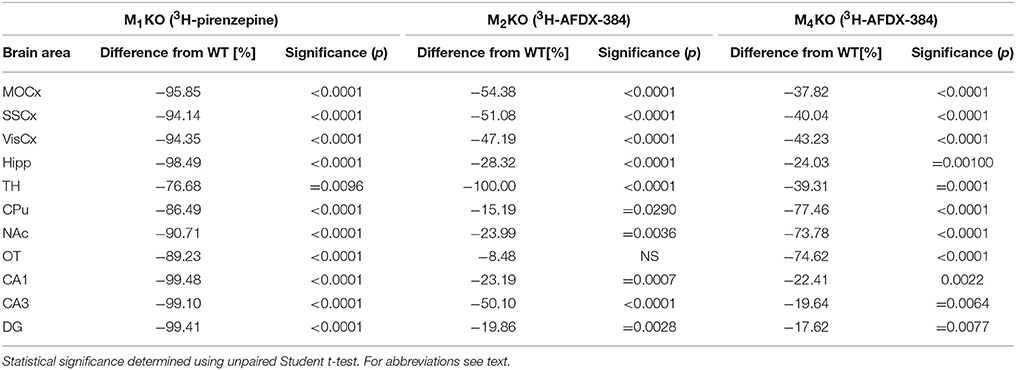
Table 2. Percentage difference in 3H-pirenzepine binding in M1KO, and in 3H-AFDX-384 to M2KO, and M4KO animals to WT animals.
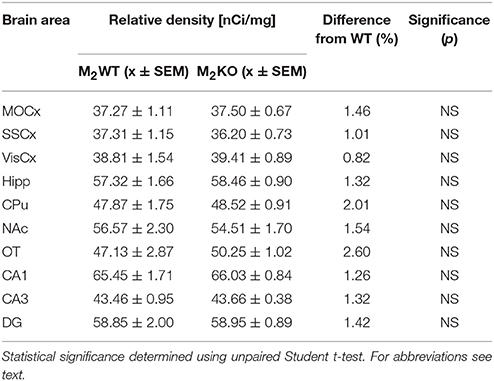
Table 3. Binding and percentage difference in 3H-pirenzepine binding in M2KO animals and WT animals.
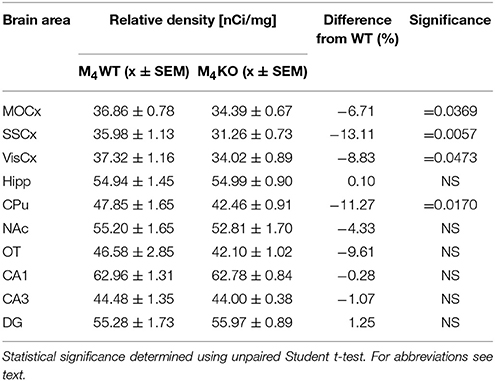
Table 4. Binding and percentage difference in 3H-pirenzepine binding in M4KO animals and WT animals.
The Selectivity of 3H-AFDX-384 Toward M2MR
In contrast to the above mentioned experiment, 3H-AFDX-384 did not show selectivity toward M2MR. If 3H-AFDX-384 was highly selective toward M2MR, the binding of 3H-AFDX-384 would be barely visible in M2 KO mice, as they do not express M2MR. As it can be deduced from Figure 1 and Table 2, the binding reduction in many brain areas varied between 20 and 50%, with exception of pons, medulla oblongata and TH, where uniform population of M2MR was found. Olfactory tubercle is the only area, in which the binding of 3H-AFDX-384 was completely preserved.
The Selectivity of 3H-AFDX-384 Toward M4MR
Similar to the experiment in M2 KO mice, the binding in M4 KO mice showed only limited selectivity of 3H-AFDX-384 toward this MR subtype. However, 3H-AFDX-384 had similar selectivity to M4MR and M2MR (see Figure 1 and Table 2). A decrease in 3H-AFDX-384 binding depends on the brain area analyzed. This experiment also verified that pons and medulla oblongata express only M2MR which can be deduced from the unchanged binding in M4 KO mice. However, thalamus was not confirmed as an area with only M2MR. Representative autoradiograms for M1, M2 and M4 WT and KO mice are shown in Figure 2.
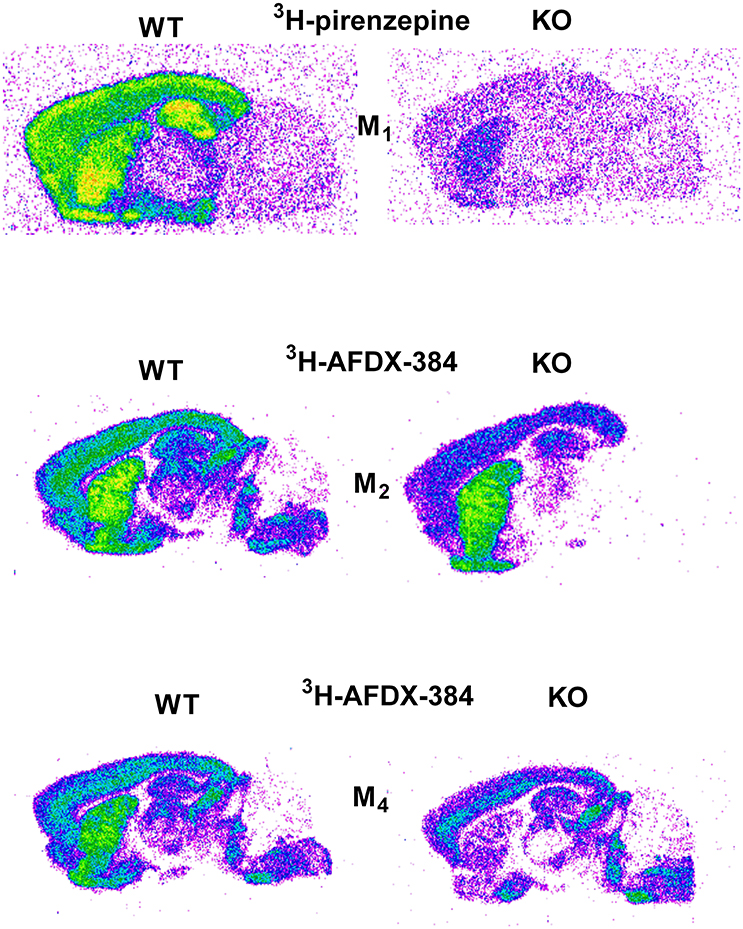
Figure 2. Representative autoradiograms in WT and KO mice: 3H-pirenzepine binding in M1 WT and KO mice (top), 3H-AFDX 384 binding in M2 WT and KO mice (middle), and 3H-AFDX 384 binding in M4 WT and KO mice (bottom).
Binding of 3H-AFDX-384 in the Presence of M4MR Antagonist PD102807
The effort to block M4MR using antagonist PD102807 was unsuccessful as demonstrated in M2 KO mice and M4 KO mice. When the slices were incubated with 1 μmol/l of PD102807, we have recorded 100% inhibition of residual binding in M2 KO mice (see Table 5). However, PD102807 also dose-dependently reduced 3H-AFDX-384 binding in M4 KO mice, suggesting limited selectivity toward M4MR.
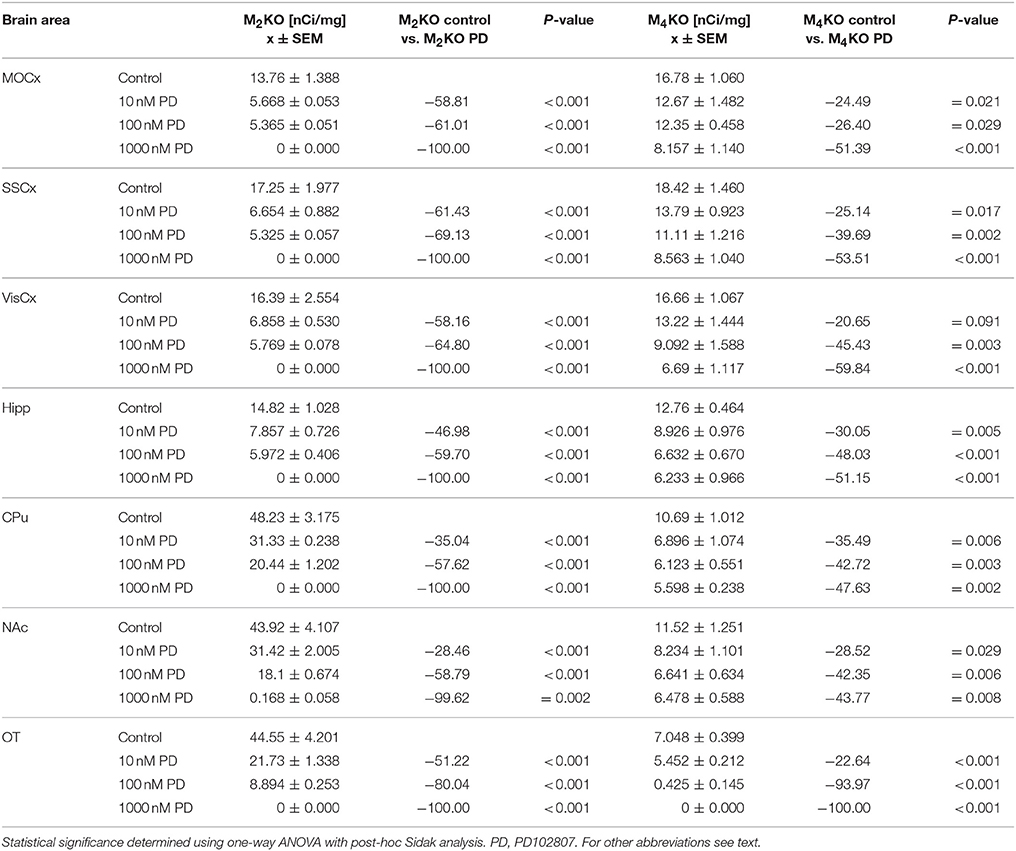
Table 5. Binding and percentage difference in 3H-AFDX 384 binding with PD102807 in M2KO, and M4KO animals vs. control (no PD102807).
Binding of 3H-AFDX-384 in the Presence of M4MR Specific Toxin MT3
Similar to PD102807, MT3 toxin was unable to block M4MR. This can be seen in competitions binding experiment with increasing concentration of MT3 toxin (Table 6). MT3 toxin was unable to completely block residual 3H-AFDX-384 binding in M2 KO mice. Moreover, MT3 also dose-dependently reduced 3H-AFDX-384 binding in M4 KO mice.
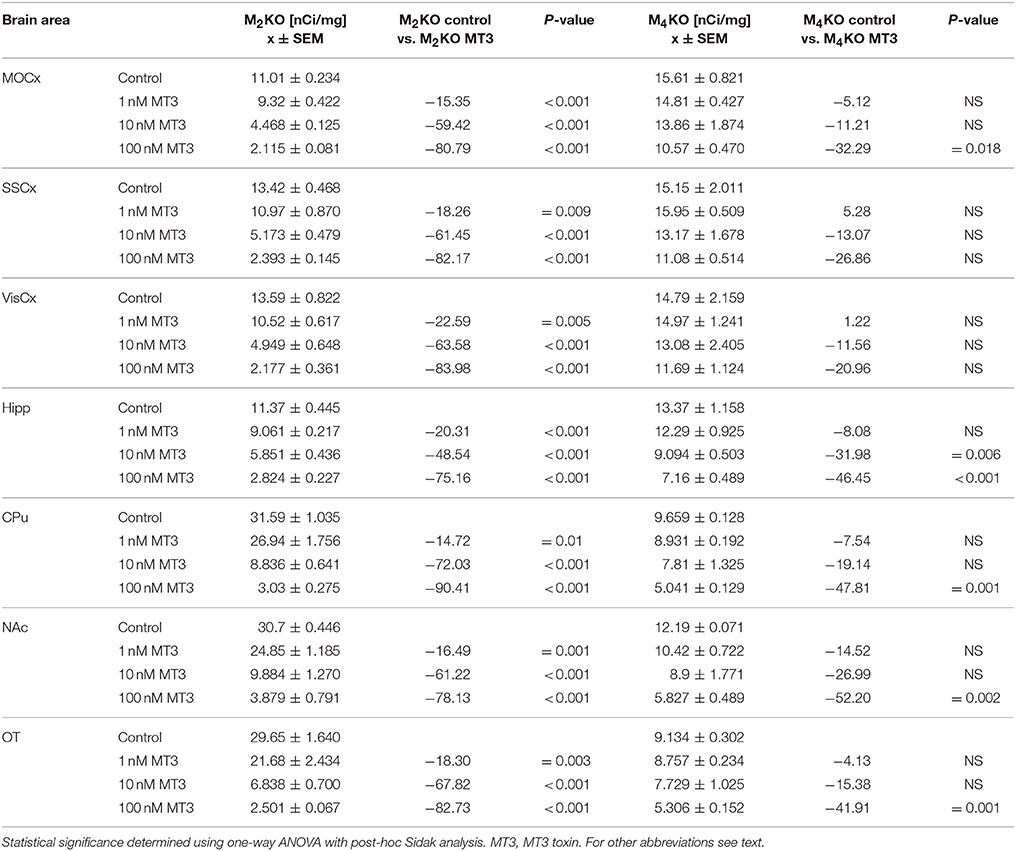
Table 6. Binding and percentage difference in 3H-AFDX 384 binding with MT3 toxin in M2KO, and M4KO animals vs. control (no MT3 toxin).
Binding of 3H-AFDX-384 in the Presence of Pirenzepine and M1MR Specific Toxin MT7 in WT and M4 KO Mice
Incubation with pirenzepine (Table 7) showed dose-dependent reduction of 3H-AFDX-384 binding both in WT and M4 KO mice. Irreversible, and very specific toxin MT7 also dose-dependently reduced 3H-AFDX-384 binding in both WT and M4 KO mice (see Table 8).
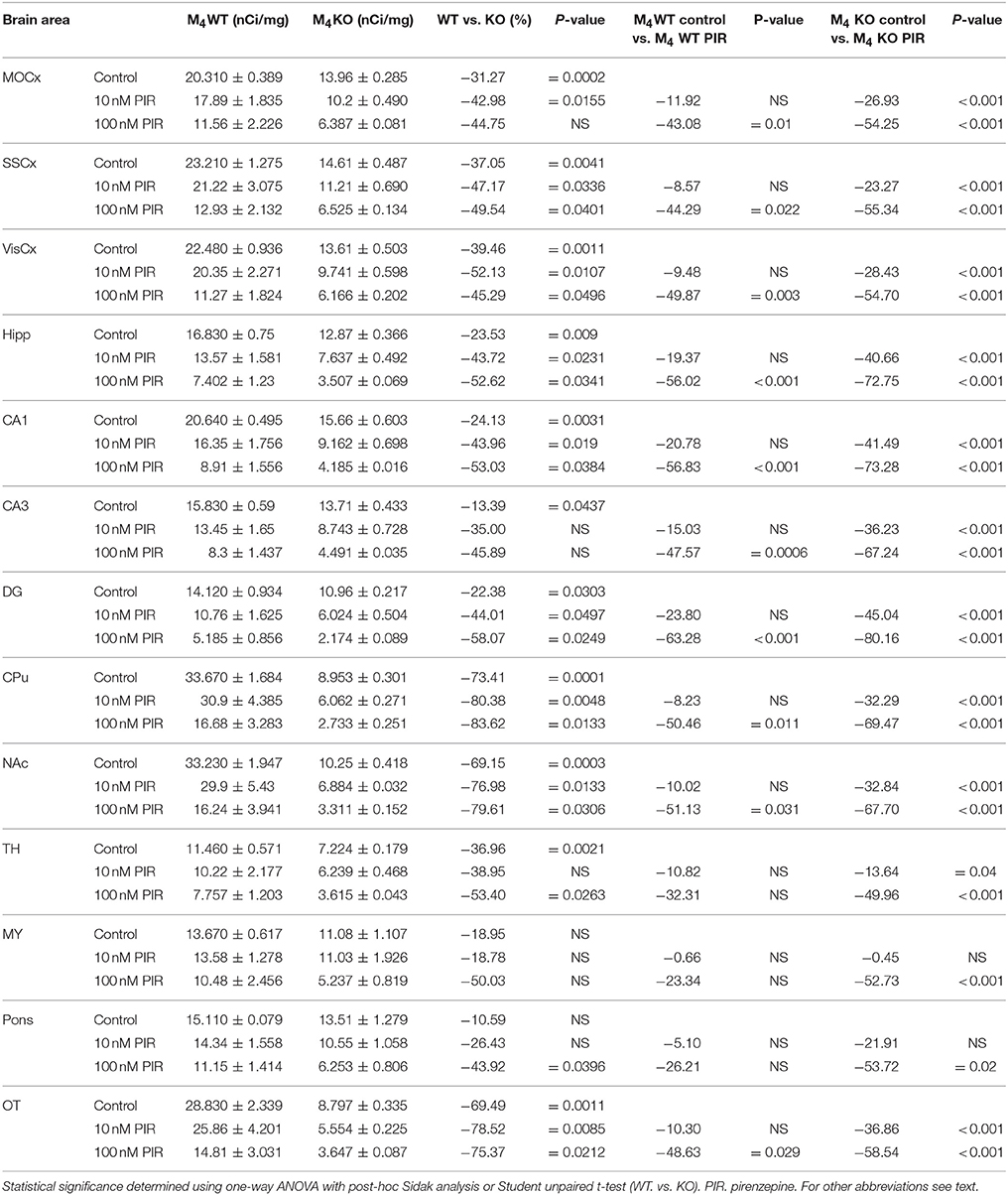
Table 7. Binding and percentage difference in 3H-AFDX 384 binding with pirenzepine in M4WT and M4KO animals vs. control (no pirenzepine).
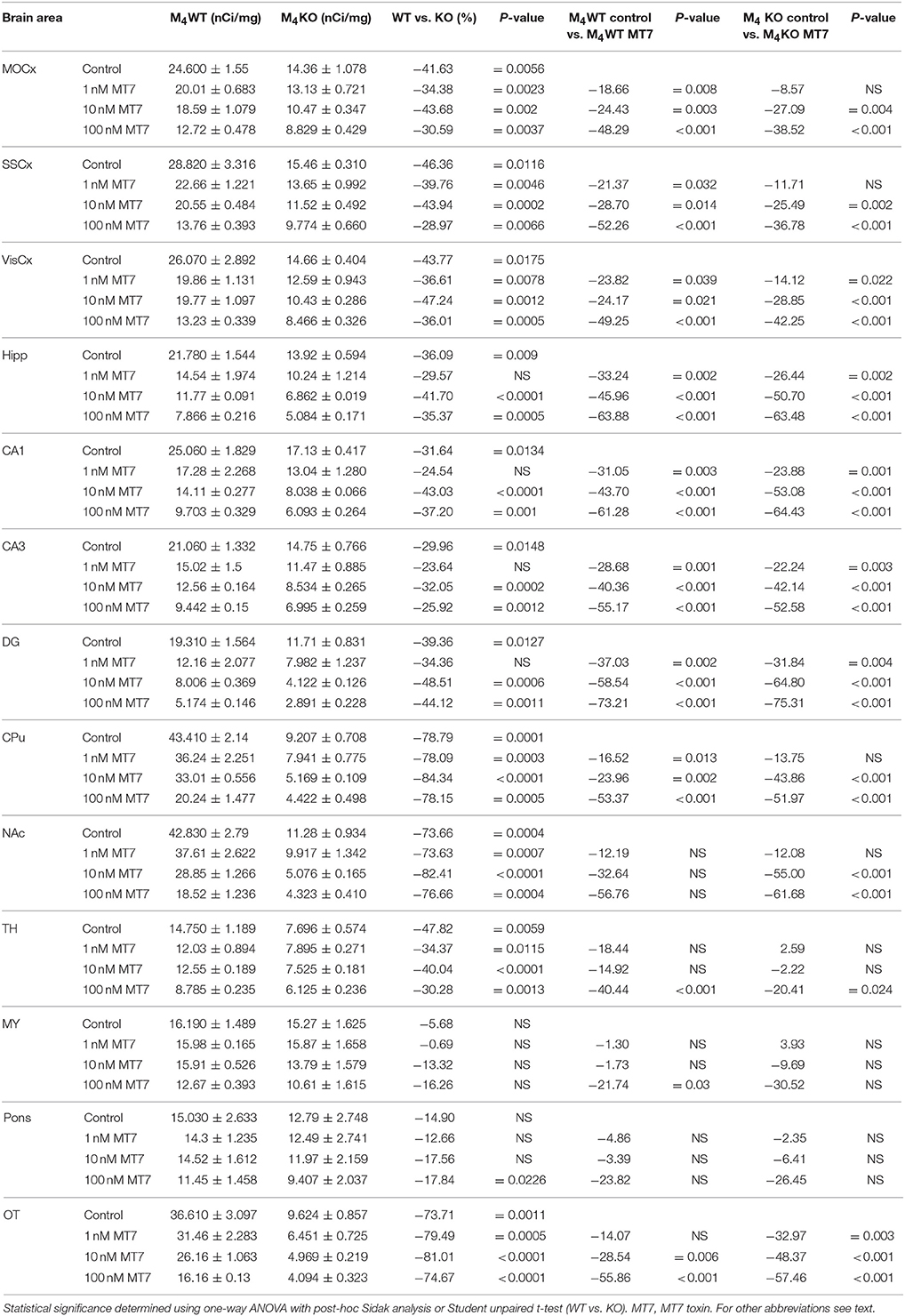
Table 8. Binding and percentage difference in 3H-AFDX 384 binding with MT7 toxin in M4WT, and M4KO animals vs. control (no MT7 toxin).
Binding of 3H-QNB in WT and M1 KO, M2 KO, and M4 KO Mice
M1 KO mice
Binding decrease in M1 KO mice (Figure 3, top) showed high densities of M1 MR in almost all brain areas. In cortical structures, there was approximately 60% of M1 MR (64, 58, and 61% in MoCx, SSCx, and VisCx, respectively). The highest density of M1 MR was found in hippocampus (91%) and in hippocampal regions (CA1: 89%, CA3 88%, dentate gyrus 98%, respectively). In striatum, there was 37%. Similar density of M1 MR was found ventral striatum, i.e., in NAc (46%) and OT (48%).
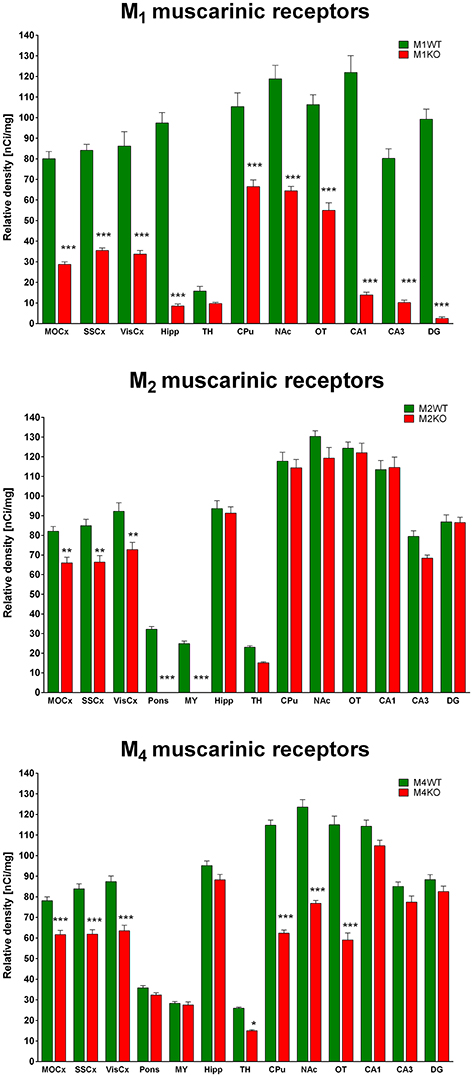
Figure 3. Changes in 3H-QNB specific binding in M1KO mice (top), M2KO mice (middle), and M4KO mice (bottom) when compared to WT animals. Non-specific binding was determined in the presence of 10 μM atropine sulfate. Ordinate: brain areas [motor cortex (MOCx), somatosensory cortex (SSCx), visual cortex (VisCx), medulla oblongate (MY), pons Varoli (pons), striatum (Caudatum-Putamen, CPu), nucleus accumbens (NAc), thalamus (TH), hippocampus (Hipp) and its specific areas CA1, CA3 and dentate gyrus (DG), olfactory tubercle (OT)], abscissa: relative density [nCi/mg]. WT, wild type; KO, knockout mice. *p < 0.05, **p < 0.01, ***p < 0.001.
M2 KO Mice
Binding decrease in M2 KO mice (Figure 3, middle) showed relatively low density of M2 MRs in MoCx, SSCx, and VisCx (20, 22, 21%, respectively). In pons and medulla oblongata, there was 100% decrease suggesting pure M2 MR population in these regions.
M4 KO Mice
The decrease in binding in M4 KO mice (Figure 3, bottom) revealed 21, 26, and 27% of M4 MRs in MoCx, SSCx, and VisCx, respectively. The population of M4 MRs is represented by 43% of total number of MRs in the thalamus. However, the muscarinic population in thalamus is low. The highest proportion of M4 MRs was found in CPu, NAc, OT, where it represents 46, 38, and 49%, respectively.
Discussion
Here we used knockout proof concept to ascertain the selectivity of well-established and commonly used autoradiography protocols for labeling of putative M2 and M1MR. Putative M2 and M1MR were labeled with 3H-AFDX-384 and 3H-pirenzepine, respectively. We compared the pattern and relative density of 3H-AFDX-384 and 3H-pirenzepine specific binding sites in WT with M1, M2, and M4 KO mice.
Our results demonstrate that 3H-pirenzepine labels predominantly, albeit not exclusively, M1MR. Thus, according to our results, 3H-pirenzepine can be used as M1MR selective ligand in brain cortex (MOCx, SSCx, VisCx) and hippocampus, in which more than 94% of 3H-pirenzepine binding sites are attributable to M1MR. In the striatum and olfactory tubercle 10–13% of 3H-pirenzepine specific binding sites correspond to another MR subtype. Apart from these limitations, 3H-pirenzepine can be considered (in concentration 5 nmol/l and in the protocol described in Methods section) as M1MR specific ligand.
To test whether 3H-pirenzepine binds also to M4MR, we performed also 3H-pirenzepine autoradiography in M4 KO mice, (see Table 1 for affinities). We assumed, that in case that 3H-pirenzepine binds M4MR, there should be decrease in 3H-pirenzepine binding in M4 KO mice. Indeed, 3H-pirenzepine binding in M4KO mice is decreased in cortical areas and in caudate putamen. This result can be explained, however by two ways. Firstly, 3H-pirenzepine binds to M4MR. Secondly, M1MR are decreased in M4 KO mice. M4MR are enriched in the striatum and expressed at modest level in cortex (Gomeza et al., 1999b). Therefore, the decrease of 3H-pirenzepine binding due to the lack of M4MR should be mostly seen in striatum and not in the cortex. We showed that there is decrease in 3H-pirenzepine binding by 11% in striatum of M4 KO mice and that the residual binding of 3H-pirenzepine in M1 KO mice is 10–13%. This suggest that indeed, in striatum 3H-pirenzepine binds also to M4MR. In contrast, there was more than 95% reduction of 3H-pirenzepine binding in cortex of M1 KO mice. The density of M4MR in the cortex is approximately three-fold lower than in the striatum, but decrease in 3H-pirenzepine binding in cortex of M4 KO mice was similar to that in striatum. Taken together, decrease in 3H-pirenzepine binding in the cortex of M4 KO mice suggests rather alteration in the density of M1 MR than binding to M4MR. Once again and taking above mentioned results into account, we can consider 3H-pirenzepine as M1MR highly specific ligand.
In contrast, 3H-AFDX-384 has poor selectivity toward M2MR and labels mixed population of MR in a brain area dependent manner. Autoradiography in M2 and M4KO mice showed that 3H-AFDX-384 binds to multiple MR populations and that this population differs between brain areas. Moreover, 3H-AFDX-384 binds to M4MR subtype in similar way as to M2MR subtype what is in contrast to opinion that 3H-AFDX-384 is M2MR preferential ligand. Our working hypothesis was that in case of 3H-AFDX-384 high selectivity toward M2MR, gene deletion of M2MR resulting in absence of M2MR protein in M2KO mice should result in complete loss of 3H-AFDX-384-specific binding. Even though 3H-AFDX-384 specific binding was reduced in M2KO mice throughout the brain, the reduction was far less than expected. Surprisingly 3H-AFDX-384 specific binding was also reduced in M4KO mice. In brain regions rich in M4MR, such as striatum the 3H-AFDX-384 specific binding was reduced more than 70%. This suggest that3H-AFDX-384 binds to M4MR subtype as well. Even though affinity of AFDX-384 is high (see Table 1), the extent of 3H-AFDX-384 binding to M4MR in vitro autoradiography is surprising. In order to increase the selectivity of 3H-AFDX-384 toward M2MR, we tried to block 3H-AFDX-384 binding to M4MR by addition of antagonist PD102807 (with pKi higher by two orders of magnitude for M4MR than for other MR subtypes: 7.3–7.4 vs. 5.2–6.7, see Table 1). Assuming the high selectivity of PD102807 for M4MR and binding of 3H-AFDX-384 also to M4MR, PD102807 should markedly reduce the residual 3H-AFDX-384 binding in M2KO mice. Indeed, addition of PD102807 into incubation medium dose-dependently reduced the residual 3H-AFDX-384 specific binding in M2KO mice, suggesting effective blocking of putative M4MR. At the highest concentration (1,000 nmol/l), there was no residual binding of 3H-AFDX-384 in M2KO mice. To test the potential binding capacity of PD102807 to other MR subtypes we performed similar experiment in M4KO mice. In case that PD102807 is selective M4MR ligand, adding PD102807 to the incubation medium should not interfere with 3H-AFDX-384 with binding capacity in M4KO mice, since there are no M4MR. However, PD102807 also dose-dependently reduced 3H-AFDX-384 specific binding in M4KO mice with no M4MR, indicating that under our experimental conditions PD102807 has poor selectivity toward M4MR. Assuming that 3H-AFDX-384 predominantly labels M2MR and M4MR and no other MR subtype our results suggest, that PD102807 binds to both M4MR and M2MR. We can therefore conclude that co-incubation with PD102807 is not a suitable protocol for M4MR binding elimination and M2MR specific binding autoradiography. Thus, we explored another specific binding antagonist—MT3 toxin from Dendroaspis angusticeps venom—with effort to eliminate binding to M4MR. This toxin is believed to block binding to M4MR with great order of magnitude difference to other MR subtypes (pKi = 8.7 (M4MR) vs. < 6 in M2, M3, M5MR and 7.1 in M1MR, see Table 1). Similarly to PD102807, MT3 toxin was unable to effectively block 3H-AFDX-384 binding to M4MR. There remained 3H-AFDX-384 binding capacity in M2 KO mice in the highest (100 nmol/l) MT3 toxin concentration which should be able to block all M4MR. Moreover, similarly to PD102807, MT3 not only dose-dependently decreased residual binding of 3H-AFDX-384 in M2 KO mice, but also in M4 KO mice. This suggest that MT3 toxin does not distinguish between M4 and M2MR, at least under our experimental conditions. Finally, we tested the hypothesis that 3H-AFDX-384 binds also to M1MR. We tested our hypotheses in M4 KO animals using two approaches: binding in the presence of pirenzepine (see Table 1 for affinities) and irreversible, and very specific toxin MT7 (with pKi = 9.8 to M1MR and pKi <6 to other muscarinic subtypes). We choose M4 KO mice in order to exclude possible binding of pirenzepine toward M4MR in striatum as discussed above. Even at 10 nM concentration, pirenzepine significantly reduced 3H-AFDX-384 specific binding in M4 KO mice, suggesting that 3H-AFDX-384 binds to a certain extent also M1MR. Moreover, irreversible, and very specific toxin MT7 showed dose-dependent reduction of 3H-AFDX-384 specific binding in M4 KO mice further supporting binding of 3H-AFDX-384 to M1MR. It is therefore possible to conclude that 3H-AFDX-384 is not suitable ligand for M2MR. Moreover, we were not successful in designing any protocol to block M4 or M1MR and increase the selectivity of 3H-AFDX-384 autoradiography of M2MR.
All five MR subtypes are highly expressed in the brain (Levey et al., 1991; Wess et al., 2003; Oki et al., 2005). The M1 and M4 MRs represent the most abundant subtypes with the highest expression in the cortex, hippocampus and striatum that can be well illustrated using 3H-QNB autoradiography in our M1 KO and M4 KO mice.
In vitro receptor autoradiography has been used for decades for mapping anatomical distribution and quantification of broad range of receptors (Manuel et al., 2015; Farar and Myslivecek, 2016). The standard autoradiography is based on equilibrium binding of radioactively labeled antagonist. The key aspect of autoradiography is thus the selectivity of radioligand toward its target. MR subtypes show often overlapping pattern of expression and most MR ligands have poor selectivity, making the discrimination of individual subtypes difficult. While there are several ligands available, which specifically label MR, they do not distinguish individual MR subtypes. In vitro heterologous expression systems have helped to describe the affinity of a broad range of antagonists toward each of the five MR (Buckley et al., 1989; Dörje et al., 1991; Dong et al., 1995). These studies have identified so-called preferential MR ligands such as AFDX-384 and pirenzepine, which are commonly used to target putative M2 and M1MR, respectively. AFDX-384 has the highest affinity toward M2MR, but also similar affinity toward M4MR (Dörje et al., 1991).
We have proofed here that autoradiography protocol for 3H-pirenzepine is suitable for M1MR detection (with limitations in CPu, NAc and OT) where pirenzepine binds also by approximately 10% to another MR subtype, likely M4MR). Thus, this ligand can be used as M1MR specific as before (e.g., Wamsley et al., 1984; McCabe et al., 1987; Farar and Myslivecek, 2016).
On the other hand, 3H-AFDX-384 is not suitable to detect M2MR as believed previously (Entzeroth and Mayer, 1990; Wolff et al., 2008; Grailhe et al., 2009). However, there are some studies that previously correctly defined 3H-AFDX-384 as M2 partially selective (Mulugeta et al., 2003) or as M2/M4 ligand (Nieves-Martinez et al., 2012).
The fact that 3H-AFDX-384 is not selective toward M2MR was predictable in the light of AFDX-384 affinity to MR subtypes (see Table 1). However, almost identical binding to M2MR and M4MR is a new finding. We also tried to find specific protocol for M2MR specific binding using different antagonists (PD102807, MT3 toxin, pirenzepine, and MT7 toxin). None of these antagonists were able to completely block other receptors. And thus it is necessary to conclude that there is no way to make the binding more specific to M2MR.
Another aspect of our data is demonstration of M1MR, M2MR, and M4MR distribution. It can be deduced from Figure 1 that M1MR are not as hugely present in the cortical structures as in some hippocampal areas (dorsal hippocampus, CA1 area, and dentate gyrus). Very low M1MR density is in the thalamus. Comparing binding in M2WT and M2KO mice we can conclude that there is relatively high density of M2MR in cortical structures, medulla oblongata, pons and thalamus. In contrast, hippocampus and striatum does not have high amount of M2MR. Striatum is, however, rich in M4MR similarly to nucleus accumbens and olfactory tubercles. Also, the density of M4MR in the cortex is relatively high. However, one should take into account that these data represent relative proportions of respective MR subtypes, since the binding was determined in radioligand concentrations around KD and the receptors were not saturated. 3H-pirenzepine has pKD around 7.9 (Watson et al., 1984), 3H-AFDX 384 has KD between 3 and 4 nmol.l−1 (Castoldi et al., 1991) what is comparable to 3H-pirenzepine. In order to verify the proportion of respective MR subtypes we have used specific MR knockouts (M1, M2, and M4 KOs) and measured a decrease in non-specific radioligand (3H-QNB) binding. This radioligand has much higher affinity to MR and pKD ranging between 10.6 and 10.8 (Peralta et al., 1987). These experiments showed the highest proportion (usually above 50%) of M1MR in virtualy all studied brain areas. M2MR take up to 20% in cortical areas and 34% in thalamus. M4MR were abundant (40% approximately) in thalamus, striatum and ventral striatum (NAc and OT), about 20% of M4MR can be found in cortical structures. This is in general agreement with previously published data (Levey et al., 1991; Wess et al., 2003; Oki et al., 2005), although we have obtained slightly different pattern of MR subtypes distribution what can be caused by the use of different radioligand. Some discrepancies between results could be attributed to the fact that not all antibodies are selective (Pradidarcheep and Michel, 2016). As referenced by Manuel et al. (2015) relatively good correlation exists between the radioligand detected receptor number and the immunolabeled receptor protein for M1 subtype.
Our results concerning the MR distribution are generally in in agreement with previous study (Oki et al., 2005) which employed different muscarinic knockouts but used non-specific radioligand (3H-NMS). However, in this study less brain areas were investigated and different method (direct radioligand binding) used. We sldo found similar pattern of MR subtype distribution as investigated using antibodies and electron microscopy (Hersch et al., 1994), immunoprecipitation (Levey et al., 1991; Tice et al., 1996) and radioligand binding (Flynn and Mash, 1993). Another study (Ferrari-Dileo et al., 1994) that used selective labeling found similar pattern of M4MR distribution as here. All five MR subtypes are highly expressed in the brain (Levey, 1996; Oki et al., 2005). The M1 and M4 MR represent the most abundant subtypes with the highest expression in the cortex, hippocampus and striatum that can be well illustrated using 3H-QNB autoradiography in our M1 KO and M4 KO mice. Previous research has shown that MRs, mostly M1 and M4 MRs, might be an interesting pharmacological target for the treatment of neurodegenerative and neuropsychiatric diseases such as Alzheimer's disease, Parkinson's disease, schizophrenia, depression and also drug abuse (Bodick et al., 1997; Scarr et al., 1999; Brady et al., 2008; Langmead et al., 2008; Bradley et al., 2017; Dall et al., 2017). Therefore, in vitro radioligand binding studies, direct and indirect, represent an important pharmacological tool to precisely characterize the involvement of specific MR subtypes in such a disease as well as to study the binding properties, affinity and efficacy of new chemical compounds with the potential to become a selective drug toward the receptor.
Conclusion
We can therefore conclude that 3H-pirenzepine showed high selectivity toward M1MR and can be used with minor limitations as M1MR specific ligand. In contrast, 3H-AFDX-384 binding sites represent different populations of MR subtypes which is brain-region-specific. This finding has to be taken into account when interpreting the binding data. Our experiments with 3H-QNB binding decrease in M1, , and M4 KO animals showed the highest proportion of M1MR in virtualy all studied brain areas. M2MR were expressed in cortical areas and in thalamus. M4MR were abundant in thalamus, striatum and ventral striatum (NAc and OT as well as in cortical structures.
Author Contributions
JM and VF contributed to the conception and design of the reported studies. PV, IK, and VF conducted all of the experiments. SF, VF, and PV analyzed the data. PV and VF contributed to the drafting and revision of the manuscript. JM wrote the manuscript in the final form. All authors approved the final version and agreed to be accountable for all aspects of the work.
Funding
This research was supported by Supported by Grant 328314 from GAUK, CZ, PROGRES Q25/1LF/2 from Charles University, CZ, and by Czech Science Foundation Grant No 17-03847S.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The reviewer AB and handling Editor declared their shared affiliation.
Acknowledgments
The authors thank Katerina Janisova for her technical assistance.
References
Aubert, I., Cécyre, D., Gauthier, S., and Quirion, R. (1992). Characterization and autoradiographic distribution of [3H]AF-DX 384 binding to putative muscarinic M2 receptors in the rat brain. Eur. J. Pharmacol. 217, 173–184. doi: 10.1016/0014-2999(92)90843-S
Bodick, N. C., Offen, W. W., Shannon, H. E., Satterwhite, J., Lucas, R., van Lier, R., et al. (1997). The selective muscarinic agonist xanomeline improves both the cognitive deficits and behavioral symptoms of Alzheimer disease. Alzheimer Dis. Assoc. Disord. 11(Suppl. 4), S16–S22.
Bradley, S. J., Bourgognon, J., Sanger, H. E., Verity, N., Mogg, A. J., White, D. J., et al. (2017). M1 muscarinic allosteric modulators slow prion neurodegeneration and restore memory loss. J. Clin. Invest. 127, 487–499. doi: 10.1172/JCI87526
Brady, A. E., Jones, C. K., Bridges, T. M., Kennedy, J. P., Thompson, A. D., Heiman, J. U., et al. (2008). Centrally active allosteric potentiators of the M4 muscarinic acetylcholine receptor reverse amphetamine-induced hyperlocomotor activity in rats. J. Pharmacol. Exp. Ther. 327, 941–953. doi: 10.1124/jpet.108.140350
Buckley, N. J., Bonner, T. I., and Brann, M. R. (1988). Localization of a family of muscarinic receptor mRNAs in rat brain. J. Neurosci. 8, 4646–4652.
Buckley, N. J., Bonner, T. I., Buckley, C. M., and Brann, M. R. (1989). Antagonist binding properties of five cloned muscarinic receptors expressed in CHO-K1 cells. Mol. Pharmacol. 35, 469–476.
Buckley, N. J., and Burnstock, G. (1986). Autoradiographic localization of peripheral M1 muscarinic receptors using [3H]pirenzepine. Brain Res. 375, 83–91. doi: 10.1016/0006-8993(86)90961-3
Bymaster, F. P., McKinzie, D. L., Felder, C. C., and Wess, J. (2003). Use of M1-M5 muscarinic receptor knockout mice as novel tools to delineate the physiological roles of the muscarinic cholinergic system. Neurochem. Res. 28, 437–442. doi: 10.1023/A:1022844517200
Castoldi, A. F., Fitzgerald, B., Manzo, L., Tonini, M., and Costa, L. G. (1991). Muscarinic M2 receptors in rat brain labeled with [3H] AF-DX 384. Res. Commun. Chem. Pathol. Pharmacol. 74, 371–374.
Cea-del Rio, C. A., Lawrence, J. J., Tricoire, L., Erdelyi, F., Szabo, G., and McBain, C. J. (2010). M3 Muscarinic acetylcholine receptor expression confers differential cholinergic modulation to neurochemically distinct hippocampal basket cell subtypes. J. Neurosci. 30, 6011–6024. doi: 10.1523/JNEUROSCI.5040-09.2010
Cortes, R., and Palacios, J. M. (1986). Muscarinic cholinergic receptor subtypes in the rat brain. I. Quantitative autoradiographic studies. Brain Res. 362, 227–238. doi: 10.1016/0006-8993(86)90448-8
Dall, C., Weikop, P., Dencker, D., Molander, A. C., Conn, P. J., Fink-Jensen, A., et al. (2017). Wörtwein, G., and Muscarinic receptor M4 positive allosteric modulators attenuate central effects of cocaine. Drug Alcohol Depend 176, 154–161. doi: 10.1016/j.drugalcdep.2017.03.014
Dong, G. Z., Kameyama, K., Rinken, A., and Haga, T. (1995). Ligand binding properties of muscarinic acetylcholine receptor subtypes (m1-m5) expressed in baculovirus-infected insect cells. J. Pharmacol. Exp. Ther. 274, 378–384.
Dörje, F., Wess, J., Lambrecht, G., Tacke, R., Mutschler, E., and Brann, M. R. (1991). Antagonist binding profiles of five cloned human muscarinic receptor subtypes. J. Pharmacol. Exp. Ther. 256, 727–733.
Eglen, R. (2012). “Overview of muscarinic receptor subtypes,” in Muscarinic Receptors, eds A. D. Fryer, A. Christopoulos, and N. M. Nathanson (Berlin; Heidelberg: Springer), 3–28.
Entzeroth, M., and Mayer, N. (1990). Labeling of rat heart muscarinic receptors using the new M2 selective antagonist [3H]AF-DX 384. Biochem. Pharmacol. 40, 1674–1676. doi: 10.1016/0006-2952(90)90473-X
Farar, V., Mohr, F., Legrand, M., d'Incamps, B. L., Cendelin, J., Leroy, J., et al. (2012). Near-complete adaptation of the PRiMA knockout to the lack of central acetylcholinesterase. J. Neurochem. 122, 1065–1080. doi: 10.1111/j.1471-4159.2012.07856.x
Farar, V., and Myslivecek, J. (2016). “Autoradiography assessment of muscarinic receptors in the central nervous system,” in Muscarinic Receptor: From Structure to Animal Models, eds J. Myslivecek and J. Jakubik (New York, NY: Springer), 159–180.
Ferrari-Dileo, G., Waelbroeck, M., Mash, D. C., and Flynn, D. D. (1994). Selective labeling and localization of the M4 (m4) muscarinic receptor subtype. Mol. Pharmacol. 46, 1028–1035.
Flynn, D. D., and Mash, D. C. (1993). Distinct kinetic binding properties of N-[3H]-methylscopolamine afford differential labeling and localization of M1, M2, and M3 muscarinic receptor subtypes in primate brain. Synapse 14, 283–296. doi: 10.1002/syn.890140406
Gomeza, J., Shannon, H., Kostenis, E., Felder, C., Zhang, L., Brodkin, J., et al. (1999a). Pronounced pharmacologic deficits in M2 muscarinic acetylcholine receptor knockout mice. Proc. Natl. Acad. Sci. U.S.A. 96, 1692–1697. doi: 10.1073/pnas.96.4.1692
Gomeza, J., Zhang, L., Kostenis, E., Felder, C., Bymaster, F., Brodkin, J., et al. (1999b). Enhancement of D1 dopamine receptor-mediated locomotor stimulation in M4 muscarinic acetylcholine receptor knockout mice. Proc. Natl. Acad. Sci. U.S.A. 96, 10483–10488. doi: 10.1073/pnas.96.18.10483
Grailhe, R., Cardona, A., Even, N., Seif, I., Changeux, J.-P., and Cloëz-Tayarani, I. (2009). Regional changes in the cholinergic system in mice lacking monoamine oxidase A. Brain Res. Bull. 78, 283–289. doi: 10.1016/j.brainresbull.2008.12.004
Hersch, S. M., Gutekunst, C. A., Rees, H. D., Heilman, C. J., and Levey, A. I. (1994). Distribution of m1-m4 muscarinic receptor proteins in the rat striatum: light and electron microscopic immunocytochemistry using subtype-specific antibodies. J. neurosci. 14, 3351–3363.
Jositsch, G., Papadakis, T., Haberberger, R. V., Wolff, M., Wess, J., and Kummer, W. (2009). Suitability of muscarinic acetylcholine receptor antibodies for immunohistochemistry evaluated on tissue sections of receptor gene-deficient mice. Naunyn-Schmiedeberg's Arch. Pharmacol. 379, 389–395. doi: 10.1007/s00210-008-0365-9
Kow, R. L., and Nathanson, N. M. (2012). Structural biology: muscarinic receptors become crystal clear. Nature 482, 480–481. doi: 10.1038/482480a
Kruse, A. C., Ring, A. M., Manglik, A., Hu, J., Hu, K., Eitel, K., et al. (2013). Activation and allosteric modulation of a muscarinic acetylcholine receptor. Nature 504, 101–106. doi: 10.1038/nature12735
Langmead, C. J., Watson, J., and Reavill, C. (2008). Muscarinic acetylcholine receptors as CNS drug targets. Pharmacol. Ther. 117, 232–243. doi: 10.1016/j.pharmthera.2007.09.009
Levey, A. I. (1996). Muscarinic acetylcholine receptor expression in memory circuits: implications for treatment of Alzheimer disease. Proc. Natl. Acad. Sci. U.S.A. 93, 13541–13546. doi: 10.1073/pnas.93.24.13541
Levey, A. I., Kitt, C. A., Simonds, W. F., Price, D. L., and Brann, M. R. (1991). Identification and localization of muscarinic acetylcholine receptor proteins in brain with subtype-specific antibodies. J. Neurosci. 11, 3218–3226.
Manuel, I., Barreda-Gómez, G., González De San Román, E., Veloso, A., Fernández, J. A., Giralt, M. T., et al. (2015). Neurotransmitter receptor localization: from autoradiography to imaging mass spectrometry. ACS Chem. Neurosci. 6, 362–373. doi: 10.1021/cn500281t
McCabe, R. T., Gibb, J. W., Wamsley, J. K., and Hanson, G. R. (1987). Autoradiographic analysis of muscarinic cholinergic and serotonergic receptor alterations following methamphetamine treatment. Brain Res. Bull. 19, 551–557. doi: 10.1016/0361-9230(87)90072-4
Mulugeta, E., Karlsson, E., Islam, A., Kalaria, R., Mangat, H., Winblad, B., et al. (2003). Loss of muscarinic M4 receptors in hippocampus of Alzheimer patients. Brain Res. 960, 259–262. doi: 10.1016/S0006-8993(02)03542-4
Myslivecek, J., Klein, M., Novakova, M., and Ricny, J. (2008). The detection of the non-M-2 muscarinic receptor subtype in the rat heart atria and ventricles. Naunyn-Schmiedebergs Arch. Pharmacol. 378, 103–116. doi: 10.1007/s00210-008-0285-8
Nieves-Martinez, E., Haynes, K., Childers, S. R., Sonntag, W. E., and Nicolle, M. M. (2012). Muscarinic receptor/G-protein coupling is reduced in the dorsomedial striatum of cognitively impaired aged rats. Behav. Brain Res. 227, 258–264. doi: 10.1016/j.bbr.2011.10.048
Oki, T., Takagi, Y., Inagaki, S., Taketo, M. M., Manabe, T., Matsui, M., et al. (2005). Quantitative analysis of binding parameters of [3H]N-methylscopolamine in central nervous system of muscarinic acetylcholine receptor knockout mice. Mol. Brain Res. 133, 6–11. doi: 10.1016/j.molbrainres.2004.09.012
Peralta, E. G., Ashkenazi, A., Winslow, J. W., Smith, D. H., Ramachandran, J., and Capon, D. J. (1987). Distinct primary structures, ligand-binding properties and tissue-specific expression of four human muscarinic acetylcholine receptors. EMBO J. 6, 3923–3929.
Pradidarcheep, W., and Michel, M. C. (2016). “Use of antibodies in the research on muscarinic receptor subtypes,” in Muscarinic Receptor: From Structure to Animal Models, eds J. Myslivecek and J. Jakubik (New York, NY: Springer), 83–94.
Reiner, C., and Nathanson, N. (2012). “Muscarinic receptor trafficking,” in Muscarinic Receptors, eds A. D. Fryer, A. Christopoulos, and N. M. Nathanson (Berlin; Heidelberg: Springer), 61–78.
Scarr, E., Dean, B., Der Zee, E. A., and Luiten, P. G. (1999). Muscarinic receptors: do they have a role in the pathology and treatment of schizophrenia? J. Neurochem. 107, 1188–1195. doi: 10.1111/j.1471-4159.2008.05711.x
Shin, J. H., Adrover, M. F., Wess, J., and Alvarez, V. A. (2015). Muscarinic regulation of dopamine and glutamate transmission in the nucleus accumbens. Proc. Natl. Acad. Sci. U.S.A. 112, 8124–8129. doi: 10.1073/pnas.1508846112
Thomsen, M., Sørensen, G., and Dencker, D. (2017). Physiological roles of CNS muscarinic receptors gained from knockout mice. Neuropharmacology. doi: 10.1016/j.neuropharm.2017.09.011. [Epub ahead of print].
Tice, M. A., Hashemi, T., Taylor, L. A., and McQuade, R. D. (1996). Distribution of muscarinic receptor subtypes in rat brain from postnatal to old age. Dev. Brain Res. 92, 70–76. doi: 10.1016/0165-3806(95)01515-9
Tien, L.-T., Fan, L.-W., Sogawa, C., Ma, T., Loh, H. H., and Ho, I.-K. (2004). Changes in acetylcholinesterase activity and muscarinic receptor bindings in μ-opioid receptor knockout mice. Mol. Brain Res. 126, 38–44. doi: 10.1016/j.molbrainres.2004.03.011
Vilaró, M. T., Palacios, J. M., and Mengod, G. (1990). Localization of m5 muscarinic receptor mRNA in rat brain examined by in situ hybridization histochemistry. Neurosci. Lett. 114, 154–159. doi: 10.1016/0304-3940(90)90064-G
Villiger, J. W., and Faull, R. L. M. (1985). Muscarinic cholinergic receptors in the human spinal cord: differential localization of [3H]pirenzepine and [3H]quinuclidinylbenzilate binding sites. Brain Res. 345, 196–199. doi: 10.1016/0006-8993(85)90854-6
Wamsley, J. K., Gehlert, D. R., Roeske, W. R., and Yamamura, H. I. (1984). Muscarinic antagonist binding site heterogeneity as evidenced by autoradiography after direct labeling with [3H]-QNB and [3H]-pirenzepine. Life Sci. 34, 1395–1402. doi: 10.1016/0024-3205(84)90012-2
Wang, Q., Wei, X., Gao, H., Li, J., Liao, J., Liu, X., et al. (2014). Simvastatin reverses the downregulation of M1/4 receptor binding in 6-hydroxydopamine-induced parkinsonian rats: the association with improvements in long-term memory. Neuroscience 267, 57–66. doi: 10.1016/j.neuroscience.2014.02.031
Watson, M., Roeske, W. R., Johnson, P. C., and Yamamura, H. I. (1984). [3H]Pirenzepine identifies putative M1 muscarinic receptors in human stellate ganglia. Brain Res. 290, 179–182. doi: 10.1016/0006-8993(84)90751-0
Weiner, D. M., Levey, A. I., and Brann, M. R. (1990). Expression of muscarinic acetylcholine and dopamine receptor mRNAs in rat basal ganglia. Proc. Natl. Acad. Sci. U.S.A. 87, 7050–7054. doi: 10.1073/pnas.87.18.7050
Wess, J., Duttaroy, A., Zhang, W., Gomeza, J., Cui, Y., Miyakawa, T., et al. (2003). M1-M5 muscarinic receptor knockout mice as novel tools to study the physiological roles of the muscarinic cholinergic system. Recept. Channels 9, 279–290. doi: 10.3109/10606820308262
Wess, J., Eglen, R. M., and Gautam, D. (2007). Muscarinic acetylcholine receptors: mutant mice provide new insights for drug development. Nat. Rev. Drug Discov. 6, 721–733. doi: 10.1038/nrd2379
Wolff, S. C., Hruska, Z., Nguyen, L., and Dohanich, G. P. (2008). Asymmetrical distributions of muscarinic receptor binding in the hippocampus of female rats. Eur. J. Pharmacol. 588, 248–250. doi: 10.1016/j.ejphar.2008.04.002
Yamamura, H. I., Wamsley, J. K., Deshmukh, P., and Roeske, W. R. (1983). Differential light microscopic autoradiographic localization of muscarinic cholinergic receptors in the brainstem and spinal cord of the rat using [3H]pirenzepine. Eur. J. Pharmacol. 91, 147–149. doi: 10.1016/0014-2999(83)90379-5
Yasuda, R. P., Ciesla, W., Flores, L. R., Wall, S. J., Li, M., Satkus, S. A., et al. (1993). Development of antisera selective for m4 and m5 muscarinic cholinergic receptors: distribution of m4 and m5 receptors in rat brain. Mol. Pharmacol. 43, 149–157.
Zavitsanou, K., Katsifis, A., Mattner, F., and Huang, X.-F. (2003). Investigation of M1//M4 muscarinic receptors in the anterior cingulate cortex in schizophrenia, bipolar disorder, and major depression disorder. Neuropsychopharmacology 29, 619–625. doi: 10.1038/sj.npp.1300367
Keywords: M1 muscarinic receptor, M2 muscarinic receptor, M4 muscarinic receptor, 3H-pirenzepine, 3H-AFDX-384, 3H-QNB, autoradiography
Citation: Valuskova P, Farar V, Forczek S, Krizova I and Myslivecek J (2018) Autoradiography of 3H-pirenzepine and 3H-AFDX-384 in Mouse Brain Regions: Possible Insights into M1, M2, and M4 Muscarinic Receptors Distribution. Front. Pharmacol. 9:124. doi: 10.3389/fphar.2018.00124
Received: 07 November 2017; Accepted: 05 February 2018;
Published: 20 February 2018.
Edited by:
Pascal Bonaventure, Janssen Research and Development, United StatesReviewed by:
Anindya Bhattacharya, Janssen Research and Development, United StatesMassimo Grilli, Università di Genova, Italy
Mengling Liu, HD Biosciences Co., Ltd., United States
Copyright © 2018 Valuskova, Farar, Forczek, Krizova and Myslivecek. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jaromir Myslivecek, am15c0BsZjEuY3VuaS5jeg==
 Paulina Valuskova1
Paulina Valuskova1 Jaromir Myslivecek
Jaromir Myslivecek