
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Pharmacol. , 01 December 2016
Sec. Pharmacogenetics and Pharmacogenomics
Volume 7 - 2016 | https://doi.org/10.3389/fphar.2016.00299
 Miles D. Thompson1,2*
Miles D. Thompson1,2* Valerie Capra3
Valerie Capra3 Mark T. Clunes4
Mark T. Clunes4 G. E. Rovati5
G. E. Rovati5 Jana Stankova6
Jana Stankova6 Mary C. Maj7
Mary C. Maj7 David L. Duffy8
David L. Duffy8Genetic variants associated with asthma pathogenesis and altered response to drug therapy are discussed. Many studies implicate polymorphisms in genes encoding the enzymes responsible for leukotriene synthesis and intracellular signaling through activation of seven transmembrane domain receptors, such as the cysteinyl leukotriene 1 (CYSLTR1) and 2 (CYSLTR2) receptors. The leukotrienes are polyunsaturated lipoxygenated eicosatetraenoic acids that exhibit a wide range of pharmacological and physiological actions. Of the three enzymes involved in the formation of the leukotrienes, arachidonate 5 lipoxygenase 5 (ALOX5), leukotriene C4 synthase (LTC4S), and leukotriene hydrolase (LTA4H) are all polymorphic. These polymorphisms often result in variable production of the CysLTs (LTC4, LTD4, and LTE4) and LTB4. Variable number tandem repeat sequences located in the Sp1-binding motif within the promotor region of the ALOX5 gene are associated with leukotriene burden and bronchoconstriction independent of asthma risk. A 444A > C SNP polymorphism in the LTC4S gene, encoding an enzyme required for the formation of a glutathione adduct at the C-6 position of the arachidonic acid backbone, is associated with severe asthma and altered response to the CYSLTR1 receptor antagonist zafirlukast. Genetic variability in the CysLT pathway may contribute additively or synergistically to altered drug responses. The 601 A > G variant of the CYSLTR2 gene, encoding the Met201Val CYSLTR2 receptor variant, is associated with atopic asthma in the general European population, where it is present at a frequency of ∼2.6%. The variant was originally found in the founder population of Tristan da Cunha, a remote island in the South Atlantic, in which the prevalence of atopy is approximately 45% and the prevalence of asthma is 36%. In vitro work showed that the atopy-associated Met201Val variant was inactivating with respect to ligand binding, Ca2+ flux and inositol phosphate generation. In addition, the CYSLTR1 gene, located at Xq13-21.1, has been associated with atopic asthma. The activating Gly300Ser CYSLTR1 variant is discussed. In addition to genetic loci, risk for asthma may be influenced by environmental factors such as smoking. The contribution of CysLT pathway gene sequence variants to atopic asthma is discussed in the context of other genes and environmental influences known to influence asthma.
Given that the prevalence of asthma may be increasing in many populations world-wide (Eder et al., 2006), it is significant that advances in treatment may result from an improved understanding of both the molecular basis of asthma and its responsiveness to therapy. Both asthma susceptibility loci, the genetic variability that confers risk for complex respiratory traits that lead to asthma, and pharmacogenetics, the genetic variability that confers responsiveness of an individual to pharmaceutical therapy, may be best modeled as independent complex traits with both genetic and environmental (GXE) contributions. Since variability in the genes encoding the proteins essential for inflammation may influence risk for many aspects of asthmatic inflammation (Drazen et al., 2000), it is to be expected that it might also increase the susceptibility to asthma independent of their pharmacogenetic relevance (Thompson et al., 2008). Here, we discuss the relative contribution of genes in the cysteinyl leukotriene (CysLT) pathway to both the etiology of asthma itself and a person’s responsiveness to leukotriene modifying (LTM) drugs.
While in some cases the genetic variants that result in disease also encode protein targets for pharmaceutical interventions, this relationship is often not consistent even with polymorphisms of a G protein-coupled receptor (GPCR) system such as the one that mediates CysLT inflammation. For example, while variability in the genes encoding drug targets such as the cysteinyl leukotriene 1 (CYSLTR1) and 2 (CYSLTR2) receptor genes may both contribute to risk for atopic asthma in some populations (Thompson et al., 2003, 2005b, 2013; Pillai et al., 2004; Brochu-Bourque et al., 2011; Yaddaden et al., 2016), only the cysteinyl leukotriene 1 (CYSLTR1) receptor is targeted by LTMs. We can only speculate, however, on the role that that variability in the cysteinyl leukotriene 2 (CYSLTR2) receptor may have in LTM pharmacogenetics since drugs targeting the CYSLTR2 receptor are not widely available.
In general, respiratory disease itself, and patient response to therapy, may be best modeled as independent complex traits with both genetic and environmental contributions. The GPCR associated with asthma, the (NPSR1) neuropeptide S receptor 1 (Laitinen et al., 2004; Kormann et al., 2005; Melén et al., 2005; Pietras et al., 2011), may represent an exception inasmuch as it also defines a potential therapeutic target. As a result, many advances in pharmacogenetics have resulted not from linkage or candidate gene studies but from Genome Wide Association Studies (GWAS). Hence, the field of pharmacogenomics has become axiomatic in the study of patient response and non-response to medication, adverse drug reactions, and optimizing drug dose for the individual.
The study of CYSLTR1 and CYSLTR2 gene variability in asthma, therefore, must be considered in the context of the genetic polymorphisms found within the synthesis pathway genes as well as genes that modify patient response to LTMs. The LTMs used to treat asthma, such as montelukast, a drug which targets cysteinyl leukotriene 1 (CYSLTR1) receptor protein, have a greater than 20% non-response rate (Noonan et al., 1998). While some of the pharmacogenetic determinants of LTM response are attributable to variability in genes that are integral (Kang et al., 2011; Mougey et al., 2013) to the signaling of cysteinyl leukotrienes (Figure 1), many true pharmacogenetic variants are located in genes that lie directly outside the pathway (Dahlin et al., 2015, 2016).
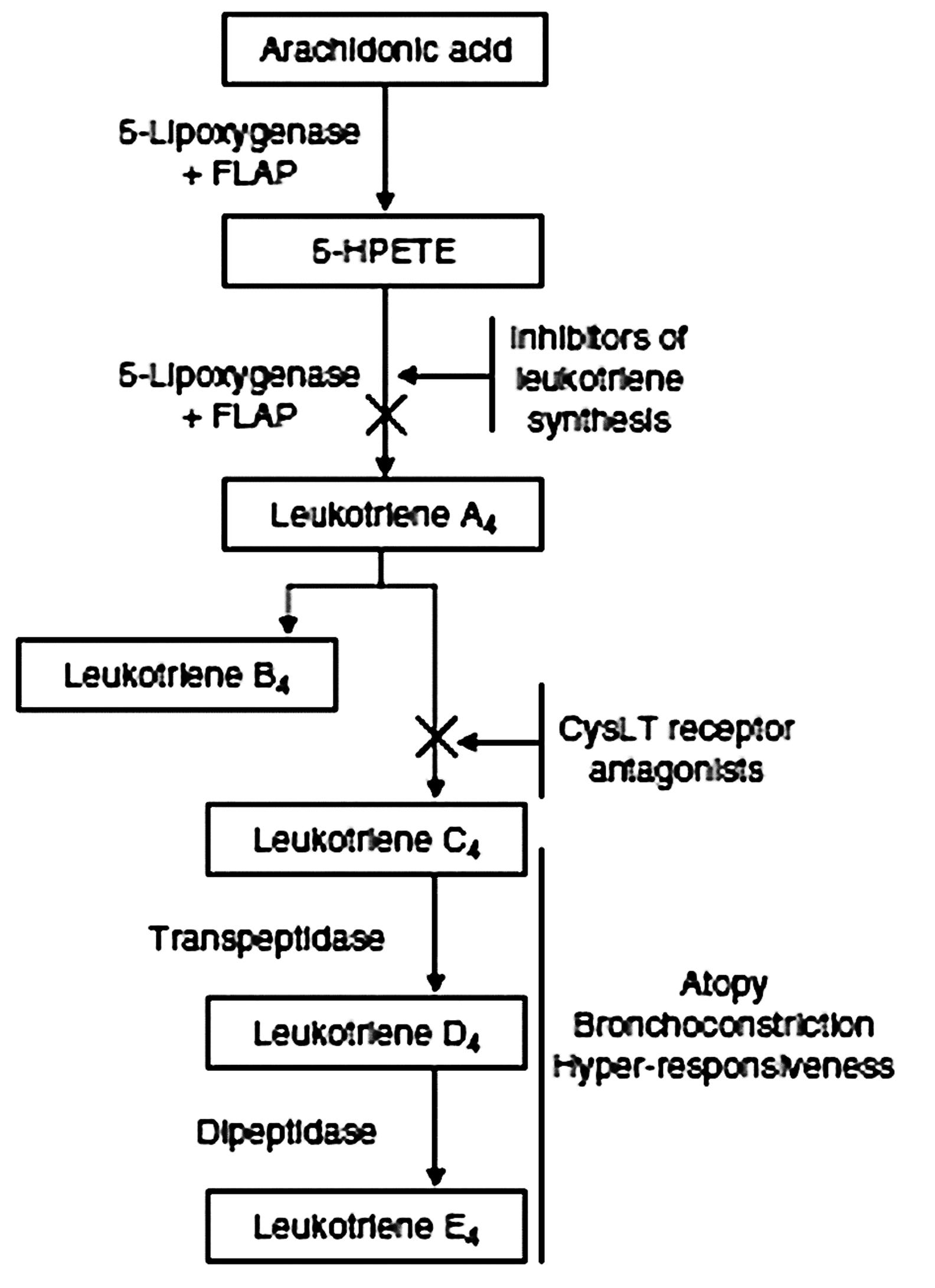
FIGURE 1. Cysteinyl leukotriene synthesis pathway. Leukotriene biosynthesis inhibitors act on either 5-lipoxygenase or its activating protein (FLAP). The class of cysteinyl leukotriene (CysLT) receptor antagonists target the CysLT receptors CYSLTR1 and CYSLTR2. 5-HPETE, 5-hydroxyperoxy-eiocosatetraenoic acid.
In order to undertake GWAS studies of complex traits, a large sample size is generally required. Thus, informative studies of the genetic basis of drug response are conducted among large samples of unrelated individuals recruited from a given population. This is especially true for asthma phenotypes studied in out-bred populations since they tend to be genetically heterogeneous (Moffatt et al., 2007; Michel et al., 2010; Sleiman et al., 2010; Ferreira et al., 2011; Melén et al., 2013). By contrast, small candidate gene studies have achieved reasonable power for detecting variants that confer risk (Marshall et al., 2013). Since data from sequencing large patient cohorts has become available, it has been possible to assess the wider relevance of some observations originally made in smaller studies.
For example, our study of atopic asthma in the Tristan da Cunha isolate provided early insight into CysLT pharmacogenetics (Thompson et al., 2006; Capra et al., 2007; Brochu-Bourque et al., 2011). In particular, the hypomorphic Met201Val CYSLTR2 variant identified on Tristan da Cunha (Thompson et al., 2003) has since been associated with atopy in large out-bred populations (Pillai et al., 2004). Selected examples of GPCR variants associated with asthma phenotypes are presented in Table 1. Tables 2 and 3 present selected variants of the CYSLTR1 and CYSLTR2 genes.
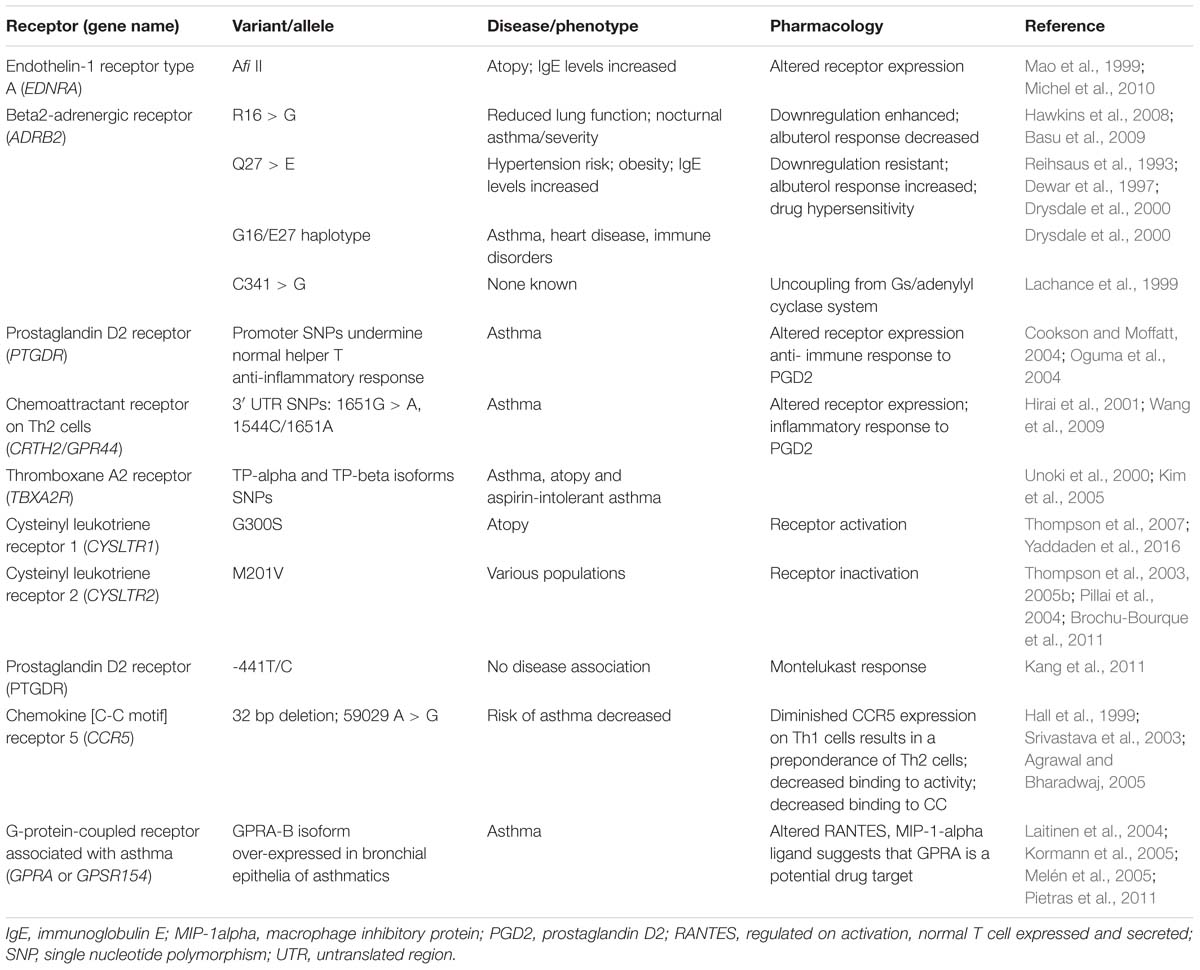
TABLE 1. Human G-protein-coupled receptor (GPCR) variants associated with asthma endophenotype or altered pharmacology.
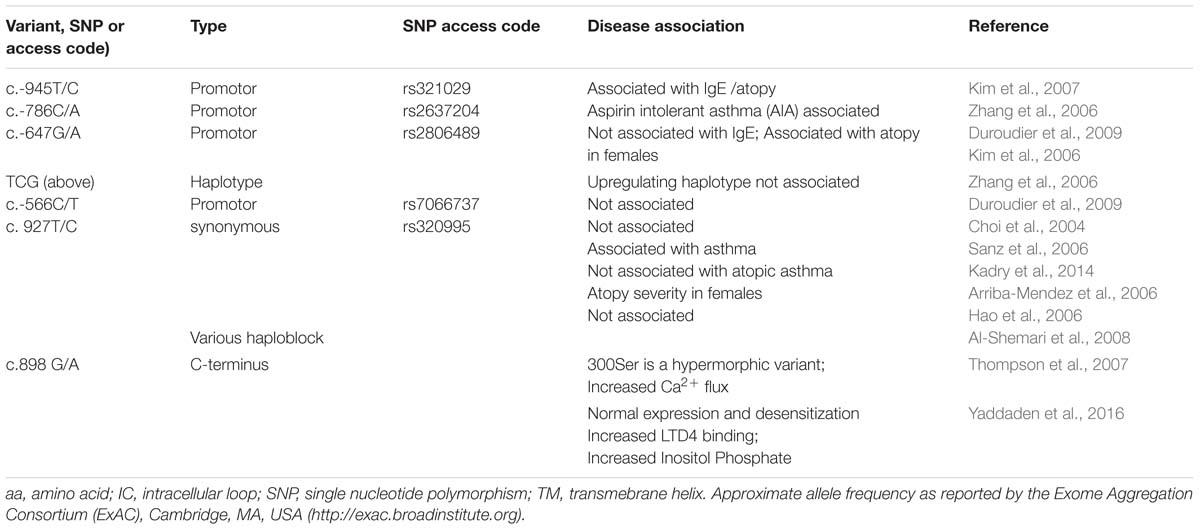
TABLE 2. CYSLTR1 receptor variants: naturally occurring polymorphisms and in vitro engineered mutants.
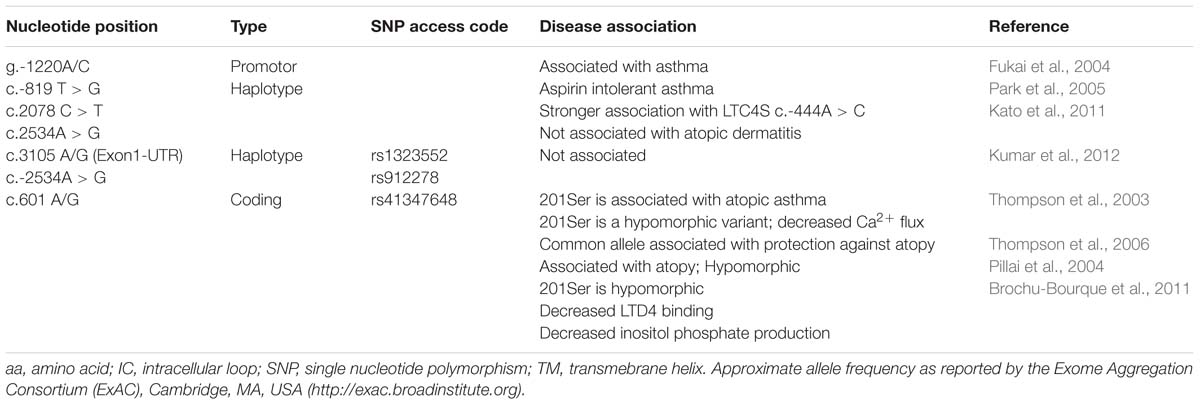
TABLE 3. CYSLTR2 receptor variants: naturally occurring polymorphisms and in vitro engineered mutants.
Studies of population isolates rely upon the fact that a founder effect limits genetic heterogeneity and results in greatly increased prevalence rates (Roberts, 1968; Zamel et al., 1996). In the Tristan da Cunha isolate, for example, there is a 45% prevalence of atopy and a 36% prevalence of asthma (Mantle and Tyrrell, 1973; Zamel et al., 1996; Camacho et al., 2011; Camacho and Cazelles, 2013). By comparison, the prevalence of asthma in most out-bred populations rarely exceeds 10%. The severity of the Tristan da Cunha atopy phenotype may be reflected in events such as the Tristan da Cunha influenza epidemic of 1971 (Mantle and Tyrrell, 1973; Zamel et al., 1996; Camacho et al., 2011; Camacho and Cazelles, 2013).
Analyzing the transmission of genetic markers in a large extended pedigree such as a population isolates, provides the opportunity to identify risk alleles originating from common ancestry (Soodyall et al., 1997, 2003). In particular, this enrichment optimizes the analysis of complex traits for gene-gene interaction effects. Even though the asthma phenotype is relatively homogenous on Tristan da Cunha, the fact that more than one of the seven founders that populated the island after 1812 were asthmatic (Levack and Levack, 2013) suggests that the disorder manifested by the islanders is influenced by complex inheritance.
The Tristan da Cunha findings (Thompson et al., 2003, 2007, 2013), therefore, must be interpreted not only in the context of a founder effect (Zamel et al., 1996; Slutsky and Zamel, 1997; Chan-Yeung et al., 1999) but also in the context of the many genes that confer risk to asthma in out-bred populations: including GPCRs variants (Table 1).
Genome Wide Association Studies have confirmed that variants of the DENND1B gene, located on chromosome 1q31 (Sleiman et al., 2010) and the ORMDL3 gene, located on chromosome 17q12 (Moffatt et al., 2007) underlie asthma pathogenesis in many populations (Ferreira et al., 2011; Melén et al., 2013). Other genes including ADAM33, CCL5, CD14, DPP10, EDN1, NPSR1, GSTP1, IL12B, IL13, IL4, IL4R, PTGDR, TNF, and VDR have also been found to be commonly associated with asthma in various study populations (Michel et al., 2010). Asthma resulting from certain genotypes has shown distinct responses to drug treatment in some instances (Berce et al., 2013). Differentiating the genetic basis of a heterogeneous disease such as asthma, therefore, creates the opportunity to tailor specific therapeutic regimens to the resulting endophenotype (Rana et al., 2001; Milligan, 2002; Thompson et al., 2012, 2014a,c).
G protein-coupled receptors are the primary drug targets for drug interventions into many forms of asthma. Many advances in therapeutics have resulted from the identification of GPCR variants that are associated with disease. Polymorphisms of the β2-adrenergic receptor (ADRB2) gene may be among the most clinically relevant with respect to drug response (Thonkham et al., 2015). Like the subsequent study of many GPCR variants, homology with the prototypical GPCR, rhodopsin (Lefkowitz, 2004), was used to predict the functional significance of sequence alterations. Many of these studies were later evaluated in vitro (Rana et al., 2001; Lefkowitz, 2004).
Early studies showed that β2-adrenergic receptor variants with altered pharmacological response are more common in patients with asthma (Taylor and Kennedy, 2001, 2002; Taylor et al., 2005; Hawkins et al., 2008) as well as congestive heart failure (Thompson et al., 2014a). More recent studies have linked polymorphisms of ADRB2 to altered drug response (Basu et al., 2009; Mathias et al., 2010). Asthma treatment with albuterol, which is targeted to ADRB2, is often associated with increased hypersensitivity (Reihsaus et al., 1993; Dewar et al., 1997; Liggett, 1997; Drysdale et al., 2000; Israel et al., 2000; Littlejohn et al., 2002). Work is currently underway to identify ADRB2 polymorphisms in populations, such as the residents of Grenada, where 30% of the children are reported to have asthma according to a modified ISAAC survey (Pearce et al., 2007; Thonkham et al., 2015).
Other candidate gene studies have identified GPCR gene variants that are associated with asthma phenotypes from various populations. In vivo and in vitro studies have determined functional roles for many GPCR variants (Rana et al., 2001), but evaluation of their physiological role in disease remains a challenge (Thompson et al., 2014b). In addition to CYSLTR1 and CYSLTR2 receptor polymorphisms (Tables 2 and 3), GPCR variants of significance to asthma pharmacogenetics are presented in Table 1. These receptors include the endothelin-1 receptor type A (EDNRA; Mao et al., 1999; Michel et al., 2010), prostaglandin D2 (PTGDR2; Cookson and Moffatt, 2004; Oguma et al., 2004), thromboxane A2 (TBXA2R; Unoki et al., 2000; Kim et al., 2005) and chemokine [C-C motif] receptor 5 (CCR5) receptors (Hall et al., 1999; Srivastava et al., 2003; Agrawal and Bharadwaj, 2005).
The cysteinyl leukotriene ligands (CysLT), including leukotriene C4 (LTC4), D4 (LTD4) and E4 (LTE4), are powerful bronchoconstrictors and pro-inflammatory mediators of atopic asthma (Samuelsson, 1983; Bisgaard, 2001). They are released by mast cells and macrophages during asthma attacks (Severien et al., 2000). The severity of an asthma attack can be influenced by the cellular concentrations of these CysLTs (Maekawa et al., 2002). These ligands regulate human airway contractions upon binding to the CysLT receptors (Metters, 1995; Takasaki et al., 2000; Figueroa et al., 2001; Kormann et al., 2005).
Polymorphic variability is found within many of the genes involved with the synthesis and signaling pathways of CysLTs. The influence of these variations to CysLT biosynthesis enzyme activity, CysLT signaling and asthma severity will be reviewed below. It is important, however, to distinguish between the effect of functional variants of risk alleles for atopy or asthma from pharmacogenetic variants that primarily influence the absorption or response to LTMs. Pharmacogenetic variants which also confer susceptibility to disease will be discussed in Section “Cysteinyl Leukotriene Receptor Polymorphisms.”
Many loci implicated in LTM pharmacogenetics, however, encode genes that are independent of CysLT-related pathways. For example, rs12422149, a common c.935G > A polymorphism in the Solute Carrier organic anion transporting polypeptide (OATP) 2B1 (SLCO2B1) was associated with absorption of and response to montelukast in humans. In particular, in vitro studies have shown that citrus juice may reduce the permeability of montelukast. Moreover, pharmacokinetic studies of montelukast co-ingested with citrus juice suggest that co-ingestion resulted in genotype specific alterations in absorption. For example, A/G heterozygotes were demonstrated to have reduced absorption relative to G/G homozygotes, independent of treatment or combined treatments (Mougey et al., 2011).
Increasingly, loci of pharmacogenetics significance have been identified by GWAS. These data underscore the fact that many loci that alter the efficacy of LTMs pharmaceuticals are not directly part of the CysLT pathway. Seminal studies by Dahlin et al. examined patient response to montelukast and the 5-LO inhibitor zileuton, over 8 weeks and (Dahlin et al., 2015) and 12 weeks (Dahlin et al., 2016), respectively, using DNA and phenotypic information from 526 and 217 people enrolled in placebo-controlled trials. This group based their initial analysis on the genome-wide genotype and phenotypic data from American Lung Association’s Asthma Clinical Research Center (ALA-ACRC) cohorts. These results showed that while rs6475448 [located in the myeloid/lymphoid or mixed-lineage leukemia (MLLT3) gene] was associated with improved montelukast response over an 8-week treatment period (Dahlin et al., 2015), rs12436663 [located in the Mitochondrial Ribonuclease P Protein 3 Precursor (MRPP3) gene, encoding a protein critical to the cleavage of tRNA molecules], was associated with worsened zileuton response over a 12-week period (Dahlin et al., 2016). In both studies, the change in Forced Expiratory Volume (ΔFEV) from baseline in response to drug was used as a measure of treatment response (Dahlin et al., 2015, 2016). Furthermore, the gene-environment (G × E) GWAS model evaluated in the 12-week study identified an association between rs517020 (located in the glycosyltransferase 1 domain containing 1 (GLT1D1) gene) with worsening responses to both montelukast and zileuton (Dahlin et al., 2016). These findings implicate loci in LTM response that, by contrast with leukotriene candidate genes, are not intuitively related to LTM therapeutic responsiveness (Dahlin et al., 2015, 2016).
Next, we will review the polymorphisms in the genes encoding portions of the CysLT pathway implicated in asthma pathogenesis and treatment (Figure 1) before discussing the receptor polymorphisms per se.
Leukotrienes are fatty acid-derived mediators which are divided in two main biologically active subgroups, LTB4 and the cysteine-conjugated leukotrienes (LTC4, LTD4, and LTE4). The arachidonate 5-lipoxygenase (ALOX5) is required for the synthesis of the unstable epoxide precursor LTA4 (Figure 1), on which acts the leukotriene A4 hydrolase (LTA4H) to originate the proinflammatory chemoattractant LTB4 or the leukotriene C4 synthase (LTC4S) to conjugate the tripeptide glutathione and form LTC4; LTD4 and LTE4 are then formed by subsequent removal of the terminal amino acid of the tripeptide. The genes which encode for the leukotriene synthesis enzymes all have polymorphisms that have been investigated in asthma endophenotypes (Capra et al., 2007; Tantisira and Drazen, 2009).
The polymorphic region of the ALOX5 gene contains a functionally important variable number tandem repeat (VNTR) sequence (rs59439148). The VNTR of the SP1-binding motif of the ALOX5 promoter (In et al., 1997) is likely to determine the expression levels of ALOX5 (In et al., 1997; Silverman et al., 1998). An early study found that patients with no wild type (WT) allele at the ALOX5 promoter locus have a diminished response to treatment with the 5-LO inhibitor, ABT-761, which targets ALOX5 (Drazen et al., 1999b). Though this polymorphism may not predict asthma risk (Sayers et al., 2003), it may be a significant determinant of the leukotriene burden associated with bronchoconstriction (Holgate et al., 1996; Drazen et al., 1999a). More recently, ALOX5 polymorphisms have been associated with increased cysteinyl leukotriene production as well as reduced lung function in children with poorly controlled asthma (Mougey et al., 2013). In this study, 270 children, 6- to 17-years old, with poorly controlled asthma were enrolled in a 6-month clinical trial. A smaller cohort of 137 patients was examined in order to test the association of the ALOX5 promoter with asthma outcomes using both additive and recessive genetic models. Mougey et al. reported that between 14.8% (40/270) and 28% (38/135) of African Americans were homozygous for variant alleles of rs59439148. These results suggest that the ALOX5 promoter contributes to increased urinary LTE4 levels, reduced lung function and potentially worse asthma control. ALOX5 promoter variants may, therefore, be a risk factor for worse asthma outcomes.
The ALOX5 variants that confer susceptibility to asthma, therefore, may be of pharmacogenetic significance if an inadequate quantity of ALOX5 protein is available as a drug target (Lima et al., 2006; Telleria et al., 2008; Tantisira et al., 2009). Evidence that g.20C > T (rs4987105) and g.213411G > A (rs4986832) are associated with improved forced expiratory volume (FEV1) in response to montelukast, suggests the clinical relevance of the ALOX5 polymorphisms (Klotsman et al., 2007). These studies suggest that the 15% or so of individuals who carry these SNPs may have a heightened response to montelukast that reflects their endogenously excessive levels of CysLTs.
There is evidence that the gene encoding LTC4S, the enzyme that adducts glutathione at the C-6 position of the arachidonic acid backbone (Lam and Austen, 2000), is also polymorphic. The promoter of the LTC4S gene contains a -444A > C SNP which is transcriptionally active. However, the activity of this polymorphic promoter is reported to cause alterations in CysLT production which are associated with severe asthma (Sampson et al., 2000; Kedda et al., 2004). This relationship has been confirmed by meta-analysis (Zhang et al., 2012).
The variant may be of pharmacogenetic significance. Subjects who were homozygous for the WT LTC4S -444A allele had a lower response to the CYSLTR1 antagonist zafirlukast, compared with CC or CA genotypes (Sampson et al., 2000). Two recent studies report the profound physiological and functional significance of this polymorphism. The first study found that, compared with corn oil, a combination of borage and echium seed oils may improve airflow obstruction in mild to moderate asthmatics who carry the variant allele in the LTC4S gene (A-444C; Kazani et al., 2014). The second group that investigated urinary leukotrienes reported that leukotriene E4 levels may be higher in patients with the LTC4 synthase -444 A/C polymorphism (Kadry et al., 2014).
The efficacy of LTMs such as Montelukast are influenced my many polymorphisms in addition to -444A/C LTC4 polymorphisms. For example, the -441T/C polymorphism of the prostaglandin D2 receptor (PTGDR) gene is associated with treatment responsiveness to montelukast. In particular, when LTA release was examined by exercise challenge in 100 asthmatic children prescribed 5 mg/kg Montelukast, it was found that minor alleles of the PTGDR -441T/C and LTC4S -444A/C polymorphisms were associated with eosinophil count in atopic asthmatic children. While the LTC4S -444A/C and PTGDR -441T/C were not associated with the susceptibility for asthma, there was a significant association between responsiveness to montelukast and the PTGDR polymorphism. These pharmacogenetic data suggest that therapies that target the PTGDR may be useful for modulating the responsiveness to LTRAs (Kang et al., 2011).
These observations suggest that the genes encoding the CysLT pathway (Figure 1), may contribute additively or synergistically to altered drug responses or asthma endophenotypes. This phenomenon was previously suggested by Park et al. (2005). This group reported that the LTC4S -444A > C [c.-444A > C] gene polymorphism, in conjunction with the rare c.-819T > G, c.2078C > T or c.2534A > G alleles of the CYSLTR1 gene, was associated with aspirin intolerant asthma (Park et al., 2005). Al-Shemari et al. (2008) initially showed that SNPs of the ALOX5, CYSLTR1, and ALOX5 genes were found to be associated with allergic rhinosinusitis. However, the association was diminished upon correction for multiple testing. This may have resulted because the study lacked the power to detect polymorphisms conferring a relative risk of 2.0 or less to the phenotype: reflecting the fact that individual risk factors for complex disease can be quite small (Al-Shemari et al., 2008). However, while an association of the LTC4S -1072G/A, rs377694, and rs730012A/C haplotype with asthma was reported, no evidence of an interaction with the CYSLTR1 gene was identified (Kumar et al., 2012).
Study of genetic variability in the genes conferring response to LTMs, including those encoding the CysLT pathway (Figure 1), therefore, should be subjected to GWAS analysis since this method is well-suited to assessing the addictive or multiplicative contribution of genes complex traits.
There are two distinct genes which encode the two CysLT receptors: the cysteinyl leukotriene 1 (CYSLTR1) and 2 (CYSLTR2) receptors genes. These two receptor proteins exhibit variable tissue expression, however, both are expressed in lung macrophages and in airway smooth muscle (ASM; Metters, 1995; Lynch et al., 1999; Heise et al., 2000; Takasaki et al., 2000; Figueroa et al., 2001; Mita et al., 2001). Knock-out mouse studies have given insight into the pathobiology mediated by the CysLTs (Kanaoka et al., 2001). CYSLTR2 was revealed to have a major role in chronic inflammation associated with fibrosis (Beller et al., 2004a). By contrast, CYSLTR1 was found to be associated with acute constriction of smooth muscle and anti-inflammatory counteraction of chronic injury (Beller et al., 2004b). Biochemical studies have also shown that these two receptors are functionally distinct. Although, CYSLTR1 has its highest affinity for LTD4 (Lynch et al., 1999; Woszczek et al., 2005), CYSLTR2 binds LTC4 and LTD4 with equal affinity (Heise et al., 2000; Maekawa et al., 2001); a phenomenon that may define the specificity of the acute and chronic activity of the CysLTs.
As with other GPCR genes, variants of the cysteinyl leukotriene receptors (CysLT) include single nucleotide polymorphisms and insertion or deletions that alter GPCR expression or function (Balasubramanian et al., 2005). Early evidence of the possible significance of CysLT receptor variability was observed in mice: in which the CYSLTR1 transcript has been shown to undergo alternative splicing that alters the function of the protein (Maekawa et al., 2001). From 2001 to the present, a number of groups have been investigating the contribution of variability of the two CysLT receptors to atopic asthma and response to pharmacological interventions. Selected CysLT receptor polymorphisms are presented in Tables 2 and 3.
Human polymorphisms, including coding variants as well as has functional promotor haplotypes that either increase or decrease the expression of either receptor are presented in Tables 2 and 3. The epidemiology and in vitro studies discussed below suggest that functional receptor variants are associated with asthma endophenotypes and altered pharmacological response once environmental factors such as smoking were controlled for (Thompson et al., 2014a). The disturbances include the alteration of: ligand binding; G-protein coupling; dimerization; desensitization; receptor trafficking to the cell surface as well as receptor mRNA expression (Rana et al., 2001; Thompson et al., 2014a).
Early studies of the effect of amino acid substitutions in the CYSLTR1 and CYSLTR2 receptor genes in atopic asthma were based on a personalized medicine hypothesis (Thompson et al., 2005a, 2006, 2014b; Capra et al., 2007). The LTM class of drugs has been found to be ineffective in greater than 20% of patients (Noonan et al., 1998). We originally hypothesized that genetic polymorphisms influenced the efficacy of drugs that act at the CYSLTR1 receptor, such as the high-affinity antagonist ligands (e.g., montelukast, pranlukast, zafirlukast; Reiss et al., 1996; Grossman et al., 1997; Suissa et al., 1997; Obase et al., 2001). Further, we proposed that even though the CYSLTR2 receptor is not a current drug target, knowledge of the polymorphisms of this potential drug target maybe clinically valuable to refractory patients in the future.
We discuss these findings in the context of the body of cysteinyl leukotriene receptor pharmacogenetics that has since been established. Selected CYSLTR1 and CYSLTR2 polymorphisms are presented in Tables 2 and 3.
Four independent groups (Thompson et al., 2003, 2013) have identified significant associations between the CYSLTR2 gene loci and atopy (Fukai et al., 2004; Pillai et al., 2004; Park et al., 2005; Thompson et al., 2013). The amino acid alignment (Figure 2) shows the location of CYSLTR2 variant with respect to the transmembrane (TM) domains. The CYSLTR2 receptor is shown schematically in Figure 3.
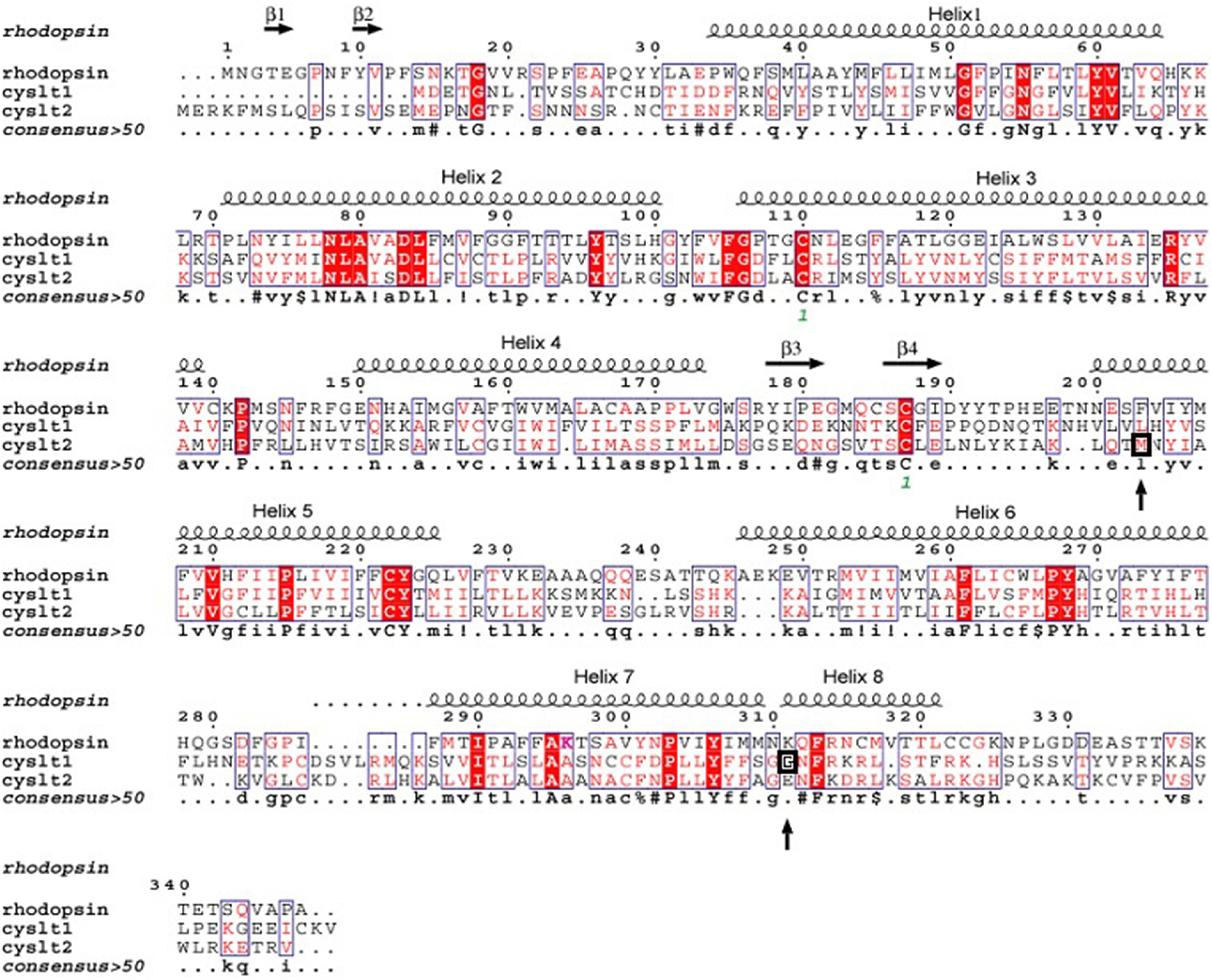
FIGURE 2. Alignment of the protein structure of the cysteinyl leukotriene 1 (CYSLTR1) and 2 (CYSLTR2) receptors in relation to rhodopsin. The amino acids conserved between these family A receptors are shown. The consensus is greater than 50%. These data formed the basis of the model predicting the CYSLTR1 and CYSLTR2 transmembrane domains (helices 1–7), the four β-sheets, and the putative cysteinyl leukotriene-binding domain. The amino acid variants that are associated with atopy or asthma, the G300S CYSLTR1variant, and the M201V CYSLTR2 variant are each boxed and noted with arrows (adapted from Thompson et al., 2014a).
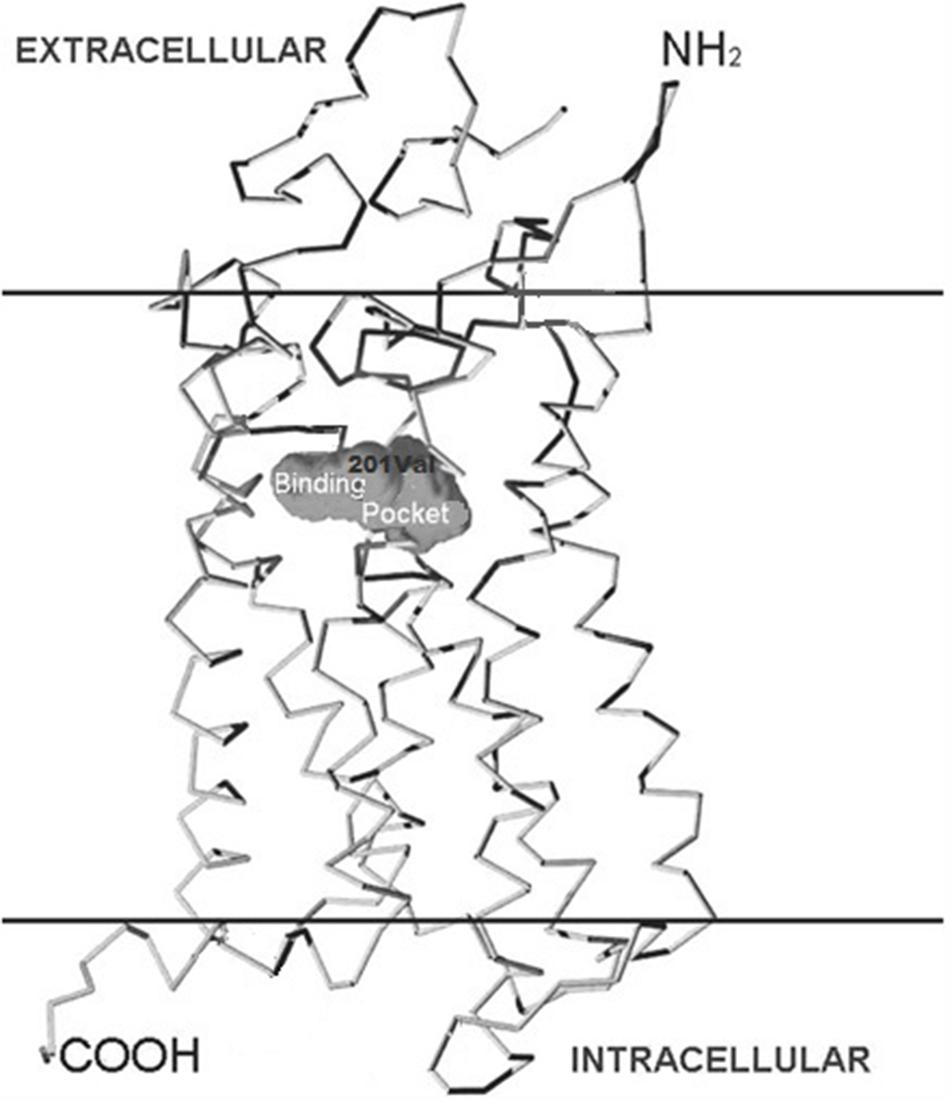
FIGURE 3. Structure of the cysteinyl leukotriene 2 (CYSLTR2) receptor and variants. The positions of the transmembrane (TM)-spanning domains of the CysLT receptor, the putative binding pocket, and the Met201Val amino acid substitution are shown in relation to the cutaway plasma membrane.
The 610 A > G variant which encodes the Met201Val CYSLTR2 receptor variant was originally identified in the Tristan da Cunha population has also been reported by Pillai et al. (2004). A less remarkable Ser236Leu variant, which was originally identified in the Tristan population (Thompson et al., 2003), may be present in as many as 10% of persons of African descent according to the Exome Aggregation Consortium, ExAC1. Though the Ser236Leu variant has not been examined in asthmatics of African origin, it has been seen compounded with Met201Val. This may be important for Tristanian asthmatics identified to be compound heterozygous for the Met201Val and Ser236Leu variants of CYSLTR2.
The Met201Val variant that was found to be associated with atopic asthma on Tristan da Cunha (Table 3) has subsequently been reported to have frequency of between 2.6% in the general population according to the Exome Aggregation Consortium (ExAc)1. The essentially inactivating character of the Met201Val the polymorphism (Thompson et al., 2003), to be discussed in below, has been confirmed by the group of Pillai et al. (2004) Their data show that Met201Val is associated with asthma phenotypes in a large population genetics study. Definitive studies using SNPs that saturate CYSLTR2 may be necessary before these findings are placed in context, however, since association studies with non-coding SNPs or haplotypes built from only a few SNPs (Kato et al., 2011; Kumar et al., 2012) are unlikely to be definitive.
The presence of SNPs in both the CYSLTR1 and CYSLTR2 genes may increase the risk for atopic asthma (Tables 2 and 3). Since the CYSLTR1is the target for anti-asthma drugs such as montelukast, the CYSLTR1 SNPs may provide an opportunity to examine the amino acid residues necessary to the structure-activity relationships that define drug action. The CYSLTR1 gene, located at Xq13-21.1, has been examined in atopic asthma (Table 2). CYSLTR1 variants, such as the 927 T > C variant (rs320995), have been associated with asthma (Hong et al., 2009) in out-bred populations. While this SNP was also noted in the Tristan da Cunha population (Thompson et al., 2013), it has not been replicated in all studies of atopic asthma (Kadry et al., 2014). A nucleotide substitution at 898G > A resulting, in vitro, in a functional Gly300Ser substitution (Thompson et al., 2007) is discussed below.
Taken together, many studies suggest that transcriptionally active CYSLTR1 variants influence the onset and severity of asthma associated endophenotypes such as atopy (Sokolowska et al., 2009). The significance of CYSLTR1 variants can be interpreted in the context of a large study of asthmatics born in Great Britain during 1958. This study, however, did not identify coding variants associated with atopy or asthma (Duroudier et al., 2009). Among those variants examined in vitro, only the 206Ser variant has been found by the Exome Aggregation Consortium1. Our ongoing GWAS study of Tristan da Cunha will ultimately resolve the questions raised by the pioneering work we undertook in this population.
We can hypothesize that epistasis involving inheritance of variants of both receptors contributes to the phenotype. The putative co-inheritance of CYSLTR1 and CYSLTR2 variants suggest a mode of asthma inheritance that is the result of epistasis. Proof of principal may be found in data showing that variants of two genes encoding other GPCR pathway proteins appear to be implicated in asthma pathogenesis. For example, it is likely that an interaction between variant forms of NPSR1 with variants of the retinoid receptor-related orphan receptor alpha (RORA) nuclear repressor gene may contribute to a variety of asthma phenotypes (Acevedo et al., 2013). Similarly, the contribution of CYSLTR1 and CYSLTR2 alleles may contribute additively or synergistically to asthma in association with other risk alleles and environmental factors.
The associations of the CYSLTR1 and CYSLTR2 genes with atopy and asthma provided the rationale for studying the functional consequences of these variants with respect to agonist and their possible contribution to the atopy phenotype (Thompson et al., 2003, 2007, 2013). By contrast, a putative high affinity LTE4 receptor (Kanaoka et al., 2013), GPR99 (also known as an oxoglutarate receptor) may not be involved since the actions of LTD4 can account for the phenotype at the cognate CYSLTR1 and CYSLTR2 receptors. The CysLT receptor CYSLTR2 variants are discussed in the context of their potential to influence LTM response.
The 601 A > G variant that is associated with atopic asthma in both the Tristan da Cunha (Thompson et al., 2003) and European populations (Pillai et al., 2004) encodes the Met201Val variant of transmembrane domain five (TMD5; Figure 3). GPCR models suggest this variant creates a mildly deleterious hypomorphic protein (Bromberg and Rost, 2007; Kazius et al., 2008; Bromberg et al., 2009; Vroling et al., 2011; Rana et al., 2012; Thompson et al., 2014a). Our work has recently confirmed the variant to be hypomorphic (Brochu-Bourque et al., 2011). The CYSLTR2 inactivating mutation results in reduced ligand binding, Ca2+ flux (Figures 4D and 5B) and inositol phosphate (IP) generation (Figure 6; Thompson et al., 2003; Pillai et al., 2004; Brochu-Bourque et al., 2011).
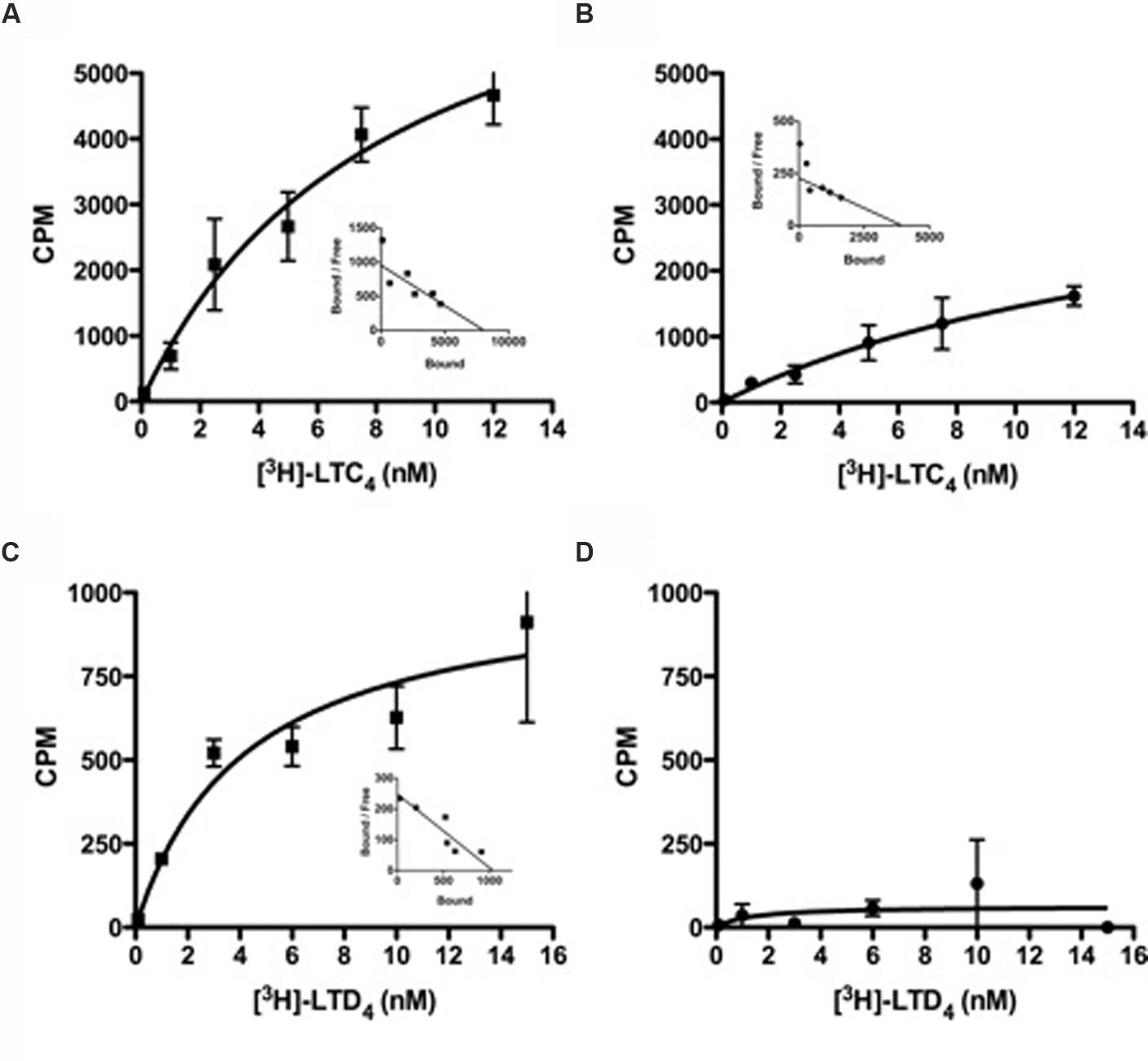
FIGURE 4. Radioligand binding studies of LTC4 or LTD4 with CYSLTR2 WT or CYSLTR2 Met201Val cells. (A,B) Saturation binding isotherms for [3H]LTC4 to CYSLTR2 WT (A) or CYSLTR2 Met201Val (B). Radioligand binding assay was performed with increasing amounts of [3H]LTC4 (0.1–12 nM) in the presence or absence of unlabeled ligand. (C,D) Saturation binding isotherms for [3H]LTD4 to CYSLTR2 WT (C) or CYSLTR2 Met201Val (D). Radioligand binding assay was performed with increasing amounts of [3H]LTD4 (0.1–15 nM) in the presence or absence of unlabeled ligand. The calculated specific binding was analyzed by non-linear transformation using Prism (Graph Pad Software, Inc.) to generate Kd and Bmax values. Saturation curves are representative of four independent experiments. Insets: Scatchard representation of the specific binding isotherm (adapted from Brochu-Bourque et al., 2011).
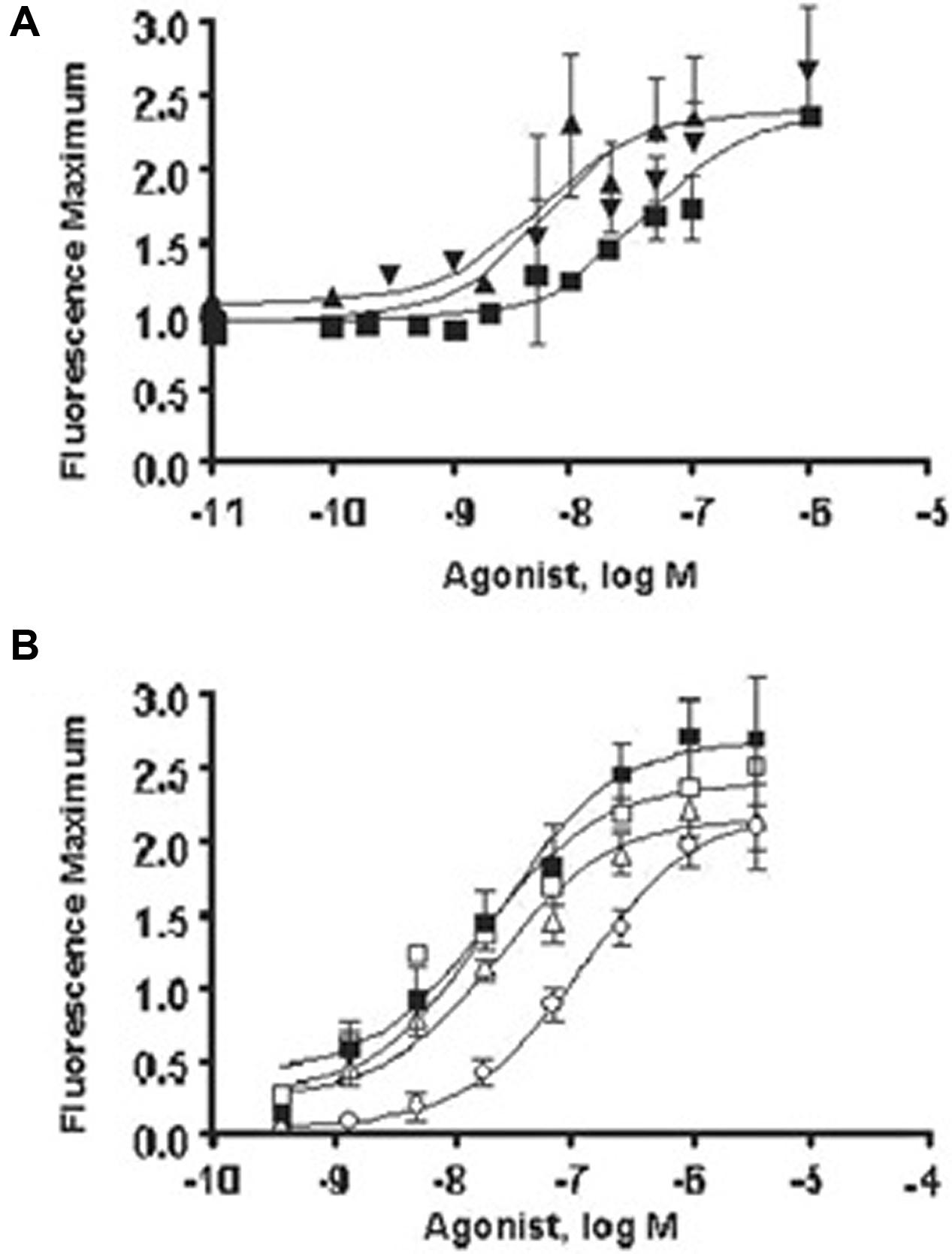
FIGURE 5. Summary of the in vitro effects of Gly300Ser cysteinyl leukotriene 1 (CYSLTR1) receptor and Met201Val on CYSLTR2 receptor signaling compared with wild type (WT). (A) Cysteinyl leukotriene D (LTD) concentration-response curve for CysLT receptors in transfected cells. Inositol triphosphate (InsP) generation assay of the variants and WT forms of the CYSLTR1 receptor. both 300Ser and 206Ser variants’ EC50 were significantly different from WT. The concentrations of LTD required to produce the InsP effect were much higher than those used in the [Ca2+] assay shown in (B), in which calcium flux was assayed for the variants and WT and variant forms of the CYSLTR1 receptor challenged with LTD. The resulting changes in intracellular calcium concentrations were measured as fluorescence maximum. For LTD4, the Met201Val variant (○) had a significantly greater EC50 compared to WT (■), while the Ser236Leu(Δ) and Ala293Gly/Arg316Lys (®) variants were not different. However, the Ala293Gly/Arg316Lys variant showed decreased efficacy (V; adapted from Thompson et al., 2014a).
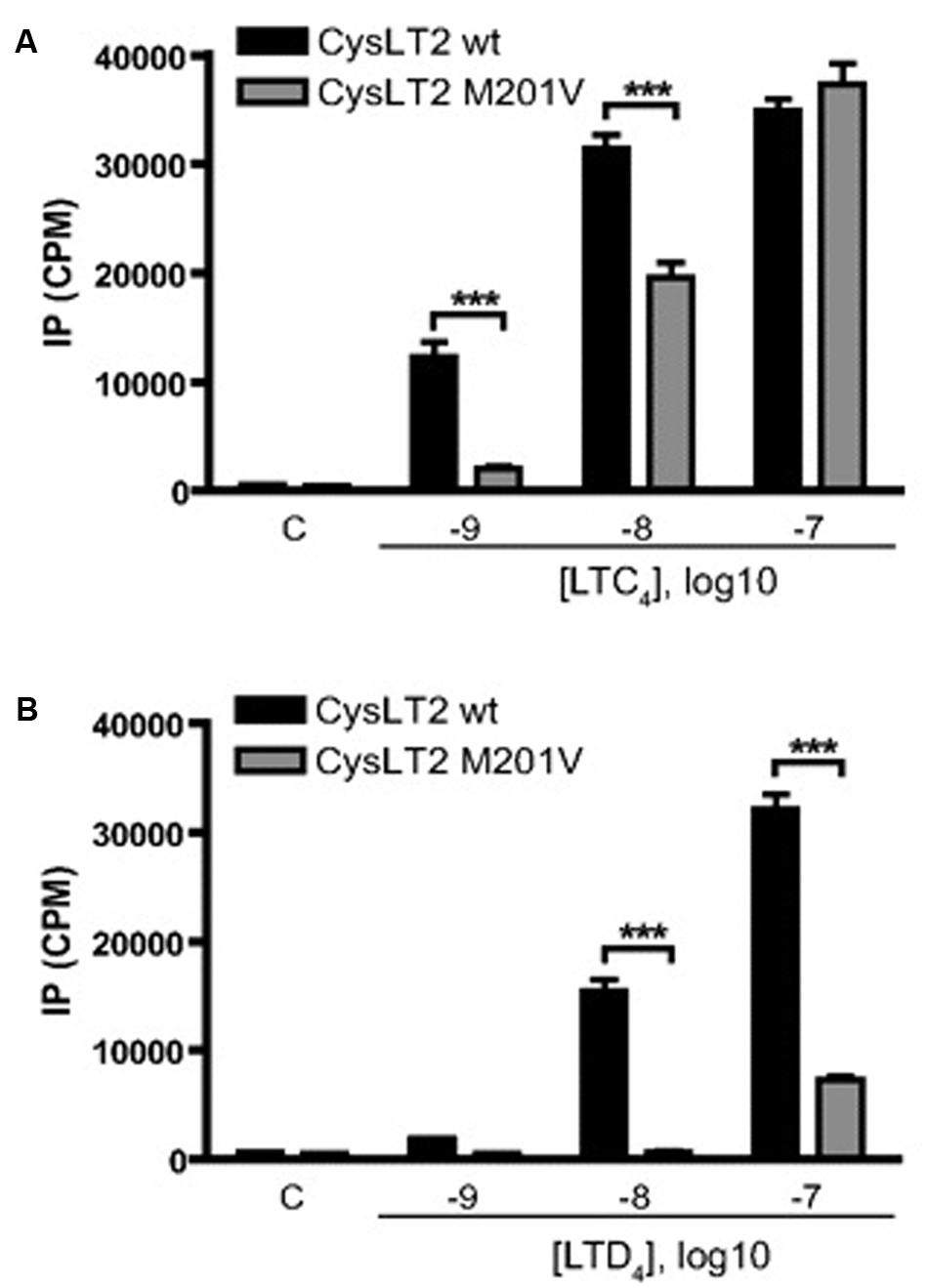
FIGURE 6. Effect of CysLTs on IP production by CYSLTR2 WT (black bars) or CYSLTR2 Met201Val (gray bars) cells. Cells were labeled with [3H]myo-inositol for 16 h, and then stimulated for 45 min with EtOH and LTC4 (A) or LTD4 (B) at the indicated concentrations before measurement of radioactivity. Results are presented as cpm of inositol phosphate production. Data are expressed as mean ± SE of 15 (A) or 9 (B) independent experiments. Lane C, control using EtOH (100 nM). ∗∗∗p < 0.001, using two-way ANOVA with Bonferroni post-tests (adapted from Brochu-Bourque et al., 2011).
In general, the effects of Met201Val were more pronounced with respect to LTD4 compared with LTC4. This effect is compounded by deficits in ligand binding that were again more significant for LTD4 than LTC4 (Figure 4). The fact that LTD4 release is more commonly associated with asthma severity, may explain why this variant is clinically relevant. The fact that the CYSLTR2 variant is both relatively common and associated with asthma in out-bred populations suggests that the variant is of relevance to the general population.
Since the CYSLTR1 Gly300Ser variant has not been reported in the general population, our functional analysis of the Gly300Ser variant will be especially informative if the CYSLTR1 gene is implicated in disease pathogenesis. Our in vitro study of the Gly300Ser variant (Figures 7 and 8; Yaddaden et al., 2016) provided among the first structural insights into CysLT receptors using site directed mutagenesis. This represents a significant advance in understanding the relatively under-studied GPCR receptors for the cysteinyl leukotrienes.
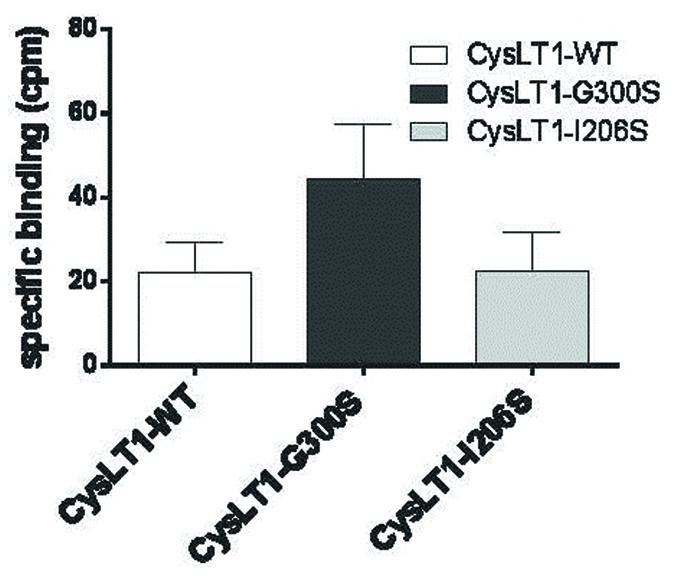
FIGURE 7. Binding of LTD4 to the CYSLTR1-WT receptor and its variants in transiently transfected HEK293 cells. Forty-eight hours post-transfection, 3H-LTD4 was added to the cells for 1 h, in the presence or absence of non-radioactive LTD4. Following several washes, the radioactivity was measured in a beta-counter (n = 3). Specific binding was calculated by subtracting the non-specific binding (in presence of competition from radioactive LTD4) from the total binding measured (Yaddaden et al., 2016).
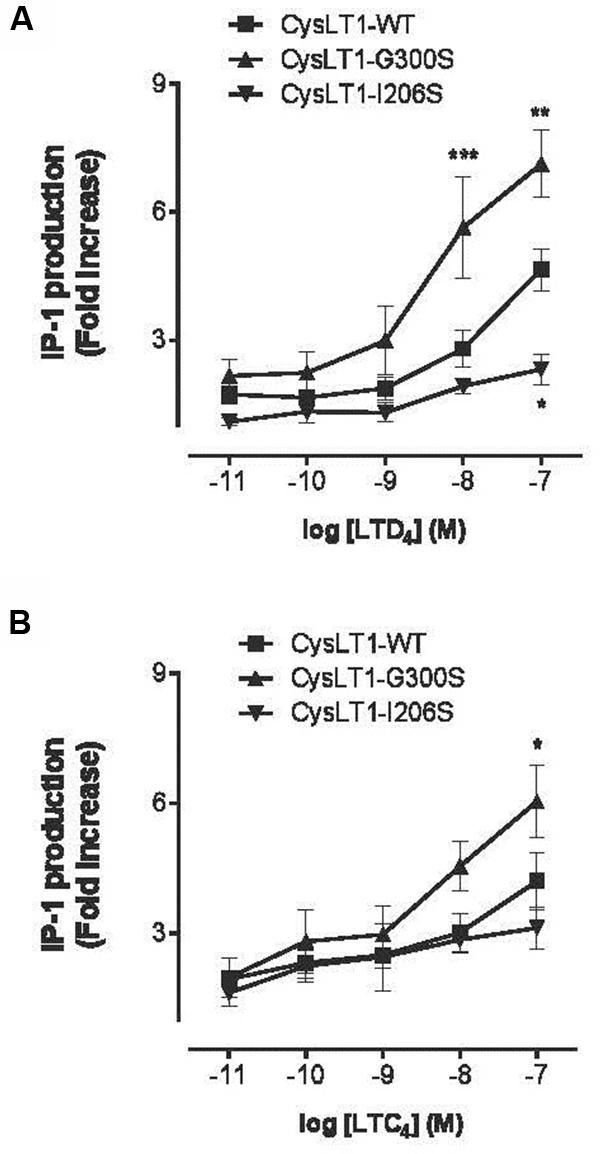
FIGURE 8. Inositol phosphate production in CYSLTR1-transfected cells. Forty-eight hours post-transfection, cells were stimulated for 30 min with LTD4 or LTC4 at 0.01–100 nM. IP-1 production was measured using an antibody against IP-1 and a fluorescent FRET IP-1 conjugate competitor. (A) IP-1 production by CYSLTR1-transfected HEK-293 cells in response to LTD4 (B) and to LTC4. Two-way ANOVA analysis; n = 6; ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001 (adapted from Yaddaden et al., 2016).
When the engineered Gly300Ser variant receptor was expressed it in COS-7 cells, the variant was more sensitive to the LTD4 agonist (Figures 7 and 8). Binding properties of the variant (measured by Bmax and Kd) indicates that the CYSLTR1 variant has higher affinity for ligand than the WT (Figure 7) by comparison with the CYSLTR2 polymorphism that has less affinity for ligand than the WT (Figure 4). By contrast, the CYSLTR1 206Ser variant, originally identified in the Tristan da Cunha population, and later found at low frequencies in atopics, asthmatics and control subjects throughout the world, was not found to be functional (Thompson et al., 2007; Yaddaden et al., 2016).
In fact, while ligand binding (Figure 7), IP generation (Figure 8) and Ca2+ flux (Figure 5A) was reduced compared to CYSLTR1 WT, the expression of the CYSLTR1 Gly300Ser variant receptors (Figure 9) and their desensitization in response to agonist was found to be normal (Thompson et al., 2007). These data suggest that CYSLTR1 CYSLTR1 antagonists, such as montelukast, may correct a putative elevation in CYSLTR1 signaling in atopic asthma.
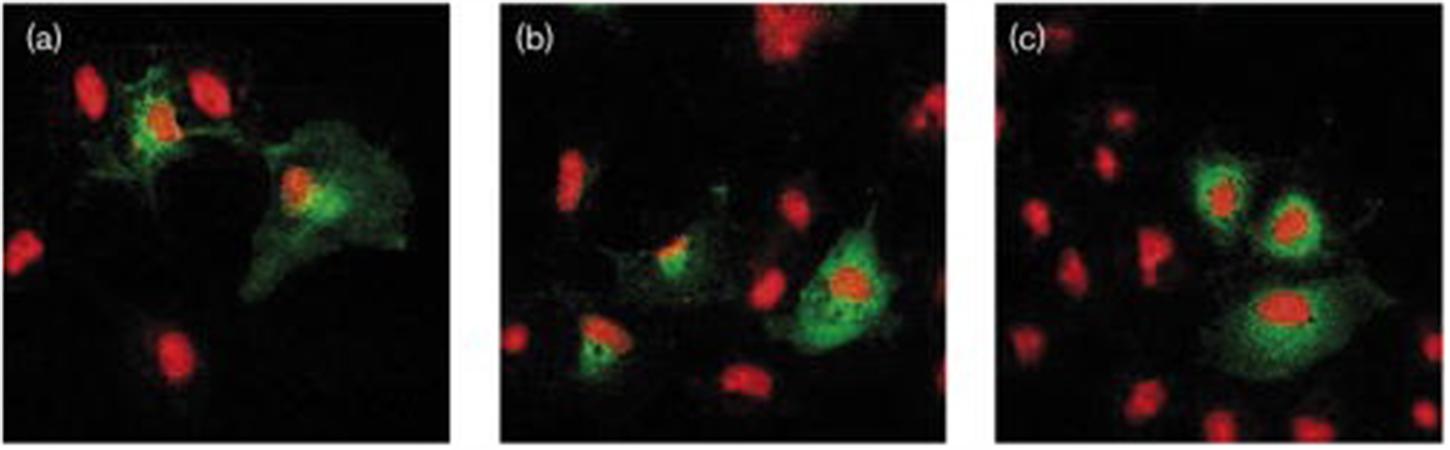
FIGURE 9. Confocal microscopy imaging of cysteinyl-leukotriene 1 (CYSLTR1) WT and variant receptors in transiently transfected cells. Cells were fixed and immunofluorescently stained with anti-CYSLTR1R antibody (green) followed by Alexa 488 goat anti-rabbit antibody (red) as described under Methods section. Superimposed images of immunoreactivity of (a) WT CYSLTR1 receptor, (b) Gly300Ser CYSLTR1 receptor variant or (c) Ile206Ser CYSLTR1 receptor variant, in basal condition. One representative experiment is shown (adapted from Thompson et al., 2007).
The relative location of variants of the receptors is shown in an alignment of each CysLT receptor (Figure 2) that was constructed with respect to the transmembrane-spanning and the putative binding pocket of the receptors. The data suggests that the putative CysLT binding site is partially determined by the integrity of the transmembrane domains. The abnormal but opposite pharmacology of variants of the receptors, causing increased potency of LTD4 at the Gly300Ser CYSLTR1 receptor variant (located in the intracellular portion of TMD7) and decreased potency of LTD4 at the Met201Val CYSLTR2 receptor variant (located in the extracellular portion of TMD5) contributes important in vitro data on the structural significance of the specific amino acid residues in these GPCRs.
It has been proposed that macrophages and eosinophils attenuate the strength of CysLT-mediated signaling by limiting the formation of CYSLTR1 receptor homodimers and/or controlling their surface expression. With respect to MAP Kinase signals, it seems possible that WT CYSLTR2 receptors interact with WT CYSLTR1 receptors to inhibit functional responses of WT CYSLTR1 receptors. In mast cells, ligand activation of the WT CYSLTR2 receptor results in reduced signaling from ligand activated CYSLTR1 WT receptors. Although, the CYSLTR1 and CYSLTR2 receptor interaction is best modeled with respect to mitogenic signals (Jiang et al., 2007), it is possible that more rapid processes such as IP generation or calcium flux may be analogous. We hypothesize that a CYSLTR1/CYSLTR2 “counterbalance” may regulate CysLT-dependent functions in immune and inflammatory responses. This balance may be disrupted by the Met201Val variant of CYSLTR2 if the LTD4 dependent signal is necessary to balance the LTD4 ligand-dependent CYSLTR1 signal. This effect may be amplified in people who also carry a CYSLTR1 variant.
The CYSLTR2 receptor may be important to the pharmacology of CYSLTR1 pharmaceuticals if, like many GPCRs, these receptors form functional heterodimers with unique pharmacological properties. Co-expression of variant receptors may alter CysLT signaling. While evidence for a functional interaction between CYSLTR1 and CYSLTR2 receptors in mast cells seems likely (Jiang et al., 2007), the functional consequence of putative heterodimers, or other functional interactions formed between variants, remains to be fully demonstrated. The fact that deletion of either the CYSLTR1 or CYSLTR2 receptors totally eliminates CysLT-mediated cutaneous edema (Lynch et al., 1999), however, suggests that a CYSLTR1/CYSLTR2 interaction may regulate some responses to CysLT mediators. It may be possible to test the hypothesis that CYSLTR2 negatively regulates CYSLTR1 signaling by examining the signaling consequences of co-expressing CYSLTR1 variants with the 201Val CYSLTR2 variant.
Central to this narrative is a discussion of how variability in the genes encoding the proteins essential for CysLT signaling may influence risk for inflammation that is associated with asthma, cognizant of the fact that variability these genes may also increase susceptibility to asthma independent of their potential LTM pharmacogenetic relevance. Asthma susceptibility loci, which confer risk for the complex respiratory traits that lead to asthma, and pharmacogenetic loci, which confer the traits that define the responsiveness of an individual to pharmaceutical therapy, are presented in the context of the linkage analysis, candidate gene studies and/or GWAS approaches used to identify them. In most cases, these loci can be modeled as independent complex traits with both genetic and environmental (GXE) contributions. Therefore, we review the relative contribution of genes in the CysLT pathway to both the etiology of asthma itself and a person’s responsiveness to LTM drugs.
Common genetic variants (e.g., SNPs) typically contribute effects of as little as 1% to a complex phenotype described in a given population. As a result, GWAS can be used to confirm the significance of the cysteinyl leukotriene gene variants to atopic asthma on Tristan da Cunha. While it is possible that affected Tristanians may inherit a variety of other genetic risk factors for atopy and/or asthma, including potential ETS-2 and ETS-3 variability (Baron et al., 2002; Silverman et al., 2002; Wu et al., 2008), we present evidence that suggests that the CYSLTR2 601 A > G variant is associated with atopic asthma. The contribution of other known atopy variants, such as those in the DENND1B gene, located on chromosome 1q31 (Sleiman et al., 2010) and the ORMDL3 gene, located on chromosome 17q12 (Moffatt et al., 2007) to the asthma phenotype is also possible. Like other asthma susceptibility loci, the CYSLTR2 601 A > G variant has been shown to contribute to asthma in large out-bred populations (Pillai et al., 2004).
While the CYSLTR2 variant was originally identified in 13% of the Tristan da Cunha population (Thompson et al., 2003) it is only present in approximately 2.6% in the European population. In both instances, the variant has been associated with atopic asthma (Thompson et al., 2003; Pillai et al., 2004). A large study of CYSLTR1 variants in asthmatics born in Great Britain during 1958 did not identify the 898 G > A variant (Duroudier et al., 2009) nor is it found in over 60,000 exomes by the Exome Aggregation Consortium1. The 898 G > A CYSLTR1 is, however, among the best studied coding variants of a receptor for any LTM drug.
By contrast with asthma susceptibility loci, in most cases the pharmacogenetic determinants of drug response represent a distinct category of risk loci that may not influence disease onset directly. For example, the pharmacogenetic variability that influences LTM response can be attributable to genes that are both integral (Kang et al., 2011; Mougey et al., 2013; Figure 1), or lie outside (Dahlin et al., 2015, 2016) CysLT signaling pathways. In this context, CysLT receptor variants have provided insight into the role of GPCR variants as genetic risk factors for disease, altered drug response, or ADRs (see Table 1) and may represent reagents necessary for refined LTM drug discovery and/or personalized medical interventions (Spiegel and Weinstein, 2004). The significance of the CysLT receptor variants to both the development of asthma and LTM response, however, is best evaluated in the context of other polymorphisms that influence the phenotype. For example, the enzymes involved in the formation of the leukotrienes (Figure 1) often result in variable production of the CysLTs (LTC4, LTD4, and LTE4) and LTB4. The many examples discussed include, the VNTR sequences located in the Sp1-binding motif within the promotor region of the ALOX5 gene that are associated with both LTM response, leukotriene burden and bronchoconstriction (Drazen et al., 1999a), and the 444A > C SNP polymorphism in the LTC4S gene that is associated with severe asthma, leukotriene burden and altered response to the CYSLTR1 LTMs (Sampson et al., 2000; Kadry et al., 2014; Kazani et al., 2014). Genetic variability in CysLT pathway may contribute additively or synergistically to altered drug responses.
Since very large GWAS studies are needed to detect polymorphisms conferring a relative risk of 2.0 or less to the phenotype (Al-Shemari et al., 2008), results suggesting that CTSLT1R variants may interact with other polymorphic variants can be difficult to replicate (Kumar et al., 2012). This may reflect the fact that LTM targets can be quite diverse. For example, the fact that PTGDR polymorphism has been significantly associated with responsiveness to montelukast may confound earlier studies of LTM pharmacogenetics. Indeed, LTM therapies may be developed that selectively target the PTGDR since this receptor may be a useful way to modulate responsiveness to LTRA (Kang et al., 2011).
Many genes that are associated with altered efficacy of LTMs pharmaceuticals, however, are not directly part of the CysLT pathway. For example, variation in the SLCO2B1 gene has associated with altered absorption of montelukast administered in the presence of citrus. These data suggest that genetic factors influence pharmacokinestics of LTM response (Mougey et al., 2011).
GWAS studies of LTM response in people enrolled in placebo-controlled trials have also been undertaken. Dahlin et al. (2015, 2016) examined LTM response using DNA and phenotypic information from placebo-controlled trials from the American Lung Association’s Asthma Clinical Research Center (ALA-ACRC) cohorts. SNPs that both favored [rs6475448, located in the myeloid/lymphoid or mixed-lineage leukemia (MLLT3) gene; Dahlin et al., 2015], or worsened [rs12436663, located in the Mitochondrial Ribonuclease P Protein 3 Precursor (MRPP3) gene], rs517020 [located in the glycosyltransferase 1 domain containing 1 (GLT1D1) gene] LTM response have been identified (Dahlin et al., 2016). These findings implicate loci in LTM response that, by contrast with CysLT receptor genes or the biosynthesis genes (Figure 1), are not intuitively related to LTM therapeutic responsiveness (Dahlin et al., 2015, 2016).
As a result, it is necessary that a GWAS approach is used to confirm the significance of the putative contribution of cysteinyl leukotriene gene variants to asthma and/or drug response in the context of other asthma susceptibility genes and genes that influence LTM pharmacogenetics. This strategy might we reveal novel genotype–phenotype correlations; as we did for the melanocortin 1 receptor (MC1R) variants (Beaumont et al., 2007).
With respect to Tristan da Cunha, this analysis is likely to identify variety of other genetic risk factors for atopic asthma and/or LTM drug response. These include loci associated with asthma in other populations such as the DENND1B gene, located on chromosome 1q31 (Sleiman et al., 2010). Exclusion of the ORMDL3 gene, located on chromosome 17q12 (Moffatt et al., 2007), may also be possible. In addition, loci already implicated in the Tristan phenotype such as ETS-2 and ETS-3 (Baron et al., 2002; Silverman et al., 2002; Wu et al., 2008) and the better documented CYSLTR2, can be evaluated in the context of those loci, such as CYSLTR1, that may have pharmacogenetic significance.
All authors listed, have made substantial, direct and intellectual contribution to the work, and approved it for publication.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Funding was provided by The Scottish Rite Charitable Foundation of Canada; The Canadian Institutes of Health Research (CIHR); The National Science and Engineering Research Council (NSERC) and an Italy-Canada International Scientific Cooperation Grant (ID 16755). We thank Saint George’s University School of Medicine for supporting this study.
Acevedo, N., Sääf, A., Söderhäll, C., Melén, E., Mandelin, J., Pietras, C. O., et al. (2013). Interaction between retinoid acid receptor-related orphan receptor alpha (RORA) and neuropeptide S receptor 1 (NPSR1) in asthma. PLoS ONE 8:e60111. doi: 10.1371/journal.pone.0060111
Agrawal, D. K., and Bharadwaj, A. (2005). Allergic airway inflammation. Curr. Allergy Asthma Rep. 5, 142–148. doi: 10.1007/s11882-005-0088-7
Al-Shemari, H., Bossé, Y., Hudson, T. J., Cabaluna, M., Duval, M., Lemire, M., et al. (2008). Influence of leukotriene gene polymorphisms on chronic rhinosinusitis. BMC Med. Genet. 26:21. doi: 10.1186/1471-2350-9-21
Arriba-Mendez, S., Sanz, C., Isidoro-Garcia, M., Davild, I., Laffond, E., Horeno, E., et al. (2006). 927T>C polymorphism of the cysteinyl-leukotriene type-1 receptor (CYSLTR1) gene in children with asthma and atopic dermatitis. Pediatr. Allergy Immunol. 17, 323–328. doi: 10.1111/j.1399-3038.2006.00416.x
Balasubramanian, S., Xia, Y., Freinkman, E., and Gerstein, M. (2005). Sequence variation in G-protein-coupled receptors: analysis of single nucleotide polymorphisms. Nucleic Acids Res. 33, 1710–1721. doi: 10.1093/nar/gki311
Baron, R. M., Palmer, L. J., Tantisira, K., Gabriel, S., Sonna, L. A., Le, L., et al. (2002). DNA sequence variants in epithelium-specific ETS-2 and ETS-3 are not associated with asthma. Am. J. Respir. Crit. Care. Med. 166, 927–932. doi: 10.1164/rccm.200201-048OC
Basu, K., Palmer, C. N., Tavendale, R., Lipworth, B. J., and Mukhopadhyay, S. (2009). Adrenergic beta(2)-receptor genotype predisposes to exacerbations in steroid-treated asthmatic patients taking frequent albuterol or salmeterol. J. Allergy Clin. Immunol. 124:1188-94.e3. doi: 10.1016/j.jaci.2009.07.043
Beaumont, K. A., Shekar, S. N., Newton, R. A., James, M. R., Stow, J. L., Duffy, D. L., et al. (2007). Receptor function, dominant negative activity and phenotype correlations for MC1R variant alleles. Hum. Mol. Genet. 16, 2249–2260. doi: 10.1093/hmg/ddm177
Beller, T. C., Friend, D. S., Maekawa, A., Lam, B. K., Austen, K. F., and Kanaoka, Y. (2004a). Cysteinyl leukotriene 1 receptor controls the severity of chronic pulmonary inflammation and fibrosis. Proc. Natl. Acad. Sci. U.S.A. 101, 3047–3052. doi: 10.1073/pnas.0400235101
Beller, T. C., Maekawa, A., Friend, D. S., Austen, K. F., and Kanaoka, Y. (2004b). Targeted gene disruption reveals the role of the cysteinyl leukotriene 2 receptor in increased vascular permeability and in a bleomycin-induced pulmonary fibrosis in mice. J. Biol. Chem. 279, 46129–46134. doi: 10.1074/jbc.M407057200
Berce, V., Kozmus, C. E., and Potočnik, U. (2013). Association among ORMDL3 gene expression, 17q21 polymorphism and response to treatment with inhaled corticosteroids in children with asthma. Pharmacogenomics J. 13, 523–529. doi: 10.1038/tpj.2012.36
Bisgaard, H. (2001). Pathophysiology of the cysteinyl leukotrienes and effects of leukotriene receptor antagonists in asthma. Allergy 56, 7–11. doi: 10.1034/j.1398-9995.56.s66.2.x
Brochu-Bourque, A., Véronneau, S., Rola-Pleszczynski, M., and Stankova, J. (2011). Differential signaling defects associated with the M201V polymorphism in the cysteinyl leukotriene type 2 receptor. J. Pharmacol. Exp. Ther. 336, 431–439. doi: 10.1124/jpet.110.172411
Bromberg, Y., Overton, J., Vaisse, C., Leibel, R. L., and Rost, B. (2009). In silico mutagenesis: a case study of the melanocortin 4 receptor. FASEB J. 23, 3059–3069. doi: 10.1096/fj.08-127530
Bromberg, Y., and Rost, B. (2007). SNAP: predict effect of non-synonymous polymorphisms on function. Nucleic Acids Res. 35, 3823–3835. doi: 10.1093/nar/gkm238
Camacho, A., Ballesteros, S., Graham, A. L., Carrat, F., Ratmann, O., and Cazelles, B. (2011). Explaining rapid reinfections in multiple-wave influenza outbreaks: Tristan da Cunha 1971 epidemic as a case study. Proc. Biol. Sci. 278, 3635–3643. doi: 10.1098/rspb.2011.0300
Camacho, A., and Cazelles, B. (2013). Does homologous reinfection drive multiple-wave influenza outbreaks? Accounting for immunodynamics in epidemiological models. Epidemics 5, 187–196. doi: 10.1016/j.epidem.2013.09.003
Capra, V., Thompson, M. D., Sala, A., Cole, D. E., Folco, G., and Rovati, G. E. (2007). Cysteinyl-leukotrienes and their receptors in asthma and other inflammatory diseases: critical update and emerging trends. Med. Res. Rev. 27, 469–527. doi: 10.1002/med.20071
Chan-Yeung, M., McClean, P. A., Sandell, P. R., Slutsky, A. S., and Zamel, N. (1999). Sensitization to cat without direct exposure to cats. Clin. Exp. Allergy 29, 762–765. doi: 10.1046/j.1365-2222.1999.00597.x
Choi, J. H., Park, H. S., Oh, H. B., Lee, J. H., Suh, Y. J., Park, C. S., et al. (2004). Leukotriene-related gene polymorphisms in ASA-intolerant asthma: an association with a haplotype of 5-lipoxygenase. Hum. Genet. 114, 337–344. doi: 10.1007/s00439-004-1082-1
Cookson, W., and Moffatt, M. (2004). Making sense of asthma genes. N. Engl. J. Med. 351, 1794–1796. doi: 10.1056/NEJMe048232
Dahlin, A., Litonjua, A., Irvin, C. G., Peters, S. P., Lima, J. J., Kubo, M., et al. (2016). Genome-wide association study of leukotriene modifier response in asthma. Pharmacogenomics J. 16, 151–157. doi: 10.1038/tpj.2015.34
Dahlin, A., Litonjua, A., Lima, J. J., Tamari, M., Kubo, M., Irvin, C. G., et al. (2015). Genome-wide association study identifies novel pharmacogenomic loci for therapeutic response to montelukast in asthma. PLoS ONE 10:e0129385. doi: 10.1371/journal.pone.0129385
Dewar, J. C., Wilkinson, J., Wheatley, A., Thomas, N. S., Doull, I., Morton, N., et al. (1997). The glutamine 27 beta(2)-adrenoceptor polymorphism is associated with elevated IgE levels in asthmatic families. J. Allergy Clin. Immunol. 100, 261–265. doi: 10.1016/S0091-6749(97)70234-3
Drazen, J. M., Israel, E., and O’Byrne, P. M. (1999a). Treatment of asthma with drugs modifying the leukotriene pathway. N. Engl. J. Med. 340, 197–206. doi: 10.1056/NEJM199901213400306
Drazen, J. M., Silverman, E. K., and Lee, T. H. (2000). Heterogeneity of therapeutic responses in asthma. Br. Med. Bull. 56, 1054–1070.
Drazen, J. M., Yandava, C. N., Dubé, L., Szczerback, N., Hippensteel, R., Pillari, A., et al. (1999b). Pharmacogenetic association between ALOX5 promoter genotype and the response to anti-asthma treatment. Nat. Genet. 22, 168–170. doi: 10.1038/9680
Drysdale, C. M., McGraw, D. W., Stack, C. B., Stephens, J. C., Judson, R. S., Nandabalan, K., et al. (2000). Complex promoter and coding region beta(2)-adrenergic receptor haplotypes alter receptor expression and predict in vivo responsiveness. Proc. Natl. Acad. Sci. U.S.A. 97, 10483–10488. doi: 10.1073/pnas.97.19.10483
Duroudier, N. P., Strachan, D. P., Blakey, J. D., and Hall, I. P. (2009). Association of the cysteinyl leukotriene receptor 1 gene with atopy in the British 1958 birth cohort. J. Allergy Clin. Immunol. 124, 566–572. doi: 10.1016/j.jaci.2009.06.004
Eder, W., Ege, M. J., and von Mutius, E. (2006). The asthma epidemic. N. Engl. J. Med. 355, 2226–2235. doi: 10.1056/NEJMra054308
Ferreira, M. A., McRae, A. F., Medland, S. E., Nyholt, D. R., Gordon, S. D., Wright, M. J., et al. (2011). Association between ORMDL3, IL1RL1 and a deletion on chromosome 17q21 with asthma risk in Australia. Eur. J. Hum. Genet. 19, 458–464. doi: 10.1038/ejhg.2010.191
Figueroa, D. J., Breyer, R. M., Defoe, S. K., Kargman, S., Daugherty, B. L., Waldburger, K., et al. (2001). Expression of the cysteinyl leukotriene 1 receptor in normal human lung and peripheral blood leukocytes. Am. J. Respir. Crit. Care Med. 163, 226–233. doi: 10.1164/ajrccm.163.1.2003101
Fukai, H., Ogasawara, Y., Migita, O., Koga, M., Ichikawa, K., Shibasaki, M., et al. (2004). Association between a polymorphism in cysteinyl leukotriene receptor 2 on chromosome 13q14 and atopic asthma. Pharmacogenetics 14, 683–690. doi: 10.1097/00008571-200410000-00006
Grossman, J., Faiferman, I., Dubb, J. W., Tompson, D. J., Busse, W., Bronsky, E., et al. (1997). Results of the first U.S. double-blind, placebo-controlled, multicenter clinical study in asthma with pranlukast, a novel leukotriene receptor antagonist. J. Asthma 34, 321–328. doi: 10.3109/02770909709067222
Hall, I. P., Wheatley, A., Christie, G., McDougall, C., Hubbard, R., and Helms, P. J. (1999). Association of CCR5 del 32 with reduced risk of asthma. Lancet 354, 1264–1265. doi: 10.1016/S0140-6736(99)03425-X
Hao, L., Sayers, I., Cakebread, J. A., Barton, S. J., Beghé, B., Holgate, S. T., et al. (2006). The cysteinyl-leukotriene type 1 receptor polymorphism 927T/C is associated with atopy severity but not with asthma. Clin. Exp. Allergy 36, 735–741. doi: 10.1111/j.1365-2222.2006.02511.x
Hawkins, G. A., Weiss, S. T., and Bleecker, E. R. (2008). Clinical consequences of ADRbeta2 polymorphisms. Pharmacogenomics 9, 349–358. doi: 10.2217/14622416.9.3.349
Heise, C. E., O’Dowd, B. F., Figueroa, D. J., Sawyer, N., Nguyen, T., Im, D. S., et al. (2000). Characterization of the human cysteinyl leukotriene 2 receptor. J. Biol. Chem. 275, 30531–30536. doi: 10.1074/jbc.M003490200
Hirai, H., Tanaka, K., Yoshie, O., Ogawa, K., Kenmotsu, K., Takamori, Y., et al. (2001). Prostaglandin D2 selectively induces chemotaxis in T helper type 2 cells, eosinophils, and basophils via seven-transmembrane receptor CRTH2. J. Exp. Med. 193, 255–261. doi: 10.1084/jem.193.2.255
Holgate, S. T., Bradding, P., and Sampson, A. P. (1996). Leukotriene antagonists and synthesis inhibitors: new directions in asthma therapy. J. Allergy Clin. Immunol. 98, 1–13. doi: 10.1016/S0091-6749(96)70220-8
Hong, X., Zhou, H., Tsai, H. J., Wang, X., Liu, X., Wang, B., et al. (2009). Cysteinyl leukotriene receptor 1 gene variation and risk of asthma. Eur. Respir. J. 33, 42–48. doi: 10.1183/09031936.00057708
In, K. H., Asano, K., Beier, D., Grobholz, J., Finn, P. W., Silverman, E. K., et al. (1997). Naturally occurring mutations in the human 5-lipoxygenase gene promoter that modify transcription factor binding and reporter gene transcription. J. Clin. Invest. 99, 1130–1137. doi: 10.1172/JCI119241
Israel, E., Drazen, J. M., Liggett, S. B., Boushey, H. A., Cherniack, R. M., Chinchilli, V. M., et al. (2000). The effect of polymorphisms of the beta(2)-adrenergic receptor on the response to regular use of albuterol in asthma. Am. J. Respir. Crit. Care Med. 162, 75–80. doi: 10.1164/ajrccm.162.1.9907092
Jiang, Y., Borrelli, L. A., Kanaoka, Y., Bacskai, B. J., and Boyce, J. A. (2007). CysLT2 receptors interact with CysLT1 receptors and down-modulate cysteinyl leukotriene dependent mitogenic responses of mast cells. Blood 110, 3263–3270. doi: 10.1182/blood-2007-07-100453
Kadry, H. M., Atta, A. H., Sultan, M. K., and El-Baz, N. A. (2014). Leukotriene C4 synthase and leukotriene receptor-1 genes polymorphism among atopic asthmatic patients. Am. J. Immunol. 10, 63–72. doi: 10.3844/ajisp.2014.63.72
Kanaoka, Y., Maekawa, A., and Austen, K. F. (2013). Identification of GPR99 protein as a potential third cysteinyl leukotriene receptor with a preference for leukotriene E4 ligand. J. Biol. Chem. 288, 10967–10972. doi: 10.1074/jbc.C113.453704
Kanaoka, Y., Maekawa, A., Penrose, J. F., Austen, K. F., and Lam, B. K. (2001). Attenuated zymosan-induced peritoneal vascular permeability and IgE dependent passive cutaneous anaphylaxis in mice lacking leukotriene C-4 synthase. J. Biol. Chem. 276, 22608–22613. doi: 10.1074/jbc.M103562200
Kang, M. J., Kwon, J. W., Kim, B. J., Yu, J., Choi, W. A., Shin, Y. J., et al. (2011). Polymorphisms of the PTGDR and LTC4S influence responsiveness to leukotriene receptor antagonists in Korean children with asthma. J. Hum. Genet. 56, 284–289. doi: 10.1038/jhg.2011.3
Kato, T., Saeki, H., Tsunemi, Y., Shibata, S., Sekiya, T., Nakamura, K., et al. (2011). Cysteinyl leukotriene receptor 2 gene polymorphism -1220 A/C is not associated with atopic dermatitis or psoriasis vulgaris in Japanese patients. J. Dermatol. 38, 497–499. doi: 10.1111/j.1346-8138.2010.01001.x
Kazani, S., Arm, J. P., Boyce, J., Chhay, H., Dutile, S., Wechsler, M. E., et al. (2014). LTC4 synthase polymorphism modifies efficacy of botanical seed oil combination in asthma. Springerplus 3, 661. doi: 10.1186/2193-1801-3-661
Kazius, J., Wurdinger, K., van Iterson, M., Kok, J., Bäck, T., and Ijzerman, A. P. (2008). GPCR NaVa database: natural variants in human G protein-coupled receptors. Hum. Mutat. 29, 39–44. doi: 10.1002/humu.20638
Kedda, M. A., Shi, J., Duffy, D., Phelps, S., Yang, I., O’Hara, K., et al. (2004). Characterization of two polymorphisms in the leukotriene C4 synthase gene in an Australian population of subjects with mild, moderate, and severe asthma. J. Allergy Clin. Immunol. 113, 889–895. doi: 10.1016/j.jaci.2004.02.008
Kim, S. H., Choi, J. H., Park, H. S., Holloway, J. W., Lee, S. K., Park, C. S., et al. (2005). Association of thromboxane A receptor gene polymorphism with the phenotype of acetyl salicylic acid-intolerant asthma. Clin. Exp. Allergy 35, 585–590. doi: 10.1111/j.1365-2222.2005.02220.x
Kim, S. H., Oh, J. M., Kim, Y. S., Palmer, L. J., Suh, C. H., Nahm, D. H., et al. (2006). Cysteinyl leukotriene receptor 1 promoter polymorphism is associated with aspirin-intolerant asthma in males. Clin. Exp. Allergy 36, 433–439. doi: 10.1111/j.1365-2222.2006.02457.x
Kim, S. H., Yang, E. M., Park, H. J., Ye, Y. M., Lee, H. Y., and Park, H. S. (2007). Differential contribution of the CysLTR1 in patients with aspirin hypersensitivity. J. Clin. Immunol. 27, 613–619. doi: 10.1007/s10875-007-9115-x
Klotsman, M., York, T. P., Pillai, S. G., Vargas-Irwin, C., Sharma, S. S., van den Oord, E. J., et al. (2007). Pharmacogenetics of the 5-lipoxygenase biosynthetic pathway and variable clinical response to montelukast. Pharmacogenet. Genomics 17, 189–196. doi: 10.1097/FPC.0b013e3280120043
Kormann, M. S., Carr, D., Klopp, N., Illig, T., Leupold, W., Fritzsch, C., et al. (2005). G-protein-coupled receptor polymorphisms are associated with asthma in a large German population. Am. J. Respir. Crit. Care Med. 171, 1358–1362. doi: 10.1164/rccm.200410-1312OC
Kumar, A., Sharma, S., Agrawal, A., and Ghosh, B. (2012). Association of the -1072G/A polymorphism in the LTC4S gene with asthma in an Indian population. Int. Arch. Allergy Immunol. 159, 271–277. doi: 10.1159/000336675
Lachance, M., Ethier, N., Wolbring, G., Schnetkamp, P. P., and Hébert, T. E. (1999). Stable association of G proteins with β2AR is independent of the state of receptor activation. Cell Signal. 11, 523–533. doi: 10.1016/S0898-6568(99)00024-8
Laitinen, T., Polvi, A., Rydman, P., Vendelin, J., Pulkkinen, V., Salmikangas, P., et al. (2004). Characterization of a common susceptibility locus for asthma-related traits. Science 304, 300–304. doi: 10.1126/science.1090010
Lam, B. K., and Austen, K. F. (2000). Leukotriene C-4 synthase: a pivotal enzyme in the biosynthe- sis of the cysteinyl leukotrienes. Am. J. Respir. Crit. Care Med. 161, S16–S19. doi: 10.1164/ajrccm.161.supplement_1.ltta-4
Lefkowitz, R. J. (2004). Historical review: a brief history and personal retrospective of seven-transmembrane receptors. Trends. Pharmacol. Sci. 25, 413–422. doi: 10.1016/j.tips.2004.06.006
Levack, P. A., and Levack, I. D. (2013). Medical practice on Tristan da Cunha–the remotest island community in the world. J. R. Coll. Physicians Edinb. 43, 290–293. doi: 10.4997/JRCPE.2013.402
Liggett, S. B. (1997). Polymorphisms of the beta2-adrenergic receptor and asthma. Am. J. Respir. Crit. Care Med. 156(4 Pt. 2), S156–S162. doi: 10.1164/ajrccm.156.4.12tac-15
Lima, J. J., Zhang, S., Grant, A., Shao, L., Tantisira, K. G., Allayee, H., et al. (2006). Influence of leukotriene pathway polymorphisms on response to montelukast in asthma. Am. J. Respir. Crit. Care Med. 173, 379–385. doi: 10.1164/rccm.200509-1412OC
Littlejohn, M. D., Taylor, D. R., Miller, A. L., and Kennedy, M. A. (2002). Determination of beta2-adrenergic receptor (ADRB2) haplotypes by a multiplexed polymerase chain reaction assay. Hum. Mutat. 2:479. doi: 10.1002/humu.9091
Lynch, K. R., O’Neill, G. P., Liu, Q., Im, D. S., Sawyer, N., Metters, K. M., et al. (1999). Characterization of the human cysteinyl leukotriene CysLT1 receptor. Nature 399, 789–793. doi: 10.1038/21658
Maekawa, A., Austen, K. F., and Kanaoka, Y. (2002). Targeted gene disruption reveals the role of cysteinyl leukotriene 1 receptor in the enhanced vascular permeability of mice undergoing acute inflammatory responses. J. Biol. Chem. 277, 20820–20824. doi: 10.1074/jbc.M203163200
Maekawa, A., Kanaoka, Y., Lam, B. K., and Austen, K. F. (2001). Identification in mice of two isoforms of the cysteinyl leukotriene 1 receptor that result from alternative splicing. Proc. Natl. Acad. Sci. U.S.A. 98, 2256–2261. doi: 10.1073/pnas.041624398
Mantle, J., and Tyrrell, D. A. (1973). An epidemic of influenza on Tristan da Cunha. J. Hyg. (Lond). 71, 89–95. doi: 10.1017/S0022172400046246
Mao, X. Q., Gao, P. S., Roberts, M. H., Enomoto, T., Kawai, M., Sasaki, S., et al. (1999). Variants of endothelin-1 and its receptors in atopic asthma. Biochem. Biophys. Res. Commun. 262, 259–262. doi: 10.1006/bbrc.1999.1158
Marshall, S. L., Guennel, T., Kohler, J., Man, M., and Fossceco, S. (2013). Estimating heritability in pharmacogenetic studies. Pharmacogenomics 14, 369–377. doi: 10.2217/pgs.13.20
Mathias, R. A., Grant, A. V., Rafaels, N., Hand, T., Gao, L., Vergara, C., et al. (2010). A genome-wide association study on African-ancestry populations for asthma. J. Allergy Clin. Immunol. 125, 336–346. doi: 10.1016/j.jaci.2009.08.031
Melén, E., Bruce, S., Doekes, G., Kabesch, M., Laitinen, T., Lauener, R., et al. (2005). Haplotypes of G protein-coupled receptor 154 are associated with childhood allergy and asthma. Am. J. Respir. Crit. Care Med. 171, 1089–1095. doi: 10.1164/rccm.200410-1317OC
Melén, E., Granell, R., Kogevinas, M., Strachan, D., Gonzalez, J. R., Wjst, M., et al. (2013). Genome-wide association study of body mass index in 23 000 individuals with and without asthma. Clin. Exp. Allergy 43, 463–474. doi: 10.1111/cea.12054
Metters, K. M. (1995). Leukotriene receptors. J. Lipid Mediat. Cell Signal. 12, 413–427. doi: 10.1016/0929-7855(95)00027-N
Michel, S., Liang, L., Depner, M., Klopp, N., Ruether, A., Kumar, A., et al. (2010). Unifying candidate gene and GWAS approaches in asthma. PLoS ONE 12:e13894. doi: 10.1371/journal.pone.0013894
Milligan, G. (2002). Strategies to identify ligands for orphan G-protein-coupled receptors. Biochem. Soc. Trans. 30, 789–793. doi: 10.1042/bst0300789
Mita, H., Hasegawa, M., Saito, H., and Akiyama, K. (2001). Levels of cysteinyl leukotriene receptor mRNA in human peripheral leucocytes: significantly higher expression of cysteinyl leukotriene receptor 2 mRNA in eosinophils. Clin. Exp. Allergy 31, 1714–1723. doi: 10.1046/j.1365-2222.2001.01184.x
Moffatt, M. F., Kabesch, M., Liang, L., Dixon, A. L., Strachan, D., Heath, S., et al. (2007). Genetic variants regulating ORMDL3 expression contribute to the risk of childhood asthma. Nature 448, 470–473. doi: 10.1038/nature06014
Mougey, E., Lang, J. E., Allayee, H., Teague, W. G., Dozor, A. J., Wise, R. A., et al. (2013). ALOX5 polymorphism associates with increased leukotriene production and reduced lung function and asthma control in children with poorly controlled asthma. Clin. Exp. Allergy 2013, 512–520. doi: 10.1111/cea.12076
Mougey, E. B., Lang, J. E., Wen, X., and Lima, J. J. (2011). Effect of citrus juice and SLCO2B1 genotype on the pharmacokinetics of montelukast. J. Clin. Pharmacol. 51, 751–760. doi: 10.1177/0091270010374472
Noonan, M. J., Chervinsky, P., Brandon, M., Zhang, J., Kundu, S., McBurney, J., et al. (1998). Montelukast, a potent leukotriene receptor antagonist, causes dose-related improvements in chronic asthma. Eur. Respir. J. 11, 1232–1239. doi: 10.1183/09031936.98.11061232
Obase, Y., Shimoda, T., Tomari, S., Mitsuta, K., Fukushima, C., Kawano, T., et al. (2001). Effects of pranlukast on aspirin-induced bronchoconstriction: differences in chemical mediators between aspirin-intolerant and tolerant asthmatic patients. Ann. Allergy. Asthma Immunol. 87, 74–79. doi: 10.1016/S1081-1206(10)62328-9
Oguma, T., Palmer, L. J., Birben, E., Sonna, L. A., Asano, K., and Lilly, C. M. (2004). Role of prostanoid DP receptor variants in susceptibility to asthma. N. Engl. J. Med. 351, 1752–1763. doi: 10.1056/NEJMoa031785
Park, J. S., Chang, H. S., Park, C. S., Lee, J. H., Lee, Y. M., Choi, J. H., et al. (2005). Association analysis of cysteinyl-leukotriene receptor 2 (CYSLTR2) polymorphisms with aspirin intolerance in asthmatics. Pharmacogenet. Genomics 15, 483–492. doi: 10.1097/01.fpc.0000166456.84905.a0
Pearce, N., Ait-Khaled, N., Beasley, R., Mallol, J., Keil, U., Mitchell, E., et al. (2007). Worldwide trends in the prevalence of asthma symptoms: phase III of the International Study of Asthma and Allergies in Childhood (ISAAC). Thorax 2007, 758–766. doi: 10.1136/thx.2006.070169
Pietras, C. O., Vendelin, J., Anedda, F., Bruce, S., Adner, M., Sundman, L., et al. (2011). The asthma candidate gene NPSR1 mediates isoform specific downstream signalling. BMC Pulm. Med. 11:39. doi: 10.1186/1471-2466-11-39
Pillai, S. G., Cousens, D. J., Barnes, A. A., Buckley, P. T., Chiano, M. N., Hosking, L. K., et al. (2004). A coding polymorphism in the CYSLT2 receptor with reduced affinity to LTD4 is associated with asthma. Pharmacogenetics 14, 627–633. doi: 10.1097/00008571-200409000-00007
Rana, B. K., Bourne, P. E., and Insel, P. A. (2012). Receptor databases and computational websites for ligand binding. Methods Mol. Biol. 897, 1–13. doi: 10.1007/978-1-61779-909-9_1
Rana, B. K., Shiina, T., and Insel, P. A. (2001). Genetic variations and polymorphisms of G protein-coupled receptors: functional and therapeutic implications. Annu. Rev. Pharmacol. Toxicol. 41, 593–624. doi: 10.1146/annurev.pharmtox.41.1.593
Reihsaus, E., Innis, M., MacIntyre, N., and Liggett, S. B. (1993). Mutations in the gene encoding for the beta-2-adrenergic receptor in normal and asthmatic subjects. Am. J. Respir. Cell Mol. Biol. 8, 334–339. doi: 10.1165/ajrcmb/8.3.334
Reiss, T. F., Altman, L. C., Chervinsky, P., Bewtra, A., Stricker, W. E., Noonan, G. P., et al. (1996). Effects of montelukast (MK-0476), a new potent cysteinyl leukotriene (LDT(4)) receptor antagonist, in patients with chronic asthma. J. Allergy Clin. Immunol. 98, 528–534. doi: 10.1016/S0091-6749(96)70086-6
Roberts, D. F. (1968). Genetic effects of population size reduction. Nature 220, 1084–1088. doi: 10.1038/2201084a0
Sampson, A. P., Siddiqui, S., Buchanan, D., Howarth, P., Holgate, S., Holloway, J., et al. (2000). Variant LTC4 synthase allele modifies cysteinyl leukotriene synthesis in eosinophils and predicts clinical response to zafirlukast. Thorax 55(Suppl. 2), S28–S31. doi: 10.1136/thorax.55.suppl_2.S28
Samuelsson, B. (1983). Leukotrienes: mediators of immediate hypersensitivity reactions and inflammation. Science 220, 568–575. doi: 10.1126/science.6301011
Sanz, C., Isidro-García, M., Dávila, I., Moreno, E., Laffond, E., and Lorente, F. (2006). Analysis of 927T> C CYSLTRI and -444A > C LTC4S polymorphisms in patients with asthma. J. Investig. Allergol. Clin. Immunol. 16, 331–337.
Sayers, I., Barton, S., Rorke, S., Sawyer, J., Peng, Q., Beghé, B., et al. (2003). Promoter polymorphism in the 5-lipoxygenase (ALOX5) and 5-lipoxygenase-activating protein (ALOX5AP) genes. Clin. Exp. Allergy 33, 1103–1110. doi: 10.1046/j.1365-2222.2003.01733.x
Severien, C., Artlich, A., Jonas, S., and Becher, G. (2000). Urinary excretion of leukotriene E-4 and eosinophil protein X in children with atopic asthma. Eur. Respir. J. 16, 588–592. doi: 10.1034/j.1399-3003.2000.16d03.x
Silverman, E. S., Baron, R. M., Palmer, L. J., Le, L., Hallock, A., Subramaniam, V., et al. (2002). Constitutive and cytokine-induced expression of the ETS transcription factor ESE-3 in the lung. Am. J. Respir. Cell Mol. Biol. 27, 697–704. doi: 10.1165/rcmb.2002-0011OC
Silverman, E. S., Du, J., De, Sanctis G. T., Rådmark, O., Samuelsson, B., Drazen, J. M., et al. (1998). Egr-1 and Sp1 interact functionally withreceptor with the 5-lipoxygenase promoter and its naturally occurring mutants. Am. J. Respir. Cell Mol. Biol. 19, 316–323. doi: 10.1165/ajrcmb.19.2.3154
Sleiman, P. M., Flory, J., Imielinski, M., Bradfield, J. P., Annaiah, K., Willis-Owen, S. A., et al. (2010). Variants of DENND1B associated with asthma in children. N. Engl. J. Med. 362, 36–44. doi: 10.1056/NEJMoa0901867
Slutsky, A. S., and Zamel, N. (1997). Genetics of asthma: the University of Toronto Program. University of Toronto Genetics of Asthma Research Group. Am. J. Respir. Crit. Care Med. 156, S130–S132. doi: 10.1164/ajrccm.156.4.12tac-10
Sokolowska, M., Wodz-Naskiewicz, K., Cieslak, M., Seta, K., Bednarek, A. K., and Pawliczak, R. (2009). Variable expression of cysteinyl leukotriene type I receptor splice variants in asthmatic females with different promoter haplotypes. BMC Immunol. 10:63. doi: 10.1186/1471-2172-10-63
Soodyall, H., Jenkins, T., Mukherjee, A., du Toit, E., Roberts, D. F., and Stoneking, M. (1997). The founding mitochondrial DNA lineages of Tristan da Cunha Islanders. Am. J. Phys. Anthropol. 104, 157–166.
Soodyall, H., Nebel, A., Morar, B., and Jenkins, T. (2003). Genealogy and genes: tracing the founding fathers of Tristan da Cunha. Eur. J. Hum. Genet. 11, 705–709. doi: 10.1038/sj.ejhg.5201022
Spiegel, A. M., and Weinstein, L. S. (2004). Inherited diseases involving G proteins and G protein-coupled receptors. Annu. Rev. Med. 55, 27–39. doi: 10.1146/annurev.med.55.091902.103843
Srivastava, P., Helms, P. J., Stewart, D., Main, M., and Russell, G. (2003). Association of CCR5 Delta 32 with reduced risk of childhood but not adult asthma. Thorax 58, 222–226. doi: 10.1136/thorax.58.3.222
Suissa, S., Dennis, R., Ernst, P., Sheehy, O., and Wood-Dauphinee, S. (1997). Effectiveness of the leukotriene receptor antagonist zafirlukast for mild-to-moderate asthma—a randomized, double-blind, placebo-controlled trial. Ann. Intern. Med. 126, 177–183. doi: 10.7326/0003-4819-126-3-199702010-00001
Takasaki, J., Kamohara, M., Matsumoto, M., Saito, T., Sugimoto, T., Ohishi, T., et al. (2000). The molecular characterization and tissue distribution of the human cysteinyl leukotriene CysLT(2) receptor. Biochem. Biophys. Res. Commun. 274, 316–322. doi: 10.1006/bbrc.2000.3140
Tantisira, K. G., and Drazen, J. M. (2009). Genetics and pharmacogenetics of the leukotriene pathway. J. Allergy Clin. Immunol. 124, 422–427. doi: 10.1016/j.jaci.2009.06.035
Tantisira, K. G., Lima, J., Sylvia, J., Klanderman, B., and Weiss, S. T. (2009). 5-Lipoxygenase pharmacogenetics in asthma: overlap with Cys-leukotriene receptor antagonist loci. Pharmacogenet. Genomics 19, 244–247. doi: 10.1097/FPC.0b013e328326e0b1
Taylor, D. R., Epton, M. J., Kennedy, M. A., Smith, A. D., Iles, S., Miller, A. L., et al. (2005). Bronchodilator response in relation to beta2-adrenoceptor haplotype in patients with asthma. Am. J. Respir. Crit. Care Med. 172, 700–703. doi: 10.1164/rccm.200501-092OC
Taylor, D. R., and Kennedy, M. A. (2001). Genetic variation of the beta(2)-adrenoceptor: its functional and clinical importance in bronchial asthma. Am. J. Pharmacogenomics 1, 165–174. doi: 10.2165/00129785-200101030-00002
Taylor, D. R., and Kennedy, M. A. (2002). Beta-adrenergic receptor polymorphisms and drug responses in asthma. Pharmacogenomics 3, 173–184. doi: 10.1517/14622416.3.2.173
Telleria, J. J., Blanco-Quiros, A., Varillas, D., Armentia, A., Fernandez-Carvajal, I., Jesus, Alonso M, et al. (2008). ALOX5 promoter genotype and response to montelukast in moderate persistent asthma. Respir. Med. 102, 857–861. doi: 10.1016/j.rmed.2008.01.011
Thompson, M. D., Burnham, W. M., and Cole, D. E. (2005a). The G protein-coupled receptors: pharmacogenetics and disease. Crit. Rev. Clin. Lab. Sci. 42, 311–392. doi: 10.1080/10408360591001895
Thompson, M. D., Capra, V., and Stankova, J. (2013). Cysteinyl leukotriene 1 and cysteinyl leukotriene 2 receptors are associated with atopic asthma in a founder population. Eur. J. Hum. Genet. 21(Suppl. 2), 405 (P16.114).
Thompson, M. D., Capra, V., Takasaki, J., Maresca, G., Rovati, G. E., Slutsky, A. S., et al. (2007). A functional G300S variant of the cysteinyl leukotriene 1 receptor is associated with atopy in a Tristan da Cunha isolate. Pharmacogenet. Genomics 17, 539–549. doi: 10.1097/FPC.0b013e328012d0bf
Thompson, M. D., Cole, D. E., Capra, V., Siminovitch, K. A., Rovati, G. E., Burnham, W. M., et al. (2014a). Pharmacogenetics of the G protein-coupled receptors. Methods Mol. Biol. 1175, 189–242. doi: 10.1007/978-1-4939-0956-8_9
Thompson, M. D., Cole, D. E., Jose, P. A., and Chidiac, P. (2014b). G protein-coupled receptor accessory proteins and signaling: pharmacogenomic insights. Methods Mol. Biol. 2014, 121–152. doi: 10.1007/978-1-4939-0956-8_7.157
Thompson, M. D., Gravesande, K. S. V., Galczenski, H., Burnham, W. M., Siminovitch, K. A., Zamel, N., et al. (2003). A cysteinyl leukotriene 2 receptor variant is associated with atopy in the population of Tristan da Cunha. Pharmacogenetics 13, 641–649. doi: 10.1097/00008571-200310000-00008
Thompson, M. D., Noble-Topham, S., Percy, M. E., Andrade, D. M., and Ebers, G. C. (2012). Chromosome 1p36 in migraine with aura: association study of the 5HT(1D) locus. Neuroreport 23, 45–48. doi: 10.1097/WNR.0b013e32834e5af3
Thompson, M. D., Siminovitch, K. A., and Cole, D. E. (2008). G protein-coupled receptor pharmacogenetics. Methods Mol. Biol. 448, 139–185. doi: 10.1007/978-1-59745-205-2_8
Thompson, M. D., Takasaki, J., Capra, V., Rovati, G. E., Siminovitch, K. A., Burnham, W. M., et al. (2006). G protein-coupled receptors and asthma endophenotypes: the cysteinyl leukotriene system in perspective. Mol. Diagn. Ther. 10, 353–366. doi: 10.1007/BF03256212
Thompson, M. D., Xhaard, H., Sakurai, T., Rainero, I., and Kukkonen, J. P. (2014c). OX1 and OX2 orexin/hypocretin receptor pharmacogenetics. Front. Neurosci. 8:57. doi: 10.3389/fnins.2014.00057
Thompson, M. D., Zamel, N., Slutsky, A. S., Lilly, C. M., Burnham, W. M., Siminovitch, K. A., et al. (2005b). Genetic variants in both cysteinyl leukotriene 1 and 2 receptors are independently associated with atopy and asthma in a Tristan da Cunha population. Eur. J. Hum. Genet. 13:1066.
Thonkham, D., Tran, J., Clunes, M. T., and Brahim, F. (2015). Prevalence and severity of asthmatic symptoms in Grenadian School children: the Grenada National Asthma Survey. BMJ Open 5:e008557. doi: 10.1136/bmjopen-2015-008557
Unoki, M., Furuta, S., Onouchi, Y., Watanabe, O., Doi, S., Fujiwara, H., et al. (2000). Association studies of 33 single nucleotide polymorphisms (SNPs) in 29 candidate genes for bronchial asthma: positive association a T924C polymorphism in the thromboxane receptor gene. Hum. Genet. 106, A440–A446. doi: 10.1007/s004390000267
Vroling, B., Sanders, M., Baakman, C., Borrmann, A., Verhoeven, S., Klomp, J., et al. (2011). GPCRDB: information system for G protein-coupled receptors. Nucleic Acids Res. 39, D309–D319. doi: 10.1093/nar/gkq1009
Wang, J., Xu, Y., Zhao, H., Sui, H., Liang, H., and Jiang, X. (2009). Genetic variations in chemoattractant receptor expressed on Th2 cells (CRTH2) is associated with asthma susceptibility in Chinese children. Mol. Biol. Rep. 36, 1549–1553. doi: 10.1007/s11033-008-9349-6
Woszczek, G., Pawliczak, R., Qi, H. Y., Nagineni, S., Alsaaty, S., Logun, C., et al. (2005). Functional characterization of human cysteinyl leukotriene 1 receptor gene structure. J. Immunol. 175, 5152–5159. doi: 10.4049/jimmunol.175.8.5152
Wu, J., Duan, R., Cao, H., Field, D., Newnham, C. M., Koehler, D. R., et al. (2008). Regulation of epithelium-specific Ets-like factors ESE-1 and ESE-3 in airway epithelial cells: potential roles in airway inflammation. Cell Res. 18, 649–663. doi: 10.1038/cr.2008.57
Yaddaden, L., Véronneau, S., Thompson, M. D., Rola-Pleszczynski, M., and Stankova, J. (2016). Cellular signalling of cysteinyl leukotriene type 1 receptor variants CysLT1-G300S and CysLT1-I206S. Prostaglandins Leukot. Essent. Fatty Acids 105, 1–8. doi: 10.1016/j.plefa.2015.12.004
Zamel, N., McClean, P. A., Sandel, P. R., Siminovitch, K. A., and Slutsky, A. S. (1996). Asthma on Tristan da Cunha: looking for the genetic link. The University of Toronto Genetics of Asthma Research Group. Am. J. Respir. Crit. Care Med. 153, 1902–1906. doi: 10.1164/ajrccm.153.6.8665053
Zhang, J., Migita, O., Koga, M., Shibasaki, M., Arinami, T., and Noguchi, E. (2006). Determination of structure and transcriptional regulation of CYSLTR1 and an association study with asthma and rhinitis. Pediatr. Allergy Immunol. 17, 242–249. doi: 10.1111/j.1399-3038.2005.00347.x
Keywords: montelukast, pharmacogenetics, pharmacogenomics, cysteinyl leukotriene 1 (CYSLTR1), cysteinyl leukotriene 2 (CYSLTR2), epistasis, epigenetics, Tristan da Cunha
Citation: Thompson MD, Capra V, Clunes MT, Rovati GE, Stankova J, Maj MC and Duffy DL (2016) Cysteinyl Leukotrienes Pathway Genes, Atopic Asthma and Drug Response: From Population Isolates to Large Genome-Wide Association Studies. Front. Pharmacol. 7:299. doi: 10.3389/fphar.2016.00299
Received: 04 June 2015; Accepted: 24 August 2016;
Published: 01 December 2016.
Edited by:
Martin A. Kennedy, University of Otago, New ZealandReviewed by:
Martin C. Michel, Johannes Gutenberg University, GermanyCopyright © 2016 Thompson, Capra, Clunes, Rovati, Stankova, Maj and Duffy. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Miles D. Thompson, bWlsZXNkLnRob21wc29uQG91dGxvb2suY29t
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.