- 1Department of Pathology and Immunology, Baylor College of Medicine, Houston, TX, USA
- 2Department of Pathology, Texas Children’s Hospital, Houston, TX, USA
Beneficial microbes are responsible for the synthesis of nutrients and metabolites that are likely important for the maintenance of mammalian health. Many nutrients and metabolites derived from the gut microbiota by luminal conversion have been implicated in the development, homeostasis and function of innate and adaptive immunity. These factors clearly suggest that intestinal microbiota may influence host immunity via microbial metabolite-dependent mechanisms. We describe how intestinal microbes including probiotics generate microbial metabolites that modulate mucosal and systemic immunity.
Introduction
The mammalian gastrointestinal tract, site for digestion and nutrition absorption harbors commensal microbiota, a population composed of 1000–5000 different bacterial species. Metagenomics of the Human Intestinal Tract (MetaHit) project containing 249 newly sequenced samples with 1,018 previously sequenced samples were combined to create a cohort from three continents. From this the integrated gene catalog (IGC) comprising 9,879,896 genes were established. The catalog includes close-to-complete sets of genes for most gut microbes. Analyses of a group of samples from Chinese and Danish individuals using IGC revealed country-specific gut microbial signatures. This expanded catalog should facilitate quantitative characterization of metagenomic, metatranscriptomic, and metaproteomic data from the gut microbiome to understand its variation across populations in human health and disease (Qin et al., 2010; Ferreira et al., 2014; Li et al., 2014). Recent studies show that changes in the commensal bacterial composition are linked to various metabolic and inflammatory diseases including inflammatory bowel disease (IBD; Sokol et al., 2008), obesity and type 2 diabetes (Everard et al., 2013; Dao et al., 2015), allergy (Berni Canani et al., 2015), and colorectal cancer (Swidsinski et al., 1998). These interrelationships provoke multiple fundamental questions regarding the cellular and molecular pathways through which commensal microbiota regulates mammalian gene expression and influence a wide range of clinically important diseased complications. The intestinal microbiota affects host physiology in many ways such as influencing the maturation of the immune response and fortifying the intestinal barrier against pathogenic bacteria. Importantly, intestinal microbes are potential regulators of digestion converting a wide range of non-digestible carbohydrates to short chain fatty acids (SCFA), which can be absorbed by the host and used as energy sources (Sharma et al., 2010; Becker et al., 2011).
Dysregulation of intestinal immune response by commensal microbiota plays an important role in the onset and development of different immune-mediated disorders (Wohlgemuth et al., 2009; Feng et al., 2010). For example, the presence of Akkermansia muciniphila, commensal mucin degrader, has been shown to exacerbate Salmonella Typhimurium infection by worsening intestinal inflammation, increasing macrophage infiltration and elevating proinflammatory cytokines in gnotobiotic mice (Ganesh et al., 2013). Flagellin-detecting toll like receptor 5 (TLR5) knockout mice colonized with adherent-invasive Escherichia coli (AIEC) during microbiota acquisition drove chronic colitis. AIEC instigates chronic inflammation by increasing microbiota levels of LPS and flagellin (Chassaing et al., 2014). Recent findings described how commensals are recognized by the intestinal innate immune system and how individual species can influence specific modules of the innate and adaptive immunity. Germ-free mice were shown to have fewer and smaller Peyer patches, exhibit a local defect or absence of TH1, TH17, and TREG cells, and their intestinal epithelia express lower amounts of TLRs and MHC class II, as compared with mice that have been exposed to normal microbiota (commensals). Similarly, symbiosis factor polysaccharide A (produced by Bacteroides fragilis) can induce TREG cells and suppress TH17 cells via engagement of TLR2 on CD4+ T cells (Round et al., 2011). Similarly, another human commensal Faecalibacterium prausnitzii suppresses IL-8 production and NF-κB signaling in response to inflammatory secretion of IL-1β (Sokol et al., 2008). Altogether, recent evidence has provided insights into immune-mediated mechanisms in metabolic disorders (Borchers et al., 2009). Taken all the findings together, existing data argues for the need to probe the microbiome for new strategies for immunomodulation, either by enhancing (immunodeficiency) or by suppressing (allergy) host immunity. Microbial metabolites and nutrients derived from beneficial bacteria in the intestine via luminal conversion may modulate host immunity and profoundly affect mammalian biology of the “holobiont.”
Changes in Microbial Diversity and Treatment with Probiotics
Recent studies in rodents show that inflammation and/or infection is correlated with changes in bacterial composition (Packey and Sartor, 2009; Saulnier et al., 2011; Pflughoeft and Versalovic, 2012; Ganesh et al., 2013). Molecular techniques are clarifying changes in the composition of the mucosal associated and fecal microbiota in patients with IBD esp., ulcerative colitis (UC), and Crohn’s diseases (CD) together with widely expanding previous culture based studies. Patients with UC and CD have decreased complexity of commensal microbiota revealed by examining DNA libraries (Frank et al., 2007). More specifically, members of the phyla Bacteroidetes and Firmicutes are decreased in CD and UC patients (Backhed et al., 2005). A member of the family Firmicutes, F. prausnitzii was reduced in the patients with CD and this was confirmed and associated with increased risk of post-resection recurrence of ileal CD (Frank et al., 2007; Sokol et al., 2008; Swidsinski et al., 2008). In vitro peripheral blood mononuclear cell stimulation by F. prausnitzii decreased pro-inflammatory cytokines IL-12 and IFN-γ and stimulated secretion of anti-inflammatory cytokine IL-10. Oral administration of live F. prausnitzii or its supernatant reduced the inflammation severity by TNBS and corrected the associated dysbiosis (Baumgart et al., 2007). However, the abundance of E. coli is increased in IBD patients (Figure 1; Kotlowski et al., 2007). Similarly, the mucosal E. coli numbers in situ correlates with the severity of ileal disease and invasive E. coli are restricted to inflamed mucosa. Finally, fecal and mucosal associated microbial communities of UC and CD patients are consistently less diverse with increased instability. Commensal non-pathogenic bacteria can cause colitis in host with immunomodulatory and mucosal barrier deficits. Interleukin (IL)-10–/– germ-free mice colonized with Enterococcus faecalis and/or invasive E. coli, showed aggressive TH1/TH17-mediated colitis within 3 weeks but this was not observed in the WT mice. LPS from microbes were detected by dendritic cells (DCs). DCs play an important role through antigen presentation via TLRs in linking between the innate and adaptive immunity (McKenna et al., 2005). DCs are the initial cells to synthesize IL-12 under well characterized microbial stimulants of the cytokines. IL-12 selectively promotes the differentiation of Th1 CD4+ cells upon stimulation with antigens (de Jong et al., 2002). Th1 cell-mediated immune response leads to the paradigm of T-helper cell differentiation in which IL-12 cytokine mediated activation of STAT4 and is critical for generation of Th1 cells (Kaplan et al., 1998). IL-12 mediated immune response is dependent upon the presence of CD4+ and CD8+ T lymphocytes and upon the production of IFN-γ finally causing cell-mediated adaptive immunity (Figure 1; Kim et al., 2007). However, certain class of bacteria like probiotic bacterium, Bifidobacterium breve increased IL-10 secretion Tr-1 cells in the colon and inhibits inflammation (Jeon et al., 2012). Introducing such beneficial strains in an unhealthy intestinal environment will potentially be a novel therapeutic strategy.
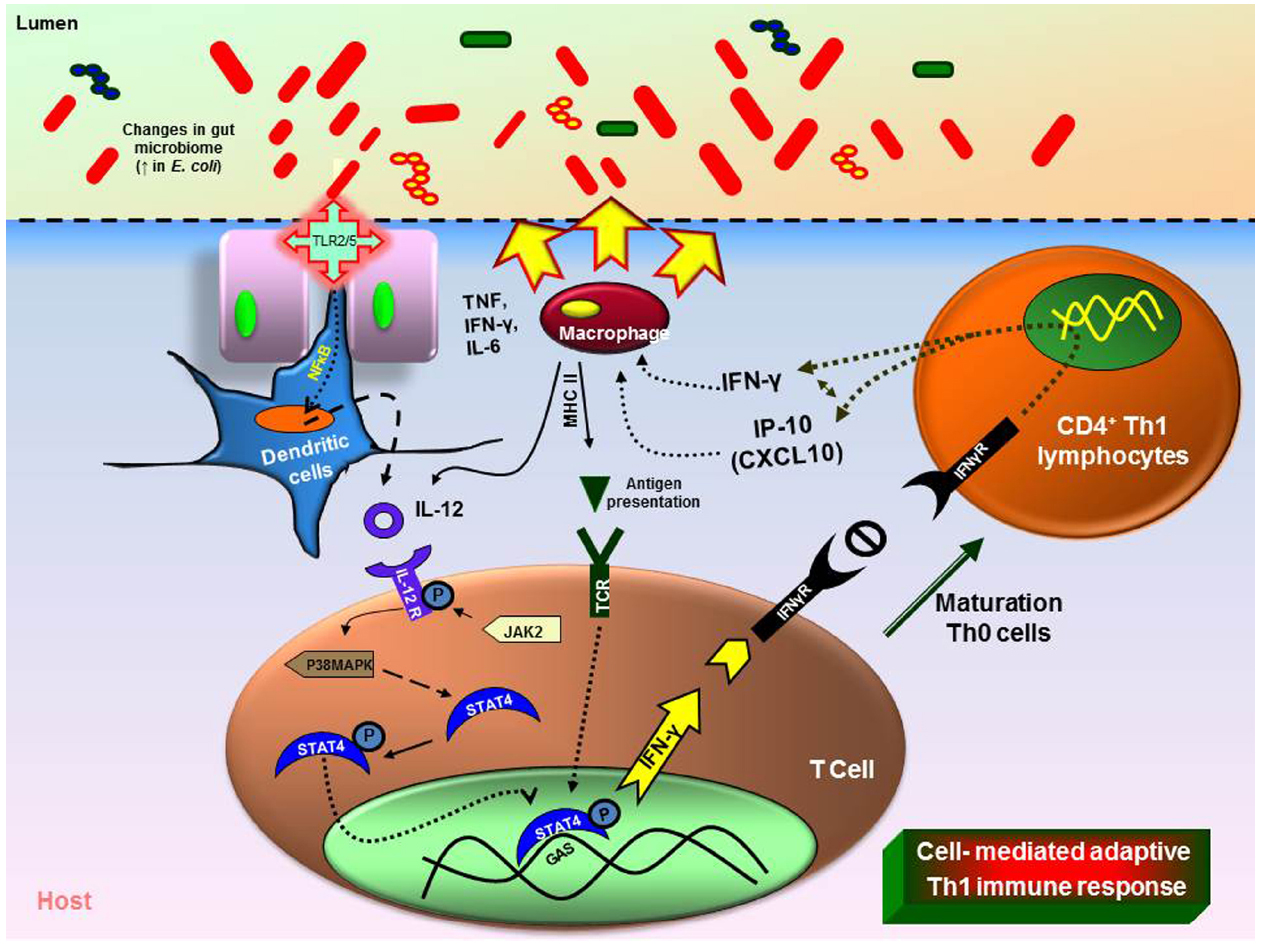
Figure 1. Immune responses triggered by changes in the gut microbiome. Intestinal inflammation in the UC or CD leads to dysbiosis (imbalance microbiota). Overgrowth of enteropathogenic bacteria causing increased activation of toll-like receptors (TLR) 2 or 4. This causes the activation and translocation of nuclear factor kappa B (NFκB) and causes secretion of pro-inflammatory cytokine interleukin (IL)-12. Increased IL-12 causes T-helper (Th) Th1/Th2 immune response with increase in tumor necrosis factor (TNF), IL-6, interferon gamma (IFN-γ). The dysbiosis leads to increase in immune cells (macrophages, neutrophils) at the infected site causing severe inflammation (MHCII—major histocompatibility complex).
Most importantly, metabolites produced by intestinal microbiota have direct effects on the host mucosa. Commensal bacterial fermentation of non-digestible fiber leads to increased luminal bioavailability of SCFAs like butyrate, acetate, fumarate, and propionate (Cummings and Macfarlane, 1997). Bacterial metabolites such as butyrate serve as potential energy sources for colonic epithelial cells, whereas other fermentation by-products like hydrogen sulfide (HS), nitric oxide (NO) and proteases produced by subsets of commensals may enhance histopathology. Butyrate metabolism by colonic epithelial cells might be suppressed by HS/NO metabolites, resulting in starvation of colonocytes and yielding histopathology similar to that of UC (Roediger et al., 1993; Packey and Sartor, 2009; Cain and Karpa, 2011). The butyrate producing probiotic bacterium Clostridium butyricum MIYAIRI 588, increase the butyrate availability in the presence of fibrous diet (Weng et al., 2015). Intracellular butyrate and propionate (but not acetate) has been shown to inhibit the activity of histone deacetylases (HDACs) in colonocytes and immune cells, which promotes the hyperacetylation of histones, in addition to some transcription factors and proteins that are involved in signal transduction. This has multiple consequences for gene expression and cellular differentiation, including the down-regulation of pro-inflammatory cytokines, such as IL-6 and IL-12, in colonic macrophages and is also known to inhibit colorectal cancer (Louis et al., 2014). Similarly, pretreatment of Helicobacter pylori-induced gastric ulcers with C. butyricum in mice showed significantly reduced numbers of mucosal lesions with decreased quantities of proinflammatory cytokines (Wang et al., 2015). Probiotics may provide beneficial functions into the GI tract which might enhance the functionality of the existing commensal communities. Probiotics may also affect the composition of the intestinal microbiota by providing colonization resistance and competition for nutrients or production of pathogenic inhibitors and modulates intestinal immune response.
Probiotics possess the ability to transiently colonize the gut (Valeur et al., 2004; Ukibe et al., 2015; Vieira et al., 2015) and facilitating proliferation of commensal microbes, while enhancing microbial diversity (Sherman et al., 2009). Probiotics are known to exert antimicrobial effects as a front line of defense against the luminal pathogens. For example, some probiotics are known to elaborate some microbial products known as bacteriocins. These probiotic factors can inhibit the growth and virulence of enteric bacterial pathogens (Corr et al., 2007). Bifidobacterium animalis subsp. lactis (B. lactis), Streptococcus thermophilus, two different strains of Lactobacillus delbrueckii subsp and L. lactis subsp in fermented milk were used to determine the impact of microbes in a mouse model of IBD. The findings show that B. lactis containing fermented milk decreased cecal pH, altered SCFA concentrations, increased the relative quantities of lactate- and butyrate-consuming bacteria, and reduced intestinal inflammation scores (Veiga et al., 2010). In addition, lactic-acid producing bacteria are known to exert antimicrobial effects on pathogens by reducing the pH of the microenvironment in the lumen of the GI tract (Fayol-Messaoudi et al., 2005). Probiotics or their metabolites reduced the secretion of immunomodulation molecule autoinducer-2 by the pathogenic E. coli, which results in reduced gene expression contained in the locus of enterocyte effacement (Pathogenicity Island) which is critical for mediating intimate bacterial binding to the host cell surfaces, called attachment and effacing lesion (Mack et al., 1999; Russell et al., 2007). Lactobacillus plantarum has been shown to have the capacity to enhance the production and secretion of mucins esp. MUC2 and MUC3 from the human intestinal epithelial cells (Mack et al., 1999), which improves the epithelial barrier function (Corfield et al., 1992, 2000). Similarly, bacteria and their by-products may have direct effect on the betterment of host health.
Luminal Conversion of Dietary Components by the Intestinal Microbiota
Human diet may have a direct impact on the intestinal microbiota which ultimately leads to the changes in the microbiota composition. These changes have been recently validated using mouse model experiments. Mice subjected to the high fat diet in obese mice showed major changes in microbial composition with an increased proportion of the phylum Firmicutes and decreased proportion of Bacteroidetes. In particular, species like Clostridium ramosum was correlated with increased body weight (Fleissner et al., 2010; Woting et al., 2014). Vitamins, amino acids or dietary fibers with the diet are assimilated and converted into other metabolites in the lumen by intestinal microbiota. Some of the products of these bio-chemical conversions were SCFA, biogenic amines (such as histamine) or other amino acid derived metabolites like serotonin or gamma-aminobutyric acid (GABA; Bravo et al., 2011; Figure 2) which may have beneficial effect on host health (Hemarajata and Versalovic, 2013; Hemarajata et al., 2013). Serotonin is a neurotransmitter, biochemically derived from tryptophan (Best et al., 2010). Bifidobacterium infantis colonization in rats modulated the bioavailability of tryptophan by yielding increased concentrations of tryptophan in plasma, reduced 5-HIAA (hydroxyindoleacetic acid) concentrations in the frontal cortex, and diminished quantities of 3,4-dihydroxyphenylacetic acid (DOPAC) in the amygdaloid cortex (Desbonnet et al., 2008). Gut microbial populations in SPF mice modulated brain development by contributing to suppressed expression of postsynaptic density protein (PSD)-95 and synaptophysin in the striatum compared to germ-free mice (Diaz Heijtz et al., 2011). Treatment with Bifidobacterium species resulted in normalization of the immune response, reversal of behavioral deficits, and restoration of basal noradrenaline concentrations in the brainstem, thereby alleviating depression of the CNS (Desbonnet et al., 2010). In addition, orally gavaged BALB/c mice with Lactobacillus rhamnosus (JB-1) reduced GABAAα2 gene expression in the prefrontal cortex and amygdala, but increased GABAAα2 gene expression in the hippocampus. These findings provide evidence that Lactobacillus strains regulate emotions, behavior and central GABA receptor expression (Bravo et al., 2011). Intestinal microbiota may modulate the bioavailability of tryptophan in the intestine, and may in turn influence availability of neurotransmitters such as serotonin in the host. Non-digestible carbohydrates can be fermented in the lumen resulting in production of SCFAs such as lactate, formate, acetate, propionate, butyrate and valerate (Blaut, 2013). These metabolically active SCFAs are involved in various biological processes as an energy source in intestinal epithelial cell proliferation (Astbury and Corfe, 2012; Fung et al., 2012; Matthews et al., 2012). Additionally, fermentation of prebiotic carbohydrates such as inulin and fructo-oligosaccharides has been shown to increase the proportion of beneficial microbes like Bifidobacterium spp. and Lactobacillus spp. in the obese mice and was negatively correlated with serum entoxin levels (Salazar et al., 2014). Consumption of western diet showed increased level of plasma LPS concentration and this was correlated with increased changes in microbiota composition (Cani et al., 2013; Everard and Cani, 2013; Everard et al., 2013). Moreover, a recent study shows that dietary plant lignans were converted to estradiol like metabolite enterodiol and enterolactone by intestinal bacteria in germ-free rats colonized with lignan-converting consortium, such as Clostridium saccharogumia, Blautia producta, Eggerthella lenta, and Lactonifactor longoviformis. The produced enterolignans suppressed tumor number and tumor cell proliferation in hormone related cancer (Mabrok et al., 2012).
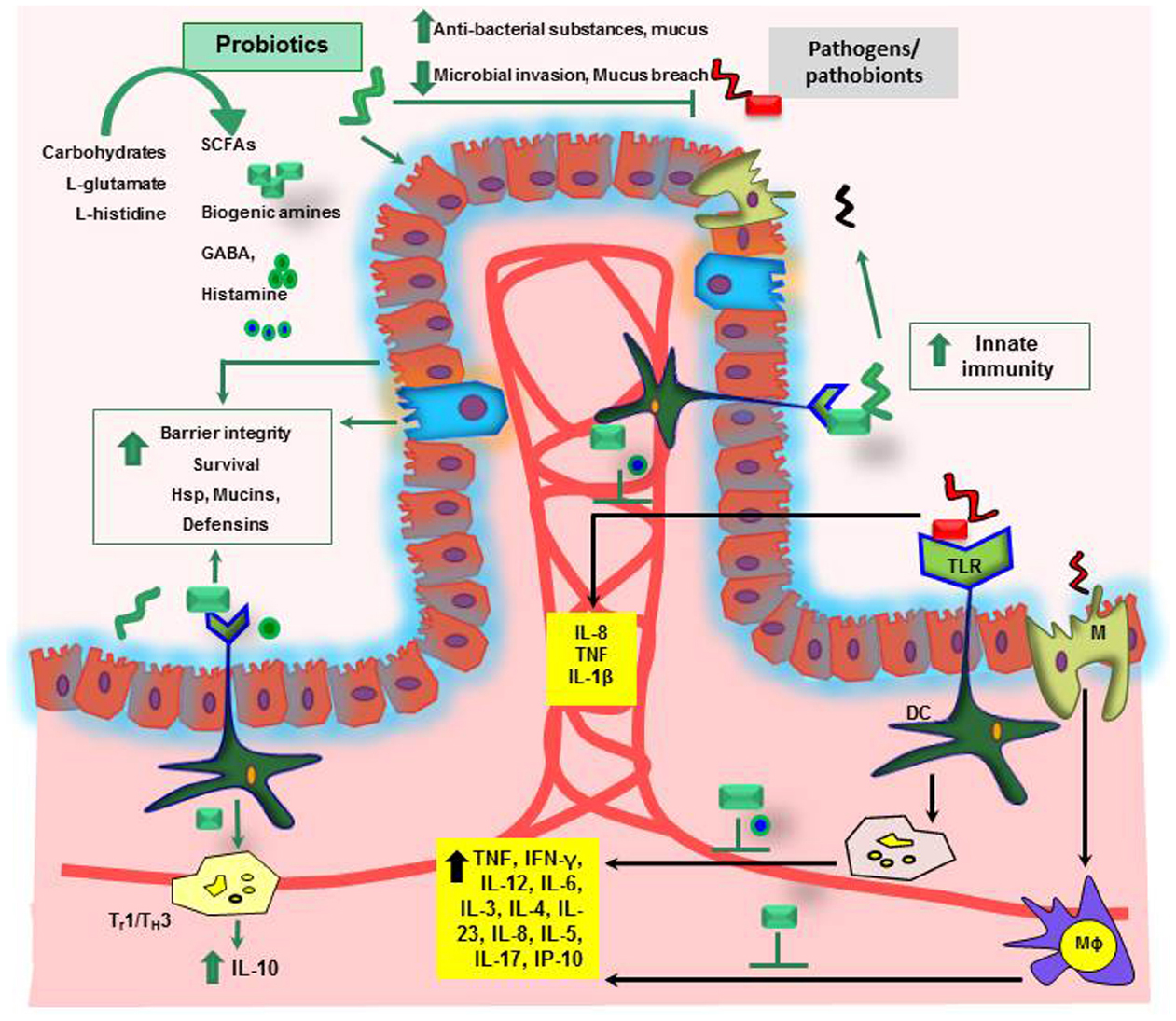
Figure 2. Mechanisms of probiosis in the gastrointestinal tract. SCFAs, short chain fatty acids; GABA, gamma-aminobutyric acid; Hsp, heat shock proteins; IL, interleukin; TNF, tumor necrosis factor; Th, T-helper; IFN-γ, interferon gamma; Mϕ, Macrophage; DC, dendritic cell; M, microfold cells.
The secondary plant metabolites, glucosinolates from Brassica vegetables, were converted to isothiocyanates (glucosinolate derivative) and were measured in urine, luminal contents and plasma of mice (Budnowski et al., 2013). In addition, glucosinolates and their derivatives have been shown to reduce AOM/DSS induced colon carcinogenesis in mice (Lippmann et al., 2014). For example, Bacteroides thetaiotaomicron isolated from human fecal sample can convert glucosinolates into isothiocyanates, measured in luminal contents of rats (Elfoul et al., 2001; Krul et al., 2002), and these compounds potentially suppress lung cancer cell metastasis by inhibiting cell survival signaling molecules Akt and NFκB activation in human lung large cell carcinoma (Wu et al., 2010b). Similarly, isoflavones have been implicated in the prevention of hormone-dependent and age related diseases, including cancer (Birt et al., 2001; Scalbert et al., 2005; Geller and Studee, 2006; Usui, 2006). Intestinal bacteria, e.g., Slackia isoflavoniconvertens, play an important role in the metabolism of isoflavones, daidzein and genistein to equol (Chang and Nair, 1995; Rafii et al., 2003; Matthies et al., 2008, 2012). Based on the structural similarities of these bacterial by-products with estrogens, they bind to estrogen receptors and thus may prevent cancer progression (Matthies et al., 2008; Lepri et al., 2014).
Immunomodulation by Probiotics
Probiotics (beneficial microbes) are frequently, though not necessarily be a commensal bacteria. Probiotics are defined as “beneficial live micro-organisms which when administrated in adequate amounts confer beneficial effects on the host health” (Mack et al., 1999; Peran et al., 2006; Borchers et al., 2009; Ganesh et al., 2012; Isolauri et al., 2012; Klaenhammer et al., 2012; Thomas et al., 2012; Morelli and Capurso, 2012; Arena et al., 2014; Dylag et al., 2014; Galdeano et al., 2015; Ki et al., 2014; Repa et al., 2014; Sah et al., 2014; Sanders et al., 2014). Most known probiotics until now are either lactobacilli or bifidobacteria representatives of which are normal inhabitants of the gastro-intestinal (GI) tract (Blum et al., 2002; Wohlgemuth et al., 2009). Recently, animal experiments and human studies suggest that therapeutic manipulation of the balance between beneficial and detrimental intestinal bacterial species can influence health and disease (Fitzpatrick, 2013). The known mechanisms of probiosis include manipulation of intestinal microbial communities, suppression of pathogens, immunomodulation, activation of anti-apoptotic genes in human or mouse intestinal epithelial cells from cytokine induced apoptosis, differentiation and fortification of the intestinal barrier (Thomas and Versalovic, 2010). For example, simultaneous treatment with probiotic Streptococcus thermophilus ATCC19258 and Lactobacillus acidophilus ATCC 4356, prevent invasion of entero-invasive E. coli and enhance the intestinal epithelial barrier function by amplifying the phosphorylation of occludin and ZO-1 together with a reduction of pro-inflammatory responses in vitro (Resta-Lenert and Barrett, 2003). Another similar study also demonstrated that application of probiotic E. coli NISSLE (EcN) is able to mediate up-regulation of ZO-1 expression in murine IECs and confer protection from the Dextran sodium sulphate (DSS) colitis-associated increase in mucosal permeability to mice luminal substances (Ukena et al., 2007).
Loss of tolerance to the patient’s own commensal microbiota has been implicated in the development of IBD (Wu et al., 2010a). Use of probiotics, to shift the existing microbiota balance in favor of protective microbial species and to treat IBD, has been extensively reviewed (Ochoa-Reparaz et al., 2009). The ability of some probiotics to synthesize bacteriocins (Awaisheh et al., 2013) or to induce the secretion of antibacterial cryptidins by Paneth cells (Hooper et al., 2003; Ayabe et al., 2004) could account for such changes in microbiota composition or even for the protection against pathogenic bacteria. In addition to the effects mediated by bacteria–bacteria interactions, probiotics may have a direct effect on host physiology. In the inflamed gut, the down-regulation of pro-inflammatory cytokines by probiotics may be an important factor for the observed improvement of symptoms (Figure 2; Ma et al., 2004). For example, Lactobacillus casei DN-114001 treatment increases the number of CD4+FoxP3+ regulatory T cells in mesenteric lymph nodes (mLN), decreases the production of the pro-inflammatory cytokines TNF-α and IFN-γ, changes the gut microbiota composition and prevents DSS induced colitis in BALB/c mice (Zakostelska et al., 2011). However, only few molecular mechanisms underlying probiotic action have so far been identified. Activation of TLR9 by bacterial DNA has been proposed as one possible mechanism of a probiotic-mediated amelioration of experimental colitis (Rachmilewitz et al., 2004). TLRs belong to highly conserved receptors of the innate immune system. TLR activation results in the translocation of the nuclear factor NFκB into the cell nucleus triggering transcription of immunorelevant genes (Cario and Podolsky, 2005). In addition, L. casei inhibits post-transcription of pro-inflammatory interferon γ-induced protein 10 (IP-10) in intestinal epithelial cells of colitic IL-10 knock-out mice (Hormannsperger et al., 2009).
An intact intestinal epithelial cell layer is of utmost importance for preventing the uncontrolled intrusion of pathogenic bacteria. However, pathogenic bacteria are capable of compromising the integrity of the epithelium by disrupting the tight junctions between epithelial cells (Berkes et al., 2003). Bacterial factors improving epithelial integrity have been identified for the probiotic Lactobacillus GG. This strain produces two soluble proteins (p40 and p75) which protect epithelial cells from apoptosis and thereby increase mucosal integrity. The secreted proteins activate anti-apoptotic protein kinase B (PKB/Akt) in a phosphatidylinositol-3′-kinase (PI3K)-dependent pathway and inhibit the pro-apoptotic p38/mitogen-activated protein kinase (MAPK; Yan et al., 2007).
Similarly, the biogenic amine, histamine, produced by decarboxylation of amino acid L-histidine by histidine decaxboxylase gene cluster (hdc) in Lactobacillus reuteri ATCC 6475 showed immunomodulatory effects by suppressing TNF production in myeloid progenitor cell lines (Figure 3) whereas the L. reuteri lacking hdc gene cluster was unable to suppress the pro-inflammatory cytokine TNF. The bacterial derived histamine binds to and activates histamine receptor H2 (HRH2) and there by inhibits MEK/ERK MAPK signaling pathway and presumably suppress TNF transcription and Ap-1 translocation (Thomas et al., 2012). These findings clearly demonstrate that bacterial interactions directly or indirectly have an impact on host physiology. Therefore, in the current review we mainly focused on the different beneficial bacteria and their metabolites on immunoregulation of the host.
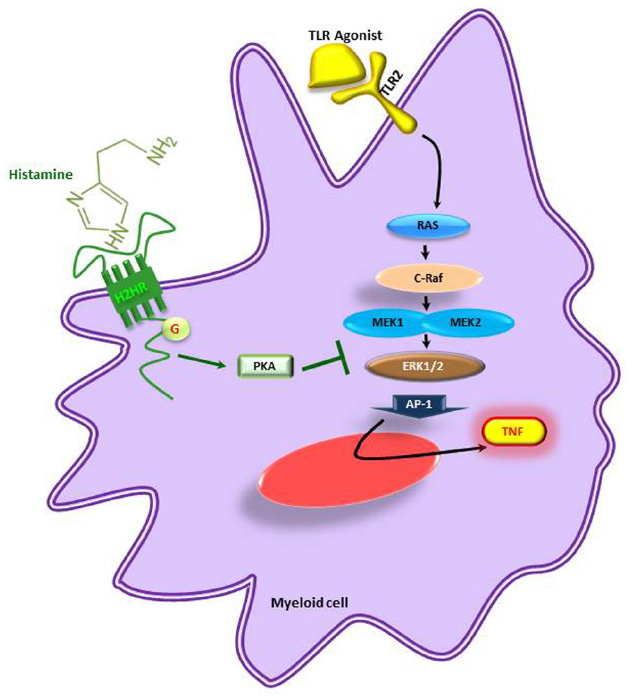
Figure 3. Microbe-derived histamine mediated suppression of pro-inflammatory cytokines. TNF is suppressed by inhibition of the MEK/ERK pathway in myeloid cells. H2HR, histamine receptor 2; PKA, activated protein kinase A; TNF, tumor necrosis factor; TLR, toll-like receptor. Adapted from Thomas et al. (2012).
Conclusion
In the presented review we demonstrated how probiotic bacteria or their metabolites regulate immunomodulatory effects on the host health. Probiotics have been proposed as preventive and therapeutic measures in order to restore the healthy microbiota composition and function of the GI tract. Additionally restoring the current balance is very important because the commensal bacteria are important source of vitamins, amino acids and lipid homeostasis and alternation in the levels of these metabolites might have an influence on the immune system (Brestoff and Artis, 2013). Therefore, therapeutic manipulations of intestinal bacteria by selectively altering the beneficial versus detrimental species by probiotics and or prebiotics administration could reverse the inflammatory responses and restore mucosal homeostasis. Future challenges include interrogations of molecular mechanisms through nutrients and beneficial bacterial metabolites, regulate immune response and linking the commensal bacteria-beneficial probiotic bacteria-metabolite-immune system axis in the content of health and diseases, may provide useful insights for the development of improved, preventive and therapeutically cost-effective and non-toxic approaches to treating different disorders mainly IBD.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The work was supported in part by research support from the National Institutes of Health (R01 AT004326, R01 DK065075, U01 CA 170930, and UH3 DK 083990). We also acknowledge the support of the NIH (P30 DK56338) for the Texas Medical Center Digestive Diseases Center.
References
Arena, M. P., Russo, P., Capozzi, V., Lopez, P., Fiocco, D., and Spano, G. (2014). Probiotic abilities of riboflavin-overproducing Lactobacillus strains: a novel promising application of probiotics. Appl. Microbiol. Biotechnol. 98, 7569–7581. doi: 10.1007/s00253-014-5837-x
Astbury, S. M., and Corfe, B. M. (2012). Uptake and metabolism of the short-chain fatty acid butyrate, a critical review of the literature. Curr. Drug Metab. 13, 815–821. doi: 10.2174/138920012800840428
Awaisheh, S. S., Al-Nabulsi, A. A., Osaili, T. M., Ibrahim, S., and Holley, R. (2013). Inhibition of Cronobacter sakazakii by heat labile bacteriocins produced by probiotic LAB isolated from healthy infants. J. Food Sci. 78, M1416–M1420. doi: 10.1111/1750-3841.12209
Ayabe, T., Ashida, T., Kohgo, Y., and Kono, T. (2004). The role of Paneth cells and their antimicrobial peptides in innate host defense. Trends Microbiol. 12, 394–398. doi: 10.1016/j.tim.2004.06.007
Backhed, F., Ley, R. E., Sonnenburg, J. L., Peterson, D. A., and Gordon, J. I. (2005). Host-bacterial mutualism in the human intestine. Science 307, 1915–1920. doi: 10.1126/science.1104816
Baumgart, M., Dogan, B., Rishniw, M., Weitzman, G., Bosworth, B., Yantiss, R., et al. (2007). Culture independent analysis of ileal mucosa reveals a selective increase in invasive Escherichia coli of novel phylogeny relative to depletion of Clostridiales in Crohn’s disease involving the ileum. ISME J. 1, 403–418. doi: 10.1038/ismej.2007.52
Becker, N., Kunath, J., Loh, G., and Blaut, M. (2011). Human intestinal microbiota: characterization of a simplified and stable gnotobiotic rat model. Gut Microbes 2, 25–33. doi: 10.4161/gmic.2.1.14651
Berkes, J., Viswanathan, V. K., Savkovic, S. D., and Hecht, G. (2003). Intestinal epithelial responses to enteric pathogens: effects on the tight junction barrier, ion transport, and inflammation. Gut 52, 439–451. doi: 10.1136/gut.52.3.439
Berni Canani, R., Sangwan, N., Stefka, A. T., Nocerino, R., Paparo, L., Aitoro, R., et al. (2015). Lactobacillus rhamnosus GG-supplemented formula expands butyrate-producing bacterial strains in food allergic infants. ISME J. 1–9. doi: 10.1038/ismej.2015.151 [Epub ahead of print].
Best, J., Nijhout, H. F., and Reed, M. (2010). Serotonin synthesis, release and reuptake in terminals: a mathematical model. Theor. Biol. Med. Model. 7, 34. doi: 10.1186/1742-4682-7-34
Birt, D. F., Hendrich, S., and Wang, W. (2001). Dietary agents in cancer prevention: flavonoids and isoflavonoids. Pharmacol. Ther. 90, 157–177. doi: 10.1016/S0163-7258(01)00137-1
Blaut, M. (2013). Ecology and physiology of the intestinal tract. Curr. Top. Microbiol. Immunol. 358, 247–272. doi: 10.1007/82_2011_192
Blum, S., Haller, D., Pfeifer, A., and Schiffrin, E. J. (2002). Probiotics and immune response. Clin. Rev. Allergy Immunol. 22, 287–309. doi: 10.1385/CRIAI:22:3:287
Borchers, A. T., Selmi, C., Meyers, F. J., Keen, C. L., and Gershwin, M. E. (2009). Probiotics and immunity. J. Gastroenterol. 44, 26–46. doi: 10.1007/s00535-008-2296-0
Bravo, J. A., Forsythe, P., Chew, M. V., Escaravage, E., Savignac, H. M., Dinan, T. G., et al. (2011). Ingestion of Lactobacillus strain regulates emotional behavior and central GABA receptor expression in a mouse via the vagus nerve. Proc. Natl. Acad. Sci. U.S.A. 108, 16050–16055. doi: 10.1073/pnas.1102999108
Brestoff, J. R., and Artis, D. (2013). Commensal bacteria at the interface of host metabolism and the immune system. Nat. Immunol. 14, 676–684. doi: 10.1038/ni.2640
Budnowski, J., Hanschen, F. S., Lehmann, C., Haack, M., Brigelius-Flohe, R., Kroh, L. W., et al. (2013). A derivatization method for the simultaneous detection of glucosinolates and isothiocyanates in biological samples. Anal. Biochem. 441, 199–207. doi: 10.1016/j.ab.2013.07.002
Cain, A. M., and Karpa, K. D. (2011). Clinical utility of probiotics in inflammatory bowel disease. Altern. Ther. Health Med. 17, 72–79.
Cani, P. D., Everard, A., and Duparc, T. (2013). Gut microbiota, enteroendocrine functions and metabolism. Curr. Opin. Pharmacol. 13, 935–940. doi: 10.1016/j.coph.2013.09.008
Cario, E., and Podolsky, D. K. (2005). Intestinal epithelial TOLLerance versus inTOLLerance of commensals. Mol. Immunol. 42, 887–893. doi: 10.1016/j.molimm.2004.12.002
Chang, Y. C., and Nair, M. G. (1995). Metabolism of daidzein and genistein by intestinal bacteria. J. Nat. Prod. 58, 1892–1896. doi: 10.1021/np50126a014
Chassaing, B., Koren, O., Carvalho, F. A., Ley, R. E., and Gewirtz, A. T. (2014). AIEC pathobiont instigates chronic colitis in susceptible hosts by altering microbiota composition. Gut 63, 1069–1080. doi: 10.1136/gutjnl-2013-304909
Corfield, A. P., Myerscough, N., Longman, R., Sylvester, P., Arul, S., and Pignatelli, M. (2000). Mucins and mucosal protection in the gastrointestinal tract: new prospects for mucins in the pathology of gastrointestinal disease. Gut 47, 589–594. doi: 10.1136/gut.47.4.589
Corfield, A. P., Wagner, S. A., Clamp, J. R., Kriaris, M. S., and Hoskins, L. C. (1992). Mucin degradation in the human colon—production of sialidase, sialate O-acetylesterase, N-acetylneuraminate lyase, arylesterase, and glycosulfatase activities by strains of fecal bacteria. Infect. Immun. 60, 3971–3978.
Corr, S. C., Li, Y., Riedel, C. U., O’Toole, P. W., Hill, C., and Gahan, C. G. (2007). Bacteriocin production as a mechanism for the antiinfective activity of Lactobacillus salivarius UCC118. Proc. Natl. Acad. Sci. U.S.A. 104, 7617–7621. doi: 10.1073/pnas.0700440104
Cummings, J. H., and Macfarlane, G. T. (1997). Role of intestinal bacteria in nutrient metabolism. JPEN J. Parenter. Enteral Nutr. 21, 357–365. doi: 10.1177/0148607197021006357
Dao, M. C., Everard, A., Aron-Wisnewsky, J., Sokolovska, N., Prifti, E., Verger, E. O., et al. (2015). Akkermansia muciniphila and improved metabolic health during a dietary intervention in obesity: relationship with gut microbiome richness and ecology. Gut doi: 10.1136/gutjnl-2014-308778 [Epub ahead of print].
de Jong, E. C., Vieira, P. L., Kalinski, P., Schuitemaker, J. H., Tanaka, Y., Wierenga, E. A., et al. (2002). Microbial compounds selectively induce Th1 cell-promoting or Th2 cell-promoting dendritic cells in vitro with diverse Th cell-polarizing signals. J. Immunol. 168, 1704–1709. doi: 10.4049/jimmunol.168.4.1704
Desbonnet, L., Garrett, L., Clarke, G., Bienenstock, J., and Dinan, T. G. (2008). The probiotic Bifidobacteria infantis: an assessment of potential antidepressant properties in the rat. J. Psychiatr. Res. 43, 164–174. doi: 10.1016/j.jpsychires.2008.03.009
Desbonnet, L., Garrett, L., Clarke, G., Kiely, B., Cryan, J. F., and Dinan, T. G. (2010). Effects of the probiotic Bifidobacterium infantis in the maternal separation model of depression. Neuroscience 170, 1179–1188. doi: 10.1016/j.neuroscience.2010.08.005
Diaz Heijtz, R., Wang, S., Anuar, F., Qian, Y., Bjorkholm, B., Samuelsson, A., et al. (2011). Normal gut microbiota modulates brain development and behavior. Proc. Natl. Acad. Sci. U.S.A. 108, 3047–3052. doi: 10.1073/pnas.1010529108
Dylag, K., Hubalewska-Mazgaj, M., Surmiak, M., Szmyd, J., and Brzozowski, T. (2014). Probiotics in the mechanism of protection against gut inflammation and therapy of gastrointestinal disorders. Curr. Pharm. Des. 20, 1149–1155. doi: 10.2174/13816128113199990422
Elfoul, L., Rabot, S., Khelifa, N., Quinsac, A., Duguay, A., and Rimbault, A. (2001). Formation of allyl isothiocyanate from sinigrin in the digestive tract of rats monoassociated with a human colonic strain of Bacteroides thetaiotaomicron. FEMS Microbiol. Lett. 197, 99–103. doi: 10.1111/j.1574-6968.2001.tb10589.x
Everard, A., Belzer, C., Geurts, L., Ouwerkerk, J. P., Druart, C., Bindels, L. B., et al. (2013). Cross-talk between Akkermansia muciniphila and intestinal epithelium controls diet-induced obesity. Proc. Natl. Acad. Sci. U.S.A. 110, 9066–9071. doi: 10.1073/pnas.1219451110
Everard, A., and Cani, P. D. (2013). Diabetes, obesity and gut microbiota. Best Pract. Res. Clin. Gastroenterol. 27, 73–83. doi: 10.1016/j.bpg.2013.03.007
Fayol-Messaoudi, D., Berger, C. N., Coconnier-Polter, M. H., Lievin-Le Moal, V., and Servin, A. L. (2005). pH-, Lactic acid-, and non-lactic acid-dependent activities of probiotic Lactobacilli against Salmonella enterica Serovar Typhimurium. Appl. Environ. Microbiol. 71, 6008–6013. doi: 10.1128/AEM.71.10.6008-6013.2005
Feng, T., Wang, L., Schoeb, T. R., Elson, C. O., and Cong, Y. (2010). Microbiota innate stimulation is a prerequisite for T cell spontaneous proliferation and induction of experimental colitis. J. Exp. Med. 207, 1321–1332. doi: 10.1084/jem.20092253
Ferreira, C. M., Vieira, A. T., Vinolo, M. A., Oliveira, F. A., Curi, R., and Martins Fdos, S. (2014). The central role of the gut microbiota in chronic inflammatory diseases. J. Immunol. Res. 2014, 689492. doi: 10.1155/2014/689492
Fitzpatrick, L. R. (2013). Probiotics for the treatment of Clostridium difficile associated disease. World J. Gastrointest. Pathophysiol. 4, 47–52. doi: 10.4291/wjgp.v4.i3.47
Fleissner, C. K., Huebel, N., Abd El-Bary, M. M., Loh, G., Klaus, S., and Blaut, M. (2010). Absence of intestinal microbiota does not protect mice from diet-induced obesity. Br. J. Nutr. 104, 919–929. doi: 10.1017/S0007114510001303
Frank, D. N., St Amand, A. L., Feldman, R. A., Boedeker, E. C., Harpaz, N., and Pace, N. R. (2007). Molecular-phylogenetic characterization of microbial community imbalances in human inflammatory bowel diseases. Proc. Natl. Acad. Sci. U.S.A. 104, 13780–13785. doi: 10.1073/pnas.0706625104
Fung, K. Y., Cosgrove, L., Lockett, T., Head, R., and Topping, D. L. (2012). A review of the potential mechanisms for the lowering of colorectal oncogenesis by butyrate. Br. J. Nutr. 108, 820–831. doi: 10.1017/S0007114512001948
Galdeano, C. M., Nuñez, I. N., Carmuega, E., de LeBlanc, A. D., and Perdigón, G. (2015). Role of probiotics and functional foods in health: gut immune stimulation by two probiotic strains and a potential probiotic yoghurt. Endocr. Metab. Immune Disord. Drug Targets 15, 37–45.
Ganesh, B. P., Klopfleisch, R., Loh, G., and Blaut, M. (2013). Commensal Akkermansia muciniphila exacerbates gut inflammation in Salmonella Typhimurium-infected gnotobiotic mice. PLoS ONE 8:e74963. doi: 10.1371/journal.pone.0074963
Ganesh, B. P., Richter, J. F., Blaut, M., and Loh, G. (2012). Enterococcus faecium NCIMB 10415 does not protect interleukin-10 knock-out mice from chronic gut inflammation. Benef. Microbes 3, 43–50. doi: 10.3920/BM2011.0050
Geller, S. E., and Studee, L. (2006). Soy and red clover for mid-life and aging. Climacteric 9, 245–263. doi: 10.1080/13697130600736934
Hemarajata, P., Gao, C., Pflughoeft, K. J., Thomas, C. M., Saulnier, D. M., Spinler, J. K., et al. (2013). Lactobacillus reuteri-specific immunoregulatory gene rsiR modulates histamine production and immunomodulation by Lactobacillus reuteri. J. Bacteriol. 195, 5567–5576. doi: 10.1128/JB.00261-13
Hemarajata, P., and Versalovic, J. (2013). Effects of probiotics on gut microbiota: mechanisms of intestinal immunomodulation and neuromodulation. Therap. Adv. Gastroenterol. 6, 39–51. doi: 10.1177/1756283X12459294
Hooper, L. V., Stappenbeck, T. S., Hong, C. V., and Gordon, J. I. (2003). Angiogenins: a new class of microbicidal proteins involved in innate immunity. Nat. Immunol. 4, 269–273. doi: 10.1038/ni888
Hormannsperger, G., Clavel, T., Hoffmann, M., Reiff, C., Kelly, D., Loh, G., et al. (2009). Post-translational inhibition of IP-10 secretion in IEC by probiotic bacteria: impact on chronic inflammation. PLoS ONE 4:e4365. doi: 10.1371/journal.pone.0004365
Isolauri, E., Rautava, S., and Salminen, S. (2012). Probiotics in the development and treatment of allergic disease. Gastroenterol. Clin. North Am. 41, 747–762. doi: 10.1016/j.gtc.2012.08.007
Jeon, S. G., Kayama, H., Ueda, Y., Takahashi, T., Asahara, T., Tsuji, H., et al. (2012). Probiotic Bifidobacterium breve induces IL-10-producing Tr1 cells in the colon. PLoS Pathog. 8:e1002714. doi: 10.1371/journal.ppat.1002714
Kaplan, M. H., Wurster, A. L., and Grusby, M. J. (1998). A signal transducer and activator of transcription (Stat)4-independent pathway for the development of T helper type 1 cells. J. Exp. Med. 188, 1191–1196. doi: 10.1084/jem.188.6.1191
Ki, Y., Kim, W., Cho, H., Ahn, K., Choi, Y., and Kim, D. (2014). The effect of probiotics for preventing radiation-induced morphological changes in intestinal mucosa of rats. J. Korean Med. Sci. 29, 1372–1378. doi: 10.3346/jkms.2014.29.10.1372
Kim, S. C., Tonkonogy, S. L., Karrasch, T., Jobin, C., and Sartor, R. B. (2007). Dual-association of gnotobiotic IL-10–/– mice with 2 nonpathogenic commensal bacteria induces aggressive pancolitis. Inflamm. Bowel Dis. 13, 1457–1466. doi: 10.1002/ibd.20246
Klaenhammer, T. R., Kleerebezem, M., Kopp, M. V., and Rescigno, M. (2012). The impact of probiotics and prebiotics on the immune system. Nat. Rev. Immunol. 12, 728–734. doi: 10.1038/nri3312
Kotlowski, R., Bernstein, C. N., Sepehri, S., and Krause, D. O. (2007). High prevalence of Escherichia coli belonging to the B2+D phylogenetic group in inflammatory bowel disease. Gut 56, 669–675. doi: 10.1136/gut.2006.099796
Krul, C., Humblot, C., Philippe, C., Vermeulen, M., van Nuenen, M., Havenaar, R., et al. (2002). Metabolism of sinigrin (2-propenyl glucosinolate) by the human colonic microflora in a dynamic in vitro large-intestinal model. Carcinogenesis 23, 1009–1016. doi: 10.1093/carcin/23.6.1009
Lepri, S. R., Zanelatto, L. C., da Silva, P. B., Sartori, D., Ribeiro, L. R., and Mantovani, M. S. (2014). Effects of genistein and daidzein on cell proliferation kinetics in HT29 colon cancer cells: the expression of CTNNBIP1 (beta-catenin), APC (adenomatous polyposis coli) and BIRC5 (survivin). Hum. Cell 27, 78–84. doi: 10.1007/s13577-012-0051-6
Li, J., Jia, H., Cai, X., Zhong, H., Feng, Q., Sunagawa, S., et al. (2014). An integrated catalog of reference genes in the human gut microbiome. Nat. Biotechnol. 32, 834–841. doi: 10.1038/nbt.2942
Lippmann, D., Lehmann, C., Florian, S., Barknowitz, G., Haack, M., Mewis, I., et al. (2014). Glucosinolates from pak choi and broccoli induce enzymes and inhibit inflammation and colon cancer differently. Food Funct. 5, 1073–1081. doi: 10.1039/c3fo60676g
Louis, P., Hold, G. L., and Flint, H. J. (2014). The gut microbiota, bacterial metabolites and colorectal cancer. Nat. Rev. Microbiol. 12, 661–672. doi: 10.1038/nrmicro3344
Ma, D., Forsythe, P., and Bienenstock, J. (2004). Live Lactobacillus reuteri is essential for the inhibitory effect on tumor necrosis factor alpha-induced interleukin-8 expression. Infect. Immun. 72, 5308–5314. doi: 10.1128/IAI.72.9.5308-5314.2004
Mabrok, H. B., Klopfleisch, R., Ghanem, K. Z., Clavel, T., Blaut, M., and Loh, G. (2012). Lignan transformation by gut bacteria lowers tumor burden in a gnotobiotic rat model of breast cancer. Carcinogenesis 33, 203–208. doi: 10.1093/carcin/bgr256
Mack, D. R., Michail, S., Wei, S., McDougall, L., and Hollingsworth, M. A. (1999). Probiotics inhibit enteropathogenic E. coli adherence in vitro by inducing intestinal mucin gene expression. Am. J. Physiol. 276(4 Pt 1), G941–G950.
Matthews, G. M., Howarth, G. S., and Butler, R. N. (2012). Short-chain fatty acids induce apoptosis in colon cancer cells associated with changes to intracellular redox state and glucose metabolism. Chemotherapy 58, 102–109. doi: 10.1159/000335672
Matthies, A., Clavel, T., Gutschow, M., Engst, W., Haller, D., Blaut, M., et al. (2008). Conversion of daidzein and genistein by an anaerobic bacterium newly isolated from the mouse intestine. Appl. Environ. Microbiol. 74, 4847–4852. doi: 10.1128/AEM.00555-08
Matthies, A., Loh, G., Blaut, M., and Braune, A. (2012). Daidzein and genistein are converted to equol and 5-hydroxy-equol by human intestinal Slackia isoflavoniconvertens in gnotobiotic rats. J. Nutr. 142, 40–46. doi: 10.3945/jn.111.148247
McKenna, K., Beignon, A. S., and Bhardwaj, N. (2005). Plasmacytoid dendritic cells: linking innate and adaptive immunity. J. Virol. 79, 17–27. doi: 10.1128/JVI.79.1.17-27.2005
Morelli, L., and Capurso, L. (2012). FAO/WHO Guidelines on probiotics 10 years later FOREWORD. J. Clin. Gastroenterol. 46, S1–S2. doi: 10.1097/MCG.0b013e318269fdd5
Ochoa-Reparaz, J., Mielcarz, D. W., Ditrio, L. E., Burroughs, A. R., Foureau, D. M., Haque-Begum, S., et al. (2009). Role of gut commensal microflora in the development of experimental autoimmune encephalomyelitis. J. Immunol. 183, 6041–6050. doi: 10.4049/jimmunol.0900747
Packey, C. D., and Sartor, R. B. (2009). Commensal bacteria, traditional and opportunistic pathogens, dysbiosis and bacterial killing in inflammatory bowel diseases. Curr. Opin. Infect. Dis. 22, 292–301. doi: 10.1097/QCO.0b013e32832a8a5d
Peran, L., Camuesco, D., Comalada, M., Nieto, A., Concha, A., Adrio, J. L., et al. (2006). Lactobacillus fermentum, a probiotic capable to release glutathione, prevents colonic inflammation in the TNBS model of rat colitis. Int. J. Colorectal. Dis. 21, 737–746. doi: 10.1007/s00384-005-0773-y
Pflughoeft, K. J., and Versalovic, J. (2012). Human microbiome in health and disease. Annu. Rev. Pathol. 7, 99–122. doi: 10.1146/annurev-pathol-011811-132421
Qin, J., Li, R., Raes, J., Arumugam, M., Burgdorf, K. S., Manichanh, C., et al. (2010). A human gut microbial gene catalogue established by metagenomic sequencing. Nature 464, 59–65. doi: 10.1038/nature08821
Rachmilewitz, D., Katakura, K., Karmeli, F., Hayashi, T., Reinus, C., Rudensky, B., et al. (2004). Toll-like receptor 9 signaling mediates the anti-inflammatory effects of probiotics in murine experimental colitis. Gastroenterology 126, 520–528. doi: 10.1053/j.gastro.2003.11.019
Rafii, F., Davis, C., Park, M., Heinze, T. M., and Beger, R. D. (2003). Variations in metabolism of the soy isoflavonoid daidzein by human intestinal microfloras from different individuals. Arch. Microbiol. 180, 11–16. doi: 10.1007/s00203-003-0551-6
Repa, A., Thanhaeuser, M., Endress, D., Weber, M., Kreissl, A., Binder, C., et al. (2014). Probiotics (Lactobacillus acidophilus and Bifidobacterium bifidum) prevent NEC in VLBW infants fed breast milk but not formula. Pediatr. Res. 77, 381–388. doi: 10.1038/pr.2014.192
Resta-Lenert, S., and Barrett, K. E. (2003). Live probiotics protect intestinal epithelial cells from the effects of infection with enteroinvasive Escherichia coli (EIEC). Gut 52, 988–997. doi: 10.1136/gut.52.7.988
Roediger, W. E., Duncan, A., Kapaniris, O., and Millard, S. (1993). Reducing sulfur compounds of the colon impair colonocyte nutrition: implications for ulcerative colitis. Gastroenterology 104, 802–809.
Round, J. L., Lee, S. M., Li, J., Tran, G., Jabri, B., Chatila, T. A., et al. (2011). The Toll-like receptor 2 pathway establishes colonization by a commensal of the human microbiota. Science 332, 974–977. doi: 10.1126/science.1206095
Russell, R. M., Sharp, F. C., Rasko, D. A., and Sperandio, V. (2007). QseA and GrlR/GrlA regulation of the locus of enterocyte effacement genes in enterohemorrhagic Escherichia coli. J. Bacteriol. 189, 5387–5392. doi: 10.1128/JB.00553-07
Sah, B. N., Vasiljevic, T., McKechnie, S., and Donkor, O. N. (2014). Effect of probiotics on antioxidant and antimutagenic activities of crude peptide extract from yogurt. Food Chem. 156, 264–270. doi: 10.1016/j.foodchem.2014.01.105
Salazar, N., Dewulf, E. M., Neyrinck, A. M., Bindels, L. B., Cani, P. D., Mahillon, J., et al. (2014). Inulin-type fructans modulate intestinal Bifidobacterium species populations and decrease fecal short-chain fatty acids in obese women. Clin. Nutr. 34, 501–507. doi: 10.1016/j.clnu.2014.06.001
Sanders, M. E., Lenoir-Wijnkoop, I., Salminen, S., Merenstein, D. J., Gibson, G. R., Petschow, B. W., et al. (2014). Probiotics and prebiotics: prospects for public health and nutritional recommendations. Ann. N. Y. Acad. Sci. 1309, 19–29. doi: 10.1111/nyas.12377
Saulnier, D. M., Santos, F., Roos, S., Mistretta, T. A., Spinler, J. K., Molenaar, D., et al. (2011). Exploring metabolic pathway reconstruction and genome-wide expression profiling in Lactobacillus reuteri to define functional probiotic features. PLoS ONE 6:e18783. doi: 10.1371/journal.pone.0018783
Scalbert, A., Manach, C., Morand, C., Remesy, C., and Jimenez, L. (2005). Dietary polyphenols and the prevention of diseases. Crit. Rev. Food Sci. Nutr. 45, 287–306. doi: 10.1080/1040869059096
Sharma, R., Young, C., and Neu, J. (2010). Molecular modulation of intestinal epithelial barrier: contribution of microbiota. J. Biomed. Biotechnol. 2010, 305879. doi: 10.1155/2010/305879
Sherman, P. M., Ossa, J. C., and Johnson-Henry, K. (2009). Unraveling mechanisms of action of probiotics. Nutr. Clin. Pract. 24, 10–14. doi: 10.1177/0884533608329231
Sokol, H., Pigneur, B., Watterlot, L., Lakhdari, O., Bermudez-Humaran, L. G., Gratadoux, J. J., et al. (2008). Faecalibacterium prausnitzii is an anti-inflammatory commensal bacterium identified by gut microbiota analysis of Crohn disease patients. Proc. Natl. Acad. Sci. U.S.A. 105, 16731–16736. doi: 10.1073/pnas.0804812105
Swidsinski, A., Khilkin, M., Kerjaschki, D., Schreiber, S., Ortner, M., Weber, J., et al. (1998). Association between intraepithelial Escherichia coli and colorectal cancer. Gastroenterology 115, 281–286. doi: 10.1016/S0016-5085(98)70194-5
Swidsinski, A., Loening-Baucke, V., Vaneechoutte, M., and Doerffel, Y. (2008). Active Crohn’s disease and ulcerative colitis can be specifically diagnosed and monitored based on the biostructure of the fecal flora. Inflamm. Bowel Dis. 14, 147–161. doi: 10.1002/ibd.20330
Thomas, C. M., Hong, T., van Pijkeren, J. P., Hemarajata, P., Trinh, D. V., Hu, W., et al. (2012). Histamine derived from probiotic Lactobacillus reuteri suppresses TNF via modulation of PKA and ERK signaling. PLoS ONE 7:e31951. doi: 10.1371/journal.pone.0031951
Thomas, C. M., and Versalovic, J. (2010). Probiotics-host communication: modulation of signaling pathways in the intestine. Gut Microbes 1, 148–163. doi: 10.4161/gmic.1.3.11712
Ukena, S. N., Singh, A., Dringenberg, U., Engelhardt, R., Seidler, U., Hansen, W., et al. (2007). Probiotic Escherichia coli Nissle 1917 inhibits leaky gut by enhancing mucosal integrity. PLoS ONE 2:e1308. doi: 10.1371/journal.pone.0001308
Ukibe, K., Miyoshi, M., and Kadooka, Y. (2015). Administration of Lactobacillus gasseri SBT2055 suppresses macrophage infiltration into adipose tissue in diet-induced obese mice. Br. J. Nutr. 114, 1180–1187. doi: 10.1017/S0007114515002627
Usui, T. (2006). Pharmaceutical prospects of phytoestrogens. Endocr. J. 53, 7–20. doi: 10.1507/endocrj.53.7
Valeur, N., Engel, P., Carbajal, N., Connolly, E., and Ladefoged, K. (2004). Colonization and immunomodulation by Lactobacillus reuteri ATCC 55730 in the human gastrointestinal tract. Appl. Environ. Microbiol. 70, 1176–1181. doi: 10.1128/AEM.70.2.1176-1181.2004
Veiga, P., Gallini, C. A., Beal, C., Michaud, M., Delaney, M. L., DuBois, A., et al. (2010). Bifidobacterium animalis subsp. lactis fermented milk product reduces inflammation by altering a niche for colitogenic microbes. Proc. Natl. Acad. Sci. U.S.A. 107, 18132–18137. doi: 10.1073/pnas.1011737107
Vieira, A. T., Galvao, I., Amaral, F. A., Teixeira, M. M., Nicoli, J. R., and Martins, F. S. (2015). Oral treatment with Bifidobacterium longum 51A reduced inflammation in a murine experimental model of gout. Benef. Microbes doi: 10.3920/BM2015.0015 [Epub ahead of print].
Wang, F. Y., Liu, J. M., Luo, H. H., Liu, A. H., and Jiang, Y. (2015). Potential protective effects of Clostridium butyricum on experimental gastric ulcers in mice. World J. Gastroenterol. 21, 8340–8351. doi: 10.3748/wjg.v21.i27.8340
Weng, H., Endo, K., Li, J., Kito, N., and Iwai, N. (2015). Induction of peroxisomes by butyrate-producing probiotics. PLoS ONE 10:e0117851. doi: 10.1371/journal.pone.0117851
Wohlgemuth, S., Haller, D., Blaut, M., and Loh, G. (2009). Reduced microbial diversity and high numbers of one single Escherichia coli strain in the intestine of colitic mice. Environ. Microbiol. 11, 1562–1571. doi: 10.1111/j.1462-2920.2009.01883.x
Woting, A., Pfeiffer, N., Loh, G., Klaus, S., and Blaut, M. (2014). Clostridium ramosum promotes high-fat diet-induced obesity in gnotobiotic mouse models. mBio 5, e01530-14. doi: 10.1128/mBio.01530-14
Wu, H. J., Ivanov, I. I., Darce, J., Hattori, K., Shima, T., Umesaki, Y., et al. (2010a). Gut-residing segmented filamentous bacteria drive autoimmune arthritis via T helper 17 cells. Immunity 32, 815–827. doi: 10.1016/j.immuni.2010.06.001
Wu, X., Zhu, Y., Yan, H., Liu, B., Li, Y., Zhou, Q., et al. (2010b). Isothiocyanates induce oxidative stress and suppress the metastasis potential of human non-small cell lung cancer cells. BMC Cancer 10:269. doi: 10.1186/1471-2407-10-269
Yan, F., Cao, H., Cover, T. L., Whitehead, R., Washington, M. K., and Polk, D. B. (2007). Soluble proteins produced by probiotic bacteria regulate intestinal epithelial cell survival and growth. Gastroenterology 132, 562–575. doi: 10.1053/j.gastro.2006.11.022
Zakostelska, Z., Kverka, M., Klimesova, K., Rossmann, P., Mrazek, J., Kopecny, J., et al. (2011). Lysate of probiotic Lactobacillus casei DN-114 001 ameliorates colitis by strengthening the gut barrier function and changing the gut microenvironment. PLoS ONE 6:e27961. doi: 10.1371/journal.pone.0027961
Keywords: probiotics, metabolites, commensal bacteria, immunomodulation, diet, dietary compounds, microbiome
Citation: Ganesh BP and Versalovic J (2015) Luminal Conversion and Immunoregulation by Probiotics. Front. Pharmacol. 6:269. doi: 10.3389/fphar.2015.00269
Received: 21 July 2015; Accepted: 26 October 2015;
Published: 12 November 2015.
Edited by:
Irene Lenoir-Wijnkoop, Utrecht University, NetherlandsReviewed by:
Domenico Criscuolo, Genovax, ItalyJean Fioramonti, Institut National de la Recherche Agronomique, France
Copyright © 2015 Ganesh and Versalovic. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: James Versalovic, amFtZXN2QGJjbS5lZHU=
 Bhanu Priya Ganesh
Bhanu Priya Ganesh