- 1 Dipartimento di Patologia Generale, Seconda Università degli Studi di Napoli, Napoli, Italy
- 2 National Centre of Research, Institute of Genetics and Biophysics, CNR-IGB, Napoli, Italy
Sirtuins represent a promising new class of conserved histone deacetylases, originally identified in yeast. The activity of the sirtuin (SirT) family – made up of seven members (SirT1-7) – is NAD+ dependent. Sirtuins target a wide range of cellular proteins in nucleus, cytoplasm, and mitochondria for post-translational modification by acetylation (SirT1, 2, 3, and 5) or ADP-ribosylation (SirT4 and 6). Sirtuins regulate responses to stress and ensure that damaged DNA is not propagated, thus contrasting the accumulation of mutations. To date, sirtuins have emerged as potential therapeutic targets for treatment of human pathologies such as metabolic, cardiovascular and neurodegenerative diseases, and cancer. SirT1 is the founding member of this class of enzymes and is currently the best known of the group. SirT1 acts in various cellular processes, deacetylating both chromatin and non-histone proteins, and its role in cancer and aging has been extensively studied. SirT1 may play a critical role in tumor initiation and progression as well as drug resistance by blocking senescence and apoptosis, and by promoting cell growth and angiogenesis. Recently, growing interest in sirtuin modulation has led to the discovery and characterization of small molecules able to modify sirtuin activity. The present review highlights SirT mechanism(s) of action and deregulation in cancer, focusing on the therapeutic potential of SirT modulators both in cancer prevention and treatment.
Introduction
Epigenetic modifications play an important role in regulating gene expression, lifespan, and tumorigenesis. In particular, acetylation regulates a wide variety of cellular functions. Class III histone deacetylases (HDACs), also known as sirtuins, are the silent information regulator 2 (Sir2) family of proteins. Unlike other HDACs, sirtuins are NAD+-dependent deacetylases, not modulated by inhibitors of class I, II, and IV HDACs. The deacetylase activity of sirtuins is controlled by the cellular [NAD+]/[NADH] ratio, where NAD+ works as an activator, while nicotinamide and NADH act as inhibitors. Expressed from bacteria to humans (Vaquero, 2009), sirtuins seem to preferentially target non-histone proteins, although little is currently known about target specificity and selectivity. Two reactions may be catalyzed by sirtuins: deacetylation and ADP-ribosylation. In both reactions, the cleavage of NAD+ is the initial chemical step. Sirtuins (SirT1, SirT2, SirT3, SirT5, and SirT7) catalyze a deacetylation reaction on lysine residues of target proteins, using NAD+ as cofactor and releasing nicotinamide with the production of 2′-O-acetyl-ADP ribose. In contrast, SirT4 and SirT6 catalyze an ADP-ribosylation reaction, in which ADP-ribosyl moiety is transferred to the substrate (Yamamoto et al., 2007).
Catalytic Domain of Sirtuins
Each member of the sirtuin family contains a highly conserved core domain consisting of an NAD+ binding site and a catalytic domain (Alcaín and Villalba, 2009). Human SirT2 consists of two globular domains: one large and one small. In the large domain, with a Rossmann fold, six β-strands form a parallel β-sheet and six α-helices. The small domain is composed of a helical and a zinc-binding module. The active site is located at the interface between the large and small domains, with a binding site for NAD+. The NAD+-binding pocket can be divided into three spatially distinct regions: the A site, with affinity for adenine–ribose, the B site, with affinity for nicotinamide–ribose, and the C site, which binds NAD+. In the presence of an acetyl-lysine substrate, the NAD+-bound B site can undergo a conformational change, bringing nicotinamide in proximity to the C site, where it can be cleaved. The ADP ribose product returns to the B site, allowing deacetylation. The C site is the binding site for free nicotinamide. At high concentrations it may occupy the site and block the conformational change of NAD+ (North and Verdin, 2004).
Sirtuin Family
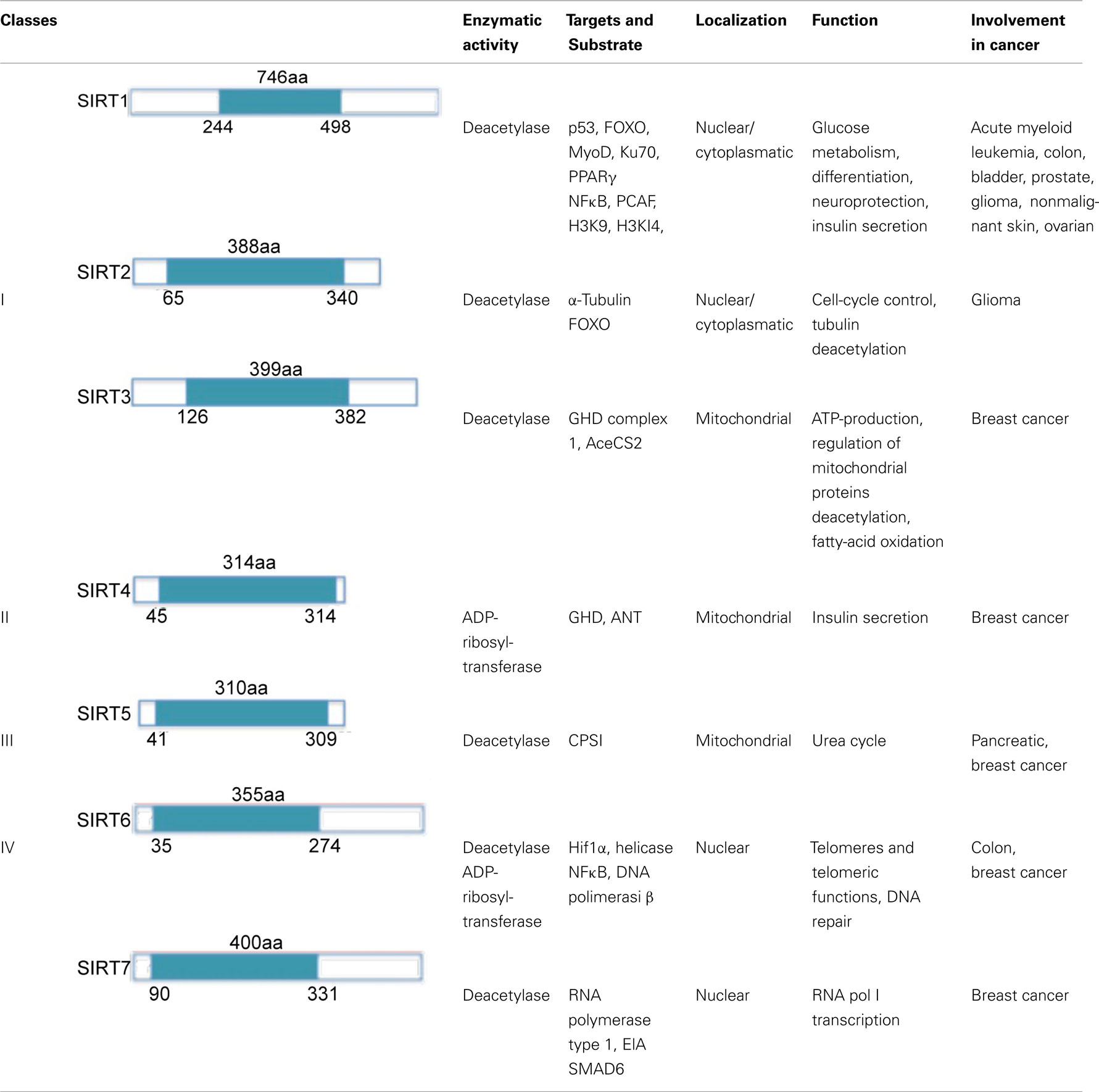
The mammalian sirtuin family comprises seven proteins (SirT1-7). These proteins differ in cellular localization, activity, and function, and are subdivided into four classes (Alcaín and Villalba, 2009; Table 1). SirT1, the closest to yeast Sir2 in terms of sequence, is a proto-member of the family. SirT1 mediates heterochromatin formation through deacetylation of K26 on histone H1, K9 on histone H3, and K16 on histone H4. SirT1 is also involved in the deacetylation of non-histone proteins, which can be divided into three groups: (i) transcription factors (p53, FOXO, E2F1, BCL6, p73, Rb, and others); (ii) DNA repair proteins; (iii) signaling factors. Due to its ability to deacetylate a variety of substrates, SirT1 is involved in a broad range of physiological functions, including control of gene expression, metabolism, tumorigenesis, and aging.
Human SirT2 co-localizes with microtubules in cytoplasm. Thus, knockdown of SirT2 results in tubulin hyperacetylation. SirT2 is also able to bind FOXO3a, reducing its acetylation levels.
SirT3 is localized in mitochondrial inner membrane, and is activated by caloric restriction. SirT4 is also mitochondrial, but no NAD+-dependent deacetylase activity has so far been detected for this sirtuin. SirT4 uses NAD+ for ADP-ribosylation of glutamate dehydrogenase, resulting in inhibition of insulin secretion and enzymatic activity of β-cells. The third mitochondrial sirtuin, SirT5, is localized in the inter-membrane space and can deacetylate cytochrome c. The last two sirtuins, SirT6 and SirT7, have recently been characterized. SirT6 exerts an NAD+-dependent histone H3 K9 deacetylase action able to modulate telomeric chromatin functions (Alcaín and Villalba, 2009). SirT6 has a role in genome stability. In mammalian cells subjected to oxidative stress, SirT6 is recruited on DNA double-strand breaks (DSB) and stimulates repair through both homologous and non-homologous end-joining recombination. Moreover, some results indicate that SirT6 is physically associated with poly [adenosine diphosphate (ADP)-ribose] polymerase 1 (PARP1). The amount of SirT6–PARP1 complexes increase after DNA damage. PARP1 is mono-ADP-ribosylated on lysine residue K521, thus increasing PARP1 activity and enhancing DSB repair under oxidative stress (Mao et al., 2011). SirT7 is a predominantly nucleolar protein associated with active rRNA genes (rDNA), where it interacts with RNA polymerase I and histones (Alcaín and Villalba, 2009). SirT7 does not seem to have any NAD+-dependent activity. Other functional characteristics of SirT6 and SirT7 remain to be further elucidated.
Some sirtuins may relocalize depending on cell or tissue type, developmental stage, stress condition, and metabolic status, suggesting that their localization is important for regulating function (McGuinness et al., 2011).
Sirtuins and Genome Stability
Some sirtuins (SirT1, 2, 3, 6) are linked to chromatin regulation, as they are responsible for the regulation of two histone post-translational modifications crucial for chromatin structure: acetylation of K16 and K9 on histone H4 and H3 respectively (McGuinness et al., 2011).
SirT1 is responsible for heterochromatin formation by a deacetylation process. Histone deacetylation can in turn facilitate histone methylation, thus enhancing global transcriptional repression. SirT1 has no effect on H3K4 methylation (a marker of open chromatin) and promotes H3K9 tri-methylation through its involvement with the histone methyltransferase Suv39H1 (Liu et al., 2009; Bosch-Presegue and Vaquero, 2011).
SirT2 and SirT3 are responsible for regulation of H4K16 acetylation. Deacetylation occurs specifically during cell-cycle in G2/M transition, in particular when SirT2 is shuttled to the nucleus (Shogren-Knaak et al., 2006). The mechanism of SirT3 is more complex. Although SirT3 is localized in mitochondria, its feeble nuclear expression is probably responsible for deacetylation of H4K16ac and H3K9ac in some settings (McGuinness et al., 2011). The finding that SirT3 knockdown does not lead to global H4K16 hyperacetylation suggests that its action occurs in limited regions of the genome.
Recently, SirT6 has been attributed with deacetylase activity (McGuinness et al., 2011), since it modulates acetylation of K9 and K56 on histone H3.
Sirtuins and Cancer
Interest in sirtuins has grown in the last decade, mainly because of their critical role in different biological processes, such as regulation of gene expression, control of metabolic processes, apoptosis and cell survival, DNA repair, development, neuro-protection, and inflammation. Sirtuins control many vital functions and are involved in several pathologies such as metabolic diseases, neurodegenerative disorders, and cancer.
SirT1 has been shown to be significantly up-regulated in different types of cancer including acute myeloid leukemia (AML), prostate, colon, and skin cancers.
Given that only SirT1 and SirT2 seem to control levels of H4K16 acetylation in many settings, and that only SirT1 is proven to be altered in certain cancers, H4K16ac loss in cancer has been correlated with SirT1 deregulated activity.
Nevertheless, SirT1 seems to play contradictory roles, both as tumor suppressor or tumor promoter (Deng, 2009; Bosch-Presegue and Vaquero, 2011). The initial evidence that SirT1 acts as tumor promoter derives from its repressive effect on tumor suppressor p53 (Deng, 2009). By interacting physically with p53, SirT1 blocks its functions through deacetylation at the C-terminal K382 residue (Vaziri et al., 2001). Moreover, SirT1 overexpression in cancer represses p53-dependent cell-cycle arrest and apoptosis in response to DNA damage and oxidative stress. DNA damage-induced acetylation of p53 leads to its activation. In response to damage, SirT1 binds to and deacetylates p53, thus reducing its functional and transcriptional activities. Overexpression of SirT1 disrupts p53-dependent pathways, resulting in a significant reduction in the cell’s ability to respond to stress and DNA damage. In contrast, the inhibition of SirT1 potentiates p53-dependent apoptosis (Deng, 2009; Rahman and Islam, 2011). These findings underline the involvement of SirT1 in tumorigenesis, given that SirT1 overexpression may increase the risk of cancer in mammals by inhibiting p53 and potentially other tumor suppressor genes, including FOXO family members, p73, Rb, and several others.
In contrast, several studies have suggested that SirT1 also has a tumor suppressor role. Decreased SirT1 levels have been reported for glioma, bladder, prostate, and ovarian cancers. Furthermore, some studies have suggested that SirT1 overexpression in APC−/+ mice reduces rather than increases colon cancer formation (Firestein et al., 2008). This action seems to be caused by SirT1 deacetylation of β-catenin, which promotes cytoplasmic localization of the nuclear-localized oncogenic form of β-catenin. In addition, in SirT1-deficient mice, embryos die at middle gestation stages, displaying increased acetylation of H3K9 and H4K16, reduced chromosome condensation and aberrant mitosis (Firestein et al., 2008; Deng, 2009). SirT1 deficiency also causes reduced DNA double-strand break repair and radiation sensitivity (Wang et al., 2008). It has also been suggested that SirT1 plays a role as tumor suppressor by regulating c-Myc. c-Myc binds to and induces SirT1. This interaction results in decreased c-Myc stability, compromising its transformational capability. These data provide strong evidence for a tumor suppressor role of SirT1 in these settings. Importantly, it still seems possible that the expression level and activity of SirT1 may modulate a delicate balance between suppression and promotion of oncogenesis, thus displaying dual dependence on its spatial and temporal distribution and stage of tumorigenesis (Bosch-Presegue and Vaquero, 2011). A potential tumor suppressor role has also been proposed for the other human sirtuins (McGuinness et al., 2011). This hypothesis is supported by several findings such as the reduction of SirT2 in a large number of human brain tumor cell lines, and its involvement in cell-cycle progression. SirT3 is the only mitochondrial sirtuin implicated in tumorigenesis. Its reduction in several cancers leads to an increase in ROS (reactive oxygen species) production, which results in enhanced tumor growth (Kim et al., 2010). SirT5 overexpression has been found in a study of pancreatic cancer (Ouaïssi et al., 2008). Recently, a role for SirT6 and SirT7 in tumorigenesis has also been proposed. SirT6 might be involved as a result of its control of the NFκB pathway and DNA double-strand repair. SirT7, whose expression inversely correlates with the tumorigenic potential in several murine cell lines (Vakhrusheva et al., 2008), displays increased expression levels in breast cancer (Ashraf et al., 2006).
Following these rationals, tumors with deregulated expression or function of sirtuins may benefit from an approach based on the use of SirT inhibitors.
Sirtuin and Aging Diseases
Sirtuins are believed to be involved in metabolic diseases, neurodegeneration, and aging.
It is well known that overexpression of Sir2 (or its orthologs) can extend organism lifespan in a wide range of lower eukaryotes (Bosch-Presegue and Vaquero, 2011). Sir2 function is often correlated to caloric restriction. The link between the role of sirtuins, caloric restriction, and longevity was first described in S. cerevisiae. In yeast, caloric restriction leads to increased replicative lifespan. Lifespan extension has not been observed in yeast lacking Sir2 (Lin et al., 2000). Currently, the role of sirtuins in the regulation of mammalian lifespan is still unclear. However, considering that sirtuins are an evolutionary conserved family of proteins, it is fair to speculate on their role in the modulation of aging-related processes in higher organisms (Westphal et al., 2007; Brooks and How, 2009). In humans, the aging process is associated with telomere erosion. SirT1 and SirT6 have a role in both the maintenance of telomeres and their function, as well as the aging process. Recent studies have demonstrated that reduction or removal of SirT6 results in telomere dysfunction and end-to-end chromosomal fusions. Symptoms displayed in the absence of SirT6 are similar to those caused by a premature aging disease, known as Werner’s syndrome. To date, little is known about other sirtuins, but no evidence suggests their involvement with telomere function, formation, and stability (Westphal et al., 2007; McGuinness et al., 2011). Recently, SirT1 has been shown to promote replicative senescence in response to chronic oxidative stress, by upregulation of the p19ARF senescence regulator, which in turn positively regulates p53 via inhibition of MDM2, a protein mediating p53 degradation (Chua et al., 2005). This effect seems to be in contrast to Sir2 function in yeast, which extends replicative lifespan.
Many studies have demonstrated that the pathogenesis of neurodegenerative diseases includes broad changes and involvement of multiple biochemical pathways. Sirtuins play a critical role in several models of neurodegenerative diseases. Many reports support the role of SirT1 in axonal protection from damage in animal models of Wallerian degenerative disease (Parkinson’s disease; Araki et al., 2004; McGuinness et al., 2011; Rahman and Islam, 2011). SirT1 overexpression also protects against Alzheimer’s and Huntington’s disease as well as amyotrophic lateral sclerosis. In models of Huntington’s disease, SirT1 activation reduces cell death by inhibition of NFκB signaling. Alzheimer’s disease is also related to sirtuin action given that SirT1 overexpression in the brain of mice reduces β-Amyloid production and the formation of plaques. The β-Amyloid peptide generates protein aggregates (plaques) in the brain of patients (Outiero et al., 2008). Recent studies also suggest that SirT1 may benefit learning and memory by activating the gene for brain-derived neurotrophic factor (Gao et al., 2010).
Sirtuin Modulators
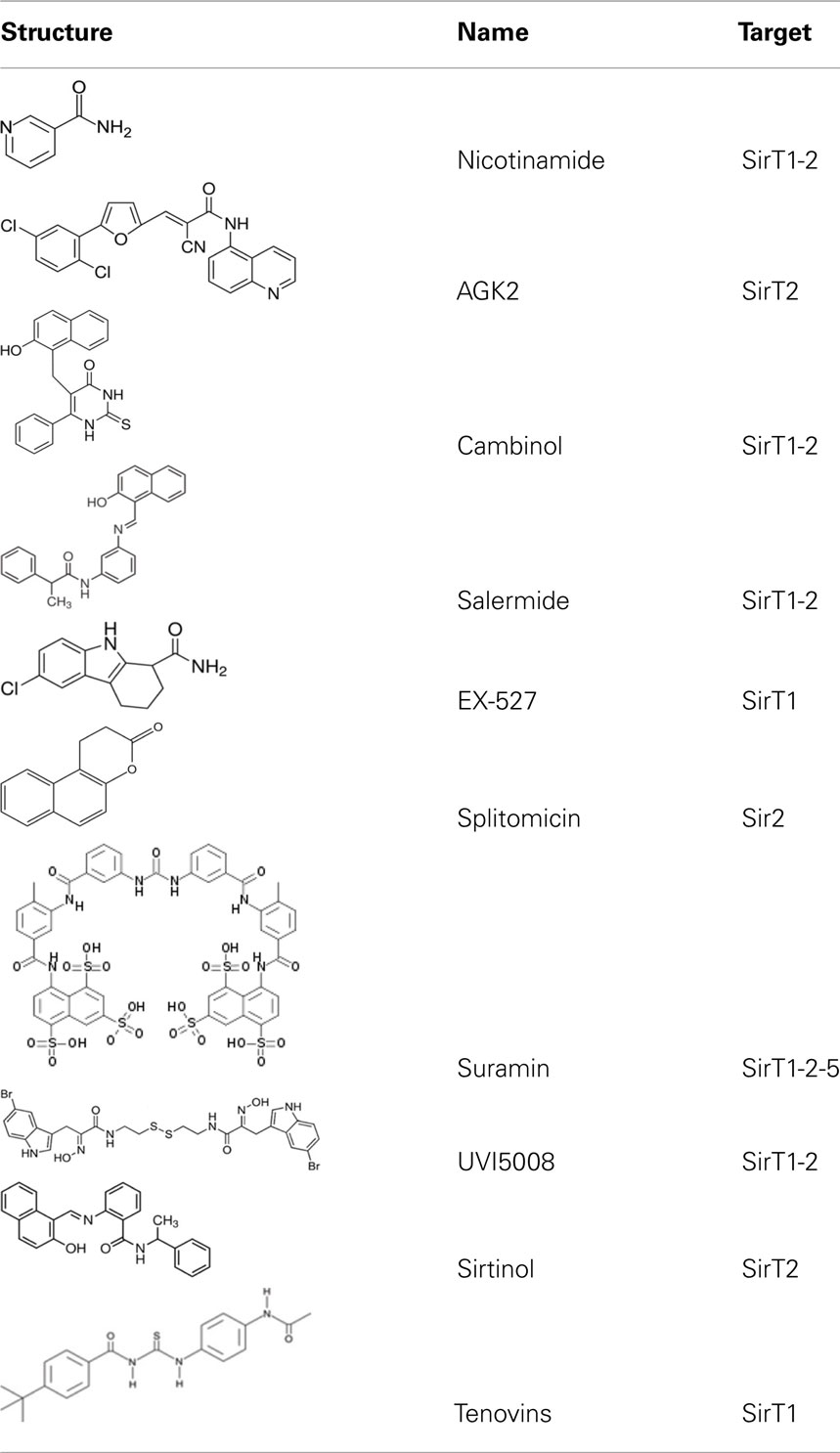
Sirtuins play a key role in several pathologies. Recently, a great deal of research interest has been focused on the identification of small chemical compounds that modulate these proteins. To date, many inhibitors of sirtuins have been proposed for therapy against neurodegenerative diseases and cancer (Alcaín and Villalba, 2009). In recent years a number of inhibitors have been discovered and characterized. In addition to nicotinamide, the physiologic sirtuin inhibitor, some specific inhibitors have been proposed, including splitomicin and its analogs, tenovins, AGK2, sirtinol, suramin, the indole derivative EX-257, cambinol, salermide, and UVI5008 (Table 2).
Splitomicin and its derivatives were discovered in a screening for inhibitors of yeast Sir2. While splitomicin did not act very efficiently against human sirtuins, a series of its analogs with a different orientation of the β-phenyl group were developed and characterized for their activity on SirT2 (Bedalv et al., 2001; Neugebauer et al., 2008). Sirtinol is mainly an inhibitor of the yeast Sir2 and human SirT2 (Grozinger et al., 2001). Its inhibitory effect on growth has been described in human breast, lung, and prostate cancer cells, though using a high micromolar range.
Another potent and selective inhibitor of SirT2 is AGK2 with IC50 of 3.5 μM. It was demonstrated that the inhibition of SirT2 protects against dopaminergic cell death in a Drosophila model of Parkinson’s disease (Outeiro et al., 2007).
Suramin is a potent inhibitor of many sirtuins: SirT1 with IC50 of 297 nM, SirT2 with IC50 of 1.15 μM, and SirT5 with IC50 of 22 μM. However, its reported neurotoxicity limits therapeutic use (Schuetz et al., 2007; Trapp et al., 2007).
Cambinol and salermide (Heltweg et al., 2006; Lara et al., 2009) are reported as inhibitors of SirT1 and SirT2. Cambinol shows the most potent activity against Burkitt lymphoma cell lines by a mechanism involving BCL6 acetylation. Salermide induces apoptosis in cancer cells in a p53-independent manner. The indole derivative, EX-527, is a cell-permeable, selective inhibitor of SirT1 (IC50 = 98 nM). Treatment with EX-527 dramatically increases acetylation at K382 of p53 after DNA damage induction in primary human mammary epithelial cells and several cell lines (Solomon et al., 2006). The direct interaction between SirT1 and p53 highlights potential SirT1 involvement in p53 functions, including differentiation, DNA repair, and aging. Currently, EX-527 is in phase 1 clinical trials for the treatment of Huntington’s disease. In preclinical studies this inhibitor was shown to reduce neuronal death.
The only SirT inhibitors that have been tested in cancer mice models are cambinol, tenovins, and UVI5008. Tenovins, a family of small-molecule inhibitors, are able to inhibit SirT1 at single-digit micromolar concentration and prevent tumor growth in vivo in a p53-dependent manner as single agents. Tenovins have a potential therapeutic interest for the development of new drugs that act against tumor cells overexpressing SirT1 (Lain et al., 2008).
UVI5008 has recently been reported as a novel epigenetic modifier able to inhibit simultaneously histone deacetylases, sirtuins, and DNA methyltransferases. This compound induces cancer cell-selective death in several human models. Importantly, UVI5008 action is independent of p53, Bcl-2-modifying factor (BMF), and TNF-related apoptosis-inducing ligand (TRAIL), affecting the growth of tumor cells deficient or mutated for these factors. This finding potentiates its application in therapy by overcoming potential drug resistance limits. The simultaneous modulation of three classes of epigenetic enzymes offers a promising strategy for applying a single drug exerting multiple-interventions against cancer (Nebbioso et al., 2011).
Phenol derivates, including quercetin, piceatannol, and resveratrol, were shown to have SirT1-activating properties. The most potent activator of the three is resveratrol, a polyphenol found in grapes, and grape products. However, its activity is still debated. Given that the activation of SirT1 by resveratrol requires the use of peptides conjugated with a non-physiological fluorophore and that no activation is observed when peptides lacking this fluorophore are used, other technical approaches are necessary to establish its effective modulation of SirT1 (Borra et al., 2005).
Conclusion and Perspectives
Sirtuins influence several cellular processes. Growing evidence has recently underlined their involvement in many diseases. Although SirT1 has been extensively researched a better understanding of its involvement in pathogenesis is required. Many questions remain to be addressed, such as the role of the other sirtuins and their function. Further research is needed to provide clearer perspectives. The development of potent sirtuin modulators may revert the disease process and possibly extend healthy human lifespan.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We wish to dedicate this review article to the memory of A. Scognamiglio, who struggled against cancer until the very end. This work was supported by the Italian Association for Cancer Research (AIRC), EU 7th frame programs (Aposys contract no. 200767; Blueprint contract no. 28251); the Ministero dell’Istruzione Università e Ricerca (PRIN no. 2009PX2T2E).
References
Araki, T., Sasaki, Y., and Milbrandt, J. (2004). Increased nuclear NAD biosynthesis and SIRT1 activation prevent axonal degeneration. Science 305, 1010–1013.
Ashraf, N., Zino, S., Macintyre, A., Kingsmore, D., Payne, A. P., George, W. D., and Shiels, P. G. (2006). Altered sirtuin expression is associated with node-positive breast cancer. Br. J. Cancer 95, 1056–1061.
Bedalv, A., Gatbonton, T., Irvine, W. P., Gottschling, D. E., and Simon, J. A. (2001). Identification of a small molecule inhibitor of Sir2p. Proc. Natl. Acad. Sci. U.S.A. 15113–15118.
Borra, M. T., Smith, B. C., and Denu, J. M. (2005). Mechanism of human SIRT1 activation by resveratrol. J. Biol. Chem. 280, 17187–17195.
Bosch-Presegue, L., and Vaquero, A. (2011). The dual role of sirtuins in cancer. Genes Cancer 2, 648–662.
Brooks, C. L., and How, W. G. (2009). Does SIRT1 affect metabolism, senescence and cancer? Nat. Rev. Cancer 9, 123–128.
Chua, K. F., Mostoslavsky, R., Lombard, D. B., Pang, W. W., Saito, S., Franco, S., Kaushal, D., Cheng, H. L., Fischer, M. R., Stokes, N., Murphy, M. M., Appella, E., and Alt, F. W. (2005). Mammalian SIRT1 limits replicative life span in response to chronic genotoxic stress. Cell Metab. 67–76.
Deng, C. X. (2009). SirT1, is it a tumor promoter or tumor suppressor? Int. J. Biol. Sci. 5, 147–152.
Firestein, R., Blander, G., and Michan, S. (2008). The sirt1 deacetylase suppress intestinal tumorigenesis and colon cancer growth. PLoS ONE 3, e2020. doi: 10.1371/journal.pone.0002020
Gao, J., Wang, W. Y., Mao, Y. W., Gräff, J., Guan, J. S., Pan, L., Mak, G., Kim, D., Su, S. C., and Tsai, L. H. (2010). A novel pathway regulates memory and plasticity via SIRT1 and miR-134. Nature 466, 1105–1109.
Grozinger, C. M., Chao, E. D., Blackwell, H. E., Moazed, D., and Schreiber, S. L. (2001). Identification of a class of small molecule inhibitors of the sirtuin family of NAD-dependent deacetylases by phenotypic screening. J. Biol. Chem. 19, 38837–38843.
Heltweg, B., Gatbonton, T., Schuler, A. D., Posakony, J., Li, H., Goehle, S., Kollipara, R., Depinho, R. A., Gu, Y., Simon, J. A., and Bedalov, A. (2006). Antitumor activity of a small-molecule inhibitor of human silent information regulator 2 enzymes. Cancer Res. 66, 4368–4377.
Kim, H. S., Patel, K., Muldoon-Jacobs, K., Bisht, K. S., Aykin-Burns, N., Pennington, J. D., Van der Meer, R., Nguyen, P., Savage, J., Owens, K. M., Vassilopoulos, A., Ozden, O., Park, S. H., Singh, K. K., Abdulkadir, S. A., Spitz, D. R., Deng, C. X., and Gius, D. (2010). SIRT3 is a mitochondria-localized tumor suppressor required for maintenance of mitochondrial integrity and metabolism during stress. Cancer Cell 19, 41–52.
Lain, S., Hollick, J. J., Campbell, J., Staples, O. D., Higgins, M., Aoubala, M., McCarthy, A., Appleyard, V., Murray, K. E., Baker, L., Thompson, A., Mathers, J., Holland, S. J., Stark, M. J., Pass, G., Woods, J., Lane, D. P., and Westwood, N. J. (2008). Discovery, in vivo activity, and mechanism of action of a small-molecule p53 activator. Cancer Cell 13, 454–463.
Lara, E., Mai, A., Calvanese, V., Altucci, L., Lopez-Nieva, P., Martinez-Chantar, M. L., Varela-Rey, M., Rotili, D., Nebbioso, A., Ropero, S., Montoya, G., Oyarzabal, J., Velasco, S., Serrano, M., Witt, M., Villar-Garea, A., Imhof, A., Mato, J. M., Esteller, M., and Fraga, M. F. (2009). Salermide, a sirtuin inhibitor with a strong cancer-specific proapoptotic effect. Oncogene 12, 781–791.
Lin, S. J., Defossez, P. A., and Guarente, L. (2000). Requirement of NAD and Sir2 for life-span extension by calorie restriction in Saccharomyces cerevisiae. Science 289, 2126–2128.
Liu, T., Liu, P. Y., and Marshall, G. M. (2009). The critical role of the class III histone deacetylase SIRT1 in cancer. Cancer Res. 69, 1702–1705.
Mao, Z., Hine, C., Tian, X., Van Meter, M., Au, M., Vaidya, A., Seluanov, A., and Gorbunova, V. (2011). SirT6 promotes DNA repair under stress by activating PARP1. Science 332 1443–1446.
McGuinness, D., McGuinness, D. H., McCaul, J. A., and Shiels, P. G. (2011). Sirtuins, bioageing and cancer. J. Aging Res. 2011, 235754.
Nebbioso, A., Pereira, R., Khanwalkar, H., Matarese, F., García-Rodríguez, J., Miceli, M., Logie, C., Kedinger, V., Ferrara, F., Stunnenberg, H. G., de Lera, A. R., Gronemeyer, H., and Altucci, L. (2011). Death receptor pathway activation, and increase of ROS production by the triple epigenetic inhibitor, UVI5008. Mol. Cancer Ther. 10, 2394–2404.
Neugebauer, R. C., Uchiechowska, U., Meier, R., Hruby, H., Valkov, V., Verdin, E., Sippl, W., and Jung, M. (2008). Structure-activity studies on splitomicin derivatives as sirtuin inhibitors and computational prediction of binding mode. J. Med. Chem. 13, 1203–1213.
North, B. J., and Verdin, E. (2004). Sirtuins: Sir2-related NAD-dependent protein deacetylases. Genome Biol. 5, 224–228.
Ouaïssi, M., Sielezneff, I., Silvestre, R., Sastre, B., Bernard, J. P., Lafontaine, J. S., Payan, M. J., Dahan, L., Pirrò, N., Seitz, J. F., Mas, E., Lombardo, D., and Ouaissi, A. (2008). High histone deacetylase 7 (HDAC7) expression is significantly associated with adenocarcinomas of the pancreas. Ann. Surg. Oncol. 15, 2318–2328.
Outeiro, T. F., Kontopoulos, E., Altmann, S. M., Kufareva, I., Strathearn, K. E., Amore, A. M., Volk, C. B., Maxwell, M. M., Rochet, J. C., McLean, P. J., Young, A. B., Abagyan, R., Feany, M. B., Hyman, B. T., and Kazantsev, A. (2007). Sirtuin 2 inhibitors rescue alpha-synuclein-mediated toxicity in models of Parkinson’s disease. Science 27, 516–519.
Outiero, T. F., Marques, O., and Kazantev, A. (2008). Therapeutic role of sirtuins in neurodegenerative disease. Biochim. Biophys. Acta 363–369.
Rahman, S., and Islam, R. (2011). Mammalian Sirt1: insight on its biological functions. Cell Commun. Signal 9, 11.
Schuetz, A., Min, J., Antoshenko, T., Wang, C. L., Allali-Hassani, A., Dong, A., Loppnau, P., Vedadi, M., Bochkarev, A., Sternglanz, R., and Plotnikov, A. N. (2007). Structural basis of inhibition of the human NAD+-dependent deacetylase SIRT5 by suramin. Structure 15, 377–389.
Shogren-Knaak, M., Ishii, H., Sun, J. M., Pazin, M. J., Davie, J. R., and Peterson, C. L. (2006). Histone H4k16 acetylation controls cromatine structure and protein interaction. Science 311, 844–847.
Solomon, J. M., Pasupuleti, R., Xu, L., McDonagh, T., Curtis, R., Di Stefano, P. S., and Huber, L. J. (2006). Inhibition of SIRT1 catalytic activity increases p53 acetylation but does not alter cell survival following DNA damage. Mol. Cell. Biol. 26, 28–38.
Trapp, J., Meier, R., Hongwiset, D., Kassack, M. U., Sippl, W., and Jung, M. (2007). Structure-activity studies on suramin analogues as inhibitors of NAD+-dependent histone deacetylases (sirtuins). ChemMedChem 2, 1419–1431.
Vakhrusheva, O., Braeuer, D., Liu, Z., Broun, T., and Bober, E. (2008). Sirt7-dependent inhibition of cell growth and proliferation might be instrumental to mediate tissue integrity during aging. J. Physiol. Pharmacol. 59 (Suppl. 9), 201–212.
Vaquero, A. (2009). The conserved role of sirtuins in chromatin regulation. Int. J. Dev. Biol. 53, 303–322.
Vaziri, H., Dessain, S. K., Ng Eaton, E., Imai, S. I., Frye, R. A., Pandita, T. K., Guarente, L., and Weinberg, R. A. (2001). hSIR2(SIRT1) functions as an NAD-dependent p53 deacetylase. Cell 107, 149–159.
Wang, R. H., Sengupta, K., Li, C., Kim, H. S., Cao, L., Xiao, C., Kim, S., Xu, X., Zheng, Y., Chilton, B., Jia, R., Zheng, Z. M., Appella, E., Wang, X. W., Ried, T., and Deng, C. X. (2008). Impaired DNA damage response, genome instability, and tumorigenesis in SIRT1 mutant mice. Cancer Cell 312–323.
Westphal, C. H., Dipp, M. A., and Guarente, L. (2007). A therapeutic role for sirtuins in diseases of aging? Trends Biochem. Sci. 32, 555–560.
Keywords: sirtuins, SirT inhibitors, cancer, human diseases, epigenetics
Citation: Carafa V, Nebbioso A and Altucci L (2012) Sirtuins and disease: the road ahead. Front. Pharmacol. 3:4. doi: 10.3389/fphar.2012.00004
Received: 13 December 2011; Paper pending published: 01 January 2012;
Accepted: 09 January 2012; Published online: 31 January 2012.
Edited by:
Aleksey G. Kazantsev, Harvard Medical School and Massachusetts General Hospital, USAReviewed by:
Antonello Mai, Sapienza University of Rome, ItalyHendrik Stunnenberg, Radboud University, Netherlands
Copyright: © 2012 Carafa, Nebbioso and Altucci. This is an open-access article distributed under the terms of the Creative Commons Attribution Non Commercial License, which permits non-commercial use, distribution, and reproduction in other forums, provided the original authors and source are credited.
*Correspondence: Lucia Altucci, Dipartimento di Patologia Generale, Seconda Università degli Studi di Napoli, Vico L.de Crecchio 7, Napoli 80138, Italy. e-mail: lucia.altucci@unina2.it; lucia@altucci.com