
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
BRIEF RESEARCH REPORT article
Front. Pediatr., 10 June 2024
Sec. Pediatric Gastroenterology, Hepatology and Nutrition
Volume 12 - 2024 | https://doi.org/10.3389/fped.2024.1421051
This article is part of the Research TopicInsights in Pediatric Gastroenterology, Hepatology & NutritionView all 7 articles
 Jia Xu1
Jia Xu1 Rebbeca M. Duar2
Rebbeca M. Duar2 Baoling Quah1
Baoling Quah1 Min Gong1
Min Gong1 Felicia Tin1
Felicia Tin1 Penny Chan1,3
Penny Chan1,3 Choon Kiat Sim1
Choon Kiat Sim1 Kok Hian Tan4,5
Kok Hian Tan4,5 Yap Seng Chong1,6
Yap Seng Chong1,6 Peter D. Gluckman1,7
Peter D. Gluckman1,7 Steven A. Frese8
Steven A. Frese8 David Kyle2
David Kyle2 Neerja Karnani1,3,9*
Neerja Karnani1,3,9*
Background: The loss of ancestral microbes, or the “disappearing microbiota hypothesis” has been proposed to play a critical role in the rise of inflammatory and immune diseases in developed nations. The effect of this loss is most consequential during early-life, as initial colonizers of the newborn gut contribute significantly to the development of the immune system.
Methods: In this longitudinal study (day 3, week 3, and month 3 post-birth) of infants of Asian ancestry born in Singapore, we studied how generational immigration status and common perinatal factors affect bifidobacteria and Bifidobacterium longum subsp. infantis (B. infantis) colonization. Cohort registry identifier: NCT01174875.
Results: Our findings show that first-generation migratory status, perinatal antibiotics usage, and cesarean section birth, significantly influenced the abundance and acquisition of bifidobacteria in the infant gut. Most importantly, 95.6% of the infants surveyed in this study had undetectable B. infantis, an early and beneficial colonizer of infant gut due to its ability to metabolize the wide variety of human milk oligosaccharides present in breastmilk and its ability to shape the development of a healthy immune system. A comparative analysis of B. infantis in 12 countries by their GDP per capita showed a remarkably low prevalence of this microbe in advanced economies, especially Singapore.
Conclusion: This study provides new insights into infant gut microbiota colonization, showing the impact of generational immigration on early-life gut microbiota acquisition. It also warrants the need to closely monitor the declining prevalence of beneficial microbes such as B. infantis in developed nations and its potential link to increasing autoimmune and allergic diseases.
Assembly of the gut microbiota during early life plays a critical role in shaping life-long immune and metabolic health. Among the first long-term colonizing gut microbes, bifidobacteria are reported to play a significant role in priming the immune system and to be protective against susceptibility to diverse diseases later in life (1, 2). However, modern lifestyle and medical practices such as the increase in the rates of cesarean section delivery, widespread use of antibiotics, and reduced breastfeeding duration are thought to have resulted in disparate rates of infants with a bifidobacteria-dominated gut microbiota between countries. Healthy infants from developing countries are more often colonized at higher abundance with bifidobacteria compared to infants from industrialized nations (3–8), where the rates of metabolic and immune-related diseases are rising (9).
Dominance of Bifidobacterium spp. in the gut is driven largely by the presence of human milk oligosaccharides (HMOs), which are complex carbohydrates that constitute the third most abundant component of breastmilk after lactose and lipids (10). Bifidobacterium longum subsp. infantis is unique among the bifidobacteria as it contains a large repertoire of genes encoding glycosidases and oligosaccharide transporters required to metabolize HMOs (11, 12). Furthermore, accumulating evidence suggests that B. infantis benefits the infant through immunoregulation of CD4+ T cells implicated in autoimmune and allergic diseases in children (13), preventing inflammation (14, 15), and improving gut barrier function (16, 17).
The highest prevalence of infant colonization with B. infantis has been reported in developing nations and in populations living traditional, agrarian lifestyles where breastfeeding rates are high (18–20). On the contrary, in high-income industrialized nations with a history of interrupted or low rates of breastfeeding, B. infantis colonization of infants is extremely rare (6–8, 20, 21). Recent findings have now shown that an initial high carriage of B. infantis in the gut of the breastfed infant provides a profound, durable colonization resistance (22–24). In its absence, populations of endotoxin-producing taxa, which harbour virulence and antibiotic resistance-related genes persist despite exclusive breastfeeding (23, 25), and several studies have documented evidence of high levels of enteric inflammation within the first 60 days of life among infants lacking high levels of bifidobacteria (13, 14). Retrospective studies have shown that such an early chronic enteric inflammation is associated with a dysfunction of the immune system that presents as a blunting of vaccine responses (18, 26) and increased prevalence of autoimmune disorders such as atopic dermatitis, food allergies, asthma, and Type I Diabetes (27–29).
Singapore is a high-income developed nation with high immigration and representation from three ethnic groups, Chinese (76.9%), Malays (14.6%) and Asian Indians (6.4%). Singapore has also shown an increase in the incidence of inflammatory disorders such as atopic dermatitis and allergic rhinitis in children (30, 31), affecting quality of life and raising the financial burden for the affected families and the health care system. Given the known connection between the microbiome in early life and immune health, we evaluated factors influencing the abundance and acquisition of bifidobacteria as well as the prevalence of B. infantis in infants from the GUSTO (Growing Up in Singapore Towards healthy Outcomes) birth cohort in Singapore.
Infants involved in this study were part of the GUSTO birth cohort which was designed to investigate the developmental origins of health and disease (DOHaD) (32). Briefly, the GUSTO cohort recruited 1,237 pregnant women at their 1st trimesters (aged 18 and above) from two major public maternity hospitals in Singapore from 2009 to 2010 and continues to follow-up their children until now (Clinicaltrials.gov registration no. NCT01174875). All the recruited women are homogeneous Chinese, Malay, or Indian ethnic ancestry. Ethnic approvals were obtained from the SingHealth Centralised Institutional Review Board (Reference 2009/280/D) and the National Healthcare Group Domain Specific Review Board (Reference D/09/021). All participants provided the written informed consent. Written informed consent was obtained from mothers of all infants who participated in the GUSTO study. Approval for the study was granted by the ethics boards of both KK Women's and Children's Hospital (KKH) and National University Hospital (NUH), which are the Centralized Institute Review Board (CIRB) and the Domain Specific Review Board (DSRB), respectively.
To assess the factors that could influence the early colonization of bifidobacteria and prevalence of B. infantis in Singaporean infants, we included infants with fecal samples available longitudinally at 3 time points (day 3, week 3 and month 3 post-birth) from 3 ethnicities (Chinese, Malay, and Indian) in the current study. For this, seventy-five infants per ethnicity per time point were selected (Supplementary Table S1). All the 75 Chinese and Malay infants and 62 Indian infants had fecal samples longitudinally at 3 time points. We randomly selected some Indian infants with fecal samples available at any 2 time points or either day 3 or Month 3 to ensure all the 3 ethnicities had the same sample size of 75 at each time point. Altogether, 675 fecal samples were used in this study. General characteristics of infants included in this study are summarized in Supplementary Table S2.
DNA extraction was performed using the QIAmp Powerfecal Pro DNA Kit (Qiagen, Hilden, Germany) as per the manufacturer's protocol with minor modifications. Briefly, approximately 0.25 g of infant fecal sample was suspended in 800 μl of CD1 solution in a PowerBead Pro tube. Samples were vortexed and bead-beaten twice for 5 min each in the TissueLyser II (Qiagen, Hilden, Germany) at 25 Hz. After the removal of inhibitors, DNA was purified through a spin column and eluted with 75 μl of pre-warmed 37°C solution C6, following which a second elution was performed to maximize the DNA yield. DNA concentration was determined by using Quant-iT PicoGreen dsDNA Assay Kit (Life Technologies, Carlsbad, California, USA).
The quantification of Real-time q-PCR was carried out using QuantStudio 6 Flex Real-Time PCR System (Applied Biosystems, Carlsbad, USA). Primers and probes (Supplementary Table S3) targeting Bifidobacterium spp (33). and B. infantis (34) were purchased from Integrated DNA Technologies (IDT, Singapore). Standard curves were generated using genomic DNA from B. infantis ATCC 15697. The standard DNA was diluted in a 10-fold serial dilution from 106 to 101 copies/reaction. For quality control, R2 value of the standard curve was >99%, and the coefficient of variation for duplications was <5%.
Reactions for the qPCR of bifidobacteria were carried out in a final volume of 25 μl containing 1× Fast SYBR green PCR mastermix (Bio-Rad, USA), 12.5 µM of forward/reverse primer, 20 ng of template DNA. The thermocycler setting consisted of an initial activation of the polymerase at 95°C for 5 min, followed by 40 cycles of 95°C for 30 s, 62°C for 20 s and 72°C for 40 s.
Reactions for the qPCR of B. infantis were carried out in a final volume of 15 μl containing 1× TaqMan Fast PCR mastermix (Applied Biosystems, Carlsbad, USA), 10 μM of forward/reverse primer, 5 μM probe and 2 ng of template DNA. The thermocycler setting consisted of an initial activation of the polymerase at 95°C for 30 s, followed by 40 cycles of 95°C for 30 s and 60°C for 20 s.
Normality tests for the abundance of bifidobacteria were performed using the Shapiro–Wilk test. As the data were not normally distributed, non-parametric tests were applied for all statistical analyses. For comparisons across different time points, Friedman's test was utilized, followed by Dunn's test for post-hoc pairwise comparisons. In the univariate analysis assessing the effects of phenotypic variables on the early colonization of bifidobacteria, the Kruskal–Wallis test was applied for variables with more than two categories, and the Mann–Whitney test was used for binary variables. For the multivariate analysis, rank-based regression models were employed to adjust for the mutual influences of significant factors identified in the univariate analysis (35). This adjustment was performed using the “Rfit” package in R version 4.3.0. The Chi-square test was employed to explore pairwise relationships between categorical phenotypic factors. All P-values, particularly those from multiple comparisons, were adjusted using the Bonferroni correction method to control for type I errors.
To assess the factors influencing the early colonization of bifidobacteria in Singaporean infants, bifidobacteria abundance was assessed in fecal samples of all study subjects at all 3 time points (day 3, week 3 and month 3 post-birth) and across all 3 ethnicities (Chinese, Malay and Indian; n = 75 for each ethnicity, 675 samples in total). Bifidobacteria abundance at each time point and within each ethnic group did not show a normal distribution (Shapiro–Wilk test, P-value < 0.0001, Supplementary Figure S1). Over time, there was a significant increase in bifidobacteria abundance with the highest mean abundance observed at the 3-month timepoint (Figure 1A and Supplementary Table S4).
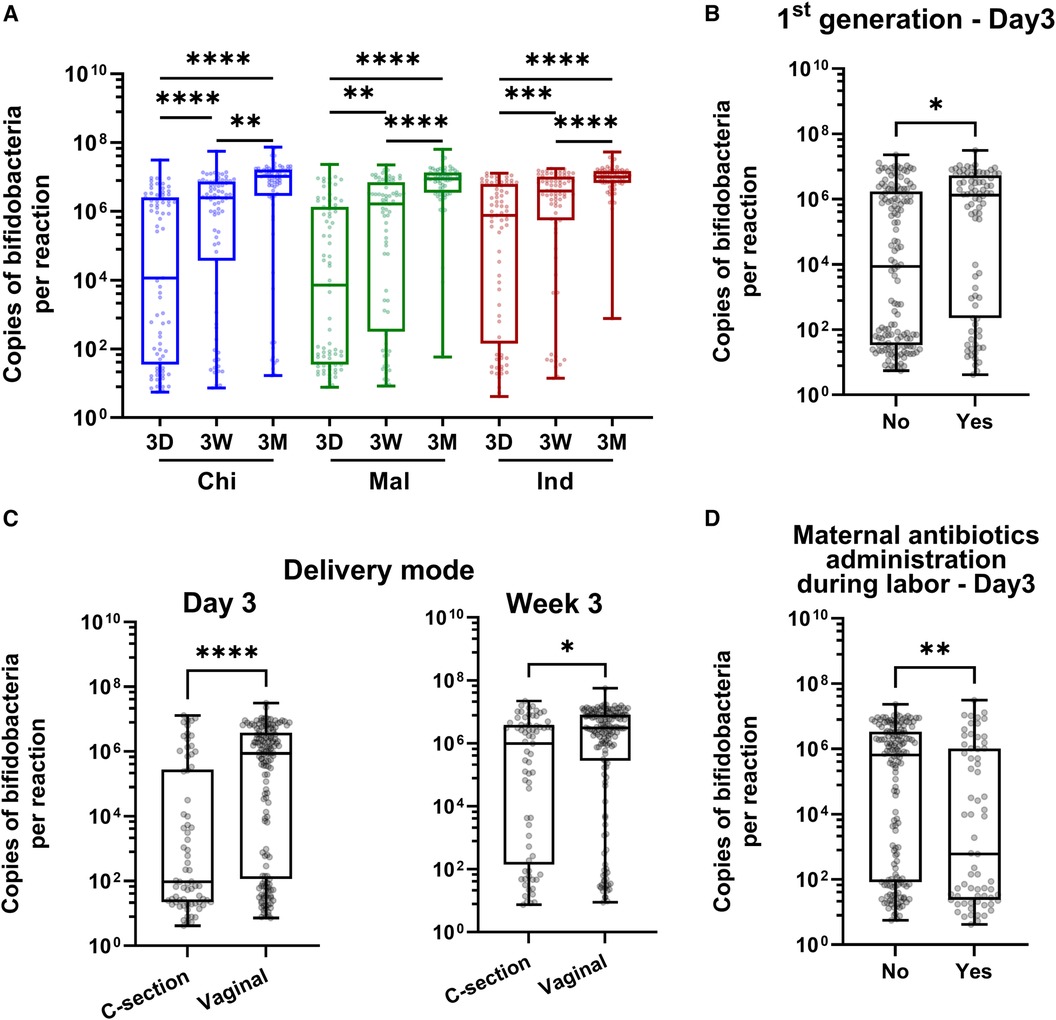
Figure 1. Abundance of bifidobacteria in infant gut in 3 Asian ethnic groups and its association with immigration status and clinical variables. (A) Longitudinal assessment of bifidobacteria abundance in infant fecal samples collected at 3 days (3D), 3 weeks (3W) and 3 months (3M) post-birth in Chinese (Chi), Malay (Mal) and Indian (Ind) ethnic groups. (B–D) Factors significantly associated with early colonization of bifidobacteria: First-generation status (B), delivery mode (C), and maternal antibiotics administration during labor (D). FDR corrected P-values were calculated using Dunn's post-hoc test (A), and rank-based regression adjusting for covariates followed by Bonferroni correction for multiple comparisons (B–D), where **** = P < 0.0001, *** = P < 0.001, ** = P < 0.01, * =P < 0.05 (A). N is 225 for 3D, 3W and 3M respectively.
Ethnicity, first generation immigration, delivery mode, and maternal administration of antibiotics during labor significantly influenced the colonization of bifidobacteria in the univariate test (Table 1). Ethnicity and first-generation status were significantly correlated with each other (Supplementary Table S5 and Supplementary Figure S2). As ethnicity, first generation status, delivery mode, and maternal antibiotics administration during labor demonstrated significant effects in the univariate analysis, they were further examined in the multivariate analysis, adjusting for the effects of the other three factors (Table 1 and Figures 1B–D). Upon mutual adjustment, associations of first-generation status, delivery mode and maternal antibiotics administration during labor with bifidobacteria abundance at Day 3 post-birth remained significant. Additionally, the effect of caesarean-section lasted until 3 weeks post-birth (Table 1 and Figures 1B–D). Therefore, long term migratory status i.e., non-first generation, birth by caesarean-section, and maternal administration of antibiotics during labor delayed the colonization of bifidobacteria in infant gut.
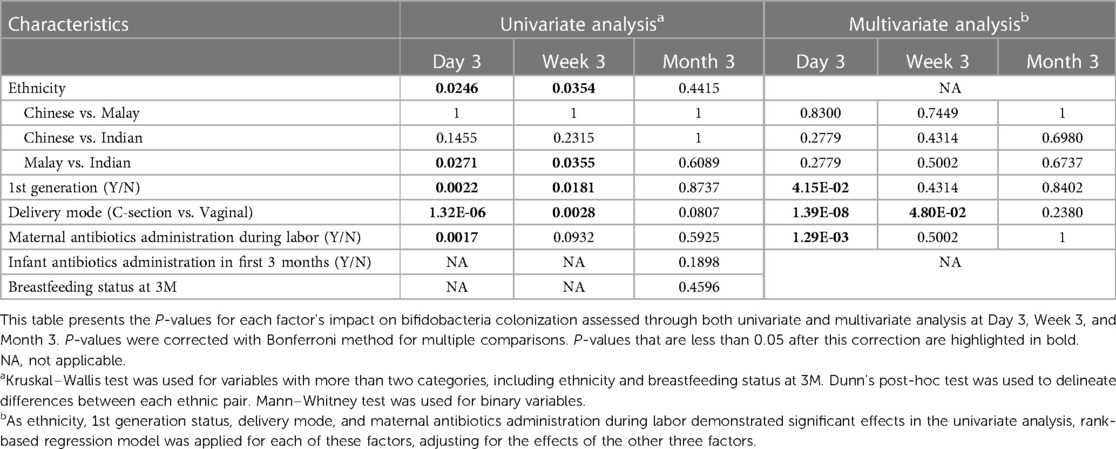
Table 1. Univariate and multivariate analysis of factors influencing early colonization of bifidobacteria over time.
B. infantis was detected in 10/225 (4.4%) infants, and its carriage was not ethnicity dependent (Supplementary Table S6). Among the 10 subjects containing B. infantis, 3 were Chinese, 5 Malay, and 2 Indian. Only 1 subject (Chinese) had B. infantis at all three time points. The relative abundance of B. infantis was extremely low in the total bifidobacteria pool ranging from 0.00001 to 6.9% (Figure 2 and Supplementary Table S7). There was no consistent trend between the prevalence of B. infantis and factors such as antibiotics administration, mode of delivery, or duration of breastfeeding (Supplementary Table S8).
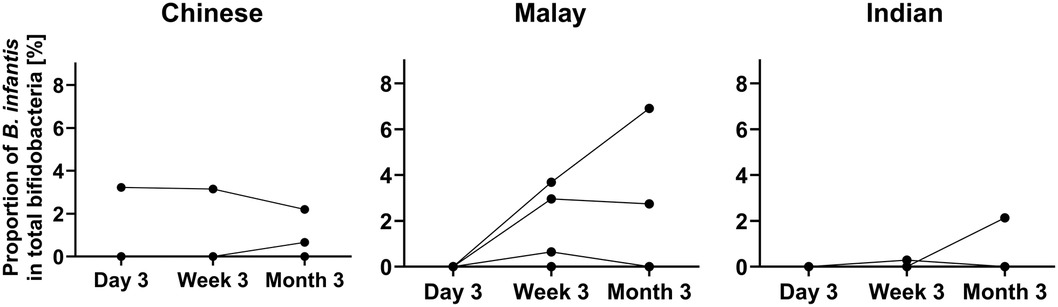
Figure 2. The proportion of B. infantis in total bifidobacteria in 10 subjects over time. N = 3 for Chinese, 5 for Malay, and 2 for Indian.
Comparison of these results with the published datasets employing several different techniques from 11 countries identified a wide variation in the prevalence of B. infantis among infant populations, with trends to high (>80%), low (<25% and >5%), or very low (<5%) prevalence (Figure 3A and Supplementary Table S9). Infants from Gambia and Bangladesh are reported to have the highest prevalence of B. infantis (>80%). Infants from mainland China, Russia, Estonia, Switzerland, Finland, United States and Australia report the prevalence of B. infantis to be moderate (between 23% to 6.2%). The lowest prevalence (<5%) of B. infantis was reported in Germany, Austria and Singapore (Supplementary Table S9). A further analysis comparing the economic status of these countries showed a significant negative correlation between GDP per capita of the country and the prevalence of B. infantis (Spearman correlation, P = 0.0208, R = −0.6550; Figure 3B and Supplementary Table S9).
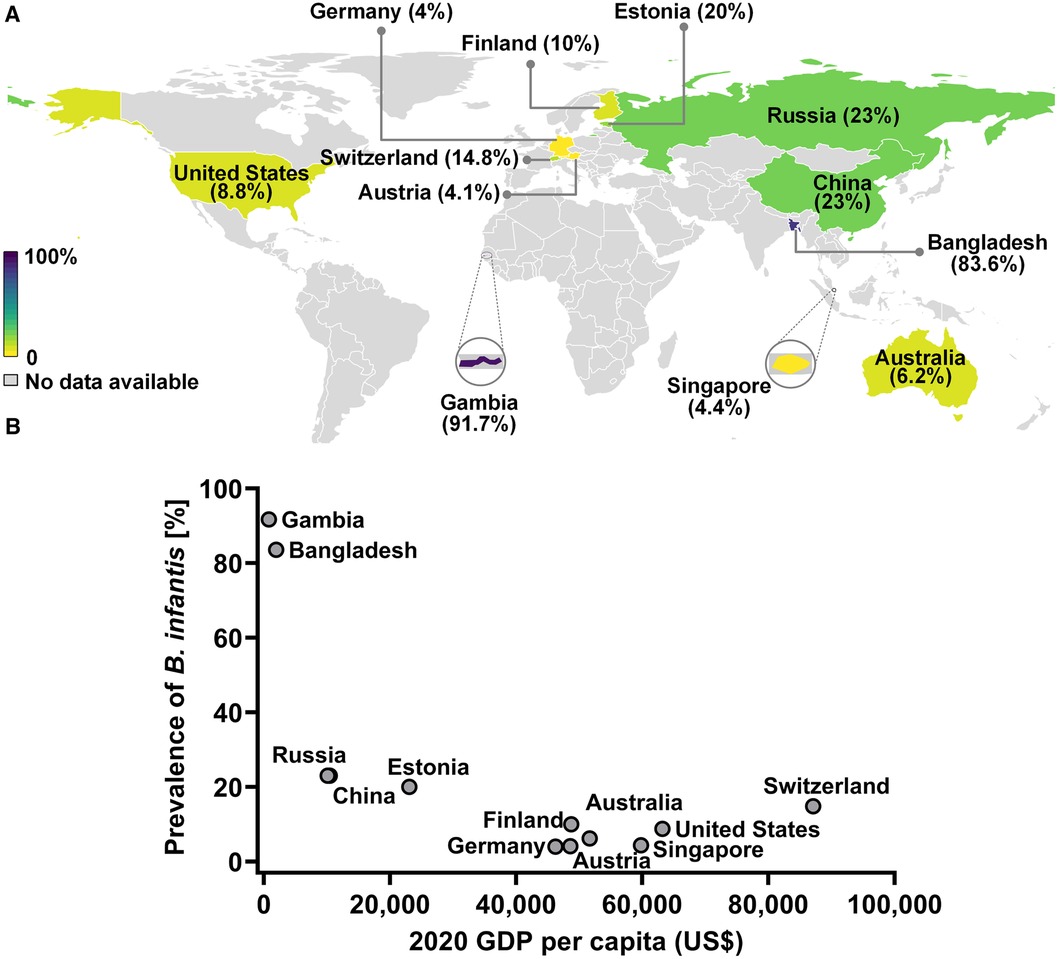
Figure 3. The prevalence of B. infantis in infant gut in developing and developed countries. (A) World map representation of B. infantis prevalence. (B) Negative correlation between the prevalence of B. infantis and the respective country's economic status as ascertained by 2020 GDP data from The World Bank (https://data.worldbank.org/indicator/NY.GDP.PCAP.CD?end=2020&name_desc=false&start=2000&view=chart).
In this study we identified the factors influencing colonization of bifidobacteria and the prevalence of B. infantis in first 3 days to 3 months of life among infants born in Singapore, a high-income, developed country. We observed use of antibiotics and cesarean section delivery delayed the colonization of bifidobacteria in the infant gut, which is consistent with previous findings (36–39). Generational immigration status was also a significant factor affecting colonization by bifidobacteria. Specifically, not being of first-generation immigration to Singapore delayed the acquisition of bifidobacteria. Similar findings have been described in adults, where immigration from developing to a developed, high-income country has a significant impact on the composition and function of microbiome (40, 41).
The reduced relative abundance of bifidobacteria among infants has been correlated to enteric inflammation and increased risk of chronic diseases as well as reduced vaccine response (13, 14, 18, 21, 26). Recent reports have suggested that loss of Bifidobacterium spp. in the infant born in developed countries is linked to increased incidence of allergic and autoimmune diseases (20) while carriage of B. infantis appears to associated with reduced risk (42). Vangay et al. tracked the gut microbiome of subjects living in China and Thailand before and after immigration to the US, and observed immigration from a non-Western country to the US decreased the gut microbial diversity, increased the ratio of Bacteroides/Prevotella, and reduced the capacity to degrade plant-derived complex carbohydrates (40). In our study, we observed a relatively higher abundance of bifidobacteria in the gut of first-generation infants born in Singapore. As the disparity in the levels of bifidobacteria in 1st generation infants was observed only at the Day 3 timepoint, our results suggest a delay in the acquisition of this microbial taxa in the non-first generation infants. Since the newborns acquire their first microbes from their environment, it's possible that modern lifestyles and practices (e.g., industrialization, urbanization, maternal diet, maternal antibiotic exposure, and other lifestyle differences) may have contributed to the delayed colonization of bifidobacteria in non-first-generation infants.
B. infantis, the only bifidobacteria species containing all the genes required to metabolize HMOs in human milk, was found at a very low prevalence (<5%) among Singaporean infants. This is relevant as the presence of HMO-utilization genes in the infant gut is correlated with inflammation and immune system development (13). Interestingly, the prevalence of B. infantis in Singapore was similar to what has been reported for other high-income developed nations. A comparative analysis of B. infantis in 12 countries by their GDP per capita showed a remarkably low prevalence of this microbe in advanced economies. High-income nations have also shown an increased burden of inflammatory diseases and Singapore is no exception to this trend. Incidence of allergic rhinitis in school-going children in Singapore is 40% (31), and is linked to other comorbidities including asthma, atopic dermatitis/eczema, allergic conjunctivitis and chronic sinusitis and chronic otitis media with effusion. Notably, the carriage of B. infantis was not ethnicity specific, indicating generic effects of urbanization and modern medical practices on infant gut microbiome.
Previous studies suggested that the colonization of B. infantis is associated with breastfeeding rates and antibiotic use (8). However, in our study, we did not find any consistent trends between the presence of B. infantis and factors such as antibiotics use, mode of delivery, or duration of breastfeeding. This could either be due to the small sample size in this study or that B. infantis has already diminished over generations in Singapore. Notably, there is evidence that bifidobacteria abundance (inclusive of B. infantis) appears to have declined from historic levels among other high-income countries (43).
This study establishes a foundational understanding of bifidobacteria colonization patterns in Singaporean infants, suggesting the significant influences of generational immigration and modern medical practices. Notably, it highlights the remarkably low presence of B. infantis in developed nations. We will expand our analysis to explore the diversity and abundance of different bifidobacteria species across the entire GUSTO cohort in early life, providing a more comprehensive and longitudinal view of the Bifidobacterium spp. that can colonize the newborn intestine. Furthermore, we intend to correlate the abundance and diversity of Bifidobacterium spp. with key phenotypic outcomes observed within the cohort, such as metabolic and mental health adversities, as well as allergic disorders. Understanding these relationships will further elucidate the role of early microbial colonization in the developmental origins of health and disease, paving the way for targeted interventions and proactive healthcare strategies. This approach not only deepens our understanding of microbial influences on early childhood development but also provides potential pathways for enhancing public health outcomes through precision medicine.
In conclusion, our study shows that immigration status and modern medical practices may influence the rates of infant colonization by bifidobacteria and sustainability of beneficial microbes such as B. infantis in infants born in high income developed nations, although the full health implications for infants born in high income developed nations remains unclear. Given the importance of proper microbiome assembly in early life for the development of the immune system, and the known immunomodulatory role of metabolites produced by B. infantis (13, 44), closer attention should be placed on the connection between the low prevalence of this important infant gut symbiont and the rise of autoimmune and allergic diseases.
The clinical and demographics data supporting the conclusions of this article are available upon request from the GUSTO cohort committee (https://gustodatavault.sg/policies). The qPCR data supporting the conclusions of this article is included in the Supplementary Material.
The studies involving humans were approved by KK Women’s and Children’s Hospital (KKH) and National University Hospital (NUH), which are the Centralized Institute Review Board (CIRB) and the Domain Specific Review Board (DSRB). The studies were conducted in accordance with the local legislation and institutional requirements. Written informed consent for participation in this study was provided by the participants’ legal guardians/next of kin.
JX: Data curation, Formal Analysis, Investigation, Validation, Visualization, Writing – original draft, Writing – review & editing. RMD: Investigation, Writing – original draft, Writing – review & editing, Validation. BQ: Data curation, Investigation, Methodology, Writing – original draft. MG: Data curation, Investigation, Methodology, Writing – review & editing. FT: Data curation, Investigation, Methodology, Writing – review & editing. PC: Investigation, Writing – review & editing. CKS: Investigation, Writing – review & editing. KHT: Writing – review & editing, Resources. YSC: Funding acquisition, Writing – review & editing, Resources. PDG: Supervision, Writing – review & editing. SAF: Investigation, Writing – review & editing. DK: Investigation, Writing – review & editing. NK: Conceptualization, Funding acquisition, Supervision, Writing – review & editing.
The author(s) declare financial support was received for the research, authorship, and/or publication of this article.
The authors declare that this study received funding from Infinant Health, Inc.; the National Research Foundation (NRF) under the Open Fund-Large Collaborative Grant (OF-LCG; MOH-000504) administered by the Singapore Ministry of Health’s National Medical Research Council (NMRC) and the Agency for Science, Technology and Research (A*STAR). In RIE2025, GUSTO is supported by funding from the NRF’s Human Health and Potential (HHP) Domain, under the Human Potential Program.
We would like to thank the participants and the GUSTO study group which includes Allan Sheppard, Amutha Chinnadurai, Anne Eng Neo Goh, Anne Rifkin-Graboi, Anqi Qiu, Arijit Biswas, Bee Wah Lee, Birit F. P. Broekman, Boon Long Quah, Borys Shuter, Chai Kiat Chng, Cheryl Ngo, Choon Looi Bong, Christiani Jeyakumar Henry, Cornelia Yin Ing Chee, Yam Thiam Daniel Goh, Doris Fok, Fabian Yap, George Seow Heong Yeo, Helen Chen, Hugo P. S. van Bever, Iliana Magiati, Inez Bik Yun Wong, Ivy Yee-Man Lau, Jeevesh Kapur, Jenny L. Richmond, Jerry Kok Yen Chan, Joanna D. Holbrook, Joshua J. Gooley, Keith M. Godfrey, Kenneth Kwek, Kok Hian Tan, Krishnamoorthy Niduvaje, Leher Singh, Lin Lin Su, Lourdes Mary Daniel, Lynette P. Shek, Marielle V. Fortier, Mark Hanson, Mary Foong-Fong Chong, Mary Rauff, Mei Chien Chua, Michael Meaney, Mya Thway Tint, Neerja Karnani, Ngee Lek, Oon Hoe Teoh, P. C. Wong, Peter D. Gluckman, Pratibha Agarwal, Rob M. van Dam, Salome A. Rebello, Seang-Mei Saw, Shang Chee Chong, Shirong Cai, Shu-E Soh, Sok Bee Lim, Chin-Ying Stephen Hsu, Victor Samuel Rajadurai, Walter Stunkel, Wee Meng Han, Wei Wei Pang, Yap-Seng Chong, Yin Bun Cheung, Yiong Huak Chan and Yung Seng Lee.
RMD, SAF and DK were employed by the company Infinant Health, Inc. (previously known as Evolve BioSystems). YSC and NK are part of an academic consortium that has received research funding from Abbott Nutrition, Nestec, and Danone.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fped.2024.1421051/full#supplementary-material
1. Kalliomaki M, Collado MC, Salminen S, Isolauri E. Early differences in fecal microbiota composition in children may predict overweight. Am J Clin Nutr. (2008) 87(3):534–8. doi: 10.1093/ajcn/87.3.534
2. Sjogren YM, Jenmalm MC, Bottcher MF, Bjorksten B, Sverremark-Ekstrom E. Altered early infant gut microbiota in children developing allergy up to 5 years of age. Clin Exp Allergy. (2009) 39(4):518–26. doi: 10.1111/j.1365-2222.2008.03156.x
3. Grzeskowiak L, Collado MC, Mangani C, Maleta K, Laitinen K, Ashorn P, et al. Distinct gut microbiota in southeastern African and northern European infants. J Pediatr Gastroenterol Nutr. (2012) 54(6):812–6. doi: 10.1097/MPG.0b013e318249039c
4. Tannock GW, Lee PS, Wong KH, Lawley B. Why don't all infants have bifidobacteria in their stool? Front Microbiol. (2016) 7:834. doi: 10.3389/fmicb.2016.00834
5. Lawley B, Otal A, Moloney-Geany K, Diana A, Houghton L, Heath AM, et al. Fecal microbiotas of Indonesian and New Zealand children differ in complexity and bifidobacterial taxa during the first year of life. Appl Environ Microbiol. (2019) 85:19. doi: 10.1128/AEM.01105-19
6. Casaburi G, Duar RM, Brown H, Mitchell RD, Kazi S, Chew S, et al. Metagenomic insights of the infant microbiome community structure and function across multiple sites in the United States. Sci Rep. (2021) 11(1):1472. doi: 10.1038/s41598-020-80583-9
7. Olm MR, Dahan D, Carter MM, Merrill BD, Yu FB, Jain S, et al. Robust variation in infant gut microbiome assembly across a spectrum of lifestyles. Science. (2022) 376(6598):1220–3. doi: 10.1126/science.abj2972
8. Taft DH, Lewis ZT, Nguyen N, Ho S, Masarweh C, Dunne-Castagna V, et al. Bifidobacterium Species colonization in infancy: a global cross-sectional comparison by population history of breastfeeding. Nutrients. (2022) 14:7. doi: 10.3390/nu14071423
9. Sonnenburg ED, Sonnenburg JL. The ancestral and industrialized gut microbiota and implications for human health. Nat Rev Microbiol. (2019) 17(6):383–90. doi: 10.1038/s41579-019-0191-8
10. Nijman RM, Liu Y, Bunyatratchata A, Smilowitz JT, Stahl B, Barile D. Characterization and quantification of oligosaccharides in human milk and infant formula. J Agric Food Chem. (2018) 66(26):6851–9. doi: 10.1021/acs.jafc.8b01515
11. Asakuma S, Hatakeyama E, Urashima T, Yoshida E, Katayama T, Yamamoto K, et al. Physiology of consumption of human milk oligosaccharides by infant gut-associated bifidobacteria. J Biol Chem. (2011) 286(40):34583–92. doi: 10.1074/jbc.M111.248138
12. Sakanaka M, Gotoh A, Yoshida K, Odamaki T, Koguchi H, Xiao JZ, et al. Varied pathways of infant gut-associated Bifidobacterium to assimilate human milk oligosaccharides: prevalence of the gene set and its correlation with bifidobacteria-rich microbiota formation. Nutrients. (2019) 12:1. doi: 10.3390/nu12010071
13. Henrick BM, Rodriguez L, Lakshmikanth T, Pou C, Henckel E, Arzoomand A, et al. Bifidobacteria-mediated immune system imprinting early in life. Cell. (2021) 184(15):3884–3898.e11. doi: 10.1016/j.cell.2021.05.030
14. Henrick BM, Chew S, Casaburi G, Brown HK, Frese SA, Zhou Y, et al. Colonization by B. infantis EVC001 modulates enteric inflammation in exclusively breastfed infants. Pediatr Res. (2019) 86(6):749–57. doi: 10.1038/s41390-019-0533-2
15. Meng D, Sommella E, Salviati E, Campiglia P, Ganguli K, Djebali K, et al. Indole-3-lactic acid, a metabolite of tryptophan, secreted by Bifidobacterium longum subspecies infantis is anti-inflammatory in the immature intestine. Pediatr Res. (2020) 88(2):209–17. doi: 10.1038/s41390-019-0740-x
16. Guo S, Gillingham T, Guo Y, Meng D, Zhu W, Walker WA, et al. Secretions of Bifidobacterium infantis and Lactobacillus acidophilus protect intestinal epithelial barrier function. J Pediatr Gastroenterol Nutr. (2017) 64(3):404–12. doi: 10.1097/MPG.0000000000001310
17. Karav S, Casaburi G, Frese SA. Reduced colonic mucin degradation in breastfed infants colonized by Bifidobacterium longum subsp. infantis EVC001. FEBS Open Bio. (2018) 8(10):1649–57. doi: 10.1002/2211-5463.12516
18. Huda MN, Lewis Z, Kalanetra KM, Rashid M, Ahmad SM, Raqib R, et al. Stool microbiota and vaccine responses of infants. Pediatrics. (2014) 134(2):e362–372. doi: 10.1542/peds.2013-3937
19. Davis JC, Lewis ZT, Krishnan S, Bernstein RM, Moore SE, Prentice AM, et al. Growth and morbidity of gambian infants are influenced by maternal milk oligosaccharides and infant gut microbiota. Sci Rep. (2017) 7:40466. doi: 10.1038/srep40466
20. Seppo AE, Bu K, Jumabaeva M, Thakar J, Choudhury RA, Yonemitsu C, et al. Infant gut microbiome is enriched with Bifidobacterium longum ssp. infantis in old order mennonites with traditional farming lifestyle. Allergy. (2021) 76(11):3489–503. doi: 10.1111/all.14877
21. Vatanen T, Plichta DR, Somani J, Munch PC, Arthur TD, Hall AB, et al. Genomic variation and strain-specific functional adaptation in the human gut microbiome during early life. Nat Microbiol. (2019) 4(3):470–9. doi: 10.1038/s41564-018-0321-5
22. Frese SA, Hutton AA, Contreras LN, Shaw CA, Palumbo MC, Casaburi G, et al. Persistence of supplemented Bifidobacterium longum subsp. infantis EVC001 in breastfed infants. mSphere. (2017) 2(6). doi: 10.1128/mSphere.00501-17
23. Casaburi G, Frese SA. Colonization of breastfed infants by Bifidobacterium longum subsp. infantis EVC001 reduces virulence gene abundance. Hum Microbiome J. (2018) 9:7–10. doi: 10.1016/j.humic.2018.05.001
24. Duar RM, Kyle D, Casaburi G. Colonization resistance in the infant gut: the role of B. infantis in reducing pH and preventing pathogen growth. High Throughput. (2020) 9(2). doi: 10.3390/ht9020007
25. Casaburi G, Duar RM, Vance DP, Mitchell R, Contreras L, Frese SA, et al. Early-life gut microbiome modulation reduces the abundance of antibiotic-resistant bacteria. Antimicrob Resist Infect Control. (2019) 8:131. doi: 10.1186/s13756-019-0583-6
26. Huda MN, Ahmad SM, Alam MJ, Khanam A, Kalanetra KM, Taft DH, et al. Bifidobacterium abundance in early infancy and vaccine response at 2 years of age. Pediatrics. (2019) 143:2. doi: 10.1542/peds.2018-1489
27. Orivuori L, Mustonen K, de Goffau MC, Hakala S, Paasela M, Roduit C, et al. High level of fecal calprotectin at age 2 months as a marker of intestinal inflammation predicts atopic dermatitis and asthma by age 6. Clin Exp Allergy. (2015) 45(5):928–39. doi: 10.1111/cea.12522
28. Arrieta MC, Arevalo A, Stiemsma L, Dimitriu P, Chico ME, Loor S, et al. Associations between infant fungal and bacterial dysbiosis and childhood atopic wheeze in a nonindustrialized setting. J Allergy Clin Immunol. (2018) 142(2):424–434.e10. doi: 10.1016/j.jaci.2017.08.041
29. Insel R, Knip M. Prospects for primary prevention of type 1 diabetes by restoring a disappearing microbe. Pediatr Diabetes. (2018) 19(8):1400–6. doi: 10.1111/pedi.12756
30. Tan TN, Lim DL, Lee BW, Van Bever HP. Prevalence of allergy-related symptoms in Singaporean children in the second year of life. Pediatr Allergy Immunol. (2005) 16(2):151–6. doi: 10.1111/j.1399-3038.2005.00242.x
31. Siow JK, Alshaikh NA, Balakrishnan A, Chan KO, Chao SS, Goh LG, et al. Ministry of health clinical practice guidelines: management of rhinosinusitis and allergic rhinitis. Singapore Med J. (2010) 51(3):190–7.20428739
32. Soh SE, Tint MT, Gluckman PD, Godfrey KM, Rifkin-Graboi A, Chan YH, et al. Cohort profile: growing up in Singapore towards healthy outcomes (GUSTO) birth cohort study. Int J Epidemiol. (2014) 43(5):1401–9. doi: 10.1093/ije/dyt125
33. Satokari RM, Vaughan EE, Akkermans AD, Saarela M, de Vos WM. Bifidobacterial diversity in human feces detected by genus-specific PCR and denaturing gradient gel electrophoresis. Appl Environ Microbiol. (2001) 67(2):504–13. doi: 10.1128/AEM.67.2.504-513.2001
34. Lawley B, Munro K, Hughes A, Hodgkinson AJ, Prosser CG, Lowry D, et al. Differentiation of Bifidobacterium longum subspecies longum and infantis by quantitative PCR using functional gene targets. PeerJ. (2017) 5:e3375. doi: 10.7717/peerj.3375
35. McKean JW, Kloke JD. Rfit: rank-based estimation for linear models. R J. (2012) 4(2):57–64. doi: 10.32614/RJ-2012-014
36. Azad MB, Konya T, Maughan H, Guttman DS, Field CJ, Chari RS, et al. Gut microbiota of healthy Canadian infants: profiles by mode of delivery and infant diet at 4 months. CMAJ. (2013) 185(5):385–94. doi: 10.1503/cmaj.121189
37. Backhed F, Roswall J, Peng Y, Feng Q, Jia H, Kovatcheva-Datchary P, et al. Dynamics and stabilization of the human gut microbiome during the first year of life. Cell Host Microbe. (2015) 17(5):690–703. doi: 10.1016/j.chom.2015.04.004
38. Dominguez-Bello MG, De Jesus-Laboy KM, Shen N, Cox LM, Amir A, Gonzalez A, et al. Partial restoration of the microbiota of cesarean-born infants via vaginal microbial transfer. Nat Med. (2016) 22(3):250–3. doi: 10.1038/nm.4039
39. Xu J, Lawley B, Wong G, Otal A, Chen L, Ying TJ, et al. Ethnic diversity in infant gut microbiota is apparent before the introduction of complementary diets. Gut Microbes. (2020) 11(5):1362–73. doi: 10.1080/19490976.2020.1756150
40. Vangay P, Johnson AJ, Ward TL, Al-Ghalith GA, Shields-Cutler RR, Hillmann BM, et al. US immigration westernizes the human gut microbiome. Cell. (2018) 175(4):962–972.e10. doi: 10.1016/j.cell.2018.10.029
41. Copeland JK, Chao G, Vanderhout S, Acton E, Wang PW, Benchimol EI, et al. The impact of migration on the gut metagenome of South Asian Canadians. Gut Microbes. (2021) 13(1):1–29. doi: 10.1080/19490976.2021.1902705
42. Dai DLY, Petersen C, Hoskinson C, Del Bel KL, Becker AB, Moraes TJ, et al. Breastfeeding enrichment of B. longum subsp. infantis mitigates the effect of antibiotics on the microbiota and childhood asthma risk. Med. (2023) 4(2):92–112.e5. doi: 10.1016/j.medj.2022.12.002
43. Henrick BM, Hutton AA, Palumbo MC, Casaburi G, Mitchell RD, Underwood MA, et al. Elevated fecal pH indicates a profound change in the breastfed infant gut microbiome due to reduction of Bifidobacterium over the past century. mSphere. (2018) 3(2). doi: 10.1128/mSphere.00041-18
Keywords: Bifidobacterium longum subsp. infantis, infant, ethnicity, immigration, delivery mode, antibiotics, gut microbiome, GUSTO
Citation: Xu J, Duar RM, Quah B, Gong M, Tin F, Chan P, Sim CK, Tan KH, Chong YS, Gluckman PD, Frese SA, Kyle D and Karnani N (2024) Delayed colonization of Bifidobacterium spp. and low prevalence of B. infantis among infants of Asian ancestry born in Singapore: insights from the GUSTO cohort study. Front. Pediatr. 12:1421051. doi: 10.3389/fped.2024.1421051
Received: 21 April 2024; Accepted: 29 May 2024;
Published: 10 June 2024.
Edited by:
Andrew S. Day, University of Otago, New ZealandReviewed by:
Marta Mangifesta, Consultant, Salt Lake City, United States© 2024 Xu, Duar, Quah, Gong, Tin, Chan, Sim, Tan, Chong, Gluckman, Frese, Kyle and Karnani. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Neerja Karnani, bmVlcmphX2thcm5hbmlAYmlpLmEtc3Rhci5lZHUuc2c=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.