- 1Department of Pathobiological Sciences, University of Wisconsin School of Veterinary Medicine, Madison, WI, United States
- 2Department of Pediatrics, University of Wisconsin School of Medicine and Public Health, Madison, WI, United States
- 3The Kasiisi Project, Fort Portal, Uganda
Introduction: Respiratory illness is the most common childhood disease globally, especially in developing countries. Previous studies have detected viruses in approximately 70-80% of respiratory illnesses.
Methods: In a prospective cohort study of 234 young children (ages 3-11 years) and 30 adults (ages 22-51 years) in rural Western Uganda sampled monthly from May 2019 to August 2021, only 24.2% of nasopharyngeal swabs collected during symptomatic disease had viruses detectable by multiplex PCR diagnostics and metagenomic sequencing. In the remaining 75.8% of swabs from symptomatic participants, we measured detection rates of respiratory bacteria Haemophilus influenzae, Moraxella catarrhalis, and Streptococcus pneumoniae by quantitative PCR.
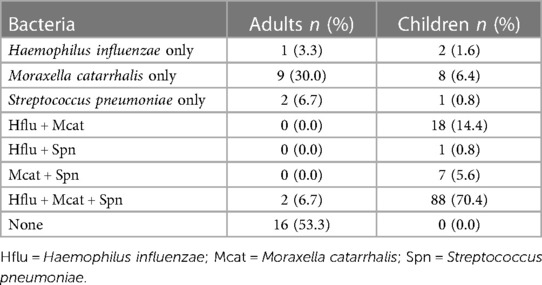
Results: 100% of children tested positive for at least one bacterial species. Detection rates were 87.2%, 96.8%, and 77.6% in children and 10.0%, 36.7%, and 13.3% for adults for H. influenzae, M. catarrhalis, and S. pneumoniae, respectively. In children, 20.8% and 70.4% were coinfected with two and three pathogens, respectively, and in adults 6.7% were coinfected with three pathogens but none were coinfected with two. Detection of any of the three pathogens was not associated with season or respiratory symptoms severity, although parsing detection status by symptoms was challenged by children experiencing symptoms in 80.3% of monthly samplings, whereas adults only reported symptoms 26.6% of the time. Pathobiont colonization in children in Western Uganda was significantly more frequent than in children living in high-income countries, including in a study of age-matched US children that utilized identical diagnostic methods. Detection rates were, however, comparable to rates in children living in other Sub-Saharan African countries.
Discussion: Overall, our results demonstrate that nonviral colds contribute significantly to respiratory disease burden among children in rural Uganda and that high rates of respiratory pathobiont colonization may play a role. These conclusions have implications for respiratory health interventions in the area, such as increasing childhood immunization rates and decreasing air pollutant exposure.
1 Introduction
Respiratory illness is the most common childhood disease globally, especially in low- and middle-income countries (LMICs) (1). Seminal studies detected viruses in 70%–80% of respiratory illnesses, but these studies sampled populations in affluent locations, primarily the US (2, 3) or European countries (4). However, these studies lack generalizability to children in LMICs, where the burden of bacterial disease is much higher and uptake of conjugate vaccines for respiratory pathogens Streptococcus pneumoniae (pneumococcus, PCV) and Haemophilus influenzae type b (Hib) is much lower (5). There are currently few comparable data from Sub-Saharan Africa, which vary by location and methodology. In a study of Ugandan children under five years admitted to a hospital for febrile respiratory illness, 80.0% of nasopharyngeal swabs tested positive for a virus by metagenomics (6), whereas only 48.8% and 45.7% tested PCR-positive among children of the same age admitted to intensive care units in Mozambique (7) and South Africa (8), respectively. Because most people do not seek medical care for relatively mild respiratory illnesses, particularly where health care access is limited, such hospital-based studies may not accurately capture prevalence rates of viral and nonviral colds. We sought to characterize respiratory illness etiologies in rural Western Uganda through a community-based longitudinal study.
We conducted a prospective cohort study of young children and adults in rural communities in Kabarole District, Western Uganda (9). Each month from May 2019 to August 2021, we collected paired respiratory symptoms surveys and nasopharyngeal swabs by visiting participants at their schools, homes, and workplaces. We first examined whether respiratory symptoms were caused by common respiratory pathogens using a multiplex PCR panel that detects 17 viruses and three bacteria. We measured frequencies of both viral and nonviral respiratory disease and compared them to elsewhere (2–4, 10).
We then measured CXC chemokine ligand 10 (CXCL10) in nasopharyngeal swabs. Induced by interferon-γ, CXCL10 plays an important role in both innate and adaptive antiviral immune responses by inducing chemotaxis of NK cells, macrophages, dendritic cells, and T lymphocytes to sites of infection (11) and polarizing Th1 cells (12). Elevated CXCL10 levels can indicate active viral infection (13–15). We therefore examined individuals with high CXCL10 levels using metagenomics to investigate rare or novel viruses unrepresented on our multiplex PCR panel.
Next, we measured carriage of respiratory pathobionts (pathogenic bacteria that first colonize the upper respiratory tract as commensals) Haemophilus influenzae, Moraxella catarrhalis, and Streptococcus pneumoniae. These pathobionts often cause acute otitis media, sinusitis, and pneumonia in children (16) as well as exacerbations of chronic obstructive pulmonary disease in adults (17, 18). Pathogenic microbiota overgrowth generally follows acute insult from either respiratory viral infections or air pollutants. Detection rates and densities increase after respiratory virus infection by inducing cellular receptors used by bacteria for adhesion (19, 20) and disrupting epithelial barrier function (21). Bacterial components can activate Toll-like receptors, prompting the release of large amounts of inflammatory cytokines, resulting in increased risk of wheezing illnesses and asthma exacerbations (10, 22, 23). Additionally, environmental and household air pollution can increase levels of these pathobionts, especially in many parts of Sub-Saharan Africa (e.g., South Africa (24), Ghana (25), and Malawi (26)) where annual fine particulate matter (PM2.5) concentrations exceed WHO standards by as much as 10-fold (27). To contrast, we compared respiratory pathobiont prevalence among children in this study with prevalence among age-matched, suburban children from a US cohort study that utilized the same diagnostic methods (10), allowing for direct comparison of these two populations.
Fortuitously, the study occurred before, during, and after the emergence COVID-19 when public health measures (e.g., closure of schools and businesses, prohibition of travel internationally or between districts in-country, mandatory mask-wearing, etc.) were instituted, allowing us to capture the changing dynamics of respiratory disease during a time of intense public health measures with the goal of reducing respiratory disease transmission. Our results shed new light on nonviral respiratory illnesses in Sub-Saharan Africa.
2 Methods
2.1 Study site, subjects, and sample collection
The design, methods, and study population for this study have been previously reported in detail (9). Briefly, we conducted a prospective cohort study between May 2019 and August 2021 in rural Western Uganda. Household characteristics relevant for respiratory disease risk in this region include 7.4% adult tobacco smoking prevalence (28), 89% use of solid biomass cooking fuel (charcoal) (29), 44% living in dwellings made with permanent wall materials (30), and 40% access to adequate indoor ventilation (29). 69% of the local population rely on subsistence farming for their livelihoods (30). After obtaining written informed consent from adult participants and parents of child participants as well as assent from children >8 years old, we enrolled 234 children (ages 3–11) and 30 adults (ages 22–51), some of whom were parents of the child participants. Each month, trained nurses collected monthly nasopharyngeal swabs and respiratory symptoms scores (9) from all participants at their schools, homes, or workplaces. After Uganda instated national lockdown for COVID-19 on March 20, 2020, we obtained permission from study participants and the Ugandan government to continue sampling adult participants and their children (n = 31) at their homes with strict biosafety precautions to protect participants and study team personnel. Although primary schools in Uganda did not reopen until January 2022, some lockdown restrictions began to loosen in October 2020 (e.g., opening businesses, major roads, and the international airport), a period we denote “late lockdown.” De-identification of participant data required for institutional ethics approval precluded collecting demographic data beyond age ranges for each cohort.
2.2 Viral diagnostics
We tested nasopharyngeal swabs using the NxTAG Respiratory Pathogen Panel (RPP) (Luminex Corporation, Austin, TX, USA) as previously described (31, 32). Immediately upon sample collection, Dacron swabs were placed in RNAlater preservation buffer (Thermo Fisher Scientific, Waltham, MA, USA) and stored at −20°C until shipment on dry ice to Madison, Wisconsin, facilitating molecular analysis. Nucleic acids were extracted as previously described using the NucliSENS EasyMag kit (bioMérieux, Marcy-l'Étoile, France) (31). The RPP tests for influenza viruses A and B, rhinovirus/enterovirus, adenovirus, parainfluenza viruses 1–4, coronaviruses (CoV NL63, CoV 229E, CoV HKU1, CoV OC43, and SARS-CoV-2), respiratory syncytial viruses A and B, metapneumovirus, human bocavirus, and the bacterial targets Chlamydophila pneumoniae, Mycoplasma pneumoniae, and Legionella pneumophilia. Sensitivity and specificity vary by pathogen but on average are approximately 95% and 99%, respectively (33).
2.3 Quantification of CXCL10 and respiratory bacteria
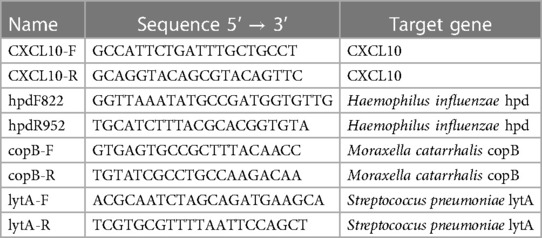
We measured CXCL10 mRNA levels via quantitative PCR (qPCR) in a representative subset of swabs (n = 232) to interrogate potential cryptic viral infections in samples that tested PCR-negative as described elsewhere (15) using specific primers (Table 1). Levels of H. influenzae, M. catarrhalis, and S. pneumoniae were measured in a subset of PCR-negative swabs from adults (n = 30) and children aged 3–6 years (n = 125) as described elsewhere (34–36) using specific primers (Table 1). CXCL10 and bacteria qPCR reactions were performed in 25 µl volumes consisting of 13.8 µl POWER SYBR Green PCR Master Mix (Thermo Fisher Scientific), 100 µM each primer, PCR-grade water, and 2 µl cDNA. Thermal cycling parameters consisted of an initial incubation of 50°C for 2 min and 95°C for 10 min, followed by 40 cycles of 95°C for 15 s and 60°C for 1 min. The qPCR assay was performed using a CFX96 Touch Real-Time PCR Detection System (Bio-Rad, Hercules, CA, USA).
2.4 Metagenomic sequencing and bioinformatics
Metagenomic sequencing was used to identify viruses in 24 nasopharyngeal swabs from children who displayed moderate to severe respiratory symptoms (symptoms scores = 6–16) but tested virus-negative by RPP, using previously described methods (37–40). Briefly, Dacron swab tips were homogenized with 50 μl RNAlater suspension and 350 μl Hanks' Balanced Salt Solution (HBSS) and centrifuged to clarify. The supernatant was treated with nucleases to digest nucleic acids not encapsidated within virions (41). Nucleic acids were extracted using the QIamp MinElute Virus Spin Kit (Qiagen, Hilden, Germany), with carrier RNA omitted. RNA was converted to double-stranded cDNA with the Superscript IV system (Thermo Fisher), which was purified using Agencourt AMPure XP beads (Beckman Coulter, Brea, CA, USA) as previously described (37–40). Genomic libraries were prepared using the Illumina Nextera XT kit (Illumina, San Diego, CA, USA) and sequenced on an Illumina MiSeq instrument using 300 × 300 cycle paired-end (V3) chemistry.
Sequences of low quality (Phred score <30) and short length (<50 bp) were trimmed and sequences matching known contaminants and host DNA were discarded using CLC Genomics Workbench v. 20.0.4 (Qiagen, Hilden, Germany). Remaining reads were then subjected to de novo assembly using the metaviral option in SPAdes v. 3.15.2 (42). The resultant contiguous sequences (contigs) were compared to viruses in NCBI databases at both the nucleotide and amino acid levels using the BLASTn and BLASTx algorithms, respectively (43, 44).
2.5 Comparison of nonviral colds and bacteria colonization with suburban US children
Symptoms status, viral infection status, and bacteria colonization status were compared to data collected as part of the 2006–2008 RhinoGen study described in detail elsewhere (10, 22, 45). Three hundred eight children (166 with asthma and 142 without asthma) aged 4–12 years living in Madison, Wisconsin, a suburban college town in the US, were enrolled in the study. Children provided weekly nasal lavage samples (45), which have been shown to yield similar rates of bacterial detection as nasopharyngeal swabs (22). Methods utilized for viral diagnostics and qPCR for H. influenzae, M. catarrhalis, and S. pneumoniae were identical to those used in this study (10), allowing for direct comparison. Only age-matched children (4–6 years) were included (n = 289).
2.6 Statistical analysis
Parametric model assumptions were assessed with Shapiro-Wilk tests for verification of normality and with Levene's test for verification of homogeneity of variances. For power analysis for the bacteria assays, we utilized the functions cohensD and pwr.t.test in R (46) on a pilot subset of samples (n = 31), aiming for a significance level of 0.05 and a power of 0.8. Symptoms status, viral infection, and age class were compared to bacteria colonization by X2 test or Fisher's exact test for association. Presence of nonviral colds or bacteria colonization were compared to calendar month with ANOVA or Kruskal-Wallis test. Levels of CXCL10 or bacteria, measured by qPCR Ct values, were compared to age class or symptoms status by Mann-Whitney U test. A two-sided p-value of less than 0.05 was regarded as statistically significant.
3 Results
In total, we collected 2,047 symptoms scores (534 scores from 30 adults and 1,513 scores from 234 children) and 1,989 nasopharyngeal swabs from 264 individuals (538 swabs from adults and 1,451 swabs from children) from May 2019 through August 2021. We obtained 1,976 paired swabs and symptoms scores.
3.1 Nonviral colds
If participants reported experiencing any respiratory symptoms, their nasopharyngeal swabs were 2.3 times more likely to test PCR-positive for at least one of 17 common respiratory viruses than if they experienced no symptoms (Fisher's exact test, p < 0.001) (Figure 1). However, samples which tested PCR-negative but were collected from people experiencing respiratory symptoms (“S+/V-”; n = 993) accounted for 50.3% of the total sample set (n = 1,976 paired swabs with symptoms scores) and 75.8% of all swabs collected during symptomatic disease (n = 1,310) (Figure 1). The proportion of S+/V- swabs decreased significantly during the most stringent period of lockdown (March 2020–September 2020; 6.0%) compared to the pre-pandemic (May 2019–February 2020; 23.6%) or late lockdown (October 2020–August 2021; 18.7%) periods (one-way ANOVA with Tukey HSD, pairwise p = 0.004 pre-pandemic vs. March–September 2020, pairwise p = 0.03 March–September 2020 vs. October 2020–August 2021) (Figure 2).
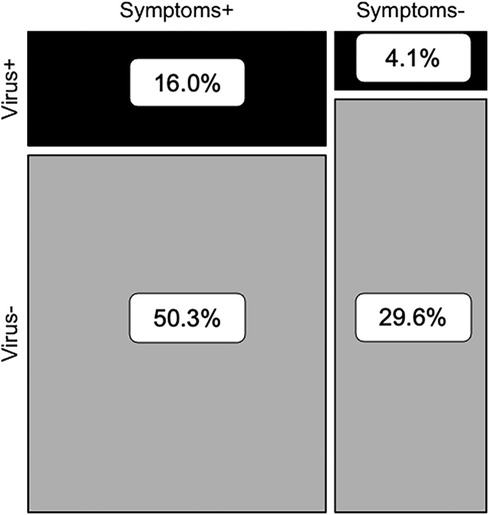
Figure 1. Contingency analysis of presence/absence of viral respiratory pathogens and respiratory symptoms (n = 1,976 paired nasopharyngeal swabs and symptoms scores).
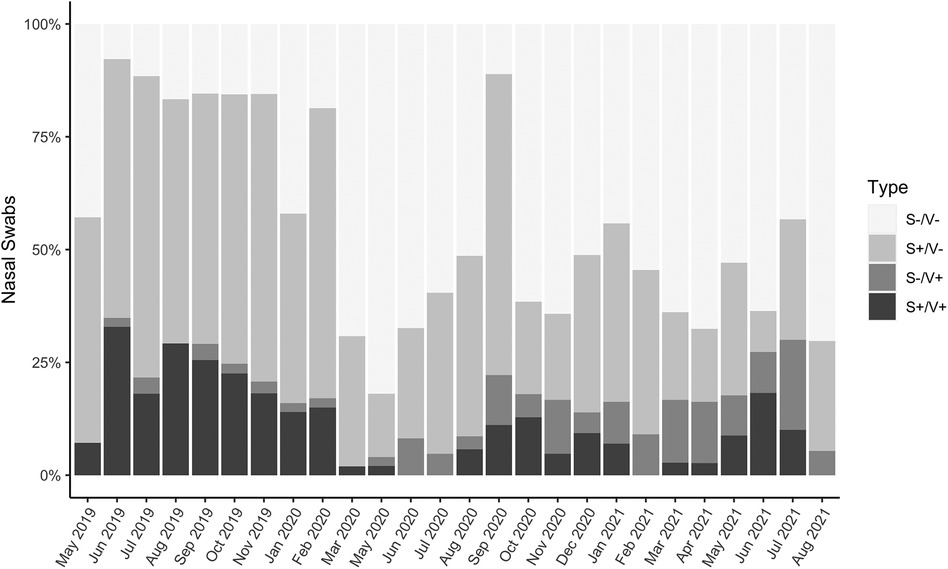
Figure 2. Proportions of symptoms-negative/virus-negative (S-/V-; healthy), symptoms-positive/virus-negative (S+/V-; nonviral cold), symptoms-negative/virus-positive (S-/V+; asymptomatic infection), and symptoms-positive/virus-positive (S+/V+; symptomatic infection) samples, May 2019-August 2021 (n = 1,976 paired nasopharyngeal swabs and symptoms scores). There were no samplings in December 2019 or April 2020.
3.2 CXCL10 expression
CXCL10 expression levels were higher in virus-infected people than virus-negative, as determined by multiplex PCR (Mann-Whitney U test, p < 0.0001) (Figure 3A). People experiencing moderate to severe cold symptoms (symptoms scores > 4), regardless of etiology, had higher CXCL10 levels than people experiencing no cold symptoms (symptoms scores = 0) (Mann-Whitney U test, p = 0.032) (Figure 3B). Experiencing cold symptoms with a PCR-positive viral infection (S+/V+) was associated with increased CXCL10 levels compared to S+/V- cases (Kruskal-Wallis test with Dunn's multiple comparison, p < 0.0001) and S-/V- cases (Kruskal-Wallis test with Dunn's multiple comparison, p = 0.0001) (Figure 3C).
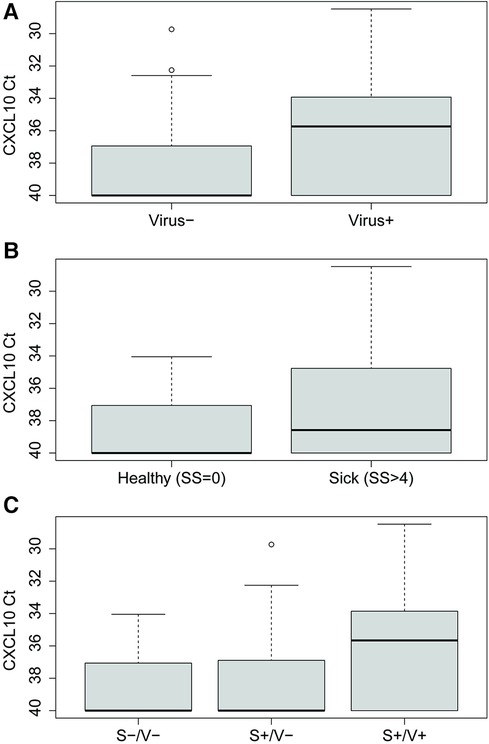
Figure 3. CXCL10 expression levels quantified by Ct values for (A) virus-positive (n = 100) vs. virus-negative (n = 132) people, as diagnosed by multiplex PCR panel, (B) no symptoms (n = 23) vs. moderate-to-severe symptoms (n = 209), as measured by symptoms scores (SS), and (C) symptoms-negative/virus-negative (S-/V-) (n = 23), symptoms-positive/virus-negative (S+/V-) (n = 109), and symptoms-positive/virus-positive (S+/V+) (n = 100) people.
3.3 Metagenomic sequencing
Following quality trimming and in silico subtraction of host and known contaminant sequences from the sequenced S+/V- samples, we retained a total of 26,756,296 reads with an average length of 141.5 bp for analysis. No reads mapped to genomes of mammalian viruses, whereas 100% of reads matched phage, bacteria, or fungi (data not shown).
3.4 Bacterial detection in children and adults
Prevalence of respiratory pathobionts H. influenzae, M. catarrhalis, and S. pneumoniae detected in nasopharyngeal swabs was higher in children than adults (87.2% vs. 20.0%, X2 = 167.7, p < 0.0001) (Table 2). Levels were also significantly higher in children than adults for each bacterial species (Mann-Whitney U test, p < 0.0001 for each species) (Figure 4). Of the three species, M. catarrhalis was detected with the highest frequencies (X2 = 16.6, p = 0.0002) (Table 2) and levels (Kruskal-Wallis with Dunn's multiple comparison, p < 0.0001) (Figure 4).
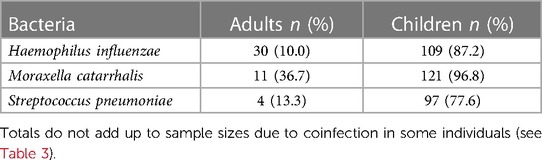
Table 2. Prevalence of respiratory pathobionts Haemophilus influenzae, Moraxella catarrhalis, and Streptococcus pneumoniae detected in nasopharyngeal swabs of virus-negative adults (n = 30) and children (n = 125).
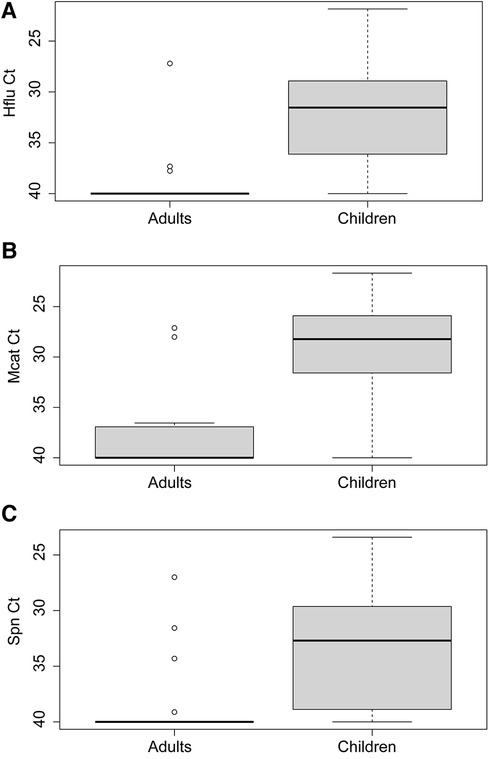
Figure 4. Levels of (A) Haemophilus influenzae (Hflu), (B) Moraxella catarrhalis (Mcat), and (C) Streptococcus pneumoniae (Spn) in nasopharyngeal swabs of virus-negative adults (n = 30) and children (n = 125).
Coinfection was much more likely in children than in adults (Table 3). Coinfection was also more common than monoinfection in children, whereas the opposite was true for adults. The most common colonization status in children was infection with all three pathobionts. No children tested negative for all three pathobionts, whereas a triple-negative was the most common colonization status in adults.
Colonization with any of the three bacterial species was not associated with increased symptoms scores in either children or adults (Supplementary Table S1). Positive cases also were not associated with sampling month for any of the three bacteria (Kruskal-Wallis test, all pairwise comparisons p > 0.05) (Supplementary Figure S1).
3.5 Comparison of nonviral cold prevalence and bacteria colonization with US suburban children
Age-matched children living in a suburban US community (see Methods) were just as likely as rural Ugandan children from this study to have PCR-positive viral infections when they experienced respiratory symptoms (OR: 2.2 vs. 2.3; see Figure 1). However, the Ugandan children had frequencies of nonviral colds two times higher than the US children (50.2% vs. 25.3% of samples). Prevalence of all three bacterial species was significantly higher in the Ugandan children (Hflu: X2 = 151.0, p < 0.0001; Mcat: X2 = 154.2, p < 0.0001; Spn: X2 = 17.4, p < 0.0001) than in the virus-negative US children (Table 4). While nearly a third (32.9%) of the virus-negative US children were PCR-negative for all three pathobionts, no Ugandan children similarly tested triple-negative (Table 5). If the US children tested positive for any bacteria, the most commonly detected was S. pneumoniae (25.3%), whereas S. pneumoniae only was the least common detection (0.8%) in the Ugandan children (Table 5).
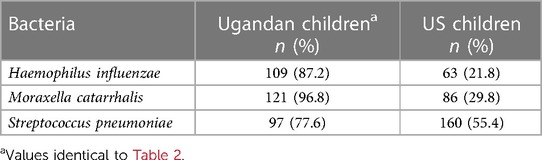
Table 4. Prevalence of respiratory pathobionts Haemophilus influenzae, Moraxella catarrhalis, and Streptococcus pneumoniae detected in nasal samples of age-matched, virus-negative Ugandan children (n = 125) and US children (n = 289).
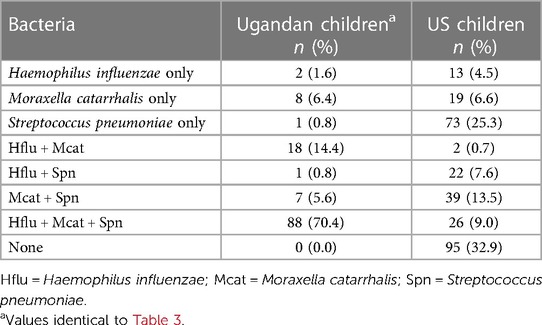
Table 5. Respiratory pathobiont coinfection in nasal samples of age-matched, virus-negative Ugandan children (n = 125) and US children (n = 289).
4 Discussion
In this study of respiratory illness in rural Western Uganda, people were 2.3 times more likely to test PCR-positive for at least one of 17 common respiratory viruses if they were experiencing respiratory symptoms than if they felt healthy. However, 50.2% of all nasopharyngeal swabs, including 75.8% of swabs collected during symptomatic disease, tested PCR-negative for respiratory viruses (S+/V-), which was confirmed by metagenomic sequencing for a subset of samples with high symptoms scores. We then tested S+/V- samples for the respiratory pathobionts H. influenzae, M. catarrhalis, and S. pneumoniae and found that all child participants tested positive for at least one bacterial species, with coinfections of all three species as the most common presentation (70.4% of swabs). These children had much higher prevalence rates and levels of these bacteria than adults as well as much higher prevalence rates compared to age-matched children living in the suburban US community of Madison, Wisconsin. Adults tested positive for each of the three pathobionts, although detection rates in adults were much lower than in children.
The prevalence of nonviral colds was much higher in our study (50.2%) in rural Western Uganda than recorded elsewhere in the world. Nonviral colds accounted for 22% of samplings in the Tecumseh study, a seminal respiratory disease cohort study performed in the US in the 1960s (2, 47), as well as 31% of samplings in Finland in the 1990s (4). Despite using the same diagnostic methods as in this study, nonviral colds only accounted for 25.3% of samplings among age-matched children in Madison, Wisconsin, in the RhinoGen study (10). It has been argued that purportedly nonviral colds may be caused by viruses yet to be identified (48), especially after the discoveries of human metapneumovirus (49) and human bocavirus (50) in the past 20 years. However, we did not find any novel viruses in these samples using metagenomic DNA sequencing, a technique our group has used extensively to identify novel infectious agents in a variety of host species (38, 51, 52).
On average, S+/V- samples had lower CXCL10 expression than S+/V+ samples, concurring with current understanding that CXCL10 expression is increased significantly during inflammation induced by viral infection (11), including by respiratory viruses such as rhinovirus (53), respiratory syncytial virus (54), and coronaviruses (55). However, CXCL10 expression is not always specific to viral infection, as evidenced by the S+/V- samples that also had higher CXCL10 expression than S-/V- samples, despite testing negative for viruses by multiplex PCR panel and metagenomic sequencing. Indeed, CXCL10 expression has been demonstrated to increase in response to infection with bacterial and protozoal infections common in Uganda, such as tuberculosis (56), scrub typhus (57), malaria (58), and leishmaniasis (59), as well as in noninfectious inflammatory disorders, such as asthma (60) and chronic obstructive pulmonary disorder (COPD) (61). We therefore explored nonviral causes of S+/V- illnesses in our data set.
Carriage of respiratory pathobionts H. influenzae, M. catarrhalis, and S. pneumoniae was ubiquitous among young children experiencing nonviral colds in our study; 100.0% of S+/V- swabs from children tested positive for at least one species. Although the goal of the current study was to elucidate causes of nonviral colds, previous studies have shown that pathobiont carriage increases with respiratory viral infections (10, 22) and data for this population in future studies would be a valuable comparison. For each bacterial species, detection rates were much higher than those among children in Madison, Wisconsin, using the same qPCR methods as this study (10). There is growing evidence that pathobiont carriage rates among children differ geographically (62) and by socioeconomic status (63). For example, rates were similar to those measures elsewhere in Sub-Saharan Africa among Gambian neonates (64) as well as HIV-positive children living in Tanzania (65) and Ethiopia (66). Seasonality did not appear to affect pathobiont carriage, as has been reported in other studies (67–69), in which rates were inversely proportional to temperature with a peak during winter months. However, these studies all took place in temperate regions of the US where seasonality is more pronounced. It is possible that patterns of respiratory pathobiont colonization are more variable in tropical regions, similar to patterns described for respiratory viruses (70, 71).
Pathobiont colonization was not associated with increased symptoms severity, in contrast to previous studies (16, 22). Because children in this study reported experiencing respiratory symptoms during 80.3% of samplings and adults reported symptoms only 26.6% of the time (9), it is possible we did not have the statistical power to differentiate symptoms status by colonization status. In fact, we assayed every sample from children with no symptoms and adults with at least moderate symptoms (symptoms scores > 4) and we still failed to detect significant differences between these groups and the more common symptomatic children and asymptomatic adults, respectively. Mechanisms explaining the observed association between pathobiont colonization and respiratory symptom severity in our study cohorts therefore remain unknown. Nonetheless, high pathobiont detection rates in early childhood have well known associations with clinically significant conditions, such as acute otitis media, sinusitis, pneumonia, and asthma (16). Children in this population may be at increased risk for these conditions. Future studies should investigate pathobiont diversity measures (e.g., alpha and beta diversity) and Th1 cytokine pathways beyond CXCL10 to further characterize the relationship between colonization with these bacteria and airway inflammation.
Although adults had much lower frequencies of respiratory pathobiont colonization than children, these frequencies, especially for M. catarrhalis, were still higher than those recorded elsewhere, including among healthy adults in England (72) and adults with COPD in the US (17). In fact, M. catarrhalis, H. influenzae, and S. pneumoniae cause approximately half of COPD exacerbations among adults (18). Chronic bacterial colonization in the respiratory tract leads to sloughing of highly immunogenic cell wall antigens that leads to the hallmark airway inflammation of COPD (18). COPD is a growing problem across Sub-Saharan Africa and is expected to overtake HIV/AIDS as the leading cause of death in this region by 2030 (73). In previous study in Uganda, place of residence (rural vs. urban) was the most significant determinant of COPD diagnosis, with COPD being more prevalent in rural areas (74). Asymptomatic carriage of these bacteria, however, is common among adults (18) and children (75, 76). Thus, PCR positivity should not be used exclusively to diagnose COPD in study participants, although we suspect it may contribute to the high prevalence of nonviral respiratory illnesses.
Other potential causes of nonviral colds in children and adults include allergic rhinitis, asthma, or air pollutant exposure. Respiratory allergies can also provoke non-infectious nasal and chest symptoms in children, but is an unlikely explanation for symptoms in Uganda, where the prevalence of allergic rhinitis is <5% (77). Evidence from other studies suggests that asthma may be underdiagnosed in Uganda, both in young children (78) and adults (79). Critically, however, both indoor and outdoor air pollution in Western Uganda regularly exceed levels deemed unsafe by the WHO by four to six times (80). Annual mean PM2.5 concentrations in the Western Uganda region measure in the top quintile for the country (27). Smoke and carbon monoxide from indoor charcoal-fueled cookstoves used in the area may contribute to respiratory disease symptoms (29), including asthma in children (81) and COPD exacerbations in adults (73). This association is not unique to Uganda or Sub-Saharan Africa. In urban areas of the US, spikes in air pollutants similarly can produce upper and lower respiratory illnesses in children (82). Air pollution also exacerbates COPD in adults all over the world (83). We advocate for mitigating exposure to non-infectious respiratory disease factors that are prevalent in rural Uganda, such as indoor biomass smoke and outdoor air pollution from vehicles, industrial and agricultural practices, or fires (84).
In summary, our findings demonstrate that people living in rural Western Uganda experience high frequencies of nonviral colds. Although we detected high detection rates of respiratory bacteria, especially in children, carriage was not associated with increased cold symptoms severity. However, this may be due to the low number of children not reporting any respiratory symptoms in the study. Our data do not support treatment of these bacteria as a way to reduce the frequency and severity of nonviral colds in this population. Although there is evidence that azithromycin reduces wheezing illness in babies and preschool-aged children (85, 86), suggesting a relationship between wheezing and airway bacteria dysbiosis (87), serious risks include antimicrobial resistance, killing healthy microbes, drug costs, and potential side effects. Antibiotic therapy therefore is not advised in uncomplicated colds.
Vaccinations against these pathobionts are either already in use for S. pneumoniae (PCV13) (88) or in development for M. catarrhalis and non-typeable H. influenzae (89). Since they were first licensed in 2000, pneumococcal conjugate vaccines have altered population-level detection rates of non-vaccine type S. pneumoniae (90), H. influenzae (91, 92), and M. catarrhalis (91). Although the current study is limited by a lack of individual participant health and demographic information, such as age and immunization status, due to required data de-identification, differential immunization rates between the US and Uganda could explain the differences in pathobiont colonization we observed between the two populations compared here. Indeed, childhood immunization rates are currently “suboptimal” in Uganda (93). For example, Uganda introduced PCV10 in 2013, later than neighboring Kenya (2011), Tanzania (2013) or Rwanda (2010), after experiencing funding challenges that hindered vaccine rollout and health worker training (94, 95). In July 2023, the official Ugandan government estimate for full three-dose PCV coverage was 90% [at the time of the RhinoGen study, PCV coverage in the US was 93% after introduction in 2000 (96)], but the Ugandan figure was disputed by the WHO and UNICEF estimates of national immunization coverage (WUENIC) because no nationally representative household survey had been conducted in the previous five years (97). Closing this gap in vaccine uptake presents an opportunity for increasing protection against respiratory illnesses and their sequelae in Ugandan children.
Incidentally, our study occurred before, during, and after the emergence of COVID-19 in Uganda. We documented that the proportion of S+/V- samples decreased sharply during the most stringent part of COVID-19 lockdown in Uganda from March to September 2020, mirroring a drop in respiratory viral illnesses observed around the world during the same period (98). The decrease in nonviral colds we observed may have been associated with nonpharmaceutical interventions relied upon before the introduction of COVID-19 vaccines that would have protected mask-wearers from inhaling noxious particulate matter (99) and children staying home from school from pathobiont transmission (100). Lockdown had many deleterious social and economic effects across Sub-Saharan Africa due to lost social safety nets, daily wages, and educational opportunities (101–103). However, our findings support the fact that there were significant improvements to respiratory health in rural Uganda, particularly among children. Therefore, future public health policy measures should build upon these gains by exploring strategies that are more sustainable in the long-term. For example, based on the results of this study, the local government health office has changed its clinical response to respiratory disease in our study population by reducing overcrowding in primary school classrooms and laying cement over dirt floors to avoid particulate inhalation. We hope this study continues to serve as a model of how such research can have clinical implications and result in direct, meaningful, and specific changes in practice.
Data availability statement
The datasets presented in this article are not readily available because of research ethics requirements to protect study participants' health information privacy. Requests to access the datasets should be directed to the corresponding author (tony.goldberg@wisc.edu).
Ethics statement
The studies involving humans were approved by the Makerere University School of Health Sciences Research & Ethics Committee and the University of Wisconsin-Madison Institutional Review Board. The studies were conducted in accordance with the local legislation and institutional requirements. All human biological samples and survey data were collected with institutional approval from Makerere University (2018-077) and the Uganda National Council for Science and Technology (NS 657) under the guiding principles of the World Medical Association Declaration of Helsinki. De-identified data were analyzed with institutional approval from the University of Wisconsin-Madison (2019-0229-CR003). All subjects participated voluntarily. Informed consent was obtained from adult participants and parents of child participants (below 18 years old). Assent was obtained from children over eight years old. All informed consent/assent conversations were conducted in study participants' native language, Rutooro, by native speakers.
Author contributions
TW: Conceptualization, Data curation, Formal Analysis, Investigation, Methodology, Visualization, Writing – original draft, Writing – review & editing. TP: Investigation, Writing – review & editing. PT: Investigation, Project administration, Writing – review & editing. ST: Investigation, Writing – review & editing. ER: Funding acquisition, Supervision, Writing – review & editing. JG: Conceptualization, Funding acquisition, Resources, Supervision, Writing – review & editing. TG: Conceptualization, Funding acquisition, Resources, Supervision, Writing – review & editing.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article.
Morris Animal Foundation (D21ZO-044), Disney Conservation Fund, Arcus Foundation (G-PGM-2011-3337), One Health Commission/Georgia Aquarium Dr. Gregory D. Bossart Memorial One Health Scholarship, University of Wisconsin-Madison Global Health Institute Graduate Student Research Award, University of Wisconsin-Madison Research Forward.
Acknowledgments
We thank the Uganda National Council for Science and Technology and Makerere University College of Health Sciences for granting research approval in Uganda. We are immensely grateful to the Kibale Forest Schools Program staff for their assistance with study coordination. We also thank Eishika Dissanayake and Yury Bochkov of the University of Wisconsin Hospital & Clinics for providing qPCR primers and assistance with the qPCR protocols.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher's note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fped.2024.1379131/full#supplementary-material
References
1. Jin X, Ren J, Li R, Gao Y, Ya Zhang H, Li J, et al. Global burden of upper respiratory infections in 204 countries and territories, from 1990 to 2019. EClin Med. (2021) 37:100986. doi: 10.1016/j.eclinm.2021.100986
2. Monto AS, Sullivan KM. Acute respiratory illness in the community. Frequency of illness and the agents involved. Epidemiol Infect. Cambridge University Press. (1993) 110(1):145. doi: 10.1017/S0950268800050779
3. Byington CL, Ampofo K, Stockmann C, Adler FR, Herbener A, Miller T, et al. Community surveillance of respiratory viruses among families in the Utah better identification of germs-longitudinal viral epidemiology (BIG-LoVE) study. Clin Infect Dis. Oxford Academic. (2015) 61(8):1217–24. doi: 10.1093/CID/CIV486
4. Mäkelä MJ, Puhakka T, Ruuskanen O, Leinonen M, Saikku P, Kimpimäki M, Blomqvist S, Hyypiä T, et al. Viruses and bacteria in the etiology of the common cold. J Clin Microbiol. American Society for Microbiology (ASM). (1998) 36(2):539. doi: 10.1128/JCM.36.2.539-542.1998
5. Zar HJ, Polack FP. Childhood pneumonia: the role of viruses. Thorax. (2015) 70(9):811–2. doi: 10.1136/thoraxjnl-2015-207320
6. Ramesh Akshaya, Nakielny Sara, Hsu Jennifer, Kyohere Mary, Byaruhanga Oswald, de Bourcy Bourcy, et al. Metagenomic next-generation sequencing of samples from pediatric febrile illness in Tororo, Uganda. PLoS One. Public Library of Science. (2019) 14(6):e0218318. doi: 10.1371/JOURNAL.PONE.0218318
7. O Callaghan-Gordo Cristina, Bassat Quique, Morais Luis, Díez-Padrisa Núria, Machevo Sónia, Nhampossa Tacilta, et al. Etiology and epidemiology of viral pneumonia among hospitalized children in rural Mozambique: a malaria endemic area with high prevalence of human immunodeficiency virus. Pediatr Infect Dis J. Lippincott Williams and Wilkins. (2011) 30(1):39–44. doi: 10.1097/INF.0B013E3181F232FE
8. Famoroti T, Sibanda W, Ndung’u T. Prevalence and seasonality of common viral respiratory pathogens, including cytomegalovirus in children, between 0 and 5 years of age in KwaZulu-natal, an HIV endemic province in South Africa. BMC Pediatr. (2018) 18(1):240. doi: 10.1186/s12887-018-1222-8
9. Weary Taylor E, Tusiime Patrick, Tuhaise Shamilah, Mandujano Reyes, Juan Francisco, Ross Elizabeth, Gern James E, et al. Respiratory disease patterns in rural Western Uganda, 2019–2022. Front Pediatr. (2024) 12:e1336009. doi: 10.3389/fped.2024.1336009
10. Bashir Hiba, Grindle Kristine, Vrtis Rose, Vang Fue, Kang Teresa, Salazar Lisa, et al. Association of rhinovirus species with common cold and asthma symptoms and bacterial pathogens. J Allergy Clin Immunol. Mosby Inc. (2018) 141(2):822–824.e9. doi: 10.1016/j.jaci.2017.09.027
11. Liu Mingli, Guo Shanchun, Hibbert Jacqueline M, Jain Vidhan, Singh Neeru, Wilson Nana O, et al. CXCL10/IP-10 In infectious diseases pathogenesis and potential therapeutic implications. Cytokine Growth Factor Rev. Elsevier. (2011) 22(3):121. doi: 10.1016/j.cytogfr.2011.06.001
12. Lee AJ, Ashkar AA. The dual nature of type I and type II interferons. Front Immunol. Frontiers Media S.A. (2018) 9(SEP):403701. doi: 10.3389/FIMMU.2018.02061/BIBTEX
13. Trifilo Matthew J, Montalto-Morrison Cynthia, Stiles Linda N, Hurst Kelley R, Hardison Jenny L, Manning Jerry E, et al. CXC chemokine ligand 10 controls viral infection in the central nervous system: evidence for a role in innate immune response through recruitment and activation of natural killer cells. J Virol. American Society for Microbiology (ASM). (2004) 78(2):585–94. doi: 10.1128/JVI.78.2.585-594.2004
14. Cheng Vincent CC, Lau Susanna KP, Woo Patrick CY, Yuen Kwok Yung. Severe acute respiratory syndrome coronavirus as an agent of emerging and reemerging infection. Clin Microbiol Rev. American Society for Microbiology (ASM). (2007) 20(4):660–94. doi: 10.1128/CMR.00023-07
15. Bochkov Yury A, Busse William W, Brockman-Schneider Rebecca A, Evans Michael D, Jarjour Nizar N, McCrae Christopher, et al. Budesonide and formoterol effects on rhinovirus replication and epithelial cell cytokine responses. Respir Res. BioMed Central. (2013) 14(1):1–9. doi: 10.1186/1465-9921-14-98/FIGURES/4
16. Chapman Timothy J, Morris Matthew C, Xu Lei, Pichichero Michael E Nasopharyngeal colonization with pathobionts is associated with susceptibility to respiratory illnesses in young children. PLoS One. Public Library of Science. (2020) 15(12):e0243942. doi: 10.1371/JOURNAL.PONE.0243942
17. Murphy Timothy F, Brauer Aimee L, Grant Brydon JB, Sethi Sanjay. Moraxella catarrhalis in chronic obstructive pulmonary disease: burden of disease and immune response. Am J Respir Crit Care Med. American Thoracic Society. (2005) 172(2):195. doi: 10.1164/RCCM.200412-1747OC
18. Murphy TF, Parameswaran GI. Moraxella catarrhalis, a human respiratory tract pathogen. Clin Infect Dis. Oxford Academic. (2009) 49(1):124–31. doi: 10.1086/599375/2/49-1-124-TBL003.GIF
19. Avadhanula Vasanthi, Rodriguez Carina A, Devincenzo John P, Wang Yan, Webby Richard J, Ulett Glen C, et al. Respiratory viruses augment the adhesion of bacterial pathogens to respiratory epithelium in a viral species- and cell type-dependent manner. J Virol. American Society for Microbiology. (2006) 80(4):1629–36. doi: 10.1128/JVI.80.4.1629-1636.2006/ASSET/BC8F4DE9-4D24-4CBF-BE91-30F9FC3CC78E/ASSETS/GRAPHIC/ZJV0040674110005.JPEG
20. Dissanayake Eishika, Brockman-Schneider Rebecca A, Stubbendieck Reed M, Helling Britney A, Zhang Zhumin, Bochkov Yury A, et al. Rhinovirus increases Moraxella catarrhalis adhesion to the respiratory epithelium. Front Cell Infect Microbiol. Frontiers Media S.A. (2023) 12:1060748. doi: 10.3389/FCIMB.2022.1060748/FULL
21. Looi K, Buckley AG, Rigby PJ, Garratt LW, Iosifidis T, Zosky GR, et al. Effects of human rhinovirus on epithelial barrier integrity and function in children with asthma. Clin Exp Allergy. John Wiley & Sons, Ltd. (2018) 48(5):513–24. doi: 10.1111/CEA.13097
22. Kloepfer Kirsten M, Lee Wai Ming, Pappas Tressa E, Kang Theresa J, Vrtis Rose F, Evans Michael D, et al. Detection of pathogenic bacteria during rhinovirus infection is associated with increased respiratory symptoms and asthma exacerbations. J Allergy Clin Immunol. Mosby Inc. (2014) 133(5):1301–1307.e3. doi: 10.1016/j.jaci.2014.02.030
23. Teo Shu Mei, Mok Danny, Pham Kym, Kusel Merci, Serralha Michael, Troy Niamh, et al. The infant nasopharyngeal microbiome impacts severity of lower respiratory infection and risk of asthma development. Cell Host Microbe. (2015) 17(5):704–15. doi: 10.1016/j.chom.2015.03.008
24. Vanker Aneesa, Nduru Polite M, Barnett Whitney, Dube Felix S, Sly Peter D, Gie Robert P, et al. Indoor air pollution and tobacco smoke exposure: impact on nasopharyngeal bacterial carriage in mothers and infants in an African birth cohort study. ERJ Open Res. European Respiratory Society. (2019) 5(1):00052-2018. doi: 10.1183/23120541.00052-2018
25. Carrión Daniel, Kaali Seyram, Kinney Patrick L, Owusu-Agyei Seth, Chillrud Steven, Yawson Abena K, et al. Examining the relationship between household air pollution and infant microbial nasal carriage in a Ghanaian cohort. Environ Int. Pergamon. (2019) 133:105150. doi: 10.1016/J.ENVINT.2019.105150
26. Dherani Mukesh K, Pope Daniel, Tafatatha Terence, Heinsbroek Ellen, Chartier Ryan, Mwalukomo Thandie, et al. Association between household air pollution and nasopharyngeal pneumococcal carriage in Malawian infants (MSCAPE): a nested, prospective, observational study. Lancet Glob Health. Elsevier. (2022) 10(2):e246–56. doi: 10.1016/S2214-109X(21)00405-8
27. Clarke Kayan, Ash Kevin, Coker Eric S, Sabo-Attwood Tara, Bainomugisha Engineer A social vulnerability index for air pollution and its spatially varying relationship to PM2.5 in Uganda. Atmosphere (Basel). MDPI. (2022) 13(8):1169. doi: 10.3390/ATMOS13081169/S1
28. Kabwama Steven Ndugwa, Ndyanabangi Sheila, Mutungi Gerald, Wesonga Ronald, Bahendeka Silver K, Guwatudde David. Tobacco use and associated factors among adults in Uganda: findings from a nationwide survey. Tob Induc Dis. The International Society for the Prevention of Tobacco Induced Diseases. (2016) 14:1. doi: 10.1186/S12971-016-0093-8
29. Kansiime Winnifred K, Mugambe Richard K, Atusingwize Edwinah, Wafula Solomon T, Nsereko Vincent, Ssekamatte Tonny, et al. Use of biomass fuels predicts indoor particulate matter and carbon monoxide concentrations; evidence from an informal urban settlement in fort portal city, Uganda. BMC public Health. BioMed Central Ltd. (2022) 22(1):1723. doi: 10.1186/s12889-022-14015-w
30. Uganda Bureau of Statistics. National Population and Housing Census 2014 Area Specific Profiles: Kabarole District. Kampala, Uganda (2017). Available online at: https://www.ubos.org/wp-content/uploads/publications/2014CensusProfiles/KABAROLE.pdf (Accessed April 18, 2024).
31. Scully Erik J, Basnet Sarmi, Wrangham Richard W, Muller Martin N, Otali Emily, Hyeroba David, et al. Lethal respiratory disease associated with human rhinovirus C in wild chimpanzees, Uganda, 2013. Emerg Infect Dis. (2018) 24(2):267–74. doi: 10.3201/eid2402.170778
32. Negrey Jacob D, Reddy Rachna B, Scully Erik J, Phillips-Garcia Sarah, Owens Leah A, Langergraber Kevin E, et al. Simultaneous outbreaks of respiratory disease in wild chimpanzees caused by distinct viruses of human origin. Emerg Microbes Infect. NLM (Medline). (2019) 8(1):139–49. doi: 10.1080/22221751.2018.1563456
33. Bochkov Yury A, Grindle Kristine, Vang Fue, Evans Michael D, Gern James E. Improved molecular typing assay for rhinovirus species A, B, and C. J Clin Microbiol. (2014) 52(7):2461–71. doi: 10.1128/JCM.00075-14
34. Greiner Oliver, Day Philip JR, Altwegg Martin, Nadal David. Quantitative detection of Moraxella catarrhalis in nasopharyngeal secretions by real-time PCR. J Clin Microbiol. American Society for Microbiology (ASM). (2003) 41(4):1386. doi: 10.1128/JCM.41.4.1386-1390.2003
35. Wang Xin, Mair Raydel, Hatcher Cynthia, Theodore M, Jordan Edmond K, Wu Henry M, et al. Detection of bacterial pathogens in Mongolia meningitis surveillance with a new real-time PCR assay to detect Haemophilus influenzae. Int J Med Microbiol. Urban & Fischer. (2011) 301(4):303–9. doi: 10.1016/J.IJMM.2010.11.004
36. Wessels Els, Schelfaut Jacqueline JG, Bernards Alexandra T, Claas Eric CJ. Evaluation of several biochemical and molecular techniques for identification of Streptococcus pneumoniae and Streptococcus pseudopneumoniae and their detection in respiratory samples. J Clin Microbiol. American Society for Microbiology1752 N St., N.W., Washington, DC. (2012) 50(4):1171–7. doi: 10.1128/JCM.06609-11/SUPPL_FILE/JCM-JCM06609-11-S01.PDF
37. Goldberg Tony L, Bennett Andrew J, Kityo Robert, Kuhn Jens H, Chapman Colin A. Kanyawara virus: a novel rhabdovirus infecting newly discovered nycteribiid bat flies infesting previously unknown pteropodid bats in Uganda. Sci Rep. (2017) 7(1):5287. doi: 10.1038/s41598-017-05236-w
38. Goldberg Tony L, Sibley SD, Pinkerton ME, Dunn CD, Long LJ, White LC. Multidecade mortality and a homolog of hepatitis C virus in bald eagles (Haliaeetus leucocephalus), the national bird of the USA. Sci Rep. (2019) 9:14953. doi: 10.1038/s41598-019-50580-8
39. Toohey-Kurth K, Sibley SD, Goldberg TL. Metagenomic assessment of adventitious viruses in commercial bovine sera. Biologicals. Academic Press. (2017) 47:64–8. doi: 10.1016/j.biologicals.2016.10.009
40. Negrey Jacob D, Mitani John C, Wrangham Richard W, Otali Emily, Reddy Rachna B, Pappas Tressa E, et al. Viruses associated with ill health in wild chimpanzees. Am J Primatol. John Wiley and Sons Inc. (2022) 84(2):e23358. doi: 10.1002/AJP.23358
41. Allander Tobias, Emerson Suzanne U, Engle Ronald E, Purcell Robert H, Bukh Jens. A virus discovery method incorporating DNase treatment and its application to the identification of two bovine parvovirus species’, proceedings of the National Academy of Sciences. Proc Natl Acad Sci USA. (2001) 98(20):11609–14. doi: 10.1073/pnas.211424698
42. Bankevich Anton, Nurk Sergey, Antipov Dmitry, Gurevich Alexey A, Dvorkin Mikhail, Kulikov Alexander S, et al. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol. Mary Ann Liebert Inc. (2012) 19(5):455–77. doi: 10.1089/cmb.2012.0021
43. Altschul Stephen F, Gish Warren, Miller Webb, Myers Eugene W, Lipman David J. Basic local alignment search tool. J Mol Biol. Elsevier BV. (1990) 215(3):403–10. doi: 10.1016/s0022-2836(05)80360-2
44. Gish W, States DJ. Identification of protein coding regions by database similarity search. Nat Genet. Springer Science and Business Media LLC. (1993) 3(3):266–72. doi: 10.1038/ng0393-266
45. Olenec Jaime P, Kim Woo Kyung, Lee Wai-Ming, Vang Fue, Pappas Tressa E, Salazar Lisa EP, et al. Weekly monitoring of children with asthma for infections and illness during common cold seasons. J Allergy Clin Immunol. Elsevier. (2010) 125(5):1001–1006.e1. doi: 10.1016/j.jaci.2010.01.059
46. Cohen J. Statistical Power Analysis for the Behavioral Sciences. 2nd ed Hillsdale, NJ: Lawrence Erlbaum (1988).
47. Monto AS, Ullman BM. Acute respiratory illness in an American community: the tecumseh study. JAMA. (1974) 227(2):164–9. doi: 10.1001/jama.1974.03230150016004
48. Heikkinen T, Järvinen A. The common cold. Lancet. Elsevier. (2003) 361(9351):51. doi: 10.1016/S0140-6736(03)12162-9
49. van den Hoogen Bernadette G, de Jong Jan C, Groen Jan, Kuiken Thijs, de Groot Groot, Fouchier Ron AM, et al. A newly discovered human pneumovirus isolated from young children with respiratory tract disease. Nat Med. Nature Publishing Group. (2001) 7(6):719. doi: 10.1038/89098
50. Allander Tobias, Tammi Martti T, Eriksson Margareta, Bjerkner Annelie, Tiveljung-Lindell Annika, Andersson Björn. Cloning of a human parvovirus by molecular screening of respiratory tract samples. Proc Natl Acad Sci U S A. National Academy of Sciences. (2005) 102(36):12891–6. doi: 10.1073/pnas.0504666102
51. Bennett Andrew J, Paskey Adrian C, Ebinger Arnt, Pfaff Florian, Priemer Grit, Höper Dirk, et al. Relatives of rubella virus in diverse mammals. Nature. Nature Publishing Group. (2020) 586(7829):424–8. doi: 10.1038/s41586-020-2812-9
52. Owens Leah A, Colitti Barbara, Hirji Ismail, Pizarro Andrea, Jaffe Jenny E, Moittié Sophie, et al. A Sarcina bacterium linked to lethal disease in sanctuary chimpanzees in Sierra Leone. Nat Commun. Nature Publishing Group. (2021) 12(1):1–16. doi: 10.1038/s41467-021-21012-x
53. Basnet Sarmila, Mohanty Chitrasen, Bochkov Yury A, Brockman-Schneider Rebecca A, Kendziorski Christina, Gern James E. Rhinovirus C causes heterogeneous infection and gene expression in airway epithelial cell subsets. Mucosal Immunol. (2023) 16(4):386–98. doi: 10.1016/j.mucimm.2023.01.008
54. Dyer Kimberly D, Percopo Caroline M, Fischer Elizabeth R, Gabryszewski Stanislaw J, Rosenberg Helene F. Pneumoviruses infect eosinophils and elicit MyD88-dependent release of chemoattractant cytokines and interleukin-6. Blood. United States. (2009) 114(13):2649–56. doi: 10.1182/blood-2009-01-199497
55. Loo Su-Ling, Wark Peter AB, Esneau Camille, Nichol Kristy S, Hsu Alan CY, Bartlett Nathan W. The pathophysiology of COVID-19 and SARS-CoV-2 infection human coronaviruses 229E and OC43 replicate and induce distinct antiviral responses in differentiated primary human bronchial epithelial cells. Am J Physiol Lung Cell Mol Physiol. (2020) 319:926–31. doi: 10.1152/ajplung.00374.2020.-The
56. Biraro Irene Andia, Kimuda Simon, Egesa Moses, Cose Stephen, Webb Emily L, Joloba Moses, et al. The use of interferon gamma inducible protein 10 as a potential biomarker in the diagnosis of latent Tuberculosis infection in Uganda. PLoS One. United States. (2016) 11(1):e0146098. doi: 10.1371/journal.pone.0146098
57. de Fost Fost, Chierakul Wirongrong, Pimda Kriangsak, Dondorp Arjen M, White Nicholas J, Van der Poll Tom. Activation of cytotoxic lymphocytes in patients with scrub typhus. Am J Trop Med Hyg. United States. (2005) 72(4):465–7. doi: 10.4269/ajtmh.2005.72.465
58. Armah Henry B, Wilson Nana O, Sarfo Bismark Y, Powell Michael D, Bond Vincent C, Anderson Winston, et al. Cerebrospinal fluid and serum biomarkers of cerebral malaria mortality in Ghanaian children. Malar J. England. (2007) 6:147. doi: 10.1186/1475-2875-6-147
59. Vasquez RE, Xin L, Soong L. Effects of CXCL10 on dendritic cell and CD4+ T-cell functions during leishmania amazonensis infection. Infect Immun. United States. (2008) 76(1):161–9. doi: 10.1128/IAI.00825-07
60. Wark Peter AB, Bucchieri Fabio, Johnston Sebastian L, Gibson Peter G, Hamilton Lynnsey, Mimica Joanna, et al. IFN-γ-induced protein 10 is a novel biomarker of rhinovirus-induced asthma exacerbations. J Allergy Clin Immunol. Elsevier. (2007) 120(3):586–93. doi: 10.1016/j.jaci.2007.04.046
61. Saetta Marina, Mariani Margherita, Panina-Bordignon Paola, Turato Graziella, Buonsanti Cecilia, Baraldo Simonetta, et al. Increased expression of the chemokine receptor CXCR3 and its ligand CXCL10 in peripheral airways of smokers with chronic obstructive pulmonary disease. Am J Respir Crit Care Med. United States. (2002) 165(10):1404–9. doi: 10.1164/rccm.2107139
62. Vieceli Tarsila, Tejada Sofia, Martinez-Reviejo Raquel, Pumarola Tomas, Schrenzel Jacques, Waterer Grant W, et al. Impact of air pollution on respiratory microbiome: a narrative review. Intensive and Critical Care Nursing. Churchill Livingstone. (2023) 74:103336. doi: 10.1016/j.iccn.2022.103336
63. Gern James E, Pappas Tressa, Visness Cynthia M, Jaffee Katy F, Lemanske Robert F, Togias Alkis, et al. Comparison of the etiology of viral respiratory illnesses in inner-city and suburban infants. J Infect Dis. Oxford Academic. (2012) 206(9):1342–9. doi: 10.1093/INFDIS/JIS504
64. Kwambana Brenda A, Barer Michael R, Bottomley Christian, Adegbola Richard A, Antonio Martin. Early acquisition and high nasopharyngeal co-colonisation by Streptococcus pneumoniae and three respiratory pathogens amongst gambian new-borns and infants. BMC Infect Dis. BioMed Central. (2011) 11(1):1–8. doi: 10.1186/1471-2334-11-175/FIGURES/3
65. Anthony Laura, Meehan Andrea, Amos Ben, Mtove George, Mjema Julius, Malahiyo Rajabu, et al. Nasopharyngeal carriage of Streptococcus pneumoniae: prevalence and risk factors in HIV-positive children in Tanzania. Int J Infect Dis. Elsevier. (2012) 16(10):e753–7. doi: 10.1016/j.ijid.2012.05.1037
66. Mulu Wondemagegn, Yizengaw Endalew, Alemu Megbaru, Mekonnen Daniel, Hailu Derese, Ketemaw Kassaw, et al. Pharyngeal colonization and drug resistance profiles of Moraxella catarrrhalis, Streptococcus pneumoniae, Staphylococcus aureus, and Haemophilus influenzae among HIV infected children attending ART clinic of felegehiwot referral hospital, Ethiopia. PLoS One. Edited by R. Borrow. PLOS. (2018) 13(5):e0196722. doi: 10.1371/journal.pone.0196722
67. Gray BM, Turner ME, Dillon HC. Epidemiologic studies of Streptococcus pneumoniae in infants: the effects of season and age on pneumococcal acquisition and carriage in the first 24 months of life. Am J Epidemiol. Oxford Academic. (1982) 116(4):692–703. doi: 10.1093/oxfordjournals.aje.a113452
68. Collazos J, Miguel J, Ayarza R. Moraxella catarrhalis bacteremic pneumonia in adults: two cases and review of the literature’, European journal of clinical microbiology & infectious diseases. Eur J Clin Microbiol Infect Dis. (1992) 11(3):237–40. doi: 10.1007/BF02098086
69. Dowell Scott F, Whitney Cynthia G, Wright Carolyn, Rose Charles E, Schuchat Anne. Seasonal patterns of invasive pneumococcal disease—volume 9, number 5—may 2003—emerging infectious diseases journal—cDC. Emerg Infect Dis. Centers for Disease Control and Prevention (CDC). (2003) 9(5):573–9. doi: 10.3201/EID0905.020556
70. Li You, Reeves Rachel M, Wang Xin, Bassat Quique, Brooks W, Abdullah Cohen Cheryl, et al. Global patterns in monthly activity of influenza virus, respiratory syncytial virus, parainfluenza virus, and metapneumovirus: a systematic analysis. Lancet Glob Health. Elsevier Ltd. (2019) 7(8):e1031–45. doi: 10.1016/S2214-109X(19)30264-5/ATTACHMENT/A7CC1C2A-404D-46A0-805F-710D19CC8314/MMC2.MP4
71. Suryadevara M, Domachowske JB. Epidemiology and seasonality of childhood respiratory syncytial virus infections in the tropics. Viruses. Multidisciplinary Digital Publishing Institute (MDPI). (2021) 13(4):696. doi: 10.3390/v13040696
72. Morris Denise E, Osman Karen L, Cleary David W, Clarke Stuart C. The characterization of Moraxella catarrhalis carried in the general population. Microbial Genomics. Microbiology Society. (2022) 8(5):000820. doi: 10.1099/MGEN.0.000820/CITE/REFWORKS
73. Adeloye Davies, Basquill Catriona, Papana Angeliki, Chan Kit Yee, Rudan Igor, Campbell Harry. An estimate of the prevalence of COPD in Africa: a systematic analysis. COPD. Taylor & Francis. (2015) 12(1):71–81. doi: 10.3109/15412555.2014.908834
74. Siddharthan Trishul, Grigsby Matthew, Morgan Brooks, Kalyesubula Robert, Wise Robert A, Kirenga Bruce, et al. Prevalence of chronic respiratory disease in urban and rural Uganda. Bull W H O. Switzerland. (2019) 97(5):318–27. doi: 10.2471/BLT.18.216523
75. Verhaegh Suzanne JC, Streefland André, Dewnarain Joy K, Farrell David J, van Belkum Belkum, Hays John P. Colonization of healthy children by Moraxella catarrhalis is characterized by genotype heterogeneity, virulence gene diversity and co-colonization with Haemophilus influenzae. Microbiology. Microbiology (Reading). (2011) 157(1):169–78. doi: 10.1099/mic.0.042929-0
76. Maleki A, Mirnaseri Z, Kouhsari E, Taherikalani M, Pakzad I, Mohammadi J, et al. Asymptomatic carriers of Neisseria meningitidis and Moraxella catarrhalis in healthy children. New Microbes New Infect. Elsevier. (2020) 36:100691. doi: 10.1016/J.NMNI.2020.100691
77. Lule Swaib A, Mpairwe Harriet, Nampijja Margaret, Akello Florence, Kabagenyi Joyce, Namara Benigna, et al. Life-course of atopy and allergy-related disease events in tropical Sub-Saharan Africa: a birth cohort study. Pediatr Allergy Immunol. (2017) 28(4):377–83. doi: 10.1111/pai.12719
78. Nantanda Rebecca, Tumwine James K, Ndeezi Grace, Ostergaard Marianne S. Asthma and pneumonia among children less than five years with acute respiratory symptoms in mulago hospital, Uganda: evidence of under-diagnosis of asthma. PLoS One. Public Library of Science. (2013) 8(11):e81562. doi: 10.1371/JOURNAL.PONE.0081562
79. Kirenga Bruce J, de Jong Jong, Katagira Winceslaus, Kasozi Samuel, Mugenyi Levicatus, Boezen Marike, et al. Prevalence and factors associated with asthma among adolescents and adults in Uganda: a general population based survey. BMC Public Health. BioMed Central Ltd. (2019) 19(1):1–9. doi: 10.1186/S12889-019-6562-2/TABLES/3
80. Okure Deo, Ssematimba Joel, Sserunjogi Richard, Gracia Nancy Lozano, Soppelsa Maria Edisa, Bainomugisha Engineer. Characterization of ambient air quality in selected urban areas in Uganda using low-cost sensing and measurement technologies. Environ Sci Technol. American Chemical Society. (2022) 56(6):3324–39. doi: 10.1021/acs.est.1c01443
81. Masekela R, Vanker A. Lung health in children in sub-saharan Africa: addressing the need for cleaner air. Int J Environ Res Public Health. Multidisciplinary Digital Publishing Institute (MDPI). (2020) 17(17):6178. doi: 10.3390/ijerph17176178
82. Altman Matthew C, Kattan Meyer, O Connor George T, Murphy Ryan C, Whalen Elizabeth, LeBeau Petra, et al. Associations between outdoor air pollutants and non-viral asthma exacerbations and airway inflammatory responses in children and adolescents living in urban areas in the USA: a retrospective secondary analysis. The Lancet Planetary Health. Elsevier B.V. (2023) 7(1):e33–44. doi: 10.1016/S2542-5196(22)00302-3
83. Duan RR, Hao K, Yang T. Air pollution and chronic obstructive pulmonary disease. Chron Dis Transl Med. Chinese Medical Association. (2020) 6(4):260. doi: 10.1016/J.CDTM.2020.05.004
84. Bauer Susanne E, Im Ulas, Mezuman Keren, Gao Chloe Y. Desert dust, industrialization, and agricultural fires: health impacts of outdoor air pollution in Africa. J Geophys Res Atmos. Blackwell Publishing Ltd. (2019) 124(7):4104–20. doi: 10.1029/2018JD029336
85. Stokholm Jakob, Chawes Bo L, Vissing Nadja H, Bjarnadóttir Elín, Pedersen Tine M, Vinding Rebecca K, et al. Azithromycin for episodes with asthma-like symptoms in young children aged 1–3 years: a randomised, double-blind, placebo-controlled trial. Lancet. Respiratory medicine. Lancet Respir Med. (2016) 4(1):19–26. doi: 10.1016/S2213-2600(15)00500-7
86. Thorsen Jonathan, Stokholm Jakob, Rasmussen Morten Arendt, Mortensen Martin Steen, Brejnrod Asker Daniel, Hjelmsø Mathis, et al. The airway microbiota modulates effect of azithromycin treatment for episodes of recurrent asthma-like symptoms in preschool children: a randomized clinical trial’, American journal of respiratory and critical care medicine. Am J Respir Crit Care Med. (2021) 204(2):149–58. doi: 10.1164/RCCM.202008-3226OC
87. Durack J, Boushey HA, Lynch SV. Airway microbiota and the implications of dysbiosis in asthma’, current allergy and asthma reports. Curr Allergy Asthma Rep. (2016) 16(8):52. doi: 10.1007/S11882-016-0631-8
88. Daniels CC, Rogers PD, Shelton CM. A review of pneumococcal vaccines: current polysaccharide vaccine recommendations and future protein antigens. J Pediatr Pharmacol Ther. Pediatric Pharmacology Advocacy Group. (2016) 21(1):27. doi: 10.5863/1551-6776-21.1.27
89. Andreas Stefan, Testa Marco, Boyer Laurent, Brusselle Guy, Janssens Wim, Kerwin Edward, et al. Non-typeable Haemophilus influenzae–Moraxella catarrhalis vaccine for the prevention of exacerbations in chronic obstructive pulmonary disease: a multicentre, randomised, placebo-controlled, observer-blinded, proof-of-concept, phase 2b trial. Lancet Respir Med. NLM (Medline). (2022) 10(5):435–46. doi: 10.1016/S2213-2600(21)00502-6
90. Kaur R, Casey JR, Pichichero ME. Emerging Streptococcus pneumoniae strains colonizing the nasopharynx in children after 13-valent pneumococcal conjugate vaccination in comparison to the 7-valent era, 2006–2015. Pediatr Infect Dis J. Lippincott Williams and Wilkins. (2016) 35(8):901–6. doi: 10.1097/INF.0000000000001206
91. Revai Krystal, McCormick David P, Patel Janak, Grady James J, Saeed Kokab, Chonmaitree Tasnee. Effect of pneumococcal conjugate vaccine on nasopharyngeal bacterial colonization during acute otitis media’, pediatrics. Pediatrics. (2006) 117(5):1823–9. doi: 10.1542/PEDS.2005-1983
92. Cleary David W, Devine Vanessa T, Morris Denise E, Osman Karen L, Gladstone Rebecca A, Bentley Stephen D, et al. Pneumococcal vaccine impacts on the population genomics of non-typeable haemophilus influenzae. Microb Genom. Microbiology Society. (2018) 4(9):e000209. doi: 10.1099/MGEN.0.000209/CITE/REFWORKS
93. Okello Gerald, Izudi Jonathan, Ampeire Immaculate, Nghania Frehd, Dochez Carine, Hens Niel. Two decades of regional trends in vaccination completion and coverage among children aged 12–23 months: an analysis of the Uganda demographic health survey data from 1995 to 2016. BMC Health Serv Res. BioMed Central Ltd. (2022) 22(1):1–17. doi: 10.1186/S12913-021-07443-8/TABLES/4
94. GAVI. GAVI Full Country Evaluation 2013: Process evaluation of pneumococcal vaccine introduction in Mozambique, Uganda, and Zambia (2014).
95. Olayinka F, Ewald L, Steinglass R. Beyond new vaccine introduction: the uptake of pneumococcal conjugate vaccine in the African region. Pan Afr Med J. African Field Epidemiology Network. (2017) 27(Suppl 3):3. doi: 10.11604/pamj.supp.2017.27.3.11531
96. WHO. Pneumococcal vaccination coverage: United States, License: CC BY-NC-SA 3.0 IGO (2023). Available online at: https://immunizationdata.who.int/pages/coverage/pcv.html?CODE=USA&ANTIGEN=PCV3&YEAR= (accessed July 20, 2023).
97. WHO and UNICEF. Uganda, WHO and UNICEF estimates of immunization coverage: 2022 version (2023). Available online at: https://data.unicef.org/wp-content/uploads/cp/immunisation/uga.pdf (accessed July 20, 2023).
98. Chow EJ, Uyeki TM, Chu HY. The effects of the COVID-19 pandemic on community respiratory virus activity. Nat Rev Microbiol. Nature Publishing Group. (2022) 21:195–210. doi: 10.1038/s41579-022-00807-9
99. Pacitto A, Amato F, Salmatonidis A, Moreno T, Alastuey A, Reche C, et al. Effectiveness of commercial face masks to reduce personal PM exposure. Sci Total Environ. Elsevier. (2019) 650:1582–90. doi: 10.1016/J.SCITOTENV.2018.09.109
100. Sá-Leão Raquel, Nunes Sónia, Brito-Avô António, Alves Carla R, Carriço João A, Saldanha Joana, et al. High rates of transmission of and colonization by Streptococcus pneumoniae and Haemophilus influenzae within a day care center revealed in a longitudinal study. J Clin Microbiol. American Society for Microbiology. (2008) 46(1):225–34. doi: 10.1128/JCM.01551-07/ASSET/34A23013-443E-4851-94BC-065266CB36C5/ASSETS/GRAPHIC/ZJM0010878440002.JPEG
101. Schewitz Ivan Anton, Zar Heather J, Masekela Refiloe, Gordon Stephen, Ozoh Obianuju, Kagima Jacqueline, et al. Unintended consequences of the COVID-19 pandemic in Africa. J Pan Afr Thorac Soc. (2020) 1:3. doi: 10.25259/JPATS_6_2020
102. Anyanwu JC, Salami AO. The impact of COVID-19 on African economies: an introduction. Afr Dev Rev. John Wiley & Sons, Ltd. (2021) 33(S1):S1–S16. doi: 10.1111/1467-8268.12531
103. Muchena D. Southern Africa: Homes become dangerous place for women and girls during COVID-19 lockdown. Amnesty International (2021). Available online at: https://www.amnesty.org/en/latest/press-release/2021/02/southern-africa-homes-become-dangerous-place-for-women-and-girls-during-covid19-lockdown/ (accessed July 21, 2023).
Keywords: respiratory disease, pediatrics, Uganda, COVID-19, asthma, COPD, air pollution
Citation: Weary TE, Pappas T, Tusiime P, Tuhaise S, Ross E, Gern JE and Goldberg TL (2024) High frequencies of nonviral colds and respiratory bacteria colonization among children in rural Western Uganda. Front. Pediatr. 12:1379131. doi: 10.3389/fped.2024.1379131
Received: 30 January 2024; Accepted: 19 April 2024;
Published: 2 May 2024.
Edited by:
George A. Syrogiannopoulos, University of Thessaly, GreeceReviewed by:
Mohammad Bagher Rahmati, Hormozgan University of Medical Sciences, IranMalik Aydin, Witten/Herdecke University, Germany
Ville Peltola, Turku University Hospital, Finland
© 2024 Weary, Pappas, Tusiime, Tuhaise, Ross, Gern and Goldberg. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Tony L. Goldberg dG9ueS5nb2xkYmVyZ0B3aXNjLmVkdQ==
 Taylor E. Weary
Taylor E. Weary Tressa Pappas2
Tressa Pappas2 James E. Gern
James E. Gern