
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Pediatr. , 12 October 2022
Sec. Pediatric Pulmonology
Volume 10 - 2022 | https://doi.org/10.3389/fped.2022.959439
This article is part of the Research Topic Translational Research in Pediatric Respiratory Diseases - From Bench to Bedside View all 6 articles
Objective: CLCA1 is a secreted protein with protease activity, and its expression is associated with inflammatory airway diseases. This study aimed to investigate the role of CLCA1 and IL-13 in pediatric asthma.
Methods: In asthmatic and healthy children, the correlation between CLCA1 expression and blood IL-4, and IL-13 levels were investigated by serological analyses such as RT-qPCR and ELISA. The effects on the activity and apoptosis of bronchial epithelial cells following IL-13 stimulation were explored in vitro by the CCK-8 assay and flow cytometry, respectively. CLCA1 siRNA was used to knock down the expression level of bronchial epithelial cells and the effect of IL-13 stimulation on these cells was assessed by the CCK-8 assay and flow cytometry.
Results: CLCA1, IL-4, and IL-13 were highly expressed in the serum of children with asthma. CLCA1 expression was highly correlated to serum IL-13. IL-13 stimulation reduced the activity of bronchial epithelial cells in vitro and promoted apoptosis. Lastly, knockdown of CLCA1 rescued the IL-13-induced decrease in activity and apoptosis.
Conclusion: CLCA1 is highly expressed in children with asthma and mediates the contributory effect of IL-13 on the occurrence and development of pediatric asthma.
Worldwide, pediatric asthma is the most common childhood chronic lower respiratory disease (1). Pediatric asthma begins early in life and may progress or resolve over time. Approximately, 50% of children with asthma become asymptomatic over time. However, some asthma symptoms may persist throughout the life of the child. In atopic and severe cases, asthma has a disproportionate impact on a patient's quality of life (2). Pediatric asthma differs from adult asthma due to the lack of mature respiratory and immune systems in children, lack of good evidence, and difficulty in establishing a diagnosis to provide medication (3). However, genetic differences are hypothesized to play a role in the onset and progression of pediatric asthma.
Calcium-activated chloride channel regulator (CLCA) proteins are a family of secreted self-cleaving and zinc-dependent metalloproteases that activate calcium-dependent chloride currents in mammalian cells (4, 5). CLCA1 is the most studied member of the CLCA family and mainly plays an important role in human respiratory diseases. Furthermore, previous studies have shown that CLCA1 is an important secretory mediator in asthma, chronic obstructive pulmonary disease, cystic fibrosis, and other diseases that exhibit increased mucus production (6, 7). In addition to regulating airway mucus secretion, CLCA1 may also be involved in the regulation of tissue inflammation during the innate immune response through the regulation of cytokine and chemokine production (8). The secreted form of CLCA1 acts as a signaling ligand that activates U-937 cells, a human monocytic cell line, as well as primary cultured porcine alveolar macrophages in a dose-dependent manner and increases proinflammatory interleukin (IL)-1β, IL-6, IL- 8, and tumor necrosis factor-α (TNF-α) expression, which act as pleiotropic factors in lung inflammation [24349445]. Therefore, CLCA1 is considered to have an important role in respiratory diseases; however, its role in pediatric asthma has not been elucidated.
Additionally, type 2 asthma, the most common form of asthma, is characterized by airway and blood eosinophilia, of which the type 2 cytokine, IL-13, is the main biomarker (9). In childhood, type 2 asthma is usually caused by allergic sensitization and exposure to inhaled allergens. This sensitization is driven by allergen-specific CD4+ Th2 cells and is evidenced by the presence of allergen-specific IgE in serum. Additionally, multiple studies have reported evidence to show that Th2 cell-related cytokines produced by CD4+ T cells are associated with asthma (10–12). In Guangdong, Wu et al. reported that the IL-13 promoter polymorphisms are important contributors to childhood asthma. Furthermore, the authors showed that the mutant T allele is associated with asthma and can increase serum IgE levels (13). Additionally, in Xinjiang Uygur, several studies have found that IL-4 and IL-13 gene polymorphisms are associated with childhood asthma (14). The IL-13 gene is located on human chromosome 5q23-q31 and consists of 4 exons and 3 introns, encoding a 132 amino acid protein (15). IL-13 is adjacent to the IL-4, IL-3, IL-5, IL-9, and GM-CSF genes, forming a cytokine gene cluster. As stated above, IL-13 is mainly secreted by activated CD4+ T cells (Th2), and its biological function is to inhibit the release of inflammatory cytokines and chemical factors from monocytes, induce B cell proliferation and differentiation, promote IgE synthesis and the production of several adhesion molecules that are expressed in endothelial cells (16).
In this paper, we investigated the expression levels of CLCA1, IL-4, and IL-13 in pediatric asthma. We found that the expression of CLCA1 and IL-13 were positively correlated in pediatric asthma relative to healthy controls. At the same time, in vitro IL-13 stimulation reduced the activity of bronchial epithelial cells and promoted apoptosis. Knockdown of CLCA1 alleviated the IL-13-induced decrease in activity and apoptosis of bronchial epithelial cells. Together, these results demonstrate that CLCA1 may be a potential therapeutic target in pediatric asthma.
In this study, 34 patients (age 2–7 years old) diagnosed with asthma were selected from individuals with pediatric asthma (Ningbo Women's and Children's Hospital). All patients were examined and diagnosed as asthma by clinicians according to the clinical guidelines of the Global Initiative for Asthma (GINA, 2019). In addition, 35 individuals with an age range of 2–7 years (matched with patients) were selected as normal controls. The individuals in the normal control group showed no signs or symptoms of asthma. As shown in Supplementary materials.This study was approved by the hospital ethics committee and all patients signed informed consent.
The GSE103166 dataset was retrieved from the GEO database (https://www.ncbi.nlm.nih.gov/geo/). Differentially expressed mRNA transcripts were identified using the limma package deployed in R (version: 3.40.2). “Adjusted p < 0.05 and log2 (fold change) > 1 or log2(fold change) < −1” was defined as a threshold mRNA differential expression screen.
Bronchial epithelial cell lines (BEAS-2B cells) were purchased from the American Type Culture Collection (ATCC; Manassas, VA, USA). The cells were cultured in RPMI-1640 medium (Hyclone, USA) supplemented with 10% fetal bovine serum (Gibco, USA). The cells were cultured in a humidified atmosphere at 37 °C supplemented with 5% CO2. For in vitro stimulation experiments, the cells were treated with recombinant IL-13 (2 ng/ml, Thermo Fisher Scientific, USA). CLCA1 siRNA (Tsingke, China) and control RNA were transfected into BEAS-2B cells in logarithmic growth phase.Transfections were performed using Lipofectamine 3,000 transfection reagent (Invitrogen, USA) according to the manufacturer's protocol.
Peripheral venous blood and upper serum were taken, and the serum IL-4 and IL-13 levels of the subjects were detected by ELISA in strict accordance with the instructions of the ELISA kit (Lianke, China), and the absorbance value at 450 nm wavelength was measured by a microplate reader. Results The standard curve was drawn to calculate the levels of IL-4 and IL-13.
Total RNA was extracted from the blood and cells using the TRIzol reagent (Invitrogen, USA) following the manufacturer's protocol. The concentration and purity of RNA were detected by a trace nucleic acid protein analyzer. To reverse transcribe the RNA, 500 ng RNA was used to synthesize cDNA using the TaKaRa Reverse Transcription Kit (Takara, Japan). For qRT-PCR, the reaction was carried out according to the instructions of the TaKaRa fluorescence quantitative PCR detection kit (Takara, Japan) using the newly synthesized cDNA as a template. The experiment was repeated three times and GAPDH was used as an internal reference. The forward primer sequence for CLCA1 was CGTCAAATACTCCCCATCGT (5′ to 3′) and the reverse primer sequence was GCTGATGTTCTGGTTGCTGA (5′ to 3′). The forward primer sequence for GAPDH was ACCCACTCCTCCACCTTTGAC (5′ to 3′) and the reverse primer sequence was TGTTGCTGTAGCCAAATTCGTT (5′ to 3′). The relative expression levels of CLCA1 were assessed using the 2-ΔΔCt method.
BEAS-2B cells in the logarithmic growth phase were diluted and spread at a volume of 200 μl per well in a 96-well plate. Each group consisted of 3 wells. At approximately 70% confluency, the cells were transfected with siRNA was added. After culturing for 0, 24, 48, 72, and 48 h, 10 μl of CCK-8 solution (Dojindo, Japan) was added to each well, and the cells were incubated at 37 °C for 2 h in the dark. Thereafter, absorbance was measured at 450 nm in a microplate reader (Labsystems, Finland).
Statistical analysis was performed using GraphPad Prism 6 software. Data are presented as mean ± SD and all experiments were performed in triplicate. The chi-square test was used to assess the relationship between normal children and clinical characteristics of children with asthma. Differences between the two groups were analyzed using Student's t-test. p < 0.05 or p < 0.01 indicated statistical significance.
The clinical characteristics of patients and normal controls are summarized in Table 1. Hypersensitivity C-reactive protein (p < 0.0209), Immunoglobulin IgE (p < 0.0003), Hemoglobin (p < 0.0001), White blood cell count (p < 0.0001), Neutrophil percentage (p < 0.0001) and Lymphocytes percentage (p < 0.0001) levels were significantly increased.
To identify differentially expressed genes in asthmatic patients vs. healthy control, pediatric asthma mRNA microarray data (Accession No. GSE67472) was extracted from the GEO database. Bioinformatics analysis was to annotate, normalize and analyze the differential expression of the data. The generated cluster heatmaps indicated that many mRNAs were differentially expressed in the airway epithelial tissue of asthmatic vs. healthy controls (Figure 1A). Additionally, the generated volcano plots highlighted significant differentially expressed genes between the asthmatic and healthy control groups. Of these significantly differentially expressed genes, 12 were upregulated and 7 were downregulated. CLCA1 exhibited the greatest fold difference between the asthma children and healthy controls and was therefore selected as the candidate gene for our study (Figure 1B).
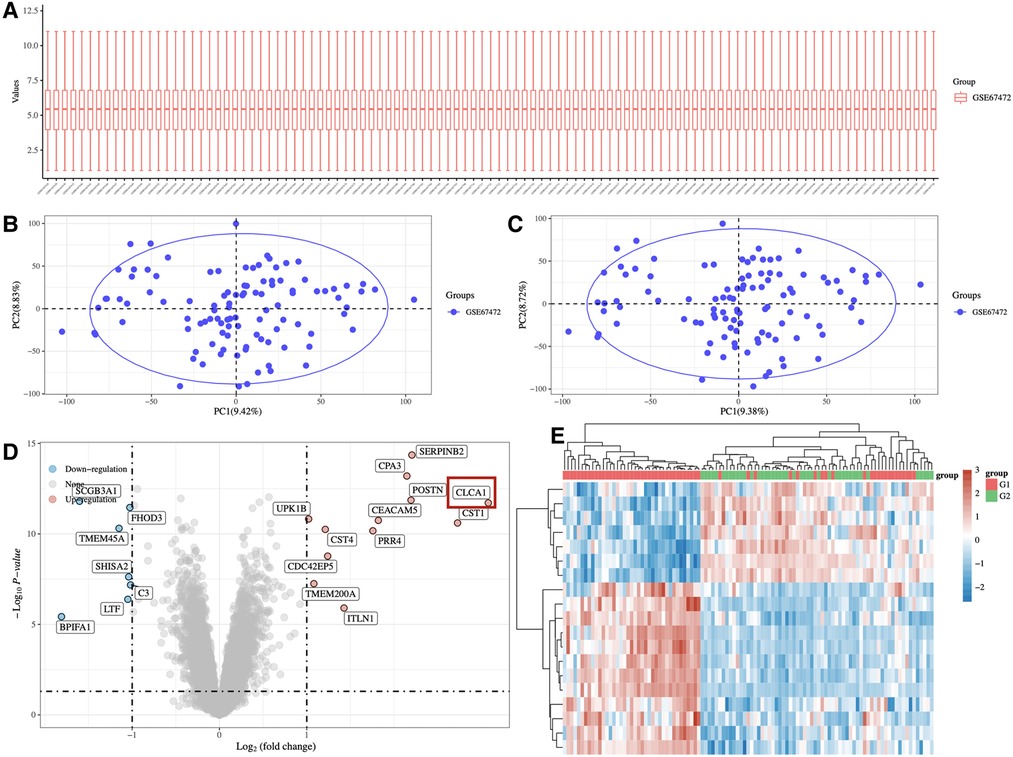
Figure 1. Profile of mRNA in the airway epithelium of asthma patients and normal people. (A) Heatmap depicting differentially expressed mRNAs in the airway epithelial tissues of 65 asthma patients and 43 normal airway epithelial tissues. (B) Volcano plot depicting the fold change values and p-values of differentially expressed genes.
Bioinformatic analysis showed that CLCA1 is highly expressed in asthmatic children. In agreement with these data, qRT-PCR analysis healthy children and children with asthma confirmed that CLCA1 is significantly expressed in the blood of children with asthma (Figure 2).
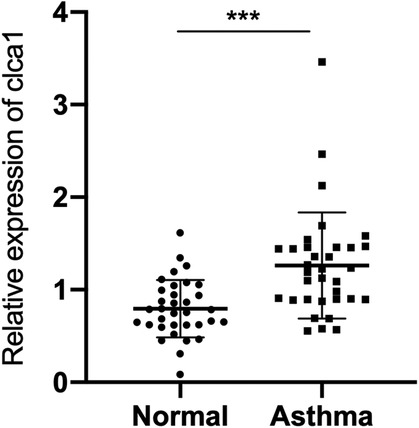
Figure 2. CLCA1 is highly expressed in the serum of children with asthma. RT-qPCR analysis of CLCA1 expression in airway epithelial tissues of 34 pairs of healthy children and children with asthma. ***p < 0.001.
Pediatric asthma is a chronic inflammatory airway disease associated with type 2 cytokines, IL-4, and IL-13 (17). Therefore, the levels of IL-4 and IL-13 were investigated in the blood of healthy children and children with asthma using ELISA. The concentration of IL-4 and IL-13 was significantly increased in the blood of pediatric asthma patients relative to the healthy control group (Figure 3A,B).
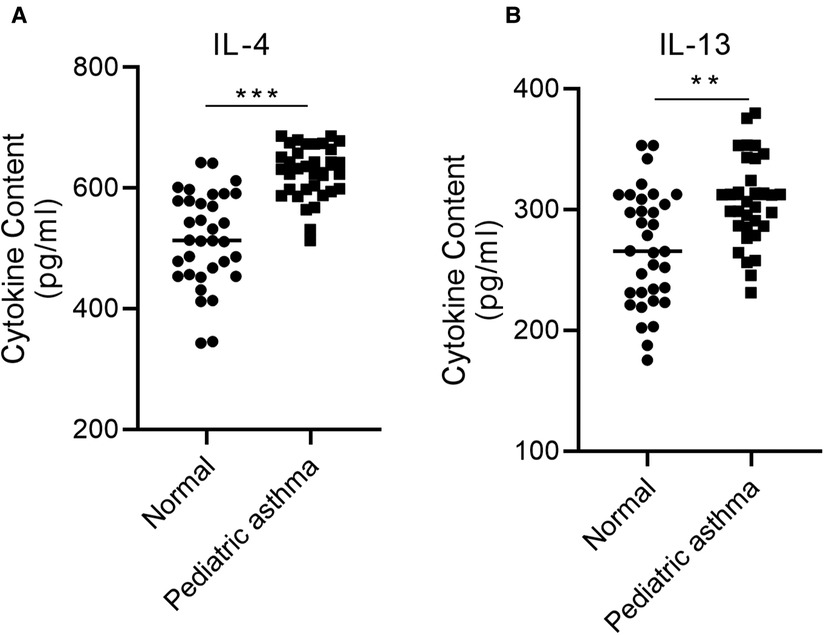
Figure 3. IL-4 and IL-13 are highly expressed in the serum of children with asthma. The concentration of (A) IL-4 and (B) IL-13 in the blood of asthmatic children relative to healthy controls was assessed via ELISA. **p < 0.01, ***p < 0.001.
Based on the ELSA data, we hypothesized that the expression level of CLCA1 in the blood may be related to IL-4 and IL-13 secretion. To determine whether the expression level of CLCA1 is correlated with blood IL-4 and IL-13 concentration, a correlation analysis of CLCA1 expression and serum IL-4 and IL-13 was performed. CLCA1 expression was weakly correlated with secreted IL-4 (Figure 4A), and highly correlated with secreted IL-13 (Figure 4B).
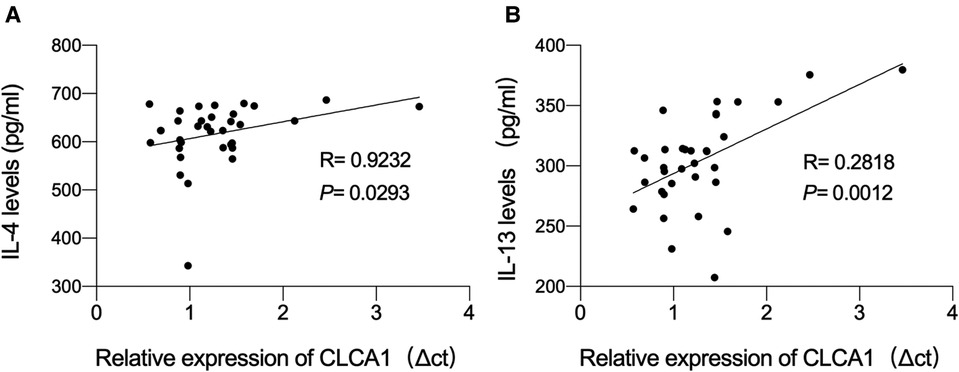
Figure 4. CLCA1 expression level is highly correlated with IL-13 content. (A,B) Person correlation analysis of the correlation of CLCA1 and IL-4 expression and the correlation of CLCA1 and IL-13 expression in the blood of healthy children and asthmatic children.
To validate the role of IL-13 in pediatric asthma, bronchial epithelial cells (BEAS-2B cells) were stimulated with IL-13 to mimic asthma in vitro. IL-13 stimulation reduced the viability of BEAS-2B cells (Figure 5A). Additionally, flow cytometric analysis revealed that IL-13 stimulation promoted BEAS-2B apoptosis (Figure 5B).
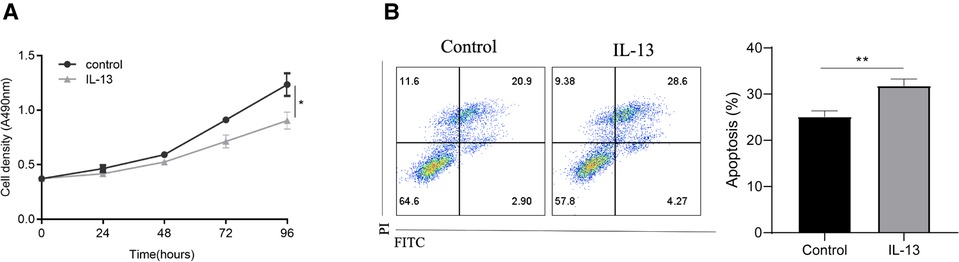
Figure 5. IL-13 stimulation enhances CLCA1 expression and downregulates bronchial epithelial cell function in vitro. BEAS-2B cells were stimulated with IL-13. Following stimulation, (A) cell proliferation was assessed using the CCK-8 assay and (B) apoptotic cells were assessed via flow cytometry. *p < 0.05, **p < 0.01.
As in vitro IL-13 stimulation reduced the activity of bronchial epithelial cells and promoted apoptosis, we next aimed to determine the specific effect of CLCA1. To achieve this, CLCA1 was knocked down in BEAS-2B cells using siRNA. The knocked-down cells were then stimulated with IL-13 (Figure 6A). CLCA1 knockdown rescued the IL-13-induced reduction in bronchial epithelial cell viability (Figure 6B) as well as apoptosis (Figure 6C).
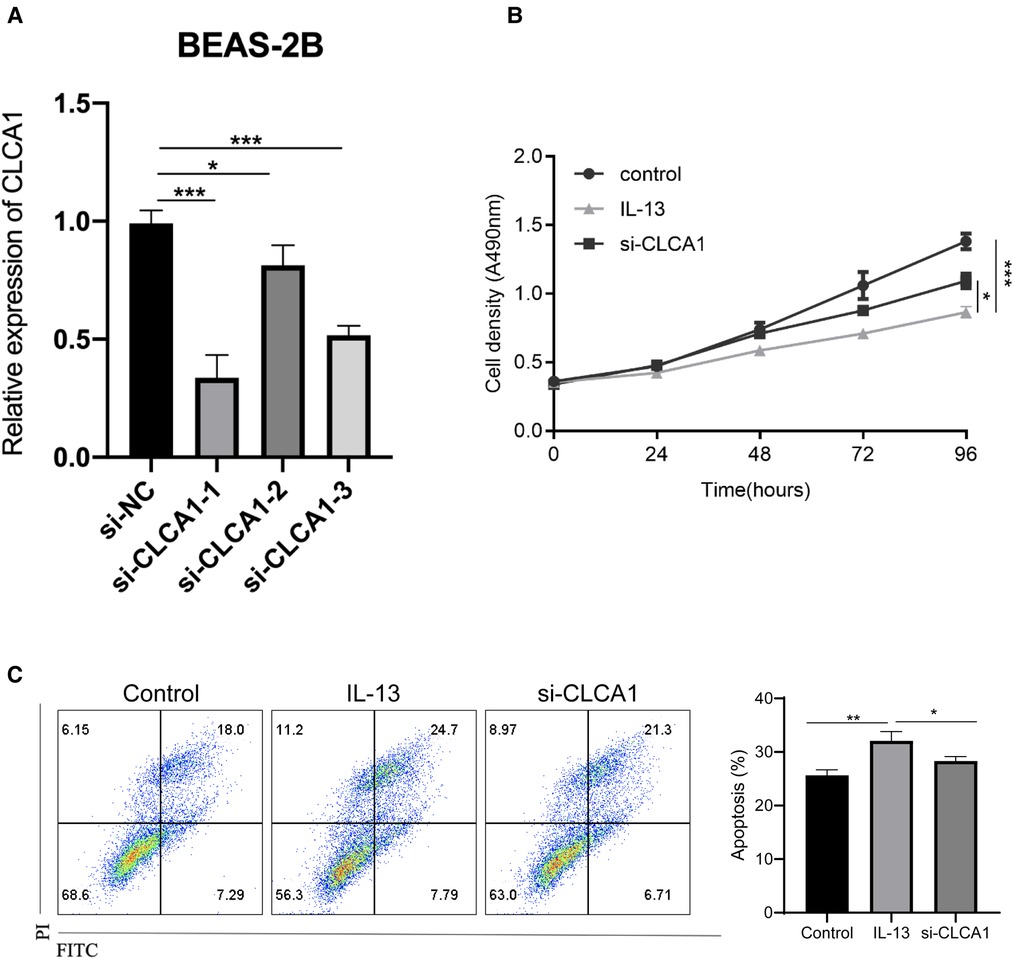
Figure 6. Knockdown of CLCA1 attenuates IL-13-induced bronchial epithelial cell activity and reduces cell apoptosis. CLCA1 knocked down BEAS-2B cells were stimulated with IL-13 and their (A,B) cell proliferation ability was assessed using the CCK-8 assay. (C) Flow cytometry was used to detect apoptotic cells.*p < 0.05,***p < 0.001.
CLCA1 is hypothesized to be widely involved in the pathogenesis of inflammatory airway diseases such as asthma. CLCA1 was bioinformatically identified as an aberrantly expressed gene in pediatric asthma. Here, we confirm that CLCA1 was indeed significantly overexpressed in the blood of asthmatic children relative to healthy controls. Additionally, elevated levels of IL-4 and IL-13 were detected in the serum of children with asthma, and IL-13 was positively correlated with the expression of CLCA1. In vitro IL-13 stimulation significantly reduced the viability of lung endothelial cells (BEAS-2B) and promoted apoptosis. Interestingly, CLCA1 knockdown reversed the IL-13-induced damage to BEAS-2B cells. Together, these results suggest that CLCA1 mediates the regulatory effect of IL-13 on pediatric asthma.
Early identification and intervention are key strategies for controlling childhood asthma. However, novel strategies that allow for the prevention of childhood asthma would be even more important. At present, many studies indicate that genetic factors are an important factor contributing to pediatric asthma (18, 19). In German children, polymorphisms in variants of the homeobox transcription factor gene, HLX1, were correlated with an increased risk of childhood asthma (18). Additionally, a study of gene variants in T-cell-specific TBX21, a factor that induces TH1 differentiation and blocks TH2 commitment together with HLX1, suggests that TBX21 polymorphisms contribute to asthma, possibly through altered TBX21 promoter activity (19). Together, these studies suggest that a better understanding of the functional effects of gene-related polymorphisms could help identify key pathways in childhood asthma development.
Several previous studies suggest that respiratory infections may be associated with childhood asthma. In the National Health and Nutrition Examination Survey, Gergen et al. examined the relationship between total IgE levels and asthma. Those authors noted that total IgE levels were associated with asthma only in persons with positive results for at least 1 allergen-specific IgE (20). In addition, cytokines appear to be strongly associated with childhood asthma. Previous studies have shown that the IL-26 secretion can be used as a biomarker for childhood asthma (21). Additionally, IL-10 and IL-1β gene polymorphisms are associated with allergic asthma in children (22, 23) and the association between IL-13 polymorphisms and susceptibility to childhood asthma has been extensively studied (24–26). Consistent with these findings, our experimental results show that IL-13 has a significant correlation with pediatric asthma.
Studies have shown that CLCA1 plays an important role in a mouse model of dextran sulfate sodium-induced colitis by the modulation of early immune responses through altered cytokine secretion (27). Furthermore, CLCA1 deficiency resulted in decreased cytokine expression and decreased leukocyte recruitment in an acute pneumonia mouse model (28). Finally, CLCA1 has been shown to activate macrophages in vitro. Consistent with our findings, previous studies have shown that, in normal human bronchial epithelial cells, IL-13 receptor activation leads to STAT6 activation and subsequent induction of CLCA1 gene expression (29). Furthermore, we demonstrated that the expression of IL-13 is highly correlated with CLCA1 in pediatric asthma. Together, these data indicate that CLCA1 may play an important role in pediatric asthma.
While this study suggests that CLCA1-mediated IL-13 has a role in the regulation of pediatric asthma, several aspects remain to be improved. Firstly, a limited number of serological samples from asthmatics patients were assessed. In future studies, the number of included asthmatic patients that should ideally be representative of different global sociodemographic populations should be increased to ensure sufficient power to identify robust biological phenomena. Secondly, the regulatory effects of CLCA1 and IL-13 on airway epithelial cell function in cell lines were only assessed in vitro. Therefore, the role of CLCA1 and IL-13 should be robustly verified in animal models. Finally, CLCA1 is associated with inflammatory airway disease; however, its role in pediatric asthma had not been reported. Whilst this study implicates the potential role of CLCA1 in pediatric asthma, additional in-depth investigations into the biological mechanism are still required.
In conclusion, this study shows that the expression of CLCA1 and IL-13 was positively correlated in pediatric asthma, and that knockdown of CLCA1 attenuated the IL-13-induced decreased activity and apoptosis of bronchial epithelial cells. These findings provide new research insights and elucidate potential therapeutic targets for pediatric asthma.
Publicly available datasets were analyzed in this study. This data can be found here: GSE103166; https://www.ncbi.nlm.nih.gov/geo/.
The studies involving human participants were reviewed and approved by Ethics Committee of Ningbo Women's and Children's Hospital. Written informed consent to participate in this study was provided by the participants’ legal guardian/next of kin.
QY and YX conceived and designed this study, performed the experiment, analyzed the data, and wrote the manuscript. YX, QY and LC participated in discussion of the results and offered scientific advice. JC provided part of the samples. DJ, PR, and QY supervised the study. QY reviewed and revised this manuscript. All authors contributed to the article and approved the submitted version.
This study was supported by Ningbo Natural Science Foundation Project (NO.202003N4276): Ningbo Clinical Research Center for Children's Health and Diseases (NO.2019A21002): Ningbo Leading Medical / Health Discipline, Project Number: (2022-B17;2010-S04).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
1. Avery C, Perrin EM, Lang JE. Updates to the pediatrics asthma management guidelines. JAMA Pediatr. (2021) 175:966–7. doi: 10.1001/jamapediatrics.2021.1494
2. Devonshire AL, Kumar R. Pediatric asthma: principles and treatment. Allergy Asthma Proc. (2019) 40:389–92. doi: 10.2500/aap.2019.40.4254
3. Papadopoulos NG, Arakawa H, Carlsen KH, Custovic A, Gern J, Lemanske R, et al. International consensus on (ICON) pediatric asthma. Allergy. (2012) 67:976–97. doi: 10.1111/j.1398-9995.2012.02865.x
4. Cunningham SA, Awayda MA, Bubien JK, Ismailov II, Arrate MP, Berdiev BK, et al. Cloning of an epithelial chloride channel from bovine trachea. J Biol Chem. (1995) 270:31016–26. doi: 10.1074/jbc.270.52.31016
5. Yurtsever Z, Rabanal MS, Randolph DT, Scheaffer SM, Roswit WT, Alevy YG, et al. Self-cleavage of human CLCA1 protein by a novel internal metalloprotease domain controls calcium-activated chloride channel activation. J Biol Chem. (2012) 287:42138–49. doi: 10.1074/jbc.M112.410282
6. Kamada F, Suzuki Y, Shao W, Tamari M, Hasegawa K, Hirota T, et al. Association of the hCLCA1 gene with childhood and adult asthma. Genes Immun. (2004) 5:540–7. doi: 10.1038/sj.gene.6364124
7. Hegab AE, Sakamoto T, Uchida Y, Nomura A, Ishii Y, Morishima Y, et al. CLCA1 Gene polymorphisms in chronic obstructive pulmonary disease. J Med Genet. (2004) 41:e27. doi: 10.1136/jmg.2003.012484
8. Liu CL, Shi GP. Calcium-activated chloride channel regulator 1 (CLCA1): more than a regulator of chloride transport and mucus production. World Allergy Organ J. (2019) 12:100077. doi: 10.1016/j.waojou.2019.100077
9. Jia G, Erickson RW, Choy DF, Mosesova S, Wu LC, Solberg OD, et al. Periostin is a systemic biomarker of eosinophilic airway inflammation in asthmatic patients. J Allergy Clin Immunol. (2012) 130:647–654.e610. doi: 10.1016/j.jaci.2012.06.025
10. Demenais F, Jeannin PM, Barnes KC, Cookson WOC, Altmüller J, Ang W, et al. Multiancestry association study identifies new asthma risk loci that colocalize with immune-cell enhancer marks. Nat Genet. (2018) 50:42–53. doi: 10.1038/s41588-017-0014-7
11. Seumois G, Chavez L, Gerasimova A, Lienhard M, Omran N, Kalinke L, et al. Epigenomic analysis of primary human T cells reveals enhancers associated with TH2 memory cell differentiation and asthma susceptibility. Nat Immunol. (2014) 15:777–88. doi: 10.1038/ni.2937
12. Yang IV, Pedersen BS, Liu A, O'Connor GT, Teach SJ, Kattan M, et al. DNA methylation and childhood asthma in the inner city. J Allergy Clin Immunol. (2015) 136:69–80. doi: 10.1016/j.jaci.2015.01.025
13. Wu B, Liu JL, Chen M, Deng RQ, Wu D. Correlation of interleukin-13 gene - 1112c/T polymorphism with asthma and total plasma IgE levels. Zhonghua Jie He He Hu Xi Za Zhi. (2004) 27:668–71.16200868
14. Zhang JH, Zhang M, Wang YN, Zhang XY. Correlation between IL-4 and IL-13 gene polymorphisms and asthma in uygur children in Xinjiang. Exp Ther Med. (2019) 17:1374–82. doi: 10.3892/etm.2018.7096
15. McKenzie AN, Culpepper JA, Malefyt RDW, Brière F, Punnonen J, Aversa G, et al. Interleukin 13, a T-cell-derived cytokine that regulates human monocyte and B-cell function. Proc Natl Acad Sci. (1993) 90:3735–9. doi: 10.1073/pnas.90.8.3735
16. Bieber T. Interleukin-13: targeting an underestimated cytokine in atopic dermatitis. Allergy. (2020) 75:54–62. doi: 10.1111/all.13954
17. Lambrecht BN, Hammad H, Fahy JV. The cytokines of asthma. Immunity. (2019) 50:975–91. doi: 10.1016/j.immuni.2019.03.018
18. Suttner K, Ruoss I, Rosenstiel P, Depner M, Pinto LA, Schedel M, et al. HLX1 Gene variants influence the development of childhood asthma. J Allergy Clin Immunol. (2009) 123:82–88.e86. doi: 10.1016/j.jaci.2008.09.047
19. Suttner K, Rosenstiel P, Depner M, Schedel M, Pinto LA, Ruether A, et al. TBX21 Gene variants increase childhood asthma risk in combination with HLX1 variants. J Allergy Clin Immunol. (2009) 123:1062–8, 1068.e1061-1068. doi: 10.1016/j.jaci.2009.02.025
20. Gergen PJ, Arbes SJ Jr., Calatroni A, Mitchell HE, Zeldin DC. Total IgE levels and asthma prevalence in the US population: results from the national health and nutrition examination survey 2005–2006. J Allergy Clin Immunol. (2009) 124:447–53. doi: 10.1016/j.jaci.2009.06.011
21. Konradsen JR, Nordlund B, Levänen B, Hedlin G, Linden A. The cytokine interleukin-26 as a biomarker in pediatric asthma. Respir Res. (2016) 17:32. doi: 10.1186/s12931-016-0351-6
22. Sobkowiak P, Banaszak IW, Kowalewska M, Wasilewska E, Langwiński W, Kycler Z, et al. Interleukin 1β polymorphism and serum level are associated with pediatric asthma. Pediatr Pulmonol. (2017) 52:1565–71. doi: 10.1002/ppul.23893
23. Zhong M, Xu Z. Correlations of IL-10 gene polymorphisms with infantile asthma. Panminerva Med. (2021) 63:389–91. doi: 10.23736/s0031-0808.19.03698-x
24. Leung TF, Tang NL, Chan IH, Li AM, Ha G, Lam CW. A polymorphism in the coding region of interleukin-13 gene is associated with atopy but not asthma in Chinese children. Clin Exp Allergy. (2001) 31:1515–21. doi: 10.1046/j.1365-2222.2001.01212.x
25. Kabesch M, Schedel M, Carr D, Woitsch B, Fritzsch C, Weiland S, et al. IL-4/IL-13 pathway genetics strongly influence serum IgE levels and childhood asthma. J Allergy Clin Immunol. (2006) 117:269–74. doi: 10.1016/j.jaci.2005.10.024
26. Kang MJ, Lee SY, Kim HB, Yu J, Kim B, Choi W, et al. Association of IL-13 polymorphisms with leukotriene receptor antagonist drug responsiveness in Korean children with exercise-induced bronchoconstriction. Pharmacogenet Genomics. (2008) 18:551–8. doi: 10.1097/FPC.0b013e3282fe94c5
27. Erickson NA, Mundhenk L, Giovannini S, Glauben R, Heimesaat MM, Gruber AD. Role of goblet cell protein CLCA1 in murine DSS colitis. J Inflammation. (2016) 13:5. doi: 10.1186/s12950-016-0113-8
28. Erickson NA, Dietert K, Enders J, Glauben R, Nouailles G, Gruber AD, et al. Soluble mucus component CLCA1 modulates expression of leukotactic cytokines and BPIFA1 in murine alveolar macrophages but not in bone marrow-derived macrophages. Histochem Cell Biol. (2018) 149:619–33. doi: 10.1007/s00418-018-1664-y
Keywords: CLCA1, pediatric asthma, IL-13, IL-4, in vitro
Citation: Xu Y, Cao L, Chen J, Jiang D, Ruan P and Ye Q (2022) CLCA1 mediates the regulatory effect of IL-13 on pediatric asthma. Front. Pediatr. 10:959439. doi: 10.3389/fped.2022.959439
Received: 1 June 2022; Accepted: 21 September 2022;
Published: 12 October 2022.
Edited by:
Mauricio Tomas Caballero, Consejo Nacional de Investigaciones Científicas y Técnicas (CONICET), ArgentinaReviewed by:
Yuchen Liu, Shenzhen University, China© 2022 Xu, Cao, Chen, Jiang, Ruan and Ye. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Qinsong Ye Zmx5LTExNUAxNjMuY29t
Specialty Section: This article was submitted to Pediatric Pulmonology, a section of the journal Frontiers in Pediatrics
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.