
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Pediatr., 30 March 2022
Sec. Pediatric Neurology
Volume 10 - 2022 | https://doi.org/10.3389/fped.2022.865181
Background: Nuclear factor I B (NFIB) plays an important role in regulating the transcription of multiple biological processes. Mutations in NFIB cause intellectual disability and macrocephaly. However, studies on abnormal brain and lung development caused by NFIB mutations are lacking.
Methods: In the present study, we enrolled a fetus with brain malformation and lung lobulation defects from China. Whole-exome sequencing (WES) was performed to detect the candidate genes and Sanger sequencing was performed for mutational analysis.
Results: After data filtering and bioinformatics prediction, a novel non-sense mutation of NFIB (NM_001190737:c.870C > A;p.Tyr290*) was identified in the fetus. This variant was predicted to produce a truncated NFIB protein because of a premature stop codon and was absent in 200 healthy controls.
Conclusion: To the best of our knowledge, this is the first case of brain malformation and lung lobulation defects caused by a NFIB variant in Asia. These findings contribute to genetic diagnosis and family counseling and expand our understanding of NFIB mutations as well as brain and lung maturation.
The nuclear factor I (NFI) family was first described that took part in the replication of Adenovirus DNA (1). As a family of transcription factors, NFIs were later found to be required for the transcriptional regulation, particularly during development of each organ system (2). In humans and most vertebrates, there are four NFI family members: NFIA, NFIB, NFIC, and NFIX. All of them share a highly conserved DNA-binding and dimerization domain at their N-terminal (3).
As one of the important members of NFI family, NFIB can bind DNA and plays a critical role in the development of multiple organ systems (4), including the central nervous system (5–7), lungs (8), and mammary glands (9). Among those, NFIB is a site-specific DNA-binding protein that plays an important role in increasing chromatin accessibility and regulating the transcription of multiple biological processes. NFIB is closely related to different cancer types. An analysis of small cell lung cancer cells by Denny et al. revealed that high expression of Nfib altered the chromatin state globally to promote cancer metastasis (10). Moreover, Zilli et al. discovered that overexpression of NFIB was sufficient to enhance primary mammary tumor growth and promote lung metastatic colonization via increased Ero1l/ERO1A expression (11). Recent studies have shown that mutations in NFIB are associated with intellectual disability and macrocephaly (12–14). However, additional reports of NFIB-related cases remain lacking.
Here, we report an aborted fetus with brain malformation and lung lobulation defects and a novel non-sense mutation in NFIB (NM_001190737:c.870C > A;p.Tyr290*) using whole-exome sequencing (WES).
The Review Board of Shenzhen Longgang District Maternity and Child Healthcare Hospital approved this study. The study was performed in accordance with the principles outlined in the Declaration of Helsinki in the ethics subsection. The parents provided written informed consent to participate in the study. Amniotic fluid was obtained from a mother undergoing amniocentesis. Blood was collected from the parents of the fetus.
Genomic DNA was extracted using a DNeasy Blood & Tissue Kit (Qiagen, Valencia, CA, United States). The main part of WES was performed at Guangdong Women’s and Children’s Hospital. The filtering strategies used conformed to those outlined in our previous study (15). Briefly, after preliminary quality control for the data, variants within intronic, intergenic and untranslated regions (UTRs), synonymous single nucleotide variants (SNVs), as well as variants with an alternative allele frequency more than 1% in the public databases [1000 Genomes, dbSNP144, YH database, and Genome Aggregation Database (gnomAD)], were firstly removed before further analysis. Then variants predicted by SIFT,1 PolyPhen-2,2 and MutationTaster3 as “Disease-causing,” were retained.
Sanger sequencing was performed to confirm the potential causative variants in the family. Primer pairs were designed using the PrimerQuest Tool from IDT4; the PCR primer sequences are available upon request.
Increased nuchal translucency (NT) (4.8 mm) was detected in a fetus (13 weeks and 5 days) using ultrasonography (>2.5 mm could be considered as increased NT) (Figure 1A). Karyotyping and copy number variation analysis of amniotic fluid from the mother revealed no abnormalities (Supplementary Figure 1). Level III ultrasonography also showed no obvious abnormalities. To find the real cause of NT increased, the parents agreed to perform exome sequencing. Subsequent WES revealed a novel heterozygous non-sense mutation of NFIB (NM_001190737:c.870C > A;p.Tyr290*) in the fetus, which was verified by Sanger sequencing (Figure 1B). This mutation was absent in the parents. This novel non-sense mutation of NFIB (p.Tyr290*) resulted in a premature stop codon at position 290 in exon 6 of the NFIB gene; it was also absent in a local cohort comprising 200 control cases (15), and in the dbSNP, Exome Variant Server databases,5 and gnomAD. Meanwhile, all results from multiple bioinformatics software (MutationTaster, PolyPhen-2, and SIFT) indicated that the variant is “disease-causing.” According to the American College of Medical Genetics (ACMG) guidelines, the variant of NFIB (c.870C > A;p.Tyr290*) can be classified as Pathogenic, which meets the following criteria from the ACMG guidelines: PVS1 (null variant with evidence supporting for disease mechanism), PS2 (de novo variant), PM1 (functional domain variation), PM2 (absent from controls in the gnomAD), and PP3 (multiple lines of computational evidence showed the variant as “Disease-causing”).
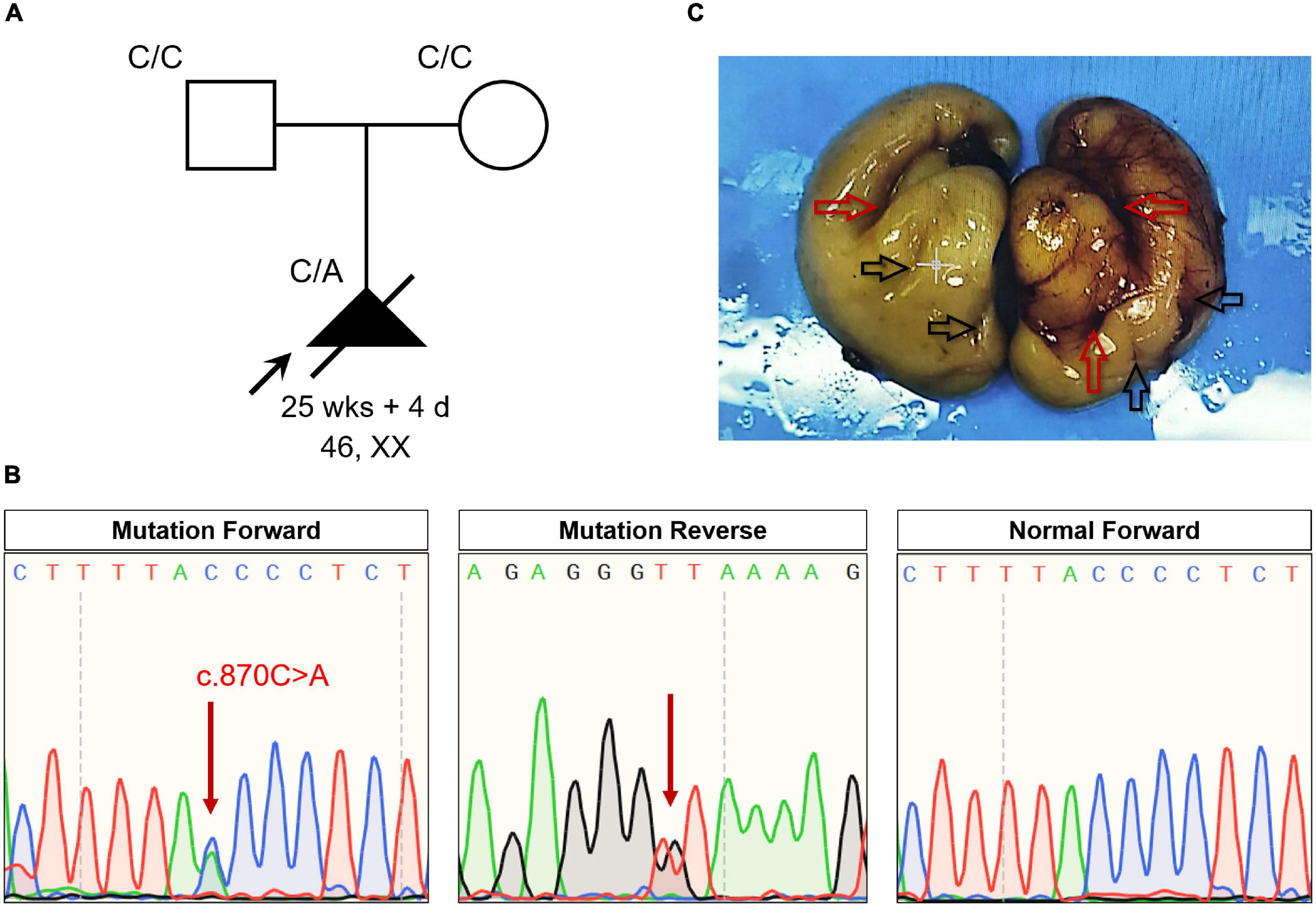
Figure 1. (A) The pedigree of this family. Black indicates the affected fetus with lissencephaly and lung lobulation defects. white circles/squares are unaffected. Arrow indicates the proband. (B) Sequencing results of the NFIB mutation. Sequence chromatograms indicate the heterozygosity of the NFIB non-sense mutation (NM_001190737:c.870C > A;p.Tyr290*) in the fetus and in a normal patient. Red arrow indicates the mutation. (C) Anatomical appearance of the cerebrum from the aborted fetus. Red arrows indicate the fissures of the brain. Black arrows indicate the brain sulci.
Mutations in NFIB are known to be pathogenic for intellectual disability and macrocephaly, as well as pulmonary dysplasia in humans and mice (12). Therefore, the family decided to end their pregnancy by abortion at 25 weeks and 4 days. Anatomical findings showed that fetal body length and other developmental parameters were consistent with those at 26 weeks of gestation. The head circumference of the fetus was 220 mm (−2 to −3 SD); weight of cerebrum was 110 g, cerebellum was 8 g. However, there was no obvious structure of the gyrus and sulcus in the cerebral cortex (especially the left side of the brain) of the fetus (Figure 1C). Meanwhile, incomplete lobulation between the upper and middle lobes of the right lung was detected. Only a 1.0 cm of fissure was found on the right lung of the fetus. Abnormalities including biliary atresia and telecanthus (20 mm) were also observed in the fetus.
The NFI family genes share a highly conserved DNA-binding domain; these genes regulate cell proliferation and differentiation of multiple organ systems. However, there are few studies on abnormal brain and lung development caused by NFIB deficiency. Currently, only 12 missense/non-sense and 2 small deletion/insertion variants of NFIB have been recorded in the Human Gene Mutation Database (HGMD; data retrieved in January 2022) (Figure 2). Here, we identified a novel non-sense NFIB variant (NM_001190737:c.870C > A;p.Tyr290*) in an aborted fetus. The variant was de novo (absent in parents) and resulted in premature termination by introducing a stop codon at the site of mutation. Our findings demonstrate the first case of abnormal brain development attributed to NFIB mutation (NM_001190737:c.870C > A;p.Tyr290*) in Asia, which expands our understanding of NFIB mutations and the associated physical development.
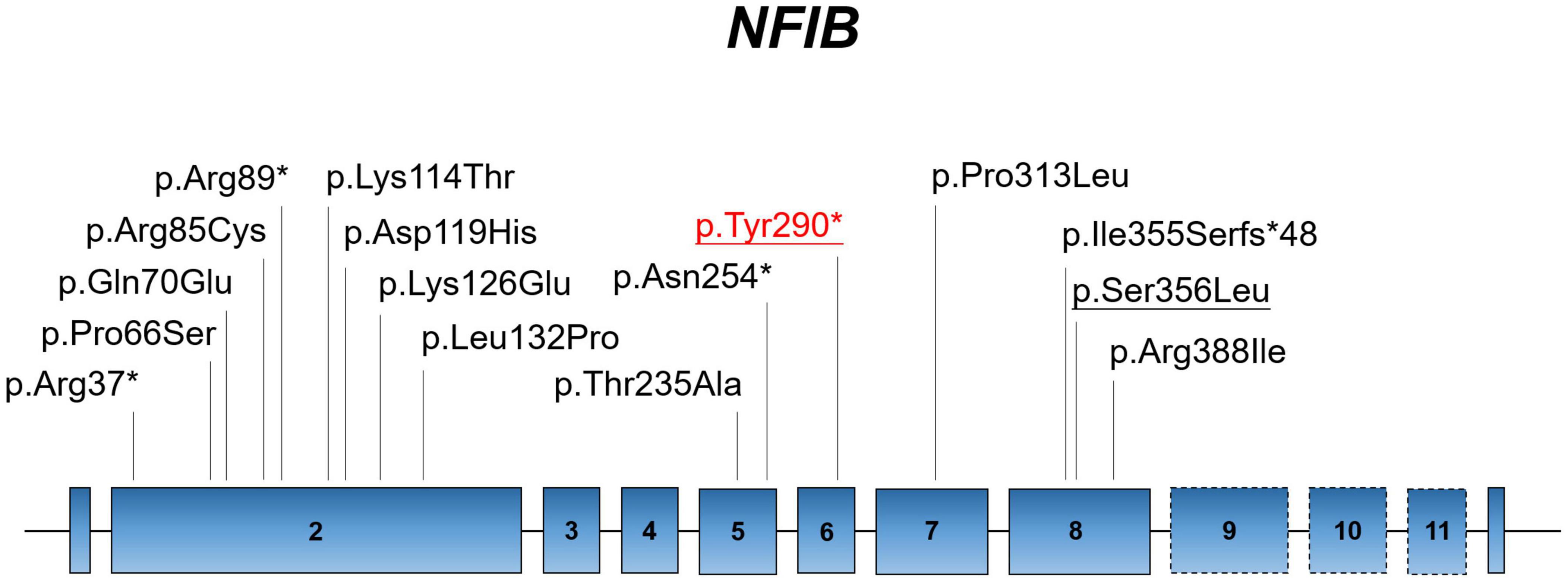
Figure 2. Localization of known mutations on the linear topology of NFIB. Red labeling represents the mutation identified in the present study. Underlined labels symbolize the fetal case.
The case presented here manifested no obvious structure of the gyrus and sulcus in the cerebral cortex of the fetus, suggesting lissencephaly or brain developmental retardation (Figure 1C). Lissencephaly, which literally means “smooth brain,” was first described in 1914 (16, 17). Patients with lissencephaly also manifest intellectual disabilities, developmental delays, seizures, and muscle spasms (18, 19). In the present study, a novel non-sense NFIB variant (NM_001190737:c.870C > A;p.Tyr290*) was identified by using WES. Mutations in NFIB are known to cause disability and macrocephaly. However, lissencephaly is often accompanied by microcephaly instead of macrocephaly (20). Considering the fetus in our case was aborted at 25 weeks and 4 days, most of the gyri and sulci on the brain surface were still under development, the reason is more likely to be retardation in brain development caused by this NFIB mutation. Nevertheless, more in-depth evidences were needed to draw a definite conclusion.
Studies on Nfib-deficient mice have shown that loss of Nfib results in perinatal lethality due to lung maturation defects along with severe callosal agenesis and forebrain defects (21). Even Nfib hemizygous mice show reduced lung maturation and decreased survival (22, 23). Dorsal telencephalon-specific Nfib conditional knockout mice show an obvious increase in cortical size (12), which may explain the macrocephaly observed in NFIB-deficient patients. These results were partially consistent with our findings of brain malformation and right lung lobulation defects in a fetus, possibly as a result of a non-sense variant (p.Tyr290*) of NFIB. Our study is also the first report of lung maturation defects caused by NFIB deficiency in humans. Notably, the NFIB (c.870C > A;p.Tyr290*) variant detected in the present study has not been reported previously and is therefore a novel variant.
We used WES to explore genetic lesions in a fetus with unexplained brain malformation and lung lobulation defects and identified a novel non-sense mutation of NFIB (NM_001190737:c.870C > A;p.Tyr290*). Our study thus contributes to insights for the genetic diagnosis and family counseling, and expands our understanding of NFIB mutations in association with brain and lung maturation.
The datasets for this article are not publicly available due to concerns regarding participant/patient anonymity. Requests to access the datasets should be directed to the corresponding author.
The studies involving human participants were reviewed and approved by the Review Board of Shenzhen Longgang District Maternity and Child Healthcare Hospital. Written informed consent to participate in this study was provided by the participants’ legal guardian/next of kin.
RX conceived and directed the project. HH, LW, HW, and HP collected the data and information. HH, YD, and JJ analyzed and interpreted the data. HH and JJ wrote the manuscript with the help of all the other authors.
This study was supported by the National Natural Science Foundation of China (81970403), China Postdoctoral Science Foundation (2020TQ0363 and 2020M682598), the Natural Science Foundation of Hunan, China (2021JJ40992), and the Youth Science Foundation of Xiangya Hospital (2021Q11).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
We thank the family for their participation in this study.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fped.2022.865181/full#supplementary-material
1. Hay RT. Origin of adenovirus DNA replication. Role of the nuclear factor I binding site in vivo. J Mol Biol. (1985) 186:129–36. doi: 10.1016/0022-2836(85)90263-3
2. Gronostajski RM. Roles of the NFI/CTF gene family in transcription and development. Gene. (2000) 249:31–45. doi: 10.1016/s0378-1119(00)00140-2
3. Chen KS, Lim J, Richards LJ, Bunt J. The convergent roles of the nuclear factor I transcription factors in development and cancer. Cancer Lett. (2017) 410:124–38. doi: 10.1016/j.canlet.2017.09.015
4. Adam RC, Yang H, Ge Y, Infarinato NR, Gur-Cohen S, Miao Y, et al. NFI transcription factors provide chromatin access to maintain stem cell identity while preventing unintended lineage fate choices. Nat Cell Biol. (2020) 22:640–50. doi: 10.1038/s41556-020-0513-0
5. Kilpatrick DL, Wang W, Gronostajski R, Litwack ED. Nuclear factor I and cerebellar granule neuron development: an intrinsic-extrinsic interplay. Cerebellum. (2012) 11:41–9. doi: 10.1007/s12311-010-0227-0
6. Zenker M, Bunt J, Schanze I, Schanze D, Piper M, Priolo M, et al. Variants in nuclear factor I genes influence growth and development. Am J Med Genet C Semin Med Genet. (2019) 181:611–26. doi: 10.1002/ajmg.c.31747
7. Clark BS, Stein-O’Brien GL, Shiau F, Cannon GH, Davis-Marcisak E, Sherman T, et al. Single-Cell RNA-Seq analysis of retinal development identifies NFI factors as regulating mitotic exit and Late-Born cell specification. Neuron. (2019) 102:1111–26. doi: 10.1016/j.neuron.2019.04.010
8. Hsu YC, Osinski J, Campbell CE, Litwack ED, Wang D, Liu S, et al. Mesenchymal nuclear factor I B regulates cell proliferation and epithelial differentiation during lung maturation. Dev Biol. (2011) 354:242–52. doi: 10.1016/j.ydbio.2011.04.002
9. Murtagh J, Martin F, Gronostajski RM. The Nuclear Factor I (NFI) gene family in mammary gland development and function. J Mammary Gland Biol Neoplasia. (2003) 8:241–54. doi: 10.1023/a:1025909109843
10. Denny SK, Yang D, Chuang CH, Brady JJ, Lim JS, Gruner BM, et al. Nfib promotes metastasis through a widespread increase in chromatin accessibility. Cell. (2016) 166:328–42. doi: 10.1016/j.cell.2016.05.052
11. Zilli F, Marques RP, Auf DMP, Jehanno C, Sethi A, Coissieux MM, et al. The NFIB-ERO1A axis promotes breast cancer metastatic colonization of disseminated tumour cells. Embo Mol Med. (2021) 13:e13162. doi: 10.15252/emmm.202013162
12. Schanze I, Bunt J, Lim J, Schanze D, Dean RJ, Alders M, et al. NFIB haploinsufficiency is associated with intellectual disability and macrocephaly. Am J Hum Genet. (2018) 103:752–68. doi: 10.1016/j.ajhg.2018.10.006
13. Barrus K, Rego S, Yip T, Martin PM, Glen OA, Van Ziffle J, et al. The expanding spectrum of NFIB-associated phenotypes in a diverse patient population-A report of two new patients. Am J Med Genet A. (2020) 182:2959–63. doi: 10.1002/ajmg.a.61852
14. Rao A, Goel H. Pathogenic nonsense variant in NFIB in another patient with dysmorphism, autism spectrum disorder, agenesis of the corpus callosum, and intellectual disability. Eur J Med Genet. (2020) 63:104092. doi: 10.1016/j.ejmg.2020.104092
15. Huang H, Chen Y, Jin J, Du R, Tang K, Fan L, et al. CSRP3, p.Arg122*, is responsible for hypertrophic cardiomyopathy in a Chinese family. J Gene Med. (2022) 24:e3390. doi: 10.1002/jgm.3390
16. Juric-Sekhar G, Hevner RF. Malformations of cerebral cortex development: molecules and mechanisms. Annu Rev Pathol. (2019) 14:293–318. doi: 10.1146/annurev-pathmechdis-012418-012927
17. Di Donato N, Chiari S, Mirzaa GM, Aldinger K, Parrini E, Olds C, et al. Lissencephaly: expanded imaging and clinical classification. Am J Med Genet A. (2017) 173:1473–88. doi: 10.1002/ajmg.a.38245
18. Koenig M, Dobyns WB, Di Donato N. Lissencephaly: update on diagnostics and clinical management. Eur J Paediatr Neurol. (2021) 35:147–52. doi: 10.1016/j.ejpn.2021.09.013
19. Di Donato N, Timms AE, Aldinger KA, Mirzaa GM, Bennett JT, Collins S, et al. Analysis of 17 genes detects mutations in 81% of 811 patients with lissencephaly. Genet Med. (2018) 20:1354–64. doi: 10.1038/gim.2018.8
20. Mochida GH. Genetics and biology of microcephaly and lissencephaly. Semin Pediatr Neurol. (2009) 16:120–6. doi: 10.1016/j.spen.2009.07.001
21. Steele-Perkins G, Plachez C, Butz KG, Yang G, Bachurski CJ, Kinsman SL, et al. The transcription factor gene Nfib is essential for both lung maturation and brain development. Mol Cell Biol. (2005) 25:685–98. doi: 10.1128/MCB.25.2.685-698.2005
22. Grunder A, Ebel TT, Mallo M, Schwarzkopf G, Shimizu T, Sippel AE, et al. Nuclear factor I-B (Nfib) deficient mice have severe lung hypoplasia. Mech Dev. (2002) 112:69–77. doi: 10.1016/s0925-4773(01)00640-2
Keywords: brain malformation, non-sense, mutation, NFIB, lung lobulation defects, lissencephaly
Citation: Huang H, Jin J, Wu L, Wu H, Pi H, Dong Y and Xiang R (2022) A de novo Non-sense Nuclear Factor I B Mutation (p.Tyr290*) Is Responsible for Brain Malformation and Lung Lobulation Defects. Front. Pediatr. 10:865181. doi: 10.3389/fped.2022.865181
Received: 29 January 2022; Accepted: 07 March 2022;
Published: 30 March 2022.
Edited by:
Alberto Spalice, Sapienza University of Rome, ItalyReviewed by:
Louise Bicknell, University of Otago, New ZealandCopyright © 2022 Huang, Jin, Wu, Wu, Pi, Dong and Xiang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Rong Xiang, c2hpcmxlc21pbGVAY3N1LmVkdS5jbg==
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.