- 1The Second School of Clinical Medicine, Southern Medical University, Guangzhou, Guangdong, China
- 2Department of Pediatric Surgery, Zhujiang Hospital, Southern Medical University, Guangzhou, Guangdong, China
Background: There are numerous published studies on the association between RET polymorphisms and susceptibility to Hirschsprung disease (HSCR). However, some of the results are inconsistent and the studies were conducted with small sample sizes. Therefore, we performed a meta-analysis to clarify the relationship.
Methods: Relevant data were retrieved from PubMed, Web of Science, Cochrane Library, EMBASE, CNKI, and Google Scholar according to PRISMA guidelines. Odds ratios (OR) were calculated to assess susceptibility to HSCR. Meanwhile, heterogeneity and publication bias were also calculated by R software package (version 4.2.1). The protocol was published in PROSPERO (CRD42022348940).
Results: A total of 12 studies were included in the meta-analysis and comprised 12 studies on the RET polymorphism rs2435357 (1,939 subjects and 3,613 controls) and 7 studies on the RET polymorphism rs2506030 (1,849 patients with HSCR and 3,054 controls). The analysis revealed that rs2435357 [A vs. G: odds ratio (OR) = 3.842, 95% confidence interval (CI) 2.829–5.220; AA vs. GG: OR = 2.597, 95% CI 1.499–4.501; AA + AG vs. GG: OR = 6.789, 95% CI 3.0711–14.9973; AA vs. AG + GG: OR = 8.156, 95%CI 5.429–12.253] and rs2506030 (A vs. G: OR = 0.519, 95% CI 0.469–0.573; AA vs. GG: OR = 0.543, 95% CI 0.474–0.623; AA + AG vs. GG: OR = 0.410, 95% CI 0.360–0.468; AA vs. AG + GG: OR = 0.361, 95%CI 0.292–0.447) were significantly associated with susceptibility to HSCR.
Conclusions: The polymorphisms rs2435357 and rs2506030 in the RET may be related to susceptibility to HSCR, of which rs2435357 (T > C) is the causal locus and rs2506030 (A > G) is the protective locus.
Systematic Review Registration: https://www.crd.york.ac.uk/prospero/, identifier:CRD42022348940.
Introduction
Hirschsprung disease (HSCR) is a very common developmental deformity of the digestive tract with a global incidence of approximately 1 case per 5,000 live births (1). The highest incidence is in Asian populations (approximately 2.8 cases per 10,000 live births) (2). HSCR is more common in males compared with females, with a ratio of approximately 4:1 (3). The main etiology of HSCR is the dysfunction of ectodermal neural crest cells in the migration process, resulting in a lack of ganglion cells in the myenteric nerve plexus of the distal intestinal wall (4), which causes abnormal intestinal function and morphology (5). Based on the extent of aganglionosis, HSCR is classified into the total colon aganglionic type (TCA: 5%), the long segment type (L-HSCR: 15%), and the short segment type (S-HSCR: 80%) (6). Delayed meconium excretion occurs in the majority of neonatal HSCR cases, and patients present with malnutrition and developmental delay as they get older. Delayed treatment can affect a child's development and can even be life threatening.
There is increasing evidence that genetic factors play an important role in the pathogenesis of HSCR (2, 7, 8). With the extensive application of high-throughput sequencing and genotyping technology, genome-wide association studies (GWAS) are facilitating the discovery of susceptibility genes for a variety of complex diseases, including HSCR (9).
The gene RET is the main susceptible gene in HSCR (10). RET is located on the long arm of chromosome 10, consists of 21 exons, and encodes a tyrosine kinase receptor superfamily protein containing 1,114 amino acids. RET is activated by ligands of the glial cell line derived neurotrophic factor (GDNF) family and participates in the proliferation, migration and differentiation of enteric neural crest cells (ENCCs) (11, 12). Abnormal expression of RET gene leads to abnormal colonization of ENCCs in the intestinal tract, which promotes apoptosis of ENCCs and causes the occurrence of HSCR (13).
A large number of case-control studies have investigated the association between single nucleotide polymorphisms (SNPs) and HSCR susceptibility (14–16). However, some SNPs still lack a corresponding meta-analysis. RET gene-related mutations are associated with the pathogenesis of HSCR (17). Rs2435357 and rs2506030 have been found to be closely related to HSCR (18). Rs2435357 (T > C) is the causal locus, while rs2506030 (A > G) is the protective locus. Numerous studies on the susceptibility analysis of HSCR with rs2435357 and rs2506030 have been published. The existing meta-analysis data of rs2435357 need to be updated, and there is no meta-analysis of rs2506030 and the results of its association with HSCR susceptibility are controversial. Therefore, revealing the degree of association between rs2435357 and rs2506030 with HSCR susceptibility via meta-analysis will provide the possibility to further study the pathogenesis of HSCR.
Methods
This systematic review and meta-analysis study was performed in accordance with the guidelines of the Preferred Reporting Items for Systematic Reviews and Meta-Analyses (PRISMA). The protocol was published in PROSPERO (https://www.crd.york.ac.uk/prospero/, CRD42022348940).
Literature search strategy
The retrieval work was independently completed by two researchers through PubMed, Web of Science, Cochrane Library, EMBASE, Chinese National Knowledge Infrastructure (CNKI), and Google Scholar. Search terms were [(HSCR) or (Hirschsprung) or (HD) or (congenital megacolon)] and [(ret) or (rs2435357) or (rs2506030)], and relevant data were manually searched according to specified standards. All studies considered potentially relevant were read in their entirety and subsequently selected for inclusion in the current study. Any differences arising therein were resolved after discussion with a third researcher. Finally, the selected studies were summarized.
Inclusion and exclusion criteria
Criteria were developed for the screening process to ensure the stability of the included data. Inclusion criteria: (1) analyzing the relationship between rs2435357 or rs2506030 and HSCR susceptibility, (2) cohort or case-control study, and (3) data for each genotype could be collected. Exclusion criteria: (1) the genotype frequency of the control group in the study did not meet the Hardy–Weinberg equilibrium (HWE, P < 0.01), 2) review articles, (3) animal experiments, (4) studies with duplicate data, and (5) severe data loss.
Data collection
Data were extracted from articles that met the inclusion criteria for analysis. This included the year of publication, region, author, HWE (P < 0.01), sample size, genotypic data from the cases and controls, genotypic classification, and sample source.
Quality assessment
Two independent reviewers assessed and scored the quality of the selected studies using the Newcastle–Ottawa scale (NOS) (19). The NOS scale assessed the quality of the study through eight items in three dimensions: exposure, study population selection, and comparability. The highest possible NOS score was 9, and a score of 6 points or above was considered high-quality research.
Statistical analysis
Meta-package and Meta-for package in R Studio were used for relevant data analysis. Odds ratios (ORs) and corresponding confidence intervals (CIs) were analyzed using the data collected to quantify the relationship between the two SNPs (rs2435357 and rs2506030) and susceptibility to HSCR. During analysis, the uppercase letters “A” and “T” were identified as the mutant alleles for rs2435357 and rs2506030, respectively, while the uppercase letters “G” and “C” were defined as wild-type alleles. The HWE for each study was assessed by Chi-square test (20). Additionally, to determine the integrity of the analysis, five genetic models were selected as the main content of data analysis; these models were allele model, dominant model, recessive model, heterozygous model, and homozygous model. Subsequently, the Q test was used to assess the heterogeneity of the data included in the study. A P value >0.1 indicated that the data included in the study were quite heterogeneous and suitable for the use of a random effects model; otherwise, a fixed effects model was used (21). Because heterogeneity of meta-analysis can be derived from race, genotyping method, and sample source (22–24), the data included in the study were further classified according to race and genotyping method, and then subgroup analysis was performed to identify the source of the heterogeneity. In addition, the sensitivity of the overall meta-analysis was assessed by deleting data for each single study (25).
Results
Study characteristics
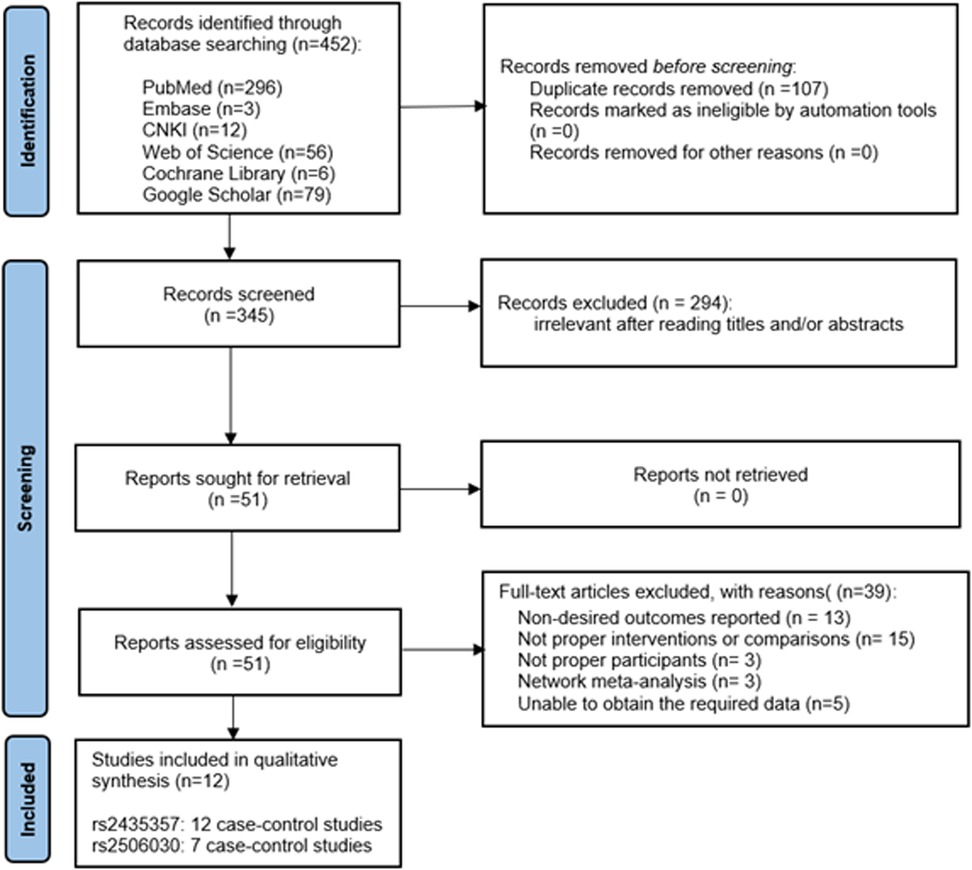
A total of 452 studies were retrieved from PubMed, EMBASE, Web of Science and Cochrane Library, Google Scholar, and CNKI. Among them, 107 repetitive articles were excluded and 294 articles that did not meet the inclusion criteria were excluded according to the title and abstract. Subsequently, a further 39 articles were excluded after analysis of the full text of the remaining 51 articles. Ultimately, 12 articles were included in the meta-analysis. Of the 12 selected studies, 7 investigated both rs2435357 and rs2506030. A PRISMA flowchart of the literature retrieval strategy is shown in Figure 1.
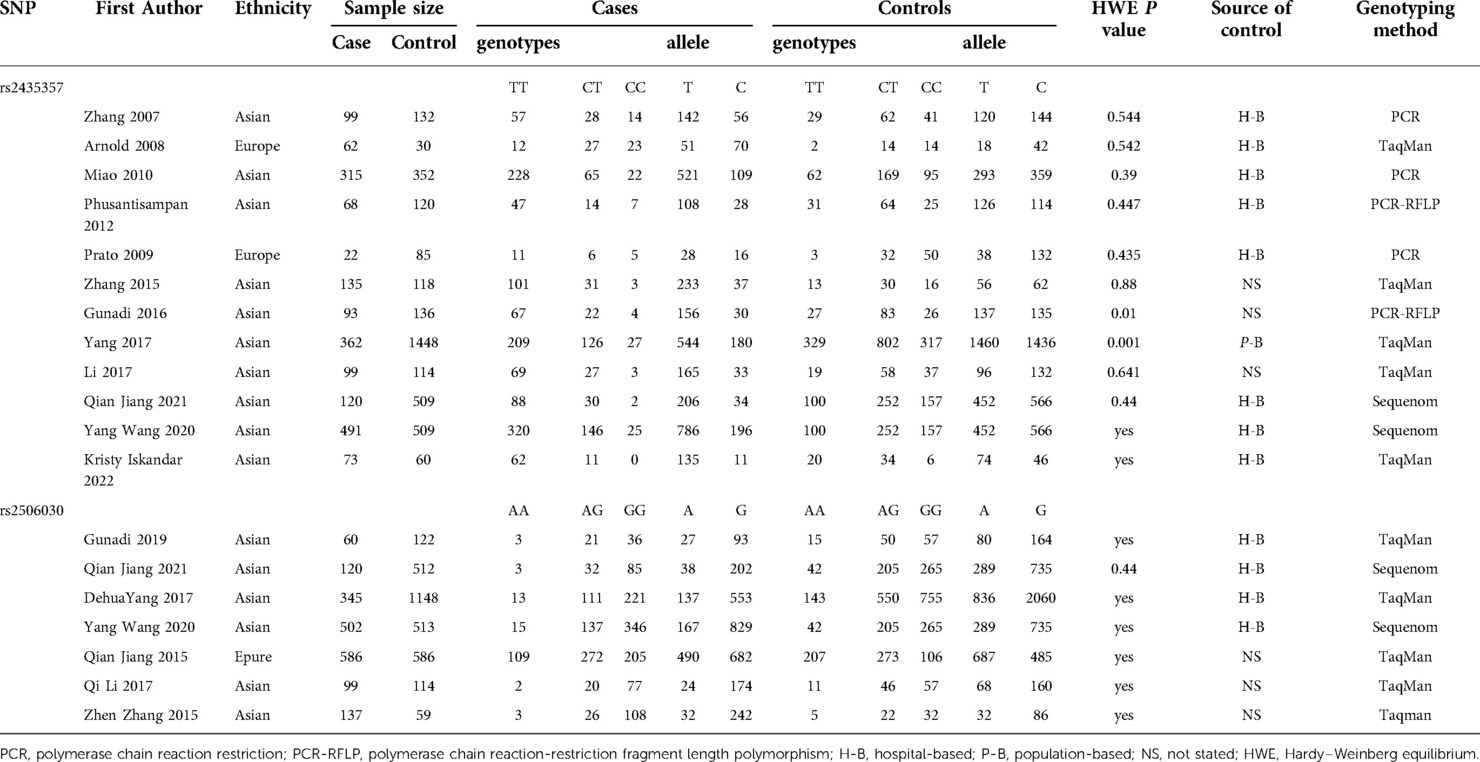
There were 12 studies on rs2435357 (3, 17, 26–35) and 7 studies on rs2506030 (17, 27, 28, 31, 33–36) included in the meta-analysis. The general characteristics of the enrolled studies are presented in Table 1. Two studies were conducted in Europe (26, 30) and 10 in Asia. Only one study explored the long segment type and the short segment type of HSCR separately (28). Sample sizes ranged from 107 to 1810. Genotypic classification methods were TaqMan, Sequenom, PCR-RFLP, and PCR. Most studies were HWE balanced and three studies did not state HWE (17, 27, 28). Eight studies were hospital-based, one was population-based, and three did not state their source of controls.
Research quality
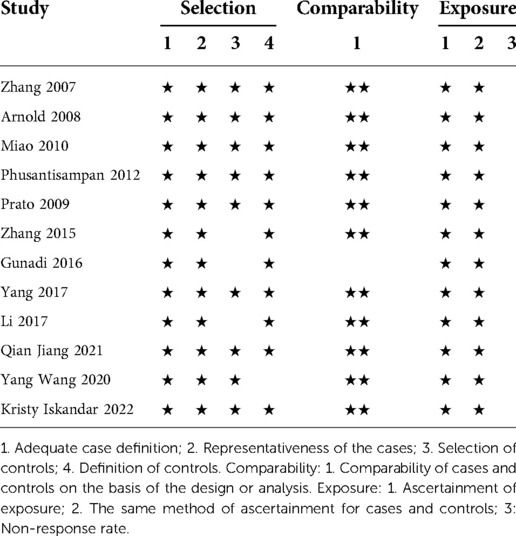
All studies included in the meta-analysis were of high quality with NOS scores between 6 and 8 (Table 2). Furthermore, all the studies had a good definition and representation of cases, and all clearly described the investigation and evaluation methods. In each study, the genetic typing method of the controls and the cases was the same. However, the selection scores of the control groups were not high, and 25% of the studies did not score on this item.
Rs2435357
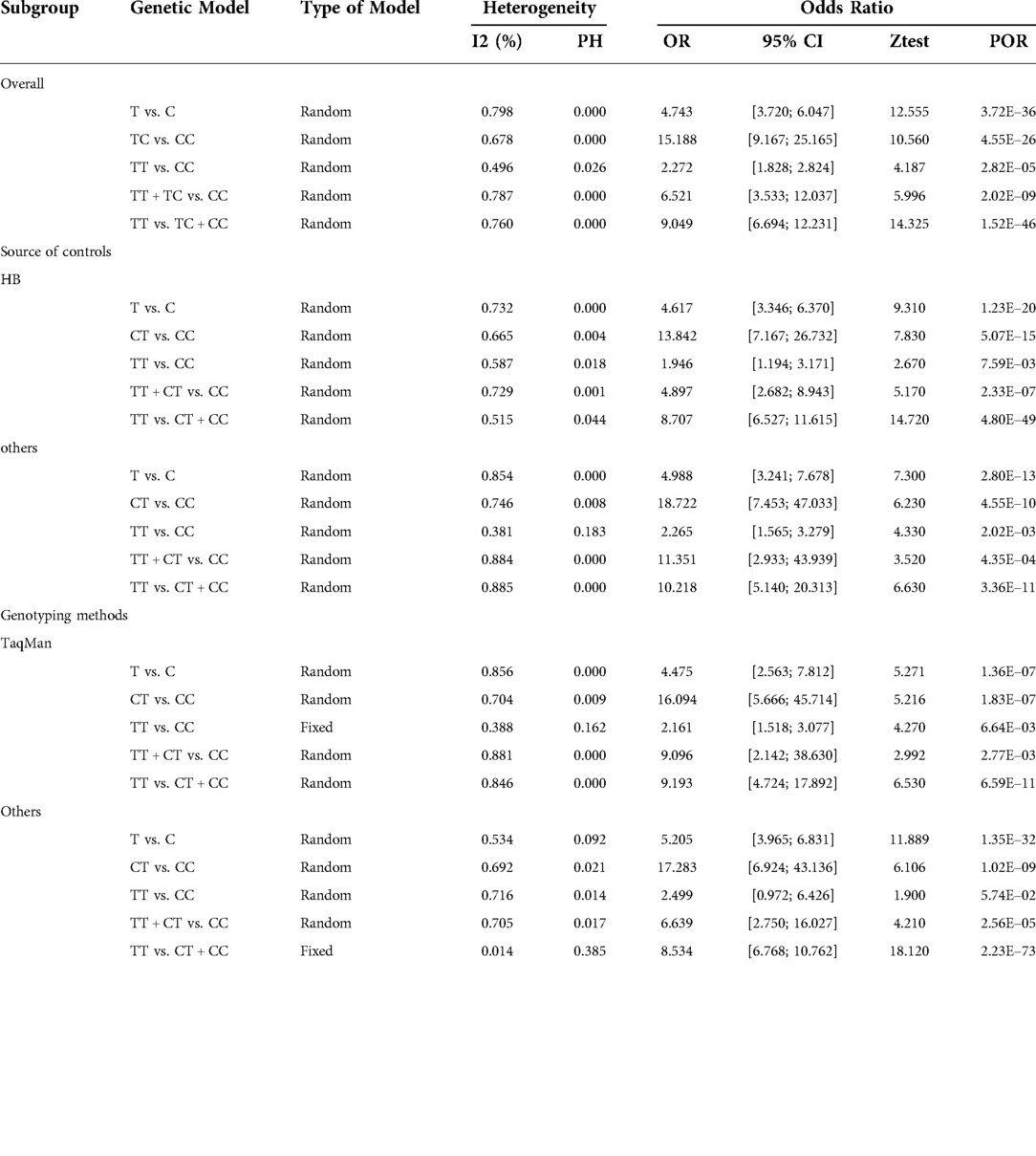
The meta-analysis results of the association between the rs2435357 polymorphism of the RET and the susceptibility risk of HSCR are presented in Figure 2. A total of 12 studies were summarized, and 1,939 cases and 3,613 controls were included in the data analysis. The rs2435357 polymorphism was significantly associated with HSCR susceptibility risk (T vs. C: OR = 4.743, 95% CI 3.720–6.047, P = 3.72e-36, I2 = 79.8%, P = 0.000). In addition, the following allelic models were also significantly associated with HSCR susceptibility: homozygous (TT vs. CC: OR = 2.272, 95% CI 1.828–2.824, P = 2.82e-5, I2 = 68.0%, P = 0.026); dominant (TT + CT vs. CC: OR = 6.521, 95% CI 3.533–12.037, P = 2.21e-06, I2 = 78.7%, P = 0.000); and recessive (TT + CT + CC: OR = 9.046, 95% CI 6.694–12.231, P = 1.52e-46, I2 = 76.0%, P = 0.000).
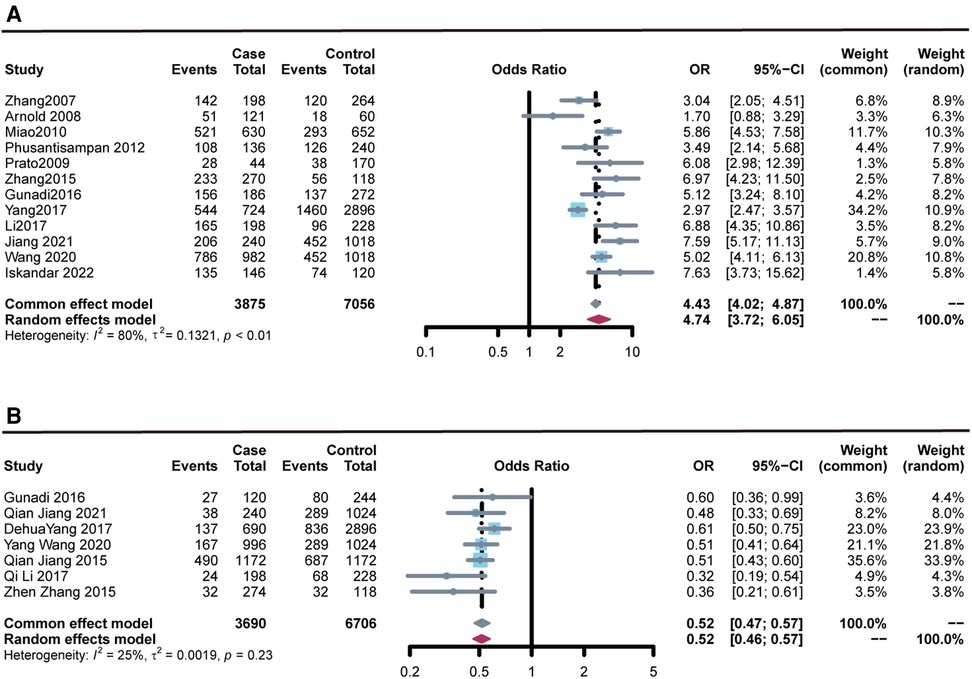
Figure 2. Forest plots of the association of rs2435357 and rs2506030 polymorphisms of the RET with susceptibility to HSCR. (A) rs2435357 (allele model); (B) rs2506030 (allele model).
Stratified analysis by ethnicity revealed that the rs2435357 polymorphism was significantly associated with HSCR risk in Asia: allele mode (T vs. C: OR = 4.992, 95% CI 3.956–6.298, P = 7.43e-42, I2 = 80.2%, P = 0.000); homozygous model (TT vs. CC: OR = 2.335, 95% CI 1.537–3.548, P = 7.06e-5, I2 = 55.1%, P = 0.018); heterozygous model (TT vs. CT: OR = 15.655, 95% CI 9.185–26.681, P = 4.89e-24, I2 = 70.2%, P = 0.000); dominant model (TT + CT vs. CC: OR = 7.855, 95% CI 4.028–15.320, P = 1.47e-09, I2 = 79.1%, P = 0.000); recessive model (TT + CT + CC: OR = 8.956, 95% CI 6.596–12.161, P = 8.20e-45, I2 = 78.4%, P = 0.000). Subgroup analysis by genotyping method and source of controls also revealed there was association between the rs2435357 polymorphism and HSCR risk in TaqMan, PCR, and Hospital-Based (H-B) subgroups. Results for other subgroups are presented in Table 3.
Rs2506030
Data were collected from seven studies, comprising 1,849 cases with HSCR and 3,054 controls. The meta-analysis results (Figure 2) revealed that the rs2506030 polymorphism was significantly associated with susceptibility to HSCR (allele: A vs. G: OR = 0.519, 95% CI 0.469–0.572, P = 2.21E-33, I2 = 25.5%, and P = 0.234). There was also sufficient evidence that the following allelic models were associated with HSCR susceptibility: homozygous (AA vs. GG: OR = 0.543, 95% CI 0.474–0.623, P = 7.00e-13, I2 = 29.8%, P = 0.200); dominant (AA + AG vs. GG: OR = 0.410, 95% CI 0.360–0.468, P = 2.49e-40, I2 = 0.00%, P = 0.541); and recessive (AA vs. AG + GG: OR = 0.361, 95% CI 0.292–0.447, P = 7.54e-17, I2 = 0.00%, P = 0.803).
Subgroup analyses were performed by ethnicity, source of controls, and genotyping method. The association between RET gene polymorphism and HSCR susceptibility was significant in the TaqMan subgroup (A vs. G: OR = 0.525, 95% CI 0.467–0.591, P = 3.92e-11, I2 = 49.0%, P = 0.098; AA vs. GG: OR = 0.565, 95% CI 0.477–0.670, P = 5.51e-6, I2 = 49.6%, P = 0.094; AG vs. GG: OR = 0.274, 95% CI 0.208–0.361, P = 5.68e-21, I2 = 0.00, P = 0.851; AA + AG vs. GG: OR = 0.385, 95% CI 0.327–0.453, P = 2.36e-30, I2 = 0.00%, P = 0.507; AA vs. AG + GG: OR = 0.368, 95% CI 0.292–0.465, P = 2.16e-10, I2 = 0.00%, P = 0.593). In addition, there was an association between rs2506030 polymorphism and HSCR risk in the Hospital-Based (H-B) subgroup. Results of the subgroup analysis of sample source and genotyping method are shown in Table 4.
Sensitivity analysis
Sensitivity analysis calculates the overall correlation coefficient obtained by removing a group of studied data respectively, and then quantifies the difference between each correlation coefficient to obtain the stability of the whole data and the model. The robustness of the meta-analysis was assessed by comparing the results of analyses before and after the removal of one study. The processed results were not significantly different from the previous results, thus the data and model have high robustness (Figure 3).

Figure 3. Forest plots of the sensitivity of the meta-analysis. (A) rs2435357 (allele model); (B) rs2506030 (allele model).
Publication bias
Harbord's test was performed to assess the possible publication bias of included studies. The results showed that in the meta-analysis of rs2435357, five genetic models showed no publication bias: allele mode (T vs. C: P = 0.395); homozygous model (TT vs. CC: P = 0.216); heterozygous model (TT vs. CT: P = 0.898, Figure 4A); dominant model (TT + CT vs. CC: P = 0.902); recessive model (TT + CT + CC: P = 0.287). In addition, in the meta-analysis of rs2506030, no publication bias found: allele: A vs. G: P = 0.225); homozygous (AA vs. GG: P = 0.470); heterozygous model (AA vs. AG: P = 0.172); dominant model (AA + AG vs. GG: P = 0.732, Figure 4B); and recessive model (AA vs. AG + GG: P = 0.070).
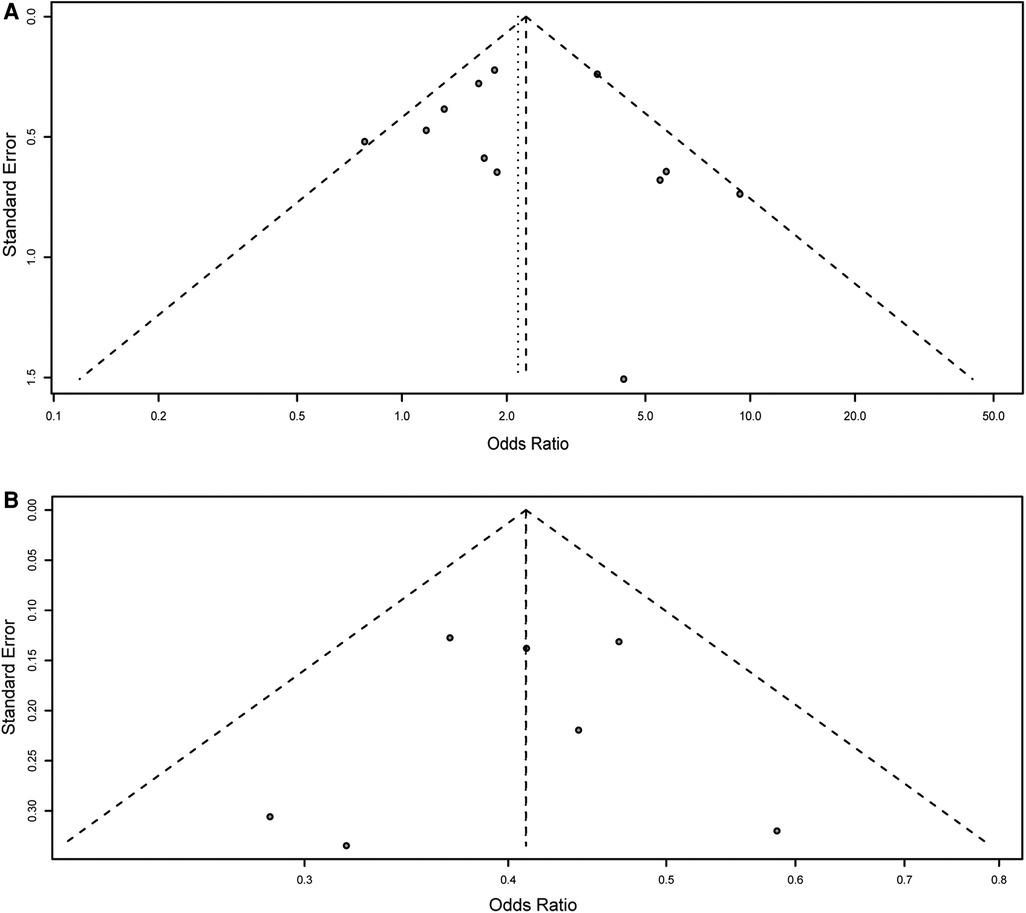
Figure 4. Funnel plot for the detection of the publication bias for association of rs2435357 and rs2506030 polymorphisms of the RET gene with HSCR risk. (A) rs2435357 (heterozygous model); (B) rs2506030 (dominant model).
Discussion
RET gene is located on the long arm of chromosome 10, contains 21 exons, and encodes receptor tyrosine kinase transmembrane protein. RET protein has three different subtypes, named RET51, RET43, and RET9. Moreover, Bhattarai et al. (37) showed that interactions between RET protein and its ligands can control the survival, migration, proliferation, differentiation, and maturation of vagal and sacral neural crest cells. Nagy et al. (38) demonstrated that excessive stimulation of RET al. one could lead to HSCR. Mutations in the RET are currently detected in approximately half of familial aggregate patients with HSCR and in 7% to 35% of patients with sporadic HSCR (39–41). Using the strategy of combining gene-gene interaction analysis with case-control study, Wang et al. demonstrated for the first time that the genetic markers of RET, ARHGEF3, and CTNNAL1 and the related genetic interaction network can change the susceptibility risk of HSCR in the Han population (33). In addition, there is sufficient evidence that RET plays an important role in other developmental and metabolic disorders (42).
The results of the present study are generally consistent with previous meta-analyses of RET polymorphisms and susceptibility to HSCR. Rs2435357 has been the focus of attention since Liang et al. (43) conducted the first meta-analysis to explore the association between RET polymorphisms and HSCR susceptibility in 2014. However, due to various limitations, previous meta-analyses for RET polymorphisms and HSCR susceptibility are problematic to differing degrees in that the sample sizes are small and the analysis indexes are shallow. To obtain accurate and effective data on the potential association, a meta-analysis with larger samples and analysis at a deeper level was needed.
Of the two SNPs included in our meta-analysis, rs2435357 is currently the most studied and the most significantly associated with HSCR susceptibility. rs2506030 is an association point for a ∼125-kb upstream of RET, identified by Jiang et al. (Manuscript in Review at the American Journal of Human Genetics) in a single GWAS using cases of HSCR of European origin (EA) from five different countries (US, Italy, France, Spain, and the Netherlands). Three SNPs—rs2435357, rs2506030, and rs2506004—were previously reported to be potentially unrelated to HSCR susceptibility (17). The results of the current study confirmed a significant association between two genetic polymorphisms—rs2435357 and rs2506030—and susceptibility to HSCR. Interestingly, not all gene models were related to HSCR risk in ethnicity subgroups and genotyping subgroups in the present study, which suggested that ethnicity and genotyping methods may influence the association of genetic polymorphisms with HSCR susceptibility. In addition, all study data were found to be from Europe and Asia during the analysis, and no studies from America and Africa meeting the inclusion criteria were available to evaluate the association between RET and HSCR susceptibility through meta-analysis. One of the two SNPs included in this study was a causal locus, whereas the other was a protective locus. The OR value of rs2435357 (T > C) is 3.842, suggesting that rs2435357 promotes the incidence of HSCR; while the OR value of rs2506030 (A > G) is 0.519, indicating that rs2506030 (A > G) has a positive effect on HSCR. Previous studies have found that rs2435357 and rs2506030 do not have linkage disequilibrium (44), implying that the effects of these two SNPs can be enhanced or inhabited by each other. Therefore, the purpose of further research can be achieved by detecting the mutation of rs2435357 and rs2506030.
However, the study also has defects. The first is that the sample size of the study is still small, which may affect the final results. Second, the results of this study may not be applicable to cases from other regions since the cases included in this study were only from Asia and Europe. Third, only published study data were included in the meta-analysis, which could cause publication bias. In addition, most of the articles included in the study lacked data on gender, age, and pathological type of HSCR of the subjects, therefore hierarchical analysis was performed on these covariates. Finally, HSCR is a combination of environmental and genetic aspects that cannot be comprehensively analyzed in a meta-analysis due to limitations in the study data.
Conclusion
This study revealed the relationship between two polymorphisms of RET—rs2435357 and rs2506030 and HSCR susceptibility, with rs2506030 exhibiting the most significant association with HSCR susceptibility. In the future, larger samples and more comprehensive analysis will more fully determine their association. Moreover, the related findings of the association between RET gene polymorphisms and HSCR susceptibility provide novel suggestions for HSCR pathogenesis research.
Data availability statement
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author/s.
Author contributions
JM: preparation manuscript, statistics, writing and finalizing the manuscript. YZ, XL, GL: preparation of data and tools. YL, ZH, CL: reviewed the manuscript. WK: guide the project, and revise the manuscript. All authors contributed to the article and approved the submitted version.
Funding
This study was supported by the Natural Science Foundation of Guangdong (2019A1515011086) and The Innovation and Entrepreneurship Training Program for College Students (202212121229).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher's note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
1. Heuckeroth RO. Hirschsprung disease—integrating basic science and clinical medicine to improve outcomes. Nat Rev Gastroenterol Hepatol. (2018) 15:152–67. doi: 10.1038/nrgastro.2017.149.
2. Amiel J, Sproat-Emison E, Garcia-Barcelo M, Lantieri F, Burzynski G, Borrego S, et al. Hirschsprung disease, associated syndromes and genetics: a review. J Med Genet. (2008) 45:1–14. doi: 10.1136/jmg.2007.053959.
3. Phusantisampan T, Sangkhathat S, Phongdara A, Chiengkriwate P, Patrapinyokul S, Mahasirimongkol S. Association of genetic polymorphisms in the RET-protooncogene and NRG1 with hirschsprung disease in Thai patients. J Hum Genet. (2012) 57:286–93. doi: 10.1038/jhg.2012.18.
4. Mueller JL, Goldstein AM. The science of hirschsprung disease: what we know and where we are headed. Semin Pediatr Surg. (2022) 31:151157. doi: 10.1016/j.sempedsurg.2022.151157.
5. Núñez-Torres R, Fernández RM, Acosta MJ, Enguix-Riego Mdel V, Marbá M, Carlos de Agustín J, et al. Comprehensive analysis of RET common and rare variants in a series of spanish hirschsprung patients confirms a synergistic effect of both kinds of events. BMC Med Genet. (2011) 12:138. doi: 10.1186/1471-2350-12-138.
6. Smigiel R, Lebioda A, Patkowski D, Czernik J, Dobosz T, Pesz K, et al. Single nucleotide polymorphisms in the RET gene and their correlations with hirschsprung disease phenotype. J Appl Genet. (2006) 47:261–7. doi: 10.1007/bf03194634.
7. Butler Tjaden NE, Trainor PA. The developmental etiology and pathogenesis of hirschsprung disease. Transl Res. (2013) 162:1–15. doi: 10.1016/j.trsl.2013.03.001.
8. Wallace AS, Anderson RB. Genetic interactions and modifier genes in hirschsprung's disease. World J Gastroenterol. (2011) 17:4937–44. doi: 10.3748/wjg.v17.i45.4937.
9. Tang CS, Gui H, Kapoor A, Kim JH, Luzón-Toro B, Pelet A, et al. Trans-ethnic meta-analysis of genome-wide association studies for hirschsprung disease. Hum Mol Genet. (2016) 25:5265–75. doi: 10.1093/hmg/ddw333.
10. Alves MM, Sribudiani Y, Brouwer RW, Amiel J, Antiñolo G, Borrego S, et al. Contribution of rare and common variants determine complex diseases-hirschsprung disease as a model. Dev Biol. (2013) 382:320–9. doi: 10.1016/j.ydbio.2013.05.019.
11. Burzynski GM, Nolte IM, Bronda A, Bos KK, Osinga J, Plaza Menacho I, et al. Identifying candidate hirschsprung disease-associated RET variants. Am J Hum Genet. (2005) 76:850–8. doi: 10.1086/429589.
12. Iwashita T, Kurokawa K, Qiao S, Murakami H, Asai N, Kawai K, et al. Functional analysis of RET with hirschsprung mutations affecting its kinase domain. Gastroenterol. (2001) 121:24–33. doi: 10.1053/gast.2001.25515.
13. Karim A, Tang CS, Tam PK. The emerging genetic landscape of hirschsprung disease and its potential clinical applications. Front Pediatr. (2021) 9:638093. doi: 10.3389/fped.2021.638093.
14. Amooee A, Lookzadeh MH, Mirjalili SR, Miresmaeili SM, Aghili K, Zare-Shehneh M, et al. ASSOCIATION OF RS2435357 AND RS1800858 POLYMORPHISMS IN RET PROTO-ONCOGENE WITH HIRSCHSPRUNG DISEASE: SYSTEMATIC REVIEW AND META-ANALYSIS. Arq Bras Cir Dig. (2019) 32:e1448. doi: 10.1590/0102-672020190001e1448.
15. Bahrami R, Shajari A, Aflatoonian M, Noorishadkam M, Akbarian-Bafghi MJ, Morovati-Sharifabad M, et al. Association of REarranged during transfection (RET) c.73 + 9277T > C and c.135G > a polymorphisms with susceptibility to hirschsprung disease: a systematic review and meta-analysis. Fetal Pediatr Pathol. (2020) 39:476–90. doi: 10.1080/15513815.2019.1672225.
16. Tomuschat C, Puri P. RET Gene is a major risk factor for hirschsprung's disease: a meta-analysis. Pediatr Surg Int. (2015) 31:701–10. doi: 10.1007/s00383-015-3731-y.
17. Xia RP, Zhao F, Ma TD, Zou CJ, Xu G, Zhou CG. Circ-ITCH overexpression promoted cell proliferation and migration in hirschsprung disease through miR-146b-5p/RET axis. Pediatr Res. (2022. doi: 10.1038/s41390-021-01860-5.
18. Zhang Z, Jiang Q, Li Q, Cheng W, Qiao G, Xiao P, et al. Genotyping analysis of 3 RET polymorphisms demonstrates low somatic mutation rate in Chinese hirschsprung disease patients. Int J Clin Exp Pathol. (2015) 8:5528–34.
19. Stang A. Critical evaluation of the Newcastle-Ottawa scale for the assessment of the quality of nonrandomized studies in meta-analyses. Eur J Epidemiol. (2010) 25:603–5. doi: 10.1007/s10654-010-9491-z.
20. Salanti G, Amountza G, Ntzani EE, Ioannidis JP. Hardy-Weinberg equilibrium in genetic association studies: an empirical evaluation of reporting, deviations, and power. Eur J Hum Genet. (2005) 13:840–8. doi: 10.1038/sj.ejhg.5201410.
21. DerSimonian R, Laird N. Meta-analysis in clinical trials. Control Clin Trials. (1986) 7:177–88. doi: 10.1016/0197-2456(86)90046-2.
22. Namazi A, Forat-Yazdi M, Jafari MA, Foroughi E, Farahnak S, Nasiri R, et al. Association between polymorphisms of ERCC5 gene and susceptibility to gastric cancer: a systematic review and meta-analysis. Asian Pac J Cancer Prev. (2017) 18:2611–7. doi: 10.22034/apjcp.2017.18.10.2611.
23. Forat-Yazdi M, Jafari M, Kargar S, Abolbaghaei SM, Nasiri R, Farahnak S, et al. Association between SULT1A1 Arg213His (rs9282861) polymorphism and risk of breast cancer: a systematic review and meta-analysis. J Res Health Sci. (2017) 17:e00396.
24. Aslebahar F, Neamatzadeh H, Meibodi B, Karimi-Zarchi M, Tabatabaei RS, Noori-Shadkam M, et al. Association of tumor necrosis factor-α (TNF-α) -308G > A and -238G > A polymorphisms with recurrent pregnancy loss risk: a meta-analysis. Int J Fertil Steril. (2019) 12:284–92. doi: 10.22074/ijfs.2019.5454.
25. Thabane L, Mbuagbaw L, Zhang S, Samaan Z, Marcucci M, Ye C, et al. A tutorial on sensitivity analyses in clinical trials: the what, why, when and how. BMC Med Res Methodol. (2013) 13:92. doi: 10.1186/1471-2288-13-92.
26. Arnold S, Pelet A, Amiel J, Borrego S, Hofstra R, Tam P, et al. Interaction between a chromosome 10 RET enhancer and chromosome 21 in the down syndrome-hirschsprung disease association. Hum Mutat. (2009) 30:771–5. doi: 10.1002/humu.20944.
27. Gunadi DA, Iskandar K, Makhmudi A, Rochadi . Accuracy of polymerase chain reaction-restriction fragment length polymorphism for RET rs2435357 genotyping as hirschsprung risk. J Surg Res. (2016) 203:91–4. doi: 10.1016/j.jss.2016.02.039
28. Li Q, Zhang Z, Diao M, Gan L, Cheng W, Xiao P, et al. Cumulative risk impact of RET, SEMA3, and NRG1 polymorphisms associated with hirschsprung disease in han Chinese. J Pediatr Gastroenterol Nutr. (2017) 64:385–90. doi: 10.1097/mpg.0000000000001263.
29. Miao X, Leon TY, Ngan ES, So MT, Yuan ZW, Lui VC, et al. Reduced RET expression in gut tissue of individuals carrying risk alleles of hirschsprung's disease. Hum Mol Genet. (2010) 19:1461–7. doi: 10.1093/hmg/ddq020.
30. Prato AP, Musso M, Ceccherini I, Mattioli G, Giunta C, Ghiggeri GM, et al. Hirschsprung disease and congenital anomalies of the kidney and urinary tract (CAKUT): a novel syndromic association. Med (Baltimore). (2009) 88:83–90. doi: 10.1097/MD.0b013e31819cf5da.
31. Yang D, Yang J, Li S, Jiang M, Cao G, Yang L, et al. Effects of RET, NRG1 and NRG3 polymorphisms in a Chinese population with hirschsprung disease. Sci Rep. (2017) 7:43222. doi: 10.1038/srep43222.
32. Zhang XN, Zhou MN, Qiu YQ, Ding SP, Qi M, Li JC. Genetic analysis of RET, EDNRB, and EDN3 genes and three SNPs in MCS+9.7 in Chinese patients with isolated hirschsprung disease. Biochem Genet. (2007) 45:523–7. doi: 10.1007/s10528-007-9093-y.
33. Wang Y, Jiang Q, Cai H, Xu Z, Wu W, Gu B, et al. Genetic variants in RET, ARHGEF3 and CTNNAL1, and relevant interaction networks, contribute to the risk of hirschsprung disease. Aging (Albany NY). (2020) 12:4379–93. doi: 10.18632/aging.102891.
34. Iskandar K, Simanjaya S, Indrawan T, Kalim AS, Marcellus HD, et al. Is there any mosaicism in REarranged during transfection variant in hirschsprung disease's patients? Front Pediatr. (2022) 10:842820. doi: 10.3389/fped.2022.842820.
35. Jiang Q, Arnold S, Heanue T, Kilambi KP, Doan B, Kapoor A, et al. Functional loss of semaphorin 3C and/or semaphorin 3D and their epistatic interaction with ret are critical to hirschsprung disease liability. Am J Hum Genet. (2015) 96:581–96. doi: 10.1016/j.ajhg.2015.02.014.
36. Jiang Q, Wang Y, Gao Y, Wang H, Zhang Z, Li Q, et al. RET Compound inheritance in Chinese patients with hirschsprung disease: lack of penetrance from insufficient gene dysfunction. Hum Genet. (2021) 140:813–25. doi: 10.1007/s00439-020-02247-y.
37. Bhattarai C, Poudel PP, Ghosh A, Kalthur SG. The RET gene encodes RET protein, which triggers intracellular signaling pathways for enteric neurogenesis, and RET mutation results in hirschsprung's disease. AIMS Neurosci. (2022) 9:128–49. doi: 10.3934/Neuroscience.2022008.
38. Nagy N, Guyer RA, Hotta R, Zhang D, Newgreen DF, Halasy V, et al. RET Overactivation leads to concurrent hirschsprung disease and intestinal ganglioneuromas. Development. (2020) 147. doi: 10.1242/dev.190900.
39. Seri M, Yin L, Barone V, Bolino A, Celli I, Bocciardi R, et al. Frequency of RET mutations in long- and short-segment hirschsprung disease. Hum Mutat. (1997) 9:243–9. 10.1002/(sici)1098-1004(1997)9:3<243::Aid-humu5 > 3.0.Co;2-8.
40. Attié T, Pelet A, Edery P, Eng C, Mulligan LM, Amiel J, et al. Diversity of RET proto-oncogene mutations in familial and sporadic hirschsprung disease. Hum Mol Genet. (1995) 4:1381–6. doi: 10.1093/hmg/4.8.1381.
41. Angrist M, Bolk S, Thiel B, Puffenberger EG, Hofstra RM, Buys CH, et al. Mutation analysis of the RET receptor tyrosine kinase in hirschsprung disease. Hum Mol Genet. (1995) 4:821–30. doi: 10.1093/hmg/4.5.821.
42. Takahashi M. RET Receptor signaling: function in development, metabolic disease, and cancer. Proc Jpn Acad Ser B Phys Biol Sci. (2022) 98:112–25. doi: 10.2183/pjab.98.008.
43. Liang CM, Ji DM, Yuan X, Ren LL, Shen J, Zhang HY. RET And PHOX2B genetic polymorphisms and hirschsprung's disease susceptibility: a meta-analysis. PLoS One. (2014) 9:e90091. doi: 10.1371/journal.pone.0090091.
Keywords: hirschsprung disease, meta-analysis, susceptibility, single nucleotide polymorphism, RET
Citation: Mu J, Zhang Y, Liao G, Li X, Luo Y, Huang Z, Luo C and Wu K (2022) Association of rs2435357 and rs2506030 polymorphisms in RET with susceptibility to hirschsprung disease: A systematic review and meta-analysis. Front. Pediatr. 10:1030933. doi: 10.3389/fped.2022.1030933
Received: 29 August 2022; Accepted: 27 September 2022;
Published: 17 October 2022.
Edited by:
Jiexiong Feng, Huazhong University of Science and Technology, ChinaReviewed by:
Huaxiang Zhao, Xi'an Jiaotong University, ChinaFatih Akbiyik, Yuksek Ihtisas University, Turkey
© 2022 Mu, Zhang, Liao, Li, Luo, Huang, Luo and Wu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Kai Wu d3VrYWlAc211LmVkdS5jbg==
Specialty Section: This article was submitted to Pediatric Surgery, a section of the journal Frontiers in Pediatrics
 Jianhua Mu
Jianhua Mu Yuxi Zhang
Yuxi Zhang Guoying Liao
Guoying Liao Xinxin Li1
Xinxin Li1 Kai Wu
Kai Wu