
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Pediatr., 25 August 2021
Sec. Pediatric Neurology
Volume 9 - 2021 | https://doi.org/10.3389/fped.2021.718704
This article is part of the Research TopicNeurodevelopment and Preterm BirthView all 6 articles
 Xiaohua Dong1,2*†
Xiaohua Dong1,2*† Jing Zhao1†
Jing Zhao1† Yinyin Shen1†
Yinyin Shen1† Qing Sun1†
Qing Sun1† Xiaohui Wu2
Xiaohui Wu2 Yanqing Zhu2
Yanqing Zhu2 Lingling Yu2*
Lingling Yu2* Yingmin Zhao2*
Yingmin Zhao2*Neonatal hypoxic–ischemic encephalopathy (HIE) is a common neurological disorder triggered by perinatal cerebral ischemia and hypoxia. Accumulating evidence has shown that peptides have neuroprotective effects in nerve injury. However, the function of endogenous peptides in the pathogenesis of HIE has not been studied. In the present study, a comparative peptidomic profile was performed in the serum of the human umbilical cord blood with HIE (three patients) and the control group (three health control) by liquid chromatography–mass spectrometry (LC-MS). Our study demonstrated that a total of 49 peptides derived from 25 precursor proteins were differentially expressed in the serum of HIE compared with normal controls, including 33 upregulated peptides and 16 downregulated peptides. Each of the differentially expressed peptides has specific characteristics, including pI, Mw, and cleavage pattern. Gene ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) analyses indicated that the precursor proteins of differentially expressed peptides participate in the different biological process. Moreover, among the 49 differentially expressed peptides, 21 peptides were identified from the fibrinogen chain family, which plays a role in neurological diseases, suggesting that these peptides may play an important role in maintaining brain health. In conclusion, our results showed a comparative peptidomic profile from human umbilical cord blood of HIE patients and normal controls. These dysregulated peptides may have potentially important functions in umbilical cord blood with HIE and may be involved in the pathogenesis of the HIE.
Neonatal hypoxic–ischemic encephalopathy (HIE) is the major cause of morbidity and mortality in neonates, mainly because less oxygen and glucose are delivered to the brain (1, 2). Asphyxia is the most common pathway leading to HIE, and it can occur before, during, or after delivery due to complicating factors (3, 4). In high-income countries, the incidence rate of HIE in neonates is 0.8 to 2 per thousand. The prognosis of HIE is closely related to the clinical grade of encephalopathy (5). Approximately 50% of moderate HIE is associated with long-term nerve injury, including death, epilepsy, neurodevelopmental disability, and intellectual disability (6).
The cellular pathophysiology of hypoxic–ischemic brain injury is complicated and mainly initiates the apoptotic pathway caused by secondary energy failure (3, 7). Hypothermia therapy is considered to be the most effective method for the treatment of moderate and severe HIE by decreasing the brain's metabolic rate and reducing neurological sequelae without adverse effects (8–12). The majority of the studies have illustrated that therapeutic hypothermia must start within 6 h after birth to improve the neurological function of the survivors (13, 14). HIE is a rapidly developing encephalopathy, and the neurologic examination of infants will change rapidly over time. Routine clinical investigation and instrument examination have great risk for the early and accurate diagnosis of HIE (15). Therefore, early and accurate diagnosis of newborns with moderate and severe HIE is an urgent problem to be solved.
Peptidomics is the quantitative analysis of large numbers of low-molecular-weight polypeptides that are intermediate products of protein hydrolysis in living cells (16). Accumulated studies indicate that peptides are involved in various biological processes and are considered to be used in either diagnosis or further treatment (17–19). However, the peptide spectrum in cord blood serum of neonates with HIE has not been elucidated. In this study, we identified a total of 49 peptides that are differentially expressed in the serum of umbilical cord blood of neonates with HIE compared with healthy controls. These differentially expressed peptides can be used as rapid screening biomarkers for HIE and may be involved in the pathogenesis of HIE.
Serum samples were collected from the umbilical cord blood of the neonates at the Jingjiang People's Hospital affiliated to Yangzhou University from October 2019 to February 2020. The serum samples were divided into two groups according to clinical diagnosis: the HIE group (n = 3) and normal controls (n = 3). The clinical features of HIE and control samples are listed in Table 1. The diagnosis of HIE was confirmed by regular examination: an Apgar score of 2–6 for 1 min and <8 after first 5 min after birth was indicated as HIE. This study was approved by the Medical Ethics Committee of Jingjiang People's Hospital in China [approval no. (2020)38].
To a 300-μl serum sample, 100% TCA (trichloroacetic acid) was added to the final concentration of 20%, and then an equal volume of CHL3 (trichloromethane) was added. The sample was then mixed thoroughly and placed on ice for 1 h. The sample was then centrifuged for 10 min at 150 × g at 4°C, and to the supernatant 10 μl of water and 10 μl of methanol were added, after which the sample was mixed thoroughly and centrifuged for an additional 10 min at 150 × g at 4°C. To the supernatant, 1 M TEAB was added to four times the volume of the supernatant, adjusting the pH to 2–3.
Twenty milligrams of C18 column material was activated with 1 ml of methanol, vortexed, and centrifuged, and the supernatant was discarded. One milliliter of 0.1% FA (formic acid) was added for acidification, followed by vortexing and centrifugation, and the supernatant was discarded. The sample was eluted with 1 ml of 0.1% FA plus 3% can (acetonitrile) three times, and the supernatant was discarded. Then, 800 μl of ACN and 200 μl of 0.1% FA were added and mixed for 30 min, and the supernatant was taken. Peptides were quantitated with the BCA method and were then freeze-dried.
Equal amounts of peptide were dissolved in 0.5 M TEAB and labeled according to the instructions of the iTRAQ-8 kit (SCIEX, Framingham, MA, USA). The labeled peptides were mixed, and the peptide samples were separated using an Ultimate 30 HPLC system (Dionex, Thermo Scientific, Waltham, MA, USA). The column used was a C18 (Durashell, 5 μm, 100 Å, 4.6 × 250 mm). The peptides were separated by increasing the ACN concentration under alkaline conditions. The flow rate was 1 ml/min, and fractions were collected every minute. A total of 42 secondary fractions were collected and combined into 12 components. The combined components were desalted on a Strata-X column and vacuum-dried.
A TripleTOF 560 and an MS system (SCIEX, USA) were used for MS data acquisition. The peptide samples were dissolved in 2% AC/0.1% FA, added to a C18 capture column (5 μm, 10 μm × 20 mm), and eluted on a C18 analytical column (3 μm, 75 μm × 150 mm) at 30 nl/min for 90 min. For information-dependent acquisition (IDA), the mass spectrometer (MS) was scanned with 250 ms of ion accumulation time, and the MS2 spectra of 30 precursor ions were collected with 50 ms of ion accumulation time. The MS1 spectra were collected in the range of 350–1,500 m/z, and the MS2 spectra were collected in the range of 100–1,500 m/z. The dynamic removal time of precursor ions was 15 s.
ProteinPilot (V4.5) software was used for protein identification (unused score ≥ 1.3, conf ≥ 95%) and quantification of iTRAQ-labeled proteomics. Peptides with a fold change ≥ 1.2 or ≤ 0.83 with a Student's test p ≤ 0.05 were selected as differentially expressed peptides.
The ProParm tool (https://web.expasy.org/protparam/) was used to calculate the isoelectric point (pI) and Mw of each peptide. The GO and KEGG pathways were predicted using websites http://geneontology.org/ and http://www.genome.jp/kegg/ or http://www.kegg.jp/. The interaction network function of the identified peptide precursor proteins was analyzed by STRING (https://string-db.org, Version 11.0). PeptideRanker (http://distilldeep.ucd.ie/PeptideRanker/) was used to predict bioactive peptides (20).
Data were analyzed using the SPSS 20.0 software package with an independent-sample t-test for comparisons between two groups. A p < 0.05 was indicated as a statistically significant difference.
A total of 198 peptides derived from 62 precursor proteins were identified and quantified by performing iTRAQ in the serum of the human umbilical cord samples of the HIE newborns and normal-born babies. By analyzing the general properties of the identified serum peptides, we found that the lengths of most peptides ranged from 8 to 18 amino acids (Figure 1A). In addition, the relationship between the molecular weight (Mw) and the isoelectric point (PI) was also investigated, and the most distributed PI 3.0–PI 5.0 peptides had Mws between 1.0 and 2.0 kDa, whereas the most distributed PI 7.0–PI 9.0 peptides had Mws between 0.5 and 1.5 kDa (Figure 1B). Furthermore, the cleaved amino acids from the precursor proteins for the preceding peptides had a certain pattern (Figure 1C). Alanine (A) and glycine (G) were the high-frequency amino acid break sites of the protein precursor groups.
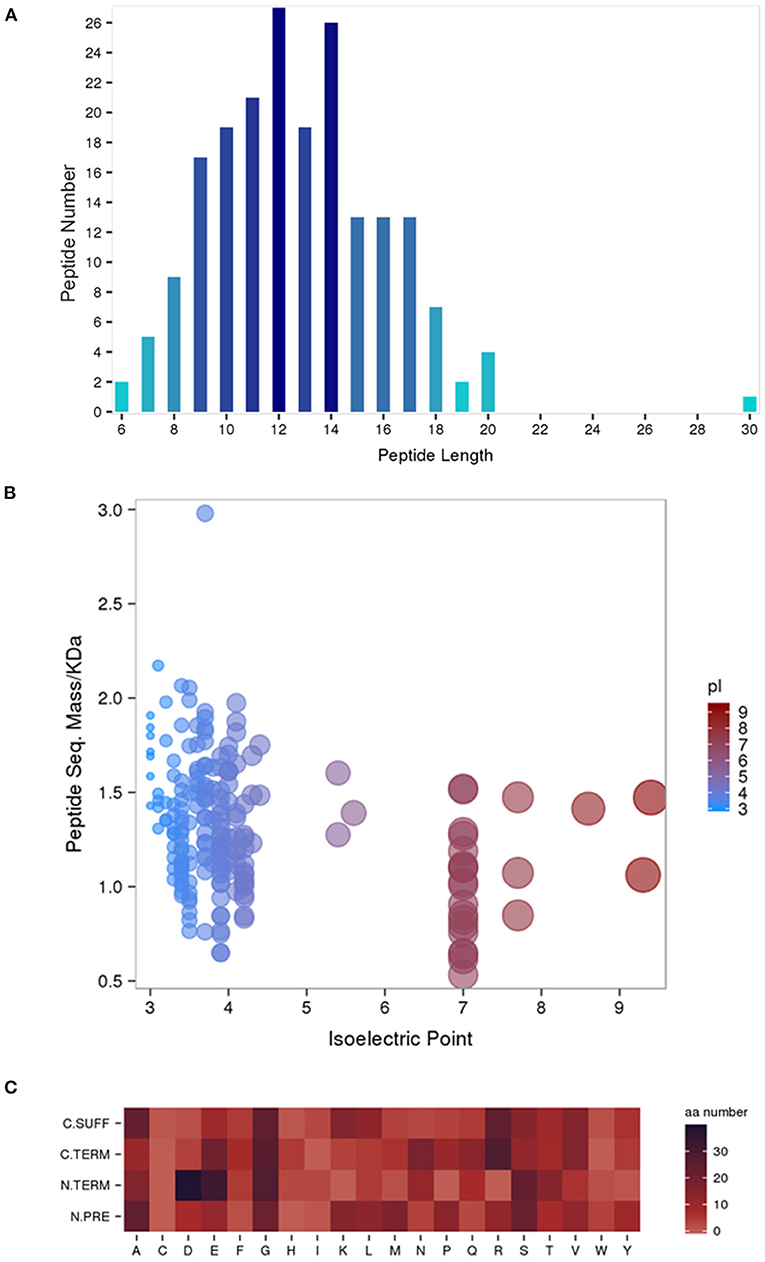
Figure 1. General properties of the identified serum peptides by iTRAQ. Peptide lengths (A). The molecular weight (Mw) vs. isoelectric point (PI) distribution of the identified total peptide segments (B). Heat map of the amino acid distribution of the cleavage sites of the precursor proteins (C). N. PRE represents the former amino acid at the N-terminus of the peptide, N. TERM represents the amino acid at the N-terminus of the peptide, (C) TERM represents the amino acid at the C-terminus of the peptide, and C. SUFF represents the latter amino acid at the C-terminus of the peptide.
A subset of 49 peptides were found to be significantly differentially expressed (p < 0.05) in the HIE group compared with the normal group, representing 41.4% of the total identified peptidomes. These 49 peptides included 33 upregulated peptides and 16 downregulated peptides (FDR > 1.2 or FDR <0.8), which were derived from 25 precursor proteins. Hierarchical clustering (Figure 2A) and volcano mapping (Figure 2B) showed these differentially expressed peptides between the two groups of newborn HIE and normal-born babies. The sequences of some differentially expressed peptides in the HIE group compared with the control group are shown in Table 2.
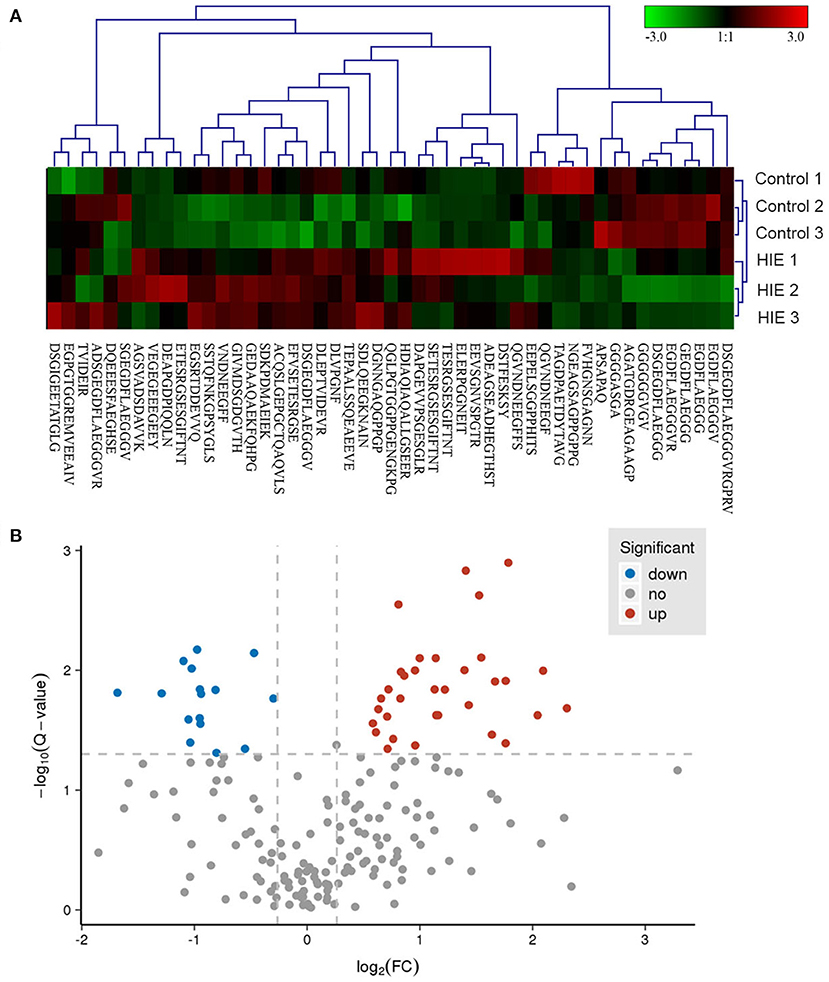
Figure 2. Hierarchical clustering and volcano map of differentially expressed peptides. Hierarchical clustering of differentially expressed peptides (A). Volcano map of differentially expressed peptides (B).
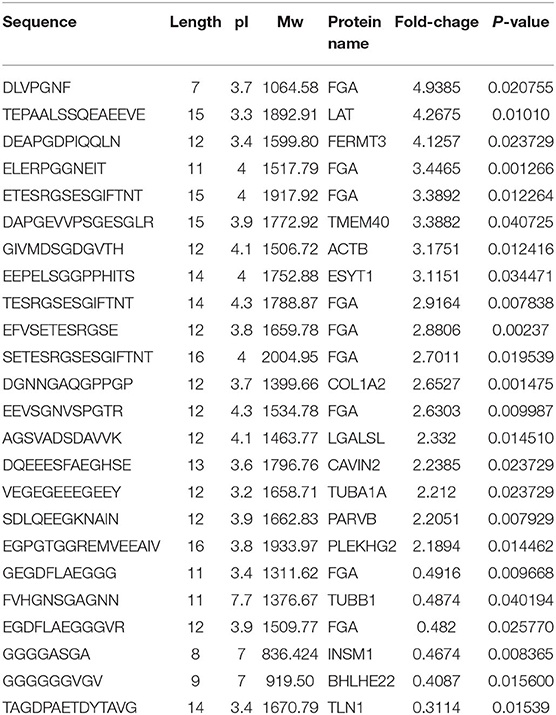
Table 2. Part peptides of differentially expressed in HIE serum compared with control (Foldchange >2 or < 0.5).
We first analyzed the general characteristics of the differentially expressed peptides between HIE umbilical cord serum and relative control umbilical cord serum. The distribution of Mw, pI, and Mw vs. pI of the differentially expressed peptides was analyzed (Figures 3A–C). We found that the distribution of Mw, pI, and Mw vs. pI was in a large-scale range. As shown in Figure 3, the Mws of most differentially expressed peptides were ranged from 1,200 to 1,800 D, whereas the pIs were mostly distributed in the ranges of 3–5 and 7–8.
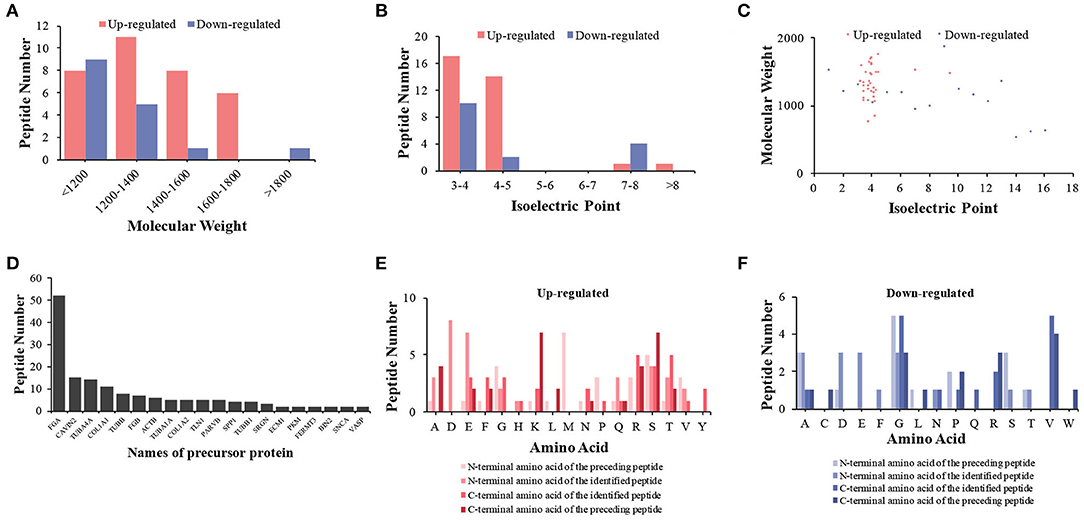
Figure 3. Features and cleavage patterns of differentially expressed peptides. Molecular weights of upregulated and downregulated peptides (A). Isoelectric points of upregulated and downregulated peptides (B). Scatter plot of isoelectric point vs. molecular weight in upregulated and downregulated peptides (C). Peptide numbers for each protein precursor (D). Distribution of the cleavage sites in upregulated peptides (E). Distribution of the cleavage sites in downregulated peptides (F).
Peptides are generated by the protein degradation or restricted proteolysis from their precursor protein (21). According to the inconsistency of peptide breakage sites, these precursor proteins can produce multiple peptides. We found 20 precursor proteins containing more than one peptide in our samples (Figure 3D). We further investigated the cleavage patterns of all differentially expressed peptides to determine the possible functional changes of those peptides. The results showed that among the upregulated peptides, the commonly used cleavage sites for the preceding peptide of the N-terminal amino acid of the preceding peptide, the N-terminal amino acid of the identified peptide, the C-terminal amino acid of the preceding peptide, and the C-terminal amino acid of the identified peptide were methionine (M), lysine (K) or serine (S), aspartic acid (D), and arginine (R) or threonine (T), respectively (Figure 3E). Among the downregulated peptides, the most frequent cleavage sites were glycine (G), alanine (A)/aspartic acid (D)/glutamic acid (E) or glycine (G), glycine (G) or valine (V), and valine (V) (Figure 3F). The bioactivity of peptides could be predicted with the cleavage sites and sequences of the peptides.
To determine the potential function of the differentially expressed peptides and corresponding precursor proteins, GO and pathway analyses were performed. The cellular components, molecular functions, biological processes, and putatively involved pathways of these peptides and precursor proteins were investigated by enrichment analysis. Regarding cellular components, organelle part, membrane part, intracellular organelle, cell part, and cell were the dominant enriched subcategories (Figure 4A). For molecular function, structural molecule activity and enzyme binding were the most highly enriched subcategories (Figure 4B). For biological processes, wound healing, response to wounding, response to stress, regulation of biological quality, and biological regulation were the most highly enriched subcategories (Figure 4C). Pathway analysis indicated that the main precursor proteins were participated in the regulation of the actin cytoskeleton, phagosome, gap junction, and focal adhesion (Figure 4D).
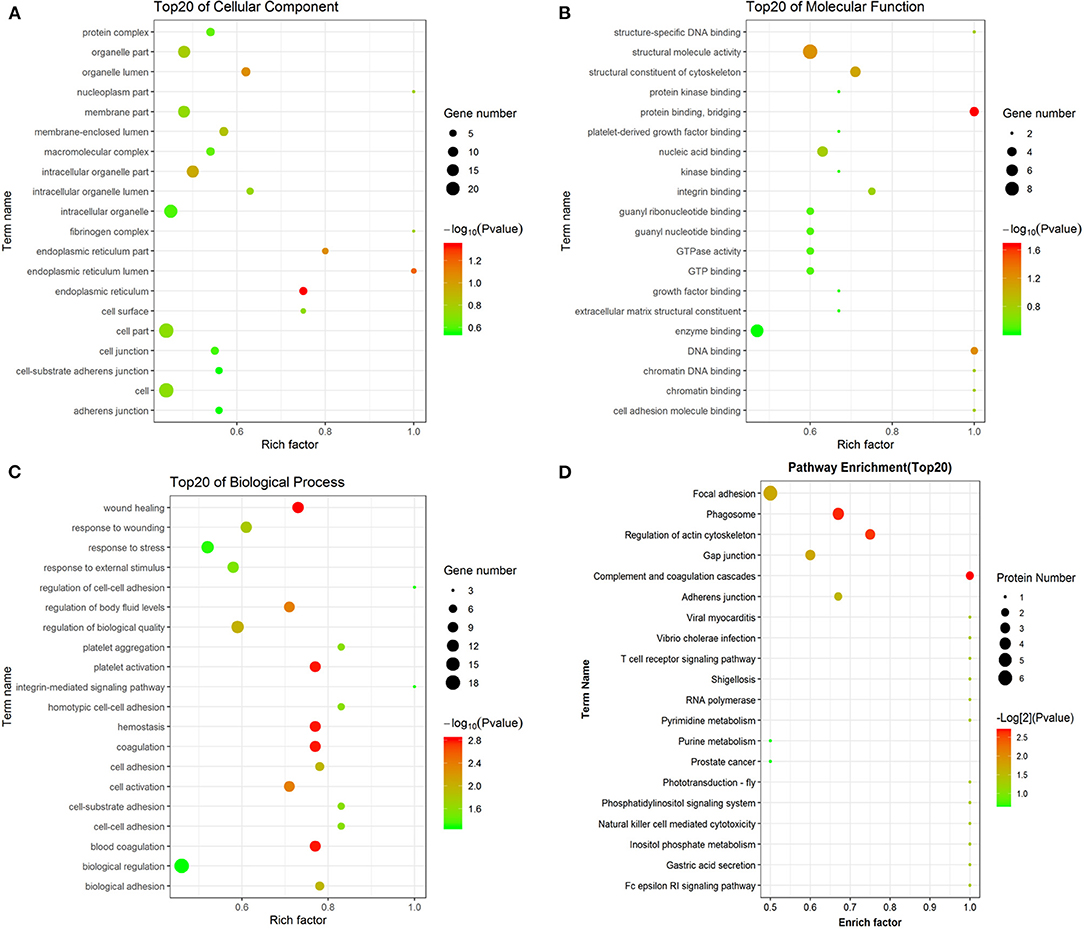
Figure 4. GO and pathway analyses of precursor proteins of differentially expressed peptides. The cellular component categories (A). The molecular function categories (B). The biological process categories (C). Pathway analysis (D).
Interaction network analysis by STRING was performed to uncover potential interactions between differentially expressed peptides and relative precursor proteins. The results suggested that 14 were related to each other (Figure 5). Furthermore, we predicted the bioactivity of the differentially excreted peptides by PeptideRanker and found that there were 15 peptides with probabilities of bioactivity higher than 0.50 that came from five precursor proteins (Table 3). Among these five precursor proteins, fibrinogen alpha chain (FGA), collagen type 1 alpha 2 chain (COL1A2), collagen type III alpha 1 chain (COL3A1), and talin1 (TLN1) interact with each other (Figure 5).
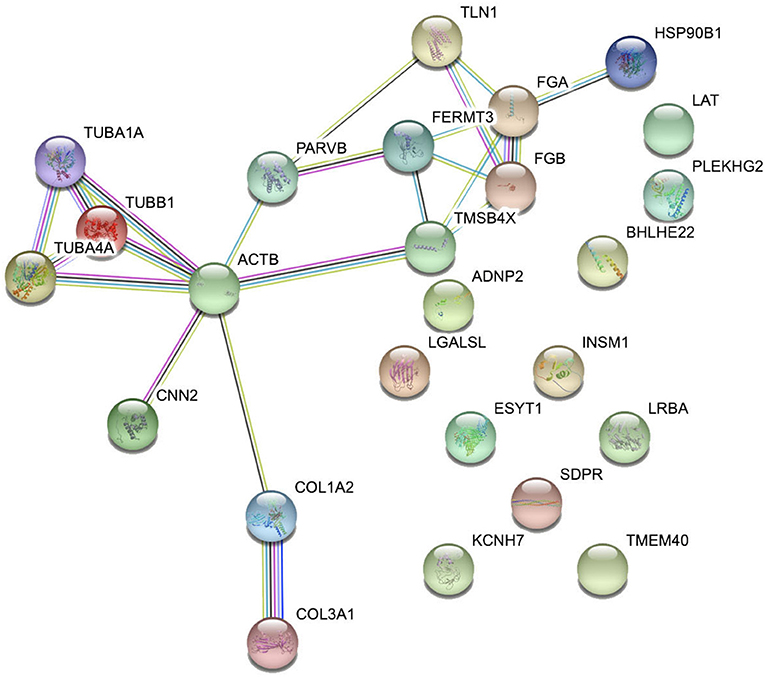
Figure 5. Interaction network analysis of precursor proteins of differentially expressed peptides according to STRING.
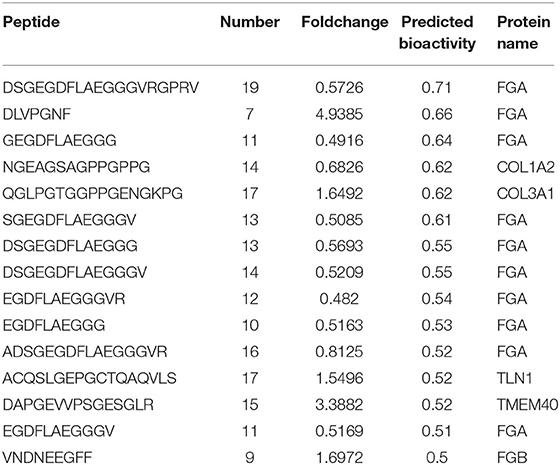
Table 3. Predicted the bioactivity of the part differentially excreted peptides by peptideranker (bioactivity > 0.50).
In the current study, we screened and identified serum peptide expression in neonatal umbilical cord blood and compared the different peptide profiles in the serum of umbilical cord blood of neonates with HIE and normal controls. This study was the first to identify potential endogenous functional peptides that may be involved in the progression of HIE and served as novel diagnostic biomarkers of HIE.
To investigate the pathogenesis of HIE, there are several reports on studying RNA (including miRNA, lncRNA, and circRNA) and protein profiling using blood samples from HIE patients (22–24). Zhu et al. (25) reported differential proteomic profiles in neonate blood with mild, moderate, severe, or without HIE and identified a total of 51 commonly differentially expressed proteins between HIE and healthy controls (25). Peptides are naturally generated in the body through protein degradation or restricted proteolysis (21). More than 13,000 natural peptides have been identified, and these usually play important roles in human physiology (26). Approximately 60 FDA-approved therapeutic peptides are on the market, and over 140 peptide-based drugs are currently in clinical development (27). It has been recognized that peptides play a vital role in the maintenance of central nervous system function. These include tridecapeptide neurotensin, cholecystokinin, neuropeptide Y, and amylin (28). In addition to their therapeutic effect, peptides can also be used as biomarkers for disease diagnosis (29). Here, we demonstrated that 49 peptides were differentially expressed in the serum of umbilical cord blood of the HIE group compared with the control group by performed LC-MS/MS. Thirty-three upregulated peptides and 16 downregulated peptides which were derived from 25 precursor proteins. These differential peptides suggest that they may be new biomarkers for the early diagnosis of HIE.
Most notably, 21 differentially expressed peptides were derived from the fibrinogen chain family in a total of 49 identified differentially expressed peptides. Among these peptides, 18 peptides were derived from the fibrinogen alpha chain (FGA), and three peptides were generated from the fibrinogen beta chain (FGB). Fibrinogen is an important blood coagulation protein and participates in numerous brain pathologies, subarachnoid hemorrhage, multiple sclerosis, and Alzheimer's disease (30–33). Fibrinogen inhibits oligodendrocyte progenitor cell (OPC) differentiation and myelination by activating the bone morphogenetic protein (BMP) signaling pathway (34). In addition, fibrinogen promotes cognitive deficits by CD11b/CD18-mediated microglial activation of Alzheimer's disease (AD) pathogenesis (35). Furthermore, fibrinogen is a potential biomarker for multiple sclerosis and Alzheimer's disease (36, 37). Neurons and astrocytes have been reported to have the ability to express FGA and FGB, which indicates that the FGA- and FGB-derived peptides may be involved in the HIE pathological processes and may have implications for the early diagnosis of HIE.
In conclusion, peptidomic analysis was performed by LC-MS and used to detect differentially expressed peptides between neonates with HIE and controls. The present study is the first to examine the potential relationship between peptides and HIE in newborns. In addition, GO and KEGG pathway analyses were used to identify the potential functions of the precursor proteins. This study provides a new view for further understanding the progress of HIE and may provide a new biomarker for the detection of HIE. However, the relevance of those peptides in umbilical cord blood to HIE need to be further investigated.
The original contributions presented in the study are included in the article/supplementary material, further inquiries can be directed to the corresponding author/s.
The studies involving human participants were reviewed and approved by the Medical Ethics Committee of Jingjiang People's Hospital in China [approval no. (2020)38]. Written informed consent to participate in this study was provided by the participants' legal guardian/next of kin.
XD, LY, and YZha designed the project and edited the manuscript. YS and QS wrote the manuscript. JZ collected the clinical data and revised the manuscript. XW and YZhu participated in the design of this study. All authors contributed to the article and approved the submitted version.
This work was supported by the following funds: the National Natural Science Foundation of China (81801503), the Natural Science Foundation of the Jiangsu Province of China (BK20180286), the Sixth People's Hospital of Shanghai Medical Group Projects of China (ly202002), the Changning District Committee of Science and Technology of China (CNKW2020Y07), and the Taizhou Science and Technology Support Plan (Social Development) project (SSF20200345).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
1. Verklan MT. The chilling details: hypoxic-ischemic encephalopathy. J Perinat Neonatal Nurs. (2009) 23:59–68; quiz 69–70. doi: 10.1097/01.JPN.0000346221.48202.7e
2. Qin X, Cheng J, Zhong Y, Mahgoub OK, Akter F, Fan Y, et al. Mechanism and Treatment Related to Oxidative Stress in Neonatal Hypoxic-Ischemic Encephalopathy. Front Mol Neurosci. (2019) 12:88. doi: 10.3389/fnmol.2019.00088
3. Valkounova I, Maresova D, Trojan S. Perinatal complication: hypoxic-ischemic encephalopathy. Sb Lek. (2001) 102:455–63.
4. Rainaldi MA, Perlman JM. Pathophysiology of Birth Asphyxia. Clin Perinatol. (2016) 43:409–22. doi: 10.1016/j.clp.2016.04.002
5. Sarnat HB, Sarnat MS. Neonatal encephalopathy following fetal distress. A clinical and electroencephalographic study. Arch Neurol. (1976) 33:696–705. doi: 10.1001/archneur.1976.00500100030012
6. Lee AC, Kozuki N, Blencowe H, Vos T, Bahalim A, Darmstadt GL, et al. Intrapartum-related neonatal encephalopathy incidence and impairment at regional and global levels for 2010 with trends from 1990. Pediatr Res. (2013) 74(Suppl. 1):50–72 doi: 10.1038/pr.2013.206
7. Calvert JW, Zhang JH. Pathophysiology of an hypoxic-ischemic insult during the perinatal period. Neurol Res. (2005) 27:246–60. doi: 10.1179/016164105X25216
8. Jacobs SE, Berg M, Hunt R, Tarnow-Mordi WO, Inder TE, Davis PG. Cooling for newborns with hypoxic ischaemic encephalopathy. Cochrane Database Syst Rev. (2013) 2013:CD003311. doi: 10.1002/14651858.CD003311.pub3
9. Edwards AD, Yue X, Squier MV, Thoresen M, Cady EB, Penrice J, et al. Specific inhibition of apoptosis after cerebral hypoxia-ischaemia by moderate post-insult hypothermia. Biochem Biophys Res Commun. (1995) 217:1193–9. doi: 10.1006/bbrc.1995.2895
10. Azzopardi DV, Strohm B, Edwards AD, Dyet L, Halliday HL, Juszczak E, et al. Moderate hypothermia to treat perinatal asphyxial encephalopathy. N Engl J Med. (2009) 361:1349–58. doi: 10.1056/NEJMoa0900854
11. Azzopardi D, Strohm B, Marlow N, Brocklehurst P, Deierl A, Eddama O, et al. Effects of hypothermia for perinatal asphyxia on childhood outcomes. N Engl J Med. (2014) 371:140–9. doi: 10.1056/NEJMoa1315788
12. Gunn AJ, Laptook AR, Robertson NJ, Barks JD, Thoresen M, Wassink G, et al. Therapeutic hypothermia translates from ancient history in to practice. Pediatr Res. (2017) 81:202–9. doi: 10.1038/pr.2016.198
13. Silveira RC, Procianoy RS. Hypothermia therapy for newborns with hypoxic ischemic encephalopathy. J Pediatr (Rio J). (2015) 91:S78–83. doi: 10.1016/j.jped.2015.07.004
14. Jia W, Lei X, Dong W, Li Q. Benefits of starting hypothermia treatment within 6 h vs. 6-12 h in newborns with moderate neonatal hypoxic-ischemic encephalopathy. BMC Pediatr. (2018) 18:50 doi: 10.1186/s12887-018-1013-2
15. Martinello K, Hart AR, Yap S, Mitra S, Robertson NJ. Management and investigation of neonatal encephalopathy: 2017 update. Arch Dis Child Fetal Neonatal Ed. (2017) 102:F346–58. doi: 10.1136/archdischild-2015-309639
16. Apostolopoulos V, Bojarska J, Chai TT, Elnagdy S, Kaczmarek K, Matsoukas J, et al. A global review on short peptides: frontiers and perspectives. Molecules. (2021) 26:430. doi: 10.3390/molecules26020430
17. Hu Y, Wang J, Zhou Y, Xie H, Yan X, Chu X, et al. Peptidomics analysis of umbilical cord blood reveals potential preclinical biomarkers for neonatal respiratory distress syndrome. Life Sci. (2019) 236:116737. doi: 10.1016/j.lfs.2019.116737
18. Baig MH, Ahmad K, Rabbani G, Choi I. Use of Peptides for the Management of Alzheimer's Disease: Diagnosis and Inhibition. Front Aging Neurosci. (2018) 10:21. doi: 10.3389/fnagi.2018.00021
19. Mahendru S, Roy K, Kukreti S. Peptide Biomarkers: Exploring the Diagnostic Aspect. Curr Protein Pept Sci. (2017) 18:914–9. doi: 10.2174/1389203717666160724203746
20. Mooney C, Haslam NJ, Pollastri G, Shields DC. Towards the improved discovery and design of functional peptides: common features of diverse classes permit generalized prediction of bioactivity. PLoS ONE. (2012) 7:e45012. doi: 10.1371/journal.pone.0045012
21. Shahinian H, Tholen S, Schilling O. Proteomic identification of protease cleavage sites: cell-biological and biomedical applications. Expert Rev Proteomics. (2013) 10:421–33. doi: 10.1586/14789450.2013.841547
22. Looney AM, Walsh BH, Moloney G, Grenham S, Fagan A, O'Keeffe GW, et al. Downregulation of umbilical cord blood levels of miR-374a in neonatal hypoxic ischemic encephalopathy. J Pediatr. (2015) 167:269–73 e262. doi: 10.1016/j.jpeds.2015.04.060
23. Dong X, Zhao Y, Huang Y, Yu L, Yang X, Gao F. Analysis of long noncoding RNA expression profiles in the whole blood of neonates with hypoxic-ischemic encephalopathy. J Cell Biochem. (2018) 120:8499–509. doi: 10.1002/jcb.28138
24. Dong X, Zhuang S, Huang Y, Yang X, Fu Y, Yu L, et al. Expression profile of circular RNAs in the peripheral blood of neonates with hypoxicischemic encephalopathy. Mol Med Rep. (2020) 22:87–96. doi: 10.3892/mmr.2020.11091
25. Zhu Y, Yun Y, Jin M, Li G, Li H, Miao P, et al. Identification of novel biomarkers for neonatal hypoxic-ischemic encephalopathy using iTRAQ. Ital J Pediatr. (2020) 46:67. doi: 10.1186/s13052-020-00822-7
26. Yuan Y. Mechanisms inspired targeting peptides. Adv Exp Med Biol. (2020) 1248:531–46. doi: 10.1007/978-981-15-3266-5_21
27. Fosgerau K, Hoffmann T. Peptide therapeutics: current status and future directions. Drug Discov Today. (2015) 20:122–8. doi: 10.1016/j.drudis.2014.10.003
28. McGonigle P. Peptide therapeutics for CNS indications. Biochem Pharmacol. (2012) 83:559–66. doi: 10.1016/j.bcp.2011.10.014
29. Akashi YJ, Springer J, Lainscak M, Anker SD. Atrial natriuretic peptide and related peptides. Clin Chem Lab Med. (2007) 45:1259–67. doi: 10.1515/CCLM.2007.274
30. Shi X, Ohta Y, Liu X, Shang J, Morihara R, Nakano Y, et al. Chronic cerebral hypoperfusion activates the coagulation and complement cascades in Alzheimer's disease mice. Neuroscience. (2019) 416:126–36. doi: 10.1016/j.neuroscience.2019.07.050
31. Bijak M, Olejnik A, Rokita B, Morel A, Dziedzic A, Miller E, et al. Increased level of fibrinogen chains in the proteome of blood platelets in secondary progressive multiple sclerosis patients. J Cell Mol Med. (2019) 23:3476–82. doi: 10.1111/jcmm.14244
32. Golanov EV, Sharpe MA, Regnier-Golanov AS, Del Zoppo GJ, Baskin DS, Britz GW. Fibrinogen Chains Intrinsic to the Brain. Front Neurosci. (2019) 13:541. doi: 10.3389/fnins.2019.00541
33. Petersen MA, Ryu JK, Akassoglou K. Fibrinogen in neurological diseases: mechanisms, imaging and therapeutics. Nat Rev Neurosci. (2018) 19:283–301. doi: 10.1038/nrn.2018.13
34. Petersen MA, Ryu JK, Chang KJ, Etxeberria A, Bardehle S, Mendiola AS, et al. Fibrinogen activates BMP signaling in oligodendrocyte progenitor cells and inhibits remyelination after vascular damage. Neuron. (2017) 96:1003–12 e1007. doi: 10.1016/j.neuron.2017.10.008
35. Merlini M, Rafalski VA, Rios Coronado PE, Gill TM, Ellisman M, Muthukumar G, et al. Fibrinogen induces microglia-mediated spine elimination and cognitive impairment in an Alzheimer's disease model. Neuron. (2019) 101:1099–108 e1096. doi: 10.1016/j.neuron.2019.01.014
36. Ahmad U, Frederiksen JL. Fibrinogen: a potential biomarker for predicting disease severity in multiple sclerosis. Mult Scler Relat Disord. (2020) 46:102509. doi: 10.1016/j.msard.2020.102509
Keywords: hypoxic-ischemic encephalopathy, neonates, peptides, peptidomic, KEGG
Citation: Dong X, Zhao J, Shen Y, Sun Q, Wu X, Zhu Y, Yu L and Zhao Y (2021) Peptidomic Analysis of Neonate Umbilical Cord Blood for the Identification of Endogenous Peptides Involved in Hypoxic–Ischemic Encephalopathy. Front. Pediatr. 9:718704. doi: 10.3389/fped.2021.718704
Received: 01 June 2021; Accepted: 23 July 2021;
Published: 25 August 2021.
Edited by:
Serafina Perrone, University of Parma, ItalyReviewed by:
Diego Iacono, Biomedical Research Institute of New Jersey, United StatesCopyright © 2021 Dong, Zhao, Shen, Sun, Wu, Zhu, Yu and Zhao. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xiaohua Dong, bmpmaXNoeGlhb2h1YUAxNjMuY29t; Lingling Yu, eGlhb2ZpYWh5dUAxNjMuY29t; Yingmin Zhao, eXlmNzEwNTI1QDE2My5jb20=
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.