
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
MINI REVIEW article
Front. Pediatr., 14 April 2021
Sec. Pediatric Nephrology
Volume 9 - 2021 | https://doi.org/10.3389/fped.2021.632248
This article is part of the Research TopicAcute Kidney Injury: It’s Not Just Acute, and It’s Not Just the KidneysView all 10 articles
Sepsis is a leading cause of morbidity and mortality in critically ill children, and acute kidney injury (AKI) is a frequent complication that confers an increased risk for poor outcomes. Despite the documented consequences of sepsis-associated AKI (SA-AKI), no effective disease-modifying therapies have been identified to date. As such, the only treatment options for these patients remain prevention and supportive care, both of which rely on the ability to promptly and accurately identify at risk and affected individuals. To achieve these goals, a variety of biomarkers have been investigated to help augment our currently limited predictive and diagnostic strategies for SA-AKI, however, these have had variable success in pediatric sepsis. In this mini-review, we will briefly outline the current use of biomarkers for SA-AKI, and propose a new framework for biomarker discovery and utilization that considers the individual patient's sepsis inflammatory response. Now recognized to be a key driver in the complex pathophysiology of SA-AKI, understanding the dysregulated host immune response to sepsis is a growing area of research that can and should be leveraged to improve the prediction and diagnosis of SA-AKI, while also potentially identifying novel therapeutic targets. Reframing SA-AKI in this manner – as a direct consequence of the individual patient's sepsis inflammatory response – will facilitate a precision medicine approach to its management, something that is required to move the care of this consequential disorder forward.
Sepsis is common in the pediatric intensive care unit (PICU), accounting for 75,000 hospitalizations annually in the United States (1). Children with sepsis suffer substantial morbidity and mortality, and those risks are further increased by the co-incidence of acute kidney injury (AKI), a frequent complication of pediatric sepsis (2, 3). Impacting almost half of critically ill children who meet criteria for severe sepsis (4), sepsis-associated AKI (SA-AKI) has been associated with poor outcomes including prolonged lengths of stay, higher mortality, and increased healthcare costs (3, 5, 6). Unfortunately, despite the burden it imposes on health outcomes, there are currently no effective disease-modifying therapies for SA-AKI once present (7, 8). As a result, therapeutic approaches are centered on prevention and supportive care, including employment of renal protective measures and timely initiation of renal replacement therapy, if indicated (9, 10).
A primary limitation to the successful development of novel therapies to treat SA-AKI is an incomplete understanding of the pathophysiology. Historical perspectives considered SA-AKI to be the result of renal hypoperfusion during the shock state, leading to acute tubular necrosis (11). However, the modern pathophysiologic model of SA-AKI recognizes a much more complex and heterogeneous disease – a combination of a dysregulated host inflammatory response to infection, altered microcirculatory blood flow, and metabolic derangements that ultimately lead to cell cycle arrest and apoptosis of renal tubular epithelial cells (12, 13). Difficulties in treatment development encountered due to this complexity are further exacerbated by the limited diagnostic strategies for SA-AKI, as it is now well-established that serum creatinine and urine output – the current gold standards for diagnosis – are fraught with issues, particularly among patients with sepsis (14). Indeed, the limitations of these tools are highlighted by the recent 23rd Acute Dialysis Quality Initiative recommendations, which suggest incorporating tubular injury biomarker status into the definition of AKI, when available (15). Taken together, the lack of precision diagnostics for SA-AKI, coupled with a limited understanding of the individual heterogeneous pathophysiology, have prevented advances in therapy beyond our current standards of prevention and supportive care (16).
With these issues in mind, it is not surprising that there is a growing interest in identifying biomarkers for SA-AKI. In addition to the clear need for validated injury biomarkers to improve diagnostic precision once present, biomarkers that allow for early identification of patients at risk for severe, persistent SA-AKI and those that reflect the patient-specific underlying pathophysiology are needed, as they might allow for prompt implementation of renal protective strategies, identification of biologically important targets for development of novel therapies, and provide a mechanism for enrichment of future clinical trials (17). Given the complexity of SA-AKI outlined above, it is unlikely that one biomarker measured at one moment in time will be able to achieve these goals, and this reality should inform our approach to identifying and employing biomarkers for SA-AKI.
This mini-review proposes a new framework for the discovery and utilization of biomarkers for SA-AKI. The foundational premise is that the pathophysiology of SA-AKI is directly tied to an individual's unique sepsis-related inflammatory response, and thus the diagnostic and treatment approach to SA-AKI may be different from other forms of AKI. Within this framework, we will briefly describe the current state of biomarkers for SA-AKI and discuss their limitations. We will then evaluate how biomarkers have been employed for the identification of individual sepsis molecular signatures, and how these may be leveraged in SA-AKI. Ultimately, if biomarkers can be biologically linked to the dysregulated inflammatory response to sepsis, then a precision medicine approach to the diagnosis and treatment of SA-AKI can be utilized to improve patient outcomes.
To date, a variety of biomarkers for SA-AKI have been studied. The biological underpinnings of these biomarkers vary, and include direct markers of tubular injury, regulatory proteins responsible for promoting cell cycle arrest, and more recently, proteins involved in the inflammatory cascade induced by sepsis (18, 19). Thus, far, SA-AKI research utilizing biomarkers has been limited to improving diagnostic and predictive capacity, most with modest success (18). Importantly, there have been no interventional trials to date utilizing biomarkers to initiate disease-specific therapy in SA-AKI. An overview of the biomarkers that have been most widely investigated in SA-AKI is included in Table 1.While the purpose of this review is not to cover these previously studied biomarkers of SA-AKI in detail, two deserve more in depth discussion.
As the most widely studied biomarker of AKI, NGAL – a protein produced by the injured nephron that can be measured in both urine and serum – has also been studied extensively as a biomarker of SA-AKI (37). While NGAL has been shown to successfully identify patients with AKI secondary to a variety of etiologies (38–40), its utility in sepsis is less clear (41–44). This is in large part due to an increase in systemic NGAL production – namely by neutrophils and the liver – as part of the inflammatory response to infection, independent of injury to nephrons (20). The consequence of this lack of kidney-specific production of NGAL has been modest performance when utilized for diagnosis and prediction of SA-AKI, often with high sensitivity but poor specificity (Table 1) (21, 22). Unfortunately, difficulty disentangling the fraction of NGAL elevation that is attributable to AKI, vs. a more generic systemic inflammatory response among patients with sepsis, likely limits its utility as a single biomarker for the diagnosis of SA-AKI, although more study is warranted.
The induction of cell cycle arrest in renal tubular epithelial cells plays an important role in the early pathophysiology of all forms of AKI (11). Consequently, the expression of cell cycle arrest proteins tissue inhibitor of metalloproteinase-2 (TIMP-2) and insulin-like growth factor-binding protein 7 (IGFBP7) have been shown to be increased in renal tubular cells in response to stress or injury (18, 25). The combination of TIMP-2 and IGFBP7 for the prediction of AKI in high risk patients has been examined in several landmark studies (Table 1) (26, 28, 45), and is now approved by the U.S. Food and Drug Administration for critically ill adults with one or more risk factors for AKI, including sepsis (25). In adults with sepsis, this tool (known as NephroCheck®) demonstrated an area under the receiver operating curve (AUROC) of 0.84 for the prediction of SA-AKI, and its predictive performance significantly improved via the addition of a clinical prediction model (AUROC of 0.94) (27). The use of NephroCheck® to assess the impact of directed implementation of standardized renal protection strategies compared to standard of care in patients with septic shock will be assessed in the upcoming Limiting AKI Progression in Sepsis (LAPIS) Trial (NCT04434209) (46). Unfortunately, this tool has not been studied robustly nor been validated in children.
As noted above and in Table 1, there are several limitations to the use of these biomarkers in pediatric SA-AKI. First and foremost, the data for their use in pediatric sepsis is scare, and this is especially problematic given a growing host of literature to suggest fundamental differences in the sepsis inflammatory response – and thus, the risk of SA-AKI – based on age (47–50). Furthermore, the biologic action of many of these biomarkers appear to be non-specific to sepsis (38, 51, 52), thereby providing no information regarding the patient's underlying inflammatory state, which is likely necessary to identify effective, patient-specific therapies for SA-AKI. Taken together, these realities suggest that additional approaches to biomarker discovery and utilization is required.
As outlined above, sepsis is a complex syndrome that stems from a dysregulated host immune response to an infectious trigger, and is a leading cause of death and disability in critically ill children (53). Given these consequences, substantial resources have been focused on improving the care of patients with sepsis, however, these efforts have failed to produce meaningful therapeutic advances beyond the mainstays of supportive care and antibiotics (54). Failures are undoubtedly tied to the heterogeneity of the disease expression on the individual patient level (17). As such, attempts to resolve this heterogeneity by identifying the sepsis molecular signature of a patient are becoming more common, as successful strategies for doing so could allow for more targeted employment of therapies (55–61).
This concept of separating a heterogeneous group of patients into more homogenous subgroups to guide management is termed enrichment, a fundamental tenant of precision medicine (62). Prognostic enrichment refers to selecting a subgroup of patients who share a similar likelihood of suffering an outcome of interest, such as mortality, while predictive enrichment selects a subgroup who are more likely to respond to a particular therapy based on underlying biology (63). This general concept, and how it may be employed to direct a precision medicine approach to SA-AKI therapy, is depicted in Figure 1.
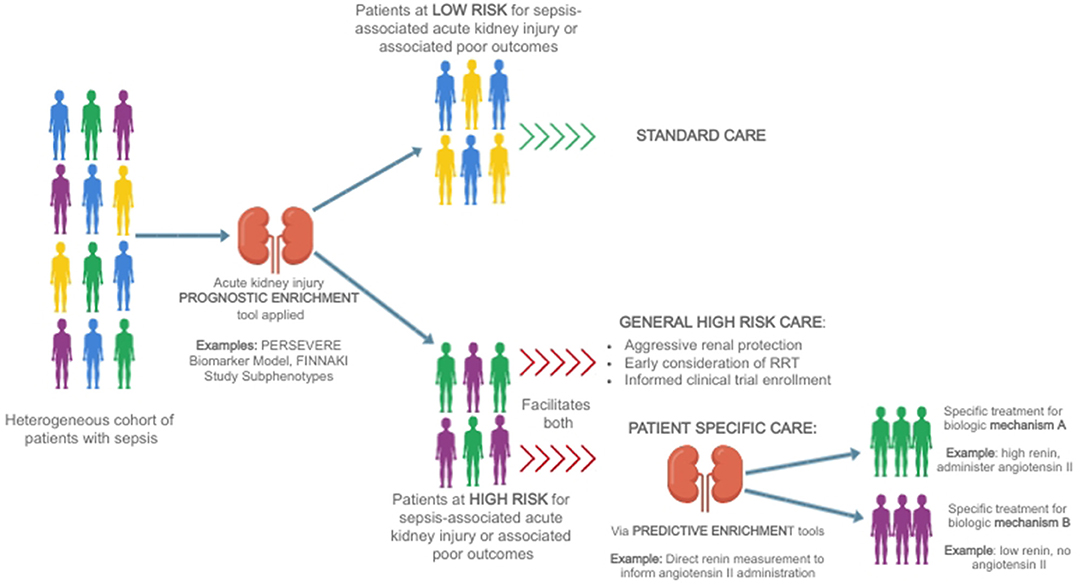
Figure 1. A heterogeneous group of patients with sepsis is divided first using a prognostic enrichment tool into those at high and those at low risk of sepsis-associated acute kidney injury [for example, the PERSEVERE Biomarker Model (64)] or associated poor outcomes such as delayed renal recovery or mortality [for example, FINNAKI Study subphenotypes (65)]. Patients who are at low risk may be treated with standard therapy, while patients at high-risk may receive more aggressive care aimed at renal protection, earlier consideration of renal support in the form of renal replacement therapy, and be considered for informed enrollment in clinical trials aimed at identifying therapies for sepsis-associated acute kidney injury. Ultimately, the goal is to utilize predictive enrichment tools to further subdivide patients on the basis of biology [for example, using direct renin levels to inform the use of angiotensin II (66)], allowing for the implementation of patient-specific therapies.
In pediatric sepsis, prognostic enrichment strategies have been used to develop a set of serum biomarkers – known as the Pediatric Sepsis Biomarker Risk Model (PERSEVERE) – capable of reliably assigning baseline risk of 28-day mortality (67). This model incorporates five serum protein biomarkers measured in the first 24 h of septic shock that were originally identified utilizing discovery-oriented genome-wide profiling of children with septic shock (68, 69), and then narrowed further using classification and regression tree (CART) modeling for estimation of baseline mortality risk (67). An updated version of the model (PERSEVERE-II) incorporates platelet count, and has been recently prospectively validated for the prediction of mortality (70). Similarly, predictive enrichment has also been utilized to subgroup patients based on gene expression, which led to the identification of two distinct endotypes that may require different treatment approaches (58). For example, one particular endotype – endotype A – is associated with increased repression of genes that regulate adaptive immunity and glucocorticoid receptor signaling, and patients with this endotype have demonstrated an increased mortality rate when treated with corticosteroids (71, 72). This association between endotype A and poor outcome in response to corticosteroids was recently corroborated among adults with septic shock (73). Given our current understanding of the significant role that the host inflammatory response plays in the pathophysiology of SA-AKI (12, 13), it is reasonable to consider leveraging this pediatric sepsis enrichment work to improve the care of SA-AKI, a similarly heterogeneous disorder (11, 12).
A precision medicine approach to SA-AKI will require both prognostic enrichment tools to identify high risk patients early and accurately, and predictive enrichment tools to deliver the right treatment to the right patient. Biomarkers play an important role in achieving these goals, however, we believe that a shift to include biomarkers of the dysregulated immune response to infection is prudent. Such a shift will also require a reframing of AKI in sepsis, recognizing that it is not simply “associated” with sepsis (as suggested by the term SA-AKI), but a disease state that is induced by the host inflammatory response. In this section, we will outline the current application of precision medicine to the study of SA-AKI within this framework, and highlight the remaining critical knowledge gaps.
The first step to improving outcomes for patients with SA-AKI is early identification of those at highest risk. While sepsis is perhaps the most significant risk factor for AKI in critically ill patients, a significant proportion of patients with septic shock do not develop AKI. Therefore, further delineation of an individual's risk profile via the development of prognostic enrichment tools is required. To date, few validated prognostic enrichment strategies for SA-AKI that incorporate sepsis-specific biomarkers exist, as outlined below.
Leveraging work done in the more advanced field of sepsis precision medicine, researchers have utilized “omic” technologies (notably genomics, transcriptomics and proteomics) to identify patients at high risk for persistent SA-AKI (10, 64, 74). Using microarray technology to study SA-AKI related transcriptomics, one group retrospectively identified 21 candidate biomarkers for the prediction of SA-AKI based on the upregulation of mRNA gene probes in patients with persistence of severe SA-AKI at day 7 of septic shock (74). The expression pattern of these 21 upregulated genes were shown to predict the presence of this severe, persistent form of SA-AKI with high sensitivity (98%) and reasonable specificity (80%) (74). Results from this work informed a second study in which the protein products of five of the aforementioned 21 genes–elastase 2 (ELA2), fibroblast growth factor 13, matrix metalloproteinase 8 (MMP8), olfactomedin 4 (OFM4), and proteinase 3 (PRTN3) – were incorporated into a new CART-derived model to predict the presence of SA-AKI at day 3 of septic shock (10). The test characteristics of this model in the derivation cohort were robust, with an AUROC of 0.95; when tested in a validation cohort, the predictive capacity of the model remained reasonable with an AUROC of 0.82, which was superior to knowledge of AKI stage by serum creatinine on the day of septic shock development alone (AUROC 0.73) (10). Using a similar approach, a more recent study utilized the PERSEVERE biomarkers and AKI stage by serum creatinine on the day of admission to develop a model for prediction of severe SA-AKI at day 3. This model had similarly impressive test characteristics, with an AUROC of 0.95 (64). Unfortunately, while these models represent potentially feasible prognostic enrichment tools for SA-AKI, they have not yet been prospectively validated nor utilized to inform patient care, which represent areas of future study.
Another strategy incorporating biomarkers that has been utilized for prognostic enrichment in SA-AKI is latent class analysis (LCA). This approach allows for the incorporation of multiple variables– including comorbidities, clinical data and biomarkers – to allow for the identification of potential subphenotypes of heterogeneous disease states. Using this methodology, a recent post-hoc analysis of the FINNAKI Study described two subphenotypes of critically ill patients with SA-AKI who have significantly different rates of mortality and renal recovery (65). Patients categorized as subphenotype 2 – which was associated with increased mortality and decreased short-term renal recovery – demonstrated elevations in biomarkers associated with endothelial dysfunction and an overall increased inflammatory state. Interestingly, four of the significantly upregulated inflammatory biomarkers in subphenotype 2 (ELA2, OFM4, MMP8 and PRTN3) overlapped with the above mentioned AKI prediction model derived by Wong and colleagues (10). Using a similar approach, a second group also identified two SA-AKI subphenotypes (AKI-SP1 and AKI-SP2) via the application of LCA to a panel of 29 clinical and biomarker variables (75). This study similarly showed decreased survival and renal recovery in patients with upregulation of biomarkers associated with endothelial dysfunction and inflammation, although the included biomarkers differed. While these LCA-driven studies identified high-risk subphenotypes of patients already known to have SA-AKI, they represent potentially viable prognostic enrichment tools, specifically to help delineate patients most likely to benefit from enrollment in clinical trials, as well as from potentially high-risk and high-resource utilizing therapies such as renal replacement therapy (RRT).
The identification of predictive enrichment tools for SA-AKI–those that provide insight into the underlying pathophysiology and thereby reveal potential treatment strategies–remains an elusive goal. Predictive enrichment tools are particularly helpful in heterogeneous disease states, as they may identify subphenotypes of patients who might benefit from a specific, biologically-based therapy. While Figure 1 outlines an ideal circumstance in which predictive enrichment occurs in an identified high risk subset of patients, it is important to note that the development and use of predictive enrichment tools does not necessarily rely on the availability of reliable prognostic enrichment strategies. However, the identification of effective predictive enrichment tools requires a deep understanding of the patient-specific pathophysiology, which remains a significant barrier in SA-AKI.
Thus far, the only proposed predictive enrichment strategy that is clinically feasible was elucidated via a series of post-hoc analyses of the Angiotensin II for the Treatment of High-Output Shock (ATHOS-3), a clinical trial of adults with vasodilatory shock treated with angiotensin II (76). In these studies, the authors were able to demonstrate that patients who were treated with angiotensin II had improved 28-day survival and earlier discontinuation of RRT (77), and that these advantages were best seen in patients who had higher serum renin levels prior to angiotensin II administration, suggestive of sepsis-induced angiotensin converting enzyme deficiency (and thus angiotensin II deficiency) in the setting of endothelial injury (66). From these findings, the authors postulated that administration of exogenous angiotensin II to patients with vasodilatory shock may be beneficial beyond simply increasing blood pressure, as it was also demonstrated to normalize high renin levels, which have been known to be proinflammatory (66). Given that serum renin levels can be easily measured, this example of predictive enrichment can and should be applied prospectively in future studies examining the effect of angiotensin II on mitigation of SA-AKI.
SA-AKI is a common and consequential diagnosis in critically ill children, yet successful diagnostic and treatment strategies remain unacceptably scarce. In order to improve the care of patients with SA-AKI, researchers must move toward a precision medicine approach that considers the heterogeneity of the disease on the individual patient level. While biomarkers will undoubtedly play an important role in these endeavors, the complex pathophysiology of SA-AKI requires that we consider the use of biomarkers specific to the individual sepsis inflammatory response, a key driver of renal injury in these patients. To do this, researchers must leverage and build upon existing sepsis precision medicine work, facilitating the development of prognostic and predictive enrichment tools that could advance the care of SA-AKI beyond prevention and renal support. A necessary and feasible first step in this process is the development and validation of reliable tools for the prediction of patients at highest risk for SA-AKI, as such a tool could facilitate the implementation of early and aggressive renal protection strategies, and perhaps more importantly in pediatrics, reduce the number of patients needed to study by informing enrollment in clinical trials aimed at identifying disease-modifying therapies. While the use of individual patient biology-driven therapies via predictive enrichment remains an elusive goal, reframing SA-AKI as a heterogeneous disease that will likely require an individualized approach to therapy is an important first step that should inform future research.
JO, NS, and HW were responsible for writing and editing this mini-review. All authors approved the final version for submission.
This work was funded by National Institutes of Health; R35 GM126943 (HW).
The Cincinnati Children's Research Foundation has submitted a provisional patent application for the sepsis-associated acute kidney injury prediction strategy discussed in this review article. NS and HW were named as co-inventors in the patent application.
The remaining author declares that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
1. Hartman ME., Linde-Zwirble WT, Angus DC, Watson RS. Trends in the epidemiology of pediatric severe sepsis. Pediatr Crit Care Med. (2013) 14:686–93. doi: 10.1097/PCC0b013e3182917fad
2. Weiss SL, Fitzgerald JC, Pappachan J, Wheeler D., Jaramillo-Bustamante JC, Salloo A, et al. Global epidemiology of pediatric severe sepsis: the sepsis prevalence, outcomes, and therapies study. Am J Respir Crit Care Med. (2015) 191:1147–57. doi: 10.1164/rccm201412-2323OC
3. Fitzgerald JC, Basu R, Akcan-Arikan A, Izquierdo LM, Piñeres Olave BE, Hassinger AB, et al. Acute kidney injury in pediatric severe sepsis, an independent risk factor for death and new disability. Crit Care Med. (2016) 44:2241–50. doi: 10.1097/CCM0000000000002007
4. Fitzgerald JC, Ross ME, Thomas NJ, Weiss SL, Balamuth F, Anderson AH. Risk factors and inpatient outcomes associated with acute kidney injury at pediatric severe sepsis presentation. Pediatric Nephrology. (2018) 33:1781–90. doi: 10.1007/s00467-018-3981-8
5. Stanski NL, Cvijanovich NZ, Fitzgerald JC, Bigham MT, Wong HR, Weiss SL, et al. Severe acute kidney injury is independently associated with mortality in children with septic shock. Intensive Care Med. (2020) 46:1050–1. doi: 10.1007/s00134-020-05940-8
6. Angus DC, Linde-Zwirble WT, Lidicker J, Clermont G, Carcillo J, Pinsky MR. Epidemiology of severe sepsis in the United States: analysis of incidence, outcome, and associated costs of care. Crit Care Med. (2001) 29:1303–10. doi: 10.1097/00003246-200107000-00002
7. Alobaidi R, Basu RK, Goldstein SL, Bagshaw SM. Sepsis-associated acute kidney injury. Semin Nephrol. (2015) 35:2–11. doi: 10.1016/j.semnephrol.2015.01002
8. Godin M, Murray P, Mehta RL. Clinical approach to the patient with AKI and sepsis. Semin Nephrol. (2015) 35:12–22. doi: 10.1016/j.semnephrol.2015.01003
9. Bellomo R, Kellum JA, Ronco C, Wald R, Martensson J, Maiden M, et al. Acute kidney injury in sepsis. Intensive Care Med. (2017) 43:816–28. doi: 10.1007/s00134-017-4755-7
10. Wong HR, Cvijanovich NZ, Anas N, Allen GL, Thomas NJ, Bigham MT, et al. A multibiomarker-based model for estimating the risk of septic acute kidney injury. Crit Care Med. (2015) 43:1646–53. doi: 10.1097/CCM0000000000001079
11. Gomez H, Ince C, De Backer D, Pickkers P, Payen D, Hotchkiss J, et al. A unified theory of sepsis-induced acute kidney injury: inflammation, microcirculatory dysfunction, bioenergetics, and the tubular cell adaptation to injury. Shock. (2014) 41:3–11. doi: 10.1097/SHK0000000000000052
12. Fani F, Regolisti G, Delsante M, Cantaluppi V, Castellano G, Gesualdo L, et al. Recent advances in the pathogenetic mechanisms of sepsis-associated acute kidney injury. J Nephrol. (2018) 31:351–9. doi: 10.1007/s40620-017-0452-4
13. Emlet DR, Shaw AD, Kellum JA. Sepsis-associated AKI: epithelial cell dysfunction. Semin Nephrol. (2015) 35:85–95. doi: 10.1016/j.semnephrol.2015.01009
14. Doi K, Yuen PST, Eisner C, Hu X, Leelahavanichkul A, Schnermann J, et al. Reduced production of creatinine limits its use as marker of kidney injury in sepsis. J Am Soc Nephrol. (2009) 20:1217–21. doi: 10.1681/ASN2008060617
15. Ostermann M, Zarbock A, Goldstein S, Kashani K, Macedo E, Murugan R, et al. Recommendations on acute kidney injury biomarkers from the acute disease quality initiative consensus conference: a consensus statement. JAMA Netw Open. (2020) 3:e2019209. doi: 10.1001/jamanetworkopen.202019209
16. Peerapornratana S, Manrique-Caballero CL, Gómez H, Kellum JA. Acute kidney injury from sepsis: current concepts, epidemiology, pathophysiology, prevention and treatment. Kidney Int. (2019) 96:1083–99. doi: 10.1016/j.kint.2019.05026
17. Stanski NL, Wong HR. Prognostic and predictive enrichment in sepsis. Nat Rev Nephrol. (2020) 16:20–31. doi: 10.1038/s41581-019-0199-3
18. Wang K, Xie S, Xiao K, Yan P, He W, Xie L. Biomarkers of sepsis-induced acute kidney injury. Biomed Res Int. (2018) 2018:6937947. doi: 10.1155/2018/6937947
19. Umbro I, Gentile G, Tinti F, Muiesan P, Mitterhofer AP. Recent advances in pathophysiology and biomarkers of sepsis-induced acute kidney injury. J Infect. (2016) 72:131–42. doi: 10.1016/j.jinf.2015.11008
20. Singer E, Markó L, Paragas N, Barasch J, Dragun D, Müller DN, et al. Neutrophil gelatinase-associated lipocalin: pathophysiology and clinical applications. Acta Physiol. (2013) 207:663–72. doi: 10.1111/apha12054
21. Wheeler DS, Devarajan P, Ma Q, Harmon K, Monaco M, Cvijanovich N, et al. Serum neutrophil gelatinase-associated lipocalin (NGAL) as a marker of acute kidney injury in critically ill children with septic shock. Crit Care Med. (2008) 36:1297–303. doi: 10.1097/CCM0b013e318169245a
22. Zhang A, Cai Y, Wang P-F, Qu J-N, Luo Z-C, Chen X-D, et al. Diagnosis and prognosis of neutrophil gelatinase-associated lipocalin for acute kidney injury with sepsis: a systematic review and meta-analysis. Crit Care. (2016) 20:41. doi: 10.1186/s13054-016-1212-x
23. Waanders F, van Timmeren MM, Stegeman CA, Bakker SJL, van Goor H. Kidney injury molecule-1 in renal disease. J Pathol. (2010) 220:7–16. doi: 10.1002/path2642
24. Tu Y, Wang H, Sun R, Ni Y, Ma L, Xv F, et al. Urinary netrin-1 and KIM-1 as early biomarkers for septic acute kidney injury. Renal Failure. (2014) 36:1559–63. doi: 10.3109/0886022X.2014949764
25. Vijayan A, Faubel S, Askenazi DJ, Cerda J, Fissell WH, Heung M, et al. Clinical use of the urine biomarker [TIMP-2] × [IGFBP7] for acute kidney injury risk assessment. Am J Kidney Dis. (2016) 68:19–28. doi: 10.1053/j.ajkd.2015.12033
26. Kashani K, Al-Khafaji A, Ardiles T, Artigas A, Bagshaw SM, Bell M, et al. Discovery and validation of cell cycle arrest biomarkers in human acute kidney injury. Crit Care. (2013) 17:R25. doi: 10.1186/cc12503
27. Honore PM, Nguyen HB, Gong M, Chawla LS, Bagshaw SM, Artigas A, et al. Urinary tissue inhibitor of metalloproteinase-2 and insulin-like growth factor-binding protein 7 for risk stratification of acute kidney injury in patients with sepsis. Crit Care Med. (2016) 44:1851–60. doi: 10.1097/CCM0000000000001827
28. Meersch M, Schmidt C, Van Aken H, Rossaint J, Görlich D, Stege D, et al. Validation of cell-cycle arrest biomarkers for acute kidney injury after pediatric cardiac surgery. PLoS ONE. (2014) 9:e110865. doi: 10.1371/journal.pone0110865
29. Bouchon A, Facchetti F, Weigand MA, Colonna M. TREM-1 amplifies inflammation and is a crucial mediator of septic shock. Nature. (2001) 410:1103–7. doi: 10.1038/35074114
30. Gibot S, Cravoisy A, Kolopp-Sarda M-N, Béné M-C, Faure G, Bollaert P-E, et al. Time-course of sTREM (soluble triggering receptor expressed on myeloid cells)-1, procalcitonin, and C-reactive protein plasma concentrations during sepsis. Crit Care Med. (2005) 33:792–6. doi: 10.1097/01.CCM.0000159089.164624A
31. Zhang J, She D, Feng D, Jia Y, Xie L. Dynamic changes of serum soluble triggering receptor expressed on myeloid cells-1 (sTREM-1) reflect sepsis severity and can predict prognosis: a prospective study. BMC Infect Dis. (2011) 11:53. doi: 10.1186/1471-2334-11-53
32. Su L, Xie L, Liu D. Urine sTREM-1 may be a valuable biomarker in diagnosis and prognosis of sepsis-associated acute kidney injury. Crit Care. (2015) 19:281. doi: 10.1186/s13054-015-0998-2
33. Dai X, Zeng Z, Fu C, Zhang S, Cai Y, Chen Z. Diagnostic value of neutrophil gelatinase-associated lipocalin, cystatin C, and soluble triggering receptor expressed on myeloid cells-1 in critically ill patients with sepsis-associated acute kidney injury. Crit Care. (2015) 19:1–10. doi: 10.1186/s13054-015-0941-6
34. Su L, Feng L, Zhang J, Xiao Y, Jia Y, Yan P, et al. Diagnostic value of urine sTREM-1 for sepsis and relevant acute kidney injuries: a prospective study. Crit Care. (2011) 15:R250. doi: 10.1186/cc10508
35. Dinarello C, Novick D, Kim S, Kaplanski G. Interleukin-18 and IL-18 binding protein. Front Immunol. (2013) 4:289. doi: 10.3389/fimmu.201300289
36. Parikh CR, Abraham E, Ancukiewicz M, Edelstein CL. Urine IL-18 is an early diagnostic marker for acute kidney injury and predicts mortality in the intensive care unit. JASN. (2005) 16:3046–52. doi: 10.1681/ASN2005030236
37. Kim S, Kim H-J, Ahn H-S, Song JY, Um T-H, Cho C-R, et al. Is plasma neutrophil gelatinase-associated lipocalin a predictive biomarker for acute kidney injury in sepsis patients? A systematic review and meta-analysis. J Crit Care. (2016) 33:213–23. doi: 10.1016/j.jcrc.2016.02014
38. Haase M, Bellomo R, Devarajan P, Schlattmann P, Haase-Fielitz A. Accuracy of neutrophil gelatinase-associated lipocalin (NGAL) in diagnosis and prognosis in acute kidney injury: a systematic review and meta-analysis. Am J Kidney Dis. (2009) 54:1012–24. doi: 10.1053/j.ajkd.2009.07020
39. Mishra J, Dent C, Tarabishi R, Mitsnefes MM, Ma Q, Kelly C, et al. Neutrophil gelatinase-associated lipocalin (NGAL) as a biomarker for acute renal injury after cardiac surgery. Lancet. (2005) 365:1231–8. doi: 10.1016/S0140-6736(05)74811-X
40. Zappitelli M, Washburn KK, Arikan AA, Loftis L, Ma Q, Devarajan P, et al. Urine neutrophil gelatinase-associated lipocalin is an early marker of acute kidney injury in critically ill children: a prospective cohort study. Crit Care. (2007) 11:R84. doi: 10.1186/cc6089
41. Mårtensson J, Bell M, Oldner A, Xu S, Venge P, Martling C-R. Neutrophil gelatinase-associated lipocalin in adult septic patients with and without acute kidney injury. Intensive Care Med. (2010) 36:1333–40. doi: 10.1007/s00134-010-1887-4
42. Bagshaw SM, Bennett M, Haase M, Haase-Fielitz A, Egi M, Morimatsu H, et al. Plasma and urine neutrophil gelatinase-associated lipocalin in septic versus non-septic acute kidney injury in critical illness. Intensive Care Med. (2010) 36:452–61. doi: 10.1007/s00134-009-1724-9
43. Törnblom S, Nisula S, Petäjä L, Vaara ST, Haapio M, Pesonen E, et al. Urine NGAL as a biomarker for septic AKI: a critical appraisal of clinical utility—data from the observational FINNAKI study. Ann Intensive Care. (2020) 10:51. doi: 10.1186/s13613-020-00667-7
44. Wang B, Chen G, Zhang J, Xue J, Cao Y, Wu Y. Increased neutrophil gelatinase-associated lipocalin is associated with mortality and multiple organ dysfunction syndrome in severe sepsis and septic shock. Shock. (2015) 44:234–8. doi: 10.1097/SHK0000000000000408
45. Hoste EAJ, McCullough PA, Kashani K, Chawla LS, Joannidis M, Shaw AD, et al. Derivation and validation of cutoffs for clinical use of cell cycle arrest biomarkers. Nephrol Dial Transplant. (2014) 29:2054–61. doi: 10.1093/ndt/gfu292
46. BioMérieux. Limiting AKI Progression In Sepsis (LAPIS): A Phase 4, Multicenter, Randomized Controlled Trial of Biomarker-guided Delivery of Kidney-sparing Care Measures in Sepsis Subjects at Risk of Developing AKI. clinicaltrials.gov (2020). Available online at: https://clinicaltrials.gov/ct2/show/NCT04434209 (accessed September 22, 2020).
47. Wynn J, Cornell TT, Wong HR, Shanley TP, Wheeler DS. The host response to sepsis and developmental impact. Pediatrics. (2010) 125:1031–41. doi: 10.1542/peds2009-3301
48. Raymond SL, López MC, Baker HV, Larson SD, Efron PA, Sweeney TE, et al. Unique transcriptomic response to sepsis is observed among patients of different age groups. PLoS ONE. (2017) 12:e0184159. doi: 10.1371/journal.pone0184159
49. Gentile LF, Nacionales DC, Lopez MC, Vanzant E, Cuenca A, Cuenca AG, et al. Protective immunity and defects in the neonatal and elderly immune response to sepsis. J Immunol. (2014) 192:3156–65. doi: 10.4049/jimmunol1301726
50. Atreya MR, Whitacre BE, Cvijanovich NZ, Bigham MT, Thomas NJ, Schwarz AJ, et al. Proprotein Convertase subtilisin/kexin type 9 loss-of-function is detrimental to the juvenile host with septic shock. Crit Care Med. (2020) 48:1513–20. doi: 10.1097/CCM0000000000004487
51. Shao X, Tian L, Xu W, Zhang Z, Wang C, Qi C, et al. Diagnostic value of urinary kidney injury molecule 1 for acute kidney injury: a meta-analysis. PLoS ONE. (2014) 9:e84131. doi: 10.1371/journal.pone0084131
52. Ramesh G, Kwon O, Ahn K. Netrin-1: a novel universal biomarker of human kidney injury. Transplant Proc. (2010) 42:1519–22. doi: 10.1016/j.transproceed.2009.11040
53. Weiss SL, Peters MJ, Alhazzani W, Agus MSD, Flori HR, Inwald DP, et al. Surviving sepsis campaign international guidelines for the management of septic shock and sepsis-associated organ dysfunction in children. Intensive Care Med. (2020) 46:10–67. doi: 10.1007/s00134-019-05878-6
54. Marshall JC. Why have clinical trials in sepsis failed? Trends Mol Med. (2014) 20:195–203. doi: 10.1016/j.molmed.2014.01007
55. Pimienta G, Heithoff DM, Rosa-Campos A, Tran M, Esko JD, Mahan MJ, et al. Plasma proteome signature of sepsis: a functionally connected protein network. PROTEOMICS. (2019) 19:1800389. doi: 10.1002/pmic201800389
56. Feng A, Rice AD, Zhang Y, Kelly GT, Zhou T, Wang T. S1PR1-associated molecular signature predicts survival in patients with sepsis. Shock. (2020) 53:284–92. doi: 10.1097/SHK0000000000001376
57. Sweeney TE, Wynn JL, Cernada M, Serna E, Wong HR, Baker HV, et al. Validation of the sepsis metascore for diagnosis of neonatal sepsis. J Pediatric Infect Dis Soc. (2018) 7:129–35. doi: 10.1093/jpids/pix021
58. Wong HR, Cvijanovich N, Lin R, Allen GL, Thomas NJ, Willson DF, et al. Identification of pediatric septic shock subclasses based on genome-wide expression profiling. BMC Med. (2009) 7:34. doi: 10.1186/1741-7015-7-34
59. Scicluna BP, van Vught LA, Zwinderman AH, Wiewel MA, Davenport EE, Burnham KL, et al. Classification of patients with sepsis according to blood genomic endotype: a prospective cohort study. Lancet Respir Med. (2017) 5:816–26. doi: 10.1016/S2213-2600(17)30294-1
60. Sweeney TE, Perumal TM, Henao R, Nichols M, Howrylak JA, Choi AM, et al. A community approach to mortality prediction in sepsis via gene expression analysis. Nat Commun. (2018) 9:694. doi: 10.1038/s41467-018-03078-2
61. Davenport EE, Burnham KL, Radhakrishnan J, Humburg P, Hutton P, Mills TC, et al. Genomic landscape of the individual host response and outcomes in sepsis: a prospective cohort study. Lancet Respir Med. (2016) 4:259–71. doi: 10.1016/S2213-2600(16)00046-1
62. Wong HR, Lindsell CJ. An enrichment strategy for sepsis clinical trials. Shock. (2016) 46:632–4. doi: 10.1097/SHK0000000000000693
63. Atreya MR, Wong HR. Precision medicine in pediatric sepsis. Curr Opin Pediatr. (2019) 31:322–7. doi: 10.1097/MOP0000000000000753
64. Stanski NL, Stenson EK, Cvijanovich NZ, Weiss SL, Fitzgerald JC, Bigham MT, et al. Persevere biomarkers predict severe acute kidney injury and renal recovery in pediatric septic shock. Am J Respir Crit Care Med. (2020) 201:848–55. doi: 10.1164/rccm201911-2187OC
65. Wiersema R, Jukarainen S, Vaara ST, Poukkanen M, Lakkisto P, Wong H, et al. Two subphenotypes of septic acute kidney injury are associated with different 90-day mortality and renal recovery. Crit Care. (2020) 24:150. doi: 10.1186/s13054-020-02866-x
66. Bellomo R, Forni LG, Busse LW, McCurdy MT, Ham KR, Boldt DW, et al. Renin and survival in patients given angiotensin ii for catecholamine-resistant vasodilatory shock. Am J Respir Crit Care Med. (2020)rccm.201911-2172OC. doi: 10.1164/rccm201911-2172OC
67. Wong HR, Salisbury S, Xiao Q, Cvijanovich NZ, Hall M, Allen GL, et al. The pediatric sepsis biomarker risk model. Crit Care. (2012) 16:R174. doi: 10.1186/cc11652
68. Kaplan JM, Wong HR. Biomarker discovery and development in pediatric critical care medicine. Pediatr Crit Care Med. (2011) 12:165–73. doi: 10.1097/PCC0b013e3181e28876
69. Standage SW, Wong HR. Biomarkers for pediatric sepsis and septic shock. Expert Rev Anti Infect Ther. (2011) 9:71–9. doi: 10.1586/eri.10154
70. Wong HR, Caldwell JT, Cvijanovich NZ, Weiss SL, Fitzgerald JC, Bigham MT, et al. Prospective clinical testing and experimental validation of the pediatric sepsis biomarker risk model. Sci Transl Med. (2019) 11:eaax9000. doi: 10.1126/scitranslmedaax9000
71. Wong HR, Cvijanovich NZ, Allen GL, Thomas NJ, Freishtat RJ, Anas N, et al. Validation of a gene expression-based subclassification strategy for pediatric septic shock. Crit Care Med. (2011) 39:2511–7. doi: 10.1097/CCM0b013e3182257675
72. Wong HR, Cvijanovich NZ, Anas N, Allen GL, Thomas NJ, Bigham MT, et al. Developing a clinically feasible personalized medicine approach to pediatric septic shock. Am J Respir Crit Care Med. (2015) 191:309–15. doi: 10.1164/rccm201410-1864OC
73. Wong HR, Hart KW, Lindsell CJ, Sweeney TE. External corroboration that corticosteroids may be harmful to septic shock endotype A patients. Crit Care Med. (2020) 49:e98–e101. doi: 10.1097/CCM0000000000004709
74. Basu RK, Standage SW, Cvijanovich NZ, Allen GL, Thomas NJ, Freishtat RJ, et al. Identification of candidate serum biomarkers for severe septic shock-associated kidney injury via microarray. Crit Care. (2011) 15:1–11. doi: 10.1186/cc10554
75. Bhatraju PK, Zelnick LR, Herting J, Katz R, Mikacenic C, Kosamo S, et al. Identification of acute kidney injury subphenotypes with differing molecular signatures and responses to vasopressin therapy. Am J Respir Crit Care Med. (2019) 199:863–72. doi: 10.1164/rccm201807-1346OC
76. Khanna A, English SW, Wang XS, Ham K, Tumlin J, Szerlip H, et al. Angiotensin II for the treatment of vasodilatory shock. N Engl J Med. (2017) 377:419–30. doi: 10.1056/NEJMoa1704154
Keywords: sepsis, acute kidney injury, biomarkers, precision medicine, enrichment
Citation: Odum JD, Wong HR and Stanski NL (2021) A Precision Medicine Approach to Biomarker Utilization in Pediatric Sepsis-Associated Acute Kidney Injury. Front. Pediatr. 9:632248. doi: 10.3389/fped.2021.632248
Received: 22 November 2020; Accepted: 22 March 2021;
Published: 14 April 2021.
Edited by:
Katja Michelle Gist, Children's Hospital Colorado, United StatesReviewed by:
Emily Lauren Joyce, Rainbow Babies & Children's Hospital, United StatesCopyright © 2021 Odum, Wong and Stanski. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Natalja L. Stanski, bmF0YWxqYS5zdGFuc2tpQGNjaG1jLm9yZw==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.