- 1Department of Experimental Medicine, University of British Columbia, Vancouver, BC, Canada
- 2Precision Vaccines Program, Division of Infectious Diseases, Boston Children's Hospital, Boston, MA, United States
- 3Harvard Medical School, Boston, MA, United States
- 4Division of Medicine Critical Care, Boston Children's Hospital, Harvard Medical School, Boston, MA, United States
- 5Institute for Medical Immunology, Université libre de Bruxelles, Brussels, Belgium
Over the past decade, there has been a growing awareness of the vital role of the microbiome in the function of the immune system. Recently, several studies have demonstrated a relationship between the composition of the microbiome and the vaccine-specific immune response. As a result of these findings, the administration of probiotics has been proposed as a means of boosting vaccine-specific immunity. Early results have so far been highly inconsistent, with little evidence of sustained benefit. To date, a precise determination of the aspects of the microbiome that impact immunity is still lacking, and the mechanisms of action are also unknown. Further investigations into these questions are necessary to effectively manipulate the microbiome for the purpose of boosting immunity and enhancing vaccine-specific responses in infants. In this review, we summarize recent studies aimed at altering the neonatal gut microbiome to enhance vaccine responses and highlight gaps in knowledge and understanding. We also discuss research strategies aimed at filling these gaps and developing potential therapeutic interventions.
Introduction
Following the relative sterility of the intrauterine environment, the microbiome is seeded from the earliest postnatal microbial exposures. The early life maturation of the microbiome also coincides with the period when the immune system is undergoing rapid development and is most susceptible to perturbations in the microbial environment. This early period of microbial and immunological maturation coincides with the timing of the primary series of immunizations. Thus, these early influences have the potential to have significant long-term effects on the development of vaccine-specific immunity.
The Evolution of the Infant Microbiome
Bacterial diversity increases over the first year of life (1) and by 2 years of age much of the individual variability of the infant microbiota has converged into a relatively stable profile that resembles that of the typical adult (2–4). The dominant phyla in the intestinal microbiome of a healthy adult are Firmicutes and Bacteroidetes, while in the infant gut, members of the phylum Actinobacteria often predominate (5). During this early period of rapid intestinal colonization, the relative abundance and diversity within each phyla may be influenced by a multitude of factors. Principal among these are mode of delivery, mode of feeding, environmental, and genetics (3).
Mode of Delivery
The long-standing view that the in utero environment is sterile has been challenged in recent years by studies reporting evidence of prenatal microbial colonization resulting from exposure to microbial communities of the placenta and amniotic fluid (6). In a carefully controlled study of 537 placentas, de Goffau et al. found no evidence for the existence of a placental microbiome (7). Further, a comprehensive review of the literature found that the existence of a fetal microbiome is not supported (8).
Within the first few hours of life, infants are exposed to multiple microbial microenvironments that may contribute to the infant's own microbiome, including those of the maternal gut, vagina, and skin. The mode of delivery represents a major early microbial exposure event that shapes the development of the infant microbiome, and therefore has implications for health in infancy and childhood.
The gut microbiome of vaginally delivered infants more closely resembles their mothers' vaginal microbiome, while newborns born by cesarean section have an increased prevalence of skin and environmental microbes (9, 10). Compared to vaginally born infants, intestinal colonization by Bacteroides spp. was impaired in infants born by cesarean section (9, 11–13). Vaginal delivery also favored colonization with members of the phylum Actinobacteria (9, 14, 15). Maternal dietary factors also influence the infant microbiome, although this effect varies by mode of delivery (16).
In contrast, the presence of opportunistic microorganisms was more prevalent in infants born by cesarean, including Clostridium difficile (12). Cesarean section delivery is also associated with reduced diversity of the infant gut microbiota over the first 3 months of life (17). By 6 months of age diversity within the 4 most common phyla (Bacteroidetes, Proteobacteria, Firmicutes, and Actinobacteria), did not vary by mode of delivery (17). The distortions observed in the infant microbiome associated with mode of delivery typically disappear in the first year of life (9, 11, 14, 18, 19).
Mode of Feeding
Mode of feeding also has a significant influence on the development of the infant gut microbiome (20), which in turn differs in variety and abundance in infants who are formula fed compared to breastfed. The intestinal microbiome of the mother influences the development of the infant via breastfeeding (21) and an overlap is observed between the composition of the infant stool microbiome and that of both the maternal gut and breastmilk (22). Over the first month of life, a quarter of the bacteria in the stool of primarily breastfed infants was derived from breastmilk, the microbiome of which is dominated by members of the phylum Proteobacteria (1). Areolar skin, which is predominantly colonized by members of the phylum Firmicutes (1), is another source of microbial exposure distinguished by mode of feeding. Firmicutes is one of the two dominant phyla in a healthy adult (5), therefore exposure of infants to maternal areolar skin bacteria via breastfeeding affects the overall infant microbiome composition.
Breastfeeding is also associated with an increased abundance of probiotic organisms, including Lactobacillus and Bifidobacterium (9), the latter of which is typically the dominant organism in breast-fed infants (23). In contrast, formula feeding is associated with reductions in probiotic microbes including Lactobacillus and Bifidobacterium (3) as well as an expansion of Bacteroides (12, 24) and potentially enteropathogenic species including Escherichia coli and C. difficile (3, 12). Consequently, lack of breastfeeding has been associated with childhood health effects similar to those associated with cesarean deliveries.
Human milk oligosaccharides (HMO) and extracellular vehicles (EV) found in breast milk also influences the development of the infant microbiome. HMO are soluble complex carbohydrates which are indigestible to the infant. They function as a prebiotic, binding pathogenic bacteria, and modulating the intestinal immune response to promote beneficial bacteria (25), such as Bifidobacterium. While EVs carry mRNA, miRNA, cytosolic, and membrane proteins, and are implicated within cell-to-cell signaling further developing the infant microbiome (25).
Environmental Factors
The definition of environmental effects on the infant microbiome covers a broad category that involves both intrauterine exposures (such as maternal disease, maternal diet, and maternal medications), and the postnatal environment (such as geography, and diet); these have been previously reviewed in references (26, 27). Although an important contributor to the development of the infant microbiome, environmental factors such as geography are less susceptible to manipulation.
Exposure to medications such as antibiotics, whether due to maternal use or post-natal treatment, is an area that can be clinically adjusted based on the potential to significantly disrupt the intestinal microbiota. Studies have shown that antibiotic usage during pregnancy disrupts the intestinal microbiota of infants in both mice (28, 29) and humans (30). Infant antibiotic treatment was associated with reduced quantity of Bacteroides and Bifidobacteria (5, 12), while the effects of maternal intrapartum antibiotic prophylaxis (IAP) were most pronounced in the reduction of Bacteroidetes, as measured at 3 months of age (19). The role of antibiotics in the intestinal microbiome is complex and multi-dimensional. Antibiotics may restore balance through the elimination of pathogenic microbes, but not without affecting commensal organisms.
Genetic Influences
One of the earliest influences or determinants of what the microbiome looks like is attributable to the influence of host genetics. The relationship between genes and the human microbiome was recently reviewed (31–33). In brief, twin studies show that there are heritable taxa, most notably from the family Christensenellaceae (phylum Firmicutes), and heritability ranges from 0.1 to 0.3 (31) but can be as high as 0.67 based on sibling studies (34). Furthermore, certain genotypes or single nucleotide polymorphisms (SNP) may result in altered diet or altered immune system function. For example, the lactase gene (which confer lactose intolerance) is associated with abundance of Bifidobacteria and even more importantly, there is the interplay of genes associated with complex diseases like obesity and diabetes that play a role, although less so in the infant phase. Consequently, the interaction between genetics and other factors in the infant, including the immune system, results in a distinct composition of the microbiome.
The Microbiome and the Immune System
The role of the intestinal microbiome in the modulation of immune responses is evidenced by the association between dysbiosis and immune-mediated diseases including diabetes (35, 36), Crohn's disease (37), rheumatoid arthritis (38), multiple sclerosis (39), inflammatory bowel disease (40), allergies (41), colorectal cancer (42), hypertension (43), and artherosclerosis (44). However, the mechanism(s) underlying these associations are still largely unknown, and much remains to be determined about how microbial dysbiosis during infancy impacts on the development of the immune system (29). In this review, we highlight how studies involving germ-free mice and probiotics administration have furthered our understanding of the interplay between the microbiome and immune development and/or function.
Insights From Germ-Free Mice
Experiments with germ-free mice have demonstrated the importance of the gut microbiome in healthy immune development. Infant mice lacking a microbiome demonstrated reduced quantity and function of lymphoid cells (45). These findings are reflected in human studies. Colonization of the infant intestine with Bacteroides fragilis at 1 month of age was associated with increased maturation of IgA-secreting cells at 2 months of age (46), suggesting that the microbiome plays a role in the priming of the infant immune system (47).
The lack of a mature microbiome in infancy contributes to a bias toward a T-helper (Th) cell 2 response (48). In germ-free mice, the Th1 response is reduced and the Th1/Th2 balance is skewed toward a Th2 response (49). However, the Th1 response can be restored by a bacterial polysaccharide (PSA) associated with Bacteroides fragilis (50). Colonization of germ-free mice with segmented filamentous bacteria was associated with the induction of Th17 cells (51). Additionally, adaptive immunity to pathogens can be altered both directly and indirectly by interaction with specific commensal microbes (48, 52). These responses can include inducing the differentiation of IgA-producing B cells, as well as the expansion of Th17 and T-regulatory (Treg) cells (48, 52).
Furthermore, although the majority of studies conducted thus far have investigated the effect of the microbiome on the immune system, there is also evidence demonstrating that the immune system facilitates the maintenance of factors essential to the host-microbe relationship (53). The lack of TLR5 expression in the gut epithelium of neonatal mice influenced the composition of the intestinal microbiome including increased representation of Bacteroidetes and Clostridia taxa relative to wildtype (54). It has also been demonstrated that polymorphisms in both innate and adaptive immune pathways influence microbial composition in mice (55).
Insights From Administration of Probiotics
Further mechanistic evidence may be derived from studies that have attempted to enhance immune responses through the administration of probiotics. Infants who received supplementation with Lactobacillus acidophilus for the first 6 months of life demonstrated reduced IL-10 responses to tetanus toxoid (TT) vaccine (56). Supplementation with Lactobacillus acidophilus for the first 6 months of life had no effect on innate immune responses (56); however, in response to polyclonal stimulation with SEB, production of IL-5 and TGF-beta increased in the probiotic group (56). In a group randomly assigned a 6-month course of probiotics, endotoxin-induced IL-6 levels were significantly reduced compared to controls, while no difference was detected in IFN-y production (57).
There is also evidence that Bifidobacteria has a role in the maturation of the infant immune system, and one way to alter the gut microbiome is via the addition to formula. Infants given formula supplemented with Bifidobacterium longum BB536 had increased Bifidobacteria and a higher Bifidobacteria/Enterobacteriaceae ratio at 2 and 4 months of age, as well as a higher number of IFN-γ secreting cells and an increased ratio of IFN-γ/IL-4 secreting cells at 7 months of age, indicating that BB536 may contribute to the bias toward a pro-Th1 response (58, 59). Although outside the scope of this review, probiotics use in the treatment of antibiotic-associated diarrhea in pediatrics has some utility (59, 60); however, results are strain specific and evidence is lacking. There are also risks that should be considered such as the potential that translocation of bacteria in probiotics may cause adverse health effects.
Finally, there is significant inter-individual variability (61) of the microbiome in humans. Together with genetic diversity, this presents a challenge in the investigation of host–microbe interactions (62). Therefore, the clinical utility of probiotic supplementation, especially in younger individuals, requires additional studies targeting risks and benefits.
The Microbiome and Vaccines
Given the associations between the microbiome and the function and composition of the immune system, the potential implications for vaccine responsiveness have been an area of interest. There is evidence implicating the host intestinal microbiome in the development of vaccine-specific immunity, although the mechanisms by which the microbiome modulates vaccine immune response are largely undetermined.
In addition to influencing microbial composition, the role of TLR5 in the microbiome also influences vaccine responses. A study in mice showed that TLR5-mediated sensing of flagellin that was produced by the gut microbiota was necessary for antibody responses to the influenza vaccine, as mice deficient for TLR5 had substantially impaired responses to the vaccine at day 7 and 14 post-vaccination (63), while germ-free (GF) or antibiotic-treated mice had significantly impaired antibody responses to the influenza vaccine (62–64). These data suggest that the gut microbiota enhances systemic vaccine responses but may suppress oral vaccine responses (65).
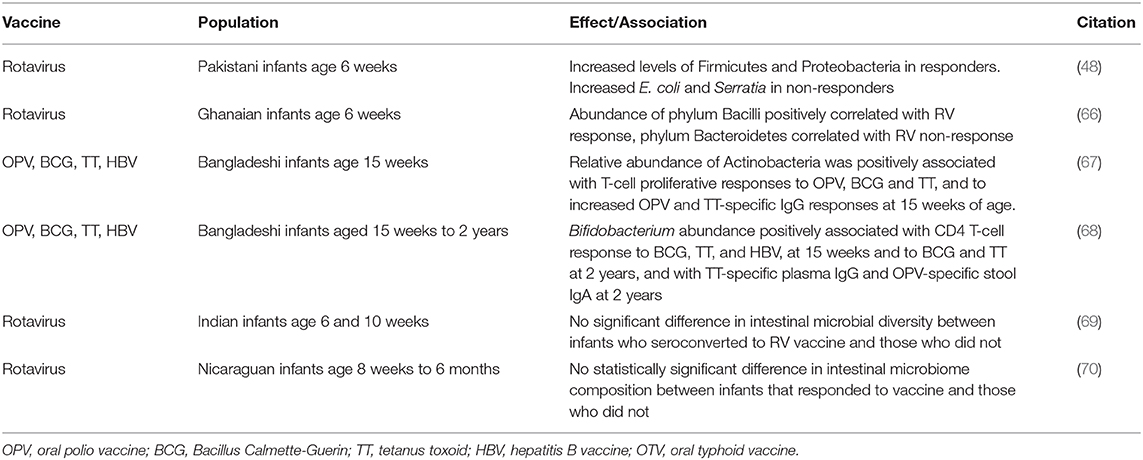
While several studies have reported an association between the composition of the infant intestinal microbiome and the quality of their response to vaccines (Table 1), the findings to date have been inconsistent. Furthermore, studies aimed at the effect of intramuscular or subcutaneous vaccines on the microbiome are still lacking.
Effects of Dysbiosis on Vaccine Responses
The infant immune response is skewed toward a Th2 response dominated by IL-4, IL-5, and IL-10 cytokine production (71) and Th2 cells are associated with the development of allergies (72). Because the infant intestinal microbiome is uniquely susceptible to disruption, deficits or disruptions in microbial communities that biases toward a Th2-type response (48, 49, 73) could be related to the association between intestinal dysbiosis and allergy development (41). On the other hand, colonization with certain bacteria (i.e., Bifidobacteria) can shift the balance toward a Th1 response (58).
Dysbiosis may also be induced via antibiotics as shown in both human and mice studies. Patterns of bacterial colonization due to gestational age and may be in part due to early antibiotic exposure in the premature infant secondary to treatment for infections (i.e., sepsis, necrotizing enterocolitis) (74). Antibiotic-induced dysbiosis in early-life resulted in reduced specific IgG responses to five different vaccines, although following fecal transfer of commensal microbiota, immunity was restored (28). Additionally, infant gut dysbiosis induced by maternal antibiotic usage was associated with reduced adaptive antiviral immunity in response to infection with Vaccinia-OVA (29). In contrast, antibiotic administration did not impaired vaccine responses in adult mice (28) and did not influence anti-rotavirus (RV) IgA titres in adult humans (52). These findings support the hypothesis that in cases of dysbiosis, vaccine responses can be boosted through manipulation of the microbiome in infants (not necessarily in adults), although details such as the duration of therapy required to adequately restore disruptions to the infant microbiome still need to be determined.
Finally, impaired vaccine responsiveness has been reported in low-income settings compared to wealthier regions (75–77). These regions often experience conditions of reduced sanitation and a correspondingly high burden of enteric pathogens (3), which may result in increased dysbiosis. It has been suggested that this excessive pathogenic burden could contribute to the reduced efficacy of oral vaccines that is often observed in the developing world (78, 79). Evidence from a Zimbabwean cohort supports this hypothesis, as improvements in sanitation were associated with improved responsiveness to the rotavirus (RV) vaccine (80).
Effects of Probiotics on Vaccine Responses
Given that the intestinal microbiome of infants is still developing and is characterized by low diversity and rapid change (27), it has been surmised that the infant microbiome may be much more susceptible to influence from external factors such as probiotic supplementation (3). While probiotics have produced beneficial effects in the treatment of some autoimmune conditions (81, 82), studies have also investigated the use of probiotics and prebiotics as adjuvants to enhance vaccine responses, the results of which have been thoroughly reviewed elsewhere (3, 83). Thus far, the results have been highly inconsistent in infant populations, with variations between studies in the type, dose, and duration of probiotic supplementation further complicating any conclusions that may be drawn from them regarding the utility of probiotics as vaccine adjuvants.
At 4 months of age, infants given enhanced formula to promote growth of intestinal Bifidobacteria displayed an enhanced polio-specific response following vaccination and a positive correlation was observed between total Bifidobacteria as a percentage of intestinal flora and titres of anti-poliovirus IgA (47). Infants given Lactobacillus rhamnosus GG (LGG) had an increased rate of rotavirus IgA seroconversion (84), while infants given a cereal supplemented with Lactobacillus paracasei F19 (LF19), from 4 to 13 months of age, demonstrated an increase in immune responses to diphtheria following vaccination, with no difference in Hib or tetanus-specific IgG levels (85). A combination of four probiotic strains was associated with increased seroconversion rates in response to vaccination with Hib, but failed to elicit a significantly enhanced response to either tetanus or diphtheria (86). Additionally, probiotic supplementation appeared to positively influence anti-HBsAg responses when the vaccine series included two monovalent vaccines and concluded with a quadrivalent vaccine at 6 months, although this trend did not reach statistical significance; however, no effect was observed when a 3-dose monovalent series of HBV was administered (87).
In contrast, a few studies have even reported a negative association between probiotics and vaccine responses. Maternal supplementation with Lactobacillus rhamnosus GG (LGG) was associated with a reduction in infant antibody responses to tetanus, Hib, and pneumococcus vaccines (88), and Bangladeshi children who received a 4-week course of BBG-01 had reduced levels of cholera toxin subunit B-specific IgA levels compared to controls (89). The administration of probiotics was associated with an increase in secretory IgA in stool samples of children at 5 months of age (90); however, this did not translate to improved clinical outcomes, as infants exposed to probiotics reported a higher frequency of mucosal infections up to 2 years of age, despite both groups being able to clear infections at comparable rates (90).
Just as the bacterial composition of the intestine can influence the response to oral vaccines, it has also been proposed that the administration of oral vaccines can cause a perturbation that would be reflected in alterations to the microbiome following vaccination (91). However, few studies have investigated this possibility to date. These conflicting trends confirm that a more thorough understanding of the interaction between the intestinal microbiome and the development of vaccine-specific immunity is necessary before probiotic supplementation can be developed as an effective adjuvant to improve vaccine responses.
Methodologies for Analysis of the Microbiome
While the most commonly analyzed samples are derived from stool, tissue biopsies allow for actual visualization relative to tissue structure as well as evaluation of adherence microbes. Methods used to study the human microbiome have been previously described (92–94) and involve using DNA-based methods (metagenomics), RNA-based approaches (metatranscriptomics), protein-based approaches (metaproteomics), and metabolite-based approaches (metabolomics).
Conclusion and Future Research Directions
The infant microbiome is a rapidly evolving and dynamic environment that is determined and influenced by a variety of internal and external factors. This dynamic microbial environment is integral in modulating the infant's immune function and composition, which in turn has implications for vaccine specific responsiveness. A great deal more research is required to establish a definitive connection between microbial composition and vaccine responsiveness. From the limited data available, the interaction between microbiome and vaccine response varies by vaccine and may also be influenced by other factors known to influence both vaccine responsiveness and microbial composition including age, gender, and route of administration.
Additionally, it has been hypothesized that the composition of the intestinal microbiome as it relates to the development of vaccine-specific immunity may be modulated by probiotic supplementation. Direct modulation of the intestinal microbiome would appear to favor the role of probiotics in influencing the response to oral vaccines; however, the emerging role of the microbiome in the function of systemic immunity suggests probiotics may benefit the response to parenteral vaccines as well. Furthermore, each individual vaccine has the potential to modulate the microbiome such that the vaccination schedule of each country influences the resulting microbiome due to bidirectional interactions between microbiome and vaccines.
Consequently, a better understanding of the factors that enhance or impede the immunological, mucosal, and microbial protection is required. There is a further need to understand the microbiome-immune interplay, and how one influences the other. To accomplish this, we recommend (1) longitudinal prospective cohort studies in infants across a broad geographical and socioeconomic spectrum to define the ontogeny of the microbiome ontogeny, (2) utilization of both in-vivo, ex-vivo and in-vitro methods to better understand how alterations in the microbiome affects the microbiome-immune interplay (either due to intentional manipulation via administration of an agent such as a probiotic or dysbiosis secondary to disease), (3) a standardized approach to evaluate the microbiome in order to allow comparisons among different cohorts (i.e., 16S rRNA, metagenomics, metatranscriptomics, and metabolomics) (94, 95), and (4) a systematic integration of data from various molecular approaches to define the dominant microbial profile at different stages of life and how they change over time.
We believe the above strategies will not only improve the gaps in understanding the microbiome-immune system-vaccine interplay but could also help decrease mortality and morbidity in the pediatric population by developing potential therapeutic interventions and enhancing immunogenicity.
Author Contributions
CR, OO, and KS conceived the manuscript, co-wrote and reviewed the content. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by funds from the Boston Children's Hospital Global Health Program.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The authors would like to acknowledge the support of the Global Health Program at Boston Children's Hospital, Harvard Medical School, Boston, MA, USA.
Abbreviations
BCG, Bacillus Calmette-Guerin; DNA, deoxyribonucleic acid; HBV, hepatitis B vaccine; HBsAg, hepatitis B surface antigen; Hib, Haemophilus influenzae type B vaccine; IFN, interferon; Ig, immunoglobulin; IL, interleukin; OPV, oral polio vaccine; OTV, oral typhoid vaccine; RNA, ribonucleic acid; RV, rotavirus vaccine; SEB, staphylococcal enterotoxin B; Th, T-helper; TLR, Toll-like receptor; and TT, tetanus toxoid.
References
1. Pannaraj PS, Li F, Cerini C, Bender JM, Yang S, Rollie A, et al. Association between breast milk bacterial communities and establishment and development of the infant gut microbiome. JAMA Pediatr. (2017) 171:647–54. doi: 10.1001/jamapediatrics.2017.0378
2. Dogra S, Sakwinska O, Soh SE, Ngom-Bru C, Bruck WM, Berger B, et al. Rate of establishing the gut microbiota in infancy has consequences for future health. Gut Microbes. (2015) 6:321–5. doi: 10.1080/19490976.2015.1078051
3. Valdez Y, Brown EM, Finlay BB. Influence of the microbiota on vaccine effectiveness. Trends Immunol. (2014) 35:526–37. doi: 10.1016/j.it.2014.07.003
4. Palmer C, Bik EM, DiGiulio DB, Relman DA, Brown PO. Development of the human infant intestinal microbiota. PLoS Biol. (2007) 5:e177. doi: 10.1371/journal.pbio.0050177
5. Turroni F, Milani C, Duranti S, Lugli GA, Bernasconi S, Margolles A, et al. The infant gut microbiome as a microbial organ influencing host well-being. Ital J Pediatr. (2020) 46:16. doi: 10.1186/s13052-020-0781-0
6. Collado MC, Rautava S, Aakko J, Isolauri E, Salminen S. Human gut colonisation may be initiated in utero by distinct microbial communities in the placenta and amniotic fluid. Sci Rep. (2016) 6:23129. doi: 10.1038/srep23129
7. de Goffau MC, Lager S, Sovio U, Gaccioli F, Cook E, Peacock SJ, et al. Human placenta has no microbiome but can contain potential pathogens. Nature. (2019) 572:329–34. doi: 10.1038/s41586-019-1451-5
8. Perez-Munoz ME, Arrieta MC, Ramer-Tait AE, Walter J. A critical assessment of the “sterile womb” and “in utero colonization” hypotheses: implications for research on the pioneer infant microbiome. Microbiome. (2017) 5:48. doi: 10.1186/s40168-017-0268-4
9. Backhed F, Roswall J, Peng Y, Feng Q, Jia H, Kovatcheva-Datchary P, et al. Dynamics and stabilization of the human gut microbiome during the first year of life. Cell Host Microbe. (2015) 17:852. doi: 10.1016/j.chom.2015.05.012
10. Dominguez-Bello MG, Costello EK, Contreras M, Magris M, Hidalgo G, Fierer N, et al. Delivery mode shapes the acquisition and structure of the initial microbiota across multiple body habitats in newborns. Proc Natl Acad Sci USA. (2010) 107:11971–5. doi: 10.1073/pnas.1002601107
11. Hill CJ, Lynch DB, Murphy K, Ulaszewska M, Jeffery IB, O'Shea CA, et al. Evolution of gut microbiota composition from birth to 24 weeks in the INFANTMET Cohort. Microbiome. (2017) 5:4. doi: 10.1186/s40168-016-0213-y
12. Penders J, Thijs C, Vink C, Stelma FF, Snijders B, Kummeling I, et al. Factors influencing the composition of the intestinal microbiota in early infancy. Pediatrics. (2006) 118:511–21. doi: 10.1542/peds.2005-2824
13. Gronlund MM, Lehtonen OP, Eerola E, Kero P. Fecal microflora in healthy infants born by different methods of delivery: permanent changes in intestinal flora after cesarean delivery. J Pediatr Gastroenterol Nutr. (1999) 28:19–25. doi: 10.1097/00005176-199901000-00007
14. Hill CJ, Lynch DB, Murphy K, Ulaszewska M, Jeffery IB, O'Shea CA, et al. Erratum to: evolution of gut microbiota composition from birth to 24 weeks in the INFANTMET Cohort. Microbiome. (2017) 5:21. doi: 10.1186/s40168-017-0240-3
15. Biasucci G, Rubini M, Riboni S, Morelli L, Bessi E, Retetangos C. Mode of delivery affects the bacterial community in the newborn gut. Early Hum Dev. (2010) 86(Suppl. 1):13–5. doi: 10.1016/j.earlhumdev.2010.01.004
16. Lundgren SN, Madan JC, Emond JA, Morrison HG, Christensen BC, Karagas MR, et al. Maternal diet during pregnancy is related with the infant stool microbiome in a delivery mode-dependent manner. Microbiome. (2018) 6:109. doi: 10.1186/s40168-018-0490-8
17. Rutayisire E, Huang K, Liu Y, Tao F. The mode of delivery affects the diversity and colonization pattern of the gut microbiota during the first year of infants' life: a systematic review. BMC Gastroenterol. (2016) 16:86. doi: 10.1186/s12876-016-0498-0
18. Chu DM, Ma J, Prince AL, Antony KM, Seferovic MD, Aagaard KM. Maturation of the infant microbiome community structure and function across multiple body sites and in relation to mode of delivery. Nat Med. (2017) 23:314–26. doi: 10.1038/nm.4272
19. Azad MB, Konya T, Persaud RR, Guttman DS, Chari RS, Field CJ, et al. Impact of maternal intrapartum antibiotics, method of birth and breastfeeding on gut microbiota during the first year of life: a prospective cohort study. BJOG. (2016) 123:983–93. doi: 10.1111/1471-0528.13601
20. Martin V, Maldonado-Barragan A, Moles L, Rodriguez-Banos M, Campo RD, Fernandez L, et al. Sharing of bacterial strains between breast milk and infant feces. J Hum Lact. (2012) 28:36–44. doi: 10.1177/0890334411424729
21. Jost T, Lacroix C, Braegger CP, Rochat F, Chassard C. Vertical mother-neonate transfer of maternal gut bacteria via breastfeeding. Environ Microbiol. (2014) 16:2891–904. doi: 10.1111/1462-2920.12238
22. Schanche M, Avershina E, Dotterud C, Oien T, Storro O, Johnsen R, et al. High-resolution analyses of overlap in the microbiota between mothers and their children. Curr Microbiol. (2015) 71:283–90. doi: 10.1007/s00284-015-0843-5
23. Yoshioka H, Iseki K, Fujita K. Development and differences of intestinal flora in the neonatal period in breast-fed and bottle-fed infants. Pediatrics. (1983) 72:317–21.
24. Harmsen HJ, Wildeboer-Veloo AC, Raangs GC, Wagendorp AA, Klijn N, Bindels JG, et al. Analysis of intestinal flora development in breast-fed and formula-fed infants by using molecular identification and detection methods. J Pediatr Gastroenterol Nutr. (2000) 30:61–7. doi: 10.1097/00005176-200001000-00019
25. Le Doare K, Holder B, Bassett A, Pannaraj PS. Mother's milk: a purposeful contribution to the development of the infant microbiota and immunity. Front Immunol. (2018) 9:361. doi: 10.3389/fimmu.2018.00361
26. Amenyogbe N, Kollmann TR, Ben-Othman R. Early-life host-microbiome interphase: the key frontier for immune development. Front Pediatr. (2017) 5:111. doi: 10.3389/fped.2017.00111
27. Milani C, Duranti S, Bottacini F, Casey E, Turroni F, Mahony J, et al. The first microbial colonizers of the human gut: composition, activities, and health implications of the infant gut microbiota. Microbiol Mol Biol Rev. (2017) 81:e00036-17. doi: 10.1128/MMBR.00036-17
28. Lynn MA, Tumes DJ, Choo JM, Sribnaia A, Blake SJ, Leong LEX, et al. Early-life antibiotic-driven dysbiosis leads to dysregulated vaccine immune responses in mice. Cell Host Microbe. (2018) 23:653–60.e5. doi: 10.1016/j.chom.2018.04.009
29. Gonzalez-Perez G, Hicks AL, Tekieli TM, Radens CM, Williams BL, Lamouse-Smith ES. Maternal antibiotic treatment impacts development of the neonatal intestinal microbiome and antiviral immunity. J Immunol. (2016) 196:3768–79. doi: 10.4049/jimmunol.1502322
30. Tanaka S, Kobayashi T, Songjinda P, Tateyama A, Tsubouchi M, Kiyohara C, et al. Influence of antibiotic exposure in the early postnatal period on the development of intestinal microbiota. FEMS Immunol Med Microbiol. (2009) 56:80–7. doi: 10.1111/j.1574-695X.2009.00553.x
31. Cahana I, Iraqi FA. Impact of host genetics on gut microbiome: take-home lessons from human and mouse studies. Animal Model Exp Med. (2020) 3:229–36. doi: 10.1002/ame2.12134
32. Goodrich JK, Davenport ER, Clark AG, Ley RE. The relationship between the human genome and microbiome comes into view. Annu Rev Genet. (2017) 51:413–33. doi: 10.1146/annurev-genet-110711-155532
33. Kurilshikov A, Wijmenga C, Fu J, Zhernakova A. host genetics and gut microbiome: challenges and perspectives. Trends Immunol. (2017) 38:633–47. doi: 10.1016/j.it.2017.06.003
34. Turpin W, Espin-Garcia O, Xu W, Silverberg MS, Kevans D, Smith MI, et al. Association of host genome with intestinal microbial composition in a large healthy cohort. Nat Genet. (2016) 48:1413–7. doi: 10.1038/ng.3693
35. Hara N, Alkanani AK, Ir D, Robertson CE, Wagner BD, Frank DN, et al. The role of the intestinal microbiota in type 1 diabetes. Clin Immunol. (2013) 146:112–9. doi: 10.1016/j.clim.2012.12.001
36. Knip M, Siljander H. The role of the intestinal microbiota in type 1 diabetes mellitus. Nat Rev Endocrinol. (2016) 12:154–67. doi: 10.1038/nrendo.2015.218
37. Khanna S, Raffals LE. The microbiome in Crohn's disease: role in pathogenesis and role of microbiome replacement therapies. Gastroenterol Clin North Am. (2017) 46:481–92. doi: 10.1016/j.gtc.2017.05.004
38. Maeda Y, Kurakawa T, Umemoto E, Motooka D, Ito Y, Gotoh K, et al. Dysbiosis contributes to arthritis development via activation of autoreactive T cells in the intestine. Arthritis Rheumatol. (2016) 68:2646–61. doi: 10.1002/art.39783
39. Shahi SK, Freedman SN, Mangalam AK. Gut microbiome in multiple sclerosis: the players involved and the roles they play. Gut Microbes. (2017) 8:607–15. doi: 10.1080/19490976.2017.1349041
40. Fava F, Danese S. Intestinal microbiota in inflammatory bowel disease: friend of foe? World J Gastroenterol. (2011) 17:557–66. doi: 10.3748/wjg.v17.i5.557
41. Chua HH, Chou HC, Tung YL, Chiang BL, Liao CC, Liu HH, et al. Intestinal dysbiosis featuring abundance of Ruminococcus gnavus associates with allergic diseases in infants. Gastroenterology. (2018) 154:154–67. doi: 10.1053/j.gastro.2017.09.006
42. Shen XG, Wang C, Li Y, Wang L, Zhou B, Xu B, et al. Downregulation of caspase-9 is a frequent event in patients with stage II colorectal cancer and correlates with poor clinical outcome. Colorectal Dis. (2010) 12:1213–8. doi: 10.1111/j.1463-1318.2009.02009.x
43. Yang T, Santisteban MM, Rodriguez V, Li E, Ahmari N, Carvajal JM, et al. Gut dysbiosis is linked to hypertension. Hypertension. (2015) 65:1331–40. doi: 10.1161/HYPERTENSIONAHA.115.05315
44. Jie Z, Xia H, Zhong SL, Feng Q, Li S, Liang S, et al. The gut microbiome in atherosclerotic cardiovascular disease. Nat Commun. (2017) 8:845. doi: 10.1038/s41467-017-00900-1
45. Tibbs TN, Lopez LR, Arthur JC. The influence of the microbiota on immune development, chronic inflammation, and cancer in the context of aging. Microb Cell. (2019) 6:324–34. doi: 10.15698/mic2019.08.685
46. Gronlund MM, Arvilommi H, Kero P, Lehtonen OP, Isolauri E. Importance of intestinal colonisation in the maturation of humoral immunity in early infancy: a prospective follow up study of healthy infants aged 0–6 months. Arch Dis Child Fetal Neonatal Ed. (2000) 83:F186–92. doi: 10.1136/fn.83.3.F186
47. Mullie C, Yazourh A, Thibault H, Odou MF, Singer E, Kalach N, et al. Increased poliovirus-specific intestinal antibody response coincides with promotion of Bifidobacterium longum-infantis and Bifidobacterium breve in infants: a randomized, double-blind, placebo-controlled trial. Pediatr Res. (2004) 56:791–5. doi: 10.1203/01.PDR.0000141955.47550.A0
48. Harris V, Ali A, Fuentes S, Korpela K, Kazi M, Tate J, et al. Rotavirus vaccine response correlates with the infant gut microbiota composition in Pakistan. Gut Microbes. (2018) 9:93–101. doi: 10.1080/19490976.2017.1376162
49. Wu HJ, Wu E. The role of gut microbiota in immune homeostasis and autoimmunity. Gut Microbes. (2012) 3:4–14. doi: 10.4161/gmic.19320
50. Mazmanian SK, Liu CH, Tzianabos AO, Kasper DL. An immunomodulatory molecule of symbiotic bacteria directs maturation of the host immune system. Cell. (2005) 122:107–18. doi: 10.1016/j.cell.2005.05.007
51. Ivanov II, Atarashi K, Manel N, Brodie EL, Shima T, Karaoz U, et al. Induction of intestinal Th17 cells by segmented filamentous bacteria. Cell. (2009) 139:485–98. doi: 10.1016/j.cell.2009.09.033
52. Harris VC, Haak BW, Handley SA, Jiang B, Velasquez DE, Hykes BL Jr, et al. Effect of antibiotic-mediated microbiome modulation on rotavirus vaccine immunogenicity: a human, randomized-control proof-of-concept trial. Cell Host Microbe. (2018) 24:197–207 e4. doi: 10.1016/j.chom.2018.07.005
53. Zheng D, Liwinski T, Elinav E. Interaction between microbiota and immunity in health and disease. Cell Res. (2020) 30:492–506. doi: 10.1038/s41422-020-0332-7
54. Fulde M, Sommer F, Chassaing B, van Vorst K, Dupont A, Hensel M, et al. Neonatal selection by Toll-like receptor 5 influences long-term gut microbiota composition. Nature. (2018) 560:489–93. doi: 10.1038/s41586-018-0395-5
55. Khan AA, Yurkovetskiy L, O'Grady K, Pickard JM, de Pooter R, Antonopoulos DA, et al. Polymorphic immune mechanisms regulate commensal repertoire. Cell Rep. (2019) 29:541–50.e4. doi: 10.1016/j.celrep.2019.09.010
56. Taylor AL, Hale J, Wiltschut J, Lehmann H, Dunstan JA, Prescott SL. Effects of probiotic supplementation for the first 6 months of life on allergen- and vaccine-specific immune responses. Clin Exp Allergy. (2006) 36:1227–35. doi: 10.1111/j.1365-2222.2006.02553.x
57. Adler Sorensen C, Fuglsang E, Jorgensen CS, Laursen RP, Larnkjaer A, Molgaard C, et al. Probiotics and the immunological response to infant vaccinations; a double-blind randomized controlled trial. Clin Microbiol Infect. (2019) 25:511 e1-e7. doi: 10.1016/j.cmi.2018.07.031
58. Wu BB, Yang Y, Xu X, Wang WP. Effects of Bifidobacterium supplementation on intestinal microbiota composition and the immune response in healthy infants. World J Pediatr. (2016) 12:177–82. doi: 10.1007/s12519-015-0025-3
59. Szajewska H, Canani RB, Guarino A, Hojsak I, Indrio F, Kolacek S, et al. Probiotics for the prevention of antibiotic-associated diarrhea in children. J Pediatr Gastroenterol Nutr. (2016) 62:495–506. doi: 10.1097/MPG.0000000000001081
60. Szajewska H, Ruszczynski M, Radzikowski A. Probiotics in the prevention of antibiotic-associated diarrhea in children: a meta-analysis of randomized controlled trials. J Pediatr. (2006) 149:367–72. doi: 10.1016/j.jpeds.2006.04.053
61. Schirmer M, Smeekens SP, Vlamakis H, Jaeger M, Oosting M, Franzosa EA, et al. Linking the human gut microbiome to inflammatory cytokine production capacity. Cell. (2016) 167:1897. doi: 10.1016/j.cell.2016.11.046
62. Ciabattini A, Olivieri R, Lazzeri E, Medaglini D. Role of the microbiota in the modulation of vaccine immune responses. Front Microbiol. (2019) 10:1305. doi: 10.3389/fmicb.2019.01305
63. Oh JZ, Ravindran R, Chassaing B, Carvalho FA, Maddur MS, Bower M, et al. TLR5-mediated sensing of gut microbiota is necessary for antibody responses to seasonal influenza vaccination. Immunity. (2014) 41:478–92. doi: 10.1016/j.immuni.2014.08.009
64. Nakaya HI, Wrammert J, Lee EK, Racioppi L, Marie-Kunze S, Haining WN, et al. Systems biology of vaccination for seasonal influenza in humans. Nat Immunol. (2011) 12:786–95. doi: 10.1038/ni.2067
65. Lynn DJ, Pulendran B. The potential of the microbiota to influence vaccine responses. J Leukoc Biol. (2018) 103:225–31. doi: 10.1189/jlb.5MR0617-216R
66. Harris VC, Armah G, Fuentes S, Korpela KE, Parashar U, Victor JC, et al. Significant correlation between the infant gut microbiome and rotavirus vaccine response in rural Ghana. J Infect Dis. (2017) 215:34–41. doi: 10.1093/infdis/jiw518
67. Huda MN, Lewis Z, Kalanetra KM, Rashid M, Ahmad SM, Raqib R, et al. Stool microbiota and vaccine responses of infants. Pediatrics. (2014) 134:e362–72. doi: 10.1542/peds.2013-3937
68. Huda MN, Ahmad SM, Alam MJ, Khanam A, Kalanetra KM, Taft DH, et al. Bifidobacterium abundance in early infancy and vaccine response at 2 years of age. Pediatrics. (2019) 143(2). doi: 10.1542/peds.2018-1489
69. Parker EPK, Praharaj I, Zekavati A, Lazarus RP, Giri S, Operario DJ, et al. Influence of the intestinal microbiota on the immunogenicity of oral rotavirus vaccine given to infants in south India. Vaccine. (2018) 36:264–72. doi: 10.1016/j.vaccine.2017.11.031
70. Fix J, Chandrashekhar K, Perez J, Bucardo F, Hudgens MG, Yuan L, et al. Association between gut microbiome composition and rotavirus vaccine response among Nicaraguan infants. Am J Trop Med Hyg. (2020) 102:213–9. doi: 10.4269/ajtmh.19-0355
71. Rosenthal KS, Zimmerman DH. Vaccines: all things considered. Clin Vaccine Immunol. (2006) 13:821–9. doi: 10.1128/CVI.00152-06
72. Maggi E. The TH1/TH2 paradigm in allergy. Immunotechnology. (1998) 3:233–44. doi: 10.1016/S1380-2933(97)10005-7
73. Oyama N, Sudo N, Sogawa H, Kubo C. Antibiotic use during infancy promotes a shift in the T(H)1/T(H)2 balance toward T(H)2-dominant immunity in mice. J Allergy Clin Immunol. (2001) 107:153–9. doi: 10.1067/mai.2001.111142
74. Baranowski JR, Claud EC. Necrotizing enterocolitis and the preterm infant microbiome. Adv Exp Med Biol. (2019) 1125:25–36. doi: 10.1007/5584_2018_313
75. Jiang V, Jiang B, Tate J, Parashar UD, Patel MM. Performance of rotavirus vaccines in developed and developing countries. Hum Vaccin. (2010) 6:532–42. doi: 10.4161/hv.6.7.11278
76. Grassly NC, Jafari H, Bahl S, Durrani S, Wenger J, Sutter RW, et al. Mucosal immunity after vaccination with monovalent and trivalent oral poliovirus vaccine in India. J Infect Dis. (2009) 200:794–801. doi: 10.1086/605330
77. Hallander HO, Paniagua M, Espinoza F, Askelof P, Corrales E, Ringman M, et al. Calibrated serological techniques demonstrate significant different serum response rates to an oral killed cholera vaccine between Swedish and Nicaraguan children. Vaccine. (2002) 21:138–45. doi: 10.1016/S0264-410X(02)00348-1
78. Magwira CA, Taylor MB. Composition of gut microbiota and its influence on the immunogenicity of oral rotavirus vaccines. Vaccine. (2018) 36:3427–33. doi: 10.1016/j.vaccine.2018.04.091
79. Levine MM. Immunogenicity and efficacy of oral vaccines in developing countries: lessons from a live cholera vaccine. BMC Biol. (2010) 8:129. doi: 10.1186/1741-7007-8-129
80. Church JA, Rukobo S, Govha M, Lee B, Carmolli MP, Chasekwa B, et al. The impact of improved water, sanitation, and hygiene on oral rotavirus vaccine immunogenicity in Zimbabwean infants: substudy of a cluster-randomized trial. Clin Infect Dis. (2019) 69:2074–81. doi: 10.1093/cid/ciz140
81. Zamani B, Golkar HR, Farshbaf S, Emadi-Baygi M, Tajabadi-Ebrahimi M, Jafari P, et al. Clinical and metabolic response to probiotic supplementation in patients with rheumatoid arthritis: a randomized, double-blind, placebo-controlled trial. Int J Rheum Dis. (2016) 19:869–79. doi: 10.1111/1756-185X.12888
82. Asemi Z, Zare Z, Shakeri H, Sabihi SS, Esmaillzadeh A. Effect of multispecies probiotic supplements on metabolic profiles, hs-CRP, and oxidative stress in patients with type 2 diabetes. Ann Nutr Metab. (2013) 63:1–9. doi: 10.1159/000349922
83. Zimmermann P, Curtis N. The influence of probiotics on vaccine responses—a systematic review. Vaccine. (2018) 36:207–13. doi: 10.1016/j.vaccine.2017.08.069
84. Isolauri E, Joensuu J, Suomalainen H, Luomala M, Vesikari T. Improved immunogenicity of oral D x RRV reassortant rotavirus vaccine by Lactobacillus casei GG. Vaccine. (1995) 13:310–2. doi: 10.1016/0264-410X(95)93319-5
85. West CE, Gothefors L, Granstrom M, Kayhty H, Hammarstrom ML, Hernell O. Effects of feeding probiotics during weaning on infections and antibody responses to diphtheria, tetanus and Hib vaccines. Pediatr Allergy Immunol. (2008) 19:53–60. doi: 10.1111/j.1399-3038.2007.00583.x
86. Kukkonen K, Nieminen T, Poussa T, Savilahti E, Kuitunen M. Effect of probiotics on vaccine antibody responses in infancy—a randomized placebo-controlled double-blind trial. Pediatr Allergy Immunol. (2006) 17:416–21. doi: 10.1111/j.1399-3038.2006.00420.x
87. Soh SE, Ong DQ, Gerez I, Zhang X, Chollate P, Shek LP, et al. Effect of probiotic supplementation in the first 6 months of life on specific antibody responses to infant Hepatitis B vaccination. Vaccine. (2010) 28:2577–9. doi: 10.1016/j.vaccine.2010.01.020
88. Licciardi PV, Ismail IH, Balloch A, Mui M, Hoe E, Lamb K, et al. Maternal supplementation with LGG reduces vaccine-specific immune responses in infants at high-risk of developing allergic disease. Front Immunol. (2013) 4:381. doi: 10.3389/fimmu.2013.00381
89. Matsuda F, Chowdhury MI, Saha A, Asahara T, Nomoto K, Tarique AA, et al. Evaluation of a probiotics, Bifidobacterium breve BBG-01, for enhancement of immunogenicity of an oral inactivated cholera vaccine and safety: a randomized, double-blind, placebo-controlled trial in Bangladeshi children under 5 years of age. Vaccine. (2011) 29:1855–8. doi: 10.1016/j.vaccine.2010.12.133
90. Quin C, Estaki M, Vollman DM, Barnett JA, Gill SK, Gibson DL. Probiotic supplementation and associated infant gut microbiome and health: a cautionary retrospective clinical comparison. Sci Rep. (2018) 8:8283. doi: 10.1038/s41598-018-26423-3
91. Eloe-Fadrosh EA, McArthur MA, Seekatz AM, Drabek EF, Rasko DA, Sztein MB, et al. Impact of oral typhoid vaccination on the human gut microbiota and correlations with s. Typhi-specific immunological responses. PLoS ONE. (2013) 8:e62026. doi: 10.1371/journal.pone.0062026
92. Allaband C, McDonald D, Vazquez-Baeza Y, Minich JJ, Tripathi A, Brenner DA, et al. Microbiome 101: studying, analyzing, and interpreting gut microbiome data for clinicians. Clin Gastroenterol Hepatol. (2019) 17:218–30. doi: 10.1016/j.cgh.2018.09.017
93. Turnbaugh PJ, Ley RE, Hamady M, Fraser-Liggett CM, Knight R, Gordon JI. The human microbiome project. Nature. (2007) 449:804–10. doi: 10.1038/nature06244
94. Environmental Chemicals, the Human Microbiome, and Health Risk: A Research Strategy. The National Academies Collection: Reports funded by National Institutes of Health. Washington, DC: National Academy of Sciences (2017).
Keywords: microbiome, vaccines, infant microbiome, immune system, maternal microbiome
Citation: Ruck CE, Odumade OA and Smolen KK (2020) Vaccine Interactions With the Infant Microbiome: Do They Define Health and Disease? Front. Pediatr. 8:565368. doi: 10.3389/fped.2020.565368
Received: 25 May 2020; Accepted: 05 November 2020;
Published: 26 November 2020.
Edited by:
Vassiliki Papaevangelou, National and Kapodistrian University of Athens, GreeceReviewed by:
Yuying Liu, University of Texas Health Science Center at Houston, United StatesAntonio Condino-Neto, University of São Paulo, Brazil
Copyright © 2020 Ruck, Odumade and Smolen. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Kinga K. Smolen, a2luZ2Euc21vbGVuQGNoaWxkcmVucy5oYXJ2YXJkLmVkdQ==
 Candice E. Ruck
Candice E. Ruck Oludare A. Odumade
Oludare A. Odumade Kinga K. Smolen
Kinga K. Smolen