- 1Symbiome Inc., San Francisco, CA, United States
- 2Division of Gastroenterology, Department of Medicine, University of California, San Francisco, San Francisco, CA, United States
- 3School of Medical and Health Sciences, Edith Cowan University, Joondalup, WA, Australia
- 4WA Human Microbiome Collaboration Centre, School of Molecular and Life Sciences, Curtin University, Perth, WA, Australia
Human gastrointestinal and respiratory tracts are colonized by diverse polymicrobial communities shortly after birth, which are continuously molded by environmental exposure. The development of the resident microbiota in early life is a critical factor in the maturation of a healthy immune system. Disturbances to the intricate relationship between environmental exposure and maturation of the infant microbiome have been increasingly identified as a potential contributor to a range of childhood diseases. This review details recent evidence that implicates the contribution of gut and airway microbiome to pediatric respiratory health.
Introduction
The human superorganism has coevolved with a wide variety of microbial species that inhabit the human body in an assortment of body-site-specific consortia (1–4). These resident microbes support many functions in the human body, including the metabolism of complex carbohydrates. Fermentation of carbohydrates leads to, amongst other products, anti-inflammatory, anti-proliferative short-chain fatty acids (SCFAs) that represent an essential energy source for gastrointestinal (GI) epithelial cells (5–7). Other microbial-derived bioactive metabolites include essential vitamins (8, 9), hormones (10, 11), and neurotransmitters (12, 13). Resident microbes also modulate drug absorption (14) and the efficacy of vaccines (15).
Of equal importance is the role of the microbiome in protection against pathogenic organisms via competitive colonization (16–18), in addition to microbial regulation of the development and subsequent modulation of local and systemic, innate and adaptive immune function (19–21). This is particularly applicable to the evolving early-life microbiome, which has been recognized as having a strong influence on long-term health from childhood through to adulthood (22).
The early-life human microbiota is evident shortly after birth at multiple body sites, and it continues to assemble, evolve and mature throughout childhood, continuously shaped by environmental exposures. Early exposures to environmental toxicants, livestock, and pet ownership, along with birth delivery by cesarean section (vs. vaginal birth), exclusive formula (rather than breast) feeding, and antimicrobial administration, have all been implicated in shaping the developing microbiome in children (23–29). Disturbances to the intricate relationship between environmental exposure and the maturation of the microbiome have been increasingly identified as an enhancer in the development of a range of childhood diseases including those affecting the airways (30–34). This review aims to summarize the most recent and compelling evidence that implicates the contribution of gut and airway microbiome in modulating pediatric respiratory health. The focus of this review is on acute respiratory illness, recurrent wheeze, and asthma, with the omission of cystic fibrosis, which has recently been reviewed (35–37).
The Origin of Gut and Airway Microbiota at Birth
Although there is emerging evidence of perinatal microbial exposure, based on the identification of microbial communities in the peri- (38) and post-natal meconium (39–41), the vast proportion of microbial colonization of the human body takes place postpartum. According to studies examining infant microbiome development at multiple body sites, colonization with niche-specific microbial assemblages is evident in the first few weeks following delivery (26, 40). Pioneer microbial colonizers of the infant gut have been tracked to multiple maternal body sites in vaginally delivered infants, with strains originating from the maternal gut accounting for a substantial proportion of the overall microbial abundance in the newborn gut, followed by maternal vaginal, oral and skin microbial reservoirs (40). The gut microbiome of infants delivered by Cesarean section was mostly devoid of the maternal gut commensals, particularly of the genus Bacteroides (27, 42), and was more likely to include maternal skin and oral microbes, as well as bacteria found in the delivery room (42–45).
Significantly less is known about the origin of the initial airway microbial colonizers, as no studies to-date have mapped a direct species transfer from the mother to the infant. However, the presence of microbial species common to multiple maternal body sites has been observed in the oral microbiota of newborns (40), with these pioneer communities also influenced by the delivery mode (46). Since the microbiota of the lungs is thought to originate primarily from the dispersion of microbes from the oral, and to a much lesser extent the nasal cavity (2, 3, 47), the former likely serves as the route of initial microbial transmission for the lower airways at birth. What is certain, however, is that the nascent microbiome, both in the intestinal and airway tracts continues to acquire microbes from maternal sources and their surrounding environment including other family members and caretakers in the first few years of life (40, 48). Maternal health, reflected in the composition of her microbiome, in addition to other environmental factors, may be necessary for the appropriate assembly of nascent microbial communities in infancy. Improving our understanding of which key microbial species are required during this early developmental window for the geneses of health would aid the ongoing efforts of developing efficacious microbiota-targeted therapeutics as a potential preventative strategy in mitigating the onset of disease. This is an enormous task and, without a doubt, the Holy Grail of pediatric microbiome research to date.
Microbiome Maturation—the Influence of Environment Over Genetics and What it Means for Respiratory Health
The microbial composition is unique for each individual irrespective of body site, even in the first few days of life. These nascent communities continue to diversify shaped by environmental exposure as the gut and airway microbiota assemble and mature (39, 49, 50). The infant microbial ecosystem assembly follows body-site specific yet coordinated trajectories (51), resulting in distinct microbial biogeography at each body site, with a personalized unique microbial fingerprint for each individual (52).
The uniqueness of individual microbial assemblies could be partly attributable to host genetics, with several studies linking genetics to microbiota composition in a healthy population (53, 54). Other more recent studies of >1,000 participants found the impact of genetics on the microbiota to be less influential (55, 56), pointing to environmental factors as the main contributors to inter-person microbiome variability in healthy individuals. Although genetically related individuals tend to harbor more similar microbiota (27, 57, 58), this relative similarity is likely due to shared environmental exposures and cohabitation, which facilitates microbial transfer between family members (59, 60). This concept is supported by an observation of gut microbiome divergence in monozygotic twins after living apart (60).
Environmental exposures appear to play a pivotal role in the early microbial acquisition and community succession in infancy. Exposures such as the aforementioned cesarean section delivery (27, 29, 42, 44, 61–63), peri- and post-natal antibiotic use (64, 65), gestational age (66), maternal health (26, 39, 67, 68), and early diet (42, 62, 63, 67), have all been associated with variability observed in the composition and successional trajectories of the gut microbiota in the early years. A recent study assessing the effect of delivery mode on the gut microbiota maturation of 120 newborns in the first year of life, independent of intrapartum antibiotics, showed enrichment of Bifidobacterium and a reduction of Enterococcus and Klebsiella spp. in vaginally delivered infants, who exhibited a more stable microbiota development (29). Mode of delivery, infant feeding, and antibiotic use have also been shown to alter the upper respiratory microbiota maturation in infancy (49, 69). Infants born by vaginal birth were quicker to acquire Corynebacterium and Dolosigranulum spp. in their nasopharyngeal microbiota than their caesarian-born counterparts. The acquisition of these species coincided with enhanced microbial community stability and fewer number of respiratory tract infections in the first year of life (69, 70).
The environment surrounding the infant serves as a natural microbial source that can colonize different body sites and potentially modulate immune maturation and tolerance. Oral (23) or intranasal (71) installation of house-dust from low-risk for asthma households protected mice against airway allergen challenge in experimental models of asthma. Gut microbiota of mice gavaged with house dust from homes with dogs was enriched for Lactobacillus johnsonii (23). Oral supplementation with this species reduced allergic airway sensitization to cockroach allergen, reducing airway levels of pro-inflammatory cytokines IL-4, IL-5, and IL-13 (23). This response was mediated by enhanced levels of immunomodulatory fatty acids, including docosahexaenoic acid (72). Similarly, nasal administration of farm-derived bacteria, either Lactococcus lactis, Acinetobacter lwoffii (73), or Staphylococcus sciuri (74), resulted in decreased hallmarks of T helper type 2 (Th2) driven allergic airway inflammation. Results from these mouse-model-based experiments are consistent with the observation that children who grow up with dogs or are exposed to farms and livestock are less likely to develop atopy and childhood asthma (75) and have distinct microbiota in infancy (24, 76–78). However, it should be noted that only a limited number of microorganisms from the environment are ecologically adapted for the successful and persistent colonization of a mammalian host. Colonizing germ-free mice with microorganisms present in diverse environmental samples resulted in a small fraction of these surviving in the mouse gut. Most of the surviving organisms were replaced by more adapted mouse or human-derived microbial strains (79).
The acquisition and appropriate development of the infant microbiome appear to be important in establishing a healthy host-microbiome symbiosis, and disruption of this harmonious relationship has been associated with childhood respiratory diseases. For instance, delayed gut microbiota maturation (39, 68) and decreased microbial diversity (80, 81) in the first year of life has been observed in infants at higher risk for asthma development. Whereas, disruption of microbial niche specificity along the respiratory tract (2, 3), together with an influx of oral commensals in the nasopharynx, appears to precede the development of respiratory tract infections (82). An improved understanding of the intricate interplay between early life microbiota acquisition, environmental microbial engraftment, and immune conditioning is needed to elucidate the microbial impact on the etiology of childhood respiratory disease.
Microbiota Development and the Immune System
Microbial assembly in infancy appears to be vital in establishing appropriate local and systemic innate and adaptive immune functions (83). A recent study elegantly demonstrated that exposure to a specific subset of intestinal microbiota enriched in riboflavin-synthesizing bacteria in the first few weeks of life is necessary for appropriate mucosal-associated invariant T (MAIT) cells development in the skin, lungs and small intestine of mice (84). Conversely, colonization later in life failed to promote MAIT cell development within tissues, indicating that exposure to specific microbes must occur during an early-life window for correct immune priming.
The long-term effect of the appropriate microbial priming of the host on immune system development is particularly evident from studies of germ-free (GF) mice. These animals have a significantly underdeveloped immune system with a lack of regulatory gut CD4+CD8αα+double-positive intraepithelial T lymphocytes (85), possess fewer T regulatory cells (86), display a thinner gut (87), and lung (88) mucus lining and have reduced expression of immunoglobulin A (89) necessary for clearance of pathogenic microbes from the gut lumen (90). Similar immune dysregulation is also observed in neonatal mice following exposure to broad-spectrum antibiotics (91, 92). Neonatal mice exposed to a single dose of macrolide antibiotic soon after birth exhibited persistent alterations in their gut microbiota composition, ileal gene expression, intestinal T-cell populations and significant reduction of fecal IgA levels. These features were not observed in neonatal GF or conventional adult mice receiving the same treatment (91). Absence of microbial colonization of the lungs appears to have a less profound effect on the subset of localized immune cells (88), except iNKT cells which are present at higher levels in both lungs and gut of GF mice (93) and dissipate following microbiota-transfer from conventional (non-GF) animals.
Exposure of neonatal mice to broad-spectrum antibiotics is known to diminish microbial diversity and leads to an exacerbation of allergen-induced airway inflammation (94, 95). Specifically, antibiotic treatment reduced the number of SCFA producing gut bacteria, leading to a subsequent reduction in systemic levels of immunomodulatory SCFAs (5, 95, 96). Conversely, supplementation of mice with SCFA or increasing dietary fiber ameliorated allergic airway inflammation in sensitized animals (5, 95). This resulted in reprogramming of hematopoiesis and subsequent seeding of the lungs by dendritic cells with high phagocytic capacity but an impaired ability to promote Th2 cell effector function or to transport inhaled aeroallergens to lung draining nodes. Airway microbiota has also been shown to influence the development of allergic airway inflammation. Immediately after birth, neonatal mice exhibit enhanced Th2 cell inflammation and airway hyperresponsiveness following exposure to house dust mite aeroallergens. However, as the airway microbiome matured, the exacerbated response to aeroallergen diminished (97). The absence of airway colonization during this early postpartum window resulted in sustained susceptibility to allergic inflammation through adulthood. Supporting these murine observations are pediatric epidemiological studies focused on the etiology of childhood disease, which implicate early-life antibiotic administration as a risk factor for childhood respiratory disease development (98, 99). Collectively, these observations provide strong evidence for the establishment of commensal microbial communities early in life as a critical factor for healthy immune development, ensuring appropriate control, and a potential reduction in cases of airway inflammation.
Airway Microbiome and Respiratory Health
Evidence implicating perturbations to the composition and function of the airway microbiota in pediatric respiratory disease has grown substantially in the last decade (30, 31, 34, 100). Unlike studies of the enteric microbiota, which are primarily based on profiling the highly abundant fecal microbial communities, studies of the airway microbiota present more of a challenge.
Challenges Associated With Studying the Airway Microbiome
Challenges of studying the airway microbiome arise from a relatively low microbial burden typically recovered from airway samples and the distinct composition of microbes present in samples collected using different methods and the respiratory tract compartment being sampled, making it difficult to compare key findings between independent studies. The airway extends from the nasal opening to the alveoli of the lungs, with each compartment providing a distinct microenvironment for microbial colonization. Ideally, samples for microbiome studies addressing associations with pulmonary disease should be obtained at a site where the inflammatory processes contributing to respiratory symptoms occur, and for most pulmonary conditions, these manifest in the lower airways. Unfortunately, sampling the lower airways, particularly in children who cannot expectorate, requires invasive bronchoscopy. This procedure is poorly suited for studies involving infants, healthy children, and large-scale studies due to the need for anesthesia and specialized procedural expertise. As a result, extensive studies of lower airway microbiome in young children are lacking. Most clinical studies focused on the airway microbiota in children are based on the non-invasive samples of the upper respiratory tract (e.g., nasopharyngeal/nasal swabs and nasal wash/lavage); these will form the main focus of discussion in this review.
The upper airway is a poor surrogate for the lower airway microbiota (2, 3, 47, 101–104). Compared to the lower airways, the upper airways (both nasal and nasopharyngeal compartments) harbor less complex microbiota, comprising of distinct microbial communities (3, 47, 103, 104). Microbial composition of the lower airway is more reflective of the oral microbiota (2, 3, 47, 105, 106). However, it should be noted that a loss of microbial topography along the respiratory tract have been observed in individuals with respiratory disease (3, 107), with nasal microbiota contributing a portion of respiratory disease-associated bacterial taxa to the lower airways (3, 47, 103). The exact mechanisms of interaction between the upper and lower airway microbiota in respiratory disease remain unclear. However, there is growing evidence for microbe-microbe interactions and microbe-host interactions (3, 104) within and across various compartments of the respiratory tract.
Dissimilar microbiota was also observed between sampling methods within the same compartment of the respiratory tract, such as those described in bronchial brush vs. bronchial alveolar lavage (108), and nasal brush vs. nasal wash samples (109). This is also the case for the two most representative anatomical sites of the upper airway microbiome studies, the nares and nasopharynx, which comprise of distinct microbial assemblies with considerable compositional overlap (110). Interpretation of airway microbiota studies should, therefore, be carried out with caution, delineating not only the different compartments of the respiratory tract but also based on sample collection procedure. Despite the aforementioned limitations, there has been tremendous recent progress in uncovering the role of the developing upper airway microbiome in modulating and improving respiratory health.
Upper Airway Microbiome and Acute Respiratory Illness
Several extensive studies have characterized the upper respiratory tract microbiome in children as being most frequently dominated by one of the following six bacterial genera—Moraxella, Streptococcus, Corynebacterium, Staphylococcus, Haemophilus, and Alloiococcus [annotated as Dolosigranulum in some databases] (50, 107, 111–114). These distinct bacterial microbiota profiles differentially relate to respiratory illness (Figure 1). In a study of 234 children at high risk for atopy, early colonization of the nasopharynx with Haemophilus, Streptococcus or Moraxella was found to be strongly associated with acute respiratory illness (ARI), including lower respiratory illness (LRI) in the first 5 years of life (50). Conversely, the relative abundance of Staphylococcus, Corynebacterium, and Alloiococcus in the upper airways of infants under 2 years of age were negatively associated with ARIs. The incidence rate of LRIs was highest in children with an early nasal Moraxella-dominant profile and lowest in those with a Corynebacterium-dominant microbiota profile (113). In a different matched case-control study of 307 children hospitalized with LRIs and 154 age-matched controls, nasopharyngeal microbiota profiles dominated by Haemophilus influenzae and Streptococcus pneumoniae were significantly associated with LRIs whereas those dominated by Moraxella catarrhalis/nonliquefaciens or by Corynebacterium propinquum and Dolosigranulum pigrum were related to relatively stable health (107). Although most ARI events involved viral pathogens, shifts in the microbiota toward dominance by one of the pathogenic bacterial genera preceded the detection of viral pathogens and acute respiratory symptoms (50). Risk of severe respiratory tract illness was significantly increased when rhinovirus (RV) or respiratory syncytial virus (RSV) were detected concurrently with nasal Moraxella, Streptococcus, or Haemophilus (111, 112, 115, 116). Similarly, in children with LRIs caused by respiratory syncytial virus (RSV), the relative abundance of H. influenzae and S. pneumoniae in the nasopharynx was strongly associated with increased inflammation (117–119) characterized by overexpression of genes linked to neutrophil/macrophage activation and signaling (117). Young adults with nasal Moraxella-dominated microbiome cluster exhibited the most increase in the concentration of inflammatory markers and the highest viral load during experimental RV infection (120). Conversely, among children hospitalized for bronchiolitis, those harboring Haemophilus dominant nasopharyngeal and nasal microbiota had increased odds for intensive care treatment and an extended hospital stay, compared to those with Moraxella-dominated microbiome profiles (110, 121). Together these observations suggest that bacterial colonization may increase susceptibility to and amplify the host innate immune response to viral respiratory pathogens, thus modulating the severity of ARIs.
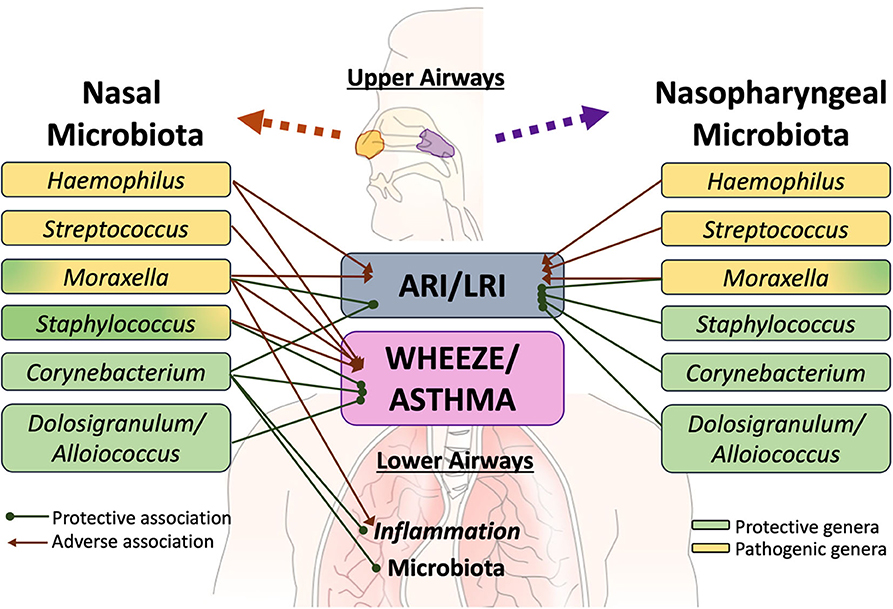
Figure 1. Specific members of the upper airway microbiota have been differentially linked to acute respiratory illness (ARI), including lower respiratory illness (LRI), recurrent wheeze, asthma risk and exacerbation. Although limited, there is some evidence that interactions exist between at least some members of the upper airway microbiota with those colonizing the lower airways and with the pulmonary immune system. However, further work is required to elucidate these links.
Upper Airway Microbiome in Children With Recurrent Wheeze and Asthma
The same three opportunistic respiratory pathogens (i.e., Streptococcus; Haemophilus, and Moraxella) have also been implicated in increasing the risk for recurrent wheeze (50, 111), asthma development and exacerbation during childhood (69, 115, 122). In a recent study of 842 infants hospitalized for bronchiolitis, increased nasal Moraxella and Streptococcus relative abundance was associated with a higher risk of recurrent wheezing by age 3 years (123). In another study of 413 school children with asthma, those that experienced at least one exacerbation irrespective of RV infection were more likely to possess Moraxella-dominated nasal microbiota composition (112) and elevated eosinophil cationic protein concentration in nasal secretions (a marker for activated eosinophils)—suggesting that such microbial communities may promote asthma exacerbation in the absence of RV. In support of this were in vitro observations of enhanced epithelial damage and gene expression of the pro-inflammatory cytokines IL-8 and IL-33 following exposure of human alveolar epithelial cells to cell-free products of nasal Moraxella isolates. Additionally, Moraxella-dominated nasal communities were found to be more stable over time, and children who persistently exhibited this microbial signature in their longitudinal samples were more likely to have viral asthma exacerbations and a higher number of ARIs (112). Relatedly, the relative abundance of nasal Moraxella was positively correlated with systemic and lower airway eosinophilia and bronchial pro-inflammatory cytokine levels in adult subjects with asthma (3). Conversely, children with Alloiococcus, Corynebacterium, and Staphylococcus-dominated nasal microbiota had decreased risk of respiratory virus detection and asthma exacerbation (112). These findings agree with a similar study of 214 children with asthma, in whom nasal microbiota co-dominated by Corynebacterium and Dolosigranulum was associated with better asthma control with fewer exacerbations (114) than those with nasal microbiota dominated by more pathogenic bacteria (i.e., Moraxella, Staphylococcus- specifically S. aureus-and Streptococcus). Additionally, loss of asthma control was accompanied by a shift in nasal bacterial communities from Corynebacterium/Dolosigranulum-dominated to those dominated by Moraxella. This transition was associated with the highest risk of exacerbation compared to the other combinations (114). During periods of uncontrolled asthma, the relative abundance of nasal Corynebacterium was associated with a lower risk of progression to severe exacerbation, suggesting that members of this genera may modulate airway inflammation in asthma (114). This appears to be the case in adult asthmatics whose nasal microbiota are less frequently dominated by members of this genus compared to healthy controls and in whom levels of bronchial inflammation inversely associated with the relative abundance of nasal Corynebacterium (3). Asthmatic subjects with Corynebacterium-dominated nasal microbiota exhibited a lower relative abundance of asthma-associated bronchial microbial taxa (3), possibly explaining reduced inflammation observed in these subjects. Similarly, the relative abundance of nasal Corynebacterium associated with lower transcription of inflammatory gene expression in the nares of children (104) and showed the highest associations with bronchial genera compared to other nasal taxa. Collectively these observations highlight the need and importance to better understand the fluctuation of microbial assemblies in the upper airways overtime and mechanisms by which these contribute to childhood respiratory disease.
A Synopsis on the Key Upper Airway Bacterial Players in Respiratory Health
Overall there is consensus on a beneficial role of Corynebacterium, Alloiococcus/Dolosigranulum, and coagulase-negative Staphylococcus (CoNS; i.e., other than S. aureus) species in moderating airway inflammation. Most studies summarized above agree that the loss of these commensals is associated with enhanced pro-inflammatory immune activation. Competitive colonization is a plausible way by which upper airway commensals protect against pathogen colonization and overgrowth. For instance, Corynebacterium has been shown to inhibit the growth of S. pneumoniae (124) and S. aureus (125, 126) by releasing the antibacterial free fatty acids that may prevent nasal colonization with the pathogenic organisms. Similarly, nasal commensal CoNS S. epidermidis triggers antimicrobial peptide production in the nasal epithelium, providing it a competitive advantage over S. aureus and M. catarrhalis leading to decreased inflammation in vivo (127). Additionally, S. epidermidis has been shown to enhance interferon-λ-dependent immunity against viral influenza resulting in the suppression of viral replication in the nasal mucosa (128). Collectively these observations indicate that from an ecological perspective, a microbiome at equilibrium may resist colonization with pathogenic bacteria and that this microbial stability is important for the maintenance of a healthy airway.
However, when it comes to pathogenic bacteria and associations with respiratory disease, there are currently a lot of unknowns, particularly regarding Moraxella. Understanding species and strain-specific differences between Moraxella spp., colonizing distinct compartments of the airway, by incorporating more granular approaches such as metagenomics into future studies will likely reveal whether more pathogenic strains are prevalent in certain cohorts. A strain-specific granularity in airway microbiome studies would also uncover whether certain pathogenic types of Moraxella spp., are able to descend deeper into the respiratory tract or remain localized in the nasopharynx. The co-occurrence of microorganisms also needs to be considered in future studies of the respiratory microbiota since microbes do not exist in isolation but in complex communities where members influence the virulence potential of opportunistic pathogens. This is seen with co-occurring Moraxella which attenuated the positive association of Alloiococcus with ARI in older children (50); the observation of Moraxella co-occurring with other members of this genus in asthmatic children likely enhancing their virulence potential (112); or the distinct dynamics of associations reported between microbial members in the upper airways of asthmatic and healthy children colonized with the same dominant microbial genera (104). Such studies may help to explain the discrepancy concerning members of genus Moraxella observed in pediatric studies summarized above. Improving our understanding of the microbe-microbe associations within and across the airway compartments and relating them to microbe-host mucosal and systemic immune interactions in multidisciplinary studies will help uncover the mechanisms by which opportunistic respiratory pathogens contribute to various respiratory illnesses in children.
Gut Microbiome and Respiratory Health
The gut is the most densely colonized organ of the human body harboring a diverse range of microbial symbionts, including bacteria, archaea, protozoa, and fungi (129–133). In recent years the importance of the bidirectional crosstalk along the gut-lung axis has been increasingly recognized as a contributor to respiratory health, although the mechanisms of these interactions remain poorly elucidated (134, 135). Unsurprisingly, respiratory diseases are often accompanied by gastrointestinal (GI) comorbidities and vice versa. Patients with obstructive pulmonary disease, for example, have increased intestinal permeability during severe acute exacerbations (136) and are 2–3 times more likely to be diagnosed with irritable bowel disease (137), whereas impaired pulmonary function is prevalent in patients with chronic GI disease (138–140). In infants, Bacteroides-dominated fecal microbiota was associated with a higher likelihood of being hospitalized for bronchiolitis (141). Although this does not directly support the role of the microbiome in the gut-lung crosstalk, the associative evidence is strong.
Further evidence for the gut-lung axis crosstalk comes from studies implicating early-life gut microbiome perturbations, characterized by loss of commensal Bifidobacteria, Lachnospira, Faecalibacteria, and Akkermansia (39, 68, 80, 81) in risk for childhood asthma development. Loss of these enteric commensals was accompanied by depletion of fecal acetate, anti-inflammatory polyunsaturated fatty acids and breast milk oligosaccharides, all known to influence gut epithelial colonization of infants at high risk for asthma (39, 80, 81). Mice born to dames gavaged with feces from high-risk infants exhibited exacerbated allergic lung inflammation in an experimental model of asthma, which was alleviated upon the supplementation of dames with bacterial taxa depleted in the microbiota of high-risk infants (80). This suggests a causal role of these early-life enteric bacteria in preventing airway inflammation. Furthermore, soluble products of the gut microbiota from high-risk infants induced Th2 cell expansion, increased IL-4 expression, and decreased regulatory T cell populations ex vivo, the latter attributed in part to elevated fecal levels of the oxylipin 12,13-diHOME (81). This oxylipin was initially observed in airways of adult asthmatics following a bronchial challenge with birch pollen (142). The link between pulmonary inflammation and elevated fecal levels of 12,13-diHOME was found to be related to reprogrammed dendritic cell activity and a reduction in the number of pulmonary Treg cells (143). In the infant gut, three epoxide hydrolase genes (EH), responsible for the production of the lipokine were encoded by Bifidobacterium bifidum and Enterococcus faecalis. Either elevated concentration of 12,13-diHOME or increased abundance of the EH genes was found to significantly increase the probability of developing childhood allergies, eczema, and asthma (143). Future research is needed to determine the role of the gut environment in the expression of these bacterial genes and how it relates to increased pulmonary inflammation in asthma. This will be important as B. bifidum is a widely used probiotic for infants, and certain strains encoding EHs may not be suitable for all infants or conditions. It should also be noted that 12,13-diHOME has a profound role in brown adipose tissue activation and is negatively correlated with body-mass index and insulin resistance in mice (144). It remains unclear why this lipokine seems to have different effects in distinct mammalian tissues/systems and what role the time of exposure to elevated levels plays in the development of various pathologies.
Gut Metabolome and Infant Diet
Infant diet is the earliest, well-established microbial selection pressure, where exclusive breastfeeding is known to select for distinct microbiota compared to formula-feeding (145). Breastfeeding has also shown to mitigate against LRI in a study of 5,322 children in whom breastfeeding for 6 months was associated with a lower incidence of LRI up to 4 years compared to children who had never been breastfed (146). The timing of solid food introduction and its nutritional composition have also been shown to significantly alter the gut microbiome in infants (147). The impact of different dietary habits on shaping the developing gut microbiota has been demonstrated in a comparative study of European and African children aged 1–6 years (148). A significantly higher intake of fiber with low animal protein and fat consumption in the rural African diet promoted enrichment of Prevotella and Xylanibacter compared to that of Bacteroides observed in the western children, leading to significantly increased levels of bacteria-derived fecal anti-inflammatory SCFA (148, 149). The microbial capability of SCFA production by fermenting complex carbohydrates is also evident in infants before weaning (150, 151), highlighting the potential of the infant gut microbiome to ferment complex carbohydrates beyond inulin, fructo- and galacto-oligosaccharides currently used in infant formula.
Harvesting enteric microbial capacity to produce SCFAs via nutritional intervention represents an attractive avenue for gut microbial modulation as a preventative strategy for respiratory disease development (152). As demonstrated in several studies, SCFAs promote intestinal-epithelial integrity leading to reduced inflammation locally in the gut as well as in the respiratory tract (5, 95, 153–155). The mechanisms linking microbial-derived SCFAs and effects on the respiratory tract are just beginning to be elucidated; however, there is no doubt that this relationship is multifaceted (135). Mechanisms of SCFAs mode of action on modulating immune responses have been linked to G protein-coupled receptors and inhibition of histone deacetylase activity (135, 154). It is essential to keep in mind that SCFA effects are not only dependent on their availability, concentration and affinity to receptors but also on the expression of various transporter molecules and downstream effectors in distinct cell types (135). Whether dietary interventions aimed at modulating gut microbiota prove effective in preventing respiratory disease in children remains to be determined.
Gut Interventions to Improve Respiratory Health
To date, although some promising results have been seen from trials using symbiotics (probiotics/prebiotics or a combination) in reducing the rate of pediatric respiratory tract infections (156, 157), this had not been the case for attempts at preventing atopic asthma (158–161). Encouragingly, infant gut microbiota composition, metabolic function, and host-immune interaction have been shown to be susceptible to modulation by a single Lactobacillus rhamnosus strain administered to high risk for asthma infants from birth once-daily for 6 months (39). The positive effect of the probiotic on the microbiome was not sustained following cessation of supplementation, suggesting the need for earlier intervention or use of a multispecies probiotic supplement consisting of species more adapted to the neonatal gut environment to achieve long term-term efficacy (32, 39). Overall, evidence implicating gut microbial alterations in respiratory disease development is rapidly building. More mechanistic studies are needed to improve the understanding of underlying mechanisms driving microbe-microbe and microbe-host interactions locally in the gut in parallel with these along the gut-lung axis before targeted interventions will likely be shown to have clinical efficacy.
Summary and Concluding Remarks
Convincing evidence from both murine and human studies implicates perturbations to the composition and function of airway and gut microbiota in pediatric respiratory disease. Disruptions to the developmental assembly of the microbiota maturation have long-lasting consequences manifesting in an enhanced response to viral or allergen exposure and consequently, respiratory disease. Despite tremendous progress in uncovering underlying microbial mechanisms responsible for respiratory disease development or exacerbation, the microbiome field is in a nascent state, and many knowledge gaps and opportunities for improved understanding remain. An integrative systems biology approach linking all the members of the microbiota (bacteria, fungal and viral) in the respiratory and gastrointestinal compartments to host immune function is required to elucidate specific microbial mechanisms that govern respiratory disease susceptibility.
Author Contributions
JD and CC conceived and wrote the review article. All authors contributed to the article and approved the submitted version.
Funding
CC is supported by the Telethon – Perth Children's Research Fund from the WA Department of Health.
Conflict of Interest
JD is employed by a skincare company Symbiome Inc.
The remaining author declares that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
1. Caporaso JG, Lauber CL, Costello EK, Berg-Lyons D, Gonzalez A, Stombaugh J, et al. Moving pictures of the human microbiome. Genome Biol. (2011) 12:R50. doi: 10.1186/gb-2011-12-5-r50
2. Bassis CM, Erb-Downward JR, Dickson RP, Freeman CM, Schmidt TM, Young VB, et al. Analysis of the upper respiratory tract microbiotas as the source of the lung and gastric microbiotas in healthy individuals. MBio. (2015) 6:1–10. doi: 10.1128/mBio.00037-15
3. Durack J, Huang YJ, Nariya S, Christian LS, Mark Ansel K, Beigelman A, et al. Bacterial biogeography of adult airways in atopic asthma. Microbiome. (2018) 6:1–16. doi: 10.1186/s40168-018-0487-3
4. Gilbert JA, Blaser MJ, Caporaso JG, Jansson JK, Lynch SV, Knight R. Current understanding of the human microbiome. Nat Med. (2018) 24:392–400. doi: 10.1038/nm.4517
5. Trompette A, Gollwitzer ES, Yadava K, Sichelstiel AK, Sprenger N, Ngom-Bru C, et al. Gut microbiota metabolism of dietary fiber influences allergic airway disease and hematopoiesis. Nat Med. (2014) 20:159–66. doi: 10.1038/nm.3444
6. Kelly CJ, Zheng L, Campbell EL, Saeedi B, Scholz CC, Bayless AJ, et al. Crosstalk between microbiota-derived short-chain fatty acids and intestinal epithelial HIF augments tissue barrier function. Cell Host Microbe. (2015) 17:662–71. doi: 10.1016/j.chom.2015.03.005
7. Zheng L, Kelly CJ, Battista KD, Schaefer R, Lanis JM, Alexeev EE, et al. Microbial-derived butyrate promotes epithelial barrier function through IL-10 receptor–dependent repression of claudin-2. J Immunol. (2017) 199:2976–84. doi: 10.4049/jimmunol.1700105
8. Magnúsdóttir S, Ravcheev D, De Crécy-Lagard V, Thiele I. Systematic genome assessment of B-vitamin biosynthesis suggests cooperation among gut microbes. Front Genet. (2015) 6:148. doi: 10.3389/fgene.2015.00148
9. Karl JP, Meydani M, Barnett JB, Vanegas SM, Barger K, Fu X, et al. Fecal concentrations of bacterially derived vitamin K forms are associated with gut microbiota composition but not plasma or fecal cytokine concentrations in healthy adults. Am J Clin Nutr. (2017) 106:1052–61. doi: 10.3945/ajcn.117.155424
10. Kunc M, Gabrych A, Witkowski JM. Microbiome impact on metabolism and function of sex, thyroid, growth and parathyroid hormones. Acta Biochim Pol. (2016) 63:189–201. doi: 10.18388/abp.2015_1093
11. Yan J, Herzog JW, Tsang K, Brennan CA, Bower MA, Garrett WS, et al. Gut microbiota induce IGF-1 and promote bone formation and growth. Proc Natl Acad Sci USA. (2016) 113:E7554–63. doi: 10.1073/pnas.1607235113
12. Yano JM, Yu K, Donaldson GP, Shastri GG, Ann P, Ma L, et al. Indigenous bacteria from the gut microbiota regulate host serotonin biosynthesis. Cell. (2015) 161:264–76. doi: 10.1016/j.cell.2015.02.047
13. Romano KA, Martinez-del Campo A, Kasahara K, Chittim CL, Vivas EI, Amador-Noguez D, et al. Metabolic, epigenetic, and transgenerational effects of gut bacterial choline consumption. Cell Host Microbe. (2017) 22:279–90.e7. doi: 10.1016/j.chom.2017.07.021
14. Spanogiannopoulos P, Bess EN, Carmody RN, Turnbaugh PJ. The microbial pharmacists within us: a metagenomic view of xenobiotic metabolism. Nat Rev Microbiol. (2016) 14:273–87. doi: 10.1038/nrmicro.2016.17
15. Vlasova AN, Takanashi S, Miyazaki A, Rajashekara G, Saif LJ. How the gut microbiome regulates host immune responses to viral vaccines. Curr Opin Virol. (2019) 37:16–25. doi: 10.1016/j.coviro.2019.05.001
16. Caballero S, Kim S, Carter RA, Leiner IM, Sušac B, Miller L, et al. Cooperating commensals restore colonization resistance to vancomycin-resistant Enterococcus faecium. Cell Host Microbe. (2017) 21:592–602.e4. doi: 10.1016/j.chom.2017.04.002
17. Nakatsuji T, Chen TH, Narala S, Chun KA, Two AM, Yun T, et al. Antimicrobials from human skin commensal bacteria protect against Staphylococcus aureus and are deficient in atopic dermatitis. Sci Transl Med. (2017) 9:1–12. doi: 10.1126/scitranslmed.aah4680
18. Leech JM, Dhariwala MO, Lowe MM, Chu K, Merana GR, Cornuot C, et al. Toxin-triggered interleukin-1 receptor signaling enables early-life discrimination of pathogenic versus commensal skin bacteria. Cell Host Microbe. (2019) 26:795–809.e5. doi: 10.1016/j.chom.2019.10.007
19. Levy M, Thaiss CA, Zeevi D, Dohnalová L, Zilberman-Schapira G, Mahdi JA, et al. Microbiota-modulated metabolites shape the intestinal microenvironment by regulating NLRP6 inflammasome signaling. Cell. (2015) 163:1428–43. doi: 10.1016/j.cell.2015.10.048
20. Schirmer M, Smeekens SP, Vlamakis H, Jaeger M, Oosting M, Franzosa EA, et al. Linking the human gut microbiome to inflammatory cytokine production capacity. Cell. (2016) 167:1125–36.e8. doi: 10.1016/j.cell.2016.10.020
21. Al Nabhani Z, Eberl G. Imprinting of the immune system by the microbiota early in life. Mucosal Immunol. (2020) 13:183–9. doi: 10.1038/s41385-020-0257-y
22. Stiemsma LT, Michels KB. The Role of the microbiome in the developmental origins of health and disease. Pediatrics. (2018) 141:e20172437. doi: 10.1542/peds.2017-2437
23. Fujimura KE, Demoor T, Rauch M, Faruqi AA, Jang S, Johnson CC, et al. House dust exposure mediates gut microbiome Lactobacillus enrichment and airway immune defense against allergens and virus infection. Proc Natl Acad Sci USA. (2014) 111:805–10. doi: 10.1073/pnas.1310750111
24. Levin AM, Sitarik AR, Havstad SL, Fujimura KE, Wegienka G, Cassidy-Bushrow AE, et al. Joint effects of pregnancy, sociocultural, and environmental factors on early life gut microbiome structure and diversity. Sci Rep. (2016) 6:1–16. doi: 10.1038/srep31775
25. Ahmadizar F, Vijverberg SJH, Arets HGM, de Boer A, Turner S, Devereux G, et al. Early life antibiotic use and the risk of asthma and asthma exacerbations in children. Pediatr Allergy Immunol. (2017) 28:430–7. doi: 10.1111/pai.12725
26. Chu DM, Ma J, Prince AL, Antony KM, Seferovic MD, Aagaard KM. Maturation of the infant microbiome community structure and function across multiple body sites and in relation to mode of delivery. Nat Med. (2017) 23:314–26. doi: 10.1038/nm.4272
27. Korpela K, Costea P, Coelho LP, Kandels-Lewis S, Willemsen G, Boomsma DI, et al. Selective maternal seeding and environment shape the human gut microbiome. Genome Res. (2018) 28:561–8. doi: 10.1101/gr.233940.117
28. O'Connor GT, Lynch SV, Bloomberg GR, Kattan M, Wood RA, Gergen PJ, et al. Early-life home environment and risk of asthma among inner-city children. J Allergy Clin Immunol. (2018) 141:1468–75. doi: 10.1016/j.jaci.2017.06.040
29. Reyman M, van Houten MA, van Baarle D, Bosch AATM, Man WH, Chu MLJN, et al. Impact of delivery mode-associated gut microbiota dynamics on health in the first year of life. Nat Commun. (2019) 10:4997. doi: 10.1038/s41467-019-13014-7
30. Durack J, Boushey HA, Lynch SV. Airway microbiota and the implications of dysbiosis in asthma. Curr Allergy Asthma Rep. (2016) 16:52. doi: 10.1007/s11882-016-0631-8
31. Abdel-Aziz MI, Vijverberg SJH, Neerincx AH, Kraneveld AD, Maitland-van der Zee AH. The crosstalk between microbiome and asthma: exploring associations and challenges. Clin Exp Allergy. (2019) 49:1067–86. doi: 10.1111/cea.13444
32. Durack J, Lynch SV. The gut microbiome: relationships with disease and opportunities for therapy. J Exp Med. (2019) 216:20–40. doi: 10.1084/jem.20180448
33. Frati F, Salvatori C, Incorvaia C, Bellucci A, Di Cara G, Marcucci F, et al. The role of the microbiome in asthma: the gut–lung axis. Int J Mol Sci. (2019) 20:1–12. doi: 10.3390/ijms20010123
34. Kumpitsch C, Koskinen K, Schöpf V, Moissl-Eichinger C. The microbiome of the upper respiratory tract in health and disease. BMC Biol. (2019) 17:87. doi: 10.1186/s12915-019-0703-z
35. Bevivino A, Bacci G, Drevinek P, Nelson MT, Hoffman L, Mengoni A. Deciphering the ecology of cystic fibrosis bacterial communities: towards systems-level integration. Trends Mol Med. (2019) 25:1110–22. doi: 10.1016/j.molmed.2019.07.008
36. Héry-Arnaud G, Boutin S, Cuthbertson L, Elborn SJ, Tunney MM. The lung and gut microbiome: what has to be taken into consideration for cystic fibrosis? J Cyst Fibros. (2019) 18:13–21. doi: 10.1016/j.jcf.2018.11.003
37. Françoise A, Héry-Arnaud G. The microbiome in cystic fibrosis pulmonary disease. Genes. (2020) 11:536. doi: 10.3390/genes11050536
38. Rackaityte E, Halkias J, Fukui EM, Mendoza VF, Hayzelden C, Crawford ED, et al. Viable bacterial colonization is highly limited in the human intestine in utero. Nat Med. (2020) 26:599–607. doi: 10.1038/s41591-020-0761-3
39. Durack J, Kimes NE, Lin DL, Rauch M, McKean M, McCauley K, et al. Delayed gut microbiota development in high-risk for asthma infants is temporarily modifiable by Lactobacillus supplementation. Nat Commun. (2018) 9:707. doi: 10.1038/s41467-018-03157-4
40. Ferretti P, Pasolli E, Tett A, Asnicar F, Gorfer V, Fedi S, et al. Mother-to-infant microbial transmission from different body sites shapes the developing infant gut microbiome. Cell Host Microbe. (2018) 24:133–45.e5. doi: 10.1016/j.chom.2018.06.005
41. Stinson LF, Boyce MC, Payne MS, Keelan JA. The not-so-sterile womb: evidence that the human fetus is exposed to bacteria prior to birth. Front Microbiol. (2019) 10:1124. doi: 10.3389/fmicb.2019.01124
42. Bäckhed F, Roswall J, Peng Y, Feng Q, Jia H, Kovatcheva-Datchary P, et al. Dynamics and stabilization of the human gut microbiome during the first year of life. Cell Host Microbe. (2015) 17:690–703. doi: 10.1016/j.chom.2015.04.004
43. Dominguez-Bello MG, Costello EK, Contreras M, Magris M, Hidalgo G, Fierer N, et al. Delivery mode shapes the acquisition and structure of the initial microbiota across multiple body habitats in newborns. Proc Natl Acad Sci USA. (2010) 107:11971–5. doi: 10.1073/pnas.1002601107
44. Wampach L, Heintz-Buschart A, Fritz JV, Ramiro-Garcia J, Habier J, Herold M, et al. Birth mode is associated with earliest strain-conferred gut microbiome functions and immunostimulatory potential. Nat Commun. (2018) 9:5091. doi: 10.1038/s41467-018-07631-x
45. Shao Y, Forster SC, Tsaliki E, Vervier K, Strang A, Simpson N, et al. Stunted microbiota and opportunistic pathogen colonization in caesarean-section birth. Nature. (2019) 574:117–21. doi: 10.1038/s41586-019-1560-1
46. Li H, Wang J, Wu L, Luo J, Liang X, Xiao B, et al. The impacts of delivery mode on infant's oral microflora. Sci Rep. (2018) 8:1–6. doi: 10.1038/s41598-018-30397-7
47. Marsh RL, Kaestli M, Chang AB, Binks MJ, Pope CE, Hoffman LR, et al. The microbiota in bronchoalveolar lavage from young children with chronic lung disease includes taxa present in both the oropharynx and nasopharynx. Microbiome. (2016) 4:1–18. doi: 10.1186/s40168-016-0182-1
48. Yassour M, Jason E, Hogstrom LJ, Arthur TD, Tripathi S, Siljander H, et al. Strain-level analysis of mother-to-child bacterial transmission during the first few months of life. Cell Host Microbe. (2018) 24:146–54.e4. doi: 10.1016/j.chom.2018.06.007
49. Ta LDH, Yap GC, Tay CJX, Lim ASM, Huang CH, Chu CW, et al. Establishment of the nasal microbiota in the first 18 months of life: correlation with early-onset rhinitis and wheezing. J Allergy Clin Immunol. (2018) 142:86–95. doi: 10.1016/j.jaci.2018.01.032
50. Teo SM, Tang HHF, Mok D, Judd LM, Watts SC, Pham K, et al. Airway microbiota dynamics uncover a critical window for interplay of pathogenic bacteria and allergy in childhood respiratory disease. Cell Host Microbe. (2018) 24:341–52.e5. doi: 10.1016/j.chom.2018.08.005
51. Grier A, McDavid A, Wang B, Qiu X, Java J, Bandyopadhyay S, et al. Neonatal gut and respiratory microbiota: coordinated development through time and space. Microbiome. (2018) 6:1–19. doi: 10.1186/s40168-018-0566-5
52. Franzosa EA, Huang K, Meadow JF, Gevers D, Lemon KP, Bohannan BJM, et al. Identifying personal microbiomes using metagenomic codes. Proc Natl Acad Sci USA. (2015) 112:E2930–8. doi: 10.1073/pnas.1423854112
53. Blekhman R, Goodrich JK, Huang K, Sun Q, Bukowski R, Bell JT, et al. Host genetic variation impacts microbiome composition across human body sites. Genome Biol. (2015) 16:1–12. doi: 10.1186/s13059-015-0759-1
54. Turpin W, Espin-Garcia O, Xu W, Silverberg MS, Kevans D, Smith MI, et al. Association of host genome with intestinal microbial composition in a large healthy cohort. Nat Genet. (2016) 48:1413–7. doi: 10.1038/ng.3693
55. Rothschild D, Weissbrod O, Barkan E, Kurilshikov A, Korem T, Zeevi D, et al. Environment dominates over host genetics in shaping human gut microbiota. Nature. (2018) 555:210–5. doi: 10.1038/nature25973
56. Scepanovic P, Hodel F, Mondot S, Partula V, Byrd A, Hammer C, et al. A comprehensive assessment of demographic, environmental, and host genetic associations with gut microbiome diversity in healthy individuals. Microbiome. (2019) 7:1–15. doi: 10.1186/s40168-019-0747-x
57. Goodrich JK, Waters JL, Poole AC, Sutter JL, Koren O, Blekhman R, et al. Human genetics shape the gut microbiome. Cell. (2014) 159:789–99. doi: 10.1016/j.cell.2014.09.053
58. Schloss PD, Iverson KD, Petrosino JF, Schloss SJ. The dynamics of a family's gut microbiota reveal variations on a theme. Microbiome. (2014) 2:1–13. doi: 10.1186/2049-2618-2-25
59. Lax S, Smith DP, Hampton-Marcell J, Owens SM, Handley KM, Scott NM, et al. Longitudinal analysis of microbial interaction between humans and the indoor environment. Science. (2014) 345:1048–52. doi: 10.1126/science.1254529
60. Xie H, Guo R, Zhong H, Feng Q, Lan Z, Qin B, et al. Shotgun metagenomics of 250 adult twins reveals genetic and environmental impacts on the gut microbiome. Cell Syst. (2016) 3:572–84.e3. doi: 10.1016/j.cels.2016.10.004
61. Jakobsson HE, Abrahamsson TR, Jenmalm MC, Harris K, Quince C, Jernberg C, et al. Decreased gut microbiota diversity, delayed Bacteroidetes colonisation and reduced Th1 responses in infants delivered by caesarean section. Gut. (2014) 63:559–66. doi: 10.1136/gutjnl-2012-303249
62. Bokulich NA, Chung J, Battaglia T, Henderson N, Jay M, Li H, et al. Antibiotics, birth mode, and diet shape microbiome maturation during early life. Sci Transl Med. (2016) 8:1–14. doi: 10.1126/scitranslmed.aad7121
63. Stokholm J, Thorsen J, Chawes BL, Schjørring S, Krogfelt KA, Bønnelykke K, et al. Cesarean section changes neonatal gut colonization. J Allergy Clin Immunol. (2016) 138:881–9.e2. doi: 10.1016/j.jaci.2016.01.028
64. Yassour M, Vatanen T, Siljander H, Hämäläinen AM, Härkönen T, Ryhänen SJ, et al. Natural history of the infant gut microbiome and impact of antibiotic treatment on bacterial strain diversity and stability. Sci Transl Med. (2016) 8:343ra81. doi: 10.1126/scitranslmed.aad0917
65. Tapiainen T, Koivusaari P, Brinkac L, Lorenzi HA, Salo J, Renko M, et al. Impact of intrapartum and postnatal antibiotics on the gut microbiome and emergence of antimicrobial resistance in infants. Sci Rep. (2019) 9:1–11. doi: 10.1038/s41598-019-46964-5
66. Fouhy F, Watkins C, Hill CJ, O'Shea CA, Nagle B, Dempsey EM, et al. Perinatal factors affect the gut microbiota up to four years after birth. Nat Commun. (2019) 10:1–10. doi: 10.1038/s41467-019-09252-4
67. Baumann-Dudenhoeffer AM, D'Souza AW, Tarr PI, Warner BB, Dantas G. Infant diet and maternal gestational weight gain predict early metabolic maturation of gut microbiomes. Nat Med. (2018) 24:1822–9. doi: 10.1038/s41591-018-0216-2
68. Stokholm J, Blaser MJ, Thorsen J, Rasmussen MA, Waage J, Vinding RK, et al. Maturation of the gut microbiome and risk of asthma in childhood. Nat Commun. (2018) 9:1–10. doi: 10.1038/s41467-017-02573-2
69. Bosch AATM, Piters WAAS, van Houten MA, Chu MLJN, Biesbroek G, Kool J, et al. Maturation of the infant respiratory microbiota, environmental drivers, health consequences. Am J Respir Crit Care Med. (2017) 196:1582–90. doi: 10.1164/rccm.201703-0554OC
70. Biesbroek G, Tsivtsivadze E, Sanders EAM, Montijn R, Veenhoven RH, Keijser BJF, et al. Early respiratory microbiota composition determines bacterial succession patterns and respiratory health in children. Am J Respir Crit Care Med. (2014) 190:1283–92. doi: 10.1164/rccm.201407-1240OC
71. Stein MM, Hrusch CL, Gozdz J, Igartua C, Pivniouk V, Murray SE, et al. Innate immunity and asthma risk in amish and hutterite farm children. N Engl J Med. (2016) 375:411–21. doi: 10.1056/NEJMoa1508749
72. Fonseca W, Lucey K, Jang S, Fujimura KE, Rasky A, Ting HA, et al. Lactobacillus johnsonii supplementation attenuates respiratory viral infection via metabolic reprogramming and immune cell modulation. Mucosal Immunol. (2017) 10:1569–80. doi: 10.1038/mi.2017.13
73. Debarry J, Garn H, Hanuszkiewicz A, Dickgreber N, Blümer N, von Mutius E, et al. Acinetobacter lwoffii and Lactococcus lactis strains isolated from farm cowsheds possess strong allergy-protective properties. J Allergy Clin Immunol. (2007) 119:1514–21. doi: 10.1016/j.jaci.2007.03.023
74. Hagner S, Harb H, Zhao M, Stein K, Holst O, Ege MJ, et al. Farm-derived gram-positive bacterium Staphylococcus sciuri W620 prevents asthma phenotype in HDM- and OVA-exposed mice. Allergy Eur J Allergy Clin Immunol. (2013) 68:322–9. doi: 10.1111/all.12094
75. Fall T, Lundholm C, Örtqvist AK, Fall K, Fang F, Hedhammar Å, et al. Early exposure to dogs and farm animals and the risk of childhood asthma. JAMA Pediatr. (2015) 169:e153219. doi: 10.1001/jamapediatrics.2015.3219
76. Tun HM, Konya T, Takaro TK, Brook JR, Chari R, Field CJ, et al. Exposure to household furry pets influences the gut microbiota of infants at 3-4 months following various birth scenarios. Microbiome. (2017) 5:1–14. doi: 10.1186/s40168-017-0254-x
77. Dhakal S, Wang L, Antony L, Rank J, Bernardo P, Ghimire S, et al. Amish. (rural) vs. Non-amish. (urban) infant fecal microbiotas are highly diverse and their transplantation lead to differences in mucosal immune maturation in a humanized germfree piglet model. Front Immunol. (2019) 10:1509. doi: 10.3389/fimmu.2019.01509
78. Kirjavainen PV, Karvonen AM, Adams RI, Täubel M, Roponen M, Tuoresmäki P, et al. Farm-like indoor microbiota in non-farm homes protects children from asthma development. Nat Med. (2019) 25:1089–95. doi: 10.1038/s41591-019-0469-4
79. Seedorf H, Griffin NW, Ridaura VK, Reyes A, Cheng J, Rey FE, et al. Bacteria from diverse habitats colonize and compete in the mouse gut. Cell. (2014) 159:253–66. doi: 10.1016/j.cell.2014.09.008
80. Arrieta MC, Stiemsma LT, Dimitriu PA, Thorson L, Russell S, Yurist-Doutsch S, et al. Early infancy microbial and metabolic alterations affect risk of childhood asthma. Sci Transl Med. (2015) 7:307ra152. doi: 10.1126/scitranslmed.aab2271
81. Fujimura KE, Sitarik AR, Havstad S, Lin DL, Levan S, Fadrosh D, et al. Neonatal gut microbiota associates with childhood multisensitized atopy and T cell differentiation. Nat Med. (2016) 22:1187–91. doi: 10.1038/nm.4176
82. Man WH, Clerc M, de Steenhuijsen Piters WAA, van Houten MA, Chu MLJN, Kool J, et al. Loss of microbial topography between oral and nasopharyngeal microbiota and development of respiratory infections early in life. Am J Respir Crit Care Med. (2019) 200:760–70. doi: 10.1164/rccm.201810-1993OC
83. Torow N, Hornef MW. The neonatal window of opportunity: setting the stage for life-long host-microbial interaction and immune homeostasis. J Immunol. (2017) 198:557–63. doi: 10.4049/jimmunol.1601253
84. Constantinides MG, Link VM, Tamoutounour S, Wong AC, Perez-Chaparro PJ, Han S-J, et al. MAIT cells are imprinted by the microbiota in early life and promote tissue repair. Science. (2019) 366:eaax6624. doi: 10.1126/science.aax6624
85. Cervantes-Barragan L, Chai JN, Tianero MD, Di Luccia B, Ahern PP, Merriman J, et al. Lactobacillus reuteri induces gut intraepithelial CD4+CD8αα+ T cells. Science. (2017) 357:806–10. doi: 10.1126/science.aah5825
86. Ohnmacht C, Park JH, Cording S, Wing JB, Atarashi K, Obata Y, et al. The microbiota regulates type 2 immunity through RORγt+ T cells. Science. (2015) 349:989–93. doi: 10.1126/science.aac4263
87. Wrzosek L, Miquel S, Noordine ML, Bouet S, Chevalier-Curt MJ, Robert V, et al. Bacteroides thetaiotaomicron and Faecalibacterium prausnitzii influence the production of mucus glycans and the development of goblet cells in the colonic epithelium of a gnotobiotic model rodent. BMC Biol. (2013) 11:61. doi: 10.1186/1741-7007-11-61
88. Remot A, Descamps D, Noordine ML, Boukadiri A, Mathieu E, Robert V, et al. Bacteria isolated from lung modulate asthma susceptibility in mice. ISME J. (2017) 11:1061–74. doi: 10.1038/ismej.2016.181
89. Hapfelmeier S, Lawson MAE, Slack E, Kirundi JK, Stoel M, Heikenwalder M, et al. Reversible microbial colonization of germ-free mice reveals the dynamics of IgA immune responses. Science. (2010) 328:1705–9. doi: 10.1126/science.1188454
90. Moor K, Diard M, Sellin ME, Felmy B, Wotzka SY, Toska A, et al. High-avidity IgA protects the intestine by enchaining growing bacteria. Nature. (2017) 544:498–502. doi: 10.1038/nature22058
91. Ruiz VE, Battaglia T, Kurtz ZD, Bijnens L, Ou A, Engstrand I, et al. A single early-in-life macrolide course has lasting effects on murine microbial network topology and immunity. Nat Commun. (2017) 8:518. doi: 10.1038/s41467-017-00531-6
92. Lynn MA, Tumes DJ, Choo JM, Sribnaia A, Blake SJ, Leong LEX, et al. Early-life antibiotic-driven dysbiosis leads to dysregulated vaccine immune responses in mice. Cell Host Microbe. (2018) 23:653–60.e5. doi: 10.1016/j.chom.2018.04.009
93. Olszak T, An D, Zeissig S, Vera MP, Richter J, Franke A, et al. Microbial exposure during early life has persistent effects on natural killer T cell function. Science. (2012) 336:489–93. doi: 10.1126/science.1219328
94. Russell SL, Gold MJ, Hartmann M, Willing BP, Thorson L, Wlodarska M, et al. Early life antibiotic-driven changes in microbiota enhance susceptibility to allergic asthma. EMBO Rep. (2012) 13:440–7. doi: 10.1038/embor.2012.32
95. Cait A, Hughes MR, Antignano F, Cait J, Dimitriu PA, Maas KR, et al. Microbiome-driven allergic lung inflammation is ameliorated by short-chain fatty acids. Mucosal Immunol. (2018) 11:785–95. doi: 10.1038/mi.2017.75
96. Corrêa-Oliveira R, Fachi JL, Vieira A, Sato FT, Vinolo MAR. Regulation of immune cell function by short-chain fatty acids. Clin Transl Immunol. (2016) 5:1–8. doi: 10.1038/cti.2016.17
97. Gollwitzer ES, Saglani S, Trompette A, Yadava K, Sherburn R, McCoy KD, et al. Lung microbiota promotes tolerance to allergens in neonates via PD-L1. Nat Med. (2014) 20:642–7. doi: 10.1038/nm.3568
98. Pitter G, Ludvigsson JF, Romor P, Zanier L, Zanotti R, Simonato L, et al. Antibiotic exposure in the first year of life and later treated asthma, a population based birth cohort study of 143,000 children. Eur J Epidemiol. (2016) 31:85–94. doi: 10.1007/s10654-015-0038-1
99. Loewen K, Monchka B, Mahmud SM, Jong G, Azad MB. Prenatal antibiotic exposure and childhood asthma: a population-based study. Eur Respir J. (2018) 52:1702070. doi: 10.1183/13993003.02070-2017
100. Dinwiddie DL, Denson JL, Kennedy JL. Role of the airway microbiome in respiratory infections and asthma in children. Pediatr Allergy Immunol Pulmonol. (2018) 31:236–40. doi: 10.1089/ped.2018.0958
101. Prevaes SMPJ, De Steenhuijsen Piters WAA, De Winter-De Groot KM, Janssens HM, Tramper-Stranders GA, Chu MLJN, et al. Concordance between upper and lower airway microbiota in infants with cystic fibrosis. Eur Respir J. (2017) 49:1602235. doi: 10.1183/13993003.02235-2016
102. An SQ, Warris A, Turner S. Microbiome characteristics of induced sputum compared to bronchial fluid and upper airway samples. Pediatr Pulmonol. (2018) 53:921–8. doi: 10.1002/ppul.24037
103. Kloepfer KM, Deschamp AR, Ross SE, Peterson-Carmichael SL, Hemmerich CM, Rusch DB, et al. In children, the microbiota of the nasopharynx and bronchoalveolar lavage fluid are both similar and different. Pediatr Pulmonol. (2018) 53:475–82. doi: 10.1002/ppul.23953
104. Chun Y, Do A, Grishina G, Grishin A, Fang G, Rose S, et al. Integrative study of the upper and lower airway microbiome and transcriptome in asthma. JCI Insight. (2020) 5:1–16. doi: 10.1172/jci.insight.133707
105. Dickson RP, Erb-Downward JR, Freeman CM, Mccloskey L, Falkowski NR, Huffnagle GB. Bacterial topography of the healthyhuman lower respiratory tract. MBio. (2017) 8:1–12. doi: 10.1128/mBio.02287-16
106. Ahmed B, Cox MJ, Cuthbertson L, James PL, Cookson WOC, Davies JC, et al. Comparison of the upper and lower airway microbiota in children with chronic lung diseases. PLoS ONE. (2018) 13:1156. doi: 10.1371/journal.pone.0201156
107. Man WH, van Houten MA, Mérelle ME, Vlieger AM, Chu MLJN, Jansen NJG, et al. Bacterial and viral respiratory tract microbiota and host characteristics in children with lower respiratory tract infections: a matched case-control study. Lancet Respir Med. (2019) 7:417–26. doi: 10.1016/S2213-2600(18)30449-1
108. Denner DR, Sangwan N, Becker JB, Hogarth DK, Oldham J, Castillo J, et al. Corticosteroid therapy and airflow obstruction influence the bronchial microbiome, which is distinct from that of bronchoalveolar lavage in asthmatic airways. J Allergy Clin Immunol. (2016) 137:1398–1405.e3. doi: 10.1016/j.jaci.2015.10.017
109. Pérez-Losada M, Crandall KA, Freishtat RJ. Two sampling methods yield distinct microbial signatures in the nasopharynges of asthmatic children. Microbiome. (2016) 4:1–6. doi: 10.1186/s40168-016-0170-5
110. Luna PN, Hasegawa K, Ajami NJ, Espinola JA, Henke DM, Petrosino JF, et al. The association between anterior nares and nasopharyngeal microbiota in infants hospitalized for bronchiolitis. Microbiome. (2018) 6:1–14. doi: 10.1186/s40168-017-0385-0
111. Teo SM, Mok D, Pham K, Kusel M, Serralha M, Troy N, et al. The infant nasopharyngeal microbiome impacts severity of lower respiratory infection and risk of asthma development. Cell Host Microbe. (2015) 17:704–15. doi: 10.1016/j.chom.2015.03.008
112. McCauley K, Durack J, Valladares R, Fadrosh DW, Lin DL, Calatroni A, et al. Distinct nasal airway bacterial microbiotas differentially relate to exacerbation in pediatric patients with asthma. J Allergy Clin Immunol. (2019) 144, 1187–97. doi: 10.1016/j.jaci.2019.05.035
113. Toivonen L, Hasegawa K, Waris M, Ajami NJ, Petrosino JF, Camargo CA, et al. Early nasal microbiota and acute respiratory infections during the first years of life. Thorax. (2019) 74:592–9. doi: 10.1136/thoraxjnl-2018-212629
114. Zhou Y, Jackson D, Bacharier LB, Mauger D, Boushey H, Castro M, et al. The upper-airway microbiota and loss of asthma control among asthmatic children. Nat Commun. (2019) 10:1–10. doi: 10.1038/s41467-019-13698-x
115. Kloepfer KM, Lee WM, Pappas TE, Kang TJ, Vrtis RF, Evans MD, et al. Detection of pathogenic bacteria during rhinovirus infection is associated with increased respiratory symptoms and asthma exacerbations. J Allergy Clin Immunol. (2014) 133:1301–7.e3. doi: 10.1016/j.jaci.2014.02.030
116. Sonawane AR, Tian L, Chu CY, Qiu X, Wang L, Holden-Wiltse J, et al. Microbiome-transcriptome interactions related to severity of respiratory syncytial virus infection. Sci Rep. (2019) 9:1–14. doi: 10.1038/s41598-019-50217-w
117. De Steenhuijsen Piters WAA, Heinonen S, Hasrat R, Bunsow E, Smith B, Suarez-Arrabal MC, et al. Nasopharyngeal microbiota, host transcriptome, and disease severity in children with respiratory syncytial virus infection. Am J Respir Crit Care Med. (2016) 194:1104–15. doi: 10.1164/rccm.201602-0220OC
118. Brealey JC, Chappell KJ, Galbraith S, Fantino E, Gaydon J, Tozer S, et al. Streptococcus pneumoniae colonization of the nasopharynx is associated with increased severity during respiratory syncytial virus infection in young children. Respirology. (2018) 23:220–7. doi: 10.1111/resp.13179
119. Ederveen THA, Ferwerda G, Ahout IM, Vissers M, de Groot R, Boekhorst J, et al. Haemophilus is overrepresented in the nasopharynx of infants hospitalized with RSV infection and associated with increased viral load and enhanced mucosal CXCL8 responses. Microbiome. (2018) 6:1–13. doi: 10.1186/s40168-017-0395-y
120. Lehtinen MJ, Hibberd AA, Männikkö S, Yeung N, Kauko T, Forssten S, et al. Nasal microbiota clusters associate with inflammatory response, viral load, and symptom severity in experimental rhinovirus challenge. Sci Rep. (2018) 8:1–12. doi: 10.1038/s41598-018-29793-w
121. Hasegawa K, Mansbach JM, Ajami NJ, Espinola JA, Henke DM, Petrosino JF, et al. Association of nasopharyngeal microbiota profiles with bronchiolitis severity in infants hospitalised for bronchiolitis. Eur Respir J. (2016) 48:1329–39. doi: 10.1183/13993003.00152-2016
122. Depner M, Ege MJ, Cox MJ, Dwyer S, Walker AW, Birzele LT, et al. Bacterial microbiota of the upper respiratory tract and childhood asthma. J Allergy Clin Immunol. (2017) 139:826–34.e13. doi: 10.1016/j.jaci.2016.05.050
123. Mansbach JM, Luna PN, Shaw CA, Hasegawa K, Petrosino JF, Piedra PA, et al. Increased Moraxella and Streptococcus species abundance after severe bronchiolitis is associated with recurrent wheezing. J Allergy Clin Immunol. (2020) 145:518–527.e8. doi: 10.1016/j.jaci.2019.10.034
124. Bomar L, Brugger SD, Yost BH, Davies SS, Lemon P. Corynebacterium accolens releases antipneumococcal free fatty acids from human nostril and skin surface triacylglycerols. MBio. (2016) 7:1–13. doi: 10.1128/mBio.01725-15
125. Yan M, Pamp SJ, Fukuyama J, Hwang PH, Cho DY, Holmes S, et al. Nasal microenvironments and interspecific interactions influence nasal microbiota complexity and S. aureus carriage. Cell Host Microbe. (2013) 14:631–40. doi: 10.1016/j.chom.2013.11.005
126. Hardy BL, Dickey SW, Plaut RD, Riggins DP, Stibitz S, Otto M, et al. Corynebacterium pseudodiphtheriticum exploits Staphylococcus aureus virulence components in a novel polymicrobial defense strategy. MBio. (2019) 10:1–24. doi: 10.1128/mBio.02491-18
127. Liu Q, Liu Q, Meng H, Lv H, Liu Y, Liu J, et al. Staphylococcus epidermidis contributes to healthy maturation of the nasal microbiome by stimulating antimicrobial peptide production. Cell Host Microbe. (2019) 27:68–78.e5. doi: 10.1016/j.chom.2019.11.003
128. Kim HJ, Jo A, Jeon YJ, An S, Lee KM, Yoon SS, et al. Nasal commensal Staphylococcus epidermidis enhances interferon-λ-dependent immunity against influenza virus. Microbiome. (2019) 7:1–12. doi: 10.1186/s40168-019-0691-9
129. Sender R, Fuchs S, Milo R. Are we really vastly outnumbered? Revisiting the ratio of bacterial to host cells in humans. Cell. (2016) 164:337–40. doi: 10.1016/j.cell.2016.01.013
130. Huseyin CE, O'Toole PW, Cotter PD, Scanlan PD. Forgotten fungi-the gut mycobiome in human health and disease. FEMS Microbiol Rev. (2017) 41:479–511. doi: 10.1093/femsre/fuw047
131. Koskinen K, Pausan MR, Perras AK, Beck M, Bang C, Mora M, et al. First insights into the diverse human archaeome: specific detection of archaea in the gastrointestinal tract, lung, and nose and on skin. MBio. (2017) 8:1–17. doi: 10.1128/mBio.00824-17
132. Nash AK, Auchtung TA, Wong MC, Smith DP, Gesell JR, Ross MC, et al. The gut mycobiome of the Human Microbiome Project healthy cohort. Microbiome. (2017) 5:153. doi: 10.1186/s40168-017-0373-4
133. Lokmer A, Cian A, Froment A, Gantois N, Viscogliosi E, Chabé M, et al. Use of shotgun metagenomics for the identification of protozoa in the gut microbiota of healthy individuals from worldwide populations with various industrialization levels. PLoS ONE. (2019) 14:e0211139. doi: 10.1371/journal.pone.0211139
134. Budden KF, Gellatly SL, Wood DLA, Cooper MA, Morrison M, Hugenholtz P, et al. Emerging pathogenic links between microbiota and the gut-lung axis. Nat Rev Microbiol. (2017) 15:55–63. doi: 10.1038/nrmicro.2016.142
135. Dang AT, Marsland BJ. Microbes, metabolites, and the gut–lung axis. Mucosal Immunol. (2019) 12:843–50. doi: 10.1038/s41385-019-0160-6
136. Sprooten RTM, Lenaerts K, Braeken DCW, Grimbergen I, Rutten EP, Wouters EFM, et al. Increased small intestinal permeability during severe acute exacerbations of COPD. Respiration. (2018) 95:334–42. doi: 10.1159/000485935
137. Keely S, Talley NJ, Hansbro PM. Pulmonary-intestinal cross-talk in mucosal inflammatory disease. Mucosal Immunol. (2012) 5:7–18. doi: 10.1038/mi.2011.55
138. Roussos A, Koursarakos P, Patsopoulos D, Gerogianni I, Philippou N. Increased prevalence of irritable bowel syndrome in patients with bronchial asthma. Respir Med. (2003) 97:75–9. doi: 10.1053/rmed.2001.1409
139. Rutten EPA, Lenaerts K, Buurman WA, Wouters EFM. Disturbed intestinal integrity in patients with COPD : effects of activities of daily living. Chest. (2014) 145:245–52. doi: 10.1378/chest.13-0584
140. Deshmukh F, Vasudevan A, Mengalie E. Association between irritable bowel syndrome and asthma: a meta-analysis and systematic review. Ann Gastroenterol. (2019) 32:570–7. doi: 10.20524/aog.2019.0426
141. Hasegawa K, Linnemann RW, Mansbach JM, Ajami NJ, Espinola JA, Petrosino JF, et al. The fecal microbiota profile and bronchiolitis in infants. Pediatrics. (2016) 138:e20160218. doi: 10.1542/peds.2016-0218
142. Lundström SL, Yang J, Källberg HJ, Thunberg S, Gafvelin G, Haeggström JZ, et al. Allergic asthmatics show divergent lipid mediator profiles from healthy controls both at baseline and following birch pollen provocation. PLoS ONE. (2012) 7:e33780. doi: 10.1371/journal.pone.0033780
143. Levan SR, Stamnes KA, Lin DL, Panzer AR, Fukui E, Mccauley K, et al. Elevated fecal 12,13-diHOME concentration in neonates at high risk for asthma is produced by gut bacteria and impedes immune tolerance. Nat Microbiol. (2019) 4:2020. doi: 10.1038/s41564-019-0574-7
144. Lynes MD, Leiria LO, Lundh M, Bartelt A, Shamsi F, Huang TL, et al. The cold-induced lipokine 12,13-diHOME promotes fatty acid transport into brown adipose tissue. Nat Med. (2017) 23:631–7. doi: 10.1038/nm.4297
145. Ho NT, Li F, Lee-Sarwar KA, Tun HM, Brown BP, Pannaraj PS, et al. Meta-analysis of effects of exclusive breastfeeding on infant gut microbiota across populations. Nat Commun. (2018) 9:4169. doi: 10.1038/s41467-018-06473-x
146. Tromp I, Jong JK, De Raat H, Jaddoe V, Franco O, Hofman A, et al. Breastfeeding and the risk of respiratory tract infections after infancy: the generation R study. PLoS ONE. (2017) 12:1–12:e1272763. doi: 10.1371/journal.pone.0172763
147. McKeen S, Young W, Mullaney J, Fraser K, McNabb WC, Roy NC. Infant complementary feeding of prebiotics for the microbiome and immunity. Nutrients. (2019) 11:25–33. doi: 10.3390/nu11020364
148. De Filippo C, Cavalieri D, Di Paola M, Ramazzotti M, Poullet JB, Massart S, et al. Impact of diet in shaping gut microbiota revealed by a comparative study in children from Europe and rural Africa. Proc Natl Acad Sci USA. (2010) 107:14691–6. doi: 10.1073/pnas.1005963107
149. Chen T, Long W, Zhang C, Liu S, Zhao L, Hamaker BR. Fiber-utilizing capacity varies in Prevotella- versus Bacteroides-dominated gut microbiota. Sci Rep. (2017) 7:1–7. doi: 10.1038/s41598-017-02995-4
150. Gopalsamy G, Mortimer E, Greenfield P, Bird AR, Young GP, Christophersen CT. Resistant starch is actively fermented by infant faecal microbiota and increases microbial diversity. Nutrients. (2019) 11:1–16. doi: 10.3390/nu11061345
151. Wang Y, Mortimer EK, Katundu KGH, Kalanga N, Leong LEX, Gopalsamy GL, et al. The capacity of the fecal microbiota from Malawian infants to ferment resistant starch. Front Microbiol. (2019) 10:1459. doi: 10.3389/fmicb.2019.01459
152. Donovan SM. Introduction to the special focus issue on the impact of diet on gut microbiota composition and function and future opportunities for nutritional modulation of the gut microbiome to improve human health. Gut Microbes. (2017) 8:75–81. doi: 10.1080/19490976.2017.1299309
153. Fukuda S, Toh H, Hase K, Oshima K, Nakanishi Y, Yoshimura K, et al. Bifidobacteria can protect from enteropathogenic infection through production of acetate. Nature. (2011) 469:543–7. doi: 10.1038/nature09646
154. Thorburn AN, McKenzie CI, Shen S, Stanley D, MacIa L, Mason LJ, et al. Evidence that asthma is a developmental origin disease influenced by maternal diet and bacterial metabolites. Nat Commun. (2015) 6:7320. doi: 10.1038/ncomms8320
155. Chiu CY, Cheng ML, Chiang MH, Kuo YL, Tsai MH, Chiu CC, et al. Gut microbial-derived butyrate is inversely associated with IgE responses to allergens in childhood asthma. Pediatr Allergy Immunol. (2019) 30:689–97. doi: 10.1111/pai.13096
156. Li KL, Wang BZ, Li ZP, Li YL, Liang JJ. Alterations of intestinal flora and the effects of probiotics in children with recurrent respiratory tract infection. World J Pediatr. (2019) 15:255–61. doi: 10.1007/s12519-019-00248-0
157. Chan CKY, Tao J, Chan OS, Li H-B, Pang H. Preventing respiratory tract infections by synbiotic interventions: a systematic review and meta-analysis of randomized controlled trials. Adv Nutr. (2020) 11:979–88. doi: 10.1093/advances/nmaa003
158. Azad MB, Coneys GJ, Kozyrskyj AL, Field CJ, Ramsey CD, Becker AB, et al. Probiotic supplementation during pregnancy or infancy for the prevention of asthma and wheeze: systematic review and meta-Analysis. BMJ. (2013) 347:f6471. doi: 10.1136/bmj.f6471
159. Cuello-Garcia CA, Brozek JL, Fiocchi A, Pawankar R, Yepes-Nuñez JJ, Terracciano L, et al. Probiotics for the prevention of allergy: a systematic review and meta-analysis of randomized controlled trials. J Allergy Clin Immunol. (2015) 136:952–61. doi: 10.1016/j.jaci.2015.04.031
160. Zuccotti G, Meneghin F, Aceti A, Barone G, Callegari ML, Di Mauro A, et al. Probiotics for prevention of atopic diseases in infants: systematic review and meta-analysis. Allergy Eur J Allergy Clin Immunol. (2015) 70:1356–71. doi: 10.1111/all.12700
Keywords: gut microbiome, microbiome development, asthma, acute respiratory disease, respiratory microbiome
Citation: Durack J and Christophersen CT (2020) Human Respiratory and Gut Microbiomes—Do They Really Contribute to Respiratory Health? Front. Pediatr. 8:528. doi: 10.3389/fped.2020.00528
Received: 18 March 2020; Accepted: 24 July 2020;
Published: 03 September 2020.
Edited by:
Bruce K. Rubin, Children's Hospital of Richmond at VCU, United StatesReviewed by:
Andre Schultz, Perth Children's Hospital, AustraliaLeah Cuthbertson, Imperial College London, United Kingdom
Copyright © 2020 Durack and Christophersen. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Juliana Durack, anVsaWFAc3ltYmlvbWUuY29t
 Juliana Durack
Juliana Durack Claus T. Christophersen
Claus T. Christophersen