- 1Division of Neonatology, Department of Pediatric, The Affiliated Hospital of Inner Mongolia Medical University, Hohhot, China
- 2Clinical Medical Research Center of the Affiliated Hospital, Inner Mongolia Medical University, Hohhot, China
Aim: To study the relationship between rs1059057 polymorphism of pulmonary surfactant protein A1 (SP-A1) and respiratory distress syndrome (RDS) in Mongolian very premature infants.
Methods: Applying the strategy of case-control study, 120 Mongolian RDS very premature infants (58 males and 62 females) in the western part of Inner Mongolia were selected as the case group, and 120 subjects of non-RDS very premature infants (56 males and 64 females) with the same nationality, same sex and similar gestational age were used as the control group. The single nucleotide polymorphism (SNP) site rs1059057 of SP-A1 was genotyped using polymerase chain reaction-single strand conformational polymorphism (PCR-SSCP).
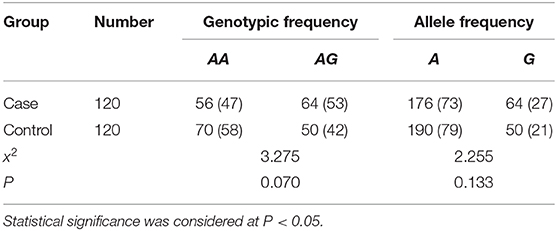
Results: Two genotypes, A/G and A/A, were detected at the SP-A1 rs1059057 locus in the western part of Inner Mongolia. In the case group, the frequencies of two genotypes were 53 and 47%, and the frequencies of A allele and G allele were 73 and 27%, respectively. In the control group, the frequencies of the two genotypes were 42 and 58%, and the frequencies of A allele and G allele were 79 and 21%, respectively. There was no significant difference in the genotype frequency of SP-A1 (rs1059057) locus between the case group and the control group (X2 = 3.275, P > 0.05), and no significant difference in allele frequency between the case group and the control group (X2 = 2.255, P > 0.05).
Conclusion: The genotypes and allele frequencies of SP-A1 (rs1059057) locus were not associated with the incidence of RDS in Mongolian very premature infants in western Inner Mongolia.
Introduction
Neonatal respiratory distress syndrome (NRDS), also known as hyaline membrane disease, is mainly due to the lack of pulmonary surfactant (PS), which leads to an increase in alveolar wall surface tension and decreased pulmonary compliance, and initiates newborn's sexual dyspnea shortly after birth, even clinical syndromes of respiratory failure (1). NRDS often occurs in premature infants, especially in premature infants within 34 weeks (2). The probability of NRDS in premature infants with gestational age within 28 weeks is up to 80% (3, 4). At present, international and domestic medical technologies are developing rapidly, and the levels of medical treatment in neonatal ward are also constantly improving. The survival problem of NRDS children has been basically solved, yet prognosis and treatment have remained difficult. Therefore, the respiratory tract management problems of children with NRDS should not be underestimated, we still need to continue to strive our efforts on them.
Pulmonary surfactant protein-A1 is a kind of alveolar cell surfactant protein synthesized and released by type II alveolar epithelium. The causes of NRDS are very complicated and there are many different opinions nowdays, but many studies have found that the lack and abnormal changes of SP-A1 are the main cause of NRDS (5–7). Studies have demonstrated that the incidence of NRDS may be related to SP-A gene polymorphism (8, 9). Through the cDNA sequence analysis of SP-A1, it is confirmed that SP-A1 has four alleles (6A, 6A2, 6A3, and 6A4), and different gene mutations can make the expression of SP-A1 abnormal, which leads to the occurrence of respiratory diseases (10). It is worth noting that there are some differences in the relationship between SP-A gene polymorphism and diseases among different regions, races, and ethnic groups (11). Therefore, our team detected the polymorphism of SP-A1 gene locus in Mongolian very premature infants in western Inner Mongolia to explore its role in NRDS etiology. This paper mainly introduces the relationship between the gene polymorphism rs1059057 of SP-A1 and the RDS of Mongolian very premature infants, in order to provide help for the rescue of Mongolian very premature infants in the western part of Inner Mongolia.
Subjects and Methods
Subjects
Applying the strategy of case-control study, one hundred and twenty Mongolian RDS very premature infants (58 males and 62 females) who were hospitalized in the department of neonatal pediatrics in our hospital from January 2012 to January 2019 were selected as the case group. The selection criteria were as follows: ➀ the immediate family members have lived in Mongolians in the western part of Inner Mongolia for at least three generations. ➁ The sex ratio was roughly balanced, the birth weight was 0.138~ 0.184 kg, and the birth weight was 28+3 weeks ≤ the gestational age <32 weeks. ➂ In accordance with the diagnostic criteria of RDS issued in Europe (4). One hundred and twenty Mongolian non-RDS very early infants (56 males and 64 females) who were hospitalized in the department of neonatal pediatrics in our hospital with the same period, race, sex, and gestational age were selected as the control group. The subjects were selected according to the following criteria: ➀ the immediate family members have lived in Mongolians in the western part of Inner Mongolia for at least three generations. ➁ The sex ratio was roughly balanced, the birth weight was 0.142~0.190 kg, 29+1 week ≤ the gestational age <32 weeks. ➂ The common chest X-ray showed no pulmonary inflammation and RDS, and the blood routine and C-reactive protein examination showed no obvious infection.
The following subjects were excluded: ➀ congenital or genetic metabolic diseases; ➁ laboratory examination showed severe infection; ➂ severe history of intrauterine or postnatal asphyxia; ➃ gestational diabetes mellitus; ➄ other diseases that may be accompanied by respiratory symptoms; ➅ there are other diseases and related factors that may affect the experimental results.
Materials and Reagents
Blood Genome DNA extraction Kit and the main regents of PCR were from Sangon Bioengineering Co., Ltd (Shanghai, China). 6 × DNA Loading Dye and DNA Ladder Mix (100–10,000 bp) were from ThemoFisher (R0611 and SM0332). SanPrep column PCR product purification kit was also from Sangon Bioengineering Co., Ltd (Shanghai, China). Main reagent of DNA sequencing were from ThermoFisher (Applied Biosystems™).
Sample Collection and Processing
The venous blood of Mongolian RDS very premature infants and non-RDS very premature infants in western Inner Mongolia was collected and stored at −80°C. At the same time, the clinical data of gestational age, sex, and birth weight of very premature infants in the experimental group were collected.
Extraction and Detection of Genome DNA From Samples
The sample DNA was extracted strictly according to the instructions of genomic DNA extraction kit (Sangon Bioengineering Co., Ltd, Shanghai) and the extracted DNA was stored at −20°C. DNA quality detection: ➀ Five microliter of DNA solution was loaded to a 1% agarose gel and the gel was run in 1xTAE at 120 V. A single clear band indicates the extracted DNA to be intact and of sufficient concentration for a PCR reaction. ➁ The concentration and purity were detected by spectrophotometer, and 1 μL DNA solution was loaded to Nanodrop to determine the OD values. A value of OD260/280 between 1.7 and 2.0 demonstrates a reliable quality of the extracted DNA.
SP-A1 Gene Polymorphism
We selected rs1059057 of SP-A1 as our study SNP, according to the following reasons: ➀ SNP database Genbank (http://www.Ncbi.Gov/genBank) provides a number of SNP research sites located in the first functional gene (SP-A1) of SP-A, in which the site studied in this paper is numbered as rs1059057 in the genebank dbSNP database. ➁ Global minor allele frequency (GMAF) of rs1059057 is 0.08(>5%), and the minimum allele >5% meets the basic conditions for SNP selecting. ➂ Located in exon 6 of this gene coding region.
PCR Amplification
➀ Template: zero point five to one microliter of blood containing anticoagulant (EDTAK2) was directly added to 20 μL PCR reaction system. ➁ Paraffin-embedded tissue samples: a single 10 μm paraffin section was treated with 50–200 μL PCR reaction buffer containing 0.2 mg/ml protease K. The volume of buffer was proportional to the size of tissue section. The sample was bathed at 60°C for 1 h, and then inactivated at 98°C for 10 min. After cooling, the sample was centrifuged (16,000 × g, 2 min) and the supernatant was transferred to a new tube. One to two microliter of the supernatant was used as the template in a 20 μL PCR reaction. ➂ PCR reaction conditions: pre-denaturation at 95°C for 3 min; denaturation at 94°C for 30 s; annealing at 55–60°C for 25–30 s; extension at 72°C for 30–50 s; 35 cycles followed by repair-extension at 72°C for 5–8 min. ➃ PCR reaction: one to two microliter of template DNA at 20–50 ng/μL; forward primer 10 μM, 2 μL; reverse primer10 μM, 2 μL; dNTP (mix) 10 mM, 2 μL; 10 × Taq Buffer (with MgCl2), 5 μL Taq enzyme 5 U/μL, 0.5 μL; Add ddH2O to 50 μL.
Genotyping of DNA Samples
The rs1059057 locus of SP-A1 gene was genotyped by PCR-SSCP method. The results were compared with the normal sequences of Gen Bank gene pool and analyzed by sequence analysis software.
Statistical Analysis
The data were analyzed by SPSS 22.0 statistical software. The sex, mode of delivery, and regularity of lung maturation in the two groups were tested by X2-test. Birth weight and gestational age were tested by t-test, and the ratio of gene polymorphism rs1059057 in SP-A1 was tested by X2-test. Statistical significance was considered at P < 0.05. A power calculation on the G*Power program was also performed, based on Cohen's method. When an effect size index of 0.2 (corresponding to “weak to moderate” gene effect) was used the present sample size revealed a >93% power for detection of significant association (a < 0.05).
Results
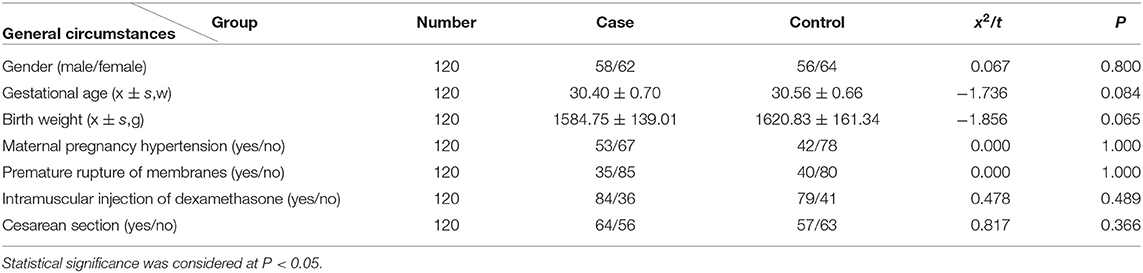
No significant difference in sex, gestational age, birth weight, mode of birth, cesarean section, and regular lung maturation was observed between the case and the control groups (P > 0.05) (Table 1).
Two genotypes of A/G and A/A were detected at SP-A1 rs1059057 locus in both the case and control group. In the case group, the frequencies of the two genotypes were 53 and 47%, and the frequencies of A allele and G allele were 73 and 27%, respectively. In the control group, the frequencies of the two genotypes were 42 and 58%, and the frequencies of A allele and G allele were 79 and 21%, respectively. No significant difference was observed in the genotype frequency of SP-A1 (rs1059057) locus between the case and the control groups (X2 = 3.275, P > 0.05). No significant difference was observed neither in the allele frequencies between the case and the control groups (X2 = 2.255, P > 0.05) (Table 2).
Discussion
Alveolar surfactant (PS) is a complex composed of lipids and special proteins synthesized and secreted by type II alveolar epithelial cells. It contains four protein components, SP-A, SP-B, SP-C, and SP-D, which play different roles based on functional and structural differences. They can not only reduce alveolar surface tension, but also participate in innate immunity (12). The lack of pulmonary surfactant can lead to the increase of alveolar surface tension and alveolar rupture, and affect the ventilation function of lung tissue, which has been considered to be the main cause of NRDS by previous studies, especially for very early births of younger gestational age (13). SP-A is a protein encoded by the SFTPA1 gene on the long arm of chromosome 10, which is a member of the C-lectin subfamily (14, 15) and play an important role in regulating the homeostasis of pulmonary surfactants and preventing the invasion of respiratory pathogens by binding lipids and carbohydrates on the surface of microorganisms (16). SP-A is one of the most abundant surface active proteins secreted by type II alveolar epithelial cells, and its deficiency affects the normal function of pulmonary and the metabolism of alveolar surfactant (17). SP-A can participate in the inflammatory response of lung tissue by binding to pathogens (18). Meanwhile SP-A can also reflect the injury of alveolar epithelial cells (19). SP-A1 protein is one of the subtypes of SP-A protein, the deficiency or structural changes of which lead to abnormal alveolar function and affect the normal function of lung tissue. It was found that mice lacking only SP-A did not develop RDS after full-term birth (20), while mice with the SP-A gene knocked out were more likely to develop pulmonary infection (21). It can be inferred that RDS in very premature infants may occur under the dual effects of decreased lung function and pulmonary inflammation caused by lack of SP-A.
A large number of studies have found that SP-A gene polymorphism is related to the occurrence of RDS in premature infants. For example, Jo et al. found that 1A0 variants and homozygous 1A0/1A0 genotypes of SP-A2 gene had protective effects on RDS (22). The haplotype (6A2/1A0) of SP-A1 may be closely related to the occurrence of RDS in an independent population, but this risk is limited only to very premature infants (23). SP-A1 haplotype 6A4 is a susceptible factor for RDS in late Greek preterm infants (24). In a study on the United States population, it was found that some SP-A alleles/haplotypes were susceptible factors of RDS, such as (1A0, 6A2, 1A0/6A2), and some SP-A alleles/haplotypes were protective factors of RDS, such as (1A5, 6A4, 1A5/6A4) (25); However, the results obtained in the population study in South Korea were the opposite (22). A Dutch study of twin fetuses found that their haplotypes were not associated with the occurrence of RDS (26). Chang et al. found that the polymorphism of SP-A (+186A/G) gene was closely related to the occurrence of RDS in premature infants (27). In addition, our previous studies found that the genotype and allele frequencies of SP-A1 (SNP) locus (rs1059047, and rs1136450) were not associated with the occurrence of RDS in Mongolian premature infants, and the haploid 6A2 of SP-A1 allele was the susceptible gene of RDS in Mongolian premature infants, and haploid 6A was the protective gene (28). Thus, it can be seen that the association between SP-A gene polymorphism and the occurrence of RDS is affected by race or regional environment.
The main purpose of this study is to investigate the relationship between SP-A1 rs1059057 locus gene polymorphism and Mongolian very premature infants' RDS. Our results showed that there was no significant association between the frequencies of genotypes and alleles of SP-A1 rs1059057 locus and the incidence of RDS in Mongolian very preterm infants, which is consistent with the results of Dutch twin study (26) and our previous study on SP-A1 (SNP) locus (rs1059047, rs1136450) (28). However, many previous studies also showed that SP-A gene polymorphism was related to the occurrence of RDS in premature infants (23–25, 27), which is inconsistent with our results. Two points are especially worthy of notice. The first is that the populations are different in those studies. The samples selected in our study are very premature Mongolian infants in the western part of Inner Mongolia, and the experimental results may be influenced by the stratification of the population, regional environment, ethnic groups, lifestyle, and other factors. Another point is that the technical approaches and sample size are different. The samples selected in this study were very premature Mongolian infants in western Inner Mongolia, the sample size was relatively small, and the time span of collecting samples was long; therefore, the genotyping method of PCR-SSCP used in the early stage of our research was used throughout the study for the consistency of experimental methods and conditions, which may also have a certain impact on the experimental results.
In a word, in this study we investigated the relationship between the rs1059057 gene polymorphism of SP-A1 and the RDS of Mongolian very premature infants in western Inner Mongolia, and found that the gene polymorphism of SP-A1 (rs1059057) was not related to the incidence of RDS in Mongolian very premature infants in western Inner Mongolia. Of course, it is necessary to increase the sample size and adopt more advanced and more sensitive detection methods to verify our current results in the future.
Data Availability Statement
All datasets generated for this study are included in the article/supplementary material.
Ethics Statement
The studies involving human participants were reviewed and approved by the Medical Ethics Committee of the affiliated Hospital of Inner Mongolia Medical University. Written informed consent to participate in this study was provided by the participants' legal guardian/next of kin.
Author Contributions
HM, CA, CL, YayZ, YanZ, and CX has made contributions to the research, design, revision and finalization of manuscripts. XW and YuZ contributed to the analysis and interpretation of the data and the drafting of the manuscript.
Funding
This work was supported by National Natural Science Foundation of China (81260107), Project of Natural Science Foundation of Inner Mongolia Autonomous region (2015MS (LH) 0810; 2011MS2011), Project of Youth Innovation Fund of Inner Mongolia Medical University (YKD2017QNCX062).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
1. Reuter S, Moser C, Baack M. Respiratory distress in the newborn. Pediatr Rev. (2014) 35:417–28; quiz: 429. doi: 10.1542/pir.35-10-417M
2. Li W, Chang L, Liu W, Rong Z, Cai B. Retrospective analysis of 86 cases of neonatal severe acute respiratory distress syndrome. Chin J Emerg Med. (2015) 24:258–62. doi: 10.3760/cma.j.issn.1671-0282.2015.03.006
3. Sweet DG, Carnielli V, Greisen G, Hallman M, Ozek E, Plavka R, et al. European consensus guidelines on the management of respiratory distress syndrome - 2016 update. Neonatology. (2017) 111:107–25. doi: 10.1159/000448985
4. Sweet DG, Carnielli V, Greisen G, Hallman M, Ozek E, Te Pas A, et al. European consensus guidelines on the management of respiratory distress syndrome - 2019 update. Neonatology. (2019) 115:432–51. doi: 10.1159/000499361
5. Alkan S, Ozer EA, Ilhan O, Sutcuoglu S, Tatli M. Surfactant treatment for neonatal respiratory disorders other than respiratory distress syndrome. J Matern Fetal Neonatal Med. (2015) 28:131–3. doi: 10.3109/14767058.2014.906575
6. Lopez-Rodriguez E, Pascual A, Arroyo R, Floros J, Perez-Gil J. Human pulmonary surfactant protein SP-A1 provides maximal efficiency of lung interfacial films. Biophys J. (2016) 111:524–36. doi: 10.1016/j.bpj.2016.06.025
7. Gupta A, Zheng SL. Genetic disorders of surfactant protein dysfunction: when to consider and how to investigate. Arch Dis Child. (2017) 102:84–90. doi: 10.1136/archdischild-2012-303143
8. Somaschini M, Nogee LM, Sassi I, Danhaive O, Presi S, Boldrini R, et al. Unexplained neonatal respiratory distress due to congenital surfactant deficiency. J Pediatr. (2007) 150:649–53, 653.e1. doi: 10.1016/j.jpeds.2007.03.008
9. Zhai L, Wu H, Wei K, Zhao S, Jiang H. Study on SP-A gene polymorphism in neonatal respiratory distress syndrome. Chin J Contemp Pediatr. (2008) 10:295–8.
10. Silveyra P, Floros J. Genetic complexity of the human surfactant-associated proteins SP-A1 and SP-A2. Gene. (2013) 531:126–32. doi: 10.1016/j.gene.2012.09.111
11. Phua J, Badia JR, Adhikari NKJ, Friedrich JO, Fowler RA, Singh JM, et al. Has mortality from acute respiratory distress syndrome decreased over time?: A systematic review. Am J Respir Crit Care Med. (2009) 179:220–7. doi: 10.1164/rccm.200805-722OC
12. Hallman M, Haataja R. Surfactant protein polymorphisms and neonatal lung disease. Semin Perinatol. (2006) 30:350–61. doi: 10.1053/j.semperi.2006.09.002
13. Hallman M, Arjomaa P, Mizumoto M, Akino T. Surfactant proteins in the diagnosis of fetal lung maturity. I. Predictive accuracy of the 35 kD protein, the lecithin/sphingomyelin ratio, and phosphatidylglycerol. Am J Obstet Gynecol. (1988) 158:531–5. doi: 10.1016/0002-9378(88)90019-1
14. Gjerstorff M, Dueholm B, Bendixen C, Holmskov U, Hansen S. Assignment of the surfactant protein A gene (SFTPA) to bovine chromosome 28q1.8–>q1.9 by radiation hybrid mapping. Cytogenet Genome Res. (2004) 106:142. doi: 10.1159/000078575
15. Nathan N, Taytard J, Duquesnoy P, Thouvenin G, Corvol H, Amselem S. Surfactant protein A: a key player in lung homeostasis. Int J Biochem Cell Biol. (2016) 81:151–5. doi: 10.1016/j.biocel.2016.11.003
16. Nayak A, Dodagatta-Marri E, Tsolaki AG, Kishore U. An insight into the diverse roles of surfactant proteins, SP-A and SP-D in innate and adaptive immunity. Front Immunol. (2012) 3:31. doi: 10.3389/fimmu.2012.00131
17. Kingma PS, Whitsett JA. In defense of the lung: surfactant protein A and surfactant protein D. Curr Opin Pharmacol. (2006) 6:277–83. doi: 10.1016/j.coph.2006.02.003
18. Thorenoor N, Umstead TM, Zhang X, Phelps DS, Floros J. Survival of surfactant protein-A1 and SP-A2 transgenic mice after infection, exhibits sex-, gene-, and variant specific differences; treatment with surfactant protein improves survival. Front Immunol. (2018) 9:2404. doi: 10.3389/fimmu.2018.02404
19. Ye S, Li Q, Yuan S, Shu H, Yuan Y. Restrictive fluid resuscitation leads to better oxygenation than non-restrictive fluid resuscitation in piglets with pulmonary or extrapulmonary acute respiratory distress syndrome. Med Sci Monit. (2015) 21:2008–20. doi: 10.12659/MSM.892734
20. Wright JR. Immunoregulatory functions of surfactant proteins. Nat Rev Immunol. (2005) 5:58–68. doi: 10.1165/rcmb.2004-0003OC
21. McCormack FX, Whitsett JA. The pulmonary collectins, SP-A and SP-D, orchestrate innate immunity in the lung. J Clin Invest. (2002) 109:707–12. doi: 10.1172/JCI15293
22. Jo HS, Cho S-I, Chang YH, Kim BI, Choi J-H. Surfactant protein A associated with respiratory distress syndrome in Korean preterm infants: evidence of ethnic difference. Neonatology. (2013) 103:44–7. doi: 10.1159/000342498
23. Rämet M, Haataja R, Marttila R, Floros J, Hallman M. Association between the surfactant protein A (SP-A) gene locus and respiratory-distress syndrome in the Finnish population. Am J Hum Genet. (2000) 66:1569–79. doi: 10.1086/302906
24. Tsitoura MEI, Stavrou EF, Maraziotis IA, Sarafidis K, Athanassiadou A, Dimitriou G. Surfactant protein A and B gene polymorphisms and risk of respiratory distress syndrome in late-preterm neonates. PLoS ONE. (2016) 11:e0166516. doi: 10.1371/journal.pone.0166516
25. Floros J, Fan R, Matthews A, DiAngelo S, Luo J, Nielsen H, et al. Family-based transmission disequilibrium test (TDT) and case-control association studies reveal surfactant protein A (SP-A) susceptibility alleles for respiratory distress syndrome (RDS) and possible race differences. Clin Genet. (2001) 60:178–87. doi: 10.1034/j.1399-0004.2001.600303.x
26. Marttila R, Haataja R, Rämet M, Pokela M-L, Tammela O, Hallman M. Surfactant protein A gene locus and respiratory distress syndrome in Finnish premature twin pairs. Ann Med. (2003) 35:344–52. doi: 10.1080/07853890310006389
27. Chang H-Y, Li F, Li F-S, Zheng C-Z, Lei Y-Z, Wang J. Genetic polymorphisms of SP-A, SP-B, and SP-D and risk of respiratory distress syndrome in preterm neonates. Med Sci Monit. (2016) 22:5091–100. doi: 10.12659/MSM.898553
Keywords: respiratory distress syndrome (RDS), pulmonary surfactant protein A1 (SP-A1), gene polymorphism, Mongolian very premature infants, respiratory tract management
Citation: Wang X, Zhang Y, Mei H, An C, Liu C, Zhang Y, Zhang Y and Xin C (2020) Study on the Relationship Between Respiratory Distress Syndrome and SP-A1 (rs1059057) Gene Polymorphism in Mongolian Very Premature Infants. Front. Pediatr. 8:81. doi: 10.3389/fped.2020.00081
Received: 10 November 2019; Accepted: 18 February 2020;
Published: 17 March 2020.
Edited by:
Yuan Shi, The Children's Hospital of Chongqing Medical University, ChinaReviewed by:
Jianhua Fu, ShengJing Hospital of China Medical University, ChinaLi Wang, Daping Hospital, China
Copyright © 2020 Wang, Zhang, Mei, An, Liu, Zhang, Zhang and Xin. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Hua Mei, bWVpaHVheWFuaUBzaW5hLmNvbQ==; Caiyan An, YWN5XzE5OTlAMTYzLmNvbQ==
 Xiaoli Wang
Xiaoli Wang Yuheng Zhang1
Yuheng Zhang1 Caiyan An
Caiyan An