
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Pediatr. , 27 February 2019
Sec. Pediatric Immunology
Volume 7 - 2019 | https://doi.org/10.3389/fped.2019.00047
This article is part of the Research Topic Human Milk Composition and Health Outcomes in Children View all 13 articles
Evidence is accumulating that demonstrates the importance of the gut microbiota in health and diseases such as allergy. Recent studies emphasize the importance of the “window of opportunity” in early life, during which interventions altering the gut microbiota induce long-term effects. The neonate's gut microbiota composition and metabolism could therefore play an essential role in allergic disease risk. Breastfeeding shapes the gut microbiota in early life, both directly by exposure of the neonate to the milk microbiota and indirectly, via maternal milk factors that affect bacterial growth and metabolism such as human milk oligosaccharides, secretory IgA, and anti-microbial factors. The potential of breastmilk to modulate the offspring's early gut microbiota is a promising tool for allergy prevention. Here, we will review the existing evidence demonstrating the impact of breastfeeding on shaping the neonate's gut microbiota and highlight the potential of this strategy for allergy prevention.
Most of the human body is heavily colonized by all kinds of microorganisms, including bacteria, viruses, fungi, protozoa, and parasites. The largest amount of microorganisms are found within the colon, although the presence in other parts of the gastrointestinal tract cannot be neglected (1, 2). The development of the human gut microbiome is a highly complex process (1). The order and timing by which the gut is colonized early in life has a lasting impact on the microbiome and contributes largely to the variation in microbiota observed between individuals (3). Over the first 3 years of life, the microbiota evolves from relatively simple but rapidly increasing in diversity to an adult state that is more complex and more stable (1). In addition to mode of delivery and antibiotic exposure, nutrition is a key factor in shaping the early microbiota composition and function [as reviewed in Tamburini et al. (4)]. Besides the important role of the gut microbiota in nutrient and bile acid metabolism and the production of vitamins (5), colonization and signaling by microbes plays a pivotal role in gut mucosal immunity as well as systemic immunity. The development of the microbiota ecology parallels that of the gut mucosal immune system. Accumulating evidence is showing that perturbations in the gut microbiota in early life, while the immune system is still developing, can have long-lasting effects on local and systemic immune health.
To date, a clear protective effect of breastmilk on allergy development has not been demonstrated (6–11). However, breastmilk contains factors that can affect key players in allergy development such as gut barrier function, the gut microbiota and oral tolerance induction (12). Therefore, we propose that the modulation of breastmilk composition could be a promising tool for allergy prevention. Here we will review the existing evidence demonstrating the impact of breastfeeding on the neonate's gut microbiota and highlight the potential of shaping the neonate's gut microbiome through breastmilk to decrease allergic disease risk. A literature search was performed in PubMed on original studies and review articles addressing (1) the association of the early life gut microbiota with allergic outcomes (2) immunological mechanisms for the early life gut microbiota to affect allergy development and (3) how breastfeeding shapes the gut microbiota. The primary focus of the review were recent studies addressing these topics in the early life window.
Epidemiological and animal studies have linked perturbations in the infant gut microbiota, when the immune system matures and the gut is colonized with microbiota, with disease risk later in life (4, 13). This highlights the existence of a window of opportunity for disease prevention, including atopic disease, which matches the period in life of breastfeeding. Germ-free (GF) mice have elevated levels of serum immunoglobulin (Ig)E because B cells undergo more isotype class switching to IgE in Peyer's patches and mesenteric lymph nodes. Microbial colonization of GF pups starting between birth and 1 week after weaning, completely inhibits this induction of IgE if a diverse microbiota is used. This implies the need for a critical level of microbial diversity following birth to prevent IgE induction (14). Colonization of GF mice with gut microbiota in early life, but not in adults, is also sufficient to protect from mucosal invariant natural killer T (iNKT) cell accumulation in the lung and allergic airway inflammation (15). Antibiotic administration exacerbated allergic airway inflammation, reduced FoxP3+ regulatory T cells (Treg) in the colon and increased serum IgE only when treatment was initiated in early life (16). In humans, a link between gut microbial dysbiosis in the first 100 days of life and an increased risk of asthma was demonstrated (17). Also other studies have shown an association between the early intestinal microbiota and the risk of allergic sensitization (18–20). All provide evidence of a link between early gut microbiota dysbiosis and an increased risk to develop allergic disease.
It is still very preliminary to define a “healthy” or “normal” human gut microbiota. The most recent studies define a healthy microbiota as highly diverse, i.e., above 600.000 bacterial genes (21). Nutrition is certainly one way to reach such diversity (2, 22). In addition to diversity there is a core microbiota which is common the most human guts and which has been proposed as a “must have“-set of bacteria (23, 24). Their precise role still needs to be clearly defined but they are mostly involved in the control of inflammation and innate immunity. Although much less details are known in early life, more and more studies try to determine associations between alterations in neonatal microbiota and disease outcome. Microbiota richness, diversity, and composition all may play a role in shaping of the immune response by the gut microbiota. Gut microbial alterations are not limited to shifts in the abundance of certain microbes, they also include alterations in microbiota metabolism and changes in the production of microbial-derived metabolites such as short-chain fatty acids (SCFA) (25, 26).
Bisgaard et al. (18) demonstrated the inverse association between the early gut bacterial diversity and the risk of allergic sensitization, but not asthma or atopic dermatitis (18). However, another study showed that infants with cow's milk allergy had an increased diversity of the gut microbiota and an altered microbial composition dominated by Lachnospiraceae as compared to non-allergic infants (27). In a Chinese cohort, 20 key food allergy-associated bacterial genera, but no difference in fecal microbiota diversity, were observed. The specific microbiota signature detected, could distinguish between IgE-mediated and non-IgE-mediated food allergic infants. The genus Clostridium sensu stricto significantly correlated with antigen-specific IgE in infants with food allergy (28). A low gut microbiota richness, overrepresentation of Enterobacteriaceae and underrepresentation of Bacteroidaceae in early infancy were associated with food sensitization in a subset of the Canadian Healthy Infant Longitudinal Development (CHILD) study (20). In the same cohort, Canadian infants at risk of asthma showed a reduction in the relative abundance of the bacterial genera Lachnospira, Veillonella, Faecalibacterium, and Rothia in early life and had lower fecal concentrations of the SCFA acetate (17, 29). A causal role of these bacterial taxa was demonstrated in mouse experiments (17). The impact of microbial dysbiosis at 3 months of age was further confirmed in a non-industrialized population in rural Ecuador (30). Interestingly, different bacterial taxa were involved compared to Canadian infants. Some fecal fungal taxa were altered too and genes involved in carbohydrate and taurine metabolism were highly altered (30). Another birth cohort showed that neonates with a relatively lower abundance of bacteria such as Bifidobacterium, Akkermansia, and Faecalibacterium, along with higher abundance of the fungi Candida and Rhodotorula and pro-inflammatory fecal metabolites, had the highest risk of childhood atopy and asthma (31). Russian children at low risk for the development of allergic disease had higher proportions of Bifidobacterium, whereas Finnish and Estonian children with a higher risk of allergies had increased abundance of Bacteroides (32). Furthermore, early colonization with Lactobacilli was shown to decrease the risk of allergy (19) while early colonization with Staphyloccocus aureus and Clostridium difficile characterizes infants developing allergy later in life (33–35). Colonization with Escherichia coli was associated with IgE-mediated eczema (36, 37). However, a study using early administration of E. coli as probiotic strategy found a reduction in allergy development, pointing toward strain-specific effects of E. coli (38). Recently it was also reported that the kinetic of development of the gut microbiome during the first year of life affects the risk of childhood asthma in children from asthmatic mothers. One-year-old children with an immature microbial composition had an increased risk of asthma at age 5 years compared to children with mature microbiota (39).
To induce tolerance at mucosal surfaces, antigens are taken up by dendritic cells (DC) which migrate to the lymph nodes where the local production of factors like transforming growth factor beta (TGF-β) induces the differentiation of naïve T cells to antigen-specific Treg (40). Here, we will summarize the current observations in early life specifically, as this coincides with the period of breastfeeding, which demonstrate an effect of the microbiota on the maturation of the immune system (Figure 1). Various studies have demonstrated a role of the microbiota in early life on the development of FoxP3+ Treg. Ex vivo culturing of human adult peripheral T cells with sterile fecal water from children at high risk of developing atopic disease, reduced the percentage of FoxP3+ Treg cells (31). Neonatal colonization with a specific strain of the commensal E. coli lead to oral tolerance failure. It reduced tolerogenic DC and subsequently Treg populations (41). On the other hand, neonatal enrichment of mice with Clostridium species from human indigenous microbiota resulted in higher numbers of colonic FoxP3+ Treg in adulthood, likely induced by intestinal epithelial cell-secreted TGF-β, and lower allergy risk (42). Another study demonstrated the pivotal role of early life colonization with Bacteroides fragilis expressing polysaccharide A (PSA) for iNKT cell inhibition and Treg development in the intestine (43). Colonizing adult mice did not have this effect (43). Another study has emphasized a role for the gut microbiota in the modulation of IL-22 secretion and gut barrier function. Colonization of young mice with Clostridia induced IL-22 production by group 3 innate lymphoid cells (ILC3) and T helper 17 cells in the intestinal lamina propria. IL-22 was critical for sensitization to food allergen as it induces antimicrobial peptide production by Paneth cells and mucus production by goblet cells to strengthen the gut barrier. This prevents the transfer of dietary antigen across the barrier and therefore allergic sensitization (44). Recent studies have also linked the kynurenine pathway, involved in the breakdown of tryptophan by host cells, with the gut microbiota and allergy (45). In host immune and epithelial cells the enzyme indoleamine 2,3-dioxygenase (IDO) metabolizes tryptophan to kynurenine and downstream products (46). IDO can become activated in response to allergen-induced immune activation (45). Kynurenines regulate immune homeostasis and exhibit tolerogenic effects by causing T cell anergy and apoptosis and induce the generation of Treg, leading to attenuation of allergic responses (45). The gut microbiota plays an important role in stimulating IDO activity, as demonstrated in GF mice (46, 47), making this pathway dependent on early life gut microbiota development (45). Targeting the gut microbiota to modulate tryptophan metabolism could therefore have the potential to prevent allergic disease (45).
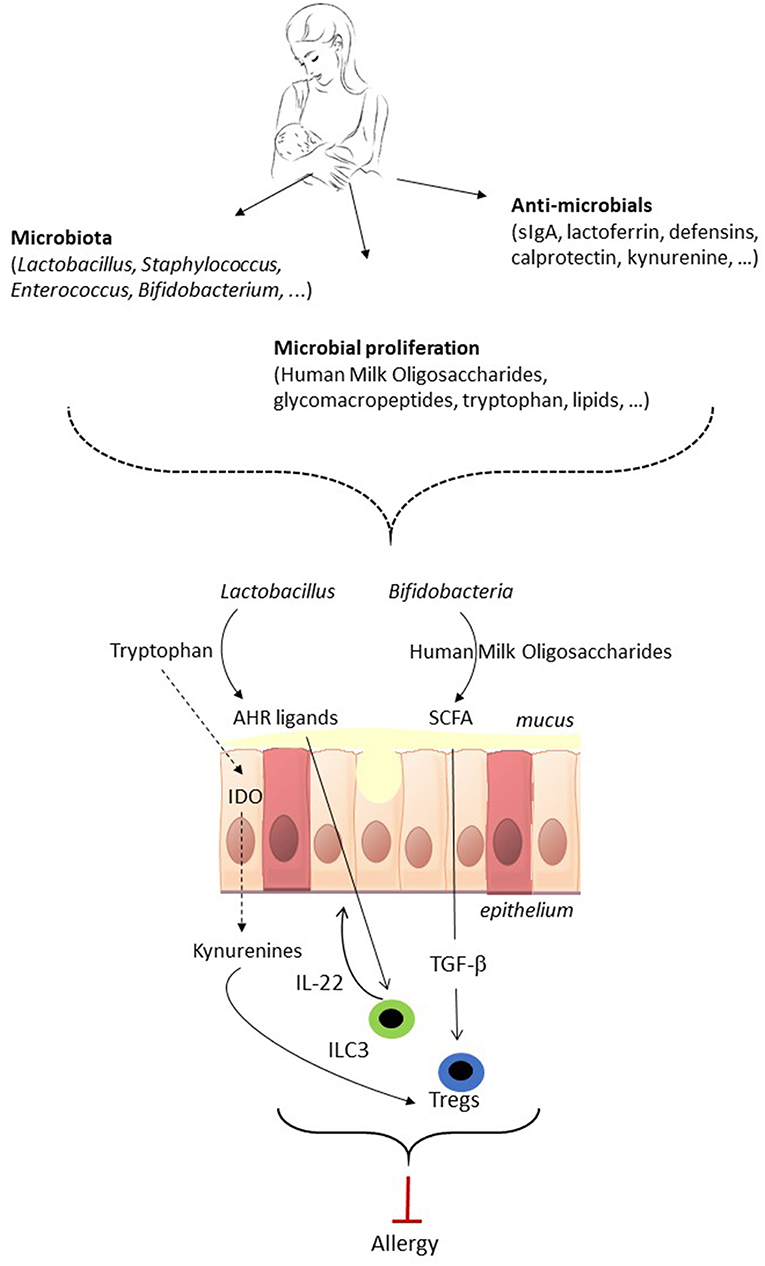
Figure 1. The potential of breastmilk to prevent allergic disease by shaping of the neonatal gut microbiota. Breastmilk contains microbes as well as factors that indirectly shape the gut microbiota of the neonate. Breastmilk can direct the early microbiota composition, i.e., favor the growth of Bifidobacteria and Lactobacillus, and affect microbiota metabolic function, which subsequently can impact on immune development and maturation. The gut microbiota in early life impacts on immune maturation via microorganism-associated molecular patterns (MAMPs) signaling (not shown in the figure) and via microbiota metabolites such as short-chain fatty acids (SCFA) and aryl hydrocarbon receptor (AHR) ligands. The gut microbiota of the neonate can direct the immune system toward allergy prevention via the induction of FoxP3+ regulatory T cells (Treg), which contribute to oral tolerance induction, and IL-22 production by group 3 innate lymphoid cell (ILC3) which strengthens the gut barrier. As a result, shaping of the infant's gut microbiota by breastmilk has the potential to direct the immune system toward allergy prevention. TGF-β, Transforming Growth Factor beta; IDO, indoleamine 2,3-dioxygenase.
The gut microbiota exerts direct effects on the immune system through microorganism-associated molecular pattern (MAMP) signaling. Bacterial as well as fungal and viral molecular patterns such as lipopolysaccharides (LPS), flagellin, peptidoglycans (PG), formyl peptides, and unique nucleic acid structures are sensed by pattern recognition receptors (PRR) including membrane bound Toll-like receptors (TLR) and cytoplasmic NOD-like receptors (NLR) (25).
LPS binds to a complex of membranous TLR4, CD14 and myeloid differentiation protein 2 (MD-2) or circulating sCD14 receptors. TLR4 is the receptor transmitting the MAMP's signal inside the cell to trigger genes of inflammation. LPS are complex molecules with a lipid A part containing different fatty acids esterified to a glucosamine structure. It can possess 4, 5, or 6 aliphatic chains of different length including cyclic fatty acids specific from bacteria. Hexa-acylated LPS, such as seen in E. coli, is considered highly inflammatory whereas LPS with a lower number of acylated fatty acids, such as those from the Porphyromonadaceae family, are less inflammatory or even anti-inflammatory (48). Variation in LPS immunogenicity can play a role in immune education and driving allergic disease. High exposure to Bacteroides-derived penta- or tetra-acylated LPS, which has immune inhibitory properties, was demonstrated in infants with high risk of allergic disease. This form of LPS does not induce endotoxin tolerance, leading to inflammatory responses later in life (32, 49). A similar observation was reported for PGs which can be pro- or anti-inflammatory according to the structure and molecular weight of the molecule. In addition, PG can bind to different TLRs, particularly TLR2, as well as the intracellular NLR NOD1 and 2, with different outcome (50, 51). Furthermore, TLR signaling by bacterial products induces a tolerogenic environment and Treg expansion in the intestine (52). These studies demonstrate that MAMPs can engage innate and adaptive immunity through the stimulation of different TLRs and NLRs. This suggests that the gut microbiota ecology at birth directs the specific crosstalk between MAMPs and the immune system and educates both innate and adaptive immunity. This could result in protection against the development of allergic diseases.
Besides the direct effects of gut microbes on the immune system, the production of metabolites by the fermentation of dietary fiber and other complex macronutrients that escape digestion in the small intestine, plays an important role. Molecules produced or derived from bacterial metabolism are drivers of cellular host functions and notably the intestinal immune, epithelial, vascular, and neural systems (53). Bacterial metabolites can induce epigenetic changes such as chromatin modifications that allow the microbiota to exert long-lasting effects on immunity (54). Metabolites of microbial origin include choline metabolites, vitamins, and phenolic derivatives (53). Primary bile acids can be transformed into secondary bile acids by the metabolism of bacteria, which can differently activate the bile acid receptors farnesoid X receptor (FXR) and G protein-coupled bile acid receptor (TGR5) (55). Specific commensal bacteria, especially Lactobacilli, metabolize the essential amino acid tryptophan into indole derivatives that can bind to aryl hydrocarbon receptors (AHR) expressed by immune and epithelial cells. AHR signaling is important for ILC3 activation and intestinal barrier function (25). The role of AHR ligands on immune development during the neonatal phase specifically is largely unclear to date. Branched-chain fatty acids such as valerate, isobutyrate, and isovalerate are derived from bacterial amino acid metabolism. SCFA, predominantly butyrate, acetate and propionate, are produced upon breakdown of dietary fiber. Lactate and succinate are intermediate metabolites in the production of SCFA, but can exert immune modulating effects themselves too (56, 57). SCFA are an important energy source for intestinal epithelial cells. Furthermore, SCFA signal through G protein couples receptors such as GPR43, GPR41, and GPR109A present on epithelial and immune cells, and inhibition of histone deacetylases (25, 58). In adult mice it has been demonstrated that SCFA can enhance the intestinal barrier function, induce tolerogenic DC and promote anti-inflammatory Treg in the colon, all contributing to immune tolerance (25). In the lung SCFA impair the ability of DC to promote a T helper 2 response and reduce ILC2 proliferation and function (59, 60). Data on the effects of SCFA on the maturation of the gut and immune system in the neonate is currently largely lacking. However, there is limited evidence that higher acetate levels in infants might assist in the protection against allergic disease (17).
Recent studies suggest that microbial transfer from the mother to the fetus already occurs in utero. Microbes have been detected in the placenta, amniotic fluid, fetal membrane, umbilical cord blood, and meconium (4, 61). However, the first major exposure of the neonate to microbes happens during birth and is highly dependent on the mode of delivery (1, 4). Besides birth mode and/or antibiotics use just before or after delivery, early nutrition is a key factor directing the early microbiota composition and function as it provides nutrients for bacterial growth and dictates their production of metabolites (1, 2, 22). Recently, a large, multi-center study confirmed that breastfeeding status was the most significant factor associated with microbiome structure in early life (62). The first bacteria to establish in the neonatal gut are mostly aerobic or facultative anaerobic bacteria such as enterobacteria, enterococci, and staphylococci. During their growth they consume oxygen allowing the rise of anaerobic bacteria including bifidobacteria (63). The gut microbiome in breastfed infants is usually dominated by bifidobacteria and Lactobacillus species, while formula-fed infants harbor a more diverse gut microbiota that resembles that of older children (22, 64). Relatively small amounts of formula supplementation of breastfed infants, only during the first days of life, already resulted in shifts in microbiota composition (65). In addition to shaping microbiota composition, early feeding practice affects microbiota metabolism. The microbiomes of newborns and young infants are enriched in genes required for the degradation of sugars from breastmilk (human milk oligosaccharides, HMOs) (22). Compared with formula-fed children, breastfed infants have lower absolute concentrations of fecal SCFA, potentially due to the less diverse microbiota, and higher concentrations of lactate. However, the relative proportion of acetate was higher in exclusively breastfed children (56). The introduction of solid foods changes the metabolic function of the gut bacteria as genes involved in the degradation of sugars from breastmilk are less needed and utilized. Instead, the microbiota adapts to the available energy sources and functionally matures to be able to degrade complex sugars and starch found in solid food (22). Interestingly, the microbiota composition in African and European infants is very similar until the introduction of solid foods, indicating the dominant role of diet over other variables in shaping the microbial composition of the gut in early life (66).
Breastmilk provides the neonate with its own microbiota as well as prebiotic, immunological and other microbiota-shaping compounds that indirectly can alter colonization patterns in the neonate (Figure 1). Therefore, a varied composition of breastmilk could be considered as a selective bioactor to reach gut microbiota diversity and hence good health.
Breastmilk contains 102-104 viable bacteria per mL (67), and thereby can directly affect the establishment of the neonatal microbiota (68). Lactobacillus, Staphylococcus, Enterococcus, and Bifidobacterium are transferred through breastfeeding (67). Milk bacterial communities are complex and vary between individuals. The breastmilk microbiota also evolves over the period of breastfeeding. Colostrum microbiota has a higher diversity than mature milk (69). In colostrum, Staphylococcus, lactic acid bacteria and Streptococcus are the most abundant (69). After 1 month, the Staphylococcus abundance is dramatically reduced, while the lactic acid bacteria are still highly abundant (68, 69). The maturation of the breastmilk microbiota happens in parallel with the evolution of the neonate's microbiota. As soon as 3–4 days after birth, the gut microbiota of infants begins to resemble the colostrum microbiota (61), followed by a gut microbiota rich in bifidobacteria and lactobacilli (22).
The origin of bacteria in breastmilk is under debate. Some suggest that human milk bacteria are derived from the maternal skin as some bacterial phyla that are common in human milk, such as Staphylococcus, are usually present on adult skin (67). It has also been demonstrated that during suckling breastmilk flows back into the mammary ducts (67, 70), which provides a route for bacteria found in the infants oral cavity to enter the mammary gland (67, 69). However, most studies propose that the translocation of maternal gut bacteria to the mammary gland is the major pathway (2, 64). DC and macrophages can sample live commensal bacteria from the gut lumen and keep them in the mesenteric lymph nodes. From there, the bacteria can circulate to other locations in the body, including the mammary glands (67). Mothers having a cesarean section, show a more diverse milk microbiota with reduced frequency of bifidobacteria as compared to mothers after a vaginal delivery. This effect is most pronounced for infants from women undergoing an elective cesarean-section, suggesting that signals related to labor affect bacterial transfer to the mammary glands (69).
HMOs are structurally complex sugars unique to human breastmilk. They are indigestible and do not provide energy for the infant but serve as prebiotics, which are substrates for fermentation processes by intestinal microbes, inducing the growth or activity of beneficial bacteria (71). HMOs are highly abundant in human milk but absent in most formula nutrition and are believed to play a major role in the differences between the gut microbiota in breast- vs. formula-fed infants. HMO composition in maternal milk is regulated by genetic fucosyltransferase-2 (FUT2) secretor status and other factors including lactation stage, maternal health and ethnicity (72). HMOs act as antiadhesive agents that inhibit pathogen adhesion to mucosal surfaces, preventing colonization and as antimicrobials by preventing proliferation of certain bacteria (72, 73). Furthermore, HMO favor Bifidobacterium growth and are their preferred substrates for the production of SCFA and lactate in infancy (56, 74, 75). Bifidobacterium predominance in the stool is the main characteristic of breastfed infants (68) and the higher relative proportion of acetate in breastfed compared to formula-fed infants may be due to the absence of HMO in formula (56). Infants receiving breastmilk from non-secretor mothers, who lack the functional FUT2 enzyme, show a delay in the establishment of bifidobacteria highlighting the need for maternal milk HMOs in bifidobacteria growth (76). A recent study further demonstrates that HMOs in breastmilk can also modulate the transcriptional activity of certain bacteria such as B. fragilis, rather than modifying their relative abundance (26). Furthermore, another commensal, E. coli, that cannot directly degrade HMOs, benefits indirectly by consuming metabolites produced by B. fragilis upon HMOs degradation (26). An association between HMO profiles, but not individual HMOs, and food sensitization has been demonstrated in 1-year-old infants (77). A small clinical study further found that infants receiving maternal milk with low concentrations of the HMO lacto-N-fucopentaose III were more likely to develop cow's milk allergy (76). A likely explanation, besides the possibility of direct effects of HMO on immune cells, is the prebiotic modulation of the gut microbiota by HMOs influencing immune development and food sensitization (77).
Casein glycomacropeptide (GMP) is a small glycoconjugated peptide present in human milk. GMP has a prebiotic effect on bifidobacteria and lactic acid bacteria (78).
Secretory IgA (sIgA) are also important factors in shaping the gut microbiota in the neonate. IgA-producing plasma cells in the mammary gland originate from the maternal gut. The specificity of sIgA in breastmilk is therefore dictated by maternal exposure to pathogenic enteric bacteria and by commensal bacteria in the maternal gut. Maternal IgA produced in the mammary gland, are transported across the epithelial cells into the milk and acquire the secretory part from epithelial cells (79). As newborns produce only low levels of sIgA, breastmilk-derived sIgA prevent expansion and penetration of pathogenic bacteria while their intestinal immune system is developing (80, 81). IgA also has functions beyond pathogen exclusion. Experiments in mice using IgA-deficient dams demonstrated a long-lasting role of breastmilk-derived sIgA in shaping of the gut microbiota. Taxa from the family Lachnospiraceae were upregulated in the absence of sIgA from breastmilk (82). Milk sIgA are also necessary for the prevention of excessive expansion of pro-inflammatory segmented filamentous bacteria (SFB) by coating these bacteria (83). A recent birth cohort showed that IgA recognition patterns differed between healthy and allergic children. This was already visible at 1 month of age, when IgA is predominantly maternally derived in breastfed children. Interestingly, mainly butyrate-producing gut commensals such as Faecalibacterium were IgA free in children with allergic symptoms (84). IgA can also modulate the production of metabolites by the microbiota, as recently shown in an adult mouse model (85). In this study it was demonstrated that IgA has the capacity to modulate bacterial composition, gene expression, and metabolic function of the gut microbiota by antigen-independent binding to intestinal bacteria (85).
Antimicrobial factors in breastmilk have the potential to shape the microbiota. Breastmilk is the main source of lactoferrin for the infant and protects them from bacterial invasion by sequestering iron from bacterial pathogens and direct interaction with bacteria (79). The ability to protect the infant against pathogenic microorganisms helps the development of a beneficial microbiota (79). The amount of fecal bifidobacteria and lactobacilli in newborns positively correlated with fecal lactoferrin (86). This suggests that lactoferrin promotes specific microbial composition and might be critical for the microbiota to develop in early life. High levels of calprotectin (heterodimer of calcium binding proteins S100A8 and S100A9) also demonstrate antimicrobial properties that can be attributed to its metal ion chelation capacity. This results in growth inhibition of especially manganese sensitive bacteria such as S. aureus and group B streptococci. After birth, calprotectin concentrations are high in human milk (87). Defensins in breastmilk also have antimicrobial activity against common neonatal pathogens (88) and have been shown to affect the intestinal microbiota (89).
Tryptophan is a precursor of a large number of metabolites, including endogenous metabolites such as kynurenine and bacterial metabolites such as indole derivatives (46, 47). Tryptophan and its metabolites are present in breastmilk (90, 91) and can have profound effects on the gut microbial composition, metabolism and function in the infant (47). Kynurenines have antimicrobial properties, which can directly impact on the gut microbiota (47). AHR ligands in breastmilk originate from the maternal diet as well as from the maternal microbiota. Maternal milk immunoglobulins help in the transfer of these metabolites to the neonate (91). The AHR signaling pathway is also able to influence microbiota composition (92).
Factors of the innate immune response that are present in breastmilk, but not infant formula, include soluble TLR2, TLR4 and their co-receptors CD14 and MD2, which are involved in binding LPS (89, 93, 94). These factors probably contribute to the composition of luminal and enterocyte surface bacteria (89, 95).
Lipids impact on the gut microbiota ecology either directly by feeding some bacteria or indirectly by triggering the host to secrete hormones and bile acids (96). Bile acids are major regulators of the gut microbiota (55). They are detergents for bacteria explaining, at least in part, the low bacterial count in the duodenum where bile acids are mostly released. Human milk fatty acid profiles were associated with the relative abundance of five taxa (Bacteroides, Enterobacteriaceae, Veillonella, Streptococcus, and Clostridium) in the gut microbiota of breastfed neonates (97).
The potential of breastmilk to alter the offspring's early gut microbiota is a promising tool for immune education and allergy prevention (Figure 1). This requires identifying (1) what a beneficial microbiota for allergy prevention in our modern environment is (2) which factors in breastmilk are necessary to guide the establishment of such a beneficial microbiota and (3) how to enrich breastmilk with the required factors. There have been major advances in the recent years in the identification of a beneficial microbiota for allergic disease prevention. In particular, the necessity of a diverse gut microbiota and the importance of the metabolism of the microbiota have been highlighted. The role of the breastmilk microbiota and HMOs in shaping of the neonatal gut microbiota has become more and more evident recently. There is however still poor knowledge on the possibilities to modulate breastmilk factors involved in the microbiota shaping. The administration of probiotics to lactating mothers has led to inconsistent results regarding the possibility to change the milk microbiota (98, 99) and there is currently no clue on the possibility to modify HMOs content in breastmilk. There is a need for randomized intervention trials that study the allergy preventive effects of supplementation of lactating mothers with milk-modulating factors such as pre- and probiotics, to shape the infants microbiota and subsequently program the immune response.
LvdE and VV proposed the topic to be covered in the manuscript and wrote the main content with the contribution of JG and RB.
This work was supported by the University of Western Australia and by the Larsson-Rosenquist Foundation.
JG is employed by Danone, a company producing products for infant nutrition.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
1. Gensollen T, Iyer SS, Kasper DL, Blumberg RS. How colonization by microbiota in early life shapes the immune system. Science (2016) 352:539–44. doi: 10.1126/science.aad9378
2. van Best N, Hornef MW, Savelkoul PH, Penders J. On the origin of species: factors shaping the establishment of infant's gut microbiota. Birth Defects Res C Embryo Today (2015) 105:240–51. doi: 10.1002/bdrc.21113
3. Martinez I, Maldonado-Gomez MX, Gomes-Neto JC, Kittana H, Ding H, Schmaltz R, et al. Experimental evaluation of the importance of colonization history in early-life gut microbiota assembly. Elife (2018) 7:e36521. doi: 10.7554/eLife.36521
4. Tamburini S, Shen N, Wu HC, Clemente JC. The microbiome in early life: implications for health outcomes. Nat Med. (2016) 22:713–22. doi: 10.1038/nm.4142
5. Sekirov I, Russell SL, Antunes LC, Finlay BB. Gut microbiota in health and disease. Physiol Rev. (2010) 90:859–904. doi: 10.1152/physrev.00045.2009
6. van Odijk J, Kull I, Borres MP, Brandtzaeg P, Edberg U, Hanson LA, et al. Breastfeeding and allergic disease: a multidisciplinary review of the literature (1966–2001) on the mode of early feeding in infancy and its impact on later atopic manifestations. Allergy (2003) 58:833–43. doi: 10.1034/j.1398-9995.2003.00264.x
7. Gdalevich M, Mimouni D, Mimouni M. Breast-feeding and the risk of bronchial asthma in childhood: a systematic review with meta-analysis of prospective studies. J Pediatr. (2001) 139:261–6. doi: 10.1067/mpd.2001.117006
8. Ip S, Chung M, Raman G, Chew P, Magula N, DeVine D, et al. Breastfeeding and maternal and infant health outcomes in developed countries. Evid Rep Technol Assess (2007) 1–186.
9. Dogaru CM, Nyffenegger D, Pescatore AM, Spycher BD, Kuehni CE. Breastfeeding and childhood asthma: systematic review and meta-analysis. Am J Epidemiol. (2014) 179:1153–67. doi: 10.1093/aje/kwu072
10. Victora CG BR, Barros AJ, França GVA, Horton S, Krasevec J, Murch S, et al. Breastfeeding in the 21st century: epidemiology, mechanisms, and lifelong effect. Lancet (2016) 387:475–90. doi: 10.1016/S0140-6736(15)01024-7
11. Lodge CJ, Tan DJ, Lau MX, Dai X, Tham R, Lowe AJ, et al. Breastfeeding and asthma and allergies: a systematic review and meta-analysis. Acta Paediatr. (2015) 104:38–53. doi: 10.1111/apa.13132
12. Munblit D, Verhasselt V. Allergy prevention by breastfeeding: possible mechanisms and evidence from human cohorts. Curr Opin Allergy Clin Immunol. (2016) 16:427–33. doi: 10.1097/ACI.0000000000000303
13. Renz H, Adkins BD, Bartfeld S, Blumberg RS, Farber DL, Garssen J, et al. The neonatal window of opportunity-early priming for life. J Allergy Clin Immunol. (2018) 141:1212–4. doi: 10.1016/j.jaci.2017.11.019
14. Cahenzli J, Koller Y, Wyss M, Geuking MB, McCoy KD. Intestinal microbial diversity during early-life colonization shapes long-term IgE levels. Cell Host Microbe (2013) 14:559–70. doi: 10.1016/j.chom.2013.10.004
15. Olszak T, An D, Zeissig S, Vera MP, Richter J, Franke A, et al. Microbial exposure during early life has persistent effects on natural killer T cell function. Science (2012) 336:489–93. doi: 10.1126/science.1219328
16. Russell SL, Gold MJ, Hartmann M, Willing BP, Thorson L, Wlodarska M, et al. Early life antibiotic-driven changes in microbiota enhance susceptibility to allergic asthma. EMBO Rep. (2012) 13:440–7. doi: 10.1038/embor.2012.32
17. Arrieta MC, Stiemsma LT, Dimitriu PA, Thorson L, Russell S, Yurist-Doutsch S, et al. Early infancy microbial and metabolic alterations affect risk of childhood asthma. Sci Transl Med. (2015) 7:307ra152. doi: 10.1126/scitranslmed.aab2271
18. Bisgaard H, Li N, Bonnelykke K, Chawes BL, Skov T, Paludan-Muller G, et al. Reduced diversity of the intestinal microbiota during infancy is associated with increased risk of allergic disease at school age. J Allergy Clin Immunol. (2011) 128:646–52 e1-5. doi: 10.1016/j.jaci.2011.04.060
19. Johansson MA, Sjogren YM, Persson JO, Nilsson C, Sverremark-Ekstrom E. Early colonization with a group of Lactobacilli decreases the risk for allergy at five years of age despite allergic heredity. PLoS ONE (2011) 6:e23031. doi: 10.1371/journal.pone.0023031
20. Azad MB, Konya T, Guttman DS, Field CJ, Sears MR, HayGlass KT, et al. Infant gut microbiota and food sensitization: associations in the first year of life. Clin Exp Allergy (2015) 45:632–43. doi: 10.1111/cea.12487
21. Le Chatelier E, Nielsen T, Qin J, Prifti E, Hildebrand F, Falony G, et al. Richness of human gut microbiome correlates with metabolic markers. Nature (2013) 500:541–6. doi: 10.1038/nature12506
22. Backhed F, Roswall J, Peng Y, Feng Q, Jia H, Kovatcheva-Datchary P, et al. Dynamics and stabilization of the human gut microbiome during the first year of life. Cell Host Microbe (2015) 17:852. doi: 10.1016/j.chom.2015.05.012
23. Arumugam M, Raes J, Pelletier E, Le Paslier D, Yamada T, Mende DR, et al. Enterotypes of the human gut microbiome. Nature (2011) 473:174–80. doi: 10.1038/nature09944
24. Qin J, Li R, Raes J, Arumugam M, Burgdorf KS, Manichanh C, et al. A human gut microbial gene catalogue established by metagenomic sequencing. Nature (2010) 464:59–65. doi: 10.1038/nature08821
25. Rooks MG, Garrett WS. Gut microbiota, metabolites and host immunity. Nat Rev Immunol. (2016) 16:341–52. doi: 10.1038/nri.2016.42
26. Charbonneau MR, O'Donnell D, Blanton LV, Totten SM, Davis JC, Barratt MJ, et al. Sialylated milk oligosaccharides promote microbiota-dependent growth in models of infant undernutrition. Cell (2016) 164:859–71. doi: 10.1016/j.cell.2016.01.024
27. Berni Canani R, Sangwan N, Stefka AT, Nocerino R, Paparo L, Aitoro R, et al. Lactobacillus rhamnosus GG-supplemented formula expands butyrate-producing bacterial strains in food allergic infants. ISME J. (2016) 10:742–50. doi: 10.1038/ismej.2015.151
28. Ling Z, Li Z, Liu X, Cheng Y, Luo Y, Tong X, et al. Altered fecal microbiota composition associated with food allergy in infants. Appl Environ Microbiol. (2014) 80:2546–54. doi: 10.1128/AEM.00003-14
29. Stiemsma LT, Arrieta MC, Dimitriu PA, Cheng J, Thorson L, Lefebvre DL, et al. Shifts in Lachnospira and Clostridium sp. in the 3-month stool microbiome are associated with preschool age asthma. Clin Sci. (2016) 130:2199–207. doi: 10.1042/CS20160349
30. Arrieta MC, Arevalo A, Stiemsma L, Dimitriu P, Chico ME, Loor S, et al. Associations between infant fungal and bacterial dysbiosis and childhood atopic wheeze in a nonindustrialized setting. J Allergy Clin Immunol. (2018) 142:424–34 e10. doi: 10.1016/j.jaci.2017.08.041
31. Fujimura KE, Sitarik AR, Havstad S, Lin DL, Levan S, Fadrosh D, et al. Neonatal gut microbiota associates with childhood multisensitized atopy and T cell differentiation. Nat Med. (2016) 22:1187–91. doi: 10.1038/nm.4176
32. Vatanen T, Kostic AD, d'Hennezel E, Siljander H, Franzosa EA, Yassour M, et al. Variation in microbiome LPS immunogenicity contributes to autoimmunity in humans. Cell (2016) 165:1551. doi: 10.1016/j.cell.2016.05.056
33. Bjorksten B, Sepp E, Julge K, Voor T, Mikelsaar M. Allergy development and the intestinal microflora during the first year of life. J Allergy Clin Immunol. (2001) 108:516–20. doi: 10.1067/mai.2001.118130
34. Kalliomaki M, Kirjavainen P, Eerola E, Kero P, Salminen S, Isolauri E. Distinct patterns of neonatal gut microflora in infants in whom atopy was and was not developing. J Allergy Clin Immunol. (2001) 107:129–34. doi: 10.1067/mai.2001.111237
35. Penders J, Thijs C, van den Brandt PA, Kummeling I, Snijders B, Stelma F, et al. Gut microbiota composition and development of atopic manifestations in infancy: the KOALA Birth Cohort Study. Gut (2007) 56:661–7. doi: 10.1136/gut.2006.100164
36. Penders J, Thijs C, Vink C, Stelma FF, Snijders B, Kummeling I, et al. Factors influencing the composition of the intestinal microbiota in early infancy. Pediatrics (2006) 118:511–21. doi: 10.1542/peds.2005-2824
37. Kirjavainen PV, Arvola T, Salminen SJ, Isolauri E. Aberrant composition of gut microbiota of allergic infants: a target of bifidobacterial therapy at weaning? Gut (2002) 51:51–5. doi: 10.1136/gut.51.1.51
38. Lodinova-Zadnikova R, Cukrowska B, Tlaskalova-Hogenova H. Oral administration of probiotic Escherichia coli after birth reduces frequency of allergies and repeated infections later in life (after 10 and 20 years). Int Arch Allergy Immunol. (2003) 131:209–11. doi: 10.1159/000071488
39. Stokholm J, Blaser MJ, Thorsen J, Rasmussen MA, Waage J, Vinding RK, et al. Maturation of the gut microbiome and risk of asthma in childhood. Nat Commun. (2018) 9:141. doi: 10.1038/s41467-017-02573-2
40. Pabst O, Mowat AM. Oral tolerance to food protein. Mucosal Immunol. (2012) 5:232–9. doi: 10.1038/mi.2012.4
41. Secher T, Payros D, Brehin C, Boury M, Watrin C, Gillet M, et al. Oral tolerance failure upon neonatal gut colonization with Escherichia coli producing the genotoxin colibactin. Infect Immun. (2015) 83:2420–9. doi: 10.1128/IAI.00064-15
42. Atarashi K, Tanoue T, Oshima K, Suda W, Nagano Y, Nishikawa H, et al. Treg induction by a rationally selected mixture of Clostridia strains from the human microbiota. Nature (2013) 500:232–6. doi: 10.1038/nature12331
43. An D, Oh SF, Olszak T, Neves JF, Avci FY, Erturk-Hasdemir D, et al. Sphingolipids from a symbiotic microbe regulate homeostasis of host intestinal natural killer T cells. Cell (2014) 156:123–33. doi: 10.1016/j.cell.2013.11.042
44. Stefka AT, Feehley T, Tripathi P, Qiu J, McCoy K, Mazmanian SK, et al. Commensal bacteria protect against food allergen sensitization. Proc Nat Acad Sci USA. (2014) 111:13145–50. doi: 10.1073/pnas.1412008111
45. Van der Leek AP, Yanishevsky Y, Kozyrskyj AL. The kynurenine pathway as a novel link between allergy and the gut microbiome. Front Immunol. (2017) 8:1374. doi: 10.3389/fimmu.2017.01374
46. Agus A, Planchais J, Sokol H. Gut microbiota regulation of tryptophan metabolism in health and disease. Cell Host Microbe (2018) 23:716–24. doi: 10.1016/j.chom.2018.05.003
47. Gao J, Xu K, Liu H, Liu G, Bai M, Peng C, et al. Impact of the gut microbiota on intestinal immunity mediated by tryptophan metabolism. Front Cell Infect Microbiol. (2018) 8:13. doi: 10.3389/fcimb.2018.00013
48. Coats SR, Pham TT, Bainbridge BW, Reife RA, Darveau RP. MD-2 mediates the ability of tetra-acylated and penta-acylated lipopolysaccharides to antagonize Escherichia coli lipopolysaccharide at the TLR4 signaling complex. J Immunol. (2005) 175:4490–8. doi: 10.4049/jimmunol.175.7.4490
49. Feehley T, Belda-Ferre P, Nagler CR. What's LPS got to do with it? a role for gut LPS variants in driving autoimmune and allergic disease. Cell Host Microbe (2016) 19:572–4. doi: 10.1016/j.chom.2016.04.025
50. Denou E, Lolmede K, Garidou L, Pomie C, Chabo C, Lau TC, et al. Defective NOD2 peptidoglycan sensing promotes diet-induced inflammation, dysbiosis, and insulin resistance. EMBO Mol Med. (2015) 7:259–74. doi: 10.15252/emmm.201404169
51. Amar J, Chabo C, Waget A, Klopp P, Vachoux C, Bermudez-Humaran LG, et al. Intestinal mucosal adherence and translocation of commensal bacteria at the early onset of type 2 diabetes: molecular mechanisms and probiotic treatment. EMBO Mol Med. (2011) 3:559–72. doi: 10.1002/emmm.201100159
52. Plunkett CH, Nagler CR. The influence of the microbiome on allergic sensitization to food. J Immunol. (2017) 198:581–9. doi: 10.4049/jimmunol.1601266
53. Nicholson JK, Holmes E, Kinross J, Burcelin R, Gibson G, Jia W, et al. Host-gut microbiota metabolic interactions. Science (2012) 336:1262–7. doi: 10.1126/science.1223813
54. Amenyogbe N, Kollmann TR, Ben-Othman R. Early-life host-microbiome interphase: the key frontier for immune development. Front Pediatr. (2017) 5:111. doi: 10.3389/fped.2017.00111
55. Baars A, Oosting A, Knol J, Garssen J, van Bergenhenegouwen J. The gut microbiota as a therapeutic target in IBD and metabolic disease: a role for the bile acid receptors FXR and TGR5. Microorganisms (2015) 3:641–66. doi: 10.3390/microorganisms3040641
56. Bridgman SL, Azad MB, Field CJ, Haqq AM, Becker AB, Mandhane PJ, et al. Fecal short-chain fatty acid variations by breastfeeding status in infants at 4 months: differences in relative versus absolute concentrations. Front Nutr. (2017) 4:11. doi: 10.3389/fnut.2017.00011
57. Iraporda C, Errea A, Romanin DE, Cayet D, Pereyra E, Pignataro O, et al. Lactate and short chain fatty acids produced by microbial fermentation downregulate proinflammatory responses in intestinal epithelial cells and myeloid cells. Immunobiology (2015) 220:1161–9. doi: 10.1016/j.imbio.2015.06.004
58. van den Elsen LW, Poyntz HC, Weyrich LS, Young W, Forbes-Blom EE. Embracing the gut microbiota: the new frontier for inflammatory and infectious diseases. Clin Transl Immunol. (2017) 6:e125. doi: 10.1038/cti.2016.91
59. Trompette A, Gollwitzer ES, Yadava K, Sichelstiel AK, Sprenger N, Ngom-Bru C, et al. Gut microbiota metabolism of dietary fiber influences allergic airway disease and hematopoiesis. Nature medicine (2014) 20:159–66. doi: 10.1038/nm.3444
60. Thio CL, Chi PY, Lai AC, Chang YJ. Regulation of type 2 innate lymphoid cell-dependent airway hyperreactivity by butyrate. J Allergy Clin Immunol. (2018) 142:1867–83.e12 doi: 10.1016/j.jaci.2018.02.032
61. Collado MC, Rautava S, Aakko J, Isolauri E, Salminen S. Human gut colonisation may be initiated in utero by distinct microbial communities in the placenta and amniotic fluid. Sci Rep. (2016) 6:23129. doi: 10.1038/srep23129
62. Stewart CJ, Ajami NJ, O'Brien JL, Hutchinson DS, Smith DP, Wong MC, et al. Temporal development of the gut microbiome in early childhood from the TEDDY study. Nature (2018) 562:583–8. doi: 10.1038/s41586-018-0617-x
63. Le Huerou-Luron I, Blat S, Boudry G. Breast- v. formula-feeding: impacts on the digestive tract and immediate and long-term health effects. Nutr Res Rev. (2010) 23:23–36. doi: 10.1017/S0954422410000065
64. Rautava S. Early microbial contact, the breast milk microbiome and child health. J Dev Orig Health Dis. (2016) 7:5–14. doi: 10.1017/S2040174415001233
65. Forbes JD, Azad MB, Vehling L, Tun HM, Konya TB, Guttman DS, et al. Association of exposure to formula in the hospital and subsequent infant feeding practices with gut microbiota and risk of overweight in the first year of life. JAMA Pediatr. (2018) 172:e181161. doi: 10.1001/jamapediatrics.2018.1161
66. De Filippo C, Cavalieri D, Di Paola M, Ramazzotti M, Poullet JB, Massart S, et al. Impact of diet in shaping gut microbiota revealed by a comparative study in children from Europe and rural Africa. Proc Nat Acad Sci USA. (2010) 107:14691–6. doi: 10.1073/pnas.1005963107
67. Fernandez L, Langa S, Martin V, Maldonado A, Jimenez E, Martin R, et al. The human milk microbiota: origin and potential roles in health and disease. Pharmacol Res. (2013) 69:1–10. doi: 10.1016/j.phrs.2012.09.001
68. Murphy K, Curley D, O'Callaghan TF, O'Shea CA, Dempsey EM, O'Toole PW, et al. The composition of human milk and infant faecal microbiota over the first three months of life: a pilot study. Sci Rep. (2017) 7:40597. doi: 10.1038/srep40597
69. Cabrera-Rubio R, Collado MC, Laitinen K, Salminen S, Isolauri E, Mira A. The human milk microbiome changes over lactation and is shaped by maternal weight and mode of delivery. Am J Clin. Nutr. (2012) 96:544–51. doi: 10.3945/ajcn.112.037382
70. Ramsay DT, Kent JC, Owens RA, Hartmann PE. Ultrasound imaging of milk ejection in the breast of lactating women. Pediatrics (2004) 113:361–7. doi: 10.1542/peds.113.2.361
71. Roberfroid M. Prebiotics: the concept revisited. J Nutr. (2007) 137(3 Suppl. 2):830S−7S. doi: 10.1093/jn/137.3.830S
72. Bode L. The functional biology of human milk oligosaccharides. Early Hum Dev. (2015) 91:619–22. doi: 10.1016/j.earlhumdev.2015.09.001
73. Bode L. Human milk oligosaccharides: every baby needs a sugar mama. Glycobiology (2012) 22:1147–62. doi: 10.1093/glycob/cws074
74. Zivkovic AM, German JB, Lebrilla CB, Mills DA. Human milk glycobiome and its impact on the infant gastrointestinal microbiota. Proc Nat Acad Sci USA. (2011) 108 (Suppl, 1):4653–8. doi: 10.1073/pnas.1000083107
75. Ayechu-Muruzabal V, van Stigt AH, Mank M, Willemsen LEM, Stahl B, Garssen J, et al. Diversity of human milk oligosaccharides and effects on early life immune development. Front Pediatr. (2018) 6:239. doi: 10.3389/fped.2018.00239
76. Seppo AE, Autran CA, Bode L, Jarvinen KM. Human milk oligosaccharides and development of cow's milk allergy in infants. J Allergy Clin Immunol. (2017) 139:708–11 e5. doi: 10.1016/j.jaci.2016.08.031
77. Miliku K, Robertson B, Sharma AK, Subbarao P, Becker AB, Mandhane PJ, et al. Human milk oligosaccharide profiles and food sensitization among infants in the CHILD Study. Allergy (2018) 73:2070–3 doi: 10.1111/all.13476
78. Pacheco AR, Barile D, Underwood MA, Mills DA. The impact of the milk glycobiome on the neonate gut microbiota. Annu Rev Anim Biosci. (2015) 3:419–45. doi: 10.1146/annurev-animal-022114-111112
79. Demmelmair H, Prell C, Timby N, Lonnerdal B. Benefits of lactoferrin, osteopontin and milk fat globule membranes for infants. Nutrients (2017) 9:E817. doi: 10.3390/nu9080817
80. Kaetzel CS. Cooperativity among secretory IgA, the polymeric immunoglobulin receptor, and the gut microbiota promotes host-microbial mutualism. Immunol Lett. (2014) 162(2 Pt. A):10–21. doi: 10.1016/j.imlet.2014.05.008
81. Brandtzaeg P. Mucosal immunity: integration between mother and the breast-fed infant. Vaccine (2003) 21:3382–8. doi: 10.1016/S0264-410X(03)00338-4
82. Rogier EW, Frantz AL, Bruno ME, Wedlund L, Cohen DA, Stromberg AJ, et al. Secretory antibodies in breast milk promote long-term intestinal homeostasis by regulating the gut microbiota and host gene expression. Proc Nat Acad Sci USA. (2014) 111:3074–9. doi: 10.1073/pnas.1315792111
83. Jiang HQ, Bos NA, Cebra JJ. Timing, localization, and persistence of colonization by segmented filamentous bacteria in the neonatal mouse gut depend on immune status of mothers and pups. Infect Immun. (2001) 69:3611–7. doi: 10.1128/IAI.69.6.3611-3617.2001
84. Dzidic M, Abrahamsson TR, Artacho A, Bjorksten B, Collado MC, Mira A, et al. Aberrant IgA responses to the gut microbiota during infancy precede asthma and allergy development. J Allergy Clin Immunol. (2017) 139:1017–25 e14. doi: 10.1016/j.jaci.2016.06.047
85. Nakajima A, Vogelzang A, Maruya M, Miyajima M, Murata M, Son A, et al. IgA regulates the composition and metabolic function of gut microbiota by promoting symbiosis between bacteria. J Exp Med. (2018) 215:2019–34. doi: 10.1084/jem.20180427
86. Mastromarino P, Capobianco D, Campagna G, Laforgia N, Drimaco P, Dileone A, et al. Correlation between lactoferrin and beneficial microbiota in breast milk and infant's feces. Biometals (2014) 27:1077–86. doi: 10.1007/s10534-014-9762-3
87. Pirr S, Richter M, Fehlhaber B, Pagel J, Hartel C, Roth J, et al. High amounts of S100-alarmins confer antimicrobial activity on human breast milk targeting pathogens relevant in neonatal sepsis. Front Immunol. (2017) 8:1822. doi: 10.3389/fimmu.2017.01822
88. Trend S, Strunk T, Hibbert J, Kok CH, Zhang G, Doherty DA, et al. Antimicrobial protein and Peptide concentrations and activity in human breast milk consumed by preterm infants at risk of late-onset neonatal sepsis. PLoS One (2015) 10:e0117038. doi: 10.1371/journal.pone.0117038
89. Walker WA, Iyengar RS. Breast milk, microbiota, and intestinal immune homeostasis. Pediatr Res. (2015) 77:220–8. doi: 10.1038/pr.2014.160
90. Gomez-Gallego C, Morales JM, Monleon D, du Toit E, Kumar H, Linderborg KM, et al. Human breast milk NMR metabolomic profile across specific geographical locations and its association with the milk microbiota. Nutrients (2018) 10:E1355. doi: 10.3390/nu10101355
91. Gomez de Aguero M, Ganal-Vonarburg SC, Fuhrer T, Rupp S, Uchimura Y, Li H, et al. The maternal microbiota drives early postnatal innate immune development. Science (2016) 351:1296–302. doi: 10.1126/science.aad2571
92. Korecka A, Dona A, Lahiri S, Tett AJ, Al-Asmakh M, Braniste V, et al. Bidirectional communication between the Aryl hydrocarbon receptor (AhR) and the microbiome tunes host metabolism. NPJ Biofilms Microbiomes (2016) 2:16014. doi: 10.1038/npjbiofilms.2016.14
93. Labeta MO, Vidal K, Nores JE, Arias M, Vita N, Morgan BP, et al. Innate recognition of bacteria in human milk is mediated by a milk-derived highly expressed pattern recognition receptor, soluble CD14. J Exp Med. (2000) 191:1807–12. doi: 10.1084/jem.191.10.1807
94. LeBouder E, Rey-Nores JE, Rushmere NK, Grigorov M, Lawn SD, Affolter M, et al. Soluble forms of Toll-like receptor (TLR)2 capable of modulating TLR2 signaling are present in human plasma and breast milk. J Immunol. (2003) 171:6680–9. doi: 10.4049/jimmunol.171.12.6680
95. LeBouder E, Rey-Nores JE, Raby AC, Affolter M, Vidal K, Thornton CA, et al. Modulation of neonatal microbial recognition: TLR-mediated innate immune responses are specifically and differentially modulated by human milk. J Immunol. (2006) 176:3742–52. doi: 10.4049/jimmunol.176.6.3742
96. Ojeda P, Bobe A, Dolan K, Leone V, Martinez K. Nutritional modulation of gut microbiota - the impact on metabolic disease pathophysiology. J Nutr Biochem. (2016) 28:191–200. doi: 10.1016/j.jnutbio.2015.08.013
97. Jiang T, Liu B, Li J, Dong X, Lin M, Zhang M, et al. Association between sn-2 fatty acid profiles of breast milk and development of the infant intestinal microbiome. Food Funct. (2018) 9:1028–37. doi: 10.1039/C7FO00088J
98. Wickens K, Barthow C, Mitchell EA, Stanley TV, Purdie G, Rowden J, et al. Maternal supplementation alone with Lactobacillus rhamnosus HN001 during pregnancy and breastfeeding does not reduce infant eczema. Pediatr Allergy Immunol. (2018) 29:296–302. doi: 10.1111/pai.12874
Keywords: breastmilk, allergy, gut microbiota, neonate, prevention
Citation: van den Elsen LWJ, Garssen J, Burcelin R and Verhasselt V (2019) Shaping the Gut Microbiota by Breastfeeding: The Gateway to Allergy Prevention? Front. Pediatr. 7:47. doi: 10.3389/fped.2019.00047
Received: 05 October 2018; Accepted: 04 February 2019;
Published: 27 February 2019.
Edited by:
George N. Konstantinou, 424 General Military Hospital, GreeceReviewed by:
Klaus Tenbrock, RWTH Aachen Universität, GermanyCopyright © 2019 van den Elsen, Garssen, Burcelin and Verhasselt. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Lieke W. J. van den Elsen, bGlla2UudmFuZGVuZWxzZW5AdXdhLmVkdS5hdQ==
Valerie Verhasselt, dmFsZXJpZS52ZXJoYXNzZWx0QHV3YS5lZHUuYXU=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.